Abstract
Pathogens usurp host pathways to generate a permissive environment for their propagation. The current spread of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection presents the urgent need to understand the complex pathogen–host interplay for effective control of the virus. SARS-CoV-2 reorganizes the host cytoskeleton for efficient cell entry and controls host transcriptional processes to support viral protein translation. The virus also dysregulates innate cellular defenses using various structural and nonstructural proteins. This results in substantial but delayed hyperinflammation alongside a weakened interferon (IFN) response. We provide an overview of SARS-CoV-2 and its uniquely aggressive life cycle and discuss the interactions of various viral proteins with host signaling pathways. We also address the functional changes in SARS-CoV-2 proteins, relative to SARS-CoV. Our comprehensive assessment of host signaling in SARS-CoV-2 pathogenesis provides some complex yet important strategic clues for the development of novel therapeutics against this rapidly emerging worldwide crisis.
Keywords: SARS-CoV-2, COVID-19, cell signaling, viral proteins, immune response
Introduction to SARS-CoV-2
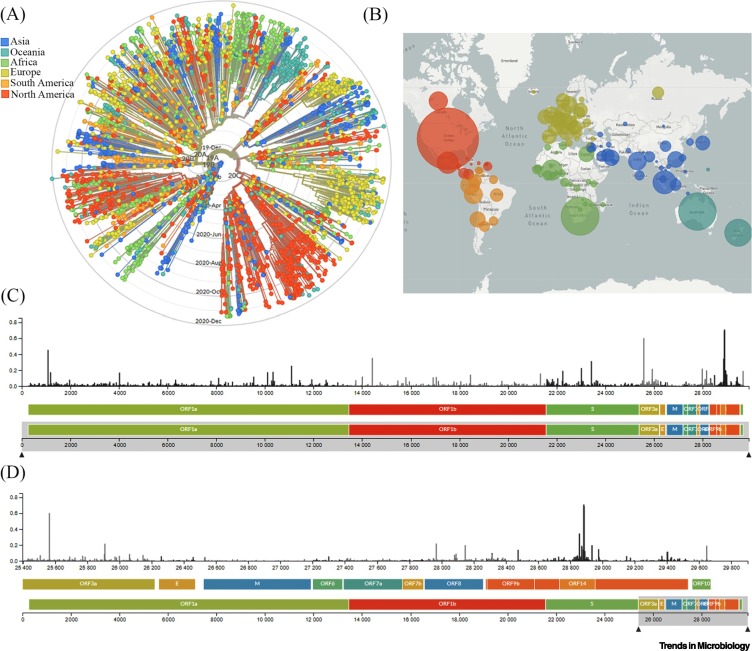
Coronaviruses are single-stranded RNA (ssRNA) viruses belonging to the family Coronaviridae. Viruses of this family have caused three major outbreaks worldwide: severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, SARS-CoV-2 in 2019, and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 with smaller outbreaks in 2015 and 2018. In 2019, a major pandemic of SARS-CoV-2 emerged in Wuhan, China, and subsequently spread across the globe, with cases being confirmed in virtually all countries and infecting about 73 million people with over 1 627 046 deaths recorded as of November 30, 2020 (https://www.worldometers.info/). The spread, evolution, and the phylogenetic analysis of SARS-CoV-2 sequences are helping researchers to investigate the mutation patterns relative to differential pathogenicity and worldwide spread of the virus (Figure 1 ) [1]. Despite strong epidemiological evidence, the host–virus molecular interactions are poorly studied, which limits efficient pandemic control.
Figure 1.
Global Genomic Epidemiology of Novel Coronavirus Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
(A) Phylogenetic analysis of 3485 genomes of SARS-CoV-2 sequences globally from December 2019 to December 2020. The different genetic variants of circulating SARS-CoV-2 are grouped into five clades defined by specific signature mutations showing their global distribution on the time scale. The clades 19A and 19B dominated the early outbreak in Wuhan and represent a higher proportion in Asia. Clades 20A, 20B, and 20C dominate in Europe and North America. (B) Geographical distribution of genomes. Each circle is centered on an individual country. The color indicates the region, and the size (area) of the circle represents the number of genomes from that country. (C) A ‘diversity’ panel that shows the novel coronavirus genome, its genes, and sites of amino acid mutations. (D) Subsection of subfigure (C) highlighting the mutation pattern in the 25 400–29 800 bp range of the genome. Apart from the spike (S) region, the genomic regions of open reading frame (ORF)14, ORF9b, ORF8, and ORF3a appear to be highly variable between clinical isolates of SARS-CoV-2. Source: latest global SARS-CoV-2 updated daily at https://nextstrain.org/sars-cov-2. Abbreviations: E, envelope; M, membrane.
Both SARS-CoV and SARS-CoV-2 belong to the β subfamily of coronaviruses. SARS-CoV-2 consists of 29 different proteins, including 16 nonstructural proteins: proteases, RNA-dependent RNA polymerases, a helicase, nuclease, and methyltransferase encoded by 14 open reading frames (ORFs) (Table 1 , Key Table) [2]. Structural proteins and nine putative accessory factors are encoded by 13 ORFs [2]. SARS-CoV-2 shares about 79% genome similarity with SARS-CoV and 50% with MERS-CoV [3]. Epidemiological data suggest that SARS-CoV-2 has the highest transmission rate but the smallest mortality rate (2.3%) when compared with SARS-CoV (9.6%) or MERS-CoV (35%). The higher transmission and pathogenicity of the SARS-CoV-2 could be attributed to the genetic alterations in different structural and nonstructural proteins. To identify potential molecular targets of SARS-CoV-2, and thus generate novel therapeutics against this devastating virus, it is important to understand the mechanisms by which the virus usurps host cell pathways and functions to cause disease. Here we discuss some of the genomic variations in viral proteins and contemplate their roles in the modulation of host cell signaling.
Table 1.
