Abstract
Several reports have highlighted the contributions of host factors such as age, gender and co-morbidities such as diabetes, hypertension and coronary heart disease in determining COVID-19 disease severity. However, inspite of initial efforts at understanding the contributions of SARS-CoV-2 variants, most were unable to delineate causality. Hence, in this study we re-visited the contributions of different clades of viruses (G, GR and GH) along with other attributes in explaining the disparity in mortality rates among countries. A total of 26,642 high quality SARS-CoV-2 sequences were included and the A23,403G (S:D614G) variant was found to be in linkage disequilibrium with C14,408 U (RdRp: P323L). Linear regression analyses revealed increase in age [Odds ratio: 1.055 (p-value 0.000358)] and higher frequency of clade-G viruses [Odds ratio: 1.029(p-value 0.000135)] could explain 37.43% of the differences in mortality rates across the 58 countries (Multiple R-squared: 0.3743). Next, Machine-Learning algorithms LogitBoost and AdaboostM1 were applied to determine whether countries belonging to high/low mortality groups could be classified using the same attributes and accurate classification was achieved in 70.69% and 62.07% of the countries, respectively. Further, evolutionary analyses of the Indian viral population (n = 662) were carried out. Allele frequency spectrum, nucleotide diversity (π) values and negative Tajima's D values across ORFs were indicative of population expansion. Network analysis revealed the presence of two major clusters of viral haplotypes, namely, clade-G and a variant of clade L [Lv] having the RdRp:A97V amino acid change. Clade-G genomes were found to be evolving more rapidly and were also found in higher proportions in three states with highest mortality rates namely, Gujarat, Madhya Pradesh and West Bengal. Thus, the findings of this study and results from in vitro studies highlighting the role of these variants in increasing transmissibility and altering response to antivirals reflect the role of viral factors in disease prognosis.
Keywords: SARS-CoV-2, Clade-G, COVID-19 mortality, Phylogenetic networks, India
1. Introduction
Coronavirus disease 2019 (COVID-19) outbreak was first reported in December 2019 from Wuhan, China and since then has spread across the globe causing >5,00,000 deaths worldwide (Lu et al., 2020; WHO, 2020). The causal agent, SARS-CoV-2 is a positive strand RNA virus speculated to be of animal origin which has been found to cluster among 6 major clades circulating across countries. The first viral whole genome (RNA) sequence information was published on 5th of January 2020 (Wu et al., 2020) and so far >2,68,000 SARS-CoV-2 sequences have been submitted from across the world to Global Initiative on Sharing All Influenza Data (GISAID) (GISAID, 2020). However, a report from Shanghai, China describing viral variants from patients visiting a clinic between January and February 2020 were unable to find any association between viral haplotypes and enhanced viral dissemination or disease severity. However, clinical data from these patients were instrumental in identifying host predictive factors such as reduced CD4+ and CD8+ T cell counts with elevated IL-6 and IL-8 levels (Zhang et al., 2020a). Further, age, gender and comorbidities such as hypertension, diabetes and coronary heart disease have also been implicated in disease severity. The daily cumulative indices which are indicative of rapid increase in infected individual numbers have also been found to result in higher deaths in different countries due to health-care resource constraints (Al-Tawfiq et al., 2020; Ji et al., 2020; Zhou et al., 2020).
World-wide, spread of SARS-CoV-2 has greatly been influenced by social distancing measures and lockdown of cities undertaken by countries to control the pandemic. However, the differences in mortality rates given the incidence rates could not completely be explained by all the variables mentioned earlier.
Hence, in this report we revisit the role of viral genome variations found across the globe and explore if viral haplotype changes in combination with known host factors such as age could be used to explain the variability that we see in mortality rates given the incidence rates across the globe. Further, we also make an effort at elucidating the evolutionary trajectories of the Indian viral population and its distribution profiles to see if the world-wide patterns are being replicated in India as well.
2. Materials and methods
2.1. Information on age, SARS-CoV-2 clade information and cumulative COVID-19 incidence, death rates across countries
Age and SARS-CoV-2 clade information were obtained from the GSAID database on the 11th of June 2020. Countries that did not have age information or did not have more than three complete, high coverage sequence submissions were excluded from the analyses. Information on cumulative COVID-19 incidence and deaths were collected from the WHO database on the same day. The ratio of cumulative death given the cumulative incidence was used as a variable to determine mortality rates.
2.2. Viral sequence information
A total of 26,642 high quality SARS-CoV-2 sequences belonging to 59 countries were downloaded on the 11th of June 2020 from the GSAID database (Supplementary table S1). This included countries from Asia (n = 19), Africa (n = 5), Europe (n = 26), North America (n = 3), South America (n = 5) and Oceania (n = 1).
