Graphical abstract
Keywords: SARS-CoV-2, COVID-19, Paper-based, Biosensor, IL-6, Smartphone
Abstract
Decentralizing COVID-19 care reduces contagions and affords a better use of hospital resources. We introduce biosensors aimed at detecting severe cases of COVID-19 in decentralized healthcare settings. They consist of a paper immunosensor interfaced with a smartphone. The immunosensors have been designed to generate intense colorimetric signals when the sample contains ultralow concentrations of IL-6, which has been proposed as a prognosis biomarker of COVID-19. This is achieved by combining a paper-based signal amplification mechanism with polymer-filled reservoirs for dispensing antibody-decorated nanoparticles and a bespoken app for color quantification. With this design we achieved a low limit of detection (LOD) of 10−3 pg mL-1 and semi-quantitative measurements in a wide dynamic range between 10−3 and 102 pg mL-1 in PBS. The assay time is under 10 min. The low LOD allowed us to dilute blood samples and detect IL-6 with an LOD of 1.3 pg mL-1 and a dynamic range up to 102 pg mL-1. Following this protocol, we were able to stratify COVID-19 patients according to different blood levels of IL-6. We also report on the detection of IL-6 in respiratory samples (bronchial aspirate, BAS) from COVID-19 patients. The test could be easily adapted to detect other cytokines such as TNF-α and IL-8 by changing the antibodies decorating the nanoparticles accordingly. The ability of detecting cytokines in blood and respiratory samples paves the way for monitoring local inflammation in the lungs as well as systemic inflammation levels in the body.
1. Introduction
The SARS-CoV-2 pandemic has rapidly spread worldwide with enormous social and economic repercussions [1]. Patients may be asymptomatic, have mild flu-like symptoms, or progress to severe pneumonia [2]. Previous studies on SARS-CoV and MERS-CoV show that infection of epithelial lung cells provokes a strong local inflammatory response characterized by high levels of cytokines and chemokines [3]. When these pro-inflammatory factors reach the bloodstream they stimulate the release of immature granulocytes in the bone marrow, which exacerbates the inflammation [4]. The resulting hyperinflammatory syndrome or “cytokine storm” is believed to be responsible for many severe cases of COVID-19. IL-6 has been extensively used for prognosticating severe cases of COVID-19. Mild cases show serum IL-6 levels between 5.1 and 18.8 pg mL−1, whereas in moderate/severe cases this value increases to 22.5–198 pg mL-1 [[5], [6], [7], [8], [9], [10]]. Stratifying patients according to severity is crucial in order to optimize COVID-19 management. During the first peak of the pandemic, saturation of hospitals forced healthcare providers to decentralize COVID-19 care. Even after “flattening the contagion curve” many mild cases of COVID-19 are cared for at home in order to reduce the risk of contagion [11]. Patients with severe symptoms are rapidly transferred to hospitals, where they receive specialized care to prevent poor outcomes. Biosensors for detecting elevated cytokine levels at the point of care could help physicians stratify patients according to their prognosis [12]. The challenge in designing such devices is that they would need to be highly portable, easy to manipulate, and require minimal infrastructure to ensure an easy implementation in a decentralized health care strategy [12,13].
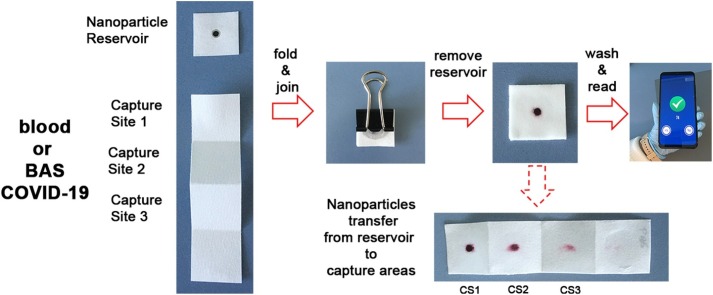
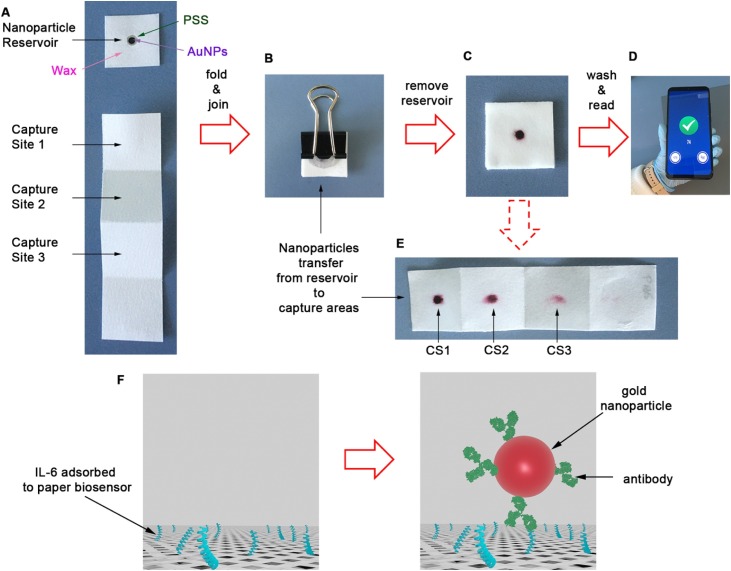
In this manuscript we meet this challenge with mobile biosensors that measure levels of IL-6 in blood and respiratory samples from COVID-19 patients in less than 10 min. In order to meet the urgent need for rapid testing we collated previous discoveries into a new design that meets the needs for decentralized COVID-19 diagnostics. It consists of devices that are entirely made of paper and comprised of two parts: a paper square containing a nanoparticle reservoir and a paper strip for target capture (Fig. 1 A). The nanoparticle reservoir is made by adding polystyrene sulfonate (PSS) to a region of the paper. This prevents irreversible interactions between antibody-decorated nanoparticles and the cellulose substrate [14]. The reservoir dimensions are defined by a hydrophobic barrier made of paraffin. The paper strip has three capture sites (CS) for amplifying colorimetric signals without adding additional analytical steps [15]. To detect IL-6, a drop of sample is added to each capture site and quickly dried. The paper is then folded and soaked with a blocking solution. Subsequently, the reservoir is pressed on top of the folded strip with the aid of a clamp for 5 min (Fig. 1B). During this time, nanoparticles are transferred vertically from the reservoir to the three capture sites (Fig. 1E), where the antibodies recognize IL-6 specifically. After removing the reservoir, a colored spot appears whose pixel intensity