Key Table. Overview of the Common Names and Currently Known Functions of the SARS-CoV-2 Viral Proteinsa
| Structural protein | Function | Refs |
|---|---|---|
| Spike protein | Viral entry – binds to ACE2 receptor and heparan sulfate | [25,94] |
| Envelope protein | Virion structure | [3] |
| Membrane protein | Virion structure | [3] |
| Nucleocapsid | Houses genome, interferes with host translation and cell cycle, regulates cytoskeleton organization | [30,99] |
| Nonstructural protein | ||
| NSP1 | Inhibits host translation and the IFN response | [33,35] |
| NSP2 | Disrupts cell cycle progression and apoptosis, possibly regulates actin assembly | [37,38] |
| NSP3 | Facilitates transcription of the viral genome, arrests cell cycle in the G0/G1 phase, contains PLpro which antagonizes host immune responses and frees NSPs 1–3 | [40,95,97] |
| NSP4 | Participates in membrane rearrangement | [45] |
| NSP5 | Contains the main protease, regulates epigenetic and gene expression, influences ER and mitochondrial functioning | [2,48,50] |
| NSP6 | Restricts host autophagosome expansion | [52] |
| NSP7 | Complexes with NSP8 to form RNA primase | [53] |
| NSP8 | Complexes with NSP7 to form RNA primase | [53] |
| NSP9 | RNA-binding phosphatase | [56] |
| NSP10 | Activation cofactor for NSP14 and NSP16 | [60,61] |
| NSP11 | Unknown | [98] |
| NSP12 | RdRp – complexes with NSP7 and NSP8 to replicate viral genome | [65] |
| NSP13 | Helicase | [69,70] |
| NSP14 | 3′-5′-exoribonuclease, interacts with replication-transcription complex | [74] |
| NSP15 | Endoribonuclease, IFN inhibitor, and modifies host cell cycle progression | [76,79] |
| ORF3a | Ion channel protein, influences cytokine responses | [81,82] |
| ORF6 | Inhibits antiviral IFN response | [84] |
| ORF7a | Suppresses STAT1 phosphorylation, inhibits type 1 IFN response | [8] |
| ORF7b | Suppresses STAT1 and STAT2 phosphorylation, inhibits type 1 IFN response | [8] |
| ORF8 | Inhibits antiviral IFN response, immune evasion virulence factor | [78] |
The proteins are divided between structural and nonstructural types. The functions and associated references are provided for each protein.
The Viral Life Cycle
The life cycle of SARS-CoV-2 begins with the priming of the viral spike (S) protein by host proteases or the transmembrane protease serine 2 (TMPRSS2) [4] to ensure fusion of the viral envelope with the host cell membrane and entry of the viral genome into the cell [5]. Alternatively, SARS-CoV-2 virions can be endocytosed by the cell after attaching to the angiotensin-converting enzyme 2 (ACE2) receptor. Once internalized, host enzyme cathepsin L can cleave the S protein that results in a similar release of the viral genome to the cell [5].
Once inside the cell, the positive-sense ssRNA genome becomes translated using host ribosomes. The primary viral gene ORF1ab codes for the polyproteins pp1a and pp1ab which autocleaves the papain-like protease (PLpro) and 3C-like protease (3CL-pro) from itself [6]. PLpro further processes the viral polyprotein and releases nonstructural proteins (NSPs) 1–3, while 3CL-pro releases the remaining NSPs 4–16 [6]. Certain SARS-CoV-2 NSPs form an RNA replication complex, such as the RNA-dependent RNA polymerase (RdRp) NSP12 and the helicase NSP13 [7]. NSPs 10, 13, 14, and 16 participate in mRNA capping, and NSPs 10, 14, and 15 proofread the nascent genome [7]. While the genome is replicating, multiple viral proteins – such as NSP1, NSP3, NSPs 12-14, ORF3, ORF6, and ORF7a/b – antagonize host immune responses by targeting the innate immune system, in particular the type 1 interferon (IFN) pathway [8,9]. However, the virus activates certain host Toll-like receptors (TLRs) during infection, which stimulates the release of numerous proinflammatory cytokines, including interleukin (IL)-1β, IL-2, IL-6, IL-7, granulocyte-colony stimulating factor (GCSF), IFN-γ, and tumor necrosis factor-alpha (TNF-α) [10., 11., 12.]. Analysis of peripheral blood from severely infected patients revealed decreased CD4 and CD8 T cell counts, but the fractions of hyperactivated T cells increased [13]. Furthermore, the concentrations of T helper (Th)17 cells and perforin and granulysin-expressing CD8 T cells were enhanced, both of which contribute to hyperinflammation [13]. Host Th17 responses are major contributors to cytokine storms, a hallmark of severe coronavirus disease 2019 (COVID-19) infections [14].
Subsequently, the viral structural proteins and the viral genome assemble into the virion, which travels to the host cell surface using vesicles and is released via lysosomal, Arl8b-dependent exocytosis [3,15]. The host endoplasmic reticulum (ER) chaperone GRP78/BiP is cotrafficked with the virion and released during this process as well [15]. Alternatively, immature virions use the nucleocapsid (N) protein to localize to a glycosylated envelope (E) protein at the cell surface [16]. This interaction allows for a reorientation of the virion, followed by budding from the host cell [16]. Finally, the destruction of host organelles can lead to the release of lysosomal contents, triggering cell death and the release of viral progeny [17].
