Significance
The rotation of the Earth around its own axis creates daily changes in the environment for virtually all living organisms. To anticipate and adapt to those changes, mammals possess an evolutionarily conserved circadian clock that controls most aspects of physiology. Using a previously undescribed analysis tool, we studied the impact of the circadian clock and its underlying feeding rhythms on hepatic gene expression. Our analysis shows that the loss of feeding rhythms in clock-disrupted animals is an important component of their phenotype. Finally, we were able to decipher the specific role of feeding rhythms, the circadian clock, and its controlled output of PARbZip transcription factors in the regulation of liver rhythmic gene expression.
Keywords: circadian clock, feeding–fasting cycle, liver metabolism, transcriptomics, differential rhythmicity analysis
Abstract
The circadian clock and feeding rhythms are both important regulators of rhythmic gene expression in the liver. To further dissect the respective contributions of feeding and the clock, we analyzed differential rhythmicity of liver tissue samples across several conditions. We developed a statistical method tailored to compare rhythmic liver messenger RNA (mRNA) expression in mouse knockout models of multiple clock genes, as well as PARbZip output transcription factors (Hlf/Dbp/Tef). Mice were exposed to ad libitum or night-restricted feeding under regular light–dark cycles. During ad libitum feeding, genetic ablation of the core clock attenuated rhythmic-feeding patterns, which could be restored by the night-restricted feeding regimen. High-amplitude mRNA expression rhythms in wild-type livers were driven by the circadian clock, but rhythmic feeding also contributed to rhythmic gene expression, albeit with significantly lower amplitudes. We observed that Bmal1 and Cry1/2 knockouts differed in their residual rhythmic gene expression. Differences in mean expression levels between wild types and knockouts correlated with rhythmic gene expression in wild type. Surprisingly, in PARbZip knockout mice, the mean expression levels of PARbZip targets were more strongly impacted than their rhythms, potentially due to the rhythmic activity of the D-box–repressor NFIL3. Genes that lost rhythmicity in PARbZip knockouts were identified to be indirect targets. Our findings provide insights into the diurnal transcriptome in mouse liver as we identified the differential contributions of several core clock regulators. In addition, we gained more insights on the specific effects of the feeding–fasting cycle.
Almost all organisms are subjected to daily changes in their environment with light–dark cycles that are caused by Earth’s rotation around its own axis. To anticipate these changes, organisms possess an evolutionarily conserved endogenous oscillator, the circadian clock, that drives daily rhythms in behavior and physiology with a 24-h period (1, 2). In mammals, the circadian clock is hierarchically organized, with a master pacemaker located in the bilateral hypothalamic superchiasmatic nuclei (SCN) and peripheral clocks present in virtually all organs, including the liver (3, 4). The SCN clock receives light via the retina and transmits this information to the peripheral clocks via direct nervous connections or controlled secretion of humoral factors. On a molecular level, the circadian clock consists of interconnected transcriptional and translational negative-feedback loops of so-called clock genes (5). The core oscillator loop consists of a positive limb in which BMAL1 (named also ARNTL) and its heterodimerization partners CLOCK and NPAS2 promote gene expression of several clock target genes via E-box motifs. These include Period (Per) and Cryptochrome (Cry), factors of the negative limb of the core loop, which then in turn inhibit the transcriptional activity of BMAL1. In addition to this core loop, another crucial loop exists in which BMAL1 heterodimers target RORα, RORγ, REV-ERBα (also named NR1D1), and REV-ERBβ (also named NR1D2) regulate expression of Bmal1 and its heterodimeric partners by binding to response elements (RORE) present in their promotors (6, 7). Furthermore, the circadian clock controls the expression of proline and acidic amino acid-rich basic leucine zipper (PARbZip) transcription factors DBP, HLF, and TEF and their repressive counterpart NFIL3 (named also E4BP4) factors. In vivo experiments in mice indicate that these transcription factors are not directly involved in the circadian clock machinery but mediate circadian clock output pathways via D-box elements (8, 9). To date, the relative contribution of each loop of the molecular network to rhythmic gene expression has been unclear.
In the liver, this oscillatory network regulates systems-wide rhythmic gene expression programs (10), including components of fundamental metabolic pathways (11). The cell-autonomous liver clock directly regulates genes involved in the regulation of glucose metabolism or xenobiotic detoxification through the positive loop and BMAL1 (12, 13). In addition, the PER and CRY proteins of the negative loop indirectly regulate lipid and glucose metabolism through interaction with nuclear receptors and cellular signaling (14–17). Nevertheless, the circadian clock is not the only driver of hepatic rhythmic gene expression. The SCN synchronize the peripheral clocks, including the liver clock, but also regulate systemic cues, such as locomotor activity, body temperature, and feeding and drinking behavior (18, 19). There is increasing evidence that daily food intake has a major impact on rhythmic gene expression and liver physiology. Specifically, feeding during the night, the activity phase of nocturnal animals like mice, increases the rhythmic amplitudes of liver physiology (20), while feeding during the resting phase inverts the temporal pattern of liver gene expression (21, 22). Constant feeding suppresses a large fraction of rhythmic gene expression in the liver (21, 23, 24). Nevertheless, the interplay of feeding cycles and the cell-autonomous clock on rhythmic gene expression in the liver is not fully understood.
A major reason for this gap of knowledge is the lack of comprehensive transcriptome datasets and analysis methods to investigate differential rhythmic gene expression in clock-disrupted mouse models and under varying feeding regimens. Here, we generated murine transcriptome datasets to systematically dissect the contribution of the molecular clock network and the role of natural feeding cycles on the temporal liver transcriptome. We performed transcriptome analyses of multiple circadian clock gene knockout (KO) models. This included the KO of the positive limb (Bmal1 KO) or the negative limb (Cry1/2 KO) of the core clock loop under ad libitum (AL) and night restricted-feeding (NRF) regimens. In addition, we assessed the role of the PARbZip-mediated clock output pathway using Hlf/Dbp/Tef KO mice. Moreover, we have implemented a statistical framework to assess differential rhythmicity and mean gene expression across multiple conditions. Our work defines the respective contributions of different components of the circadian clock network and of natural feeding cycles on rhythmic gene expression.
Results
A Statistical Framework to Detect Differential Rhythmicity across Many Conditions.
While an increasing number of statistical methods allow the analysis of rhythmicity from time-series gene expression analysis, algorithms to assess differential rhythmicity across multiple conditions remain scarce (25, 26). Previously, we have used harmonic regression and model selection to assess differential rhythmicity between conditions in various settings (27–33). The key idea is that rhythmicity parameters (amplitude and phase) for each condition can be shared across subsets of conditions, leading to a combinatorial set of possible models for each gene (SI Appendix, Fig. S1 A and B). By assigning each gene (probabilistically) to one of these models (Fig. 1A), this method provides a powerful alternative to common approaches based on intersecting the identified sets from the individual conditions, which is typically sensitive to thresholds. However, the method had to be customized for each specific setting and was not implemented into a flexible and accessible statistical package. We have now extended this method to detect and estimate changes in the parameters amplitude (log2 fold-change peak-to-trough), phase (time of peak), and mean expression levels for datasets with two or more conditions. Moreover, the models are now tailored to the noise properties of RNA-sequencing (RNA-Seq) data. The method was named dryR (for Differential Rhythmicity Analysis in R) and is available as an R package (https://github.com/naef-lab/dryR).
