Abstract
Marine bacterium Rhodococcus sp. NJ-530 has developed several ultraviolet (UV) adaptive characteristics for survival and growth in extreme Antarctic environment. Rhodococcus sp. NJ-530 DNA photolyase encoded by a 1146 bp photolyase-homologous region (phr) was identified in genome. Quantitative real-time PCR demonstrated that transcriptional levels of phr were highly up-regulated by ultraviolet-B (UV-B) radiation (90 μW·cm−2) and increased to a maximum of 149.17-fold after exposure for 20 min. According to the results of SDS-PAGE and western blot, PHR was effectively induced by isopropyl-β-d-1-thiogalactopyranoside (IPTG) at the genetically engineered BL21(DE3)-pET-32a( +)-phr construct under the condition of 15 °C for 16 h and 37 °C for 4 h. In terms of in vivo activity, compared with a phr-defective E. coli strain, phr-transformed E. coli exhibited higher survival rate under high UV-B intensity of 90 μW·cm−2. Meanwhile, the purified PHR, with blue light, presented obvious photorepair activity toward UV-induced DNA damage in vitro assays. To sum up, studying the mechanisms of Rhodococcus sp. NJ-530 photolyase is of great interest to understand the adaptation of polar bacteria to high UV radiation, and such data present important therapeutic value for further UV-induced human skin and genetic damage diseases.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02660-8.
Keywords: Antarctica, Rhodococcus sp. NJ-530, Photolyase, DNA damage, DNA repair, Photorepair
Introduction
Normally, ozone layer functions as an ultraviolet (UV, 280–400 nm) radiation barrier to absorb almost all ultraviolet-C (UV-C, 100–280 nm) radiation and approximately 90% of ultraviolet-B (UV-B, 280–315 nm) radiation (Hu and Adar 2017; Sinha and Hader 2002). It is unfortunate that the ozone layer has been severely damaged and depleted due to pollution and the use of fluoridated compounds, especially in Antarctica (Bais et al. 2015). The increased UV levels exert increasingly adverse effects on molecules such as DNA, protein and lipids in bacteria, fungi, plants, animals and even humans (Dahms and Lee 2010; Gill et al. 2015; Jans et al. 2005; Roldan-Arjona and Ariza 2009).
UV radiation has the oxidizing activity in triggering oxidative stress in organisms by generate reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), hydroxyl radicals (·OH), superoxide anion radicals (·O2−) and singlet oxygen (1O2) (Subedi et al. 2017). What is more, genotoxic UV light mainly damages organismic DNA in two ways: formation of cis-syn cyclobutane pyrimidine dimers (CPDs) and pyrimidine–pyrimidone (6–4) photoproducts [(6–4) PPs] (Kageyama and Waditee-Sirisattha 2019; Sancar 2004; Sinha and Hader 2002). CPDs are the major type of UV-induced dimers, and the spatial configuration of both photoproducts is altered compared to the DNA double-helix structure (Dehez et al. 2017). Unfortunately, if left unrepaired, the DNA damage may result in mutations or even cell death in organisms (Hader and Sinha 2005; Lee et al. 2018; Rastogi et al. 2010). In addition, UV-induced DNA damage causes a series of skin problems, such as inflammation, immunosuppression, and even skin cancer (Armstrong and Kricker 2001; Pfeifer and Besaratinia 2012). The protection of skin with traditional sunscreens is inherently prophylactic, while sunscreens containing DNA photolyase (EC 4.1.99.3) can repair DNA damage caused by UV-B (Berardesca et al. 2012). According to recent studies, topical use of photolyase is very effective in protecting human skin and serves as both a preventive measure and a method for treating UV-induced damage to human skin (Carducci et al. 2015; Laino et al. 2015; Megna et al. 2017; Purohit et al. 2016; Puviani et al. 2013; Stege et al. 2000). Photolyase, which belongs to CPF (cryptochrome (CRY) and photolyase family), specifically binds to CPDs or (6–4) PPs to repair UV-induced DNA damage using the energy from visible light photons (Stege et al. 2000). However, a single kind of photolyase cannot bind to both products. Thus, CPD photolyase can only repair CPDs, while 6–4 photolyase only repair (6–4) PPs.
The molecular mechanism of DNA photolyase has been extensively explored, but reports of photolyase in Antarctic microorganisms are still limited. To the best of our knowledge, Antarctic photolyases were found in bacteria Pseudomonas, Hymenobacter and Sphingomonas (Marizcurrena et al. 2019, 2017), unicellular green alga Chlamydomonas sp. ICE-L (Li et al. 2015), and several zooplankton (Isely et al. 2009; Malloy et al. 1997; Rocco et al. 2002). What is more, scientists also found photolyase gene in Antarctic bacterium Hymenobacter nivis P3T and moss Pohlia nutans, but their functional characterization and verification had not been carried out yet (Li et al. 2019; Terashima et al. 2019). Therefore, in this study, the actinomycete Rhodococcus sp. NJ-530 from Antarctica was selected as experimental model to study DNA photolyase in view of extreme UV adaptability. We successfully cloned and expressed the photolyase-homologous region (PHR) from Rhodococcus sp. NJ-530 genome, which establishes a foundation for studying UV radiation response in certain Rhodococcus strains.
