Abstract
Background: There are two highly conserved thyroid hormone (triiodothyronine [T3]) receptor (TR) genes, TRα and TRβ, in all vertebrates, and the expression of TRα but not TRβ is activated earlier than T3 synthesis during development. In human, high levels of T3 are present during the several months around birth, and T3 deficiency during this period causes severe developmental abnormalities including skeletal and intestinal defects. It is, however, difficult to study this period in mammals as the embryos and neonates depend on maternal supply of nutrients for survival. However, Xenopus tropicalis undergoes a T3-dependent metamorphosis, which drastically changes essentially every organ in a tadpole. Of interest is intestinal remodeling, which involves near complete degeneration of the larval epithelium through apoptosis. Concurrently, adult intestinal stem cells are formed de novo and subsequently give rise to the self-renewing adult epithelial system, resembling intestinal maturation around birth in mammals. We have previously demonstrated that T3 signaling is essential for the formation of adult intestinal stem cells during metamorphosis.
Methods: We studied the function of endogenous TRα in the tadpole intestine by using knockout animals and RNA-seq analysis.
Results: We observed that removing endogenous TRα caused defects in intestinal remodeling, including drastically reduced larval epithelial cell death and adult intestinal stem cell proliferation. Using RNA-seq on intestinal RNA from premetamorphic wild-type and TRα-knockout tadpoles treated with or without T3 for one day, before any detectable T3-induced cell death and stem cell formation in the tadpole intestine, we identified more than 1500 genes, which were regulated by T3 treatment of the wild-type but not TRα-knockout tadpoles. Gene Ontology and biological pathway analyses revealed that surprisingly, these TRα-regulated genes were highly enriched with cell cycle-related genes, in addition to genes related to stem cells and apoptosis.
Conclusions: Our findings suggest that TRα-mediated T3 activation of the cell cycle program is involved in larval epithelial cell death and adult epithelial stem cell development during intestinal remodeling.
Keywords: metamorphosis, Xenopus tropicalis, thyroid hormone receptor, RNA-seq, intestine, stem cells, apoptosis
Introduction
The synthesis of thyroid hormone (triiodothyronine [T3]) and its release for blood circulation are regulated by the hypothalamus–pituitary–thyroid axis through thyrotropin-releasing hormone and thyrotropin (1). T3 affects nearly all aspects of biology including postembryonic development, a period around birth when plasma T3 level peaks, and adult metabolism in vertebrates. Expectedly, T3 deficiency during postembryonic development causes severe developmental problems including the defects in maturation of the brain and intestine (2,3).
T3 receptors (TRs) are transcription factors, which belong to the superfamily of nuclear hormone receptors (4). TRs bind to T3 response elements, mainly as heterodimers with 9-cis-retinoic acid receptors, which are also members of the nuclear hormone receptor superfamily, to regulate gene transcription by recruiting different cofactors in a T3-dependent manner (4–20). TRs repress target gene expression in the absence of T3 by recruiting corepressors such as nuclear receptor corepressor and silencing mediator of retinoid and thyroid hormone receptor. When T3 binds to TR, the expression of these T3-target genes is increased by liganded TR due to the recruitment of coactivators such as P300 and steroid receptor coactivators (4,5,7,11–20).
There are two TR genes, TRα and TRβ, in all vertebrates. In mammals, the two TR genes produce three T3 binding isoforms, TRα1, TRβ1, and TRβ2, and another one incapable of binding to T3, TRα2, due to alternative splicing, with differing tissue distributions (1). Numerous transgenic, knockin, and knockout mouse models have been developed and revealed many roles of TRs during development and in adult organ physiology/pathology. TRα1 is the predominant TR in the intestine, bone, muscle, heart, and the central nervous system, suggesting that TRα regulates the development of these organs. TRβ1 is mainly expressed in the inner ear, retina, and liver, while TRβ2 is predominantly expressed in the hypothalamus and pituitary to control T3 levels (3,21). Additionally, TRα is expressed earlier than T3 synthesis during vertebrate development (22,23), suggesting that TRα regulates T3 target genes to affect early development. However, it is difficult to investigate the roles of TRs during postembryonic development in vertebrates as the embryos and neonates depend on maternal supply for survival.
We have been using amphibian metamorphosis as a model to study TR function during vertebrate development (7,18,24–26). Metamorphosis in anurans such as Xenopus tropicalis resembles postembryonic development in mammals (18,24), making it a useful model to study T3 action and adult organ development in vertebrates. During metamorphosis, many organs are drastically remodeled to the adult form with distinct morphology. In particular, the tadpole intestine is a simple tubular structure with a single epithelial fold (27,28). During metamorphosis, the larval epithelial cells undergo apoptosis and adult intestinal stem cells are formed de novo, which subsequently proliferate and differentiate to form a multiply folded adult epithelium surrounded by thick layers of connective tissues and muscles (28–34). This offers a unique opportunity to study how T3 regulates adult stem cell development.
As in mammals, TRα is highly expressed earlier than TRβ and before the maturation of the thyroid gland and the onset of metamorphosis at stage 54 in X. tropicalis (9,22,35–40). TRβ has little expression during premetamorphosis but is strongly activated during metamorphosis. Thus, TRα is likely to have a more important role during premetamorphosis and early metamorphic period, which is supported by recent TRα and TRβ knockout studies in X. tropicalis (41–52). How TRα or TRβ affects X. tropicalis development remains unclear. Here, we have carried out RNA-seq analyses of the intestine in wild-type and TRα-knockout X. tropicalis tadpoles treated with or without T3 and discovered interesting gene regulation pathways controlled by TRα that underlie larval epithelial cell death and adult stem cell development during metamorphosis.
Materials and Methods
Animal rearing and genotyping
Wild-type adult X. tropicalis were purchased from Nasco Co. (Fort Atkinson, WI) or raised in the laboratory. Tadpoles were staged according to the description for Xenopus laevis (53). All animal care and treatments were performed as approved by the Animal Use and Care Committee of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health. Premetamorphic X. tropicalis tadpoles at stage 54 were treated with 10 nM T3 at 25°C for up to three days.
Sexually mature TRα heterozygous (+/−) mutant X. tropicalis frogs (46) were mated to produce TRα homozygous (−/−) tadpoles. Briefly, a TRα+/− male and a TRα+/− female were injected with 20 U of human chorionic gonadotropin (hCG; Novarel, Tarrytown, NY) one day before egg laying for priming. Next day, 200 U of hCG was injected into each frog before natural mating. After reaching stage 45 (onset of feeding), the resulting tadpoles were moved into 9 or 3 L tanks equipped with a water circulating system at 26°C under a 15:9 hour light:dark cycle. Tadpoles at stage 54 were anesthetized with ice-cold water, and tail tip was clipped and lysed in 20 μL of QuickExtract (Epicentre, Madison, WI) at 55°C for 1 to 2 hours. Genotyping was performed on the tail tip DNA by using polymerase chain reaction (PCR): 2 μL of DNA extraction solution was added to each PCR containing 10 μL of GoTaq Green Master Mix (Promega, Madison, WI) and 0.5 μL of 20 μM primers. PCR cycling conditions were 94°C for 30 seconds, 68°C for 60 seconds, and 72°C for 180 seconds for 35 cycles. The forward and reverse primers were: wild-type TRα forward, 5′-AGCTATCTGGACAAAGACGAGCCG-3′, mutant TRα forward, 5′-ACATCCCCAGCTATCCCCAGCTATG-3′, reverse, 5′-GCAAACTTTTTGGCTCAGAGGCCAC-3′ (46).
Histological analysis
Tadpoles at stage 54 were randomly selected and treated with 10 nM T3 for 0–3 days at 25°C as indicated. After treatment, the tadpole was genotyped, and the intestine was isolated. The intestine length was measured and normalized against body weight. The intestines were then fixed in 4% paraformaldehyde in phosphate-buffered saline for one day and processed with a tissue processor (Excelsior AS Tissue Processor; Thermo Fisher Scientific, Waltham, MA). Next, the intestines were embedded in paraffin, cut into 5 μm sections, and placed on slides. After deparaffinizing, the tissue sections were stained with methyl green-pyronin Y (MGPY; Muto, Tokyo, Japan) as described (33). For detecting proliferating cells, 1.25 μL of 10 mg/mL of 5-ethynyl-2′-deoxyuridine (EdU) was injected into tadpoles, and 30 minutes later, the tadpoles were euthanized (54). Tissue sections cut at 5 μm were prepared as described above, and EdU was detected with the Click-iT Plus EdU Alexa Fluor 594 Imaging kit (Invitrogen, Carlsbad, CA). Apoptotic cells were detected by using terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) in situ cell death detection kit (Roche Diagnostics, Indianapolis, IN). Nuclei were then stained with DAPI (4′,6-diamidino-2-phenylindole). EdU-positive cells and apoptotic cells in epithelial region were measured by using the ImageJ software (National Institutes of Health, Bethesda, MD).