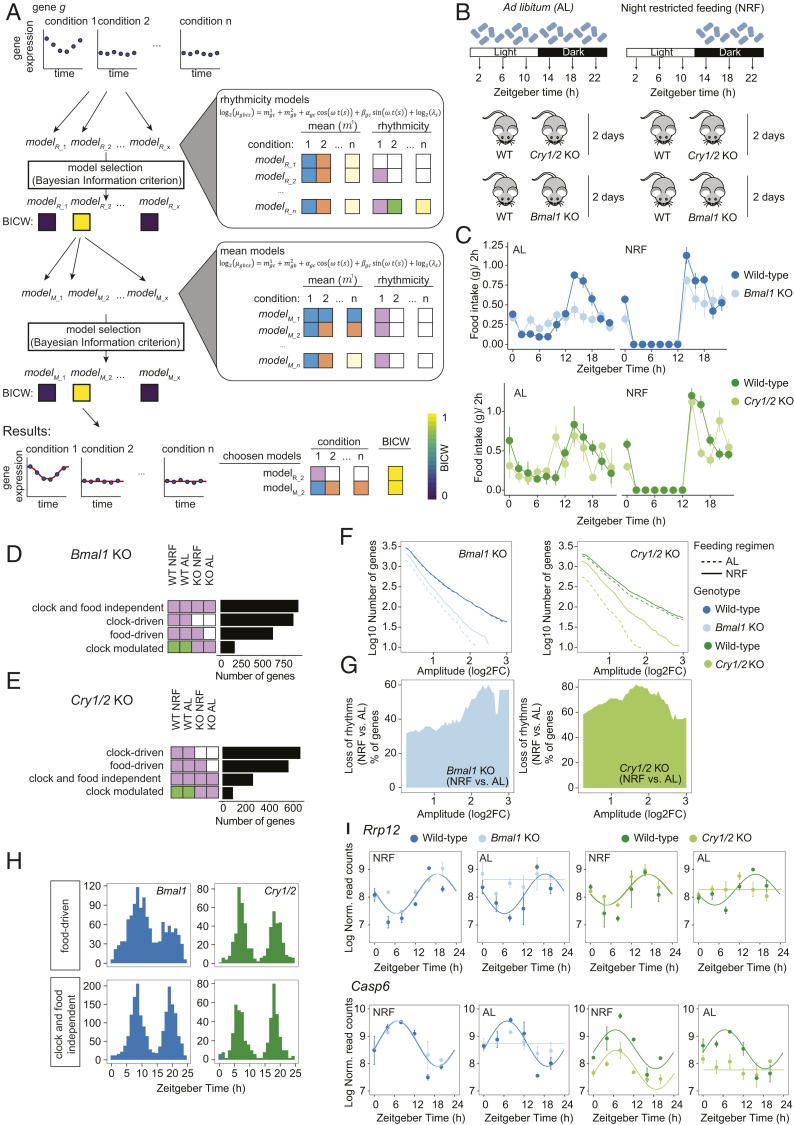
Fig. 1.
Regulation of liver rhythmic gene expression by the circadian clock and feeding rhythms analyzed with dryR. (A) Schematic illustrating the workflow of the statistical framework used in dryR. BICW is color-coded as indicated in the ramp (Lower Right). The same color for the remaining boxes indicates shared mean levels and rhythmic parameters between the indicated conditions for mean and rhythmic models, respectively. (B) Experimental design. (C) Rhythmic feeding in WT, Bmal1 KO, and Cry1/2 KO mice under AL and NRF. The Zeitgebertime (ZT) defines the timing of entrainment by light (ZT0 lights on; ZT12: lights off). (D and E) Number of genes classified in rhythmic models in Bmal1 KO (D) and Cry1/2 KO (E). White indicates no rhythm detected, and the same color indicates shared rhythmic parameters (amplitudes and phase) between the indicated conditions. (F) Cumulative number of rhythmic genes in the liver of WT, Bmal1 KO, and Cry1/2 KO in function of minimal amplitude. NRF partially restores rhythmicity in KOs. (G) Relative number of genes that lose rhythmicity under the AL compared with NRF regimen in KO in function of minimal amplitude. (H) Phase distribution of rhythmic genes classified as food-driven and clock- and food-independent. (I) Temporal expression pattern of Rrp12 and Casp6.
To systematically assess the performance of dryR and compare it with existing rhythmicity analysis methods, we simulated rhythmic count data for four conditions with a commonly used sampling interval of 4 h and two replicates. We first tested goodness of fit of dryR and detected overall good fits for phase, mean, and amplitude with higher noise at lower mean counts (SI Appendix, Fig. S1C). In a second step, we used simulations to test the reliability of detecting rhythmicity in four conditions and benchmarked it to published rhythm-detection methods (SI Appendix, Fig. S1D). The number of correctly detected rhythms across all four conditions was in the same range as with best-performing other algorithms, for which each condition was tested individually. Moreover, dryR performed reliably across a broad spectrum of conditions, including varying sampling intervals, missing samples, and numbers of replicates (SI Appendix, Fig. S1E). Next, we compared the ability of dryR to detect phase shifts and changes in amplitude with methods designed for differential rhythmicity analysis between two conditions of temporal data: CircaCompare (34), Detection of Differential Rhythmicity (35), and LimoRhyde (36). To this end, we simulated rhythmic data with either a 4-h phase shift or a doubling in amplitude between two conditions. dryR reliably performs the calling of differential rhythmicity with comparable specificity and sensitivity as the best-performing algorithms across variable sampling intervals or changing numbers of replicates and missing samples (SI Appendix, Fig. S1 F–I). Thus, the performance of dryR is comparable to other methods for detecting rhythmicity or differential rhythmicity, but the advantage of dryR is that it combines rhythmicity detection, differential gene expression, and rhythmicity analysis for two or more conditions.
Clock-Disrupted Mice Exhibit Attenuated Feeding Rhythms, Dampening Rhythmic Liver Gene Expression.
The fasting–feeding cycle is an important driver of rhythmic gene expression in the mouse liver (21, 23, 24). To distinguish the contribution of natural feeding cycles (food intake occurs largely during the night in wild-type [WT] animals with free access to food [AL]), and of the circadian clock to rhythmic gene expression, we kept Cry1/2 and Bmal1 KO mice and WT controls under a 12-h light–dark cycle under AL feeding (Fig. 1B). Compared with WT animals, both Bmal1 and Cry1/2 KOs lost their feeding rhythms under AL feeding (Fig. 1C). In parallel, to control for feeding rhythms, we conducted the same experiment with restricted access to food during the active phase (NRF) (Fig. 1B). As expected, feeding rhythms comparable to WT AL were found in the KOs under NRF (Fig. 1C).
We then investigated temporal gene expression in the liver of these mice using RNA-Seq (sampled every 4 h, two independent replicates) and analyzed the results by dryR. The vast majority of circadian core clock genes lost rhythmicity in Bmal1 and Cry1/2 KOs irrespective of the offered feeding regimen (SI Appendix, Figs. S2A and S3 and Dataset S1), except for Per2, consistent with previous work (37). Moreover, of all transcripts assigned to a model with identical rhythms in WT AL and NRF (Fig. 1 D and E), a comparable proportion was detected to be regulated by the circadian clock (clock-driven) in both Bmal1 KO (30%; Fig. 1D) and Cry1/2 KO (37%; Fig. 1E). Accordingly, these transcripts lost rhythmicity in both KO AL and KO NRF. We next used the KO mice under both feeding regimens to identify transcript rhythms that depend on a natural feeding–fasting cycle. In WT animals, AL and NRF showed similar numbers of cyclic genes across the entire range of amplitudes, with nearly 100 transcripts with amplitudes above 8-fold in both genotypes (Fig. 1F). In contrast, at least 32% and 54% of rhythmic transcripts under NRF lost rhythmicity in the absence of rhythmic food intake (AL) in Bmal1 and Cry1/2 KOs, respectively, across the entire range of amplitude (Fig. 1 F and G). Among those, transcripts assigned to the food-driven model comprised 22% (Bmal1 KO; Fig. 1D) and 31% (Cry1/2 KO; Fig. 1E) of all rhythmic genes. These transcripts shared amplitude and phase across three conditions (WT AL, WT NRF, and KO NRF) but lost rhythmic expression in the absence of rhythmic food intake (KO AL) (Fig. 1 D and E and SI Appendix, Fig. S2B). These feeding–fasting-driven rhythmic genes (food-driven) exhibited a bimodal phase distribution (Fig. 1H and SI Appendix, Fig. S2C) and on average 36% lower amplitudes (SI Appendix, Fig. S2D) compared with clock-driven or clock-modulated genes. As exemplified by Rrp12 (night phase) and Casp6 (light phase) (Fig. 1I), they lost rhythmicity in the KOs under AL feeding, while they retained rhythms under NRF conditions. Of all rhythmic genes, 5% (Bmal1 KO) and 6% (Cry1/2 KO) were regulated by feeding rhythms but showed different rhythmic parameters (i.e., amplitude and/or phase) in the KO, suggesting that feeding rhythms and the clock participate in their regulation (clock-modulated; Fig. 1 D and E and SI Appendix, Fig. S2B). Strikingly, 32% (Bmal1 KO) and 14% (Cry1/2 KO) of rhythmic genes exhibited consistent rhythmicity (identical phases and amplitudes) across all four tested conditions (clock- and food-independent; Fig. 1 D and E and SI Appendix, Fig. S2 B–D). Clock- and food-independent rhythms also exhibited a biphasic phase pattern (Fig. 1H) and showed amplitudes comparable to (Bmal1 KO) or lower (Cry1/2 KO) than those of food-driven transcripts (SI Appendix, Fig. S2 B–D). For example, Cirbp and Hsp90ab1 keep rhythmic messenger RNA (mRNA) levels in all conditions (SI Appendix, Fig. S2E). These findings suggest that under a light–dark cycle, additional clock-independent systemic signals other than feeding drive low-amplitude rhythmic gene expression in the liver.
