Abstract
Simple Summary
Complex traits that require observations over multiple time points for the same individual are called longitudinal traits. Understanding the genetic architecture of beef cattle growth cannot be limited simply to a genome-wide association study (GWAS) for body weight at any specific ages, but should be extended to a more general purpose by considering the longitudinal weight–age using a growth curve approach. We compared three nonlinear models that described the body weight data of Chinese Simmental beef cattle. The parameters of the suitable model were treated as phenotypes of single-trait GWAS and multi-trait GWAS. We identified 87 significant single nucleotide polymorphisms (SNPs) associated with body weight. Many candidate genes associated with body weight were identified which may be useful for exploring the full genetic architecture underlying growth and development traits in livestock.
Abstract
The objective of the present study was to perform a genome-wide association study (GWAS) for growth curve parameters using nonlinear models that fit original weight–age records. In this study, data from 808 Chinese Simmental beef cattle that were weighed at 0, 6, 12, and 18 months of age were used to fit the growth curve. The Gompertz model showed the highest coefficient of determination (R2 = 0.954). The parameters’ mature body weight (A), time-scale parameter (b), and maturity rate (K) were treated as phenotypes for single-trait GWAS and multi-trait GWAS. In total, 9, 49, and 7 significant SNPs associated with A, b, and K were identified by single-trait GWAS; 22 significant single nucleotide polymorphisms (SNPs) were identified by multi-trait GWAS. Among them, we observed several candidate genes, including PLIN3, KCNS3, TMCO1, PRKAG3, ANGPTL2, IGF-1, SHISA9, and STK3, which were previously reported to associate with growth and development. Further research for these candidate genes may be useful for exploring the full genetic architecture underlying growth and development traits in livestock.
Keywords: longitudinal data, growth curve model, single-trait GWAS, multi-trait GWAS, Chinese Simmental beef cattle
1. Introduction
With improved quality of life in China, the demand for meat, particularly beef, is increasing [1]. The Simmental breed accounts for more than 70% of the total number of beef cattle in China [2]. Therefore, it is necessary to analyze the genetic mechanism of the growth traits in beef cattle production [3].
Genome-wide association study (GWAS) and genomic selection (GS) are powerful statistical tools that can broadly identify candidate genes with significant single nucleotide polymorphisms (SNPs) involved in production traits [4,5], growth traits [6,7] and fertility traits [8,9]. However, current research mainly focuses on single data records, such as birth weight, weaning weight, and weight before slaughter [3,10,11]. Complex traits that require observations over multiple time points for the same individual are called longitudinal traits. Compared with single data records, longitudinal data can better describe the growth and production of livestock and poultry [12,13]. The fitting growth curve model is one of the most common strategies for such data [14]. Different models [15] provide a few parameters for people to show the regularity of weight gain in livestock, such as mature body weight (A), time-scale parameter (b), and maturity rate (K), which might reflect the influence of genetic impacts on body weight. In the current study, parameters (such as A and K) were considered as phenotypes of the mixed linear model, and quantitative trait loci affecting the growth curve were identified by GWAS. In addition, Das et al. [16] proposed a series of methods based on random regression models. Previous research has demonstrated that these methods are more sophisticated and flexible [17], but they did not provide biologically-interpretable parameters, such as A and K, which are usually required in daily breeding management.
Generally, a quantitative trait locus (QTL), which affects complex traits, may affect multiple traits [18]. Therefore, the genetic correlations between the parameter estimates (mainly A and K) should be considered. These correlations may be attributed to SNPs that have pleiotropic effects on multiple traits. Therefore, multiple trait GWAS (multi-trait GWAS) is more reasonable in this study [18,19].
In this study, body weights of 808 Chinese Simmental beef cattle at four stages of growth were used to fit the growth curve. The best fitting growth curve parameters were treated as phenotypes for single-trait GWAS and multi-trait GWAS. The aim of our study was to comprehensively analyze candidate genes and QTL regions associated with growth traits by two GWAS methods. We also undertook post-GWAS analyses to identify and prioritize annotated genes within detected genomic regions using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Our findings offer valuable insights for exploring the full genetic architecture underlying growth and development traits in livestock.
2. Materials and Methods
2.1. Resource Population and Phenotypes Collection
All animals used in the study were treated following the guidelines established by the Council of China Animal Welfare. Protocols of the experiments were approved by the Science Research Department of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China (approval number: RNL09/07). The training population consisted of 808 Chinese Simmental beef cattle established in Ulgai, Xilingole League, Inner Mongolia of China. Body weight at four growth stages (0, 6, 12, and 18 months after birth) were measured for each individual. Since fixed effects were related to body weight and not to growth curve parameters, original body weight data at each age were pre-adjusted for fixed effects (breed, year, and month of birth) by a generalized linear model (GLM). The descriptive statistics of the pre-adjusted phenotypic data are presented in Table 1.
Table 1.
The descriptive statistics of body weight for Chinese Simmental beef cattle.
| Age (Month) | Max (kg) | Min (kg) | Mean (kg) | Standard Deviation (SD) |
|---|---|---|---|---|
| 0 | 55.20 | 25.00 | 38.79 | 6.21 |
| 6 | 326.00 | 107.00 | 208.68 | 39.48 |
| 12 | 561.00 | 242.00 | 398.70 | 56.21 |
| 18 | 739.00 | 346.00 | 520.10 | 73.18 |
2.2. Genotyping and Quality Control
Genomic DNA was isolated from blood samples using the TIANamp Blood DNA Kit (Tiangen Biotech Co.Ltd., Beijing, China) and DNA quality was acceptable when the A260/A280 ratio was between 1.8 and 2.0. In total, 808 cattle were genotyped using Illumina BovineHD Beadchip (Illumina Inc., San Diego, CA, USA). Before statistical analysis, SNPs were pre-processed by PLINK v1.07 [20]. Duplicated SNPs were also removed. Finally, 671,192 SNPs on 29 autosomal chromosomes with an average distance of 3 kb were generated for the analysis. SNPs were deleted according to the following standards, including minor allele frequency (<0.01), SNP call rate (<0.05), and Hardy–Weinberg equilibrium values (p < 1 × 10−6).
2.3. Growth Curve Fitting
Three of the most widely used nonlinear models (Table 2) to describe animal growth curves—Gompertz, Logistic, and Brody—were fitted for each animal using the iterative nonlinear least-squares method via the Gauss–Newton [21] algorithm implemented in SAS 9.4. In the function, A is the mature body weight, which means the ultimate body weight of an individual; b is the time-scale parameter, which means the time for an individual to reach its maximum growth rate; K is the maturity rate, which means the rate at which an individual approaches its mature body weight (A).
Table 2.
Growth curve model. A is the mature body weight, b is the time-scale parameter, K is the maturity rate, W is the observed body weight, t is the growth time, and e is the natural logarithm.
| Model | Function |
|---|---|
| Gompertz | W = Aexp(−bexp−Kt) |
| Logistic | W = A(1 + bexp−Kt)−1 |
| Brody | W = A(1 − bexp−Kt) |
The coefficient of determination R2 [22] was used to evaluate the fitting effect of the growth curve model. The expression is as follows:
| (1) |
where W represents the observation of body weight, represents the estimated body weight of the growth curve model, and is the average value of body weight.
The genetic correlation of A, b, and K was also calculated, which used the following formula:
| (2) |
where is the additive genetic variance of trait 1, is the additive genetic variance of trait 2, and is the covariance of additive effects.
