Graphical abstract
Keywords: SARS-CoV, SARS-CoV-2, MERS-CoV, Medicinal chemistry, Molecular modeling
Abstract
Severe respiratory infections were highlighted in the SARS-CoV outbreak in 2002, as well as MERS-CoV, in 2012. Recently, the novel CoV (COVID-19) has led to severe respiratory damage to humans and deaths in Asia, Europe, and Americas, which allowed the WHO to declare the pandemic state. Notwithstanding all impacts caused by Coronaviruses, it is evident that the development of new antiviral agents is an unmet need. In this review, we provide a complete compilation of all potential antiviral agents targeting macromolecular structures from these Coronaviruses (Coronaviridae), providing a medicinal chemistry viewpoint that could be useful for designing new therapeutic agents.
1. Introduction
Coronaviruses are enveloped viruses grouped into the Coronaviridae family (subfamily: Coronavirinae) and order Nidovirales. 1 The subfamily has four genera, Alpha-, Beta-, Gamma- and - Deltacoronavirus. Coronaviruses are known to infect bats, rodents, and can be transmitted to other mammals (Alpha- and Betacoronavirus) and birds (Gamma- and Deltacoronavirus).2 Alpha- and Betacoronavirus can infect humans and cause mild or severe respiratory syndromes - Human Coronaviruses (HCoV).2 Two severe respiratory syndromes epidemics were previously reported, being Severe Acute Respiratory Syndrome-coronavirus (SARS-CoV) in 2002,3 and Middle East Respiratory Syndrome-coronavirus (MERS-CoV) in 2012.4 In December 2019, in Wuhan, China, new cases of severe pneumonia were reported, which was later associated with the emergence of a novel coronavirus, initially called 2019-nCoV (2019 novel coronavirus).5 Taxonomic and phylogenetic analyzes of it pointed similarities with the SARS-CoV prototype, renaming it as SARS-CoV-2.6 Gradually, new cases have been reported in Asia, Europe, and Americas, quickly reaching a global scale and leading the World Health Organization (WHO) to declare the pandemic situation in March 2020.7
Medicinal chemistry strategies aim to develop new candidates to progress through several standard levels in the drug development process, in which apply a broad variety of tools involving numerous related disciplines. Normally, these levels include (i) the selection of a biological target of medicinal interest and identifying initial hits and/or lead compounds; (ii) compounds’ design and development of an efficient structure–activity relationship (SAR) analysis in the lead-optimization process; (iii) application of in silico methods; (iv) screening of synthesized compounds performing in vitro biological assays; and (v) pharmacokinetics and in vivo studies.8 In recent years, the term “one-target-one-disease” has been growing, mainly due to the emergence of several diseases with high complexity.9 In this context, the discovery, biological evaluation, and optimization of active small molecules is the core of the drug discovery process. So far, this approach has allowed to discover potential lead compounds from natural and synthetic sources, as well as combinatorial compounds libraries.10 These last have been applied for both drug lead discovery and optimization,11, 12, 13 since several methods have been able to search diverse chemical libraries containing a wide range of natural, synthetic small-molecules (organics), and (non)peptidomimetic analogs.14 Notwithstanding these facts, computer-aided drug design (CADD) uses in silico methods that were developed to facilitate the identification of new active molecules by applying rigorous steps in drug discovery and development workflows, such as the creation of virtual libraries, molecular docking, and in silico screening.15, 16, 17 Additionally, it quickly identifies potential targets, optimizes the research, and avoids unnecessary expenses.18, 19 Among the CADD approaches, structure-based drug design (SBDD) is a very effective alternative since it allows that drug-like compounds to be designed based on the structure of a macromolecular target.20, 21 In SBDD, virtual chemical libraries containing millions of molecules and structural databases are utilized to verify the ability of ligands to interact with the target.21 Then, a potential drug-like is designed and synthesized considering its biological target in order to ensure that the new molecule acts specifically on the target chosen.22 Still, ADMET (absorption, distribution, metabolism, excretion, and toxicity) filters can be applied to enhance the probability of obtaining hits or leads with good drug-like properties.23, 24 Along with in silico methods increase, drug repurposing has emerged as an interesting alternative to re-discover (repurpose) known drug against new diseases. Considering this, Kumar et al. (2019)25 and Pillaiyar et al. (2020)26 have reported perspectives about the recent advances and challenges in drug repurposing for different diseases, including dengue fever, cancer, and central nervous system (CNS) disorders. By using drug repurposing, several recent studies have used this approach to identify potentially effective treatments for SARS-CoV-2 (COVID-19) pandemic.27, 28, 29, 30, 31, 32, 33, 34, 35, 36
In addition, clinical trials with adult patients applying different COVID-19 therapeutic strategies are being carried out around the world, as previously reviewed by Wang and coworkers (2020).37 The most common treatments under antiviral research used drugs such as remdesivir, ribavirin, favipiravir, kaletra (a compound preparation with lopinavir and ritonavir), chloroquine-hydroxychloroquine, interferon-α, and arbidol. Despite researchers’ efforts, no treatment or drugs has been really effective, pointing to a necessary search for new drugs, vaccines or peptide inhibitors.37 Recently, an important review study containing inhibitors for SARS-, MERS- and others human coronaviruses has been reported. In general, these compounds block the membrane fusion stage or the co-receptor interaction, providing valuable tools in the race against SARS-CoV-2.38
Deeming the aforementioned background, in this review, we initially provide information concerning biological aspects from Coronaviruses (CoV), such as the viral cycle of infection and proteins’ organization. Thereafter, a deep, recent, and complete compilation of all the most relevant researches involving the development of potential antiviral agents against macromolecular targets from SARS-CoV, MERS-CoV, SARS-CoV-2 (COVID-19), and HCoV-229E, as well. Hereafter, we discussed these works in detail, including chemical structures (from natural and synthetic origins), viral strains, and molecular docking studies (showing covalent, hydrophobic, and hydrogen-bonding interactions). Subsequently, we provide a table compiling all inhibitors targeting CoV biological structures found in the literature. Finally, we believe that this ‘piece of knowledge’ represents a novel useful scientific source for designing new inhibitors as promising agents to fight against Coronavirus outbreaks.
2. Human coronaviruses (HCoV)
Alpha- and Betacoronavirus can infect the human respiratory tract, ranges from mild to more serious illness that can lead to death.2 Previously, six coronaviruses had been identified as capable of infecting humans, being HCov-OC43, HCoV-229E, HCoV-NL63, HKU1, SARS-CoV, and MERS-CoV.4, 39, 40, 41, 42, 43 The first epidemic of coronavirus took place between 2002 and 2003, caused by SARS-CoV, where 8,096 people contracted the virus and had an outcome of 774 deaths, exhibiting a 9% mortality rate.43, 44 Ten years later, MERS-CoV emerges in 2012,4 and the cases have progressed slowly and, despite the epidemic situation regressed, cases are still reported. By November 2019, a total of 2,494 cases and 858 deaths were reported, reaching a mortality rate of 35%.45
It is considered that coronaviruses transmission origin can occur from a reservoir host to an intermediate host until finally infect humans. Genetic sequencing analyzes pointed out the origin of SARS-CoV, MERS-CoV, HCoV-NL63, and HCoV-229E from bats, while HCoV-OC43 and HKU1 are originated from rodents.46, 47, 48, 49, 50, 51, 52
The seventh coronavirus to infect humans was identified in December 2019, named SARS-CoV-2, causing a severe acute respiratory syndrome (SARS) and spreading the current pandemic scenario.5 The infection was namely COVID-19 (Coronavirus Disease 2019) and there was reported 13,150,645 cases and 574,464 deaths worldwide on July 15, 2020.53 There are no vaccines to either HCoV, therefore, this review will focus on the three types of Betacoronavirus that have demonstrated major impacts on global health (SARS-CoV, MERS-CoV, and SARS-CoV-2) and their possible targets in medicinal chemistry viewpoint.
2.1. SARS-CoV, MERS-CoV, and SARS-CoV-2
The first coronavirus epidemic emerged from an outbreak in Guangdong province, China, in November 2002 caused by SARS-CoV and was controlled in mid of July 2003, by isolating infected people.3, 43 At the time, it was characterized the transmission through direct contact with infected people or with contaminated fomites related to droplets and aerosols released by sick individuals.43 The virus infects airway epithelial cells that can worsen and become a pneumonia characterized as a SARS.43 Normally, the infection is characterized by fever, dyspnea, lymphopenia and, in some rare cases, diarrhea and other gastrointestinal symptoms, which may occur the active replication of the virus in the small and large intestine, which suggests a possible fecal-oral transmission.43, 54 Also, a prolonged clotting time and elevated liver enzymes were observed.43 Still, SARS-CoV is able to infect macrophages and dendritic cells, inducing the pro-inflammatory cytokines and chemokines releasing, which play an important role in the disease progression.55, 56 However, the exact mechanism involved in pulmonary consolidation and severity remains uncertain.57
After the SARS-CoV epidemic in 2002–2003, MERS-CoV appeared in Jeddah, Saudi Arabia in 2012,58 causing infections in the lower respiratory system similar to those caused by SARS-CoV, and also with a similar transmission.4, 58, 59, 60, 61 Phylogenetic analyzes concluded that the MERS-CoV genome has almost identical proximity to the virus found in dromedaries, an animal widely used in the Middle East and, therefore, has direct contact with humans.62, 63, 64 Symptoms are also similar to those of SARS-CoV, and in more severe cases, there is progression to an acute syndrome of respiratory distress, septic shock, multiple organ failure, and death.4, 59, 60, 61 MERS-CoV induces the activation of the innate immune response, through the interferon I pathway, and as a mode of evading the immune system, the virus produces proteins that block the signaling of IFN and NF-κB pathways thus increasing viral replication and pathogenicity.65, 66, 67
Seven years later, in December 2019, a new coronavirus caused severe cases of pneumonia in the city of Wuhan, China,5 and triggered the second pandemic of the 21st century, after the swine flu caused by the Influenza A (H1N1) virus in 2009.68 SARS-CoV-2 (named after phylogenetic analyzes6) has been the aim of molecular, immunological and pharmacological studies to understand the viral mechanisms, and what treatments would be viable, in addition to the rational development of vaccines. The virus transmission mode is person-to-person through respiratory droplets, as well as contact with contaminated fomites, in which the main symptoms are equal to SARS-CoV and MERS-CoV. The secretion of cytokines in COVID-19 has also been reported, as well as in SARS and MERS, and may lead to the so-called “Cytokine Storm Syndrome” presenting high-levels of analytes such as IL-6, MCP-1, VEGF, and IL-8, among others, contributing to hypotension and pulmonary dysfunction.69
There are still no specific antivirals or effective vaccines for coronaviruses. Then, the only effective methods to control the disease are to avoid contamination promoting social distancing, reinforcement of hygiene procedures, and masks utilization. As COVID-19 is an ongoing pandemic caused by a highly contagious virus,53 the search for new effective antiviral drugs is extremely essential.
3. Viral structure and genome organization of HCoV
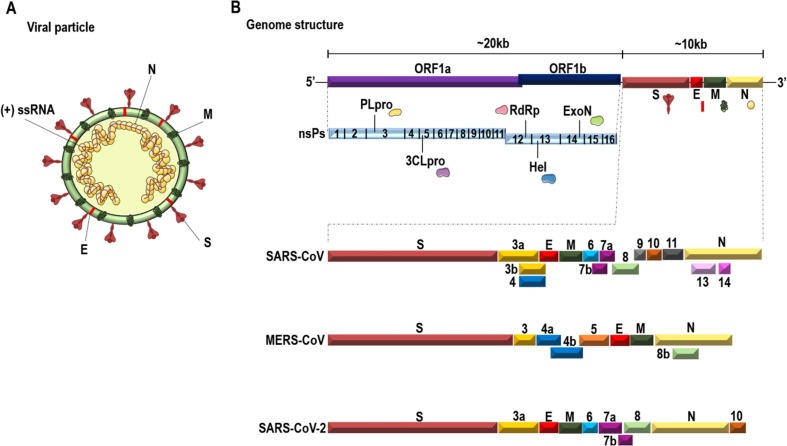
Virion organization revealed by cryo-electron microscopy (Cryo-EM) showed coronaviruses as spherical viruses with approximately 80 to 125 nm with a particular feature of protruding oligomers (Spike protein) projected on its surface, presenting a crown-like appearance and giving rise to its name.1, 70 Three other proteins make up the viral structure, namely membrane protein (M), envelope protein (E), and nucleocapsid protein (N). Within the viral membrane, the helical nucleocapsid, composed by the N protein, can induce a cell cycle delay, assists viral RNA during replication, and be complexed with the genetic material (Fig. 1 A).1, 71
Fig. 1.
A. Schematic representation of the HCoVs viral structure and genome organization. HCoVs are spherical viruses, with oligomeric Spike proteins (S) that protrude on their surface, and interact with the cell receptors. The surface has also the lipid bilayer from the host cell with the membrane (M), the envelope (E) proteins, and the (+) ssRNA packed with the nucleocapsid protein (N) on a helical shape. B. Organization of the SARS-CoV, MERS-CoV, and SARS-CoV-2 genome. The HCoVs genome is a (+) ssRNA of ~30 kb encoding non- and structural proteins. The 5′ end encodes the ORFs 1a and 1b giving rise to the pp1a and pp1ab, which is cleaved into non-structural proteins. The 3′ end encodes the structural proteins that will be assembled into the viral particle. The figure shows a schematic representation of the SARS-CoV, MERS-CoV and SARS-CoV-2 genome, with an emphasis on structural and accessory proteins, highlighting the similarities and differences among them. Figure based on complete genome sequences from Genbank: AY274119.3, NC_019843.3, and NC_045512.2.
The coronavirus (CoV) genome is a single-stranded, positive-sense RNA of approximately 30 kb, with a 5́cap and a 3́poly (A) tail (Fig. 1B). The 5′ end shows the replicase gene encoding two Open Reading Frames (ORFs) 1a and 1b with an overlapping. The ORF1ab encode two polyproteins named pp1a (ORF1a) and pp1ab (ORF1a and ORF1b).1, 57 These polyproteins are then proteolytically cleaved, generating 16 non-structural proteins (nsP’s). The 3′ end of the genome encodes structural and accessory proteins interspersed within them. Both ends have untranslated regions (UTRs) that play a role in RNA viral transcription, replication, and synthesis. The 5′ end UTR has stem-loop structures that are necessary for these processes.1, 57
4. The replication cycle of HCoV
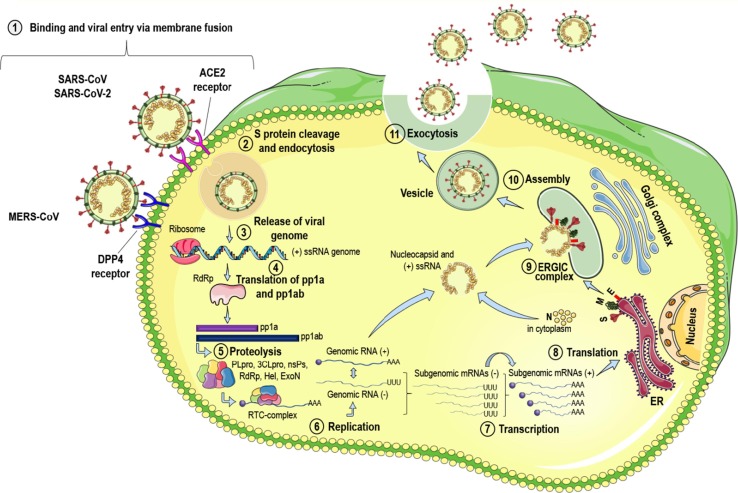
The initial contact with the host organism occurs through mucous membranes from the nose, mouth, and eyes. The coronaviruses bind the RBD domain from Spike protein (S) to the host cell, recognizing the human angiotensin-converting enzyme 2 (ACE2) receptor in infections by SARS-CoV and SARS-CoV-2,72, 73 and the dipeptidyl peptidase 4 (DPP4) receptor in infections caused by MERS-CoV74 (Fig. 2 ). This interaction ends up causing a conformational rearrangement resulting in the fusion between the viral membrane and the host cell membrane, after the cleavage of S protein at its S2 site and then releasing the nucleocapsid within the cell cytoplasm.1, 57, 75 Once inside the cell, the 5′ end from the RNA has the ORFs 1a and 1b translated into polyproteins pp1a and pp1ab, processed later into the nsP1-11 and nsP1-16, respectively.1, 57, 76 To encode the two polyproteins, the virus uses a stop codon sequence (5′-UUUAAAC-3′) and a pseudoknot RNA that causes a −1 ribosomal frameshifting upstream of the ORF 1a stop codon, allowing the continuous reading to ORF1b. Thus, the pseudoknot structure is unrolled until it finds the stop sequence in ORF1a, preventing continuous ribosome elongation, extending the conversion to ORF 1b and resulting in the translation of pp1ab.1, 57, 77
Fig. 2.
