Abstract
In the ongoing coronavirus disease 2019 (COVID-19) pandemic, there remain unanswered questions regarding the nature and significance of the humoral immune response toward other coronavirus infections. Here, we investigate the cross-reactivity of antibodies raised against the first severe acute respiratory syndrome coronavirus (SARS-CoV) for their reactivity toward SARS-CoV-2. We extensively characterize a selection of 10 antibodies covering all of the SARS-CoV structural proteins: spike, membrane, nucleocapsid, and envelope. Although nearly all of the examined SARS-CoV antibodies display some level of reactivity to SARS-CoV-2, we find only partial cross-neutralization for the spike antibodies. The implications of our work are two-fold. First, we establish a set of antibodies with known reactivity to both SARS-CoV and SARS-CoV-2, which will allow further study of both viruses. Second, we provide empirical evidence of the high propensity for antibody cross-reactivity between distinct strains of human coronaviruses, which is critical information for designing diagnostic and vaccine strategies for COVID-19.
Keywords: CSARS-CoV-2, antibody, cross-reactivity, spike, nucleocapsid, envelope membrane, COVID-19
Graphical Abstract
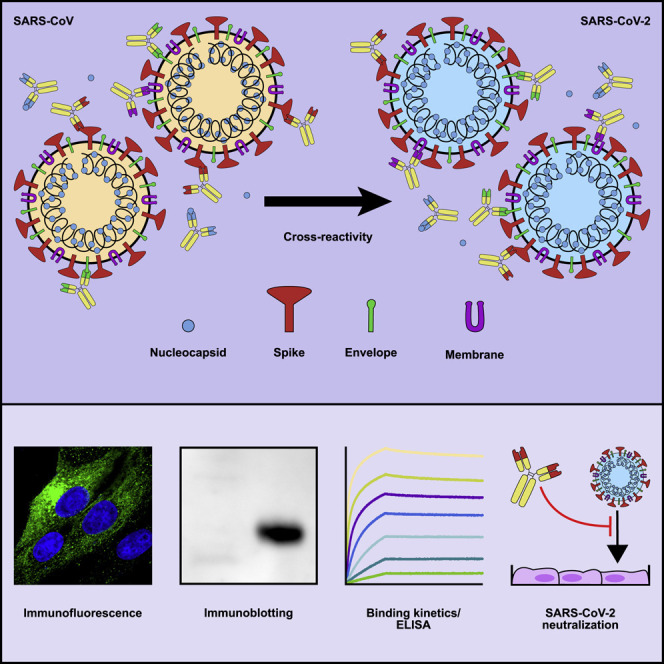
Bates et al. extensively examine the cross-reactivity of selected monoclonal antibodies raised against SARS-CoV structural proteins with SARS-CoV-2. Although most SARS-CoV antibodies display remarkable cross-reactivity toward SARS-CoV-2 proteins, the spike antibodies of SARS-CoV show minimal cross-neutralization of live SARS-CoV-2 virus.
Introduction
The recent emergence of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019 has led to an ongoing worldwide coronavirus disease 2019 (COVID-19) pandemic and public health crisis (Zhu et al., 2020). At the time of writing, there are over 65 million confirmed infections and 1.5 million fatalities worldwide (Dong et al., 2020). SARS-CoV-2 has been designated as a strain of the same species as the original SARS coronavirus (SARS-CoV) because of a high degree of sequence similarity (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). SARS-CoV-2 falls within the family Coronaviridae and can be further subcategorized as a Betacoronavirus of lineage B (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). There is an urgent need for tools to study this novel CoV, as part of the effort to quickly and safely develop vaccines and treatments. One avenue that merits exploration is the repurposing of reagents that were developed for use with SARS-CoV, because many are both extremely effective and commercially available.
CoVs are enveloped, positive-sense, single-stranded RNA viruses with exceptionally large genomes of up to 32 kb on a single RNA molecule. The genomes of most CoVs, including SARS-CoV-2, contain two large open reading frames (which collectively code for 16 nonstructural proteins) in addition to several other open reading frames that are individually responsible for expression of four structural proteins (spike [S], nucleocapsid [N], membrane [M], and envelope [E]) and nine accessory proteins (Khailany et al., 2020). Coronaviridae are a large and diverse family of viruses, with several genera further divided into several lineages, and human (HCoV) and animal CoVs are intermixed within each of these categories (Forni et al., 2017). Of the HCoVs, the SARS-CoVs are most closely related to the lineage C beta-CoV MERS, followed by the lineage A beta-CoVs HCoV-HKU1 and HCoV-OC43, and then the alpha CoVs HCoV-NL63 and HCoV-229E. The lineage A beta-CoVs and the alpha-CoVs are distributed worldwide with seroprevalence exceeding 90% in some studies, although they cause relatively mild disease compared with the rarer acute respiratory syndrome CoVs (Killerby et al., 2018; Severance et al., 2008).
The four SARS-CoV-2 structural proteins are critical for shaping the physical form of the virion, but most available information about them has been extrapolated from other CoVs. Generally, the CoV M protein is involved in shaping the viral envelope membrane (Neuman et al., 2011), the N protein complexes with the viral RNA (McBride et al., 2014), the S protein mediates receptor recognition and membrane fusion (Li, 2016; Zhou et al., 2020), and the E protein contributes to the structure of the viral envelope (Schoeman and Fielding, 2019). Furthermore, several of these CoV structural proteins have been shown to have intracellular functions unrelated to their role as structural proteins (McBride et al., 2014). There are limits to the utility of extrapolation; it is known, for example, that the topology of the CoV E protein varies dramatically among various viruses (Schoeman and Fielding, 2019), and the differences among the receptor binding domains (RBDs) of the S protein can be dramatic. Therefore, tools to interrogate the specific functions of each of the SARS-CoV-2 structural proteins would be of immense and immediate use.
CoV-specific antibodies are one type of tool used in such studies. Antibodies against the SARS-CoV-2 structural proteins could be used as reagents in microscopy and western blotting, as structural tools to probe functional epitopes, and even as antiviral therapies. The protein that produces the greatest SARS-CoV-2-specific antibody response in humans is the viral S protein (Ahmed et al., 2020), but it is known that antibodies are produced against the N, M, and E proteins as well (Ahmed et al., 2020; Severance et al., 2008). Because SARS-CoV and SARS-CoV-2 are such markedly similar viruses, as discussed below, it is reasonable to assume that there may be some cross-reactivity between SARS-CoV antibodies against their cognate SARS-CoV-2 structural proteins, and, indeed, there is already some evidence that this is the case (Lv et al., 2020; Tian et al., 2020; Wrapp et al., 2020; Yuan et al., 2020).
SARS-CoV and SARS-CoV-2 S proteins share 76% amino acid sequence homology, and both rely on cellular angiotensin-converting enzyme 2 (ACE2) as an attachment receptor, as well as the TMPRSS2 protease for priming (Hoffmann et al., 2020). Recent reports have identified cross-reactive antibodies that bind to the S protein of both SARS-CoV and SARS-CoV-2; however, no such cross-reactive antibodies have been identified for the remaining structural proteins (Lv et al., 2020; Tian et al., 2020; Wrapp et al., 2020; Yuan et al., 2020). A non-human-primate model of SARS-CoV-2 DNA vaccination found that a polyclonal antibody response to S alone is sufficient to protect from SARS-CoV-2 challenge, similar to results from a human S-only vaccine trial for SARS-CoV (Martin et al., 2008; Yu et al., 2020). Additionally, convalescent plasma from recovered COVID-19 cases has been broadly shown to reduce mortality of individuals with serious disease (Sullivan and Roback, 2020; Zhang et al., 2020). The sequence similarities between SARS-CoV and CoV-2 N, M, and E proteins are high, at 91%, 90%, and 95%, respectively, making it likely that any individual antibody may be cross-reactive. Indeed, there are reports of human antibodies against the S, N, and M proteins for which the epitopes are identical between SARS-CoV and SARS-CoV-2, further supporting the possibility of cross-reactivity, although none has been experimentally verified (Ahmed et al., 2020).