Materials and methods
Bacterial strain and culture method
Rhodococcus sp. NJ-530 was isolated from floating ice near the Zhongshan Research Station of Antarctica (69° S, 77° E) during China’s 22nd Antarctic Expedition in 2001. The complete genome of Rhodococcus sp. NJ-530 was sequenced and submitted to National Center for Biotechnology Information (NCBI) website under the accession Number of NZ_CP034152.1. The sample was stored at the Key Laboratory of Marine Bioactive Substances, First Institute of Oceanography, Ministry of Natural Resources, China. Cultures were incubated in 2216E medium at 25 °C.
Bioinformatic analysis
The open-reading frame (ORF) and homology of phr were analyzed with ORF Finder and BLASTX, respectively, from NCBI website. Nucleotide sequence of phr was translated into a protein sequence (http://searchlauncher.bcm.tmc.edu/seq-util/Options/sixframe.html). Then, theoretical isoelectric point (pI) and molecular weight (Mw) of the photolyase were calculated using Compute pI/Mw tool in ExPASy (http://web.expasy.org/protparam). Predictprotein (https://www.predictprotein.org/) and Swiss-Model were used to predict its function and structure (Waterhouse et al. 2018). Phylogenetic tree was built using neighbor-joining method and evaluated using a bootstrap analysis method with 1000 replicates by MEGA 6.0 software.
phr cloning and plasmid constructs
EasyPure® Bacteria Genomic DNA Kit (TransGen Biotech, Beijing, China) was used to extracted Rhodococcus sp. NJ-530 genomic DNA. Specific primers for phr (Table 1) were designed to amplify phr with EasyTaq® DNA polymerase (TransGen Biotech, Beijing, China) using Rhodococcus sp. NJ-530 genomic DNA as the template. PCR procedure was as follows: 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 90 s, and a final step of 10 min at 72 °C. The desired PCR products were then separated by electrophoresis in a 1.0% agarose gel and purified using an EasyPure® PCR Purification Kit (TransGen Biotech, China). After inserted into PMD19T vector, the purified phr was maintained in Escherichia coli strain DH5α and finally sequenced (Sangon, China) to confirm its correctness.
Table 1.
Primers used in phr cloning and expression
| Primers | Sequence (5′-3′) |
|---|---|
| Primers for phr cloning | |
| phr-F | CGGACAGACGTTGGGT |
| phr-R | TCAGCCTTTGATCTCGC |
| Primers for phr transcriptional expression | |
| phr-qF | CGCAACTGGTGGACGGTGAC |
| phr-qR | CTTCAACGAGTGGACGGCCTTC |
| 16S rRNA-qF | TGCGAGACCGTGAGGTGGAG |
| 16S rRNA-qR | GCGATTACTAGCGACTCCGACTTC |
| GAPDH-qF | CGACGGCAGCCAGAACATCATC |
| GAPDH-qR | GCGTGGATCGTGGTCATCAGAC |
| recA-qF | CACCACCGTCATCTTCATCAACCAG |
| recA-qR | CGACGGCAGCCAGAACATCATC |
| Ldh-qF | GACCAACCTCGCCATCGGATTG |
| Ldh-qR | CGTGGGAAGGGACAGGCAAAC |
| rpoB-qF | GTCGTTCAAGGTGCTCCTCAAGG |
| rpoB-qR | TCGTCGCCGTCCGCCATC |
| PGK-qF | GCGAGACCAGCAAGGACGAAG |
| PGK-qR | ACGCCTGCTTACGGTGAACAAC |
Quantitative real-time PCR analysis of phr expression
Rhodococcus sp. NJ-530 cells were cultured to exponential growth phase for the analysis of phr transcription levels. Three biological replicates of cells in exponential growth phase were irradiated with UV-B at intensities of 0, 30, and 90 μW·cm−2 for different culture times (0, 10, 20, 40, 90 and 150 min) to investigate how phr was expressed at the transcriptional level in response to UV radiation. Meanwhile, OD600 value in different UV-B conditions was measured to draw the growth curve. RNA, extracted by TransZol (TransGen Biotech, China) from 10 mL of cells, was observed by electrophoresis in a 1.0% agarose gel and detected with NanoVue (GE Healthcare) (He et al. 2019). cDNA was obtained from high quality RNA with PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time) (Takara, Dalian).
