SUMMARY
Immune cells’ metabolism influences their differentiation and functions. Given that a complex interplay of environmental factors within the tumor microenvironment (TME) can have a profound impact on the metabolic activities of various immune, stromal, and tumor cell types, there is emerging interest to advance understanding of these diverse metabolic phenotypes in the TME. Discussing various cell-extrinsic contributions to the metabolic activities of immune cells and recent technical advances in experimental systems and metabolic profiling technologies, we propose future directions to better understand how immune cells meet their metabolic demands in the TME, which can be leveraged for therapeutic benefit.
Keywords: metabolism, immunometabolism, tumor microenvironment, immunology, in vitro modeling, physiologic media, metabolomics, stable isotope tracing
INTRODUCTION
Metabolic conditions in the tumor microenvironment (TME) are influenced by many factors, including gradients of nutrient and oxygen levels, tissue vascularization, heterocellular interactions, and systemic metabolism. However, much of the research used to define principles of tumor cell metabolism have relied heavily on cultured cells, which cannot fully recapitulate the metabolic conditions of tumor metabolism in vivo, especially tumor cell-extrinsic factors such as nutrient availability and immune infiltration. To improve the modeling capacity of experimental systems used to study the TME, various cell-intrinsic and environmental inputs should be considered. However, as the TME contains sundry cell types with diverse and flexible metabolic programming, precise investigation of the metabolic demands, adaptations, and interactions among various immune and cancer cells is complex. Recent discussion of such intercellular interactions has been presented in detail by a number of excellent reviews (Bader et al., 2020; Lau and Vander Heiden, 2020). Here, we first highlight how immune cell metabolism and function can be influenced by the TME, and then discuss emerging technical advances in experimental modeling and metabolomics that may be exploited in future studies to further our understanding of immune cell metabolism in the complex context of the TME.
1. ENVIRONMENT SHAPES IMMUNE CELL METABOLISM AND FUNCTION
1.1. Nutrient Availability
Much of the work in immunometabolism to date has focused on how cells concurrently and differentially utilize nutrients to satisfy metabolic demands associated with effector functions and differentiation states. Indeed, the metabolic activities of different immune cell types, particularly lymphocytes, has been extensively reviewed elsewhere (Buck et al., 2017; Lau and Vander Heiden, 2020). The metabolic program of normal resting cells primarily serves to meet the bioenergetic requirements of maintaining homeostatic processes that require ATP. In contrast, proliferating cells—both normal and malignant—not only need to generate energy to support cell growth, but must also meet various anabolic demands of macromolecule biosynthesis (e.g., proteins, nucleotides, lipids) and cellular redox homeostasis (Cantor and Sabatini, 2012; Kong and Chandel, 2018; O’Sullivan et al., 2019; Pearce et al., 2013; Sugiura and Rathmell, 2018). A central metabolic program associated with activation of innate (dendritic cells (DCs), macrophages, neutrophils) and adaptive (CD8+ and CD4+ T cells, B cells) immune cell is aerobic glycolysis, also commonly known as the Warburg effect (Lunt and Vander Heiden, 2011; Warburg, 1956). Increasing glucose uptake and glycolysis not only provides precursors for proliferation, such as glucose-dependent serine production for nucleotide synthesis (Ma et al., 2019), but also supports IFN-γ production (Cham and Gajewski, 2005; Chang et al., 2013a). For instance, T helper (Th) cells are highly glycolytic (Michalek et al., 2011; Shi et al., 2011), relying on the glucose transporter GLUT1 for inflammatory cell (i.e., Th1 and Th17) effector function, and GLUT1-mediated glycolysis in regulatory T (Treg) cells regulates the balance between proliferation and suppressive capacity (Gerriets et al., 2016). However, this may pose a challenge for tumor-infiltrating immune cells given that glucose availability can vary in the TME (Siska et al., 2017; Sullivan et al., 2019). Specifically, there is evidence that glucose limitation in TME can compromise effector functions of tumor infiltrating lymphocytes (TILs) and the antitumor effects of natural killer (NK) cells (Chang et al., 2013b, 2015; Ho et al., 2015).
In addition to glucose, the availability of different amino acids can also influence immune cell metabolism. For example, T cells and macrophages may catabolize glutamine to fuel ATP production (Blagih et al., 2015; Carr et al., 2010; Jha et al., 2015). Yet, blunting glutamine catabolism can promote both increased CD8+ TIL effector function and tumor regression, owing to differential demands for glutamine utilization among tumor cells and TILs (Leone et al., 2019). Given the metabolic variability among cell types in the TME, the targeted inhibition of metabolic enzymes may drive distinct cell-specific consequences. For instance, either genetic or pharmacologic blockade of glutaminase (GLS)—which metabolizes glutamine into glutamate—has been shown to impair proliferation and activation of CD4+ and CD8+ T cells with little effect on cytokine production. The same blockade can also suppress the differentiation of CD4+ Th17 cells (Johnson et al., 2018). In addition, supraphysiologic arginine levels provided by the synthetic cell culture medium RPMI 1640 can blunt cytokine production mediated by T cells but increase their survival (Geiger et al., 2016). Tryptophan depletion also can reportedly hinder T-cell function through activation of the integrated stress response (Munn et al., 2005). Further, the extracellular availability of both serine (Labuschagne et al., 2014; Ma et al., 2017; Ron-Harel et al., 2016) and methionine (Gao et al., 2019; Roy et al., 2020; Sinclair et al., 2019) can influence the proliferation of tumor cells and T lymphocytes through effects linked to one-carbon metabolism. In T cells, as the folate and methionine cycles are uncoupled and serine-derived carbon does not transit to the methionine cycle, methionine may be the exclusive source of the methyl moiety in S-adenosylmethionine (SAM) and, by extension, an essential amino acid for methylation reactions (Roy et al., 2020). In addition, others have recently demonstrated that alanine availability can affect protein synthesis during the early stages of T cell activation (Ron-Harel et al., 2019). Interestingly, genes that encode amino acid transporters are amongst the most upregulated genes upon T cell activation (Howden et al., 2019), suggesting that amino acid exchange is a critical aspect of immune regulation in the TME. Together, these results highlight that amino acid availability can have distinct cell-specific effects on immune cell metabolism and function, perhaps suggesting the potential to target amino acid metabolism of different immune subtypes in the treatment of human disease.
Purported waste products of cellular metabolism—including lactate, kynurenine, and adenosine—can also exert immunomodulatory effects in the TME. For example, whereas lactate can suppress the cytolytic capacity of CD8+ effector cells (Brand et al., 2016; Calcinotto et al., 2012; Fischer et al., 2007), it may instead be used by Treg cells to support metabolic demands (Angelin et al., 2017; Wang et al., 2020). Further, lactate accumulation in a murine model of arthritis induces upregulation of the monocarboxylate transporter SLC5A12 in CD4+ T cells, a transporter class similarly upregulated in activated T cells. This leads to elevated levels of intracellular acetyl-CoA and citrate, as well as decreased T cell motility (Pucino et al., 2019). Others have also demonstrated that lactate generated by melanoma cells can diminish immune surveillance by T cells and NK cells by suppressing nuclear factor of activated T cells (NFAT)-dependent IFN-γ production (Brand et al., 2016). Further, recent evidence also indicates that lactate can be used to fuel the TCA cycle in both normal tissues and certain lung cancers (Faubert et al., 2017; Hui et al., 2017), suggesting that lactate may promote immune evasion in the TME by supporting demands of tumor growth (Angelin et al., 2017). In addition, elevated levels of kynurenine, a downstream product of tryptophan catabolism, can induce immunosuppression in T cells (Fallarino et al., 2006; Mezrich et al., 2010; Opitz et al., 2011). Remarkably, Triplett and colleagues recently demonstrated that the enzyme-mediated depletion of systemic kynurenine could increase the tumor infiltration and proliferation of CD8+ lymphocytes (Triplett et al., 2018). Lastly, the accumulation of adenosine, a breakdown product of nucleotide metabolism, effectively reduces the cytotoxic effects mediated by T and NK cells, but enhances the activation of immunoregulatory M2 macrophages (Csóka et al., 2012; Huang et al., 1997).