is directly related to the concentration of IL-6 in the sample. Since filter paper is semi-transparent when wet, and the biosensors generated a colorimetric signal in each capture site, the colored spot on the folded paper is the sum of the three independent signals generated at each capture site [15]. This collective, amplified signal is then quantified with a previously developed smartphone app for densitometry at the point of need [16]. The origami-enabled signal amplification mechanism achieves a limit of detection of 10−3 pg mL-1, which is one of the lowest reported to date (Table S1) [[16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39]]. Thanks to this low limit of detection in PBS we were able to dilute patient samples for detecting elevated levels of IL-6 without performing any additional purification steps. This was accomplished in both blood and respiratory samples, thus paving the way for using the proposed biosensors for monitoring systemic and local inflammation. Being able to detect high levels of IL-6 in different biological matrices, along with the rapid turnaround time and highly portable detection system, makes our biosensors ideal for the decentralized monitoring of COVID-19 patients both at home and in hospitals.
Fig. 1.
Photographs of the paper immunosensor and key analytical steps for detecting IL-6; (A) The biosensors consists of a piece of paper containing antibody-decorated nanoparticles, and a paper strip with three capture sites (CS); (B) After adding a drop of sample in each capture site and drying it, the paper is folded; (C) After removing the reservoir, a colored spot appears whose pixel intensity is the sum of the colorimetric signal generated in each capture site; (D) After washing away excess reagents the colorimetric signal is measured with a previously developed smartphone app within seconds; (E) Unfolding the paper strip in (C) reveals that nanoparticles are transferred vertically to the 3 binding sites; (F) Schematic representation of biomolecular interactions; IL-6 is adsorbed onto the paper substrate and subsequently detected by antibody-decorated gold nanoparticles. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Experimental Section
2.1. Materials
Whatman filter paper #41 and #1 was obtained from GE Healthcare Life Sciences. Wax Paper Squares (6″) was purchased from NORPRO. Gold (III) chloride hydrate, sodium citrate tribasic dihydrate, poly (ethylene glycol) 2-mercaptoethyl ether acetic acid (thiol-PEG-acid) 2100, N-Hydroxysulfosuccinimide sodium salt (sulfo-NHS), N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDAC; EDC), O-(2-biotinyl-aminoethyl) polyethylene glycol 3000 (biotin-PEG), poly (sodium 4-styrenesulfonate, average Mw ≈ 70.000) 30 % solution, Tween-20, 2-ethanesulfonic acid (MES) and recombinant human IL-6 (I2786) were obtained from Sigma Aldrich. Rabbit anti-IL-6 polyclonal antibody (ab6672) was purchased from Abcam. Albumin from bovine serum (BSA, protease free) was obtained from VWR Chemicals. PBS refers to phosphate buffered saline pH 7.4. PBST refers to PBS supplemented with 0.1 % Tween-20. RT refers to room temperature.
2.2. Antibody-decorated gold nanoparticles
Gold nanoparticles with a diameter of ca. 40 nm were obtained with the citrate-reducing method as previously described [16]. The nanoparticles were subsequently modified with antibodies adapting a published protocol. In brief, the citrate around the nanoparticles was substituted for thiol-PEG-acid. Then, the carboxylated moieties were transformed into reactive sulfo-NHS esters via the addition of EDC (1 mg) and sulfo-NHS (2 mg) in 0.5 M MES buffer pH 5.5 for 30 min. Then 200 μL of the nanoparticles were centrifuged at 8000 rpm for 6 min and rabbit anti-IL-6 polyclonal antibody (1 mg mL−1) was added overnight. Then 200 μL of a blocking solution containing glycine (0.1 M) and BSA (10 mg mL−1) in phosphate buffer (0.1 M, pH 7.0) was added for at least 30 min to cap any remaining sulfo-NHS ester groups. Subsequently, nanoprobes were washed 5 times with PBST (6000 rpm, 6 min). Finally, the pellet was resuspended in 100 μL of PBST ([Au] ≈ 250 mM).
2.3. Fabrication of origami biosensors
For the reservoir, Whatman paper #1 was cut into 2 × 2 cm squares, which were then printed with wax with the following procedure. First, wax paper was cut into 2 × 2 cm squares. A 5 × 5 mm circle was removed from the center with a hole puncher. The remaining wax was aligned with the Whatman paper and the paraffin was transferred by pressing a hot iron against it for 5 s. The procedure was repeated on the other side of the paper to completely seal it. Then, a nanoparticle-reservoir was fabricated in the unwaxed paper area by adding 3 μL of 30 % PSS and letting it dry at room temperature (RT) followed by adding 1 μL of antibody-decorated nanoparticles and letting it dry at RT. Paper strips for analyte capture consisted of pieces of Whatman paper #41 cut into 2 × 8 cm strips as shown in Fig. 1A.