Clock- and System-Driven Biological Functions of the Rhythmic Liver Transcriptome.
To further distinguish the specific contribution of the core clock oscillator from rhythmic feeding on rhythmic gene expression in the mouse liver, we focused our analysis on Bmal1 and Cry1/2 KO animals kept under NRF. Using dryR, genes were grouped according to their temporal expression patterns into five categories (system-driven, clock-driven, clock-modulated, Cry1/2 KO-specific, Bmal1 KO-specific) (Fig. 2 A and B and Dataset S2). Of all rhythmic genes in WT mice, 33% were independent of a functional clock and remained rhythmic in both KOs and, thus, were named system-driven. Expectedly, these gene promoters lacked binding of circadian clock transcription factors (SI Appendix, Fig. S4A). System-driven genes turned out to exhibit lower amplitudes compared with clock-driven or clock-modulated genes (Fig. 2C and SI Appendix, Fig. S4 B and C) and showed a bimodal phase distribution (Fig. 2D and SI Appendix, Fig. S4B). In a temporal resolved-enrichment analysis, these system-driven genes can be assigned to mRNA translation and ribosome biogenesis (“ribosome biogenesis,” “nucleolus,” and “de novo posttranslational folding”), known to be regulated by systemic signals (32), and metabolism of food (“cellular lipid metabolic process” and “response to insulin”) (Fig. 2E). Moreover, immune function (“BCR B-cell”) was also enriched for system-driven genes, which is likely a consequence of the rhythmic circulation of B cells (38). In comparison, clock-driven genes, which lost rhythmicity in the KO mice, represented 20% of all rhythmic genes in the mouse liver (Fig. 2A). These genes had generally a high amplitude (Fig. 2C and SI Appendix, Fig. S4 B and C). Interestingly, this gene set showed a homogenous distribution of peak times (Fig. 2D and SI Appendix, Fig. S4B). Clock-driven genes could be assigned, besides their evident role in the circadian clock oscillator, to a function in metabolism of xenobiotics (“xenobiotic metabolism” and “biotransformation”), a process previously implicated with the circadian clock (39). Surprisingly, we also found a hitherto unknown clock-driven function in ion transport (“Na2+ transmembrane transporter activity”) (Fig. 2E). Like the clock-driven genes, the set of clock-modulated genes was targeted by the circadian core clock factors but displayed an altered rhythmic oscillation in the absence of a functional clock (7%; Fig. 2A and SI Appendix, Fig. S4A). These genes shifted from a pattern resembling clock-driven genes that show a homogeneous phase distribution, to a bimodal phase distribution in clock-disrupted animals with lower amplitude, a pattern similar to what was observed for system-driven genes (Fig. 2D and SI Appendix, Fig. S4 B and D). Clock-modulated genes appeared to play a role for the immune system and interferon response (“IRF target genes”), known as a rhythmic process (40) and the modulation of food-related insulin response (“insulin signalling”) (Fig. 2E). Together, our observations suggest that systemic signals generate bimodal and low-amplitude rhythmicity in gene expression in the mouse liver, whereas the circadian clock drives gene expression rhythms with high amplitudes covering all phases.
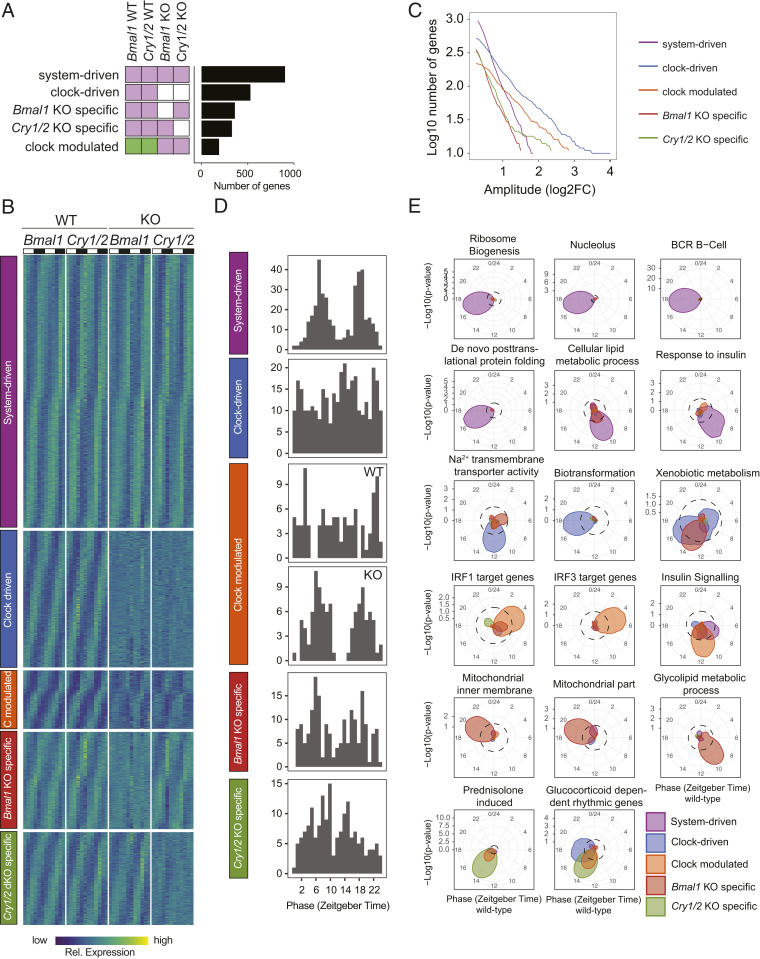
Fig. 2.
Systemic cues and the circadian clock equally shape rhythmic liver gene expression. (A) Number of genes classified in rhythmic models in Bmal1 KO and Cry1/2 KO under NRF. White indicates no rhythm detected, and the same color indicates shared rhythmic parameters (amplitudes and phase) between samples. (B) Heat maps of normalized rhythmic mRNA levels in the liver of WT, Bmal1 KO, and Cry1/2 KO mice. Genes were classified as system-driven, clock-driven, Bmal1 KO-specific, Cry1/2 KO-specific, and clock-modulated. (C) Cumulative number of genes classified in the indicated rhythmic model in function of minimal amplitude. (D) Phase distribution of indicated models. (E) Examples of functional enrichment around the clock. Enrichment of the indicated functional terms is represented by the radial coordinate at the indicated time point. P values were calculated by comparing the genes within a sliding window of 4 h with all expressed genes.
Differential Rhythmicity between Bmal1 and Cry1/2 KO Models in Liver Gene Expression.
The transcriptional circuitry of the mouse circadian clock is a complex network of positive- and negative-feedback loops. Our dataset containing KOs for Bmal1 and Cry1/2 allows the comparison of the effects of the core transcriptional activator BMAL1 and its repressors CRY1 and CRY2. Therefore, we continued to characterize the differential effects on rhythmic liver gene expression in these KOs under NRF in 12-h light–dark cycles. We detected genes that lost rhythmicity only in one of the KOs (i.e., rhythms that were lost in either Bmal1 KO or Cry1/2 KO; Fig. 2 A–D). The amplitudes of these rhythmic transcript were overall lower than clock-driven and comparable to system-driven transcripts (SI Appendix, Fig. S4C). These findings suggest that systemic signals blunted specifically in Bmal1 or Cry1/2 KO can modulate rhythmic gene expression. Indeed, glucocorticoid responsive genes (“Prednisolone induced” and “Glucocorticoid dependent rhythmic genes”) are more likely to lose rhythm specifically in Cry1/2 KO (Fig. 2E), which might reflect the direct repression of the glucocorticoid receptor by CRY1 and CRY2 (16). Conversely, loss of rhythms related to mitochondrial activity and metabolism of glycolipids were more specific to the deletion of Bmal1 (Fig. 2E).
We further analyzed genes showing a phase shift in the two KO models. Interestingly, in Cry1/2 KO mice under NRF, a subset of rhythmic genes exhibited a phase advance of ∼4 h compared with WT (SI Appendix, Fig. S5A). In comparison, Bmal1 KO mice did not show such biased phases (SI Appendix, Fig. S5A). To identify potential transcription factors involved in this phase shift, we performed an untargeted ChIP-enrichment analysis (Dataset S2). This revealed that CREB- and PPARα-binding sites are significantly enriched in genes with a phase differences in the two KO models. While CREB activity has been shown to be phase-delayed in Bmal1 KO mice (41), PPARα potentially mediates the phase advance in Cry1/2 KO mice. Indeed, PPARα target genes were enriched not only in clock-driven genes but also in genes showing altered rhythms in both KOs (SI Appendix, Fig. S5B). Moreover, only in Cry1/2 KO, PPARα target genes showed a phase advanced of 4 h (SI Appendix, Fig. S5A), as exemplified by Pnpla2, Lpin2, and Cpt1a (SI Appendix, Fig. S5C). While expression of Pparα was slightly decreased in Bmal1 KO, as previously shown (42), we did not detect any rhythmicity changes of Pparα mRNA expression in Cry1/2 KO (SI Appendix, Fig. S5D). However, nuclear PPARα protein levels were phase-advanced (SI Appendix, Fig. S5E) based on published nuclear proteomics data (43). Together, the Bmal1 and Cry1/2 KOs show both shared effects on clock-driven genes expression as well as distinct differences in rhythmic gene expression in the liver.