If cross-reactivity with SARS-CoV-2 is a common feature of SARS-CoV antibodies, then many recovered SARS-CoV patients may still possess SARS-CoV-2 reactive antibodies; antibody responses were shown to remain at high levels for at least 12 years according to a recent preprint (Guo et al., 2020). Although sequence conservation is lower for more common HCoVs, their high prevalence may lead to widespread antibodies with cross-reactivity to SARS-CoV-2. Furthermore, antibodies promoting antibody-dependent cellular phagocytosis have been shown to assist in elimination of SARS-CoV infection, showing that cross-reactive antibodies need not be neutralizing to play a productive role in resolution of CoV infection (Yasui et al., 2014).
This report characterizes a series of SARS-CoV monoclonal antibodies for cross-reactivity, experimental utility, and neutralization of the live SARS-CoV-2 virus. Information about how antibodies from different CoV infections interact is critical for several reasons. It is an important factor to consider during the design of antibody-based CoV tests, particularly for those as closely related as SARS-CoV and SARS-CoV-2. New treatments for SARS-CoV-2 that interact with a patient’s immune system will also need to take into account the prevalence of cross-reactive antibodies as a result of previous CoV infections. Further, information about the basic biology of this novel virus will be critical in developing such tailored treatments, and cross-reactive antibodies could be extremely useful in such studies.
Results
Sequence similarities of the structural proteins of HCoVs
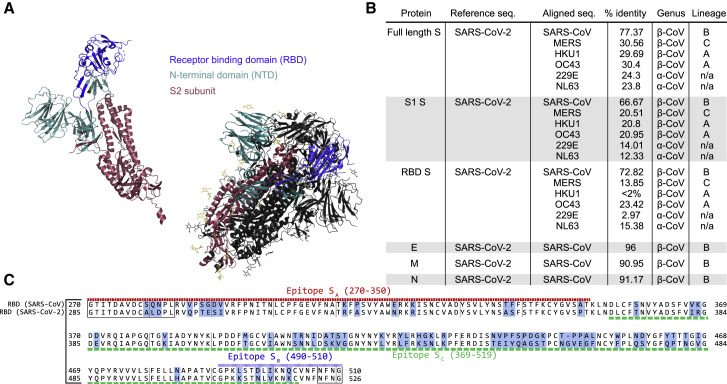
To begin to evaluate structural potential for cross-reactivity, we compared the amino acid sequences of each SARS-CoV-2 structural protein with the homologous protein from the other HCoVs (Figure 1 A). We first looked at the amino acid homology among the S proteins of the common HCoVs and found that other human beta-CoVs (MERS-CoV, HCoV-HKU1, and HCoV-OC43) show only about 30% similarity to the SARS-CoV-2 S protein, and human alpha-CoVs (HCoV-229E and HCoV-NL63) show only about 24% similarity to SARS-CoV-2 S protein. The S protein of the original SARS-CoV, however, is much more closely related, showing 77% similarity between SARS-CoV and SARS-CoV-2, which lends support to the idea that anti-SARS-CoV S antibodies could be cross-reactive with the SARS-CoV-2 S protein. The E, M, and N protein sequences show striking similarity between SARS-CoV and SARS-CoV-2; they are 96%, 91%, and 91% similar, respectively (Figure 1A).
Figure 1.
Sequence similarity between SARS-CoV and SARS-CoV-2
(A) Similarity scores for each of the SARS-CoV-2 structural proteins compared with SARS-CoV and other common coronaviruses. Similarity in the S protein is substantially lower than for the other structural proteins.
(B) Sequence alignment of the receptor binding domain (RBD) of SARS-CoV and SARS-CoV-2. Regions of difference are highlighted in blue, while the epitopes of the antibodies used in this study are underlined according to their designations in Table 1. The boxed regions fall outside of the canonical RBD sequence but are included because of overlap with the above epitope regions.
(C) Crystal structure of the S protein color coded by domain both as monomer and in the functional homotrimeric form in which one of the monomers is colored, while the other two are shown in white. As shown, the NTD and RBD compose the majority of the S1 region.
See also Figure S1.
The Biodefense and Emerging Infections (BEI) Research Resources Repository has available several types of antibodies and immune sera against each of the structural SARS-CoV proteins, as well as whole virus (summarized in Table 1 ). Eight of these are mouse monoclonal antibodies (240C, 341C, 540C, 154C, 472C, 19C, 283C, 42C) of either the IgM, IgG2a, or IgG1 class, recognizing either the SARS-CoV E, M, N, or S proteins. Of these, only two are neutralizing, 341C and 540C (Tripp et al., 2005). There are also polyclonal rabbit sera against the SARS-CoV S protein and an anti-S monoclonal human IgG1 antibody (CR3022) isolated from a SARS-CoV patient (ter Meulen et al., 2006), all of which are neutralizing.
Table 1.
SARS-CoV antibodies utilized by this study
| Antibody | Reference | Protein specificity | Species | Class | Neutralization of SARS-CoV | Epitope |
|---|---|---|---|---|---|---|
| 240C | Tripp et al. (2005) | S | mouse | IgG2a | no | SA (490–510) |
| 341C | Tripp et al. (2005) | S | mouse | IgG2a | yes | SA (490–510) |
| 540C | Tripp et al. (2005) | S | mouse | IgG2a | yes | SA (490–510) |
| 154C | Tripp et al. (2005) | S | mouse | IgM | no | SB (270–350) |
| CR3022 | (ter Meulen et al., 2006) (GenBank: DQ168569 and DQ168570) | S | human | IgG1 | yes | SC (369–519) |
| NRC-772 | Made by BEI | S | rabbit | serum | yes | — |
| 472C | Tripp et al. (2005) | E | mouse | IgM | no | |
| 19C | Tripp et al. (2005) | M | mouse | IgM | no | |
| 283C | Tripp et al. (2005) | M | mouse | IgG1 | no | |
| 42C | Tripp et al. (2005) | N | mouse | IgM | no |
Although antibodies that recognize each of the structural proteins are of interest as experimental tools, antibodies that recognize the S protein are particularly so because of their potential to neutralize infectious virus. Structural information about the specific biochemical interactions between S-specific antibodies and the S protein is of great value. For the anti-S monoclonal antibodies (through BEI Resources) described in Table 1, the epitopes can be traced to one of three regions of the RBD. Whereas 240C, 341C, and 540C all bind within a region at the end of the RBD (epitope SA) (Tripp et al., 2005), the 154C antibody binds to a region at the beginning of the RBD (epitope SB); and the human monoclonal antibody CR3022 binds to specific residues in a broad region in the middle of the RBD (epitope SC) (Yuan et al., 2020). These epitopes are indicated in Figure 1B, along with the alignment of the SARS-CoV and SARS-CoV-2 RBDs. Although not identical, these regions do show some level of similarity between the two virus strains. The three-dimensional structure of the S protein in both monomeric and the functional trimeric form is displayed to illustrate the general accessibility of each portion of the protein (Figure 1C).
