Abstract
Purpose
To generate and assess an algorithm combining eye tracking and speech recognition to extract brain lesion location labels automatically for deep learning (DL).
Materials and Methods
In this retrospective study, 700 two-dimensional brain tumor MRI scans from the Brain Tumor Segmentation database were clinically interpreted. For each image, a single radiologist dictated a standard phrase describing the lesion into a microphone, simulating clinical interpretation. Eye-tracking data were recorded simultaneously. Using speech recognition, gaze points corresponding to each lesion were obtained. Lesion locations were used to train a keypoint detection convolutional neural network to find new lesions. A network was trained to localize lesions for an independent test set of 85 images. The statistical measure to evaluate our method was percent accuracy.
Results
Eye tracking with speech recognition was 92% accurate in labeling lesion locations from the training dataset, thereby demonstrating that fully simulated interpretation can yield reliable tumor location labels. These labels became those that were used to train the DL network. The detection network trained on these labels predicted lesion location of a separate testing set with 85% accuracy.
Conclusion
The DL network was able to locate brain tumors on the basis of training data that were labeled automatically from simulated clinical image interpretation.
© RSNA, 2020
Summary
A deep learning (DL) model trained with eye-tracking data combined with speech-recognition data can automatically extract labeled data from clinical interpretations for DL, producing a large volume of labeled data to accelerate DL in radiology.
Key Points
■ Combining eye tracking and speech recognition, brain tumors were localized with 92% accuracy, which represents the accuracy in identifying and localizing the lesions in training-set data.
■ A keypoint detection deep learning (DL) model trained with these locations achieved 85% accuracy in detecting and locating new lesions in a test dataset.
■ Using the proposed method, expert-labeled data for DL can be extracted automatically from standard-of-care clinical interpretations and thus can provide expansive quantities of data for DL without additional effort by radiologists.
Introduction
Obtaining appropriately annotated data in sufficient quantities for effective deep learning (DL) is costly, tedious, time-consuming, and often impractical (1–4). For example, annotating 1000 images has been estimated to require an entire month of full-time work by two expert radiologists (2). Hence, what is lacking and needed for all of radiology artificial intelligence is an automated method to obtain and annotate radiologic data, during normal workflow, from every clinical study that is performed, with no additional work by the radiologists.
To accelerate DL in radiology, a variety of annotation software tools have been used for object segmentation, such as ITKSnap (5) and 3D Slicer (6–8). More recently, dedicated software tools for artificial intelligence–intended annotation have emerged, such as MD.ai (9), XNAT (10), and ePad (11). However, these tools all require manual annotation of lesions by mouse clicking and/or dragging (4), which is tedious and time-consuming. Another approach being employed to annotate images is crowdsourcing, currently offered by numerous online services (12–14). However, this inevitably risks compromising patients’ protected health information, despite state-of-the-art de-identification software (15–17). Additionally, it generally employs nonexpert and/or nonradiologist users, often leading to incorrect labeling (18) and requires additional time and effort by imaging experts to improve quality control (19). Additionally, all of this needs to be performed in the research setting, which may slow algorithm development because of the busy clinical schedules of most radiologists who may participate in the research.
Although radiologists in clinical practice often measure lesions, in many cases they do not measure all lesions, let alone all structures (ie, muscles, organs), while interpreting a scan. However, all radiologists do physically look at the specific structures they are analyzing and describing. Eye-tracking technology allows a computer to know precisely where, when, and for how long a radiologist is looking at a location within an image. Because eye tracking provides this information without impeding the radiologist’s viewing of a monitor, it can automatically harness expert-labeled imaging data during routine clinical interpretation. The vital link to harnessing eye tracking for image labeling is to use the other activity that radiologists perform as they view images: they dictate what they see, usually just when they see it, by using a microphone or dictaphone. The audio input is processed in real time by speech-recognition software to generate a report. On the basis of our collective clinical experience, we surmise that in the majority of cases, radiologists fixate on structures while describing them. By recording the time at which a keyword indicating a structure is spoken, we can combine voice-dictation and eye-tracking data to produce labeled images during clinical imaging interpretation, without added input from the radiologist.
Thus, in this study, we sought proof of concept that an algorithm combining eye tracking and speech recognition can extract lesion location labels automatically for DL.
