Abstract
Effective treatment choices to the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) are limited because of the absence of effective target-based therapeutics. The main object of the current research was to estimate the antiviral activity of cannabinoids (CBDs) against the human coronavirus SARS-CoV-2. In the presented research work, we performed in silico and in vitro experiments to aid the sighting of lead CBDs for treating the viral infections of SARS-CoV-2. Virtual screening was carried out for interactions between 32 CBDs and the SARS-CoV-2 Mpro enzyme. Afterward, in vitro antiviral activity was carried out of five CBDs molecules against SARS-CoV-2. Interestingly, among them, two CBDs molecules namely Δ9 -tetrahydrocannabinol (IC50 = 10.25 μM) and cannabidiol (IC50 = 7.91 μM) were observed to be more potent antiviral molecules against SARS-CoV-2 compared to the reference drugs lopinavir, chloroquine, and remdesivir (IC50 ranges of 8.16–13.15 μM). These molecules were found to have stable conformations with the active binding pocket of the SARS-CoV-2 Mpro by molecular dynamic simulation and density functional theory. Our findings suggest cannabidiol and Δ9 -tetrahydrocannabinol are possible drugs against human coronavirus that might be used in combination or with other drug molecules to treat COVID-19 patients.
Keywords: Cannabinols, In vitro antiviral assay, SARS-CoV-2 and Mpro enzyme
1. Introduction
The COVID-19 epidemic has caused in a global health emergency [1] and has spread to 213 countries and is uncontrollable so far. Coronaviruses are tiny (diameter, 65 to 125 nm) and their nucleic materials range in length from 2 to 32 kbs, and these viruses are widely distributed in humans and other mammals [[2], [3], [4]]. COVID-19 has an extensive variety of symptoms and signs and can cause respiratory and gastrointestinal symptoms or death. It shares 79% genetic similarity with human SARS-CoV [5,6], and interestingly, all acknowledged sequences of coronavirus share ≥98% similarity with the bat coronavirus RaTG13 [7,8]. Alpha (α), gamma (γ), beta (β), and delta (δ) are used to represent subcategories of the coronavirus family [7]. SARS-CoV was firstly reported in Foshan, China in November 2002, and it was transported to Hong Kong from where it spread worldwide. Later, in April 2012, MERS-CoV occurred in Jordan, causing persistent endemics worldwide. SARS-CoV-2, MERS-CoV, and SARS-CoV infect mainly the lower respiratory tract and produce pneumonia. Besides, human coronaviruses NL63, 229E, OC43, and HKU1 typically are accompanying by minor symptoms involving the infection of the upper tract of the respiratory [9,10]. SARS-CoV-2, MERS-CoV, and SARS-CoV commonly cause lung cell injury because these human coronaviruses have been evolutionally acquiring the capability to encode various proteins which allow them to evade the host immune system. Furthermore, these proteins attack and over-activate more inflammatory and immune cell via hyper-activation and massive section, and induce the cytokine storm, resulting severe injury of infected tissues [11,12].
The SARS-CoV-2 genome encodes ~25 proteins, which are required by the virus to infect humans and replicate. One of them, spike (S) protein initiates virus infection by recognizing and binding with angiotensin-converting enzyme-2 in the human lung cells [[13], [14], [15]]. Two proteases cleave viral and human proteins, and RNA polymerase synthesizes viral RNA and viral RNA-cleaving endoribonuclease. SARS-CoV-2 MPro (main protease) also known as 3CLPro plays a major role in the lifecycle of human SARS-CoV-2 [[16], [17], [18]]. SARS-CoV-2 Mpro and papain-like protease process translated viral RNA polyproteins. The molecular mass of Mpro is 33,797 Da. SARS-CoV-2 Mpro recognizes and acts remarkably at fewer than 11 cleavage sites of Leu-Gln↓ (Ser, Ala, Gly) of the polyprotein replicase 1ab. The activated Mpro is a homodimer containing two proteomes and a noncanonical Cys-His dyad situated in the cleft between the domain I and II. Mpro is preserved among coronavirus and the substates of Mpro in various coronavirus have several mutual features. SARS-CoV-2 Mpro generates 12 non-structural proteins (Nsp4 and Nsp16) through the cleaves of the viral polyproteins, including RNA-dependent RNA polymerase (RdRp, Nsp12), and the helicase (Nsp13) [19]. Therefore, SARS-CoV-2 Mpro is viewed as the best molecular target to block coronavirus replication [[19], [20], [21]]. Because comparable cleavage specificity has not been yet reported for human proteases, the inhibition of SARS-CoV-2 Mpro is unlikely to cause any toxic effect to humans. The SARS-CoV-2 Mpro 3D cystography X-ray structure has recently been resolved (PDB ID: 6Y2F [19]), and this provides a means of discovering its inhibitors [19,22].
Currently, it was observed that inflammatory molecule levels (e.g. C reactive protein and pro-inflammatory cytokines, IL-1β, IL-6, IL-7, IL-8, IL-9, IL-10, fibroblast growth factor, IFN, granulocyte-colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, tumor necrosis factor, macrophage inflammatory protein 1 α, and vascular endothelial growth factor) are elevated in the cells of the lung of the COVID-19 patients. The presence of these proinflammatory cytokines in the lung is linked with acute respiratory distress, severe injury, and death [23,24].
Viral replication is accompanying by immune activation and inflammation in the host, and it has been well documented that immune cells (e.g. B cells, CD8 lymphocytes, natural killer cells, monocytes, CD4 lymphocytes, and neutrophils) highly express the levels of cannabinoid receptor type 2 (CB-2) in lung, liver, nasal epithelium, spleen, thymus, kidney, and brain [25]. CB-2 is best known for its immunosuppressive and apoptotic effects, increasing anti-inflammatory cytokine levels, reducing pro-inflammatory cytokine production, and encouraging regulatory expression of T cells [26,27]. Furthermore, the CB-2 receptor inhibits inflammatory processes and macrophage migration [28] and offers a healing drug target for reducing immune-associated pathological processes in viral associated infections [25]. Evidence indicates CB-2 receptors influence immunomodulatory progression, and coagulative, inflammatory, and the levels of cytokine in the lung. Also, the role of CB-2 against the inflammation had been studied by Mechoulam et al. that CBDs act as an inverse agonist at the CB2 receptor, partially explaining its anti-inflammatory properties [29,30]. Another similar study was reported by Jieting et al. which revealed the anti-inflammatory role of the CB-2 receptor in the lung to reduce the ischemic-reperfusion injury in mice. Moreover, proinflammatory cytokines are elevated in COVID-19 patients, which suggests that CB-2 agonists maybe play important role in the COVID-19 infected patient to regulate the inflammation in lung cells. Also, the well-known role of SARS-CoV-2 Mpro in the progression of viral development, the antagonist against it may stop the role in the progression of viral. Thus, CB-2 and SARS-CoV-2 Mpro can be considered therapeutic targets for the SARS-CoV-2 infected patient.
Based on the effectiveness's of CBDs to reduce inflammation by acting as agonists of CB-2 receptor [31,32], and the importance of SARS-CoV-2 Mpro in the SARS-CoV-2 lifecycle to the translation of viral RNA [33,34], we anticipated that CBDs might be useful for treating COVID-19 patients by inhibiting SARS-CoV-2 Mpro as well as binding with CB-2 receptor as an agonist to reduce inflammation and lung injury.
In the present research, we screened 32 CBDs and evaluated their binding affinities with SARS-CoV-2 Mpro using AUTODOCK and VINA. Lead molecules were estimated for their conformational stability, ligand reactivity, and stability with SARS-CoV-2 Mpro. Five CBDs, namely, Δ9-tetrahydrocannabinolic acid (Δ9-THCA), Δ9-tetrahydrocannabinol (Δ9-THC), cannabinol (CBN), cannabidiol acid (CBDA), and cannabidiol (CBD) (Table 1 ) were subjected to further in vitro antiviral assays to predict the abilities of these five CBDs to inhibit human SARS-CoV-2. While the possible therapeutic use of CBDs for COVID-19 patients has been proposed recently [35], it is the first report that provides actual targets and possible mechanisms of CBDs against the SARS-CoV-2. Privileged safety and current in vitro antiviral results of CBDs collectively supported that CBD and Δ9-THC might work as dual-acting for treating the human coronavirus infections.
