Abstract
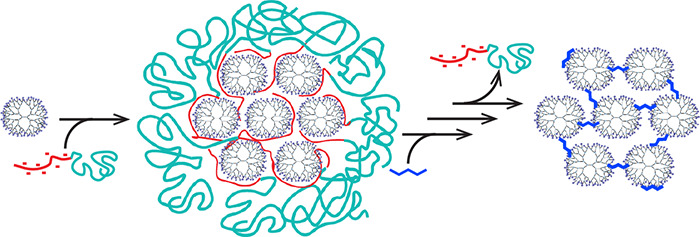
A versatile method is presented to form dendrimer superstructures by exploiting coacervate-core micelles as a template to confine and organize the hyperbranched macromolecules. First, complex coacervate-core micelles are formed from negative–neutral block copolymers and positively charged polyamidoamine dendrimers. The dendrimers inside the micellar core are then covalently cross-linked with each other upon addition of glutaraldehyde. After removal of the block copolymer from the assembly by increasing the salt concentration, consecutively, the formed Schiff bases cross-linking the dendrimers are reduced to amines, followed by a final dialysis step. This leads to well-defined covalently cross-linked nanostructures, coined dendroids, with a size of around 30 nm in diameter and a molecular weight of approximately 2.5 MDa. By incorporating dendrimer-encapsulated gold nanoparticles (AuDENs) into the micelle template strategy, the aggregation number of dendrimers inside the dendroids is determined by counting the nanoparticles in TEM micrographs. Furthermore, TEM performed at different tilt angles and AFM analysis corroborate formation of stable, covalently linked three-dimensional structures. Reconstruction of the TEM tilt series results in a tomogram further illustrating the 3D distribution of the gold nanoparticles, and hence the individual dendrimers, in the nanostructure. These dendroids appear to have a hard, poorly compressible core and a relatively soft outside. The versatility of the hierarchical building up of the supermolecules is illustrated by the controlled and synchronous incorporation of empty dendrimers and AuDENs into a single hybrid dendroid structure. The presented strategy allows for the preparation of a variety of classes of supermolecules, depending on the type of micellar-core macromolecule, e.g., dendrimer, cross-linker, and nanoparticles, used. Considering the broad interest in dendrimers as well as micelles in a plethora of research areas, e.g., (targeted) drug delivery, biomedical imaging, theragnostics, and catalysis, there is a great potential for dendroids and related classes of covalently linked macromolecules, viz., supermolecules.
Keywords: dendrimers, supermolecules, covalent tethers, micelle template, nanostructures, TEM tomography
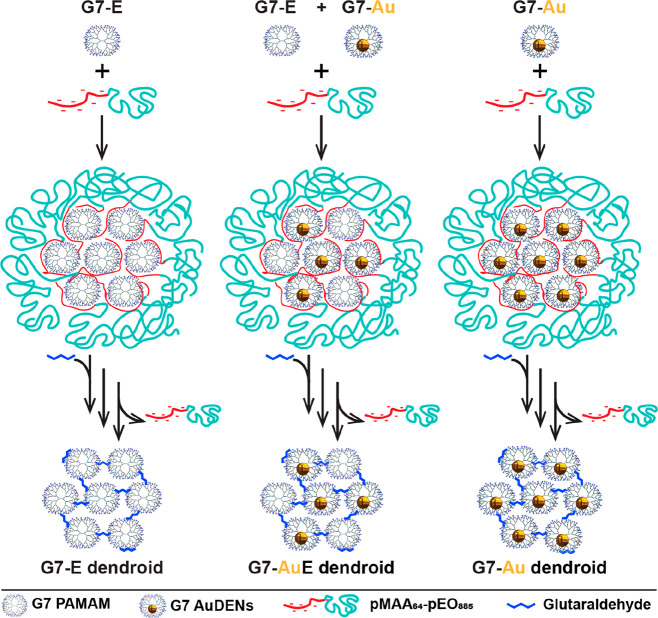
Mastering the design and manufacturing of molecular superstructures is one of the major driving forces for synthetic chemists; in fact, a plethora of self-assembly strategies has fulfilled the promise to create nanoscale superstructures, exploiting mainly noncovalent interactions of (macro)molecules, metal ions, and nanoparticles.1−3 Exploring geometric constraints in macromolecular building blocks has led to great control in 1D, 2D, and 3D structures.4,5 Micelles for example, and in particular polyion complex or complex coacervate-core micelles, PICs and C3Ms, respectively, are well studied self-assembled nanostructures, and their formation and breakdown are often controllable.6−9 To improve the stability of such structures against high salt concentrations and large pH ranges, chemical cross-linking of micellar subcomponents has been explored.8 A challenge that remains is the design of strategies for discrete synthesis of nanostructures of a single type of macromolecular building blocks that are tethered by covalent bonds and the consecutive characterization of such structures.10 Here we present a promising template strategy to build up discrete and well-defined supermacromolecular structures in the size range of 30 nm and a molecular weight of about 2.5 MDa. We start from coacervate-core dendrimicelles, in which the charge interactions of the respective polymers result in assembling a controlled number of dendrimers in close proximity to each other, to allow formation and isolation of covalently cross-linked discrete dendrimer clusters, coined dendroids.
Dendrimers, with polyamidoamines (PAMAMs) being the most widely studied example, are highly symmetrical, hyperbranched polymers with a controlled shape, size, and high density of functional end groups.11−13 Furthermore, it is possible to functionalize the outside end groups14,15 or encapsulate small molecules, e.g., drugs, or nanoparticles inside the voids of the dendrimers,16−19 making them ideal building blocks for the formation of nanostructures.20 The synthesis of PAMAM dendrimers, up to generation 10 with a size of about 14 nm, is well established. However, the traditional techniques of iterative reaction and purification have considerable disadvantages for manufacturing the higher generation structures, such as a large number of steps required during synthesis and the dense packing of end groups, all leading to polydisperse and less well-defined compounds.21 Therefore, Tomalia and other groups introduced different approaches to scale up dendrimers, forming clusters by self-assembling or cross-linking dendrimers in solution.21−29 Most of these methods, however, offer limited control of size, aggregation number, or stability. We envisioned an alternative way of controlled nanocluster formation with a template method, which allows for good control of morphology, size, and structure of the resulting nanomaterial;30 dendrimer-based micelles would then lead to the formation of covalently linked dendrimer superstructures.
Micelles provide a promising platform for the template method and have been used, for example, for the fabrication of hollow nanospheres and nanocages.31,32 The groups of Crooks and Kataoka already studied the encapsulation of single dendrimers and dendrimer zinc porphyrins in micelles, respectively.33,34 More recently, our group developed the concept of PAMAM dendrimicelles, formed by coassembly of negatively charged carboxylic acid-terminated dendrimers and oppositely charged poly(vinylpyridine)-poly(ethylene oxide) diblock copolymers or positively charged amine-terminated PAMAM dendrimers and oppositely charged poly(methacrylic acid)-poly(ethylene oxide) diblock copolymers.35,36 The aggregation number of dendrimers in the micellar core can be tuned by varying the dendrimer generation36 or the pH.37 Furthermore, nanoparticles have been proven to reside inside the PAMAM dendrimers,19,38,39 providing the possibility to localize and monitor the otherwise poorly visible dendrimers in transmission electron microscopy (TEM). In fact, recently we have shown that it is also possible to encapsulate dendrimer-encapsulated gold dendrimers (AuDENs) into dendrimicelles,35,40−42 providing a straightforward determination of the micellar aggregation numbers.