Replication cycle of HCoV. The virus enters the host cell by binding protein S to the ACE2 receptor (SARS-CoV and SARS-CoV-2) or the DPP4 receptor (MERS-CoV), leading the viral membrane fusion with the cell membrane host (1). Fusion occurs because of S protein cleavage, allowing entry through the endosomal pathway (2). The viral RNA is released into the cell cytoplasm (3) and the pp1a and pp1ab polyproteins (4) are translated, which will be cleaved by the proteases of the RTC complex (5), synthesizing the (+) and (−) RNAs (6). From (−) gRNA, sgRNAs will be discontinuously transcribed until act as mRNAs (7) and finally being translated into the structural proteins (8) that will be transferred to the ERGIC intermediate complex (9). The N proteins then bind to the (+) gRNA in the cytoplasm and are assembled with structural proteins in the ERGIC complex (10). Vesicles containing virion are transported to the plasma membrane and released via exocytosis (11) in the extracellular space, thus infecting neighboring cells.
The ORF1a from SARS-CoV, SARS-CoV-2, and MERS-CoV has the PLpro (papain-like protease) domain within the nsP3, responsible for the processing of mature replicase proteins. Also, ORF1a presents another domain composed of a serine-type protease, 3CLpro (chymotrypsin-like picornavirus 3C-like cysteine protease or main protease (Mpro)) within nsP5.78, 79, 80 While PLpro does the cleavage at the limits between nsP1-2, nsP2-3, and nsP3-4, 3CLpro is responsible for 11 other remaining cleavage events.1, 57, 78 The nsP’s are organized in the replicase-transcriptase complex (RTC) and have other domains and enzymatic functions. For example, in ORF1b there are nsP’s encoding the RNA polymerase-dependent RNA domain (RdRp), RNA helicase, exoribonuclease, and a ribose dependent on 2′-O-methyltransferase activity.1, 57 All of these nsP’s with specific activities contribute to the construction of an environment that enables the synthesis of RNA, the replication and transcription of subgenomic RNAs (sgRNAs), and genomic RNAs (gRNA)57, 81 (Fig. 2).
The replication initiates with the synthesis of a negative polarity viral RNA that produces both gRNA and sgRNAs, which works as mRNA for the translation of structural and accessory proteins. From the RTC complex, there is the transcription of subgenomic mRNAs with 3′-coterminal end and gRNA, which have in common a leader sequence derived from the 5′ end of the genome. From there, the proteins are translated, and E and M proteins are translocated to the Endoplasmic Reticulum-Golgi intermediate compartment (ERGIC), where it is believed to be the site of assembly, budding, and transport of virions.1, 57, 82 The nucleocapsid proteins pack the genomic RNA to form the nucleocapsids on helical structures. The N protein then interacts with the E and M proteins, producing the virus-like particles (VLPs) that incorporate the S proteins. Mature virions are transported to the cell surface inside vesicles and are released into the extracellular space, allowing the interaction and infection of neighboring cells (Fig. 2).1, 57
5. Druggable targets from HCoVs
5.1. Spike glycoprotein (also named as S protein)
The CoV spike protein (S) is a type I transmembrane glycoprotein playing roles in viral attachment, fusion, and cell entry as well as tissue tropism.83 S proteins assemble into trimers protruding from the viral surface forming the crown-like appearance.84 It has two functional subunits named S1 (responsible for binding to the cell receptors) and S2 (acts in the fusion of the viral and cellular membranes).83, 84, 85, 86, 87
The S1 subunit forms the protein globular head and presents the receptor-binding domain (RBD) responsible for virus interaction with the host cellular receptors (e.g. ACE2 for SARS-CoV and SARS-CoV-2 or DPP4 for MERS-CoV),88, 73, 74 thus allowing the viral entry into the targeted cell. The receptor-binding S1 subunit of CoV S proteins has two domains, the N-terminal and C-terminal domains, either them can act as RBDs.90 The Cryo-EM structure of SARS-CoV-2 S trimer was performed87, 89 and a furin cleavage site at the S1/S2 boundary was identified which is known to be processed during biogenesis.87
Recently, it was identified that RBD in SARS-CoV-2 S protein can bound strongly to both human and bat ACE2 receptors, and interestingly a higher binding affinity was detected for SARS-CoV-2 RBD than to SARS-CoV.89, 91 The crystal structure of the SARS-CoV-2 RBD bounded to the ACE2 was elucidated and is nearly identical to that of the SARS-CoV RBD.92 The SARS-CoV-2 entry requires cellular proteins to process S protein helping the infection. In this way, S protein is primed by cellular serine protease TMPRSS293 and a furin preactivation of the SARS-CoV-2 S protein-enhanced virus entry into some cell types.94 Similarly to the SARS-CoV,95 the cellular serine protease TMPRSS2 was pointed as a possible antiviral target for entry step of SARS-CoV-2.96
The C-terminal S coronavirus S2 subunit forms a stalk-like structure anchored in the membrane during the virus fusion to the host cell membrane and presents heptad repeats (HRs) motifs.97 SARS-CoV and SARS-CoV-2 have an 89.8% sequence identity in their S2 subunits.97 After the binding of RBD at S1 subunit to the ACE2 receptor on the host cell, the membrane distal 1 (HR1) and membrane-proximal 2 (HR2) heptad repeat domains in the S2 subunit interact with each other, forming a six-helix bundle (6-HB) fusion core thus bringing viral and cellular membranes into proximity for fusion and infection.97, 98 Interestingly, it was shown that SARS-CoV-2 has a much higher capacity of membrane fusion than SARS-CoV.97
5.1.1. S protein inhibitors
The attachment of SARS-CoV spike (S) protein to cellular Angiotensin-Converting Enzyme 2 (ACE2) is the first step in SARS-CoV infection. Several reports indicated that blocking the S protein attachment to cellular receptors could prevent the virus entry. Thus, the SARS-CoV S protein becomes an attractive target for the development of anti-SARS drugs.99
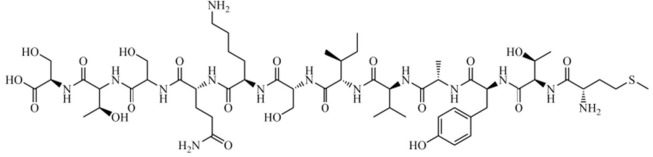
Small-peptide-derived structures of HR regions from the SARS-CoV S protein have been shown to inhibit SARS-CoV infection by interfering in SARS-CoV fusion upon target cells.100, 101, 102, 103 Encouraged by these results, Ho et al. (2006)99 synthesized 14 small synthetic peptide derivatives from the S protein and screened all of them against the S protein attachment to the ACE2, and upon the S-protein-pseudotyped retrovirus infectivity, as well. Among these, the peptide SP-10 (residues 668–679 - STSQKSIVAYTM) (Fig. 3 ) was identified as the most promising protein–protein interaction (PPI) inhibitor, exhibiting 90% inhibition at 10 nmol concentration and blocking this interaction in a dose-dependent manner, with an IC50 value of 0.0018 μM (1.8 nM). The inhibitory effect of SP-10 on SARS-CoV S protein and Vero E6 cells was analyzed by Biotinylated Enzyme-Linked Immunosorbent Assay (ELISA), Immunofluorescence assay (IFA), and S-protein-pseudotyped retrovirus infectivity. Vero E6 cells were treated with BSA, Biotin-labeled S protein or SP-10/biotin-labeled S protein mixture. Moreover, the results indicated that SP-10 was capable of blocking the attachment of S protein to Vero cells. Additionally, it was investigated the possible domains involved in receptor interactions to identify biologically active peptides using peptide-scanning method, which involved overlapping peptides of 12 residues covering additional amino acids on both the N- and C-terminals from the SP-10 structure. The inhibitory effect of small overlapping peptides on the SARS-CoV S protein and ACE2 interaction was analyzed by competitive biotinylated ELISA. The receptor-binding regions from the SARS-CoV S protein have been defined in different studies.104, 105, 106 It was suggested that the region comprising residues 660–683 from the SARS-CoV S protein may interacts with ACE2. Therefore, SP-10 was the first small-peptide designed as a PPI inhibitor targeting SARS-CoV S protein attachment to the ACE2. According to these authors, it could be developed as an anti-SARS-CoV agent for the treatment of SARS-CoV infection.99
Fig. 3.
Chemical structure of SP-10 (peptide sequence: STSQKSIVAYTM), a small-peptide-derived from the SARS-CoV S protein.
Finally, another peptide-derived inhibitor (comprising the sequence: EEQAKTFLDKFNHEAEDLFYQSSGLGKGDFR) of SARS-CoV S protein is shown in Table 1 .
Table 1.
Inhibitors targeting Coronaviruses found in the literature.
| Structure | Source | Organism | IC50 | Target | PDB - Interactions/(H-bond) | Ref. |
|---|---|---|---|---|---|---|
| Spike (S) protein | ||||||
| EEQAKTFLDKFNHEAEDLFYQSSGLGKGDFR | Synthetic | SARS-CoV | 0.1 µM | S protein | Not revealeda | 209 |
| Chymotrypsin-like cysteine protease (3CLpro) | ||||||
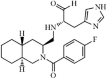 |
Synthetic | SARS-CoV | 57 µM | 3CLpro | (PDB: 4TWW) - Cys145, His41, Met49, Met165, Asp187, His163(H), Phe140, Leu141, and Glu166 | 210 |
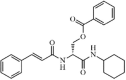 |
Synthetic | SARS-CoV | 30 µM | 3CLpro | (PDB: 3AW1) - Cys145(H) and Gln189(H) | 211 |
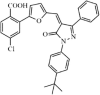 |
Synthetic | SARS-CoV | 5.8 µM | 3CLpro | (PDB: 2ALV) - Gly143(H), Ser144(H), Cys145(H), His163(H), His41(H), Leu27, Met49, and Gln189 | 212 |
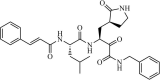 |
Synthetic | HCoV-NL63 | 1.08 µM | 3CLpro | (PDB: 6FV2) - His41(H), *Gly142(H), Phe139(H), and Glu166(H). | 213 |
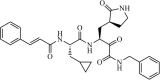 |
Synthetic | SARS-CoV | 0.24 µM | 3CLpro | (PDB: 5N5O) - Met49, Met165, Asp187, Gln189(H), and Thr190 | 213 |
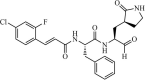 |
Synthetic | MERS-CoV | 1.7 µM | 3CLpro | (PDB: 4RSP) - Glu166(H), Glu169(H), Gln192(H), Cys148 (Covalent), and His41 | 214 |
| SARS-CoV | 0.2 µM | 3CLpro | Not revealeda | 214 | ||
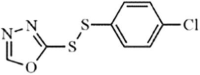 |
Synthetic | SARS-CoV | 0.51 µM | 3CLpro | (PDB: 2AMD) - Phe140, Leu141, His163, Met165, Glu166, His172, Asn142(H), Gly143(H), and Cys145(H) | 215 |
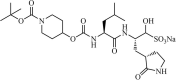 |
Synthetic | MERS-CoV | 0.4 µM | 3CLpro | (PDB: 5WKL) - Cys148, Gln192(H), *Gln167(H), Glu169(H), His41(H), *His166(H), and *Phe143(H) | 216 |
| SARS-CoV | 5.1 µM | 3CLpro | Not revealeda | 216 | ||
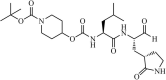 |
Synthetic | MERS-CoV | 0.6 µM | 3CLpro | Not revealeda | 216 |
| SARS-CoV | 2.1 µM | 3CLpro | Not revealeda | 216 | ||
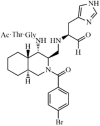 |
Synthetic | SARS-CoV | 26 µM | 3CLpro | (PDB: 4TWW) - Cys145, His163(H), Phe140, Leu141, and Glu166 | 217 |
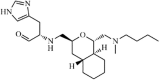 |
Synthetic | SARS-CoV | 95 µM | 3CLpro | Not revealeda | 218 |
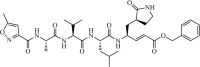 |
Synthetic | MERS-CoV | 0.28 µM | 3CLpro | (PDB: 1UK3) - Cys148 (Covalent), *His166(H), and His175(H) | 219, 220 |
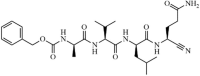 |
Synthetic | SARS-CoV | 4.6 µM | 3CLpro | (PDB: 3VB5) - Cys145(Covalent), Pro168, Thr190(H), Glu166(H), Gln189(H), His164(H), Phe140(H), and His163(H) | 221 |
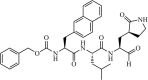 |
Synthetic | SARS-CoV | 0.23 µM | 3CLpro | Not revealeda | 222 |
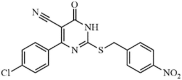 |
Synthetic | SARS-CoV | 6.1 µM | 3CLpro | (PDB: 1UK4) - Glu166(H), Gly143(H), Cys145(H), Met49, and Gln189 | 223 |
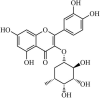 |
Synthetic | SARS-CoV | 24.1 µM | 3CLpro | Not revealeda | 224 |
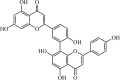 |
Natural | SARS-CoV | 8.3 µM | 3CLpro | (PDB: 2Z3E) - Leu141(H), His163(H), Val186(H), Gln189(H), and Gln192(H), | 225 |
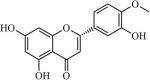 |
Natural | SARS-CoV | 8.3 µM | 3CLpro | Not revealeda | 226 |
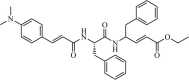 |
Synthetic | SARS-CoV | 1.0 µM | 3CLpro | Not revealeda | 227 |
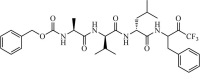 |
Synthetic | SARS-CoV | 10.0 µM | 3CLpro | (PDB: 1UK4) - His41, Met49, Phe140, Gly143(H), Cys145(H), His163, Met165, Glu166(H), Pro168, and Gln189(H) | 228 |
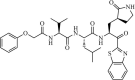 |
Synthetic | SARS-CoV | 1.7 µM | 3CLpro | Not revealeda | 229 |
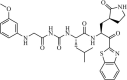 |
Synthetic | SARS-CoV | 10.0 µM | 3CLpro | (PDB: 1WOF) - Cys145, Ser144(H), His163(H), His164(H), Glu166(H), and Gln189(H) | 230 |
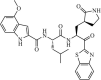 |
Synthetic | SARS-CoV | 0.74 µM | 3CLpro | (PDB: 1WOF) - Cys145, Ser144(H), His163(H), His164(H), Glu166(H), and Gln189(H) | 231 |
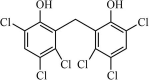 |
Synthetic | SARS-CoV | 5.0 µM | 3CLpro | (PDB: 1UK4) - Glu166(H), His163(H), Cys145(H), Ser144(H), Asn142(H), Phe140(H), Thr26(H), and His41 | 232 |
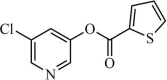 |
Synthetic | SARS-CoV | 0.5 µM | 3CLpro | Not revealeda | 233 |
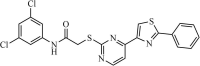 |
Synthetic | SARS-CoV | 3.0 µM | 3CLpro | Not revealeda | 234 |
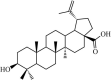 |
Natural | SARS-CoV | 10.0 µM | 3CLpro | Not revealeda | 235 |
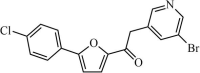 |
Synthetic | SARS-CoV | 13.0 µM | 3CLpro | Not revealeda | 236 |
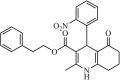 |
Synthetic | SARS-CoV | 8.9 µM | 3CLpro | Not revealeda | 237 |
| SARS-CoV | 2.5 µM | 3CLpro | Not revealeda | 238 | ||
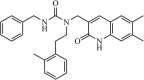 |
