Abstract
Patients suffering from Coronavirus disease 2019 (COVID-19) can develop neurological sequelae, such as headache and neuroinflammatory or cerebrovascular disease. These conditions—termed here as Neuro-COVID—are more frequent in patients with severe COVID-19. To understand the etiology of these neurological sequelae, we utilized single-cell sequencing and examined the immune cell profiles from the cerebrospinal fluid (CSF) of Neuro-COVID patients compared with patients with non-inflammatory and autoimmune neurological diseases or with viral encephalitis. The CSF of Neuro-COVID patients exhibited an expansion of dedifferentiated monocytes and of exhausted CD4+ T cells. Neuro-COVID CSF leukocytes featured an enriched interferon signature; however, this was less pronounced than in viral encephalitis. Repertoire analysis revealed broad clonal T cell expansion and curtailed interferon response in severe compared with mild Neuro-COVID patients. Collectively, our findings document the CSF immune compartment in Neuro-COVID patients and suggest compromised antiviral responses in this setting.
Keywords: transcriptomics, single-cell RNA sequencing, cerebrospinal fluid, COVID-19, SARS-CoV-2, neurological manifestation, Neuro-COVID, T cell exhaustion, interferon-stimulated genes
Graphical Abstract
Highlights
-
•
Single-cell atlas of cerebrospinal fluid in Neuro-COVID and controls
-
•
Expansion of dedifferentiated monocytes and exhausted CD4+ T cells in Neuro-COVID
-
•
Less pronounced interferon signature in Neuro-COVID than in viral encephalitis
-
•
Curtailed interferon-response in severe Neuro-COVID
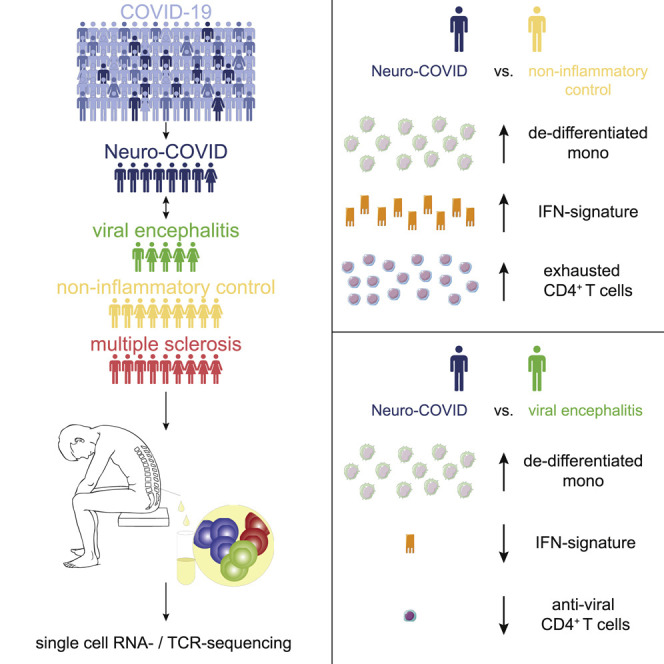
Neurological manifestations associated with COVID-19 (Neuro-COVID) are recognized but under-studied. Using single-cell transcriptomics, Heming et al. identify expansion of dedifferentiated monocytes and exhausted CD4+ T cells in the cerebral spinal fluid of Neuro-COVID patients. This work provides a basis for improved understanding of the neurological sequelae associated with COVID-19.
Introduction
Patients suffering from coronavirus disease 2019 (COVID-19), the global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), can develop acute or long-term neurological sequelae, henceforth collectively termed “Neuro-COVID” (Ellul et al., 2020). Diverse neurological signs and symptoms have been associated with COVID-19, ranging from mild symptoms (e.g., anosmia, ageusia, headache, dizziness) to severe complications, such as seizures, encephalitis, ischemic stroke, and intracerebral hemorrhage (Mao et al., 2020; Romero-Sánchez et al., 2020; Varatharaj et al., 2020). Consensus criteria for the diagnosis and classification of the severity of Neuro-COVID have been proposed (Fotuhi et al., 2020; Paterson et al., 2020). The prevalence of Neuro-COVID varies considerably between individual studies ranging from 4.1% (Xiong et al., 2020) to 57.4% (Romero-Sánchez et al., 2020) and even 84% in COVID-19 with acute respiratory distress syndrome (ARDS) (Helms et al., 2020). The heterogeneity might be due to variable definitions of Neuro-COVID (Pezzini and Padovani, 2020) or selection bias, as most evidence stems from retrospective studies of hospitalized patients (Helms et al., 2020; Mao et al., 2020; Romero-Sánchez et al., 2020; Xiong et al., 2020). Most of the specific neurological signs and symptoms, with the exception of anosmia and ageusia, are more prevalent in severe COVID-19 infection and severely affected patients will be hospitalized more frequently. During infection with the new SARS-CoV-2, the prevalence of neurological manifestations ranges orders of magnitude higher than previous reports of infections with the related SARS-CoV (0.09%) and Middle East respiratory syndrome coronavirus (MERS-CoV) (0.36%) (Ellul et al., 2020). Neurological sequelae associated with SARS-CoV-2 infection thus constitute an unexpectedly frequent health burden and the underlying mechanisms remain poorly defined.
Three potential mechanisms of Neuro-COVID have been proposed: (1) non-specific complications of systemic disease for example because of oxygen deprivation, sepsis, or hyperpyrexia; (2) direct viral damage to cells of the nervous system because of neurotropism of SARS-CoV-2; and (3) indirect damage to the nervous system because of an infection-triggered excessive and detrimental immune activation (Ellul et al., 2020). These suspected mechanisms are not mutually exclusive and might coexist in individual patients. In support of direct neuro-tropism, previous studies have shown that SARS-CoV-2 can enter the nervous system by crossing the neural-mucosal interface in olfactory mucosa by binding to ACE2 (Hoffmann et al., 2020; Zhou et al., 2020). The virus subsequently spreads along neuroanatomical structures and penetrates respiratory and cardiovascular control centers in the medulla oblongata (Meinhardt et al., 2020). SARS-CoV-2 was accordingly detected in the brains of 53% of brain autopsy specimens with enrichment in cranial nerves originating from the lower brainstem (Matschke et al., 2020). In support of detrimental immunity, the same study provided neuropathological evidence of microglia activation and in 79% of the analyzed patients infiltration of CD8+ T cells in the brain and meninges (Matschke et al., 2020). However, autopsy and brain biopsy materials are rarely available from patients and will naturally be restricted to very severely affected patients. Analyzing a more readily available biomaterial in Neuro-COVID is thus preferable.
Cerebrospinal fluid (CSF) forms a unique immune compartment that surrounds the central nervous system (CNS) and can be sampled from patients to facilitate the diagnosis and understanding of neurological diseases (Ransohoff and Engelhardt, 2012). The noncellular fraction of CSF represents mainly an ultrafiltrate of serum, while the composition of leukocytes in the CSF is tightly controlled and differs substantially from blood (Ransohoff and Engelhardt, 2012). For example, while CD4+ T cells are the most abundant cell type in CSF, myeloid and B cells are reduced compared with blood (Han et al., 2014; Kowarik et al., 2014). Applying modern technologies, such as single-cell transcriptomics to the CSF, multiplies the potential of the CSF to decipher the pathogenesis of neurological diseases (Meyer Zu Hörste et al., 2020) as previously exemplified in multiple sclerosis and Alzheimer’s disease (Gate et al., 2020; Ramesh et al., 2020; Schafflick et al., 2020).
Previous studies have analyzed CSF in Neuro-COVID using established techniques, such as cell counting and protein characterization. Most studies identified a normal cell count, except when the presentation was encephalitis, and normal or mildly elevated protein (Espíndola et al., 2020), yet a detailed leukocyte characterization in CSF is lacking. Notably, SARS-CoV-2 RNA was undetectable in the CSF in nearly all studies, and its detection was equally rare in pulmonary COVID-19 (2 out of 578 patients, see Destras et al., 2020) and in Neuro-COVID (0 out of 30 patients, see Neumann et al., 2020; 0 out of 31, see Bellon et al., 2020; 1 out of 37, see Kremer et al., 2020). This suggests indirect mechanisms primarily causing neurological sequelae of COVID-19 patients. However, these studies provide a low dimensional characterization of CSF in Neuro-COVID and lack unbiased techniques.
In contrast, high-resolution data are available from the blood and bronchoalveolar fluid (BALF) in pulmonary COVID-19. Recent single-cell RNA sequencing (scRNA-seq) studies have revealed complex immune dysregulation in the blood with a severity-specific pattern of myeloid cells and clonally expanded CD8+ T cells in pulmonary COVID-19 (Schulte-Schrepping et al., 2020; Zhu et al., 2020). Whether similar or distinct changes occur in the CSF in Neuro-COVID, remains unknown.
Here, we therefore constructed an unbiased single-cell transcriptional atlas of CSF leukocytes in patients with Neuro-COVID and infectious, autoimmune, and non-inflammatory controls. We identified an increase of dedifferentiated monocytes and exhausted T cells in the CSF, which was specific to Neuro-COVID. An interferon (IFN) response in Neuro-COVID was present but attenuated compared with viral encephalitis. Severe Neuro-COVID exhibited a broad clonal T cell expansion and a decreased IFN response. Thus, leukocytes exhibit disease-specific signs of local immune overactivation, despite the absence of SARS-CoV-2 in the CSF. We thereby provide further evidence for immune-mediated mechanisms in Neuro-COVID.
Results
Single-Cell Atlas of Cerebrospinal Fluid Leukocytes in Neuro-COVID Compared with Controls Including Viral Encephalitis
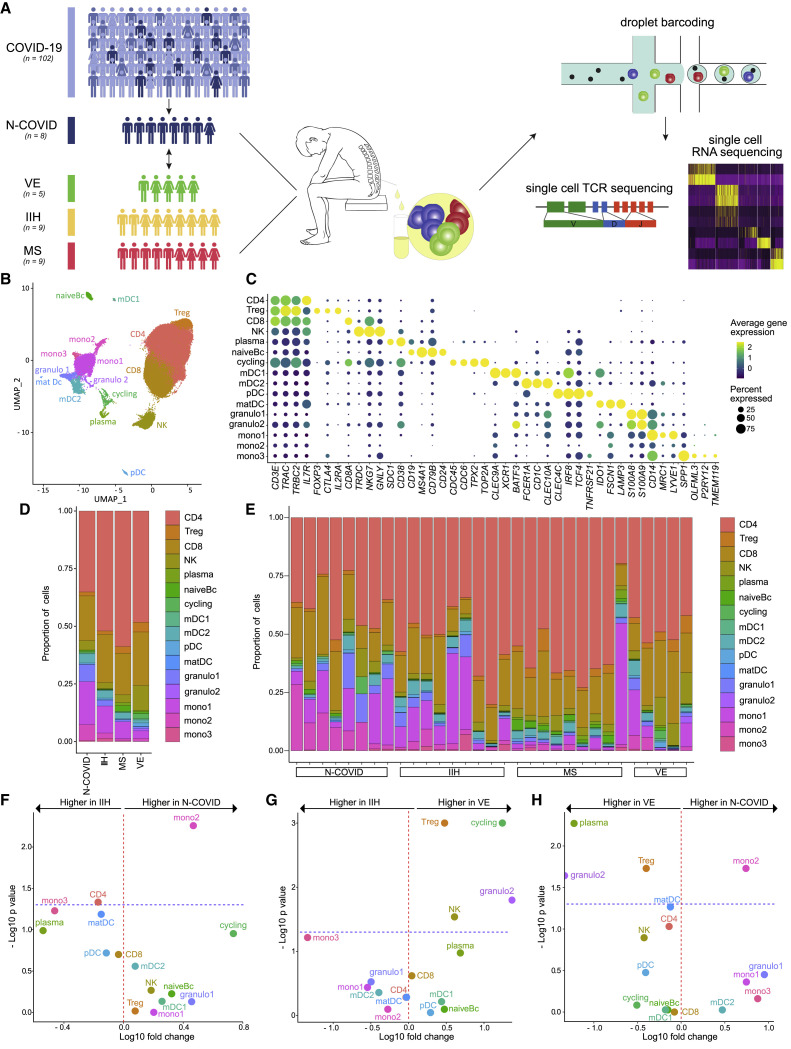
We successfully collected CSF leukocytes from eight Neuro-COVID patients for scRNA-seq analysis (see STAR Methods) (Figure 1 A; Table S1). Two of these patients had severe acute respiratory distress syndrome (ARDS), necessitating mechanical ventilation, while six had mild-to-moderate pulmonary COVID-19 manifestation (Table S1). Three patients (two of them with ARDS) had severe neurological signs/symptoms, including seizures, flaccid paresis, or stroke. Five patients showed mild neurological manifestations, including headache, dizziness, anosmia/ageusia, and cognitive impairment. In line with previous reports (Bellon et al., 2020; Destras et al., 2020; Espíndola et al., 2020; Neumann et al., 2020), we did not detect SARS-CoV-2 in the CSF of any patient using PCR. Additionally, we did not identify any cells expressing SARS-CoV-2 RNA with single-cell transcriptomics (see below and STAR Methods). We then collected previously published scRNA-seq data from controls with idiopathic intracranial hypertension (IIH) (n = 4), a non-inflammatory condition characterized by excess CSF, and with the brain autoimmune disease multiple sclerosis (MS) in relapse (n = 4) (Schafflick et al., 2020). To improve statistical power, we recruited additional patients with IIH (n = 5), active MS (n = 5), and viral encephalitis (VE) (n = 5) and performed scRNA-seq of CSF cells (Figure 1A; Table S1).
Figure 1.
