Abstract
The emergence and spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in significant global morbidity, mortality, and societal disruption. A better understanding of virus-host interactions may potentiate therapeutic insights toward limiting this infection. Here we investigated the dynamics of the systemic response to SARS-CoV-2 in hamsters by histological analysis and transcriptional profiling. Infection resulted in consistently high levels of virus in the upper and lower respiratory tracts and sporadic occurrence in other distal tissues. A longitudinal cohort revealed a wave of inflammation, including a type I interferon (IFN-I) response, that was evident in all tissues regardless of viral presence but was insufficient to prevent disease progression. Bolstering the antiviral response with intranasal administration of recombinant IFN-I reduced viral disease, prevented transmission, and lowered inflammation in vivo. This study defines the systemic host response to SARS-CoV-2 infection and supports use of intranasal IFN-I as an effective means of early treatment.
Keywords: COVID-19, IFN-I, mRNA-seq, pandemic, hamster, intranasal, therapeutic, prophylactic, cytokine, transcriptomics
Graphical abstract
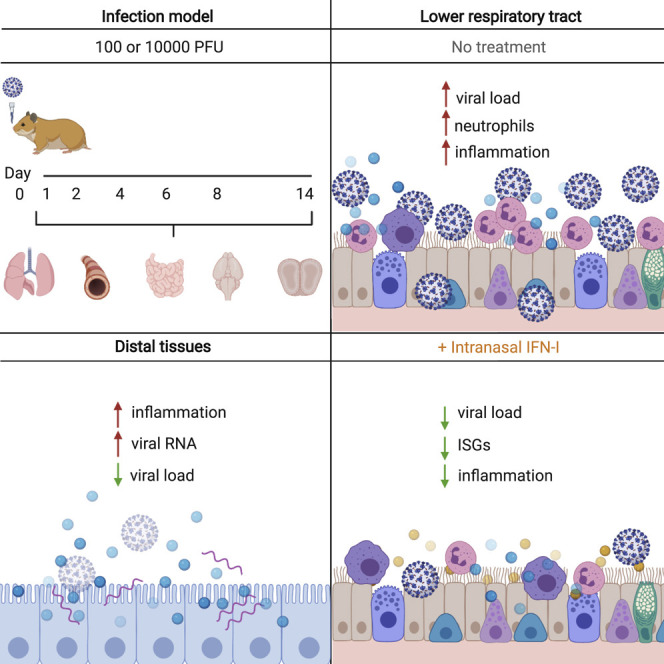
The host response to SARS-CoV-2 results in significant inflammation. To understand this biology, Hoagland et al. utilize infected hamsters to elucidate transcriptional footprints across tissues longitudinally, showing an inflammatory response beyond the site of acute replication. Local administration of IFN-I reduces virus load and improves immune infiltrate.
Introduction
The emergence and global spread of a virus is by no means a unique event in recorded history. Smallpox, measles, human immunodeficiency virus, and other viral pandemics have had profound consequences for global health and international economies (Eisinger and Fauci, 2018). Four major influenza A virus pandemics in the last century and seasonal epidemics have accounted for significant morbidity and mortality (Krammer et al., 2018). Countless numbers of emerging and re-emerging pathogens, including dengue, Ebola, Nipah, and Zika viruses, have caused global outbreaks as the climate changes and zoonotic incidence of transmission increases. As evidenced by the societal disruption caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), we remain inadequately prepared for these disease outbreaks despite the many advances of biomedical research.
All organisms have the capacity to respond to virus infection. CRISPR-based antiviral defenses of archaea and bacteria work through acquisition of viral genetic material by the host that is then utilized as a template to guide a nuclease to other copies of the virus genome, causing its degradation (Barrangou et al., 2007). Similarly, in plants, arthropods, and worms, Dicer proteins process double-stranded RNA from viruses into small genetic fragments that enable the RNA-induced silencing complex to bind and cleave virus-specific transcripts (Guo et al., 2019). In contrast to these sequence-specific genetic defenses, vertebrates utilize a relatively non-specific pathogen detection surveillance system as a first-line defense to inhibit virus replication (tenOever, 2016). In vertebrates, cellular recognition of virus infection culminates in transcriptional induction of a family of cytokines, type I and III interferons (IFN-I and IFN-III, respectively) that initiate a cascade of events that ultimately renders cells less permissive to virus infection (Lazear et al., 2019; Mesev et al., 2019; Park and Iwasaki, 2020).
Transcriptional induction of IFNs requires cellular recognition of the pathogen through engagement of pattern recognition receptors (PRRs) (Janeway and Medzhitov, 2002). Engagement of these PRRs results in activation of two families of transcription factors: nuclear factor κB (NF-κB) and the IFN regulatory factors (IRFs) (Maniatis et al., 1998). Co-activation of NF-κB and members of the IRF family (IRF1, IRF3, and IRF7) causes transcriptional activation of IFN beta (IFNB), an IFN-I, which is then transcribed, translated, and secreted, signaling in an autocrine and paracrine manner to invoke an antiviral state. IFN-I binding to its cognate cell surface receptor induces activation of a transcription factor complex called IFN-stimulated gene factor 3 (ISGF3), composed of STAT1, STAT2, and IRF9 (Stark et al., 2018). ISGF3 is responsible for enhanced expression of at least 150 genes that have direct antiviral effects (Schneider et al., 2014; Schoggins and Rice, 2011). This early IFN-I response limits virus replication and cell-to-cell spread and aids in coordinating the pathogen-specific adaptive immune response and development of immune memory. Viruses have co-evolved to specifically block the IFN-I response, precisely because of their critical roles in viral clearance (García-Sastre, 2017). This is evident for all viruses that cause significant morbidity, including smallpox, measles, influenza, mumps, rubella, and Ebola. Indeed, SARS-CoV-2, the etiological agent for the coronavirus disease 2019 (COVID-19) pandemic, encodes in its genome several IFN-I antagonists (Konno et al., 2020; Lei et al., 2020; Miorin et al., 2020; Thoms et al., 2020; Xia et al., 2020). An interesting feature of SARS-CoV-2 infection is that the virus modulates the immune response in a way that suppresses the IFN-I system but induces and sustains high levels of chemokine mRNAs, resulting in an imbalanced host response (Blanco-Melo et al., 2020; Qin et al., 2020; Zheng et al., 2020b). This juxtaposition of low IFN-I and high chemokines may be responsible for enhanced infiltration of neutrophils and monocytes to the respiratory tract, resulting in COVID-19 (Coperchini et al., 2020; Zheng et al., 2020a).
Here we sought to comprehensively characterize the host response to SARS-CoV-2 by tracking disease systemically from initial exposure to the virus to better understand the biology of COVID-19. SARS-CoV-2 infection resulted in a wave of inflammation that culminated in a systemic response that included distal organs, such as the brain and gastrointestinal (GI) tract. Lung pathology after SARS-CoV-2 infection in hamsters was similar to what has been described among people with COVID-19 and demonstrated lasting transcriptional changes that extended even after the virus had been cleared (Carfì et al., 2020; Weerahandi et al., 2020). Last, administration of intranasal IFN-I reduced the viral load and tissue damage, suggesting that this approach may be an effective early intervention for respiratory disease caused by SARS-CoV-2.
Results
SARS-CoV-2 drives an imbalanced antiviral response
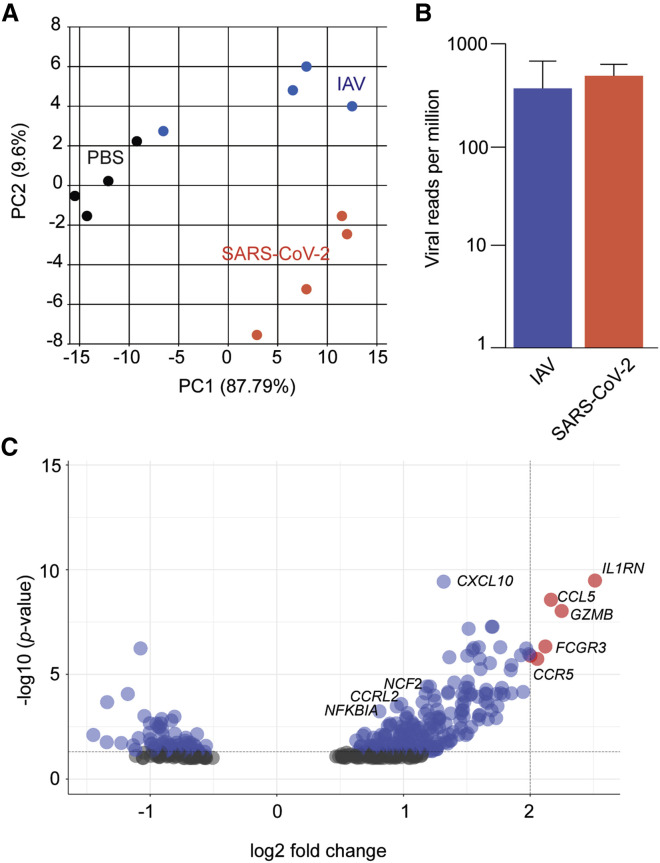
To assess the relative host response to SARS-CoV-2, we infected golden hamsters (Mesocricetus auratus) with the pandemic H1N1 influenza A virus (IAV) (A/California/04/2009) or a clinical isolate of SARS-CoV-2 (USA-WA1/2020). Five days after infection, total lungs were collected and analyzed by mRNA sequencing (mRNA-seq). Principal-component analysis demonstrated distinct and consistent transcriptional signatures between mock infection (PBS) and these two respiratory viruses (Figure 1 A). Aligning captured reads from each animal with the host and viral genomes demonstrated comparable viral loads of IAV and SARS-CoV-2 at the time of analysis, suggesting that any differences in the host response were due to virus biology (Figure 1B). To identify the unique aspects of the host response to SARS-CoV-2, we identified differentially expressed genes (DEGs), comparing IAV and SARS-CoV-2-infected cohorts, and assessed all host genes with a log2 fold change greater than 2 to identify enriched biological processes (Figure 1C; Table S1). SARS-CoV-2 induced a greater inflammatory signature than IAV, as exemplified by high mRNA levels of Ccl5, Gzmb, and Il1rn, consistent with what has been observed in people with COVID-19 and a ferret model (Blanco-Melo et al., 2020). Moreover, expression of the neutrophil markers Fcgr3, Ccrl2, and Cf2 were increased significantly in response to SARS-CoV-2-infected hamsters compared with IAV infection, consistent with what has been reported with COVID-19 autopsies (Table S1; Skendros et al., 2020).
Figure 1.
The transcriptional signatures from SARS-CoV-2- and IAV-infected hamster lungs differ
(A) Principal-component analysis of mRNA-seq samples derived from total lungs of hamsters (n = 4/cohort) treated with IN PBS, IAV (A/Cal/04/2009), or SARS-CoV-2 (USA-WA1/2020), collected 5 days after treatment, and aligned to virus and hamster reference genomes.
(B) Reads per million of SARS-CoV-2 and IAV from (A) relative to host mRNA. Error bars indicate standard error of the mean.