Materials and Methods
Image Selection and Data Source
This retrospective study did not employ any human participant data other than the publicly available Brain Tumor Segmentation (BraTS) database (20) and hence did not require institutional review board oversight. The study was conducted in compliance with the Health Insurance Portability and Accountability Act.
We used the BraTS 2016 public brain tumor database (20), which consists of MRI scans of 220 patients with high-grade glioma and 54 patients with low-grade glioma. From this database, we examined the skull-stripped T1-weighted postcontrast images of high-grade gliomas. We vertically stacked the image volumes, for a total of 8003 two-dimensional (2D) sections. We then randomly scrambled the image sections. We used the first 700 2D sections for the eye-tracking experiment and as our training set. These images were written to a Microsoft PowerPoint file, one image for each page, for display during the eye-tracking experiments. We set aside the next 100 as testing-set images. However, 15 were of poor quality or did not contain a discernible tumor (eg, in the extreme cranial or caudal aspect of the original image volume) and were discarded, leaving a total of 85 testing-set images.
Extraction of Labels with Eye Tracking and Speech Recognition
The 700 BraTS images (sections) were viewed by a single neuroradiologist (J.N.S., 2 years of experience), and one of three keywords—“tumor,” “mass,” or “lesion”—was spoken into a microphone as part of a standard phrase so as to simulate the typical process of dictating a report while examining images (Fig 1). Standard phrases were of the form “There is a tumor/lesion/mass in the right/left frontal/parietal/temporal lobe/cerebellum.” The use of standard phrases was meant to embed the keywords within a larger description and to make the interpretation of 700 images more fluid.
Figure 1:
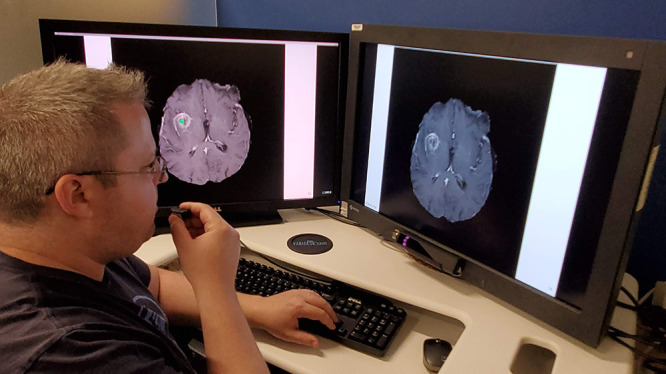
Eye-tracking setup. The user is looking at the monitor on the right while dictating into a microphone in his right hand. The gaze position appears on the left monitor as a small green dot.
As in our prior work, we performed the eye-tracking experiments and simulated interpretations using the Fovio Eye Tracker remote eye-tracker system (Seeing Machines, Canberra, Australia) with gaze data collected using the EyeWorks Suite (version 3.12). Gaze data were acquired at a rate of 60 Hz on a Dell Precision T3600 (Windows 7, Intel Xeon central processing unit E5-1603 at 2.80 GHz with 128 GB of random access memory). Images were presented from a PowerPoint (Microsoft, Redmond, Wash) presentation on a 30-inch EIZO liquid crystal display monitor. Before the experiments, a nine-point calibration procedure in EyeWorks Record was required. The Fovio remote system was again situated 2 cm beneath the bottom of the viewing screen and at a 26° angle with respect to the monitor.
Following calibration, the user completed the task of interpreting each 2D image section as a new slide in the PowerPoint presentation. A single neuroradiologist (J.N.S.) viewed the images and dictated for the simulated interpretation. Each lesion was interpreted as a new image and lesion. All images were used, even those that were clearly degraded or lacking adequate brain parenchyma.
A screen-captured Windows Media video–format video file with continuous recording of time points was generated. The video displayed user gaze position as a function of time overlaid on the images, which was essentially a video of the user’s gaze during the entire eye-tracking process. The entire experiment interpreting the 700 training-set images produced 313 904 gaze points. The gaze points were spaced on the order of 10 msec apart in time, and the total time of the experiment was roughly 1 hour 25 minutes. The radiologist was aware that gaze points were being recorded on a separate monitor but was unable to view that monitor during the experiments.
