Abstract
Simple Summary
The present study investigated the emergence, antimicrobial susceptibility profiles, serotypes, and genotypes of Salmonella enterica from hatcheries and their upstream breeder farms to determine the occurrence of antimicrobial-resistant (AMR) Salmonella enterica contamination in hatcheries and its dissemination in an integrated broiler chicken operation. The hatcheries showed a high prevalence of Salmonella isolates with high antimicrobial resistance and no susceptible isolate. The AMR isolates from hatcheries originating from breeder farms could disseminate to the final retail market along the broiler chicken supply chain. Hatcheries play a more important role than breeder farms in the occurrence and spread of Salmonella in the broiler production chain. The emergence of AMR Salmonella in hatcheries may be due to the horizontal spread of resistant isolates rather than the first contamination of susceptible Salmonella and acquisition of resistance. Therefore, Salmonella control in hatcheries, particularly its horizontal transmission, is important.
Abstract
Positive identification rates of Salmonella enterica in hatcheries and upstream breeder farms were 16.4% (36/220) and 3.0% (6/200), respectively. Among the Salmonella serovars identified in the hatcheries, S. enterica ser. Albany (17/36, 47.2%) was the most prevalent, followed by the serovars S. enterica ser. Montevideo (11/36, 30.6%) and S. enterica ser. Senftenberg (5/36, 13.9%), which were also predominant. Thirty-six isolates showed resistance to at least one antimicrobial tested, of which 52.8% (n = 19) were multidrug resistant (MDR). Thirty-three isolates (enrofloxacin, MIC ≥ 0.25) showed point mutations in the gyrA and parC genes. One isolate, S. enterica ser. Virchow, carrying the blaCTX-M-15 gene from the breeder farm was ceftiofur resistant. Pulsed-field gel electrophoresis (PFGE) showed that 52.0% S. enterica ser. Montevideo and 29.6% S. enterica ser. Albany isolates sourced from the downstream of hatcheries along the broiler chicken supply chain carried the same PFGE types as those of the hatcheries. Thus, the hatcheries showed a high prevalence of Salmonella isolates with high antimicrobial resistance and no susceptible isolate. The AMR isolates from hatcheries originating from breeder farms could disseminate to the final retail market along the broiler chicken supply chain. The emergence of AMR Salmonella in hatcheries may be due to the horizontal spread of resistant isolates. Therefore, Salmonella control in hatcheries, particularly its horizontal transmission, is important.
Keywords: Salmonella enterica, antimicrobial resistance, hatchery, integrated broiler chicken operation, transmission
1. Introduction
Salmonellosis, caused by Salmonella enterica (S. enterica), is among the most frequently reported foodborne bacterial diseases [1]. Contaminated poultry and its products are a major source of motile Salmonellae causing salmonellosis in human worldwide [2,3]. In South Korea, Salmonella is the leading (23%) cause of bacterial foodborne poisoning. The annual production of chicken meat, the second-largest source of animal protein, was 957,000 metric tons in 2019, indicating an increase of about 1.6% since 2018 [4,5]. Among them, broilers represented 77% of slaughtered chickens in 2018. Contamination of poultry may occur throughout the broiler production chain, and the potential risk for contamination at each stage has been identified [6].
Humans are likely to be exposed to antimicrobial-resistant (AMR) Salmonella, which results from the use of antimicrobials in animals, through contaminated food, thus, leading in a health threat [7]. In recent years, increases in the emergence and spread of AMR Salmonella, particularly multidrug-resistant (MDR) Salmonella, in humans and animals have been reported worldwide, making it a global challenge [8,9,10]. Therefore, fluoroquinolones (FQs) and third generation cephalosporins (3GC) have become critically important for treating salmonellosis in humans [11]. Thus, Salmonella resistant to FQs and 3GC frequently arise in animals with easy dissemination across the food chain [12]. The dissemination of AMR Salmonella through the food chain, particularly through chickens, has important implications for the failure of salmonellosis treatment, thus, creating an increased risk to public health by the spread of AMR Salmonella via chickens [13].
It is essential to inhibit microorganisms in the broiler chicken supply chain to produce hygienic chicken meat. Epidemiological studies have clearly shown the transmission pathway of S. enterica and its serovars associated with poultry and the challenges posed by their virulence and antimicrobial resistance profiles [14]. Consequently, this pathogen has become one of the main targets for the implementation of control strategies along the poultry production chain. Breeding flocks, hatcheries, rearing farms, and slaughter plants are all recognized as critical focal points for managing the risk associated with salmonellae. A hatchery plays an important role in collecting hatching eggs from the upper breeder farm and selling newly hatched chicks to a commercial broiler farm. Some Salmonella serovars can persist in hatcheries longer than others, probably due to their ability to form biofilms [15]. Hazard Analysis and Critical Control Point (HACCP) has been applied to poultry farms (including broiler and breeder farms) and chicken slaughterhouses in Korea. However, it has not been applied to hatcheries, thus, warranting a systematic investigation and evaluation of hatchery hygiene, which has not yet been performed [16].
Salmonella can be introduced into hatcheries by horizontal and vertical transmission routes. The newly hatched chicks are more susceptible to Salmonella infection than older birds; as their intestinal flora and immune system are immature, they may become infected with a challenge of 10–100 Salmonella cells [17]. A high prevalence of Salmonella in one-day-old chickens from hatcheries has been previously reported [18]. Salmonella contamination in a hatchery can produce poor-quality chicks, resulting in a decreased feed conversion rate, increased mortality, and poor flock uniformity [16]. Moreover, the prevalence of Salmonella in hatcheries is related to Salmonella prevalence in derived meat products during processing [19]. Prevention of Salmonella contamination in chicken products requires detailed knowledge of the major sources of contamination. The critical role of a hatchery in disseminating Salmonella to commercial broiler farms and possibly exposing breeder flocks to contamination on egg trays, trolleys, and vehicles has also been previously reported [20,21,22]. Most of these works have focused on the potential for cross-contamination and infection caused by Salmonella in chicks during incubation. Considering that most integrated companies show vertical integration in Korea with numerous potential sources of Salmonella contaminants in this system, Salmonella control in integrated broiler chicken operations is complicated [23].
It is necessary to investigate the occurrence and antimicrobial resistance of Salmonella in the poultry production chain as it may aid the optimization of HACCP strategies and reduce the incidence of salmonellosis in humans. In recent years, several reports describing Salmonella prevalence in the integrated broiler supply chain in Korea have been published [19,24,25,26]. However, studies focusing on the dissemination or tracing of the AMR Salmonella along an integrated broiler chicken operation are limited. The dissemination of AMR Salmonella in the broiler farm, slaughterhouse, and its downstream retail markets has been previously described, possibly contributing to the original dissemination of AMR Salmonella to retail chicken meat [6,27]. The high prevalence of AMR Salmonella colonized in newly hatched chicks in broiler farms has emphasized upstream breeder farm and hatchery as the sources of AMR Salmonella in broiler chickens. The purposes of the present study were to identify AMR Salmonella occurrence in hatcheries and their upstream breeder farms and reveal the dissemination in a vertically integrated broiler chicken operation in South Korea.
2. Materials and Methods
2.1. Sample Collection
There were no vulnerable populations involved, and no endangered species were used in the experiments. No chickens were killed; cloacal swab samples were taken by a veterinarian with prior consent of the farm managers. The present study did not require ethical approval.
From September 2015 to August 2016, 420 fresh samples were collected from 44 hatcheries and their upstream 25 breeder farms. The sampling was done in two parts as follows:
-
(i)
A total of 220 cloacal swab test samples were collected from 44 hatcheries. Twenty-five cloacal swab samples were randomly collected from the entire area of each hatchery, and samples from five chicks were pooled into one test sample (S, n = 5).
-
(ii)
A total of 200 test samples, including 125 cloacal swab samples and 75 litter samples, were collected from 25 breeder farms. Twenty-five cloacal swab samples and fifteen litter samples were randomly collected from the entire areas of each breeder farm, and five samples obtained from the similar area were pooled into one test sample. Finally, cloacal swabs (S, n = 5) and litter (L, n = 3) were collected from each farm.
