Abstract
Purpose
The current study aims to determine the molecular mechanisms of diabetic retinopathy (DR) using the protein-protein interactome and metabolome map. We examined the protein network of novel biomarkers of DR for direct (physical) and indirect (functional) interactions using clinical target proteins in different models.
Methods
We used proteomic tools including 2-dimensional gel electrophoresis, mass spectrometry analysis, and database search for biomarker identification using in vivo murine and human model of diabetic retinopathy and in vitro model of oxidative stress. For the protein interactome and metabolome mapping, various bioinformatic tools that include STRING and OmicsNet were used.
Results
We uncovered new diabetic biomarkers including prohibitin (PHB), dynamin 1, microtubule-actin crosslinking factor 1, Toll-like receptor (TLR 7), complement activation, as well as hypothetical proteins that include a disintegrin and metalloproteinase (ADAM18), vimentin III, and calcium-binding C2 domain-containing phospholipid-binding switch (CAC2PBS) using a proteomic approach. Proteome networks of protein interactions with diabetic biomarkers were established using known DR-related proteome data. DR metabolites were interconnected to establish the metabolome map. Our results showed that mitochondrial protein interactions were changed during hyperglycemic conditions in the streptozotocin-treated murine model and diabetic human tissue.
Conclusions
Our interactome mapping suggests that mitochondrial dysfunction could be tightly linked to various phases of DR pathogenesis including altered visual cycle, cytoskeletal remodeling, altered lipid concentration, inflammation, PHB depletion, tubulin phosphorylation, and altered energy metabolism. The protein-metabolite interactions in the current network demonstrate the etiology of retinal degeneration and suggest the potential therapeutic approach to treat DR.
Electronic supplementary material
The online version of this article (10.1007/s40200-020-00570-9) contains supplementary material, which is available to authorized users.
Keywords: Diabetic retinopathy, Interactome, Metabolome, Prohibitin, Mitochondria, Tubulin, Proteomics, ADAM18, CAC2PBS
Introduction
Diabetic retinopathy (DR) is the complication of diabetes mainly diagnosed as the pathological alterations in the microvasculature of the retina. DR mechanisms include abnormal vascular capillaries due to blood vessel leakage, blockade, and the development of new, fragile blood vessels [1–5]. The impaired vascular capillary results in bleeding, fluid leaking, inflammation, and apoptosis that would eventually lead to photoreceptor cell death. Before the vascular symptom, complicated pathological changes occur at the molecular, cellular, and tissue levels during the progression of diabetic retinopathy. For example, early DR stage is associated with synaptic protein changes, neurodegeneration, retinal pigment epithelium (RPE) apoptosis, and thinning of the inner nuclear layer [6–8]. DR involves mitochondrial dysfunction, increased oxidative stress, increased advanced glycation end product, and apoptosis of photoreceptor cells [9–12]. Currently, clinical detection of these changes is challenging and the molecular mechanism of DR is not fully elucidated [5, 13, 14].
DR pathogenesis involves activation of the polyol pathway, excessive generation of reactive oxygen species, VEGF activation, neovascularization, and blood-retinal barrier (BRB) damage [15–18]. However, there has been no comprehensive understanding of DR pathogenesis on the molecular level, where information on mitochondrial dysfunction, metabolite interactions, and the crosstalk between metabolome-proteome is lacking. We hypothesize that molecular mechanisms of DR could be revealed using the DR-specific protein-protein interactome and metabolome mapping through all known proteome in diabetes. Previously, we have studied apoptotic signaling in the retina and retinal pigment epithelium under oxidative stress using proteomic tools [19–33]. We built the protein interactome based on phosphoproteomics experiments to map the molecular interactions in age-related macular degeneration (AMD) [25, 26, 32, 33].
In this study, we determined the DR interactome and metabolome map using the proteomic and bioinformatic tools to unveil molecular mechanisms including amino acids synthesis, energy balance, retinoid binding, insulin secretion, and insulin resistance. Using the metabolome map, we determined whether specific amino acids, carbohydrate, ATP/ADP, pH, and glycolysis/citric acid cycles are the distinctive components in the DR mechanism.
Our protein interactome suggests that DR mechanisms could be connected to the altered visual cycle, mitochondrial dysfunction, inflammation, altered lipid concentration, altered energy metabolism, tubulin phosphorylation, and specific DR biomarkers including prohibitin. The interactome suggests that new diabetic biomarkers including a disintegrin and metalloproteinase (ADAM18), calcium-binding C2 domain-containing phospholipid-binding switch (CAC2PBS), vimentin variant (vimentin III), tubulin, and prohibitin, may employ the anti-apoptotic or tumor suppressor effects through changes in their expression levels as well as post-translational modifications. Further, the whole protein interactions were divided into different subgroups based on their functional roles by the clustering algorithm. The obtained clusters showed the networking with the rhodopsin cycle, angiogenesis, lipid metabolism, and apoptotic pathways. The protein-protein interaction (PPI) network dissects the molecular mechanism of the multifactorial complex DR to understand retinal degeneration on the molecular level and eventually provides a therapeutic guide for DR treatment.
Materials and methods
In vitro study of prohibitin signaling in DR pathogenesis
For in vitro experiments, retinal pigment epithelial cells (ARPE-19) were obtained from ATCC (Manassas, VA). Retinal progenitor cells were generously donated by Dr. Harold J. Sheeldo at the University of North Texas. ARPE-19 cells were cultured in a 5% CO2 incubator at 37 °C in 100 mm dishes (Nalge Nunc International, Naperville, IL) using Dulbecco’s modified Eagle’s medium (DMEM) with fetal bovine serum (10%) and penicillin/streptomycin (1%). Confluent cells were trypsinized (5–7 min at 37 °C) using a trypsin-EDTA buffer (0.1%), then centrifugation at 300 × g for 7 min. Cells (eight to nine passages) were grown to confluence for 2–4 days and then were treated with H2O2 (200 µM), intense light (7000–10,000 lx, 1–24 h), or constant light (48 h, 700 lx). Cells were washed using Modified Dulbecco’s PBS and lysed by IP lysis buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, and protease inhibitor cocktail (0 oC, pH 7.4, 5 min) with periodic sonication (3 × 5 min), and centrifugation (10 min, 13,000 × g). The proteins were equilibrated with Laemmli sample buffer (5X, 5% β-mercaptoethanol), separated using SDS-PAGE, and visualized using Coomassie blue (Pierce, IL) or silver staining.
In vivo study of prohibitin signaling in DR pathogenesis
Male Wistar rats (250–350 g/rat, eight weeks-old, Charles River Laboratories) were randomly assigned to control or diabetic groups. All procedures were in agreement with the EU Directive 2010/63/EU, NIH guideline, and ARVO guideline for animal experiments. Diabetes was introduced with an injection (IP) of streptozotocin (STZ in 10 mM sodium citrate, 65 mg/kg, pH 4.5, Sigma, St. Louis, MO, USA). Blood glucose levels exceeding 250 mg/dl were considered as hyperglycemic and confirmed two days after STZ injection with a glucometer (Elite, Bayer, Portugal). Before euthanization, blood was collected for glucose analysis and animals were weighed. Age-matched controls and diabetic rats were anesthetized with halothane and sacrificed, 2 and 8 weeks after the onset of diabetes.
Retinal protein extraction
The diabetic and age-matched control eyes were enucleated and placed in phosphate-buffered saline (2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, pH 7.4, 4 ºC). Retinas were dissected and lysed in RIPA buffer (0.1% SDS, 1 mM DTT, 50 mM Tris–HCl, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 0.5% DOC, pH 7.4) supplemented with complete protease inhibitor cocktail tablets and phosphatase inhibitors (10 mM NaF and 1 mM Na3VO4). Protein extracts were sonicated, centrifuged, and collected in the supernatant (10 min, 16,000 x g, 4 ºC). All proteins in the retina were analyzed using 1D and 2D SDS-PAGE, western blot, and mass spectrometry. New proteins in STZ-treated rats were further studied using database search and BLAST analysis (Table 1).
Table 1.
Proteomic analysis of diabetic retina of STZ-treated rat. Proteins were isolated and separated by 2D SDS-PAGE, followed by mass spectrometry analysis. Four hypothetical/unknown proteins were uncovered as not reported proteins previously (a-d). Gene/protein name, model, isoelectric point (pI), molecular weight, and accession number are presented.
| Protein Identified | Gene Name | Model | Nominal Mass (Da) | pl | Accession Number | |
|---|---|---|---|---|---|---|
| 1 | Adenylate cyclase type 4 | ADCY4 | Rattus norvegicus | 118799 | 8.15 | gi|117788 |
| 2 | Hypothetical/unknowna | - | Rattus norvegicus | 27756 | 5.37 | gi|149057175 |
| 3 | Myogenic regulatory factor 2 | MRF2 | Rattus norvegicus | 12630 | 9.75 | gi|159162479 |
| 4 | SH3 domain-binding protein 1 | SH3BP1 | Rattus norvegicus | 74852 | 6.16 | gi|285002227 |
| 5 | TCRB protein | TCRB | Rattus norvegicus | 35227 | 8.78 | gi|165971024 |
| 6 | Hypothetical/unknownb | ADAM18 | Rattus norvegicus | 135097 | 8.91 | gi|1820960860 |
| 7 | Hypothetical/unknownc | VIMIII | Homo sapiens | 47488 | 5.09 | gi|16552261 |
| 8 | Vinculin | VCL | Rattus norvegicus | 116615 | 5.83 | gi|157822133 |
| 9 | Prohibitin | PHB | Rattus norvegicus | 29820 | 5.57 | gi|13937353 |
| 10 | Toll-like Receptor 7 | TLR7 | Rattus norvegicus | 121299 | 6.38 | gi|124245106 |
| 11 | Hypothetical/unknownd | FAM81B | Rattus norvegicus | 48781 | 6.78 | gi|1046827298 |
| 12 | Thioredoxin doamin containing protein 17 | TXNDC17 | Rattus norvegicus | 14092 | 4.85 | gi|157786640 |
| 13 | Insulin like growth factor 2 receptor A | IGF2R | Rattus norvegicus | 174358 | 5.45 | gi|149027461 |
| 14 | Junction plakoglobin | JUP | Rattus norvegicus | 81801 | 5.75 | gi|41350891 |
| 15 | Microtubule-actin cross linking factor 1 | MACF1 | Rattus norvegicus | 830743 | 5.29 | gi|402534525 |
| 16 | Dynamin-1like protein | DNM1L | Rattus norvegicus | 83908 | 6.64 | gi|2425052 |
| 17 | E3 ubiquitin-protein ligase RNF135 | RNF135 | Rattus norvegicus | 46012 | 5.53 | gi|58865598 |
| 18 | Regulator of G-protein signalling 3 CRA_d | RGS3 | Rattus norvegicus | 68401 | 8.54 | gi|149059607 |
aPutative calcium-binding C2 phospholipid-binding switch (CAC2PBS), bA disintegrin and metalloproteinase (ADAM18), cVimentin variant III, dProtein family 81 (FAM81B)
Immunoblotting analysis
Proteins were denatured in the buffer (10% SDS, 30% glycerol, 0.5 M Tris, 0.6 M DTT, 0.012% bromophenol blue, 6x sample buffer) and heated for 5 min at 95 ºC. Proteins were separated by SDS-PAGE using ingredient gel (8–16%). Proteins were transferred into PVDF membranes (Millipore, Billerica, MA, USA) and the membrane was blocked using low-fat milk (5%) in Tris-buffered saline (TBS-T, 20 mM Tris-HCl, 137 mM NaCl, 0.1% Tween- 20, pH 7.6, 1 h, 25 oC). The membrane was incubated with primary antibody (rabbit polyclonal, Genemed Synthesis, San Antonio, TX) overnight at 4 ºC. The membrane was washed (TBS-T, 0.5% low-fat milk, 1 h) and incubated with the anti-mouse or anti-goat alkaline phosphatase-linked IgG secondary antibody (1:10,000, GE Healthcare, Buckinghamshire, UK, or 1:10,000 dilution, anti-rabbit, Agrisera, Vännäs, Sweden) in TBS-T (1% low-fat milk, 1 h, 25 oC). The membrane was washed (TBS-T, 0.5% low-fat milk, 1 h) and proteins were visualized using the enhanced chemifluorescence (ECF, GE Healthcare). Proteins were detected using an imaging system (Typhoon FLA 9000, GE Healthcare) and analyzed quantitatively by Image Quant 5.0 software (Molecular Dynamics, Inc., Sunnyvale. CA, USA). The membrane was reprobed for β-actin immunoreactivity (1:5,000, Sigma), β-III tubulin (1: 5,000, Covance), and an HRP-conjugated secondary antibody (1:7,000, anti-mouse, Santa Cruz Biotechnology, Santa Cruz, CA) as loading/positive controls.