(C) Volcano plot comparing DEGs following SARS-CoV-2 versus IAV infection as described in (A) (gray, not significant; blue, significant; red, significant and log2 fold change > 2). Genes are called by their human ortholog.
See also Table S1.
When evaluating these data, it also became apparent that, like many immune-related genes, Ifnb1 was not annotated in the golden hamster genome. Given this, we next performed a de novo build on these mRNA-seq datasets and also a BLASTn search for conserved Ifnb1 genomic sequences shared by other rodents. We identified a transcript that was 85% similar to Ifnb1 at the nucleotide level with the Chinese hamster (Cricetulus griseus) (Figures S1A and S1B). At the protein level, this transcript was more closely related to Ifnb1 from the white-footed mouse (Peromyscus leucopus), showing a 65% identity match (Figure S1C). To ensure that this gene was functional, we cloned the cDNA, expressed it in golden hamster fibroblasts (BHK-21 cells), and demonstrated that transfection and transfer of supernatant resulted in robust induction of host IFN-stimulated genes (ISGs) in hamster- but not human-derived cell lines (Figures S1D and S1E). When aligning the Mesocricetus auratus IFN beta 1 transcript (MaIfnb1), we found no significant induction in SARS-CoV-2- or IAV-infected animals (Figure S1F). These data suggest that SARS-CoV-2 induces high chemokine expression in the context of a muted IFN-I response.
SARS-CoV-2 infection can be initiated by varying virus inocula and routes of entry
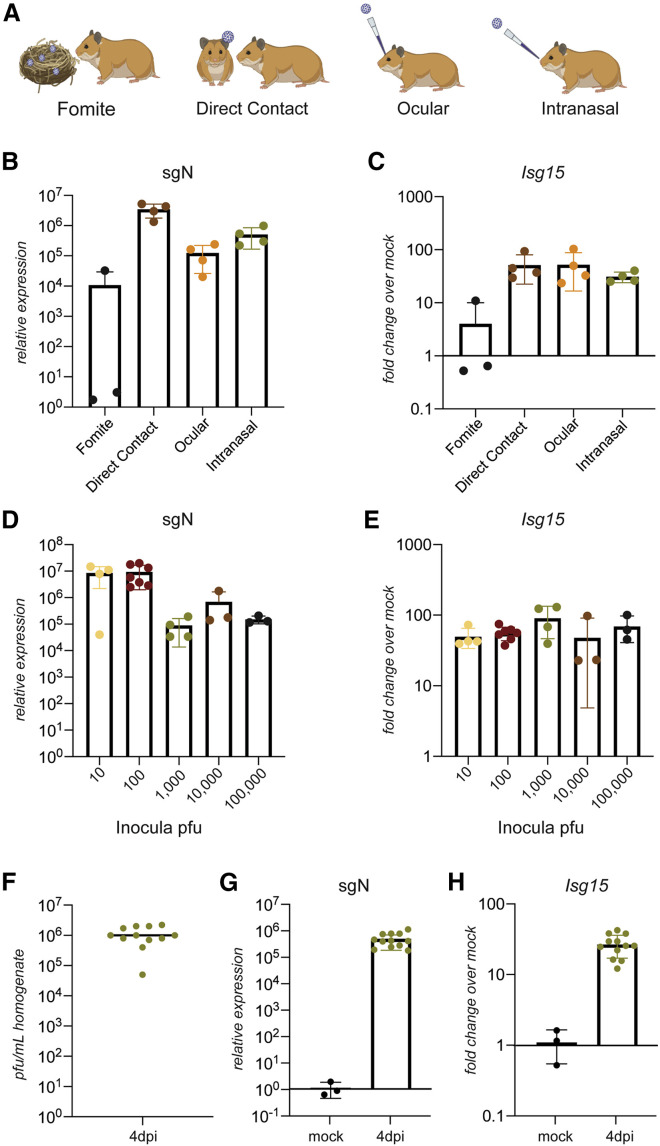
To examine how different routes of infection influence disease progression, we exposed naive hamsters to SARS-CoV-2-contaminated fomites, infected animals, or direct inoculation via the eye or nose (Figure 2 A). First, we verified that our viral stocks of SARS-CoV-2 maintained the furin cleavage site in the spike protein. Mutations at this site, which have been commonly demonstrated to occur during passaging in Vero E6 cells, have been associated with in vivo attenuation (Liu et al., 2020). SARS-CoV-2 levels from infected hamsters were initially measured by real-time qRT-PCR of the subgenomic nucleocapsid (N) transcript (sgN) in the lower respiratory tract. Fomite transmission was the least efficient means of infection, whereas direct inoculation and contact transmission resulted in consistent virus detection in the lungs on day 4 after infection (Figure 2B). Moreover, all transmission routes, including ocular infection, showed evidence of a host respiratory response with elevated levels of Isg15 and the pro-inflammatory chemokine C-X-C motif chemokine ligand 11 (Cxcl11) in the lungs (Figures 2C and S2A).
Figure 2.
SARS-CoV-2 infects the respiratory tract and activates the innate immune response in hamsters
(A) Tested modes of transmission: fomite transmission using dirty nesting from a cage of an infected hamster, direct contact with an infected individual hamster, and direct inoculation ocularly or IN.
(B and C) Nucleoprotein subgenomic RNA (sgRNA) (sgN) and (C) Isg15 real-time qRT-PCR of RNA extracted from hamster lungs after exposure to virus via fomites in dirty cages, direct contact with an infected hamster, eye drops (60,000 PFUs), or IN droplets (1,000 PFUs) of SARS-CoV-2, shown as fold change of transcript abundance compared with uninfected controls (n = 4; fomite transmission, n = 3).
(D and E) sgN and (E) Isg15 real-time qRT-PCR of RNA extracted from hamster lungs 2 days after infection with 10, 100, 1,000, 10,000, or 100,000 PFUs of SARS-CoV-2, shown as fold change of transcript abundance compared with uninfected controls (n = 4 for 10 and 1,000 PFUs, n = 6 for 100 PFUs, and n = 3 for 10,000 and 100,000 PFUs).
(F) Virus load determined by plaque assay from lung homogenates derived from lung tissue from golden hamsters infected IN with 1,000 PFUs collected 4 days after infection and homogenized in PBS (n = 12).
(G and H) sgN and (H) Isg15 real-time qRT-PCR of RNA extracted from pelleted lung homogenate from (F), shown as fold change of transcript abundance compared with uninfected controls.
Data are represented as mean, and the error bars indicate standard deviation. See also Figure S2.
Given the capacity to precisely control the inoculum and timing of an infection when the virus is administered intranasally, we chose this route of infection for all subsequent experiments. In contrast to the ferret model, where 105 plaque-forming units (PFUs) of SARS-CoV-2 were required to initiate a host response that was confined to the upper respiratory tract (Blanco-Melo et al., 2020), 10 PFUs were sufficient to establish a lower respiratory tract infection in hamsters (Figure 2D). Despite inducing a similar host response, as measured by Isg15 and Irf7, increasing the inoculum doses to as high as 105 PFUs not only failed to increase overall replication but tended to result in lower levels of sgN and nsp14 (Figures 2D, 2E, S2B, and S2C). To determine the reliability of intranasal infection in this outbred animal model, we infected 12 hamsters and found that lung viral titers, as measured by plaque assay, were consistently 106 PFUs 4 days after infection, with just a single outlier (Figure 2F). Reproducibility was corroborated further by real-time qRT-PCR for sgN and a concomitant host response, as measured by Isg15 mRNA (Figures 2G and 2H).
Progressive SARS-CoV-2 infection in hamsters results in lung pathology
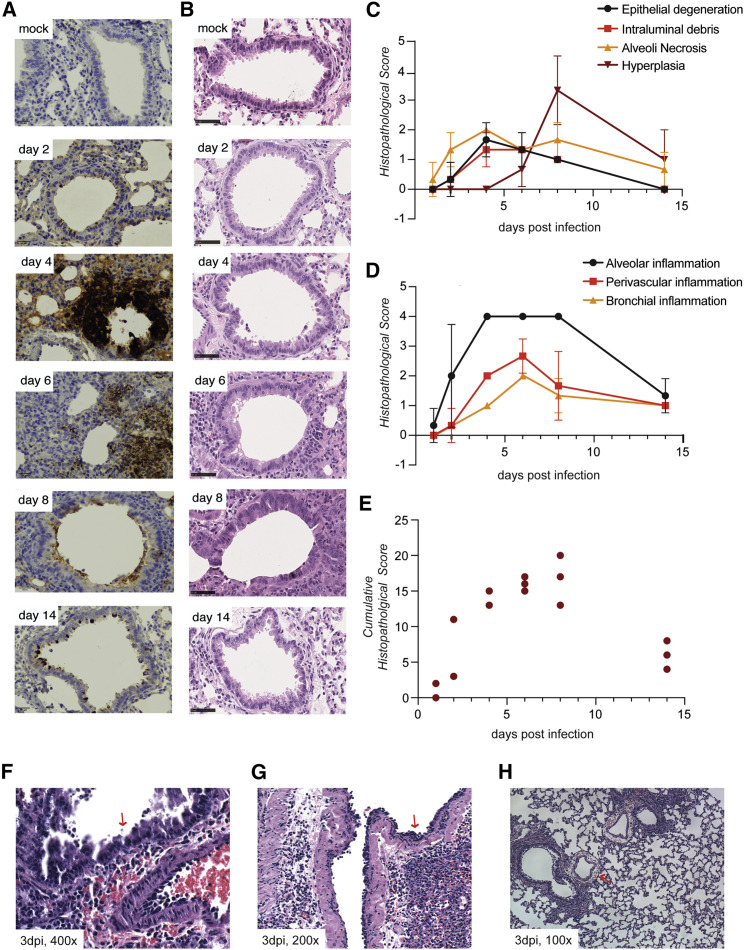
To minimize any confounding factors that may arise with a non-physiological viral challenge, we assessed lung pathology in response to inoculation with 100 PFUs. Lungs were harvested on days 2, 4, 6, 8, and 14 after infection, and tissue sections were stained with an antibody for viral N and hematoxylin and eosin (H&E) (Figures 3A and 3B). The most robust N protein expression appeared 4 days after infection and occurred predominantly in epithelial cells comprising the air interface of the lung (Figure 3A). Despite N protein levels diminishing dramatically beyond 4 days after infection, viral protein was still evident at 14 days, consistent with pathological assessment of the H&E-stained tissue sections. In agreement with earlier reports, epithelial degeneration in bronchioles and alveolar necrosis were apparent, as well as type II pneumocyte hyperplasia and cytopathy, peaking 4 and 8 days after infection, respectively (Figure 3C; Imai et al., 2020). There was also evidence of alveolar, perivascular, and bronchial inflammation, peaking between 4 and 8 days after infection (Figures 3D and S3A). Inflammation, as defined by cumulative histopathological score, was maximal on day 8 after infection (Figure 3E; Table S2). Moreover, atypical adenomatous hyperplasia of alveoli was observed 14 days after infection, even after the majority of inflammation had subsided (Figures 3B and S3B; Table S2). Further analysis of H&E staining of lung tissue on day 3 after infection revealed several distinct phenomena. We observed apoptosis in the bronchial epithelium as well as nested accumulations of neutrophils in the bronchioles of infected hamsters (Figures 3F and 3G). Furthermore, severe vascular edema was observed, even in sites of the lung with relatively little inflammation (Figure 3H). These data highlight the value of this small animal model, in which an unmodified clinical strain of SARS-CoV-2 causes progressive infection in the lower respiratory tract, ultimately resulting in severe pneumonia as observed in people with COVID-19.
