Summary
Background
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces an antibody response targeting multiple antigens that changes over time. This study aims to take advantage of this complexity to develop more accurate serological diagnostics.
Methods
A multiplex serological assay was developed to measure IgG and IgM antibody responses to seven SARS-CoV-2 spike or nucleoprotein antigens, two antigens for the nucleoproteins of the 229E and NL63 seasonal coronaviruses, and three non-coronavirus antigens. Antibodies were measured in serum samples collected up to 39 days after symptom onset from 215 adults in four French hospitals (53 patients and 162 health-care workers) with quantitative RT-PCR-confirmed SARS-CoV-2 infection, and negative control serum samples collected from healthy adult blood donors before the start of the SARS-CoV-2 epidemic (335 samples from France, Thailand, and Peru). Machine learning classifiers were trained with the multiplex data to classify individuals with previous SARS-CoV-2 infection, with the best classification performance displayed by a random forests algorithm. A Bayesian mathematical model of antibody kinetics informed by prior information from other coronaviruses was used to estimate time-varying antibody responses and assess the sensitivity and classification performance of serological diagnostics during the first year following symptom onset. A statistical estimator is presented that can provide estimates of seroprevalence in very low-transmission settings.
Findings
IgG antibody responses to trimeric spike protein (Stri) identified individuals with previous SARS-CoV-2 infection with 91·6% (95% CI 87·5–94·5) sensitivity and 99·1% (97·4–99·7) specificity. Using a serological signature of IgG and IgM to multiple antigens, it was possible to identify infected individuals with 98·8% (96·5–99·6) sensitivity and 99·3% (97·6–99·8) specificity. Informed by existing data from other coronaviruses, we estimate that 1 year after infection, a monoplex assay with optimal anti-Stri IgG cutoff has 88·7% (95% credible interval 63·4–97·4) sensitivity and that a four-antigen multiplex assay can increase sensitivity to 96·4% (80·9–100·0). When applied to population-level serological surveys, statistical analysis of multiplex data allows estimation of seroprevalence levels less than 2%, below the false-positivity rate of many other assays.
Interpretation
Serological signatures based on antibody responses to multiple antigens can provide accurate and robust serological classification of individuals with previous SARS-CoV-2 infection. This provides potential solutions to two pressing challenges for SARS-CoV-2 serological surveillance: classifying individuals who were infected more than 6 months ago and measuring seroprevalence in serological surveys in very low-transmission settings.
Funding
European Research Council. Fondation pour la Recherche Médicale. Institut Pasteur Task Force COVID-19.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing COVID-19 emerged in Wuhan, China, in December, 2019. Since then, it has spread rapidly, with confirmed cases being recorded in nearly every country in the world. The presence of viral infection can be directly detected via quantitative RT-PCR (RT-qPCR) on samples from nasopharyngeal or throat swabs.1, 2 In many countries, neither mild cases nor asymptomatic cases will be tested by RT-qPCR (unless they are direct contacts of known cases), and even among tested individuals with SARS-CoV-2 infection, many might have a negative result at time of testing due to low viral load. While not suitable for diagnosis of clinical cases, serology is a promising tool for identifying individuals with previous infection by detecting antibodies generated in response to SARS-CoV-2 infection. However, the utility of serological testing depends on the kinetics of the anti-SARS-CoV-2 antibody response during and after infection.
Research in context.
Evidence before this study
We searched PubMed on July 29, 2020, with no limitations, using the terms (“SARS-CoV-2” OR “COVID-19”) AND (“antibody” OR “serology”) AND “multiplex”. Our search revealed eight publications, two of which described multiplex immunoassays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using Luminex technology. Given the fast-paced generation of new evidence, we next turned to the preprint literature and searched medRxiv on the same date with the same search terms, where we found 78 publications. In total, we identified more than 20 studies using multiplex immunoassays to classify previous infection with SARS-CoV-2, evidence of a rapidly moving and dynamic field. Several studies described diagnostic tests with multiplex combinations of antigens with more than 90% sensitivity and more than 99% specificity validated on up to 400 samples. A common limitation of many previous studies is insufficient diversity in panels of samples tested in terms of location and symptom severity.
Added value of this study
Our work advances on previous work through the use of large numbers of serum samples from different panels and antigens, as well as the use of statistically robust cross-validation of classification algorithms. This combination of multiplex assays and algorithms allows us to accurately identify previously infected individuals with 98·8% (95% CI 96·5–99·6%) sensitivity and 99·3% (97·6–99·8) specificity. In addition to classifying serum samples from recently infected individuals, we present new analytic tools for two key applications of multiplex serological assays for SARS-CoV-2 surveillance. First, by analysing our data with a mathematical model of antibody kinetics, we show how multiplex assays can be used to provide more accurate classification after antibody levels have started to decay. Second, through analysis of quantitative measurements of multiple antibody responses with statistical algorithms, we show how serosurveillance can be optimised for a range of different transmission settings, most notably in low-transmission settings.
Implications of all the available evidence
Multiplex assays are set to be a key tool for serological surveillance of SARS-CoV-2, especially as the epidemic progresses and the epidemiology becomes more complex. Many assays have already shown high levels of accuracy at classifying recently infected individuals when antibody levels are still high. The challenge now turns to how to optimally use the high-dimensional data generated by these assays to allow surveillance of the SARS-CoV-2 epidemic as it progresses.
An individual is seropositive to a pathogen if they have detectable antibodies specific for that pathogen. In practice, serological assays are used to measure antibody responses in blood samples. However, individuals who have never been infected with the target pathogen might have non-zero antibody responses due to cross-reactivity with other pathogens or background assay noise. To account for this, defining seropositivity is equivalent to determining whether the measured antibody response is greater or lower than some pdefined cutoff value.
