Abstract
The rapid identification of pathogenic microorganisms plays a crucial role in the timely diagnosis and treatment strategies during a global pandemic, especially in resource-limited area. Herein, we present a sensitive biosensor strategy depended on botulinum neurotoxin type A light chain (BoNT/A LC) activated complex assay (BACA). BoNT/A LC, the surrogate of BoNT/A which embodying the most potent biological poisons, could serve as an ultrasensitive signal reporter with high signal-to-noise ratio to avoid common strong background response, poor stability and low intensity of current biosensor methods. A nanoparticle hybridization system, involving specific binding probes that recognize pathogenic 16S rRNAs or SARS-CoV-2 gene site, was developed to measure double-stranded biotinylated target DNA containing a single-stranded overhang using Fluorescence Resonance Energy Transfer (FRET)-based assay and colorimetric method. The method is validated widely by six different bacteria strains and severe acute respiratory related coronavirus 2 (SARS-CoV-2) nucleic acid, demonstrating a single cell or 1 aM nucleic acid detecting sensitivity. This detection strategy offers a solution for general applications and has a great prospect to be a simple instrument-free colorimetric tool, especially when facing public health emergency.
Keywords: Bacterial diagnostic, SARS-CoV-2, Botulinum toxin, Fluorescence resonance energy transfer, Colorimetric detection, Biosensors
1. Introduction
Pathogenic microorganisms pose a massive threat to public health safety (Sun et al., 2020; Zhou et al., 2020). The rapid diagnosis is vital for diseases diagnosis and treatment to contain the epidemic, especially pandemic. However, traditional culture-based methods are usually time-consuming and tedious (Forne et al., 2000; Guan et al., 2013). Currently, novel biosensors based diagnostic strategies have been developed with an improved sensitivity and accuracy (Ren et al., 2019; Song et al., 2019; Yoo and Lee, 2016; Zhang et al., 2020). Those sensors include (Hong et al., 2020; Wan et al., 2018; Wang and Alocilja, 2015; Wu et al., 2015) enzyme-based reporters [horseradish peroxidase (HRP) (Wen et al., 2016), alkaline phosphatase (ALP) (Han et al., 2020), and catalase (de la Rica and Stevens, 2012)], fluorescent molecule-based reporters [Fluorescein probes (Cardoso et al., 2011) and Molecular Beacons (Cao et al., 2014)] and nanomaterial-based reporters [magneto-DNA nanoparticle system (Chung et al., 2013), Gold nanoparticles (Yang et al., 2020)]. Although enzyme-based reporters shortened analysis time in many experiments, their sensitivity is limited due to strong background response (Seo et al., 2015). The functionalized fluorescent molecule-based reporters provide an excellent way as a rapid on-site tool for pathogenic diagnostics. However, their poor stability and low intensity restrict the further utilization (Tsourkas et al., 2002). Most nanomaterial-based reporters require complex operations and specialized training (Naja et al., 2007). Thus, there is an urgent need for a sensitive signal reporter with a high signal-to-noise ratio that can be used in the general pathogen detection.
Currently, Corona Virus Disease 2019 (COVID-19) caused by SARS-COV-2 virus has become a global pandemic, with over 1,730,000 confirmed deaths and 1,730,000 confirmed cases as of December 2020. Gold-standard assays using real-time reverse transcriptase–polymerase chain reaction (RT-PCR) of patient samples, such as blood, urine, stool and nasopharyngeal swabs, have been employed to confirm the presence of SARS-COV-2. But they are not portable and require complicated laboratory manipulations. Although serology tests using lateral flow assay (LFA) is rapid and need minimal infrastructure, their application may not be suitable for the confirmation of acute COVID-19, due to the long window stage (1–2 weeks) for patients to obtain a detectable antibody level. To accelerate clinical diagnostic testing for SARS-COV-2, a novel diagnostic test for the virus should be developed in a user friendly way (Fabiani et al., 2021; Miripour et al., 2020; Seo et al., 2020).
Botulinum toxin (BoNT), produced by Clostridium botulinum and related strains, has attracted significant attention in bioanalytical chemistry due to their extraordinary neurotoxic and ultra strong catalytic characters. It functions by cleaving Soluble NSF Attachment Protein Receptor (SNARE) required for neurotransmitter release. Depending on the protein structure and sequence differences, BoNTs can be classified into eight types (A-H) of (Barash and Arnon, 2014; Chen and Barbieri, 2006; Dennis et al., 2000). Among them, BoNT/A is the most toxic serotype which causes severe symptoms via specifically cleaving synaptosome-associated protein of 25 kDa (SNAP-25) (Blasi et al., 1993). It is the most poisonous substance known: a single gram of the toxin can kill one million people (Dhaked et al., 2010). The light chain of BoNT/A (BoNT/A LC) is a surrogate of BoNT/A and retains the same catalytic activity (Caglic et al., 2014). So far, there has been no report for utilization of BoNTs as signal reporters in analytical chemistry.
