Abstract
Ethnopharmacological relevance
Since the occurrence of coronavirus disease 2019 (COVID-19) in Wuhan, China in December 2019, COVID-19 has been quickly spreading out to other provinces and countries. Considering that traditional Chinese medicine (TCM) played an important role during outbreak of SARS and H1N1, finding potential alternative approaches for COVID-19 treatment is necessary before vaccines are developed. According to previous studies, Maxing Shigan decoction (MXSGD) present a prominent antivirus effect and is often used to treat pulmonary diseases. Furthermore, we collected 115 open prescriptions for COVID-19 therapy from the National Health Commission, State Administration of TCM and other organizations, MXSGD was identified as the key formula. However, the underlying molecular mechanism of MXSGD against COVID-19 is still unknown.
Aim of the study
The present study aimed to evaluate the therapeutic mechanism of MXSGD against COVID-19 by network pharmacology and in vitro experiment verification, and screen the potential components which could bind to key targets of COVID-19 via molecular docking method.
Materials and methods
Multiple open-source databases related to TCM or compounds were employed to screen active ingredients and potential targets of MXSGD. Network pharmacology analysis methods were used to initially predict the antivirus and anti-inflammatory effects of MXSGD against COVID-19. IL-6 induced rat lung epithelial type Ⅱ cells (RLE-6TN) damage was established to explore the anti-inflammatory damage activity of MXSGD. After MXSGD intervention, the expression level of related proteins and their phosphorylation in the IL-6 mediated JAK-STAT signaling pathway were detected by Western blot. Molecular docking technique was used to further identify the potential substances which could bind to three key targets (ACE2, Mpro and RdRp) of COVID-19.
Results
In this study, 105 active ingredients and 1025 candidate targets were selected for MXSGD, 83 overlapping targets related to MXSGD and COVID-19 were identified, and the protein-protein interaction (PPI) network of MXSGD against COVID-19 was constructed. According to the results of biological enrichment analysis, 63 significant KEGG pathways were enriched, and most of them were related to signal transduction, immune system and virus infection. Furthermore, according the relationship between signal pathways, we confirmed MXSGD could effectively inhibit IL-6 mediated JAK-STAT signal pathway related protein expression level, decreased the protein expression levels of p-JAK2, p-STAT3, Bax and Caspase 3, and increased the protein expression level of Bcl-2, thereby inhibiting RLE-6TN cells damage. In addition, according to the LibDock scores screening results, the components with strong potential affinity (Top 10) with ACE2, Mpro and RdRp are mainly from glycyrrhiza uralensis (Chinese name: Gancao) and semen armeniacae amarum (Chinese name: Kuxingren). Among them, amygdalin was selected as the optimal candidate component bind to all three key targets, and euchrenone, glycyrrhizin, and glycyrol also exhibited superior affinity interactions with ACE2, Mpro and RdRp, respectively.
Conclusion
This work explained the positive characteristics of multi-component, multi-target, and multi-approach intervention with MXSGD in combating COVID-19, and preliminary revealed the antiviral and anti-inflammatory pharmacodynamic substances and mechanism of MXSGD, which might provide insights into the vital role of TCM in the prevention and treatment of COVID-19.
Keywords: Novel coronavirus, Traditional Chinese medicine, Maxing Shigan decoction, Antiviral, Anti-inflammatory, In vitro experiment
Graphical abstract
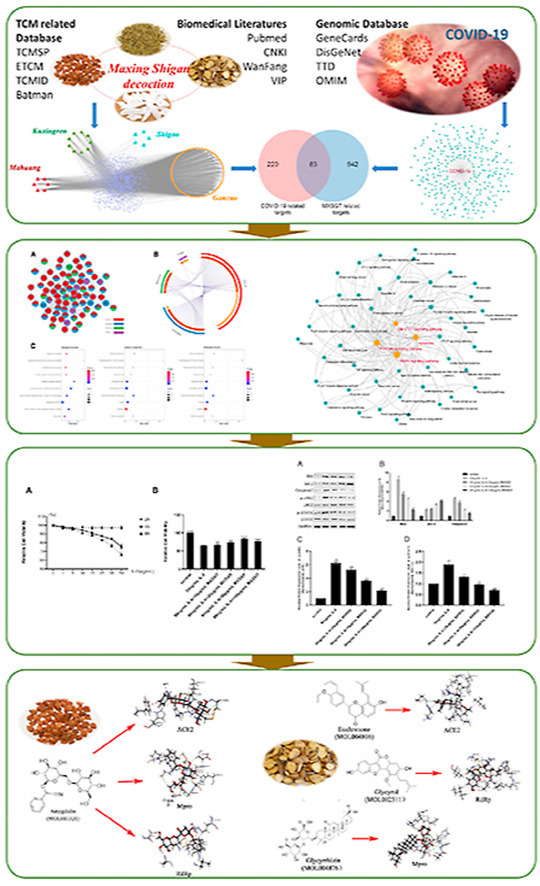
Abbreviation
- COVID-19
coronavirus disease 2019
- MXSGD
Maxing Shigan decoction
- ACE2
angiotensin-converting enzyme II
- Mpro
M protease
- RdRp
RNA-derived RNA polymerase
- TCM
traditional Chinese medicine;
- OB
Oral bioavailability
- DL
Drug-likeness
- Caco-2
intestinal epithelial permeability
- PPI
protein-protein interaction
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MF
molecular function
- BP
biological process
- CC
cellular component
1. Introduction
Since the outbreak of coronavirus disease 2019 (COVID-19) in Wuhan, China in December 2019, the novel coronavirus has been quickly spreading out to other provinces (Munster et al., 2020; Wu et al., 2020). With the increasing incidence of confirmed cases, corresponding spread control policies and emergency actions are taking place, such as "Diagnosis and Treatment Program for novel coronavirus pneumonitis (1st to 7th editions)" issued by the National Health Committee and the State Administration of traditional Chinese medicine (TCM). At present, China has been successful in combating COVID-19 and maintaining a massive infection control of the epidemic, TCM has been demonstrated to be effective in improving patients' immune systems and health conditions in the fight against COVID-19.
Maxing Shigan decoction (MXSGD), composed of four kinds of Chinese medicine, including Ephedra sinica (ehedraceae, Chinese name: Mahuang), Semen armeniacae amarum (rosaceae, Chinese name: Kuxingren), Gypsum Fibrosum (sulfates, Chinese name: Shigao) and Glycyrrhiza uralensis (leguminosae, Chinese name: Gancao), is a classic prescription for treating lung diseases. In previous four large-scale epidemics in China since 1949, including COVID-19, many TCM prescriptions were formed on the basis of MXSGD. The results of a series of case studies suggest that modified MXSGD combined with the western medicine routine treatment has a remarkable clinical curative effect on common COVID-19 without obvious damage of liver and kidney function (Qu et al., 2020). According a report by State Administration of TCM, the three formulas and three medicines have proved to be effective in treating COVID-19. It is worth noting that 4 of the 6 effective TCM recipes, including Lianhuaqingwen capsule/granule, Qingfei Paidu decoction, Huashi Baidu formula, and Xuanfei Baidu granule, were both formulated based on MXSGD, and the the efficacy and safety of which had been confirmed by multiple clinical studies (Hu et al., 2020; Xin et al., 2020). From the perspective of syndrome differentiation and treatment, MXSGD is suitable for COVID-19 patients with superficial exogenous pathogens on the surface and phlegm-heat in the lung. The key point of TCM syndrome differentiation is the number of thin yellow veins with moss for fever, asthma and cough. Mahuang has the effect of effuse sweat and resolve exterior, diffuse lung and calm asthma, disinhibit water and disperse edema. Kuxingren is effective in treating of cough and asthma accompanied by stuffiness in the chest and profuse expectoration. Shigao is effective in febrile diseases due to exogenous affection with high fever and dire thirst, asthma and cough caused by heat in the lung. Gancao mainly used to modulate the pharmaceutical activity of different herbal medicines, and it can also moisten lung and relieve cough. Four Chinese medicines cooperate with each other to play the main effect of ventilating lung, and relieving cough and asthma. Above all, it is reasonable to believe that MXSGD is the core combination of most clinically effective formulations and has application value. However, the mechanism of MXSGD in the treatment of COVID-19 is still unclear.
