Abstract
MicroRNA dysregulation contributes to malignant progression, dissemination, and profound treatment resistance in multiple cancers. miR-449a is recognized as a tumor suppresser. However, the roles of miR-449a in lung cancer initiation and progression are largely unknown. Our study aims to investigate the roles and underlying mechanism of miR-449a in modulating sensitivity to ionizing radiation (IR) in non-small cell lung cancer (NSCLC). Lung cancer cells were transfected with miR-449a mimics or negative control and exposed to IR; the levels of target protein, glycolysis, cell viability, apoptosis, and DNA damage were examined. miR-449a was suppressed in lung cancer tissues and cancer cells (A549 and H1299). IR exposure significantly increased the expression of miR-449a in A549 cells at doses ranging from 4 to 8 Gy at 24 h, whereas no significant change in miR-449a was found in H1299 cells after IR. When A549 cells were exposed to IR at a dose of 8 Gy, the miR-449a level only had an acute increase within 12 h. miR-449a restoration dramatically suppressed IR-induced cell apoptosis and DNA damage in both A549 and H1299 cells. Bioinformatics analysis indicated that lactate dehydrogenase A (LDHA) was a potential target of miR-449a. miR-449a mimic transfection substantially suppressed the LDHA expression and production of lactate catalyzed by LDHA as well as glucose uptake. We confirmed that miR-449a could bind to the 3′-UTR of LDHA mRNA using luciferase reporter assay. LDHA siRNA-transfected cells showed enhanced cell apoptosis and DNA damage in response to IR exposure. miR-449a upregulation enhanced IR sensitivity of lung cancer cells by suppressing LDHA and the subsequent glycolysis.
Key words: Non-small cell lung cancer (NSCLC), Ionizing radiation (IR), miR-449a, Lactate dehydrogenase A (LDHA), Glycolysis
INTRODUCTION
Lung cancer ranks among the most prevalent and lethal malignant tumors around the world. The majority of lung cancers is non-small cell lung cancer (NSCLC), which is characterized by rapid progression, high metastasis risk, and profound treatment resistance. NSCLC manifests with four main histologic types, including adenocarcinoma, squamous cell carcinoma, large cell carcinoma, and mixed histologies1. To date, platinum-based chemotherapy with concurrent radiotherapy comprised the first-line strategy for locally advanced NSCLC. However, despite optimal chemoradiotherapy, approximately 40% of patients suffer recurrent disease and treatment failure within the irradiated areas, which contributes to a dismal 15%–25% survival rate at 5 years2. The cellular sensitivity of cancers to ionizing radiation (IR) is dependent on an integrated molecular network, either by activation or inhibition of those molecules that account for DNA repair, metabolism, survival, and signal transduction regulation3. Therefore, identification and getting insight into the mechanisms of the molecular targets for radiosensitization are of importance for lung cancer therapy. MicroRNAs (miRNAs) are a class of short noncoding RNAs with approximately 20 to 25 nucleotides that regulate gene expression posttranscriptionally by binding to a complementary sequence in the 3′-untranslated region (3′-UTR) of target mRNAs4. miRNAs function by either degrading the targeted mRNA or structurally suppressing their transcriptional activities. Emerging evidence shows that multiple miRNAs participate in various fundamental biological processes, including cell proliferation, apoptosis, differentiation, and metabolism. Moreover, change in both circulating and tissue miRNA expression profiles was documented in NSCLC patients in comparison with healthy subjects5,6. Several plasma miRNAs were identified to be effective in predicting radiosensitivity, such as hsa-miR-98-5p, hsa-miR-302e, hsa-miR-495-3p, and hsa-miR-6137. miR-449a is expressed at a low level in several cancer cell lines, and solid tumors such as gastric cancer, bladder cancer, and NSCLC4,7,8. It was reported that miR-34b/c overexpression increased radiosensitivity even at low doses of radiation in NSCLC cells9. miR-34b/c and miR-449a are two functionally related miRNA clusters10. However, the potential roles of miR-449a in overcoming radiotherapy resistance and the underlying mechanism in NSCLC remain unknown.
Reprogrammed cellular metabolism is a hallmark of cancer for rapid growth and proliferation11. Aerobic glycolysis represents a common phenomenon in cancer cells to consume more glucose to produce less energy and glycolytic intermediates that are required for accelerated tumor growth regardless of their oxygen status. Therefore, the alternative energy metabolism strategy utilized by cancer cells highlights a potential pathway for cancer therapy. Lactate dehydrogenase A (LDHA) is a step-control enzyme that plays critical roles in the modulation of glycolysis in cancer cells. Emerging evidence has documented that LDHA was upregulated and correlated with the clinicopathologic features and prognosis of patients with NSCLC12. Moreover, Zhou et al. reported that [18F]fluorodeoxyglucose ([18F]FDG) accumulation in primary lung adenocarcinomas was correlated with the expression of LDHA, and knockdown of LDHA inhibited cellular proliferation and increased drug sensitivity of cancer cells1. However, the epigenetic regulation of LDHA expression and its role in regulating radiosensitivity are not completely understood. In the current study, we emphasize the role of miR-449a in regulating radiotherapy sensitivity in NSCLC and then investigate the possible regulatory mechanism of LDHA by miR-449a, which will be utilized as a new strategy for overcoming radioresistance in patients with NSCLC.