Inspired by its catalytic properties for the cleavage of SNAP-25, we established pathogenic and SARS-COV-2 detection technique based on BoNT/A LC activated complex assay (BACA). Combined with bacterial multiple 16S rRNA strands and magnetic enrichment, BACA enabled to significantly improve the detection sensitivity. In this assay, the amplified bacterial/viral targets were captured by streptavidin magnetic beads (MB) and followed by recruitment of BoNT/A LC modified gold nanoparticle detection probes (BNPs) decorated via engineered gold binding peptide (GBP)-tagged BoNT/A LC. After magnetic separation, the generated BoNT/A LC functionalized target-probe nanocomplexes activated the fluorescence quenching substrates to produce fluorescence signals. Meanwhile, the target phenotypes could be further determined through a colorimetric biosensor. In this work, BACA demonstrated high specificity and excellent sensitivity. It has been used for bacteria detection and rapid detection of SARS-CoV-2.
2. Material and methods
2.1. Preparation of tag overhung targets
Escherichia coli (CICC 10412), Eberthella typhi (CICC 21483), Pseudomonas aeruginosa (CICC 21625), Staphylococcus aureus (CICC 10384), Streptococcus pyogenes (CICC 10373) and Enterococcus faecalis (CICC 10396) were provided from the China Center of Industrial Culture Collection (CICC). The 16S rRNA sequences of 6 different bacterial genus types were downloaded from NCBI database and aligned to choose corresponding unique regions (50–80 nucleotides in length) as detection target sequences using MegAlign software (DNAStar). Each pathogenic RNA was extracted using Bacterial RNA Kit (Omega) and cDNA was obtained using PrimeScript II 1st Strand cDNA Synthesis Kit (Takara). The double-stranded biotinylated target DNA containing a single-stranded TAG (TAGCGCGACCATAGTGAAGAAATA) overhang was then amplified by DreamTaq™ Hot Start DNA Polymerase (Thermo) with a TAG-containing forward primer and a 5’ biotinylated reverse primer through universal PCR (Deshpande and White, 2012).
While for SARS-CoV-2 (Genbank Accession NO.MN908947.3), the recombinant plasmid containing full-length N gene was obtained from Tiandz Biotech as our positive COVID-19 mimic target due to clinic and laboratory limitations. The published primer sets designed by Chinese Center For Disease Control And Prevention (CDC) were used in this work. BACA was tested to reveal its potential for rapid and effective detection of SARS-CoV-2 nucleic acid. All primers and probes (Table S1) were synthesized by Sangon Biotech and primer specificity was analyzed by agarose gel electrophoresis.
2.2. Probe conjugations
To fabricate BoNT/A LC modified gold nanoparticle detection probes (BNP), 20 μL of 100 μM thiol-DNA in 30 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Sigma-Aldrich) buffer, 2 μL of 0.2 M NaH2PO4 at pH 5.2, and 2 μL of 30 mM TCEP were mixed and reacted at 37 °C for 1 h followed by 176 μL of 10 mM PB at pH 7.4. The freshly prepared oligonucleotides were put into gold nanoparticles (1 OD/1 mL) and were incubated at a 37 °C. Then, 10 mM PB buffer containing 2 M NaCl was gradually added to the solution every 30 min until a final concentration of 500 mM was reached. Then the reaction mixture was incubated in overnight. To remove extra thiol-DNA, the DNA-conjugated gold nanoparticles (DNP) were washed 3 times with PB buffer. The number of oligonucleotides that successfully modified onto the DNP was quantified using the Qubit® ssDNA Assay Kit (Invitrogen by Thermo Fisher Scientific, USA). BNP probes were fabricated as follow: the DNA-modified DNPs (10 nM, 0.2 mL) were reacted with excess GBP-tagged BoNT/A LC (0.3 μM, 0.2 mL) for 0.5 h at 37 °C. Then it was collected and resuspended with reaction buffer (50 mM HEPES, pH 7.4, 500 mM NaCl, 0.05% TWEEN20) for later usage. And the amount of BoNT/A LC conjugated onto the BNP was estimated using the SNAPtide (FITC/DABCYL, List Biological Laboratories, Campbell, CA).
2.3. BACA assay for bacteria and SARS-CoV-2 nucleic acid detection
To gather the amplified tag overhung targets, 30 μL of streptavidin magnetic beads (2 mg/mL, New Englands Biolabs) was added to 40 μL of PCR amplicons and incubated for 30 min at 37 °C followed by magnetic separation and washing with PB buffer. Subsequently, 70 μL of BNP probes that dually decorated with anti-TAG sequences and engineered GBP-tagged BoNT/A LC were added. After magnetic separation, the target-probe complexes were washed with reaction buffer (50 mM HEPES, pH 7.4, 500 mM NaCl, 0.05% TWEEN20) for two times and finally suspended with cleavage buffer (50 mM HEPES, pH 7.4, 2.3 M TMAO, 0.05% TWEEN20). The BoNT/A substrate SNAPtide (FITC/DABCYL) was then added to the assay with a final concentration of 5 μM. Reactions (10 μL, 384-well microplate format) were incubated in a fluorescence plate reader (BioTek H1 microplate reader) for up to 180 min at 37 °C with fluorescence read every 5 min (SNAPtide reporter = λex: 490 nm; λem: 523 nm) as a fluorometric biosensor. Meanwhile, the color change of supernatant could be observed to the naked eyes or measured by UV–vis spectrophotometer at 520 nm.
