Abstract
Dopaminergic (DA) neurons derived from human pluripotent stem cells (hPSCs) represent a renewable and available source of cells useful for understanding development, developing disease models, and stem-cell therapies for Parkinson’s disease (PD). To assess the utility of stem cell cultures as an in vitro model system of human DA neurogenesis, we performed high-throughput transcriptional profiling of ~20,000 ventral midbrain (VM)-patterned stem cells at different stages of maturation using droplet-based single-cell RNA sequencing (scRNAseq). Using this dataset, we defined the cellular composition of human VM cultures at different timepoints and found high purity DA progenitor formation at an early stage of differentiation. DA neurons sharing similar molecular identities to those found in authentic DA neurons derived from human fetal VM were the major cell type after two months in culture. We also developed a bioinformatic pipeline that provided a comprehensive long noncoding RNA landscape based on temporal and cell-type specificity, which may contribute to unraveling the intricate regulatory network of coding and noncoding genes in DA neuron differentiation. Our findings serve as a valuable resource to elucidate the molecular steps of development, maturation, and function of human DA neurons, and to identify novel candidate coding and noncoding genes driving specification of progenitors into functionally mature DA neurons.
Keywords: human pluripotent stem cells, dopamine neuron differentiation, single-cell RNA sequencing
1. Introduction
Dopaminergic (DA) neurons in the ventral midbrain (VM) are essential for controlling key functions such as control of voluntary movement, reward processing, and working memory. Dysfunction of DA neurons in the ventral tegmental area (VTA), which projects into corticolimbic structures, is associated with the development of neuropsychiatric disorders, drug addiction, and depression while degeneration of DA neurons in the substantia nigra compacta (SNc) is the main pathology in Parkinson’s disease (PD). Over the last three decades, scientific endeavors have focused on designing a novel cell-based therapy for PD via replacement of lost cells with new healthy DA neurons [1]. Major efforts have thus focused on generating ventral midbrain DA neurons from human pluripotent stem cells (hPSCs) for use in disease models and diagnostics [2] as well as in cell-based therapies for PD [3,4]. It is now possible to obtain mature and functional DA neurons from hPSCs for disease modeling and therapeutics [5,6,7], and these 2D culture systems could potentially also be used as stem cell models of human VM DA neurogenesis.
We previously reported a standardized differentiation approach based on floor-plate transition with the addition of dual-SMAD inhibition (Noggin and SB431542) combined with i) sonic hedgehog (SHH) to induce differentiation into ventral neural fates and ii) glycogen synthase kinase 3 inhibitor (GSK3i) to progressively pattern hPSCs toward a caudal VM progenitor phenotype [8]. Our protocol also included the timed delivery of fibroblast growth factor 8b (FGF8b) to drive a more fine-tuned control of rostro-caudal patterning of VM progenitors, which subsequently gave rise to mature DA neurons [9,10].
In this study, we assessed the potential of long-term in vitro differentiation as a stem-cell model of human DA neuron specification and maturation. We used 10× single-cell RNA sequencing (scRNAseq) to perform high-throughput transcriptional profiling of a large number of hPSCs (19,841) at different developmental stages of VM patterning and differentiation. To better dissect the DA developmental program, we examined the cellular composition of VM-patterned stem-cell cultures at the progenitor state, and after one and two months, when functionally mature DA neurons have been formed. Based on single-cell data obtained at different timepoints of DAgenesis, we reconstructed developmental trajectories able to deduce the origin and timing of cell type appearance during VM differentiation. The advent of single-cell resolution technologies has brought unprecedented insights into the complexity and diversity of cell types during brain development [11,12]. However, most scRNAseq studies of neural differentiation currently focus on coding genes, failing to consider the vast amount of noncoding RNA. Long noncoding RNAs (lncRNAs) are a class of noncoding transcripts longer than 200 nucleotides that have emerged as key transcriptional and posttranscriptional regulators acting at multiple levels of gene expression [13,14,15,16]. We therefore developed a bioinformatic pipeline for profiling the expression of lncRNA and obtained a single-cell landscape of these noncoding genes during DA differentiation in the 10× Genomics dataset by mapping their temporal and cell-type-specific expression.
Together, our findings constitute a valuable resource that may help identify both candidate lncRNAs and coding genes involved in regulatory mechanisms during DA neurogenesis. This single-cell dataset will serve as a powerful tool to elucidate human DA neuron development, maturation, and function, and will contribute to establishing more refined DA differentiation protocols.
2. Materials and Methods
2.1. hPSC Culture and 2D Differentiation
Undifferentiated RC17 (Roslin Cells, #hPSCreg RCe021-A) were maintained on 0.5 μg/cm2 Lam-521 (BioLamina, #LN-521)-coated plates in iPS Brew 4 medium (Miltenyi, #130-104-368) until the start of differentiation. They were passaged with 0.5mM EDTA (ethylenediaminetetraacetic acid) roughly every 7 days (with 10µM Y-27632 and seeding density 2500 cells/cm2) or when becoming confluent. The cells were differentiated into 2D VM-patterned progenitors using our good manufacturing practice (GMP)-grade protocol [9]. On day 0 of the differentiation the cells were detached with EDTA and seeded (10,000 cells/cm2) onto Lam-111 (BioLamina, #LN-111)-coated plates (1 μg/cm2) in N2 medium with SB431542 (10 μM), Noggin (100 ng/mL), Shh-C24II (300 ng/mL), CHIR99021 (0.9 μM), and Y-27632 (10 μM). Media was changed on days 2, 4, 7. On day 9 the media was changed to either N2 medium with FGF8b (fibroblast growth factor 8b, 100 ng/mL) or solely N2 medium. The cells were detached with accutase on day 11 and replated (800,000 cells/cm2) on Lam-111-coated plates (1 μg/cm2) in B27 medium with BDNF (brain-derived neurotrophic factor, 20 ng/mL), AA (ascorbic acid, 0.2 mM), and Y-27632 (10 μM), either with or without FGF8b (100 ng/mL). Media was changed on day 14. On day 16 the cells were either taken for analysis or replated for terminal differentiation. For long-term culture the cells were detached with accutase and replated at a lower density (155,000 cells/cm2) on Lam-111-coated plates in B27 medium with BDNF (20 ng/mL), AA (0.2 mM), GDNF (glial cell line-derived neurotrophic factor, 10 ng/mL) + db-cAMP (dibutyryl cyclic adenosine monophosphate, 500 μM), DAPT (notch inhibitor, 1 μM), and Y-27632 (10 μM). Media was changed every 2–3 days until the end of the experiment. For the long-term cultures the cells were plated on Lam-111 with double concentration (2 μg/cm2) and after day 25 only 50–75% of the media was changed in order to minimize the risk of detachment.
2.2. scRNA-Seq Analysis
Cell suspensions were loaded into a 10× Genomics Chromium Single Cell System (10× Genomics) and libraries were generated using version 3 chemistry according to the manufacturer’s instructions. Libraries were sequenced on Illumina NextSeq500 (400 million reads flow cells) using the recommended read length. Sequencing data was first pre-processed through the Cell Ranger pipeline (10× Genomics, Cellranger count v2) with default parameters (expect-cells set to number of cells added to 10× system), aligned to GrCH38 (v3.1.0) and resulting matrix files were used for subsequent bioinformatic analysis. Seurat (version 3.1.1 and R version 3.6.1, R, Vienna, Austria) was utilized for downstream analysis. Batch effects were removed using the Harmony algorithm (1.0), treating individual 10× runs as a batch. Cells with at least 200 detected genes were retained and the data was normalized to transcript copies per 10,000, and log-normalized to reduce sequencing depth variability. For visualization and clustering, manifolds were calculated using UMAP methods (RunUMAP, Seurat) on 20 precomputed principal components. Clusters were identified by calculating a shared-neighbor graph and then defined (FindClusters, Seurat) with a resolution of 0.2. Identification of differentially expressed genes between clusters was carried out using the default Wilcoxon rank sum test (Seurat). For pseudotime and trajectory analysis Slingshot (1.4) was adopted. For comparison with fetal data Seurats LabelTransfer was used with fetal data as reference object projected the PCA structure onto the data.
