Highlights
-
•
The highly pathogenic MERS-CoV, SARS-CoV and SARS-CoV-2 cause acute illness and are often fatal.
-
•
MERS-CoV, SARS-CoV and SARS-CoV-2 tend to infect the respiratory tract and can cause acute respiratory syndrome.
-
•
SARS-related mortality rate is relatively higher than influenza.
-
•
CT scans and RT-PCR are the most promising diagnostic tools for COVID-19 diagnosis.
Keywords: MERS-CoV, SARS-CoV, SARS-CoV-2, Point-of-Care Diagnostics, Biosensors
Abstract
The highly pathogenic MERS-CoV, SARS-CoV and SARS-CoV-2 cause acute respiratory syndrome and are often fatal. These new viruses pose major problems to global health in general and primarily to infection control and public health services. Accurate and selective assessment of MERS-CoV, SARS-CoV and SARS-CoV-2 would assist in the effective diagnosis of infected individual, offer clinical guidance and aid in assessing clinical outcomes. In this mini-review, we review the literature on various aspects, including the history and diversity of SARS-CoV-2, SARS-CoV and MERS-CoV, their detection methods in effective clinical diagnosis, clinical assessment of COVID-19, safety guidelines recommended by World Health Organization and legal regulations. This review article also deals with existing challenges and difficulties in the clinical diagnosis of SARS-CoV-2. Developing alternative diagnostic platforms by spotting the shortcomings of the existing point-of-care diagnostic devices would be useful in preventing future outbreaks.
1. Introduction
A number of potentially fatal viruses have arose in recently [1], [2]. In addition to raising major global public health problems, they have been able to cause massive human mortality [3], [4]. Its global epidemic can pose a risk everywhere because of contemporary life and extensive travel of people and goods [5], [6], [7]. Recently, 3 new viruses have been implicated in severe acute illness, namely Severe Acute Respiratory Syndrome-Corona-Virus (SARS-CoV), Severe Acute Respiratory Syndrome-Corona-Virus 2 (SARS-CoV-2) and Middle East Respiratory Syndrome-Corona-Virus (MERS-CoV) [8], [9], [10], [11], [12]. At present, the SARS-CoV-2 epidemic has disrupted global supply chains, production, tourism, trade and commerce. The automotive industry was especially severely affected. As a result of travel bans in various countries, restauranteurs, travel agencies and public transport service providers have been badly affected [8], [9], [10], [11], [12]. Tourism-dependent countries such as Malaysia and Thailand are indeed struggling financially as scared visitors opt not to venture out. On the other hand, stock markets responded extremely strongly to the pandemic of the SARS-CoV-2 virus [10], [11], [12]. Most companies in Asian countries have implemented stringent guidelines and restrictions to reduce the risks posed by the COVID-19 pandemic to an appropriate level through a confluence of management control and prevention measures [13], [14], [15], [16].
SARS-CoV-2, SARS-CoV and MERS-CoV cause acute illness and are often fatal. Even though lung injury was being reported at all age groups, in certain cases, including the older people or those with co-occurring diseases, the above-foresaid viruses seem to more probable to cause multi-organ failure, acute respiratory distress syndrome and intestinal pneumonia, which are vulnerable to heart failure and high mortality rates [10], [11], [12], [13], [14]. Due to the high mortality rate, it has a twin negative effect: the huge responsibility on the pubic healthcare system, including certain healthcare professionals and the prescribing physician as well as fear of outsourcing one or more of them amongst the public [13], [14], [15], [16].
Usually, the virus reservoir is animal which include chimpanzees, bats or camels. In addition to animal-to-human transmission, human-to-human transmission of SARS-CoV, SARS-CoV-2 and MERS-CoV has been reported, typically from a highly infectious patient to a member of the clinical team, including other hospitalized patients [17], [18], [19], [20], [21]. Although so far no particular therapy has been suggested for its management, supportive therapy appears to have worked better. Antiviral vaccines are currently under development [22], [23], [24]. These new viruses pose major problems to global health in general and primarily to infection control and public health services [25], [26], [27]. In the current scenario, extensive awareness of in-depth education about SARS-CoV-2, SARS-CoV and MERS-CoV and multi-directional care between healthcare professionals and hospitalized patients infected with the above-mentioned viruses can help reduce the spread of these viruses [28], [29], [30]. It is therefore necessary to have an adequate knowledge of its reservoir, its distribution method and the best accessible management as well as preventive measures [31], [32], [33]. Closer monitoring and awareness remain a primary concern for doctors as well as health officials alike [34], [35]. The community spread of the viral infection emphasizes the significance of the clinical diagnosis of illness caused by SARS-CoV-2, SARS-CoV and MERS-CoV. In compliance with the guidelines provided by the Centers for Disease Control and Prevention, the clinicians must organize screening tests for viral infections in patients with nearby health professionals [26], [27], [28], [29], [30]. Traditionally, real-time reverse transcription-polymerase chain reaction (RT-PCR) is the most extensively employed diagnostic tool for diagnosing viral infection caused by SARS-CoV-2, SARS-CoV and MERS-CoV, while viral cultures are not approved [27], [28], [29], [30]. In addition, the viral infection can be identified from a combined effect of symptoms, health issues and a chest computed tomography (CT) scan displaying the characteristics of pneumonia [32], [33], [34], [35]. In this article we have reviewed the literature on various aspects, including the history and diversity of SARS-CoV-2, SARS-CoV and MERS-CoV, their detection methods in effective clinical diagnosis (see Scheme 1 ), diagnostic challenges, safety guidelines recommended by World Health Organization (WHO) and legal regulations. This review article also deals with existing challenges and difficulties in the clinical diagnosis of SARS-CoV-2.
Scheme 1.
Schematic representation of the use of point-of-care diagnostics in the diagnosis of SARS-CoV, MERS-CoV and SARS-CoV-2.
2. History of coronavirus
As of December 2019, few groups of unknown patients with pneumonia were affected by a new coronavirus in China [1]. Temporarily, the WHO coined the new coronavirus as 2019 novel coronavirus (2019-nCoV) on January 7, 2020, unexpectedly came into our sight [2]. Later, the virus was consequently renamed SARS-CoV-2. The SARS-CoV-2 caused disease was named coronavirus disease 2019 (COVID-19) [3], [13], [14]. As of April 11, 2020, more than 1,735,719 patients have been verified positive by nucleic acid screening in China and 209 other countries, and it led to a death rate of 106,558 owing to acute respiratory failure or other respiratory-related complications. Furthermore, more than 392,901 patients all over the world have recovered from coronavirus disease. And, those who recovered from coronavirus disease were confirmed negative by nucleic acid testing [3], [13], [14].
In China, previous outbreaks of evolving infectious diseases have negatively affected the blood supply [1], [2], [3]. But, the safety of the transfusion recipient must be also taken into account, despite the fact that the evolving infection is a respiratory disease. Recent studies suggested that after onset of symptoms, viral ribonucleic acid (RNA) can be identified from serum or plasma of patients infected with SARS-CoV-2, SARS-CoV and MERS-CoV [2], [3], [4], [5]. Identification of viral RNA through polymerase chain reaction, however, is not analogous to identification of intact infectious virus [3], [4], [5], [6], [7]. Though WHO reported in 2003 that almost no instances of SARS-CoV had been recorded owing to the transfusion of blood products, there seemed to be a possible risk of transfusion of SARS-CoV [1], [2], [3], [4]. With increasing asymptomatic infection cases observed among COVID-19 infected patients, blood safety during transfusion is of great importance.
3. Diversity of coronavirus
In addition, coronaviruses are divided into 4, namely δ-CoVs, γ-CoVs, β-CoVs and α-CoVs, amongst which β-CoVs and α-CoVs can infect mammals, while δ-CoVs and γ-CoVs can infect birds [13], [14], [15], [16], [17]. Up to now, 7 coronaviruses have been found to be transmittable through human contact. Among them, HCoV-HKU1, HCoV-NL63, HCoV-OC43 and HCoV-229E cause upper respiratory disease [13], [14], [15], [16], [17]. Sometimes these viruses can cause more serious illnesses in humans, especially in the elderly. Since the 1960s, HCoV-OC43 and HCoV-229E were well known. Subsequently, SARS-CoV, HCoV-NL63 and HCoV-HKU1 were came into attention in 2003, 2004 and 2005, respectively [1], [2], [3], [4], [5]. MERS-CoV, separated in 2012, is analogous to SARS-CoV. Both MERS-CoV and SARS-CoV tend to infect the lower respiratory tract and can potentially cause acute respiratory syndrome.
4. SARS-CoV
Initially, SARS corona (SARS-CoV) appears as a Flu-like illness that advances to respiratory failure, pneumonia and, in certain instances, death [36], [37], [38], [39], [40]. SARS-related mortality rate is relatively higher than influenza or other common infections of the respiratory tract [40], [41], [42], [43], [44]. In 2003, SARS-CoV is identified. SARS-CoV is believed to be an animal virus from an unidentified animal reservoir, possibly bats, which has often spread to several other animals and has been infected for the first time in humans in Guangdong province in southern China in 2002 [44], [45], [46], [47], [48]. In March 2003, the viral infection spread rapidly in Canada, Singapore, Vietnam, Hong Kong and Bejing [49], [50], [51], [52], [53]. An outbreak of SARS significantly impacted 26 nations and led to over 8000 cases in 2003. There have been no documented cases of SARS in almost any part of the world since 2004 [54], [55], [56], [57]. Since then, a small percentage of cases have occurred due to laboratory accidents, or plausibly, transmission from animals to humans. SARS-CoV is predominantly transmitted from one person to another [57], [58], [59].
Auspiciously, it has been shown that the hospitalized patients infected with SARS-CoV are not infectious during the period of incubation [60], [61], [62]. It usually occurs primarily after fourteen days of illness, which is correlated with the peak excretion of virus in faeces and respiratory secretions [60], [61], [62], [63]. Many cases of transmission from human to human occurred in the medical environment, in the unavailability of appropriate precautionary measures to control infection. The implementation of good practices to control SARS-CoV viral infection has ended the international epidemic [63], [64], [65], [66].
The symptoms of SARS-CoV viral infection may include shivering, diarrhoea, headache, myalgia, malaise and fever [56], [57], [58], [59], [60]. No group of symptoms has been reported to date to demonstrate that they are specific to a diagnosis of SARS. Even though fever is perhaps the most commonly reported symptom, it is often absent on preliminary assessment, particularly patients with immunosuppression and in the elderly [62], [63], [64], [65], [66], [67]. In most cases, especially during the first or second week of SARS-CoV viral infection, diarrhoea, shortness of breath and cough are present. Extreme cases frequently develop swiftly, advancing to respiratory distress and necessitating intensive treatment [64], [65], [66], [67], [68], [69].
Accurate and selective assessment of SARS-CoV would assist in the effective diagnosis of infected individual, offer clinical guidance and aid in assessing clinical outcomes (see Table 1 ). Until now, specific 41-base SARS-CoV double-stranded DNA (dsDNA) genome, single biotinylated deoxyribonucleic acid (DNA)/streptavidin-coated sensor chip, S protein/human angiotensin-converting enzyme 2, SARS-CoV specific antibody/protein A and Feline coronavirus (FIP) type I anti-viral antiserum were the reported bio-recognition elements widely used to detect SARS-CoV [70], [71], [72], [73], [74].
Table 1.
