Abstract
Our aim was to identify the differentially expressed genes (DEGs) in peripheral blood mononuclear cells (PBMC) of Parkinson’s disease (PD) patients and healthy controls by microarray technology and analysis of related molecular pathways by functional annotation. Thirty PD patients and 30 controls were enrolled. Agilent Human 8X60 K Oligo Microarray was used for gene level expression identification. Gene ontology and pathway enrichment analyses were used for functional annotation of DEGs. Protein–protein interaction analyses were performed with STRING. Expression levels of randomly selected DEGs were quantified by real time quantitative polymerase chain reaction (RT-PCR) for validation. Flow cytometry was done to determine frequency of regulatory T cells (Tregs) in PBMC. A total of 361 DEGs (143 upregulated and 218 downregulated) were identified after GeneSpring analysis. DEGs were involved in 28 biological processes, 12 cellular components and 26 molecular functions. Pathway analyses demonstrated that upregulated genes mainly enriched in p53 (CASP3, TSC2, ATR, MDM4, CCNG1) and PI3K/Akt (IL2RA, IL4R, TSC2, VEGFA, PKN2, PIK3CA, ITGA4, BCL2L11) signaling pathways. TP53 and PIK3CA were identified as most significant hub proteins. Expression profiles obtained by RT-PCR were consistent with microarray findings. PD patients showed increased proportions of CD49d+ Tregs, which correlated with disability scores. Survival pathway genes were upregulated putatively to compensate neuronal degeneration. Bioinformatics analysis showed an association between survival and inflammation genes. Increased CD49d+ Treg ratios might signify the effort of the immune system to suppress ongoing neuroinflammation.
Subject terms: Neuroimmunology, Transcriptomics
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease after Alzheimer’s disease and affects 1% of the population above 60 year-old1. Accumulation of α-synuclein in intracellular deposits named Lewy bodies and loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) are the core pathological features of the disease2. Depletion of dopamine causes dysfunction of basal ganglia and leads to the classical motor symptoms of PD including bradykinesia, rigidity, resting tremor and postural instability. PD is also associated with numerous non-motor symptoms that might precede motor symptoms3. Although diagnosis of PD mainly relies on clinical findings, moderate to severe dopaminergic neuron loss occurs before the onset of motor symptoms4. Thus, it is crucial to diagnose the disease at early stages and develop disease-modifying treatments that will reduce neurodegeneration. Understanding of cellular mechanisms leading to loss of dopaminergic neurons might provide a basis for identification of diagnostic biomarkers and improvement of new therapeutic strategies for PD.
Etiopathogenesis of PD is not fully understood but environmental factors like pesticides, toxins, metals, head injury and certain drugs have received great attention for years5. However recent studies revealed that genetic susceptibility has a key role in PD pathogenesis and complex interactions of genetic and environmental factors are necessary for development of PD6. Monogenic forms of PD constitute < 10% of all cases but identification of these genes provides insight into molecular mechanisms underlying the disease7. Several studies established that oxidative stress, mitochondrial dysfunction and impairment of protein homeostasis contribute to disease mechanisms8. Neuroinflammation also seems to play a role in PD, but whether it induces harmful effects due to release of proinflammatory cytokines or has a protective role in terms of clearance of extracellular debris and production of trophic factors has not yet been established9. Evidence of CD4+ T cell infiltration in postmortem studies of PD brain specimens and increased expression of inflammatory cytokines have strengthened the idea that inflammation could be a prominent feature of PD10,11.
Microarray technology allows comparison of numerous gene expression profiles between healthy subjects and patients and thus provides a common strategy for analysis of neurodegenerative diseases. Several microarray studies have been carried out using both brain tissue and peripheral blood to date and gene expression profiles of PD have been established12–17. Blood-based gene expression analyses revealed altered expression of genes associated with ubiquitination/proteasomal system, mitochondrial function, oxidation and metabolism in PD patients as compared to controls15. Blood transcriptomics of drug-naïve sporadic PD showed increased expression of genes that are involved in leukocyte activation and epigenetic alterations that regulate chromatin remodelling16. Comparison of PD patients with LRRK2 mutation idiopathic PD cases and controls revealed altered expression of genes that are associated with Akt signaling pathway and B cell differentiation18. Tumor necrosis factor (TNF) signaling pathway was also found to be a key pathway involved in PD pathogenesis19. Analyses of differentially expressed gene profiles in experimental mouse model of MPTP induced PD revealed increased expression of systemic inflammation and programmed cell death genes20 Zhang et al. reported decreased expression of complex I transcripts and increased expression of heat shock proteins and some anti-apoptotic gene groups in PD patients21. Durrenberger et al. showed upregulation of P2X7 and NOS pathways thereby emphasizing that extracellular ATP and reactive astrocytes are responsible of microglial activation and subsequent release of proinflammatory cytokines that contribute to dopaminergic cell death22. In another microarray study, expression levels of neuro-immune signaling related transcripts were found to be increased in nucleated blood cells of PD patients23. Overall, these results indicate importance of neuroinflammation in PD pathogenesis.
To bring light to possible pathological mechanisms underlying PD and discover biomarkers associated with progression of motor symptoms in PD, we identified differentially expressed genes through microarray and transcriptome studies. Our results pinpointed expression alterations particularly in inflammation and survival genes. Notably, peripheral blood ratios of a subset of regulatory T cells (Tregs) showed association with altered disability scores of PD.
Materials and methods
Patient selection
Thirty consecutive idiopathic PD patients who were diagnosed by movement disorders specialists according to the United Kingdom Parkinson’s Disease Society Brain Bank Criteria and 30 controls with no prior history of neurological and inflammatory disease were enrolled (Table 2). All patients were under dopaminergic drug treatment. Exclusion criteria were presence of accompanying inflammatory or autoimmune diseases and being under immunosuppressive treatment. Included patients were evaluated by a neurologist, and each patient received the Unified Parkinson Disease Rating Scale (UPDRS) and the Hoehn and Yahr (H&Y) Scale.
Table 2.
Clinical and demographical characteristics of participants.
| PD (n = 30) | HC(n = 30) | p value | |
|---|---|---|---|
| Gender (female/male) | 10/20 | 11/19 | 0.518 |
| Age (mean ± SD) | 56.51 ± 1.40 | 51.18 ± 1.33 | 0.007 |
| Age at disease onset, (mean ± SD) | 49.36 ± 9.17 | ||
| Disease duration, years (mean ± SD) | 6.91 ± 3.32 | ||
| UPDRS total (mean ± SD) | 42.45 ± 19.02 | ||
| UPDRS I | 6.57 ± 4.15 | ||
| UPDRS II | 11.09 ± 8.36 | ||
| UPDRS III | 23.57 ± 14.34 | ||
| UPDRS IV | 1.85 ± 5.48 | ||
| H&Y scale | 2.12 ± 0.60 |
H&Y Scale Hoehn and Yahr Scale, HC healthy controls, UPDRS :Unified Parkinson's Disease Rating Scale, PD Parkinson’s disease.
Microarray expression profiling and DEGs screening
Eighteen PD patients (12 males, mean age ± SD was 58.28 ± 9.45) and 18 control subjects (15 males, mean age ± SD was 48.54 ± 8.22) from the original group were enrolled for transcriptome studies. Mean years of disease duration of patients ± SD was 6.87 ± 4.08, mean total UPDRS scores ± SD was 34.94 ± 18.09, mean UPDRS III score ± SD was 17.50 ± 12.08 and mean H&Y score ± SD was 2.05 ± 0.64. Total RNA was extracted from whole blood samples by using Qiagen RNeasy (CatNo: 74104) kit according to the manufacturer’s protocol. Assessment of total RNA quality was performed by the Agilent 2100 Expert Software that automatically provides RNA Integrity Number (RIN). The minimum RIN number was defined as 7. Agilent Human 8 × 60 K Oligo Microarray has been used for gene level expression identification by manufacturer’s protocol (Agilent Technologies, Palo Alto, CA). Cyanine-3 was used to label targets, after the labeling, the samples were purified by using Absolutely RNA Nanoprep Kit (Agilent) and purified RNA samples were hybridized with Hybridization Kit (Agilent) at 65 °C for 17 h. After washing steps, array slide was put into the Agilent slide holder. After generating the microarray scan images, .tif images were extracted by using the Feature Extraction Software (FE) 4.0.1.21. After normalization by using Robust Multi-array Average (RMA) method, differentially expressed genes (DEGs) between PD and healthy controls were analyzed by Genespring. The significance level was set at p < 0.05 and log2 fold change > 2 for identifying DEGs.
Gene ontology and pathway enrichment analyses
Gene ontology analysis (GO) provides functionally annotation of DEGs within a biological context with respect of biological processes, cellular components and molecular functions24. Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) pathway database was used for analyzing gene functions and to link this data with molecular pathways25. GO terms and KEGG pathway enrichment analyses for DEGs were performed by using the database for annotation, visualization and integrated discovery (DAVID) online database (DAVID Bioinformatics Resources 6.8).
Protein–protein interaction
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://www.string-db.org/) is a database that identifies known and predicted interactions between proteins26. Selected pathways were filtered into the DEG protein–protein interaction (PPI) network complex containing nodes and edges with parameters including a minimum required interaction score > 0.4 (medium confidence). Only query proteins were displayed. Hub genes, highly interconnected with nodes in a module, have been considered functionally significant. A hub is a node that has a higher degree than other nodes in the graph. In our study, hub genes were defined by module connectivity. Furthermore, we uploaded all genes in the hub module to the STRING database to construct PPI to screen hub nodes in PPI network. We defined genes with the node connectivity > 2 (total edges/total nodes) as the hub nodes in PPI network. Additionally, the genes with p value less than 0.05 were identified as real hub genes.