2.3. LncRNA Quantification
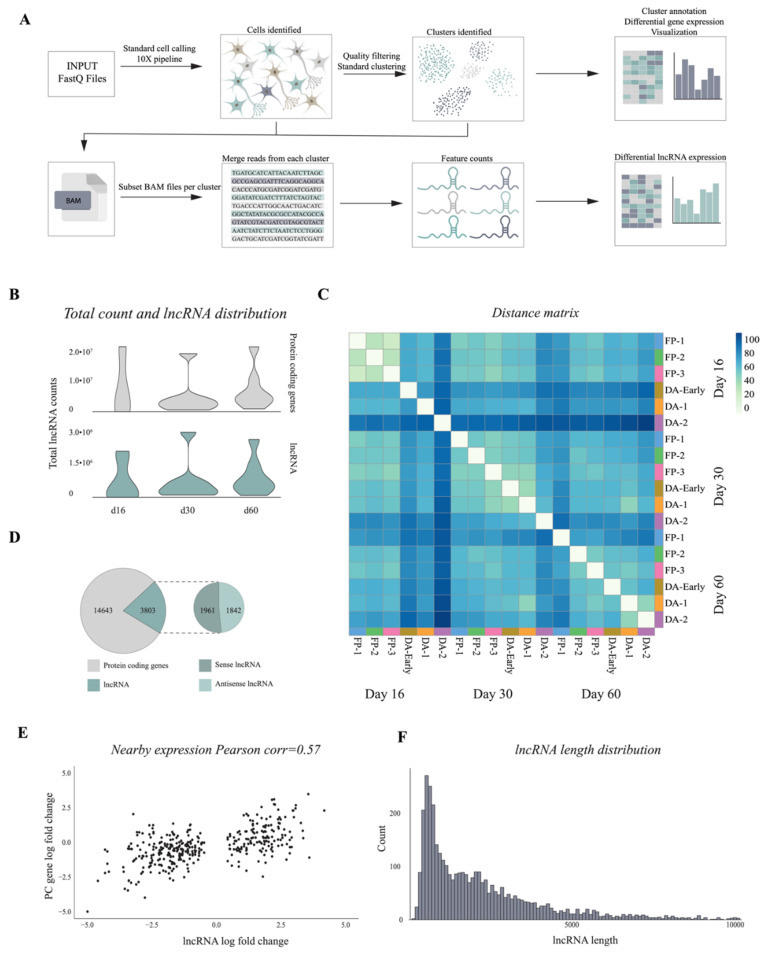
Cell populations identified on the basis of protein-coding genes were used to quantify lncRNA from each cell type across three time points. The expression of lncRNAs was analyzed by extracting cell barcodes for all clusters using Seurat function WhichCells and the original .bam files obtained from the Cellranger pipeline were used to subset aligned files for each cluster (subset-bam tool provided by 10×. Each cluster-specific .bam file was then used as input to count LncRNA expression using FeatureCounts (forward strand). The LncRNA annotation file was downloaded from Gencode (release 36, assembly GRCh38). Differential lncRNA expression analysis was performed using DeSeq2 for both cell types (DA neuron and floor-plate progenitors (FP)) and time points (days 16 and 60) as design (p adj < 0.01). This tool creates a general linear model, assuming a negative binomial distribution. LncRNA that overlapped within a 5kb distance to a protein-coding gene were defined as intragenic. Nearby correlation analysis was performed computing Pearson correlation coefficient using change in protein coding gene expression and lncRNA expression. All data visualization was performed using tidyverse (V1.3), ggplot2 (V3.3.0), and genomic ranges (V1.40.0) running on R (V. 4.0).
2.4. qRT-PCR
Total RNAs were isolated on day of analysis using the RNeasy Micro Kit (QIAGEN#74004) according to manufacturer instructions. Approximately 1 μg of RNA was reverse transcribed using Maxima First Strand cDNA Synthesis Kit (Thermo Fisher 2#K1642Invitrogen). cDNA was prepared together with SYBR Green Master mix (Roche#04887352001) using the Bravo instrument (Agilent) and analyzed by quantitative PCR on a LightCycler 480 II instrument (Roche) using a 2-step protocol with a 95 °C, 0.5 min denaturation step followed by a 60 °C, 1 min annealing/elongation step for 40 cycles in total. All quantitative RT-PCR (qRT-PCR) samples were run in technical triplicates, analyzed with the ΔΔCt-method, normalized against the two housekeeping genes ACTB (actin-beta) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and results are given as a fold change of expression over undifferentiated hPSCs. Details and list of primers are reported in Supplementary Table S1.
2.5. Electrophysiology
Prior to recording, cells on coverslips were transferred to a recording chamber containing Krebs solution gassed with 95% O2 and 5% CO2 at RT and exchanged every 20 min during recordings. The standard solution was composed of (in mM): 119 NaCl, 2.5 KCl, 1.3 MgSO4, 2.5 CaCl2, 25 glucose, and 26 NaHCO3. For recordings, a Multiclamp 700B Microelectrode Amplifier (Molecular Devices) was used together with borosilicate glass pipettes (3–7 MOhm) filled with the following intracellular solution (in mM): 122.5 potassium gluconate, 12.5 KCl, 0.2 EGTA (egtazic acid), 10 HEPES (N-2-hydroxyethylpiperazine-N-ethanesulfonic acid), 2 MgATP, 0.3 Na3GTP, and 8 NaCl adjusted to pH 7.3 with KOH, as previously described. Data acquisition was performed with pCLAMP 10.2 software (Molecular Devices, San Jose, CA, United States); current was filtered at 0.1 kHz and digitized at 2 kHz. Cells with neuronal morphology and round cell body were selected for recordings. Resting membrane potentials were monitored immediately after breaking-in in current-clamp mode. Thereafter, cells were kept at a membrane potential of −60 mV to −80 mV, and 500 ms currents were injected from −85 pA to +165 pA with 20 pA increments to induce action potentials. For inward sodium and delayed rectifying potassium current measurements, cells were clamped at −70 mV and voltage-depolarizing steps were delivered for 100 ms at 10 mV increments.
2.6. Immunocytochemistry
The cells were washed with PBS and fixed in 4% paraformaldehyde solution for 15 min at room temperature (RT) prior to staining. The cells were pre-incubated in a blocking solution containing 0.1 M PBS with potassium (KPBS) +0.1% Triton +5% serum (of secondary antibody host species) for 1–3 h before the primary antibody solution was added.
The cells were incubated with the primary antibodies overnight at 4 °C and the following day they were washed with KPBS before adding the secondary antibody solution containing fluorophore-conjugated antibodies (1:200, Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and DAPI (4′,6-diamidino-2-phenylindole) (1:500). The cells were incubated with the secondary antibodies for 2 h at RT and finally washed with KPBS. Primary antibodies used were: rabbit anti-TH(tyrosine hydroxylase) (1:1000, AB152, Merck Millipore, Burlington, MA, USA), chicken anti-MAP2 (1:10,000, ab5392, Abcam, Cambridge, United Kingdom), mouse anti-Ki67 (1: 500, ab550609, BD Biosciences, San Jose, CA, USA), mouse anti-FOXA2 (1:1000, ab101060, Santa Cruz, Dallas, TX, USA), rabbit anti-LMX1A (1:1000, AB10533, Merck Millipore), goat anti-OTX2 (1:2000, AF1979, R&D Systems, Minneapolis, MN, USA), rabbit anti-TAU (1:2000, A0024, DAKO, Glostrup, Denmark), rabbit anti-GIRK2 (G-protein-regulated inward-rectifier potassium channel 2) (1:500, APC006, Alomone Labs, Jerusalem, Israel), rabbit anti-AADC (aromatic L-amino acid decarboxylase) (1:500, Merck Millipore AB1519), rabbit anti-CALB (calbindin) (1:200, cb38, Swant Inc., Marly, Fribourg, Switzerland), and mouse anti-GFAP (glial fibrillary acidic protein) (1:200 SMI 21, BioLegend San Diego, CA, USA).
2.7. Microscopy
Fluorescent images were captured using a Leica DMI6000B widefield microscope with a Leica 12V 100W Lamphouse 11,504,103 BZ: 00 light source. The images were either taken with a 10× objective (HC PL FLUOTAR 10×, NA = 0.30, immersion = DRY) or a 20× objective (HC FLUOTAR L 20x, NA = 0.4, immersion = DRY). The channels and filters used were DAPI (EX=excitation (nm), EM= emissions (nm); EX: 320–400, EM: 430-510), Cy2 (EX: 430–510, EM: 475–575), Cy3 (EX: 485–585, EM: 535-685), and Cy5 (EX: 560–680, EM: 625–775). Image acquisition software was Leica LAS X and images were processed using Adobe Photoshop CC 2020 (Adobe, San Jose, CA, USA). Any adjustments were applied equally across the entire image, and without the loss of any information. All brightfield images were taken with an Olympus CKX53 with a 4x objective (UPlanFL N 4×/0.13 NA, immersion = DRY) and the software used was OLYMPUS cellSens Standard V1.18.