2.4. Detection of IL-6
First, 10 μL of sample was added to the first three squares of the paper strip (CS1, 2, and 3 in Fig. 1A) and dried with a hair drier for 30 s. Then, the strip was folded and 1 mL of PBS-BSA was added. Immediately afterwards, the nanoparticle reservoir was placed onto the strip and held with the aid of a clamp for 5 min. The reservoir was then peeled off and the paper strip was washed five times with 1 mL of PBST. The resulting colorimetric signal was measured immediately afterwards with the mobile app [16] Details about the development of the mobile app have been published elsewhere. [16] The app guides the user to establish a controlled distance and angle between the camera of the Huawei POT-LX1smartphone and the assay by using a virtual frame on the screen that matches the shape of the paper biosensor. After pressing “Set” the app automatically finds the region of interest, calculates the pixel intensity and subtracts it from the background. Uneven light conditions are also automatically corrected. The app stops taking measurements if there are any changes in imaging conditions. This informs the user that the smartphone needs to be realigned, at which point the measurements resume. The data set is evaluated after 50 valid measurements have been acquired. The resulting colorimetric signal S is the highest for black (255) and the lowest for white (0).
Spiked blood samples and bronchial aspirate (BAS) samples were obtained and processed as follows. A blood sample from a healthy donor was spiked with IL-6 by adding 1 μL of a solution 10 times more concentrated to 9 μL of blood. The final concentration of added IL-6 was the one reported in Fig. 3B. The samples were then diluted 1000 times with PBS and tested as above. BAS samples were first liquefied by adding 100 μL of 0.3 M H2O2 to 10 mg of samples for 1 min. The IL-6 concentration of these samples was determined with an in-house ELISA (Fig. S1). A liquefied BAS sample containing less than 3 pg mL−1 IL-6 (below the limit of detection of ELISA) was spiked with the same procedure detailed for blood samples. Samples were diluted 10 times with PBS before applying them to the paper biosensor.
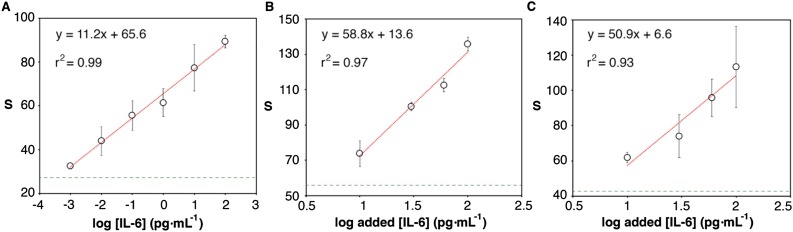
Fig. 3.
Detection of IL-6 in PBS (A), or spiked into blood (B), or bronchial aspirate (BAS, (C)). Horizontal green dotted lines represent the signal above 3 times the standard deviation of the blank. Error bars are the standard deviation (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Blood samples from COVID-19 patients were obtained from the Immunology Department at Son Espases University Hospital (Ethics Committee Protocol IB 4169/20 PI). The IL-6 concentration was determined with a DIASource Immunoassay ELISA kit. Informed consent was obtained from all participants. BAS samples from patients included in this study were provided by Biobank IdISBa and CIBERES Pulmonary Biobank Consortium, a network currently formed by twelve tertiary Spanish hospitals integrated in the Spanish National Biobanks Network. They were processed following standard procedures with the appropriate approval of the Ethics and Scientific Committees and with the collaboration of the healthcare services of the Hospital Universitario Son Espases and Hospital Son Llàtzer.
3. Results and discussion
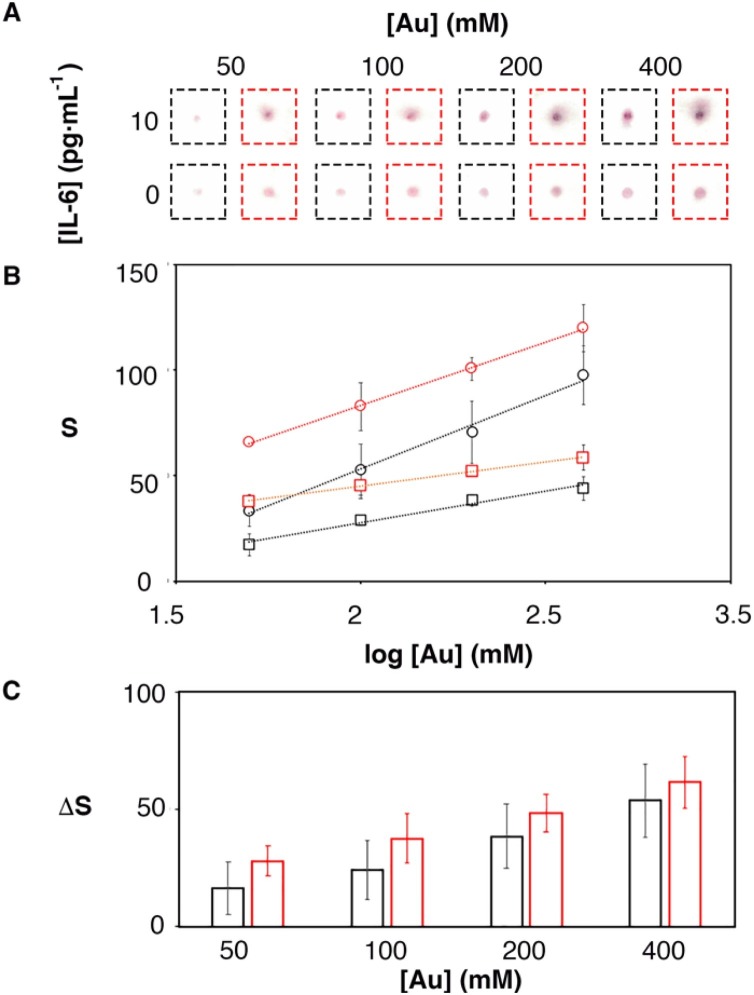
The biosensors shown in Fig. 1 incorporate a signal amplification mechanism based on adding the colorimetric signals generated in multiple capture sites. This concept has already been tested for amplifying signals in paper immunosensors, although not for the detection of IL-6 [15]. To test the impact of implementing this design in the IL-6 biosensors, samples containing 0 or 10 pg mL−1 IL-6 were tested with biosensors modified with nanoparticles at different concentrations. The colorimetric signal was measured with two methods. The first method calculated the pixel intensity only in the capture site CS1. The second method involved folding the paper to measure the collective signal from CS1, CS2 and CS3. Fig. 2 Ashows scanned images for the resulting assays. As the concentration of gold nanoprobes increased the color in the spot also increased. The color in the folded samples (highlighted in red) was more intense than in the unfolded ones. This is because in the folded sample, the signal from the three capture sites is cumulative [15]. Colored spots were more intense in samples containing IL-6, which indicated that the main contribution to the signal was the specific recognition of IL-6 by antibody-decorated nanoparticles. Fig. 2B summarizes the results obtained after quantifying the color intensity in 3 