2.4. Single-Trait GWAS
After selecting the nonlinear model which best fit the body weight data, the parameters A, b, and K were used in GWAS. Firstly, we used principal component analysis (PCA) and obtained the kinship matrix using the package GAPIT [23] (Genomic Association and Prediction Integrated Tool) (http://www.maizegenetics.net/gapit) in R software (R 3.6.1). The following mixed linear model was considered:
| y = Ws + Xβ + Zμ + e | (3) |
where y represents the vector of observations from the three phenotypes (A, b, K estimates) for each individual; s is the SNP effects vector; W is a matrix of SNP genotype indicators, which were coded as 0, 1, and 2 corresponding to the three genotypes AA, AB, and BB; μ is a vector of these polygenic effects with an assumed N (0, Kσ2) distribution, where σ2 is the polygenic variance and K is a marker inferred kinship matrix; X is an incidence matrix for β, and β is a non-genetic vector of fixed effects only including principal component effects (the top three eigenvectors of the Q matrix). The other fixed effects were not included at this step, since they were already considered before fitting nonlinear functions (see pre-adjustment for fixed effects in Section 2.1). Z is an incidence matrix for μ; while e is a vector for random residual errors with a putative N (0, ) distribution, where is the residual variance.
The false discovery rate (FDR) was used to determine the threshold values for single-trait GWAS and multi-trait GWAS. The FDR was set as 0.05, and the threshold p-value was calculated as follows:
| p = FDR × n/m | (4) |
where n is the number of p < 0.05 in the results and m is the total number of SNPs [24].
2.5. Multi-Trait GWAS
The multi-trait GWAS was conducted to detect pleiotropic SNPs for the parameters A, b, and K. The model was a Chi-squared distribution which was calculated for each SNP using the following formula [25]:
| (5) |
| (6) |
where is a 3 × 1 vector of the t-values for ith SNP obtained from single-trait GWAS; is the estimate of v; is the corresponding variance which can be obtained by the compressed mixed linear model (CMLM); is the transpose of the vector ; is the inverse of the 3 × 3 correlation matrix between traits, which was calculated using the qualified SNPs.
2.6. Gene Function Annotation
We explored the biological mechanism of significant SNPs based on the interpretability of the gene functions related to the relevant SNPs. To select the candidate genes based on the physical location of SNPs, the BioMart module of Ensembl was used to match the significant SNPs with the UMD Bostaurus 3.1 (http://www.animalgenome.org). Then we confirmed the biological function of related genes by the genome databanks National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/), and the genes associated with growth and development traits were screened out. GO and KEGG pathways were used to annotate the main biological functions, metabolic pathways, and signal transduction pathways involved in differentially expressed genes.
3. Results
3.1. Growth Curve Fitting
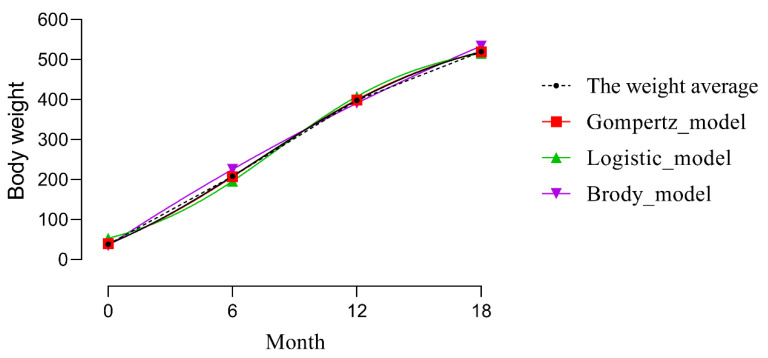
Models are shown in Table 3, and the three growth curves are plotted in Figure 1. The R2 values for the Gompertz, Logistic, and Brody models were 0.954, 0.951, and 0.951, respectively; the Gompertz model showed the best goodness of fit. Figure 1 shows four growth curves of the Gompertz model, Logistic model, Brody model, and the weight average. The curves representing the Gompertz model and average body weight overlap almost completely, while the other curves have some deviation. Therefore, the parameters of the Gompertz model were selected as phenotypes of GWAS.
Table 3.
Estimated values of the growth curve model.
| Parameter | Models | ||
|---|---|---|---|
| Gompertz | Logistic | Brody | |
| A | 617.900 | 551.000 | 1458.500 |
| b | 2.740 | 9.304 | 0.976 |
| K | 0.153 | 0.273 | 0.024 |
| R2 | 0.954 | 0.951 | 0.951 |
Figure 1.
Plot of growth curve models.
The correlation coefficients were 0.087 (A and b), –0.578 (A and K), and 0.369 (b and K) respectively, which showed that A and K have a strong negative correlation.
3.2. Principal Components Analysis (PCA)
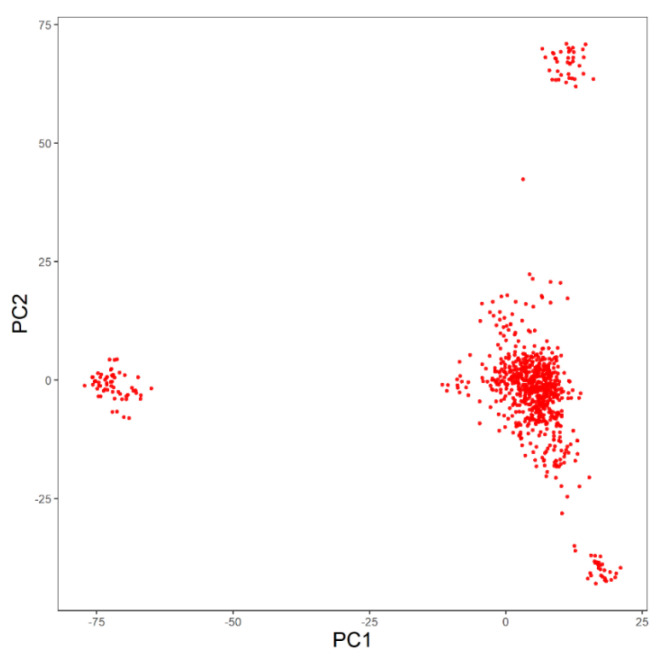
The population stratification of the Simmental beef cattle population based on the PCA is shown in Figure 2. The population was divided into five separate clusters, demonstrating an obvious stratification in the reference population. The majority of individuals are located in the lower right corner, while a small number of individuals are distributed in other regions. Therefore, the first two principal components are selected as covariables to eliminate the influence of population stratification on correlation analysis.
Figure 2.
Population structure identified by principal components analysis. PC stands for the principal components of principal components analysis.
