Abstract
DNA ligase I (LIG1) joins DNA strand breaks during DNA replication and repair transactions and contributes to genome integrity. The mutations (P529L, E566K, R641L and R771W) in LIG1 gene are described in patients with LIG1-deficiency syndrome that exhibit immunodeficiency. LIG1 senses 3’-DNA ends with a mismatch or oxidative DNA base inserted by a repair DNA polymerase. However, the ligation efficiency of the LIG1 variants for DNA polymerase-promoted mutagenesis products with 3’-DNA mismatches or 8-oxo-2’-deoxyguanosine (8-oxodG) remains undefined. Here, we report that R641L and R771W fail in the ligation of nicked DNA with 3’-8-oxodG, leading to an accumulation of 5’-AMP-DNA intermediates in vitro. Moreover, we found that the presence of all possible 12 non-canonical base pairs variously impacts the ligation efficiency by P529L and R771W depending on the architecture at the DNA end, whereas E566K exhibits no activity against all substrates tested. Our results contribute to the understanding of the substrate specificity and mismatch discrimination of LIG1 for mutagenic repair intermediates and the effect of non-synonymous mutations on ligase fidelity.
Introduction
Exposure to environmental agents and endogenous sources of mutagens can generate chemical modifications to the structure of DNA resulting in the formation of DNA lesions, such as an abasic or apurinic/apyrimidinic (AP) site and an oxidised base (8-oxo-2’-deoxyguanosine, 8-oxodG) (1,2). These lesions are mutagenic and can block DNA replication and repair leading to many human diseases, such as cancer (3,4). DNA ligase I (LIG1) contributes to genome stability by sealing processed Okazaki fragments during DNA replication and catalysing the ultimate ligation step during most DNA repair transactions (5–8). LIG1 protein contains a C-terminal region that includes three conserved domains: (i) adenylation domain (AdD), (ii) the oligonucleotide/oligosaccharide binding-fold domain (OBD) and (iii) the less conserved DNA-binding domain (DBD) that encircles duplex DNA as revealed by crystal structures(9,10). Studies have shown that the cellular functions of LIG1 are mediated through its non-catalytic N-terminal domain that contains the nuclear localisation signal and participates in protein–protein interactions (11). For example, LIG1 interacts with proliferating cell nuclear antigen and replication factor C during DNA replication, and a physical interaction between DNA polymerase (pol) β and LIG1 during base excision repair (BER) has also been reported (12–14).
LIG1-deficiency disease and Bloom Syndrome (BS), which is caused by the mutations in BLM gene, have been mistaken for each other in the past because they exhibit a range of overlapping clinical phenotypes (15,16). In both cases, a predisposition for cancer, stunted growth, photosensitivity and a high amount of sister chromatid exchange have been reported (17,18). The mutations that have been identified in the BS patients truncate the RecQ-like helicase (BLM) or result in its impaired function related to DNA replication and repair of DNA double-strand breaks (19). The majority of the patients with LIG1-deficiency syndrome have been described with symptoms of immunodeficiency (20–25). The inherited mutations in the LIG1 gene that have been identified are located in the C-terminal domain of the protein (P529L, E566K, R641L and R771W) and presumably impact the enzyme activity variously, excluding the truncated polypeptide (Thr415*) that lacks the catalytic region altogether (26,27). The first female individual case of human LIG1 deficiency was reported in 1992, and she was anaemic at birth, with subsequent growth retardation and developmental delay (22,23). This discovery was followed by the identification of five patients carrying biallelic inherited mutations in the LIG1 gene that exhibited a variety of clinical symptoms, such as hypogammaglobulinemia, severe eczema, adenovirus and urinary tract infections (24–26). In the present study, we investigated all of these LIG1 gene variant mutations identified in the patients (Supplementary Table 1).
LIG1 completes DNA repair that requires high-fidelity DNA synthesis by DNA polymerases, which discriminate against the incorporation of incorrect nucleotides to varying degrees based on base pairing and/or sugar identity (28). Successful ligation relies on the formation of a Watson–Crick base pair between 5’-phosphate (P) and 3’-hydoxyl (OH) termini of the resulting nicked product following DNA polymerase-mediated correct nucleotide insertion (14). Replicative DNA polymerases achieve high fidelity of DNA synthesis and exhibit proofreading activity that is responsible for the removal of incorrectly incorporated nucleotides from the primer terminus (29). However, most of the repair DNA polymerases, such as the X-family members of pol β, pol λ and pol μ, lack 3’-5’ proofreading exonuclease activity and incorporate a single nucleotide into the gap DNA with moderate fidelity; their intrinsic fidelity is much lower than that of polymerases involved in DNA replication (30,31). Furthermore, it has been shown that the mismatch recognition and error-detection mechanisms by a repair DNA polymerase are compromised by the mutagenic incorporation of 8-oxodG opposite adenine, mimicking a cognate base pair (32,33). In our previous studies, we reported the extent to which human DNA ligases I and IV discriminate 3’-ends, including a mismatch (deoxyribonucleotides or ribonucleotides) or oxidative DNA base damage (8-oxodG) during BER or double-strand break repair, respectively (34–42).
The insertion of 8-oxodG or a mismatched base by a DNA polymerase leads to the formation of an intermediate that is a poor substrate for a DNA ligase (14). In the presence of a damaged or modified DNA end, the ligation reaction fails and an abortive 5’-adenylate (AMP) product is formed (34–37). DNA intermediates with a 5’-AMP block and a 3’-damaged or mismatched end could serve as mutagenic repair intermediates that may trigger harmful nuclease activities, cytotoxicity, double-strand break formation in replicating DNA and, ultimately, genomic instability (38–42). However, we lack a full understanding of the mechanism of DNA-end recognition and substrate specificity of LIG1. Moreover, the impact of an impaired LIG1 activity caused by the disease-associated mutations in the LIG1 gene on the ligation of mutagenic repair intermediates formed after DNA polymerase-promoted insertion of an oxidative DNA damage or a mismatched base remains entirely unknown.
In the present study, we investigated the ligation efficiency of pre-inserted 8-oxodG and all possible 12 non-canonical base pairings at the 3’-end of nicked DNA substrates by LIG1 variants (P529L, R641L, E566K and R771W) in vitro relative to the wild-type enzyme. Our findings demonstrate that the ligase activity of the variants R641L and R771W for 3’-8-oxodG substrates was significantly compromised in comparison with those of wild type and P529L. Homology modelling suggests a potential impact of the R641L and R771W mutations on the structure/function of LIG1 in complex with nicked DNA containing 8-oxodG. Moreover, we found that the LIG1 disease variants P529L and R771W exhibit diverse ligation efficiency for the 12 non-canonical base pairings at the 3’-end of nicked DNA, while E566K has no activity against all DNA substrates tested in this study.
Materials and methods
Preparation of DNA substrates
Oligodeoxyribonucleotides with and without a 6-carboxyfluorescein (FAM) label were obtained from Integrated DNA Technologies. The nicked DNA substrates containing template bases A, T, G or C and 3’-pre-inserted bases (dA, dT, dG or dC) or 8-oxodG were prepared as described previously (36–42) and used for the ligation assays (Supplementary Table 2).
Preparation of DNA LIG1 mutants
The plasmid constructs of (Δ261) DNA LIG1 variants (P529L, E566K, R641L and R771W) were cloned into pET-24b (Novagen) expression vector by site-directed mutagenesis using the wild-type DNA LIG1 construct (40). The coding sequences of all mutants were confirmed by DNA sequencing.