Table 1.
Molecular binding affinities of CBDs with SARS-CoV-2 Mpro, where common residues of the amino acid are indicated by bold font. Δ9-THCA, Δ9-THC, CBN, CBD, and CBDA are the tested compounds and α-ketoamide 13b (PC) is a positive control.
2. Materials and methods
2.1. SARS-CoV-2 Mpro protein and ligands collection
SARS-CoV-2 Mpro protein crystal structure (PDB: 6LU7) has been retrieved from RCSB protein database (https://www.rcsb.org/) at a resolution of 2.16 Å [36]. The binding active pocket was predicted by the CASTp server (http://sts.bioe.uic.edu/) for computational based virtual screening and compared to previous studies [36]. The protein preparation wizards' approach has been used to prepare and refine the targeted protein by Schrodinger suite. Excess co-crystallizing water molecules were deleted, and the required hydrogens were added to the complex structure of the protein. The 2D structures of 32 cannabinoids were downloaded in a simulation description format (SDF) from PubChem (https://pubchem.ncbi.nlm.nih.gov/).
2.2. Preparation of ligands and structure-based virtual molecule screening
CBDs were prepared using LigPrep tools in Schrodinger's suite and optimized for minimum energy using a density functional theory (DFT) approach [37,38]. Bond orders and conformations were refined and minimized using OPLS 2005 force field [39]. These prepared ligand libraries were exposed to structure-based virtual screening with the CASTp server predicted active binding pocket of SARS-CoV-2 Mpro [40]. Also, the CASTp server predicted active binding site of SARS-CoV-2 Mpro was again re-confirmed by VINA random 25 runs between CBDs and SARS-CoV-2 Mpro (Fig. S1). To the virtual screening, drug molecules were treated as rigid entities, while the receptor was treated as a flexible entity [41,42]. To ensure result reliability, validity, and reproducibility, molecular docking was performed by two different docking methods, namely AUTODOCK [43] and VINA [44]. Also, binding energies and interactions between CBDs and SARS-CoV-2 Mpro were estimated by a computational approach [43]. To visualize interactions CBDs and the protein, BIOVIA Discovery Studio Visualizer was used.
2.3. Molecular dynamic (MD) simulation and energy calculation
For access the docking conformations complexes stabilities of the five CBDs, Δ9-THCA, Δ9-THC, CBN, CBD, and CBDA with SARS-CoV-2 Mpro, molecular dynamic simulations were carried out in explicit water solution using YASARA dynamic software [45] using a SAMSUNG Intel(R) Core (TM) i5-CPU, 4GB RAM system, Windows 7 enterprise version (64). Best docking binding conformations of CBDs complexes were subjected to evaluate their binding confirmation stabilities with SARS-CoV-2 Mpro. The simulation cells of complexes were defined using periodic cell boundaries and filled with an explicit water solvent at 0.997 g/L (density). A periodic simulation cell boundary size of X = 61.30 A°, Y = 82.84 A°, Z = 52.14 A° was built around the entire system. Chloride and sodium ions were arbitrarily placed to achieve charge neutrality. Pka values were only predicted for the sidechains of Asp, His, Glu, and Lys residues [46]. The AMBER14 molecular dynamic force field was selected for MD simulation under physiological conditions 298 K, 0.9% NaCl, and pH 7.4 [47]. System energies were minimized initially by steepest descent minimization and this was followed as previously described method [48]. More than 20 ns run molecular dynamic simulations were performed at constant temperature and pressure and MD trajectories were saved for 250 ps for further analysis. These MD trajectories were subjected to analysis by the YASARA template file (“md_analysis.mcr”). Binding energy conformations of complexes were analyzed using another YASARA template file (“md_analyzebindenergy.mcr”) [49]. Average conformation stabilities of models were estimated from simulations and root means square deviations (RMSDs). As per the YASARA manual, free binding energies were calculated without the involvement of the entropy term, as follows. YASARA provides positive binding energies, and therefore, more positive energies indicate favorable binding with a receptor in the selected force field, while negative binding energies indicate weak binding. Trajectory analysis data obtained by MD simulation were represented graphically using SigmaPlot 10.0. CBD/SARS-CoV-2 Mpro binding conformations were visualized using Discovery Studio visualization software.
2.4. Structural stabilities of CBDs as determined by DFT
Frontier molecular orbitals (FMOs) namely lowest unoccupied molecular orbitals (LUMOs), and highest occupied molecular orbitals (HOMOs) of CBDs were analyzed by density functional theory (DFT). The LUMOs and HOMOs of molecules Δ9-THCA, Δ9-THC, CBN, CBD, and CBDA were determined. HOMO-LUMO energy gaps for CBD molecules were calculated by the given equation;
| (1) |
Chemical potentials (μ) and chemical hardnesses (η) were calculated using the energies associated with HOMOs and LUMOs.
| (2) |
| (3) |
Electronegativity (χ) and electrophilicity (ω) were calculated using ionization potentials (I), which are generally defined as negative EHOMO values, and electron affinities (A) was defined to be equal to negative ELUMO values.
| (4) |
2.5. Extraction and identification of CBDs extraction
Preparative HPLC was performed on a LC-Forte/R (YMC, Japan) equipped with an ultra-violet (UV) detector (220 nm) using a Phenomenex Luna C18 column (250 × 21.2 mm, 10 μm), whereas semi-preparative LC was conducted system using (Gilson, USA) a refractive index (RI) detector and a Phenomenex Luna C18(2) column (250 × 10 mm, 5 μm). NMR spectra were recorded on a Varian Superconducting FT-NMR System (500 and 125 MHz for 1H and 13C, respectively) in chloroform-d. Chemical shifts of proton and carbon spectra were obtained in chloroform-d were reported, peaks are reported residual solvent peaks at 7.26 ppm and 77.0 ppm, respectively. Ultra-high-performance liquid chromatography (UPLC) ESI mass spectrometry was carried out on a Shimadzu LCMS-2020 system. Chongsam (Korean hemp, Cannabis sativa L.) was collected from the association (Andong city, Gyeongsangbuk-do, South Korea) in accord with assignment/transfer approval processes (approval No. 1564) stipulated by the Korean Ministry of Food and Drug Safety and the Seoul Regional Food and Drug Administration.
Chongsam leaves were harvested in July 2019, naturally dried, finely cut, and 10 g was extracted twice with ethanol (200 mL) at the temperature of the room and filtered. The ethanolic extract (1.64 g) was then suspended in water and afterward successively partitioned versus normal hexane, which yielded 720 mg of residue. Silica open column chromatography (Merck, 230–400 mesh, 2.0 × 10.0 cm ID) was carried out by using a hexane: ethyl acetate (F1–10:0, F2–25:1, F3–10:1, and F4–0:10; each 200 mL) stepwise gradient. The F2 (187 mg) fraction was subjected to preparative HPLC (Phenomenex Luna C18 column; 250 × 21.2 mm, 10 μm) eluted using a water (A) and MeCN (B) gradient system (70 to 85% MeCN over 60 min) at 10 mL/min flow rate using a 220 nm UV detector to yield four subfractions (a–d). Further purification of each sub-fraction was performed by semi-preparative HPLC (Phenomenex Luna C18 (2); 250 × 10 mm, 5 μm) using 70 to 85% MeCN as an eluant at 4 mL/min flow rate to afford pure compounds Δ9-THCA (17.0 mg), Δ9-THC (48 mg), CBN (1.1 mg), and CBD (1.9 mg) (Fig. S3). Fraction F3 (35 mg) was subjected to preparative HPLC (Phenomenex Luna C18 column; 250 × 21.2 mm, 10 μm) and gradient eluted with water (A) and MeCN (B) at 65 to 80% MeCN over 60 min at 10 mL/min flow rate using a 220 nm UV detector to yield one sub-fraction (e). This was purified by semi-preparative HPLC (Phenomenex Luna C18 (2); 250 × 10 mm, 5 μm) using a 65 to 80% MeCN gradient at a flow rate of 4 mL/min to afford pure CBDA (7.9 mg). The five isolated molecules were analyzed by comparing NMR spectra with those previously reported (supplementary) [50,51].