To convert from internal coordinates of the eye-tracking display to those of the image matrix, we had to account for monitor’s size and resolution. The monitor used was 1600 × 1598 pixels, so that the y-coordinate of the gaze data had to be scaled as 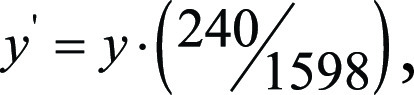 where y is in the screen-captured coordinates and y´ is rescaled into the coordinates of the original 2D BraTS image sections, which are sized at 240 × 240 pixels. The x-coordinate scaling is similar, although we had to account for a 426-pixel offset on the left side of the monitor because of unmatched aspect ratios of the PowerPoint display and the monitor:
where y is in the screen-captured coordinates and y´ is rescaled into the coordinates of the original 2D BraTS image sections, which are sized at 240 × 240 pixels. The x-coordinate scaling is similar, although we had to account for a 426-pixel offset on the left side of the monitor because of unmatched aspect ratios of the PowerPoint display and the monitor: 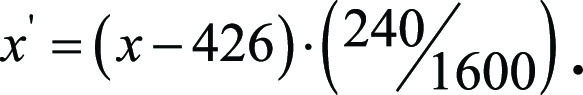
During simulated interpretations, as gaze data were acquired, we simultaneously recorded the entire dictation session by a microphone as a waveform audio file with a size 907.2 MB. Then we simulated clinical speech-recognition software–style voice interpretation. To do so, we employed the Google Cloud Speech-to-Text Application Programming Interface, which allowed us to generate a spreadsheet file specifying the image number, keyword spoken for that image, and beginning and end time for the keyword. It also generated a text file containing the full dictation, which contained 7673 words, or roughly 11 words per image.
More specifically, the waveform audio file was uploaded into the Google Cloud Platform, and then the key for the corresponding account and location of the file in the Google Cloud “bucket” was specified in a Google Colab Python file. Beginning and end times for keywords were obtained using the “start_time” and “end_time” attributes in the Google Python module “speech_v1.” Then the corresponding timestamps were matched to the times in the gaze-point spreadsheet by selecting the gaze time points closest to those in the speech-recognition spreadsheet. We used the Pandas library in Python for spreadsheet import, manipulation, and export. In this manner, we obtained all of the gaze points between the beginning of speaking each keyword and the end of the articulation. We then extracted the mean of these gaze positions for each lesion. The approach is illustrated for a sample image with a few representative gaze points in Figure 2.
Figure 2:
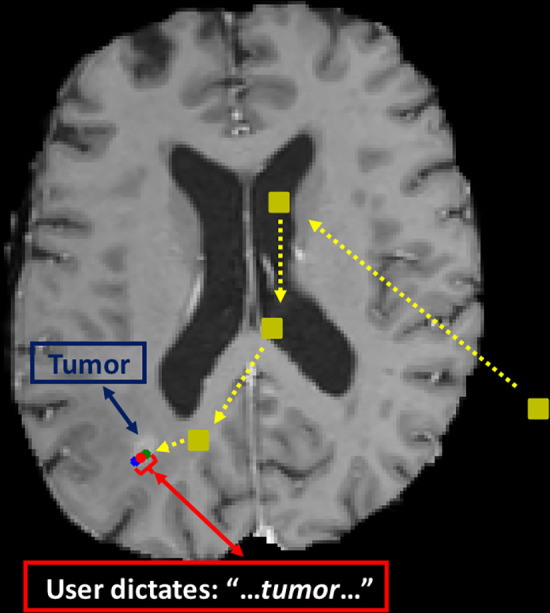
Example gaze plot. Initial points viewed are shown as yellow squares. The gaze position corresponding to the beginning of speaking the keyword is the green dot, and that for the end of the word is the blue dot. We take the average point (red dot) as the extracted label.