Changes in Mean mRNA Levels in Bmal1 and Cry1/2 KOs Predict Amplitude and Distinct Expression Phases.
Most circadian transcriptome studies across several conditions (altered feeding, KOs) have focused on rhythmic gene expression but have not investigated changes in mean mRNA levels. Therefore, we used the dryR framework to detect patterns in mean mRNA levels (Fig. 1A) and learn about circadian clock-regulated gene expression. Overall, more genes were differentially expressed in Bmal1 KO than in Cry1/2 KO mice (Fig. 3A). Consistent with the molecular function of BMAL1 as a positive regulator of transcription and CRY1/2 as a repressor in the core clock loop, more genes were down-regulated (compared with up-regulated) in Bmal1 KO and vice versa in Cry1/2 KO mice (Fig. 3B), indicating that the direct effects of those transcription regulators are dominant.
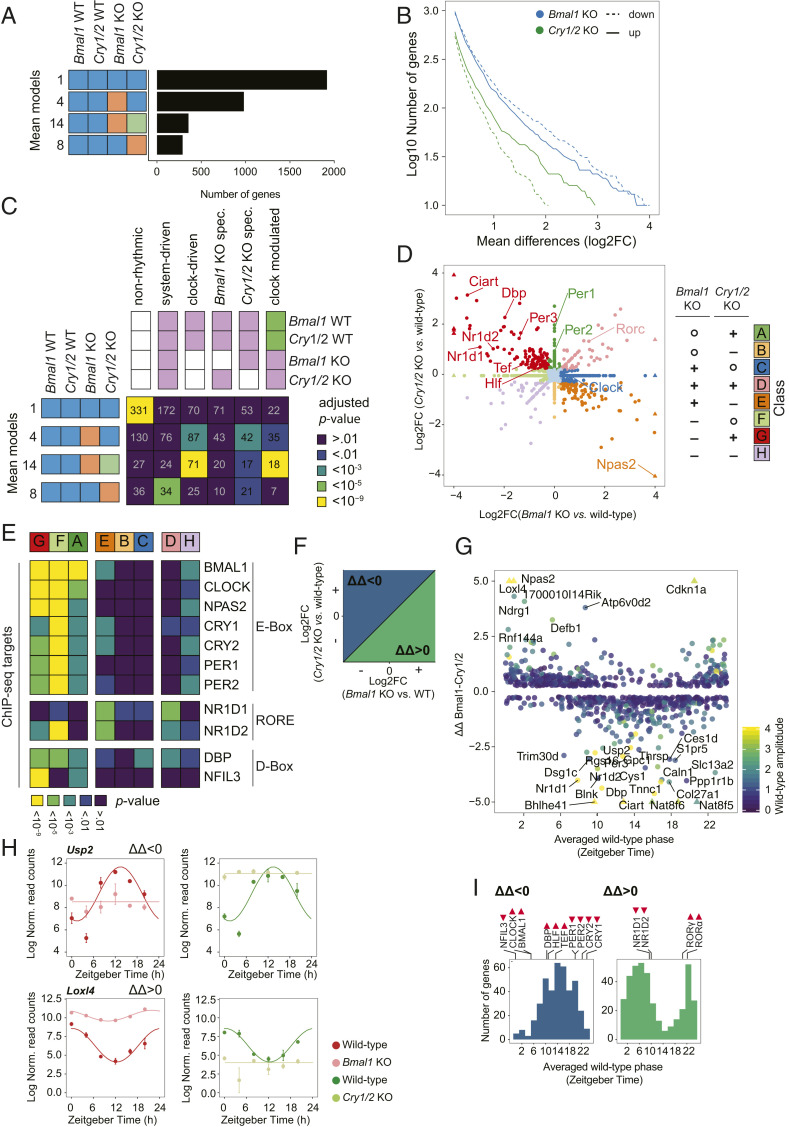
Fig. 3.
Differential mean expression between KOs of the positive and negative limb can predict rhythmic parameters of gene expression. (A) Number of genes classified in mean models under NRF in Bmal1 KO and Cry1/2 KO. The same color indicates shared mean expression levels between samples. (B) Cumulative number of genes that are up-regulated and down-regulated in mean expression with a log2 fold-change larger than the value on the x axis between the indicated KO and WT. (C) Number of genes corresponding to the indicated mean and rhythm model. Genes that show constant mean expression levels in both KO models are more likely to display no rhythmicity. Genes with differences in mean levels in Bmal1 KO or Cry1/2 KO are more likely to also be rhythmically expressed. (D) Scatterplot of log2 fold-changes in mean levels in Bmal1 KO vs. Cry1/2 KO. The color represents classes that group genes according to their differential expression pattern in the two KOs (+, up-regulation; −, down-regulation; o, not differentially expressed). (E) Enrichment analysis of clock gene targets for differentially expressed genes in the indicated class. (F) Schematic of log2 fold-changes in mean levels in Bmal1 KO/WT vs. Cry1/2 KO/WT to indicate the meaning of a positive or negative ΔΔBmal1/Cry1/2 (ΔΔ). (G) Scatterplot of phase in WT mice in function of the difference of log2 fold-changes in mean levels of Bmal1 KO vs. WT and log2 fold-changes in mean levels of Cry1/2 KO vs. WT (ΔΔBmal1/Cry1/2). (H) Example of rhythmic genes expression patterns with differing mean levels. (I) Phase histogram of genes with a positive or negative ΔΔBmal1/Cry1/2. Time of peak (▲) or trough (▼) of clock protein levels in liver nuclei (43) are indicated as triangles above the histograms.
Next, we elucidated the relationships between changes in mean levels in the KOs and rhythmicity in liver gene expression. Genes that shared their mean expression across all four conditions were more likely nonrhythmic. Conversely, genes that showed differential mean levels between KO and WT were enriched among clock-driven and clock-modulated rhythmic genes (Fig. 3C). We next compared mean gene expression changes in both KOs and grouped genes accordingly (Fig. 3D). Target genes of E-box– or D-box–binding core clock and clock output proteins (i.e., BMAL1, CLOCK, NPAS2, CRY1/2, PER1/2, DBP, and NFIL3) were down-regulated in Bmal1 KO mice and derepressed in Cry1/2 KOs (Fig. 3 D and E). On the other hand, genes regulated by the BMAL1-regulated repressors NR1D1 and NR1D2 showed opposite mean differences (Fig. 3 D and E). Interestingly, among core clock and clock-related genes, only Npas2 and Rorγ showed atypical patterns. Npas2 was the only clock gene that showed a clear up-regulation in expression in Cry1/2 KO and a down-regulation in Bmal1 KO (Fig. 3D and SI Appendix, Fig. S3), further supporting the idea that Npas2 is a NR1D1/2 and ROR target (44). Surprisingly, the mean levels of Rorγ were up-regulated in both KOs (Fig. 3D and SI Appendix, Fig. S3). Our observation points to a complex feedback regulation in the circadian oscillatory core loop, as supported by others (45). The reported regulation of Rorγ by both NR1D1/2 and BMAL1 might play a role in this complex regulation (46).
Finally, we established how quantitative differences in mean level between KO and WT correlated with rhythmic parameters such as phase and amplitude. To this end, we calculated a double-difference score (ΔΔBmal1/Cry1/2) summarizing the effects of changes in both Cry1/2 KO and Bmal1 KO (Fig. 3F). The absolute ΔΔBmal1/Cry1/2 value correlated with amplitude, while genes expressed at different phases showed a bias in positive or negative values (Fig. 3G). For example, genes expressed near ZT14 showed predominantly negative values. Conversely, the phase distributions differed depending on the ΔΔBmal1/Cry1/2. Indeed mRNA levels of genes with a negative ΔΔBmal1/Cry1/2 (e.g., Usp2) were peaking around ZT14 (Fig. 3 G–I), fitting to the reported activity (9, 47) and nuclear protein levels of E-box and D-box transcriptional factors (43). Genes with a positive ΔΔBmal1/Cry1/2 (e.g., Loxl4) exhibited phases reminiscent of RORα/γ and NR1D1/2 target genes (Fig. 3 G–I). Overall, opposing transcriptional changes in Bmal1 KO and Cry1/2 KO correlate with rhythmicity, higher amplitudes, and preferred peak phases.
