Abstract
Purpose
To determine the influence of preprocessing on the repeatability and redundancy of radiomics features extracted using a popular open-source radiomics software package in a scan-rescan glioblastoma MRI study.
Materials and Methods
In this study, a secondary analysis of T2-weighted fluid-attenuated inversion recovery (FLAIR) and T1-weighted postcontrast images from 48 patients (mean age, 56 years [range, 22–77 years]) diagnosed with glioblastoma were included from two prospective studies (ClinicalTrials.gov NCT00662506 [2009–2011] and NCT00756106 [2008–2011]). All patients underwent two baseline scans 2–6 days apart using identical imaging protocols on 3-T MRI systems. No treatment occurred between scan and rescan, and tumors were essentially unchanged visually. Radiomic features were extracted by using PyRadiomics (https://pyradiomics.readthedocs.io/) under varying conditions, including normalization strategies and intensity quantization. Subsequently, intraclass correlation coefficients were determined between feature values of the scan and rescan.
Results
Shape features showed a higher repeatability than intensity (adjusted P < .001) and texture features (adjusted P < .001) for both T2-weighted FLAIR and T1-weighted postcontrast images. Normalization improved the overlap between the region of interest intensity histograms of scan and rescan (adjusted P < .001 for both T2-weighted FLAIR and T1-weighted postcontrast images), except in scans where brain extraction fails. As such, normalization significantly improves the repeatability of intensity features from T2-weighted FLAIR scans (adjusted P = .003 [z score normalization] and adjusted P = .002 [histogram matching]). The use of a relative intensity binning strategy as opposed to default absolute intensity binning reduces correlation between gray-level co-occurrence matrix features after normalization.
Conclusion
Both normalization and intensity quantization have an effect on the level of repeatability and redundancy of features, emphasizing the importance of both accurate reporting of methodology in radiomics articles and understanding the limitations of choices made in pipeline design.
Supplemental material is available for this article.
© RSNA, 2020
See also the commentary by Tiwari and Verma in this issue.
Summary
Radiomic feature extraction from MRI can be highly variable, and although preprocessing can improve the repeatability of these features, there is a lack of consistency in performance improvement across feature types and sequences; identification of repeatable and informative features should be a prerequisite in radiomics studies.
Key Points
■ Intensity and texture (gray-level co-occurrence matrix) features from MRI show low repeatability on a scan-rescan dataset of patients with glioblastoma using the default settings for feature extraction.
■ Normalization can improve the overlap between the region of interest intensity histograms of scan and rescan.
■ Intensity quantization settings must be chosen carefully; lower numbers of quantization bins result in higher correlation between texture features, indicating higher redundancy.
Introduction
In recent years, radiology has shifted toward more quantitative analysis of imaging to aid medical decision making. Among the most prominent techniques leading this shift is radiomics, which is defined by the extraction of quantitative features from a region of interest (ROI) on medical images (1). These quantitative descriptors can then be used to build predictive models for clinical variables, such as molecular markers, treatment response, and prognosis (2). This approach has the advantage that, in comparison to biopsies, features can reflect the full diversity of the ROI and factors such as tumor heterogeneity can be captured more easily and in a noninvasive manner (3,4). However, if these models are used for patient stratification with potential treatment decisions based on their predicted outcomes, the features used must fulfill two criteria: repeatability and reproducibility. Repeatability refers to “variability of the quantitative image biomarker when repeated measurements are acquired on the same experimental unit under identical or nearly identical conditions” to determine the measurement error (5). Reproducibility refers to “variability in the quantitative image biomarker measurements associated with using the imaging instrument in real-world clinical settings,” such as different settings of a software package attempting to identify and separate measurement errors from the reproducibility conditions (5).
Both repeatability and reproducibility of radiomic features have been described to be sensitive to various factors, such as image acquisition, resolution, reconstruction, preprocessing, and the software package used to extract them (6). Most published studies have described repeatability on CT (2,6–8). In contrast to CT, absolute voxel intensities on MRI do not have tissue-specific values, and changing signal intensities can leave tissue contrast unaltered (9). Therefore, intensity normalization might be needed to correct for these changes in intensity to make features comparable between and within patients, especially when scanned under slightly different conditions (8).
Relatively few studies have examined the repeatability and reproducibility of radiomic features at contrast-enhanced MRI (11–13). One potential reason is the challenge with test-retest studies that require the use of contrast agents. Of the few studies that have examined this topic, very little has been reported on the underlying causes for the lack of robustness of features. In a recent study on the repeatability of radiomic features for small prostate tumors, Schwier et al (11) showed that different features extracted with different MRI sequences might require different settings to increase their repeatability. However, this study only evaluated the effect of preprocessing and feature extraction configurations on the intraclass correlation coefficient (ICC) for features extracted from a small ROI and for a relatively small dataset of 15 patients. In our study, we build on this previous work, examining repeatability and feature redundancy in a unique scan-rescan dataset of patients with newly diagnosed glioblastoma, with the aim of understanding some of the reasons for the lack of repeatability of radiomic features.