Immunocytochemistry
Retinal cells were rinsed using the PBS buffer and fixed (4% sucrose, 4% PFA, 10 min, 25 oC) and permeabilized (PBS, 1% Triton X-100, 10 min). Cells were incubated with the buffer (5% FBS, 0.2% Tween-20 PBS, 20 min) to prevent non-specific antibody binding. Cells were incubated with the primary antibody (2 h, 25 oC), rinsed (PBS), and incubated with the secondary antibody for 1 h. The cell nuclei were visualized using DAPI staining (1:5,000). The coverslips were mounted on glass slides using Dako Fluorescence mounting medium (Dako, Denmark). Cells were visualized using a laser scanning confocal microscope LSM 710 META (Zeiss, Germany) and were analyzed quantitatively using ImageJ software (version 1.53a).
Mass spectrometry analysis
The excised piece from protein bands/spots (1D/2D) were cut into 1 mm2 cubes. The Coomassie-stained or silver-stained gel pieces were incubated using a Coomassie destaining buffer (200 µL of 50% MeCN in 25 mM NH4HCO3, pH 8.0, 20 min, 25 oC) or silver destaining buffer containing 30 mM potassium ferricyanide (50%) and 100 mM sodium thiosulfate (50%). The gels were dehydrated using acetonitrile (200 µL) and dried in vacuum (Speed Vac, Savant, Holbrook, NY). Proteins were reduced (100 mM NH4HCO3, 10 mM DTT, 56 ºC, 30 min), alkylated (100 mM NH4HCO3, 55 mM iodoacetamide, 20 min, 25 oC, dark) and digested (13 ng/µL sequencing-grade trypsin, Promega, 10 mM NH4HCO3, 10% MeCN, 37 oC, overnight).
The peptides were enriched using a buffer (50 µL of 50% MeCN in NH4HCO3, 5% formic acid, 20 min, 37 ºC). Dried peptides were dissolved in a mass spectrometry sample buffer (5–10 µL, 75% MeCN in NH4HCO3, 1% trifluoroacetic acid). The organic matrix (alpha-cyano-4-hydroxycinnamic acid, 5 mg/mL, MW 189.04, Sigma-Aldrich, St. Louis, MO) was freshly dissolved in a matrix buffer (50% MeCN, 50% NH4HCO3, 1% trifluoroacetic acid) and centrifuged (13,000 x g, 5 min). The matrix-peptides (0.5 µL) were spotted onto the MALDI plate (Ground steel, Bruker Daltonics, Germany). The peptide mass was analyzed in 800–3000 Da (Flex MALDI-TOF mass spectrometer, Bruker Daltonics, Germany) using 70–75% laser intensity and 100–300 shots. All spectra were calibrated using trypsin peptides (842.5099, 2211.105 Da) and were analyzed using Flex analysis software. Peptides were identified using the Mascot software (Matrix Science) and proteins were searched using NCBI/SwissProt database by the following criteria: 50–300 ppm mass tolerance, zero missed cleavage, carbamidomethyl cysteine, methionine oxidation, and 4 minimum peptide match. Protein identification was validated based on protein sequence coverage, a number of matched peptides, MOWSE score, and tandem mass amino acid sequence. MOWSE score is expressed as -10logP as a probability value to compute the composite probability P.
Cholesterol treatment in vitro
Cholesterol (Matheson Coleman & Bell) stock solution was prepared in 95% ethanol to a final concentration of 10 mg/ml. β-cyclodextrin (Sigma) was dissolved in media and filtered before applied to cells. While applied to cells, all the chemicals were dissolved in FBS free media, and cells were incubated for 12 h with fresh media before the treatment.
Construction of PPI network
We identified more than 1000 proteins as proteins in the retina, RPE, and vitreous (Table 2). The primary dataset for normal proteins in the eye was collected by performing a keyword search against SwissProt (http://www.expasy.ch/spot/), HPRD (http://hprd.org), and Gene Ontology (GO) database (http://www.geneontology.org). When gene ontology information was unavailable, we used TargetP (http://www.cbs.dtu.dk/service/TargetP/) and Psort (http://psort.ims.u-tokyo.ac.jp/) algorithms that predict the subcellular localization of a given protein in the retina to filter out false positives. Only proteins that were predicted to be retinal proteins by all algorithms were included as the positive control list.
The interactome was established using PPI software STRING 11. (http//:string-db.org/) by adding 1000 identified normal retinal proteins in the query. Protein interactions were presented using eight categories including neighborhood (green), gene fusion (red), co-occurrence (dark blue), co-expression (black), binding experiment (purple), database (blue), text mining (lime), and homology (cyan). Proteins in the retina were validated using the databases (https://sph.uth.edu/retnet/; RPGeNet 2.0; RD5000 DB; RETINOBASE) as well as the cross-search from the PubMed database and Google Scholar in January 2019 for the terms “proteins in the eye”, “proteomics, retina”, and “retina proteins” in the title, abstract, and keywords. Retinal proteins were collected based on their accession number, gene name, protein name, and type of model as selection criteria. All proteins in the retina, RPE, and the vitreous were validated using the Uniprot Knowledgebase (UniProtKB).
For diabetic retinopathy proteins, we collected > 500 interacting proteins that are identified in the diabetic retinopathy pathogenesis (Table 3). The proteins were searched using the Database of Interacting Proteins (DIP), Human Protein Reference Database (HPRD), European Molecular Biology Laboratory (EMBL)-European Bioinformatics Institute (EBI) and protein sequences of these genes were obtained using FASTA (protein and DNA sequence alignment software package) from NCBI (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/). The PubMed database and Google scholar were used for validation of retinal proteins and proteomic study in the retina.
For metabolomics study, all known metabolites in DR were analyzed using bioinformatics tools including OmicsNet software (https://www.omicsnet.ca/). OmicsNet is a visual network system to create a network relationship among genes, proteins, miRNAs, metabolites, or transcription factors in three-dimensional (3D) space. A list of molecules in DR was obtained via OmicsNet. More than 100 metabolites in DR were connected in purine metabolism, nitrogen metabolism, aerobic glycolysis, mitochondrial metabolites, and the TCA cycle (Table 4).
The metabolites were validated using their KEGG (Kyoto Encyclopedia of Genes and Genomes) and PubChem identification number (https://pubchem.ncbi.nlm.nih.gov/rest/pug). The metabolites were inserted into the OmicsNet.ca software to generate a comprehensive metabolome map. For the detailed mechanistic study, DR-specific 121 metabolites were selected and mapped. The metabolome map was used to validate our protein interactome to uncover any hidden interactions and molecular mechanisms. Further, the proteins obtained from the metabolome map were used to generate the DR metabolite-protein interactome map reversely to elucidate metabolite-related protein network using STRING software.
DR interactome and metabolome map
STRING (http://string-db.org/) was used to establish the protein-protein interaction map. The interactions of Homo sapiens were selected and the interaction network was constructed using the ‘‘add more interactors’’ option in the STRING database. STRING used various protein-interacting tools including physical protein interaction, co-expression, gene fusion, neighborhood, co-occurrence, databases, and text mining. The interactome map was a comprehensive network of protein interactions associated with DR generated from all available sources including physical interactions and pathway databases. The search strategy involved the terms “diabetic retinopathy,” “diabetic eye disease,” “metabolomics,” “metabonomics,” “metabolites,” “mass spectroscopy,” and “proteomics”. All metabolomics literature was searched, reviewed, and relevant metabolites were presented in Table 4.
Assigning accession number
An accession number is a unique identifier given to a DNA or protein sequence record to allow for tracking of different versions in a single data repository. In the current study, the GI number of the nucleotide sequence was assigned in the tables.
Statistical analysis
Values are presented as the mean of three independent experiments. Two-group comparisons were analyzed by the 2-tailed t-test. Multiple comparisons were evaluated by ANOVA and Tukey or Dunnet tests, as appropriate. P < 0.05 is considered statistically significant.
Results
New biomarkers in DR pathogenesis
To determine whether early biomarkers in hyperglycemic models play crucial roles in the progression of DR, we introduced a system-wide, unbiased, and high-throughput approach to examine protein changes using a proteomic tool and protein interaction mapping. Proteomic approaches, including retinal protein collection from STZ-induced diabetic rats, 2D gel electrophoresis, and protein identification by mass spectrometry, were used to determine protein identification and modifications in DR.
New DR-related biomarkers found in our proteomic experiments are presented in Table 1. Four hypothetical proteins were further analyzed using bioinformatics tools including BLAST search and post-translational modification study. Four unknown proteins were calcium-binding C2 domain-containing phospholipid-binding switch (CAC2PBS), a disintegrin and metalloproteinase (ADAM18), vimentin variant III, and FAM81B. In addition to these hypothetical proteins identified by mass spectrometry analysis, our proteomics data suggested that new biomarkers of DR include prohibitin (PHB), vinculin (VCL), tubulin beta III (TUBB3), E3 ubiquitin-protein ligase (RNF135), microtubule-actin crosslinking factor 1 (MACF1), TCRB protein (TCRB), toll-like receptor 7 precursor (TLR7), insulin-like growth factor 2 receptor (IGF2R), and myogenic regulatory factor 2 (MRF2). The identities of DR-specific proteins were presented by accession number, pI value, nominal mass, and their gene name (Table 1).
DR specific interactome map
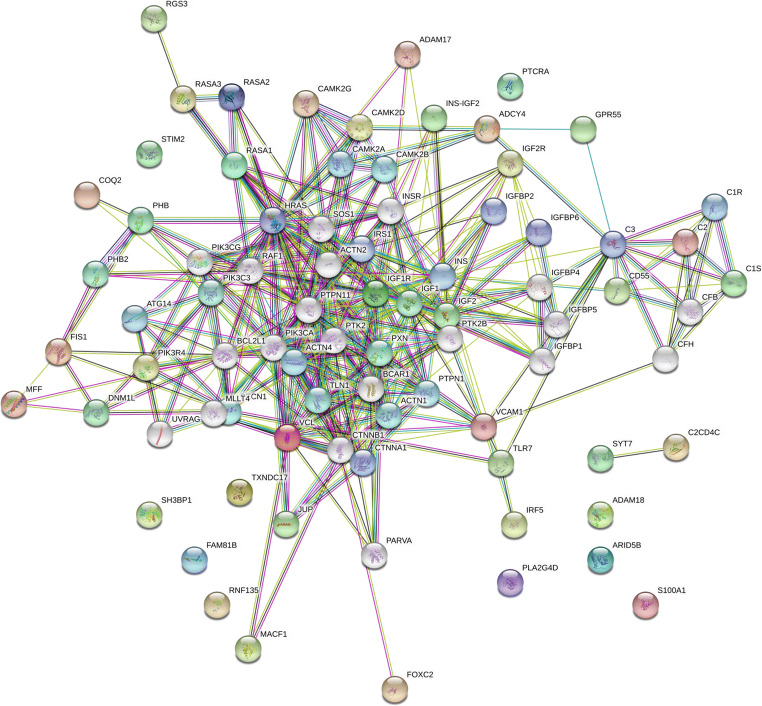
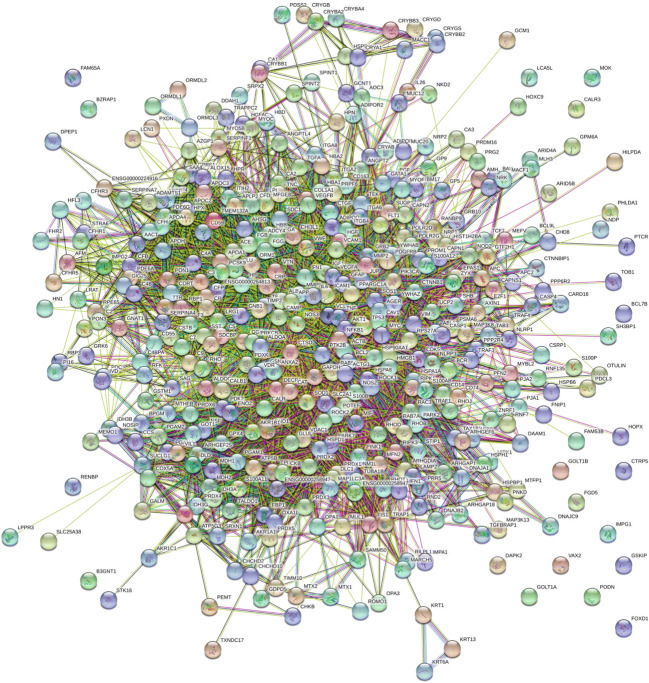
DR specific protein interactome map was established using new biomarkers uncovered in the current study (ADAM18, FAM81B, PHB, TXNDC17, ADCY4, VCL, DNM1L, RNF135, CAC2PBS, SH3BP1, MRF2, TLR7, TCRB, MACF1, JUP, and IGF2). These DR biomarkers may provide a foundation to determine potential target proteins implicated in DR pathogenesis. The DR specific protein interactome was connected using proteomics data from DR tissues in vivo murine models. Our initial interactome map using new biomarkers suggests that mitochondrial protein trafficking, crystalline aggregation, ubiquitin-based protein degradation, and complement activation network could occur in DR pathogenesis as presented in Fig. 1. The interactome suggests the new candidates including ADAM17 and mechanisms in DR including mitochondrial dysfunction, oxidative stress, cytoskeletal remodeling by microtubule, actin filament, intermediate filament (vimentin), and inflammation switch (complement activation).
