Abstract
The superfamily 1 helicase nonstructural protein 13 (nsp13) is required for SARS-CoV-2 replication. The mechanism and regulation of nsp13 has not been explored at the single-molecule level. Specifically, force-dependent unwinding experiments have yet to be performed for any coronavirus helicase. Here, using optical tweezers, we find that nsp13 unwinding frequency, processivity, and velocity increase substantially when a destabilizing force is applied to the RNA substrate. These results, along with bulk assays, depict nsp13 as an intrinsically weak helicase that can be activated >50-fold by piconewton forces. Such force-dependent behavior contrasts the known behavior of other viral monomeric helicases, such as hepatitis C virus NS3, and instead draws stronger parallels to ring-shaped helicases. Our findings suggest that mechanoregulation, which may be provided by a directly bound RNA-dependent RNA polymerase, enables on-demand helicase activity on the relevant polynucleotide substrate during viral replication.
Significance
SARS-CoV-2 relies on its helicase, nonstructural protein 13 (nsp13) for viral replication. Nsp13 in SARS-Cov-2 has >99% sequence identity to its ortholog in SARS-CoV-1, suggesting a conserved helicase mechanism across coronaviridae. However, this mechanism has not been investigated at single-molecule detail as necessary to compare it with those of other well-studied viral helicases. Here, using optical tweezers assays, we show that the catalytic efficiency of nsp13 RNA unwinding activity is increased >50-fold by applied forces. This strong force dependence is phenomenologically similar to bacteriophage helicases, but contrasts the behavior of hepatitis C NS3 helicase. Overall, our results indicate that nsp13 may rely on external forces, potentially provided by viral polymerase, for its activity in the context of viral replication.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-stranded RNA coronavirus responsible for the coronavirus disease 2019 (COVID-19) pandemic (1,2). Its genome encodes 16 nonstructural proteins, including the RNA-dependent RNA polymerase nonstructural protein 12 (RdRP) and the helicase nonstructural protein 13 (nsp13) (3). Nsp13 forms a complex with RdRP and is essential for viral replication (4,5). Nsp13 from SARS-CoV-2 is a superfamily 1 helicase and is >99% similar to nsp13 from severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) (also referred to as SARS-CoV), differing by only a single amino acid (V570I) (6). This high level of conservation indicates that nsp13 from SARS-CoV-1 and -CoV-2 have a shared unwinding mechanism. It also indicates that antiviral therapeutics targeting the helicase would be effective toward SARS-CoV-1 and -CoV-2 nsp13 and potentially on helicases from future coronaviridae. Despite this therapeutic potential, the detailed mechanism and regulation of replicative helicases in coronaviridae in general remain poorly studied.
Previous work on SARS-CoV-1 nsp13 established, using bulk assays, a 5′-3′ directionality of unwinding activity (7). In addition, nsp13 has been shown to unwind both RNA and DNA substrates with approximately equal initial rates despite the virus having an RNA genome (8,9). The efficiency of nsp13 unwindase activity is low compared with other helicases, but may be enhanced through different mechanisms such as by cooperativity or by interaction with the viral RdRp (9, 10, 11), an interaction for which structural evidence has been recently obtained (5). Nonetheless, the potential mechanisms underlying nsp13 regulation have not been explored.
Mechanical forces offer potential means for the regulation of viral helicase activation. Some viral helicases, such as hexameric T4 bacteriophage gp41 and T7 bacteriophage gp4, exhibit enhanced unwinding velocities when destabilizing forces are applied to their polynucleotide substrates (12,13). Other viral helicases, such as the monomeric hepatitis C virus (HCV) nonstructural protein 3 (NS3), show little change in unwinding velocity when forces are applied to their substrate (14). However, force-dependent measurements have yet to be performed for any coronavirus helicase.
In this work, we investigate the mechanism of SARS-CoV-2 nsp13-driven polynucleotide unwinding activity using both bulk and single-molecule approaches. Using bulk assays, we find that nsp13 unwinds both DNA and RNA with weak intrinsic activity. Using optical tweezers, we find that applying piconewton forces to the nucleotide substrate increases the event frequency, unwinding velocity, and processivity of nsp13, leading to a >50-fold enhancement of catalytic activity. We directly observe the stepping behavior of nsp13, estimating one base pair (bp) unwound per ATP. Overall, our results provide a biochemical and biophysical characterization of the SARS-CoV-2 nsp13 helicase and suggest that mechanoregulation may be a sufficient means of activating its unwinding activity in vivo.
Materials and methods
Cloning
The nsp13 plasmid was generated as previously described (5). Briefly, the gene was synthesized and cloned into the pet28(a)+ vector using NdeI and XhoI restriction sites by GenScript Biotech (Piscataway, NJ). We then performed mutagenesis using the QuikChange Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) to insert an HRV-3C (PreScission; APExBIO Technology, Houston, TX) protease cleavage site and remove the thrombin cleavage site.
Protein expression and purification
The nsp13 was expressed and purified as reported previously with minimal changes (5). Briefly, the plasmid expressing the SARS-CoV-2 nsp13 helicase (SARS-CoV-2-nsp13 Pet28a or SARS-CoV-2-nsp13 Pet28a-PreScission; APExBIO Technology) was transformed in BL21 Rosetta cells, and 4–6 L of culture were grown and induced at 37 and 18°C. Cell lysate was subjected to Ni-NTA affinity, cleaved overnight with PreScission protease (APExBIO Technology), and then further subjected to cation-exchange and size-exclusion chromatography. The affinity tag-free recombinant protein was eluted in sizing buffer (25 mM HEPES (pH 7.0), 250 mM KCl, 1 mM MgCl2, 5% glycerol, and 1 mM tris(2-carboxyethyl)phosphine (TCEP)), flash frozen in liquid N2, and stored at −80°C. A full protocol can be found in the Supporting Materials and Methods.
Differential scanning fluorimetry
These experiments were carried out on a C1000 Touch Thermal cycler CFX-96 instrument (GE Healthcare, Chicago, IL). Recombinant constructs were diluted to a final concentration of 4 μM in differential scanning fluorimetry (DSF) buffer (25 mM HEPES-KOH (pH 7.5), 25 mM KCl, 5 mM MgCl2, 1 mM TCEP, and 0.005% Triton X-100 (Sigma-Aldrich, St. Louis, MO)) and assayed in a 96-well plate (Hard-Shell HSP9665; Bio-Rad Laboratories, Hercules, CA). Ligands were added to a final concentration of 1 mM. SYPRO Orange fluorescent dye (S5692, excitation 490 nm and emission 590 nm; Sigma-Aldrich) was used at 1:500 dilution. The temperature was linearly increased with a step of 0.5°C for 55 min, from 25 to 95°C, and fluorescence readings were taken at each interval. Melting temperatures were estimated as the minimal value of the first derivative of the fluorescence versus temperature curves.