Protein purifications
The recombinant human DNA LIG1 (Δ261) mutant proteins (P529L, E566K, R641L and R771W) were purified as described previously (36–42). Briefly, proteins were overexpressed in Rosetta (DE3) pLysS Escherichia coli cells (Millipore Sigma) grown in Terrific Broth media with kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml) at 37°C. Once the Optical Density reached 1.0, expression was induced with 0.5 mM Isopropyl ß-D-1-thiogalactopyranoside and continued overnight at 16°C. After centrifugation at 5000× rpm for 15 min, cells were lysed in lysis buffer (50 mM Tris-HCl pH 7.0, 500 mM NaCl, 20 mM imidazole, 5% glycerol, 1 mM Phenylmethylsulfonyl fluoride and one EDTA-free protease inhibitor cocktail tablet) by sonication at 4°C. The lysate was clarified by centrifugation at 16 000× rpm for 1 h. The cell lysis solution was then filter-clarified and loaded onto a HisTrap HP column (GE Health Sciences) that was equilibrated with binding buffer A (50 mM Tris-HCl pH 7.0, 500 mM NaCl, 20 mM imidazole and 5% glycerol). The column was washed with binding buffer A, followed by buffer B (50 mM Tris-HCl pH 7.0, 500 mM NaCl, 35 mM imidazole and 5% glycerol). The protein was eluted with an increasing imidazole gradient (0–500 mM) in elution buffer A (50 mM Tris-HCl pH 7.0, 500 mM NaCl, 500 mM imidazole and 5% glycerol) at 4°C. The collected fractions containing the LIG1 protein were then loaded onto a Superdex 200 10/300 (GE Health Sciences) column and eluted in elution buffer B (50 mM Tris-HCl pH 7.0, 500 mM NaCl, 5% glycerol and 1 mM Dithiothreitol (DTT)). The protein was further purified by Resource Q (GE Health Sciences) and finally by HiTrap Heparin (GE Health Sciences) columns with binding buffer B (20 mM Tris-HCl pH 7.0, 50 mM NaCl and 5% glycerol) and then with elution buffer C (20 mM Tris-HCl pH 7.0, 1 M NaCl and 5% glycerol). Protein quality was evaluated onto 10% Sodium dodecyl sulfate polyacrylamide gel electrophoresis, and the protein concentration was measured using absorbance at 280 nm. The final proteins were stored in storage buffer (20 mM Tris-HCl pH 7.0, 300 mM NaCl and 5% glycerol) in aliquots at −80°C.
Structure modelling of DNA LIG1 variants
Structure analysis of DNA LIG1 variants was performed based on the solved crystal structure of human DNA LIG1 bound to adenylated DNA containing an 8-oxodG:A base pair (PDB:6P0E) using the Coot software (43,44). All structural images were drawn using PyMOL (http://www.pymol.org/).
DNA ligation assays
The ligation assays were performed as described previously (36–42). Briefly, the reaction was performed in a mixture containing 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 100 µg/ml Bovine serum albumin, 10% glycerol and the indicated nicked DNA substrate (500 nM) in a final volume of 10 µl. The ligation assays were initiated with the addition of wild-type or one of the DNA LIG1 variants: P529L, E566K, R641L or R771W (100 nM). The reaction mixture was incubated at 37°C and stopped at the time points indicated in the figure legends. The reaction products were then quenched with an equal amount of gel loading buffer (95% formamide, 20 mM ethylenediaminetetraacetic acid, 0.02% bromophenol blue and 0.02% xylene cyanol) and separated by electrophoresis on an 18% denaturing polyacrylamide gel. The gels were scanned with a Typhoon PhosphorImager (Amersham Typhoon RGB), and the data were analysed using ImageQuant software.
Results
Ligation of repair intermediates with 3’-8-oxodG by LIG1 variants
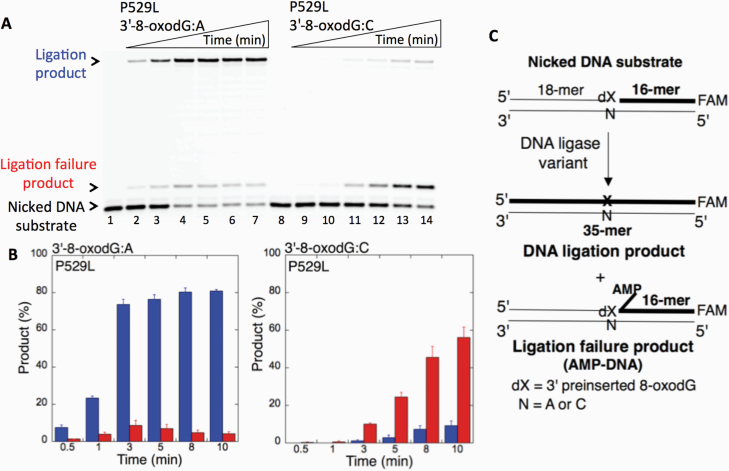
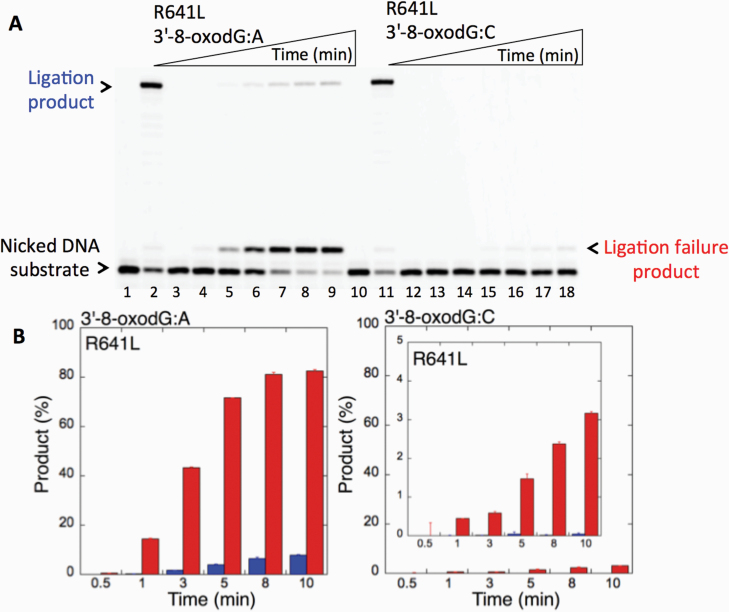
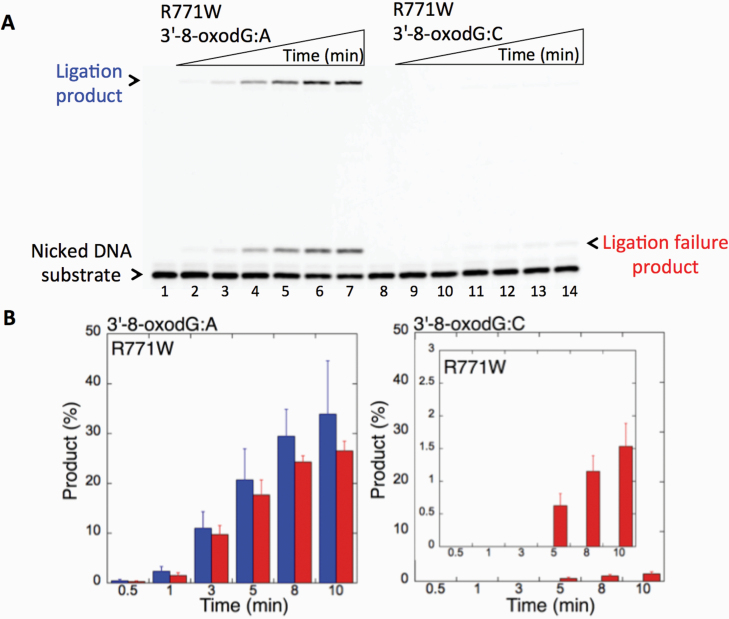
We first examined the impact of LIG1 disease mutations (P529L, R641L and R771W) on the ligation fidelity of nicked DNA substrates with a 3’-pre-inserted 8-oxodG opposite A or C in vitro (Figure 1C). LIG1 variant P529L can ligate the DNA substrate with 3’-8-oxodG:A (Figure 1A, Lanes 2–7) more efficiently than that of nicked DNA with 3’-8-oxodG:C (Figure 1A, Lanes 9–14). However, we obtained more ligation failure products in the reaction with the 3’-8-oxodG:C substrate (Figure 1B). LIG1 variant R641L exhibited very low activity on both DNA substrates (3’-8-oxodG:A and 3’-8-oxodG:C) compared to the ligation of nicked DNA with undamaged ends (Figure 2A, Lanes 2 and 11). The R641L mutant failed in the presence of the 3’-8-oxodG:A substrate (Figure 2A, Lanes 4–9) and yielded a higher amount of 5’-AMP-DNA intermediates (Figure 2B) than obtained with the nicked 3’-8-oxodG:C DNA (Figure 2A, Lanes 13–18). The ligation reactions containing the 3’-8-oxodG:A substrate and the LIG1 variant R771W were accompanied by an accumulation of AMP-DNA intermediates (Figure 3A, Lanes 2–7, and Figure 3B). Similar to R641L (Figure 2), R771W was not active on the 3’-8-oxodG:C substrate (Figure 3A, Lanes 9–14), and a very low amount of ligation failure products were observed (Figure 3B). We did not observe any ligation products by the LIG1 variant E566K (Supplementary Figure 1A).