Analytical response characteristics of various proposed sensors used for the detection of severe acute respiratory syndrome associated coronavirus.
| Analytical method | Modifier | Target | Detection limit | Assay time | Detection range | Sensitivity | References |
|---|---|---|---|---|---|---|---|
| Micro-cantilever | Feline coronavirus (FIP) type I anti-viral antiserum |
SARS-CoV | 0.1 µg ml−1 | <1 h | 0.1–100 µg ml−1 | – | [70] |
| Surface Plasmon Resonance | SARS-CoV specific antibody/protein A | SARS-CoV antigen | – | – | – | 1.66 × 104 PFU mL−1 | [72] |
| Flexural plate wave | S protein/human angiotensin-converting enzyme 2 | SARS-CoV | – | 60 s | – | – | [73] |
| Surface Plasmon Resonance | Single biotinylated DNA/streptavidin-coated sensor chip | SARS-CoV | – | – | – | – | [71] |
| AlGaN/GaN high electron mobility transistor | specific 41-base SARS-CoV double-stranded DNA (dsDNA) genome | SARS-CoV N protein | 0.003 nM | – | 0.003–3000 nM | – | [74] |
Microcantilevers have evolved since the late 1990s as innovative platforms for sensing biomolecules with on-chip circuitry and excellent sensitivity [70]. The enhanced selectivity of microcantilevers can be accomplished by tailoring the surface of microcantilevers with an engineered biorecognition element capable of binding chemical or biological species of interest. Microcantilevers have the peculiar characteristic of bending under the effect of unequal stresses when the analyte of interest adsorbs to one side of the cantilever. Velankani et al. [70] fabricated an efficient sensing element by modifying the surface of the microcantilever with feline coronavirus (FIP) type I anti-viral antiserum for the detection of SARS-CoV. When the surface engineered microcantilever was exposed to FIP type I virus positive sample, Velankani et al. [70] observed the bending of the microcantilever, asserting the adsorption of FIP type I virus on the surface of the microcantilever. The limit of detection of the developed sensing element was 0.1 µg mL−1. Furthermore, the assay time of the proposed method was less than 1 h [70]. However, this analytical approach has its own limitations. The existence of narrow laser beam in a biological specimen can lead to significant thermal management complexities resulting in erroneous readings.
Recently, surface plasmon resonance (SPR) has arisen as one of the most promising diagnostic (the term diagnostic refers techniques for diagnosing diseases or other problems, while diagnosis is the process of assessing the illness or condition that describes a person’s signs and symptoms) tools in many important fields including food monitoring, drug, biotechnology, environmental monitoring and medicine because of its ability to measure biomolecular interactions without labelling [71]. Conventionally, SPR is extensively employed in protein-protein, RNA-protein and DNA-protein interactions [71]. Nevertheless, RNAs are synthesized with a restricted length for each measurement and purified by HPLC, which involves regular replacement of the chips and makes them expensive. Hence, Yang et al. [71] suggested an alternative approach (one-chip-for-all strategy) based on SPR analysis in which the single biotinylated DNA/streptavidin-coated sensor chip was used as a sensing platform to study the interactions between RNA and SARS-CoV N protein. The estimated dissociation constant of the SARS-CoV N protein – target RNA complex was 4.60 ± 0.3 nM. The proposed diagnostic tool was run at high temperature (37 °C) to ensure swift RNA hybridization. However, the key downside of this method was the increase in noise signal due to the high operating temperature. In order to further improve the sensitivity of SARS-CoV sensor, Wang et al. [72] constructed a novel SPR biosensor which is capable of monitoring reaction sites as well as reference simultaneously in a single flow cell. Wang et al. [72] detected SARS-CoV antigen with the aid of two reference biochips. The sensing element was fabricated by immobilizing SARS-CoV antibody on the surface of one chip, while SARS-CoV antibody was immobilized on the surface of second chip via protein A. Relatively small signal variations occurred when the former was employed to detect SARS-CoV antigen. On the other hand, the second chip showed a noticeable detection signal with a small shift in the reference signal. The fabricated biochip exhibited an enhanced sensitivity of 1.66 × 104 PFU mL−1. Also, the ability of the biochip to distinguish the adsorption of impurities in the analyte was also reported. Although the proposed biochip has improved sensitivity, the detection of SARS-CoV antigen based on SPR has limitations, since it could not clearly differentiate between non-specific and specific interactions with the surface of the sensing element.
In recent times, one-port- and two-port- based interdigitated transducers have received immense attention among researchers in the construction of acoustic wave sensors [73]. In the fabrication of acoustic wave sensors, one port interdigitated transducers are extensively used to conduct surfaces in one path, which in-turn helps to establish resonant cavity to assess the change in resonant frequency. However, designing a vital resonator with a wider bandwidth for biosensor is very difficult [73]. On the other hand, acoustic wave sensors based on two-port interdigitated transducers positioned at opposite sides of a split could easily determine the change in frequency of signal associated to chemical and biological species, delay as well as the attenuation in biosensing applications. Hence, Chang et al. [73] suggested a novel approach of fabricating and integrating the miniaturize system with two port flexural plate sensor for the construction of portable biosensor to detect SARS-CoV in human blood samples. When the immobilized human angiotensin-converting enzyme 2 binds to SARS-CoV through S protein, phase delay has occurred which could provide information on quantitative detection of SARS-CoV in human blood samples [73]. One major limitation of this type of biosensor is the distribution of acoustic energy throughout the surface of the flexible plate wave sensor chip which reduces sensitivity.
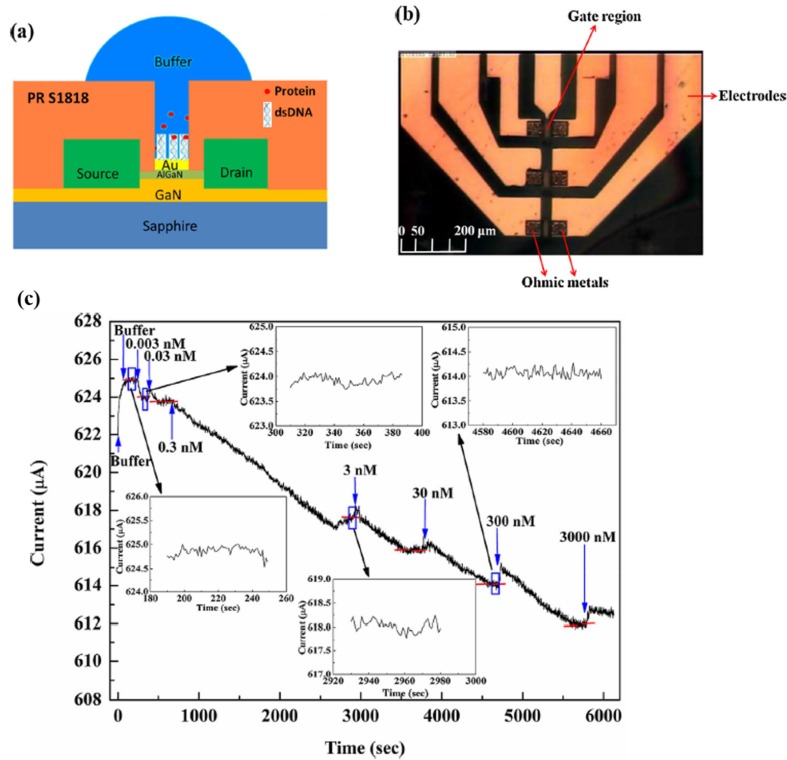
An alternative approach for improving the analytical response characteristics of SARS-CoV biosensor is to increase its sensitivity by enhancing the binding affinity between nucleocapsid protein (N protein) of SARS-CoV and SARS-CoV double-stranded DNA (dsDNA) genome (see Fig. 1 ) [2]. To improve the binding affinity between SARS-CoV dsDNA genome and C-terminal domain of SARS-CoV nucleocapsid protein, Hsu et al. [74] utilized high electron mobility transistors fabricated with two different materials including aluminium gallium nitride and gallium nitride. Hsu et al. [74] correlated the binding affinity between SARS-CoV dsDNA genome and C-terminal domain of SARS-CoV nucleocapsid protein with disassociation constant for the quantitative SARS-CoV detection. Hsu et al. [74] observed that there were at least two binding sites on the SARS-CoV dsDNA genome, which facilitate the binding of nucleotide – protein interaction. Their corresponding disassociation constants were estimated as 0.052 nM and 51.24 nM. It has been found to be transmittable through indirect or direct contact with animals infected with MERS-CoV. In addition, estimation of different IFN-α4 mRNA concentrations with the aid of RT-qPCR was also reported (see Fig. 2 ) [8]. Although high electron mobility transistors based on aluminium gallium nitride and gallium nitride facilitate the binding of nucleotide – protein interaction, the surface potential affects the piezoelectric-induced carrier density in the channel of high electron mobility transistors, leading to changes in the drain current.
Fig. 1.
Investigation of binding affinity between nucleocapsid protein (N protein) of SARS-CoV and SARS-CoV double-stranded DNA (dsDNA) genome using transistors. (a) Schematics of the N protein sensor, (b) plan-view photography of the sensor and (c) real-time detection of the N protein from 0.003 nM to 3000 nM at constant bias of 350 mV. Figures modified from Ref. [2] and used with permission.
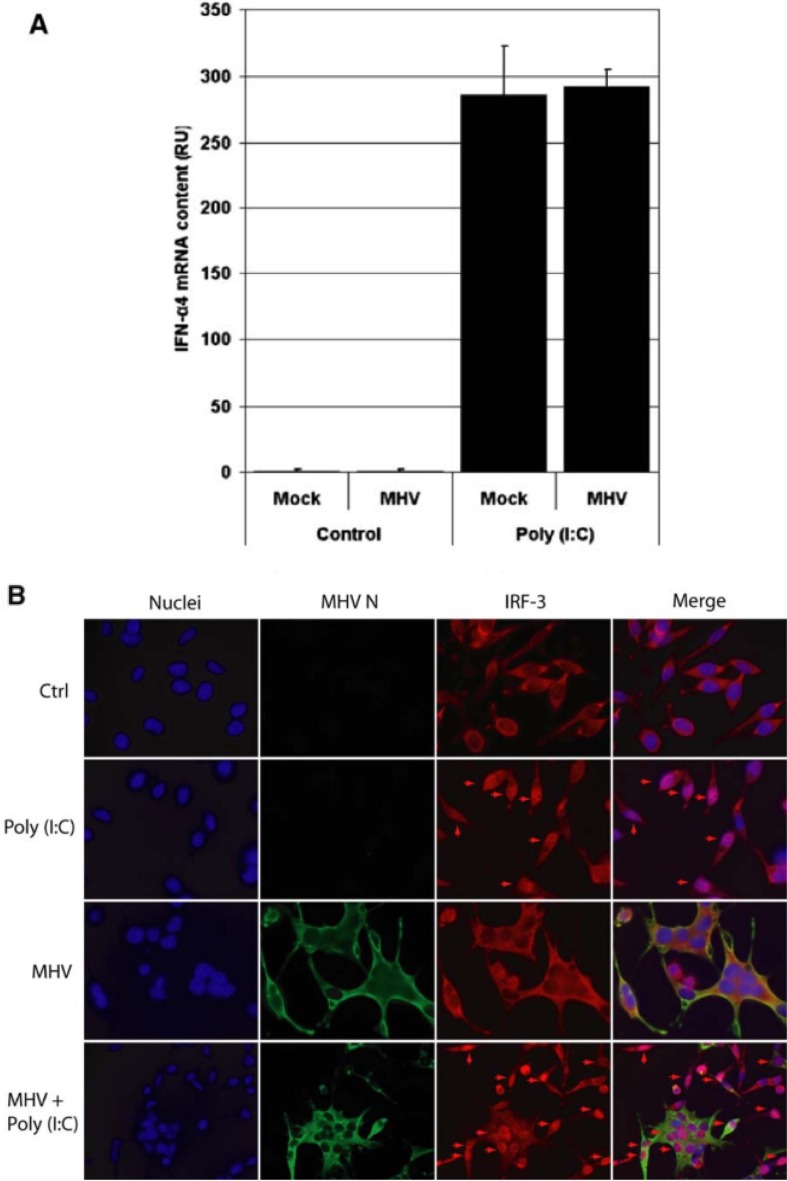
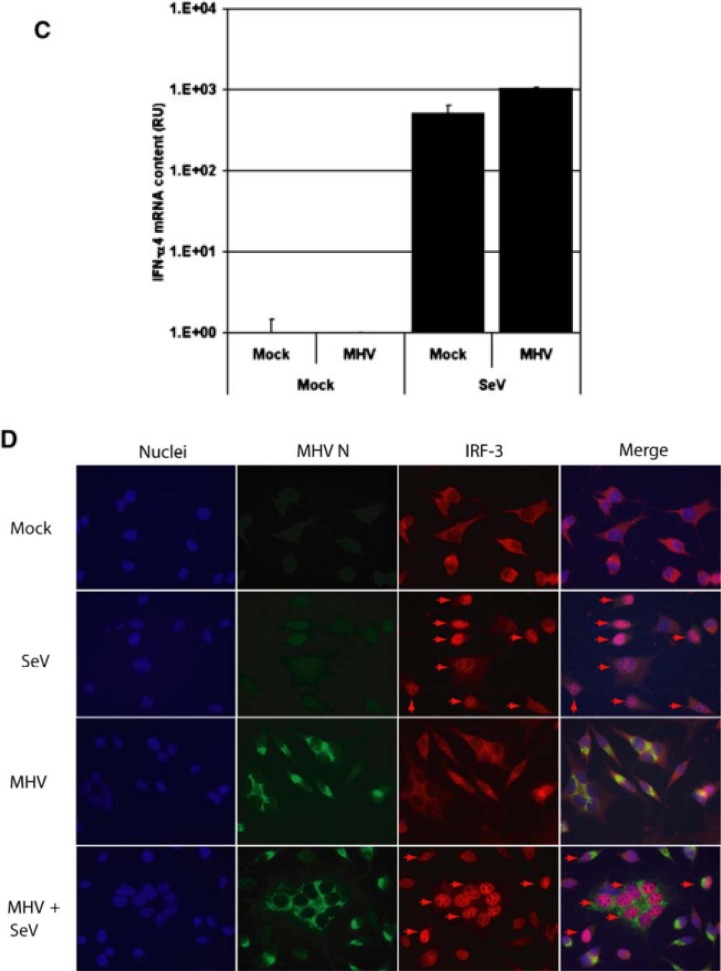
Fig. 2.
