Abstract
The potent HIV-1 capsid inhibitor GS-6207 is an investigational principal component of long-acting antiretroviral therapy. Here we show that GS-6207 inhibits HIV-1 by stabilizing and thereby preventing functional disassembly of the capsid shell in infected cells. X-ray crystallography, cryo-electron microscopy, and hydrogen-deuterium exchange experiments reveal that GS-6207 tightly binds two adjoining capsid subunits and promotes distal intra- and inter-hexamer interactions that strikingly stabilize the curved capsid lattice. In addition, GS-6207 interferes with capsid binding to the cellular HIV-1 cofactors Nup153 and CPSF6 that mediate viral nuclear import and direct integration into gene-rich regions of chromatin. These findings elucidate structural insights into the multimodal, potent antiviral activity of GS-6207 and provide a means for rationally developing second-generation therapies.
One sentence summary:
The potent HIV-1 inhibitor GS-6207 binds and stabilizes curved capsid assemblies.
Long-acting antiretroviral therapy would substantially improve the care of people living with HIV and mitigate a number of challenges including the necessity of daily administration of current HIV medications, suboptimal treatment adherence, and emergence of drug resistance. GS-6207 (Lenacapavir, Gilead Sciences) is the first-in-class long-acting ultra-potent HIV capsid (CA) inhibitor. Recently completed phase 1 clinical trials (NCT03739866) have suggested a six-month dosing interval may be possible. Based on these results GS-6207 has advanced into phase 2/3 clinical trials (NCT04143594/NCT04150068). Initial mechanistic studies with GS-CA1, an archetypal predecessor of GS-6207, revealed its multi-stage mechanism of antiviral action (1). GS-CA1 potently (EC50 = 87 pM) inhibited early steps of HIV-1 replication, and also exhibited a second, less potent (EC50 = 240 pM) antiviral activity during virus egress. Molecular modeling studies predicted that both GS-CA1 and GS-6207 bind to the hydrophobic pocket formed by two adjoining CA subunits within the hexamer (2). HIV-1 genotyping, after selection in cell culture in the presence of the inhibitor, identified a number of CA mutations positioned near the potential inhibitor binding site that conferred substantial resistance to GS-CA1 (1). However, the structural and mechanistic bases for how this class of compounds binds and alters biological functions of HIV-1 CA remain unclear.
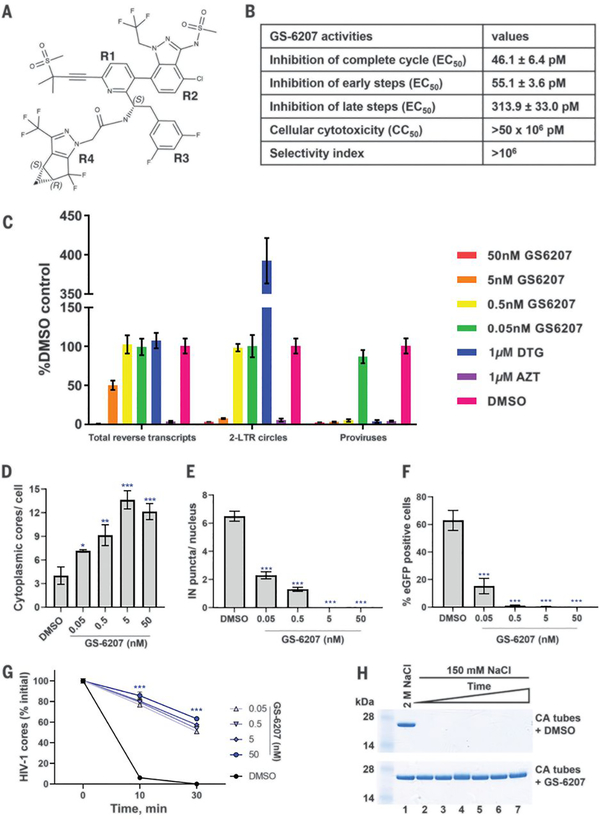
We synthesized GS-6207 (Fig. 1A) and examined its antiviral activities. GS-6207 inhibited HIV-1 replication in peripheral blood mononuclear cells (PBMCs) and various cell lines with EC50 values in the range of ~12 to 314 pM (Fig. 1B and Table S1). PBMCs and MT4 T cells were fully viable in the presence of 50 μM (highest concentration tested) GS-6207, indicating a selectivity index of >106 (Fig. 1B). GS-6207 exhibited higher potency during early (EC50 of ~55 pM) versus late (EC50 of ~314 pM) steps of HIV-1 replication (Fig. 1B and Table S1). Our subsequent efforts focused on understanding the structural and mechanistic bases for inhibition of incoming HIV-1 by GS-6207.