3.3. Summary of Results by Two GWAS Methods
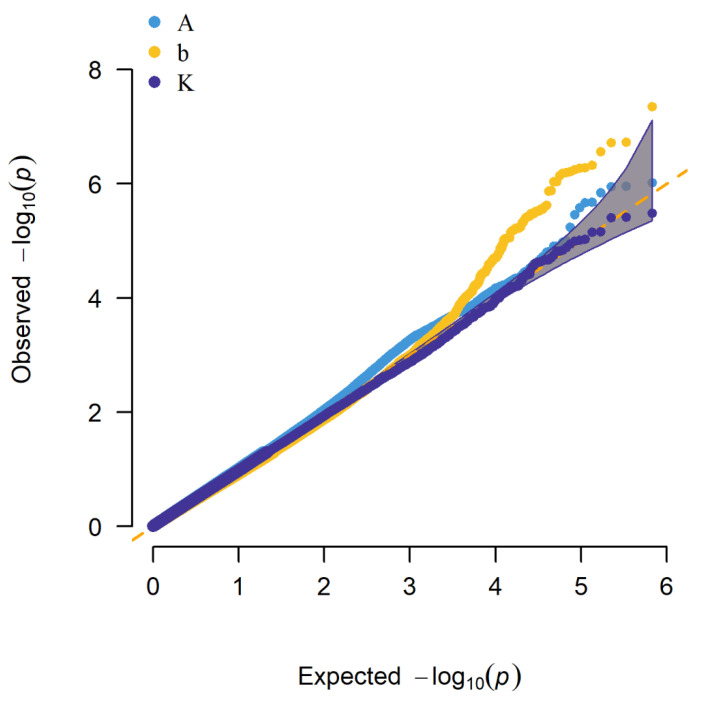
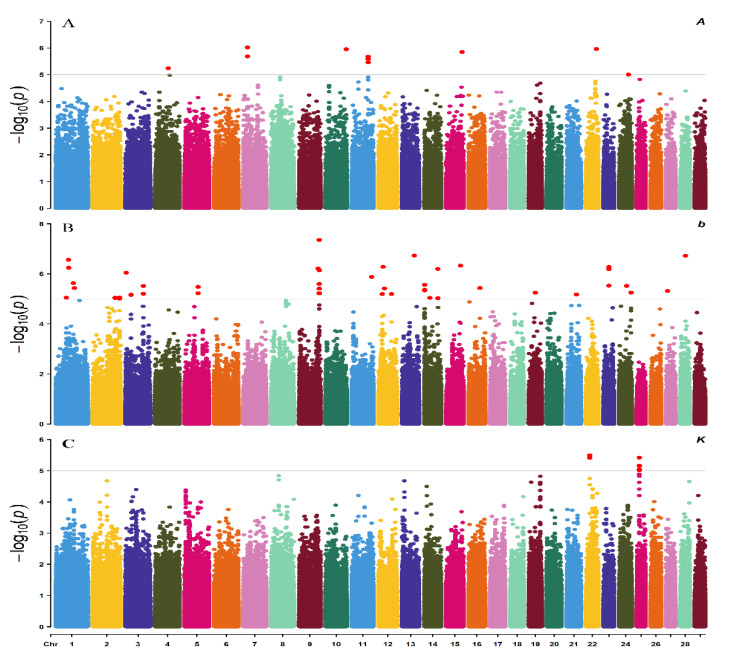
The quantile-quantile (Q-Q) plots and Manhattan plots of single-trait GWAS are shown in Figure 3 and Figure 4. Most points were near the diagonal line in quantile-quantile (Q-Q) plots because the population structure was considered in the GWAS function. The Q-Q plots suggested that there was no inflation or systematic bias in this research. There were nine significant SNPs for mature body weight (A) in the Manhattan plots of single-trait GWAS. The nine SNPs were located on bos taurus autosomes (BTA) 4, 7, 10, 11, 15, and 22, and the locus with the lowest p-value was located at 20,500,709 bp on BTA 7 (Figure 4A). The 49 significant SNPs were shown for time-scale parameter (b), and the SNP with the lowest p-value was located at 98,989,710 bp on BTA 9. For the maturity rate (K), Manhattan plot indicated seven significant loci which were located on BTA 22 and 25, and the SNP with the lowest p-value was located at 18,694,612 bp on BTA 22. We observed several associated genes involved in growth and development, including PLIN3, KCNS3, TMCO1, ANGPTL2, IGF-1, ASPH, ALPL, GRM7, and SHISA9. All results are shown in Table 4.
Figure 3.
Quantile-quantile (Q-Q) plots of single-trait genome-wide association study (GWAS) for A, b, and K.
Figure 4.
Manhattan plots of GWAS for A, b, and K. (A) stands for the Manhattan plot of parameter A; (B) stands for the Manhattan plot of parameter b; (C) stands for the Manhattan plot of parameter K.
Table 4.
The results of single-trait GWAS and multi-trait GWAS.
| Trait | SNP | BTA | Position | Distance | Gene | p-Value |
|---|---|---|---|---|---|---|
| A | ARS-BFGL-NGS-14531 | 7 | 20,500,709 | 6291 | PLIN3 | 9.55 × 10−7 |
| BovineHD2200014587 | 22 | 51,133,487 | within (intronic) | BSN | 1.10 × 10−6 | |
| BovineHD1000029459 | 10 | 101,577,026 | within (intronic) | TTC8 | 1.12 × 10−6 | |
| BovineHD1500022754 | 15 | 78,218,321 | within (intronic) | C15H11ofF49 | 1.42 × 10−6 | |
| BovineHD0700005699 | 7 | 20461 012 | within (intronic) | UHRF1 | 2.08 × 10−6 | |
| BovineHD1100023174 | 11 | 80,858,593 | 14,458 | KCNS3 | 2.13 × 10−6 | |
| BovineHD1100023180 | 11 | 80,883,741 | 39,606 | KCNS3 | 2.61 × 10−6 | |
| BovineHD1100023175 | 11 | 80,860,546 | 16,411 | KCNS3 | 3.46 × 10−6 | |
| Hapmap36353-SCAFFOLD29708_3468 | 4 | 64,923,141 | 62,596 | PDE1C | 5.78 × 10−6 | |
| b | BovineHD0900028514 | 9 | 98,989,710 | within (exonic) | PRKN | 4.43 × 10−8 |
| BovineHD1300017399 | 13 | 60,669,478 | 15,189 | RSPO4 | 1.86 × 10−7 | |
| BTB-00981633 | 28 | 24,967,427 | within (intronic) | DNA2 | 1.90 × 10−7 | |
| BovineHD1500020257 | 15 | 70,169,617 | 1,074,228 | LRRC4C | 4.73 × 10−7 | |
| BovineHD1200006711 | 12 | 22,401,586 | 317,236 | COG6 | 5.26 × 10−7 | |
| BovineHD2300007448 | 23 | 27,217,994 | within (intronic) | SKIV2L | 5.33 × 10−7 | |
| BovineHD0900026419 | 9 | 93,361,299 | 12,446 | NOX3 | 6.17 × 10−7 | |
| BovineHD1400018901 | 14 | 67,713,519 | within (intronic) | STK3 | 6.38 × 10−7 | |
| BovineHD2300007441 | 23 | 27,195,210 | within (intronic) | C4A | 6.58 × 10−7 | |
| BovineHD0900028515 | 9 | 98,990,425 | within | PRKN | 7.26 × 10−7 | |
| BovineHD0300000940 | 3 | 3,186,646 | within (intronic) | TMCO1 | 9.11 × 10−7 | |
| BovineHD0300000941 | 3 | 3,189,462 | within (intronic) | TMCO1 | 9.11 × 10−7 | |
| BovineHD1100028458 | 11 | 97,919,703 | 60,177 | ANGPTL2 | 1.33 × 10−6 | |
| BovineHD1100028450 | 11 | 97,903,021 | 43,495 | ANGPTL2 | 1.34 × 10−6 | |
| BovineHD0100024671 | 1 | 86,573,589 | 93,224 | DNAJC19 | 2.37 × 10−6 | |
| BovineHD0900028520 | 9 | 99,001,573 | within (exonic) | PRKN | 2.54 × 10−6 | |
| BovineHD1400000353 | 14 | 2,382,595 | within (intronic) | ZC3H3 | 2.76 × 10−6 | |
| BovineHD1400000354 | 14 | 2,384,748 | within (intronic) | ZC3H3 | 2.76 × 10−6 | |
| BovineHD2300007455 | 23 | 27,227,600 | within (intronic) | CFB | 3.00 × 10−6 | |
| BovineHD2400010016 | 24 | 36,578,137 | 458,512 | ADCYAP1 | 3.03 × 10−6 | |
| BovineHD0300025174 | 3 | 87,908,532 | 16,189 | MYSM1 | 3.07 × 10−6 | |
| BovineHD0500018625 | 5 | 66,594,318 | within (intronic) | IGF-1 | 3.34 × 10−6 | |
| BovineHD0500018629 | 5 | 66,609,814 | 5314 | IGF-1 | 3.34 × 10−6 | |
| BovineHD0500018633 | 5 | 66,624,481 | 19,981 | IGF-1 | 3.34 × 10−6 | |
| BovineHD0100026284 | 1 | 92,441,255 | 1,184,964 | NLGN1 | 3.72 × 10−6 | |
| BovineHD1200008652 | 12 | 29,267,967 | within (exonic) | RXFP2 | 3.85 × 10−6 | |