2.8. Statistical Analysis
All data are expressed as mean ± standard error of the mean (SEM). A Shapiro–Wilk normality test was used to assess the normality of the distribution and parametric or nonparametric tests were performed accordingly. Statistical analyses were conducted using GraphPad Prism 8.0 (GraphPad, San Diego, CA, USA).
2.9. Code Availability
Code for bioinformatics pipeline used in this analysis is available at GitHub https://github.com/davhg96/SC_lncRNA.git.
3. Results
3.1. hPSCs Are Patterned into Functionally Mature DA Neurons
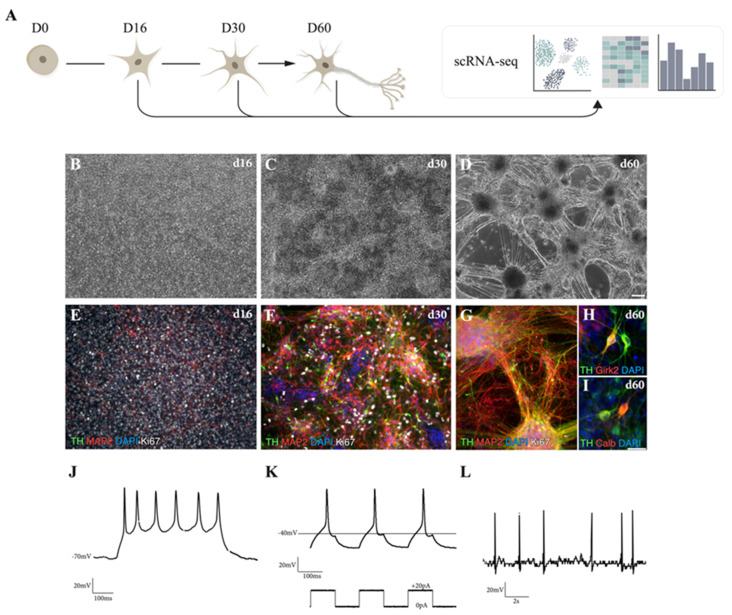
We used a well-established differentiation protocol [9] that relies on dual-SMAD inhibition for neuralization, combined with exposure to the ventralizing secreted factor SHH and to GSK3i for caudalization in order to efficiently drive hPSC differentiation into DA progenitors and subsequently into mature VM DA neurons for single-cell sequencing (Figure 1A). This protocol results in the formation of a homogeneous VM progenitor population after 16 days, as assessed by immunostaining for FOXA2, LMX1a, and OTX2 (Figure 1B,E and Supplementary Figure S1A,B). Subsequent differentiation (Figure 1C,D) led to downregulation of the progenitor markers SOX2 and SHH1, and upregulation of the DA transcripts NURR1 and TH (Supplementary Figure S1E and Supplementary Table S1). The progressive formation of DA neurons was confirmed by staining for TH/MAP2, TAU/MAP2 and AADC/MAP2 (Figure 1F,G and Supplementary Figure S1C,D), showing an increase in morphological complexity of cultures over time as DA progenitors exited from cell cycle and differentiated into postmitotic DA neurons. Immunocytochemistry for Ki67 showed that cells were highly proliferative at days 16 and 30, but that proliferation was minimal at day 60 (Figure 1E–G). At this later timepoint, DA neurons had acquired a subtype identity, expressing GIRK2 (Figure 1H) and CALB (Figure 1I), and had reached functional maturity, as confirmed by whole-cell patch-clamp electrophysiological recordings (Figure 1J–L). Patched cells (n = 5) exhibited functional properties such as hyperpolarized resting membrane potentials (−47.12 mV) and presence of inward sodium (Na+)-outward delayed-rectifier potassium (K+) currents (Supplementary Figure S1F,G). Neuronal function was confirmed by the ability to fire multiple induced-action potentials (APs) upon current injections (n = 4/5) (Figure 1J) with majority of the cells (n = 4/5) showing rebound APs after brief depolarization (Figure 1K), characteristic of midbrain DA neurons in vitro [17]. Furthermore, some cells (n = 3/5) displayed the ability to fire APs without current injection, as shown by spontaneous firing (Figure 1L), indicative of functional neuronal maturation.
Figure 1.
VM-patterned hPSC differentiation generates functionally mature dopaminergic (DA) neurons. (A) Schematic overview of the experimental design. (B–D) Representative bright-field images of ventral midbrain (VM) differentiation cultures at different time points (16, 30, and 60 days). Scale bars, 100 µm. (E–G) Immunofluorescence staining of tyrosine hydroxylase (TH), MAP2, and Ki67 at days 16, 30, and 60. Scale bars, 100 µm. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). (H) Immunofluorescence staining of DA markers TH/GIRK2. (I) TH/calbindin (CALB) at day 60. Scale bars, 25 µm. Nuclei were stained with DAPI. (J–L) Electrophysiological assessment of DA neuron-rich cultures using patch-clamp analysis. (J) Cells analyzed at day 60 displayed induced action potentials. (K) Induced action potentials upon brief depolarization. (L) Spontaneous firing characteristic of DA neurons.
3.2. scRNAseq Reveals Cell-Type Specificity and Developmental Trajectories During VM Differentiation
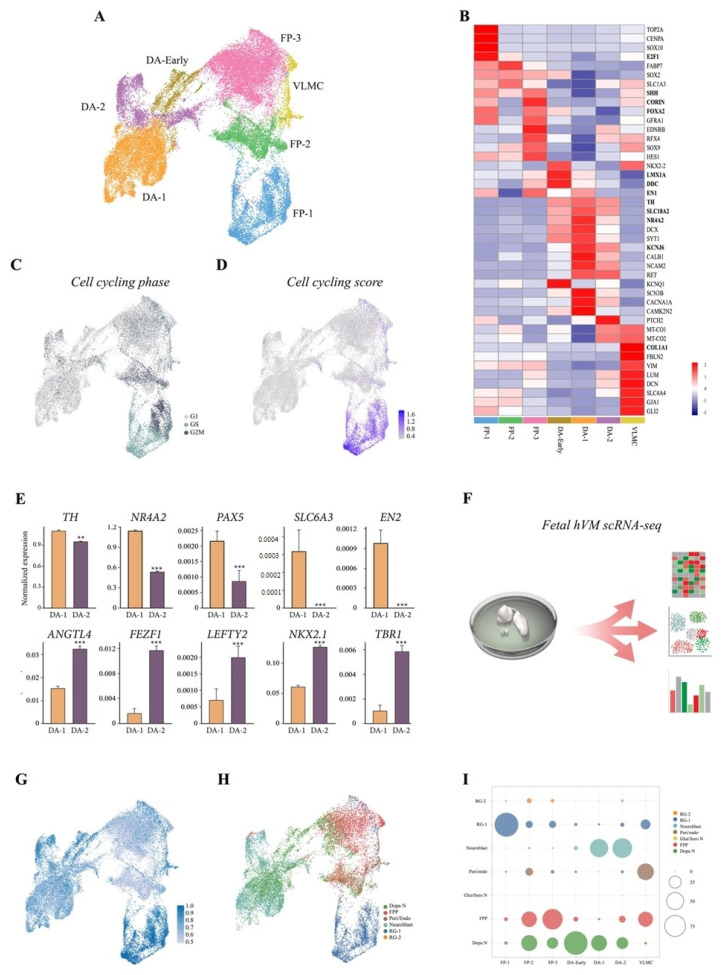
We next performed a 10× Genomics droplet-based single-cell time course transcriptomic analysis at days 16, 30, and 60 of differentiation (Figure 1A and Supplementary Figure S2C), and a total of 19,841 cells were retained for analysis following quality control (QC). Uniform manifold approximation and projection (UMAP) and graph-based clustering assigned the majority of cells to either a floor plate or a DA progenitor/neuron identity in the integrated dataset (Figure 2A,B). A few cells with features of vascular leptomeningal cells (VLMCs), a new cell type associated with vasculature in the brain [18], but none with a glia cell signature were detected (Figure 2A,B). These findings were confirmed at protein level by immunocytochemistry (Supplementary Figure S2A,B). Cell cycle analysis corroborated immunostaining data (Figure 1E–G), with 33%, 6%, and <1% of cells in active cell cycle at day 15, 30, and 60, respectively (Figure 2C,D). A large population of floor-plate cells was also detected, and further analysis of cell cycle showed that the major segregation within this population was due to cycling genes (Figure 2B–D). UMAP distinguished three clusters, which we named FP-1, FP-2, and FP-3 (Figure 2A). We then identified the most highly differentially expressed genes using the Wilcoxon rank sum test for each cell cluster. FP-1 and FP-2 shared key molecular features of radial glial (RG) cells, characterized by expression of SOX2, PLP1, EDNRB, and SOX9 (Figure 2B and Supplementary Figure S2D,E). FP-1 differed primarily from FP-2 in that it included cycling cells with a highly proliferative signature (TOP2A and CENPA), as visualized by UMAP cell cycle score (S.Score and G2M.Score) as well as in expression of the pro-neural gene SOX2 and the chromatin-associated gene E2F1(Figure 2B–D and Supplementary Figure S2D,E). The FP-3 cluster was instead highly enriched by expression of the morphogens SHH, CORIN, and WNt5A as well as FOXA2, LMX1A, and OTX2, which define the VM floor plate (Figure 2B and Supplementary Figure S2D,E). Furthermore, the scRNAseq data revealed the absence of pluripotency-associated genes (NANOG and OCT4) as well as forebrain (FOXG1 and SIX6) and hindbrain (HOXA2 and GBX2) cells, showing efficient VM patterning (Supplementary Figure S2F).