Preparation of DNA and RNA duplex substrates
All oligos used for the preparation of DNA and RNA substrates were purchased from Integrated DNA Technologies (Coralville, IA). The 5′ overhang DNA substrate with fluorescent dye and quencher labels (hereafter DNA substrate) was prepared by mixing Oligo1 (5′-TTTTTTTTTTCTGATGTTAGCAGCTTCG-BHQ2-3′) and Oligo2 (5′-TAMRA-CGAAGCTGCTAACATCAG-3′) in a 1.2:1 molar ratio and annealed by heating at 95°C for 5 min in a temperature-controlled aluminum block, followed by cooling at a rate of 1°C/min to 25°C. The DNA substrate lacking quencher label was prepared by mixing unlabeled Oligo1 and Oligo2 in a 1.2:1 molar ratio and annealing as above. The 5′ overhang RNA substrate with fluorescent dye and quencher labels (hereafter RNA substrate) was prepared by mixing Oligo3 (5′-UUUUUUUUUUCUGAUGUUAGCAGCUUCG-BHQ2-3′) and Oligo4 (5′-TAMRA-CGAAGCUGCUAACAUCAG-3′) in a 1.2:1 molar ratio and annealed by heating at 75°C for 5 min in a temperature-controlled aluminum block, followed by cooling the sample at a rate of 1°C/min to 25°C. The RNA substrate lacking quencher label was prepared by mixing unlabeled Oligo3 and Oligo4 in a 1.2:1 molar ratio and annealing as per the RNA substrate. Capture oligos, complementary to the duplexed portion of the quenching strands, have the sequence 5′-CGAAGCTGCTAACATCAG-3′ and 5′-CGAAGCUGCUAACAUCAG-3′ for DNA and RNA substrates, respectively.
Fluorescence anisotropy binding assay
For fluorescence anisotropy studies, nsp13 (34 μM, in buffer containing 20 mM HEPES (pH = 7.5), 5 mM MgCl2, 150 mM KCl, 1 mM TCEP, and 3% glycerol) and partial duplex DNA/RNA (as prepared above, in assay buffer) were diluted 10× into assay buffer (20 mM HEPES (pH = 7.5), 5 mM MgCl2, 1 mM TCEP, and 0.005% Triton X-100) with KCl added to a final concentration of 25 mM (20 μL final reaction volume). Fluorescence anisotropy was measured after incubating for 10 min at room temperature using a Synergy Neo2 plate reader (532/25 nm excitation filter, 590/35 nm emission filter; Biotek, Winooski, VT) in dual photomultiplier tube mode. Gain values on the two photomultiplier tubes (reading parallel and perpendicular emission) were automatically scaled to give equal counts from a fluorophore-only control well, and the G factor was automatically adjusted.
Bulk helicase activity assays
Initial velocities of nsp13 helicase activity were measured by mixing recombinant nsp13 (10 nM) with partial duplex DNA or RNA substrates (0–6 μM) in the presence of ATP (2 mM) and capture oligo (8 μM DNA or 12 μM RNA) in 24-μL reaction volumes using solid black, 384-well NBS microplates (Corning, Corning, NY). The nsp13 was prepared as a 2× stock solution in helicase buffer (20 mM HEPES (pH 7.5), 40 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM TCEP, 0.5 mM glutathione, 0.01% Triton X-100, and 0.1 mg/mL bovine serum albumin), and 12 μL was added to reaction wells. DNA/RNA substrates were prepared as 4× stock solutions in helicase buffer followed by two-fold serial dilutions in helicase buffer. Substrates (6 μL) were added to reaction wells containing nsp13 and incubated for 10 min. ATP/capture oligos were prepared as a combined 4× stock solution in helicase buffer, and 6 μL were added to assay wells to initiate the reactions. Control reactions included wells with no enzyme and no ATP added. Substrate unwinding was measured by continuously recording the fluorescence (excitation: 544 nm and emission: 590 nm) on a Synergy Neo2 plate reader (Biotek) for 30 min in 15-s intervals. Relative fluorescence units (RFU) were converted to units of molarity with standard curves generated from DNA Oligo2 and RNA Oligo4. Enzyme parameters KM, kcat, and Hill coefficients were determined by fitting the plot of initial rates (first ∼150 s) against the substrate concentration to the Michaelis-Menten equation or the Hill equation using Prism v.6.0 (GraphPad Software).
Inhibition assays were performed by combining 4× nsp13/substrate/capture oligo (6-μL) stock solution in assay buffer with twofold ADP:AlF3 (prepared as a 2× 1:2 molar ratio stock) serial dilutions (12 μL) in helicase buffer in reaction wells. Reactions were initiated by the addition of a 4× ATP solution in the helicase buffer. Final concentrations were 10 nM nsp13, 0.5 μM DNA or RNA substrate, corresponding to 2.5 μM capture oligo, 2 mM ATP, and 0–500 μM ADP:AlF3. Control reactions containing no ATP and no ADP:AlF3 were included in the assays. Data were fitted using a sigmoidal dose dependent equation:
using Prism v.6.0 (GraphPad Software).
See Supporting Material and Methods for variable enzyme and salt concentration assay details and gel-based helicase assay details.
Preparation of substrate for optical tweezers assay
The substrate consists of two 1.5-kbp-long biotinylated double-stranded DNA (dsDNA) handles annealed to an RNA hairpin containing a 180-bp stem and a tetraloop. A 20-nt single-stranded region at the 5′ flank of the hairpin serves as the nsp13 loading site (with a sequence of 5′-CAGAAAGAAAAACCGGAUCC-3′). The hairpin sequence was made by in vitro transcription of a DNA template used previously (15) using the MEGAscript T7 Transcription Kit (Thermo Fisher Scientific, Waltham, MA). RNA was then purified using the MEGAclear Kit (Thermo Fisher Scientific). The DNA handles were generated by PCR using modified primers to generate the overhang for annealing to the hairpin-containing RNA. The forward primer for the 5′ overhang handle contains an inverted base that terminates Phusion DNA polymerase (with a sequence of 5′-CAACCATGAGCACGAATCCTAAACCT/ilnvdT/GCATAACCCCTTGGGGCCTCTAAACG-3′). The forward primer for the 3′ overhang handle contains inverted bases at its 5′ end that terminate Taq polymerase (with a sequence of 5′-/5InvdG//iInvdC//iInvdA//iInvdA//iInvdA//iInvdT//iInvdC//iInvdT//iInvdC//iInvdC//iInvdG//iInvdG//iInvdG//iInvdG//iInvdT//iInvdT//iInvdC//iInvdC//iInvdC//iInvdC//iInvdA//iInvdA//iInvdT//iInvdA//iInvdC//iInvdG/TAGTCTAGAGAATTCATTGCGTTCTGTACA/3ddC/-3′). The reverse primers for the 5′ and 3′ overhang handles contain a biotin at their respective 5′ ends for bead attachment.
The annealed complex containing the 5′ overhang handle and the RNA hairpin was assembled by mixing 1 μM 5′ overhang handles and 400 nM RNA hairpin in a total volume 20 μL of TL buffer (50 mM HEPES (pH 7.5), 55 mM KOAc, 6 mM Mg(OAc)2, and 1 mM dithiothreitol) and incubated at 65°C for 90 s and cooled to 4°C. A 10-μL reaction containing 3.5 μL of annealed complex and 1.6 μL of 0.5% streptavidin-coated polystyrene beads (2-μm diameter; Spherotech, Lake Forest, IL) was incubated on ice for 30 min. Then, the mixture was diluted with 1 mL TL buffer. A separate reaction containing 50-nM 3′ overhang handles and 1.6 μL of beads was incubated and diluted as above.