Figure 3.
SARS-CoV-2 infection results in severe lower respiratory tract pathology
(A and B) Representative images of golden hamster lung tissue collected 2, 4, 6, 8, and 14 days after infection (100 PFUs) and immunohistochemistry with (A) N-specific antibody and (B) H&E (scale bar, 50 μm, n = 3).
(C and D) Average values of histopathological scores, assessed by a certified pathologist for each of the observations, corresponding to the time points in (A) and (B) (n = 3). Error bars indicate standard deviation.
(E) Cumulative clinical scores for nine different histopathological assessments (Table S2). Each point represents the score for one animal.
(F and G) Representative images of (F) apoptosis in the bronchial epithelium (400× magnification), (G) accumulation of neutrophils (200× magnification), and (H) severe vascular edema (100× magnification) in hamster lungs collected 3 days after SARS-CoV-2 infection (100 PFU).
The host IFN-I response to SARS-CoV-2 infection is insufficient to prevent spread to the lower respiratory tract
We next examined the transcriptional response in the respiratory tract of hamsters infected with 100 or 10,000 PFUs, respectively, over the same time course during which we had observed progressive lung pathology. Total RNA from lungs was assayed by real-time qRT-PCR for sgN and host Isg15 levels on the indicated days after infection (Figures S4A and S4B). In contrast to the rise, plateau, and clearance of subgenomic viral replication, which was maximal between days 2 and 4 after infection following challenge with 100 PFUs, a 10,000-PFU challenge immediately resulted in high RNA levels (Figure S4A). This was followed by a general decline of viral transcripts through day 6 after infection (Figure S4A). Furthermore, there was an increase and decline of the host response to infection with the lower-dose inoculum of virus, as measured by Isg15 levels, that mirrored the kinetics of viral replication. For the higher-dose virus inoculum, Isg15 levels remained elevated through day 8 after infection, after viral RNA was largely cleared (Figure S4B). These data suggest that the amount of viral inoculum can influence the host response to infection and should be considered when analyzing data that are extrapolated to human disease.
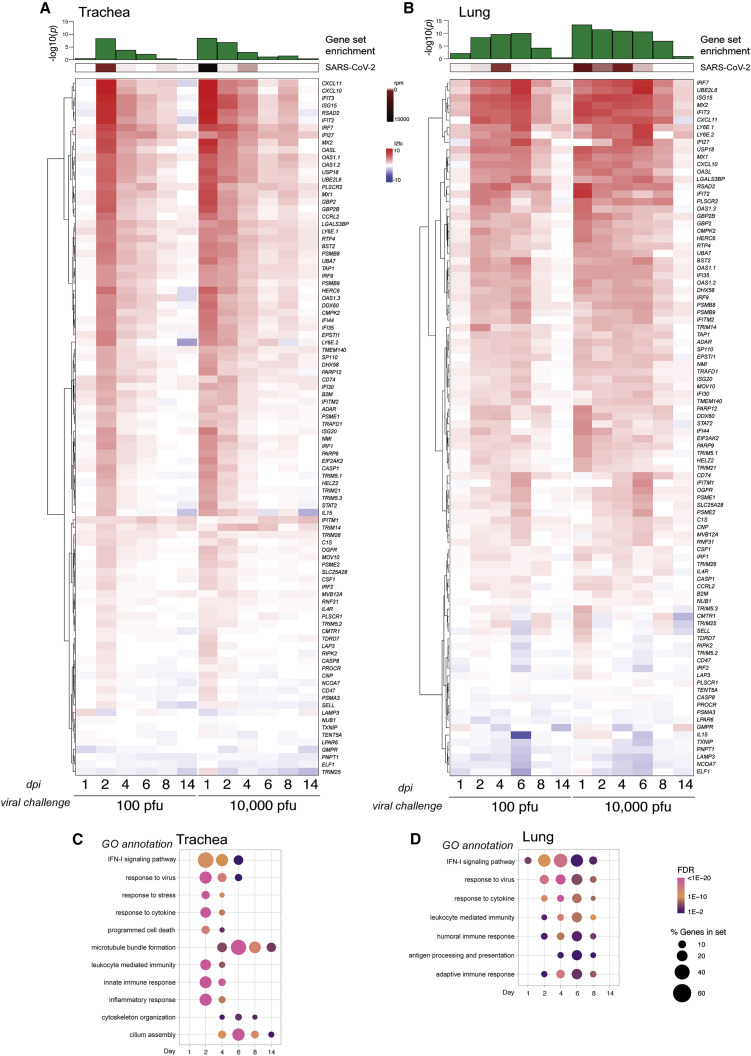
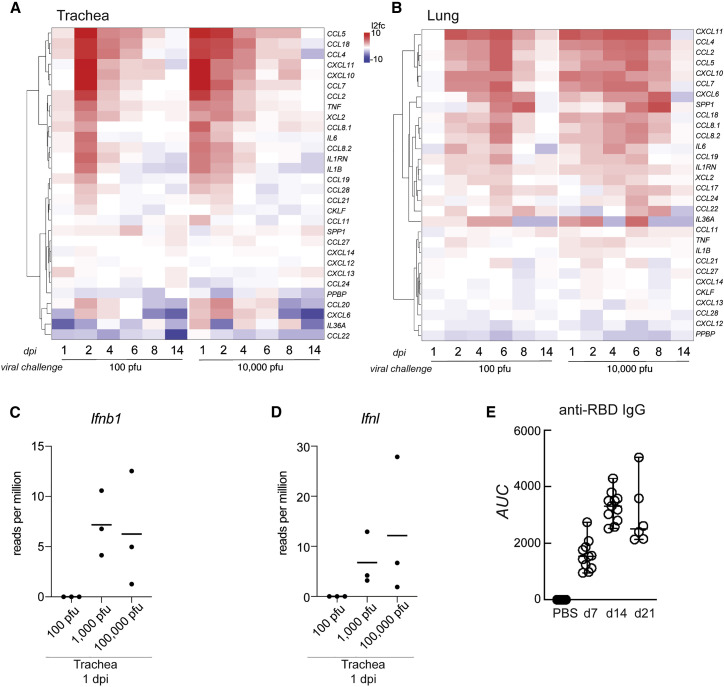
To further characterize the full antiviral response to infection, we performed mRNA-seq on a longitudinal study and assessed the levels of a curated list of ISGs in the trachea and lungs on the indicated days with the same low and high viral challenge doses (Figures 4 A, 4B, and S4C). The host response to SARS-CoV-2 correlated with viral load, as defined by virus reads per million, in the trachea (Figure 4A). However, despite elevated transcription of many ISGs, the host response was insufficient to contain virus replication in the upper respiratory tract, culminating in a high viral load in the lower respiratory tract on day 4 after infection (Figure 4B). This response to the virus was different when the viral challenge was increased by 2 orders of magnitude, where we observed no delay in ISG production in the trachea or the lungs (Figures 4A and 4B). Regardless, the host response was unable to restrict virus propagation. These data also demonstrate that, irrespective of inoculum dose, SARS-CoV-2 infection appears to limit induction of many of the classical ISGs, in agreement with different studies (Konno et al., 2020; Lei et al., 2020; Lucas et al., 2020; Thoms et al., 2020; Miorin et al., 2020).
Figure 4.
The upper and lower respiratory tract IFN-I transcriptional signatures
(A and B) Differential gene expression of a curated list of ISGs calculated from bulk mRNA-seq from (A) tracheae and (B) lungs of hamsters infected with SARS-CoV-2 for the times and inocula indicated compared with uninfected controls. The heatmaps represent the log2 fold change of each gene indicated on the right (human ortholog) at the time points below. Averages of reads per million (rpm) mapping to the SARS-CoV-2 genome in both tissues throughout the time course are shown above each heatmap (in brown), and the statistical significance of the enrichment of the gene list in the total number of DEGs (in green) is shown as −log10(adjusted p value) (control group, n = 5; each experimental group, n = 3).
(C and D) Gene enrichment analysis of all DEGs (log2 fold change > 1, false discovery rate [FDR] < 0.05) increased during infection for (C) the trachea and (D) the lungs, as depicted for the time course of infection with 100 PFU. Each dot indicates a statistically significant representation of the category of genes listed on the left at each time point. The size of the dot indicates the percentage of enriched genes from each GO annotation set, and the color represents its FDR.
See also Figure S4.
In addition to inducing an IFN-I response, infected cells activate other transcriptional programs aimed at reducing viral load. DEGs from the trachea and lungs were enriched for transcripts involved in an inflammatory response, which included recruitment of the adaptive immune system, as determined by enrichment of relevant Gene Ontology (GO) annotations (Figures 4C and 4D; Table S3). In the trachea, induction of genes associated with a cellular response to a virus, to stress, and cytokines mirrored virus levels, generally peaking 2–6 days after infection and dissipating thereafter (Figure 4C). Gene enrichment analyses suggested significant changes to cellular composition, as denoted by induction of transcripts involved in cilium assembly, microtubule bundle formation, and cytoskeleton organization, consistent with the H&E staining, which suggested that SARS-CoV-2 may cause loss of cilia or ciliated cells (Figures 4C and S4D). Similar gene-enriched pathways were observed in infected lungs, although these tended to be sustained for the first 8 days of disease progression (Figures 4B and 4D). By day 14, SARS-CoV-2 infection in the trachea and the lungs was resolved, as measured by lack of DEGs beyond regeneration of cilia (Figures 4C and 4D; Table S3). These findings outline the transcriptional response to disease progression in SARS-CoV-2-infected hamsters and may be used as a resource to better understand the molecular biology underlying the antiviral response in people with COVID-19.