Fig. 1. Multimodal mechanism of action of GS-6207.
(A) Chemical structure of GS-6207. (B) Antiviral activities and cytotoxicity of GS-6207 (also see Table S1). (C) Effects of GS-6207 on formation of total reverse transcripts, 2-LTR circles and proviruses. Error bars indicate SD for three independent experiments. (D) Effect of GS-6207 on the number of post-fusion HIV-1 cores in the cytoplasm (also see Fig. S2). (E) Inhibition of nuclear import of HIV-1 (see Fig. S3A). (F) Effect of GS-6207 on HIV-1 infectivity (see Fig. S3B). (G) GS-6207 increases the stability of isolated HIV-1 cores in vitro (see Fig. S4). Error bars in (D-G) represent SEM from 4 fields of view for a representative experiment out of 2 independent experiments (***p < 0.0001). (H) Effects of GS-6207 on the stability of recombinant CA tubes. Only pelleted fractions of CA from each reaction are shown. CA tubes were assembled in 2 M NaCl in the absence (upper image) or presence of GS-6207 (lower image) and then either directly pelleted (lane 1) or exposed to low ionic strength (150 mM NaCl) buffer for increasing periods of time (0, 1, 4, 24, 48 and 96 h shown in lanes 2–7) and then pelleted.
To dissect HIV-1 post-entry infection steps targeted by GS-6207 we monitored viral DNA intermediates, including total reverse transcripts, 2-long terminal repeat (LTR) circles (a surrogate for nuclear import), and integrated proviruses (the viral copy DNA incorporated into the host cell DNA) (Fig. 1C). In parallel, we examined the effects of GS-6207 on viral DNA levels in the cytoplasm and nuclei of infected cells (Fig. S1). In control experiments, the reverse transcriptase (RT) inhibitor azidothymidine (AZT) impaired viral DNA synthesis, whereas the integrase (IN) inhibitor dolutegravir (DTG) specifically blocked integration as evidenced by marked reduction of proviral DNA and increased levels of 2-LTR circles. In contrast, GS-6207 affected multiple sequential steps of virus ingress in a dose dependent manner. At a comparatively high concentration (50 nM), GS-6207 effectively inhibited reverse transcription. At pharmacologically relevant concentration (5 nM) (3), the inhibitor partly impaired viral DNA synthesis and effectively blocked formation of 2-LTR circles and integrated HIV-1 DNA. In line with these results (Fig. 1C), 5 nM GS-6207 markedly reduced viral DNA levels in both the cytoplasm and nucleus (Fig. S1). At tenfold lower concentration (0.5 nM), GS-6207 inhibited integration without detectably affecting reverse transcription. Although 0.5 nM GS-6207 and 1 μM DTG similarly inhibited integration, the former failed to increase 2-LTR circle formation likely due to concomitant inhibition of nuclear import (Fig. 1C). Results of cellular fractionation indeed support this interpretation of the population-specific PCR assays (Fig. S1). Compared with the DMSO control, 0.5 nM GS-6207 increased and decreased viral DNA levels in the cytoplasm and nucleus, respectively (Fig. S1). The multi-step inhibition, which depends on the concentration of GS-6207, is likely due to the inhibitor affecting the multifaceted roles of CA during virus ingress (4).