To form a tether, a bead conjugated to the 5′ overhang handle complex was immobilized on a pipette via suction and brought into close proximity to a second bead conjugated to 3′ overhang handles held in an optical trap. Upon hybridization of the 3′ overhang handle with the 5′-overhang-handle-RNA-hairpin complex, a tether was formed.
Optical tweezers measurements
All single-molecule measurements were made using an optical tweezers instrument (“MiniTweezers”; Tweezerman, Port Washington, NY) (16). The sample buffer consisted of 20 mM HEPES (pH 7.5), 20 mM KCl, 5 mM MgSO4, 1 mM TCEP, 0.1 mg/mL bovine serum albumin, and 0.005% Triton X-100. At the start of each experiment, a tether was formed in the sample chamber, and its force-extension behavior was obtained by moving the optically trapped bead at a speed of 70 nm/s away from the second bead held in the pipette. Once a single tether was confirmed by the shape of the force-extension curve, nsp13 (0.1 nM) and ATP (1 mM unless otherwise stated) were injected into the chamber, and unwinding data were collected in a force-feedback mode to maintain a constant tension in the tethered polynucleotide substrate (instrument response time ∼5-10 ms). Data were collected at 200 Hz. All experiments were conducted at 23 ± 1°C.
Single-molecule data were analyzed in MATLAB (The MathWorks, Natick, MA). The raw readout in nanometers was converted to bps by using the extensible worm-like chain model with the following parameters (17): persistence length 1 nm, contour length 0.59 nm per RNA nucleotide, and stretch modulus 1000 pN. Velocity was determined by linear fitting to the time-distance data (from the start to the end of a processive event). Processivity was determined as the maximal distance traveled before detachment. Event frequency was determined by counting the number of independent unwinding events divided by the total recording time for a given tether.
Step-size experiments were run as above but with 2 μM ATP present and 18 pN constant force. Time series data (200 Hz) were filtered using a five-data-point nonoverlapping median boxcar and fitted using the tDetector step-finding algorithm, which iteratively searches for statistically significant change points in time series data using a two-sample t-test (18,19). An α-value (significance level for t-test) of 10−7 was used. The gamma distribution parameters for the dwell-time data were estimated using the gamfit function in MATLAB (MathWorks).
Results
Biochemical characterization of SARS-CoV-2 nsp13
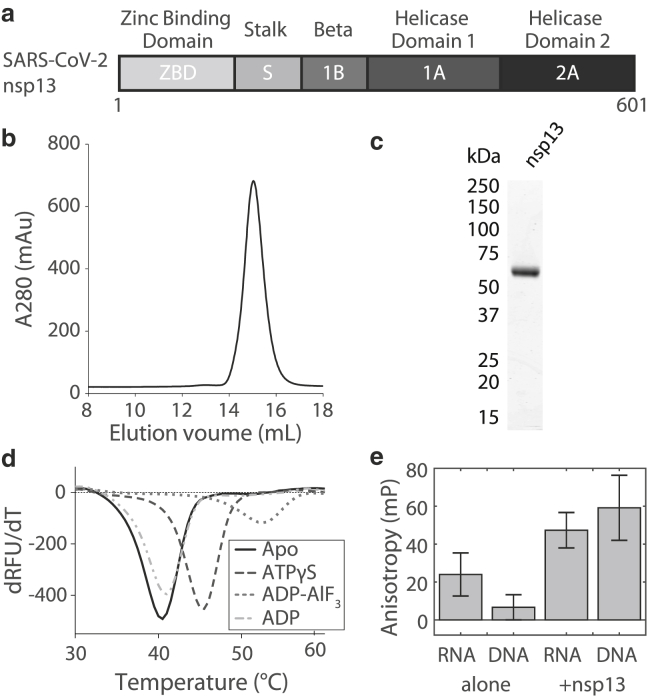
The SARS-CoV-2 nsp13 helicase consists of an N-terminal zinc-binding domain followed by stalk and β-domains (S and 1B domains, respectively) and, finally, the catalytic RecA1 and RecA2 (A1 and A2) helicase domains (Fig. 1 a). Previous structural and biochemical data on SARS-CoV-1 and MERS-CoV (4,20) have identified the zinc-binding domain and S domains as important for structural stability of the enzyme. The A1 and A2 domains regulate ATP hydrolysis and, along with the 1B domain, are proposed to regulate oligonucleotide binding.
Figure 1.
Biochemical characterization of nsp13. (a) Diagram showing the domain architecture of SARS-CoV-2 nsp13. (b) Size-exclusion chromatography (Superdex 200 Increase) of purified nsp13. (c) SDS-PAGE gel (Coomassie stain; Abcam, Cambridge, UK) showing nsp13 purity. (d) Differential scanning fluorimetry (DSF) of SARS-CoV-2 nsp13 in the absence or presence of ADP or analogs (1 mM). Melting temperatures are the following: APO, 40.5°C; ATPγS, 45.5°C; ADP-AlF3, 52.5°C; and ADP, 41.0°C for n = 2 independent measurements from two separate protein preparations. (e) Fluorescence anisotropy measurements for fluorescent DNA and RNA partial duplexes (10 nM) with and without nsp13 (3.4 μM) added. Data shown as mean ± standard deviation (SD) for n = 3 independent measurements from two separate protein preparations.
Biochemical studies of SARS-CoV-1 nsp13 indicated that a GST-tag, which has the potential to induce dimerization, could substantially alter activity (>280-fold) compared with a His6-tag (9). However, the positively charged His6-tag may also artificially influence polynucleotide binding. Therefore, for our analyses, we used recombinant, tag-free, full-length SARS-CoV-2 nsp13 (hereafter, nsp13), which we have reported recently (5). Briefly, the protein was subjected to Ni-NTA affinity, cation-exchange (CaptoS HP; Cytiva, Marlborough, MA) and size-exclusion (S200 increase) chromatography. The inclusion of an ion exchange step reduced DNA contamination and resulted in monodisperse peak off of the gel filtration column at 15 mL (Fig. 1 b) that was >95% pure by SDS-PAGE (Fig. 1 c).
To examine the nucleotide binding by nsp13, we used DSF (21). We found that nsp13 alone (apo) had a melting temperature of 40.5°C and that the addition of ATPγS or ADP-AlF3 (1 mM) shifted the melting temperature by ∼5–12°C (Fig. 1 d). These shifts indicate stabilization via ligand binding and are consistent with recent structural data showing ADP-AlF3 binding within the nsp13 active site (Fig. 1 d; (5)). No significant shift was observed in the presence of ADP (1 mM) (Fig. 1 d).