Fig. 1.
The interactome of diabetic retinopathy specific proteins. DR specific proteins in vivo were determined using proteomic tools including 2D SDS-PAGE and mass spectrometry. DR specific protein interactome was established using protein-protein interaction (PPI) map software STRING 11.0 (http://string-db.org/) by adding newly identified 20 DR specific proteins in the query. Protein interactions were presented using eight categories, including homology (cyan), neighborhood (green), co-occurrence (dark blue), gene fusion (red), co-expression (black), binding experiments (purple), databases (blue), and text mining (lime). DR specific interactome was determined by genomic context, high-throughput experiments, co-expression, and previous publications in PubMed
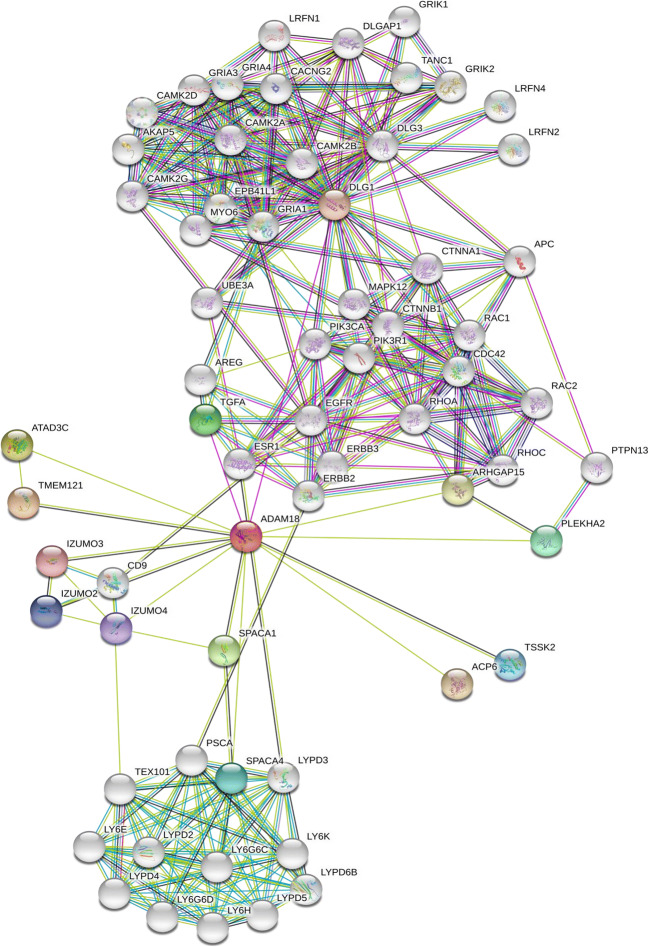
The interactions of ADAM18 as a new DR biomarker were further established by adding a seed ‘‘ADAM18’’ only in the query (Fig. 2) and increase their first and second shell interactions to 50 with a lower threshold. The ADAM18 interactome suggests that ADAM18 may contribute to specific dual phosphorylation signal including EGFR, PI3K, APC, RAC1/2, and RHO through the direct TGFA interaction, as well as calcium signal through calcium/calmodulin-dependent kinase IIα/β (CAMK2A/B) and calcium channel (CACNG2) as shown in Fig. 2. It is of interest that ADAM18 may interact indirectly with the PI3K catalytic subunit (PIK3CA, p110) and the regulatory subunit (PIK3R1, p85) separately through an estrogen receptor (ESR1). Phosphorylation reactions by PI3K p110, PI3K p85, EGFR are relayed to RAC1, CDC42, and RHOA molecules. ADAM18 is also related to calcium reactions by direct interaction with DLG1 that may control CAMK and a MYO protein network. ADAM18-UBE3A interaction demonstrated the ubiquitin-related pathway through ubiquitin ligase E3A.
Fig. 2.
ADAM18 interactome. One of four hypothetical proteins (ADAM18, CAC2PBS, vimentin variant III, FAM81B) was further analyzed using bioinformatics tools including BLAST and functional domain search. ADAM18 interactome was established using PPI map software STRING 11.0 (http://string-db.org/) by adding a seed (ADAM18) in the query and applied 50/50 first and second shell interactions. The ADAM18 interactome shows a functional, indirect association and direct physical interactions. ADAM18 interactions in DR are derived from eight sources, including neighborhood, gene fusion, co-occurrence, high-throughput interaction experiments, and published knowledge (PubMed). ADAM18-UBE3A interaction shows a potential ubiquitin signaling by ubiquitin ligase E3A. Our ADAM18 interactome implies that phosphorylation signaling through ADAM18 may go through ADAM18-TGFA-EGFR-RHOA for one direction, as well as ADAM18-ESR1-PI3K-RAC1-CDC42-RAC2 pathway for the other direction
Prohibitin signaling in DR pathogenesis
Previously, we tested the hypothesis of whether prohibitin (PHB) signaling could be altered in the oxidative stress model [20, 26, 29, 31]. The current experiments demonstrated that mitochondrial PHB was decreased during hyperglycemic conditions in the streptozotocin-treated murine models and diabetic human tissue. Furthermore, we examined the hypothesis that PHB depletion could lead to mitochondrial disruption using the increased/decreased cholesterol model [34, 35].
First, we examined the PHB expression in the retina of the DR models to understand oxidative damage and mitochondrial dysfunction in hyperglycemia and aging. Our results showed that the PHB level was changed during hyperglycemic conditions in the streptozotocin-treated murine model and diabetic human tissue (Fig. 3).
Fig. 3.
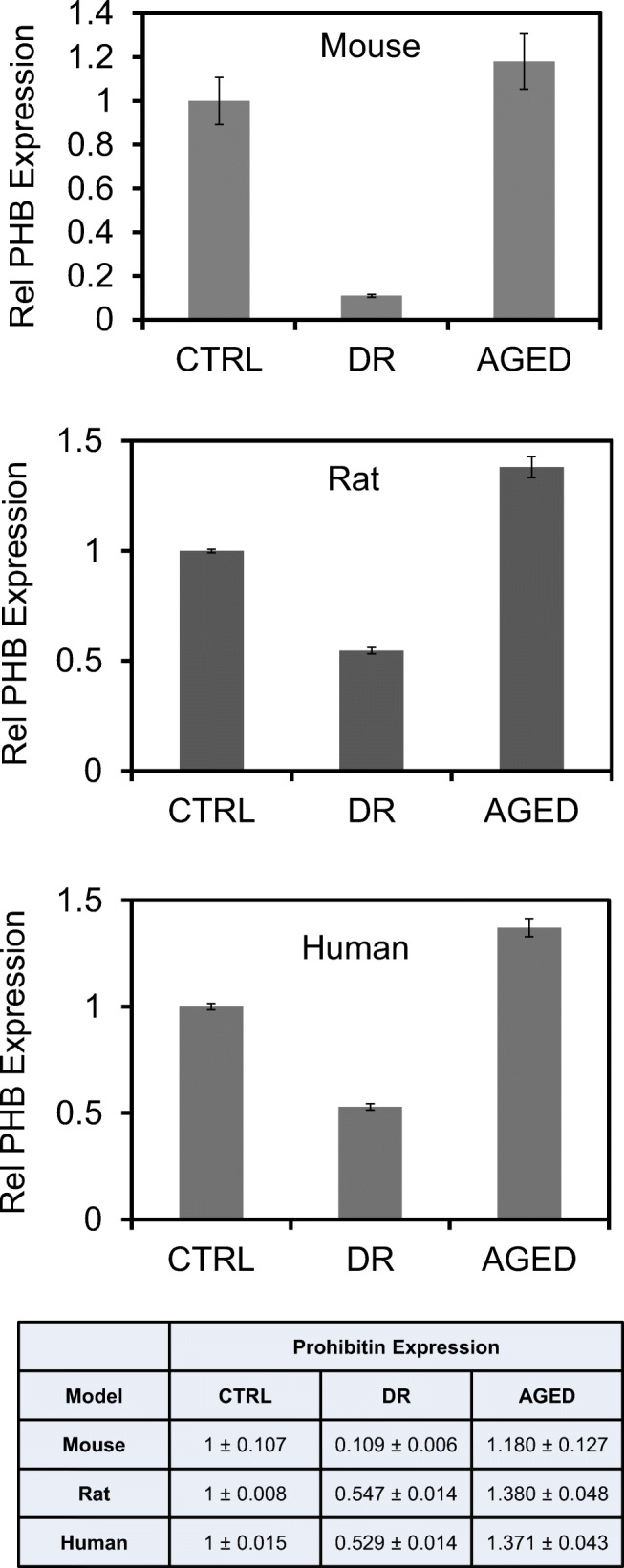
Prohibitin as the DR biomarker. Prohibitin expression in control, diabetic retinopathy (DR), and aged retina (n = 09) for biological triplicate, and technical triplicate were analyzed quantitatively using SDS-PAGE and western blotting. The beta-actin blot was used as a loading control. (A) Prohibitin expression was analyzed quantitatively in diabetic, aged, and control mouse retina. (B) Prohibitin expression was analyzed quantitatively in diabetic, aged, and control rat retina. (C) Prohibitin expression was analyzed quantitatively in diabetic retinopathy (DR), aged, and control retina in post-mortem human tissue. Statistical comparisons between means were performed by a 2-tailed t-test. A p-value of ≤ 0.05 was considered significant statistically (p > 0.05 ns, not significant; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001)
Tubulin βIII analysis in DR
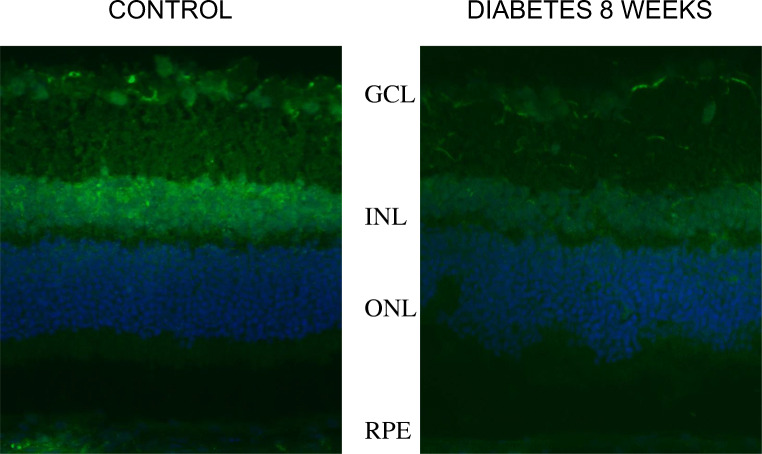
Next, we analyzed cytoskeletal proteins including actin, tubulin, and vimentin in the rat retina to determine the function of cytoskeletal polymers after 8 weeks of diabetes based on our proteomics data. Our results revealed that diabetes does not induce widespread retinal degeneration after 8 weeks, however, tubulin βIII was significantly decreased, especially in the GCL, INL, and RPE layer (Fig. 4). These alterations suggest that mitochondrial trafficking could be altered by actin, tubulin, and vimentin polymers under diabetes leading to cytoskeletal and mitochondrial remodeling [22, 32].
Fig. 4.
β-tubulin III as the DR biomarker. β-tubulin III in rat retinas was analyzed using immunohistochemistry. The ganglion cell layer (GCL)), the inner nuclear layer (INL), the outer nuclear layer (ONL), and the RPE are presented. β-tubulin III is abundant in the GCL, INL, and RPE layer. Our data revealed that diabetes does not induce widespread retinal degeneration after 8 weeks, however, the protein level of beta-tubulin III was altered significantly
Control interactome map in the retina
Next, the control retina map was established to connect more than 1000 retinal proteins using STRING 11.0 software and analyzed protein interactions involved in normal reactions/non-disease conditions in the retina (Supplement Table 2). Control proteins could be involved in inflammation, lipid metabolism, energy metabolism, retinoid binding, oxidative stress, apoptosis, transcription, and signal regulation (Fig. 5). Protein interactions of the PI3K-AKT pathway, WNT signaling, actin cytoskeleton, apoptosis, p53 pathway, VEGF pathway, and HIF-1 signaling are noticed in the control map (Fig. 5, Supplement Fig. 1A and H).
Fig. 5.