Materials and Methods
Study Population
This is a secondary analysis of prospectively collected data from two clinical trials (ClinicalTrials.gov ID NCT00662506 and NCT00756106) at Massachusetts General Hospital and Dana-Farber Cancer Institute (14). All patients underwent the same imaging protocol, and both studies were approved by the institutional review board. A total of 54 adult patients (mean age, 57 years [age range, 22–77 years]; 33 men, 21 women) were included in the initial evaluation. Patients received either chemoradiation with cediranib (NCT00662506) or standard chemoradiation (NCT00756106). In addition to standard eligibility criteria, all patients were required to have a contrast-enhancing tumor of at least 1 cm in diameter. Patients underwent two pretreatment scans 2–6 days apart (mean, 3.7 days apart). Patients for whom both scans were not available were excluded from this study, resulting in a cohort size of 48 (mean age, 56 years [age range, 22–77 years]; 27 men, 21 women). The patients received no treatment between scan and rescan. None of the tumors had clinically significant changes between scans, as measured by the change in contrast-enhancing tumor volume or fluid-attenuated inversion recovery (FLAIR) hyperintensity.
MRI Scans
Scan and rescan images were acquired by using an identical imaging protocol and were obtained with the same model of 3.0-T MRI system (TimTrio; Siemens Medical Solutions, Malvern, Pa) at the same research institution. A total of 40 of 48 patients underwent both scan and rescan using identical MRI scanners. To improve scan-to-scan reproducibility, AutoAlign (Siemens) was used to ensure automatic alignment of the slice positions in a standard reproducible way for each scan. Further analysis was limited to axial T1-weighted postcontrast and axial T2-weighted FLAIR sequences. Axial T2-weighted FLAIR images were acquired with a repetition time of 10 000 msec, an echo time of 70 msec, 5-mm section thickness, 1-mm intersection gap, 0.43-mm in-plane resolution, 23 sections, and a 512 × 512 matrix. Axial T1-weighted postcontrast images were obtained after the injection of a bolus of 0.1 mmol per kilogram of body weight of Magnevist (Bayer Healthcare, Warrendale, Pa) with repetition time of 600 msec, an echo time of 12 msec, 5-mm section thickness, 1-mm intersection gap, 0.43-mm in-plane resolution, 23 sections, and a 512 × 512 matrix.
Segmentation, Annotation, and Preprocessing
Segmentations of enhancing lesions on T1-weighted postcontrast sequences and areas of T2 abnormality on T2-weighted FLAIR sequences were performed by expert raters (E.R.G., neuro-oncologist with 12 years of experience; M.C.P., neuroradiologist with 11 years of experience) blinded to patient identity, order of scans, and patient treatment status. Both scans of each patient were annotated by the same rater. After segmentation, each patient’s T1-weighted postcontrast sequences were registered to corresponding T2-weighted FLAIR sequences using the BRAINSfit module in 3D Slicer (https://www.slicer.org/) (15,16). The N4 bias-correction algorithm was applied to all images using the Nipype (Neuroimaging in Python: Pipelines and Interfaces) Python package (version 1.1.7; http:nipy.org/nipype) (17). Whole-brain extraction was performed on T1-weighted postcontrast images using the ROBEX (RObust Brain EXtraction, https://www.nitrc.org/projects/robex) system (18), and the resulting brain mask was applied to T2-weighted FLAIR images.
Normalization of input images was performed as part of the feature extraction (built-in z score normalization) or by using a histogram-matching technique as a separate step before feature extraction. The built-in normalization normalizes each input volume such that the mean of the voxel intensity distribution is centered at zero with unit variance (z score normalization). Histogram matching of the non-ROI region is a common normalization technique in radiomics (19). In our study, we implemented histogram matching using the method described by Nyúl and Udupa (20), in which a piecewise linear transformation is applied such that the histogram of a source image is matched to that of a chosen reference image. A randomly chosen patient was used as reference to which the histograms of all other patients were matched.
In addition to the aforementioned manual masks, we derived union masks of both visits by registering the rescan to the scan and taking the union of both masks separately for the enhancing tumor ROI on T1-weighted postcontrast images and total tumor ROI on T2-weighted FLAIR images. For feature extraction, these masks were then registered back to the nonregistered images.
