Abstract
Since human coronavirus (HCoVs) was first described in the 1960s, seven strains of respiratory human coronaviruses have emerged and caused human infections. After the emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), a pneumonia outbreak of coronavirus disease 2019 (COVID-19) caused by a novel coronavirus (SARS-CoV-2) has represented a pandemic threat to global public health in the 21st century. Without effectively prophylactic and therapeutic strategies including vaccines and antiviral drugs, these three coronaviruses have caused severe respiratory syndrome and high case-fatality rates around the world. In this review, we detail the emergence event, origin and reservoirs of all HCoVs, compare the differences with regard to structure and receptor usage, and summarize therapeutic strategies for COVID-19 that cause severe pneumonia and global pandemic.
Keywords: Human coronavirus, Reservoirs, Gene structure, Receptor usage, Therapeutic strategies
1. Introduction
Over the past 20 years, three previously unknown coronaviruses, SARS-CoV, MERS-CoV and SARS-CoV-2, have emerged [[1], [2], [3]]. The outbreak of COVID-19, caused by SARS-CoV-2, becomes a global public health emergency after the 1918H1N1 influenza pandemic [4]. Since the outbreak of COVID-19 later in 2019 in Wuhan city of China, the coronavirus has spread to 213 countries, areas or territories with nearly 3,000,000 confirmed cases and over 200,000 deaths all over the world as of April 27, 2020. While the other HCoVs induce mild upper respiratory diseases, the three highly pathogenic viruses attack the lower respiratory system in humans [5,6]. The current study demonstrates that the novel coronavirus emerged in 2019 is more transmissible than SARS-CoV in 2002 [7]. The lack of effective therapeutic strategies for COVID-19, which causes global pandemic and has high morbidity and mortality rates, highlights the need for vaccines and antiviral drugs [8]. In this review of the seven human coronaviruses, we detail the emergence events, summarize the origin and evolution, highlight the biological features comprised of gene structures, protein organizations, receptor usage and cell entry. At last, we discuss current knowledge of prophylactic and therapeutic strategies including vaccines development and drugs discovery for these three highly pathogenic coronaviruses and expect to bring valuable countermeasures against COVID-19 and novel coronaviruses emerged in the future.
Coronaviruses (CoVs) are enveloped viruses with a single-strand, positive-sense RNA genome approximately 26–32 kb in size, which is the largest known genome among all RNA viruses. The term ‘coronavirus’ refers to the appearance of CoV virions considering the protruding spike proteins on their surface that look like a crown under electron microscopy (“corona” means crown). The first coronavirus is an infectious bronchitis virus and was isolated from chicken embryos in 19,37 [9], along with subsequent viral isolations in rodents, domestic animals, and humans. Coronaviruses have been identified in many mammalian animals including humans and avian species and can induce various severe diseases involving respiratory, gastrointestinal, enteric, and neurological systems.
Coronaviruses belong to the subfamily Coronavirinae in the family Coronaviridae and the order Nidovirales. Coronaviridae are further subdivided phylogenetically into four genera Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. Alphacoronaviruses and Betacoronaviruses are found in mammals, whereas Gammacoronaviruses and Deltacoronaviruses are primarily found in birds. International Committee of Taxonomy of Viruses (ICTV) in 2018 claimed that Betacoronavirus lineage was reclassified into five subgenera, namely Embecovirus, Sarbecovirus, Merbecovirus, Nobecovirus, and Hibecovirus. HCoV-229 E in the subgenus Duvinacovirus and HCoV-NL63 in the subgenus Setracovirus belong to the Alphacoronavirus genus. HCoV-HKU1 and OC43 in the subgenus Embecovirus, MERS-CoV in the subgenus Merbecovirus, SARS-CoV and SARS-CoV-2 in the subgenus Sarbecovirus belong to the genus Betacoronavirus [10,11] (Fig. 1 ).
Fig. 1.
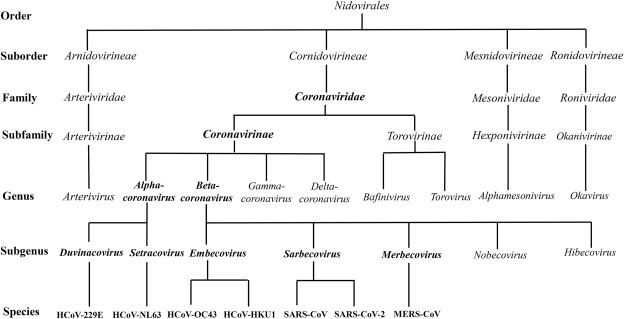
The taxonomy of the order Nidovirales. HCoV, human coronavirus; MERS, Middle East respiratory syndrome; SARS, severe acute respiratory syndrome.
2. Emergence and identification of HCoVs
The first human coronavirus was identified in 1965 in humans infected with the common cold [12]. The aetiology of the common cold was considered to be a bacterium before Doches et al. found that the viruses were likely causative agents in the 1930s (Fig. 2 ). By 1965, a substantial proportion of colds seemed not to be caused by known myxoviruses such as influenza and parainfluenza viruses. Strain B814 was the first human coronavirus isolated from a patient with a cold but it was lost in the laboratory.
Fig. 2.
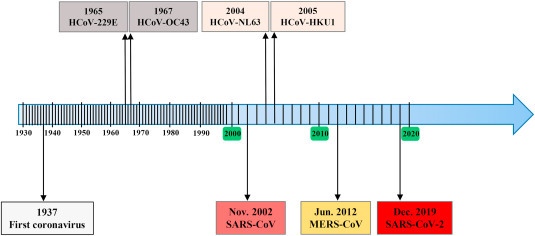
Timeline for the emergency of human coronaviruses. HCoV, human coronavirus; MERS, Middle East respiratory syndrome; SARS, severe acute respiratory syndrome.
2.1. HCoV-229E and OC43
HCoV-229 E was harvested from the respiratory tract in patients with the common cold by Hamre and Procknow in 1965 [13]. HCoV-OC43 was isolated from a nasopharyngeal washing patient by Mclntosh and his colleagues in 1967 [14]. HCoV-229 E and OC43 respectively survived in WI-38 lung fibroblast cells and human embryonic cell culture. Having been subsequently detected worldwide, these two coronaviruses present the same clinical symptoms including headache, sneezing, runny nose, and sore throat but rarely infections of the lower respiratory tract [5]. These infections are usually self-limiting and require no specific treatment or therapy.
2.2. HCoV-NL63 and HKU1
HCoV-NL63 was identified from the nasopharyngeal aspirate of an infant with bronchiolitis, fever, coryza, and conjunctivitis in Amsterdam, Netherlands in 2004 [15]. The patient had mild respiratory tract infection symptoms like the common cold (fever, cough, and rhinorrhea) [16]. HCoV-HKU1 was first identified from the nasopharyngeal aspirate in a Hong Kong patient with bronchiolitis and pneumonia in 2005 [17]. It’s challenging to study HKU1 considering that HKU1 viruses cannot be propagated in continuous cell lines. In 2013, Dijkman et al. isolated HKU1 viruses from clinical specimens by using primary, differentiated human tracheal bronchial epithelial cells cultured at an air-liquid interface [18]. HCoV-NL63 and HKU1 are widespread globally and associated with upper respiratory tract infections (URTI) including fever, running nose, and cough. Furthermore, they also cause mild to serious lower respiratory tract infections (LRTI) in infants and children with bronchiolitis and pneumonia [19].
2.3. SARS
Severe acute respiratory syndrome (SARS) first emerged in Foshan City, Guangdong, China in November 2002 (https://www.who.int/ith/diseases/sars/en/). In April 2003, SARS-CoV, a novel coronavirus which could cause an unusual epidemic of atypical pneumonia, was identified under global containment effort [1,20]. Being highly transmissible among human, SARS rapidly spread to other provinces in China causing significant outbreaks and then to other countries in Southeast Asia including Singapore and Vietnam in only 9 months [[21], [22], [23]]. SARS-CoV infected patients present early systemic symptoms with malaise, headache, myalgia, fever, dry cough, and shortness of breath, followed by respiratory distress generally and develop severe pneumonia by week 1–2 of illness, which may result in death. By July 2003, SARS-CoV had caused more than 8000 laboratory confirmed cases and 780 deaths with a 9.6% case fatality rate (CFR) in 29 countries (https://www.cdc.gov/sars/surveillance/absence.html). The first epidemic of SARS ended in July 2003, as announced by the World Health Organization (WHO) (www.who.int/csr/don/2003_07_05/en). Next year, five additional SARS cases from zoonotic transmission emerged without human SARS cases detected in Guangzhou City, China [24]. However, four SARS infected cases occurred with neither mortality nor further transmission [25].