Here, we present how dendrimicelles can be used to fabricate dendrimer-based nanoclusters based on the template method depicted in Scheme 1. The dendrimers in the micellar core are covalently cross-linked, and subsequently the block copolymer is removed, which results in well-defined nanoclusters, dendroids, consisting of the dendrimers previously present in the micellar core. More precisely, we use amine-terminated PAMAM dendrimer generation 7, which we cross-link with glutaraldehyde, a widely used cross-linker for macromolecules containing amine groups.28,43,44 Glutaraldehyde is able to react with the primary amines in the dendrimer structure to selectively cross-link the dendrimers inside the micellar core. By increasing the NaCl concentration the block copolymer is removed from the cross-linked dendrimer structure. In order to prevent hydrolysis of the cross-linker, the formed imine bonds are subsequently reduced by sodium borohydride to yield stable secondary amines. The formation of dendrimicelles and dendroids has been monitored with dynamic light scattering (DLS) and atomic force microscopy (AFM). The resulting nanostructure is a supermolecule of about 2.5 MDa consisting of about 21 tethered single building blocks, i.e., generation 7 dendrimers. We also show that AuDENs and a mixture of AuDENs and empty dendrimers can be encapsulated in dendrimicelles, consecutively forming completely filled or half-filled DENdroids, respectively. In fact, the gold nanoparticles allow for a straightforward characterization of the hybrid structures by normal and cryo transmission electron microscopy, including TEM tomography.
Scheme 1. Scheme of the formation of dendroids based on generation 7 based PAMAM dendrimers. Left column: Dendrimicelles are made by mixing dendrimers with a pMAA64-pEO885 block copolymer at stoichiometric charge ratio. After micelle formation, glutaraldehyde is added to cross-link the dendrimers in the micellar core. Upon increasing the ionic strength to a NaCl concentration of 0.8 M, the block copolymer dissociates from the core, and dendroids are formed. The imide bonds are subsequently reduced by sodium borohydride, and the dendroids are isolated via dialysis. Right column: When using dendrimer-encapsulated gold nanoparticles (AuDENs) with a pMAA64-pEO885 block copolymer at stoichiometric charge ratio, dendroids are formed where the number of gold particles is equal to the aggregation number of the dendrimers in the micelles. Middle column: Dendroids with different loadings of AuDENs are readily obtained by mixing empty dendrimers and Au-DENs at the start of the procedure. Note: The charges on the periphery of the dendrimers (positive at pH 7) and inside micelles are omitted for clarity reasons.
Results and Discussion
The assembly of dendrimicelles and the following dendroid formation, via cross-linking and consecutive reduction of the imines, and isolation were monitored by DLS. Next, the influence of different ratios of cross-linking agents on the dendroid formation was investigated. Characterization of the dendroids by AFM was used to prove the 3D shape and hence covalent cross-linking of the dendrimers. The number of dendrimers present in the supermolecules was determined exploiting gold nanoparticles residing inside the dendrimers, with TEM allowing straightforward counting to obtain the aggregation number. The versatility of the template methodology was further extended by mixing in different dendrimers, i.e., empty and Au-NP filled, resulting in hybrid structures. Finally, the 3D shape of the dendroids has been visualized using variable-angle TEM and tomography, completing the structural characterization. Below, these various aspects are discussed in more detail.
Formation of Dendroids
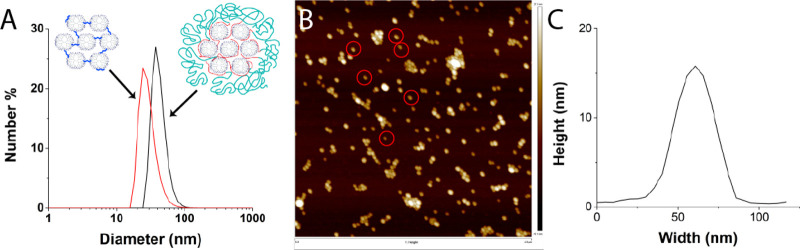
Following the strategy described in Scheme 1 (left column), a series of dendroids was prepared. Based on previous experience with dendrimicelles,35,36 generation-7 amine-terminated PAMAM dendrimers (G7) were used; in particular, G7 allows controlled dendrimer-encapsulated nanoparticle formation and avoids dendrimer-stabilized nanoparticle formation.16−18 Dendrimicelles were formed upon mixing aqueous solutions of dendrimers and poly(methacrylic acid)-poly(ethylene oxide), pMAA64-pEO885, as an anionic-neutral diblock copolymer. Micelle formation was done at pH 7, as both the PAMAM and the polymethacrylic acid subpart are oppositely charged at this pH. The neutral poly(ethylene oxide) subpart of the block copolymer assures stability of the formed coacervate nanostructure by forming a corona around the dendrimer core. Dendrimicelle formation was confirmed by DLS showing a hydrodynamic diameter of 41–48 nm (Figures 1A, S1).
Figure 1.
(A) DLS number-averaged plot of dendrimicelles (black) and the resulting dendroids after purification with dialysis (red). (B) AFM image of G7-E-based dendroids. (C) Corresponding average height profile of dendroids indicated with a red circle in B.
After dendrimicelle formation, glutaraldehyde was added to cross-link the dendrimers inside the micellar core. A similar approach was successfully used by Maggi etal. to form chitosan nanogels inside micelle cores.45 DLS measurements show that the size of the dendrimicelles decreases a few (1–3) nanometers upon addition of glutaraldehyde, indicating that possibly a few block copolymers are released from the micelles, or the chemical cross-linking might lead to a minor contraction or reorganization of the core (Figure S1). The micelles are still intact after addition of the cross-linker, in fact many amine end groups are still available for the complex coacervate interaction and potentially later can be used for consecutive functionalization of the dendroids. After addition of NaCl to a final concentration of 0.8 M, the observed hydrodynamic radius by DLS decreased by about 10 nm (Figures 1A, S1). In previous research the thickness of the micellar corona was estimated to be around 7 nm42 using cryo-TEM, so the here-observed decrease in size illustrates the removal of the block copolymer from the complex, and only the dendrimer core is seen. The removal of the block copolymer was monitored by fluorescence emission using a fluorescently labeled block copolymer (Figures S3, S4). The fluorescence emission decreased dramatically after dialysis for 3 days, showing that the block copolymer was successfully removed from the dendroid sample (Figure S5). A control experiment, in which the cross-linking step was omitted, showed micelles of 46 nm diameter, which after addition of NaCl increased dramatically in size, indicating swelling and/or formation of worm-like structures due to charge screening (Figure S2). This observation corroborates that 0.8 M is above the critical salt concentration of dendrimicelles, determined to be ∼0.5 M in previous research.35
For reduction of the imines, formed in the cross-linking step, to the more stable secondary amines and concomitant reduction of possibly residual free (glutar-) aldehydes to alcohol groups, 10 molar equivalents of sodium borohydride with respect to the cross-linker were added to the dendroid solution. After NaBH4 addition, the size observed with DLS did not change, indicating that the reduction step does not influence the structure of the dendroids (Figure S1). The hydrodynamic radius of the dendroids after purification with dialysis is 29–35 nm. The dendroid size is similar to the micellar core size of DENdrimicelles as determined with cryo-TEM, vide infra. Interestingly, the packing fraction of DENs inside the micellar core was previously estimated to be about 0.9,42 leading to the hypothesis of the close proximity of the dendrimers in the micelle core to allow for efficient cross-linking. In addition, the size of the core dendrimer aggregates (which remains about the same after removal of the block copolymer when cross-linked with the short cross-linker) proves in fact that the dendrimers are densely packed in the core of dendrimicelles. We speculate that the reciprocal dendrimer proximity and other properties of the dendroids could be simply tuned in the future by using cross-linkers with different lengths and structure.
