Peripartum cardiomyopathy (PPCM) is a rare form of heart failure occurring in the last trimester of pregnancy or in the first months after delivery.1 The diagnosis is based on exclusion criteria, and specific biomarkers remain unidentified.1 Proposed mechanisms of PPCM pathophysiology are based on various mouse models harboring cardiac-specific genetic defects,2,3 which demonstrated typical PPCM characteristics, but mutations in these genes remain to be associated with PPCM in patients.4 This study aims to identify aberrant pathways in cardiomyocytes obtained from induced pluripotent stem cells (iPSC) derived from patients with PPCM.
Two patients with PPCM with acute heart failure (diagnosed within weeks after delivery) have been included in the study. Patients A and B were aged 37 and 28 years with a left ventricular ejection fraction of 17% and 45% and elevated levels of N-terminal pro-B-type natriuretic peptide 1309 ng/L and 2743 ng/L at presentation, respectively. Patient A was compared with her sister and patient B with her mother in paired analyses; both controls were healthy, had a normal echocardiogram, and have had multiple uncomplicated deliveries. Patients were screened for genetic variations in 61 genes associated with cardiomyopathies, but none was identified. All participants provided written informed consent. This study was approved by the local Medical Ethical Committee (METc:2014.104).
Pregnancy-associated wall stress was simulated in vitro by applying cyclic mechanical stretch to the iPSC-derived cardiomyocytes (iPSC-CM; Figure [A]). RNA sequencing analysis of these conditions resulted in a total of 3316 differentially expressed genes (Figure [B]). We identified 95 common differentially expressed genes in healthy mechanically stretched iPSC-CM and in both static and stretched PPCM iPSC-CM compared with healthy static iPSC-CM (designated as the intersection of interest in yellow, Figure [B]). Therefore, a stretch-induced stress response was observed in all mechanically stretched cells, but also in static PPCM iPSC-CM. Relative expression levels of these 95 genes were visualized in a heat map (Figure [C]), and a pathway overrepresentation analysis was performed. Predicted gene ontology terms were organized into superclusters (Figure [D]). Gene ontology terms pertained mostly to lipid metabolism as part of the fatty alcohol biosynthesis supercluster of gene ontology terms, and 28 of 95 differentially expressed genes were associated to metabolic processes (depicted in red, Figure [C]). In silico transcription factor enrichment analysis identified key transcription factors SREBP1 (sterol regulatory element-binding transcription factor 1), NFY (nuclear transcription factor Y), SP1 (Sp1 transcription factor), MAX (MYC-associated factor X), and CEBPB (CCAAT enhancer-binding protein beta).
Figure.
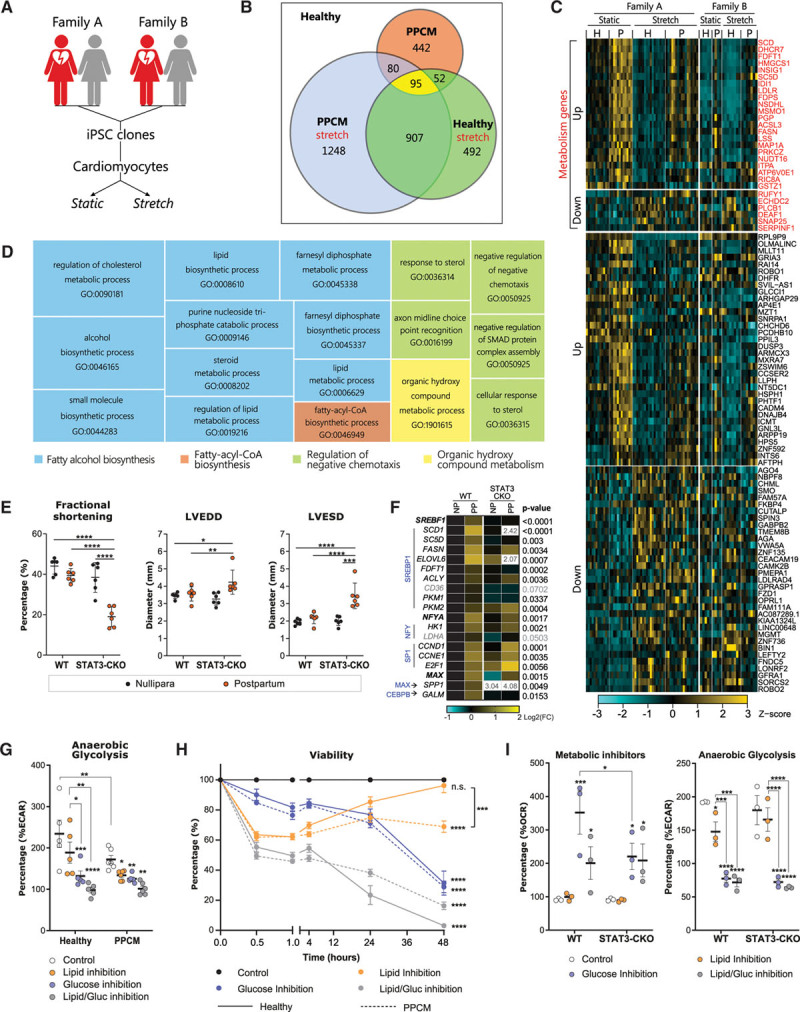
Pathways related to lipid metabolism were aberrantly regulated in PPCM-specific iPSC-derived cardiomyocytes. A, Schematic overview of the experimental setup. Multiple iPSC clones were generated from patient A (4 clones), patient B (2 clones), and both controls (2 clones from each). Each clone has been differentiated to cardiomyocytes at least 3 times. Cardiomyocytes have been subjected to cyclic equiaxial mechanical stretch (15% elongation at 1 Hz for 48 hours) to mimic pregnancy-related hemodynamic stress. B, Venn diagram denoting the number of differentially expressed genes (DEGs) in stretched healthy cardiomyocytes, static and stretched cardiomyocytes derived from patients with PPCM, and reciprocal intersections compared with static healthy cardiomyocytes (background). The yellow intersection of interest marks the overlapping set of 95 DEGs in cardiomyocytes from stretched healthy, static PPCM, and stretched PPCM conditions. C, A heatmap showing expression patterns of the 95 genes at the intersection of interest. Cyan and yellow indicate reduced and increased gene expression, respectively. Genes associated with metabolic processes have been grouped (red) in addition