2.2. Isolation and Identification of Salmonella
Samples were collected in sterile plastic conical tubes (50 mL; SPL Life Sciences Co., Ltd., Seoul, Korea) and stored under refrigeration in the laboratory until analysis, which was performed within 48 h of arrival. Salmonellae were isolated using previously described standard methods [28]. The DNA of Salmonella-positive colonies, extracted by the boiling method, was further tested by polymerase chain reaction (PCR) using the Salmonella-specific gene (invA) [29]. All strains were serotyped per the Kauffmann–White scheme using slide agglutination with O and H antigen-specific sera (BD Difco, Sparks, MD, USA and Denka Seiken Co., Ltd., Tokyo, Japan) [30].
2.3. Antimicrobial Susceptibility Test
The minimum inhibitory concentrations (MICs) of the 16 antimicrobials were determined using the KRNV4F Sensititre panel (TREK Diagnostic Systems, Incheon, Korea). The MIC of enrofloxacin was determined using the agar dilution method. Escherichia coli (ATCC 25922) was used as the quality control strain. The susceptibility breakpoints of most antimicrobials were interpreted based on the Clinical and Laboratory Standard Institute (CLSI) guidelines [31], whereas those of the antimicrobials used for the animals, including ceftiofur, enrofloxacin, and florfenicol, were interpreted based on the CLSI standards document VET-01 [32]. The CLSI criteria were not available for streptomycin or neomycin, for which we used other references [7,33] (see Supplementary Materials, Table S1). Salmonella isolates, resistant to at least three antimicrobial classes, were considered to be MDR.
2.4. Molecular Characterization of Resistance
For determining the molecular characterization of quinolone resistance genes, the quinolone-resistant isolates were further detected on plasmid-mediated quinolone resistance (PMQR) genes by PCR and sequencing. These genes included qnrA, qnrB, qnrS, qnrD, qepA, oqxA, and aac(6′)-lb-cr and mutations in the quinolone-resistance determining region (QRDR), gyrA, and parC genes, as previously described [34]. Positive controls were used in all PCR reactions. PCR products were purified using the QIAquick Gel Extraction kit (Qiagen, Hilden, Germany) and were further sequenced directly (SolGent Co., Ltd., Daejeon, Korea) for sequence analysis and aligned using the BLAST program (www.ncbi.nlm.nih.gov/BLAST/). The inferred amino acid sequences of the QRDR-encoding genes were compared with the corresponding regions of the reference strain S. Typhimurium LT2 (GenBank accession no. AE006468).
For detecting the molecular characterization of resistance to 3GC, isolates exhibiting extended-spectrum β-lactamase/ampicillin-class C (ESBL/AmpC) phenotypes were further screened by PCR, as previously described [35].
2.5. Pulsed-Field Gel Electrophoresis and BioNumerics Analysis
The Salmonella isolates (n = 38) selected from the hatcheries and upstream breeder farms were genotyped using pulsed-field gel electrophoresis (PFGE) following the protocols of the Centers for Disease Control and Prevention available on PulseNet (www.pulsenetinternational.org) with certain modifications, as described previously [28]. Among them, S. enterica ser. Albany (n = 17), S. enterica ser. Montevideo (n = 9), and S. enterica ser. Virchow (n = 2) were selected to further compare the genotypic relatedness of isolates with relevant serovars (S. enterica ser. Montevideo, n = 100; S. enterica ser. Albany, n = 71; and S. enterica ser. Virchow, n = 25) from downstream of the integrated broiler chicken operation, including broiler farms, slaughterhouse, and retail markets (in our previous studies) [6,27].
2.6. Statistical Analysis
The Chi-square test was used to test for significant differences in the rates of Salmonella isolation among hatcheries and breeder farms and p-values less than 0.05 were considered to be statistically significant. The software SPSS (version 19.0; IBM Co., Armonk, NY, USA) was used for statistical analysis.
3. Results
3.1. Prevalence and Serovars of Salmonella
A total of 36 isolates (16.4%) from the samples obtained from hatcheries and six isolates (3.0%) obtained from their upstream breeder farms were positive for Salmonella (Table S2). There was a significant difference (p < 0.05) in isolation rates among the breeder farm and hatchery; however, there were no significant differences in isolation rates among the cloacal swabs samples (1/125, 0.8%) and the litter samples (5/75, 6.7%) in the breeder farms. Three of 25 (12.0%) breeder farms were positive for S. enterica; eighteen of 44 hatcheries (40.9%) were positive for Salmonella. The prevalence of S. enterica per hatchery and in breeder farms is shown in Table S3. Five serovars were identified among the 42 Salmonella-positive isolates (Table 1). The most common serotype recovered from the hatcheries was S. enterica ser. Albany (17 isolates, 47.2%), followed by S. enterica ser. Montevideo (11 isolates, 30.6%) and S. enterica ser. Senftenberg (five isolates, 13.9%). Only one isolate with S. enterica ser. Omuna was detected, and two isolates (5.6%) marked as “untypable” could not be assigned to specific serotypes. The serotypes recovered from breeder farms were S. enterica ser. Montevideo (3 isolates, 50%) and S. enterica ser. Virchow (three isolates, 50%).
Table 1.
Serotype distribution of Salmonella enterica in hatcheries and their upstream breeder farms.
| Serovar (Serogroup) | Sampling Site, n a (%) | ||
|---|---|---|---|
| Hatchery | Breeder Farm | Total | |
| Albany (C2–C3) | 17 (47.2) | 0 | 17 (40.5) |
| Montevideo (C1) | 11 (30.6) | 3 (50.0) | 14 (33.3) |
| Senftenberg (E4) | 5 (13.9) | 0 | 5 (11.9) |
| Virchow (C1) | 0 | 3 (50.0) | 3 (7.1) |
| Omuna (C1) | 1 (2.8) | 0 | 1 (2.4) |
| Untypable | 2 (5.6) | 0 | 2 (4.8) |
| Total | 36 | 6 | 42 |
an, number of isolates.
3.2. Antimicrobial Susceptibility Analysis
All 36 isolates from hatcheries were resistant to at least one antimicrobial; 19 of these isolates (52.8%) were MDR (Table 2). Among these 19 MDR isolates were all isolates of S. enterica ser. Albany (n = 17) and the only isolate of S. enterica ser. Omuna (n = 1) and untypable (n = 1); no isolate of S. enterica ser. Senftenberg or S. enterica ser. Montevideo was MDR. Isolates of S. enterica ser. Albany with resistance to NAL (17/17, 100.0%), TET (17/17, 100.0%), AMP (17/17, 100.0%), SXT (17/17, 100.0%), and CHL (17/17, 100.0%) were the most prevalent, followed by those resistant to STR (11/17, 64.7%) and FFN (3/17, 17.6%). Isolates of S. enterica ser. Montevideo with resistance to NAL (11/11, 100.0%) were the most prevalent, followed by the one resistant to SXT (1/11, 9.1%). Isolates of S. enterica ser. Senftenberg with resistance to NAL (5/5, 100.0%) were the most prevalent. Among all isolates from hatcheries, 50.0% (18/36) and 47.2% (17/36) showed intermediate resistance to CIP and enrofloxacin (ENR), respectively. All 36 isolates from the hatcheries were susceptible to the four antimicrobials NEO, GEN, CEP, and XNL. Six antimicrobial resistance profiles were observed among Salmonella isolates from the hatcheries; the antimicrobial resistance profile NAL (13/36, 36.1%) was the most prevalent antimicrobial resistance profile, followed by NAL-STR-TET-SXT-AMP-CHL (12/36, 33.3%) (Table 3). The isolates of S. enterica ser. Albany were all resistant to ≥5 antimicrobials. Antimicrobial resistance profiles of NAL-STR-TET-SXT-AMP-CHL (n = 11), NAL-TET-SXT-AMP-CHL (n = 3), and NAL-TET-SXT-AMP-CHL-FFN (n = 3) were confirmed. Five of six isolates from breeder farms were MDR.
Table 2.