The most fundamental measure of antibody level is via concentration in a sample (eg, in units of μg/mL); however, a measurement in terms of molecular mass per volume is usually impossible to obtain. Instead, a range of assays can provide measurements that are positively associated with the true antibody concentration—eg, an optical density from an ELISA or a median fluorescent intensity (MFI) from a Luminex microsphere assay. In contrast to the continuous measurement of antibody response provided by laboratory-based research assays, most point-of-care serological tests provide a binary outcome: seronegative or seropositive. There are several commercially available tests for detecting SARS-CoV-2 antibody responses, which are being catalogued by FIND Diagnostics.3 These tests are typically based on lateral flow assays mounted in plastic cartridges that detect antibodies in small-volume blood samples. A key feature of many rapid tests is that they are dependent on the choice of seropositivity cutoff, and there could be substantial misclassification for antibody levels close to this cutoff.
Antibody levels are not constant and change over time. The early kinetics of the antibody response to SARS-CoV-2 have been well documented, with a rapid rise in antibody levels occurring 5–15 days after symptom onset leading to seroconversion (depending on the choice of cutoff).1, 4 Assuming the long-term kinetics of the SARS-CoV-2 antibody response are similar to other pathogens,5, 6, 7 we expect to observe a biphasic pattern of decay, with rapid decay in the first 3–6 months after infection, followed by a slower rate of decay. Notably, this decay pattern might lead to seroreversion, whereby previously seropositive individuals revert to being seronegative. If a serological test with an inappropriately high choice of cutoff is used for SARS-CoV-2 serological surveys, there is a major risk that seroreversion could lead to previously infected individuals testing seronegative.8
The antibody response generated after SARS-CoV-2 infection is diverse, consisting of multiple isotypes targeting several proteins on the virus including the spike protein (and its receptor-binding domain [RBD]) and nucleoprotein.9 This diversity of biomarkers provides both a challenge and an opportunity for diagnostics research. The challenge lies in selecting appropriate biomarkers and choosing between the increasing number of commercial assays, many of which have not been extensively validated and might produce conflicting results. The opportunity is that with multiple biomarkers, it is possible to generate a serological signature of infection that is robust to how antibody levels change over time,10, 11, 12, 13 rather than relying on classification of seropositive individuals using a single cutoff antibody level.
In this analysis, we apply mathematical models of antibody kinetics and machine learning classifiers to identify serological signatures of SARS-CoV-2 infection generated using multiplex assays, which can provide accurate serological diagnosis that is robust to the waning of antibody levels over time.
Methods
Study population and samples
We analysed 97 serum samples from 53 adult patients admitted to two hospitals with COVID-19 in Paris between Jan 24 and April 1, 2020, and 162 serum samples from 162 health-care workers in two hospitals in Strasbourg with RT-qPCR-confirmed SARS-CoV-2 infection taken between April 6 and April 7, 2020, who had mostly mild symptoms (table).14, 15, 16 Samples were collected up to 39 days after symptom onset. Four patients from Hôpital Bichat provided 6–15 samples in total. 35 patients from Hôpital Cochin provided one sample each and 14 patients provided two samples each. Negative control samples were selected from panels of pre-epidemic cohorts (before December, 2019) with ethical approval for broad antibody testing. We chose a panel representative of the target population (177 serum samples from healthy adult French blood donors), plus two geographically diverse panels (68 plasma samples from healthy adult donors from the Thai Red Cross and 90 serum samples from Peruvian healthy adult donors). All samples underwent a viral inactivation protocol by heating at 56°C for 30 min. The potential effect of the viral inactivation protocol on the measurement of antibody levels was assessed using serum positive for anti-malaria antibodies. IgG and IgM antibody levels were measured in matched samples before and after the inactivation protocol. The viral inactivation protocol did not affect measured IgG or IgM levels (data not shown).
Table.
Panels of samples
| Number of participants | Number of samples | Age, years* |
Symptom severity |
||
|---|---|---|---|---|---|
| Mild | Severe | ||||
| SARS-CoV-2 cases confirmed by RT-qPCR | |||||
| Hôpital Bichat (Paris, France) | 4 | 34 | 39 (31–80) | 0 | 4 |
| Hôpital Cochin (Paris, France) | 49 | 63 | 56 (26–79) | 27 | 22 |
| Nouvel Hôpital Civil and Hôpital de Haute Pierre (Strasbourg, France) | 162 | 162 | 32 (20–65) | 160 | 2 |
| Pre-epidemic seronegative controls | |||||
| Thai Red Cross | 68 | 68 | >18 | .. | .. |
| Peruvian donors | 90 | 90 | >18 | .. | .. |
| France blood donors (Établissement Français du Sang) | 177 | 177 | >18 | .. | .. |
Positive control serum samples are from patients with RT-qPCR confirmed SARS-CoV-2 infection. RT-qPCR=quantitative RT-PCR. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
Age is presented as median (range) for SARS-CoV-2 cases.
Serum samples were biobanked at the Clinical Investigation and Access to BioResources platform at Institut Pasteur (Paris, France). Samples were obtained from consenting individuals through the CORSER study (NCT04325646), directed by Institut Pasteur and approved by the Comité de Protection des Personnes Ile de France III, and the French COVID cohort (NCT04262921), sponsored by Inserm and approved by the Comité de Protection des Personnes Ile de France VI. Sample collection in Hôpital Cochin was approved by the Research Ethics Commission of Necker-Cochin Hospital. Samples from French blood donors were approved for use by Etablissement Français du Sang (Lille, France). Use of the Peruvian negative controls was approved by the Institutional Ethics Committee from the Universidad Peruana Cayetano Heredia (SIDISI 100873). The Human Research Ethics Committee at the Walter and Eliza Hall Institute of Medical Research and the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University, Thailand, approved the use of the Thai negative control samples. Informed written consent was obtained from all participants or their next of kin.
Serological assays
We optimised a 12-plex assay for detecting IgG and IgM antibody responses against seven SARS-CoV-2 antigens (two nucleoprotein constructs, five spike proteins) and one nucleoprotein for each of seasonal coronaviruses NL63 and 229E. Three antigens from other viruses were included (influenza A [H1N1] virus, adenovirus type 40, and rubella virus), for which a large part of the population is expected to be seropositive due to vaccination or natural infection and hence serve as internal controls. Antigens were supplied by Institut Pasteur or Native Antigen (Oxford, UK). Antigen names, expression systems, suppliers, and catalogue numbers are provided in the appendix (p 4)). All proteins were coupled to magnetic beads as described elsewhere.17 The masses of proteins coupled to beads were optimised to generate a log-linear standard curve with a pool of positive serum prepared from patients with RT-qPCR-confirmed SARS-CoV-2 infection.
