Graphical abstract
Keywords: SARS-CoV2, COVID-19, Immune response, Immunomodulation, Cytokines
Abstract
Emerging infectious diseases always pose a threat to humans along with plant and animal life. SARS-CoV2 is the recently emerged viral infection that originated from Wuhan city of the Republic of China in December 2019. Now, it has become a pandemic. Currently, SARS-CoV2 has infected more than 27.74 million people worldwide, and taken 901,928 human lives. It was named first ‘WH 1 Human CoV’ and later changed to 2019 novel CoV (2019-nCoV). Scientists have established it as a zoonotic viral disease emerged from Chinese horseshoe bats, which do not develop a severe infection. For example, Rhinolophus Chinese horseshoe bats harboring severe acute respiratory syndrome-related coronavirus (SARSr-CoV) or SARSr-Rh-BatCoV appear healthy and clear the virus within 2–4 months period. The article introduces first the concept of EIDs and some past EIDs, which have affected human life. Next section discusses mysteries regarding SARS-CoV2 origin, its evolution, and human transfer. Third section describes COVID-19 clinical symptoms and factors affecting susceptibility or resistance. The fourth section introduces the SARS-CoV2 entry in the host cell, its replication, and the establishment of productive infection. Section five describes the host’s immune response associated with asymptomatic, symptomatic, mild to moderate, and severe COVID-19. The subsequent seventh and eighth sections mention the immune status in COVID-19 convalescent patients and re-emergence of COVID-19 in them. Thereafter, the eighth section describes viral strategies to hijack the host antiviral immune response and generate the “cytokine storm”. The ninth section describes about transgenic humane ACE2 (hACE2) receptor expressing mice to study immunity, drugs, and vaccines. The article ends with the development of different immunomodulatory and immunotherapeutics strategies, including vaccines waiting for their approval in humans as prophylaxis or treatment measures.
1. Introduction
Emerging infectious diseases (EIDs, comprise diseases originating due to pathogenic microbes and have recently originated and entered in a given population for the first time or an infectious disease that has occurred previously, but increased incidence, impact and/or geographic location has already occurred or expected to increase in future with a given time period) always remain a threat to human life along with animals and plants on the planet [1]. The phrase “emerging diseases” has been in use in the scientific community since the early 1960s. However, the emergence of outbreaks of genital herpes (in 1970s, caused by herpes simplex virus -2 or HSV-2) and human immunodeficiency virus-1 (HIV-1) infection causing acquired immunodeficiency syndrome (AIDS, discovered in 1981 in patients having unusual cancer (Kaposi’s sarcoma) and opportunistic infections (pneumonia caused by Pneumocystis carinii), and the HIV-1 was first identified in 1983 by Luc Montagnier’s team at the Pasteur Institute in Paris) raised the concern about EIDs and the use of the term became popular worldwide [2], [3], [4], [5]. Luc Montagnier and Françoise Barré-Sinoussi from his team won the 2008 Nobel Prize in physiology and medicine for isolating and characterizing the HIV-1 for the first time.
In 2004, Woolhouse and Gaunt have noted four major characteristics to describe most EIDs: (1) causal organism is a RNA virus, (2) disease is caused by the pathogen having a non-human or animal reservoir, (3) pathogen having a wide host range, and (4) have a potential for human-to-human transmission [6]. The details of factors, including pathogen-derived, environment derived, human derived, political, and other factors (bioterrorism and technology) have been described in detail somewhere else [7]. The emergence of Ebola virus (although first discovered in 1947 near Ebola river, Democratic Republic of Congo) infection in 2013–2015 in West Africa that started in a rural setting of south-eastern Guinea, spread to urban area and within weeks crossed the border, and became a global epidemic in months [8]. The discovery of Bombali virus (BOMV) in African free-tailed bats in Sierra Leone [(little free-tailed (Chaerephon pumilus) and Angolan free-tailed (Mops condylurus)] indicates the bat origin of the Ebola virus [9]. Zika virus (a Flavivirus from Aedes aegypti, discovered in Uganda in 1947) infection outbreak occurred more recently in 2013–2014 in French Polynesia, in May 2015 in Brazil, which moved to Americas in 2016 infecting more than twenty countries of the continent [10], [11].
The 1918 influenza pandemic caused by the H1N1 influenza virus with genes of avian origin comprises the largest epidemic of the last or 20th century with an infection of more than 500 million people. The number of deaths associated with H1N1 influenza worldwide comprise approximately 50 million with ≥675,000 death in the USA alone [12], [13]. However, the exact origin of H1N1 pandemic is unknown [14]. The 1957–1958 global pandemic due to H2N2 influenza virus (emerged in the East Asia and triggered Asian flu) killed 1.1 million people around the world and the USA alone had 116,000 deaths [15], [16]. The 1968 H3N2 influenza A pandemic emerged in September 1968 in the USA killing over 1 million people worldwide and about 100, 000 in the USA [12]. In 1997 the first human infection with avian H5N1 was reported in Hong Kong. On 15th April 2009, a new H1N1 influenza virus detected in California, USA and world health organization (WHO) on 11th June 2009 declared new H1N1 pandemic. According the center for disease control (CDC) at least 1 million people infected with 2009 H1N1 pandemic in the USA.
Bats serve as crucial animal reservoirs for EIDs (Marburg virus infection, Nipah virus infection, Rabies virus infection, Ebola virus infections, severe acute respiratory syndrome or SARS, and Middle Eastern respiratory syndrome or MERS) in humans [17], [18]. SARS originated from Guangdong district in China in year 2002–2003 (spread to thirty countries worldwide) due to SARS-CoV and MERS first reported in a hospital in Jeddah, Saudi Arabia with the death of the patient in 2012 due to MERS-CoV that spread to twenty seven countries worldwide causing an epidemic are [19], [20]. Both, these viruses belong to the Coronaviridae family and are betacoronaviruses (βCoVs). The emergence of SARS-CoV2 infection in the December 2019, in the Wuhan city of Hubei district of the Republic of China has led to the modern day pandemic infecting more than 27.74 million people around the world [21]. These EIDs are very challenging in terms of identifying their primary or secondary reservoirs, control, and treatment. The SARS-CoV2 has been categorized as a Group 4 virus on the basis of occupational hazards due to the non-availability of effective prophylaxis and antiviral treatment [22]. Hence, it becomes essential to study the SARS-CoV2 emergence, infection strategies, associated immune response, and factors controlling the pathogenesis to design and develop immune-based therapeutics against COVID-19.
2. Unsolved mystery of SARS-CoV2 origin and infection
The SARS-CoV2 infection is named as corona virus disease-19 (COVID-19) by the WHO. SARS-CoV2 is most closely related to a group of SARS-like coronaviruses (SARS-LCoVs, i.e. bat-SL-CoVZC45 and bat-SL-CoVZXC21) with the 89.1% similarity in the nucleotide sequence (nts). SARS-CoV2 protein sequence has also a high degree of similarity with bat-SL-CoVZXC21 and SARS-CoV [23]. SARS-LCoVs (bat-SL-CoVZC45 and bat-SL-CoVZXC21) discovered in bats in the eastern Zhoushan of eastern China belong to βCoV genus and the subgenus Sarbecovirus [24], [25]. Of note, severe acute respiratory syndrome-related coronavirus (SARSr-CoV) in Rhinolophus Chinese horseshoe bats (SARSr-Rh-BatCoV) causes an acute, but a self-limiting infection, which serve as a primary reservoir for recombination between strains from different geographical locations within a reachable foraging range [26]. This needs to establish for SARS-CoV2 evolution. SARS-CoV2 is closely related to SARS-CoV, but is also distinct from SARS-CoV [27]. For example, SARS-CoV2 has a higher binding affinity for ACE2 receptor than SARS-CoV, and the monoclonal antibodies (mAbs) raised against SARS-CoV receptor binding domain (RBD) do not bind effectively to SARS-CoV2 S-protein RBD [28]. This indicates the limited cross-reactivity of mAbs raised against SARS-CoV S-protein RBD with SARS-CoV2 S-protein RBD. Hence, the bioinformatics tools-based prediction for the RBDs of SARS-CoV and SARS-CoV2 as epitopes and the generation of B cell immune response needs further investigations. The two novel bat CoVs (RsSHC014 and Rs3367) from Chinese horseshoe bats (family: Rhinolophidae) in Yunnan, China are more closely related to SARS-CoV in their RBD of the spike (S) protein [29]. The isolated live SL-CoV (bat SL-CoV-WIV1) from bat faecal samples in Vero E6 cells have 99.9% sequence identity to Rs3367, and use ACE2 receptors to enter the cell and cause infection in humans, civets, and Chinese horseshoe bats [29].
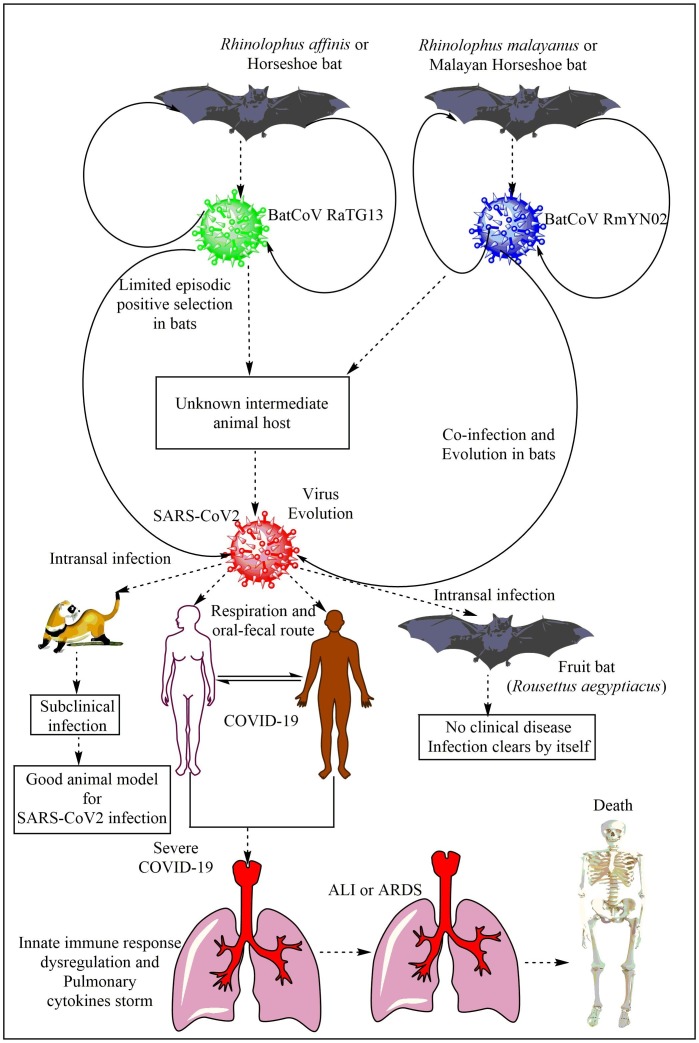
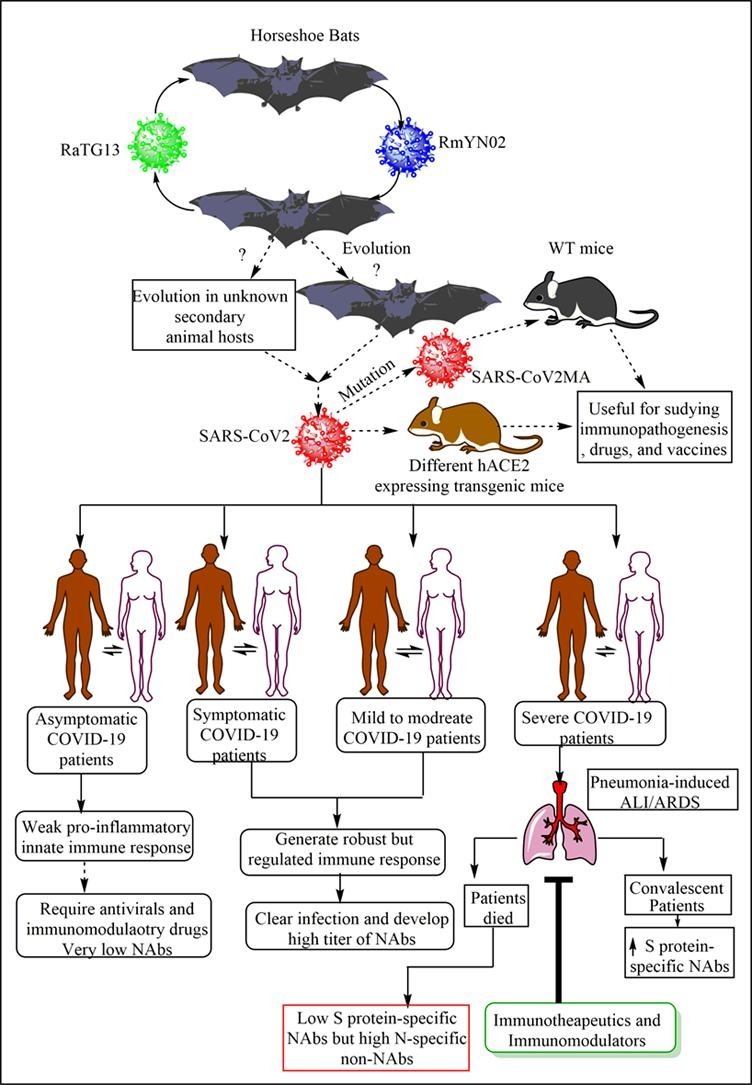
A bat CoV RaTG13, a SARS-related CoV (SARS-rCoV)] found in Rhinolophus affinis from Yunnan province shares 96.2% overall genome sequence identity with the SARS-CoV2 (Fig. 1 ) [30]. And, this 4% difference between the genome sequences of RaTG13-CoV and SARS-CoV2 may involve decades of genetic evolution, which may have occurred during the passage and survival in the secondary host that may have spread the infection in humans. However, SARS-CoV2 may not be directly evolved from the bat RaTG13-CoV due to the difference in the amino-acid (AA) sequence divergence in the RBD, despite angiotensin converting enzyme 2 (ACE2) receptor usage by both to enter the cell for initiating the infection [31]. Also, the RaTG13-CoV binds to the ACE2 receptor in human cells without spike (S) protein cleavage into S1 and S2 due to the absence of furin cleavage site in its RBD [32]. Metagenomic analysis of 227 samples obtained from the bats (Rhinolophus malayanus or Malayan Horseshoe bat) of the Yunnan province of the China between May and October 2019 has identified another bat-derived CoV called RmYN02 (Fig. 1) [33]. RmYN02 shares 93.3% nts identity with SARS-CoV-2 at the scale of the complete virus genome and 97.2% identity in the lab gene (21,285 nucleotides), indicating it as the closest relative of SARS-CoV2 reported to date [33]. RmYN02 exhibits far lower sequence identity to SARS-CoV2 in the S gene (nucleotide 71.8%, AA 72.9%), compared to 97.4% AA similarity between RaTG13 and SARS-CoV2. However, RmYN02 exhibits a low AA sequence identity (only 62.4%) to SARS-CoV2 in its RBD, and may not bind to the human ACE2 to cause infection. A similarity plot, called Simplot has also showed that the RmYN02 is more similar to the SARS-CoV2 than RaTG13 in most genome regions [33]. Of note, the multiple AAs insertion at the junction site of the S1 and S2 subunits of the S protein characterized the RmYN02. Hence, insertion of AAs may occur naturally in βCoVs to which SARS-CoV2 belongs. Thus we know that the COVID-19 is of bat origin but we do not have its confirmatory proof. We do not have a confirmatory report for the transmission of SARS-CoV2 from bats to humans.
Fig. 1.
Origin of SARS-CoV2 infection or COVID-19 in humans. The BatCoV RaTG13 and RmYN02 are most closely related to SARS-CoV2. However, they are not able to cause infection in their natural form. So different theories have emerged, including the use of secondary animal hosts (which are not confirmed yet) by the virus to evolve there to cause infection to humans. However, another theory suggest the RaTG13 and RmYN02 evolved or undergone positive episodic selection in horseshoe bats. Fruit bats and ferret can also get the SARS-CoV2 infection through intranasal route. However, fruit bats do not develop visible symptoms and clear the infection, whereas ferrets can serve an animal model for asymptomatic SARS-CoV2 infection of the humans as they develop subclinical symptoms. The severe infection with SARS-CoV2 in humans causes severe pneumonia and the associated ALI/ARDS through generating the cytokine storm to cause death of the patient.
Some researchers have also suggested its transmission to humans from snakes [Bungarus multicinctus (the many-banded krait) and Naja atra (the Chinese cobra)] and pangolins via serving as secondary or reservoir hosts [34], [35], [36]. However, we do not have any evidence of SARS-CoV2 transmission from snakes to humans. SARS-CoV-2-like CoV (named Pangolin-CoV) isolated from dead Malayan pangolins is 91.02% and 90.55% identical to SARS-CoV2 and BatCoV RaTG13 at the whole genome level [36]. Another pangolin CoV, called pangolin-CoV2020 isolated from three sick Malayan pangolins does not provide the evidence of direct emergence of SARS-CoV2 from it [37]. Also, all the pangolin βCoVs isolated to date have no polybasic (furin-like) S1//S2 cleavage site in their S protein like SARS-CoV2 [38]. Both, Pangolin-CoV and RaTG13 have lost their putative furin recognition sequence motif at S1/S2 cleavage site, which is present in SARS-CoV2 [36]. Only SARS-CoV2 has an unique peptide called PRRA, which may be involved in the proteolytic cleavage of the spike protein by cellular proteases to impact host range and transmissibility and pangolin CoVs lack it [39]. Hence, the RatTG13-CoV is more closer to SARS-CoV2 than CoVs obtained from two pangolin samples (SRR10168377 and SRR10168378) [39]. Thus, hypotheses supporting genetic recombination, convergence, and adaptation for the SARS-CoV2 evolution from these known sources of virus are still the hot topic for debate and need further investigation.
A recent study has contrasted the role of positive selection and recombination in the Sarbecoviruses in horseshoe bats to SARS-CoV-2 evolution in humans [40]. The incidence of some positive selection in SARS-CoV2 may be attributed to recombination and sequencing artefacts. The purifying selection is also weaker in SARS-CoV2 than the Sarbecoviruses in horseshoe bats, but the purifying episodic selection are present in the bat virus lineage from SARS-CoV2 has emerged [40]. The bat virus RmYN02 has a recombinant CpG content in its S protein that indicates coinfection and evolution in bats without the involvement of other species (Fig. 1). This study suggests that non-human progenitor of SARS-CoV-2 is capable of human-human transmission as a result of its natural evolution in bats [40]. Another study indicates the divergence of SARS-CoV-2 from BatCoV RaTG13 through limited episodic positive selection process (Fig. 1) [41]. Thus natural selection occurred in bat CoVs, which infected humans through SARS-CoV2, but not in humans. Of note, SARS-CoV2 infects horseshoe bats intestinal epithelial cell (IEC) organoid in vitro and also replicates there [42]. However, in vivo infection of SARS-CoV2 infection in the horseshoe bats will be interesting to study as the natural presence of SARS-CoV2 and clinical infection or symptoms in horseshoe still needs experimental investigations.
A phylogenetic and phylodynamic analysis of SARS-CoV2 has indicated the time to the most recent common ancestor (TMRCA) and evolutionary rate of SARS-CoV-2 to be 12 November 2019 (95% BCI: 11 October 2019 and 09 December 2019) and 9.90 × 10−4 substitutions per site per year (95% BCI: 6.29 × 10−4–1.35 × 10−3) [43]. Another study based on Bayesian time-scaled phylogenetic analysis using the tip-dating method has estimated the TMRCA and evolutionary rate for SARS-CoV2 ranges from 22 to 24 November 2019 and 1.19 to 1.31 × 10−3 substitutions per site per year [39]. Another phylogenetic dating study has indicated that divergence dates between SARS-CoV2 and the bat sarbecovirus (RaTG13) reservoirs comprise as 1948 (95% highest posterior density (HPD): 1879–1999), 1969 (95% HPD: 1930–2000) and 1982 (95% HPD: 1948–2009) [44]. This study indicates that the lineage giving rise to the current SARS-CoV2 has been circulating without any notice in bats for decades. Thus horseshoe bats are the primary reservoirs for SARS-CoV2 lineage, and SARS-CoV2 and RaTG13 may have single ancestral lineage. A recent study has indicated that SARS-CoV2 can also infect fruit bats (Rousettus aegyptiacus) through intranasal route, who develop transient infection without any clinical sign and may serve as reservoir host (Fig. 1) [45]. Oral and fecal shedding of the virus has also been reported from infected fruit bats. Of note, despite the virus detection by RT-qPCR, no viral antigen (Ag) is detectable in the lungs of these fruit bats. Ferrets develop SARS-CoV2 subclinical infection resembling to asymptomatic human patients showing efficient virus replication in the nasal cavity and infection transmission (Fig. 1). Ferrets have also not shown viral Ag in their lungs, but it is seen in their nasal cavity, nasal respiratory and olfactory epithelium, and olfactory epithelium of the vomero-nasal organ. Thus ferrets may prove beneficial animal model for SARS-CoV2 infection and testing the efficacy of vaccines or antiviral drugs (Fig. 1). The intranasal inoculation of the SARS-CoV2 does not produce infection in pigs and chickens, and they remain healthy and SARS-CoV2 free [45]. The ACE2 in chickens has alteration in its three AAs residues out of five AAs ((Lys31Glu, Glu35Arg, and Met82Arg), which does not support the SARS-CoV2 binding and entry in the host cells.
We still do not know the exact origin of the SARS-CoV2 and it has created a panic around the whole world by causing overwhelming hospitalization of the affected people, death, fear, psychological stress, and social stigma in both COVID-19 infected and non-infected people, and financial crisis in the major economies of the world. Despite not knowing the exact source of its transmission to humans, we know that it is an enveloped βCoV of Coronaviridae family with single stranded (ss), positive sense RNA genome (30 kb long) encoding four structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), which is foreign to the immune system. It has 11 genes in total with 11 open reading frames (ORFs,) described somewhere else [46]. For example, ORF1ab and ORF2 (S protein), ORF3a and ORF4 (E protein), ORF5 (M protein), ORF6, ORF7a, ORF7b, ORF8, and ORF9 (N protein), and ORF10 codes for ORF10 protein [46]. ORF7 is not essential for SARS-CoV2 replication in vitro [47]. A SARS-CoV2 variant with 382-nucleotide deletion (Δ382) truncates the ORF7b, which deletes ORF8-transcription-reegulatory sequence to eliminate ORF8 transcription has caused infection in January and February 2020 in Singapore [48]. Thus, this variant has successfully caused infection in the early phase of the epidemic in Singapore, but it disappeared after March 2020. Travellers returning to Taiwan from Wuhan, China in early February 2020 also had the identical SARS-CoV2 Δ382 variant, whereas SARS-CoV2 different variants with different deletion in the ORF8 have been reported in Australia (Δ138), Bangladesh (Δ345), and Spain (Δ62) [48]. The SARS-CoV2 Δ382 variant is associated with the milder phenotype of COVID-19 with less pro-inflammatory cytokine response and less incidence of hypoxia in patients [48]. Hence, ORF8 inhibition in severe COVID-19 patients may prove helpful to decrease the severity and mortality among patients.
Out of 4 structural proteins, S protein serves as a major protective antigen, which elicits highly potent neutralizing antibodies (NAbs), 16 non-structural proteins (Nsp1-Nsp16), and many other accessory proteins also impact host immune response [46], [49]. Of note, S proteins of SARS-CoV and SARS-CoV2 share 75% homology in AA sequence [47]. Table 1 mentions the functions of different SARS-CoV2 proteins. The emergence of SARS-CoV-2 variant carrying the S protein AA change D614G (an A-to-G nucleotide mutation at position 23,403 in the Wuhan reference strain that appeared first in the Europe) has become most prevalent in this global pandemic of COVID-19 and its recurrent pattern (G614) has increased at multiple geographic levels, including national, regional, and municipal [50]. The D614-G614 transition has occurred asynchronously in different regions worldwide with a starting from the Europe to North America and Oceania, and then to Asia [50]. The viral shift from D614 to G614 has also occurred in even local epidemics (Wales, Nottingham, Spain, New South Wales, China, Japan, Hong Kong, and Thailand) with established D614 infection, indicating the fitness advantage of G614.
Table 1.
Different proteins of SARS-CoV2 and their function in virus life cycle (replication, assembly, and pathogenesis).
| SARS-CoV2 proteins | Functions |
|---|---|
|
Expressed as polyprotein comprising of 16 Nsps (mentioned below) |
|
Binds to host 40S ribosome and promotes host cell RNA degradation, Blocks RIG-1-dependent antiviral immune response |
|
Binds to host cell prohibitin 1 and 2 (PHB1 and PHB2) to disrupt host cell environment |
|
|
|
Binds to Nsp3 and helps in virus replication |
|
Cleaves at 11 sites of (3C-like proteinase) NSP polyprotein |
|
|
|
Forms complex with Nsp8 and Nsp12 to yield NSP8 RNA polymerase activity |
|
|
|
Interacts with DEAD-box RNA helicase 5 (DDX5) of host cell to increase viral replication |
|
Activates NSP14 and NSP16, which are methyltransferases |
|
Unknown |
|
RNA-dependent RNA polymerase, forms complex with Nsp7 and Nsp8 |
|
|
|
Exerts 3′-5′ exoribonucleaseactivity and N7-methyltransferase activity |
|
|
|
Acts as a 2′‑O‑Ribose‑Methyltransferase and methylates the 2′-hydroxy group of adenine using S-adenosylmethionine (SAM) as the methyl source |
|
Recognizes and binds to ACE2 receptor of the host cells to enter for replication and productive infection |
|
|
|
|
|
|
|
Interacts with Nsp8 to promote RNA polymerase activity |
|
Accessory protein acting as a type 1 trans-membrane protein |
|
Accessory protein, localizes to Golgi apparatus, may help in virus assembly |
|
Comprises of ORF8a and ORF8b, binds to IRF association domain (IAD) of IRF3 via ORF8b to inhibit type 1 IFNs release |
|
Interacts with NLRX1 and inhibits RLR and NLRP3-mediated innate immune response, but promotes TLR signaling-dependent pro-inflammatory immune response |
|
Unknown |
|
|
The G614 variant grows to a higher titre as a pseudovirion and in infected individuals it is linked with low RT-PCR cycle threshold, suggesting its higher levels in the upper respiratory tract, but no association with disease severity [50]. Thus G614-bearing viruses have significantly higher infectious titers (2.6- to 9.3-fold increase) than their D614 counterparts as G614 increase the S protein stability and membrane incorporation preventing S1 shedding [50], [51]. Of note, S protein with G614 does not bind ACE2 more efficiently than S protein having D614 [51]. Hence, SARS-CoV2 with S protein having G614 transmit more easily due to the increased S protein stability without any change in the tendency to infect the host cell as compared to the virus with S protein having D614 [50], [51]. This is because D614G mutation does not affect TMPRSS2 activity required for virus entry along with ACE2. Hence, this study indicates changes crucial for understanding the mechanisms of virus infection, including the continuing surveillance of S protein mutations to design immunological interventions. This is because S protein is responsible for establishing COVID-19 infection and NAbs developing against S protein RBDs provide protection to the host. Of note, SARS-CoV2 sequence has a very low diversity, but still natural selection may act upon rare but favourable mutations [52]. SARS-CoV2 has some evidence of seasonal waning and the persistence of current pandemic may enable immunologically relevant mutations in the virus [53]. For example, antigenic drift has been seen in influenza viruses and SARS-CoV, but not seen in SARS-CoV2 yet. However, SARSCoV2 can acquire mutations for its fitness and immune escape [50]. This study needs further investigation. Thus along with different viral proteins and their variants, different host genetic factors controlling immune response play a significant role in SARS-CoV2 and other related CoVs infections [54]. Hence, host immune system plays a significant role in the pathogenesis and outcome of the infection, including the determination of the host resistance and susceptibility to the infection that will be discussed in later sections.
3. Clinical picture of SARS-CoV2 infection or COVID-19, including in pregnant women and pregnancy outcome
The symptoms of COVID-19 in adult patients include fever, dry cough, malaise, myalgia, vomiting, diarrhea, and abdominal pain. Severe COVID-19 patients also show neurological symptoms (headache, loss of taste and smell, and vision impairment etc.), described in detail somewhere else [19], [55], [56]. Ageusia (loss of taste) occurs specifically to the COVID-19 patients as compared to other flu-like upper respiratory infections [57]. Loss of taste in severe COVID-19 patients may be explained on the basis of ACE2 receptors expression on tongue epithelial cells (TECs), which can be infected and may cause pro-inflammatory damage due to increased pro-inflammatory cytokines. Anosmia (loss of smell) in patients with COVID-19 may be correlated to the ACE2 expression in the nasal epithelia and their infection with SARS-CoV2 (Fig. 2 ). SARS-CoV2-mediated ultrastructural remodeling in both apical and basal sides of human airway epithelial cells (HAECs) of nasal and bronchial origin occurs at 48 h post infection [58]. The increase in the virus replication and virus number in HAECs reduces the epithelial integrity at 48 h post infection [58]. The nasal HAECs at 48 h post infection contain high amount of Nsp14 RNA. Also, the SARS-CoV2 may infect ACE2 and TMPRSS2 expressing supporting cells and vascular pericytes of the olfactory epithelium and bulb causing loss of smell (Fig. 2) [59]. However, neurons of olfactory epithelium and bulb do not express ACE2. The loss of smell occurs in response to the increase in the pro-inflammatory cytokines, including TNF-α in the olfactory epithelium [60]. However, IL-1β levels remain same in the control and infected patients with anosmia. Neuroleptic malignant syndrome (NMS) has also been reported in severe COVID-19 patient [61]. NMS is characterized by severe rigidity, tremor, fever, altered mental status, autonomic dysfunction, and elevated serum creatinine phosphokinase and white blood cell (WBC) count [62].
Fig. 2.
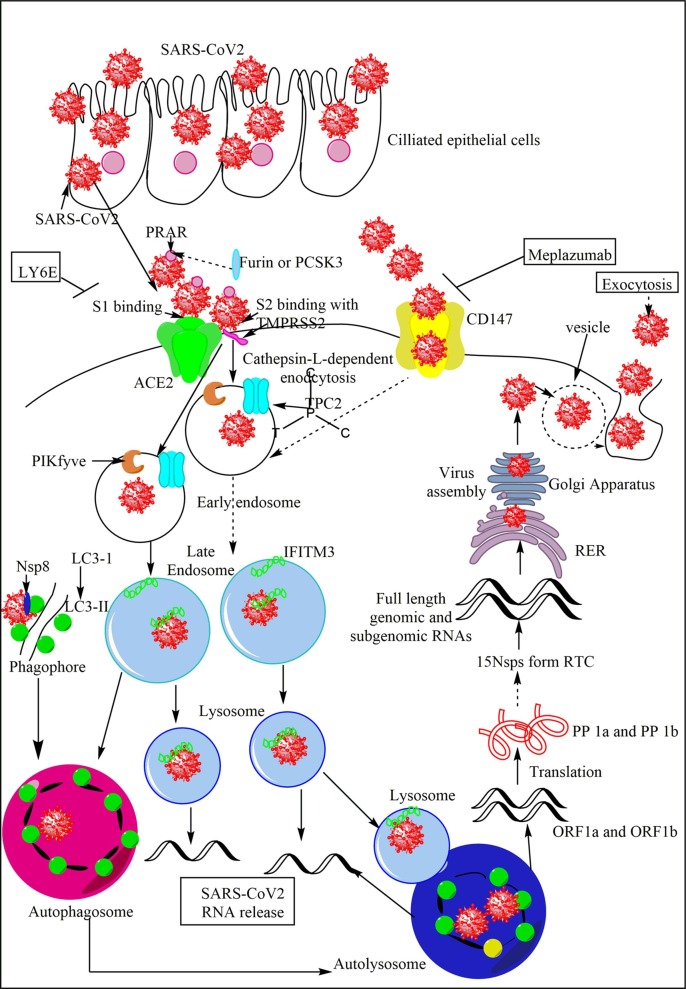
Entry, replication, and productive infection establishment by the SARS-CoV2 in humans. The SARS-CoV2 enters and infects the ciliated epithelial cells (nasal or pulmonary epithelial cells primarily) via the interaction of its S protein with the ACE2 receptors of the host cells. The interaction is mediated by S1 protein that generates due to cleavage of S protein into S1 and S2 by the furin or PCSK3. The furin acts on the PRAR, a dibasic furin binding site at the S protein. The S2 protein bins to the TMPRSS, which further increases the SARS-CoV2 entry in the host cell. SARS-CoV2 also enters the cell through CD147 as the mAb, meplazumab against it inhibits the severity and symptoms of COVID-19. SARS-CoV2 and ACE2 interaction promotes its cellular endosomal entry via cathepsin-L-dependent endocytosis. TPC2 on early endosome and lysosome, and PIKfyve on early endosome also play a crucial role in virus entry in the endosome via maintaining endolysosomal homeostasis. The IFITM3 on endosomes fuse with the SARS-CoV2 to enhance its trafficking to the lysosomes. The endosomes fuse with lysosomes to form endolysosomes or autolysosomes containing the virion. On the other hand, the interaction between LC3-II of the phagophore and Nsp8 forms the autophagosome containing virus. The autophagosome fuses with lysosomes to form autophagolysosomes or autolysosomes. The ssRNA of ORF1a and ORF1b release in the cytosol and translate into polyprotein 1a (PP1a) and PP1b synthesizing 15Nsps, which form RTC. The RTC forms full length genomic and subgenomic RNAs, which form the virion that via endoplasmic reticulum (ER) to Golgi apparatus ERGIC. The virion in the vesicle comes out form the cell via exocytosis.
Pneumonia is one of the major symptoms of the COVID-19 causing acute lung injury (ALI) or acute respiratory distress syndrome (ARDS) to put the patient on the respiratory support system (ventilator). Chest radiographs of some patients show ground-glass lung changes, normal or lower than average WBC count, and platelet counts, hypoxemia, along with kidney and liver dysfunction [19]. The gastrointestinal symptoms (vomiting, diarrhea, nausea, and abdominal discomfort) occur in some COVID-19 patients first, before fever and cough indicating its feco-oral transmission [63]. The differentiated human gut enterocytes, including ileal absorptive enterocytes highly express ACE2 receptor and TMPRSS2 for SARS-CoV2 entry and promoting the productive infection and viral replication (Fig. 2) [64], [65]. Along with TMPRSS2, TMPRSS4 also helps in the SARS-CoV2 infection of the intestinal epithelial cells (IECs) [66]. A recent study has shown the SARS-CoV2 infection of human IECs [42]. The virus replicates in IECs and has been detected in the stool samples of COVID-19 patients with diarrhea [42]. However, the SARS-CoV2 virus released in the lumen of the colon becomes inactive in response to the human colonic fluid as infectious virus is absent in the stool samples of the patients [66]. Hence, the first appearance of gastrointestinal symptoms in patients without fever and cough indicates the oral transmission of the SARS-CoV2.
Male patients with COVID-19 show higher mortality rate as compared to their counterpart females, and the case fatality rate (CFR) in males is 1.7 times higher than that of females independent of age [67], [68], [69], [70]. In addition to the increased severity and mortality among males, they also comprise high (71.4%) recovery from severe COVID-19 [71]. The laboratory findings have suggested that male patients with moderate COVID-19 have higher plasma levels of CCL5 than female patients [72]. These males also show higher levels of non-classical monocytes (CD14−CD16+) in circulation than females and females show higher numbers of intermediate monocytes (CD14+CD16+). Hence, these males have increased number of CD14−CD16+ non-classical monocytes and CCL5, which are significantly correlated, but lower number of T cells in circulation as compared to female COVID-19 patients having increased number of intermediated monocytes. Female COVID-19 patients show a higher number of CD38+HLA-DR+ activated T cells (Both CD4+ and CD8+T cells) than male patients [72]. Also, females have higher number of TIM-3 and PD-1 positive exhausted or terminally differentiated CD4+ and CD8+T cells than male patients. This gender-based difference in T cells is most robust for CD8+T cells. However, this study has drawback regarding healthy donors (HDs) who were not age, body mass index (BMI), and underlying risk factors matched, but it has suggested the immunological difference between male and female may indicate the higher severity of the infection in males. Future studies will help to explore sex-based immunological differences in SARS-CoV2 pathogenesis.
Even the patient with blood group A are at high risk of developing severe COVID-19 in comparison to others and people with blood group O are more resistant to develop COVID-19 [73]. One can hypothesize the presence of anti-B antibodies (Abs) in patients with blood group A may be enhancing the virus immune escape or entry in the cells for its replication and productive infection. Whereas, people with the blood group O have both anti-A and anti-B Abs, which may be neutralizing the SARS-CoV2 due to cross-reactivity, we do not know yet now. Hence, this will be interesting to study in detail. We do not know the cause of anti-A Ab origin, but the anti-B Abs originate as a result to immunity to gut microbiota [74], [75]. The similar pattern (blood group O people were resistant to it and blood group A people were most susceptible) has also been observed in SARS-CoV infection-induced severe acute respiratory syndrome (SARS) 2002–2003 epidemic [76]. This is also true for malaria, for example, people with blood group A are more susceptible to severe malaria and people with blood group O are resistant to it due to the reduced rosetting (a virulent Plasmodium falciparum phenomenon associated with severe malaria [77], [78]. Hence, the relationship between COVID-19 infection and its severity with the person’s blood group is an interesting research direction.