PARbZip Target Genes Remain Rhythmic with Lower Mean Expression in the Liver of Hlf/Dbp/Tef KO Mice.
To interfere with further regulators of rhythmic gene expression, we extended our analysis to D-box transcriptional regulators (HLF, DBP, and TEF) that mediate outputs of the circadian clock (39, 48). To this end, Hlf/Dbp/Tef KO and WT mice were exposed to a light–dark cycle under an AL feeding regimen (Fig. 4A). Hlf/Dbp/Tef KOs exhibited an intact circadian locomotor activity, as previously shown (48), and conserved feeding rhythms comparable to those in WT under AL (Fig. 4B), unlike the core clock KO animals (Fig. 1C). Liver transcriptome analysis using dryR (sampled every 4 h, two independent replicates) showed that the lack of the circadian clock output regulators HLF, DBP, and TEF had little effect on the rhythmic expression patterns of circadian clock genes and showed only minor effects on the mean expression level of a few circadian clock genes (SI Appendix, Fig. S6 A and B and Dataset S3). Overall, the number of rhythmic transcripts in Hlf/Dbp/Tef KOs was 18% lower than in WT mice, uniformly across the entire range of amplitudes (Fig. 4C and SI Appendix, Fig. S6C). Specifically, amplitude and phase of most oscillating genes in WT (59%) were unaltered in Hlf/Dbp/Tef KOs while 28% of rhythmic genes lost rhythmicity in the KO and depended on the presence of PARbZip regulators (Fig. 4 D and E and SI Appendix, Fig. S6D); 10% of all rhythmic genes were oscillating only in Hlf/Dbp/Tef KOs (Fig. 4 D and E and SI Appendix, Fig. S6D). Unaltered rhythmic genes were not only genes showing promoter-bound E-box and RORE transcriptional regulators but unexpectedly also by D-box transcriptional regulators such as DBP and NFIL3 (Fig. 4F). Conversely, rhythmic genes that lost rhythms in PARbZip-deficient mice were not enriched in direct targets of D-box transcriptional regulators (Fig. 4F), hinting at indirect effects.
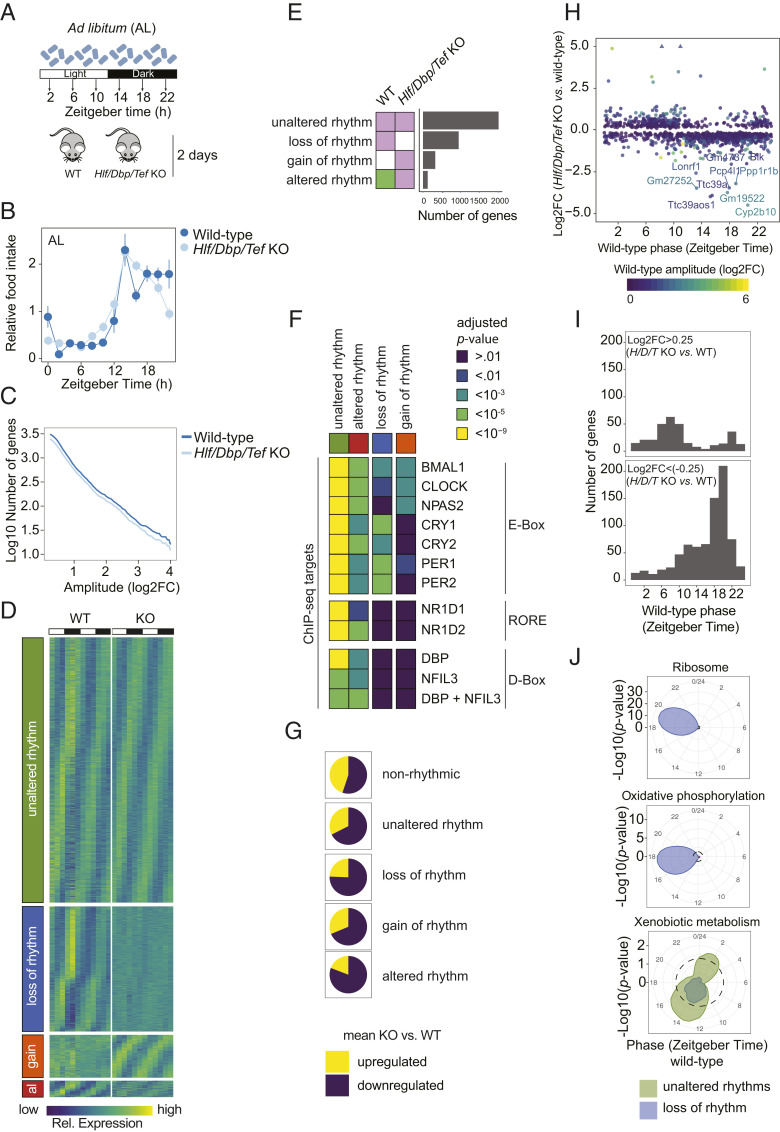
Fig. 4.
PARbZip target genes have reduced expression in KO animals but maintain rhythmicity. (A) Experimental design. (B) Rhythmic-feeding patterns in WT and Hlf/Dbp/Tef KO mice under an AL feeding regimen. (C) Cumulative number of genes classified in the indicated rhythmic model in function of minimal amplitude. (D) Heat maps of normalized rhythmic mRNA levels in the liver of WT and Hlf/Dbp/Tef KO mice. Genes were classified according to their temporal expression pattern in Hlf/Dbp/Tef KO compared with WT mice: unaltered rhythm, loss of rhythm, gain of rhythm (gain), and altered rhythm (al). (E) Number of genes classified in models according to their hepatic temporal gene expression pattern in Hlf/Dbp/Tef KO and WT mice. White indicates no rhythm detected, and the same color indicates shared rhythmic parameters (i.e., amplitudes and phase) between the indicated conditions. (F) Enrichment analysis of clock gene targets for differentially expressed genes in the indicated class (for details, see D). (G) Ratio of mean differential gene expression in the liver of the indicated rhythm model. (H) Scatterplot of phase in WT mice in function of log2 fold-changes in mean levels in Hlf/Dbp/Tef KO vs. WT mice. (I) Phase distribution of genes that are up-regulated (Top) or down-regulated (Bottom) in their mean expression in Hlf/Dbp/Tef KO compared with WT mice. (J) Enrichment of the indicated functional terms is represented by the radial coordinate at the indicated time point. P values were calculated by comparing the genes within a sliding window of 4 h with all expressed genes.
We next used dryR to assess differential mean level changes in the Hlf/Dbp/Tef KOs (SI Appendix, Fig. S6 E and F). Genes with a loss of rhythm, but surprisingly also those with an unaltered rhythm, were more often down-regulated than up-regulated in Hlf/Dbp/Tef KOs (Fig. 4G). Many of the strongly down-regulated rhythmic genes exhibited a peak in phase at ZT20, the phase typically shown by PARbZip target genes, whereas strongly up-regulated genes rather peaked in the opposite phase (Fig. 4 H and I).
We next checked which biological functions were overrepresented in the rhythmic mRNAs with altered mean expression level in Hlf/Dbp/Tef KOs. Major parts of the xenobiotic metabolic network were down-regulated but kept their rhythmic gene expression in Hlf/Dbp/Tef KOs (Fig. 4J and SI Appendix, Fig. S7 A and B). Some bona fide direct PARbZip target genes associated with xenobiotic metabolism (e.g., Alas, Por, Car [Nr1i3]) were down-regulated but maintained their rhythmic expression patterns in the KO (SI Appendix, Fig. S7A). Moreover, targets of the xenobiotic nuclear receptor CAR (NR1I3) also showed a similar expression pattern (e.g., Ces1d, Cyp2b10, Akl4) (SI Appendix, Fig. S7A). The second xenobiotic receptor PXR (NR1I2) and its target genes tended to keep rhythmic expression patterns in Hlf/Dbp/Tef KOs as well but showed a decreased expression level (SI Appendix, Fig. S7 A and B). The bile acid receptor FXR (NR1H4) is the only nuclear receptor that showed a robust loss of rhythms in Hlf/Dbp/Tef KOs and so did members of its target cascade, including Nr0b2 (Shp) and Cyp7a1 (SI Appendix, Fig. S7A), consistent with published results (49).