independent experiments. In this Figure, signals increase when the paper was folded in agreement with the observations in Fig. 2A. In Fig. 2C the specific signal obtained after subtracting the blank from the 10 pg mL−1 sample was plotted as a function of the concentration of nanoprobes. In this plot, the specific signal was always higher when the paper was folded. Furthermore, the standard deviation of three independent experiments was smaller when the paper was folded than when measuring a single capture site. These results confirm that the proposed method enhances specific colorimetric signals in paper immunosensors without increasing variability. The method only required adding three drops of sample instead of one. No additional reagents or analytical steps were required, making it useful for amplifying signals in rapid diagnostic tests.
Fig. 2.
Colorimetric signal obtained from samples containing 0 or 10 pg mL−1 when using 3 capture sites (CS) and the paper is folded so that the 3 signals add up (red), or unfolded and measuring only CS1 (black); (A) Photographs of the assays; (B) colorimetric signal for 10 (dots) and 0 (squares) pg·mL−1; (C) Specific signal after subtracting 0 from 10 pg·mL−1; Error bars are the standard deviation (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
After testing the signal amplification mechanism, we calibrated the biosensors with solutions containing different concentrations of recombinant human IL-6 in PBS. In Fig. 3A, the biosensors generated dose-dependent signals that increased linearly with the logarithm of the concentration of IL-6 in the range between 10−3 and 100 pg mL-1. The limit of detection, expressed as the assayed sample that yields a signal above three times the standard deviation of the blank (99 % confidence), was 10−3 pg mL-1. This is one of the lowest limits of detection reported for IL-6, and it was achieved with a fast turnaround time under 10 min (Table S1).
However, the proposed biosensors may have a drawback when detecting IL-6 in real matrices. With our device, the analyte is captured by drying the sample in the paper. While this expedites the detection of IL-6, biomolecules such as proteins and mucins present in real matrices may also physically adsorb to the paper and hinder the capture of IL-6. They may also interact non-specifically with the antibody-nanoparticles, interfering with the detection of IL-6. Blood samples are also highly colored, which can be problematic for colorimetric signal generation methods. Fortunately, the biosensors have an exceptionally low limit of detection that allows for the dilution of samples prior to testing, therefore diluting color interferents from the matrix as well. It has been proposed that healthy individuals have IL-6 levels between 0.5 and 6 pg mL−1 [40]. Diluting samples 103 times should still enable detecting IL-6 above 6 pg mL−1 (0.006 pg mL-1 in the diluted sample), while reducing matrix effects. With this in mind, we spiked blood samples from a healthy donor with IL-6 in the concentration range between 10 and 100 pg mL−1. Then we diluted these samples 1000 times with PBS and tested them with our biosensors. In Fig. 3B, the biosensor yielded a linear response with the logarithm of the concentration of added IL-6 in the concentration range proposed. These experiments demonstrate that the biosensors can detect IL-6 in blood in the relevant range for COVID-19 care [41]. According to the standard addition method, the basal IL-6 concentration can be estimated in Fig. 3b when Y = 0. This yields a value of 0.6 pg mL−1, which fits well with the IL-6 levels in a healthy individual. The limit of detection obtained by substituting Sblank + 3σ in the linear fitting yields a value of 0.7 pg mL−1. Since this this has been added to basal levels in the sample, the final limit of detection in blood is 1.3 pg mL−1. Taking into consideration that the sample was diluted 103 times, this limit of detection in blood agrees well with the value obtained in PBS (10-3 pg mL−1, Fig. 3a), which validates our biosensors for detecting abnormal levels of IL-6 (above 6 pg mL−1, highest value for healthy individuals) in blood samples. Finally, we tested whether our biosensors could detect IL-6 in spiked respiratory samples (bronchial aspirate, BAS). To this end, liquefied BAS samples were diluted 10 times with PBS and tested with the same procedure shown above. In Fig. 3C, all samples yield a dose-dependent signal above three times the standard deviation of the matrix signal. These results indicate that our biosensors could be used for monitoring variations in cytokine levels in respiratory samples from COVID-19 patients as well.