Antibodies of the SARS-CoV structural proteins show cross-reactivities with SARS-CoV-2 by microscopy
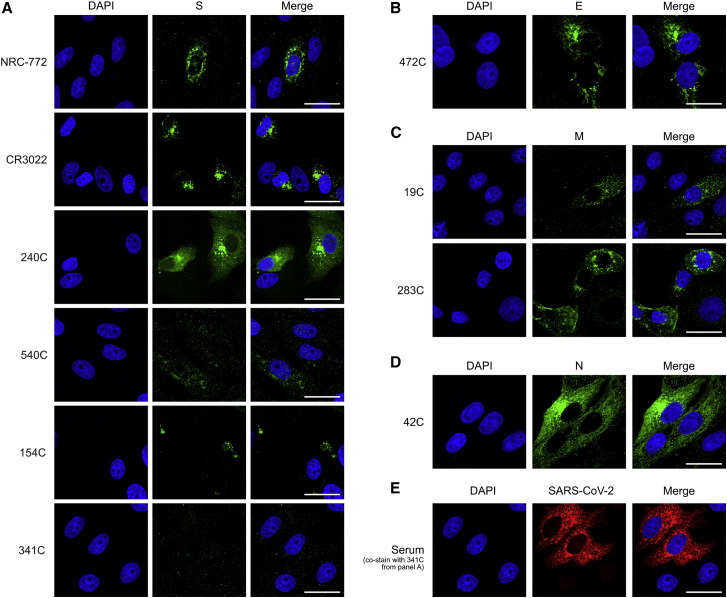
To assess SARS-CoV antibodies against SARS-CoV-2, we first performed immunofluorescence (IF) staining of Vero E6 cells infected with live SARS-CoV-2 virus (Figure 2 ). The S-specific antibodies NRC-772, CR3022, and 240C all showed strong staining, whereas 540C and 154C showed weak staining. Antibody 341C showed no staining. The E-specific (472C), M-specific (19C and 283C), and N-specific (42C) antibodies all displayed robust staining. We confirmed the presence of SARS-CoV-2-infected cells by co-staining with human convalescent serum, which demonstrates that negative 341C staining is not due to a lack of infection. To further validate the utility of these antibodies for IF, we performed staining of 293T cells transiently transfected with Strep-tagged constructs of each of the individual SARS-CoV-2 structural proteins (Gordon et al., 2020). We compared the staining of the strep-tag within each structural protein in IF against that of the experimental antibodies, finding that the staining pattern of a majority of these antibodies is detectable, with some being highly similar to the strep-tag antibody (Figure S2). These results match our findings for the live SARS-CoV-2 infection; however, 42C (N-specific) showed markedly reduced staining of transiently transfected cells. Together, these antibodies provide complete coverage of SARS-CoV-2 structural proteins, showing their utility for SARS-CoV-2 experiments involving microscopy.
Figure 2.
Immunofluorescence of SARS-CoV-2 structural proteins using SARS-CoV antibodies
(A–D) Representative immunofluorescence images of Vero cells infected with SARS-CoV-2. 24 h post-infection, cells were fixed and stained with the listed SARS-CoV antibodies (green): (A) spike, (B) envelope, (C) membrane, and (D) nucleocapsid.
(E) 341C was co-stained with human convalescent serum (red) to confirm the presence of infected cells. Scale bars, 30 μm. DAPI (blue) was used to visualize cell nuclei.
See also Figure S2.
Antibodies of the SARS-CoV structural proteins show cross-reactivities with SARS-CoV-2 by immunoblotting
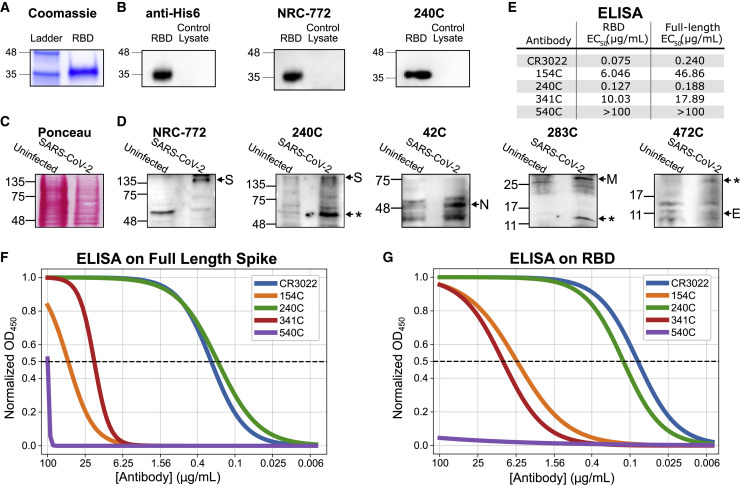
We next evaluated these antibodies by western blot. His6-tagged RBD from SARS-CoV-2 was produced in HEK293 cells and purified by Ni-NTA chromatography (Figure S3A). The purified RBD was then used for a western blot with each of the mouse monoclonal antibodies (Figures 3A and 3B). Anti-His6 antibody demonstrates high purity of the RBD protein. The staining produced by each experimental antibody was compared with lysate from untransfected 293T cells to assess background. Of these antibodies, 240C and NR-772 produced strong signal with little background, whereas the other monoclonal antibodies (CR3022, 154C, 341C, and 540C) did not produce detectable signal.
Figure 3.
Biochemical characterization of SARS-CoV antibodies for their cross-reactivity with SARS-CoV-2 proteins
Characterization of the S-specific antibodies by western blot and ELISA.
(A) Coomassie stain of in-house-purified His6-tagged RBD protein produced in HEK293-F suspension cells and purified by Ni-NTA chromatography.
(B) Western blot of purified RBD with anti-His6 antibody and SARS-CoV S-specific antibodies.
(C) Ponceau stain of SARS-CoV-2-infected and uninfected Vero E6 cell lysate.
(D) Western blot of SARS-CoV-2-infected lysate probed with SARS-CoV structural protein-specific monoclonal antibodies. Shown are representative images of two to three independent experiments.
(E) Summary table of observed EC50 values from both sets of ELISAs.
(F) ELISA on purified full-length spike coated at 2 μg/mL.
(G) ELISA on purified RBD coated at 2 μg/mL. n = 3 (each done in triplicates). Asterisks indicate an expected alternate band.
See also Figures S3 and S4.
We also performed western blots on SARS-CoV-2 (Isolate USA-WA1/2020)-infected and uninfected Vero E6 cell lysates. Probing with human convalescent serum revealed bands at the expected size for each of the structural SARS-CoV-2 proteins: S, N, M, and E (Figure S3D). The 42C, 540C, NRC-772, 240C, and 283C antibodies each developed bands unique to the SARS-CoV-2-infected samples; however, not all of the bands were at the expected molecular weight. Previous reports have shown that SARS-CoV-2 proteins, including S, N, M, and E, all produce bands at several different molecular weights when expressed exogenously in HEK293T cells, and it is not surprising that these bands also exist in our blots (Gordon et al., 2020). What is unexpected is that for S monoclonal antibodies 240C, 540C, and 283C, we detect the lower molecular weight band (∼50 kDa) and not the band at the expected molecular weight (Figure S3). This could be because of masking of the epitope by glycosylation absent from the truncated protein or specific recognition of proteolytically cleaved peptides. For instance, proteolytic cleavage of the S protein is known to be important for proper maturation of SARS-CoV-2 particles (Hoffmann et al., 2020; Walls et al., 2020). The N monoclonal antibody 42C is able to detect a band at the correct molecular weight (∼46 kDa); however, there is a moderate level of background (Figure 3D). The M monoclonal antibody 283C detects a band at the expected molecular weight (∼25 kDa) but also detects a lower molecular weight band similar to what has been shown in previous reports that showed that the M protein is particularly prone to proteolytic degradation in western blots, as well as a high molecular weight smear (Gordon et al., 2020). The M monoclonal antibody 19C shows similar staining, but weaker, and both display a high level of background staining. The E monoclonal antibody 472C displays a weak band at the expected molecular weight (∼8 kDa), but the intensity is similar to that of background, so a positive determination cannot be made.
S antibodies show cross-reactivity in binding
The fact that the S glycoprotein is responsible for virus binding and entry into host cells makes it an attractive target for antibody generation because some of these antibodies may be neutralizing. Because of the potential functional role for these antibodies, and because of the number of different antibody clones, we decided to examine the S-protein-specific antibodies more thoroughly.