2.4. MERS
A 60-year-old man presented a fatal pneumonia and acute renal failure and died in June of 2012 in Saudi Arabia. MERS-CoV, as a novel coronavirus, was identified from his sputum of lungs [2]. MERS-CoV caused diseases, ranging from asymptomatic to atypical pneumonia along with renal failure and gastrointestinal symptoms, are often fatal [26]. A typical infection caused by MERS-CoV first starts from fever, cough, and shortness of breath to pneumonia and dyspnea rapidly. As of January 15, 2020, 2506 totally confirmed cases of MERS, and 862 related deaths with a CFR of 34.4% were reported worldwide (https://www.who.int/csr/don/31-january-2020-mers-united-arab-emirates/en/). The majority of these cases were reported from Saudi Arabia with 2102 cases (CFR = 37.1%) according to the WHO. Because MERS-CoV does not transmit easily between humans unless there is close contact, lots of MERS cases were independent clusters and confined to the countries in the Middle East, especially in KSA (approximately 80%). Outside of this region, 27 countries have reported cases of MERS in African, Asia and Europe and the USA in person who went to the Middle East or was in contact with those who did.
2.5. COVID-19
On December 12, 2019, 27 cases (41 cases revised subsequently) of unknown cause of pneumonia was firstly reported by Wuhan Municipal Health Commission in Wuhan, Hubei Province, China [3]. Actually, the date of symptom onset of the first known case is December 1, 2019 basing on retrospective analyses of clinical data later [27]. A novel coronavirus was identified through unbiased sequencing of bronchoalveolar-lavage fluid from patients on December 21, 2019 [6] and the WHO was informed of the cases cluster on December 31, 2019. Twenty seven of 41 patients had a history of direct exposure to the Huanan seafood wholesale market where a number of non-aquatic animals such as poultry, birds, snakes, and other wildlife animals were on sale before the outbreak [27,28]. Studies have demonstrated person-to-person transmission via droplets or in contact with a patient directly [29]. Eight children were reported to be persistently positive testing on rectal swabs, suggesting the potential for fecal–oral transmission [30]. Typical symptoms of these infections include fever, dyspnea, muscle ache, dry cough, sore throat, and diarrhea. Disease onset includes bilateral pneumonia, multiple mottling and ground-glass opacity by transverse chest x-ray and CT images [31]. On January 8, 2020, a novel coronavirus was isolated from a Wuhan pneumonia patient and two days later the complete viral genome sequence were determined along with first fatal case reported [32]. The virus was tentatively named by WHO as the 2019 novel coronavirus (2019-nCoV) on January 12, 2020, eventually recognized by the Coronavirus Study Group of ICTV based on phylogeny, taxonomy and established practice as SARS-CoV-2 [33], which infected disease was named COVID-19 meanwhile by WHO on February 11, 2020. COVID-19 outbreak coincides with the Chinese Spring Festival, during which about 3 billion trips were made through China, with 15 million trips happening in Wuhan in 2020. As April 27, 2020, at total of 84,341 laboratory confirmed cases have been detected in China, including 4643 deaths (CRF = 5.5%). Infections in medical workers of more than 3000 cases and family clusters were also reported [34]. However, the number of COVID-19 cases was reversed, controlling the confirmed cases to 29,839 with the Wuhan travel ban and the national emergency response, 96% fewer than expected 744,000 cases in the absence of interventions by an epidemic model analysis [35]. SARS-CoV-2 is more infectious than the other two highly pathogenic coronaviruses, and can be transmitted in asymptomatic or presymptomatic infections. To date, SARS-CoV-2 has rapidly spread to 213 countries/territories/areas worldwide. Almost 3,000,000 confirmed cases and more than 200,000 deaths have been counted in the world, which is from situation report of WHO. Given how far the virus has spread and its devastating global impacts, WHO has declared the COVID-19 outbreak a global pandemic.
3. Origins and reservoirs of HCoVs
Bats are ancient and diverse mammals with the second largest number of species. Over 1000 species of bats belong to the Chiroptera order and traditionally include two suborders: the megabats and the microbats, which are mostly frugivorous and insectivorous respectively. Recently the classification is revised: Yinpterochiroptera and Yangochiroptera. Bats are recognized to be possible reservoirs to a wide variety of viruses, which could infect humans and domestic animal species [36]. These viruses include coronaviruses, henipaviruses, filoviruses, lyssaviruses and several highly pathogenic ones, such as Hendra virus, Nipah virus and Ebola virus [[37], [38], [39], [40]]. Bats have been recognized as the natural reservoirs of many human coronaviruses [41] (Table 1 ).
Table 1.
Comparison of host, genome features, receptor-binding site, and receptor of human coronaviruses.
| HCoV | Host |
Genome features |
Receptor-binding sites |
Receptor |
||||
|---|---|---|---|---|---|---|---|---|
| Natural | Intermediate | Size (nt) | GC (%) | RBD | RBM | Protein Determinant | Protein Determinant | |
| HCoV-229 E | Bats | Camelids | 27,240 | 38 | 417–547 | – | APN | |
| HCoV-NL63 | Bats | Unknown | 27,553 | 34 | 476–616 | – | --ACE2 | |
| HCoV-HKU1 | Rodents | Unknown | 29,926 | 32 | 15–302 | – | Unknown | 9-O-Ac-Sia |
| HCoV-OC43 | Rodents | Cattle | 30,738 | 37 | 15–302 | – | Unknown | 9-O-Ac-Sia |
| MERS-CoV | Bats | Camels | 30,119 | 41 | 367–606 | 484–567 | DPP4 | |
| SARS-CoV | Bats | Civets | 29,727 | 41 | 306–527 | 424–494 | ACE2 | |
| SARS-CoV-2 | Bats | Pangolins? | 29,903 | 38 | 333–527 | 438–506 | ACE2 | |
3.1. HCoV-229E, NL63, OC43 and HKU1
Firstly, natural reservoirs of HCoV-229 E and NL63 were recently found in African bats. HCoV-229 E-like viruses, whose RNA-dependent RNA polymerases fragment had a 92% identity with HCoV-229 E, were found in Hipposideros bats [42] and the intermediate reservoirs of HCoV-229 E are likely camelids [43]. HCoV-NL63-like viruses have been found in Triaenops afer bats [44,45]. Unlike Alphacoronavirus genus and other Betacoronavirus subgenus, HCoV-OC43 and HKU1 from the subgenus Embecovirus seem more likely to originate from rodents [46,47]. Furthermore, agriculturally important animals include cattle or swine are thought to be intermediate hosts of HCoV-OC43 [48,49].
3.2. SARS-CoV
SARS-CoV was initially identified in Himalayan palm civets (Pagkuma larvata), raccoon dogs (Nyctereutes procyonoides) and Chinese ferret badgers (Melogale moschata) considering that early SARS cases contacted with animals from live animal markets in Guangdong, China [50]. However, large-scale epidemiological research soon revealed civets were not the natural hosts of SARS-CoV, which was not found in the wild or domestic civets without live animal market exposure [40,51]. In 2005, novel SARS-related coronaviruses (SARSr-CoVs), known as SARS-like coronaviruses (SL-CoVs) were found in horseshoe bats (Rhinolophus genus) in China. Genome sequences of these bat SL-CoVs manifested nearly 90% identity to human or civet SARS-CoV [40,52]. In addition, RsSHC014 and Rs3367, as two novel SL-CoVs found in 2013, were found in horseshoe bats [53]. SARS-CoV is possible origin from recombination of different SL-CoVs because of frequent RNA recombination within coronaviruses. So far, most bat SL-CoVs were found in a variety of horseshoe bats species (genus Rhinolophus of suborder Yinpterochiroptera) [50]. Bats are now generally thought to be natural reservoirs and civets and other small carnivore are intermediate hosts of SARS-CoV.
3.3. MERS-CoV
Most early MERS infected patients were independent clusters and only appear in the Middle East countries, especially in Saudi Arabia. Some cases were reported to contact with dromedary camels, a major livestock species in the Middle East [26,54]. The whole genomic sequences of human MERS-CoVs and camels in the Middle East have a 99% identity [[55], [56], [57]]. Moreover, serum samples collected from camels in the Middle East, Africa and Asia were positive for antibodies against MERS-CoV [56,58]. There’s a lot of evidence that camels are the primary zoonotic hosts for MERS-CoV, however, subsequent work suggests bats are the ancestral reservoir host [[59], [60], [61]]. Based on genomic sequence analysis, MERS-CoV and bat CoVs belong to the Merbecovirus subgenus, which have been detected from a variety of bats in suborder Yangochiroptera of Betacoronavirus genus [62]. Bat coronaviruses (BatCoVs), including HKU4 and HKU5 prior to the emergence of MERS-CoV, were discovered in various bat species that belong to the Vespertilionidae family [63]. Since the emergency of MERS, MERS-related coronaviruses (MERSr-CoVs) were identified in various bat species and countries [[64], [65], [66]]. Taken together, dromedary camels may have important roles as intermediate reservoirs for person to person transmission and bats are likely the ancestral hosts for MERS-CoV.