Antimicrobial resistance of Salmonella enterica isolated from hatcheries and their upstream breeder farms a.
| Serovar | n | No. (%) of Isolates Resistant to Antimicrobials | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL | CIP | ENR | NEO | GEN | STR | TET | AMC | CEP | FOX | XNL | AMP | SXT | COL | FFN | CHL | MDR | ||
| Hatchery | ||||||||||||||||||
| Albany | 17 | 17 (100.0) | 0 | 0 | 0 | 0 | 11 (64.7) | 17 (100.0) | 0 | 0 | 0 | 0 | 17 (100.0) | 17 (100.0) | 0 | 3 (17.6) | 17 (100.0) | 17 (100.0) |
| Montevideo | 11 | 11 (100.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (9.1) | 0 | 0 | 0 | 0 |
| Senftenberg | 5 | 5 (100.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Omuna | 1 | 1 (100.0) | 0 | 0 | 0 | 0 | 1 (100.0) | 1 (100.0) | 0 | 0 | 0 | 0 | 1 (100.0) | 1 (100.0) | 0 | 0 | 1 (100.0) | 1 (100.0) |
| Untypable | 2 | 1 (50.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (50.0) | 0 | 1 (50.0) | 1 (50.0) | 1 (50.0) | 0 | 0 | 1 (50.0) |
| Subtotal | 36 | 35 (97.2) | 0 | 0 | 0 | 0 | 15 (41.7) | 18 (50.0) | 0 | 0 | 1 (2.8) | 0 | 19 (52.8) | 20 (55.6) | 1 (2.8) | 3 (8.3) | 18 (50.0) | 19 (52.8) |
| Breeder farm | ||||||||||||||||||
| Montevideo | 3 | 3 (100.0) | 0 | 0 | 3 (100.0) | 0 | 3 (100.0) | 3 (100.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (100.0) |
| Virchow | 3 | 2 (66.7) | 0 | 0 | 3 (100.0) | 0 | 2 (66.7) | 3 (100.0) | 0 | 0 | 0 | 2 (66.7) | 2 (66.7) | 0 | 0 | 0 | 0 | 2 (66.7) |
| Subtotal | 6 | 5 (83.3) | 0 | 0 | 6 (100.0) | 0 | 5 (83.3) | 6 (100.0) | 0 | 0 | 0 | 2 (33.3) | 2 (33.3) | 0 | 0 | 0 | 0 | 5 (83.3) |
| Total | 42 | 40 (95.2) | 0 | 0 | 6 (14.3) | 0 | 17 (40.5) | 24 (57.1) | 0 | 0 | 1 (2.4) | 2 (4.8) | 21 (50.0) | 23 (54.8) | 1 (2.4) | 3 (7.1) | 18 (42.9) | 24 (57.1) |
a NAL, nalidixic acid; CIP, ciprofloxacin; ENR, enrofloxacin; NEO, neomycin; GEN, gentamicin; STR, streptomycin; TET, tetracycline; AMC, amoxicillin/clavulanic acid; CEP, cephalexin; FOX, cefoxitin; XNL, ceftiofur; AMP, ampicillin; SXT, trimethoprim/sulfamethoxazole; COL, colistin; FFN, florfenicol; CHL, chloramphenicol; MDR, multiple drug resistance; n, number of isolates.
Table 3.
Antimicrobial resistance profile of Salmonella isolates from hatcheries and their upstream breeder farm.
| No. | Antimicrobial Resistance Profile a | Hatchery (n b = 36) | Breeder Farm (n = 6) | ||
|---|---|---|---|---|---|
| n (%) | Serovars (n) | n (%) | Serovars (n) | ||
| 1 | NAL | 13 (36.1) | Montevideo (7), Senftenberg (5), untypable (1) | 0 | - |
| 2 | NAL-STR-TET-SXT-AMP-CHL | 12 (33.3) | Albany (11), Omuna (1) | 0 | - |
| 3 | NAL-SXT | 4 (11.1) | Montevideo (4) | 0 | - |
| 4 | NAL-TET-SXT-AMP-CHL | 3 (8.3) | Albany (3) | 0 | - |
| 5 | NAL-TET-SXT-AMP-CHL-FFN | 3 (8.3) | Albany (3) | 0 | - |
| 6 | SXT-AMP-FOX-COL | 1 (2.7) | Untypable (1) | 0 | - |
| 7 | NAL-NEO-STR-TET | 0 | - | 3 (50.0) | Montevideo (3) |
| 8 | NAL-NEO-STR-TET-AMP-XNL | 0 | - | 1 (16.7) | Virchow (1) |
| 9 | NEO-STR-TET | 0 | - | 1 (16.7) | Virchow (1) |
| 10 | NAL-NEO-TET-AMP-XNL | 0 | - | 1 (16.7) | Virchow (1) |
NAL, nalidixic acid; NEO, neomycin; STR, streptomycin; TET, tetracycline; FOX, cefoxitin; XNL, ceftiofur; AMP, ampicillin; SXT, trimethoprim/sulfamethoxazole; COL, colistin; FFN, florfenicol; CHL, chloramphenicol; n, number of isolates.
3.3. Quinolone-Resistance Determining Region (QRDR) Mutations, Plasmid-Mediated QuinoloneRresistance (PMQR), and Extended-Spectrum β-Lactamase (ESBL)-Producing Isolates
The prevalence of PMQR and QRDR mutations among Salmonella isolates (ENR, MIC ≥ 0.25) is shown in Table 4. PMQR genes were not observed in any test isolate; all test isolates from hatcheries (n = 33) showed single point mutations in gyrA and parC genes. Point mutations with Ser-83-Tyr, Ser-83-Phe, and Asp-87-Gly were found in the gyrA gene; point mutations with Tyr-57-Ser and Tyr-57-Thy were found in the parC gene. Of the two Salmonella isolates from breeder farm with resistance to XNL (MICs ≥ 8), one isolate carried an ESBL gene, which was blaCTX-M-15.
Table 4.
Prevalence of plasmid-mediated quinolone resistance (PMQR) and quinolone-resistance determining region (QRDR) mutations among Salmonella (enrofloxacin (ENR), minimum inhibitory concentrations (MIC) ≥ 0.25) isolated from hatcheries and their upstream breeder farms.
| Patterns | ENR MIC (µg/mL) | PMQR | QRDR Mutations | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gyrA | parC | No. of Isolates | ||||||||
| Ser-83-Tyr | Ser-83-Phe | Ser-87-Gly | Tyr-57-Ser | Tyr-57-Thy | Hatchery | Breeder Farm | Total | |||
| P1 | 0.25−0.50 | □ | □ | □ | ■ | ■ | □ | 15 | 3 | 18 |
| P2 | 0.25−0.50 | □ | □ | ■ | □ | □ | ■ | 9 | 1 | 10 |
| P3 | 0.25−0.50 | □ | □ | ■ | □ | ■ | □ | 8 | 0 | 8 |
| P4 | 0.25 | □ | ■ | □ | □ | ■ | □ | 1 | 0 | 1 |
| P5 | 0.50 | □ | □ | □ | □ | ■ | □ | 0 | 1 | 1 |
| n, % | 0 | 1 (2.6) | 18 (47.4) | 18 (47.4) | 37 (97.4) | 1 (2.6) | 33 | 5 | 38 | |
n, indicates the number of isolates; % indicates the percentage; ■/□, indicate the presence/absence, respectively, of designated gene and mutation in each Salmonella isolate tested.
