Significance
Phytochemical diversity affects plant fitness and is the source of numerous medicines. Despite this, we know remarkably little about how phytochemical diversity is distributed across the plant kingdom and the environment. To address this challenge, we coupled untargeted metabolomics on 416 grassland vascular plant species across Switzerland with phylogenetic information, species distribution modelling, and ensemble machine learning to construct a framework for comprehensively predicting landscape-scale phytochemical diversity of both known and currently unclassified molecules. We show that phytochemicals diversity and identity can be predicted in the landscape as a function of phylogenetic information, as well as of climatic, topographic, and edaphic factors. We demonstrate that it is thus possible to map hotspots of phytochemical diversity and phytochemical endemism across the landscape.
Keywords: plant secondary metabolites, landscape ecology, diversity hotspots, chemical ecology, alpine habitat
Abstract
To cope with environmental challenges, plants produce a wide diversity of phytochemicals, which are also the source of numerous medicines. Despite decades of research in chemical ecology, we still lack an understanding of the organization of plant chemical diversity across species and ecosystems. To address this challenge, we hypothesized that molecular diversity is not only related to species diversity, but also constrained by trophic, climatic, and topographical factors. We screened the metabolome of 416 vascular plant species encompassing the entire alpine elevation range and four alpine bioclimatic regions in order to characterize their phytochemical diversity. We show that by coupling phylogenetic information, topographic, edaphic, and climatic variables, we predict phytochemical diversity, and its inherent composition, of plant communities throughout landscape. Spatial mapping of phytochemical diversity further revealed that plant assemblages found in low to midelevation habitats, with more alkaline soils, possessed greater phytochemical diversity, whereas alpine habitats possessed higher phytochemical endemism. Altogether, we present a general tool that can be used for predicting hotspots of phytochemical diversity in the landscape, independently of plant species taxonomic identity. Such an approach offers promising perspectives in both drug discovery programs and conservation efforts worldwide.
Phytochemical diversity describes the richness and abundance of the specialized metabolites produced by vegetation. It is a key aspect of plant functional diversity and, thus, affects plant fitness (1), ecosystem functioning (2), and services to humankind (3). Despite its relevance, chemical ecologists still struggle to understand both the evolutionary origin of phytochemical diversity and its variation across ecosystems (4). Only a small fraction of the >300,000 currently described phytochemicals (5) has been ascribed to a known ecosystem function or process (6). This is because most identification work has been undertaken on model organisms, such as crop plants (7), and because drug discovery programs have so far been based on prior ethno-medicinal knowledge or random sampling, rather than systematic sampling from the tree of life (8) or guided by ecologically relevant information (9). The ability to better predict the presence and diversity of phytochemicals of interest from phylogenetic information, or from specific environments or habitat types, could uncover the full spectrum and function of phytochemicals in the landscape while also orienting drug discovery research (10). Moreover, documenting landscape variability in phytochemical diversity is particularly important in the context of land use change, which is causing losses of plants that possess a yet-unknown value to medicine and science (11).
The plant metabolome includes both primary functions, expected to be conserved across species, and specialized functions, associated to specific lineages or environments (1). Thus, phytochemical variation in the landscape is expected to arise from a combination of evolutionary (12, 13) and ecological (14, 15) constraints. From a macroevolutionary standpoint, some classes of phytochemical compounds are specific to plant clades (e.g., glucosinolates in Brassicaceae, or tropane alkaloids in Solanales; ref. 16). Such lineage-dependent variation is thought to be driven by chemical defense innovations followed by coevolutionary dynamics with herbivores (17, 18). In particular, the escape-and-radiate model (13) predicts that plant lineages diversify by creating novel, more potent, or complex chemical mixtures in response to biotic pressure (19). Therefore, plant lineages that have experienced more evolutionary split events are predicted to have evolved higher levels of phytochemical diversity (13). From an ecological perspective, phytochemical diversity is expected to be the result of plant adaptation to abiotic and biotic conditions, both of which vary along ecological gradients in landscapes (2, 20). For example, habitats that impose constraints on plant growth, such as cold and resource-poor environments, may be expected to drive selection toward potent chemical defense mechanisms that reduce tissue loss (21). At the same time, it is well established that herbivores and pathogens can promote divergent selection between plant congeners, leading to increasing chemical dissimilarity (22). As such, species relatedness alone is a poor predictor of site-level phytochemical diversity.
Here, we questioned whether phytochemical diversity can be predicted from the phylogenetic and ecological heterogeneity observed in the landscape. We hypothesized that phytochemical diversity is not only related to local plant species diversity but is also constrained by other ecological factors, especially trophic, climatic, edaphic, and topographic variation. We developed a methodological framework involving: 1) comprehensive sampling of plant species along ecological transects that cover the entire range of regional vegetation ecological boundaries; 2) assessing species-level phytochemical composition and combining it with species distribution models (SDMs) for extrapolating phytochemical diversity across the landscape based on species occurrences; 3) extracting climatic and topographical variables associated with each unique molecule observed across all species to build molecular distribution models (MDMs); and 4) projecting phytochemical diversity and composition across the landscape based on these MDMs (SI Appendix, Fig. S1). Here, we consider phytochemical diversity both as the richness of clustered metabolic features and the presence/absence of the families of compounds they represent. We expected that phytochemical diversity values compiled from the projected MDMs would better explain plot-level phytochemical diversity and composition than phytochemical diversity calculated from plant species composition alone.