Radiomics Software
Radiomics features were extracted using the PyRadiomics open-source Python package (version 2.1.0; https://pyradiomics.readthedocs.io/) (7). Features for the scan and rescan were extracted separately from both T1-weighted postcontrast and T2-weighted FLAIR images. Whenever indicated, the package default image normalization was applied to brain-extracted images as part of the feature extraction process (z score normalization), and all features defined as default by PyRadiomics were extracted from three-dimensional tumor volumes. We limited our analysis of texture features to features derived from gray-level co-occurrence matrices (GLCMs) and excluded the following features from further analysis: compactness1, compactness2, and spherical disproportion are perfectly correlated with sphericity; and homogeneity1 and homogeneity2 are directly correlated with inverse difference moment. For each experimental setting and sequence (T1-weighted postcontrast and T2-weighted FLAIR) we extracted 13 shape, 17 intensity, and 23 texture features.
Statistical Analysis
For each feature extracted from both T1-weighted postcontrast and T2-weighted FLAIR sequences, we calculated the ICC between the feature value extracted from the scan and rescan over the sample of 48 patients. We used a two-way model of the ICC (unit, single; type, consistency; 95% CI) as implemented in the R statistical software (version 3.5.2; R Foundation for Statistical Computing) “IRR” package (version 0.84). Features were then grouped into shape (describing size and shape), intensity, and texture features, as proposed by Kalpathy-Cramer et al (21) and following the image biomarker standardization initiative classes for feature groups for further analysis (22). To determine association between features, we calculated the pairwise Spearman correlation coefficient between features for all patients (one scan) and took the absolute value to reflect the strength of the correlation. For comparison of the ROI intensity distributions between scan and rescan, we chose the maximum range of voxel values of both images and divided it into 100 bins. These bins were then used to derive the intensity histograms for both visits. We used these histograms to calculate the Jensen-Shannon divergence (JSD) between visits. On the basis of the Kullback-Leibler divergence, the JSD has the advantage of being both symmetric and an unbiased measure of the similarity between two probability distributions (23). Statistical significance between feature groups was assessed using a Kruskal-Wallis test followed by post hoc pairwise Dunn multiple-comparisons tests with Bonferroni correction to determine the relationship between the individual means. Analysis of statistical differences between normalization approaches was performed with the paired Wilcoxon test with respect to the chosen baseline (no normalization) and Bonferroni correction for multiple comparisons. The significance threshold for adjusted P values was .05. Statistical analysis was performed using R statistical software (version 3.5.2).
Results
Repeatability of Feature Extraction from Unnormalized MRI
First, we examined the repeatability of shape, intensity, and texture features using the PyRadiomics default settings (no normalization, intensity quantization with constant bin width set to 10). Figure 1 shows the distribution of the ICC scores for each feature group for both sequences. The ICC is computed based on the full study population. For both sequences, purely segmentation-dependent features in the shape group are highly repeatable between the scan and rescan, with a median ICC of 0.98 (range, 0.88–0.99) for T2-weighted FLAIR images and 0.96 (range, 0.78–0.98) for T1-weighted postcontrast images. Features in the intensity and texture feature groups, which depend on voxel intensity values, show low ICCs and high variability in the ICCs within the groups for both sequences, with median ICC values for T2-weighted FLAIR and T1-weighted postcontrast images of 0.60 (range, 0.38–0.84) and 0.71 (range, 0.36–0.83), respectively, for intensity, and 0.68 (range, 0.10–0.94) and 0.78 (range, 0.48–0.86), respectively, for texture features. We observed differences in the ICC distribution for T2-weighted FLAIR and T1-weighted postcontrast images, respectively, between shape and intensity (P < .001 and < .001, adjusted for three comparisons) and shape and texture features (P < .001 and < .001, adjusted for three comparisons) on the pairwise Dunn test. Accordingly, we assessed how the repeatability of features in the intensity and texture groups that are calculated based on voxel intensities can be improved.
Figure 1:
Distribution of intraclass correlation coefficient (ICC) values per feature group under default feature extraction settings. Each boxplot represents the distribution of one radiomics feature group (shape, intensity, texture) between scan and rescan for the cohort of 48 patients. A, T2-weighted fluid-attenuated inversion recovery (T2W-FLAIR). B, T1-weighted (T1W) postcontrast. Features were extracted from nonnormalized images using the PyRadiomics default settings (no normalization, constant bin width for intensity quantization).
Effect of Normalization on the Intensity Distribution and Intensity Quantization on Within-Scan Feature Correlation
Influence of normalization on the ROI intensity distribution.—Intensity features describe the distribution of voxel intensity values in the segmented region. GLCM features are computed on the basis of the GLCM, which represents the relationships of the voxel intensities of neighboring voxels in the ROI. Before we studied the repeatability of intensity and GLCM features, we first assessed the effect of normalization on the ROI intensity histogram of both the scan and the rescan and voxel intensity quantization on the correlation between GLCM features.