Validation of expression levels of DEGs
Expression levels of DEGs were confirmed by real time quantitative polymerase chain reaction (RT-PCR) studies for randomly selected 5 genes (PRKACB, TSC2, GATA2, PI3KCA, CASP3). Venous blood samples were collected in 10 ml EDTA containing tubes that were obtained from 8 randomly selected PD patients (3 males, mean age ± SD was 59.00 ± 3.62) and 8 age and sex matched healthy controls (4 males, mean age ± SD was 60.00 ± 3.46). Mean years of disease duration ± SD of patients was 6.86 ± 3.34, mean total UPDRS scores ± SD was 39.25 ± 22.64, mean UPDRS III score ± SD was 22.75 ± 17.17, mean H&Y score ± SD was 2.25 ± 0.65. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by using Ficoll density gradient centrifugation and cryopreserved within FBS and 10% DMSO at liquid nitrogen until assayed. RNA was isolated from frozen PBMCs by using RNA isolation commercial kit (Jena Bioscience, Total RNA Purification Kit, PP-210L). Purity assessment was determined by O.D. ratios of 260/280 and 260/230 and concentrations of isolated RNAs were detected with Thermo Scientific Nanodrop 2000. cDNA was synthesized with a transcription kit (Jena Bioscience, PCR511) from 2 µl RNA with the concentration of 100 ng/ml by using Bio-Rad Thermal Cycler according to the manufacturer’s protocol. The quantitative real time PCR (qRT-PCR) assay was performed on an Agilent Stratagene 3005P system. Each reaction was run in 10 μl reaction mixture containing 1 μl of cDNA (50 ng/μl), 10 pmol/µl of forward and reverse primer sets for each specific gene (Table 1) by using Sybr green method (PCR master kit, Jena Bioscience, qPCR GreenMaster with UNG/lowROX kit, PCR306). Beta-actin (ACTB) was used as a normalization reference gene for quantitating the relative levels of gene expressions. Relative gene expressions were analyzed by 2−ΔΔCt method27.
Table 1.
Primers for real time PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| CASP3 | ATTGTGGAATTGATGCGTGA | GGCAGGCCTGAATAATGAAA |
| PRKACB | GCCACGACAGATTGGATTG | TCCAGAGCCTCTAAACTTTGGT |
| ACTB | AACCGCGAGAAGATGACCCA | GGATAGCACAGCCTGGATAGCA |
| GATA2 | CCGCCACATCCATCCTAGCAG | TGCAGACGGCAACGGC |
| TSC2 | AGCTCACGGAAACCTGTCTG | AGGAGACCTCTTCGGGACAG |
| PI3KCA | TCAAAGGATTGGGCACTTTT | GCCTCGACTTGCCTATTCAG |
Immunophenotyping by flow cytometry
To validate the findings of microarray study, a flow cytometry study was conducted in order to determine peripheral immune cell phenotype in PD patients. Frozen PBMCs obtained from 20 PD patients (mean age ± SD was 55.36 ± 5.41 mean age, 12 males) and 20 age and sex matched healthy controls (p = 0.111 and p = 0.744 respectively) from the original group (mean age ± SD 53.55 ± 7.63, 13 males) (8 PD and 8 controls were selected from subjects that were enrolled in RT-PCR study). Mean years of disease duration ± SD of patients was 7.17 ± 2.73, mean total UPDRS scores ± SD was 45.15 ± 19.04, mean UPDRS III score ± SD was 26.15 ± 14.63 mean H&Y score ± SD was 2.1 ± 0.55. Frozen PBMCs were used for extracellular labeling, anti-human monoclonal antibodies applied as followed: CD3-FITC, CD16/CD56-PE, CD45-PerCP, CD19-APC (Beckton Dickens (BD)-MultitestTM, Franklin Lakes, NJ, USA), CD3-Alexa Fluor 700, CD4-PerCP, CD8-APC-Cy7, CD25-APC, CD127-PE-Cy7, CD49d-PE-Cy5 (all from BD) and incubated for 20 min in the dark at room temperature. For intracellular IL-10 labeling at least 106 PBMCs were incubated with 0.25 μg/ml Phorbol-12-myristate-13-acetate (PMA) (AdipoGen, San Diego, CA, USA), 1 μg/ml ionomycin (Santa Cruz, Dallas, TX, USA), and 10 μg/ml brefeldin (Ebioscience, Santa Clara, CA,USA) in complete medium (RPMI 1640 enriched with 10% fetal bovine serum, 1% nonessential amino acids, 1%L-glutamine, 1%Na-pyruvate, 1% minimum essential medium vitamin, 1% penicillin–streptomycin; Gibco, Waltham, MA, USA) for 16 h at 37 °C and %5 CO2. Then, PBMCs were permeabilized by 1 ml of fixation and permeabilization buffer (FoxP3 Staining Buffer Set, eBioscience) and incubated at 4 °C for an hour on ice. After washing with fixation buffer, PBMCs were stained with anti-IL-10-PE antibody (BD), and incubated at 4 °C for an hour on ice. Experiments were utilized in Novocyte (ACEA Bioscience, Inc.) and data were analyzed by using NovoExpress and Flowjo software. The results were finally expressed as percentage of positive cells of the parent (%).
Statistical analyses
Categorical variables between groups were compared by chi-square test, Student t test was used for comparison of parametric variables and Mann–Whitney U for non-parametric variables. Pearson correlation was used to determine correlation between percentages of immune cell subtypes, expression levels of genes and clinical findings. A p value of < 0.05 was considered statistically significant. Statistical analyses were conducted using Prism (GraphPad Software, Inc) or SPSS software (IBM).
Ethics approval and consent to participate
Written informed consent was obtained from all participants and study was approved by Istanbul University, Istanbul Faculty of Medicine Ethical Committee and conformed to the provisions of the Declaration of Helsinki.
Results
Clinical features and demographical characteristics of subjects are presented in Table 2.
Analysis of DEGs
The boxplots for normalized gene expression data represent a good normalization (Supplementary Figure 1). After statistical assessment a total of 361 DEGs (143 upregulated and 218 downregulated) were identified between PD patients and controls (Supplementary Table .1). Hierarchical clustering and heat map analysis with 361 DEGs were performed for validation (Supplementary Figure 2).
Functional annotation
GO analysis revealed that DEGs were involved in 28 biological processes, 12 cellular components and 26 molecular functions (Table 3).
Table 3.
GO analysis of DEGs.
| GO-ID | Term | Count | p value |
|---|---|---|---|
| Biological process (BP) | |||
| GO:0006334 | Nucleosome assembly | 12 | 1.80E−05 |
| GO:0006351 | Transcription, DNA-templated | 61 | 1.50E−04 |
| GO:0043484 | Regulation of RNA splicing | 6 | 2.00E−04 |
| GO:0008380 | RNA splicing | 12 | 2.00E−04 |
| GO:0071456 | Cellular response to hypoxia | 8 | 0.002 |
| GO:0048536 | Spleen development | 5 | 0.005 |
| GO:0098609 | Cell–cell adhesion | 13 | 0.006 |
| GO:0006397 | Mrna processing | 10 | 0.008 |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 24 | 0.012 |
| GO:0051028 | mRNA transport | 5 | 0.012 |
| GO:0045814 | Negative regulation of gene expression, epigenetic | 5 | 0.015 |
| GO:0045931 | Positive regulation of mitotic cell cycle | 4 | 0.016 |
| GO:0045214 | Sarcomere organization | 4 | 0.018 |