RNA extraction, cDNA synthesis, and quantitative reverse-transcription PCR
Isolation of total RNA was performed with RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). Reverse transcription was carried out with QuantiTect® Reverse Transcription Kit (Qiagen). Total RNA (500 ng) was reversed transcribed into cDNA in a 20 μL reaction including 2 μL of gDNA wipeout buffer, 4 μL of 5 × RT buffer, 1 μL of RT Primer Mix, and 1 μL of Quantiscript Reverse Transcriptase. The mixture was incubated at 42°C for 15 minutes and then at 95°C for 3 minutes. It was then diluted 10-fold in sterile water. Two microliters of the cDNA solution was added to 18 μL of quantitative PCR mixture containing 10 μL of 2 × SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and 0.5 μL of 20 μM primers. Quantitative reverse-transcription PCR (qRT-PCR) was performed in triplicates by using the Step One Plus Real-Time PCR System (Applied Biosystems). The sequences of the primers are described in Supplementary Table S1. The ribosomal protein L8 gene (rpl8) (55) was used as a house keeping gene for normalization, and the gene expression analysis was performed at least twice, with similar results.
RNA-seq analysis
Total RNA was extracted from the intestine of wild-type and TRα−/− tadpoles treated with or without T3 for 18 hours. The mRNA was purified from total RNA by using poly-T oligo-attached magnetic beads and chemically fragmented. The integrity of RNA (RIN) was determined using Agilent Bioanalyzer 2100 system (Agilent Technologies, CA) with RIN >8.0. First-strand cDNA was synthesized by using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H-). Second-strand cDNA synthesis was subsequently performed by using DNA Polymerase I and RNase H. The cDNA libraries were generated by using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA). The libraries were sequenced on the Illumina HiSeq 2000 platform to obtain 100 nt paired-end reads by the Molecular Genomics Core, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health. The demultiplexed and adapter removed short reads were mapped to NCBI Xenopus tropicalis genome assembly, Xenbase Xenopus tropicalis Genome assembly (v9.1), and Ensembl Xenopus tropicalis Genome (JGI 4.2) with STAR software (version 2.6.1c), and reads counts for each gene/exon were obtained with featureCounts tool of Subread software (version 1.6.3). R Bioconductor DESeq2 package was used for gene differential expression analysis.
Gene Ontology and pathway analysis
To identify significantly enriched biological themes and functional groups, Gene Ontology (GO) and pathway analyses were performed by using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 program (56,57). Ensembl Xenopus ID were changed into human ID to increase the hit count (58). The GO analysis was used to identify enriched biological terms by using “biological process” subontology of GO (GO:BP) GO terms defined by the Gene Ontology Consortium (59). We further analyzed genes related to “stem cell regulation” (GO:0048864, GO:0036335, GO:0019827, GO:0072089, GO:0072091, and GO:2000648), “cell proliferation” (GO:0008283), and “apoptotic process” (GO:1904019 and GO:0006915).
Statistical analysis
All graphs show one representative experiment of at least three. The results were analyzed using the 4-Step Excel Statistics software application (OMS Publishing, Inc., Tokorozawa, Saitama, Japan).
Results
Lack of TRα suppresses intestinal remodeling during T3-induced metamorphosis in X. tropicalis
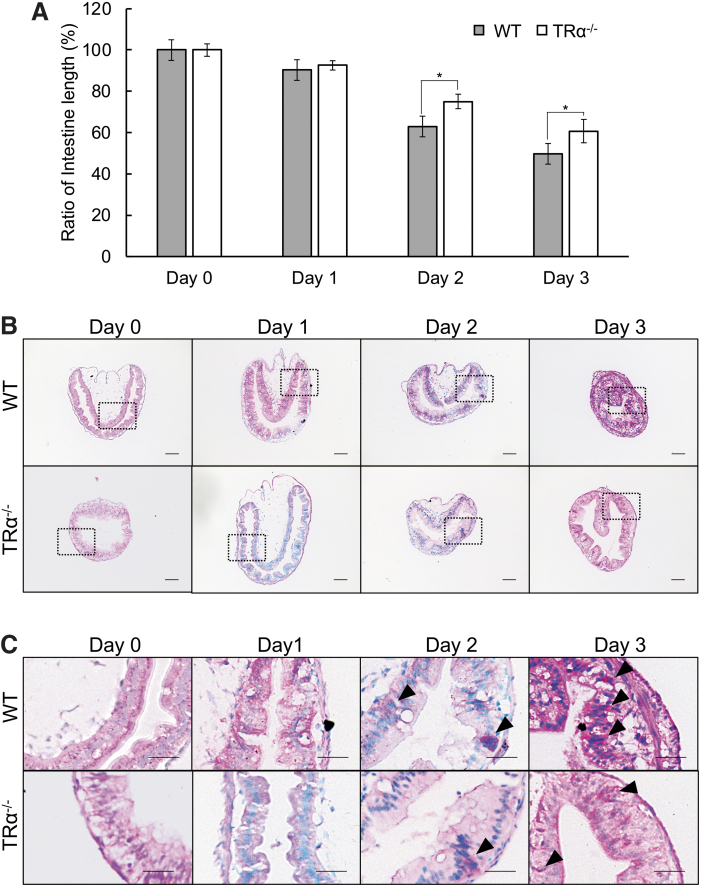
To investigate the role of TRα during intestinal remodeling, wild-type and TRα−/− tadpoles at stage 54 were treated with T3 for three days. Intestinal length is known to be reduced by as much as 80–90% during natural metamorphosis (27,28). Consistently, the relative intestinal length, normalized against body weight, was reduced in the T3-treated wild-type animals (Fig. 1A). The reduction in the relative intestinal length in TRα−/− tadpoles was significantly less than that of wild-type tadpoles after two or three days of T3 treatment (Fig. 1A and Supplementary Fig. S1A, B), suggesting reduced/delayed response to T3 treatment (note that during natural development, intestinal length gradually increases from stage 54 to stage 58, two to three weeks from stage 54, and then begins to shorten) (53,60). In addition, the intestine at stage 54 is made of mainly a monolayer of larval epithelial cells surrounded by little connective tissue and muscles, and the larval epithelium degenerates during metamorphosis and is replaced by a multiple-folded epithelium with elaborate connective tissue at the end of metamorphosis (27). We therefore investigated intestinal morphology during T3-induced metamorphosis by using MGPY staining, which stains DNA blue and RNA red (33,61). Since the stem cells have RNA-rich cytoplasm and form clusters that are stained strongly red, MGPY can easily detect proliferating adult intestinal stem cells during metamorphosis (27). After three days of T3 treatment, the intestinal epithelium in wild-type tadpoles started to form multiple folds with numerous clusters of proliferating stem cells (Fig. 1B, C). The intestinal epithelium in TRα−/− tadpoles changed little during T3 treatment, remaining largely a monolayer (Fig. 1B, C). Thus, TRα is required for the initiation of intestinal remodeling during T3-induced metamorphosis, consistent with the observed delayed in intestinal remodeling in TRα−/− tadpoles during natural metamorphosis (41,46).
FIG. 1.
TRα knockout inhibits intestinal remodeling during T3-induced metamorphosis. (A) The relative intestinal length shortens less in the TRα−/− tadpoles than the wild-type ones after two to three days of T3 treatment. Tadpoles at stage 54 were treated with 10 nM T3 for up to three days. Intestinal length was measured from bile duct junction to colon and normalized against body weight. The relative intestinal length was calculated as a % of that in the control animals (0 day) for each genotype, respectively. Each group included more than five tadpoles. Note that the animal weight or intestinal length change little within three days around stage 54 without T3 treatment (53). Each bar represents the mean + S.E. The asterisk (*) indicates a significant difference between TRα−/− tadpoles and wild-type tadpoles (p < 0.05). (B, C) The morphological changes induced by T3 are inhibited/delayed in the intestine of TRα−/− tadpoles. Cross sections of the intestine from TRα−/− and wild-type tadpoles treated with T3 were stained with MGPY, which stains DNA blue and RNA red (B) with the boxed regions shown at higher magnification in (C). The DNA was stained blue while the RNA was stained red. Note that there were more epithelial cells in clusters stained by MGPY after two to three days of T3 treatment of the wild-type animals (black arrowheads). The connective tissue in the wild-type intestine was also thicker (more developed), especially after three days of T3 treatment. Scale bar, 20 μm (B) and 10 μm (C). The experiment was repeated twice with similar results. MGPY, methyl green-pyronin Y; S.E., standard error; T3, triiodothyronine; WT, wild type.
