Significance
Fruit aroma influences herbivory and food choice by humans, ultimately affecting seed dispersal and plant reproductive success. Despite the significance of scent, our understanding of the biosynthesis of odor-active volatiles is incomplete. Herein, we detail a plant pathway that uses pyruvate and acetyl-CoA to form citramalic acid and, through a series of recursive reactions that bypass regulation at threonine deaminase, enables 1-C α-ketoacid elongation and synthesis of isoleucine and straight and branched chain esters. The initiating enzyme, citramalate synthase, is a neofunctionalized form of 2-isopropylmalate synthase that is insensitive to feedback inhibition. Engagement of the “citramalate pathway” in ripening fruit provides for an elevated and persistent production of isoleucine and volatile esters as fruit tissues ripen, age, and senesce.
Keywords: fruit, citramalate, esters, isoleucine, ripening
Abstract
A plant pathway that initiates with the formation of citramalate from pyruvate and acetyl-CoA by citramalate synthase (CMS) is shown to contribute to the synthesis of α-ketoacids and important odor-active esters in apple (Malus × domestica) fruit. Microarray screening led to the discovery of a gene with high amino acid similarity to 2-isopropylmalate synthase (IPMS). However, functional analysis of recombinant protein revealed its substrate preference differed substantially from IPMS and was more typical of CMS. MdCMS also lacked the regulatory region present in MdIPMS and was not sensitive to feedback inhibition. 13C-acetate feeding of apple tissue labeled citramalate and α-ketoacids in a manner consistent with the presence of the citramalate pathway, labeling both straight- and branched-chain esters. Analysis of genomic DNA (gDNA) revealed the presence of two nearly identical alleles in “Jonagold” fruit (MdCMS_1 and MdCMS_2), differing by two nonsynonymous single-nucleotide polymorphisms (SNPs). The mature proteins differed only at amino acid 387, possessing either glutamine387 (MdCMS_1) or glutamate387 (MdCMS_2). Glutamate387 was associated with near complete loss of activity. MdCMS expression was fruit-specific, increasing severalfold during ripening. The translated protein product was detected in ripe fruit. Transient expression of MdCMS_1 in Nicotiana benthamiana induced the accumulation of high levels of citramalate, whereas MdCMS_2 did not. Domesticated apple lines with MdCMS isozymes containing only glutamate387 produced a very low proportion of 2-methylbutanol- and 2-methylbutanoate (2MB) and 1-propanol and propanoate (PROP) esters. The citramalate pathway, previously only described in microorganisms, is shown to function in ripening apple and contribute to isoleucine and 2MB and PROP ester biosynthesis without feedback regulation.
Esters are aroma impact compounds produced by many fruits and contribute notably to the sensory quality of apple (Malus × domestica) fruit, accounting for 80 to 95% of the total volatiles emitted (1). The esters hexyl acetate, butyl acetate, and 2-methylbutyl acetate are abundantly produced and considered to confer typical apple aroma characteristics (2, 3), which are perceived as “fruity” and “floral.” Volatile esters produced in apple fruit are largely composed of either straight-chain (SC) or branched-chain (BC) alkyl (alcohol-derived) and alkanoate (acid-derived) groups, which typically possess one to eight carbons (1). The final step of ester formation is the condensation of an alcohol and a CoA thioester by alcohol acyltransferase (AAT) (4). Surprisingly, despite the importance of aroma in fruit consumption, the biochemistry of ester formation is poorly understood.
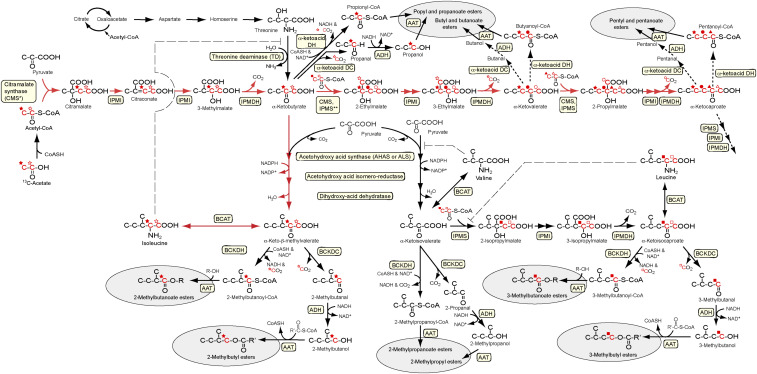
It has been suggested that ester precursors are produced primarily by degradative processes and that BC ester precursors, in particular, are derived from branched-chain amino acid (BCAA) degradation (5–9). In apples, isoleucine accumulates during apple fruit ripening, but valine and leucine do not (10–12). Correspondingly, esters related to isoleucine metabolism predominate, while those from valine can be detected only occasionally and usually at low levels, and no esters are produced from the leucine pathway (9, 13, 14). In plants, isoleucine is normally synthesized from threonine based on evidence for autotrophy in Nicotiana plumbaginifolia (15, 16). Threonine is deaminated to α-ketobutyrate by threonine deaminase (TD) (17), and α-ketobutyrate is subsequently metabolized to α-keto-β-methylvalerate, the isoleucine precursor, by three enzymes (Fig. 1). These same three enzymes form α-ketoisovalerate from pyruvate in the synthesis pathway for valine. Leucine synthesis begins with the valine precursor α-ketoisovalerate to form the leucine precursor α-ketoisocaproate.
Fig. 1.
Branched-chain amino acid metabolism, proposed citramalate pathway, and routes to ester biosynthesis. Citramalate-dependent pathway (in red) and its contribution to straight- and branched-chain ester biosynthesis. Adapted from Sugimoto et al. (40). The long dashed lines indicate feedback inhibition. The short dashed lines indicate not all reactions shown. Abbreviations: AAT, alcohol acyl-CoA transferase; ADH, alcohol dehydrogenase; BCAT, branched-chain amino transferase; BCKDC, branched-chain α-ketoacid decarboxylase; BCKDH, branched-chain α-ketoacid dehydrogenase; CMS, citramalate synthase; IPMDH, 3-isopropylmalate dehydrogenase; IPMI, 2-isopropylmalate isomerase; IPMS, 2-isopropylmalate synthase. An asterisk indicates the gene has been previously found in bacteria, but not in plants. The double asterisk indicates activity previously given the trivial name of 2-ethylmalate synthase (77). Hydrogens in carbon–hydrogen bonds are not shown. Carbons derived from the C-1 and C-2 positions of acetyl-CoA are, respectively, indicated with open and solid symbols adjacent to the carbon atoms.
The final reaction in the synthesis of isoleucine, valine, and leucine involves branched-chain aminotransferase (BCAT), which catalyzes a freely reversible reaction (Fig. 1). BC esters can be produced from exogenously supplied BCAAs, but also by the application of BC α-ketoacids (α-KEAs) (6). Given that BC α-KEAs are in approximate equilibrium with their respective BCAAs (18), it may be reasonable to expect that, for apple, the pool of isoleucine roughly mirrors the pool of its respective BC α-KEA. Therefore, the accumulation of isoleucine in apples during ripening may well be an indication of the content of its precursor, α-keto-β-methylvalerate. Furthermore, α-keto-β-methylvalerate is ultimately the direct precursor to the BC ester 2-methylbutyl acetate, an important aroma impact compound for apple (1).
Biosynthesis of all three BCAAs is responsive to feedback regulation. TD is inhibited by isoleucine, although this inhibition is antagonized by valine; acetohydroxyacid synthase is principally inhibited by valine and leucine; and 2-isopropylmalate synthase (IPMS) is inhibited by leucine (19–21). Given that isoleucine biosynthesis is under feedback regulation, the explanation for the exclusive accumulation of this amino acid in ripening apple fruit is not obvious. Sugimoto et al. (12) used this evidence to propose the existence of an alternative pathway for α-ketobutyrate formation in ripening apple fruit, whose first step involves the formation of citramalate.
The citramalate pathway has been described in several strains of bacteria for isoleucine biosynthesis (22–25). In this pathway, acetyl-CoA and pyruvate are substrates for the formation of citramalate by citramalate synthase (CMS) (Fig. 1). Several bacteria form (R)-citramalate, whereas yeast and apple form (S)-citramalate (26, 27). CMS is closely related to IPMS, which belongs to an acyltransferase family (EC 2.3.3). The acyltransferase family also includes citrate synthase, homocitrate synthase, malate synthase, and methylthioalkylmalate synthase (MAM). Each differs in substrate specificity, preferring, respectively, oxaloacetate, α-ketoglutarate, glyoxylate, α-ketoisovalerate, and various ω-methylthio-α-ketoalkanoates (28, 29).
In Leptospira interrogans, LiCMS (UniProtKB-Q8F3Q1) protein has a sequence similar to Mycobacterium tuberculosis MtIPMS, but unlike IPMS, its activity is specific to pyruvate as the α-KEA substrate (30, 31). In Arabidopsis, four genes in the IPMS family (IPMS1 [At1g18500], IPMS2 [At1g74040], MAM1 [At5g23010], and MAM3 [At5g23020]) have been characterized (28, 32, 33). The amino acid sequence identity is ∼60% between AtIPMS and AtMAM proteins (32) and the most significant difference is the presence of an additional 130-aa sequence in the C-terminal region in AtIPMS. This domain, called the “R-region,” is involved in leucine feedback inhibition in the yeast IPMS protein (LEU4) (34). AtIPMS and LiCMS enzymes are inhibited by leucine (32) and isoleucine (35), respectively; however, the lack of the R-region in AtMAM eliminates leucine feedback inhibition (36).
CMS proteins in Methanococcus jannaschii (UniProtKB-Q58787) and L. interrogans have been characterized for their activity and specificity (30, 37). In yeast, CMS activity is evident in both Saccharomyces cerevisiae (38) and Saccharomyces carlsbergensis (26), but no nucleotide or amino acid sequence for CMS has been identified as yet in the yeast genome database. In plants, Kroumova and Wagner (39) reported the involvement of one-carbon fatty acid biosynthesis (1-C FAB) in the formation of sugar esters in some (e.g., tobacco [Nicotiana tabacum] and petunia [Petunia × hybrida]), but not all, members of the Solanaceae and suggested that an α-KEA elongation pathway initiated by the condensation of acetyl-CoA and pyruvate enables 1-C FAB. However, there has been no molecular characterization of the entry point into the pathway via CMS, for example, in these or other plant species, nor, in fact, in any eukaryote.