3.4. SARS-CoV-2
COVID-19, an emerging coronavirus infection induced by SARS-CoV-2, may originate in Huanan seafood wholesale market. About a month later after COVID-19 outbreak, Chinese scientists published new coronavirus genome sequence information, revealing that the virus exhibited an 89.1% identity to SL-CoVs which originated from bats. Phylogenetic analysis suggested that the novel coronavirus clustered with members of the subgenus Sarbecovirus of genus Betacoronavirus , including the SARS-CoV and SL-CoVs from bats [32]. Meanwhile, another research group obtained whole genome sequences from five patients and found that the virus shares a 96.2% identity to a bat coronavirus, RaTG13, which was discovered from horseshoe bats (Rhinolophus affinis) in Yunnan Province [67]. However, the ~4% difference means 17% divergence at the neutral sites because the genomic average dS value is 0.17, suggesting the large divergence between SARS-CoV-2 and RaTG13 [68]. Furthermore, the receptor binding domain (RBD) of SARS-CoV-2 is only ~85% identical and only share one of six critical residues with RBD of RaTG13 [69]. SARS-CoV-2 had 79.5% sequence similarity to SARS-CoV and formed a distinct lineage compared with SL-CoVs based on phylogenetic tree [67]. SARS-CoV-2 contains a polybasic cleavage site insertion between the two subunits of the spike protein, which is also observed in RmYN02 sampled in another Rhinolophus bat from Yunnan province [67,70]. It cannot be proved that emergence of SARS-CoV-2 comes from a recent recombination event [71], but recombination may have happened before 2009, when the SARS-CoV-2 ancestors in bats first acquired genetic characteristics of SARS by incorporation of a SL-CoV RBD [72].
Bats are thought to be likely native reservoirs for SARS-CoV-2, but bat-derived coronavirus rarely infect humans directly without an intermediate host considering the contradictions that COVID-19 emerged during the hibernation of bats. It seems to be likely that the novel virus is probably transmitted to humans by another intermediate host, as previously described coronaviruses. Pangolins are likely the intermediate host of the SARS-CoV-2 by full-length genome sequence comparison with coronaviruses from Malayan pangolins. The spike protein of the pangolin-CoV, which is responsible for receptor binding domain, is virtually identical to that of SARS-CoV-2, with one amino acid difference [73,74]. Metagenomic sequencing identified a novel pangolin-CoV, which was isolated from SL-CoVs positive Malayan pangolins during 2017–2018, with approximately 85.5%–92.4% similarity to SARS-CoV-2 [75]. Prior to the emergence of COVID-19, SL-CoVs had been detected in two dead Malayan pangolins with a frothy liquid in lung [76]. The order Pholidota includes the eight living species of pangolins worldwide (Manis. javanica, Manis. pentadactyla, Manis. crassicaudata. Manis. culionensis. Manis. tricuspis, Manis. tetradactyla, Manis. gigantea and Manis. temminckii) [77]. All the eight pangolin species are regarded as critically endangered because of the huge demand for people in food and medicines. The Huanan seafood wholesale market where wild animals including Malayan pangolins were sold, was initially thought to be the origin of COVID-19, considering that SARS-CoV-2 was identified in initial patients in contact with the market. Therefore, pangolins could be one of the potential intermediate reservoirs for SARS-CoV-2. Further research is needed to confirm pangolins as an intermediate host and find other potential intermediate reservoirs.
4. Genome structure and organization
Coronaviruses particles consist of four or five structural proteins along with various minor components including nonstructural proteins (nsp). Human coronaviruses have a similar genome structure and protein organization, including an open reading frame ORF1a/b to encode 16 nonstructural proteins (nsp1 through nsp16). The large overlapping polyproteins, ORF1a and ORF1b, commonly referring to pp1a and pp1b, comprise approximately 2/3 of the genome and encode the replicase polyprotein. These polyproteins are cleaved by papain-like cysteine protease (PLpro, resides within nsp3) and 3C-like serine protease (3CLpro, also known as main protease Mpro, resides within nsp5) to produce nsps, including RNA-dependent RNA polymerases (RdRp, resides within nsp12) and helicase (Hel, resides within nsp13) shown in Fig. 3 . The remaining 1/3 of the genome mainly encodes four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N), and some also encode a hemagglutinin esterase (HE) protein which are encoded by other ORFs at the 3′ polyadenylated (Fig. 3a). The HE protein is a glycoprotein with neuraminate O-acetyl-esterase activity and the active site FGDS, is present downstream to ORF1a/b and upstream to S gene. The S protein is responsible for the characteristic crown-like appearance because of the spike present on the surface of CoVs and the most variable sequences for binding receptor and entry to the host cells. The E protein is a small hydrophobic integral membrane protein and critical for virus assembly. The M protein is associated with the envelope in all coronaviruses to induce membrane curvature. The N protein is a nonspecific RNA-binding protein that forms the ribonucleocapsid with viral genomic RNA [[78], [79], [80]].
Fig. 3.
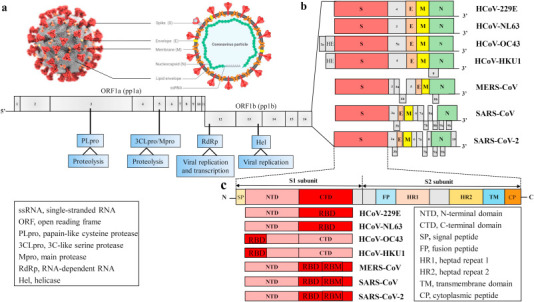
Genomes and structures of human coronaviruses. (a) The ultrastructural morphology of coronavirus on the left illustration (the Centers for Disease Control and Prevention, CDC) and right virus particle. (b) The genomes of HCoVs contain a single-stranded, positive-sense RNA (ssRNA) genome of 27–32 kb in size. The 5′-terminal ORF1a/b within two-thirds of the genome encodes two large polyproteins 1a (pp1a) and pp1b. These polyproteins are cleaved by PLpro and 3CLpro, also known as Mpro, to produce non-structural proteins (nsps), including RdRp and Hel, which are important enzymes involved in the transcription and replication. The 3′ one-third of genome encodes four structural proteins: Spike (S), Membrane (M), Envelope (E) and Nucleocapsid (N), which are essential for virus-cell receptor binding and virion assembly, and other non-structural proteins along with a set of accessory proteins unique to each virus species. Some coronaviruses express an additional structural protein, hemagglutinin-esterase (HE). (c) The S protein of HCoVs consists of S1 subunit and S2 subunit. The S1 region contains an NTD and a CTD (also referred to as the RBD), whereas the S2 region includes a TM region, FP, HR1 and HR2. The HE glycoprotein consists of two functional domains: a corresponding sialate O-acetylesterase domain and an O-acetylated sialic acid binding domain. HCoV-OC43 and HKU1 bind 9-O-Ac-Sia via NTD of S1 subunit and mediate sialate-O-acetylesterase activity by RDE domain of HE protein. Other coronaviruses mediate attachment to the cellular receptors by RBD (CTD) in S1 region. Herein, we compare and contrast genome structures of seven identified HCoVs strains HCoV-229 E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, MERS-CoV, SARS-CoV and SARS-CoV-2.
4.1. HCoV-229E and OC43
The RNA genome of HCoV-229 E is about 27,240 nucleotides, with a poly(A) tail. The GC content is nearly 38%. The laboratory strains of HCoV-229 E have eight putative protein-coding genes with the characteristic gene order ORF1a, ORF1b, HE, S, ORF4a, ORF4b, E, M, and N, while there are eight in the subsequent clinical strains present as an intact ORF4 [[81], [82], [83]] (Fig. 3b). For HCoV-229 E clinical strain, transcription regulatory sequence (TRS) motif is present in 3′ end of the leader sequence and upstream of each structural gene. The TRS core structure is UCUCAACU for leader, S and E gene, UCUAAACU for the M and N gene, and UCAACU for ORF4 gene. The first complete genome sequence of the HCoV-OC43 comprises 30,738 nucleotides, with a poly(A) tail. The GC content is 37%, with the characteristic gene order ORF1a, ORF1b, NS2a, HE, S, NS5a, E, M, and N [48,84] (Fig. 3b). HE protein reinforces the ability of HCoV-OC43 to infect because of its acetyl-esterase activity [85]. The TRS core structure is UCUAAAC for leader, NS2a and S gene, UCUUAAG for the NS5a and E gene, UUAAAC for HE gene, UCCAAAC for M gene and UCUAAA for N gene [86].