MATERIALS AND METHODS
Tissue Sample Collection and Cell Cultures
Twenty-two NSCLC tissue samples and matched noncancerous normal tissue samples were surgically obtained in our hospital from March 2014 to April 2015. None of the patients received any chemotherapy or radiation therapy before surgery. Tissue samples were immediately snap frozen in liquid nitrogen until RNA extraction. In addition, serum samples were collected from NSCLC patients who were sensitive (n = 15) or resistant (n = 10) to radiotherapy in our department from August 2015. Informed consent was obtained from all patients, and the current work was approved by the Ethics Committee of Shaanxi Provincial Tumor Hospital.
Human lung adenocarcinoma cells A549 and NCI-H1299 were obtained from the ATCC (Manassas, VA, USA) and cultured in RPMI-1640 media containing 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) at 37°C in humidified atmosphere with 5% CO2. Normal human bronchial epithelial (HBE) cells were purchased from Sciencell (Carlsbad, CA, USA) and grown in specialty medium (Sciencell) at 37°C in a humidified atmosphere with 5% CO2. The lung adenocarcinoma cells were plated in 25-cm2 polystyrene flasks 24 h prior to exposure of γ-ray irradiation and then were grown to subconfluent monolayers. Then the irradiation of the cell monolayers was performed using a Gamma Cell 40 Exactor (Nordion International, Kanata, Ontario, Canada) at a dose rate of 2.4 Gy/min at increasing doses of 2, 4, 8, and 12 Gy.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA and total miRNA were extracted from NSCLC tissue samples and matched noncancerous normal tissue samples, as well as in A549 and H1299 cells γ-ray irradiated or not using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) based on the manufacturer’s protocols. For determining the miR-449a level after stimulation with IR, 1 μg of total RNA was reverse transcribed using One-Step PrimeScript® miRNA cDNA Synthesis Kit (Takara, Dalian, P.R. China) for 60 min at 37°C and for 5 s at 85°C. The subsequent qRT-PCRs were performed using the SYBR® Premix Ex TaqTM II (Perfect Real Time; Takara), with each reaction system containing 20 ng of templates and specific primers for miR-449a and U6. The expression of miR-124 was normalized based on the internal U6b using the 2−ΔΔCt method. Moreover, for the analysis of the mRNA levels of LDHA, the purified RNA (1.5 μg) was used to generate the first-strand cDNA using SuperScript® III Reverse Transcriptase (Invitrogen). The resultant cDNA was amplificated using a SYBR Green Real-Time PCR Master Mix Kit (Takara Biotechnology) according to the manufacturer’s instructions. Each PCR was performed in triplicate. LDHA primers (forward: 5′-CAT CCT GGG CTA TTG GAC TCT-3′; reverse: 5′-TGT CCC AAA ATG CAA GGA ACA-3′) were used with GAPDH primers (forward: 5′-GCC AAA AGG GTC ATC ATC TC-3′; reverse: 5′-GTA GAG GCA GGG ATG ATG TTC-3′) as the internal control. The relative fold change in expression with respect to a control sample was calculated by the 2−ΔΔCt method. All real-time PCR assays were performed with the Bio-Rad IQTM5 Multicolour Real-Time PCR Detection System.
Western Blot
Total cellular proteins were extracted using RIPA lysis buffer supplemented with a proteinase inhibitor cocktail, and the concentration of proteins in the centrifuged lysis buffer was determined using a commercially available BCA Assay Kit (Bio-Rad, Hercules, CA, USA). Total protein (20 μg) in the supernatant was mixed with 2× SDS loading buffer and then loaded onto a 10–12% precast Bis-Tris gel (Invitrogen). After electrophoresis, the separated proteins were transferred to PVDF (polyvinylidene difluoride) membranes (Millipore, Billerica, MA, USA) and blocked with 5% skim milk powder at room temperature for 60 min. Then the membranes were incubated with 1.0 μg/ml primary antibodies against LDHA, γ-H2AX, and β-actin (Cell Signaling Technology, Beverly, MA, USA) overnight at 4°C, respectively. Afterward, the membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies (1:1,000 dilution) for 60 min at room temperature. The target proteins were indicated using ECL Western blotting detection reagents (ECL New England Biolabs, Beverly, MA, USA). Quantification of the target proteins was performed using a Bio Image Intelligent Quantifier 1-D (Version 2.2.1; Nihon-BioImage Ltd., Tokyo, Japan) with β-actin used as an internal control.
Oligonucleotide Transfection
miR-449 mimics and the matched control were purchased from RiboBio (Guangzhou, P.R. China). LDHA siRNA and the corresponding negative control siRNA were obtained from Invitrogen. Cell transfection of miR-449a mimics and LDHA siRNA (50 nM) as well as their control oligonucleotides was performed using the Lipofectamine-based protocol (Invitrogen). The expression levels of miR-449a and LDHA in the cultured cells were evaluated 48 h after transfection.