We assessed the binding of the S-protein-specific antibodies to both the full-length SARS-CoV-2 S protein and the purified RBD by ELISA (Figures 3F, 3G, and S4; summarized in Figure 3E). The CR3022 and 240C antibodies showed strong binding to both the full-length S and the RBD (EC50 75 and 127 ng/mL, respectively, to the RBD). 154C and 341C showed weak but detectable binding (6.046 and 10.03 μg/mL, respectively, to the RBD), whereas 540C did not demonstrate binding at all. The trend for these antibodies is generally similar to what was seen in the previous studies, where the antibodies were tested against recombinant SARS-CoV S protein (Tripp et al., 2005). The original report of CR3022 did not perform a direct ELISA for us to compare with our results; however, our data agree with studies of CR3022 on SARS-CoV-2 showing that it binds strongly to both full-length S and the RBD (Yuan et al., 2020). Although CR3022 appears to be the strongest binder to RBD, 240C is marginally better on the full-length S protein.
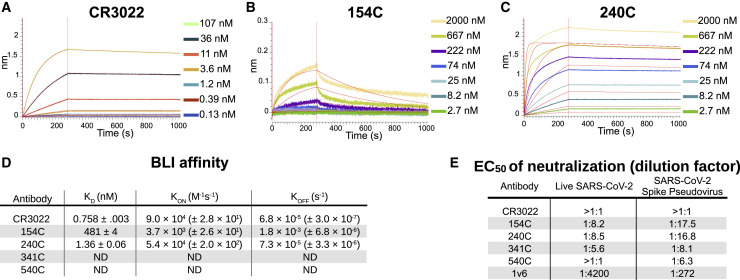
To assess the binding kinetics of the antibody-RBD interaction in more detail, we measured the antibody-epitope interactions using biolayer interferometry (BLI). The three monoclonal antibodies that showed the strongest binding with the ELISA displayed high affinity for the SARS-CoV-2 RBD: CR3022 showed the strongest binding with a calculated KD of 758 pM (Figure 4 A), while 240C demonstrated a 1.36 nM KD (Figure 4B) and 154C a 481 nM KD (Figure 4C). As summarized in Figure 4D, these antibodies showed fast-on/slow-off kinetics in agreement with a previous report of CR3022 binding kinetics on RBD (Yuan et al., 2020). The other antibodies we tested displayed no measurable binding at the highest concentration used (Figure S5). Importantly, BLI does not account for the avidity of these antibodies, and it is likely that the interaction of each epitope/paratope pair is substantially lower than that of the intact antibody; however, the intact antibody more closely resembles the interaction that is likely to occur in most in vitro assays, or indeed in vivo. Our KD is substantially lower than reported in Tian et al. (2020); however, this is likely due to differences in the reagents used. Tian et al. (2020) expressed their RBD in E. coli, preventing glycosylation, while our RBD was produced in mammalian cells. Additionally, their CR3022 was produced as a single-chain variable fragment (scFv) in E. coli, which would contain only a single paratope and may fold differently from our full CR3022 antibody, which was produced in a plant expression system, is bivalent, and contains intact constant domains.
Figure 4.
Binding kinetics and functional testing of Spike-specific antibodies against the RBD of the SARS-CoV-2
(A–C) Biolayer interferometry curves for CR3022, 154C, and 240C with 3-fold dilutions. Streptavidin biosensors were coated with biotinylated RBD, then blocked with 1 μM D-Biotin in kinetics buffer. Negative binding curves for 341C and 540C shown in Figure S5. Curve fitting was performed using 1:1 binding model in ForteBio Analysis HT 10.0 software.
(D) Summary of quantified binding kinetics of Spike monoclonal antibodies from BLI experiment.
(E) Neutralization assay 50% neutralization values against live SARS-CoV-2 by focus-forming assay and SARS-CoV-2 spike pseudotyped lentivirus by fluorescence microscopy. EC50 values represent triplicate experiments (n = 3). 1v6 is positive control from COVID-19 patient convalescent serum collected at day 14. The concentration of all monoclonal antibody stocks was 1 mg/mL. 154C and 240C showed only partial neutralization at the highest concentration tested (1:10 dilution), whereas 341C, 540C, and CR3022 failed to reliably neutralize pseudotyped virus at this dilution.
See also Figures S5 and S6.
S antibodies of SARS-CoV show limited cross-neutralization of SARS-CoV-2
Finally, we assessed the neutralizing capabilities of these S protein-specific monoclonal antibodies. We set up a neutralization assay using a Lentivirus GFP-reporter pseudotyped with the SARS-CoV-2 S protein (Crawford et al., 2020). Neutralization was assessed by quantitative fluorescent microscopy, using the area of GFP expression compared with that of an antibody-untreated control. Serial dilutions of antibodies were used to generate neutralization curves and estimate the antibody concentration necessary for 50% neutralization. This readout was used because the monoclonal antibodies displayed only partial neutralization at the highest concentration used in our assay. To validate our assay, we used human convalescent serum from a SARS-CoV-2-positive patient. This anti-serum demonstrated 50% neutralization at a dilution of 1:270 (Figure 4E). Consistent with a previous report, CR3022 failed to show any neutralization at 100 μg/mL despite its potent binding in every other assay (Yuan et al., 2020). 154C and 240C both showed partial neutralization, with a 50% reduction in GFP area at 57.8 and 61.3 μg/mL, respectively. Consistent with the BLI results, 341C and 540C did not show substantial neutralization. We were surprised to see 154C perform the best in this assay, particularly because the original report of these antibodies on SARS-CoV showed 341 and 540 as the only antibodies with neutralizing capabilities. One unique aspect of 154C is that it is the only IgM antibody from this selection of S-specific antibodies; however, it is not clear how this might affect neutralization (Yuan et al., 2020).
To further validate our pseudotyped lentivirus neutralization data, we set up focus-forming assay (FFA)-based neutralization studies using live SARS-CoV-2 (Isolate USA-WA1/2020) as previously described (Case et al., 2020). The virus was titrated such that each well received 30 plaque-forming units (PFUs)/well, which was pre-incubated for 1 h with antibody dilutions starting at 1:10 down to 1:1,280. The results from this assay were broadly similar with those seen in the pseudovirus neutralization assay, with 240C and 154C showing partial neutralization, and 341C, 540C, and CR3022 showing minimal neutralization (Figures 4E and S6). The human convalescent serum from a SARS-CoV-2 patient (1v6) performed better in the FFA, whereas the monoclonal antibodies each performed slightly less well than in the pseudotype neutralization assay. The reasons for this variation may be because of the substantial differences between the design of these two assays, including the cell type, virus type and quantity, and detection method. Despite these differences, the similar neutralizing trends in both assays show limited cross-neutralization of SARS-CoV-2 by the S monoclonal antibodies of SARS-CoV.
Summary of cross-reactivity of SARS-CoV structural protein-specific antibodies to SARS-CoV-2 structural proteins in various assays
The utility of each of the antibodies used in this study has been summarized in Table 2 . In particular, the S protein-specific 240C performed well in every assay we performed, excluding neutralization. In contrast, 540C showed no detectable binding in any of our assays. The other S protein-specific monoclonal antibodies 154C, 341C, and CR3022 showed mixed utility in different assays (Table 2). The rabbit polyclonal antibody NRC-772 also worked in every assay in which it was tested; however, polyclonal sera is limited to experiments where structural information about particular epitopes is not important due to the unknown admixture of the contained antibody clones. The antibodies against E, M, and N demonstrated utility in IF and showed some success in western blots in the case of the 42C and 283C antibodies (Table 2). Further studies could explore these antibodies in greater detail by producing purified E, M, and N proteins for use in biochemical assays, such as the ones we used to characterize the S protein-specific antibodies in this report.
Table 2.