2.5.1. Analysis of cannabinoids
An analytical reversed-phase Shimadzu Nexera X2 UHPLC system comprised of a solvent degassing unit (DGU–20A), binary pump (LC–30AD), autosampler (SIL–30AC), system controller unit (CBM–20A), photodiode array detector (SPD–M20A), and column oven unit (CTO–20AC) was used for qualitative and or quantitative analysis. Electrospray ionization (ESI)-mass spectrometry (MS) (Shimadzu LCMS-2020 system) was used for qualitative analysis. A Phenomenex Luna Omega polar C18 column (150 × 2.1 mm, 1.6 μm) was used. The mobile phase contained the binary gradient of solvent A (water) and solvent B (MeCN), as follows, initially; 70% B, 10.0 min; 85% B, 11.0 min; 95% B, and 15.0 min; 70% B. The flow rate was established to 0.3 mL/min and 220 nm detection wavelength was used.
2.6. In vitro estimations of the antiviral potencies of CBDs against SARS-CoV-2
The antiviral potencies of CBDs were investigated as per the previously described method [52]. The images of confocal microscopy of viral N protein and cell nuclei were analyzed using image mining (IM) software. A dose-response curve (DRC) was made for individual compounds. From the American Type Culture Collection (ATCC CCL-81), Vero cells were obtained for in vitro drug screening assays. Cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM; Welgene) containing, 10% heat-inactivated fetal bovine serum (FBS) and 1× antibiotic-antimycotic solution (Gibco) in a 5% CO2 atmosphere at 37 °C. Virus SARS-CoV-2 (βCoV/KOR/KCDC03/2020) was given by the Korea Centers for Disease Control and Prevention (KCDC), and the virus was propagated in Vero cells. The viral titers were estimated by subjecting Vero cells to plaque assays. Institute Pasteur Korea supported this research work, which was carried in accord with rules issued through the Korea National Institute of Health (KNIH), South Korea, using level-3 of biosafety control procedures in laboratories approved for use by the KCDC.
2.6.1. Drug and reagents
Isolated CBDs were used, and lopinavir (LPV; S1380) was purchased from SelleckChem and remdesivir (HY- 104077) from MedChemExpress, and chloroquine was obtained from Sigma-Aldrich. Chloroquine was mixed in Dulbecco's Phosphate-Buffered Saline (DPBS; Welgene). For in vitro studies, all reagents were well mixed in DMSO. The Anti-SARS-CoV-2 N protein antibody was obtained from Sino Biological Inc. (China), while Hoechst-33342 and Alexa Fluor-488 goat anti-rabbit IgG (H + L) secondary antibody were bought from the Molecular Probes. Also, paraformaldehyde (PFA) (32% aqueous solution) and normal goat serum were purchased from Vector Laboratories, Inc. (Burlingame, CA) and Electron Microscopy Sciences (Hatfield, PA), respectively.
2.6.2. Dose-response curve analysis as estimated by immunofluorescence
DRC analysis was carried out as previously described protocol[52]. Briefly, Vero cells were sown (1.2 × 104 cells/well) in DMEM containing 2% FBS in black and 1× Antibiotic-Antimycotic solution (Gibco), 384-well, μClear plates (Greiner Bio-One), and allowed to stand for 24 h, then, cells were treated with compounds at the conc. ranging 0.05–100 μM, in duplicate. Nine-point DRCs were generated. For viral infection, plates were moved into the BSL-3 containment facility and SARS-CoV-2 was added at MOI of 0.0125. Later, cells were fixed at 24 hpi with 4% PFA and examined using an immunofluorescence Operetta (Perkin Elmer). Images acquired using in-house software was used to estimate infection ratios and cell numbers, and antiviral activity was normalized versus negative (0.5% DMSO) and positive (mock) controls. DRCs were fitted using the following sigmoidal dose-response model using: Y = Bottom + (Top − Bottom) / (1 + (IC50 / X)Hillslope), using Prism or 7XLfit 4 Software. Finally, IC50 values were analyzed from the normalized activity dataset-fitted curves. IC50 and CC50 values were also analyzed in duplicate, and the quality of each assay was verified using the Z'-factors and coefficients of variation.
3. Results
3.1. Molecular docking of CBDs with SARS-CoV-2 Mpro and mapping of their molecular interactions
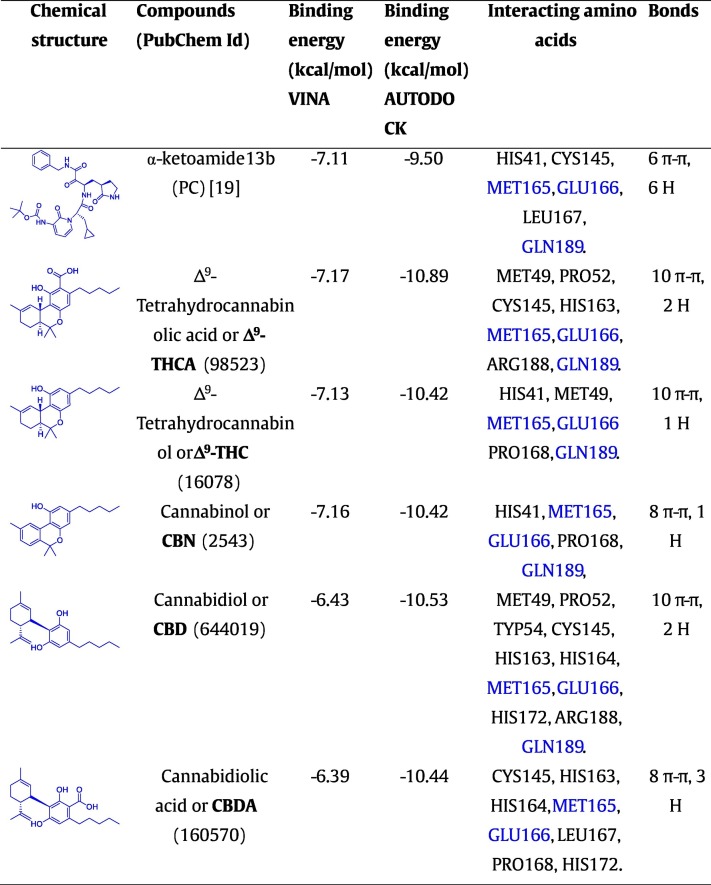
To estimate and explore the interactions between CBDs and SARS-CoV-2 Mpro, we carried out the virtual screening of 32 known CBDs (Table S1). The predicted active pocket of SARS-CoV-2 Mpro was identified by the CASTp server which was similar to the privileged reported binding pocket [19]. As a result, SARS-CoV-2 Mpro (domain-I) was assigned as the binding active pocket for CBDs docking (Fig. S1), which is similar to earlier studies of α-ketoamide 13b binding [19]. All CBDs were estimated for their binding with SARS-CoV-2 Mpro, showing the binding affinities range between −5.67 to −7.62, kcal/mol (VINA) (Table S1). Redocking of the 32 CBDs was carried out by AUTODOCK, and all 32 showed binding energies between −8.40 and −10.89 kcal/mol (Table S1). Based on molecular docking confirmation stabilities, and interaction with residues of SARS-CoV-2 Mpro, five CBDs namely Δ9-THCA, Δ9-THC, CBN, CBD, and CBDA showed stable conformation and remarkable binding interactions with SARS-CoV-2 Mpro (Figs. 1 and S2) where Δ9-THCA, CBN, and CBDA had binding affinities of −10.89, −10.42, and −10.44 kcal/mol, respectively, while Δ9-THC and CBD showed molecular binding energies of −10.42 and −10.53 kcal/mol, respectively, compared to positive control (α-ketoamide 13b) binding energy of ˗9.50 kcal/mol in AUTODOCK. The interaction between Δ9-THC and SARS-CoV-2 Mpro involved one hydrogen bond and ten π-π bonds interaction residues (Fig. 1). CBD-SARS-CoV-2 Mpro complex showed two hydrogen and ten π-π interactions with various amino acid residues, while Δ9-THCA, CBN, and CBDA showed the two, one, three hydrogen bindings and ten, eight, and eight π-π interactions with amino acid residues, respectively. Importantly, it was observed that all five hits (Δ9-THCA, Δ9-THC, CBN, CBD and, CBDA) and the positive control (α-ketoamide 13b) that bound to SARS-CoV-2 Mpro interacted with similar common amino acids as GLN189, MET165, and GLU166, (Tables 1 and S1) therefore, it can be suggested that these common interacting residues may be required for binding with the active pocket of SARS-CoV-2 Mpro to produce the inhibitory effect.