TRα knockout inhibits T3-induced intestinal stem cell proliferation during metamorphosis in X. tropicalis
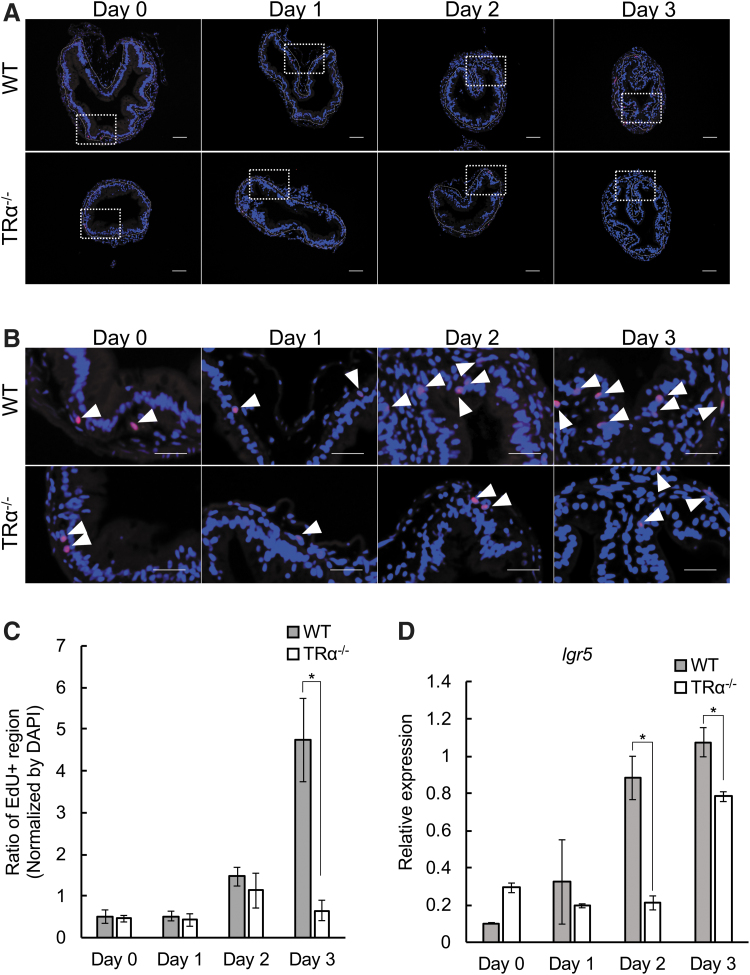
We have previously demonstrated that proliferating intestinal epithelial cells during T3-induced metamorphosis can be labeled with EdU and also express high levels of the adult intestinal stem cell marker Lgr-5 (33,62). To directly investigate whether TRα knockout affected epithelial stem cell proliferation, the tadpoles treated with or without T3 as above were injected with EdU 30 minutes before being sacrificed. Staining for EdU showed that cell proliferation gradually increased during T3 treatment in the wild-type intestine, but this increase was absent in TRα−/− tadpoles (Fig. 2A–C), indicating that TRα is important for intestinal stem cell proliferation during metamorphosis. Additionally, the expression of the stem cell marker gene lgr5 was increased, as expected, during this period in the wild-type tadpole intestine, while this upregulation was inhibited/delayed in TRα−/− tadpoles (Fig. 2D), supporting a role of TRα in regulating intestinal stem cell proliferation during metamorphosis.
FIG. 2.
TRα knockout inhibits T3-induced epithelial cell proliferation during metamorphosis. (A) Proliferating cells in the TRα−/− intestine after three days of T3 treatment are reduced compared with those in the wild-type intestine. The intestinal sections from stage 54 tadpoles treated with 10 nM T3 for up to three days were double-stained with EdU for cell proliferation (red) and DAPI for DNA (blue). The dotted white lines depict the epithelium–mesenchyme boundary. Scale bar indicates 20 μm. (B) The boxed regions in (A) are shown at a higher magnification. White arrowheads denote EdU-positive cells. Scale bar indicates 10 μm. (C) Quantification of cell proliferation as detected in (A, B). The EdU-positive areas (red) in the epithelium were quantified with the ImageJ software and normalized against the total cellular area in the epithelium as determined by DAPI staining. The asterisk (*) indicates a significant difference between the TRα−/− tadpoles and wild-type tadpoles (p < 0.05). Experiment was repeated twice, and each bar represents the mean + S.E. (D) The expression of the adult intestinal stem cell marker lgr5 is reduced in the TRα−/− intestine. Total intestinal RNA from wild-type and knockout stage 54 tadpoles treated with 10 nM T3 for up to 0–3 days was analyzed by RT-PCR for the expression of lgr5. Note that lgr5 was upregulated by T3 after two to three days of T3 treatment in wild-type animals, and this upregulation was delayed/reduced in the knockout tadpoles. DAPI, 4′,6-diamidino-2-phenylindole; EdU, 5-ethynyl-2′-deoxyuridine; RT-PCR, reverse transcription polymerase chain reaction.
TRα is required for T3-induced larval epithelial apoptosis during metamorphosis in X. tropicalis
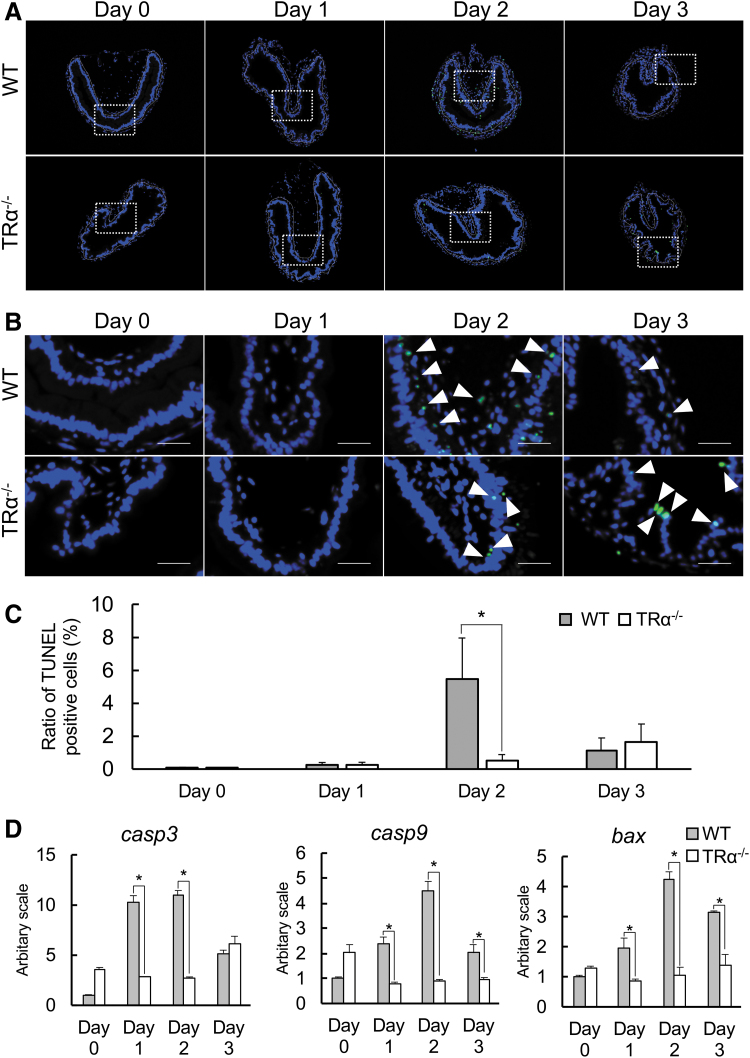
The vast majority of larval epithelial cells undergo apoptosis in response to rising T3 levels during intestinal metamorphosis (28,63). Thus, we next investigated whether knocking out TRα affected larval epithelial degeneration during T3-induced metamorphosis by using TUNEL labeling. As expected, little apoptotic signals were present in the epithelium of intestine in wild-type or TRα−/− tadpoles after 0 and 1 day of T3 treatment. After two days of T3 treatment, high levels of apoptotic signals were present only in the intestinal epithelium of wild-type but not knockout tadpoles (Fig. 3A–C). Cell death was largely complete after three days of treatment (Fig. 3C) as the epithelium was now mostly proliferating adult stem cells (Fig. 2). Interestingly, little cell death was detected in the knockout tadpoles even after three days of treatment (Fig. 3C), suggesting that TRα was essential for T3-induced cell death during the treatment. Consistently, when we investigated intestinal expression of apoptotic factors caspase3 (casp3), caspase9 (casp9), and Bcl-2-associated X protein (bax) (29), we observed that T3 induced the upregulation of these genes in the wild-type tadpoles, whereas the induction was abolished in TRα-knockout tadpoles (Fig. 3D).
FIG. 3.
T3-induced larval epithelial cell death is reduced in TRα−/− tadpoles. (A) Apoptotic cells in the TRα−/− intestine are reduced during T3-induced metamorphosis. The cross sections of the intestine from stage 54 tadpoles treated with 10 nM T3 for 0–3 days were double-stained with TUNEL for apoptotic cells and DAPI for DNA. The dotted white lines depict the epithelium–mesenchyme boundary. Scale bar indicates 20 μm. (B) The boxed regions in (A) are shown at a higher magnification. White arrowheads denote TUNEL-positive cells. Scale bar indicates 10 μm. (C) Quantification of apoptosis as detected in (A, B). The TUNEL-positive cells (green) in the epithelium were counted with the ImageJ software and normalized against the total cells in the epithelium as determined by DAPI staining. Note that apoptotic cell number reached the peak after two days of T3 treatment in wild-type tadpoles, while it was still low in the TRα−/− intestine even after T3 treatment for three days. The asterisk (*) indicates a significant difference between the TRα−/− tadpoles and wild-type tadpoles (p < 0.05). Experiment was repeated three times, and each bar represents the mean + S.E. (D) The mRNA levels of apoptosis-regulatory genes in the intestine are significantly induced by T3 treatment of wild-type but not TRα−/− premetamorphic tadpoles. Total intestinal RNA was isolated from wild-type and knockout stage 54 tadpoles treated with 10 nM T3 for up to 0–3 days. The mRNA levels of caspase3 (casp3), caspase9 (casp9), and Bcl-2-associated X protein (bax) were determined by real-time PCR and shown as relative levels to those in the intestine of wild-type animals at stage 54. Note that the wild-type intestine had much higher levels of casp3, casp9, and bax after one and two days of T3 treatment compared with the knockout one. The asterisk (*) indicates a significant difference between TRα−/− and wild-type tadpoles (p < 0.05). Experiment was repeated twice, and each bar represents the mean + S.E. TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling.