| GO:0043486 | Histone exchange | 3 | 0.018 |
| GO:0060968 | Regulation of gene silencing | 3 | 0.018 |
| GO:0006974 | Cellular response to DNA damage stimulus | 10 | 0.019 |
| GO:0060325 | Face morphogenesis | 4 | 0.019 |
| GO:0006278 | RNA-dependent DNA biosynthetic process | 3 | 0.021 |
| GO:0045893 | Positive regulation of transcription, DNA-templated | 18 | 0.021 |
| GO:0007160 | Matrix adhesion | 6 | 0.029 |
| GO:0051726 | Regulation of cell cycle | 7 | 0.032 |
| GO:0080182 | Histone H3-K4 trimethylation | 3 | 0.033 |
| GO:0018105 | Peptidyl-serine phosphorylation | 7 | 0.033 |
| GO:0000183 | Chromatin silencing at rdna | 4 | 0.034 |
| GO:0018117 | Protein adenylylation | 2 | 0.038 |
| GO:0036303 | Lymph vessel morphogenesis | 2 | 0.038 |
| GO:0035574 | Histone H4-K20 demethylation | 2 | 0.038 |
| GO:0043488 | Regulation of mrna stability | 6 | 0.048 |
| Cellular component (CC) | |||
| GO:0005654 | Nucleoplasm | 91 | 4.80E−08 |
| GO:0005634 | Nucleus | 148 | 5.30E−08 |
| GO:0000786 | Nucleosome | 9 | 3.70E−04 |
| GO:0005737 | Cytoplasm | 125 | 8.70E−04 |
| GO:0005720 | Nuclear heterochromatin | 4 | 0.008 |
| GO:0005913 | Cell–cell adherens junction | 14 | 0.008 |
| GO:0019013 | Viral nucleocapsid | 4 | 0.015 |
| GO:0070062 | Extracellular exosome | 66 | 0.035 |
| GO:0002944 | Cyclin K-CDK12 complex | 2 | 0.037 |
| GO:0043234 | Protein complex | 14 | 0.045 |
| GO:0005694 | Chromosome | 6 | 0.046 |
| GO:0000788 | Nuclear nucleosome | 4 | 0.048 |
| Molecular function (MF) | |||
| GO:0044822 | Poly(A) RNA binding | 51 | 2.40E−08 |
| GO:0005515 | Protein binding | 202 | 1.60E−04 |
| GO:0042393 | Histone binding | 10 | 5.80E−04 |
| GO:0016779 | Nucleotidyltransferase activity | 5 | 0.002 |
| GO:0031492 | Nucleosomal DNA binding | 6 | 0.002 |
| GO:0003677 | DNA binding | 50 | 0.002 |
| GO:0003723 | RNA binding | 22 | 0.002 |
| GO:0004674 | Protein serine/threonine kinase activity | 17 | 0.003 |
| GO:0004693 | Cyclin-dependent protein serine/threonine kinase activity | 5 | 0.004 |
| GO:0003720 | Telomerase activity | 3 | 0.010 |
| GO:0031625 | Ubiquitin protein ligase binding | 13 | 0.010 |
| GO:0098641 | Cadherin binding involved in cell–cell adhesion | 13 | 0.010 |
| GO:0001968 | Fibronectin binding | 4 | 0.013 |
| GO:0003714 | Transcription corepressor activity | 10 | 0.017 |
| GO:0000166 | Nucleotide binding | 14 | 0.017 |
| GO:0003682 | Chromatin binding | 15 | 0.019 |
| GO:0004672 | Protein kinase activity | 14 | 0.022 |
| GO:0003676 | Nucleic acid binding | 29 | 0.025 |
| GO:0004864 | Protein phosphatase inhibitor activity | 4 | 0.025 |
| GO:0005524 | ATP binding | 40 | 0.031 |
| GO:0070733 | Protein adenylyltransferase activity | 2 | 0.038 |
| GO:0035575 | Histone demethylase activity (H4-K20 specific) | 2 | 0.038 |
| GO:0097493 | Structural molecule activity conferring elasticity | 2 | 0.038 |
| GO:0003779 | Actin binding | 11 | 0.043 |
| GO:0008307 | Structural constituent of muscle | 4 | 0.046 |
| GO:0001046 | Core promoter sequence-specific DNA binding | 4 | 0.049 |
KEGG pathway analysis revealed that upregulated genes were mainly enriched in p53 (CASP3, TSC2, ATR, MDM4, CCNG1) and PI3K/Akt (IL2RA, IL4R, TSC2, VEGFA, PKN2, PIK3CA, ITGA4, BCL2L11) signaling pathways and downregulated genes were enriched in leukocyte transendothelial migration pathway (ACTG1, RAPGEF3, JAM3, MYL9, PTPN11) (Tables 4 and 5). Thus, several significantly altered genes were involved in survival and inflammation processes.
Table 4.
Pathway analysis of upregulated genes in PD.
| Category | Term | Genes | Count | p value |
|---|---|---|---|---|
| KEGG_PATHWAY | hsa04115:p53 signaling pathway | CASP3, TSC2, ATR, MDM4, CCNG1 | 5 | 0.0030 |
| REACTOME_PATHWAY | R-HSA-211227:R-HSA-211227 (Activation of DNA fragmentation factor) | HIST1H1E, CASP3, HIST1H1D | 3 | 0.0085 |
| REACTOME_PATHWAY | R-HSA-1221632:R-HSA-1221632 (Meiotic synapsis) | SYNE2, HIST2H2BE, HIST2H2AC, SUN1, ATR | 5 | 0.0107 |
| REACTOME_PATHWAY | R-HSA-212436:R-HSA-212436 (Generic Transcription Pathway) | ZNF419, ZNF558, ZNF692, ZNF160, ZNF141, ZNF550, ZIK1, ZNF480, ZNF615, ZNF37A | 10 | 0.0160 |
| KEGG_PATHWAY | hsa04151:PI3K-Akt signaling pathway | IL2RA, IL4R, TSC2, VEGFA, PKN2, PIK3CA, ITGA4, BCL2L11 | 8 | 0.0338 |
| KEGG_PATHWAY | hsa05414:Dilated cardiomyopathy | PRKACB, ITGA4, TPM2, TTN | 4 | 0.0392 |
| KEGG_PATHWAY | hsa05206:MicroRNAs in cancer | DNMT3A, CASP3, VEGFA, ZEB2, MDM4, CCNG1, BCL2L11 | 7 | 0.0416 |
| REACTOME_PATHWAY | R-HSA-606279:R-HSA-606279 (Deposition of new CENPA-containing nucleosomes at the centromere) | HIST2H2BE, HIST2H2AC, CENPC, CENPT | 4 | 0.0468 |
| KEGG_PATHWAY | hsa03015:mRNA surveillance pathway | SMG5, SMG1, PABPC1L, NXT2 | 4 | 0.0479 |
Table 5.
Pathway analysis of downregulated genes in PD.
| Category | Term | Genes | Count | p-value |
|---|---|---|---|---|
| KEGG_PATHWAY | ptr04670:leukocyte transendothelial migration | ACTG1, RAPGEF3, JAM3, MYL9, PTPN11 | 5 | 0.0116 |
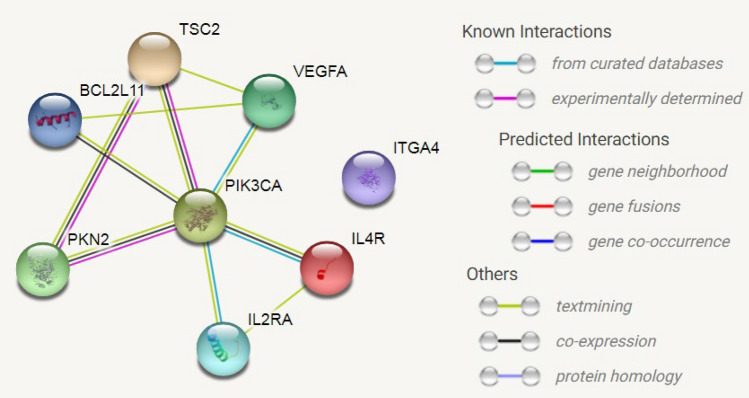
PPI network construction
PPI network of proteins that were encoded by most significantly upregulated DEGs were involved in p53 and PI3K/Akt signaling pathways. TP53 and PIK3CA were identified to be most significant hub proteins (Figs. 1 and 2).
Figure 1.
Protein–protein interaction (PPI) network for upregulated DEGs in PI3K/Akt pathway. Nodes represent proteins and lines represent interactions of proteins in networks.
Figure 2.
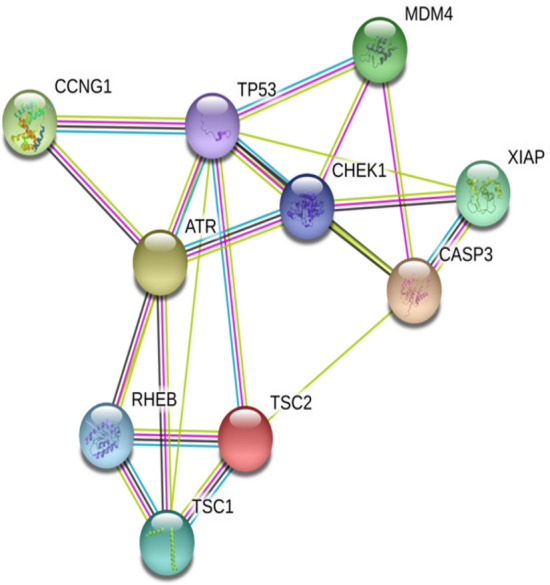
Protein–protein interaction (PPI) network for upregulated DEGs in p53 pathway. Nodes represent proteins and lines represent interactions of proteins in networks.
Validation of gene expressions with RT-PCR
For validation of the microarray data, expression levels of 5 genes (PRKACB, TSC2, GATA2, PI3KCA, CASP3) that were randomly selected among the upregulated DEGs were quantified by RT-PCR analysis using peripheral blood samples of randomly selected 8 PD patients and 8 healthy controls. In consistency with microarray data, expression levels of selected genes were elevated in PD patients compared to healthy controls (Table 6). There was no correlation between expression levels of selected genes and clinical findings of PD patients such as age, age of disease onset, disease duration, UPDRS and H&Y scale scores (data not shown).
Table 6.
Fold chance values of RT-PCR and microarray data.
| Q-RT-PCR FC* | Microarray FC | |
|---|---|---|
| PRKACB | 1.951 | 1.725 |
| TSC2 | 1.191 | 1.615 |
| GATA2 | 1.042 | 1.657 |
| PI3KCA | 2.188 | 1.156 |
| CASP3 | 1.579 | 1.348 |
Q-RT-PCR quantitative real time polymerase chain reaction, FC fold change.
*Note that FC was assessed by the 2−ΔΔCt method for Q-RT-PCR.