Comparison of calibration plots in Fig. 3 reveals that experiments performed in real matrices yield higher sensitivity (higher slope of the calibration plot). That is, the presence of a diluted biologic matrix facilitates the detection of IL-6 compared to the same experiments performed in PBS. The basal concentration of IL-6 in the samples is a constant value added to the concentration spiked into the sample, and therefore cannot be responsible for this increase in sensitivity. We hypothesized that biological components in the matrix that remain in the paper after drying could facilitate antibody-antigen interactions, for example by providing a native environment that preserves IL-6 conformation. The most abundant protein in blood is human serum albumin (HSA), which is a present at a concentration of around 40 mg mL−1. To test whether the presence of albumin in the sample could increase the sensitivity of our biosensors, we repeated the calibration experiments, but instead of diluting recombinant IL-6 with PBS we diluted it with PBS supplemented with 40 μg mL−1 bovine serum albumin (BSA). BSA is very similar to HSA in its amino acid composition (76 % homology, [42]), and 40 μg mL−1 is the approximate concentration after diluting the sample 103 times. In Fig. S2, the slope of the calibration plot increases from 11.2 in PBS to 22.4 in PBS-BSA. These experiments demonstrate that the presence of excess proteins in the sample increases the sensitivity of the proposed biosensors. Furthermore, comparison of the same tests measured with the mobile app or via image analysis with ImageJ yielded a higher slope with the app (22.4 vs 16.3), which shows that the quantification method also helps increase the sensitivity of the assay (Fig. S2). To summarize, the high sensitivity and low limit of detection of our tests originate from the paper-based signal amplification mechanism (Fig. 2), the composition of diluted biological matrices (Figs. 3 and S2), and the utilization of a bespoken app for color quantification (Fig. S2). While our tests are semi-quantitative compared to ELISA (Fig. S1) their sensitivity in real matrices may be high enough for detecting high levels of IL-6 in patient samples at the point of care. This is explored below.
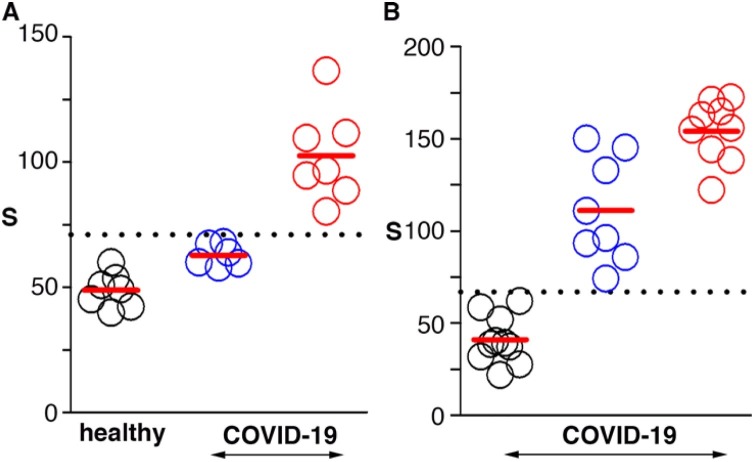
Next we tested whether the proposed biosensors could be used for detecting high levels of IL-6 in a panel of blood and BAS samples from patients, which were also analyzed with the gold standard (ELISA) in order to quantify IL-6. In Fig. 4 A, patient samples containing IL-6 above 17 pg mL−1 yielded signals statistically different when compared to blood samples from healthy donors (Kruskall-Wallis test, p < 0.001). This threshold value was in agreement with the cut-off value proposed in the literature to prognosticate severe COVID-19 as well as to guide specific anti-inflammatory treatments [43,44]. Fig. 4B summarizes the results obtained from analyzing BAS samples. BAS is usually obtained from intubated patients, and therefore we were not able to compare our results with healthy donors. Instead, we used samples containing IL-6 below 10 pg mL−1 in order to establish a cut-off value. In Fig. 4B, samples containing IL-6 with a concentration of 10 pg mL−1 or higher can be differentiated from those containing undetectable levels by ELISA (Kruskall-Wallis test, p < 0.001). However, it was not possible to distinguish among samples containing intermediate (10−100 pg mL−1) and high (above 100 pg mL−1) levels of IL-6 with the proposed biosensors. To the best of our knowledge no clinical study correlating the levels of IL-6 in respiratory samples with patient severity has yet been published in the context of COVID-19. It should be noted that the decentralized detection of IL-6 in respiratory samples would require sputum rather than BAS, since BAS is usually obtained from intubated patients. While the experiments shown here demonstrate the possibility of measuring IL-6 in samples from the lower respiratory tract, further studies should be performed for determining appropriate cut-off values in sputum samples.
Fig. 4.