MTT Assay
The cell proliferation capacity of the lung cancer cells in response to IR with or without miR-449a mimic transfection was determined using MTT assays. In brief, A549 and H1299 cells were grown at a density of 5,000 cells/well in 96-well plates and cultured at 37°C for 24 h and then transfected with miR-449a mimics and the negative control. The cells were then incubated for another 48 h and were radiated, trypsinized, and plated in six-well cell culture dishes in triplicate. The MTT assay was performed according to the manufacturer’s instructions (Molecular Probes, Eugene, OR, USA) after incubation for 48 h. The absorbance values were determined at 570 nm using a Spectra Max 250 spectrophotometer (VersaMax; Molecular Devices, Sunnyvale, CA, USA).
Colony Formation Assays
Cells transfected with miR-449a mimics or negative control were incubated for 48 h and then were subjected to irradiation. Afterward, the cells were incubated at 37°C in 5% CO2 for 14 days. The percentage of surviving cells was calculated by comparing the number of colonies formed in the nonirradiated control cultures.
Cell Apoptosis
Lung cancer cells were plated in six-well plates at 3 × 105 cells per well and were transfected with miR-449a mimics or LDHA siRNA followed by IR exposure. Cells were harvested after 48 h and then were stained with FITC-conjugated annexin V and propidium iodide (PI; BD Pharmingen, San Diego, CA, USA) based on the manufacturer’s instructions. The stained apoptotic cells were analyzed by flow cytometry (Accuri C6; BD Biosciences, San Diego, CA, USA).
Immunofluorescence Analysis of Intranuclear Focus Formation
Cancer cells transfected with oligonucleotides were plated on glass coverslips and exposed to indicated irradiation and were then fixed with 4% paraformaldehyde for 15 min. After blocking with 3% BSA/PBS containing Tween 20, cells were incubated overnight with primary antibody against γ-H2AX (ab22551; Abcam, Cambridge, UK). The cells were then washed twice with PBS and incubated with secondary antibodies for 30 min. Images were viewed and acquired using a 60× oil objective lens on a Zeiss fluorescence microscope controlled by AxioVision 4.8 software.
Determination of Glucose Uptake and Lactate Production
The IR-exposed lung cancer cells were starved for 3 h in glucose-free medium and then were incubated with 0.1 mM 2-deoxy-d-glucose (2-DG) (Sigma-Aldrich, St. Louis, MO, USA) containing 10 μCi/ml 2-deoxy-d-[2,6-3H] glucose (Sigma) for 10 min at 37°C. The reaction was terminated by washing three times with ice-cold PBS containing 10 mM d-glucose. Afterward, the cells were lysed, and radioactivity taken up by the cells was measured by a liquid scintillation counter. Lactate production was measured by the lactate assay kit (Biovision, Mountain View, CA, USA) based on the manufacturer’s instructions. The results were normalized based on total cellular protein amounts.
Luciferase Report Assay
Dual-luciferase report assay was performed to evaluate whether miR-449a could bind to the 3′-UTR of the LDHA gene. In brief, A549 cells were plated in a 96-well plate and then were transfected with 50 nM miR-449a or its corresponding negative control miRNA. The cells were then cotransfected with 0.2 mg/ml of wild-type or mutant 3′-UTR of LDHA gene vectors. After incubation for 48 h, luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Firefly luciferase activity was then normalized to the corresponding Renilla luciferase activity.
Statistical Analysis
Statistical analyses of data were performed by one-way analysis of variance followed by Fisher’s PSLD post hoc test using SPSS 22.0 software (IBM, Chicago, IL, USA). The results shown are the mean ± SEM. A value of p < 0.05 was considered statistically significant.
RESULTS
Expression of miR-449a in Lung Cancer Tissues and IR-Exposed Lung Cancer Cells
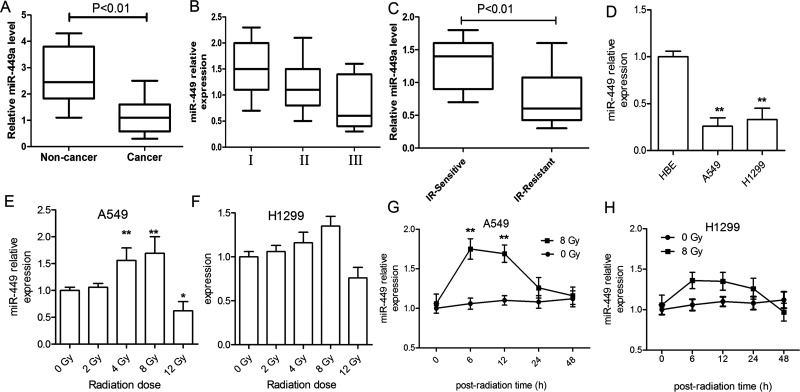
We first evaluated the miR-449a levels in lung cancer tissue samples and matched noncancerous samples using qRT-PCR. It was shown that miR-449a decreased to a low level in cancerous tissues compared to the matched noncancerous tissues (p < 0.01), which gradually decreased with the progression of the tumor stages, although no significant differences were indicated (p > 0.05) (Fig. 1A and B). Additionally, we detected the serum miR-449a levels in IR treatment-sensitive patients who had stable or decreased lung cancer and in IR-resistant patients who had progressed lung cancer. It was demonstrated that IR treatment-sensitive patients had higher levels of miR-449a than IR-resistant patients (p < 0.01) (Fig. 1C). Moreover, we detected miR-449a in normal HBE cells and lung cancer cells (A549 and H1299) using qRT-PCR. It was shown that miR-449a was significantly suppressed in lung cancer cells compared to normal HBE cells (p < 0.01) (Fig. 1D). Furthermore, the expression of miR-449a in response to various doses of γ-ray radiation ranging from 2 to 12 Gy at 12 h was determined in A549 and H1299 cells. The results demonstrated that miR-449a was significantly induced in A549 cells at 4- and 8-Gy IR, which had a sharp decrease after exposure to 12-Gy IR (Fig. 1E). However, we found that no statistical increase in miR-449a was observed in response to IR in H1299 cells at each irradiation dose (Fig. 1F). In addition, we detected the change in miR-449a expression after 8-Gy IR within 48 h in the two cell lines. It was shown that miR-449a had an acute increase within 12 h following IR exposure to 8 Gy in both A549 and H1299 cells to a different extent. However, its expression decreased 24 h after IR (Fig. 1G and H).