Flow cytometry analyses
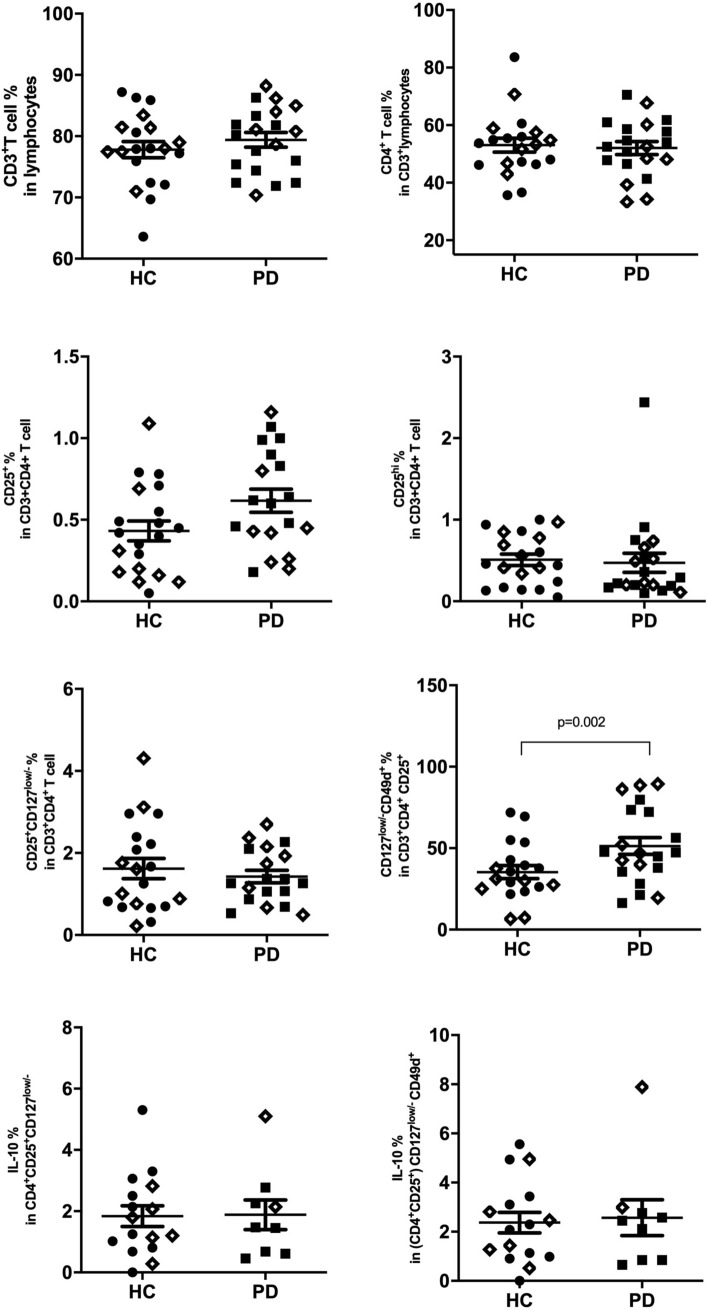
An important finding of the microarray data was enhanced expression of Treg-associated surface expression factors IL2RA (CD25) and CD49d (Suppl. Table 1). To validate the alteration of Treg populations in PD, peripheral blood frequencies of immune cell subsets were assessed by flow cytometry. The gating strategy has been displayed in Supplementary Figure 3. Frequencies of CD3+ T cells (79.40 ± 1.201 vs. 77.81 ± 1.331), CD4+ helper T cells (52.06 ± 2.259 vs. 53.05 ± 2.430), CD8+ cytotoxic T cells (33.57 ± 1.875 vs. 31.39 ± 1.932), CD19+ B cells (5.805 ± 0.4360 vs. 5.651 ± 0.5065) and CD16/56+ NK cells (11.06 ± 1.17 vs. 14.31 ± 1.12) were comparable between PD and control groups. Percentage of CD3+CD4+CD25+ T cells and CD3+CD8+CD25+ T cells were slightly increased in PD group without attaining statistical significance. Frequency of Tregs, which were defined as CD4+CD25+CD127low T cells28, showed no differences among patient and control groups. For further evaluation of effects of CD49 expression on suppressive capacity of Tregs, subpopulations of CD49d positive and negative cells were evaluated within CD4+CD25+CD127low Tregs. Frequency of CD49d+ Tregs were significantly higher in PD patients than in controls (51.33 ± 5.187 vs 35.33 ± 4.035, p = 0.002). To assess functional immunosuppressive capacity of Tregs, we compared frequencies of IL-10 producing cells within CD4+CD25+CD127low and CD4+CD25+CD127low/-CD49d+ subtypes between PD and controls. There were no differences between two groups in terms of frequencies of IL-10 producing cells for both subtypes (Fig. 3). Proportions of CD4+CD25+CD127lowCD49d+ Tregs were found to be negatively correlated with total UPDRS score in PD patients (p = 0.028, r = -0.491) (Fig. 4). There was no correlation between CD49d+ Treg frequencies versus age or disease duration.
Figure 3.
Frequencies of T cell subsets are shown on y axis for PD and control groups, symbol ◊ represents subjects enrolled in both RT-PCR and flow cytometry studies.
Figure 4.
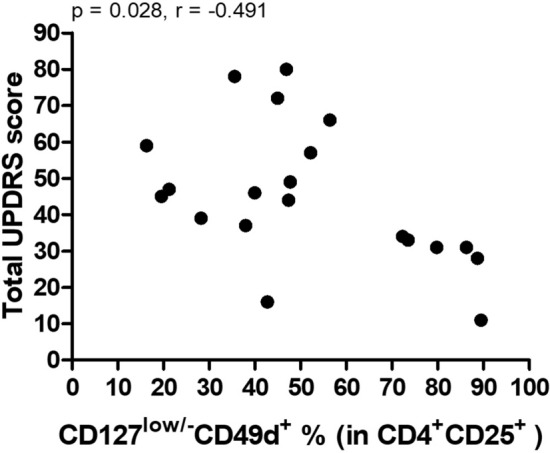
Correlation plot between total UPDRS score and proportion of CD4+CD25+CD127lowCD49d+ Tregs.
Discussion
PD is an important health care problem that causes serious impairment in daily life quality especially in elderly population. The disease can only be treated symptomatically with the current treatment strategies and the degenerative process cannot be prevented. An important reason for this is that significant loss of neurons has already occurred at the stage of diagnosis. Improved understanding of the pathophysiology of PD and the mechanisms underlying neuronal loss are crucial for early diagnosis and development of disease-specific treatments. Our results revealed that genes that are most differentially expressed between PD patients and healthy controls are particularly associated with survival pathways and inflammation. p53 and PI3K/Akt pathways were the most profoundly affected survival-associated pathways.
Apoptosis is crucially involved in physiological and pathological conditions such as oxidative stress and severe DNA damage. Genotoxic stress in the cell is recognized by ataxia telangiectasia mutated (ATM) kinase and ATM- and Rad3-related (ATR) kinase proteins, which provide phosphorylation of murine double minute gene 2 (MDM2), a ubiquitin-ligase that enhances p53 degradation29,30. This posttranslational change causes inactivation of MDM2 while activating p5331. Activated p53 stops the cell cycle in order to eliminate DNA damage. In cases that the DNA damage cannot be corrected, p53 initiates programmed cell death. p53-induced pro-apoptotic genes cause release of cytochrome-c from the mitochondrial membrane. Cytochrome-c also interacts with p53-induced APAF-1 protein to activate the caspase cascade32. p53 suppresses the Akt/mTOR pathway both by enhancing tuberous sclerosis complex 2 (TSC2) transcription and inducing synthesis of PTEN protein that degrades PIP333,34. In postmortem studies and experimental models of PD, apoptosis has been shown as an important mediator of dopaminergic cell death35. p53, an important tumor suppressor gene, plays a critical role in the process of apoptosis by transcriptional control of the genes associated with cell death and Bcl2 family interaction36. SNpc of PD brains shows increased p53 expression37, deletion of DJ1 causes p53-mediated loss of dopaminergic neurons in experimental models, p53 inhibition has a neuroprotective effect and p53 activates transcription of the alpha synuclein gene (33–34). Thus, overexpression of the p53 pathway might be one of the essential factors involved in the pathogenesis of sporadic PD.
PI3K-Akt signaling pathway is a highly conserved cascade that promotes cell survival and proliferation and plays important role in synaptic plasticity. Numerous growth factors and cytokines carry out their intracellular effects via PI3K pathway38. In our study, PPI analysis showed that phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PI3KCA) protein has a central role in this network. The PI3K enzyme catalyzes the synthesis of an important secondary messenger phosphatidylinositol (3,4,5)-trisphosphate (PIP3) molecule from the phosphatidylinositol 4,5-bisphosphate (PIP2). PIP3 activates phosphorylation of the serine threonine kinase Akt39. mTORC1, one of the downstream proteins of this signaling pathway, is a protein kinase complex that acts as a sensor for metabolic status of the cell and plays an important role in decision-making in the direction of survival or apoptosis40. Akt eliminates inhibition of mTORC1 by phosphorylating the TSC2 protein, which has an inhibitory effect on mTORC1. Activation of mTORC1 increases anabolic activity in the cell and suppresses autophagy. Defects in this pathway have been associated with neurodegeneration in diseases such as Alzheimer and Huntington disease40–42. In an experimental study, TSC2 expression was shown to be increased in response to synuclein accumulation in the transgenic mouse model, but no significant difference was found between TSC2 expression levels in the brain of human PD cases and controls43. In another study, it has been shown that proteins regulating autophagy prevent the ubiquitination of TSC2 in the neurotoxin-induced animal model and cause autophagy in dopaminergic neurons through this mechanism44. Activation of autophagic mechanisms may protect dopaminergic neurons from synuclein toxicity by increasing synuclein degradation45. In our study TSC2 gene expression was increased in the PD group and this upregulation might be due to the effort to activate the autophagy process and could be a therapeutic target, especially in the early stage of the disease.