Quantitative real-time PCR (qRT-PCR) was performed with an ABI StepOnePlusTM Real-Time PCR System (Applied Biosystems, USA) and SYBR Premix Ex Taq™ II (TaKaRa Biotech Co., Dalian, China). Following conditions were used for qRT-PCR: 95 °C for 30 s, 40 cycles of 95 °C for 5 s, 58 °C for 11 s, and 72 °C for 30 s, and a final step of 5 min at 72 °C. Housekeeping genes of 16S ribosomal RNA (16S rRNA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phosphoglycerate kinase (PGK), recombinase A (recA), L-lactate dehydrogenase (Ldh), and DNA-directed RNA polymerase subunit beta (rpoB) were selected as internal control genes. Gene-specific primers used for qRT-PCR are shown in Table 1. Then, experimental data were further analyzed using Ct (2−ΔΔCT) model (Livak and Schmittgen 2001). Meanwhile, Ct raw data outputs were analyzed on EXCEL-based program BestKeeper and with geNorm program.
Codon optimization analysis and plasmid constructs
The codon optimization analysis (amino acid sequence invariance) was performed on the basis of codon adaptation index (CAI), GC content, restriction sites and cis-acting elements. In addition, Trx and His tags were added to the optimized phr in following order: ATG-Trx-His, tag-KpnI-TEV, protease, site-phr-Stop, codon-HindIII (GenScript Biotech Corp, Nanjing, China). The final sequence containing phr was inserted into expression vector pET-32a( +), which was then transformed to E. coli BL21 (DE3) by screening and a positive clone with high expression was identified. The sequence of final constructed recombinant strain BL21(DE3)-pET-32a( +)-phr was verified again and then preserved for subsequent PHR expression.
PHR expression, purification and detection
BL21(DE3)-pET-32a( +)-phr was inoculated into a 1-L Erlenmeyer flask containing 300 mL of liquid LB medium and 100 μg·mL−1 ampicillin, followed by incubation at 37 °C with shaking at 220 rpm. When OD600 reached 0.6 ~ 0.8, isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, and the culture was incubated at 15 °C for 16 h or at 37 °C for 4 h with shaking at 150 rpm to induce PHR expression, after which cells were collected by centrifugation. BL21(DE3)-pET-32a( +) without IPTG was used as a control.
The collected bacterial cells were suspended in lysis buffer 1 (150 mM NaCl, 0.5 mM Phenylmethanesulfonyl fluoride and 25 mM Tris, pH 8.0), disrupted by ultrasonication (JY88-IIN, Ningbo Scientz Biotechnology, China), and then centrifuged at 12,000×g for 20 min at 4 °C. At this point, supernatant and cell sediments in cell lysate were collected. Cell sediments were resuspended with lysis buffer 2 (7 M guanidine hydrochloride, 50 mM Tris, 300 mM NaCl, 0.1% Triton X-100, pH 8.0), and then further purified with Ni–NTA resin (TaKaRa BIO INC., Beijing, China) to harvest PHR protein. After renatured via a dialysis bag of 25 kDa in dialysis solution (50 mM Tris, 300 mM NaCl, pH 8.0) at 4 °C, purified protein were further concentrated to reconstituted for functional analysis. Protein expression and detection were determined using Mass Spectrometry (MS) analysis (Sangon, China), SDS-PAGE and western blot. The primary antibody used for western blot was an anti-His antibody (GenScript Biotech Corp, Nanjing, China).
In vivo photorepairing activity of PHR
Depending on pretest, BL21(DE3)-pET-32a( +) and BL21(DE3)-pET-32a( +)-phr in logarithmic phase were 105 fold times diluted and then steaked on LB agar plates. Experimental groups consisted of BL21(DE3)-pET-32a( +)-phr irradiated with UV-B (90 μW·cm−2) for 0, 1, 2, 5, 10, 20, or 30 min and then recovered under visible light (80 μmol·m−2 s−1) for 30 min. Control groups consisted of BL21(DE3)-pET-32a( +) subjected to the same conditions. The two groups of Petri dishes were cultured at 37 °C overnight. Survival rate was based on the number of monoclonal colonies surviving in each group under UV-B irradiation compared with survival rate without UV-B exposure: survival rate (%) = (N/Ntotal) × 100% (N: numbers of monoclonal colonies that survived under UV-B irradiation; Ntotal: numbers of monoclonal colonies without UV-B). Three parallel experiments were performed for each set of conditions.
In vitro photorepairing activity of PHR
An oligo (dT)16 (′5-TTT TTT TTT TTT TTT T-3′) solution was exposed to 100 μW·cm−2 of UV-B lamps (Beijing Normal University, Beijing, China) for 9 h to obtain CPDs substrates. OD260 values for oligo (dT)16 were measured at 0, 3, 5.5, 7.5 and 9 min. Meanwhile, repair buffer (50 mM Tris–HCl, 100 mM NaCl, 1 mM EDTA, 10% glycerol, and 1 mM dithiothreitol, pH 7.6) was prepared. In vitro photorepairing activity of PHR was tested (total 300 μL) by adding 1 μM purified PHR to prepared 200 μL repair buffer and 5 μM UV-oligo (dT)16. The mixture was placed in the dark for 15 min, and then exposed to blue light (450 nm, 10 nm FWHM, 50 μmol·m−2 s−1). OD260 value of the initial UV-oligo (dT)16 and repair buffer was set as 0. Then, OD260 values were measured at 0, 2, 10, 40, 60, and 100 min to evaluate UV-induced DNA damage repair activity of the purified PHR. In the control group, purified PHR was replaced by bovine serum albumin (BSA) (Li et al. 2015; Mu et al. 2005).