Optimizing Cross-Linking and Stability
To optimize the core cross-linking, 15 different ratios of dendrimers to glutaraldehyde were tested, from 1:10 to 1:500 (Table S1). When using the ratios below and including 1:30, an increase in size after addition of NaCl is observed, suggesting that the core cross-linking degree is not sufficient to form well-defined dendroids. Most likely smaller networks form, and/or predominantly intramolecular cross-linking in single dendrimers occurs. The procedures with higher glutaraldehyde ratios above and including 1:40 showed the earlier-mentioned characteristic decrease in size by ∼10 nm upon NaCl addition, indicative for the dendroid formation. For the experiments described above and below, unless otherwise stated, we chose the ratio 1:60 to be sure that cross-linking worked well, yet to have still many amine end groups available in the final nanostructure. Generation 7 PAMAM dendrimers have, theoretically, 512 terminal primary amine groups, and glutaraldehyde has two reactive aldehyde groups. Therefore, a ratio of 1:60 corresponds to a maximum functionalization degree of about 25% of the number of dendrimer end groups.
As imine bonds are prone to hydrolysis, we decided to incorporate a reduction step with sodium borohydride. Interestingly, while monitoring the size of the dendroids by DLS prepared with and without the reduction step, we did not observe a difference over the time course of 14 days. Due to the multitude of cross-linking groups, in combination with the reversible nature of the Schiff base formation and hydrolysis, the imine-based dendroids show a notable kinetic stability. We do believe that this allows for designs in which the slow degradation of the dendroids into individual dendrimers can be tuned with cross-linker ratio and type, with potential applications where slow release of components is of relevance, such as biomedical diagnostics and therapeutics.
AFM of Dendroids
Figure 1B shows the AFM image of dendroids, and in Figure 1C the average height profile of a selected number of dendroids is provided; the ratio of dendrimer to glutaraldehyde in the preparation here was 1:100. The average height observed is 16 nm, and the width is 31 nm at mid height (and 56 nm at the bottom). We here will focus on the height alone and will discuss the width of the dendroids more in detail in the TEM discussion below. A single G7 PAMAM dendrimer has a diameter in solution of about 8 nm, but the height of a single G7-PAMAM as measured by AFM is about 2 nm (Figure S6A,B).46 The height of dendrimicelles measured by AFM is about 5 nm, showing that these micelles are not stable in the dried measuring conditions (Figure S6C,D). Therefore, the observed height of 16 nm for the dendroids excludes the formation of a 2D, flat, array of cross-linked dendrimers, but corroborates a 3D superstructure of multiple covalently tethered dendrimers. Moreover, considering the compressibility of a single G7-PAMAM, as can be deduced from the 2 nm height measured with AFM,46 it is reasonable to assume the cross-linking in the dendroids results in an overall stiffening of the individual dendrimer components. Whereas a single G7 PAMAM is compressed by a factor of 4, comparing the hydrodynamic radius to the AFM height, the dendroids have a compressibility of only a factor of 2. Clearly the cross-linking results in the stiffening and reduced compressibility of the individual dendrimers, yet the dendroid as a whole is still somewhat compressed in the vertical direction when dried on a mica surface. It is likely that flattening occurs to dendrimers when part of a dendroid structure, but clearly much less than in individual dendrimers and also less than in tecto core–shell dendrimers.47 A micelle core is a dynamic environment of different macromolecular subcomponents, and cross-linking of about 21, vide infra, dendrimers can occur in different ways. Although higher generations such as G7 are considered to be spherically shaped particles in solution, it is unlikely the dendrimers are organized in a close-packed sphere organization. Besides the dynamic nature of the coacervate micelle core, one also has to consider the presence of the block copolymer in the micelle core. Therefore, it is probable that the dendrimers are not all perfectly three-dimensionally cross-linked to more dendrimers, but occasionally also only to one or two neighboring dendrimers. This will allow for a 3D structure with certain compressibility, as corroborated also by TEM, vide infra.
DENdroids Based on Dendrimer-Encapsulated Au-Nanoparticles
For more extensive characterization, micelles with generation 7 based dendrimer encapsulated gold nanoparticles (AuDENs) were formed. The G7-Au256 DENs were characterized with UV–vis and TEM and had a size of 1.5 ± 0.5 nm (Figure S7). Upon mixing a block copolymer with AuDENs (G7-Au micelles), DENdrimicelles are formed with a 46 nm diameter determined with DLS, similar to the above-described dendrimicelles (Figure S8A). In addition to the overall size, from cryo-TEM micrographs also the micelle-core size is readily determined to be 26 ± 3 nm (Figure S9A,B).35 Most informative, the aggregation number of dendrimers in micelles is deduced by simply counting the AuDENs per micelle in the cryo-TEM pictures, as only one Au-nanoparticle is encapsulated per dendrimer.16 The G7-Au-based DENdrimicelles contain 22 ± 4 AuNPs per micelle (Figure S9C).