Fig. 1.
3D conformations of ligand-protein interaction of Δ9-THCA, Δ9-THC, CBN, CBD, CBDA, and α-ketoamide 13b (positive control) with SARS-CoV-2 Mpro. Dotted lines of green colour show hydrogen bonds between the SARS-CoV-2 Mpro residues and ligands.
3.2. Absorption, distribution, metabolism, and excretion (ADME) profiles of CBDs
All optimized CBDs were subjected to analysis of their ADME profiles using Schrodinger software. Accordingly, Lipinski's rule, all CBDs had an acceptable range of molecular weights, Lipinski's violation, n-OHNH donors, n-OH acceptors, and n-ROTB (Table S2), indicating these CBDs are likely to exhibit good absorption profiles and pharmacological effects.
3.3. Stability confirmation of CBDs estimated through density functional theory (DFT)
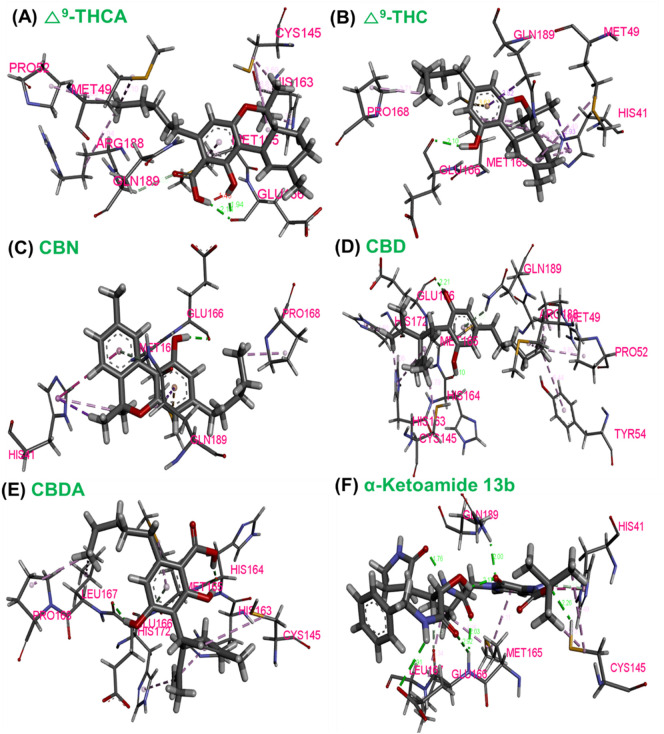
DFT approach was applied to calculate the energy gaps between HOMO and LUMO for the five CDBs. HOMO orbitals were located at 2, 2-dimethylchroman-5-ol and methyl-1′, 2′, 3′, 4′-tetrahydro-[1,1′-biphenyl]-2, 6-diol ring motifs of Δ9-THC, and CBD, respectively (Fig. 2 ). The energy gap was observed to be slightly high for compounds Δ9-THC, and CBD than CBDA. However, the LUMOs of these CBDs were detected at the 3-propyl-6a, 7, 8, 10a-tetrahydro-6H-benzo[c]chromen-1-ol and (S)-4, 5′-dimethyl-1′, 2′, 3′, 4′-tetrahydro-[1, 1′-biphenyl]-2, 6 -diol motif of Δ9-THC, and CBD, respectively (Fig. 2). While HOMO orbitals of Δ9-THCA, CBN, and CBDA were located at chromene with the carboxylic acid group, chromene fused cyclic ring with an aliphatic side chain and, dihydroxyl benzoic acid motif, respectively. It is well documented that a molecule with a small frontier energy orbital gap is chemically reactive, highly polarized, and has low kinetic stability, while a higher HOMO to LUMO gap between frontier orbitals confers stability (Table 2 ) [53]. Compounds Δ9-THC and CBD had a slightly higher HOMO to LUMO energy gap than Δ9-THCA, CBN, and CBDA, which indicated that Δ9-THC and CBD have a good stability profile. Chemical hardness (η) is defined as resistance to distortion of the electron cloud during chemical processes and represents compound stability [54]. Interestingly the η values of Δ9-THC and CBD were higher than Δ9-THCA, CBN, and CBDA, showing the good stability of Δ9-THC and CBD.
Fig. 2.
Molecular geometries with charges and atomic numbers for A. Δ9-THCA, C.Δ9-THC, E. CBN, G. CBD, and I. CBDA. Frontier molecular orbitals used to explore the HOMO-LUMO of optimized B. Δ9-THCA, D. Δ9-THC, F. CBN, H. CBD, and J. CBDA were calculated using the DFT approach of Gaussian program package.
Table 2.
DFT calculation results of CBDs for quantum chemical parameters.
| Quantum chemical parameters (eV unit) | Δ9-THCA | Δ9-THC | CBN | CBD | CBDA |
|---|---|---|---|---|---|
| HOMO | −6.07 | −5.74 | −5.81 | −5.91 | −6.16 |
| LUMO | −0.78 | −0.10 | −0.95 | −0.11 | −0.91 |
| Energy gap (ΔE) | 5.29 | 5.63 | 4.85 | 5.80 | 5.25 |
| Chemical potential (μ) | −3.42 | −2.92 | −3.38 | −3.01 | −3.53 |
| Global hardness (η) | 2.64 | 2.82 | 2.42 | 2.90 | 2.62 |
| Ionization potential (I) | 6.07 | 5.74 | 5.81 | 5.91 | 6.16 |
| Electrophilicity (ω) | 2.21 | 1.51 | 2.37 | 1.56 | 2.37 |
3.4. Estimation of in vitro antiviral effects of CBDs on SARS-CoV-2
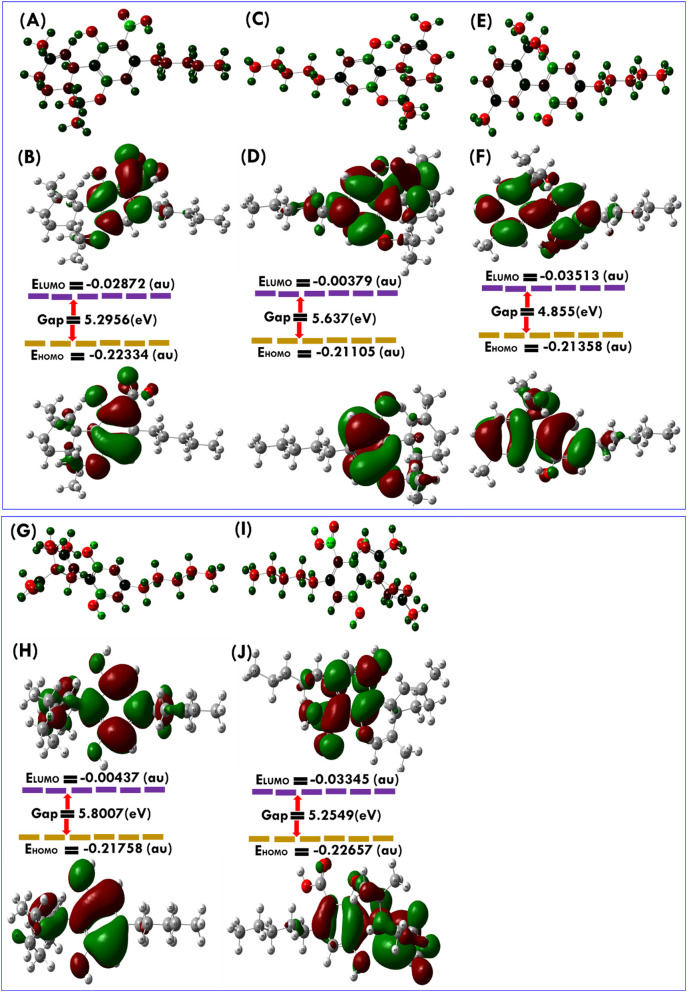
Based on our virtual screening results with SARS-CoV-2, five CBDs were assigned to be assessed in vitro antiviral assay against the SARS-CoV-2. These five CBDs, namely for Δ9-THCA, Δ9-THC, CBN, CBD, and CBDA (Table 1) were successfully isolated and purified (Fig. S3) and their in vitro antiviral activities were estimated using Vero cells as previously reported [52]. Using in-house Image Mining software, the confocal microscope images of viral N protein and cell nuclei were evaluated, and DRC was produced for individual CBDs (Fig. 3 ). In this experiment, chloroquine, ramdesivir, and lopinavir were used as standard drugs; they have IC50 of 9.78, 8.17, and 13.16 μM, respectively, which is matched with previously reported studies [52]. Of the five CBDs, Δ9-THC, and CBD showed potent antiviral activity against SARS-CoV-2 with IC50 of 10.25 and 7.91 μM, respectively, while for Δ9-THCA, CBN, and CBDA showed moderate antiviral activity with IC50 of 13.17, 11.07, and 37.61 μM, respectively. The most active compound CBD exhibited higher inhibition of SARS-CoV-2 virus at concentration 12.50 μM (99.19%) and a Vero cell survival ratio was 97.46% (Fig. 3). Also, it was noticed that the host cell survival ratio suddenly decreased at a concentration of 50 μM. Therefore, 12.50 μM of CBD was deemed safe for inhibiting SARS-CoV-2 without causing any cytotoxicity, in vitro. A similar concentration-dependent inhibition pattern was also observed for Δ9-THCA.