The transplacental transmission or vertical transmission of COVID-19 from mother to the offspring has also been reported along with the infection in the placenta (syncytiotrophoblasts) and the macrophage infiltration [79], [80], [81], [82], [83], [84], [85]. The pregnant mother shows a high degree of viremia and the neonate born also, following the placental infection [82]. However, possibility of vertical transmission of SARS-CoV2 from a mildly symptomatic pregnant woman has also been indicated in a study [83]. Even a case of second trimester miscarriage of the pregnancy in a woman with COVID-19 has also been reported due to the transfer of the SARS-CoV2 the fetus through infected placenta [86]. Hence, SARS-CoV2 infection may transfer vertically to the new born from the infected mother. The patients dying with severe COVID-19 more commonly show dyspnoea, chest tightness, consciousness disorders, leucocytosis, and lymphopenia. COVID-19 patients also have dysregulated serum or plasma biochemical markers of the acute infection and inflammation (alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine, creatine kinase (CK), lactate dehydrogenase (LDH), ferritin, cardiac troponin I, N-terminal pro-brain natriuretic peptide, and D-dimer (increases due to increased plasmin levels and activity during hyperfibrinolysis) [19], [71], [87], [88]. Severe-COVID-19 patients have high serum ferritin levels [71]. The patients with COVID-19 also show an increased biliverdin (an oxidized form of bilirubin) plasma levels, indicating increase oxidative stress during disease, which decreases in the recovered patients [89]. Plasma 5-hydroxy-tryptophan (5-HT) levels also increase in the COVID-19 patients. Thus patients dying due to severe COVID-19 develop ARDS and sepsis along with other complications, including type 1 respiratory failure, acute cardiac injury, heart failure, alkalosis, hyperkalemia, acute kidney injury (AKI), thromboembolic complications, and hypoxic encephalopathy [71], [87]. For example, patients with severe or critical COVID-19 have a higher acute physiology and clinical health evaluation (APACHE) III score than moderate COVID-19 subjects [71].
The high APACHE III score in severe COVID-19 patients directly correlates with the increased age. On the other hand, children and infants do not develop severe COVID-19 [90]. For example, children younger than 10 years of age have accounted for only 1% of the total COVID-19 cases [91]. The author has described in details regarding the decreased susceptibility of children to severe COVID-19 over adults [19]. Females, young children, and patients with blood group O are more resistant to severe COVID-19. And, age, sex, pregnancy status, and blood group impact the immunological status of the host along with the existence of previous chronic diseases, including obesity, hyperlipidemia, hypertension, and other heart diseases, diabetes, cancer, lung diseases, including asthma, chronic obstructive pulmonary disease (COPD), cystic fibrosis, smoking habits etc. [19], [71], [92], [93]. People with impaired fasting glucose levels, type 1 and 2 diabetes mellitus develop severe COVID-19, require more hospitalization, and exhibit higher mortality than people with normal blood glucose level [94], [95], [96], [97]. For example, people with underlying pulmonary diseases and depending on their smoking status (current or former smokers) have high incidence of COVID-19 [71]. Immune status of the healthy host significantly varies from patients with senile life style and chronic inflammatory and metabolic diseases. Hence, the immune status of the host plays a crucial role in the SARS-CoV2 infection and its severity, described in detail in the following sections.
4. SARS-CoV2 entry in the different cell types and the infection pathogenesis
The S protein of the SARS-CoV2 recognizes ACE2 receptor of the host cells to enter. ACE2 is expressed on the nasal epithelia, airway epithelia, cornea, gastrointestinal tract (GIT), gall bladder, cholangiocytes (epithelial cells lining the bile duct), urinary tract, testes, cardio-vascular system, endothelial cells, oral mucosal epithelial cells (OMECs), including TECs [19], [98], [99], [100], [101], [102], [103]. In humans, respiratory epithelia lining the nasal epithelia contain low amount of ACE2, squamous epithelium lining oropharyngeal tonsillar tissue also express ACE2, and progressively reduced ACE2 expression occurs in the lower airway regions, which culminates to the minimum level in alveolar region [47]. Nasal epithelia express higher ACE2 levels than bronchial epithelial tissues, but it has a low TMPRSS2 expression [47]. The nasal goblet secretory cells co-express ACE2 and TMPRSS2 [65]. However, TMPRSS2 expression is higher in whole respiratory tract than ACE2.
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) also express ACE2 receptor through which SARS-CoV2 enter and replicate in cardiomyocytes [104]. The increased viral replication in cardiomyocytes and cytopathic effects induce their apoptotic death via caspase 3 (CASP3) activation, and cardiomyocytes cease to beat after 72 h of SARS-CoV2 infection [104]. The SARS-CoV2 infection to cardiomyocytes decreases ACE2 expression, but increases the CXCL2 expression along with different interleukins (ILs, IL-1β and IL-11), and antiviral protein called 2′-5′-oligoadenylate synthetase 3 (OAS3), which is induced by IFNs, and are activated by viral dsRNA [104]. The IL-11 is crucial for cardiovascular fibrosis and acts through binding to IL-11 receptor expressed on cardiac fibroblasts to activate non-canonical, ERK-dependent autocrine signalling to synthesize fibrogenic protein [105]. The OAS3 activates antiviral enzyme RNase L that inhibits viral and cellular RNAs restricting protein synthesis, and viral replication to prevent the infection spread [106]. However, SARS-CoV2 inhibits IFN generation so it should be novel to check the OAS3 at protein level in cardiomyocytes and other infected cells, including immune cells. The SARS-CoV2 infection in cardiomyocytes also reduces cardiomyocyte marker genes (TNNT2, encodes cardiac muscle troponin T and TNNC1, encodes troponin C1) and mitochondrial gene responsible for oxidative phosphorylation (OXPHOS) called creatine kinase M2 (CKMT2) [104]. Hence, SARS-CoV2 infection in cardiomyocytes changes their gene expression program regulating metabolism and immune response.
The epithelial cells of renal system (kidney epithelial cells (KECs), renal tubular cells (RTCs)) and male reproductive system (Leydig cells, seminal vesicles, and cells of seminiferous tubules, including spermatids) also express ACE2 receptor [19], [107], [108], [109]. In human female reproductive system ovaries, decidua, and uterus (epithelial cells) express ACE2 receptor [109], [110], [111]. Hence, these organs may also be infected with SARS-CoV2. A recent study has shown the negligible co-transcription of ACE2 and TMPRSS2 in human placenta throughout the pregnancy that minimally expresses the canonical cell-entry mediators for SARS-CoV-2 [112]. However, CD133+CD34+Lin−CD45− cells in human umbilical cord blood (UCB), classified as hematopoietic stem cells (HSCs) and endothelial progenitor cells (EPCs) express ACE2 and TMPRSS2 [113]. The cells called very small embryonic-like stem cells (VSELs) and HSCs interact with SARS-CoV2 through ACE2 receptor and activate NLRP3 inflammasome [113]. A higher ACE2 expression on OMECs explains feco-oral transmission of COVID-19 [19], [114]. Even human immune cells (monocytes, B cells, and T cells are susceptible to the active SARS-CoV2 infection and show virus replication [115]. The immunohistochemical analysis of lungs from severe COVID-19 patients has shown the presence of SARS-CoV2 infected immune cells. Also, the whole peripheral blood monocytes (PBMCs) get infected with SARS-CoV2 in vitro.
The ACE2 (a metallopeptidase with molecular weight 110 kDa) serves as a binding receptor and entry site of the SARS-CoV2 causing COVID-19 (Fig. 2) [116], [117]. Of note, SARS-CoV2 does not recognize mouse and rat ACE2. The S1 proteins of SARS-CoV2 bind to the ACE2 and its enzymatic activity cleaves S protein into S1 and S2 (Fig. 2). The S1 binds to the ACE2 and S2 activated by the host cell serine protease transmembrane protein serine 2 (TMPRSS2) induces membrane fusion to enter the cell through the cathepsin-L-dependent pathway (Fig. 2) [118], [119]. Thus, TMRSS2 enhances the virus entry in the cells and the infection spread in the lung alveolar cells. The presence of a multi-basic cleavage site (cleaved by host furin) in the S protein at S1/S2 site of the SARS-CoV2 is essential for infecting human lung epithelial cells or HAECs (Fig. 2) [120]. Hence, both TMPRSS2 and furin enhance the virus entry and replication in the cell, and cytopathology of SARS-CoV2 [47]. A mammalian two-pore channel, TPC2 (two-pore channel 2, a cation channel for endocytic membrane trafficking) in intracellular endosomes and lysosomes, and the kinase PIKfyve (Phosphatidylinositol 3-phosphate 5-kinase or Fab1 in yeast) synthesizing PI(3,5)P2 in early endosome required for endolysosomal homeostasis plays a significant role in SARS-CoV2 entry (Fig. 2) [121]. PIKfyve inhibition dysregulates endolysosomal functions, endosomal homeostasis, and autophagy [122], [123], [124], [125]. The SARS-CoV-2 receptor binding motif (RBM) of RBD plays a crucial role in S protein interaction with ACE2 [32]. RBM, which contains all residues interacting with ACE2 is present in the carboxy-terminal of the RBD [126]. Hence, RBD comprises of a core and the RBM [32]. The mutations in the RBD of S protein of SARS-CoV2 may enhance or decrease their binding capacity or affinity for ACE2. However, there is no evidence that these ACE2-affinity enhancing mutations may have been selected in current SARS-CoV-2 pandemic isolates [127]. Most AA mutations to the RBD are deleterious for RBD expression and activity that is ACE2 binding, but substantial number of mutations are well tolerated and even enhance ACE2 binding, including the one at ACE interface residues, which vary across SARS-CoV-related viruses [127]. The details of SARS-CoV2 S protein (RBM of the RBD) interaction with ACE2 has been discussed by the author somewhere else [19].
All cell types (including epithelial cells and immune cells) express CD147 that also serves as a potential SARS-CoV-2 receptor, suggesting another route for SARS-CoV2 infection [128], [129], [130]. The PBMCs isolated from children highly express CD147, and adolescents and adults express it at lower level [130]. Also, the CD147 expression increases with BMI and older age in the circulating immune cells, but not in barrier cells, including epithelial cells of the lungs [130]. Thus, older people and conditions with increased BMI (obesity, diabetes, and hypertension) are more prone to develop COVID-19. CD147 is slightly highly expressed in the human bronchial epithelial cells (HBECs) of chronic obstructive pulmonary disease (COPD) [130]. Azithromycin treatment in hospitalized COVID-19 patients decreases the viral load via blocking the SARS-CoV2 interaction with CD147 [131]. Meplazumab (a humanized mAb raised against CD147) has successfully treated COVID-19-associated pneumonia and along with improving the lymphopenia and C-RP levels (Fig. 2) [132].
Certain cytokines, including IL-1β and IFN-β increase the ACE2 expression in the alveolar epithelial cells (AECs), but they decrease the TMPRRS2 expression [47]. IL-13 (a crucial Th2 cytokine, which has anti-inflammatory action) increased in the lungs of asthmatic patients also decreases the ACE2 expression in AECs [47]. However, HBECs isolated from asthma patients show a higher TMPRSS2 expression raising the possibility of increased S protein cleavage into S1 and S2 [130]. Cigarette smoking increases the ACE2 expression in HBECs [130]. Hence, patients with previous pulmonary disorders (cystic fibrosis (CF), tuberculosis, viral infections (influenza, SARS or MERS), and asthma) may show different outcome of the COVID-19 than people not previously exposed to these infections or not having CF or asthma. The lymphocyte antigen 6 complex locus E (LY6E), an interferon-stimulated gene (ISG) restricts the SARS-CoV2 entry in the cell via S protein (Fig. 2) [133], [134], [135]. The increased TMRSS2 expression or amphotericin treatment neutralizing the interferon-induced transmembrane protein 3 (IFITM3)-mediated restriction of SARS-CoV entry do not affect the LY6E-mediated SARS-CoV2 entry restriction [133]. The IFITM3 levels increase in lung epithelial cells during COVID-19 [136]. IFITM3 fusion with the virus particles in the endosome enhances virus trafficking to lysosomes (Fig. 2) [137]. S-plamitoylation plays a crucial role in the IFITM3-mediated virus trafficking to the lysosome. A profound IFITM3 expression arrests internalized virus particles within endocytic vesicles and prevents the virus release in the cytosol [138], [139], [140].
COVID-19 patients have clusters of T cells and monocytes characterized by interferon-response genes (ISGs) such as IFN-α inducible protein 27 (IFI27 or p27, a hydrophobic mitochondrial protein regulating apoptosis and congenital immunity), IFITM3, or ISG15 [141]. The microtubule associated protein 1B light chain 3-II (LC3-II)-mediated internalization of SARS-CoV2 in the phagophore and, then into autophagosome also occurs [142]. The Nsp8 co-localizes with LC3-II in the phagophore [143]. The conversion of microtubule-associated protein 1B light chain 3-1 (LC3-I) to LC3-II involves the autophagy (Atg) protein complex (Atg4 + Atg3 + Atg7). The autophagosomes and lysosomes containing the virus fuse to form autolysosomes. The autophagy genes, including Atg5, Atg12, and LC3-I upregulate during SARS-CoV2 infection [144]. Atg5−/− mice show an aberrant CoV replication [145]. The CoV Nsp6 protein via LC3 fusion also induces the omegasome (The ER domains containing double FYVE-domain containing protein (DFCP1), which recruits ULK1, Atg14, Atg5, and LC3 proteins) formation, which matures into autophagosome that delivers LC3 and the CoV in the autophagolysosomes via lysosome fusion [146]. Hence, along with endosomes and lysosomes, autophagy also plays a crucial role in the SARS-CoV2 replication and infection process. Thus with the release and uncoating of viral nucleocapsid in the cytosol, the CoV replication starts with the ORF1a and ORF1b translation into polyprotein (PP) 1a and PP 1b (Fig. 2) [147].
The downstream of translation of ORF1a and ORF1b occurs through ribosomal frameshift mechanism. Subsequently, PP 1a and PP 1b are cleaved into at least 15 Nsps, which form a replication-transcription complex (RTC) (Fig. 2) [147]. Following replicase-polymerase assembly, the full-length positive strand of genomic RNA transcribes to form a full-length negative-strand template to synthesize new genomic RNAs and overlapping subgenomic negative-strand templates (Fig. 2) [147]. These subgenomic RNAs then transcribe and translate to form structural and accessory proteins [147]. The assembly of virion occurs once the helical nucleocapsid containing the genomic RNA interacts with other viral structural proteins (S, E, and M proteins) [147]. The CoV assembly completes with the budding of the helical nucleocapsid through membranes early in the secretory pathway from the endoplasmic reticulum to the Golgi intermediate compartment (ERGIC) (Fig. 2). Host proteins do not take part in this phase of the infection cycle.
The viral M protein interacts with various structural proteins, including S and E protein to assemble into a mature CoV. The M protein, a central player of CoV assembly, turns cellular membranes into workshops for virus, and host factors get together to synthesize new virus particles [148]. The interaction between M and structural proteins (E protein) generates the scaffold of virion envelope to induce the budding and release of the M protein-modified membrane with S protein assembled into spikes in the virus envelope [148], [149], [150]. Thereafter, the CoV moves to the vesicle for the exocytosis. The exocytosed virus is ready to infect new host cells. However, the detailed discussion of human CoV (hCoV), including SARS-CoV2 replication process and virion assembly is beyond the scope of the current review and has been discussed in detail somewhere else [151], [152], [153], [154]. More recently, a transition model from Nsp7-Nsp8 hexadecameric primase complex to the Nsp12-Nsp7-Nsp8 polymerase complex has also been proposed to understand CoV transcription and replication machinery [155].
The SARS-CoV2 infection of the host epithelial cell (Caco-2 cells) changes the phosphoproteome within 24 h post infection and main fractions of phosphopeptides, include modified serines (86.4%), threonine (13.4%), and tyrosine (0.2%) [156]. SARS-CoV2 infection of the host cells reshapes their splicing machinery and inhibition of splicing by pladeinolide B decreases SARS-CoV2 pathogenic effects. The SARS-CoV2 infection of the host epithelial cells increases growth factor receptor (GFR) signaling and phosphorylation of the whole pathway, including related components of cytoskeleton remodelling and receptor endocytosis [156]. The GFR signaling, amongst others, activates 1) the RAF/MEK/ERK MAPK signaling cascade and 2) integrates [via phosphoinositide 3 kinase (PI3K) and protein kinase B (AKT)] with mTORC1 signaling to regulating proliferation [156]. The PI3KB inhibitors (pictilisib and omipalisib) inhibit the SARS-CoV2 replication. Both, RAF inhibitor sorafenib and the dual RAF/MEK inhibitor RO5126766 decrease the cytopathic effect of the virus and decrease its replication. The same inhibitory effects of the GFR inhibition have been observed in another cell line (UKF-RC-2 cells) infected with SARS-CoV2. Hence, activation of GFR signaling during SARS-CoV2 infection increases its replication in host epithelial cells.
5. Immunology of the SARS-CoV2 infection or the COVID-19
The immune system plays a crucial role in the outcome of the SARS-CoV2 infection. The data from 63 hospitals involved in treating COVID-19 patients in the UK has identified a core peripheral blood immune signature indicating the enrichment with SARS-CoV2-specific Abs, plasmablasts, cytokines (IL-6, IL-8, and IL-10), chemokine (CXCL10 or IFN-γ-induced protein 10 or IP-10), highly cycling T cells and CD8+T cells with exhaustion phenotype (expressing T-cell immunoglobulin mucin-3 (TIM-3) and programmed cell death-1 or PD-1), altered B cells compartment, and depletion of αβ and γδT cells [157]. The increased plasmablasts correlate with the COVID-19 severity, but the dynamic B cell biology essentially contributes to the immunophenotype of all COVID-19 patients [157]. Also, the increased circulating IL-6 and IL-10 levels correlate well with the severity along with the chemokine IP-10 that is also highest in severe COVID-19 patients [157]. Both, IP-10 and IFN dynamics do not correlate concomitant C-reactive protein (CRP) measurements. People, who are q-PCR positive for SARS-CoV2 infection develop IgG anti-N, IgM anti-N, IgG anti-S1, and IgA anti-S1 Abs, which increase over two months after q-PCR diagnosis and remain at plateau over the next two months [158]. IgM anti-N Abs increase in concentration rapidly soon after COVID-19 diagnosis and they become undetectable after two months [158]. IgA anti-S1 Abs start to decrease after 1 month of diagnosis but remain detectable thereafter. IgG anti-N and anti-S1 antibody levels increase during the first 6 weeks after diagnosis and their number increases with the severity of the disease. Hence, the potency and the severity of immune response determines the following phenotype among COVID-19 patients: (1) Asymptomatic, (2) mild, (3) moderate, and (4) severe cases of COVID-19.
5.1. The immune response in asymptomatic COVID-19 patients
Asymptomatic COVID-19 cases cannot be differentiated on the basis of anatomy and extent of infection from symptomatic cases of the disease as 50% of asymptomatic COVID-19 patients show radiographic abnormalities, including ground-glass opacities on chest CT scans [159], [160]. Another study had shown approximately 50% patients of 634 people screened for COVID-19 were asymptomatic in Yokoyama, Japan, in a cruise ship hosting 3711 people [161]. Hence, immunological investigations become a crucial laboratory-based diagnosis to differentiate asymptomatic patients from symptomatic ones. Asymptomatic individuals show mild clinical and laboratory indications, and mild lung lesions indicating a low pro-inflammatory innate immune response generation than the symptomatic individuals (Fig. 3 ) [162]. For example, SARS-CoV2 positive asymptomatic individuals induce a weaker pro-inflammatory tissue damaging innate immune response (very low levels of eighteen pro- and anti-inflammatory cytokines) and the median virus shedding (the time period between first and last nasopharyngeal swab) duration among this group is nineteen days (interquartile range (IQR), 15–26 days) [163].
Fig. 3.
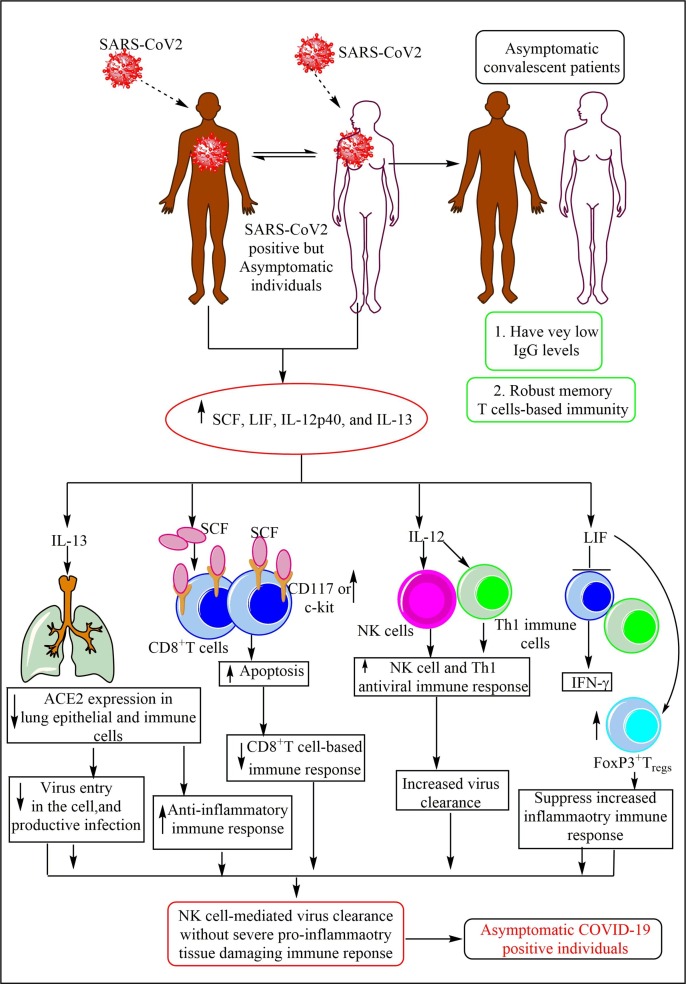
The immune response in the SARS-CoV2 positive asymptomatic patients. Asymptomatic patients are SARS-CoV2 infected but do not show the signs of the COVID-19. They produce a higher amount of IL-13, SCF, IL-12, and LIF. IL-13 decreases the ACE2 expression on pulmonary epithelial cells (PECs) and other immune cells therefore a decreased virus entry and replication occurs without the productive infection. SCF increases the apoptosis among CD8+ cytotoxic T cells that decrease cytotoxic and pro-inflammatory action against the virus. IL-12 increases the NK cell and T cell antiviral immunity. LIF inhibits the IFN-γ production and induces FoxP3+Tregs causing immunosuppression. Hence, a decrease in the virus entry in the cell, generation of the weak pro-inflammatory immune response, and the activation of NK and Th1 cells-mediated anitviral action generate the asymptomatic phenotype in SARS-CoV2 infected people. These asymptomatic patients at their convalescent phase show very low amount of neutralizing IgG Abs.
Asymptomatic individuals have higher circulating stem cell factor (SCF), IL-13, IL-12p40 (a subunit of IL-12), and leukemia inhibitory factor (LIF) levels than healthy controls (Fig. 3). The increase in IL-12 levels increase the antiviral immune response of NK cells and Th1 cells (direct cytotoxic action and IFN-γ generation) during early phase of the immune response (Fig. 3) [164], [165]. IL-12 inhibits early viral replication through activating NK cell cytotoxic action and IFN-γ release, but is not essential to clear the infection in case of influenza virus infection [165]. Mice deficient in IL-12 are defective in IFN-γ production and show impaired Th1 immune response without affecting cytotoxic T lymphocyte (CTL) generation [166]. However, IL-12 is not an essential cytokine for the generation of polarized Th1 cell cytokine expression and associated immune response generation during viral infections, including mouse hepatitis virus (MHV, a murine coronavirus) [167], [168]. Antigen presenting cells (APCs), including macrophages and DCs secrete IL-12 in response to the viral infection [169]. The IL-12 production may antagonize Th2 cell differentiation and inhibits the class switching to IgE to a greater extent than to other Immunoglobulins (Igs, IgM and IgGs) [170]. Hence, early IL-12 induction from APCs in asymptomatic patients helps in the SARS-CoV2 clearance through activating NK and Th1 cell-mediated cytotoxic action and antiviral IFN-γ release. However, the exact role of IL-12 during SARS-CoV2 action should be studied in different transgenic mice or ferrets that can develop asymptomatic COVID-19 resembling humans. The increased IL-13 level is associated with the decreased ACE2 expression in lungs indicating that in asymptomatic individuals SARS-CoV2 tendency to infect the cell via ACE2 receptor decreases for a productive infection (Fig. 3). However, CD147-dependent SARS-CoV2 infection of the host cells remains unchanged. Thus a less severe SARS-CoV2 infection without severe pro-inflammatory immune reaction may develop in these people as indicated by their asymptomatic phenotype. Also, IL-13 cytokine induces macrophage polarization towards anti-inflammatory alternatively activated macrophages (AAMs, which secrete anti-inflammatory IL-10) or M2 macrophages and suppresses the pro-inflammatory cytokines (IL-6, IL-1, TNF-α, and IL-8) secretion [171], [172]. On the other hand, IL-13 has a little positive effect in inducing IFN-γ from large granular lymphocytes (LGLs, which have large azurophilic granules in their cytoplasm), which include NK cells and CD8+ cytotoxic T cells [171]. However, the increased SCF levels may induce apoptotic cell death of CD8+T cells expressing the c-kit or CD117 (the ligand for SCF) in the asymptomatic patients (Fig. 3) [173]. Hence, CD8+T cell-mediated antiviral immune response (direct cytotoxicity and IFN-γ production) decreases in asymptomatic SARS-CoV2 positive people (Fig. 3). Higher the CD117 or c-kit expression on CD8+T cells, lesser is their proliferation and differentiation, but higher is their apoptotic death.
The magnitude of the CD117 on CD8+T cells inversely depends on the strength of the stimuli. In asymptomatic patients, decreased SARS-CoV2 infection of CD8+T cells occurs due to the decreased ACE2 expression that increases the CD117 expression over them (Fig. 3). Thus, CD8+T cells response is weaker in asymptomatic patients of COVID-19. Also, the SCF-c-kit (CD117) interaction maintains gut immune homeostasis and thus asymptomatic SARS-CoV2 people may not be showing gastrointestinal disturbance, an area remains to explore [174]. LIF also suppresses the IFN-γ release from T cells, promotes FoxP3+ regulatory T (Tregs) and Th2 cell proliferation, suppresses Th17 cells, and exerts host protective anti-inflammatory effects opposite to the pro-inflammatory cytokine, IL-6 (Fig. 3) [175], [176], [177], [178]. Although, LIF belongs to IL-6 cytokine family. These immunological effects may lead to the asymptomatic phenotype in certain people infected with SARS-CoV2. Thus elevated IL-12 promote direct cytotoxic action of NK cells and antiviral IFN-γ release, and elevated IL-13 and LIF levels suppress increased viral invasion to the host cells and generate anti-inflammatory immune response (AAMs activation) to protect the tissue damage, generation of Tregs further suppress pro-inflammatory tissue damaging immune response. Hence, infection gets cleared in asymptomatic patients without the generation of severe pro-inflammatory immune response causing severe harm. Further studies in animal models will put light on immune response associated with asymptomatic phenotype in patients.
Also, the asymptomatic individuals show very low levels of SARS-CoV2 S protein-specific IgG Abs as compared to acute symptomatic COVID-19 patients [159], [163]. Even convalescent asymptomatic individuals show reduction in the SARS-CoV2 NAbs and IgGs during the early convalescent phase as compared to the symptomatic convalescent patients. For example, higher number (40%) of asymptomatic convalescent patients become seronegative for SARS-CoV2-specific IgGs in the early convalescent phase (8 weeks after their hospital discharge), whereas only 12.9% symptomatic convalescent patients become seronegative for SARS-CoV2-specific Abs during this period [163]. However, convalescent asymptomatic COVID-19 patients develop a robust memory T cell-based immunity [179]. This may be due to less cytotoxic CD8+T cell response in these patients and exposure to the virus for longer duration may induce CD8+ memory T cells precursors expressing lymphoid homing molecules (CCR7, L-selectin) and homeostatic cytokine receptors (IL-7α, IL-2/IL-15β) [180], [181]. The CD4+T cell help for CD8+T cell primary response depends on the degree of inflammation as asymptomatic COVID-19 patients do not develop severe inflammation, they do not require CD4+T cells for the process [182]. However, primary CD8+T cell immune response occurring without the help of CD4+T cells may be of a good quality, but their ability to make good quality secondary immune response fades away. Hence, it will be better to see the effectiveness of CD8+ memory T cells in terms of its durability and strength with time. Further studies are required in the field to explore the generation of CD8+T memory cells in asymptomatic convalescent COVID-19 patients.
5.2. The immune response in symptomatic COVID-19 patients
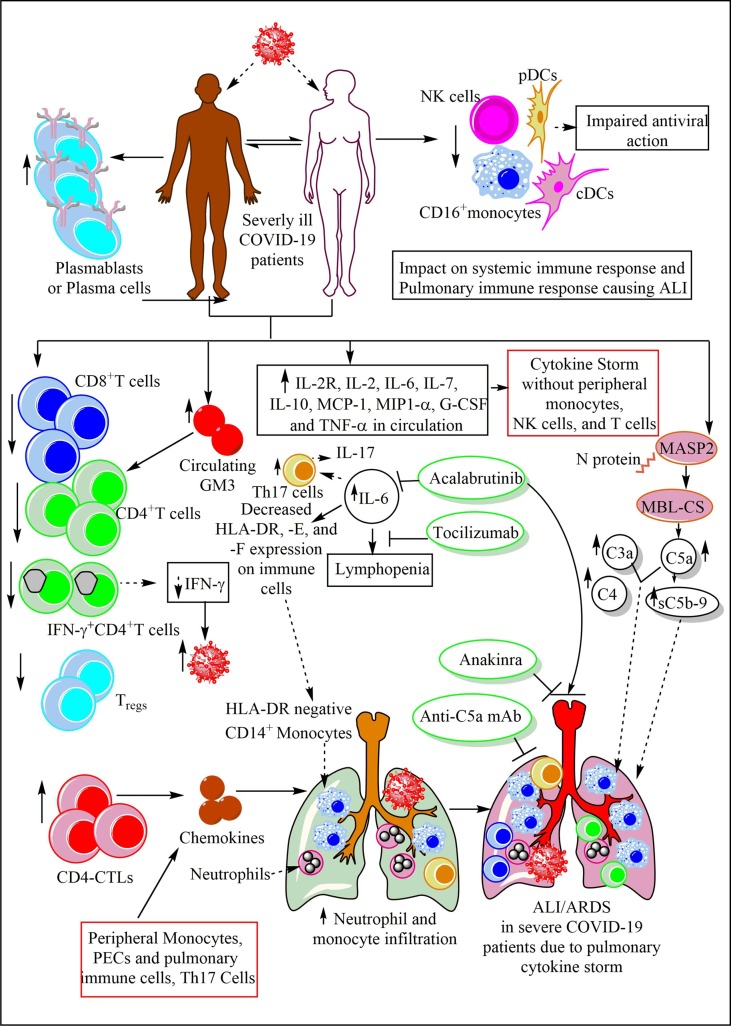
The symptomatic COVID-19 patients have higher IgG levels as compared to asymptomatic patients and their IgGs also stays for longer duration during convalescent stage than the asymptomatic convalescent patient (Fig. 3) [163]. The symptomatic patients have increased levels of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), macrophage colony-stimulating factor (M-CSF), growth-regulated oncogene-α (GRO-α), granulocyte colony-stimulating factor (G-CSF), and IL-6 [163]. GRO-α belongs to the chemokine ligand family and is also called CXCL1, it increase the neutrophil infiltration in the infected target organ through CXCR2, a G-protein coupled chemokine receptor (Fig. 4 ) [183]. GRO-α supports luminal and sub-endothelial cell neutrophil crawling to get in the target organ (Fig. 4) [184]. GRO-α is a 73 AA 8-kDa protein that resembles GRO-β/CXCL2, GRO-γ/CXCL3, and IL-8 [185]. GRO-α is 48% identical to IL-8 at the AA level and shares similar functions [186]. GRO-α modulates immune cell migration and angiogenesis. GRO-α production increases the virus (HIV-1) replication and survival in the infected macrophages and T cells to increase the disease severity [187]. Also, it activates ORF74 of the Kaposi's sarcoma-associated herpesvirus (KSHV) to induce angiogenic phenotype of Kaposi’s sarcoma lesions via activating JNK/SAPK and p38MAPK signaling as induced by inflammatory cytokines capable of activating the angiogenesis [188], [189]. Hence, it should be checked in context to SARS-CoV2 infection as both (HIV-1 and SARS-CoV2) are enveloped ssRNA viruses. GRO-α is also called melanoma growth stimulatory activity (MGSA) as it was first discovered in malignant melanoma cells as an autocrine growth factor [190].
Fig. 4.
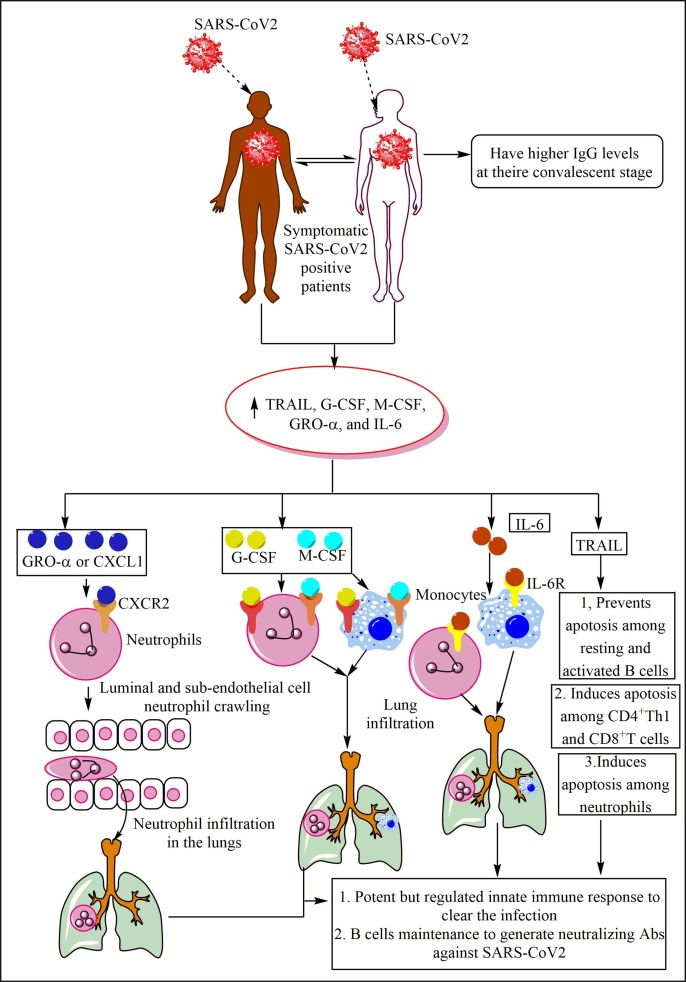
The immune response in the COVID-19 symptomatic patients. The symptomatic patients produce a high amount of GRO-α or CXCL1, which increase neutrophil infiltration in the lung via enhancing luminal and sub-endothelial cell neutrophil crawling. The increased G-CSF and M-CSF in these patients also increase neutrophil and monocyte infiltration in the lungs and their survival and differentiation. The increased IL-6 levels also increase the neutrophil and monocyte infiltration in the lungs. The increased TRAIL levels prevent the resting and active B cell apoptosis, induce apoptosis among CD4+T and CD8+T cells, and neutrophils to prevent the exaggeration of inflammation and the lung injury. These patients have higher neutralizing Abs in their convalescent stage than asymptomatic individuals.
G-CSF and M-CSF are required for neutrophil and monocyte development, maturation, and infiltration in the target organ to clear the infection and induce pro-inflammatory immune response (Fig. 4) [191]. On the other hand, elevated levels of TRAIL can promote apoptosis among primary plasma cells generated in a T cell-dependent immune response [192]. However, resting and activated B cells are resistant to TRAIL-dependent apoptosis despite the comparable TRAIL and death receptors (DRs, DR5 and DR4) expression (Fig. 4) [192]. On the other hand, TRAIL also induces apoptosis among immature DCs, although mature DCs are resistant to it [193]. TRAIL also induces apoptosis among differentiating CD4+Th1 cells and CD8+T cells generated in the absence of CD4+T cells (Fig. 4) [194]. TRAIL also induces cell cycle arrest in G2/M phase among CD8+T cells, rather inducing their apoptosis [195].
TRAIL may also induce apoptosis among human neutrophils to prevent exaggerated inflammation [196]. It will comprise a novel approach to measure the circulating serum/plasma and bronchoalveolar lavage fluid (BALF) levels of these factors (CXCL1, TRAIL, G-CSF, and M-CSF) in patients who have recovered from mild symptomatic stage or have progressed to moderate or severe COVID-19. We can interpret that in symptomatic COVID-19 patients the pro-inflammatory and antiviral innate immune response generation prevents its exaggeration to severe COVID-19 phenotype. Also, as the neutrophils are not resistant to the apoptotic death so they do not cause profound bystander tissue damage as observed during sepsis-induced acute lung injury (ALI) [197]. The prevention of apoptotic cell death among resting and activated B cells plays a crucial in the protective IgG generation, which is maintained at higher levels in the convalescent patients as compared to asymptomatic convalescent patients. The presence of Abs in these patients is the indicator of presence of Abs secreting plasma cells generated in the absence of T cell-dependent immune response. Hence, the Abs generated do not have longevity and their plasma cells reside within extra-follicular region of secondary lymphoid organs (SLOs), including spleen [198]. Further studies are required in the field.