Other members of the PI3K pathway that are upregulated in PD patients are also profoundly associated with the disease. Akt regulates the expression of Bcl-2 like 11 (BCL2L11) through phosphorylation of forkhead phosphorylation of transcription factors (FOXO)46. BCL2L11, also known as Bim (Bcl 2-interacting mediator of cell death), is a potent pro-apoptotic protein that can bind anti-apoptotic proteins with high affinity47. In the MPTP-induced PD mouse model, Bim expression is increased regionally in the midbrain and remains high during dopaminergic neuron death48. VEGFA is the biologically most active isoform of VEGF growth factor family that potently promotes proliferation and migration of vascular endothelial cells and controls vascular permeability 49,50. Although VEGFA is primarily identified as an angiogenesis factor, recent studies have implied neurotrophic, neuroprotective and chemoattractant roles, as well51. Upregulated expression of VEGF gene has been reported in reactive astrocytes within substantia nigra of PD patients52. This upregulation is likely to have occurred as a compensatory mechanism to enhance neurogenesis and to maintain microcirculation.
In this study, we found increased expression levels of several inflammation-related genes. Proteomics analysis suggested that upregulation of at least some of the inflammation genes (e.g. IL-2RA and IL-4R) was by virtue of the enhanced expression of the PI3K/Akt pathway, which is intimately associated with inflammatory pathways. Early evidence of involvement of inflammation in PD pathogenesis has come from the studies that revealed presence of human leukocyte antigen DR (HLA-DR) positive reactive microglia in SNc of PD patients. Subsequent studies showed that increased expression of proinflammatory cytokines released from activated microglia contributes to the degeneration of dopaminergic neurons and activation of nuclear factor kappa B (NF-κB) pathway in PD patients and experimental models of PD53–57. Consistent with these findings increased expression of genes encoding proinflammatory cytokines has been found in SN of PD patients58.
Inflammatory factors may display both hazardous and neuroprotective roles in PD. As an example, IL-4, an anti-inflammatory cytokine, may both suppress microglial activity and modulate neurogenesis59,60. In the lipopolysaccharide-treated rat model, increased endogenous IL-4 synthesis caused elevation of pro-inflammatory cytokines of microglial origin thereby contributing to degeneration of vulnerable dopaminergic neurons61. By contrast, infiltrating IL-4 producing CD4+ T cells in the CNS have been shown to improve neuronal survival and achieve axonal healing through the Akt signaling pathway62. Similarly, in experimental autoimmune encephalitis models, externally administered IL-4 has been shown to improve axonal damage via IL-4 receptor (IL-4R)-mediated signaling pathway63. While PD is typified with Th1-type inflammation64, IL-4 may shift the balance in favor of Th2-type inflammation and thus may regulate neuroinflammation in PD. IL-4 also prevents neuronal death and induces neuronal growth through binding IL-4 receptor alpha (IL4-Rα) on neurons62. Thus overexpression of IL4-R in PD might be a compensating countermeasure to suppress the ongoing neuroinflammation.
Although the role of the peripheral immune system in PD-related neuroinflammation has not yet been entirely elucidated, infiltrating CD4+ and CD8+ T lymphocytes have been suggested to contribute to neurodegeneration65. CD3+ T cells, which have been found in close proximity to glial cells in brains of DLB patients and α-synuclein transgenic mice, have been suggested to participate in activation of glial cells66. In vitro studies suggested that CD4+/CD25− effector T cells promote microglial activation and contribute to neurodegeneration while CD4+/CD25+ Tregs might be inhibiting microgliosis67. Furthermore, adoptive transfer of CD3 activated Tregs to mice with MPTP-induced PD has provided greater than 90% protection of the nigrostriatal system68. It is thought that dysfunction of Tregs may contribute to PD pathogenesis by enhancement of neuroinflammation or by impairment of tolerance to neuronal antigens69. However, the underlying mechanisms of decreased effectiveness of regulatory T cells in PD patients have not been elucidated.
IL2RA (CD25) gene encodes the alpha chain of the receptor complex of IL-2, an important cytokine associated with immune homeostasis and self-tolerance. IL-2 acts as a growth factor for T cells70. CD25+ Tregs suppress the inflammatory response and autoimmunity presumably by competing for and consuming the IL-2 cytokine. The effector CD4+ T cell apoptosis induced by Treg cells through cytokine deprivation has been shown to require the presence of BIM proapoptotic protein71, which was upregulated in our study. Dopaminergic cell damage induced by 1-methyl-4-phenylpyridinium (MPP+) is prevented by Tregs. Suppression of microglial oxidative stress and inflammation by Tregs might likely be responsible for this protective mechanism.
There are controversial results for proportion of Tregs in PD. While in some studies Tregs were found to be increased in PD, some other researchers reported that Tregs were decreased or not altered in PD patients72–74. Saunder et al. found that Tregs from PD patients had reduced ability to suppress effector T cells while proliferative capacity of CD4+ T cells did not change. Besides, α4β1+ CD4+ T cells were elevated slightly but not significantly in PD patients75. α4β1 integrin is an adhesion molecule that facilitates the migration of immune system cells into the central nervous system. The alpha chain of α4β1 integrin is encoded by ITGA4 (CD49d) gene. The ability of CD49d+ Tregs to suppress effector T cells was found to be weaker compared to CD49d negative T cells in vitro76. In our study, CD49d expression was increased in PD and also PD patients displayed higher frequencies of CD49d+ Tregs in the peripheral blood. Notably, PD patients with higher frequencies of CD49d+ Tregs showed trends towards displaying lower UPDRS scores, indicating a neuroprotective role of this Treg subset in PD presumably through suppression of infiltrating T cells in the brain. These results could also be explained by reduction of CD49d+ Tregs by increased age and disease duration. However, no correlation could be found between these factors ruling out this assumption.
It is known that neuroinflammation induced by activation of astrocytes and microglia with subsequent dysfunction of endotelhial cells contributes to PD pathogenesis. Besides central inflammation, peripheral proinflammatory profile with altered CD4+/CD8+ T cells ratios and Treg cells might facilitate neurodegeneration77. Several transcriptome studies that were performed in different platforms with different tissues have highlighted the importance of inflammatory pathways in PD pathogenesis78. These studies have also shown that even different regions of PD brain may show altered immune gene expression profiles79. Highly diverse DEG profiles have been obtained in previous transcriptome studies that were recently conducted with peripheral blood samples of sporadic PD patients with a special emphasis on immune system-related genes (Table 7)16,18,19,23,81–93. Possible reasons underlying this variability could be the heterogeneous physiopathology of sporadic PD, different inclusion criteria and clinical confounders (such as early onset, treatment status and exercise), different expression detection methods (microarray or RNA-seq) and diverse enrichment analysis methods. In general, expression levels of inflammatory factors are increased in the peripheral blood of PD patients. Nevertheless, essential T lymphocyte activation genes have also been found to be decreased in the reports of Jiang et al., Chi et al. and Calligaris et al.16,83,84. It is tempting to speculate that this may be attributed to the increased Treg activity, which was demonstrated by our study. In addition, Calligaris et al. and Mutez et al. reported an alteration in the signaling pathway of IL-4R (a receptor highly abundant on Tregs), while we found altered expression of the IL-4R16,85. The fact that the recruited patients in the Calligaris study were not under treatment, unlike our patient cohort, suggests that the altered expression of this pathway cannot be attributed to dopaminergic therapy. Another resemblance with our findings was altered expression of cytokine receptor (e.g. IL-18R, IL-2RA) and cell adhesion molecule (e.g. ICAM1) gene expression levels in studies reported by Kurvits et al., Hu et al. and Tan et al.19,81,82. Furthermore, altered expression levels of Treg related genes were highlighted by Mutez et al. and Pinho et al.85,93. Grünblatt et al. reported increased expression of integrin, alpha M (ITGAM) in substantia nigra of sporadic PD patients80 By contrast, immune-related genes that were significantly altered in our study and not reported in previous studies were VEGFA and ITGA4.
Table 7.
Comparison of the recently performed peripheral blood transcriptome studies of PD patients with our study.