Fig. 3.
Dose-response curve analysis of three control drugs, lopinavir, chloroquine, and remdesivir, and the five CBDs, Δ9-THCA, Δ9-THC, CBN, CBD, and CBDA. The blue circles represent the inhibitory concentrations against SARS-CoV-2 infection (%) and the red squares represent Vero cell viability (%). Means ± SD was calculated from the results of duplicate experiments.
3.5. Conformation of stability of CBDs-SARS-CoV-2 Mpro complexes by molecular dynamic (MD) simulation
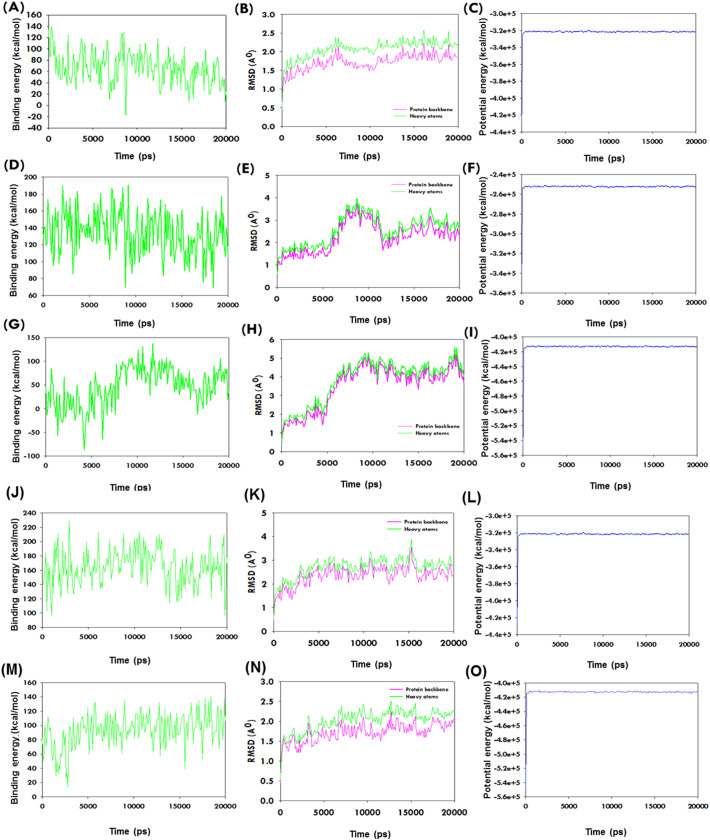
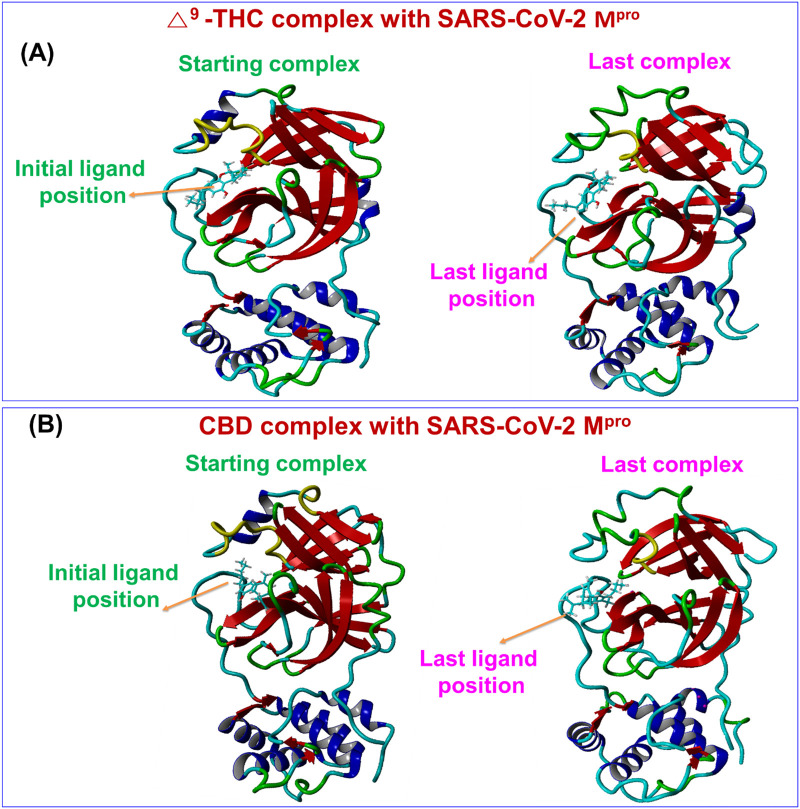
The motions and conformation stabilities of CBDs-SARS-CoV-2 Mpro complexes were further evaluated under solution conditions by MD simulation. Complexes with high binding energy were allowed to the estimation of amino acid interactions over time. Generated trajectories were analyzed to determine ligand binding energies, root-mean-square deviation (RMSD), and total potential energies of ligand-receptor complexes over time 20 ns. The average binding energies of the Δ9-THC and CBD were 164.73 and 93.10 kcal/mol, respectively. These positive binding energies as per the assigned forcefield suggested that CBDs bind strongly with SARS-CoV-2 Mpro throughout MD runs. Δ9-THC-SARS-CoV-2 Mpro and CBD-SARS-CoV-2 Mpro complexes exhibited the equilibrium of trajectories after 1500 and 500 ps respectively, indicated complex stability during a 20 ns MD run. Interestingly, the RMSD of the complex of SARS-CoV-2 Mpro with Δ9-THC, and CBD, respectively did not show additional fluctuation during MD simulation runs (Fig. 4 ). The stability profiles of SARS-CoV-2 Mpro with Δ9-THCA, CBN, and CBDA complexes are represented in Fig. 4 in MD simulation, where CBN showed the stability at 1300 ps and Δ9-THCA showed at 1100 ps (Fig. 4). Besides, compound CBDA exhibited slightly higher fluctuation in RMSD of backbone and heavy atoms. These observations showed CBDA-SARS-CoV-2 Mpro complex is slightly less stable than Δ9-THC-SARS-CoV-2 Mpro and CBD-SARS-CoV-2 Mpro complexes. The initial and last poses of Δ9-THC and CBD to the SARS-CoV-2 Mpro binding pocket did not show any conformational changes in the complex structures over 20 ns (Fig. 5 ). The potential energies of Δ9-THC-SARS-CoV-2 Mpro and CBD-SARS-CoV-2 Mpro complexes exhibited a linear trajectory. Hence, RMSDs and binding energies confirmed the stabilities of complexes Δ9-THC, and CBD with SARS-CoV-2 Mpro during the 20 ns MD simulation.
Fig. 4.
