Abstract
RBMS3, a gene encoding a glycine-rich RNA-binding protein, belongs to the family of c-Myc gene single-strand binding proteins (MSSP). Recently, several reports have provided evidence that RBMS3 was deregulated in a diverse range of solid tumors and played a critical role in tumor progression. However, it remains unclear whether RBMS3 inhibits the progression of human breast cancer. Thus, the aim of this study was to investigate the role of RBMS3 in breast cancer and explore the underlying mechanism in breast cancer progression. Our results showed, for the first time, that the expression of RBMS3 at both the mRNA and protein levels was significantly downregulated in human breast cancer tissues and cell lines. In addition, RBMS3 overexpression dramatically suppressed the proliferation, migration, and invasion of breast cancer cells in vitro and attenuated tumor growth in vivo. Furthermore, we observed that RBMS3 greatly inhibited the protein expression of β-catenin, cyclin D1, and c-Myc in breast cancer cells. In summary, we have shown that RBMS3 inhibited the proliferation and tumorigenesis of breast cancer cells, at least in part, through inactivation of the Wnt/β-catenin signaling pathway. Thus, RBMS3 may be a potential treatment target for breast cancer.
Key words: RBMS3, Breast cancer, Metastasis, Tumorigenesis, Wnt/β-catenin signaling
INTRODUCTION
Breast cancer is one of the most common malignant tumors in women around the world1,2. Despite recent advances in early detection and multidisciplinary therapy, about 30% of patients with breast cancer suffer from uncontrolled metastasis and recurrence, resulting in a 5-year decrease in survival3–5. Thus, it is urgent that we identify new therapeutic molecular targets and therapeutic strategies for the prevention and treatment of breast cancer.
Members of the c-Myc gene single-strand binding proteins (MSSP) family are single-strand DNA-binding proteins that cooperate with the Myc protein to regulate cell proliferation, cell cycle progression, apoptosis, DNA replication, and gene transcription6,7. RBMS3, a gene encoding a glycine-rich RNA-binding protein, belongs to the MSSP family. It was first identified by screening a human fibroblast cDNA library with a labeled DNA fragment derived from α2 (I) collagen promoter Box 5A8. One study reported that RBMS3 was involved in liver fibrosis, and ectopic expression of RBMS3 induced the expression of Prx1 and collagen α1 (I) in hepatic stellate cells (HSCs)9. In addition, previous studies demonstrated that RBMS3 may act as a tumor suppressor gene in several types of cancer10–12. A study by Li et al. confirmed that the expression of RBMS3 mRNA and protein levels was greatly downregulated in esophageal squamous cell carcinoma (ESCC), and overexpression of RBMS3 effectively suppressed the ESCC cell growth rate, soft agar colony formation, and tumor formation in nude mice11. However, it remains unclear whether RBMS3 inhibits the progression of human breast cancer. Thus, the aim of this study was to investigate the role of RBMS3 in breast cancer and explore the underlying mechanism in breast cancer progression.
MATERIALS AND METHODS
Tissue Collection
Human breast cancer tissues were obtained from patients with breast cancer who underwent tumor resection between 2014 and 2015 at the Department of Oncology, The Second Xiangya Hospital of Central South University (P.R. China). The samples were immediately immersed in liquid nitrogen. The study was approved by the institutional ethics committee at Huaihe Hospital of Henan University, and written informed consent was obtained from all patients included in this study.
Cell Culture
Human breast cancer cell lines MCF-7, MDA-MB-231, and BT-474, as well as normal human breast epithelial cell line (NBEC), were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen), 100 U/ml penicillin (Gibco), and 100 U/ml streptomycin (Gibco) at 37°C in a humidified atmosphere containing 5% CO2.
Construction of Plasmids and Transfection
The full-length RBMS3 cDNA was amplified and subcloned into pcDNA3.1 (Invitrogen), whereas the empty vector pcDNA3.1 was used as the control. MCF-7 cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The transfection efficiency was confirmed by quantitative real-time (qRT)-PCR and Western blot.