L-AC2 cells were infected with MHV at MOI 10. At 1 h p.i., cells were infected with 4 µg poly(I:C). At 8 h p.i. total RNA was isolated and reverse transcribed using random hexamers. (A) IFN-α4 mRNA concentration was determined using specific RT-qPCR and (B) endogenous IRF3 localization and MHV nucleocapsid protein were detected in IFA with specific poly- and monoclonal antibodies, respectively. Nuclear IRF3 is indicated with arrows. Subsequently, L-ACE2 cells were seeded on coverslips in 35-mm wells and infected with SeV for 30 min. Next, cells were infected with MHV at MOI 5. At 8.5 h.p.i. cells on coverslips were fixed in 3% paraformaldehyde. From the remainder of the cells, total RNA was isolated and reverse transcribed using random hexamers. (C) IFN-α4 mRNA concentration was determined using specific RT-qPCR and (D) endogenous IRF3 and MHV nucleocapsid protein were detected in IFA with specific poly- and monoclonal antibodies, respectively. Nuclear IRF3 is indicated with arrows. Figures modified from Ref. [8] and used with permission.
In the early phases of SARS-CoV, protease inhibitors including ribavirin together with ritonavir and lopinavir are used to treat infected patients in antiviral therapy, but the impact of certain steroids and interferon on the control of SARS-CoV infection in patients is not well established and requires further study. Antiviral vaccines and other antiviral therapies are currently under development.
5. MERS-CoV
MERS-CoV, a single-stranded RNA virus, is regarded a lethal virus which often binds to the DPP4 receptor and enters its host cell. Since January 2016, WHO estimated that 1638 people were infected owing to MERS-CoV [75], [76], [77], [78], [79]. About 35% of people diagnosed with MERS-CoV were reported dead. This could be an overestimation of the actual mortality rate, considering that current monitoring systems may ignore mild cases of MERS-CoV. The MERS-CoV was detected in numerous countries including European region, Korea and gulf region with an apparently high mortality rate. Though MERS-CoV is taxonomically analogous to SARS-CoV, it is genetically different from SARS-CoV. MERS-CoV was first confirmed in 2012 Saudi Arabia from a human who fell infected with a flu-like respiratory disease. Subsequently, MERS-CoV was separated from the lung of MERS-CoV infected patient belonging to Egypt. Besides Saudi Arabia, MERS-CoV cases have been confirmed in almost 20 countries including Philippines, Thailand, China, the United States, South Korea, the United Kingdom, Austria, Bangladesh, Algeria, Oman, Turkey, Kuwait, the United Arab Emirates, Egypt, Qatar, and Jordan.
A traveller from Saudi Arabia who visited the United States on May 2, 2014 was diagnosed with MERS-CoV and was the first MERS-CoV case identified in the United States. After 9 days of first instance, the second case of MERS-CoV was reported on May 11, 2014, who is also a traveller from Saudi Arabia. Subsequently, in the Republic of Korea, the first instance of MERS-CoV was identified on May 20, 2015 and the same was reported to WHO on that day, in Korea so far, 185 people have been infected due to MERS-CoV, including 36 deaths. Numerous studies on MERS-CoV have shown that MERS-CoV is a virus transmitted from infected camels to humans. The transmission of MERS-CoV from one infected person to another person is plausible, but few transmissions have been reported between family members residing in the same residence. Even though transmission from human to human is minimal, it persists from patients to healthcare professionals. While most of the reports of MERS-CoV cases have been ascribed to the person-to-person infections in clinical environments, available scientific data strongly suggest that camels may be the source of MERS-CoV infection to the humans. Nevertheless, the primary roles of camels in MERS-CoV transmission and the specific modalities of its transmission remain obscure. When there is no direct interaction between MERS-CoV infected patient and other healthy people, MERS-CoV doesn’t appear to be easily transmitted.
The common symptoms of MERS-CoV involve shortness of breath, cough and fever. In addition, diarrhoea, pneumonia and gastrointestinal-associated illness symptoms have also been reported. Many clinically confirmed cases of diagnosis of MERS-CoV are documented without symptoms, which implies that they show practically no clinical symptoms, but after laboratory examination, they are positive for MERS-CoV infection. Many of these symptomless instances were found after intense assessment of a lab setting-confirmed case. The MERS-CoV usually causes progressively severe illness in the elderly and individuals with debilitated immune systems.
The field of research for MERS-CoV monitoring is a major and positive advancement in the international in vitro diagnostic industries. Early detection seems to be the only way to control the outbreak, which would be a challenge using conventional analytical techniques due to expensive equipment, the need for professionals and low data quality. On the other hand, polymerase chain reaction (PCR) is being used as a promising diagnostic tool for the detection of ultralow levels of human viral pathogens with enhanced specificity and sensitivity. In particular, individuals who are deemed to be infected with MERS-CoV based on their clinical signs are often diagnosed with RT-PCR. Nevertheless, it requires a well-established laboratory and a skilled workforce to operate expensive equipment. Hence, point-of-care testing techniques are essential to fill such gaps by miniaturising the size of device and minimizing the device’s cost and thereby offering fast, simple-to-use diagnostics without extensive training. For instance, Kim et al. [75] proposed a novel diagnostic kit for the detection of MERS-CoV in human blood samples in which double stranded DNA immobilized on self-assembly shielded AuNPs acts as a sensing element for MERS-CoV sensing. The proposed sensor could verify the presence of MERS-CoV in human blood samples based on the change in localized SPR and the alteration of colour of AuNPs when subjected to UV–vis wavelength range. The fabricated sensing element can detect 1 pmol µL−1 of 30 bp MERS-CoV and is also suitable for the on-site diagnosis of several infectious diseases, in particular in environments with limited resources. The main advantage of the proposed analytical approach is that more detailed information can be obtained, as the method uses only double stranded DNA and self-assembled shielded AuNPs.
Integrating biotechnology with nanotechnology plays a significant role in therapeutic interventions, especially in the fabrication of nanobiosensors for the diagnosis of viral infections in human nasal fluid samples. Among various approaches, immunosensors based on interdigitated electrodes have become an attractive choice owing to their superior sensitivity, inexpensive, simplicity of use and potential for miniaturization. Recently, Layqah et al. [76] developed an electrochemical biosensor for the detection of MERS-CoV in human nasal fluid samples by immobilizing AuNPs on the surface of interdigitated carbon electrodes. The sensing mechanism behind the electrochemical detection of MERS-CoV was that there was an indirect competition between the recombinant spike protein S1 of MERS-CoV present in the nasal fluid samples and the immobilized MERS-CoV antibody. The fabricated interdigitated carbon electrode exhibited good linearity in the range of 0.001–100 ng mL−1 with a low detection limit of 1 pg mL−1. The limitation of this approach is the complexity of precise control of the geometry and position of the carbon electrodes.
Alternatively, DNA hydrogel formation by isothermal amplification of complementary targets (DhITACT) has been extensively employed for the early diagnosis of human viral pathogens. However, clogging of microchannels occurs owing to the large number of amplified DNA strands, which limits its sensitivity and detection limit. To resolve the prevailing shortcomings of the conventional DhITACT system and to establish an effective diagnostic platform for testing hospitalized patients infected with MERS-CoV, the conventional DhITACT system must be improved to achieve enhanced sensitivity and reduced assay evaluation period. For instance, Jung et al. [79] proposed an advanced DhITACT system based on fluorescence detection for an effective and rapid diagnosis of MERS-CoV, in which the sensitivity of the DhITACT system was improved by 100 folds when monitored under a UV lamp. Jung et al. [79] fabricated DhITACT system employing chemical vapour deposition technique by depositing functional polymers on stainless steel mesh, followed by the immobilization of DNA primers and hybridization of DNA templates on microfluidic channels. The diagnostic device proposed for the detection of MERS-CoV was very simple and inexpensive compared to other diagnostic platforms currently in use. However, its key drawbacks are overlapping signals owing to the autofluorescence of biomolecules and the existence of background signals when the fabricated DhITACT system is deployed in highly scattering media.
Paper based analytical devices (PADs) are the fast growing technology that has received tremendous interest due to their ease of use, low cost, portability and disposability. So far, PADs have been widely employed as an effective sensing platform for the detection of ultra-low levels of biomolecules in human blood serum samples. Owing to these advantages, Teengam et al. [77] coupled PADs with colorimetric assay for the detection of MERS-CoV by immobilizing pyrrolidinyl peptide nucleic acid (acpcPNA) on the aggregated AgNPs. The sensing mechanism of the proposed point-of-care diagnostic device was that the dispersion of silver nanoparticles resulting from the formation of acpcPNA-DNA complex when subjecting the fabricated sensor to the human blood samples obtained from patients infected with MERS-CoV. Furthermore, one step amplification of viral RNA and rapid diagnosis of MERS-CoV in infected patients using bio-optical sensor have also been reported (see Fig. 3 ) [78]. Despite the remarkable functionalities of paper-based devices, there are certain drawbacks regarding the reliability and simultaneous detection of multiple human viral pathogens.
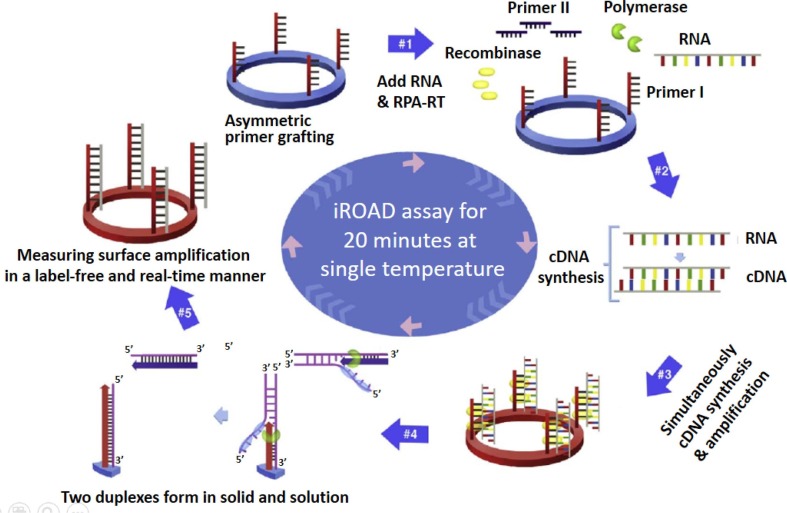
Fig. 3.
Schematic representation of the principle of an isothermal, rapid and label-free one-step RNA amplification/detection (iROAD) assay. First, preparation of the iROAD chip through the primers (forward) grafting on the optical sensor would be needed for a ready-to-use viral RNA detection assay (#1). Then, the mixture containing recombinase polymerase amplification-reverse transcription (RPA-RT) reagents, reverse primers, and extracted RNA is added into the reaction chip (#2). During the isothermal reaction, complementary DNA (cDNA) is synthesized from the RNA template via RNA-RT kit (#3). Thereafter, recombinase/primer complexes bind to double-stranded target cDNA and facilitate strand exchange at a constant temperature. After the displaced strand forms a D-loop by gp32 (sky blue), the immobilized primers are extended by polymerase (light green) on the surface of the silicon microring resonator (#4). The formation of two duplexes is caused by the amplification of the solid and the solution. The exponential RNA amplification after the reverse transcription based on the asymmetric assay is achieved by the repletion of the process (#5). The amplification and detection of the target is simultaneously monitored by measuring the wavelength shift on an optical sensor for 20 min. Figures modified from Ref. [78] and used with permission.