Host Signaling during SARS-CoV-2 Infection
During infection, viruses tend to exploit host machinery to create a permissive host microenvironment. A phosphoproteomic analysis of SARS-CoV-2-infected Vero cells revealed regulation of 97 cellular kinases [18]. The activated kinases include some of those involved in p38-mediated signaling and guanosine monophosphate-dependent protein kinases. Downregulated kinases include those involved in cell growth, the cell cycle, and cytoskeleton regulators. The most highly activated transcription factors, such as nuclear factor-κB (NF-ĸB), monocyte-specific enhancer factor 2C, and c-Jun, are downstream of the p38/MAPK (mitogen activated protein kinase) pathway which is known to mediate stimulation of proinflammatory cytokines, including IL-6, TNF-α, CCL2, CCL20, and CXCL8 [18]. Additionally, cell cycle arrest at the S/G2 transition is highly correlated with productive SARS-CoV-2 infection, whereas M phase progression is negatively correlated. S/G2 arrest during infection may be due to a dysregulation of cyclin-dependent kinase 2 (CDK2) activity during viral infection [18]. Studies have found atypical, delayed induction of host immune response to SARS-CoV-2 infection which favors virus replication in the absence of type I and III IFN responses and simultaneously induces high levels of chemokines [19,20]. An integrated immune analysis on a cohort of 50 COVID-19 patients with various levels of disease severity has shown that NF-κB partially drives the inflammation. Such inflammatory responses are the outcome of increased TNF-α and IL-6 signaling [19]. Recent work shows that TNF-α and IFN-γ induce inflammatory cell death and tissue damage through extensive crosstalk among different modes of cell death, including pyroptosis, apoptosis, and necroptosis, together termed PANoptosis [21]. Treatment with a combination of TNF-α and IFN-γ was found to activate the Janus kinase (JAK)/STAT1(signal transducer and activator of transcription 1)/IRF1 (interferon regulatory factor 1) axis, leading to nitric oxide production that drives caspase-8/FADD-mediated PANoptosis. Interestingly, TNF-α and IFN-γ together induce a SARS-CoV-2-like cytokine storm in mice, which can be reversed using PANoptosis inhibitors. Furthermore, treatment of SARS-CoV-2-infected mice with neutralizing antibodies against TNF-α and IFN-γ protects mice from death. This emphasizes the clinical significance of cytokine-mediated inflammatory cell death signaling pathways.
Structural Proteins
SARS-CoV-2 encodes five structural proteins: (i) spike, (ii) envelope, (iii) membrane, and (iv) nucleocapsid.
Spike Protein
The SARS-CoV-2 S protein mainly functions in viral entry. Mechanistically, the S protein binds to human heparan sulfate (HS) and ACE2 using its receptor-binding domain (RBD) and is activated by human proteases, but, unlike SARS-CoV, cell entry of SARS-CoV-2 is less dependent on host proteases as it is preactivated by the pro-protein convertase furin [22]. The rate of transmission of SARS-CoV-2 could be attributed to structural differences in the highly variable RBD of its S proteins (Box 1 ). Unlike many of its other proteins, the S protein of SARS-CoV-2 amplifies the activation of IFN-β [9]. A preprint by Hsu et al. suggests that the SARS-CoV-2 S protein contributes to host hyperinflammatory responses via its binding to the ACE2 receptor [23]. This interaction stimulates the MAPK–NF-ĸB axis and results in the release of proinflammatory cytokines such as TNF-α and CCL2. MyD88 and TRIF are critical adaptor molecules for this signaling pathway to function. Furthermore, downstream signaling from the S–ACE2 interaction increases ER stress, causing an unfolded protein response and activation of NF-ĸB. While these results are from a preprint, and should be viewed with some caution, an older study found that the SARS-CoV S protein induced the activation of NF-ĸB signaling as well [24]. The induction of inflammatory cytokines by the S protein may contribute to the cytokine storm and the associated epithelial damage and organ injuries observed in COVID-19 patients.
Box 1. Spike Protein Mutations in SARS-CoV-2.
Structurally, in addition to 12 nucleotide insertions, amino acid mutations of the RBD of SARS-CoV-2 include L455, F486, Q493, S494, N501, and Y505. These modifications result in an increase in 3-O-linked glycans around the binding site and thus higher binding affinity for the human receptor ACE2. A variant carrying amino acid change D614G has been recently implicated to increase infectivity of SARS-CoV-2 [95].
Alt-text: Box 1
In addition to ACE2, an increasing body of evidence points to the importance of cell surface HS in the initial cellular attachment and viral entry [25]. The higher binding affinity of S protein (Kd 55 nM) to HS compared with RBD (Kd 1 μM) suggests that heparin, an HS mimetic, or soluble HS, can be used to block viral entry in the host cell [26]. HS is a complex glycosaminoglycan found ubiquitously on human cells, with documented involvement in key cellular processes of inflammation, cellular communication, and microbial invasion [27,28]. Recent evidence for the robust proviral actions of the HS-degrading enzyme heparanase (HPSE) across viral families make HS a promising system for the study of SARS-CoV-2 viral entry [29].
Envelope and Membrane Proteins
Envelope (E) and Membrane (M) proteins constitute important components of the SARS-CoV-2 virus envelope. Sequence comparison shows that these SARS-CoV-2 proteins are highly similar to those of SARS-CoV [9]. Interestingly, E and M proteins are nearly identical among bat and pangolin coronaviruses. The M protein also independently inhibits the activation of IFN-β, while the E protein stimulates the transcription of interferon-stimulated genes (ISGs) [9].
Nucleocapsid (ORF9/9a) Protein
The nucleocapsid (N) competes with host protein synthesis by dephosphorylating the translational regulator ribonucleoprotein LARP1, which lets it bind to untranslated regions of mRNA. Interestingly, overexpression of the SARS-CoV N protein upregulates casein kinase 2 activity, which induces actin polymerization and thereby regulates cytoskeleton organization, a process commonly associated with viral infection [30]. The SARS-CoV N protein also inhibits S phase progression by inhibiting CDK2, either by direct binding of N protein or by downregulating protein levels of CDK2, cyclin E, and cyclin A [31]. Further information on ORF9 isoforms is given in Box 2 .
Box 2. Functions of ORF9b and ORF9c.
Functions of these two proteins remain elusive, with little information available in the literature to date. In SARS-CoV, ORF9b suppresses type I IFN signaling by severing a link between mitochondrial antiviral-signaling protein (MAVS) and downstream mediators TRAF3 and TRAF6 [96]. Protein expression of dynamin-related protein (DRP1), which is involved in physiological mitochondrial fission, was found to be diminished with overexpression of ORF9b, resulting in mitochondrial elongation [96]. While recent investigation on this protein has been limited, initial work directed at understanding host protein interactions of viral proteins suggests that SARS-CoV-2 ORF9b may influence cellular proliferation by binding to multiple MAP kinases, while ORF9c binds to several proteins involved in electron transport [2]. Interestingly, SARS-CoV-2 ORF9c may not be a true open reading frame or may not be required for viral replication, as multiple clinical isolates analyzed contained premature stop codons in the ORF sequence [2].