The voxel values for MRI are not normalized, and there are no tissue-specific intensity ranges, so features based on voxel intensities showed great variability in ICC between scan and rescan. Therefore, we first studied the effect of normalization on the intensity distribution of the segmented tumor region (ROI intensity histogram) by comparing the voxel intensity histograms between scan and rescan before turning to the repeatability of intensity features. We used (a) the built-in normalization (z score normalization over all voxels in the input volume) and (b) histogram matching to a reference case. The overlap between histograms was measured by the JSD between the ROI intensity histogram of the scan and rescan and the effect of normalization as change in JSD before and after normalization for all 48 patients.
For our study population, both normalization techniques (z score and histogram normalization of brain-extracted images) significantly improved the similarity between the histograms, as measured by JSD on T2-weighted FLAIR and T1-weighted postcontrast images (paired Wilcoxon test against the not-normalized baseline without comparisons between the normalized groups, adjusted P values for two comparisons, z score and histogram matching, respectively, of P < .001 and P < .001 on T2-weighted FLAIR images and P = .002 and P = .03 on T1-weighted postcontrast images). Figure 2, A and C, shows the ROI intensity histograms for both scans before normalization (column 1) and the change in overlap between the histograms owing to normalization (columns 2 and 3). Figure 2, A, illustrates the effect of normalization techniques for a representative case.
Figure 2:
Effect of normalization on the region of interest (ROI) intensity histograms. Intensity histograms of the ROI segmentations from the scan (blue) and rescan (orange) of representative cases on both T2-weighted fluid-attenuated inversion recovery (T2W-FLAIR) and T1-weighted (T1W) postcontrast sequences of, A, a representative case and, C, a failure case. The first column shows ROI intensity histograms without preprocessing; the second column, after brain extraction and normalization via histogram matching; and the third column, after brain extraction and z score normalization. The overlap between the histograms is quantified by Jensen-Shannon divergence (JSD). B, D, Axial sections from the T2-weighted FLAIR and T1-weighted postcontrast scan and rescan after brain extraction of the corresponding cases, A, C, respectively.
In some cases, normalization caused an increase in JSD between scan and rescan instead of the expected decrease. Failure cases were defined as cases for which normalization resulted in an increase in JSD for both T1-weighted postcontrast and T2-weighted FLAIR sequences (this analysis was constrained to z score normalization). Six of 48 patients’ scans were identified as failure cases. Visual assessment of these cases revealed that for all of them, the brain extraction step was not performed properly. We identified two patterns: (a) either too aggressive or (b) total or partial failed brain extraction (leaving either the full skull or parts of the skull behind). The latter mode of failure is illustrated in Figure 2, C and D, with representative axial slices of T2-weighted FLAIR and T1-weighted postcontrast scan and rescan (Fig 2, D), illustrating the brain extraction failure patterns.
We therefore additionally examined the JSD distributions of images that were normalized without previous brain extraction. As shown in Figure 3, the JSD values of the scan and rescan ROI intensity histograms of images normalized without previous brain extraction were not significantly different from brain-extracted and normalized images.
Figure 3:
Jensen-Shannon divergence (JSD) distributions with and without brain extraction. Distribution of the JSD between the region of interest intensity histograms of the scan and rescan for the entire cohort using T2-weighted fluid-attenuated inversion recovery (T2W-FLAIR) (left) and T1-weighted (T1W) postcontrast (right) for not-normalized, z score–normalized, and histogram-matched images, each with (blue) and without (orange) brain extraction performed before normalization. For each normalization approach (no normalization, z score normalization, histogram-matched), the absence of brain extraction before normalization did not have a significant effect on the JSD.
Influence of voxel intensity quantization on feature correlation.—In a manner similar to how intensity features describe the distribution of intensity values in the ROI, GLCM features describe the GLCM. For computation of the GLCM, intensity values first need to be quantized into discrete intensity ranges. This quantization step can be performed using either a defined bin width (absolute binning) or a preset number of bins (relative binning) adapted to the range of intensity values in the ROI. Assuming the user has normalized the intensities, using the default intensity quantization settings as implemented in PyRadiomics (constant bin width set to 10) results in nonsensical binning for the computation of the GLCM (ie, all voxel intensities are placed into only two bins, as this choice of bin width is too coarse for the existing range of voxel values following normalization). Texture features calculated based on this GLCM do not capture the true variability in image intensity that is present within the images. This setting results in extremely high correlations between texture features (mean Spearman correlation coefficient for all features, 0.95) on T1-weighted postcontrast images. By explicitly specifying the number of bins (relative binning) rather than a fixed bin width, the aforementioned effect can be avoided. With increasing numbers of bins and quantization levels, the overall correlation between texture features decreases (mean Spearman correlation coefficient for all GLCM features, 0.52 [five bins], 0.47 [64 bins], and 0.43 [256 bins]; T1-weighted postcontrast imaging), without an adverse effect on the repeatability of these features (Fig E1 [supplement]). The same effect can be observed on T2-weighted FLAIR images. On the basis of these results, for data reported in the following sections, we did not use the constant bin width setting; rather, we explicitly set the number of intensity value bins for intensity quantization to 256.