Figure 1.
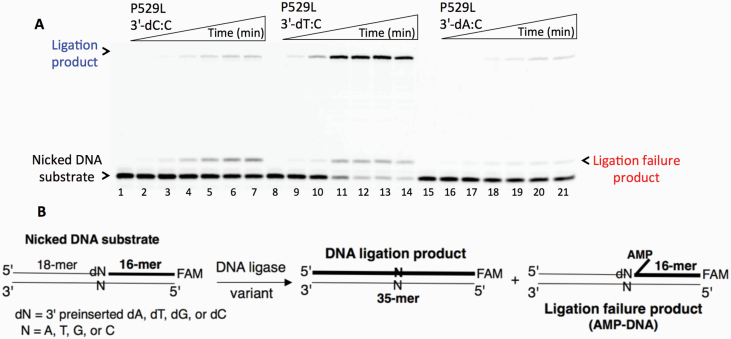
Ligation of the repair intermediate with 3’-8-oxodG by LIG1 variant P529L. (A) Lanes 1 and 8 are the negative enzyme controls of the nicked DNA substrates with 3’-8-oxodG opposite templates A and C, respectively. Lanes 2–7 and 9–14 show the reaction products obtained for 3’-8-oxodG:A and 3’-8-oxodG:C substrates, respectively, at Time points 0.5, 1, 3, 5, 8 and 10 min. (B) The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data are presented as the averages from three independent experiments ± SDs. (C) Illustrations of the nicked DNA substrate with 3’-8-oxodG and the products observed in the ligation reaction.
Figure 2.
Ligation of the repair intermediate with 3’-8-oxodG by LIG1 variant R641L. (A) Lanes 1 and 10 are the negative enzyme controls of the nicked DNA substrates with 3’-dT opposite template A and 3’-dC opposite template G, respectively. Lanes 2 and 11 show the reaction products obtained for 3’-dT:A and 3’-dC:G, respectively. Lanes 3 and 12 are the negative enzyme controls of the nicked DNA substrates with 3’-8-oxodG opposite templates A and C, respectively. Lanes 4–9 and 13–18 show the reaction products obtained for 3’-8-oxodG:A and 3’-8-oxodG:C substrates, respectively, at Time points 0.5, 1, 3, 5, 8 and 10 min. (B) The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data are presented as the averages from three independent experiments ± SDs.
Figure 3.
Ligation of the repair intermediate with 3’-8-oxodG by LIG1 variant R771W. (A) Lanes 1 and 8 are the negative enzyme controls of the nicked DNA substrates with 3’-8-oxodG opposite templates A and C, respectively. Lanes 2–7 and 9–14 show the reaction products obtained for 3’-8-oxodG:A and 3’-8-oxodG:C substrates, respectively, at Time points 0.5, 1, 3, 5, 8 and 10 min. (B) The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data are presented as the averages from three independent experiments ± SDs.
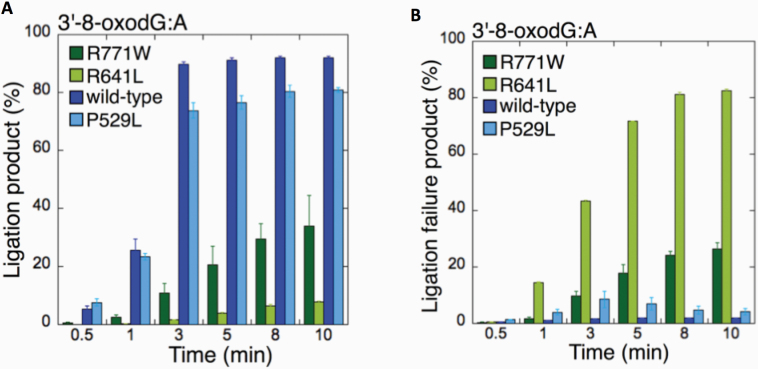
We then compared the wild-type enzyme (Supplementary Figure 1B) to the different LIGI variants (Figures 1–3) for the 3’-8-oxodG:A substrate as we obtained products for all ligases using this approach (Figure 4). The comparisons revealed that there was no significant difference in ligation efficiency between wild type and P529L, while R641L and R771W were compromised in their ability to complete the ligation reaction (Figure 4A). Indeed, we found a significant increase (~40–80- and ~10-30-fold for R641L and R771W, respectively) in the amount of ligation failure products when compared to wild-type LIG1 (Figure 4B).
Figure 4.
Comparisons for the ligation of 3’-8-oxodG:A substrate by the wild-type and LIG1 variants. (A, B) The graphs show the time-dependent changes in the ligation (A) and ligation failure (B) products for the wild-type and LIG1 variants P529L, R641L and R771W. The data are presented as the averages from three independent experiments ± SDs.
Structure and function analysis of LIG1 variants
To gain insight into the possible structure-function mechanisms underlying the defects, we performed homology modelling to determine the impact of the LIG1 disease mutations (P529L, E566K, R641L and R771W) on ligase activity (Figures 1–4) using the previously solved crystal structure of human LIG1 (PDB: 6P0E) in complex with DNA containing 8-oxodG:A (44).