Synthetic | SARS-CoV | 17.2 µM | 3CLpro | Not revealeda | 239 |
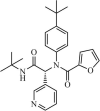 |
Synthetic | SARS-CoV | 1.5 µM | 3CLpro | Not revealeda | 240 |
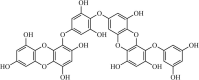 |
Natural | SARS-CoV | 2.7 µM | 3CLpro | (PDB: 2ZU5) - Thr190(H), His163(H), Ser144(H), His41(H), and Cys145(H) | 241 |
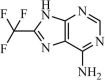 |
Synthetic | SARS-CoV | 13.9 µM | 3CLpro | Not revealeda | 242 |
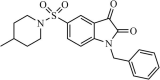 |
Synthetic | SARS-CoV | 1.04 µM | 3CLpro | (PDB: 1UK4) - Met49, Leu141, Asn142, Gly143(H), Ser144, Cys145(H), Met165, Arg188, Gln189, and Gln192 | 243 |
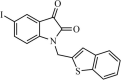 |
Synthetic | SARS-CoV | 0.95 µM | 3CLpro | (PDB: 1UK4) - Gly143(H), Cys145(H), His41(H), His164, Phe140, His163, and Met165 | 244 |
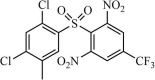 |
Synthetic | SARS-CoV | 0.3 µM | 3CLpro | (PDB: 1UK4) - Met49, His41(H), Pro39, Leu27, Cys145, His164, Met165, Leu167, Gln192, and Gln189 | 245 |
| Synthetic | SARS-CoV | 0.06 µM | 3CLpro | (PDB: 2A5A) - Glu189, Met49, His41, Cys145, Gly143(H), His163(H), Phe140, His172, and Glu166 | 246 | |
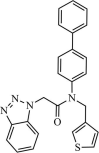 |
Synthetic | SARS-CoV | 0.051 µM | 3CLpro | Not revealeda | 247 |
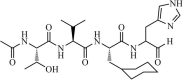 |
Synthetic | SARS-CoV | 0.098 µM | 3CLpro | (PDB: 3ATW) - Thr90(H), Glu166(H), His163(H), Phe140, Leu141, Asn142, His41, Met49, Met165, Asp187, and Cys145(H). | 248 |
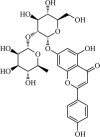 |
Natural | SARS-CoV | 27.5 µM | 3CLpro | (PDB: 4WY3) - Gln166(H), Leu167(H), Phe140(H), Gln189(H), Asn142(H), Thr26(H), and Thr24(H). | 249 |
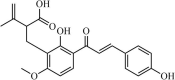 |
Natural | SARS-CoV | 11.4 µM | 3CLpro | (PDB: 2ZU5) - His163(H), Ser144(H), and Cys145(H) | 250 |
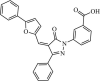 |
Synthetic | MERS-CoV | 5.8 µM | 3CLpro | (PDB: 4YLU) - His41, Ser147, Glu169(H), Leu170, Val193, and Gln195 | 212 |
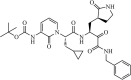 |
Synthetic | SARS-CoV2 | 0.67 µM | 3CLpro | (PDB: 6Y2F) - His41(H), Gly143(H), Cys145(H), Ser144(H), Phe140(H), Glu166(H), and His163(H) | 127 |
| SARS-CoV | 0.90 µM | 3CLpro | Not revealeda | 127 | ||
| MERS-CoV | 0.58 µM | 3CLpro | Not revealeda | 127 | ||
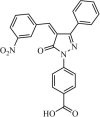 |
Synthetic | SARS-CoV | 9.6 µM | 3CLpro | (PDB: 1UK4) - Glu166(H), Gly143, Gln192, Met49, Arg188, and Gln189 | 251 |
| Papain-like protease (PLpro) | ||||||
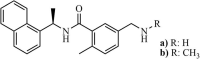 |
Synthetic | SARS-CoV | (a) 0.46 µM and (b) 1.3 µM | PLpro | (PDB: 3E9S) - Tyr269, Gln270, and Asp165 | 252 |
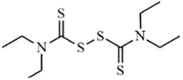 |
Synthetic | SARS-CoV | 14.2 µM | PLpro | (PDB: 4M0W) - Cys271 | 253 |
| MERS-CoV | 22.7 µM | PLpro | Not revealeda | 253 | ||
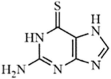 |
SARS-CoV | 5.0 µM | PLpro | Not revealeda | 254 | |
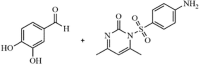 |
Synthetic | MERS-CoV | 6.6 µM | PLpro | (PDB: 4RF1) - Tyr279(H), Ser167(H), Pro163, Asp164, Asp165, *Gly248, *Thr249, Pro250, *Phe269, Glu273, Ala275, Val276, Gly277, and Thr308 | 255 |
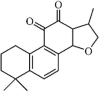 |
Natural | SARS-CoV | 0.8 µM | PLpro | Not revealeda | 256 |
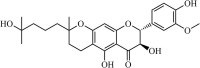 |
Natural | SARS-CoV | 5.0 µM | PLpro | Not revealeda | 257 |
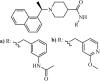 |
Synthetic | SARS-CoV | (a) 0.39 µM and (b) 0.35 µM | PLpro | (PDB: 3 MJ5) - Cys112, *Leu163, Asp165, Pro248, Pro249, Tyr265, Tyr269, Gln270, Tyr274, Thr302, and Asp303 | 258 |
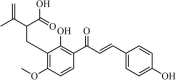 |
Natural | SARS-CoV | 1.2 µM | PLpro | (PDB: 3 MJ5) - His176(H) and His172(H) | 250 |
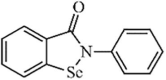 |
Synthetic | SARS-CoV | 8.45 µM | PLpro | (PDB: 2FE8) - His290, Asp287, His273, Ala289, Lys106(H), Cys112(Covalent), and Trp107(H). | 259b |
| SARS-CoV2 | 2.26 µM | PLpro | (PDB: 6W9C) - *Leu289, Ala288, Asp286, Lys105, Tyr268, *Trp106, His272, and *Cys111(Covalent). | 259b | ||
| NTPase/Helicase (nsP13) | ||||||
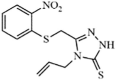 |
Synthetic | SARS-CoV | 6 µM | nsP13 | Not revealeda | 260 |
| 12 µM 50 µM 5.9 µM |
Y277AnsP13 K508AnsP13 WTnsP13 |
(homology model) Tyr277, Arg507, and Lys508. | 261 | |||
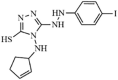 |
Synthetic | MERS-CoV | 2.5 µM | nsP13 | (homology model) Tyr7, Tyr159, Arg163, and Tyr171. | 261, 262 |
([Bi(L5) (H2O) (ClO4)3], L5 = 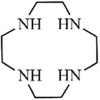
|
Synthetic | SARS-CoV | 5.0 µM | nsP13 | Not revealeda | 263 |
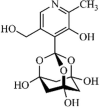 |
Synthetic | SARS-CoV | 3.0 µM | nsP13 | Not revealeda | 264 |
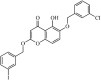 |
Synthetic | SARS-CoV | 11.0 µM | nsP13 | Not revealeda | 265 |
| Guanine-N7-Methyltransferase (also named as N7-MTase or nsP14) | ||||||
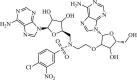 |
Synthetic | SARS-CoV | 0.6 µM | nsP14 | (PDB: 5C8T) - Trp385, Phe401, Tyr420, Phe426, Phe506, Cys387, Pro335, Val290, Trp292, Ile332, Phe367, Ala353, Val389, Asn386 (H), Arg310 (H), Gly333 (H), Ile338 (H), Lys336 (H), His424 (H), and Asp352 (H). | 266 |
*: Divergent residue labeling due to the difference between the SARS-CoV, SARS-CoV-2, and MERS-CoV proteins’ structures. a: The authors have not performed in silico studies. b: Preprint study published online at BioRxiv, doi: 10.1101/2020.05.17.100768, available at: https://www.biorxiv.org/content/10.1101/2020.05.17.100768v1.
5.2. Envelope protein (also named as E protein)
The envelope protein (E) is the smallest structural protein that plays roles in the viral assembly, budding, virion release, and pathogenesis.82, 107, 108 Its structure has a short hydrophilic N-terminal, a large hydrophobic (TMD), and a hydrophilic C-terminal domain.82, 109 For the SARS-CoV, E protein was mainly detected in the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), and only a small portion is incorporated into the virion envelope.110
The C-terminal SARS-CoV E domain interacts with the PALS1, a tight junction-associated protein, and alters tight junction formation and epithelial morphogenesis in mammalian cells.111 Besides, the SARS-CoV E protein forms ion conductive pores in lipid bilayers,112 exhibiting an important function in the virus-host interaction. Also, it was shown that the activity of SARS-CoV E protein ion channel contributes to virus pathogenesis and it was required for inflammasome activation in infected-mice.113
The SARS-CoV E protein-induced apoptosis in Jurkat T-cells and interacted with antiapoptotic protein BcL-xL suggesting a novel mechanism of T-cell apoptosis observed in SARS infection.114 In another study, it was reported that the E protein PDZ-binding motif involved in protein–protein interactions has a major role in SARS-CoV virulence using the cellular protein syntenin as a mediator of p38 MAPK induced inflammation.115
5.2.1. E protein inhibitors
Expressed in SARS- and MERS-CoV, and most recently described in SARS-CoV-2,116 envelope (E) protein is a small membrane protein responsible for forming ion channels and is directly related to the viral infection, replication, dissemination through host tissues and modulation of the immune response.82, 117 Notwithstanding these functions, E protein has been considered as an important target in the development of selective inhibitors against CoV.118
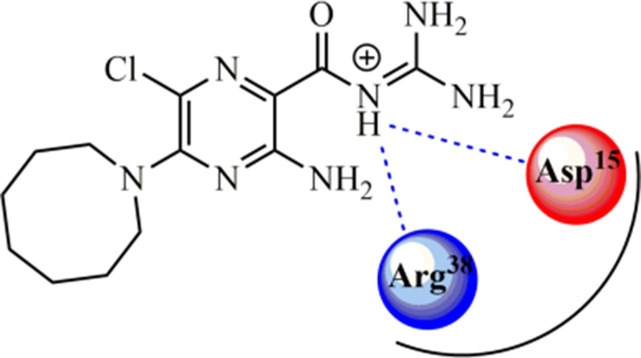
Among the classes of active compounds against E protein,118 acylguanidines (AG) standout for having several antiviral potential agents, where BIT225 is the first derivative from this class in clinical trials against hepatitis C (HCV) and immunodeficiency type 1 (HIV1) viruses,119 as well as AG derivatives from zanamivir and oseltamivir, potentially actives against influenza infections.120 Concerning this chemical class, Wilson et al. (2006)121 performed a study involving the hexamethylene amiloride (HMA, Fig. 4 ), an acylguanidine analogous to the amiloride drug, where a peptide corresponding to the E protein sequences from three coronaviruses in the GenBank database was synthetized. Thereafter, it was observed that the HMA significantly inhibited the conductance of HCoV-229E E protein ion channel (−70 mV) at 100 µM concentration. The authors reported its inhibitory activity with an EC50 value of 1.34 µM upon the HCoV-229E replication, as well as from the mouse hepatitis virus (MHV), with an EC50 value of 3.91 µM.
Fig. 4.
Hexamethylene amiloride (HMA) inhibitor of E protein from HCoV-229E. Hydrogen-bonding interactions are shown as blue dotted lines.
Posteriorly, Pervushin et al. (2009)122 synthesized a peptide (sequence: E8TGTLIVNSVLLFLAFVVFLLVTLAILTALR-NH2) corresponding to the transmembrane domain of SARS-CoV E protein to evaluate the binding mode with the HMA. By using Nuclear Overhauser Effect (NOE) in NMR analysis, it was revealed that HMA has two binding sites in the SARS-CoV E protein, being one near to the Asp15 residue and another at Arg38. Besides, they also revealed that HMA nitrogen 5 is found in the protonated state (NH+) and bound to the ion channel, in which it is stabilized by hydrogen-bonding interactions with Asp15 and Arg38 amino acid residues, as a signal at 10.7 ppm (for Asp15) in the 1H NMR spectrum.
5.3. Chymotrypsin-like picornavirus 3C-like protease (3CLpro) (also named as main protease (M pro), or 3C)
The 3CLpro (also called Mpro) is a protease (corresponding to nsP5) highly conserved in HCoVs, and as well as PLpro, is responsible for the cleavage of the polyproteins pp1a and pp1ab playing an essential role in the viral replication.123, 124 This protease cleaves the polyprotein at 11 conserved sites involving the amino acid sequence Leu-Gln/Ser-Ala-Gly, which cleavage site is between the Gln and Ser.123 Although the cleavage pattern of 3CLpro seems to be conserved in SARS-CoV and SARS-CoV-2,123, 125 additional mechanisms are required to the polyprotein processing in the MERS-CoV, such as dimerization of the substrate-binding site to obtain a mature dimer.126
The coronavirus 3CLpro was studied, revealing three domains. The N-terminal 3CLpro I and II domains are antiparallel β-sheets with 6 to 13 β-ribbons forming a chymotrypsin-like structure.123, 125 The catalytic binding site is located between these two domains and a loop structure links domain II to the C-terminal domain III, comprising a dyad of conserved residues (cysteine and histidine) both in SARS-CoV and SARS-CoV-2.123, 127 Domain III consists of a globular cluster of five helices associated with the protein dimerization due interactions between these domains of each monomer. Also, domain III has been associated with the proteolytic activity of 3CLpro.123, 125, 126
5.3.1. 3CLpro inhibitors
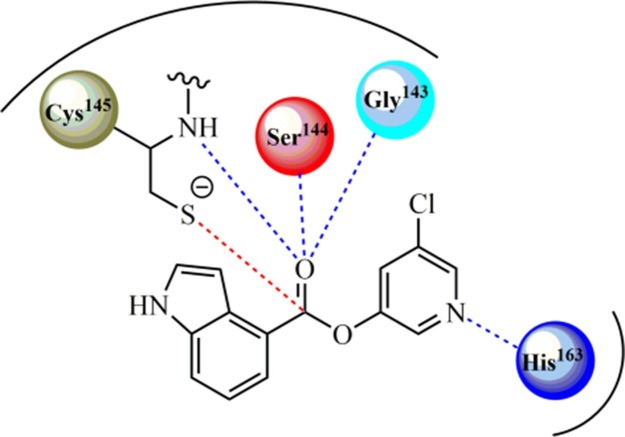
Ghosh et al. (2008)128 reported the design and biological evaluation of a series of 5-chloropyridine ester-derived inhibitors against chymotrypsin-like protease (3CLpro) from SARS-CoV. This protease is crucial for viral replication, along with papain-like protease (PLpro), constituting important targets in the search for selective and effective antiviral inhibitors.129, 130, 131 Deeming this information, 11 chloropyridine ester derivatives were synthesized and biologically evaluated. Then, the indole-derived (Fig. 5 ) was found to be a covalent inhibitor, confirmed by MALDI-TOF analysis. Thereafter, it demonstrated to be the most promising inhibitor against SARS-CoV 3CLpro, with an IC50 value of 0.03 µM. Furthermore, it was verified that this compound presented activity against infected cells with the virus, exhibiting an EC50 value of 6.9 µM.128
Fig. 5.
Promising covalent inhibitor of 3CLpro from SARS-CoV. Covalent and hydrogen-bonding interactions are shown as red and blue dotted lines, respectively.
Computational studies of molecular docking by using GOLD® v. 3.2 software were carried out involving this target (PDB ID: 2HOB). It was suggested that the critical residues for the interaction between the indole-derived inhibitor and the protease involve a carbonyl group at position 4. Finally, the authors described that the distance between the carbon dioxide and the sulfur atom from the Cys145 residue prior to the nucleophilic attack is about 2.8 Å. Also, the distance between the nitrogen from the chloropyridinyl ring (as a H-acceptor group) and His163 residue is about 2.4 Å. Furthermore, the carbonyl oxygen atom is located between three backbone nitrogens, forming three hydrogen-bonding interactions with Cys145 (NH···O, 2.3 Å), Ser144 (NH···O, 2.4 Å), and Gly143 (NH···O, 2.8 Å) residues, increasing the stability of the complex and favoring the nucleophilic attack by Cys145 residue (Fig. 5). In this context, Ghosh et al. (2008)128 reported that in the covalently modified state after the aforementioned nucleophilic attack (PDB ID: 2V6N), the indole ring is placed into the S1 pocket, where the chloropyridinyl ring was before the nucleophilic attack, suggesting a dynamic enzymatic reorganization. Still, heterocycles indole and imidazole (from the His41 residue) are within a distance of 4 Å, suggesting that these rings have their initial position modified by the covalent reaction progress, reinforcing the aforementioned enzymatic reorganization.
Finally, diverse 3CLpro inhibitors from CoV have been reported in the literature and they can be found in Table 1.