3.4. Correlations among Salmonella Isolates from Hatcheries and Downstream Stages along an Integrated Broiler Chicken Operation Based on Genotypic Characteristics
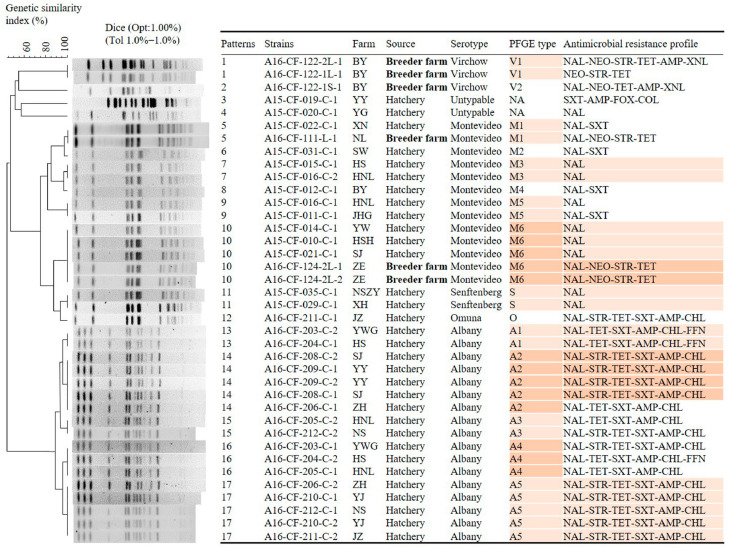
A total of 38 selected isolates were subdivided into 17 PFGE types with 100% similarity (Figure 1). Different serotypes were detected on the same farms. S. enterica ser. Montevideo and S. enterica ser. Virchow isolates were identified from farm BY, S. enterica ser. Albany and S. enterica ser. Montevideo from farm SJ, and S. enterica ser. Omuna and S. enterica ser. Albany isolates from farm JZ. Isolates, including 17 S. enterica ser. Albany and 13 S. enterica ser. Montevideo, were classified into five and six PFGE types, respectively, based on characterization with PFGE using the restriction enzyme XbaI. Among these, one PFGE type (M1) was shared among breeder farms and hatchery isolates. Isolates of the same PFGE type were observed in different hatchery farms (type M3 in farms HS and HNL; type M5 in farms HNL and JHG; type M6 in farms YW, HSH, SJ, and ZE; type S in farms NSZY and XH; type A1 in farms YWG and HS; type A2 in farms YY, SJ, and ZH; type A3 in farms HNL and NS; type A4 in farms YWG, HS, and HNL; and type A5 in farms ZH, YJ, NS, and JZ). Conversely, isolates of different PFGE types were observed in the same farms (types M3, M5, A3, and A4 in farm HNL; types A1 and A4 in farms YWG and HS; and types A2 and A5 in farm ZH).
Figure 1.
Dendrograms of the pulsed-field gel electrophoresis (PFGE) types of the 38 selected Salmonella isolates recovered from the hatchery and upstream breeder farms. These isolates and their association with source and antimicrobial resistance profiles are shown. Backgrounds with the same-colored bars (orange) indicate samples with the same PFGE patterns and antimicrobial resistance profiles; bold and non-bold indicate samples with different source.
The 224 isolates of S. enterica (S. enterica ser. Montevideo, n = 109; S. enterica ser. Albany, n = 88; and S. enterica ser. Virchow, n = 27) from the integrated broiler chicken operation can be divided into 50 PFGE types, including 30, 11, and 9 PFGE types in S. enterica ser. Montevideo, S. enterica ser. Albany, and S. enterica ser. Virchow isolates, respectively (Figure S1). The distribution of PFGE types of Salmonella isolates from different stages along the chicken production chain is shown in Table 5. Eleven PFGE types of Salmonella isolates from hatcheries were detected. Among these, two (18.2%) PFGE types (SM-3 and SM-7) were found consistent with upstream breeder farms and eight (72.7%) PFGE types (SM-3, SM-7, SM-11, SM-12, and SM-13; SA-9, SA-10, and SA-11) were consistent with downstream stages, including broiler farms (SM-3, SM-7, SM-11, SM-12, and SM-13), slaughterhouses (SM-7, SM-12, SA-10, and SA-11), and retail markets (SM-7, SM-12, SA-9, SA-10, and SA-11). Among these PFGE types, SM-7 was found in isolates from breeder farms, hatcheries, broiler farms, slaughterhouses, and retail markets (Figure S1a). Furthermore, the types SA-10 and SA-11 were also found in isolates from hatcheries, slaughterhouses, and retail markets (Figure S1b); no identical PFGE type was found in isolates of S. enterica ser. Virchow from breeder farms and the downstream stages (Figure S1c). The 52 (52.0%, 52/100) S. enterica ser. Montevideo isolates procured from the downstream of the hatchery stage carried the same PFGE types as those of the hatchery, and the most prevalent PFGE types were SM-7 and SM-12, accounting for 25.0% (25/100) and 14.0% (14/100) of the isolates, respectively. Similarly, the 21 (29.6%, 21/71) S. enterica ser. Albany isolates procured from the downstream of the hatchery stage carried the same PFGE types as those of the hatchery, and the second most prevalent PFGE type SA-11 accounted for 19.7% (14/71) of the isolates (Table 6 and Figure S2). The prevalence of overlapped PFGE types in S. enterica ser. Montevideo (52/100, 52.0%) is significantly higher (p = 0.003) than that of in S. enterica ser. Albany (21/71, 29.6%); the prevalence of the most prevalent overlapped PFGE types in each serotype (SM-7 of S. enterica ser. Montevideo and SA-11 of S. enterica ser. Albany, respectively) is significantly higher than that of non-overlapped PFGE types (SM-6 (or SM-15, 22, 26, 28) and SA-4, respectively) (Table S4).
Table 5.
The distribution of PFGE type of Salmonella isolates from different stages along the chicken production chain.
| Serovar | PFGE Type | Sampling Site | ||||
|---|---|---|---|---|---|---|
| Breeder Farm | Hatchery | Broiler Farm | Slaughterhouse | Retail Market | ||
| Montevideo | SM | 3 b, 7 | 3, 7, 11, 12, 13, 14 | 3, 5, 6, 7, 9, 10, 11, 12, 13, 15, 16, 17, 18, 20, 24, 25, 26, 27, 28, 29, 30 | 1, 2, 4, 7, 8, 10, 12, 21, 23 | 7, 12, 19, 22 |
| Albany | SA | - a | 6, 8, 9, 10, 11 | - | 1, 2, 3, 4, 5, 7, 10, 11 | 1, 2, 4, 5, 9, 10, 11 |
| Virchow | SV | 8, 9 | - | 2 | 2, 3, 4, 6, 7 | 1, 2, 4, 5 |
a No isolate of the target serotype; b number indicating the PFGE type, as shown in Figure S1.
Table 6.
The prevalence of Salmonella isolates from downstream of the hatchery stage along the integrated broiler chicken operation, with overlapped PFGE types with the hatchery stage *.
| Serovar | Overlap or Not | PFGE Type | Downstream (Broiler Farm, Slaughterhouse, Retail Market) | |
|---|---|---|---|---|
| Number of Isolates/per PFGE Type | Percentage (%) | |||
| Montevideo | Overlap | SM-7 | 25 | 25.0 |
| SM-12 | 14 | 14.0 | ||
| SM-13 | 5 | 5.0 | ||
| SM-3, 11 | 4 | 4.0 | ||
| Subtotal | 52 | 52.0 | ||
| Non-overlap | SM-6, 15, 22, 26, 28 | 4 | 4.0 | |
| SM-27 | 3 | 3.0 | ||
| SM-1, 5, 10, 16, 19, 23, 29 | 2 | 2.0 | ||
| SM-2, 4, 8, 9, 17, 18, 20, 21, 24, 25, 30 | 1 | 1.0 | ||
| Subtotal | 48 | 48.0 | ||
| Total | 100 | 100.0 | ||
| Albany | Overlap | SA-11 | 14 | 19.7 |
| SA-10 | 4 | 5.6 | ||
| SA-9 | 3 | 4.2 | ||
| Subtotal | 21 | 29.6 | ||
| Non-overlap | SA-4 | 24 | 33.8 | |
| SA-7 | 9 | 12.7 | ||
| SA-1 | 8 | 11.3 | ||
| SA-5 | 4 | 5.6 | ||
| SA-2 | 3 | 4.2 | ||
| SA-3 | 2 | 2.8 | ||
| Subtotal | 50 | 70.4 | ||
| Total | 71 | 100.0 | ||
| Virchow | Non-overlap | SV-2 | 11 | 44.0 |
| SV-4 | 6 | 24.0 | ||
| SV-1 | 4 | 16.0 | ||
| SV-3, 5, 6, 7 | 1 | 4.0 | ||
| Total | 25 | 100.0 | ||
* This table was converted to a Venn diagram to illustrate the distribution of PFGE types in Figure S2.