More formally, we defined the set of all time points during eye tracking and simulated image interpretation, in increments of 0.1 second, as 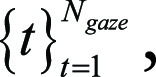 where Ngaze is the total number of time points during the entire eye tracking simulation. Then the set of gaze points
where Ngaze is the total number of time points during the entire eye tracking simulation. Then the set of gaze points 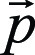 during eye tracking and simulated image interpretation was given by using
during eye tracking and simulated image interpretation was given by using 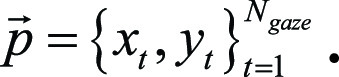 We also defined the set of time points at the beginning or end of stating any one of the keywords (“tumor,” “mass,” or “lesion”) by using
We also defined the set of time points at the beginning or end of stating any one of the keywords (“tumor,” “mass,” or “lesion”) by using 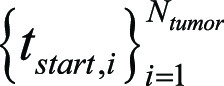 and
and 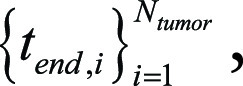 respectively. Here,
respectively. Here, 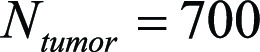 is the total number of lesions. Then the gaze-point times at (closest to) the beginning and end of keyword utterances were given by using
is the total number of lesions. Then the gaze-point times at (closest to) the beginning and end of keyword utterances were given by using 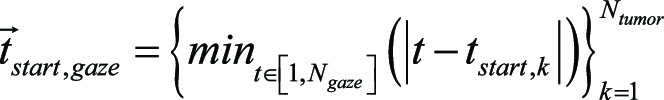 and
and 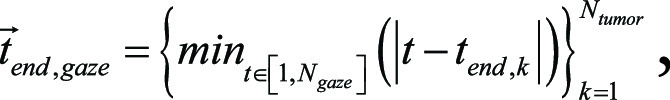 respectively. Next, for tumor i, we took the mean gaze point as
respectively. Next, for tumor i, we took the mean gaze point as 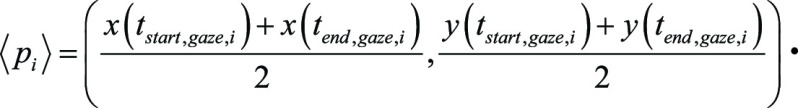
The speech-recognition software accurately detected the keyword for 647 out of the 700 lesions (92%). The start and end times for the 53 images in which the keyword was not detected were entered into the spreadsheet manually. This was done by watching the gaze-point screen-captured video recording also containing an audio recording. These video and audio files were obtained simultaneously with the audio-only waveform audio file, with a Windows Media video file of size 209.7 MB. Audio from the waveform audio file and not from the Windows Media video was used for speech recognition because of sound quality and compatibility with the Google speech-recognition software.
Convolutional Neural Network
We trained a keypoint detection convolutional neural network (CNN) on the 700 (x, y) positions. Our network used four convolution-rectified linear unit-pooling layers followed by two fully connected dense layers, with a two node–dense layer for the output (x, y) network predictions, which were then scaled by a sigmoid activation function. The algorithm architecture was adapted from notebooks used by contestants in the 2018 Kaggle Facial Keypoints Detection Competition (21). Some of the major differences were that whereas the facial keypoint detection task required identifying 15 facial landmarks, we only sought to detect one corresponding to a point within the lesion; hence, we used the two node–dense layer. Additionally, the level of detail in the competition’s images exceeded that of our BraTS images, so it was appropriate to use fewer layers and filters for our application.
Backpropagation employed the Adam optimizer with a learning rate of 0.001, the loss function being the mean average error. Of the 700 images and eye-tracking point labels, 630 were used for training, and the remaining 70 were used for validation. We trained the CNN with a batch size of 20 for 50 epochs. All training was performed in the Google Colab environment with their tensor processing unit, Python 3.7 (https://www.python.org/), and TensorFlow 2.0 (https://www.tensorflow.org/). The training time was roughly 42 minutes.
Statistical Analysis
The simple metric of percent accuracy was calculated in Python 3.7. For accuracy of lesion localization in the training and testing sets, the denominators were the respective set sizes, 700 and 85. Percent accuracy was the percentage of images for which the predicted keypoint was within the bounding box of the user-annotated lesion mask. Bounding boxes were computed from the hand-annotated masks, which were traced in MATLAB version 2016a (MathWorks, Natick, Mass). Then, in Python, a short function was used to calculate the minimum and maximum x and y values of the masks, which formed the edges of the bounding boxes.