Figure 1.
Expression of miR-449a level in lung cancerous tissues and cells. (A) Reverse transcription polymerase chain reaction (RT-PCR) evaluation of miR-449a expression in non-small cell lung cancer (NSCLC) tissue samples and matched noncancerous normal tissue samples (n = 22). (B) Expression of miR-449a in NSCLC tissue samples at different stages (I: n = 6, II: n = 11, III: n = 5). (C) Levels of serum miR-449a in ionizing radiation (IR)-sensitive (n = 15) and IR-resistant (n = 12) patients. (D) Expression of miR-449a in normal human bronchial epithelial (HBE) cells and lung cancer cells. **p < 0.01, compared to HBE. (E, F) Change of miR-449a expression at 48 h of culture in response to increasing doses of IR at 2, 4, 8, and 12 Gy in A549 and H1299 cells. **p < 0.01, *p < 0.05, compared to 0 Gy. (G, H) Change in miR-449a expression within 48 h in A549 and H1299 cells after IR (8 Gy) treatment. **p < 0.01, compared to 0 Gy. All experiments were repeated at least three times.
miR-449a Restoration Profoundly Inhibited Lung Cancer Cell Viability Exposed to IR
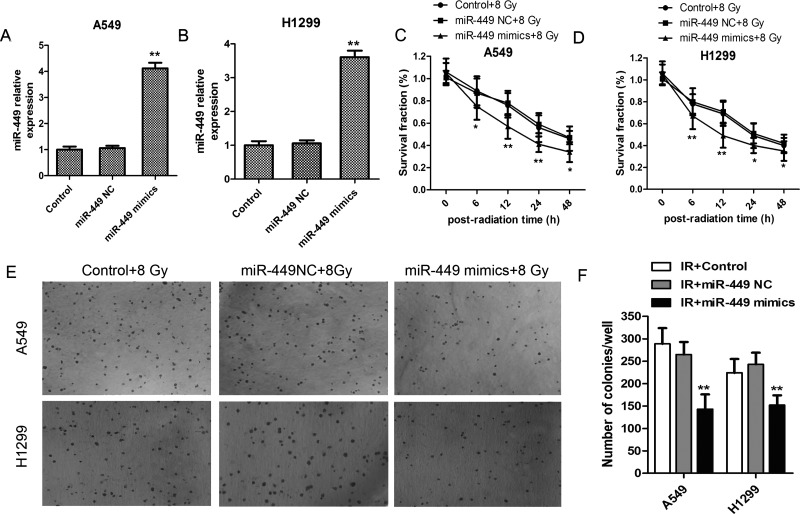
We supposed that IR-induced acute elevation of miR-449a may play a role in inhibiting lung cancer cell viability. Therefore, we transfected A549 and H1299 cells with miR-449a mimics to elevate its expression approximately fourfold in A549 and H1299 cells as shown in Figure 2A and B. Afterward, the miR-449a mimics/NC-transfected cells were exposed to 8-Gy IR, and cell viability was determined using the MTT assay. It was confirmed that miR-449a upregulation dramatically suppressed the growth of lung cancer cells in both A549 and H1299 cells (Fig. 2C and D). In addition, miR-449a upregulation profoundly inhibited colony formation of A549 and H1299 cells injured by IR (Fig. 2E and F).