3. Results and discussion
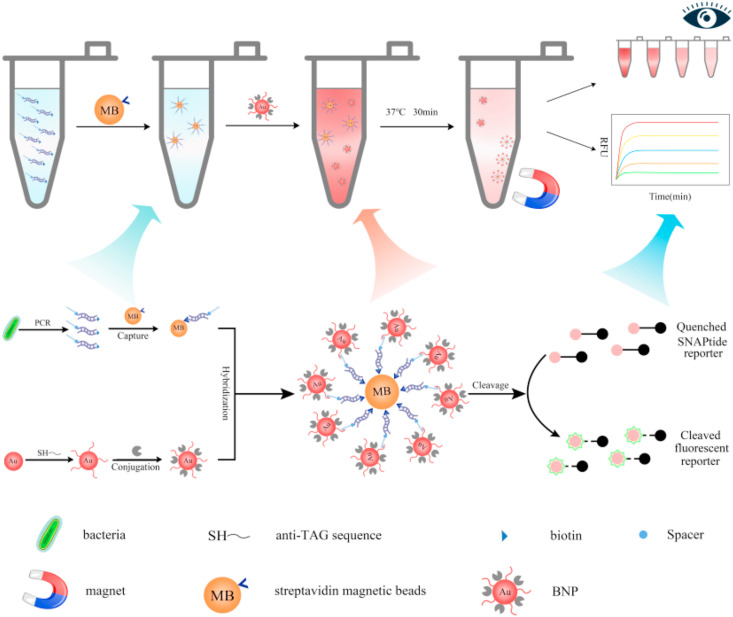
Motivated by the increasing severe situation of pathogenic outbreaks, we draw here the evolution and validation of BACA assays to sense these pathogenic nucleic acids. The strategy of BACA for bacterial and viral detection is illustrated in Fig. 1 . The advantage of BACA is the design of BoNT/A LC catalysis reaction by addition of SNAPtide substrate to amplify the signal of FRET-based assay as a biosensing signal mechanism (Bagramyan et al., 2008; Lucas et al., 2015; Simpson, 2003). In detail, the double-stranded biotinylated target DNA containing a single-stranded TAG overhang was firstly obtained by RT-PCR using a TAG-containing forward primer and a 5’ biotinylated reverse primer. Taking advantage of streptavidin magnetic beads, the sequence-specific targets were separated, and the overhanging tag allowed hybridization to anti-TAG probe sequence that modified on BNPs which had already been conjugated to BoNT/A LC. The specific binding of the double-stranded biotinylated target via sandwich hybridization reaction would result in the presence of engineered BoNT/A LC via magnetic separation. Sequentially, the engineered BoNT/A LC in the sandwich complexes cleaved the SNAPtide substrate to generate fluorescence signals. The signal of BACA was directly correlated with amplified target derivative as a fluorometric biosensor. Meanwhile, the color change of the assay combined with magnetic separation could be observed by the naked eyes without any additional instruments or complicated statistical analysis or by UV–vis spectra. Thus, this technique could not only dramatically decrease test cost, but also provide quick and precise diagnosis, making it well fit for utilization in source-limit area.
Fig. 1.
BoNT/A LC activated complex assay (BACA) for bacterial or virus diagnosis. The target nucleic acid was amplified through universal polymerase chain reaction (PCR). Sequence-specific amplicons were then captured by streptavidin magnetic beads (MB) followed by recruitment of BoNT/A LC and anti-TAG sequence dually modified BNPs, forming magnetic nanocomplexes. Fluorescent signals generated upon the SNAPtide cleavage activity of BoNT/A LC on the magnetic nanocomplexes. Moreover, the color changes of the assay could be observed with naked eyes or UV–vis spectra. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To verify the key step of BoNT/A LC-catalysis reaction of BACA, the engineered BoNT/A LC was synthesized, and its activity was confirmed. We initially found that plasmid pTIG-Trx-BoNT-ALC without ribosome binding site (RBS) between the thioredoxin gene (trxA) and BoNT/A LC gene could not be expressed in the E. coli cells. Plasmid pTIG-Trx-RBS-BoNT-ALC was then reconstructed based on pTIG-Trx-BoNT-ALC to obtain the signal reporter of BACA, GBP-tagged BoNT/A LC (Fig. S1a-S1e and S2). Trx-RBS sequence was amplified and subcloned into pTIG-Trx-BoNT-ALC to form the new recombinant plasmid pTIG-Trx-RBS-BoNT-ALC. The activity of BoNT-ALC was confirmed using SNAPtide FRET-based assay and SDS-PAGE analysis method (Fig. S1f and S1g).