TRα is required for T3 regulation of genes in the cell cycle and extracellular matrix pathway during the early phase of intestinal remodeling
Having shown that TRα is required for T3-induced intestinal metamorphosis, we next investigated gene regulation network controlled by TRα by using RNA-seq analysis (Supplementary Fig. 2A). We focused on the early gene expression program, that is, after only 18 hours of T3 treatment when little morphological changes had occurred in the intestine (Fig. 1). We isolated total RNA from the intestine of wild-type and TRα−/− tadpoles at stages 52–54 treated with T3 for 18 hours and validated the success of the T3 treatment by analyzing the expression of known T3 target genes such as TRβ (thrb) and TH/bzip (thibz) (Supplementary Fig. S2B). The RNA was then subjected to sequencing on the Illumina HiSeq 2000 platform to identify differentially expressed genes. All sequencing data sets have been submitted to GEO under accession No. GSM4551285 to GSM4551296.
To increase the hit count for global gene expression analysis, the orthologs of human gene were used for analysis of GO terms and pathways as reported previously (58). To do so, we first evaluated the raw RNA-seq data against the genome databases NCBI, Ensembl, and Xenbase, and we found that Ensembl database yielded the most identifiable genes after conversion to human gene ID (Supplementary Fig. S2C).
To confirm the reliability of RNA-seq results, selected genes, including three genes upregulated by T3 in both the wild-type and TRα−/− intestine, and three genes upregulated by T3 only in the wild-type intestine, were analyzed by qRT-PCR on independent RNA samples. The expression patterns by qRT-PCR were consistent with the RNA-seq results (Supplementary Fig. S3A, B).
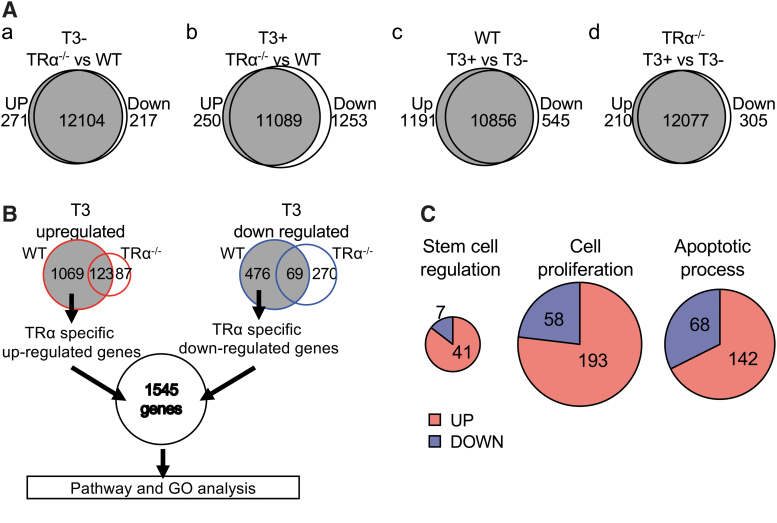
By comparing the global gene expression profiles between the wild-type and TRα−/− intestines from tadpoles with or without T3 treatment (Fig. 4A and Supplementary Table S2), we found that only 3.8% of genes were upregulated or downregulated in the intestine due to TRα knockout in the premetamorphic tadpoles at stage 54 without T3 treatment (Fig. 4A-a). However, ∼10% of genes had lower expression levels in the TRα−/− intestine compared with the wild-type ones after T3 treatment (Fig. 4A-b), suggesting that TRα−/− is important for mediating the effects of T3.
FIG. 4.
RNA-seq analysis reveals that the upregulation of most of the T3-induced genes in the WT intestine is blocked in the TRα−/− intestine. (A) Venn diagram analysis for genes upregulated or downregulated by T3 or TRα knockout (twofold or more and padj <0.02). (a) Two hundred seventy-one genes were upregulated and 217 genes downregulated in the TRα−/− intestine compared with the WT intestine at stage 54. (b) Two hundred fifty genes were upregulated and 1253 genes downregulated in the T3-treated TRα−/− intestine compared with the T3-treated WT intestine. (c) One thousand one hundred ninety-two genes were upregulated and 545 genes downregulated in the intestine by T3 treatment of WT tadpoles. (d) Two hundred ten genes were upregulated and 305 genes downregulated in the intestine by T3 treatment of TRα−/− tadpoles. (B) Venn diagram comparison of T3 upregulated (left) and downregulated (right) genes in the WT and TRα−/− intestine reveals 1545 genes regulated by T3 treatment of WT but not TRα−/− tadpoles (i.e., TRα target genes). Left: 1069 genes were upregulated by T3 only in the WT intestine, 87 genes were upregulated by T3 only in the TRα−/− intestine, and 123 genes were upregulated in both the WT intestine and TRα−/− intestine. Right: 476 genes were downregulated by T3 only in the WT intestine, 270 genes were downregulated by T3 only in the TRα−/− intestine, and 69 genes were downregulated in both the WT intestine and TRα−/− intestine. TRα-specific upregulated (1069) and downregulated (476) genes were combined as TRα-specific T3-regulated genes and subjected to GO and pathway analysis. (C) Three of the significantly enriched GO terms among the TRα-specific T3-regulated genes: stem cell regulation, cell proliferation, and apoptotic process. Note that most of the regulated genes in these GO terms (41, 193, and 142 genes, respectively) were upregulated by T3 in the WT intestine. GO, gene ontology.
When we compared the gene expression profiles of the genes in the intestine of tadpoles with or without T3 treatment, we identified 1192 genes upregulated and 545 downregulated by at least twofolds or more after T3 treatment in the intestine of wild-type tadpoles (Fig. 4A-c). In the TRα-knockout animals, T3 treatment only upregulated 210 genes and downregulated 305 genes (Fig. 4A-d), indicating that the knockout had a much broader effect on T3 upregulated genes than the downregulated ones.
Of the T3-upregulated genes in the wild-type intestine, 123 genes were also upregulated in the TRα−/− intestine, indicating that TRα was required for the upregulation of the other 1069 genes in the wild-type intestine (Fig. 4B). Among the downregulated genes, 475 were found to require TRα (Fig. 4B). Together, we thus found that 1545 genes were regulated by TRα in the wild-type intestine (Fig. 4B and Supplementary Table S3). Among them, we found 48, 251, and 210 genes belonging to GO terms “stem cell regulation,” “cell proliferation,” and “apoptotic process,” respectively (Fig. 4C and Supplementary Table S4), most of which were upregulated by TRα (Fig. 4C). These are consistent with the major events, larval epithelial cell death and adult epithelial proliferation that take place during intestinal metamorphosis.