4.2. HCoV-NL63 and HKU1
The HCoV-NL63 genome encompasses 27,553 nucleotides, with a poly(A) tail. The GC content is 34%, very low GC contents among all HCoVs. The genome order is ORF1a, ORF1b, S, ORF3, E, M, and N. Similar to HCoV-229 E, only one ORF protein is located between the S and E genes [87,88]. It was reported that the accessory gene was structural N-glycosylated and virion-incorporated for cell infection [89]. The TRS core sequence of the virus is defined as AACUAAA for the most of ORFs except the E gene for which the TRS is 5′-AACUAUA-3′ [88,90]. The RNA genome of HCoV-HKU1 is about 29,926 nucleotides, with a poly(A) tail. HCoV-HKU1 has the lowest GC contents (32%) among all HCoVs. Similar to the genome organization of HCoV-OC43, the genome order is ORF1a, ORF1b, HE, S, ORF4, E, M, N and ORF8 [87,91] (Fig. 3b). The TRS core sequence of HCoV-HKU1 is 5′-AAUCUAAAC-3′ located upstream of all the ORFs except the E gene which may share the same TRS with ORF417. Like HCoV-OC43, HE protein of HCoV-HKU1 may have an acetyl-esterase activity as receptor-destroying enzymes (RDE) by using sialic acids [92].
4.3. SARS-CoV
The RNA genome of SARS-CoV encompasses 29,727 nucleotides, and the GC content is 41%, with a poly(A). The characteristic gene order of SARS-CoV is ORF1a, ORF1b, S, ORF3a, ORF3b, E, M, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, N, ORF9a and ORF9b [93,94]. The exact function of these nonstructural accessory proteins is unclear. The ORF8 protein sequences of SARS-CoV isolated from early-phase patients is full length [25]. However, The ORF8 derived from the mid- and late-phase SARS-CoV infected patients contains ORF8a and 8 b because of a 29-nucleotide deletion, which leads the split of complete ORF8 into putative ORF8a and 8 b [95,96]. Recently, Lau et al. revealed that the ORF8 protein of SARS-CoV, which was likely acquired from SL-CoVs by recombination, may be responsible for animal-to-human transmission [97]. The N-glycosylation sites for ORF3a and ORF8b and O-glycosylation sites for ORF3a and ORF9b were predicted using the NetNGlyc server [98]. A study found a putative TRS core leader sequence is 5′-CUAAAC-3′ excluding ORF1a and ORF1b [93]. Another group reported that 5′-AAACGAAC-3′ (genomic nucleotides 65 to 72), was present upstream of ORF1a and five other ORFs but not including ORF1b and E [94]. Soon, Thiel and colleagues isolated a conserved sequence (5′-ACGAAC-3′) located in front of nine predicted ORFs (ORF1a, S, ORF3a, E, M, ORF6, ORF7a, ORF8a and N) [99,100].
4.4. MERS-CoV
The RNA genome of MERS-CoV is about 30,119 nucleotides in length and the GC content is 41%, with a poly(A) tail. The characteristic gene order of SARS-CoV is ORF1a, ORF1b, S, ORF3, ORF4a, ORF4b, ORF5, E, M, ORF8b, and N [101,102]. Although the exact function of the accessory proteins including ORF3, ORF4a, ORF4b, ORF5 and ORF8b, is still unclear, these proteins have been reported to play an important role in viral replication. Deletion-mutants of ORF3, ORF4a, ORF4b, or ORF5 are attenuated for MERS-CoV replication in vitro [103,104]. Further studies suggest that these accessory proteins are involved in evading the host immune system, such as ORF4a/4 b and ORF5 proteins as strong IFN antagonists [105,106]. The TRS core (5′-AACGAA-3′) are located at the 3′ end of the leader sequence and at different positions upstream of genes in the genomic 3′-proximal domain of MERS-CoV [107].
4.5. SARS-CoV-2
The genome of SARS-CoV-2 is nearly 30,000 nucleotides, and the GC content is about 38%, with a poly(A). The characteristic gene order is ORF1a, ORF1b, S, ORF3a/3 b, E, M, ORF6, ORF7a/7 b, ORF8, N, ORF9a, ORF9b and ORF1032,[[108], [109], [110]] (Fig. 3b). As with SARS-CoV, SARS-CoV-2 has a predicted ORF8 gene, without known functional domain or motif, located between the M and N genes. But, the ORF8 genome sequences are more similar to those of bat SARSr-CoVs ZXC21 and ZC45 than ORF8 derived from SARS-CoV [111]. A high conservation was observed for structural proteins E, M and A across the Betacoronavirus genus, while accessory proteins ORF8 seem to have much stricter evolutionary constraints [112]. The ORF8 protein has a high possibility to form a protein with an alpha-helix, following with a beta-sheet containing six strands based on a secondary structure prediction [111]. The TRS core structure of SARS-CoV-2 is CUAAAC for ORF1a/b, S, M, ORF8, N and ORF10 gene, ACGAAC for the ORF3, E, ORF6 and ORF7 [113]. To date, high-resolution crystal structures of SARS-CoV-2 proteins, including PLpro, Mpro, RdRp and S proteins, have been determined by research groups from all over the world (Suppl Table 1).
5. Receptor recognition and cell entry
The S glycoprotein, which is responsible for the protruding “spikes” like a crown under electron microscopy, binds receptor of host cells and determines the species tropism. The S protein consists of two functionally regions: the S1 unit contains an N-terminal domain (NTD) and a C-terminal domain (CTD, namely RBD), whereas the S2 unit includes a transmembrane region, fusion peptide (FP), and heptad repeats (HR1 and HR2) [114,115] (Fig. 3c). The S proteins mediate attachment to the cellular receptors by RBD domain in S1 region and subsequent fusion of the virus-cell membrane by transmembrane fusion domain in S2 region [116,117]. The difference in receptor usage among these coronaviruses is attributed to diversity in local architecture and receptor-binding site accessibility from S protein [118,119].
5.1. APN for HCoV-229 E and ACE2 for HCoV-NL63
Aminopeptidase N (APN) is a type II glycoprotein that belongs to the family of membrane-bound metalloproteases. The APN glycoprotein is expressed in a variety of tissues, including epithelial cells from the renal proximal tubules, intestinal brush border and the respiratory tract, cells of the monocytic and granulocytic lineage, synaptic membranes of the central nervous system [120]. Human aminopeptidase N (hAPN/CD13/ANPEP) is reported to be a receptor for HCoV-229 E [121]. The RBD of HCoV-229 E spike protein is located between residues 417 and 547, [122] (Table 1). Although HCoV-229 E has been proved to take an endosomal pathway for cell entry [123], the virus preferred cell-surface TMPRSS2 to endosomal cathepsin to enter cell [124]. Angiotensin converting enzyme 2 (ACE2) is a type I transmembrane protein that belongs to the angiotensin-converting enzyme family. ACE2 protein is expressed in various human organs, including lung alveolar epithelial cells, pneumocytes and enterocytes of the small intestine, and arterial and venous endothelial cells and smooth muscle cells [125,126]. HCoV-NL63 is found to employ ACE2 as a receptor for cellular entry in 2002 [127]. The minimal RBD of the HCoV-NL63 S protein is located between residues 476 and 616,[128] (Table 1). HCoV-NL63 was observed to enter the cell by clathrin-mediated endocytosis [129].
5.2. 9-O-Ac-Sia for HCoV-OC43 and HKU1
Sialic acids (Sias) are acidic nine-carbon sugars that commonly cap the glycan chains of cell surface glycoproteins and glycolipids. 9-O-acetylated sialic acid (9-O-Ac-Sia), as an attachment receptor determinant, can be carried by sialoglycan-based receptors [130]. The S protein of HCoV-OC43 and HKU1 employ 9-O-Ac-Sia residues on glycoproteins to initiate the infection of host cells [131,132]. The HE protein consists of two functional units: a corresponding sialate O-acetylesterase domain and an O-acetylated sialic acid binding domain [133]. However, the HE protein of these two coronaviruses has not exhibited O-acetylated sialic acid binding activity [132]. In contrast, the HE protein possesses an acetyl-esterase activity recognized by the S protein, which plays a role as an RDE [85,134]. In conclusion, HCoV-OC43 and HKU1 early utilize 9-O-Ac-Sia as a receptor determinant by the S protein to initiate the infection of host cells and late mediates sialate-O-acetylesterase RDE activity by the HE protein. Furthermore, the S protein of HCoV-OC43 and HKU1 was recently reported to bind 9-O-Ac-Sia via a domain A of S1 subunit (S1A) [135,136]. Similar to HCoV-229 E, OC43 and HKU1 also liked the cell-surface TMRRSS2 rather than endosomal cathepsins for cell entry [124].