Inflammation in response to SARS-CoV-2 follows infection from the upper to the lower respiratory tract
SARS-CoV-2 infection is capable of disrupting aspects of IFN-I biology but can also lead to hyperinflammation (Manson et al., 2020) Given this, we generated a curated list of inflammatory cytokines and compared the same transcriptional datasets from our longitudinal study (Figures 5A and 5B). Consistent with our earlier data, infection initiated with a high inoculum dose resulted in rapid induction of host inflammatory cytokines, in contrast to the response accompanying a low-PFU challenge. Furthermore, the peak of the host response in the trachea preceded that observed in the lungs by approximately 4 days (Figures 5A and 5B). For example, the most robust induction of host genes in response to SARS-CoV-2 was chemokines, which are responsible for orchestrating lung immune cell infiltration and the respiratory distress that is the hallmark of COVID-19 (Tay et al., 2020; Vabret et al., 2020). In response to a viral challenge of 100 PFU, the levels of Ccl4 and Ccl5 peaked on day 2 in the trachea, in contrast to the lungs, where peak levels were not achieved until day 6 (Figures 5A, 5B, and S5A). The potent induction of these chemokines, together with Cxcl10 and Cxc11, suggested recruitment of activated T cells and macrophages to the respiratory tract. This was supported by an observed increase in Xcl2 transcripts that likely originated from T cells or natural killer (NK) cells in the tissue in addition to increased expression of Cd8a, Cd4, and Cd3g (Figures 5A, 5B, S5B, and S5C). Furthermore, attraction of macrophages was suggested by increased expression of Ccl2, Ccl7, Ccl8, and Itgam (Figure S5C). In addition, cytokine and chemokine expression levels were not always equivalent in the upper and lower respiratory tract. For example, Tnf and Il1b were induced to high levels in the upper respiratory tract but generally absent in the lower airways. In contrast, Il36a, Cxcl6, and Ccl22 reached higher levels of expression in the lungs compared with the trachea (Figures 5A and 5B). These differences likely reflect the heterogenous composition of these tissues and differences in immune cell subsets that were recruited to these sites, given that overall viral levels were comparable in these airway compartments at different times during infection.
Figure 5.
The chemokine expression profile of the upper and lower respiratory tract in infected golden hamsters
(A and B) Differential gene expression of a curated list of chemokines and cytokines calculated from bulk mRNA-seq from (A) the trachea and (B) the lungs of hamsters infected with SARS-CoV-2 for the times and dosages indicated compared with uninfected controls. The heatmaps represent the log2 fold change of each gene indicated on the right (human ortholog) at the time points below (control group, n = 5; each experimental group, n = 3).
(C and D) Mean number of rpm from RNA-seq mapping to (C) Ifnb or (D) Ifnl in the trachea 1 day after infection (n = 3).
(E) Mean IgG antibody titers specific to the spike protein in 9- to 10-week-old hamsters and controls (n = 8) 7 (n = 11), 14 (n = 11), and 21 (n = 6) days after infection, measured by ELISA and depicted as area under the curve (AUC).
Error bars indicate standard deviation. See also Figure S5.
Aligning these longitudinal RNA-seq data with Ifnl and Ifnb1 revealed that, at a high virus inoculation dose, detectable levels of both IFNs were only present in the trachea 1 day after infection, with no reads for either transcript in the lungs or any other tissue at any time after infection (Figures 5C and 5D). Comparable with the ISG profile, cytokine expression also decreased by day 14, which correlated with the peak of virus spike protein-specific immunoglobulin G (IgG) antibodies in serum, shown previously to elicit protection against re-infection (Figures 5A, 5B, and 5E; Imai et al., 2020; Tostanoski et al., 2020).
SARS-CoV-2 infection causes systemic inflammation in distal tissues
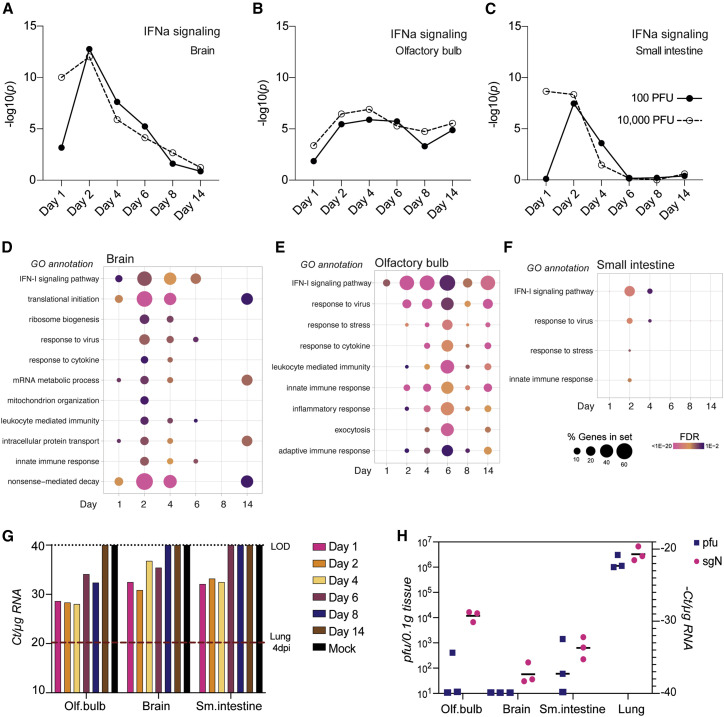
Given the extensive viral replication and consequent host response to SARS-CoV-2 infection observed in hamsters, we next examined tissues distal to the respiratory tract. Specifically, we examined SARS-CoV-2 infection in the olfactory bulb, the four lobes of the brain (the frontal, parietal, temporal, and occipital lobes, here denoted simply as “brain”), and the small intestine but found inconsistent evidence of virus replication, as measured by viral reads per million, which were orders of magnitude lower than in the respiratory tract (Figures 4 and S6A). However, despite the low levels of viral reads in tissues outside of the lungs, a strong antiviral transcriptional response was evident, as measured by enrichment score for IFN-I signaling (Figures 6 A–6C). In brain tissue, the proportion of DEGs was highest 2 days after infection, which correlated with the peak viral load observed in the upper respiratory tract (Figures 6D and S6B). Genes associated with response to virus, response to cytokines, IFN signaling, and innate immunity appeared prior to peak differential gene expression in the lower respiratory tract, suggesting that inflammation, or possibly infection (led by chemokine spread or virus), spread bi-directionally from the trachea. Additional gene signatures in the brain included those associated with translation initiation, nonsense-mediated decay, and mRNA metabolism, suggesting a distinct transcriptional response that, in some cases, was sustained for the duration of the experiment. Consistent with evidence of inflammation in the absence of consistent infection, we observed the most sustained IFN response in the olfactory bulb (Figures 6B, 6E, and S6B). In addition to classical ISGs, gene signatures for a stress response and an adaptive immune response were evident, which may account for the acute olfactory impairment (anosmia) reported in mild and severe cases of COVID-19 in humans and hamsters (Cantuti-Castelvetri et al., 2020; Carignan et al., 2020; Imai et al., 2020). The most modest inflammatory response in the assessed tissues was in the small intestine, where a mild antiviral gene signature was observed that correlated with the time of peak viral load in the respiratory tract; similar etiologies have been reported in a subset of people with COVID-19 (Yang and Tu, 2020).
Figure 6.
Sites distal to direct SARS-CoV-2 replication display significant inflammation
(A–C) The statistical significance of the enrichment of a curated list of ISGs among the total number of DEGs is shown as –log10(adjusted p value) above each time point from transcriptome analysis of total RNA extracted from (A) the brain, (B) the olfactory bulb, or (C) the small intestine (all groups n = 3; small intestine 10,000 PFUs day 8, n = 2).
(D and E) Gene enrichment analysis of all DEGs (log2 fold change > 1, FDR < 0.05) that increased during infection in (D) the brain, (E) the olfactory bulb, and (F) the small intestine are depicted for the time course of infection with 100 PFU. Each dot indicates a statistically significant representation of the category of genes listed on the left at each time point. The size of the dot indicates the percentage of enriched genes from each GO annotation set, and the color represents its FDR (n = 3).
(G) sgN levels represented by real-time qRT-PCR Ct score per 1 μg RNA from the olfactory bulb, brain, and small intestine at the indicated time points after infection with 100 PFUs SARS-CoV-2. RNA samples of each organ and corresponding time point were pooled before analysis (n = 3 per condition). The red dotted line indicates the Ct value of a sample corresponding to lungs collected from a hamster 4 days after infection.
(H) sgN levels represented by real-time qRT-PCR (−)Ct score per 1 μg RNA and PFUs per 100 mg of analyzed tissue homogenate (n = 3 per condition). Horizontal bars indicate mean.
Because many of the transcriptional signatures observed would require detection of pathogen-associated molecular patterns (PAMPs), we next performed more sensitive assays to determine whether low levels of virus replication could be detected, as reported by others (Imai et al., 2020; Tostanoski et al., 2020). We subjected the same RNA from the mRNA-seq dataset to real-time qRT-PCR for sgN. Using this assay, we were able to amplify a PCR product after fewer than 40 cycles in olfactory bulb, brain, and small intestine on days 1, 2, and 4 after infection (Figure 6G). Moreover, sgN transcripts could be detected as far out as 8 days after infection in the olfactory bulb, suggesting the presence of replicating virus, albeit at 1/200th of the level of the lungs 4 days after infection. To corroborate these results, we assessed each aforementioned tissue for infectious particles by plaque assay of tissue homogenate 3 days after infection. We found that sgN transcript levels did not consistently correlate with infectious particles at the level of detection allowed by the plaque assay (Figure 6H). For example, in the olfactory bulb, ∼400 PFUs per 0.1 mg of tissue were detected only in a single animal, but sgN remained consistent at a cycle threshold (Ct) score of ∼28 (Figure 6H). Together with the apoptosis observed in the bronchial epithelium and vascular edema in the lungs of infected hamsters, these data suggest that dissemination of virus-derived PAMPs may be responsible for the systemic inflammation observed across tissues (Figures 3F, 3H, and 6). Further studies will be needed to substantiate these results, but if these data accurately reflect the inconsistencies of tissue tropism and inflammation in people with COVID-19, then they could point to the underpinnings of diverse clinical manifestations.