We considered the following two scenarios to account for the observed inhibitions of viral DNA replication intermediates: i) GS-6207 could adversely affect functional disassembly of the CA shell through stabilizing or destabilizing its architecture, which in turn would adversely affect reverse transcription, nuclear import and integration; ii) GS-6207 could interfere with CA interactions with cognate cellular co-factors needed for nuclear import, and/or trafficking of pre-integration complexes inside the nucleus to preferred sites of integration.
To examine these possibilities, we imaged the effects of GS-6207 on incoming HIV-1 by using single particle detection of virus cores co-labeled with CypA-DsRed (a marker for CA) and INmNG (IN fused to NeonGreen protein) (Fig. S2) (5). GS-6207 substantially increased levels of virus cores in the cytoplasm suggesting a stabilizing effect of the inhibitor (Fig. 1D). Conversely, GS-6207 inhibited the formation of IN puncta in the nucleus with concomitant inhibition of HIV-1 infection (Fig. 1E and 1F and S3). These findings indicate that GS-6207 stabilizes virus cores, leading to their accumulation in the cytoplasm and preventing nuclear import.
To explore the stabilizing role of the inhibitor on the CA shell, we conducted in vitro assays with isolated HIV-1 particles (6). In the absence of inhibitor, virus cores fully dissociated within 30 min, whereas pM concentrations of GS-6207 markedly enhanced the stability of native cores (Fig. 1G and S4). Next, we tested effects of GS-6207 on tubular assemblies made in the presence of 2 M NaCl (7) (Fig. 1H). The preassembled tubes dissociated immediately upon their exposure to a buffer containing 150 mM NaCl (<1 min, see lane 2 in Fig. 1H). In sharp contrast, addition of GS-6207 to preassembled CA tubes rendered these tubular assemblies highly resistant to low ionic strength (150 mM NaCl) conditions. Strikingly, in the presence of GS-6207, tubular CA assemblies remained stable even after 96 h of incubation under physiologically relevant conditions (Fig. 1H). The stabilizing effects correlated with a GS-6207 to CA ratio of ~1:1 (Fig. S5).
We tested the effects of the cellular CA binding partner CypA on GS-6207 activities. As expected (8, 9), the addition of increasing concentrations of CypA resulted in effective disassembly of the pre-formed CA tubes in the absence of the inhibitor (Fig. S6). In sharp contrast, GS-6207 stabilized CA tubes remained intact in the presence of CypA (Fig. S6). Furthermore, GS-6207 antiviral activities remained unaffected by depletion or overexpression of CypA in Jurkat and MT4 T cells (Fig. S7).
Next, we examined whether GS-6207 affects CA interactions with known cellular cofactors Nup153 and CPSF6 needed for nuclear import (10, 11). GS-6207 substantially reduced binding of cellular Nup153 and CPSF6 to preassembled CA tubes (Fig. S8). Because CPSF6 is also known to regulate integration site selectivity (12, 13), we tested whether GS-6207 influences sites of HIV-1 integration. The inhibitor substantially reduced integration in gene dense regions and conversely, enhanced integration in lamina-associated domains (LADs) (Fig. S9). These GS-6207 mediated effects on integration targeting mimicked the CPSF6 depletion phenotype. However, the extent of inhibitor induced changes was less than those seen with CPSF6 knockout, suggesting that GS-6207 may not fully displace the cellular cofactor. Taken together, our mechanistic studies reveal striking stabilizing effects of GS-6207 on viral cores coupled with the ability of the inhibitor to also interfere with CA binding to cognate cellular cofactors CPSF6 and Nup153.