To examine nsp13 binding to RNA or DNA, we performed fluorescence anisotropy measurements. We designed fluorescently tagged dsDNA and double-stranded RNA (dsRNA) substrates containing an 18-bp duplex region with a 10-nucleotide 5′ overhang. A 10-nucleotide overhang likely allows only one copy of nsp13 to bind at a time (10). The fluorescence anisotropy of these substrates (10 nM) was measured both in the absence and presence of nsp13 (3.4 μM) in ATP-free buffer. We measured an increase in fluorescence anisotropy for both DNA and RNA, indicating that nsp13 can bind both of these substrates (Fig. 1 e). Together, these data indicate that recombinant tag-free nsp13 binds nucleotide analogs and can bind both RNA or DNA in the absence of nucleotide.
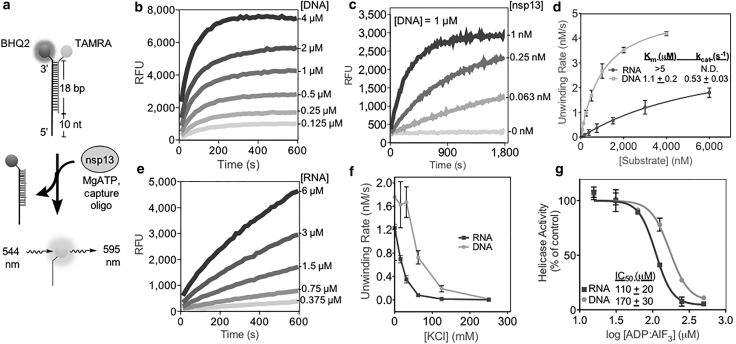
Bulk measurements of nsp13-driven DNA and RNA unwinding
To examine the helicase activity of SARS-CoV-2 nsp13, we utilized a bulk fluorescence-based assay (22). Briefly, we generated the DNA and RNA substrates described above but with the addition of a quencher on one of the unlabeled strands such that fluorescence signal is suppressed while in the duplex but not when the strands are dissociated (Fig. 2 a). The inclusion of a capture DNA oligonucleotide was required for clear changes in fluorescence signal in this assay (Fig. S1). The nsp13 (10 nM) unwound DNA in the presence of ATP (2 mM), as revealed by an increase in the fluorescence signal (Fig. 2 b). This activity was also observed using a gel-based assay (Fig. S2 a). We found that helicase activity could be readily observed at nsp13 concentrations below 100 pM in the presence of DNA (1 μM) (Fig. 2 c). We next measured the initial reaction velocity of nsp13 over a range of DNA concentrations. Fitting these data to the Michaelis-Menten equation revealed a kcat of 0.53 ± 0.03 s−1 (or 9.5 ± 0.5 bp-s−1 given the 18-bp duplex) and a KM of 1.1 ± 0.2 μM for DNA (fit ± 95% confidence intervals (CIs); Fig. 2 d). This rate of DNA unwinding is similar to that reported for HCV NS3 helicase in a fluorescence-based helicase assay (23). We determined a Hill coefficient of ∼1, indicating a lack of cooperativity in nsp13-driven unwinding for our substrates.
Figure 2.
The nsp13 is a DNA and RNA unwindase. (a) Diagram of fluorescence-based helicase assay using a partial duplex oligonucleotide substrate with 10-nt overhang. Unwinding of the duplex results in a fluorescence signal. (b) Nsp13 helicase activity (10 nM) as a function of time with increasing DNA substrate concentrations. (c) Helicase activity as a function of time at constant DNA (1 μM) and increasing nsp13 concentrations. (d) Plot of initial substrate unwinding velocities versus substrate concentration (0–4 μM for DNA and 0–6 μM for RNA). Data points show the mean ± SD for n = 3 measurements. Fitting the data points to the Michaelis-Menten model revealed KM and kcat values for DNA unwinding (inset; fit ± 95% confidence intervals (CI)). (e) Nsp13 helicase activity (10 nM) as a function of time with increasing RNA substrate concentrations. (f) Initial DNA unwinding rates at different KCl concentrations for DNA and RNA substrates. Data points show the mean ± SD for n = 3 measurements. (g) Dose-response curves of nsp13 helicase activity in the presence of ATP (2 mM) and an increasing concentration of ADP-AlF3 (0–500 μM). Data points show the mean ± SD for n = 3 measurements. IC50 values were determined by fitting the data to the Hill equation (inset).
We next examined nsp13 helicase activity using an RNA duplex with an analogous sequence. As with DNA, we observed an RNA concentration-dependent increase in fluorescence signal (Fig. 2 e). Compared with DNA, we observed that the unwinding rate for the RNA substrate was ∼2–3-fold lower and that the initial rate profile for RNA remained linear even at the highest substrate concentration (6 μM) tested (Fig. 2, d and e). Therefore, we can only estimate the lower bound of KM to be ∼5 μM for the RNA substrate used here.
To examine experimental factors that can alter helicase activity, we next repeated the bulk unwinding assay in buffers with different ionic strengths. DNA unwinding was faster than RNA unwinding (between 1.5- and 8-fold) at all salt concentrations tested (Fig. 2 f). The nsp13 activity was highest when no KCl was added to the assay buffer; increasing the salt concentration led to a decrease in the duplex unwinding rate, with no measurable activity at ≥250 mM KCl. A similar salt dependence of nsp13 helicase activity was observed in gel-based assays (Fig. S2 b). The ionic strength dependence of nsp13 helicase activity could be due to multiple factors that are not mutually exclusive, including changes in protein binding and differences in stability of the duplex substrates. Finally, ADP-AlF3 inhibited both nsp13-dependent RNA and DNA (500 nM) unwinding with similar IC50 values (Fig. 2 g). Overall, our data suggest that nsp13 possesses 5′-3′ helicase activity, albeit with a weak KM for the specific duplexes we have tested, and unwinds the DNA substrate more readily than the RNA substrate.
Single-molecule measurements of nsp13-driven RNA unwinding
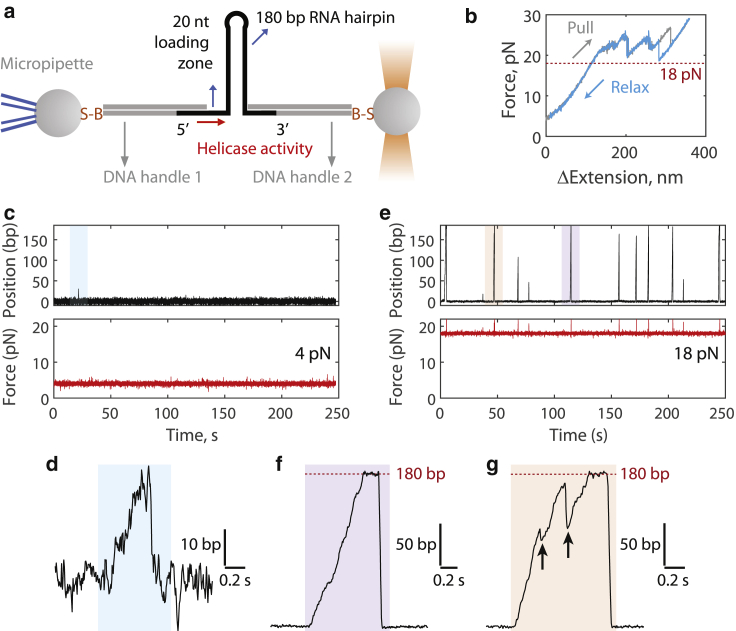
To further investigate nsp13 helicase activity, we designed an optical trap assay to directly visualize RNA unwinding at the single-molecule level. Briefly, an RNA hairpin including a 20-nucleotide 5′ loading zone, a 180-bp duplex region, and a tetraloop was annealed to two 1.5-kbp dsDNA handles, which were conjugated to polystyrene beads (see Materials and Methods; Figs. 3 a and S3). One bead was held in a micropipette, whereas the other was held in an optical trap and manipulated to apply a specified amount of force to the tether. A single tether between the two beads was ensured by measuring the force-extension curve of each tether (Fig. 3 b). Under 20 pN, the hairpin remained stably folded by itself for the duration of the experiments. Nsp13 was then injected into the sample chamber, and RNA unwinding was monitored by the changes in the optical trap position required to hold a constant force. The nsp13-catalyzed unwinding of the hairpin would result in an increase in the distance between the two beads.