Two separate assays were used for measuring IgG and IgM antibodies (appendix p 2). For each assay, samples were run on 96 well plates, containing two blanks (only beads, no serum) and a standard curve prepared from two-fold serial dilutions (1:50 to 1:25 600) of a pool of positive controls. Plates were read using a Luminex MAGPIX system (Austin, TX, USA) and MFI was used for analysis. A five-parameter logistic curve was used to convert MFI to antibody dilution, relative to the standard curve performed on the same plate to account for interassay variations. The multiplex immunoassay was validated by checking that the MFIs obtained were well correlated with those obtained in monoplex (only one conjugated bead type per well). For non-SARS-CoV-2 antigens, MFI data were used for the analysis. The IgG and IgM assays were run on all samples shown in the table. Analytic validation of the assay was implemented by comparison with data from two other immunoassays, S-Flow and pseudo-neutralisation, on matched sample sets from the Strasbourg health-care workers. Further details of assay validation are provided in the appendix (pp 2–3, 5).
Diagnostic performance
For antibody responses to a single antigen, diagnostic sensitivity was defined as the proportion of individuals with RT-qPCR-confirmed SARS-CoV-2 infection with antibody levels above a given seropositivity cutoff. For assessment of classification performance, samples taken from individuals fewer than 10 days after symptom onset were excluded, because antibody levels have often not yet increased during this time period. Diagnostic specificity was defined as the proportion of negative controls (with no history of SARS-CoV-2 infection) with antibody levels below a given seropositivity cutoff. This trade-off between sensitivity and specificity was evaluated using receiver operating characteristic (ROC) analysis.
Antibody responses to multiple antigens can be combined to identify individuals with previous SARS-CoV-2 infection using statistical classification or machine learning algorithms.10 A random forests algorithm was chosen due to its superior classification performance over statistical classifiers such as logistic regression (appendix p 9). Classification algorithms were implemented in R (version 3.4.3). Uncertainty in sensitivity and specificity is quantified in three ways: (i) binomial CIs calculated using Wilson's method; (ii) 1000-fold repeat cross-validation with a training set comprising two thirds of the data and a disjoint testing set comprising a third of the data; and (iii) cross-panel validation with algorithms trained and tested on disjoint panels of data (appendix p 12).
Differences in measured antibody responses were assessed using a two-sided t test. Correlations between measured antibody responses were assessed using Spearman's correlation. Differences in classification performance were assessed using McNemar's test.
Mathematical model of antibody kinetics
SARS-CoV-2 antibody kinetics during the first year after symptom onset are described using a previously published mathematical model of the immunological processes underlying the generation and waning of antibody responses following infection.5 The existing model is adapted to account for the frequent data available in the first weeks of infection, as follows:
where B denotes the level of B lymphocytes, b is the rate of differentiation of B lymphocytes into antibody secreting plasma cells, Ps denotes the level of short-lived plasma cells with rate of decay cs, Pl denotes the level of long-lived plasma cells with rate of decay cl, A denotes antibody levels, ρ is the proportion of plasma cells that are short lived, g is the rate of generation of antibodies (IgG or IgM) from plasma cells, and r is the rate of decay of antibody molecules (appendix p 15).
Statistical inference was implemented within a mixed-effects framework allowing for characterisation of the kinetics within each individual while also describing the population-level patterns. On the population level, both the mean and variation in antibody kinetics are accounted for. Models were fitted in a Bayesian framework using Markov chain Monte Carlo methods with priors informed by estimates from long-term studies of antibody kinetics following infection with other coronaviruses (appendix pp 13–14). Predicted antibody levels are presented as posterior medians with 95% credible intervals (CrIs).
Serological surveillance
Imperfect diagnostic assays can cause bias in seroprevalence estimates. This can be accounted for by statistically adjusting for known values of sensitivity and specificity. A ROC curve obtained from a training dataset consisting of positive and negative samples is described by a sequence of estimated sensitivities and specificities {E(sensi), E(speci)}, where E denotes an estimator. N-fold cross-validation generates N samples of sensitivity {sensi, … sensi,N} for each estimated specificity E(speci) and N samples of specificity {speci,1, … , speci,N} for each estimated sensitivity E(sensi). Following a previously outlined approach,18, 19 for each pair i of sensitivity and specificity, we obtain N estimates of the measured seroprevalence Mi in a scenario with true seroprevalence T as follows:
The point estimates of sensitivity and specificity can be used to calculate an adjusted estimate of true seroprevalence for each of the N estimates of the measured seroprevalence Mi:
with E(Ti,n) = 0 if Mi,n <1E(sensi). Both the measured seroprevalence Mi and the estimated true seroprevalence E(Ti) are summarised as medians with 95% ranges. We calculate the expected relative error as
Role of the funding source
The funder of the study played no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data in the study and the corresponding author had final responsibility for the decision to submit for publication.
Results
For all 14 SARS-CoV-2 biomarkers (seven antigens, IgG and IgM for both), measured responses were significantly higher in samples with RT-qPCR-confirmed infection than in negative control samples (appendix p 5; two-sided t test p<0·0001).