Summary of reagent quality in assays
| Antibody | Protein target | Immunofluorescence | ELISA | Western blot | Biolayer interferometry | Neutralization |
|---|---|---|---|---|---|---|
| 240C | S | +++ | +++ | +++ | +++ | partial |
| 154C | S | − | ++ | − | + | partial |
| 341C | S | − | ++ | − | − | − |
| 540C | S | + | − | ++ | − | − |
| CR3022 | S | ++ | +++ | − | +++ | − |
| NRC-772 | S | +++ | N/D | +++ | +++ | N/D |
| 42C | N | +++ | N/D | ++ | N/D | N/D |
| 427C | E | ++ | N/D | − | N/D | N/D |
| 19C | M | ++ | N/D | + | N/D | N/D |
| 283C | M | +++ | N/D | ++ | N/D | N/D |
Discussion
Our results demonstrate measurable cross-reactivity from a majority of the SARS-CoV structural protein-targeted antibodies that we evaluated against SARS-CoV-2 S, N, M, and E proteins. These tools can be readily obtained from BEI Resources and utilized by labs to study the properties of untagged SARS-CoV-2 structural proteins. These antibodies can serve the unmet need for more resources enabling the study of SARS-CoV-2. It is critical to understand the basic biology of SARS-CoV-2 in order to inform efforts toward improved diagnostics and treatments. Further, information about cross-reactivity of antibodies between SARS-CoV and SARS-CoV-2 may assist bioinformaticians in developing computational tools for predicting cross-reactivity of other antibodies, or even guiding rational design of improved CoV antibodies and small-molecule therapeutics.
We have shown that these publicly available antibodies are of potential use in several different types of assays with SARS-CoV-2 proteins. We found that several of these SARS-CoV structural protein antibodies demonstrated good staining in IF of SARS-CoV-2-infected cells and in an overexpression system (240C, NRC-772, and CR3022 against S; 42C against N; 283C against M; 472C against E). The anti-S antibodies, 240C and NRC-772, also give clear signal in western blot with minimal background. Several S antibodies show potent binding to full-length S and the RBD by ELISA, as well as binding to the RBD by BLI (240C, CR3022, and NRC-772). This wide range of uses substantially broadens our ability to investigate the biochemical properties of SARS-CoV-2 structural proteins.
The neutralization experiments we performed showed that antibodies that were previously shown to be neutralizing against SARS-CoV were actually less likely to be strongly cross-reactive with SARS-CoV-2. This may be because of a phenomenon well described among rapidly evolving viruses, such as HIV and influenza, wherein neutralizing antibodies are more likely to bind to highly variable epitopes lying on the host-interacting surfaces of the viral proteins (Pauthner and Hangartner, 2020; Sicca et al., 2018). It is likely that the specific amino acid substitutions present in the RBD of the SARS-CoV-2 S protein compared with that of SARS-CoV were selected for, in part, because of their ability to avoid binding by existing SARS-CoV antibodies among the wild animal populations from whence SARS-CoV-2 emerged. It is then, perhaps, unsurprising that 240C and 154C retained partial neutralizing ability, whereas 341C and 540C seem to have lost their capacity to neutralize when faced with SARS-CoV-2. There are ongoing efforts to determine the evolutionary forces that are shaping the continued change of the SARS-CoV-2 S protein in response to more widespread antibody-based immunity in the worldwide population; it may even be possible to anticipate mutations that could give rise to more virulent strains (Starr et al., 2020).
Although these antibodies only partially neutralized a SARS-CoV-2 model infection, they are still of interest for their potential to elucidate the structure and function of their protein targets. Antibodies have been critical tools in structure determination and in the mapping of proteins’ functional regions. Having a wide array of antibodies that recognize varying epitopes is of great help in this endeavor. Additionally, with the current dearth of knowledge regarding the life cycle and pathogenesis of SARS-CoV-2, particularly regarding the understudied M, N, and E proteins, we believe that these antibodies could be used in experiments to better understand the nuances of their functions beyond their obvious structural roles.
Our results also speak to the high proportion of SARS-CoV antibodies that display substantial cross-reactivity to SARS-CoV-2 structural proteins. Anecdotal evidence supports the efficacy of convalescent plasma treatment for COVID-19, indicating that cross-reactive antibodies generated during previous CoV infections may prove beneficial for emerging CoV infections (Duan et al., 2020). Another recent study found that following recovery from infection with SARS-CoV-2, patients expressed increased levels of antibodies capable of binding to peptides from more distantly related HCoVs, such as HCoV-OC43 and HCoV-229E (Shrock et al., 2020). Conversely, studies of COVID-19 patients have found neutralizing antibody titers to be directly proportional to disease severity, suggesting a more complicated relationship between antibodies and COVID-19 (Long et al., 2020; Wu et al., 2020). Some have hypothesized that this may be because of high concentrations of virus and neutralizing antibodies acting together to drive greater immune pathology (Jacobs, 2020; Zohar and Alter, 2020). A better understanding of the functions of individual antibody isotypes against different antigenic targets will be critical to predicting the utility of a particular antibody against SARS-CoV-2.
Further studies could also investigate possible cooperation between antibodies recognizing different epitopes, especially because CR3022 neutralization was shown to have synergy with another anti-S antibody that recognized a different epitope on the protein (ter Meulen et al., 2006). A recent study (Chi et al., 2020), for example, characterized a neutralizing monoclonal antibody that did not bind the RBD at all, and instead recognized an epitope in the NTD of the S protein. Knowledge about the variety of vulnerable epitopes, and possible synergy between antibodies that target them, brings us ever closer to being able to design and deploy effective therapeutics and vaccines in this time of urgent need.