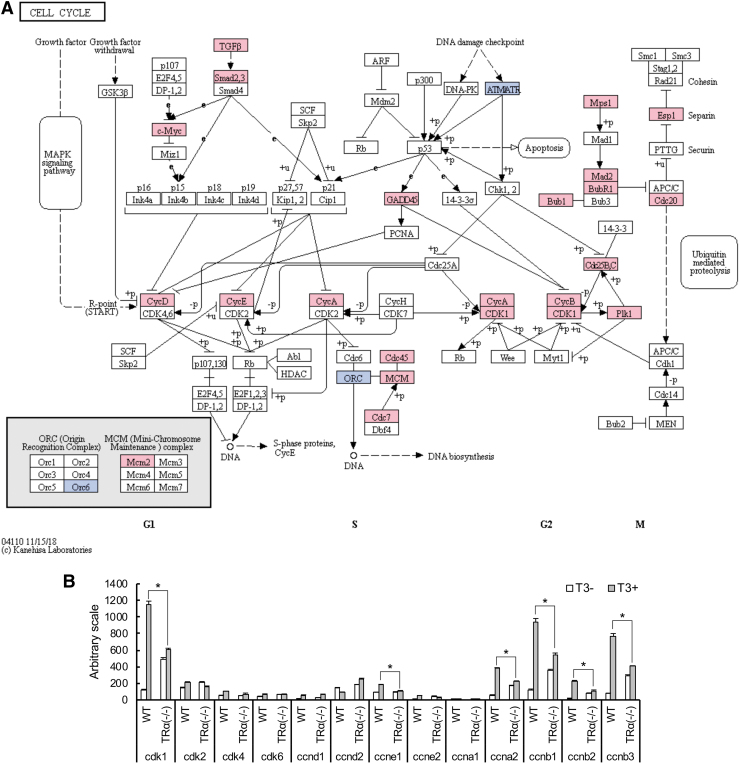
GO analyses of 1545 genes regulated by TRα revealed significant enrichment for genes in the translation, cell division and extracellular matrix (ECM)-related GO terms (Table 1 and Supplementary Table S5). Similarly, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses showed enrichment of pathways such as those related to ECM–receptor interaction and cell cycle (Table 2). Interestingly, under both analyses, “stem cell regulation” and “apoptotic process” were not among the significantly enriched GO terms or KEGG pathways (Tables 1 and 2 and Supplementary Table S5). This likely reflects the fact that apoptosis and stem cell formation/proliferation take place much later, only after two to three days of T3 treatment. Thus, many of the genes involved in these processes were not affected after the 18 hour T3 treatment used for the RNA-seq analysis. Surprisingly, many cell cycle-related GO terms were among the top significantly enriched GO terms (Table 1 and Supplementary Table S5). Similarly, cell cycle pathway was among the enriched KEGG pathways (Table 2). Of interest, in the cell cycle pathway, 23 genes were upregulated while only 3 genes were downregulated by T3 among the TRα-regulated genes (Fig. 5A). In addition, it is known that cell cycle progression is mainly regulated by cyclin-dependent kinases (CDKs) and cyclins. We observed that cdk1 (cdk1), cyclin E1 (ccne1), cyclin E2 (ccne2), cyclin A2 (ccna2), cyclin B1 (ccnb1), cyclin B2 (ccnb2), and cyclin B3 (ccnb3) were upregulated by more than twofold in the wild-type intestine but not in the knockout tadpole intestine after T3 treatment (Fig. 5B). Thus, an important role of TRα is to activate cell cycle genes to promote cell cycle progression during intestinal metamorphosis.
Table 1.
Top Significantly Enriched Gene Ontology Terms Among Genes Regulated by TRα
| No. | GO ID | Term | Count | p | FDR |
|---|---|---|---|---|---|
| 1 | GO:6614 | SRP-dependent cotranslational protein targeting to membrane | 45 | <0.001 | <0.001 |
| 2 | GO:19083 | Viral transcription | 47 | <0.001 | <0.001 |
| 3 | GO:6413 | Translational initiation | 51 | <0.001 | <0.001 |
| 4 | GO:184 | Nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 45 | <0.001 | <0.001 |
| 5 | GO:6412 | Translation | 60 | <0.001 | <0.001 |
| 6 | GO:6364 | rRNA processing | 53 | <0.001 | <0.001 |
| 7 | GO:51301 | Cell division | 71 | <0.001 | <0.001 |
| 8 | GO:9813 | Flavonoid biosynthetic process | 14 | <0.001 | <0.001 |
| 9 | GO:7067 | Mitotic nuclear division | 54 | <0.001 | <0.001 |
| 10 | GO:30574 | Collagen catabolic process | 24 | <0.001 | <0.001 |
| 11 | GO:7155 | Cell adhesion | 81 | <0.001 | <0.001 |
| 12 | GO:52696 | Flavonoid glucuronidation | 14 | <0.001 | <0.001 |
| 13 | GO:7062 | Sister chromatid cohesion | 30 | <0.001 | <0.001 |
| 14 | GO:2227 | Innate immune response in mucosa | 14 | <0.001 | <0.001 |
| 15 | GO:30198 | Extracellular matrix organization | 41 | <0.001 | <0.001 |
| 16 | GO:30199 | Collagen fibril organization | 16 | <0.001 | <0.001 |
| 17 | GO:52695 | Cellular glucuronidation | 10 | <0.001 | 0.004 |
| 18 | GO:2181 | Cytoplasmic translation | 12 | <0.001 | 0.006 |
| 19 | GO:19731 | Antibacterial humoral response | 15 | <0.001 | 0.028 |
| 20 | GO:19433 | Triglyceride catabolic process | 11 | <0.001 | 0.050 |
FDR, false discovery rate; GO, Gene Ontology.
Table 2.
Significantly Enriched Kyoto Encyclopedia of Genes and Genomes Pathways Among Genes Regulated by TRα
| No. | GO ID | Term | Count | p | Benjamini | FDR |
|---|---|---|---|---|---|---|
| 1 | hsa03010 | Ribosome | 50 | <0.001 | <0.001 | <0.001 |
| 2 | hsa04974 | Protein digestion and absorption | 32 | <0.001 | <0.001 | <0.001 |
| 3 | hsa04512 | ECM–receptor interaction | 28 | <0.001 | <0.001 | <0.001 |
| 4 | hsa00053 | Ascorbate and aldarate metabolism | 15 | <0.001 | <0.001 | <0.001 |
| 5 | hsa00830 | Retinol metabolism | 23 | <0.001 | <0.001 | <0.001 |
| 6 | hsa00983 | Drug metabolism—other enzymes | 19 | <0.001 | <0.001 | <0.001 |
| 7 | hsa00040 | Pentose and glucuronate interconversions | 16 | <0.001 | <0.001 | <0.001 |
| 8 | hsa00860 | Porphyrin and chlorophyll metabolism | 17 | <0.001 | <0.001 | <0.001 |
| 9 | hsa00140 | Steroid hormone biosynthesis | 20 | <0.001 | <0.001 | 0.001 |
| 10 | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 22 | <0.001 | <0.001 | 0.004 |
| 11 | hsa05204 | Chemical carcinogenesis | 22 | <0.001 | <0.001 | 0.016 |
| 12 | hsa00982 | Drug metabolism—cytochrome P450 | 19 | <0.001 | 0.001 | 0.060 |
| 13 | hsa05322 | Systemic lupus erythematosus | 29 | <0.001 | 0.001 | 0.066 |
| 14 | hsa04510 | Focal adhesion | 36 | <0.001 | 0.010 | 0.642 |
| 15 | hsa04110 | Cell cycle | 25 | <0.001 | 0.010 | 0.742 |
| 16 | hsa04972 | Pancreatic secretion | 18 | 0.006 | 0.103 | 8.049 |
| 17 | hsa00790 | Folate biosynthesis | 6 | 0.008 | 0.115 | 9.490 |
| 18 | hsa00601 | Glycosphingolipid biosynthesis—lacto and neolacto series | 8 | 0.009 | 0.131 | 11.441 |
| 19 | hsa04978 | Mineral absorption | 11 | 0.010 | 0.136 | 12.522 |
| 20 | hsa05203 | Viral carcinogenesis | 31 | 0.011 | 0.146 | 14.051 |
| 21 | hsa01100 | Metabolic pathways | 140 | 0.013 | 0.154 | 15.582 |
| 22 | hsa05146 | Amoebiasis | 18 | 0.022 | 0.246 | 25.829 |
| 23 | hsa05222 | Small cell lung cancer | 15 | 0.029 | 0.300 | 32.598 |
| 24 | hsa05150 | Staphylococcus aureus infection | 11 | 0.030 | 0.292 | 32.836 |
| 25 | hsa04914 | Progesterone-mediated oocyte maturation | 15 | 0.035 | 0.325 | 37.653 |
| 26 | hsa00100 | Steroid biosynthesis | 6 | 0.036 | 0.322 | 38.451 |
| 27 | hsa04973 | Carbohydrate digestion and absorption | 9 | 0.042 | 0.356 | 43.555 |
ECM, extracellular matrix.
FIG. 5.
Suppression of cell cycle gene expression during intestinal remodeling in TR−/− tadpoles. (A) Mapping of TRα-specific T3-regulated genes to cell cycle pathway (KEGG). Genes regulated by T3 for at least 2.0-fold, after 18 hours of T3 treatment of wild-type but not TRα−/− tadpoles were mapped onto the KEGG pathway for the cell cycle. Pink boxes indicate upregulation, and blue boxes indicate downregulation. Note that most of regulated genes were upregulated by T3. (B) Bar graph representation of the expression of cell cycle genes from RNA-seq. Note that cdk1 (cdk1), cyclin A2 (ccna2), cyclin B1 (ccnb1), cyclin B2 (ccnb2), and cyclin B3 (ccnb3) were upregulated by T3 only in the wild-type intestine. The asterisk (*) indicates a significant difference between the TRα−/− and wild type (FDR <0.05). FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes.