Based on current understanding of COVID-19, the novel coronavirus and the SARS-coronavirus share central biological properties which can guide risk assessment and intervention (Lu et al., 2020). As with SARS-CoV, the spike (S) protein of the COVID-19 can bind to angiotensin-converting enzyme II (ACE2), a cellular entry receptor, and employ the transmembrane protease serine 2 (TMPRSS2) to facilitate viral entry into host target cells (Kuba et al., 2005; Wan et al., 2020b). The coronavirus M protease (Mpro) and RNA-derived RNA polymerase (RdRp) are two additional key targets for drug screening against COVID-19(Liu et al., 2020a, Liu et al., 2020b; Shi et al., 2020).
According to the characteristics of the multicomponent and multitarget of Chinese herbal medicine, network pharmacology may be a new and well-documented method to find some meaningful information. Through network pharmacology, we can mine drugs and disease targets from massive data, and understand the mechanisms and pathways between them(Li et al., 2013; Hao et al., 2014).At present, the application scope of network pharmacology is expanding, including exploring the basic pharmacological action and mechanism of TCM recipes on diseases (Zheng et al., 2020), analyzing the theory of TCM and exploring the development and application of Chinese herbal medicine (Zhang et al., 2019a; Luan et al., 2020). Therefore, in this study we used network pharmacology to screen active ingredients and potential targets of MXSGD, and to reveal its potential medicinal substance and mechanism in antivirus and anti-inflammatory. Molecular docking was used to further identify the potential substances which could bind to three key targets of COVID-19 (ACE2, Mpro and RdRp).
2. Materials and methods
2.1. Screening of active components and targets of MXSGD
The four Chinese medicine ingredients of MXSGD were obtained from the TCMSP (http://tcmspw.com/tcmsp.php, accessed January 2020) (Ru et al., 2014), ETCM (http://www.tcmip.cn/ETCM/index.php/Home/Index/, accessed January 2020)(Xu et al., 2019), TCMID (http://119.3.41.228:8000/tcmid/, accessed January 2020)(Huang et al., 2018), and Batman (http://bionet.ncpsb.org/batman-tcm/, accessed January 2020)(Liu et al., 2016) databases. In addition, the databases of Pubmed, CNKI, WanFang, and VIP were used to supplement any other omitted components. The structures of chemical components were obtained from PubChem (https://www.ncbi.nlm.nih.gov/, accessed January 2020). Many aspects of drug-like properties can be quantified through physico-chemical indices and these, in turn, can be calculated in Chinese medicine compound. Criteria such as the "lipinski rule of five" is a rule of thumb that describes the drugability of a determinate molecule, which can be used to filter libraries and remove molecules with predicted poor drug-like properties (Lipinski et al., 2001). TCMSP provides these parameters, and currently the most commonly used screening indicators in network pharmacological analysis are oral bioavailability (OB), drug-likeness (DL) and intestinal epithelial permeability (Caco-2). OB is an important index for objective evaluation of drug absorption and determines the success of clinical drug trials (Sietsema, 1989). Drug-likeness DL is a property that is a comprehensive reflection of the pharmacokinetic characteristics of compounds administered to the human body (Proudfoot, 2002). Accurate evaluation of DL is helpful for screening desirable compounds and improving the hit rate of drug candidates. Therefore, these three important parameters (OB ≥ 30%, DL ≥ 0.18, Caco-2 ≥-0.4) were used for the bioactive components screening (Li et al., 2015).
Based on previous experience and considering the recognition of target prediction method, we chose the ligand-based prediction technology for target prediction (Yu et al., 2012; Wu et al., 2016). First, we used the two common chemical component databases of TCMSP and PubChem as the main sources of structure information on chemical components. From this we obtained detailed information about the bioactive components of MXSGD, including canonical SMILES, molecular structures, and their "mol2" files. Second, Swiss Target Prediction (http://www.swisstargetprediction.ch/, accessed January 2020) and Pharm-Mapper (http://lilab.ecust.edu.cn/pharmmapper/index.php, accessed January 2020) were used to predict target protein names of bioactive components depending on the chemical similarities and pharmacophore models of its components (Gfeller et al., 2014; Wang et al., 2017). Finally, the targets were further mapped to UniProt Database (https://www.uniprot.org/, accessed February 2020) to normalize their corresponding nomenclatures and transform into corresponding gene names, with the species limited to "Homo sapiens".
2.2. Identification of common targets of MXSGD in COVID-19
"Coronavirus infection" and "coronavirus pneumonia" were applied as search terms to find COVID-19 related targets. Four databases were employed for biological information accumulation, including GeneCards (https://www.genecards.org/, accessed January 2020) (Rebhan et al., 1997), DisGeNET (http://www.disgenet.org/, accessed January 2020) (Pinero et al., 2017), TTD (http://db.idrblab.net/ttd/, accessed January 2020) (Chen et al., 2002), and OMIM (https://omim.org/)(Amberger et al., 2019). All of the COVID-19 related targets were normalized with Uniprot ID, using the Bioconductor package of R software (Li et al., 2018) to remove duplication.
A list of target genes of compounds and COVID-19 were prepared based on the preceding steps. The genes overlapping these two sets were filtered with R 3.6.1 software using the Venn Diagram package (Chen and Boutros, 2011). Common genes were considered as potential targets of MXSGD against COVID-19, further analyzed and visualized using Cytoscape 3.6.0 software (Shannon et al., 2003).
2.3. Protein-protein interaction (PPI) network construction and bioinformatic analysis
Proteins connected within a PPI network are likely to collaborate (e.g., form a signal transduction pathway or molecular complex) to perform various related biological processes (De Las and Fontanillo, 2010). Therefore, the potential targets of MXSGD against COVID-19 were submitted to the STRING database (https://string-db.org/) to obtain a PPI network (Szklarczyk et al., 2017), and underwent visual management with Cytoscape 3.6.0. We explored the underlying mechanisms of these common targets with gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using DAVID (https://david.ncifcrf.gov, v6.8) (Huang et al., 2009) to perform functional annotation and enrichment analysis, and performed meta-analysis for different herbs with Metascape (https://metascape.org/gp/index.html) (Yingyao et al., 2019). Finally, the functional categories were identified and ranked by p-values, and those GO terms and KEGG pathways with p-value ≤ 0.05 and FDR ≤0.05 were recognized as significant.