Alt-text: Box 2
Nonstructural Proteins
SARS-CoV-2 encodes 16 NSPs, which cooperate and form complexes to execute their functions. Here, we provide an overview of the role of these NSPs in the viral life cycle along with certain accessory ORF proteins expressed by the virus. Given the relative scarcity of peer-reviewed studies on SARS-CoV-2, we offer an overview of these proteins in both SARS-CoV and SARS-CoV-2 as their sequence similarities suggest that they could play analogous functions. However, information on SARS-CoV must be recognized as such, and caution must be exercised when it is applied to SARS-CoV-2.
NSP1
SARS-CoV-2 NSP1 shares 85% sequence identity with SARS-CoV and helps to neutralize the host response to infection. When HEK293T cells are stimulated with IFN-α, SARS-CoV-2 NSP1 inhibits phosphorylation of STAT1, thereby suppressing downstream IFN-α-mediated signaling. In addition, NSP1 also abolishes IFN-β production in HEK293T cells [9,32]. The mechanism by which this occurs is less clear as NSP1 likely inhibits proteins both upstream and downstream of IRF3. The mutations in NSP1 found in SARS-CoV and MERS-CoV appear to improve its ability to inhibit type 1 IFN responses [8].
Multiple roles have been discovered for NSP1 of SARS-CoV which may apply to its variant in SARS-CoV-2. SARS-CoV NSP1 blocks the host translation process by associating with the 40S subunit of the ribosome [33]. NSP1 also recruits a host endonuclease to create breaks in the 5′-UTR (untranslated region) region of host mRNA sequences, which are then degraded in an Xrn1-mediated manner [34]. Interestingly, viral mRNAs have a leader sequence in the 5′-region that confers protection against endonucleolytic cleavage and promotes the selective translation of viral proteins [35]. Mutations in SARS-CoV NSP1 jeopardize its ability to block the host IFN response, suggesting that NSP1 interacts with interferon response factor (IRF)-3, IRF7, and c-Jun signaling [36]. Both SARS-CoV and SARS-CoV-2 NSP1 function to negatively regulate the host immune response.
NSP2
NSP2 consists of 638 amino acids and shares only 68% sequence homology with its counterpart in SARS-CoV. One mutation in the endosome-associated-protein-like domain has been suggested to contribute to the increased contagiousness of SARS-CoV-2 [37]. NSP2 is one of the few SARS-CoV-2 proteins which significantly accelerates the activation of IFN-β [9]. While not confirmed in SARS-CoV-2, SARS-CoV NSP2 interacts with host proteins prohibitin 1 and 2 to disrupt cell cycle progression and apoptosis [38]. NSP2 has been shown to interact with the Wiskott–Aldrich syndrome protein and scar homology (WASH) complex component strumpellin [2]. Strumpellin is known to regulate actin assembly on intracellular vesicles, which may help in viral egress [2,39]. Considering the differences in sequence homology of NSP2 in SARS-CoV-2 compared with SARS-CoV, the protein merits additional research.
NSP3
NSP3 is a large, multidomain protein that plays versatile roles in the SARS-CoV-2 life cycle. SARS-CoV-2 NSP3 delays the activation of IFN-β, allowing more time for the virus to replicate safely [9]. However, the SARS-CoV NSP3 was a much more potent inhibitor of the IFN response and could deubiquitinate proteins more efficiently as well [40]. NSP3 contains a papain-like protease (PLpro) domain that cleaves the replicase polyprotein and frees NSP1, NSP2, and NSP3 [6]. The SARS-CoV PLpro was demonstrated to inhibit many components of the innate immune response, particularly by blocking type I IFN signaling through interaction with the stimulator of interferon genes (STING)–TRAF3 (TNF receptor associated factor 3)–TBK1 (Tank-binding kinase 1) complex and degradation of p53 [41,42]. SARS-CoV PLpro also impedes TLR7-mediated cytokine induction via the removal of Lys63-linked ubiquitin bound to TRAF3 and TRAF6 [43]. A macrodomain of NSP3, termed the X domain, forms an essential component of the coronavirus replication complex. The resulting protein from the X domain also inhibits type I IFN, ISGs, and cytokine responses by removing ADP-ribose from target proteins [44]. ADP-ribosylated proteins can stimulate a variety of immune responses, and many viruses have been found to encode proteins capable of removing ADP-ribose from host proteins and evading host immunity [44]. Given these results, SARS-CoV-2 PLpro may interact with the STING and TLR7 signaling pathways to dysregulate the host immune response.
NSP4
NSP4 is one of the viral proteins released during the cleavage of the viral polyproteins pp1a and pp1ab [45]. While there is a scarcity of information of SARS-CoV-2 NSP4, it may have similar roles as its SARS-CoV variant as they share nearly 80% sequence similarity [9]. Structural modeling of the SARS-CoV protein suggests that it has multiple transmembrane domains [46]. Early after its release, it becomes N-glycosylated upon interaction with the ER membrane. Two domains located in a luminal loop of SARS-CoV NSP4 interact with NSP3 to translocate from the ER to the perinuclear region [47]. SARS-CoV NSP4 (amino acid residues H120 and F120) participates in membrane rearrangement to facilitate the viral replication and transcription complex (RTC) formation.
NSP5
NSP5 is the main protease of SARS-CoV-2, with 3CL-pro being its putative functional domain [2]. The protein shows 99% and 96% similarity with BAT-SL-CovZXC21 and SARS-CoV respectively [48]. NSP5 acts as an epigenetic and gene expression regulator and can also influence the functions of the ER and mitochondria. In a protein interactome study, SARS-CoV-2 NSP5 interacted with histone deacetylase 2 (HDAC2), likely restricting its nuclear translocation to inhibit innate antiviral immunity and resulting in enhanced ACE2-mediated cell entry [2,49,50]. Studies also show that catalytically inactive NSP5 interacts with tRNA methyltransferase, which dimethylates guanosine on nuclear and mitochondrial tRNAs [2]. SARS-CoV-2 NSP5 may mediate the cleavage of TRMT1, making it lose its zinc-finger domain and nuclear localization sequence [2,51]. In addition, gene ontology data predict involvement of NSP5_C145A in regulation of the cell membrane matrix, cell death, response to oxidative stress, and sensory perception of mechanical stimuli.