Although previous works have suggested that catabolic pathways are primarily responsible for ester biosynthesis (5–8), the lack of supportive molecular and biochemical data suggests that a reassessment of this conceptual model is appropriate. The objective of this work was to evaluate whether the citramalate pathway operates in specialized plant organs like apple fruit and whether it contributes to the synthesis of BC and SC esters. Herein, we identify and characterize two MdCMS alleles and their translated protein isomers, demonstrate the presence of an active citramalate pathway in apple that includes α-KEA elongation, and link these elements to ester biosynthesis. Our work builds upon that of Hulme (27), who originally identified citramalate from plant (apple) extracts, and confirms the hypothesis of Sugimoto et al. (12, 40) regarding the existence of a citramalate pathway in plants that, in apple, contributes to ester biosynthesis.
Materials and Methods
Isotope Feeding Study with 13C Acetic Acid.
We studied the incorporation of 13C-labeled acetate (1-[13C] or 2-[13C] or 1,2-[13C2]) into esters and other metabolites synthesized by peel discs of “Jonagold” apple fruit. Methanol was added to the incubation solution to enhance the synthesis of methyl esters, which are normally present at extremely low levels in apple fruit (40). Thus, 13C-labeled methyl esters could be considered largely as being synthesized de novo during the experimental run. The incorporation of 13C into soluble intermediates and headspace volatiles was analyzed, respectively, by liquid (LC) and gas chromatography (GC) coupled with mass spectrometry (MS) as described in SI Appendix, Materials and Methods. Isotopologs of headspace volatiles and soluble acids were quantified by integrating chromatogram peaks for molecular or unique ions. Mass isotopolog (M+1 to M+5) enrichment was calculated relative to the unlabeled mass fraction (M). The expected position of the isotopic carbon from labeled acetate in the various compounds of interest is predicted in the proposed pathway (Fig. 1).
Developmental Changes in MdCMS, MdIPMS1, and MdIPMS2 Expression and Citramalate Content.
To determine the developmental patterns of gene expression and citramalate content, eight developmental stages of Jonagold apple fruit were selected. Developmental stages represent multiple time points from the early preclimacteric through ripening and senescence over a period of 81 d at room temperature as previously described (40) and further detailed in SI Appendix, Materials and Methods.
Plant material, volatile and metabolite analysis, RNA isolation, and microarray printing, design, labeling, and statistical analysis.
For each developmental stage, 20 apples were randomly chosen and their internal ethylene content was used to select those for further analysis (RNA isolation, gene expression, aroma profile, protein, and metabolite) as described in SI Appendix, Materials and Methods. Ripening-related genes were identified by untargeted screening using custom microarrays created from “Mutsu” apple as described in SI Appendix, Materials and Methods. The initial tentative identity for citramalate synthase was IPMS during the gene screening, but the designation MdCMS was adopted following protein functional analysis and is used hereafter.
Determination of mRNA transcript levels by RT-PCR.
The expression of MdCMS (a mixture of alleles MdCMS_1 and _2), MdIPMS1, MdIPMS2, and 18S ribosomal RNA (18s rRNA) was measured on fruit, leaf, root, and stem tissues from Jonagold apple trees using semiquantitative RT-PCR analysis as described in SI Appendix, Materials and Methods using primers listed in SI Appendix, Table S1.
Citramalate and 2-isopropylmalate analysis.
Citramalate and 2-isopropylmalate content was determined for fruit skin tissues using a MS (Quattro Premier XE; Waters Corporation) coupled to an ultraperformance LC (Acquity; Waters Corporation). MS and chromatographic identification and quantification protocols identical to those previously published (40). Data were collected and analyzed with proprietary software (MassLynx 4.1 and QuanLynx; Waters).
MdCMS and MdIPMS Cloning, Identification, and Sequencing from Jonagold.
Total RNA from Jonagold fruit tissues was used to identify and clone MdCMS_1 and _2, and MdIPMS1 and 2 sequences. Identification and sequence determination and assembly of mRNA and genomic DNA, and construction of cDNA are described in SI Appendix, Materials and Methods and using primer sequences listed in SI Appendix, Table S2.
MdCMS Allelic Composition and Association with Volatile Profile.
Genomic DNA was extracted from 99 apple lines and the nucleotide identity for a SNP (C or G) at base 2488 of MdCMS, responsible for a nonsynonymous amino acid substitution (respectively, glutamine387 or glutamate387), was determined to learn whether the line was homozygous or heterozygous for MdCMS_1 and _2 as described in SI Appendix, Materials and Methods and using primer sequences listed in SI Appendix, Table S3. The volatile profile for ripe fruit from these lines (40) was used to associate 1-propanol– and propanoate-derived (PROP) and the branched chain 2-methylbutanol- and 2-methylbutanoate-derived (2MB) ester phenotype with MdCMS allelotype.
Expression of MdCMS and MdIPMS in Escherichia coli and Protein Purification.
Coding sequence analysis was as described in SI Appendix, Materials and Methods. Open reading frame (ORF) with or without the putative chloroplast transit peptide of gel-purified MdCMS_1 and _2 and MdIPMS1 and 2 PCR products were cloned directly into the PET101/D-TOPO (Invitrogen) vector, mobilized into BL21(DE) E. coli cells, and protein produced and purified as described in SI Appendix, Materials and Methods.
MdCMS and MdIPMS Protein Characterization.
Enzyme assay and kinetics for MdCMS and MdIPMS.
Activity and substrate preference assays for MdCMS_1 and _2 and MdIPMS1 and 2 were performed using a 5,5-dithio-bis-(2-nitrobenzoic acid) (DTNB) endpoint assay as described by de Kraker et al. (32) with modifications (SI Appendix, Materials and Methods) for 12 α-KEAs (Table 1). Enzyme kinetics to determine Km and Vmax values for different α-KEA substrates and acetyl-CoA were performed as described in SI Appendix, Materials and Methods.
Table 1.
Activity of citramalate synthase (MdCMS_1 and MdCMS_2) and 2-isopropylmalate synthase (MdIPMS1 and 2) proteins and activity relative to use of pyruvate (pyr) as a substrate under saturating substrate conditions using the DTNB endpoint assays described in SI Appendix, Materials and Methods
| MdCMS_1 activity | MdCMS_2 activity | MdIPMS1 activity | MdIPMS2 activity | |||||
| Substrate | μmol⋅min−1⋅g−1 | Relative to pyr, % | μmol⋅min−1⋅g−1 | Relative to pyr, % | μmol⋅min−1⋅g−1 | Relative to pyr, % | μmol⋅min−1⋅g−1 | Relative to pyr, % |
| α-Ketobutyrate | 485 ± 135 | 199 | 1.3 ± 1.9 | 5 | 260 ± 50 | 2,910 | 189 ± 32 | 3,060 |
| α-Ketoisovalerate | 32 ± 22 | 12 | 1.7 ± 1.3 | 6 | 824 ± 247 | 8,810 | 617 ± 47 | 9,720 |
| α-Keto-β-methylvalerate | 0 | 0 | 1.3 ± 1.9 | 5 | 13 ± 2.0 | 143 | 8 ± 11 | 70 |
| α-Ketoisocaproate | 1.8 ± 2.6 | 1 | 2.0 ± 2.8 | 7 | 1 ± 1.0 | 13 | 2.3 ± 3.2 | 20 |
| α-Ketohexanoic acid | 0 | 0 | 1.0 ± 1.4 | 3 | 6 ± 1.6 | 77 | 2.3 ± 3.2 | 20 |
| α-Ketooctanoic acid | 6.4 ± 9 | 2 | 1.7 ± 2.4 | 6 | 6 ± 8.9 | 113 | 2.3 ± 3.2 | 20 |
| Oxaloacetate | 89 ± 41 | 35 | 8.2 ± 1.5 | 27 | 9 ± 0.8 | 104 | 15.9 ± 0.0 | 245 |
| Glyoxylate | 1.8 ± 2.6 | 1 | 0.7 ± 0.9 | 2 | 8 ± 5.4 | 73 | 0 | 0 |
| α-Ketoglutarate | 0 | 0 | 1.7 ± 2.4 | 6 | 1 ± 1.7 | 7 | 1.1 ± 1.6 | 10 |
| α-Keto-γ-(methylthio)butyric acid | 0 | 0 | 0 | 0 | 10 ± 2.4 | 111 | 0 | 0 |
| Pyruvate | 245 ± 82 | 100 | 30.1 ± 1.2 | 100 | 11 ± 7.7 | 100 | 8.0 ± 4.8 | 100 |
| α-Ketovaleric acid | 104 ± 35 | 42 | 1.0 ± 1.4 | 3 | 391 ± 92 | 4,290 | 276 ± 53 | 4,500 |
The absorbance was corrected by subtracting the background of the identical enzyme assay mixture without α-ketoacids. Experiments were repeated two to three times for each substrate/enzyme combination.
Determination of pH and amino acid feedback regulation.
The optimum pH range and feedback regulation for MdCMS_1, MdIPMS1 and 2 by BCAAs and threonine were determined as described in SI Appendix, Materials and Methods. The impact of pH on activity was determined using several buffers spanning pH 5.5 to 10.5. To determine inhibition by BCAAs, activity was measured following addition of valine, leucine, isoleucine, and threonine at concentrations ranging 0 to 10 mM. Pyruvate was used as the α-keto substrate for MdCMS_1 and _2, and α-ketoisovalerate was used for MdIPMS1 and 2.
Subcellular localization of MdCMS and MdIPMS.