5.4. Papain-like protease (also named as PLpro)
The PLpro is a cysteine protease encoded by nsP3 protein that recognizes the consensus cleavage sequence LXGG at the amino-terminal of replicase products in the cell cytosol during the virus replication. The tetrapeptide motif is found between nsP1/2, nsP2/3, and nsP3/4 proteins releasing the nsP1-2-3 from the viral polyprotein.78, 132
Additionally, the PLpro enzymatic activity is also characterized as the deubiquitinating activity of cellular proteins, possibly providing a favoring environment for intracellular viral replication. PLpro molecular structure resembles deubiquitinating enzymes (DUBs) responsible for cleaving both Ubiquitin and UBL-ISG15, which are important cellular modifiers that are covalently attached to target proteins by a peptide bond. Like DUBs, PLpro acts on this isopeptide bond cleaving viral proteins during the viral replication process.132, 133, 134
Also, it is known that due to the deubiquitinating activity, PLpro can inhibit the production of cytokines and chemokines having a significant role in the innate immune response against viral infection antagonizing IFN pathway.135
The molecular structure and function of PLpro from SARS-CoV, MERS-CoV, and SARS-CoV-2 is very similar, with slight conformational differences. The monomer consists of four domains: ubiquitin-like (UBL), the thumb, the palm, and the fingers.136, 137 Previously, some studies showed that PLpro domains are conserved in all coronaviruses, and the same was observed with SARS-CoV-2.138 The catalytic core has a Zn-ribbon domain with four cysteine residues, demonstrated in HCoV-229E to be important for enzyme activity.139 The PLpro hydrophobic domain, important to the processing of the nsP3/4 site, is glycosylated, mediating an intracellular membrane association. It is hypothesized that the anchoring of PLpro may be important to the assemble of replication complex during the virus replication.78
5.4.1. PLpro inhibitors
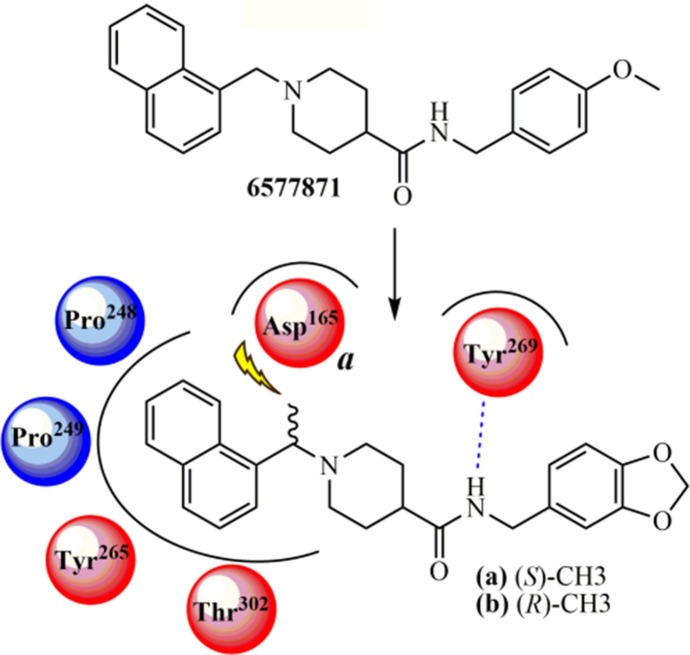
Ghosh et al. (2010)140 developed a new lead compound belonging to the class of piperidine carboxamide from a chemical library using high-throughput screening (HTS). The compound 6577871 (Fig. 6 ) exhibited an IC50 value of 59 μM. Also, studies involving lead optimization and structure–activity relationship (SAR) analyses were performed to provide improved inhibitors targeting PLpro and antiviral activity against infected Vero E6 cells with SARS-CoV. The (S)-Me (a) (IC50 = 0.56 μM for PLpro; and EC50 = 9.1 μM for antiviral assays) and (R)-Me (b) (IC50 = 0.32 μM for PLpro; and EC50 = 9.1 μM for antiviral assays) derivatives were the most potent compounds in this series (Fig. 6), showing equivalent enzymatic inhibition and antiviral activity. From the structure–activity relationship (SAR) analysis, the authors established that the introduction of a benzodioxolane ring led to more strong inhibitory activity than its precursor compound (6577871). Also, 1-(naphthyl)ethylamides are more potent than 2-naphthyl derivatives, and it was observed a preference for (R)-methyl enantiomers. The X-ray structure of complex for b-PLpro from SARS-CoV was determined in a 2.6 Å resolution and a model for a-PLpro complex revealed molecular features associated with interactions, in which it was shown a unique binding mode into the PLpro structure. In silico studies demonstrated that the activity of this series of compounds is not associated with stereoisomerism, in contrast to other compounds already synthesized. Superposing the structure of compound a on the crystal structure of the b-PLpro complex, the authors observed that there is an inversion of the piperidine ring between compounds a and b binding modes, which allows the naphthyl rings of both isomers to be accommodated at the active site, in a very similar orientation. Finally, the flexible piperidine ring acts as a spacer (or linker) group that enables the carboxamide (CONH) from both a and b compounds to perform hydrogen-bonding interactions with the backbone carbonyl oxygen at the Tyr269 residue, thereby retaining the potency of both enantiomers.
Fig. 6.
Chemical structures of piperidine carboxamide analogs with activity against PLpro from SARS-CoV. Hydrogen-bonding interaction is shown as a blue dotted line. Bumping collision is represented by a yellow-ray for the compound a.
The crystal structure of the b-PLpro complex confirmed the presence of water molecules conserved between the apoenzyme and compound b. Water molecules conserved into the pocket, located between residues Asp165, Asp303, and Thr302, preventing naphthyl rings from occupying this pocket, while water nearby residues such as Leu163 and Lys158 prevent the forthcoming of the benzodioxolane ring to residue Lys158. The naphthyl ring is placed into the hydrophobic pocket formed by Pro248, Pro249, Tyr265, Tyr269, and Thr302 amino acid residues. Based on these interactions, it was observed that a gem-dimethyl substitution at the same position of methyl from the a or b inhibitors decreases the freedom degree around the carbon atom and locks the compound in a conformation in which one methyl group exhibits a bumping collision with the side chain of Asp165 residue when compared to the (R)-Me substituted analog (b). Therefore, SAR analysis, systematic modification guided by ligands-PLpro complex X-ray crystallography, and subsequent molecular modeling resulted in a potent inhibitor (b), with an IC50 value of 320 nM and antiviral activity with an EC50 value of 9.1 μM upon SARS-CoV-infected Vero E6 cells.140
Lastly, other PLpro inhibitors from CoV have been reported in the literature and these compounds are shown in Table 1.
5.5. RNA-dependent RNA polymerase (also named as nsP12)
The enzymatic complex of HCoVs presents an RNA-dependent RNA polymerase (RdRp, also named nsP12) as a catalytic subunit for the synthesis of a negative-sense RNA, gRNA and sgRNA being a central component of coronaviral replication and transcription machinery. This protein has two domains: the N-terminal with nidovirus RdRp-associated nucleotidyltransferase (NiRAN) activity and the C-terminal canonic RdRp domain, connected by an interface domain.141, 142, 143
The RdRp adopts the conserved structure of viral polymerase family, of a cupped right hand composed of the fingers, palm and thumb subdomains in the C-terminal region, catalyzing the viral RNA during the replication and transcription cycle.144 The RdRp catalyzes the formation of phosphodiester bonds between ribonucleotides from an RNA template in the presence of divalent metal ions and thus synthesizing the negative-sense RNA, gRNA, and sgRNA.143
It was showed that SARS-coronavirus RdRp (nsP12) needs to interact with nsP7 and nsP8 as a tripartite complex to activate its capability to replicate long RNA.145 Recently, the Cryo-EM structure of SARS-CoV-2 nsP12 in complex with the cofactors nsP7 and nsP8 and a newly β-hairpin domain at its N-terminal region was reported.146 For the MERS-CoV, it was demonstrated that nsP8 and nsP12 form an active complex.147
5.5.1. RdRp (nsP12) promising inhibitors
Since RdRp (nsP12) is involved in the replication-transcription process in CoV, as well as in the production of genomic RNA, this target represents an interesting alternative for the development of new drug candidates in search of an effective pharmacotherapy against SARS-CoV-2.148, 149, 150, 151 Thus, recent researches have been developed, especially involving in silico methods, as well as drug repurposing,131, 152, 153 in order to inhibit the viral replication cycle and, consequently, reducing the infection.154, 155
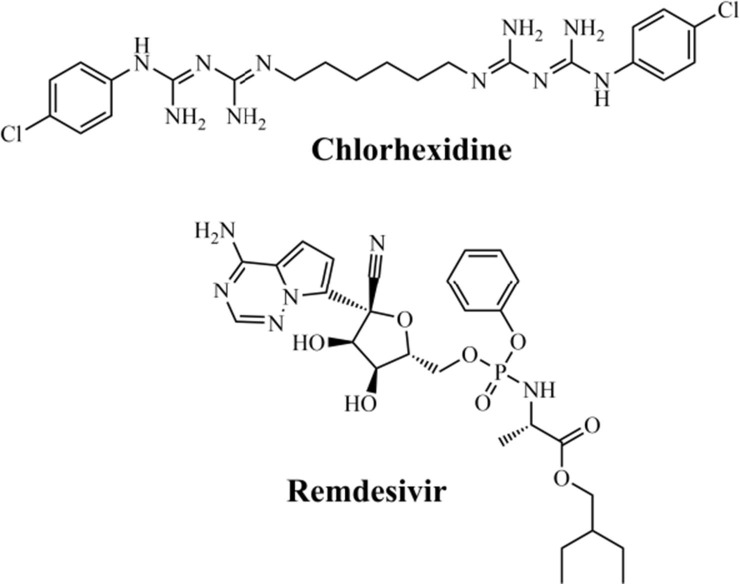
Recently, in a study performed by Choudhury et al. (2020),156 molecular docking was used to analyze 30 FDA-approved drugs at the active site of the RdRp enzyme (PDB ID: 6M71) from SARS-CoV-2. Then, the authors demonstrated that the drugs chlorhexidine and remdesivir (Fig. 7 ) presented the best docking score values, being −132.84 and −114.46, respectively. Based on these values, the authors conclude that chlorhexidine was considered as the best inhibitor in the study, while the remdesivir was found to be the best among the listed antiviral drugs.
Fig. 7.
Promising FDA-approved drugs virtually screened against RdRp from SARS-CoV-2.
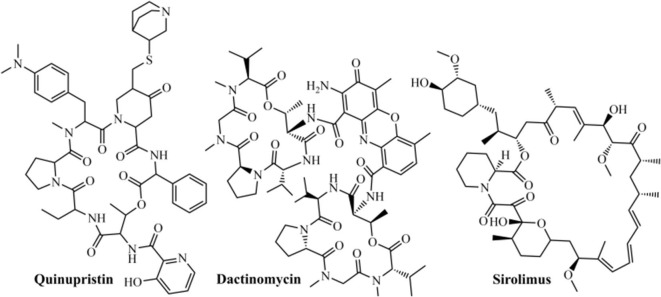
Posteriorly, Pokhrel et al. (2020)157 performed a virtual screening of 1,930 FDA-approved drugs forward RdRp enzyme (PDB ID: 6M71) from SARS-CoV-2, utilizing the Chemoinformatic Tools and Databases, and also the AutoDock Vina to perform docking (by using ensemble, rigid, and flexible methods). In this study, the authors observed that quinupristin (Fig. 8 ), an antimicrobial active against Gram-positive bacteria, was the most promising ligand in all three types of docking studies, exhibiting a docking score of −12.3 Kcal/mol. Moreover, dactinomycin and sirolimus antibiotics (Fig. 8) also demonstrated encouraging results, with scores of −12.2 Kcal/mol for both, where these results could be used as a basis for the search for RdRp from SARS-CoV-2 pandemic.
Fig. 8.
Promising antibiotic inhibitors against RdRp from SARS-CoV-2.
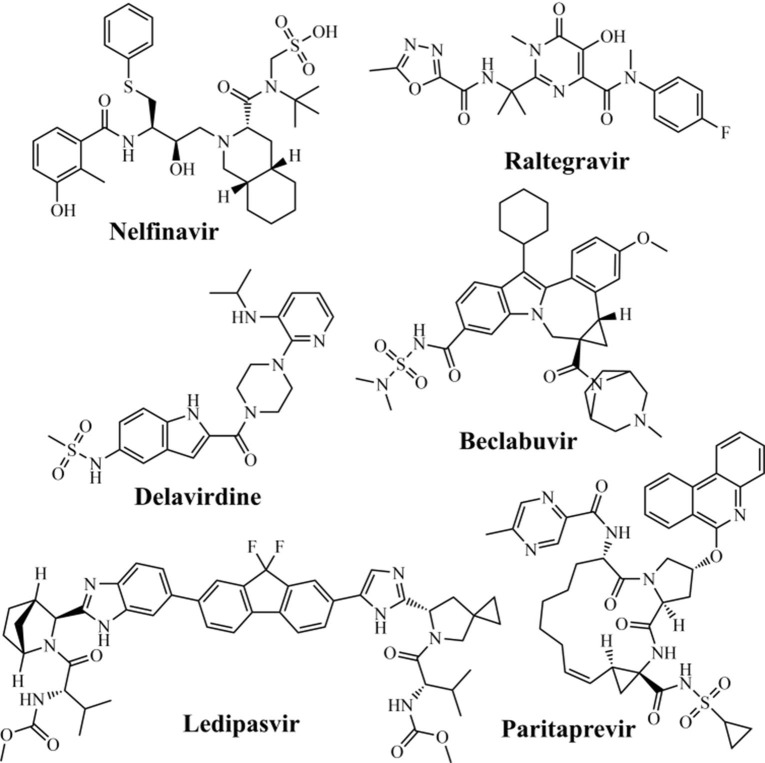
Searching for repurposing drugs against RdRp, Beg et al. (2020)158 performed a study based on molecular homology of RdRp from SARS-CoV-2 (PDB ID: 6NUR), using the SWISS-MODEL server, as well as molecular docking in the AutoDock Tools software. Then, the authors analyzed 74 different antiviral drugs, including HCV, HIV, human cytomegalovirus (HCMV), herpes simplex virus (HSV), human papillomavirus (HPV), varicella-zoster virus (VZV), and influenza virus. It was observed that the anti-HIV nelfinavir (−8.8 Kcal/mol), raltegravir (−8.7 Kcal/mol), and delavirdine (−8.5 Kcal/mol) drugs (Fig. 9 ), as well as anti-HCV paritaprevir (−10 Kcal/mol), beclabuvir (−10 Kcal/mol), and ledipasvir (−10 Kcal/mol) (Fig. 9) drugs were to be the most promising molecules against SARS-CoV-2 RdRp.
Fig. 9.
Anti-HIV and -HCV promising drugs screened against SARS-CoV-2 RdRp.
Among anti-HIV drugs, the main amino acid residues involved in hydrogen-bonding interactions were identified as Ala576, Asn582, and Ser650 for delavirdine; Arg444, Asp509, Asp514, and Arg515 for nelfinavir; and Asp343, Arg444, Asp514, Thr571, Ser573, and Asn582 for raltegravir. Concerning anti-HCV drugs, hydrogen-bonding interactions were observed with Ser392, Asn398, and Asn434 for beclabuvir; Lys391, Arg446, Arg460, and Asp514 for ledipasvir; and Ser392, Asn398, and Lys402 for paritaprevir.
Finally, the aforementioned studies reported the search for RdRp inhibitors by using techniques of drug repurposing, in which promising compounds have been identified that could be useful against SARS-CoV-2 in the future. Furthermore, this compilation will serve as a basis to accelerate the search for promising and effective compounds to inhibit RdRp from SARS-CoV-2.
5.6. NTPase/Helicase (also named as nsP13)
Helicases are classified into six superfamilies (SFs) and those from the Nidovirales order belong to the SF1 superfamily and the Upf1 family (SF1B), characterized by moving in the 5′ → 3′ direction along the nucleic acid chain.143, 159 In coronaviruses, the SF1 helicase domain (HEL1) is located in the C-terminal region of the nsP13, from a cleavage product of pp1ab replicase. The N-terminal region of nsP13 contains a multinuclear zinc-binding domain (ZBD), which is one of the most conserved domains of the Nidovirales order. Structurally, the CoV HEL1 domain is formed by two RecA-like domains, named 1A and 2A.143, 160
For the SARS-CoV nsP13, both RNA and DNA duplex-unwinding activities were detected allowing the efficient strand separation of extended regions of double-stranded RNA and DNA in a 5′-to-3′direction. Besides, both (deoxy)nucleoside triphosphatase (dNTPase) and RNA-5′-triphosphatase activities were also reported being the SARS-CoV nsP13. Subcellular localization showed nsP13 on membranes from the endoplasmic reticulum, apparently the site of SARS-CoV RNA synthesis.161 The biochemical characterization of MERS-CoV nsP13 showed a dsRNA-unwinding activity with the helicase requiring more ATP for the optimal unwinding of RNA substrates with short 5′ loading strands.162 Recently, the NTPase and RNA helicase activities were reported for SARS-CoV-2 nsP13 and that bismuth salts can inhibit both these activities in a dose-dependent manner.163 There is speculation that SARS-CoV nsP13 may has a function in RNA capping because of the demonstrated RNA triphosphatase TPase activity, although further studies are necessary.164
5.6.1. NTPase/Helicase (nsP13) inhibitors
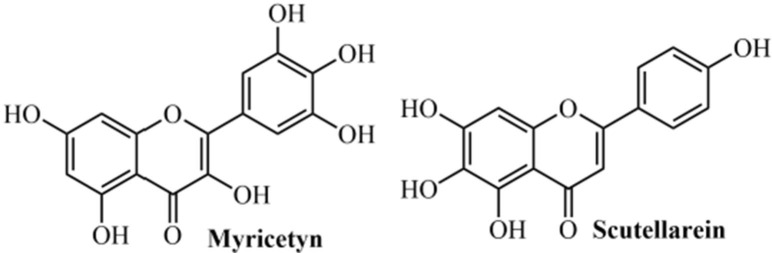
Keum & Jeong (2012)165 evaluated the viral helicase as a potential target for developing chemical inhibitors against SARS-CoV. In this study, they analyzed the full genome sequence from SARS-CoV and verified that 2/3 of the SARS-CoV genome consists of viral replicase genes, which encode 16 non-structural proteins (nsP’s). Among these, RNA-dependent RNA-polymerase (named as nsP12 or RdRp) and NTPase/helicase (nsP13) are very important for viral replication. The SARS-CoV NTPase/helicase consists of 601 amino acids obtained from the replicase region,166, 167 and analyses of the amino acid sequence suggest that the helicase is divided into two separate domains. The first is a metal-binding domain (MBD) located at the N-terminus and, the second, a helicase domain (Hel).168 The SARS-CoV helicase uses ATP and dATP as preferred energy sources.161, 169 The authors conducted in vitro biochemical experiments to find natural compounds that might suppress either the DNA unwinding or the ATPase activity of the SARS-CoV NTPase/helicase. It was observed that none of the 64 compounds evaluated interfered with the DNA unwinding activity of the nsP13 protein. However, the flavonoid myricetin (IC50 = 2.71 µM) and flavone scutellarein (IC50 = 0.86 µM) strongly inhibited the ATPase activity of nsP13 from the SARS-CoV (Fig. 10 ). Nevertheless, other investigations are still required since it is unclear how myricetin and scutellarein suppress the ATPase activity.