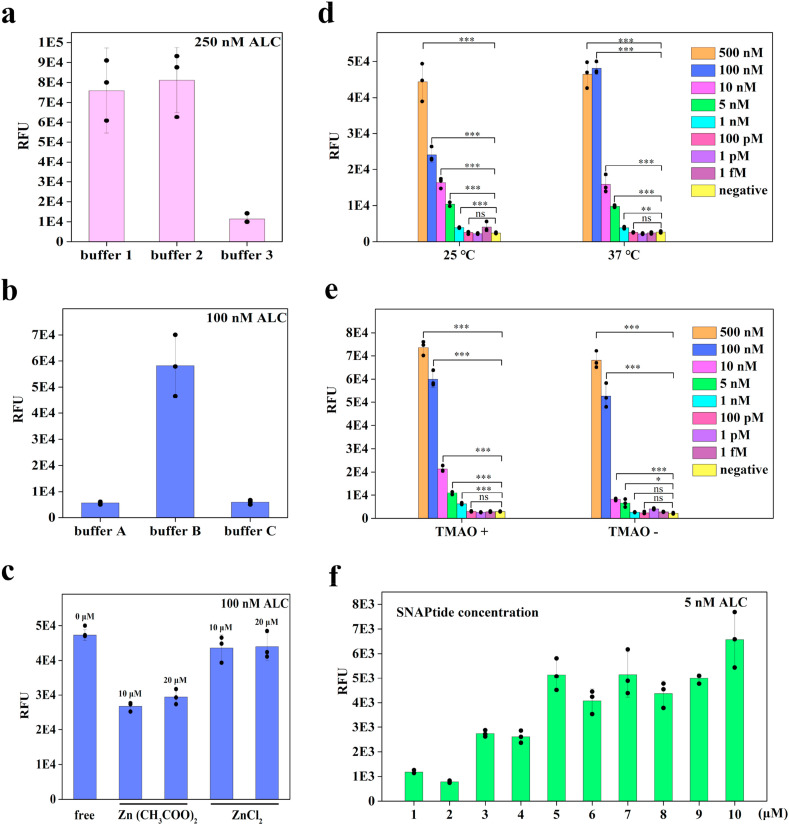
To optimize BoNT/A LC cleavage activity and to achieve highest fluorescent signal, we tested both reactions of SNAP25 substrate cleavage and SNAPtide fluorescence assay in a series of cleavage buffers (HEPES and Tris-HCl) containing two Zn2+ types at different cleavage reaction temperatures (25 °C and 37 °C). Purified BoNT/A LC protein was diluted with three different dilution buffers (PB and HEPES) before added to the cleavage reaction buffer. And fluorescence signals were measured with or without 2.3 M trimethylamino oxide (TMAO) to verify its improvement on the cleavage activity of BoNT/A LC. Then, we selected cost effective SNAPtide concentration by adding a series of SNAPtide concentrations (1–10 μM) to the FRET detection assay. As shown in Fig. 2 and S3, when BoNT/A LC in cleavage buffer 1 with TMAO (2.3 M) at 37 °C, the real-time fluorescence spectra of BoNT/A LC-based assay had the strongest response. Therefore, SNAPtide FRET-based substrate was an appropriate reporter for sensitive biosensing assay.
Fig. 2.
BoNT/A LC optimization. BoNT/A LC cleavage activity with different cleavage buffers (a) (cleavage buffer 1: 50 mM HEPES, 0.05% TWEEN20, pH 7.4; cleavage buffer 2: 50 mM HEPES, 20 μM ZnCl2, 0.05% TWEEN20, pH 7.4; cleavage buffer 3: 10 mM Tris-HCl with 20 mM NaCl, pH 7.6), different dilution buffers (b) (dilution buffer A: 10 mM PB buffer at pH 7.4; dilution buffer B: 50 mM HEPES, 0.05% TWEEN20, pH 7.4; dilution buffer C: 50 mM HEPES, 200 mM NaCl, pH 7.6) and cleavage activity with 10 μM/20 μM Zn(CH3COO)2 or 10 μM/20 μM ZnCl2 or without metal ions (c). BoNT/A LC cleavage activities were detected at different temperatures (25 °C or 37 °C) (d), TMAO in cleavage buffer (e) and different SNAPtide concentrations ranged from 1 to 10 μM (f).