Influence of Normalization on the Repeatability of Intensity and Texture Features
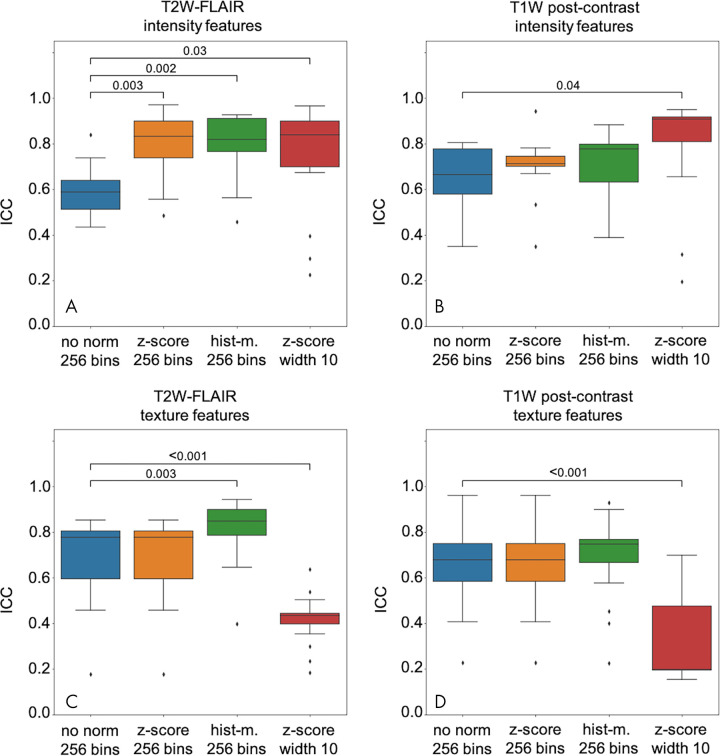
As described previously, the application of z score normalization and histogram matching improved the overlap between the ROI intensity histograms of the scan and rescan. Furthermore, the previous results highlight the importance of an appropriate binning strategy. Building on these results, we examined the influence of the normalization on the repeatability of intensity and texture features between scan and rescan using relative binning with 256 bins for intensity quantization for features extracted from not normalized, z score normalized, and histogram-matched scans. For comparison, we also included the ICC data computed on features extracted using z score normalization in combination with the default absolute intensity quantization setting (constant bin width, 10). The effect of the choice of the normalization technique on the ICC between both scans for intensity and texture features is presented in Figure 4 (top row, intensity; bottom row, texture features). The ICCs of single features are shown in Figure E2 (supplement) for intensity and Figure E3 (supplement) for texture.
Figure 4:
Distribution of intensity and texture intraclass correlation coefficient (ICC) values under different conditions. ICC for, A, B, intensity and, C, D, texture features extracted from T2-weighted fluid-attenuated inversion recovery (T2W FLAIR) (left) and T1-weighted (T1W) postcontrast (right) using either z score normalization (z-score) or histogram matching (hist-m.) compared with features extracted from not-normalized (no norm) images. Significant differences in the feature group mean ICC between feature extraction strategies (paired Wilcoxon test) are indicated with brackets.
Intensity Features
While both normalization techniques lead to an improved overlap between the ROI intensity histograms of scan and rescan for both T2-weighted FLAIR and T1-weighted postcontrast sequences, the effect of normalization on the repeatability of intensity features varies between the sequences (Fig 4, A and B). On T2-weighted FLAIR images (Fig 4, A), both z score normalization and histogram matching improved the repeatability of intensity features with respect to the not-normalized baseline (relative binning with 256 bins; paired Wilcoxon test against the not-normalized baseline without comparisons between the normalized groups; adjusted P values for three comparisons, P = .003 [z score normalization] and P = .002 [histogram matching]). On T1-weighted postcontrast images, however, neither z score normalization nor histogram matching resulted in a significant effect on the ICC of intensity features between scan and rescan (Fig 4, B).