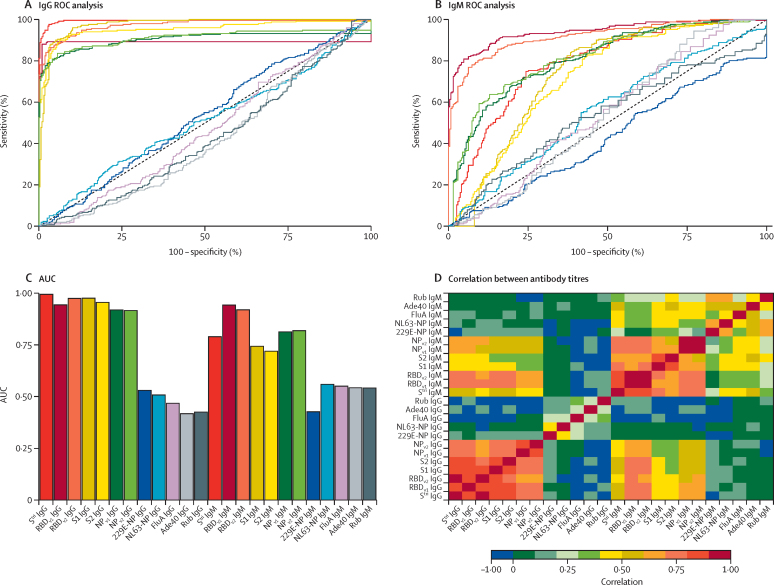
The trade-off between sensitivity and specificity obtained by varying the cutoff for seropositivity was investigated using a ROC curve (figure 1A, B) and summarised using area under the ROC curve (figure 1C). Depending on the characteristics of the desired diagnostic test, different targets for sensitivity and specificity can be considered. The results of three targets are summarised in the appendix (p 11)): high sensitivity target enforcing sensitivity greater than 99%, balanced sensitivity and specificity where both are approximately equal, and high specificity target enforcing specificity greater than 99%. Focusing on the high specificity target, anti-trimeric spike (Stri) IgG was the best-performing biomarker with 99·1% specificity (95% CI 97·4–99·7) and 91·6% sensitivity (87·5–94·5). Anti-Stri IgG provided significantly better classification than all other biomarkers (appendix p 8; McNemar's test p<10−7). There was significant correlation between antibody responses against all SARS-CoV-2 antigens, but no significant correlation between antibody responses to SARS-CoV-2 and the seasonal coronaviruses 229E and NL63 (figure 1D).
Figure 1.
Anti-SARS-CoV-2 antibody responses
ROC curves for IgG antibodies (A) and IgM antibodies (B) obtained by varying the cutoff for seropositivity. Colours correspond to antibodies against different antigens, as shown in panel C. (C) AUC for individual biomarkers. (D) Spearman's correlation between measured antibody responses. Ade40=adenovirus type 40 hexon (capsid). AUC=area under the ROC curve. FluA=influenza A virus (H1N1) haemagglutinin recombinant antigen. NL63-NP=human coronavirus NL63 NP. NP=SARS-CoV-2 nucleoprotein. ROC=receiver operating characteristic. RBD=SARS-CoV-2 spike glycoprotein receptor-binding domain. Rub=rubella virus-like particles (spike glycoprotein E1, spike glycoprotein E2, and capsid protein). SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. Stri=SARS-CoV-2 trimeric spike protein. S1=SARS-CoV-2 spike glycoprotein (S1 domain). S2=SARS-CoV-2 spike glycoprotein (S2 domain). 229E-NP=human coronavirus 229E NP.
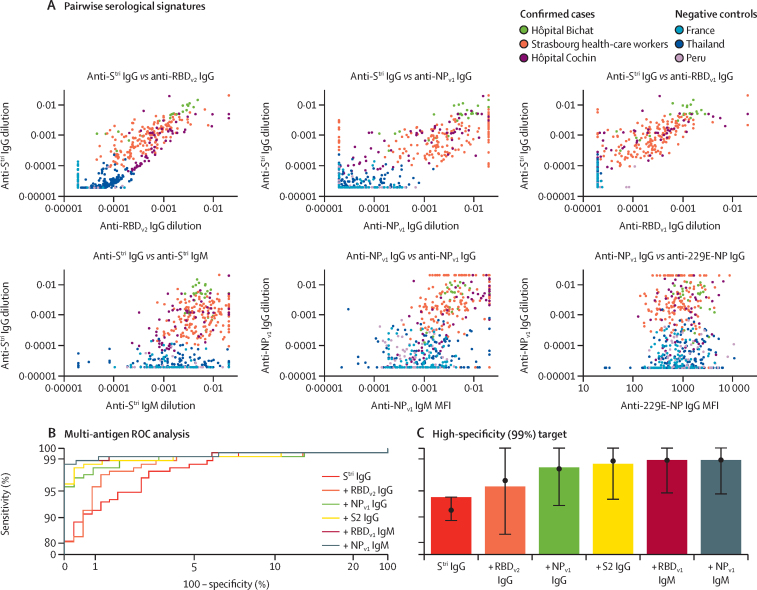
With 24 biomarkers, there are 156 pairwise comparisons. Figure 2A provides an overview of six pairwise comparisons of antibody responses. The data are noisy, highly correlated, and high dimensional (although only two dimensions are depicted here). We refer to the pattern of antibody responses in multiple dimensions as the serological signature. For all plots of SARS-CoV-2 biomarkers, there are two distinct clusters: antibody responses from negative control samples in the blue cluster in the bottom left, and antibody responses from serum samples from individuals with RT-qPCR-confirmed SARS-CoV-2 infection clustered in the centre and top right.
Figure 2.
Serological signatures of SARS-CoV-2 infection
(A) Pairwise combinations of antibody responses. Each point denotes a measured antibody response from a sample from Hôpital Bichat (n=34), health-care workers from Nouvel Hôpital Civil and Hôpital de Haute Pierre in Strasbourg (n=162), and Hôpital Cochin (n=63). Negative control samples are included from Thailand (n=68), Peru (n=90), and French blood donors (n=177). (B) ROC curves for multiple biomarker classifiers generated using a random forests algorithm. Biomarkers are added sequentially. The axes have been rescaled to better differentiate between high values of sensitivity and specificity. (C) For a high specificity target (>99%), sensitivity increases with additional biomarkers, added sequentially. Sensitivity was estimated using a random forests classifier. Points and whiskers denote the median and 95% CIs from repeat cross-validation. MFI= median fluorescent intensity. NP=SARS-CoV-2 nucleoprotein. RBD=SARS-CoV-2 spike glycoprotein receptor-binding domain. ROC=receiver operating characteristic. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. Stri=SARS-CoV-2 trimeric spike protein. S2=SARS-CoV-2 spike glycoprotein (S2 domain). 229E-NP=human coronavirus 229E NP.