Figure 3.
Single-molecule measurement of nsp13 helicase activity. (a) Geometry of the single-molecule hairpin unwinding assay. DNA handles separate the RNA hairpin from the beads (connected by the biotin-streptavidin linkage, B-S). A 20 nucleotide (nt) region allows single nsp13 molecules to bind and begin 5′-3′ helicase activity. The diagram is not to scale. The full construct design is shown in Fig. S3. (b) Representative force-extension curve of a single tether (no nsp13 present). Above 20 pN, external force begins to open the RNA hairpin. (c) Representative example trace at 4 pN of external force in the presence of nsp13. Only one event is observed in ∼250 s. (d) Zoomed-in blue region from (c). (e) Representative example trace at 18 pN of external force in the presence of nsp13. Numerous events are observed in ∼250 s. (f) Zoomed-in purple region from (e). (g) Zoomed-in orange region from (e). Arrows denote slippage or dissociation events. The red dotted line denotes 180 bp, the total length of the RNA hairpin, in (f) and (g). To see this figure in color, go online.
We first made measurements at low applied forces (4 pN) (Fig. 3 c). At this force, the unwinding activity of nsp13 (100 pM) was consistent with the low catalytic efficiency measured in the bulk assays (Fig. 2). The long time period between the unwinding events relative to the durations of these events suggest that each unwinding is likely driven by a single helicase enzyme (12). RNA unwinding events by nsp13 were rare and short, typically terminating after a few tens of bps, indicating poor processivity (Fig. 3, c and d). We next made measurements at 18 pN, the highest applied force feasible without directly starting to open the hairpin (Fig. 3 b). We observed three major differences at 18 pN (Fig. 3 e) in comparison with 4 pN: 1) unwinding events occurred more frequently at the same nsp13 concentration, 2) unwinding events became longer in distance, and 3) unwinding appeared to be much faster. In some instances, nsp13 unwound the entire 180-bp hairpin (Fig. 3 f). Interestingly, when nsp13 reached the end of the hairpin, we observed that the hairpin remained fully open for an extended period (Fig. 3, f and g). This observation contrasts previous results for T4 gp41 and DnaB helicases, which continue to translocate past the hairpin loop, leading to rezipping of the hairpin (Fig. S3 d; (12,13,24)). The dataset of n = 27 “pause” durations at 180 bp under 18 pN was exponentially distributed with a characteristic time of ∼0.26 s (Fig. S3 e). The fact that we observed pausing rather than rezipping (motions with negative slope) possibly indicates that a kinetic barrier precludes nsp13 from switching from unwinding to single-stranded translocation activities. Similar end-of-hairpin pausing was recently observed for the RNA helicase Mtr4p (25). An alternative explanation, which we do not believe is likely under our experimental conditions, is that pauses were caused by the collective action of multiple nsp13 molecules.
At 18 pN, we also observed events in which very rapid (>1 kbp-s−1) backward motions interspersed RNA unwinding (Fig. 3 g, black arrows; observed in ∼33% of the recorded traces at 18 pN). These events may correspond to backward slippage by single nsp13 molecules or to dissociation of the leading nsp13 and subsequent action of a trailing nsp13, which is possible when multiple nsp13 molecules bind in succession. Based on comparing the unwinding event frequency (∼1 event per 25 s) with the duration between backward motions (∼0.2 s), we favor the former interpretation. Furthermore, both the unwinding velocity and processivity decreased with reduced ATP (Fig. S4), indicating that nsp13 may detach from the RNA while waiting for ATP.
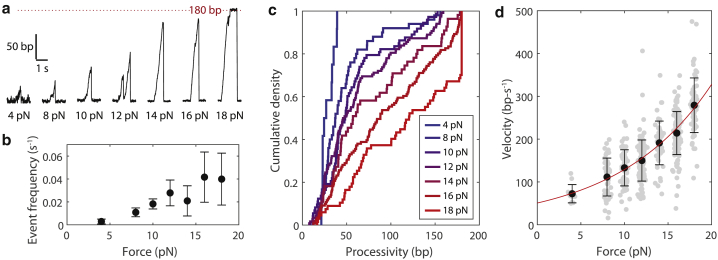
The nsp13 event frequency, processivity, and velocity are enhanced by applied force
To quantify the force dependence of nsp13 helicase activity, we made single-molecule measurements of RNA unwinding at multiple applied forces between 4 and 18 pN (Fig. 4 a). We first measured the event frequency or the number of independent unwinding events per unit time. The event frequency increased ∼14-fold between 4 and 18 pN (Fig. 4 b). Assuming a simple model in which there is no rate-limiting step to initiate unwinding, the event frequency is proportional to the on rate of nsp13 binding to RNA, meaning that applied force on the RNA substrate may increase the nsp13 on rate ∼14-fold. The low event frequency extrapolated to zero force is consistent with the weak KM for RNA substrates measured in bulk assays (Fig. 2 d). An alternative explanation is that applied force decreases the energy barrier to initiate unwinding events and that the data in Fig. 4 b represent an increased fraction of nsp13-binding events that successfully initiate unwinding of the RNA hairpin before dissociating.
Figure 4.
Force-dependent activation of nsp13 helicase activity. (a) Example traces of nsp13 unwinding hairpin RNA at 4, 8, 10, 12, 14, 16, and 18 pN force. (b) The number of unwinding initiation events per second at various forces. Data are shown as mean ± SD for n = 5–10 independent RNA hairpins. (c) Distributions of measured processivities for nsp13 at various forces (n = 12, 50, 73, 118, 55, 121, and 67 events for 4, 8, 10, 12, 14, 16, and 18 pN applied force, respectively). 180 bp is the total length of the RNA hairpin. (d) The measured velocity of nsp13 at various forces (n = 12, 50, 73, 118, 55, 121, and 67 events for 4, 8, 10, 12, 14, 16, and 18 pN applied force, respectively). Each measurement is shown as a gray dot slightly spaced out in force for visibility. Black data points show mean ± SD. Data were fitted to an exponential function v(F) = v0exp(F/Fe) (red line), revealing a y-intercept (v0) of 51 ± 9 bp and a rate term (Fe) of 10.4 ± 1.3 pN (fitted values ± 95% CI). To see this figure in color, go online.