Fig. 10.
Natural compounds able to inhibit the NTPase/helicase from SARS-CoV.
Finally, other inhibitors of NTPase/helicase (nsP13) from CoV are displayed in Table 1.
5.7. Guanine-N7-Methyltransferase (also named as N7-MTase or nsP14)
The nsP14 has both exoribonuclease (ExoN) and guanine-N7-methyltransferase (N7-MTase) activities and both roles are important for viral replication and transcription. Coronaviruses replicate in the cytoplasm and have evolved strategies to cap their RNAs, with several nsP’s involved in this process. The SARS-CoV nsP14 was identified as a cap guanine-N7-methyltransferase (N7-MTase) producing the cap-0 structure (m7GpppN), from the family of S-adenosyl-l-methionine (SAM)-dependent methyltransferases.170 Several critical residues for the nsP14 methyltransferase activity on GTP were identified including F73, R84, W86, R310, D331, G333, P335, Y368, C414, and C416.171 The SARS-CoV nsP14 C-terminal N7-MTase domain has a noncanonical MTase fold with a β-sheet insertion and a peripheral zinc finger.172
Interestingly, the association of nsP10 with nsP14 stimulates the ExoN activity while does not affect the N7-MTase activity.164, 173 In this way, one molecule of nsP10 interacts with N-terminal ExoN domain of nsP14 to stabilize it and stimulate its activity, and the presence of two zinc fingers are crucial for this nsP14 function.172
In contrast with nsP14 MTase, SARS-CoV nsP16 also presented MTase methylation activity, in a sequence-specific manner to m7GpppA-RNA. Crystal structure showed an nsP16/nsP10 complex, where nsP16 binds to substrates m7GpppA-RNA and SAM cofactor with the assistance of nsP10.164
5.7.1. N7-MTase (nsP14) inhibitors
Natural products from plants, animals, and microorganisms have been used to treat diseases since the beginning of human life.174, 175 Among diverse bacteria of medicinal interest, Streptomyces species (Gram-positive bacteria) have been widely reported as chassis organisms suitable for the development of bioactive molecules,176, 177, 178 constituting the main resource of antibiotics for clinical use.179, 180
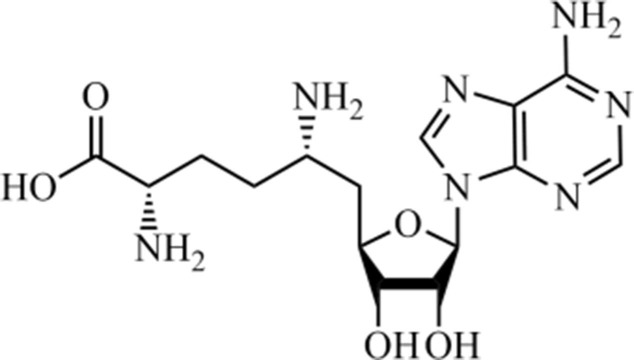
Sinefungin (Fig. 11 ) constitutes an important amino acid-containing nucleoside, obtained from Streptomyces griseolus and S. incarnatus species,181 structurally related to S-adenosyl-methionine (SAM) and S-adenosyl-L-homocysteine (SAH), thus constituting an anticancer agent against metastasis in lung and primary breast tumors.182, 183, 184 Recently, promising antiviral activity against the Zika virus (ZIKV),185, 186 and Feline herpesvirus type 1 (FHV-1).187 In this context, Sun et al. (2014)188 described the activity of this natural product against SARS-CoV, exhibiting a promising activity at 1.68 µM concentration. Moreover, it acts as an inhibitor of the enzyme guanine-N 7-methyltransferase, which has been identified as a promising target for the development of new antiviral drugs against SARS-CoV since it inhibits the RNA-processing enzyme.
Fig. 11.
Sinefungin, a pan-inhibitor against SAM-dependent methyltransferases.
The aforementioned authors identified this inhibitor by developing a genetic system as a high-throughput enzymatic activity assay platform, involving 3,000 natural product extracts as candidates for N 7-MTases inhibitors. These extracts were obtained from the natural microbial product library at Hubei Biopesticide Engineering Research Center (HBERC), in which secondary metabolites were isolated using classical methods. Additionally, it was concluded that sinefungin represents a broad-spectrum inhibitor, since it has shown activity upon N 7-MTase enzymes from different CoV species, such as murine coronavirus (MHV), transmissible gastroenteritis coronavirus (TGEV), and infectious bronchitis virus (IBV), with IC50 values of 1.89, 1.67, and 1.9 µM, respectively. Unfortunately, it does not constitute an ideal viral inhibitor since it has a poor selectivity index (SI) of 0.32, due to its results towards human N 7-MTase (IC50 = 0.55 nM) and SARS-CoV N 7-MTase (IC50 = 1.68 µM). In contrast, it contributes as a potential scaffold for designing new selective N7-MTases inhibitors, in which it could be also used against SARS-CoV-2.
In a study developed by Aouadi et al. (2017)189 was described a high-throughput N 7-MTase assay based on Homogenous Time-Resolved Fluorescence (HTRF®), where they identified 20 potential compounds that inhibit SARS-CoV N 7-MTase. Therefore, 2,000 compounds obtained from the Prestwick Chemical Library®, including 1,280 small molecules (FDA- and EMA-approved drugs), 320 natural products, and 400 pyridazine-derived compounds were also screened in this study. Among these compounds, 20 best molecules were identified as potential antiviral agents. Lastly, the natural product, a polyphenol, demonstrated to be the most promising inhibitor targeting the SARS-CoV nsP14 (Fig. 12 ), with an IC50 value of 0.019 µM.
Fig. 12.
Most promising inhibitor against guanine-N7-methyltransferase from SARS-CoV.
Furthermore, the authors also reported that sinefungin has shown inhibition <0.05 µM against human mRNA cap guanine-N7-methyltransferase (RNMT), which characterizes it has poor selectivity because of its low selectivity index (SI) value (<2.63). This low SI value could be associated with the structural homology between the SARS-CoV N 7-MTase and human RNMT forms, which limits the possibility of discovering specific inhibitors for this viral target.
Finally, another inhibitor of SARS-CoV N7-MTase is presented in Table 1.
5.8. Nucleocapsid protein (also named as N protein)
The nucleocapsid protein (N) is a multifunctional RNA-binding protein having pivotal roles in the viral replication cycle as viral core formation, viral assembly, virus budding/envelope formation, and mRNA replication/gRNA synthesis.190 This protein binds and packs the viral RNA genome into a helical nucleocapsid structure called ribonucleoprotein (RNP) complex. Also, the coronavirus N protein has several roles in the cellular response including chaperone activity, cell cycle regulation, stress response, viral pathogenesis/immune system interference, and signal transduction.190
N protein structure has an N-terminal RNA-binding domain (NTD) responsible for RNA binding, a central Ser/Arg(SR)-rich linker intrinsically disordered, and a C-terminal dimerization domain (CTD).190, 191 The N-terminal domain of the SARS-CoV N protein has five-strand β-sheet and single-stranded RNAs bind to the protein surface at the junction between positively charged β-hairpin and the core structure.191 The crystallographic structure of the SARS-CoV N protein CTD showed a dimer with extensive interactions between the two subunits thus suggesting that the N protein is not stable in the monomeric form.192
Recently, the crystallographic structure of the N-terminal RNA binding domain of SARS-CoV-2 N protein was determined and a unique potential hydrophobic RNA binding pocket alongside the β-sheet core was detected.193 Also, the biochemical characterization of expressed SARS-CoV-2 N protein detected the protein as a large dimer in solution by CTD-CTD interaction.194
5.8.1. N Protein inhibitors
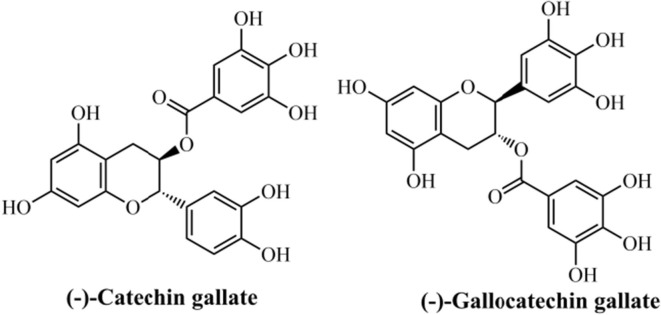
Among the CoV structural proteins, the nucleocapsid (N) protein seems to be a promising target for designing therapeutic drugs with antiviral activity, since it plays essential function on the virus particle assembly.195 The mouse CoV N protein is a major pathological marker in the host cell, in which could lead to the upregulation of proinflammatory cytokines, block innate immune response, and also cause induce host cell apoptosis.196 Some studies have reported the utilization of polyphenolic compounds with therapeutic properties, acting as antioxidants and anticancer, as well as antiviral and antimicrobial agents.197, 198, 199, 200 Both flavonoids (−)-catechin gallate and (−)-gallocatechin gallate demonstrate medicinal interest, due to their antioxidant effects with possible application for different disorders,201 including cancer.202 Also, (−)-catechin gallate has demonstrated promising effects in the treatment of HIV,203, 204 as well as against prostatic,205 brain,206 and other types of cancers.207
Deeming these natural compounds, Roh et al. (2012)208 developed a new approach for the inhibitor screening forwards SARS-CoV N protein by using a quantum dots-conjugated oligonucleotide system with broad application in imaging analysis on a biochip. From a high-throughput screening (HTS) strategy using an optical nanoparticle-based RNA oligonucleotide, it was identified the inhibitory effects of (−)-catechin gallate and (−)-gallocatechin gallate (Fig. 13 ) upon SARS-CoV N protein. The biological evaluation revealed that both flavonoids exhibit high inhibition activity in a concentrated manner against SARS-CoV N protein. These compounds, at 0.005 µg/mL or more, concentration-dependently attenuated the binding affinity as evidenced by quantum dots-RNA oligonucleotide on the biochip. This concentration was found to be the IC50 value for both natural products. Regarding this, the authors have concluded that a new property for (−)-catechin gallate and (−)-gallocatechin gallate has been identified, and also their approach was found to be promising for drug discovery. Besides, no information about molecular interactions from in silico studies were provided by the authors. Finally, the authors’ technique could be mainly characterized by low-cost, high-sensibility, fast result, low labor-effort, compatibility for miniaturization, allowing which it could be applicable for discovering inhibitors in screenings against other types of diseases.
Fig. 13.
Flavonoids with activity against SARS-CoV N protein.
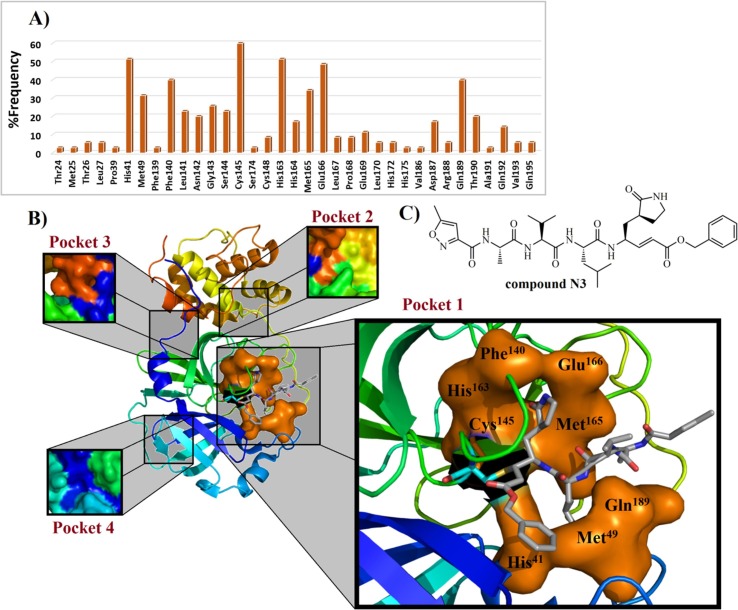
Based on Table 1, it is observed that Han et al. (2006)209 have developed a peptide sequence (containing 31 amino acids) capable of inhibiting the S protein from SARS-CoV, with an IC50 value of 0.1 µM. In general, it is possible to verify that 3CLpro inhibitors are more broadly studied in the medicinal chemistry of CoV, followed by PLpro inhibitors. In this context, we performed a complete analysis of the interactions associated with 3CLpro inhibitors found in the literature (IC50 values ranging from 97 to 0.051 µM). Then, it was revealed that hydrophobic contacts are present in 49.09% of all interactions. Additionally, hydrogen-bonding interactions are observed for 49.54% of compounds, being the Cys145 (11.1%), His163 (12.96%), and Glu166 (14.81%) residues most present in interactions between ligands and SARS-CoV 3CLpro. In some studies, the Glu166 residue has been replaced with His166 residue,216, 219 because the authors have used MERS-CoV 3CLpro (PDB ID: 4RSP) and SARS-like human β-Coronaviruses 2c EMC/2012 (HCoV-EMC), respectively. In general, MERS-CoV 3CLpro is also a chymotrypsin-like cysteine protease that has a catalytic dyad constituted of Cys148 and His41, along with an extended binding site.219, 267, 268, 269, 270 This target exhibits a rigorous primary substrate specificity for a P 1 Gln residue.271 Also, a strong affinity for a P 2 Leu residue has been identified. Finally, P 3 residue side chain is oriented towards the hydrophobic P 4 residue, such as Ala.216, 271 In contrast, some studies have suggested that SARS-CoV 3CLpro has a catalytic dyad Cys145-His41, similarly to the SARS-CoV-2 (Cys145 and His41), HCoV 3CLpro (Cys144 and His41), and TGEV 3CLpro (Cys145 and His41).125, 272 Some works have suggested that cysteine residue from catalytic dyad could act as a nucleophile, being Cys145 from SARS-CoV221 and SARS-CoV-2 3CLpro,273 and Cys148 from MERS-CoV 3CLpro.214, 219 Then, considering all interactions reported for 3CLpro inhibitors into Table 1, it was built a graphic containing the frequency of residues (Fig. 14 A). This type of graphic has been used by our research team to identify fingerprints of residues from a cysteine protease.274 Regarding the Fig. 14A, it is possible to observe that eight amino acids are more frequently associated with higher than 30% of interactions between ligands and 3CLpro target, being His41 (51.4%), Met49 (31.4%), Phe140 (40%), Cys145 (60%), His163 (51.4%), Met165 (34.2%), Glu166 (48.5%), and Gln189 (40%) residues from protomer A. Moreover, the cysteine (Cys145-148) residue performs covalent bonding modes in only 1.37% of interactions. All these residues were used to build a tridimensional active site structure (Fig. 14B) in complex with the inhibitor N3 (Fig. 14C), containing only the most important residues, as previously discussed. It was verified that all these amino acids are comprised into binding pocket 1 from SARS-CoV-2 3CLpro, as described by Shi et al. (2020).275 Notwithstanding these observations, it is evident that most of the active compounds found in the literature preferably bind into the pocket 1. Recently, Harcourt et al. (2004)78 verified that the PLpro from SARS-CoV-2 is a crucial viral enzyme and a potential target for designing inhibitors. Additionally, SARS-CoV PLpro has a catalytic triad constituted of Cys112, His273, and Asp287. Also, the side chain of Trp107 that is placed into the oxyanion hole participates in the stabilization of the negatively charged tetrahedral transition state from the intermediate structure.132, 258, 276 In the SARS-CoV-2 PLpro structure, the catalytic triad is composed of Cys111, His272, and Asp286 residues. Besides, the auxiliary amino acid for stabilization of the negatively charged intermediate is Trp106 residue.259 However, this stabilizing function could be shared with other amino acids, such as Tyr268, Ala289, and Leu298 residues.259 Lastly, these different residues labeling is associated with the X-ray crystallographic structure used by the authors (PDB ID: 6W9C).277 Regarding the PLpro inhibitors (IC50 values ranging from 24.4 to 0.35 µM) in Table 1, these compounds commonly interact with Asp165, Tyr269 (from SARS-CoV), Gln270, Cys111 (SARS-CoV-2), and Cys112 (from SARS-CoV). Helicases are proteins responsible for catalyzing duplex oligonucleotides separation (double-stranded RNA-dsRNA) into single strands (ssRNA), using energy from ATP hydrolysis.278 Previously, a triazole-derived inhibitor, named SSYA10-001, was found to be capable of inhibiting nsP13 Y277A, nsP13 K508A, and WT SARS-CoV nsP13, exhibiting IC50 values of 12, 50, and 5.9 µM, respectively.261 Also, it was revealed that SSYA10-001 interacts with Tyr277, Arg507, and Lys208 residues from SARS- and MERS-CoV nsP13 targets.260, 261 Still, concerning NTPase/helicase (nsP13) inhibitors (IC50 values varying from 11 to 2.5 µM), a triazole derivative recently synthesized by Zaher et al. (2020)262 exhibited activity against MERS-CoV nsP13 (IC50 = 2.5 µM), where it was investigated by using in silico studies. As a result, it was observed that there are only hydrophobic interactions involving Tyr7, Arg163, Tyr159, and Tyr171 amino acid residues from MERS-CoV nsP13 (PDB ID: 5WWP).279 N7- and 2′-O-methylation of the viral RNA cap are critical steps for viral infections since their inhibition leads to a decrease in the synthesis of viral proteins and helps the virus elimination by stimulation of the immune response.280 Then, MTases are considered as attractive targets for drug design and discovery,188, 189 where the Phe426 residue is considered as the largest influence in nsP14 activity.281 S-adenosyl methionine (SAM) and RNA binding sites of N7-MTase nsP14 present 95% sequence identity, when comparing SARS-CoV with SARS-CoV-2 organisms.282 In this context, nucleoside SAM analogs could target nsP14 from both viruses. A potent SARS-CoV nsP14 inhibitor was synthesized by Ahmed-Belkacem et al. (2020),266 in which it demonstrated an IC50 value of 0.6 µM. Additionally, it was verified that this compound is able to perform seven hydrogen-bonding interactions, with Arg310, Gly333, Lys336, Ile338, Asp352, Asn386, and His424.