The classification performance with random forests algorithms of multiplex combinations of antibody responses is shown with ROC curves in figure 2B. Sequentially including data from additional biomarkers leads to significant improvements in classification performance, albeit with diminishing returns in sensitivity (figure 2C; appendix pp 10–11). For example, for the high-specificity target, with a single biomarker (anti-Stri IgG) we can achieve 91·6% (95% CI 87·5–94·5) sensitivity. Including anti-RBDv2 IgG increases sensitivity to 95·6% (92·3–97·5). Combinations of five to six biomarkers provide 98·8% (96·5–99·6) sensitivity with 99·3% (97·6–99·8) specificity (figure 2C; appendix pp 10–11).
For individuals with RT-qPCR-confirmed SARS-CoV-2 infection, the model-predicted IgG antibody response to SARS-CoV-2 shows a biphasic pattern of waning for all antigens, with a first rapid phase between 1 and 3 months after symptom onset, followed by a slower rate of waning (appendix p 19). The percentage reduction in antibody level after 1 year was mostly determined by prior information and estimated to be 47·1% (95% CrI 17·5–90·3) for anti-Stri IgG antibodies, with comparable estimates for other antigens (appendix p 21). Sensitivity was assessed using the seropositivity cutoff based on a high-specificity target (>99%). For all antigens considered, we predict that there will be a reduction in sensitivity over time, although there is a large degree of uncertainty (appendix p 19).
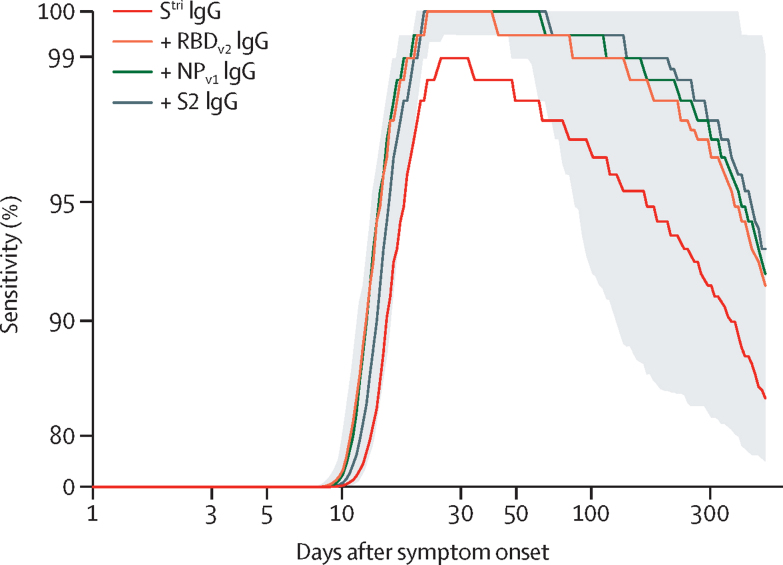
We predict that multiplex diagnostic tests will have higher classification performance for longer durations of time than single antigen tests. In particular, we predicted that the sensitivity based on anti-Stri IgG antibody responses after 1 year will be 88·7% (95% CrI 63·4–97·4) and that the sensitivity of a four-antigen multiplex classifier after 1 year will be 96·4% (80·9–100·0; figure 3; appendix pp 21).
Figure 3.
Model-predicted sensitivity over time
Proportion of 215 individuals (patients and health-care workers) with RT-qPCR-confirmed SARS-CoV-2 infection testing seropositive over time. A random forests algorithm was used for classification of multiple antigen multiplex data, with antigens added sequentially. The grey shaded region shows the 95% credible interval for the four-antigen multiplex classifier (black line). The x-axis is on a log scale and the y-axis has been rescaled to better differentiate between high values of sensitivity. NP=SARS-CoV-2 nucleoprotein. RBD=SARS-CoV-2 spike glycoprotein receptor-binding domain. RT-qPCR=quantitative RT-PCR. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. Stri=SARS-CoV-2 trimeric spike protein. S2=SARS-CoV-2 spike glycoprotein (S2 domain).
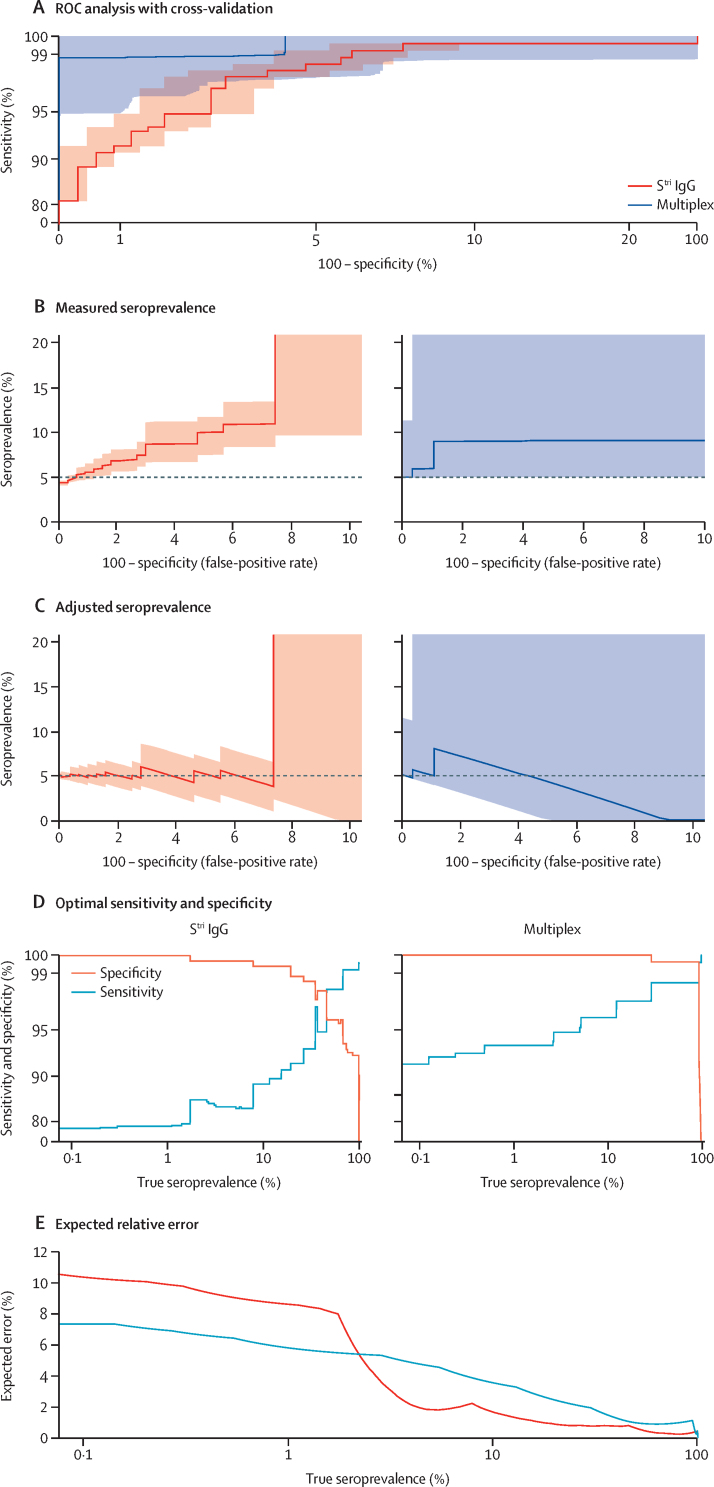
For serological diagnosis of individual samples, the aim is typically to optimise sensitivity while enforcing high specificity (>99%). A serological assay that accurately classifies individual samples will also perform well at estimating seroprevalence in populations. However, an assay optimised for individual-level classification is not necessarily optimal for population-level surveillance where the aim is to obtain accurate estimates of true seroprevalence. Figure 4 shows how imperfect diagnostic sensitivity and specificity can cause bias in measured seroprevalence, and how this can be statistically adjusted for. The expected error in adjusted seroprevalence can be minimised by selecting optimal values of sensitivity and specificity. Figure 4A presents ROC curves for a monoplex anti-Stri IgG assay and a multiplex assay using six biomarkers, with quantification of uncertainty via repeat cross-validation (appendix p 11). In an epidemiological scenario with true seroprevalence of 5%, the measured seroprevalence will depend on the assay sensitivity and false-positive rate (ie, 1 – specificity; figure 4B). For a high false-positive rate, the measured seroprevalence overestimates the true seroprevalence. Applying a statistical correction to account for imperfect sensitivity and specificity, we can obtain more accurate estimates of seroprevalence (figure 4C). For both the monoplex and multiplex serological assays, the adjusted estimates are not accurate when the false-positive rate exceeds the true prevalence.