Figure 2.
Single-cell RNA sequencing (ScRNAseq) captures distinct cell-type identities during human pluripotent stem cell DA neuron differentiation. (A) Uniform manifold approximation and projection (UMAP) plot embeddings of 19,841 cells from VM cultures at day 16, 30, and 60. Cell-type assignments are indicated. (B) Heat map showing expression levels of selected genes for each cluster. Values are given as standard deviations relative to average expression across all clusters. (C) UMAP embeddings showing the predicted cell cycle phase. (D) Cell-cycling score (S.Score + G2M.Score) calculated using Seurat CellCycleScoring function. (E) Bar plot of DA-1 and DA-2 clusters showing expression of meso/diencephalic markers. Data represent mean ± SEM, ** p < 0.01, *** p < 0.001. (F) Schematic overview of the experimental design. (G) UMAP embeddings of prediction score for human fetal VM cells from three separate fetuses (6, 8, and 11 weeks post-conception) vs. hPSC VM-derived culture cell types. (H) Predicted cell types using fetal derived cell types as reference. (I) Relative overlapping quantification (% cells overlapping) of human fetal midbrain vs. hPSC VM culture datasets.
We also identified a large neuronal cluster (8989 cells) with increased expression of MAP2, DCX, POU2F2, and RBFIX1 (all upregulated with an adjusted p-value <10−30 compared to FPs) (Figure 2B and Supplementary Figure S2C–E). This neuronal cluster was further refined into three subclusters, which we called DA-early, DA-1, and DA-2 (Figure 2A and Supplementary Figure S2C). DA-early differed mainly from the DA-1 and DA-2 clusters in its retained expression of the floor-plate markers LMX1A and FOXA2, but was starting to gain additional expression of the neuronal precursor markers DCX and SYT1, and the late DA progenitor markers Nurr1 and DDC (Figure 2B and Supplementary Figure S2D). DA-1 and DA-2 subclusters both expressed TH (Figure 2B,C). DA-1 (orange in Figure 2A and Supplementary Figure S2C) was characterized by key markers of VM DA neurons such as TH, NR4A2, PAX5, SLC6A3, and EN2 (Figure 2E), while the smaller DA-2 cluster (purple in Figure 2A) displayed lower or no expression of most DA-related genes (Figure 2E). This latter cluster was instead enriched for non-DA genes such as ANGLT4, FEZF1, LEFTY2, NKX2.1, and TBR1 (Figure 2E).
We next compared the molecular identity of the cells in our 2D VM cultures to a newly generated single-cell dataset of human fetal VM from fetuses dissected 6–11 weeks post conception using the Seurats data projection method in order to assess how well the in vitro stem-cell system captures human VM development (Figure 2F). The fetal dataset comprised 23,438 cells, including ~12,400 progenitors and almost 4000 cells classified as DA neurons (Birtele et al) [19]. Overall prediction quality between the 2D VM-patterned stem-cell culture and fetal VM was high, with a median prediction score of 0.67. Accuracy was higher for the FP-1 and the neuronal clusters, and lower for FP-2 (Figure 2G). Focusing on the two DA clusters in 2D culture revealed 43% of the cells in DA-1 and 25% in DA-2 predicted to be in the fetal DA neuron cluster (median prediction score 0.70 and 0.60, respectively) (Figure 2H,I). However, 56% of DA cells in DA-1 and 68% in DA-2 were also predicted to be fetal neuroblasts, suggesting that not all DA neurons fully mature under 2D culture conditions (Figure 2H,I).
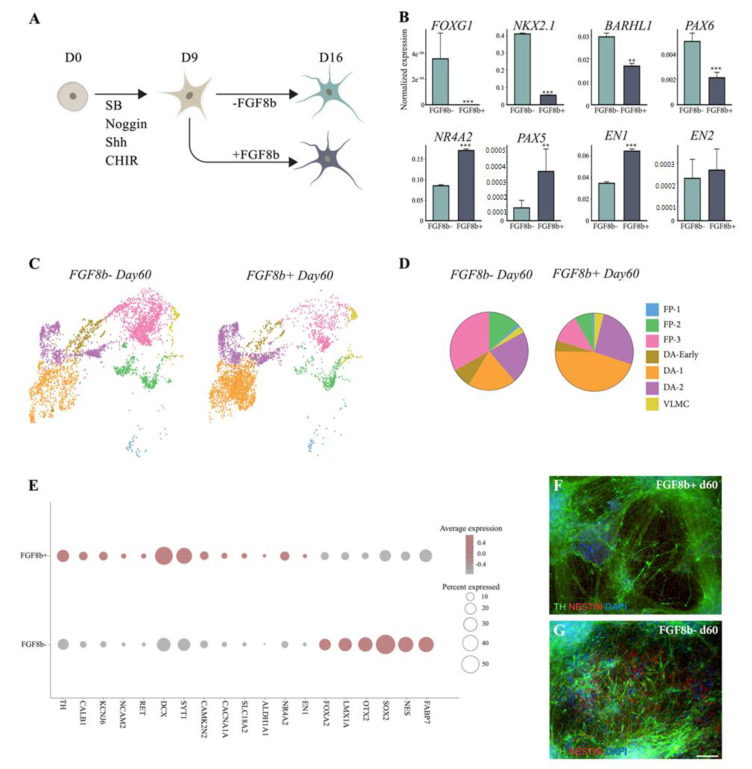
The fine-tuned balance of patterning factors is crucial for ensuring homogeneous VM cell commitment. We previously showed that addition of FGF8b to VM progenitors at days 9–16 resulted in more robust and precise DA neuron formation after transplantation in a PD rat model [8,9]. In this study, we compared the cellular composition of terminally differentiated VM cultures patterned with or without FGF8b at single-cell level (Figure 3A). scRNAseq showed that the absence of FGF8b correlated with ectopic expression of rostral meso/diencephalic markers (BARHL1, NKX2.1, PAX6, and FOXG1) accompanied by downregulation of caudal VM markers (PAX5, EN1, and NR4A2) (Figure 3B).
Figure 3.
Fibroblast growth factor 8 (FGF8b) facilitates progenitor transition into mature DA neurons. (A) Schematic overview of the experimental design. (B) Bar plots showing expression of rostral and caudal mesencephalic markers with and without FGF8b treatment at 2 months from single-cell dataset. Data represent mean ± SEM, ** p < 0.01, *** p < 0.001. (C) UMAP embeddings showing cells from 60-day VM culture with and without FGF8b colored by cell types. (D) Pie chart showing cell-type proportion with and without FGF8b at 2 months. (E) Dot plot showing percent of cells expressing early and late DA markers in VM culture with and without FGF8b at 2 months. (F) Immunofluorescence staining of TH/NESTIN with and (G) without FGF8b treatment at 2 months VM culture. Scale bars, 100 µm. Nuclei were stained with DAPI.
Further investigation of the cellular composition of VM culture with and without FGF8b at day 60 showed that in this 2D model, the timed and dosed delivery of FGF8b also resulted in an increased yield of the DA neuron population (Figure 3C,D).
Interestingly, after two months, over twice the number of cells were found in the DA-1 cluster cultured under FGF8b+ conditions than in the FGF8b- culture (45% vs. 20%, respectively) (Figure 3D). In contrast, a larger DA-early and floor-plate clusters were found in the FGF8b- (DA-early, 8%; FP clusters, 47%) than in the FGF8b+ (DA-early, 4%; floor plate, 20%) (Figure 3C,D) cultures, pointing to the fact that a considerable number of cells with immature identities were still present in 2D cultures grown without FG8b. Single-cell analysis (Figure 3E) and immunohistochemistry of early and late DA markers also confirmed that the absence of FGF8b affected the recapitulation of more mature human DA neurons (Figure 3F,G).