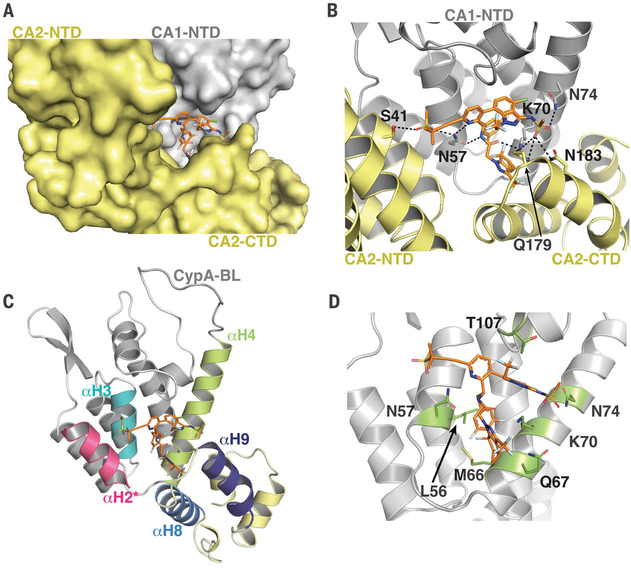
To understand the structural basis for GS-6207 interaction with CA, we solved a co-crystal structure of the inhibitor bound to a pre-stabilized CAA14C/E45C/W184A/M185A hexamer (14) (Fig. 2 and Table S2). The high-resolution structure (2.22 Å) revealed that GS-6207 binds in the hydrophobic pocket formed by two adjacent CA subunits (Fig. 2A and Fig. S10) with a stoichiometry of six GS-6207 compounds bound per each CAA14C/E45C/W184A/M185A hexamer. GS-6207 makes extensive Van der Waals and hydrogen bonding interactions with CA1-NTD (the N-terminal domain of CA subunit 1), CA2-CTD (the C-terminal domain of CA subunit 2) and CA2-NTD. Two ring systems R3 and R4 primarily drive the Van der Waals interactions with CA1-NTD and CA2-CTD (Fig. S10). R1 and R2 also provide additional interactions with CA1-NTD and CA2-NTD. GS-6207 establishes a hydrogen bonding network with the side chains of N57, K70 and N74 of CA1-NTD, S41 of CA2-NTD, and Q179 and N183 of CA2-CTD (Fig. 2B, S10 and Table S3).
Fig. 2. Structural basis for GS-6207 interaction with CA hexamer.
(A) X-ray crystal structure of GS-6207 (orange) bound to the pre-stabilized CAA14C/E45C/W184A/M185A hexamer (PDB ID: 6VKV). GS-6207 binds at the pocket formed by two adjoining CA subunits CA1 (light grey) and CA2 (pale yellow). Relative positioning of CA1-NTD, CA2-NTD, and CA2-CTD are indicated. (B) Cartoon representation of the structure indicating GS-6207’s interactions with the two subunits that form the binding pocket, CA1 and CA2. Hydrogen bonds are denoted by black dashed lines. (C) The main helices (αH2*, αH3, αH4, αH8 and αH9) that interact with GS-6207 are indicated. (D) Reported resistance mutations (green) for GS-CA1 (1) are shown in the context of GS-6207 bound to CA1-NTD.
The interacting helices that predominantly form the GS-6207 binding pocket include αH3 and αH4 from CA1-NTD, αH8 and αH9 from CA2-CTD, and αH2* from CA2-NTD (Fig. 2C). Particularly noteworthy is that GS-6207 strongly influences the conformation and relative positioning of αH9 of CA2-CTD with respect to αH4 of CA1-NTD. For comparison, αH9 is seen to exhibit substantial conformational variation in the absence or presence of different cellular protein partners bound to CAA14C/E45C/W184A/M185A or native CA hexamers (Fig. S11–20).
Previously reported resistant mutations to predecessor compound GS-CA1 are within close proximity of the GS-6207 binding site (Fig. 2D). M66 is a key constituent of the hydrophobic pocket and forms strong Van der Waals interactions with rings R3 and R4. The M66I substitution had the most profound effects on loss of GS-6207 potency, reducing activity by more than four orders of magnitude (Table S4). N57S, Q67H, K70A, and N74D substitutions, which are expected to adversely affect direct interactions of CA with GS-6207, reduced potency by ~60, ~10, ~45, and 14-fold, respectively (Table S4). Consistent with a previous report (1), infectivity of the M66I mutant virus, which conferred the greatest extent of GS-6207 resistance, was markedly compromised (Fig. S21). Infectivity of N57S and K70A mutant viruses, which exhibited substantial resistance to the inhibitor, were severely and considerably reduced, respectively. Q67H and N74D, which exhibited lower levels of resistance, displayed WT HIV-1 infectivity (Fig. S21).