| Authors | Patient selection criteria | Method | Enrichment analyses | Number of samples | Most significant DEGs related with immune system | Most significant pathways related with immune system | Comment |
|---|---|---|---|---|---|---|---|
| Kurvits et al.81 | Idiopathic PD with standard medical treatment | Whole blood RNA-Seq | KEGG | 12 PD, 12 HC | IL18R1 (upregulated) | Immune system related pathways were not specified | IL18R1 is an interleukin receptor linked with proinflammatory responses |
| Hu et al.82 | Idiopathic PD without any immune system disorders and under standard medical treatment | RNA-Seq from blood leukocytes before and after exercise | KEGG | 21 PD | CCL5, CD28, CD3E, IL23A, IL2RA, PRKCQ, TMIGD2, TNFSF9, TNFRSF18 ZAP70 (downregulated after exercise) |
T-cell receptor signaling pathway, cytokine receptor-signaling pathway and primary immunodeficiency pathway were highly enriched in downregulated genes after exercise |
Exercise can modulate the abnormal immunity in patients with PD through T-cell-related functions |
| Jiang et al.83 | Early-stage idiopathic PD | Microarray (Affymetrix) | KEGG, CYTOSCAPE | 255 PD, 255 HC | TNFSF14 (Upregulated), IL23A (Downregulated) | TNFSF14 was enriched in the pathways associated with immunoregulation | TNFSF14 stimulates the proliferation of T cells. IL23A is involved in Th17-type immune response |
| Tan et al.19 | Drug-naïve sporadic PD | Microarray (Affymetrix) | KEGG, STRING | 40 PD, 19 HC | CD40, GATA3 (upregulated) ICAM1 (downregulated) | Chemokine signaling pathway, Cytokine–cytokine receptor interaction, Cell adhesion molecules, NF-kappa B signaling pathway | ICAM1, other cell adhesion molecules and chemokines are involved in persistent neuroinflammation. GATA3 is a regulator of T-cell development |
| Chi et al.84 | Early-stage idiopathic PD | Microarray (Affymetrix) | KEGG | 50 PD, 22 HC | HLADQB1, IFI27 (downregulated) | The downregulated DEGs were enriched in the immune response | HLADQB1 is related with T cell activation |
| Calligaris et al.16 | Drug-naïve sporadic PD | Microarray (Affymetrix) | GO, DAVID, GSEA | 40 PD, 20 HC | TCF3, DOCK10, MAN1C1 (downregulated) | Lymphocyte activation, leukocyte activation | TCF3 is involved in development and differentiation of B and T cells. DOCK10 is involved in IL-4 receptor signaling. MAN1C1 is a key protein required for innate immunity activation |
| Mutez et al.85 | Familial PD with LRRK2 mutation | Microarray (Agilent) | Ingenuity Pathways Analysis | 10 familial PD, 7 HC, 1 pooled HC | Immune related genes were not specified | TGF-β signaling, IL-10 signaling, IL-4 signaling, leukocyte extravasation signaling, toll-like receptor signaling, IL-6 signaling | CD4+CD25+Foxp3 regulatory T cells modulate microglial inflammation through IL-10- and TGF-β-mediated mechanisms |
| Infante et al.86 | PD with LRRK2 mutation and idiopathic PD | RNA-Seq | KEGG | 20 familial PD, 20 idiopathic PD, 20 HC | Immune related genes were not specified | Complement cascade, cell adhesion molecules | The complement system is an effector arm of the innate immune response, which may play role in PD pathogenesis |
| Infante et al.18 | PD with LRRK2 mutation and idiopathic PD | RNA-Seq | Not performed | 20 familial PD, 20 asymptomatic carriers of the mutation, 20 idiopathic PD, 20 controls, 7 PD before and after initiating dopaminergic therapy | FCER2 (downregulated in PD) | Not specified | FCER2 is a key molecule for B-cell activation and growth and is involved in oxidative stress mediated damage |
| Soreq et al.23 | Early stage PD | Microarray (Affymetrix) | BiNGO | 50 PD, 22 HC, 33 NC | CR1, FCGRT CLEC4E, IGSF6, interleukins and their receptors, MEFV, IRF4 upregulated compared to HC; TNFSF14, TNFRSF9, TNFRSF10C upregulated compared to NC | IL-4 biosynthesis | Transcription and immune signaling-related transcripts are distinctly expressed in nucleated blood cells early in PD progress |
| Soreq et al.87 | PD patients pre- and post-deep brain stimulation | Exon array | Post-hoc GO analyses | 7 PD pre-DBS, 7 PD post-DBS (stim-ON), 7 PD post -DBS (stim-OFF), 6 HC | Immune related genes were not specified | IL-8 receptor activity, IL-1 receptor activity, Th1 and Th2 immune response, B cell differentiation, cytokine production | The DBS stimulus induced more leucocyte transcriptional change than the disease itself. This was reversed by DBS stimulation |
| Soreq et al.88 | Advanced PD | Exon array | Matlab | 7 PD, 6 HC | Immune related genes were not specified | NF-B cascade and immune response | PD patients’ blood leukocytes exhibit alternative splicing of immune response related genes |
| Alieva et al.89 | Untreated PD at early stage | Microarray (pooled RNA) Illumina Human HT-12 v4.0 | DAVID | 4 PD, 4 HC | Immune related genes were not specified | Immune system process, Defense response, Response to cytokine stimulus, Positive regulation of I-kappaB kinase/NF-kappaB signaling | Immune system activation may play an important role in the development of PD |
| Karlsson et al.90 | Patients with clinically defined PD, patients with de novo PD | Microarray Illumina Human HT-12 v4.0 | Ingenuity Pathway Analysis | 79 PD,75 HC | Immune related genes were not specified | Inflammatory response pathways | Changes in gene expression in the brain due to PD can also be found in blood and could be used as molecular biomarker for early detection of PD |
| Kobo et al.91 | PD patients with GBA mutation | Exon array, Affymetrix | DAVID | 59 PD with GBA mutation, 59 HC | BANK1, CD180, CD19, CD22, CD72, CD791, FCRL1, FCRL2, FCRL3,IGHD, IGHM downregulated | Immune response, B cell receptor signaling pathway | The down-regulation of B cell-related genes are observed in PD-GBA patients and correlate with disease progression |
| Li et al.92 | Early-stage idiopathic PD | Microarray, Affymetrix | Ingenuity pathways analysis | 50 PD, 22 HC | Immune related genes were not specified | T cell receptor signaling, iCOS-iCOSL signaling in T helper cells, CD28 signaling in T helper cells, PI3K signaling in B lymphocytes, B cell receptor signaling | mTOR signaling and CD28 signaling in T helper cells were able to accurately classify PD and healthy control samples |
| Pinho et al.93 | PD with slow and rapid progression | Microarray, Affymetrix | DAVID | 35 PD with slow progression and 35 PD with rapid progression | FOXP1 downregulated in rapid progressive patients | Immune function pathways | Decreased FOXP1 expression may result in increased levels of pro-inflammatory mediators |
| Karaaslan et al | Idiopathic PD with standard medical treatment | Microarray (Agilent) | KEGG, STRING | 18 PD, 18 HC | IL2RA, IL4R, ITGA4 (CD49d) | Immune system related pathways were not specified | Upragulated genes were associated with Treg function, increased expression of ITGA4 (CD49d) might be the explanation of dysregulated Treg functions in PD |
PD Parkinson’s disease, HC healthy control, NC neurological control, BINGO biological networks gene oncology, KEGG Kyoto Encyclopedia of Genes and Genomes, DAVI: database for annotation, visualization and integrated discovery, STRING search tool for the retrieval of interacting genes/proteins, GO gene ontology, GBA glucosidase beta acid.
To further understand the mechanism, by which CD49d+ Tregs might be suppressing neuroinflammation in PD, we assessed Tregs with anti-inflammatory IL-10 production and could not find difference between PD patients and controls. Nevertheless, Tregs might be operating through TGF-β rather than IL-10 production in PD and this assertion should be further investigated in future studies. Another limitation of our study in this context was not assessing pro-inflammatory cytokine-producing T cell subtypes and thus the inflammation status of our PD patients and suppression capacity of CD49d+ Tregs could not be duly investigated. Other limitations of our study were the low sample size for both gene expression analyses and validation studies. Although there were numerous gene expression studies with larger cohorts that revealed possible role of immune system in the pathogenesis of PD, the data we have obtained with a limited number of samples may provide a basis for discussing Treg-PD relationship in functional terms.
Conclusion
In brief, we detected altered expression levels of genes involved in survival, apoptosis and inflammation related pathways in PD patients. We also found clues indicating the functionality of a subset of Tregs. Our results emphasize once again the complex interactions between neuronal apoptosis and inflammation in neurodegeneration. The exaggerated proinflammatory responses and/or insufficient anti-inflammatory mechanisms may result in the loss of vulnerable dopaminergic neurons. The increased detection of CD49d expression, which is expected to be low in Tregs with high suppressive capacity, suggested that dysfunctional Tregs may participate in PD pathogenesis. Nevertheless, increased CD49 expression may be a factor that positively affects the prognosis of the disease in the advanced stages through as yet unknown mechanisms.
Supplementary Information
Acknowledgements
We thank all participants.
Abbreviations
- DEGs
Differentially expressed genes
- PD
Parkinson’s disease
- RT-PCR
Real time polymerase chain reaction
- Tregs
Regulatory T cells
- SNpc
Substantia nigra pars compacta
- UPDRS
Unified Parkinson disease rating scale
- H&Y Scale
Hoehn and Yahr scale
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DAVID
Database for annotation, visualization and integrated discovery
- STRING
Search tool for the retrieval of interacting genes/proteins
- ATM
Ataxia telangiectasia mutated
- ATR
ATM- and Rad3-related (ATR)
- MDM2
Murine double minute gene 2
- PI3KCA
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- PIP3
Phosphatidylinositol (3,4,5)-trisphosphate
- PIP2
Phosphatidylinositol 4,5-bisphosphate
- TSC2
Tuberous sclerosis complex 2
- BCL2L11
Bcl-2 like 11
- FOXO
Forkhead phosphorylation of transcription factors
- MPP +
1-Methyl-4-phenylpyridinium
Author contributions
C.I.K., E.T., A.H.E., C.U. conceived and designed the experiments. B.B., H.A.H. and Z.K. collected the data. Z.K., E.S., V.Y., C.U. and O.T.K. performed the experiments. O.T.K., V.Y., C.I.K. and E.T. analyzed the experimental data. Z.K., C.I.K. and E.T. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Istanbul University Research Fund (project number BAP-34229/27642).
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-81961-7.