Quantitative Real-Time (qRT)-PCR
Total RNA was extracted from breast cancer tissues and cells using TRIzol reagent (Invitrogen) and reverse transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA, USA). PCR analysis was performed on the Applied Biosystems 7500 Sequence Detection system (ABI) using SYBR-Green PCR Mix (Invitrogen). The following primers were used for RT-PCR: RBMS3, 5′-CGCGGATCCTACCCCCAGTACACTTA-3′ (sense) and 5′-CCGGAATTCTTACTTAGACTGTTGGTA-3′ (antisense); β-actin, 5′-CCGTGAAAAGATGACCCAGATC-3′ (sense) and 5′-CACAGCCTGGATGGCTACGT-3′ (antisense). The relative RBMS3 mRNA expression was calculated using the comparative cycle threshold method for relative quantitation and normalized to β-actin levels.
Western Blot Analysis
For Western blot, breast cancer tissues or cells were lysed in RIPA lysis buffer (Invitrogen). Thirty micrograms of protein per sample was separated by 10% SDS-PAGE and transferred to a PVDF membrane (Millipore, Boston, MA, USA). After blocking with 5% skim milk, the membranes were incubated with primary antibodies overnight at 4°C. The following antibodies were used: anti-RBMS3, anti-E-cadherin, anti-vimentin, anti-β-catenin, anti-c-Myc, anti-cyclin D1, and anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Horseradish peroxidase (HRP)-labeled secondary antibodies (1:2,000 dilution; Santa Cruz Biotechnology) were then added for incubation for 1 h at room temperature. Blots were visualized with the use of an enhanced chemiluminescence kit (Amersham Biosciences Inc., Piscataway, NJ, USA).
Cell Proliferation Assay
Cell proliferation was evaluated using the 3-(4.5-methylthiozol-2yl)-2.5-diphenyltetrazolium bromide (MTT) assay. MCF-7 cells (1 × 105 cells/well) transfected with RBMS3-pcDNA3.1 or pcDNA3.1 were seeded onto 96-well plates and then cultured at 24-h intervals for 4 days. Then 20 μl of 5 mg/ml MTT solution (Sigma-Aldrich, St. Louis, MO, USA) was added to each well and incubated for 4 h at 37°C. Subsequently, the supernatant was removed, and the formazan crystals were dissolved in 150 μl of dimethyl sulfoxide (DMSO; Sigma). The optical density of each well was assessed at a wavelength of 570 nm using a microplate reader (Model 550; Bio-Rad, Hercules, CA, USA).
Migration and Invasion Assays
For cell migration assay, infected MCF-7 cells (1 × 105 cells/well) in 200 μl of serum-free DMEM were added to the upper compartment with 8-μm pores (BD Biosciences, Eugene, OR, USA). DMEM (500 μl) containing 10% FBS was then added to the lower chamber. After 24 h of incubation, the cells on the lower surface of the filter were fixed with 3.7% formaldehyde and then stained with 1% crystal violet for 30 min. The average number of migrated cells was counted in five random fields per well under a light microscope (magnification: 100×). The methods utilized in the invasion assays were similar to those used in the cell migration assays except that the inserts were precoated with 20 μg of Matrigel.
Tumorigenicity Assay in Xenograft Tumor Model
Animal experiments were carried out according to the protocols approved by The Animal Care and Use Committee of the Second Xiangya Hospital of Central South University. Six-week-old female Balb/c athymic nude mice were subcutaneously injected in the right flank with 1 × 106 RBMS3-overexpressing cells in 0.1 ml of PBS (n = 8 each group). The dimensions of xenograft nodules were callipered every 1 week for 4 successive weeks. The volume of tumor was calculated using the following equation: V = L × W 2 (V = volume, L = length, W = width). Four weeks later, the animals were sacrificed, and the xenografts were isolated and weighed.
Statistical Analysis
Statistical analysis was carried out with the SPSS statistical software package (Version 13.0; SPSS Inc., Chicago, IL, USA). Data were presented as mean ± SD. The statistical significance of the difference was analyzed by ANOVA and post hoc Dunnett’s test. A value of p < 0.05 was considered statistically significant.