Subtypes of Monocytic Cells Increased in Neuro-COVID in the CSF
(A) Scheme illustrating the study design; 33 out of 102 patients (32%) with COVID-19 infection had neurological manifestations (N-COVID). Cerebrospinal fluid (CSF) was collected from 12 of these patients for clinical reasons and CSF leukocytes from 8 patients were successfully analyzed by single-cell RNA sequencing (scRNA-seq) and single-cell T cell receptor (TCR) sequencing. CSF leukocytes from patients with viral encephalitis (VE), idiopathic intracranial hypertension (IIH), and relapsing-remitting multiple sclerosis (MS) were used as controls.
(B) UMAP plot showing 16 color-coded cell clusters of 80,919 raw single-cell transcriptomes from CSF cells from N-COVID (n = 8), IIH (n = 9), MS (n = 9), and VE (n = 5) patients.
(C) Marker genes of cell clusters are shown. Color encodes average gene expression; dot size represents percentage of cells expressing the gene.
(D and E) Proportions of cells split by diagnosis (D) or individual patient (E).
(F–H) Changes of cluster abundances in N-COVID (n = 8) versus IIH (n = 9) (F), VE (n = 5) versus IIH (n = 9) (G), and N-COVID (n = 8) versus VE (n = 5) (H). Logarithmic fold change of cluster abundance is plotted against negative logarithmic p value (two-sided Wilcoxon’s rank-sum test). The horizontal dashed line represents the significance threshold (p = 0.05).
Abbreviations: CD4, CD4+ T cells; Treg, regulatory T cells; CD8, CD8+ T cells; NK, NK cells; plasma, plasma cells; naiveBc, naive B cells; mDC, myeloid dendritic cells; pDC, plasmacytoid dendritic cells; matDC, mature dendritic cells; granulo, granulocytes; mono, monocytes.
Basic CSF analysis (e.g., cell numbers, protein) yielded an increase of CSF leukocyte counts and protein concentration in VE compared with all other patients (Figure S1A). Blood-CSF-barrier disruption (BCBD) was more prevalent in Neuro-COVID and VE patients than in MS and IIH. CSF of MS patients exhibited intrathecal immunoglobulin synthesis, as expected (Filippi et al., 2018) (Figure S1A). Even though basic CSF parameters thus showed disease-consistent changes, they failed to sufficiently explain neurological involvement in COVID-19 infection.
We therefore merged all available scRNA-seq data of Neuro-COVID and control patients (IIH, MS, VE), which then encompassed 80,919 total single-cell transcriptomes with 2,610 ± 504 SEM average cells per sample and 1,027 ± 85 SEM median genes detected per cell (Table S1). This dataset considerably exceeds existing scRNA-seq studies from the CSF (Esaulova et al., 2020; Schafflick et al., 2020). We performed unbiased cell clustering (Figure 1B) and annotated the resulting 16 clusters based on the expression of marker genes (Figure 1C; Table S2).
Three T cell clusters (CD3E, TRAC, TRBC2) were classified into CD4+ T cells (CD4: IL7R), regulatory T cells (Treg: FOXP3, CTLA4, IL2RA) and CD8+ T cells clusters (CD8: CD8A) (Figure 1C). NK cells (NK: NKG7, GNLY) clustered together with γδ T cells (TRDC) (Figure 1C). B lineage clusters separated into plasma cells (plasma: SDC1/CD138, CD38) and naive B cells (naiveBc: CD19, MS4A1/CD20, CD24) (Figure 1C). One cluster with inconclusive lineage assignment showed expression of canonical proliferation-associated cell cycle genes (cycling: CDC45, TPX2, TOP2A) (Figure 1C). We identified four dendritic cell (DC) clusters that represented myeloid DC type I (mDC1: CLEC9A, XCR1, BATF3), type II (mDC2: CLEC10A, CD1C, FCER1A), plasmacytoid DC (pDC: CLEC4C, IRF8, TCF4/E2-2), and mature DC (matDC: IDO1, FSC1, LAMP3) (Figure 1C). Further myeloid-lineage clusters included granulocytes (granulo1/2: S100A8, S100A9) and three monocyte-like clusters (mono1–3) (Figure 1C). The mono1 cluster preferentially expressed CNS-border-associated macrophage genes (mono1: MRC1, LYVE1), and the mono3 cluster expressed known microglia-associated genes (mono3: SPP1, OLFML3, P2RY12, TMEM119) (Figure 1C), which were previously observed in CSF-derived leukocytes (Esaulova et al., 2020). Non-manual cluster annotation supported our cluster labeling (Figure S1B). We thus replicated the known composition and phenotype of hematopoietic lineages in the CSF (Esaulova et al., 2020; Farhadian et al., 2018; Ramesh et al., 2020; Schafflick et al., 2020).
Neuro-COVID Cerebrospinal Fluid Features an Expansion of Dedifferentiated Monocytes
Next, we systematically compared the CSF cluster composition between disease conditions (Figures 1D–1H). To validate our approach, we first compared MS with IIH. In accordance with previous studies in the CSF (Han et al., 2014; Schafflick et al., 2020), MS patients featured a significant increase of naiveBc, plasma, and Treg clusters (Figure S1C). Next, we compared Neuro-COVID with controls. We found a significant expansion of the mono2 cluster in Neuro-COVID compared with IIH (Figure 1F). In contrast, in patients with VE, Treg, cycling, NK, and granulo2 clusters expanded, but the mono2 cluster did not change considerably (Figure 1G). When directly compared with VE, Neuro-COVID accordingly featured a specific increase of the mono2 cluster while Treg, granulo2, and plasma clusters were more abundant in VE (Figure 1H). Neuro-COVID thus induced a specific pattern of leukocyte changes in the CSF - preferentially affecting a subset of monocytes.
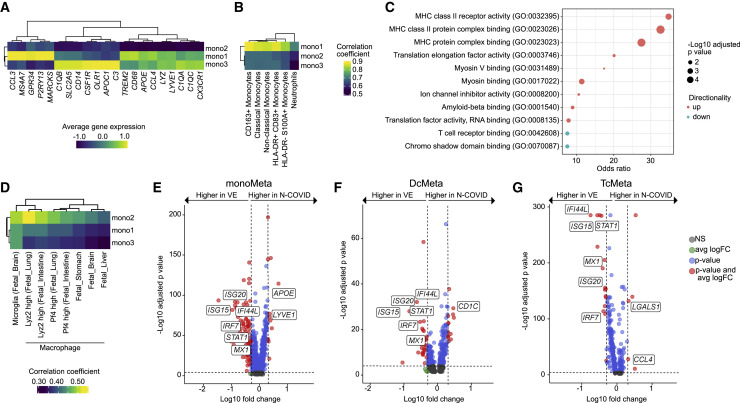
We aimed to better characterize the mono2 cluster. In contrast to the mono1 and mono3 clusters, mono2 exhibited reduction of pan-monocytic markers (CD14, LYZ, CD68) and markers of microglia and border-associated macrophages (e.g., CX3CR1, LYVE1, APOE, TREM2) (Figure 2 A; Table S2). Mono2 also showed an overall lower expression of a previously identified microglia gene set (Sankowski et al., 2019) than the mono1 and mono3 clusters (Figure S1D; Table S2). We compared our monocytic clusters in CSF with previously published COVID-19-associated monocytes in blood (Schulte-Schrepping et al., 2020) but did not identify overlap with a specific subset (Figure 2B). Compared with all other clusters, mono2 featured enhanced antigen-presenting characteristics in pathway enrichment analysis (Figure 2C; Table S2). In addition, mono2 partially resembled developmental macrophages in comparison with data from the Mouse Cell Atlas (Han et al., 2018) (Figure 2D), potentially indicating dedifferentiation. Pseudotime analysis revealed trajectories within monocytic clusters likely originating from the mono2 cluster (Figure S1E). Neuro-COVID thus features monocytes in the CSF with a partially developmental and antigen-presenting phenotype.
Figure 2.
Neuro-COVID Displays a Reduced Interferon Response Compared with Viral Encephalitis
(A) Average gene expression of monocytes, microglia, and border-associated macrophages markers in mono1, mono2, and mono3 clusters.
(B) Correlation coefficients between gene expression of monocytic clusters of this study and of Schulte-Schrepping et al. (Schulte-Schrepping et al., 2020).
(C) Top: enriched GO terms of molecular functions in the mono2 cluster compared with all other clusters. Size encodes the significance and color indicates whether the term was enriched in genes with increased or decreased expression.
(D) Correlation coefficients between gene expression of monocytic clusters of this study and clusters of the Mouse Cell Atlas (Han et al., 2018) containing “fetal” in their cluster annotation.
(E–G) Differentially expressed (DE) genes in cell type meta clusters in N-COVID versus viral encephalitis (VE) patients in the monoMeta (E), DCMeta (F), and TcMeta (G) clusters. See Figure S2 for meta cluster definition. Logarithmic fold change is plotted against negative logarithmic adjusted p value. The horizontal dashed line represents an adjusted p value of 0.0001 and the vertical dashed lines display a common logarithmic fold change of 0.3. Selected transcripts are labeled.
Neuro-COVID Results in a Less Pronounced IFN-Signature than in Viral Encephalitis
Next, we sought to identify Neuro-COVID-specific transcriptional changes. In scRNA-seq, differential expression (DE) analysis tends to perform poorly in clusters with low cell numbers (Wang et al., 2019). We therefore merged clusters into three gross “meta-clusters” of T/NK (TcMeta), DC (DCMeta), and monocytes/granulocyte (monoMeta) cell types (Figure S2A). The B lineage and the cycling clusters were removed because of low total cell numbers. The monoMeta cluster increased expression of some IFN-driven transcripts (IFITM3, IFI27, IFNGR2, IL18), including CCAAT/enhancer-binding proteins (CEBPA, CEBPB) in Neuro-COVID compared with IIH controls (Table S3). The TcMeta cluster also increased expression of IFN-regulated transcripts (PRF1, XCL1, ETS1) when Neuro-COVID samples were compared with IIH controls (Table S3). In a direct DE analysis against VE, however, we found that multiple canonical IFN-signaling transcripts (e.g., STAT1, IRF7, MX1, ISG15) were expressed considerably higher in VE than in Neuro-COVID across all meta-clusters (Figures 2E–2G; Table S3). In Neuro-COVID, CSF leukocytes might thus not elicit a comparably strong antiviral response as in direct VE. The DCMeta and monoMeta clusters instead showed higher expression of homeostatic microglia/border-associated macrophage transcripts (e.g., LYVE1, APOE, CD1C) in Neuro-COVID than in VE (Figures 2E and 2F). In the same comparison, the TcMeta cluster expressed some trafficking (CCL4) and immunosuppressive transcripts (LGALS1) (Figure 2G). We thus identified a Neuro-COVID-specific transcriptional response in CSF leukocytes clearly distinct from conditions with direct viral damage to the CNS.
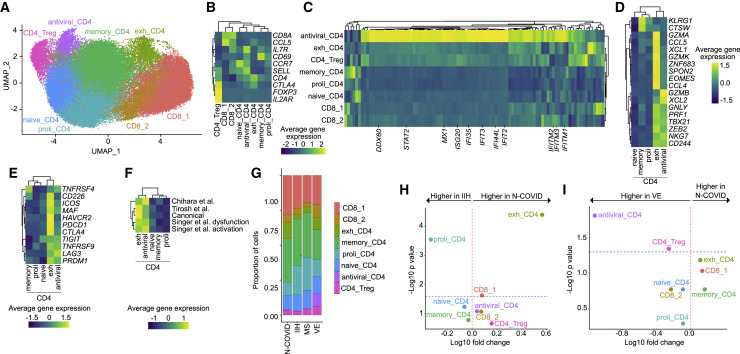
Exhausted CD4+ T cells Are Increased in the Cerebrospinal Fluid in Neuro-COVID
T cell subsets are transcriptionally similar and tend to require targeted analytical approaches (Schafflick et al., 2020). We therefore subclustered all T cells (Figure 3 A). This returned the following subclusters: two CD8+ T cell clusters (CD8_1, CD8_2: CD8A, CCL5), regulatory CD4+ T cells (Treg: CTLA4, FOXP3, IL2RA), naive CD4+ T cells (naive_CD4: CCR7, SELL), proliferating CD4+ T cells (proli_CD4: CCR7, ribosomal genes), and memory-like CD4+ T cells (memory_CD4: IL7R, CD69) (Figure 3B; Table S4). In addition, we identified one CD4+ T cell cluster with strong induction of IFN-stimulated genes (antiviral_CD4: e.g., IFITM3, IFI44L, MX1; Figure 3C; Table S4). A further cluster, which we named exhausted CD4+ T cells (exh_CD4), expressed a partial cytotoxicity signature (e.g., GZMA, GZMK, CCL5, XCL1; Figure 3D) (Patil et al., 2018) and canonical T cell exhaustion transcripts (exh_CD4: PDCD1, ICOS, CTLA4, HAVCR2/TIM3, CD226; Figure 3E). Additional previously published exhaustion signatures (Chihara et al., 2018; Singer et al., 2016; Tirosh et al., 2016) were also expressed by the exh_CD4 cluster (Figure 3F), including previously identified co-inhibitory genes modules (Chihara et al., 2018) transcriptionally controlled by c-MAF (IL10, IL12RB1, PLEKHF1, ALCAM) and Blimp1/PRDM1 (PLEKHO2, MAP3K5, RUNX2) (Figure S2B).
Figure 3.
Exhausted CD4+ T Cells Expand in the CSF in Neuro-COVID
(A) UMAP plot depicting 8 color-coded T cell subclusters from Neuro-COVID (N-COVID; n = 8), 9 idiopathic intracranial hypertension (IIH) (n = 9), multiple sclerosis (MS) (n = 9), and viral encephalitis (VE) (n = 5) patients; in total 61,642 single-cell transcriptomes.