The control interactome of the retina. Normal eye proteins are presented in the interactome map. Control interactome map was established using PPI software STRING version 11.0 (http://string-db.org/) by adding over 1000 identified eye proteins in the query. The interactome shows direct physical interaction as well as indirect functional association. Physical and functional interactions include high throughput interaction experiment, genomic context, published knowledge in PubMed and MiPs, and conserved co-expression. The control map revealed the following; PI3K-AKT pathway, Wnt pathway, p53 pathway, actin cytoskeletal pathway, VEGF pathway, apoptosis, HIF-1 signaling (Supplement Fig. 1A and H)
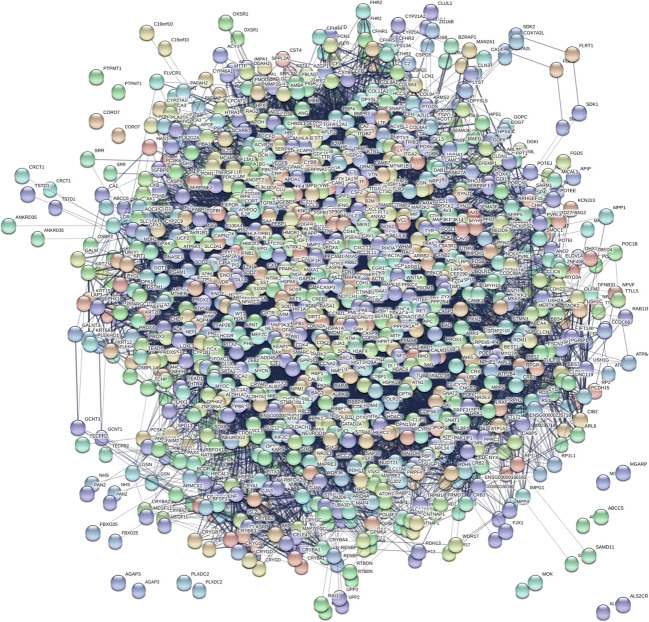
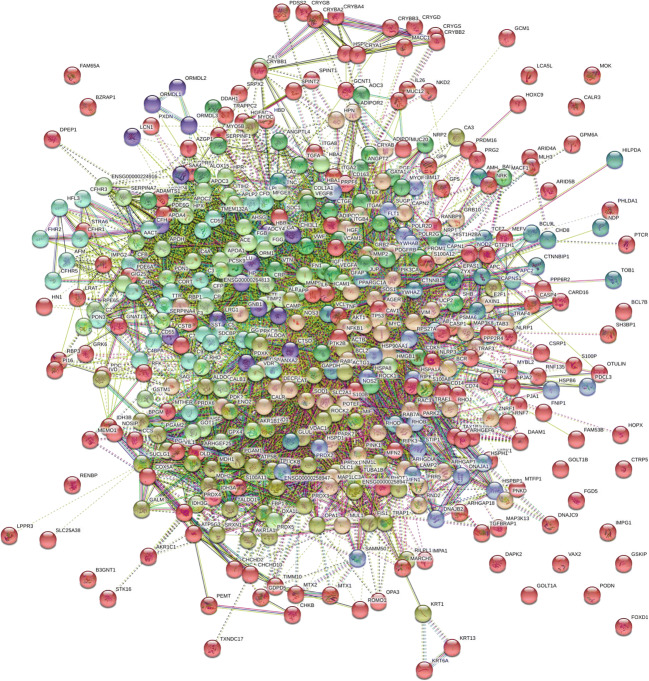
DR interactome map
The comprehensive DR interactome was established using 500 DR-specific proteins from the current study and known DR proteins using STRING software (Fig. 6, Supplement Table 3). Our results demonstrated that these interacting proteins in DR, when upregulated or downregulated under various stress conditions, may give influence to the visual cycle, energy metabolism, rhodopsin cycle, mitochondrial structure, and complement activation that can potentiate insulin depletion, apoptotic process, and oxidative stress.
Fig. 6.
The protein interactome in DR. Potential biomarkers are presented in the interaction map. Over 500 proteins found in DR were added in STRING software version 11.0 and determined protein interactions. The current interactome integrates interaction data from the above sources for a large number of organisms. The DR interactome suggests that phosphorylation signaling in DR could be initiated under hyperglycemia. Oxidative stress, mitochondria dysfunctions, inflammation, apoptosis, cytoskeletal remodeling, altered lipid metabolism, and altered energy metabolism are noticed (Supplement Fig. 2A and K)
The analysis of DR and control map interaction revealed that protein signaling in DR could be initiated from the oxidative stress biomarkers. The PPI map supported the idea that interacting proteins under oxidative stress could be involved in apoptosis in the RPE. The current study identified altered apoptosis as the major pathological pathway determined by the DR network. Further, the DR interactome could identify the clinical target of inhibition for the anti-apoptotic and anti-angiogenic networks (Fig. 6, Supplement Fig. 2A and K).
DR interactome map with nine clusters
The DR-specific proteins in Table 3 were divided into several groups based on their functional regulatory roles by k-means clustering algorithms. Grouping the protein interactions into clusters further confirmed the sub-network on their functional roles including the rhodopsin cycle, inflammation, angiogenesis, oxidative stress, lipid, energy metabolism, apoptosis, mitochondria organization, and retinoid binding (Fig. 7). The k-means algorithm was essential due to their unsupervised clustering method based on the adjacent matrix that determines which molecules go to the group of pre-specified criteria. Most of the interactors were connected by multiple lines suggesting interactions derived from more than one source of information. On segregating the network based on k-means clustering, the members within each cluster were observed to be highly interconnected, reflecting a functional association indicating crosstalk between networks.
Fig. 7.
Clustering of DR interactome network into 9 clusters. Our interactors were connected by multiple lines suggesting that multiple interactions are derived from more than one source of information
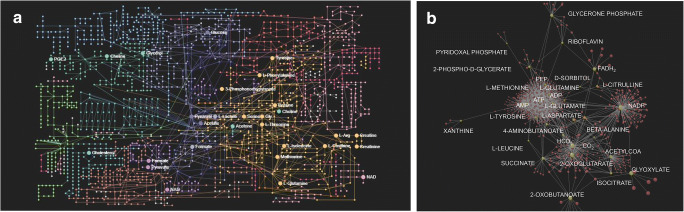
DR metabolome mapping
To confirm our DR interactome, the metabolome map of all known DR metabolites was established using various bioinformatic tools including OmicsNet software (Fig. 8A). Over 100 metabolites collected from the database and the literature and specific 121 metabolites related to DR were mapped to identify metabolites-proteome interactions (Supplement Table 4). The DR metabolome map showed that molecules were interconnected to the TCA cycle, purine metabolites, xanthine, amino acid, nitrogen metabolism, glycolysis, and mitochondrial reaction in DR.
Fig. 8.
DR metabolome mapping. A The metabolome map was established using OmicsNet software by adding 121 metabolites in the query. These metabolites carry protein interactions on their edges. Metabolites in the retina including amino acids, oligonucleotides, carbohydrates, ketones, aldehydes, amines, and lipids are shown on the map. The metabolome mapping identified low molecular weight metabolites in DR samples. B The metabolome map was dissected into the smaller-scale map for clarification. Forty-three metabolites are shown in the metabolome map. The dissected metabolome map suggests that energy balance (ATP/ADP, ADP/AMP), reducing power (NADPH/NADP+, FADH2), amino acid metabolism (methionine, tyrosine, leucine, aspartate, glutamine, citrulline, beta-alanine, 4-aminobutanoate), purine degradation (xanthine), vitamins (pyridoxal phosphate, riboflavin), glycolysis (PEP, 2-phosphoglycerate, glycerone phosphate), hyperglycemia (sorbitol), citric acid cycle (acetyl CoA, succinate, 2-oxoglutarate, glyoxylate), and pH (HCO3−, CO2) could be altered as the potential pathogenic mechanisms in DR
The metabolome map was dissected into the smaller-scale map using 43 metabolites for clarification (Fig. 8B). The second metabolome map demonstrated that potential DR mechanisms may include energy balance (ATP/ADP, ADP/AMP ratio), reducing power (NADPH/NADP+, FADH2), altered amino acid metabolism (methionine, tyrosine, leucine, aspartate, glutamine, citrulline/nitric oxide, beta-alanine, 4-aminobutanoate, glutamic acid), purine degradation (xanthine), vitamins (pyridoxal phosphate, riboflavin), glycolysis (PEP, 2-phosphoglycerate, glycerone phosphate), hyperglycemia (sorbitol), citric acid cycle (acetyl CoA, succinate, 2-oxoglutarate, glyoxylate), and cellular pH regulation (HCO3−, CO2).
Reverse interactome map from the metabolome-proteome interactions
The second DR interactome was established reversely using STRING software and proteins from the metabolome map (Supplement Table 5). Proteins were identified from the metabolome-proteome interactions in the metabolome map. The new DR interactome from the metabolome map suggests that a positive correlation may exist between the metabolites in energy balance and proteins in diabetes. Subsequently, the metabolite-protein interaction was validated again using the second DR interactome and the metabolome-proteome analysis suggesting that direct and indirect networks exist between DR-related metabolites and specific proteins in the retina (Fig. 9).
Fig. 9.
The second interactome network obtained from the metabolome map. Proteins obtained from the metabolome map were added in STRING software and determined the protein interactions to confirm the original DR interactome. The metabolites were specific amino acids, carbohydrates, ATP/ADP/AMP, and lipids that are closely related to insulin resistance and secretion. The second interactome confirmed the first DR interactome and suggests a correlation between the metabolites and DR biomarkers
Dissection of DR map and its interacting pathway in DR pathogenesis
Based on our proteomic data, DR interactome, and DR metabolome map, the potential DR mechanism was proposed. The pathway information is vital for the quantitative analysis of cellular remodeling of the biological system. Mitochondria have been implicated in the origin of DR because of the essential role in energy balance, lipid reactions, and the control of apoptotic pathways. Activation of complement pathways can initiate and accelerate apoptosis, thrombosis, and leukostasis, and these reactions are significant steps in the development of DR. Mitochondrial dysfunction, oxidative stress, altered retinoid metabolism, altered cytoskeleton, altered lipid metabolism, energy balance (ATP/ADP), angiogenesis, apoptosis, and inflammation were also implicated (Fig. 10).
Fig. 10.
Mechanistic dissection using the DR interactome. Based on our proteomic data, the potential DR mechanism is proposed as follows: (1) oxidative stress; (2) hyperglycemia; (3) mitochondrial dysfunction; (4) complement activation; (5) apoptosis; (6) angiogenesis; (7) inflammation; (8) lipid metabolism; (9) energy metabolism; (10) cytoskeletal remodeling; (11) vascular permeability; (12) altered retinoid metabolism; (13) altered phosphorylations by Tyr-dependent kinase, Ser/Thr-dependent kinase, and calcium-dependent kinase
Discussion
New biomarkers in DR
Diabetic retinopathy is recognized as a metabolic disease and the predominant cause of blindness among adults throughout the world [14]. The pathogenesis of DR is related to hyperglycemia-induced unwavering energy disturbances, inflammation, and oxidative stress [4, 15]. It is an unarguable fact that DR also occurs in some patients whose blood glucose level is well-controlled suggesting that other pathogenic factors may contribute to the phenomenon [14, 36–39]. The direct and indirect protein interactions constitute a functional association of an essential survival complex in the retina. The assembly of all known and unknown proteins, as well as the genetic association in the cell, result in a protein network of proteome-wide functional connectivity. Thus, the protein network is an ideal scaffold for data integration, visualization, and biomarker discovery. An in-depth understanding of the molecular mechanism of the disease requires knowledge of all functional integration between the expressed proteins and metabolites.
Based on our proteomics data, DR specific protein interactome map revealed the new biomarkers as hypothetical proteins that have not been reported in the database except their gene sequence in DR pathogenesis (Fig. 1; Table 1).
One of the identified hypothetical molecules in the database is calcium-binding C2 domain-containing phospholipid-binding switch protein (CAC2PBS) [40]. C2 domain proteins are mainly signal transducers including protein kinase C (single C2 domain) and membrane trafficking proteins including synaptotagmins (multiple C2 domains). C2 domains regulate the respective protein function by forming the Ca2+-dependent or Ca2+-independent phospholipid complex.
The other unknown protein II in proteomics data was identified as ADAM18 by mass spectrometry analysis and BLAST search [41–43]. ADAM18 contains a unique N-terminus sequence that has no homologous proteins found in the BLAST search using the NCBInr database, however, the gene shows the similarity as a disintegrin and metalloproteinase with thrombospondin domain (ADAMTS) [44].
The next unknown protein III was identified as vimentin variant III that encodes a type III intermediate filament [45–49]. Intermediate filaments, along with microtubules and actin microfilaments, make up the cytoskeleton. This protein is involved in neuritogenesis and cholesterol transport as well as cell attachment, migration, and phosphorylation-dependent polymerization.
Finally, the fourth unknown protein was identified as FAM81B-like protein. Not much information has been reported on this protein until recently [50, 51]. FAM81B contains a DNA binding domain and was reported as a chromosome segregation protein.
The ADAM18 interactome
The ADAM18 interactome indicates that ADAM18 phosphorylations may go through a dual-direction including ADAM18-TGFA-EGFR-RHOA as one direction, in addition to ADAM18-ESR1-PI3K-RAC1-CDC42-RAC2 pathway as the alternative direction. RAC1 is a precursor of NOX2 signaling and CDC42 is involved in insulin secretion. Overall ADAM18 network may propose a dual phosphorylation mechanism of Ser/Thr kinase-based phosphorylation (CAMK) and Tyr kinase (PI3K) signaling at the same time to increase the number of controlling reactions through specific kinases [43, 46].