5.3. The immune response in mild to moderate COVID-19 patients
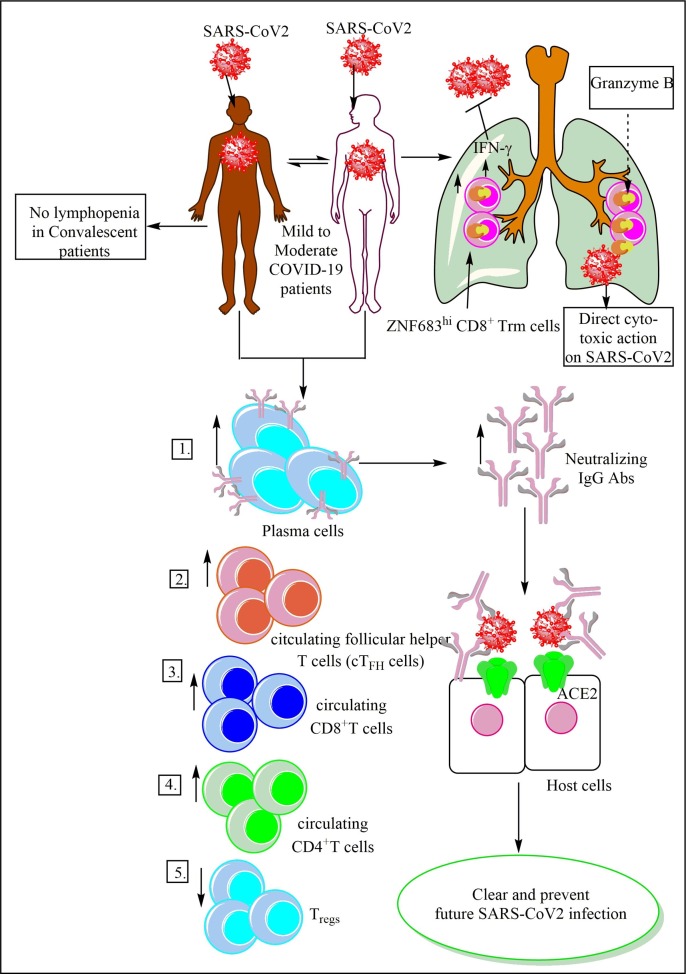
The mild cases of COVID-19 show lower body temperature and their body temperature normalizes faster as compared to patients with severe COVID-19 [199]. They have the median viral shedding duration of fourteen days [163]. The patients with mild COVID-19 also have viral shedding restricted to the respiratory tract and lack detectable virus RNA 10 days after the disease onset as indicated [200]. The neutrophils infiltrated in the lungs of mild COVID-19 patients execute their antiviral action (type 1IFN production) in an immunologically ‘controlled’ manner under the regulation of fully differentiated Th17, Th1, CD8+T resident memory (TRMs), and partially exhausted T (TEX) cells [201]. The dying apoptotic neutrophils are efficiently cleared through efferocytosis (phagocytic clearance of dead cells) by macrophages, indicating the mechanisms controlling exaggerated inflammation and tissue repair in these patients [201], [202], [203]. For example, mild to moderate COVID-19 patients before symptomatic recovery (on day 7th of the infection) that persists for at least seven days following the convalescence show higher levels of CD3–CD19+CD27hiCD38hi antibody-secreting cells (ASCs), CD4+CXCR5+ICOS+PD-1+ circulating follicular helper T cells (cTFH cells), activated CD38+HLA-DR+ CD4+T, and CD38+HLA-DR+ CD8+T cells, along with IgM and IgG antibodies binding to SARS-CoV2 in their peripheral blood samples (Fig. 5 ) [204]. The IgM levels are very low in mild COVID-19 patients as compared to the severe cases of COVID-19 [200]. The increase in cTFH cells correlates well with the increase in IL-6 as it inhibits the IL-2Rβ (CD122) upregulation via preventing the STAT5 association with the Il2rb locus to allow germinal center (GC)-TFH cells for receiving sustained T cell receptor (TCR) signaling and producing IL-2 without initiating a TCR/IL-2 inhibitory feedback loop [205].
Fig. 5.
The immune response in the mild to moderate COVID-19 patients. These patients show an increase in Ab secreting plasma cells, cTFH cells, CD4+T cells, and CD8+T cells with a decrease in the Tregs in the circulation. These patients have residential memory T cells (Trems) in their lungs, which exert a direct cytotoxic action on SARS-CoV2 via granzyme B. The neutralizing Abs protect the host form the severe infection via virus clearance and also provide future protection to the host. These patient during convalescent stage do not show lymphopenia and exhibit a higher level of neutralizing Abs that asymptomatic and symptomatic patients.
The human B cell compartment in COVID-19 patients alters rapidly with the early recruitment of B cells expressing a limited subset of variable heavy (VH) chain genes, and extensive polyclonal IgG and IgA Abs activation without a significant somatic hypermutation (SHM) [206]. However, COVID-19 individuals show a broad range of SHMs [207]. The high SHM pattern is associated with clinically more severe pattern of the COVID-19 and B cell receptors (BCRs) with high SHM do not show clonal expansion [207]. The clusters of related BCR sequences in Ab-positive COVID-19 repertoires spanning different individuals, independently of one another, show a very similar B cell response as indicated by the preferential usage of Ig heavy chain variable region 3 (IGHV3) subfamily genes, mostly rearranged with Ig heavy chain joining region 4 (IGHJ4) or IGHJ6 gene segments [207]. The majority of COVID-19-specific IGHV3 BCR clusters have unmutated BCR sequences. This suggests that the patients with effective immune response have a large pool of naïve B cells. Expansion of B cell clones and highly convergent Abs with highly similar sequences across SARS-CoV2 infected patients highlights their stereotyped naïve nature against the infection [206]. Recently, polyclonal IgGs and Fabs recognizing SRAS-CoV2 S protein have been isolated from convalescent COVID-19 patients, which differ in their binding to the RBD epitopes, recognizing α- and β-CoVs, and contribution to avidity to increased binding/neutralization of IgGs against Fabs [208]. Also, the polyclonal plasma Fabs recognize both S1A and RBD epitopes on SARS-CoV2 S protein, including the epitope that blocks ACE2 receptor binding. Even these IgGs do not get affected with identified S protein mutations of the SARS-CoV2 [208]. Hence, the characterization of these recurrent polyclonal IgGs against SARS-CoV2 provide us a criteria to evaluating vaccine elicited Abs.
The neutralizing Abs showing minimum somatic mutation and the limited clonal expansion recognize epitopes of the RBD and inhibit the SARS-CoV2 S-ACE2 receptor interaction through neutralizing the SARS-CoV2 (Fig. 5) [209]. SARS-CoV2 NAbs develop within 10–15 days post infection and their titers well correlate with S-protein binding Abs targeting S1, RBD, and S2 regions [210]. The elderly and middle-aged patients have high levels of plasma NAbs and SARS-CoV2-S protein-specific Abs than younger patients. Whereas, the non-NAbs generated within first seven days of the infection target epitopes outside the RBD of the S protein to disrupt the interaction between ACE2- SARS-CoV2-S protein without undergoing the considerable maturation [209]. Hence, both neutralizing and non-neutralizing Abs have a potential for immunotherapeutics. However, convalescent patients with mild COVID-19 rapidly loss protective Abs against SARS-CoV2, which is more rapid than the SARS-CoV (serum IgG Abs were present even after 1 year (3 years) but IgM Abs lost after 6 months) [163], [211], [212], [213].
Another study has shown the low frequency of NAbs in COVID-19 convalescent patients in addition to the presence of rare, but recurring RBD-specific Abs with a potent antiviral activity [214]. The production and level of anti-RBD IgG Abs moderately correlate with the age of the patient and the severity of infection. Whereas, anti-RBD IgM Abs do not correlate with the duration of symptoms and the timing of sample collection [214]. Of note, women show less anti-RBD and anti-spike (S) IgG titers than male patients independent of age, severity of symptoms, timing of sample collection relative to the onset or duration of symptoms [214], [215]. The IgG titers are greater than either IgM or IgA for S1, full length S, and S-RBD in the overall COVID-19 convalescent patients and 80% patients show NAbs [215]. Some mild COVID-19 patients show very low undetectable NAbs levels indicating that they have T cell-mediated immunity to protect against the virus [210].
The mild COVID-19 patients have increased level of ZNF683hi CD8+ Trm (resident memory T) cells in their BALF (Fig. 5) [129]. The CD8+Trm memory cell formation occurs through the extracellular adenosine triphosphate (eATP) recognition via purinergic receptor P2X 7 (P2RX7) expressed on CD8+T cells, which enhances their responsiveness to the transforming growth factor-β (TGF-β), an essential step for tissue residency of memory T cells [216]. The long term maintenance of CD8+Trms requires sustained P2RX7 signaling at barrier sites, including respiratory tract or lungs. Thus only healthy individuals with healthy mitochondria will have CD8+Trms for longer duration, which could clear the related infection in near future. The zinc finger protein 683 (ZNF863 or Hobit) is a homolog of B lymphocyte-induced maturation protein-1 (Blimp-1) in T cells. The quiescent and long-lived effector-type CD8+T cells highly express ZNF863 or Hobit [217]. The T-bet and IFN-γ expression within the effector CD8+ T-cell population highly correlate with the Hobit expression. Hence, T-bet and Hobit, both are crucial for IFN-γ production by the effector memory CD8+T cells (Fig. 5) [217].
The Hobit or ZNF683 maintain cytotoxicity (via regulating granzyme B) in a deployment-ready mode in Trm [218]. Blimp-1 controls the generation of these CD8+Trm cells in the lungs [219]. However, in mature human NK cells, ZNF683 or Hobit negatively regulates IFN-γ production by mature human NK cells [220]. Therefore, the ZNF683 expression in human NK cells isolated from COVID-19 should also be explored as severe COVID-19 patients fail to produce IFN-γ. Thus, patients with mild COVID-19 are efficient in clearing the virus due to the presence of high number of ZNF683hi CD8+ Trm cells, which are capable of exerting direct and immediate cytotoxic action on the SARS-CoV2 (Fig. 5). Of note, some (20–28%) HDs with no prior history of SARS-CoV2 infection also have SARS-CoV2 reactive memory T cells directed against S or M protein, but not against nucleocapsid (N protein) [179], [221], [222]. This indicates the induction of SARS-CoV2 cross-reactive T cells in response to the common cold. And these SARS-CoV2 reactive T cells in the unexposed population may help to clear the infection via causing mild or moderate infection.
The COVID-19 recovered patients show the presence of neutralizing IgG antibody and majority of recovered patients also have anti-RBD IgM and IgA antibodies [222], [223], [224]. However, convalescent COVID-19 patients do not have expanded plasmablasts [71]. The frequency of plasmablasts in individuals with severe COVID-19 does not correlate with age, since days onset of the COVID-19 symptoms, APACHE III score, and frequency of CD4+ cTFHs [71]. The CD4+cTFHs generated from SARS-CoV2 S protein-specific CD4+T cells and M or N-specific CD4+T cells skew towards Th1 or Th1/Th17 cells [179], [207]. The presence of RBD-specific IgGs has been detected in COVID-19 patients six days post PCR confirmation of the infection [224]. These IgG Abs show class switching to IgG1 and IgG3 subclass, and no IgG2 and IgG4 have been detected [224]. Hence, NAbs production and IgG class-switching occur early during acute COVID-19. The plasmablast (express Ki-67 and low levels of CXCR5) frequency does not correlate with the anti-RBD IgM or IgG levels, but a positive correlation between the anti-RBD-specific IgM and IgG levels, and time since the onset of the infection in moderate and severe COVID-19 patients occurs [71]. The S protein-specific CD4+ T cell responses correlate well with the magnitude of the anti-spike RBD IgG titers, while the non-spike SARS-CoV-2-specific CD4+ T cell response does not correlate well with anti-spike RBD IgG titers. The anti-RBD IgM and IgA production depends on the generation of S protein-specific CD4+T cells immune response, but the non-spike SARS-CoV-2-specific CD4+ T cell response is independent of anti-spike RBD IgG titer [222]. Hence, the Ab, CD4+, and CD8+ T cell responses specific to S protein of SARS-CoV-2 generally correlate well, but less strongly correlate with circulating pro-inflammatory cytokines (IL-6, IL-10, and TNF-α) [179].
The T cell repertoire increase their diversity and richness above the level of convalescent HDs [207]. The COVID-19 patients show a skewed usage of T cell receptor β chain variable and joining (TRBV/J) gene. More than 80% COVID-19 patients have SARS-CoV2 S protein-specific CD4+T cells [225]. The majority of the CD4+ T cell response in COVID-19 cases is directed against highly expressed SARS-CoV-2 ORFs S, M, and N proteins. On an average, these Ags account for 27%, 21%, and 11% of the total CD4+ T cell response, respectively [222]. Most COVID-19 patients also generate SARS-CoV-2 Nsp3, Nsp4, and ORF8 specific CD4+ T cells, which account for ~5% of the total CD4+T cell response each. Whereas E, ORF6, hypothetical ORF10, and Nsp1 are all small Ags, and remain unrecognized by CD4+T cells. On the other hand, the SARS-CoV2 unexposed control group have about 23–35% of CD4+T cells against S protein of SARS-CoV2, indicating the widely distributed cross-reactivity with SARS-CoV2 [222], [225]. However, these S protein-reactive CD4+T cells from HDs target primarily to C-terminal epitopes indicating their higher homology to S glycoproteins of human endemic coronaviruses (HCoVOC43 and HCoV229E) compared to N-terminal epitopes [225].
The immunodominance pattern of SARS-CoV-2 CD8+ T cells comprises as, with spike protein accounts for ~26% of the reactivity, and N accounts for ~12%. For other proteins or antigens, it comprises as M (22%), Nsp6 (15%), ORF8 (10%), and ORF3a (7%). The less CD8+T cells are present in uninfected control group (HDs) showing cross-reactivity with SARS-CoV2. The recovered patients show slightly increased CD3+ cells as compared to the control non-exposed group. There is no significant difference between CD4+ and CD8+ T cells in both (recovered and control) groups [222]. However, CD19+ cells decrease in recovered patients as compared to the control group, but CD3–CD19– cells or CD14+CD16– monocytes (classical monocytes) remain same in both groups [71], [222]. Even the total portion of monocytes (CD14+ and HLA-DR+) and monocytes depending on CD14 and CD16 expression [classical (CD14+CD16–) monocytes, intermediated CD14+CD16+) monocytes, and non-classical monocytes (CD14−CD16–)] do not alter significantly in control, mild, severe, and COVID-19 recovered patients in the peripheral circulation [71]. The convalescent patients do not show the signs of lymphopenia (Fig. 5).
5.4. The immune response in severely ill COVID-19 patients
In severely infected COVID-19 patients the absolute number of T cells, CD4+T, and CD8+T cells decreases (Fig. 6 ) [71], [179], [226], [227], [228], [229]. The severity of COVID-19 increases with the older age and comorbidity as these patients have higher number of SARS-CoV2-specific CD4+T cells, harbor higher IL-2 levels, and their IFN-γ producing cells (NK, NKT, γδT, CD4+Th1, and CD8+ cytotoxic T, and CD11ahiFcγRIIIhi B cells, also called IFN-γ producing B cells) decrease [230], [231], [232]. The diminished frequencies of membrane protein reactive IFNγ+ T cells (CD4+ and CD8+T cells) in severe COVID-19 patients admitted in the intensive care unit (ICU) well correlate with APACHE II score of the severity. Hence, age and comorbidities (hypertension, diabetes, chronic respiratory diseases) predetermine the SARS-CoV2-specific T cell immune response. The decrease in the T cells can be suggested as a result of their apoptotic cell death as indicated by increased apoptosis marker (Annexin V) upon infection with the SARS-CoV 2 [115], [201]. Total T cell count, including CD8+ T cells or CD4+ T cells decreasing below 800, 300, or 400/μL is associated with the mortality among severe COVID-19 patients [229]. The CXCR5+CD38+CD8+T cells significantly decrease in the peripheral blood of severe COVID-19 patients, but their number increases in the lungs correlating with the severity of the pneumonia [233]. The T cell number in circulation is inversely correlated with the systemic IL-6, TNF-α, and IL-10 levels. Hence, COVID-19 convalescent patients with low IL-6, TNF-α, and IL-10 levels have T cell numbers equal to healthy controls. Also, IFN-γ expressing CD4+T and CD8+T cells get lower in severe cases as compared to mild or moderate cases (Fig. 6) [227], [234]. The CD38+ and HLA-DR+CD38+ memory CD4+ and CD8+T cells increase in severe COVID-19 patients as compared to healthy controls [71]. Severe COVID-19 patients also have higher frequency of PD-1+ CD4+ T memory cells but not CD8+T cells depending on the age, than healthy controls.
Fig. 6.
The immune response in the severe COVID-19 patients. These sever COVID-19 patients show a decrease in the CD4+T cells, CD8+T cells, IFN-γ+CD4+T cells, and Tregs. Due to decreased IFN-γ levels the viral load increases. The increase in the circulating GM3 is well correlated with the decreased CD4+T cell number in the circulation. The increased CD4-CTLs along with peripheral monocytes, pulmonary epithelial cells (PECs) and pulmonary Th17 cells secrete various chemokines, which increase the neutrophil and monocyte infiltration in the lung. The increased accumulation of neutrophils, monocytes, CD4+T cells, and Th17 cells induce pulmonary cytokine storm to cause ALI/ ARDS. The systemic cytokine also generates but does not involve monocytes, NK cells, and T cells. The increased IL-6 levels cause lymphopenia, increase Th17 cells secreting pro-inflammatory cytokines (IL-17, IL-22 etc.). IL-6 also decreases HLA-DR expression on immune cells, including monocytes, which migrate to the lungs. The HLA-E and HLA-F expression on immune cells also decreases. Tocilizumab inhibits IL-6-mediated immune response to protect against COVID-19-induced pneumonia and ALI. Acalabrutinib, a BTK inhibitor also inhibits IL-6 production and the associated lymphopenia and other severe COVID-19 symptoms, including ALI. The CS activation also play a crucial role in sever COVID-19. The increased C5a, C3a, C4, and membrane attack complex (MAC) levels well correlate with COVID-19 severity and ALI. The anti-C5a mAb protects against severe COVID-19 and ALI. Anakinra, an IL-1R antagonist also inhibits neutrophils and monocytes infiltration in the lung to protect against severe COVID-19-induced ALI. These patients show increased levels of plasma cells than mild or moderate COVID-19 patients. A decrease in the systemic NK cells, pDCs, cDCs (CD1c+ and CD141+DCs), and CD16+ monocytes occurs systemically causing an impaired antiviral immune response.
The increased PD-1 and TIM-3 expression indicates their exhaustion and terminal differentiation [229], [235]. However, PD-1 expression is more pronounced on CD4+T cell subsets than CD8+T cells, and TIM-3 is more expressed on CD8+T cells and CD4+T cell subsets correlating with a strong downregulation of the metabolically active ecto-5′-nucleotidase CD73 [207]. Another study has shown that the memory CD8+T cells in severe CVID-19 patients express CD38, HLA-DR, Ki-67, and PD-1 [179]. The CD4+T effector memory cells (CD4+TEM) show lower expression of genes encoding T cells receptor (TCR) signaling components and highly express HIF-1α as an adaptation to the hypoxia or dysoxia (abnormal tissue oxygen utilization) during severe COVID-19 [157]. On the other hand, CD8+TEMs highly express cytolysis-associated genes NCR1 (encoding NKp46), FASLG (encodes Fas Ligand or CD95-L, also called Tumor necrosis factor ligand superfamily member 6 or TNFSF6), several GZM (encode granzymes) genes and PRF1 (encoding perforin), and overexpression of TNFRSF10B (encoding TRAIL-receptor) and caspase 3 (CASP3) indicates their increased susceptibility to apoptotic cell death than the CD8+TEMs of healthy controls [157].
Both, CD4+ and CD8+T cells highly express B and T lymphocyte attenuator (BTLA), which is responsible for increased mortality during sepsis via dysregulating innate immune response and lymphocyte apoptotic death [207], [236], [237]. However, the CD4+/CD8+T cell ratio in COVID-19 patients is higher than HDs indicating that CD8+T cell loss is higher than CD4+T cells [207], [230], [234]. The CD8+T cells from severe COVID-19 patients also show lower IL-2 expression than HDs. These patients also show conserved T central memory (Tcm CD45RA+CCR7+) cells, but naive CD4+T cells and T effector memory (TEM, CDR45+RA−CCR7−) cells decrease as compared to HDs [234]. The number of senescent or exhausted CD8+T cells increase in severe COVID-19 patients as compared to HDs. Both, moderate and severe COVID-19 patients have reduced CD4+CD25+CD127low and CD45RA+Tregs (Fig. 6) [226], [238]. Also, the patients with acute COVID-19 show delayed RBD- and nucleocapsid protein (NP)-specific T cell responses in the first three weeks of the first appearance of symptoms [228]. The decrease in the systemic CD4+T cell count in severe COVID-19 patients negatively correlates with the plasma monosialodihexosyl ganglioside (GM3) levels as it increases with severity of the disease and CD4+T cells decrease with increasing severity of COVID-19 (Fig. 6) [89]. Thus exosomes containing GM3 may participate in the severe COVID-19 via directly targeting systemic CD4+T cells. However, severe COVID-19 patients have increased number of CD4+ cytotoxic lymphocytes (CD4-CTLs) or cytotoxic T helper cells than mild cases [238]. These cytotoxic T helper cells may explain the high level of IFN-γ at early admission of the patients, which decreases through the recovery. The increased IFN-γ stimulates the CD62L+ eosinophil expansion from day 2 to day 6 after admission, which are reminiscent of lung resident neutrophils [199]. This transient expansion of CD62L+ eosinophils occurs just prior to the development of ALI in severe COVID-19 patients around 1 week after admission. Hence, an early IFN-γ and CD62L+ axis should also be investigated in the ALI associated with severe COVID-19. These CD4-CTLs highly express chemokines for myeloid cell (monocytes, macrophages, neutrophils, and DCs) infiltration in the lungs (Fig. 6) [238].
The infiltrated myeloid cells are responsible for the pro-inflammatory cytokine generation and inflammatory lung damage among severe COVID-19 patients (Fig. 6). All acute COVID-19 patients show specific CD4+T cells against, S, M, and N proteins of SARS-CoV2 [230]. Of note, males have more CD4+T cells against SARS-CoV2 N proteins than female patients, which comprise lower frequencies of IFNγ+ and IFNγ+TNFα+ bifunctional T cells than those specific for S- and M-protein [230]. The SARS-CoV-2-specific CD4+ and CD8+ T cells in 100% and 80% of COVID-19 patients with ARDS have been observed, which induce a strong pro-inflammatory immune response against SARS-CoV2 S protein, and also release Th1 and Th17 cytokines (Fig. 6) [221], [222]. The high circulating and lung-specific IL-6 promotes the differentiation of Th17 cell, which secrete various pro-inflammatory cytokines, including IL-17, IL-21, and IL-22, and induce neutrophil and monocyte infiltration in the lungs via releasing CXCL8 (Fig. 6) [239], [240]. The actively proliferating CD8+T cells in acute moderate and severe COVID-19 patients are defined by the expression of Ki-67 and are CCR7−CD27+CD28+CD45−CD127− [179]. However, these patients during convalescent stage do not contain these SARS-CoV2-specific CD8+T cells because they skew towards the TEMs, which are CCR7+CD127+CD45RA+/−TCF1+, and their number increases with number of days without infection and symptoms [179]. The granzyme B (GrB) of these CCR7+CD127+CD45RA+/−TCF1+ TEMs decreases with the increase in infection and symptoms free days. Thus, severe SARS-CoV2 infection causes the emergence of stem-like memory SARS-CoV2-specific CD8+T cells. The CD8+T cells and γδT cells comprise the most affected T cell population in severe COVID-19 patients [157]. The frequencies of TCRα/β- and γ/δ-positive T cells do not differ between COVID-19 patients and HDs along with frequencies of Tregs and TFHs [234] Also, Vγ9Vδ2 T cells (non-conventional T cells, which can sense phosphorylated metabolites or antigens (pAgs) generated through mevalonate pathway of host cells) also show a severe decrease in COVID-19 patients, and this loss shifts the compartment composition to Vδ+T cells [157].
The total B cell count remains normal or increases in active COVID-19 patients [207]. However, both moderate and severe COVID-19 patients have low number of CD21+CD27+ B cells in the circulation as compared to HDs [71]. On the other hand, COVID-19 recovered patients and HDs have same amount of CD21+CD27+ B cells. Both, moderate and Severe COVID-19 patients have a higher number of CD21−CD27− B cells than HDs. The moderate COVID-19 patients also have lower levels of CD11c+CD21−CD27− B cells than HDs. B cell activating factor (BAFF) is higher in active COVID-19 patient’s plasma than HDs, but a proliferation inducing ligand (APRIL, supports long lived plasma cells in its microenvironment) is higher only in the plasma of COVID-19 recovered patients [207]. Also, secretory variants of immune checkpoint molecules, including soluble TIM-3 (sTIM-3), lymphocyte activation gene-3 (sLAG-3), sCD25, and sGalectin-9 are higher in active COVID-19 patients, which get lower in convalescent patients. Severe COVID-19 patients and recovered patients have higher levels of SARS-CoV2-RBD-specific IgM and IgG Abs independent of plasmablasts frequency [71]. The severe COVID-19 is associated with oligoclonal expansion of B cells with Abs having long and divergent complementarity determining region 3 (CDR3) sequences.
The patients with severe COVID-19 also have higher serum levels of IL-2R (sCD25), IL-6, IL-10, and TNF-α than mild or moderate cases (Fig. 6) [207], [226], [227]. IL-13 levels are low in both active and convalescent COVID-19 patients [207]. The profound circulating IL-6 levels cause low HLA-DR expression on immune cells (macrophages, B cells, DCs, and NK cells) and lymphopenia (due to profound CD4+T, CD3−CD19+ (B cells), and CD3−CD56+ NK cell depletion) in patients with COVID-19-induced severe respiratory failure (SRF) due to its sustained production and hyper-inflammation (Fig. 6) [141], [234], [241]. Among CD3+ cells, CD4+, CD8+, and CD56+ (NKT) cells significantly reduce in circulation [234]. The decrease in the HLA-DR expression on CD14+ monocytes is responsible for COVID-19 associated SRF (Fig. 6). The increased serum IL-6 levels in severe COVID-19 patients and the impairment in antiviral cytokine production by CD4+T cells, CD8+T cells, and NK cells well correlate, and these patients have less GrA positive NK cells [234], [242]. The IL-6 responsive genes, including IL-6R, signal transducer and activator of transcription 3 (STAT3), and suppressor of cytokine signaling 3 (SOCS3) increase with the severity of COVID-19, indicating that high IL-6 levels play a crucial role in the dysregulated immune response [243].
The TNF-α-induced gene TNFSF10 increase also indicates its role in inflammation. CD8+T cells and NK cells in patients with high serum IL-6 level requiring intensive care admission also show less cytotoxic action against SARS-CoV2 [234], [242]. Tocilizumab, an IL-6 inhibitor treatment in plasma-enriched cells increases the HLA-DR expression on immune cells in vitro, and in vivo it reverses the lymphopenia and restores NK cell cytotoxicity potential (restores GrA and perforin) in severe COVID-19 patients (Fig. 6) [234], [241], [242]. The intravenous tocilizumab, a recombinant humanised mAb of the IgG1 class against both soluble and membrane-bound IL-6 receptor (IL-6R) (8 mg/kg bodyweight (up to a maximum of 800 mg) in two infusions, 12 hrs apart or subcutaneous (162 mg administered in two simultaneous doses, one in each thigh) decreases the risk of mechanical ventilation and death in severe COVID-19 patients with pneumonia [244]. Tocilizumab also decreases circulating CRP levels, indicating its potential inhibitory action on IL-6-mediated pro-inflammatory activity [234]. However, 13% of the 179 patients treated with tocilizumab have developed new infection in the study. Hence, the tocilizumab should be used with a caution. Recently, Roche’s Actemra/RoActemra (tocilizumab) has failed to meet the primary endpoint of improved clinical status in the Phase III COVACTA trial in hospitalized adult patients with severe COVID-19-induced pneumonia and ALI. The high IL-6 levels during severe COVID-19 well correlates with the auto phosphorylation of Bruton’s tyrosine kinase (BTK) in macrophages [245]. The BTK inhibitor, acalabrutinib treatment improves lung oxygenation within 1–3 days in severe COVID-19 without any toxicity. This treatment also decreases inflammatory markers (C-RP and IL-6) and normalizes lymphopenia (Fig. 6) [245]. Thus BTK inhibitor may serve as an immunomodulatory agent during severe COVID-19. However, Sanofi and Regeneron’s the phase 3 clinical trial of the IL-6 inhibitor sarilumab [Kevzara, used in rheumatoid arthritis (RA)] has also failed.
Severe COVID-19 patients show increase in circulating IL-2, IL-7, G-CSF, IP-10, MCP-1 or CCL2, MIP1-α, and TNF-α, indicating the cytokine storm generation responsible for severe COVID-19 induced acute lung injury (ALI) and multiorgan failure (Fig. 6) [246], [247]. Additionally, tumor necrosis factor superfamily 14 (TNFSF14) protein, (also called LIGHT or CD258, which binds to the herpes virus entry mediator (HVEM) or decoy receptor 3 (DR3) or lymphotoxin β receptor), extracellular newly identified RAGE (receptor for advanced glycation end product) binding protein (EN-RAGE or S100A12, which serves as a marker for coronary heart disease (CHD) and promotes sepsis-induced ALI/ARDS), and oncostatin-M (OS-M, an IL-6 regulator) also increase systemically in severe COVID-19 patients [141], [248], [249]. The increased EN-RAGE expression occurs specifically in monocytes of COVID-19 patients without inducing pro-inflammatory cytokines, which well correlates with its higher levels in the plasma. The severe COVID-19 also induces the systemic releases of gut or pulmonary microbial products, including bacterial DNA or lipopolysaccharide (LPS), which well correlates with plasma IL-6, MCP-3, TNF-α, EN-RAGE, TNFSF14, and OS-M levels [141]. Thus, pro-inflammatory damage in lugs or gut to the commensal bacteria may release their microbial-associated molecular pattern (MAMPs) to further enhance the cytokine storm.
The coagulation factor III (F3) variant allele rs72729504 has a very strong association with COVID-19 severity as these people are prone to form more fibrin fragment D-dimers (most widely used clinical marker for blood coagulation), and increased D-dimer levels (>1µg/ml) are associated poor outcome in COVID-19 patients [250], [251], [252]. The lead common variant rs12029080 associated with higher D-dimer levels has also shown a great association with severe COVID-19 [251]. Anakinra (a 17 kD recombinant, non-glycosylated human IL-1 receptor antagonist (hIL-1ra) with a half-life of about 3–4 h and good safety profile) seems to be promising in patients with COVID-19-induced cytokine storm as indicated by improve clinical and laboratory findings (Fig. 6) [253]. Another study has shown the safety (without serious side effects) of the anakinra use in severe COVID-19 patients to decrease the mechanical ventilation and death [254]. Anakinra is safe to use as an immunomodulatory agent with remdesivir (an antiviral drug) in severe COVID-19 patients [255]. Remdesivir should be used with caution as a randomised, double-blind, placebo-controlled, multicentre trial and the compassionate use of remdesivir have indicated that it does not significantly improves clinical symptoms among adult patients with severe COVID-19 [256], [257].
IL-1β is absent in all severe and moderate cases of COVID-19 patients requiring ICU admission, indicating that Caspase 1 (CASP1) and NLRP3 inflammasome activation to release mature IL-1β does not occurs at severe stages of COVID-19 requiring ICU admission and ventilator support for oxygen [258], [259]. Hence, inflammasomes are not crucial in the induction of COVID-19-induced cytokine storm at later stages. Authors has mentioned the details of IL-1β during COVID-19 somewhere else [19]. IL-1α in circulation has also not been detected in severe COVID-19 patients [243]. Severe or critical COVID-19 patients show IL-10 levels in the circulation but they do not have IFN-γ. The increased circulating furin during severe COVID-19 may increase TNF-α expression in macrophages during COVID-19 via activating TNF-α Converting Enzyme (TACE) required to release soluble TNF-α from its membrane-bound precursor [260]. Hence circulating furin may also play crucial role in the severity of COVID-19 that is discussed somewhere else [19].
The complement system (CS) robustly comes in action during SARS-CoV2 infection [251]. Patient with both moderate and severe COVID-19 have higher plasma levels of C5a and sC5b-9 (Fig. 6) [261]. Of note, sC5b-9 or membrane attack complex (MAC) levels are also higher in those patients having normal C5a level due to its faster clearance. Thus sC5b-9 may serve as a crucial marker for in vivo complement activation that may cause pulmonary damage and endothelial damage during severe COVID-19. The N protein of SARS-CoV2 activates the mannose-binding lectin (MBL) pathway of the CS through MBL-associated serine protease 2 (MASP-2) binding that activates MAC causing inflammatory tissue damage (Fig. 6) [262]. COVID-19 patients with lung injury show high MBL, MASP-2, C3, C4a, C4d, and C5b-9 deposition even in the vascular endothelium [263]. The damage to the vascular endothelium is confirmed by the presence of higher levels of von Willebrand factor (vWF) released from endothelial cell’s Weibel-Palade bodies in severe COVID-19 patients [264], [265]. The treatment with anti-C5a monoclonal Ab (mAb) suppresses COVID-19-induced ALI (Fig. 6) [262]. Thus pro-inflammatory cytokines and CS play crucial roles in the COVID-19 severity and associated ALI. Severe COVID-19 patients show an increase in neutrophil extracellular traps (NETs), tissue factor (TF) activity, and sC5b-9 (MAC) levels in the plasma [266]. The thrombin or NETosis inhibition or the C5aR blockage attenuates platelet-mediated NET-induced thrombogenicity [266].
The C3 inhibition with compstatin Cp40 disrupts TF expression in neutrophils, and thus the NET, TF, and platelet-mediated thrombus formation. Thus the CS, neutrophils (NETs and TF) play crucial role in immunothrombosis and ALI. Men (between 20 and 50 years of age) have higher C3 and C4 levels in their plasma and cerebrospinal fluid (CSF), which may also explain the high mortality rate in males than females of their counterpart age due to the higher inflammatory and tissue damaging activity of CS during severe COVID-19 [267]. For example, C3–/− mice infected with SARS-CoV show protection against protection against inflammatory ALI (Fig. 6) [268]. The complement genes C1S and C1R also upregulate during SARS-CoV2 infection, whereas the C1q/TNF-related protein 6, which inhibits complement activation downregulates during SARS-CoV2 infection [144]. Hence, CS activation in COVID-19 patients may also exaggerated inflammatory cascade, and the ALI. Role of complement proteins in neurological symptoms among COVID-19 patients should also be studied due to the higher levels of C3 and C4 in the CSF males than females.
Patients with severe COVID-19 show loss of different innate immune cells (γδT cells, plasmacytoid dendritic cells (pDC), conventional DCs (cDCs are CD1c+ and CD141+DCs), CD16+ monocytes, CD8+ mucosal-associated invariant T (MAIT) cells, innate lymphoid cells (ILCs), and NK cells) [71], [269]. However, CD11c+CD1c− DCs become predominant in circulation [157]. The patients with severe COVID-19 have an increased plasma calprotectin (S100A8/S100A9) level, which correlates with the loss of non-classical CD14lowCD16high monocytes and the accumulation of HLA-DRlow classical monocytes [270]. COVID-19 patients with ARDS show a significant decrease in cDCs, CD16+ monocytes, and NK cells in the peripheral circulation (Fig. 6). The acute COVID-19 significantly reduces DCs and their function and cDC/pDC ratio increase in these patients [228]. The NK cells also show impaired antiviral function due to the reduced intracellular expression of CD107a, Ksp37, GrB, and granulysin, and IFN-γ, TNF-α, and chemokine production (Fig. 6) [269], [271]. The increase in NKG2A expression on NK cells in COVID-19 patients decreases their cytotoxic action via binding to HLA-E [271]. The genes responsible for inhibitory receptors (LAG3 or CD223, an immune checkpoint receptor, and TIM-3 or Hepatitis A virus cellular receptor 2 (HAVCR2) also upregulate in NK cells isolated from severe COVID-19 patients [269]. The increased LAG3 and TIM3 expression on human NK cells during severe COVID-19 impairs their function, indicating their exhaustion [272], [273], [274]. The expression of T cell immunoreceptor with Ig and immunoreceptor tyrosine-based inhibition motif domain (TIGIT), an inhibitory receptor on NK cells also downregulates [207]. TIGIT binds to CD155 expressed on DCs, T cells, B cells, and macrophages, thus NK cells do not interact with these cells during severe COVID-19. The TIGIT interaction with CD155 on DCs stimulates them to release IL-10, but inhibits the IL-12 release promoting tolerogenic DCs, which downregulate T cell responses [275]. Hence, NK cell-mediated tolerogenic DC formation and T cell function inhibition do not occur in COVID-19 patients. The NF-κB activation plays a crucial role in the decreased systemic type 1 IFN and the increased production pro-inflammatory cytokines (IL-6, IL-8, and TNF-α) in severe COVID-19 patients (Fig. 6) [243], [276].
Severe COVID-19 patients with ARDS show a significantly higher number of plasmablasts, indicating the generation of the robust B cell immune response. The severity of COVID-19 well correlates with IgG immune response [277], [278]. The increase in neutrophil to lymphocyte ratio (NLR) and IgG response (NLRhiIgGhi) well correlates with the COVID-19 severity. These patients have higher pro-inflammatory cytokines levels (IL-2, IL-6, and IL-10) and decreased CD4+T cell number than NLRlowIgGlow patients. Older (more than 50 years of age) COVID-19 patients with an NLR ≥ 3.13 develop critical COVID-19 [279]. The high NLR can be used as prognostic biomarker for the COVID-19 severity and organ failure [71]. Additionally, severe COVID-19 patients also show a high neutrophil: T cell ratio (NTR) that directly correlates with NLR [71]. The high NLR and NTR well correlate with APACHE III score of severe COVID-19 patients. This indicates that neutrophils infiltrated in the lungs of severe COVID-19 patients are resistant to apoptotic death, similarly to neutrophils infiltrated in lungs of sepsis patients [197]. Whereas, T cells during severe COVID-19 due to apoptosis as mentioned previously. Of note, the COVID-19 severity and NLR correlation is independent of age and vascular comorbidity, but NTR and COVID-19 severity depends on the age and vascular comorbidity [71].