Tobacco (N. tabacum cv. Samson) was used for transient expression assays. The MdCMS_1 and two MdIPMS vectors were generated as described in SI Appendix, Materials and Methods and were spliced into pEarleyGate101 (35S-Gateway-YFP-HA tag-OCS 3′; ABRC stock no. CD3-683) and mobilized into Agrobacterium tumefaciens strain EHA105 as described in SI Appendix, Materials and Methods. MdCMS- or MdIPMS-pEarleyGate 101 in Agrobacterium was syringe-infiltrated into leaves and the infiltrated areas were analyzed after 3 d by confocal microscopy as described by Reumann et al. (41).
Transient expression in Nicotiana benthamiana.
N. benthamiana plants were transfected with Agrobacterium tumefaciens strain EHA105 and induced to transiently express MdCMS_1 and _2 and MdIPMS2. The quantity of citramalate, leucine, isoleucine, threonine, and valine was determined by GC/MS relative to wild-type, mock, and empty vector constructs 5 d after infiltration as described in SI Appendix, Materials and Methods.
Yeast complementation.
A TD knockout was created in the yeast strain YMRX-3B (leu4Δ and leu9Δ, kindly donated by Enrico Casalone, Università di Chieti, Chieti, Italy) to prevent isoleucine synthesis and test MdCMS function by complementation as described in SI Appendix, Materials and Methods. Oligonucleotide primers to create the TD knockout line are listed in SI Appendix, Table S4.
Identification of IPMS and CMS in Rosaceae.
MdCMS_1, and MdIPMS1 and 2 protein sequences lacking predicted chloroplast targeting peptides were used as a query for tblastn searches against Rosaceae nucleotide databases (https://www.rosaceae.org/). Default parameters were used: e-value threshold: 0.001; word size: 3; max matches in a query range: 0; scoring matrix: PAM30; gap costs: existence:7 extension:2. Results are listed in SI Appendix, Table S5.
Identification of MdCMS Protein In Situ.
Purified recombinant MdCMS_1 protein was separated using two-dimensional (2D) electrophoresis/SDS-PAGE, and fragment peptides from MdCMS_1 were identified by a quadrupole time-of-flight LC/MS instrument (XeVo; Waters). To confirm the presence of MdCMS protein in apple fruit, MdCMS protein fragments were sought in protein preparations from ripening “Golden Delicious” (Malus × domestica Borkh.) apple fruits. The detailed protein preparations and identification procedures are described in SI Appendix, Materials and Methods.
Protein Sequence Alignment and Phylogenetic Tree.
Protein sequence alignment and phylogenetic tree were developed for CMS and IPMS as they related to bacterial CMS and eukaryotic IPMS and MAM. Additional protein sequence comparisons were also performed for CMS and IPMS in members of the Rosaceae family. Procedures for analysis are as described in SI Appendix, Materials and Methods.
Results
13C-Acetate Feeding.
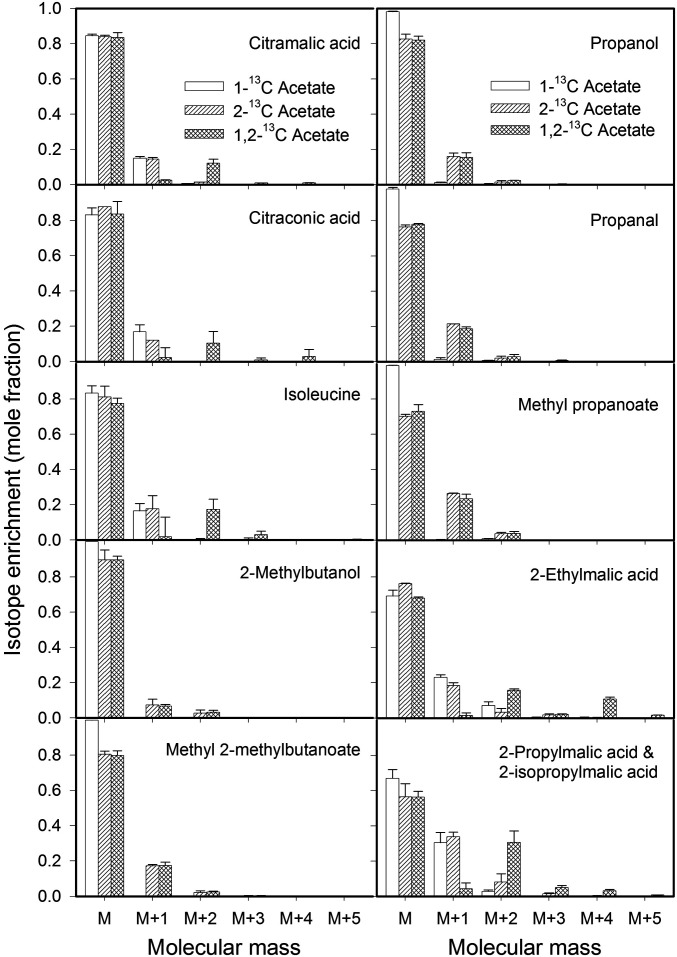
Previous work in plant and nonplant systems (22–24, 41–45) was used to predict differential labeling of citramalate and other metabolites in the citramalate pathway from 1-[13C], 2-[13C], and 1,2-[13C2]-acetate feeding (Fig. 1). Consistent with the proposed pathway, the labeled carbon from 13C-labeled acetate was incorporated into citramalate, citraconate, isoleucine, 2-ethylmalate, 2-propylmalate (and/or 2-isopropylmalate), 1-propanal, 1-propanol, methyl propanoate, methyl 2-methylbutanoate, and 2-methylbutanol (Fig. 2). Citramalate was distinguishable from 2-hydroxyglutarate as a derivatized analyte based on retention time and mass spectra. As predicted in the proposed pathway, the molecular mass for citramalate, citraconate, isoleucine, 2-ethylmalate, and 2-propylmalate and/or 2-isopropylmalate increased by 1 Da when fed with 1-[13C] or 2-[13C]-acetate and increased mass by 2 Da when fed with 1,2-[13C2] acetate. Also as predicted for 1-propanal, 1-propanol, methyl propanoate, methyl 2-methylbutanoate, and 2-methylbutanol, the mole fraction of M+1 was enriched more when fed 2-[13C] acetate than 1-[13C] acetate.
Fig. 2.
Mass isotopolog distribution of acids, alcohols, aldehydes, and esters from apple discs fed with 1-[13C] acetate, 2-[13C] acetate, and 1,2-[13C2] acetate. The isotope distribution (in mole fraction) is expressed as unlabeled mass (M) and one mass unit heavier than the unlabeled mass (M+1) up to five mass units heavier (M+5) than the unlabeled compound. Except for citraconic acid analysis, where 2-[13C] acetate incorporation yielded only one sample that was quantifiable for the acid, there were two biological replicates for each data point. The vertical bars represent the SD of the mean.
1,2-[13C2] acetate incorporation into 2-ethylmalate and 2-propylmalate (and/or 2-isopropylmalate, indistinguishable since they share the same molecular mass) brought about an increase of two (M+2) and four (M+4) mass units, which, respectively, were interpreted as being indicative of one and two cycles of acetyl-CoA addition via the proposed pathway. 1,2-[13C2] acetate incorporation into M+3, M+4, and M+5 isotopologs of 2-propylmalate and/or 2-isopropylmalate, was consistent with the anticipated labeling of 2-propylmalate, but not 2-isopropylmalate. The formation of 2-ethylmalate has not been previously demonstrated in apple fruit. However, in yeast, 2-ethylmalate synthase activity has been described (46). The pattern of 1,2-[13C2] acetate incorporation into 2-ethylmalate (M+4) and 2-propylmalate (M+3, M+4, and M+5) in the current study is consistent with an active 1-C α-KEA chain-elongation process such as that described for yeast (42).
Labeling of the C-1 and C-2 positions of the butanoate portion of methyl butanoate was determined using fragment ion m/z 74, a product of McLafferty rearrangement (SI Appendix, Fig. S1). The proposed citramalate pathway predicts label (M+1 and M+2) incorporation only from the C-2 position of 2-[13C] and 1,2-[13C2] acetate, but label was detected from 1-[13C] acetate also, indicating label was derived, at least partially, through another pathway. Label in the molecular ion of 1-butanal, while low, demonstrated enrichment of M+1 by 1,2-[13C2] acetate, consistent with labeling of 1-butanol via a 1-C α-KEA elongation pathway analogous to the citramalate pathway (43). However, in that 1,2-[13C2] acetate yielded enrichment of M+4, it likely reflects incorporation via more than one pathway, perhaps via 2-C FAB.
The 13C-acetate enrichment of M+1 and M+2 in threonine was not reflective of the isoleucine labeling pattern, suggesting that the citramalate pathway, rather than threonine deamination, dominates in the formation of isoleucine in ripening apple (SI Appendix, Fig. S2).
Metabolite Analysis and CMS Gene Expression.