The impact of free fatty acids on immune cell function in the TME continues to be an active area of investigation. While early models of immune cell metabolic reprogramming proposed that glycolytic and oxidative (fueled by long-chain fatty acid oxidation (FAO)) metabolic programs were associated with distinct immune cell populations (e.g., CD8+ T effector (Teff) versus T memory (Tmem) cells, M1 versus M2 macrophages) (O’Neill and Pearce, 2016; Pearce et al., 2013), the reality has proven to be far more nuanced. Etomoxir, an inhibitor of CPT1A—an enzyme that catalyzes the rate-limiting step in mitochondrial long-chain FAO—has immunomodulatory effects on CD8+ Tmem cell differentiation and M2 macrophage polarization (Huang et al., 2014; van der Windt et al., 2012), reinforcing the notion that FAO-fueled OXPHOS supports these processes. However, genetic evidence has indicated that CPT1A is dispensable for CD8+ Teff function and Tmem generation, as well as for CD4+ Treg suppressive capacity (Raud et al., 2018). Further, the suggested influence of etomoxir on M2 macrophage polarization has been attributed to off-target effects of the compound (Divakaruni et al., 2018). Nonetheless, CD8+ T cells with elevated Cpt1a expression display a metabolic advantage in vivo (Klein Geltink et al., 2017; van der Windt et al., 2012), underscoring a role for mitochondrial metabolism in supporting optimal T cell function. Long-chain FAO may be more critical for tissue-resident immune cells; recent work has demonstrated that fatty acid import (via FABP4 and FABP5) and CPT1A-dependent lipid oxidation are required for the persistence of tissue-resident CD8+ cells in peripheral tissues (Pan et al., 2017), while the lipid transporter CD36 is required for accumulation of CD4+ Treg cells in the TME (Wang et al., 2020). Interestingly, TILs in B16 melanoma tumors display enhanced fatty acid catabolism, a phenotype attributed to low glucose levels in the TME (Zhang et al., 2017), hinting at a role for FAO in metabolically flexible immune cells in the TME.
Beyond their role as fuel sources, saturated fatty acids can also promote the production of pro-inflammatory cytokines (e.g., IL-23 during Th17 responses), whereas polyunsaturated fatty acids have been linked to anti-inflammatory cytokine production (e.g. IL-10) (Wang et al., 2015). Short chain fatty acids (SCFAs), which are largely byproducts of microbial metabolism, can act as positive regulators of CD8+ Teff function as well (Bachem et al., 2019; Balmer et al., 2016; Trompette et al., 2018). For example, butyrate—which acts as an HDAC inhibitor—can influence T cell function by mediating G-protein-coupled receptor signaling (Bachem et al., 2019). In addition, recent work by Qui and colleagues demonstrated that acetate can enhance Teff cell function by reinforcing permissive epigenetic marks (i.e., H3K27ac) and chromatin accessibility at key effector cytokine loci (e.g., Gmzc, Ifng, Tnfa) (Qiu et al., 2019). These results are consistent with the notion that the composition of the gut microbiome can influence metabolite availability and thus, patient responses to immunotherapy (reviewed in (Gopalakrishnan et al., 2018)), though specific contributions of microbiome-derived metabolites to immune function in the TME require further research.
1.2. Oxygen, pH, and Ion Concentration
Tissue oxygen (O2) concentrations are dictated by competing rates of O2 diffusion and consumption. Most tissues experience oxygen levels of 2–9% (average 40 mm Hg) (Krzywinska and Stockmann, 2018), while cells in the TME may instead experience hypoxic conditions (<2% oxygen) that are driven by disorganized vasculature and the increased metabolic rate of tumor cells (Bertout et al., 2008). Such hypoxic conditions promote the stabilization of hypoxia inducible factor-1α (HIF-1α), whose expression is associated with increased inflammatory potential in both myeloid and lymphoid cells (Palazon et al., 2014). Similar to oxygen availability, extracellular pH varies across the TME—likely in concordance with local accumulation of lactate—and extracellular acidification (pH ~5.8–6.6) can have broad effects on immune cells (Huber et al., 2017; Singer et al., 2018). Notably, small ion availability can also have dramatic effects on T cell function. Elevated sodium levels can drive increased inflammatory potential in CD4+ Th17 cells (Kleinewietfeld et al., 2013; Wu et al., 2013), and increased potassium availability can compromise nutrient uptake and effector function in CD8+ T cells (Vodnala et al., 2019).
Although Hif1a deletion has been shown not to affect metabolic reprogramming of activated T cells (Wang et al., 2011), several studies have demonstrated that hypoxic conditions can have immunomodulatory effects (Lim et al., 2020). For example, Hif1a expression in CD8+ T cells regulates their proliferation and influences their effector function by promoting a glycolytic phenotype (Finlay et al., 2012; Lum et al., 2007; Palazon et al., 2017). In murine breast tumor models, the deletion of Hif1a in CD8+ T cells can result in reduced tumor infiltration and cytotoxic function (Palazon et al., 2017), indicating that HIF-1α can serve a key role in the maintenance of Teff cell function. Further, models of adoptive cell transfer (ACT) have demonstrated that hypoxic pre-conditioning can improve the cytotoxicity and tumor clearance mediated by CD8+ T cells (Gropper et al., 2017). Notably, several studies have shown that oxygen restriction can also have immunosuppressive effects. For example, immunosuppressive (Arg1+) tumor-associated macrophages (TAMs) spatially associate with hypoxic regions in mouse breast tumors (Carmona-Fontaine et al., 2017), while Hif1a deletion in macrophages impairs both mobilization and activation (Cramer et al., 2003). In addition, TILs were absent from hypoxic regions in a murine model of fibrosarcoma (Hatfield et al., 2015), and limited oxygen availability has been associated with T cell exhaustion in a murine model of melanoma (Najjar et al., 2019). Further, HIF-1α can promote immunosuppressive characteristics of CD4+ CD25+ Tregs (Ben-Shoshan et al., 2008). Together, these studies illustrate that oxygen availability can strongly impact immune cell function in the TME.
1.3. Intercellular interactions
The diverse milieu of immune, cancer, and stromal cells present within the TME creates a dynamic environment, fostering crosstalk between cancer and immune cells that can be influenced by tumor type (Gentles et al., 2015). Advances in single cell techniques, such as mass cytometry and single-cell genomics, have revealed considerable complexity among stromal and immune cell subtypes in the TME, as well as heterogeneity in the metabolic profiles of these cells (Hartmann and Bendall, 2020; Li et al., 2017; Xiao et al., 2019). Recent work characterizing metabolic enzyme expression in T cell subsets by mass cytometry has the advantage of characterizing the metabolic potential of rare immune subsets from tumors (Hartmann et al., 2020). The major caveat with any single cell profiling approach is that metabolic enzyme expression only defines how the table is set; nutrient availability in the TME will ultimately define what meal is served. Analysis of The Cancer Genome Atlas (TCGA) datasets revealed six dominant immune signatures—wound healing, IFN-γ dominant, inflammatory, lymphocyte poor, immunologically quiet, and TGF-β dominant—associated with 33 different cancer types (Thorsson et al., 2018). The cell types that contribute to these signatures are not fully understood but likely influence anti-tumor immunity. For instance, potential nutrient competition between distinct cell populations that share overlapping metabolic demands could lead to limitations in the availability of glucose and other cellular fuels. Similarly, stromal and immune cell types can contribute to shaping the metabolic composition within the TME, thus affecting both the nutrient utilization and drug responses of neighboring cancer cells (Halbrook et al., 2018; Sousa et al., 2016).