Figure 4.
Implementation of seroprevalence surveys using monoplex (Stri IgG) and six-biomarker multiplex assays
(A) ROC analysis with cross-validated uncertainty. Solid lines represent median ROC curves and shaded regions represent 95% uncertainty intervals for specificity. The axes have been rescaled to better differentiate between high values of sensitivity and specificity. (B) In a scenario with true seroprevalence of 5%, the measured seroprevalence depends on the false-positive rate. (C) In a scenario with true seroprevalence of 5%, adjusted seroprevalence estimates are obtained by accounting for assay sensitivity and specificity. (D) Across a range of true seroprevalence values, optimal values of sensitivity and specificity can be selected to minimise the expected relative error in seroprevalence surveys. The y-axis has been rescaled to better differentiate between high values of sensitivity and specificity. (E) The expected relative error for optimal values of sensitivity and specificity. ROC=receiver operating characteristic. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. Stri=SARS-CoV-2 trimeric spike protein.
In real-life applications, however, true seroprevalence is not known a priori. For a range of seroprevalence from 0·1% to 100%, figure 4D presents values of the assay's sensitivity and specificity that have been optimised to minimise the expected relative error. For a monoplex assay based on anti-Stri IgG antibodies, if true seroprevalence is less than 20%, the relative error is minimised when we select specificity greater than 99%. When true seroprevalence is less than 2%, the relative error is minimised when specificity is 100%. For a multiplex serological assay, if true seroprevalence is less than 30%, the relative error is minimised when we implement an algorithm with specificity of 100%. When comparing the expected relative error for the Stri IgG and six-biomarker multiplex assays, the error depends on the possible values of sensitivity and specificity, as well as the uncertainty in these estimates. For true seroprevalence greater than 2%, the monoplex assay has lower error (a consequence of the lower levels of variation in the ROC curve; figure 4A, E). For true seroprevalence less than 2%, the multiplex assay has lower error, a consequence of the high levels of specificity (figure 4D, E).
Discussion
Infection with SARS-CoV-2 induces antibodies of multiple isotypes (IgG, IgM, IgA) targeting various epitopes, including spike and nucleoprotein.20 These biomarkers might exhibit distinct kinetics leading to variation in their diagnostic performance. By measuring multiple biomarkers in large numbers of individuals, it is possible to create a serological signature of previous infection,10, 11, 12, 13 leading to more accurate serological classification of individuals recently infected with SARS-CoV-2.21, 22 Although necessarily more complex than a single measured antibody response, such an approach has the potential of providing more accurate classification and being more stable over time.
IgG antibody levels to a single antigen (Stri) can classify samples from individuals previously infected with SARS-CoV-2 with 91·6% (95% CI 87·5–94·5) sensitivity and 99·1% (97·4–99·7) specificity. Measuring additional biomarkers with a multiplex assay can improve classification performance to 98·8% (96·5–99·6) sensitivity and 99·3% (97·6–99·8) specificity. A similar phenomenon is observed for serological diagnosis of HIV, where combining multiple assays can lead to improved accuracy.23 Multiplex assays provide some of the benefits of combining separate assays, but are subject to the risk that multiple biomarkers measured on the same assay are often correlated.