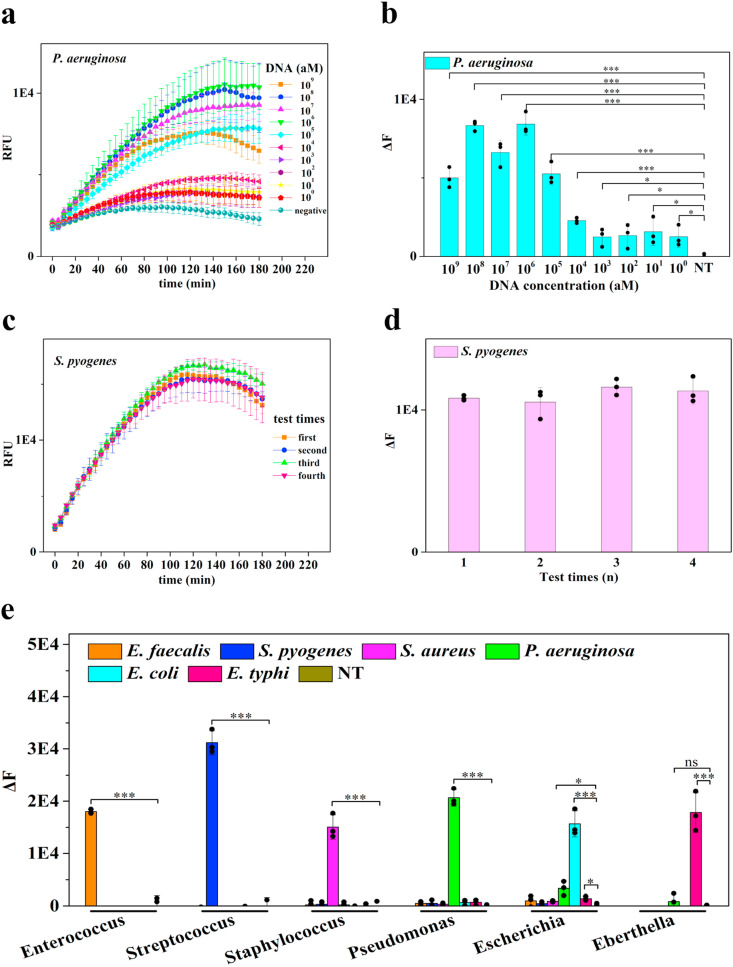
To apply BoNT/A LC to bacterial diagnostic, we focused on two important aspects: (1) building a simple fluorescent reader to offer lab-quality sensitivity and high repeatability in resource-limit conditions; (2) testing BACA specificity against bacteria and related strains. First, we used the signal generation strategy based on a sandwich hybridization assay for bacteria detection. The double-stranded biotinylated target DNA derived from bacterial 16S rRNA was amplified through conventional PCR and then separated by MBs (Fig. S4 and S5). Meanwhile, the anti-TAG probe was conjugated to DNA-conjugated gold nanoparticles (DNPs) and the binding efficiency between the probe and DNP was quantified (Fig. S6a). To optimize the BNP probes, a series of conjugation protocols were employed to screen the conditions of BoNT/A LC conjugating onto DNA-modified DNPs and the strongest binding capacity was observed under 0.3 μM BoNT/A LC with DNPs for 0.5 h (Fig. S6b). The UV–Vis spectra also verified the binding of DNPs and GBP-tagged BoNT/A LC (Fig. S6c). The BoNT/A LC-modified nanocomplexes were assembled with the target-modified MBs in a shaker-incubator. Next, we tested the performance of our method on bacterial detection after primer specificities of six different pathogens investigated (Fig. S7). P. aeruginosa was chosen as the model organism. For synthetic nucleic acids, the limit of detection (LOD) of BACA could reach to 1 aM using fluorescent readout (Fig. 3 a–b). The BACA fluorometric biosensor allowed sensitive detection of P. aeruginosa at 1 CFU/mL (Fig. S8). The LOD of BACA fluorometric biosensor of S.pyogenes reached 10 aM for DNA target and 103 CFU/mL for bacteria concentration (Fig. S8 and S9). Besides conventional amplification of bacterial multiple 16S and magnetic enrichment of BACA, the high sensitivity of BACA was mainly attributed to the specific target amplification and the catalysis activity of BoNT/A LC that one toxin molecule could cleave billions of substrate molecules in a short time. The RSD (relative standard deviation) values of intra-assay and inter-assay for target of S.pyogenes were 7.03% and 4.36%, respectively. These assays displayed excellent reproducibility of the tests through BACA fluorometric method (Fig. 3c–d). BACA fluorometric method could genotype multiple species of bacteria without cross-reactions among them (Fig. 3, Fig. 4 a). These results demonstrated that BACA was a robust and accurate way to detect bacteria.
Fig. 3.
Bacteria detection by fluorometric BACA biosensor. BoNT/A LC based fluorescent spectra (a) and signal intensity (b) for a serial dilution of synthetic P. aeruginosa DNA. The reproducibility of response in relative fluorescent intensity through BACA fluorometric method after exposure to the same target DNA in different tests (c–d). Fluorescence response for differential detection of 6 bacteria types (e) (n = 3 technical replicates, bars represent mean ± s.d. ΔF represents background subtracted fluorescence. NT represents no template control).
Fig. 4.
Specificity analysis of BACA method for bacteria detection. Bacteria species-specific target detection by fluorometric (a) and colorimetric (b) biosensor respectively. The BoNT/A substrate SNAPtide (FITC/DABCYL) was then added to the assay with a final concentration of 5 μM. Reactions were incubated in a fluorescence plate reader (BioTek H1 microplate reader) with fluorescence read every 5 min as a fluorometric biosensor. The color changes of supernatant could be observed to the naked eyes or measured by UV–vis spectrophotometer at 520 nm (n = 3 technical replicates, bars represent mean ± s.d.). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Another goal of BACA was to develop a visual readout requiring no additional device. We then studied the feasibility of BACA as a generic method using the colorimetric biosensor for detecting the presence of a series of pathogens. For model organism P. aeruginosa, the LOD of BACA colorimetric biosensor was observed for DNA target (102 aM) and bacteria (101 CFU/mL) (Fig. S8 and S10). While for S.pyogenes, the LOD was 102 aM for DNA target and 104 CFU/mL for bacteria (Fig. S8 and S9). We found that the BACA colorimetric method had a lower sensitivity than the fluorometric approach. Both the two methods displayed a high reproducibility and excellent specificity (Fig. 4b and Fig. S10). Thus, the BACA colorimetric method can be applied in point-of-care platform, which was different from crosslinking colorimetry of DNA-DNPs and salt-induced colorimetry of unmodified DNPs (Yang et al., 2020).