to up- and downregulated genes. D, Pathway overrepresentation analysis was performed on the genes at the intersection of interest, and superclusters (colored) of individual gene ontology terms (GO-terms) are displayed as a treemap. Sizes of the squares are proportionate to the level of significance of each GO-term and the superclusters. The fatty alcohol biosynthesis supercluster is markedly overrepresented. E, Cardiac function was reduced in STAT3-CKO mice after the second pregnancy and nursing period as demonstrated by reduced fractional shortening, left ventricular end diastolic diameter (LVEDD), and left ventricular end systolic diameter (LVESD). F, Key lipid metabolism-associated genes that were identified as DEGs by RNA sequencing were also found to be differentially expressed in the left ventricles of STAT3-CKO mice. Blue gene symbols mark transcription factors governing the indicated genes; bold gene symbols show transcription factors that could be detected by quantitative reverse transcription-polymerase chain reaction. White fields indicate off-scale data, which are numerically shown instead. NP indicates nullipara; PP, postpartum. Overall differences between all samples were determined per gene by 2-way ANOVA (without post hoc test). G, Oligomycin A–induced extracellular acidification rates (ECAR) were assessed by a Seahorse assay–based Mito Fuel Flex test to determine cellular responses to specific inhibition of lipid metabolism, glucose metabolism, or both simultaneously. Data are presented as change relative to respective conditions before addition of inhibitors. H, Metabolic flexibility is impaired in cardiomyocytes derived from patients with PPCM as determined by a viability assay following metabolic pathway inhibition shown after the first hour of inhibitor addition (left) and after 48 hours (right). I, Metabolic flexibility was impaired in isolated cardiomyocytes from STAT3-CKO mice as assessed by Seahorse assay (left). This effect was not observed for cytosolic anaerobic glycolysis (right). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 as determined by 2-way ANOVA followed by a Bonferroni post hoc test compared with the respective control condition unless indicated otherwise (by connecting lines). CEBPB indicates CCAAT enhancer-binding protein beta; CoA, coenzyme A; ECAR, extracellular acidification rate; iPSC, induced pluripotent stem cell; Gluc, glucose; H, healthy control; MAX, MYC-associated factor X; OCR, oxygen consumption rate; P, PPCM patient; PPCM, peripartum cardiomyopathy; SMAD, small mothers against decapentaplegic; STAT3-CKO, STAT3 gene deletion; and WT, wild-type.
To underscore the relevance of these findings, we aimed to validate our findings in an established PPCM mouse model caused by cardiac-specific STAT3 conditional knockout (STAT3-CKO).2 These STAT3-CKO mice consistently developed PPCM as demonstrated by a severely reduced cardiac function (Figure [E]). Gene expression analysis of left ventricular tissue of postpartum STAT3-CKO mice (ie, after 2 pregnancies and nursing periods) showed that key metabolic genes were also differentially regulated compared with nulliparous STAT3-CKO mice (Figure [F]).
Analysis of metabolic substrate utilization in the PPCM iPSC-CM and isolated STAT3-CKO cardiomyocytes was performed by means of a Seahorse assay–based Mito Fuel Flex test. Cytosolic anaerobic glycolysis was markedly blunted in PPCM iPSC-CM at baseline and after inhibition of lipid metabolism (Figure [G]). To assess substrate dependence, we measured cellular viability after specific inhibition of metabolic pathways (Figure [H]). Viability was rapidly decreased by inhibition of total lipid metabolism, total glucose metabolism, or both within the first hour. Interestingly, healthy iPSC-CM recovered over time, which was not observed in PPCM iPSC-CM after 48 hours. Inhibition of glucose metabolism resulted in elevated OCR in isolated wild-type cardiomyocytes, which was blunted in STAT3-CKO cardiomyocytes; this did not affect cytosolic anaerobic glycolysis (Figure [I]).