RESULTS
RBMS3 Had a Low Expression in Breast Cancer Tissues and Cell Lines
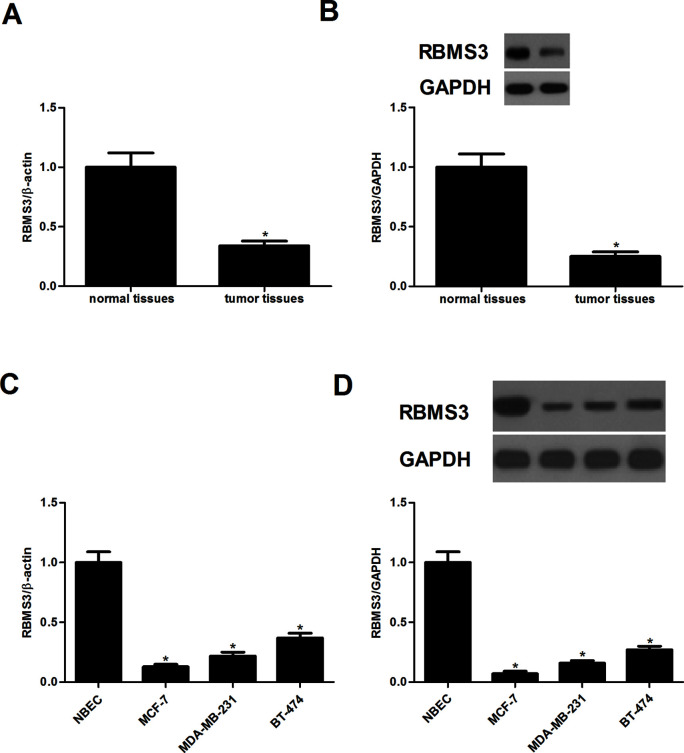
First, we examined the expression of RBMS3 in breast cancer tissues. The results of the qRT-PCR analysis indicated that the mRNA expression of RBMS3 was significantly decreased in breast cancer tissues, compared with the normal tissues (Fig. 1A). Similarly, the protein expression of RBMS3 was reduced in breast cancer tissues (Fig. 1B). Moreover, we examined the expression of RBMS3 in breast cancer cell lines. The expression of RBMS3 at both the mRNA and protein levels was obviously reduced in breast cancer cell lines (Fig. 1C and D).
Figure 1.
RBMS3 was lowly expressed in breast cancer tissues and cell lines. (A) RBMS3 mRNA expression was determined in breast cancer tissues. (B) Western blot analysis was used to examine RBMS3 protein expression in breast cancer tissues. *p < 0.05 compared to the adjacent normal tissues. All the experiments were repeated at least three times. (C, D) The expression of RBMS3 at both mRNA and protein levels was obviously reduced in breast cancer cell lines. *p < 0.05 compared to the NBEC cell line.
RBMS3 Inhibited the Proliferation of Breast Cancer Cells In Vitro
To study the role of RBMS3 in breast cancer, MCF-7 cells were transduced with RBMS3-pcDNA3.1 or pcDNA3.1. Compared with the pcDNA3.1 group, the expression of RBMS3 at both the mRNA and protein levels was obviously increased following transfection of RBMS3 in MCF-7 cells (Fig. 2A). We then evaluated the effect of RBMS3 on breast cancer cell proliferation using an MTT assay. The results showed that RBMS3 overexpression significantly inhibited the proliferation of MCF-7 cells (Fig. 2B).