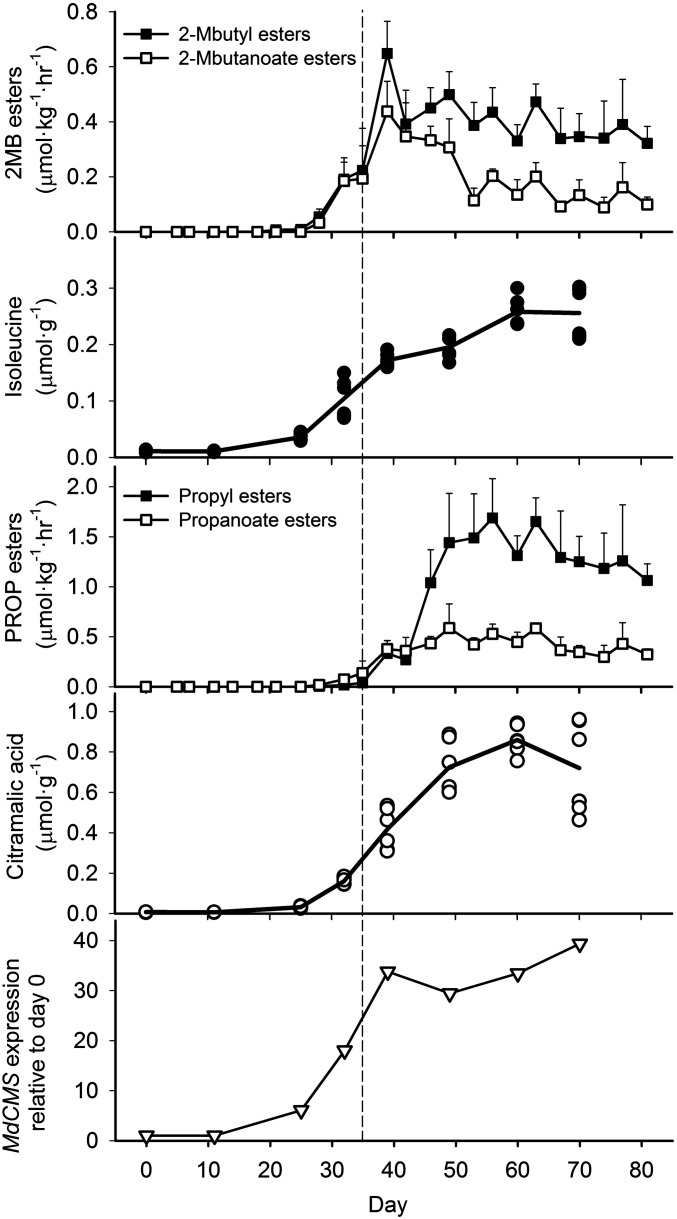
Citramalate content increased during ripening as the internal ethylene content rose above 0.1 µL·L−1 on day 25 (40) and paralleled increases in 2MB and PROP esters and isoleucine content (Fig. 3). Citramalate content was very low in unripe fruit and began to increase on day 25, increasing about 120-fold as ripening progressed. Citramalate levels remained high even during senescence.
Fig. 3.
Developmentally dependent changes in the production of branched-chain 2MB (2-methylbutanol- and 2-methylbutanoate-derived) and straight-chain PROP (1-propanol- and propanoate-derived) esters, citramalic acid, and isoleucine and in the gene expression of citramalate synthase (MdCMS) based on microarray analysis for “Jonagold” apple fruit during ripening and senescence at room temperature (21 ± 1 °C). The eight time points selected were based on the physiological stage of development (12). Symbols represent the average of four biological replications for ester analysis, three samples from each of two biological replicates (each from a pooled sample from the skin tissue of five fruit) for acid analyses, and two biological replications (each from a pooled sample from the skin tissue of five fruit) for gene expression. The vertical bars represent the SD of the mean. Expression for MdCMS is based on microarray data and is depicted relative to day 0.
MdCMS, initially annotated as IPMS, was highly expressed following the onset of fruit ripening based on microarray analysis (Fig. 3). Since leucine does not increase in ripening apple fruit (12), a large increase in IPMS expression was contraindicated. Therefore, we hypothesized that the annotation-based identification of IPMS was incorrect and that the protein may, in fact, be CMS. This interpretation had the potential to explain both isoleucine and citramalate accumulation in apple (40). The expression of MdCMS first increased on day 25, increasing further during ripening and remaining high during senescence. Semiquantitative RT-PCR analysis for MdCMS yielded an expression pattern similar to that found via microarray (SI Appendix, Fig. S3) and demonstrated that MdCMS was primarily expressed in ripening fruit (SI Appendix, Fig. S4). MdIPMS1 and MdIPMS2 differed from MdCMS in that MdIPMS1 and 2 expression in fruit was relatively constant from days 0 to 70. Expression of MdCMS increased with ripening in apple lines having high and low production of BC esters (SI Appendix, Fig. S5), suggesting that MdCMS expression alone was not responsible for the differences in ester phenotype.
MdCMS and MdIPMS Sequence Analysis.
Genomic sequence analysis of MdCMS revealed a total of eight introns within the ORF (SI Appendix, Fig. S6). Two MdCMS alleles were identified in Jonagold based on differences in length at the fourth intron and two SNPs in the coding region. The fourth intron length for MdCMS_1 was 224 bp and for MdCMS_2 was 101 bp. The MdCMS_1 and MdCMS_2 ORF nucleotide sequences differed at only two positions, bp 117 and bp 2488, yielding shifts from leucine36 and glutamine387 of MdCMS_1 to, respectively, proline36 and glutamate387 in MdCMS_2 (SI Appendix, Figs. S6 and S7). The inferred coding sequences of MdCMS_1 and _2 were 1,422 bp, about 280 bp shorter than Arabidopsis MAM genes (28). MdCMS had about 60% similarity with AtIPMS1 and 2 and AtMAM1 and 3 based on BLASTp analysis.
The two IPMS genes, MdIPMS1 and MdIPMS2, had coding sequences of 1,890 and 1,905 bp, respectively (SI Appendix, Fig. S8). These sequences were similar to those of the corresponding Arabidopsis IPMS genes, which are about 2,000 bp in length and had 93% nucleotide sequence similarity. The predicted IPMS protein sequences were only about 65% similar to MdCMS.
Alignment of MdCMS_1 and the two MdIPMS proteins with a selection of characterized plant IPMS and MAM, and LiCMS proteins revealed two shared domains containing motifs of GxGERxG and HxH[DN]D (SI Appendix, Fig. S7). The MdCMS and the MdIPMS proteins also contained chloroplast-targeting regions. MdIPMS1 and 2, like AtIPMS and LiCMS, contained an R-region reported to confer feedback inhibition by, respectively, leucine and isoleucine (32, 34, 35). However, MdCMS_1 was shorter than MdIPMS1 and 2 by ∼150 aa as it lacked the sequence corresponding to the R-region. The R-region is also lacking in AtMAMs and SlIPMS3 proteins and its omission is responsible for a lack of feedback regulation in those enzymes (29, 36). This feature likely explains the lack of inhibition by BCAAs on purified MdCMS_1 preparations (see below).
Phylogenetic tree analysis of the AA sequences sans organelle targeting and R regions showed that MdCMS clustered with the predicted pear (Pyrus communis) CMS, but was outside the plant IPMS and MAM clusters, and distant from microbial IPMS and CMS clusters (SI Appendix, Fig. S9). Within the Rosaceae family, close matches for MdCMS were only found in Pyrus species and not for the more distantly related cherry (Prunus avium), apricot (Prunus armeniaca), peach (Prunus persica), strawberry (Fragaria x ananassa), raspberry (Rubus occidentalis), and rose (Rosa chinensis) (SI Appendix, Table S5).
MdCMS Allelic Composition and Association with Volatile Profile.
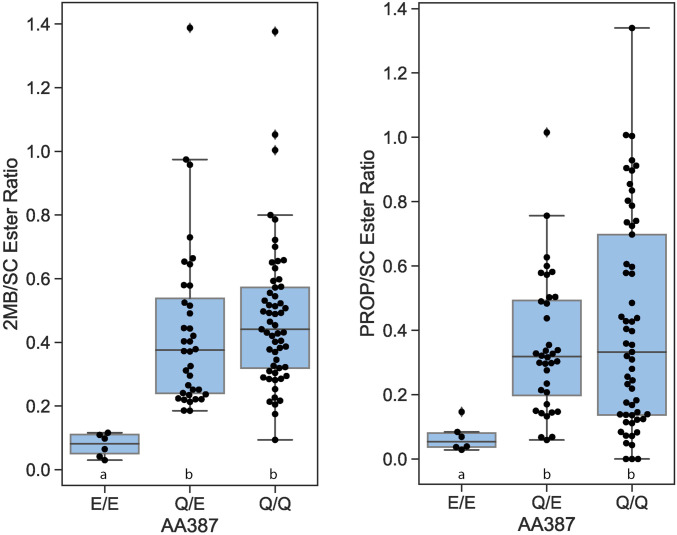
Sanger sequencing of MdCMS-specific PCR products for 99 domesticated apple lines revealed clear indications of G, mixed G and C, or C at base 2488. Respectively, this reflected MdCMS isozymes having only glutamine387 (homozygous MdCMS_1), both glutamine387 and glutamate387 (heterozygous for MdCMS_1 and _2), or only glutamate387 (homozygous MdCMS_2). Of the accessions tested, 56 lines were homozygous for glutamine387, 36 were heterozygous, and 6 lines were homozygous for glutamate387 (SI Appendix, germplasm_genotype_BCester.xls). Those lines possessing only isozymes with glutamate387 produced a much lower proportion of 2MB and PROP esters, suggesting that this allele has low or no in vivo activity (Fig. 4).
Fig. 4.
Ratio of 2-methybutyl and 2-methylbutanoate (2MB) (Left) and propyl and propanoate (PROP) (Right) ester moieties to straight-chain (SC) ester moieties for 99 apple lines classed according to the identity of amino acid 387 (AA387) for the two MdCMS isozymes, where glutamine387 (Q) is indicative of the functional MdCMS_1 and glutamate387 (E) is indicative of the nonfunctional MdCMS_2. SC esters are the sum of all alkyl and alkanoate ester elements having four or six carbons arranged in a SC. Significant differences between means (P < 0.05, least significance difference [LSD]) are denoted by different letters above the SNP designation.
MdCMS and MdIPMS Protein Characterization.
Full-length MdCMS_1 and 2 proteins with chloroplast target peptides, and MdIPMS1 and 2 proteins without chloroplast targeting regions were used for enzymatic assays as noted in SI Appendix, Materials and Methods. Predicted MdCMS and MdIPMS protein sizes of 52 and 62 kDa, respectively, were confirmed with SDS-PAGE gel analysis (SI Appendix, Fig. S10). The products of the enzyme assays for MdCMS_1 with pyruvate and MdIPMS1 with α-ketoisovalerate were verified to be citramalate and 2-isopropylmalate, respectively (SI Appendix, Fig. S11).