Fig. 14.
Frequency of residues present in ligand-3CLpro complexes (A) found in the literature and compound N3 in complex with eight most frequent amino acid residues of the pocket 1 (B) from SARS-CoV 3CLpro (PDB ID: 2HOB). In B), covalent Cys145 residue is shown as blue color, exhibiting its connectivity to Michael’s acceptor moiety. Finally, the most frequent amino acid residues from pocket 1 are demonstrated in a molecular surface overview (in orange color). In C), the Michael’s acceptor N3, a peptide-derived, which has been explored in different works, where it has been identified as a promising inhibitor of 3CLpro from MERS-CoV (IC50 = 0.28 µM),219 SARS-CoV-2 (EC50 = 16.77 µM),273 GS-WT12 (Ki = 9.0 ± 0.8 µM), WT-GPH6 (Ki = 2.3 ± 0.1 µM), and WT (Ki = 1.9 ± 0.1 µM) modified proteases.283.
6. Conclusion
Concerning all aspects aforementioned in this manuscript, it is evident that coronaviruses are viruses which account for millions of human illness worldwide, being the SARS-, MERS-, and SARS-CoV-2 the main viruses of medical interest. Currently, there are no vaccines or FDA-approved drugs to specifically treat these viral infectious diseases. Then, the development of researches involving identification and/or synthesis of inhibitors emerges as an essential approach for drug discovery, since selective antivirals targeting macromolecules from HCoVs are an unmet need. In this context, the medicinal chemistry provides tools for the development of more selective inhibitors by using computer-aided drug design (CADD) approaches, such as structure-based drug design (SBDD), in which molecular characteristics from the macromolecular target (including interactions with ligands) should be considered during the rational design of more potent and selective inhibitors. In this scenario, classical medicinal chemistry along with SBDD have resulted in promising new antiviral agents targeting HCoVs’ macromolecules, providing us important aspects about the current druggable targets from these viruses which could be used to design desirable inhibitors.
In general, it was verified that natural products have demonstrated great results in different studies, such as hexamethylene amiloride targeting E protein from HCoV-229E (IC50 = 1.34 µM, and hydrogen-bonding interactions with Asp15 and Arg38 residues), flavone scutellarein targeting nsP13 from SARS-CoV (IC50 = 0.86 µM), sinefungin targeting nsP14 from SARS-CoV (IC50 = 1.68 µM), and the flavonoids (or phenolic compounds) (−)-catechin gallate and (−)-gallocatechin gallate targeting N protein from SARS-CoV (both with IC50 = 0.005 µg/mL). Additionally, small synthetic peptides have exhibited activity against the S protein from SARS-CoV, where the best peptide sequence (STSQKSIVAYTM) displayed an IC50 value of 1.8 nM, suggesting this compound as an excellent inhibitor. However, it was not provided information about the molecular interactions by using in silico methods. Concerning 3CLpro inhibitors, it was verified that the covalent interaction between the cysteine residue from the catalytic dyad and ligands are frequently observed. The best 3CLpro, a synthetic indole analog, was able to covalently interact with Cys145 and, via hydrogen-bonding, with Gly143, Ser144, and His163 residues, at distances ranging from 2.3 to 2.8 Å. In the SARS-CoV 3CLpro inhibition assay, it demonstrated an IC50 value of 0.03 µM. Moreover, the best PLpro inhibitor, a synthetic piperidine carboxamide analog, was not able to interact with Cys residues from the catalytic triad from this target. However, it presented an IC50 value of 320 nM against SARS-CoV PLpro. Also, it was verified that this piperidine analog shows a bumping collision with the Asp165 amino acid residue when an S-methyl substituent is present. Finally, a drug repurposing study was capable of suggesting chlorhexidine (an antimicrobial drug) and remdesivir (an antiviral drug) have been highlighted as promising agents targeting nsP12 from SARS-CoV-2. However, none inhibition assay was performed upon isolated nsP12. Still, anti-HIV and HCV drugs have been also suggested as promising candidates against SARS-CoV-2 nsP12, such as nelfinavir, raltegravir, delavirdine, beclabuvir, ledipasvir, and paritaprevir.
3CLpro inhibitors have been broadly studied, allowing us to generate a %frequency graphic showing the most relevant residues observed during the ligand-target complex formation. From this, it was possible verified that these compounds preferably interact via hydrogen-bonding interactions (49.54%), where the Glu166 residue is the most frequent. Additionally, it was observed that Cys145 (from SARS-CoV) and Cys148 (MERS-CoV) residues together are verified in only 1.37% of interactions. In general, it was verified that the 3CLpro inhibitors are more frequently found into the pocket 1, being the residues His41 (51.4%), Met49 (31.4%), Phe140 (40%), Cys145 (60%), His163 (51.4%), Met165 (34.2%), Glu166 (48.5%), and Gln189 (40%) more commonly found in interactions with this macromolecular target.
Finally, we believe that the search for inhibitors targeting HCoVs macromolecular structures is in continuous development, where several studies have been released, mainly targeting 3CLpro and PLpro. However, the other targets should be considered in the drug design of new antiviral compounds. This manuscript represents a great collection of relevant studies targeting HCoV in order to develop new drugs, which could be used for guiding future studies focused on the development of inhibitors to fight against coronaviruses, including SARS-CoV-2 (COVID-19). Furthermore, it is possible suggest that the classical medicinal chemistry strategies are insufficient to quickly discover new drugs against this virus. Thus, new methodologies and technologies should be exploited to identify novel anti-SARS-CoV-2 drug candidates in a time- and cost-effective manner. Then, it is time to review the various drug discovery methods involving computer-aided protocols to seek trends and identify promising new avenues of anti-SARS-CoV-2 drug discovery.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa de Alagoas (FAPEAL) and the National Council for Scientific and Technological Development (CNPq) for their support to the Brazilian Post-Graduate Programs. Moreover, the authors also thank the Research Collaboratory for Structural Bioinformatics – Protein Data Bank to provide access to the Educational Resources and Images about HCoVs (available at: https://www.rcsb.org/news?year=2020&article=5e74d55d2d410731e9944f52&feature=true), which allowed us to elaborate the illustration for the graphical abstract. Finally, we would like to thank the Smart Medical Art server (https://smart.servier.com/) for provide graphical schemes for free utilization in academic productions.
References
- 1.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69(4):635–664. doi: 10.1128/mmbr.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo P.C.Y., Lau S.K.P., Lam C.S.F. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavi. J Virol. 2012;86(7):3995–4008. doi: 10.1128/jvi.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peiris J., Chu C., Cheng V. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Chan J.F.W., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorbalenya A.E., Baker S.C., Baric R.S. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adhanom Ghebreyesus T. WHO Director-General’s opening remarks at the media briefing on COVID-19. World Heal Organ. 2020;(March):4. [Google Scholar]
- 8.Campbell I.B., Macdonald S.J.F., Procopiou P.A. Medicinal chemistry in drug discovery in big pharma: past, present and future. Drug Discov Today. 2018;23(2):219–234. doi: 10.1016/j.drudis.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Bawa P., Pradeep P., Kumar P., Choonara Y.E., Modi G., Pillay V. Multi-target therapeutics for neuropsychiatric and neurodegenerative disorders. Drug Discov Today. 2016;21(12):1886–1914. doi: 10.1016/j.drudis.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Macarron R., Banks M.N., Bojanic D. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov. 2011;10(3):188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 11.Rasheed A., Farhat R. Combinatorial chemistry: A review. J Pharm Sci Res. 2013;4(7):2502–2516. doi: 10.13040/IJPSR.0975-8232.4(7).2502-16. [DOI] [Google Scholar]
- 12.Kennedy J.P., Williams L., Bridges T.M., Daniels R.N., Weaver D., Lindsley C.W. Application of combinatorial chemistry science on modern drug discovery. J Comb Chem. 2008;10(3):345–354. doi: 10.1021/cc700187t. [DOI] [PubMed] [Google Scholar]
- 13.Liu R., Li X., Xiao W., Lam K.S. Tumor-targeting peptides from combinatorial libraries. Adv Drug Deliv Rev. 2017;110–111:13–37. doi: 10.1016/j.addr.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu R., Li X., Lam K.S. Combinatorial chemistry in drug discovery. Curr Opin Chem Biol. 2017;38:117–126. doi: 10.1016/j.cbpa.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepherd C.A., Hopkins A.L., Navratilova I. Fragment screening by SPR and advanced application to GPCRs. Prog Biophys Mol Biol. 2014;116(2–3):113–123. doi: 10.1016/j.pbiomolbio.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Baker M. Fragment-based lead discovery grows up. Nat Rev Drug Discov. 2013;12(1):5–7. doi: 10.1038/nrd3926. [DOI] [PubMed] [Google Scholar]
- 17.Viana J de O., Félix M.B., Maia M dos S., Serafim V de L., Scotti L., Scotti M.T. Drug discovery and computational strategies in the multitarget drugs era. Brazilian J Pharm Sci. 2018;54(spe) doi: 10.1590/s2175-97902018000001010. [DOI] [Google Scholar]
- 18.Yıldırım M.A., Goh K.-I., Cusick M.E., Barabási A.-L., Vidal M. Drug—target network. Nat Biotechnol. 2007;25(10):1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 19.Lavecchia A., Giovanni C. Virtual screening strategies in drug discovery: A critical review. Curr Med Chem. 2013;20(23):2839–2860. doi: 10.2174/09298673113209990001. [DOI] [PubMed] [Google Scholar]
- 20.Wang T., Wu M.-B., Zhang R.-H. Advances in computational structure-based drug design and application in drug discovery. Curr Top Med Chem. 2015;16(9):901–916. doi: 10.2174/1568026615666150825142002. [DOI] [PubMed] [Google Scholar]
- 21.Khanna V, Ranganathan S, Petrovsky N. Rational Structure-Based Drug Design. Elsevier Ltd.; 2019. doi:10.1016/b978-0-12-809633-8.20275-6.
- 22.Roy A., Nair S., Sen N., Soni N., Madhusudhan M.S. In silico methods for design of biological therapeutics. Methods. 2017;131:33–65. doi: 10.1016/j.ymeth.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Rose S., Stevens A. Computational design strategies for combinatorial libraries. Curr Opin Chem Biol. 2003;7(3):331–339. doi: 10.1016/S1367-5931(03)00057-7. [DOI] [PubMed] [Google Scholar]
- 24.Kazmi S.R., Jun R., Yu M.-S., Jung C., Na D. In silico approaches and tools for the prediction of drug metabolism and fate: A review. Comput Biol Med. 2019;106:54–64. doi: 10.1016/j.compbiomed.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Kumar R., Harilal S., Gupta S.V. Exploring the new horizons of drug repurposing: A vital tool for turning hard work into smart work. Eur J Med Chem. 2019;182 doi: 10.1016/j.ejmech.2019.111602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillaiyar T., Meenakshisundaram S., Manickam M., Sankaranarayanan M. A medicinal chemistry perspective of drug repositioning: Recent advances and challenges in drug discovery. Eur J Med Chem. 2020;195:112275. doi: 10.1016/j.ejmech.2020.112275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar Y., Singh H., Patel C.N. In silico prediction of potential inhibitors for the Main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. J Infect Public Health. 2020 doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey A., Nikam A.N., Shreya A.B. Potential therapeutic targets for combating SARS-CoV-2: Drug repurposing, clinical trials and recent advancements. Life Sci. 2020;256:117883. doi: 10.1016/j.lfs.2020.117883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiménez-Alberto A., Ribas-Aparicio R.M., Aparicio-Ozores G., Castelán-Vega J.A. Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Comput Biol Chem. 2020;88:107325. doi: 10.1016/j.compbiolchem.2020.107325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreani J., Le Bideau M., Duflot I. In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb Pathog. 2020;145:104228. doi: 10.1016/j.micpath.2020.104228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plaze M., Attali D., Petit A.-C. Repurposing chlorpromazine to treat COVID-19: The reCoVery study. Encephale. 2020;46(3):169–172. doi: 10.1016/j.encep.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hage-Melim LI da S, Federico LB, de Oliveira NKS, et al. Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID-19. Life Sci. 2020;256:117963. doi:10.1016/j.lfs.2020.117963. [DOI] [PMC free article] [PubMed]
- 35.Fantini J., Di Scala C, Chahinian H., Yahi N. Structural and molecular modeling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020:105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251:117627. doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D., Li Z., Liu Y. An overview of the safety, clinical application and antiviral research of the COVID-19 therapeutics. J Infect Public Health. 2020 doi: 10.1016/j.jiph.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiu S, Dick A, Ju H, et al. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J Med Chem. 2020;As Soon As. doi:10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed]
- 39.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121(1):190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 40.McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci U S A. 1967;57(4):933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Der Hoek L., Pyrc K., Jebbink M.F. Identification of a new human coronavirus. Nat Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo P.C.Y., Lau S.K.P., Chu C. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79(2):884–895. doi: 10.1128/jvi.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peiris J.S.M., Lai S.T., Poon L.L.M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003 (Based on data as of the 31 December 2003.). World Health Organization.
- 45.World Health Organization W. Middle east respiratory syndrome coronavirus.
- 46.Tu C., Crameri G., Kong X. Antibodies to SARS coronavirus in civets. Emerg Infect Dis. 2004;10(12):2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kan B., Wang M., Jing H. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79(18):11892–11900. doi: 10.1128/jvi.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lau S.K.P., Woo P.C.Y., Li K.S.M. Discovery of a novel coronavirus, china rattus coronavirus HKU24, from Norway rats supports the murine origin of betacoronavirus 1 and has implications for the ancestor of betacoronavirus lineage A. J Virol. 2015;89(6):3076–3092. doi: 10.1128/jvi.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corman V.M., Baldwin H.J., Tateno A.F. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol. 2015;89(23):11858–11870. doi: 10.1128/jvi.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su S., Wong G., Shi W. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.World Health Organization. Coronavirus Disease (COVID-19) Dashboard.