Statistical analysis
The statistical tests of LSD were carried out in SPSS 19.0, and p values of < 0.05 were considered as being significant.
Results
Real-time PCR analysis of phr mRNA expression
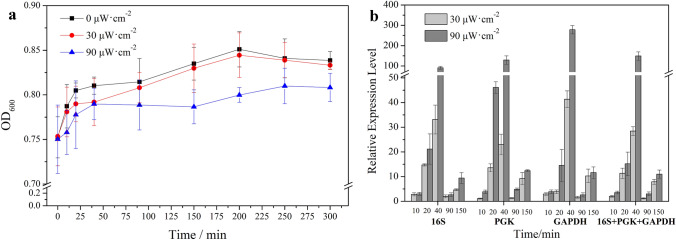
After analyzed by Ct (2−ΔΔCT) model, EXCEL-based program BestKeeper and geNorm program, 16S rRNA, GAPDH, PGK were finally suitable for normalization. Figure 1a exhibits the growth states of Rhodococcus sp. NJ-530 with (30 μW·cm−2 or 90 μW·cm−2) or without UV-B irradiation. In contrast to normal condition, UV-B irradiation inhibited Rhodococcus sp. NJ-530 growth in a dose-dependent manner. As shown in Fig. 1b, the relative mRNA expression of phr in Rhodococcus sp. NJ-530 presented different variations in response to different UV-B stresses. Transcriptional expression level of phr gradually increased, reaching maximum values (28.48 and 149.17, respectively) at 40 min under 30 μW·cm−2 and 90 μW·cm−2 UV-B irradiation. In addition, during experimental period, phr was expressed at significantly higher levels in Rhodococcus sp. NJ-530 exposed to a stronger UV-B irradiation (90 μW·cm−2) than a lower UV-B radiation (30 μW·cm−2). The expression levels then decreased but remained in a state of upregulation.
Fig. 1.
Growth curve (a) and phr transcriptional expression levels (b) of Rhodococcus sp. NJ-530 under different UV-B radiation intensities
phr cloning, bioinformatics analysis, and codon optimization
The 1146 bp length of phr predicted from the genome of Rhodococcus sp. NJ-530 was successfully amplified and verified with molecular techniques and then submitted to GenBank under accession number MH844561. phr sequence encoded a polypeptide of 381 amino acids (Supplementary Fig. 1). pI and Mw of the PHR were 5.97 and 43.06 kDa, respectively. Figure 2 depicts tertiary structure of PHR, where a DNA photolyase from E. coli was used as a template (Park et al. 1995). Phylogenetic tree of CPF (Supplementary Fig. 2) was divided into eight branches of CPD Class I PHR, CPD Class II PHR, CPD Class III PHR, Plant PHR2, (6–4) prokaryotic PHR, (6–4) eukaryotic PHR, DASH (Drosophila, Arabidopsis, Synechocystis, Homo) CRY, and Plant CRY. PHR of Rhodococcus sp. NJ-530 belongs to CPD Class I PHRs. Finally, the total number of base pairs in final modified phr (Supplementary Fig. 3) with tags was 1620, and the corresponding number of amino acids was 540. Predicted pI and Mw were 6.30 and 60.24 kDa.
Fig. 2.
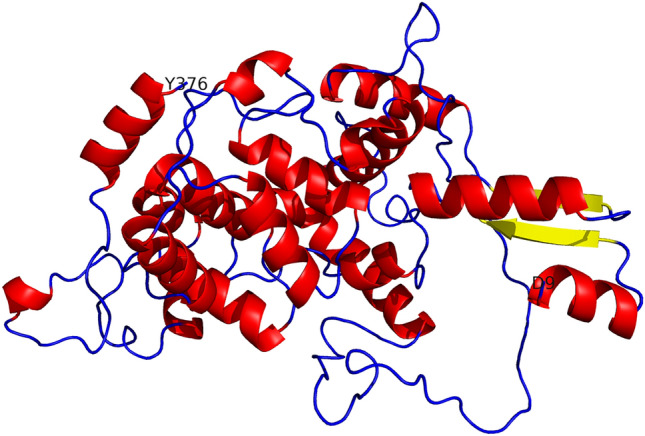
Tertiary structure of PHR from Rhodococcus sp. NJ-530. Helix was shown in red, beta sheet in yellow, and others in blue
Fig. 3.