COVID-19 patients with ARDS show significantly higher number of “immature or developing neutrophils” expressing neutrophil granule proteins (Elastase neutrophil expressed (ELANE), lactoferrin (LTF), and matrix metalloproteinase 8 (MMP8) or neutrophil collagenase), but do not express canonical neutrophil markers (CXCR2 and Fc gamma receptor 3 B (FCGR3B) or CD16b) [269]. Another study has shown the increase in immature neutrophils (CD10lowCD101−CXCR4+/−) in both blood and lungs indicating the emergency myelopoiesis [270]. Along with the increase in the neutrophils in the peripheral circulation, increase in eosinophils also occurs [71]. However, CD15 (also called Lewis × or Lex mediates neutrophils adhesion to DCs via DC-SIGN) expression decreases in neutrophils of severe COVID-19 patients as compared to HDs, but it remains unchanged for eosinophils [71], [280], [281]. This study also indicates the increase in the immature or developing neutrophils in severe COVID-19 patients, which cannot interact with DCs to induce their maturation [282]. The increase in the immature neutrophils positively correlates with the plasma calprotectin levels. Also, the peripheral blood monocytes (PBMCs) from COVID-19 patients with ARDS lack pro-inflammatory cytokine and chemokine gene expression, indicating they are not the active players of cytokine storm generation.
HLA class II expression in PBMCs, DCs, and B cells isolated from critical COVID-19 patients significantly decreases (Fig. 6). A reduced cross-talk between NK cells and DCs occurs in these critical COVID-19 patients. COVID-19 patients show decreased HLA-E (expressed by NK cells) and HLA-F on their immune cells (Fig. 6). CD16highCD56low NK cells responsible for antiviral immune function severely deplete in critical COVID-19 patients [71]. All COVID-19 patients have significantly low CD56highCD16low K cells responsible for producing IFN-γ and TNF-α [71], [269]. Clusters of proliferative lymphocytes are present in the most COVID-19 patients. For example, CD4+T cells and NK cells, which express LAG3, PD-1, and TIM-3 show exhausted phenotype in COVID-19 except CD8+T cells [243]. The exhaustion related genes (Basic leucine zipper transcription factor, ATF-like or BATF, IFN regulatory factor 4 or IRF4, and CD274 or PDL-1) express increasingly with the severity of COVID-19 [243]. Like, peripheral monocytes, T and NK cells also do not substantially express pro-inflammatory cytokine genes. Thus peripheral immune cells (monocytes, T cells, and NK cells) do not take part in the COVID-19-induced cytokine storm (Fig. 6). Although, symptomatic COVID-19 patients show an increase in granulocyte-macrophage colony-stimulating factor (GM-CSF)-producing, activated CD4+T cells and CD14+CD16+HLA-DRlow inflammatory monocytes (IMs) expressing high levels of IL-6 accelerating the inflammation (Fig. 6) [283]. These pathogenic T cells and IMs may infiltrate the lungs to enhance COVID-19-induced inflammatory lung damage and respiratory failure.
Normal human bronchial epithelial (NHBE) cells infected with SARS-CoV2 in vitro do not produce type 1 and type-III IFNs, but they produce eighty three IFN-stimulatory genes (ISGs), including those exerting direct antiviral action (IFN-induced protein with tetratricopeptide repeats (IFIT) and IFITMs) [284], [285]. Both, human nasal and bronchial AECs do not elicit specific-IFN and ISG response, and is undetectable in the first 24 h post infection [58]. However, they upregulate type I and type III IFNs (IFNB1, IFNL1, and IFNL2, -3, and -4), as well as other immunity-related genes after 24 h post infection, which peaks at 72–96 h post infection and fades away thereafter [58]. However, in vitro SARS-CoV-2 infection in NHBE cells elicits the chemokines (CCL20, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL16) and inflammatory cytokines (IL-6 and TNF-α) expression [19]. A subset of ISGs with no IFN-I or IFN-III expression in lungs of COVID-19 patients is also present [284]. The BALF samples from severe COVID-19 patients also have a higher levels of different chemokines (CCL2, CCL7, CCL8, CXCL1, CXCL2, CXCL17, and CCL11) (Fig. 6) [284], [285]. However, in nasal AECs the relative expression of genes (IL-18, IL-18R1, NF-κB2, NF-κBIA, TNFα, and TNFAIP3) associated with NF-κB and TNF-α signaling pathways upregulates at 48 h post infection and onwards, but these genes remain unaffected throughout the SARS-CoV2 infection in bronchial AECs [58]. This diverse response in nasal and bronchial AECs upon SARS-CoV2 infection highlights the induction of a distinct transcriptional immune programing between them, both in terms of kinetics and intensity. Hence, potential intrinsic differences between persons during early response to the SARS-CoV2 infection occurs in the upper and lower respiratory tracts.
The circulating IL-6, TNF-α, IL-1RA, CCL2 (monocyte chemoattractant protein 1 or MCP-1), CCL7 or MCP-3, CCL8 or MCP-2, CXCL2, CXCL8 (a classic neutrophil chemoattractant), CXCL9 (chemoattractant for NK cells), CXCL10, and CXCL16 increase in severe COVID-19 patients despite the impaired cytokine response by blood myeloid cells and CD123high pDCs (Fig. 6) [141], [284]. CXCL1 also serves as a chemoattractant for NK and T cells. High number of neutrophils, activated DCs, and activated mast cells are present in the BALF of severe COVID-19 patients with pneumonia [285]. The ficolin-1+ or FCN-1+, secreted phosphoprotein 1+ or SPP1+ or osteopontin+ or OPN+ macrophages and IMs replace alveolar macrophages (AMs) in the lungs of severe or critical COVID-19 patients as indicate by their high number in the BALF [286]. However, myeloid DCs (mDCs), pDCs and T cells become much lower in severe COVID-19 patients than moderate one. The migration of inflammatory transitional non-classical monocytes and CD1c+cDCs form the circulation to the lungs in severe COVID-19 patients occurs [233]. Another study further confirms these findings by indicating the replacement of AMs by pro-inflammatory innate immune cells (neutrophils and IMs) and the altered CD8+ cytotoxic T cell function severe COVID-19 patients [129].
The BALF from severe COVID-19 patients also have the increased NK and naïve CCR7+CD4+T cells [129]. Of note, NK cell numbers increase in the lung of severe COVID-19 patients but decrease in the circulation. The pulmonary epithelial compartment of severe COVID-19 patients shows an increase in club cells (also express TMPRSS2) and AT2 cells [129]. The lung type II pneumocytes co-express ACE2 receptors with TMPRSS2 [65]. ACE2 is a human ISG in vitro and in vivo, which gets highly expressed during SARS-CoV2 infection in humans [65]. For example, IFN-α2 and IFN-γ increase the ACE2 expression in human AECs in a dose-dependent manner [65]. This indicates that the induction of IFN-γ and type 1 IFNs during infection of AECs with SARS-CoV2 at initial stages increases the ACE2 expression by these cells and enhances their susceptibility to SARS-CoV2 infection, which aggravates the disease causing severe COVDI-19. The CD8+T and CD4+T cells only form the proliferative cell compartment and infiltrated neutrophils serve as major inducers of type 1 IFN response [129]. FCN1+ macrophages also produce type 1 IFNs but to a lesser level than infiltrated neutrophils. Hence, infiltrated neutrophils and macrophages are the only major immune cells to generate the cytokine storm in the lungs of severe COVID-19 patients.
The CD4+ T cells in the severe COVID-19 patients show a more naive phenotype, which express higher levels of IL-7R, CCR7 (regulate homing of naïve and central memory T cells via binding to CCL19 and CCL21), sphingosine 1 phosphate receptor (S1PR1, which via binding to S1P induces their trafficking, suppresses Tregs generation, Th17 cell generation, and also regulates T central memory cell and T residential memory cell formation), and lymphotoxin-β (LT-β, which induces the immune responses against limiting amounts of antigen). At the site of inflammation, the activated sphingosine kinase-1 produces a high amount of S1P that binds to S1PR1 of CD4+T cells to increases their persistence and motility in the inflammatory environment [19], [287]. However, a decrease in S1P in the blood plasma of COVID-19 patients has been observed, which increases in patients discharged from hospital relative to admission in small number of patients [89]. This indicates that in mild or moderate patients having low systemic inflammation or in severe COVID-19 patients S1P has migrated with blood circulation to the inflammatory lung environment to increase the CD4+T cells number. Hence, it will be interesting to observe S1P levels in BALF and plasma patients with COVID-19. The SPP1+ macrophages highly express inflammatory chemokines (CCL2, CCL3, CCL4, CCL7, and CCL8) and genes controlling hypoxia [heme oxygenase 1 (HMOX1) and hypoxia-inducible factor- 1α (HIF-1α)]. However, they express low levels of MHC class II (HLA-A and HLA-DQA1) and type I IFN genes (IFIT1 and 2′-5′-Oligoadenylate Synthetase 1 or OAS1, it negatively regulates type I IFN signaling and chemokine synthesis in human macrophages) [288].
The pulmonary AMs upregulate chemokines [chemokine ligand 18 (CCL18 or Pulmonary and activation-regulated chemokine (PARC), and macrophage inflammatory protein 1 β (MIP-1β or chemokine (C-C motif) ligand 4-like 2 or CCL4L2)] and the cathepsin-L and B (CTSL and CTSB) expression in the severe COVID-19 patients [19]. The SARS-CoV2 infected macrophages in severe COVID-19 patients show an increased expression chemokines, including CCL7 (MCP-3) that chemoattracts monocytes and regulates macrophage function via binding to CCR2), CCL8 or MCP2 (chemoattracts monocytes, T cells, and NK cells), and CCL18 and apolipoprotein E (APOE), and express less thousand and one kinase 1 (TAOK1, a member of Ste20-like kinases that positively regulates TLR signaling to produce IL-6 and TNF-α) [129], [289]. The increased CCL8 secretion by macrophages in the lungs of severe COVID-19 patients attracts the naïve T cells, immunosuppressive and immature DCs, and B cells (both naïve and effector) via binding to CCR8 [290]. The circulating macrophages isolated from the COVID-19 patients are bigger in size than normal monocytes and also secrete pro-inflammatory cytokines [291]. They also secrete increased pro-inflammatory cytokines (IL-6 and TNF-α). Severe COVID-19 patients exhibit the increased circulating furin levels [226]. COVID-19 patients with ARDS show lymphopenia (especially T cells) that can be explained as the high expression of furin that increases their susceptibility SARS-CoV2 infected even before becoming fully functional at an early stage of infections [19], [115]. This may lead to their apoptotic death [115]. The IL-12/STAT4 signaling pathway increasing IFN-γ secretion also upregulates the furin expression in Th1 cells [292]. Both CD4+ and CD8+T cells produce lesser amounts of IFN-γ (type-II IFN) severe COVID-19 patients [226].
6. Immune status in COVID-19 convalescent patients
The patients recovered from COVID-19 have high amount of spike (S)-specific Abs and patients died due to COVID-19 show high level of nucleocapsid (N)-specific Abs that has been validated in a large cohort [293]. Comparable naïve CD4+T cells are present in HDs and convalescent COVID-19 HDs [294]. Patient recovered from acute COVID-19 also have CD127+ effector and central memory CD4+ T-cells and CD57+ differentiated memory CD8+ T-cells and their DCs, including CD1c+, CD11c+, and pDCs also increase [199]. However, convalescent COVID-19 HDs show 2 fold decrease in the frequency of central memory CD4+T cells and 1.5 fold in CD4+ effector memory T cells [294]. There is a difference in the overall frequency of circulating TFHs between SARS-CoV2 unexposed HDs and convalescent COVID-19 HDs. The convalescent COVID-19 HDs have a higher frequency of CCR7lowPD-1+ effector-memory like circulating TFH (TFH-EM) cells, but have lower CCR7highPD-1− central-memory like circulating TFH (TFH-CM) cell frequency than controlled HDs, indicating the ongoing germinal centre (GC) response in the convalescent COVID-19 patients even in the absence of the virus [294].
The increased frequency of CXCR3+CCR6−cTFH1 cells (about 1.5-fold higher than unexposed HDs) in COVID-19 convalescents HDs shows that the prolonged production of NAbs in patients recovered from severe and moderate COVID-19. CXCR3−CCR6−cTFH2 cells also increase in COVID-19 convalescent patients. However, these convalescent patient show decrease in CXCR3−CCR6−cTFH17 cells, which provide superior help to naïve B cells to Ab production [294], [295]. Although, CXCR3+CCR6−cTFH1 and CXCR3−CCR6−cTFH2 increase in COVID-19 convalescent patients but a decrease in superior CXCR3−CCR6−cTFH17 cells may explain the no production of long term (years) Abs to protect from the reinfection. This needs an investigation and we have various transgenic human ACE2 (hACE2) receptor expressing laboratory mice on different back grounds (discussed later in the article). However, peripheral Tregs in convalescent COIVD-19 HDs are similar to unexposed HDs, but they show a decrease in the circulating CD45RA−CD127−CD25+CXCR5hiPD-1hi T follicle regulatory (TFR) cells, which are terminally differentiated cells and suppress the Ab production mediated through TFHs [294], [296]. Of note, circulating TFHs and TFRs also show memory like properties [297]. Thus this may affect the NAbs production in the COVID-19 convalescent patients by increasing the Ab production process.
The convalescent COVID-19 HDs patients show an increase in the Th2 cells, but Th1 and Th17 cells do not show any significant change as compared to the unexposed HDs [294]. However, overall expression of PD-1 is higher in T cells of COVID-19 convalescent HDs indicating either they are exhausted or ready for helping B cells to produce Abs that will not long last. Hence, it also gives another reason for no long lasting immunity by Abs in COVID-19 convalescent patients. Further studies will put light in the direction. The naïve T cells and the central memory T cells (TCMs) remain low in convalescent patients recovering from moderate COVID-19, which further decrease in severe COVID-19 convalescent patients [294]. However, effector memory T cells (TEMs) are higher in severe COVID-19 convalescent patients. The same trend is true for TFH-CM and TFH-EM in COVID-19 convalescent group (moderate and severe). The cTFH1, cTFH2 and cTFH17 cell do not significantly change depending on the COVID-19 severity, besides their overall variation to unexposed HDs [294].
Increased basophils, eosinophils, classical and non-classical monocytes, NK cells, eosinophils, and γδT-cells have also been reported in convalescent patients [199]. The circulating IL-6, MCP3, Keratin19 (KRT19), CXCL10, AREG, and IFN-γ levels also decrease in convalescent patients. Although, circulating IL-6 decrease in COVID-19 convalescent patients as compared to the active COVID-19 patients but it is still higher (4 fold) than unexposed HDs [294]. They also show higher CXCL11 (that has highest binding affinity for CXCR3) and TNF-α levels than unexposed HDs, indicating the ongoing GC process in the lymph nodes (LNs) due to presence of the SARS-CoV2 Ag in follicular DCs (FDCs), which produce these cytokines. IL-5 and IL-21 are also higher in COVID-19 convalescent patients than normal HDs [294]. Although other signature cytokines of Th1, Th2, and Th17 cells remain intact like normal HDs. The increased soluble CD40 (sCD40 shed from T cells or released from platelets) levels in convalescent patients for several weeks may predispose them to cardiovascular risk [207]. The T cell clonotypes of mild-COVID-19 recovered patients are highly diverse [207]. Hence, in convalescent COVID-19 patients the dysregulated immune response comes to an immune homeostasis with the development of Ab-dependent protective immunity that does not last long due to the absence of memory B cells and long lasting plasma cells [19]. Further studies will also put light on this topic.
7. Re-emergence of COVID-19 in convalescent patients
The re-emergence of the COVID-19 in the recovered patients in some countries has created a panic around the world. COVID-19 re-emergence may depend on the strength of immune response against primary infection. For example, severely infected patients, who have survived SARS-CoV2 infection develop CD8+TEMs and protective NAbs at highest level compared to asymptomatic and mild to moderate COVID-19 patients, so chances of them to get re-infection is lowest within in the window of 2–3 months or at least 6 months. This may be only possible in case of severe COVID-19 convalescent patients upon exposure to the new mutant SARS-CoV2 that is highly pathogenic and NAbs are not working against it or increasing their pathogenicity via antibody-dependent enhancement (ADE) of phagocytosis. This needs an exploration. However, along with environmental or community transmission, the COVID-19 may come back in asymptomatic convalescent patients in the absence of environmental or community transmission also. This may occur as result of less specificity and durability of NAbs and T memory cells produced during the first asymptomatic infeciton, and the activation of already present virus that has gone to the dormant or latent stage, which remain hidden in their hiding niches (because SARS-CoV2 can infect every cell expressing ACE2, TMPRSS2, and CD147) from the immune system, and remain unaffected to antivirals (remember we do not have an effective antiviral against SARS-CoV2, which may also help the virus to escape) unless reactivated. Only the RT-PCR of nasal swabs and blood sample are performed clinically to declare whether the patient has become COVID-19 negative or not. The inclusion of other tissue or fluid samples (urine) along with at least 2–3 month clinical follow up of the recovered patients may solve the incidence of re-emergence of COVID-19 patients. Hence, immune status of the host both, before and after the COVID-19 plays a crucial role in tracking and fighting the SARS-CoV2. However, SARS-CoV2 has evolved or developed various strategies to hide from the effective immune response or block the effective antiviral immune response in the host as discussed in the following section.
8. Hijacking or modulation of the antiviral immune response by SARS-CoV2
The cytosolic PRRs play a crucial role in the pathogen, including cytosolic virus recognition. For, example, cytosolic toll-like receptors (TLRs present in endosome, phagosome, phagolysosomes, including TLR3, TLR7, TLR8, and TLR9), retinoic acid-inducible gene-1 (RIG-1)-like receptors (RLRs), and cyclic-adenosine monophosphate (GMP)-adenosine monophosphate (AMP) synthase or cGAS recognize cytosolic nucleic acids, including different types DNAs and RNAs and play crucial role in the clearing different cytosolic pathogens (bacteria and virus) [298], [299], [300]. For, example, the recognition of viral genetic material (both DNA and RNA) by cGAS activates stimulator of interferon genes [STING, located in Golgi apparatus and endoplasmic reticulum (ER)] to synthesize NF-κB and interferon regulatory factor 3 (IRF3)-dependent cytokines and type 1 IFNs (Fig. 7 ) [301], [302], [303], [304]. Both, DNA and RNA viruses evade or block cGAS-STING signaling pathway responsible to generate NF-κB and IRF3-dependent cytokines and type 1 IFNs through different mechanisms (Fig. 7) [299], [305], [306].
Fig. 7.
Cytosolic PRRs for SARS-CoV2 for the antiviral immune response and their modulation or hijacking. TLR3 (recognize dsRNA), TLR7, and TLR8 (both recognize ssRNA) in the endosome of macrophages, pDCs, DCs, B cells, and other myeloid cells. The TLR3 activation in DCs through dsRNA stimulates downstream signaling via TRIF and TRAF. TRIF activates RIP1 to activate NF-κB and TRAF6 activates TBK1 and IKK-ε-dependent IRF3 activating IL-6, IL-12, and TNF-α and type 1 IFNs. Similarly, TLR7 and TLR8 activation through ssRNA stimulates downstream NF-κB signaling via MyD88 activation mediated IRAK-1 and IRAK-4. The TRAF6 activates p38MAPK, and TRAF3 activates IRF7 to activate pro-inflammatory cytokines and antiviral type 1IFNs. RLRs via RIG-1 and MDA5 also recognize cytosolic viral ssRNA to activate MAVS, which activates TBK1 and IKK-ε to activate IRF3 for type 1 IFN-mediated antiviral immune response. TBK1 also activates NF-κB and IRF7. Another PRR called cGAS also recognizes viral ssRNA to generate cGAMP which binds to the STING activated STING binds to the TBK1, which activates IRF3 to generate antiviral type 1 IFNs. The cGAS-STING signaling via TRAF6 also activates canonical NF-κB to generate pro-inflammatory cytokines. However, the viral proteins Nsp3 and Nsp8 via binding to TBK1 inhibit the potent type 1 IFN-mediated antiviral immune response. However, Nsp3 promotes the NF-κB-mediated tissue damaging pro-inflammatory cytokines (IL-6, IL-12, and TNF-α) via inhibiting the TLR signaling inhibitors, TLE3 and TLE4. Details are mentioned in the text.
The HMGB1, Ag presentation pathway, which may be associated with macrophage maturation and activation, activate upon cGAS-STING signaling pathway independent of type 1 IFN pathway in macrophages [307]. The cGAS-STING activation in T cells via DNA methylation activates many T cell effector signaling pathways independently of IFN, Th17 cell signaling, Th1 pathway, NF-κB, nuclear factor of activated T cells (NFAT), unfolded protein response (UPR, activates through an evolutionarily conserved UPR motif), IL-2 pathways, and cell death [307], [308]. Hence, although type 1 IFN synthesis through cGAS-STING signaling gets blocked in SARS-CoV2 infected macrophages and T cells, but it increases macrophage maturation and pro-inflammatory function, and in T cells as it promotes the Th17 cell generation and pro-inflammatory cytokine release along with their apoptotic death as seen in severe COVID-19 patients. The description of cGAS-STING signaling in inflammation and antiviral immune response in beyond the scope of present article that can be found in detail elsewhere [299], [300].
TLR3 (highly expressed on DCs) activation through downstream Toll/interleukin-1 receptor (TIR)-domain-containing adaptor-inducing interferon-β (TRIF) during SARS-CoV infection protects via regulating the NF-κB and IRF3-dependent innate immune response required to clear the infection (Fig. 7) [309]. TLR7 (highly expressed in plasmacytoid DC (pDCs) and B cells, but at low levels in epithelial cells) and TLR8 (present in myeloid cells) in endosomes also recognize ssRNA of SARS-CoV and activate myeloid differentiation factor 88 (MyD88)-dependent downstream signaling to activate mitogen activated protein kinase (MAPK) and NF-κB-dependent IL-6, IL-12, TNF-α and IFN-α release (Fig. 7) [310]. Even TLR7/TLR8 signaling in neutrophils during viral infection impairs their protective action and enhances their pro-inflammatory function [311]. Severe COVID-19 patients also show increased levels of IL-6 and TNF-α indicating the TLR7 and TLR8 signaling overactivation in macrophages and neutrophils infiltrated in the lungs. SARS-CoV2 has more ssRNA than SARS-CoV therefore it can profoundly activate TLR7/TLR8 pro-inflammatory immune response responsible for COVID-19-associated ALI and cytokine storm [312]. On the other hand, RLRs only recognize cytosolic ssRNAs and this protein family has three members, including RIG-1, melanoma differentiation associated protein 5 (MDA5), and the laboratory of genetics and physiology 2 (LGP2) (Fig. 7) [313], [314]. The activation of RLRs-dependent signaling through RIG-1 and MDA5-mediated viral RNA recognition produces type 1 IFNs to protect the host against invading virus through activating mitochondrial antiviral-signalling protein (MAVS)-dependent TANK-binding kinase 1 (TBK1)/ IκB kinase-ε (IKKε) and IRF3, IRF7, and NF-κB (Fig. 7) [315]. The details are described elsewhere [315]. Following section describes the hijacking or inhibition of these signaling pathways during SARS-CoV2 infection.
The interaction between viral Nsp13 and TANK-binding kinase 1 (TBK1) and TANK-binding kinase 1-binding protein 1 (TBKBP1/SINTBAD) inhibits the TBK1 interaction with IKKi required for an IRF-dependent type 1 IFN production downstream of cGAS-STING and RLR signaling against RNA viruses (Fig. 7) [316]. Thus SARS-CoV2 infection of innate immune cells (epithelial cells, macrophages, DCs, and NK cells) inhibits the type I IFN release to escape the lethal antiviral innate immune response. Nsp13 also interacts with multiple proteins of transducin-like Enhancer of split (TLE) that include TLE4 and TLE1 to keep going the NF-κB-dependent pro-inflammatory cytokine (IL-6, TNFα, IL-8, and IL-12) release [316]. This is because TLE proteins inhibit pro-inflammatory immune response in response to PRR activation-induced NF-κB signaling pathway through interacting with different transcription factors, including Hes1 (Hairy and enhancer of Split 1) (Fig. 7) [317], [318]. Hence, Nsp13 simultaneously blocks the type 1 IFNs release, while inducing the release of TLR3, TLR7, and TLR8-dependent pro-inflammatory cytokines (IL-6, TNF-α, IL-8, and IL-12) to induce inflammatory lung damage in severe COVID-19 patients (Table 1) through hijacking different host proteins involved in PRR signaling-induced antiviral action (Fig. 7).
Another Nsp, called Nsp15 blocks the TBK1-dependent IRF3 activation-mediated type 1 IFNs through TLR signaling pathways via binding to the RING finger protein 41 (RNF41)/ Neuregulin receptor degradation protein 1 or Nrdp1 (an E3 ubiquitin-protein ligase) (Table 1) (Fig. 7) [316], [319]. Nsp15 blocks the type 1 IFNs release during SARS-CoV2 infection at same time when Nsp13 induces the release of pro-inflammatory cytokines responsible for COVID-19-induced cytokine storm. The Nsp15 binding to Nrdp1 further enhances the MyD88-dependent TLR-7/8 signaling pathway required for NF-κB and AP-1 transcription factors induced pro-inflammatory cytokine (TNF-α, IL-6, IL-8, IL-12, and IL-10) response responsible for ‘cytokine storm’ generation during COVID-19 (Table 1) [19], [319]. The TLR signaling pathway inhibition via Nrdp1 involves K48-linked polyubiquitination-mediated MyD88 degradation. Also, the binding of Nsp15 to Nrdp1 prevents the macrophage polarization to M2 macrophages that further supports the pro-inflammatory phenotype of macrophages in COVID-19 patients causing acute lung injury (ALI) [320].
The Nrdp1 promotes pro-inflammatory M1 to anti-inflammatory M2 macrophages via Lys-63-linked ubiquitination and activating transcription factor CCAAAT/enhancer-binding protein β (C/EBPβ) to activate arginase 1 (Arg1) gene [320]. The Nsp15 binding to Nrdp1 also inhibits the TBK1 and IRF3 as Nrdp1 activates TBK1 and IRF3 signaling to synthesize type 1 IFNs. Other SARS-CoV2 proteins, including ORF3a and Nsp8 strongly bind to other E3 ubiquitin ligase (Tripartite Motif Containing 59 or TRIM59, a member of TRIM famliy) and promote TLR-mediated activation of NF-κB signaling inducing pro-inflammatory cytokine release [316]. Under normal conditions, TRIM59 binds to the evolutionarily conserved signaling intermediate in Toll pathways (ECSIT) as an adaptor protein required for the NF-κB activation via forming a complex with p65/p50 NF-κB proteins and inhibits exaggerated pro-inflammatory cytokine generation [321], [322]. ESCIT complex with TAK1 and TRAF6 also activates NF-κB and Smad1/5/8 signaling [323], [324]. On the other hand, ESCIT overexpression enhances IRF3 activation and IFN-β expression by bridging RIG-I and MDA5 to the VISA complex [325]. Mind Bomb 1 or MIB1 ubiquitinates TBK1 and NAP1 (nucleosome assembly protein 1), activates NF-κB and interferon-stimulated response element (ISRE) reporters for antiviral immune response [326].
The SARS-CoV2 ORF8 interacts with MHC-1 of the infected cells, including epithelial cells, macrophages, and DCs, and downregulate their cell surface expression via inducing their lysosomal degradation in an autophagy-dependent manner [327]. Thus, ORF8 of the SARS-CoV2 protects the virus from the direct cytotoxic action of the CD8+ cytotoxic T cells via decreasing the MHC-1-dependent Ag presentation. Hence, the SARS-CoV2 virus hijacks the MHC-1-dependent Ag presentation of the host to protect from itself from the direct killing by CD8+T cells. The SARS-CoV2 ORF9c interacts with NLRX1 [a NOD-like receptor family member, which negatively regulates TLR signaling-induced NF-κB activation via interacting with TRAF6 or IκB kinase (IKK)], impacts RLR-mediated antiviral immune response, reactive oxygen species generation, and NLRP3 inflammasome activation, thus the release of IL-1β to modulate NF-κB signaling and IκB kinase (Table 1) [316], [328], [329], [330], [331]. Hence, SARS-CoV2 infection hijacks the cytosolic cellular mechanisms responsible for generating potent type 1 IFNs through cGAS, RLR, and TLR-based mechanisms required to clear the virus and induced antiviral immune response in the adjacent cells. On the other hand, it stimulates signaling mechanisms required for generating cytokine storm, inflammatory tissue damage, and the generation of pro-inflammatory M1 macrophages. Future studies will explore other hidden mechanisms of virus hijacking of the host innate immune response proteins and to develop therapeutics to target viral proteins involved in the process.
9. Immunomodulatory and immunotherapeutic approaches for COVID-19
Currently, we do not have the SARS-CoV2-specific antiviral drugs. Gilead bioscience’s antiviral drug remdesivir (GS-5734) has emerged from a collaboration between, Gilead, the U.S. Centres for Disease Control and Prevention (CDC), and the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) [332]. Remdesivir is monophosphoramidate prodrug of an adenosine analogue with a broad antiviral spectrum (filoviruses (Ebola virus), paramyxoviruses (parainfluenza and mumps virus), pneumoviruses [respiratory syncytial virus (RSV), Dengue virus and Yellow fever virus), and coronaviruses (SARS-CoV, MERS-CoV, and SARS-CoV2)] [333], [334], [335], [336]. The antiviral action of remdesivir against RNA viruses (Ebola virus and SARS-CoV2) involves the divergent RNA-dependent RNA polymerase (RdRP) inhibition required for RNA transcription, and serves as a delayed chain terminator [332], [337]. Remdesivir (GS-5734) is a prodrug, which metabolizes within cells into an alanine metabolite (GS-704277). The alanine metabolite or GS-704277 further metabolizes into a monophosphate derivative, which ultimately transforms into an active nucleoside triphosphate derivative capable to target RdRP of the RNA viruses [155], [332]. It exerts its antiviral action against SARS-CoV2 in human nasal and bronchial AECs infected in vitro, and has been used in combination with diltiazem (a calcium channel blocker) for severe COVID-19 patients [58]. The addition of diltiazem increases the antiviral action of remdesivir via increasing the type 1 IFN production and also protects the human bronchial epithelial integrity.
The early remdesivir administration in the non-lethal COVID-19 infected rhesus macaques prevents the development of signs of pneumonia, reduces the pulmonary infiltrates as indicated on radiographs, and decreases the virus load in BALF within twelve hours of its first administration [338]. A lung necropsy after seven days of treatment has shown a significant decrease in the viral load and inflammatory lung damage. Thus, the early administration of the remdesivir in the rhesus macaques with non-lethal SARS-CoV2 infection proves clinically beneficial. Hence, the early remdesivir administration to patients with COVID-19 may prevent their progression to lethal COVID-19. The preclinical studies have shown its benefits, but in clinical settings remdesivir did not prove significantly beneficial to severe COVID-19 patients and 66% patients have shown adverse events [256]. The remdesivir has been given to serious COVID-19 patients on compassionate basis due to some clinical improvements (improved respiration), but needs further randomized clinical trials for its frequent use as it has worsened the disease in 15% patients and 13% patients died after the remdesivir treatment completion [257]. However, remdesivir treatment is superior to the placebo in adult patients despite having adverse events, and lowers the risk of respiratory tract infections therefore it is the only drug of choice for severe COVID-19 patients [339]. The USA Food and Drug Administration (FDA) has issued an emergency use authorization for remdesivir in adults and children with severe COVID-19 [340]. The remdesivir should be administered intravenously that limits its frequent use.
Ribonucleoside analogue β-d-N4-hydroxycytidine (NHC; EIDD-1931) is another broad-spectrum antiviral drug against SARS-CoV-2, MERS-CoV, SARS-CoV, and related zoonotic group 2b or 2c bat-CoVs [341]. It also highly effective against a CoV bearing resistance mutations to the remdesivir. Another advantage of ribonucleoside analog β-d-N4-hydroxycytidine over remdesivir is its good oral bioavailability as indicated in preclinical studies in animal models of SARS-CoV and MERS-CoV infection. It improves the pulmonary function, reduces virus load, and prevents weight loss in preclinical animal models [341]. However, its availability for clinical trials will take longer time. The hydroxychloroquine (HCQ) earlier suggested as an effective molecule for SARS-CoV2 has failed (even as when used as post-exposure prophylaxis within 4 days after exposure) in the clinics in all categories of COVID-19 patients [342], [343], [344], [345]. The combination of HCQ with azithromycin given to patients with mild to moderate COVID-19 had not improved their clinical status in 15 days in comparison to the standard therapy [346]. Even these patients show a prolonged corrected QT interval and elevated liver-enzyme levels upon treatment with HCQ alone or in combination with azithromycin [346]. Therefore, WHO has stopped the HCQ arm of the solidarity trial (an international clinical trial to help find an effective treatment for COVID-19, launched by WHO and partners) for COVID-19 as of 17 June 2020. Hence, currently we are lacking potent and safe antivirals against SARS-CoV2 therefore we have left with the option to prepare and strengthen our immune system to fight the disease through novel immunomodulators, immunotherapeutics (vaccines or cell-based or Abs-based immune therapies). Following sections describe the development in the direction.
A retrospective multicenter cohort study with 446 COVID-19 patients has shown the protective action of early IFN (IFN-α2b) therapy and delayed IFN therapy increases the mortality [347]. IFN-α2b and umifenovir or arbidol (antiviral drug used against influenza in Russia and China) alone or in combination reduce mortality and accelerate recovery in comparison to treatment with lopinavir/ritonavir (LPV/r) [347]. The early IFN therapy exerts both antiviral and immunomodulatory action, whereas late IFN only antiviral action [347]. The mAb, CR3022 (a neutralizing Ab obtained from SARS-CoV convalescent patient) binds tightly to the epitope distal to the SARS-CoV2 S protein RBD, which facilitates the fusion-incompetent post fusion state, and incubation of S protein with CR3022 Fab (RBD-Fab complex) destroys the prefusion trimer [348], [349], [350]. Thus this cryptic epitope in the RBD-based vaccine may serve as an immunomodulator and can be used with an Ab with a potential to block receptor attachment with the virus [348]. The CR3022 can only access the binding epitope only when at least two RBDs of the trimeric S protein are in the upright conformation and slightly rotated or in “open” conformations, but not in “closed” prefusion S structure [349]. However, CR3022 mAb does not neutralize SARS-CoV2 infection [350], [351]. The CR3022 mAb binds more tightly to the SARS-CoV epitope due to the presence of glycan, but SARS-CoV2 epitope does not have glycan [349]. However, another group has isolated and identified another monoclonal NAb called EY6A from convalescent COVID-19 patient that also binds to the SARS-CoV2 S protein RBD tightly to the epitope, which resides away from the ACE2 receptor binding site [352]. These epitope residues are key to stabilizing the prefusion S protein. The EY6A binds to the epitope that may serve as a major target for NAbs, and may serve as a therapeutic target for COVID-19.
The animal studies have shown the efficacy of murine polyclonal antibodies (PolyAbs) raised against SARS-CoV for SARS-CoV2 infection via preventing the virus entry in the host cell to establish a productive infection [353]. This shows the cross reactivity of the Abs generated against SARS-CoV to SARS-CoV2. Hence, this animal data has a potential to extrapolate to human studies. However, the sera collected from the SARS-CoV infection recovered patients only moderately protect the SARS-CoV2 S pseudovirions infected 293/hACE2 cells, but sera from COVID-19 recovered patients have shown full protection to SARS-CoV2 pseudovirions infected 293/hACE2 cells [121]. The treatment with sera of recovered COVID-19 patients to SARS-CoV S pseudovirions infected 293/hACE2 cells does not exert any effect on transduction. Hence, the sera obtained from COVID-19 and SARS recovered patient have limited cross-neutralization effect.
The in vitro treatment with the human recombinant soluble ACE2 (hrsACE2) reduces the SARS-CoV2 recovery from Vero E6 cells by the factor of 1000–5000 [354]. The hrsACE2 treatment also inhibits the SARS-CoV2 infection in human kidney and blood vessels organoids. Thus, it comprises a potential recombinant protein-based therapeutic approach at early stages of COVID-19. It has already undergone phase I and phase II clinical trials in patients with ARDS under the name GSK2586881 [355]. ACE2-Fc human IgG1 fusion protein with mutations in the catalytic domain of ACE2 (MDR504) has also a potential to use as an immunotherapeutic agent as it achieves therapeutic concentration in the lungs upon parenteral administration to neutralize SARS-CoV2 via binding to S protein [356]. ACE2-Fc human IgG1 fusion protein contains a LALA (Leu234Ala and Leu235Ala double mutation) mutation in the ACE2 catalytic domain, which abrogates its FcRγ binding, but retains its FcRN binding to prolong its half-life and achieve therapeutic concentration in the lung alveoli. This MDR504 hACE2-Fc has a great potential for pre or post-exposure prophylaxis or treatment of COVID-19. The MDR504 hACE2-Fc is effective against COVID-19 via parenteral route and has a long serum half-life of ~145 h with an excellent bioavailability in the epithelial lining fluid of the lungs [356].