Texture Features
As in the case of intensity features, normalization techniques have a different effect on both sequences. For T2-weighted FLAIR images, z score normalization did not change the ICC distribution of texture features compared with no normalization (relative intensity quantization, 256 bins), whereas histogram matching improved the repeatability (paired Wilcoxon test against the not-normalized baseline without comparisons between the normalized groups; adjusted P = .003 for three comparisons) (Fig 4, C). For T1-weighted postcontrast images, neither of the normalization techniques improved the repeatability of GLCM features (Fig 4, D). The ICC distribution of texture features extracted from z score normalized scans using the default bin width setting is presented in the fourth column in Figure 4, C and D. The coarse intensity quantization, effectively reducing the total number of bins to two for the majority of images, decreases the repeatability of GLCM features significantly on both sequences (paired Wilcoxon test, adjusted P < .001 for both T2-weighted FLAIR and T1-weighted postcontrast).
Independence of Features Extraction Repeatability of the ROI
To exclude all segmentation-dependent factors that might influence the repeatability of radiomic intensity and texture features, we extracted features using the union of the ROI of both scan and rescan. However, this approach did not produce higher ICC values for intensity and texture features (both from T2-weighted FLAIR and T1-weighted postcontrast images) than using manual masks separately defined for scan and rescan (one-sided analysis of variance). The ICC distributions are illustrated in Figure 5. This finding suggests that the low repeatability of intensity and texture features in our study is driven by differences in voxel intensities within the ROI between scan and rescan as opposed to intrarater variability in segmentations.
Figure 5:
Distribution of intensity and texture intraclass correlation coefficient (ICC) values depending on the region of interest (ROI) definition. ICC for intensity and texture features extracted from, A, T2-weighted fluid-attenuated inversion recovery (T2W FLAIR) and, B, T1-weighted (T1W) postcontrast using manual ROI masks separately outlined for scan and rescan (blue) or the union of both masks to extract features from the scan as well as rescan (orange). There is no statistically significant difference (paired Wilcoxon test) in the ICC distributions between the ROI definitions.
Discussion
In this study, we analyzed the influence of normalization (including voxel intensity quantization) on the repeatability of radiomic feature extraction from brain MRI (T2-weighted FLAIR and T1-weighted postcontrast sequences) using the open-source software package PyRadiomics for feature extraction. The high repeatability of shape features, which are computed exclusively based on the provided manual segmentations (Fig 1) indicates that the segmentations are very consistent between scan and rescan. While it has been reported that features are susceptible to variations in manual segmentations (24), we excluded this as a major driver for the low repeatability of intensity and texture features in this study based on the high consistency of the segmentations. Furthermore, using the same mask to extract radiomic features from the scan and rescan to eliminate segmentation effects did not result in an improvement in the ICC between visits. This serves as additional support to the idea that image acquisition and patient-related factors have a greater influence on the lack of radiomic feature repeatability than intrarater variability in segmentation between scan and rescan. As such, we investigated whether the repeatability of intensity and texture features can be improved by application of the appropriate normalization technique in combination with an adaptation of the intensity quantization strategy.
The dependency of intensity values in MRI on scanner properties, image acquisition, and image processing requires standardization of the image intensity to enable a comparison of features across patients (10). Accordingly, there are many approaches to normalization of medical imaging, particularly for normalization of brain MRI (25). We chose two of the most widely used normalization techniques in radiomics pipelines, which are z score normalization (built into PyRadiomics) and histogram matching to a reference case (26). The optimal intensity normalization technique is expected to result in a good overlap between the intensity histograms of scan and rescan. Both normalization techniques significantly improved overlap between the ROI intensity histograms of scan and rescan. However, we could identify cases in which, because of either too aggressive or insufficient brain extraction, normalization efforts had an adverse effect (Fig 2, C and D). This is consistent with a previous study, which showed that most commonly used brain extraction algorithms can fail in the presence of disease (27), thereby introducing another factor that can harm standardization and intensity normalization efforts.
For cohort-level analysis, we recommend manual auditing of the results of automatic brain extraction to ensure that brain extraction did not fail. Manual correction of sporadic failures might not be needed for cohort-level analyses. However, the analysis of individual scans (eg, for treatment stratification) might require manual checks of every scan to ensure appropriate brain extraction. If necessary, manual correction of the automatic brain extraction must be performed to ensure that the analysis is not impaired by flawed brain extraction and its downstream effects. We could not detect a significant difference in the JSD of normalized cases with and without brain extraction (Fig 3), but we did not examine downstream effects on feature repeatability.