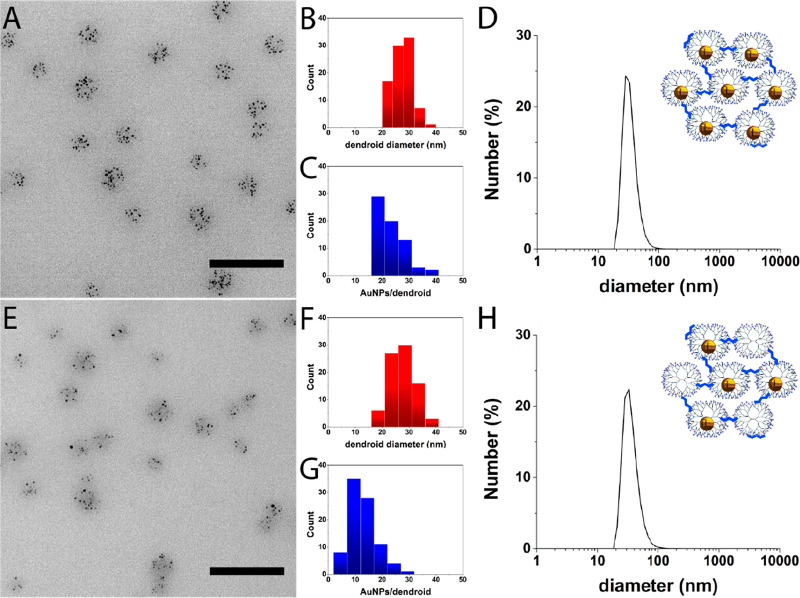
Following the procedure described above for the dendrimicelles and in Scheme 1 (right column), the DENdrimicelle cores were cross-linked with glutaraldehyde and reduced with sodium borohydride. Importantly, due to the covalent tether between the dendrimers, the DENdroids are stable to drying, in contrast to the parent DENdrimicelles, and therefore can also be characterized by normal TEM, rather than cryo-TEM. Figures 2A and S10 show representative TEM pictures of G7-Au DENdroids. The size of the DENdroids is 27 ± 3 nm (Figure 2B), similar to the parent micellar core size determined by cryo-TEM. DLS measurements show a hydrodynamic diameter of 34 nm for the DENdroids (Figure 2D), similar to the dendroids with “empty” dendrimers described above. Apparently, the packing density does not change significantly when going from the core of DENdrimicelles to DENdroids. The number of gold particles per dendroid is 21 ± 6 for the DENdroids, (Figure 2C); this corresponds well to the numbers found for the DENdrimicelles, suggesting that all DENdrimers inside a micelle are cross-linked and joined in a single supermolecule. Considering the aggregation number of 21 G7-AuDENs, the DENdroids have a total molecular weight of around 3 MDa. Taking the hydrodynamic radius, the volume of these dendroids is on the order of 10 × 103 nm3.
Figure 2.
Upper part (A–D) shows G7-Au dendroids (100% G7-Au), and lower part (E–H) shows G7-AuE dendroids (50% G7-E + 50% G7-Au). The left part shows representative TEM pictures of G7-Au (A) and G7-AuE (E) based dendroids. Scale bars represent 100 nm. The middle part shows histograms of G7-Au (B, C) and G7-AuE (F, G) based dendroids. The size of the G7-Au dendroids is 27 ± 3 nm (B, red), and the size of the G7-AuE dendroids is 28 ± 5 nm (F, red). The G7-Au and G7-AuE based dendroids contain 21 ± 6 AuNPs per dendroid (C, blue) and 12 ± 5 AuNPs per dendroid (G, blue), respectively. The right part shows number-averaged DLS plots of G7-Au (D) and G7-AuE (H) based dendroids, showing a size of about 35 nm for both dendroid types. The cartoons illustrate the type of dendroids characterized by DLS and TEM.
To prove the effectiveness of cross-linking for dendroid formation, above we described the control experiment with DLS monitoring of non-cross-linked dendrimicelles upon salt addition. We performed a similar experiment but monitoring by TEM analysis the effect when not cross-linking the core of DENdrimicelles. Figure S11 shows a TEM picture of a sample of G7-Au-based micelles, formed without a cross-linking step, after addition of NaCl to a final concentration of 0.8 M. Clearly only small clusters and individual DENs are observed. This again confirms that a salt concentration of 0.8 M is high enough to disrupt dendrimicelles and that chemical cross-linking is required to form dendroids or DENdroids.
Hybrid Den/DENdroids with Empty Dendrimers and AuDENs
To explore the versatility of hierarchically controlling dendrimer superstructures, we formed dendroids containing 50% empty dendrimers (G7-E) and 50% AuDENs (G7-Au). For this, G7-E and G7-Au dendrimers were mixed before adding the block copolymer to form dendrimicelles (Scheme 1, central column). These half-filled dendrimicelles (G7-AuE micelles) have a hydrodynamic diameter of 43 nm determined with DLS (Figure S8B). Table S2 shows the DLS data of G7-E, G7-Au, and G7-AuE micelles and corresponding dendroids. Figures 2E and S12 show TEM pictures of the resulting dendroids after cross-linking as described above. The formed dendroids have a size of 28 ± 5 and 35 nm determined with TEM and DLS, respectively (Figure 2 F,H), which again is in agreement with the G7-AuE micelle core size.35 The number of G7-Au DENs per dendroid was determined to be 12 ± 5 (Figure 2G). As this is half of the aggregation number found in DENdroids containing only G7-Au DENs, this proves that den/DENdroids were formed, i.e., with half of the dendrimers in the supermolecule filled with a Au-nanoparticle. This experiment shows that multicompartment dendroids containing different dendrimers with different encapsulations can be formed in a finely controllable way by simply mixing the different dendrimers prior to micelle formation in the desired ratio.
Tilt-Angle TEM and Tomography
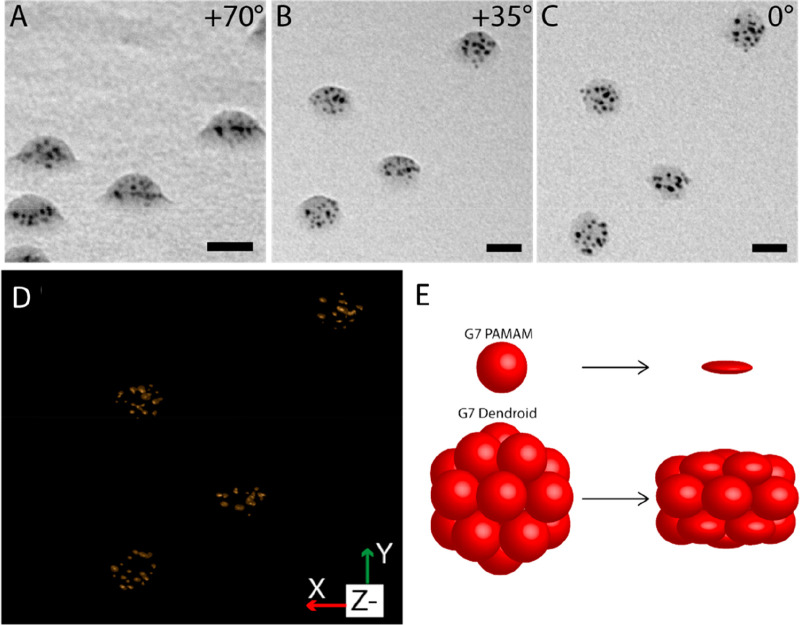
The high electron density of the AuDENs provides a stark contrast allowing more detailed TEM studies to reveal the 3D structure of the dendroids. TEM pictures of G7-Au-based DENdroids at different tilt angles were recorded, and Figure 3A–C (enlarged in Figure S13) show representative DENdroids measured at +70, +35, and 0 degree tilt angles. The pictures underline that dendroids have a clear three-dimensional structure, indicating that the cross-linking worked on the whole dendrimicelle core (see also Supplementary Movie 1: Tilt series). Similar to AFM, also here, due to the drying process, the DENdroids occur flattened with a kind of disc or dome-like shape. Considering the aggregation number of the dendrimers in the micelle core and consecutively in the dendroids, i.e., ∼21, the supermolecule consists of covalently linked dendrimers in a 3D fashion; that is not a perfect spherical shape under the TEM conditions, yet it is reasonable to assume in solution it is. This flattening also shows that the dendroids are, to a certain extent, still flexible, meaning that the cross-linking leaves some compressibility of the individual dendrimers. We reckon that this flexibility could be tuned by using different cross-linking ratios and cross-linking molecules. In order to visualize the 3D structure, we performed a tomographic reconstruction based on the TEM tilt angle series.