The plots show the stability of the interaction between the CBDs and SARS-CoV-2 Mpro complexes in MD simulation over time. (A-C) Binding energy profile, RMSD, and potential energy of Δ9-THCA-SARS-CoV-2 Mpro complex estimation during MD simulation as determined using YASARA. (D-F) Binding energy profile, RMSD, and potential energy of CBN-SARS-CoV-2 Mpro during MD simulation as determined using YASARA. (G-I) Binding energy profile, RMSD, and potential energy of CBDA- SARS-CoV-2 Mpro complex during MD simulation as determined using YASARA. (J-L) Binding energy profile, RMSD, and potential energy of Δ9-THC-SARS-CoV-2 Mpro complex during MD simulation as described using YASARA. (M-O) Binding energy profile, RMSD, and potential energy of CBD-SARS-CoV-2 Mpro complex during MD simulation as determined using YASARA.
Fig. 5.
The comparative binding position of CBDs-SARS-CoV-2 Mpro complex before and after a long run of MD simulations for (A) Δ9-THC, and (B) CBD.
4. Discussion
This study represents a preliminary effort to discover dual-acting phytochemicals capable of interacting with SARS-CoV-2 Mpro as an antagonist and CB-2 receptors in the lungs (agonist). SARS-CoV-2 Mpro is known to play a significant role in the viral replication of SARS-CoV-2, but as yet, no effective target-based vaccine or drug has been developed against COVID-19. Herein, we performed structured based screening of diverse CBDs because they directly or indirectly activate CB-2 receptor and attenuate the properties of proinflammatory cytokines. We screened 32 CBDs molecules for estimation of their interaction with the validated binding active site of SARS-CoV-2 Mpro. From the point of view of availability and cost of molecules, virtual screening has been carried out to find a smaller number of good hit molecules and reduce the risk of the matter of luck and time. Also, CBDs are the licensed product to use experimentally and these are not readily available.
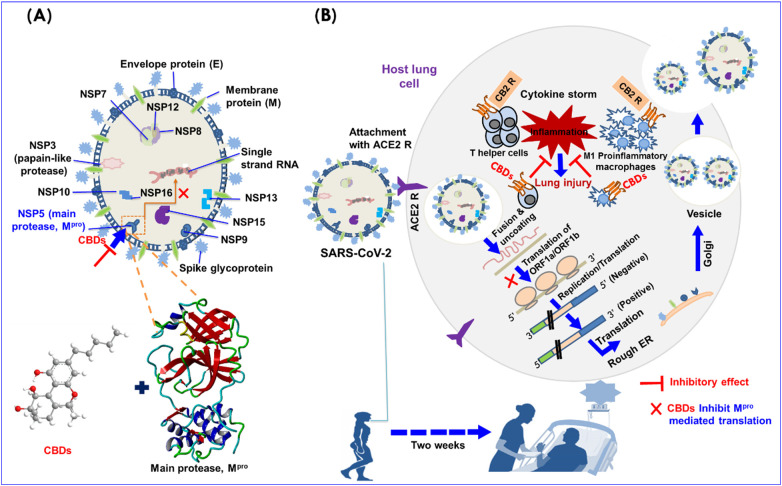
As collectively observation indicates that molecular docking, DFT, and MD simulation showed stable binding of CBDs with SARS-CoV-2 Mpro. Based on our computational study (Fig. 1) and in vitro antiviral potencies (Fig. 3) of CBD, and Δ9-THC against SARS-CoV-2 suggested that these both molecules might inhibit SARS-CoV-2 in two ways, i.e. they may bind to and inhibit SARS-CoV-2 Mpro, leads to blocking the translation procedure as well as reduce pro-inflammatory cytokines levels in lung cells by acting as agonists of CB-2 receptor (Fig. 6 ).
Fig. 6.
A. A plausible mechanism of CBDs as may act dual-acting to inhibit SARS-CoV-2: structure features of SARS-CoV-2 and its main SARS-CoV-2 Mpro binding pocket, B. SARS-CoV-2 life cycle in host lung cells is initiated by binding between viral spike glycoprotein and ACE2 cellular receptor. Spike glycoprotein facilitates viral envelope/cell fusion through the endosomal pathways, consequently, release RNA of SARS-CoV-2 and translate the RNA genome of viral by viral polyproteins replicase of 1ab and pp1a and their subsequent cleavage by the viral proteinases into small products. A sequence of subgenomic mRNAs is produced by SARS-CoV-2 Mpro, which are next translated into similar viral proteins. Further, genome RNA and viral proteins accumulate into virions in the ER and Golgi, and SARS-CoV-2 is transported in vesicles to the extracellular compartment. During this process, T-helper cells and M1 pro-inflammatory macrophages secrete interleukins and induce inflammation inside lung cells.
The structural features of SARS-CoV-2 indicate that several proteins likely spike glycoprotein (S), chymotrypsin-like main protease, SARS-CoV-2 Mpro , papain-like protease, and RNA polymerase play essential roles in the progress of SARS-CoV-2 [33]. The SARS-CoV-2 life cycle in host lung cells is initiated by binding between the S-protein of SARS-CoV-2 and ACE2 (cellular receptor) [[55], [56], [57]]. S-protein facilitates viral wrapping fusion with host cells, through an endosomal pathway, which results in the viral cell release the RNA of SARS-CoV-2 into the host cell and translates the viral genome RNA into replicase polyproteins 1ab and pp1a, are later cleaved by proteinases into small products (Fig. 6) [3,58]. SARS-CoV-2 Mpro and papain-like protease are essential for the processing of polyproteins [19]. Later, sub-genomic mRNA is formed by polymerase [59]. In this pathway, T-helper cells and M1 pro-inflammatory macrophages secrete interleukins, which cause inflammation inside lung cells [60,61]. However, the binding affinities, complex stabilities, and in vitro potencies of CBDs suggested that these CBDs inhibit SARS-CoV-2 Mpro activity and lead to blocking viral replication. Importantly, no human similar protease to SARS-CoV-2 Mpro has reported been yet, and such inhibitors (inhibitor for SARS-CoV-2 Mpro) are improbable to be toxic [19]. On the other side, CBDs trigger the stimulation of CB-2 receptor in the lung, and induce immunosuppression, apoptosis, increase anti-inflammatory cytokine levels and inhibit pro-inflammatory cytokine production and the inductions of regulatory T-cells [62].
The presence of hydrogens and diverse π-π bonds and other intramolecular interactions between the residues of amino acid of SARS-CoV-2 Mpro (Fig. 1) and the five selected CBDs suggested better conformational complex stability among all CBDs. Importantly, binding of the five CBDs, Δ9-THCA, Δ9-THC, CBN, CBD, and CBDA with the binding active site of SARS-CoV-2 Mpro had good docking scores compared to reference compound α-ketoamide with similar binding pocket of SARS-CoV-2 Mpro (Tables 1 and S1). Also, Δ9-THC and CBD showed a slightly higher HOMO-LUMO energy gap in DFT calculations, which supported that these molecules are stable with SARS-CoV-2 Mpro compared to remaining CBDs (Fig. 2). In MD simulations, Δ9-THC- or CBD-SARS-CoV-2 Mpro complexes showed better conformation stability than Δ9-THCA, CBN, and CBDA complexes (Fig. 4, Fig. 5). The above-obtained results suggested Δ9-THC or CBD bind to SARS-CoV-2 Mpro with stable conformations. For further confirmation of protein binding and affinity, the surface plasmon resonance technique or other protein binding assay is required in the next phase of the study of the research.
Moreover, the significant in vitro antiviral results against SARS-CoV-2 of Δ9-THC, and CBD compared to Δ9-THCA, CBN, and CBDA suggested their higher potency to inhibit SARS-CoV-2, but higher doses of Δ9-THC or CBD, caused cytotoxicity in the host cell, which attracts the attention regarding safety issues for human. But based on privileged safety profiles of Δ9-THC, and CBD for humans [63,64], it has been proposed subtherapeutic dosing of CBD at 0.3 mg/kg/day is effective in Crohn's disease. In humans, CBD has been tested over a wide dose range from <1 to 50 mg/kg/day [63,64]. Moreover, Δ9-THC acts as a partial agonist of CB1R and CBR2 and reportedly induces immunological and anti-inflammatory effects via activation of CB2R [65,66]. Δ9-THC is a well-known psychoactive cannabinoid and high doses of Δ9-THC cause side effects such as depolarization, paranoid delusions, the flight of ideas, disorganized thinking, and visual hallucination [67]. Also, Δ9-THC increases the anxiety when used alone, particularly at high doses under conditions of stress, while co-administration with CBD can counteract the Δ9-THC induced anxiety [66]. Thus, the safe dose of Δ9-THC for further treatment of SARS-CoV-2 infected patient is a major concern for the safety of patients.