Under physiological circumstances, maternal lipid metabolism is increased during the last trimester of pregnancy and quickly normalizes after delivery.5 This metabolic transition is greatly dependent on transcription factors governing lipid metabolism. Our results demonstrate that iPSC-CM derived from patients with PPCM showed disrupted regulation of pathways related to lipid metabolism, which was validated in an established PPCM mouse model. Furthermore, we observed impaired anaerobic glycolysis in PPCM iPSC-CM after inhibition of lipid metabolism. Healthy iPSC-CM demonstrated adequate metabolic plasticity by activation of glucose utilization in response to inhibition of lipid metabolism; this metabolic switch was blunted in PPCM iPSC-CM. Isolated wild-type cardiomyocytes immediately switched to lipid metabolism after inhibition of glucose metabolism, which was not observed in STAT3-CKO cardiomyocytes.
Although the number of patients included in this study remains a limitation, this study highlights that commonly affected pathways could be identified in 2 different families. STAT3 and PGC1α were not identified as differentially expressed genes in any condition, but these might be altered posttranscriptionally in patients with PPCM and may exacerbate PPCM progression.5 However, patients with PPCM may have mutations or epigenetic alterations in other genes, which may explain altered cardiac metabolism in PPCM iPSC-CM. Moreover, the correlation between parity and PPCM severity remains to be further investigated.
In conclusion, our data show that lipid metabolism is widely affected in PPCM iPSC-CM and highlight the potential role of metabolic regulation as a key factor for PPCM susceptibility.
Sources of Funding
This research was supported by the Dutch Heart Foundation (2012T047 to P.v.d.M.), ZonMW Clinical Fellow (90700436 to P.v.d.M.), the European Research Council (StG 715732 to P.v.d.M.), the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant (754425), the German Research Foundation (HI 842/4-3 to D.H.-K.), the DFG Clinical Research Group (KFO 311, HI 842/10-1, HI 842/10-2 to D.H.-K.; RI 2531/2-1, RI 2531/2-2 to M.R.-H.), by the State of Lower Saxony and the Volkswagen Foundation (VWZN3009 and VWZN3452 to D.H.-K. and M.R.-H.).
Disclosures
None.
Footnotes
Drs Hoes, Bomer, and Ricke-Hoch contributed equally.
Drs Hilfiker-Kleiner and van der Meer contributed equally.
Data sharing: All data and materials have been made publicly available at Array Express and can be accessed at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9053/. All cell lines used in this study have been registered in the Human Pluripotent Stem Cell Registry at https://hpscreg.eu/.
Contributor Information
Nils Bomer, Email: n.bomer@umcg.nl.
Melanie Ricke-Hoch, Email: hoch.melanie@mh-hannover.de.
Tristan V. de Jong, Email: tristan_dejong@hotmail.com.
Stefan Pietzsch, Email: pietzsch.stefan@mh-hannover.de.
Denise Hilfiker-Kleiner, Email: hilfiker.denise@mh-hannover.de.
References
- 1.Bauersachs J, König T, van der Meer P, Petrie MC, Hilfiker-Kleiner D, Mbakwem A, Hamdan R, Jackson AM, Forsyth P, de Boer RA, et al. Pathophysiology, diagnosis and management of peripartum cardiomyopathy: a position statement from the Heart Failure Association of the European Society of Cardiology Study Group on peripartum cardiomyopathy. Eur J Heart Fail. 2019;21:827–843. doi: 10.1002/ejhf.1493 [DOI] [PubMed] [Google Scholar]
- 2.Hilfiker-Kleiner D, Kaminski K, Podewski E, Bonda T, Schaefer A, Sliwa K, Forster O, Quint A, Landmesser U, Doerries C, et al. A cathepsin D-cleaved 16 kDa form of prolactin mediates postpartum cardiomyopathy. Cell. 2007;128:589–600. doi: 10.1016/j.cell.2006.12.036 [DOI] [PubMed] [Google Scholar]
- 3.Patten IS, Rana S, Shahul S, Rowe GC, Jang C, Liu L, Hacker MR, Rhee JS, Mitchell J, Mahmood F, et al. Cardiac angiogenic imbalance leads to peripartum cardiomyopathy. Nature. 2012;485:333–338. doi: 10.1038/nature11040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ware JS, Li J, Mazaika E, Yasso CM, DeSouza T, Cappola TP, Tsai EJ, Hilfiker-Kleiner D, Kamiya CA, Mazzarotto F, et al. Shared genetic predisposition in peripartum and dilated cardiomyopathies. N Engl J Med. 2016;374:233–241. doi: 10.1056/NEJMoa1505517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stapel B, Kohlhaas M, Ricke-Hoch M, Haghikia A, Erschow S, Knuuti J, Silvola JMU, Roivainen A, Saraste A, Nickel AG, et al. Low STAT3 expression sensitizes to toxic effects of β-adrenergic receptor stimulation in peripartum cardiomyopathy. Eur Heart J. 2016;38:ehw086 doi: 10.1093/eurheartj/ehw086 [DOI] [PMC free article] [PubMed] [Google Scholar]