To date, there is no effective vaccine and treatment for MERS-CoV. Successful treatment and accurate diagnosis seems to provide the basis for patient management [78], [79], [80], [81], [82], [83], [84]. Interferon, lopinavir and convalescent plasma are being used to better treat patients diagnosed with MERS-CoV [85], [86], [87], [88], [89], [90]. The most available initial treatment for MERS-CoV is potentially lopinavir and interferon. Although the use of therapeutic corticosteroid has delayed clearance of MERS-CoV, the impact of steroids on the control of MERS-CoV infection in patients is not well established [85], [86], [87], [88], [89], [90]. Furthermore, emetine dihydrochloride hydrate, anisomycin, omacetaxine mepesuccinate, IFN-β, mycophenolate, cycloheximide, cyclosporin A and ribavirin have so far been reported to treat people diagnosed with MERS-CoV [80], [81], [82], [83], [84], [85], [86], [87].
6. SARS-CoV-2
An unseen pneumonia spread swiftly in Wuhan, China, in December 2019, and the majority of the first cases were associated to source infection in a local seafood market [1], [2], [3], [4]. Subsequently, scientists sequenced and recognized a novel β-coronavirus, whose genome is nearly 86.9% similar to the earlier reported SARS-like CoV genome (MG772933.1 and bat-SL-CoVZC45) and seems to differ from MERS-CoV and SARS-CoV. The disease caused by the novel discovered coronavirus is called as COVID-19 [5], [6], [7], [8], [9], [10].
Humans with COVID-19 typically have a fever and the estimated time for incubation is within 2 weeks [11], [12], [13], [14]. Most infected individuals with SARS-CoV-2 virus suffer from chronic respiratory illnesses. The other symptoms of COVID-19 viral infection include sore throat, pains, aches, and shortness of breath. In addition, symptoms such as running nose, nausea and diarrhoea are also reported in few cases [15], [16], [17], [18], [19].
COVID-19 has the ability to wreck human health and lives, corporations, markets and ecosystems as a whole. The mining sector is not very resistant to these effects and the global recession has the potential to have serious repercussions on the industry in the short, medium and long term [1], [2], [3], [4], [5]. For instance, the mining industries in South Africa appear to be the epicentre of SARS-CoV-2. South African mining has been under surveillance for its connection to the outbreak of COVID-19. It has been found that 679 instances of COVID-19 among South African mine workers were reported among 50,000 people across the country [1], [2], [3], [4], [5]. Under constant pressure from labour unions, the mining sector of South Africa adopted strict mining safety regulations in May [1], [2], [3], [4], [5]. As a result, mines have been reopened in South Africa with a capacity of 50% to further prevent the COVID-19 outbreak. Awareness and evaluation of these negative effects for the industrial sector and their impact on economic and social development in the broad sense is a major challenge for scientific research.
Allergy sufferers are undoubtedly nervous and puzzled about what the existing COVID-19 disease outbreak could entail for them in the long and short term [6], [7], [8], [9], [10], [11], [12]. People including kids and elderly with asthma are placed in a higher risk category for COVID-19 disease, however, surprisingly, the available evidence indicated that most of these individuals were not significantly affected [6], [7], [8], [9], [10], [11], [12]. The same scenario was observed in other allergic diseases including atopic dermatitis and allergic rhinitis. This also includes children with allergies. However, there is no proof that kids with chronic allergic illnesses have a greater risk of infection with SARS-CoV-2 than healthy individuals. A persistence of the proven long-term therapy is indeed very necessary for asthmatic patients to retain control of asthma and to be better equipped for potential viral infection caused by SARS-CoV-2 [6], [7], [8], [9], [10], [11], [12]. The World Allergy Organization has framed guidelines by getting feedback from the expert team of healthcare professionals and other scientific experts to prevent panic among allergic patients during COVID-19 outbreak [6], [7], [8], [9], [10], [11], [12].
It is also reported that the elderly as well as those with underlying health issues such as cancer, chronic respiratory disease, diabetes and cardiovascular disease have a greater chance of developing serious illness [20], [21], [22], [23], [24], [25]. In particular, those 80 plus people who have never been vaccinated for pneumonia or flu, who live in facility-based long-term care services and residential care homes, are at greater risk for developing deadly diseases owing to COVID-19 [27], [28], [29], [30], [31], [32]. In addition, COVID-19 has been observed to be more lethal in senior citizens with chronic illnesses including kidney lung and heart if they are infected with HIV or have had cancer. Nearly, 15 percent of them have cough and fever, while 90 percent of people have more than one symptom. It has been urged that anyone over 65 with elevated respiratory symptoms must seek medical advice from the health care professionals [27], [28], [29], [30], [31], [32]. As most visitors to old-age homes may be carriers of the viral infection caused by SARS-CoV-2, visitors are not permitted to enter the old-age homes. Since most elderly people in old-age homes are vulnerable and intellectually disabled with numerous diseases, they are at higher risk of life - threatening illness [27], [28], [29], [30], [31], [32]. The key to decelerating transmission and combating COVID-19 virus is to create awareness of the facts that it typically causes the disease and how well it spreads. The outbreak of newly discovered corona virus began mainly via discharge from the nose or droplets of saliva once an infected individual sneezes or coughs.
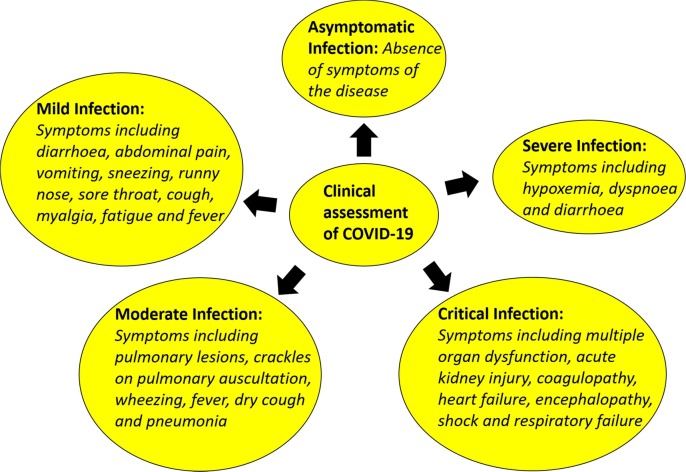
The clinical assessment of COVID-19 disease caused by SARS-CoV-2 is shown in Fig. 4 . The extent of viral infection observed in patients influences the clinical assessment of COVID-19 disease and is classified as follows,
-
•
Asymptomatic infection - absence of symptoms of the disease
-
•
Severe infection - symptoms including hypoxemia, dyspnoea and diarrhoea
-
•
Critical infection - symptoms including multiple organ dysfunction, acute kidney injury, coagulopathy, heart failure, encephalopathy, shock and respiratory failure
-
•
Moderate infection - symptoms including pulmonary lesions, crackles on pulmonary auscultation, wheezing, fever, dry cough and pneumonia
-
•
Mild infection - symptoms including diarrhoea, abdominal pain, vomiting, sneezing, runny nose, sore throat, cough, myalgia, fatigue and fever
Fig. 4.
Clinical assessment of COVID-19.
As per a prospective study of 2143 hospitalized patients identified in the registry of the China Centre for Disease Control and Prevention, 731 instances have been verified using clinical tests [27], [28], [29], [30], [31], [32], [33], [34], [35]. Of these 731 cases, 38.8% were moderately symptomatic, 50.9% were mild symptomatic and 4.4% were asymptomatic. Hardly, 5.8% of patients experienced serious or critical illness [27], [28], [29], [30], [31], [32], [33], [34], [35]. Assessing the level of COVID-19 infection is difficult because the number of overall instances, particularly mild cases, is uncertain in individuals who don’t take medication or are not being monitored.
6.1. Point-of-care immunodiagnostic tests for COVID-19
Due to the rise of COVID-19 outbreak and the scarcity of clinical laboratory-based molecular testing capacities and reagents, several diagnostic test companies have established and started to sell rapid and simple-to-use tools to enhance screening beyond laboratory settings [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. These quick and easy testing kits depend on either the recognition of SARS-CoV-2 viral proteins in respiratory samples or the identification of human antibodies present in blood or serum in response to viral infection [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. Nevertheless, they must be tested in the relevant groups and environments before such assessments can be approved. Insufficient assessments can ignore patients with an active infection or incorrectly classify patients as contracting the disease if they do not, thereby hindering the efforts of combat illness. Based on the established evidence, the WHO currently suggests the use of certain innovative immunodiagnostic point-of-care testing methods in laboratory studies. In addition, the WHO has suggested that these testing kits should be used in any other environment, as well as for clinical decision-making, until data are provided to support their use for early diagnosis [1], [2], [3], [4], [5], [6], [7], [8], [9], [10].
Rapid antigen-based screening tools identify the level of viral proteins expressed by SARS-CoV-2 in a sample from the respiratory tract of a virally infected individual [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. If the level of viral proteins expressed by SARS-CoV-2 is present in the sample at adequate amounts, it binds to specific antibodies attached to a test strip contained in a plastic case and produces a visually measurable signal, usually within 30 min [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. The identified antigens are expressed only if the virus deliberately replicates; thus, these assessments are often used to detect acute or early infection. The exact functioning of assessments depends on many factors, such as the time since the initial infection, the level of virus in the test sample obtained from an individual, and how it has been analysed, and the exact specifications of reagents in the test kits. As seen in the assessments conducted, the sensitivity of such tests could differ greatly from 34% to 80% [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. Based on the evidence, such assessments can eliminate half or more of those hospitalized with COVID-19, completely depending on the population of patient analysed. These hypotheses desperately require additional research to see if they are accurate. Furthermore, false-positive results may arise if indeed the antibodies immobilized on test strips also identify the level of viral proteins expressed by virus in the sample apart from SARS-CoV-2 [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. If some of the antigen detection methods being developed or commercially produced show acceptable efficiency, they may probably be used as a primary care tests to swiftly screen individuals who are very vulnerable to have COVID-19, minimizing or avoiding the need for a molecular confirmatory test. With the available information currently accessible, the WHO does not strongly suggest the use of antigen-detecting swift clinical care diagnostics, despite the fact that research on their clinical and prognostic efficacy is highly recommended [11], [12], [13], [14], [15], [16], [17], [18], [19], [20].
Another more popular form of rapid screening test promoted for COVID-19 is the effective screening test based on the detection of host antibodies [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32]. It is an analytical tool that confirms the prevalence of antibodies in the human blood serum of individuals suspected of being infected with SARS-CoV-2. Antibody production occurs between days and weeks, once after viral infection in healthy individuals [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32]. The efficacy of the response of antibody is based on a number of factors, including age, intake of nutrients, clinical manifestations, as well as other drugs or infections like HIV that affect the immune system. In certain persons with COVID-19, the infection verified by molecular tests (eg. RT-PCR) would have weak, delayed or missing antibody responses [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32]. Research findings indicate that most individuals do not develop antibody responses until the fourteenth day after the onset of symptoms. This implies that a detection of SARS-CoV-2 infection premised on antibody response is always plausible in the recovery phase when several prospects for medical interventions or outbreak of infectious diseases have indeed passed [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32]. Sometimes, antibody tests for COVID-19 may also interact with various other viruses, such as other human coronaviruses, and results in inaccurate tests. Assessments to diagnose antibody responses to COVID-19 in the community will be essential to enable vaccine development and to improve our assessment of the impact of infection among unrecognized persons through active-screening, monitoring activities, population attack rates and infection associated mortality rates. Nevertheless, such testing methods have limited usefulness for clinical diagnosis, as they cannot swiftly identify acute infection to notify decision necessary to evaluate the treatment regimen [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32]. Few health care professionals have employed these antibody response studies to make an early diagnosis of COVID-19 caused by SARS-CoV-2 virus in circumstances where molecular testing was unreliable. However, there was a significant epidemiological correlation to SARS-CoV-2 viral infection and associated blood samples exhibiting elevated levels of antibodies. Based on the existing documentation, the WHO does not strongly advice the use of screening tests for clinical results to detect antibodies, but encourages further work to develop its effectiveness in the fight against the disease and epidemiological research.