In endpoint DTNB assays with 12 different α-ketoacids, MdCMS_1 had the highest level of activity with α-ketobutyrate and pyruvate, the first two steps in the citramalate pathway (Table 1). This result is different from that observed for bacterial CMS proteins, which were essentially pyruvate-specific and did not have much activity with other α-KEAs tested (i.e., α-ketoisovalerate, α-ketobutyrate, and glyoxylate) (30, 31, 37). MdCMS_2 had very low overall activity and, while being highly specific for pyruvate, it had essentially no activity with other α-ketoacids evaluated, suggesting that it may not contribute to the formation of citramalate in vivo. MdIPMS1 and 2 had the highest activities with α-ketoisovalerate, α-ketovalerate, and α-ketobutyrate and had relatively low activity with pyruvate. When activity with pyruvate was used as a reference for comparing substrate preferences, the activity of MdIPMS1 and 2 for their favored substrate (α-ketoisovalerate) was roughly 9,000% that of pyruvate. The substrate preference for the two apple IPMS proteins was similar to IPMS from other species, but tended to have a lower relative activity with pyruvate (32, 47–51).
The enzyme kinetics of MdCMS_1 differed somewhat from CMS from nonplant organisms (MdCMS_2 was excluded from kinetic analysis due to its low level of activity). The Km of MdCMS_1 for pyruvate (2446 µM) was much higher than for previously reported bacterial CMS enzymes (31, 37). The Km of MdCMS_1 for acetyl-CoA was lower when pyruvate, rather than α-ketobutyrate, was used as a cosubstrate (Table 2). The Km values of MdCMS_1 and the two MdIPMS proteins for acetyl-CoA were relatively similar when α-ketobutyrate was the cosubstrate; however, MdCMS_1 had a lower Km for α-ketobutyrate than either MdIPMS1 or 2. The two MdIPMS proteins had low Km values for α-ketoisovalerate, and are highly specific for this substrate based on their catalytic efficiency, consistent with previous findings (32, 47–51). The Vmax of the reactions using α-ketobutyrate was similar for all three proteins, but those of the namesake reactions differed as much as fourfold from one another, with the Vmax of MdCMS_1 with pyruvate being lowest. The enzymatic efficiency of the three proteins was similar for α-ketobutyrate; however, MdCMS_1 had a relatively low enzyme efficiency with pyruvate compared to the efficiencies of the reaction of MdIPMS 1 and 2 with α-ketoisovalerate.
Table 2.
Kinetic parameters for citramalate synthase (MdCMS_1) and 2-isopropylmalate synthase (MdIPMS) using DTNB endpoint assay as described in SI Appendix, Materials and Methods
| Enzyme | Substrate | Km ± SE, μM | Vmax ± SE, μmol·min−1·g−1 | kcat ± SE, s−1 | kcat/Km ± SE, M−1·s−1 |
| MdCMS_1 | |||||
| Pyruvate | 2,446 ± 187 | 259 ± 27 | 0.22 ± 0.02 | 92 ± 3 | |
| Acetyl-CoA* | 6.6 ± 0.9 | 383 ± 36 | 0.33 ± 0.03 | 53,169 ± 10,928 | |
| α-Ketobutyrate | 3,559 ± 509 | 447 ± 214 | 0.39 ± 0.18 | 104 ± 37 | |
| Acetyl-CoA† | 11.3 ± 0.2 | 576 ± 75 | 0.50 ± 0.06 | 43,917 ± 4,796 | |
| MdIPMS1 | |||||
| α-Ketoisovalerate | 1,004 ± 44 | 1,059 ± 313 | 1.09 ± 0.32 | 1,106 ± 371 | |
| Acetyl-CoA‡ | 9.0 ± 1.9 | 900 ± 214 | 0.93 ± 0.22 | 102,341 ± 7,298 | |
| α-Ketobutyrate | 5,734 ± 796 | 478 ± 140 | 0.49 ± 0.14 | 82 ± 12 | |
| Acetyl-CoA† | 13.5 ± 1.6 | 288 ± 37 | 0.30 ± 0.03 | 22,083 ± 243 | |
| MdIPMS2 | |||||
| α-Ketoisovalerate | 706 ± 39 | 748 ± 89 | 0.77 ± 0.09 | 1,093 ± 91 | |
| Acetyl-CoA‡ | 8.6 ± 1.0 | 742 ± 90 | 0.77 ± 0.09 | 90,742 ± 9,753 | |
| α-Ketobutyrate | 6,287 ± 562 | 544 ± 19 | 0.56 ± 0.02 | 91 ± 11 | |
| Acetyl-CoA† | 10.4 ± 1.7 | 362 ± 61 | 0.37 ± 0.06 | 37,143 ± 6,932 |
Absorbance was corrected by subtracting the background of the identical enzyme assay mixture without α-ketoacids. Km and Vmax were determined by regression analysis of the Lineweaver–Burke plots for each substrate. There were two to three replications performed for each regression analysis.
In the presence of saturating (10 mM) pyruvate.
In the presence of saturating (10 mM) α-ketobutyrate.
In the presence of saturating (5 mM) α-ketoisovalerate.
The optimum pH range for MdCMS_1 activity was 9.0 to 9.5 and for the two MdIPMS proteins was between 8.0 and 9.0 (SI Appendix, Fig. S12). The activity of MdCMS_1 and MdIPMS enzymes was very low at pH 6.0, but gradually increased until pH 8.0 to 9.0, and then decreased sharply above pH 10.0.
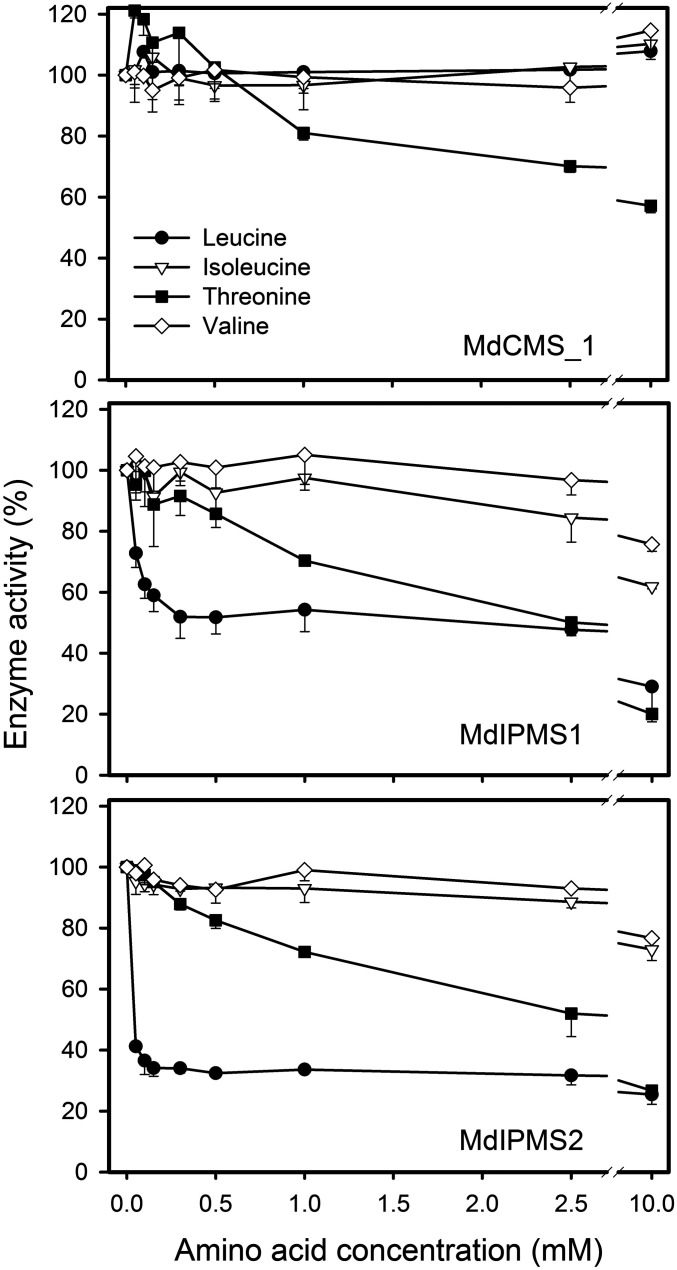
BCAA metabolism is typically regulated by feedback inhibition of the end product amino acid (19–21). However, none of the three BCAAs inhibited MdCMS_1 activity (Fig. 5). Interestingly, low threonine concentrations slightly stimulated MdCMS_1 activity and high threonine levels inhibited activity substantially (∼50%). This has not been reported for CMS in bacteria. The presence of as little as 0.05 mM leucine reduced MdIPMS1 and MdIPMS2 activities by 40% and 70%, respectively, and activity further decreased with increasing leucine concentration. MdIPMS2 was somewhat more strongly inhibited by leucine than was MdIPMS1. The concentration of leucine needed to achieve maximal inhibition of MdIPMS (0.10 to 0.3 mM) was similar to that for Neurospora (50) and yeast (48), but lower than that for Arabidopsis, which was maximally inhibited at 1 mM leucine (32). The activity of the two MdIPMS proteins was also inhibited by high concentrations of isoleucine and valine, decreasing 60 to 75%, whereas MdCMS_1 activity was not influenced. The inhibitory effect of high levels of valine and isoleucine on MdIPMS activity is also consistent with microbial IPMS enzymes (48, 50). Additionally, both MdIPMS proteins were inhibited by elevated levels of threonine, a finding not previously reported in plants or microorganisms (52). Unlike MdCMS_1, the activities of MdIPMS1 and 2 were not stimulated by low threonine concentrations.
Fig. 5.
Relative activity of MdCMS_1 and MdIPMS1 and 2 proteins as affected by branched-chain amino acids and threonine. The substrates pyruvate (10 mM) and α-ketoisovalerate (10 mM) were used for MdCMS and MdIPMS, respectively. Ellman's reagent endpoint assay was used with background correction. The vertical bars represent the SD from the mean for two replicate assays.
Transient expression of MdCMS_1 and MdIPMS1 and 2 proteins fused with YFP in tobacco indicated that these proteins are targeted to chloroplasts (SI Appendix, Fig. S13). This is consistent with previous reports for IPMS (53) from spinach (Spinacia oleracea) and AtMAM3 (33).