Intranasal administration of IFN-I limits viral replication and inflammation
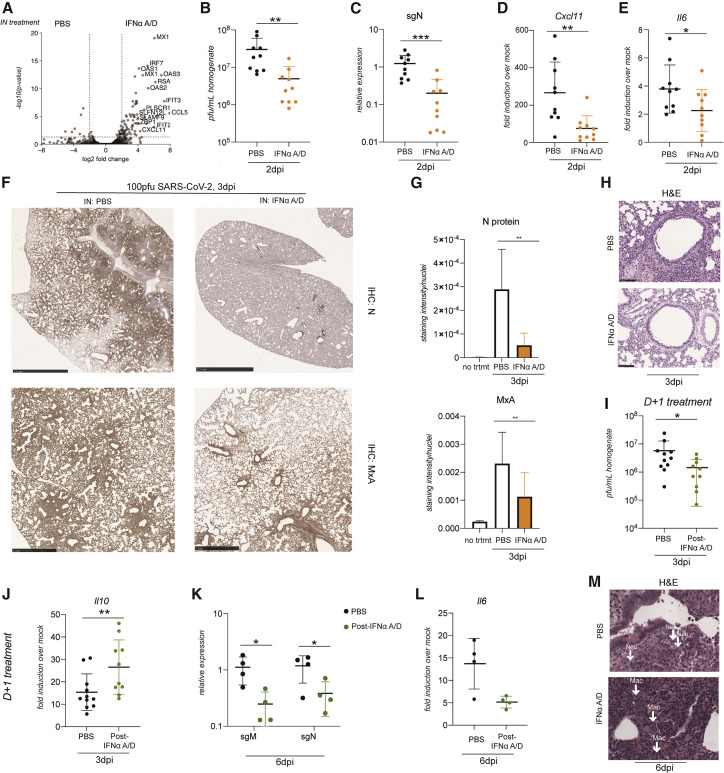
The elevated ISGs observed across all investigated tissues may reflect the capacity of IFN-I to limit tissue tropism. To determine whether IFN-I could provide a prophylactic benefit against COVID-19, we decided to test it intranasally in our hamster model. We first evaluated murine and human IFN beta alongside commercially available universal IFN (IFNα A/D) in the BHK-21 hamster cell line and found IFNα A/D to be the most potent (Figure S7A). To examine antiviral efficacy in vivo, we administered IFNα A/D to hamsters intranasally and assessed the transcriptional response in the respiratory tract by mRNA-seq and further corroborated these data by real-time qRT-PCR (Figures 7 A and S7B). A robust IFN-I response was induced, comprised of canonical ISGs, including Isg15, Mx1-2, Ifit2-3, Cxcl10-11, and Oas1-3, among others (Table S4). To ascertain whether this response was sufficient to significantly reduce the viral load in vivo, we challenged hamsters with SARS-CoV-2 24 h after intranasal (IN) PBS or IFNα A/D administration and continued daily IN treatment for the duration of the infection. We evaluated IFNα A/D- and PBS-treated animals for viral load, as measured by plaque assay, 48 h after virus challenge (Figure 7B). The viral load was reduced in hamsters receiving IFNα A/D, with a log-fold decrease in infectious virions that was also reflected in sgN RNA levels (Figure 7C). In addition to a log decrease in viral load, prophylactic IFNα A/D also reduced Cxcl11 and Il6 mRNA levels in the lungs (Figures 7D and 7E). Moreover, we performed immunohistochemistry (IHC) for SARS-CoV-2 N protein and MxA in lungs 3 days after infection, which were either treated IN with PBS or IFNα A/D daily starting 24 h before infection and quantified protein expression (see STAR methods for details; Figures 7F and 7G). N and MxA protein expression was reduced significantly. This not only corroborated the decreased viral load in hamsters treated IN with IFNα A/D but also suggested that administration of IFN-I can ultimately lower the magnitude of the innate immune response in the golden hamster model. Additionally, these lungs were also H&E stained, which revealed less inflammation in IFNα A/D-treated hamsters (Figures 7H and S7C).
Figure 7.
IFN administration reduces the viral load and lung pathology in hamsters
(A) Volcano plot showing DEGs from transcriptome analysis of RNA extracted from lungs of golden hamsters treated with 200,000 units of IN IFNα A/D for 8 h compared with mock treatment. Each dot represents a gene plotted by its log2 fold change on the x axis and the statistical significance on the y axis as −log10(p value) (n = 3 per condition). All genes with a log2 fold change of more than 4 and p value of less than 0.05 are labeled with the name of the human ortholog.
(B) Plaque assay of lung homogenate from golden hamster lungs infected with 100 PFUs of SARS-CoV-2 harvested 2 days after infection after daily IN treatment with PBS or 200,000 units of IFNα A/D starting 24 h prior to infection. n = 10 per condition from two independent experiments, p = 0.007.
(C) sgN relative expression levels by real-time qRT-PCR from lungs of hamsters infected with 100 PFUs of SARS-CoV-2 and treated IN starting 24 h before infection with PBS or IFNα A/D and collected 3 days after infection. n = 10 per condition from two independent experiments, p = 0.0007.
(D) Real-time qRT-PCR for fold induction of Cxcl11 mRNA from corresponding hamster lungs compared with mock-infected hamster lungs (n = 3). n = 10 from two independent experiments, p = 0.0039.
(E) Real-time qRT-PCR for fold induction of Il6 mRNA from corresponding hamster lungs compared with mock-infected hamster lungs (n = 3). n = 10 per condition from two independent experiments, p = 0.0476.
(F) Representative images of immunohistochemistry staining with N-specific antibody and MxA in lungs 3 days after infection with IN PBS or IFNα A/D, administered beginning 24 h before infection. N-protein scale bar, 2.5 mm; MxA scale bar, 1 mm.
(G) Protein expression quantification using ColorDeconvolution2 in ImageJ (see STAR methods for details) For N protein, n = 8 total representative images from 2 animals per treatment group, p = 0.0011; for MxA, n = 12 total representative images from 3 animals per treatment group, p = 0.0081.
(H) Representative images of H&E stains from perfused hamster lungs harvested 3 days after infection with 100 PFUs of SARS-CoV-2 and daily IN treatment with PBS or 200,000 units of IFNα A/D. Scale bar, 100 μm. The IFNα A/D representative image is of a lung in which corresponding sgN levels were lowered a log compared with PBS- and SARS-CoV-2-infected controls.
(I and J) Plaque assay and (J) real-time qRT-PCR for Il10 of lung homogenates from golden hamsters infected with 100 PFUs of SARS-CoV-2 and treated IN starting 24 h after infection with PBS or IFNα A/D and collected 3 days after infection. n = 10 per treatment group from 2 independent experiments. p = 0.0186 (I) and p = 0.0213 (J).
(K) Real-time qRT-PCR for sgM and sgN from lungs of hamsters infected with SARS-CoV-2, subsequently treated daily with IN PBS or IFNα A/D, and harvested 6 days after infection, n = 4 per treatment group; sgM, p = 0.0137; sgN, p = 0.0237.
(L) Real-time qRT-PCR for Il6 from lungs of hamsters infected with SARS-CoV-2, subsequently treated daily with IN PBS or IFNα A/D, and harvested 6 days after infection, represented as fold induction over baseline expression in mock-infected golden hamsters; n = 4 per treatment group.
(M) Representative images of H&E stains of infiltrates from lungs of hamsters infected with SARS-CoV-2, subsequently treated daily with IN PBS or IFNα A/D, and harvested 6 days after infection; 400× magnification.
“Neu” and “Mac” indicate a neutrophil or macrophage, respectively. Data are represented as mean ± standard deviation. See STAR methods for statistical tests utilized.
To determine whether IN IFNα A/D could provide a therapeutic benefit to improve disease outcome in this model, we treated animals with IFNα A/D daily, beginning 24 h after infection with SARS-CoV-2, and collected the lungs 3 and 6 days after challenge. Three days after infection, there were fewer infectious virions in IFNα A/D- compared with PBS-treated lungs, in addition to elevated levels of the anti-inflammatory cytokine interleukin-10 (Il10) (Figures 7I and 7J). Six days after infection, there were no infectious virions in the lungs in either treatment group, as measured by plaque assay, but there was less sgN and subgenomic membrane (sgM) viral RNA as well as lower Il6 mRNA levels (Figures 7K and 7L). Furthermore, analysis of H&E staining of the lungs 6 days after infection revealed that lungs treated therapeutically with IFNα A/D had an immune cell infiltrate composed more heavily of reactive macrophages and fewer neutrophils compared with their controls; increased mitosis was also observed in the bronchial epithelium, suggesting that these hamsters were closer to disease resolution (Figure 7M). In conjunction with increased Il10 expression on day 3 and decreased interleukin-6 expression on day 6, these data suggest that introduction of exogenous IN IFN-I results in a modified immune cell infiltrate, resulting in a lower viral load.
To further ascertain the prophylactic value of IFNα A/D, we applied it to a more clinically relevant transmission model where treated animals were exposed by direct contact rather than being infected IN. Animals were treated with PBS or IFNα A/D and co-caged with infected animals for 10 h (Figure S7E). In agreement with our direct infection model, IFNα A/D, given prophylactically in this transmission model, resulted in a greater than 2-log reduction of viral titers in infected animals and prevented transmission altogether in 3 of the 5 animals (Figure S7E). The latter results were further corroborated by IHC of MxA and N protein, showing IFNα A/D-mediated induction of ISGs and limited viral expression (Figures S7F and S7G). This was exemplified by the fact that overlapping low levels of MxA with N protein were present in PBS-treated animals but absent in IFNα A/D-treated hamsters (where MxA was prevalent in the absence of N staining) (Figures 7F and 7G). These data could be further corroborated by measuring the levels of sgN mRNA from total lungs (Figure S7H).
Because treatment with IFN-I may be prohibitive for various economic and geographical reasons, we next examined whether a comparable antiviral response could be achieved by IN administration of a PAMP that would induce endogenous IFN-I. The advantages of using a PAMP, such as a mimetic for double-stranded RNA (dsRNA) (poly(I:C)), include ease of manufacturing and storage as well as affordability. IN poly(I:C) administration showed antiviral activity comparable with IFNα A/D (Figure S7I).
Discussion
In response to the COVID-19 pandemic, there have been unprecedented efforts to develop therapeutic interventions in the sphere of vaccines and antivirals. Different strategies have been employed to target the SARS-CoV-2 virus, including direct inhibition of virus replication by targeting the RNA-dependent RNA polymerase, protease, and S protein and/or blocking interactions with the ACE2 entry receptor and TMPRSS2 in conjunction with further defining the intricacies of these interactions (Dieterle et al., 2020; Douangamath et al., 2020; Han et al., 2020; Sorokina et al., 2020; Tay et al., 2020). In addition, efforts are underway to repurpose US Food and Drug Administration (FDA)-approved drugs that may have therapeutic value in vivo (De Clercq and Li, 2016; Si et al., 2020). To advance general countermeasures against viral outbreaks, our strategy has been to develop broad-spectrum antiviral agents rather than pathogen-specific interventions for each emerging and re-emerging global virus outbreak. Our approach focuses on exploiting the host response to combat virus infections; namely, enhancement of the IFN-I response.
We found that the host response to SARS-CoV-2 in hamsters, although insufficient to protect against severe bronchopneumonia induced by the virus, is characterized by active IFN-I signaling. Other studies have identified a similar transcriptional response in human ACE2 (hACE2)-transgenic mice and in mice transduced with hACE2-encoding adenovirus-based vectors or by using a mouse-adapted strain of SARS-CoV-2 (Dinnon et al., 2020; Israelow et al., 2020; Sun et al., 2020; Winkler et al., 2020). Additionally, we are able to show that this response extends beyond the respiratory tract in several organs distal to the site of infection for up to 14 days. Although it has been suggested that imbalanced IFN-I is in part responsible for the severe inflammation and aberrant neutrophil infiltration, IFN-I is also necessary to control virus titers in hamsters and mice (Boudewijns et al., 2020; Sun et al., 2020). Given this effect of the IFN-I response to limit viral replication, we decided to leverage recombinant IFN-I as a prophylactic and therapeutic agent against SARS-CoV-2. IFN-I and/or IFN-III are ideal candidates for inhibiting viral infection. In addition to their pleiotropic effects in directly targeting multiple stages of the virus replication cycle, IFNs also activate an appropriate immune response to clear a virus, regardless of the pathogen (Wang and Fish, 2019). It has been demonstrated recently that IFN-I signaling, mediated by STAT2, can limit virus replication and dissemination in the hamster model, whereas IFN-III signaling appeared to be less important. Moreover, it has also been demonstrated clinically that the presence of auto-antibodies against IFNs and inborn errors of innate immunity, such as deficiencies in TLR3 and IRF7, are enriched in people with severe COVID-19 pneumonia, suggesting the potential value of exogenous IFN treatment in certain individuals whose own IFN response is impaired or obstructed (Bastard et al., 2020; Zhang et al., 2020). However, administering IFN-I systemically has proven to have very low concordance because it can cause unwanted side effects. This dynamic likely contributed to the inability of IFN-I to reduce mortality in the Solidarity Trial for COVID-19 Treatments (SOLIDARITY) (Dinnon et al., 2020; Pan et al., 2020). To avoid these potential adverse effects, we administered IFN-I locally to the respiratory tract and achieved a 2- to 3-log reduction in viral burden and kept the virus below the limit of detection in certain experimental settings. Furthermore, a recent phase 2 clinical trial of inhaled, nebulized IFN beta-1a to treat people with COVID-19 resulted in decreased time to recovery (Monk et al., 2020). This supports IN delivery of IFN-I as a potential prophylactic and therapeutic agent against COVID-19.