STAR★methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibody | ||
| Human monoclonal anti-SARS-Cov -S CR3022 | BEI Resources | NR-52392 |
| Mouse anti-SARS-CoV S Monoclonal IgM 154C | BEI Resources | NR-620 |
| Mouse anti-SARS-CoV S Monoclonal IgG2a 240C | BEI Resources | NR-616 |
| Mouse anti-SARS-CoV S Monoclonal IgG2a 341C | BEI Resources | NR-617 |
| Mouse anti-SARS-CoV S Monoclonal IgG2a 540C | BEI Resources | NR-618 |
| Mouse anti-SARS-CoV M Monoclonal IgM 19C | BEI Resources | NR-615 |
| Mouse anti-SARS-CoV M Monoclonal IgG1 283C | BEI Resources | NR-621 |
| Mouse anti-SARS-CoV E Monoclonal IgM 472C | BEI Resources | NR-614 |
| Mouse anti-SARS-CoV N Monoclonal IgM 42C | BEI Resources | NR-619 |
| Rabbit anti-SARS-CoV S polyclonal sera-hydrogel | BEI Resources | NRC-773 |
| Rabbit anti-SARS-CoV S polyclonal sera | BEI Resources | NRC-768 |
| Guinea pig anti-SARS-CoV S polyclonal sera | BEI Resources | NR-10316 |
| Mouse anti-2xStrep monoclonal antibody | Sigma Aldrich | SAB2702215 |
| Goat anti-mouse HRP IgG | R & D Systems | HAF007; RRID:AB_357234 |
| Goat anti-mouse HRP IgG | Cell Signaling | 7076; RRID:AB_330924 |
| Goat anti-mouse HRP IgM | Invitrogen | 62-6820; RRID:AB_138470 |
| Goat anti-rabbit IgG HRP | Cell Signaling | 7074; RRID:AB_2099233 |
| Donkey anti-human HRP | Sigma Aldrich | SAB3701359 |
| Goat anti-Llama IgG (H+L) HRP | Novus | NB7242; RRID:AB_10124657 |
| antiHis-HRP | Invitrogen | ma1-80218; RRID:AB_931258 |
| anti-Human-AlexaFluor488 | Invitrogen | A11013; RRID:AB_141360 |
| anti-Mouse IgG AlexaFluor555 | Invitrogen | A21422; RRID:AB_141822 |
| anti-Mouse IgM AlexaFluor488 | Invitrogen | A21042; RRID:AB_2535711 |
| anti-Rabbit FAB2 AlexaFluor555 | Invitrogen | 4413S; RRID:AB_10694110 |
| Biological samples | ||
| Human deidentified Patient Sera 1v6 | Messer Lab, Oregon Health & Science University | N/A |
| Human deidentified Patient Sera 0v1 | Messer Lab, Oregon Health & Science University | N/A |
| SARS-CoV-2 Isolate USA-WA1/2020 | BEI Resources | NR-52281 |
| Chemicals and recombinant proteins | ||
| RBD recombinant purified Sars-CoV-2 Spike | BEI Resources | NR-52306 |
| Full-length Soluble SARS-CoV-2 Spike | BEI Resources | NR-52308 |
| BSA | Gold-Bio | A-420-10 |
| FBS | VWR | 97068-085 |
| PBS | Lonza Biosciences | 17-516F |
| Trypsin | Thermo Fisher Scientific | 25200056 |
| TrueBlue | SeraCare | 5510-0030 |
| Cell lines | ||
| HEK293T | ATCC | CRL-3216 |
| HEK293T human Ace2 expressing stable cell line | BEI Resources | NR-52511 |
| Vero E6 | ATCC | VERO C1008 |
| Recombinant DNA | ||
| HDM_IDTSpike_fixK, SARS-CoV-2 plasmid | BEI Resources | NR-52514 |
| HDM_Hgpm2 | BEI Resources | NR-52517 |
| HDM_tat1b | BEI Resources | NR-52518 |
| pRC_CMV_Rev1b | BEI Resources | NR-52519 |
| pHAGE2_CMV_ZsGreen_W | BEI Resources | NR-52516 |
| pTwist-EF1alpha-nCoV-2019-S-2xStrep | Gordon et al., 2020 | N/A |
| pLVX-EF1alpha-nCoV-2019-E-IRES-Puro | Gordon et al., 2020 | N/A |
| pLVX-EF1alpha-nCoV-2019-M-IRES-Puro | Gordon et al., 2020 | N/A |
| pLVX-EF1alpha-nCoV-2019-N-IRES-Puro | Gordon et al., 2020 | N/A |
| Software | ||
| R | Rstudio https://rstudio.com/ | https://rstudio.com/ |
| Python | Python Software Foundation | https://www.python.org/ |
| T-Coffee | Notredame et al., 2000 | http://tcoffee.crg.cat/ |
| Data Analysis HT(10.0.0.48) | ForteBio | 50-5029 |
| Critical commercial assays | ||
| PureLink HiPure Plasmid Maxiprep Kit | Invitrogen | K210007 |
| ChromaLINK biotin protein labeling kit | Vector Labs | B-9007-105K |
| Streptavidin (SA) Biosensors | ForteBio | 18-5019 |
| Other | ||
| NuncSorp ELISA Plates | Thermo Fisher Scientific | 44-2404-21 |
| PVDF membrane | Thermo Fisher Scientific | 88518 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fikadu G. Tafesse (tafesse@ohsu.edu).
Materials availability
No unique reagents were generated during the course of this study.
Data and code availability
This study did not generate any unique datasets or code.
Experimental model and subject details
293T stable cell lines expressing Ace2 receptor (293T-Ace2) were a kind gift from Dr. Jesse D. Bloom from University of Washington, and described previously (Crawford et al., 2020). Wt low-passage 293T (293T-Lp), 293T-Ace2, and Vero E6 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, 10% FBS, 1% Penn-Strep, 1% NEAA) at 37C. Cells were cultured on treated T75 dishes, passaged with Trypsin at 95% confluency to avoid overcrowding.
Method details
Sequence alignment
Protein sequences were obtained from uniprot and aligned using the T-Coffee multiple sequence alignment server.
Cell transfection
Transfections were carried out in 293T cells seeded at 70%–90% cell density using Lipofectamine 3000 (ThermoFisher Scientific) as per manufacturer’s instructions. For immunofluorescence, the SARS-CoV2 structural protein plasmids pTwist-EF1alpha-nCoV-2019-S-2xStrep, pLVX-EF1alpha-nCoV-2019-E-IRES-Puro, pLVX-EF1alpha-nCoV-2019-M-IRES-Puro, or pLVX-EF1alpha-nCoV-2019-N-IRES-Puro were transfected using 2 μg of plasmid per well of a 24-well plate. Structural SARS-CoV-2 protein plasmids were a kind gift from the Krogan Lab at UCSF and are described previously (Gordon et al., 2020). For pseudotyped lentivirus production, lentivirus packaging plasmids, HDM_Hgpm2, HDM_tat1b, PRC_CMV_Rev1b, SARS_CoV-2 S plasmid HDM_IDTSpike_fixK, and LzGreen reporter plasmid pHAGE2_CMV_ZsGreen_W were transfected using 0.44 μg for packaging, 0.68 μg for S, and 2 μg for reporter plasmids per 6 cm dish. Packaging, SARS-CoV-2 S, and reporter plasmids were a kind gift from Jesse D. Bloom from University of Washington, and are described previously (Crawford et al., 2020). Transfection media was carefully removed 6 hours post transfection, and replaced with DMEM.
Pseudotyped lentivirus production
293T cells were seeded at 2 million cells/dish in 6cm TC-treated dishes. The following day, cells were transfected as described above with lentivirus packaging plasmids, SARS-CoV-2 S plasmid, and lzGreen reporter plasmid (Crawford et al., 2020). After transfection, cells were incubated at 37C for 60 hours. Viral media was harvested, filtered with 0.45 μm filter, then frozen before use. Virus transduction capability was then tittered on 293T-Ace2 cells treated with 50 μL of 5 μg/ml polybrene (Sigma-Aldritch LLC). LzGreen titer was determined by fluorescence using BZ-X700 all-in-one fluorescent microscope (Keyence), a 1:16 dilution was decided as optimal for following neutralization assays due to broad transduced foci distribution.
SARS-CoV-2 virus propagation
One tube of frozen SARS-CoV-2 (BEI Resources) was thawed and diluted 1:10 for inoculation in minimal volume onto 70% confluent Vero E6 cells. The cells were incubated for 1 hour at 37°C, rocking every 15 minutes to ensure even coverage. Additional media was added up to the manufacturer’s recommended culture volume, and the cells were incubated for 72 hours at 37°C. Supernatant was collected and spun at 3,000 × g for 5 minutes, then aliquoted for storage at −80°C.
SARS-CoV-2 infection
A 96-well plate of 50% confluent Vero cells was inoculated with 50 μL frozen SARS-CoV-2 virus stock for 1 hour at 37°C with rocking every 15 minutes. Added an additional 50 μL of fresh media and incubated for 24 hours at 37°C. Fixed plate by submerging in 4% PFA in PBS for 1 hour, then brought into BSL-1 for immunofluorescence staining.