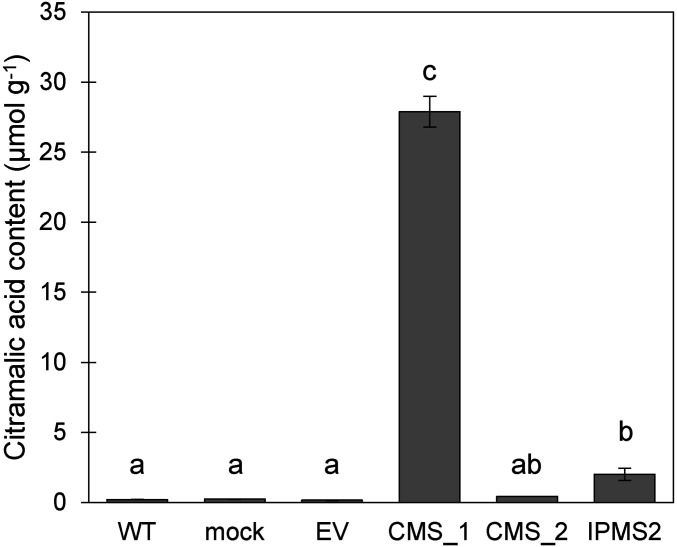
Transient expression of MdCMS_1 in N. benthamiana resulted in the production of very high levels of citramalate, but MdCMS_2 did not enhance citramalate accumulation over native levels (Fig. 6). MdIPMS2 expression led to the accumulation of 1/10th the amount of citramalate compared to MdCMS_2, reflective of its low activity with pyruvate. None of the enzymes impacted threonine, isoleucine, leucine, or valine levels (SI Appendix, Fig. S14).
Fig. 6.
Accumulation of citramalate in response to transient expression of MdCMS_1 (CMS_1), MdCMS_2 (CMS_2), and MdIPMS2 (IPMS2) in N. benthamiana. Controls include transient expression of an empty vector (EV), mock infiltration with buffer (mock), and wild type (WT). Whole transfected, mock, and WT leaves for three individual plants were sampled. Significant differences in means (P < 0.05, LSD) are denoted by different letters.
We attempted to complement a yeast strain lacking the ability to synthesize isoleucine. TD was deleted in the strain YMR-3B, which lacks LEU4 and LEU9, generating a triple-knockout YMRX-3B-TD that should have no citramalate synthase activity associated with IPMS. Unfortunately, the triple-knockout strain grew on the selective media, which lacked isoleucine, likely indicating that yeast has an additional pathway to synthesize isoleucine without the requirement of threonine.
Evidence for MdCMS_1 Translation in Apple Fruit.
Two-dimensional gels of His-tag purified MdCMS_1 products from E. coli yielded several spots with molecular mass consistent with MdCMS_1 when stained with Sypro Ruby (SI Appendix, Fig. S15). LC-MS/MS analysis of peptide fragments from indicated spots yielded sequences identical to predicted protein sequences for MdCMS_1 (SI Appendix, Table S6). Protein preparations from apple fruit yielded four peptides identical to MdCMS_1 fragments and the known CMS sequence (SI Appendix, Table S7, and highlighted and underlined, SI Appendix, Fig. S7). The detection of MdCMS protein in ripening apple confirmed that MdCMS is translated.
Discussion
Following its discovery in apple (41), citramalate has been detected in pear, banana (Musa acuminata), citrus (Citrus sinensis, Citrus limon, Citrus limettioides, Citrus paradise × Citrus reticulata, Citrus paradisi), tomato (Solanum lycopersicum), sugarbeet (Beta vulgaris), soybean (Glycine max, Glycine soja), Arabidopsis thaliana (54–61), and, in this paper, N. benthamiana. While several possible functions have been proposed for this compound, no role or roles have been demonstrated conclusively (54, 58, 62). Our study reveals a pathway for its formation and utilization as a precursor to isoleucine and aroma-active esters in ripening apple fruit.
The pattern and amount of citramalate accumulation in this study were similar to the findings of Hulme and Wooltorton (63). Concomitant increases in isoleucine, MdCMS expression, and PROP and 2MB esters, suggest a coordinated developmental process. In that apple ripening is entirely dependent upon ethylene action (10), it is likely that the induction of MdCMS and citramalate accumulation as they relate to ester biosynthesis are similarly ethylene dependent.
13C-Acetate Feeding.
The isotope enrichment pattern in response to 13C-acetate feeding of apple skin discs showed no evidence of dilution of 13C enrichment in the presumptive pathway connecting citramalate, citraconate, 2-ethylmalic acid, isoleucine, propanol, propanal, methyl propanol, and methyl 2-methylbutanoate, suggesting that this pathway may be the primary route of PROP and 2MB ester synthesis for some apple lines. This assertion is supported by the finding of Sugimoto et al. (40), who found isoleucine and PROP and 2MB ester production greatly diminished in apple lines having no increase in citramalate during ripening. The lack of label dilution in this study, in concert with previous findings of declining threonine with fruit ripening (12), argues against threonine as the primary substrate for the ripening-related increase in isoleucine and propyl and 2MB esters. Furthermore, if the label from the 1,2-[13C2] acetic acid in the feeding studies were via threonine, which originates from aspartate, it would yield a gain of more than two mass units for isoleucine (64).
Isoleucine Metabolism.
The existence of a citramalate pathway, bypassing threonine metabolism in ripening apple, would explain why isoleucine levels can increase markedly as threonine content decreases ∼90% during apple fruit ripening (10–12, 40). Normally, isoleucine inhibits its own synthesis via feedback inhibition of TD (21), which can cause an accumulation of threonine (65). The possibility of ripening-specific expression of a novel TD isozyme, insensitive to feedback inhibition, was considered. In this case, the threonine level might be expected to increase to support the observed accumulation of isoleucine. However, as noted previously, threonine levels declined during ripening (12). The labeling data, taken in conjunction with these findings, support the suggestion that synthesis of isoleucine and its derivative esters may be accomplished in part or in whole via a feedback-insensitive citramalate pathway that bypasses threonine in the synthesis of α-ketobutyrate.
The relative contributions of the TD- and CMS-dependent pathways to total α-ketobutyrate synthesis are as yet unknown. Likely, the feedback-sensitive TD pathway would meet metabolic needs associated with homeostatic plant metabolism, whereas the citramalate pathway may be more dedicated to specialized biochemistry. In this case, that would include the production of volatile metabolites at the end of life of a specialized plant reproductive organ to promote herbivory and subsequent seed dispersal (66).
The citramalic pathway, as described here, may be restricted to members of Maleae (e.g., Malus and Pyrus), since putative CMS genes were found in these and not other genera of Rosaceae. The evidence for a relatively recent genome duplication event in the Maleae tribe (67) may have provided an opportunity for specialization of IPMS. There are several instances of the development of specialized members of the IPMS family, at least two of which, MAM and IPMS3, include loss of a regulatory domain like MdCMS (28, 29, 33). Interestingly, in these instances, specialization takes the form of differing substrate preferences, producing glucosinolates in Brassicaceae (28, 33) and acylsugars in Solanaceae (29). In a study of E. coli IPMS (LeuA), Marcheschi et al. (68) identified four residues in the substrate binding pocket as important in size specificity. While these four residues are perfectly conserved in all 15 canonical IPMS proteins investigated (SI Appendix, Fig. S9), either three or four of the amino acids are altered in plant CMS and MAM proteins. In the case of MdCMS, all four have been modified (SI Appendix, Fig. S7).
Enzyme Activity.
The high activity of MdCMS_1 for pyruvate and the relatively low activity of IPMS1 and 2 for this substrate suggest that MdCMS_1 provides the entry point for carbon into the citramalate pathway. Nevertheless, the fact that MdCMS_1 and MdIPMS1 and 2 have relatively high levels of activity with α-ketobutyrate and α-ketovalerate suggests that both MdCMS and MdIPMS can carry out the chain-elongation steps in the α-KEA pathway beyond α-ketobutyrate. Label incorporation from 13C-acetate into 2-ethylmalate and 2-propylmalate in this study indicates some level of 1-C elongation of α-KEAs takes place in apple. Evaluation of the α-KEA elongation pathway in yeast demonstrated that α-ketobutyrate contributes to the production of 1-propanol, 1-butanol, and 1-pentanol via this pathway (69–71). Furthermore, in a bacterial system, the introduction of CMS activity markedly altered the metabolic profile. Transformation of E. coli with CimA from Methanococcus jannaschii enhanced 1-propanol and 1-butanol production via α-ketobutyrate by 9- and 22-fold, respectively (45).
The slightly higher catalytic efficiency of MdCMS_1 for α-ketobutyrate compared to pyruvate in apple stands in contrast to bacteria. Bacterial CMS has a relatively high specificity for pyruvate, perhaps reflecting the fact that isoleucine is synthesized exclusively via citramalate pathway in bacterial systems (72). The relatively high Km that MdCMS_1 has for pyruvate is curious. Reports of pyruvate levels in apple tissues are few; “Cox’s Orange Pippin” apple fruit extract was reported to contain about 12 µM pyruvate (73), and “Jonathan” apple fruit extract contains about 3 µM (74), well below the Km of MdCMS_1. The concentration of pyruvate in the chloroplast is unknown, however. Nevertheless, the fact that CMS uses two central metabolites of primary metabolism, acetyl-CoA and pyruvate, would suggest that even if substrates were low in concentration, they would not be limited by supply. The low catalytic efficiency of MdCMS may be important in managing an appropriate level of carbon flux through the citramalate pathway; the total carbon flux through BC esters in apple (0.6 to 1 μmol⋅kg−1⋅h−1) is roughly 1/1,000th that of respiratory CO2 production (12).