The potentially competitive environment within the TME can also induce TILs to enter an exhausted state. Exhausted CD8+ T (Tex) cells are a subpopulation of CD8+ Teff cells that exhibit reduced effector function and proliferative capacity, and can arise during chronic infection or cancer (McLane et al., 2019). Tex cells are associated with metabolic insufficiency that may be driven in part by the activation of one (or more) checkpoint inhibitor receptors, such as PD-1, LAG3, or CTLA-4 (Bengsch et al., 2016; Previte et al., 2019). Metabolic characterization of exhausted CD8+ T cells suggests that PD-1 inhibits glycolysis and supports fatty acid oxidation, resulting in the accumulation of CD4+ Tregs that can suppress the effector functions of CD8+ T cells (Bengsch et al., 2016; Patsoukis et al., 2015). Interestingly, PD-1 blockade has been shown to reverse the inhibition of glycolysis in exhausted T cells within a murine sarcoma (Chang et al., 2015). Indeed, therapeutic interventions designed to target checkpoint inhibitors have garnered extensive interest in the treatment of human cancers (Buck et al., 2017). Given the influence of tumor-immune crosstalk on anti-tumor immunity, experimental co-culture models may provide an alternative approach to monoculture strategies in order to model heterocellular interactions within the TME and how these impact immune cell responses and tumor cell sensitivity to small molecule drugs.
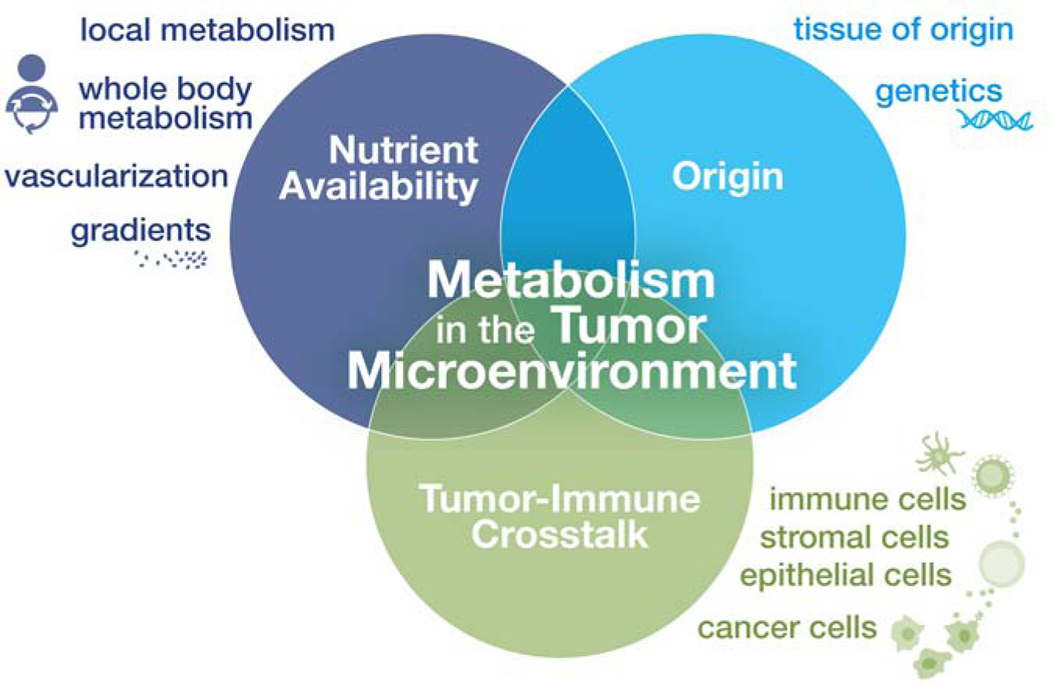
Ultimately metabolism in the TME is influenced by numerous factors that impact local nutrient availability, including cell-intrinsic metabolic programs driven by oncogenes (and complicated by genetic heterogeneity within the tumor) and cell-extrinsic factors such as tissue vascularization and nutrient and oxygen gradients within the tumor (Figure 1). Whole-body metabolic fluxes also influence tissue metabolite levels and are heavily influenced by diet and the microbiota (Ang et al., 2020; Biggs et al., 2017; Hui et al., 2020; Rooks and Garrett, 2016). Layered on top of this are tumor-stromal interactions, especially evolving tumor-immune cell interactions, that have the potential to impact the metabolic and inflammatory environment within the TME. Improved modeling of metabolism in the TME will require being mindful of environmental conditions (nutrient and oxygen levels), knowledge of tumor origin and genetic background, and tumor-immune crosstalk (Figure 1).
Figure 1: Factors influencing metabolism in the TME.
Metabolic conditions in the TME are influenced by many factors. From the perspective of the tumor, mutations in oncogenes and tumor suppressor genes can promote cell-intrinsic changes in metabolic reprogramming depending on the cancer type (“Origin”). Whole body metabolism dictates available nutrients in circulation (“Nutrient Availability”), while local nutrient availability in the TME is influenced by tumor vascularization and tissue nutrient uptake. Due in part to the high proliferation of cancer cells, nutrient and oxygen gradients can form which shape the metabolic activities of cells differently across the tumor. Nutrient utilization and signaling by many cell types present (immune, stromal, epithelial, cancer) in the TME can also modulate the metabolic conditions within the growing tumor (“Tumor-Immune Crosstalk”).
2. MODELING METABOLISM IN THE TUMOR MICROENVIRONMENT
Several recent studies have collectively established that metabolic phenotypes may vary by experimental model. For example, the use of stable isotope tracers in vivo using either mouse models or human patients has revealed that nutrient utilization by cancer and immune cells can differ from those observed using cell-based culture approaches (Davidson et al., 2016; Fan et al., 2019; Hensley et al., 2016; Ma et al., 2019). Though there are inherent caveats associated with all experimental models, an increased recognition that environmental factors contribute to cell metabolism (DelNero et al., 2018; Lyssiotis and Kimmelman, 2017; Muir et al., 2018; Singer et al., 2018; Wolpaw and Dang, 2018) has rapidly escalated efforts to improve the capacity of in vitro culture systems to more closely recapitulate conditions in human circulation or tumor-specific environments (Cantor, 2019).
2.1. Physiological media formulations
The complete medium formulations that remain the workhorses of cell culture studies across all areas of biology typically consist of a synthetic basal medium (e.g., MEM, DMEM, RPMI 1640) that poorly reflects metabolite availability in human biofluids, which is also supplemented with a largely undefined serum component (Cantor, 2019). Such media were primarily designed to promote the rapid growth of specific cell types, rather than to model in vivo biochemical conditions. Despite the extensive evidence for how biochemical conditions influence immune cell metabolism (Singer et al., 2018), most cell culture studies (Arora, 2013) are still carried out using these complete media. To address this discrepancy, there has been a recent rise in development of strategies to more closely model biochemical conditions encountered in vivo through the use of improved media formulations or systems that dynamically buffer nutrient concentrations (Birsoy et al., 2014). Here we focus on unbiased bottom-up approaches to systematically develop physiologic media that more closely recapitulate the metabolic composition of human blood.