4. Discussions
In the present study, the prevalence of S. enterica (16.4%) was higher in hatcheries than in its upstream breeder farm (3.0%), despite fumigation being routinely used during hatching (Table S2) [16]. The prevalence of Salmonella in hatcheries varied widely from operation to operation (6.78–44.9%) and may have been associated with differences in hygiene and sanitation levels of each operation and the different detection methods used in each study [25,36,37]. Herein, the Salmonella isolation rate from breeder farms was relatively lower (3.0%) than in previous studies in Korea (14.7–19.0%) and China (10.53–18.15%); however, even infected breeder flocks have been shown to cause widespread Salmonella contamination [18,25]. The comparison of our Salmonella isolation rates in cloacal swabs (1/125, 0.8%) and litter (5/75, 6.7%) samples from breeder farms with previous studies (0% and 40%, respectively) indicates that different sample types may also be a factor influencing the prevalence of Salmonella [24,38].
In total, S. enterica ser. Albany was the dominant serovar in hatcheries (Table 1). These results are consistent with our previous study on isolating AMR Salmonella isolates from chicken slaughterhouses and retail markets [6]. Similarly, studies conducted on the prevalence of Salmonella in poultries in Vietnam, Malaysia, and Myanmar showed that the predominant serovar was S. enterica ser. Albany (34.1%, 35.4%, and 38%, respectively) [39,40,41]. Contrary to our results, S. enterica ser. Hadar was the most frequently reported serovar in integrated broiler operations in Korea; the Salmonella serotype most often isolated from the hatchery was S. enterica ser. Senftenberg [16,19,25]. S. enterica ser. Albany, one of the prominent serovars in poultry that has been infecting animals and humans for several decades, may be an emerging serotype in Korea in the future [42,43]. Consistent with our results, S. enterica ser. Montevideo, S. enterica ser. Senftenberg, and S. enterica ser. Virchow were the most frequently reported serovars in the poultry industry in Korea [25,26,44]. S. Enteritidis has typically been the most common serotype responsible for Salmonella infections in poultry in Korea for several decades; however, this prevalence has decreased, and this serovar has been gradually replaced by other emerging serovars. Therefore, the predominant Salmonella serovar varies from company to company and time to time.
Antimicrobial resistance of Salmonella is a globally emerging problem of public health concern. In the present study, no susceptible isolate was found in any of the 42 isolates. Among the 16 antimicrobial agents tested, the highest resistance rate observed in the hatchery was to NAL (97.2%), followed by SXT (55.6%), AMP (52.8%), TET (50.0%), and CHL (50.0%), which is consistent with previous reports (Table 2) [6,45]. Antimicrobial resistance was detected even in isolates from hatcheries that were not treated with antimicrobials. One potential explanation is that those AMR isolates came from upstream breeder farms. Quinolones, ampicillin, and tetracyclines have been widely used for therapy, prophylaxis, and growth promotion by breeders, while sulfonamides have been used in human and veterinary medicine for 40 years [46,47]. Another potential explanation is that the hatcheries are contaminated with AMR Salmonella in the internal environment of the hatchery or the external natural environment [48]. Contrary to the increasing incidence of FQ-resistant Salmonella reported worldwide, we did not find FQ resistance in the present study. However, a marked resistance to NAL reported herein could be a matter of concern because NAL resistance has been associated with a decrease in susceptibility to FQs, which are used to treat salmonellosis in humans. The present study indicated that MDR Salmonella contamination was more widespread in the hatchery (19/36, 52.8%) (Table 2) than suggested by another report [49]. Among MDR isolates, we observed resistance to 3GC and COL, which are critically important in treating salmonellosis in humans [11]. For example, two S. enterica ser. Virchow isolates from breeder farms were resistant to XNL, with the antimicrobial resistance profiles NAL-NEO-STR-TET-AMP-XNL and NAL-NEO-TET-AMP-XNL (Table 3); one isolate (untypable) from the hatchery was resistant to COL, with the antimicrobial resistance profile SXT-AMP-FOX-COL. Colistin, as an antimicrobial substance, was used against Gram-negative bacteria. The use of colistin has been limited due to systemic toxicity. However, it has been re-introduced as a last-line option in the treatment of human infections [50]. The resistance can be transmitted to humans through the food chain. Eventually, it can lead to microbial cross-resistance and pose a threat to human health. Therefore, it is mandatory to monitor the dissemination of resistance to colistin [51].
Although PMQR has been studied and increasingly reported, QRDR mutations seem to represent the main mechanism of quinolone resistance in animal isolates [52]. Moreover, PMQR was commonly detected in Enterobacteriaceae, particularly in E. coli, and the prevalence of PMQRs in Salmonella remains extremely low [53]. This finding was consistent with our observations that the global level of FQ non-susceptibility may be mainly due to QRDR chromosomal mutations (Table 4). Our results indicated that missense mutations frequently occurred in the QRDR of gyrA and parC, which are considered to be the major quinolone resistance determinants in Salmonella [54]. In the present study, we identified QRDR point mutations in the gyrA and parC genes in all selected Salmonella isolates from hatcheries, a finding inconsistent with previous studies showing one-point mutation only in the gyrA gene as the main pattern [55,56,57]. The results imply that resistance to FQs is continuously evolving with time. The results also show that antimicrobial pressure, rather than the horizontal transmission of antimicrobial resistance genes in chickens, leads to the appearance of antimicrobial resistance; clonal dissemination seems to be a key contributing factor for increasing resistance to FQs among Salmonella in hatcheries, where antimicrobials are not applicable [58]. This finding indicates the potential risk that Salmonella isolates with mutations in gyrA and parC could naturally be maintained during hatching, even with no antimicrobial pressure. The Salmonella isolates with QRDR may be directly transmitted to the downstream broiler farms through their carrying by day-old chicks [59]. Our results also showed that one resistance gene (blaCTX-M-15), which encodes resistance to ESBL, was identified in S. enterica ser. Virchow, one of the most frequently identified serotypes in 2015–2016 [6]. There was a potential risk of ESBL-producing Salmonella isolates being transmitted to humans through contaminated poultry products [60].
Currently, PFGE is an easy and effective method to assess relatedness among Salmonella isolates from different sources [6,27]. The clonal relationship among isolates from hatcheries and their upstream breeder farms at the chromosome level was accessed using PFGE (Figure 1). There was frequent Salmonella cross-contamination among hatcheries and among hatcheries and upstream breeder farms. An identical PFGE type (M1) was shared between isolates from a hatchery and its upstream breeder farm isolates, suggesting that Salmonella contamination in hatcheries could be achieved by a direct vertical top-down transmission [36]. Isolates from different hatcheries shared the same PFGE types, indicating Salmonella cross-transmission among the hatcheries, possibly due to the sharing of the same source of eggs or trucks within the same operation [24].
To further determine and compare the genotypic relatedness of isolates from the integrated broiler chicken operation, selected isolates of the three most prevalent serotypes from breeder farms, hatcheries, broiler farms, slaughterhouses, and retail markets were analyzed by PFGE (Figure S1). A highly consistent PFGE pattern (SM-7) from different sources revealed that the AMR S. enterica clones could disseminate through the broiler chicken supply chain (Table 6 and Figure S2). The SM-7, not the main PFGE type in hatcheries, could be disseminated to the downstream stage (broiler farm) even throughout the broiler supply chain, suggesting that S. enterica ser. Montevideo could persist and is difficult to eliminate from the environment, probably due to its biofilm-producing ability [61,62]. Herein, 52% of the S. enterica ser. Montevideo and 29.6% of the S. enterica ser. Albany isolates from the downstream of the hatchery carried the same PFGE types as those of the hatchery, indicating that Salmonella contamination in hatcheries was an important source of Salmonella contamination in the integrated broiler chicken operation (Table 6 and Figure S2). From Table S4, it can be concluded that biocontrol of Salmonella contamination in hatcheries is important; and control of S. enterica ser. Montevideo isolates from hatcheries are more necessary.
