We integrated quantitative nanoscale lipid mapping with molecular dynamic simulations to define the lipid profile within caveolae. Caveolin-1 and cavin1 individually sort distinct plasma membrane lipids. Intact caveolae containing both caveolin-1 and cavin1 further generate a distinct lipid profile, with headgroup and acyl chain selectivity.
Abstract
Caveolae are specialized domains of the vertebrate cell surface with a well-defined morphology and crucial roles in cell migration and mechanoprotection. Unique compositions of proteins and lipids determine membrane architectures. The precise caveolar lipid profile and the roles of the major caveolar structural proteins, caveolins and cavins, in selectively sorting lipids have not been defined. Here, we used quantitative nanoscale lipid mapping together with molecular dynamic simulations to define the caveolar lipid profile. We show that caveolin-1 (CAV1) and cavin1 individually sort distinct plasma membrane lipids. Intact caveolar structures composed of both CAV1 and cavin1 further generate a unique lipid nano-environment. The caveolar lipid sorting capability includes selectivities for lipid headgroups and acyl chains. Because lipid headgroup metabolism and acyl chain remodeling are tightly regulated, this selective lipid sorting may allow caveolae to act as transit hubs to direct communications among lipid metabolism, vesicular trafficking, and signaling.
Introduction
Caveolae are a striking morphological feature of the plasma membrane (PM) of many vertebrate cells. Caveolae have been implicated in mechanoprotection, endocytosis, signal transduction, and lipid regulation (Cheng et al., 2015; Echarri and Del Pozo, 2015; Nassoy and Lamaze, 2012; Oh et al., 2007; Parton, 2018; Pilch and Liu, 2011). The characteristic morphology of caveolae, with a bulb connected to the PM by a highly curved neck, is generated by integral membrane proteins termed caveolins and by lipid-binding peripheral membrane proteins, the cavins (Bastiani et al., 2009; Hansen et al., 2009; Hansen et al., 2013; Hernandez et al., 2013; Hill et al., 2008; Kovtun et al., 2015; Liu et al., 2008; McMahon et al., 2009). Specifically, caveolin-1 (CAV1) and caveolin-3 (CAV3; in striated muscle) and cavin1 (or polymerase I and transcript release factor [PTRF]) are essential for caveola formation (Hill et al., 2008; Liu et al., 2008).
Another set of key caveolar components comprise the PM lipids. While biophysical studies have consistently suggested that the lateral distribution of lipids determines and/or responds to changing membrane morphology, our understanding of the lipid composition of caveolae and how this contributes to caveola formation, function, and disassembly is still relatively primitive. Early studies showed concentration of gold-labeled ganglioside-binding toxins in uncoated pits (Montesano et al., 1982; Tran et al., 1987), and these structures were subsequently identified as caveolae (Parton, 1994). The later discovery of caveolins in detergent-insoluble cholesterol and glycosphingolipid-enriched fractions isolated from cells and tissues (Dupree et al., 1993; Kurzchalia et al., 1992; Lisanti et al., 1994) suggested that caveolae represent a specialized, morphologically distinct, lipid raft domain. Other studies strengthened the links between cholesterol, glycosphingolipids, and caveolae, with caveola structure and cavin association dependent on cholesterol (Breen et al., 2012; Hill et al., 2008; Rothberg et al., 1992; Jansen et al., 2008) and caveola trafficking regulated by glycosphingolipids (Sharma et al., 2004; Shvets et al., 2015). Purification and lipid analysis of caveolar fractions isolated from adipocytes gave further insights into the specialized lipid composition of caveolae, showing enrichment of cholesterol and specific gangliosides such as GD3 but exclusion of other gangliosides (Ortegren et al., 2004).
Fractionation methods described above rely on biochemical separation of caveolae from the PM and so may not retain the 3D architectures of caveolae, where distinct caveolar curvatures may contribute to the specific lipid composition. Specialized EM techniques have been used to examine the distribution of individual lipids with respect to caveolae in situ, including phosphoinositol 4,5-bisphosphate [PtdIns(4,5)P2] that was shown to be enriched at the neck of caveolae (Fujita et al., 2009). Loss of caveolae through genetic loss or knockdown of caveolar components has significant effects on glycosphingolipid biosynthetic pathways and the cellular lipidome (Ariotti et al., 2014). In addition, knockdown or total loss of caveolar components shows similar effects to the acute disassembly of caveolae in causing alterations to the nanoscale lipid organization of the PM with significant effects on isoform-specific signaling of lipid-anchored Ras proteins (Ariotti et al., 2014). Using light microscopy and single-particle tracking, Hubert et al. (2020) demonstrated that caveolae are stabilized by sphingomyelin, while caveolar scission is promoted by cholesterol. In epithelial cells, loss of caveolae causes elevated levels of junctional PtdIns(4,5)P2 to recruit the formin FMNL2 and promote F-actin bundling (Teo et al., 2020). How these effects might be related to the lipid composition of caveolae is not known, but we have speculated that the regulated release of lipids from caveolae could couple disassembly to lipid changes (Parton et al., 2020). Testing such a model requires detailed understanding of how the caveolar lipid composition is generated.
It has been shown that various protein constituents of caveolae differentially contribute to defining the lipid composition of caveolae. Caveolins are small integral membrane proteins that bind cholesterol (Murata et al., 1995). The conserved scaffolding domain of caveolin, a highly conserved 20–amino acid region of caveolin linked to protein–protein interactions, is tightly membrane associated (Ariotti et al., 2015b) and has been shown to induce lateral segregation of phosphatidylserine (PtdSer) and PtdIns(4,5)P2 (Wanaski et al., 2003) and the formation of highly enriched cholesterol domains in liposome experiments (Epand et al., 2005). The lipid-binding properties of cavin proteins are also starting to be unraveled. In vitro cavin proteins bind PtdSer, (Gustincich et al., 1999; Hill et al., 2008; Izumi et al., 1997) and PtdIns(4,5)P2 (Kovtun et al., 2014; Tillu et al., 2018). The PtdIns(4,5)P2-binding domain of the cavin proteins has been structurally characterized and shown to reside in a highly positively charged patch within the first helical region of the protein (HR1 domain; Kovtun et al., 2014). Lysine residues in the PtdIns(4,5)P2-binding region are exposed and ubiquitinated as caveolae are disassembled to act as a sensor of the membrane association/dissociation state, but this site is not required for caveola formation (Tillu et al., 2015). Cavin1 has an additional lipid-binding domain with in vitro specificity for PtdSer composed of 11 amino acid (undecad) repeats (Tillu et al., 2018). The number of repeats varies in different species to regulate caveolar stability. Consistent with these results, functional experiments have also shown that PtdSer is vital for caveolar formation, whereas depletion of PtdIns(4,5)P2 has less-pronounced effects (Hirama et al., 2017).
Despite the wealth of biochemical and in vitro data on the lipid-binding capabilities of caveolae, we still do not know how caveolae selectively sort lipids in intact cells. More specifically, do caveolar constituents additively or synergistically contribute to distinct lipid sorting? A detailed molecular understanding of the lipid constituents of caveolae, defined both by their headgroups and acyl chains, is crucial for understanding the formation of caveolae as well as their disassembly processes that are crucial for caveola function. Moreover, caveolae can provide a paradigm for understanding how local concentrations of specific lipid species contribute to membrane morphogenesis. In this study, we combined model cellular systems with lipid depletion/rescue experiments and a quantitative ultrastructural mapping technique that has been used to define the nanoscale lipid association of Ras isoforms (Liang et al., 2019; Plowman et al., 2005; Prior et al., 2003; Zhou et al., 2014; Zhou et al., 2017; Zhou et al., 2015) to elucidate the role of individual caveolar components in the generation of the unique lipid composition of the caveolar domain and to define the role of specific lipid species in caveolar formation.
Results
A model system for de novo assembly of caveolae by caveolin and cavin1
To dissect the selective lipid sorting of each caveola-associated protein, we used MCF7 cells, which lack expression of both caveolins and cavins (McMahon et al., 2019; Gambin et al., 2014). We first expressed the basic components of caveolae, CAV1 and cavin1 (Fig. 1 A), to assemble caveolae de novo. Expression of CAV1 and cavin1 in MCF7 cells was sufficient to generate surface pits with the typical features of caveolae as judged by immunoelectron microscopy on frozen sections (Fig. 1 B), by conventional EM with a surface stain (Fig. 1 C), and by using a nanobody fused to the ascorbate peroxidase APEX2 to detect YFP-CAV1 (Fig. S1). PM sheets labeled for the expressed proteins showed coassociation of CAV1 and cavin1 with structures of caveolar morphology (Fig. 1 D), whereas cavin1 expressed alone showed a similar level of surface labeling (Fig. S2 A) but no detectable association with defined membrane domains (Fig. 1 E). In contrast to CAV1 expressed with cavin1, CAV1 expressed alone showed a heterogeneous pattern of PM labeling when localized using the nanobody–APEX system (Fig. S1).
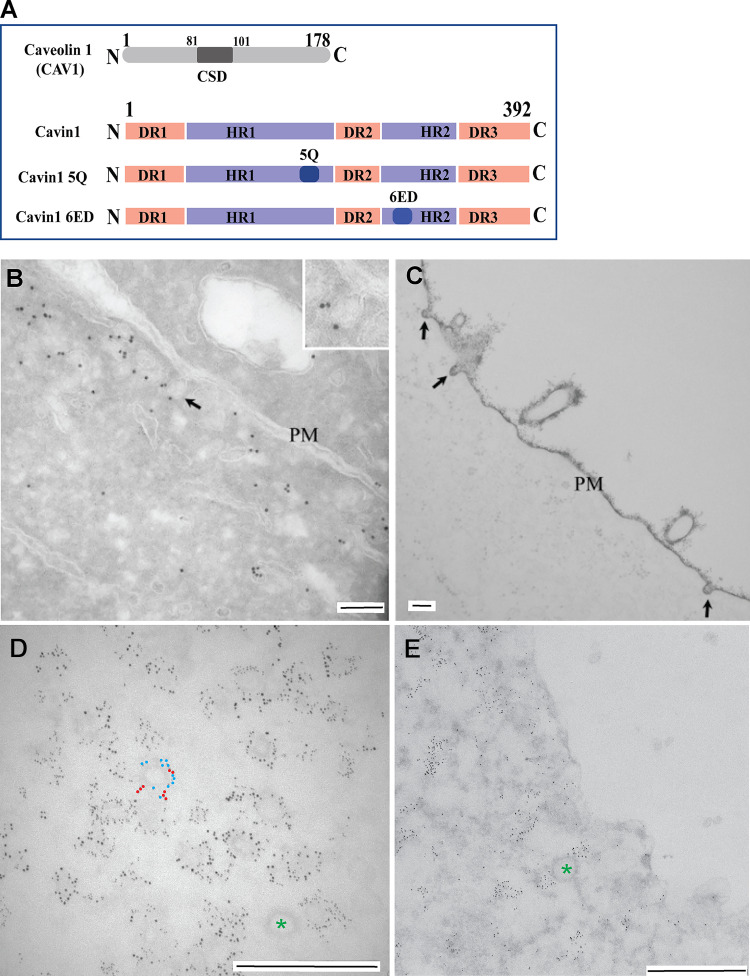
Figure 1.