Structural comparison of GS-6207 with the substantially less potent HIV-1 CA inhibitor PF74 (15–17) revealed both similarities and marked differences (Fig. S22 and Table S3). The resemblance between the two compounds is seen with respect to their interactions with CA1-NTD. Phenyl R1 and R2, and indole R3 rings of PF74 superimpose onto the indazole (R2), difluorobenzyl (R3), and cyclopenta-pyrazole (R4) rings of GS-6207, respectively. However, unlike PF74, which makes limited hydrophobic contacts with CA2-CTD, GS-6207 establishes extensive hydrogen bonding and hydrophobic interactions with adjoining CA2-NTD and CA2-CTD (Fig. 2, S10 and S22).
We also compared GS-6207 binding to known interactions of CPSF6 and Nup153 with CA hexamers (15, 18). The backbone of Nup153 aligns along R1 and R3 of GS-6207, with F1417 of Nup153 closely superimposing on the difluorobenzyl moiety (R3) of GS-6207 (Fig. S23). Similarly, there is substantial overlap between GS-6207 and the main chain of CPSF6, with F321 of CPSF6 superimposing on R3 extremely well (Fig. S24). Interestingly, binding pockets for Nup153 and CPSF6 are more open, with CTD αH9 being positioned further away from NTD αH4 than in the presence of GS-6207. In turn, the closer αH4-αH9 conformation imposed by GS-6207 creates steric clashes with Nup153 and CPSF6 (Fig. S23 and S24). Collectively, these findings provide structural explanations for displacement of Nup153 and CPSF6 by GS-6207 (Fig. S8).
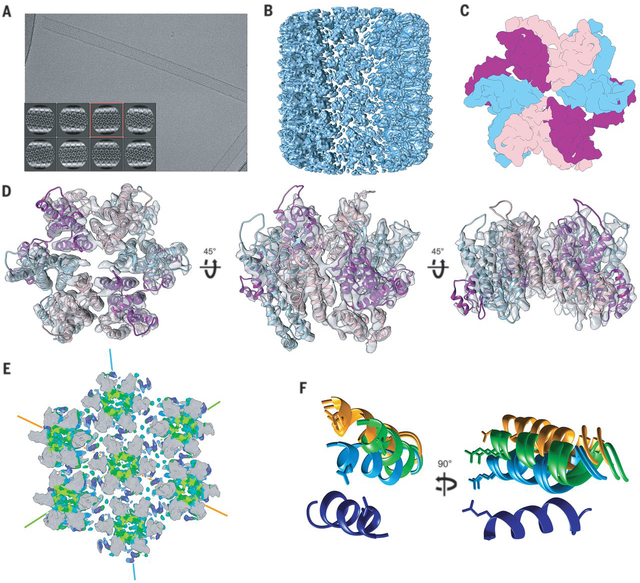
To understand the structural basis for GS-6207 interactions with curved CA assemblies we employed cryo-EM. GS-6207, but not a DMSO control, stabilized preformed tubes and resulted in well-defined tubular CA assemblies at physiological salt concentration (Fig. 3A, S25 and S26). Imaging these structures allowed us to obtain a 6.3 Å map for GS-6207 bound to A92E CA tubes (Fig. 3B, S27, S28 and Table S5; GS-6207 interacted similarly with WT and A92E CA tubes (Fig. S25), and the latter protein was successfully used for prior cryo-EM studies (8, 19)). A hexamer with pseudo two-fold symmetry characteristic of CA tubes was readily identified (Fig. 3C–D) and further refined by analyzing helical tube patches using a single particle approach (RASTR) independent of helical parameter determination (Fig. S29) (20). The mutually independent helical and RASTR approaches produced equivalent maps of a tube hexamer (Fig. S30), further validating the map’s accuracy (Fig. S31–33). Rigid body docking of individual crystallographic CA monomers in the presence of GS-6207 could account for all features in the cryo-EM hexamer, including the positions of well-defined α-helices in the CTD (Fig. 3D, S30 and S34). Density corresponding to bound GS-6207 could be identified by segmentation of the helical or RASTR cryo-EM maps (Fig. S35). Thus, we were able to obtain a model of the GS-6207 bound tube hexamers under physiologically relevant conditions.