References
- 1.de Lau LM, Breteler MM. Epidemiology of Parkinson's disease. Lancet Neurol. 2006;5:525–535. doi: 10.1016/S1474-4422(06)70471-9. [DOI] [PubMed] [Google Scholar]
- 2.Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson's disease. J. Neurol. Neurosurg. Psychiatry. 1988;51:745–752. doi: 10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalia LV, et al. Clinical correlations with Lewy body pathology in LRRK2-related Parkinson disease. JAMA Neurol. 2015;72:100–105. doi: 10.1001/jamaneurol.2014.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Y, Le W, Jankovic J. Preclinical biomarkers of Parkinson disease. Arch. Neurol. 2011;68:22–30. doi: 10.1001/archneurol.2010.321. [DOI] [PubMed] [Google Scholar]
- 5.Goldman SM. Environmental toxins and Parkinson's disease. Annu. Rev. Pharmacol. Toxicol. 2014;54:141–164. doi: 10.1146/annurev-pharmtox-011613-135937. [DOI] [PubMed] [Google Scholar]
- 6.Greenamyre JT, Hastings TG. Biomedicine. Parkinson's—Divergent causes, convergent mechanisms. Science. 2004;304:1120–1122. doi: 10.1126/science.1098966. [DOI] [PubMed] [Google Scholar]
- 7.Lesage S, Brice A. Parkinson's disease: From monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 2009;18:R48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 8.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson's disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 9.Phani S, Loike JD, Przedborski S. Neurodegeneration and inflammation in Parkinson's disease. Parkinsonism Relat. Disord. 2012;18(Suppl 1):S207–209. doi: 10.1016/S1353-8020(11)70064-5. [DOI] [PubMed] [Google Scholar]
- 10.Brochard V, et al. Infiltration of CD4+ lymphocytes into the brain contributes to neurodegeneration in a mouse model of Parkinson disease. J. Clin. Investig. 2009;119:182–192. doi: 10.1172/JCI36470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mogi M, et al. Interleukin-1 beta, interleukin-6, epidermal growth factor and transforming growth factor-alpha are elevated in the brain from parkinsonian patients. Neurosci. Lett. 1994;180:147–150. doi: 10.1016/0304-3940(94)90508-8. [DOI] [PubMed] [Google Scholar]
- 12.Hauser MA, et al. Expression profiling of substantia nigra in Parkinson disease, progressive supranuclear palsy, and frontotemporal dementia with parkinsonism. Arch. Neurol. 2005;62:917–921. doi: 10.1001/archneur.62.6.917. [DOI] [PubMed] [Google Scholar]
- 13.Moran LB, et al. Whole genome expression profiling of the medial and lateral substantia nigra in Parkinson's disease. Neurogenetics. 2006;7:1–11. doi: 10.1007/s10048-005-0020-2. [DOI] [PubMed] [Google Scholar]
- 14.Scherzer CR, et al. Molecular markers of early Parkinson's disease based on gene expression in blood. Proc. Natl. Acad. Sci. U.S.A. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamir R, et al. Analysis of blood-based gene expression in idiopathic Parkinson disease. Neurology. 2017;89:1676–1683. doi: 10.1212/WNL.0000000000004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calligaris R, et al. Blood transcriptomics of drug-naive sporadic Parkinson's disease patients. BMC Genomics. 2015;16:876. doi: 10.1186/s12864-015-2058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, et al. Genome-scale expression pattern of long non-coding RNAs in Chinese Uyghur patients with Parkinson's disease. Med. Sci. Monit. 2020;26:e925888. doi: 10.12659/MSM.925888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Infante J, et al. Identification of candidate genes for Parkinson's disease through blood transcriptome analysis in LRRK2-G2019S carriers, idiopathic cases, and controls. Neurobiol. Aging. 2015;36:1105–1109. doi: 10.1016/j.neurobiolaging.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 19.Tan C, Liu X, Chen J. Microarray analysis of the molecular mechanism involved in Parkinson's disease. Parkinsons Dis. 2018;2018:1590465. doi: 10.1155/2018/1590465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil-Martinez AL, et al. Identification of differentially expressed genes profiles in a combined mouse model of Parkinsonism and colitis. Sci. Rep. 2020;10:13147. doi: 10.1038/s41598-020-69695-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, James M, Middleton FA, Davis RL. Transcriptional analysis of multiple brain regions in Parkinson's disease supports the involvement of specific protein processing, energy metabolism, and signaling pathways, and suggests novel disease mechanisms. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;137B:5–16. doi: 10.1002/ajmg.b.30195. [DOI] [PubMed] [Google Scholar]
- 22.Durrenberger PF, et al. Inflammatory pathways in Parkinson's disease; A BNE microarray study. Parkinsons Dis. 2012;2012:214714. doi: 10.1155/2012/214714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soreq L, Israel Z, Bergman H, Soreq H. Advanced microarray analysis highlights modified neuro-immune signaling in nucleated blood cells from Parkinson's disease patients. J. Neuroimmunol. 2008;201–202:227–236. doi: 10.1016/j.jneuroim.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner M, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Liu W, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J. Exp. Med. 2006;203:1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Capetillo O, Allis CD, Nussenzweig A. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 2004;199:1671–1677. doi: 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batchelor E, Loewer A, Mock C, Lahav G. Stimulus-dependent dynamics of p53 in single cells. Mol. Syst. Biol. 2011;7:488. doi: 10.1038/msb.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu W, Feng Z, Levine AJ. The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer. 2012;3:199–208. doi: 10.1177/1947601912454734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robles AI, Bemmels NA, Foraker AB, Harris CC. APAF-1 is a transcriptional target of p53 in DNA damage-induced apoptosis. Cancer Res. 2001;61:6660–6664. [PubMed] [Google Scholar]
- 33.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Z, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 35.Tatton WG, Chalmers-Redman R, Brown D, Tatton N. Apoptosis in Parkinson's disease: Signals for neuronal degradation. Ann. Neurol. 2003;53(Suppl 3):S61–S70. doi: 10.1002/ana.10489. [DOI] [PubMed] [Google Scholar]
- 36.Levy OA, Malagelada C, Greene LA. Cell death pathways in Parkinson's disease: Proximal triggers, distal effectors, and final steps. Apoptosis. 2009;14:478–500. doi: 10.1007/s10495-008-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair VD, McNaught KS, Gonzalez-Maeso J, Sealfon SC, Olanow CW. p53 mediates nontranscriptional cell death in dopaminergic cells in response to proteasome inhibition. J. Biol. Chem. 2006;281:39550–39560. doi: 10.1074/jbc.M603950200. [DOI] [PubMed] [Google Scholar]
- 38.Liu D, Hou P, Liu Z, Wu G, Xing M. Genetic alterations in the phosphoinositide 3-kinase/Akt signaling pathway confer sensitivity of thyroid cancer cells to therapeutic targeting of Akt and mammalian target of rapamycin. Cancer Res. 2009;69:7311–7319. doi: 10.1158/0008-5472.CAN-09-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanhaesebroeck B, Vogt PK, Rommel C. PI3K: From the bench to the clinic and back. Curr. Top. Microbiol. Immunol. 2010;347:1–19. doi: 10.1007/82_2010_65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffin RJ, et al. Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer's disease pathology. J. Neurochem.. 2005;93:105–117. doi: 10.1111/j.1471-4159.2004.02949.x. [DOI] [PubMed] [Google Scholar]
- 41.Colin E, et al. Akt is altered in an animal model of Huntington's disease and in patients. Eur. J. Neurosci. 2005;21:1478–1488. doi: 10.1111/j.1460-9568.2005.03985.x. [DOI] [PubMed] [Google Scholar]
- 42.Rai SN, et al. The role of PI3K/Akt and ERK in neurodegenerative disorders. Neurotox. Res. 2019;35:775–795. doi: 10.1007/s12640-019-0003-y. [DOI] [PubMed] [Google Scholar]
- 43.Miki Y, et al. Alteration of autophagy-related proteins in peripheral blood mononuclear cells of patients with Parkinson's disease. Neurobiol. Aging. 2018;63:33–43. doi: 10.1016/j.neurobiolaging.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Ha JY, et al. Tnfaip8 l1/Oxi-beta binds to FBXW5, increasing autophagy through activation of TSC2 in a Parkinson's disease model. J. Neurochem. 2014;129:527–538. doi: 10.1111/jnc.12643. [DOI] [PubMed] [Google Scholar]
- 45.Karabiyik C, Lee MJ, Rubinsztein DC. Autophagy impairment in Parkinson's disease. Essays Biochem. 2017;61:711–720. doi: 10.1042/EBC20170023. [DOI] [PubMed] [Google Scholar]
- 46.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 47.Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perier C, et al. Two molecular pathways initiate mitochondria-dependent dopaminergic neurodegeneration in experimental Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8161–8166. doi: 10.1073/pnas.0609874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat. Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 50.Soker S, Fidder H, Neufeld G, Klagsbrun M. Characterization of novel vascular endothelial growth factor (VEGF) receptors on tumor cells that bind VEGF165 via its exon 7-encoded domain. J. Biol. Chem. 1996;271:5761–5767. doi: 10.1074/jbc.271.10.5761. [DOI] [PubMed] [Google Scholar]
- 51.Jin K, et al. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11946–11950. doi: 10.1073/pnas.182296499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wada K, et al. Expression levels of vascular endothelial growth factor and its receptors in Parkinson's disease. NeuroReport. 2006;17:705–709. doi: 10.1097/01.wnr.0000215769.71657.65. [DOI] [PubMed] [Google Scholar]