Together, these findings underscore the pivotal role of FGF8b in the induction and specification of DA progenitors into functionally mature DA neurons during VM differentiation.
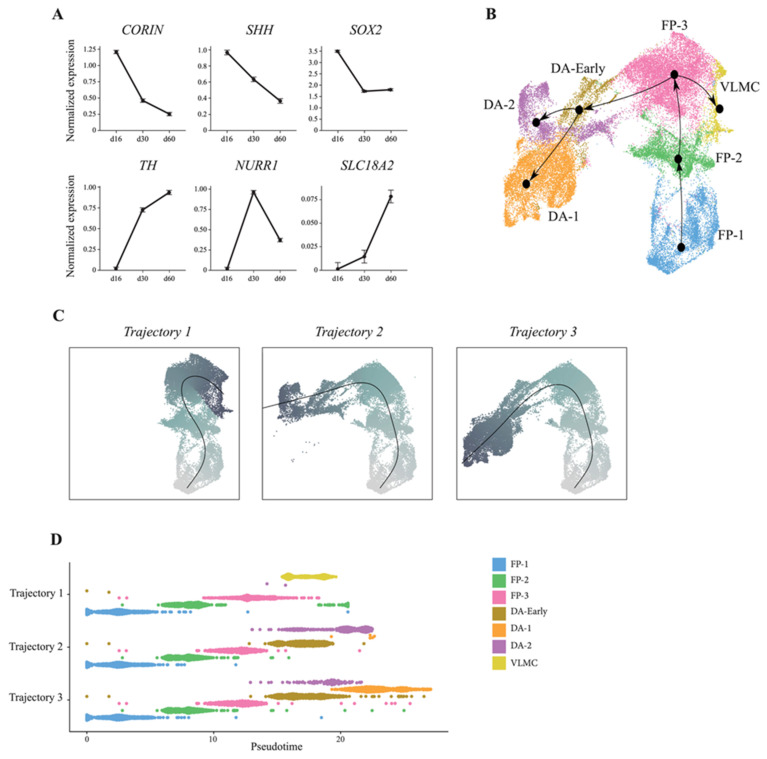
We also assessed the dynamics of DA neuron development in our stem-cell-based model. Time-based expression analysis of progenitor markers including CORIN, SHH, and SOX2 decreased from day 16 to day 60, whereas markers for maturing neurons increased over time (Figure 4A). This prompted us to exploit our a single-cell dataset to temporally reconstruct human DA neurogenesis in in vitro 2D culture using Slingshot on integrated data from all timepoints of differentiation (Figure 4B). Slingshot performs lineage inference using a cluster-based minimum spanning tree, constructing simultaneous principal curves for branching paths through the tree. Three tentative lineages were identified, all originating from the floor-plate population (Figure 4B). Specifically, the resulting plots placed the cells along a continuous path, identifying first FP-1 and then FP-2 as the parent lineage, which subsequently underwent gradual transcriptional changes projecting into three different tentative trajectories (Figure 4B,C). Two of the trajectories shared the same four progenitors (FP-1, FP-2, FP-3, and DA-early), but with DA-1 and DA-2 as terminal states, respectively (Figure 4C,D). In the third trajectory, VLMC was the terminal state, with FP-1, FP-2, and FP-3 as progenitors (Figure 4C,D). Overall, these data confirm that key molecular aspects of DA neurogenesis are recapitulated along a temporal axis in 2D VM culture following a precise developmental program, which eventually gives rise to the generation of functionally mature DA neurons. Interestingly, we also found that the VLMC and DA neuron clusters originated from the same VM floor-plate cells and did not diverge until terminally differentiated into DA neurons or VLMCs, respectively.
Figure 4.
ScRNAseq reconstructs developmental trajectories during VM-patterned hPSC differentiation. (A) Temporal expression pattern of early and late DA markers during hPSC VM differentiation (16–60 days). (B) Slingshot-based pseudotime trajectories calculated from UMAP embeddings showing combined developmental trajectories of all identified cell types and, (C) individual trajectory curves colored by pseudotime. (D), Pseudotime inference reconstruction plot showing the appearance and temporal progression of cell-types along a pseudotemporal axis.
3.3. 10× Genomics Captures a Repertoire of lncRNAs during VM Differentiation
In order to study tissue-specific expression of lncRNAs and their functional importance as critical regulators in key biological processes we developed a pipeline to examine the developmental dynamics and cell-type specificity of lncRNAs during DA neuron differentiation. The pipeline was designed to comprehensively profile differentially expressed lncRNAs at different timepoints or according to cell-type specificity based on the clusters previously defined using coding gene markers. To identify lncRNAs expressed during VM differentiation, we merged the reads from each cluster and performed lncRNA quantification in a strand-specific manner (see Materials and Methods and Figure 5A). By comparing the expression of lncRNAs and mRNAs, we found that the average number of mapped lncRNA genes at each timepoint was significantly lower than that of mRNA genes. However, no significant changes were detected from day 16 to day 60 (Figure 5B). Our pipeline detected 3802 lncRNA genes, including 1961 sense and 1842 antisense (expression base mean ≥1), and distance matrix of lncRNA expression showed a correlation between cell types and differentiation timepoints (Figure 5C,D and Supplementary Figure S3A,B). Since lncRNAs can exert either repressive or promoting activities on target genes by coordinating protein and RNA interactions both in cis (on neighboring genes) and in trans (on distant loci), we investigated their genomic distribution. Among 3803 identified lncRNA genes, the majority were intragenic (Supplementary Figure S3C). Pearson’s pairwise correlation analysis showed that lncRNAs exhibited a higher change in expression correlation with their nearest protein coding gene (<5 kb, Figure 5E). Specifically, we found a higher percentage of positive correlations between cis pairs. Length distribution showed that the majority of lncRNAs detected in this pipeline ranged from 300 to 2500 base pairs (Figure 5F). This confirms that the pipeline is able to profile lncRNAs during VM differentiation regardless of genomic location, length of transcript, or transcriptional direction.
Figure 5.
Bioinformatic pipeline detects lncRNAs from 10x Genomics single cell dataset (A) Schematic overview of pipeline designed for lncRNA quantification. (B) Count distribution of protein-coding genes and lncRNAs from each sample at three time points of hPSC DA neuron differentiation (day 16, 30, 60). (C) Heatmap of Euclidean distance across different cell types and time points (log2 normalized counts). (D) Number and distribution of protein-coding genes and lncRNAs expressed (baseMean >1), strand (sense “+”, antisense “−”). (E) Pearson correlation of change in expression of protein-coding genes and lncRNAs (±5Kb, p adj < 0.01). The x-axis shows the log fold change of the lncRNA and the y-axis the log fold change of the overlapping protein-coding gene. (F) Length distribution of expressed lncRNAs (x-axis).