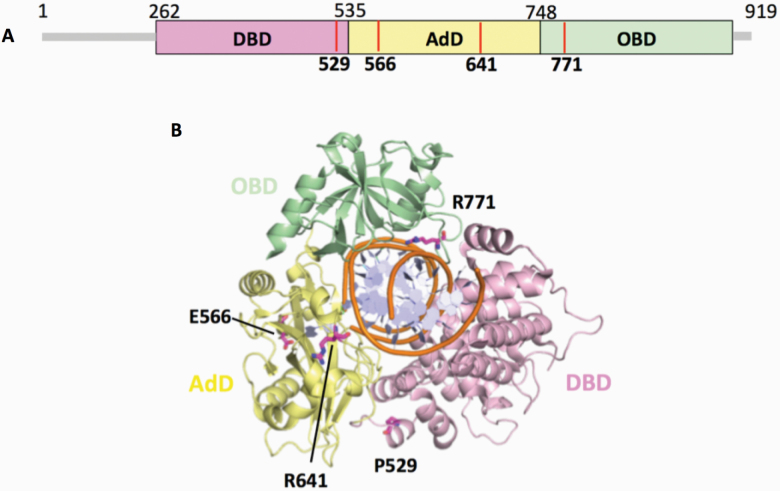
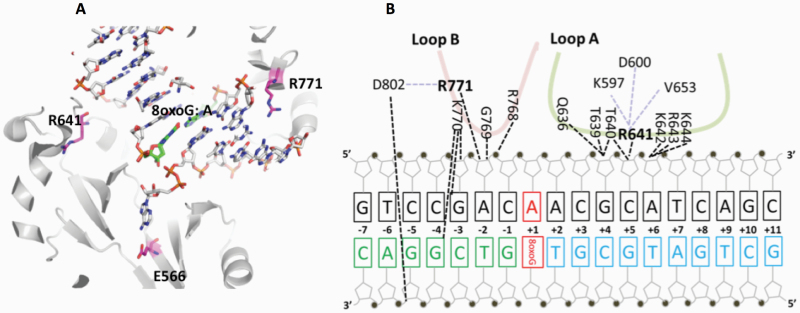
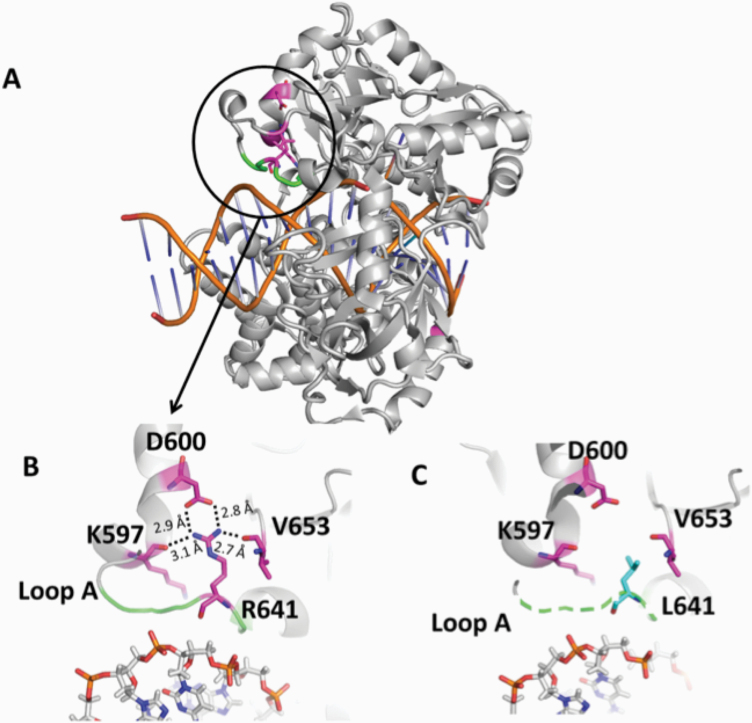
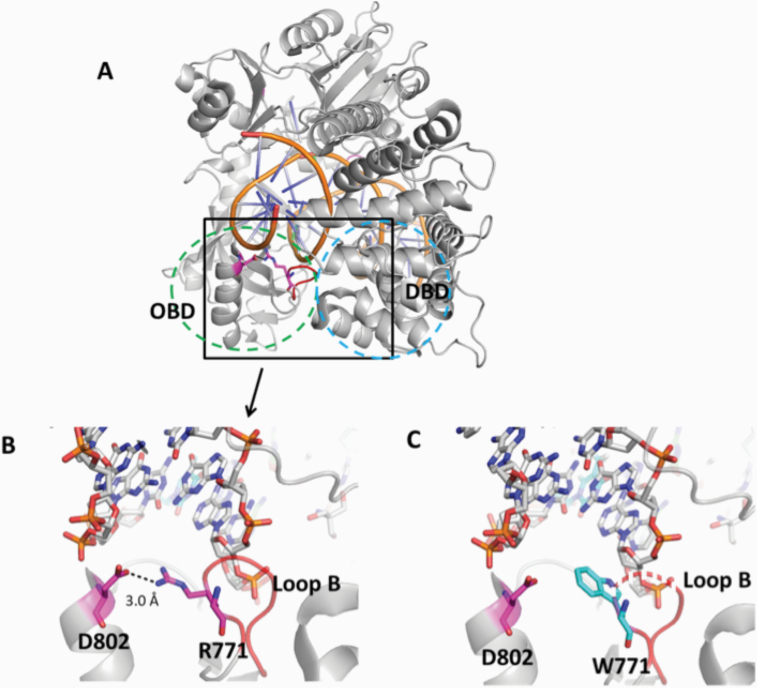
The mutations occur at amino acid residues of LIG1 that are located on different subdomains within the catalytic core of the protein (262–919 aa). E566 and R641 reside in the AdD domain, while P529 and R771 are in the DBD and OBD domains, respectively (Figure 5A). All domains encircle the nicked DNA (Figure 5B) as reported previously (10). Our structural models show that Arg641 and Arg771 are adjacent to separate DNA contact sites, namely Loops A and B, and interact with critical functional amino acid residues (Figure 6). Arg641 in Loop A makes a salt bridge with Asp600 and interacts with a carbonyl group of Lys597 and Val653 (Figure 7A and B), and the residues that are in close proximity to Arg641 (i.e. Gln636, Thr639, Thr640, Lys642, Arg643 and Lys644) interact with template DNA (Figure 6B). Arg771 directly interacts with the template DNA strand and forms a salt bond with Asp802 (Figure 8A and B). Furthermore, our models reveal template and downstream DNA interactions with Lys770 (Figure 6B). Accordingly, we hypothesise that all of these interactions stabilise the minor groove DNA conformation that enables the ligase to complete catalysis, and the replacement of Arg641 with Leu (R641L) or Arg771 with Trp (R771W) would disrupt the stabilisation of Loops A and B, confound the interactions with their interacting amino acids and, therefore, interfere with the DNA-binding capacity of LIG1 (Figures 7C and 8C).
Figure 5.
Domain architecture of LIG1 with the disease mutations. (A) Schematic view showing the domain composition of human LIG1, including the N-terminal domain (amino acids 1–261, gray), and the catalytic core (amino acids 262–919) consisting of the DBD (pink), AdD (yellow) and OBD (green). (B) LIG1 (cartoon) in complex with an adenylated nicked DNA complex (stick, orange). The amino acid residues (magenta), P529, E566, R641 and R771, that are mutated in LIG1-deficiency disease are shown as sticks.
Figure 6.
Architecture of LIG1 showing the positions of the disease-associated residues in complex with the nicked DNA containing 8-oxoG:A. (A) Ribbon diagram shows LIG1 (grey) encircling the nicked DNA (sticks) with 8-oxoG opposite template A (green). E566, R641 and R771 (magenta) are represented by sticks. (B) Nucleotide-residue contact map showing individual LIG1 residues interactions that are close proximity to R641 (loop A, green) and R771 (loop B, red) with the nicked DNA containing 8-oxoG:A.
Figure 7.
Structure model of LIG1 with R641L identified in LIG1 deficiency syndrome. (A) Ribbon diagram showing LIG1 encircling a nicked DNA duplex (orange). Arg641 (magenta) shown as sticks is located in Loop A (green). (B) Arg641 (R641) interacts with K597, D600 and V653, and (C) the replacement of Arg641 with Leu (L641) disrupts the interaction and destabilises Loop A (shown in dotted green line). All residues are shown as sticks. Positive and negative potentials are shown in blue and red, respectively.
Figure 8.
Structure model of LIG1 with R771W identified in LIG1 deficiency syndrome. (A) Ribbon diagram showing LIG1 encircling a nicked DNA duplex (orange). Arg771 (magenta) shown as sticks is located in Loop B (red) that resides in the OBD domain (green dotted circle) and interacts with the DBD (blue dotted circle). (B) Arg771 (R771) interacts with D802 (magenta) to stabilise the loop B, and (C) replacement of Arg771 with Trp (cyan, W771) disrupts this interaction and destabilises Loop B (shown in dotted red line). All residues are shown as sticks. Positive and negative potentials are shown in blue and red, respectively.