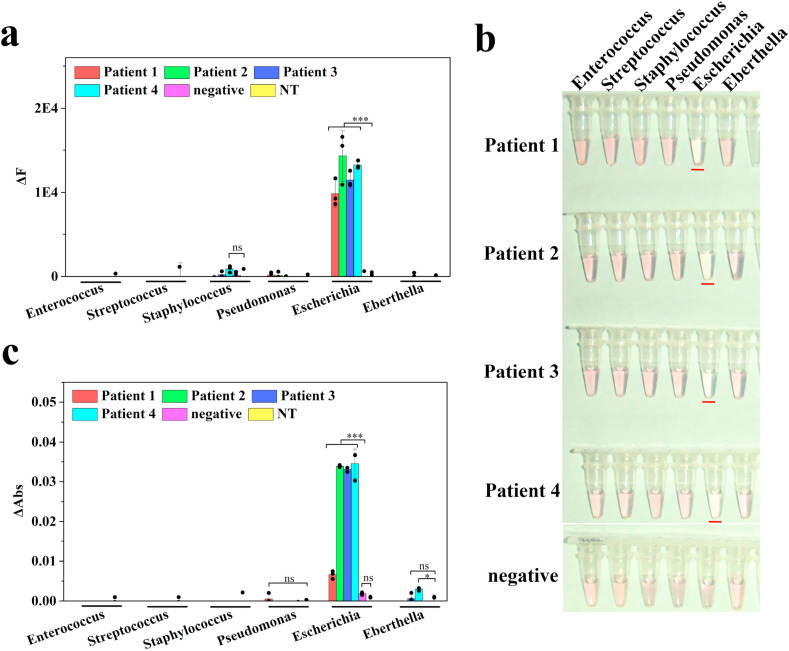
For the final validation of the BACA method, we acquired and analyzed clinical blood samples infected with pathogen. We further confirmed the possibility of the method for the diagnosis of infectious disease. Four suspected infectious samples and one healthy sample were detected by BACA fluorometric and colorimetric method. These samples were tested with BACA and qPCR method in parallel in the detecting panel. Fig. 5 displayed the results from both methods. In these results, the suspected infectious samples were identified for positive results of E.coli and one healthy sample was negative (Fig. 5 and Fig. S11), which were verified by gel electrophoresis analysis (Fig. S12). The results of BACA were well consistent with that obtained by qPCR and traditional culture technique that carried out by the hospital. Though very few patient samples were analyzed here because of the clinic and laboratory limitations, BACA performed a great potential for clinical disease diagnosis. Moreover, BACA also produced the prospect of different bacteria strain detections if additional patient samples could be supplied.
Fig. 5.
Pathogen diagnosis results of clinical blood samples using the BACA assay. Bacteria detection using fluorometric (a) and colorimetric (b and c) BACA biosensor respectively. Results are shown as mean ± s.d. The BoNT/A substrate SNAPtide (FITC/DABCYL) was then added to the assay with a final concentration of 5 μM. Reactions were incubated in a fluorescence plate reader (BioTek H1 microplate reader) with fluorescence read every 5 min as a fluorometric biosensor. The color change of supernatant could be observed to the naked eyes or measured by UV–vis spectrophotometer at 520 nm. All specimens for the tests were measured in triplicate. (NT represents no template control). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
COVID-19 was caused by SARS-CoV-2 coronavirus and had become a severe global health threat because of its person-to-person contact transmission characteristics since December 2019 (Wu et al., 2020). Currently, Real-Time PCR (RT-PCR) was applied as the gold standard for the clinical diagnosis of COVID-19, only 47–59% of the positive cases were identified using RT-PCR (Li et al., 2020). Other detection methods such as Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) (Yu et al., 2020), Reverse Transcription Recombinase Aided Amplification (RT-RAA) (Xue et al., 2020), Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) diagnostics (Broughton et al., 2020; Guo et al., 2020), Droplet Digital PCR (ddPCR) (Suo et al., 2020) and serological test (Shi et al., 2020) had been reported till now to help advance the COVID-19 diagnostic. We finally sought to demonstrate the potential of BACA for COVID-19 detection. Due to some clinic and laboratory limitations, the SARS-CoV-2 N gene plasmid was used as the mimic. As shown in Fig. 6 a and b, biotinylated target DNA of N gene could be amplified and assembled to BoNT/A LC activated complexes. The analytical sensitivity of BACA fluorometric and colorimetric methods for SARS-CoV-2 N gene detection were as low as one copy per assay (Fig. 6c and S13). Also the BACA assay only provided a positive result for SARS-CoV-2-abEN pseudovirus and no cross-reactions were observed with SARS-CoV-abEN pseudovirus, MERS-CoV-abEN pseudovirus and Human Genomic DNA as shown in Figure S14. Thus, the BACA strategy built here was undoubtedly proved to be a great choice for COVID-19 diagnostic and had a good potential to be used under resource-limited circumstances.