The outcomes of SARS-CoV-2 are heterogeneous, with the greatest dichotomy observed between young people and those of advanced age (Williamson et al., 2020). Although this disparity certainly has multiple underlying factors, numerous reports have implicated misfiring of the antiviral response as being a major contributor to the severity of disease (Blanco-Melo et al., 2020; Hadjadj et al., 2020; Lucas et al., 2020; Yao et al., 2020). As observed in cell culture and animal models, it is clear that the host response to SARS-CoV-2 results in an early aberrant IFN response juxtaposed with an overproduction of chemokines (Blanco-Melo et al., 2020; Hadjadj et al., 2020; Lucas et al., 2020). In the hamster model, SARS-CoV-2 infection results in induction of an insufficient IFN-I response as it spreads from the upper to the lower respiratory tract. Although it remains unclear whether the heightened immunity of these tissues prevents virus infection and, therefore, restricts tropism in this hamster model, this would be consistent with the fact that non-respiratory organs that express ACE2 often show an absence of virus even in severe COVID-19 cases (Ziegler et al., 2020). In any event, the combination of high pro-inflammatory cytokines and chemokines and low IFN-I levels is likely the molecular basis for the neutrophil nests observed in the lower respiratory tract. This conclusion is supported by the fact that exogenous administration of IFNα A/D reduces the neutrophil presence in the lungs.
We also observed that the inflammatory response initiated at the earliest site of replication, the upper respiratory tract, may be sufficient to induce systemic inflammation in distal tissues, where mature virions are present at vanishingly low levels if at all, despite robust detection of viral RNA. However, there have been reports of central nervous system invasion in SARS-CoV-2 PCR-positive deceased individuals (Meinhardt et al., 2020). Although the source of the inflammatory response in these distal tissues remains unclear, and whether the source of inflammation differs between cases of mild, moderate, and severe disease, it is tempting to speculate that the secretion or spillover of subgenomic material because of its abundance, perhaps bound by the N protein to form a protected ribonucleoprotein complex, may be a prevalent PAMP that is disseminated from the primary site of replication. In support of this idea, we found that detection of sgN by real-time qRT-PCR does not always correlate with detection of infectious virions. This concept warrants further study but has been suggested in the past for unrelated RNA viruses (tenOever et al., 2002, 2004).
Here we present compelling evidence that IN administration or induction of IFN-I provides protection against SARS-CoV-2 infection, whether for prophylaxis or for early therapeutic use. These data reflect the inhibitory potential of IFN-I and identify an opportunity for IN administration of IFNs as broad-spectrum antiviral agents for SARS-CoV-2 and potentially other respiratory virus infections.
Limitations of study
The transcriptional host response during early infection with SARS-CoV-2 and after viral clearance has remained largely undefined, partially because of limitations in clinical settings. Here we define this response in young golden hamsters, which are able to effectively clear the virus after the disease course. This comes with its own limitations, including a lack of representation of older animals and extrapolation to human disease. The age of the animals and severity of disease may alter the antiviral host response and/or tissue tropism of the virus. The role of IFN-I in the COVID-19 disease course has been widely debated, but clinical trials administering it IN have shown promise. This study defines early treatment of COVID-19. Further research must be performed in a lethal model to assess the value of IN IFN-I administration late in the disease course. Moreover, given the predisposition of individuals with inborn errors of innate immunity to severe COVID-19, IN IFN-I intervention may have a more striking effect in hamsters modified genetically to lack innate immune sensors such as TLR3 and/or TLR7. Future work defining the effect of IN IFN-I treatment later in the disease course and in a lethal model is warranted to further delineate the nuanced effects of bolstering IFN before, during, and/or after SARS-CoV-2 infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-SARS-CoV nucleocapsid [1C7] | Center for Therapeutic Antibody Discovery at the Icahn School of Medicine at Mount Sinai | N/A |
| Anti-MxA, clone M143 (CL143) | Millipore Sigma | Cat# MABF938; RRID: AB_2885181 |
| anti-mouse HRP (DISCOVERY OmniMap) | Roche | Cat# 760-4310, RRID: AB_2885182 |
| Goat anti-Hamster IgG (H+L) Secondary Antibody, HRP (Invitrogen) | Thermo Fisher | Cat# PA1-29626, RRID: AB_10985385 |
| Bacterial and virus strains | ||
| SARS-CoV-2 (USA-WA1/2020) | BEI Resources | Cat# NR-52281 |
| Influenza A/California/04/2009 (H1N1) | Langlois et al., 2013 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Oxoid purified agar (Thermo Scientific) | Thermo Scientific | Cat# OXLP0028B |
| Human IFN Alpha Hybrid (Universal Type I IFN, IFNα A/D) | PBL Assay Science | Cat# 11200-2 |
| Polyinosinic-polycytidylic acid sodium salt (poly I:C) | Millipore Sigma | Cat# P1530 |
| TRIzol Reagent (Invitrogen) | Thermo Scientific | Cat# 15596026 |
| Lipofectamine 2000 Transfection Reagent (Invitrogen) | Thermo Scientific | Cat# 11668019 |
| SuperScript II Reverse Transcriptase (Invitrogen) | Thermo Scientific | Cat# 18064022 |
| Oligo(dT)20 Primer (Invitrogen) | Thermo Scientific | Cat# 18418020 |
| recombinant RBD | Amanat et al., 2020 | N/A |
| Critical commercial assays | ||
| Amicon Ultra-15 centrifugal filter unit | Millipore Sigma | Cat# UFC910024 |
| FastPrep-24 5G bead beating grinder and lysis system | MP Biomedicals | Cat# 6005500 |
| Lysing Matrix A, 2 mL tube | MP Biomedicals | Cat# 6910050 |
| KAPA SYBR FAST qPCR Master Mix (2X) | Roche | KK4602 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | Cat# 20020594 |
| DNA-free DNA Removal Kit (Invitrogen) | Thermo Scientific | Cat# AM1906 |
| DISCOVERY ChromoMap DAB Kit (RUO) | Roche | Cat# 760-159 |
| Deposited data | ||
| RNA-sequencing datasets | This paper | GEO: GSE161200 |
| Mesocricetus auratus Interferon beta 1 sequence | This paper | GenBank: MW017682 |
| SARS-CoV-2/human/USA/WA-CDC-WA1/2020, complete sequence | Harcourt et al., 2020 | GenBank: MN985325.1 |
| Experimental models: cell lines | ||
| Vero E6 | ATCC | Cat# CRL-1586 |
| BHK-21 | ATCC | Cat# CCL-10 |
| A549 | ATCC | Cat# CCL-185 |
| MDCK | ATCC | Cat# CCL-34 |
| Experimental models: organisms/strains | ||
| LVG Golden Syrian hamsters | Charles River Laboratories | Strain# 049 |
| Oligonucleotides | ||
| N/A | See Table S5 for oligonucleotide sequences. | N/A |
| Recombinant DNA | ||
| pLVX-TetOne-Puro-MaIfnb1 | This paper | N/A |
| Software and algorithms | ||
| bcl2fastq | Illumina | http://basespace.illumina.com/ |
| RNA-Seq Alignment App v2.0.2 | Illumina | http://basespace.illumina.com/ |
| edgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| fry | Wu et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| STRING | Szklarczyk et al., 2019 | https://string-db.org/ |
| REVIGO | Supek et al., 2011 | http://revigo.irb.hr/ |
| gplots | CRAN | https://ggplot2.tidyverse.org/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MEGAHIT | Li et al., 2015 | https://github.com/voutcn/megahit |
| ImageJ | National Institutes of Health (USA) | https://imagej.nih.gov/ij/ |
| Color Deconvolution 2 (ImageJ plugin) | Landini et al., 2020 | https://blog.bham.ac.uk/intellimic/g-landini-software/colour-deconvolution-2/ |
| Prism v. 8.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benjamin R. tenOever (benjamin.tenoever@mssm.edu).
Materials availability
Plasmids generated in this study are available upon request.
Data and code availability
The accession number for the RNA-sequencing datasets reported in this paper is GEO: GSE161200. The accession number for the sequence of the Mesocricetus auratus Interferon Beta 1 gene annotated in this paper is GenBank: MW017682.
Experimental model and subject details
Golden Hamsters
Golden hamsters, ranging in ages from 3-5 weeks, were purchased from Charles River Laboratories and housed in a temperature-controlled environment with twelve hours of light per day at the Center for Comparative Medicine and Surgery (CCMS) at Icahn School of Medicine at Mount Sinai (New York, NY, USA). All experiments involving viral infections were carried out in a CDC/USDA-approved BSL-3 facility at CCMS and animals were transferred into the facility four days prior to onset of experiments. Same-sex litter mates were housed in pairs in ventilated cages with ad libitum access to food and water. Environmental enrichment consisted of access to gnawing blocks, extra nesting material, and a weekly fresh vegetable. All experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee at Icahn School of Medicine at Mount Sinai. During anesthesia, animals were provided thermal support to prevent loss of body heat. Male hamsters were used to generate the data in Figures 1, 2, 3, 4, 5, 6, 7A–7H, and S2–S7. Seven male hamsters and three female hamsters per group were used to generate the data in Figures 7I and 7J. One male hamster and three female hamsters per group were used to generate the data in Figures 7K–7M. For Figure 5E assessing in vivo Spike antibodies, male hamsters 8-9 weeks of age were used.
Cell lines
Vero E6 cells (kidney epithelial cells from female African green monkey, ATCC, CRL-1586), BHK-21 cells (kidney fibroblasts from male Syrian golden hamster, ATCC, CCL-10), MDCK cells (kidney epithelial cells from female dog, ATCC, CCL-34) and A549 cells (lung epithelial cells from male human, ATCC, CCL-185) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (GIBCO) supplemented with 10% FBS (Atlanta Biologicals) and 1% penicillin/streptomycin (Millipore Sigma) at 37°C in a humified atmosphere with 5% CO2. Cells tested for the presence mycoplasma bi-weekly using MycoAlert Mycoplasma Detection Kit (lonza). None of the cell lines were authenticated by an external service but were derived directly from ATCC.