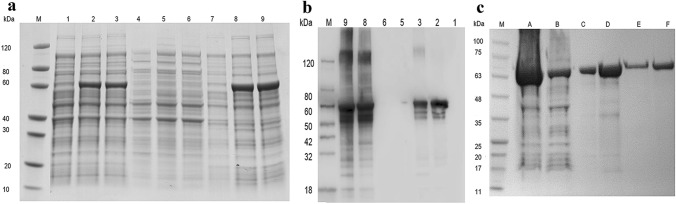
SDS-PAGE (a) and western blot (b) analysis for total protein and SDS-PAGE (c) analysis for purified PHR expressed in BL21(DE3)-pET-32a( +)-phr (Lane M: protein marker; Lane 1: cell lysate of vector pET-32a without induction; Lane 2: cell lysate with induction for 16 h at 15 °C; Lane 3: cell lysate with induction for 4 h at 37 °C; Lane 4: supernatant of cell lysate of vector pET-32a without induction; Lane 5: supernatant of cell lysate with induction for 16 h at 15 °C; Lane 6: supernatant of cell lysate with induction for 4 h at 37 °C; Lane 7: sediments of cell lysate of vector pET-32a without induction; Lane 8: sediments of cell lysate with induction for 16 h at 15 °C; Lane 9: sediments of cell lysate with induction for 4 h at 37 °C; Lane A: unpurified protein; lane B: sample effluent; lane C: 20 mM imidazole eluate; lane D: 50 mM imidazole eluate; lane E: 500 mM imidazole eluate; Lane F: 200 mM imidazole eluate)
Plasmid constructs, PHR expression, purification and in vivo photorepairing activity
Gratifyingly, as shown in Fig. 3a, b, BL21(DE3)-pET-32a( +)-phr was successfully induced to express photolyase protein in the sediments of cell lysate under incubation at 15 °C for 16 h and 37 °C for 4 h. Simultaneously, the size of target protein verified by SDS-PAGE and western blotting was consistent with the predicted. Furthermore, Fig. 3c demonstrates that after Ni–NTA resin purification, PHR was separated from protein mixture, and the 200 mM imidazole eluent showed a refined PHR product. What is more, MS analysis indicated that the coverage of purified PHR protein and target protein sequence was 27% (Supplementary Fig. 4).
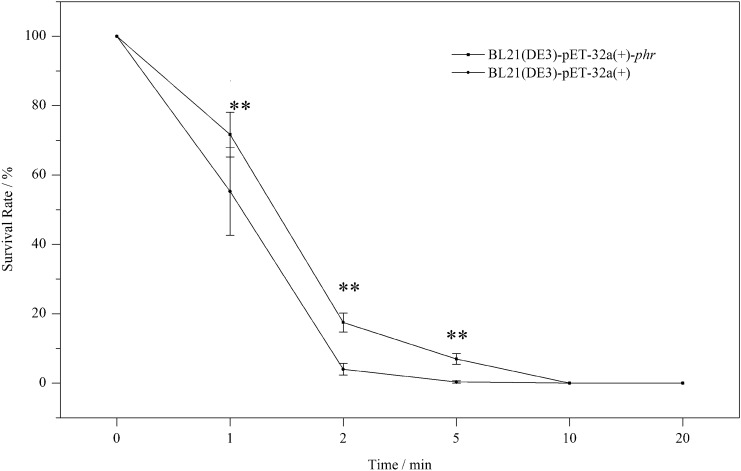
In vivo photolyase activity assays of PHR are shown in Fig. 4. As UV-B irradiation time increased, the survival rates of both BL21(DE3)-pET-32a( +)-phr and BL21(DE3)-pET-32a( +) gradually decreased. In addition, UV-B exposure of 90 μW·cm−2 for more than 10 min was completely lethal for both groups of bacteria, with mortality reaching 100%. It is worth noting that the survival rate of BL21(DE3)-pET-32a( +)-phr was higher than that of BL21(DE3)-pET-32a( +) following UV-B irradiation for up to 5 min. These preliminary results confirmed that UV-B resistance of defective E. coli was increased by exogenous phr from Rhodococcus sp. NJ-530.
Fig. 4.
Survival rate of phr-defective E. coli strain [BL21(DE3)-pET-32a( +)] and phr-transformed E. coli strain (BL21(DE3)-pET-32a( +)-phr). The method of LSD in SPSS 19.0, P < 0.05 for * and P < 0.01 for **
In vitro photorepairing activity of PHR
The results regarding in vitro photorepair activity of PHR expressed and purified from BL21(DE3)-pET-32a( +)-phr are shown in Fig. 5. In UV-oligo (dT)16 prepare phase, Fig. 5a revealed that UV-B radiation for increasing times could decrease the absorbance value of oligo(dT)16 at 260 nm from 1.21 to 0.58, which indicated that DNA damage resulted in forming CPDs. Figure 5b shows that after adding purified photolyase, damaged CPDs were largely repaired, as shown by OD260, which increased to 0.134 at 10 min, and then gradually decreased, indicating a relatively good reparation.
Fig. 5.