2.4. In vitro activity verification
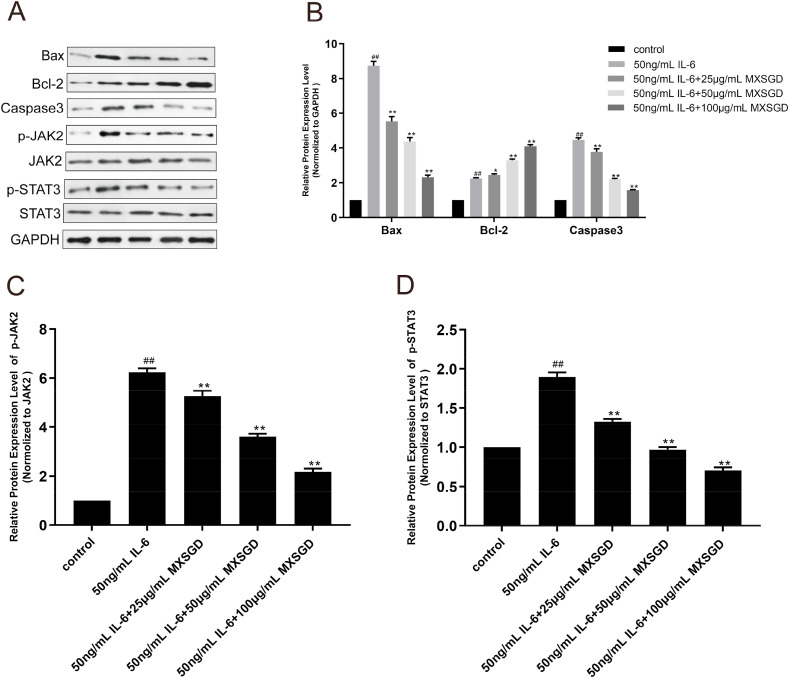
The elevated inflammatory cytokines may play a major role in the pathology of COVID-19, and induce tissue necrosis, interstitial macrophage and monocyte infiltrations in the lung, heart and gastrointestinal mucosa (Chen et al., 2020, Wan et al., 2020a; Xu et al., 2020). To block the organs damage of IL-6, one of key inflammatory cytokines, might be a potentially effective strategy for the treatment of COVID-19(Liu et al., 2020a, Liu et al., 2020b). IL-6 induced rat lung epithelial type Ⅱ cells (RLE-6TN, CRL-2300, ATCC) damage was established to explore the anti-inflammatory damage activity of MXSGD. RLE-6 TN cells were suspended in DMEM medium containing 10% fetal bovine serum, seeded in 96-well plates at a density of 5 × 10^4/mL, and incubated at 37°C and 5% CO2. After incubated for 24h and changed the basic medium overnight, RLE-6TN cells were interfered with the different concentrations of IL-6 (ab259381, Abcam). And the number of living cells was detected by MTT method at 2h, 4h, 8h, respectively, to determine the most appropriate time and concentration of IL-6 to induce RLE-6TN cell damage. The MXSGD rat medicated serum was prepared with reference to related literature (Zhang et al., 2019b). The number of living cells was detected by MTT method after administered with the different concentrations (10,20,50,100 μg/mL) of MXSGD rat medicated serum and 50 ng/mL IL-6 simultaneously, to determine the effect of MXSGD inhibiting IL-6 induced RLE-6TN cell damage. Subsequently, the expression level of related proteins and their phosphorylation, in the IL-6 mediated JAK-STAT signaling pathway were detected by Western blot. The total protein from RLE-6TN cell administered with the different concentrations (25,50,100 μg/mL) of MXSGD rat medicated serum and 50 ng/mL IL-6 simultaneously was extracted using RIPA lysis buffer, and the protein concentrations were measured with BCA protein assay kit. The protein samples were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to the polyvinylidene difluoride membrane (PVDF). And then, the PVDF membrane was blocked with 5% bovine serum album for 1 h and incubated with primary antibodies including JAK2 (ab108596, Abcam), p-JAK2 (ab32101, Abcam), STAT3 (ab68153, Abcam), p-STAT3 (ab76315, Abcam), Bax (ab32503, Abcam), Bcl-2 (ab32124, Abcam), Caspase3 (ab13847, Abcam) and GAPDH (ab9485, Abcam) at 1:1000 dilution at 4 °C overnight. After washing with 1 × TBST four times, the membranes were further incubated with horseradish peroxidase-conjugated secondary antibody IgG (ab6721,1:2000, Abcam) at room temperature for 1.5h, and washed with 1 × TBST four times. The photosensitive films were placed in an exposure box and exposed for 1 min in a dark room, and then developed and fixed. The films were taken with Lab Works™ gel imaging and analysis system to analyze the brightness value of each protein band.
2.5. Simulated molecular docking
The X-ray crystal structures of the COVID-19 related targets, including ACE2 (PDB:1R4L), Mpro (PDB: 2AMD), and RdRp (PDB: 4KHM), were downloaded from the RCSB protein data bank (http://www.pdb.org/)(Burley et al., 2018) and their resolutions were carefully examined. The semi-flexible molecular docking simulation was carried out using the program LibDock implemented in Discovery Studio 2.5 (DS 2.5, Accelrys, San Diego, USA). To evaluate the binding affinity of the components with their targets, the score was calculated using the customizable scoring function of LibDock. The LibDock scores >80% of that of the initial ligands represented a strong binding affinity of the components with their corresponding targets. A visualization of the herb-component-target interactive network was constructed using Cytoscape 3.6.0.
3. Results
3.1. Establishment of MXSGD-active components-targets network
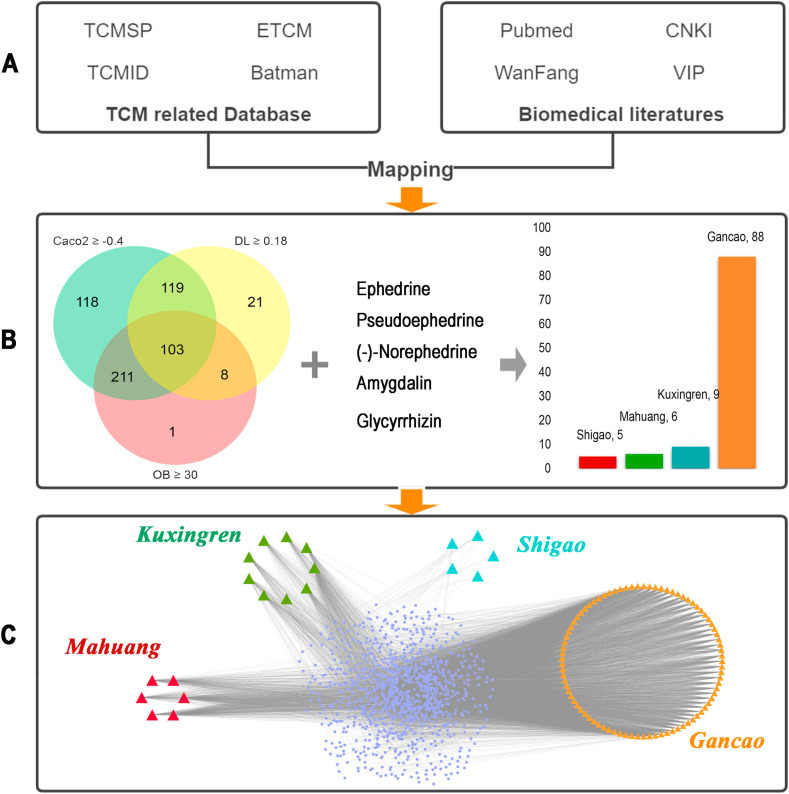
Based on TCM database and literature resources, the related components of MXSGD were integrated (Fig. 1 A). 103 bioactive components of MXSGD were collected in this study according to screening criteria. Notably, the criteria were only based on the consensus formed by the experience of previous researchers, which cannot fully reflect the authenticity of the active ingredients in relevant herbal medicine. Therefore, referring to previous studies (Miao et al., 2020; Song et al., 2016; EI-Saber et al., 2020), we retained the five most abundant, representative and effective components of related herbs, including ephedrine, pseudoephedrine, (−)-normedrine, amygdalin and glycyrrhizin. Then, 108 active components were identified and selected, including 6 from Mahuang, 9 from Kuxingren, 5 from Shigao, and 88 from Gancao (Fig. 1B, Supplementary 1). Ultimately, 105 components were deemed to be core compounds of MXSGD following removal of duplicates, and “Lipinski Rule” and screening related information were listed in Supplementary 1. 1025 potential targets were identified from the 105 bioactive components of MXSGD with ligand-based prediction strategy (Supplementary 2). The number of these targets in Mahuang, Kuxingren, Shigao, and Gancao was 234, 419, 33, and 905, respectively. The MXSGD component-targets network was constructed with Cytoscape 3.6.0 for a systematic and holistic view (Fig. 1C).
Fig. 1.