Cantor and colleagues developed human plasma-like medium (HPLM) (Cantor et al., 2017), which contains over 60 metabolites and salt ions at concentrations that represent average reported values for normal adult human plasma (Psychogios et al., 2011; Wishart et al., 2013). Through the use of HPLM, the authors discovered an example of metabolic regulation mediated by a metabolite (uric acid) that, among basal media, was uniquely defined in HPLM, and whose plasma concentrations differ by up to 10-fold between humans and mice. The authors went on to demonstrate that HPLM could influence the relative cytotoxicity induced by 5-Fluorouracil, a classic chemotherapeutic that is widely used. These results provide evidence that the influence of metabolite availability on cell physiology and drug responses need not be restricted to considerations of nutrient utilization alone, and also suggest the potential to identify additional unforeseen metabolite-drug interactions.
Through an independent and parallel approach, Vande Voorde and colleagues developed a comparable basal medium (Plasmax) designed to reflect the polar metabolite composition of human blood (Ackermann and Tardito, 2019; Voorde et al., 2019). Among the relative metabolic differences described in cultured cells, the authors reported that differential arginine availability between Plasmax and a conventional medium can influence the direction of a reaction catalyzed by argininosuccinate lyase within the urea cycle. The authors also showed that by culturing certain breast cancer cells in Plasmax supplemented with relatively low levels of serum, the availability of selenium (at concentrations defined in Advanced DMEM/F12) could influence the colony-forming capacity of these cells.
Notably, the impact of physiologic media on cultured cells is not restricted to studies of cancer metabolism. Recent work demonstrated that, compared to the historically used medium to culture normal and malignant blood cells (RPMI 1640), HPLM induced markedly different transcriptional responses in human primary T lymphocytes and improved their activation upon antigen stimulation (Leney-Greene et al., 2020). This medium-dependent influence on T cell activation was traced to the differential availability of calcium, which is provided at a 6-fold higher levels in HPLM than RPMI 1640, indicating that small ion availability should be another key consideration in future models of the TME.
The development of physiologic media need not be limited to recapitulating the metabolite composition of human plasma, particularly in considering the environment(s) encountered by cancer and immune cells within a tumor. Through metabolic characterization of tumor interstitial fluid (TIF) isolated from tissue-specific murine tumors, Sullivan and colleagues recently reported that metabolite availability in TIF can differ from matched murine plasma, and that TIF composition may be influenced by additional factors such as cancer type and tumor location (Sullivan et al., 2019). These results suggest the potential to also generate synthetic media guided by non-plasma conditions, though given the anticipated heterogeneity of intratumoral environmental contexts, such approaches will likely require careful consideration in the selection and modeling of these biofluids.
While most efforts toward modeling the defined biochemical conditions of cell-based culture models have focused on polar metabolite availability, the characterization of extracellular lipid composition in different biofluids remains an ongoing consideration. For most complete media, the major source of lipophilic species is a supplement of 10–20% serum from fetal bovine or calf. However, lipid composition can vary between serum sources and lots, thus impacting experimental reproducibility. Charcoal stripping to remove serum lipids and enhance reproducibility can also deplete critical growth factors and hormones, further complicating current strategies to incorporate lipids into culture media at defined concentrations. Given the reported role of free fatty acids as a fuel for certain cancer and immune cell populations (Röhrig and Schulze, 2016), the development of robust and accessible approaches to incorporate at least certain free fatty acids to physiologic media at defined concentrations will be an important future direction to consider.
2.2. Tumor spheroids and organoid models
While physiological culture media systems have sought to recapitulate in vivo nutrient availability, tumor spheroid and organoid models have aimed to better model other environmental factors that cells encounter in normal tissues or in the TME. Three-dimensional (3D) organoid culture models, which are designed to more closely mimic the mechanical properties of the extracellular matrix, can also affect cell metabolism and drug responses (Fujii et al., 2016). For example, colorectal cancer (CRC) spheroids used to model nutrient gradients in the TME revealed p53-dependent metabolic vulnerabilities in response to cholesterol-lowering drugs (i.e., statins), which were not otherwise observed in cells cultured as monolayers (Kaymak et al., 2020). In addition, Neal and colleagues developed an air-liquid interface (ALI) method for culturing patient-derived organoids (PDOs), which enabled the propagation of tumor cells and embedded immune cell populations (including T, B, and NK cells and CD14+ or CD68+ macrophages) for several weeks (Neal et al., 2018). Similarly, others have described the use of: a) co-cultured mouse-derived cancer organoids with autologous immune cells for the study of PD-1:PDL-1 interactions (Chakrabarti et al., 2018); b) heterotypic co-culture of CRC spheroids with immune cells to examine the infiltration and ensuing response of T and NK cells (Courau et al., 2019); c) organotypic tumor spheroids to identify associations between certain cytokines and chemokines and anti-tumor immunity (Jenkins et al., 2018); and d) co-culture of CRC-derived organoids with autologous TILs to predict cellular responses to chemoradiotherapy (Kong et al., 2018). Of note, Kong and colleagues demonstrated that antitumor immunity could be rescued by the addition of antibodies targeting PD-1, highlighting the potential utility of coculturing TILs with tumor organoids to uncover new cancer immunotherapies.
2.3. In vivo tumor characterization
One challenge of in vitro models is how best to recapitulate both the proportion of immune cell populations and their functional state within the TME. An excellent example of this is CD8+ Tex (for a comprehensive review please see (McLane et al., 2019)). Effectively recapitulating the Tex cell state in cell culture has been challenging, although new chronic stimulation methods developed to study T cell exhaustion in vitro show promise (Vardhana et al., 2020). Chronic infection models, such as LCMV Clone 13, and syngeneic tumor models remain the gold-standard for studying this cellular state. Indeed, syngeneic mouse models also remain the current workhorse model system for studying tumor-immune cell interactions, as well as for testing the efficacy of new immunotherapies (e.g., checkpoint inhibitors) (Allard et al., 2016). However, orthotopic xenograft mouse models may better recapitulate the relevant TME context (Sullivan et al., 2019). Interestingly, one recent study demonstrated that tumor infiltration by various immune cells could vary between models that used syngeneic versus orthotopic xenografts, highlighting the potential influence of tumor location on immune cell response (Zhao et al., 2017).
3. EMERGING APPROACHES FOR MODELING TUMOR-IMMUNE METABOLIC INTERACTIONS
In mice and humans, great progress has been made in the study of in vivo metabolism through the use of stable isotope-labeled nutrients in tumors or normal tissues. In some cases, these studies have also lent insights into how metabolic activity may vary between tumors in a tissue-dependent manner (Fernández-García et al., 2020; Jang et al., 2018). In contrast to typical in vitro methods, in vivo tracer studies further enable the consideration of nutrient and oxygen gradients in the context of tissue architecture, intracellular interactions, and systemic metabolism (Figure 2). For instance, recent work by Ma and colleagues used stable isotope tracer and 13C-glucose infusion techniques to track and compare glucose utilization by CD8+ T cells responding to infection in vitro and in vivo (Ma et al., 2019). This work revealed that, in contrast to their metabolic phenotypes exhibited in vitro, CD8+ T cells in vivo displayed an oxidative phenotype and used glucose carbon for anabolic demands such as nucleotide and serine biosynthesis rather than for lactate production. In another recent study, Lercher and colleagues used 13C-arginine infusion to understand how hepatic metabolism is modulated in response to viral infection (Lercher et al., 2019). Together these studies provide a potential foundation for studying the impact of local and systemic environments on T cell metabolism and protective immunity (Figure 1).