5.3. DPP4 for MERS-CoV
Dipeptidyl peptidase 4 (DPP4), which is known as CD26, is a cell surface glycoprotein of 110 kDa. As a type II transmembrane protein, DPP4 is ubiquitously expressed by a variety of cells and is cleaved of the cell membrane in a process called shedding [137]. It is reported that DPP4 is a functional receptor for the MERS-CoV [138,139]. The initial S protein RBD of MERS-CoV is located between residues 367 and 606,[140] (Table 1). The MERS-CoV S1 domain contains a core structure and an accessory subdomain receptor-binding motif (RBM, residues 484–567) (Fig. 3). The core structure is a five-stranded antiparallel β sheet (β1, β2, β3, β4 and β9) with two short α helices in the connecting loops (α1 and α2). The RBM is a four-stranded antiparallel β sheet (β5, β6, β7 and β8) [141] (Fig. 4 a and b). The flexible RBD region in MERS-CoV S proteins is located between residues 381 and 588, which is not contradictory to previous study. In addition, two states of the RBD (lying state and standing state) are captured by a high-resolution structure of the trimeric MERS-CoV S protein by cryo-EM. The dynamic RBD is facilitated to be recognized by DPP4 [142]. MERS-CoV gains entry into host cells by direct fusion at the plasma membrane by TMPRSS2. In the absence of TMPRSS2, MERS-CoV is endocytosed, and can be triggered by cathepsin L to complete viral entry [143].
Fig. 4.
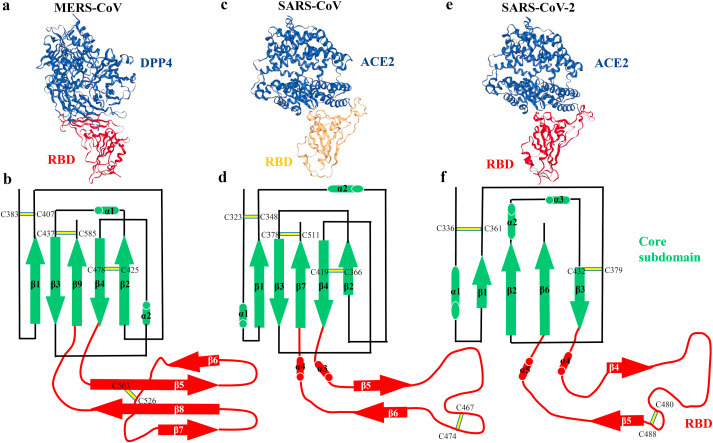
The structural comparison of RBD bound with receptor and RBD subdomains in the three coronaviruses. (a) Overall structure of RBD bound with DPP4 (PDB ID: 4KR0), and (b) schematic illustration topology of the core structure and RBM in the MERS-CoV RBD. (c) Overall structure of RBD bound with ACE2 (PDB ID: 2AJF), and (d) schematic illustration topology of the core structure and RBM in the SARS-CoV RBD. (e) Overall structure of RBD bound with ACE2 (PDB ID: 6M0J), and (f) schematic illustration topology of the core structure and RBM in the SARS-CoV-2 RBD. β strands are drawn as arrows and α helices are drawn as cylinders. The disulfide bonds are drawn as yellow sticks. The core subdomain is colored in green and the receptor-binding subdomain is colored in red.
5.4. ACE2 for SARS-CoV
SARS-CoV utilizes ACE2 as its functional entry receptor, similar to HCoV-NL63 [144]. A minimal RBD, which is located in the S1 region (residues 306–527) (Table 1), has greater affinity to bind ACE2 than does a longer fragment (residues 12–672) [145,146]. The SARS-CoV RBD also includes a core structure and an accessory subdomain RBM (residues 424–494) in direct binding ACE2145. The core subdomain is a five-stranded antiparallel β sheet (β1, β2, β3, β4 and β7) with two short α helices in the connecting loops (α1 and α2). The RBM subdomain is a two-stranded antiparallel β sheet (β5 and β6) with two short connecting α helices (α3 and α4) in the loops [141,147] (Fig. 4c and d). Particularly, two sites in RBM (residues 479 and 487), are responsible for SARS disease progression and SARS-CoV tropism. Mutations in RBM residues 479 and 487 have an effect on transmission of SARS-CoV [145,148]. A recent study reported that the flexible RBD region (residues 318–513) in SARS-CoV S protein presents two states of the RBD (lying state and standing state), similar to MERS-CoV mentioned previously [142]. A large of studies have proved that SARS-CoV use multiple pathways for host cell entry, including direct fusion with the plasma membrane by TMPRSS2, cathepsin L-mediated endocytosis, and clathrin- and caveolae-independent endocytosis [[149], [150], [151], [152]].
5.5. ACE2 for SARS-CoV-2
ACE2 is the functional surface receptor for SARS-CoV-2 that is causing the COVID-19 [153,154]. ACE2 expression in human airway epithelial cells is enhanced by interferon and influenza, which could be exploited by SARS-CoV-2 [155]. Gut enterocytes was also infected by the novel coronavirus because of ACE2 high expression in the intestinal epithelium [156]. SARS-CoV-2 has an RBD (residues 333–527) located in the S1 protein to engage ACE2, nearly identical to that of the SARS-CoV RBD (Table 1). The SARS-CoV-2 RBD also contains a core structure and an accessory subdomain RBM (residues 438–506) that interacts directly with ACE2157. The core subdomain is a twisted four-stranded antiparallel β sheet (β1, β2, β3 and β6) with two short α helices in the connecting loops (α1, α2 and α3). The RBM subdomain is a two-stranded antiparallel β sheet (β4 and β5) with two short connecting α helices (α4 and α5) in the loops [157] (Fig. 4e and f). The overall amino acid sequence identities are around 76%–78% for the S protein, around 73%–76% for the RBD, and 50%–53% for the RBM between SARS-CoV-2 and SARS-CoV [158,159]. A recent study uncovers cryo-EM structure of the full-length human ACE2, which presents two conformations (open and closed) of ACE2 [160]. SARS-CoV-2 RBD takes a more compact conformation than SARS-CoV RBD to bind ACE2, which enhances its ACE2-binding affinity [161]. Compared with SARS-CoV, ACE2 binding to SARS-CoV-2 S ectodomain has a 10- to 20-fold stronger affinity [162]. Another research group demonstrated that SARS-CoV-2 RBD binding to ACE2 displayed 4-fold higher affinity than the SARS-RBD and a mutation in ACE2 amnio acid (K353) was sufficient to abolish the binding [163]. A recent study reported that SARS-CoV-2 gained entry into cells via a new receptor CD147, suggesting the diversity of binding receptor for the virus [164]. The cellular serine protease TMPRSS2 primes SARS-CoV-2 S for entry into host cells [154]. Proprotein convertase furin preactivation of the SARS-CoV-2 S protein is helpful for cell entry and evading immune surveillance [165]. The expression of ACE2 and TMPRSS2 is higher in nasal epithelial cells than other tissues, suggesting nasal epithelial cells are the loci of initial infection [166]. Ou et al. recently revealed that SARS-CoV-2 invades hACE2 overexpressed 293 cells mainly through endocytosis [167].
6. Therapeutic strategies for HCoVs
Currently, there are no clinical treatments or prevention strategies, such as approved antiviral drugs or vaccines, for highly pathogenic coronaviruses. Severe HCoV-infected patients mainly receive supportively medical care, along with different drugs combination treatments. Several strategies have been used to treat infections with SARS-CoV, MERS-CoV and SARS-CoV-2 [168]. This part summarizes the progress of therapeutic agents and vaccines for these three highly pathogenic coronaviruses caused diseases, as described below.