Expression of CAV1, cavin1, and mutants in MCF7 cells. (A) Domain structure of CAV1, wild-type cavin1, and cavin1 mutants used in this study. DR, disordered region; HR, helical region. Cavin1 5Q and Cavin1 6ED denote amino acid substitutions in the putative PtdIns(4,5)P2–binding region of HR1 and in the UC1 domain of HR2, respectively. (B–E) MCF7 cells expressing CAV1 and cavin1 (B–D) or cavin1 alone (E). (B) Frozen section showing labeling for CAV1 in MCF7 cells coexpressing CAV1 and cavin1. Labeling is associated with membranous profiles close to the surface, some of which are connected to the PM (e.g., see arrow; shown at higher magnification in the inset). (C) Ruthenium red–labeled PM showing caveolae (arrows). (D) Sonicated PM sheet showing labeling for CAV1-mCherry (2-nm gold) and cavin1-GFP (6-nm gold) associated with approximately 60–80-nm-diameter profiles characteristic of caveolae. In panel D, the large gold (red; cavin1) and the small gold (blue; CAV1) are highlighted on one structure (raw image Fig. S2). (E) Sonicated PM sheet showing clustered labeling for Cavin1-GFP (6-nm gold). Clathrin-coated pits (green asterisks) are unlabeled. Scale bars, 100nm (B); 200 nm (C); 500 nm (D and E).
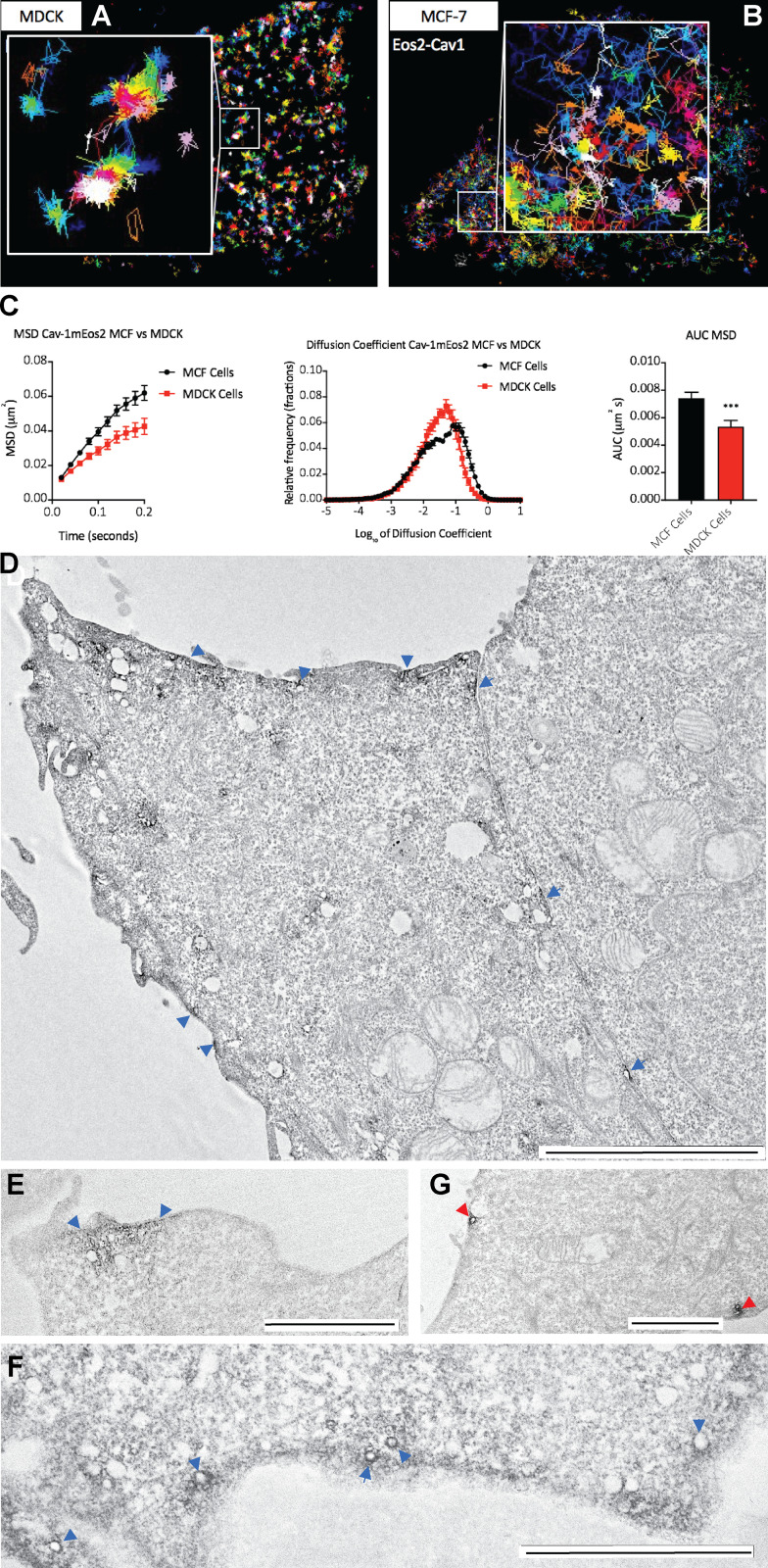
Figure S1.
Characterization of the noncaveolar pool of CAV1. (A and B) Single-molecule tracking of CAV1-mEos2 MDCK cells (A) and MCF7 cells (B). (C) Quantitative comparison of diffusion of CAV1-mEos2 in MDCK versus MCF7 cells showing changes in MSD (in micrometers squared; left), area under the MSD curve (AUC; right), and relative frequency distribution of diffusion coefficient (middle). Data are shown as mean ± SEM. Statistical significance was evaluated using one-way ANOVA, with *** indicating P < 0.05. (D–F) YFP-CAV1/APEX-nanobody expressed in MCF7 cells. Blue arrowheads indicate PM areas with high concentration of APEX staining. A range of structures are labeled, including flat areas of the PM (D), clusters of vesicles (E and F), and vesicular profiles or various diameters (F). (G) YFP-CAV1/Apex-nanobody coexpressed with cavin1 in MCF7 cells; vesicular profiles characteristic of caveolae are positive for the APEX reaction product (red arrowheads), with negligible labeling elsewhere on the PM. Scale bars, 5 µm (D); 2 µm (E–G).
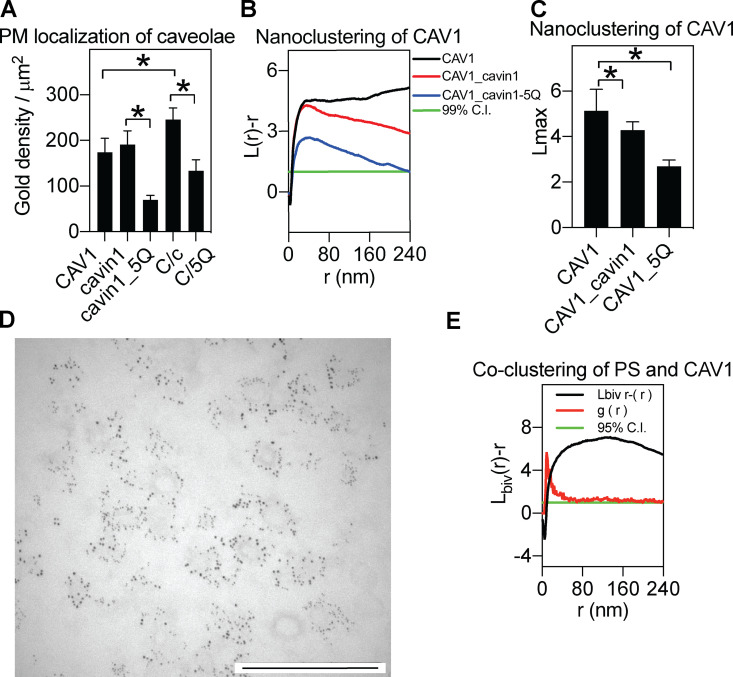
Figure S2.
Characterization of the MCF7 system. (A) Quantitative analysis of gold labeling density for CAV1, cavin1, and the cavin1.5Q mutant (5Q) or for CAV1 coexpressed with WT cavin1 (C/c) or cavin1.5Q (C/5Q). Gold density data are shown as mean ± SEM. One-way ANOVA was used to evaluate the statistical differences between constructs, with * indicating P < 0.05. (B and C) Univariate analysis of clustering of the expressed constructs is shown as raw nanoclustering curves (B) and Lmax data summary (C). In the data summary in C, data are shown as mean ± SEM. Bootstrap tests examined the statistical significance between constructs, with * indicating P < 0.05. (D) Unlabeled version of Fig. 1 D showing a sonicated plasma membrane sheet containing immunogold labeled CAV1-mCherry (2-nm gold) and Cavin1-GFP (6-nm gold). Scale bar, 500 nm. . (E) The averaged cumulative bivariate coclustering function Lbiv(r) − r, as well as the packing density function g(r), for GFP-LactC2 and RFP-CAV1 are plotted against distance r in nanometers..
We further characterized the features of this putative noncaveolar CAV1 pool. Single-molecule tracking of CAV1 tagged with monomeric Eos2 showed that CAV1 expressed alone in MCF7 cells showed greater mobility than CAV1 expressed in MDCK cells that have endogenous caveolae. Analysis of the mean square displacement (MSD) of CAV1-mEos2 mobility revealed an increase in confinement in MDCK cells as shown by the MSD curves and the area under the MSD curves (Fig. S1 C). These results are consistent with the reported effect of cavin1 knockdown on CAV1 diffusion as shown by FRAP (Hill et al., 2008) and super-resolution light microscopy (Khater et al., 2019). APEX staining for YFP-CAV1 was localized to discrete patches of the PM, in areas enriched in vesicles and tubular profiles (Fig. S1, D and E), and was also associated with pits and vesicles of varying diameter (e.g., Fig. S1 F). This pattern of staining was quite distinct from the restriction of APEX labeling to structures with caveolar morphology in cells cotransfected with cavin1 (Fig. S1 G).
Caveolae show a distinct lipid profile
Having established a system in which we can assess the surface distribution of individually expressed CAV1 and cavin1 as well as the two proteins coexpressed (caveolae), we then used immunogold EM spatial mapping to quantify the nanoscale organization of the CAV1 and cavin1 domains. This system allows an unbiased quantitative analysis of the clustering of a particular type of proteins (univariate analysis) or their association with other proteins (bivariate analysis).
For assessment of caveolar localization in cells expressing both CAV1 and cavin1, labeling of CAV1 was used as the caveolar marker. CAV1, expressed alone or with cavin1 in MCF7 cells, labeled domains of ∼40-nm radius (Fig. S2 B). The lateral univariate clustering of the gold labeling within a select 1-µm2 PM area was calculated using the Ripley’s univariate K-function analysis. The extent of nanoclustering, L(r) − r, was plotted as a function of the cluster radius, r, in nanometers (Fig. S2 B). The L(r) − r value of 1 indicates the 99% confidence interval (C.I.), the values above which indicate statistically meaningful clustering. The peak L(r) − r value is termed Lmax and summarizes nanoclustering statistics. A larger Lmax indicates more extensive nanoclustering. The spatial analysis showed a more restricted cluster size for CAV1 when expressed with cavin1 than when expressed alone (Fig. S2 B).