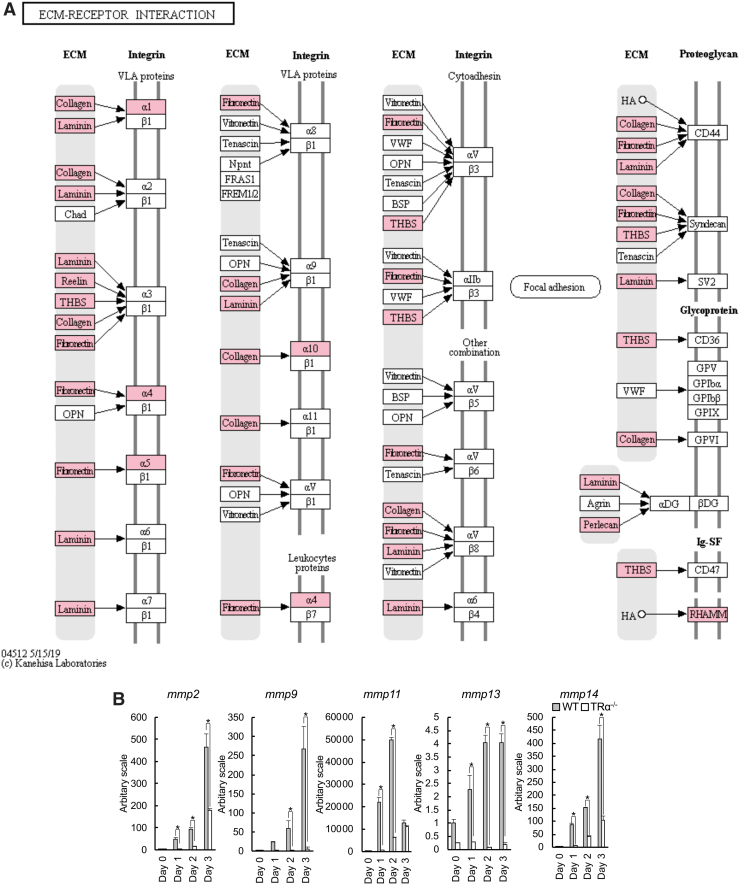
Numerous earlier studies suggest that ECM remodeling by matrix metalloproteinases (MMPs) plays a role in intestinal remodeling, especially in apoptosis of larval epithelial cells (30,64–69). Thus, perhaps not surprisingly, many genes in the ECM–receptor interaction pathway were upregulated by T3 only in the wild-type intestine but not in the TRα-knockout intestine (Fig. 6A). In particular, the expression of many MMPs such as mmp2, mmp9, mmp11, mmp13, and mmp14 was strongly induced by T3 after one day of T3 treatment and increased further after two to three days of T3 treatment of wild-type tadpoles. In TRα−/− tadpoles, their expression was not upregulated or only slightly upregulated even after three days of T3 treatment (Fig. 6B). Thus, TRα is likely critical for T3-induced larval epithelial cell death by activating ECM remodeling via MMPs.
FIG. 6.
TRα knockout causes reduction in mmp expression during intestinal remodeling. (A) Mapping of TRα-specific T3-regulated genes to the ECM–receptor interaction pathway (KEGG). Genes regulated by T3 for at least twofold after one day of T3 treatment of wild-type but not TRα−/− tadpole were mapped onto the KEGG pathway for the ECM–receptor interaction. Pink boxes indicate upregulation, and blue boxes indicate downregulation. Note that all mapped genes were upregulated by T3. (B) The mRNA level of mmp genes in the intestine is significantly increased during T3-induced metamorphosis in wild-type tadpoles but not in TRα−/− tadpoles. The mRNA levels of mmp2, mmp9, mmp11, mmp13, and mmp14 were determined by real-time PCR and shown relative to the levels in the wild-type intestine at stage 54. The wild-type intestine had much higher levels of expression of mmp2, mmp9, mmp11, mmp13, and mmp14 after one and two days of T3 treatment. The asterisk (*) indicates a significant difference between the TRα−/− tadpoles and wild-type tadpoles (p < 0.05). Experiment was repeated twice, and each bar represents the mean + S.E. ECM, extracellular matrix.
Discussion
Due to maternal dependence of mammalian embryos and neonates, it is difficult to study how T3 regulates the maturation of adult organs such as the intestine during postembryonic development in mammals. Making use of the ability to manipulate amphibian metamorphosis and taking advantages of the recent advances in genome-wide and genetic analyses of the diploid anuran X. tropicalis, we have here characterized for the first time the global gene expression changes that are controlled by TRα during early phase of intestinal metamorphosis, a model of postembryonic adult organ development in mammals. Our results have revealed that in the early phase of T3-induced metamorphosis, TRα is critical for gene regulation by T3, particularly for the T3-induced genes, in the intestine, similar to the findings on whole tadpoles (45,46). In addition, consistent with the possibility that many of the genes regulated by T3 are likely direct TR target genes, TRα knockout leads to increased levels of many of these genes at stage 54 compared with those in wild-type animals, likely due to the derepression caused by the loss of unliganded TRα at this stage. More importantly and surprisingly, we have discovered that T3 induces cell proliferation program early during intestinal metamorphosis, well before significant proliferation of the adult epithelial cells, suggesting that activation of the cell cycle program may be involved in T3-induced larval epithelial cell degeneration.
Intestinal remodeling in X. tropicalis involves apoptotic degeneration of vast majority of the larval epithelial cells and de novo formation of the adult intestinal epithelial stem cells through dedifferentiation of a small fraction of larval epithelial cells in a process controlled by T3 (28,70,71). We and others have previously demonstrated that TRα knockout alters the timing for the initiation of metamorphosis as well as the rate of metamorphic progression (41,42,44–49). Interestingly, there is also a delay in intestinal remodeling relative to the external morphological changes of the tadpole during natural metamorphosis, implicating a role of TRα in the temporal coordination of metamorphosis of various organs/tissues (41,44,46). Our studies here indicate that TRα knockout inhibits T3-induced intestinal remodeling, including drastically reduced apoptosis and adult stem cell proliferation after extended T3 treatment, demonstrating a critical role of TRα in mediating the effects of T3 during intestinal metamorphosis.
Our RNA-seq analysis of the intestine from wild-type or TRα-knockout tadpoles treated with or without T3 for 18 hours identified 1192 upregulated genes in the intestine during early phase of intestinal metamorphosis. Importantly, the vast majority of these genes (1069 genes) were not regulated by T3 in the TRα-knockout tadpole intestine. Similarly, of the 545 genes downregulated by T3 in the wild-type intestine, most were not regulated by T3 in the TRα-knockout tadpole intestine. These results demonstrate a predominant role of TRα during this early phase (the first day of T3 treatment) of intestinal metamorphosis.
Bioinformatics analyses of the genes that were regulated by T3 in the wild-type intestine but not in the TRα-knockout tadpole intestine revealed that these TRα-regulated genes were likely important for the earlier changes during intestinal metamorphosis. In particularly, the degeneration of the larval epithelium via apoptosis is perhaps the earliest and most dramatic observable morphological change during intestinal remodeling (28,33,72), with cell death easily detectable after two days of T3 treatment. The adult epithelial stem cells, which exist as clusters of proliferating cells at the climax of metamorphosis, are detected mainly after three days of T3 treatment (72). Consistently, we observed that there were more than 200 TRα-regulated apoptosis-related genes after one day of T3 treatment, with the vast majority induced by T3. Similarly, the vast majority of the stem cell-related genes were induced by T3, although only less than 50 such genes were regulated by TRα after one day of T3 treatment, likely due to the fact that stem cell formation occurs much later and thus fewer such genes are regulated early during intestinal metamorphosis.
GO and pathway analyses revealed that among the TRα-regulated genes, ECM-related GO terms and pathways were highly enriched. It is well known that ECM and its remodeling by MMPs are critical for cell fate determination (73–75). Our earlier studies have shown that ECM and MMPs affect intestinal epithelial cell fate in response to T3 (30,64–69). Our RNA-seq data here showed that numerous MMP genes, which are major components of the ECM-related GO terms and pathways, were upregulated by T3 within one day of treatment, suggesting that ECM remodeling by MMPs is a critical early step in T3-induced epithelial cell fate changes, mainly larval epithelial cell death.
Concurrently with larval epithelial cell death, adult stem cells are formed de novo, and by the climax of metamorphosis when most of the larval epithelial cells have undergone apoptosis, the proliferating adult epithelial stem cells are the predominant cells in the epithelium (28,63). Consistently, our earlier microarray analyses of gene expression changes after extended T3 treatment or during natural metamorphosis have also found that GO terms and pathways related to ECM remodeling and cell death are enriched among the regulated genes (76,77). These earlier studies have also revealed the enrichment of GO terms and pathways related to cell cycle and cell proliferation (76,77), which are likely responsible for the cell proliferation observed at the climax of metamorphosis. Consistently, the protein translation GO terms were also highly enriched among the genes during early T3-induced intestinal remodeling but not among the genes regulated near the end of metamorphosis (77), suggesting that protein translation is important for early phase of intestinal remodeling.