Additionally, viral genome sequences from Indian nationals and a few international tourists stranded in the country, both submitted from India were also included (n = 662). From a total of 662 sequences, 25 were found without state information out of which 14 were Indian citizens from Iran and 5 were Italian tourists. Additionally, 7 had been grown in Vero cells. The rest of the isolates were collected from 19 different states which included Andhra Pradesh (n = 1), Assam (n = 2), Bihar (n = 4), Delhi (n = 62), Gujarat (n = 199), Haryana (n = 4), Jammu (n = 1), Karnataka (n = 10), Kerala (n = 2), Ladakh (n = 6), Madhya Pradesh (n = 15), Maharashtra (n = 63), Odisha (n = 74), Punjab (n = 1), Rajasthan (n = 6), Tamil Nadu (n = 31), Telangana (n = 97), Uttar Pradesh (n = 5) and West Bengal (n = 47). State-wise average ages were calculated for those states that had contributed more than three viral sequences to the GISAID database.
2.3. Nucleotide alignment and variant calling
Given the initial emergence of the SARS-CoV-2 virus from Wuhan, China, alignment was carried out using the genome sequence submitted by Wu et al. (NC_045512.2) in January 2020 (Wu et al., 2020). Reference guided sequence alignment was executed with five iterations for both. Since different sequencing platforms had been used to generate SARS-CoV-2 sequence data with different error rates and filtering cut-offs so variant calling was stringently carried out with a minimum coverage of one-third genomes. Ambiguous bases found in genome sequences were considered to be unresolved for the purpose of analyses.
2.4. Measurements of diversity and deviation from neutrality for Indian sequences
Watterson's estimator (θw), nucleotide diversity (π) and Tajima's D (Tajima, 1989) for each open reading frame (ORF) was calculated using MEGA X (Kumar et al., 2018).
2.5. Phylogeny construction for Indian sequences
Phylogenetic analysis was carried out following the median-joining approach using Network 10.1.0.0 software (Bandelt and Röhl, 1999). For phylogeny construction, a variant frequency cutoff of ≥0.01 was used and a 97% cutoff for the number of sequences with resolved bases for each position to avoid spurious clustering.
2.6. Statistical analyses
Measures of linkage disequilibrium (LD) such as D′ and r2 values (Jennings, 1917) were calculated manually using observed and expected frequencies of haplotypes formed by the loci of interest. Linear regression was carried out to assess the importance of variables such as Age (average age of reported cases), World Bank income group classification (Groups 3 and 4), tests per million as on June 25, 2020 and percentage of different clades of viruses (G, GR and GH) in explaining COVID-19 mortality rates in continuous scale using IBM SPSS Statistics for Windows, version 1.0.0.1327 (IBM Corp., Armonk, N.Y., USA). and R (R Core Team, 2014). Mortality rate was defined as the ratio of cumulative deaths and cumulative incidence as on 25.06.2020.
2.7. Machine learning
Machine-Learning was used to assess whether countries belonging to high/low mortality groups could be classified based on the percentage of different clades of viruses and other attributes. For this, the Waikato Environment for Knowledge Analysis (Weka 3.8.4) (Frank et al., 2016) software suite was used. Countries with death rates above the median value of 2.8% were labeled as high death rate countries. Two major boosting algorithms AdaboostM1 (Yoav and Schapire, 1996) and LogitBoost (Friedman et al., 2000) were used in conjunction with simple decision stumps (as weak learners) and 10 iterations to predict high/low mortality groups. The mortality rate classification was based on the median COVID-19 death rate (Section 2.6). The predictor variables used were Age (average age of reported cases), Income (World Bank income group classification), Tests (tests per million as on June 25, 2020), and percentage of different clades of viruses. The accuracy of the models was further tested using 10-fold cross-validation.