6.1. Virus targeted treatments
Nucleoside analogues target the viral RdRp to block RNA synthesis and have a broad-spectrum activity against a wide range of CoVs [169,170]. Ribavirin is a guanosine analog with broad-spectrum activity against RNA viruses [171]. Although ribavirin was used to treat patients infected by SARS- and MERS- CoV, it may be associated with many side effects such as hemolytic anemia and cardiorespiratory distress at high doses [8]. So far it is unclear whether ribavirin treatment could improve the clinical outcome of COVID-19 disease. Remdesivir (GS-5734, Gilead Science) is a prodrug of the adenine derivative and broadly against MERS-CoV, SARS-CoV and Ebola virus [[172], [173], [174]]. A COVID-19 patient from the USA recovered after treatment with intravenous remdesivir [175]. Soon a study demonstrated that remdesivir combined with chloroquine effectively suppresses SARS-CoV-2 in vitro [176]. Two phase III trials have begun from February to April in 2020 [177]. Recent clinical data shown 68% severe COVID-19 patients (36/53) had an improvement in oxygen-support class after treatment with compassionate-use remdesivir [178]. In addition, the cryo-EM structure of the RdRp complexed with a template-primer RNA and remdesivir has been determined [179]. Lopinavir and ritonavir were protease inhibitors for HIV treatment initially [180]. Lopinavir-ritonavir targeted coronavirus nonstructural protein 3CLpro and had antiviral activity against MERS-CoV and SARS-CoV [181,182]. Clinical trials of lopinavir and ritonavir have been initiated in SARS-CoV-2 infected patients. However, a recent study shows that lopinavir–ritonavir treatment has not significantly accelerated clinical improvement, reduced mortality, or diminished viral RNA detectability in severe COVID-19 patients [183]. EIDD-2801, a prodrug of EIDD-1931 (NHC), is a ribonucleoside analog with broad-spectrum activity against a variety of viruses such as influenza and Ebola. Sheahan et al. recently shown that EIDD-1931 is highly active against SARS-CoV-2 in primary human airway epithelial cell cultures and SARS- or MERS-CoV infected mice [184]. To date, the crystal structures of SARS-CoV-2 RdRp and Mpro (3CLpro) have been reported, which is helpful for design of new antiviral drugs targeting RdRp and Mpro of coronaviruses [[185], [186], [187], [188]].
6.2. Host targeted treatment
The host innate interferon (IFN) response is crucial for the control of viral replication after infection, which can be augmented by the supplementation of recombinant interferons [189]. A variety of IFN types have been administrated to patients with SARS- and MERS-CoV, typically in combination with broad-spectrum antivirals such as ribavirin or lopinavir–ritonavir. IFNα was proven to be effective in a small number of cases in combination with ribavirin [190]. IFNα2a, IFNα2b and IFNβ1b, in combinations with ribavirin or lopinavir–ritonavir, were also used to treat patients with MERS-CoV [182,191,192]. However, IFNs are not recommended as first-line agents considering their side effects including anemia, fatigue and depression. Additionally, Blanco-Melo et al. recently found that interferons type I and III innate response to infection with SARS-CoV-2 is limited to inhibit virus replication [193]. Corticosteroids act as immunomodulatory agents and have an anti-inflammatory effect. Corticosteroids in combinatory therapies with ribavirin or IFN had no favorable response in patients with MERS-CoV [8]. Although IFNα1 plus corticosteroids was used to be treated SARS-CoV associated disease, it was reported that the SARS-CoV infected patient, who received prolonged treatment with corticosteroids treatment, died of aspergillosis [194,195]. Furthermore, recent clinical study does not support corticosteroid treatment for COVID-19 patients with lung injury [196].
6.3. Antibody and plasma therapy
Monoclonal antibodies (mAbs) have potentially therapeutic effects on combating highly pathogenic viral diseases, by neutralizing structural proteins on coronaviruses [197]. Convalescent plasma or other preparations (monoclonal, hyperimmune globulin) that possess neutralizing antibodies have been the most worth consideration of therapies for SARS- and MERS-CoV [198,199]. Although several studies found that mAbs targeting SARS- and MERS-CoV inhibited viral replication and ameliorated related disease in vitro or in animal models [[200], [201], [202]], large scale clinical trial involved in convalescent plasma or mAbs against SARS- and MERS-CoV are lacking [203]. Given the urgency of the 2019-nCoV outbreak and time-consuming for new interventions, it is worth to assess the existing neutralizing monoclonal antibody in the treatment of SARS- and MERS-CoV [204]. A SARS-CoV-specific human monoclonal antibody (CR3022) was first reported to have a potential binding with the RBD of SARS-CoV-2 (KD of 6.3 nM) [205]. The crystal structure of RBD complexed with CR3022 has shown that the drug targets a highly conserved epitope (Suppl Table 1). Ju et al. recently reported the isolation of over 200 RBD-specific mAbs derived from COVID-19 patients and found two potent antibodies, P2C–1F11 and P2B–2F6, out-competed ACE2 with close to 100% efficiency [206]. Meanwhile, Shi et al. also isolated two human mAbs (CA1 and CB6) from a convalescent COVID-19 patient and found the latter inhibited SARS-CoV-2 infection in rhesus macaques, suggesting that it has significant potential to serve as a therapeutic agent for COVID-19 [207]. A novel human monoclonal antibody (47D11) exhibited cross-neutralizing activity of SARS-CoV-2 and SARS-CoV by binding a conserved epitope on the spike RBD [208]. Wrapp et al. described that single-domain camelid antibodies can neutralize pathogenic Betacoronaviruses including emerging SARS-CoV-2 [209]. Hence, convalescent plasma, which is from COVID-19 patients who have recovered, and monoclonal antibody offer the possibility of prevention and therapy for emerging coronaviruses caused diseases including COVID-19 in the future [210]. Besides, a recent study demonstrated that clinical grade human recombinant soluble ACE2 can inhibit SARS-CoV-2 infection in Vero-E6 cells and human capillary and kidney organoids, suggesting a new approach to prevent SARS-CoV-2 infection [211].
6.4. Vaccines
Vaccines are considered the gold standard for highly pathogenic coronaviruses prevention and treatment. There are no currently approved vaccines for human coronaviruses caused infection, because of the requirement of many years’ efforts to develop a new vaccine [[212], [213], [214]]. Multiple vaccination strategies targeting SARS- and MERS-CoV are in the early stages, including inactivated virus vaccines, live-attenuated virus vaccines, viral vector vaccines, subunit vaccines and DNA/protein vaccines [215,216]. To date, more than 300 vaccine developments disclose the potential methodologies to treat and prevent coronavirus caused infections including COVID-19168. An animal model is necessary for vaccine development to test the effectiveness of vaccines in vivo. Bao et al. established a mouse model by using the hACE2 transgenic animal infected by SARS-CoV-2, which may facilitate the development of vaccines against the virus [217]. Another animal model has been reported that rhesus macaques was infected using SARS-CoV-2 isolated from clinical bronchoalveolar lavage fluid. Rhesus macaques infected by SARS-CoV-2 showed acute localized-to-widespread pneumonia already reported in COVID-19 patients, without obvious clinical symptoms of respiratory disease [218]. Meanwhile, another research group found that conjunctival infected-rhesus macaques presented high viral load and distribution in the nasolacrimal system [219]. Recently, Rockx et al. found SARS-CoV-2 causes COVID-19-like disease in cynomolgus macaques [220]. These animal models are especially important for testing preventive and therapeutic strategies including broad vaccines and drugs against highly pathogenic coronaviruses. Gao et al. recently developed an inactivated vaccine and found its protective immunity and neutralization activities against the novel coronavirus in rhesus macaques [221]. To date, the first COVID-19 vaccine, which is a recombinant adenovirus type-5 (Ad5) vectored COVID-19 vaccine expressing the S protein of SARS-CoV-2, underwent a phase 1 clinical trial and was found to be safe, well-tolerated and able to generate an immune response against SARS-CoV-2 in humans [222].
7. Outlook and conclusions
In the first 20 years of this century, we have witnessed three outbreaks of previously unknown highly pathogenic coronaviruses, SARS-CoV, MERS-CoV and SARS-CoV-2. COVID-19 caused by the novel coronavirus has become a global pandemic and is still continuing presently. The collected data focus on understanding the pathology and epidemiology of human coronaviruses, including pathogen transmission, the reservoir and/or intermediate hosts, and virus biology. Before the next emergency of a novel highly pathogenic coronavirus, there are many lessons still to be learned for us. For prevention, the broad-spectrum, pan-coronavirus vaccines should be developed. To prevent virus of zoonotic sources from getting into the human population, the barriers between natural reservoirs and human society must be maintained. For diagnosis, the rapid identification of the full-length genome of emerging novel coronavirus by next-generation meta-transcriptomic sequencing made it possible to design quick and accurate diagnostic assays. Zhang et al. recently developed an AI system to quickly diagnose SARS-CoV-2 by using Computed Tomography (CT) [223]. For isolation, the most effective way to prevent infections and save lives is breaking human-to-human transmission. A recent report summarizes five different non-pharmaceutical interventions, including case isolation in the home, voluntary home quarantine, social distancing of those over 70 years old, social distancing of entire population, and closure of schools and universities [224]. For therapies, with the development of high-resolution crystal structures of coronavirus proteins (Suppl Table 1) and the establishment of animal models aiming at coronaviruses, developing pan-CoV antiviral drugs against current and future emerging coronaviruses have become the ultimate therapeutic strategies.