The crystal structure of LIG1 bound to oxidatively damaged DNA uncovered fundamental conformational changes that the ligase active site must undergo in the presence of 8-oxodG:A in syn geometry (44). Our findings demonstrate that the ligation of the 3’-8-oxodG:A substrate by R641L or R771W is significantly impaired in comparison with the wild-type enzyme (Figure 4). This effect could be due to the unstable conformation of the loops and repulsive amino acid interactions (Figures 6–8) at the ligase active site, which exhibits significant structural adjustments (i.e. rearrangements of active-site amino acid residues N590 and R589) in the presence of 8-oxodG that flips out into the DNA major groove (44). We suggest that amino acid substitutions at R641 and R771 would impair the catalytic architecture of the ligase conformation that would more profoundly interfere with its DNA binding in the context of a frayed 3’-terminus (Supplementary Figure 2A and B).
The difference we observed in the ligation efficiencies of 8-oxodG:A vs. 8-oxodG:C substrates by the LIG1 variants could be due to the dual coding potentials of 8-oxodG (anti- or syn-), which use Watson–Crick to base pair with C or Hogsten edge to base pair with A, respectively (45). This variation can create two distinct structural conformations of the primer terminus backbone at the ligase active site (Supplementary Figure 2C and D). Similarly, pol β structural snapshots during an insertion of 8-oxodGTP revealed a poor active-site geometry and less insertion efficiency of 8-oxodGTP opposite C than opposite A (46). Furthermore, in our previous studies, we reported less efficient ligation of pol β 8-oxodGTP insertion products opposite C relative to A in a coupled reaction, including both DNA LIG1 and pol β in vitro (39).
Our models also show that the replacement of Pro529 with Leu (P529L) has no effect on the protein structure (Supplementary Figure 3). Accordingly, we did not obtain any significant difference in the enzyme activity for the ligation of 3’-8-oxodG substrates between the wild-type and P529L ligases (Figure 4). Furthermore, we observed an altered hydrogen bond formation with the 5’-AMP catalytic intermediate in the presence of the mutation at Glu566 (E566K) in our models (Supplementary Figure 4), as well as a complete disruption of ligase activity for E566K in comparison with the wild-type enzyme (Supplementary Figure 1A).
Specificity of LIG1 variants for the ligation of 3’-pre-inserted DNA mismatches
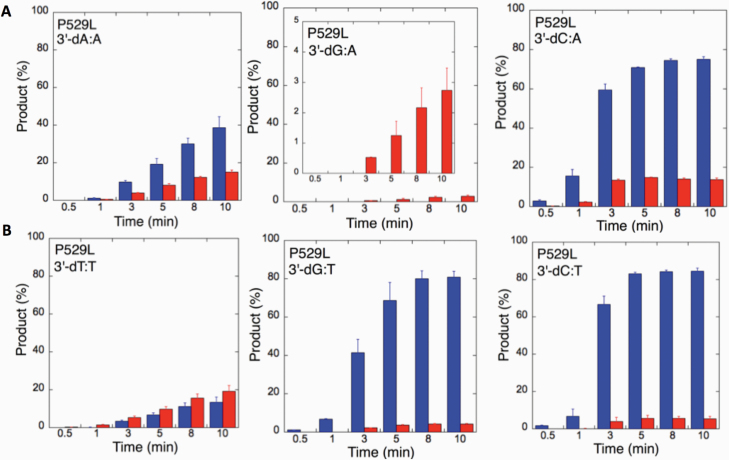
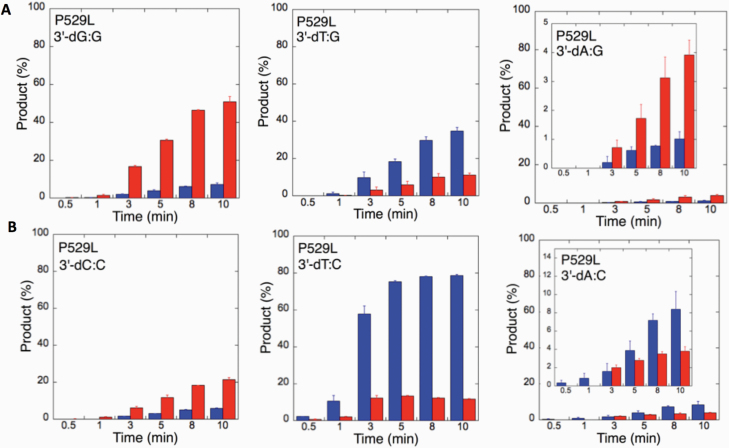
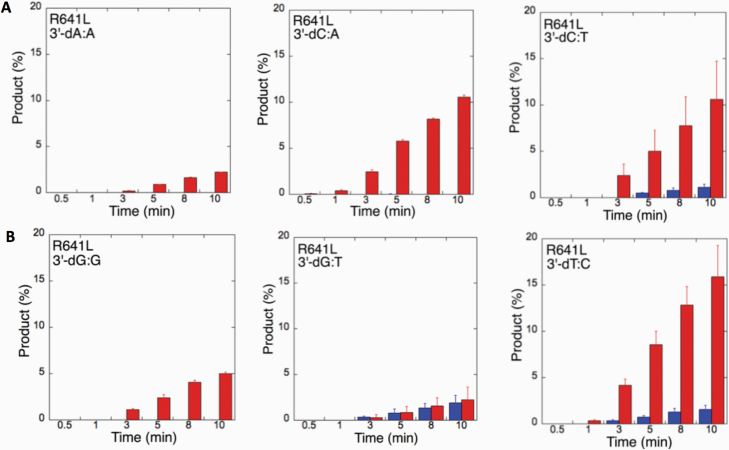
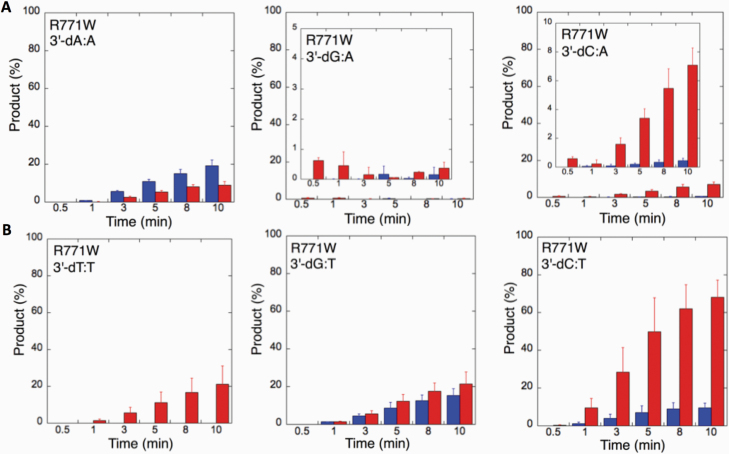
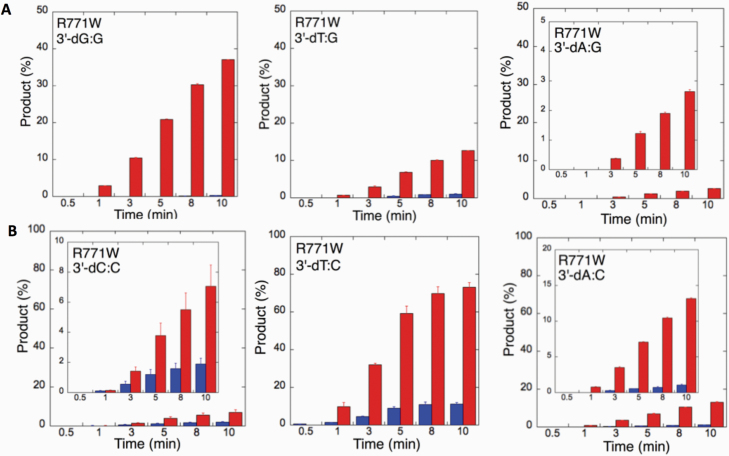
We next examined the efficiency of the LIG1 mutants for ligation of nicked DNA substrates with 3’-pre-inserted mismatches, including all possible 12 non-canonical base pairs (Figure 9B). LIG1 variant P529L exhibited ligase activity for almost all mismatch-containing DNA substrates (Figures 9A and Supplementary Figures 5–6). Specifically, we obtained a higher amount of ligation products (~60–80%) for 3’-mismatch:template bases, dC:A, dG:T, dC:T (Figure 10) and dT:C (Figure 11B) and ligation failure products (~40%) for 3’-dG:G (Figure 11A). Conversely, LIG1 variant R641L was not active on the majority of the mismatch substrates (Supplementary Figures 7–8). We observed, instead, ligation failure products only (~10%) with the nicked DNA harbouring 3’-mismatch:template bases: dA:A, dC:A, dC:T, dG:G, dG:T and dT:C (Figure 12). The LIG1 variant R771W (Supplementary Figures 9–10) yielded the highest amount of ligation failure products (~60%) for 3’-dC:T (Figure 13B) and 3’-dT:C (Figure 14B), and the amount of these products was only ~10–20% for 3’-dT:T, 3’-dG:T (Figure 13), 3’-dT:G and 3’-dA:C (Figure 14).