6.2. Well-established analytical methods for the identification of COVID-19
Until now, COVID-19 was screened and identified using CT scans, blood culture, enzyme-linked immunosorbent assay (ELISA), immune identification technology and nucleic acid testing. On the other hand, molecular approaches are better suited for reliable diagnosis than CT scan and can classify different pathogens [27], [28], [29], [30], [31], [32], [33], [34], [35]. The implementation of a molecular technique depends on the recognition of (i) the pathogen’s genomic and proteomic composition or (ii) the occurrence of alterations in protein/expression of genes in the individual, often during viral infection [20], [21], [22], [23], [24], [25], [26], [27].
Nucleic acid testing: The existing standard method for COVID-19 diagnosis is the nucleic acid test [1], [3], [15], [18], [31]. A plethora of RT-PCR diagnostic kits have been developed to diagnose the existence of SARS-CoV-2 in infected patients. RT-PCR involves two main steps to assess RNA expression levels. In the first phase, the complementary DNA strands are reverse transcripted from the RNA of SARS-CoV-2, subsequently specific regions of the complementary DNA strands are amplified [1], [3], [15], [18], [31]. Testing, optimization of assays, design of primers and probes and sequence alignment are the primary steps involved in the design process. Recently, few studies on SARS-CoV-2 have been performed to design probes and primers by analysing their genome sequences. So far, 3 regions of SARS-CoV-2 related viral genomes that would retain sequences have been identified. They are (i) nucleocapsid protein gene (N gene), (ii) envelope protein gene (E gene) and (iii) RNA dependent RNA polymerase gene (RdRP gene). Clinical studies on SARS-CoV-2 associated viral genomes indicated that the E and RdRP genes had enhanced analytical sensitivity while the N gene had relatively lower sensitivity for the detection of SARS-CoV-2 [1], [3], [15], [18], [31]. Subsequently, assay conditions are standardized prior to the PCR test, including temperature, incubation time and reagent conditions. Finally, clinical experiments must be performed in the absence and presence of SARS-CoV-2 to guarantee the measurement is accurate and to recognize experimental errors [1], [3], [15], [18], [31].
RT-PCR often uses respiratory samples for the diagnosis of COVID-19. Although samples taken from the lower respiratory tract are highly recommended for hospitalized patients infected with COVID-19, samples collected from the upper respiratory tract are mostly recommended [1], [3], [15], [18], [31]. Nasal aspirates, nasopharyngeal washes, oropharyngeal swabs and nasopharyngeal swabs are samples often collected from the upper respiratory tract. Similarly, samples that are often taken from the lower respiratory tract are tracheal aspirates, BAL fluid and sputum. The amount of SARS-CoV-2 in human blood samples relies on the days after the onset of the disease. SARS-CoV-2 can be identified more precisely in nasal swabs and sputum during the first 14 days after the onset of the illness while, the diagnosis of SARS-CoV-2 in throat swabs is inaccurate 8 days after the onset of symptoms. Due to the difference in viral loads, a negative test resulting from upper and lower respiratory samples doesn’t imply that SARS-CoV-2 is absolutely removed from the infected patient. Such shortcomings may be due to the limited amount of SARS-CoV-2 recognized in the sampled region and inappropriate sampling techniques [1], [3], [15], [18], [31].
Computed Tomography: The Hubei Province, China employed CT scans as an alternative diagnostic tool for detecting SARS-CoV-2 in hospitalized patients due to the false prediction of RT-PCR and the lack of diagnostic kits [1], [7], [16], [18]. Chest CT scan does not cut the skin or does not come into contact with the upper or lower respiratory tract, but takes multiple X-ray measurements around the patient’s chest at various angles to produce cross-sectional images [1], [7], [16], [18]. A chest CT scan could assist in speed up diagnosis and screening, particularly with the shortfalls of RT-PCR. A chest CT scan requires approximately 40 min, including 20 min for the examination and 20 min for the preparatory work [1], [7], [16], [18]. The mean radiation dose used during the chest CT scan ranged from 1 mSv to 10 mSv, depending on the part of the body tested. A low dose of radiation used in chest CT scan for the diagnosis of COVID-19 disease caused by SARS-CoV-2 is generally less than 1 mSv [1], [7], [16], [18]. With the low dose of radiation used in the chest CT scan, the probability of developing cancer from it is so minimal that it cannot be assessed accurately [1], [7], [16], [18]. Nevertheless, in many instances, the limitations involve the radiation exposure requirement and the use of a contrast dye which could pose a health risk to people and seldom cause a skin disorder called systemic nephrogenic fibrosis and kidney problems.
Radiologists analyse the images produced by several X-ray measurements to check for unusual that would result in a diagnosis. The imaging attributes of SARS-CoV-2 were versatile and relied on the phase of viral infection after the onset of symptoms [1], [7], [16], [18]. It has been reported that ground-glass opacities are observed during the initial stage of COVID-19 (approximately 4 days after the onset of symptoms). However, as SARS-CoV-2 progresses, irregular shaped paved stone pattern with ground-glass opacities are identified. Finally, consolidations of the lungs are noted in the cross-sectional images at the later stage after the onset of symptoms. Thus, consolidations of the lungs, ground-glass opacities and irregular shaped paved stone are perhaps the most common prominent features of SARS-CoV-2 that can make chest CT scan popular for diagnosing COVID-19 in infected patients [1], [7], [16], [18]. Numerous clinical trials have exhibited that CT scans have improved misdiagnosis rates and increased sensitivity by 86–98% compared to RT-PCR [1], [7], [16], [18].
In the current scenario, hospitalized patients infected with SARS-CoV-2 are detected using RT-PCR and tested for the presence of SARS-CoV-2 in patients with the aid of CT scans, yet each analytical technique has its own disadvantages [1], [2], [3], [4], [5], [6], [7]. There are several difficulties related to RT-PCR. So far, PCR reagent kits are not available to meet demand. In addition, most rural hospitals outside urban areas do not have RT-PCR facility to handle large number of samples obtained from virally infected patients. Also, RT-PCR depends on the existence of measureable SARS-CoV-2 in the obtained sample. Based on RT-PCR analysis, it is difficult to implement control measures for the asymptomatic patient infected with SARS-CoV-2. On the other hand, diagnostic platforms based on CT scans are costly, need trained personal and cannot accurately diagnose COVID-19 [1], [2], [3], [4], [5], [6], [7]. To overcome these shortcomings, further technologies should be developed for the detection of SARS-CoV-2 [5], [6], [7], [8], [9], [10], [11], [12]. On the other hand, the utilization of bronchoscopy as a screening tool for SARS-CoV-2 is not approved since the aerosol poses a significant risk to both healthcare professionals and patients [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16]. For mechanically ventilated patients whose upper respiratory specimens are negative, bronchoscopy is traditionally recommended while other screening tests could significantly improve the clinical management. Nevertheless, bronchoscopy can be suggested when medical and safety criteria are fulfilled and uncertain diagnoses are made. Optionally, nonbronchoscopic bronchoalveolar lavages and tracheal aspiration could be used in patients infected with SARS-CoV-2 to obtain respiratory samples [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16].
IgM based ELISA kit is used in many countries to qualitatively determine the presence of SARS-CoV-2 in blood serum samples, by testing COVID-19 IgM antibody [5], [6], [7], [8], [9], [10], [11], [12]. Here, microplate based enzyme immunoassay technique is employed to assess COVID-19 IgM antibody. In addition, anti-human IgM specific antibody is coated on the microplate where samples obtained from an individual infected with SARS-CoV-2 and assay controls are added along with COVID-19 specific peptide antigens are introduced. The incubation period often requires more than an hour to facilitate the antigen-antibody interaction. Subsequently, the microplate is washed several times with deionized water to eliminate the unbound protein matrix. To each microtiter wells of the microplate, a horseradish peroxidase (HRP) labelled streptavidin is introduced. If the tested sample obtained from an infected individual contains SARS-CoV-2 IgM antibody, then it forms an immunocomplex after a certain period of incubation. Afterwards, the microplate is washed several times with deionized water to remove the unbound tracer antibody. After removal of the unbound tracer antibody, the immunocomplex bound to the microtiter wells of the microplate is incubated for a certain period of time with a substrate solution of a particular concentration. Finally, spectrophotometry analyses are carried out on the microplate to determine the levels of COVID-19 IgM antibody in the tested sample obtained from an infected person [5], [6], [7], [8], [9], [10], [11], [12].
The need for effective vaccines is vital to reduce the severity of COVID-19 and to minimize human-to-human transmission of SARS-CoV-2, thereby helping to control the coronavirus epidemic [30], [31], [32], [33], [34]. A plethora of strategies are being deployed against SARS-CoV-2 for developing effective vaccines, including protein vaccines and subunit vaccines. Clinical studies are ongoing, however successful development of SARS-CoV-2 vaccines takes few months to several years [27], [28], [29], [30], [31], [32]. On the other hand, Traditional Chinese Medicine appears to have some effect in the treatment of hospitalized patients infected with SARS-CoV-2. Many pharmaceutical drugs including chloroquine phosphate, arbidol, ribavirin, lopinavir/ritonavir, IFNα-2b, teicoplanin, azhithromycin, favipiravir, hydroxychloroquine, remdesivir, are being studied in clinical laboratories to treat SARS-CoV-2 infected patients [30], [31], [32], [33], [34], [35].
6.3. Asymptomatic infection and its diagnosis
With the worldwide coronavirus epidemic, there is ample evidence that various COVID-19 infections are asymptomatic and can transmit the disease to others [91], [92], [93], [94], [95], [96], [97], [98], [99], [100]. Since most of the patients infected with SARS-CoV-2 are asymptomatic, they can only be identified by knowing their past contact information and closely observing the natural course of the COVID-19 disease. Conventionally, RT-PCR shows positive detection of nucleic acid of SARS-CoV-2 in patient samples who are asymptomatic with COVID-19, however, they have no characteristic clinical signs or symptoms and no obvious irregularities in images such as lung computed tomography [1], [2], [3], [4], [5], [6], [7], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100]. Identification of transmission route and early diagnosis of an infected individual are the significant steps to contain COVID-19. Nevertheless, most patients with asymptomatic infections do not seek emergency medical attention because there are no apparent clinical signs and insufficient knowledge of preventive steps, which further make a significant contribution to the high prevalence of COVID-19 [91], [92], [93], [94], [95], [96], [97], [98], [99], [100]. Hence, preventing and managing such a particular type of patient worldwide, who needs more special attention, is a great challenge [1], [2], [3], [4], [5], [6], [7], [8], [9], [10].
COVID-19 patients with asymptomatic infections have the same virulence as infections with symptoms. A study reported that one asymptomatic individual who had SARS-CoV-2 viral infection for a period of 19 days, after confirmation by RT-PCR, could have infected 5 people [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. These asymptomatic instances could play a significant role in the transmission of SARS-CoV-2 and thus pose a major challenge in controlling viral infections. The time interval of initial exposure to the SARS-CoV-2 to the onset of clinical symptoms is considered to be an incubation period and is usually within 14 days [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. During these 14 days, the COVID-19 patients with asymptomatic infections are likely to spread SARS-CoV-2 to healthy individuals. Recent studies have shown that the primary viral infection found in symptomatic patients was identical to that observed in asymptomatic patients, suggesting that COVID-19 patients with asymptomatic infections are capable of transmitting SARS-CoV-2, which could occur early often during infection [91], [92], [93], [94], [95], [96], [97], [98], [99], [100]. The ability of SARS-CoV-2 to infect healthy individuals primarily depends on reproductive state of the virus. On the other hand, positive nucleic acid test results of COVID-19 patients denote that the amount of SARS-CoV-2 in the human blood serum samples eventually reaches a certain maximum limit. In some instances, hyper-sensitive viral nucleic acid re-examination approaches show positive for COVID-19 patients with no apparent clinical signs or symptoms, yet these patients were not responsible for new infections. In another study, it was found that the COVID-19 patients with asymptomatic infections must be quarantined for 2 weeks [91], [92], [93], [94], [95], [96], [97], [98], [99], [100]. Further investigations are also needed to determine the infectivity duration of symptomatic and asymptomatic cases. Every screening tests employed for the early diagnosis of SARS-CoV-2 in asymptomatic cases has its own merits and demerits [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12]. In particular, specificity and selectivity of these diagnostic tool differ significantly from each other, which can lead to false positive and false negative results in patients infected with SARS-CoV-2 who have different stages of COVID-19 disease severity. The analytical response characteristics of the commercially available diagnostic tools are important for screening patients infected with SARS-CoV-2. Such information is critical in establishing control strategies to contain COVID-19 disease [91], [92], [93], [94], [95].