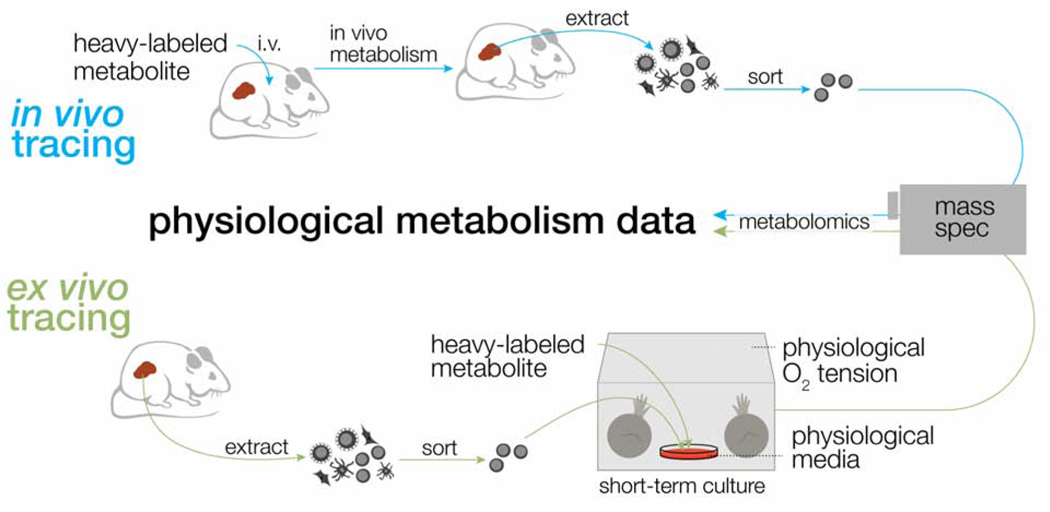
FIGURE 2: Approaches for modeling physiologic immunometabolism using stable isotope tracing.
Isotope tracers, in the form of heavy-labeled metabolites, can be administered intravenously (top) or ex vivo to sorted cell populations (bottom) to achieve accurate tracing of in vivo metabolic pathways. Ex vivo tracing (bottom), in combination with physiologic media and oxygen tension, offers a path for physiological modeling of the metabolic state of specific cell subsets in vivo that require extensive sorting after extraction.
Yet, caveats also remain for in vivo studies of immunometabolism. One challenge in using 13C tracers to study immune cell metabolism in vivo is the natural cellular heterogeneity that exists in different tissues. Given the heterocellular complexity within a tumor, the use of isotopically-labeled infusion techniques typically requires eventual isolation (e.g., via sorting or kit-based purification) of a specific cell subset. Sorting immune cell subsets after isolation takes time (30 minutes to several hours) and often involves culture in minimal buffers that promote changes in metabolite pool sizes (Llufrio et al., 2018). Although this timescale preceding metabolite extraction could be incompatible with retaining the metabolic profile of cells in vivo (Cantor, 2019; Llufrio et al., 2018), there is evidence that T cells retain their metabolic programming during short-term culture after sorting (Ma et al., 2019). Taking a different approach, sorting immune cells into distinct cell populations (i.e., Teff versus Tex cells) followed by immediate ex vivo 13C tracer analysis represents a feasible way to model immune cell metabolism in the TME, particularly when studying complex cellular heterogeneity or rare cell populations. By further incorporating the use of physiological media and more physiologically relevant oxygen tensions, ex vivo tracing experiments could potentially offer a more tangible and accessible approach for mimicking in vivo conditions (Figure 2).
Extending beyond metabolomic snapshots of isolated cells, imaging techniques such as matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometry have the potential to further elucidate spatial metabolite composition and nutrient gradients within intact tissues. For example, Fan and colleagues used an ex vivo stable isotope-resolved metabolomics (SIRM) technique and coupled histological characterization for samples resected from non-small cell lung cancer patients to investigate tissue metabolism over time (Fan et al., 2016b). Further, Hensley and colleagues infused non-small cell lung cancer patients with 13C-glucose prior to surgical resection, which revealed that tumor areas with high perfusion versus low perfusion exhibited different metabolic phenotypes (Hensley et al., 2016). Others have mapped metabolite clusters in human CRC based on 3D desorption electrospray ionization (DESI)-MS techniques (Inglese et al., 2017). Perhaps the integration of spatial metabolite profiling and high-dimensional immune cell imaging techniques (Hartmann and Bendall, 2020) will bring us closer to achieving single cell resolution of metabolism within complex tissue environments.
4. CONCLUSION
The metabolic networks of different cells are dictated by the sum and interplay of several intertwined cell-intrinsic and environmental factors. By integrating this concept with recent technical advances such as physiologic culture media, culture systems that dynamically buffer the concentrations of certain nutrients, stable isotope tracing, and in vivo metabolite imaging, it should be possible to develop new strategies that advance our understanding of immunometabolism in the TME. In addition, co-culture methods that integrate tumor organoids and peripheral blood lymphocytes (Dijkstra et al., 2018) have the potential to support more “personalized” metabolic modeling of patient TME conditions. The culture of ex vivo tissue slices (Fan et al., 2016a) or PDOs from patient-derived tumors (Neal et al., 2018) in conditions that more closely recapitulate those in the human body may enable opportunities to more closely model metabolic cross-talk between diverse cell types that reside in the TME. Therefore, contextualizing the study of immunometabolism will be important to improve our understanding of immune cell function and how such knowledge can be leveraged to treat human disease.
Acknowledgments
We thank members of the Jones laboratory for discussion and critical reading of the manuscript and Jeanie Wedberg and Michelle Minard for administrative assistance. This work was supported by grants from the NIH (K22CA225864 to JRC) and CIHR (MOP-142259 to RGJ), as well as funds from the Van Andel Institute (to RGJ).