We found that GO terms and pathways related to cell cycle and cell proliferation were highly enriched even among the TRα-regulated genes within one day of T3 treatment. This is well before significant cell proliferation during intestinal remodeling as shown in Figures 2 and 3, high levels of cell proliferation occurred only after three days of T3 treatment, after the peak of apoptotic activity at day 2 of T3 treatment. Furthermore, most of the over 250 TRα-regulated proliferation-related genes were induced by T3. It is well known that in the premetamorphic tadpole intestine, there are little connective tissue and muscles. The predominant cell type is the larval epithelial cell. Thus, for genes that are expressed in different cell types, RNA-seq reveals changes mainly in the larval epithelium, although genes highly or exclusively expressed in other tissues, such as mmp11 in the connective tissue, can also be found. In addition, most proliferating cells at the climax of intestinal metamorphosis are in the epithelium (33). Thus, the activation of the cell cycle program was most likely in the intestinal larval epithelium. Considering the fact that more than 90% of the larval intestinal epithelial cells die by apoptosis yet there is a prominent signal of proliferation genes at 18 hours of T3 treatment, this finding suggests an interesting model for T3 action during intestinal metamorphosis. That is, T3 activates the cell cycle/proliferation program in the larval epithelial cells via TRα. This, together with other signaling processes induced by T3, such as ECM-remodeling by MMPs, in turn helps the differentiated cells to choose two alternative fates: apoptosis or dedifferentiation into adult stem cells. Clearly, further studies are needed to test the interesting hypothesis that activation of the cell cycle/proliferation program in the larval epithelium is a prerequisite for larval cell death and adult stem cell development during T3-dependent intestinal metamorphosis.
Supplementary Material
Acknowledgments
We thank Nga Luu for experimental help and laboratory management and the team in the Molecular Genomics Core, NICHD, NIH, for RNA sequencing.
Authors' Contributions
Y.T. and Y.-B.S. designed the research. Y.S. and Y.T. generated the knockout animals and extracted RNA samples. H.Z. assisted with the bioinformatics. All authors participated in the article preparation and approved the final version of the article.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by the Intramural Research Program of NICHD, NIH. Yuta Tanizaki and Yuki Shibata were supported in part by Japan Society for the Promotion of Science (NIH) Fellowship.
Supplementary Material
References
- 1. Sirakov M, Plateroti M. 2011. The thyroid hormones and their nuclear receptors in the gut: from developmental biology to cancer. Biochim Biophys Acta 1812:938–946 [DOI] [PubMed] [Google Scholar]
- 2. Porterfield SP, Hendrich CE. 1993. The role of thyroid hormones in prenatal and neonatal neurological development—current perspectives. Endocr Rev 14:94–106 [DOI] [PubMed] [Google Scholar]
- 3. Flamant F, Samarut J. 2003. Thyroid hormone receptors: lessons from knockout and knock-in mutant mice. Trends Endocrinol Metab 14:85–90 [DOI] [PubMed] [Google Scholar]
- 4. Laudet V, Gronemeyer H. 2002. The Nuclear Receptor FactsBook. Academic Press, San Diego [Google Scholar]
- 5. Yen PM. 2001. Physiological and molecular basis of thyroid hormone action. Physiol Rev 81:1097–1142 [DOI] [PubMed] [Google Scholar]
- 6. Buchholz DR, Paul BD, Fu L, Shi YB. 2006. Molecular and developmental analyses of thyroid hormone receptor function in Xenopus laevis, the African clawed frog. Gen Comp Endocrinol 145:1–19 [DOI] [PubMed] [Google Scholar]
- 7. Shi YB, Matsuura K, Fujimoto K, Wen L, Fu L. 2012. Thyroid hormone receptor actions on transcription in amphibia: the roles of histone modification and chromatin disruption. Cell Biosci 2:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hsia SC, Bedadala GR, Balish MD. 2011. Effects of thyroid hormone on HSV-1 gene regulation: implications in the control of viral latency and reactivation. Cell Biosci 1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sato Y, Buchholz DR, Paul BD, Shi Y-B. 2007. A role of unliganded thyroid hormone receptor in postembryonic development in Xenopus laevis. Mech Dev 124:476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paul BD, Fu L, Buchholz DR, Shi Y-B. 2005. Coactivator recruitment is essential for liganded thyroid hormone receptor to initiate amphibian metamorphosis. Mol Cell Biol 25:5712–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burke LJ, Baniahmad A. 2000. Co-repressors 2000. FASEB J 14:1876–1888 [DOI] [PubMed] [Google Scholar]
- 12. Jones PL, Shi Y-B. 2003 N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors. In: Workman JL (ed) Current Topics in Microbiology and Immunology: Protein Complexes that Modify Chromatin. Vol. 274. Springer-Verlag, Berlin, pp. 237–268 [DOI] [PubMed] [Google Scholar]
- 13. Zhang J, Lazar MA. 2000. The mechanism of action of thyroid hormones. Annu Rev Physiol 62:439–466 [DOI] [PubMed] [Google Scholar]
- 14. Jones PL, Sachs LM, Rouse N, Wade PA, Shi YB. 2001. Multiple N-CoR complexes contain distinct histone deacetylases. J Biol Chem 276:8807–8811 [DOI] [PubMed] [Google Scholar]
- 15. McKenna NJ, O'Malley BW. 2001. Nuclear receptors, coregulators, ligands, and selective receptor modulators: making sense of the patchwork quilt. Ann N Y Acad Sci 949:3–5 [DOI] [PubMed] [Google Scholar]
- 16. Wong J, Shi YB, Wolffe AP. 1995. A role for nucleosome assembly in both silencing and activation of the Xenopus TR beta A gene by the thyroid hormone receptor. Genes Dev 9:2696–2711 [DOI] [PubMed] [Google Scholar]
- 17. Lazar MA. 1993. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev 14:184–193 [DOI] [PubMed] [Google Scholar]
- 18. Shi Y-B. 1999. Amphibian Metamorphosis: From Morphology to Molecular Biology. John Wiley & Sons, Inc., New York [Google Scholar]
- 19. Evans RM. 1988. The steroid and thyroid hormone receptor superfamily. Science 240:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai MJ, O'Malley BW. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Ann Rev Biochem 63:451–486 [DOI] [PubMed] [Google Scholar]
- 21. Jones I, Srinivas M, Ng L, Forrest D. 2003. The thyroid hormone receptor beta gene: structure and functions in the brain and sensory systems. Thyroid 13:1057–1068 [DOI] [PubMed] [Google Scholar]
- 22. Hadj-Sahraoui N, Seugnet I, Ghorbel MT, Demeneix B. 2000. Hypothyroidism prolongs mitotic activity in the post-natal mouse brain. Neurosci Lett 280:79–82 [DOI] [PubMed] [Google Scholar]
- 23. White P, Burton KA, Fowden AL, Dauncey MJ. 2001. Developmental expression analysis of thyroid hormone receptor isoforms reveals new insights into their essential functions in cardiac and skeletal muscles. FASEB J 15:1367–1376 [DOI] [PubMed] [Google Scholar]
- 24. Tata JR. 1993. Gene expression during metamorphosis: an ideal model for post-embryonic development. Bioessays 15:239–248 [DOI] [PubMed] [Google Scholar]
- 25. Brown DD, Cai L. 2007. Amphibian metamorphosis. Dev Biol 306:20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Denver RJ. 2013. Neuroendocrinology of amphibian metamorphosis. Curr Top Dev Biol 103:195–227 [DOI] [PubMed] [Google Scholar]
- 27. Sterling J, Fu L, Matsuura K, Shi Y-B. 2012. Cytological and morphological analyses reveal distinct features of intestinal development during Xenopus tropicalis metamorphosis. PLoS One 7:e47407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi Y-B, Ishizuya-Oka A. 1996. Biphasic intestinal development in amphibians: embryogensis and remodeling during metamorphosis. Curr Topics Dev Biol 32:205–235 [DOI] [PubMed] [Google Scholar]
- 29. Ishizuya-Oka A, Hasebe T, Shi YB. 2010. Apoptosis in amphibian organs during metamorphosis. Apoptosis 15:350–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Su Y, Shi Y, Stolow M, Shi Y-B. 1997. Thyroid hormone induces apoptosis in primary cell cultures of tadpole intestine: cell type specificity and effects of extracellular matrix. J Cell Biol 139:1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ishizuya-Oka A, Ueda S, Inokuchi T, Amano T, Damjanovski S, Stolow M, Shi Y-B. 2001. Thyroid hormone-induced expression of Sonic hedgehog correlates with adult epithelial development during remodeling of the Xenopus stomach and intestine. Differentiation 69:27–37 [DOI] [PubMed] [Google Scholar]
- 32. Ishizuya-Oka A, Shi YB. 2011. Evolutionary insights into postembryonic development of adult intestinal stem cells. Cell Biosci 1:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okada M, Wen L, Miller TC, Su D, Shi YB. 2015. Molecular and cytological analyses reveal distinct transformations of intestinal epithelial cells during Xenopus metamorphosis. Cell Biosci 5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun G, Shi Y-B. 2012. Thyroid hormone regulation of adult intestinal stem cell development: mechanisms and evolutionary conservations. Int J Biol Sci 8:1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shi YB. 2013. Unliganded thyroid hormone receptor regulates metamorphic timing via the recruitment of histone deacetylase complexes. Curr Top Dev Biol 105:275–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morreale de Escobar G, Calvo R, Escobar del Rey F, Obregon MJ. 1994. Thyroid hormones in tissues from fetal and adult rats. Endocrinology 134:2410–2415 [DOI] [PubMed] [Google Scholar]