Immunofluorescence
293T cells were seeded on 24-well plates containing glass coverslips coated with poly-lysine solution; 100,000 cells were seeded per well. Cells were transfected with SARS-CoV-2 structural protein plasmids as described above. After 48 hours post transfection, cells were fixed with 4% PFA in PBS. Coverslips with transfected 293T cells and the 96-well plate with SARS-CoV-2 infected Vero cells were permeabilized with 2% BSA, 0.1% Triton X-100 in PBS. Transfected cells were incubated for 3 hours at RT with the following anti-SARS-CoV structural protein monoclonal or polyclonal antibodies at a 1:250 dilution for transfected 293T cells, or 1:200 for infected Vero cells: mouse anti-SARS-CoV S monoclonal IgM 154C, mouse anti-SARS-CoV S monoclonal IgG2a 240C, mouse anti-SARS-CoV S monoclonal IgG2a 341C, mouse anti-SARS-CoV S monoclonal IgG2a 540C, mouse anti-SARS-CoV N monoclonal IgM 19C, mouse anti-SARS-CoV M monoclonal IgG1 283C, mouse anti-SARS-CoV E monoclonal IgM 472C, mouse anti-SARS-CoV N monoclonal IgM 42C, human anti-SARS-CoV S monoclonal IgG1 CR3022, and rabbit anti-SARS-CoV S polyclonal sera (BEI Resources) and mouse anti-2xStrep-tag antibody (Sigma-Aldrich). Anti-mouse IgG AF555, anti-rabbit IgG AF555, or anti-mouse IgM AF488 conjugated secondary antibodies were added at 1:500 dilution for 1 hour at RT (Invitrogen). Confocal imaging was performed with a Zeiss LSM 980 using a 63x Plan-Achromatic 1.4 NA oil immersion objective. Images were processed with Zeiss Zen Blue software. Maximum intensity z-projections were prepared in Fiji. All antibody stain images were pseudocolored for visual consistency.
Pseudovirus neutralization assay
Neutralization protocol was based on previously reported neutralization research utilizing SARS-CoV-2 S pseudotyped lentivirus (Crawford et al., 2020). 293T-Ace2 cells were seeded on tissue culture treated, poly-lysine treated 96-well plates at a density of 10,000 cells per well. Cells were allowed to grow overnight at 37°C. LzGreen SARS-COV-2 S pseudotyped lentivirus were mixed with 2-fold dilutions of the following monoclonal or polyclonal anti-SARS-CoV-2 S antibodies: mouse anti-SARS-CoV S monoclonal IgM 154C, mouse anti-SARS-CoV S monoclonal IgG2a 240C, mouse anti-SARS-CoV S monoclonal IgG2a 341C, mouse anti-SARS-CoV S monoclonal IgG2a 540C, rabbit anti-SARS-CoV S polyclonal sera, Guinea pig anti-SARS-CoV S polyclonal sera, human monoclonal anti-SARS-CoV S CR3022 (BEI Resources). Human patient sera from a SARS-CoV-2 patient was used as positive neutralization control, while virus alone was used as negative control. Sera and antibody dilutions ranged from 1:10 to 1:1048. Virus-antibody mixture was incubated at 37C for 1 hour after which virus was added to 293T-Ace2 treated with 5 μg/ml polybrene. Cells were incubated with neutralized virus for 44 hours before imaging. Cells were fixed with 4% PFA for 1 hour at RT, incubated with DAPI for 10 minutes at RT, and imaged with BZ-X700 all-in-one fluorescent microscope (Keyence). Estimated area of DAPI and GFP fluorescent pixels was calculated with built in BZ-X software (Keyence).
Focus forming assay (FFA) for live SARS-CoV-2 virus neutralization measurement
The FFA was performed as previously described (Case et al., 2020). In brief, Vero E6 cells were plated into 96 well plates at 24,000 cells/well and incubated overnight. Previously propagated SARS-CoV-2 stocks were titrated by plaque forming unit (pfu) assay and diluted to 30 pfu in 15 μL. To the virus, 15 μL of antibody dilutions were added such that the final antibody dilution was 1:10 to 1:1280 in two-fold dilutions and this was incubated at 37°C for 1 hour. All virus and antibody dilutions were prepared in Opti-MEM media with 2% FBS. 30 μL of neutralized virus was then added to the confluent Vero E6 cells and incubated for 1 hour at 37°C. 150 μL of overlay media (Opti-MEM, 2% FBS, 2% Methylcellulose) was then added to each well and incubated for 48 hours at 37°C. Following infection, the plates were fixed using formaldehyde and subsequently blocked for 30 minutes with perm buffer containing 0.1% bovine serum albumin and 0.1% saponin. SARS-CoV-2 RBD and N protein immunized alpaca polyclonal serum was used as primary antibody at 1:5,000 dilution in perm buffer, and anti-Llama-HRP secondary was used at 1:20,000 dilution. Plates were developed with TrueBlue (SeraCare) substrate and imaged with an Immunospot analyzer.
Enzyme-linked immunosorbent assay (ELISA)
ELISA plates, Nunc MaxiSorp (Invitrogen), were coated with purified recombinant SARS-COV-2 RBD domain (BIR resources, NR-52306) at 2 μg/ul in PBS. Coating was carried out overnight at 4°C. Protein was blocked in 2% BSA, 1% tween-20 in PBS for 30 minutes at RT. The following anti SARS-CoV-2 S monoclonal and polyclonal antibodies were serially diluted by 2-fold dilutions in blocking buffer: mouse anti-SARS-CoV S monoclonal IgM 154C, mouse anti-SARS-CoV S monoclonal IgG2a 240C, mouse anti-SARS-CoV S monoclonal IgG2a 341C, mouse anti-SARS-CoV S monoclonal IgG2a 540C, human monoclonal anti-SARS-CoV-S CR3022 (BEI Resources). Human patient sera from a SARS-CoV-2 patient was used as a positive control. Dilutions ranged from1:10 to 1:10480, and were incubated for 1 hour at RT. Anti-mouse HRP, and anti-human-HRP secondary antibodies were used at 1:4000 concentration in blocking buffer, and were incubated 1 hour at RT. 50 μL of TMB HRP substrate (ThermoFisher Scientific) was added, and following incubation for 10 minutes at RT, 50 μL of 2N H2SO4 was added as a stopping solution. Plate absorbance at 405nm was measured using a CLARIOstar® Plus plate fluorimeter (BMG Labtech).
RBD protein purification and biotinylation
Purified SARS-CoV-2 S-RDB protein was prepared as described previously (Stadlbauer et al., 2020). Breifly, codon optimized His-tagged RBD in pInducer-20 was used to make lentivirus in HEK293T cells which was then used to infect HEK293-F suspension cells. The suspension cells were allowed to grow for 3 days with shaking at 37°C at 8% CO2. Cell supernatant was collected, sterile filtered, and purified by Ni-NTA chromatography. The purified protein was then buffer exchanged into PBS and concentrated. For use in BLI, purified RBD was biotinylated using the ChromaLINK biotin protein labeling kit according to the manufacturer’s instructions with 5x molar equivalents of labeling reagent to achieve 1.92 biotins/protein.
Biolayer interferometry (BLI)
Streptavidin biosensors (ForteBio) were soaked in PBS for at least 30 minutes prior to starting the experiment. Biosensors were prepared with the following steps: equilibration in kinetics buffer (10 mM HEPES, 150 mM NaCl, 3mM EDTA, 0.005% Tween-20, 0.1% BSA, pH 7.5) for 300 s, loading of biotinylated RBD protein (10ug/mL) in kinetics buffer for 200 s, and blocking in 1 μM D-Biotin in kinetics buffer for 50 s. Binding was measured for seven 3-fold serial dilutions of each monoclonal antibody using the following cycle sequence: baseline for 300 s in kinetics buffer, association for 300 s with antibody diluted in kinetics buffer, dissociation for 750 s in kinetics buffer, and regeneration by 3 cycles of 20 s in 10 mM glycine pH 1.7, then 20 s in kinetics buffer. All antibodies were run against an isotype control antibody at the same concentration. Data analysis was performed using the ForteBio data analysis HT 10.0 software. Curves were reference subtracted using the isotype control and each cycle was aligned according to its baseline step. KDs were calculated using a 1:1 binding model using global fitting of association and dissociation of all antibody concentrations, excluding dilutions with response below 0.005 nm.