The reported accuracy of serological tests depends on multiple factors, most notably the validation samples used. Specificity is typically determined by pre-epidemic negative control samples, with the inclusion of greater numbers of samples providing more robust characterisation of specificity. Rather than taking large numbers of samples from a homogeneous population, we encourage the use of multiple negative control panels that are epidemiologically diverse with respect to age and location. Sensitivity is determined by positive control samples. It might be trivial to record high sensitivity when validating with samples from small numbers of individuals with severe symptoms.24 We encourage the use of multiple positive control panels that are epidemiologically diverse with respect to factors such as age, COVID-19 symptom severity, and time since symptom onset. When comparing the performance of different assays, the ideal approach is to use common serum samples. In the majority of situations where common serum samples are not available, including epidemiological information on validation samples can facilitate more effective comparison between assays.
Analysis of samples from individuals collected up to 5 months after the onset of COVID-19 symptoms indicates that antibody levels are maintained but with substantial waning.25, 26 The long-term kinetics of the antibody response to SARS-CoV-2 will not be definitively quantified until infected individuals are followed longitudinally for months and even years after RT-qPCR-confirmed infection. While these samples are being collected, mathematical models can provide important insights into how SARS-CoV-2 antibody levels might change over time. Modelling beyond the timeframe for which we have data has its limitations; however, our Bayesian approach benefits from robust quantification of uncertainty accounting for a wide range of future scenarios. Furthermore, this modelling approach provides falsifiable predictions which will allow models to be updated as new data are generated.
A limitation of this study is the absence of samples from individuals collected more than 39 days after symptom onset, and the absence of samples from individuals with RT-qPCR-confirmed asymptomatic SARS-CoV-2 infection. A further limitation is that this assay included antigens for the two alphacoronaviruses (229E, NL63) rather than the more closely related betacoronaviruses (OC43 and HKU1). Given the reported cross-reactivity between SARS-CoV-2 and OC43,27 it is possible that the inclusion of antigens for other betacoronaviruses might improve classification performance.
The simulations presented here predict that following SARS-CoV-2 infection, antibody responses will increase rapidly 1–2 weeks after symptom onset, with antibody responses peaking within 2–4 weeks. After this peak, antibody responses are predicted to decline according to a biphasic pattern, with rapid decay in the first 3–6 months followed by a slower rate of decay. Model predictions of the rise and peak of antibody response are informed by, and are consistent with, many sources of data.5, 6, 7 Model predictions of the decay of antibody responses are strongly determined by prior information on longitudinal follow-up of individuals infected with other coronaviruses.28 Under the scenario that the decay of SARS-CoV-2 antibody responses is similar to that of SARS-CoV, we would expect substantial reductions in antibody levels within the first year after infection. For the seropositivity cutoffs highlighted here, this could cause approximately 10–50% of individuals to test seronegative after 1 year, depending on the exact choice of biomarker and seropositivity cutoff.
This finding presents a potential problem for SARS-CoV-2 serological diagnostics. Most commercially available diagnostic tests compare antibody responses to a fixed seropositivity cutoff. Where these cutoffs have been validated, it is typically by comparison of serum from negative control samples collected pre-epidemic with serum from hospitalised patients in the first weeks of infection (ie, when antibody responses are likely to be at their highest).29 If we fail to account for antibody kinetics, we risk incorrectly classifying individuals with old infections (eg, >6 months) as being seronegative. This is particularly important for point-of-care rapid serological tests with fixed cutoffs, limited dynamic range, and visual evaluation. If inappropriate tests are used in seroprevalence surveys, there is a risk of substantial underestimation of the proportion of previously infected individuals.
An advantage of continuous multiplex data is that different algorithms can be applied to the same data for different epidemiological applications. We considered multiplex combination of antigens to optimise classification of individual samples against a target of maximising sensitivity given a minimum specificity of 99%. However, a test that is optimal for individual-level classification is not necessarily optimal for population-level use. A recommended target for serological assays for serosurveillance surveys is to minimise the expected error in estimated seroprevalence. For scenarios where true seroprevalence is expected to be low (<10%), we found assays with high specificity (>99%) to be optimal. Notably, this provides a potential solution to the challenge of implementing serosurveillance studies in regions where seroprevalence is expected to be lower than commonly reported false-positive rates.30 This is possible because our assay allows 100% specificity to be achieved with an accompanying reduction in sensitivity that can be statistically accounted for. In low-seroprevalence settings, there are additional challenges in collecting sufficient numbers of samples to ensure statistically robust estimates.31
The analysis presented here is based on data limited to the first 39 days after symptom onset, and the predictions we have made might subsequently be contradicted as more data become available. However, the concepts outlined here of serological signatures of SARS-CoV-2 infection generated by multiplex assays, and mathematical models of antibody kinetics, allow us to plan in advance for some of the future challenges that we might face in SARS-CoV-2 serological surveillance.
Data sharing
All data and code used for producing the results are freely available on GitHub.
Acknowledgments
Acknowledgments
This work was supported by the European Research Council (MultiSeroSurv ERC Starting Grant 852373; MTW), l'Agence Nationale de la Recherche and Fondation pour la Recherche Médicale (CorPopImm; MTW), and the Institut Pasteur International Network (CoronaSeroSurv; MTW). JR was supported by the Pasteur Paris University International PhD Program. CC was supported by the European Research Council 771813. The French COVID cohort is supported by the REACTing consortium and by the French Directorate General for Health. Darragh Duffy (Institut Pasteur, Paris) and Jérôme Hadjadj and Laura Barnabei (Institut Imagine, Paris) are thanked for their work on the Hôpital Cochin study. Dionicia Gamboa (Cayetano Heredia University, Lima) is thanked for sharing negative control samples from Peru. Jetsumon Sattabongkot (Mahidol University, Bangkok) is thanked for sharing negative control samples from Thailand. Marie-Noelle Ungeheuer and Blanca Liliana Perlaza are thanked for processing samples at the Clinical Investigation and Access to BioResources platform in Institut Pasteur. Shane Mansfield (Pierre and Marie Curie University, Paris) and Richard Davison (Heriot Watt University, Edinburgh) are thanked for their help in the solution of differential equations. We thank all patients and health-care workers who kindly agreed for samples to be used for medical research purposes.