3. Results
3.1. Patient age and the SARS-CoV-2 G-clade spike protein variant G 23403 (S: G614) explain differences in death rates across countries
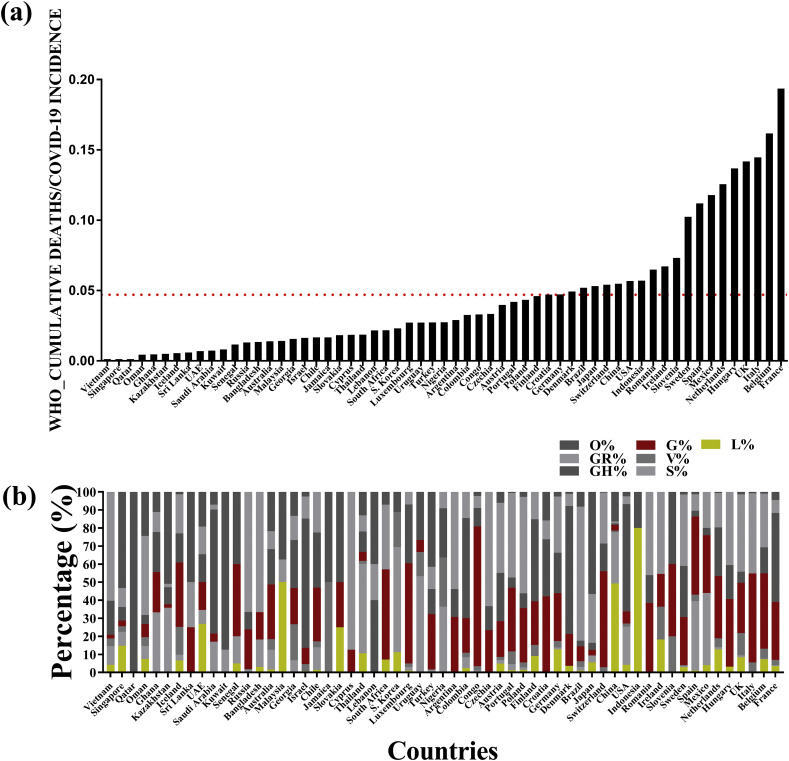
The sequences from all the 59 countries excluding India which had patient information on age and more than 3 sequence submissions were analyzed to delineate the frequency of non-synonymous changes that had been used to classify the virus into clades (Fig. 1 , Supplementary table S1-1). Comparisons were made to delineate the role all the predictors. Linear regression analysis revealed that increase in age [Odds ratio: 1.055 (p-value = 0.000358)] and the higher frequency of the G-clade viruses [Odds ratio:1.029 (p-value 0.000135)] could explain 37.43% of the differences in cumulative death rates given the cumulative incidence rates across the 58 countries (Multiple R-squared: 0.3743). Two countries had to be excluded from the analyses because of missing ‘tests per million’ values. The LogitBoost algorithm could accurately classify 41 out of 58 countries (70.69%) based on mortality rates given the attributes included in this study. Based on 10-fold CV, the AUC (Area under ROC curve) was 0.776 with a precision of 71.1%, recall value 70.7%, and F-measure of 70.7%. The AdaboostM1 algorithm could classify 36 out of 58 countries accurately (62.07%) and had an AUC value of 0.709. The final classifiers (obtained by LogitBoost and AdaboostM1) determined based on our data and ROC curves are given in the (Supplementary table S1-2, S1-3). Notably, both the classifiers used clade G percent as the most predictive attribute in the first iteration.
Fig. 1.
Comparison of the mortality rates and viral clade distribution across 59 countries across the world (a) The ratio of COVID-19 cumulative deaths to cumulative incidences for 59 countries as estimated by the World Health Organization till the 11th of June 2020. The red line indicates the average ratio. (b) The distribution of 6 different clades, namely L, S, V, G, GH, GR and O found in the same 59 countries considered for analysis.
3.2. Linkage disequilibrium between the A23403G (S: D614G) and the C14408U (NSP12:P323L) variants
Given that the RNA dependent RNA Polymerase (RdRp) holds the key to viral genome conservation, hence, we focused on understanding if there existed a non-random association between the known clade-G A23403G (S: D614G) variant and the high frequency RdRp variant C14408U (NSP12:P323L) among the clade-G viruses. Using sequence information from all the 59 countries, it was identified that both the variants was in linkage disequilibrium (LD) with D′ and r2 values of 0.9 and 0.826 respectively. It was also observed that the Asian countries had the lowest percentage of the U14408-G23403 (NSP12:L323-S:G614) haplotype (37%) with none in Qatar, South Korea, 3.47% in China and 4.17% in Malaysia in contrast to the African, European, North and South American countries where the lowest percentage was found in Uruguay at 26.67% (Supplementary table S1). The earliest G-clade virus (S: G614) with NSP12: L323 variant was identified in Zhejiang, China with a collection date of 1/24/2020 (EPI_ISL_422425) followed by its presence in Italy, France, Switzerland, England and Luxembourg by the end of February 2020 and its re-entry to China by way of individuals from Beijing with travel history to Europe in March 2020.