Figure 2.
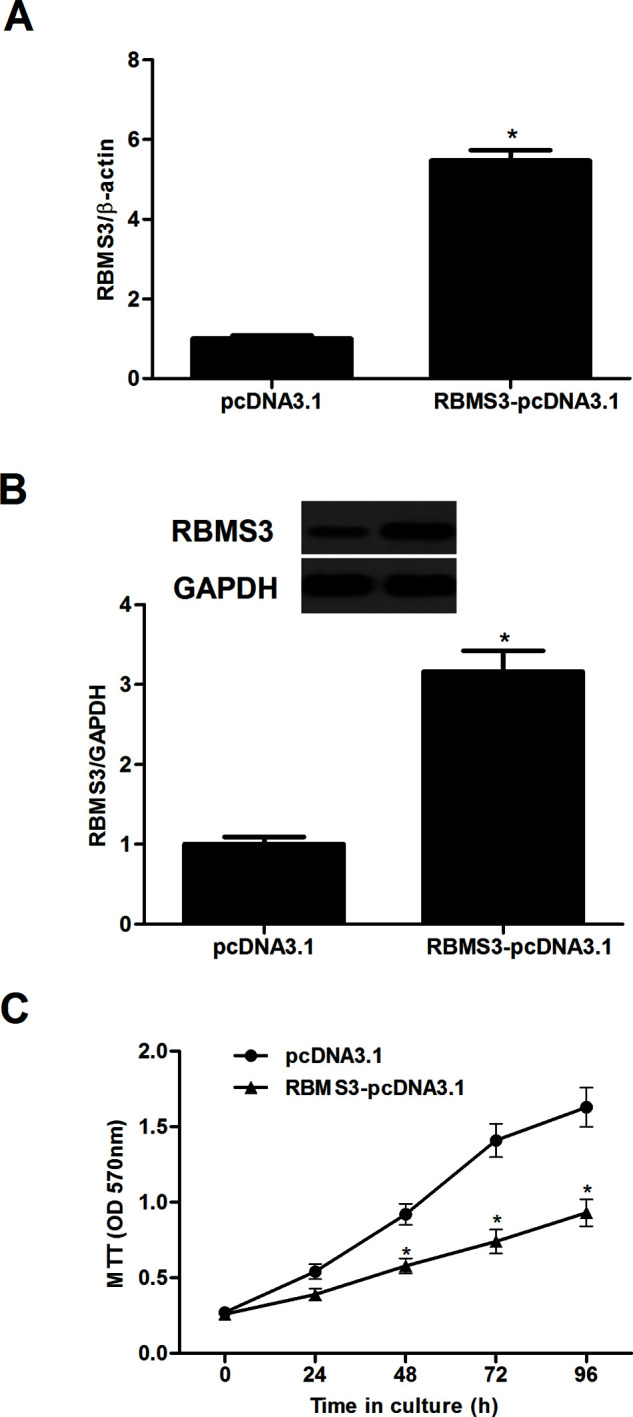
RBMS3 inhibited the proliferation of breast cancer cells in vitro. (A, B) real-time (RT)-PCR and Western blot analyses showed effective upregulation of RBMS3 following RBMS3-pcDNA3.1 transfection. (B) MCF-7 cells were transfected with RBMS3-pcDNA3.1 or pcDNA3.1. (C) The proliferating capacity of transfected cells was evaluated using an MTT assay at 0, 24, 48, 72, and 96 h subsequent to transfection. *p < 0.05 compared to the pcDNA3.1 group.
RBMS3 Inhibited the Migration and Invasion of Breast Cancer Cells
Next, we performed Transwell migration and Matrigel invasion assays to assess the effects of RBMS3 on cell migration and invasion. Overexpression of RBMS3 significantly inhibited the number of MCF-7 cells that migrated into the lower chamber, compared with the pcDNA3.1 group (Fig. 3A). The Matrigel invasion assay indicated that RBMS3 overexpression significantly repressed the invasiveness of MCF-7 cells (Fig. 3B). To investigate whether RBMS3 suppressed breast cancer cell migration and invasion by inhibiting EMT, we analyzed the expression of several EMT markers in RBMS3-overexpressing and control MCF-7 cells. RBMS3 overexpression efficiently upregulated E-cadherin expression and downregulated vimentin expression in MCF-7 cells (Fig. 3C).
Figure 3.