Regarding accuracy, if a calculated (eye tracking + speech recognition) or predicted (CNN) (x, y) coordinate lay within the bounding box, then it was counted as a true-positive result. If it fell outside of the bounding box, then it was counted as a false-negative result. Then, noting that all calculations or predictions produced a candidate (x, y) value and thus that there were no true-negative or false-positive results, we calculated accuracy according to the following: accuracy = true-positive result/(true-positive result + false-negative result) = true-positive result/700 for the training set (calculated by eye tracking + speech recognition) or true-positive result/85 for the testing-set predictions. Thus, the metric was used both for accuracy of training-set lesion localization by eye tracking plus speech recognition and accuracy of testing-set lesion predictions by the CNN.
Results
Lesion Localization with Eye Tracking and Speech Recognition
Gaze points were 92% (644 of 700) accurate, where accuracy was defined as being within the bounding box of the hand-annotated lesion mask, which was our reference standard (Fig 3). This accuracy represents the accuracy of eye tracking and speech recognition in identifying and localizing the lesions in training-set data. The other missed 8% of images consisted of near-misses, in which the gaze point was almost within the bounding box, or image sections that were either degraded or outside of the full three-dimensional context or did not manifest the lesion clearly.
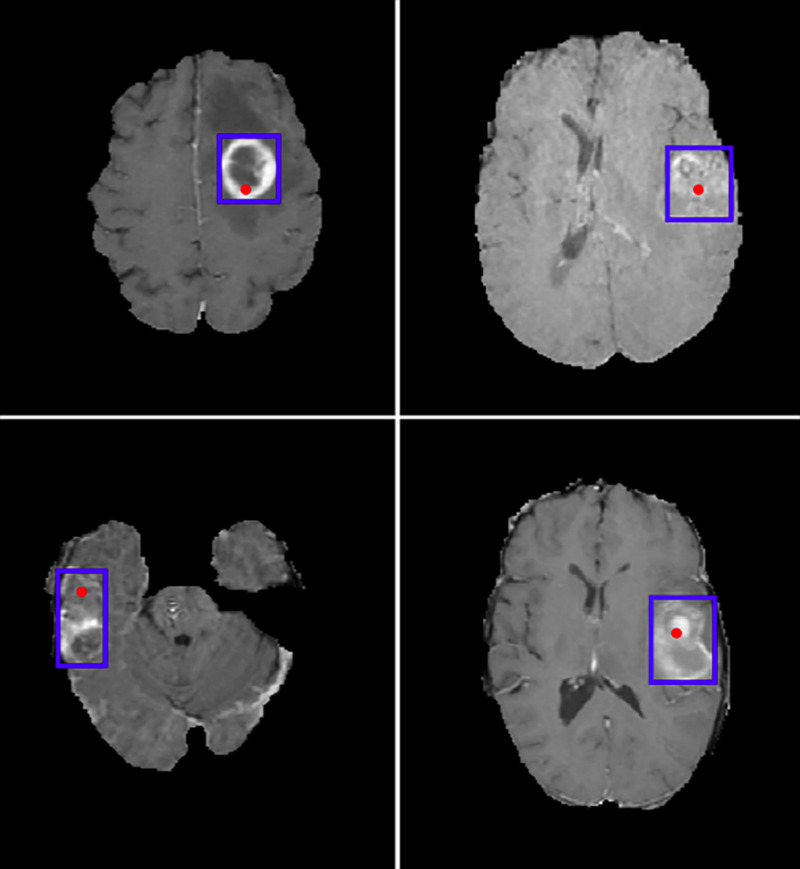
Figure 3:
Example test-set images from the Brain Tumor Segmentation database with convolutional neural network–predicted lesion location in red and bounding box of the hand-annotated mask (ground truth) in blue.
Lesion Prediction with Trained CNN
The trained CNN was able to predict the location of new lesions on test images with 85% (72 of 85) accuracy.
Discussion
In this study, we demonstrate proof of concept that an algorithm combining eye tracking and speech recognition can extract lesion location labels automatically for DL with 92% accuracy; the DL network trained from automatically labeled data was 85% accurate for predicting the location of new lesions on a test dataset. The proposed algorithm provides a method for data extraction for DL in radiology. Whereas natural language processing (which analyzes the reports) and the images themselves are available in general to be studied with DL, information about where the radiologist looked during the review of the scans and when they looked there is typically lost. Our approach recaptures that information.