3.3. In depth analyses of the variants found within the Indian SARS-CoV-2 genomes
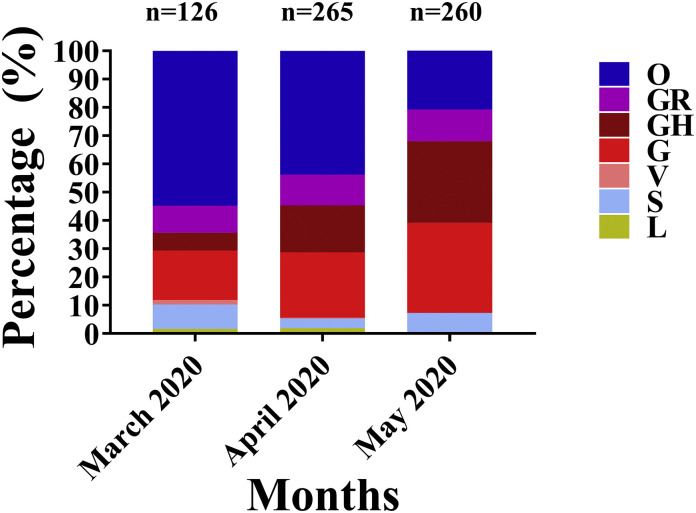
The Indian subcontinent was found to have lower morality rates given the disease incidence rates (0.028, Supplementary table S4) and the clade-G viruses formed 25.96% of the proportion of viruses that had been sequenced till 11th of June 2020. However, the clade-G virus associated with higher death rates were found to increase in frequency from 17.46% in March to 31.92% in May which formed the basis of the country-average (Fig. 2)
Fig. 2.
Month-wise distribution of viral clades in India.
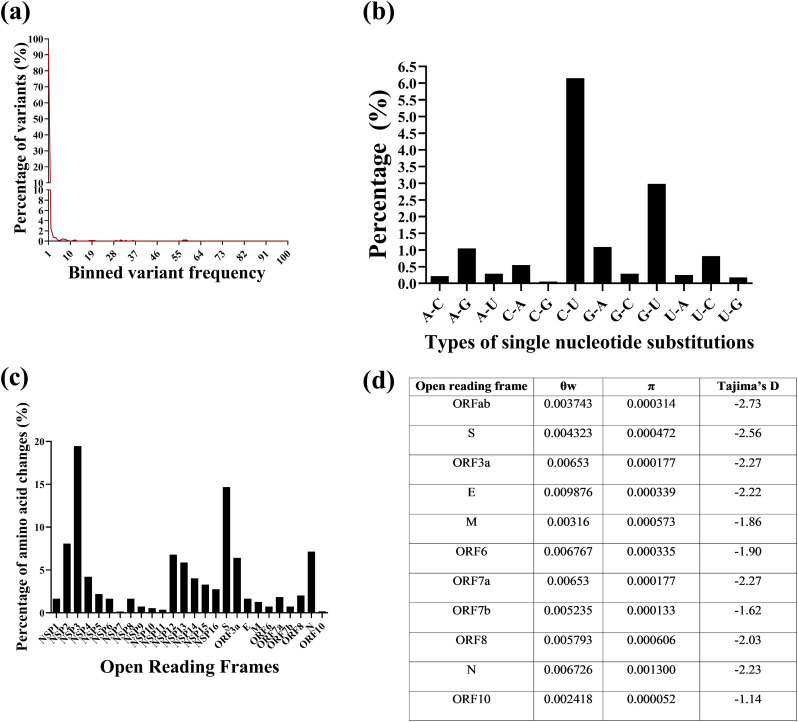
Hence, to further understand the evolutionary trajectories of these viruses, 662 Indian genome sequences and the ancestral Wuhan-Hu-1 isolate sequence (NC_045512.2) were separately aligned and a total of 887 single nucleotide variants, 21 di-nucleotide, tri-nucleotide and tetra-nucleotide substitutions and 14 insertions and deletions were identified ranging in frequencies between 0.2% to >57%. A total of 545 of these variants resulted in amino acid changes (Fig. 3)A, supplementary table S5). Among the single nucleotide variants C—U transition per C residue in the genome was highest at 6.15%, followed by G-U transversion (2.98%) (Fig. 3B, supplementary table S6). The NSP3 and S ORFs had the highest proportion of non-synonymous changes (NSP3: n = 106, 19.45%; S: n = 80, 14.68%) (Fig. 3C supplementary table S7). Given the number of variants identified, the diversities across all the ORFs were calculated. ORF10 was found to have the lowest nucleotide diversity (π) while ORF N had the highest. Overall, the nucleotide diversity (π) values were low across ORFs in comparison to the θ (Watterson's estimator) values (Fig. 3D) which was indicative of the presence of higher proportion of low frequency variants as has been described in Fig. 3A. The next objective was to determine if the patterns of diversity could be attributed to genetic drift or neutrality. Tajima's test for neutrality was applied and all the ORFs were found to have negative Tajima's D values (Fig. 3D) indicative of non-neutral evolution.