Viruses
SARS-related coronavirus 2 (SARS-CoV-2), isolate USA-WA1/2020 (NR-52281) was deposited by the Center for Disease Control and Prevention (Atlanta, GA, USA) and obtained through BEI Resources, NIAID, NIH (Bethesda, MD, USA). SARS-CoV-2 was propagated in Vero E6 cells in DMEM (GIBCO) supplemented with 2% FBS (Atlanta Biologicals), 4.5 g/L D-glucose (GIBCO), 4 mM L-glutamine (GIBCO), 10 mM non-essential amino acids (GIBCO), 1 mM sodium pyruvate (GIBCO) and 10 mM HEPES (Fisher Scientific). Virus stock was filtered by centrifugation using Amicon Ultra-15 Centrifugal filter unit (Millipore Sigma) and resuspended in viral propagation media. All infections were performed with either passage 3 or 4 SARS-CoV-2. Influenza A virus H1N1 isolate A/California/04/2009 was propagated in MDCK cells in DMEM (GIBCO) supplemented with 0.35% BSA (MP Biomedicals).
Method details
Plaque assays
Plaque assays were performed using Vero E6 cells on 12-well plates. Virus was diluted logarithmically in SARS-CoV-2 infection medium for a total volume of 200μl inoculum per well. Plates were incubated for one hour at room temperature, rocking plates every 10 minutes. An overlay of Modified Eagle Medium (GIBCO) 4 mM L-glutamine (GIBCO), 0.2% BSA (MP Biomedicals), 10 mM HEPES (Fisher Scientific), 0.12% NaHCO3, and 0.7% Oxoid agar (Thermo Scientific) was mixed and pipetted into each well. Plates were then incubated at 37°C for 48 hours and then fixed in 5% formaldehyde in PBS for 24 hours before removal from the BSL3 facility. Plaques were stained with crystal violet solution (1% crystal violet (w/v) in 20% ethanol (v/v)) for 15 minutes. Plaque assays of lung homogenates were frozen once before plaque assay was performed; plaque assays of other tissue homogenates were plaqued prior to any cryopreservation.
In vivo infections
Before intranasal infection, hamsters were anesthetized by intraperitoneal injection with 200μl of a ketamine/xylazine solution (4:1), [100mg/kg]. Virus stock was resuspended in PBS to a total volume of 100μl inoculum for each animal. Intranasal treatments with control vehicle (PBS), IFNα A/D (PBL Assay Science), and poly I:C (Millipore Sigma) were also administered in a 100μl volume resuspended in PBS. In direct contact transmission experiments, index hamsters were intranasally infected with 1000pfu of SARS-CoV-2 and caged for 24 hours. Sentinel hamsters were then co-housed with one infected hamster in a clean cage until the experimental endpoint. In fomite transmission experiments, two hamsters were infected with 1000pfu SARS-CoV-2 and housed in cages for two days. They were then removed, and a single naive hamster was moved into the cage for four days before the experimental endpoint. For ocular infections, 6000pfu of SARS-CoV-2 was administered in the eye (5μl in each eye) of anesthetized hamsters. Hamsters subjected to mRNA-Seq or histology were perfused with 60mL ice-cold PBS before harvest. A segment of the upper small intestine closest to the stomach was used for small intestine analysis. When whole brain was harvested from hamsters, the olfactory bulb was sectioned off for separate RNA extraction and analysis. The brain was cut in half longitudinally after removing hindbrain and half was used for RNA extraction and analysis.
Tissue homogenization and tissue RNA extraction
Hamster tissues were homogenized in Lysing Matrix A homogenization tubes (MP Biomedicals) for 40 s at 6.00 m/s for 2 cycles in a FastPrep-24 5G bead beating grinder and lysis system (MP Biomedicals). All tissues for mRNA-Seq analysis were homogenized directly in 1ml TRIzol (Invitrogen). If hamster lung samples were being plaqued for SARS-CoV-2 titer, they were homogenized in 1ml PBS. Homogenates were then centrifuged for five minutes at 10,000 g. Supernatant was immediately frozen for later analysis by plaque assay. The cell pellet was resuspended in TRIzol and re-homogenized as above before RNA extraction. RNA was isolated by phenol/chloroform extraction according to manufacturer’s instructions.
Quantitative real-time PCR analysis
RNA was DNase treated using DNA-free DNA removal kit (Invitrogen). 1μg of total RNA was used for each SuperScript II Reverse Transcriptase reaction with oligo d(T) primers (Invitrogen). Resultant cDNA was diluted 1:20 before real-time quantitative polymerase chain reaction (RT-qPCR) analysis. RT-qPCR reaction was performed using KAPA SYBR® FAST qPCR Master Mix (2X) Kit (Roche) on a LightCycler 480 Instrument II (Roche, Cat# 05015243001). Relative mRNA and viral RNA levels were quantified by normalizing sgRNA to actin expression and normalizing lung RNA levels to mock-infected controls.
Transcriptome analysis
1μg of total RNA was enriched for polyadenylated RNA species and prepared for short-read next-generation sequencing using the TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. Sequencing libraries were sequenced on an Illumina NextSeq 500 platform. Fastq files were generated using bcl2fastq (Illumina) and aligned to the Syrian golden hamster genome (MesAur 1.0, ensembl) using the RNA-Seq Alignment application (Basespace, Illumina). Differential gene expression was determined using the edgeR protocol in the Bioconductor package (Robinson et al., 2010). Samples from the same tissue were analyzed together. Genes that did not have a sufficiently large count in a given tissue as described in Chen et al. (2016) were excluded from the analyses. Highly expressed genes in certain samples such as genes of viral origin were taken into consideration through normalization of the library sizes to an effective library size (Robinson and Oshlack, 2010). Differential gene expression between control group and experimental groups were tested for using a negative binomial model with extended with quasi-likelihood methods (QLF) (Lun et al., 2016). All genes with a false discovery rate (FDR) < 0.05 were classified as differentially expressed genes. Statistical significance of enrichment of custom gene sets were determined using fry from the edgeR package on Bioconductor (Wu et al., 2010). Number of reads mapping to the viral genome (GenBank: MN985325.1) was performed using bowtie2 (Langmead and Salzberg, 2012). For DotPlot analyses, significantly induced genes in each sample (log2 fold change > 1, FDR < 0.05) were used as query to search for enriched biological processes (BP) using STRING (Szklarczyk et al., 2019). A reduced set of enriched Gene Ontology (GO) terms (FDR < 0.05) was obtained using REVIGO (allowed similarity = 0.5) (Supek et al., 2011). Selected enriched GO terms were visualized by Dot plot using ggplot2 (Wickham, 2016).
de novo gene annotation and verification
The identification of the interferon beta gene from golden hamsters was achieved by de novo assembly using MEGAHIT (https://github.com/voutcn/megahit; Li et al., 2015). In brief, RNA derived from the lungs of hamsters treated with poly I:C were used to generate an Illumina mRNA library. Fastq files were then inputted into MEGAHIT for de novo assembly. Assembled contigs generated as a result were subsequently used as input for a BLASTx search against human IFNB1 protein (NP_002167.1). This search resulted in a single hamster contig that showed 46% identity to human IFNB1 (see Figure S1). This gene annotation has been deposited to GenBank (accession number MW017682). The sequence was synthesized as a gBlock (IDT) and cloned into a doxycycline-inducible TetOne vector (Takara). Functionality was confirmed by transfecting the construct into BHK-21 cells cultured in normal growth medium supplemented with doxycycline 10ng/ml. Transfection was performed using Lipofectamine 2000 (Invitrogen) following manufacturer’s instructions.
Lung histology
For H&E staining, lung tissues were aseptically harvested following whole-body perfusion and fixed in 10% neutral buffered formalin for 24 hours. Tissues were then embedded in paraffin and sectioned into 8 micron thick sections onto charged slides. For H&E staining, slides were deparaffinized and rehydrated prior to staining with hematoxylin (Gill’s formula, Vector Laboratories, Cat# H3401) and eosin Y (Sigma Aldrich, Cat# E4009) according to manufacturer’s instructions. Tissues were assessed for several clinical criteria by a certified veterinary pathologist at the Icahn School of Medicine at Mount Sinai. For SARS-CoV-2 N or MxA staining, upper lobes fixed and embedded as above. Viral protein was detected using mouse monoclonal anti-SARS-CoV-2 Nucleocapsid [1C7C7] protein (a kind gift by Dr. T. Moran, Center for Therapeutic Antibody Discovery at the Icahn School of Medicine at Mount Sinai). MxA was detected using Anti-MxA, clone M143 (CL143), a mouse monoclonal antibody (EMD Millipore, MABF938). Anti-N and anti-MxA staining were conducted using the VENTANA Discovery Ultra (Roche) instrument. The anti-N antibody was diluted to 1:50 and applied manually to individual slides. The OmniMap anti-mouse HRP (Roche) was used as the secondary antibody, and the DISCOVERY OmniMap ChromoMap DAB RUO (Roche) was utilized for development. For IHC protein expression quantification, four sections of each analyzed slide were quantified using Color Deconvolution 2 (Landini et al., 2020) software for H DAB analysis by normalizing the HRP signal to the number of nuclei detected in each image, using a previously published protocol (Crowe and Yue, 2019).
Anti-RBD hamster IgG ELISA
The ELISA protocol was adapted form established protocol (Amanat et al., 2020). 96-well plates (Immulon 4 HBX, Thermo Fisher) were coated with 50μl of 2μg/ml recombinant RBD in PBS at 4°C overnight. After overnight incubation, the coating solutions were removed, and the plates were blocked with 100μl of 3% non-fat milk (americanBio) prepared in 0.1% Tween 20 containing PBS (PBS-T) for 1 hour at room temperature. To reduce the risk of containing live virus, serum samples were heated 1 hour for 56°C before use and was handled in a BSL-3 facility. Serial dilution of serum samples and dilution of secondary antibody was done in 1% non-fat milk prepared in PBS-T. After blocking, solutions were removed and 100μl of diluted serum was added following 2 hours incubation at room temperature. After the solutions were removed, the wells were washed with 250μl PBS-T three times. 100μl of 1:7500 diluted anti-Syrian hamster IgG HRP (ThermoFisher) secondary antibody was added and incubated for 1 hour at room temperature. After the solutions were removed, the wells were washed with 250μl PBS-T three times. Once the wells were completely dry, 100 μL of SIGMAFAST o-phenylenediamine dihydrochloride (Millipore Sigma) solution was added and the reaction was stopped with 30μl of 3M HCl. The optical density at 490 nm was measured and the concentrations of the antibody were analyzed by area under curve (AUC) using Prism 8 (GraphPad).