To the best of our knowledge, this is the first demonstration that automated lesion annotation for DL is possible by extracting the data directly from clinical image interpretations without added input from or effort by the radiologist. The lesion annotation can be performed in a nonobtrusive fashion that neither distracts from nor hinders patient care. Although the current work focuses on primary glial neoplasms, a notable future application of interest is brain metastases. Researchers have already used CNNs to detect and segment brain metastases (22–24). Initial success has been achieved in applying these models to radiation-therapy planning (25). However, all methods rely on the tedious and time-consuming process of hand annotation. The proposed method has potential to generate large volumes of data that could be collected from routine clinical work and used for artificial intelligence purposes.
In previous works using eye tracking for DL (20,26,27), eye tracking has been shown to accurately locate and segment lesions. Specifically, in Stember et al (27), the radiologist focused on lesion borders in 356 meningioma contrast-enhanced MR images. Then, training a U-Net CNN architecture using the resulting eye tracking–generated masks, the trained network was compared with that obtained by training on the corresponding hand-annotated masks. The average overlap between the two sets of masks, measured using the Dice similarity coefficient, was 85%. The CNNs trained on eye-tracking and hand-annotation masks were statistically equivalent to each other. The present work goes further than this previous work, making the approach clinically feasible (ie, ecologically valid). Our findings suggest that with fully simulated clinical interpretations, we can produce accurate lesion position labels using eye tracking and speech recognition. These data could then be leveraged to track lesions over time to aid in the reporting and clinical follow-up of brain metastases and to allow for the training of highly accurate and robust DL networks. The networks could in clinical practice detect and localize lesions, serving as a second reader to decrease false-negative results, particularly for small or subtle lesions.
Of importance for many DL applications is not merely locating lesions but also incorporating information about lesion shapes and sizes. Importantly, full lesion segmentation and characterization provides areas and volumes. Because we found that simulated image interpretation tends to provide points within lesions but does not actually contour shapes, this approach does not label images for DL segmentation tasks directly. However, lesion localization is widely recognized as the critical first step in bounding-box and contour prediction. Hence, we anticipate that the approach outlined here can form the foundation for transfer learning that achieves bounding-box localization by using, for instance, a faster regional CNN or YOLO (You Only Look Once) algorithm followed by segmentation by a U-Net–based architecture. Future work will incorporate these extensions.
Our voice-dictation software relied on saving the speech files, uploading them onto the Google Cloud Server, and then running our script using the Google Speech-to-Text Application Programming Interface to extract the time durations of stating the keywords. One ultimate goal is nearly real-time interpretation of speech integrated into the clinical workflow with established voice-dictation systems such as Nuance PowerScribe or M-Modal. Real-time interpretation would be an important part of actual deployment into clinical workflow for prospective research and then retrospective querying of eye-tracking and speech data to train CNNs to detect lesions or structures of interest.
Additionally, we note that normal or normal-variant structures or lesions (such as benign developmental venous anomalies or prominent perivascular spaces) are typically not studied in DL research because of their relatively prosaic clinical implications. However, DL networks to detect such structures still have the potential to expedite image interpretations by detecting such lesions and autopopulating reports. Although it is difficult to imagine research projects centered around detecting such lesions, an approach such as ours would allow for the training of these networks because radiologists often dictate or describe such lesions in clinical reports.
This study had several limitations. First, we only analyzed individual 2D image sections, as opposed to fully three-dimensional image series, to help expedite this proof-of-concept study. Using only 2D images could potentially increase the accuracy of the resulting networks because there are substantial correlations between instances of lesions across multiple sections, increasing the likelihood that radiologists will fixate on lesions directly. Second, all images were of single glial neoplasms. More general applications to lesion prediction would ideally be able to detect varying numbers and types of lesions. This is particularly true in disease states, such as brain metastases, in which a variable number of lesions are typically present. We will seek to address this by generalizing our CNN to have a recurrent “one-to-many” architecture that takes a fixed image size as the input but can output a variable number of location predictions depending on the number of lesions present.