Figure 9.
Ligation of the repair intermediate with 3’-pre-inserted mismatches opposite template C by LIG1 variant P529L. (A) Lanes 1, 8 and 15 are the negative enzyme controls of the nicked DNA substrates with 3’-dC:C, 3’-dT:C and 3’-dA:C, respectively. Lanes 2–7, 9–14 and 16–21 show the reaction products obtained at time points 0.5, 1, 3, 5, 8 and 10 min. (B) Illustrations of the nicked DNA substrate with 3’-mismatches and the products observed in the ligation reaction.
Figure 10.
Specificity of LIG1 variant P529L for the ligation of repair intermediates with 3’-pre-inserted mismatches opposite templates A and T. (A,B) The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products for 3’-pre-inserted mismatches opposite templates A (A) and T (B). The data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Supplementary Figure 5.
Figure 11.
Specificity of LIG1 variant P529L for the ligation of repair intermediates with 3’-pre-inserted mismatches opposite templates G and C. (A, B) The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products for 3’-pre-inserted mismatches opposite templates G (A) and C (B). The data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Figure 9 and Supplementary Figure 6.
Figure 12.
Specificity of LIG1 variant R641L for the ligation of repair intermediates with 3’-pre-inserted mismatches. (A, B) The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products. The data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Supplementary Figures 7 and 8.
Figure 13.
Specificity of LIG1 variant R771W for the ligation of repair intermediates with 3’-pre-inserted mismatches opposite templates A and T. (A, B) The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products for 3’-pre-inserted mismatches opposite templates A (A) and T (B). The data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Supplementary Figure 9.
Figure 14.
Specificity of R771W for the ligation of repair intermediates with 3’-pre-inserted mismatches opposite templates G and C. (A, B) The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products for 3’-pre-inserted mismatches opposite templates G (A) and C (B). The data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Supplementary Figure 10.
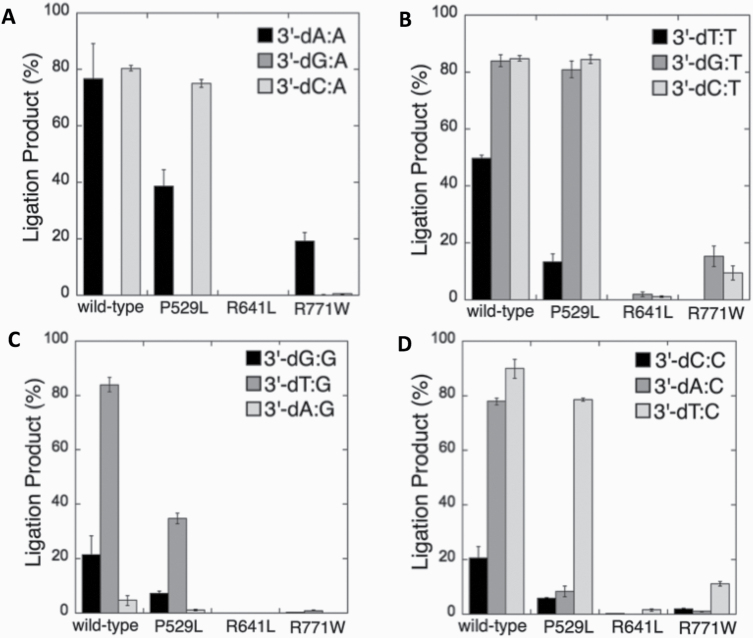
We next compared the wild-type (Supplementary Figures 11–14) and LIGI variant enzymes (Figures 10–14) for their ability to ligate nicked DNA substrates with all 3’-pre-inserted mismatches (Figure 15). Our comparisons revealed that LIG1 variant P529L shows almost the same specificity as the wild-type protein for all substrates, with a slightly less amount of ligation products (except 3’-dA:C). Nevertheless, the presence of all possible 12 non-canonical base pairs at the 3’-end variously impacts the ligation efficiency of P529L and R771W depending on the type of mismatch. Interestingly, we did not observe a comparable amount of ligation products for any of the 12 non-canonical base pairs by the LIG1 variant R641L (Figure 15). This result is consistent with the greater functional impairment of R641L for the ligation of 8-oxodG substrates relative to R771W (Figure 4A).
Figure 15.
Comparisons for the ligation of 3’-pre-inserted mismatch-containing substrates by the wild-type and LIG1 variants. (A–D) The graphs show the time-dependent changes in the amount of ligation products, and the data are presented as the averages from three independent experiments ± SDs.
Discussion
The fidelity of DNA ligation is a crucial component of the faithful replication and repair of genomic DNA (5–7). Indeed, defects in the ligation of Okazaki fragments during DNA replication has been reported to be linked to a deficiency in LIG1 activity as shown in LIG1-null mouse cells (47). Moreover, the rare human diseases with an autosomal recessive mode of inheritance, such as the LIG1-deficiency disease, are associated with greatly increased genome instability (20–27). The inherited mutations (P529L, R641L, R771W and E566K) identified in the LIG1 gene have been reported as causative factors for LIG1-deficiency disease in patients that exhibit immunodeficiency of variable severity (Supplementary Table 1). The primary fibroblast cell line 46BR and its LIG1-deficient subcell line 46BR.1G1, which expresses the individual disease mutant R771W, show an enhanced sensitivity to a broad range of DNA damage-inducing agents, including DNA alkylators (20–23). Furthermore, an elevated level of phosphorylated H2AX (a marker of DNA damage) following ionising radiation and an increased incidence of sister chromatid exchange (reflective of genome instability) have been reported in these cell lines, as well as in the lymphoblastoid B and peripheral blood T cells that express the individual disease mutant R641L (23–25). Recently, reduced repair and hypersensitivity to the DNA alkylating agent ethyl methanesulfonate have been shown for LIG1−/− HEK-293T cells expressing LIG1 R641L and R771W mutants in comparison to wild-type HEK-293T cells (26). A mouse model engineered to express the R771W mutant exhibits increased genomic instability and a predisposition to cancer (47,48). It has also been reported that steady-state LIG1 protein level is significantly higher in different types of malignant tumours compared to benign tissue samples obtained from human patients (49). Overall, the reports above highlight the essential role of LIG1 for cellular viability and the fact that defects or insufficiency in LIG1 is casually linked to genome instability and disease.