Fig. 3. Cryo-EM structure of GS-6207 stabilized CA tubes.
(A) Cryo-EM image of A92E CA tubes stabilized by GS-6207 in 150 mM NaCl. Inset shows a subset of the averages obtained by 2D clustering of tube segments. (B) 6.3 Å resolution Cryo-EM map from helical processing of GS-6207 stabilized A92E CA tubes. (C) Diagram showing the pseudo-two-fold symmetric arrangement of monomers in a tube hexamer. (D) Atomic model of a hexamer in the GS-6207 stabilized CA tube generated by rigid-body fitting of six copies of the X-ray structure of a GS-6207 bound CA monomer into the RASTR map. (E) A portion of a tube showing interactions between seven hexamers. Coloring corresponds to HDX protection levels. The cyan, green and orange lines indicate three helical directions. (F) Close up of αH9-αH9 interactions involving a central hexamer. All six H9 helices in the central hexamer (in dark blue) were superimposed. αH9 helices in neighboring hexamers are shown in cyan, green and orange, matching the coloring of helical directions in (E). The absence of true two-fold symmetry results in slight differences in the positioning of the two helices along a specific helical direction. The visible side chain is Glu180.
Comparisons of our cryo-EM structure with published cryo-EM and cryo-ET derived structures of CA hexamers from tubes and native HIV-1 particles (19, 21) reveal the principal differences in formation of curved hexameric lattices in the absence and presence of GS-6207 (S36-S41). Normally, CA CTDs move away from the adjacent NTDs to accommodate inter-hexamer contacts in the context of a curved topology (19, 21). In sharp contrast, GS-6207 strongly restricts changes in the CTD position with respect to the adjoining NTD and requirements for establishing inter-hexamer interactions on a curved surface are satisfied by repositioning of the comparatively rigid GS-6207 bound CA monomers in each hexamer (see movies S1–S4, also compare movie S5 with movies S7 and S9, and movie S6 with movies S8 and S10). Accordingly, NTD αH4 and CTD αH9 from adjacent subunits are further apart and closer together in the absence and presence of the inhibitor, respectively (Fig. S37 and S39).
To further understand how GS-6207 affects tubular CA assemblies we used hydrogen-deuterium exchange (HDX) (Fig. S42–S46). HDX experiments revealed strong protection in CA segments that directly interact with the inhibitor (Fig. S47–S48). Unexpectedly, we observed strong protection beyond the direct inhibitor binding sites. The NTDs that form the inner hexamer core and provide the binding site for IP6 (a natural cellular cofactor of CA that also stabilizes virus cores (22)) showed strong protection (Fig. S47 and S49) despite a lack of direct contacts with GS-6207. These findings suggest that GS-6207 stabilizes individual CA hexamers. This notion is further supported by thermal shift assays, which show that GS-6207 substantially increases the melting temperature of isolated CA hexamers (Fig. S50). Collectively these biochemical findings are consistent with our co-crystal structure (Fig. 2), which shows that each GS-6207 connects two adjoining monomers in a hexamer, with the binding of six inhibitors resulting in a more stable hexamer.
Strikingly, the strongest GS-6207 induced protections were seen in αH9 (Fig. S47 and S51) suggesting that the inhibitor stabilizes inter-hexamer αH9-αH9 contacts essential for curved lattice formation (Fig. 3E–F and (19, 21)). The E45A and E180A CA substitutions, which influence intra- and inter-hexameric interfaces, respectively (23–25), but do not directly interact with GS-6207 (Fig. S47), conferred partial resistance to the inhibitor (Fig. S52).