The first DR interactome showed that PHB could be involved in DR progression. Decreased PHB may modulate lipid metabolism, RPE apoptosis, nucleotide binding, and diabetic signaling through lipid-binding sequence with cardiolipin [26, 29, 31]. As an oxidative stress environment, we introduced a diabetic model to compare PHB expression in hyperglycemic, aged, and normal conditions. Our experiments demonstrated that PHB in the RPE is diminished during DR pathogenesis (Fig. 3). PHB depletion leads to mitochondrial disruption and fragmentation [29, 31–33].
Our PHB-protein binding experiments suggest that PHB may change the cytoskeleton and mitochondrial arrangement by binding to actin, tubulin, and vimentin. The beta-actin is observed to be present in both control and oxidative stress implying a general role of PHB-actin interaction on mitochondrial structure maintenance. Beta-actin has a nucleotide-binding domain that controls the flow of ions across epithelial cells and also regulates cell volume. Differential expression of PHB was observed in the post-mortem retinas of DR and aged eye as well as in the retina of mice and rats in vivo. Region-specific (macular vs. periphery) and tissue-specific (retina vs. RPE) differential expressions of PHB in the eye were observed in the post-mortem tissue. Moreover, a detailed mechanism of PHB interaction with lipid, nucleotide, and protein as shown in the previous study suggests how molecular interactions of PHB-lipid/nucleotide/protein may control the apoptotic pathway in DR and promote DR progression [21, 23, 26, 29].
Beta-tubulin is situated at the crossroad of a network of cancer pathways and epigenetic regulations that are essential for cell cycle and cell division [45, 52–56]. Although the mechanism of DR is not understood entirely, proteins involved in the cell cycle are known to be involved in DR progression. Our data suggest that the cytoskeletal proteins in the rat retina may determine the function of the mitochondrial polymer after 8 weeks of diabetes based on our proteomics data.
In the interactome map, line color indicates the type of interaction evidence, whereas line thickness indicates the strength of data reliability, and line shape indicates the mode of action. The following pathways are noticed in our control interactome map in the retina: PI3K-AKT pathway, WNT signaling, regulation of actin cytoskeleton, apoptosis, p53 pathway, VEGF network, and HIF-1 network. Frequent dysregulation of PI3K-AKT signaling is implicated in diabetes that leads to the development of small-molecule inhibitors of PI3K and AKT as potential therapeutic tools toward DR.
The normal vs. the DR interactions suggest the following aspects of DR: (1) the DR interactions indicated alterations in energy metabolisms, as well as changes in tubulin phosphorylation, mitochondrial organization, ATP-regulating reactions (ATP/ADP, ADP/AMP ratio), and reducing power (NADPH/NADP+) [57–59], (2) chaperon changes including crystalline, heat shock proteins, albumin, peroxiredoxin, thioredoxin, signified up-regulated oxidative stress in DR [60–63], (3) changes of retinoid-binding proteins, including CRABP, CRALBP, RDH, RGR, RPE65, and IRBP represent potential alterations of retinoid metabolism [64–68]. Altered expression of retinoid-binding proteins may lead to perturbed visual/rhodopsin cycle and decreased retinoid concentrations which are also suggested by cytoskeletal remodeling via actin, vimentin, tubulin phosphorylation, and angiogenic reaction modulated by VEGF and EPO [69–71], (4) altered lipid metabolism that includes cardiolipin as a mitochondrial indicator of apoptosis and cholesterol as a protein aggregator [72–74], (5) imbalance of mitochondrial stoichiometry, (6) complement activation by C3, C5, C7, C9, CFB, CFH, and CFI [16, 75]. This was observed via vitronectin and clusterin interactions, (7) cytoskeletal remodeling by microtubules, actin filament, and intermediate filament (VIM, ACT and TUBB), (8) apoptotic molecules including BAX, BAK, BAD, BCL2, pJAK2, pSTAT3, TNF, PRDX5, CLU [76–78], (9) induced inflammation markers (collagen, vitronectin, MIF, CD14, PSMA6, APOA1).
Further analysis of the interactome by grouping into 9 clusters shows indirect connections and crosstalk between specific pathways and networks. The clustering of the interactome revealed the interconnectivity between the visual cycle, inflammatory response, energy metabolism, and angiogenesis as well as apoptotic pathways.
The metabolome map demonstrated that the DR-specific metabolites including amino acid, oligonucleotides, carbohydrates, ketones, aldehydes, amines, and lipids in the retina are connected through protein bindings, which are closely related to insulin resistance and secretion.
Our metabolomics data revealed various small molecules including ATP/ADP, ADP/AMP, NADPH/NADP+, FADH2, methionine, tyrosine, leucine, aspartate, glutamine, citrulline, 4-aminobutanoate, xanthine, pyridoxal phosphate, riboflavin, PEP, 2-phosphoglycerate, glycerone phosphate, sorbitol, acetyl CoA, succinate, 2-oxoglutarate, glyoxylate, HCO3−, and CO2 as the distinctive metabolites in DR [63, 79].
Retina shows a high metabolism rate, oxygen consumption, and is considered to be particularly susceptible to hyperglycemia-related oxidative stress [80, 81]. We postulate that impairment in the metabolism of purine, arginine, methionine, tyrosine, leucine, aspartate, and glutamine, and glutamic acid as metabolic dysregulation associated with DR based on our metabolome map. The analysis of DR interactome using proteome-metabolome network reveals that there is a positive correlation between diverse phosphorylations (ATP/ADP/AMP ratio vs. Ser/Thr Kinase and Tyr kinase) and DR progression.
DR related proteins and signaling pathways in the interactome
In the diabetic retina, cells experience high glucose concentration inducing the overproduction of metabolism-related proteins while enhancing glycation and the accumulation of advanced glycation end products (AGEs) [82–85].
Under diabetic conditions, inactivation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), an enzyme that catalyzes the conversion of glyceraldehyde 3-phosphate to D-glycerate 1,3-bisphosphate, is responsible for diabetic complications, accumulation of AGE, activation of protein kinase C (PKC), and increased hexosamine pathway. Other investigations indicate that GAPDH may also be involved in glucose-induced apoptosis through translocation to the nucleus in retina Müller cells [86, 87].
The accumulation of sorbitol is originated from aldose reductase (ALR) catalyzing NADPH-dependent reduction of glucose to sorbitol. Inhibition of ALR greatly attenuates basement membrane thickening, pericyte loss, and microaneurysms in the retinal capillary. The mechanism may involve the inhibition of VEGF, decreased expression of platelet and endothelial cell adhesion molecule-1 (PECAM1), and suppression of glial activation [88–91].
Vascular homeostasis
Vascular integrity and functionality ensure the adequate blood supply and mass transfer for normal tissue physiology and stress response. The main clinical signs in DR involve high blood pressure, capillary basement thickening, microaneurysm, hemorrhage, blood-retinal barrier break-down, and neovascularization. The microvasculature consequences and blindness are the major manifestations of the end-stage in DR.
Vascular endothelial growth factor (VEGF) regulates angiogenesis and vascular permeability [18, 37]. VEGF inhibition can limit vascular leakage and inflammation. However, not all DR patients respond to anti-VEGF therapy, indicating other VEGF independent factors and pathways may induce neovascularization during DR.
Nitric oxide synthase (NOS) regulates nitric oxide (NO) concentration in the cell [92, 93]. NO maintains vascular structure by inhibiting blood clotting to prevent vascular smooth muscle cells (SMCs) from over-proliferation. NO controls basal blood flow, vascular integrity, superoxide scavenging, and protein nitration. Inducible NOS in immune cells, typically stimulated under oxidative stress, generates a high level of NO that can be cytotoxic and pro-inflammatory. NO is angiogenic through enhancing endothelial migration and inducing reorganization of extracellular matrix (ECM) proteins. NOS uncoupling caused by an insufficient substance (oxygen and arginine) abrogates NO generation property.
Angiotensin I is catalyzed to angiotensin II by angiotensin-converting enzyme (ACE) [15, 94]. The latter mainly works as a vasopressin producer and a vasoconstrictor that mediates the narrowing of the blood vessels by muscular wall contraction. Besides, it is involved in the overproduction of reactive oxygen species (ROS) and activation of nuclear factor-kappaB (NF-κB). It also stimulates VEGF, resulting in cell hypertrophy, over-proliferation, migration, and apoptosis. Clinical studies show that angiotensin II inhibition decreased the progression of DR by 50%.
Erythropoietin (EPO) is a glycosylated cytokine and growth factor that regulates blood cell production, immune reaction, and neuroprotection [8, 19, 95]. Although EPO is beneficial for diabetic patients, to reduce blood glucose levels, limit peripheral neuropathy, maintain energy metabolism, it also has significant side effects including elevated blood pressure, excessive microvascular angiogenesis, and even tumor-inducing effect.
Plexin domain-containing 2 (PLXDC2) is a tumor endothelial marker (TEM7R) and a mitogen for neural progenitors. It is highly expressed in the endothelial cells of solid tumors. In DR, it is associated with the formation of a fibrovascular membrane and may cause retinal angiogenesis [96–98].
The stress response in DR
Many pathological pathways associated with DR result in the overproduction of ROS. The increased oxidative stress further facilitates protein turnover rate, damages nucleotides, initiates immune responses, and induces growth factors including VEGF which promotes the development of diabetic complications. Antioxidant therapies have shown a promising clinical effect in slowing down the progression of DR.
Collectin superfamily (C-lectin) participates in removing damaged lipoprotein under oxidative stress. Heat shock proteins function as protein chaperones to protect immature proteins facilitating correct folding and to assist in clathrin-mediated endocytosis. Heat shock protein family A has been identified in RPE cells under oxidative stress and ischemia-reperfusion injury.
Cell adhesion and structural proteins
Retinal tissue morphology change, infiltration of immune cells, and neovascularization are associated with extracellular matrix (ECM) and cytoskeletal proteins. Changes in ECM and matrix protease make the whole tissue mechanically labile and more accessible to immune cells. Vascular basement membrane thickening involves morphological changes and apoptosis of pericytes. When pericytes are separated from the endothelial layer, blood vessel loses its structural integrity.
Integrin (α2/β1) is a collagen receptor mediating platelet adhesion in blood vessels. Different polymorphisms of α2 integrin cause different reactivity of platelet with aggregation factors, including thrombin and adenosine diphosphate. Diabetic patients have hyper-reactive platelets and micro-vascular defects [38]. In healthy blood vessels, the endothelium layer is intact, which prevents platelet from adhering to collagens underneath. Platelet in diabetic patients can be activated by the exposed collagen at the defect site. Therefore, antiplatelet therapy has been reported to slow down the progress of DR.
At the early stage of DR, there is an increase of leukocytes in the retina, which might be responsible for retinal vascular leakage, retinal capillary nonperfusion, and local inflammation. Inhibition of ICAM-1 reduced the leukocyte number, which may potentially prevent blood-retinal barrier breakdown [99–101].
Matrix metalloproteinase (MMP) is the critical enzyme for reorganizing ECM during embryonic development, reproduction, and wound healing [102, 103]. Protein expressions of MMP-1, MMP-2, MMP-3, MMP-9, and MMP-14 are enhanced in DR, which may make the capillary basement mechanically weak. With Wnt, β-catenin can travel to cell nuclear and induce T-cell factor/lymphoid enhancing factor (TCF/LEF) signaling.
The inflammatory response in DR
Our interactome suggests that apart from vascular morbidity, DR also shows characteristics consistent with chronic inflammation. Enhanced immune cells in the retina and elevated pro-inflammatory proteins result in the structural and functional changes in retina cells.
Nuclear factor κ-B (NF-κB) is a pleiotropic transcription factor associated with host defense and inflammation [104]. High glucose and AGE can induce NF-κB in retinal pericyte to enhance pericyte apoptosis. NF-κB inhibitors show anti-apoptotic effect and ameliorate diabetic nephropathy, while NF-κB activation is pro-apoptotic with increased Bax and tumor necrosis factor-alpha (TNF-α) expression in retinal pericyte.
Interleukin-1β (IL-1β) is a pro-inflammatory cytokine that can be induced in the retina during DR. IL-1β is associated with consequences including increased levels of ICAM-1 and endothelin (ET), immune cell adhesion, local inflammation, breakdown of the blood-retina barrier, and activation of glial cells [105–107]. Other similar cytokines include IL-8, tumor necrosis factor (TNF-α), and monocyte chemoattractant protein (MCP-1) were also identified in the DR retina.
Phosphorylation signaling and gene regulation
Prolonged hyperglycemia can increase the intracellular calcium level in retinal vascular endothelial cells and PKC activity [15]. For DR, PKC pathway alteration is related to insulin resistance, activation of NADPH oxidase, increased ET-1 secretion, VEGF-induced retinal endothelial permeability, smooth muscle cell apoptosis, macrophage activation and adhesion, and NO inhibition. PKC inhibitor therapy has been demonstrated to be effective to improve the visual outcome of DR patients.
Hypoxia-inducible factor-1 (HIF-1) responses under hypoxia and regulates the expression of genes for angiogenesis, cell survival, wound healing, and glucose regulation [108, 109]. In addition to hypoxia, it can also be increased by AGEs in the retina. It binds to hypoxia-responsive elements in the VEGF promoter to upregulate VEGF expression.