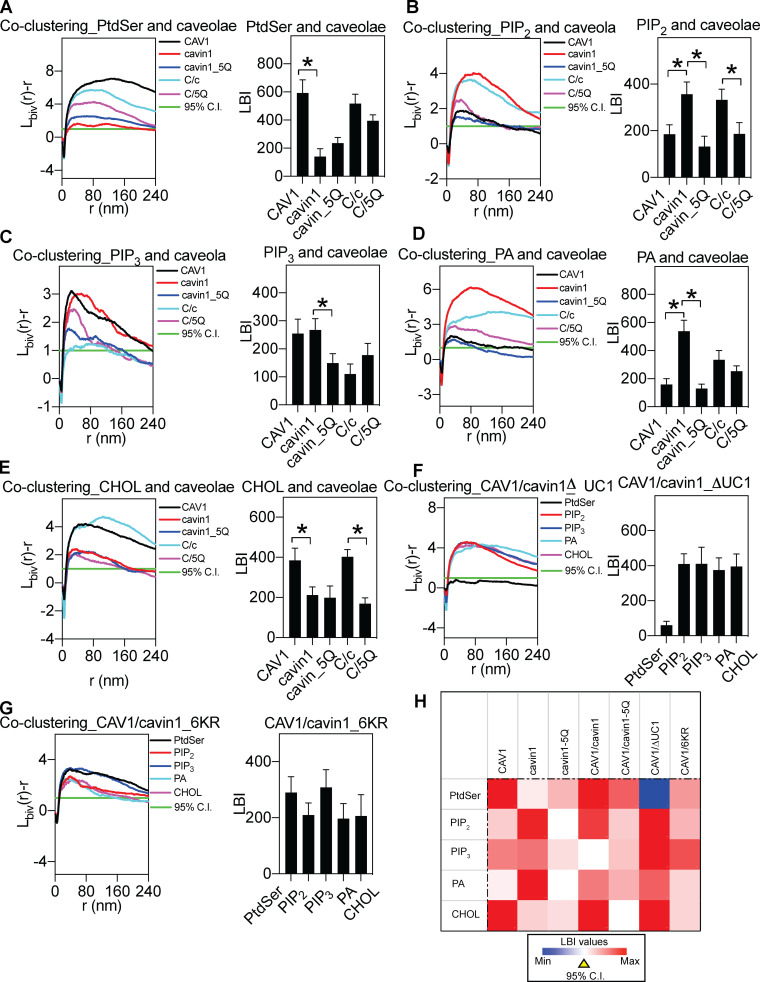
We next sought to determine the lipids colocalizing with the singly expressed proteins and the CAV1/cavin1 complex. PM sheets were prepared from MCF7 cells coexpressing RFP-labeled caveolar proteins together with a lipid-binding domain: GFP-Lact-C2, GFP-PH-PLCδ, GFP-Spo20, GFP-PH-AKT, or GFP-D4H that binds to PtdSer, PtdIns(4,5)P2, phosphatidic acid (PA), phosphoinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3], or cholesterol, respectively. PtdIns(3,4,5)P3 was also detected by a second probe, a GFP-tagged pleckstrin homology (PH) domain of ADP-ribosylation factor nucleotide-binding site opener (ARNO-PH; Fig. S3, A and B) to avoid problems with recognition of PtdIns(3,4)P2 by the PH-AKT probe. PM sheets were colabeled with anti–RFP-2nm-gold and anti–GFP-6nm-gold. The intact PM sheets were imaged using transmission EM (TEM) at a magnification of 100,000×. The lateral colocalization of the two populations of gold particles within a select 1-µm2 PM area was calculated using the Ripley’s bivariate K-function. The extent of coclustering, Lbiv(r) − r, was plotted as a function of the cluster radius, r (Fig. 2 A). The Lbiv(r) − r value of 1 indicates the 95% C.I., the values above which indicate statistically meaningful colocalization. Area under the curve between the r values of 10 and 110 nm yields L-function–bivariate integrated (LBI), which summarizes the extent of colocalization. Larger LBI values indicate more extensive colocalization, with an LBI value of 100 as the 95% C.I.
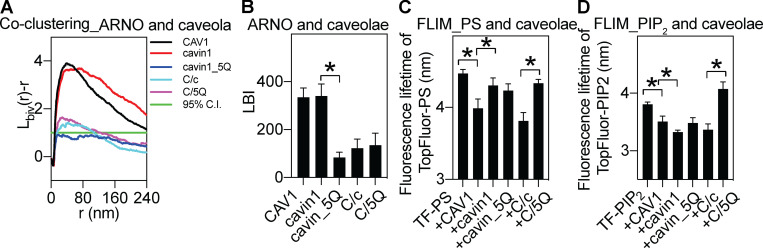
Figure S3.
Selective lipid sorting of caveolae proteins. (A) Bivariate coclustering analysis between GFP-ARNO-PH domain [specifically tagging PtdIns(3,4,5)P3] and an RFP-tagged caveolae protein on intact PM sheets of MCF7 cells. (B) LBI values (a parameter of coclustering) between GFP-ARNO-PH domain and RFP-caveolae protein are calculated from the bivariate curves shown in A. (C and D) Fluorescence lifetime of TopFluor-PtdSer (TF-PS; C) and fluorescence lifetime of TopFluor-PtdIns(4,5)P2 (TF-PIP; D) were measured in MCF7 cells expressing empty vector pC1 or an RFP-tagged caveolae protein. Data are shown as mean ± SEM, with * indicating statistical significance (P < 0.05).
Figure 2.
Lipid mapping of caveolar components expressed in a model caveola-deficient cell system. (A–E) Bivariate clustering analyses of the indicated caveolar proteins and lipid-binding probes coexpressed in MCF7 cells. 5Q, cavin1.5Q; C/c, CAV1/cavin1; C/5Q, CAV1 plus cavin1.5Q. (F and G) Bivariate analyses between the GFP-tagged lipid probes and the RFP-tagged mutants delta UC1 (F) or 6KR (G) were calculated. (H) Bivariate clustering heatmap summary. In data summary panels, data are shown as mean ± SEM. Bootstrap tests examined the statistical differences between groups, with * indicating P < 0.05.
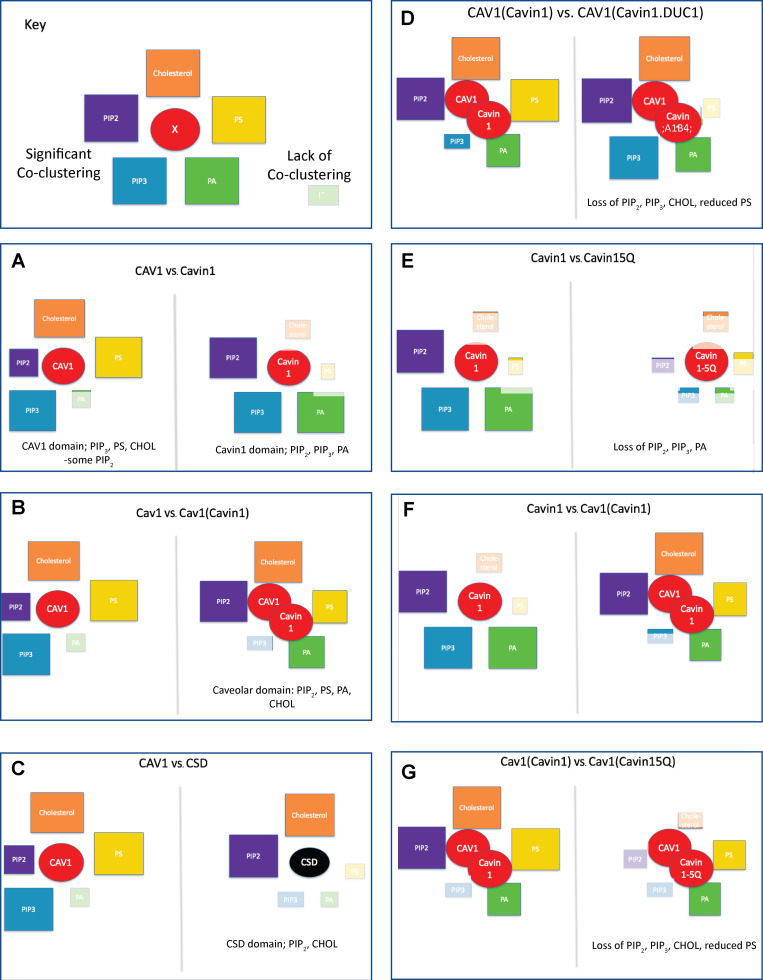
Singly expressed CAV1 showed statistically significant association with PtdSer, PtdIns(3,4,5)P3, and cholesterol (Fig. 2, A–E). In contrast, cavin1 expressed alone preferentially coclustered with PtdIns(4,5)P2, PtdIns(3,4,5)P3, and PA. This indicated that CAV1 and cavin1 sort different PM lipids. We next coexpressed CAV1 with cavin1 and monitored CAV1 association with the same set of lipids. CAV1 expressed with cavin1 shows a quantitatively distinct lipid association profile to CAV1 expressed alone; association with PtdIns(3,4,5)P3 was decreased, but association with PtdIns(4,5)P2 and PA was significantly increased (for summary schemes, see Fig. 6). We note that Lbiv(r)−r values are above the 95% C.I. at long distance (>200 nm) when the peak of the curves are large. This is likely due to the cumulative function of Ripley’s bivariate K-function. As an example shown in Fig. S2 E, the packing density g(r) for GFP-LactC2 and RFP-CAV1 shows a sharp peak at a shorter distance, while the cumulative Lbiv(r)−r curve for the same images shows a tall and broad peak. Taken together, CAV1/cavin1 alone or CAV1 and cavin1 combined each sort distinct sets of lipids.
Figure 6.
Schematic comparison of the lipid profiles associated with the indicated proteins or protein pairs (circles). Colors of square boxes indicate lipid species; the size of the box indicates the level of coclustering with the indicated proteins (faded boxes indicate statistically insignificant association).
As an independent test of the colocalization of various phospholipids and caveola components in intact cells, we performed fluorescence lifetime imaging combined with fluorescence resonance energy transfer (FLIM-FRET). Specifically, we spike-labeled MCF7 cells expressing an RFP-tagged caveolae construct with a fluorescent phospholipid such as TopFluor-PtdSer or TopFluor-PtdIns(4,5)P2. The TopFluor moiety is covalently attached to the sn-2 chains of lipids and has an excitation peak of 495 nm and an emission peak of 503 nm. FRET from the TopFluor moiety to RFP can be used to monitor association of the fluorescent lipids with coexpressed RFP-tagged proteins (Zhou et al., 2014; Zhou et al., 2017). The fluorescence lifetime of TopFluor-PtdSer was ∼4.5 ns in MCF7 cells expressing an empty vector pC1 and was decreased to ∼3.99 ns in MCF7 cells expressing RFP-CAV1 (Fig. S3 C), suggesting efficient energy transfer between the TopFluor moiety and RFP. The fluorescence lifetime of TopFluor-PtdSer was the lowest, at ∼3.8 ns, in MCF7 cells expressing RFP-tagged CAV1 and cavin1, significantly lower than cells expressing RFP-cavin1, RFP-cavin1-5Q, and a combination of RFP-CAV1 and RFP-cavin1-5Q. These FLIM data are entirely consistent with our EM measurements above. We also performed similar FLIM-FRET experiments using TopFluor-PtdIns(4,5)P2. As shown in Fig. S3 D, the changes in fluorescence lifetime values are entirely consistent with the EM data using the PtdIns(4,5)P2 probe, PH-PLCδ domain. Thus, association of fluorescent lipids with individual or coexpressed caveola proteins as monitored by FLIM-FRET shows a similar pattern to that observed with expressed lipid-binding domain proteins as measured by quantitative EM.