Third, only a single neuroradiologist (J.N.S.) participated to generate the eye-tracking and report data, as well as the hand-annotated masks. Additionally, this radiologist was also aware that eye-tracking data were being acquired during simulated interpretation; thus, the Hawthorne effect of this knowledge altering the gaze pattern and/or interpretation may be relevant. We will address this in future work by employing multiple radiology readers in more realistic clinical simulations.
A fourth limitation was that the speech-recognition program missed the keyword for 7.6% (53 of 700) of image interpretations. The Google Cloud Speech-to-Text Application Programming Interface is a general-purpose voice-recognition program and is not tailored for the lexicon of radiology. It does not undergo further specific training for a particular user’s voice and speech style, as do clinical-grade software packages. As such, speech-recognition tools for clinical practice would presumably have a lower miss rate. Nevertheless, no dictation system is perfect, and lesion keyword calls would invariably be missed in any implementation. In this work, we manually added these missed images back in. However, to be seamless, future implementation would need to leave such missed cases excluded from the training set. This would reduce the number of tagged images, but if the numbers were large enough, it would presumably be less of a hindrance. For example, let us even assume a miss rate of 10%, higher than ours of 7.6%. Extracting 10 000 clinical lesion interpretations, that would still leave 9000 labeled lesions for CNN training. Another limitation was that clinical integration will have to allow for additional computing power that is employed in parallel with that required for standard image viewing on picture archiving and communication system workstations. This would be needed, notably, for processing and recording gaze data into gaze maps. Last, it should be noted that with current eye-tracking systems, users need to maintain fairly constrained positions with respect to the display sensors to obtain consistently reliable data. In true clinical practice, radiologists move around considerably, which results in some degree of eye-tracking data loss as calibration is lost.
Future work will include incorporating the approach into the clinical workflow to obtain prospectively labeled image data to train networks from a wide variety of modalities, body parts, and lesion types. Recognizing the limitations of our study, we would plan to integrate more image types (including three-dimensional MRI volumes) and more readers. Additionally, we would anticipate that radiologists would be aware that their eye movements within the viewing monitor will be recorded, but being in a nonresearch setting, they may behave differently. This awareness influence will need to be addressed in future studies.
In conclusion, we have demonstrated how eye tracking and speech recognition can be used to extract labeled image data for DL. Although applied here in this proof-of-principle study to brain lesions, the approach is very general and could be adapted to extract any structure of interest in any imaging modality. The proposed algorithm has potential to yield high quantities of labeled image data “for free” from standard-of-care clinical interpretations.
Disclosures of Conflicts of Interest: J.N.S. disclosed no relevant relationships. H.C. disclosed no relevant relationships. D.G. disclosed no relevant relationships. N.S. disclosed no relevant relationships. R.Y. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: author paid consultant for Agios, Puma, ICON, and NordicNeuroLab; institution has grant from Agios. Other relationships: disclosed no relevant relationships. S.E. disclosed no relevant relationships. A.H. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: fMRI Consultants (purely educational entity). Other relationships: disclosed no relevant relationships. S.J. disclosed no relevant relationships. B.W. Activities related to the present article: institution receives NIH Intramural grants (work supported in part by NIH Center for Interventional Oncology and the Intramural Research Program of the NIH. Activities not related to the present article: eye tracking patents pending for imaging regarding 2D and 3D transformations. Other relationships: NIH and University of Central Florida may own intellectual property in the space; NIH and NVIDIA have a cooperative research and development agreement; NIH and Siemens have a cooperative research and development agreement; NIH and Philips have a cooperative research and development agreement. P.C. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: author has stock in Avicenna.ai and is cofounder; author received travel accommodations from Canon Medical as a consultant. Other relationships: disclosed no relevant relationships. E.K. disclosed no relevant relationships. U.B. disclosed no relevant relationships.