Results and Discussion
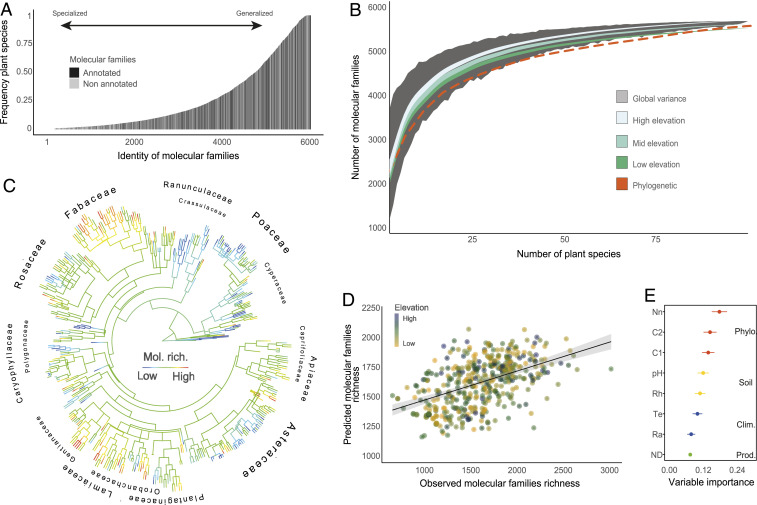
To combine phylogenetic and ecological information for predicting phytochemical diversity in the landscape, we screened the metabolomes of a representative fraction of grassland plant species occurring in the Swiss Alps (n = 416), specifically covering 55% of the total diversity of genera in Swiss grassland vegetation types (23). We sampled leaves of multiple individuals per species across six elevation gradients encompassing a range of soil conditions and four alpine bioclimatic regions (SI Appendix, Fig. S2), which we processed using UHPLC-MS (ultra-high performance liquid chromatography coupled to mass spectrometry) in untargeted fragmentation mode. By organizing the spectral dataset through molecular networking (24), the raw metabolome was processed into a functional representation of each species’ phytochemical diversity. Spectra collectively represent metabolic structures, and since those structures are tightly related to metabolites’ biological function, it is possible to approximate the functional representation of phytochemical diversity directly from the organized spectral space. This spectral space includes highly heterogeneous structures of both structurally known and unknown compounds, with reported or yet-undocumented biological function. Across the 416 vascular plant species measured, we detected more than 43,000 metabolic features encompassing 6,012 molecular families, here defined as spectrally clustered metabolic features (SI Appendix, Fig. S3). While 10% of the molecules were present in more than 80% of species, most molecular families (60%) were specialized metabolites and were produced by less than 20% of species (Fig. 1A). Furthermore, we show that a random sampling of about 100 species is sufficient to describe the full molecular family diversity displayed by all species combined (Fig. 1B). While we found a positive relationship between plant species diversity and phytochemical diversity (i.e., the number of unique molecular families; Fig. 1B, gray ribbon), we also show that this relationship was variable. Specifically, we observed that by randomly sampling 10,000 assemblages of 20 species, molecular family diversity varied 1.3-fold (Fig. 1B). This indicates that molecular family diversity, in addition to being related to plant species richness, is also constrained by plant species-specific phytochemical fingerprint. Sensitivity analyses based on null models also showed that the magnitude and trend of this relationship was dependent on the specific structure of the phytochemical diversity and the frequency of unique molecules across plant species (SI Appendix, Fig. S4). Importantly, we show that phytochemical diversity of plant assemblages is constrained by both abiotic and biotic axes (Fig. 1B), revealing that predicting molecular family richness based on plant species identity alone is biased in the absence of additional information.
Fig. 1.
Phylogenetic, abiotic, and biotic factors together predict phytochemical richness across species. (A) Frequency distribution of unique molecular families across all plant species sampled. (B) Number of unique molecular families as a function of the increasing number of species sampled. The gray shaded area represents the variance around molecular families’ prediction based on 1 million randomly sampled plant species assemblages covering 2 to 100 plant species. The different colored lines represent the average relationship for conditioned sampling based on three different elevational bands (Low = 400–900 m a.s.l.; Mid = 900–1,600 m a.s.l., and High = 1,600–3,000 m a.s.l.). Finally, the red dotted line represents the same sampling but constrained by phylogenetic proximity of species. (C) Phylogenetic tree of all plant species sampled (n = 416), where branch tips are colored by molecular family richness (low: blue, high: red). (D) Association between observed and predicted values of molecular family richness (Pearson correlation: n = 416, r = 0.49, P < 0.001) derived from an ensemble machine learning algorithm with variable selection. (E) Variable importance for the same machine learning algorithm, colored by variable class (red: phylogenetic variables [Phylo], Nn = number of intervening nodes, C1/C2 = first/second axes of cophenetic distance matrix; yellow: soil variables, pH = soil pH 95% quantile, Rh = soil moisture 5% quantile; blue: climate variables [Clim], Te = average annual temperature 95% quantile, Ra = median solar radiation 95% quantile; green: vegetation productivity [Prod], ND = mean NDVI 95% quantile).
Across the 6,012 molecular families, 40% were assigned to a known compound class or infraclass, including metabolite classes known to be related to plant–environment interactions, such as phenolic compounds, terpenes, and alkaloids (1) (SI Appendix, Fig. S5). We further confirmed that phytochemical diversity carries functional value to plants in different environments. We used the calculated phytochemical diversity for each plant species to construct experimental communities of high and low molecular family diversity. Each experimental community was built by randomly selecting 10 plant species from a pool of 50 commonly occurring species, following which five pairs of high and low phytochemical diversity communities were placed at low, mid, and high elevation along two elevational transects (transects 1 and 5 in SI Appendix, Fig. S2, respectively). Plot-level arthropod sampling across the growing season showed that experimental plots containing low molecular family diversity attracted 41% more total arthropod abundance than plots with high molecular family diversity (P = 0.02; SI Appendix, Fig. S6). These results suggest that molecular family diversity affects higher biological diversity at the ecosystem level (25) and that phytochemical diversity among plant species affects ecosystem properties.