Both normalization techniques show better repeatability for T2-weighted FLAIR images than for T1-weighted postcontrast images. One reason for this is that the repeatability of T1-weighted postcontrast scans can be complicated by variations in contrast application and the timing of image acquisition after injection, notwithstanding the controlled research conditions under which the scans used in this study were acquired. The normalization approaches used in our study do not account for these differences, as they are based on the intensity distribution of the full input volume, including the contrast-enhancing region. These findings are consistent with those in He et al (28), which showed that variability introduced by contrast enhancement can negatively affect the diagnostic performance of radiomics models on CT.
Additionally, the parameters for feature extraction, especially the choice of voxel intensity quantization, can have marked effects not just on the repeatability of radiomic feature extraction but also on the correlation between features (12,13). Features that are calculated based on binned or quantized values (eg, GLCM features) are sensitive to the choice of this setting. This is reflected in the poor ICC for texture features using a bin width of 10 on z score normalized images (Fig 4, C and D). Given the lack of standardized intensity ranges in MRI, relative binning is a more a reasonable choice, as it results in improved repeatability.
Importantly, the effects of intensity quantization require additional examination of the correlation between features. Increasing the number of histogram bins after brain extraction and normalization results in a decrease of the correlation between GLCM features within one scan, while having no adverse effect on feature repeatability (data not shown). Highly redundant features may have a negative effect on downstream predictive pipelines.
There were some limitations to our study. First, we limited the examination of texture features to GLCM features because of the popularity of these descriptors with respect to other texture features. Future studies will need to thoroughly examine other classes of texture features (eg, Laws energy, Gabor) (29,30). Second, features were extracted from two-dimensional axial sequences, and differences in slice placement can have an additional influence on the repeatability of radiomic feature extraction. Moreover, most researchers use radiomics features for some task (eg, survival analysis, disease diagnosis) to be solved via some machine learning model (eg, random forest, support vector machine classifier). In this study, we only tested for the repeatability of features. We did not test whether trained machine learning models using these radiomic descriptors are repeatable. Last, our findings and, therefore, our recommendations, may only be valid for radiomic features extracted from newly diagnosed and untreated glioblastoma as this was the use case in our study.
In summary, our findings that the optimal setting for feature extraction may vary from feature group to group (and maybe even within the separate groups) are consistent with results presented by Schwier et al (11) on the repeatability of radiomic feature extraction from MRI on a dataset of small prostate tumors. The extraction of repeatable intensity and GLCM radiomic features from MRI requires robust standardized preprocessing and careful selection of feature extraction settings. On the basis of our results, we recommend using a normalization strategy (especially for unenhanced sequences) and using relative binning strategies to account for varying intensity ranges within images. Furthermore, we recommend checking the within-scan correlations between features during feature selection and using a higher number of bins to avoid feature redundancy.
SUPPLEMENTAL FIGURES
This publication was supported by the Martinos Scholars fund to K.V.H. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Martinos Scholars fund. Research reported in this publication was supported by a training grant from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under award number 5T32EB1680 to K.C. and J.B.P. and by the National Cancer Institute (NCI) of the National Institutes of Health under award number F30CA239407 to K.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This study was supported by National Institutes of Health grants R01CA129371 to T.T.B., K23CA169021 to E.R.G., and U01 CA154601, U24 CA180927, and U24 CA180918 to J.K.C. This research was carried out in whole or in part at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies, P41EB015896, a P41 Biotechnology Resource Grant supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health.
Disclosures of Conflicts of Interest: K.V.H. disclosed no relevant relationships. J.B.P. disclosed no relevant relationships. A.L.B. disclosed no relevant relationships. K.C. disclosed no relevant relationships. P.S. disclosed no relevant relationships. J.M.B. disclosed no relevant relationships. M.C.P. disclosed no relevant relationships. T.T.B. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: received research support from Champions Biotechnology, AstraZeneca, Pfizer, and Millennium; is on the advisory board for UpToDate; is a consultant for Genomicare, Merck, NXDC, Amgen, Roche, Oxigene, Foundation Medicine, and Proximagen; provided CME lectures or material for UpToDate, Research to Practice, Oakstone Medical Publishing, and Imedex. Other relationships: disclosed no relevant relationships. E.R.G. disclosed no relevant relationships. B.R.R. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is on the advisory board for ARIA, Butterfly, DGMIF (Daegu-Gyeongbuk Medical Innovation Foundation), QMENTA, and Subtle Medical; is a consultant for Broadview Ventures, Janssen Scientific, ECRI Institute, GlaxoSmithKline, Hyperfine Research, Peking University, Wolf Greenfield, Superconducting Systems, Robins Kaplin, Millennium Pharmaceuticals, GE Healthcare, Siemens, Quinn Emanuel Trial Lawyers, Samsung, and Shenzhen Maternity and Child Health Care Hospital; is a founder of BLINKAI Technologies. Other relationships: disclosed no relevant relationships. J.K.C. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is a consultant and advisory board member for Infotech, Soft. Other relationships: disclosed no relevant relationships.