NSP6
SARS-CoV-2 NSP6 is a potent inhibitor of the host type 1 IFN response [8]. NSP6 interacts with TBK1 to suppress the phosphorylation of IRF3, which is a prerequisite step for the stimulation of IFN-β [8]. Interestingly, SARS-CoV NSP6 does not inhibit type I IFN responses [8]. Evolutionary analysis of NSP6 points to a mutation which is important for maintaining the stability of the SARS-CoV-2 protein and, in turn, contributes to its acquired role in immune inhibition [52]. However, one study has reported that SARS-CoV-2 NSP6 stimulated the promoters of the interferon-stimulated response element (ISRE) and ISG56 [9]. The NSP6 of SARS-CoV instead was reported to play an important role in the inhibition of host autophagy. It restricted the expansion of host autophagosomes by reducing the diameter of autophagosomes in infected cells [53]. NSP6 of the coronavirus infectious bronchitis virus (IBV) has been known to limit expansion of omegasomes, sites to which proteins forming the autophagosome complex are recruited [53]. Furthermore, IBV NSP6 disrupts the association of mammalian target of rapamycin (mTOR) with lysosomes [53]. Given these findings, SARS-CoV-2 NSP6 may have an uncharacterized role in impeding host autophagy as well.
NSP7 and NSP8
Both NSP7 and NSP8 are highly conserved between SARS-CoV-2 and SARS-CoV. SARS-CoV-2 NSP7 shares 98.8% sequence similarity with SARS-CoV NSP7, while SARS-CoV-2 NSP8 is 97.5% conserved, with 98.8% and 97.5% similarity, respectively. Therefore, NSP7 and NSP8 in SARS-CoV-2 likely serve similar roles, as in SARS-CoV. The SARS-CoV NSP8 is a 22-kD protein that forms a complex with NSP7 to act as an RNA primase [54]. In SARS-CoV, the NSP7+8 complex can efficiently bind to RNA and extend primers to template length [54]. Magnesium ions (as cofactors) and basic pH are required for the optimum activity of this complex [54]. The complex also indirectly facilitates replication of the viral genome. To initiate RNA replication, the RdRp NSP12 must first associate with the primase complex followed by NSP14. The SARS-CoV NSP14 is a key enzyme involved in proofreading of newly synthesized viral RNA [55]. NSP8 interacts with ribonucleoprotein 7 (LARP7), M-phase phosphoprotein 10, and cytokine-induced protein. LARP7 is an important part of the transcription assembly shown to control transcription of viral genes [18,56]. SARS-CoV-2 NSP7’s interaction with Ras homology family member A (RHOA) may contribute to downregulation of Rho GTPases and the phosphorylation of the vimentin and stathmin proteins by PAK1/2 kinase [18]. Both vimentin and stathmin are known to have a role in cytoskeletal organization. Furthermore, SARS-CoV-2 NSP7 was demonstrated to impede IFN-α signaling independently of the other NSPs [8].
NSP9
NSP9 of SARS-CoV-2 acts as an RNA-binding phosphatase and participates in the RTC [57]. It shares 97.3% sequence homology with SARS-CoV NSP9. The structure of SARS-CoV NSP9 suggests that it may stabilize viral RNA during replication and assist in RNA processing [58]. During replication, SARS-CoV NSP9 dimerizes via the binding of two α-helices with GXXXG motifs, which is a necessary step for coronavirus replication [59]. Interestingly, SARS-COV-2 NSP9 is a stimulator of the host type 1 IFN response [9].
NSP10 and NSP16
SARS-CoV-2 NSP10 is highly conserved with its counterpart in SARS-CoV [9]. SARS-CoV NSP10 forms a complex with NSP16 during the coronavirus life cycle [60]. NSP10 binds to and activates the 2-O-methyltransferase NSP16 [60]. The complex methylates the 5′ guanosine cap of the viral RNA, which helps to avoid detection from molecules that recognize host cytosolic RNA, such as RIG-I [61]. This immune evasion ability of the SARS-CoV NSP10+16 complex interferes with activation of the IFN response [60]. Loss of the NSP10+16 interaction resulted in inhibition of viral replication, suggesting that formation of the complex is an essential step in the SARS-CoV life cycle [62]. Additionally, SARS-CoV NSP10 acts as an activation cofactor in its complex with NSP14. It enables NSP14 to proofread and edit newly synthesized RNA [63]. The NSP7+8+12 complex and NSP10 may each serve regulatory roles allowing the exoribonuclease to function optimally.
NSP11
SARS-CoV-2 NSP11 is a small cleavage product resulting from the processing of pp1a by 3CL-pro [64]. Its full 13 amino acid sequence is listed as follows: SADAQSFLNGFAV [65]. In the ribosomal frameshift of ORF1b, which is required for the translation of NSP11-16, it becomes the N terminus of NSP12. No independent function has been currently described for NSP11 [64].
NSP12
NSP12 is the RdRp of SARS-CoV-2 and is essential for viral replication. It must form a polymerase complex with its cofactors NSP7 and NSP8 prior to activation [66]. NSP7+8 functions as a primase and improves the RNA-binding capacity of NSP12, which facilitates its subsequent polymerase activity to replicate the SARS-CoV-2 genome [66]. Transfection of HEK293T cells with a NSP12 plasmid prior to infection with Sendai virus significantly inhibited IFN-β activation [9]. However, NSP12 also stimulates ISRE and ISG56, suggesting that it may have a nuanced relationship with the host immune response [9]. SARS-CoV-2 NSP12 displays decreased enzymatic activity and thermostability when compared with SARS-CoV NSP12 [67]. The experimental drug remdesivir, which has gained popularity in clinical use against SARS-CoV-2, is thought to inhibit SARS-CoV-2 by binding to NSP12 [62,68]. Furthermore, Ruan et al. found eight additional compounds that demonstrate strong binding affinity to the NSP7+8+12 complex [69]. NSP12 is known to interact with ribonucleoprotein 4B, CREB regulated transcription coactivator 3, E3 ubiquitin protein ligase, ubiquitin-associated protein 2, and PLAKHA5. Many SARS-CoV-2 NSP12-interacting proteins show decreased phosphorylation upon viral protein overexpression [18].