| BovineHD0900028524 | 9 | 99,010,494 | within (exonic) | PRKN | 3.89 × 10−6 | |
| BovineHD1400000321 | 14 | 2,241,832 | 6798 | MAPK15 | 4.36 × 10−6 | |
| BovineHD1400000343 | 14 | 2,348,518 | 3233 | GSDMD | 4.68 × 10−6 | |
| BovineHD1900009534 | 19 | 32,360,589 | within (intronic) | HS3ST3A1 | 5.67 × 10−6 | |
| BovineHD0900028481 | 9 | 98,914,727 | within (exonic) | PRKN | 5.95 × 10−6 | |
| BovineHD0900028509 | 9 | 98,984,305 | within (exonic) | PRKN | 5.95 × 10−6 | |
| BovineHD0500018642 | 5 | 66,654,472 | 49,972 | IGF-1 | 6.01 × 10−6 | |
| BovineHD0900028504 | 9 | 98,967,507 | within (exonic) | PRKN | 6.05 × 10−6 | |
| BovineHD0300025183 | 3 | 87,959,712 | within (intronic) | MYSM1 | 6.27 × 10−6 | |
| BovineHD1200027060 | 12 | 64,329,068 | 1,659,351 | SLITRK5 | 6.45 × 10−6 | |
| BovineHD1200026793 | 12 | 18,310,824 | 9625 | RCBTB2 | 6.50 × 10−6 | |
| BovineHD2100014355 | 21 | 49,967,674 | 200,864 | FBXO33 | 6.69 × 10−6 | |
| BovineHD0300008509 | 3 | 26,888,743 | 28,039 | CD58 | 6.84 × 10−6 | |
| BovineHD0300008508 | 3 | 26,885,838 | 25,134 | CD58 | 6.98 × 10−6 | |
| BovineHD0200038336 | 2 | 131,809,255 | within (intronic) | ALPL | 8.81 × 10−6 | |
| BovineHD0200038337 | 2 | 131,810,815 | within (exonic) | ALPL | 8.81 × 10−6 | |
| BovineHD0200038343 | 2 | 131,820,288 | 7428 | ALPL | 8.81 × 10−6 | |
| BovineHD0200031784 | 2 | 110,303,552 | within (intronic) | EPHA4 | 9.04 × 10−6 | |
| BovineHD0100014672 | 1 | 52,227,088 | 136,783 | CCDC54 | 9.06 × 10−6 | |
| BovineHD1400008371 | 14 | 28,916,088 | 27,997 | ASPH | 9.15 × 10−6 | |
| BovineHD0200031783 | 2 | 110,302,531 | within (intronic) | EPHA4 | 9.19 × 10−6 | |
| BovineHD1400018902 | 14 | 67,716,121 | within (intronic) | STK3 | 9.42 × 10−6 | |
| BovineHD0200038341 | 2 | 131,817,068 | 4208 | ALPL | 9.84 × 10−6 | |
| K | BovineHD2200005378 | 22 | 18,694,612 | 60,070 | GRM7 | 3.24 × 10−6 |
| BovineHD2500003405 | 25 | 12,148,764 | 444,406 | SHISA9 | 3.82 × 10−6 | |
| BovineHD2200005379 | 22 | 18,697,043 | 57,639 | GRM7 | 3.89 × 10−6 | |
| BovineHD2500003397 | 25 | 12,122,951 | 418,593 | SHISA9 | 6.89 × 10−6 | |
| BovineHD2500003394 | 25 | 12,119,907 | 415,549 | SHISA9 | 7.09 × 10−6 | |
| BovineHD2500003411 | 25 | 12,164,708 | 460,350 | SHISA9 | 9.25 × 10−6 | |
| BovineHD2500003396 | 25 | 12,122,067 | 417,709 | SHISA9 | 9.70 × 10−6 | |
| Multi | BovineHD1000008269 | 10 | 25,336,507 | 11,871 | BT.86117 | 5.76 × 10−11 |
| BovineHD2300014561 | 23 | 49,948,237 | 785 | C6ORF146 | 5.11 × 10−7 | |
| BovineHD0100017897 | 1 | 63,214,855 | within (intronic) | bta-mir-2285de | 8.19 × 10−7 | |
| BovineHD1100024571 | 11 | 85,545,380 | 311,744 | TRIB2 | 9.06 × 10−7 | |
| BovineHD1900017810 | 19 | 61,961,078 | within (intronic) | ABVA10 | 1.78 × 10−6 | |
| BovineHD2200011596 | 22 | 40,545,626 | 185,054 | BT.92027 | 2.22 × 10−6 | |
| BovineHD1300005737 | 13 | 19,728,845 | 183,012 | NRP1 | 2.77 × 10−6 | |
| BovineHD1400005409 | 14 | 18,830,773 | 441,600 | BT.88023 | 2.77 × 10−6 | |
| BovineHD1400018913 | 14 | 67,761,416 | within (exonic) | STK3 | 3.41 × 10−6 | |
| BovineHD2400015566 | 24 | 54,582,317 | 107,987 | C18ORF26 | 3.89 × 10−6 | |
| Hapmap46842-BTA-57397 | 24 | 11,851,627 | 637,616 | CDH7 | 4.32 × 10−6 | |
| BovineHD1400003514 | 14 | 12,051,695 | 146,289 | GSDMC | 4.44 × 10−6 | |
| BovineHD0200034312 | 2 | 118,915,870 | within (intronic) | PSMD1 | 5.65 × 10−6 | |
| BovineHD0900014829 | 9 | 53,862,308 | 48,559 | GPR63 | 5.70 × 10−6 | |
| BovineHD2700010439 | 27 | 36,460,835 | 72,147 | KAT6A | 5.71 × 10−6 | |
| BovineHD1100023984 | 11 | 83,388,941 | 200,644 | NBAS | 6.15 × 10−6 | |
| BovineHD0900002818 | 9 | 11,192,144 | 488,896 | RIMS1 | 6.15 × 10−6 | |
| BovineHD1300006393 | 13 | 21,900,826 | within (intronic) | PLXDC2 | 6.15 × 10−6 | |
| BovineHD0200019309 | 2 | 66,721,486 | 808,880 | ACTR3 | 8.50 × 10−6 | |
| BovineHD1800012623 | 18 | 42,743,057 | 295,489 | ZNF507 | 9.33 × 10−6 | |
| BovineHD0300008523 | 3 | 26,920,280 | 59,576 | CD58 | 9.36 × 10−6 | |
| BovineHD2900005573 | 29 | 19,238,772 | 49,975 | GDPD4 | 9.53 × 10−6 |
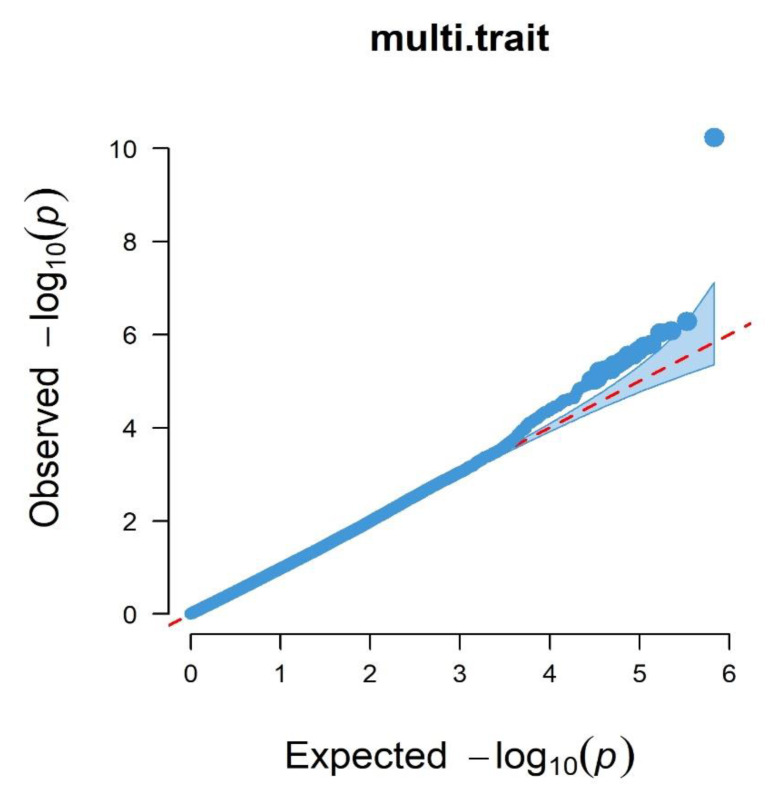
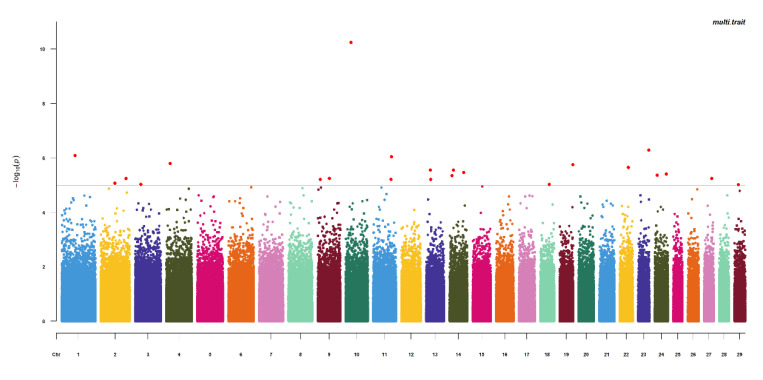
The Q-Q plot and the Manhattan plot of multi-trait GWAS are shown in Figure 5 and Figure 6. The same conclusion as the single-trait GWAS was given in the Q-Q plot of multi-trait GWAS. The 22 significant SNPs were identified. The SNP with the lowest p-value was located at 25,336,507 bp on BTA 10. We also observed several associated genes involved in growth and development which included STK3, CD58, and bta-mir-2285de. The results are shown in Table 4.