Abbreviations:
- FLAIR
- fluid-attenuated inversion recovery
- GLCM
- gray-level co-occurrence matrix
- ICC
- intraclass correlation coefficient
- JSD
- Jensen-Shannon divergence
- ROI
- region of interest
References
- 1.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48(4):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aerts HJWL, Grossmann P, Tan Y, et al. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci Rep 2016;6(1):33860 [Published correction appears in Sci Rep 2017;7:41197.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudie JD, Rauschecker AM, Bryan RN, Davatzikos C, Mohan S. Emerging Applications of Artificial Intelligence in Neuro-Oncology. Radiology 2019;290(3):607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med 2013;368(9):842–851. [DOI] [PubMed] [Google Scholar]
- 5.Raunig DL, McShane LM, Pennello G, et al. Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat Methods Med Res 2015;24(1):27–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int J Radiat Oncol Biol Phys 2018;102(4):1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garapati SS, Hadjiiski L, Cha KH, et al. Urinary bladder cancer staging in CT urography using machine learning. Med Phys 2017;44(11):5814–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madabhushi A, Udupa JK. New methods of MR image intensity standardization via generalized scale. Med Phys 2006;33(9):3426–3434. [DOI] [PubMed] [Google Scholar]
- 10.Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp 2018;2(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwier M, van Griethuysen J, Vangel MG, et al. Repeatability of Multiparametric Prostate MRI Radiomics Features. ArXiv 1807.06089 [preprint] [ICC] https://arxiv.org/abs/1807.06089. Posted 2018. Accessed January 21, 2019. [DOI] [PMC free article] [PubMed]
- 12.Molina D, Pérez-Beteta J, Martínez-González A, et al. Influence of gray level and space discretization on brain tumor heterogeneity measures obtained from magnetic resonance images. Comput Biol Med 2016;78:49–57. [DOI] [PubMed] [Google Scholar]
- 13.Duron L, Balvay D, Vande Perre S, et al. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS One 2019;14(3):e0213459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batchelor TT, Gerstner ER, Emblem KE, et al. Improved tumor oxygenation and survival in glioblastoma patients who show increased blood perfusion after cediranib and chemoradiation. Proc Natl Acad Sci U S A 2013;110(47):19059–19064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson H, Harris G, Williams K. BRAINSFit: mutual information registrations of whole-brain 3D images, using the insight toolkit. Insight J 2007. https://www.insight-journal.org/browse/publication/180/6. [Google Scholar]
- 16.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30(9):1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorgolewski K, Burns CD, Madison C, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front Neuroinform 2011;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iglesias JE, Liu CY, Thompson PM, Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 2011;30(9):1617–1634. [DOI] [PubMed] [Google Scholar]
- 19.Chen W, Liu B, Peng S, Sun J, Qiao X. Computer-Aided Grading of Gliomas Combining Automatic Segmentation and Radiomics. Int J Biomed Imaging 2018;2018:2512037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyúl LG, Udupa JK. On standardizing the MR image intensity scale. Magn Reson Med 1999;42(6):1072–1081. [DOI] [PubMed] [Google Scholar]
- 21.Kalpathy-Cramer J, Mamomov A, Zhao B, et al. Radiomics of Lung Nodules: A Multi-Institutional Study of Robustness and Agreement of Quantitative Imaging Features. Tomography 2016;2(4):430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. ArXiv 1612.07003 [preprint] http://arxiv.org/abs/1612.07003. Posted December 21, 2016. Accessed October 19, 2019.
- 23.Lin J. Divergence Measures Based on the Shannon Entropy. IEEE Trans Inf Theory 1991;37(1):145–151. [Google Scholar]
- 24.Pavic M, Bogowicz M, Würms X, et al. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol 2018;57(8):1070–1074. [DOI] [PubMed] [Google Scholar]
- 25.Shinohara RT, Sweeney EM, Goldsmith J, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin 2014;6:9–19 [Published correction appears in Neuroimage Clin 2015;7:848.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun X, Shi L, Luo Y, et al. Histogram-based normalization technique on human brain magnetic resonance images from different acquisitions. Biomed Eng Online 2015;14(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang K, Beers AL, Bai HX, et al. Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement. Neuro Oncol 2019;21(11):1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep 2016;6(1):34921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laws KI. Textured Image Segmentation. https://apps.dtic.mil/docs/citations/ADA083283. Published 1980. Accessed October 25, 2019.
- 30.Jain AK, Farrokhnia F. Unsupervised texture segmentation using Gabor filters. Pattern Recognit 1991;24(12):1167–1186. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.