The continuing epidemic threat of SARS-CoV-2 to global health told us that humans live in a ‘‘Global Village,’’ where an infectious disease emerging in any corner of the world has the potential to disseminate globally. In mind of ‘‘One Health, One World’‘, we should foster collaborative and multisectoral effects of trans-disciplines to achieve optimal health for people, animals, and the environment [225].
Acknowledgements
This work was supported by the National Natural Science Foundation of China (11902020, 31370018, 11421202, and 11827803), China Postdoctoral Science Foundation (No. 2019M660390), the National Key R&D Program of China (2017YFA0506500, 2016YFC1102203, and 2016YFC1101100), and Fundamental Research Funds for the Central Universities (ZG140S1971).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.medntd.2020.100043.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Peiris J.S.M., Phil D., Yuen K.Y., Osterhaus A.D.M.E., Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 2.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/s0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kupferschmidt K., Cohen J. Will novel virus go pandemic or be contained? Science. 2020;367:610–611. doi: 10.1126/science.367.6478.610. [DOI] [PubMed] [Google Scholar]
- 5.Su S., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang B., et al. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect Dis Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham C., Stuart H. Cultivation of the virus of infectious bronchitis of chickens in embryonated chicken eggs. Am J Vet Res. 1947;8:209–212. [PubMed] [Google Scholar]
- 10.Steffen I., Simmons G. Coronaviruses. eLS. 2015:1–9. doi: 10.1002/9780470015902.a0023611. [DOI] [Google Scholar]
- 11.Cong Y., Verlhac P., Reggiori F. The interaction between nidovirales and autophagy components. Viruses. 2017;9:182–196. doi: 10.3390/v9070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myint S. Human coronaviruses: a brief review. Rev Med Virol. 1994;4:35–46. doi: 10.1002/rmv.1980040108. [DOI] [Google Scholar]
- 13.Hamre D., Procknow J. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 14.McIntosh K., Dees J., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respirator disease. Proc Natl Acad Sci U S A. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Hoek L., Pyrc K., Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol Rev. 2006;30:760–773. doi: 10.1111/j.1574-6976.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der hoek L., et al. Identification of a new coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo P.C., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dijkman R., et al. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J Virol. 2013;87:6081–6090. doi: 10.1128/JVI.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo P.C., Lau S.K., Yip C.C., Huang Y., Yuen K.Y. More and more coronaviruses: human coronavirus HKU1. Viruses. 2009;1:57–71. doi: 10.3390/v1010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jsm P., et al. Coronavirus as a possible cause of severe acute respiratory syndrome. The Journal of Tepecik Education and Research Hospital. 2003;13:55–56. doi: 10.5222/terh.2003.26734. [DOI] [Google Scholar]
- 21.Drosten C., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 22.Ksiazek T.G., et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 23.Lee N., et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 24.Wang M., et al. SARS-CoV infection in a restaurant from palm civet. Emerg Infect Dis. 2006;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song H.-D., et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haagmans B.L., et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/s1473-3099(13)70690-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:135–143. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H. Early lessons from the frontline of the 2019-nCoV outbreak. Lancet. 2020;395:687. doi: 10.1016/s0140-6736(20)30356-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020 doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China - key questions for impact assessment. N Engl J Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 32.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coronaviridae Study Group of the International Committee on Taxonomy of, V. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5, 536-544, doi:10.1038/s41564-020-0695-z (2020). [DOI] [PMC free article] [PubMed]
- 34.Chan J.F.-W., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian H., et al. An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in China. Science. 2020;31:eabb6105. doi: 10.1126/science.abb6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan Y., Zhao K., Shi Z.L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11:210–224. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu B., Ge X., Wang L.F., Shi Z. Bat origin of human coronaviruses. Virol J. 2015;12:221–231. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee A., Kulcsar K., Misra V., Frieman M., Mossman K. Bats and coronaviruses. Viruses. 2019;11:41–56. doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 41.Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfefferle S., et al. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corman V.M., et al. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao Y., et al. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol. 2017;91 doi: 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huynh J., et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lau S.K., et al. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J Virol. 2015;89:3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W., et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vijgen L., et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vijgen L., et al. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guan Y., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 51.Tu C., et al. Antibodies to SARS coronavirus in civets. Emerg Infect Dis. 2004;10:2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lau S., et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ge X.Y., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alagaili A.N., et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5 doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Briese T., et al. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. mBio. 2014;5 doi: 10.1128/mBio.01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reusken C.B.E.M., et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/s1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perera R.A.P.M., et al. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:20574–20581. doi: 10.2807/1560-7917.ES2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- 58.Corman V.M., et al. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992-2013. Emerg Infect Dis. 2014;20:1319–1322. doi: 10.3201/eid2008.140596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Memish Z.A., et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anthony S.J., et al. Further evidence for bats as the evolutionary source of middle east respiratory syndrome Coronavirus. mBio. 2017;8 doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chastel C. Middle East respiratory syndrome (MERS): bats or dromedary, which of them is responsible? Bull Soc Pathol Exot. 2014;107:69–73. doi: 10.1007/s13149-014-0333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong A.C.P., Li X., Lau S.K.P., Woo P.C.Y. Global epidemiology of bat coronaviruses. Viruses. 2019;11 doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woo P.C., Lau S.K., Li K.S., Tsang A.K., Yuen K.Y. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg Microb Infect. 2012;1:e35. doi: 10.1038/emi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang L., et al. MERS-related betacoronavirus in Vespertilio superans bats, China. Emerg Infect Dis. 2014;20:1260–1262. doi: 10.3201/eid2007.140318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Annan A., et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu D.K.W., et al. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci U S A. 2018;115:3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang X., et al. On the origin and continuing evolution of SARS-CoV-2. National Science Review. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV2. Cell. 2020 doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coutard B., et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176:104742–104747. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paraskevis D., et al. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212–104216. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patiño-Galindo J.Á., Filip I., AlQuraishi M., Rabadan R. 2020. Recombination and convergent evolution led to the emergence of 2019 Wuhan coronavirus bioRxiv. [DOI] [Google Scholar]
- 73.Xiao K., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 74.Liu P., et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lam T.T.-Y., et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 76.Liu P., Chen W., Chen J.P. Viral metagenomics revealed sendai virus and coronavirus infection of Malayan pangolins (Manis javanica) Viruses. 2019;11:979–994. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaudin T.J., Emry R.J., Wible J.R. The phylogeny of living and extinct pangolins (mammalia, pholidota) and associated taxa: a morphology based analysis. J Mamm Evol. 2009;16:235–305. doi: 10.1007/s10914-009-9119-9. [DOI] [Google Scholar]
- 78.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dijkman R., et al. Human coronavirus 229E encodes a single ORF4 protein between the spike and the envelope genes. Virol J. 2006;3:106–114. doi: 10.1186/1743-422X-3-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thiel V., Herold J., Schelle B., Siddell S. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- 83.Farsani S.M., et al. The first complete genome sequences of clinical isolates of human coronavirus 229E. Virus Gene. 2012;45:433–439. doi: 10.1007/s11262-012-0807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lau S.K., et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Desforges M., Desjardins J., Zhang C., Talbot P.J. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J Virol. 2013;87:3097–3107. doi: 10.1128/JVI.02699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.St-Jean J.R., et al. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pyrc K., Berkhout B., van der Hoek L. The novel human coronaviruses NL63 and HKU1. J Virol. 2007;81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdul-Rasool S., Fielding B. Understanding human coronavirus HCoV-NL63. Open Virol J. 2010;4:76–84. doi: 10.2174/1874357901004010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Müller M., et al. Human coronavirus NL63 open reading frame 3 encodes a virion-incorporated N-glycosylated membrane protein. Virol J. 2010;7:6–18. doi: 10.1186/1743-422X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol J. 2004;1:7–18. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dominguez S.R., et al. Isolation, propagation, genome analysis and epidemiology of HKU1 betacoronaviruses. J Gen Virol. 2014;95:836–848. doi: 10.1099/vir.0.059832-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marra M.A., et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 94.Rota P.A., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 95.Oostra M., de Haan C.A., Rottier P.J. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J Virol. 2007;81:13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lau S.K., et al. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J Virol. 2015;89:10532–10547. doi: 10.1128/JVI.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stadler K., et al. SARS--beginning to understand a new virus. Nat Rev Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thiel V., et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 100.Snijder E.J., et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off Ffrom the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/s0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cotten M., et al. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/s0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Woo P.C., et al. Novel betacoronavirus in dromedaries of the Middle East, 2013. Emerg Infect Dis. 2014;20:560–572. doi: 10.3201/eid2004.131769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scobey T., et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Almazan F., et al. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio. 2013;4 doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang Y., et al. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Canton J., et al. MERS-CoV 4b protein interferes with the NF-kappaB-dependent innate immune response during infection. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Boheemen S., et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3 doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cui H., et al. Structural genomics and interactomics of 2019 Wuhan novel coronavirus, 2019-nCoV, indicate evolutionary conserved functional regions of viral proteins. bioRxiv. 2020 doi: 10.1101/2020.02.10.942136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taiaroa G., et al. Direct RNA sequencing and early evolution of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.05.976167. [DOI] [Google Scholar]