The caveolin scaffolding domain (CSD) contributes to the selective lipid sorting of caveolae
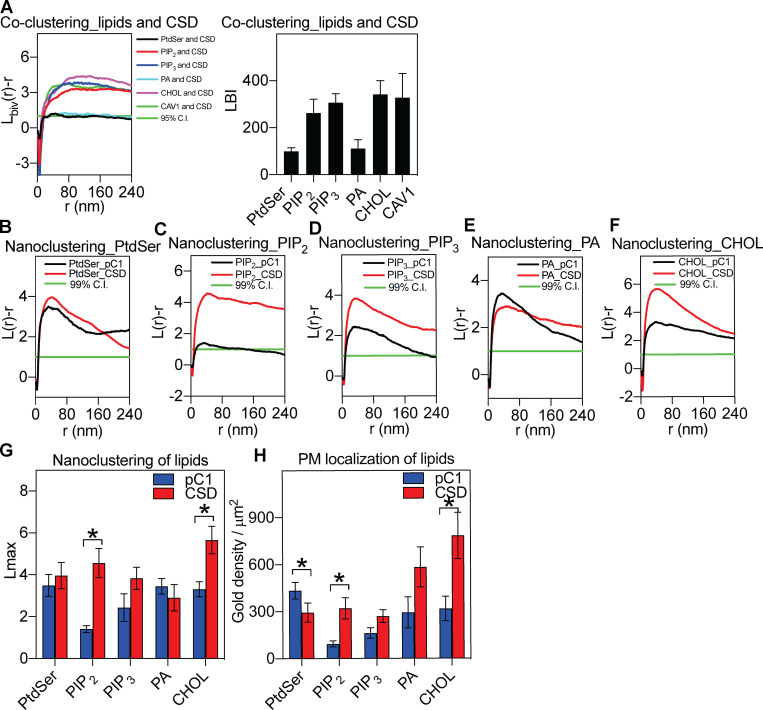
To dissect the properties of CAV1 that could dictate association with specific lipid domains, we focused on the conserved membrane proximal region of CAV1 termed the CSD (Fig. 1 A and Fig. 3). The CSD is a potent and specific inhibitor of endothelial nitric oxide synthase activity in vitro and in vivo (García-Cardeña et al., 1997; Bucci et al., 2000; Fulton et al., 2002; Yu et al., 2006) and has been reported to have membrane-binding activity (Epand et al., 2005; Wanaski et al., 2003). We first examined the specific lipid profile (probed by the GFP-tagged lipid-binding domains) of the expressed RFP-CSD in an EM-bivariate colocalization analysis. The RFP-CSD showed significant PM labeling and coclustered with PtdIns(4,5)P2, PtdIns(3,4,5)P3, and cholesterol (Fig. 3 A). Unlike full-length CAV1, however, the CSD alone showed no significant association with PtdSer (Fig. 3 A).
Figure 3.
The CSD shows a distinct lipid specificity, colocalizes with CAV1, and affects PM lipid organization. (A) Bivariate analysis: CSD coclusters with lipids and CAV1 (the left panel indicates raw data and the right panel shows the bivariate coclustering summary). (B–F) CSD affects PtdIns(4,5)P2, PtdIns(3,4,5)P3, and cholesterol nanoclustering. (G) Lipid univariate clustering summary. (H) CSD affects surface levels of PS and cholesterol. Data summaries in A, G, and H show mean ± SEM. Bootstrap tests examined the statistical differences between conditions in the spatial distribution data in A and G, with * indicating P < 0.05. For gold density in H, one-way ANOVA was used to evaluate the statistical differences, with * indicating P < 0.05.
In view of the association of the CSD with distinct lipid domains and the profound effects on signal transduction (García-Cardeña et al., 1997; Bucci et al., 2000), we hypothesized that the CSD might affect nanoscale lipid organization. To test this, the RFP-tagged CSD was coexpressed with the suite of GFP-tagged lipid markers, and their spatial distribution was quantified via the univariate spatial analysis. Expression of the CSD increased the clustering of PtdIns(4,5)P2 and cholesterol (Fig. 3, B–G). Counting the gold particles within the same 1-µm2 PM area estimates the surface density of the lipid probes. The presence of the CSD domain increased levels of PtdIns(4,5)P2 and cholesterol but caused a significant decrease in PtdSer (Fig. 3 H). These results indicate that the caveolin CSD colocalizes with a distinct set of PM lipids and can have profound effects on surface spatial organization and levels of PM lipids in distinct manners.
Cavin1 HR1 and UC1 domains contribute distinct lipid-sorting capabilities
We next focused on the other key structural protein, cavin1, and first investigated the role of the conserved PtdIns(4,5)P2-binding site in the N-terminal helical domain (HR1; residues 106–135; Kovtun et al., 2014). Substitution of five key positively charged amino acids to glutamines within the HR1 domain (5Q; Fig. 1 A) disrupts binding of purified cavin1 to PtdIns(4,5)P2 in liposome-binding assays. We therefore tested whether the coclustering of specific lipids with cavin1 or with the CAV1/cavin1 complex would be affected by the 5Q substitutions in parallel bivariate lipid mapping analysis. The 5Q substitutions in cavin1 caused a dramatic change in the association with PtdIns(4,5)P2, PtdIns(3,4,5)P3, and PA (Fig. 2, A–E). Coexpression of CAV1 and cavin1-5Q showed a decrease in association of PtdIns(4,5)P2, as well as a loss of cholesterol association, when compared with CAV1 expressed with wild-type cavin1 (heatmap in Fig. 2 H). As shown in the heatmap in Fig. 2 H and the scheme in Fig. 6, the net result of the loss of PtdIns binding through the 5Q site extended to the entire tested lipid network.
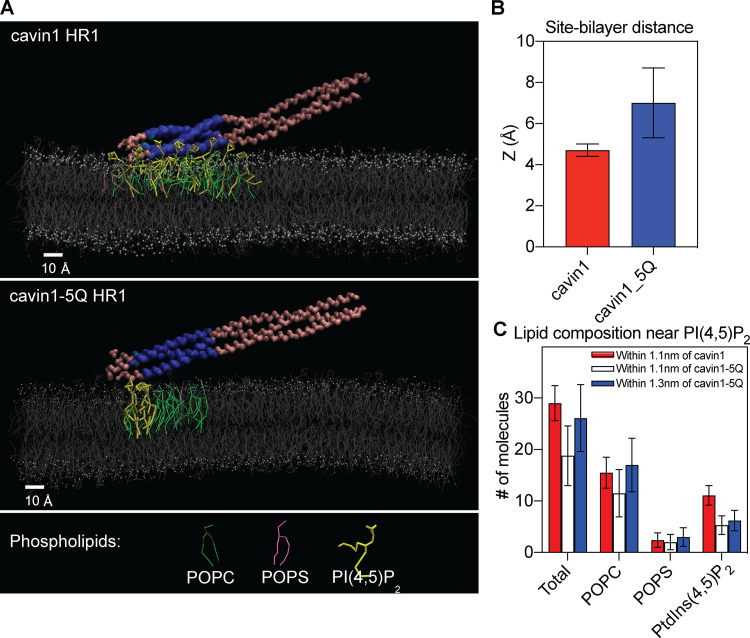
To further characterize the potential membrane lipid interactions of cavin1, we performed coarse-grained molecular dynamics simulations using the MARTINI 2.2 force field (de Jong et al., 2013; Marrink et al., 2007; Monticelli et al., 2008). The cavin1 HR1 domain trimer and its 5Q mutant were modeled using Protein Data Bank (PDB) structure 4QKV (Kovtun et al., 2014) as the initial configuration, and the interaction of each protein with a bilayer composed of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine), and PtdIns(4,5)P2 in an 80:15:5 ratio was observed over four runs of 3 µs each. In these simulations, the PtdIns(4,5)P2-binding HR1 invariably approached the bilayer within hundreds of nanoseconds, even while the interaction between the rest of the protein and the bilayer was more variable for both cavin1-HR1 and cavin1–5Q HR1 (Fig. 4 A) and settled within a few angstroms of the lipid bilayer. The proteins maintained their trimer tertiary structures, and the overall structure of the lipid bilayer was not perturbed by the approach and association of either the wild-type or mutant HR1 domain. We thus took statistics over the last 2 µs of each simulation run to quantify the interaction between the lipid bilayer and the HR1 domain. The distance between wild-type HR1 and the bilayer was consistently 4–5 Å (as the minimum distance between any protein particle and any lipid particle), while the distance between the 5Q mutant site and the bilayer was around 7 Å with considerably larger fluctuations (Fig. 4 B). This is consistent with the diminished electrostatic interactions between the 5Q mutant and the charged bilayer.
Figure 4.
Coarse-grained molecular simulations of cavin1 HR1 and cavin1–5Q HR1 interacting with a model lipid bilayer. (A) Side view of typical lipid bilayer–interacting configurations for cavin1 HR1 (top) and (bottom) cavin1–5Q HR1 (bottom). Highlighted lipids are within 1.1 nm of the cavin1 interaction site (blue) or within 1.3 nm of the cavin1-5Q interaction site (blue). (B) Minimum distance (Z in angstroms) between the closest atoms of the peptides and lipids is compared between the wild-type HR1 and the 5Q mutant. (C) Total number of lipid molecules were counted within 11 Å and 13 Å of the nearest atoms of the peptides. Data summaries in B and C are shown as mean ± SEM.
The lipid composition near either the wild-type HR1 or the 5Q mutant site was then studied by counting all lipids with particles within 11 Å of any protein particles, where the cutoff was chosen to be approximately twice the minimum distance between the wild-type HR1 and the lipid bilayer. On average, there were 11 PtdIns(4,5)P2 molecules within 11 Å of the wild-type HR1 and only five PtdIns(4,5)P2 molecules within 11 Å of the 5Q mutant (Fig. 4 C), suggesting that the wild-type cavin1 HR1 interacts strongly with PtdIns(4,5)P2 and concentrates it far above the bulk concentration (of 5% in this case) and the association of PtdIns(4,5)P2 with the 5Q mutant is correspondingly decreased. PtdSer association was similar between the wild-type and the 5Q mutant. There are more POPC molecules within 11 Å of the wild-type HR1 than the 5Q mutant, possibly attributable to the larger overall distance of the 5Q mutant from the lipid bilayer. When the lipid interaction cutoff was expanded to 13 Å, both the total number of lipids and the number of POPC molecules interacting with the 5Q mutant were similar to the wild-type, but the number of PtdIns(4,5)P2 molecules near the 5Q mutant (six on average) was still significantly lower than the wild-type. Similar trends [of PtdIns(4,5)P2 clustering near the wild-type HR1 but not the 5Q mutant] were seen when the interaction cutoff was defined as short as 7 Å, below which almost no lipids interacted with the interaction site, and as large as 15 Å, above which the bulk composition of the bilayer started to obscure interaction site-specific effects. As such, coarse-grained molecular dynamics modeling supports the observations that the cavin1 HR1 domain has a PtdIns(4,5)P2-specific interaction with membranes, and this specific interaction is significantly diminished in the 5Q mutant.
We next tested the role of a UC1 domain of cavin1 that has been shown in vitro to bind PtdSer (Tillu et al., 2018). We generated two cavin UC1 domain mutants: deletion of the entire UC1 domain (deltaUC1) and replacing lysines and arginine residues within the UC1 domain by aspartic acid and glutamate (6KR; Fig. 1 A). CAV1 was coexpressed with each cavin1 UC1 mutant in MCF7 cells for another set of lipid-mapping analysis. The deltaUC1 mutation completely ablated coclustering with PtdSer and increased coclustering with PA and PtdIns(3,4,5)P3 without affecting PtdIns(4,5)P2 and cholesterol (Fig. 2 F). Cavin1-6KR mutation also decreased PtdSer coclustering, but with less impact on other lipids than complete deletion of the UC1 domain (Fig. 2 G). Interestingly, substitution of the acidic residues in the UC1 domain also caused a reduction in cholesterol coclustering (Fig. 2 G). These results suggest that the specific lipid environment of caveolae is generated synergistically by both caveolins and by cavins. In vitro studies showing changes in PtdIns(4,5)P2 or PtdSer binding in vitro translate into remarkable changes in lipid association in cells. Moreover, loss of binding to these specific lipids has indirect effects on cholesterol association with CAV1/cavin1 domains to alter the entire lipid environment, showing a cooperativity in lipid recruitment to caveolae.