Figure 5.
Q-Q plots of multi-trait GWAS for A, b, and K.
Figure 6.
Manhattan plot of multi-trait GWAS for A, b, and K.
3.4. GO and KEGG Pathway Analysis
We found 29 KEGG pathways and 135 GO terms, and 12 pathways and 99 GO terms were significantly enriched (p < 0.05) (e.g., thiamine metabolism, circadian rhythm, protein stabilization, nephric duct morphogenesis, glycosylphosphatidylinositol (GPI)-linked ephrin receptor activity) (Table S1). Particularly, seven KEGG pathways and 14 GO terms which were related to growth and development are shown separately in Table 5, including Hippo signaling pathway—multiple species, longevity regulating pathway—multiple species, nephric duct morphogenesis, and limb morphogenesis.
Table 5.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) involved in differentially expressed genes.
| Gene Name | Term | Database | ID | DEG |
|---|---|---|---|---|
| ALPL | Hippo signaling pathway—multiple species | KEGG pathway | bta00730 | 1 |
| biomineral tissue development | Gene Ontology | GO:0031214 | 1 | |
| ANGPTL2 | angiogenesis | Gene Ontology | GO:0001525 | 1 |
| EPHA4 | Axon guidance | KEGG pathway | bta04360 | 1 |
| nephric duct morphogenesis | Gene Ontology | GO:0072178 | 1 | |
| cochlea development | Gene Ontology | GO:0090102 | 1 | |
| KAT6A | Signaling pathways regulating pluripotency of stem cells | KEGG pathway | bta04550 | 1 |
| PLIN3 | lipid storage | Gene Ontology | GO:0019915 | 1 |
| PRKAG3 | Longevity regulating pathway—multiple species | KEGG pathway | bta04213 | 1 |
| Apelin signaling pathway | KEGG pathway | bta04371 | 1 | |
| fatty acid biosynthetic process | Gene Ontology | GO:0006633 | 1 | |
| ASPH | limb morphogenesis | Gene Ontology | GO:0035108 | 1 |
| roof of mouth development | Gene Ontology | GO:0060021 | 1 | |
| ASPH, STK3 | negative regulation of cell population proliferation | Gene Ontology | GO:0008285 | 2 |
| STK3 | Hippo signaling pathway | KEGG pathway | bta04390 | 1 |
| MAPK signaling pathway | KEGG pathway | bta04010 | 1 | |
| cell differentiation involved in embryonic placenta development | Gene Ontology | GO:0060706 | 1 | |
| hepatocyte apoptotic process | Gene Ontology | GO:0097284 | 1 | |
| negative regulation of organ growth | Gene Ontology | GO:0046621 | 1 | |
| positive regulation of fat cell differentiation | Gene Ontology | GO:0045600 | 1 | |
| central nervous system development | Gene Ontology | GO:0007417 | 1 |
Note: DEG represents the number of differentially expressed genes detected in this pathway; MAPK represents the mitogen-activated protein kinase signaling pathway.
4. Discussion
4.1. Growth Curve Fitting
R2 of Gompertz model reached 0.954, which was the highest of the three models. The parameter A of the Gompertz model showed that the mature body weight of Chinese Simmental beef cattle reached 617.9 kg, which was within the normal mature weight range (600–800 kg) for the population [26]. Though the R2 of Logistic model and Brody model reached 0.951, the parameter A (551.0 and 1458.5) was inconsistent with the actual weight of Chinese Simmental beef cattle. The results indicated that the two models may not be suitable for the data in this study. Though the coefficient of determination (R2) for Logistic model and Brody model for A are the same (0.951), the estimate of A for the two models was quite different. The reason for this phenomenon may be that the function expressions of the two models are different, and the estimation methods are also different, so the models adapt to different breeds. Among the three models, the growth curve fitting by the Gompertz model with the highest R2 had well-matched performance for the actual cattle population. Therefore, the Gompertz model was chosen as the best model for Chinese Simmental beef cattle, which was the same conclusion as Liang et al. [27].
The negative relationship between parameters A and K has been reported many times [14,28,29], which suggests that individuals with smaller mature weight will gain its mature body weight at a young age. Thus, we can predict that precocious animals will not gain a high mature body weight, even if we put in the same cost (such as feed) as other individuals. The conclusion could help us reduce the cost of raising animals by learning to manage individuals separately.