3.4. Single-Cell Analysis Reveals Dynamics and Cell-Type Specificity of LncRNAs During VM Differentiation
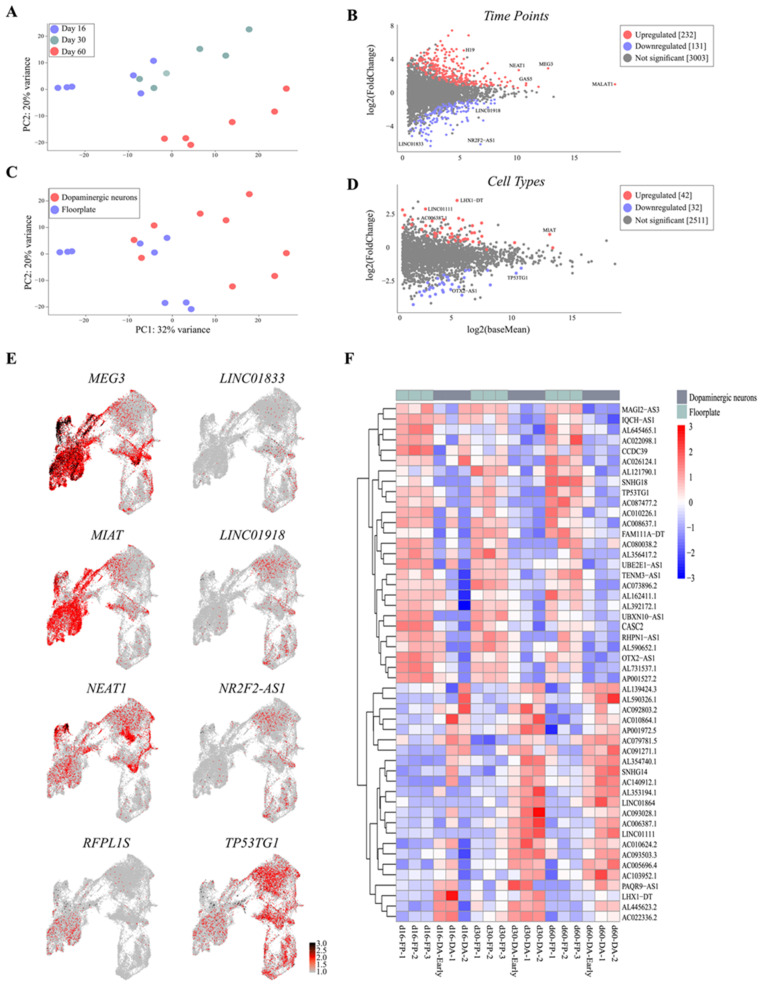
scRNAseq based on coding gene analysis distinguished cell types and defined their developmental dynamics during differentiation. We therefore investigated whether the expression of captured lncRNAs correlated to that of molecularly distinct cell clusters identified during DA neuron specification. C (PCA) clustered 456 single cells into three distinct populations, in line with the three different stages of human VM differentiation analyzed, showing that lncRNAs exhibit high stage specificity following VM developmental dynamics (Figure 6A). The MA plot showed significantly differentially expressed lncRNAs (p value adjusted < 0.01) between day 16 and day 60 (Figure 6B). As expected, during VM differentiation we found downregulated well-known lncRNAs such as NR2F2-AS1, and LHX5-AS1 were shown to play regulatory roles in neural stem-cell differentiation (Figure 6B,E and Supplementary Figure S3D) [20,21]. LINC01833 (also called SIX3os), the long noncoding opposite strand transcript of homeodomain factor Six3, known to be involved in neuronal differentiation and retinal cell specification, was also downregulated (Figure 6B,E) [22]. Captured downregulated lncNRAs also included annotated transcripts not yet associated with neurodevelopment, such as LINC01918 and AL139393.2, implicated in thyroid and medulloblastoma carcinoma, respectively [23,24], as well as SMCR2, also known as lncSREBF1, which plays a role in lipid metabolism (Figure 6B,E and Supplementary Figure S3D) [25].
Figure 6.
Catalogue of lncRNA time and cell-type specificity. (A) Principal component analysis (PCA) plot of lncRNA expression across timepoints in hPSC DA neuron differentiation. (B) MA plot of lncRNAs showing their log2 fold change and log2 baseMean expression across time points (p adj < 0.01). (C) PCA plot of lncRNA expression across cell types in hPSC DA neuron differentiation. (D) MA plot of lncRNAs showing their log2 fold change and log2 baseMean expression across cell type (p adj < 0.01). (E) Expression map of candidate lncRNAs projected on UMAP plot. (F) Expression heatmap of the top 50 most differentially expressed lncRNAs across cell types (log2 vst, p adj < 0.01).
Differentially expressed upregulated lncRNAs in long-term VM culture included the well-known nuclear paraspeckle assembly transcript 1 (NEAT1), overexpressed in the substantia nigra, as well as H19, MALAT1, and SNHG12, previously reported to be enriched at early stages of PD pathogenesis (Figure 6B,E and Supplementary Table S2) [26,27,28]. We also found upregulated expression of AC005062.1 and AL590302.1, as well as LINC02019 and the antisense SPRY4-AS1, previously associated with tumorigenesis (Supplementary Figure S3D and Supplementary Table S2) [29].
We next examined lncRNA expression based on cell-type specificity by combining single-cell data from days 16, 30, and 60. PCA revealed that lncRNAs clearly segregated into DA and floor-plate clusters, and 140 lncRNAs were found significantly differentially expressed in these two molecularly distinct cell types (Figure 6C). Consistent with these findings, we found lncRNAs associated with brain development to be upregulated in the DA cluster, such as the nuclear-localized MIAT, also known as Gomafu, reported to be expressed in postmitotic neurons (Figure 6C,D) [30]. The imprinted lncRNA Meg3 [31] (Figure 6E and Supplementary Figure S3A), expressed in the cortex, was also found enriched in DA neurons, similarly to its neighbor gene DLK1, which plays a critical role in DA specification. LINC01111, identified as an oncosuppressor in pancreatic cancer [32], and the annotated LHX1-DT and AC006387.1 transcripts were also significantly enriched in the DA population (Figure 6F and Supplementary Table S3). Noteworthy, ~80% of lncRNAs detected in the DA cell type were also found upregulated at day 60 of VM differentiation. In contrast, OTX2-AS1 was significantly enriched in the floor-plate cluster together with TP53TG1, implicated in carcinoma but not as yet associated with a neuronal phenotype [33] (Figure 6D–F and Supplementary Figure S3E). This cluster of immature cells was also characterized by SNHG18 [34], known to play an oncogenic role in glioma, LINC0052 [35], which acts as a sponge for miR-608, and the annotated transcripts AC026124.1, AL132780.2, and AC026401.3 (Figure 6D–F and Supplementary Figure S3E).
4. Discussion
In this study, we used single-cell sequencing to assess the validity and potential use of stem-cell-based differentiation to model human DA neuron development. By transcriptionally profiling almost 20,000 cells at different time points, spanning the transition from progenitor stage to functionally mature DA neurons, we were able to assess the extent to which this culture system recapitulates DA neuron development. Our findings prompt a number of important observations. Firstly, clustering analysis shows that cell types present during stem-cell differentiation are similar to those present during fetal VM development and that no aberrant cell types are found in the cultures. Secondly, the temporal appearance of different cell types corresponds to the order of progression previously observed in human fetal VM [11] and in xenograft models [10]. Thirdly, the molecular profile of the resulting DA neurons closely matches that of fetal VM-derived DA neurons. Taken together, these considerations support the use of stem-cell-based models to study aspects of human brain development.
When using the single-cell dataset to reconstruct lineage specification and DA neurogenesis, we were able to identify three distinct lineages all originating from the same floor-plate population. Two of the trajectories shared exactly the same progenitors (FP-1, FP-2, FP-3, and DA-early), but then diverged into a large DA population (DA-1) and a smaller population of neurons expressing TH but few other DA-associated markers (DA-2). This latter population also shared a much smaller overlap of genes with DA neurons in fetal VM, indicating that some cells in 2D culture underwent incomplete differentiation and/or insufficient patterning toward a VM regional fate or otherwise aberrantly express TH in non-DAergic cells as is commonly observed in in vitro cultures [36,37,38,39]. Interestingly, this lineage analysis also revealed that the VLMC and DA neuron clusters originated from the same VM floor-plate cells and did not diverge until terminal differentiation into DA neurons or VLMCs had occurred.
Rostro-caudal patterning in VM differentiation protocol can be more precisely controlled using timed delivery of FGF8b [9]. By quantifying cellular composition of cells patterened with and without FGF8 after two months in culture, we showed that FGF8b facilitates the conversion of progenitors into functionally mature DA neurons while enabling efficient differentiation toward a caudal VM pattern. These findings also underscore the need to identify and validate more markers able to predict functional maturation in both long-term culture and in vivo after transplantation.
In the past decades, large-scale genome-wide sequencing has revealed the tissue-specificity and functional relevance of lncRNAs as key regulators in neural development, refuting the paradigm of RNA as simply an intermediary between DNA and protein [40,41,42]. In this expanding view of the genomic landscape, very few scRNAseq studies have as yet provided comprehensive human lncRNA repertoires of different healthy human tissues [43,44,45,46]. Although the relatively low expression levels of lncRNAs make analysis at single-cell resolution challenging, a detailed catalog of cell-type-specific lncRNAs expressed during DA differentiation may increase our understanding of their involvement in the VM developmental program and intricate regulatory DA pathways [47,48]. Here, we exploited 3′-end high-throughput sequencing of 10× Genomics technology to capture polyadenylated noncoding genes and developed a bioinformatic pipeline that provided the first comprehensive dataset of lncRNAs showing cell-type specificity by analyzing different stages of DA neuron differentiation. 3′-end sequencing is not able to capture non polyA transcripts and is less efficient than full-length scRNAseq methodologies. Nevertheless, 10× Genomics performs high-throughput microfluidic profiling of a large number of cells. Thus, although many lncRNAs may individually have low expression levels, the extensive number of cells that is possible to sequence with this method also allows lncRNA expression to be analyzed and mapped to distinct clusters defined by the coding gene expresssion.