Figure 3.
TEM picture of G7-Au-based dendroids at tilt angles of (A) +70 degrees, (B) +35 degrees, and (C) 0 degree, showing their 3D structure. Scale bars represent 20 nm. (D) Tomographic reconstruction of the dendroids shown in A–C, showing the XY isosurface. The Supporting Information figures and movies provide more detailed data. (E) Cartoon illustrating the compressibility of G7 PAMAM dendrimers and G7-based dendroids when dried on a surface. Dendrimers and dendroids are vertically compressed by 75% and 35%, respectively.
Tomography of hybrid nanomaterials containing gold nanoparticles can severely be hampered by movement, during the measurements, of the particles or of the structure as a whole, as well as from changes to the nanoparticle size, (de)composition, and clustering.48,49 In our recent DENdrimicelle work we have also observed the various effects of prolonged and stroboscopic TEM measurements with the altering of size and distribution of the AuDENs as well as of the organic material.40 In fact, also for the dendroids we clearly observe an alteration of AuNP size and position in the tilted-angle TEM micrographs, as is evident from the full tilted angle series (Figure S13, Supplementary Movie 1). The tomographic reconstruction of the acquired TEM tilt series nevertheless resulted in an acceptable spatial resolution and provides complementary information on and insight into the basic shape, size, and nanoparticle distribution of several individual DENdroids. Figure 3D and S14 and Supplementary Movies 2 and 3 show the reconstruction of some of the DENdroids in the field of view shown in Figure 3 and based on the full tilt series. In particular the high tilt angle TEM pictures and reconstruction show that the AuNPs are distributed evenly in the space of the dendroids, indicating that the dendrimers are packed in a three-dimensional structure. Similar to what was observed in AFM, the 3D shape is clear from the tomogram, with multiple layers of dendrimers cross-linked to each other in a dome or disc structure.
Size, Density, and Compressibility
Combining the information from TEM, cryoTEM, and AFM allows for a more in-depth discussion of the overall size and compressibility of the dendroid structures. First, the average diameter of the dendroids by TEM, i.e., 27 nm (Figure 3), is practically the same as what has been determined to be the core in the parent dendrimicelles by cryo-TEM. Hence, we can conclude that the dendroids are not stretched out laterally once they are positioned on a surface for TEM analysis, which tells about the multitude of covalent cross-linking groups connecting the dendrimers together. The volume of a sphere with a radius of 27 nm encompasses 10 × 103 nm3. Taking 8 nm as the diameter for a G7 PAMAM,50 its volume is 268 nm3 and 21 dendrimers have a total volume of about 5 × 103 nm3. This implies that the volume density of dendrimers in the dendroids is about 50%.
From the X and Z projections of the reconstructed dendroids (Figure S14B,C), the height–diameter ratio of the AuNP delineated part is ∼1:2. Using a width of 22 nm, determined from Figure 3C, this leads to a height of about 12 nm as delineated by the AuDENs. Assuming the AuDENs to be positioned on average more or less in the center of the 8 nm G7 PAMAM, we can guesstimate the size of the individual dendroids from the TEM tomogram with an additional 3 nm of dendrimer on the outside of the disc structure delineated by the AuDENs. This makes the overall shape a disc or dome shape with the diameter and height being 28 and 18 nm, respectively. The compressibility (expressed as the reduction in height) is then ∼33%. Presumably, surface dendrimers have less cross-links compared to dendrimers residing in the center of the dendroid, leading to less rigidity and therefore more compressibility. The overall compressibility of dendroids is clearly strongly reduced with respect to a single dendrimer (Figure 3E). For example, a G7 PAMAM dendrimer of 8 nm diameter in solution has been measured by AFM to be only 2 nm high once prepared on a surface, implying a compressibility (expressed by the reduction in height) of 75%. Once positioned on a surface in AFM or TEM measurements, we can define a top and bottom to the dendroids; we believe the top and bottom ones can be compressed more than the dendrimers in the central layer of the dendroid. In fact, if these top/bottom dendrimers are compressed by about 50%, then they would account almost fully for the compressibility as a whole of the dendroids (see Figure 3D). In other words, these dendroids can be considered hard-core nanostructures with a relatively soft outside layer.
Conclusion
We have presented the controlled synthesis of well-defined 3D dendrimer clusters, coined dendroids, consisting of covalently linked dendrimers packed in a 3D structure that is still quite flexible but has a dramatically reduced compressibility with respect to single dendrimers. Depending on the ratios used, for most of the dendroids described here a maximum of 25% of the dendrimer end groups is participating in the cross-linking, leaving still a lot of free end groups for potential further cross-linking or functionalization in future work. Imines are known to be in equilibrium with the amine and aldehyde subcomponents and are among the archetypical reversible covalent chemistry building blocks. Our data suggest that the multitude of cross-links in the dendroids is such that the reversible covalent nature of the imine formation does not result in the disassembly of the dendroid as a whole. In other words, some cross-links might hydrolyze and re-form, but unlikely all at the same time, leading to release of dendrimers. We also showed that it is possible to encapsulate AuNPs inside the dendroids, tuning the composition of the superstructure with the mixing ratio of dendrimers and DENdrimers prior to micelle formation. The dendroids are shown to contain about 21 dendrimers in the about 30 nm sized nanostructure. Considering the number of dendrimers, the overall mass of the prepared dendroids is about 2.5 MDa and the mass of the Au-DENdroids is about 3 MDa, comparable in weight to the biggest protein known in nature, Titin,51 and encompassing a volume of ∼104 nm3. The presented strategy is versatile and provides the basis for a variety of classes of material. For instance, the aggregation number, size, and flexibility of dendroids could be tuned by using different dendrimer generations (or a mix of generations) or by varying the length and type of cross-linkers, respectively. Furthermore, different dendrimers, block copolymers, and nanoparticles can be used for the formation of dendroids. Dendroids define a highly tunable class of structures. The here-presented example based on a G7 PAMAM appears to have a hard-core soft outside structure, with a dramatically reduced compressibility with respect to the individual parent macromolecule. Further investigating the nature of the cross-linker (ratio, type) on the physical-chemical properties of the dendroids is currently of our interest. Considering the great promise of dendrimers as well as micelles in many fields, from nanomedicine to materials research, together with the hierarchically controlled strategy, this gives rise to great potential for several applications including (targeted) drug delivery, biomedical imaging, and catalysis. Finally, the micelle template strategy described here is not limited to dendrimers, and other macromolecular core-building units further expand the horizon with other classes of supermolecules.
Material and Methods
Materials
Amine-terminated polyamidoamino dendrimers generation 7 were purchased from Dendritech Inc., MI, USA, as a 5 wt % methanolic solution and used as a 2.89 mM aqueous solution based on primary amine content. pMAA64-pEO885 (Mw/Mn = 1.15) was obtained from Polymer Sources Inc., Canada, and used as a 5 mM aqueous solution based on carboxylic acid content. HAuCl4·3H2O was purchased from VWR International B.V. Glutaraldehyde 50% aqueous solution, 3-(N-morpholino)propanesulfonic acid (MOPS) sodium salt, NaBH4, NaCl, 6-aminofluorescein (AF), 1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide (EDC), 1 M HCl, and 1 M NaOH solutions were obtained from Sigma-Aldrich.