We show that functional phytochemical diversity is predictable from phylogenetic branching and ecological properties of species. We coupled the database of molecular families identified for each species with species phylogenetic information, climate-based distribution models, and machine learning to determine the best predictors of molecular family richness (i.e., the number of unique molecular families per species). We found that molecular family richness was predictable from both phylogenetic branching (Fig. 1C and SI Appendix, Fig. S7) and environmental variation combined (Fig. 1 D and E, Pearson correlation: n = 416, r = 0.49, P < 0.001, SI Appendix, Fig. S8). Specifically, we discovered a phylogenetic signal for molecular family richness (Pagel’s lambda [λ] = 0.72, P < 0.001) and found that molecular family richness increased on average by 20 molecular families with each new evolutionary split event (i.e., evolving from 60 molecular families in Equisetum telmateia with 3 split events up to 440 molecular families for Veronica teucrium with 22 split events in our phylogeny; SI Appendix, Fig. S7). Moreover, we found that molecular family richness was negatively correlated with both vegetation productivity (i.e., mean normalized difference vegetation index [NDVI], r = −0.10, P = 0.04) and soil moisture (r = −0.11, P = 0.01), but was positively correlated with solar radiation (r = 0.12, P = 0.01) and soil pH (r = 0.11, P = 0.03; SI Appendix, Fig. S8 and Fig. 1E). We thus confirm that phytochemical diversity is, at least in part, driven by functional adaptations of plants to their environment (26).
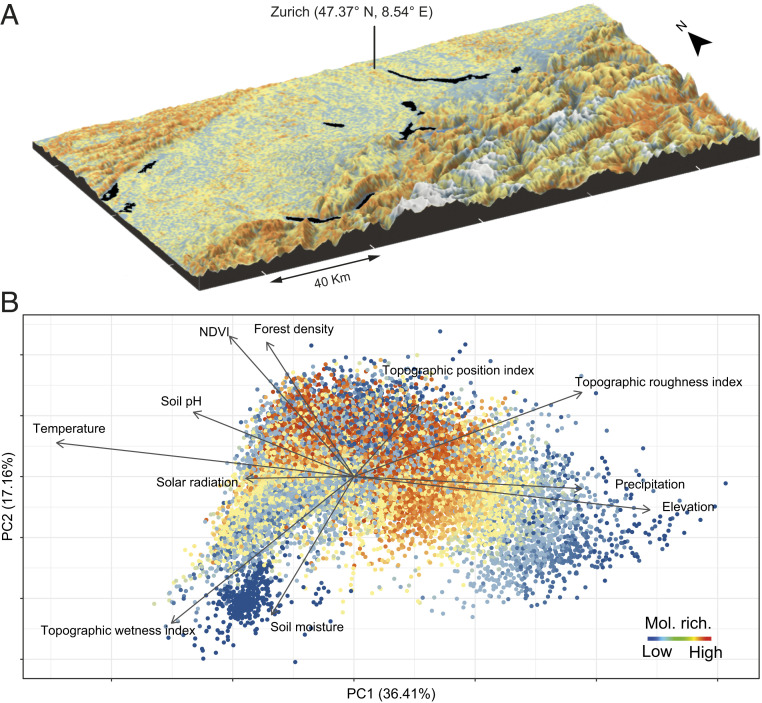
We next built MDMs by geographically mapping the distributions of the 6,012 molecular families (see workflow in SI Appendix, Fig. S1). This was achieved by combining molecular family identification for each plant species with its corresponding range map based on plant SDMs, the latter of which we predicted from climatic and topographic information (SI Appendix, Fig. S9). In doing so, we were able to predict phytochemical diversity, and its inherent composition, across the landscape (Fig. 2A). Analyses of phytochemical diversity–environment interactions revealed strong associations between phytochemical diversity and mean annual temperature (variable importance: 23%), precipitation (variable importance: 13%), NDVI (variable importance: 19%) and soil moisture (variable importance: 18%) (Fig. 2B). Specifically, habitats with more thermal energy and resources (2), but also comprising a higher risk of attack by herbivores and pathogens, such as those situated at low to mid elevation (25), displayed higher molecular family richness. The robustness of MDM predictions was assessed by two means. First, spatial predictions of phytochemical diversity from MDMs generated the same type of relationship between plant species diversity and chemical diversity as that generated from the HPLC-measured plant species chemical composition (SI Appendix, Fig. S10). We observed up to a 12-fold increase in accuracy (dependent on the number of species sampled per community) when using MDM predictions rather than plant species richness for predicting phytochemical diversity (SI Appendix, Fig. S11). Second, while molecular family richness from in situ plant communities sampled along the six transects correlated strongly with the sampled plant species richness (SI Appendix, Fig. S12; Pearson correlation between empirical and estimate values; n = 48; r = 0.87, P < 0.001), such correlation was replicated using MDMs (correlation between empirical chemical diversity and MDM-derived chemical diversity; n = 48, r = 0.64, P < 0.001), a correlation accuracy that was 17% stronger than a similar correlation based on SDMs (correlation between empirical chemical diversity and SDM-derived species richness n = 48, r = 0.53, P < 0.001). These results hence indicate that MDMs can provide robust predictions of phytochemical diversity from any location where environmental variables can be derived (Fig. 2B) (27).