Baricitinib (an oral, selective and reversible inhibitor of the Janus kinases JAK1 and JAK2 previously used in rheumatoid arthritis (RA) patient to treat exaggerated inflammatory immune response) treatment (4 mg twice daily for 2 days followed by 4 mg per day for the remaining 7 days) has shown a significant improvement in COVID-19 patients with pneumonia [357]. The baricitinib treatment significantly decreases the mortality, pro-inflammatory cytokines (IL-6, IL-1β, and TNF-α) without affecting IL-8 levels, increases circulating T (especially, CD4+T and CD3+CD4+CD45RA−CD27−TEms), and B cell numbers [357]. The increase in naïve and central memory CD4+T, and B cells also occurs with baricitinib treatment. However, baricitinib treatment does not affect absolute NK cell, neutrophil, and CD8+T cell numbers in these patients. A time-dependent decrease in senescent CD8+CD45RA+CD57+CD27−T cells along with an increase in naïve CD8+CD45RA+CD57+CD27−T cells and memory CD3+CD8+CD27+CD45RA−T cells occurs with the baricitinib treatment [357]. However, no alteration in HLA-DR+CD38+ (activated), CD3+CD8+ T cells occurs. Baricitinib treatment also significantly increases IgG Abs as compared to the control COVID-19 pneumonia patients without affecting serum IgA levels. The baricitinib treatment also decreases classical (CD14highCD16low) pro-inflammatory monocytes, but it increases non-classical monocytes (CD14lowCD16high). Hence, baricitinib has a potent anti-inflammatory and immunomodulatory effects in patients with COVID-19-induced pneumonia, and decreases the incidence of death among these patients along with standard antiviral drugs [357]. A combination of ruxolitinib (a JAK1/2 inhibitor), 10 mg/twice daily for 14 days and eculizumab (an anti-C5a complement Mab), 900 mg IV/weekly for a maximum of three weeks to severe COVID-19 patients has significantly improved respiratory symptoms, radiographic pulmonary lesions, and decreased the circulating D-dimer levels in comparison the available standard therapy [358]. The combination also turns off the dysregulated innate and adaptive immune response responsible for severe COVID-19 and associated ARDS. AMY-101 (a compstatin C3 inhibitor) in infection model has shown the blockade of IL-6 release, the major cytokine associated with severe COVID-19 and associated ARDS [359], [360]. The C3 inhibition can also inhibit the C3a and C5a generation that may block C3a-C3aR and C5a-C5aR1 interaction-mediated ALI through inhibiting the pro-inflammatory cytokines (TNF-α, and IL-6) generation and neutrophil infiltration in the lungs.
A triple antiviral therapy (IFN-β-1b, lopinavir-ritonavir, and ribavirin)-based phase II clinical trial has shown its safety and superiority to the lopinavir-ritonavir combination in alleviating the COVID-19 symptoms, shortening viral shedding, and hospital stay in patients with mild to moderate COVID-19 [361]. In this triple antiviral therapy, along with antiviral action, the IFN-β-1b also exerts immunomodulatory effects, including decrease in the pro-inflammatory cytokine releases from myeloid cells (macrophages and neutrophils) and T cells, increases Tregs production along with IL-10 and TGF-β production by B cells, and enhances the release of Th2 cytokines (IL-4, IL-5, and IL-13) [362]. Thus, it will be interesting to study the immunomodulatory action of IFN-β in COVID-19 patients. A novel lipopeptide EK1C4 through the intranasal route has shown the protection against βCoV, including SARS-CoV2 infection in mice via inhibiting the viral S protein-mediated entry of host cells through ACE2 receptor [363]. The treatment of sever COVID-19 patients over 18 years of age with mavrilimumab (an anti-GM-CSF mAb) in a single-centre prospective cohort study has shown zero mortality, great clinical improvement in terms of decrease in pneumonia-induced ALI, and early recovery from ventilators along with the fever resolution [364]. Hence, mavrilimumab has a great potential for sever-COVID-19 patients and future clinical studies will strengthen these findings for its use worldwide.
The sera or plasma obtained from COVID-19 convalescent patients is helpful for COVID-19 patients via increasing the radiological resolution, reducing the viral load, increasing the HLA-DR+ immune cells, and has improved the patient survival with less than 1% serious adverse effects in severe COVID-19 patients [204], [365], [366]. However, the prophylactic use of convalescent sera is more beneficial as compared to its use as a curative therapy [19], [366]. The use of convalescent sera as a curative therapy is most effective only soon after the diagnosis of COVID-19 [367]. COVID-19 convalescent plasma (CCP) therapy has cleared the SARS-CoV2 from 80% critical COVID-19 patients post 14 days and S- and RBD-specific IgGs increase within 3 days of the start of the therapy [368]. However, these IgGs decrease within 3 to 7 days after therapy. Of note, nucleocapsid or nucleoprotein (N)-specific Abs do not increase following the CCP therapy. Thus, immediate increase in the NAbs (IgGs) against S-protein and RBD is one of the reasons for CCP efficacy in critical or severe COVID-19 patients. Along with an increase in NAbs within three days of CCP, lymphocytes also increase significantly in this time period, which remain upregulated within 21 days post CCP therapy [368].
The CCP therapy also decreases neutrophil infiltration in the lungs and circulating CRP also decrease post CCP therapy [368]. Hence, CCP therapy has resolved the lung inflammation in 80% of the severe COVID-19 patients. The CCP therapy does not impact cardiac, renal, and liver functions except decreasing the total bilirubin levels. The IL-6, TNF-α, and IL-10 levels remain same in these patients before and after the CCP therapy [368]. Hence, the severe COVID-19 patients who respond well to CCP therapy recover from the disease and the associated ALI, clear the virus, show an increase in the lymphocytes with a significant decrease in the neutrophil infiltration in the lungs. On the other hand, non-responders to the CCP therapy show an increased CRP, LDH, B-type natriuretic peptide (produced by heart, which increases heart failure or it gets worse), urea nitrogen (indicates kidney damage), glucose, and procalcitonin (PCT) level [368]. Higher Abs in the donated plasma confer immediate immunity to the receiving patient. Thus, higher the Ab number in the donated plasma, better is the efficacy of CCP. The CCP therapy even after 2 weeks of the onset of COVID-19 symptoms is beneficial in improving the symptoms and survival in patients with severe COVID-19 [368]. Another study with small sample size (20 critical COVID-19 patients) has shown beneficial effect of CCP therapy without any adverse event, including hyper-inflammation [369]. However, the CCP therapy has its own regulatory and logistical challenges described somewhere else, should be considered [19], [370].
Recently, a research group has isolated mAbs (CA1 and CB6) from convalescent COVID-19 patients, which exert SARS-CoV2 neutralizing activity in vitro, and CB6 mAb inhibits the SARS-CoV2 infection progression when given therapeutically or prophylactically to rhesus macaques before or after infection [351]. The protective effect of CB6 mAb involves the recognition of the epitope in the RBD region of the SARS-CoV2 through its VH domain that prevents virus binding to its entry receptor ACE2. However, both VH and VL of CB6 mAb hinder the mAb-bound SARS-CoV2 to interact with ACE2 [351]. Furthermore, the LALA mutation induction in the Fc portion of CB6 mAb eliminates the Ab-dependent cellular cytotoxicity (ADCC) effect and CB6 (LALA) mAbs show protective effect in rhesus macaques infected with SARS-CoV2 without exacerbating the inflammatory tissue damage [351]. The single-domain antibodies (VHHs) isolated from llama immunized with prefusion-stabilized coronavirus spikes have shown a potential to neutralize SARS-CoV and MERS-CoV S pseudotyped viruses [371]. The crystal structure of VHHs has shown the presence of two different epitopes, but both the VHHs (SARS-CoV-specific and MERS-CoV-specific) interfere the receptor binding. However, SARS-CoV S-specific VHH cross-reacts with SARS-CoV2 S. This cross-reactive VHH neutralizes SARS-CoV2 pseudotyped viruses as a bivalent human-Ig Fc fusion [371]. Hence, these VHHs have a great potential for COVID-19 due to their less susceptibility to stearic hindrance and greater stability than larger conventional Abs [372]. Even neutralizing nanobodies (H11-D4 and H11-H4) have been developed, which bind to the SARS-CoV2 S protein RBD and block its interaction with ACE2 to neutralize the SARS-CoV infection [373]. They rely on three variable loops (CDR1, CDR2, and CDR3) to form the antigen binding site. Thus the nanobodies fused with Fc neutralize SARS-CoV2 interaction with ACE2, and show an additive effect in the presence of CR3022 [373].
The novelty of the SARS-CoV2 is the major hurdle in the treatment options for COVID-19 patients and the SARS-CoV2 infection in huge population around the world has compelled researchers to adopt different approaches to develop new or repurpose previously available antivirals and immunomodulators. Thus understanding the need of emergency of the vaccine a group has developed a pilot-scale production of PiCoVacc, a purified inactivated SARS-CoV2 vaccine capable of inducing SARS-CoV2–specific neutralizing Abs (IgG) in different laboratory animals [BALB/c mice (at 6 weeks of challenge), Wistar rats (at 7 weeks of challenge), and rhesus macaques (at 2 weeks of challenge)] [374]. These SARS-CoV2–specific neutralizing Abs generated in response to the PiCoVacc are able to neutralize ten different SARS-CoV2 strains. Hence, these NAbs have broad specificity to neutralize other SARS-CoV2 strains. However, inactivated SARS-CoV2 immunized mice produce ~30 fold less N-specific IgGs than that of Abs targeting S or RBD [374]. On the other hand humans infected with COVID-19 produce a higher amount of N-specific IgG that serves as a one of the clinical diagnosis marker of SARS-CoV2 infection, but do not provide immune protection therefore people died due to COVID-19 have high nucleocapsid or N-specific Abs [293], [375]. Hence, future studies will determine the use of this vaccine in the clinics.
Three immunizations with two different doses (3 or 6µgm/per dose) have provided a full protection to tested macaques against SARS-CoV2 infection [374]. However, this study is insufficient to rule out the induction of antibody-dependent enhancement (ADE) of the infection and studies with longer duration of observation of animals after vaccination may clarify the ADE issue. The PiCoVacc challenged animals only develop NAbs without any change in the lymphocyte subset percentage (CD3+, CD4+, and CD8+), and key cytokines (TNF-α, IL-2, IL-4, IL-5, IL-6, and IFN-γ) in comparison to sham and placebo treated group. This strategy can be used in humans in the absence of any other safer choice as this approach of vaccination has its drawbacks, including reactivation of the pathogenic virus, requirement of adjuvants, and booster doses etc. Hence, the sequence homology and bioinformatics approaches will prove useful tool to design various other novel immune-based therapeutics, including Ab and T cell-based vaccinations via identifying different conserved epitopes to overcome these hurdles (absence of cross reactivity across βCoVs and resistance in virus due to evolution) for SARS-CoV2 vaccination [23], [376]. For example, the immune epitope database and analysis resource (IEDB, a repository of epitope-related information) use has identified multiple specific regions in SARS-CoV2 with a high homology to the SARS-CoV responsible for 2002–2003 SARS epidemic and parallel bioinformatics studies have provided a priori potential for B and T cell epitopes [23]. Thus two independent experimental approaches identifying same epitope (that were not available previously for SARS-CoV2) for adaptive immune response activation indicate their high incidence for the use of vaccine candidate for SARS-CoV2 infection. For example, 12 of 17 SARS-CoV-2 T cell epitopes having ≥90% sequence identity to the SARS-CoV have been independently identified through epitope predictions depending on the SARS-CoV-2 sequences [23].
A replication competent vesicular stomatitis virus expressing a modified form of the SARS-CoV-2 spike gene in place of the native glycoprotein gene (VSV-eGFP-SARS-CoV-2) is an attenuated replication-competent vaccine candidate that generates NAbs in mice expressing humanized ACE2 upon immunization and protects these mice from pneumonia and inflammation following SARS-CoV2 infection [377], [378]. Even passive transfer of serum from immunized mice to naive mice expressing human ACE2 protects from SARS-CoV2 infection. Thus attenuated SARS-CoV2 vaccine has also been developed and tested in laboratory animals. A positive outcome in the phase 1, dose escalated, open trial, including 45 healthy individuals with the candidate vaccine mRNA-1273 encoding the stabilized prefusion SARS-CoV-2 S protein has raised a hope in the scientific community for developing effective vaccines for SARS-CoV2 [379]. The study has shown the development of S-2P specific binding IgG Abs and RBD-specific binding Abs along with the generation of NAbs in all individuals received second dose of the vaccine. Also, increasing the dose from 25 µg to 100 µg elicited the CD4+T cells based Th1 immune response and cytotoxic CD8+T cell response occurs only in group given 100 µg of the [379]. However, invited adverse reactions, including fatigue, chills, headache, and myalgia have occurred in more than 50% of people involved in the study. People receiving the first booster (higher concentration than the first vaccination) dose have shown systemic adverse events.
Another, chimpanzee adenovirus-vectored vaccine (ChAdOx1 nCoV-19, consisting of the replication-deficient simian adenovirus vector, ChAdOx1 containing the full-length S protein) of SARS-CoV-2, with a tissue plasminogen activator (tPA) leader sequence), called AZD1222-based phase 1/2 single-blind randomized trial in the UK in 18–55 years old people has also shown promising effects (a peaked spike-specific T cell immune response on day 14 and a rise in anti-spike Ab response on day 28 of intramuscular injection) [380]. All the people enrolled in the vaccine trial develop NAbs after first booster dose. Mild local and systemic immune reactions, including pain, fever, chills, myalgia, headache, and malaise develop in these people upon vaccination. Another, non-replicating adenovirus type-5 (Ad5)-vectored COVID-19 vaccine at 5 × 1010 virus particle has been found safe in healthy individuals (603 healthy adults) that induces a significant amount of RBD-specific NAbs to clear the SARS-CoV2 infection [381].
Recently, different strategies to measure the SARS-CoV2 NAbs activity involving pseudotyped [SARS-CoV-2 spike-pseudotyped, single-cycle, replication-defective human immunodeficiency virus type-1 (HIV-1)] and chimeric viruses (replication competent vesicular stomatitis virus (VSV)/SARS-CoV2 chimeric virus) have been developed [382]. These viruses may prove useful to observe the efficacy of potential SARS-CoV2 NAbs generated through different vaccines or the presence of NAbs in convalescent plasma or serum. Another group has developed a SARS-CoV-2 surrogate virus neutralization test, which identifies total immunodominant NAbs against RBD of the SARS-CoV2 S protein in an isotype- and species-independent manner [383]. This simple and rapid test blocks the interaction between ACE2 and RBD of the SARS-CoV2 through NAbs, and does not require biosafety level 3 (BSL3) facility [383]. Also, a study has identified that the IgG variable heavy chain3-53 (IGHV3-53) is the most frequently used IGHV gene to target RBD of the S protein [384]. These IGHV3-53 Abs are very potent against SARS-CoV2 RBD and show minimum affinity maturation showing their promise as a vaccine design candidate [384]. The detailed structure of IGHV3-53 Abs will prove helpful to design artificial Ags with a potential to induce NAbs. NK cell and DC-based immunotherapies should also be explored for COVID-19-associated pneumonia as antiviral and immunoregulatory function of these innate immune cells dysregulate causing severe COVID-19 infection. Hence, different immunomodulatory and vaccine-based immunotherapeutics are in pipeline for COVID-19. Their preclinical toxicity studies and clinical trials will determine their suitability in human use as vaccines or immunomodulatory drugs.
10. Laboratory mice to study SARS-CoV2 infection, immunopathogenesis, and test potent antivirals, immunomodulators, and immunotherapeutics
We cannot infect humans directly with SARS-CoV2 therefore to study its immunology and test various immunomodulators and vaccine candidates, we urgently need small animal (mice) models readily available for researchers. This is because normal laboratory mice of different strains (3-to-4-week-old BALB/c, DBA/2J, and C57BL/6), including immunocompromised ones (STAT1−/− C57BL/6, AG129 (type I and II IFN receptor-deficient), and Rag1−/− C57BL/6 (do not have mature B and T cells) do not readily get SARS-CoV2 infection due to the lack of effective and strong binding of SARS-CoV2 S protein with the mouse ACE2 (mACE2) [117], [126], [385]. Different approaches to develop transgenic laboratory mice expressing human ACE2 (hACE2) for inducing moderate to severe SARS-CoV2 infection have been utilized. Recently, transgenic mice expressing hACE2 receptor in the lungs have been developed through the intranasal instillation of the replication-defective adenoviruses encoding hACE2 (AdV-hACE2) in the BALB/c mice [385]. These mice upon SARS-CoV2 infection show high virus titer in the lungs along with low virus levels in the heart, spleen, and brain, with no virus in the serum, kidneys, and GIT [385]. This tissue distribution of SARS-CoV-2 infection may be due to the both, the delivery and expression of the AdV-hACE2, and the natural tropism of the virus [385]. However, in this transgenic model authors have not measured the mortality as seen in severe COVID-19 patients. This model is good to study COVID-19-induced pneumonia and therapeutic interventions.
Another group has developed HFH4 (FoxJ1, a gene crucial for ciliated epithelial cell differentiation) promoter driven hACE2-overexpressing transgenic mice as a model for SARS-CoV2 infection [386], [387]. These HFH4-hACE2 transgenic mice developed on C3B6 background express human ACE2 on ciliated epithelial cells of respiratory tracts (nasal, tracheal, and bronchial epithelial cells) and in the cells of central nervous system (CNS), including multiciliated ependymal cells (MECs, specialized epithelial cell lining mammalian brain ventricles) [387]. These mice are crucial as SARS-CoV2 primarily infect ciliated epithelial cells and AT2 cells or type 2 pneumocytes [47]. However, eyes, heart, and brain have also shown the presence of the viral RNA [386]. However, the SARS-CoV2 infection to these mice establishes pneumonia and ALI, but mice die with exaggerated neuroinflammation due to the viral neuroinvasion [386]. Further study utilizing whole body plethysmograph (WBE)-mediated measurement of respiratory changes has proven that mortality in these mice upon SARS-CoV2 infection occurs due to the neuroinvasion of the virus but not due to the respiratory failure [388], [389]. Hence, this approach did not develop an animal model for severe COVID-19 causing death due to the ALI or ARDS.
Another group has developed hACE2 transgenic mice on C57BL/6 background through CRISPR/Cas9 mediated gene knockin technology [390]. These hACE2 transgenic mice (both, young and older adults) develop SARS-CoV2-induced pneumonia and brain infection upon intranasal instillation of the virus. This transgenic mouse model did not show fatality and intragastric instillation of the SARS-CoV2 has also developed pneumonia [390]. Hence, this model can be used for intranasal and orally-induced SARS-CoV2 infection, and identifying novel immunomodulators and vaccine candidates. Another group has developed hACE2 transgenic mice (HB-01 background) and they have showed 6–11 months old hACE2 transgenic mice developed pneumonia whereas WT HB-01 did not [391]. Hence, this model can be used for older adults on these findings. Authors have not reported fatality in this model too, whereas OCIVD-19 is more fatal in older adults. However, it would be interesting to observe the SARS-CoV2 infection in these young transgenic mice. Another group has developed a different hACE2 transgenic mice through using Venezuelan equine encephalitis virus (VEEV) replicon expressing hACE2 with a C-terminal S-tag packed into VRPs with the help of the helper RNAs encoding VEEV capsid and envelope proteins to produce VEEV-VRP-hACE2 in both C57BL/6 and BALB/c mice [392]. These mice two days post SARS-CoV infection develop high NLR in the peripheral blood and also induce Ab production. These VEEV-VRP-hACE2 transgenic mice develop pneumonia and support viral replication in the lungs as indicated in COVID-19 patients [392]. The human mAb called CB6 has shown the protective action in these mice infected with SARS-CoV2 via clearing the infection and pulmonary lesions. Thus, VEEV-VRP-hACE2 transgenic mice are another transgenic mice shown appropriate for SARS-CoV2 mouse animal model.
Another group has developed another transgenic mice to study the diversity of the immune response seen in COVID-19 patients, these transgenic mice have cytokeratin-18 (K18) gene promoter-derived humanized ACE2 receptors (K18-hACE2) and develop signs of severe COVID-19 upon intranasal instillation of the SARS-CoV2 that spreads to the lungs and other organs (heart, kidneys, spleen, and intestinal tissues) [393]. The K18-hACE2 transgenic mice upon infection with SARS-CoV2 also shows many immunological features (neutrophil, monocyte, and T cell infiltration in the lungs, proinflammatory cytokine response, type I and II IFN signaling, and leukocyte signaling pathways) seen in COVID-19 patients. The K18-hACE2 transgenic mouse model is better. This is because the transgenic mice expressing human ACE2 in lungs only can be used only to study SARS-CoV2-induced pneumonia or ALI but the K18-hACE2 transgenic mice have human ACE2 in multiple tissues (liver, kidneys, and gastrointestinal tract) [394]. Also, K18-hACE2 transgenic mice upon intranasal SARS-CoV2 infection do not show viral RNA in the circulation and in the gastrointestinal tract till seven days post infection [393]. The tissue expression of hACE2 follows the pattern lungs ≥ colon ≥ kidneys ≥ brain. However, hACE2 in these mice declines after infection, suggesting the death hACE2 expressing cells after infection, hACE2 shedding or the receptor downregulation [393]. This transgenic SARS-CoV2 infection mouse model develops clinically observable disease in the lungs without infecting the brain. Only one of six mice show the viral RNA presence in the brain after 7 post infection days, in which infection has disseminated to the whole cortex with observable sparing of the olfactory bulb and cerebellum [393]. Thus K18-hACE2 transgenic mouse is a better animal model for studying severe-COVID-19 than HFH4-hACE2 mice developed on C3B6 background. However, one K18-hACE2 transgenic mouse out of 6 mice developed severe brain inflammation, two out of total 9 hearts showed hypereosinophilic cardiomyocytes with pyknotic nuclei, all livers were inflamed in infected animals, one kidney showed inflammatory injury and all spleens isolated form SARS-CoV2 infected K18-hACE2 transgenic mice were normal. Thus, this transgenic mouse model is also good to study COVID-19-induced multiorgan failure.
Another group has developed transgenic mice on both, BALB/c and C57BL/6 backgrounds by exogenous delivery of human ACE2 with a replication-deficient adenovirus (Ad5-hACE2) [395]. Both, these transgenic mice develop pneumonia, weight loss, and high virus load in the lungs. However, Ad5-hACE2 transgenic mice on BALB/c background show a higher weight loss (~20%), higher necrotic cell debri in lungs than transgenic mice with C57BL/6 mice background [395]. Researchers have also used Ad5-hACE2 transgenic mice for elucidating the protective role of type 1 IFNs, and STAT1, vaccine testing, CCP therapy testing, and antivirals (polyinosinic:polycytidylic acid (poly I:C) and remdesivir) [395]. Of note, the K18-hACE2 transgenic mouse has C57BL/6 background, whereas the replication-defective adenoviruses encoding human ACE2 transgenic mice has a BALB/c background. Although, both are inbred mice but show different immune response, including innate and adaptive response during infection [396], [397], [398], [399]. Hence, we need caution, while using these transgenic animal models depending on the parameters needs to studying (vaccine or immunomodulator testing or studying immune response) during SARS-CoV2 infection.
As above transgenic mice have some immunologic discrepancies in context to the COVID-19 in humans therefore another group has developed DRAGA (HLA-A2.HLA-DR4.Rag1KO.IL-2Rγc KO.NOD) mice infused with human hematopoietic stem cells from cord blood reconstituting a fully functional human immune system along with engrafting human epithelial and endothelial cells [400]. The DRAGA mice sustain SARS-CoV2 infection and develop severe-COVID-19-like symptoms, which mimic human immune response causing COVID-19-induced pathology and immunopathogenesis [400]. Hence, DRAGA mice will prove best animal model to study COVID-19 immunopathogenesis and studying the efficacy of different immunomodulators and vaccines. Advancing the one step in the direction, researchers have mutated the SARS-CoV2 to the mouse adapted SARS-CoV2MA virus that infects efficiently to younger adult BALB/c mice to cause mild to moderate infection [388]. The SARS-CoV2 does not bind to the murine ACE2 (mACE2) due to the incompatibility of SARS-CoV-2 Q498 and researchers have engineered Q498Y/P499T into the SARS-CoV-2 S gene through reverse genetics to design the recombinant virus (SARS-CoV-2MA) [47], [388]. However, this SARS-CoV2MA infects more potently and severely to older BALB/c mice (loss of pulmonary function also seen), like COVID-19 has more severe phenotype in older adults [388]. Several pro-inflammatory cytokines and chemokines increase in the lungs after 2 days post infection without an increase in the serum. Thus this virus can be used in young and older adult mice for developing mild to moderate and severe SARS-CoV2 infection resembling human COVID-19, which can be also used for immunomodulatory and vaccine candidates for COVID-19 [388]. Hence, different hACE2 transgenic mice have been developed to open the door for SARS-CoV2 biomedical research along with one mutant virus that can infect WT mice. However, caution and further studies are required.
11. Future perspectives and conclusion
The story of CoVs is not new to humans and they were first identified simultaneously in the UK and USA in early 1960s as the causal viruses for common cold and mild respiratory infections. However, they draw their attention following the emergence of SARS-CoV-induced severe acute respiratory syndrome (SARS) epidemic in 2002–2003. In 2014, another CoV-induced infection, called MERS emerged in the middle-eastern countries that spread to the UK, USA, and Canada. SARS and MERS have one thing in common that both emerged from bat CoV with different intermediate hosts. For example, masked palm civets and racoon dogs served as intermediate host for SARS and dromedary or Arabian camels for MERS-CoV. However, SARS-CoV2 has also originated from bats called horseshoe bats, but we still do not know the intermediated host from which the infection passed to humans and took the form of current pandemic. Hence, identifying the intermediate host will be beneficial to fight more specifically with the pandemic. For example, we can immunize those animals to prevent the future infection spread and also reveal the conditions, which lead to the evolution of the highly pathogenic SARS-CoV2 as the pathogenic evolution leading to the evolution of SARS-CoV2 has neither occurred in humans nor in bats as bats have closest CoV called RaTG13 [41]. However, we do not know about the common ancestor for RaTG13 and SARS-CoV2, whether that still exist, if yes in which intermediate host? Hence the chain of events leading to the SARS-CoV2 emergence and COVID-19 in humans should be discovered to efficiently deal with the virus. Studying evolution is key to investigate the emergence of biological processes, including infection and immunity.
Plenty of studies have divided patients in terms of symptoms varying form asymptomatic to mild to moderate to severe. Asymptomatic patients do not develop a robust pro-inflammatory innate immune response required to clear the infection, instead they harbor the virus, and serve as potent reservoirs and may spread infection for longer duration as compared to symptomatic individuals before recovery. Asymptomatic COVID-19 patients exhibit the NK and Th1 cells-dependent direct antiviral action agaisnt SARS-CoV2. However, active asymptomatic COVID-19 patients show an anti-inflammatory immune response (IL-13 and IL-10) and convalescent asymptomatic COVID-19 patients show weaker of Ab response that fades away very soon. They develop robust T cell memory response. The mechanisms behind this diverse immune response in asymptomatic active and convalescent COVID-19 patients should be explored. On the other hand, mild and moderate patient develop healthy and controlled immune response to clear the infection without its dysregulation as seen in severe COVID-19 patients. For example, in severe COVID-19 patients, virus hijacks the component of innate immune response (cGAS and RLR-mediated virus recognition, and the type 1 IFNs generation) which can directly recognize and kill it. But it, promotes the TLR7/TLR8-based pro-inflammatory immune response (cytokine storm), profound monocyte and neutrophil infiltration in the lungs to cause ALI or ARDS. Hence, only severe/critical COVID-19 patients lose life. Transgenic mice expressing hACE2 may serve as potential animals for studying SARS-CoV2 infection, immunity, and different drugs, including vaccines. However, a mutant SARS-CoV2MA can infect WT mice and can induce mild to moderate infection in younger mice and severe pneumonia in older mice. Thus we are entering in an era where animal model-based studies will provide unexplored immunological mechanisms.
For example, thalidomide is a potent immunomodulatory agent, and has been used in pneumonia and sepsis previously as an immunomodulatory drug [401], [402], [403]. Even a recent, clinical study has indicated its immunomodulatory and protective action in sever COVID-19 patients as an adjuvant therapy with low‐dose short‐term glucocorticoid [404]. Even currently, thalidomide is under phase II clinical trial for severe COVID-19 patients (ClinicalTrial.gov Identifier: NCT04273529). The immunomodulatory role of statins should also be studied in severe COVID-19 [405]. In a recent study, the in hospital use of statins has decreased the mortality rate in COVID-19 patients due to its immunomodulatory action that decreased IL-6 levels and neutrophil count [406]. The in vivo mechanism of thalidomide and statins in COVID-19 needs an exploration using transgenic hACE2 receptor expressing mice or mutant SARS-CoV2MA in WT mice.
12. Conclusion
Immune system has evolved to protect the host from pathogens and internal damage-associated molecular patterns (DAMPs). However, due to the novelty of the SARS-CoV2 it may be a difficult target for the human immune system causing a dysregulated immune response during COVID-19. For example, some develop asymptomatic infection and other develop severe disease causing increased mortality. Even the SARS-CoV2 has different factors or proteins, which hijack the direct antiviral defence machinery without affecting the pro-inflammatory cytokine release to induce cytokine storm. Hence, understanding the immunology of the COVID-19 or SARS-CoV2 infection is one of the several roads for the successful management of the infection. Now we have human ACE2 transgenic mice to use for immunology, immunopathogenesis, and therapeutics-based studies. Therapeutics are urgently needed to strengthen the immune system to accept the SARS-CoV2 challenge to the immune system that is “catch me if you can”.
13. Contribution
The author developed the idea, searched for the literature, wrote and compiled the manuscript, along with designing and developing the figures.
Funding
The author did not receive any funding for the current work.
Declaration of Competing Interest
The authors declare that he does not have any known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2020.106980.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurer F.D. Equine piroplasmosis–another emerging disease. J. Am. Vet. Med. Assoc. 1962;141:699–702. [PubMed] [Google Scholar]
- 3.Barré-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vézinet-Brun F., Rouzioux C., Rozenbaum W., Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 4.D.T. Fleming, G.M. McQuillan, R.E. Johnson, A.J. Nahmias, S.O. Aral, F.K. Lee, M.E. St Louis, Herpes simplex virus type 2 in the United States, 1976 to 1994, N Engl. J. Med. 337(16) (1997) 1105-11. [DOI] [PubMed]
- 5.Kaposi's sarcoma and Pneumocystis pneumonia among homosexual men--New York City and California, MMWR Morb Mortal Wkly Rep 30(25) (1981) 305-8. [PubMed]
- 6.W.M.G. E, Ecological origins of novel human pathogens, Crit. Rev. Microbiol. 33(4) (2007) 231-241. [DOI] [PubMed]
- 7.Ndow G., Ambe J.R., Tomori O. Emerging infectious diseases: a historical and scientific review. Socio-cultural Dimensions Emerging Infect Dis Africa. 2019:31–40. [Google Scholar]
- 8.Baseler L., Chertow D.S., Johnson K.M., Feldmann H., Morens D.M. The pathogenesis of Ebola virus disease. Annu. Rev. Pathol. 2017;12:387–418. doi: 10.1146/annurev-pathol-052016-100506. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein T., Anthony S.J., Gbakima A., Bird B.H., Bangura J., Tremeau-Bravard A., Belaganahalli M.N., Wells H.L., Dhanota J.K., Liang E., Grodus M., Jangra R.K., DeJesus V.A., Lasso G., Smith B.R., Jambai A., Kamara B.O., Kamara S., Bangura W., Monagin C., Shapira S., Johnson C.K., Saylors K., Rubin E.M., Chandran K., Lipkin W.I., Mazet J.A.K. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat. Microbiol. 2018;3(10):1084–1089. doi: 10.1038/s41564-018-0227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kindhauser M.K., Allen T., Frank V., Santhana R.S., Dye C. Zika: the origin and spread of a mosquito-borne virus. Bull. World Health Organ. 2016;94(9):675. doi: 10.2471/BLT.16.171082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz D.A. The origins and emergence of zika virus, the newest TORCH infection: what's old is new again. Arch. Pathol. Laboratory Med. 2016;141(1):18–25. doi: 10.5858/arpa.2016-0429-ED. [DOI] [PubMed] [Google Scholar]
- 12.Glezen W.P. Emerging infections: pandemic influenza. Epidemiol. Rev. 1996;18(1):64–76. doi: 10.1093/oxfordjournals.epirev.a017917. [DOI] [PubMed] [Google Scholar]
- 13.Schoch-Spana M. “Hospital's full-up”: the 1918 influenza pandemic. Public Health Rep. 2001;116(Suppl. 2):32–33. doi: 10.1093/phr/116.S2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilbourne E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006;12(1):9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmuir A.D. Epidemiology of Asian influenza. With special emphasis on the United States. Am. Rev. Respir Dis. 1961;83(2 Pt 2):2–14. doi: 10.1164/arrd.1961.83.2P2.2. [DOI] [PubMed] [Google Scholar]
- 16.Viboud C., Simonsen L., Fuentes R., Flores J., Miller M.A., Chowell G. Global mortality impact of the 1957–1959 influenza pandemic. J. Infect. Dis. 2016;213(5):738–745. doi: 10.1093/infdis/jiv534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brook C.E., Dobson A.P. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23(3):172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allocati N., Petrucci A.G., Di Giovanni P., Masulli M., Di Ilio C., De Laurenzi V. Bat–man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discovery. 2016;2(1):16048. doi: 10.1038/cddiscovery.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar V. Emerging human Coronavirus infections (SARS, MERS, and COVID-19): Where they are leading us. Int. Rev. Immunol. 2020 doi: 10.1080/08830185.2020.1800688. [DOI] [PubMed] [Google Scholar]
- 20.Al-Omari A., Rabaan A.A., Salih S., Al-Tawfiq J.A., Memish Z.A. MERS coronavirus outbreak: implications for emerging viral infections. Diagn. Microbiol. Infect. Dis. 2019;93(3):265–285. doi: 10.1016/j.diagmicrobio.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J.P. Unger, Comparison of COVID-19 Health Risks With Other Viral Occupational Hazards, Int. J. Health Serv. (2020) 20731420946590. [DOI] [PMC free article] [PubMed]
- 23.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host & Microbe. 2020;27(4):671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu D., Zhu C., Ai L., He T., Wang Y., Ye F., Yang L., Ding C., Zhu X., Lv R., Zhu J., Hassan B., Feng Y., Tan W., Wang C. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect. 2018;7(1):154. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau S.K., Li K.S., Huang Y., Shek C.T., Tse H., Wang M., Choi G.K., Xu H., Lam C.S., Guo R., Chan K.H., Zheng B.J., Woo P.C., Yuen K.Y. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010;84(6):2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong R., Pei S., Yin C., He R.L., Yau S.S. Analysis of the hosts and transmission paths of SARS-CoV-2 in the COVID-19 outbreak. Genes (Basel) 2020;11(6) doi: 10.3390/genes11060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.-J., Luo C.-M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.-Y., Wang L.-F., Daszak P., Shi Z.-L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.K.-S. Yuen, Z.-W. Ye, S.-Y. Fung, C.-P. Chan, D.-Y. Jin, SARS-CoV-2 and COVID-19: The most important research questions, Cell & Biosci. 10 (2020) 40-40. [DOI] [PMC free article] [PubMed]
- 32.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., Hughes A.C., Bi Y., Shi W. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30(11):2196–2203.e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S., Wei W., Cheung W.Y.-M., Li W.-J., Li L.-F., Leung G.M., Holmes E.C., Hu Y.-L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 35.Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020;92(4):433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020 doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P., Jiang J.Z., Wan X.F., Hua Y., Li L., Zhou J., Wang X., Hou F., Chen J., Zou J., Chen J. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16(5):e1008421. doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., Chaillon A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O.A. MacLean, S. Lytras, J.B. Singer, S. Weaver, S.L.K. Pond, D.L. Robertson, Evidence of significant natural selection in the evolution of SARS-CoV-2 in bats, not humans, bioRxiv (2020).
- 41.Cagliani R., Forni D., Clerici M., Sironi M. Computational inference of selection underlying the evolution of the novel coronavirus, severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94(12) doi: 10.1128/JVI.00411-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H., Cai J.-P., Yip C.C.-Y., Chan I.H.-Y., Wong K.K.-Y., Tsang O.T.-Y., Chan K.-H., Chan J.F.-W., To K.K.-W., Chen H., Yuen K.Y. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26(7):1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 43.Nie Q., Li X., Chen W., Liu D., Chen Y., Li H., Li D., Tian M., Tan W., Zai J. Phylogenetic and phylodynamic analyses of SARS-CoV-2. Virus Res. 2020;287 doi: 10.1016/j.virusres.2020.198098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 45.K. Schlottau, M. Rissmann, A. Graaf, J. Schön, J. Sehl, C. Wylezich, D. Höper, T.C. Mettenleiter, A. Balkema-Buschmann, T. Harder, C. Grund, D. Hoffmann, A. Breithaupt, M. Beer, SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study, The Lancet Microbe. [DOI] [PMC free article] [PubMed]
- 46.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein. J. 2020;39(3):198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R., Gralinski L.E., Schäfer A., Dang H., Gilmore R., Nakano S., Sun L., Fulcher M.L., Livraghi-Butrico A., Nicely N.I., Cameron M., Cameron C., Kelvin D.J., de Silva A., Margolis D.M., Markmann A., Bartelt L., Zumwalt R., Martinez F.J., Salvatore S.P., Borczuk A., Tata P.R., Sontake V., Kimple A., Jaspers I., O’Neal W.K., Randell S.H., Boucher R.C., Baric R.S. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182(2):429–446. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young B.E., Fong S.-W., Chan Y.-H., Mak T.-M., Ang L.W., Anderson D.E., Lee C.Y.-P., Amrun S.N., Lee B., Goh Y.S., Su Y.C.F., Wei W.E., Kalimuddin S., Chai L.Y.A., Pada S., Tan S.Y., Sun L., Parthasarathy P., Chen Y.Y.C., Barkham T., Lin R.T.P., Maurer-Stroh S., Leo Y.-S., Wang L.-F., Renia L., Lee V.J., Smith G.J.D., Lye D.C., Ng L.F.P. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. The Lancet. 2020;396(10251):603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A. Wu, Y. Peng, B. Huang, X. Ding, X. Wang, P. Niu, J. Meng, Z. Zhu, Z. Zhang, J. Wang, J. Sheng, L. Quan, Z. Xia, W. Tan, G. Cheng, T. Jiang, Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China, Cell Host & Microbe 27(3) (2020) 325-328. [DOI] [PMC free article] [PubMed]
- 50.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.L. Zhang, C.B. Jackson, H. Mou, A. Ojha, E.S. Rangarajan, T. Izard, M. Farzan, H. Choe, The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity, bioRxiv (2020) 2020.06.12.148726.