Detection of IL-6 in real samples; (A) blood samples from healthy donors (black) or COVID-19 patients with IL-6 levels below (blue) or above (red) 17 pg·mL−1; (B) Bronchial aspirate (BAS) samples from COVID-19 patients with IL-6 levels below 10 pg·mL−1 (black), between 10 and 100 pg·mL−1 (blue), or above 100 pg·mL−1(red). Dotted lines show the average signal plus 2 times the standard deviation of healthy donors (A) or patients with IL-6 below 3 pg·mL−1 (B). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusion
In conclusion we have introduced paper immunosensors with a mobile readout platform for the detection of IL-6 in blood and respiratory samples from COVID-19 patients. The immunosensors have a low limit of detection at 10−3 pg mL-1 in PBS. Semi-quantitative measurements can be performed in a wide dynamic range over 5 orders of magnitude due to their paper-based signal generation design. This excellent performance enables diluting real samples to reduce matrix interference. Thanks to this procedure, we were able to correctly classify blood samples from COVID-19 patients with an IL-6 content higher than 17 pg mL-1, and BAS samples above 10 pg mL-1. Higher cut-off values could be set by diluting samples to a larger extent. The biosensors could be adapted to detect other cytokines such as TNF-α and IL-8 by substituting the antibodies around the nanoparticles for others that can recognize these targets specifically. The tests are entirely made of filter paper, which makes them lightweight, easy to transport and distribute, and easy to dispose of by incineration. Our app does not require any lightbox or accessory to control photographic conditions, making it particularly useful for home testing. These features, along with the ability to identify patients with elevated IL-6 levels at low cut-off values, makes the proposed biosensors a promising tool for the decentralized management of COVID-19 patients at home or in hospitals.
Competing interests statement
A patent application describing the method for storing nanoparticles in paper reservoirs has been filed (PCT/EP2020/075013). A patent application describing the method for liquefying respiratory samples has also been filed (P202030403).
CRediT authorship contribution statement
Cristina Adrover-Jaume: Investigation, Validation, Writing - review & editing. Alejandra Alba-Patiño: Investigation, Validation, Writing - review & editing. Antonio Clemente: Investigation, Data curation, Conceptualization, Supervision, Writing - review & editing. Giulia Santopolo: Investigation, Writing - review & editing. Andreu Vaquer: Investigation, Writing - review & editing. Steven M. Russell: Software, Visualization, Writing - review & editing. Enrique Barón: Investigation, Writing - review & editing. María del Mar González del Campo: Investigation, Writing - review & editing. Joana M. Ferrer: Investigation, Writing - review & editing. María Berman-Riu: Investigation, Writing - review & editing. Mercedes García-Gasalla: Investigation, Writing - review & editing. María Aranda: Investigation. Marcio Borges: Investigation, Writing - review & editing. Roberto de la Rica: Conceptualization, Project administration, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
R.R acknowledges financial support from grants FEDER/Ministerio de Ciencia, Innovación y Universidades/Agencia Estatal de Investigación/- Proyecto CTQ2017-82432-R and FEDER/Ministerio de Ciencia, Innovación y Universidades/Agencia Estatal de Investigación/- Proyecto CTQ2017-92226-EXP. Radix (R.R.), Folium (A.C., M.G) and Talent + fellowships from IdISBa/Impost turisme sostenible/ Agència d’Estratègia Turística de les Illes Balears-Govern de les Illes Balears are reported. A.A. acknowledges a fellowship for doctoral studies from Roche (“Stop fuga de cerebros” program). E.B. acknowledges funding from the Instituto de Salud Carlos III (Sara Borrell contract). A.V. acknowledges funding from European Social Fund (ESF) through JoTReSdOS program. M. G.-G. acknowledges funding from Instituto de Salud Carlos III (project COV20/00943). We want to particularly acknowledge the patients, Biobank IdISBa and CIBERES Pulmonary Biobank Consortium (PT17/0015/0001) member of the Spanish National Biobanks Network financed by the Carlos III Health Institute, with the participation of the Units of Intensive Care, Clinical Analysis and Pulmonology of Hospital Universitario Son Espases and Hospital Son Llàtzer, for their collaboration.
Footnotes
The Multidisciplonary Sepsis Group comprises a group of clinicians and technology developers devoted to improving sepsis care. The members of the Immune Response in Human Pathology Group study the mechanisms underlying diseases of the immune system. The Infectious Diseases-HIV Group studies clinical aspects of infectious diseases such as Tuberculosis and pneumococcal infections.
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2020.129333.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Karami A., Hasani M., Jalilian F.A., Ezati R. Conventional PCR assisted single-component assembly of spherical nucleic acids for simple colorimetric detection of SARS-CoV-2. Sens. Actuators B Chem. 2020:128971. doi: 10.1016/j.snb.2020.128971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polidoro R.B., Hagan R.S., de Santis Santiago R., Schmidt N.W. Overview: systemic inflammatory response derived from lung injury caused by SARS-CoV-2 infection explains severe outcomes in COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai Q., Huang D., Ou P., Yu H., Zhu Z., Xia Z., Su Y., Ma Z., Zhang Y., Li Z., He Q., Liu L., Fu Y., Chen J. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy Eur. J. Allergy Clin. Immunol. 2020;75:1742–1752. doi: 10.1111/all.14309. [DOI] [PubMed] [Google Scholar]