Fig. 2.
Geographic mapping of phytochemical diversity. (A) A geographic representation of phytochemical diversity across ∼20,000 km2 in Switzerland. Colors are as described for B, with white and black additionally showing snowy summits and lakes, respectively. (B) PC1 and PC2 scores from a PCA of phytochemical diversity data extrapolated from a random sample of 20,000 geographic pixels (∼100-m resolution) from A. Arrows represent loadings for the environmental variables annual mean temperature (Temperature), annual precipitation sum (Precipitation), annual sum of solar radiation (Solar radiation), topographic position index (aspect, or hills versus depressions in the landscape), topographic roughness index (slope and terrain roughness), topographic wetness index, inner forest density (Forest height Q25), mean NDVI, soil moisture, and soil acidity. Point colors represent phytochemical richness (low: blue, high: red).
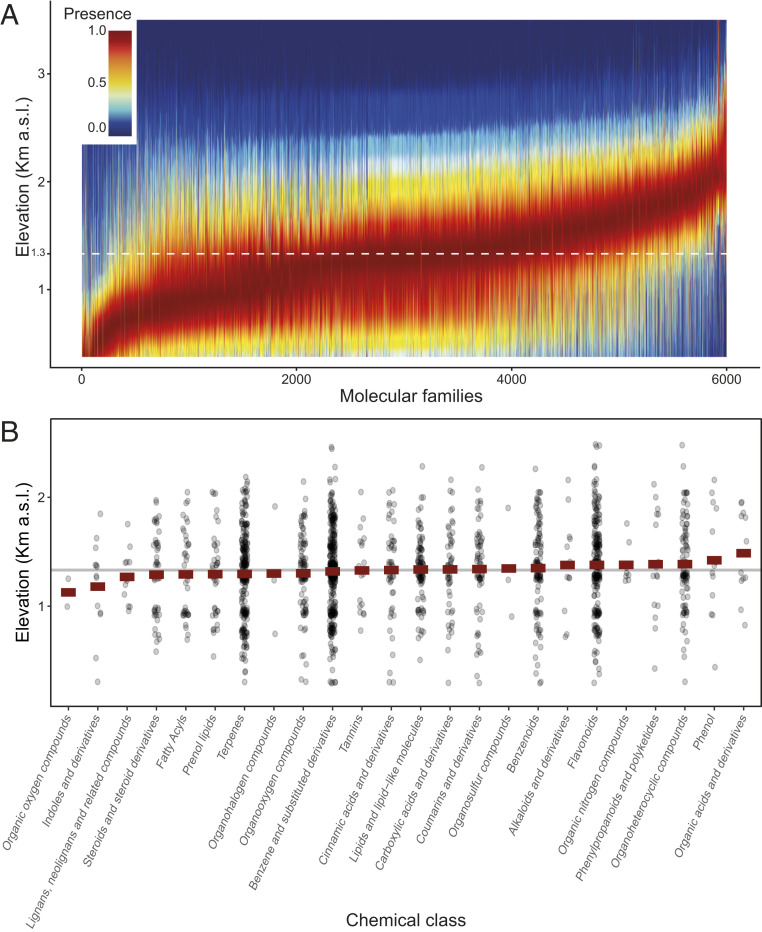
Elevation, which integrates multiple ecological variables in a single axis (28), was associated with shifts in the distributions of specific molecular families. Overall, abundance of molecular families was generally higher at mid to low elevations than at high elevation (average elevation: 1,303 m above sea level [a.s.l.]); (Fig. 3A). Nevertheless, we also detected elevation-specific distributions for different major classes of molecules (Fig. 3B). For instance, phenol-based compounds, such as anthocyanin and numerous flavonoids, were more common at high elevations, corroborating the protective role of these compounds against stressful abiotic conditions of high elevation (high ultraviolet, high wind, and freezing cold) (29). This result is independent from phylogenic constrains, since we found no phylogenetic convergence for high elevation plant assemblages (standardized effect sizes of mean pairwise distance calculated using 999 permutations = 0.03; P < 0.53; ref. 30). We further found that the average elevational range of molecular families with a restricted spread along the elevation gradient (i.e., endemic to a narrow elevation band, SI Appendix, Fig. S11) was located approximately about 300 m higher than the average elevation observed for total molecular family diversity (i.e., 1,640 m a.s.l. for endemic molecular families; SI Appendix, Fig. S11A). Taken together, these results indicate that a random sampling of a low-elevation plant assemblage would, on average, yield more phytochemical diversity than a random sampling of a high-elevation community. However, a random sampling of high elevation plants would likely result in finding unique molecules that are not present at low elevation. Such results are in line with assembly theory (31), which suggests that low elevation habitats are stable and productive, creating higher biotic pressure between species and, thus, favoring increased phytochemical diversity for protection (25, 32). Conversely, high elevation habitats, where competition and antagonistic interactions are less intense (33), but habitat heterogeneity is much larger (34), favor a general decrease of overall phytochemical diversity but select for specific molecules essential for survival in stressful environments (35) (i.e., molecular endemism).
Fig. 3.
Elevational distributions of phytochemical families. (A) Optimal distribution of 6,012 phytochemical families found in the Swiss Alps along the elevation (km a.s.l.). Distribution is based on the niche of each plant species identified through spatial modeling plus the presence of the molecule in the plant species. Colors reflect probability of occurrence (red, maximal [1]; blue, absent [0]), with the dotted white line showing the average probability of occurrence considering all molecular families. (B) Inferred major classes of metabolites ordered by average elevation optima, with the gray line representing the global median value.