We found that many lncRNAs are specific to distinct cell types associated with either progenitors or functionally mature DA neurons. Interestingly, we identified new lncRNAs potentially involved in early steps of DA differentiation, such as TP53TG1, LINC01833, and SNHG18, found specifically enriched in the floor-plate clusters, as well as two antisense transcripts of the NR2F1 and OTX2 coding genes, which play a key role in cortical development and VM floor-plate formation, respectively [49]. Our dataset also includes significantly enriched lncRNAs not previously linked to functionally mature DA cells, such as LINC01111, RFPL1S, and AL365361.1. It also confirms the expression of lncRNAs already associated with DA tissue, including MIAT, involved in DA signaling via modulation of DA transmission and neurobehavioral phenotypes [50]. Using a microfluidic approach enabled us to analyze a sufficiently large number of cells and more accurately redefine the feature expression map of lncRNAs which until now were thought to be mainly expressed in late stages of VM differentiation. NEAT1, for example, known to exert a neuroprotective role in SNc, was also found highly expressed in our floor-plate subclusters, suggesting its potential role in DA commitment [51]. The imprinted lncRNA Meg3 also displayed a particular cell-type-specific expression pattern robustly expressed in DA clusters, hinting at its involvement as a critical regulator of DA neuron specification, possibly through crosstalk with its coding neighbor gene DLK1. In this scenario, integrating single-cell transcriptomics and epigenomics technologies, such as assay for transposase-accessible chromatin using sequencing (ATAC-seq), will bring valuable insights into the complex and intricate regulatory network involving coding and noncoding genes during stem-cell DA differentiation.
Taken together, these findings provide a valuable transcriptomic dataset of coding and noncoding genes in human DA neurogenesis that may lead to the identification of novel cell types and a better definition of molecular diversity in progenitor and mature DA populations.
Acknowledgments
We thank Ulla Jarl, Marie Persson Vejgården, and Mikael Sparrenius for technical assistance; and Bengt Mattsson for his valuable help with microscopy and figure preparation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/10/1/137/s1, Figure S1: Gene expression and functional analysis of VM-patterned hPSCs, Figure S2: Cell-type identification based on scRNAseq and immunocytochemistry, Figure S3: Differentially expressed lncRNAs based on cell-type and time-point, Table S1: RT-qPCR primer list, Table S2: LncRNAs deregulated in 60 vs. 16 time points, and Table S3: LncRNAs deregulated in DA vs. FP cell-types.
Author Contributions
Conceptualization, M.P. and A.F.; methodology, M.P., A.F. and F.N.; formal analysis, F.N., A.F., E.S., P.S., M.B., Y.S. and D.H.G.; investigation, F.N., E.S., P.S., M.B., Y.S. and D.H.G.; resources, M.P. and A.F.; data curation, A.F. and F.N.; writing—original draft preparation, M.P. and A.F.; supervision, M.P. and A.F; funding acquisition, M.P. All authors have read and agreed to the published version of the manuscript.
Funding
The research leading to these results has received funding from the New York Stem Cell Foundation (MP), the European Research Council (ERC Grant Agreement no. 771427, MP), the Swedish Research Council (Grant Agreements 2016-00873 (MP)), the Swedish Parkinson Foundation (Parkinsonfonden, MP), the Swedish Brain Foundation (Hjärnfonden FO2019-0301, MP), the Strategic Research Area at Lund University MultiPark (Multidisciplinary research in Parkinson’s disease) and KAW (2018.0040). M. Parmar is a New York Stem Cell Foundation–Robertson Investigator.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barker R.A., Drouin-Ouellet J., Parmar M. Cell-based therapies for Parkinson disease—past insights and future potential. Nat. Rev. Neurol. 2015;11:492–503. doi: 10.1038/nrneurol.2015.123. [DOI] [PubMed] [Google Scholar]
- 2.Barker R.A., Parmar M., Studer L., Takahashi J. Human Trials of Stem Cell-Derived Dopamine Neurons for Parkinson’s Disease: Dawn of a New Era. Cell Stem Cell. 2017;21:569–573. doi: 10.1016/j.stem.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Parmar M. Towards stem cell based therapies for Parkinson’s disease. Development. 2018;145:dev156117. doi: 10.1242/dev.156117. [DOI] [PubMed] [Google Scholar]
- 4.Barker R.A., TRANSEURO consortium Designing stem-cell-based dopamine cell replacement trials for Parkinson’s disease. Nat. Med. 2019;25:1045–1053. doi: 10.1038/s41591-019-0507-2. [DOI] [PubMed] [Google Scholar]
- 5.Kriks S., Shim J.-W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A., et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nat. Cell Biol. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grealish S., Diguet E., Kirkeby A., Mattsson B., Heuer A., Bramoulle Y., Van Camp N., Perrier A.L., Hantraye P., Björklund A., et al. Human ESC-Derived Dopamine Neurons Show Similar Preclinical Efficacy and Potency to Fetal Neurons when Grafted in a Rat Model of Parkinson’s Disease. Cell Stem Cell. 2014;15:653–665. doi: 10.1016/j.stem.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi T., Morizane A., Doi D., Magotani H., Onoe H., Hayashi T., Mizuma H., Takara S., Takahashi R., Inoue H., et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nat. Cell Biol. 2017;548:592–596. doi: 10.1038/nature23664. [DOI] [PubMed] [Google Scholar]
- 8.Kirkeby A., Nolbrant S., Tiklova K., Heuer A., Kee N., Cardoso T., Ottosson D.R., Lelos M.J., Rifes P., Dunnett S.B., et al. Predictive Markers Guide Differentiation to Improve Graft Outcome in Clinical Translation of hESC-Based Therapy for Parkinson’s Disease. Cell Stem Cell. 2017;20:135–148. doi: 10.1016/j.stem.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nolbrant S., Heuer A., Parmar M., Kirkeby A. Generation of high-purity human ventral midbrain dopaminergic progenitors for in vitro maturation and intracerebral transplantation. Nat. Protoc. 2017;12:1962–1979. doi: 10.1038/nprot.2017.078. [DOI] [PubMed] [Google Scholar]
- 10.Tiklová K., Nolbrant S., Fiorenzano A., Björklund Åsa K., Sharma Y., Heuer A., Gillberg L., Hoban D.B., Cardoso T., Adler A.F., et al. Author Correction: Single cell transcriptomics identifies stem cell-derived graft composition in a model of Parkinson’s disease. Nat. Commun. 2020;11:1. doi: 10.1038/s41467-020-17421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., Borm L.E., Stott S.R., Toledo E.M., Villaescusa J.C., et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 2016;167:566–580.e19. doi: 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallejos C.A., Risso D., Scialdone A., Dudoit S., Marioni J.C. Normalizing single-cell RNA sequencing data: Challenges and opportunities. Nat. Methods. 2017;14:565–571. doi: 10.1038/nmeth.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The FANTOM consortium. Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., et al. The Transcriptional Landscape of the Mammalian Genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 14.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn R.A., Chang H.Y. Long Noncoding RNAs in Cell-Fate Programming and Reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiorenzano A., Pascale E., Gagliardi M., Terreri S., Papa M., Andolfi G., Galasso M., Tagliazucchi G.M., Taccioli C., Patriarca E.J., et al. An Ultraconserved Element Containing lncRNA Preserves Transcriptional Dynamics and Maintains ESC Self-Renewal. Stem Cell Rep. 2018;10:1102–1114. doi: 10.1016/j.stemcr.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grace A., Onn S. Morphology and electrophysiological properties of immunocytochemically identified rat dopamine neurons recorded in vitro. J. Neurosci. 1989;9:3463–3481. doi: 10.1523/JNEUROSCI.09-10-03463.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marques S., Zeisel A., Codeluppi S., Van Bruggen D., Falcão A.M., Xiao L., Li H., Häring M., Hochgerner H., Romanov R.A., et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birtele M., Sharma Y., Storm P., Kajtez J., Sozzi E., Nilsson F., Wahlestedt J.N., Stott S., He X.L., Mattsson B., et al. Single cell transcriptional and functional analysis of human dopamine neurons in 3D fetal ventral midbrain organoid like cultures. bioRxiv. 2020 doi: 10.1101/2020.10.01.322495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ang C.E., Ma Q., Wapinski O.L., Fan S., Flynn A.R., Lee Q.Y., Coe B.P., Onoguchi M., Olmos V.H., Do B.T., et al. The novel lncRNA lnc-NR2F1 is pro-neurogenic and mutated in human neurodevelopmental disorders. eLife. 2019;8:41770. doi: 10.7554/eLife.41770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prajapati B., Fatma M., Maddhesiya P., Sodhi M.K., Fatima M., Dargar T., Bhagat R., Seth P., Sinha S. Identification and epigenetic analysis of divergent long non-coding RNAs in multilineage differentiation of human Neural Progenitor Cells. RNA Biol. 2019;16:13–24. doi: 10.1080/15476286.2018.1553482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng S.-Y., Johnson R., Stanton L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2011;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Jin T., Shen H., Yan J., Guan M., Jin X. Identification of Long Non-Coding RNA Expression Profiles and Co-Expression Genes in Thyroid Carcinoma Based on The Cancer Genome Atlas (TCGA) Database. Med Sci. Monit. 2019;25:9752–9769. doi: 10.12659/MSM.917845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi P., Jallo G., Perera R.J. In silico analysis of long non-coding RNAs in medulloblastoma and its subgroups. Neurobiol. Dis. 2020;141:104873. doi: 10.1016/j.nbd.2020.104873. [DOI] [PubMed] [Google Scholar]