The substantially lower activity of MdCMS_2 and its high specificity for pyruvate compared to MdCMS_1 is striking in that the only amino acid sequence differences were the changes within Subdomain I (glutamine387 to glutamate387) and within the transit peptide sequence (leucine36 to proline36). The amino acid residue equivalent to MdCMS glutamine387 in IPMS, IPMS3, and MAM is conserved; it has not been previously demonstrated to be important for activity in those enzymes. Histidine386, the residue adjacent to glutamine387 in the conserved HxH[DN]D motif, however, has been implicated in acetyl-CoA binding and in catalysis (31, 75). Specifically, mutation of histidine386 to either an alanine or asparagine completely disrupts the enzymatic activity of LiCMS. These findings make it likely that mutation of glutamine387 to a negatively charged glutamate387 is sufficient to disrupt the activity of MdCMS_2.
Transient MdCMS Expression.
The accumulation of high levels of citramalate in N. benthamiana transiently expressing MdCMS_1 in the current study demonstrated that MdCMS_1 is functional in vivo. The lack of a similar boost in citramalate in response to the expression of MdCMS_2 confirms its suspected lack of functionality. Transient expression with MdIPMS2 also led to the accumulation of a small amount of citramalate, which is consistent with the small amount of citramalate synthase activity found for MdIPMS2. It is interesting that the accumulation of citramalate in N. benthamiana did not alter levels of isoleucine and may reflect a lack of activity of the 2-isopropylmalate isomerase of N. benthamiana with citramalate. The fact that apple apparently has no impediment at this step bears additional inquiry.
MdCMS_1 and MdCMS_2 Isozymes in Apple.
Knockout and complementation studies in a perennial crop like apple are difficult, expensive, and time-consuming, rendering proof-of-concept experiments impractical. However, it was possible to identify lines with no fully functional CMS by segregating those with only nonfunctional MdCMS isozymes containing glutamate387. This allelotype should be “rescued” by the presence of at least one allele of MdCMS coding for glutamine387. Of the 99 domesticated apple lines evaluated in this study, those lines containing only MdCMS isozymes with glutamate387 had a much lower proportion of 2MB and PROP esters compared to lines possessing at least one MdCMS isozyme with glutamine387. These data suggest that BC ester content in domesticated apple varieties appears, to a significant extent, to be a function of the MdCMS isoforms present. Very likely, additional diversity is brought about by variation in multiple additional factors including TD activity, ethylene sensitivity, MdCMS transcription and translation efficiency, and substrate availability.
The low frequency of the alleles with glutamate387 suggests it may be the subject of negative selection pressure. The link between MdCMS activity and the production of PROP and BC esters and their potential for impacting herbivory, including that by humans, creates a logical argument for this prospect. However, if true, it is not clear whether this is through selection in wild ancestors of modern apple cultivars or through selection via breeding programs.
Conclusion
The physiological, molecular, metabolic, enzymatic, isotopic, transgenic, and proteomic evidence presented here uniformly and consistently demonstrate the presence of a citramalate pathway in plants that, in apple fruit, contributes to the formation of aroma active esters during fruit ripening. The findings confirm the hypothesis of Sugimoto et al. (12) regarding existence of this pathway in plants and extend previous work by Kroumova et al. (76) to include the entry point into the 1-C α-KEA elongation pathway and further link this pathway to aroma formation. The citramalate pathway uses pyruvate and acetyl-CoA, products of primary metabolism, in a developmentally dependent synthetic process that is not feedback regulated. This is in contrast to previous assertions that ester biosynthesis is largely catabolic in nature. The implication is that this specialized plant organ, destined to senesce, need not catabolize existing cellular components (e.g., cell membranes) to engage in the aromatic “invitation to herbivory” that is intended to bring about consumption and seed dispersal.
Supplementary Material
Acknowledgments
A.D.J. acknowledges support from the US Department of Agriculture National Institute of Food and Agriculture, Hatch Project MICL02143 and NSF Grant DBI-0619489 for the purchase of the Quattro Premier LC/MS/MS instrument. N.S. acknowledges Dr. Jonathan Gershenzon from the Max Planck Institute for sharing his characterized clones of AtIPMS1 and 2 for our study. P.E. acknowledges the assistance of Dr. Cornelius Barry in developing transient expression assays. J.S. acknowledges Ms. Leslie Campbell Palmer for her technical assistance on proteomic analysis and support from Agriculture and Agri-Food Canada for funding by the A-Base research project (RPI 197). R.B. acknowledges support from Michigan AgBioResearch.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009988118/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Paillard N. M. M., “The flavour of apples, pears and quinces” in Food Flavours, Part C. The Flavour of Fruits, Morton I. D., Macleod A. J., Eds. (Elsevier, Amsterdam, The Netherlands, 1990), pp. 1–41. [Google Scholar]
- 2.Dimick P. S., Hoskin J. C., Review of apple flavor—state of the art. Crit. Rev. Food Sci. Nutr. 18, 387–409 (1983). [DOI] [PubMed] [Google Scholar]
- 3.Fellman J. K., Miller T. W., Mattinson D. S., Mattheis J. P., Factors that influence biosynthesis of volatile flavor compounds in apple fruits. HortScience 35, 1026–1033 (2000). [Google Scholar]
- 4.Ueda Y., Ogata K., Coenzyme A-dependent esterification of alcohols and acids in separated cells of banana pulp and its homogenate. Nippon Shokuhin Kogyo Gakkaishi 24, 624–630 (1977). [Google Scholar]
- 5.Wyllie S. G., Fellman J. K., Formation of volatile branched chain esters in bananas (Musa sapientum L.). J. Agric. Food Chem. 48, 3493–3496 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Gonda I., et al. , Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit. J. Exp. Bot. 61, 1111–1123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowan D. D., Lane H. P., Allen J. M., Fielder S., Hunt M. B., Biosynthesis of 2-methylbutyl, 2-methyl-2-butenyl, and 2-methylbutanoate esters in Red Delicious and Granny Smith apples using deuterium-labeled substrates. J. Agric. Food Chem. 44, 3276–3285 (1996). [Google Scholar]
- 8.Tressl R., Drawert F., Biogenesis of banana volatiles. J. Agric. Food Chem. 21, 560–565 (1973). [Google Scholar]
- 9.Ortiz A., Echeverría G., Graell J., Lara I., The emission of flavour-contributing volatile esters by “Golden Reinders” apples is improved after mid-term storage by postharvest calcium treatment. Postharvest Biol. Technol. 57, 114–123 (2010). [Google Scholar]
- 10.Defilippi B. G., Dandekar A. M., Kader A. A., Relationship of ethylene biosynthesis to volatile production, related enzymes, and precursor availability in apple peel and flesh tissues. J. Agric. Food Chem. 53, 3133–3141 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Nie L. C., Sun J. S., Di B., Changes in amino acid and fatty acid contents as well as activity of some related enzymes in apple fruit during aroma production. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 31, 663–667 (2005). [PubMed] [Google Scholar]
- 12.Sugimoto N., Jones A. D., Beaudry R., Changes in free amino acid content in “Jonagold” apple fruit as related to branched-chain ester production, ripening, and senescence. J. Am. Soc. Hortic. Sci. 136, 429–440 (2011). [Google Scholar]
- 13.Mattheis J. P., Buchanan D. A., Fellman J. K., Volatile compounds emitted by “Gala” apples following dynamic atmosphere storage. J. Am. Soc. Hortic. Sci. 123, 426–432 (1998). [Google Scholar]
- 14.Plotto A., McDaniel M. R., Mattheis J. P., Characterization of changes in “Gala” apple aroma during storage using OSME analysis, a gas chromatography-olfactometry technique. J. Am. Soc. Hortic. Sci. 125, 714–722 (2000). [Google Scholar]
- 15.Colau D., Negrutiu I., Van Montagu M., Hernalsteens J. P., Complementation of a threonine dehydratase-deficient Nicotiana plumbaginifolia mutant after Agrobacterium tumefaciens-mediated transfer of the Saccharomyces cerevisiae ILV1 gene. Mol. Cell. Biol. 7, 2552–2557 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidorov V., Menczel L., Maliga P., Isoleucine-requiring Nicotiana plant deficient in threonine deaminase. Nature 294, 87–88 (1981). [Google Scholar]
- 17.Binder S., Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 8, e0137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tewari Y. B., Goldberg R. N., Rozzell J. D., Thermodynamics of reactions catalysed by branched-chain-amino-acid transaminase. J. Chem. Thermodyn. 32, 1381–1398 (2000). [Google Scholar]
- 19.Eisenstein E., Cloning, expression, purification, and characterization of biosynthetic threonine deaminase from Escherichia coli. J. Biol. Chem. 266, 5801–5807 (1991). [PubMed] [Google Scholar]
- 20.Singh B. K., Shaner D. L., Biosynthesis of branched chain amino acids: From test tube to field. Plant Cell 7, 935–944 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wessel P. M., Graciet E., Douce R., Dumas R., Evidence for two distinct effector-binding sites in threonine deaminase by site-directed mutagenesis, kinetic, and binding experiments. Biochemistry 39, 15136–15143 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Gray C. T., Kornberg H. L., Enzymic formation of citramalate from acetyl-coenzyme A and pyruvate in Pseudomonas ovalis Chester, catalysed by “pyruvate transacetase.” Biochim. Biophys. Acta 42, 371–372 (1960). [DOI] [PubMed] [Google Scholar]
- 23.Hochuli M., Patzelt H., Oesterhelt D., Wüthrich K., Szyperski T., Amino acid biosynthesis in the halophilic archaeon Haloarcula hispanica. J. Bacteriol. 181, 3226–3237 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Risso C., Van Dien S. J., Orloff A., Lovley D. R., Coppi M. V., Elucidation of an alternate isoleucine biosynthesis pathway in Geobacter sulfurreducens. J. Bacteriol. 190, 2266–2274 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westfall H. N., Charon N. W., Peterson D. E., Multiple pathways for isoleucine biosynthesis in the spirochete Leptospira. J. Bacteriol. 154, 846–853 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sai T., Aida K., Uemura T., Studies on (−)-citramalic acid formation by respiration-deficient yeast mutants. V. Purification and some properties of citramalate condensing enzyme. J. Gen. Appl. Microbiol. 15, 345–363 (1969). [Google Scholar]