DNA ligases and DNA polymerases that play roles in DNA replication and repair transactions have long been prime candidates for key regulators of genome stability (28). Participation of an error-prone DNA polymerase in DNA repair processes can generate DNA nicks containing a damaged or mismatched 3’-end (14). Yet, we lack a fundamental understanding of the DNA ligation mechanism and human DNA ligase substrate specificities. The biological implications of DNA ligases and their fidelity for the ligation of mutagenic repair intermediates are critical to understand the molecular features of DNA ligation and ligase function in normal cells and in human diseases, such as cancer. As described above, cells from LIG1-deficient patients are hypersensitive to DNA damage-inducing agents that cause oxidative damage or single-base lesions that are mainly repaired by BER involving pol β nucleotide insertion and subsequent ligation by LIG1. In the present study, we comprehensively investigated the extent to which wild-type and LIG1 variants (P529L, R641L and R771W) counteract DNA polymerase-promoted mutagenesis. We elucidated the mechanism of substrate discrimination by wild-type and LIG1 variant proteins for DNA ends on the basis of base pairing (correct vs. incorrect) and oxidative DNA base damage (8-oxodG). The DNA substrates used in this study mimic the potential mutagenic repair intermediates and persistent DNA breaks that are expected to be toxic, to block transcription, or to be converted into double-strand breaks upon DNA replication.
Overall, our results revealed that functional defects in the catalytic activity of human LIG1 lead to a failure in the discrimination of damaged or mismatched DNA substrates. Consistent with our structure models, our in vitro findings indicate that two missense mutations (R641L and R771W) described in the patients with LIG1-deficiency syndrome uniquely impact ligase activity in comparison with P529L (wild-type level of activity) and E566K (lack of activity). Specifically, these two ligase variants fail in the second step of the ligation reaction, where DNA-AMP intermediates accumulate while the enzyme is transferring an AMP moiety to the 5’-P terminus of the DNA break before the last step of phosphodiester bond formation between the 3’-OH and 5’-P ends (8). Furthermore, we observed that the extent of this ligation failure is significantly sensitive to both the type of base (mismatch or damaged) and the nature of the non-synonymous mutation. Overall, LIG1 variants tested in this study are very selective for the chemical characteristics of base pairs and show different abilities to join nicks, including 3’-mismatches (12 possible non-canonical base pairs) or 8-oxodG. When comparing the genotype and clinical characteristics of the patients (Supplementary Table 1) with our in vitro ligase activity findings and structure models, we suggest that the LIG1 E566K variant with no ligase activity against all substrates tested is likely the primary driver of the phenotypes seen in the original patient who was R771W/E566K heterozygous and experienced growth retardation and died from lymphoma. For the patients with homozygous P529L and R771W mutations, our data suggest that the significantly impaired fidelity of the LIG1 R771W variant could contribute to their severe clinical phenotype, since the P529L variant protein displays normal catalytic activity. We suggest that the variable deficiencies in substrate discrimination by the LIG1 variants R771W and R641L could contribute to the phenotypic diversity of the patients carrying these mutations. Nevertheless, it is important to emphasise the overlapping functions of human LIG1 and LIG3, particularly in BER. Previous studies have demonstrated the exchange between these two human ligases during single-strand break repair in vitro and the requirement of LIG3 for the viability of LIG1-depleted mammalian cells in vivo (50). Therefore, impairment in LIGI activity could be complemented by LIG3 in cells, a hypothesis that would need to be assessed in LIG1 mutant cell lines.
DNA ligation is the last step of almost all DNA repair pathways, and DNA ligases are known as biomarkers of abnormal DNA repair (5–7). Therefore, DNA ligases are promising candidates for the development of small-molecule inhibitors for anticancer therapeutic purposes (51). Human DNA ligase inhibitors have been developed using a computer-aided rational drug design strategy based on the crystal structure of human LIG1 and have been used in preclinical investigations (51–54). For example, the LIG1-specific inhibitor L87-G17 increased cytotoxicity of MCF7 breast cancer cells to DNA-damaging agents, such as the alkylator methyl methanesulfonate and decreased colon cancer cell survival after ionising radiation with no observed effect on the sensitivity of normal cells (55,56). Our study that reports on the mechanism of DNA ligation and substrate discrimination by wild-type and disease mutants of LIG1 for potential mutagenic and toxic repair intermediates could contribute to the development of repair inhibitors towards LIG1 and improvement of current DNA ligase inhibitors to more specifically target aberrant DNA repair in cancer cells (57). However, additional structural studies are required to provide further insights into the structural basis of mutagenic ligation and to define the elements that play a crucial role in locating and engaging the mutagenic DNA lesions to optimise ligation fidelity.
Funding
This work was supported by the National Institutes of Health/National Institute of Environmental Health Sciences (grant 4R00ES026191).
Conflict of interest statement: None declared.
Supplementary Material
References
- 1. De Bont R and van Larebeke N (2004) Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis, 19, 169–185. [DOI] [PubMed] [Google Scholar]
- 2. Singh R, Kaur B, Kalina I, et al. (2007) Effects of environmental air pollution on endogenous oxidative DNA damage in humans. Mutat. Res., 620, 71–82. [DOI] [PubMed] [Google Scholar]
- 3. Nelson B C and Dizdaroglu M (2020) Implications of DNA damage and DNA repair on human diseases. Mutagenesis, 35, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chatterjee N and Walker G C (2017) Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen., 58, 235–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ellenberger T and Tomkinson A E (2008) Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem., 77, 313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Timson D J, Singleton M R and Wigley D B (2000) DNA ligases in the repair and replication of DNA. Mutat. Res., 460, 301–318. [DOI] [PubMed] [Google Scholar]
- 7. Tomkinson A E and Mackey Z B (1998) Structure and function of mammalian DNA ligases. Mutat. Res., 407, 1–9. [DOI] [PubMed] [Google Scholar]
- 8. Martin I V and MacNeill S A (2002) ATP-dependent DNA ligases. Genome Biol., 3, 3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howes T R and Tomkinson A E (2012) DNA ligase I, the replicative DNA ligase. Subcell. Biochem., 62, 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pascal J M, O’Brien P J, Tomkinson A E and Ellenberger T (2004) Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature, 432, 473–478. [DOI] [PubMed] [Google Scholar]
- 11. Tomkinson A E, Vijayakumar S, Pascal J M and Ellenberger T (2006) DNA ligases: structure, reaction mechanism, and function. Chem. Rev., 106, 687–699. [DOI] [PubMed] [Google Scholar]
- 12. Levin D S, Bai W, Yao N, O’Donnell M and Tomkinson A E (1997) An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. USA, 94, 12863–12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levin D S, Vijayakumar S, Liu X, Bermudez V P, Hurwitz J and Tomkinson A E (2004) A conserved interaction between the replicative clamp loader and DNA ligase in eukaryotes: implications for Okazaki fragment joining. J. Biol. Chem., 279, 55196–55201. [DOI] [PubMed] [Google Scholar]
- 14. Çağlayan M. (2019) Interplay between DNA polymerases and DNA ligases: influence on substrate channeling and the fidelity of DNA ligation. J. Mol. Biol., 431, 2068–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomkinson A E, Lasko D D and Lindahl T (1990) Biochemical properties of mammalian DNA ligase I and the molecular defect in Bloom’s syndrome. Prog. Clin. Biol. Res., 340A, 283–294. [PubMed] [Google Scholar]
- 16. Willis A E and Lindahl T (1987) DNA ligase I deficiency in Bloom’s syndrome. Nature, 325, 355–357. [DOI] [PubMed] [Google Scholar]