On the other hand, genes that lost rhythms in Hlf/Dbp/Tef KOs were associated with oxidative phosphorylation and ribosomes (Fig. 4J and SI Appendix, Fig. S7 C and D). Rhythmic levels of ribosome-associated genes have been linked to diurnal fluctuations of liver mass (50). We therefore tested whether a loss of liver size fluctuation in Hlf/Dbp/Tef KOs is associated with blunted rhythms in ribosome-associated genes. Indeed, Hlf/Dbp/Tef KOs lacked daily fluctuations and showed an overall larger liver mass (SI Appendix, Fig. S7E), potentially indicating that the two processes are interdependent. In conclusion, our studies revealed that disrupted PARbZip function resulted in a down-regulation of gene expression rather than a loss of rhythm of target genes. The observed loss of rhythms for some biological functions in the KO might be elicited by indirect effects such as a lack of liver size fluctuation.
NFIL3 and PARbZip Transcription Factors Cooperate to Drive Rhythmic Gene Expression in Liver.
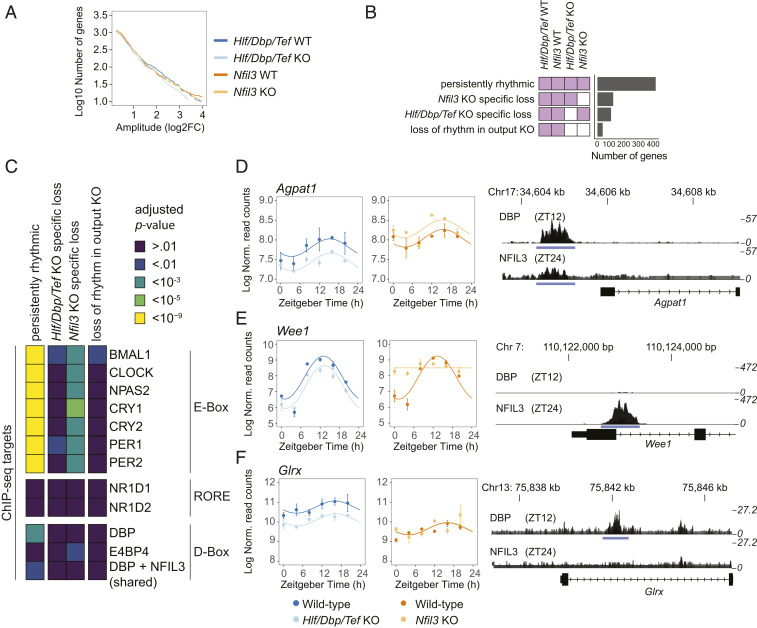
The D-Box–binding repressor NFIL3 (named also E4BP4) binds the same D-box motifs (51), and its mRNA was still rhythmically expressed in Hlf/Dbp/Tef KO mice (SI Appendix, Fig. S6A). Thus, NFIL3 potentially drives the remaining rhythmicity of PARbZip target genes in the liver of Hlf/Dbp/Tef KO mice. To test this assumption, we made use of a publicly available RNA-Seq dataset of Nfil3 KO mice fed AL and kept under a light–dark cycle (9). As in Hlf/Dbp/Tef KO mice, clock gene expression and the number of rhythmic liver genes was only slightly affected in the absence of Nfil3 (Fig. 5A and SI Appendix, Fig. S6 A and B). Correspondingly, most rhythmic genes in WT (48%) maintained their temporal expression pattern in both KOs (Fig. 5B). Considerably fewer rhythmic genes specifically lost rhythmicity in the respective KO model (13% for Hlf/Dbp/Tef KO and 15% for Nfil3 KO). Only 5% of rhythmic genes lost oscillatory expression in both KOs (Fig. 5B). These results support the hypothesis that NFIL3 can act as a driver of D-box target genes that maintain rhythmicity in Hlf/Dbp/Tef KO mice. In support of this, genes with D-box sites that are bound by DBP or both NFIL3 and DBP tended to be persistently rhythmically expressed across all KOs (Fig. 5C). An example of this scenario is the gene Agpat1 (Fig. 5D). Genes with exclusive NFIL3-bound D-boxes in their promoter region, such as Wee1, were prone to specifically lose rhythmicity in Nfil3 KO but still maintained rhythms in Hlf/Dbp/Tef KO mice (Fig. 5 C and E), further indicating that NFIL3 activity and its target genes are still rhythmic in these KO mice. However, the enrichment of DBP target genes in persistently rhythmic genes, as exemplified for Glrx (Fig. 5 C and F), also showed that NFIL3 alone might be not sufficient to maintain rhythmicity in Hlf/Dbp/Tef KO mice for some genes. These genes were still rhythmic even without binding of NFIL3 to a D-box. Together, Hlf/Dbp/Tef KO mice showed a down-regulation in mean expression levels for many bona fide PARbZip target genes, but rhythms persisted with lower mean expression levels. We hypothesize that NFIL3 is the likely driver for most of these genes with maintained rhythmicity in Hlf/Dbp/Tef KO mice. However, rhythmic NFIL3-independent genes exist, probably driven directly by the circadian clock as these genes also show direct binding by clock components (Fig. 5C).
Fig. 5.
Comparative analysis of differential liver expression in Hlf/Tef/Dbp KO and Nfil3 KO mice. (A) Cumulative number of rhythmic genes with amplitudes larger than the value on the x axis in the indicated genotype. (B) Number of genes classified in models according to their liver gene expression profile in Hlf/Dbp/Tef WT, Nfil3 WT, Hlf/Dbp/Tef KO, and Nifl3 KO mice. White indicates no rhythm detected, and the same color indicates shared rhythmic parameters (i.e., amplitudes and phase) between indicated conditions. (C) Enrichment analysis of core clock targets for genes classified into the indicated rhythmic model. Target genes have been identified using published ChIP-Seq data in mouse liver (see SI Appendix, SI Materials and Methods for details). (D–F) Gene expression over time (Left) and ChIP-Seq profiles of DBP and NFIL3 at the promotor region of Agpat1 (D), Wee1 (E), and Glrx (F) (Right) in mouse liver. ChIP-Seq profiles are derived from published datasets in mouse liver (9).
Discussion
This study provides the dryR method, implemented as an R package, to analyze rhythmicity in complex datasets comprising several conditions. We applied dryR to analyze newly generated mouse liver RNA-Seq datasets that allow genome-wide comparisons between circadian clock KOs of the positive and negative limb of the core clock loop under controlled feeding cycles. In addition, we also provided a time-resolved RNA-Seq dataset of Hlf/Dbp/Tef KO mice to examine the role of D-box–binding transcription factors for rhythmic gene expression. The analysis of these RNA-Seq datasets demonstrated the versatility of the statistical framework and also provided insights into circadian clock biology. To ease access, we made the RNA-Seq datasets and the statistical analysis available via a web application with an interactive interface (SI Appendix, Fig. S8 and https://clockprofile.epfl.ch).
Over the last two decades, many algorithms have been published to identify rhythmically transcribed genes in transcriptomic datasets designed to study circadian rhythms and related phenomena. These time-series transcriptomic datasets are often designed to compare rhythms between conditions (e.g., KO vs. WT or treated vs. untreated conditions). However, approaches designed to assess differential rhythmicity remain rare and are typically designed to test differences between two conditions (34–36). The more general dryR algorithm is a parametric model based on generalized harmonic regression and can perform rhythm detection with similar sensitivity and specificity as more specialized methods. In addition, dryR allows to simultaneously assess differential rhythmicity and differences in mean levels in more than two conditions. In contrast to other methods that use P values to indicate the presence of rhythmicity or differential rhythmicity of a certain feature, dryR provides likelihoods for a set of fitted models, which can then be converted to probabilities using established model-selection approaches, such as the Bayesian information criterion (BIC). We note that dryR is not designed to provide multiple test-corrected gene sets but is in essence a model-based clustering algorithm, where each gene is assigned to each model (Fig. 1A) probabilistically (using BIC weights). In this way, dryR can identify the most likely model for each gene expression pattern measured across different conditions and allows comparison of rhythmicity parameters such as mean, phase, and amplitude, which are shared among subsets of conditions. Moreover, although dryR was primarily designed for the analysis of RNA-Seq data, its implementation also allows analysis of rhythmic time series assuming normally distributed noise. This includes small-scale datasets, such as metabolite or body temperature measurements, and omics-scale datasets, including microarrays, metabolomics, or proteomics.