Footnotes
Declaration of Interests
J.R.C. is an inventor on a patent for HPLM (PCT/US2017/061377). RGJ is a consultant for Agios Pharmaceuticals and serves on the Scientific Advisory Board of ImmunoMet Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann T, and Tardito S. (2019). Cell culture medium formulation and its implications in cancer metabolism. Trends in Cancer 5, 329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard B, Allard D, and Stagg J. (2016). Methods to evaluate the antitumor activity of immune checkpoint inhibitors in preclinical studies In Methods in Molecular Biology, (Humana Press Inc.), pp. 159–177. [DOI] [PubMed] [Google Scholar]
- Ang QY, Alexander M, Newman JC, Tian Y, Cai J, Upadhyay V, Turnbaugh JA, Verdin E, Hall KD, Leibel RL, et al. (2020). Ketogenic Diets Alter the Gut Microbiome Resulting in Decreased Intestinal Th17 Cells. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Levine MH, Wang Z, Quinn WJ, Kopinski PK, Wang L, et al. (2017). Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 25, 1282–1293.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora M. (2013). Cell Culture Media: A Review. Mater. Methods 3. [Google Scholar]
- Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, Whitney PG, Fernandez-Ruiz D, Dähling S, Kastenmüller W, et al. (2019). Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8+ T Cells. Immunity 51, 285–297.e5. [DOI] [PubMed] [Google Scholar]
- Bader JE, Voss K, and Rathmell JC (2020). Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol. Cell 78, 1019–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer ML, Ma EH, Bantug GR, Grählert J, Pfister S, Glatter T, Jauch A, Dimeloe S, Slack E, Dehio P, et al. (2016). Memory CD8+ T Cells Require Increased Concentrations of Acetate Induced by Stress for Optimal Function. Immunity 44, 1312–1324. [DOI] [PubMed] [Google Scholar]
- Ben-Shoshan J, Maysel-Auslender S, Mor A, Keren G, and George J. (2008). Hypoxia controls CD4+ CD25+ regulatory T-cell homeostasis via hypoxia-inducible factor-1α. Eur. J. Immunol 38, 2412–2418. [DOI] [PubMed] [Google Scholar]
- Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, Stelekati E, McLane LM, Paley MA, Delgoffe GM, et al. (2016). Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8+ T Cell Exhaustion. Immunity 45, 358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertout JA, Patel SA, and Simon MC (2008). The impact of O2 availability on human cancer. Nat. Rev. Cancer 8, 967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs MB, Medlock GL, Moutinho TJ, Lees HJ, Swann JR, Kolling GL, and Papin JA (2017). Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, Wang T, Chen WW, Clish CB, and Sabatini DM (2014). Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 508, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vázquez G, Yurchenko E, Raissi TC, VanderWindt GJW, Viollet B, Pearce EL, et al. (2015). The Energy Sensor AMPK Regulates T Cell Metabolic Adaptation and Effector Responses InVivo. Immunity 42, 41–54. [DOI] [PubMed] [Google Scholar]
- Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et al. (2016). LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metab. 24, 657–671. [DOI] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, and Pearce EL (2017). Metabolic Instruction of Immunity. Cell 169, 570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcinotto A, Filipazzi P, Grioni M, Iero M, De Milito A, Ricupito A, Cova A, Canese R, Jachetti E, Rossetti M, et al. (2012). Modulation of Microenvironment Acidity Reverses Anergy in Human and Murine Tumor-Infiltrating T Lymphocytes. Cancer Res. 72, 2746–2756. [DOI] [PubMed] [Google Scholar]
- Cantor JR (2019). The Rise of Physiologic Media. Trends Cell Biol. 29, 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR, and Sabatini DM (2012). Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2, 881–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR, Abu-Remaileh M, Kanarek N, Freinkman E, Gao X, Louissaint A, Lewis CA, and Sabatini DM (2017). Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase HHS Public Access. Cell 169, 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, and Xavier JB (2017). Metabolic origins of spatial organization in the tumor microenvironment. Proc. Natl. Acad. Sci. U. S. A 114, 2934–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, and Frauwirth KA (2010). Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol 185, 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti J, Holokai L, Syu L, Steele NG, Chang J, Wang J, Ahmed S, Dlugosz A, and Zavros Y. (2018). Hedgehog signaling induces PD-L1 expression and tumor cell proliferation in gastric cancer. Oncotarget 9, 37439–37457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cham CM, and Gajewski TF (2005). Glucose Availability Regulates IFN-γ Production and p70S6 Kinase Activation in CD8 + Effector T Cells. J. Immunol [DOI] [PubMed] [Google Scholar]
- Chang C-H, Curtis JD, Maggi LB, Faubert B, Villarino AV, O’Sullivan D, Huang SC-C, van der Windt GJW, Blagih J, Qiu J, et al. (2013a). Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis. Cell 153, 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-H, Curtis JD, Maggi LB, Faubert B, Villarino AV, O’Sullivan D, Huang SC-C, van der Windt GJW, Blagih J, Qiu J, et al. (2013b). Posttranscriptional Control of T Cell Effector Function by Aerobic Glycolysis. Cell 153, 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-H, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJW, et al. (2015). Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 162, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courau T, Bonnereau J, Chicoteau J, Bottois H, Remark R, Assante Miranda L, Toubert A, Blery M, Aparicio T, Allez M, et al. (2019). Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J. Immunother. Cancer 7, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, et al. (2003). HIF-1α is essential for myeloid cell-mediated inflammation. Cell 112, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csóka B, Selmeczy Z, Koscsó B, Németh ZH, Pacher P, Murray PJ, Kepka-Lenhart D, Morris SM, Gause WC, Leibovich SJ, et al. (2012). Adenosine promotes alternative macrophage activation via A2A and A2B receptors. FASEB J. 26, 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. (2016). Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 23, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelNero P, Hopkins BD, Cantley LC, and Fischbach C. (2018). Cancer metabolism gets physical. Sci. Transl. Med 10, eaaq1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, et al. (2018). Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 174, 1586–1598.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AS, Hsieh WY, Minarrieta L, Duong TN, Kim KKO, Desousa BR, Andreyev AY, Bowman CE, Caradonna K, Dranka BP, et al. (2018). Etomoxir Inhibits Macrophage Polarization by Disrupting CoA Homeostasis. Cell Metab. 28, 490–503.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, Orabona C, Bianchi R, Belladonna ML, Volpi C, et al. (2006). The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J. Immunol 176, 6752–6761. [DOI] [PubMed] [Google Scholar]
- Fan T, Lane A, and Higashi R. (2016a). Stable Isotope Resolved Metabolomics Studies in ex vivo Tissue Slices. Bio Protoc. 6, e1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW-M, Warmoes MO, Sun Q, Song H, Turchan-Cholewo J, Martin JT, Mahan A, Higashi RM, and Lane AN (2016b). Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator β-glucan in a two-case ex vivo non-small-cell lung cancer study. Mol. Case Stud 2, a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TWM, Bruntz RC, Yang Y, Song H, Chernyavskaya Y, Deng P, Zhang Y, Shah PP, Beverly LJ, Qi Z, et al. (2019). De novo synthesis of serine and glycine fuels purine nucleotide biosynthesis in human lung cancer tissues. J. Biol. Chem 294, 13464–13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Li KYY, Cai L, Hensley CTT, Kim J, Zacharias LGG, Yang C, Do QNN, Doucette S, Burguete D, et al. (2017). Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-García J, Altea-Manzano P, Pranzini E, and Fendt SM (2020). Stable Isotopes for Tracing Mammalian-Cell Metabolism In Vivo. Trends Biochem. Sci 45, 185–20. [DOI] [PubMed] [Google Scholar]
- Finlay DK, Rosenzweig E, Sinclair LV, Carmen FC, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, and Cantrell DA (2012). PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med 209, 2441–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S, et al. (2007). Inhibitory effect of tumor cell–derived lactic acid on human T cells. Blood 109, 3812–3819. [DOI] [PubMed] [Google Scholar]
- Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, et al. (2016). A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 18, 827–838. [DOI] [PubMed] [Google Scholar]