Characterization
DLS was done on a Malvern Zetasizer Nano S equipped with a laser operating at 633 nm. The sample grids used for electron microscopy were purchased from Electron Microscopy Sciences (EMS, Hatfield, PA, USA). The grids were treated with a plasma cleaner setup (20 s at 10–1 Torr) to make them hydrophilic. For TEM measurements, samples were prepared on hydrophilic 400 mesh carbon-coated copper grids, while 200 mesh carbon-coated copper grids were used for the TEM tilt series. The TEM tilt series were acquired with SerialEM, and IMOD was used for 3D reconstruction of the dendroids from the tilt series. For cryo-TEM measurements, 400 mesh holey carbon grids were used. For cryo-TEM, after blotting, the samples were plunged into liquid ethane at about liquid nitrogen temperature by using a Vitrobot system (FEI Company). Samples for (cryo-)TEM were imaged with a 1400Plus JEOL TEM operating at 120 kV. The contrast of (cryo-)TEM pictures was adjusted by using ImageJ-win64. Atomic force microscopy images were recorded on a Bruker Multimode 5 using contact mode. NMR spectra in D2O were obtained on a Bruker Avance III spectrometer operating at 500 MHz for 1H, equipped with a 5 mm TXI probe. Fluorescence emission spectra were acquired on a Cary Eclipse spectrophotometer.
Synthesis of Dendrimer-Encapsulated Nanoparticles
G7-dendrimer-encapsulated gold nanoparticles were made following established protocols as reported before.16 Briefly, 50 μL of PAMAM G7-NH2 (5 wt % in MeOH, ∼2 mg) was evaporated under reduced pressure and afterward dissolved in 2 mL of distilled water. The pH was adjusted to ∼3 with 1 M HCl. Afterward, 1 mL of a 4.4 mM aqueous solution of HAuCl4 was added (corresponding to a dendrimer loading of 50% with respect to the number of dendrimer end groups). After stirring the solution for 10 min, 44 μL of a 1 M NaBH4 solution in 0.3 M NaOH was added (corresponding to 10 mol equiv to Au3+). The solution changed color from yellow to dark brown within seconds, indicating the reduction of Au3+ ions and formation of nanoparticles. After stirring overnight, the pH was adjusted to 7 using HCl. The resulting G7-Au were stored at 4 °C.
Dendrimicelle Formation
Dendrimicelles were formed under charge stoichiometric conditions following an established protocol.35 For this, 20 μL of an aqueous 2.89 mM PAMAM dendrimer solution (charge concentration based on surface groups) was added to 20 μL of 0.2 M MOPS buffer at pH 7. This solution was dissolved in 149 μL of water. Afterward, 11 μL of an aqueous 5 mM pMAA64-pEO885 solution (charge concentration based on −COO−) was added during sonication for 2 min. Samples were left to equilibrate overnight before characterization and core cross-linking.
Dendroid Formation from Dendrimicelles
After dendrimicelle formation, 1 mM glutaraldehyde solution was added to the micelle solution for core cross-linking in the desired ratio between PAMAM and glutaraldehyde. After an incubation at room temperature of 5 h, 50 μL of a 4 M NaCl solution was added to the cross-linked dendrimicelles to remove the block copolymer from the cross-linked dendrimer core. Addition of 10 molar equiv, with respect to glutaraldehyde, of a freshly prepared stock of 0.1 M NaBH4 in 0.1 M NaOH leads to the reduction of the imide bonds and free aldehyde groups. The resulting dendroids are purified by dialysis against a 1 M NaCl solution to remove the block copolymer and afterward against demi-water to remove the NaCl. Dialysis filters have a cutoff of 100 kDa. TEM pictures were recorded before purification with DLS.
Block Copolymer, pMAA64pEO885, Functionalization with 6-Aminofluorescein
Seven molar equivalents of 6-aminofluorescein was added to 1 mL of an aqueous 25 mM (−) solution of pMAA64pEO885. Afterward, 10 molar equiv of EDC with respect to AF was added, and the solution was mixed for several hours at room temperature covered with aluminum foil. Unreacted AF and EDC were removed by dialysis against demi-water. After purification with dialysis, the product was dried under reduced pressure and dissolved in D2O for NMR analysis. Final pMAA64pEO885-AF concentration was calculated with NMR integrals, and loading was determined to be 0.02 AF per block copolymer by using UV–vis.
Acknowledgments
Jan van Lent of the Wageningen Electron Microscopy Centre, WEMC, is thanked for his help with (cryo) electron microscopy and tomography. Anton Bunschoten is acknowledged for performing atomic force microscopy measurements and discussions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.0c09322.
UV–vis, DLS, and (cryo-)TEM characterization of G7 AuDENs, G7-E-, G7-AuE-, and G7-Au-based micelles and resulting dendroids, NMR and fluorescence data of the block copolymer, TEM tomography reconstruction and enlarged TEM pictures at different tilt angles (PDF)
Movie of TEM tilt series of G7-Au-based dendroids and of tomography reconstructions (AVI)
Movie of TEM tilt series of G7-Au-based dendroids and of tomography reconstructions (AVI)
Movie of TEM tilt series of G7-Au-based dendroids and of tomography reconstructions (AVI)
The authors declare no competing financial interest.