NSP13
NSP 13 is the SARS-CoV-2 helicase and is identical to its counterpart in SARS-CoV [48]. Therefore, information on SARS-CoV NSP13 is highly applicable to SARS-CoV-2. SARS-CoV NSP13 unwinds duplex DNA and RNA with a 5′ single-stranded tail in the 5′–3′ direction using ATP hydrolysis, and thereby plays a crucial role in viral replication [70,71]. In general, the duplex RNA-unwinding activity of SARS-CoV NSP13 is less efficient, when compared with duplex DNA unwinding [71]. However, with increased ATP concentrations, duplex RNA becomes a preferred substrate for helicase activity of SARS-CoV-2 NSP13, resulting in higher processivity in unwinding of duplex RNA compared with DNA [72]. Here, ATP causes a conformational change in helicase, diverting its affinity towards RNA. With this work, the authors conclude that ATP availability may influence the unwinding, translocation, and dissociation of SARS-CoV-2 NSP13 from its substrate (duplex DNA or RNA). Adedeji et al. further suggested that RdRp (NSP12 from SARS-CoV) enhances the catalytic efficacy of SARS-CoV NSP13 by twofold [73]. Similar to NSP6, the SARS-CoV-2 NSP13 also inhibits the activation of IFN-β in HEK293T cells by inhibiting the phosphorylation of both IRF3 and TBK1 [8,9,40]. NSP13 also inhibits IFN-α-mediated signaling in HEK293T cells [8]. Thus, SARS-CoV-2 NSP13 plays dual roles as a helicase and an inhibitor of the type 1 IFN response.
NSP14
SARS-CoV NSP14 has been shown to interact with other NSPs, like 7, 8, 10, and 12 [46,74]. Its primary role is to serve as a 3′–5′-exoribonuclease upon initial interaction with NSP10. Once active, it proofreads the viral genome and excises any errant nucleotides that it encounters. In the coronavirus murine hepatitis virus, mutations in NSP14 resulted in a 15-fold increase in genomic mutations, highlighting the essential role it plays in replication fidelity [75]. Similarly, the mutated NSP14 of SARS-CoV exhibited a 21-fold increase in genomic mutations [75]. Additionally, SARS-CoV NSP14 serves as a (guanine-N7)-methyltransferase, which can protect newly synthesized viral RNA from degradation. Its capping function requires the presence of the exoribonuclease domain, although the enzymatic activity of the domain is dispensable [76]. Finally, SARS-CoV-2 NSP14 has been shown to inhibit IFN-β production and signaling by preventing the nuclear localization of IRF3 [9,40].
SARS-CoV NSP14 can also bind to the NSP7+8+12 complex, but it lacks exoribonuclease activity when associated [55]. However, upon the addition of NSP10 to the replication complex, SARS-CoV NSP14 regains its activity [55]. Alongside NSP7, 8, and 12, NSP14 may act as another unit of a super-complex of proteins with primase, polymerase, and exoribonuclease functions.
NSP15
SARS-CoV NSP15 acts as an endoribonuclease and cleaves RNA nucleotides in the 3′ direction of uridylates [77]. The conformation of NSP15 is highly sensitive to the presence of manganese cations [78]. Mutations in SARS-CoV NSP15, including those outside of the active site, inhibit viral infection [79]. Like NSP13 and 14, SARS-CoV-2 NSP15 can also interfere with the host IFN response during infection by blocking the localization of IRF3 to the nucleus [40]. Interestingly, SARS-CoV NSP15 contains a small domain capable of binding to the retinoblastoma protein pRb [80]. NSP15 alters the distribution of pRb in Huh-7 cells, increasing the ratio of cytoplasmic to nuclear pRb [80]. The reduction of Rb expression results in changes in cell cycle progression of NSP15-transfected cells [80]. Thus, NSP15 exhibits multifaceted roles as an endoribonuclease, an IFN inhibitor, and a binding partner for pRb.
ORF3a
ORF3a, alongside ORF8a and the E protein, is an ion channel protein encoded by SARS-CoV [81]. In SARS-CoV-2, ORF3a has been demonstrated to inhibit IFN-α signaling by suppressing the phosphorylation of STAT1 [8]. Another study found that ORF3 could impede IFN-β signaling as well, making it a potent inhibitor of the type 1 IFN response [9]. Recently, ORF3a has been implicated in lysosomal deacidification [15]. Raising the pH of the lysosome allows the virus to use lysosomal organelles as vehicles to reach the plasma membrane and egress from the host cell [15]. SARS-CoV ORF3a possesses three transmembrane domains and interacts with the M and E proteins during viral assembly [82]. ORF3a also contains a PDZ-binding motif, which conceivably allows it to bind to hundreds of cellular proteins and regulate signaling [82]. SARS-CoV virions lacking ORF3a exhibit defects in replication both in vitro and in vivo [82]. Viruses which lacked the PDZ-binding motif of ORF3a replicated normally, suggesting that other domains of ORF3a may be essential for replication [82]. However, in the absence of the E protein, the PDZ-binding motif becomes essential for replication [82]. Interplay between the E protein and ORF3a exerts significant influence over the viral replication cycle. ORF3a participates in cytokine responses independently of its interactions with the E protein (Box 3 ).
Box 3. ORF3a Participates in Cytokine Storm Induction.
Independently of its interactions with the E protein, SARS-CoV ORF3a also participates in induction of the cytokine storm. In this pathway, TRAF3 acts as its key binding partner. The interaction of ORF3a and TRAF3 results in the activation of NF-ĸB, which upregulates IL-1β expression and secretion [97]. Additionally, SARS-CoV ORF3a expression leads to the ubiquitination of apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), a process mediated by TRAF3 once again [98]. Ubiquitination of ASC in NOD-, LRR-, and pyrin domain-containing protein 3 (NLRP3)-expressing cells activates caspase-1 and IL-1β maturation [98]. Due to its role in the upregulation and maturation of IL-1β, ORF3a stimulates formation of the NLRP3 inflammasome and the induction of a cytokine storm. Given these findings in SARS-CoV, the ORF3a of SARS-CoV-2 may induce cytokine storms during infection as well.