MXSGD component-targets network. (A)Two broad categories database: TCM database and literature resources. (B) 103 bioactive components screened by three criteria (red section represents the components of OB ≥30, yellow section represents DL ≥0.18, green section represents the components of Caco2 ≥-0.4), and 5 ingredients which do not meet the criteria but were included as major components of the herbal medicine, including 6 from Mahuang, 9 from Kuxingren, 5 from Shigao and 88 from Gancao. (C) Construction of MXSGD bioactive component-putative targets visual network. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Candidate targets of MXSGD against COVID-19
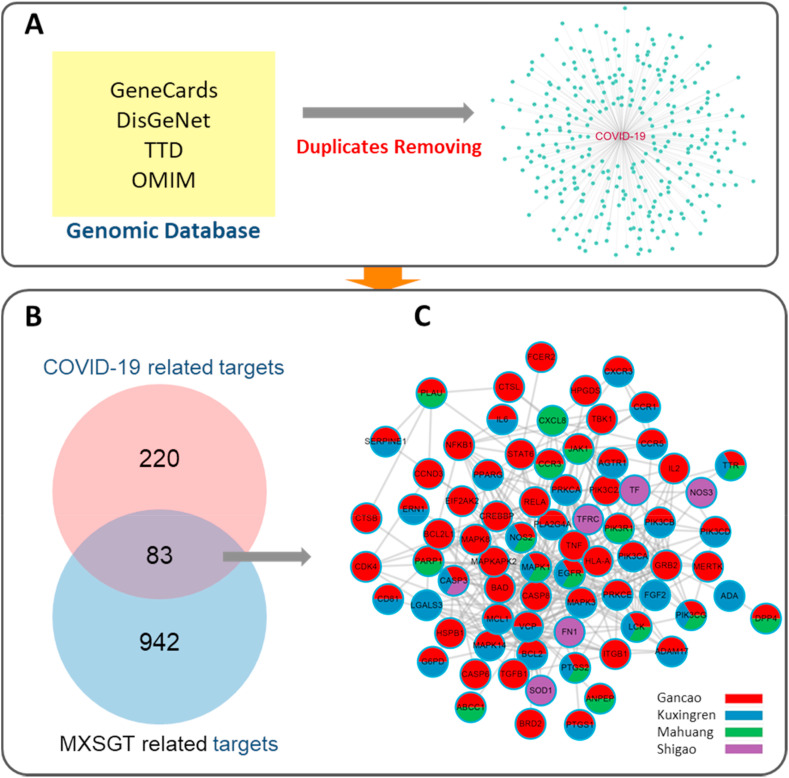
We needed to identify the overlap between the target lists of drug compounds with that of disease to determine the candidate targets of MXSGD against COVID-19. First, 303 COVID-19 related genes were collected from GeneCards, DisGeNET, TTD, and OMIM after removing duplicates and are shown in Fig. 2 A, with the information listed in Supplementary 3. Then 83 common targets of MXSGD and COVID-19 were identified through overlapping shown in Fig. 2B; the number of targets in Mahuang, Kuxingren, Shigao, and Gancao was 17, 36, 6, and 74, respectively (Supplementary 4). However, these targets were extracted from 103 components in MXSGD, and most targets were shared by two or more of the four herbals ingredients. In order to reveal the pharmacological mechanisms of MXSGD against COVID-19, the PPI network of candidate targets of MXSGD against COVID-19 was constructed and presented in Fig. 2C. The network consists of 76 nodes and 328 edges, and more than half of the nodes are derived from two or more herbs.
Fig. 2.
Identification of candidate targets of MXSGD against COVID-19. (A) Four genomic databases for screening of COVID-19 related targets, finally 303 COVID-19 related genes were collected after removing duplicates. (B) 83 candidate targets of MXSGD against COVID-19 were identified. (C) PPI network of candidate targets of MXSGD against COVID-19, where nodes are color-coded based on the identities of the gene list of different herbs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Potential mechanisms of MXSGD’s effects on COVID-19
According to biological enrichment, 160 GO terms were enriched (Supplementary 5). The top 10 terms in molecular function (MF), biological process (BP), and cellular component (CC) including 5 BP terms (lipopolysaccharide-mediated signaling pathway, inflammatory response, extrinsic apoptotic signaling pathway in absence of ligand, and intrinsic apoptotic signaling pathway in response to DNA damage), 3 CC terms (external side of plasma membrane, cytosol, and phosphatidylinositol 3-kinase complex), and the 1 MF term MAP kinase activity (p-value ≤ 0.05 and FDR ≤ 0.05). Moreover, 111 KEGG pathways were enriched and 63 of them were significant (p-value ≤ 0.05 and FDR ≤ 0.05), most of them are related to signal transduction, immune system and virus infection (Supplementary 6). Signal transduction related pathways including HIF-1 signaling pathway, TNF signaling pathway, PI3K-Akt signaling pathway, MAPK signaling pathway, JAK-STAT signaling pathway and NF-kappa B signaling pathway etc. Immune system related pathways including Toll-like receptor signaling pathway, T cell receptor signaling pathway, and NOD-like receptor signaling pathway and Chemokine signaling pathway etc. Virus infection related pathways including hepatitis B, hepatitis C, influenza A, Epstein-Barr virus, measles, and HTLV-1 infection signaling pathways. Most of the pathways are regulated by a combination of two or three herbal medicines (Supplementary 7).
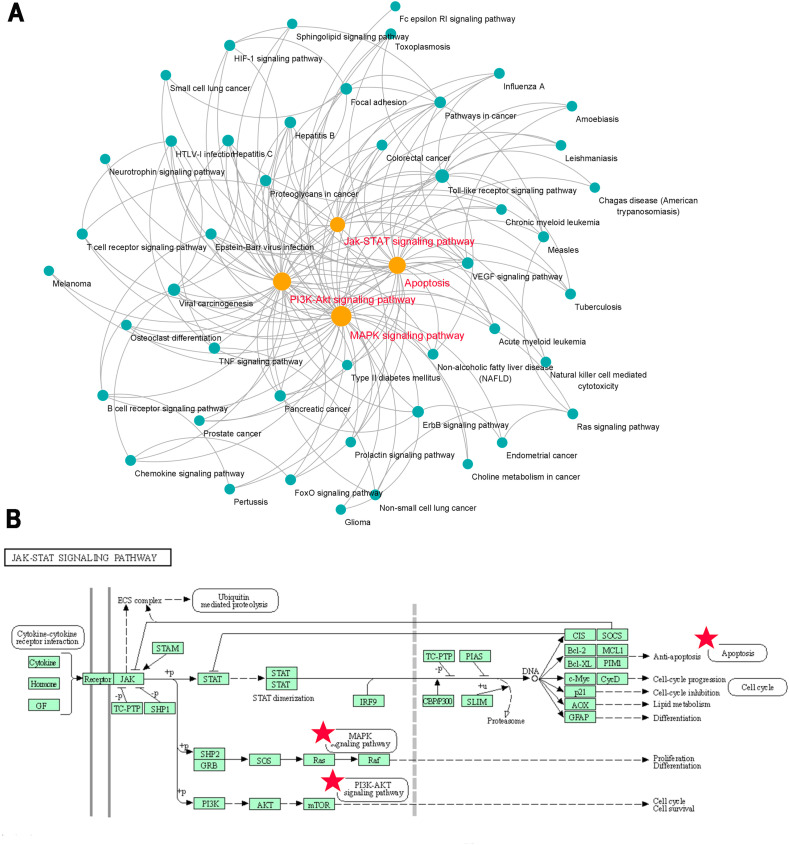
In order to clarify the interaction relationship between signal pathways and find the key roles, we established the relation network of signal pathways in Top50 according to the p-value. According Fig. 3 A, we found MAPK, PI3K-Akt, Apoptosis and Jak-STAT signaling pathway were connected more pathways in network, which indicated that changes in these pathways may affect the entire network to create a chain reaction. Furthermore, Fig. 3B shows that JAK-STAT pathway is upstream of the other 3 pathways, and this pathway is mainly influenced by Gancao and Kuxingren through Table 1 , and related targets include STAT6, PIK3CG, IL-6, CCND3, GRB2, PIK3CD, PIK3CA, JAK1, BCL2L1, IL-2. Therefore, we focused on the effect of MXSGD on Jak-STAT pathway in subsequent experimental studies.
Fig. 3.
The relevant signaling pathway network and its key role of MXSGD against COVID-19. (A) Pathway to pathway network. Each circle represents a signaling pathway, the larger the circle, the more connected pathways. (B) JAK-STAT signaling pathway affects 3 important downstream signaling pathways, including MAPK signaling pathway, PI3K-Akt signaling pathway and Apoptosis signaling pathway).
Table 1.