Figure 2.
miR-449a upregulation promoted IR sensibility in lung cancer cells. (A, B) A549 and H1299 cells were transfected with miR-449a mimics, and miR-449a level was examined. (C, D) A549 and H1299 cells were transfected with miR-449a mimics or NC and exposed to IR at 8 Gy, and cell viability within 48 h was determined using MTT assay. *p < 0.05, **p < 0.01, versus Control + 8 Gy. (E, F) Soft agar colony formation assay of A549 and H1299 cells transfected with miR-449a mimics or NC for 2 weeks (n = 3). Average number of colonies and representative images are shown. All experiments were repeated at least three times.
miR-449a Restoration Promotes IR-Induced Cell Apoptosis and DNA Damage
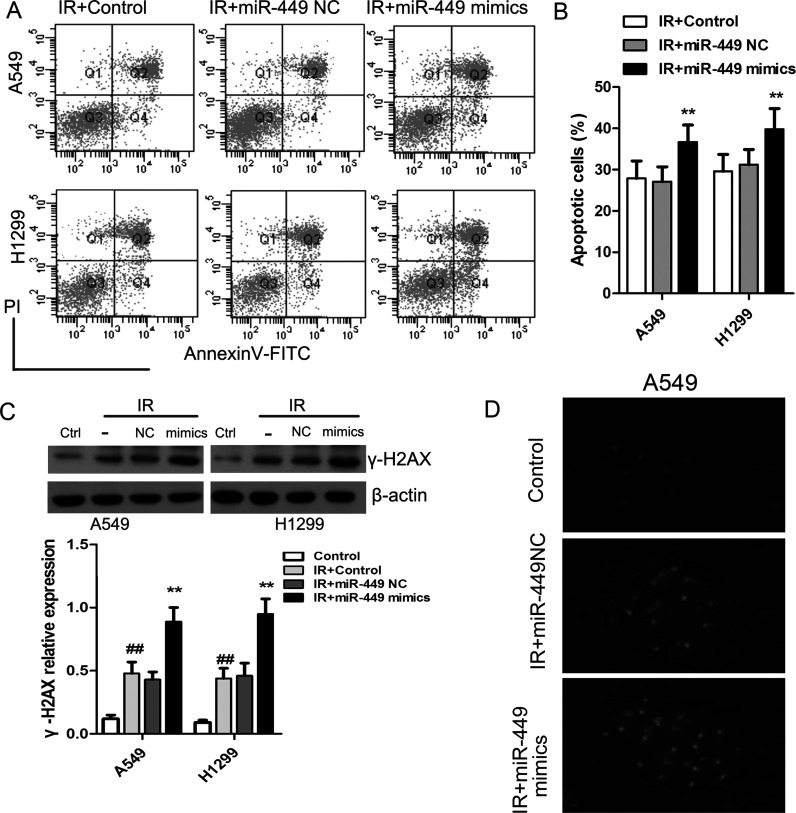
In addition, we evaluated cell apoptosis of lung cancer cells using the annexin V/PI method. The cancer cells were introduced with miR-449a mimics or the corresponding control and then were exposed to IR at 8 Gy. After 48 h of incubation, the fraction of apoptotic cells in A549 further increased to 36.7 ± 4.2% and to 39.8 ± 5.1% in H1299 cells (Fig. 3A and B). We determined the expression of γ-H2AX, a hallmark of DNA damage, using Western blot. We found that IR induced a significant increase in γ-H2AX expression compared to the non-IR-treated cells, which were profoundly corresponding by the administration of miR-449 mimics (Fig. 3C). Elevated γ-H2AX plaques were observed in IR-exposed A549 cells that were transfected with miR-449a mimics (Fig. 3D).
Figure 3.
miR-449a upregulation enhanced IR-induced cell apoptosis and DNA damage in lung cancer cells. (A, B) Cell apoptosis in IR-exposed cells in the presence or absence of miR-449 mimics. (C) Western blot analysis of the level of DNA double-strand break marker (γ-H2AX) after IR. **p < 0.01, compared to IR + miR-449 NC; ##p < 0.01, compared to Control. (D) After irradiation, A549 cells were fixed and stained with anti-γ-H2AX antibodies.
miR-449a Targeted LDHA to Suppress Glycolysis in Lung Cancer Cells
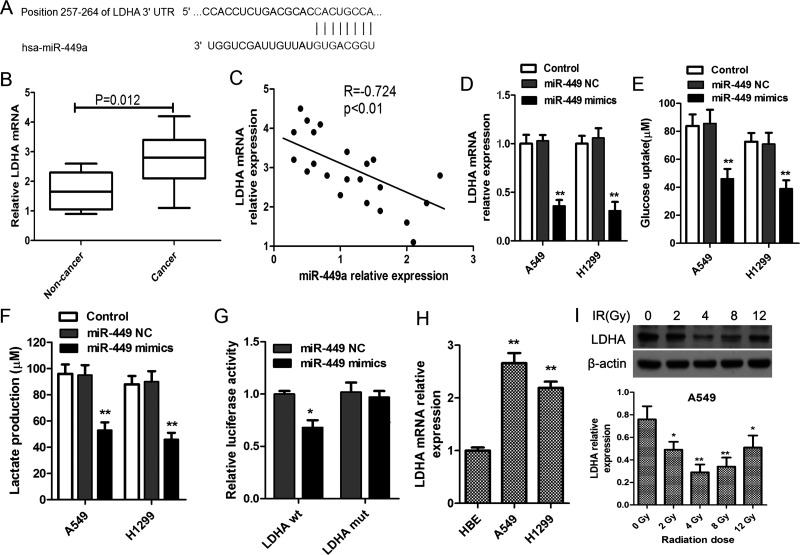
We predicted that LDHA was a potential target of miR-449a using miRDB and TargetScan network as shown in Figure 4A. We found that LDHA mRNA level in lung cancerous tissue samples was significantly higher than that in the matched noncancerous tissues (p = 0.012) (Fig. 4B). In addition, miR-449a levels in lung cancer tissues were negatively correlated to the levels of LDHA mRNA (R = −0.724, p < 0.01) (Fig. 4C). In order to examine whether miR-449a upregulation could bind to LDHA mRNA and change its expression, we determined the LDHA mRNA in miR-449a mimic- and negative control-transfected A549 and H1299 cells. The results showed that in comparison with the matched control cells, miR-449a mimics inhibited the level of LDHA in both A549 and H1299 cells (Fig. 4D). Likewise, we found that miR-449a upregulation also reduced the production of lactate and glucose uptake (Fig. 4E and F). Moreover, the luciferase report assays also confirmed that there was a binding site of miR-449a on the 3′-UTR of LDHA mRNA in A549 cells (Fig. 4G). We also examined the LDHA mRNA levels in normal bronchial epithelial cells and lung cancer cells and found that LDHA mRNA was substantially elevated in cancer cells (p < 0.05) (Fig. 4H). LDHA expression was inhibited in response to the gradually increased IR in a dose-dependent manner with a maximum inhibitory effect at 4 Gy. However, the LDHA levels had profound increase when exposed to IR at 8 and 12 Gy in comparison with that of 4 Gy (Fig. 4I).