- 37. Nagasawa T, Suzuki S, Takeda T, DeGroot LJ. 1997. Thyroid hormone receptor beta 1 expression in developing mouse limbs and face. Endocrinology 138:1276–1281 [DOI] [PubMed] [Google Scholar]
- 38. Yaoita Y, Brown DD. 1990. A correlation of thyroid hormone receptor gene expression with amphibian metamorphosis. Genes Dev 4:1917–1924 [DOI] [PubMed] [Google Scholar]
- 39. Wong J, Shi Y-B. 1995. Coordinated regulation of and transcriptional activation by Xenopus thyroid hormone and retinoid X receptors. J Biol Chem 270:18479–18483 [DOI] [PubMed] [Google Scholar]
- 40. Wang X, Matsuda H, Shi Y-B. 2008. Developmental regulation and function of thyroid hormone receptors and 9-cis retinoic acid receptors during Xenopus tropicalis metamorphosis. Endocrinology 149:5610–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi J, Ishizuya-Oka A, Buchholz DR. 2017. Growth, development, and intestinal remodeling occurs in the absence of thyroid hormone receptor alpha in tadpoles of Xenopus tropicalis. Endocrinology 158:1623–1633 [DOI] [PubMed] [Google Scholar]
- 42. Choi J, Suzuki KI, Sakuma T, Shewade L, Yamamoto T, Buchholz DR. 2015. Unliganded thyroid hormone receptor alpha regulates developmental timing via gene repression as revealed by gene disruption in Xenopus tropicalis. Endocrinology 156:735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sakane Y, Iida M, Hasebe T, Fujii S, Buchholz DR, Ishizuya-Oka A, Yamamoto T, Suzuki KT. 2018. Functional analysis of thyroid hormone receptor beta in Xenopus tropicalis founders using CRISPR-Cas. Biol Open 7:bio030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakajima K, Tazawa I, Yaoita Y. 2018. Thyroid hormone receptor alpha- and beta-knockout Xenopus tropicalis tadpoles reveal subtype-specific roles during development. Endocrinology 159:733–743 [DOI] [PubMed] [Google Scholar]
- 45. Wen L, Shi YB. 2015. Unliganded thyroid hormone receptor alpha controls developmental timing in Xenopus tropicalis. Endocrinology 156:721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wen L, Shibata Y, Su D, Fu L, Luu N, Shi Y-B. 2017. Thyroid hormone receptor α controls developmental timing and regulates the rate and coordination of tissue specific metamorphosis in Xenopus tropicalis. Endocrinology 158:1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wen L, Shi YB. 2016. Regulation of growth rate and developmental timing by Xenopus thyroid hormone receptor alpha. Dev Growth Differ 58:106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sachs LM. 2015. Unliganded thyroid hormone receptor function: amphibian metamorphosis got TALENs. Endocrinology 156:409–410 [DOI] [PubMed] [Google Scholar]
- 49. Yen PM. 2015. Unliganded TRs regulate growth and developmental timing during early embryogenesis: evidence for a dual function mechanism of TR action. Cell Biosci 5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nakajima K, Tazawa I, Shi YB. 2019. A unique role of thyroid hormone receptor beta in regulating notochord resorption during Xenopus metamorphosis. Gen Comp Endocrinol 277:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shibata Y, Tanizaki Y, Shi YB. 2020. Thyroid hormone receptor beta is critical for intestinal remodeling during Xenopus tropicalis metamorphosis. Cell Biosci 10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shibata Y, Wen L, Okada M, Shi YB. 2020. Organ-specific requirements for thyroid hormone receptor ensure temporal coordination of tissue-specific transformations and completion of Xenopus metamorphosis. Thyroid 30:300–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nieuwkoop PD, Faber J. 1965. Normal Table of Xenopus laevis. North Holland Publishing, Amsterdam [Google Scholar]
- 54. Okada M, Shi YB. 2017. Cell proliferation analysis during Xenopus metamorphosis: using 5-ethynyl-2-deoxyuridine (EdU) to stain proliferating intestinal cells. Cold Spring Harb Protoc 2017:pdb prot097717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi Y-B, Liang VC-T. 1994. Cloning and characterization of the ribosomal protein L8 gene from Xenopus laevis. Biochim Biophys Acta 1217:227–228 [DOI] [PubMed] [Google Scholar]
- 56. Huang da W, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 58. Nagasawa K, Tanizaki Y, Okui T, Watarai A, Ueda S, Kato T. 2013. Significant modulation of the hepatic proteome induced by exposure to low temperature in Xenopus laevis. Biol Open 2:1057–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dennis GJ, Sherman BT, Hosack DA, Yang H, Gao W, Lane HC, Lempicki RA. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:R60. [PubMed] [Google Scholar]
- 60. Schreiber AM, Cai L, Brown DD. 2005. Remodeling of the intestine during metamorphosis of Xenopus laevis. Proc Natl Acad Sci U S A 102:3720–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ishizuya-Oka A, Ueda S. 1996. Apoptosis and cell proliferation in the Xenopus small intestine during metamorphosis. Cell Tissue Res 286:467–476 [DOI] [PubMed] [Google Scholar]
- 62. Sun G, Hasebe T, Fujimoto K, Lu R, Fu L, Matsuda H, Kajita M, Ishizuya-Oka A, Shi YB. 2010. Spatio-temporal expression profile of stem cell-associated gene LGR5 in the intestine during thyroid hormone-dependent metamorphosis in Xenopus laevis. PLoS One 5:e13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shi Y-B, Ishizuya-Oka A. 2001. Thyroid hormone regulation of apoptotic tissue remodeling: implications from molecular analysis of amphibian metamorphosis. Prog Nucleic Acid Res Mol Biol 65:53–100 [DOI] [PubMed] [Google Scholar]
- 64. Ishizuya-Oka A, Ueda S, Shi Y-B. 1996. Transient expression of stromelysin-3 mRNA in the amphibian small intestine during metamorphosis. Cell Tissue Res 283:325–329 [DOI] [PubMed] [Google Scholar]
- 65. Fu L, Ishizuya-Oka A, Buchholz DR, Amano T, Matsuda H, Shi YB. 2005. A causative role of stromelysin-3 in extracellular matrix remodeling and epithelial apoptosis during intestinal metamorphosis in Xenopus laevis. J Biol Chem 280:27856–27865 [DOI] [PubMed] [Google Scholar]
- 66. Amano T, Kwak O, Fu L, Marshak A, Shi Y-B. 2005. The matrix metalloproteinase stromelysin-3 cleaves laminin receptor at two distinct sites between the transmembrane domain and laminin binding sequence within the extracellular domain. Cell Res 15:150–159 [DOI] [PubMed] [Google Scholar]
- 67. Amano T, Fu L, Marshak A, Kwak O, Shi YB. 2005. Spatio-temporal regulation and cleavage by matrix metalloproteinase stromelysin-3 implicate a role for laminin receptor in intestinal remodeling during Xenopus laevis metamorphosis. Dev Dyn 234:190–200 [DOI] [PubMed] [Google Scholar]
- 68. Hasebe T, Fu L, Amano T, Shi Y-B. 2007. Evidence for a cooperative role of gelatinase A and membrane type-1 matrix metalloproteinase during Xenopus laevis development. Mech Dev 124:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fu L, Hasebe T, Ishizuya-Oka A, Shi Y-B. 2007. Roles of matrix metalloproteinases and ECM remodeling during thyroid hormone-dependent intestinal metamorphosis in Xenopus laevis. Organogenesis 3:14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shi YB, Hasebe T, Fu L, Fujimoto K, Ishizuya-Oka A. 2011. The development of the adult intestinal stem cells: insights from studies on thyroid hormone-dependent amphibian metamorphosis. Cell Biosci 1:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ishizuya-Oka A, Hasebe T, Buchholz DR, Kajita M, Fu L, Shi YB. 2009. Origin of the adult intestinal stem cells induced by thyroid hormone in Xenopus laevis. FASEB J 23:2568–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Okada M, Miller TC, Wen L, Shi YB. 2017. A balance of Mad and Myc expression dictates larval cell apoptosis and adult stem cell development during Xenopus intestinal metamorphosis. Cell Death Dis 8:e2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Roskelley CD, Srebrow A, Bissell MJ. 1995. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr Opin Cell Biol 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Page-McCaw A, Ewald AJ, Werb Z. 2007. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Bio 8:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vu TH, Werb Z. 2000. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev 14:2123–2133 [DOI] [PubMed] [Google Scholar]
- 76. Buchholz DR, Heimeier RA, Das B, Washington T, Shi Y-B. 2007. Pairing morphology with gene expression in thyroid hormone-induced intestinal remodeling and identification of a core set of TH-induced genes across tadpole tissues. Dev Biol 303:576–590 [DOI] [PubMed] [Google Scholar]
- 77. Heimeier RA, Das B, Buchholz DR, Fiorentino M, Shi YB. 2010. Studies on Xenopus laevis intestine reveal biological pathways underlying vertebrate gut adaptation from embryo to adult. Genome Biol 11:R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.