Fig. 6.
Detection of SARS-CoV-2 N gene plasmid. The detection target was amplified (a) and used to fabricate complexes that leading to apparent color changes to naked eyes (b). Fluorometric detection sensitivity of SARS-CoV-2 N gene plasmid was analyzed by BACA method (c) (n = 3 technical replicates, bars represent mean ± s.d. NT represents no template control). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusions
In the current study, we have designed a universal, low-cost and easily accessible biosensor based on BoNT/A LC activated complex assay (BACA) capable of measuring and phenotyping common pathogens and SARS-CoV-2. By detecting the fluorescence signal or the color change of reaction assay, the method is fast (1–2 h), sensitive (1 aM), accurate (99%) and adaptable to many other microorganisms. Moreover, the BACA is in excellent agreement with traditional culture and qPCR methods and produces the clinic prospect. Although this method offers ultrasensitive nucleic acid diagnostic ability, an optimized version of BACA may potentially be developed in view of simple experimental procedures and minimal instrument ways demanded by people. We look forwards to this approach serving as a promising way for the early diagnosis or analysis of acute virus outbreak in point-of-care scenarios in conjunction with ready-to-use cell lysates, isothermal amplification tools and lateral flow detection.
CRediT authorship contribution statement
Fengge Song: Experimental investigation, Methodology, Data curation, Formal analysis, Writing - review & editing. Yuanyuan Shen: Investigation, Methodology. Yangdao Wei: Investigation, Methodology. Chunrong Yang: Methodology, Formal analysis. Xiaolin Ge: Formal analysis. Aimin Wang: Methodology, Formal analysis. Chaoyang Li: Methodology, Formal analysis. Yi Wan: Conceptualization, Methodology, Data curation, Formal analysis, Writing - review & editing. Jinghong Li: Conceptualization, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We gratefully acknowledge support from Key Research and the Development Program of Hainan, China (No. ZDYF(XGFY)2020002), the National Key Research and Development Program of China, China (No.2018YFD0900704), the National Natural Science Foundation of China, China (No. 41866002), Tsinghua University Initiative Scientific Research Program, China (No. 2020Z99CFZ019), Open Fund of Shandong Key Laboratory of Corrosion Science, China (KLCS201910), and Research Foundation of Hainan University, China (No. KYQD(ZR)1711).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112953.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
Multimedia component 1
References
- Bagramyan K., Barash J.R., Arnon S.S., Kalkum M. PloS One. 2008;3(4) doi: 10.1371/journal.pone.0002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash J.R., Arnon S.S. J. Infect. Dis. 2014;209(2):183–191. doi: 10.1093/infdis/jit449. [DOI] [PubMed] [Google Scholar]
- Blasi J., Chapman E.R., Link E., Binz T., Yamasaki S., Camilli P.D., Südhof T.C., Niemann H., Jahn R. Nature. 1993;365(6442):160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglic D., Krutein M.C., Bompiani K.M., Barlow D.J., Benoni G., Pelletier J.C., Reitz A.B., Lairson L.L., Houseknecht K.L., Smith G.R., Dickerson T.J. J. Med. Chem. 2014;57(3):669–676. doi: 10.1021/jm4012164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Feng C., Liu Y., Wang S., Liu F. Biosens. Bioelectron. 2014;57:133–138. doi: 10.1016/j.bios.2014.02.020. [DOI] [PubMed] [Google Scholar]
- Cardoso S.P., Patel R., Brown C., Navarrete C. Tissue Antigens. 2011;78(3):171–177. doi: 10.1111/j.1399-0039.2011.01736.x. [DOI] [PubMed] [Google Scholar]
- Chen S., Barbieri J.T. J. Biol. Chem. 2006;281(16):10906–10911. doi: 10.1074/jbc.M513032200. [DOI] [PubMed] [Google Scholar]
- Chung H.J., Castro C.M., Im H., Lee H., Weissleder R. Nat. Nanotechnol. 2013;8(5):369–375. doi: 10.1038/nnano.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rica R., Stevens M.M. Nat. Nanotechnol. 2012;7(12):821–824. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- Dennis, David T., Inglesby, Thomas V., Henderson, Donald A., Bartlett, John G., Ascher, Michael S. Jama. 2000;283(17):2281–2290. doi: 10.1001/jama.283.17.2281. [DOI] [PubMed] [Google Scholar]