- Gao X, Sanderson SM, Dai Z, Reid MA, Cooper DE, Lu M, Richie JP, Ciccarella A, Calcagnotto A, Mikhael PG, et al. (2019). Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Rieckmann JCC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al. (2016). L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. (2015). The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med 21, 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, Warmoes MO, De Cubas AA, MacIver NJ, Locasale JW, et al. (2016). Foxp3 and Toll-like receptor signaling balance T reg cell anabolic metabolism for suppression. Nat. Immunol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, and Wargo JA (2018). The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 33, 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropper Y, Feferman T, Shalit T, Salame T-M, Porat Z, and Shakhar G. (2017). Culturing CTLs under Hypoxic Conditions Enhances Their Cytolysis and Improves Their Anti-tumor Function. Cell Rep. 20, 2547–2555. [DOI] [PubMed] [Google Scholar]
- Halbrook CJ, Pasca di Magliano M, and Lyssiotis CA (2018). Tumor cross-talk networks promote growth and support immune evasion in pancreatic cancer. Am. J. Physiol. Gastrointest. Liver Physiol 315, G27–G35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FJ, and Bendall SC (2020). Immune monitoring using mass cytometry and related high-dimensional imaging approaches. Nat. Rev. Rheumatol 16, 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FJ, Mrdjen D, McCaffrey E, Glass DR, Greenwald NF, Bharadwaj A, Khair Z, Verberk SGS, Baranski A, Baskar R, et al. (2020). Single-cell metabolic profiling of human cytotoxic T cells. Nat. Biotechnol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield SM, Kjaergaard J, Lukashev D, Schreiber TH, Belikoff B, Abbott R, Sethumadhavan S, Philbrook P, Ko K, Cannici R, et al. (2015). Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci. Transl. Med 7, 277ra30–277ra30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. (2016). Metabolic Heterogeneity in Human Lung Tumors. Cell 164, 681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P-C, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui Y-C, Cui G, Micevic G, Perales JC, et al. (2015). Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 162, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden AJM, Hukelmann JL, Brenes A, Spinelli L, Sinclair LV, Lamond AI, and Cantrell DA (2019). Quantitative analysis of T cell proteomes and environmental sensors during T cell differentiation. Nat. Immunol 20, 1542–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Apasov S, Koshiba M, and Sitkovsky M. (1997). Role of A2a extracellular adenosine receptor-mediated signaling in adenosine-mediated inhibition of T-cell activation and expansion. Blood 90, 1600–1610. [PubMed] [Google Scholar]
- Huang SCC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, et al. (2014). Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol 15, 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber V, Camisaschi C, Berzi A, Ferro S, Lugini L, Triulzi T, Tuccitto A, Tagliabue E, Castelli C, and Rivoltini L. (2017). Cancer acidity: An ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin. Cancer Biol 43, 74–89. [DOI] [PubMed] [Google Scholar]
- Hui S, Ghergurovich JMJ, Morscher RRJ, Jang C, Nature XT-, 2017, undefined, Teng X, Lu W, Esparza LA, Reya T, et al. (2017). Glucose feeds the TCA cycle via circulating lactate. 551, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S, Cowan AJ, Zeng X, Yang L, TeSlaa T, Li X, Bartman C, Zhang Z, Jang C, Wang L, et al. (2020). Quantitative Fluxomics of Circulating Metabolites. Cell Metab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese P, McKenzie JS, Mroz A, Kinross J, Veselkov K, Holmes E, Takats Z, Nicholson JK, and Glen RC (2017). Deep learning and 3D-DESI imaging reveal the hidden metabolic heterogeneity of cancer. Chem. Sci 8, 3500–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C, Chen L, and Rabinowitz JD (2018). Metabolomics and Isotope Tracing. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, Bowden M, Deng J, Liu H, Miao D, et al. (2018). Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 8, 196–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AK, Huang SCC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. (2015). Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430. [DOI] [PubMed] [Google Scholar]
- Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Maseda D, Liberti MV, Paz K, Kishton RJ, et al. (2018). Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell 175, 1780–1795.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaymak I, Maier CR, Schmitz W, Campbell AD, Dankworth B, Ade CP, Walz S, Paauwe M, Kalogirou C, Marouf H, et al. (2020). Mevalonate pathway provides ubiquinone to maintain pyrimidine synthesis and survival in p53-deficient cancer cells exposed to metabolic stress. Cancer Res. 80, 189–203. [DOI] [PubMed] [Google Scholar]
- Klein Geltink RI, O’Sullivan D, Corrado M, Bremser A, Buck MD, Buescher JM, Firat E, Zhu X, Niedermann G, Caputa G, et al. (2017). Mitochondrial Priming by CD28. Cell 171, 385–397.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN, and Hafler DA (2013). Sodium chloride drives autoimmune disease by the induction of pathogenic TH 17 cells. Nature 496, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H, and Chandel NS (2018). Regulation of redox balance in cancer and T cells. J. Biol. Chem 293, 7499–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong JCH, Guerra GR, Millen RM, Roth S, Xu H, Neeson PJ, Darcy PK, Kershaw MH, Sampurno S, Malaterre J, et al. (2018). Tumor-Infiltrating Lymphocyte Function Predicts Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. JCO Precis. Oncol 1–15. [DOI] [PubMed] [Google Scholar]
- Krzywinska E, and Stockmann C. (2018). Hypoxia, metabolism and immune cell function. Biomedicines 6, E56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJF, Mackay GM, Vousden KH, and Maddocks ODK (2014). Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 7, 1248–1258. [DOI] [PubMed] [Google Scholar]
- Lau AN, and Vander Heiden MG (2020). Metabolism in the Tumor Microenvironment. Annu. Rev. Cancer Biol 4, annurev-cancerbio-030419–033333. [Google Scholar]
- Leney-Greene MA, Boddapati AK, Su HC, Cantor JR, and Lenardo MJ (2020). Human Plasma-like Medium Improves T Lymphocyte Activation. IScience 23, 100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone RD, Zhao L, Englert JM, Sun I-HI-M, Oh M-H, Sun I-HI-M, Arwood ML, Bettencourt IA, Patel CH, Wen J, et al. (2019). Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science (80-. ). 366, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher A, Bhattacharya A, Popa AM, Caldera M, Schlapansky MF, Baazim H, Agerer B, Gürtl B, Kosack L, Májek P, et al. (2019). Type I Interferon Signaling Disrupts the Hepatic Urea Cycle and Alters Systemic Metabolism to Suppress T Cell Function. Immunity 51, 1074–1087.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, et al. (2017). Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet 49, 708–718. [DOI] [PubMed] [Google Scholar]
- Lim AR, Rathmell WK, and Rathmell JC (2020). The tumor microenvironment as a metabolic barrier to effector T cells and immunotherapy. Elife 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llufrio EM, Wang L, Naser FJ, and Patti GJ (2018). Sorting cells alters their redox state and cellular metabolome. Redox Biol. 16, 381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, Simon MC, and Thompson CB (2007). The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 21, 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, and Vander Heiden MG (2011). Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu. Rev. Cell Dev. Biol 27, 441–464. [DOI] [PubMed] [Google Scholar]
- Lyssiotis CA, and Kimmelman AC (2017). Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 27, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer ML, et al. (2017). Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab. 25, 345–357. [DOI] [PubMed] [Google Scholar]
- Ma EH, Verway MJ, Johnson RM, Roy DG, Steadman M, Hayes S, Williams KS, Sheldon RD, Samborska B, Kosinski PA, et al. (2019). Metabolic Profiling Using Stable Isotope Tracing Reveals Distinct Patterns of Glucose Utilization by Physiologically Activated CD8+ T Cells. Immunity 51, 856–870.e5. [DOI] [PubMed] [Google Scholar]
- McLane LM, Abdel-Hakeem MS, and Wherry EJ (2019). CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol 37, 457–495. [DOI] [PubMed] [Google Scholar]
- Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, and Bradfield CA (2010). An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol 185, 3190–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, and Rathmell JC (2011). Cutting Edge: Distinct Glycolytic and Lipid Oxidative Metabolic Programs Are Essential for Effector and Regulatory CD4 + T Cell Subsets. J. Immunol 186, 3299–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A, Danai LV, and Vander Heiden MG (2018). Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis. Model. Mech 11, dmm035758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, Sharma MD, Baban B, Harding HP, Zhang Y, Ron D, and Mellor AL (2005). GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22, 633–642. [DOI] [PubMed] [Google Scholar]