- 53.Sawada M, Imamura K, Nagatsu T. Role of cytokines in inflammatory process in Parkinson's disease. J. Neural Transm. Suppl. 2006 doi: 10.1007/978-3-211-45295-0_57. [DOI] [PubMed] [Google Scholar]
- 54.McGeer PL, Itagaki S, Boyes BE, McGeer EG. Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson's and Alzheimer's disease brains. Neurology. 1988;38:1285–1291. doi: 10.1212/wnl.38.8.1285. [DOI] [PubMed] [Google Scholar]
- 55.Hirsch EC, Hunot S. Neuroinflammation in Parkinson's disease: A target for neuroprotection? Lancet Neurol. 2009;8:382–397. doi: 10.1016/S1474-4422(09)70062-6. [DOI] [PubMed] [Google Scholar]
- 56.Herrera AJ, Castano A, Venero JL, Cano J, Machado A. The single intranigral injection of LPS as a new model for studying the selective effects of inflammatory reactions on dopaminergic system. Neurobiol. Dis. 2000;7:429–447. doi: 10.1006/nbdi.2000.0289. [DOI] [PubMed] [Google Scholar]
- 57.Tansey MG, Goldberg MS. Neuroinflammation in Parkinson's disease: Its role in neuronal death and implications for therapeutic intervention. Neurobiol. Dis. 2010;37:510–518. doi: 10.1016/j.nbd.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duke DC, Moran LB, Pearce RKB, Graeber MB. The medial and lateral substantia nigra in Parkinson's disease: mRNA profiles associated with higher brain tissue vulnerability. Neurogenetics. 2007;8:83–94. doi: 10.1007/s10048-006-0077-6. [DOI] [PubMed] [Google Scholar]
- 59.Nam JH, et al. Interleukin-13/-4-induced oxidative stress contributes to death of hippocampal neurons in abeta1-42-treated hippocampus in vivo. Antioxid. Redox Signal. 2012;16:1369–1383. doi: 10.1089/ars.2011.4175. [DOI] [PubMed] [Google Scholar]
- 60.Kiyota T, et al. CNS expression of anti-inflammatory cytokine interleukin-4 attenuates Alzheimer's disease-like pathogenesis in APP+PS1 bigenic mice. FASEB J. 2010;24:3093–3102. doi: 10.1096/fj.10-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bok E, Cho EJ, Chung ES, Shin WH, Jin BK. Interleukin-4 contributes to degeneration of dopamine neurons in the lipopolysaccharide-treated substantia nigra in vivo. Exp. Neurobiol. 2018;27:309–319. doi: 10.5607/en.2018.27.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walsh JT, et al. MHCII-independent CD4+ T cells protect injured CNS neurons via IL-4. J. Clin. Investig. 2015;125:2547. doi: 10.1172/JCI82458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogelaar CF, et al. Fast direct neuronal signaling via the IL-4 receptor as therapeutic target in neuroinflammation. Sci. Transl. Med. 2018 doi: 10.1126/scitranslmed.aao2304. [DOI] [PubMed] [Google Scholar]
- 64.Perry VH. Innate inflammation in Parkinson's disease. Cold Spring Harb. Perspect. Med. 2012;2:a009373. doi: 10.1101/cshperspect.a009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brochard V, et al. Infiltration of CD4(+) lymphocytes into the brain contributes to neurodegeneration in a mouse model of Parkinson disease. J. Clin. Investig. 2009;119:182–192. doi: 10.1172/Jci36470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iba M, et al. Neuroinflammation is associated with infiltration of T cells in Lewy body disease and α-synuclein transgenic models. J. Neuroinflammation. 2020;17:1–14. doi: 10.1186/s12974-020-01888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reynolds AD, Stone DK, Mosley RL, Gendelman HE. Nitrated {alpha}-synuclein-induced alterations in microglial immunity are regulated by CD4+ T cell subsets. J. Immunol. 2009;182:4137–4149. doi: 10.4049/jimmunol.0803982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reynolds AD, Banerjee R, Liu J, Gendelman HE, Mosley RL. Neuroprotective activities of CD4+CD25+ regulatory T cells in an animal model of Parkinson's disease. J. Leukoc. Biol. 2007;82:1083–1094. doi: 10.1189/jlb.0507296. [DOI] [PubMed] [Google Scholar]
- 69.Kannarkat GT, Boss JM, Tansey MG. The role of innate and adaptive immunity in Parkinson's disease. J. Parkinsons Dis. 2013;3:493–514. doi: 10.3233/JPD-130250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith KA. Interleukin-2: Inception, impact, and implications. Science. 1988;240:1169–1176. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- 71.Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat. Immunol. 2007;8:1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- 72.Rosenkranz D, et al. Higher frequency of regulatory T cells in the elderly and increased suppressive activity in neurodegeneration. J. Neuroimmunol. 2007;188:117–127. doi: 10.1016/j.jneuroim.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 73.Huang Y, Liu Z, Wang XQ, Qiu YH, Peng YP. A dysfunction of CD4+ T lymphocytes in peripheral immune system of Parkinson's disease model mice. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2014;30:567–576. [PubMed] [Google Scholar]
- 74.Cen L, et al. Peripheral lymphocyte subsets as a marker of Parkinson's disease in a Chinese population. Neurosci. Bull. 2017;33:493–500. doi: 10.1007/s12264-017-0163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saunders JA, et al. CD4+ regulatory and effector/memory T cell subsets profile motor dysfunction in Parkinson's disease. J. Neuroimmune Pharmacol. 2012;7:927–938. doi: 10.1007/s11481-012-9402-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kraczyk B, Remus R, Hardt C. CD49d Treg cells with high suppressive capacity are remarkably less efficient on activated CD45RA− than on naive CD45RA+ Teff cells. Cell Physiol. Biochem. 2014;34:346–355. doi: 10.1159/000363004. [DOI] [PubMed] [Google Scholar]
- 77.Pajares M, I Roja A, Manda G, Bosca L, Cuadrado A. Inflammation in Parkinson's disease: Mechanisms and therapeutic implications. Cells. 2020 doi: 10.3390/cells9071687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borrageiro G, Haylett W, Seedat S, Kuivaniemi H, Bardien S. A review of genome-wide transcriptomics studies in Parkinson's disease. Eur. J. Neurosci. 2018;47:1–16. doi: 10.1111/ejn.13760. [DOI] [PubMed] [Google Scholar]
- 79.Lopez Gonzalez I, Garcia-Esparcia P, Llorens F, Ferrer I. Genetic and transcriptomic profiles of inflammation in neurodegenerative diseases: Alzheimer, Parkinson, Creutzfeldt–Jakob and tauopathies. Int. J. Mol. Sci. 2016;17:206. doi: 10.3390/ijms17020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grunblatt E, et al. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. J. Neural Transm. (Vienna) 2004;111:1543–1573. doi: 10.1007/s00702-004-0212-1. [DOI] [PubMed] [Google Scholar]
- 81.Kurvits L, et al. Transcriptomic profiles in Parkinson's disease. Exp. Biol. Med. (Maywood) 2020 doi: 10.1177/1535370220967325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu Y, et al. Exercise reverses dysregulation of T-cell-related function in blood leukocytes of patients with Parkinson's disease. Front. Neurol. 2019;10:1389. doi: 10.3389/fneur.2019.01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang F, Wu Q, Sun S, Bi G, Guo L. Identification of potential diagnostic biomarkers for Parkinson's disease. FEBS Open Bio. 2019;9:1460–1468. doi: 10.1002/2211-5463.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chi LM, Wang LP, Jiao D. Identification of differentially expressed genes and long noncoding RNAs associated with Parkinson's disease. Parkinsons Dis. 2019;2019:6078251. doi: 10.1155/2019/6078251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mutez E, et al. Transcriptional profile of Parkinson blood mononuclear cells with LRRK2 mutation. Neurobiol. Aging. 2011;32:1839–1848. doi: 10.1016/j.neurobiolaging.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 86.Infante J, et al. Comparative blood transcriptome analysis in idiopathic and LRRK2 G2019S-associated Parkinson's disease. Neurobiol Aging. 2016;38(214):e211–214 e215. doi: 10.1016/j.neurobiolaging.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 87.Soreq L, et al. Deep brain stimulation induces rapidly reversible transcript changes in Parkinson's leucocytes. J. Cell. Mol. Med. 2012;16:1496–1507. doi: 10.1111/j.1582-4934.2011.01444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soreq L, Bergman H, Israel Z, Soreq H. Exon arrays reveal alternative splicing aberrations in Parkinson's disease leukocytes. Neurodegener. Dis. 2012;10:203–206. doi: 10.1159/000332598. [DOI] [PubMed] [Google Scholar]
- 89.Alieva A, et al. Involvement of endocytosis and alternative splicing in the formation of the pathological process in the early stages of Parkinson's disease. Biomed. Res. Int. 2014;2014:718732. doi: 10.1155/2014/718732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karlsson MK, et al. Found in transcription: Accurate Parkinson's disease classification in peripheral blood. J. Parkinson's Dis. 2013;3:19–29. doi: 10.3233/JPD-120159. [DOI] [PubMed] [Google Scholar]
- 91.Kobo H, et al. Down-regulation of B cell-related genes in peripheral blood leukocytes of Parkinson's disease patients with and without GBA mutations. Mol. Genet. Metab. 2016;117:179–185. doi: 10.1016/j.ymgme.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 92.Li T, Tang W, Zhang L. Monte Carlo cross-validation analysis screens pathway cross-talk associated with Parkinson's disease. Neurol. Sci. 2016;37:1327–1333. doi: 10.1007/s10072-016-2595-9. [DOI] [PubMed] [Google Scholar]
- 93.Pinho R, et al. Gene expression differences in peripheral blood of Parkinson's disease patients with distinct progression profiles. PLoS ONE. 2016;11:e0157852. doi: 10.1371/journal.pone.0157852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.