- 54.Cheng V.C.C., Hung I.F.N., Tang B.S.F. Viral replication in the nasopharynx is associated with diarrhea in patients with severe acute respiratory syndrome. Clin Infect Dis. 2004;38(4):467–475. doi: 10.1086/382681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Law H.K.W., Cheung C.Y., Ng H.Y. Chemokine up-regulation in SARS-coronavirus–infected, monocyte-derived human dendritic cells. Blood. 2005;106(7):2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spiegel M., Schneider K., Weber F., Weidmann M., Hufert F.T. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J Gen Virol. 2006;87(7):1953–1960. doi: 10.1099/vir.0.81624-0. [DOI] [PubMed] [Google Scholar]
- 57.Fehr AR, Stanle, P. Coronaviruses: An overview of their replication and pathogenesis. In: Maier HJ, Bickerton E, Britton P, editors. Coronaviruses: Methods and Protocols. vol. 1282. Methods in Molecular Biology. Springer New York; 2015, pp. 1–23. doi:10.1007/978-1-4939-2438-7. [DOI] [PMC free article] [PubMed]
- 58.de Groot R.J., Baker S.C., Baric R.S. Middle east respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol. 2013;87(14):7790–7792. doi: 10.1128/jvi.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arabi Y.M., Arifi A.A., Balkhy H.H. Clinical course and outcomes of critically ill patients with middle east respiratory syndrome coronavirus infection. Ann Intern Med. 2014;160(6):389–397. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 60.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386(9997):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Senga M., Arabi Y.M., Fowler R.A. Clinical spectrum of the Middle East respiratory syndrome coronavirus (MERS-CoV) J Infect Public Health. 2017;10(2):191–194. doi: 10.1016/j.jiph.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stalin Raj V., Farag E.A.B.A., Reusken C.B.E.M. Isolation of MERS coronavirus from dromedary camel, Qatar, 2014. Emerg Infect Dis. 2014;20(8):1339–1342. doi: 10.3201/eid2008.140663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haagmans B.L., Al Dhahiry S.H.S., Reusken C.B.E.M. Middle East respiratory syndrome coronavirus in dromedary camels: An outbreak investigation. Lancet Infect Dis. 2014;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Azhar E.I., El-Kafrawy S.A., Farraj S.A. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 65.Lokugamage K.G., Narayanan K., Nakagawa K. Middle east respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the Nucleus while Sparing mRNAs of cytoplasmic origin. J Virol. 2015;89(21):10970–10981. doi: 10.1128/jvi.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol. 2014;95(PART 4):874–882. doi:10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed]
- 67.Niemeyer D., Zillinger T., Muth D. Middle east respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol. 2013;87(22):12489–12495. doi: 10.1128/jvi.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scalera N.M., Mossad S.B. The first pandemic of the 21st century: A review of the 2009 pandemic variant influenza A (H1N1) virus. Postgrad Med. 2009;121(5):43–47. doi: 10.3810/pgm.2009.09.2051. [DOI] [PubMed] [Google Scholar]
- 69.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science (80-) 2020;368(6490):473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 70.Neuman B.W., Adair B.D., Yoshioka C. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80(16):7918–7928. doi: 10.1128/jvi.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nal B., Chan C., Kien F. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S M and E. J Gen Virol. 2005;86(5):1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- 72.Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: A functional receptor for SARS coronavirus. Cell Mol Life Sci. 2004;61(21):2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raj V.S., Mou H., Smits S.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ziebuhr J. The coronavirus replicase. Curr Top Microbiol Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332(2):498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harcourt B.H., Jukneliene D., Kanjanahaluethai A. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Boheemen S., De Graaf M., Lauber C. Genomic characterization of a newly discovered coronavirus. MBio. 2012;3(6):1–9. doi: 10.1128/mBio.00473-12.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martina E., Stiefl N., Degel B. Screening of electrophilic compounds yields an aziridinyl peptide as new active-site directed SARS-CoV main protease inhibitor. Bioorganic Med Chem Lett. 2005;15(24):5365–5369. doi: 10.1016/j.bmcl.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alanagreh L., Alzoughool F., Atoum M. The human coronavirus disease covid-19: Its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9(5) doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schoeman D., Fielding B.C. Coronavirus envelope protein: Current knowledge. Virol J. 2019;16(1):1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walls A.C., Tortorici M.A., Bosch B.J. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531(7592):114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kirchdoerfer R.N., Cottrell C.A., Wang N. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531(7592):118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity Of The SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li F., Li W., Farzan M., Harrison S.C. Structural biology: Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (80-) 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 89.Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-) 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89(4):1954–1964. doi: 10.1128/jvi.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tai W., He L., Zhang X. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lan J., Ge J., Yu J. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 93.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shang J., Wan Y., Luo C. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient Activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84(24):12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li J., Zhan P., Liu X. Targeting the entry step of SARS-CoV-2: a promising therapeutic approach. Signal Transduct Target Ther. 2020;5(1):1–2. doi: 10.1038/s41392-020-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xia S., Liu M., Wang C. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bosch B.J., Martina B.E.E., van der Zee R. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci. 2004;101(22):8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ho T., Wu S., Chen J. Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2006;69(2):70–76. doi: 10.1016/j.antiviral.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu S., Xiao G., Chen Y. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic. Lancet. 2020;363(January) doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ni L., Zhu J., Zhang J., Yan M., Gao G.F., Tien P. Design of recombinant protein-based SARS-CoV entry inhibitors targeting the heptad-repeat regions of the spike protein S2 domain. Biochem Biophys Res Commun. 2005;330(1):39–45. doi: 10.1016/j.bbrc.2005.02.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu J., Xiao G., Xu Y. Following the rule: Formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem Biophys Res Commun. 2004;319(1):283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yuan K., Yi L., Chen J. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem Biophys Res Commun. 2004;319(3):746–752. doi: 10.1016/j.bbrc.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: Expression and functional characterization. Biochem Biophys Res Commun. 2003;312(4):1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Babcock G.J., Esshaki D.J., Thomas W.D., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78(9):4552–4560. doi: 10.1128/jvi.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Westerbeck J.W., Machamer C.E. A coronavirus E protein is present in two distinct pools with different effects on assembly and the secretory pathway. J Virol. 2015;89(18):9313–9323. doi: 10.1128/jvi.01237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liao Y., Yuan Q., Torres J., Tam J.P., Liu D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349(2):264–275. doi: 10.1016/j.virol.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Y., Surya W., Claudine S., Torres J. Structure of a conserved golgi complex-targeting signal in coronavirus envelope proteins. J Biol Chem. 2014;289(18):12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nieto-Torres J.L., DeDiego M.L., Álvarez E. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Teoh K.-T., Siu Y.-L., Chan W.-L. The SARS Coronavirus E Protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Nusrat A, ed. Mol Biol Cell. 2010;21(22):3838–3852. doi: 10.1091/mbc.e10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432(2):485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nieto-Torres J.L., DeDiego M.L., Verdiá-Báguena C. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5):1–19. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y., Xiong Z., Zhang S. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem J. 2005;392(1):135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML, et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. Basler CF, ed. PLoS Pathog. 2014;10(8):e1004320. doi:10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed]
- 116.Tilocca B., Soggiu A., Sanguinetti M. Immunoinformatic analysis of the SARS-CoV-2 envelope protein as a strategy to assess cross-protection against COVID-19. Microbes Infect. 2020;(xxxx) doi: 10.1016/j.micinf.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Surya W., Li Y., Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim Biophys Acta - Biomembr. 2018;1860(6):1309–1317. doi: 10.1016/j.bbamem.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Do A. ll Perspective Chemistry and Biology of SARS-CoV-2. Published online 2020:1-13. doi:10.1016/j.chempr.2020.04.023.
- 119.Gazina E.V., Petrou S. Viral targets of acylguanidines. Drug Discov Today. 2012;17(17–18):1039–1043. doi: 10.1016/j.drudis.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hsu P.H., Chiu D.C., Wu K.L. Acylguanidine derivatives of zanamivir and oseltamivir: Potential orally available prodrugs against influenza viruses. Eur J Med Chem. 2018;154:314–323. doi: 10.1016/j.ejmech.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 121.Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353(2):294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pervushin K., Tan E., Parthasarathy K. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5(7) doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science (80-) 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 124.Perlman S., Netland J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tahir ul Qamar M, Alqahtani SM, Alamri MA, Chen L-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. Published online March 2020. doi:10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed]
- 126.Tomar S., Johnston M.L., John S.E.S. Ligand-induced dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 protease (3CLpro): Implications for nsp5 regulation and the development of antivirals. J Biol Chem. 2015;290(32):19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang L., Lin D., Sun X. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science (80-) 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ghosh A.K., Gong G., Grum-Tokars V. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorganic Med Chem Lett. 2008;18(20):5684–5688. doi: 10.1016/j.bmcl.2008.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yu R, Chen L, Lan R, Shen R, Li P. Computational screening of antagonist against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int J Antimicrob Agents. 2020;2(xxxx):106012. doi:10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed]
- 131.Wu C., Liu Y., Yang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Báez-Santos YM, St. John SE, Mesecar AD. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi:10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed]
- 133.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. Deubiquitinating activity of the SARS-CoV papain-like protease. In: Advances in Experimental Medicine and Biology. Vol. 581; 2006:37–41. doi:10.1007/978-0-387-33012-9_5. [DOI] [PMC free article] [PubMed]
- 134.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79(24):15199–15208. doi: 10.1128/jvi.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J Virol. 2009;83(13):6689–6705. doi: 10.1128/jvi.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ratia K., Saikatendu K.S., Santarsiero B.D. Severe acute respiratory syndrome coronavirus papain-like-protease: Structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci U S A. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lei J., Mesters J.R., Drosten C., Anemüller S., Ma Q., Hilgenfeld R. Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features. Antiviral Res. 2014;109(1):72–82. doi: 10.1016/j.antiviral.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81(4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 139.Herold J., Siddell S.G., Gorbalenya A.E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold (Journal of Biological Chemistry (1999) 274 (14400–14405)) J Biol Chem. 1999;274(30):21490. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ghosh A.K., Takayama J., Rao K.V. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein–ligand X-ray structure and biological evaluation. J Med Chem. 2010;53(13):4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43(17):8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: Prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17(12):4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Snijder EJ, Decroly E, Ziebuhr J. The Nonstructural proteins directing coronavirus RNA synthesis and processing. In: Advances in virus research. Vol. 96. 1st ed. Elsevier Inc.; 2016, pp. 59–126. doi:10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed]
- 144.te Velthuis A.J.W., Arnold J.J., Cameron C.E., van den Worm S.H.E., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2009;38(1):203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Subissi L., Posthuma C.C., Collet A. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gao Y., Yan L., Huang Y. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (80-) 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNAdependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ganeshpurkar A., Gutti G., Singh S.K. Polymerases and their emerging roles in antiviral therapy. Viral Polymerases. 2019::1–42. doi: 10.1016/B978-0-12-815422-9.00001-2. [DOI] [Google Scholar]
- 149.Kakhki R.K., Kakhki M.K., Neshani A. COVID-19 target: A specific target for novel coronavirus detection. Gene Reports. 2020;20(May):19–21. doi: 10.1016/j.genrep.2020.100740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Venkateshan M., Muthu M., Suresh J., Ranjith Kumar R. Azafluorene derivatives as inhibitors of SARS CoV-2 RdRp: Synthesis, physicochemical, quantum chemical, modeling and molecular docking analysis. J Mol Struct. 2020;1220 doi: 10.1016/j.molstruc.2020.128741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int J Antimicrob Agents. 2020;2(xxxx):3–8. doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mirza M.U., Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Elfiky AA. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. Published online 2020:117592. doi:10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed]
- 154.Yin W, Mao C, Luan X, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science (80-). 2020;1504(June):eabc1560. doi:10.1126/science.abc1560. [DOI] [PMC free article] [PubMed]
- 155.Huang B., Ling R., Cheng Y. Characteristics and therapeutic options of the coronavirus disease 2019. Mol Ther Methods Clin Dev. 2020 doi: 10.1016/j.omtm.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Choudhury S., Moulick D., Saikia P., Mazumder M.K. Evaluating the potential of different inhibitors on RNA-dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2: A molecular modeling approach. Med J Armed Forces India. 2020;(xxxx) doi: 10.1016/j.mjafi.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pokhrel R., Chapagain P., Siltberg-Liberles J. Potential RNA-dependent RNA polymerase inhibitors as prospective therapeutics against SARS-CoV-2. J Med Microbiol. 2020:864–873. doi: 10.1099/jmm.0.001203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Beg M.A., Athar F. Anti-HIV and Anti-HCV drugs are the putative inhibitors of RNA-dependent-RNA polymerase activity of NSP12 of the SARS CoV-2 (COVID-19) Pharm Pharmacol Int J. 2020;8(3) doi: 10.15406/ppij.2020.08.00292. [DOI] [Google Scholar]
- 159.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76(1):23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 160.Lehmann K.C., Snijder E.J., Posthuma C.C., Gorbalenya A.E. What we know but do not understand about nidovirus helicases. Virus Res. 2015;202:12–32. doi: 10.1016/j.virusres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78(11):5619–5632. doi: 10.1128/jvi.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Adedeji A.O., Lazarus H. Biochemical characterization of middle east respiratory syndrome coronavirus helicase. mSphere. 2016;1(5):1–14. doi: 10.1128/msphere.00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shu T., Huang M., Wu D. SARS-coronavirus-2 Nsp13 Possesses NTPase and RNA helicase activities that can be inhibited by bismuth salts. Virol Sin. 2020;12250:1–9. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen Y., Su C., Ke M. Biochemical and structural insights into the mechanisms of sars coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7(10) doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Keum Y.S., Jeong Y.J. Development of chemical inhibitors of the SARS coronavirus: Viral helicase as a potential target. Biochem Pharmacol. 2012;84(10):1351–1358. doi: 10.1016/j.bcp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Snijder E.J., Bredenbeek P.J., Dobbe J.C. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Thiel V., Ivanov K.A., Putics Á. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84(9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 168.Bernini A., Spiga O., Venditti V. Tertiary structure prediction of SARS coronavirus helicase. Biochem Biophys Res Commun. 2006;343(4):1101–1104. doi: 10.1016/j.bbrc.2006.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tanner J.A., Watt R.M., Chai Y.-B. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J Biol Chem. 2003;278(41):39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen Y., Cai H., Pan J. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A. 2009;106(9):3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jin X., Chen Y., Sun Y. Characterization of the guanine-N7 methyltransferase activity of coronavirus nsp14 on nucleotide GTP. Virus Res. 2013;176(1–2):45–52. doi: 10.1016/j.virusres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ma Y., Wu L., Shaw N. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci U S A. 2015;112(30):9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A. 2012;109(24):9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83(3):770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 175.Xu X., Liu Y., Du G., Ledesma-Amaro R., Liu L. Microbial chassis development for natural product biosynthesis. Trends Biotechnol. 2020:1–18. doi: 10.1016/j.tibtech.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 176.Zhou Q., Ning S., Luo Y. Coordinated regulation for nature products discovery and overproduction in Streptomyces. Synth Syst Biotechnol. 2020;5(2):49–58. doi: 10.1016/j.synbio.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu R., Deng Z., Liu T. Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab Eng. 2018;50:74–84. doi: 10.1016/j.ymben.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 178.Jacob C., Weissman K.J. Unpackaging the roles of streptomyces natural products. Cell Chem Biol. 2017;24(10):1194–1195. doi: 10.1016/j.chembiol.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 179.Davison E.K., Brimble M.A. Natural product derived privileged scaffolds in drug discovery. Curr Opin Chem Biol. 2019;52:1–8. doi: 10.1016/j.cbpa.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 180.Hoefler B.C., Stubbendieck R.M., Josyula N.K., Moisan S.M., Schulze E.M., Straight P.D. A link between linearmycin biosynthesis and extracellular vesicle genesis connects specialized metabolism and bacterial membrane physiology. Cell Chem Biol. 2017;24(10):1238–1249.e7. doi: 10.1016/j.chembiol.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 181.Yin X., Zhao G., Schneller S.W. Carbocyclic sinefungin. Tetrahedron Lett. 2007;48(28):4809–4811. doi: 10.1016/j.tetlet.2007.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sala L., Franco-Valls H., Stanisavljevic J. Abrogation of myofibroblast activities in metastasis and fibrosis by methyltransferase inhibition. Int J Cancer. 2019;145(11):3064–3077. doi: 10.1002/ijc.32376. [DOI] [PubMed] [Google Scholar]