Alt-text: Box 3
ORF6
ORF6 is the least conserved protein between SARS-CoV and SARS-CoV-2, with only 66.6% sequence similarity [9]. SARS-CoV NSP8 colocalizes with ORF6 in cytoplasmic punctate structures [83]. Although ORF6 associates with NSP8, mutants lacking ORF6 demonstrate similar replication kinetics as wild-type viruses [84]. SARS-CoV ORF6 binds to karyopherin α2 and β1 to the ER/Golgi membrane, preventing them from forming a nuclear import complex [85]. Additionally, SARS-CoV ORF6 is known to interact with the NUP98/RAE complex and is correlated with increased phosphorylation of NUP98 at the S888 site. This prevents host mRNA nuclear export, resulting in inhibition of host protein synthesis [86]. The C terminus appears to be the essential region of the protein for the tethering to occur [85]. When IFN signaling is active, the loss of nuclear importation prevents STAT1 from entering the nucleus and blocks downstream STAT1 signaling [85]. In conjunction with these findings, SARS-CoV infection results in rapid production of high viral titers in STAT1 knockout mice [87]. Given the essential role of STAT1 signaling in the clearance of coronavirus infections, ORF6 plays a key role as an inhibitor of antiviral IFN signaling.
In support of these findings, recent studies have implicated SARS-CoV-2 ORF6 in the suppression of IFN-β-mediated IFN responses [8,9,40]. ORF6 binds to karyopherin α, a nuclear import factor, to prevent the translocation of IRF3 to the nucleus, which halts the subsequent production of IFN-β [8]. In addition, amino acids 53–61 of the ORF6 C terminus are sufficient for ORF6 to block the nuclear translocation of STAT1, thereby reducing the activity of the ISRE and ISG56 promoters [9].
ORF7a/b
ORF7a and 7b are accessory proteins of SARS-CoV-2. Both ORF7a and ORF7b inhibit the type 1 IFN response, but they do so through slightly different mechanisms. ORF7a inhibits the phosphorylation of STAT2, while ORF7b inhibits the phosphorylation of both STAT1 and STAT2 [8]. SARS-CoV-2 ORF7a and 7b prevent the STAT proteins from translocating to the nucleus and subsequent transcription of ISGs. Given the myriad of SARS-CoV-2 proteins which inhibit the type 1 IFN response, exogenous addition of certain IFNs may repress viral production. Prophylactic treatment of IFN-β proved sufficient to restrict viral replication in SARS-CoV-2-infected Calu-3 cells [9].
ORF8
ORF8 contains several hypervariable regions and constitutes one of the proteins that shares the smallest degree of homology with SARS-CoV [78]. ORF8 also serves as an important inhibitor of the type I IFN system. SARS-CoV-2 ORF8 overexpression results in dose-dependent inhibition of IFN-β promoter activation [88]. Its IFN-inhibitory activity could be due to downregulation of MHC-I [89]. Zhang et al. observed downregulation of human leukocyte antigen (HLA)-A2 with overexpression of ORF8 in 293T cells. SARS-CoV-2 ORF8 may serve as an immune evasion virulence factor through rapid mutation and evolution [90]. Divergent clinical isolates from various geographic regions have been identified with highly variable ORF8 and ORF 14 sequences [91., 92., 93.]. Kakhki et al. proposed that ORF8 might serve as an excellent detection target as its sequence contains stretches of nucleotides that are wholly unique to SARS-CoV-2 [94].
Concluding Remarks
Two decades following its initial outbreak, no standardized therapies for SARS-CoV are available. Therefore, a great deal of in-depth research is needed to develop effective treatments for β-coronavirus infections such as SARS-CoV-2. In this review we have provided an overview of the SARS-CoV-2 life cycle, specific details on its key proteins, and an account of the polymorphisms in the SARS-CoV-2 genome along with updated information on its global spread. The information may provide important clues about factors regulating the differential spread, high transmission, and pathogenesis of SARS-CoV-2. We have also discussed the relative differences in viral protein functions between the two SARS-CoV types. Both structural and nonstructural SARS-CoV-2 proteins play important roles in usurping cellular machinery, insights into which may be helpful to target the cellular signaling pathways to make the host microenvironment resistant to virus replication. Many SARS-CoV-2 proteins delay immune activation, which prolongs infection and allows further viral replication. This is followed by substantial IFN production, resulting in a cytokine storm that contributes to acute tissue injury. Proinflammatory viral factors are poorly studied and need to be further investigated (see Outstanding Questions). Elucidation of the crystal structures of these SARS-CoV-2 factors would likely contribute to development of safe and effective anti-COVID-19 therapies or vaccines. Collectively, it is clear that individual factors of SARS coronaviruses have diverse roles in disruption of cellular programs, and additional research focus directed in the areas outlined here may help in delineating the underlying mechanism of pathogenesis and spread in humans and will be instrumental in our quest for curative therapeutics.
Outstanding Questions.
Does the ability of the to interact with HS more strongly have a role in the high transmission rate for SARS-CoV-2? Should HS be considered an essential or accessory coreceptor for the S protein? What types of HS modification are essential for viral entry? Which viral proteins activate heparanase?
What are the structure-specific functional alterations of ORF8 and 14 which help the virus to adapt to different geographic locations? Moreover, how can these genetic variations be used to design specific immunotherapy against SARS-CoV-2?
What roles do mammalian target of rapamycin complex (mTORC)1/2 complexes play in virus protein translation and activation of immunity, and which viral proteins regulate them?
Alt-text: Outstanding Questions
Acknowledgments
This work was supported by the National Institutes of Health R01 Grants EY029426, AI139768, EY024710 (to D.S.) and a National Eye Institute (NEI) core Grant (EY001792).