The routes of Salmonella cross-contamination within the hatchery and between the hatchery and its upstream breeder farm were complicated. Salmonella was detected in the hatchery and its upstream breeder farm (represented by S. enterica ser. Montevideo) (Table 5). Two PFGE types (SM-3 and SM-7) were consistent with upstream breeder farms, indicating that vertical transfer of infection from breeding birds to progeny is an important aspect of the epidemiology of S. enterica infection within the poultry industry [24,63]. More importantly, emerging serotypes and genotypes were detected in the hatchery. The hatchery samples were contaminated with non-original Salmonella with a serotype different from those in the breeder farm (represented by S. enterica ser. Albany); the hatchery samples were contaminated with non-original Salmonella with PFGE types different from those in the breeder farm (represented by SM-11, SM-12, SM-13, and SM-14). Salmonella was absent in upstream breeder farms but present in the hatchery, suggesting that contamination can happen during hatching. Therefore, apart from the original Salmonella contamination in upstream breeder farms, there is at least one more route of Salmonella contamination in the hatchery. There are many ways in which Salmonella can enter these extensive and integrated operations and be recirculated and amplified by various routes [26]. In some cases, clonal horizontal transmission in the hatchery and on the farm during the rearing period is of greater importance and leads to the isolation of a greater variety of Salmonella serovars [64]. For instance, several risk factors for horizontal transmission have been identified, such as inadequate cleaning and disinfection of hatching houses, which leads to contamination of the downstream hatching eggs and a poor level of hygiene [64]. Usually, Salmonella infection does not interfere with hatchability, but during hatching, the organisms are widely spread in the hatcher via ducts and the fluff that is disseminated by forced ventilation [63,65]. The higher prevalence of Salmonella in a hatchery than in a relatively clean place indicates that intervention strategies must target this stage to prevent Salmonella from entering the downstream broiler farm. This approach requires that Salmonella should be detected quickly and accurately at the hatching stage before entering the broiler farm. Our study had the limitation that we did not provide direct evidence to prove the horizontal transmission of Salmonella in the hatchery. Further studies focusing on the investigation of the horizontal transmission routes of Salmonella in hatcheries are needed.
5. Conclusions
Our study found a high prevalence of Salmonella at the hatchery stage, high antimicrobial resistance, and no susceptible isolates. The AMR isolates could disseminate to the downstream stage, even to the final retail market, along the broiler chicken supply chain. Compared with breeder farms, Salmonellae in the hatchery play a more important role in their emergence and spread in the broiler production chain. The emergence of AMR Salmonella in the hatchery may be due to the horizontal spread of resistant isolates rather than the first contamination of Salmonella, and then the acquisition of resistance. Therefore, the presence of AMR Salmonella on day-old chicks in the hatchery is very likely to reduce the effectiveness of the biosafety prevention and control of Salmonella in the rearing farm. The broiler farm, with a high possibility to explore antimicrobials, could provide new or higher resistant isolates in the next stage along the broiler production chain. Using Salmonella-free chicks would be an effective way to control AMR Salmonella emergence and transmission in broiler chickens. Furthermore, the national surveillance program should be implemented in hatcheries for monitoring AMR Salmonella in chickens.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/11/1/154/s1, Figure S1: Dendrograms of the PFGE types of the selected S. enterica ser. Montevideo (a), S. enterica ser. Albany (b), and S. enterica ser. Virchow (c) isolates recovered from an integrated broiler chicken operation, Figure S2: The prevalence of Salmonella isolates from downstream of the hatchery step along the integrated broiler chicken operation, with overlap of the PFGE type with hatchery (see the details in Table 6). Backgrounds with the same-colored bars (orange) indicate samples with the same PFGE patterns and antimicrobial resistance profiles, Table S1: Antimicrobials and the range of concentration tested, Table S2: Prevalence of Salmonella enterica in hatcheries and their upstream breeder farms, Table S3: Prevalence of Salmonella enterica per hatcheries and breeder farms, Table S4: The prevalence of Salmonella isolates from downstream of the hatchery with overlap the PFGE types with the hatchery stage.
Author Contributions
K.S., M.K., and H.-K.J. contributed to conception and design of experiments; S.-Y.C. and B.W. contributed to acquisition, analysis, and interpretation of data; B.W., S.-Y.C., J.-F.Z., J.-Y.P., and Y.-J.L. drafted and/or revised the article. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through the Agriculture, Food, and Rural Affairs Convergence Technologies Program for Educating Creative Global Leader (716002-7, 320005-4) and the Animal Disease Management Technology Development Program (119059-2) funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA). In addition, this paper was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (no. 2017R1D1A1B03030883).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Centers for Disease Control and Prevention . National Salmonella Surveillance Annual Report and Appendices 2013. US Department of Health and Human Services; Atlanta, GA USA: 2016. [Google Scholar]
- 2.Li K., Ye S., Alali W.Q., Wang Y., Wang X., Xia X., Yang B. Antimicrobial susceptibility, virulence gene and pulsed-field gel electrophoresis profiles of Salmonella enterica serovar Typhimurium recovered from retail raw chickens, China. Food Control. 2017;72:36–42. doi: 10.1016/j.foodcont.2016.07.032. [DOI] [Google Scholar]
- 3.Jackson B.R., Griffin P.M., Cole D., Walsh K.A., Chai S.J. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg. Infect. Dis. 2013;19:1239. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Min K.-J., Hwang I.-G., Lee S.-H., Cho J.-I., Yoon K.-S. Determination of risk ranking of combination of potentially hazardous foods and foodborne pathogens using a risk ranger. J. Food Hyg. Saf. 2011;26:91–99. [Google Scholar]
- 5.Sunyoung Choi T.M. South Korea: Poultry and Products Annual 2019. [(accessed on 6 August 2020)]; Available online: http://agriexchange.apeda.gov.in/marketreport/Reports/Poultry_and_Products_Annual_Seoul_Korea_Republic_of_8-29-2019.pdf.
- 6.Shang K., Wei B., Jang H.-K., Kang M. Phenotypic characteristics and genotypic correlation of antimicrobial resistant (AMR) Salmonella isolates from a poultry slaughterhouse and its downstream retail markets. Food Control. 2019;100:35–45. doi: 10.1016/j.foodcont.2018.12.046. [DOI] [Google Scholar]
- 7.Centers for Disease Control . National Antimicrobial Resistance Monitoring System: Enteric Bacteria. 2012. U.S. Department of Health and Human Services, CDC; Atlanta, GA, USA: 2014. [(accessed on 21 August 2020)]. Available online: https://www.cdc.gov/narms/pdf/2012-annual-report-narms-508c.pdf. [Google Scholar]
- 8.Eller C., Simon S., Miller T., Frick J.-S., Prager R., Rabsch W., Guerra B., Werner G., Pfeifer Y. Presence of β-lactamases in extended-spectrum-cephalosporin-resistant Salmonella enterica of 30 different serovars in Germany 2005–11. J. Antimicrob. Chemother. 2013;68:1978–1981. doi: 10.1093/jac/dkt163. [DOI] [PubMed] [Google Scholar]