Association of caveolin and cavin1 requires distinct PtdSer species with unique acyl chain composition
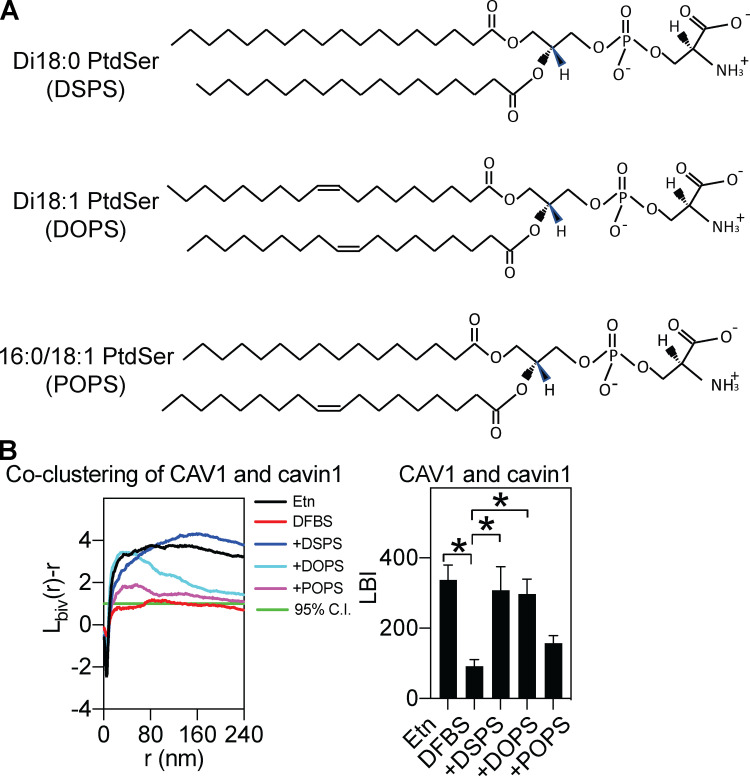
In view of the PtdSer-binding specificity via the cavin UC1 domain that is required for caveola stability and the role of PtdSer in caveola formation (Hirama et al., 2017), we next analyzed whether PtdSer levels determined CAV1/cavin1 association. PSA3 cells generate less endogenous PtdSer when grown in medium containing dialyzed FBS (DFBS), but PtdSer can be restored to near control levels by 10 µM ethanolamine (Etn) supplementation (Lee et al., 2012; Liang et al., 2019; Plowman et al., 2005; Zhou et al., 2014; Zhou et al., 2017; Zhou et al., 2015). PSA3 cells cultured under different conditions to modulate their PtdSer content were transfected with CAV1 and cavin1 for a bivariate colocalization analysis. Negligible CAV1/cavin1 association was observed in PSA3 cells depleted of PtdSer (DFBS; Fig. 5). In contrast, cells grown in Etn with near-native levels of PtdSer showed highly significant association of CAV1 and cavin1. In PtdSer-depleted PSA3 cells, we next acutely added synthetic PtdSer species (Fig. 5 A). Addback of fully saturated DSPS (1,2-distearoyl-sn-glycero-3-phospho-L-serine; di18:0) and monounsaturated DOPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine; di18:1) was highly effective in driving CAV1/cavin1 association. In contrast, the mixed chain POPS (16:0/18:1) was ineffective (Fig. 5 B). These results show that the association of caveolins and cavins is dependent on specific PtdSer acyl chain compositions.
Figure 5.
CAV1/cavin1 association is dependent on PS. (A) Structure of different synthetic PS species used in this study. (B) CAV1/cavin1 association is negligible in cells deficient in PS but is restored in cells cultured in Etn. Different PS species have distinct abilities to mediate CAV1/cavin1 association. Fully saturated DSPS and monounsaturated DOPS are effective at driving CAV1/cavin1 association, but mixed-chain POPS is ineffective. The left panel shows the raw data while the right panel shows data summaries. In the summaries, data are shown as mean ± SEM. Bootstrap tests examined the statistical differences between conditions, with * indicating P < 0.05.
Discussion
This study provides the first quantitative picture of the lipid environment associated with the cytoplasmic face of caveolae in situ (for schematic summary, see Fig. 6). CAV1 and cavin1 in this model system generate caveolae significantly enriched in PtdIns(4,5)P2, PtdSer, and cholesterol. This environment is quantitatively distinct from that seen with expression of CAV1 alone [PtdSer, cholesterol, PtdIns(3,4,5)P3, lower PtdIns(4,5)P2] or cavin1 alone [PtdIns(3,4,5)P3, PtdIns(4,5)P2, PA]. Although we have tested a cohort of anionic lipids, our findings show that the selective lipid sorting by caveolae is clearly mediated by more than electrostatics alone. This is indeed supported by our spatial analysis testing different PtdSer species with the same charged headgroup but distinct acyl chains. Thus, caveolae are dependent on the local lipid environment established by a complex set of interactions involving both surface charges of lipid headgroups and packing integrity of their acyl chains. This is also consistent with the mechanical nature of caveolae, where the structural integrity of caveolae depends on membrane tension.
Through a combination of mutational and coexpression studies, we can start to further define the mechanisms and molecular determinants involved in generating these distinct lipid profiles. The CSD has been extensively characterized as an inhibitor of signal transduction pathways in vitro and in vivo (García-Cardeña et al., 1997; Bucci et al., 2000), but the mechanisms involved remain controversial (Byrne et al., 2012; Collins et al., 2012). The isolated domain has lipid-binding activity (Wanaski et al., 2003; Epand et al., 2005). In the full-length protein, the CSD has been suggested to be at least partially buried within the membrane of caveolae (Ariotti et al., 2015b). In mammalian cells, the CSD region of CAV1 is accessible to antibodies within the Golgi complex but not in caveolae unless cholesterol is depleted from the PM (Pol et al., 2005). We now show that the CSD itself has the ability to cocluster with cholesterol and PtdIns(4,5)P2 when expressed in isolation. Moreover, expression of the CSD has a striking effect on the organization of the PM lipids, increasing the nanoclustering and surface levels of cholesterol and PtdIns(4,5)P2 while significantly decreasing the surface PtdSer levels. These distinct lipid-sorting capabilities may impact numerous signaling pathways.
We also show that two distinct lipid-binding sites in cavin1 contribute to the selective lipid sorting of caveolae. The widespread changes in the coclustered lipids induced by changing these sites, which confer specific lipid-binding activity in vitro, emphasize that the lipid composition of the domain is not generated by single binding interactions but cooperative interactions between multiple lipid-binding interfaces and membrane biophysical properties. This is also validated in our molecular dynamics simulations. These findings are consistent with studies on other lipid-binding proteins, such as the BAR domain protein Bin1 and other phosphoinositol (PI)-interacting proteins (Picas et al., 2014) showing that, rather than 1:1 lipid interactions, a PtdIns(4,5)P2-binding domain of higher stoichiometry is generated by localized electrostatic interactions with the protein. This generates a PtdIns(4,5)P2 domain with higher capacity to bind other PI-binding proteins (in the above example, dynamin2). A similar principle can apply to caveolae; the high concentration of CAV1 and cavin1, both of which form oligomers, interacts with lipids through multiple interaction interfaces in a cooperative fashion to generate a unique lipid profile. Loss of just one of these interfaces, in the case of PtdIns(4,5)P2-binding site, can change the entire complement of associated lipids. Moreover, the role of the CAV1/cavin1 complex in generating the lipid domain is not simply through increased association of specific lipids; for example, comparison of PtdIns(3,4,5)P3 coclustering across the different combinations of expressed caveolar proteins shows that only with WT CAV1/cavin1 is there a complete exclusion (negligible coclustering with CAV1) of PtdIns(3,4,5)P3 (Fig. 2 and Fig. 6). This finding again highlights that lipid sorting by caveolae is not exclusively mediated by headgroup electrostatic interactions. In addition to the extensive surface charges carried by PtdIns(3,4,5)P3, the overall packing characteristics of PtdIns(3,4,5)P3 lipids also contribute to how well these highly charged lipids can pack within caveolae. This concept is supported by our findings using different PtdSer species. Despite having the same charged headgroup, these different PtdSer species display distinct abilities to mediate packing of caveolae components.
Here, we have focused on the lipid domain generated in the cytoplasmic leaflet of caveolae by cavins and caveolins. However, we suggest that the well-documented transbilayer coupling between lipids such as PtdSer and extracellular leaflet lipids (Raghupathy et al., 2015) can contribute to the generation of the unique glycosphingolipid composition of caveolae.
These findings point to a highly cooperative process of caveola formation involving caveolin, cavin, and lipids. This is further supported by analysis of the lipid requirements for CAV1/cavin association, focusing on PtdSer. We now show that CAV1/cavin1 association is dependent on specific PtdSer species in intact PM sheets of MCF7 cells. In vitro validation using purified caveola components reconstituted in synthetic liposomes of different lipid constituents will be needed to further resolve the mechanisms involved. Protein–protein interactions can contribute to this association, but in this model system, this is not sufficient to generate the CAV1/cavin1 domain required for caveola formation. We envisage a coincidence detection mechanism in which multiple low-affinity interactions, protein–lipid and lipid–lipid, contribute to formation of the caveolar domain. Analogous to BAR domain association with PtdIns(4,5)P2, the arrangement of the membrane lipids embedded in the bilayer and the lipid-binding interfaces of the oligomeric protein complexes may be crucial. Only fully saturated DSPS (di18:0) and monounsaturated DOPS (di18:1), but not mixed chain POPS (16:0/18:1), were compatible with CAV1/cavin1 association, implicating the lipid’s fatty acyl chains in facilitating these interactions.
In mammalian cells, the mixed-chain PtdSer species comprise the majority of the PtdSer species. In particular, POPS is highly abundant at ∼40% of all PtdSer species, whereas the saturated PtdSer species is much less abundant. However, this does not mean these saturated PtdSer species are less biologically important. For instance, PtdIns(4,5)P2 comprises <1% of total PM lipid content but plays critical biological roles and binds to a wide variety of membrane-associating proteins. Thus, locally concentrated populations of a less abundant saturated PtdSer species within caveolae should contribute to the structural integrity of membrane architectures. It has been proposed that different PtdSer species vary in their response to changes in PM curvature (Liang et al., 2019). Whether lipid packing geometry can contribute to caveolar architecture must await testing in a reconstituted liposome system using purified caveolae components.