Fig. 3.
Description of SARS-CoV-2 variants, the gene diversities and the results of the test for neutrality among the Indian viral population. (a) The frequencies of variants binned on the basis of frequency. (b) The types of single nucleotide changes incurred by the viral genomes (c) The percentage of amino acid changes per open reading frame out of a total of 388 non-synonymous changes that were observed. (d) The diversity and Tajima's D values across open reading frames.
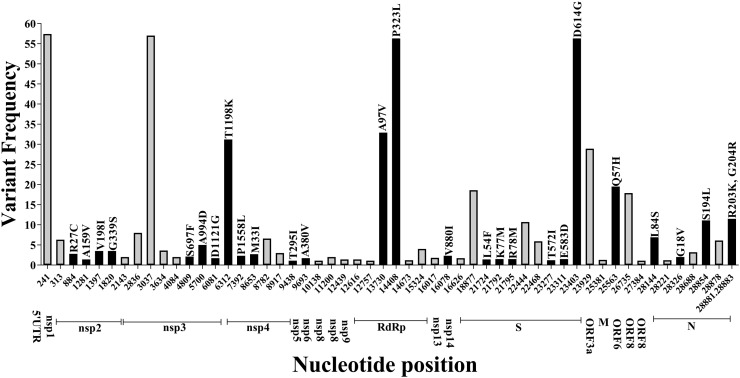
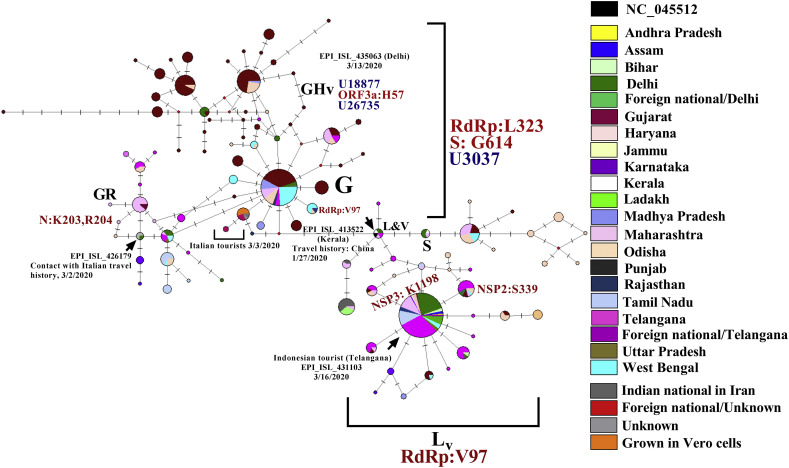
To trace the ancestries of the Indian viral isolates, network analysis was carried out using the Hamming distances of variants present at ≥1% frequency among the genomes (Supplementary table S8). The haplotypes were generated stringently using a total of 53 single nucleotide variants and a tri-nucleotide substitution at positions 28,881–28,883GGG-AAC together resulting in 27 amino acid changes among the 663 Indian SARS-CoV-2 genome sequences (Fig. 4 , Supplementary table S9). A total of 101 nodes with 101 distinct haplotypes were discovered using 663 genome sequences. The network appearance was as expected from an ongoing pandemic with the presence of ancestral viral haplotypes existing along with newly mutated genomes. There were two major clusters of haplotypes that were found to have emerged from the ancestral Wuhan-Hu-1 virus (clade L); the first identified to be belonging to a variant of the clade L which has been annotated here as Lv (n = 208) and harbored the RNA dependent RNA polymerase (RdRp) protein [C13,730 U,(A97V)] from which a sub-cluster (n = 126) was formed harboring an additional non-synonymous change in the NSP3 protein [C6312A (T1198K)] (Fig. 5 ). The second cluster was formed by the clade G viruses (n = 173) which differed at three loci resulting in one amino acid change each in the RdRp, S protein [C3037U, C14,408 U (RdRp: P323L), A23,403G (S:D614G)] known to be in LD across the world. The first clade G viruses to be sequenced from India were isolated on the 3rd of March from Italian tourists (Accession IDs: EPI_ISL_420543, EPI_ISL_420545, EPI_ISL_420547, EPI_ISL_420549, EPI_ISL_420551 and EPI_ISL_420553) and were clade G variants harboring an additional amino acid change in the NSP3 protein [C4,809U (S697F)] followed by an Indian from Iran and two contacts of Indians with travel history to Italy (EPI_ISL_424362, EPI_ISL_424364, EPI_ISL_424365). Two sub-clusters were observed evolving from clade G; GH variant mentioned here as GHv with the variants C18877U, G25563U (ORF3a:Q57H), C26735U having multiple evolving branches and GR with the tri-nucleotide GGG-AAC substitution at positions 28,881–28,883 resulting in two amino acid changes R203K, G204R in the N protein (Fig. 4). Additional synonymous and non-synonymous variants among both the clusters resulted in the 101 haplotypes (supplementary table S9). Reticulations were also observed which could happen because of parallel mutations or homoplasy (Fig. 5).