Effect of purified PHR on photoreparation of UV-induced DNA damage. a The absorbance at 260 nm of oligo (dT)16 irradiated with 100 μW·cm−2 UV-B. b CPDs photorepairing activity of PHR from Rhodococcus sp. NJ-530. BSA was set as control
Discussion
Photolyase/cryptochrome genes have been identified in animals, plants, bacteria, fungi and human cells over the past few decades (Kobayashi et al. 1998; Landry et al. 1997; Li et al. 2015; Malhotra et al. 1994; Tagua et al. 2015; Todo et al. 1996). Since the first report of E. coli photolyase, bacterial photolyases have been extensively studied (Sancar et al. 1984). Rhodococcus sp. NJ-530 was isolated from Antarctic ice water and belongs to the actinomycetes. Bioinformatics analysis showed that PHR from Rhodococcus sp. NJ-530 has high similarity to E. coli photolyase. Considering that CPDs account for approximately 70% of DNA lesions (Bennett et al. 2003), studying CPD photolyase has become much more important in terms of practical significance and application value. This paper reports the cloning, expression, and in vivo and in vitro activity analyses of the unique photolyase gene phr from Rhodococcus sp. NJ-530, which have not previously been reported.
DNA repair by photolyases can be summarized as three photoinduced processes: photoinitiation, photoreduction, and photoreactivation (Kim and Sundin 2001; Zhang et al. 2017). The activity of photolyase must be stimulated by light, which implies an essential requirement for photoreceptors. CPD PHRs belong to three groups: Class I (mostly in unicellular organisms), Class II (unicellular and multicellular organisms), and Class III (plants) (Ozturk 2017). In this research, results showed PHR from Rhodococcus sp. NJ-530 belonged to CPD Class I PHR.
Photolyase plays an irreplaceable role in physiological adaptation mechanisms of Rhodococcus sp. NJ-530 to adapt to high-intensity UV radiation in Antarctica. qRT-PCR analysis reflected the expression of phr in response to different intensities of UV-B radiation. Based on the results, transcriptional expression of phr was induced by stronger UV-B, which increased the expression of phr so that it was expressed at very high levels when exposed to high-intensity UV-B (90 μW·cm−2). Especially at 40 min, the relative expression level of phr was up to 149.17 times. Diurnal change in CPD photorepair activity, which is presumably regulated by transcript level of phr, may play an important role in minimizing growth inhibition due to UV-B irradiation. These research findings were consistent with previous results presented by other researchers (Kihara et al. 2004; Li et al. 2015; Takahashi et al. 2002). Compared with 40 min, phr expression level at 90 and 150 min were both decreased but still in an up-regulated level. The reason for decreasing may be related to Rhodococcus sp. NJ-530 passed logarithmic growth phase to stationary phase after 90 min (Fig. 1a).
Given the existence of codon usage bias in different organisms and tandem rare codons found in native genes that reduce the efficiency of translation or even disengage the translational machinery, a codon optimization analysis was performed to achieve expression of PHR in large quantities (Dever 2002). GC content and unfavorable peaks were optimized to prolong the half-life of mRNA. In addition, stem-loop structures were disrupted in consideration of impacting ribosomal binding and stability of mRNA. Finally, after all these attempts, PHR enzyme was expressed and purified from BL21(DE3)-pET-32a( +)-phr after induction with 0.5 mM IPTG at 15 °C for 16 h or 37 °C for 4 h.
Photolyase is important for the reparation of UV-induced gene damage and the prevention of further lethal biological effects. First, UV-resistance ability was detected in two E. coli strains, BL21(DE3)-pET-32a( +)-phr and BL21(DE3)-pET-32a( +). The survival rate of former strain was much higher than that of the latter, indicating that phr played important roles in UV radiation endurance in vivo. Finally, purified PHR was further used to explore photoreactivation activity of photolyase. Given that CPD photolyase can repair CPD damage induced by UV light, that UV-oligo (dT)16 showed a trend of increasing over time in the presence of PHR, indicated that PHR exhibited effective photoreactivation activity. In addition, survival rate was significantly different at 1, 2 and 5 min, and increased by 0.4-, 3.25- and 19-folds, respectively. However, its enzyme activity was not maintained for long. Perhaps in vitro photoremediation reaction lacks a complete, recyclable electron-transfer mechanism (Liu et al. 2015; Zhang et al. 2017). All the findings in this study provide a theoretical reference for photolyase drug development in the hopes of preventing gene damage and even human skin cancer, which enriched the exploitation and utilization of Antarctic marine resources.
Conclusion
The prevention of UV-induced DNA damage and the ability to maintain stability and integrity of genetic material are essential for living organisms, particularly organisms living in areas with strong UVR. Therefore, we first isolated a strain of Rhodococcus sp. NJ-530 from Antarctic ice to explore the repair activity of photolyase. In this study, we expressed and purified PHR in E. coli (DE3). Simultaneously, in vitro and in vivo analysis showed considerable UV-induced DNA damage repair activities of purified PHR, which provides further insight supporting the use of photolyase in the treatment of photoimmunology and photocarcinogenesis.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Supplementary Fig. 1 Protein functional sites prediction of the amino acids sequence of Rhodococcus sp. NJ-530 PHR (TIF 73 KB)
Supplementary file2 Supplementary Fig. 2 Phylogenetic tree of CPF. PHR from Rhodococcus sp. NJ-530, shown in red box, belongs to CPD Class I PHRs (TIF 3408 KB)
Supplementary file3 Supplementary Fig. 3 Gene sequence of phr from Rhodococcus sp. NJ-530, before and after codon optimization for the next PHR expression (TIF 1723 KB)
Supplementary file4 Supplementary Fig. 4 Protein sequence of PHR from Rhodococcus sp. NJ-530. Peptides matched the results of mass spectrometry (MS) shown in bold red (TIF 356 KB)
Author contributions
Conceptualization: CQ and JM; methodology: YH, CQ and JM; formal analysis and investigation: YH, LZ; writing—original draft preparation: YH; writing—review and editing: YH; funding acquisition: CQ and JM; supervision: CQ and JM.