We next measured the processivity or total length of RNA unwound during each event. The processivity increased monotonically from what was observed at 4 pN, in which no event was greater than 50 bp long, to 18 pN, in which ∼35% of the nsp13 molecules unwound the entire 180-bp duplex region (Fig. 4 c). At the lower forces measured (4–14 pN) in which events were not truncated by the limited length of the RNA hairpin, distributions of measured processivities could be fitted to a single exponential, indicating that for this given RNA hairpin, nsp13 has a constant probability of detaching as it unwinds (i.e., does not preferentially detach at a sequence-specific position) (Fig. S5).
We next measured the velocity of nsp13 in response to applied force on the RNA substrate. Remarkably, we found that the velocity increased exponentially with force, increasing from ∼70 bp-s−1 at 4 pN to ∼280 bp-s−1 at 18 pN (Fig. 4 d). Velocity is proportional to kcat, meaning that applied force increases the kcat ∼4-fold. We can also estimate the nsp13 off rate as the velocity divided by the processivity. Because both of these values approximately increase in proportion over the measured force range (Figs. 4 c and S5), we estimate the nsp13 off rate to be force independent at ∼3 s−1. We note that this value (obtained using 8–14 pN data, for which processivity was low enough to be quantified; Fig. S5) is in agreement with the inverse of the pause duration of nsp13 at the 180-bp position under the 18-pN load (∼4 s−1; Fig. S3 e). Coupling the fourfold change in the kcat with the 14-fold change in the KM, we estimate that increasing the applied force from 4–18 pN increases the catalytic efficiency >50-fold. Fitting the velocity data to an exponential function yielded an extrapolated zero-force velocity of ∼50 bp-s−1 (Fig. 4 d). Overall, we find that applying a destabilizing force to the RNA hairpin enhances nsp13 unwinding frequency, processivity, and velocity.
The nsp13 unwinds RNA in discrete steps at low ATP
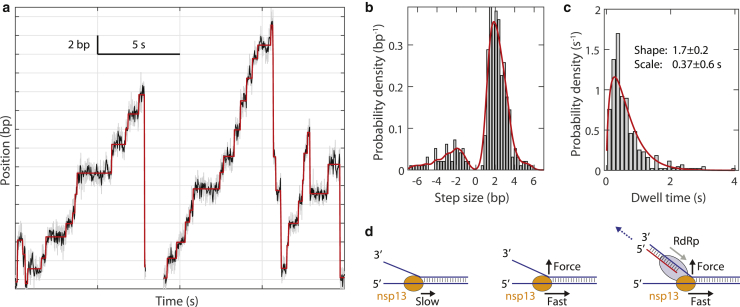
To gain further insight into the nsp13 mechanochemical cycle, we measured nsp13 unwinding activity at very low ATP concentrations (2 μM). This ATP concentration is ∼50-fold lower than the KM (Fig. S4 c), and thus, we expected to find fast unwinding motions separated by relatively long dwell times in which the enzyme waits for ATP. We indeed observed step-like motion underlying ∼5 bp-s−1 unwinding (Fig. 5 a). Forward steps were on the order of 1–3 bp and were mixed with backward steps (of similar magnitude) and larger backward motions (>3 bp). The large backward motions may represent a diffusive slip by a single nsp13 molecule or the dissociation of a leading nsp13 followed by unwinding by a trailing enzyme. We favor the former interpretation because the latter would require multiple nsp13 molecules to be within 10 nt of each other (e.g., Fig. 5 a, right trace).
Figure 5.
Stepping analysis for nsp13. (a) Example stepping data (two separate traces) for nsp13 at 18 pN force and 2 μM ATP. Shown are raw data (gray, 200 Hz) and median-filtered data (black, 40 Hz) along with output of tDetector step-finding algorithm (red). (b) Distribution of step sizes for n = 344 steps (from 29 traces). The kernel-density-smoothing function for the distribution (red) has a maximum at 1.9 and −1.9 bp. (c) Distribution of dwell times preceding each step (n = 344). Fit to the gamma distribution is shown in red, with fitted parameters (fit ± 95% CI) shown inset. (d) Diagram showing that application of destabilizing force enhances the velocity of nsp13. RdRp and nsp13 form a complex and may move in the same direction (in the configuration shown). In a plausible scenario, if the red strand is elongated while nsp13 is stationary (holding the bottom strand in place) and waiting to step, the product may be pushed outward (blue dotted arrow), leading to a destabilizing force. To see this figure in color, go online.
To quantify nsp13 stepping behavior, we collected many traces and analyzed them using the tDetector step-finding algorithm (18,19). An example step-finding output is shown in red in Fig. 5 a. A histogram of measured step sizes (n = 344 steps) is shown in Fig. 5 b. The peak step size was 2 bp. Because of the limited resolution of our assay, it is possible that smaller steps exist but are missed in this analysis.
Backward steps of various sizes were frequent (66 out of 344 measured steps, ∼19%), and the average dwell times preceding a forward and backward step were approximately equal (0.63 ± 0.03 and 0.62 ± 0.07 s, respectively; mean ± standard error of the mean for N = 278 and 66). Similar backstepping has been observed for other helicases, and it is expected that the backstep frequency should increase at low ATP concentrations (26,27). Analyzing successive steps, we measured the probability of two forward steps in a row to be 0.68 ± 0.24 (215 out of N = 315 measurements, error propagated from Poisson counting error) and two negative steps in a row to be 0.08 ± 0.04 (25 out of 315), roughly equal to the calculated probability if each step is treated as being independent (0.812 = 0.65 and 0.192 = 0.04). Furthermore, the probability of a forward step after a backward step (0.11 ± 0.05, 34 out of 315) was approximately equal to the probability of a backward step after a forward step (0.13 ± 0.06, 41 out of 315). Taken altogether, these results indicate that step type (forward or backward) is uncorrelated with dwell time and unconditional on the previous step type; such statistics are consistent with forward and backward steps being generated stochastically from the same kinetic state (28).
We next compiled the distribution of all measured dwell times preceding a step (n = 344; Fig. 5 c). The dwell-time data were fitted to the gamma distribution using maximal likelihood estimation, returning a shape factor of 1.7 ± 0.2 (fit ± 95% CI). This result is consistent with two approximately equal rate-limiting hidden steps preceding each measured unwinding or rewinding motion (28,29). Given that the peak step size was 2 bp (Fig. 5 b) and that ATP binding is the rate-limiting step in these experiments, we interpret these results to mean that two ATP binding events drive each two-bp motion; under this interpretation, one ATP is needed for each bp unwound. Other viral helicases, including NS3 (30,31) and T7 gp4 (32) helicases, have a similar step size and also hydrolyze one ATP per bp traveled.