Supplementary Material
References
- Whitesides G. M.; Grzybowski B. Self-Assembly at All Scales. Science 2002, 295 (5564), 2418–2421. 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- Zeng X. B.; Ungar G.; Liu Y. S.; Percec V.; Dulcey S. E.; Hobbs J. K. Supramolecular Dendritic Liquid Quasicrystals. Nature 2004, 428 (6979), 157–160. 10.1038/nature02368. [DOI] [PubMed] [Google Scholar]
- Chakrabarty R.; Mukherjee P. S.; Stang P. J. Supramolecular Coordination: Self-Assembly of Finite Two- and Three-Dimensional Ensembles. Chem. Rev. 2011, 111 (11), 6810–6918. 10.1021/cr200077m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel N.; Retsch M.; Fustin C. A.; del Campo A.; Jonas U. Advances in Colloidal Assembly: The Design of Structure and Hierarchy in Two and Three Dimensions. Chem. Rev. 2015, 115 (13), 6265–6311. 10.1021/cr400081d. [DOI] [PubMed] [Google Scholar]
- Ling X. Y.; Reinhoudt D. N.; Huskens J. From Supramolecular Chemistry to Nanotechnology: Assembly of 3D Nanostructures. Pure Appl. Chem. 2009, 81 (12), 2225–2233. 10.1351/PAC-CON-09-07-04. [DOI] [Google Scholar]
- Cabral H.; Miyata K.; Osada K.; Kataoka K. Block Copolymer Micelles in Nanomedicine Applications. Chem. Rev. 2018, 118 (14), 6844–6892. 10.1021/acs.chemrev.8b00199. [DOI] [PubMed] [Google Scholar]
- Kataoka K.; Harada A.; Nagasaki Y. Block Copolymer Micelles for Drug Delivery: Design, Characterization and Biological Significance. Adv. Drug Delivery Rev. 2001, 47 (1), 113–131. 10.1016/S0169-409X(00)00124-1. [DOI] [PubMed] [Google Scholar]
- Talelli M.; Barz M.; Rijcken C. J. F.; Kiessling F.; Hennink W. E.; Lammers T. Core-Crosslinked Polymeric Micelles: Principles, Preparation, Biomedical Applications and Clinical Translation. Nano Today 2015, 10 (1), 93–117. 10.1016/j.nantod.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voets I. K.; de Keizer A.; Cohen Stuart M. A. Complex Coacervate Core Micelles. Adv. Colloid Interface Sci. 2009, 147–148, 300–18. 10.1016/j.cis.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Grill L.; Dyer M.; Lafferentz L.; Persson M.; Peters M. V.; Hecht S. Nano-Architectures by Covalent Assembly of Molecular Building Blocks. Nat. Nanotechnol. 2007, 2 (11), 687–691. 10.1038/nnano.2007.346. [DOI] [PubMed] [Google Scholar]
- Astruc D.; Boisselier E.; Ornelas C. Dendrimers Designed for Functions: From Physical, Photophysical, and Supramolecular Properties to Applications in Sensing, Catalysis, Molecular Electronics, Photonics, and Nanomedicine. Chem. Rev. 2010, 110 (4), 1857–1959. 10.1021/cr900327d. [DOI] [PubMed] [Google Scholar]
- Bosman A. W.; Janssen H. M.; Meijer E. W. About Dendrimers: Structure, Physical Properties, and Applications. Chem. Rev. 1999, 99 (7), 1665–1688. 10.1021/cr970069y. [DOI] [PubMed] [Google Scholar]
- Tomalia D. A. Birth of a New Macromolecular Architecture: Dendrimers as Quantized Building Blocks for Nanoscale Synthetic Polymer Chemistry. Prog. Polym. Sci. 2005, 30 (3–4), 294–324. 10.1016/j.progpolymsci.2005.01.007. [DOI] [Google Scholar]
- Ruggi A.; Beekman C.; Wasserberg D.; Subramaniam V.; Reinhoudt D. N.; van Leeuwen F. W. B.; Velders A. H. Dendritic Ruthenium(II)-Based Dyes Tuneable for Diagnostic or Therapeutic Applications. Chem. - Eur. J. 2011, 17 (2), 464–467. 10.1002/chem.201002514. [DOI] [PubMed] [Google Scholar]
- Trinchi A.; Muster T. H. A Review of Surface Functionalized Amine Terminated Dendrimers for Application in Biological and Molecular Sensing. Supramol. Chem. 2007, 19 (7), 431–445. 10.1080/10610270601120363. [DOI] [Google Scholar]
- Gröhn F.; Bauer B. J.; Akpalu Y. A.; Jackson C. L.; Amis E. J. Dendrimer Templates for the Formation of Gold Nanoclusters. Macromolecules 2000, 33 (16), 6042–6050. 10.1021/ma000149v. [DOI] [Google Scholar]
- Scott R. W. J.; Wilson O. M.; Crooks R. M. Synthesis, Characterization, and Applications of Dendrimer-Encapsulated Nanoparticles. J. Phys. Chem. B 2005, 109 (2), 692–704. 10.1021/jp0469665. [DOI] [PubMed] [Google Scholar]
- Wilson O. M.; Scott R. W. J.; Garcia-Martinez J. C.; Crooks R. M. Synthesis, Characterization, and Structure-Selective Extraction of 1–3-nm Diameter AuAg Dendrimer-Encapsulated Bimetallic Nanoparticles. J. Am. Chem. Soc. 2005, 127 (3), 1015–1024. 10.1021/ja045224m. [DOI] [PubMed] [Google Scholar]
- Zhao M. Q.; Sun L.; Crooks R. M. Preparation of Cu Nanoclusters within Dendrimer Templates. J. Am. Chem. Soc. 1998, 120 (19), 4877–4878. 10.1021/ja980438n. [DOI] [Google Scholar]
- Sun H. J.; Zhang S. D.; Percec V. From Structure to Function via Complex Supramolecular Dendrimer Systems. Chem. Soc. Rev. 2015, 44 (12), 3900–3923. 10.1039/C4CS00249K. [DOI] [PubMed] [Google Scholar]
- Uppuluri S.; Swanson D. R.; Piehler L. T.; Li J.; Hagnauer G. L.; Tomalia D. A. Core-Shell Tecto(Dendrimers): I. Synthesis and Characterization of Saturated Shell Models. Adv. Mater. 2000, 12 (11), 796–800. . [DOI] [Google Scholar]
- Cheng Z. L.; Thorek D. L. J.; Tsourkas A. Gadolinium-Conjugated Dendrimer Nanoclusters as a Tumor-Targeted T-1 Magnetic Resonance Imaging Contrast Agent. Angew. Chem., Int. Ed. 2010, 49 (2), 346–350. 10.1002/anie.200905133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. S.; Mecke A.; Orr B. G.; Holl M. M. B.; Baker J. R. DNA-Directed Synthesis of Generation 7 and 5 PAMAM Dendrimer Nanoclusters. Nano Lett. 2004, 4 (3), 391–397. 10.1021/nl0343497. [DOI] [Google Scholar]
- Huang C. H.; Nwe K.; Al Zaki A.; Brechbiel M. W.; Tsourkas A. Biodegradable Polydisulfide Dendrimer Nanoclusters as MRI Contrast Agents. ACS Nano 2012, 6 (11), 9416–9424. 10.1021/nn304160p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. M.; Wang H.; Yang W. J.; Cheng Y. Y. Disulfide Cross-Linked Low Generation Dendrimers with High Gene Transfection Efficacy, Low Cytotoxicity, and Low Cost. J. Am. Chem. Soc. 2012, 134 (42), 17680–17687. 10.1021/ja307290j. [DOI] [PubMed] [Google Scholar]
- Song C.; Xiao Y. C.; Ouyang Z. J.; Shen M. W.; Shi X. Y. Efficient Co-Delivery of microRNA 21 Inhibitor and Doxorubicin to Cancer Cells Using Core-Shell Tecto Dendrimers Formed via Supramolecular Host-Guest Assembly. J. Mater. Chem. B 2020, 8 (14), 2768–2774. 10.1039/D0TB00346H. [DOI] [PubMed] [Google Scholar]