Fig. 4.
Distribution of nucleotide variants on the basis of which Indian SARS-CoV-2 ancestries were derived with variant frequencies and associated amino acid changes. The black bars represent the non-synonymous variants with the amino acid changes mentioned above and the coding regions are indicated below.
Fig. 5.
Phylogenetic network of 662 SARS-CoV-2 viral genomes isolated from Indians and foreign nationals present in India. Each circle represents a haplotype and the diameter is proportional to the number of genomes belonging to each haplotype. Each notch on a horizontal line represents a differentiating variant and the lengths of the connecting lines are proportional to the number of variants. The colors indicate the different states from which specimens were collected. The arrow indicates the node containing the ancestral NC_0145512 viral haplotype and the sequence ID indicate the first sequenced viral genome from a symptomatic individual with travel history to Wuhan, China. A total of 53 variants were used to construct the haplotypes and the nucleotide and amino acid differences in comparison with NC_045512 among the major clades Lv(RdRp:A97V), G, GHv [C18887U, C26735U] and GR with the 48,881-48,883(GGG-AAC, N:R203K, G204R) variants are mentioned. The details of the index patient infected with each clade of the virus in the Indian context are mentioned with isolation dates.
There were 17 variants [C1707U, C6310A, U8022G, G11083U, A8026U, G11083U, G12685U, A15435G, C19524U, A21550C, A21551U, A24389U, G24390C, U24622C, C28311U, G29742U, A29827U, A29830U] that could not be included in the network analysis because of the presence of unresolved bases at multiple viral genome sequences and the G26144U (ORF3a:G251V) variant that segregates clade V was excluded because of allele frequency cutoff of 1%.
3.4. State-wise distribution of clade-G viruses and the U14408-G23403 (NSP12:L323-S: G614) haplotypes
In February 2020, the clade-G and GR viruses were first reported from India among Italian tourists and a contact of an Indian national with travel history to Italy. This was followed by the identification of a clade-GH virus in an individual from Delhi in mid-February 2020 (Fig. 5). All the Indian clade-G viruses were found to harbor the U14,408 (RdRp: L323) variant. In states with the highest number of deaths given the number of confirmed cases like Madhya Pradesh (4.21%), West Bengal (4.40%) and Gujarat (6.28%) (Supplementary table S10), reported higher rates of clade-G viruses at 100% (n = 10), 72.34% (n = 34) and 36.18% (n = 72) with average age counts of 35 (±18.8), 45(±18.4) and 44.8(±18.7) years respectively. Additionally, total of 6 out of 34 (17.7%) and one isolate out of 14 (7.1%) clade-G viruses from West Bengal and Odisha respectively harbored the U13,730 (RdRp:V97) variant. This variant which originated among the clade-L viruses was first reported in a clade-G isolate in the USA [EPI_ ISL_431080, 3/2/2020] and the first from India was reported in an individual from West Bengal who got infected while travelling [EPI_ ISL_455644, 4/2/2020]. The isolates from the states of Assam (n = 2) and Ladakh (n = 6, average age = 26.4) which had lowest COVID-19 deaths (0.22% and 0.42%) did not harbor the clade-G viruses however, the number of isolates were few.
4. Discussion
In this report, for the first time our results highlight that both high age and SARS-CoV-2 clade-G viral infections can explain 37.43% of the observed variability in cumulative mortality rates across 58 countries. Further, a machine learning algorithm trained on some of these broad attributes including percent of clade-G infections has reasonably good (>70%) accuracy to classify countries into high/low mortality groups. The clade-G viruses were first reported from the Zhejiang and the Sichuan provinces in China (EPI_ISL_422425, EPI_ISL_451345) with a collection date of 1/24/2020 followed by its spread to Bavaria, Germany by way of a Chinese citizen visiting the city for work at the end of January 2020 and in line with our findings a recent report on a few European countries with high death rates have also highlighted the presence of clade-G viruses (Eaaswarkhanth et al., 2020). Similarly, an earlier report on clinical outcome from Sheffield, England have also demonstrated the G614 mutation to be associated with higher viral loads (Korber et al., 2020).