- 52.Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., Vogels C.B., Brito A.F., Alpert T., Muyombwe A. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020 doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.S.T. Sehra, J.D. Salciccioli, D.J. Wiebe, S. Fundin, J.F. Baker Maximum daily temperature, precipitation, ultra-violet light and rates of transmission of SARS-Cov-2 in the United States. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. LoPresti, D.B. Beck, P. Duggal, D.A.T. Cummings, B.D. Solomon, The Role of Host Genetic Factors in Coronavirus Susceptibility: Review of Animal and Systematic Review of Human Literature, medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 55.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., Miao X., Li Y., Hu B. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan. China, JAMA Neurology. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Pallanti, Importance of SARs-Cov-2 anosmia: From phenomenology to neurobiology, Compr Psychiatry 100 (2020) 152184. [DOI] [PMC free article] [PubMed]
- 57.Vaira L.A., Salzano G., Fois A.G., Piombino P., De Riu G. Potential pathogenesis of ageusia and anosmia in COVID-19 patients. Int. Forum Allergy Rhinol. 2020 doi: 10.1002/alr.22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., Fouret J., Dubois J., Gaymard A., Lescure F.-X., Dulière V., Brun P., Constant S., Poissy J., Lina B., Yazdanpanah Y., Terrier O., Rosa-Calatrava M. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep. Med. 2020;1(4) doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D. Brann, T. Tsukahara, C. Weinreb, M. Lipovsek, K. Van den Berge, B. Gong, R. Chance, I. Macaulay, H.-j. Chou, R. Fletcher, D. Das, K. Street, H.R. de Bezieux, Y.-G. Choi, D. Risso, S. Dudoit, E. Purdom, J. Mill, R.A. Hachem, H. Matsunami, D. Logan, B. Goldstein, M. Grubb, J. Ngai, S.R. Datta, Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia, bioRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 60.Torabi A., Mohammadbagheri E., Akbari Dilmaghani N., Bayat A.H., Fathi M., Vakili K., Alizadeh R., Rezaeimirghaed O., Hajiesmaeili M., Ramezani M., Simani L., Aliaghaei A. Proinflammatory cytokines in the olfactory mucosa result in COVID-19 induced anosmia. ACS Chem. Neurosci. 2020;11(13):1909–1913. doi: 10.1021/acschemneuro.0c00249. [DOI] [PubMed] [Google Scholar]
- 61.Kajani R., Apramian A., Vega A., Ubhayakar N., Xu P., Liu A. Neuroleptic malignant syndrome in a COVID-19 patient. Brain Behav. Immun. 2020 doi: 10.1016/j.bbi.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pelonero A.L., Levenson J.L., Pandurangi A.K. Neuroleptic malignant syndrome: a review. Psychiatric Services. 1998;49(9):1163–1172. doi: 10.1176/ps.49.9.1163. [DOI] [PubMed] [Google Scholar]
- 63.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential Fecal–oral transmission. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J. SARS-CoV-2 Receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zang R., Castro M.F.G., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5(47):eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin J.-M., Bai P., He W., Wu F., Liu X.-F., Han D.-M., Liu S., Yang J.-K. Gender differences in patients with COVID-19: focus on severity and mortality. Front. Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.H.a.d.G. Peckham, M. Nina, Raine, Charles and Radziszewska, Anna and Ciurtin, Coziana and Wedderburn, Lucy R. and Rosser, Elizabeth C. and Deakin, Claire T. and Webb, Kate., Sex-Bias in COVID-19: A Meta-Analysis and Review of Sex Differences in Disease and Immunity. or Lancet Infect Dis (2020). [DOI] [PMC free article] [PubMed]
- 69.N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, Y. Qiu, J. Wang, Y. Liu, Y. Wei, J.a. Xia, T. Yu, X. Zhang, L. Zhang, Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study, The Lancet 395(10223) (2020) 507-513. [DOI] [PMC free article] [PubMed]
- 70.Scully E.P., Haverfield J., Ursin R.L., Tannenbaum C., Klein S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., Kuthuru O., Apostolidis S.A., Bershaw L., Dougherty J., Greenplate A.R., Pattekar A., Kim J., Han N., Gouma S., Weirick M.E., Arevalo C.P., Bolton M.J., Goodwin E.C., Anderson E.M., Hensley S.E., Jones T.K., Mangalmurti N.S., Luning Prak E.T., Wherry E.J., Meyer N.J., Betts M.R. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5(49):eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.T. Takahashi, M.K. Ellingson, P. Wong, B. Israelow, C. Lucas, J. Klein, J. Silva, T. Mao, J.E. Oh, M. Tokuyama, P. Lu, A. Venkataraman, A. Park, F. Liu, A. Meir, J. Sun, E.Y. Wang, A. Casanovas-Massana, A.L. Wyllie, C.B.F. Vogels, R. Earnest, S. Lapidus, I.M. Ott, A.J. Moore, K. Anastasio, M.H. Askenase, M. Batsu, H. Beatty, S. Bermejo, S. Bickerton, K. Brower, M.L. Bucklin, S. Cahill, M. Campbell, Y. Cao, E. Courchaine, R. Datta, G. DeIuliis, B. Geng, L. Glick, R. Handoko, C. Kalinich, W. Khoury-Hanold, D. Kim, L. Knaggs, M. Kuang, E. Kudo, J. Lim, M. Linehan, A. Lu-Culligan, A.A. Malik, A. Martin, I. Matos, D. McDonald, M. Minasyan, S. Mohanty, M.C. Muenker, N. Naushad, A. Nelson, J. Nouws, M. Nunez-Smith, A. Obaid, I. Ott, H.-J. Park, X. Peng, M. Petrone, S. Prophet, H. Rahming, T. Rice, K.-A. Rose, L. Sewanan, L. Sharma, D. Shepard, E. Silva, M. Simonov, M. Smolgovsky, E. Song, N. Sonnert, Y. Strong, C. Todeasa, J. Valdez, S. Velazquez, P. Vijayakumar, H. Wang, A. Watkins, E.B. White, Y. Yang, A. Shaw, J.B. Fournier, C.D. Odio, S. Farhadian, C. Dela Cruz, N.D. Grubaugh, W.L. Schulz, A.M. Ring, A.I. Ko, S.B. Omer, A. Iwasaki, I.r.t. Yale, Sex differences in immune responses that underlie COVID-19 disease outcomes, Nature (2020). [DOI] [PMC free article] [PubMed]
- 73.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., Asselta R., Grimsrud M.M., Milani C., Aziz F., Kässens J., May S., Wendorff M., Wienbrandt L., Uellendahl-Werth F., Zheng T., Yi X., de Pablo R., Chercoles A.G., Palom A., Garcia-Fernandez A.E., Rodriguez-Frias F., Zanella A., Bandera A., Protti A., Aghemo A., Lleo A., Biondi A., Caballero-Garralda A., Gori A., Tanck A., Carreras Nolla A., Latiano A., Fracanzani A.L., Peschuck A., Julià A., Pesenti A., Voza A., Jiménez D., Mateos B., Nafria Jimenez B., Quereda C., Paccapelo C., Gassner C., Angelini C., Cea C., Solier A., Pestaña D., Muñiz-Diaz E., Sandoval E., Paraboschi E.M., Navas E., García Sánchez F., Ceriotti F., Martinelli-Boneschi F., Peyvandi F., Blasi F., Téllez L., Blanco-Grau A., Hemmrich-Stanisak G., Grasselli G., Costantino G., Cardamone G., Foti G., Aneli S., Kurihara H., ElAbd H., My I., Galván-Femenia I., Martín J., Erdmann J., Ferrusquía-Acosta J., Garcia-Etxebarria K., Izquierdo-Sanchez L., Bettini L.R., Sumoy L., Terranova L., Moreira L., Santoro L., Scudeller L., Mesonero F., Roade L., Rühlemann M.C., Schaefer M., Carrabba M., Riveiro-Barciela M., Figuera Basso M.E., Valsecchi M.G., Hernandez-Tejero M., Acosta-Herrera M., D'Angiò M., Baldini M., Cazzaniga M., Schulzky M., Cecconi M., Wittig M., Ciccarelli M., Rodríguez-Gandía M., Bocciolone M., Miozzo M., Montano N., Braun N., Sacchi N., Martínez N., Özer O., Palmieri O., Faverio P., Preatoni P., Bonfanti P., Omodei P., Tentorio P., Castro P., Rodrigues P.M., Blandino Ortiz A., de Cid R., Ferrer R., Gualtierotti R., Nieto R., Goerg S., Badalamenti S., Marsal S., Matullo G., Pelusi S., Juzenas S., Aliberti S., Monzani V., Moreno V., Wesse T., Lenz T.L., Pumarola T., Rimoldi V., Bosari S., Albrecht W., Peter W., Romero-Gómez M., D'Amato M., Duga S., Banales J.M., Hov J.R., Folseraas T., Valenti L., Franke A., Karlsen T.H. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Galili U., Mandrell R.E., Hamadeh R.M., Shohet S.B., Griffiss J.M. Interaction between human natural anti-alpha-galactosyl immunoglobulin G and bacteria of the human flora. Infect. Immun. 1988;56(7):1730–1737. doi: 10.1128/iai.56.7.1730-1737.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Branch D.R. Anti-A and anti-B: what are they and where do they come from? Transfusion. 2015;55(Suppl. 2):S74–S79. doi: 10.1111/trf.13087. [DOI] [PubMed] [Google Scholar]
- 76.Cheng Y., Cheng G., Chui C.H., Lau F.Y., Chan P.K., Ng M.H., Sung J.J., Wong R.S. ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA. 2005;293(12):1450–1451. doi: 10.1001/jama.293.12.1450-c. [DOI] [PubMed] [Google Scholar]
- 77.Rowe J.A., Handel I.G., Thera M.A., Deans A.M., Lyke K.E., Koné A., Diallo D.A., Raza A., Kai O., Marsh K., Plowe C.V., Doumbo O.K., Moulds J.M. Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc Natl Acad Sci U S A. 2007;104(44):17471–17476. doi: 10.1073/pnas.0705390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goel S., Palmkvist M., Moll K., Joannin N., Lara P., Akhouri R.R., Moradi N., Öjemalm K., Westman M., Angeletti D., Kjellin H., Lehtiö J., Blixt O., Ideström L., Gahmberg C.G., Storry J.R., Hult A.K., Olsson M.L., von Heijne G., Nilsson I., Wahlgren M. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat. Med. 2015;21(4):314–317. doi: 10.1038/nm.3812. [DOI] [PubMed] [Google Scholar]
- 79.Di Mascio D., Khalil A., Saccone G., Rizzo G., Buca D., Liberati M., Vecchiet J., Nappi L., Scambia G., Berghella V., D'Antonio F. Outcome of Coronavirus spectrum infections (SARS, MERS, COVID 1–19) during pregnancy: a systematic review and meta-analysis. Am J Obstet Gynecol MFM. 2020;2(2):100107. doi: 10.1016/j.ajogmf.2020.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Juan J., Gil M.M., Rong Z., Zhang Y., Yang H., Poon L.C. Effects of coronavirus disease 2019 (COVID-19) on maternal, perinatal and neonatal outcomes: a systematic review. Ultrasound Obstet. Gynecol. 2020 doi: 10.1002/uog.22088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., Li J., Zhao D., Xu D., Gong Q., Liao J., Yang H., Hou W., Zhang Y. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. The Lancet. 2020;395(10226):809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vivanti A.J., Vauloup-Fellous C., Prevot S., Zupan V., Suffee C., Do Cao J., Benachi A., De Luca D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020;11(1):3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kulkarni R., Rajput U., Dawre R., Valvi C., Nagpal R., Magdum N., Vankar H., Sonkawade N., Das A., Vartak S., Joshi S., Varma S., Karyakarte R., Bhosale R., Kinikar A. Early-onset symptomatic neonatal COVID-19 infection with high probability of vertical transmission. Infection. 2020 doi: 10.1007/s15010-020-01493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.J. Sisman, M.A. Jaleel, W. Moreno, V. Rajaram, R.R.J. Collins, R.C. Savani, D. Rakheja, A.S. Evans, INTRAUTERINE TRANSMISSION OF SARS-COV-2 INFECTION IN A PRETERM INFANT, The Pediatric Infectious Disease Journal Online First (9000). [DOI] [PubMed]
- 85.Hosier H., Farhadian S.F., Morotti R.A., Deshmukh U., Lu-Culligan A., Campbell K.H., Yasumoto Y., Vogels C.B.F., Casanovas-Massana A., Vijayakumar P., Geng B., Odio C.D., Fournier J., Brito A.F., Fauver J.R., Liu F., Alpert T., Tal R., Szigeti-Buck K., Perincheri S., Larsen C., Gariepy A.M., Aguilar G., Fardelmann K.L., Harigopal M., Taylor H.S., Pettker C.M., Wyllie A.L., Cruz C.D., Ring A.M., Grubaugh N.D., Ko A.I., Horvath T.L., Iwasaki A., Reddy U.M., Lipkind H.S. SARS–CoV-2 infection of the placenta. J. Clin. Invest. 2020;130(9):4947–4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baud D., Greub G., Favre G., Gengler C., Jaton K., Dubruc E., Pomar L. Second-trimester miscarriage in a pregnant woman with SARS-CoV-2 infection. JAMA. 2020;323(21):2198–2200. doi: 10.1001/jama.2020.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ji H.-L., Zhao R., Matalon S., Matthay M.A. Elevated Plasmin(ogen) as a common risk factor for COVID-19 susceptibility. Physiol. Rev. 2020;100(3):1065–1075. doi: 10.1152/physrev.00013.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song J.W., Lam S.M., Fan X., Cao W.J., Wang S.Y., Tian H., Chua G.H., Zhang C., Meng F.P., Xu Z., Fu J.L., Huang L., Xia P., Yang T., Zhang S., Li B., Jiang T.J., Wang R., Wang Z., Shi M., Zhang J.Y., Wang F.S., Shui G. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020 doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kelvin A.A., Halperin S. COVID-19 in children: the link in the transmission chain. Lancet. Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 92.Akhtar H., Patel C., Abuelgasim E., Harky A. COVID-19 (SARS-CoV-2) infection in pregnancy: a systematic review. Gynecol. Obstet. Invest. 2020 doi: 10.1159/000509290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martinelli I., Ferrazzi E., Ciavarella A., Erra R., Iurlaro E., Ossola M., Lombardi A., Blasi F., Mosca F., Peyvandi F. Pulmonary embolism in a young pregnant woman with COVID-19. Thromb. Res. 2020;191:36–37. doi: 10.1016/j.thromres.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu L., She Z.G., Cheng X., Qin J.J., Zhang X.J., Cai J., Lei F., Wang H., Xie J., Wang W., Li H., Zhang P., Song X., Chen X., Xiang M., Zhang C., Bai L., Xiang D., Chen M.M., Liu Y., Yan Y., Liu M., Mao W., Zou J., Liu L., Chen G., Luo P., Xiao B., Zhang C., Zhang Z., Lu Z., Wang J., Lu H., Xia X., Wang D., Liao X., Peng G., Ye P., Yang J., Yuan Y., Huang X., Guo J., Zhang B.H., Li H. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing Type 2 diabetes. Cell Metab. 2020;31(6):1068–1077.e3. doi: 10.1016/j.cmet.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J., Kong W., Xia P., Xu Y., Li L., Li Q., Yang L., Wei Q., Wang H., Li H., Zheng J., Sun H., Xia W., Liu G., Zhong X., Qiu K., Li Y., Wang H., Wang Y., Song X., Liu H., Xiong S., Liu Y., Cui Z., Hu Y., Chen L., Pan A., Zeng T. Impaired fasting glucose and diabetes are related to higher risks of complications and mortality among patients with coronavirus disease 2019. Front Endocrinol (Lausanne) 2020;11:525. doi: 10.3389/fendo.2020.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu J., Huang J., Zhu G., Wang Q., Lv Q., Huang Y., Yu Y., Si X., Yi H., Wang C., Liu Y., Xiao H., Zhou Q., Liu X., Yang D., Guan X., Li Y., Peng S., Sung J., Xiao H. Elevation of blood glucose level predicts worse outcomes in hospitalized patients with COVID-19: a retrospective cohort study. BMJ Open Diabetes Res. Care. 2020;8(1) doi: 10.1136/bmjdrc-2020-001476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barron E., Bakhai C., Kar P., Weaver A., Bradley D., Ismail H., Knighton P., Holman N., Khunti K., Sattar N., Wareham N.J., Young B., Valabhji J. Associations of type 1 and type 2 diabetes with COVID-19-related mortality in England: a whole-population study. Lancet Diabetes Endocrinol. 2020 doi: 10.1016/S2213-8587(20)30272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo J., Huang Z., Lin L., Lv J. Coronavirus Disease 2019 (COVID-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 infection. J. Am. Heart Assoc. 2020;9(7):e016219. doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12(1):8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.H. Zhang, Z. Kang, H. Gong, D. Xu, J. Wang, Z. Li, X. Cui, J. Xiao, T. Meng, W. Zhou, J. Liu, H. Xu, The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes, bioRxiv (2020) 2020.01.30.927806.
- 101.X. Chai, L. Hu, Y. Zhang, W. Han, Z. Lu, A. Ke, J. Zhou, G. Shi, N. Fang, J. Fan, J. Cai, J. Fan, F. Lan, Specific ACE2 Expression in Cholangiocytes May Cause Liver Damage After 2019-nCoV Infection, bioRxiv (2020) 2020.02.03.931766.
- 102.W. Sungnak, N. Huang, C. Bécavin, M. Berg, R. Queen, M. Litvinukova, C. Talavera-López, H. Maatz, D. Reichart, F. Sampaziotis, K.B. Worlock, M. Yoshida, J.L. Barnes, N.E. Banovich, P. Barbry, A. Brazma, J. Collin, T.J. Desai, T.E. Duong, O. Eickelberg, C. Falk, M. Farzan, I. Glass, R.K. Gupta, M. Haniffa, P. Horvath, N. Hubner, D. Hung, N. Kaminski, M. Krasnow, J.A. Kropski, M. Kuhnemund, M. Lako, H. Lee, S. Leroy, S. Linnarson, J. Lundeberg, K.B. Meyer, Z. Miao, A.V. Misharin, M.C. Nawijn, M.Z. Nikolic, M. Noseda, J. Ordovas-Montanes, G.Y. Oudit, D. Pe’er, J. Powell, S. Quake, J. Rajagopal, P.R. Tata, E.L. Rawlins, A. Regev, P.A. Reyfman, O. Rozenblatt-Rosen, K. Saeb-Parsy, C. Samakovlis, H.B. Schiller, J.L. Schultze, M.A. Seibold, C.E. Seidman, J.G. Seidman, A.K. Shalek, D. Shepherd, J. Spence, A. Spira, X. Sun, S.A. Teichmann, F.J. Theis, A.M. Tsankov, L. Vallier, M. van den Berge, J. Whitsett, R. Xavier, Y. Xu, L.-E. Zaragosi, D. Zerti, H. Zhang, K. Zhang, M. Rojas, F. Figueiredo, H.C.A.L.B. Network, SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes, Nat Med 26(5) (2020) 681-687. [DOI] [PMC free article] [PubMed]
- 103.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14(2):185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sharma A., Garcia G., Jr., Wang Y., Plummer J.T., Morizono K., Arumugaswami V., Svendsen C.N. Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Rep. Med. 2020;1(4) doi: 10.1016/j.xcrm.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schafer S., Viswanathan S., Widjaja A.A., Lim W.-W., Moreno-Moral A., DeLaughter D.M., Ng B., Patone G., Chow K., Khin E., Tan J., Chothani S.P., Ye L., Rackham O.J.L., Ko N.S.J., Sahib N.E., Pua C.J., Zhen N.T.G., Xie C., Wang M., Maatz H., Lim S., Saar K., Blachut S., Petretto E., Schmidt S., Putoczki T., Guimarães-Camboa N., Wakimoto H., van Heesch S., Sigmundsson K., Lim S.L., Soon J.L., Chao V.T.T., Chua Y.L., Tan T.E., Evans S.M., Loh Y.J., Jamal M.H., Ong K.K., Chua K.C., Ong B.-H., Chakaramakkil M.J., Seidman J.G., Seidman C.E., Hubner N., Sin K.Y.K., Cook S.A. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552(7683):110–115. doi: 10.1038/nature24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Y., Banerjee S., Wang Y., Goldstein S.A., Dong B., Gaughan C., Silverman R.H., Weiss S.R. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. 2016;113(8):2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.C. Fan, K. Li, Y. Ding, W.L. Lu, J. Wang, ACE2 Expression in Kidney and Testis May Cause Kidney and Testis Damage After 2019-nCoV Infection, medRxiv (2020) 2020.02.12.20022418.
- 108.Rabb H. Kidney diseases in the time of COVID-19: major challenges to patient care. J. Clin. Invest. 2020;130(6):2749–2751. doi: 10.1172/JCI138871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.J.W. Zhang, Y.; Wang, R.; Lu, K.; Tu, M.; Guo, H.; Xie, W.; Qin, Z.; Li, S.; Zhu, P.; Wang, X., Bioinformatic Analysis Reveals That the Reproductive System is Potentially at Risk from SARS-CoV-2., Preprints (2020).
- 110.Vaz-Silva J., Carneiro M.M., Ferreira M.C., Pinheiro S.V., Silva D.A., Silva-Filho A.L., Witz C.A., Reis A.M., Santos R.A., Reis F.M. The vasoactive peptide angiotensin-(1–7), its receptor Mas and the angiotensin-converting enzyme type 2 are expressed in the human endometrium. Reprod. Sci. 2009;16(3):247–256. doi: 10.1177/1933719108327593. [DOI] [PubMed] [Google Scholar]
- 111.Reis F.M., Bouissou D.R., Pereira V.M., Camargos A.F., dos Reis A.M., Santos R.A. Angiotensin-(1–7), its receptor Mas, and the angiotensin-converting enzyme type 2 are expressed in the human ovary. Fertil. Steril. 2011;95(1):176–181. doi: 10.1016/j.fertnstert.2010.06.060. [DOI] [PubMed] [Google Scholar]
- 112.R. Pique-Regi, R. Romero, A.L. Tarca, F. Luca, Y. Xu, A. Alazizi, Y. Leng, C.-D. Hsu, N. Gomez-Lopez, Does the human placenta express the canonical cell entry mediators for SARS-CoV-2?, eLife 9 (2020) e58716. [DOI] [PMC free article] [PubMed]
- 113.Ratajczak M.Z., Bujko K., Ciechanowicz A., Sielatycka K., Cymer M., Marlicz W., Kucia M. SARS-CoV-2 entry receptor ACE2 is expressed on very small CD45(-) precursors of hematopoietic and endothelial cells and in response to virus spike protein activates the Nlrp3 inflammasome. Stem Cell Rev. Rep. 2020:1–12. doi: 10.1007/s12015-020-10010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-L., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.M.C. Pontelli, I.A. Castro, R.B. Martins, F.P. Veras, L.L. Serra, D.C. Nascimento, R.S. Cardoso, R. Rosales, T.M. Lima, J.P. Souza, D.B. Caetité, M.H.F. de Lima, J.T. Kawahisa, M.C. Giannini, L.P. Bonjorno, M.I.F. Lopes, S.S. Batah, L. Siyuan, R.L. Assad, S.C.L. Almeida, F.R. Oliveira, M.N. Benatti, L.L.F. Pontes, R.C. Santana, F.C. Vilar, M.A. Martins, T.M. Cunha, R.T. Calado, J.C. Alves-Filho, D.S. Zamboni, A. Fabro, P. Louzada-Junior, R.D.R. Oliveira, F.Q. Cunha, E. Arruda, Infection of human lymphomononuclear cells by SARS-CoV-2, bioRxiv (2020) 2020.07.28.225912.
- 116.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7):e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82(17):8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(4):779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ferguson C.J., Lenk G.M., Meisler M.H. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 2009;18(24):4868–4878. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rutherford A.C., Traer C., Wassmer T., Pattni K., Bujny M.V., Carlton J.G., Stenmark H., Cullen P.J. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 2006;119(Pt 19):3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.de Lartigue J., Polson H., Feldman M., Shokat K., Tooze S.A., Urbé S., Clague M.J. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10(7):883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shisheva A. PIKfyve: Partners, significance, debates and paradoxes. Cell Biol. Int. 2008;32(6):591–604. doi: 10.1016/j.cellbi.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020 doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.K. Wang, W. Chen, Y.-S. Zhou, J.-Q. Lian, Z. Zhang, P. Du, L. Gong, Y. Zhang, H.-Y. Cui, J.-J. Geng, B. Wang, X.-X. Sun, C.-F. Wang, X. Yang, P. Lin, Y.-Q. Deng, D. Wei, X.-M. Yang, Y.-M. Zhu, K. Zhang, Z.-H. Zheng, J.-L. Miao, T. Guo, Y. Shi, J. Zhang, L. Fu, Q.-Y. Wang, H. Bian, P. Zhu, Z.-N. Chen, SARS-CoV-2 invades host cells via a novel route: CD147-spike protein, bioRxiv (2020) 2020.03.14.988345.
- 129.Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., Deczkowska A., Zhang S., Schwikowski B., Zhang Z., Amit I. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181(7):1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Radzikowska U., Ding M., Tan G., Zhakparov D., Peng Y., Wawrzyniak P., Wang M., Li S., Morita H., Altunbulakli C., Reiger M., Neumann A.U., Lunjani N., Traidl-Hoffmann C., Nadeau K., O’Mahony L., Akdis C.A., Sokolowska M. Distribution of ACE2, CD147, CD26 and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy n/a(n/a) 2020 doi: 10.1111/all.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev. Rep. 2020;16(3):434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.H. Bian, Z.-H. Zheng, D. Wei, Z. Zhang, W.-Z. Kang, C.-Q. Hao, K. Dong, W. Kang, J.-L. Xia, J.-L. Miao, R.-H. Xie, B. Wang, X.-X. Sun, X.-M. Yang, P. Lin, J.-J. Geng, K. Wang, H.-Y. Cui, K. Zhang, X.-C. Chen, H. Tang, H. Du, N. Yao, S.-S. Liu, L.-N. Liu, Z. Zhang, Z.-W. Gao, G. Nan, Q.-Y. Wang, J.-Q. Lian, Z.-N. Chen, P. Zhu, Meplazumab treats COVID-19 pneumonia: an open-labelled, concurrent controlled add-on clinical trial, medRxiv (2020) 2020.03.21.20040691.
- 133.X. Zhao, S. Zheng, D. Chen, M. Zheng, X. Li, G. Li, H. Lin, J. Chang, H. Zeng, J.-T. Guo, LY6E Restricts the Entry of Human Coronaviruses, including the currently pandemic SARS-CoV-2, bioRxiv (2020) 2020.04.02.021469. [DOI] [PMC free article] [PubMed]
- 134.Lee P.Y., Wang J.-X., Parisini E., Dascher C.C., Nigrovic P.A. Ly6 family proteins in neutrophil biology. J. Leukoc. Biol. 2013;94(4):585–594. doi: 10.1189/jlb.0113014. [DOI] [PubMed] [Google Scholar]
- 135.Upadhyay G. Emerging role of lymphocyte antigen-6 family of genes in cancer and immune cells. Front. Immunol. 2019;10(819) doi: 10.3389/fimmu.2019.00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hachim M.Y., Al Heialy S., Hachim I.Y., Halwani R., Senok A.C., Maghazachi A.A., Hamid Q. Interferon-induced transmembrane protein (IFITM3) is upregulated explicitly in SARS-CoV-2 infected lung epithelial cells. Front. Immunol. 2020;11:1372. doi: 10.3389/fimmu.2020.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Spence J.S., He R., Hoffmann H.-H., Das T., Thinon E., Rice C.M., Peng T., Chandran K., Hang H.C. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 2019;15(3):259–268. doi: 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Feeley E.M., Sims J.S., John S.P., Chin C.R., Pertel T., Chen L.-M., Gaiha G.D., Ryan B.J., Donis R.O., Elledge S.J. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10):e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wee Y.S., Roundy K.M., Weis J.J., Weis J.H. Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immunity. 2012;18(6):834–845. doi: 10.1177/1753425912443392. [DOI] [PubMed] [Google Scholar]
- 140.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., Wagh D., Coller J., Pellegrini K.L., Kazmin D., Alaaeddine G., Leung W.S., Chan J.M.C., Chik T.S.H., Choi C.Y.C., Huerta C., Paine McCullough M., Lv H., Anderson E., Edupuganti S., Upadhyay A.A., Bosinger S.E., Maecker H.T., Khatri P., Rouphael N., Peiris M., Pulendran B. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020:eabc6261. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Maier H.J., Britton P. Involvement of autophagy in coronavirus replication. Viruses. 2012;4(12):3440–3451. doi: 10.3390/v4123440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78(18):9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.K. Singh, Y.-C. Chen, J.T. Judy, F. Seifuddin, I. Tunc, M. Pirooznia, Network Analysis and Transcriptome Profiling Identify Autophagic and Mitochondrial Dysfunctions in SARS-CoV-2 Infection, bioRxiv : the preprint server for biology (2020) 2020.05.13.092536. [DOI] [PMC free article] [PubMed]
- 145.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279(11):10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7(11):1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4(3) doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S.M., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82(22):11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Luo H., Wu D., Shen C., Chen K., Shen X., Jiang H. Severe acute respiratory syndrome coronavirus membrane protein interacts with nucleocapsid protein mostly through their carboxyl termini by electrostatic attraction. Int. J. Biochem. Cell Biol. 2006;38(4):589–599. doi: 10.1016/j.biocel.2005.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sevajol M., Subissi L., Decroly E., Canard B., Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Neuman B.W., Angelini M.M., Buchmeier M.J. Does form meet function in the coronavirus replicative organelle? Trends Microbiol. 2014;22(11):642–647. doi: 10.1016/j.tim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 154.P. V’kovski, H. Al-Mulla, V. Thiel, B.W. Neuman, New insights on the role of paired membrane structures in coronavirus replication, Virus Research 202 (2015) 33-40. [DOI] [PMC free article] [PubMed]
- 155.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182(2):417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.K. Klann, D. Bojkova, G. Tascher, S. Ciesek, C. Münch, J. Cinatl, Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication, Molecular Cell. [DOI] [PMC free article] [PubMed]
- 157.Laing A.G., Lorenc A., del Molino I., del Barrio A., Das M., Fish L., Monin M., Muñoz-Ruiz D.R., McKenzie T.S., Hayday I., Francos-Quijorna S., Kamdar M., Joseph D., Davies R., Davis A., Jennings I., Zlatareva P., Vantourout Y., Wu V., Sofra F., Cano M., Greco E., Theodoridis J., Freedman S., Gee J.N.E., Chan S., Ryan E., Bugallo-Blanco P., Peterson K., Kisand L., Haljasmägi L., Chadli P., Moingeon L., Martinez B., Merrick K., Bisnauthsing K., Brooks M.A.A., Ibrahim J., Mason F. Lopez, Gomez K., Babalola S., Abdul-Jawad J., Cason C., Mant J., Seow C., Graham K.J., Doores F. Di, Rosa J., Edgeworth M., Shankar-Hari A.C. Hayday. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020 doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 158.Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F., Thorsteinsdottir B., Kristjansdottir S., Birgisdottir K., Kristinsdottir A.M., Sigurdsson M.I., Arnadottir G.A., Ivarsdottir E.V., Andresdottir M., Jonsson F., Agustsdottir A.B., Berglund J., Eiriksdottir B., Fridriksdottir R., Gardarsdottir E.E., Gottfredsson M., Gretarsdottir O.S., Gudmundsdottir S., Gudmundsson K.R., Gunnarsdottir T.R., Gylfason A., Helgason A., Jensson B.O., Jonasdottir A., Jonsson H., Kristjansson T., Kristinsson K.G., Magnusdottir D.N., Magnusson O.T., Olafsdottir L.B., Rognvaldsson S., le Roux L., Sigmundsdottir G., Sigurdsson A., Sveinbjornsson G., Sveinsdottir K.E., Sveinsdottir M., Thorarensen E.A., Thorbjornsson B., Thordardottir M., Saemundsdottir J., Kristjansson S.H., Josefsdottir K.S., Masson G., Georgsson G., Kristjansson M., Moller A., Palsson R., Gudnason T., Thorsteinsdottir U., Jonsdottir I., Sulem P., Stefansson K. Humoral immune response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hung I.F.-N., Cheng V.C.-C., Li X., Tam A.R., Hung D.L.-L., Chiu K.H.-Y., Yip C.C.-Y., Cai J.-P., Ho D.T.-Y., Wong S.-C. SARS-CoV-2 shedding and seroconversion among passengers quarantined after disembarking a cruise ship: a case series. Lancet. Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tabata S., Imai K., Kawano S., Ikeda M., Kodama T., Miyoshi K., Obinata H., Mimura S., Kodera T., Kitagaki M. Clinical characteristics of COVID-19 in 104 people with SARS-CoV-2 infection on the Diamond Princess cruise ship: a retrospective analysis. Lancet. Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25(10):2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ma Y., Xu Q.-N., Wang F.-L., Ma X.-M., Wang X.-Y., Zhang X.-G., Zhang Z.-F. Characteristics of asymptomatic patients with SARS-CoV-2 infection in Jinan. China, Microbes Infect. 2020;22(4):212–217. doi: 10.1016/j.micinf.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020 doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 164.Orange J.S., Wolf S.F., Biron C.A. Effects of IL-12 on the response and susceptibility to experimental viral infections. J. Immunol. 1994;152(3):1253–1264. [PubMed] [Google Scholar]
- 165.Monteiro J.M., Harvey C., Trinchieri G. Role of Interleukin-12 in Primary Influenza Virus Infection. J. Virol. 1998;72(6):4825–4831. doi: 10.1128/jvi.72.6.4825-4831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Magram J., Connaughton S.E., Warrier R.R., Carvajal D.M., Wu C.Y., Ferrante J., Stewart C., Sarmiento U., Faherty D.A., Gately M.K. IL-12-deficient mice are defective in IFN gamma production and type 1 cytokine responses. Immunity. 1996;4(5):471–481. doi: 10.1016/s1074-7613(00)80413-6. [DOI] [PubMed] [Google Scholar]
- 167.Schijns V.E., Haagmans B.L., Wierda C.M., Kruithof B., Heijnen I.A., Alber G., Horzinek M.C. Mice lacking IL-12 develop polarized Th1 cells during viral infection. J. Immunol. 1998;160(8):3958–3964. [PubMed] [Google Scholar]
- 168.Oxenius A., Karrer U., Zinkernagel R.M., Hengartner H. IL-12 is not required for induction of type 1 cytokine responses in viral infections. J. Immunol. 1999;162(2):965–973. [PubMed] [Google Scholar]
- 169.Komastu T., Ireland D.D., Reiss C.S. IL-12 and viral infections. Cytokine Growth Factor Rev. 1998;9(3–4):277–285. doi: 10.1016/S1359-6101(98)00017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Morris S.C., Madden K.B., Adamovicz J.J., Gause W.C., Hubbard B.R., Gately M.K., Finkelman F.D. Effects of IL-12 on in vivo cytokine gene expression and Ig isotype selection. J. Immunol. 1994;152(3):1047–1056. [PubMed] [Google Scholar]
- 171.Minty A., Chalon P., Derocq J.M., Dumont X., Guillemot J.C., Kaghad M., Labit C., Leplatois P., Liauzun P., Miloux B., Minty C., Casellas P., Loison G., Lupker J., Shire D., Ferrara P., Caput D. lnterleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362(6417):248–250. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 172.Junttila I.S. Tuning the cytokine responses: an update on Interleukin (IL)-4 and IL-13 receptor complexes. Front. Immunol. 2018;9(888) doi: 10.3389/fimmu.2018.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Frumento G., Zuo J., Verma K., Croft W., Ramagiri P., Chen F.E., Moss P. CD117 (c-Kit) is expressed during CD8+ T Cell priming and stratifies sensitivity to apoptosis according to strength of TCR engagement. Front. Immunol. 2019;10(468) doi: 10.3389/fimmu.2019.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Puddington L., Olson S., Lefrançois L. Interactions between stem cell factor and c-Kit are required for intestinal immune system homeostasis. Immunity. 1994;1(9):733–739. doi: 10.1016/s1074-7613(94)80015-4. [DOI] [PubMed] [Google Scholar]
- 175.Metcalfe S.M. LIF in the regulation of T-cell fate and as a potential therapeutic, Genes &. Immunity. 2011;12(3):157–168. doi: 10.1038/gene.2011.9. [DOI] [PubMed] [Google Scholar]
- 176.Janssens K., Van Den Haute C., Baekelandt V., Van Horssen J., Van Wijmeersch B., Stinissen P., Hendriks J.J.A., Slaets H., Hellings N. Leukemia inhibitory factor shifts the immune balance towards regulatory T cells. J. Neuroimmunol. 2014;275(1):155. doi: 10.1016/j.bbi.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 177.West N.R. Coordination of immune-stroma crosstalk by IL-6 family cytokines. Front. Immunol. 2019;10(1093) doi: 10.3389/fimmu.2019.01093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.S.M. Davis, L.A. Collier, E.D. Winford, C.C. Leonardo, C.T. Ajmo, Jr., E.A. Foran, T.J. Kopper, J.C. Gensel, K.R. Pennypacker, Leukemia inhibitory factor modulates the peripheral immune response in a rat model of emergent large vessel occlusion, J. Neuroinflammation 15(1) (2018) 288-288. [DOI] [PMC free article] [PubMed]
- 179.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Wullimann D.J., Kammann T., Emgård J., Parrot T., Folkesson E., Rooyackers O., Eriksson L.I., Henter J.-I., Sönnerborg A., Allander T., Albert J., Nielsen M., Klingström J., Gredmark-Russ S., Björkström N.K., Sandberg J.K., Price D.A., Ljunggren H.-G., Aleman S., Buggert M. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.S.S. Cush, K.M. Anderson, D.H. Ravneberg, J.L. Weslow-Schmidt, E. Flaño, Memory generation and maintenance of CD8+ T cell function during viral persistence, J. Immunol. (Baltimore, Md. : 1950) 179(1) (2007) 141-153. [DOI] [PMC free article] [PubMed]
- 181.Kaech S.M., Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naïve cells. Nat. Immunol. 2001;2(5):415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bevan M.J. Helping the CD8+ T-cell response. Nat. Rev. Immunol. 2004;4(8):595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- 183.S. Lian, X. Zhai, X. Wang, H. Zhu, S. Zhang, W. Wang, Z. Wang, J. Huang, Elevated expression of growth-regulated oncogene-alpha in tumor and stromal cells predicts unfavorable prognosis in pancreatic cancer, Medicine (Baltimore) 95(30) (2016) e4328-e4328. [DOI] [PMC free article] [PubMed]
- 184.Girbl T., Lenn T., Perez L., Rolas L., Barkaway A., Thiriot A., Del Fresno C., Lynam E., Hub E., Thelen M., Graham G., Alon R., Sancho D., von Andrian U.H., Voisin M.B., Rot A., Nourshargh S. Distinct compartmentalization of the chemokines CXCL1 and CXCL2 and the atypical receptor ACKR1 determine discrete stages of neutrophil diapedesis. Immunity. 2018;49(6):1062–1076.e6. doi: 10.1016/j.immuni.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zlotnik A., Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36(5):705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Baggiolini M., Dewald B., Moser B. Interleukin-8 and related chemotactic cytokines–CXC and CC chemokines. Adv. Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 187.Lane B.R., Strieter R.M., Coffey M.J., Markovitz D.M. Human Immunodeficiency Virus Type 1 (HIV-1)-induced GRO-α production stimulates HIV-1 replication in macrophages and T lymphocytes. J. Virol. 2001;75(13):5812–5822. doi: 10.1128/JVI.75.13.5812-5822.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lane B.R., Liu J., Bock P.J., Schols D., Coffey M.J., Strieter R.M., Polverini P.J., Markovitz D.M. Interleukin-8 and growth-regulated oncogene alpha mediate angiogenesis in Kaposi's Sarcoma. J. Virol. 2002;76(22):11570–11583. doi: 10.1128/JVI.76.22.11570-11583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bais C., Santomasso B., Coso O., Arvanitakis L., Raaka E.G., Gutkind J.S., Asch A.S., Cesarman E., Gershengorn M.C., Mesri E.A. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391(6662):86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 190.Richmond A., Thomas H.G. Purification of melanoma growth stimulatory activity. J. Cell. Physiol. 1986;129(3):375–384. doi: 10.1002/jcp.1041290316. [DOI] [PubMed] [Google Scholar]
- 191.Hamilton J.A. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008;8(7):533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 192.Ursini-Siegel J., Zhang W., Altmeyer A., Hatada E.N., Do R.K.G., Yagita H., Chen-Kiang S. TRAIL/Apo-2 ligand induces primary plasma cell apoptosis. J. Immunol. 2002;169(10):5505–5513. doi: 10.4049/jimmunol.169.10.5505. [DOI] [PubMed] [Google Scholar]
- 193.Leverkus M., Walczak H., McLellan A., Fries H.-W., Terbeck G., Bröcker E.-B., Kämpgen E. Maturation of dendritic cells leads to up-regulation of cellular FLICE-inhibitory protein and concomitant down-regulation of death ligand–mediated apoptosis. Blood J. Am. Soc. Hematol. 2000;96(7):2628–2631. [PubMed] [Google Scholar]
- 194.Janssen E.M., Droin N.M., Lemmens E.E., Pinkoski M.J., Bensinger S.J., Ehst B.D., Griffith T.S., Green D.R., Schoenberger S.P. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434(7029):88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- 195.Bosque A., Pardo J., Martínez-Lorenzo M.J., Lasierra P., Larrad L., Marzo I., Naval J., Anel A. Human CD8+ T cell blasts are more sensitive than CD4+ T cell blasts to regulation by APO2L/TRAIL. Eur. J. Immunol. 2005;35(6):1812–1821. doi: 10.1002/eji.200526046. [DOI] [PubMed] [Google Scholar]
- 196.Renshaw S.A., Parmar J.S., Singleton V., Rowe S.J., Dockrell D.H., Dower S.K., Bingle C.D., Chilvers E.R., Whyte M.K.B. Acceleration of human neutrophil apoptosis by TRAIL. J. Immunol. 2003;170(2):1027–1033. doi: 10.4049/jimmunol.170.2.1027. [DOI] [PubMed] [Google Scholar]
- 197.Kumar V. Pulmonary innate immune response determines the outcome of inflammation during pneumonia and sepsis-associated acute lung injury. Front. Immunol. 2020;11(1722) doi: 10.3389/fimmu.2020.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.O'Connor B.P., Gleeson M.W., Noelle R.J., Erickson L.D. The rise and fall of long-lived humoral immunity: terminal differentiation of plasma cells in health and disease. Immunol. Rev. 2003;194:61–76. doi: 10.1034/j.1600-065x.2003.00055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rodriguez L., Pekkarinen P.T., Lakshmikanth T., Tan Z., Consiglio C.R., Pou C., Chen Y., Mugabo C.H., Nguyen N.A., Nowlan K., Strandin T., Levanov L., Mikes J., Wang J., Kantele A., Hepojoki J., Vapalahti O., Heinonen S., Kekäläinen E., Brodin P. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Reports Medicine. 2020;1(5) doi: 10.1016/j.xcrm.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., Song T., Alshukairi A.N., Chen R., Zhang Z., Gan M., Zhu A., Huang Y., Luo L., Mok C.K.P., Al Gethamy M.M., Tan H., Li Z., Huang X., Li F., Sun J., Zhang Y., Wen L., Li Y., Chen Z., Zhuang Z., Zhuo J., Chen C., Kuang L., Wang J., Lv H., Jiang Y., Li M., Lin Y., Deng Y., Tang L., Liang J., Huang J., Perlman S., Zhong N., Zhao J., Malik Peiris J.S., Li Y., Zhao J. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130(10) doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.E. Wauters, P. Van Mol, A.D. Garg, S. Jansen, Y. Van Herck, L. Vanderbeke, A. Bassez, B. Boeckx, B. Malengier-Devlies, A. Timmerman, T. Van Brussel, T. Van Buyten, R. Schepers, E. Heylen, D. Dauwe, C. Dooms, J. Gunst, G. Hermans, P. Meersseman, D. Testelmans, J. Yserbyt, P. Matthys, S. Tejpar, J. Neyts, J. Wauters, J. Qian, D. Lambrechts, Discriminating Mild from Critical COVID-19 by Innate and Adaptive Immune Single-cell Profiling of Bronchoalveolar Lavages, bioRxiv (2020) 2020.07.09.196519.