- 27.Hulme A. C., The isolation of l-citramalic acid from the peel of the apple fruit. Biochim. Biophys. Acta 14, 36–43 (1954). [DOI] [PubMed] [Google Scholar]
- 28.Textor S., et al. , Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: Recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 218, 1026–1035 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Ning J., et al. , A feedback-insensitive isopropylmalate synthase affects acylsugar composition in cultivated and wild tomato. Plant Physiol. 169, 1821–1835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H., et al. , Isoleucine biosynthesis in Leptospira interrogans serotype lai strain 56601 proceeds via a threonine-independent pathway. J. Bacteriol. 186, 5400–5409 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J., et al. , Molecular basis of the substrate specificity and the catalytic mechanism of citramalate synthase from Leptospira interrogans. Biochem. J. 415, 45–56 (2008). [DOI] [PubMed] [Google Scholar]
- 32.de Kraker J.-W., Luck K., Textor S., Tokuhisa J. G., Gershenzon J., Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol. 143, 970–986 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Textor S., de Kraker J. W., Hause B., Gershenzon J., Tokuhisa J. G., MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 144, 60–71 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavalieri D., et al. , Trifluoroleucine resistance and regulation of alpha-isopropyl malate synthase in Saccharomyces cerevisiae. Mol. Gen. Genet. 261, 152–160 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Zhang P., et al. , Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans. Biochem. J. 421, 133–143 (2009). [DOI] [PubMed] [Google Scholar]
- 36.de Kraker J.-W., Gershenzon J., From amino acid to glucosinolate biosynthesis: Protein sequence changes in the evolution of methylthioalkylmalate synthase in Arabidopsis. Plant Cell 23, 38–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howell D. M., Xu H., White R. H., (R)-citramalate synthase in methanogenic archaea. J. Bacteriol. 181, 331–333 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Losada M., Canovas J. L., Ruiz Amil M., Oxaloacetate, citramalate and glutamate formation from pyruvate in baker’s yeast. Biochem. Z. 340, 60–74 (1964). [PubMed] [Google Scholar]
- 39.Kroumova A. B., Wagner G. J., Different elongation pathways in the biosynthesis of acyl groups of trichome exudate sugar esters from various solanaceous plants. Planta 216, 1013–1021 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Sugimoto N., Forsline P., Beaudry R., Volatile profiles of members of the USDA Geneva Malus core collection: Utility in evaluation of a hypothesized biosynthetic pathway for esters derived from 2-methylbutanoate and 2-methylbutan-1-ol. J. Agric. Food Chem. 63, 2106–2116 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Reumann S., et al. , In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150, 125–143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekiel I., Smith I. C. P., Sprott G. D., Biosynthesis of isoleucine in methanogenic bacteria: A carbon-13 NMR study. Biochemistry 23, 1683–1687 (1984). [Google Scholar]
- 43.Feng X., et al. , Characterization of the central metabolic pathways in Thermoanaerobacter sp. strain X514 via isotopomer-assisted metabolite analysis. Appl. Environ. Microbiol. 75, 5001–5008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nesbakken T., Kolsaker P., Ormerod J., Mechanism of biosynthesis of 2-oxo-3-methylvalerate in Chlorobium vibrioforme. J. Bacteriol. 170, 3287–3290 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atsumi S., Liao J. C., Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl. Environ. Microbiol. 74, 7802–7808 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strassman M., Ceci L. N., A study of acetyl-CoA condensation with α-keto acids. Arch. Biochem. Biophys. 119, 420–428 (1967). [DOI] [PubMed] [Google Scholar]
- 47.Kohlhaw G. B., α-Isopropylmalate synthase from yeast. Methods Enzymol. 166, 414–423 (1988). [DOI] [PubMed] [Google Scholar]
- 48.Ulm E. H., Böhme R., Kohlhaw G., α-Isopropylmalate synthase from yeast: Purification, kinetic studies, and effect of ligands on stability. J. Bacteriol. 110, 1118–1126 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Carvalho L. P., Blanchard J. S., Kinetic and chemical mechanism of α-isopropylmalate synthase from Mycobacterium tuberculosis. Biochemistry 45, 8988–8999 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webster R. E., Gross S. R., The α-isopropylmalate synthetase of Neurospora. I. The kinetics and end product control of α-isopropylmalate synthetase function. Biochemistry 4, 2309–2318 (1965). [Google Scholar]
- 51.Kohlhaw G., Leary T. R., Umbarger H. E., α-Isopropylmalate synthase from Salmonella typhimurium. Purification and properties. J. Biol. Chem. 244, 2218–2225 (1969). [PubMed] [Google Scholar]
- 52.Wiegel J., Schlegel H. G., Leucine biosynthesis: Effect of branched-chain amino acids and threonine on α-isopropylmalate synthase activity from aerobic and anaerobic microorganisms. Biochem. Syst. Ecol. 5, 169–176 (1977). [Google Scholar]
- 53.Hagelstein P., Schultz G., Leucine synthesis in spinach chloroplasts: Partial characterization of 2-isopropylmalate synthase. Biol. Chem. Hoppe Seyler 374, 1105–1108 (1993). [DOI] [PubMed] [Google Scholar]
- 54.Degu A., et al. , Inhibition of aconitase in citrus fruit callus results in a metabolic shift towards amino acid biosynthesis. Planta 234, 501–513 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Fraser P. D., et al. , Manipulation of phytoene levels in tomato fruit: Effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19, 3194–3211 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudell D. R., Mattheis J. P., Curry E. A., Prestorage ultraviolet-white light irradiation alters apple peel metabolome. J. Agric. Food Chem. 56, 1138–1147 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Ulrich R., “Organic acids” in The Biochemistry of Fruits and Their Products, Hulme A. C., Ed. (Academic Press, London, 1970), pp. 89–118. [Google Scholar]
- 58.Khorassani R., et al. , Citramalic acid and salicylic acid in sugar beet root exudates solubilize soil phosphorus. BMC Plant Biol. 11, 121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiehn O., Kopka J., Trethewey R. N., Willmitzer L., Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 72, 3573–3580 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Oms-Oliu G., et al. , Metabolic characterization of tomato fruit during preharvest development, ripening, and postharvest shelf-life. Postharvest Biol. Technol. 62, 7–16 (2011). [Google Scholar]
- 61.Zhang J., Yang D., Li M., Shi L., Metabolic profiles reveal changes in wild and cultivated soybean seedling leaves under salt stress. PLoS One 11, e0159622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noro S., Kudo N., Kitsuwa T., Differences in sugars and organic acids between red and yellow apple cultivars at time of coloring, and effect of citramalic acid on development of anthocyanin. Hortic. J. 57, 381–389 (1988). [Google Scholar]
- 63.Hulme A. C., Wooltorton L. S. C., Determination and isolation of the non-volatile acids of pome fruits and a study of acid changes in apples during storage. J. Sci. Food Agric. 9, 150–158 (1958). [Google Scholar]
- 64.Jolchine G., Le metabolisme de l’acide acetique dans les feuilles de Bryophyllum diagremontianum Berger. La genese des acides organiques par differentes reactions de condensation du radical acetyle, dans les chloroplastes et dans la fraction cellulaire non chloroplastique [in French]. Bull. Soc. Chim. Biol. (Paris) 44, 337–364 (1962).14452175 [Google Scholar]
- 65.Martinez-Force E., Benitez T., Amino acid overproduction and catabolic pathway regulation in Saccharomyces cerevisiae. Biotechnol. Prog. 10, 372–376 (1994). [Google Scholar]
- 66.Cipollini M., Secondary metabolites of vertebrate-dispersed fruits: Evidence for adaptive functions. Rev. Chil. Hist. Nat. 73, 421–440 (2000). [Google Scholar]
- 67.Xiang Y., et al. , Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol. 34, 262–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marcheschi R. J., et al. , A synthetic recursive “+1” pathway for carbon chain elongation. ACS Chem. Biol. 7, 689–697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guymon J. F., Ingraham J. L., Crowell E. A., The formation of n-propyl alcohol by Saccharomyces cerevisiae. Arch. Biochem. Biophys. 95, 163–168 (1961). [DOI] [PubMed] [Google Scholar]
- 70.Vollbrecht D., Three pathways of isoleucine biosynthesis in mutant strains of Saccharomyces cerevisiae. Biochim. Biophys. Acta 362, 382–389 (1974). [DOI] [PubMed] [Google Scholar]
- 71.Ingraham J. L., Guymon J. F., Crowell E. A., The pathway of formation of n-butyl and n-amyl alcohols by a mutant strain of Saccharomyces cerevisiae. Arch. Biochem. Biophys. 95, 169–175 (1961). [DOI] [PubMed] [Google Scholar]
- 72.Zou Y., Guo X., Picardeau M., Xu H., Zhao G., A comprehensive survey on isoleucine biosynthesis pathways in seven epidemic Leptospira interrogans reference strains of China. FEMS Microbiol. Lett. 269, 90–96 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Hulme A. C., Smith W. H., Wooltorton L. S. C., Biochemical changes associated with the development of low-temperature breakdown in apples. J. Sci. Food Agric. 15, 303–307 (1964). [Google Scholar]
- 74.Wills R. B. H., McGlasson W. B., Changes in the organic acids of Jonathan apples during cool storage in relation to the development of breakdown. Phytochemistry 7, 733–739 (1968). [Google Scholar]
- 75.Kumar R., et al. , Molecular basis of the evolution of methylthioalkylmalate synthase and the diversity of methionine-derived glucosinolates. Plant Cell 31, 1633–1647 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kroumova A. B., Xie Z., Wagner G. J., A pathway for the biosynthesis of straight and branched, odd- and even-length, medium-chain fatty acids in plants. Proc. Natl. Acad. Sci. U.S.A. 91, 11437–11441 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Canovas J. L., Ruiz-Amil M., Losada M., Condensation of α-ketobutyrate and acetyl-CoA in baker’s yeast. Arch. Mikrobiol. 50, 164–170 (1965). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.