Feeding behavior in most mammals follows a pronounced circadian rhythmicity pattern with food mostly consumed during the activity phase (1). Mice are nocturnal animals and consume around 70% of the food during the night (52, 53). In line with others (12, 21, 54, 55), we observed that under a light–dark cycle, the absence of a functional molecular clock dampens rhythms in food intake under an AL paradigm. The loss of a functional SCN master clock in Bmal1 and Cry1/2 KO mice is likely the main reason for the observed alterations (56, 57). We have seen that these damped rhythms in turn change temporal gene expression in mouse liver. Thus, investigations of transcriptomes in clock-disrupted animal models should be performed under controlled feeding conditions to avoid confounding effects. This conclusion is not restricted to studies with clock-disrupted animal models, as many conditions alter temporal feeding patterns, including high-fat diet (58), calorie restriction (59), stress (60), and age (61).
Restoring the natural feeding rhythms of KO animals by NRF revealed that 33% of rhythmic genes were system-driven and that fewer genes (20%) were direct targets of the cell-autonomous molecular liver clock. Day-restricted feeding is insufficient to completely restore the loss of rhythmic gene expression caused by blunted feeding rhythms in clock-disrupted KO mice (21). In line with our findings, arrhythmic feeding in WT mice blunted the oscillation of the majority of rhythmic liver genes (23, 24). Conspicuously, clock-dependent rhythms had significantly higher amplitudes, which is consistent with our previous analysis of Bmal1 KOs (27). The number of feeding-dependent rhythmic genes that we observed in our study was higher than in previous reports where 90% of rhythmic genes in the liver are dependent on the cell-autonomous hepatic molecular clock (13, 37). Conversely, the restoration of the cell-autonomous liver clock in Bmal1 KO mice has been recently shown to rescue only 10% of rhythmic gene expression in the liver (62). This suggests that systemic rhythmic signals generated by hepatocytes can potentially have an impact on the clock in other tissues that, in turn, can feedback on the hepatocyte clock. In that context, clock disruption in hepatocytes reportedly alters rhythmic gene expression in nearby endothelial and Kupffer cells (63). It will be interesting to explore how cell-type–specific circadian clocks communicate with each other in complex tissues.
We observed that both the circadian clock and natural feeding rhythms are key drivers of rhythmic gene expression in the liver. Nevertheless, under a light–dark cycle, a subset of rhythmic liver genes oscillated even in the absence of a functional clock and rhythmic food intake. This observation suggested that clock-independent signals can drive the remaining rhythmicity of those genes, likely involving the SCN. The SCN can receive light signals via the retinohypothalamic tract (64) and, thus, keep a rhythmic activity under a light–dark cycle. Indeed, under a light–dark cycle, both Cry1/2 KO and Bmal1 KO mice reportedly exhibit rhythmic locomotor activity, which is a known output of the SCN (56, 57). Moreover, the ablation of the SCN attenuates rhythmicity of the liver transcriptome under constant darkness (65). We found that a rhythmic gene cluster of PPARα targets (SI Appendix, Fig. S5) and the mTOR signaling pathway (66) show a Cry1/2 KO-specific phase advance that follows the reported advance in SCN activity in Cry1/2 (67). This further supports the idea that SCN-borne signals drive the remaining liver rhythms in the absence of a functional circadian clock and natural feeding cycles. It will be interesting to examine how the SCN conveys rhythmicity to the liver transcriptome. Body temperature cycles might be one route. Indeed, rhythmic body temperature is conserved under constant feeding (24). In addition, we have seen that the temperature-sensitive transcript Cirbp (68) and the heat shock protein pathway (69) exhibit rhythmic expression profiles that were conserved across NRF and AL feeding regimens even in the absence of a functional clock. Another possible route is the release of glucocorticoids by the adrenal gland that is also dependent on SCN-borne cues (70, 71), and glucocorticoids have been shown to mediate rhythmic gene expression (29, 72, 73).
The analysis of gene expression in Hlf/Dbp/Tef KO and Nfil3 KO mice revealed that these clock output factors are involved in the transcriptional regulation of a limited number of genes involved in xenobiotic and lipid metabolism, as already described (39, 74). Surprisingly, PARbZip inactivation has a higher impact on mean expression level than on rhythmicity of the target genes (SI Appendix, Fig. S7F). This effect on expression levels revisits the role of PARbZip transcription factors in the regulatory landscape of circadian clock-regulated gene expression, identifying factors modulating mean expression levels instead of rhythmicity alone. As an example, while BMAL1 is able to regulate several aspects of xenobiotic detoxification (13), the PARbZip factors amplify this regulation and are required for normal liver detoxification (39). Another interesting finding is the role of the PARbZip factors in the regulation of the bile acid–FXR pathway, which loses rhythmicity in KO mice. While a direct PARbZip regulation has been suggested for example for Cyp7a1 (75), we did not detect binding of PARbZip transcription factors near the promoters of this gene in published ChIP-Seq datasets (9). The entire detoxification pathway is also regulated by bile acids (76). Bile acid elimination is controlled by the xenobiotic receptors PXR and CAR (77). Therefore, an indirect regulation through the deficient rhythmic elimination of bile acids is an alternative mechanism.
While our study has focused on the transcriptional regulation of circadian physiology, posttranscriptional (78) and posttranslational (79) regulation is equally important. Thus, future omics-scale investigations on the posttranscriptional and posttranslational level would be desirable and could provide more mechanistic insights into the role of feeding cycles and the circadian clock network on liver physiology. Overall, combining a powerful analysis framework with a comprehensive dataset of several clock KO models under controlled feeding regimens provides an integrated view on how the clock network and feeding cues work together in concert to drive rhythmic gene expression in the liver.
Materials and Methods
Details about the described procedures and data analysis are available at SI Appendix, SI Materials and Methods.
Mouse Experiments.
Male mice were kept under a 12-h light–12-h dark cycle. All mice used in the experiments were between 9 to 14 wk old. The generation of Bmal1 and Cry1/2 KO mice have been previously described (57, 66). Four days before sacrifice, Bmal1 and Cry1/2 KO mice had either free access to chow food (ad libitum [AL]) or only during the dark phase (night restricted feeding [NRF]). The generation of Hlf/Dbp/Tef KO mice has been already described (48). These animals were fed AL as feeding rhythms were not different from WT siblings (Fig. 4B). Mice were killed, and livers were dissected, snap-frozen in liquid nitrogen, and stored at −80 °C until further processing. The studies were conducted in accordance with the regulations of the veterinary office of the Canton of Vaud (experiments conducted at Nestlé Research) and Geneva (experiments conducted at University of Geneva).
Food-Intake Measurements.
Bmal1 and Cry1/2 KO mice were kept single-housed in phenotyping cages (TSE Systems) under a 12-h light/12-h dark cycle with an NRF and AL regimen as previously described (80). Food intake was measured every 15 min. Feeding frequency for Hlf/Dbp/Tef KO mice and WT littermates was measured as previously described (81).
RNA Extraction and RNA-Seq.
RNA extraction was performed as described (82). Libraries were prepared using the TruSeq Stranded mRNA Library Prep kit (Illumina) and sequenced on a HiSeq2500 or NextSeq500 (Illumina).
Supplementary Material
Acknowledgments
We thank Ueli Schibler for the utilization of PARbZip KO mouse data and Gijsbertus van der Horst for providing Cry1/2 KO mice. We thank José-Luis Sanchez-Garcia for technical support and Jake Yeung for providing insightful discussions. This work was supported by the European Research Council through individual Starting Grant ERC-2010-StG-260988 (to F.G.). F.G. receives support from the Institute for Molecular Bioscience, The University of Queensland. F.N. is supported by Swiss National Science Foundation Grant 310030_173079 and the Ecole Polytechnique Fédérale de Lausanne (EPFL). We thank Frederic Raymond and Sylviane Metairon from Nestlé Research and Bastien Mangeat and Lionel Ponsonnet from the Gene Expression Core Facility at EPFL for high-throughput sequencing.
Footnotes
Competing interest statement: B.D.W., C.G., F.A., E.M, A.C., and F.G. were employees of Société des Produits Nestlé SA.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015803118/-/DCSupplemental.
Data Availability.
Raw files and technical details about the RNA-Seq data have been deposited in NCBI's Gene Expression Omnibus (83) and are accessible through GEO Series accession nos. GSE135898 and GSE135875. The DryR code is available in GitHub athttps://github.com/naef-lab/dryR. Results and visualization are available at https://clockprofile.epfl.ch.