- Song C.; Shen M. W.; Rodrigues J.; Mignani S.; Majoral J. P.; Shi X. Y.. Superstructured Poly(amidoamine) Dendrimer-Based Nanoconstructs as Platforms for Cancer Nanomedicine: A Concise Review. Coord. Chem. Rev. 2020, 421, 213463. 10.1016/j.ccr.2020.213463. [DOI] [Google Scholar]
- Khopade A. J.; Mohwald H. Statistical Megamer Morphologies and Materials from PAMAM Dendrimers. Macromol. Rapid Commun. 2005, 26 (6), 445–449. 10.1002/marc.200400523. [DOI] [Google Scholar]
- Tomioka N.; Takasu D.; Takahashi T.; Aida T. Electrostatic Assembly of Dendrimer Electrolytes: Negatively and Positively Charged Dendrimer Porphyrins. Angew. Chem., Int. Ed. 1998, 37 (11), 1531–1534. . [DOI] [PubMed] [Google Scholar]
- Xie Y. D.; Kocaefe D.; Chen C. Y.; Kocaefe Y.. Review of Research on Template Methods in Preparation of Nanomaterials. J. Nanomater. 2016, 2016, 1. 10.1155/2016/2302595. [DOI] [Google Scholar]
- Bastakoti B. P.; Munkaila S.; Guragain S. Micelles Template for the Synthesis of Hollow Nickel Phosphate Nanospheres. Mater. Lett. 2019, 251, 34–36. 10.1016/j.matlet.2019.05.034. [DOI] [Google Scholar]
- Huang H. Y.; Remsen E. E.; Kowalewski T.; Wooley K. L. Nanocages Derived from Shell Cross-Linked Micelle Templates. J. Am. Chem. Soc. 1999, 121 (15), 3805–3806. 10.1021/ja983610w. [DOI] [Google Scholar]
- Stapert H. R.; Nishiyama N.; Jiang D. L.; Aida T.; Kataoka K. Polyion Complex Micelles Encapsulating Light-Harvesting Ionic Dendrimer Zinc Porphyrins. Langmuir 2000, 16 (21), 8182–8188. 10.1021/la000423e. [DOI] [Google Scholar]
- Chechik V.; Zhao M. Q.; Crooks R. M. Self-Assembled Inverted Micelles Prepared from a Dendrimer Template: Phase Transfer of Encapsulated Guests. J. Am. Chem. Soc. 1999, 121 (20), 4910–4911. 10.1021/ja990445r. [DOI] [Google Scholar]
- ten Hove J. B.; Wang J.; van Leeuwen F. W. B.; Velders A. H.. Dendrimer-Encapsulated Nanoparticle-Core Micelles as Modular Strategy for Particle-in-a-Box-in-a-Box Nanostructures. Nanoscale 2017, 9, 18619. 10.1039/C7NR06773A. [DOI] [PubMed] [Google Scholar]
- Wang J.; Voets I. K.; Fokkink R.; van der Gucht J.; Velders A. H. Controlling the Number of Dendrimers in Dendrimicelle Nanoconjugates from 1 to More than 100. Soft Matter 2014, 10 (37), 7337–7345. 10.1039/C4SM01143K. [DOI] [PubMed] [Google Scholar]
- Wang J.; Lei L.; Voets I. K.; Cohen Stuart M. A.; Velders A. H. Dendrimicelles with pH-Controlled Aggregation Number of Core-Dendrimers and Stability. Soft Matter 2020, 16 (34), 7893–7897. 10.1039/D0SM00458H. [DOI] [PubMed] [Google Scholar]
- Gomez M. V.; Guerra J.; Velders A. H.; Crooks R. M. NMR Characterization of Fourth-Generation PAMAM Dendrimers in the Presence and Absence of Palladium Dendrimer-Encapsulated Nanoparticles. J. Am. Chem. Soc. 2009, 131 (1), 341–350. 10.1021/ja807488d. [DOI] [PubMed] [Google Scholar]
- Gomez M. V.; Guerra J.; Myers V. S.; Crooks R. M.; Velders A. H. Nanoparticle Size Determination by H-1 NMR Spectroscopy. J. Am. Chem. Soc. 2009, 131 (41), 14634–14635. 10.1021/ja9065442. [DOI] [PubMed] [Google Scholar]
- ten Hove J. B.; van Leeuwen F. W. B.; Velders A. H. Manipulating and Monitoring Nanoparticles in Micellar Thin Film Superstructures. Nat. Commun. 2018, 9 (1), 5207. 10.1038/s41467-018-07568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove J. B.; van Oosterom M. N.; van Leeuwen F. W. B.; Velders A. H. Nanoparticles Reveal Extreme Size-Sorting and Morphologies in Complex Coacervate Superstructures. Sci. Rep. 2018, 8 (1), 13820. 10.1038/s41598-018-32240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove J. B.; Wang J.; van Oosterom M. N.; van Leeuwen F. W. B.; Velders A. H. Size-Sorting and Pattern Formation of Nanoparticle-Loaded Micellar Superstructures in Biconcave Thin Films. ACS Nano 2017, 11, 11225–11231. 10.1021/acsnano.7b05541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Hernandez J.; Babin J.; Zappone B.; Lecommandoux S. Preparation of Shell Cross-Linked Nano-Objects from Hybrid-Peptide Block Copolymers. Biomacromolecules 2005, 6 (4), 2213–2220. 10.1021/bm0501499. [DOI] [PubMed] [Google Scholar]
- Migneault I.; Dartiguenave C.; Bertrand M. J.; Waldron K. C. Glutaraldehyde: Behavior in Aqueous Solution, Reaction with Proteins, and Application to Enzyme Crosslinking. BioTechniques 2004, 37 (5), 790–802. 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- Maggi F.; Ciccarelli S.; Diociaiuti M.; Casciardi S.; Masci G. Chitosan Nanogels by Template Chemical Cross-Linking in Polyion Complex Micelle Nanoreactors. Biomacromolecules 2011, 12 (10), 3499–3507. 10.1021/bm201015h. [DOI] [PubMed] [Google Scholar]
- Li J.; Piehler L. T.; Qin D.; Baker J. R.; Tomalia D. A.; Meier D. J. Visualization and Characterization of Poly(amidoamine) Dendrimers by Atomic Force Microscopy. Langmuir 2000, 16 (13), 5613–5616. 10.1021/la000035c. [DOI] [Google Scholar]
- Li J.; Swanson D. R.; Qin D.; Brothers H. M.; Piehler L. T.; Tomalia D.; Meier D. J. Characterizations of Core-Shell Tecto-(dendrimer) Molecules by Tapping Mode Atomic Force Microscopy. Langmuir 1999, 15 (21), 7347–7350. 10.1021/la990517s. [DOI] [Google Scholar]
- Ercius P.; Alaidi O.; Rames M. J.; Ren G. Electron Tomography: A Three-Dimensional Analytic Tool for Hard and Soft Materials Research. Adv. Mater. 2015, 27 (38), 5638–5663. 10.1002/adma.201501015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azcarate J. C.; Fonticelli M. H.; Zelaya E. Radiation Damage Mechanisms of Monolayer-Protected Nanoparticles via TEM Analysis. J. Phys. Chem. C 2017, 121 (46), 26108–26116. 10.1021/acs.jpcc.7b08525. [DOI] [Google Scholar]
- Jackson C. L.; Chanzy H. D.; Booy F. P.; Drake B. J.; Tomalia D. A.; Bauer B. J.; Amis E. J. Visualization of Dendrimer Molecules by Transmission Electron Microscopy (TEM): Staining Methods and Cryo-TEM of Vitrified Solutions. Macromolecules 1998, 31 (18), 6259–6265. 10.1021/ma9806155. [DOI] [Google Scholar]
- Labeit S.; Kolmerer B. Titins - Giant Proteins in Charge of Muscle Ultrastructure and Elasticity. Science 1995, 270 (5234), 293–296. 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.