- 202.Kumar V. Phagocytosis: phenotypically simple yet a mechanistically complex process. Int. Rev. Immunol. 2020;39(3):118–150. doi: 10.1080/08830185.2020.1732958. [DOI] [PubMed] [Google Scholar]
- 203.Boada-Romero E., Martinez J., Heckmann B.L., Green D.R. The clearance of dead cells by efferocytosis. Nat. Rev. Mol. Cell Biol. 2020;21(7):398–414. doi: 10.1038/s41580-020-0232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B., Tong S.Y.C., Lewin S.R., Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Papillion A., Powell M.D., Chisolm D.A., Bachus H., Fuller M.J., Weinmann A.S., Villarino A., O’Shea J.J., León B., Oestreich K.J., Ballesteros-Tato A. Inhibition of IL-2 responsiveness by IL-6 is required for the generation of GC-T<sub>FH</sub> cells. Sci. Immunol. 2019;4(39):eaaw7636. doi: 10.1126/sciimmunol.aaw7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.S.C.A. Nielsen, F. Yang, R.A. Hoh, K.J.L. Jackson, K. Roeltgen, J.-Y. Lee, A. Rustagi, A.J. Rogers, A.E. Powell, P.S. Kim, T.T. Wang, B. Pinsky, C.A. Blish, S.D. Boyd, B cell clonal expansion and convergent antibody responses to SARS-CoV-2, Res Sq (2020) rs.3.rs-27220. [DOI] [PMC free article] [PubMed]
- 207.Schultheiß C., Paschold L., Simnica D., Mohme M., Willscher E., von Wenserski L., Scholz R., Wieters I., Dahlke C., Tolosa E., Sedding D.G., Ciesek S., Addo M., Binder M. Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53(2):442–455.e4. doi: 10.1016/j.immuni.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., Lorenzi J.C.C., Finkin S., Hägglöf T., Hurley A., Millard K.G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P.D., Caskey M., Robbiani D.F., Nussenzweig M.C., Bjorkman P.J. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.E. Seydoux, L.J. Homad, A.J. MacCamy, K.R. Parks, N.K. Hurlburt, M.F. Jennewein, N.R. Akins, A.B. Stuart, Y.-H. Wan, J. Feng, R.E. Whaley, S. Singh, M. Boeckh, K.W. Cohen, M.J. McElrath, J.A. Englund, H.Y. Chu, M. Pancera, A.T. McGuire, L. Stamatatos, Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation, Immunity. [DOI] [PMC free article] [PubMed]
- 210.F. Wu, A. Wang, M. Liu, Q. Wang, J. Chen, S. Xia, Y. Ling, Y. Zhang, J. Xun, L. Lu, S. Jiang, H. Lu, Y. Wen, J. Huang, Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications, medRxiv (2020) 2020.03.30.20047365.
- 211.Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chang S.-C., Wang J.-T., Huang L.-M., Chen Y.-C., Fang C.-T., Sheng W.-H., Wang J.-L., Yu C.-J., Yang P.-C. Longitudinal analysis of Severe Acute Respiratory Syndrome (SARS) coronavirus-specific antibody in SARS patients. Clin. Diagn. Lab. Immunol. 2005;12(12):1455–1457. doi: 10.1128/CDLI.12.12.1455-1457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cao W.-C., Liu W., Zhang P.-H., Zhang F., Richardus J.H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 2007;357(11):1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 214.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.-H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020 doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Klein S.L., Pekosz A., Park H.-S., Ursin R.L., Shapiro J.R., Benner S.E., Littlefield K., Kumar S., Naik H.M., Betenbaugh M., Shrestha R., Wu A.A., Hughes R.M., Burgess I., Caturegli P., Laeyendecker O., Quinn T.C., Sullivan D.J., Shoham S., Redd A.D., Bloch E.M., Casadevall A., Tobian A.A.R. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J. Clin. Invest. 2020 doi: 10.1172/JCI142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Borges da Silva H., Peng C., Wang H., Wanhainen K.M., Ma C., Lopez S., Khoruts A., Zhang N., Jameson S.C. Sensing of ATP via the purinergic receptor P2RX7 promotes CD8<sup>+</sup> Trm cell generation by enhancing their sensitivity to the cytokine TGF-β. Immunity. 2020;53(1):158–171.e6. doi: 10.1016/j.immuni.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vieira Braga F.A., Hertoghs K.M., Kragten N.A., Doody G.M., Barnes N.A., Remmerswaal E.B., Hsiao C.C., Moerland P.D., Wouters D., Derks I.A., van Stijn A., Demkes M., Hamann J., Eldering E., Nolte M.A., Tooze R.M., ten Berge I.J., van Gisbergen K.P., van Lier R.A. Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. Eur. J. Immunol. 2015;45(10):2945–2958. doi: 10.1002/eji.201545650. [DOI] [PubMed] [Google Scholar]
- 218.Kragten N.A.M., Behr F.M., Vieira Braga F.A., Remmerswaal E.B.M., Wesselink T.H., Oja A.E., Hombrink P., Kallies A., van Lier R.A.W., Stark R., van Gisbergen K. Blimp-1 induces and Hobit maintains the cytotoxic mediator granzyme B in CD8 T cells. Eur. J. Immunol. 2018;48(10):1644–1662. doi: 10.1002/eji.201847771. [DOI] [PubMed] [Google Scholar]
- 219.Behr F.M., Kragten N.A.M., Wesselink T.H., Nota B., van Lier R.A.W., Amsen D., Stark R., Hombrink P., van Gisbergen K. Blimp-1 rather than Hobit drives the formation of tissue-resident memory CD8(+) T cells in the lungs. Front. Immunol. 2019;10:400. doi: 10.3389/fimmu.2019.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Post M., Cuapio A., Osl M., Lehmann D., Resch U., Davies D.M., Bilban M., Schlechta B., Eppel W., Nathwani A., Stoiber D., Spanholtz J., Casanova E., Hofer E. The Transcription factor ZNF683/HOBIT regulates human NK-cell development. Front. Immunol. 2017;8(535) doi: 10.3389/fimmu.2017.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., Haagmans B.L., de Swart R.L., Sette A., de Vries R.D. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ni L., Ye F., Cheng M.-L., Feng Y., Deng Y.-Q., Zhao H., Wei P., Ge J., Gou M., Li X., Sun L., Cao T., Wang P., Zhou C., Zhang R., Liang P., Guo H., Wang X., Qin C.-F., Chen F., Dong C. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., Vanderheiden A., Nyhoff L., Davis C.W., Adekunle O., Affer M., Sherman M., Reynolds S., Verkerke H.P., Alter D.N., Guarner J., Bryksin J., Horwath M.C., Arthur C.M., Saakadze N., Smith G.H., Edupuganti S., Scherer E.M., Hellmeister K., Cheng A., Morales J.A., Neish A.S., Stowell S.R., Frank F., Ortlund E., Anderson E.J., Menachery V.D., Rouphael N., Mehta A.K., Stephens D.S., Ahmed R., Roback J.D., Wrammert J. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 2020;1(3) doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Röhmel J., Voigt S., Schmitz J., Miltenyi S., Demuth I., Müller M.A., Hocke A., Witzenrath M., Suttorp N., Kern F., Reimer U., Wenschuh H., Drosten C., Corman V.M., Giesecke-Thiel C., Sander L.E., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020 doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 226.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Pedersen S.F., Ho Y.C. SARS-CoV-2: a storm is raging. J. Clin. Invest. 2020 doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhou R., Kai-Wang To K., Wong Y.-C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.-Y., Tsz-Kan Lau T., Yeung P., Chan W.-M., Ka-Lun Wu A., Lung K.-C., Tak-Yin Tsang O., Leung W.-S., Fan-Ngai Hung I., Yuen K.-Y., Chen Z. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020 doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11(827) doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sattler A., Angermair S., Stockmann H., Heim K.M., Khadzhynov D., Treskatsch S., Halleck F., Kreis M.E., Kotsch K. SARS-CoV-2 specific T-cell responses and correlations with COVID-19 patient predisposition. J. Clin. Invest. 2020 doi: 10.1172/JCI140965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Bao Y., Liu X., Han C., Xu S., Xie B., Zhang Q., Gu Y., Hou J., Qian L., Qian C., Han H., Cao X. Identification of IFN-γ-producing innate B cells. Cell Res. 2014;24(2):161–176. doi: 10.1038/cr.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Frucht D.M., Fukao T., Bogdan C., Schindler H., O'Shea J.J., Koyasu S. IFN-γ production by antigen-presenting cells: mechanisms emerge. Trends Immunol. 2001;22(10):556–560. doi: 10.1016/s1471-4906(01)02005-1. [DOI] [PubMed] [Google Scholar]
- 233.I. Sánchez-Cerrillo, P. Landete, B. Aldave, S. Sánchez-Alonso, A. Sánchez-Azofra, A. Marcos-Jiménez, E. Ávalos, A. Alcaraz-Serna, I. de los Santos, T. Mateu-Albero, L. Esparcia, C. López-Sanz, P. Martínez-Fleta, L. Gabrie, L. del Campo Guerola, H. de la Fuente, M.J. Calzada, I. González-Álvaro, A. Alfranca, F. Sánchez-Madrid, C. Muñoz-Calleja, J.B. Soriano, J. Ancochea, E. Martín-Gayo, COVID-19 severity associates with pulmonary redistribution of CD1c+ DC and inflammatory transitional and nonclassical monocytes, J Clin Invest (2020). [DOI] [PMC free article] [PubMed]
- 234.Mazzoni A., Salvati L., Maggi L., Capone M., Vanni A., Spinicci M., Mencarini J., Caporale R., Peruzzi B., Antonelli A., Trotta M., Zammarchi L., Ciani L., Gori L., Lazzeri C., Matucci A., Vultaggio A., Rossi O., Almerigogna F., Parronchi P., Fontanari P., Lavorini F., Peris A., Rossolini G.M., Bartoloni A., Romagnani S., Liotta F., Annunziato F., Cosmi L. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest. 2020;130(9):4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wolf Y., Anderson A.C., Kuchroo V.K. TIM3 comes of age as an inhibitory receptor. Nat. Rev. Immunol. 2020;20(3):173–185. doi: 10.1038/s41577-019-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shubin N.J., Chung C.S., Heffernan D.S., Irwin L.R., Monaghan S.F., Ayala A. BTLA expression contributes to septic morbidity and mortality by inducing innate inflammatory cell dysfunction. J. Leukoc. Biol. 2012;92(3):593–603. doi: 10.1189/jlb.1211641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Shubin N.J., Monaghan S.F., Heffernan D.S., Chung C.-S., Ayala A. B and T lymphocyte attenuator expression on CD4+ T-cells associates with sepsis and subsequent infections in ICU patients. Crit. Care. 2013;17(6):R276. doi: 10.1186/cc13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.B.J. Meckiff, C. Ramírez-Suástegui, V. Fajardo, S.J. Chee, A. Kusnadi, H. Simon, A. Grifoni, E. Pelosi, D. Weiskopf, A. Sette, F. Ay, G. Seumois, C.H. Ottensmeier, P. Vijayanand, Single-cell transcriptomic analysis of SARS-CoV-2 reactive CD4+T cells, bioRxiv (2020) 2020.06.12.148916.
- 239.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 2008;8(5):337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 240.Timoteo R.P., da Silva M.V., Miguel C.B., Silva D.A.A., Catarino J.D.S., Rodrigues Junior V., Sales-Campos H., Freire Oliveira C.J. Th1/Th17-related cytokines and chemokines and their implications in the pathogenesis of pemphigus vulgaris. Mediators Inflamm. 2017:7151285. doi: 10.1155/2017/7151285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host & Microbe. 2020;27(6):992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Mazzoni A., Salvati L., Maggi L., Capone M., Vanni A., Spinicci M., Mencarini J., Caporale R., Peruzzi B., Antonelli A., Trotta M., Zammarchi L., Ciani L., Gori L., Lazzeri C., Matucci A., Vultaggio A., Rossi O., Almerigogna F., Parronchi P., Fontanari P., Lavorini F., Peris A., Rossolini G.M., Bartoloni A., Romagnani S., Liotta F., Annunziato F., Cosmi L. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest. 2020;130(9) doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.-A., Merkling S.H., Treluyer J.-M., Veyer D., Mouthon L., Blanc C., Tharaux P.-L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.G. Guaraldi, M. Meschiari, A. Cozzi-Lepri, J. Milic, R. Tonelli, M. Menozzi, E. Franceschini, G. Cuomo, G. Orlando, V. Borghi, A. Santoro, M. Di Gaetano, C. Puzzolante, F. Carli, A. Bedini, L. Corradi, R. Fantini, I. Castaniere, L. Tabbì, M. Girardis, S. Tedeschi, M. Giannella, M. Bartoletti, R. Pascale, G. Dolci, L. Brugioni, A. Pietrangelo, A. Cossarizza, F. Pea, E. Clini, C. Salvarani, M. Massari, P.L. Viale, C. Mussini, Tocilizumab in patients with severe COVID-19: a retrospective cohort study, The Lancet Rheumatology. [DOI] [PMC free article] [PubMed]
- 245.Roschewski M., Lionakis M.S., Sharman J.P., Roswarski J., Goy A., Monticelli M.A., Roshon M., Wrzesinski S.H., Desai J.V., Zarakas M.A., Collen J., Rose K.M., Hamdy A., Izumi R., Wright G.W., Chung K.K., Baselga J., Staudt L.M., Wilson W.H. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci. Immunol. 2020;5(48):eabd0110. doi: 10.1126/sciimmunol.abd0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020;53(1):19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ligthart S., Sedaghat S., Ikram M.A., Hofman A., Franco O.H., Dehghan A. EN-RAGE: a novel inflammatory marker for incident coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 2014;34(12):2695–2699. doi: 10.1161/ATVBAHA.114.304306. [DOI] [PubMed] [Google Scholar]
- 249.Z. zhang, N. Han, Y. Shen, S100A12 promotes inflammation and cell apoptosis in sepsis-induced ARDS via activation of NLRP3 inflammasome signaling, Molecular Immunol. 122 (2020) 38-48. [DOI] [PubMed]
- 250.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., May B., De Freitas J.K., Glicksberg B.S., Mason C.E., Tatonetti N.P., Shapira S.D. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 2020 doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.L. Smith Nicholas, E. Huffman Jennifer, P. Strachan David, J. Huang, A. Dehghan, S. Trompet, M. Lopez Lorna, S.-Y. Shin, J. Baumert, V. Vitart, C. Bis Joshua, H. Wild Sarah, A. Rumley, Q. Yang, G. Uitterlinden Andre, J. Stott David, G. Davies, M. Carter Angela, B. Thorand, O. Polašek, B. McKnight, H. Campbell, R. Rudnicka Alicja, M.-H. Chen, M. Buckley Brendan, E. Harris Sarah, A. Peters, D. Pulanic, T. Lumley, J.M. de Craen Anton, C. Liewald David, C. Gieger, S. Campbell, I. Ford, J. Gow Alan, M. Luciano, J. Porteous David, X. Guo, N. Sattar, A. Tenesa, M. Cushman, P.E. Slagboom, M. Visscher Peter, D. Spector Tim, T. Illig, I. Rudan, G. Bovill Edwin, F. Wright Alan, L. McArdle Wendy, G. Tofler, A. Hofman, G.J. Westendorp Rudi, M. Starr John, J. Grant Peter, M. Karakas, D. Hastie Nicholas, M. Psaty Bruce, F. Wilson James, D.O. Lowe Gordon, J. O'Donnell Christopher, C.M. Witteman Jacqueline, J.W. Jukema, J. Deary Ian, N. Soranzo, W. Koenig, C. Hayward, Genetic Predictors of Fibrin D-Dimer Levels in Healthy Adults, Circulation 123(17) (2011) 1864-1872. [DOI] [PMC free article] [PubMed]
- 253.G. Dimopoulos, Q. de Mast, N. Markou, M. Theodorakopoulou, A. Komnos, M. Mouktaroudi, M.G. Netea, T. Spyridopoulos, R.J. Verheggen, J. Hoogerwerf, A. Lachana, F.L. van de Veerdonk, E.J. Giamarellos-Bourboulis, Favorable Anakinra Responses in Severe Covid-19 Patients with Secondary Hemophagocytic Lymphohistiocytosis, Cell Host & Microbe (2020). [DOI] [PMC free article] [PubMed]
- 254.Huet T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I., Sacco E., Naccache J.-M., Bézie Y., Laplanche S., Le Berre A., Le Pavec J., Salmeron S., Emmerich J., Mourad J.-J., Chatellier G., Hayem G. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2(7):e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Franzetti M., Pozzetti U., Carugati M., Pandolfo A., Molteni C., Faccioli P., Castaldo G., Longoni E., Ormas V., Iemoli E., Piconi S. Interleukin-1 receptor antagonist anakinra in association with remdesivir in severe Coronavirus disease 2019: a case report. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X., Nicastri E., Oda R., Yo K., Quiros-Roldan E., Studemeister A., Redinski J., Ahmed S., Bernett J., Chelliah D., Chen D., Chihara S., Cohen S.H., Cunningham J., D'Arminio Monforte A., Ismail S., Kato H., Lapadula G., L'Her E., Maeno T., Majumder S., Massari M., Mora-Rillo M., Mutoh Y., Nguyen D., Verweij E., Zoufaly A., Osinusi A.O., DeZure A., Zhao Y., Zhong L., Chokkalingam A., Elboudwarej E., Telep L., Timbs L., Henne I., Sellers S., Cao H., Tan S.K., Winterbourne L., Desai P., Mera R., Gaggar A., Myers R.P., Brainard D.M., Childs R., Flanigan T. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zhao C., Zhao W. NLRP3 inflammasome—a key player in antiviral responses. Front. Immunol. 2020;11:211. doi: 10.3389/fimmu.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Chen I.Y., Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015;23(1):55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 260.Cordova Z.M., Grönholm A., Kytölä V., Taverniti V., Hämäläinen S., Aittomäki S., Niininen W., Junttila I., Ylipää A., Nykter M., Pesu M. Myeloid cell expressed proprotein convertase FURIN attenuates inflammation. Oncotarget. 2016;7(34):54392–54404. doi: 10.18632/oncotarget.11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.M. Cugno, P.L. Meroni, R. Gualtierotti, S. Griffini, E. Grovetti, A. Torri, M. Panigada, S. Aliberti, F. Blasi, F. Tedesco, F. Peyvandi, Complement activation in patients with COVID-19: A novel therapeutic target, Journal of Allergy and Clinical Immunology. [DOI] [PMC free article] [PubMed]
- 262.T. Gao, M. Hu, X. Zhang, H. Li, L. Zhu, H. Liu, Q. Dong, Z. Zhang, Z. Wang, Y. Hu, Y. Fu, Y. Jin, K. Li, S. Zhao, Y. Xiao, S. Luo, L. Li, L. Zhao, J. Liu, H. Zhao, Y. Liu, W. Yang, J. Peng, X. Chen, P. Li, Y. Liu, Y. Xie, J. Song, L. Zhang, Q. Ma, X. Bian, W. Chen, X. Liu, Q. Mao, C. Cao, Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation, medRxiv (2020) 2020.03.29.20041962.
- 263.Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.R. Escher, N. Breakey, B. Lämmle, Severe COVID-19 infection associated with endothelial activation, Thromb Res 190 (2020) 62-62. [DOI] [PMC free article] [PubMed]
- 265.Panigada M., Bottino N., Tagliabue P., Grasselli G., Novembrino C., Chantarangkul V., Pesenti A., Peyvandi F., Tripodi A. Hypercoagulability of COVID-19 patients in intensive care unit: a report of thromboelastography findings and other parameters of hemostasis. J. Thromb. Haemost. 2020;18(7):1738–1742. doi: 10.1111/jth.14850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Skendros P., Mitsios A., Chrysanthopoulou A., Mastellos D.C., Metallidis S., Rafailidis P., Ntinopoulou M., Sertaridou E., Tsironidou V., Tsigalou C., Tektonidou M.G., Konstantinidis T., Papagoras C., Mitroulis I., Germanidis G., Lambris J.D., Ritis K. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 2020 doi: 10.1172/JCI141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Kamitaki N., Sekar A., Handsaker R.E., de Rivera H., Tooley K., Morris D.L., Taylor K.E., Whelan C.W., Tombleson P., Loohuis L.M.O., Ripke S., Neale B.M., Corvin A., Walters J.T.R., Farh K.-H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H., Pers T.H., Agartz I., Agerbo E., Albus M., Alexander M., Amin F., Bacanu S.A., Begemann M., Belliveau R.A., Bene J., Bergen S.E., Bevilacqua E., Bigdeli T.B., Black D.W., Bruggeman R., Buccola N.G., Buckner R.L., Byerley W., Cahn W., Cai G., Cairns M.J., Campion D., Cantor R.M., Carr V.J., Carrera N., Catts S.V., Chambert K.D., Chan R.C.K., Chen R.Y.L., Chen E.Y.H., Cheng W., Cheung E.F.C., Chong S.A., Cloninger C.R., Cohen D., Cohen N., Cormican P., Craddock N., Crespo-Facorro B., Crowley J.J., Curtis D., Davidson M., Davis K.L., Degenhardt F., Del Favero J., DeLisi L.E., Demontis D., Dikeos D., Dinan T., Djurovic S., Donohoe G., Drapeau E., Duan J., Dudbridge F., Durmishi N., Eichhammer P., Eriksson J., Escott-Price V., Essioux L., Fanous A.H., Farrell M.S., Frank J., Franke L., Freedman R., Freimer N.B., Friedl M., Friedman J.I., Fromer M., Genovese G., Georgieva L., Gershon E.S., Giegling I., Giusti-Rodríguez P., Godard S., Goldstein J.I., Golimbet V., Gopal S., Gratten J., de Haan L., Mitjans M., Hamshere M.L., Hansen M., Hansen T., Haroutunian V., Hartmann A.M., Henskens F.A., Herms S., Hirschhorn J.N., Hoffmann P., Hofman A., Hollegaard M.V., Hougaard D.M., Ikeda M., Joa I., Julià A., Kahn R.S., Kalaydjieva L., Karachanak-Yankova S., Karjalainen J., Kavanagh D., Keller M.C., Kelly B.J., Kennedy J.L., Khrunin A., Kim Y., Klovins J., Knowles J.A., Konte B., Kucinskas V., Kucinskiene Z.A., Kuzelova-Ptackova H., Kähler A.K., Laurent C., Keong J.L.C., Lee S.H., Legge S.E., Lerer B., Li M., Li T., Liang K.-Y., Lieberman J., Limborska S., Loughland C.M., Jan L., Lönnqvist J., Macek M., Magnusson P.K.E., Maher B.S., Maier W., Mallet J., Marsal S., Mattheisen M., Mattingsdal M., McCarley R.W., McDonald C., McIntosh A.M., Meier S., Meijer C.J., Melegh B., Melle I., Mesholam-Gately R.I., Metspalu A., Michie P.T., Milani L., Milanova V., Mokrab Y., Morris D.W., Mors O., Murphy K.C., Murray R.M., Myin-Germeys I., Müller-Myhsok B., Nelis M., Nenadic I., Nertney D.A., Nestadt G., Nicodemus K.K., Nikitina-Zake L., Nisenbaum L., Nordin A., O’Callaghan E., O’Dushlaine C., O’Neill F.A., Oh S.-Y., Olincy A., Olsen L., Van Os J., Pantelis C., Papadimitriou G.N., Steixner A.A., Parkhomenko E., Pato M.T., Paunio T., Pejovic-Milovancevic M., Perkins D.O., Pietiläinen O., Pimm J., Pocklington A.J., Powell J., Price A., Pulver A.E., Purcell S.M., Quested D., Rasmussen H.B., Reichenberg A., Reimers M.A., Richards A.L., Roffman J.L., Roussos P., Ruderfer D.M., Salomaa V., Sanders A.R., Schall U., Schubert C.R., Schulze T.G., Schwab S.G., Scolnick E.M., Scott R.J., Seidman L.J., Shi J., Sigurdsson E., Silagadze T., Silverman J.M., Sim K., Slominsky P., Smoller J.W., So H.-C., Spencer C.C.A., Stahl E.A., Stefansson H., Steinberg S., Stogmann E., Straub R.E., Strengman E., Strohmaier J., Stroup T.S., Subramaniam M., Suvisaari J., Svrakic D.M., Szatkiewicz J.P., Söderman E., Thirumalai S., Toncheva D., Tooney P.A., Tosato S., Veijola J., Waddington J., Walsh D., Wang D., Wang Q., Webb B.T., Weiser M., Wildenauer D.B., Williams N.M., Williams S., Witt S.H., Wolen A.R., Wong E.H.M., Wormley B.K., Wu J.Q., Xi H.S., Zai C.C., Zheng X., Zimprich F., Wray N.R., Stefansson K., Visscher P.M., Adolfsson R., Andreassen O.A., Blackwood D.H.R., Bramon E., Buxbaum J.D., Børglum A.D., Cichon S., Darvasi A., Domenici E., Ehrenreich H., Esko T., Gejman P.V., Gill M., Gurling H., Hultman C.M., Iwata N., Jablensky A.V., Jönsson E.G., Kendler K.S., Kirov G., Knight J., Lencz T., Levinson D.F., Li Q.S., Liu J., Malhotra A.K., McCarroll S.A., McQuillin A., Moran J.L., Mortensen C. P.B. Schizophrenia Working Group of the Psychiatric Genomics, C. Psychosis Endophenotypes International, C. Wellcome Trust Case-Control, Complement genes contribute sex-biased vulnerability in diverse disorders. Nature. 2020;582(7813):577–581. doi: 10.1038/s41586-020-2277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Gralinski L.E., Sheahan T.P., Morrison T.E., Menachery V.D., Jensen K., Leist S.R., Whitmore A., Heise M.T., Baric R.S. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9(5) doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.A. Silvin, N. Chapuis, G. Dunsmore, A.-G. Goubet, A. Dubuisson, L. Derosa, C. Almire, C. Hénon, O. Kosmider, N. Droin, P. Rameau, C. Catelain, A. Alfaro, C. Dussiau, C. Friedrich, E. Sourdeau, N. Marin, T.-A. Szwebel, D. Cantin, L. Mouthon, D. Borderie, M. Deloger, D. Bredel, S. Mouraud, D. Drubay, M. Andrieu, A.-S. Lhonneur, V. Saada, A. Stoclin, C. Willekens, F. Pommeret, F. Griscelli, L.G. Ng, Z. Zhang, P. Bost, I. Amit, F. Barlesi, A. Marabelle, F. Pène, B. Gachot, F. André, L. Zitvogel, F. Ginhoux, M. Fontenay, E. Solary, Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19, Cell. [DOI] [PMC free article] [PubMed]
- 271.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Byun H.J., Jung W.W., Lee D.S., Kim S., Kim S.J., Park C.G., Chung H.Y., Chun T. Proliferation of activated CD1d-restricted NKT cells is down-modulated by lymphocyte activation gene-3 signaling via cell cycle arrest in S phase. Cell Biol. Int. 2007;31(3):257–262. doi: 10.1016/j.cellbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 273.da Silva I.P., Gallois A., Jimenez-Baranda S., Khan S., Anderson A.C., Kuchroo V.K., Osman I., Bhardwaj N. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 Blockade. Cancer Immunol. Research. 2014;2(5):410–422. doi: 10.1158/2326-6066.CIR-13-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Khan M., Arooj S., Wang H. NK cell-based immune checkpoint inhibition. Front. Immunol. 2020;11:167. doi: 10.3389/fimmu.2020.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Harjunpää H., Guillerey C. TIGIT as an emerging immune checkpoint. Clin. Exp. Immunol. 2020;200(2):108–119. doi: 10.1111/cei.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.C. Gruber, Impaired interferon signature in severe COVID-19, Nature Reviews Immunology 20(6) (2020) 353-353. [DOI] [PMC free article] [PubMed]
- 277.Zhang B., Zhou X., Zhu C., Song Y., Feng F., Qiu Y., Feng J., Jia Q., Song Q., Zhu B., Wang J. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and IgG level predicts disease severity and outcome for patients with COVID-19. Front. Mol. Biosci. 2020;7(157) doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R., Song M., Wang L., Zhang W., Han B., Yang L., Wang X., Zhou G., Zhang T., Li B., Wang Y., Chen Z., Wang X. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. Journal of Translational Medicine. 2020;18(1):206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.van Gisbergen K.P.J.M., Ludwig I.S., Geijtenbeek T.B.H., van Kooyk Y. Interactions of DC-SIGN with Mac-1 and CEACAM1 regulate contact between dendritic cells and neutrophils. FEBS Lett. 2005;579(27):6159–6168. doi: 10.1016/j.febslet.2005.09.089. [DOI] [PubMed] [Google Scholar]
- 281.van Gisbergen K.P., Sanchez-Hernandez M., Geijtenbeek T.B., van Kooyk Y. Neutrophils mediate immune modulation of dendritic cells through glycosylation-dependent interactions between Mac-1 and DC-SIGN. J. Exp. Med. 2005;201(8):1281–1292. doi: 10.1084/jem.20041276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Kumar V., Sharma A. Neutrophils: Cinderella of innate immune system. Int. Immunopharmacol. 2010;10(11):1325–1334. doi: 10.1016/j.intimp.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 283.Y. Zhou, B. Fu, X. Zheng, D. Wang, C. Zhao, Y. qi, R. Sun, Z. Tian, X. Xu, H. Wei, Aberrant pathogenic GM-CSF<sup>+</sup> T cells and inflammatory CD14<sup>+</sup>CD16<sup>+</sup> monocytes in severe pulmonary syndrome patients of a new coronavirus, bioRxiv (2020) 2020.02.12.945576.
- 284.D. Blanco-Melo, B.E. Nilsson-Payant, W.C. Liu, S. Uhl, D. Hoagland, R. Møller, T.X. Jordan, K. Oishi, M. Panis, D. Sachs, T.T. Wang, R.E. Schwartz, J.K. Lim, R.A. Albrecht, B.R. tenOever, Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19, Cell 181(5) (2020) 1036-1045.e9. [DOI] [PMC free article] [PubMed]
- 285.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27(6):883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 287.Jaigirdar S.A., Benson R.A., Elmesmari A., Kurowska-Stolarska M.S., McInnes I.B., Garside P., MacLeod M.K.L. Sphingosine-1-phosphate promotes the persistence of activated CD4 T cells in inflamed sites. Front. Immunol. 2017;8(1627) doi: 10.3389/fimmu.2017.01627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Lee W.-B., Choi W.Y., Lee D.-H., Shim H., Kim-Ha J., Kim Y.-J. OAS1 and OAS3 negatively regulate the expression of chemokines and interferon-responsive genes in human macrophages. BMB Rep. 2019;52(2):133–138. doi: 10.5483/BMBRep.2019.52.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Zhu L., Yu Q., Gao P., Liu Q., Luo X., Jiang G., Ji R., Yang R., Ma X., Xu J., Yuan H., Zhou J., An H. TAOK1 positively regulates TLR4-induced inflammatory responses by promoting ERK1/2 activation in macrophages. Mol. Immunol. 2020;122:124–131. doi: 10.1016/j.molimm.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 290.Schutyser E., Richmond A., Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J. Leukoc. Biol. 2005;78(1):14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.D. Zhang, R. Guo, L. Lei, H. Liu, Y. Wang, Y. Wang, T. Dai, T. Zhang, Y. Lai, J. Wang, Z. Liu, A. He, M. O'Dwyer, J. Hu, COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome, medRxiv (2020) 2020.03.24.20042655.