Although there are few studies about growth curves in Chinese Simmental beef cattle, some authors have concluded that the Gompertz model provides the best fit for body weight of beef cattle. Zainaguli et al. [30] used four common models (Logistic, Gompertz, Brody, and Bertallanffy) to fit the weight growth curves of 344 Xinjiang Brown cattle. The Gompertz model showed the best fit for the population. Liang et al. [27] compared four growth curve models (Logistic, Gompertz, Brody, and Bertallanffy) fitted to body weight of Simmental beef cattle and concluded that the Gompertz model was superior to the other models.
4.2. GWAS, GO, and KEGG Pathway Analysis
We performed single-trait GWAS and multi-trait GWAS for the body weight trait of Chinese Simmental beef cattle. A great number of genes involved in growth and development were identified by each method. The reason for this phenomenon may be the limited dataset. However, since most growth and development traits are controlled by multiple genes [31], the genes associated with growth and development identified by separate GWAS cannot be ignored. Single-trait GWAS and multi-trait GWAS have their specific advantages in the identification of distinct loci. For example, compared to the meta-analysis GWAS, the single-population GWAS was more powerful for the identification of SNPs [32], whereas multi-trait GWAS has the advantage of increasing statistical power and identifying pleiotropic loci [33,34,35]. Therefore, it should be noted that multi-trait GWAS cannot replace single-trait GWAS, rather it was complementary to single-trait GWAS. Thus, combining single-trait GWAS and multi-trait GWAS methods was expected to markedly improve the analysis of the genetic mechanism of the body weight traits for Chinese Simmental beef cattle.
Single-trait GWAS: For mature body weight (A), the significant locus ARS-BFGL-NGS-14531 which has the lowest p-value was near PLIN3 (perilipin 3). PLIN3 is an important regulator of adipogenesis and triglyceride storage [36], and PLIN3 functions are intertwined with the lipogenic pathways implicated in sebaceous lipogeneses, such as desaturation and triglyceride synthesis [37]; three significant SNPs were near KCNS3 (potassium voltage-gated channel modifier subfamily S member 3) which was proven to be significantly associated with the percent body fat (%BF) [38]. For time-scale parameter (b), two SNPs were within TMCO1 (transmembrane and coiled-coil domains 1), which may affect muscle development because of the significant relationship with PRKAG3 (protein kinase AMP-activated non-catalytic subunit gamma 3); two significant SNPs were near ANGPTL2 (angiopoietin like 2). A study showed that ANGPTL2 may be used as a new type of adipocyte factor [39]. One SNP was located in CFB (complement factor B), which has been identified as related to the total number of piglets born (TNB) and reproductive traits [40]; four significant SNPs were near or within IGF-1 (insulin-like growth factor 1). IGF-1 and its signaling pathway play a primary role in normal growth and aging [41,42]. The locus BovineHD1400008371 was near ASPH (aspartate beta-hydroxylase), which is involved in regulating the growth and development of beef cattle carcass [43]; four significant SNPs were near or within ALPL (alkaline phosphatase, biomineralization associated). A study showed that the expression level of ALPL in white blood cells of obese people is significantly higher than that of lean people, indicating that ALPL may be related to the production of fat [44]; two significant loci were within EPHA4 (EPH receptor A4) which was one of the potential candidate genes for growth trait of pigs [45]. Seven significant loci of maturity rate (K) were concentrated on chromosomes 22 and 25; two associated genes GRM7 (glutamate metabotropic receptor 7) and SHISA9 (shisa family member 9) were found and SHISA9 was highly correlated with growth and development [46].
Multi-trait GWAS: CD58 (CD58 molecule) and STK3 (serine/threonine kinase 3) were found by both methods. Two SNPs (BovineHD1400018901 and BovineHD1400018902) from single-trait GWAS and one SNP (BovineHD1400018913) from multi-trait GWAS were near STK3, which is also named MST2 (macrophage stimulating 2). MST1 (macrophage stimulating 1) and MST2 were central to the Hippo signaling pathway in mammals, which enabled the dynamic regulation of tissue homeostasis in animal development [47]. One significant SNP (BovineHD0100017897) was within bta-mir-2285de which might be an important regulator of bovine mammary lipogenesis and metabolism [48]. The locus BovineHD2700010439 was near KAT6A (lysine acetyltransferase 6A), and it has been shown to be significantly associated with growth retardation [49]. One significant SNP(BovineHD1100023984) was near NBAS (NBAS subunit of NRZ tethering complex) which was significantly associated with bone development [50].
GO and KEGG pathway: There are also some candidate genes which are closely related to growth and development found by GO and KEGG pathway. For example, STK3 was involved in more than one GO and KEGG pathway, including Hippo signaling pathway—multiple species, Hippo signaling pathway, MAPK signaling pathway, cell differentiation involved in embryonic placenta development [51], hepatocyte apoptotic process, negative regulation of organ growth, negative regulation of cell population proliferation, positive regulation of fat cell differentiation, and central nervous system development, which suggests that STK3 may be closely related to cell proliferation and differentiation [51], organ growth and development, and nervous system development [52]. ASPH was involved in limb morphogenesis and roof of mouth development, which suggests that ASPH is significantly associated with body development [43]. ANGPTL2 was involved in angiogenesis, which suggests that ANGPTL2 is closely related to the development of individuals [39].
5. Conclusions
In conclusion, the three growth curve models were used to fit the body weight data of Chinese Simmental beef cattle. The parameters of the Gompertz model with the best fitting effect are the phenotypes of GWAS. A total of 65 significant SNPs from single-trait GWAS and 22 SNPs from multi-trait GWAS were found. Seven KEGG pathways and 14 GO terms, which were related to growth and development, were also identified. Several candidate genes that were significantly associated with growth and development traits were observed, including PLIN3, KCNS3, TMCO1, ANGPTL2, CFB, IGF-1, ALPL EPHA4, SHISA9, STK3, and bta-mir-2285de. The role of associated genes in growth and development was also discussed. Further research for these candidate genes may be useful for exploring the full genetic architecture underlying growth and development traits in livestock.
Acknowledgments
The authors would like to thank all staff at the cattle experimental unit in Beijing and Ulagai for animal care and sample collection.
Abbreviations
The following abbreviations are used in this manuscript:
| GWAS | genome-wide association study |
| GS | genomic selection |
| SNP | single nucleotide polymorphism |
| QTL | quantitative trait loci |
| FDR | false discovery rate |
| PCA | principal components analysis |
| BTA | Bos taurus autosomes |
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/11/1/192/s1, Table S1: Gene Ontology (GO) and KEGG pathway analysis.
Author Contributions
Data curation, X.D., B.A., and L.D.; formal analysis, X.D., T.C., and B.-G.Y.; funding acquisition, H.G.; investigation, L.X., L.Z., J.L., G.E, and H.G.; methodology, X.D. and H.G.; software, X.D., L.D., and M.L.; supervision, G.E and H.G.; writing—original draft, X.D.; writing—review and editing, B.A., G.E, and H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funds from the National Natural Science Foundations of China (31872975) and the Program of National Beef Cattle and Yak Industrial Technology System (CARS-37). Cattle Breeding Innovative Research Team supported statistical analysis and writing of the paper.
Institutional Review Board Statement
Ethics approval: All animals used in the study were treated following the guidelines established by the Council of China Animal Welfare. Protocols of the experiments were approved by the Science Research Department of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China (approval number: RNL09/07).