- 6.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang Xiaoyun, Chen H., Yu H., Zhang Xiaoping, Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020 doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han H., Ma Q., Li C., Liu R., Zhao L., Wang W., Zhang P., Liu X., Gao G., Liu F., Jiang Y., Cheng X., Zhu C., Xia Y. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon J.-S., Kim J.Y., Kim M.-C., Park S.Y., Kim B.-N., Bae S., Cha H.H., Jung J., Kim M.-J., Lee M.J., Choi S.-H., Chung J.-W., Shin E.-C., Kim S.-H. Factors of severity in patients with COVID-19: cytokine/chemokine concentrations, viral load, and antibody responses. Am. J. Trop. Med. Hyg. 2020;103:2412–2418. doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D., Cuapio A., Ask E.H., Hull R.M., Haroun-Izquierdo A., Schaffer M., Klingström J., Folkesson E., Buggert M., Sandberg J.K., Eriksson L.I., Rooyackers O., Ljunggren H.G., Malmberg K.J., Michaëlsson J., Marquardt N., Hammer Q., Strålin K., Björkström N.K. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5:eabd6832. doi: 10.1126/SCIIMMUNOL.ABD6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Luo S., Shen Y., Li M., Zhang Z., Dong Y., Zhou H., Lin L., Guo W., Kang Z., Xing L., Li J., Ye H., Gui W., Hu Yi, Yuan M., Han S., Zhu R., Ye Y., Wang L., Jiang X., Zhao H., Zheng X., Fan H., Zhao L., Hu Yu, Hu D. Multiple enzyme release, inflammation storm and hypercoagulability are prominent indicators for disease progression in COVID-19: a multi-centered, correlation study with CT imaging score. Lancet Infect. Dis. 2020 doi: 10.2139/ssrn.3544837. [DOI] [Google Scholar]
- 11.Hollander J.E., Carr B.G. Virtually perfect? Telemedicine for Covid-19. N. Engl. J. Med. 2020;382:1679–1681. doi: 10.1056/NEJMp2003539. [DOI] [PubMed] [Google Scholar]
- 12.Russell S.M., Alba-Patiño A., Barón E., Borges M., Gonzalez-Freire M., De La Rica R. Biosensors for managing the COVID-19 cytokine storm: challenges ahead. ACS Sens. 2020;5:1506–1513. doi: 10.1021/acssensors.0c00979. [DOI] [PubMed] [Google Scholar]
- 13.Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alba-Patiño A., Adrover-Jaume C., de la Rica R. Nanoparticle reservoirs for paper-only immunosensors. ACS Sens. 2020;5:147–153. doi: 10.1021/acssensors.9b01937. [DOI] [PubMed] [Google Scholar]
- 15.Alba-Patiño A., Russell S.M., de la Rica R. Origami-enabled signal amplification for paper-based colorimetric biosensors. Sens. Actuators B Chem. 2018;273:951–954. doi: 10.1016/j.snb.2018.07.019. [DOI] [Google Scholar]
- 16.Alba-Patiño A., Russell S.M., Borges M., Pazos-Perez N., Alvarez-Puebla R.A., de la Rica R. Nanoparticle-based mobile biosensors for the rapid detection of Sepsis biomarkers in whole blood. Nanosc. Adv. 2020;2:253–1260. doi: 10.1039/d0na00026d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aydın E.B. Highly sensitive impedimetric immunosensor for determination of interleukin 6 as a cancer biomarker by using conjugated polymer containing epoxy side groups modified disposable ITO electrode. Talanta. 2020;215 doi: 10.1016/j.talanta.2020.120909. [DOI] [PubMed] [Google Scholar]
- 18.Aydın E.B., Aydın M., Sezgintürk M.K. The development of an ultra-sensitive electrochemical immunosensor using a PPyr-NHS functionalized disposable ITO sheet for the detection of interleukin 6 in real human serums. New J. Chem. 2020 doi: 10.1039/d0nj03183f. [DOI] [Google Scholar]
- 19.Borse V., Srivastava R. Fluorescence lateral flow immunoassay based point-of-care nanodiagnostics for orthopedic implant-associated infection. Sens. Actuators B Chem. 2019;280:24–33. doi: 10.1016/j.snb.2018.10.034. [DOI] [Google Scholar]
- 20.Chen H., Li J., Zhang X., Li X., Yao M., Zheng G. Automated in vivo nanosensing of breath-borne protein biomarkers. Nano Lett. 2018;18:4716–4726. doi: 10.1021/acs.nanolett.8b01070. [DOI] [PubMed] [Google Scholar]
- 21.Chen P., Chung M.T., McHugh W., Nidetz R., Li Y., Fu J., Cornell T.T., Shanley T.P., Kurabayashi K. Multiplex serum cytokine immunoassay using nanoplasmonic biosensor microarrays. ACS Nano. 2015;9:4173–4181. doi: 10.1021/acsnano.5b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diacci C., Berto M., Di Lauro M., Bianchini E., Pinti M., Simon D.T., Biscarini F., Bortolotti C.A. Label-free detection of interleukin-6 using electrolyte gated organic field effect transistors. Biointerphases. 2017;12 doi: 10.1116/1.4997760. [DOI] [PubMed] [Google Scholar]
- 23.Fabri-Faja N., Calvo-Lozano O., Dey P., Terborg R.A., Estevez M.C., Belushkin A., Yesilköy F., Duempelmann L., Altug H., Pruneri V., Lechuga L.M. Early sepsis diagnosis via protein and miRNA biomarkers using a novel point-of-care photonic biosensor. Anal. Chim. Acta. 2019;1077:232–242. doi: 10.1016/j.aca.2019.05.038. [DOI] [PubMed] [Google Scholar]
- 24.Gentili D., D’Angelo P., Militano F., Mazzei R., Poerio T., Brucale M., Tarabella G., Bonetti S., Marasso S.L., Cocuzza M., Giorno L., Iannotta S., Cavallini M. Integration of organic electrochemical transistors and immuno-affinity membranes for label-free detection of interleukin-6 in the physiological concentration range through antibody-antigen recognition. J. Mater. Chem. B. 2018;6:5400–5406. doi: 10.1039/c8tb01697f. [DOI] [PubMed] [Google Scholar]