Contributors
MTW implemented the analysis and wrote the first draft of the report. MTW and IM conceived the study. JR, SPel, CC, SHM, CD, and TB processed samples. NN analysed the data. RL designed the laboratory protocols. AM, MB, SPet, and FD generated proteins. SK, BT, SF-K, JdS, and AF wrote study protocols and coordinated sample collection. MTW, JR, and SPel have verified the underlying data.
Declaration of interests
MTW, IM, JR, SPel, MB, and SPet are inventors on provisional patent PCT/US 63/057.471 on a serological antibody-based diagnostics of SARS-COV-2 infection. TB is a coinventor on provisional patent PCT/US 63/020,063 entitled “S-Flow: a FACS-based assay for serological analysis of SARS-CoV-2 infection” submitted by Institut Pasteur. SK reports personal fees from Accelerate Diagnostics, Astellas, Merck Sharp & Dohme, Pfizer, and Menarini, and grants and personal fees from bioMérieux, outside of the submitted work. All other authors declare no competing interests.
Supplementary Material
References
- 1.To KK, Tsang OT, Leung WS. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siordia JA., Jr Epidemiology and clinical features of COVID-19: a review of current literature. J Clin Virol. 2020;127 doi: 10.1016/j.jcv.2020.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Foundation for Innovative New Diagnostics SARS-CoV-2 diagnostics: performance data. https://www.finddx.org/covid-19/dx-data
- 4.Borremans B, Gamble A, Prager KC. Quantifying antibody kinetics and RNA detection during early-phase SARS-CoV-2 infection by time since symptom onset. eLife. 2020;9 doi: 10.7554/eLife.60122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White MT, Griffin JT, Akpogheneta O. Dynamics of the antibody response to Plasmodium falciparum infection in African children. J Infect Dis. 2014;210:1115–1122. doi: 10.1093/infdis/jiu219. [DOI] [PubMed] [Google Scholar]
- 6.Teunis PF, van Eijkeren JC, de Graaf WF, Marinović AB, Kretzschmar ME. Linking the seroresponse to infection to within-host heterogeneity in antibody production. Epidemics. 2016;16:33–39. doi: 10.1016/j.epidem.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Andraud M, Lejeune O, Musoro JZ, Ogunjimi B, Beutels P, Hens N. Living on three time scales: the dynamics of plasma cell and antibody populations illustrated for hepatitis a virus. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams ER, Anand R, Andersson MI. Antibody testing for COVID-19: a report from the National COVID Scientific Advisory Panel. Wellcome Open Res. 2020;5:139. doi: 10.12688/wellcomeopenres.15927.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hachim A, Kavian N, Cohen CA. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- 10.Longley RJ, White MT, Takashima E. Development and validation of serological markers for detecting recent Plasmodium vivax infection. Nat Med. 2020;26:741–749. doi: 10.1038/s41591-020-0841-4. [DOI] [PubMed] [Google Scholar]
- 11.Azman AS, Lessler J, Luquero FJ. Estimating cholera incidence with cross-sectional serology. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aau6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helb DA, Tetteh KK, Felgner PL. Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc Natl Acad Sci USA. 2015;112:E4438–E4447. doi: 10.1073/pnas.1501705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perraut R, Varela ML, Loucoubar C. Serological signatures of declining exposure following intensification of integrated malaria control in two rural Senegalese communities. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadjadj J, Yatim N, Barnabei L. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescure FX, Bouadma L, Nguyen D. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fafi-Kremer S, Bruel T, Madec Y. Serologic responses to SARS-CoV-2 infection among hospital staff with mild disease in eastern France. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longley RJ, França CT, White MT. Asymptomatic Plasmodium vivax infections induce robust IgG responses to multiple blood-stage proteins in a low-transmission region of western Thailand. Malar J. 2017;16:178. doi: 10.1186/s12936-017-1826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diggle PJ. Estimating prevalence using an imperfect test. Epi Res International. 2011;2011 [Google Scholar]
- 19.Gelman A, Carpenter B. Bayesian analysis of tests with unknown specificity and sensitivity. J Roy Stat Soc C. 2020;69:1269–1283. doi: 10.1111/rssc.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grzelak L, Temmam S, Planchais C. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer AS, Jones FK, Nodoushania A. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv. 2020 doi: 10.1101/2020.07.18.20155374. published online July 20. (preprint) [DOI] [Google Scholar]
- 22.Becker M, Strengert M, Junker D. Going beyond clinical routine in SARS-CoV-2 antibody testing—a multiplex coronavirus antibody test for the evaluation of cross-reactivity to endemic coronavirus antigens (version 2) medRxiv. 2020 doi: 10.1101/2020.07.17.20156000. published online Aug 18. (preprint) [DOI] [Google Scholar]
- 23.WHO . World Health Organization; Geneva: 2011. When and how to use assays for recent infection to estimate HIV incidence at a population level. [Google Scholar]
- 24.Takahashi S, Greenhouse B, Rodriguez-Barraquer I. Are seroprevalence estimates for severe acute respiratory syndrome coronavirus 2 biased? J Inf Dis. 2020;222:1772–1775. doi: 10.1093/infdis/jiaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudbjartsson DF, Norddahl GL, Melsted P. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheatley AK, Juno JA, Wang JJ. Evolution of immunity to SARS-CoV-2 (version 2) medRxiv. 2020 doi: 10.1101/2020.09.09.20191205. published online Sept 11. (preprint) [DOI] [Google Scholar]
- 27.Huang AT, Garcia-Carreras B, Hitchings MDT. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11 doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long Q, Deng H, Chen J. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020 doi: 10.1101/2020.03.18.20038018. published online March 20. (preprint) [DOI] [Google Scholar]
- 30.Bryant JE, Azman AS, Ferrari MJ. Serology for SARS-CoV-2: Apprehensions, opportunities, and the path forward. Sci Immunol. 2020;5:47. doi: 10.1126/sciimmunol.abc6347. [DOI] [PubMed] [Google Scholar]
- 31.Larremore DB, Fosdick BK, Bubar KM. Estimating SARS-CoV-2 seroprevalence and epidemiological parameters with uncertainty from serological surveys (version 2) medRxiv. 2020 doi: 10.1101/2020.04.15.20067066. published online June 22. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and code used for producing the results are freely available on GitHub.