Discussion
In this work, we show that SARS-CoV-2 nsp13 has weak intrinsic helicase activity that can be stimulated >50-fold by externally provided forces. We also provide direct measurements of the nsp13 step size. These results provide insight into the mechanism and regulation of nsp13, enabling a deeper molecular understanding of the processes underlying SARS-CoV-2 replication.
Comparing bulk and single-molecule results, we find the interesting trend that nsp13 velocity depends on substrate stability. dsRNA is more stable than dsDNA, each of which are more stable than a hairpin RNA duplex (33); we estimate velocities of ≤5, 10, and 50 bp-s−1 on these substrates, respectively (Figs. 2 d and 4 d). Furthermore, destabilizing the RNA hairpin by force increases the velocity of nsp13 (Fig. 4 d). Decreases in processivity correlated with decreases in velocity across data sets (Figs. 2 d, 4 d, and S4). Thus, the weaker KM (which depends on the processivity relative to the duplex length) of nsp13 on dsRNA relative to dsDNA (Fig. 2 d) may be due to differences in substrate stability.
Although the in vivo velocity of nsp13 has not yet been determined, the Escherichia coli replication complex travels at ∼1000 bp-s−1 (34,35). We thus speculate that nsp13 moves much faster than its basal unwinding velocity during viral replication. Our data suggest that nsp13 RNA helicase activity can be stimulated >50-fold by forces in the 10–20-pN regime that act on the nucleic acid substrate. Although the degree of activation may change if force is applied in different geometries, and more force would be needed to destabilize a truly double-stranded substrate as opposed to a hairpin (24,36), we nonetheless propose that forces in this range can activate nsp13 in vivo. E. coli RNA polymerase and T7 viral RNA polymerase can each provide ∼20 pN of force (37,38). Based on recent structural data showing that nsp13 forms a complex with RdRp (5), it is possible that asynchronous action of the RdRp on the same duplex may provide the destabilizing force necessary for force activation (Fig. 5 d). Consistently, existing bulk kinetic data show that RdRP enhances nsp13 helicase activity for SARS-CoV-1 (9). Moreover, there are numerous instances of polymerases activating helicases in other viral systems, including Zika (39), T7 (13,34,40), and T4 (41, 42, 43). We note that there are multiple alternative mechanisms by which RdRp may activate nsp13. RdRp may more directly apply force to nsp13 rather than to the RNA substrate. It is also possible that, like T7 gp4, binding to RdRp directly alters the unwinding behavior of nsp13 (13). Finally, it is possible that, similar to the T4 primosome complex, RdRP activates nsp13 by increasing its binding affinity to the replication fork (44,45). Further single-molecule assays, including the entire RdRp-nsp13 holo-complex, will be needed to investigate these possibilities.
Replicative helicases from the six superfamilies (46) fall into two broad classes: ring-shaped hexameric helicases and nonhexameric helicases (47,48). Broadly, two mechanisms have been proposed for helicases: an active mechanism in which the helicase directly destabilizes the DNA or RNA substrate and a passive mechanism in which the helicase opportunistically advances upon the spontaneous opening of base pairs (35,49,50). These mechanisms are not mutually exclusive, and a continuum between them exists (51). The nonhexameric superfamily 2 helicase HCV NS3 and the nonhexameric superfamily 1 DNA repair helicase UvrD are proposed to be highly active helicases, each having unwinding velocities that do not increase in response to destabilizing forces (14,52). We show here that the unwindase activity of nsp13, a nonhexameric helicase (5), more closely matches that of ring-shaped bacteriophage DNA helicases such as T4 gp41 (passive) and T7 gp4 (weakly active), each of which show a strong force-dependent velocity (12,13). Although we cannot definitively determine the degree of passiveness for nsp13 with these data, a purely passive helicase is expected to have an exponential force-velocity relationship, and the more active the helicase (i.e., the stronger the interaction potential), the more the force-velocity curve deviates from an exponential (26). Interestingly, the force-activated velocity of nsp13 more closely matches that of hexameric helicases than it does other nonhexameric helicases. We note the possibility that nsp13 may shift to a stronger active mechanism under different conditions; T7 gp4 has been proposed to do so when bound to its polymerase (13), and DnaB helicase does so under certain DNA-pulling geometries (24). Further work, including assays on dsRNA forks in which force can be applied to either strand, is needed to explore these possibilities in detail.
Potent force activation of nsp13 unwindase activity potentially enables on-demand regulation. This is likely important to viral RNA genome stability because promiscuous unwinding would expose it to degradation. A default low-activity state for the helicase also maintains the integrity of the viral replication machinery and prevents dissociation (53). The nsp13 in SARS-CoV-2 is >99% similar by sequence identity to its counterpart in SARS-CoV-1 (6), suggesting that force regulation of replicative helicase activity is likely conserved across coronaviridae that are evolutionarily proximal to the causative agent of COVID-19.
The high conservation of nsp13 helicases across coronaviridae suggests that therapeutic agents targeting this enzyme could be developed to treat COVID-19 and other diseases. Targeting viral helicases in general has been challenging (54,55). However, recent successes in targeting different ATPases (56, 57, 58) suggests that selective and potent inhibitors of nsp13 could be developed. The bulk and single-molecule assay platforms reported here may be leveraged in future drug discovery efforts and in detailed mechanistic investigation of nsp13 inhibition.
Author contributions
K.J.M. and X.C. performed single-molecule assays and processed the data. P.M.M.S. performed bulk helicase assays. M.G. expressed and purified proteins with aid from K.J.M. and S.E.W. M.G. and P.M.M.S. performed DSF assays. K.J.M. performed fluorescence anisotropy assays. A.A. provided the key reagents. S.L. and T.M.K. supervised the research. K.J.M., P.M.M.S., M.G., S.L., and T.M.K. wrote the manuscript with input from all authors.
Acknowledgments
The authors thank members of the Kapoor Lab as well as the Seth Darst and Elizabeth Campbell Labs for useful discussions. The authors are also grateful to the Pels Center for Biochemistry and Structural Biology for support.
T.M.K. is grateful to the National Institutes of Health, National Institute of General Medical Sciences for funding (R35 GM130234). S.L. is supported by the Robertson Foundation, Sinsheimer Foundation, and National Institutes of Health (DP2HG010510). K.J.M. is supported by a National Cancer Institute K00 Fellowship (K00CA223018).
Editor: Ahmet Yildiz.
Footnotes
Keith J. Mickolajczyk, Patrick M. M. Shelton, Michael Grasso, and Xiaocong Cao contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2020.11.2276.
Contributor Information
Shixin Liu, Email: shixinliu@rockefeller.edu.
Tarun M. Kapoor, Email: kapoor@rockefeller.edu.