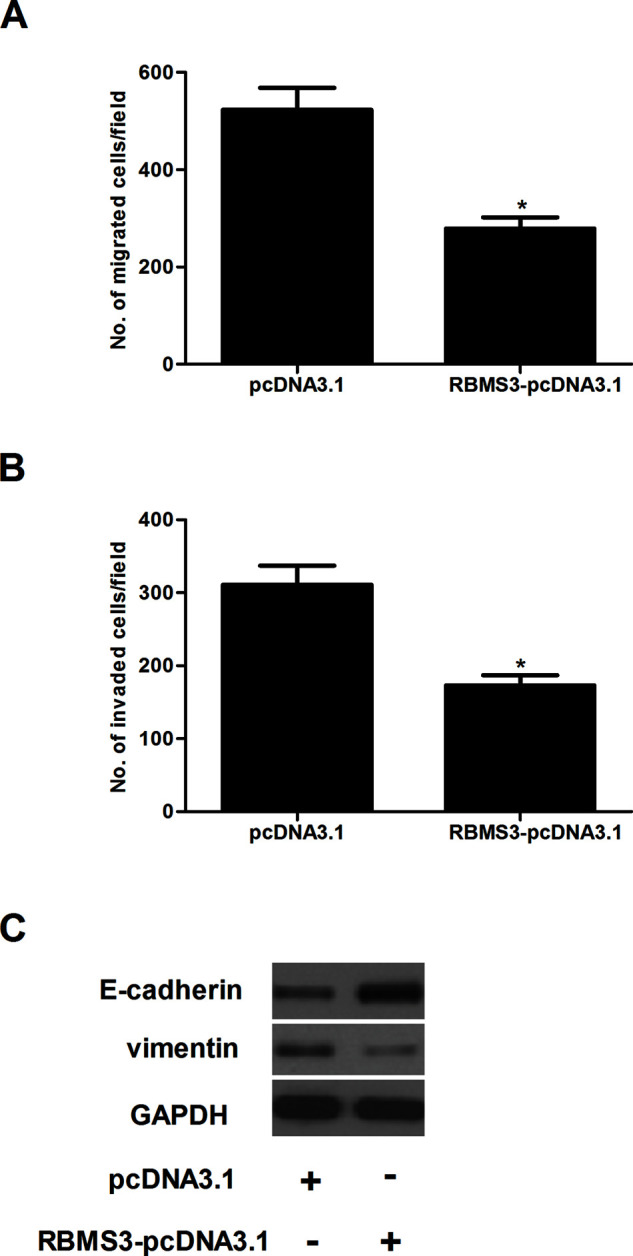
RBMS3 inhibited the migration and invasion of breast cancer cells. MCF-7 cells were transfected with RBMS3-pcDNA3.1 or pcDNA3.1. (A) Cell migration was detected using the Transwell migration assay. (B) Cell invasion was detected using a Matrigel invasion assay. (C) The expression of E-cadherin and vimentin was evaluated by Western blot. The relative expression of each gene was normalized to the housekeeping gene GAPDH. *p < 0.05 compared to the pcDNA3.1 group.
RBMS3 Prevented the Wnt/β-Catenin Pathway in Breast Cancer Cells
The Wnt/β-catenin pathway plays an important role in tumor initiation, progression, and metastasis. Thus, we investigated whether RBMS3 regulates the Wnt/β-catenin pathway to inhibit breast cancer progression. RBMS3 overexpression significantly downregulated the protein expression of β-catenin in MCF-7 cells, compared with the pcDNA3.1 group (Fig. 4). It was the same for the downstream target genes, such as c-Myc and cyclin D1.
Figure 4.
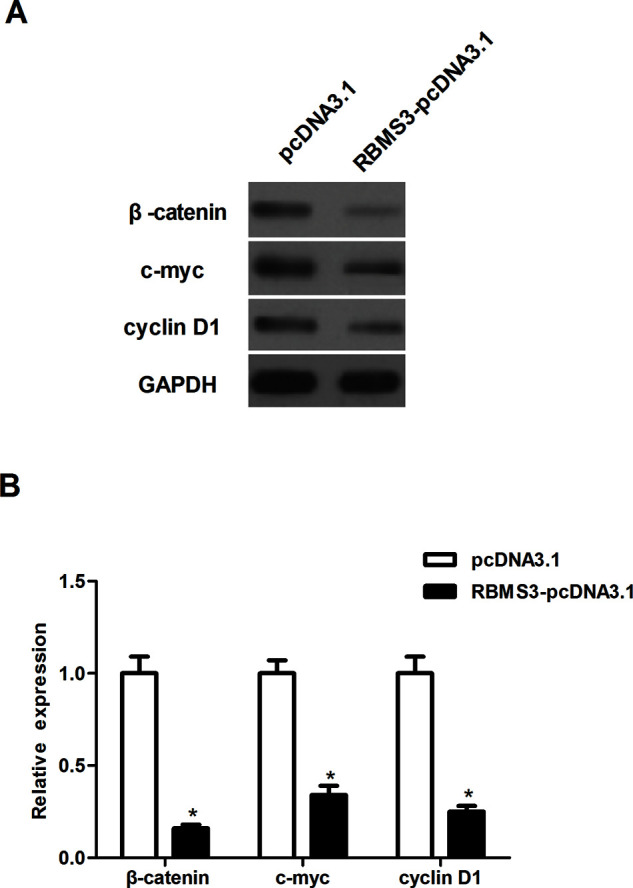
RBMS3 prevented the Wnt/β-catenin pathway in breast cancer cells. MCF-7 cells were transfected with RBMS3-pcDNA3.1 or pcDNA3.1. (A) The expression of β-catenin, c-Myc, and cyclin D1 was evaluated by Western blot. The relative expression of each gene was normalized to the housekeeping gene GAPDH. (B) Quantification analysis of relative protein levels of β-catenin, c-Myc, and cyclin D1 was performed using Gel-Pro Analyzer version 4.0 software. *p < 0.05 compared to the pcDNA3.1 group.