- Deshpande A., White P.S. Expert Rev. Mol. Diagn. 2012;12(6):645–659. doi: 10.1586/erm.12.60. [DOI] [PubMed] [Google Scholar]
- Dhaked R.K., Singh M.K., Singh P., Gupta P. Indian J. Med. Res. 2010;132(5):489–503. [PMC free article] [PubMed] [Google Scholar]
- Fabiani L., Saroglia M., Galata G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forne M., Dominguez J., Fernandez-Banares F., Lite J., Viver J.M. J. Gastroenterol. 2000;95(9):2200–2205. doi: 10.1111/j.1572-0241.2000.02303.x. [DOI] [PubMed] [Google Scholar]
- Guan Z.P., Jiang Y., Gao F., Zhang L., Zhou G.H., Guan Z.J. Eur. Food Res. Technol. 2013;237(4):627–637. [Google Scholar]
- Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R.R., Zhou Q., Li W. Cell Discov. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Chen J., Li Z., Chen H., Qiu H. Biosens. Bioelectron. 2020;148:111811. doi: 10.1016/j.bios.2019.111811. [DOI] [PubMed] [Google Scholar]
- Hong F., Ma D., Wu K., Mina L.A., Luiten R.C., Liu Y., Yan H., Green A.A. Cell. 2020;180(5):1018–1032. doi: 10.1016/j.cell.2020.02.011. e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Debruyne D.N., Spencer J., Kapoor V., Liu L.Y., Zhou B., Lee L., Feigelman R., Burdon G., Liu J., Oliva A., Borcherding A., Tan H., Urban A.E., Liu G., Liu Z. bioRxiv. 2020 doi: 10.1101/2020.03.12.988246. 2020.03.12.988246. [DOI] [Google Scholar]
- Lucas A.D., Prima M.A.D., Hitchins V.M. Applied Biosafety. 2015;20(2):104–109. [Google Scholar]
- Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naja G., Bouvrette P., Hrapovic S., Luong J.H. Analyst. 2007;132(7):679–686. doi: 10.1039/b701160a. [DOI] [PubMed] [Google Scholar]
- Ren X., Zhang K., Deng R., Li J. Inside Chem. 2019;5(10):2571–2592. [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Seo S.H., Lee Y.R., Ho Jeon J., Hwang Y.R., Park P.G., Ahn D.R., Han K.C., Rhie G.E., Hong K.J. Biosens. Bioelectron. 2015;64:69–73. doi: 10.1016/j.bios.2014.08.038. [DOI] [PubMed] [Google Scholar]
- Shi J., Han D., Zhang R., Li J., Zhang R. Clin. Chem. 2020;66(8):1030–1046. doi: 10.1093/clinchem/hvaa122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L.L. Annu. Rev. Pharmacol. Toxicol. 2003;44(1):167–193. doi: 10.1146/annurev.pharmtox.44.101802.121554. [DOI] [PubMed] [Google Scholar]
- Song F., Deng R., Liu H., Wang A., Ma C., Wei Y., Cui X., Wan Y., Li J. Anal. Chem. 2019;91(21):14043–14048. doi: 10.1021/acs.analchem.9b03717. [DOI] [PubMed] [Google Scholar]
- Sun P., Lu X., Xu C., Sun W., Pan B. J. Med. Virol. 2020;92(6):548–551. doi: 10.1002/jmv.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Chen G., Lan K., Chen Y. Emerg. Microb. Infect. 2020;9(1):1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsourkas A., Behlke M.A., Bao G. Nucleic Acids Res. 2002;30(23):5168–5174. [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Song F., Wang G., Liu H., An M., Wang A., Wu X., Ma C., Wang N. Anal. Chem. 2018;90(16):9853–9858. doi: 10.1021/acs.analchem.8b01933. [DOI] [PubMed] [Google Scholar]
- Wang Y., Alocilja E.C. J. Biol. Eng. 2015;9(1):16. doi: 10.1186/s13036-015-0014-z. -16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Wang L., Xu L., Li L., Ren S., Cao C., Jia N., Aldalbahi A., Song S., Shi J. Analyst. 2016;141(18):5304–5310. doi: 10.1039/c6an01435f. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Wang Y., Duan N., Ma H., Wang Z. J. Agric. Food Chem. 2015;63(35):7849–7854. doi: 10.1021/acs.jafc.5b03224. [DOI] [PubMed] [Google Scholar]
- Xue G., Li S., Zhang W., Du B., Cui J., Yan C., Huang L., Chen L., Zhao L., Sun Y., Li N., Zhao H., Feng Y., Wang Z., Liu S., Zhang Q., Xie X., Liu D., Yao H., Yuan J. Anal. Chem. 2020;92(14):9699–9705. doi: 10.1021/acs.analchem.0c01032. [DOI] [PubMed] [Google Scholar]
- Yang T., Luo Z., Tian Y., Qian C., Duan Y. TrAC Trends Anal. Chem. (Reference Ed.) 2020;124:115795. [Google Scholar]
- Yoo S.M., Lee S.Y. Trends Biotechnol. 2016;34(1):7–25. doi: 10.1016/j.tibtech.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.-H., Yin X. Clin. Chem. 2020;66(7):975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Qin S., Wu S., Liang Y., Li J. Chem. Sci. 2020;11(25):6352–6361. doi: 10.1039/d0sc01353f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1