In vitro studies have highlighted that incorporation of this variant stabilizes the S protein leading to greater incorporation in virus-like particles which has been speculated to have contributed to the transmission efficiency of this haplotype (Zhang et al., 2020b). Further, given that the hACE2 gene is interferon inducible in the airway epithelium (Chua et al., 2020) and infection results in increase in ACE2 production, a virus with higher proportions of S protein could mount higher inflammatory response associated with poor prognosis. However, this variant might not be contributing alone. Earlier reports on antivirals used in treatment which target the RdRp protein such as Remdesivir, Filibuvir etc. have shown these antivirals binding to the RdRp-NSP7-NSP8 complex through a hydrophobic groove involving amino acid residues F324, F325, F326 of the RdRp protein which is just next to the P323L variant site with potential to alter response to treatment with these antivirals which is in strong LD with the A23,403G (S:D614G) variant (Pachetti et al., 2020; Ruan et al., 2020).
Given the pattern of virulence observed across the world, an attempt was also made to decipher the pattern of evolution among the Indian isolates to infer the ancestries of the viral isolates and build SARS-CoV-2 infection paths as has been done before (Forster et al., 2020) with special emphasis on the clade-G viruses. While comparing the 662 Indian SARS-CoV-2 genomes, a number of nucleotide variants or segregating sites were identified, however, the nucleotide diversity values (π) were indicative of an excess of low frequency variants. This could be because of recent population expansions as has been observed in H1N1 populations involved in outbreaks and epidemics (Martinez-Hernandez et al., 2010) and the uniform negative Tajima's D values across all the SARS-CoV-2 ORFs could also be attributed to it and this finding has remained unchanged with increase in viral sequences numbers (Bhattacharjee and Pandit, 2020).The clade G (S:G614) viruses with the C14,408U (RdRp: P323L) variant were found to incur more number of variations leading to the emergence of a number of sub-clusters of viruses with increased branch lengths in comparison to the clade Lv viruses including clade GHv and GR. These clade G viruses and its sub- clusters were also found in more numbers in states where higher mortality rates were recorded [Gujarat (72/199, Madhya Pradesh (10/10) and West Bengal (34/47)]. Earlier reports have also attributed the occurrence of higher numbers of mutations to the presence of C14,408 U (RdRp: P323L) variant occurring in the interaction domain of RdRp thus impairing protein-protein interactions with NSP7, NSP8 and NSP14 resulting in altered proofreading and processivity as has been speculated in earlier reports (Chand and Azad, 2020; Pachetti et al., 2020).
These implications about the clade-G viruses need to be further tested using comprehensive clinical data and genomic data from all the countries, however, information on the presence of viral variants might prove to be useful in diagnostics and in decision making during treatment. This idea is reinforced with the recent emergence and rapid spread of the B.1.1.7 variant with multiple additional spike protein mutations [deletion 69–70 (diagnostic failure), deletion145, N501Y (increased hACE2 binding affinity), A570D, P681H (Furin cleavage site), T716I, S982A, D1118H) on the background of the G614 mutation (Kupferschmidt, 2020).
The following is the supplementary data related to this article.
S1 table: Details of all the sequences included in the study. S2 table: Detailed results of LogitBoost algorithm. S3 table: Detailed results of AdaBoostM1 algorithm. S3 table: Country-wise values of variables including frequencies of clade-G viruses and 14,408–23,403 haplotypes. S4 table: Description all the variants identified in this study among the Indian viral population. S5 table: Proportions of different types of single nucleotide variants observed among the Indian viral isolates. S6 table: Distribution of amino acid changes across open reading frames in the Indian viral isolates. S7 table: Features of the variants included in the construction of viral haplotypes. S8 table: Indian viral haplotype information. S9 table: Official state-wise numbers of confirmed COVID-19 cases and deaths from 19 Indian states as updated on 11th June 2020.
Declaration Competing Interest
None declared.
Funding
B.B was supported by the Ramanujan fellowship funded by the Department of Science and Technology, Government of India.
Author Contributions
Conceptualization, B.B.; data curation, B.B.; data analysis, S.B. & B.B, writing-original draft, B.B. & B.P.; writing- review and editing, B.B., B.P. & S.B. All three authors approved the manuscript.