- 9.Djeffal S., Bakour S., Mamache B., Elgroud R., Agabou A., Chabou S., Hireche S., Bouaziz O., Rahal K., Rolain J.-M. Prevalence and clonal relationship of ESBL-producing Salmonella strains from humans and poultry in northeastern Algeria. BMC Vet. Res. 2017;13:132. doi: 10.1186/s12917-017-1050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindermann D., Gopinath G., Chase H., Negrete F., Althaus D., Zurfluh K., Tall B.D., Stephan R., Nüesch-Inderbinen M. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010–2015: Poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front. Microbiol. 2017;8:1322. doi: 10.3389/fmicb.2017.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization . Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human Use. WHO; Geneva, Switzerland: 2017. [Google Scholar]
- 12.Eguale T., Birungi J., Asrat D., Njahira M.N., Njuguna J., Gebreyes W.A., Gunn J.S., Djikeng A., Engidawork E. Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in central Ethiopia. Antimicrob. Resist. Infect. Control. 2017;6:13. doi: 10.1186/s13756-017-0171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youn S.Y., Jeong O.M., Choi B.K., Jung S.C., Kang M.S. Comparison of the antimicrobial and sanitizer resistance of Salmonella isolates from chicken slaughter processes in Korea. J. Food Sci. 2017;82:711–717. doi: 10.1111/1750-3841.13630. [DOI] [PubMed] [Google Scholar]
- 14.Racicot M., Comeau G., Tremblay A., Quessy S., Cereno T., Charron-Langlois M., Venne D., Hébert G., Vaillancourt J.P., Fravalo P. Identification and selection of food safety-related risk factors to be included in the Canadian Food Inspection Agency’s Establishment-based Risk Assessment model for Hatcheries. Zoonoses Public Health. 2020;67:14–24. doi: 10.1111/zph.12650. [DOI] [PubMed] [Google Scholar]
- 15.Mueller-Doblies D., Clouting C., Davies R. Investigations of the distribution and persistence of Salmonella and ciprofloxacin-resistant Escherichia coli in turkey hatcheries in the UK. Zoonoses Public Health. 2013;60:296–303. doi: 10.1111/j.1863-2378.2012.01524.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim J., Kim K. Hatchery hygiene evaluation by microbiological examination of hatchery samples. Poult. Sci. 2010;89:1389–1398. doi: 10.3382/ps.2010-00661. [DOI] [PubMed] [Google Scholar]
- 17.Cox N., Bailey J., Blankenship L., Meinersmann R., Stern N., McHan F. Research note: Fifty percent colonization dose for Salmonella Typhimurium administered orally and intracloacally to young broiler chicks. Poult. Sci. 1990;69:1809–1812. doi: 10.3382/ps.0691809. [DOI] [PubMed] [Google Scholar]
- 18.Fei X., Yin K., Yin C., Hu Y., Li J., Zhou Z., Tian Y., Geng S., Chen X., Pan Z. Analyses of prevalence and molecular typing reveal the spread of antimicrobial-resistant Salmonella infection across two breeder chicken farms. Poult. Sci. 2018;97:4374–4383. doi: 10.3382/ps/pey305. [DOI] [PubMed] [Google Scholar]
- 19.Choi S.W., Ha J.S., Kim B.Y., Lee D.H., Park J.K., Youn H.N., Hong Y.H., Lee S.B., Lee J.B., Park S.Y., et al. Prevalence and characterization of Salmonella species in entire steps of a single integrated broiler supply chain in Korea. Poult. Sci. 2014;93:1251–1257. doi: 10.3382/ps.2013-03558. [DOI] [PubMed] [Google Scholar]
- 20.Cox N., Bailey J., Berrang M. Bactericidal treatment of hatching eggs I. Chemical immersion treatments and Salmonella. J. Appl. Poult. Res. 1998;7:347–350. doi: 10.1093/japr/7.4.347. [DOI] [Google Scholar]
- 21.Davies R., Breslin M. Environmental contamination and detection of Salmonella enterica serovar enteritidis in laying flocks. Vet. Rec. 2001;149:699–704. doi: 10.1136/vr.149.23.699. [DOI] [PubMed] [Google Scholar]
- 22.Davies R., Nicholas R., McLaren I., Corkish J., Lanning D., Wray C. Bacteriological and serological investigation of persistent Salmonella Enteritidis infection in an integrated poultry organisation. Vet. Microbiol. 1997;58:277–293. doi: 10.1016/S0378-1135(97)00157-0. [DOI] [PubMed] [Google Scholar]
- 23.Bailey J., Stern N., Fedorka-Cray P., Craven S., Cox N., Cosby D., Ladely S., Musgrove M. Sources and movement of Salmonella through integrated poultry operations: A multistate epidemiological investigation. J. Food Prot. 2001;64:1690–1697. doi: 10.4315/0362-028X-64.11.1690. [DOI] [PubMed] [Google Scholar]
- 24.Kim A., Lee Y.J., Kang M.S., Kwag S.I., Cho J.K. Dissemination and tracking of Salmonella spp. in integrated broiler operation. J. Vet. Sci. 2007;8:155–161. doi: 10.4142/jvs.2007.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha J.S., Seo K.W., Kim Y.B., Kang M.S., Song C.-S., Lee Y.J. Prevalence and characterization of Salmonella in two integrated broiler operations in Korea. Irish Vet. J. 2018;71:1–9. doi: 10.1186/s13620-018-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim M.S., Lim T.H., Jang J.H., Lee D.H., Kim B.Y., Kwon J.H., Choi S.W., Noh J.Y., Hong Y.H., Lee S.B., et al. Prevalence and antimicrobial resistance of Salmonella species isolated from chicken meats produced by different integrated broiler operations in Korea. Poult. Sci. 2012;91:2370–2375. doi: 10.3382/ps.2012-02357. [DOI] [PubMed] [Google Scholar]
- 27.Shang K., Wei B., Kang M. Distribution and dissemination of antimicrobial-resistant Salmonella in broiler farms with or without enrofloxacin use. BMC Vet. Res. 2018;14:257. doi: 10.1186/s12917-018-1590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International Organization for Standardization . Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. European Committee for Standardization; Geneva, Switzerland: 2017. [Google Scholar]
- 29.Shanmugasamy M., Velayutham T., Rajeswar J. Inv A gene specific PCR for detection of Salmonella from broilers. Vet. World. 2011;4:562. doi: 10.5455/vetworld.2011.562-564. [DOI] [Google Scholar]
- 30.Ewing W. Edwards and Ewing’s Identification of Enterobacteriaceae. Elsevier Science Publishing Co. Inc.; Amsterdam, The Netherlands: 1986. [Google Scholar]
- 31.Clinical Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing, Approved Standard M100-S30. CLSI Wayne; Chester County, PA, USA: 2020. 30th Informational Supplement. [Google Scholar]
- 32.Clinical Laboratory Standards Institute (CLSI) Approved Standard Vet01-A4. CLSI Wayne; Chester County, PA, USA: 2018. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. [Google Scholar]
- 33.Ojo O.E., Ogunyinka O.G., Agbaje M., Okuboye J.O., Kehinde O.O., Oyekunle M.A. Antibiogram of Enterobacteriaceae isolated from free-range chickens in Abeokuta, Nigeria. Vet. Arhiv. 2012;82:577–589. [Google Scholar]
- 34.Jeong H.S., Kim J.A., Shin J.H., Chang C.L., Jeong J., Cho J.-H., Kim M.-N., Kim S., Kim Y.R., Lee C.H. Prevalence of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes in Salmonella isolated from 12 tertiary-care hospitals in Korea. Microb. Drug Resist. 2011;17:551–557. doi: 10.1089/mdr.2011.0095. [DOI] [PubMed] [Google Scholar]
- 35.Dierikx C., van Essen-Zandbergen A., Veldman K., Smith H., Mevius D. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 2010;145:273–278. doi: 10.1016/j.vetmic.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 36.Liljebjelke K.A., Hofacre C.L., Liu T., White D.G., Ayers S., Young S., Maurer J.J. Vertical and horizontal transmission of Salmonella within integrated broiler production system. Foodborne Pathog. Dis. 2005;2:90–102. doi: 10.1089/fpd.2005.2.90. [DOI] [PubMed] [Google Scholar]
- 37.Fei X., He X., Guo R., Yin C., Geng H., Wu K., Yin K., Geng S., Pan Z., Li Q. Analysis of prevalence and CRISPR typing reveals persistent antimicrobial-resistant Salmonella infection across chicken breeder farm production stages. Food Control. 2017;77:102–109. doi: 10.1016/j.foodcont.2017.01.023. [DOI] [Google Scholar]