- 25.Muret K., Désert C., Lagoutte L., Boutin M., Gondret F., Zerjal T., Lagarrigue S. Long noncoding RNAs in lipid metabolism: Literature review and conservation analysis across species. BMC Genom. 2019;20:1–18. doi: 10.1186/s12864-019-6093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyu Y., Bai L., Qin C. Long noncoding RNAs in neurodevelopment and Parkinson’s disease. Anim. Model. Exp. Med. 2019;2:239–251. doi: 10.1002/ame2.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simchovitz A., Hanan M., Niederhoffer N., Madrer N., Yayon N., Bennett E.R., Greenberg D.S., Kadener S., Soreq H. NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J. 2019;33:11223–11234. doi: 10.1096/fj.201900830R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L., Zhuang Y., Zhao X., Li X.-K. Long Non-coding RNA in Neuronal Development and Neurological Disorders. Front. Genet. 2019;9:744. doi: 10.3389/fgene.2018.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X.L., Subramanian M., Jones M.F., Chaudhary R., Singh D.K., Zong X., Gryder B., Sindri S., Mo M., Schetter A., et al. Long Noncoding RNA PURPL Suppresses Basal p53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell Rep. 2017;20:2408–2423. doi: 10.1016/j.celrep.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun C., Huang L., Li Z., Leng K., Xu Y., Jiang X.-M., Cui Y. Long non-coding RNA MIAT in development and disease: A new player in an old game. J. Biomed. Sci. 2018;25:1–7. doi: 10.1186/s12929-018-0427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan H., Rao J., Yuan J., Gao L., Huang W., Zhao L., Ren J. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate ischemic neuronal death by targeting miR-21/PDCD4 signaling pathway. Cell Death Dis. 2017;8:1–15. doi: 10.1038/s41419-017-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan S., Shen M., Zhou M., Shi X., He R., Yin T., Wang M., Guo X., Qin R.Y. Long noncoding RNA LINC01111 suppresses pancreatic cancer aggressiveness by regulating DUSP1 expression via microRNA-3924. Cell Death Dis. 2019;10:1–16. doi: 10.1038/s41419-019-2123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Lagares A., Crujeiras A.B., Lopez-Serra P., Soler M., Setien F., Goyal A., Sandoval J., Hashimoto Y., Martinez-Cardús A., Gomez A., et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc. Natl. Acad. Sci. USA. 2016;113:E7535–E7544. doi: 10.1073/pnas.1608585113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng R., Yao Q., Li X., Xu B. Long Noncoding Ribonucleic Acid SNHG18 Promotes Glioma Cell Motility via Disruption of α-Enolase Nucleocytoplasmic Transport. Front. Genet. 2019;10:1140. doi: 10.3389/fgene.2019.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang T., Zhang Y., Tang S., Wang Y. Long non-coding RNA LINC00052 regulates miR-608/EGFR axis to promote progression of head and neck squamous cell carcinoma. Exp. Mol. Pathol. 2019;111:104321. doi: 10.1016/j.yexmp.2019.104321. [DOI] [PubMed] [Google Scholar]
- 36.Cooper O., Hargus G., Deleidi M., Blak A., Osborn T., Marlow E., Lee K., Levy A., Pérez-Torres E., Yow A., et al. Differentiation of human ES and Parkinson’s disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid. Mol. Cell. Neurosci. 2010;45:258–266. doi: 10.1016/j.mcn.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fasano C.A., Chambers S.M., Lee G., Tomishima M.J., Studer L. Efficient Derivation of Functional Floor Plate Tissue from Human Embryonic Stem Cells. Cell Stem Cell. 2010;6:336–347. doi: 10.1016/j.stem.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Kirkeby A., Parmar M. Building authentic midbrain dopaminergic neurons from stem cells—Lessons from development. Transl. Neurosci. 2012;3:314–319. doi: 10.2478/s13380-012-0041-x. [DOI] [Google Scholar]
- 39.Daadi M.M., Weiss S. Generation of Tyrosine Hydroxylase-Producing Neurons from Precursors of the Embryonic and Adult Forebrain. J. Neurosci. 1999;19:4484–4497. doi: 10.1523/JNEUROSCI.19-11-04484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K.C., Chang H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiorenzano A., Pascale E., Patriarca E.J., Minchiotti G., Fico A. LncRNAs and PRC2: Coupled Partners in Embryonic Stem Cells. Epigenomes. 2019;3:14. doi: 10.3390/epigenomes3030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grassi D.A., Brattås P.L., Jönsson M.E., Atacho D., Karlsson O., Nolbrant S., Parmar M., Jakobsson J. Profiling of lincRNAs in human pluripotent stem cell derived forebrain neural progenitor cells. Heliyon. 2020;6:e03067. doi: 10.1016/j.heliyon.2019.e03067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S.J., Nowakowski T.J., Pollen A.A., Lui J.H., Horlbeck M.A., Attenello F.J., He D., Weissman J.S., Kriegstein A.R., Diaz A., et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:1–17. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alessio E., Buson L., Chemello F., Peggion C., Grespi F., Martini P., Massimino M.L., Pacchioni B., Millino C., Romualdi C., et al. Single cell analysis reveals the involvement of the long non-coding RNA Pvt1 in the modulation of muscle atrophy and mitochondrial network. Nucleic Acids Res. 2019;47:1653–1670. doi: 10.1093/nar/gkz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang S., Cheng S.-J., Ren L.-C., Wang Q., Kang Y.-J., Ding Y., Hou M., Yang X., Lin Y., Liang N., et al. An expanded landscape of human long noncoding RNA. Nucleic Acids Res. 2019;47:7842–7856. doi: 10.1093/nar/gkz621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J., Xu J., Zhang L., Liu S., Ma Y., Wen X., Hao J., Li Z., Ni Y., Li X., et al. Combined Single-Cell Profiling of lncRNAs and Functional Screening Reveals that H19 Is Pivotal for Embryonic Hematopoietic Stem Cell Development. Cell Stem Cell. 2019;24:285–298.e5. doi: 10.1016/j.stem.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 47.Jönsson M.E., Wahlestedt J.N., Åkerblom M., Kirkeby A., Malmevik J., Brattås P.L., Jakobsson J., Parmar M. Comprehensive analysis of microRNA expression in regionalized human neural progenitor cells reveals microRNA-10 as a caudalizing factor. Development. 2015;142:3166–3177. doi: 10.1242/dev.122747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fico A., Fiorenzano A., Pascale E., Patriarca E.J., Minchiotti G. Long non-coding RNA in stem cell pluripotency and lineage commitment: Functions and evolutionary conservation. Cell. Mol. Life Sci. 2019;76:1459–1471. doi: 10.1007/s00018-018-3000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vernay B., Koch M., Vaccarino F., Briscoe J., Simeone A., Kageyama R., Ang S.-L. Otx2 Regulates Subtype Specification and Neurogenesis in the Midbrain. J. Neurosci. 2005;25:4856–4867. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ip J.Y., Sone M., Nashiki C., Pan Q., Kitaichi K., Yanaka K., Abe T., Takao K., Miyakawa T., Blencowe B.J., et al. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci. Rep. 2016;6:27204. doi: 10.1038/srep27204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai L., Tu L., Li T., Yang X., Ren Y., Gu R., Zhang Q., Yao H., Qu X., Wang Q., et al. Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease through the inhibition of the PI3K/Akt signaling pathway. Int. Immunopharmacol. 2019;75:105734. doi: 10.1016/j.intimp.2019.105734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in supplementary material.