(B) Average gene expression of canonical T cell markers in T cell subclusters.
(C) Average gene expression of interferon-stimulated genes obtained from Wilk et al. (Wilk et al., 2020) in T cell subclusters. The complete list of genes is provided in Table S4.
(D) Average gene expression of cytotoxicity markers in CD4+ clusters obtained from Patil et al. (Patil et al., 2018).
(E) Average gene expression of canonical T cell exhaustion markers in CD4+ clusters.
(F) Average gene expression of T cell exhaustion gene sets from Singer et al. (Singer et al., 2016), Tirosh et al. (Tirosh et al., 2016), Chihara et al. (Chihara et al., 2018), and canonical exhaustion markers in CD4+ T cell clusters. The complete gene sets are provided in Table S4.
(G) Proportions of T cells split by diagnosis.
(H and I) Changes of T cell subcluster abundances in Neuro-COVID (n = 8) versus IIH (n = 9) (H) and Neuro-COVID (n = 8) versus VE (n = 5) (I). Logarithmic fold change of cluster abundance is plotted against negative logarithmic p value (two-sided Wilcoxon’s rank-sum test). The horizontal dashed line represents the significance threshold (p = 0.05).
Abbreviations: proli_CD4, proliferating CD4+ T cells; memory_CD4, memory-like CD4+ T cells; exh_CD4, exhausted CD4+ T cells; CD8, CD8+ T cells; CD4_Treg, regulatory CD4+ T cells.
Human CD4+ T cells are poorly classified by clustering (Schafflick et al., 2020). We therefore applied cell set enrichment analysis (CSEA) to identify cluster-independent compositional changes (Schafflick et al., 2020). T cells resembling IL-10−CD8+ and IL-10+CD8+ cells in murine coronavirus encephalitis (Trandem et al., 2011), and resembling PD-1 expressing T cells, associated with T cell dysfunction (GSE26495), were enriched in Neuro-COVID compared to VE (Table S4).
When abundances between disease conditions were compared, Neuro-COVID patients featured a significant expansion of the exh_CD4 and reduction of the proli_CD4 cluster compared to IIH (Figures 3G and 3H). This indicates that T cell-driven antiviral immunity in the CSF might be inefficient or “exhausted” in Neuro-COVID, although this could be confounded by the time of sampling (Table S1). When Neuro-COVID and VE were directly compared, the antiviral_CD4 and, to a lesser extent, Treg clusters characterized VE (Figure 3I). The relative lack of antiviral IFN-producing CD4+ T cells might suggest an ineffective immune response in the CSF of Neuro-COVID patients but could also reflect differences in study recruitment kinetics. Because VE features a strong antiviral immune response in the CSF, the antiviral response in Neuro-COVID might also be simply weaker, without necessarily being ineffective.
Severely Affected Neuro-COVID Patients Show Curtailed IFN Responses
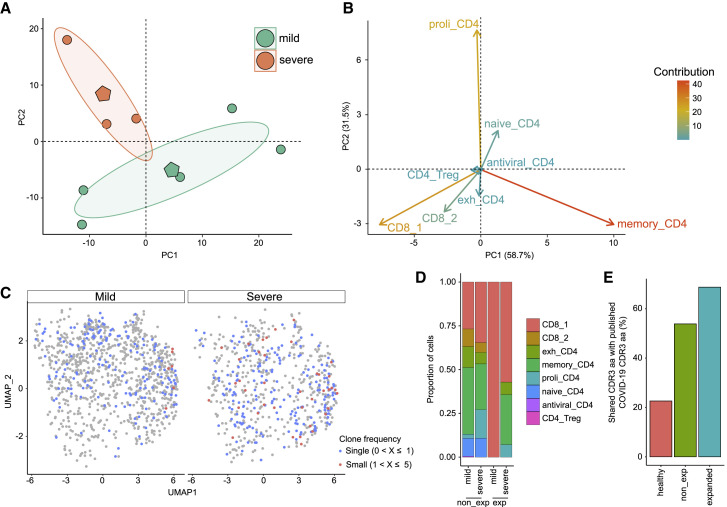
Previous studies reported severity-dependent changes in the blood during pulmonary COVID-19 infection (Lee et al., 2020; Schulte-Schrepping et al., 2020; Silvin et al., 2020; Wilk et al., 2020). We tested for similar changes in Neuro-COVID by categorizing the Neuro-COVID patients based on their neurological signs and symptoms into mild/moderate (n = 5; mean age 69 years) and severe (n = 3; mean age 64 years) cases (Table S1). We then performed principal component analysis (PCA) of the Neuro-COVID cluster abundances, thus plotting their relative scRNA-seq cluster abundance into a two-dimensional space. In contrast to the general clusters (Figure S2C), the T cell subclusters clearly distinguished between mild and severe Neuro-COVID (Figure 4 A). Abundance of the proli_CD4 cluster characterized severely affected Neuro-COVID, while memory_CD4 was a hallmark of mildly affected Neuro-COVID (Figure 4B). In a direct mild versus severe comparison, no cluster proportions passed our significance threshold (Figures S2D–S2G). Severe Neuro-COVID showed a non-significant trend toward increased naiveBc and proli_CD4 (Figures S2E and S2G). Most likely, our study was not sufficiently powered to achieve significance in this direct subgroup comparison. In accordance with an underpowered study size, the increase of BCBD in severe Neuro-COVID reported previously (Alexopoulos et al., 2020) also did not reach significance in our severe versus mild comparison (Figure S2H).
Figure 4.
Broad Clonal T Cell Expansion in Severe Neuro-COVID
(A) Principal component analysis (PCA) of T cell subcluster abundances in Neuro-COVID patients categorized by neurological severity (mild, n = 5; severe, n = 3). Each circle represents one patient, the polygons represent the group means. The ellipses around the group mean represent the confidence regions.
(B) Individual variables of the PCA. The contribution of each variable is color-coded.
(C) UMAP plot of Neuro-COVID (n = 8) T cell subclusters show clone frequency split by neurological disease severity.
(D) Proportions of Neuro-COVID T cells split by clone frequency and neurological disease severity (mild, n = 5; severe, n = 3).
(E) Overlap of complementarity-defining region (CDR) 3 amino acid (aa) sequences of healthy clones from a public repository (Corrie et al., 2018), non-expanded (non_exp), and expanded clones from Neuro-COVID with published CDR3 aa sequences from seven COVID-19 studies retrieved from the same repository (see STAR Methods).
Abbreviations: Dim, dimension; proli_CD4, proliferating CD4+ T cells; memory_CD4, memory-like CD4+ T cells; exh_CD4, exhausted CD4+ T cells; CD8, CD8+ T cells; CD4_Treg, regulatory CD4+ T cells; exp, expanded; non_exp, non-expanded.
We next investigated severity-dependent transcriptional differences in the meta clusters. Expression of several IFN-associated transcripts (MX1, IFNGR1, IRF1) was reduced in the TcMeta and monoMeta clusters in severe compared to mild Neuro-COVID (Table S4). Concurrently, the expression of several class I (HLA-B, HLA-C) and class II (HLA-DRB1, HLA-DQB1, HLA-DQA1) genes was reduced in TcMeta and monoMeta in severe disease (Table S4). In addition, expression of trafficking molecules (CXCR4, ITGB1, ITGA4) was decreased in TcMeta in severe versus mild Neuro-COVID (Table S4). Transcriptionally, we thus found signs of potentially impaired antiviral responses in the CSF of severely affected Neuro-COVID patients.
Severe Neuro-COVID Patients Exhibit a Broad Clonal T Cell Expansion
Finally, we reconstructed single-cell T cell receptor (TCR) sequence information from Neuro-COVID samples (see STAR Methods; Table S2). We successfully identified any α- or β-chain clonotype in 25% of all T cells. In mild Neuro-COVID, expanded clonotypes (defined as ≥2 identified clones) aggregated in the CD8_1 cluster, while severe Neuro-COVID showed expanded clones across multiple T cell clusters (Figures 4C and 4D). In accordance, bronchoalveolar CD8+ T cells had shown more pronounced transcriptional heterogeneity in severe infection than in moderate pulmonary COVID-19 (Liao et al., 2020). We then compared the complementarity-defining region (CDR) 3 amino acid (aa) sequences with published CDR3 aa information from seven COVID-19 studies in a public repository (>180 million clonotypes) (Corrie et al., 2018). We found larger overlap in the expanded (69%) than in the non-expanded (54%) clonotypes with CDR3 aa sequences of COVID-19 patients from the repository. In contrast, healthy controls from COVID-19 studies of the same repository showed only 23% overlap with COVID-19 patients based on their CDR3 aa sequences (Figure 4E). This provides evidence that virus-associated adaptive immunity might be shared between CSF and periphery and across individuals in COVID-19 infection.
Neuro-COVID-Associated Changes Are Partially Maintained when Controlling for Confounders
We systematically compared our observations in CSF with published scRNA-seq studies from leukocytes in the blood and BALF to facilitate putting our findings into context (Table S5).
Next, we aimed to control for potential biases in our study. After tentative removal of one Neuro-COVID patient with comorbid MS, core results, including the expansion of the mono2 and exh_CD4 clusters in Neuro-COVID, remained unchanged (Figures S3A–S3H)—arguing against a relevant distortion of our observations caused by this patient.
Cohorts in our study are imbalanced with regard to age and sex, because MS and IIH primarily manifest in young females (Filippi et al., 2018; Mollan et al., 2019), while severe COVID-19 infection and Neuro-COVID predominate in aged males (Karagiannidis et al., 2020; Mao et al., 2020). To account for this, we performed regression analysis using age, sex, and diagnosis as predictors of cluster proportions. In the Neuro-COVID versus IIH comparison, the increase of the mono2 cluster did not reach significance (p = 0.12) (Table S5). In the same comparison, the increase of the exh_CD4 cluster remained significant (p = 0.029) when controlling for age and sex as covariates (Table S5). Neuro-COVID-associated changes in the CSF are thus partially maintained if controlled for confounders.
Discussion
In this single-cell transcriptomics study, we identified specific immune alterations in the CSF of Neuro-COVID patients featuring an increase of dedifferentiated monocytes and exhausted T cells. We observed an IFN response in Neuro-COVID that was attenuated compared with viral encephalitis. Severe Neuro-COVID exhibited a broad clonal T cell expansion and curtailed IFN response compared with mild Neuro-COVD. Thus, our study potentially indicates a compromised antiviral response and points toward immune-mediated mechanisms contributing to Neuro-COVID. Our observations could in fact represent only the “tip-of-the-iceberg” of Neuro-COVID associated alterations because this study had limited statistical power.
In line with our findings in the CSF, several studies reported an increase of specific monocyte lineage cells in the blood of pulmonary COVID-19 patients which were classified as proinflammatory macrophages (Chua et al., 2020; Liao et al., 2020), HLA-DRlo classical monocytes (Silvin et al., 2020), or CD14+ classical monocytes (Fan et al., 2020; Lee et al., 2020). Similarly, T cell exhaustion has been described in the blood of COVID-19 patients, especially in severe cases (De Biasi et al., 2020; Zheng et al., 2020a, 2020b). Another hallmark of COVID-19 in blood was an IFN-dominated transcriptional response (Huang et al., 2020; Wilk et al., 2020; Zhang et al., 2020), which was reduced in severe COVID-19 (Blanco-Melo et al., 2020; Hadjadj et al., 2020) and compared with influenza virus (Lee et al., 2020). In line with those results, we observed attenuated IFN response in the CSF of severe Neuro-COVID and compared with viral encephalitis. Inversely, many studies reported an expansion of plasma cells and plasmablasts (Bernardes et al., 2020; Zhang et al., 2020; Zhu et al., 2020) that we did not observe in the CSF. Immune alterations may thus be partially shared between compartments and partially compartment specific. Considering the tightly controlled leukocyte composition in CSF differing from blood (Ransohoff and Engelhardt, 2012), synonymous alterations between compartments are notable. We speculate that Neuro-COVID does not simply represent a “side-effect” of pulmonary COVID-19 but displays CSF-specific mechanisms in support of immune-mediated mechanisms in Neuro-COVID.
Exhausted or dysfunctional T cells arise because of repetitive over-stimulation during chronic infection and in the tumor micro-milieu. They are characterized by loss of effector functions, high expression of co-inhibitory receptors, and a specific transcriptional program (Chihara et al., 2018; McLane et al., 2019; Singer et al., 2016; Tirosh et al., 2016). Checkpoint inhibitor treatment can “reinvigorate” exhausted T cells. This treatment has revolutionized the therapy of multiple malignancies (Robert, 2020) but has also been investigated in chronic viral infections (Wykes and Lewin, 2018), including infections with HIV (Kaufmann et al., 2007), influenza A virus (Rutigliano et al., 2014), and notably also in a mouse model of coronavirus (Karnam et al., 2012). Whether this could represent a therapeutic avenue in some instances of Neuro-COVID remains to be investigated, especially by confirming exhaustion in functional assays and understanding immune mechanisms in early versus late stages of Neuro-COVID.
Our study also articulates hypotheses to be investigated in future clinical studies. Because detection of SARS-CoV-2 RNA in the CSF is extremely rare (Neumann et al., 2020; Bellon et al., 2020; Kremer et al., 2020) and the diagnosis of Neuro-COVID often relies on subjective symptoms, one could envision flow cytometric detection of a specific immune profile, e.g., dedifferentiated monocytes and exhausted T cells, in the CSF to substantiate and standardize the definition of Neuro-COVID. Using CSF analysis for the diagnosis of Neuro-COVID would first require testing CSF from patients with COVID-19 infectious without neurological involvement in the future to define the specificity of our observations. Our findings could also gain predictive potential. It could be tested whether CSF analysis at the onset of Neuro-COVID can predict outcome or the severity of incipient Neuro-COVID. Studies with longitudinal clinical follow-up will be required to address this question. Additionally, the effects of immune responses may be stage-specific in Neuro-COVID as observed in the multi-stage kinetics in pulmonary COVID-19 (Mann et al., 2020; Tian et al., 2020).