Abbreviations:
- BraTS
- Brain Tumor Segmentation
- CNN
- convolutional neural network
- DL
- deep learning
- 2D
- two-dimensional
References
- 1.Kohli M, Prevedello LM, Filice RW, Geis JR. Implementing machine learning in radiology practice and research. AJR Am J Roentgenol 2017;208(4):754–760. [DOI] [PubMed] [Google Scholar]
- 2.Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: a primer for radiologists. RadioGraphics 2017;37(7):2113–2131. [DOI] [PubMed] [Google Scholar]
- 3.European Society of Radiology (ESR) . What the radiologist should know about artificial intelligence - an ESR white paper. Insights Imaging 2019;10(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang A, Tam R, Cadrin-Chenevert A, et al. Canadian Association of Radiologists white paper on artificial intelligence in radiology. Can Assoc Radiol J 2018;69(2):120–135. [DOI] [PubMed] [Google Scholar]
- 5.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 2006;31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- 6.Kikinis R, Pieper S. 3D Slicer as a tool for interactive brain tumor segmentation. In: Proceedings of the 2011 annual international conference of the IEEE Engineering in Medicine and Biology Society. Piscataway, NJ: Institute of Electrical and Electronics Engineers, 2011; 6982–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.3D Slicer website. https://www.slicer.org/. Accessed July 25, 2019.
- 8.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30(9):1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MD.ai website. https://www.md.ai. Accessed July 25, 2019.
- 10.XNAT website. https://www.xnat.org/. Accessed July 25, 2019.
- 11.Rubin DL, Ugur Akdogan M, Altindag C, Alkim E. ePAD: an image annotation and analysis platform for quantitative imaging. Tomography 2019;5(1):170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amazon Mechanical Turk. Amazon website. https://www.mturk.com/. Accessed July 25, 2019.
- 13.Lionbridge website. https://lionbridge.ai/. Accessed July 25, 2019.
- 14.fiverr website. https://www.fiverr.com/. Accessed July 25, 2019.
- 15.Langlotz CP, Allen B, Erickson BJ, et al. A Roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/The Academy workshop. Radiology 2019;291(3):781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geis JR, Brady AP, Wu CC, et al. Ethics of artificial intelligence in radiology: summary of the joint European and North American multisociety statement. Radiology 2019;293(2):436–440. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y, Heider P, Meystre S. Ensemble-based methods to improve de-identification of electronic health record narratives. AMIA Annu Symp Proc 2018;2018:663–672. [PMC free article] [PubMed] [Google Scholar]
- 18.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer 2018;18(8):500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ørting S, Doyle A, van Hilten MHA, et al. A survey of crowdsourcing in medical image analysis. ArXiv 1902.09159 [preprint] https://arxiv.org/abs/1902.09159. Posted February 25, 2019.
- 20.Khosravan N, Celik H, Turkbey B, Jones EC, Wood B, Bagci U. A collaborative computer aided diagnosis (C-CAD) system with eye-tracking, sparse attentional model, and deep learning. Med Image Anal 2019;51:101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Facial keypoints detection. Kaggle website. https://www.kaggle.com/c/facial-keypoints-detection. Accessed March 5, 2020.
- 22.Liu Y, Stojadinovic S, Hrycushko B, et al. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One 2017;12(10):e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med 2018;95:43–54. [DOI] [PubMed] [Google Scholar]
- 24.Dikici E, Ryu JL, Demirer M, et al. Automated brain metastases detection framework for T1-weighted contrast-enhanced 3D MRI. ArXiv 1908.04701 [preprint] https://arxiv.org/abs/1908.04701. Posted August 13, 2019. [DOI] [PubMed]
- 25.Shirokikh B, Dalechina A, Shevtsov A, et al. Deep learning for brain tumor segmentation in radiosurgery: prospective clinical evaluation. ArXiv 1909.02799 [preprint] https://arxiv.org/abs/1909.02799. Posted September 6, 2019.
- 26.Khosravan N, Celik H, Turkbey B, et al. Gaze2Segment: a pilot study for integrating eye-tracking technology into medical image segmentation. In: Müller H, Kelm BM, Arbel T, et al., eds. Medical computer vision and Bayesian and graphical models for biomedical imaging. BAMBI 2016, MCV 2016. Vol 10081, Lecture Notes in Computer Science. Cham, Switzerland: Springer, 2016; 94–104. [Google Scholar]
- 27.Stember JN, Celik H, Krupinski E, et al. Eye tracking for deep learning segmentation using convolutional neural networks. J Digit Imaging 2019;32(4):597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]