- 38.Hurd H., McKean J., Griffith R., Rostagno M. Estimation of the Salmonella enterica prevalence in finishing swine. Epidemiol. Infect. 2004;132:127–135. doi: 10.1017/S0950268803001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moe A.Z., Paulsen P., Pichpol D., Fries R., Irsigler H., Baumann M.P., Oo K.N. Prevalence and antimicrobial resistance of Salmonella isolates from chicken carcasses in retail markets in Yangon, Myanmar. J. Food Prot. 2017;80:947–951. doi: 10.4315/0362-028X.JFP-16-407. [DOI] [PubMed] [Google Scholar]
- 40.Nidaullah H., Abirami N., Shamila-Syuhada A.K., Chuah L.-O., Nurul H., Tan T.P., Abidin F.W.Z., Rusul G. Prevalence of Salmonella in poultry processing environments in wet markets in Penang and Perlis, Malaysia. Vet. World. 2017;10:286. doi: 10.14202/vetworld.2017.286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ta Y.T., Nguyen T.T., To P.B., Pham D.X., Le H.T.H., Thi G.N., Alali W.Q., Walls I., Doyle M.P. Quantification, serovars, and antibiotic resistance of Salmonella isolated from retail raw chicken meat in Vietnam. J. Food Prot. 2014;77:57–66. doi: 10.4315/0362-028X.JFP-13-221. [DOI] [PubMed] [Google Scholar]
- 42.Chen M., Wang S., Hwang W., Tsai S., Hsih Y., Chiou C., Tsen H. Contamination of Salmonella Schwarzengrund cells in chicken meat from traditional marketplaces in Taiwan and comparison of their antibiograms with those of the human isolates. Poult. Sci. 2010;89:359–365. doi: 10.3382/ps.2009-00001. [DOI] [PubMed] [Google Scholar]
- 43.Chiu L.-H., Chiu C.-H., Horn Y.-M., Chiou C.-S., Lee C.-Y., Yeh C.-M., Yu C.-Y., Wu C.-P., Chang C.-C., Chu C. Characterization of 13 multi-drug resistant Salmonella serovars from different broiler chickens associated with those of human isolates. BMC Microbiol. 2010;10:86. doi: 10.1186/1471-2180-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Na S.H., Moon D.C., Kang H.Y., Song H.-J., Kim S.-J., Choi J.-H., Yoon J.W., Yoon S.-S., Lim S.-K. Molecular characteristics of extended-spectrum β-lactamase/AmpC-producing Salmonella enterica serovar Virchow isolated from food-producing animals during 2010–2017 in South Korea. Int. J. Food Microbiol. 2020;322:108572. doi: 10.1016/j.ijfoodmicro.2020.108572. [DOI] [PubMed] [Google Scholar]
- 45.Sodagari H.R., Mashak Z., Ghadimianazar A. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from retail chicken meat and giblets in Iran. J. Infect. Dev. Ctries. 2015;9:463–469. doi: 10.3855/jidc.5945. [DOI] [PubMed] [Google Scholar]
- 46.Cheong H.J., Lee Y.J., Hwang I.S., Kee S.Y., Cheong H.W., Song J.Y., Kim J.M., Park Y.H., Jung J.-H., Kim W.J. Characteristics of non-typhoidal Salmonella isolates from human and broiler-chickens in southwestern Seoul, Korea. J. Korean Med. Sci. 2007;22:773–778. doi: 10.3346/jkms.2007.22.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poroś-Głuchowska J., Markiewicz Z. Antimicrobial resistance of Listeria monocytogenes. Acta Microbiol. Pol. 2003;52:113–129. [PubMed] [Google Scholar]
- 48.Wilkins M., Bidol S., Boulton M., Stobierski M., Massey J., Robinson-Dunn B. Human salmonellosis associated with young poultry from a contaminated hatchery in Michigan and the resulting public health interventions, 1999 and 2000. Epidemiol. Infect. 2002;129:19–27. doi: 10.1017/S0950268802007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren X., Li M., Xu C., Cui K., Feng Z., Fu Y., Zhang J., Liao M. Prevalence and molecular characterization of Salmonella enterica isolates throughout an integrated broiler supply chain in China. Epidemiol. Infect. 2016;144:2989–2999. doi: 10.1017/S0950268816001515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun S., Negrea A., Rhen M., Andersson D.I. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 2009;53:2298–2305. doi: 10.1128/AAC.01016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J., Wang F., Jin H., Hu J., Yuan Z., Shi W., Yang X., Meng J., Xu X. Laboratory monitoring of bacterial gastroenteric pathogens Salmonella and Shigella in Shanghai, China 2006–2012. Epidemiol. Infect. 2015;143:478–485. doi: 10.1017/S0950268814001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hordijk J., Veldman K., Dierikx C., van Essen-Zandbergen A., Wagenaar J.A., Mevius D. Prevalence and characteristics of quinolone resistance in Escherichia coli in veal calves. Vet. Microbiol. 2012;156:136–142. doi: 10.1016/j.vetmic.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 53.Lin D., Chen K., Chan E.W.-C., Chen S. Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Sci. Rep. 2015;5:1–8. doi: 10.1038/srep14754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui M., Zhang P., Li J., Chengtao S., Wu C. Prevalence and characterization of fluoroquinolone resistant Salmonella isolated from an integrated broiler chicken supply chain. Front. Microbiol. 2019;10:1865. doi: 10.3389/fmicb.2019.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamang M.D., Nam H.-M., Kim A., Lee H.-S., Kim T.-S., Kim M.-J., Jang G.-C., Jung S.-C., Lim S.-K. Prevalence and mechanisms of quinolone resistance among selected nontyphoid Salmonella isolated from food animals and humans in Korea. Foodborne Pathog. Dis. 2011;8:1199–1206. doi: 10.1089/fpd.2011.0899. [DOI] [PubMed] [Google Scholar]
- 56.Bae D.-H., Dessie H.K., Baek H.-J., Kim S.-G., Lee H.-S., Lee Y.-J. Prevalence and characteristics of Salmonella spp. isolated from poultry slaughterhouses in Korea. J. Vet. Med. Sci. 2013:13–0093. doi: 10.1292/jvms.13-0093. [DOI] [PubMed] [Google Scholar]
- 57.Kim K.-Y., Park J.-H., Kwak H.-S., Woo G.-J. Characterization of the quinolone resistance mechanism in foodborne Salmonella isolates with high nalidixic acid resistance. Int. J. Food Microbiol. 2011;146:52–56. doi: 10.1016/j.ijfoodmicro.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 58.Deshpande L.M., Sader H.S., Debbia E., Nicoletti G., Fadda G., Jones R.N. Emergence and epidemiology of fluoroquinolone-resistant Streptococcus pneumoniae strains from Italy: Report from the SENTRY Antimicrobial Surveillance Program (2001–2004) Diagn. Microbiol. Infect. Dis. 2006;54:157–164. doi: 10.1016/j.diagmicrobio.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 59.Baker S., Duy P.T., Nga T.V.T., Dung T.T.N., Phat V.V., Chau T.T., Turner A.K., Farrar J., Boni M.F. Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. Elife. 2013;2:e01229. doi: 10.7554/eLife.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J.S., Yun Y.-S., Kim S.J., Jeon S.-E., Lee D.-Y., Chung G.T., Yoo C.-K., Kim J., Group P.K.W. Rapid emergence and clonal dissemination of CTX-M-15–producing Salmonella enterica serotype Virchow, South Korea. Emerg. Infect. Dis. 2016;22:68. doi: 10.3201/eid2201.151220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prunić B., Milanov D., Velhner M., Pajić M., Pavlović L., Mišić D. Clonal persistence of Salmonella enterica serovars Montevideo, Tennessee, and Infantis in feed factories. J. Infect. Dev. Ctries. 2016;10:662–666. doi: 10.3855/jidc.7313. [DOI] [PubMed] [Google Scholar]
- 62.Nesse L.L., Nordby K., Heir E., Bergsjoe B., Vardund T., Nygaard H., Holstad G. Molecular analyses of Salmonella enterica isolates from fish feed factories and fish feed ingredients. Appl. Environ. Microbiol. 2003;69:1075–1081. doi: 10.1128/AEM.69.2.1075-1081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cason J., Cox N., Bailey J. Transmission of Salmonella Typhimurium during hatching of broiler chicks. Avian Dis. 1994:583–588. doi: 10.2307/1592082. [DOI] [PubMed] [Google Scholar]
- 64.Heyndrickx M., Vandekerchove D., Herman L., Rollier I., Grijspeerdt K., De Zutter L. Routes for Salmonella contamination of poultry meat: Epidemiological study from hatchery to slaughterhouse. Epidemiol. Infect. 2002;129:253–265. doi: 10.1017/S0950268802007380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey J., Cox N., Berrang M. Hatchery-acquired salmonellae in broiler chicks. Poult. Sci. 1994;73:1153–1157. doi: 10.3382/ps.0731153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.