A previous unpublished study used scRNA-seq to investigate CSF leukocytes in Neuro-COVID, yet with a different design (Song et al., 2020). The authors analyzed CSF compared with blood but did not provide autoimmune or encephalitis controls. Non-inflammatory controls in that study were mostly (8 out of 11 patients, 72%) derived from another preceding study, which could cause systematic bias (Song et al., 2020). In accordance with our results, the authors observed a compartmentalized immune response within the CSF, featuring activation of T cells, interleukin (IL)-12-associated immune cell activation and induction of IFN-stimulated genes (Song et al., 2020). The authors reported an expansion of B cells in the CSF in Neuro-COVID, which did not reach significance in our IIH (0.27%) versus Neuro-COVID (0.56%) comparison (p = 0.59). Patients in the Song et al. study were sampled on average 12.5 days after hospital admission versus 17.25 days in our study. Observations may thus also reflect different stages of COVID-19 infection. Moreover, the authors identified anti-SARS-CoV-2 antibodies in the CSF with widespread cross-reactivity against anti-neural and anti-glia epitopes, while SARS-CoV-2 RNA was consistently undetectable in the CSF (Song et al., 2020). Autoantibodies of unknown relevance had been detected in the CSF in Neuro-COVID in another study (Franke et al., 2020).
In summary, our study delineated local immune mechanisms and, in synergy with other studies, lends support to immune-mediated mechanisms contributing to neurological sequelae of COVID-19 patients. Our findings set the basis for better understanding and potentially diagnosing Neuro-COVID in the future.
Limitations of the Study
There are limitations to our study. CSF cell numbers were considerably lower in Neuro-COVID than in other conditions, and we did not include COVID-19 patients without neurological manifestations as controls because of ethical concerns in performing an invasive lumbar puncture for only scientific reasons in these patients. Moreover, our study lacks patient-matched blood data so that we cannot draw conclusions about CSF-specificity of immune alterations solely based on our data. The time between sample collection and clinical presentation varied among patients and among groups. Our findings may thus be biased, especially because of the known stage-dependent immune alterations of COVID-19 (Mann et al., 2020; Tian et al., 2020). Finally, neurological signs and symptoms in Neuro-COVID might not only depend on the observed CSF changes, but also on systemic factors, such as oxygen or serum cytokine levels.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Neuro-COVID patients | Department of Neurology, University Hospital Essen, Germany | N/A |
| IIH patients as controls | Department of Neurology with Institute of Translational Neurology, University Hospital Münster, Münster, Germany | N/A |
| MS patients as controls | Department of Neurology with Institute of Translational Neurology, University Hospital Münster, Münster, Germany | N/A |
| VE patients as controls | Department of Neurology with Institute of Translational Neurology, University Hospital Münster, Münster, Germany | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| X-Vivo15 media | Lonza | cat#BE02-060F |
| T4 Polynucleotide Kinase | New England Biolabs | cat#M0201S |
| T4 DNA Ligase | New England Biolabs | cat#M0202S |
| RecJF | New England Biolabs | cat#M0264S |
| Lambda Exonuclease | New England Biolabs | cat#M0262S |
| Critical Commercial Assays | ||
| Chromium Next GEM Single Cell 3′ Library & Gel Bead Kit v2 | 10x Genomics | cat#CG00052 |
| Chromium Next GEM Single Cell 3′ Library & Gel Bead Kit v3 | 10x Genomics | cat#CG000185 |
| Chromium Next GEM Single Cell 3′ Library & Gel Bead Kit v3.1 | 10x Genomics | cat#CG000204 |
| Chromium Next GEM Single Cell 5′ Library & Gel Bead Kit v1 | 10x Genomics | cat#CG000186 |
| Chromium Next GEM Single Cell 5′ Library & Gel Bead Kit v1.1 | 10x Genomics | cat#CG009207 |
| Chromium Single Cell VDJ Enrichment Kit, Human T Cell | 10x Genomics | cat#CG000086 |
| AMPure XP beads | Beckman Coulter | cat#A63880 |
| MagNA Pure 96 DNA and Viral NA Small Volume Kit | Roche | cat#06543588001 |
| MagNA Pure 96 DNA and Viral NA Large Volume Kit | Roche | cat#06374891001 |
| RealStar SARS-CoV-2 RT-PCR Kit 1.0 | Altona Diagnostics | cat#821015 |
| anti-SARS-CoV-2 IgG ELISA | Euroimmun | cat#EI2606-9601G |
| Bio-Plex Pro Human Cytokine IL-1α Singleplex Set | Bio-Rad | cat#171B6001M |
| Bio-Plex Pro Human Cytokine TNF-α Singleplex Set | Bio-Rad | cat#171B5026M |
| Bio-Plex Pro Human Cytokine IL-6 Singleplex Set | Bio-Rad | cat#171B5006M |
| Bio-Plex Pro Human Cytokine IL-8/CXCL8 Singleplex Set | Bio-Rad | cat#171B5008M |
| RealStar HHV6 PCR Kit 1.0 | Altona Diagnostics | cat#311013 |
| RealStar HSV PCR Kit 1.0 | Altona Diagnostics | cat#061013 |
| RealStar VZV PCR Kit 1.0 | Altona Diagnostics | cat#071003 |
| HSV 1 HSV2 VZV R-gene Kit | Biomerieux | cat#69-004B |
| Artus CMV QS-RGQ Kit | QIAGEN | cat#4503363 |
| Artus EBV QS-RGQ Kit | QIAGEN | cat#4501363 |
| SERION ELISA classic FSME/TBE IgG | Virion/Serion | cat#ESR112G |
| SERION ELISA classic FSME/TBE IgM | Virion/Serion | cat#ESR112M |
| Deposited Data | ||
| scRNA-seq from patients with Neuro-COVID, multiple sclerosis, idiopathic intracranial hypertension and viral encephalitis, see Table S1 | This Paper | GSE163005 |
| scTCR from patients with Neuro-COVID, multiple sclerosis, idiopathic intracranial hypertension and viral encephalitis, see Table S1 | This Paper | GSE163005 |
| interactive version of the entire dataset | This Paper | http://covid.mheming.de/ |
| Supplemental Tables | Mendeley Dataset | https://data.mendeley.com/datasets/5mt97xcyyw/2 |
| Software and Algorithms | ||
| R v4.0 | R Core Team | https://www.r-project.org |
| R factoextra package v1.0 | Alboukadel Kassambara and Fabian Mundt | https://github.com/kassambara/factoextra |
| R EnhancedVolcano package v1.6 | Kevin Blighe, Sharmila Rana and Myles Lewis | https://doi.org/10.18129/B9.bioc.EnhancedVolcano |
| R scRepertoire package v1.2 | Nick Borcherding | http://www.bioconductor.org/packages/release/bioc/html/scRepertoire.html |
| R ggpubr package v4.0 | Alboukadel Kassambara | https://github.com/cran/ggpubr |
| R rstatix package v0.6 | Alboukadel Kassambara | https://github.com/kassambara/rstatix |
| R ggplot2 package v3.3 | Hadley Wickham | https://www.rdocumentation.org/packages/ggplot2 |
| R pheatmap package v1.0 | Raivo Kolde | https://www.rdocumentation.org/packages/pheatmap/versions/1.0.10/topics/pheatmap |
| R enrichR package v2.1 | Wajid Jawaid | https://www.rdocumentation.org/packages/enrichR/versions/2.1 |
| R Seurat package v3.2 | Satijalab | https://github.com/satijalab/seurat |
| R harmony package v1.0 | Ilya Korsunsky, Nghia Millard, Jean Fan, Kamil Slowikowski and Soumya Raychaudhuri | https://github.com/immunogenomics/harmony |
| R clustifyr package v1.1 | Rui Fu, Austin Gillen, Ryan M. Sheridan, Chengzhe Tian, Michelle Daya, Yue Hao, Jay R. Hesselberth and Kent A. Riemondy | https://rnabioco.github.io/clustifyr/ |
| R slingshot package v1.6 | Kelly Street, Davide Risso, Russell B. Fletcher, Diya Das and John Ngai, Nir Yosef, Elizabeth Purdom and Sandrine Dudoit | https://doi.org/10.18129/B9.bioc.slingshot |
| cellranger v3.1.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| AIRR Data Commons API | AIRR Community | https://docs.airr-community.org/en/stable/api/adc_api.html |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gerd Meyer zu Hörste (gerd.meyerzuhoerste@ukmuenster.de).
Materials Availability
All unique reagents generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
Raw sequencing data are available in the Gene Expression Omnibus (GEO) repository (GSE163005). An interactive version of the entire dataset using cerebroApp (Hillje et al., 2020) is available at: http://covid.mheming.de. We followed the official tutorial of the packages listed, no custom code was generated. Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/5mt97xcyyw.2.
Experimental Model and Subject Details
Neuro-COVID Subjects
Cerebrospinal fluid (CSF) was collected from eight patients classified as Neuro-COVID admitted to the University Hospital Essen, Germany between May and June 2020. All patients were tested positive for SARS-CoV-2 by reverse transcription polymerase chain reaction (PCR) of nasopharyngeal swabs and developed neurological signs/symptoms between 10 days prior to and 20 days after the diagnostic PCR was performed (Table S1). PCR for SARS-CoV-2 in CSF was performed in all patients and was negative in all patients. Seven patients were male and the age ranged from 53 to 82 years (Table S1). Based on pulmonary and systemic signs/symptoms, disease severity was classified into 3 categories as described (Buonsenso et al., 2020).
-
1
Mild disease: asymptomatic infection or mild respiratory signs/symptoms (e.g., fever, sore throat, pharyngeal congestion) without any radiographic abnormalities and no septic presentation
-
2
Moderate disease: clinical signs of mild pneumonia (e.g., fever, dry or productive cough, abnormal breath sounds on auscultation) without signs of hypoxemia (e.g., shortness of breath)
-
3
Severe disease: mild or moderate disease and signs of disease progression (e.g., respiratory failure requiring mechanical ventilation, septic shock or organ dysfunction requiring intensive care unit-monitoring)
The severity of neurological signs/symptoms in COVID-19 infected patients was classified as follows: Neuro-COVID severity 1 was defined by mild signs/symptoms (e.g., headache, dizziness, anosmia, ageusia), severity 2 by moderate signs/symptoms (e.g., fatigue, mono/para/quadriparesis), and severity 3 by severe neurological manifestations (e.g., seizures, stroke, cognitive impairment or muscle weakness) (Fotuhi et al., 2020). To account for the relatively small sample size, we merged Neuro-COVID patients in stage 1 and 2 and refer to these patients as ‘mild’ Neuro-COVID throughout the manuscript. Patients in stage 3 are termed ‘severe’ Neuro-COVID. None of these patients had received any immunomodulatory treatment previously (Table S1).
Control Subjects
In total, we included data from 20 control patients in our study (Table S1). All control patients were treated in inpatient or outpatient clinics of the Department of Neurology with Institute of Translational Neurology at the University Clinic Münster, Germany between 2017 and 2020. Out of these 20 control patient data, 8 had been published previously (Schafflick et al., 2020). Previously published scRNA-seq data from controls with idiopathic intracranial hypertension (IIH) (n = 4) and relapsing-remitting multiple sclerosis (MS) (n = 4) in active relapse were used (Schafflick et al., 2020). Additionally, we recruited five new treatment-naive patients with relapsing remitting (RR)MS in active relapse leading to the first diagnosis and IIH, respectively, and 5 patients with VE (Table S1). The detailed inclusion and exclusion criteria have been described previously (Schafflick et al., 2020). Briefly, all MS patients were included at first relapse leading to the diagnosis of disease, met the revised McDonald criteria (Thompson et al., 2018) and were treatment-naive. An extensive diagnostic workup was performed in order to exclude differential diagnoses (Schafflick et al., 2020). Patients with a questionable diagnosis, secondary progressive (SP)MS and any immunomodulatory therapy were excluded (Schafflick et al., 2020). The Expanded Disability Status Scale (EDSS) was used to assess disease severity (Kurtzke, 1983).
Patients with IIH were diagnosed based on the diagnostic criteria of IIH (Friedman and Jacobson, 2002). These included signs/symptoms of an increased intracranial pressure, elevated opening pressure (above 200 mmH2O) on lumbar puncture, normal routine CSF studies and no structural cause of intracranial hypertension. Patients with VE were included, if they showed clinical signs of encephalitis (e.g., headache, seizure, psychiatric symptoms, altered mental status and focal neurologic abnormalities) and alterations of the CSF that were compatible with the diagnosis (Table S1). Specific viruses (Herpes simplex virus-1/2 (HSV-1/HSV-2), Cytomegalovirus (CMV), Ebstein-Barr virus (EBV), Varicella zoster virus (VZV), Human herpesvirus-6 (HHV-6) and Enterovirus were tested by PCR in all VE patients. HSV-1 and VZV could be detected in the CSF of one patient, respectively. Screening for serum antibodies to the Tick-borne encephalitis (TBE) virus was performed by ELISA and was positive in one patient. CSF and serum were tested by Treponema pallidum hemagglutination assay to exclude Treponema pallidum infection. ELISA and immunoblotting were performed to detect Borrelia burgdorferi. Screening for the following antibodies was performed using immunofluorescence assays (IFA) and immunoblotting to exclude autoimmune encephalitis: Anti-Hu, Ri, ANNA-3, Yo, TR/DNER, Ma/Ta, GAD65, Amphiphysin, NMDA receptor, GABA-b receptor, GABA-a receptor, LGI1, CASPR2, ZIC4, DPPX, glycine receptor, mGluR1, mGluR5, RhoGTPase activating protein, Recoverin, GluDR2, Flotilin, Homer3, Neurochondrin, AMPA receptor.