- 25.Hao Z., Pan Y., Shao W., Lin Q., Zhao X. Graphene-based fully integrated portable nanosensing system for on-line detection of cytokine biomarkers in saliva. Biosens. Bioelectron. 2019;134:16–23. doi: 10.1016/j.bios.2019.03.053. [DOI] [PubMed] [Google Scholar]
- 26.Huang D., Ying H., Jiang D., Liu F., Tian Y., Du C., Zhang L., Pu X. Rapid and sensitive detection of interleukin-6 in serum via time-resolved lateral flow immunoassay. Anal. Biochem. 2020;588 doi: 10.1016/j.ab.2019.113468. [DOI] [PubMed] [Google Scholar]
- 27.Khosravi F., Loeian S.M., Panchapakesan B. Ultrasensitive label-free sensing of IL-6 based on PASE functionalized carbon nanotube micro-arrays with RNA-aptamers as molecular recognition elements. Biosensors. 2017;7 doi: 10.3390/bios7020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Militano F., Poerio T., Mazzei R., Salerno S., De Bartolo L., Giorno L. Development of biohybrid immuno-selective membranes for target antigen recognition. Biosens. Bioelectron. 2017;92:54–60. doi: 10.1016/j.bios.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Nie R., Huang J., Xu X., Yang L. A portable pencil-like immunosensor for point-of-care testing of inflammatory biomarkers. Anal. Bioanal. Chem. 2020;412:3231–3239. doi: 10.1007/s00216-020-02582-z. [DOI] [PubMed] [Google Scholar]
- 30.Russell C., Ward A.C., Vezza V., Hoskisson P., Alcorn D., Steenson D.P., Corrigan D.K. Development of a needle shaped microelectrode for electrochemical detection of the sepsis biomarker interleukin-6 (IL-6) in real time. Biosens. Bioelectron. 2019;126:806–814. doi: 10.1016/j.bios.2018.11.053. [DOI] [PubMed] [Google Scholar]
- 31.Sabaté J., Henry O.Y.F., Jolly P., Ingber D.E. An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 2019;14:1143–1149. doi: 10.1038/s41565-019-0566-z. [DOI] [PubMed] [Google Scholar]
- 32.Tang C.K., Vaze A., Shen M., Rusling J.F. High-throughput electrochemical microfluidic immunoarray for multiplexed detection of cancer biomarker proteins. ACS Sens. 2016;1:1036–1043. doi: 10.1021/acssensors.6b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tertis M., Ionut P., Bogdan D., Suciu M., Graur F., Cristea C. Biosensors and Bioelectronics Impedimetric aptasensor for the label-free and selective detection of Interleukin-6 for colorectal cancer screening. Biosens. Bioelectron. 2019;137:123–132. doi: 10.1016/j.bios.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Sun J., Hou Y., Zhang C., Li D., Li H., Yang M., Fan C., Sun B. A SERS-based lateral flow assay biosensor for quantitative and ultrasensitive detection of interleukin-6 in unprocessed whole blood. Biosens. Bioelectron. 2019;141 doi: 10.1016/j.bios.2019.111432. [DOI] [PubMed] [Google Scholar]
- 35.Wei H., Ni S., Cao C., Yang G., Liu G. Graphene oxide signal reporter based multifunctional immunosensing platform for amperometric profiling of multiple cytokines in serum. ACS Sens. 2018;3:1553–1561. doi: 10.1021/acssensors.8b00365. [DOI] [PubMed] [Google Scholar]
- 36.Wu J., Chen Y., Yang Mingzhu, Wang Y., Zhang C., Yang Mo, Sun J., Xie M., Jiang X. Streptavidin-biotin-peroxidase nanocomplex-amplified microfluidics immunoassays for simultaneous detection of inflammatory biomarkers. Anal. Chim. Acta. 2017;982:138–147. doi: 10.1016/j.aca.2017.05.031. [DOI] [PubMed] [Google Scholar]
- 37.Yin B., Zheng W., Dong M., Yu W., Chen Y., Joo S.W., Jiang X. An enzyme-mediated competitive colorimetric sensor based on Au@Ag bimetallic nanoparticles for highly sensitive detection of disease biomarkers. Analyst. 2017;142:2954–2960. doi: 10.1039/c7an00779e. [DOI] [PubMed] [Google Scholar]
- 38.Zhang K., Liu G., Goldys E.M. Robust immunosensing system based on biotin-streptavidin coupling for spatially localized femtogram mL−1 level detection of interleukin-6. Biosens. Bioelectron. 2018;102:80–86. doi: 10.1016/j.bios.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J., He J., Verano M., Brimmo A.T., Glia A., Qasaimeh M.A., Chen P., Aleman J.O., Chen W. An integrated adipose-tissue-on-chip nanoplasmonic biosensing platform for investigating obesity-associated inflammation. Lab Chip. 2018;18:3550–3560. doi: 10.1039/c8lc00605a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo H.J., Park K.K., Han S.S., Chung W.Y., Son M.W., Kim W.B., Surh Y.J. Cytokine serum levels in soft tissue sarcoma patients: correlations with clinico-pathological features and prognosis. Int. J. Cancer. 2002;100:463–471. doi: 10.1002/ijc.10496. [DOI] [PubMed] [Google Scholar]
- 41.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., Marron T.U., Xie H., Patel M., Tuballes K., Van Oekelen O., Rahman A., Kovatch P., Aberg J.A., Schadt E., Jagannath S., Mazumdar M., Charney A.W., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang B.X., Kim H.-Y., Dass C. Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:1237–1247. doi: 10.1016/j.jasms.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Fu B., Xu X., Wei H. Why tocilizumab could be an effective treatment for severe COVID-19? J. Transl. Med. 2020;18:164. doi: 10.1186/s12967-020-02339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in COVID-19: a single center experience. J. Med. Virol. 2020 doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.