Figure 4.
miR-449a targets lactate dehydrogenase A (LDHA) to suppress its expression. (A) LDHA mRNA levels in NSCLC tissue samples and matched noncancerous normal tissue samples (n = 22). (B) Bioinformatic prediction of the binding site of miR-449a on the 3′-untranslated regions (3′-UTRs) of LDHA. (C) Correlations of miR-449a and LDHA mRNA levels in NSCLC tissues. (D) A549 and H1299 cells were transfected with miR-449a mimics or negative control oligonucleotide, and miR-449a levels were evaluated using qRT-PCR after 24 h. Glucose uptake (E) and lactate production (F) in lung cancer cells in response to miR-449a upregulation by miR-449a mimics. *p < 0.05, **p < 0.01, versus control. (G) Luciferase assay of the binding of miR-449a on LDHA. A549 cells were cotransfected with miR-449a mimics, NC oligonucleotide, and a luciferase reporter containing LDHA 3′-UTR (LDHA wt) or mutant constructs of LDHA (LDHA mut). Then relative luciferase activity was determined. *p < 0.01, versus miR-449 NC. (H) LDHA mRNA levels in normal lung cell HBE and lung cancer cells (A549 and H1299). **p < 0.01, versus HBE. (I) A549 cells were exposed to escalating doses of radiation ranging from 2 to 12 Gy, and the expression of LDHA was detected using Western blot after incubation for 24 h. **p < 0.01, *p < 0.05, compared to 0 Gy. All experiments were repeated at least in triplicate.
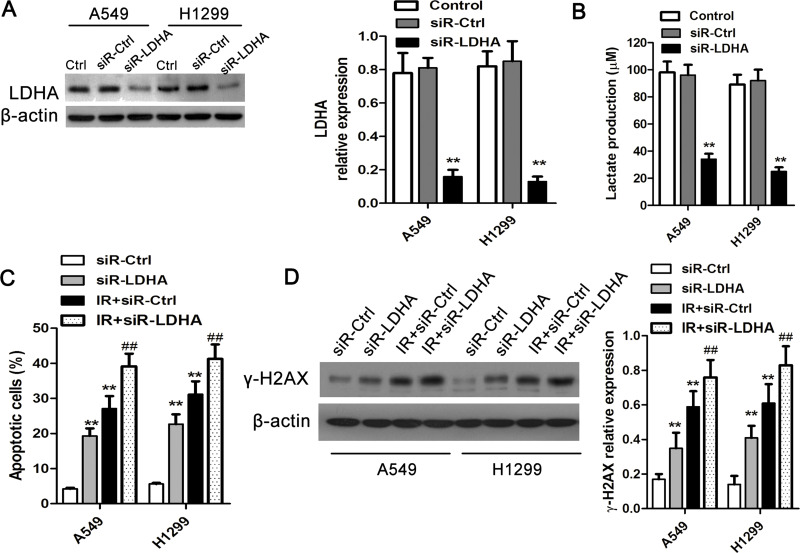
LDHA Was Involved in the Regulation of IR Sensitivity
To investigate the role of LDHA in the regulation of IR sensitivity of lung cancer cells, LDHA siRNA and its matched control siRNA were introduced into lung cancer cells (Fig. 5A) and then were exposed to IR at 8 Gy. Cellular DNA damage and apoptosis were evaluated after 24 h. It was shown that LDHA siRNA significantly suppressed the expression of LDHA along with the production of lactate (Fig. 5B). Moreover, after LDHA interference had significant inhibitory effects on cell apoptosis in A549 and H1299 cells without irradiation. In addition, the transfection of LDHA siRNA profoundly elevated the ratio of apoptotic cells following IR insults (Fig. 5C). Likewise, we found that LDHA siRNA markedly promoted the presentation of γ-H2AX in lung cancer cells in response to IR exposure (Fig. 5D).
Figure 5.