6.4. Role of diagnostics in the testing strategy of COVID-19
Research findings indicate that any individual suspicious of COVID-19 who tests RT-PCR negative must be separated and tested thoroughly 24 h later. After 2 successive negative nucleic acid tests with no evidence of disease, patients who are completely recovered from SARS-CoV-2 viral infection can be discharged from hospital [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]. Nevertheless, if the prevalence of SARS-CoV-2 in these individuals cannot be reliably determined based on clinical signs, then they should live in isolation and monitored 24 h until the SARS-CoV-2 is ruled out or confirmed [25], [26], [27], [28], [29], [30]. Any cases of positive RT-PCR would result in the collective admission and treatment of the patient depending on the extent of the disease. The Italian recommendations referred to Chest X-ray (CXR) as a reliable first-line radiological test in combination with RT-PCR for the surveillance and rapid diagnosis of suspicious COVID-19 patients [31], [32], [33], [34], [35]. Although CT and CXR chest attributes are prevalent in SARS-CoV-2 infected patients and the protocols are relatively easy and fast to achieve results, CXR is not recommended as a screening tool for the first-line alone because it seems to lack specificity. On the other hand, RT-PCR is used as an alternative screening tool to test patients with fever, decreased white cell counts and CT abnormalities for confirmation. Conversely, chest CT and CXR has been recommended for unstable or stable symptomatic individuals as per the Italian guidelines, but have not been taken into consideration for asymptomatic individuals [31], [32], [33], [34], [35]. Numerous studies in both repeated steps and the implementation of various technologies have greatly improved the diagnostic specificity. In a variety of recommendations, the clinical knowledge acquired for the diagnosis of COVID-19 and the control of infections with identical clinical symptoms was considered to be crucial [17], [18], [19], [20], [21], [22], [23], [24]. The way in which the assessments can be integrated to facilitate diagnosis remains unclear, particularly in those who are RT-PCR negative. To overcome this issue, nucleic acids from numerous locations of the specimen can be detected with the help of nucleic acid tests to increase the reliability of the diagnostic test. In addition, chemiluminescence, ELISA and immune-chromatography based serum antibody testing for the early diagnosis of COVID-19 diseases have also been studied to improve the robustness of the diagnostic test [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12]. Although more effort is required in this area, multi-site testing in SARS-CoV-2 affected individuals at the same time can enhance sensitivity and decrease false-negative outcomes.
7. Safety guidelines recommended by World Health Organization
-
•
Regular hand washing with disinfectant-based hand sterilizing solution or alcohol and hot water can help prevent the spread of disease
-
•
Gloves must be of disposable type and can be used for handling biological fluids of the infected individuals
-
•
If you want to live in the same room with someone who is infected with SARS, then you must wear a surgical mask to cover both the mouth and nose
-
•
Eyeglasses can also be worn to protect a person from the spread of viral infection
-
•
Warm water and soap must be utilized to clean the knives, forks, spoons, clothes, bedsheets and blankets of a person infected with SARS
-
•
Discharge from the nose or droplets of saliva an infected individual sneezes or coughs
-
•
Disinfection of exteriors which have been tainted either with discharge from the nose or droplets of saliva when an infected individual sneezes or coughs
-
•
Take all infection prevention measures for at least 10 days, even when the patient is fully recovered and has not shown any signs of the disease
8. Legal regulations
The emergence of SARS-CoV-2 pandemic continues to pose unparalleled regulatory and other legal challenges [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110]. Law and order play a vital role in combating COVID-19 disease outbreak. As countries across the world are working hard to control SARS-CoV-2 virus, numerous social challenges are arising day by day, which necessitate legislative and rational responses [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110]. Owing to the recent WHO statement on the transmission of SARS-CoV-2 which causes COVID-19 in humans, numerous nations have enforced public health policies related to epidemics and pandemics. The national lockdown has emerged as an essential part of the government’s plan to counter the COVID-19 pandemic in many countries [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110]. With companies closed, contracts revoked, scheduled prolonged and supply chains affected, this activity generated significant economic losses in organized sectors. On the other hand, the unorganized sector has experienced a significant breakdown without any legal intervention. While the self-quarantine has enabled to control the transmission of SARS-CoV-2 causing COVID-19, legislative and regulatory investigations of this procedure have evaded inspection so far [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110]. Quarantine is a social exclusion in which individuals subjected to an infectious disease are placed for a certain period of time in order to limit the further transmission of viral infection. As most individual are currently in self-quarantine, it is therefore imperative that national governments should develop contractual jurisdiction to effectively adopt and enforce public health legislation. In 2005, the WHO set out international regulatory guidelines to tackle global health emergencies. The ultimate aim of the International Health Regulation is to monitor the spread of diseases within the countries and to provide basic necessities, foods, commodities of healthcare goods and staff. In addition to that, the Global Health Security Index, a collaborative project initiated by the Johns Hopkins Center for Health Security and the Nuclear Threat Initiative, assesses health safety across 195 countries to resolve outbreaks of infectious diseases that could give rise to international pandemics and epidemics [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110]. As a whole, the Global Health Security Index recognizes significant deficiencies in a country’s potential to suppress, identify and respond to public health crises; substantial dipartites in health care systems; susceptibility to environmental, socio-economic and political uncertainties that may complicate awareness and preparedness plans to outbreaks; and failure to adapt to international standards. In India, the number of confirmed COVID-19 cases is rising day by day [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110]. Despite the general standards of cleanliness, social practice of living in a group and under-resourced public health care system could have the worst possible impact on India [111], [112], [113], [114], [115]. As the number of confirmed COVID-19 cases increasing day by day, on March 12, 2020, the central government of India enacted an Act to adopt adequate steps to control the SARS-CoV-2 pandemic [116], [117], [118], [119], [120]. This urged all union territories and states to impose Section 2 of the Act, which permits the authorities to enact special steps to enforce regulations on deadly infectious diseases. On the other hand, numerous legal and regulatory necessities have been implemented for a commercial organization to assess and equip itself to adapt to this situation. In certain countries, any individual susceptible of transmitting viral infection of any life-threatening disease is liable to fines or even 24 months imprisonment at times [120], [121], [122], [123], [124].
9. Conclusion
Precise and accurate diagnosis aids control transmission of the disease in the early stages of the outbreak. However, most of the diagnostics tools used for the diagnosis of SARS consume time for analysis and require sophisticated laboratory with trained personal. Therefore, the development of portable point-of-care diagnostics that can diagnose SARS in infected patients in seconds is the need of the hour. The next step being worked on is to develop 3-D printed microfluidic diagnostic devices to analyse multiple samples obtained from SARS infected patients with high throughput. Integrating nanotechnology with microfluidic diagnostic systems seems to be an appealing option for the fabrication of micron-sized components in a single platform, making the device portable and cheaper. In the absence of effective vaccines and treatments, the only way to reduce the COVID-19 epidemic is to identify the infected persons with the aid of nano-biosensors and isolate them from other healthy individuals. In particular, diagnosis of SARS in densely populated countries requires low-cost point-of-care diagnostic kits that must be addressed as one of the current challenges. Currently, there are only two well established analytical techniques for diagnosing COVID-19, including CT scans and RT-PCR, however they are expensive and not suitable for point-of-care diagnosis in resource-limited settings. Therefore, it is crucial to find an alternative diagnostic platforms by recognizing the shortcomings of the existing point-of-care diagnostic devices. In addition, these point-of-care diagnostic tools must be easy to use and mass-produced at low cost in large scale so that it can be accessed by a common man in resource-limited settings. We also believe that the integration of point-of-care diagnostic kits with smart phones will ease diagnostic process by automating readings and repositories.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Mrs. Madeshwari Ezhilan thanks Council of Scientific and Industrial Research, New Delhi, India, for the financial support (09/1095(0044)/19-EMR-I). We also acknowledge SASTRA Deemed University, Thanjavur, India for extending infrastructure support to carry out the study.
References
- 1.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Diagnosis of the Coronavirus disease (COVID-19):rRT-PCR or CT? Eur. J. Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu Y.R., Kang Y.W., Fang J.Y., Lee G.Y., Chyi J.I., Chang C.K. Investigation of C-terminal domain of SARS nucleocapsid protein–Duplex DNA interaction using transistors and binding-site models. Sens. Actuators B: Chem. 2014;193:334–339. doi: 10.1016/j.snb.2013.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong Q., Li Z., Shen X., Xu K., Shen Y., Fang Q., Chen F., Liang T. CT imaging features of patients with different clinical types of coronavirus disease 2019 (COVID-19) J. Zhejiang Univ. Med. Sci. 2020;49 doi: 10.3785/j.issn.1008-9292.2020.03.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.L.M. Pandey, Design of engineered surfaces for prospective detection of Sars-CoV-2 using quartz crystal microbalance based techniques, Expert Rev. Proteom. (2020) https://doi.org/10.1080/14789450.2020.1794831. [DOI] [PubMed]
- 6.K. Priyadarshi, V.L. Nag, S.P. Kombade, R.S. Gadepalli, S. Misra, K. Singh, Molecular diagnosis of COVID-19: an update and review, Ann. Natl. Acad. Med. Sci. n.d.;(EFirst). https://doi.org/10.1055/s-0040-1713836.