LibDock scores of the candidate components in TCHMs for COVID-19 related targets.
| Target | Compound ID | Molecule name | Libdock score | Source |
|---|---|---|---|---|
| ACE2 | MOL001320 | Amygdalin | 167.60 | Kuxingren |
| MOL002372 | Spinacen | 147.10 | Kuxingren | |
| MOL004806 | Euchrenone | 146.38 | Gancao | |
| MOL005001 | Gancaonin H | 143.41 | Gancao | |
| MOL004885 | Licoisoflavanone | 141.71 | Gancao | |
| MOL004989 | 6-prenylated eriodictyol | 139.18 | Gancao | |
| MOL005008 | Glycyrrhiza flavonol A | 138.45 | Gancao | |
| MOL004905 | Glyuranolide | 138.35 | Gancao | |
| MOL004856 | Gancaonin A | 134.73 | Gancao | |
| MOL004857 | Gancaonin B | 133.78 | Gancao | |
| Mpro | MOL001320 | Amygdalin | 164.17 | Kuxingren |
| MOL004876 | Glycyrrhizin | 159.87 | Gancao | |
| MOL002372 | Spinacen | 152.57 | Kuxingren | |
| MOL004935 | Sigmoidin-B | 149.57 | Gancao | |
| MOL004806 | Euchrenone | 148.04 | Gancao | |
| MOL005001 | Gancaonin H | 146.33 | Gancao | |
| MOL004805 | Shinflavanone | 143.99 | Gancao | |
| MOL004949 | Isolicoflavonol | 142.35 | Gancao | |
| MOL005007 | Glyasperins M | 140.04 | Gancao | |
| MOL000497 | Licochalcone a | 139.55 | Gancao | |
| RdRp | MOL001320 | Amygdalin | 144.05 | Kuxingren |
| MOL002311 | Glycvrol | 142.96 | Gancao | |
| MOL004935 | Sigmoidin B | 140.41 | Gancao | |
| MOL005008 | Glycyrrhiza flavonol A | 139.96 | Gancao | |
| MOL004948 | Isoglycyrol | 139.29 | Gancao | |
| MOL002372 | Spinacen | 139.21 | Kuxingren | |
| MOL004857 | Gancaonin B | 138.63 | Gancao | |
| MOL004904 | licopyranocoumarin | 137.38 | Gancao | |
| MOL004863 | Gancaonin L | 135.91 | Gancao | |
| MOL004980 | Inflacoumarin A | 135.79 | Gancao |
3.4. In vitro activity verification
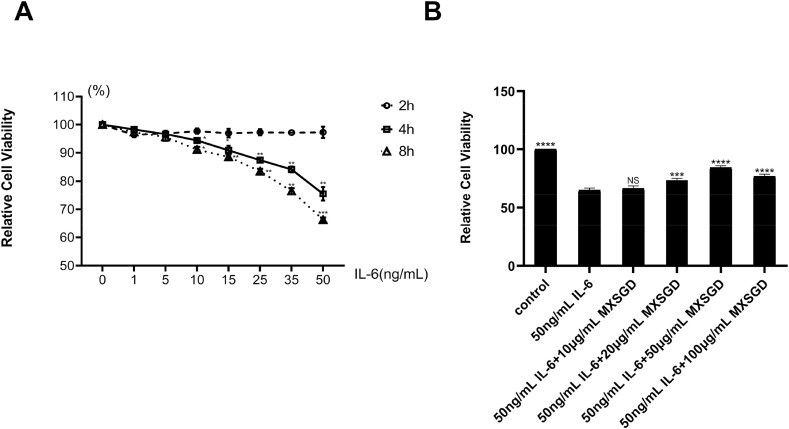
As shown in Fig. 4 A, with the increase of IL-6 concentration and the extension of the intervention time, the survival rate of RLE-6TN cells gradually decreases, especially after 50 ng/mL IL-6 intervention for 8h, which indicated that IL-6 could cause RLE-6TN cells damage. After the different concentrations of MXSGD rat medicated serum treatment, the survival rate of RLE-6TN cells was significantly increased in a dose-dependent manner compared with 50 ng/mL IL-6 (Fig. 4B), which indicated that MXSGD could effectively inhibit IL-6 induced RLE-6TN cells damage. The Western blot results found that after the different concentrations of MXSGD rat medicated serum treatment, the protein expression levels of p-JAK2, p-STAT3, Bax and Caspase 3 significantly decreased, while the protein expression level of Bcl-2 significantly increased in a dose-dependent manner (Fig. 5 ). It indicated that MXSGD could effectively inhibit IL-6 mediated JAK-STAT signal pathway related protein expression level, thereby inhibiting RLE-6TN cells damage.
Fig. 4.
the survival rate of RLE-6TN cells after treated with IL-6 and MXSGD. (A) the survival rate of RLE-6TN cells after treated with the different concentrations of IL-6 for 2h, 4h and 8h (Compared with 0 ng/mL IL-6, *p<0.05, **p <0.01, ***p<0.001). (B) the survival rate of RLE-6TN cells after treated with the different concentrations of MXSGD and 50 ng/mL IL-6 for 8h (Compared with IL-6 group, ***p<0.001, ****p<0.0001).
Fig. 5.
the expression level of related proteins and their phosphorylation in the IL-6 mediated JAK-STAT signaling pathway after MXSGD treatment. (A) After MXSGD and IL-6 treatment, the expression level of related proteins and their phosphorylation in the IL-6 mediated JAK-STAT signaling pathway examined by Western blotting assay. (B) The expression level of Bax, Bcl-2 and Caspase 3 after treated with the different concentrations of MXSGD and 50 ng/mL IL-6. (C) The expression level of p-JAK2 after treated with the different concentrations of MXSGD and 50 ng/mL IL-6. (D) The expression level of p-STAT3 after treated with the different concentrations of MXSGD and 50 ng/mL IL-6 (Compared with control group, ##p <0.01. Compared with 50 ng/mL IL-6, *p<0.05, **p <0.01).
3.5. Molecular docking simulation and binding site analysis for candidate components
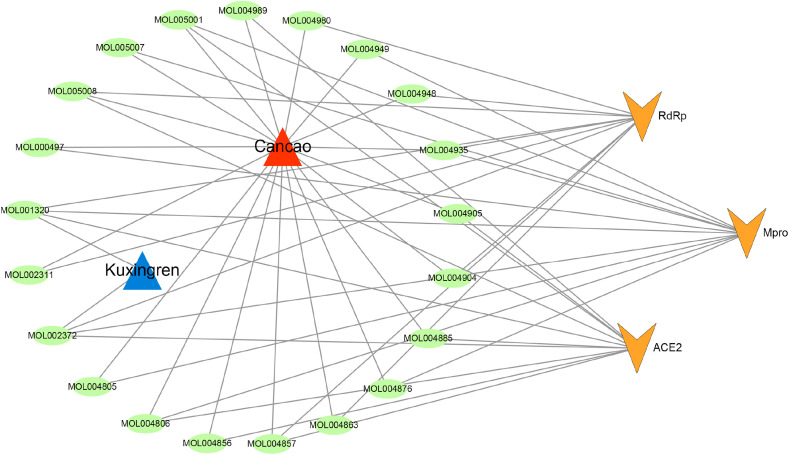
Since the main component of Shigao is hydrous calcium sulfate (CaSO4·2H2O), we focused on Mahuang, Kuxingren and Gancao for indepth analysis. As shown in Table 1, according to the LibDock scores screening results, the components with strong potential affinity (Top 10) with ACE2, Mpro and RdRp are mainly from Kuxingren and Gancao. A visualization of the herb-component-target interactive network is presented in Fig. 6 , and indicates that the multiple components in Gancao and Kuxingren could exert anti-coronavirus activity by acting on multiple targets.
Fig. 6.