CBD controls the stimulation of the immune not only in the human immunodeficiency virus but also in post-Ebola syndrome when administered orally at 10–20 mg/kg and 1.7–10 mg/kg daily, respectively [68]. Notably, the Therapeutic Goods Administration of the Australian Department of Health recently (April 2020) [69] recommended that low dose CBD can be administered orally or by inhaler or vaporization for the treatment of insomnia and anxiety secondary to post-traumatic stress disorder at 25–40 mg/day for a 10-year-old child and 50 to 75 mg/day for adults [70,71]. CBD has been also administered at 1 mg/kg/day for the management of local and systemic chronic pain [72,73]. Thus, it appears CBD is safe at up to 60 mg per day. Furthermore, it has been described that a single oral dose of CBD at 100 mg, or a combination of 10.8 mg THC and 10 mg CBD was safe and tolerable in the healthy volunteers [74,75]. Based on these above privileged preclinical studies on these two cannabinoids (CBD and Δ9-THC) for their safety profiles for the management of various diseases and our encouraging in vitro antiviral potencies of CBD and Δ9-THC (Fig. 3) against SARS-CoV-2, it can be suggested that CBD and Δ9-THC may attract additional research communities to estimate their significance clinically for the treatment of human coronavirus affected patients.
5. Conclusion
In summary, this report demonstrates the antiviral potencies of CBD and Δ9-THC against SARS-CoV-2. Based on privileged safety index CBD and Δ9-THC in human and their current in vitro potencies against SARS-CoV-2, it can be concluded that these compounds are potential antiviral molecules towards SARS-CoV-2 and may have worked as dual-acting against SARS-CoV-2, not only block the viral translation procedure by inhibiting SARS-CoV-2 Mpro but also reduce pro-inflammatory cytokines levels in lung cells by acting as agonists of CB-2 receptor. The successful in vitro work here of CBD and Δ9-THC lays the framework for their application in human clinical trials for the treatment of human coronavirus infections. Thus, CBD and Δ9-THC may be used in combination or with other drugs to treat COVID-19 patients.
Funding
This research work was supported by the Korea Institute of Science and Technology (KIST) institutional program (Project No. 2Z06260 and 2V08900), by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through the Innovational Food Technology Development Program, funded by Korean Ministry of Agriculture, Food and Rural Affairs (MAFRA)(119034-3), and the Priority Research Centers Program through the National Research Foundation (NRF) of Korea funded by the Korean Ministry of Education (2014R1A6A1031189).
CRediT authorship contribution statement
VR, JGP, K-YC, JH, and JL designed the study and wrote the manuscript. PC, TK, and JH isolated CBD compounds. All authors read and approved the final manuscript.
Declaration of competing interest
The authors have no conflict of interest to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2020.12.020.
Appendix A. Supplementary data
Supplementary material
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong H., Rogers J.A., Xu S. American Association for the Advancement of Science. 2020. Continuous on-body sensing for the COVID-19 pandemic: Gaps and opportunities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cárdenas-Conejo Y., Liñan-Rico A., García-Rodríguez D.A., Centeno-Leija S., Serrano-Posada H. An exclusive 42 amino acid signature in pp1ab protein provides insights into the evolutive history of the 2019 novel human-pathogenic coronavirus (SARS-CoV-2) J. Med. Virol. 2020;92:688–692. doi: 10.1002/jmv.25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong M.C., Javornik Cregeen S.J., Ajami N.J., Petrosino J.F. 2020. Evidence of Recombination in Coronaviruses Implicating Pangolin Origins of nCoV-2019, bioRxiv : The Preprint Server for Biology. (2020.2002.2007.939207) [Google Scholar]
- 9.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Resp. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman H., Carter I., Basile K., Donovan L., Kumar S., Tran T., Ko D., Alderson S., Sivaruban T., Eden J.-S. Interpret with caution: an evaluation of the commercial AusDiagnostics versus in-house developed assays for the detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;104374 doi: 10.1016/j.jcv.2020.104374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gralinski L.E., Baric R.S. Molecular pathology of emerging coronavirus infections. J. Pathol. 2015;235:185–195. doi: 10.1002/path.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respirat. Res. 2020;21:1–14. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao B., Olvera de la Cruz M. Enhanced binding of SARS-CoV-2 spike protein to receptor by distal polybasic cleavage sites. ACS Nano. 2020;14:10616–10623. doi: 10.1021/acsnano.0c04798. [DOI] [PubMed] [Google Scholar]
- 15.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H. 2020. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor, bioRxiv. (Online version) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 17.Moitra P., Alafeef M., Dighe K., Frieman M., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped Plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parvez M., Alam S., Karim M., Hasan M., Jaman J., Karim Z., Tahsin T., Hasan M., Hosen M.J. 2020. Prediction of Potential Inhibitors for RNA-Dependent RNA Polymerase of SARS-CoV-2 Using Comprehensive Drug Repurposing and Molecular Docking Approach, arXiv preprint arXiv:2004.07086. Online version. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Khafaji K., AL-DuhaidahawiL D., Tok T. Taskin. Using integrated computational approaches to identify safe and rapid treatment for SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1764392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson B.S., Laloraya M. Cytokine storm in COVID-19 patients transforms to a cytokine super cyclone in patients with risk factors. Cytokine Growth Factor Rev. 2020;54:32–42. doi: 10.1016/j.cytogfr.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kronbichler A., Effenberger M., Eisenhut M., Lee K.H., Shin J.I. Seven recommendations to rescue the patients and reduce the mortality from COVID-19 infection: an immunological point of view. Autoimmun. Rev. 2020;19 doi: 10.1016/j.autrev.2020.102570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi F., Tortora C., Argenziano M., Di Paola A., Punzo F. Cannabinoid receptor type 2: a possible target in SARS-CoV-2 (CoV-19) infection? Int. J. Mol. Sci. 2020;21:3809. doi: 10.3390/ijms21113809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rieder S.A., Chauhan A., Singh U., Nagarkatti M., Nagarkatti P. Cannabinoid-induced apoptosis in immune cells as a pathway to immunosuppression. Immunobiology. 2010;215:598–605. doi: 10.1016/j.imbio.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jean-Gilles L., Braitch M., Latif M.L., Aram J., Fahey A.J., Edwards L.J., Robins R.A., Tanasescu R., Tighe P.J., Gran B. Effects of pro-inflammatory cytokines on cannabinoid CB 1 and CB 2 receptors in immune cells. Acta Physiol. 2015;214:63–74. doi: 10.1111/apha.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guindon J., Hohmann A. Cannabinoid CB2 receptors: a therapeutic target for the treatment of inflammatory and neuropathic pain. Br. J. Pharmacol. 2008;153:319–334. doi: 10.1038/sj.bjp.0707531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribeiro A., Almeida V., Costola-de-Souza C., Ferraz-de-Paula V., Pinheiro M., Vitoretti L., Gimenes-Junior J., Akamine A., Crippa J., Tavares-de-Lima W. Cannabidiol improves lung function and inflammation in mice submitted to LPS-induced acute lung injury. Immunopharmacol. Immunotoxicol. 2015;37:35–41. doi: 10.3109/08923973.2014.976794. [DOI] [PubMed] [Google Scholar]
- 30.Mechoulam R., Peters M., Murillo-Rodriguez E., Hanuš L.O. Cannabidiol–recent advances. Chem. Biodivers. 2007;4:1678–1692. doi: 10.1002/cbdv.200790147. [DOI] [PubMed] [Google Scholar]