Funding
This study was funded by the National Key Research and Development Program of China (2018YFD0900705), the China Ocean Mineral Resources R&D Association (DY135-B2-14), the Natural Science Foundation of China (41576187), the Natural Science Foundation of Shandong (ZR2019BD023), Key Research and Development Program of Shandong Province (2018GHY115034, 2018GHY115039), Ningbo Public Service Platform for High-Value Utilization of Marine Biological Resources (NBHY-2017-P2), and Youth Fund Project of Shandong Natural Science Foundation (ZR2017QD008).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Changfeng Qu, Email: cfqu@fio.org.cn.
Jinlai Miao, Email: miaojinlai@fio.org.cn.
References
- Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J Photochem Photobiol B. 2001;63:8–18. doi: 10.1016/S1011-1344(01)00198-1. [DOI] [PubMed] [Google Scholar]
- Bais AF, McKenzie RL, Bernhard G, Aucamp PJ, Ilyas M, Madronich S, Tourpali K. Ozone depletion and climate change: impacts on UV radiation. Photochem Photobiol Sci Off J Eur Photochem Assoc Eur Soc Photobiol. 2015;14:19–52. doi: 10.1039/c4pp90032d. [DOI] [PubMed] [Google Scholar]
- Bennett CJ, Webb M, Willer DO, Evans DH. Genetic and phylogenetic characterization of the type II cyclobutane pyrimidine dimer photolyases encoded by Leporipoxviruses. Virology. 2003;315:10–19. doi: 10.1016/S0042-6822(03)00512-9. [DOI] [PubMed] [Google Scholar]
- Berardesca E, Bertona M, Altabas K, Altabas V, Emanuele E. Reduced ultraviolet-induced DNA damage and apoptosis in human skin with topical application of a photolyase-containing DNA repair enzyme cream: clues to skin cancer prevention. Mol Med Rep. 2012;5:570–574. doi: 10.3892/mmr.2011.673. [DOI] [PubMed] [Google Scholar]
- Carducci M, Pavone PS, De Marco G, Lovati S, Altabas V, Altabas K, Emanuele E. Comparative effects of sunscreens alone vs sunscreens plus DNA repair enzymes in patients with actinic keratosis: clinical and molecular findings from a 6-month. Randomized Clin Study J Drugs Dermatol JDD. 2015;14:986–990. [PubMed] [Google Scholar]
- Dahms HU, Lee JS. UV radiation in marine ectotherms: molecular effects and responses. Aquat Toxicol (Amsterdam, Netherlands) 2010;97:3–14. doi: 10.1016/j.aquatox.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Dehez F, Gattuso H, Bignon E, Morell C, Dumont E, Monari A. Conformational polymorphism or structural invariance in DNA photoinduced lesions: implications for repair rates. Nucl Acids Res. 2017;45:3654–3662. doi: 10.1093/nar/gkx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/S0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Gill SS, Anjum NA, Gill R, Jha M, Tuteja N. DNA damage and repair in plants under ultraviolet and ionizing radiations. Sci World J. 2015;2015:250158. doi: 10.1155/2015/250158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hader DP, Sinha RP. Solar ultraviolet radiation-induced DNA damage in aquatic organisms: potential environmental impact. Mutat Res. 2005;571:221–233. doi: 10.1016/j.mrfmmm.2004.11.017. [DOI] [PubMed] [Google Scholar]
- He Y, et al. Molecular cloning and functional analysis of a Delta(12)-fatty acid desaturase from the Antarctic microalga Chlamydomonas sp. ICE-L 3 Biotech. 2019;9:328. doi: 10.1007/s13205-019-1858-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Adar S. The cartography of UV-induced DNA damage formation and DNA repair. Photochem Photobiol. 2017;93:199–206. doi: 10.1111/php.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isely N, Lamare M, Marshall C, Barker M. Expression of the DNA repair enzyme, photolyase, in developmental tissues and larvae, and in response to ambient UV-R in the Antarctic sea urchin Sterechinus neumayeri. Photochem photobiol. 2009;85:1168–1176. doi: 10.1111/j.1751-1097.2009.00566.x. [DOI] [PubMed] [Google Scholar]