- 110.Wu A., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chan J.F., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kirchdoerfer R.N., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alsaadi E., Jones I. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Walls A.C., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gallagher T., Perlman S. Public health: broad reception for coronavirus. Nature. 2013;495:176–177. doi: 10.1038/495176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Walls A.C., et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kolb A., Hegyi A., Siddell S. Identification of residues critical for the human coronavirus 229E receptor function of human aminopeptidase N. J Gen Virol. 1997;78:2795–2802. doi: 10.1099/0022-1317-78-11-2795. [DOI] [PubMed] [Google Scholar]
- 121.Yeager C., et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bonavia A., Zelus B.D., Wentworth D.E., Talbot P.J., Holmes K.V. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J Virol. 2003;77:2530–2538. doi: 10.1128/jvi.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J Virol. 2017;91 doi: 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hamming I., et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kuba K., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hofmann H., et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lin H.X., et al. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD-ACE2 receptor interaction. J Gen Virol. 2008;89:1015–1024. doi: 10.1099/vir.0.83331-0. [DOI] [PubMed] [Google Scholar]
- 129.Milewska A., et al. Entry of human coronavirus NL63 into the cell. J Virol. 2017;92 doi: 10.1128/JVI.01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Baumann A.M., et al. 9-O-Acetylation of sialic acids is catalysed by CASD1 via a covalent acetyl-enzyme intermediate. Nat Commun. 2015;6:7673–7685. doi: 10.1038/ncomms8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci U S A. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang X., et al. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zeng Q., Langereis M., Vliet A., Huizinga E., de Groot R. Structure of coronavirus hemagglutinin-esterase offers insight into Corona- and Influenza virus evolution. Proc Natl Acad Sci U S A. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bakkers M.J., et al. Betacoronavirus adaptation to humans involved progressive loss of hemagglutinin-esterase lectin activity. Cell Host Microbe. 2017;21:356–366. doi: 10.1016/j.chom.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hulswit R.J.G., et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tortorici M.A., et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lambeir A.M., Durinx C., Scharpe S., De Meester I. Dipeptidyl-peptidase IV from bench to bedside: an update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Crit Rev Clin Lab Sci. 2003;40:209–294. doi: 10.1080/713609354. [DOI] [PubMed] [Google Scholar]
- 138.Raj V.S., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Letko M., et al. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 2018;24:1730–1737. doi: 10.1016/j.celrep.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lu G., et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang N., et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yuan Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092–15101. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Qing E., Hantak M.P., Galpalli G.G., Gallagher T. Evaluating MERS-CoV entry pathways. Methods Mol Biol. 2019;(2020):9–20. doi: 10.1007/978-1-0716-0211-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li W., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li F., Li W., Farzan M., Harrison S. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 146.Wong S.K., Li W., Moore M.J., Choe H., Farzan M.A. 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Du L., et al. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Qu X.X., et al. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J Biol Chem. 2005;280:29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hofmann H., Pohlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Matsuyama S., et al. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang I.C., et al. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang H., et al. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ziegler C.G.K., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020 doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lamers M.M., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. eabc 1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 158.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yan R., et al. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020 doi: 10.1126/science.abb2762. eabb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang Q., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang K., et al. 2020. SARS-CoV-2 invades host cells via a novel route CD147-spike protein bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shang J., et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020 doi: 10.1073/pnas.2003138117. 202003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sungnak W., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020 doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ou X., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620–1632. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu C., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020 doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Clercq E.D. Antiviral activity spectrum and target of action of different classes of nucleoside analogues. Nucleosides and Nucleotides. 1994;13:1271–1295. doi: 10.1080/15257779408012151. [DOI] [Google Scholar]
- 170.De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Graci J.D., Cameron C.E. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol. 2006;16:37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Menachery V.D., et al. SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sheahan T.P., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sheahan T.P., et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:222–236. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Holshue M.L., et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang, M. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30, 269-271, doi:10.1038/s41422-020-0282-0 (2020). [DOI] [PMC free article] [PubMed]
- 177.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 178.Grein J., et al. Compassionate use of remdesivir for patients with severe covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yin W., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. eabc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Walmsley S., et al. Lopinavir–ritonavir versus nelfinavir for the initial treatment of HIV infection. N Engl J Med. 2002;346:2039–2046. doi: 10.1056/NEJMoa012354. [DOI] [PubMed] [Google Scholar]
- 181.Chu C.M., et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chan J.F., et al. Treatment with lopinavir/ritonavir or interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cao B., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe COVID-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sheahan T.P., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020 doi: 10.1126/scitranslmed.abb5883. eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gao Y., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jin Z., et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 187.Zhang L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 doi: 10.1126/science.abb3405. eabb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dai W., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020 doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Menachery V.D., et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5 doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhao Z., et al. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol. 2003;52:715–720. doi: 10.1099/jmm.0.05320-0. [DOI] [PubMed] [Google Scholar]
- 191.Omrani A.S., et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis. 2014;14:1090–1095. doi: 10.1016/s1473-3099(14)70920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Falzarano D., et al. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Blanco-Melo D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Loutfy M.R., et al. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. J Am Med Assoc. 2003;290:3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- 195.Wang H., et al. Fatal aspergillosis in a patient with SARS who was treated with corticosteroids. N Engl J Med. 2003;349:507–508. doi: 10.1056/NEJM200307313490519. [DOI] [PubMed] [Google Scholar]
- 196.Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473–475. doi: 10.1016/s0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Totura A.L., Bavari S. Broad-spectrum coronavirus antiviral drug discovery. Expert Opin Drug Discov. 2019;14:397–412. doi: 10.1080/17460441.2019.1581171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Soo Y., et al. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin Microbiol Infect. 2004;10:676–678. doi: 10.1111/j.1469-0691.2004.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Arabi Y.M., et al. Feasibility of using convalescent plasma immunotherapy for MERS-CoV infection, Saudi Arabia. Emerg Infect Dis. 2016;22:1554–1561. doi: 10.3201/eid2209.151164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhu Z., et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Jiang L., et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med. 2014;6:234ra259. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 202.Pascal K.E., et al. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci U S A. 2015;112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.de Wit E., van Doremalen N., Falzarano D., Munster V.J., Sars, Mers Recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Shanmugaraj B., Siriwattananon K., Wangkanont K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pac J Allergy Immunol. 2020 doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 205.Tian X., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microb Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ju B., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 207.Shi R., et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 208.Wang C., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wrapp D., et al. Structural basis for potent neutralization of betacoronaviruses. Cell. 2020 doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020 doi: 10.1016/s1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Monteil V., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Piot P., et al. Immunization: vital progress, unfinished agenda. Nature. 2019;575:119–129. doi: 10.1038/s41586-019-1656-7. [DOI] [PubMed] [Google Scholar]
- 213.Lu S. Timely development of vaccines against SARS-CoV-2. Emerg Microb Infect. 2020;9:542–544. doi: 10.1080/22221751.2020.1737580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang N., Tang J., Lu L., Jiang S., Du L. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res. 2015;202:151–159. doi: 10.1016/j.virusres.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wang L., et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun. 2015;6:7712–7723. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bao L., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 218.Shan C., et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Res. 2020 doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Deng W., et al. Rhesus macaques can be effectively infected with SARS-CoV-2 via ocular conjunctival route. bioRxiv. 2020 doi: 10.1101/2020.03.13.990036. [DOI] [Google Scholar]
- 220.Rockx B., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020 doi: 10.1126/science.abb7314. eabb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gao Q., et al. Rapid development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhu F.-C., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020 doi: 10.1016/s0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang K., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements and prognosis of COVID-19 pneumonia using computed Tomography. Cell. 2020 doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ferguson N.M., et al. Impact of non-pharmaceutical interventions (NPIs) to reduce COVID19 mortality and healthcare demand. Imperial College COVID-19 Response Team. 2020 doi: 10.25561/77482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lebov J., et al. A framework for One Health research. One Health. 2017;3:44–50. doi: 10.1016/j.onehlt.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