Exclusion Criteria
Exclusion criteria for all control patients were: pregnancy and breastfeeding, patients under 18 years, severe concomitant infectious or autoimmune disease and a red blood cell count over 200/μl in CSF.
Methods Details
Ethics Statements
All patients gave written informed consent to sample collection and data analysis. The study was approved by the local ethics committee in Essen (Ethics Committee of the University Duesburg-Essen; reference number 20-284-BO) and in Münster, Germany (Ethics Committee of the Board of Physicians of the Region Westfalen-Lippe and of the Westfälische Wilhelms-University Münster; reference number 2015-522-f-S).
Diagnostic Procedures and CSF Analysis
Lumbar puncture was performed between day 0 and day 40 (Neuro-COVID patients), on day 0 or 1 (MS patients), between day 0 and 7 (IIH patients) and between day 0 and 7 (patients with VE) after admission to the hospital. CSF was processed within an h to ensure optimal sample quality. CSF cells were counted manually in a Fuchs-Rosenthal chamber, total protein and intrathecal immunoglobulin concentrations were assessed by nephelometry. Concentration of protein and immunoglobulins in serum and CSF were compared and a Reiber scheme was created to evaluate the integrity of the blood-CSF-barrier (BCBD), quantified by the ratio between CSF albumin and serum albumin. Oligoclonal bands (OCB) were detected by isoelectric focusing and silver nitrate staining. Electroencephalography, using a standard 10-20-EEG system, was recorded in seven Neuro-COVID cases and abnormalities (generalized or focal slowing and epileptiform activity) were classified by a board certified neurologist (Table S1). Imaging studies (either CT or MRI head) were performed in 24 out of 27 included patients to identify structural abnormalities (Table S1).
Single-Cell RNA Sequencing
CSF was processed for scRNA-seq as described previously (Schafflick et al., 2020). Briefly, CSF samples were collected into round bottom polypropylene tubes and then centrifuged for 10 min at 300 x g. The CSF supernatant was removed, and CSF cells were resuspended in 5 mL of X-Vivo15 media (Lonza) and stored at 4°C. The samples were then transported to the laboratory of the Department of Neurology with Institute of Translational Neurology at the University Clinic Münster, Germany for scRNA-seq. Transport time of samples collected in Münster was 10-30 min. Transport time of samples collected in Essen was < 60 min. Cell suspensions were then centrifuged again for 5 min at 400 x g and then resuspended in 40 μL of X-Vivo15 media (Lonza). Out of this volume, 6 μL were used for cell counting and the remaining volume was entirely used for scRNA-seq.
Single-cell suspensions were then loaded onto the Chromium Single Cell Controller using the Chromium Single Cell 3′ or 5′ Library & Gel Bead Kit (both from 10X Genomics) with different versions of chemistry (details see Table S1). Sample processing and library preparation was performed according to manufacturer’s instructions using AMPure beads (Beckman Coulter). Single-cell immune receptor repertoires were reconstructed from Neuro-COVID samples (Table S1). Sequencing was carried out commercially on an Illumina Nextseq500 with either 26-8-0-57 or 26-8-0-132 read setup, or an Illumina Novaseq 6000 with 150-8-8-150 read setup (for details see Table S1). Processing of sequencing data was performed with the cellranger pipeline v3.1.0 (10X Genomics) according to the manufacturer’s instructions. Briefly, subsequent read alignments and transcript counting was done individually for each sample using the cellranger count pipeline with standard parameters. To detect SARS-CoV-2 positive CFS cells, the cellranger count pipeline was additionally performed with a customized reference genome constructed by cellranger mkref that included the SARS-CoV-2 genome (NCBI Reference Sequence: NC_045512.2) added to the pre-built human GRCh38 reference. The cellranger aggr pipeline was employed to aggregate samples with mapping normalization to leave each sample with similar sequencing depth per cell. The total and per sample cell numbers are listed in Table S1.
Single-Cell TCR-Sequencing
Four Neuro-COVID samples (Table S1) were processed with the Chromium Single Cell 5′ Library & Gel Bead Kit and barcoded, full-length VDJ segments from T cells were enriched from first-strand cDNA via PCR amplification with the Chromium Single Cell VDJ Enrichment Kit, Human T Cell (from 10X Genomics) following the manufacturer’s instructions. Further library preparation was performed using the Chromium Single Cell 5′ Library Construction Kit (10X Genomics). Sequencing was carried out on an Illumina Nextseq550 with 26-8-0-132 read setup (for details see Table S1). Raw data was processed by the cellranger pipeline v3.1 (10X Genomics) according to the manufacturer’s instructions.
Three Neuro-COVID samples (Table S1) were processed with the Chromium Single Cell 3′ Library & Gel Bead Kit (10x Genomics) and we developed a novel method to sequence antigen receptor information from 3′ scRNA-seq libraries. The method allows shortening the constant region of antigen receptors during enrichment, while maintaining their cell barcode and unique molecular identifier (UMI) information attached to the 3′ of the cDNA molecules. In summary, the method involves self-circulating the cDNA library, enriching the VDJ region and re-linearizing. Each step maintains the VDJ region together with the cell barcode and UMI in the same molecule. Primer Poly A is used as a 5′ race primer and two T cell receptor (TCR) reverse primer pools were synthesized from Eurofins according to sequences of reverse primers of 10x Genomics Chromium Single Cell V(D)J Enrichment Kit, Human T Cell, PN-1000005 (Sequences and concentration see Table S2). All primers were synthesized by Eurofins Genomics; the primers are listed in Table S2. For circularization, cDNA generated from 10x Genomics Chromium Single Cell V(D)J Enrichment Kit, Human T Cell, PN-1000005v3 were end-phosphated with the T4 Polynucleotide Kinase (New England Biolabs), and purified by 0.6x Ampure XP beads (Beckman Coulter). Then 1,000 units of T4 DNA Ligase (New England Biolabs) were added to self-circularize the phosphated cDNA at 16°C for 16 h. Subsequently, 0.7x Ampure XP beads were used to purify. Remaining linear DNA was digested by 0.9 units/μl RecJf and 0.1 units/μl Lambda Exonuclease (both from New England Biolabs). Circularized cDNA libraries were purified by 0.7x Ampure XP beads. A 5′ race nested PCR enrichment was performed to enrich the TCR variable region. A size selection was done by 0.5x - 0.8 x Ampure XP beads. Then, the nested PCR products were phosphated, circulated and linear digested again as above. A PCR with primers read1 and TSO was used to re-linearize the circulated library. PCR products were purified by 0.5x - 0.8 x Ampure XP beads (Beckman Coulter) and libraries were prepared from them using the Chromium Single Cell 3′ Library Kit v3 (10x Genomics). Sequencing was carried out on an Illumina Nextseq550 with 26-8-0-132 read setup (for details see Table S1). In order to analyze them with cellranger pipelines from 10x Genomics, we converted read 2 into reverse-complement and replaced the cell barcode white list of the Chromium Next GEM Single Cell 5′ Library & Gel Bead Kit v1.1 with the one of Chromium Next GEM Single Cell 3′ Library & Gel Bead Kit v3 in the folder /cellranger-3.1.0/cellranger-cs/3.1.0/lib/python/cellranger/barcodes/ of the cellranger pipeline. Processing of modified sequencing data was performed with the modified cellranger VDJ pipeline v3.1.0 (10X Genomics) according to the manufacturer’s instructions.
Data Analysis of Single-Cell RNA Sequencing
Downstream analysis of scRNA-seq datasets was performed using Seurat v3.2 (Stuart et al., 2019) and R v4.0 as described previously (Wolbert et al., 2020). Each sample was filtered individually to remove cell doublets and low-quality cells with few genes (< 200) high genes (> 1200-6000) or high mitochondrial percentages (5%–20%). To account for technical noise, data were normalized using logarithmic transformation normalization, highly variable genes were identified, and data were scaled taking into account mitochondrial percentages. Principal component analysis (PCA) was performed using highly variable genes. Based on the elbow plot, we used the first 40 principal components for further analysis. To cluster the cells, we used the ‘FindNeighbors’ (based on KNN graphs) and the ‘FindCluster’ (based on Louvain method; resolution 0.3) functions in Seurat. Batch effects were taken into account using Harmony (Korsunsky et al., 2019), which project cells into a shared embedding. To visualize cells in a two-dimensional dataset we performed UMAP. We annotated clusters based on known marker genes or automated with the clustifyr (Fu et al., 2020) package based on Monaco et al. (Monaco et al., 2019). One cluster with 921 cells that showed high percentage of mitochondrial transcripts (4.9%; mean mitochondrial percentage of all clusters 2.5%) and low UMI counts (864; mean UMI count of all clusters 5403) was removed because it most likely represents dying cells. We subsetted the T cell clusters and re-clustered these subclusters with a higher resolution (0.5). DotPlots and FeaturePlots were created using the internal visualization functions of Seurat. Volcano plots of cell cluster abundance were created using ggplot2 v3.3 using unpaired two-sided t test in R. Heatmaps were created with pheatmap v1.0 and data were clustered using complete linkage with Euclidean distance measure. Significant cluster abundance changes were adjusted for age and sex employing a linear regression analysis in R and including age and size as predictors. Comparisons of our scRNA-seq data with the Mouse Cell Atlas (MCA) (Han et al., 2018) and Schrepping et al. (Schulte-Schrepping et al., 2020) were carried out using clustifyr and were based on the official cluster annotations. PCA of cluster abundances was performed and visualized with the R package factoextra.
Differentially expressed (DE) genes were determined with the FindMarker function in Seurat with the Wilcoxon rank sum test with a threshold of adjusted p value (based on Bonferroni correction) of 0.05 and a minimum of 10 cells per cluster. Volcano plots of DE genes were created with the R package EnhancedVolcano. To perform gene set enrichment analysis, we used the Enrichr tool (Kuleshov et al., 2016). and the GO Molecular Function 2018 reference dataset.
We performed pseudotime of the mono1, mono2 and mono3 clusters using Slingshot (Street et al., 2018). Slingshot was performed with the UMAP embeddings and cluster annotations of the Seurat analysis (see above).
Single-Cell Immune Repertoire Analysis
Single-cell T cell receptor sequencing (scTCR) data were analyzed using the R package scRepertoire v1.2 (Borcherding et al., 2020) following the official vignette. Clonotypes were removed if any cell barcode had more than 2 immune receptor chains. scTCR data were merged with scRNA-seq data of the T cell clusters only based on the cell barcodes. TCR clones that matched barcodes of cells that were not located in T cell clusters (based on our previous annotations using Seurat) were removed. TCR were categorized into expanded (defined as ≥2 identified clones) and non-expanded clones (clone frequency 1). Published immune repertoire data of COVID-19 were retrieved from iReceptor (Corrie et al., 2018) using the AIRR Data Commons API. Repertoires with diagnosis ID DOID:0080600 (COVID-19) were queried from http://covid19-1.ireceptor.org/airr/v1/ and http://covid19-2.ireceptor.org/airr/v1/ comprising 7 studies at the time of access: Nielsen et al. (Nielsen et al., 2020), Minervina et al. (Minervina et al., 2020), Galson et al. (Galson et al., 2020), Liao et al. (Liao et al., 2020), Schultheiß et al. (Schultheiß et al., 2020), Shomuradova et al. (Shomuradova et al., 2020) and Alsoussi et al. (Alsoussi et al., 2020). In a next step, CDR3 amino acid (aa) sequences were retrieved from these patients resulting in 181,969,096 sequences. We determined the overlap of CDR3 aa sequences from our Neuro-COVID clones with these sequences. As a negative control we retrieved repertoires by filtering for “Control (Healthy)” study group description from http://covid19-1.ireceptor.org/airr/v1/ and http://covid19-2.ireceptor.org/airr/v1/. We then downloaded all CDR3 aa sequences from these patients resulting in 2,156,752 sequences. Finally, we calculated the overlap of CDR3 aa sequences from these healthy controls with those of COVID-19 patients.
Cell Set Enrichment Analysis (CSEA)
CSEA was performed as previously described (Schafflick et al., 2020). Briefly, we used the gene-cell expression matrix, labels for the input cells, and the gene set of interest as input for CSEA. Using the genes in the specified signature, CSEA computed the score for each cell by dividing the log gene expression values by the mean expression across all cells, taking the sum across all genes, and normalizing the resulting values between 0 and 1. Then, the cells were rank-ordered based on the computed score, and the Enrichment Score (ES) was computed using the ranked cell scores as previously described (Schafflick et al., 2020). To determine the p value of the CSEA ES, we selected a new set of genes and re-compute the cell scores 100 times and then computed the probability that the maximum ES computed with the original gene signature was greater than the ES computed with the permuted gene sets. The random gene set was selected by finding the same number of genes as in the original gene set and finding the top 100 genes that had the closest mean expression to each gene in the original signature set and then randomly sampling one of those 100 genes. We also computed the p values for the enrichment in IIH as the negative enrichment p values in order to select for gene signatures that were only enriched in Neuro-COVID cells not IIH cells. We corrected for multiple testing using the Benjamini-Hochberg procedure to generate the corrected p values for both positive and negative enrichment. We filtered the result based on the following criteria: the corrected p value of the true signature set is smaller than 0.05 and the corrected p value of the control signature set is > 0.05.