Pliability of intra- and inter-hexameric interactions is essential for both proper assembly of the CA shell during virion maturation and its subsequent disassembly during virus ingress (8, 23). GS-6207 disrupts this delicately balanced interplay by rigidifying the CTD conformation, and stabilizing both intra- and αH9-αH9 inter-hexamer interactions (Fig. 2, 3 and S47). These findings provide structural clues as to how GS-6207 inhibits functional disassembly of virus cores and blocks incoming HIV-1 in infected cells (Fig. 1). Taken together, our study elucidates the structural and mechanistic bases for the multimodal, potent antiviral activity of GS-6207 and provide a platform for rationally developing improved long-acting therapies.
We note that during the revision of the present manuscript an article describing clinical targeting of HIV CA by GS-6207, which also includes synthesis of the inhibitor and a crystal structure of GS-6207 bound to CA hexamer (Fig. S53), was published (26).
Supplementary Material
Acknowledgements:
We are grateful to Stephanie Rebensburg, Pratibha Koneru and other members of the participating laboratories for their help with data analysis and valuable suggestions.
Funding: This work was supported by NIH grants R01 AI062520 and R01 AI143649 (to M.K.), U54 AI150472 (to M.K., P.G., A.C.F., G.B.M., and A.N.E.), R01 AI129862 to G.B.M., R01 AI052014 (to A.N.E.), R01 AI77344 and DP1 DA043915 (to E.M.P).
Footnotes
Competing Interests: A.N.E. declares fees from ViiV Healthcare Co. for work unrelated to this project. No other authors declare competing interests.
Data availability:
All data is available in the manuscript or the supplementary materials. The co-crystal structure and cryo-EM derived atomic model are deposited into the PDB under the accession numbers: 6VKV and 6VWS, respectively. The EM maps are deposited into the EMDB under accession codes: EMD-21423 and EMD-21424. DNA sequences for integration site mapping were deposited in the National Center for Biotechnology Sequence Read Archive (NCBI SRA) under accession code PRJNA608802.
References and Notes:
- 1.Yant SR et al. , A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model. Nat. Med. 25, 1377–1384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh K et al. , GS-CA Compounds: First-In-Class HIV-1 Capsid Inhibitors Covering Multiple Grounds. Front. Microbiol. 10, 1227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daar E et al. , in Conference on Retroviruses and Opportunistic Infections (Boston, USA, 2020). [Google Scholar]
- 4.Yamashita M, Engelman AN, Capsid-Dependent Host Factors in HIV-1 Infection. Trends Microbiol. 25, 741–755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis AC, Marin M, Shi J, Aiken C, Melikyan GB, Time-Resolved Imaging of Single HIV-1 Uncoating In Vitro and in Living Cells. PLoS Pathog. 12, e1005709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis AC, Melikyan GB, Single HIV-1 Imaging Reveals Progression of Infection through CA-Dependent Steps of Docking at the Nuclear Pore, Uncoating, and Nuclear Transport. Cell Host Microbe 23, 536–548 e536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrlich LS, Agresta BE, Carter CA, Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66, 4874–4883 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C et al. , Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun. 7, 10714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grattinger M et al. , In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology 257, 247–260 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Bejarano DA et al. , HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matreyek KA, Engelman A, The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 85, 7818–7827 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Achuthan V et al. , Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe 24, 392–404 e398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sowd GA et al. , A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U.S.A 113, E1054–1063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pornillos O et al. , X-ray structures of the hexameric building block of the HIV capsid. Cell 137, 1282–1292 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price AJ et al. , Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS Pathog. 10, e1004459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya A et al. , Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. U.S.A 111, 18625–18630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito A et al. , Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74. J. Virol. 