Calcium/calmodulin-dependent protein kinase IV (CaMKIV) is an insulin-regulating protein [71, 110]. Insulin transcription is enhanced by an elevated intracellular calcium level, which is regulated by activating transcription factor-2 (ATF-2), interacting with the cAMP-responsive elements of insulin. With CaMKIV, this binding causes elevated insulin transcription, while with CaMK inhibitor, this enhanced effect is eliminated. Therefore, it is suggested that ATF-2 regulates insulin gene expression, which is dependent on the concentration of calcium and CaMKIV.
The current study tried to dissect the mechanism using the protein-metabolite interactome implying physiological, genetic, and metabolic reactions that can lead to DR pathogenesis.
The interactome shows new diabetic biomarkers in DR that include a disintegrin and metalloproteinase (ADAM18), calcium-binding C2 domain-containing phospholipid-binding switch (CAC2PBS), vimentin variant (vimentin III), and FAM81B as hypothetical proteins, as well as tubulin and prohibitin as the anti-apoptotic or angiogenic-related molecules. The protein interactome and metabolome in this study reveal the molecular mechanism to propose a clinical application toward DR treatment [111–117].
Electronic supplementary material
Dissection of the retina control map showing several pathways, including PI3K, WNT, cytoskeleton, apoptosis, p53, VEGF, HIF-1, and cholesterol mechanism. A. PI3K-AKT signaling, B. WNT network, C. Actin cytoskeletal network, D. Apoptosis, E. p53 network, F. VEGF pathway, G. HIF-1 network, H. Cholesterol metabolism. (PDF 5027 kb)
Mechanistic dissection of the interactome network leading to the pathogenesis of diabetic retinopathy, A. Apoptosis, B. Angiogenesis, C. Inflammation network, D. Retinoid metabolism, E. Vascular permeability/occlusion, F. Energy metabolism network, G. Lipid metabolism, H. Cytoskeleton organization, I. Mitochondria dysfunction, J. Hyperglycemia, K. Oxidative stress. (PDF 7678 kb)
Identified proteins in the retina as the control. A total of 1090 proteins were identified as biological, cellular, and molecular functional proteins which serve as the control proteins in the retina. Control interactome map was established using bioinformatics tools including PPI software STRING (http://string-db.org/) by adding 1104 identified normal eye proteins in the query. (XLSX 111 kb)
DR biomarkers. DR-specific proteins were identified from our proteomics study, protein database, and proteomics literature. These proteins were considered DR biomarkers and used in bioinformatics analysis. DR-specific interactome map was established using STRING to find the probable interacting partners of the proteins associated with DR. These potential DR biomarkers are presented in the interaction map and mechanistic dissection was presented in Supplement Figures 2A-2K. (XLSX 35 kb)
Metabolites associated with DR pathogenesis. A total of 121 metabolites were collected from the literature and databases including KEGG and PubChem. The metabolome map was established using bioinformatics tools including OmicsNet software by adding 121 metabolites in the query. (XLSX 19 kb)
Reverse interactome mapping from the metabolome map. A total of 115 proteins related to DR were identified as the DR-specific metabolome-proteome interactions from the nodes of metabolites in the metabolome map. Proteins obtained from the metabolome-proteome interactions were added in STRING software and determined the protein interactions. (XLSX 14 kb)
Funding information
The current research was supported in part by Research Assistantship and Teaching Assistantship from the American University of Nigeria.
Compliance with ethical standards
Conflict of interest
None declared.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pusparajah P, Lee LH, Kadir KA. Molecular markers of diabetic retinopathy: Potential screening tool of the future? Front Physiol. 2016;7:1–19. doi: 10.3389/fphys.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.VanGuilder HD, Bixler GV, Kutzler L, Brucklacher RM, Bronson SK, Kimball SR, Freeman WM. Multi-modal proteomic analysis of retinal protein expression alterations in a rat model of diabetic retinopathy. PLoS One. 2011;6:e16271. doi: 10.1371/journal.pone.0016271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuo JZ, Wong TY, Rotter JI, Wiggs JL. Challenges in elucidating the genetics of diabetic retinopathy. JAMA Ophthalmol. 2014;132:96–107. doi: 10.1001/jamaophthalmol.2013.5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang J, Kern TS. Inflammation in diabetic retinopathy. Prog Retin Eye Res. 2011;30:343–58. doi: 10.1016/j.preteyeres.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrovič D. Candidate genes for proliferative diabetic retinopathy. Biomed Res Int 2013;2013:1–9. 10.1155/2013/540416. [DOI] [PMC free article] [PubMed]
- 6.Ly A, Scheerer MF, Zukunft S, Muschet C, Merl J, Adamski J, Hrabě M, De Angelis S, Neschen SM, Hauck M, Ueffing Retinal proteome alterations in a mouse model of type 2 diabetes. Diabetologia. 2014;57:192–203. doi: 10.1007/s00125-013-3070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decanini A, Karunadharma PR, Nordgaard CL, Feng X, Olsen TW, Ferrington DA. Human retinal pigment epithelium proteome changes in early diabetes. Diabetologia. 2008;51:1051–61. doi: 10.1007/s00125-008-0991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah SS, Tsang SH, Mahajan VB. Erythropoetin receptor expression in the human diabetic retina. BMC Res Notes. 2009;2:234. doi: 10.1186/1756-0500-2-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caprara C, Grimm C. From oxygen to erythropoietin: relevance of hypoxia for retinal development, health and disease. Prog Retin Eye Res. 2012;31:89–119. doi: 10.1016/j.preteyeres.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Schrier SA, Falk MJ. Mitochondrial disorders and the eye. Curr Opin Ophthalmol. 2011;22:325–31. doi: 10.1097/ICU.0b013e328349419d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra M, Kowluru RA. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Investig Ophthalmol Vis Sci. 2015;56:5133–42. doi: 10.1167/iovs.15-16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajala A, Gupta VK, Anderson RE, Rajala RVS. Light activation of the insulin receptor regulates mitochondrial hexokinase. A possible mechanism of retinal neuroprotection. Mitochondrion. 2013;13:566–76. doi: 10.1016/j.mito.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fort PE, Freeman WM, Losiewicz MK, Singh RSJ, Gardner TW. The Retinal Proteome in Experimental Diabetic Retinopathy. Mol Cell Proteomics. 2009;8:767–79. doi: 10.1074/mcp.M800326-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Csősz É, Deák E, Kalló G, Csutak A, Tőzsér J. Diabetic retinopathy: Proteomic approaches to help the differential diagnosis and to understand the underlying molecular mechanisms. J Proteomics. 2017;150:351–8. doi: 10.1016/j.jprot.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 15.Cunha-Vaz JG. Pathophysiology of diabetic retinopathy. Br J Ophthalmol. 1978;62:351–5. doi: 10.1136/bjo.62.6.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dagher Z, Park YS, Asnaghi V, Hoehn T, Gerhardinger C, Lorenzi M. Studies of rat and human retinas predict a role for the polyol pathway in human diabetic retinopathy. Diabetes. 2004;53:2404–11. doi: 10.2337/diabetes.53.9.2404. [DOI] [PubMed] [Google Scholar]
- 17.Kowluru RA, Mishra M. Oxidative stress, mitochondrial damage and diabetic retinopathy. Biochim Biophys Acta Mol Basis Dis. 2015 doi: 10.1016/j.bbadis.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Antonetti DA, Barber AJ, Hollinger LA, Wolpert EB, Gardner TW. Vascular endothelial growth factor induces rapid phosphorylation of tight junction proteins occludin and zonula occluden 1. J Biol Chem. 1999;274:23463–7. doi: 10.1074/jbc.274.33.23463. [DOI] [PubMed] [Google Scholar]
- 19.Chung H, Lee H, Lamoke F, Hrushesky WJM, Wood P, Jahng WJ. Neuroprotective role of erythropoietin by antiapoptosis in the retina. J Neurosci Res. 2009;87:2365–74. doi: 10.1002/jnr.22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnouk H, Lee H, Zhang R, Chung H, Hunt RC, Jahng WJ. Early biosignature of oxidative stress in the retinal pigment epithelium. J Proteomics. 2011;74:254–61. doi: 10.1016/j.jprot.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R, Hrushesky WJM, Wood P, Lee SH, Hunt RC, Jahng WJ. Melatonin reprogrammes proteomic profile in light-exposed retina in vivo. Int J Biol Macromol. 2010;47:255–60. doi: 10.1016/j.ijbiomac.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sripathi SR, Prigge CL, Elledge B, He W, Offor J, Gutsaeva DR, Jahng WJ. Melatonin modulates prohibitin and cytoskeleton in the retinal pigment epithelium. Int J Sci Eng Res. 2017;8:502–6. doi: 10.14299/ijser.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshua M, Okere C, Sylvester O, Yahaya M, Precious O, Dluya T, Um J-Y, Neksumi M, Boyd J, Vincent-Tyndall J, Choo D-W, Gutsaeva DR, Jahng WJ. Disruption of angiogenesis by anthocyanin-rich extracts of hibiscus sabdariffa. Int J Sci Eng Res. 2017;8:299–307. doi: 10.14299/ijser.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jahng WJ. New Biomarkers in the Retina and RPE Under Oxidative Stress, in: Adio A, editor, Ocul. Dis., IntechOpen, 2012: pp. 121–155. 10.5772/48785.
- 25.He W, Sripathi SR, Joshua M, Zhang R, Tosin F, Ambrose P, Gutsaeva DR, Jahng WJ. Mechanistic dissection of macular degeneration using the phosphorylation interactome, in: Lo G, Giudice, editors, Vis. Impair. Blind., IntechOpen, 2020. 10.5772/intechopen.83016.