These results not only have implications for understanding the formation of caveolae but also for their function. Although we focused mostly on anionic phospholipids here, our findings strongly suggest that the selective lipid sorting by caveolae is not simply electrostatic. This is more evident in our testing of different PtdSer species with the same charged headgroup but distinct acyl chains. This is consistent with the flattening of caveolae in response to membrane tension changes (Sinha et al., 2011) or other stresses (McMahon et al., 2019) that has emerged as a crucial aspect of caveolar biology. Our results suggest that the caveola domain is stabilized by multiple synergistic low-affinity interactions, dependent on specific lipids rather than stable protein–protein interactions and can therefore be considered a metastable domain poised to disassemble. Loss of caveolae through genetic ablation or treatments that cause acute flattening induces changes in the nanoscale lipid organization of the PM (Ariotti et al., 2014). Key lysine residues that form the PtdIns binding site of cavin1 become ubiquitinated upon caveola disassembly, suggesting their decreased interaction with PtdIns(4,5)P2 and changing the local lipid environment as cavins dissociate into the cytosol. By comparing the properties of CAV1 expressed alone with CAV1/cavin1 coexpressed together, we showed that noncaveolar CAV1, recently termed the CAV1 scaffold (Khater et al., 2019), diffuses more rapidly in the absence of cavin1, as monitored by single-molecule tracking, and has a distinct complement of associated lipids. CAV1 domains are enriched in PtdIns(3,4,5)P3 and are relatively depleted of PtdIns(4,5)P2 and PA (Fig. 2) compared with the CAV1/cavin1 (caveolar) domain. Caveola disassembly may thus release particular lipid types into the bulk membrane and influence the properties of the bulk membrane, which may contribute to the modulation of Ras signaling (Ariotti et al., 2014) or actin organization at the junctional caveolae enriched with PtdIns(4,5)P2 (Teo et al., 2020). In view of the high concentration of cavin proteins associated with each caveola (estimated as 50–80; Gambin et al., 2014) and the proposed number of cavin-associated PtdIns(4,5)P2 molecules per cavin trimer (Fig. 4), as suggested from our coarse-grain simulations, disassembly of caveolae could change the accessibility of a considerable pool of PI lipids. Defining the lipid components that associate with caveolae and the mechanisms that dictate their association with, and dissociation from, caveolae will be crucial to understanding how caveolae function but will also provide general insights into the interactions between protein complexes and the hundreds of membrane lipid species required for cellular function.
Materials and methods
Cell culture and transfection
MCF7 and Madin-Darby canine kidney (MDCK) cells were obtained from American Type Culture Collection and were regularly mycoplasma tested. MCF7 cells were checked by Western blotting for lack of CAV1 (note that unlike the line used here, some strains of MCF7 cells do express low levels of CAV1). MDCK cells were grown in DMEM/F-12 (Life Technologies) supplemented with 10% FBS and 2 mM L-glutamine. MCF7 cells were grown in DMEM supplemented with 10% FBS and 2 mM L-glutamine. CAV1 and cavin1 were cloned as described in Hill et al. (2008). The CSD construct (Chaudhary et al., 2014) and cavin constructs (Kovtun et al., 2014; Tillu et al., 2018) were described previously. Tagged constructs were transfected using Lipofectamine 2000 reagent (Life Technologies) following the manufacturer’s instructions using a 1:3 ratio of DNA:Lipofectamine. All transient transfections were performed with Lipofectamine 2000 per the manufacturer’s instruction. PSA3 cells were a generous gift from Dr. Hiroyuki Arai at the University of Tokyo, Tokyo, Japan. PSA3 cells were cultured in DMEM containing 10% DFBS without/with 10 µM Etn for 72 h before experiments.
Acute exogenous PtdSer addback
Synthetic PtdSer species and fluorescent TopFluor-PtdSer or TopFluor-PtdIns(4,5)P2 were purchased from Avanti Polar Lipids, Inc., dissolved in chloroform, and kept in nitrogen at −20°C. The appropriate amount of lipid/chloroform solution was transferred to a glass vial (for a final working concentration of 10 µM) using a Hamilton syringe. Chloroform was evaporated by purging with nitrogen and was kept under vacuum overnight in the dark to eliminate residual chloroform. The dried PtdSer film was rehydrated in DMEM containing 10% DFBS and sonicated for 20 min in a bath sonicator. For TopFluor-PtdSer and TopFluor-PtdIns(4,5)P2, the lipid film was rehydrated in ice-cold phosphate buffer solution containing 3% BSA before sonication.
EM
Standard TEM using ruthenium red and immunoelectron microscopy on frozen sections using antibodies to CAV1 followed by 10 nm protein A-gold were performed as described previously (Hill et al., 2008). Sonication of MCF7 cells to generate PM lawns was performed exactly as described (Gambin et al., 2014). Briefly, cells were plated on poly-L-lysine–coated 35-mm plastic dishes, and then a probe sonicator was used to generate basal PM sheets adhered to the substratum. Cells were then fixed and labeled before embedding in resin and sectioning close to the substratum. Localization of YFP-CAV1 using a nanobody-based APEX2 approach was as described previously (Ariotti et al., 2015a).
Quantitative spatial analysis
Univariate analysis
The univariate analysis calculates the spatial distribution of a single population of gold nanoparticles within a select PM area (Zhou et al., 2017; Zhou et al., 2015). The GFP-tagged proteins/peptides of interest on intact PM sheets were attached to copper EM grids, fixed with 4% PFA and 0.1% glutaraldehyde, and immunolabeled with 4.5 nm gold coupled to anti-GFP antibody and negative-stained with uranyl acetate. Gold nanoparticles were imaged with TEM at 100,000× magnification. ImageJ was then used to assign x, y coordinates for each gold particle. The spatial distribution of gold particles within a selected 1-µm2 area was calculated using Ripley’s K-function, which tests a null hypothesis that all points in the analyzed area are randomly distributed:
| (1) |
and
| (2) |
where K(r) describes the univariate K-function for the total number of gold particles, denoted as n, in a selected area of A; r denotes the distance between 1 and 240 nm with an increment of 1 nm. || · || characterizes Euclidean distance, with 1(·) as the indicator function. We define 1(·) = 1 if ||xi − xj|| ≤ r and 1(·) = 0 if ||xi − xj|| > r. We incorporate a parameter of wij−1 to achieve an unbiased edge correction. For a circle with xi at the center and ||xi − xj|| as the radius, wij−1 defines the proportion of the circumference of the circle. L(r) − r is a linear transformation of K(r) and is normalized against the 99% C.I. calculated from Monte Carlo simulations. A L(r) − r value of 0 describes a complete random pattern of particles distribution, while a L(r) − r value above the 99% C.I. of 1 illustrates a statistically meaningful clustering pattern. For each condition, at least 15 PM sheets were imaged and analyzed. Statistical significance between different conditions was evaluated by comparing our calculated distribution patterns against 1,000 bootstrap samples in bootstrap tests (Zhou et al., 2017; Zhou et al., 2015).
Bivariate coclustering analysis
Coclustering between two different-sized gold particles was calculated using the bivariate K-function coclustering analysis (Zhou et al., 2017; Zhou et al., 2015). The GFP- and RFP-tagged proteins on intact PM sheets attached to EM grids were coimmunolabeled with 2-nm gold linked to anti-RFP antibody and 6-nm gold linked to anti-GFP antibody, respectively. As above, the x, y coordinates of gold particles were assigned using ImageJ. Bivariate K-function analysis then calculated the colocalization between 6- and 2-nm gold populations. The null hypothesis of this bivariate analysis is that the two gold populations spatially separate from each other (Eqs. 3–6):
| (3) |
| (4) |
| (5) |
and
| (6) |
where Kbiv(r) denotes the bivariate estimator containing two distinct bivariate K-functions: Kbs(r) calculates the distribution of all the 6-nm gold particles (b = big gold) with respect to each 2-nm small gold (s = small gold), and Ksb(r) calculates the distribution of all the 2-nm gold with respect to each 6-nm gold. The total number of 6-nm big gold is termed nb, while the total number of 2-nm small gold is ns. Other notations for Eqs. 3–6 still follow the same description as in Eq. 1 and Eq. 2. Lbiv(r) − r is a linear transformation of Kbiv(r) and is further normalized against the 95% C.I. An Lbiv(r) − r value of 0 indicates lateral separation between 6-nm/2-nm gold particles. On the other hand, Lbiv(r) − r values above the 95% C.I. of 1 indicate statistically meaningful colocalization.
To better summarize the colocalization data, we integrated the Lbiv(r) − r curves within a fixed range 10 < r < 110 nm and termed the parameter as bivariate Lbiv(r) − r integrated (or LBI):
| (7) |
For each test, at least 15 PM sheets were imaged and analyzed. The same bootstrap tests used above in the univariate analysis were also used here to evaluate the statistical significance between tests (Zhou et al., 2017; Zhou et al., 2015).
Single-molecule tracking and quantitation
Single-particle tracking photoactivated localization microscopy of MCF and MDCK cells transfected with Cav1-mEos2 was performed on the Roper Scientific total internal reflection fluorescence (TIRF) microscope equipped with an iLas2 double-laser illuminator (Roper Technologies), a CFI Apo TIRF 100× (1.49 NA) objective (Nikon), and an Evolve512 delta EMCCD camera (Photometrics). Images were acquired using Metamorph software (version 7.78; Molecular Devices) at 50 Hz, and 16,000 frames were acquired per cell. A 405-nm laser was used to photoconvert mEos2, with simultaneous 561-nm exposure to excite the photoconverted mEos2. For stochastic photoconversion of mEos2 molecules, a low amount (3–5%) of 405-nm laser and 75–80% of 561-nm laser was used. Data analysis was performed as previously described (Bademosi et al., 2017; Kasula et al., 2016) using PALM-Tracer, a plugin in Metamorph software (Molecular Devices).
FLIM-FRET
For spike-labeling of TopFluor-PtdSer or TopFluor-PtdIns(4,5)P2, MCF7 cells expressing empty vector pC1 or RFP-tagged caveolae proteins were washed with ice-cold PBS containing 3% BSA three times. Cells were then incubated in ice-cold PBS/3%BSA containing TopFluor-tagged lipid at 4°C for 10 min. After aspirating off the lipid/PBS suspension, cells were washed with ice-cold PBS/3% BSA three times before incubation with medium containing 10% FBS for 1 h at 37°C.
MCF7 cells spike-labeled with TopFluor-lipid were washed with PBS two times and incubated in 10 mM NH4Cl for 10 min. Following 2× PBS washing, MCF7 cells were then incubated in 4% PFA for 30 min and then washed with PBD 4× and double-distilled water 4×. Fixed cells were then mounted on glass slides for imaging. The spike-labeled MCF7 cells were imaged using a wide-field microscope at 60× Plan-Apo oil immersion objective with 1.4 NA. Fluorophore TopFluor was excited using a sinusoidally simulated, modulating 3-W 497-nm light-emitting diode at 40 MHz, which is part of a FLIM unit attached to the wide-field microscope. Three independent experiments were performed, and one-way ANOVA was used to evaluate the statistical significance between groups.
Molecular simulations
The interaction between cavin1-HR1 and lipid bilayers was modeled using the MARTINI 2.2 force field (de Jong et al., 2013; Marrink et al., 2007; Monticelli et al., 2008). The Research Collaboratory for Structural Bioinformatics PDB structure ID 4QKV was used as the starting point for the cavin1-HR1 structure, with the 5Q point mutations performed using the Mutate Residue tool in Visual Molecular Dynamics (Humphrey et al., 1996). Both cavin1-HR1 and cavin1-HR1-5Q atomistic structures were then coarse-grained using the martinize.py script (de Jong et al., 2013) with default parameters. Using the insane.py script (Wassenaar et al., 2015), the resulting coarse-grained protein structures were horizontally aligned and placed 4 nm away from a coarse-grained 80:15:5 POPC:POPS:PtdIns(4,5)P2 lipid bilayer with 840 lipids per leaflet. The simulation unit cell was cubic with initial size 24 × 24 × 20 nm3.
All simulations were performed in GROMACS 2019.3 (Abraham et al., 2015) using standard MARTINI 2.2 parameters (de Jong et al., 2016; with van der Waals Lennard–Jones potentials shifted to zero at a cutoff of 1.1 nm, and long-range Coulomb interactions treated with a reaction field with a cutoff of 1.1 nm and dielectric constant εr = 15). Each of four runs began with 10,000 steps of energy minimization, followed by 0.5 ns of NVT equilibration and 1 ns of NPT equilibration, using a time step of 20 fs, V-rescale thermostat at 300K with time constant 1 ps and (for the second equilibration) Berendsen semi-isotropic barostat at 1 bar with time constant 4 ps, and compressibility 4.5 × 10−5 bar−1. The production run was then performed for 3 μs, using a time step of 30 fs, V-rescale thermostat at 300K with time constant 1 ps and Parrinello-Rahman semi-isotropic barostat at 1 bar with time constant 12 ps, and compressibility 3.0 × 10−4 bar−1.