90, 5808–5823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AJ et al. , CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog. 8, e1002896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao G et al. , Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randolph PS, Stagg SM, Reconstruction of Average Subtracted Tubular Regions (RASTR) enables structure determination of tubular filaments by cryo-EM. J. Struct. Biol. 4, 100023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattei S, Glass B, Hagen WJ, Krausslich HG, Briggs JA, The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354, 1434–1437 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Dick RA et al. , Inositol phosphates are assembly co-factors for HIV-1. Nature 560, 509–512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.del Alamo M, Mateu MG, Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J. Mol. Biol. 345, 893–906 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Shi J, Zhou J, Shah VB, Aiken C, Whitby K, Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol. 85, 542–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forshey BM, von Schwedler U, Sundquist WI, Aiken C, Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76, 5667–5677 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Link JO et al. , Clinical targeting of HIV capsid protein with a long-acting small molecule. Nature, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chanput W, Mes JJ, Wichers HJ, THP-1 cell line: an in vitro cell model for immune modulation approach. Int. Immunopharmacol. 23, 37–45 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Connor RI, Chen BK, Choe S, Landau NR, Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206, 935–944 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Naldini L et al. , In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Koneru PC et al. , HIV-1 integrase tetramers are the antiviral target of pyridine-based allosteric integrase inhibitors. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A et al. , A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase. PLoS Pathog. 10, e1004171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Pizzato M et al. , A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J. Virol. Methods 156, 1–7 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Feng L et al. , The Competitive Interplay between Allosteric HIV-1 Integrase Inhibitor BI/D and LEDGF/p75 during the Early Stage of HIV-1 Replication Adversely Affects Inhibitor Potency. ACS Chem. Biol. 11, 1313–1321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serrao E, Cherepanov P, Engelman AN, Amplification, Next-generation Sequencing, and Genomic DNA Mapping of Retroviral Integration Sites. J. Vis. Exp, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson-Daniels J et al. , Dominant Negative MA-CA Fusion Protein Is Incorporated into HIV-1 Cores and Inhibits Nuclear Entry of Viral Preintegration Complexes. J. Virol. 93, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung M et al. , Large-scale functional purification of recombinant HIV-1 capsid. PLoS One 8, e58035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pornillos O, Ganser-Pornillos BK, Banumathi S, Hua Y, Yeager M, Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J. Mol. Biol. 401, 985–995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nettleship JE, Brown J, Groves MR, Geerlof A, Methods for protein characterization by mass spectrometry, thermal shift (ThermoFluor) assay, and multiangle or static light scattering. Methods Mol. Biol. 426, 299–318 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Kessl JJ et al. , A multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 287, 16801–16811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W, XDS. Acta Crystallogr. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ, Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Vagin A, Teplyakov A, An approach to multi-copy search in molecular replacement. Acta Crystallogr. D 56, 1622–1624 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M, HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zivanov J et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Chalmers MJ et al. , Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, Smith DL, Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pascal BD et al. , HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keppel TR, Weis DD, Mapping residual structure in intrinsically disordered proteins at residue resolution using millisecond hydrogen/deuterium exchange and residue averaging. J. Am. Soc. Mass Spectrom. 26, 547–554 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Graupe M et al. , in International Search Report and Writen Opinion Isued inPCT/ US2017/047416, dated Oct.27, 2017, 10pages., U. S. Patent, Ed (Gilead Science, Inc.,, United States, 2018).
- 56.Liu GC, Cogan DA, Ellman JA, Catalytic asymmetric synthesis of tert-butanesulfinamide. Application to the asymmetric synthesis of amines. J. Am. Chem. Soc. 119, 9913–9914 (1997). [Google Scholar]
- 57.Krissinel E, Henrick K, Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the manuscript or the supplementary materials. The co-crystal structure and cryo-EM derived atomic model are deposited into the PDB under the accession numbers: 6VKV and 6VWS, respectively. The EM maps are deposited into the EMDB under accession codes: EMD-21423 and EMD-21424. DNA sequences for integration site mapping were deposited in the National Center for Biotechnology Sequence Read Archive (NCBI SRA) under accession code PRJNA608802.