Acknowledgments
The authors acknowledge the submitters of coronavirus sequence data to the GISAID database, the database managers, developers and scientists associated with GISAID.
References
- Al-Tawfiq J.A., Leonardi R., Fasoli G., Rigamonti D. Prevalence and fatality rates of COVID-19: what are the reasons for the wide variations worldwide? Travel med. Infect. Dis. Ther. 2020;35:101711. doi: 10.1016/j.tmaid.2020.101711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H.-J., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee B., Pandit B. 2020. Phylogenetic Clustering of the Indian SARS-CoV-2 Genomes Reveals the Presence of Distinct 1clades of Viral Haplotypes among States. [DOI] [Google Scholar]
- Chand G.B., Azad G.K. 2020. Identification of Novel Mutations in RNA-Dependent RNA Polymerases of SARS-CoV-2 and their Implications on its Protein Structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thurmann L., Kurth F., Volker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Muller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.E., Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Eaaswarkhanth M., Al Madhoun A., Al-Mulla F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int. J. Infect. Dis. 2020;96:459–460. doi: 10.1016/j.ijid.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank X.E., Hall M.A., Witten I.H. Fourth ed. Morgan Kaufmann; 2016. The WEKA Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”. [Google Scholar]
- Friedman Y.J., Hastie T., Tibshirani R. Additive logistic regression: a statistical view of boosting (with discussion and a rejoinder by the authors) Ann. Stat. 2000;28(2):337–407. [Google Scholar]
- GISAID 2020. https://www.gisaid.org/
- Jennings H.S. The numerical results of diverse systems of breeding, with respect to two pairs of characters, linked or independent, with special relation to the effects of linkage. Genetics. 1917;2:97–154. doi: 10.1093/genetics/2.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Ma Z., Peppelenbosch M.P., Pan Q. Potential association between COVID-19 mortality and health-care resource availability. Lancet Glob. Health. 2020;8 doi: 10.1016/S2214-109X(20)30068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B.F.W., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E.E., Bhattacharya T., Parker M.D., Partridge D.G., Evans C.M., de Silva T., on behalf of the Sheffield COVID-19 Genomics Group, LaBranche C.C., Montefiori D.C. 2020. Spike Mutation Pipeline Reveals the Emergence of a More Transmissible form of SARS-CoV-2. [DOI] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. 2020. Mutant Coronavirus in the United Kingdom Sets off Alarms, but its Importance Remains Unclear. [DOI] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hernandez F., Jiminez-Gonzalez D.E., Martinez-Flores A., Villalobos-Castillejos G., Vaughan G., Kawa-Karasik S., Flisser A., Maravilla P., Romero-Valdovinos M. What happened after the initial global spread of pandemic human influenza virus A (H1N1)? A population genetics approach. Virol. J. 2010;7:196. doi: 10.1186/1743-422X-7-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing. Austria. URL; Vienna: 2014. R: A language and environment for statistical computing.http://www.R-project.org/ [Google Scholar]
- Ruan Z., Liu C., Guo Y., He Z., Huang X., Jia X., Yang T. 2020. Potential Inhibitors Targeting RNA-Dependent RNA Polymerase Activity (NSP12) of SARS-CoV-2. (Preprints 2020030024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2020. (https://covid19.who.int/). WHO.
- Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoav Z.F., Schapire R.E. 1996. Experiments with a New Boosting Algorithm. Machine Learning: Proceedings of the Thirteenth International Conference; pp. 148–156. [Google Scholar]
- Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S., Chen J., Wang W., Chen B., Jiang L., Yu S., Lu J., Wang J., Xu M., Yuan Z., Zhang Q., Zhao G., Wang S., Chen S., Lu H. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. 2020. The D614G Mutation in the SARS-CoV-2 Spike Protein Reduces S1 Shedding and Increases Infectivity. [DOI] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 table: Details of all the sequences included in the study. S2 table: Detailed results of LogitBoost algorithm. S3 table: Detailed results of AdaBoostM1 algorithm. S3 table: Country-wise values of variables including frequencies of clade-G viruses and 14,408–23,403 haplotypes. S4 table: Description all the variants identified in this study among the Indian viral population. S5 table: Proportions of different types of single nucleotide variants observed among the Indian viral isolates. S6 table: Distribution of amino acid changes across open reading frames in the Indian viral isolates. S7 table: Features of the variants included in the construction of viral haplotypes. S8 table: Indian viral haplotype information. S9 table: Official state-wise numbers of confirmed COVID-19 cases and deaths from 19 Indian states as updated on 11th June 2020.