- 17. Chan J Y, Becker F F, German J and Ray J H (1987) Altered DNA ligase I activity in Bloom’s syndrome cells. Nature, 325, 357–359. [DOI] [PubMed] [Google Scholar]
- 18. Lehmann A R, Willis A E, Broughton B C, James M R, Steingrimsdottir H, Harcourt S A, Arlett C F and Lindahl T (1988) Relation between the human fibroblast strain 46BR and cell lines representative of Bloom’s syndrome. Cancer Res., 48, 6343–6347. [PubMed] [Google Scholar]
- 19. Arora H, Chacon A H, Choudhary S, McLeod M P, Meshkov L, Nouri K and Izakovic J (2014) Bloom syndrome. Int. J. Dermatol., 53, 798–802. [DOI] [PubMed] [Google Scholar]
- 20. Barnes D E, Tomkinson A E, Lehmann A R, Webster A D and Lindahl T (1992) Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell, 69, 495–503. [DOI] [PubMed] [Google Scholar]
- 21. Webster A D, Barnes D E, Arlett C F, Lehmann A R and Lindahl T (1992) Growth retardation and immunodeficiency in a patient with mutations in the DNA ligase I gene. Lancet, 339, 1508–1509. [DOI] [PubMed] [Google Scholar]
- 22. Teo I A, Arlett C F, Harcourt S A, Priestley A and Broughton B C (1983) Multiple hypersensitivity to mutagens in a cell strain (46BR) derived from a patient with immuno-deficiencies. Mutat. Res., 107, 371–386. [DOI] [PubMed] [Google Scholar]
- 23. Henderson L M, Arlett C F, Harcourt S A, Lehmann A R and Broughton B C (1985) Cells from immunodeficient patient (46BR) with a defect in DNA ligation are hypomutable but hypersensitive to the induction of sister chromatid exchanges. Proc. Natl. Acad. Sci. USA, 82, 2044–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prigent C, Satoh M S, Daly G, Barnes D E and Lindahl T (1994) Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol. Cell. Biol., 14, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teo I A, Broughton B C, Day R S, James M R, Karran P, Mayne L V and Lehmann A R (1983) A biochemical defect in the repair of alkylated DNA in cells from an immunodeficient patient (46BR). Carcinogenesis, 4, 559–564. [DOI] [PubMed] [Google Scholar]
- 26. Maffucci P, Chavez J, Jurkiw T J, et al. (2018) Biallelic mutations in DNA ligase 1 underlie a spectrum of immune deficiencies. J. Clin. Invest., 128, 5489–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tomkinson A E, Naila T and Bhandari S K (2019) Altered DNA ligase activity in human disease. Mutagenesis, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Showalter A K, Lamarche B J, Bakhtina M, Su M I, Tang K H and Tsai M D (2006) Mechanistic comparison of high-fidelity and error-prone DNA polymerases and ligases involved in DNA repair. Chem. Rev., 106, 340–360. [DOI] [PubMed] [Google Scholar]
- 29. Kunkel T A. (2009) Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol., 74, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ahn J, Kraynov V S, Zhong X, Werneburg B G and Tsai M D (1998) DNA polymerase beta: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochem. J., 331(Pt 1), 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamtich J and Sweasy J B (2010) DNA polymerase family X: function, structure, and cellular roles. Biochim. Biophys. Acta, 1804, 1136–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hsu G W, Ober M, Carell T and Beese L S (2004) Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature, 431, 217–221. [DOI] [PubMed] [Google Scholar]
- 33. Batra V K, Beard W A, Hou E W, Pedersen L C, Prasad R and Wilson S H (2010) Mutagenic conformation of 8-oxo-7,8-dihydro-2’-dGTP in the confines of a DNA polymerase active site. Nat. Struct. Mol. Biol., 17, 889–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cağlayan M, Batra V K, Sassa A, Prasad R and Wilson S H (2014) Role of polymerase β in complementing aprataxin deficiency during abasic-site base excision repair. Nat. Struct. Mol. Biol., 21, 497–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Çağlayan M, Horton J K, Prasad R and Wilson S H (2015) Complementation of aprataxin deficiency by base excision repair enzymes. Nucleic Acids Res., 43, 2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Çaglayan M, Prasad R, Krasich R, et al. (2017) Complementation of aprataxin deficiency by base excision repair enzymes in mitochondrial extracts. Nucleic Acids Res., 45, 10079–10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Çağlayan M and Wilson S H (2015) Oxidant and environmental toxicant-induced effects compromise DNA ligation during base excision DNA repair. DNA Repair (Amst)., 35, 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Çaglayan M and Wilson S H (2017) Role of DNA polymerase β oxidized nucleotide insertion in DNA ligation failure. J. Radiat. Res., 58, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Çağlayan M, Horton J K, Dai D P, Stefanick D F and Wilson S H (2017) Oxidized nucleotide insertion by pol β confounds ligation during base excision repair. Nat. Commun., 8, 14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Çağlayan M. (2020) The ligation of pol β mismatch insertion products governs the formation of promutagenic base excision DNA repair intermediates. Nucleic Acids Res., 48, 3708–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caglayan M and Wilson S H (2018) Pol μ dGTP mismatch insertion opposite T coupled with ligation reveals a promutagenic DNA intermediate during double strand break repair. Nat. Comm., 9, 4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Çağlayan M. (2020) Pol μ ribonucleotide insertion opposite 8-oxodG facilitates the ligation of premutagenic DNA repair intermediate. Sci. Rep., 10, 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr., 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 44. Tumbale P P, Jurkiw T J, Schellenberg M J, Riccio A A, O’Brien P J and Williams R S (2019) Two-tiered enforcement of high-fidelity DNA ligation. Nat. Commun., 10, 5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brieba L G, Eichman B F, Kokoska R J, Doublié S, Kunkel T A and Ellenberger T (2004) Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J., 23, 3452–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Freudenthal B D, Beard W A, Perera L, Shock D D, Kim T, Schlick T and Wilson S H (2015) Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature, 517, 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harrison C, Ketchen A M, Redhead N J, O’Sullivan M J and Melton D W (2002) Replication failure, genome instability, and increased cancer susceptibility in mice with a point mutation in the DNA ligase I gene. Cancer Res., 62, 4065–4074. [PubMed] [Google Scholar]
- 48. Bentley D J, Harrison C, Ketchen A M, Redhead N J, Samuel K, Waterfall M, Ansell J D and Melton D W (2002) DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J. Cell Sci., 115, 1551–1561. [DOI] [PubMed] [Google Scholar]
- 49. Sun D, Urrabaz R, Nguyen M, Marty J, Stringer S, Cruz E, Medina-Gundrum L and Weitman S (2001) Elevated expression of DNA ligase I in human cancers. Clin. Cancer Res., 7, 4143–4148. [PubMed] [Google Scholar]
- 50. Le Chalony C, Hoffschir F, Gauthier L R, Gross J, Biard D S, Boussin F D and Pennaneach V (2012) Partial complementation of a DNA ligase I deficiency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells. Cell. Mol. Life Sci., 69, 2933–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tomkinson A E, Howes T R and Wiest N E (2013) DNA ligases as therapeutic targets. Transl. Cancer Res., 2, 1219. [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong S, Chen X, Zhu X, et al. (2008) Identification and validation of human DNA ligase inhibitors using computer-aided drug design. J. Med. Chem., 51, 4553–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson G M, MacKerell A D Jr and Tomkinson A E (2008) Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res., 68, 3169–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Howes T R L, Sallmyr A, Brooks R, Greco G E, Jones D E, Matsumoto Y and Tomkinson A E (2017) Structure-activity relationships among DNA ligase inhibitors: characterization of a selective uncompetitive DNA ligase I inhibitor. DNA Repair (Amst)., 60, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jahagirdar D. (2018) Combinatorial use of DNA ligase inhibitor L189 and temozolomide potentiates cell growth arrest in HeLa. Curr. Cancer Ther. Rev., 14, 1–11. [Google Scholar]
- 56. Srivastava M, Nambiar M, Sharma S, et al. (2012) An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell, 151, 1474–1487. [DOI] [PubMed] [Google Scholar]
- 57. Kelley M R, Logsdon D and Fishel M L (2014) Targeting DNA repair pathways for cancer treatment: what’s new? Future Oncol., 10, 1215–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.