Quantification and Statistical Analysis
Statistical analysis was performed using R v4.0. Boxplots were created with the ggpubr package. When comparing one categorical variable with two groups (severe versus mild Neuro-COVID) with one continuous parameter (e.g., cells, protein, IL-6, IL-8) we used Wilcoxon’s rank-sum test (two-sided). When comparing one categorical parameter with multiple groups (Neuro-COVID, MS, IIH, VE) with a continuous parameter, we performed Kruskal-Wallis with Dunn test as a post hoc test (rstatix package in R). To compare a categorical parameter with two groups (severe versus mild Neuro-COVID) with another categorical parameter with two groups (OCB, BBBD) we used Fisher’s exact test (two-sided). In the case of one categorical parameter with multiple groups (Neuro-COVID, MS, IIH, VE) with another categorial, we used the Freeman-Halton extension of Fisher’s exact test. For pairwise comparisons we performed Fisher’s exact test (two-sided) with multiple testing adjustments using Benjamini-Hochberg’s method (rstatix package). The significance level alpha was set at 0.05.
Systematic Meta-Analysis
For the meta-analysis-like literature review, we systematically searched the databases PubMed, MedRxiv, and BioRxiv until December 2020 using the MeSH terms “Coronavirus,” “Coronavirus Infections/virology,” “Coronavirus Infections/immunology,” “Coronavirus Infections/pathology,” “RNA-Seq/methods,” “Sequence Analysis,” “RNA/ methods,” “Single-Cell Analysis,” and “Sequencing.” We included original research articles assessing the impact of COVID-19 on immune cell populations by analyzing samples obtained from SARS-CoV-2 positive patients with genome-wide single-cell transcriptomics techniques, such as 10x Genomics or drop-seq.
Additional Resources
In addition to the deposition of the raw sequencing data on GEO, we provide an interactive platform of the entire dataset using cerebroApp (Hillje et al., 2020) at http://covid.mheming.de/. The platform includes analysis information, shows key analytic results, such as UMAP visualization, marker genes and cluster abundances and allows interactive exploration of the results based on custom genes and gene sets.
Acknowledgments
We are deeply indebted to the patients for their participation. We thank Carolin Lewe and Marc Hennies for excellent technical assistance and Dr. Chenling Xu for help with data analysis. G.M.z.H. was supported by grants from the Deutsche Forschungsgemeinschaft (DFG; grant number ME4050/4-1) and DFG grant ME4050/8-1, under the frame of E-Rare-3, the ERA-Net for Research on Rare Diseases; and from the Ministerium für Innovation, Wissenschaft und Forschung (MIWF) des Landes Nordrhein-Westfalen. This study was supported by the DFG through Sonderforschungsbereich Collaborative Research Center Transregio (CRC-TR) 128, projects A09 (to H.W.) and B03 (to G.M.z.H.). The study was also supported by the Foundation University Hospital Essen (M.S.). H.W. received project-related research support from Biogen. O.W. was supported by an unrestricted grant of the Rudolf-Ackermann-Stiftung (Stiftung für Klinische Infektiologie). The graphical abstract contains images from Servier Medical Art (https://smart.servier.com, licensed under a Create Commons Attribution 3.0 Unported License).
Author Contributions
G.M.z.H. and M.S. conceived and supervised the study. A.K.M., M.F., F.S., O.W., T.B., U.D., and M.S. recruited COVID-19 patients. S.R., A.-L.B., and M. Hartlehnert performed data acquisition. X.L. and I.-N.L. carried out sequencing experiments. M. Heming., A.S., N.Y., and X.L. performed data analysis. C.K. and H.W. co-supervised the study. G.M.z.H. and M. Heming. wrote the manuscript. All authors critically revised the manuscript.
Declaration of Interests
A patent application covering the method for reconstructing TCR information from 3′ libraries has been requested under the filing number LU101949 by X.L. and G.M.z.H. The remaining authors declare no competing interests.
Published: December 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.12.011.
Supplemental Information
Sheet 1 named patient info: Patient characteristics. Ab - antibody; AF - atrial fibrillation; BCBD - blood-CSF-barrier dysfunction; BPH: benign prostatic hyperplasia; CAD: coronary artery disease; C - COVID-19; CIM/CIP - critical illness myopathy/polyneuropathy; cn - cranial nerve; COPD - Chronic obstructive pulmonary disease; CRC - colorectal cancer; CRP - C-reactive protein; CTS - Carpal tunnel syndrome; EDSS - Expanded Disability Status Scale; GERD - gastresophageal reflux disease; Ig - immunoglobulin; IIH - intracranial hypertension; IL - interleukin; LP - lumbar puncture; MS - multiple sclerosis; NSCLC - non-small cell lung cancer; OCB - oligoclonal band; OSA - obstructive sleep apnea; PAD - peripheral arterial disease; PCT - procalcitonin; PNP - polyneuropathy; S/p - status post; RBC - red blood cell; TBI - traumatic brain injury; TIA - transient ischemic attack; VE - viral encephalitis; vvECMO - veno-venous extracorporeal membrane oxygenation; WBC - white blood cell count; VZV - Varicella zoster virus; HSV - Herpes simplex virus; Sheet 2 named scRNA-seq: Technical information on single-cell RNA sequencing. Please note that the columns ‘cells’ and ‘median genes per cell’ refer to the raw cellranger output, while the columns ‘final cells’ and ‘final median genes per cell’ refer to the final Seurat object after filtering low-quality cells and removing the low quality-cluster (see Methods). Sheet 3 named scTCR-seq: Technical information on single-cell T cell receptor sequencing. TCR - T cell receptor sequencing
Sheet 1 named Marker genes: Top marker genes of all clusters. Average logFC threshold was set to 0.25 and minimal percentage of expressing genes in either of the two populations to 0.25. All genes with an adjusted p value above 0.05 were removed. Only genes with a positive average log fold change were retained. avg_logFC - natural log fold change of the average expression between the corresponding cluster and all remaining clusters; pct.1 - percentage of cells with the gene detected in the corresponding cluster pct.2 - percentage of cells with the gene detected in all remaining clusters; p_val_adj - adjusted p value (based on Bonferroni method). Sheet 2 named DE mono2 versus mono1 and mono3: Differentially expressed genes of mono2 versus mono1 and mono3. Average logFC threshold was set to 0.1 and minimal percentage of expressing genes in either of the two populations to 0.1. All genes with an adjusted p value above 0.05 were removed. A positive average logFC value shows a higher expression in the mono2 cluster. All mitochondrial and ribosomal genes were removed. avg_logFC - natural log fold change of the average expression between mono2 and mono1/3; pct.1 - percentage of cells with the gene detected mono2; pct.2 - percentage of cells with the gene detected in mono1 and mono3 clusters; p_val_adj - adjusted p value (based on Bonferroni method). Sheet 3 named TCR Primer: Technical information of TCR sequencing. TCR - T cell receptor; R - a purine base (arginine (A) or guanine (G); Y - a pyrimidine base (cytosine (C) or thymidine (T); B - a base of either C, G or T; 4 step PCR was used in TCR Repertoire enrichment. 2 annealing steps (63°C and 57°C) were used due to the annealing temperatures’ difference between PolyA primer and TCR rev primer. Temperature was defined with degree Celsius (°C) and duration time in hh:mm:ss. Sheet 4 named Microglia marker: Microglia markers from Sankowski et al. (Sankowski et al., 2019) used for the gene score feature plot (Figure S1D). Sheet 5 named Enrichment analysis of DE genes: Enrichment analysis of significant DE genes of mono2 cluster using EnrichR tool in GO Molecular Function 2018 dataset. Mitochondrial and ribosomal DE genes were removed in advance.
Sheets 1-6: DE genes in Neuro-COVID versus IIH (sheet 1-3) and Neuro-COVID versus VE (sheet 4-6) clusters in TcMeta (sheet 1 & 4), DCMeta (sheet 2 & 5) and monoMeta (sheet 3 & 6) were identified with the FindMarkers function in Seurat. Average logFC threshold was set to 0.1 and minimal percentage of expressing genes in either of the two populations to 0.1. All genes with an adjusted p value above 0.05 were removed. A positive average logFC value shows a higher expression in Neuro-COVID, a negative average logFC value shows a higher expression in IIH / VE. All mitochondrial and ribosomal genes were removed. avg_logFC - natural log fold change of the average expression between the corresponding cluster in Neuro-COVID and the corresponding cluster in IIH / VE; pct.1 - percentage of cells with the gene detected in the corresponding cluster in Neuro-COVID; pct.2 - percentage of cells with the gene detected in the corresponding cluster in IIH / VE; p_val_adj - adjusted p value (based on Bonferroni method).
Sheet 1 named Marker genes T subclusters: Marker genes of the T cell subclusters. Average logFC threshold was set to 0.1 and minimal percentage of expressing genes in either of the two populations to 0.1. All genes with an adjusted p value above 0.05 were removed. Only genes with a positive average log fold change were retained. avg_logFC - natural log fold change of the average expression between the corresponding cluster and all remaining clusters; pct.1 - percentage of cells with the gene detected in the corresponding cluster; pct.2 - percentage of cells with the gene detected in all remaining clusters; p_val_adj - adjusted p value (based on Bonferroni method). Sheet 2 named Marker genes exhaustion and ISG: T cell exhaustion and interferon-stimulated gene (ISG) signatures. Interferon-stimulated genes (ISG) from Wilk et al. (Wilk et al., 2020) that were used for the ISG signature heatmap in Figure 3C and markers of T cell exhaustion from Singer et al. (Singer et al., 2016) (Dysfunction and Activation module), Chihara et al. (Chihara et al., 2018) and Tirosh et al. (Tirosh et al., 2016) that were used for the T cell exhaustion signature in Figure 3E and Figure S2. Sheet 3-4: CSEA Neuro-COVID versus IIH (sheet 3) and Neuro-COVID versus VE (sheet 4) in CD4+ T cells. Summary of the CSEA enrichment score, the total number of cells in the leading edge, which drive the enrichment of the specific signature, the enrichment in the leading edge that demonstrates how enriched the leading edge is for Neuro-COVID cells compared to the total cell population, the corrected p value, which is the p value for positive enrichment (Neuro-COVID versus IIH/VE) and the negative p value, which is the corrected p value for negative enrichment (IIH/VE versus Neuro-COVID). Sheet 5-7: DE genes in severe versus mild Neuro-COVID in TcMeta (sheet 5), DCMeta (sheet 6) and monoMeta (sheet 7) clusters. Average logFC threshold was set to 0.1 and minimal percentage of expressing genes in either of the two populations to 0.1. All genes with an adjusted p value above 0.05 were removed. A positive average logFC value shows a higher expression in severe Neuro-COVID, a negative average logFC value shows a higher expression in mild Neuro-COVID. All mitochondrial and ribosomal genes were removed. avg_logFC - natural log fold change of the average expression between the corresponding cluster in severe Neuro-COVID and the corresponding cluster in mild Neuro-COVID; pct.1 - percentage of cells with the gene detected in the corresponding cluster in severe Neuro-COVID; pct.2 - percentage of cells with the gene detected in mild Neuro-COVID; p_val_adj - adjusted p value (based on Bonferroni method).
Sheet 1 named Meta analysis: Summary of 20 single-cell RNA-sequencing studies of COVID-19. Changes of cluster abundance are listed for pulmonary COVID-19 versus healthy control, severe versus mild COVID-19 and COVID-19 versus inflammatory controls (e.g., influenza). BALF - bronchoalveolar fluid, CCL - CC-chemokine ligand, CCL4L2 - C-C Motif Chemokine Ligand 4 Like 2, CLEC9A - C-Type Lectin Domain Containing 9A, cMono - classical monocytes, CTL -cytotoxic T lymphocyte, CTLA 4 - cytotoxic T-lymphocyte-associated Protein 4, CTSB - Cathepsin B, CTSL - Cathepsin L, CXCL - C-X-C motif chemokine, DC - dendritic cell, EM - effector memory, FCN - Ficolin, FU - follow-up, GZM - granzyme, hi - high, HIF1A - Hypoxia Inducible Factor 1 Subunit Alpha, HLA - human leukocyte antigen, lo - low, HMOX1 - Heme Oxygenase 1, ICOS - inducible T cell costimulator, IFI27 - Interferon Alpha Inducible Protein 27, IFITM3 - Interferon Induced Transmembrane Protein 3, IFN - interferon, IL - interleukin, IL1RN - Interleukin 1 Receptor Antagonist, iMono - intermediate monocytes, ISG - Interferon-stimulated gene, ITGAE - Integrin Subunit Alpha E, KLRB - Killer Cell Lectin Like Receptor, MAIT - Mucosal associated invariant T cells, LTB - lymphotoxin β, Mac - macrophages, MAPK - mitogen-activated protein kinase, mDC - myeloid dendritic cell, MK - megakaryocytes, Mono - monocyte, n/a - not available, ncMono - non-classical monocytes, NK - natural killer cell, NRG - neuregulin, OAS1 - Oligoadenylate Synthetase 1, pDC - plasmacytoid dendritic cell, PRF - perforin, S1PR1 - Sphingosine-1-Phosphate Receptor 1, Tcm - T central memory cell, TFH - T follicular helper cell, TH - T helper, TEM / Tem - T effector memory cell, TM - T memory cell, TNF - tumor necrosis factor, Treg - regulatory T cell, TTE - terminal effector, XCL - X-C Motif Chemokine Ligand, ZNF683 - Zinc Finger Protein 683. Sheet 2 named Linear regression. Linear regression analysis. Regression was performed for all significant cluster abundances changes with age, sex and diagnosis as predictors.