- 26.Lee H, Arnouk H, Sripathi S, Chen P, Zhang R, Bartoli M, Hunt RC, Hrushesky WJM, Chung H, Lee SH, Jahng WJ. Prohibitin as an oxidative stress biomarker in the eye. Int J Biol Macromol. 2010;47:685–90. doi: 10.1016/j.ijbiomac.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H, Chung H, Lee SH, Jahng WJ. Light-induced phosphorylation of crystallins in the retinal pigment epithelium. Int J Biol Macromol. 2011;48:194–201. doi: 10.1016/j.ijbiomac.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Chung H, Arnouk H, Lamoke F, Hunt RC, Hrushesky WJM, Wood PA, Lee SH, Jahng WJ. Cleavage of the retinal pigment epithelium-specific protein RPE65 under oxidative stress. Int J Biol Macromol. 2010;47:104–8. doi: 10.1016/j.ijbiomac.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sripathi SR, He W, Atkinson CL, Smith JJ, Liu Z, Elledge BM, Jahng WJ. Mitochondrial-nuclear communication by prohibitin shuttling under oxidative stress. Biochemistry. 2011;50:8342–51. doi: 10.1021/bi2008933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sripathi SR, He W, Um J, Moser T, Dehnbostel S, Kindt K, Goldman J, Frost MC, Jahng WJ. Nitric oxide leads to cytoskeletal reorganization in the retinal pigment epithelium under oxidative stress. Adv Biosci Biotechnol. 2012;03:1167–78. doi: 10.4236/abb.2012.38143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sripathi SR, Sylvester O, He W, Moser T, Um J, Lamoke F, Ramakrishna W, Bernstein PS, Bartoli M, Jahng WJ. Prohibitin as the molecular binding switch in the retinal pigment epithelium. Protein J. 2016;35:1–16. doi: 10.1007/s10930-015-9641-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sripathi SR, He W, Sylvester O, Neksumi M, Um J-Y, Dluya T, Bernstein PS, Jahng WJ. Altered cytoskeleton as a mitochondrial decay signature in the retinal pigment epithelium. Protein J. 2016;35:179–92. doi: 10.1007/s10930-016-9659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sripathi S, He W, Prigge CL, Sylvester O, Um J-Y, Powell FL, Neksumi M, Bernstein PS, Choo D-W, Bartoli M, Gutsaeva DR, Jahng WJ. Interactome mapping guided by tissue-specific phosphorylation in age-related macular degeneration. Int J Sci Eng Res. 2017;8:680–98. doi: 10.14299/ijser.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponce J, Brea D, Carrascal M, Guirao VV, Degregorio-Rocasolano N, Sobrino TT, Castillo JJ, Davalos A, Gasull T, Dávalos A, Gasull T. The effect of simvastatin on the proteome of detergent-resistant membrane domains: decreases of specific proteins previously related to cytoskeleton regulation, calcium homeostasis and cell fate. Proteomics. 2010;10:1954–65. doi: 10.1002/pmic.200900055. [DOI] [PubMed] [Google Scholar]
- 35.Dong P, Flores J, Pelton K, Solomon KR. Prohibitin is a cholesterol-sensitive regulator of cell cycle transit. J Cell Biochem. 2010;111:1367–74. doi: 10.1002/jcb.22865. [DOI] [PubMed] [Google Scholar]
- 36.Kowluru RA, Santos JM, Zhong Q. Sirt1, A negative regulator of matrix metalloproteinase-9 in diabetic retinopathy. Investig Ophthalmol Vis Sci. 2014;55:5653–60. doi: 10.1167/iovs.14-14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pusparajah P, Lee LH, Kadir KA. Molecular markers of diabetic retinopathy: Potential screening tool of the future? Front Physiol. 2016 doi: 10.3389/fphys.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liyanage VRB, Jarmasz JS, Murugeshan N, Bigio MRD, Rastegar M, Davie DNA., Jr modifications: Function and applications in normal and disease states. Biology (Basel) 2014;3(4):670–723. doi: 10.3390/biology3040670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin J, Min H, Kim SJ, Oh S, Kim K, Yu HG, Park T, Kim Y. Development of diagnostic biomarkers for detecting diabetic retinopathy at early stages using quantitative proteomics. J Diabetes Res. 2016;2016:6571976. doi: 10.1155/2016/6571976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: Structural and functional diversity. Protein Sci. 1996;5:2375–90. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venhoranta H, Bauersachs S, Taponen J, Lohi H, Taira T, Andersson M, Kind A, Schnieke A, Flisikowski K. Fetal growth restriction caused by MIMT1 deletion alters brain transcriptome in cattle. Int J Dev Neurosci. 2013;31:463–7. doi: 10.1016/j.ijdevneu.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Zou B, Liu X, Gong Y, Cai C, Li P, Xing S, Pokhrel B, Zhang B, Li J. A novel 12-marker panel of cancer-associated fibroblasts involved in progression of hepatocellular carcinoma. Cancer Manag Res. 2018;10:5303–11. doi: 10.2147/CMAR.S176152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu R, Cheng M, Lu T, Yang N, Ye S, Pan Y-H, Hong T, Dang S, Zhang W. A disintegrin and metalloproteinase with thrombospondin motifs 18 deficiency leads to visceral adiposity and associated metabolic syndrome in mice. Am J Pathol. 2018;188:461–73. doi: 10.1016/j.ajpath.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 44.Shalaby L, Thounaojam M, Tawfik A, Li J, Hussein K, Jahng WJ, Al-Shabrawey M, Kwok HF, Bartoli M, Gutsaeva D. Role of endothelial ADAM17 in early vascular changes associated with diabetic retinopathy. J Clin Med. 2020;9:400. doi: 10.3390/jcm9020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brockhaus K, Melkonyan H, Prokosch-Willing V, Liu H, Thanos S. Alterations in tight- and adherens-junction proteins related to glaucoma mimicked in the organotypically cultivated mouse retina under elevated pressure. Investig Opthalmol Vis Sci. 2020;61:46. doi: 10.1167/iovs.61.3.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nita M, Grzybowski A, Ascaso FJ, Huerva V. Age-related macular degeneration in the aspect of chronic low-grade inflammation (Pathophysiological paraInflammation) Mediat Inflamm. 2014;2014:1–10. doi: 10.1155/2014/930671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livne-Bar I, Lam S, Chan D, Guo X, Askar I, Nahirnyj A, Flanagan JG, Sivak JM. Pharmacologic inhibition of reactive gliosis blocks TNF-α-mediated neuronal apoptosis. Cell Death Dis. 2016 doi: 10.1038/cddis.2016.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arsenijevic Y, Taverney N, Kostic C, Tekaya M, Riva F, Zografos L, Schorderet D, Munier F. Non-neural regions of the adult human eye: A potential source of neurons? Investig Ophthalmol Vis Sci. 2003 doi: 10.1167/iovs.02-0267. [DOI] [PubMed] [Google Scholar]
- 49.Shinoda K, Hirakata A, Hida T, Yamaguchi Y, Fukuda M, Maekawa S, Azuma N, Ultrastructural and immunohistochemical findings in five patients with vitreomacular traction syndrome, Retina. 2000. 10.1097/00006982-200003000-00011. [DOI] [PubMed]
- 50.Viewer GD. Fam81b family with sequence similarity 81, member B [Rattus norvegicus (Norway rat)]. 2019;3:1–5.
- 51.Ramirez-Ardila DE, Ruigrok-Ritstier K, Helmijr JC, Look MP, van Laere S, Dirix L, Berns EMJJ, Jansen MPHM. LRG1 mRNA expression in breast cancer associates with PIK3CA genotype and with aromatase inhibitor therapy outcome. Mol Oncol. 2016;10:1363–73. doi: 10.1016/j.molonc.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 53.Eckmiller MS. Renewal of the ciliary axoneme in cone outer segments of the retina of Xenopus laevis. Cell Tissue Res. 1996;285:165–9. doi: 10.1007/s004410050632. [DOI] [PubMed] [Google Scholar]
- 54.Gong J, Sagiv O, Cai H, Tsang SH, Del LV, Priore Effects of extracellular matrix and neighboring cells on induction of human embryonic stem cells into retinal or retinal pigment epithelial progenitors. Exp Eye Res. 2008;86:957–65. doi: 10.1016/j.exer.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldenberg-Cohen N, Avraham-Lubin B-CR, Sadikov T, Goldstein RS, Askenasy N. Primitive stem cells derived from bone marrow express glial and neuronal markers and support revascularization in injured retina exposed to ischemic and mechanical damage. Stem Cells Dev. 2012;21:1488–500. doi: 10.1089/scd.2011.0366. [DOI] [PubMed] [Google Scholar]
- 56.Struebing FL, Lee RK, Williams RW, Geisert EE. Genetic networks in mouse retinal ganglion cells. Front Genet. 2016;7:169. doi: 10.3389/fgene.2016.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loukovaara S, Sandholm J, Aalto K, Liukkonen J, Jalkanen S, Yegutkin GG. Deregulation of ocular nucleotide homeostasis in patients with diabetic retinopathy. J Mol Med. 2017;95:193–204. doi: 10.1007/s00109-016-1472-6. [DOI] [PubMed] [Google Scholar]
- 58.Loukovaara S, Sahanne S, Jalkanen S, Yegutkin GG. Increased intravitreal adenosine 5′-triphosphate, adenosine 5′-diphosphate and adenosine 5′-monophosphate levels in patients with proliferative diabetic retinopathy. Acta Ophthalmol. 2015;93:67–73. doi: 10.1111/aos.12507. [DOI] [PubMed] [Google Scholar]
- 59.Du Y, Veenstra A, Palczewski K, Kern TS. Photoreceptor cells are major contributors to diabetes-induced oxidative stress and local inflammation in the retina. Proc Natl Acad Sci. 2013;110:16586–16591. doi: 10.1073/pnas.1314575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlotterer A, Kolibabka M, Lin J, Acunman K, Dietrichá N, Sticht C, Fleming T, Nawroth P, Hammes H-P. Methylglyoxal induces retinopathy-type lesions in the absence of hyperglycemia: studies in a rat model. FASEB J. 2019;33:4141–53. doi: 10.1096/fj.201801146RR. [DOI] [PubMed] [Google Scholar]
- 61.Acute FPE Complications C. Diabetes. 2011;60:A133–95. 10.2337/db11-478-715.
- 62.Zamora DO, Riviere M, Choi D, Pan Y, Planck SR, Rosenbaum JT, David LL, Smith JR. Proteomic profiling of human retinal and choroidal endothelial cells reveals molecular heterogeneity related to tissue of origin. Mol Vis. 2007;13:2058–65. http://www.ncbi.nlm.nih.gov/pubmed/18079679. [PubMed]
- 63.Devi TS, Yumnamcha T, Yao F, Somayajulu M, Kowluru RA, Singh LP. TXNIP mediates high glucose-induced mitophagic flux and lysosome enlargement in human retinal pigment epithelial cells. Biol Open. 2019;8:bio038521. doi: 10.1242/bio.038521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamane K, Minamoto A, Yamashita H, Takamura H, Miyamoto-Myoken Y, Yoshizato K, Nabetani T, Tsugita A, Mishima HK. Proteome analysis of human vitreous proteins. Mol Cell Proteomics. 2003;2:1177–87. doi: 10.1074/mcp.M300038-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Garcia-Ramírez M, Hernández C, Villarroel M, Canals F, Alonso MA, Fortuny R, Masmiquel L, Navarro A, García-Arumí J, Simó R. Interphotoreceptor retinoid-binding protein (IRBP) is downregulated at early stages of diabetic retinopathy. Diabetologia. 2009;52:2633–41. doi: 10.1007/s00125-009-1548-8. [DOI] [PubMed] [Google Scholar]
- 66.Minamoto A, Yamane K, Yokoyama T. Proteomics of Vitreous Fluid. In: Proteomics Hum Body Fluids. Totowa: Humana Press; 2007. p. 495–507. 10.1007/978-1-59745-432-2_23.
- 67.Kang M-K, Lee E-J, Kim Y-H, Kim D, Oh H, Kim S-I, Kang Y-H. Chrysin ameliorates malfunction of retinoid visual cycle through blocking activation of AGE-RAGE-ER stress in glucose-stimulated retinal pigment epithelial cells and diabetic eyes. Nutrients. 2018;10:1046. doi: 10.3390/nu10081046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldstein AS, Witte ON. A plethora of progenitors in the post-natal prostate. EMBO Rep. 2012 doi: 10.1038/embor.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fisher JW. Landmark advances in the development of erythropoietin. Exp Biol Med (Maywood) 2010;235:1398–411. doi: 10.1258/ebm.2010.010137. [DOI] [PubMed] [Google Scholar]
- 70.Wu T, Handa JT, Gottsch JD. Light-induced oxidative stress in choroidal endothelial cells in mice. Invest Ophthalmol Vis Sci. 2005;46:1117–23. doi: 10.1167/iovs.04-0517. [DOI] [PubMed] [Google Scholar]
- 71.Jarajapu YPR, Cai J, Yan Y, Li Calzi S, Kielczewski JL, Hu P, Shaw LC, Firth SM, Chan-Ling T, Boulton ME, Baxter RC, Grant MB. Protection of blood retinal barrier and systemic vasculature by insulin-like growth factor binding protein-3. PLoS One. 2012;7:e39398. doi: 10.1371/journal.pone.0039398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreira PI, Santos MS, Moreno AM, Proenca T, Seica R, Oliveira CR. Effect of streptozotocin-induced diabetes on rat brain mitochondria. J Neuroendocrinol. 2004;16:32–8. doi: 10.1111/j.1365-2826.2004.01107.x. [DOI] [PubMed] [Google Scholar]
- 73.Alam NM, Mills WC, Wong AA, Douglas RM, Szeto HH, Prusky GT. A mitochondrial therapeutic reverses visual decline in mouse models of diabetes. Dis Model Mech. 2015;8:701–710. doi: 10.1242/dmm.020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gargiulo P, Goldberg J, Romani B, Schiaffini R, Ciampalini P, Faulk WP, McIntyre JA. Qualitative and quantitative studies of autoantibodies to phospholipids in diabetes mellitus. Clin Exp Immunol. 1999;118:30–4. doi: 10.1046/j.1365-2249.1999.01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerl VB, Bohl J, Pitz S, Stoffelns B, Pfeiffer N, Bhakdi S. Extensive deposits of complement C3d and C5b-9 in the choriocapillaris of eyes of patients with diabetic retinopathy. Investig Ophthalmol Vis Sci. 2002;43:1104–8. https://iovs.arvojournals.org/article.aspx?articleid=2200162. [PubMed]