Protein-bilayer distances and lipid occupancies were calculated using GROMACS tools gmx mindist and gmx select, respectively. Distances were measured as the minimum between any protein particle and any lipid particle, while occupancies were measured by counting all lipids with at least one particle within the occupancy cutoff of any protein particle.
Online supplemental material
Our supplemental materials include three supplemental figures. Specifically, in Fig. S1, we used single-molecule tracking and EM to validate the structures of caveolae. These data further support our findings shown in Fig. 1 in the main text. In Fig. S2, we used EM-spatial analysis to quantify the lipid composition of caveolae. These data further support our findings in the main Fig. 2. In Fig. S3, we applied EM spatial mapping and FLIM to further validate the lipid sorting data illustrated in our main Fig. 2.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia (grants APP1140064 and APP1150083 and fellowship APP1156489 to R.G. Parton and GNT1120381 and GNT1155794 [fellowship to F.A. Meunier]). B.M. Collins is supported by a National Health and Medical Research Council Senior Research Fellowship (APP1136021). N. Ariotti is supported by a National Health and Medical Research Council grant (APP1102730). R.G. Parton is supported by the Australian Research Council Centre of Excellence in Convergent Bio-Nano Science and Technology. Y. Zhou and J.F. Hancock are supported by National Institutes of Health grant R01 GM124233. Computational resources for the coarse-grained molecular dynamics simulations were provided by the Research Computing Centre at the University of Queensland.
The authors declare no competing financial interests.
Author contributions: Y. Zhou: designed and performed experiments, analyzed data, wrote and edited manuscript. N. Ariotti: designed and performed experiments, analyzed data, edited manuscript. J. Rae: designed and performed experiments and analyzed data. H. Liang: performed experiments and analyzed data. V. Tillu: performed experiments and analyzed data. S. Tee: performed experiments, analyzed data, edited manuscript. M. Bastiani: performed experiments and analyzed data. A.T. Bademosi: performed experiments and analyzed data. B.M. Collins: designed experiments, analyzed data, edited manuscript. F.A. Meunier: designed experiments, analyzed data, wrote and edited manuscript. J.F. Hancock: wrote and edited manuscript. R.G. Parton: designed and performed experiments, analyzed data, wrote and edited manuscript.
References
- Abraham, M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1-2:19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Ariotti, N., Fernández-Rojo M.A., Zhou Y., Hill M.M., Rodkey T.L., Inder K.L., Tanner L.B., Wenk M.R., Hancock J.F., and Parton R.G.. 2014. Caveolae regulate the nanoscale organization of the plasma membrane to remotely control Ras signaling. J. Cell Biol. 204:777–792. 10.1083/jcb.201307055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti, N., Hall T.E., Rae J., Ferguson C., McMahon K.A., Martel N., Webb R.E., Webb R.I., Teasdale R.D., and Parton R.G.. 2015a. Modular Detection of GFP-Labeled Proteins for Rapid Screening by Electron Microscopy in Cells and Organisms. Dev. Cell. 35:513–525. 10.1016/j.devcel.2015.10.016 [DOI] [PubMed] [Google Scholar]
- Ariotti, N., Rae J., Leneva N., Ferguson C., Loo D., Okano S., Hill M.M., Walser P., Collins B.M., and Parton R.G.. 2015b. Molecular characterization of caveolin-induced membrane curvature. J. Biol. Chem. 290:24875–24890. 10.1074/jbc.M115.644336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademosi, A.T., Lauwers E., Padmanabhan P., Odierna L., Chai Y.J., Papadopulos A., Goodhill G.J., Verstreken P., van Swinderen B., and Meunier F.A.. 2017. In vivo single-molecule imaging of syntaxin1A reveals polyphosphoinositide- and activity-dependent trapping in presynaptic nanoclusters. Nat. Commun. 8:14492. 10.1038/ncomms14492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani, M., Liu L., Hill M.M., Jedrychowski M.P., Nixon S.J., Lo H.P., Abankwa D., Luetterforst R., Fernandez-Rojo M., Breen M.R., et al. 2009. MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J. Cell Biol. 185:1259–1273. 10.1083/jcb.200903053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen, M.R., Camps M., Carvalho-Simoes F., Zorzano A., and Pilch P.F.. 2012. Cholesterol depletion in adipocytes causes caveolae collapse concomitant with proteosomal degradation of cavin-2 in a switch-like fashion. PLoS One. 7:e34516. 10.1371/journal.pone.0034516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci, M., Gratton J.P., Rudic R.D., Acevedo L., Roviezzo F., Cirino G., and Sessa W.C.. 2000. In vivo delivery of the caveolin-1 scaffolding domain inhibits nitric oxide synthesis and reduces inflammation. Nat. Med. 6:1362–1367. 10.1038/82176 [DOI] [PubMed] [Google Scholar]
- Byrne, D.P., Dart C., and Rigden D.J.. 2012. Evaluating caveolin interactions: do proteins interact with the caveolin scaffolding domain through a widespread aromatic residue-rich motif? PLoS One. 7:e44879. 10.1371/journal.pone.0044879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary, N., Gomez G.A., Howes M.T., Lo H.P., McMahon K.A., Rae J.A., Schieber N.L., Hill M.M., Gaus K., Yap A.S., and Parton R.G.. 2014. Endocytic crosstalk: cavins, caveolins, and caveolae regulate clathrin-independent endocytosis. PLoS Biol. 12:e1001832. 10.1371/journal.pbio.1001832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J.P., Mendoza-Topaz C., Howard G., Chadwick J., Shvets E., Cowburn A.S., Dunmore B.J., Crosby A., Morrell N.W., and Nichols B.J.. 2015. Caveolae protect endothelial cells from membrane rupture during increased cardiac output. J. Cell Biol. 211:53–61. 10.1083/jcb.201504042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, B.M., Davis M.J., Hancock J.F., and Parton R.G.. 2012. Structure-based reassessment of the caveolin signaling model: do caveolae regulate signaling through caveolin-protein interactions? Dev. Cell. 23:11–20. 10.1016/j.devcel.2012.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, D.H., Singh G., Bennett W.F., Arnarez C., Wassenaar T.A., Schäfer L.V., Periole X., Tieleman D.P., and Marrink S.J.. 2013. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J. Chem. Theory Comput. 9:687–697. 10.1021/ct300646g [DOI] [PubMed] [Google Scholar]
- de Jong, D.H., Bauokina S., Ingolfsson H.I., and Marrink S.J.. 2016. Martini straight: Boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 199:1–7. 10.1016/j.cpc.2015.09.014 [DOI] [Google Scholar]
- Dupree, P., Parton R.G., Raposo G., Kurzchalia T.V., and Simons K.. 1993. Caveolae and sorting in the trans-Golgi network of epithelial cells. EMBO J. 12:1597–1605. 10.1002/j.1460-2075.1993.tb05804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echarri, A., and Del Pozo M.A.. 2015. Caveolae - mechanosensitive membrane invaginations linked to actin filaments. J. Cell Sci. 128:2747–2758. 10.1242/jcs.153940 [DOI] [PubMed] [Google Scholar]
- Epand, R.M., Sayer B.G., and Epand R.F.. 2005. Caveolin scaffolding region and cholesterol-rich domains in membranes. J. Mol. Biol. 345:339–350. 10.1016/j.jmb.2004.10.064 [DOI] [PubMed] [Google Scholar]
- Fujita, A., Cheng J., Tauchi-Sato K., Takenawa T., and Fujimoto T.. 2009. A distinct pool of phosphatidylinositol 4,5-bisphosphate in caveolae revealed by a nanoscale labeling technique. Proc. Natl. Acad. Sci. USA. 106:9256–9261. 10.1073/pnas.0900216106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton, D., Fontana J., Sowa G., Gratton J.P., Lin M., Li K.X., Michell B., Kemp B.E., Rodman D., and Sessa W.C.. 2002. Localization of endothelial nitric-oxide synthase phosphorylated on serine 1179 and nitric oxide in Golgi and plasma membrane defines the existence of two pools of active enzyme. J. Biol. Chem. 277:4277–4284. 10.1074/jbc.M106302200 [DOI] [PubMed] [Google Scholar]
- Gambin, Y., Ariotti N., McMahon K.A., Bastiani M., Sierecki E., Kovtun O., Polinkovsky M.E., Magenau A., Jung W., Okano S., et al. 2014. Single-molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae. eLife. 3:e01434. 10.7554/eLife.01434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cardeña, G., Martasek P., Masters B.S., Skidd P.M., Couet J., Li S., Lisanti M.P., and Sessa W.C.. 1997. Dissecting the interaction between nitric oxide synthase (NOS) and caveolin. Functional significance of the nos caveolin binding domain in vivo. J. Biol. Chem. 272:25437–25440. 10.1074/jbc.272.41.25437 [DOI] [PubMed] [Google Scholar]
- Gustincich, S., Vatta P., Goruppi S., Wolf M., Saccone S., Della Valle G., Baggiolini M., and Schneider C.. 1999. The human serum deprivation response gene (SDPR) maps to 2q32-q33 and codes for a phosphatidylserine-binding protein. Genomics. 57:120–129. 10.1006/geno.1998.5733 [DOI] [PubMed] [Google Scholar]
- Hansen, C.G., Bright N.A., Howard G., and Nichols B.J.. 2009. SDPR induces membrane curvature and functions in the formation of caveolae. Nat. Cell Biol. 11:807–814. 10.1038/ncb1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, C.G., Shvets E., Howard G., Riento K., and Nichols B.J.. 2013. Deletion of cavin genes reveals tissue-specific mechanisms for morphogenesis of endothelial caveolae. Nat. Commun. 4:1831. 10.1038/ncomms2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, V.J., Weng J., Ly P., Pompey S., Dong H., Mishra L., Schwarz M., Anderson R.G., and Michaely P.. 2013. Cavin-3 dictates the balance between ERK and Akt signaling. eLife. 2:e00905. 10.7554/eLife.00905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, M.M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S.J., Walser P., Abankwa D., Oorschot V.M., Martin S., et al. 2008. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 132:113–124. 10.1016/j.cell.2007.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirama, T., Das R., Yang Y., Ferguson C., Won A., Yip C.M., Kay J.G., Grinstein S., Parton R.G., and Fairn G.D.. 2017. Phosphatidylserine dictates the assembly and dynamics of caveolae in the plasma membrane. J. Biol. Chem. 292:14292–14307. 10.1074/jbc.M117.791400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert, M., Larsson E., Vegesna N.V.G., Ahnlund M., Johansson A.I., Moodie L.W., and Lundmark R.. 2020. Lipid accumulation controls the balance between surface connection and scission of caveolae. eLife. 9:e55038. 10.7554/eLife.55038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38: 27–28. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Izumi, Y., Hirai S., Tamai Y., Fujise-Matsuoka A., Nishimura Y., and Ohno S.. 1997. A protein kinase Cdelta-binding protein SRBC whose expression is induced by serum starvation. J. Biol. Chem. 272:7381–7389. 10.1074/jbc.272.11.7381 [DOI] [PubMed] [Google Scholar]
- Jansen, M., Pietiaïnen V.M., Pölönen H., Rasilainen L., Koivusalo M., Ruotsalainen U., Jokitalo E., and Ikonen E.. 2008. Cholesterol substitution increases the structural heterogeneity of caveolae. J. Biol. Chem. 283:14610–14618. 10.1074/jbc.M710355200 [DOI] [PubMed] [Google Scholar]