Quantification and statistical analysis
Statistical significance was determined by Student’s t test or Mann-Whitney test when data points did not follow normal distribution. In the case of three experimental groups, one-way ANOVA was applied, and the p-value was corrected for multiple comparisons using Dunnett’s test. Statistical tests were performed with Prism 8.0 (GraphPad) software. For all RNA sequencing analyses, statistical significance of differential gene expression between groups was determined through the edgeR pipeline (see transcriptome analysis, methods section). For all data, a p-value < 0.05 or a false discovery rate < 0.05 was considered to be statistically significant. Statistical details for each experiment including statistical tests applied, p-values and number of replicates can be found in the figure legends.
Acknowledgments
This work was funded by the Marc Haas Foundation and DARPA's] PREPARE program (HR0011-20-2-0040). The views, opinions, and/or findings expressed are those of the author and should not interpreted as representing the official views or policies of the Department of Defense or the U.S. government. D.B.-M. is an Open Philanthropy Fellow of the Life Sciences Research Foundation. We would like to thank Carlos Franco and Lenny Martinez, supervisors of the ISMMS animal facility, for all of their help.
Author contributions
Animal work, experimental design, and subsequent assays were performed by R.M., D.A.H., and K.O. IgG titers were determined by S.H. Illumina libraries were prepared by M.P. and R.M. J.F., I.G., and S.A.U. performed additional animal work and real-time qRT-PCR follow up. D.B.-M., D.S., and R.M. performed all computational work pertaining to mRNA-seq data. J.K.L. and K.A. performed the histological evaluation. R.M., D.A.H., and B.R.t. wrote the paper.
Declaration of interests
The authors declare no competing interests.
Published: January 29, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.01.017.
Supplemental information
All statistically significantly differentially expressed genes between SARS-CoV-2- and IAV-infected hamster lungs. Genes are shown as ensembl gene ID next to known golden hamster gene name and human orthologs. Expression of each gene is shown as mean count, log2 fold change and adjusted p-value (padj).
Differential gene expression for all genes expressed in each tissue indicated compared to uninfected control tissues. Genes are shown as ensembl gene ID together with known human ortholog. Expression is indicated as log2 fold change (logFC) and false discovery rate (FDR). Each sheet contains gene expression information for one tissue from animals infected with either 100pfu (L) or 10,000pfu (H).
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., Van Weyenbergh J., De Keyzer C., Bervoets L., Sharma S., et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carfì A., Bernabei R., Landi F., Gemelli Against COVID-19 Post-Acute Care Study Group Persistent Symptoms in Patients After Acute COVID-19. JAMA. 2020;324:603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carignan A., Valiquette L., Grenier C., Musonera J.B., Nkengurutse D., Marcil-Héguy A., Vettese K., Marcoux D., Valiquette C., Xiong W.T., et al. Anosmia and dysgeusia associated with SARS-CoV-2 infection: an age-matched case-control study. CMAJ. 2020;192:E702–E707. doi: 10.1503/cmaj.200869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lun A.T., Smyth G.K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe A.R., Yue W. Semi-quantitative Determination of Protein Expression using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Biol. Protoc. 2019;9:e3465. doi: 10.21769/BioProtoc.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E., Li G. Approved Antiviral Drugs over the Past 50 Years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M.E., Haslwanter D., Bortz R.H., 3rd, Wirchnianski A.S., Lasso G., Vergnolle O., Abbasi S.A., Fels J.M., Laudermilch E., Florez C., et al. A Replication-Competent Vesicular Stomatitis Virus for Studies of SARS-CoV-2 Spike-Mediated Cell Entry and Its Inhibition. Cell Host Microbe. 2020;28:486–496.e6. doi: 10.1016/j.chom.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Ábrányi-Balogh P., et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger R.W., Fauci A.S. Ending the HIV/AIDS Pandemic. Emerg. Infect. Dis. 2018;24:413–416. doi: 10.3201/eid2403.171797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Li Y., Ding S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019;19:31–44. doi: 10.1038/s41577-018-0071-x. [DOI] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., et al. Identification of SARS-CoV-2 Inhibitors using Lung and Colonic Organoids. Nature. 2020;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C.A., Jr., Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Nakagawa S., Sato K., USFQ-COVID19 Consortium SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C., Palese P., Shaw M.L., Treanor J., Webster R.G., García-Sastre A. Influenza. Nat. Rev. Dis. Primers. 2018;4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini G., Martinelli G., Piccinini F. Colour Deconvolution - stain unmixing in histological imaging. Bioinformatics. 2020:btaa847. doi: 10.1093/bioinformatics/btaa847. [DOI] [PubMed] [Google Scholar]
- Langlois R.A., Albrecht R.A., Kimble B., Sutton T., Shapiro J.S., Finch C., Angel M., Chua M.A., Gonzalez-Reiche A.S., Xu K., et al. MicroRNA-based strategy to mitigate the risk of gain-of-function influenza studies. Nat. Biotechnol. 2013;31:844–847. doi: 10.1038/nbt.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Schoggins J.W., Diamond M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Liu Z., Zheng H., Lin H., Li M., Yuan R., Peng J., Xiong Q., Sun J., Li B., Wu J., et al. Identification of Common Deletions in the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020;94 doi: 10.1128/JVI.00790-20. e00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T., Chen Y., Smyth G.K. It’s DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol. Biol. 2016;1418:391–416. doi: 10.1007/978-1-4939-3578-9_19. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Falvo J.V., Kim T.H., Kim T.K., Lin C.H., Parekh B.S., Wathelet M.G. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- Manson J.J., Crooks C., Naja M., Ledlie A., Goulden B., Liddle T., Khan E., Mehta P., Martin-Gutierrez L., Waddington K.E., et al. COVID-19-associated hyperinflammation and escalation of patient care: a retrospective longitudinal cohort study. Lancet Rheumatol. 2020;2:E594–E602. doi: 10.1016/S2665-9913(20)30275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt J., Radke J., Dittmayer C., Franz J., Thomas C., Mothes R., Laue M., Schneider J., Brunink S., Greuel S., et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2020 doi: 10.1038/s41593-020-00758-5. Published online November 30, 2020. [DOI] [PubMed] [Google Scholar]
- Mesev E.V., LeDesma R.A., Ploss A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019;4:914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M., Gabbay F.J., Davies D.E., Holgate S.T., Ho L.P., et al. Inhaled Interferon Beta COVID-19 Study Group Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30511-7. S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Peto R., Karim Q.A., Alejandria M., Henao-Restrepo A.M., García C.H., Kieny M.-P., Malekzadeh R., Murthy S., Preziosi M.-P., et al. Repurposed antiviral drugs for COVID-19 –interim WHO SOLIDARITY trial results. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2023184. Published online December 2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A., Iwasaki A. Type I and Type III Interferons - Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si L., Bai H., Rodas M., Cao W., Oh C.Y., Jiang A., Moller R., Hoagland D., Oishi K., Horiuchi S., et al. Human organ chip-enabled pipeline to rapidly repurpose therapeutics during viral pandemics. bioRxiv. 2020 doi: 10.1101/2020.04.13.039917. [DOI] [Google Scholar]
- Skendros P., Mitsios A., Chrysanthopoulou A., Mastellos D.C., Metallidis S., Rafailidis P., Ntinopoulou M., Sertaridou E., Tsironidou V., Tsigalou C., et al. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 2020;130:6151–6157. doi: 10.1172/JCI141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokina M., M C Teixeira J., Barrera-Vilarmau S., Paschke R., Papasotiriou I., Rodrigues J.P.G.L.M., Kastritis P.L. Structural models of human ACE2 variants with SARS-CoV-2 Spike protein for structure-based drug design. Sci. Data. 2020;7:309. doi: 10.1038/s41597-020-00652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Cheon H., Wang Y. Responses to Cytokines and Interferons that Depend upon JAKs and STATs. Cold Spring Harb. Perspect. Biol. 2018;10:a028555. doi: 10.1101/cshperspect.a028555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L., Liu D., Huang J., He J., Zhu A., Zhao J., et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell. 2020;182:734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever B.R. The Evolution of Antiviral Defense Systems. Cell Host Microbe. 2016;19:142–149. doi: 10.1016/j.chom.2016.01.006. [DOI] [PubMed] [Google Scholar]
- tenOever B.R., Servant M.J., Grandvaux N., Lin R., Hiscott J. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 2002;76:3659–3669. doi: 10.1128/JVI.76.8.3659-3669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever B.R., Sharma S., Zou W., Sun Q., Grandvaux N., Julkunen I., Hemmi H., Yamamoto M., Akira S., Yeh W.C., et al. Activation of TBK1 and IKKvarepsilon kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 2004;78:10636–10649. doi: 10.1128/JVI.78.19.10636-10649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hedge S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.X., Fish E.N. Global virus outbreaks: Interferons as 1st responders. Semin. Immunol. 2019;43:101300. doi: 10.1016/j.smim.2019.101300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerahandi H., Hochman K.A., Simon E., Blaum C., Chodosh J., Duan E., Garry K., Kahan T., Karmen-Tuohy S., Karpel H., et al. Post-discharge health status and symptoms in patients with severe COVID-19. J. Gen. Intern. Med. 2020:1–8. doi: 10.1007/s11606-020-06338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Lim E., Vaillant F., Asselin-Labat M.L., Visvader J.E., Smyth G.K. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Tu L. Implications of gastrointestinal manifestations of COVID-19. Lancet Gastroenterol. Hepatol. 2020;5:629–630. doi: 10.1016/S2468-1253(20)30132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Bora S.A., Parimon T., Zaman T., Friedman O.A., Palatinus J.A., Surapaneni N.S., Matusov Y.P., Cerro Chiang G., Kassar A.G., et al. Cell type-specific immune dysregulation in severely ill COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.07.23.20161182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. COVID-STORM Clinicians. COVID Clinicians. Imagine COVID Group. French COVID Cohort Study Group. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort. NIAID-USUHS/TAGC COVID Immunity Group Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Williams E.P., Malireddi R.K.S., Karki R., Banoth B., Burton A., Webby R., Channappanavar R., Jonsson C.B., Kanneganti T.D. Impaired NLRP3 inflammasome activation/pyroptosis leads to robust inflammatory cell death via caspase-8/RIPK3 during coronavirus infection. J. Biol. Chem. 2020;295:14040–14052. doi: 10.1074/jbc.RA120.015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. HCA Lung Biological Network. Electronic address. lung-network@humancellatlas.org. HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All statistically significantly differentially expressed genes between SARS-CoV-2- and IAV-infected hamster lungs. Genes are shown as ensembl gene ID next to known golden hamster gene name and human orthologs. Expression of each gene is shown as mean count, log2 fold change and adjusted p-value (padj).
Differential gene expression for all genes expressed in each tissue indicated compared to uninfected control tissues. Genes are shown as ensembl gene ID together with known human ortholog. Expression is indicated as log2 fold change (logFC) and false discovery rate (FDR). Each sheet contains gene expression information for one tissue from animals infected with either 100pfu (L) or 10,000pfu (H).
Data Availability Statement
The accession number for the RNA-sequencing datasets reported in this paper is GEO: GSE161200. The accession number for the sequence of the Mesocricetus auratus Interferon Beta 1 gene annotated in this paper is GenBank: MW017682.