A visualization of herb-component-target interactive network. Red and blue triangles, green ellipses and orange arrows represent herbs, candidate components and COVID-19 related targets, respectively. The edges represent the mutual relations among the herbs, ingredients and targets. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
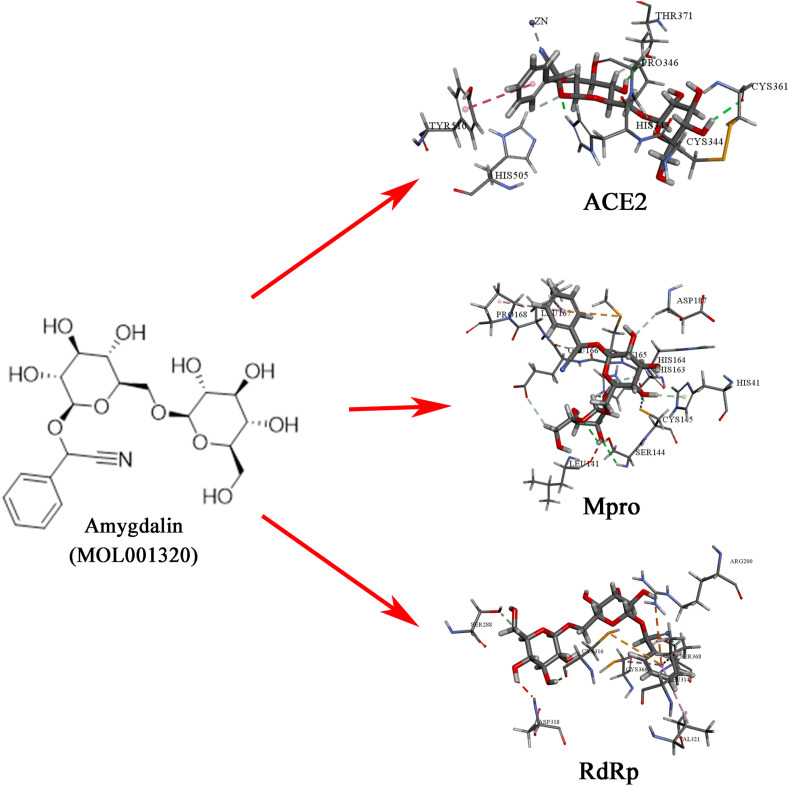
In order to reveal the interaction between the candidate compounds and COVID-19 related targets, the binding sites of the compounds and targets were further analyzed. According to the LibDock scores of the compounds and targets in Supplementary 8, amygdalin was selected as the optimal candidate component for ACE2, Mpro, and RdRp. The binding atoms, distance, and interaction category of amygdalin with COVID-19 related targets are presented in Table 2 . There were 7 hydrogen bonds (HIS345, THR371, CYS361, PRO346, HIS505, THR371 and CYS344) and 1 hydrophobic interaction (TYR510) formed between amygdalin and ACE2, 9 hydrogen bonds (SER144, CYS145, HIS163, ASP187, GLU166, HIS164, GLU166 and HIS41) and 2 hydrophobic interactions (LEU167, PRO168) between amygdalin and Mpro, and 4 hydrogen bonds (CYS316, SER368, CYS316 and SER288), 1 electrostatic (ARG200), and 3 hydrophobic (LEU314, VAL321 and CYS366) interactions between amygdalin and RdRp (Fig. 7 ). The results indicate that amygdalin could be a potential inhibitor for ACE2, Mpro, and RdRp, and may be promising as a potential anti-coronavirus drug.
Table 2.
Binding atoms, distance, and interaction category of amygdalin with COVID-19 related targets.
| Targets | Binding atoms | Distance | Interaction category | Types |
|---|---|---|---|---|
| ACE2 | A:HIS345:HE2 - MOL001320:O10 | 1.84229 | Hydrogen Bond | Conventional Hydrogen Bond |
| MOL001320:H45 - A:THR371:OG1 | 2.48164 | Hydrogen Bond | Conventional Hydrogen Bond | |
| MOL001320:H55 - A:CYS361:O | 2.40125 | Hydrogen Bond | Conventional Hydrogen Bond | |
| A:PRO346:HD2 - MOL001320:O30 | 2.45426 | Hydrogen Bond | Carbon Hydrogen Bond | |
| A:HIS505:HE1 - MOL001320:O10 | 2.72189 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H44 - A:THR371:OG1 | 2.85535 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H54 - A:CYS344:O | 2.43107 | Hydrogen Bond | Carbon Hydrogen Bond | |
| A:TYR510 - MOL001320 | 4.9534 | Hydrophobic | Pi-Pi T-shaped | |
| Mpro | A:SER144:HN - MOL001320:O26 | 2.76804 | Hydrogen Bond | Conventional Hydrogen Bond |
| A:CYS145:SG - MOL001320:O17 | 3.71570 | Hydrogen Bond | Conventional Hydrogen Bond | |
| A:HIS163:HE2 - MOL001320:O24 | 2.11830 | Hydrogen Bond | Conventional Hydrogen Bond | |
| A:ASP187:HA - MOL001320:O13 | 2.68742 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H38 - A:GLU166:O | 2.50247 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H42 - A:HIS164:O | 2.69184 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H46 - A:HIS164:O | 2.63320 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H58 - A:GLU166:OE1 | 2.84674 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H45 - A:HIS41 | 2.32425 | Hydrogen Bond | Pi-Donor Hydrogen Bond | |
| MOL001320 - A:LEU167 | 5.19132 | Hydrophobic | Pi-Alkyl | |
| MOL001320 - A:PRO168 | 5.19354 | Hydrophobic | Pi-Alkyl | |
| RdRp | MOL001320:H53 - A:CYS316:O | 1.85071 | Hydrogen Bond | Conventional Hydrogen Bond |
| A:SER368:HB2 - MOL001320:N9 | 2.47321 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H50 - A:CYS316:O | 2.81425 | Hydrogen Bond | Carbon Hydrogen Bond | |
| MOL001320:H57 - A:SER288:OG | 2.41247 | Hydrogen Bond | Carbon Hydrogen Bond | |
| A:ARG200:NH1 - MOL001320 | 4.72350 | Electrostatic | Pi-Cation | |
| MOL001320 - A:LEU314 | 4.49061 | Hydrophobic | Pi-Alkyl | |
| MOL001320 - A:VAL321 | 5.08548 | Hydrophobic | Pi-Alkyl | |
| MOL001320 - A:CYS366 | 4.78123 | Hydrophobic | Pi-Alkyl |
Fig. 7.
The chemical structure of amygdalin and the binding sites with ACE2, Mpro, and RdRp.
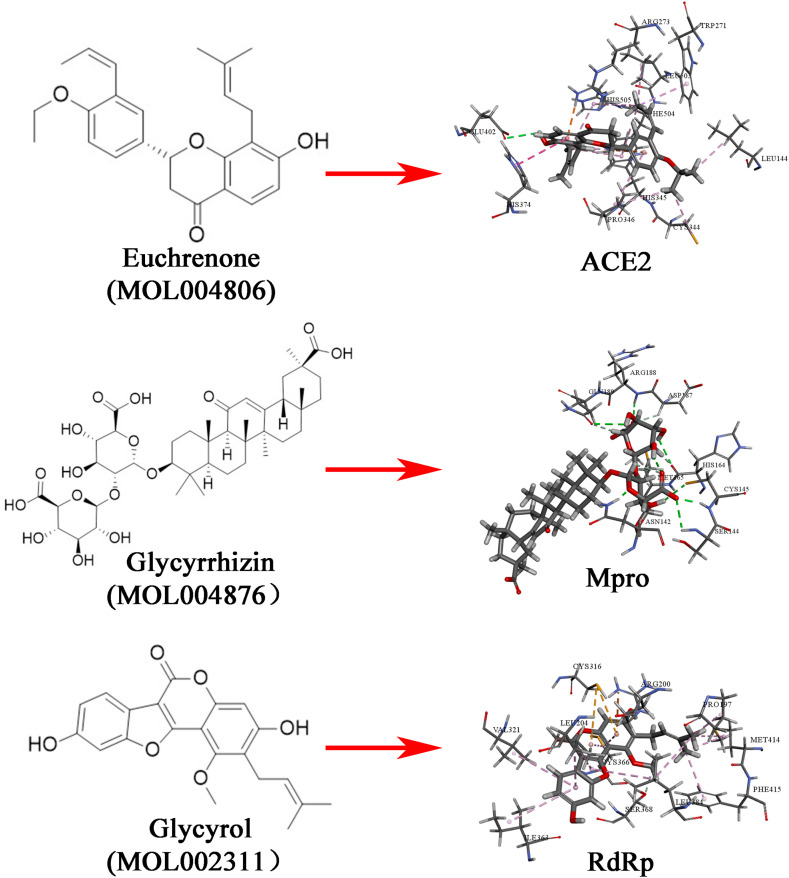
Euchrenone, glycyrrhizin, and glycyrol, three candidate components of Gancao, also exhibited affinity interactions with ACE2, Mpro, and RdRp, respectively. As shown in Fig. 7, Fig. 8 hydrogen bonds (HIS345, GLU402, and ARG273), 2 electrostatic interactions (ARG273 and HIS345), and 12 hydrophobic interactions (HIS374, LEU503, LEU144, CYS344, PRO346, TRP271, HIS345, PHE504, HIS505 and PRO3464) formed between euchrenone and ACE2, 11 hydrogen bonds (ASN142, SER144, CYS145, ARG188, CYS145, HIS164, GLN189, MET165, ASP187, HIS164 and GLN189) formed between glycyrrhizin and Mpro, and 1 hydrogen bond (SER368), 1 electrostatic interaction (ARG200), and 11 hydrophobic interactions (MET414, PRO197, LEU384, MET414, PHE415, CYS366, LEU204, LEU384, LEU204, VAL321 and ILE363) formed between glycyrol and RdRp. The results indicate that a variety of chemical components, such as triterpenoid saponins, flavones and their glycosides from Gaocao, one of the more frequently used herbs in TCM, could be promising as potential anti-coronavirus drugs and are worthy of further research and development.