To date, studies addressing variation of phytochemical diversity across taxa have focused on specific classes of metabolites, such as terpenes (36), cardenolides (17), or glucosinolates (37). Further studies have addressed the entire phytochemical makeup of coexisting plant species within specific genera, such as Inga (22), or Piper (25), but they have not considered how the metabolome changes along environmental clines. By coupling a suite of latest generation methodologies in computational metabolomics and spatial modeling, we provide untargeted, multispecies evidence that phytochemical diversity is constrained not only by phylogenetic history (13), but also by environmental variation (38). We thus propose that phytochemical families exist within a determined environmental domain, akin to the species ecological niche concept (39), which makes them spatially predictable.
In summary, we show that plant phytochemical diversity in plant species results from the sum of many specialized molecular families on top of more widespread molecules, and that such patterns of phytochemical diversity are predictable across species and ecosystems (Fig. 2). We show that molecular endemism is generally strong at higher elevations (SI Appendix, Fig. S13), which could explain why plants from alpine regions of the world have historically provided a large number of medicines (40). This is important because alpine habitats are highly vulnerable to climate change (41), which could lead to the disappearance of currently uncharacterized phytochemicals. Altogether, we show that variation in biotic and abiotic factors, at least within the boundaries of the mid-European continental climate, can be used to predict which molecules, broad classes of molecules, or even molecular functions are likely to occur at a given location, thus improving our capacity to seek molecules of interest over entire landscapes (10). By mapping landscape-level phytochemical diversity, we ultimately provide an approach for orienting the discovery of bioactive molecules outside well-established biodiversity hotspots (i.e., chemodiversity hotspots), and for prioritizing areas for future conservation efforts in light of climate and land use change.
Methods
Field Sampling and Plant Species.
We aimed to establish a representative metabolomics dataset for the most frequent plant species spanning most of the ecological conditions for Switzerland. To this end, we sampled 48 vegetation plots supporting grassland communities across six transects of the Swiss Alps every 200 m of elevation (SI Appendix, Fig. S2). The six elevation transects spanned all the elevation bands and the four bioclimatic regions of the Central European Alps and included both calcareous and acidic soil types (SI Appendix, Fig. S2). We sampled ∼80% of all vascular plant species found in the vegetation plots. For each species, we sampled between 2 and 10 of the youngest fully expanded leaves from a total of two to three individuals. Plants were all sampled during peak flowering season across the different bands of the elevational gradient in order to mitigate potential phenological and ontogenetic factors. Leaves were stored at 4 °C for less than 5 h before being dried at 40 °C for 5 d and ground to powder using a MM400 Retch TissueLyser (Qiagen).
Environmental Variables.
We selected a set of 10 environmental variables to describe the main ecological gradients in the region: annual mean temperature, annual precipitation sum, annual sum of solar radiation, topographic position index, topographic roughness index, topographic wetness index, inner forest density (Forest height Q25), mean NDVI, soil moisture, and soil pH. Predictors were not strongly correlated (Pearson correlations |r| < 0.59) and were selected due to being established descriptors of plant distributions in Switzerland (42). See SI Appendix, Supplementary Methods, for description of environmental variables calculations.
Metabolome Analysis by Mass Spectrometry and Chemical Data Processing.
We performed untargeted metabolomics analyses to estimate phytochemical diversity of all plant species collected in the field (n = 416 species). We extracted 20 mg of each of the dry ground tissue with 0.5 mL of extraction solvent (MeOH: MilliQ water: formic acid; 80:19.5:0.5). The mixture was thoroughly homogenized for 3 min at 30 Hz using glass beads and centrifuged for 3 min at 14,000 rpm. The supernatant was transferred to a chromatographic vial, and a volume of 2.5 μL was injected into an Acquity UPLC C18 column (50 mm × 2.1 mm, 1.7 μm; Waters). We analyzed the sample via UHPLC-quadrupole time‐of‐flight mass spectrometry (QTOFMS) using an Acquity UPLC coupled to a Synapt G2 MS (Waters). We used a binary solvent system consisting of H2O and acetonitrile, both supplemented with 0.05% formic acid. The chromatographic separation was carried out at a flow rate of 0.6 mL/min at a temperature of 40 °C using a linear gradient of 2–100% acetonitrile in 6.0 min. MS detection was done in positive electrospray ionization over a mass range of 85–1,200 Da. Data were acquired in the data-independent acquisition (DIA) mode, in which all precursor ions from the full mass range are fragmented to yield MS/MS spectra. Quality control samples made of a pool of all samples were run at the beginning of each batch and after every 30 injections. The MS source was cleaned before each of the five batches running over 5 d.
Molecular Networking and Annotation.
We used MS-DIAL (43) for peak detection and assignment of the parent mass to each of the fragmented spectra of the DIA data. The output of MS-DIAL was implemented in the Global Natural Products Social (GNPS) to cluster the MS/MS spectra into compound families based on their cosine similarity and molecular networking (24, 44) (SI Appendix, Fig. S3, MS/MS data and molecular network can be accessed through GNPS in the webpage: https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp). Each feature was annotated by spectral matching against an in silico spectral database version of the Dictionary of Natural Product according to a previously described methodology (45). The ClassyFire taxonomy was employed (46) to estimate the consensus classes, or superclasses, of the molecular families. Despite the presence of partially convoluted spectra (as typically obtained in DIA) and the presence of degenerate features (adducts, dimers), the clusters of the molecular network appeared as an efficient proxy to evaluate the chemical diversity of this wide spectral dataset. Output feature data were combined and averaged per plant species. To obtain an index of expression for each molecular family per plant species, we summed the scaled peak height within species and molecular families. To estimate chemical diversity, we summed the number of molecular families present in each plant species. Because MS/MS spectra were clustered by their fragmentation profile, the total number of molecular families reflects the maximal potential functional chemical diversity as analyzed by the mass spectrometer.