References
- 1.Hadfield J., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu D., et al. Vaccines and therapies in development for SARS-CoV-2 infections. J. Clin. Med. 2020;9:1885. doi: 10.3390/jcm9061885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmons G., et al. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du L., et al. The spike protein of SARS-CoV – a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romano M., et al. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia H., et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. (Cambridge) 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei X., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conti P., et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34:327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 11.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y., et al. CT manifestations of two cases of 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295:208–209. doi: 10.1148/radiol.2020200280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh S., et al. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535.E14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poduri R., et al. Drugs targeting various stages of the SARS-CoV-2 life cycle: Exploring promising drugs for the treatment of Covid-19. Cell. Signal. 2020;74:109721. doi: 10.1016/j.cellsig.2020.109721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- 18.Bouhaddou M. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadjadj J., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco-Melo D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karki R., et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell (Cambridge) 2020:31542–31547. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang J., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu A., et al. SARS-CoV-2 Spike protein promotes hyper-inflammatory response that can be ameliorated by Spike-antagonistic peptide and FDA-approved ER stress and MAP kinase inhibitors in vitro. bioRxiv. 2020 doi: 10.1101/2020.09.30.317818. Published online October 1, 2020. [DOI] [Google Scholar]
- 24.Dosch S.F., et al. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142:19–27. doi: 10.1016/j.virusres.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L., et al. SARS-CoV-2 spike protein binds heparan sulfate in a length- and sequence-dependent manner. bioRxiv. 2020 doi: 10.1101/2020.05.10.087288. Published online May 10, 2020. [DOI] [Google Scholar]
- 26.Clausen T.M., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057.E15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esko J.D., Lindahl U. Molecular diversity of heparan sulfate. J. Clin. Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park P.J., Shukla D. Role of heparan sulfate in ocular diseases. Exp. Eye Res. 2013;110:1–9. doi: 10.1016/j.exer.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakkar N., et al. Emerging roles of heparanase in viral pathogenesis. Pathogens. 2017;6:43. doi: 10.3390/pathogens6030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mp T., et al. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surjit M., et al. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia H., et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong S., et al. A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019-nCoV) J. Med. Virol. 2020;92:1542–1548. doi: 10.1002/jmv.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narayanan K., et al. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C., et al. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wathelet M.G., et al. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angeletti S., et al. COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92:584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornillez-Ty C.T., et al. Severe acute respiratory syndrome coronavirus nonstructural protein 2 Interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seaman M.N.J., et al. Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol. 2013;23:522–528. doi: 10.1016/j.tcb.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuen C., et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X., et al. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan L., et al. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290:3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S., et al. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016;17:678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fehr A.R., et al. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7 doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oostra M., et al. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graham R.L., et al. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133:88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakai Y., et al. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan J.F., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinto B.G.G., et al. ACE2 Expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. J. Infect. Dis. 2020;222:556–563. doi: 10.1093/infdis/jiaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu P., et al. NOS1 inhibits the interferon response of cancer cells by S-nitrosylation of HDAC2. J. Exp. Clin. Cancer Res. 2019;38:483. doi: 10.1186/s13046-019-1448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dewe J.M., et al. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol. Cell. Biol. 2017;37 doi: 10.1128/MCB.00214-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benvenuto D., et al. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020;81:e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cottam E.M., et al. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.te Velthuis Aartjan J.W., et al. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subissi L., et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. PNAS. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mbonye U.R., et al. Phosphorylation of HEXIM1 at Tyr271 and Tyr274 promotes release of P-TEFb from the 7SK snRNP complex and enhances proviral HIV gene expression. Proteomics. 2015;15:2078–2086. doi: 10.1002/pmic.201500038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Astuti I. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Egloff M., et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. PNAS. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miknis Z.J., et al. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 2009;83:3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouvet M., et al. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., et al. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015;89:8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mirza M.U., Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J. Pharm. Anal. 2020;10:320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma Y., et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. PNAS. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naqvi A.A.T., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hillen H.S., et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 67.Peng Q., et al. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Q., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruan Z., et al. SARS-CoV-2 and SARS-CoV: Virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12) J. Med. Virol. 2021;93:389–400. doi: 10.1002/jmv.26222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seybert A., et al. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5'-to-3' polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanner J.A., et al. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jang K., et al. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci. Rep. 2020;10:4481. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adedeji A.O., et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang C., et al. The SARS coronavirus nucleocapsid protein – forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eckerle L.D., et al. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y., et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhardwaj K., et al. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen S., et al. Extended ORF8 gene region is valuable in the epidemiological investigation of severe acute respiratory syndrome-similar coronavirus. J. Infect. Dis. 2020;222:223–233. doi: 10.1093/infdis/jiaa278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang H., et al. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhardwaj K., et al. The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. J. Virol. 2012;86:4294–4304. doi: 10.1128/JVI.07012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Castano-Rodriguez C., et al. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9:2325. doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan X., et al. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar P., et al. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366:293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yount B., et al. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frieman M., Baric R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol. Mol. Biol. Rev. 2008;72:672–685. doi: 10.1128/MMBR.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krull S., et al. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29:1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hogan R.J., et al. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 2004;78:11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li J., et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv. 2020 doi: 10.1101/2020.05.24.111823. Published online May 24, 2020. [DOI] [Google Scholar]
- 90.Tan Y., et al. Novel immunoglobulin domain proteins provide insights into evolution and pathogenesis of SARS-CoV-2-related viruses. mBio. 2020;11 doi: 10.1128/mBio.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gong Y., et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microbes Infect. 2020;9:1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Muth D., et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8:15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Su Y.C.F., et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11 doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kakhki R.K., et al. COVID-19 target: A specific target for novel coronavirus detection. Gene Rep. 2020;20:100740. doi: 10.1016/j.genrep.2020.100740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi C., et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kanzawa N., et al. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-κB activation. FEBS Lett. 2006;580:6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Siu K., et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim C. SARS-CoV-2 evolutionary adaptation toward host entry and recognition of receptor O-acetyl sialylation in virus–host interaction. Int. J. Mol. Sci. 2020;21:4549. doi: 10.3390/ijms21124549. [DOI] [PMC free article] [PubMed] [Google Scholar]