- 76.Liu X, Mameza MG, Lee YS, Eseonu CI, Yu C-R, Kang Derwent JJ, Egwuagu CE. Suppressors of cytokine-signaling proteins induce insulin resistance in the retina and promote survival of retinal cells. Diabetes. 2008;57:1651–1658. doi: 10.2337/db07-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.del Olmo-Aguado S, Núñez-Álvarez C, Osborne NN. Blue light action on mitochondria leads to cell death by necroptosis. Neurochem Res. 2016;41:2324–35. doi: 10.1007/s11064-016-1946-5. [DOI] [PubMed] [Google Scholar]
- 78.Ren X, Lu H, Wang N, Zhang C, Ji Y, Cui S, Dong Y, Yang K, Du M, Diao F, Kong L. Thioredoxin is implicated in the anti-apoptotic effects of grape seed proanthocyanidin extract during hyperglycemia. Mol Med Rep. 2017;16:7731–7. doi: 10.3892/mmr.2017.7508. [DOI] [PubMed] [Google Scholar]
- 79.Kim SS, Utsunomiya H, Koski J, Wu BM, Cima MJ, Sohn J, Mukai K, Griffith LG, Vacanti JP. Survival and function of hepatocytes on a novel three-dimensional synthetic biodegradable polymer scaffold with an intrinsic network of channels. Ann Surg. 1998;228:8–13. doi: 10.1097/00000658-199807000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pang C, Jia L, Jiang S, Liu W, Hou X, Zuo Y, Gu H, Bao Y, Wu Q, Xiang K, Gao X, Jia W. Determination of diabetic retinopathy prevalence and associated risk factors in Chinese diabetic and pre-diabetic subjects: Shanghai diabetic complications study. Diabetes Metab Res Rev. 2012;28:276–83. doi: 10.1002/dmrr.1307. [DOI] [PubMed] [Google Scholar]
- 81.Sheetz MJ. Molecular understanding of hyperglycemia’s adverse effects for diabetic complications. JAMA. 2002;288:2579. doi: 10.1001/jama.288.20.2579. [DOI] [PubMed] [Google Scholar]
- 82.Popova EA, Mironova RS, Odjakova MK. Non-enzymatic glycosylation and deglycating enzymes. Biotechnol Biotechnol Equip. 2010;24:1928–35. doi: 10.2478/V10133-010-0066-7. [DOI] [Google Scholar]
- 83.Pacher P, Szabó C. Role of poly(ADP-Ribose) polymerase-1 activation in the pathogenesis of diabetic complications: endothelial dysfunction, as a common underlying theme. Antioxid Redox Signal. 2005;7:1568–80. doi: 10.1089/ars.2005.7.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kiss L, Szabó C. The pathogenesis of diabetic complications: the role of DNA injury and poly(ADP-ribose) polymerase activation in peroxynitrite-mediated cytotoxicity. Mem Inst Oswaldo Cruz. 2005;100:29–37. doi: 10.1590/S0074-02762005000900007. [DOI] [PubMed] [Google Scholar]
- 85.Warboys CM, Fraser PA. Hyperglycemia attenuates acute permeability response to advanced glycation end products in retinal microvasculature. Microvasc Res. 2010;80:174–6. doi: 10.1016/j.mvr.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 86.Lind KR, Ball KK, Cruz NF, Dienel GA. The unfolded protein response to endoplasmic reticulum stress in cultured astrocytes and rat brain during experimental diabetes. Neurochem Int. 2013;62:784–95. doi: 10.1016/j.neuint.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eljarrat-Binstock E, Raiskup F, Stepensky D, Domb AJ, Frucht-Pery J. Delivery of gentamicin to the rabbit eye by drug-loaded hydrogel iontophoresis. Investig Opthalmology Vis Sci. 2004;45:2543. doi: 10.1167/iovs.03-1294. [DOI] [PubMed] [Google Scholar]
- 88.Huang SP, Palla S, Ruzycki P, Varma RA, Harter T, Reddy GB, Petrash JM. Aldo-keto reductases in the eye. J Ophthalmol. 2010;2010:1–6. doi: 10.1155/2010/521204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawanishi K, Ueda H, Moriyasu M. Aldose reductase inhibitors from the nature. Curr Med Chem. 2003;10:1353–74. doi: 10.2174/0929867033457304. [DOI] [PubMed] [Google Scholar]
- 90.Kumar H, Shah A, Sobhia ME. Novel insights into the structural requirements for the design of selective and specific aldose reductase inhibitors. J Mol Model. 2012;18:1791–9. doi: 10.1007/s00894-011-1195-0. [DOI] [PubMed] [Google Scholar]
- 91.Veeresham C, Swetha E, Rao AR, Asres K. Vitro and in vivo aldose reductase inhibitory activity of standardized extracts and the major constituent of andrographis paniculata. Phyther Res. 2013;27:412–6. doi: 10.1002/ptr.4722. [DOI] [PubMed] [Google Scholar]
- 92.Zhan X, Du Y, Crabb JS, Gu X, Kern TS, Crabb JW. Targets of tyrosine nitration in diabetic rat retina. Mol Cell Proteomics. 2008;7:864–74. doi: 10.1074/mcp.M700417-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Toda N, Nakanishi-Toda M. Nitric oxide: ocular blood flow, glaucoma, and diabetic retinopathy. Prog Retin Eye Res. 2007;26:205–38. doi: 10.1016/j.preteyeres.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 94.Gao B-B, Phipps JA, Bursell D, Clermont AC, Feener EP. Angiotensin AT1 receptor antagonism ameliorates murine retinal proteome changes induced by diabetes. J Proteome Res. 2009;8:5541–9. doi: 10.1021/pr9006415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J, Wu Y, Jin Y, Ji F, Sinclair SH, Luo Y, Xu G, Lu L, Dai W, Yanoff M, Li W, Xu G-T. Intravitreal injection of erythropoietin protects both retinal vascular and neuronal cells in early diabetes. Invest Ophthalmol Vis Sci. 2008;49:732–42. doi: 10.1167/iovs.07-0721. [DOI] [PubMed] [Google Scholar]
- 96.Huang Y-C, Lin J-M, Lin H-J, Chen C-C, Chen S-Y, Tsai C-H, Tsai F-J. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. 2011;118:642–8. doi: 10.1016/j.ophtha.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 97.Qin G, Mallik S, Mitra R, Li A, Jia P, Eischen CM, Zhao Z. MicroRNA and transcription factor co-regulatory networks and subtype classification of seminoma and non-seminoma in testicular germ cell tumors. Sci Rep. 2020;10:852. doi: 10.1038/s41598-020-57834-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miller SFC, Summerhurst K, Rünker AE, Kerjan G, Friedel RH, Chédotal A, Murphy P, Mitchell KJ. Expression of Plxdc2/TEM7R in the developing nervous system of the mouse. Gene Expr Patterns. 2007;7:635–44. doi: 10.1016/j.modgep.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 99.Perrone L, Devi TS, Hosoya K, Terasaki T, Singh LP. Thioredoxin interacting protein (TXNIP) induces inflammation through chromatin modification in retinal capillary endothelial cells under diabetic conditions. J Cell Physiol. 2009;221:262–72. doi: 10.1002/jcp.21852. [DOI] [PubMed] [Google Scholar]
- 100.Abhary S, Hewitt AW, Burdon KP, Craig JE. A systematic meta-analysis of genetic association studies for diabetic retinopathy. Diabetes. 2009;58:2137–47. doi: 10.2337/db09-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simões MJ, Lobo C, Egas C, Nunes S, Carmona S, Costa M, Duarte T, Ribeiro L, Faro C, Cunha-Vaz JG. Genetic variants in ICAM1, PPARGC1A and MTHFR are potentially associated with different phenotypes of diabetic retinopathy. Ophthalmologica. 2014;232:156–62. doi: 10.1159/000365229. [DOI] [PubMed] [Google Scholar]
- 102.Kowluru RA. Role of matrix metalloproteinase-9 in the development of diabetic retinopathy and its regulation by H-Ras. Investig Opthalmology Vis Sci. 2010;51:4320. doi: 10.1167/iovs.09-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mishra M, Flaga J, Kowluru RA. Molecular mechanism of transcriptional regulation of matrix metalloproteinase-9 in diabetic retinopathy. J Cell Physiol. 2016;231:1709–18. doi: 10.1002/jcp.25268. [DOI] [PubMed] [Google Scholar]
- 104.Kinnunen K, Puustjarvi T, Terasvirta M, Nurmenniemi P, Heikura T, Laidinen S, Paavonen T, Uusitalo H, Yla-Herttuala S. Differences in retinal neovascular tissue and vitreous humour in patients with type 1 and type 2 diabetes. Br J Ophthalmol. 2009;93:1109–15. doi: 10.1136/bjo.2008.148841. [DOI] [PubMed] [Google Scholar]
- 105.GUSTAVSSON C, AGARDH C-D, HAGERT P, AGARDH E. Inflammatory markers in nondiabetic and diabetic rat retinas exposed to ischemia followed by reperfusion. Retina. 2008;28:645–52. doi: 10.1097/IAE.0b013e31815ec32d. [DOI] [PubMed] [Google Scholar]
- 106.Azcutia V, Abu-Taha M, Romacho T, Vázquez-Bella M, Matesanz N, Luscinskas FW, Rodríguez-Mañas L, Sanz MJ, Sánchez-Ferrer CF, Peiró C. Inflammation determines the pro-adhesive properties of high extracellular d-glucose in human endothelial cells in vitro and rat microvessels in vivo. PLoS One. 2010;5:e10091. doi: 10.1371/journal.pone.0010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie M, Hu A, Luo Y, Sun W, Hu X, Tang S. Interleukin-4 and melatonin ameliorate high glucose and interleukin-1β stimulated inflammatory reaction in human retinal endothelial cells and retinal pigment epithelial cells. Mol Vis. 2014;20:921–8. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4077596/. [PMC free article] [PubMed]
- 108.Arjamaa O, Nikinmaa M. Oxygen-dependent diseases in the retina: role of hypoxia-inducible factors. Exp Eye Res. 2006;83:473–83. doi: 10.1016/j.exer.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 109.Loukovaara S, Koivunen P, Inglés M, Escobar J, Vento M, Andersson S. Elevated protein carbonyl and HIF-1α levels in eyes with proliferative diabetic retinopathy. Acta Ophthalmol. 2014;92:323–7. doi: 10.1111/aos.12186. [DOI] [PubMed] [Google Scholar]
- 110.Abu-El-Asrar AM, Dralands L, Missotten L, Al-Jadaan I, Geboes K. Expression of apoptosis markers in the retinas of human subjects with diabetes. Invest Ophthalmol Vis Sci. 2004;45:2760–6. doi: 10.1167/iovs.03-1392. [DOI] [PubMed] [Google Scholar]
- 111.Shevchenko AV, Prokofiev VF, Konenkov VI, Klimontov VV, Tyan NV, Chernykh DV, Trunov AN, Chernykh VV. Polymorphisms of extracellular connective tissue remodeling proteinases and MMP2, MMP3, MMP9 genes, and neoangigenesis VEGF gene in retinal microangiopathy in the patients with type 2 diabetes mellitus. Med Immunol. 2019;21:441–50. doi: 10.15789/1563-0625-2019-3-441-450. [DOI] [Google Scholar]
- 112.Karumanchi DK, Gaillard ER, Dillon J. Early diagnosis of diabetes through the eye. Photochem Photobiol. 2015;91:1497–504. doi: 10.1111/php.12524. [DOI] [PubMed] [Google Scholar]
- 113.Lyons TJ, Basu A. Biomarkers in diabetes: hemoglobin A1c, vascular and tissue markers. Transl Res. 2012;159:303–12. doi: 10.1016/j.trsl.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Laíns I, Gantner M, Murinello S, Lasky-Su JA, Miller JW, Friedlander M, Husain D. Metabolomics in the study of retinal health and disease. Prog Retin Eye Res. 2019;69:57–79. doi: 10.1016/j.preteyeres.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 115.Rhee SY, Jung ES, Park HM, Jeong SJ, Kim K, Chon S, Yu S-Y, Woo J-T, Lee CH. Plasma glutamine and glutamic acid are potential biomarkers for predicting diabetic retinopathy. Metabolomics. 2018;14:89. doi: 10.1007/s11306-018-1383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Paris LP, Johnson CH, Aguilar E, Usui Y, Cho K, Hoang LT, Feitelberg D, Benton HP, Westenskow PD, Kurihara T, Trombley J, Tsubota K, Ueda S, Wakabayashi Y, Patti GJ, Ivanisevic J, Siuzdak G, Friedlander M. Global metabolomics reveals metabolic dysregulation in ischemic retinopathy. Metabolomics. 2016;12:15. doi: 10.1007/s11306-015-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haines NR, Manoharan N, Olson JL, D’Alessandro A, Reisz JA. Metabolomics Analysis of human vitreous in diabetic retinopathy and rhegmatogenous retinal detachment. J Proteome Res. 2018;17:2421–7. doi: 10.1021/acs.jproteome.8b00169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dissection of the retina control map showing several pathways, including PI3K, WNT, cytoskeleton, apoptosis, p53, VEGF, HIF-1, and cholesterol mechanism. A. PI3K-AKT signaling, B. WNT network, C. Actin cytoskeletal network, D. Apoptosis, E. p53 network, F. VEGF pathway, G. HIF-1 network, H. Cholesterol metabolism. (PDF 5027 kb)
Mechanistic dissection of the interactome network leading to the pathogenesis of diabetic retinopathy, A. Apoptosis, B. Angiogenesis, C. Inflammation network, D. Retinoid metabolism, E. Vascular permeability/occlusion, F. Energy metabolism network, G. Lipid metabolism, H. Cytoskeleton organization, I. Mitochondria dysfunction, J. Hyperglycemia, K. Oxidative stress. (PDF 7678 kb)
Identified proteins in the retina as the control. A total of 1090 proteins were identified as biological, cellular, and molecular functional proteins which serve as the control proteins in the retina. Control interactome map was established using bioinformatics tools including PPI software STRING (http://string-db.org/) by adding 1104 identified normal eye proteins in the query. (XLSX 111 kb)
DR biomarkers. DR-specific proteins were identified from our proteomics study, protein database, and proteomics literature. These proteins were considered DR biomarkers and used in bioinformatics analysis. DR-specific interactome map was established using STRING to find the probable interacting partners of the proteins associated with DR. These potential DR biomarkers are presented in the interaction map and mechanistic dissection was presented in Supplement Figures 2A-2K. (XLSX 35 kb)
Metabolites associated with DR pathogenesis. A total of 121 metabolites were collected from the literature and databases including KEGG and PubChem. The metabolome map was established using bioinformatics tools including OmicsNet software by adding 121 metabolites in the query. (XLSX 19 kb)
Reverse interactome mapping from the metabolome map. A total of 115 proteins related to DR were identified as the DR-specific metabolome-proteome interactions from the nodes of metabolites in the metabolome map. Proteins obtained from the metabolome-proteome interactions were added in STRING software and determined the protein interactions. (XLSX 14 kb)