References
- Alexopoulos H., Magira E., Bitzogli K., Kafasi N., Vlachoyiannopoulos P., Tzioufas A., Kotanidou A., Dalakas M.C. Anti-SARS-CoV-2 antibodies in the CSF, blood-brain barrier dysfunction, and neurological outcome: Studies in 8 stuporous and comatose patients. Neurol. Neuroimmunol. Neuroinflamm. 2020;7:e893. doi: 10.1212/NXI.0000000000000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon M., Schweblin C., Lambeng N., Cherpillod P., Vazquez J., Lalive P.H., Schibler M., Deffert C. Cerebrospinal fluid features in SARS-CoV-2 RT-PCR positive patients. Clin. Infect. Dis. 2020:ciaa1165. doi: 10.1093/cid/ciaa1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes J.P., Mishra N., Tran F., Bahmer T., Best L., Blase J.I., Bordoni D., Franzenburg J., Geisen U., Josephs-Spaulding J., HCA Lung Biological Network. Deutsche COVID-19 Omics Initiative (DeCOI) Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity. 2020;53:1296–1314.e9. doi: 10.1016/j.immuni.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherding N., Bormann N.L., Kraus G. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res. 2020;9:47. doi: 10.12688/f1000research.22139.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonsenso D., Parri N., De Rose C., Valentini P., Gemelli-pediatric COVID-19 team Toward a clinically based classification of disease severity for paediatric COVID-19. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30396-0. S1473-3099(20)30396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara N., Madi A., Kondo T., Zhang H., Acharya N., Singer M., Nyman J., Marjanovic N.D., Kowalczyk M.S., Wang C. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 2018;558:454–459. doi: 10.1038/s41586-018-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Corrie B.D., Marthandan N., Zimonja B., Jaglale J., Zhou Y., Barr E., Knoetze N., Breden F.M.W., Christley S., Scott J.K. iReceptor: A platform for querying and analyzing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol. Rev. 2018;284:24–41. doi: 10.1111/imr.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destras G., Bal A., Escuret V., Morfin F., Lina B., Josset L., COVID-Diagnosis HCL Study Group Systematic SARS-CoV-2 screening in cerebrospinal fluid during the COVID-19 pandemic. Lancet Microbe. 2020;1:e149. doi: 10.1016/S2666-5247(20)30066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellul M.A., Benjamin L., Singh B., Lant S., Michael B.D., Easton A., Kneen R., Defres S., Sejvar J., Solomon T. Neurological associations of COVID-19. Lancet Neurol. 2020;19:767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaulova E., Cantoni C., Shchukina I., Zaitsev K., Bucelli R.C., Wu G.F., Artyomov M.N., Cross A.H., Edelson B.T. Single-cell RNA-seq analysis of human CSF microglia and myeloid cells in neuroinflammation. Neurol. Neuroimmunol. Neuroinflamm. 2020;7:e732. doi: 10.1212/NXI.0000000000000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espíndola O.M., Siqueira M., Soares C.N., Lima M.A.S.D., Leite A.C.C.B., Araujo A.Q.C., Brandão C.O., Silva M.T.T. Patients with COVID-19 and neurological manifestations show undetectable SARS-CoV-2 RNA levels in the cerebrospinal fluid. Int. J. Infect. Dis. 2020;96:567–569. doi: 10.1016/j.ijid.2020.05.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Chi X., Ma W., Zhong S., Dong Y., Zhou W., Ding W., Fan H., Yin C., Zuo Z. Single-cell RNA-seq and V(D)J profiling of immune cells in COVID-19 patients. MedRxiv. 2020 doi: 10.1101/2020.05.24.20101238. [DOI] [Google Scholar]
- Farhadian S.F., Mehta S.S., Zografou C., Robertson K., Price R.W., Pappalardo J., Chiarella J., Hafler D.A., Spudich S.S. Single-cell RNA sequencing reveals microglia-like cells in cerebrospinal fluid during virologically suppressed HIV. JCI Insight. 2018;3:121718. doi: 10.1172/jci.insight.121718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi M., Bar-Or A., Piehl F., Preziosa P., Solari A., Vukusic S., Rocca M.A. Multiple sclerosis. Nat. Rev. Dis. Primers. 2018;4:43. doi: 10.1038/s41572-018-0041-4. [DOI] [PubMed] [Google Scholar]
- Fotuhi M., Mian A., Meysami S., Raji C.A. Neurobiology of COVID-19. J. Alzheimers Dis. 2020;76:3–19. doi: 10.3233/JAD-200581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke C., Ferse C., Kreye J., Reincke M., Sanchez-Sendin E., Rocco A., Steinbrenner M., Angermair S., Treskatsch S., Zickler D. High frequency of cerebrospinal fluid autoantibodies in COVID-19 patients with neurological symptoms. MedRxiv. 2020 doi: 10.1101/2020.07.01.20143214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D.I., Jacobson D.M. Diagnostic criteria for idiopathic intracranial hypertension. Neurology. 2002;59:1492–1495. doi: 10.1212/01.wnl.0000029570.69134.1b. [DOI] [PubMed] [Google Scholar]
- Fu R., Gillen A.E., Sheridan R.M., Tian C., Daya M., Hao Y., Hesselberth J.R., Riemondy K.A. clustifyr: an R package for automated single-cell RNA sequencing cluster classification. F1000Res. 2020;9:223. doi: 10.12688/f1000research.22969.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galson J.D., Schaetzle S., Bashford-Rogers R.J.M., Raybould M.I.J., Kovaltsuk A., Kilpatrick G.J., Minter R., Finch D.K., Dias J., James L. Deep sequencing of B cell receptor repertoires from COVID-19 patients reveals strong convergent immune signatures. bioRxiv. 2020 doi: 10.1101/2020.05.20.106294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gate D., Saligrama N., Leventhal O., Yang A.C., Unger M.S., Middeldorp J., Chen K., Lehallier B., Channappa D., De Los Santos M.B. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature. 2020;577:399–404. doi: 10.1038/s41586-019-1895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Lin Y.C., Wu T., Salgado A.D., Mexhitaj I., Wuest S.C., Romm E., Ohayon J., Goldbach-Mansky R., Vanderver A. Comprehensive immunophenotyping of cerebrospinal fluid cells in patients with neuroimmunological diseases. J. Immunol. 2014;192:2551–2563. doi: 10.4049/jimmunol.1302884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M. Neurologic Features in Severe SARS-CoV-2 Infection. N. Engl. J. Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillje R., Pelicci P.G., Luzi L. Cerebro: interactive visualization of scRNA-seq data. Bioinformatics. 2020;36:2311–2313. doi: 10.1093/bioinformatics/btz877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Shi Y., Gong B., Jiang L., Liu X., Yang J., Tang J., You C., Jiang Q., Long B. Blood single cell immune profiling reveals the interferon-MAPK pathway mediated adaptive immune response for COVID-19. MedRxiv. 2020 doi: 10.1101/2020.03.15.20033472. [DOI] [Google Scholar]
- Karagiannidis C., Mostert C., Hentschker C., Voshaar T., Malzahn J., Schillinger G., Klauber J., Janssens U., Marx G., Weber-Carstens S. Case characteristics, resource use, and outcomes of 10 021 patients with COVID-19 admitted to 920 German hospitals: an observational study. Lancet Respir. Med. 2020;8:853–862. doi: 10.1016/S2213-2600(20)30316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnam G., Rygiel T.P., Raaben M., Grinwis G.C.M., Coenjaerts F.E., Ressing M.E., Rottier P.J.M., de Haan C.A.M., Meyaard L. CD200 receptor controls sex-specific TLR7 responses to viral infection. PLoS Pathog. 2012;8:e1002710. doi: 10.1371/journal.ppat.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann D.E., Kavanagh D.G., Pereyra F., Zaunders J.J., Mackey E.W., Miura T., Palmer S., Brockman M., Rathod A., Piechocka-Trocha A. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat. Immunol. 2007;8:1246–1254. doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.-R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarik M.C., Grummel V., Wemlinger S., Buck D., Weber M.S., Berthele A., Hemmer B. Immune cell subtyping in the cerebrospinal fluid of patients with neurological diseases. J. Neurol. 2014;261:130–143. doi: 10.1007/s00415-013-7145-2. [DOI] [PubMed] [Google Scholar]
- Kremer S., Lersy F., de Sèze J., Ferré J.-C., Maamar A., Carsin-Nicol B., Collange O., Bonneville F., Adam G., Martin-Blondel G., SFNR-COVID Group Brain MRI Findings in Severe COVID-19: A Retrospective Observational Study. Radiology. 2020;297:E242–E251. doi: 10.1148/radiol.2020202222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw377. W90-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzke J.F. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.-S. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Mann E.R., Menon M., Knight S.B., Konkel J.E., Jagger C., Shaw T.N., Krishnan S., Rattray M., Ustianowski A., Bakerly N.D., NIHR Respiratory TRC. CIRCO Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19. Sci. Immunol. 2020;5:eabd6197. doi: 10.1126/sciimmunol.abd6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschke J., Lütgehetmann M., Hagel C., Sperhake J.P., Schröder A.S., Edler C., Mushumba H., Fitzek A., Allweiss L., Dandri M. Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. 2020;19:919–929. doi: 10.1016/S1474-4422(20)30308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane L.M., Abdel-Hakeem M.S., Wherry E.J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- Meinhardt J., Radke J., Dittmayer C., Franz J., Thomas C., Mothes R., Laue M., Schneider J., Brünink S., Greuel S. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2020 doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- Meyer Zu Hörste G., Gross C.C., Klotz L., Schwab N., Wiendl H. Next-Generation Neuroimmunology: New Technologies to Understand Central Nervous System Autoimmunity. Trends Immunol. 2020;41:341–354. doi: 10.1016/j.it.2020.02.005. [DOI] [PubMed] [Google Scholar]
- Minervina A.A., Komech E.A., Titov A., Bensouda Koraichi M., Rosati E., Mamedov I.Z., Franke A., Efimov G.A., Chudakov D.M., Mora T. Longitudinal high-throughput TCR repertoire profiling reveals the dynamics of T cell memory formation after mild COVID-19 infection. bioRxiv. 2020 doi: 10.1101/2020.05.18.100545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollan S.P., Aguiar M., Evison F., Frew E., Sinclair A.J. The expanding burden of idiopathic intracranial hypertension. Eye (Lond.) 2019;33:478–485. doi: 10.1038/s41433-018-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco G., Lee B., Xu W., Mustafah S., Hwang Y.Y., Carré C., Burdin N., Visan L., Ceccarelli M., Poidinger M. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep. 2019;26:1627–1640.e7. doi: 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B., Schmidbauer M.L., Dimitriadis K., Otto S., Knier B., Niesen W.-D., Hosp J.A., Günther A., Lindemann S., Nagy G., PANDEMIC and the IGNITE study groups Cerebrospinal fluid findings in COVID-19 patients with neurological symptoms. J. Neurol. Sci. 2020;418:117090. doi: 10.1016/j.jns.2020.117090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.C.A., Yang F., Hoh R.A., Jackson K.J.L., Roeltgen K., Lee J.-Y., Rustagi A., Rogers A.J., Powell A.E., Kim P.S. B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Res Sq. 2020 doi: 10.1016/j.chom.2020.09.002. rs.3.rs-27220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson R.W., Brown R.L., Benjamin L., Nortley R., Wiethoff S., Bharucha T., Jayaseelan D.L., Kumar G., Raftopoulos R.E., Zambreanu L. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain. 2020;143:3104–3120. doi: 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V.S., Madrigal A., Schmiedel B.J., Clarke J., O’Rourke P., de Silva A.D., Harris E., Peters B., Seumois G., Weiskopf D. Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol. 2018;3:eaan8664. doi: 10.1126/sciimmunol.aan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzini A., Padovani A. Lifting the mask on neurological manifestations of COVID-19. Nat. Rev. Neurol. 2020;16:636–644. doi: 10.1038/s41582-020-0398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A., Schubert R.D., Greenfield A.L., Dandekar R., Loudermilk R., Sabatino J.J., Jr., Koelzer M.T., Tran E.B., Koshal K., Kim K., University of California, San Francisco MS-EPIC Team A pathogenic and clonally expanded B cell transcriptome in active multiple sclerosis. Proc. Natl. Acad. Sci. USA. 2020;117:22932–22943. doi: 10.1073/pnas.2008523117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff R.M., Engelhardt B. The anatomical and cellular basis of immune surveillance in the central nervous system. Nat. Rev. Immunol. 2012;12:623–635. doi: 10.1038/nri3265. [DOI] [PubMed] [Google Scholar]
- Robert C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020;11:3801. doi: 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Sánchez C.M., Díaz-Maroto I., Fernández-Díaz E., Sánchez-Larsen Á., Layos-Romero A., García-García J., González E., Redondo-Peñas I., Perona-Moratalla A.B., Del Valle-Pérez J.A. Neurologic manifestations in hospitalized patients with COVID-19: The ALBACOVID registry. Neurology. 2020;95:e1060–e1070. doi: 10.1212/WNL.0000000000009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutigliano J.A., Sharma S., Morris M.Y., Oguin T.H., 3rd, McClaren J.L., Doherty P.C., Thomas P.G. Highly pathological influenza A virus infection is associated with augmented expression of PD-1 by functionally compromised virus-specific CD8+ T cells. J. Virol. 2014;88:1636–1651. doi: 10.1128/JVI.02851-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankowski R., Böttcher C., Masuda T., Geirsdottir L., Sagar, Sindram E., Seredenina T., Muhs A., Scheiwe C., Shah M.J. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 2019;22:2098–2110. doi: 10.1038/s41593-019-0532-y. [DOI] [PubMed] [Google Scholar]