Fig. 8.
The chemical structures of three components of Gancao and their binding sites with ACE2, Mpro, and RdRp.
4. Discussion
As a newly emerged acute respiratory infectious disease, COVID-19 has been well controlled in China, but it is still spreading in many countries around the world. Effect interventions, such as vaccines, monoclonal antibodies, oligonucleotide-based therapies, peptides, interferon therapies, and small-molecule drugs, are likely to require months to years to develop (Liu et al., 2020a, Liu et al., 2020b; Li et al., 2020). Since the advantage of TCM intervention in complex diseases lies in its focus on the recovery of overall function, it is urgent and reasonable that the effective complementary and alternative treatment strategies from the perspective of TCM should be explored for the prevention and control of the current epidemic. The concept of network pharmacology is consistent with TCM theory, and it is suitable for exploring the mechanisms of prescriptions composed of multiple ingredients. Therefore, we applied network pharmacology combined with simulated molecular docking analysis to examine the mechanism of MXSGD against COVID-19, as well as important components with good binding ability to COVID-19 targets.
In this study, 83 potential targets of MXSGD against COVID-19 were extracted from 103 components of MXSGD and most targets were shared by two or more herbs. The results suggest that several herbs and most compounds from MXSGD affect multiple targets. For example, (−)-norephedrine, pseudoephedrine, amygdalin, butanedioic acid, glycyrrhizin, and gancaonin B acted on 7, 5,10, 13, 11, and 17 COVID-19 related targets, respectively. Although the number of putative targets in each single herb was different, the overlapping targets between different herbs were numerous. In other words, multiple compounds from MXSGD may have the same target, providing synergistic effects. In combination with analysis of enrichment of GO and KEGG pathways, we believe that the mechanism of action of MXSGD against COVID-19 is closely related to antivirus, anti-inflammatory and immune regulation. Especially, JAK-STAT signaling pathways may be the key pathways for MXSGD against COVID-19.
Taking full advantage of these opportunities requires a better understanding of the mechanisms involved in disease. The latest research confirmed that several inflammatory cytokines that were involved in CRS and correlate with adverse clinical outcomes in COVID-19 employ a distinct intracellular signaling pathway mediated by Janus kinases (JAKs), and suggested that JAK-STAT signaling might be an excellent therapeutic target for the development of much needed therapies for COVID-19(Luo et al., 2020). The potential role of JAK inhibitors in treating patients with COVID-19-associated CRS is an area of active investigation with multiple ongoing clinical trials. Considering the global urgency of alleviating the COVID-19 pandemic, inhibiting JAK seems to be an attractive treatment option for developing much-needed therapies. In addition, the results from a previous study indicated that MXSGD may regulate the PI3K/AKT signaling pathway against influenza virus A/WSN/33 (H1N1), with broad-spectrum inhibitory activity against different strains of human influenza A viruses, including clinical oseltamivir-resistant isolates and an H1N1pdm strain (Hsieh et al., 2012b). PI3K/AKT signaling pathway plays many roles in viral infection, such as through its anti-apoptotic function during viral entry, and PI3K inhibitors cause accumulation of influenza virus particles on the cell surface, unable to move to the endosome (Wong et al., 2005; Ehrhardt et al., 2006). Combined with our research findings, MXSGD might have an advantage in regulating JAK-STAT and PI3K/AKT signaling pathway, and mainly dependent on Gancao and Kuxingren. Furthermore, through in vitro experiments, we confirmed MXSGD could effectively inhibit IL-6 mediated JAK-STAT signal pathway related protein expression level, decreased the protein expression levels of p-JAK2, p-STAT3, Bax and Caspase 3, and increased the protein expression level of Bcl-2, thereby inhibiting RLE-6TN cells damage.
Through the above analysis, we believed that MXSGD had certain antiviral effects, so we further discovered the effective components which could bind to the key targets of COVID-19 by simulating molecular docking. In this study, several components of Kuxingren and Gancao presented excellent binding ability, which was consistent with the above discussion. Our results suggested that euchrenone, glycyrrhizin, and glycyrol, three candidate components in Gancao, exhibited superior affinity interactions with ACE2, Mpro, and RdRp, respectively. Papers and reports on the antiviral effects of Gancao had now become relatively widespread, especially concerning the component glycyrrhizin. It was reported that glycyrrhizin had the best potential activity in inhibiting replication of the two clinical isolates of coronavirus (FFM-1 and FFM-2) from SARS patients (Cinatl et al., 2003). Furthermore, studies indicate that glycyrrhizin could exhibit direct anti-HBV effects, improving liver dysfunction for chronic hepatitis B patients and the immune status of HBV (Sun et al., 2019). In contrast, the antiviral potential of Kuxingren has not been aroused much attention, and relevant reports are insufficient. However, in our research, amygdalin, one of the main active components of Kuxingren, was selected as the optimal candidate component for ACE2, Mpro and RdRp. which might be promising as a potential anti-coronavirus drug.
In summary, this study combined network pharmacology and simulated molecular docking method to analyze the pharmacodynamic material basis and mechanism of MXSGD in treating COVID-19, and the in vitro experiment verified our prediction results to a certain extent. Although Gancao and Kuxingren may have a more prominent role in treating novel coronavirus, each TCM prescription is a complex system and has multiple targets and links in curing disease. From the viewpoint of prescription and compatibility, Mahuang and Shigao may have the effect of relieving clinical symptoms of COVID-19 patients, such as reducing fever, promoting sweat excretion, and relieving cough, etc. However, the experimental verification and more proof is necessary to further confirm.
5. Conclusion
This work explained the positive characteristics of multi-component, multi-target, and multi-approach intervention with MXSGD in combating COVID-19, and preliminary revealed the antiviral and anti-inflammatory pharmacodynamic substances and mechanism of MXSGD, which might provide insights into the vital role of TCM in the prevention and treatment of COVID-19.
Declaration of competing interest
The authors declare no conflict of interest, financial or otherwise.
Acknowledgments
This work was supported by the National Key Research and Development Project of China (2018YFC1704106 and 2018YFC1704100), National Natural Science Foundation of China (81630080, 81703931 and 81903792) and China Postdoctoral Science Foundation Grant (2019M650600).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jep.2021.113854.
Author's contributions
Yuan Li and Fuhao Chu performed main analysis and drafted the manuscript. Xia Ding and Xiaohong Gu designed the research. Ping Li, Nadia Johnson and Tao Li helped for introduction and discussion. Yan Wang, Rongxian An, Dantong Wu, Jiena Chen and Zeqi Su assisted in the data cleaning and preparation. All authors wrote, read, and approved the manuscript.