RBMS3 Attenuated Tumor Growth in Mouse Xenograft Models
To examine the role of RBMS3 in tumor growth in vivo, we established mouse xenograft models with RBMS3-overexpressing MCF-7 cells and control cells by subcutaneous injection. The results demonstrated that the volume and weight of tumors derived from RBMS3-overexpressing MCF-7 cells were drastically reduced compared to those of tumors generated by control cells (Fig. 5A and B).
Figure 5.
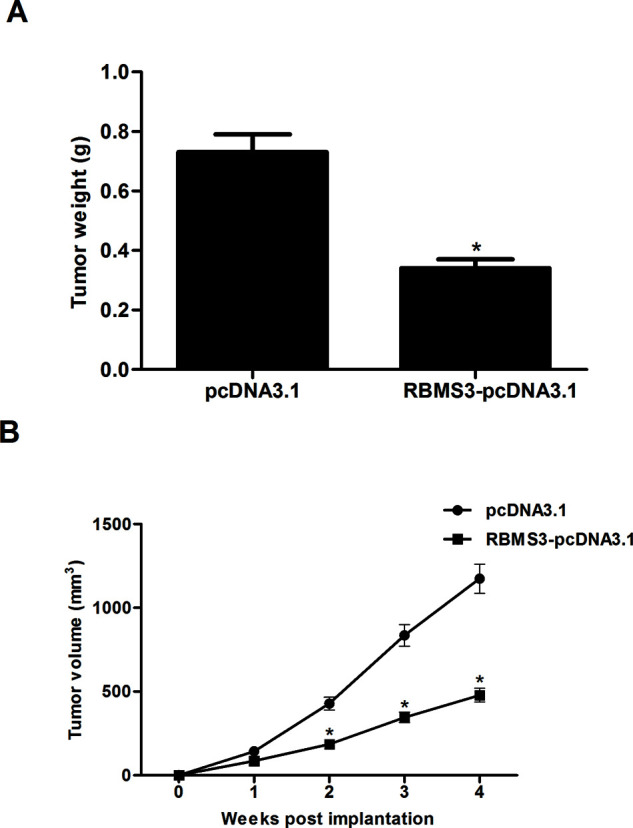
RBMS3 attenuated tumor growth in mouse xenograft models. Tumor xenotransplant mouse model was established with the injection of RBMS3-overexpressing MCF-7 cells and control cells. (A) Four weeks later, the animals were sacrificed, and the xenografts were isolated and weighed. (B) The volume of tumors was monitored every 1 week. *p < 0.05 compared to the pcDNA3.1 group.
DISCUSSION
In the present study we showed, for the first time, that the expression of RBMS3 at both the mRNA and protein levels was significantly downregulated in human breast cancer tissues and cell lines. In addition, RBMS3 overexpression dramatically suppressed the proliferation, migration, and invasion of breast cancer cells in vitro and attenuated tumor growth in vivo. Furthermore, we observed that RBMS3 greatly inhibited the protein expression of β-catenin, cyclin D1, and c-Myc in breast cancer cells.
Several reports have provided evidence that RBMS3 was deregulated in a diverse range of solid tumors and played a critical role in tumor progression. Zhang et al. confirmed that the expression of RBMS3 was remarkably lower in gastric cancer tissues than that in the matched normal tissues13. Another study showed that RBMS3 expression was downregulated in nasopharyngeal carcinoma cell lines and tissues. Upregulation of RBMS3 inhibited the proliferation of nasopharyngeal carcinoma cells14. Consistent with the above data, in the present study we observed that the expression of RBMS3 was obviously downregulated in human breast cancer tissues and cell lines. RBMS3 overexpression significantly suppressed the proliferation of breast cancer cells in vitro and attenuated tumorigenicity in vivo in a nude mouse model. These data imply that RBMS3 as a tumor suppressor may play an important role in development and progression of breast cancer.