- Kasula, R., Chai Y.J., Bademosi A.T., Harper C.B., Gormal R.S., Morrow I.C., Hosy E., Collins B.M., Choquet D., Papadopulos A., and Meunier F.A.. 2016. The Munc18-1 domain 3a hinge-loop controls syntaxin-1A nanodomain assembly and engagement with the SNARE complex during secretory vesicle priming. J. Cell Biol. 214:847–858. 10.1083/jcb.201508118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khater, I.M., Liu Q., Chou K.C., Hamarneh G., and Nabi I.R.. 2019. Super-resolution modularity analysis shows polyhedral caveolin-1 oligomers combine to form scaffolds and caveolae. Sci. Rep. 9:9888. 10.1038/s41598-019-46174-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, O., Tillu V.A., Jung W., Leneva N., Ariotti N., Chaudhary N., Mandyam R.A., Ferguson C., Morgan G.P., Johnston W.A., et al. 2014. Structural insights into the organization of the cavin membrane coat complex. Dev. Cell. 31:405–419. 10.1016/j.devcel.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Kovtun, O., Tillu V.A., Ariotti N., Parton R.G., and Collins B.M.. 2015. Cavin family proteins and the assembly of caveolae. J. Cell Sci. 128:1269–1278. 10.1242/jcs.167866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia, T.V., Dupree P., Parton R.G., Kellner R., Virta H., Lehnert M., and Simons K.. 1992. VIP21, a 21-kD membrane protein is an integral component of trans-Golgi-network-derived transport vesicles. J. Cell Biol. 118:1003–1014. 10.1083/jcb.118.5.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Uchida Y., Emoto K., Umeda M., Kuge O., Taguchi T., and Arai H.. 2012. Impaired retrograde membrane traffic through endosomes in a mutant CHO cell defective in phosphatidylserine synthesis. Genes Cells. 17:728–736. 10.1111/j.1365-2443.2012.01622.x [DOI] [PubMed] [Google Scholar]
- Liang, H., Mu H., Jean-Francois F., Lakshman B., Sarkar-Banerjee S., Zhuang Y., Zeng Y., Gao W., Zaske A.M., Nissley D.V., et al. 2019. Membrane curvature sensing of the lipid-anchored K-Ras small GTPase. Life Sci. Alliance. 2:e201900343. 10.26508/lsa.201900343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti, M.P., Scherer P.E., Vidugiriene J., Tang Z., Hermanowski-Vosatka A., Tu Y.H., Cook R.F., and Sargiacomo M.. 1994. Characterization of caveolin-rich membrane domains isolated from an endothelial-rich source: implications for human disease. J. Cell Biol. 126:111–126. 10.1083/jcb.126.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Brown D., McKee M., Lebrasseur N.K., Yang D., Albrecht K.H., Ravid K., and Pilch P.F.. 2008. Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab. 8:310–317. 10.1016/j.cmet.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink, S.J., Risselada H.J., Yefimov S., Tieleman D.P., and de Vries A.H.. 2007. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 111:7812–7824. 10.1021/jp071097f [DOI] [PubMed] [Google Scholar]
- McMahon, K.A., Zajicek H., Li W.P., Peyton M.J., Minna J.D., Hernandez V.J., Luby-Phelps K., and Anderson R.G.. 2009. SRBC/cavin-3 is a caveolin adapter protein that regulates caveolae function. EMBO J. 28:1001–1015. 10.1038/emboj.2009.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, K.A., Wu Y., Gambin Y., Sierecki E., Tillu V.A., Hall T., Martel N., Okano S., Moradi S.V., Ruelcke J.E., et al. 2019. Identification of intracellular cavin target proteins reveals cavin-PP1alpha interactions regulate apoptosis. Nat. Commun. 10:3279. 10.1038/s41467-019-11111-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano, R., Roth J., Robert A., and Orci L.. 1982. Non-coated membrane invaginations are involved in binding and internalization of cholera and tetanus toxins. Nature. 296:651–653. 10.1038/296651a0 [DOI] [PubMed] [Google Scholar]
- Monticelli, L., Kandasamy S.K., Periole X., Larson R.G., Tieleman D.P., and Marrink S.J.. 2008. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 4:819–834. 10.1021/ct700324x [DOI] [PubMed] [Google Scholar]
- Murata, M., Peränen J., Schreiner R., Wieland F., Kurzchalia T.V., and Simons K.. 1995. VIP21/caveolin is a cholesterol-binding protein. Proc. Natl. Acad. Sci. USA. 92:10339–10343. 10.1073/pnas.92.22.10339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassoy, P., and Lamaze C.. 2012. Stressing caveolae new role in cell mechanics. Trends Cell Biol. 22:381–389. 10.1016/j.tcb.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Oh, P., Borgström P., Witkiewicz H., Li Y., Borgström B.J., Chrastina A., Iwata K., Zinn K.R., Baldwin R., Testa J.E., and Schnitzer J.E.. 2007. Live dynamic imaging of caveolae pumping targeted antibody rapidly and specifically across endothelium in the lung. Nat. Biotechnol. 25:327–337. 10.1038/nbt1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortegren, U., Karlsson M., Blazic N., Blomqvist M., Nystrom F.H., Gustavsson J., Fredman P., and Strålfors P.. 2004. Lipids and glycosphingolipids in caveolae and surrounding plasma membrane of primary rat adipocytes. Eur. J. Biochem. 271:2028–2036. 10.1111/j.1432-1033.2004.04117.x [DOI] [PubMed] [Google Scholar]
- Parton, R.G. 1994. Ultrastructural localization of gangliosides; GM1 is concentrated in caveolae. J. Histochem. Cytochem. 42:155–166. 10.1177/42.2.8288861 [DOI] [PubMed] [Google Scholar]
- Parton, R.G. 2018. Caveolae: Structure, Function, and Relationship to Disease. Annu. Rev. Cell Dev. Biol. 34:111–136. 10.1146/annurev-cellbio-100617-062737 [DOI] [PubMed] [Google Scholar]
- Parton, R.G., Kozlov M.M., and Ariotti N.. 2020. Caveolae and lipid sorting: Shaping the cellular response to stress. J. Cell Biol. 219:e201905071. 10.1083/jcb.201905071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picas, L., Viaud J., Schauer K., Vanni S., Hnia K., Fraisier V., Roux A., Bassereau P., Gaits-Iacovoni F., Payrastre B., et al. 2014. BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat. Commun. 5:5647. 10.1038/ncomms6647 [DOI] [PubMed] [Google Scholar]
- Pilch, P.F., and Liu L.. 2011. Fat caves: caveolae, lipid trafficking and lipid metabolism in adipocytes. Trends Endocrinol. Metab. 22:318–324. 10.1016/j.tem.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman, S.J., Muncke C., Parton R.G., and Hancock J.F.. 2005. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 102:15500–15505. 10.1073/pnas.0504114102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, A., Martin S., Fernández M.A., Ingelmo-Torres M., Ferguson C., Enrich C., and Parton R.G.. 2005. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol. Biol. Cell. 16:2091–2105. 10.1091/mbc.e04-08-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior, I.A., Muncke C., Parton R.G., and Hancock J.F.. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160:165–170. 10.1083/jcb.200209091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghupathy, R., Anilkumar A.A., Polley A., Singh P.P., Yadav M., Johnson C., Suryawanshi S., Saikam V., Sawant S.D., Panda A., et al. 2015. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell. 161:581–594. 10.1016/j.cell.2015.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg, K.G., Heuser J.E., Donzell W.C., Ying Y.S., Glenney J.R., and Anderson R.G.. 1992. Caveolin, a protein component of caveolae membrane coats. Cell. 68:673–682. 10.1016/0092-8674(92)90143-Z [DOI] [PubMed] [Google Scholar]
- Sharma, D.K., Brown J.C., Choudhury A., Peterson T.E., Holicky E., Marks D.L., Simari R., Parton R.G., and Pagano R.E.. 2004. Selective stimulation of caveolar endocytosis by glycosphingolipids and cholesterol. Mol. Biol. Cell. 15:3114–3122. 10.1091/mbc.e04-03-0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvets, E., Bitsikas V., Howard G., Hansen C.G., and Nichols B.J.. 2015. Dynamic caveolae exclude bulk membrane proteins and are required for sorting of excess glycosphingolipids. Nat. Commun. 6:6867. 10.1038/ncomms7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, B., Köster D., Ruez R., Gonnord P., Bastiani M., Abankwa D., Stan R.V., Butler-Browne G., Vedie B., Johannes L., et al. 2011. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 144:402–413. 10.1016/j.cell.2010.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, J.L., Gomez G.A., Weeratunga S., Davies E.M., Noordstra I., Budnar S., Katsuno-Kambe H., McGrath M.J., Verma S., Tomatis V., et al. 2020. Caveolae control contractile tension for epithelia to eliminate tumor cells. Dev. Cell. 54:75–91.e7. 10.1016/j.devcel.2020.05.002 [DOI] [PubMed] [Google Scholar]
- Tillu, V.A., Kovtun O., McMahon K.A., Collins B.M., and Parton R.G.. 2015. A phosphoinositide-binding cluster in cavin1 acts as a molecular sensor for cavin1 degradation. Mol. Biol. Cell. 26:3561–3569. 10.1091/mbc.E15-06-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillu, V.A., Lim Y.W., Kovtun O., Mureev S., Ferguson C., Bastiani M., McMahon K.A., Lo H.P., Hall T.E., Alexandrov K., et al. 2018. A variable undecad repeat domain in cavin1 regulates caveola formation and stability. EMBO Rep. 19:e45775. 10.15252/embr.201845775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, D., Carpentier J.L., Sawano F., Gorden P., and Orci L.. 1987. Ligands internalized through coated or noncoated invaginations follow a common intracellular pathway. Proc. Natl. Acad. Sci. USA. 84:7957–7961. 10.1073/pnas.84.22.7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanaski, S.P., Ng B.K., and Glaser M.. 2003. Caveolin scaffolding region and the membrane binding region of SRC form lateral membrane domains. Biochemistry. 42:42–56. 10.1021/bi012097n [DOI] [PubMed] [Google Scholar]
- Wassenaar, T.A., Ingólfsson H.I., Böckmann R.A., Tieleman D.P., and Marrink S.J.. 2015. Computational Lipidomics with insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theory Comput. 11:2144–2155. 10.1021/acs.jctc.5b00209 [DOI] [PubMed] [Google Scholar]
- Yu, J., Bergaya S., Murata T., Alp I.F., Bauer M.P., Lin M.I., Drab M., Kurzchalia T.V., Stan R.V., and Sessa W.C.. 2006. Direct evidence for the role of caveolin-1 and caveolae in mechanotransduction and remodeling of blood vessels. J. Clin. Invest. 116:1284–1291. 10.1172/JCI27100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Liang H., Rodkey T., Ariotti N., Parton R.G., and Hancock J.F.. 2014. Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol. Cell. Biol. 34:862–876. 10.1128/MCB.01227-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Wong C.O., Cho K.J., van der Hoeven D., Liang H., Thakur D.P., Luo J., Babic M., Zinsmaier K.E., Zhu M.X., et al. 2015. SIGNAL TRANSDUCTION. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science. 349:873–876. 10.1126/science.aaa5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Prakash P., Liang H., Cho K.J., Gorfe A.A., and Hancock J.F.. 2017. Lipid-Sorting Specificity Encoded in K-Ras Membrane Anchor Regulates Signal Output. Cell. 168:239–251.e16. 10.1016/j.cell.2016.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]