- 7.Wang K., Kang S., Tian R., Zhang X., Wang Y. Imaging manifestations and diagnostic value of chest CT of coronavirus disease 2019 (COVID-19) in the Xiaogan area. Clin. Radiol. 2020;75:341–347. doi: 10.1016/j.crad.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vessteeg G.A., Bredenbeek P.J., Worm S.H.E., Spaan W.J.M. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virol. 2007;361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falzone L., Musso N., Gattuso G., Bongiorno D., Palermo I.C., Scalia G. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020;46(3):957–964. doi: 10.3892/ijmm.2020.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 11.Loforte A., Gliozzi G., Martin Suarez S., Pacini D. Contributory role of positron emission tomography in a left ventricular assist device recipient at the time of COVID-19 pandemic. ASAIO J. 2020:599–602. doi: 10.1097/MAT.0000000000001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge C., Li M., Li M., Peyghan A.A. Au-decorated BN nanotube as a breath analyser for potential medical applications. J. Mol. Liq. 2020;312 doi: 10.1016/j.molliq.2020.113454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan F., Ghaffar A., Khan N., Cho S.H. An overview of signal processing techniques for remote health monitoring using impulse radio UWB transceiver. Sensors (Switzerland) 2020;20(9) doi: 10.3390/s20092479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.X. Bemtgen, K. Krüger, A. Supady, D. Duerschmied, D. Schibilsky, F. Bamberg, et al., First successful treatment of coronavirus disease 2019 induced refractory cardiogenic plus vasoplegic shock by combination of percutaneous ventricular assist device and extracorporeal membrane oxygenation: a case report, ASAIO J. (2020) 607–609. https://doi.org/10.1097/MAT.0000000000001178. [DOI] [PMC free article] [PubMed]
- 15.Lv D.F., Ying Q.M., Weng Y.S., Shen C.B., Chu J.G., Kong J.P., Sun D.H., Gao X., Weng X.B., Chen X.Q. Dynamic change process of target genes by RT-PCR testing of SARS-CoV-2 during the course of a coronavirus disease 2019 patient. Clin. Chim. Acta. 2020;506:172–175. doi: 10.1016/j.cca.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y.H., Dong J.H., An W.M., Lv X.Y., Yin X.P., Zhang J.Z., Dong L., Ma X., Zhang H.J., Gao B.L. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. J. Infect. 2020;80:394–400. doi: 10.1016/j.jinf.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 18.Li D., Wang D., Dong J., Wang N., Huang H., Xu H., Xia C. False negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning based ct diagnosis and insights from two cases. Korean J. Radiol. 2020;21:505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cordes A.K., Heim A. Rapid random access detection of novel SARS-Coronavirus-2 (SARS-CoV-2, previously 2019-nCoV) using an open access protocol for the Panther Fusion. J. Clinic Virol. 2020;125:104305. doi: 10.1016/j.jcv.2020.104305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghorbel O., Ayedi R., Ben Chikha H., Shehin O., Frikha M. Design of a smart medical bracelet prototype for COVID-19 based on wireless sensor networks. Int. J. Adv. Trends Comput. Sci. Eng. 2020;9(3):2684–2688. doi: 10.30534/ijatcse/2020/30932020. [DOI] [Google Scholar]
- 21.Rane K.P. Design and development of low cost humanoid robot with thermal temperature scanner for COVID-19 virus preliminary identification. Int. J. Adv. Trends Comput. Sci. Eng. 2020;9(3):3485–3493. doi: 10.30534/ijatcse/2020/153932020. [DOI] [Google Scholar]
- 22.Chen H.-Y., Chen A., Chen C. Investigation of the impact of infrared sensors on core body temperature monitoring by comparing measurement sites. Sensors (Switzerland) 2020;20(10) doi: 10.3390/s20102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahmoudi M. Emerging biomolecular testing to assess risk of mortality from COVID-19 Infection. Mol. Pharm. 2020 doi: 10.1021/acs.molpharmaceut.0c00371. [DOI] [PubMed] [Google Scholar]
- 24.Kumar R., Nagpal S., Kaushik S., Mendiratta S. COVID-19 diagnostic approaches: different roads to the same destination. Virus Dis. 2020;31(2):97–105. doi: 10.1007/s13337-020-00599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adhikari S.P., Meng S., Wu Y.J., Mao Y.P., Ye R.X., Wang Q.Z., Sun C., Sylvia S., Rozelle S., Raat H., Zhou H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-190) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9:29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rong X.M., Yang L., Chu H.D., Fan M. Effect of delay in diagnosis on transmission of COVID-19. Math. Biosci. Eng: MBE. 2020;17:2725–2740. doi: 10.3934/mbe.2020149. [DOI] [PubMed] [Google Scholar]
- 27.Poncette A.S., Mosch L., Spies C., Schmieding M., Schiefenhövel F., Krampe H. Improvements in patient monitoring in the intensive care unit: survey study. J. Med. Internet Res. 2020;22(6):e19091. doi: 10.2196/19091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y., Yang G.D., Feng K., Huang H., Yun Y.X., Mou X.Y., Wang L.F. Clinical features and chest CT findings of coronavirus disease 2019 in infants and young children. Chinese J. Contemp. Pediatr. 2020;22:215–220. doi: 10.7499/j.issn.1008-8830.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szunerits S., Saada T.N., Meziane D., Boukherroub R. Magneto-optical nanostructures for viral sensing. Nanomaterials. 2020;10(7):1–12. doi: 10.3390/nano10071271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.P. Pokhrel, C. Hu, H. Mao, Detecting the coronavirus (COVID-19). ACS Sensors https://doi.org/10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed]
- 31.Freire-Paspuel B., Vega-Marino P.A., Velez A., Cruz M., Perez F., Garcia B.M.A. Evaluation of Viasure SARS-CoV-2 RT-qPCR kit (CerTest Biotec) using CDC FDA EUA RT-qPCR kit as a gold standard. MedRxiv. 2020 doi: 10.1101/2020.06.29.20131367. [DOI] [Google Scholar]
- 32.Gao J., Tian Z., Yang X. Breakthrough; chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:1047. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 33.Greiwe J., Nyenhuis S.M. Wearable technology and how this can be implemented into clinical practice. Curr. Allergy Asthma Rep. 2020;20(8) doi: 10.1007/s11882-020-00927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dharavath B., Yadav N., Desai S., Sunder R., Mishra R., Ketkar M. A one-step, one-tube real-time RT-PCR based assay with an automated analysis for detection of SARS-CoV-2. Heliyon. 2020;6(7) doi: 10.1016/j.heliyon.2020.e04405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mujawar M.A., Gohel H., Bhardwaj S.K., Srinivasan S., Hickman N., Kaushik A. Nano-enabled biosensing systems for intelligent healthcare: towards COVID-19 management. Mater. Today Chem. 2020;17 doi: 10.1016/j.mtchem.2020.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu R.W.K., Jin Y., Chung G.T.Y., Lui W.-B., Chan A.T.C., Lim W. Automated extraction protocol for quantification of SARS-coronavirus RNA in serum: an evaluation study. BMC Infect. Dis. 2006;6 doi: 10.1186/1471-2334-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Wu Y.S., Tang Y.P., Li M. Detection of SARS-associated coronavirus N protein by time-resolved fluoroimmunoassay. Di Yi Jun Yi Da Xue Xue Bao. 2005;25(4):429–431. 434. [PubMed] [Google Scholar]
- 38.Meyer B., Drosten C., Muller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L.-L., Hu Z.-H., Wang H.-L., Han X., Deng F. Evaluation of sensitivities and specificities of SARS-CoV detection by real-time quantitative reverse transcription-PCR assays. Virol. Sin. 2009;24(3):187–193. doi: 10.1007/s12250-009-3021-8. [DOI] [Google Scholar]
- 40.Hu Q.H., Liu X.L., Zhuang Z.X., Shi X.L., He Y., Liu J. Modified molecular beacon-based dual real-time PCR for detection of SARS virus and its application. Wei Sheng Yan Jiu. 2005;34(4):416–418. [PubMed] [Google Scholar]
- 41.Liang F.Y., Lin L.C., Ying T.H., Yao C.W., Tang T.K., Chen Y.W., Hou M.H. Immunoreactivity characterization of the three structural regions of the human coronavirus OC43 nucleocapsid protein by Western blot: Implications for the diagnosis of coronavirus infection. J. Virol. Methods. 2013;187:413–420. doi: 10.1016/j.jviromet.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H.H. Sunwoo, A. Palaniyappan, A. Ganguly, P.K. Bhatnagar, D. Das, A.O.S. EI-kadi, M.R. Suresh, Quantitative and sensitive detection of the SARS-CoV spike protein using bispecific monoclonal antibody-based enzyme-linked immunoassay, J. Virol. Methods 187 (2013) 72–78. [DOI] [PMC free article] [PubMed]
- 43.Liu X.E., Li J., Li Y.H., Wang L., Li T., Lu H.Y. Comparison among four kits in detection of anti-SARS-CoV IgG in sera of SARS patients. Zhonghua Liu Xing Bing Xue Za Zhi. 2004;25(6):514–516. [PubMed] [Google Scholar]
- 44.Liu Y., Wang Z., Deng Y.J., Ni A.P., Ma C., Wen J. Reliability of detecting SARS-CoV antibody for diagnosis of SARS. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003;25(5):539–541. [PubMed] [Google Scholar]
- 45.Zhou X., Liu D., Zhong R., Dai Z., Wu D., Wang H. Determination of SARS-coronavirus by a microfluidic chip system. Electrophoresis. 2004;25(17):3032–3039. doi: 10.1002/elps.200305966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drosten C., Chiu L.-L., Panning M., Leong H.N., Preiser W., Tam J.S. Evaluation of advanced reverse transcription-PCR assays and an alternative PCR target region for detection of severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol. 2004;42(5):2043–2047. doi: 10.1128/JCM.42.5.2043-2047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roh C., Jo S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011;86:1475–1479. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J., Wang D., Li Y., Zhao Q., Huang A., Zheng J., Chen W. SARS coronavirus nucleocapsid protein monoclonal antibodies developed using a prokaryotic expressed protein. Hybridoma. 2011;30:481–485. doi: 10.1089/hyb.2011.0028. [DOI] [PubMed] [Google Scholar]
- 49.Hadjinicolaou A.V., Farcas G.A., Demetriou V.L., Mazzulli T., Poutanen S.M., Willey B.M. Development of molecular-beacon-based multi-alleic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): a general methodology for detecting rapidly mutating viruses. Arch. Virol. 2011;156:671–680. doi: 10.1007/s00705-010-0906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrich A., Mahony J., Chong S., Broukhanski G., Gharabaghi F., Johnson G. Multicenter comparison of nucleic acid extraction methods for detection of severe acute respiratory syndrome coronavirus RNA in stool specimens. J. Clin. Microbiol. 2006;44(8):2681–2688. doi: 10.1128/JCM.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guan M., Chen H.Y., Foo S.Y., Tan Y.-J., Goh P.-Y., Wee S.H. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 2004;11(2):287–291. doi: 10.1128/CDLI.11.2.287-291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahony J.B., Petrich A., Louie L., Song X., Chong S., Smieja M. Performance and cost evaluation of one commercial and six in-house conventional and real-time reverse transcription-PCR assays for detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42(4):1471–1476. doi: 10.1128/JCM.42.4.1471-1476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He Q., Manopo I., Lu L., Leung B.P., Chng H.H., Ling A.E. Novel immunofluorescence assay using recombinant nucleocapsid-spike fusion protein as antigen to detect antibodies against severe acute respiratory syndrome coronavirus. Clin. Diagn. Lab. Immunol. 2005;12(2):321–328. doi: 10.1128/CDLI.12.2.321-328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hourfar M.K., Roth W.K., Seifried E., Schmidt M. Comparison of two real-time quantitative assays for detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42(5):2094–2100. doi: 10.1128/JCM.42.5.2094-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang J.C., Chang Y.F., Chen K.H., Su L.C., Lee C.W., Chen C.C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009;25:320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S., Lu D., Zhang M., Che J., Yin Z., Zhang S. Double-antigen sandwich ELISA for detection of antibodies to SARS-associated coronavirus in human serum. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24(8):549–553. doi: 10.1007/s10096-005-1378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan J.F., Li Y., Zhang H.J., Zhou P., Ke Z.H., Zhang Y.Z., Shi Z.L. Indirect enzyme-linked immunosorbent assay based on the nucleocapsid protein of SARS-like coronaviruses. Virol. Sin. 2009;24:146–151. [Google Scholar]
- 58.Manopo I., Lu L., He Q., Chee L.L., Chan S.-W., Kwang J. Evaluation of a safe and sensitive Spike protein-based immunofluorescence assay for the detection of antibody responses to SARS-CoV. J. Immunol. Methods. 2005;296(1–2):37–44. doi: 10.1016/j.jim.2004.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren Y., Ding H.G., Wu Q.F., Chen W.J., Chen D., Bao Z.Y. Detection of SARS-CoV RNA in stool samples of SARS patients by nest RT-PCR and its clinical value. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003;25(3):368–371. [PubMed] [Google Scholar]
- 60.Zhou Z.T., Jiang W.L., Yuan Z.H., Shen F., Qu D., Gu S.M. Detection associated SARS-associated coronavirus infection by reverse transcriptase-polymerase chain reaction, indirect immunofluorescence assay and enzyme-linked immunosorbent assay. World Scientific Publishing Co., Recent works on Microbes and infection in China: Selected from the J. Microbes Infect. China. 2009:117–125. [Google Scholar]