- 183.Cao H., Li L., Deying Y. Recent progress in histone methyltransferase (G9a) inhibitors as anticancer agents. Eur J Med Chem. 2019;179:537–546. doi: 10.1016/j.ejmech.2019.06.072. [DOI] [PubMed] [Google Scholar]
- 184.Raghavendra N.K., Rao D.N. Exogenous AdoMet and its analogue sinefungin differentially influence DNA cleavage by R. EcoP15I - Usefulness in SAGE. Biochem Biophys Res Commun. 2005;334(3):803–811. doi: 10.1016/j.bbrc.2005.06.171. [DOI] [PubMed] [Google Scholar]
- 185.Hercik K., Brynda J., Nencka R., Boura E. Structural basis of Zika virus methyltransferase inhibition by sinefungin. Arch Virol. 2017;162(7):2091–2096. doi: 10.1007/s00705-017-3345-x. [DOI] [PubMed] [Google Scholar]
- 186.Tao Z., Cao R., Yan Y. Design, synthesis and in vitro anti-Zika virus evaluation of novel Sinefungin derivatives. Eur J Med Chem. 2018;157:994–1004. doi: 10.1016/j.ejmech.2018.08.057. [DOI] [PubMed] [Google Scholar]
- 187.Kuroda Y., Yamagata H., Nemoto M., Inagaki K., Tamura T., Maeda K. Antiviral effect of sinefungin on in vitro growth of feline herpesvirus type 1. J Antibiot (Tokyo) 2019;72(12):981–985. doi: 10.1038/s41429-019-0234-4. [DOI] [PubMed] [Google Scholar]
- 188.Sun Y., Wang Z., Tao J. Yeast-based assays for the high-throughput screening of inhibitors of coronavirus RNA cap guanine-N7-methyltransferase. Antiviral Res. 2014;104(1):156–164. doi: 10.1016/j.antiviral.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Aouadi W., Eydoux C., Coutard B. Toward the identification of viral cap-methyltransferase inhibitors by fluorescence screening assay. Antiviral Res. 2017;144:330–339. doi: 10.1016/j.antiviral.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Huang Q., Yu L., Petros A.M. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43(20):6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 192.Yu I.-M., Oldham M.L., Zhang J., Chen J. Crystal Structure of the Severe Acute Respiratory Syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J Biol Chem. 2006;281(25):17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kang S., Yang M., Hong Z. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;(xxx) doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zeng W., Liu G., Ma H. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hatakeyama S., Matsuoka Y., Ueshiba H. Dissection and identification of regions required to form pseudoparticles by the interaction between the nucleocapsid (N) and membrane (M) proteins of SARS coronavirus. Virology. 2008;380(1):99–108. doi: 10.1016/j.virol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hurst K.R., Kuo L., Koetzner C.A., Ye R., Hsue B., Masters P.S. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J Virol. 2005;79(21):13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Galeotti F., Barile E., Curir P., Dolci M., Lanzotti V. Flavonoids from carnation (Dianthus caryophyllus) and their antifungal activity. Phytochem Lett. 2008;1(1):44–48. doi: 10.1016/j.phytol.2007.10.001. [DOI] [Google Scholar]
- 198.Yamamoto Y., Gaynor R.B. Therapeutic potential of inhibition of the NF-κB pathway in the treatment of inflammation and cancer. J Clin Invest. 2001;107(2):135–142. doi: 10.1172/JCI11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cushnie T.P.T., Lamb A.J. Antimicrobial activity of flavonoids. Int J Antimicrob Agents. 2005;26(5):343–356. doi: 10.1016/j.ijantimicag.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Spencer J.P.E. Flavonoids: modulators of brain function? Br J Nutr. 2008;99(E-S1):ES60–ES77. doi: 10.1017/S0007114508965776. [DOI] [PubMed] [Google Scholar]
- 201.Katiyar S., Elmets C., Katiyar S. Green tea and skin cancer: photoimmunology, angiogenesis and DNA repair. J Nutr Biochem. 2007;18(5):287–296. doi: 10.1016/j.jnutbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 202.Pyrko P., Schonthal A.H., Hofman F.M., Chen T.C., Lee A.S. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67(20):9809–9816. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 203.Williamson M., McCormick T., Nance C., Shearer W. Epigallocatechin gallate, the main polyphenol in green tea, binds to the T-cell receptor, CD4: Potential for HIV-1 therapy. J Allergy Clin Immunol. 2006;118(6):1369–1374. doi: 10.1016/j.jaci.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 204.Hamza A., Zhan C.-G. How Can (−)-Epigallocatechin Gallate from Green Tea Prevent HIV-1 Infection? Mechanistic Insights from Computational Modeling and the Implication for Rational Design of Anti-HIV-1 Entry Inhibitors. J Phys Chem B. 2006;110(6):2910–2917. doi: 10.1021/jp0550762. [DOI] [PubMed] [Google Scholar]
- 205.Hsieh T.-C., Wu J.M. Targeting CWR22Rv1 prostate cancer cell proliferation and gene expression by combinations of the phytochemicals EGCG, genistein and quercetin. Anticancer Res. 2009;29(10):4025–4032. [PMC free article] [PubMed] [Google Scholar]
- 206.Das A., Banik N.L., Ray S.K. Flavonoids activated caspases for apoptosis in human glioblastoma T98G and U87MG cells but not in human normal astrocytes. Cancer. 2009 doi: 10.1002/cncr.24699. NA–NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Philips B.J., Coyle C.H., Morrisroe S.N., Chancellor M.B., Yoshimura N. Induction of apoptosis in human bladder cancer cells by green tea catechins. Biomed Res. 2009;30(4):207–215. doi: 10.2220/biomedres.30.207. [DOI] [PubMed] [Google Scholar]
- 208.Roh C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int J Nanomed Radiat Technol Inst. 2012;7:2173–2179. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Han D.P., Penn-Nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350(1):15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shimamoto Y., Hattori Y., Kobayashi K. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorganic Med Chem. 2015;23(4):876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Konno H., Wakabayashi M., Takanuma D., Saito Y., Akaji K. Design and synthesis of a series of serine derivatives as small molecule inhibitors of the SARS coronavirus 3CL protease. Bioorganic Med Chem. 2016;24(6):1241–1254. doi: 10.1016/j.bmc.2016.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kumar V, Tan K, Wang Y, Lin S, Liang P. Bioorganic & Medicinal Chemistry Identification , synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. 2016;24:3035–3042. [DOI] [PMC free article] [PubMed]
- 213.Zhang L., Lin D., Kusov Y. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem. 2020 doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 214.Kumar V., Shin J.S., Shie J.J. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antiviral Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang L., Bao B.B., Song G.Q. Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical synthesis, biological evaluation, molecular docking and 3D-QSAR study. Eur J Med Chem. 2017;137:450–461. doi: 10.1016/j.ejmech.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Galasiti Kankanamalage A.C., Kim Y., Damalanka V.C. Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element. Eur J Med Chem. 2018;150:334–346. doi: 10.1016/j.ejmech.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ohnishi K., Hattori Y., Kobayashi K., Akaji K. Evaluation of a non-prime site substituent and warheads combined with a decahydroisoquinolin scaffold as a SARS 3CL protease inhibitor. Bioorganic Med Chem. 2019;27(2):425–435. doi: 10.1016/j.bmc.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yoshizawa S ichiro, Hattori Y, Kobayashi K, Akaji K. Evaluation of an octahydroisochromene scaffold used as a novel SARS 3CL protease inhibitor. Bioorganic Med Chem. 2020;28(4):115273. doi:10.1016/j.bmc.2019.115273. [DOI] [PMC free article] [PubMed]
- 219.Ren Z., Yan L., Zhang N. The newly emerged SARS-Like coronavirus HCoV-EMC also has an “Achilles’’ heel“: Current effective inhibitor targeting a 3C-like protease”. Protein Cell. 2013;4(4):248–250. doi: 10.1007/s13238-013-2841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yang H., Xie W., Xue X. Design of wide-spectrum inhibitors targeting coronavirus main proteases. Bjorkman P, ed. PLoS Biol. 2005;3(10):e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chuck C.P., Chen C., Ke Z., Chi-Cheong Wan D., Chow H.F., Wong K.B. Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. Eur J Med Chem. 2013;59:1–6. doi: 10.1016/j.ejmech.2012.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Prior A.M., Kim Y., Weerasekara S. Design, synthesis, and bioevaluation of viral 3C and 3C-like protease inhibitors. Bioorganic Med Chem Lett. 2013;23(23):6317–6320. doi: 10.1016/j.bmcl.2013.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ramajayam R., Tan K.-P., Liu H.-G., Liang P.-H. Synthesis, docking studies, and evaluation of pyrimidines as inhibitors of SARS-CoV 3CL protease. Bioorg Med Chem Lett. 2010;20(12):3569–3572. doi: 10.1016/j.bmcl.2010.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Chen L., Li J., Luo C. Binding interaction of quercetin-3-β-galactoside and its synthetic derivatives with SARS-CoV 3CLpro: Structure–activity relationship studies reveal salient pharmacophore features. Bioorg Med Chem. 2006;14(24):8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ryu Y.B., Jeong H.J., Kim J.H. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorganic Med Chem. 2010;18(22):7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lin C.-W., Tsai F.-J., Tsai C.-H. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res. 2005;68(1):36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Shie J.J., Fang J.M., Kuo T.H. Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic α, β-unsaturated esters. Bioorganic Med Chem. 2005;13(17):5240–5252. doi: 10.1016/j.bmc.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shao Y.-M., Yang W.-B., Kuo T.-H. Design, synthesis, and evaluation of trifluoromethyl ketones as inhibitors of SARS-CoV 3CL protease. Bioorg Med Chem. 2008;16(8):4652–4660. doi: 10.1016/j.bmc.2008.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Konno S., Thanigaimalai P., Yamamoto T. Design and synthesis of new tripeptide-type SARS-CoV 3CL protease inhibitors containing an electrophilic arylketone moiety. Bioorg Med Chem. 2013;21(2):412–424. doi: 10.1016/j.bmc.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Thanigaimalai P., Konno S., Yamamoto T. Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: Structure–activity relationship study. Eur J Med Chem. 2013;65:436–447. doi: 10.1016/j.ejmech.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Thanigaimalai P., Konno S., Yamamoto T. Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: Design, synthesis, biological evaluation, and docking studies. Eur J Med Chem. 2013;68:372–384. doi: 10.1016/j.ejmech.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Liu Y.-C., Huang V., Chao T.-C. Screening of drugs by FRET analysis identifies inhibitors of SARS-CoV 3CL protease. Biochem Biophys Res Commun. 2005;333(1):194–199. doi: 10.1016/j.bbrc.2005.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Blanchard J.E., Elowe N.H., Huitema C. High-Throughput Screening Identifies Inhibitors of the SARS Coronavirus Main Proteinase. Chem Biol. 2004;11(10):1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tsai K.-C., Chen S.-Y., Liang P.-H. Discovery of a novel family of SARS-CoV protease inhibitors by virtual screening and 3D-QSAR studies. J Med Chem. 2006;49(12):3485–3495. doi: 10.1021/jm050852f. [DOI] [PubMed] [Google Scholar]
- 235.Wen C.-C., Kuo Y.-H., Jan J.-T. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem. 2007;50(17):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 236.Zhang J., Huitema C., Niu C. Aryl methylene ketones and fluorinated methylene ketones as reversible inhibitors for severe acute respiratory syndrome (SARS) 3C-like proteinase. Bioorg Chem. 2008;36(5):229–240. doi: 10.1016/j.bioorg.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ge F., Xiong S., Sen Lin F, Zhang Z.P., Zhang X.E. High-throughput assay using a GFP-expressing replicon for SARS-CoV drug discovery. Antiviral Res. 2008;80(2):107–113. doi: 10.1016/j.antiviral.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kao R.Y., Tsui W.H.W., Lee T.S.W. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem Biol. 2004;11(9):1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mukherjee P., Desai P., Ross L., White E.L., Avery M.A. Structure-based virtual screening against SARS-3CLpro to identify novel non-peptidic hits. Bioorganic Med Chem. 2008;16(7):4138–4149. doi: 10.1016/j.bmc.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jacobs J., Grum-Tokars V., Zhou Y. Discovery, synthesis, and structure-based optimization of a series of N -(tert -Butyl)-2-(N -arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL. J Med Chem. 2013;56(2):534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Park J.Y., Kim J.H., Kwon J.M. Dieckol, a SARS-CoV 3CLpro inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorganic Med Chem. 2013;21(13):3730–3737. doi: 10.1016/j.bmc.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lee H., Mittal A., Patel K. Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual and high-throughput screenings. Bioorganic Med Chem. 2014;22(1):167–177. doi: 10.1016/j.bmc.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Liu W., Zhu H.-M., Niu G.-J. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg Med Chem. 2014;22(1):292–302. doi: 10.1016/j.bmc.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chen L.R., Wang Y.C., Yi W.L. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorganic Med Chem Lett. 2005;15(12):3058–3062. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Lu I.-L., Mahindroo N., Liang P.-H. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J Med Chem. 2006;49(17):5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- 246.Niu C., Yin J., Zhang J., Vederas J.C., James M.N.G. Molecular docking identifies the binding of 3-chloropyridine moieties specifically to the S1 pocket of SARS-CoV Mpro. Bioorganic Med Chem. 2008;16(1):293–302. doi: 10.1016/j.bmc.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Turlington M., Chun A., Tomar S. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: Identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorganic Med Chem Lett. 2013;23(22):6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Akaji K., Konno H., Mitsui H. Structure-based design, synthesis, and evaluation of peptide-mimetic SARS 3CL protease inhibitors. J Med Chem. 2011;54(23):7962–7973. doi: 10.1021/jm200870n. [DOI] [PubMed] [Google Scholar]
- 249.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. 2020;35(1):145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Park J.Y., Ko J.A., Kim D.W. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J Enzyme Inhib Med Chem. 2016;31(1):23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 251.Ramajayam R., Tan K.P., Liu H.G., Liang P.H. Synthesis and evaluation of pyrazolone compounds as SARS-coronavirus 3C-like protease inhibitors. Bioorganic Med Chem. 2010;18(22):7849–7854. doi: 10.1016/j.bmc.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ghosh A.K., Takayama J., Aubin Y. Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome−coronavirus papain-like protease. J Med Chem. 2009;52(16):5228–5240. doi: 10.1021/jm900611t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lin M.H., Moses D.C., Hsieh C.H. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Res. 2018;150(November 2017):155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Chou C.Y., Chien C.H., Han Y.S. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem Pharmacol. 2008;75(8):1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lee H., Ren J., Pesavento R.P. Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via high-throughput screening and molecular modeling. Bioorganic Med Chem. 2019;27(10):1981–1989. doi: 10.1016/j.bmc.2019.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Park J.Y., Kim J.H., Kim Y.M. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorganic Med Chem. 2012;20(19):5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Cho JK, Curtis-Long MJ, Lee KH, et al. Corrigendum to “Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa” [Bioorg. Med. Chem. 21 (2013) 3051–3057]. Bioorg Med Chem. 2013;21(22):7229. doi:10.1016/j.bmc.2013.09.005. [DOI] [PMC free article] [PubMed]
- 258.Báez-Santos Y.M., Barraza S.J., Wilson M.W. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J Med Chem. 2014;57(6):2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Węglarz-Tomczak E, Tomczak JM, Talma M, Brul S. Ebselen as a highly active inhibitor of PLProCoV2. bioRxiv. Published online 2020:2020.05.17.100768. doi:10.1101/2020.05.17.100768.
- 260.Adedeji A.O., Singh K., Calcaterra N.E. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob Agents Chemother. 2012;56(9):4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Adedeji A.O., Singh K., Kassim A. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and middle east respiratory syndrome coronaviruses. Antimicrob Agents Chemother. 2014;58(8):4894–4898. doi: 10.1128/AAC.02994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zaher N.H., Mostafa M.I., Altaher A.Y. Design, synthesis and molecular docking of novel triazole derivatives as potential CoV helicase inhibitors. Acta Pharm. 2020;70(2):145–159. doi: 10.2478/acph-2020-0024. [DOI] [PubMed] [Google Scholar]
- 263.Yang N., Tanner J.A., Wang Z. Inhibition of SARS coronavirus helicase by bismuth complexes. Chem Commun. 2007;42:4413–4415. doi: 10.1039/b709515e. [DOI] [PubMed] [Google Scholar]
- 264.Tanner J.A., Zheng B.J., Zhou J. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem Biol. 2005;12(3):303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kim M.K., Yu M.S., Park H.R. 2,6-Bis-arylmethyloxy-5-hydroxychromones with antiviral activity against both hepatitis C virus (HCV) and SARS-associated coronavirus (SCV) Eur J Med Chem. 2011;46(11):5698–5704. doi: 10.1016/j.ejmech.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ahmed-Belkacem R., Sutto-Ortiz P., Guiraud M. Synthesis of adenine dinucleosides SAM analogs as specific inhibitors of SARS-CoV nsp14 RNA cap guanine-N7-methyltransferase. Eur J Med Chem. 2020;201:112557. doi: 10.1016/j.ejmech.2020.112557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Needle D., Lountos G.T., Waugh D.S. Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity. Acta Crystallogr Sect D Biol Crystallogr. 2015;71(5):1102–1111. doi: 10.1107/S1399004715003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wu A., Wang Y., Zeng C. Prediction and biochemical analysis of putative cleavage sites of the 3C-like protease of Middle East respiratory syndrome coronavirus. Virus Res. 2015;208:56–65. doi: 10.1016/j.virusres.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lee H., Lei H., Santarsiero B.D. Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV. ACS Chem Biol. 2015;10(6):1456–1465. doi: 10.1021/cb500917m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27(2):157–162. doi: 10.1016/S0006-291X(67)80055-X. [DOI] [PubMed] [Google Scholar]
- 272.Yang H., Yang M., Ding Y. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Jin Z., Du X., Xu Y. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 274.da Silva-Junior E.F., Barcellos Franca P.H., Ribeiro F.F. Molecular docking studies applied to a dataset of cruzain inhibitors. Curr Comput Aided Drug Des. 2017;14(1):68–78. doi: 10.2174/1573409913666170519112758. [DOI] [PubMed] [Google Scholar]
- 275.Shi Y., Zhang X., Mu K. D3Targets-2019-nCoV: a webserver for predicting drug targets and for multi-target and multi-site based virtual screening against COVID-19. Acta Pharm Sin B. 2020 doi: 10.1016/j.apsb.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ratia K, Kilianski A, Baez-Santos YM, Baker SC, Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. Rey FA, ed. PLoS Pathog. 2014;10(5):e1004113. doi:10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed]
- 277.Osipiuk J., Jedrzejczak R., Tesar C. The crystal structure of papain-like protease of SARS CoV-2. Cent Struct Genomics Infect Dis. 2020 doi: 10.2210/pdb6w9c/pdb. [DOI] [Google Scholar]
- 278.Frick D., Lam A. Understanding helicases as a means of virus control. Curr Pharm Des. 2006;12(11):1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hao W, Wojdyla JA, Zhao R, et al. Crystal structure of Middle East respiratory syndrome coronavirus helicase. Fremont DH, ed. PLOS Pathog. 2017;13(6):e1006474. doi:10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed]
- 280.Becares M., Pascual-Iglesias A., Nogales A., Sola I., Enjuanes L., Zuñiga S. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. Perlman S, ed. J Virol. 2016;90(11):5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Ma Y., Wu L., Shaw N. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci. 2015;112(30):9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Srinivasan S., Cui H., Gao Z. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12(4):360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Xue X., Yang H., Shen W. Production of authentic SARS-CoV Mpro with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J Mol Biol. 2007;366(3):965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]