Metastasis is a major feature in aggressive breast cancer. The prognosis of breast cancer patients is often poor due to metastasis, especially for those diagnosed at advanced stages15,16. EMT is considered to be an important early step in cancer progression and metastasis. During tumor development, epithelial cells undergo dynamic cytoskeletal rearrangement and lose cell adhesion and epithelial components (such as E-cadherin) while acquiring mesenchymal and migratory phenotypes17. Our findings demonstrated that RBMS3 overexpression efficiently suppressed breast cancer cell migration and invasion capacity, as well as inhibited the EMT phenotype, which was accompanied with increased E-cadherin and decreased vimentin expression in MCF-7 cells. These results suggest that RBMS3 functions as a novel suppressor of EMT and tumor metastasis in breast cancer.
The Wnt/β-catenin signaling pathway plays a critical role in regulating cancer cell proliferation, survival, cell cycle, metastasis, and apoptosis18–20. Aberrant activation of the Wnt/β-catenin signaling is well documented to be closely associated with carcinogenesis in breast cancer21–23. β-Catenin is a key component of the Wnt signaling cascade, and accumulation of β-catenin is a hallmark of Wnt signaling activation24. Cyclin D is an established target of Wnt signaling, and elevated cyclin D levels are characteristic of advanced breast cancer25. In addition, the Wnt/β-catenin signaling pathway was involved in the regulation of EMT in breast cancer cells26. The current study revealed that overexpression of RBMS3 decreased the expression of β-catenin and its target genes such as c-Myc and cyclin D1 in breast cancer cells, which is consistent with results of a previous study showing that RBMS3 downregulated the expression of MMP2 and β-catenin in nasopharyngeal carcinoma cells14. These data strongly suggest that RBMS3 inhibited tumor growth and migration of breast cancer cells in vitro and in vivo, at least in part, through inactivation of the Wnt/β-catenin signaling pathway.
In summary, we have shown that RBMS3 inhibited the proliferation and metastasis of breast cancer cells, at least in part, through inactivation of the Wnt/β-catenin signaling pathway. Thus, RBMS3 may be a potential treatment target for breast cancer.
REFERENCES
- 1. Desantis CE, Fedewa SA, Goding SA, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42. [DOI] [PubMed] [Google Scholar]
- 2. Beral V. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet 2007;369:1703–10. [DOI] [PubMed] [Google Scholar]
- 3. Bickell NA, Wang JJ, Oluwole S, Schrag D, Godfrey H, Hiotis K, Mendez J, Guth AA. Missed opportunities: Racial disparities in adjuvant breast cancer treatment. J Clin Oncol. 2006;24:1357–62. [DOI] [PubMed] [Google Scholar]
- 4. Moseley AL, Carati CJ, Piller NB. A systematic review of common conservative therapies for arm lymphoedema secondary to breast cancer treatment. Ann Oncol. 2007;18:639–46. [DOI] [PubMed] [Google Scholar]
- 5. Enger SM, Thwin SS, Buist DS, Field T, Frost F, Geiger AM, Lash TL, Prout M, Yood MU, Wei F. Breast cancer treatment among older women in integrated health care settings. J Clin Oncol. 2011;24:4377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Niki T, Izumi S, Saëgusa Y, Taira T, Takai T, Iguchi-Ariga SMM, Ariga H. MSSP promotes ras/myc cooperative cell transforming activity by binding to c-Myc. Genes Cells 2000;5:127–41. [DOI] [PubMed] [Google Scholar]
- 7. Fujimoto M, Matsumoto K, Iguchi-Ariga SM, Ariga H. Disruption of MSSP, c-myc single-strand binding protein, leads to embryonic lethality in some homozygous mice. Genes Cells 2001;6:1067–75. [DOI] [PubMed] [Google Scholar]
- 8. Penkov D, Ni R, Else C, Piñol-Roma S, Ramirez F, Tanaka S. Cloning of a human gene closely related to the genes coding for the c-myc single-strand binding proteins. Gene 2000;243:27–36. [DOI] [PubMed] [Google Scholar]