Data availability statement
The data used to support the findings of our study are included within the article, or within the Supplementary Material. For additional information or questions, contact the corresponding author.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Amberger J.S., Bocchini C.A., Scott A.F., Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019;47(D1):D1038–D1043. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K., Berman H.M., Christie C., Duarte J.M., Feng Z., Westbrook J., Young J., Zardecki C. RCSB Protein Data Bank: sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education. Protein Sci. 2018;27(1):316–330. doi: 10.1002/pro.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Boutros P.C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ji Z.L., Chen Y.Z. TTD: therapeutic target database. Nucleic Acids Res. 2002;30(1):412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las R.J., Fontanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 2010;6(6) doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- eEhrhardt C., Marjuki H., Wolff T., Nurnberg B., Planz O., Pleschka S., Ludwig S. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol. 2006;8:1336–1348. doi: 10.1111/j.1462-5822.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- El-Saber Batiha G., Magdy Beshbishy A., El-Mleeh A., Abdel-Daim M.M. Prasad devkota, H., traditional uses, bioactive chemical constituents, and pharmacological and toxicological activities of Glycyrrhiza glabra L. (fabaceae) Biomolecules. 2020;10(3):352. doi: 10.3390/biom10030352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller D., Grosdidier A., Wirth M., Daina A., Michielin O., Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42:W32–W38. doi: 10.1093/nar/gku293. Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao da C., Xiao P.G. Network pharmacology: a Rosetta Stone for traditional Chinese medicine. Drug Dev Res. 2014;75(5):299–312. doi: 10.1002/ddr.21214. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hu K., Guan W.J., Bi Y., Zhang W., Li L., Zhang B., Liu Q., Song Y., Li X., Duan Z., Zheng Q., Yang Z., Liang J., Han M., Ruan L., Wu C., Zhang Y., Jia Z.H., Zhong N.S. Efficacy and safety of Lianhuaqingwen capsules, a repurposed Chinese herb, in patients with coronavirus disease 2019: a multicenter, prospective, randomized controlled trial. Phytomedicine. Phytomedicine. 2020:153242. doi: 10.1016/j.phymed.2020.153242. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., Wen C. Tcmid 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 2018;46(D1):D1117–D1120. doi: 10.1093/nar/gkx1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li M., Zhou Z., Guan X., Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020;111:102452. doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Clercq E.D. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;111(102452) doi: 10.1038/d41573-020-00016-0. https://www.nature.com/articles/d41573-020-00016-0#ref-CR1 [DOI] [PubMed] [Google Scholar]
- Li J., Zhao P., Li Y., Tian Y., Wang Y. Systems pharmacology-based dissection of mechanisms of Chinese medicinal formula Bufei Yishen as an effective treatment for chronic obstructive pulmonary disease. Sci. Rep. 2015;5:15290. doi: 10.1038/srep15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Qu H., Wang S., Wei J., Zhang L., Ma R., Lu J., Zhu J., Zhong W.D., Jia Z. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34:2515–2517. doi: 10.1093/bioinformatics/bty124. [DOI] [PubMed] [Google Scholar]
- Li S., Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin. J. Nat. Med. 2013;11(2):110–120. doi: 10.1016/S1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Liu W., Morse J.S., Lalonde T., Xu S. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Guo F., Wang Y., Li C., Zhang X., Li H., Diao L., Gu J., Wang W., Li D., He F. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci. Rep. 2016;6:21146. doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan X., Zhang L.J., Li X.Q., Rahman K., Zhang H., Chen H.Z., Zhang W.D. Compound-based Chinese medicine formula: from discovery to compatibility mechanism. J. Ethnopharmacol. 2020;254:112687. doi: 10.1016/j.jep.2020.112687. [DOI] [PubMed] [Google Scholar]
- Miao S.M., Zhang Q., Bi X.B., Cui J.L., Wang M.L. A review of the phytochemistry and pharmacological activities of Ephedra herb. Chin. J. Nat. Med. 2020;18(5):321–344. doi: 10.1016/S1875-5364(20)30040-6. [DOI] [PubMed] [Google Scholar]
- Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China - key questions for impact assessment. N. Engl. J. Med. 2020;382(8):692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot J.R. Drugs, leads, and drug-likeness: an analysis of some recently launched drugs. Bioorg. Med. Chem. Lett. 2002;12(12):1647–1650. doi: 10.1016/s0960-894x(02)00244-5. [DOI] [PubMed] [Google Scholar]
- Qu Y.F., Fang W., Jing Y.Z., Qin C., Niu X.C., Zhang N., Zhang J.R. Forty cases of common COVID-19 treated with modified ephedra and apricot kernel and Gypsum and licorice decoction combined with western medicine routine treatment. Henan.Tradit. Chin. Med. 2020;40(5):666–669. [Google Scholar]
- Rebhan M., Chalifa-Caspi V., Prilusky J., Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13(4):163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Li P., Guo Z., Tao W., Yang Y., Xu X., Li Y., Wang Y., Yang L. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Ye H.Q., Deng C.L., Li R., Zhang B., Gong P. A nucleobase-binding pocket in a viral RNA-dependent RNA polymerase contributes to elongation complex stability. Nucleic Acids Res. 2020;48(3):1392–1405. doi: 10.1093/nar/gkz1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sietsema W.K. The absolute oral bioavailability of selected drugs. Int. J. Clin. Pharmacol. Ther. Toxicol. 1989;27(4):179–211. [PubMed] [Google Scholar]
- Song S., Ma Q., Tang Q., Chen F., Xing X., Guo Y., Guo S., Tan X., Luo J. Stereoselective metabolism of amygdalin-based study of detoxification of semen armeniacae amarum in the herba ephedrae-semen armeniacae amarum herb pair. J. Ethnopharmacol. 2016;179:356–366. doi: 10.1016/j.jep.2015.12.019. [DOI] [PubMed] [Google Scholar]
- Sun Z.G., Zhao T.T., Lu N., Yang Y.A., Zhu H.L. Research progress of glycyrrhizic acid on antiviral activity. Mini Rev. Med. Chem. 2019;19(10):826–832. doi: 10.2174/1389557519666190119111125. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S.X., Yi Q.J., Fan S.B., Lv J.L., Zhang X.X., Guo L., Lang C.H., Xiao Q., Xiao K.H., Yi Z.J., Qiang M., Xiang J.L., Zhang B.S., Chen Y.P. 2020. Characteristics of Lymphocyte Subsets and Cytokines in Peripheral Blood of 123 Hospitalized Patients with 2019 Novel Coronavirus Pneumonia (NCP) (Preptint) MedRxiv 2020.02.10. [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;94(7):e00127. doi: 10.1128/JVI.00127-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shen Y., Wang S., Li S., Zhang W., Liu X., Lai L., Pei J., Li H. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45(W1):W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W.R., Chen Y.Y., Yang S.M., Chen Y.L., Horng J.T. Phosphorylation of PI3K/Akt and MAPK/ERK in an early entry step of enterovirus 71. Life Sci. 2015;78(1):82–90. doi: 10.1016/j.lfs.2005.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.W., Lu L., Liang S.W., Chen C., Wang S.M. Application of drug-target prediction technology in network pharmacology of traditional Chinese medicine. Zhongguo Zhong Yao Za Zhi. 2016;41:377–382. doi: 10.4268/cjcmm20160303. [DOI] [PubMed] [Google Scholar]
- Wu P., Hao X., Lau E., Wong J.Y., Leung K., Wu J.T., Cowling B.J., Leung G.M. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveill. 2020;25(3):2000044. doi: 10.2807/1560-7917.ES.2020.25.3.2000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin S., Cheng X., Zhu B., Liao X., Yang F., Song L., Shi Y., Guan X., Su R., Wang J., Xing L., Xu X., Jin L., Liu Y., Zhou W., Zhang D., Liang L., Yu Y., Yu R. Clinical retrospective study on the efficacy of Qingfei Paidu decoction combined with Western medicine for COVID-19 treatment. Biomed. Pharmacother. 2020;129:110500. doi: 10.1016/j.biopha.2020.110500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.Y., Zhang Y.Q., Liu Z.M., Chen T., Lv C.Y., Tang S.H., Zhang X.B., Zhang W., Li Z.Y., Zhou R.R., Yang H.J., Wang X.J., Huang L.Q. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47(D1):D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J. Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet. Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingyao Zhou, Bin Zhou, Lars Pache, Max Chang, Alireza Hadj, Hadj, Olga Tanaseichuk, Christopher Benner, Sumit K., Sumit Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Chen J., Xu X., Li Y., Zhao H., Fang Y., Li X., Zhou W., Wang W., Wang Y. A systematic prediction of multiple drug-target interactions from chemical, genomic, and pharmacological data. PloS One. 2012;7(5) doi: 10.1371/journal.pone.0037608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Huai Y., Miao Z., Qian A., Wang Y. Systems pharmacology for investigation of the mechanisms of action of traditional Chinese medicine in drug discovery. Front. Pharmacol. 2019;10:743. doi: 10.3389/fphar.2019.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Baak J.P., Li S., Xiao W., Ren H., Yang H., Gan Y., Wen C. Network pharmacology analysis of the therapeutic mechanisms of the traditional Chinese herbal formula Lian Hua Qing Wen in Corona virus disease 2019 (COVID-19), gives fundamental support to the clinical use of LHQW. Phytomedicine. 2020;79:153336. doi: 10.1016/j.phymed.2020.153336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., He G.L., Lu F.G., Li L., Zhang B., Dai B., Wei K., Chen S.Q., Ning Y., Hu J., Wu T. Mechanism research of anti influenza virus of ephedra decocted earlier Maxing Shigan Decoction from the expression level of IFN-α/β protein mediated by TLR7/8. China.J. Tradit.Chin. Med.Pharm. 2019;34(3):1188–1193. 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of our study are included within the article, or within the Supplementary Material. For additional information or questions, contact the corresponding author.