- 31.Turcotte C., Blanchet M.-R., Laviolette M., Flamand N. The CB 2 receptor and its role as a regulator of inflammation. Cell. Mol. Life Sci. 2016;73:4449–4470. doi: 10.1007/s00018-016-2300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bie B., Wu J., Foss J.F., Naguib M. An overview of the cannabinoid type 2 (CB2) receptor system and its therapeutic potential. Curr. Opin. Anaesthesiol. 2018;31:407. doi: 10.1097/ACO.0000000000000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks J.M., Smith J.C. How to discover antiviral drugs quickly. N. Engl. J. Med. 2020;382:2261–2264. doi: 10.1056/NEJMcibr2007042. [DOI] [PubMed] [Google Scholar]
- 34.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Micr. 2020 doi: 10.1016/j.chom.2020.03.002. Online version. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill K.P. Cannabinoids and the coronavirus. Cannabis Cannabinoid Res. 2020;5:118–120. doi: 10.1089/can.2020.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 37.Schrödinger L. Schrödinger, LLC; New York, NY: 2016. Schrödinger Suite. [Google Scholar]
- 38.Parr R.G., Yang W. Density functional approach to the frontier-electron theory of chemical reactivity. J. Am. Chem. Soc. 1984;106:4049–4050. [Google Scholar]
- 39.Shivakumar D., Williams J., Wu Y., Damm W., Shelley J., Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput. 2010;6:1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- 40.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F., Xiao X., Cheng W., Yang W., Yu P., Song Z., Yarov-Yarovoy V., Zheng J. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol. 2015;11:518–524. doi: 10.1038/nchembio.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S.-Y., Zou X. Advances and challenges in protein-ligand docking. Int. J. Mol. Sci. 2010;11:3016–3034. doi: 10.3390/ijms11083016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krieger E., Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krieger E., Dunbrack R.L., Hooft R.W., Krieger B. Comput. Drug Discov. Des. Springer; 2012. Assignment of protonation states in proteins and ligands: Combining pK a prediction with hydrogen bonding network optimization; pp. 405–421. [DOI] [PubMed] [Google Scholar]
- 47.Dickson C.J., Madej B.D., Skjevik Å.A., Betz R.M., Teigen K., Gould I.R., Walker R.C. Lipid14: the amber lipid force field. J. Chem. Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piotto S., Sessa L., Iannelli P., Concilio S. Computational study on human sphingomyelin synthase 1 (hSMS1) Biochim. Biophys. Acta Biomembr. 2017;1859:1517–1525. doi: 10.1016/j.bbamem.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Krieger E., Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi Y.H., Hazekamp A., Peltenburg-Looman A.M., Frédérich M., Erkelens C., Lefeber A.W., Verpoorte R. NMR assignments of the major cannabinoids and cannabiflavonoids isolated from flowers of Cannabis sativa. Phytochem Anal: Int. J. Plant Chem. Biochem. Tech. 2004;15:345–354. doi: 10.1002/pca.787. [DOI] [PubMed] [Google Scholar]
- 51.Vaillancourt V., Albizati K.F. A one-step method for the. alpha.-arylation of camphor. Synthesis of (-)-cannabidiol and (-)-cannabidiol dimethyl ether. J. Org. Chem. 1992;57:3627–3631. [Google Scholar]
- 52.Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. e00819–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynam M.M., Kuty M., Damborsky J., Koca J., Adriaens P. Molecular orbital calculations to describe microbial reductive dechlorination of polychlorinated dioxins. Environ. Toxicol. Chem: Int. J. 1998;17:988–997. [Google Scholar]
- 54.Mineva T., Sicilia E., Russo N. Density-functional approach to hardness evaluation and its use in the study of the maximum hardness principle. J. Am. Chem. Soc. 1998;120:9053–9058. [Google Scholar]
- 55.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burton D.R., Walker L.M. Rational vaccine design in the time of COVID-19. Cell Host Microbe. 2020;27:695–698. doi: 10.1016/j.chom.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu T.Y., Frieman M., Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat. Nanotech. 2020;15:247–249. doi: 10.1038/s41565-020-0674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ortiz-Prado E., Simbaña-Rivera K., Gómez-Barreno L., Rubio-Neira M., Guaman L.P., Kyriakidis N.C., Muslin C., Jaramillo A.M.G., Barba-Ostria C., Cevallos-Robalino D. Clinical, molecular and epidemiological characterization of the SARS-CoV2 virus and the coronavirus disease 2019 (COVID-19), a comprehensive literature review. Diagn. Micr. Infec. Dis. 2020;98 doi: 10.1016/j.diagmicrobio.2020.115094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Citti C., Linciano P., Russo F., Luongo L., Iannotta M., Maione S., Laganà A., Capriotti A.L., Forni F., Vandelli M.A. A novel phytocannabinoid isolated from Cannabis sativa L. with an in vivo cannabimimetic activity higher than Δ 9-tetrahydrocannabinol: Δ 9-Tetrahydrocannabiphorol. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-56785-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Millar S.A., Stone N., Bellman Z., Yates A., England T., O'Sullivan S. A systematic review of cannabidiol dosing in clinical populations. Br. J. Clin. Pharmacol. 2019;85:1888–1900. doi: 10.1111/bcp.14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Costiniuk C.T., Saneei Z., Routy J.-P., Margolese S., Mandarino E., Singer J., Lebouché B., Cox J., Szabo J., Brouillette M.-J. Oral cannabinoids in people living with HIV on effective antiretroviral therapy: CTN PT028—study protocol for a pilot randomised trial to assess safety, tolerability and effect on immune activation. BMJ Open. 2019:9. doi: 10.1136/bmjopen-2018-024793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pertwee R. The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: Δ9-tetrahydrocannabinol, cannabidiol and Δ9-tetrahydrocannabivarin. Br. J. Pharmacol. 2008;153:199–215. doi: 10.1038/sj.bjp.0707442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boggs D.L., Nguyen J.D., Morgenson D., Taffe M.A., Ranganathan M. Clinical and preclinical evidence for functional interactions of cannabidiol and Δ 9-tetrahydrocannabinol. Neuropsychopharmacology. 2018;43:142–154. doi: 10.1038/npp.2017.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sherif M., Radhakrishnan R., D’Souza D.C., Ranganathan M. Human laboratory studies on cannabinoids and psychosis. Biol. Psych. 2016;79:526–538. doi: 10.1016/j.biopsych.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 68.Naftali T., Mechulam R., Marii A., Gabay G., Stein A., Bronshtain M., Laish I., Benjaminov F., Konikoff F.M. Low-dose cannabidiol is safe but not effective in the treatment for Crohn’s disease, a randomized controlled trial. Digest. Dis. Sci. 2017;62:1615–1620. doi: 10.1007/s10620-017-4540-z. [DOI] [PubMed] [Google Scholar]
- 69.D.O.H.T.G.A.O.A.G.P.A. report Cannabidiol and Sports Performance: A Narrative Review of Relevant Evidence and Recommendations for Future Research. https://www.tga.gov.au [DOI] [PMC free article] [PubMed]
- 70.Zuardi A.W. Cannabidiol: from an inactive cannabinoid to a drug with wide spectrum of action. Braz. J. Psychiat. 2008;30:271–280. doi: 10.1590/s1516-44462008000300015. [DOI] [PubMed] [Google Scholar]
- 71.Shannon S., Opila-Lehman J. Effectiveness of cannabidiol oil for pediatric anxiety and insomnia as part of posttraumatic stress disorder: a case report. Perm. J. 2016:20. doi: 10.7812/TPP/16-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Atsmon J., Heffetz D., Deutsch L., Deutsch F., Sacks H. Single-dose pharmacokinetics of oral cannabidiol following administration of PTL101: a new formulation based on gelatin matrix pellets technology. Clin. Pharmacol. Drug Dev. 2018;7:751–758. doi: 10.1002/cpdd.408. [DOI] [PubMed] [Google Scholar]
- 73.Fasinu P.S., Phillips S., ElSohly M.A., Walker L.A. Current status and prospects for cannabidiol preparations as new therapeutic agents. Pharmacother: J Human Pharmacol. Drug Ther. 2016;36:781–796. doi: 10.1002/phar.1780. [DOI] [PubMed] [Google Scholar]
- 74.Qian Y., Gurley B.J., Markowitz J.S. The potential for pharmacokinetic interactions between cannabis products and conventional medications. J. Clin. Psychopharmacol. 2019;39:462–471. doi: 10.1097/JCP.0000000000001089. [DOI] [PubMed] [Google Scholar]
- 75.W.H.O Expert Committee on Drug Dependence, Thirty-Ninth Meeting, Geneva. https://www.who.int/medicines/access/controlledsubstances/5.2_CBD.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material