- 292.Pesu M., Muul L., Kanno Y., O'Shea J.J. Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood. 2006;108(3):983–985. doi: 10.1182/blood-2005-09-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.C. Atyeo, S. Fischinger, T. Zohar, M.D. Slein, J. Burke, C. Loos, D.J. McCulloch, K.L. Newman, C. Wolf, J. Yu, K. Shuey, J. Feldman, B.M. Hauser, T. Caradonna, A. Schmidt, T.J. Suscovich, C. Linde, Y. Cai, D. Barouch, E.T. Ryan, R.C. Charles, D. Lauffenburger, H. Chu, G. Alter, Distinct early serological signatures track with SARS-CoV-2 survival, Immunity. [DOI] [PMC free article] [PubMed]
- 294.Gong F., Dai Y., Zheng T., Cheng L., Zhao D., Wang H., Liu M., Pei H., Jin T., Yu D., Zhou P. Peripheral CD4+ T cell subsets and antibody response in COVID-19 convalescent individuals. J. Clin. Invest. 2020 doi: 10.1172/JCI141054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Morita R., Schmitt N., Bentebibel S.E., Ranganathan R., Bourdery L., Zurawski G., Foucat E., Dullaers M., Oh S., Sabzghabaei N., Lavecchio E.M., Punaro M., Pascual V., Banchereau J., Ueno H. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34(1):108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Sage P.T., Ron-Harel N., Juneja V.R., Sen D.R., Maleri S., Sungnak W., Kuchroo V.K., Haining W.N., Chevrier N., Haigis M., Sharpe A.H. Suppression by TFR cells leads to durable and selective inhibition of B cell effector function. Nat. Immunol. 2016;17(12):1436–1446. doi: 10.1038/ni.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sage P.T., Alvarez D., Godec J., von Andrian U.H., Sharpe A.H. Circulating T follicular regulatory and helper cells have memory-like properties. J. Clin. Invest. 2014;124(12):5191–5204. doi: 10.1172/JCI76861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.K. V., Toll-like receptors in immunity and inflammatory diseases: Past, present, and future, Int. Immunopharmacol. 59 (2018) 391-412. [DOI] [PMC free article] [PubMed]
- 299.Ahn J., Barber G.N. STING signaling and host defense against microbial infection. Experimental Mol. Med. 2019;51(12):1–10. doi: 10.1038/s12276-019-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kumar V. A STING to inflammation and autoimmunity. J. Leukoc. Biol. 2019;106(1):171–185. doi: 10.1002/JLB.4MIR1018-397RR. [DOI] [PubMed] [Google Scholar]
- 301.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Li X.D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341(6152):1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Gao D., Wu J., Wu Y.-T., Du F., Aroh C., Yan N., Sun L., Chen Z.J. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341(6148):903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ni G., Ma Z., Damania B. cGAS and STING: at the intersection of DNA and RNA virus-sensing networks. PLoS Pathog. 2018;14(8):e1007148. doi: 10.1371/journal.ppat.1007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Eaglesham J.B., Kranzusch P.J. Conserved strategies for pathogen evasion of cGAS–STING immunity. Curr. Opin. Immunol. 2020;66:27–34. doi: 10.1016/j.coi.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wu J., Dobbs N., Yang K., Yan N. Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity. 2020;53(1):115–126.e5. doi: 10.1016/j.immuni.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Wu J., Chen Y.-J., Dobbs N., Sakai T., Liou J., Miner J.J., Yan N. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell deathSTING-mediated ER stress primes T cell death. J. Exp. Med. 2019;216(4):867–883. doi: 10.1084/jem.20182192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Totura A.L., Whitmore A., Agnihothram S., Schäfer A., Katze M.G., Heise M.T., Baric R.S. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus. Infection. mBio 2015,;6(3):e00638–e715. doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Li Y., Chen M., Cao H., Zhu Y., Zheng J., Zhou H. Extraordinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response. Microbes Infect. 2013;15(2):88–95. doi: 10.1016/j.micinf.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Tang F.S., Van Ly D., Spann K., Reading P.C., Burgess J.K., Hartl D., Baines K.J., Oliver B.G. Differential neutrophil activation in viral infections: Enhanced TLR-7/8-mediated CXCL8 release in asthma. Respirology. 2016;21(1):172–179. doi: 10.1111/resp.12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Moreno-Eutimio M.A., López-Macías C., Pastelin-Palacios R. Bioinformatic analysis and identification of single-stranded RNA sequences recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. Microbes Infection. 2020;22(4):226–229. doi: 10.1016/j.micinf.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.G. Hartmann, Nucleic acid immunity, Advances in immunology, Elsevier, 2017, pp. 121-169. [DOI] [PMC free article] [PubMed]
- 314.D. Goubau, S. Deddouche, C.R. e Sousa, Cytosolic sensing of viruses, Immunity 38(5) (2013) 855-869. [DOI] [PMC free article] [PubMed]
- 315.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Zhang X., Li X., Ning F., Shang Y., Hu X. TLE4 acts as a corepressor of Hes1 to inhibit inflammatory responses in macrophages. Protein Cell. 2019;10(4):300–305. doi: 10.1007/s13238-018-0554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ramasamy S., Saez B., Mukhopadhyay S., Ding D., Ahmed A.M., Chen X., Pucci F., Yamin R., Wang J., Pittet M.J., Kelleher C.M., Scadden D.T., Sweetser D.A. Tle1 tumor suppressor negatively regulates inflammation in vivo and modulates NF-κB inflammatory pathway. Proc. Natl. Acad. Sci. U S A. 2016;113(7):1871–1876. doi: 10.1073/pnas.1511380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wang C., Chen T., Zhang J., Yang M., Li N., Xu X., Cao X. The E3 ubiquitin ligase Nrdp1 'preferentially' promotes TLR-mediated production of type I interferon. Nat. Immunol. 2009;10(7):744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 320.Ye S., Xu H., Jin J., Yang M., Wang C., Yu Y., Cao X. The E3 ubiquitin ligase neuregulin receptor degradation protein 1 (Nrdp1) promotes M2 macrophage polarization by ubiquitinating and activating transcription factor CCAAT/enhancer-binding Protein β (C/EBPβ) J. Biol. Chem. 2012;287(32):26740–26748. doi: 10.1074/jbc.M112.383265. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 321.Kondo T., Watanabe M., Hatakeyama S. TRIM59 interacts with ECSIT and negatively regulates NF-κB and IRF-3/7-mediated signal pathways. Biochem. Biophys. Res. Commun. 2012;422(3):501–507. doi: 10.1016/j.bbrc.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 322.Jin Z., Zhu Z., Liu S., Hou Y., Tang M., Zhu P., Tian Y., Li D., Yan D., Zhu X. TRIM59 protects mice from sepsis by regulating inflammation and phagocytosis in macrophages. Front. Immunol. 2020;11(263) doi: 10.3389/fimmu.2020.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Kopp E., Medzhitov R., Carothers J., Xiao C., Douglas I., Janeway C.A., Ghosh S. ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes Dev. 1999;13(16):2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Wi S.M., Moon G., Kim J., Kim S.T., Shim J.H., Chun E., Lee K.Y. TAK1-ECSIT-TRAF6 complex plays a key role in the TLR4 signal to activate NF-κB. J. Biol. Chem. 2014;289(51):35205–35214. doi: 10.1074/jbc.M114.597187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Lei C.Q., Zhang Y., Li M., Jiang L.Q., Zhong B., Kim Y.H., Shu H.B. ECSIT bridges RIG-I-like receptors to VISA in signaling events of innate antiviral responses. J. Innate Immun. 2015;7(2):153–164. doi: 10.1159/000365971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Kumagai Y., Akira S. Mind bomb proteins in the antiviral arsenal. Immunity. 2011;35(3):320–322. doi: 10.1016/j.immuni.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 327.Y. Zhang, J. Zhang, Y. Chen, B. Luo, Y. Yuan, F. Huang, T. Yang, F. Yu, J. Liu, B. Liu, Z. Song, J. Chen, T. Pan, X. Zhang, Y. Li, R. Li, W. Huang, F. Xiao, H. Zhang, The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I, bioRxiv (2020) 2020.05.24.111823.
- 328.Xia X., Cui J., Wang H.Y., Zhu L., Matsueda S., Wang Q., Yang X., Hong J., Songyang Z., Chen Z.J., Wang R.F. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity. 2011;34(6):843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Allen I.C., Moore C.B., Schneider M., Lei Y., Davis B.K., Scull M.A., Gris D., Roney K.E., Zimmermann A.G., Bowzard J.B., Ranjan P., Monroe K.M., Pickles R.J., Sambhara S., Ting J.P. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity. 2011;34(6):854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Nagai-Singer M.A., Morrison H.A., Allen I.C. NLRX1 is a multifaceted and enigmatic regulator of immune system function. Front. Immunol. 2019;10:2419. doi: 10.3389/fimmu.2019.02419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Hung S.C., Huang P.R., Almeida-da-Silva C.L.C., Atanasova K.R., Yilmaz O., Ojcius D.M. NLRX1 modulates differentially NLRP3 inflammasome activation and NF-κB signaling during Fusobacterium nucleatum infection. Microbes Infect. 2018;20(9–10):615–625. doi: 10.1016/j.micinf.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Cho A., Saunders O.L., Butler T., Zhang L., Xu J., Vela J.E., Feng J.Y., Ray A.S., Kim C.U. Synthesis and antiviral activity of a series of 1'-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg. Med. Chem. Lett. 2012;22(8):2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11(4) doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.B.N. Williamson, F. Feldmann, B. Schwarz, K. Meade-White, D.P. Porter, J. Schulz, N.v. Doremalen, I. Leighton, C.K. Yinda, L. Pérez-Pérez, A. Okumura, J. Lovaglio, P.W. Hanley, G. Saturday, C.M. Bosio, S. Anzick, K. Barbian, T. Cihlar, C. Martens, D.P. Scott, V.J. Munster, E.d. Wit, Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2, bioRxiv (2020) 2020.04.15.043166. [DOI] [PMC free article] [PubMed]
- 339.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.-D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of Covid-19 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.R. Dolin, M.S. Hirsch, Remdesivir — an important first step, New England J. Med. (2020). [DOI] [PMC free article] [PubMed]
- 341.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541) doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery. 2020;6(1):16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Tang W., Cao Z., Han M., Wang Z., Chen J., Sun W., Wu Y., Xiao W., Liu S., Chen E., Chen W., Wang X., Yang J., Lin J., Zhao Q., Yan Y., Xie Z., Li D., Yang Y., Liu L., Qu J., Ning G., Shi G., Xie Q. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 2020;369:m1849. doi: 10.1136/bmj.m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., Skipper C.P., Nascene A.A., Nicol M.R., Abassi M., Engen N.W., Cheng M.P., LaBar D., Lother S.A., MacKenzie L.J., Drobot G., Marten N., Zarychanski R., Kelly L.E., Schwartz I.S., McDonald E.G., Rajasingham R., Lee T.C., Hullsiek K.H. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.W. Tang, Z. Cao, M. Han, Z. Wang, J. Chen, W. Sun, Y. Wu, W. Xiao, S. Liu, E. Chen, W. Chen, X. Wang, J. Yang, J. Lin, Q. Zhao, Y. Yan, Z. Xie, D. Li, Y. Yang, L. Liu, J. Qu, G. Ning, G. Shi, Q. Xie, Hydroxychloroquine in patients with COVID-19: an open-label, randomized, controlled trial, medRxiv (2020) 2020.04.10.20060558.
- 346.A.B. Cavalcanti, F.G. Zampieri, R.G. Rosa, L.C.P. Azevedo, V.C. Veiga, A. Avezum, L.P. Damiani, A. Marcadenti, L. Kawano-Dourado, T. Lisboa, D.L.M. Junqueira, P.G.M. de Barros e Silva, L. Tramujas, E.O. Abreu-Silva, L.N. Laranjeira, A.T. Soares, L.S. Echenique, A.J. Pereira, F.G.R. Freitas, O.C.E. Gebara, V.C.S. Dantas, R.H.M. Furtado, E.P. Milan, N.A. Golin, F.F. Cardoso, I.S. Maia, C.R. Hoffmann Filho, A.P.M. Kormann, R.B. Amazonas, M.F. Bocchi de Oliveira, A. Serpa-Neto, M. Falavigna, R.D. Lopes, F.R. Machado, O. Berwanger, Hydroxychloroquine with or without Azithromycin in Mild-to-Moderate Covid-19, New England Journal of Medicine (2020).
- 347.N. Wang, Y. Zhan, L. Zhu, Z. Hou, F. Liu, P. Song, F. Qiu, X. Wang, X. Zou, D. Wan, X. Qian, S. Wang, Y. Guo, H. Yu, M. Cui, G. Tong, Y. Xu, Z. Zheng, Y. Lu, P. Hong, Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients, Cell Host Microbe. [DOI] [PMC free article] [PubMed]
- 348.Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., Tan T.K., Rijal P., Coombes N., Bewley K.R., Tree J.A., Radecke J., Paterson N.G., Supasa P., Mongkolsapaya J., Screaton G.R., Carroll M., Townsend A., Fry E.E., Owens R.J., Stuart D.I. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host & Microbe. 2020 doi: 10.1016/j.chom.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.M.G. Joyce, R.S. Sankhala, W.H. Chen, M. Choe, H. Bai, A. Hajduczki, L. Yan, S.L. Sterling, C.E. Peterson, E.C. Green, C. Smith, N. de Val, M. Amare, P. Scott, E.D. Laing, C.C. Broder, M. Rolland, N.L. Michael, K. Modjarrad, A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein, bioRxiv (2020).
- 351.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.-S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 352.Zhou D., Duyvesteyn H.M.E., Chen C.P., Huang C.G., Chen T.H., Shih S.R., Lin Y.C., Cheng C.Y., Cheng S.H., Huang Y.C., Lin T.Y., Ma C., Huo J., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., Tan T.K., Rijal P., Donat R.F., Godwin K., Buttigieg K.R., Tree J.A., Radecke J., Paterson N.G., Supasa P., Mongkolsapaya J., Screaton G.R., Carroll M.W., Gilbert-Jaramillo J., Knight M.L., James W., Owens R.J., Naismith J.H., Townsend A.R., Fry E.E., Zhao Y., Ren J., Stuart D.I., Huang K.A. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020 doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 353.A.C. Walls, Y.-J. Park, M.A. Tortorici, A. Wall, A.T. McGuire, D. Veesler, Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell. [DOI] [PMC free article] [PubMed]
- 354.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Khan A., Benthin C., Zeno B., Albertson T.E., Boyd J., Christie J.D., Hall R., Poirier G., Ronco J.J., Tidswell M., Hardes K., Powley W.M., Wright T.J., Siederer S.K., Fairman D.A., Lipson D.A., Bayliffe A.I., Lazaar A.L. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care. 2017;21(1):234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.N. Iwanaga, L. Cooper, L. Rong, B. Beddingfield, J. Crabtree, R.A. Tripp, J.K. Kolls, Novel ACE2-IgG1 fusions with improved activity against SARS-CoV2, bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 357.Bronte V., Ugel S., Tinazzi E., Vella A., De Sanctis F., Canè S., Batani V., Trovato R., Fiore A., Petrova V., Hofer F., Barouni R.M., Musiu C., Caligola S., Pinton L., Torroni L., Polati E., Donadello K., Friso S., Pizzolo F., Iezzi M., Facciotti F., Pelicci P.G., Righetti D., Bazzoni P., Rampudda M., Comel A.C., Mosaner W., Lunardi C., Olivieri O. Baricitinib restrains the immune dysregulation in severe COVID-19 patients. J Clin Invest. 2020 doi: 10.1172/JCI141772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Giudice V., Pagliano P., Vatrella A., Masullo A., Poto S., Polverino B.M., Gammaldi R., Maglio A., Sellitto C., Vitale C., Serio B., Cuffa B., Borrelli A., Vecchione C., Filippelli A., Selleri C. Combination of ruxolitinib and eculizumab for treatment of severe SARS-CoV-2-related acute respiratory distress syndrome: a controlled study. Front. Pharmacol. 2020;11:857. doi: 10.3389/fphar.2020.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Mastellos D.C., Ricklin D., Lambris J.D. Clinical promise of next-generation complement therapeutics. Nat. Rev. Drug Discovery. 2019;18(9):707–729. doi: 10.1038/s41573-019-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Risitano A.M., Mastellos D.C., Huber-Lang M., Yancopoulou D., Garlanda C., Ciceri F., Lambris J.D. Complement as a target in COVID-19? Nat. Rev. Immunol. 2020;20(6):343–344. doi: 10.1038/s41577-020-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Hung I.F.-N., Lung K.-C., Tso E.Y.-K., Liu R., Chung T.W.-H., Chu M.-Y., Ng Y.-Y., Lo J., Chan J., Tam A.R., Shum H.-P., Chan V., Wu A.K.-L., Sin K.-M., Leung W.-S., Law W.-L., Lung D.C., Sin S., Yeung P., Yip C.C.-Y., Zhang R.R., Fung A.Y.-F., Yan E.Y.-W., Leung K.-H., Ip J.D., Chu A.W.-H., Chan W.-M., Ng A.C.-K., Lee R., Fung K., Yeung A., Wu T.-C., Chan J.W.-M., Yan W.-W., Chan W.-M., Chan J.F.-W., Lie A.K.-W., Tsang O.T.-Y., Cheng V.C.-C., Que T.-L., Lau C.-S., Chan K.-H., To K.K.-W., Yuen K.-Y. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. The Lancet. 2020;395(10238):1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Kasper L.H., Reder A.T. Immunomodulatory activity of interferon-beta. Ann. Clin. Transl. Neurol. 2014;1(8):622–631. doi: 10.1002/acn3.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.G. De Luca, G. Cavalli, C. Campochiaro, E. Della-Torre, P. Angelillo, A. Tomelleri, N. Boffini, S. Tentori, F. Mette, N. Farina, P. Rovere-Querini, A. Ruggeri, T. D'Aliberti, P. Scarpellini, G. Landoni, F. De Cobelli, J.F. Paolini, A. Zangrillo, M. Tresoldi, B.C. Trapnell, F. Ciceri, L. Dagna, GM-CSF blockade with mavrilimumab in severe COVID-19 pneumonia and systemic hyperinflammation: a single-centre, prospective cohort study, The Lancet Rheumatology. [DOI] [PMC free article] [PubMed]
- 365.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Joyner M.J., Wright R.S., Fairweather D., Senefeld J.W., Bruno K.A., Klassen S.A., Carter R.E., Klompas A.M., Wiggins C.C., Shepherd J.R.A., Rea R.F., Whelan E.R., Clayburn A.J., Spiegel M.R., Johnson P.W., Lesser E.R., Baker S.E., Larson K.F., Ripoll J.G., Andersen K.J., Hodge D.O., Kunze K.L., Buras M.R., Vogt M.N.P., Herasevich V., Dennis J.J., Regimbal R.J., Bauer P.R., Blair J.E., Van Buskirk C.M., Winters J.L., Stubbs J.R., Paneth N.S., Verdun N.C., Marks P., Casadevall A. Early safety indicators of COVID-19 convalescent plasma in 5000 patients. J. Clin. Invest. 2020;130(9):4791–4797. doi: 10.1172/JCI140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Casadevall A., Pirofski L.-A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130(4):1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Xia X., Li K., Wu L., Wang Z., Zhu M., Huang B., Li J., Wang Z., Wu W., Wu M., Li W., Li L., Cai Y., Bosco B., Zhong A., Liu X., Lv T., Gan Z., Chen G., Pan Y., Liu C., Zhang K., Xu X., Wang C., Wang Q. Improved clinical symptoms and mortality among patients with severe or critical COVID-19 after convalescent plasma transfusion. Blood. 2020;136(6):755–759. doi: 10.1182/blood.2020007079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Hegerova L., Gooley T.A., Sweerus K.A., Maree C., Bailey N., Bailey M., Dunleavy V., Patel K., Alcorn K., Haley R., Johnsen J.M., Konkle B.A., Lahti A.C., Alexander M.L., Goldman J.D., Lipke A., Lim S.-J., Sullivan M.D., Pauk J.S., Pagel J.M. Use of convalescent plasma in hospitalized patients with COVID-19: case series. Blood. 2020;136(6):759–762. doi: 10.1182/blood.2020006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L., van Buskirk C., Grossman B.J., Joyner M., Henderson J.P., Pekosz A., Lau B., Wesolowski A., Katz L., Shan H., Auwaerter P.G., Thomas D., Sullivan D.J., Paneth N., Gehrie E., Spitalnik S., Hod E., Pollack L., Nicholson W.T., Pirofski L.-A., Bailey J.A., Tobian A.A.R. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 2020;130(6):2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181(5):1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Forsman A., Beirnaert E., Aasa-Chapman M.M., Hoorelbeke B., Hijazi K., Koh W., Tack V., Szynol A., Kelly C., McKnight A., Verrips T., de Haard H., Weiss R.A. Llama antibody fragments with cross-subtype human immunodeficiency virus type 1 (HIV-1)-neutralizing properties and high affinity for HIV-1 gp120. J. Virol. 2008;82(24):12069–12081. doi: 10.1128/JVI.01379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020 doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 374.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Liu L., Liu W., Zheng Y., Jiang X., Kou G., Ding J., Wang Q., Huang Q., Ding Y., Ni W., Wu W., Tang S., Tan L., Hu Z., Xu W., Zhang Y., Zhang B., Tang Z., Zhang X., Li H., Rao Z., Jiang H., Ren X., Wang S., Zheng S. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect. 2020;22(4):206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Tilocca B., Soggiu A., Sanguinetti M., Babini G., De Maio F., Britti D., Zecconi A., Bonizzi L., Urbani A., Roncada P. Immunoinformatic analysis of the SARS-CoV-2 envelope protein as a strategy to assess cross-protection against COVID-19. Microbes Infect. 2020;22(4):182–187. doi: 10.1016/j.micinf.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Case J.B., Rothlauf P.W., Chen R.E., Kafai N.M., Fox J.M., Smith B., Shrihari S., McCune B.T., Harvey I.B., Keeler S.P., Bloyet L.-M., Zhao H., Ma M., Adams L.J., Winkler E.S., Holtzman M.J., Fremont D.H., Whelan S.P.J., Diamond M.S. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.J.B. Case, P.W. Rothlauf, R.E. Chen, Z. Liu, H. Zhao, A.S. Kim, L.-M. Bloyet, Q. Zeng, S. Tahan, L. Droit, M.X.G. Ilagan, M.A. Tartell, G. Amarasinghe, J.P. Henderson, S. Miersch, M. Ustav, S. Sidhu, H.W. Virgin, D. Wang, S. Ding, D. Corti, E.S. Theel, D.H. Fremont, M.S. Diamond, S.P.J. Whelan, Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2, Cell Host Microbe (2020). [DOI] [PMC free article] [PubMed]
- 379.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.P.M. Folegatti, K.J. Ewer, P.K. Aley, B. Angus, S. Becker, S. Belij-Rammerstorfer, D. Bellamy, S. Bibi, M. Bittaye, E.A. Clutterbuck, C. Dold, S.N. Faust, A. Finn, A.L. Flaxman, B. Hallis, P. Heath, D. Jenkin, R. Lazarus, R. Makinson, A.M. Minassian, K.M. Pollock, M. Ramasamy, H. Robinson, M. Snape, R. Tarrant, M. Voysey, C. Green, A.D. Douglas, A.V.S. Hill, T. Lambe, S.C. Gilbert, A.J. Pollard, J. Aboagye, K. Adams, A. Ali, E. Allen, J.L. Allison, R. Anslow, E.H. Arbe-Barnes, G. Babbage, K. Baillie, M. Baker, N. Baker, P. Baker, I. Baleanu, J. Ballaminut, E. Barnes, J. Barrett, L. Bates, A. Batten, K. Beadon, R. Beckley, E. Berrie, L. Berry, A. Beveridge, K.R. Bewley, E.M. Bijker, T. Bingham, L. Blackwell, C.L. Blundell, E. Bolam, E. Boland, N. Borthwick, T. Bower, A. Boyd, T. Brenner, P.D. Bright, C. Brown-O'Sullivan, E. Brunt, J. Burbage, S. Burge, K.R. Buttigieg, N. Byard, I. Cabera Puig, A. Calvert, S. Camara, M. Cao, F. Cappuccini, M. Carr, M.W. Carroll, V. Carter, K. Cathie, R.J. Challis, S. Charlton, I. Chelysheva, J.-S. Cho, P. Cicconi, L. Cifuentes, H. Clark, E. Clark, T. Cole, R. Colin-Jones, C.P. Conlon, A. Cook, N.S. Coombes, R. Cooper, C.A. Cosgrove, K. Coy, W.E.M. Crocker, C.J. Cunningham, B.E. Damratoski, L. Dando, M.S. Datoo, H. Davies, H. De Graaf, T. Demissie, C. Di Maso, I. Dietrich, T. Dong, F.R. Donnellan, N. Douglas, C. Downing, J. Drake, R. Drake-Brockman, R.E. Drury, S.J. Dunachie, N.J. Edwards, F.D.L. Edwards, C.J. Edwards, S.C. Elias, M.J. Elmore, K.R.W. Emary, M.R. English, S. Fagerbrink, S. Felle, S. Feng, S. Field, C. Fixmer, C. Fletcher, K.J. Ford, J. Fowler, P. Fox, E. Francis, J. Frater, J. Furze, M. Fuskova, E. Galiza, D. Gbesemete, C. Gilbride, K. Godwin, G. Gorini, L. Goulston, C. Grabau, L. Gracie, Z. Gray, L.B. Guthrie, M. Hackett, S. Halwe, E. Hamilton, J. Hamlyn, B. Hanumunthadu, I. Harding, S.A. Harris, A. Harris, D. Harrison, C. Harrison, T.C. Hart, L. Haskell, S. Hawkins, I. Head, J.A. Henry, J. Hill, S.H.C. Hodgson, M.M. Hou, E. Howe, N. Howell, C. Hutlin, S. Ikram, C. Isitt, P. Iveson, S. Jackson, F. Jackson, S.W. James, M. Jenkins, E. Jones, K. Jones, C.E. Jones, B. Jones, R. Kailath, K. Karampatsas, J. Keen, S. Kelly, D. Kelly, D. Kerr, S. Kerridge, L. Khan, U. Khan, A. Killen, J. Kinch, T.B. King, L. King, J. King, L. Kingham-Page, P. Klenerman, F. Knapper, J.C. Knight, D. Knott, S. Koleva, A. Kupke, C.W. Larkworthy, J.P.J. Larwood, A. Laskey, A.M. Lawrie, A. Lee, K.Y. Ngan Lee, E.A. Lees, H. Legge, A. Lelliott, N.-M. Lemm, A.M. Lias, A. Linder, S. Lipworth, X. Liu, S. Liu, R. Lopez Ramon, M. Lwin, F. Mabesa, M. Madhavan, G. Mallett, K. Mansatta, I. Marcal, S. Marinou, E. Marlow, J.L. Marshall, J. Martin, J. McEwan, L. McInroy, G. Meddaugh, A.J. Mentzer, N. Mirtorabi, M. Moore, E. Moran, E. Morey, V. Morgan, S.J. Morris, H. Morrison, G. Morshead, R. Morter, Y.F. Mujadidi, J. Muller, T. Munera-Huertas, C. Munro, A. Munro, S. Murphy, V.J. Munster, P. Mweu, A. Noé, F.L. Nugent, E. Nuthall, K. O'Brien, D. O'Connor, B. Oguti, J.L. Oliver, C. Oliveira, P.J. O'Reilly, M. Osborn, P. Osborne, C. Owen, D. Owens, N. Owino, M. Pacurar, K. Parker, H. Parracho, M. Patrick-Smith, V. Payne, J. Pearce, Y. Peng, M.P. Peralta Alvarez, J. Perring, K. Pfafferott, D. Pipini, E. Plested, H. Pluess-Hall, K. Pollock, I. Poulton, L. Presland, S. Provstgaard-Morys, D. Pulido, K. Radia, F. Ramos Lopez, J. Rand, H. Ratcliffe, T. Rawlinson, S. Rhead, A. Riddell, A.J. Ritchie, H. Roberts, J. Robson, S. Roche, C. Rohde, C.S. Rollier, R. Romani, I. Rudiansyah, S. Saich, S. Sajjad, S. Salvador, L. Sanchez Riera, H. Sanders, K. Sanders, S. Sapaun, C. Sayce, E. Schofield, G. Screaton, B. Selby, C. Semple, H.R. Sharpe, I. Shaik, A. Shea, H. Shelton, S. Silk, L. Silva-Reyes, D.T. Skelly, H. Smee, C.C. Smith, D.J. Smith, R. Song, A.J. Spencer, E. Stafford, A. Steele, E. Stefanova, L. Stockdale, A. Szigeti, A. Tahiri-Alaoui, M. Tait, H. Talbot, R. Tanner, I.J. Taylor, V. Taylor, R. Te Water Naude, N. Thakur, Y. Themistocleous, A. Themistocleous, M. Thomas, T.M. Thomas, A. Thompson, S. Thomson-Hill, J. Tomlins, S. Tonks, J. Towner, N. Tran, J.A. Tree, A. Truby, K. Turkentine, C. Turner, N. Turner, S. Turner, T. Tuthill, M. Ulaszewska, R. Varughese, N. Van Doremalen, K. Veighey, M.K. Verheul, I. Vichos, E. Vitale, L. Walker, M.E.E. Watson, B. Welham, J. Wheat, C. White, R. White, A.T. Worth, D. Wright, S. Wright, X.L. Yao, Y. Yau, Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial, The Lancet 396(10249) (2020) 467-478. [DOI] [PMC free article] [PubMed]
- 381.Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., Li J.-X., Yang B.-F., Wang L., Wang W.-J., Wu S.-P., Wang Z., Wu X.-H., Xu J.-J., Zhang Z., Jia S.-Y., Wang B.-S., Hu Y., Liu J.-J., Zhang J., Qian X.-A., Li Q., Pan H.-X., Jiang H.-D., Deng P., Gou J.-B., Wang X.-W., Wang X.-H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. The Lancet. 2020;396(10249):479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.-H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Caskey M., Robbiani D.F., Nussenzweig M.C., Rice C.M., Hatziioannou T., Bieniasz P.D. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217(11) doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R., Tan Y.-J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.-F. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 384.Yuan M., Liu H., Wu N.C., Lee C.-C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., Rogers T.F., Landais E., Sok D., Jardine J.G., Burton D.R., Wilson I.A. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020:eabd2321. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., Turner J.S., Schmitz A.J., Lei T., Shrihari S., Keeler S.P., Fremont D.H., Greco S., McCray P.B., Perlman S., Holtzman M.J., Ellebedy A.H., Diamond M.S. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182(3):744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Jiang R.-D., Liu M.-Q., Chen Y., Shan C., Zhou Y.-W., Shen X.-R., Li Q., Zhang L., Zhu Y., Si H.-R., Wang Q., Min J., Wang X., Zhang W., Li B., Zhang H.-J., Baric R.S., Zhou P., Yang X.-L., Shi Z.-L. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182(1):P50–P85. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Ostrowski L.E., Hutchins J.R., Zakel K., O'Neal W.K. Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol. Ther. 2003;8(4):637–645. doi: 10.1016/s1525-0016(03)00221-1. [DOI] [PubMed] [Google Scholar]
- 388.Dinnon K.H., Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Hou Y.J., Adams L.E., Gully K.L., Brown A.J., Huang E., Bryant M.D., Choong I.C., Glenn J.S., Gralinski L.E., Sheahan T.P., Baric R.S. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020 doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Menachery V.D., Gralinski L.E., Baric R.S., Ferris M.T. New metrics for evaluating viral respiratory pathogenesis. PLoS ONE. 2015;10(6):e0131451. doi: 10.1371/journal.pone.0131451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Sun S.H., Chen Q., Gu H.J., Yang G., Wang Y.X., Huang X.Y., Liu S.S., Zhang N.N., Li X.F., Xiong R., Guo Y., Deng Y.Q., Huang W.J., Liu Q., Liu Q.M., Shen Y.L., Zhou Y., Yang X., Zhao T.Y., Fan C.F., Zhou Y.S., Qin C.F., Wang Y.C. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28(1):124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao L., Liu P., Zhao L., Ye F., Wang H., Zhou W., Zhu N., Zhen W., Yu H., Zhang X., Guo L., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu H., Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q., Wu G., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 392.Zhang Y.N., Li X.D., Zhang Z.R., Zhang H.Q., Li N., Liu J., Li J.Q., Zhang H.J., Wang Z.J., Shen S., Shi Z.L., Wei H.P., Yuan Z.M., Ye H.Q., Zhang B. A mouse model for SARS-CoV-2 infection by exogenous delivery of hACE2 using alphavirus replicon particles. Cell Res. 2020:1–3. doi: 10.1038/s41422-020-00405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., Ritter J.H., Kang L.-I., Dort S., Robichaud A., Head R., Holtzman M.J., Diamond M.S. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020 doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.McCray P.B., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81(2):813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Sun J., Zhuang Z., Zheng J., Li K., Wong R.L.-Y., Liu D., Huang J., He J., Zhu A., Zhao J., Li X., Xi Y., Chen R., Alshukairi A.N., Chen Z., Zhang Z., Chen C., Huang X., Li F., Lai X., Chen D., Wen L., Zhuo J., Zhang Y., Wang Y., Huang S., Dai J., Shi Y., Zheng K., Leidinger M.R., Chen J., Li Y., Zhong N., Meyerholz D.K., McCray P.B., Perlman S., Zhao J. Generation of a broadly useful model for COVID-19 pathogenesis. Vaccination Treat. Cell. 2020;182(3):734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Wakeham J., Wang J., Xing Z. Genetically determined disparate innate and adaptive cell-mediated immune responses to pulmonary Mycobacterium bovis BCG infection in C57BL/6 and BALB/c mice. Infect. Immun. 2000;68(12):6946–6953. doi: 10.1128/iai.68.12.6946-6953.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Morokata T., Ishikawa J., Ida K., Yamada T. C57BL/6 mice are more susceptible to antigen-induced pulmonary eosinophilia than BALB/c mice, irrespective of systemic T helper 1/T helper 2 responses. Immunology. 1999;98(3):345–351. doi: 10.1046/j.1365-2567.1999.00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Preston J.A., Beagley K.W., Gibson P.G., Hansbro P.M. Genetic background affects susceptibility in nonfatal pneumococcal bronchopneumonia. Eur. Respir. J. 2004;23(2):224–231. doi: 10.1183/09031936.03.00081403. [DOI] [PubMed] [Google Scholar]
- 399.Fransen F., Zagato E., Mazzini E., Fosso B., Manzari C., El Aidy S., Chiavelli A., D'Erchia A.M., Sethi M.K., Pabst O., Marzano M., Moretti S., Romani L., Penna G., Pesole G., Rescigno M. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity. 2015;43(3):527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 400.T.D. Brumeanu, P. Vir, S. Shashikumar, A.F. Karim, S. Kar, K.K. Chung, K.P. Pratt, S. Casares, A Human-Immune-System mouse model for COVID-19 research (DRAGA mouse: HLA-A2.HLA-DR4.Rag1KO.IL-2Rγc KO.NOD), bioRxiv (2020).
- 401.Kumar V., Harjai K., Chhibber S. A combination of thalidomide and augmentin protects BALB/c mice suffering from Klebsiella pneumoniae B5055-Induced Sepsis. J. Chemother. 2009;21(2):159–164. doi: 10.1179/joc.2009.21.2.159. [DOI] [PubMed] [Google Scholar]
- 402.Kumar V., Chhibber S. Anti-inflammatory effect of thalidomide alone or in combination with augmentin in Klebsiella pneumoniae B5055 induced acute lung infection in BALB/c mice. Eur. J. Pharmacol. 2008;592(1–3):146–150. doi: 10.1016/j.ejphar.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 403.Kumar V., Harjai K., Chhibber S. Thalidomide treatment modulates macrophage pro-inflammatory function and cytokine levels in Klebsiella pneumoniae B5055 induced pneumonia in BALB/c mice. Int. Immunopharmacol. 2010;10(7):777–783. doi: 10.1016/j.intimp.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 404.Chen C., Qi F., Shi K., Li Y., Li J., Chen Y., Pan J., Zhou T., Lin X., Zhang J., Luo Y., Li X., Xia J. Thalidomide combined with low-dose short-term glucocorticoid in the treatment of critical Coronavirus Disease 2019. Clin. Transl. Med. 2020 doi: 10.1002/ctm2.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Rodrigues-Diez R.R., Tejera-Muñoz A., Marquez-Exposito L., Rayego-Mateos S., Sanchez L.S., Marchant V., Santamaria L.T., Ramos A.M., Ortiz A., Egido J., Ruiz-Ortega M. Statins: could an old friend help the fight against COVID-19? Br. J. Pharmacol. 2020 doi: 10.1111/bph.15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Zhang X.-J., Qin J.-J., Cheng X., Shen L., Zhao Y.-C., Yuan Y., Lei F., Chen M.-M., Yang H., Bai L., Song X., Lin L., Xia M., Zhou F., Zhou J., She Z.-G., Zhu L., Ma X., Xu Q., Ye P., Chen G., Liu L., Mao W., Yan Y., Xiao B., Lu Z., Peng G., Liu M., Yang J., Yang L., Zhang C., Lu H., Xia X., Wang D., Liao X., Wei X., Zhang B.-H., Zhang X., Yang J., Zhao G.-N., Zhang P., Liu P.P., Loomba R., Ji Y.-X., Xia J., Wang Y., Cai J., Guo J., Li H. In-hospital use of statins is associated with a reduced risk of mortality among individuals with COVID-19. Cell Metab. 2020;32(2):176–187.e4. doi: 10.1016/j.cmet.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.