- 61.Chui L., Drebot M., Andonov A., Petrich A., Glushek M., Mahony J. Comparison of 9 different PCR primers for the rapid detection of severe acute respiratory syndrome coronavirus using 2 RNA extraction methods. Diagn. Microbiol. Infect. Dis. 2005;53(1):47–55. doi: 10.1016/j.diagmicrobio.2005.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J.H., Kang M., Park E., Chung D.R., Kim J., Hwang E.S. A simple and multiplex loop-mediated isothermal amplification (LAMP) assay for rapid detection of SARS-CoV. Biochip J. 2019;13:341–351. doi: 10.1007/s13206-019-3404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu X., Cheng G., Di B., Yin A., He Y., Wang M. Establishment of a fluorescent polymerase chain reaction method for the detection of the SARS-associated coronavirus and its clinical application. Chin. Med. J. (Engl) 2003;116(7):988–990. [PubMed] [Google Scholar]
- 64.Shin G.C., Chung Y.S., Kim I.S., Cho H.W., Kang C. Preparation and characterization of a novel monoclonal antibody specific to severe acute respiratory syndrome-coronavirus nucleocapsid protein. Virus Res. 2006;122(1–2):109–118. doi: 10.1016/j.virusres.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J.H., Ma S.X., Ouyang X.L., Wang H.B., Yu Y., Li X.J. Determination and comparison of anti-SARS antibody in children and adults. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2004;12(2):217–219. [PubMed] [Google Scholar]
- 66.Lupia T., Scabini S., Mornese Pinna S., Di Perri G., De Rosa F.G., Corcione S. 2019 novel coronavirus (2019-nCoV) outbreak challenge: a new challenge. J. Global Antimicrob. Resist. 2020;21:22–27. doi: 10.1016/j.jgar.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gralinski L.E., Sheahan T.P., Morrison T.E., Menachery V.D., Jensen K., Leist S.R. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9:e01753–e1818. doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin M.H., Moses D.C., Hsieh C.H., Cheng S.C., Chen Y.H., Sun C.Y., Chou C.Y. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Res. 2018;150:155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Velanki S., Ji H.F. Detection of feline coronavirus using microcantilever sensors. Meas. Sci. Technol. 2006;17:2964–2968. doi: 10.1088/0957-0233/17/11/015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Y., Wang Q., Guo D. A novel strategy for analyzing RNA-protein interactions by surface plasmon resonance biosensor. Mol. Biotechnol. 2008;40:87–93. doi: 10.1007/s12033-008-9066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y.J., Cui D.F., Deng W., Qin C., Wang J.B., Cai H.Y. Research on the detection of SARS-CoV antigen with reference SPR biosensor. Chinese J. Sens. Actuators. 2007;20:2151–2154. [Google Scholar]
- 73.Chang W.Y., Sung P.H., Chu C.H., Shih C.J., Lin Y.C. Phase detection of the two-port FPW sensor for biosensing. IEE Sens. J. 2008;8:501–507. doi: 10.1109/IEMBS.2005.1616467. [DOI] [PubMed] [Google Scholar]
- 74.Hsu Y.R., Lee G.Y., Chyi J.I., Chang C.K., Huang C.C., Hsu C.P. Investigation of the binding affinity of C-terminal domain of SARS coronavirus nucleocapsid protein to nucleotide using AlGaN/GaN high electron mobility transistors. Proc. IEEE Sens. 2012 6411277. [Google Scholar]
- 75.Kim H., Park M., Hwang J., Kim J.H., Chung D.R., Lee K.S., Kang M. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sens. 2019;4:1306–1312. doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- 76.Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tee-ngam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB and HPV oligonucleotides. Anal. Chem. 2017;89:5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koo B., Jin C.E., Lee T.Y., Lee J.H., Park M.K., Sung H. An isothermal, label-free, and rapid one-step RNA amplification/detection assay for diagnosis of respiratory viral infections. Biosens. Bioelectron. 2017;90:187–194. doi: 10.1016/j.bios.2016.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jung Y., You J.B., Choi B.R., Kim J.S., Lee H.K., Jang B. A highly sensitive molecular detection platform for robust and facile diagnosis of Middle East respiratory syndrome (MERS) corona virus. Adv. Healthcare Mater. 2016;5:2168–2173. doi: 10.1002/adhm.201600334. [DOI] [PubMed] [Google Scholar]
- 80.Lu X., Whitaker B., Sakthivel S.K.K., Kamili S., Rose L.E., Lowe L. Real-time reverse transcription-pcr assay panel for middle east respiratory syndrome coronavirus. J. Clin. Microbiol. 2014;52(1):67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corman V.M., Ölschläger S., Wendtner C.-M., Drexler J.F., Hess M., Drosten C. Performance and clinical validation of the RealStar® MERS-CoV Kit for detection of Middle East respiratory syndrome coronavirus RNA. J. Clin. Virol. 2014;60(2):168–171. doi: 10.1016/j.jcv.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Go Y.Y., Kim Y.-S., Cheon S., Nam S., Ku K.B., Kim M. Evaluation and clinical validation of two field-deployable reverse transcription-insulated isothermal PCR assays for the detection of the Middle East respiratory syndrome-coronavirus. J. Mol. Diagnost. 2017;19(6):817–827. doi: 10.1016/j.jmoldx.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hashemzadeh M.S., Rasouli R., Zahraei B., Izadi M., Tat M., Saadat S.H. Development of dual TaqMan based one-step rRT-PCR assay panel for rapid and accurate diagnostic test of MERS-CoV: a novel human coronavirus, ahead of Hajj pilgrimage. Iran Red Crescent Med. J. 2016;18(11) doi: 10.5812/ircmj.23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huh H.J., Kim J.-Y., Kwon H.J., Yun S.A., Lee M.-K., Ki C.-S. Performance evaluation of the PowerChek MERS (upE & ORF1a) real-time PCR kit for the detection of middle east respiratory syndrome coronavirus RNA. Ann. Lab Med. 2017;37(6):494–498. doi: 10.3343/alm.2017.37.6.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee J.-S., Ahn J.S., Yu B.S., Cho S.I., Kim M.J., Choi J.M. Evaluation of a real-time reverse transcription-PCR (RT-PCR) assay for detection of middle east respiratory syndrome coronavirus (MERS-COV) in clinical samples from an outbreak in South Korea in 2015. J. Clin. Microbiol. 2017;55(8):2554–2555. doi: 10.1128/JCM.00667-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim M.-N., Ko Y.J., Seong M.-W., Kim J.-S., Shin B.-M., Sung H. Analytical and clinical validation of six commercial middle east respiratory syndrome coronavirus RNA detection kits based on real-Time reverse-Transcription PCR. Ann Lab Med. 2016;36(5):450–456. doi: 10.3343/alm.2016.36.5.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hecht L.-S., Jurado-Jimenez A., Hess M., Halas H.E., Bochenek G., Mohammed H. Verification and diagnostic evaluation of the RealStar® Middle East respiratory syndrome coronavirus (N gene) reverse transcription-PCR kit 1.0. Future Microbiol. 2019;14(11):941–948. doi: 10.2217/fmb-2019-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fung J., Lau S.K.P., Woo P.C.Y. Antigen capture enzyme-linked immunosorbent assay for detecting middle east respiratory syndrome coronavirus in humans. Methods Mol. Biol. 2020;2099:89–97. doi: 10.1007/978-1-0716-0211-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee K., Ko H.L., Lee E.-Y., Park H.-J., Kim Y.S., Kim Y.-S. Development of a diagnostic system for detection of specific antibodies and antigens against Middle East respiratory syndrome coronavirus. Microbiol. Immunol. 2018;62(9):574–584. doi: 10.1111/1348-0421.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li J., Mao N.-Y., Zhang C., Yang M.-J., Wang M., Xu W.-B. The development of a GeXP-based multiplex reverse transcription-PCR assay for simultaneous detection of sixteen human respiratory virus types/subtypes. BMC Infect. Dis. 2012;12 doi: 10.1186/1471-2334-12-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reijans M., Dingemans G., Klaassen C.H., Meis J.F., Keijdener J., Mulders B. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J. Clin. Microbiol. 2008;46(4):1232–1240. doi: 10.1128/JCM.02294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pierce V.M., Elkan M., Leet M., McGowan K.L., Hodinka R.L. Comparison of the idaho technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J. Clin. Microbiol. 2012;50(2):364–371. doi: 10.1128/JCM.05996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pabbaraju K., Wong S., Tokaryk K.L., Fonseca K., Drews S.J. Comparison of the luminex xTAG respiratory viral panel with xTAG respiratory viral panel fast for diagnosis of respiratory virus infections. J. Clin. Microbiol. 2011;49(5):1738–1744. doi: 10.1128/JCM.02090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang N., Wang L., Deng X., Liang R., Su M., He C. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anderson T.P., Werno A.M., Barratt K., Mahagamasekera P., Murdoch D.R., Jennings L.C. Comparison of four multiplex PCR assays for the detection of viral pathogens in respiratory specimens. J. Virol. Methods. 2013;191(2):118–121. doi: 10.1016/j.jviromet.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xie C., Jiang L., Huang G., Pu H., Gong B., Lin H. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoo S.J., Kuak E.Y., Shin B.M. Detection of 12 respiratory viruses with two-set multiplex reverse transcriptase-PCR assay using a dual priming oligonucleotide system. Korean J. Lab. Med. 2007;27(6):420–427. doi: 10.3343/kjlm.2007.27.6.420. [DOI] [PubMed] [Google Scholar]
- 100.Chen K.-F., Rothman R.E., Ramachandran P., Blyn L., Sampath R., Ecker D.J. Rapid identification viruses from nasal pharyngeal aspirates in acute viral respiratory infections by RT-PCR and electrospray ionization mass spectrometry. J. Virol. Methods. 2011;173(1):60–66. doi: 10.1016/j.jviromet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Timani K.A., Ye L., Ye L., Zhu Y., Wu Z., Gong Z. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J. Clin. Virol. 2004;30(4):309–312. doi: 10.1016/j.jcv.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nalla A.K., Casto A.M., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.R. Konrad, U. Eberle, A. Dangel, B. Treis, A. Berger, K. Bengs, et al., Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020, Eurosurveillance (2020) https://doi.org/10.2807/1560-7917.ES.2020.25.9.2000173. [DOI] [PMC free article] [PubMed]
- 104.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu J., Liu J., Li S., Peng Z., Xiao Z., Wang X. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mei X., Lee H.-C., Diao K.-Y., Huang M., Lin B., Liu C. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jääskeläinen A.J., Kekäläinen E., Kallio-Kokko H., Mannonen L., Kortela E., Vapalahti O. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Eurosurveillance. 2020;25(18) doi: 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.P.B. Van Kasteren, B. van der Veer, S. van den Brink, L. Wijsman, J. de Jonge, A. van den Brandt, et al., Comparison of seven commercial RT-PCR diagnostic kits for COVID-19, J. Clin. Virol. 128 (2020) https://doi.org/10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed]
- 109.Zainol Rashid Z., Othman S.N., Abdul Samat M.N., Ali U.K., Wong K.K. Diagnostic performance of COVID-19 serology assays. Malays. J. Pathol. 2020;42(1):13–21. [PubMed] [Google Scholar]
- 110.Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26(7):771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cui Z., Chang H., Wang H., Lim B., Hsu C.-C., Yu Y. Development of a rapid test kit for SARS-CoV-2: an example of product design. Bio-Des. Manuf. 2020;3(2):83–86. doi: 10.1007/s42242-020-00075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shen L., Huang F., Chen X., Xiong Z., Yang X., Li H. Diagnostic efficacy of three test kits for SARS-CoV-2 nucleic acid detection. J. Zhejiang Univ. Med. Sci. 2020;49(2):185–190. doi: 10.3785/j.issn.1008-9292.2020.03.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.A.I. Khan, J.L. Shah, M.M. Bhat, CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images, Comput. Methods Programs Biomed. 196 (2020) https://doi.org/10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed]
- 114.Mahapatra S., Chandra P. Clinically practiced and commercially viable nanobio engineered analytical methods for COVID-19 diagnosis. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fukumoto T., Iwasaki S., Fujisawa S., Hayasaka K., Sato K., Oguri S. Efficacy of a novel SARS-CoV-2 detection kit without RNA extraction and purification. Int. J. Infect. Dis. 2020;98:16–17. doi: 10.1016/j.ijid.2020.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stanley K. AusDiagnostics SARS-CoV-2 kits shown to be more sensitive than reference laboratory test. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yip C.C.-Y., Sridhar S., Cheng A.K.-W., Leung K.-H., Choi G.K.-Y., Chen J.H.-K. Evaluation of the commercially available LightMix® Modular E-gene kit using clinical and proficiency testing specimens for SARS-CoV-2 detection. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kitagawa Y., Orihara Y., Kawamura R., Imai K., Sakai J., Tarumoto N. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Blairon L., Wilmet A., Beukinga I., Tré-Hardy M. Implementation of rapid SARS-CoV-2 antigenic testing in a laboratory without access to molecular methods: Experiences of a general hospital. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.T. Mahmud, M.A. Rahman, S.A. Fattah, CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization, Comput. Biol. Med. 122 (2020) https://doi.org/10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed]
- 122.Li D., Zhang M., Lin D., Hu Y., Wen Z., Liu S. Evaluation of the sensitivity and specificity of ELISA kits for the SARS-CoV-2 diagnosis. J. Shenzhen Univ. Sci. Eng. 2020;37(3):224–226. doi: 10.3724/SP.J.1249.2020.03224. [DOI] [Google Scholar]
- 123.Sapkal G., Shete-Aich A., Jain R., Yadav P., Sarkale P., Lakra R. Development of indigenous IgG ELISA for the detection of anti-SARS-CoV-2 IgG. Indian J. Med. Res. 2020;151(5):444–449. doi: 10.4103/ijmr.IJMR_2232_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tan M., Ou J., Huang Y., Li Y., Mai J., Ma C. Performance evaluation and clinical application of three antibody test kits for novel coronavirus. Chinese J. Microbiol. Immunol. 2020;40(4):250–255. doi: 10.3760/cma.j.cn112309-20200303-00097. [DOI] [Google Scholar]