- 9. Fritz D, Stefanovic B. RNA-binding protein RBMS3 is expressed in activated hepatic stellate cells and liver fibrosis and increases expression of transcription factor Prx1. J Mol Biol. 2007;371:585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang YN, Liu Y, Meng Q, Li X, Wang F, Yao G, Wang L, Fu S, Tong D. RBMS3 is a tumor suppressor gene that acts as a favorable prognostic marker in lung squamous cell carcinoma. Med Oncol. 2015;32:1–9. [DOI] [PubMed] [Google Scholar]
- 11. Li Y, Chen L, Nie CJ, Zeng TT, Liu H, Mao X, Qin Y, Zhu YH, Fu L, Guan XY. Downregulation of RBMS3 is associated with poor prognosis in esophageal squamous cell carcinoma. Cancer Res. 2011;71:6106–15. [DOI] [PubMed] [Google Scholar]
- 12. Chen J, Fu L, Zhang LY, Kwong DL, Yan L, Guan XY. Tumor suppressor genes on frequently deleted chromosome 3p in nasopharyngeal carcinoma. Chin J Cancer 2012;31:215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang T, Wu Y, Fang Z, Yan Q, Zhang S, Sun R, Khaliq J, Li Y. Low expression of RBMS3 and SFRP1 are associated with poor prognosis in patients with gastric cancer. Am J Cancer Res. 2016;6:2679–89. [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J, Kwong DL, Zhu CL, Chen LL, Dong SS, Zhang LY, Tian J, Qi CB, Cao TT, Wong AM. RBMS3 at 3p24 inhibits nasopharyngeal carcinoma development via inhibiting cell proliferation, angiogenesis, and inducing apoptosis. PLoS One 2012;7:e44636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manni A, Washington S, Griffith JW, Verderame MF, Mauger D, Demers LM, Samant RS, Welch DR. Influence of polyamines on in vitro and in vivo features of aggressive and metastatic behavior by human breast cancer cells. Clin Exp Metastasis 2002;19:95–105. [DOI] [PubMed] [Google Scholar]
- 16. Guo BH, Feng Y, Zhang R, Xu LH, Li MZ, Kung HF, Song LB, Zeng MS. Bmi-1 promotes invasion and metastasis, and its elevated expression is correlated with an advanced stage of breast cancer. Mol Cancer 2011;10:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalluri R. EMT: When epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119:1417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shan S, Lv Q, Zhao Y, Liu C, Sun Y, Xi K, Xiao J, Li C. Wnt/β-catenin pathway is required for epithelial to mesenchymal transition in CXCL12 over expressed breast cancer cells. Int J Clin Exp Pathol. 2015;8:12357–67. [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Jin K, Pelt GWV, Dam HV, Yu X, Mesker WE, Dijke PT, Zhou F, Zhang L. c-Myb enhances breast cancer invasion and metastasis through the Wnt/β-catenin/Axin2 pathway. Cancer Res. 2016;76:3364–75. [DOI] [PubMed] [Google Scholar]
- 20. Song L, Liu D, He J, Wang X, Dai Z, Yang Z, Kang H, Wang B. SOX1 inhibits breast cancer cell growth and invasion through suppressing the Wnt/β-catenin signaling pathway. APMIS 2016;124:547–55. [DOI] [PubMed] [Google Scholar]
- 21. Ma F, Zhang J, Zhong L, Wang L, Liu Y, Wang Y, Peng L, Guo B. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/β-catenin signaling. Gene 2014;535:191–7. [DOI] [PubMed] [Google Scholar]
- 22. Yin X, Xiang T, Li L L, Su X, Shu X, Luo X, Huang J, Yuan Y, Peng W, Oberst M. DACT1, an antagonist to Wnt/β-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. 2013;15:7039–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song L, Liu D, Wang B, He J, Zhang S, Dai Z, Ma X, Wang X. miR-494 suppresses the progression of breast cancer in vitro by targeting CXCR4 through the Wnt/β-catenin signaling pathway. Oncol Rep. 2015;34:525–31. [DOI] [PubMed] [Google Scholar]
- 24. Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell 2012;149:1192–205. [DOI] [PubMed] [Google Scholar]
- 25. Wang Z, Li B, Zhou L, Yu S, Su Z, Song J, Sun Q, Sha O, Wang X, Jiang W. Prodigiosin inhibits Wnt/β-catenin signaling and exerts anticancer activity in breast cancer cells. Proc Natl Acad Sci USA 2016;113:13150–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma X, Yan W, Dai Z, Gao X, Ma Y, Xu Q, Jiang J, Zhang S. Baicalein suppresses metastasis of breast cancer cells by inhibiting EMT via downregulation of SATB1 and Wnt/β-catenin pathway. Drug Des Dev Ther. 2016;10:1419–41. [DOI] [PMC free article] [PubMed] [Google Scholar]