Data Availability Statement
We confirm that all raw data underlying our findings are publicly available without restriction. Data is available from the Dryad Digital Repository: doi: https://doi.org/10.5061/dryad.4qc06.
Conflicts of Interest
The authors declare that they have no competing interests. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mao Y., Hopkins D.L., Zhang Y., Luo X. Consumption patterns and consumer attitudes to beef and sheep meat in China. Am. J. Food Nutr. 2016;4:30–39. [Google Scholar]
- 2.An B., Xu L., Xia J., Wang X., Miao J., Chang T., Song M., Ni J., Xu L., Zhang L. Multiple association analysis of loci and candidate genes that regulate body size at three growth stages in Simmental beef cattle. BMC Genet. 2020;21:1–11. doi: 10.1186/s12863-020-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W., Xu L., Gao H., Wu Y., Gao X., Zhang L., Zhu B., Song Y., Bao J., Li J., et al. Detection of candidate genes for growth and carcass traits using genome-wide association strategy in Chinese Simmental beef cattle. Anim. Prod. Sci. 2018;58:224–233. doi: 10.1071/AN16165. [DOI] [Google Scholar]
- 4.Jiang L., Liu J., Sun D., Ma P., Ding X., Yu Y., Zhang Q. Genome wide association studies for milk pro-duction traits in Chinese Holstein population. PLoS ONE. 2010;5:e13661. doi: 10.1371/journal.pone.0013661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meredith B.K., Kearney F.J., Finlay E.K., Bradley D.G., Fahey A.G., Berry D.P., Lynn D.J. Ge-nome-wide associations for milk production and somatic cell score in Holstein-Friesian cattle in Ireland. BMC Genet. 2012;13:21. doi: 10.1186/1471-2156-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buzanskas M.E., Grossi D.A., Ventura R.V., Schenkel F.S., Sargolzaei M., Meirelles S.L., Mokry F.B., Higa R.H., Mudadu M.A., Da Silva M.V.G.B., et al. Genome-Wide Association for Growth Traits in Canchim Beef Cattle. PLoS ONE. 2014;9:e94802. doi: 10.1371/journal.pone.0094802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin B., Bao W.-J., Wu Z.-Q., Xia X.-H. In Situ Monitoring of Protein Adsorption on a Nanoparticulated Gold Film by Attenuated Total Reflection Surface-Enhanced Infrared Absorption Spectroscopy. Langmuir. 2012;28:9460–9465. doi: 10.1021/la300819u. [DOI] [PubMed] [Google Scholar]
- 8.Huang W., Kirkpatrick B.W., Rosa G.J.M., Khatib H. A genome-wide association study using selective DNA pooling identifies candidate markers for fertility in Holstein cattle. Anim. Genet. 2010;41:570–578. doi: 10.1111/j.1365-2052.2010.02046.x. [DOI] [PubMed] [Google Scholar]
- 9.Sahana G., Guldbrandtsen B., Bendixen C., Lund M. Genome-wide association mapping for female fertility traits in Danish and Swedish Holstein cattle. Anim. Genet. 2010;41:579–588. doi: 10.1111/j.1365-2052.2010.02064.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Miao J., Chang T., Xia J., An B., Li Y., Xu L., Zhang L., Gao X., Li J., et al. Evaluation of GBLUP, BayesB and elastic net for genomic prediction in Chinese Simmental beef cattle. PLoS ONE. 2019;14:e0210442. doi: 10.1371/journal.pone.0210442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu B., Guo P., Wang Z., Zhang W., Chen Y., Zhang L., Gao H., Wang Z., Gao X., Xu L., et al. Accuracies of genomic prediction for twenty economically important traits in Chinese Simmental beef cattle. Anim. Genet. 2019;50:634–643. doi: 10.1111/age.12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning C., Wang D., Zheng X., Zhang Q., Zhang S., Mrode R., Liu J.-F. Eigen decomposition expedites longitudinal genome-wide association studies for milk production traits in Chinese Holstein. Genet. Sel. Evol. 2018;50:1–10. doi: 10.1186/s12711-018-0383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Oliveira H.R., Lourenco D., Masuda Y., Misztal I., Tsuruta S., Jamrozik J., Brito L., Silva F., Cant J., Schenkel F. Single-step genome-wide association for longitudinal traits of Canadian Ayrshire, Holstein, and Jersey dairy cattle. J. Dairy Sci. 2019;102:9995–10011. doi: 10.3168/jds.2019-16821. [DOI] [PubMed] [Google Scholar]
- 14.Crispim A.C., Kelly M.J., Guimaraes S.E., Fonseca e Silva F., Fortes M.R., Wenceslau R.R., Moore S. Multi-Trait GWAS and New Candidate Genes Annotation for Growth Curve Parameters in Brahman Cattle. PLoS ONE. 2015;10:e0139906. doi: 10.1371/journal.pone.0139906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.France J., Kebreab E. Mathematical Modelling in Animal Nutrition. CABI; Wallingford, UK: 2008. [Google Scholar]
- 16.Das K., Li J., Wang Z., Tong C., Fu G., Li Y., Xu M., Ahn K., Mauger D., Li R., et al. A dynamic model for genome-wide association studies. Qual. Life Res. 2011;129:629–639. doi: 10.1007/s00439-011-0960-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ning C., Kang H., Zhou L., Wang D., Wang H., Wang A., Fu J., Zhang S., Liu J.-F. Performance Gains in Genome-Wide Association Studies for Longitudinal Traits via Modeling Time-varied effects. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-00638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolormaa S., Pryce J.E., Reverter A., Zhang Y., Barendse W., Kemper K., Tier B., Savin K., Hayes B.J., Goddard M.E. A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle. PLoS Genet. 2014;10:e1004198. doi: 10.1371/journal.pgen.1004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortes M., Kemper K., Sasazaki S., Reverter A., Pryce J., Barendse W., Bunch R., McCulloch R., Harrison B., Bolormaa S. Evidence for pleiotropism and recent selection in the PLAG 1 region in Australian Beef cattle. Anim. Genet. 2013;44:636–647. doi: 10.1111/age.12075. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., De Bakker P.I.W., Daly M.J., et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakushinskii A.B. The problem of the convergence of the iteratively regularized Gauss-Newton method. Comput. Math. Math. Phys. 1992;32:1353–1359. [Google Scholar]
- 22.Spiess A.-N., Neumeyer N. An evaluation of R 2 as an inadequate measure for nonlinear models in pharmacological and biochemical research: A Monte Carlo approach. BMC Pharmacol. 2010;10:6. doi: 10.1186/1471-2210-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipka A.E., Tian F., Wang Q., Peiffer J., Li M., Bradbury P.J., Gore M.A., Buckler E.S., Zhang Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to mul-tiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995;57:289–300. [Google Scholar]
- 25.Bolormaa S., Neto L.P., Zhang Y., Bunch R., Harrison B., Goddard M., Barendse W. A genome-wide as-sociation study of meat and carcass traits in Australian cattle. J. Anim. Sci. 2011;89:2297–2309. doi: 10.2527/jas.2010-3138. [DOI] [PubMed] [Google Scholar]