Supporting material
References
- 1.Wu F., Zhao S., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F.-W., Kok K.-H., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia Z., Yan L., Rao Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J., Malone B., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frick D.N., Virdi R.S., Silvaggi N.R. Molecular basis for ADP-ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry. 2020;59:2608–2615. doi: 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner J.A., Watt R.M., Huang J.-D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov K.A., Thiel V., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adedeji A.O., Marchand B., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee N.-R., Kwon H.-M., Kim D.-E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang K.-J., Jeong S., Kim D.-E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci. Rep. 2020;10:4481. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lionnet T., Spiering M.M., Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson D.S., Bai L., Wang M.D. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumont S., Cheng W., Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai V.P., Frank F., Bustamante C. Co-temporal force and fluorescence measurements reveal a ribosomal gear shift mechanism of translation regulation by structured mRNAs. Mol. Cell. 2019;75:1007–1019.e5. doi: 10.1016/j.molcel.2019.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith S.B., Cui Y., Bustamante C. [7] Optical-trap force transducer that operates by direct measurement of light momentum. Methods in Enzymology. 2003;361:134–162. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- 17.Liphardt J., Onoa B., Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y., Deffenbaugh N.C., Hancock W.O. Molecular counting by photobleaching in protein complexes with many subunits: best practices and application to the cellulose synthesis complex. Mol. Biol. Cell. 2014;25:3630–3642. doi: 10.1091/mbc.E14-06-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mickolajczyk K.J., Deffenbaugh N.C., Hancock W.O. Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proc. Natl. Acad. Sci. USA. 2015;112:E7186–E7193. doi: 10.1073/pnas.1517638112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao W., Wojdyla J.A., Cui S. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017;13:e1006474. doi: 10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 22.Özeş A.R., Feoktistova K., Fraser C.S. Real-time fluorescence assays to monitor duplex unwinding and ATPase activities of helicases. Nat. Protoc. 2014;9:1645–1661. doi: 10.1038/nprot.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter D.J., Short S.A., Consler T.G. Product release is the major contributor to kcat for the hepatitis C virus helicase-catalyzed strand separation of short duplex DNA. J. Biol. Chem. 1998;273:18906–18914. doi: 10.1074/jbc.273.30.18906. [DOI] [PubMed] [Google Scholar]
- 24.Ribeck N., Kaplan D.L., Saleh O.A. DnaB helicase activity is modulated by DNA geometry and force. Biophys. J. 2010;99:2170–2179. doi: 10.1016/j.bpj.2010.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick E.M., Srinivasan S., Comstock M.J. The RNA helicase Mtr4p is a duplex-sensing translocase. Nat. Chem. Biol. 2017;13:99–104. doi: 10.1038/nchembio.2234. [DOI] [PubMed] [Google Scholar]
- 26.Chakrabarti S., Jarzynski C., Thirumalai D. Processivity, velocity, and universal characteristics of nucleic acid unwinding by helicases. Biophys. J. 2019;117:867–879. doi: 10.1016/j.bpj.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi Z., Pugh R.A., Chemla Y.R. Sequence-dependent base pair stepping dynamics in XPD helicase unwinding. eLife. 2013;2:e00334. doi: 10.7554/eLife.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moffitt J.R., Chemla Y.R., Bustamante C. Methods in statistical kinetics. Methods Enzymol. 2010;475:221–257. doi: 10.1016/S0076-6879(10)75010-2. [DOI] [PubMed] [Google Scholar]
- 29.Yildiz A., Tomishige M., Selvin P.R. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 30.Cheng W., Arunajadai S.G., Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myong S., Bruno M.M., Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Syed S., Pandey M., Ha T. Single-molecule fluorescence reveals the unwinding stepping mechanism of replicative helicase. Cell Rep. 2014;6:1037–1045. doi: 10.1016/j.celrep.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesnik E.A., Freier S.M. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995;34:10807–10815. doi: 10.1021/bi00034a013. [DOI] [PubMed] [Google Scholar]
- 34.Ha T. Need for speed: mechanical regulation of a replicative helicase. Cell. 2007;129:1249–1250. doi: 10.1016/j.cell.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Manosas M., Xi X.G., Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribeck N., Saleh O.A. DNA unwinding by ring-shaped T4 helicase gp41 is hindered by tension on the occluded strand. PLoS One. 2013;8:e79237. doi: 10.1371/journal.pone.0079237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M.D., Schnitzer M.J., Block S.M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 38.Thomen P., Lopez P.J., Heslot F. T7 RNA polymerase studied by force measurements varying cofactor concentration. Biophys. J. 2008;95:2423–2433. doi: 10.1529/biophysj.107.125096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu S., Ci Y., Shi L. Zika virus NS3 is a canonical RNA helicase stimulated by NS5 RNA polymerase. Nucleic Acids Res. 2019;47:8693–8707. doi: 10.1093/nar/gkz650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stano N.M., Jeong Y.-J., Patel S.S. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong F., Weitzel S.E., von Hippel P.H. A coupled complex of T4 DNA replication helicase (gp41) and polymerase (gp43) can perform rapid and processive DNA strand-displacement synthesis. Proc. Natl. Acad. Sci. USA. 1996;93:14456–14461. doi: 10.1073/pnas.93.25.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delagoutte E., von Hippel P.H. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry. 2001;40:4459–4477. doi: 10.1021/bi001306l. [DOI] [PubMed] [Google Scholar]
- 43.Manosas M., Spiering M.M., Benkovic S.J. Collaborative coupling between polymerase and helicase for leading-strand synthesis. Nucleic Acids Res. 2012;40:6187–6198. doi: 10.1093/nar/gks254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jose D., Weitzel S.E., von Hippel P.H. Breathing fluctuations in position-specific DNA base pairs are involved in regulating helicase movement into the replication fork. Proc. Natl. Acad. Sci. USA. 2012;109:14428–14433. doi: 10.1073/pnas.1212929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee W., Jose D., von Hippel P.H. A single-molecule view of the assembly pathway, subunit stoichiometry, and unwinding activity of the bacteriophage T4 primosome (helicase-primase) complex. Biochemistry. 2013;52:3157–3170. doi: 10.1021/bi400231s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 47.Patel S.S., Picha K.M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 48.Lohman T.M., Tomko E.J., Wu C.G. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 49.Pincus D.L., Chakrabarti S., Thirumalai D. Helicase processivity and not the unwinding velocity exhibits universal increase with force. Biophys. J. 2015;109:220–230. doi: 10.1016/j.bpj.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amaratunga M., Lohman T.M. Escherichia coli rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 51.Pyle A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 52.Sun B., Wei K.-J., Xi X.G. Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO J. 2008;27:3279–3287. doi: 10.1038/emboj.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Hippel P.H., Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 54.Shadrick W.R., Ndjomou J., Frick D.N. Discovering new medicines targeting helicases: challenges and recent progress. J. Biomol. Screen. 2013;18:761–781. doi: 10.1177/1087057113482586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta A., Brosh R.M., Jr. New insights into DNA helicases as druggable targets for cancer therapy. Front. Mol. Biosci. 2018;5:59. doi: 10.3389/fmolb.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleymann G., Fischer R., Rübsamen-Waigmann H. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 57.Crute J.J., Grygon C.A., Cordingley M.G. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 58.Cupido T., Pisa R., Kapoor T.M. Designing a chemical inhibitor for the AAA protein spastin using active site mutations. Nat. Chem. Biol. 2019;15:444–452. doi: 10.1038/s41589-019-0225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.