References
- 1.Atger F., Mauvoisin D., Weger B., Gobet C., Gachon F., Regulation of mammalian physiology by interconnected circadian and feeding rhythms. Front. Endocrinol. (Lausanne) 8, 42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rijo-Ferreira F., Takahashi J. S., Genomics of circadian rhythms in health and disease. Genome Med. 11, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astiz M., Heyde I., Oster H., Mechanisms of communication in the mammalian circadian timing system. Int. J. Mol. Sci. 20, 343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schibler U., et al. , Clock-talk: Interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb. Symp. Quant. Biol. 80, 223–232 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preitner N., et al. , The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Sato T. K., et al. , A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Gachon F., Physiological function of PARbZip circadian clock-controlled transcription factors. Ann. Med. 39, 562–571 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Yoshitane H., et al. , Functional D-box sequences reset the circadian clock and drive mRNA rhythms. Commun. Biol. 2, 300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mermet J., Yeung J., Naef F., Systems chronobiology: Global analysis of gene regulation in a 24-hour periodic world. Cold Spring Harb. Perspect. Biol. 9, a028720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinke H., Asher G., Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 20, 227–241 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Lamia K. A., Storch K. F., Weitz C. J., Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson B. P., et al. , Hepatocyte circadian clock controls acetaminophen bioactivation through NADPH-cytochrome P450 oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 111, 18757–18762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang E. E., et al. , Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 16, 1152–1156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmutz I., Ripperger J. A., Baeriswyl-Aebischer S., Albrecht U., The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 24, 345–357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamia K. A., et al. , Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimaldi B., et al. , PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 12, 509–520 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai K., Nishio T., Nakagawa H., Nakamura S., Fukuda Y., Effect of bilateral lesions of the suprachiasmatic nuclei on the circadian rhythm of food-intake. Brain Res. 142, 384–389 (1978). [DOI] [PubMed] [Google Scholar]
- 19.Stephan F. K., Zucker I., Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U.S.A. 69, 1583–1586 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatori M., et al. , Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vollmers C., et al. , Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 21453–21458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damiola F., et al. , Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mange F. et al.; CycliX Consortium , Diurnal regulation of RNA polymerase III transcription is under the control of both the feeding-fasting response and the circadian clock. Genome Res. 27, 973–984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenwell B. J., et al. , Rhythmic food intake drives rhythmic gene expression more potently than the hepatic circadian clock in mice. Cell Rep. 27, 649.e645–657.e645 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Hughes M. E., et al. , Guidelines for genome-scale analysis of biological rhythms. J. Biol. Rhythms 32, 380–393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeung J., Naef F., Rhythms of the genome: Circadian dynamics from chromatin topology, tissue-specific gene expression, to behavior. Trends Genet. 34, 915–926 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Yeung J., et al. , Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res. 28, 182–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weger B. D., et al. , The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab. 29, 362.e8–382.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weger B. D., et al. , Extensive regulation of diurnal transcription and metabolism by glucocorticoids. PLoS Genet. 12, e1006512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cederroth C. R., et al. , Circadian regulation of cochlear sensitivity to noise by circulating glucocorticoids. Curr. Biol. 29, 2477–2487.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrenko V., et al. , Pancreatic α- and β-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes Dev. 31, 383–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atger F., et al. , Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. U.S.A. 112, E6579–E6588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulido R. S., et al. , Neuronal activity regulates blood-brain barrier efflux transport through endothelial circadian genes. Neuron 108, 937–952.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons R., Parsons R., Garner N., Oster H., Rawashdeh O., CircaCompare: A method to estimate and statistically support differences in mesor, amplitude and phase, between circadian rhythms. Bioinformatics 36, 1208–1212 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Thaben P. F., Westermark P. O., Differential rhythmicity: Detecting altered rhythmicity in biological data. Bioinformatics 32, 2800–2808 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Singer J. M., Hughey J. J., LimoRhyde: A flexible approach for differential analysis of rhythmic transcriptome data. J. Biol. Rhythms 34, 5–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornmann B., Schaad O., Bujard H., Takahashi J. S., Schibler U., System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Druzd D., et al. , Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity 46, 120–132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gachon F., Olela F. F., Schaad O., Descombes P., Schibler U., The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4, 25–36 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., et al. , A proteomics landscape of circadian clock in mouse liver. Nat. Commun. 9, 1553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobel J. A. et al.; CycliX consortium , Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLoS Biol. 15, e2001069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canaple L., et al. , Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol. Endocrinol. 20, 1715–1727 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Wang J., et al. , Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 25, 102–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crumbley C., Wang Y., Kojetin D. J., Burris T. P., Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J. Biol. Chem. 285, 35386–35392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baggs J. E., et al. , Network features of the mammalian circadian clock. PLoS Biol. 7, e52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho H., et al. , Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koike N., et al. , Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gachon F., et al. , The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 18, 1397–1412 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiang J. Y., Kimmel R., Weinberger C., Stroup D., Farnesoid X receptor responds to bile acids and represses cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J. Biol. Chem. 275, 10918–10924 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Sinturel F., et al. , Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169, 651.e614–663.e614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitsui S., Yamaguchi S., Matsuo T., Ishida Y., Okamura H., Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 15, 995–1006 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurokawa M., Akino K., Kanda K., A new apparatus for studying feeding and drinking in the mouse. Physiol. Behav. 70, 105–112 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Gannon K. S., Smith J. C., Henderson R., Hendrick P., A system for studying the microstructure of ingestive behavior in mice. Physiol. Behav. 51, 515–521 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Adamovich Y., et al. , Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 19, 319–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turek F. W., et al. , Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bunger M. K., et al. , Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Horst G. T., et al. , Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Kohsaka A., et al. , High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Acosta-Rodriguez V. A., de Groot M. H. M., Rijo-Ferreira F., Green C. B., Takahashi J. S., Mice under caloric restriction self-impose a temporal restriction of food intake as revealed by an automated feeder system. Cell Metab. 26, 267–277.e262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang S. Z., Eiden L. E., Activation of the HPA axis and depression of feeding behavior induced by restraint stress are separately regulated by PACAPergic neurotransmission in the mouse. Stress 19, 374–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamada C., Mogami S., Hattori T., Psychological stress exposure to aged mice causes abnormal feeding patterns with changes in the bout number. Aging (Albany NY) 9, 2269–2287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koronowski K. B., et al. , Defining the independence of the liver circadian clock. Cell 177, 1448.e14–1462.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guan D., et al. , The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science 369, 1388–1394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hastings M. H., Maywood E. S., Brancaccio M., Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Akhtar R. A., et al. , Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12, 540–550 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Jouffe C., et al. , The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11, e1001455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Albus H., et al. , Cryptochrome-deficient mice lack circadian electrical activity in the suprachiasmatic nuclei. Curr. Biol. 12, 1130–1133 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Morf J., et al. , Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science 338, 379–383 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Reinke H., et al. , Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 22, 331–345 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buijs R. M., et al. , Anatomical and functional demonstration of a multisynaptic suprachiasmatic nucleus adrenal (cortex) pathway. Eur. J. Neurosci. 11, 1535–1544 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Ishida A., et al. , Light activates the adrenal gland: Timing of gene expression and glucocorticoid release. Cell Metab. 2, 297–307 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Oishi K., et al. , Genome-wide expression analysis reveals 100 adrenal gland-dependent circadian genes in the mouse liver. DNA Res. 12, 191–202 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Reddy A. B., et al. , Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology 45, 1478–1488 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Gachon F., et al. , Proline- and acidic amino acid-rich basic leucine zipper proteins modulate peroxisome proliferator-activated receptor alpha (PPARalpha) activity. Proc. Natl. Acad. Sci. U.S.A. 108, 4794–4799 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lavery D. J., Schibler U., Circadian transcription of the cholesterol 7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 7, 1871–1884 (1993). [DOI] [PubMed] [Google Scholar]
- 76.Goodwin B., et al. , A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 (2000). [DOI] [PubMed] [Google Scholar]
- 77.Wagner M., et al. , CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology 42, 420–430 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Wang J., et al. , Circadian clock-dependent and -independent posttranscriptional regulation underlies temporal mRNA accumulation in mouse liver. Proc. Natl. Acad. Sci. U.S.A. 115, E1916–E1925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mauvoisin D., Gachon F., Proteomics in circadian biology. J. Mol. Biol. 432, 3565–3577 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Jouffe C., et al. , Perturbed rhythmic activation of signaling pathways in mice deficient for Sterol Carrier Protein 2-dependent diurnal lipid transport and metabolism. Sci. Rep. 6, 24631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown S. A., Zumbrunn G., Fleury-Olela F., Preitner N., Schibler U., Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr. Biol. 12, 1574–1583 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Schmidt E. E., Schibler U., Cell size regulation, a mechanism that controls cellular RNA accumulation: Consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J. Cell Biol. 128, 467–483 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw files and technical details about the RNA-Seq data have been deposited in NCBI's Gene Expression Omnibus (83) and are accessible through GEO Series accession nos. GSE135898 and GSE135875. The DryR code is available in GitHub athttps://github.com/naef-lab/dryR. Results and visualization are available at https://clockprofile.epfl.ch.