LDHA inhibition enhances IR sensitivity in lung cancer cells. (A) LDHA siRNA and its control siRNA were transfected into A549 and H1299 cells, and the expression of LDHA was determined 48 h after the transfections using Western blot. (B) Lactate production after LDHA inhibition in lung cancer cells was examined 48 h after transfection. **p < 0.01, compared to control. (C) Cell apoptosis in A549 and H1299 cells after LDHA inhibition with or without IR exposure at 48 h. (D) DNA damages by IR exposure in lung cancer cells transfected with LDHA siRNA or control siRNA. **p < 0.01, compared to siR-Ctrl; ##p < 0.01, compared to IR-siR-Ctrl. All experiments were repeated at least three times.
DISCUSSION
Radioresistance is a major obstacle and challenge for NSCLC treatment, and it is associated with poor prognosis in NSCLC patients. Our study demonstrated that miR-449a upregulation could enhance the sensitivity of lung cancer cells to IR by targeting LDHA, which in turn leads to the modulation of glucose metabolism by suppressing glycolysis and promoting cell apoptosis in lung cancer.
Emerging data showed that miR-449a was frequently downregulated in various cancers, including lung cancer, breast cancer, papillary thyroid carcinoma, hepatocellular carcinoma, among others4,13,14. miR-449a belongs to the p53-responsive miRNA family and shares similar structure and function with the miR-34 family. miR-449a was reported to play important roles not only in ensuring proper normal cell functions but also in avoiding cancer, marking a close link between cell differentiation and tumor suppression15. Moreover, miR-449 family members are potent inducers of cell death and cell cycle arrest16. Luo et al. identified that miR-449a was significantly downregulated in NSCLC tissues and cell lines, which promoted cell apoptosis by binding to and inhibiting c-Met4. In addition, Jeon and his colleagues demonstrated that miR-449a could inhibit the expression of histone deacetylases (HDACs) and may represent a potential therapeutic candidate in patients with primary lung cancers17. In our study, we also noticed that miR-449a was less expressed in lung cancer cells, which had an acute increase in lung cancer cells after IR stimulation, which may be an adaptive response to DNA damage induced by IR. However, this acute response might be insufficient to reverse the occurrence of radiotherapy resistance and affect cell fate. Similarly, previous evidence revealed that radiation acted as an environmental stimuli occurring at the mature miRNA level, and it induced rapid miRNA activation of miR-34 through 5′-end phosphorylation18. However, whether miR-449a upregulation employs the same mechanism as miR-34 in response to IR in lung cancer cells needs further studies.
LDHA is a step-control enzyme involved in cell glucose metabolism by converting pyruvate to lactate in an anaerobic condition, which is well acknowledged as anaerobic glycolysis. It is confirmed that abnormal glycolysis frequently occurs in cancer cells in spite of sufficient oxygen supply, termed “Warburg effects.” Studies have demonstrated that aerobic glycolysis is a hallmark of cancer cell metabolism, including lung cancers. The aerobic glycolysis reactions meet the high demand for amino acids, fatty acids, and nucleotides for rapid cancer proliferation and distant metastasis, although not for efficient production of adenosine triphosphate (ATP). Therefore, targeting glycolysis in lung cancer cells may highlight a promising strategy for diagnosis and treatment. As a typical modulator of glucose metabolism in cancer cells, the widely upregulated LDHA was indicated to be involved not only in the initiation but also in the tumor maintenance and progression of lung cancer12. A recent clinical study proved that LDHA was correlated with the clinicopathologic features and prognosis of NSCLC patients1. However, the role of LDHA in response to the treatment of radiation therapy in lung cancer cells is not completely understood. Our study found that LDHA expression and activity were suppressed following a mild dose of IR within 24 h. However, this inhibition was discounted against the profound increased dose of IR. We supposed that the DNA damage repair system may work to fight against IR insults. Recent compelling studies showed that targeting LDHA combined with radiotherapy could increase radiosensitivity in prostate cancer cells, indicating that LDHA was an ideal therapeutic target to develop combination therapy for overcoming radioresistance19,20. In the current study, LDHA was one of the direct targets of miR-449a as reflected by the luciferase reporter assay. Moreover, exogenous genetic increase in miR-449a could reinforce the efficiency of IR treatment, suggesting that miR-449a and γ-radiation synergistically promoted cell death. However, whether the impact of miR-449a on cell survival could be replicated in other cells and cancers still requires additional studies.
In conclusion, our study demonstrated that exogenous miR-449a could enhance the sensitivity of lung cancer cell to IR treatment by suppressing LDHA expression and activity, which in turn led to the inhibition of cell glycolysis and cell survival in lung cancers. Our study highlights an alternative approach to overcome radioresistance in cancers. However, further studies concerning the potential roles of miR-449a in other NSCLC cell lines and the underlying mechanism in response to radiation therapy are needed.