Predictive Model of Chemical Diversity at the Species Level.
To explore the structure of phytochemical diversity across our 416 plant species, we first estimated the distribution of each molecular family across all plant species. Additionally, we performed rarefaction curve analyses by, 1) simulating 1,000,000 randomly sampled plant species assemblages covering 2 to 100 plant species (gray area in Fig. 1B), 2) splitting the 416 plant species into three groups (Low = 400–900 m a.s.l.; Mid = 900–1,600 m a.s.l., High = 1,600–3,000 m a.s.l.) based on the elevation optima of each species, which was obtained from the calculated SDMs (n = 100,000 species assemblages per elevational group in Fig. 1B, green lines), and 3) constraining the sampling according to the minimal phylogenetic distance from the initially randomly sampled plant species (Fig. 1B, dashed red line). To obtain the phylogenetic relationship of the 416 plant species, we pruned the DaPhnE 0.1 supertree of the European vascular plant species (47). We then predicted the chemical diversity combining species phylogenetic information and environmental conditions. We measured the phylogenic signal for phytochemical diversity across species using Pagel’s lambda (phylosig in the phytools package; ref. 48). A lambda value of 1 indicates phylogenetic conservatism consistent with the tree topology and a random walk model (i.e., chemical diversity similarity is directly proportional to the extent of shared evolutionary history). A lambda value of 0 indicates phylogenetic independence. To test the escalation of chemical diversity hypothesis (13), we regressed the number of splitting nodes (distRoot in the adephylo package; ref. 49) against the chemical diversity of each plant species (SI Appendix, Fig. S7). From the phylogenetic tree, we computed the pairwise cophenetic distance for each species and projected it on two dimensions using multidimensional scaling. These two metrics of phylogenetic similarity were then used with the number of splitting nodes as phylogenetic variables. To test for the relationship between species-level chemical differences, phylogenetic and environmental variables extracted from the species-distribution models, we calculated a correlation matrix with all pairwise combinations (SI Appendix, Fig. S8). To explore the predictability of species-level phytochemical diversity from evolutionary and environmental variables, we performed a machine learning ensemble model composed of Random Forest (Ranger packages; ref. 50), recursive partitioning (Rpart package; ref. 51) and Extreme Gradient Boosting (Xgboost package; ref. 52). To build the predictive model we first performed variable selection based on rank importance [vip package (53)]. The variables that ranked in the top 10 in at least two of the three models were retained, i.e., number of nodes, cophenetic Dimension 1 and 2, NDVI quantile 0.95, soil moisture quantile 0.05, soil pH quantile 0.95, annual temperature average quantile 0.95, solar radiation median. For all species, we predicted phytochemical diversity from a model trained recursively on a dataset excluding the target species. The correlation between the predicted and observed phytochemical diversity values indicates an ability to forecast phytochemical diversity at the species level. Finally, we estimated variable importance for the ensemble learning model (vimp package; ref. 54).
Geographic Mapping of Phytochemical Diversity.
We modelized the distribution of the 6,012 chemical families across the alpine landscape by combining the 416 species metabolomics dataset and SDMs. First, we mapped the potential distribution of all the 416 plant species at a ∼100-m spatial grid resolution using SDMs (55) (see SI Appendix for SDM construction). To represent the probability of a specific molecular family being present in the landscape (i.e., the environmental conditions corresponding to each spatial grid pixel), we summed the value of expression index related to that molecular family for all the plant species present in the pixel grid that produced the molecule. All the data were scaled between 0 and 1 within a molecular family to allow interfamily comparison. We then extracted the probability of the presence of molecular families from 20,000 randomly chosen spatial points, which we related to corresponding environmental variables. We next coupled this dataset with a machine learning algorithm (extreme gradient boosting model XGBoost; ref. 56) to build MDMs based on the environmental variables found to be related to each molecular family. MDMs were trained on 70% of the data and validated on the remaining 30% (mean validation accuracy = 0.94). This training process was also used to compute the importance of each environmental factor from the 6,012 MDMs. We projected the probability of presence of all the 6,012 molecular families on 20,000 km2 of Switzerland using the produced MDMs. By stacking the 6,012 projection maps, we estimated an index of local molecular diversity (sum of potentially present molecular families) at a scale of 100 m2 resolution and used it for the geographic mapping of phytochemical diversity. By using the value of the predicted phytochemical diversity index in the 20,000 randomly chosen locations, we next built an environmental principal component analysis (PCA) to describe the phytochemical diversity distribution across the multivariate environmental space.
To validate the MDM approach for predicting phytochemical diversity across the landscape, we first correlated plant species richness extracted from the SDMs with the number of unique molecular families (based on field-collected chemical composition) from 50,000 randomly chosen locations. We used this dataset to build a predictive model of phytochemical diversity based on Gradient Boost Model (GBM). Second, we correlated plant species richness from the SDMs and the MDM-based predictions of phytochemical diversity from the same locations. Finally, we compared the accuracy of our MDMs predictions versus the GBM predictions.