- Schafflick D., Xu C.A., Hartlehnert M., Cole M., Schulte-Mecklenbeck A., Lautwein T., Wolbert J., Heming M., Meuth S.G., Kuhlmann T. Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat. Commun. 2020;11:247. doi: 10.1038/s41467-019-14118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., Deutsche COVID-19 OMICS Initiative (DeCOI) Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiß C., Paschold L., Simnica D., Mohme M., Willscher E., von Wenserski L., Scholz R., Wieters I., Dahlke C., Tolosa E. Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease. Immunity. 2020;53:442–455.e4. doi: 10.1016/j.immuni.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomuradova A.S., Vagida M.S., Sheetikov S.A., Zornikova K.V., Kiryukhin D., Titov A., Peshkova I.O., Khmelevskaya A., Dianov D.V., Malasheva M. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T-cell receptors. Immunity. 2020;53:1245–1257.e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Wang C., Cong L., Marjanovic N.D., Kowalczyk M.S., Zhang H., Nyman J., Sakuishi K., Kurtulus S., Gennert D. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell. 2016;166:1500–1511.e9. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E., Bartley C.M., Chow R.D., Ngo T.T., Jiang R., Zamecnik C.R., Dandekar R., Loudermilk R.P., Dai Y., Feimei L. Exploratory neuroimmune profiling identifies CNS-specific alterations in COVID-19 patients with neurological involvement. bioRxiv. 2020 2020.09.11.293464. [Google Scholar]
- Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J., Banwell B.L., Barkhof F., Carroll W.M., Coetzee T., Comi G., Correale J., Fazekas F., Filippi M., Freedman M.S. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17:162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- Tian W., Zhang N., Jin R., Feng Y., Wang S., Gao S., Gao R., Wu G., Tian D., Tan W. Immune suppression in the early stage of COVID-19 disease. Nat. Commun. 2020;11:5859. doi: 10.1038/s41467-020-19706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trandem K., Zhao J., Fleming E., Perlman S. Highly activated cytotoxic CD8 T cells express protective IL-10 at the peak of coronavirus-induced encephalitis. J. Immunol. 2011;186:3642–3652. doi: 10.4049/jimmunol.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varatharaj A., Thomas N., Ellul M.A., Davies N.W.S., Pollak T.A., Tenorio E.L., Sultan M., Easton A., Breen G., Zandi M., CoroNerve Study Group Neurological and neuropsychiatric complications of COVID-19 in 153 patients: a UK-wide surveillance study. Lancet Psychiatry. 2020;7:875–882. doi: 10.1016/S2215-0366(20)30287-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Li B., Nelson C.E., Nabavi S. Comparative analysis of differential gene expression analysis tools for single-cell RNA sequencing data. BMC Bioinformatics. 2019;20:40. doi: 10.1186/s12859-019-2599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbert J., Li X., Heming M., Mausberg A.K., Akkermann D., Frydrychowicz C., Fledrich R., Groeneweg L., Schulz C., Stettner M. Redefining the heterogeneity of peripheral nerve cells in health and autoimmunity. Proc. Natl. Acad. Sci. USA. 2020;117:9466–9476. doi: 10.1073/pnas.1912139117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykes M.N., Lewin S.R. Immune checkpoint blockade in infectious diseases. Nat. Rev. Immunol. 2018;18:91–104. doi: 10.1038/nri.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Mu J., Guo J., Lu L., Liu D., Luo J., Li N., Liu J., Yang D., Gao H. New onset neurologic events in people with COVID-19 in 3 regions in China. Neurology. 2020;95:e1479–e1487. doi: 10.1212/WNL.0000000000010034. [DOI] [PubMed] [Google Scholar]
- Zhang J.-Y., Wang X.-M., Xing X., Xu Z., Zhang C., Song J.-W., Fan X., Xia P., Fu J.-L., Wang S.-Y. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Yang P., Zhao Y., Zhuang Z., Wang Z., Song R., Zhang J., Liu C., Gao Q., Xu Q. Single-Cell Sequencing of Peripheral Mononuclear Cells Reveals Distinct Immune Response Landscapes of COVID-19 and Influenza Patients. Immunity. 2020;53:685–696.e3. doi: 10.1016/j.immuni.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet 1 named patient info: Patient characteristics. Ab - antibody; AF - atrial fibrillation; BCBD - blood-CSF-barrier dysfunction; BPH: benign prostatic hyperplasia; CAD: coronary artery disease; C - COVID-19; CIM/CIP - critical illness myopathy/polyneuropathy; cn - cranial nerve; COPD - Chronic obstructive pulmonary disease; CRC - colorectal cancer; CRP - C-reactive protein; CTS - Carpal tunnel syndrome; EDSS - Expanded Disability Status Scale; GERD - gastresophageal reflux disease; Ig - immunoglobulin; IIH - intracranial hypertension; IL - interleukin; LP - lumbar puncture; MS - multiple sclerosis; NSCLC - non-small cell lung cancer; OCB - oligoclonal band; OSA - obstructive sleep apnea; PAD - peripheral arterial disease; PCT - procalcitonin; PNP - polyneuropathy; S/p - status post; RBC - red blood cell; TBI - traumatic brain injury; TIA - transient ischemic attack; VE - viral encephalitis; vvECMO - veno-venous extracorporeal membrane oxygenation; WBC - white blood cell count; VZV - Varicella zoster virus; HSV - Herpes simplex virus; Sheet 2 named scRNA-seq: Technical information on single-cell RNA sequencing. Please note that the columns ‘cells’ and ‘median genes per cell’ refer to the raw cellranger output, while the columns ‘final cells’ and ‘final median genes per cell’ refer to the final Seurat object after filtering low-quality cells and removing the low quality-cluster (see Methods). Sheet 3 named scTCR-seq: Technical information on single-cell T cell receptor sequencing. TCR - T cell receptor sequencing
Sheet 1 named Marker genes: Top marker genes of all clusters. Average logFC threshold was set to 0.25 and minimal percentage of expressing genes in either of the two populations to 0.25. All genes with an adjusted p value above 0.05 were removed. Only genes with a positive average log fold change were retained. avg_logFC - natural log fold change of the average expression between the corresponding cluster and all remaining clusters; pct.1 - percentage of cells with the gene detected in the corresponding cluster pct.2 - percentage of cells with the gene detected in all remaining clusters; p_val_adj - adjusted p value (based on Bonferroni method). Sheet 2 named DE mono2 versus mono1 and mono3: Differentially expressed genes of mono2 versus mono1 and mono3. Average logFC threshold was set to 0.1 and minimal percentage of expressing genes in either of the two populations to 0.1. All genes with an adjusted p value above 0.05 were removed. A positive average logFC value shows a higher expression in the mono2 cluster. All mitochondrial and ribosomal genes were removed. avg_logFC - natural log fold change of the average expression between mono2 and mono1/3; pct.1 - percentage of cells with the gene detected mono2; pct.2 - percentage of cells with the gene detected in mono1 and mono3 clusters; p_val_adj - adjusted p value (based on Bonferroni method). Sheet 3 named TCR Primer: Technical information of TCR sequencing. TCR - T cell receptor; R - a purine base (arginine (A) or guanine (G); Y - a pyrimidine base (cytosine (C) or thymidine (T); B - a base of either C, G or T; 4 step PCR was used in TCR Repertoire enrichment. 2 annealing steps (63°C and 57°C) were used due to the annealing temperatures’ difference between PolyA primer and TCR rev primer. Temperature was defined with degree Celsius (°C) and duration time in hh:mm:ss. Sheet 4 named Microglia marker: Microglia markers from Sankowski et al. (Sankowski et al., 2019) used for the gene score feature plot (Figure S1D). Sheet 5 named Enrichment analysis of DE genes: Enrichment analysis of significant DE genes of mono2 cluster using EnrichR tool in GO Molecular Function 2018 dataset. Mitochondrial and ribosomal DE genes were removed in advance.
Sheets 1-6: DE genes in Neuro-COVID versus IIH (sheet 1-3) and Neuro-COVID versus VE (sheet 4-6) clusters in TcMeta (sheet 1 & 4), DCMeta (sheet 2 & 5) and monoMeta (sheet 3 & 6) were identified with the FindMarkers function in Seurat. Average logFC threshold was set to 0.1 and minimal percentage of expressing genes in either of the two populations to 0.1. All genes with an adjusted p value above 0.05 were removed. A positive average logFC value shows a higher expression in Neuro-COVID, a negative average logFC value shows a higher expression in IIH / VE. All mitochondrial and ribosomal genes were removed. avg_logFC - natural log fold change of the average expression between the corresponding cluster in Neuro-COVID and the corresponding cluster in IIH / VE; pct.1 - percentage of cells with the gene detected in the corresponding cluster in Neuro-COVID; pct.2 - percentage of cells with the gene detected in the corresponding cluster in IIH / VE; p_val_adj - adjusted p value (based on Bonferroni method).
Sheet 1 named Marker genes T subclusters: Marker genes of the T cell subclusters. Average logFC threshold was set to 0.1 and minimal percentage of expressing genes in either of the two populations to 0.1. All genes with an adjusted p value above 0.05 were removed. Only genes with a positive average log fold change were retained. avg_logFC - natural log fold change of the average expression between the corresponding cluster and all remaining clusters; pct.1 - percentage of cells with the gene detected in the corresponding cluster; pct.2 - percentage of cells with the gene detected in all remaining clusters; p_val_adj - adjusted p value (based on Bonferroni method). Sheet 2 named Marker genes exhaustion and ISG: T cell exhaustion and interferon-stimulated gene (ISG) signatures. Interferon-stimulated genes (ISG) from Wilk et al. (Wilk et al., 2020) that were used for the ISG signature heatmap in Figure 3C and markers of T cell exhaustion from Singer et al. (Singer et al., 2016) (Dysfunction and Activation module), Chihara et al. (Chihara et al., 2018) and Tirosh et al. (Tirosh et al., 2016) that were used for the T cell exhaustion signature in Figure 3E and Figure S2. Sheet 3-4: CSEA Neuro-COVID versus IIH (sheet 3) and Neuro-COVID versus VE (sheet 4) in CD4+ T cells. Summary of the CSEA enrichment score, the total number of cells in the leading edge, which drive the enrichment of the specific signature, the enrichment in the leading edge that demonstrates how enriched the leading edge is for Neuro-COVID cells compared to the total cell population, the corrected p value, which is the p value for positive enrichment (Neuro-COVID versus IIH/VE) and the negative p value, which is the corrected p value for negative enrichment (IIH/VE versus Neuro-COVID). Sheet 5-7: DE genes in severe versus mild Neuro-COVID in TcMeta (sheet 5), DCMeta (sheet 6) and monoMeta (sheet 7) clusters. Average logFC threshold was set to 0.1 and minimal percentage of expressing genes in either of the two populations to 0.1. All genes with an adjusted p value above 0.05 were removed. A positive average logFC value shows a higher expression in severe Neuro-COVID, a negative average logFC value shows a higher expression in mild Neuro-COVID. All mitochondrial and ribosomal genes were removed. avg_logFC - natural log fold change of the average expression between the corresponding cluster in severe Neuro-COVID and the corresponding cluster in mild Neuro-COVID; pct.1 - percentage of cells with the gene detected in the corresponding cluster in severe Neuro-COVID; pct.2 - percentage of cells with the gene detected in mild Neuro-COVID; p_val_adj - adjusted p value (based on Bonferroni method).
Sheet 1 named Meta analysis: Summary of 20 single-cell RNA-sequencing studies of COVID-19. Changes of cluster abundance are listed for pulmonary COVID-19 versus healthy control, severe versus mild COVID-19 and COVID-19 versus inflammatory controls (e.g., influenza). BALF - bronchoalveolar fluid, CCL - CC-chemokine ligand, CCL4L2 - C-C Motif Chemokine Ligand 4 Like 2, CLEC9A - C-Type Lectin Domain Containing 9A, cMono - classical monocytes, CTL -cytotoxic T lymphocyte, CTLA 4 - cytotoxic T-lymphocyte-associated Protein 4, CTSB - Cathepsin B, CTSL - Cathepsin L, CXCL - C-X-C motif chemokine, DC - dendritic cell, EM - effector memory, FCN - Ficolin, FU - follow-up, GZM - granzyme, hi - high, HIF1A - Hypoxia Inducible Factor 1 Subunit Alpha, HLA - human leukocyte antigen, lo - low, HMOX1 - Heme Oxygenase 1, ICOS - inducible T cell costimulator, IFI27 - Interferon Alpha Inducible Protein 27, IFITM3 - Interferon Induced Transmembrane Protein 3, IFN - interferon, IL - interleukin, IL1RN - Interleukin 1 Receptor Antagonist, iMono - intermediate monocytes, ISG - Interferon-stimulated gene, ITGAE - Integrin Subunit Alpha E, KLRB - Killer Cell Lectin Like Receptor, MAIT - Mucosal associated invariant T cells, LTB - lymphotoxin β, Mac - macrophages, MAPK - mitogen-activated protein kinase, mDC - myeloid dendritic cell, MK - megakaryocytes, Mono - monocyte, n/a - not available, ncMono - non-classical monocytes, NK - natural killer cell, NRG - neuregulin, OAS1 - Oligoadenylate Synthetase 1, pDC - plasmacytoid dendritic cell, PRF - perforin, S1PR1 - Sphingosine-1-Phosphate Receptor 1, Tcm - T central memory cell, TFH - T follicular helper cell, TH - T helper, TEM / Tem - T effector memory cell, TM - T memory cell, TNF - tumor necrosis factor, Treg - regulatory T cell, TTE - terminal effector, XCL - X-C Motif Chemokine Ligand, ZNF683 - Zinc Finger Protein 683. Sheet 2 named Linear regression. Linear regression analysis. Regression was performed for all significant cluster abundances changes with age, sex and diagnosis as predictors.
Data Availability Statement
Raw sequencing data are available in the Gene Expression Omnibus (GEO) repository (GSE163005). An interactive version of the entire dataset using cerebroApp (Hillje et al., 2020) is available at: http://covid.mheming.de. We followed the official tutorial of the packages listed, no custom code was generated. Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/5mt97xcyyw.2.