ACKNOWLEDGMENTS
This work was supported by the Science and Technology Project of Shaanxi Province (Grant No. 2017SF-172), BIKANG Funding (Grant No. 2017BIKANGJIJIN-020), the Science and Technology Foundation of Shaanxi Province (Grant No. 2012K13-02-37), the Fundamental Research Funds for the Central Universities (Grant No. 2014gjhz11), and the Funds of the Second Affiliated School of Xi’an Jiaotong University for Young Scientists [Grant No. YJ(QN)201305]. The authors alone are responsible for the content and writing of the manuscript.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Zhou X, Chen R, Xie W, Ni Y, Liu J, Huang G. Relationship between 18F-FDG accumulation and lactate dehydrogenase a expression in lung adenocarcinomas. J Nucl Med. 2014;55(11):1766–71. [DOI] [PubMed] [Google Scholar]
- 2. Dinh TK, Fendler W, Chalubinska-Fendler J, Acharya SS, O’Leary C, Deraska PV, D’Andrea AD, Chowdhury D, Kozono D. Circulating mir-29a and mir-150 correlate with delivered dose during thoracic radiation therapy for non-small cell lung cancer. Radiation Oncol. 2016;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee JH, Choi KJ, Seo WD, Jang SY, Kim M, Lee BW, Kim JY, Kang S, Park KH, Lee YS, Bae S. Enhancement of radiation sensitivity in lung cancer cells by celastrol is mediated by inhibition of hsp90. Int J Mol Med. 2011;27(3):441–6. [DOI] [PubMed] [Google Scholar]
- 4. Luo W, Huang B, Li Z, Li H, Sun L, Zhang Q, Qiu X, Wang E. MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-met. PLoS One 2013;8(5):e64759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hou J, Meng F, Chan LW, Cho WC, Wong SC. Circulating plasma microRNAs as diagnostic markers for NSCLC. Front Genet. 2016;7:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu W, He J, Chen D, Zhang B, Xu L, Ma H, Liu X, Zhang Y, Le H. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS One 2014;9(2):e87780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen X, Xu Y, Liao X, Liao R, Zhang L, Niu K, Li T, Li D, Chen Z, Duan Y, Sun J. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol. 2016;37(9):11927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li LP, Wu WJ, Sun DY, Xie ZY, Ma YC, Zhao YG. MiR-449a and CDK6 in gastric carcinoma. Oncol Lett. 2014;8(4):1533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balca-Silva J, Sousa Neves S, Goncalves AC, Abrantes AM, Casalta-Lopes J, Botelho MF, Sarmento-Ribeiro AB, Silva HC. Effect of miR-34b overexpression on the radiosensitivity of non-small cell lung cancer cell lines. Anticancer Res. 2012;32(5):1603–9. [PubMed] [Google Scholar]
- 10. Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, Yan W. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci USA 2014;111(28):E2851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hensley CT, DeBerardinis RJ. In vivo analysis of lung cancer metabolism: Nothing like the real thing. J Clin Invest. 2015;125(2):495–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ooi AT, Gomperts BN. Molecular pathways: Targeting cellular energy metabolism in cancer via inhibition of SLC2A1 and LDHA. Clin Cancer Res. 2015;21(11):2440–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Z, Wang J, Gao R, Yang X, Zhang Y, Li J, Zhang J, Zhao X, Xi C, Lu X. Downregulation of microRNA-449 promotes migration and invasion of breast cancer cells by targeting tumor protein d52 (TPD52). Oncol Res. 2016;25(5):753–61. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Li Z, Huang X, Xu J, Su Q, Zhao J, Ma J. MiR-449 overexpression inhibits papillary thyroid carcinoma cell growth by targeting RET kinas-β-catenin signaling pathway. Int J Oncol. 2016;49(4):1629–37. [DOI] [PubMed] [Google Scholar]
- 15. Lize M, Klimke A, Dobbelstein M. MicroRNA-449 in cell fate determination. Cell Cycle 2011;10(17):2874–82. [DOI] [PubMed] [Google Scholar]
- 16. Zhou Y, Chen Q, Qin R, Zhang K, Li H. MicroRNA-449a reduces cell survival and enhances cisplatin-induced cytotoxicity via downregulation of NOTCH1 in ovarian cancer cells. Tumour Biol. 2014;35(12):12369–78. [DOI] [PubMed] [Google Scholar]
- 17. Jeon HS, Lee SY, Lee EJ, Yun SC, Cha EJ, Choi E, Na MJ, Park JY, Kang J, Son JW. Combining microRNA-449a/b with a HDAC inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer 2012;76(2):171–6. [DOI] [PubMed] [Google Scholar]
- 18. Salzman DW, Nakamura K, Nallur S, Dookwah MT, Metheetrairut C, Slack FJ, Weidhaas JB. MiR-34 activity is modulated through 5’-end phosphorylation in response to DNA damage. Nat Commun. 2016;7:10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hao J, Graham P, Chang L, Ni J, Wasinger V, Beretov J, Deng J, Duan W, Bucci J, Malouf D, Gillatt D, Li Y. Proteomic identification of the lactate dehydrogenase A in a radioresistant prostate cancer xenograft mouse model for improving radiotherapy. Oncotarget 2016;7(45):74269–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koukourakis MI, Giatromanolaki A, Panteliadou M, Pouliliou SE, Chondrou PS, Mavropoulou S, Sivridis E. Lactate dehydrogenase 5 isoenzyme overexpression defines resistance of prostate cancer to radiotherapy. Br J Cancer 2014;110(9):2217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]