- 26.Fang X., Xu S., Zhang Y. A new breed resource in China—Chinese Simmental cattle. J. Yellow Cattle Sci. 2002;5:67–69. [Google Scholar]
- 27.Liang Y., Zhu B., Jin S., Bao J., Xu L., Chen Y., Gao X., Zhang L., Gao H., Li J. The Growth Curve Fitting and the Correlation Analysis between Body Weight and Body Measurements in Chinese Simmental Beef Cattle Population. Acta Vet. Zootech. Sin. 2018;49:497–506. [Google Scholar]
- 28.Fitzhugh H.A. Analysis of Growth Curves and Strategies for Altering Their Shape. J. Anim. Sci. 1976;42:1036–1051. doi: 10.2527/jas1976.4241036x. [DOI] [PubMed] [Google Scholar]
- 29.Denise R.S.K., Brinks J.S. Genetic and Environmental Aspects of the Growth Curve Parameters in Beef Cows. J. Anim. Sci. 1985;61:1431–1440. doi: 10.2527/jas1985.6161431x. [DOI] [PubMed] [Google Scholar]
- 30.Zainaguli J., Tan R., Huang X.-X., Wang Y.-C., Rexiti A., Nuerbiye W., Cheng L.-M., Fu X.-F., Jia X.-S., Zeng L. Fitting the Weight Growth Curve of Xinjiang Brown Cattle. China Anim. Husb. Vet. Med. 2014;41:211–215. [Google Scholar]
- 31.Dekkers J.C.M. Commercial application of marker- and gene-assisted selection in livestock: Strategies and lessons. J. Anim. Sci. 2004;82 doi: 10.2527/2004.8213_supplE313x. [DOI] [PubMed] [Google Scholar]
- 32.Guo Y., Huang Y., Hou L., Ma J., Chen C., Ai H., Huang L.-S., Ren J. Genome-wide detection of genetic markers associated with growth and fatness in four pig populations using four approaches. Genet. Sel. Evol. 2017;49:21. doi: 10.1186/s12711-017-0295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bottolo L., Chadeau-Hyam M., Hastie D.I., Zeller T., Liquet B., Newcombe P., Yengo L., Wild P.S., Schillert A., Ziegler A., et al. GUESS-ing polygenic associations with multiple phenotypes using a GPU-based evolutionary stochastic search algorithm. PLoS Genet. 2013;9:e1003657. doi: 10.1371/journal.pgen.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Reilly P.F., Hoggart C.J., Pomyen Y., Calboli F.C.F., Elliott P., Järvelin M.-R., Coin L.J. MultiPhen: Joint Model of Multiple Phenotypes Can Increase Discovery in GWAS. PLoS ONE. 2012;7:e34861. doi: 10.1371/journal.pone.0034861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Covington J.D., Noland R.C., Hebert R.C., Masinter B.S., Smith S.R., Rustan A.C., Ravussin E., Bajpeyi S. Perilipin 3 Differentially Regulates Skeletal Muscle Lipid Oxidation in Active, Sedentary, and Type 2 Diabetic Males. J. Clin. Endocrinol. Metab. 2015;100:3683–3692. doi: 10.1210/JC.2014-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camera E., Dahlhoff M., Ludovici M., Zouboulis C.C., Schneider M.R. Perilipin 3 modulates specific lipogenic pathways in SZ95 sebocytes. Exp. Derm. 2014;23:759–761. doi: 10.1111/exd.12507. [DOI] [PubMed] [Google Scholar]
- 38.Costa-Urrutia P., Colistro V., Jiménez-Osorio A.S., Cárdenas-Hernández H., Solares-Tlapechco J., Ramirez-Alcántara M., Granados J., Ascencio-Montiel I.J., Rodríguez-Arellano M.E. Genome-Wide As-sociation Study of Body Mass Index and Body Fat in Mexican-Mestizo Children. Genes (Basel) 2019;10:945. doi: 10.3390/genes10110945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabia B., Andrade S., Carreira M.C., Casanueva F.F., Crujeiras A.B. A role for novel adipose tis-sue-secreted factors in obesity-related carcinogenesis. Obes. Rev. 2016;17:361–376. doi: 10.1111/obr.12377. [DOI] [PubMed] [Google Scholar]
- 40.Sato S., Kikuchi T., Uemoto Y., Mikawa S., Suzuki K. Effect of candidate gene polymorphisms on repro-ductive traits in a Large White pig population. Anim. Sci. J. 2016;87:1455–1463. doi: 10.1111/asj.12580. [DOI] [PubMed] [Google Scholar]
- 41.Frater J., Lie D., Bartlett P., McGrath J.J. Insulin-like Growth Factor 1 (IGF-1) as a marker of cognitive decline in normal ageing: A review. Ageing Res. Rev. 2018;42:14–27. doi: 10.1016/j.arr.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Argente J., Pérez-Jurado L.A. Genetic causes of proportionate short stature. Best Pr. Res. Clin. Endocrinol. Metab. 2018;32:499–522. doi: 10.1016/j.beem.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Ramayo-Caldas Y., Fortes M.R., Hudson N.J., Porto-Neto L.R., Bolormaa S., Barendse W., Kelly M., Moore S.S., Goddard M.E., Lehnert S.A., et al. A marker-derived gene network reveals the regulatory role of PPARGC1A, HNF4G, and FOXP3 in intramuscular fat deposition of beef cattle. J. Anim. Sci. 2014;92:2832–2845. doi: 10.2527/jas.2013-7484. [DOI] [PubMed] [Google Scholar]
- 44.Pan Y., Choi J.H., Shi H., Zhang L., Su S., Wang X. Discovery and Validation of a Novel Neutrophil Activation Marker Associated with Obesity. Sci. Rep. 2019;9:3433. doi: 10.1038/s41598-019-39764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Y., Fu J., Wang A. Association of EphA4 polymorphism with swine reproductive traits and mRNA ex-pression of EphA4 during embryo implantation. Mol. Biol. Rep. 2012;39:2689–2696. doi: 10.1007/s11033-011-1023-8. [DOI] [PubMed] [Google Scholar]
- 46.Wang H., Zhang L., Cao J., Wu M., Ma X., Liu Z., Liu R., Zhao F., Wei C., Du L. Genome-Wide Specific Selection in Three Domestic Sheep Breeds. PLoS ONE. 2015;10:e0128688. doi: 10.1371/journal.pone.0128688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan D. The Hippo Signaling Pathway in Development and Cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li R., Beaudoin F., Ammah A.A., Bissonnette N., Benchaar C., Zhao X., Lei C., Ibeagha-Awemu E.M. Deep sequencing shows microRNA involvement in bovine mammary gland adaptation to diets supplemented with linseed oil or safflower oil. BMC Genom. 2015;16:884. doi: 10.1186/s12864-015-1965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trinh J., Hüning I., Yüksel Z., Baalmann N., Imhoff S., Klein C., Rolfs A., Gillessen-Kaesbach G., Lohmann K. A KAT6A variant in a family with autosomal dominantly inherited microcephaly and developmental delay. J. Hum. Genet. 2018;63:997–1001. doi: 10.1038/s10038-018-0469-0. [DOI] [PubMed] [Google Scholar]
- 50.Balasubramanian M., Hurst J., Brown S., Bishop N.J., Arundel P., DeVile C., Pollitt R.C., Crooks L., Longman D., Caceres J.F., et al. Compound heterozygous variants in NBAS as a cause of atypical osteogenesis im-perfecta. Bone. 2017;94:65–74. doi: 10.1016/j.bone.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams M.D., Kerlavage A.R., Fleischmann R.D., Fuldner R.A., Bult C.J., Lee N.H., Kirkness E.F., Weinstock K.G., Gocayne J.D., White O. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–174. [PubMed] [Google Scholar]
- 52.Gaudet P., Livstone M.S., Lewis S.E., Thomas P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We confirm that all raw data underlying our findings are publicly available without restriction. Data is available from the Dryad Digital Repository: doi: https://doi.org/10.5061/dryad.4qc06.