RBD western blot
293T cells were seeded in 10 cm dishes at a density of 3.5 million cells per dish. After overnight growth, cells were transfected using lipofectamine 3000 as described above. Plasmids pTwist-EF1alpha-nCoV-2019-S-2xStrep, pLVX-EF1alpha-nCoV-2019-E-IRES-Puro, pLVX-EF1alpha-nCoV-2019-M-IRES-Puro, or pLVX-EF1alpha-nCoV-2019-N-IRES-Puro were transfected using 90 μg of DNA per 10 cm dish. Cells were scraped 48 hours post-transfection, then lysed in RIPA buffer (EMD Millipore). Cell lysates were diluted with reducing Laemmli buffer, incubated for 10 minutes at 37°C, then ran on 4%–20% Mini-PROTEAN® TGX Precast Protein Gels (BIO-RAD). Additionally, 1ug of purified recombinant S RBD-His6 was diluted in PBS and Laemmli buffer to a final volume of 20 μL and added to a 7.5% Mini-PROTEAN® TGX Precast Protein Gel (BIO-RAD). Resolved proteins were then transferred to a PVDF membrane, blocked in TBS with 2% BSA 0.1% Tween-20, then incubated with the following antibodies diluted to 1:500 in blocking buffer: mouse anti-SARS-CoV N monoclonal IgM 19C, mouse anti-SARS-CoV M monoclonal IgG1 283C, mouse anti-SARS-CoV E monoclonal IgM 472C, and mouse anti-2xStrep-tag antibody, and anti-His-HRP. Blots were stained with SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher Scientific) using an ImageQuant LAS 4000 imager (GE Life Sciences).
SARS-CoV-2 infected lysate western blot
Around 106 Vero E6 cells were infected with SARS-CoV-2 at MOI of 0.1. At 72 hours post infection, cells were washed with PBS and lysed with 8M urea + 1x RIPA buffer + 1x Laemmli buffer. The cell lysates were then removed from the BSL-3 for further analysis. An equal quantity of uninfected Vero E6 cells were processed similarly. The cell lysates were heated to 42°C for 30 minutes, then run on a 15% SDS-PAGE gel and transferred to a PVDF membrane. A sample membrane was stained with Ponceau to assess loading quantities. The remaining membranes were blocked for 30 minutes at room temperature with 2% bovine serum albumin, 1% polyvinylpyrrolidone, 0.1% tween-20 in PBS (PBS-T). 1v6 and NRC-772 were used at 1:1000 while 154C, 240C, 341C, 540C, 19C, 42C, 283C, and 472C were used at 1:100. The primary antibodies diluted in blocking buffer and incubated with the membranes at room temperature for 4 hours before being washed thrice with PBS-T. Secondary antibodies were anti-Human-HRP (SAB3701359) for 1v6 and CR3022 at 1:5000; anti-rabbit-HRP (7074) for NRC-772 at 1:5000; anti-mouse-IgG-HRP (7076) for 240C, 341C, 540C, and 283C at 1:1000; anti-mouse-IgM-HRP (62-6820) for 154C, 19C, 42C, and 472C. Blots were stained with SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher Scientific) using an ImageQuant LAS 4000 imager (GE Life Sciences).
Quantification and statistical analysis
Sequence alignments and identity scores were calculated using the T-Coffee software package via the online portal. The EC50 values for ELISA and live virus neutralization were calculated using a three-parameter logistic regression model in Python using the SciPy statistics library. Each EC50 includes data from three replicate experiments and unless otherwise noted, three technical replicates within each experiment. KD values were calculated in Fortebio Data Analysis HT software and fit to a 1:1 binding model and globally fit to both the association and dissociation curves of all concentrations with response values above 0.005 nm.
Acknowledgments
This work was supported by NIH training grant T32AI747225 on Interactions at the Microbe-Host Interface and OHSU Innovative IDEA grant 1018784. BLI data were generated on an Octet Red 384, which is made available and supported by OHSU Proteomics Shared Resource facility and equipment grant number S10OD023413. We acknowledge the support of the members of the Messer lab who performed collection of patient samples and the patients who agreed to donate samples for scientific research.
Author contributions
Conceptualization: F.G.T. and T.A.B.; methodology, formal analysis, and investigation: T.A.B., J.B.W., S.F., H.C.L., and F.G.T.; resources: W.B.M.; writing – original draft: T.A.B., J.B.W., and S.F.; writing – review and editing: all authors; visualization: T.A.B., J.B.W., S.F., H.C.L., and F.G.T.; supervision: F.G.T.; project administration: F.G.T.; fund acquisition: F.G.T.
Declaration of interests
The authors declare no competing interests.
Published: February 16, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2021.108737.
Supplemental information
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Guo Z., Duan C., Chen Z., Wang G., Lu Y., Li M., Lu J. Long-Term Persistence of IgG Antibodies in SARS-CoV Infected Healthcare Workers. medRxiv. 2020 2020.02.12.20021386. [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J.J.L. Neutralizing antibodies mediate virus-immune pathology of COVID-19. Med. Hypotheses. 2020;143:109884. doi: 10.1016/j.mehy.2020.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby M.E., Biggs H.M., Haynes A., Dahl R.M., Mustaquim D., Gerber S.I., Watson J.T. Human coronavirus circulation in the United States 2014-2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lv H., Wu N.C., Tsang O.T.-Y., Yuan M., Perera R.A.P.M., Leung W.S., So R.T.Y., Chan J.M.C., Yip G.K., Chik T.S.H. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020;31:107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D., Andrews C.A., Vogel L., Koup R.A., Roederer M., VRC 301 Study Team A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Pauthner M.G., Hangartner L. Broadly neutralizing antibodies to highly antigenically variable viruses as templates for vaccine design. In: Hangartner L., Burton D.R., editors. Vaccination Strategies Against Highly Variable Pathogens, Current Topics in Microbiology and Immunology. Springer International Publishing; 2020. pp. 31–87. [DOI] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance E.G., Bossis I., Dickerson F.B., Stallings C.R., Origoni A.E., Sullens A., Yolken R.H., Viscidi R.P. Development of a nucleocapsid-based human coronavirus immunoassay and estimates of individuals exposed to coronavirus in a U.S. metropolitan population. Clin. Vaccine Immunol. 2008;15:1805–1810. doi: 10.1128/CVI.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., MGH COVID-19 Collection & Processing Team Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicca F., Neppelenbroek S., Huckriede A. Effector mechanisms of influenza-specific antibodies: neutralization and beyond. Expert Rev. Vaccines. 2018;17:785–795. doi: 10.1080/14760584.2018.1516553. [DOI] [PubMed] [Google Scholar]
- Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan H.C., Roback J.D. Convalescent Plasma: Therapeutic Hope or Hopeless Strategy in the SARS-CoV-2 Pandemic. Transfus. Med. Rev. 2020;34:145–150. doi: 10.1016/j.tmrv.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp R.A., Haynes L.M., Moore D., Anderson B., Tamin A., Harcourt B.H., Jones L.P., Yilla M., Babcock G.J., Greenough T. Monoclonal antibodies to SARS-associated coronavirus (SARS-CoV): identification of neutralizing and antibodies reactive to S, N, M and E viral proteins. J. Virol. Methods. 2005;128:21–28. doi: 10.1016/j.jviromet.2005.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., VIB-CMB COVID-19 Response Team Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 2020.03.30.20047365. [Google Scholar]
- Yasui F., Kohara M., Kitabatake M., Nishiwaki T., Fujii H., Tateno C., Yoneda M., Morita K., Matsushima K., Koyasu S., Kai C. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology. 2014;454–455:157–168. doi: 10.1016/j.virol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Pang R., Xue X., Bao J., Ye S., Dai Y., Zheng Y., Fu Q., Hu Z., Yi Y. Anti-SARS-CoV-2 virus antibody levels in convalescent plasma of six donors who have recovered from COVID-19. Aging (Albany NY) 2020;12:6536–6542. doi: 10.18632/aging.103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T., Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 2020;20:392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.