To further validate our approach, we calculated plant assemblage-level phytochemical diversity values for the 48 experimental communities sampled established along the six elevation transects for which metabolomics data were directly available (SI Appendix, Fig. S9). We assessed the effect of elevation on community-level plant species richness and phytochemical richness (separately) using quadratic regression analyses. Next, we extracted the phytochemical diversity index at each corresponding location of the 48 vegetation plots from the MDM mapping exercise. Finally, we performed three correlations tests: 1) field-sampled chemical diversity versus field-sampled plant species richness, 2) field-sampled chemical diversity versus SDM-based plant species richness, and 3) field-sampled phytochemical diversity versus MDM-based chemical diversity.
Distribution of Molecular Families along Elevation.
As in integrative ecological axis, we plotted the probability values of presence for each molecular family obtained from MDMs along an elevation axis generated with the 20,000 randomly chosen pixels of the spatial grid. We chose to represent the elevation axis due its integrative representation of contrasted environmental conditions in the Alps (see correlation with PCA axis 1 in Fig. 2A). We used a locally polynomial quantile regression to estimate changes to the distribution of each molecular family with elevation (lprq in the Quantreg package (57), probs = 0.95). We then estimated an elevation optimum for each molecular family by extracting the elevations corresponding to the highest probability of a molecular family’s presence. Elevation optima where used to illustrate the environmental specificity of large functional classes of molecules. Finally, we identified the molecular families endemism related to elevation by filtering the quantile 0.8 of molecular families elevational range for a probability of presence corresponding to 0.5 (SI Appendix, Fig. S13).
Supplementary Material
Acknowledgments
We thank Adrien Delavallade and Maude Poirier for help during field sampling and Anurag Agrawal and Xavier Morin for providing constructive comments on an earlier version of this manuscript. This work was supported by Swiss National Science Foundation Grants 31003A-179481, 31003A-159869 (to S.R.), and 31003A-162604 (to L.P.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013344118/-/DCSupplemental.
Data Availability.
Raw data for plant species and molecular families have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.13032740).
References
- 1.Rosenthal G. A., Berenbaum M. R., Herbivores: Their Interactions with Secondary Plant Metabolites: The Chemical Participants, Rosenthal G. A., Berenbaum M. R., Eds. (Academic Press, San Diego, CA, ed. 2, 1991), vol. 1. [Google Scholar]
- 2.Hunter M. D., The Phytochemical Landscape: Linking Trophic Interactions and Nutrient Dynamics (Princeton University Press, Princeton, NJ, 2016), pp. 347. [Google Scholar]
- 3.Bruneton J., Pharmacognosy, Phytochemistry, Medicinal Plants (Lavoisier Publishing, Paris, 1995), pp. 915. [Google Scholar]
- 4.Sedio B. E., Recent breakthroughs in metabolomics promise to reveal the cryptic chemical traits that mediate plant community composition, character evolution and lineage diversification. New Phytol. 214, 952–958 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Banerjee P., et al. , Super Natural II–A database of natural products. Nucleic Acids Res. 43, D935–D939 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessler A., Kalske A., Plant secondary metabolite diversity and species interactions. Annu. Rev. Ecol. Evol. Syst. 49, 115–138 (2018). [Google Scholar]
- 7.Viant M. R., Kurland I. J., Jones M. R., Dunn W. B., How close are we to complete annotation of metabolomes? Curr. Opin. Chem. Biol. 36, 64–69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyllenhaal C., et al. , Ethnobotanical approach versus random approach in the search for new bioactive compounds: Support of a hypothesis. Pharm. Biol. 50, 30–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atanasov A. G., et al. , Honokiol: A non-adipogenic PPARγ agonist from nature. Biochim. Biophys. Acta 1830, 4813–4819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coley P. D., et al. , Using ecological criteria to design plant collection strategies for drug discovery. Front. Ecol. Environ. 1, 421–428 (2003). [Google Scholar]
- 11.Chapin F. S., 3rd, et al. , Consequences of changing biodiversity. Nature 405, 234–242 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Firn R. D., Jones C. G., Natural products–A simple model to explain chemical diversity. Nat. Prod. Rep. 20, 382–391 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Ehrlich P. R., Raven P. H., Butterflies and plants–A study in coevolution. Evolution 18, 586–608 (1964). [Google Scholar]
- 14.Coley P. D., Kursar T. A., Ecology. On tropical forests and their pests. Science 343, 35–36 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Pichersky E., Lewinsohn E., Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62, 549–566 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Wink M., Plant secondary metabolism: Diversity, function and its evolution. Nat. Prod. Commun. 3, 1205–1216 (2008). [Google Scholar]
- 17.Agrawal A. A., et al. , Evidence for adaptive radiation from a phylogenetic study of plant defenses. Proc. Natl. Acad. Sci. U.S.A. 106, 18067–18072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becerra J. X., Noge K., Venable D. L., Macroevolutionary chemical escalation in an ancient plant-herbivore arms race. Proc. Natl. Acad. Sci. U.S.A. 106, 18062–18066 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermeij G. J., On escalation. Annu. Rev. Earth Planet. Sci. 41, 1–19 (2013). [Google Scholar]
- 20.Sedio B. E., Rojas Echeverri J. C., Boya P C. A., Wright S. J., Sources of variation in foliar secondary chemistry in a tropical forest tree community. Ecology 98, 616–623 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Coley P. D., Bryant J. P., Chapin F. S. 3rd, Resource availability and plant antiherbivore defense. Science 230, 895–899 (1985). [DOI] [PubMed] [Google Scholar]