- Najjar YG, Menk AV, Sander C, Rao U, Karunamurthy A, Bhatia R, Zhai S, Kirkwood JM, and Delgoffe GM (2019). Tumor cell oxidative metabolism as a barrier to PD-1 blockade immunotherapy in melanoma. JCI Insight 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou S-HH, Salahudeen AA, Smith AR, et al. (2018). Organoid Modeling of the Tumor Immune Microenvironment. Cell 175, 1972–1988.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LAJ, and Pearce EJ (2016). Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med 213, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan D, Sanin DE, Pearce EJ, and Pearce EL (2019). Metabolic interventions in the immune response to cancer. Nat. Rev. Immunol 19, 324–335. [DOI] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, et al. (2011). An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203. [DOI] [PubMed] [Google Scholar]
- Palazon A, Goldrath AW, Nizet V, and Johnson RS (2014). HIF Transcription Factors, Inflammation, and Immunity. Immunity 41, 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazon A, Tyrakis PA, Macias D, Veliça P, Rundqvist H, Fitzpatrick S, Vojnovic N, Phan AT, Loman N, Hedenfalk I, et al. (2017). An HIF-1α/VEGF-A Axis in Cytotoxic T Cells Regulates Tumor Progression. Cancer Cell 32, 669–683.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE, et al. (2017). Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, et al. (2015). PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun 6, 6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Poffenberger MC, Chang CH, and Jones RG (2013). Fueling immunity: Insights into metabolism and lymphocyte function. Science (80-. ). 342, 1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previte DM, Martins CP, O’Connor EC, Marre ML, Coudriet GM, Beck NW, Menk AV, Wright RH, Tse HM, Delgoffe GM, et al. (2019). Lymphocyte Activation Gene-3 Maintains Mitochondrial and Metabolic Quiescence in Naive CD4 + T Cells. Cell Rep 27, 129–141.e4. [DOI] [PubMed] [Google Scholar]
- Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, et al. (2011). The human serum metabolome. PLoS One 6, e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucino V, Certo M, Bulusu V, Cucchi D, Goldmann K, Pontarini E, Haas R, Smith J, Headland SE, Blighe K, et al. (2019). Lactate Buildup at the Site of Chronic Inflammation Promotes Disease by Inducing CD4+ T Cell Metabolic Rewiring. Cell Metab. 30, 1055–1074.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Villa M, Sanin DE, Buck MD, O’Sullivan D, Ching R, Matsushita M, Grzes KM, Winkler F, Chang CH, et al. (2019). Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell Rep. 27, 2063–2074.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raud B, Roy DG, Divakaruni AS, Tarasenko TN, Franke R, Ma EH, Samborska B, Hsieh WY, Wong AH, Stüve P, et al. (2018). Etomoxir Actions on Regulatory and Memory T Cells Are Independent of Cpt1a-Mediated Fatty Acid Oxidation. Cell Metab. 28, 504–515.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig F, and Schulze A. (2016). The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749. [DOI] [PubMed] [Google Scholar]
- Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Spinelli JB, et al. (2016). Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell Metab. 24, 104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron-Harel N, Ghergurovich JM, Notarangelo G, LaFleur MW, Tsubosaka Y, Sharpe AH, Rabinowitz JD, and Haigis MC (2019). T Cell Activation Depends on Extracellular Alanine. Cell Rep. 28, 3011–3021.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks MG, and Garrett WS (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy DG, Chen J, Mamane V, Ma EH, Muhire BM, Sheldon RD, Shorstova T, Koning R, Johnson RM, Esaulova E, et al. (2020). Methionine Metabolism Shapes T Helper Cell Responses through Regulation of Epigenetic Reprogramming. Cell Metab. 31, 250–266.e9. [DOI] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, and Chi H. (2011). HIF1αdependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med 208, 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair LV, Howden AJM, Brenes A, Spinelli L, Hukelmann JL, Macintyre AN, Liu X, Thomson S, Taylor PM, Rathmell JC, et al. (2019). Antigen receptor control of methionine metabolism in T cells. Elife 8, e44210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer K, Cheng W-C, Kreutz M, Ho P-C, and Siska PJ (2018). Immunometabolism in cancer at a glance. Dis. Model. Mech 11, dmm034272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siska PJ, Beckermann KE, Mason FM, Andrejeva G, Greenplate AR, Sendor AB, Chiang YCJ, Corona AL, Gemta LF, Vincent BG, et al. (2017). Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, and Rathmell JC (2018). Metabolic Barriers to T Cell Function in Tumors. J. Immunol 200, 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MR, Danai LV, Lewis CA, Chan SH, Gui DY, Kunchok T, Dennstedt EA, Vander Heiden MG, Muir A, Heiden MGV, et al. (2019). Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife 8, e44235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T-H, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. (2018). The Immune Landscape of Cancer. Immunity 48, 812–830.e14. [Google Scholar]
- Triplett TA, Garrison KC, Marshall N, Donkor M, Blazeck J, Lamb C, Qerqez A, Dekker JD, Tanno Y, Lu W-CC, et al. (2018). Reversal of indoleamine 2,3-dioxygenasemediated cancer immune suppression by systemic kynurenine depletion with a therapeutic enzyme. Nat. Biotechnol 36, 758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, Ubags N, Fajas L, Nicod LP, and Marsland BJ (2018). Dietary Fiber Confers Protection against Flu by Shaping Ly6c− Patrolling Monocyte Hematopoiesis and CD8+ T Cell Metabolism. Immunity 48, 992–1005.e8. [DOI] [PubMed] [Google Scholar]
- Vardhana SA, Hwee MA, Berisa M, Wells DK, Yost KE, King B, Smith M, Herrera PS, Chang HY, Satpathy AT, et al. (2020). Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodnala SK, Eil R, Kishton RJ, Sukumar M, Yamamoto TN, Ha NH, Lee PH, Shin MH, Patel SJ, Yu Z, et al. (2019). T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science (80-. ). 363, eaau0135v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorde J. Vande, Ackermann T, Pfetzer N, Sumpton D, Mackay G, Kalna G, Nixon C, Blyth K, Gottlieb E, and Tardito S. (2019). Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci. Adv 5, eaau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yosef N, Gaublomme J, Wu C, Lee Y, Clish CB, Kaminski J, Xiao S, Zu Horste GM, Pawlak M, et al. (2015). CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity. Cell 163, 1413–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Franco F, Tsui YC, Xie X, Trefny MP, Zappasodi R, Mohmood SR, Fernández-García J, Tsai CH, Schulze I, et al. (2020). CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol 21, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CPP, Shi LZZ, Milasta S, Carter R, Finkelstein D, McCormick LLL, Fitzgerald P, Chi H, Munger J, et al. (2011). The Transcription Factor Myc Controls Metabolic Reprogramming upon T Lymphocyte Activation. Immunity 35, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956). On the origin of cancer cells. Science (80-. ). 123, 309–314. [DOI] [PubMed] [Google Scholar]
- van der Windt GJW, Everts B, Chang C-H, Curtis JD, Freitas TC, Amiel E, Pearce EJ, and Pearce EL (2012). Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. (2013). HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 41, D801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpaw AJ, and Dang CV (2018). Exploiting Metabolic Vulnerabilities of Cancer with Precision and Accuracy. Trends Cell Biol. 28, 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, and Kuchroo VK (2013). Induction of pathogenic TH 17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Dai Z, and Locasale JW (2019). Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun 10, 3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kurupati R, Liu L, Zhou XY, Zhang G, Hudaihed A, Filisio F, Giles-Davis W, Xu X, Karakousis GC, et al. (2017). Enhancing CD8+ T Cell Fatty Acid Catabolism within a Metabolically Challenging Tumor Microenvironment Increases the Efficacy of Melanoma Immunotherapy. Cancer Cell 32, 377–391.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li L, Starr TK, and Subramanian S. (2017). Tumor location impacts immune response in mouse models of colon cancer. Oncotarget 8, 54775–54787. [DOI] [PMC free article] [PubMed] [Google Scholar]