- 22.Kursar T. A., et al. , The evolution of antiherbivore defenses and their contribution to species coexistence in the tropical tree genus Inga. Proc. Natl. Acad. Sci. U.S.A. 106, 18073–18078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aeschimann D., Lauber K., Moser D. M., Theurillat J.-P., Flora Alpina (Haupt Berne, Switzerland, 2004). [Google Scholar]
- 24.Wang M., et al. , Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards L. A., et al. , Phytochemical diversity drives plant-insect community diversity. Proc. Natl. Acad. Sci. U.S.A. 112, 10973–10978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regos A., Gagne L., Alcaraz-Segura D., Honrado J. P., Domínguez J., Effects of species traits and environmental predictors on performance and transferability of ecological niche models. Sci. Rep. 9, 4221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holzmeyer L., et al. , Evaluation of plant sources for antiinfective lead compound discovery by correlating phylogenetic, spatial, and bioactivity data. Proc. Natl. Acad. Sci. U.S.A. 117, 12444–12451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Körner C., The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 22, 569–574 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Rasmann S., Pellissier L., Defossez E., Jactel H., Kunstler G., Climate-driven change in plant–insect interactions along elevation gradients. Funct. Ecol. 28, 46–54 (2014). [Google Scholar]
- 30.Kembel S. W., et al. , Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Sundqvist M. K., Sanders N. J., Wardle D. A., Community and ecosystem responses to elevational gradients: Processes, mechanisms, and insights for global change. Annu. Rev. Ecol. Evol. Syst. 44, 261–280 (2013). [Google Scholar]
- 32.Coley P. D., Aide T. M., “Comparison of herbivory and plant defenses in temperate and tropical broad-leaved forests” in Plant-Animal Interactions: Evolutionary Ecology in Tropical and Temperate Regions, Price P. W., Lewinsohn T. M., Fernandes G. W., Benson W. W., Eds. (Wiley, New York, NY, 1991), pp. 25–49. [Google Scholar]
- 33.Callaway R. M., et al. , Positive interactions among alpine plants increase with stress. Nature 417, 844–848 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Rasmann S., Alvarez N., Pellissier L., “The altitudinal niche-breadth hypothesis in insect-plant interactions” in Annual Plant Reviews, Volume 47, Insect-Plant Interactions, Voelckel C., Jander G., Eds. (John Wiley & Sons, Ltd, 2014), pp. 339–359. [Google Scholar]
- 35.Jacobo-Velázquez D. A., Cisneros-Zevallos L., An alternative use of horticultural crops: Stressed plants as biofactories of bioactive phenolic compounds. Agriculture 3, 596–598 (2013). [Google Scholar]
- 36.Becerra J. X., Insects on plants: Macroevolutionary chemical trends in host use. Science 276, 253–256 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Cacho N. I., Kliebenstein D. J., Strauss S. Y., Macroevolutionary patterns of glucosinolate defense and tests of defense-escalation and resource availability hypotheses. New Phytol. 208, 915–927 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Kumar S., Yadav A., Yadav M., Yadav J. P., Effect of climate change on phytochemical diversity, total phenolic content and in vitro antioxidant activity of Aloe vera (L.) Burm.f. BMC Res. Notes 10, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutchinson G. E., Concluding remarks. Cold Spring Harb. Symp. Quant. Biol. 22, 415–427 (1957). [Google Scholar]
- 40.Vitalini S., et al. , Traditional knowledge on medicinal and food plants used in Val San Giacomo (Sondrio, Italy)–An alpine ethnobotanical study. J. Ethnopharmacol. 145, 517–529 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Lamprecht A., Semenchuk P. R., Steinbauer K., Winkler M., Pauli H., Climate change leads to accelerated transformation of high-elevation vegetation in the central Alps. New Phytol. 220, 447–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Descombes P., et al. , Spatial modelling of ecological indicator values improves predictions of plant distributions in complex landscapes. Ecography 43, 1448–1463 (2020). [Google Scholar]
- 43.Tsugawa H., et al. , MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frank A. M., et al. , Clustering millions of tandem mass spectra. J. Proteome Res. 7, 113–122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allard P.-M., et al. , Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 88, 3317–3323 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Djoumbou Feunang Y., et al. , ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 8, 61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durka W., Michalski S. G., Daphne: A dated phylogeny of a large European flora for phylogenetically informed ecological analyses. Ecology 93, 2297 (2012). [Google Scholar]
- 48.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 49.Jombart T., Balloux F., Dray S., adephylo: New tools for investigating the phylogenetic signal in biological traits. Bioinformatics 26, 1907–1909 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Wright M. N., Ziegler A., ranger: A fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 77, 1–17 (2017). [Google Scholar]
- 51.Therneau T., Atkinson B., rpart: Recursive Partitioning and Regression Trees. Version 4.1-15 (2019).
- 52.Chen T., et al. , xgboost: Extreme Gradient Boosting. KDD '16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data. 785–794 (2016).
- 53.Greenwell B., Boehmke B., Gray B., Variable Importance Plots—An Introduction to the vip Package. The R Journal 12, 343–366 (2020). [Google Scholar]
- 54.Williamson B. D., Simon N., Carone M., vimp: Nonparametric Variable Importance. Version 2.1.5 (2019).
- 55.Guisan A., Zimmermann N. E., Predictive habitat distribution models in ecology. Ecol. Modell. 135, 147–186 (2000). [Google Scholar]
- 56.Chen T., Guestrin C., “XGBoost: A Scalable tree boosting system” in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (Association for Computing Machinery, San Francisco, CA, 2016), pp 785–794. [Google Scholar]
- 57.Koenker R., Quantreg: An R package for quantile regression and related methods. Version 5.75 (2004).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for plant species and molecular families have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.13032740).