- Jans J, et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol CB. 2005;15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Kageyama H, Waditee-Sirisattha R. Antioxidative, anti-inflammatory, and anti-aging properties of mycosporine-like amino acids: molecular and cellular mechanisms in the protection of skin-aging. Mar Drugs. 2019 doi: 10.3390/md17040222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara J, Moriwaki A, Matsuo N, Arase S, Honda Y. Cloning, functional characterization, and near-ultraviolet radiation-enhanced expression of a photolyase gene (PHR1) from the phytopathogenic fungus Bipolaris oryzae. Curr Genet. 2004;46:37–46. doi: 10.1007/s00294-004-0507-7. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Sundin GW. Construction and analysis of photolyase mutants of Pseudomonas aeruginosa and Pseudomonas syringae: contribution of photoreactivation, nucleotide excision repair, and mutagenic DNA repair to cell survival and mutability following exposure to UV-B radiation. Appl Environ Microbiol. 2001;67:1405–1411. doi: 10.1128/aem.67.4.1405-1411.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kanno S, Smit B, van der Horst GT, Takao M, Yasui A. Characterization of photolyase/blue-light receptor homologs in mouse and human cells. Nucl Acids Res. 1998;26:5086–5092. doi: 10.1093/nar/26.22.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laino L, Elia F, Desiderio F, Scarabello A, Sperduti I, Cota C, DiCarlo A. The efficacy of a photolyase-based device on the cancerization field: a clinical and thermographic study. J Exper Clin Cancer Res CR. 2015;34:84. doi: 10.1186/s13046-015-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Stapleton AE, Lim J, Hoffman P, Hays JB, Walbot V, Last RL. An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc Natl Acad Sci USA. 1997;94:328–332. doi: 10.1073/pnas.94.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Lee MR, Kim S, Kim JC, Park SE, Shin TY, Kim JS. Conidiogenesis-related DNA photolyase gene in Beauveria bassiana. J Invertebr Pathol. 2018;153:85–91. doi: 10.1016/j.jip.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Li C, et al. Cyclobutane pyrimidine dimers photolyase from extremophilic microalga: remarkable UVB resistance and efficient DNA damage repair. Mutat Res. 2015;773:37–42. doi: 10.1016/j.mrfmmm.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Li C, Liu S, Zhang W, Chen K, Zhang P. Transcriptional profiling and physiological analysis reveal the critical roles of ROS-scavenging system in the Antarctic moss Pohlia nutans under Ultraviolet-B radiation. Plant Physiol Biochem PPB. 2019;134:113–122. doi: 10.1016/j.plaphy.2018.10.034. [DOI] [PubMed] [Google Scholar]
- Liu Z, Wang L, Zhong D. Dynamics and mechanisms of DNA repair by photolyase. Phys Chem Chem Phys. 2015;17:11933–11949. doi: 10.1039/c4cp05286b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Malhotra K, Kim ST, Sancar A. Characterization of a medium wavelength type DNA photolyase: purification and properties of photolyase from Bacillus firmus. Biochemistry. 1994;33:8712–8718. doi: 10.1021/bi00195a012. [DOI] [PubMed] [Google Scholar]
- Malloy KD, Holman MA, Mitchell D, Detrich HW., 3rd Solar UVB-induced DNA damage and photoenzymatic DNA repair in Antarctic zooplankton. Proc Natl Acad Sci USA. 1997;94:1258–1263. doi: 10.1073/pnas.94.4.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marizcurrena JJ, Morel MA, Brana V, Morales D, Martinez-Lopez W, Castro-Sowinski S. Searching for novel photolyases in UVC-resistant Antarctic bacteria. Extremophiles. 2017;21:409–418. doi: 10.1007/s00792-016-0914-y. [DOI] [PubMed] [Google Scholar]
- Marizcurrena JJ, Martinez-Lopez W, Ma H, Lamparter T, Castro-Sowinski S. A highly efficient and cost-effective recombinant production of a bacterial photolyase from the Antarctic isolate Hymenobacter sp. UV11. Extremophiles. 2019;23:49–57. doi: 10.1007/s00792-018-1059-y. [DOI] [PubMed] [Google Scholar]
- Megna M, Lembo S, Balato N, Monfrecola G. "Active" photoprotection: sunscreens with DNA repair enzymes Giornale italiano di dermatologia e venereologia : organo ufficiale. Societa italiana di dermatologia e sifilografia. 2017;152:302–307. doi: 10.23736/s0392-0488.17.05567-5. [DOI] [PubMed] [Google Scholar]
- Mu W, Zhang D, Xu L, Luo Z, Wang Y. Activity assay of His-tagged E. coli DNA photolyase by RP-HPLC and SE-HPLC. J Biochem Biophys Methods. 2005;63:111–124. doi: 10.1016/j.jbbm.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Ozturk N. Phylogenetic and functional classification of the photolyase/cryptochrome family. Photochem Photobiol. 2017;93:104–111. doi: 10.1111/php.12676. [DOI] [PubMed] [Google Scholar]
- Park HW, Kim ST, Sancar A, Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Besaratinia A. UV wavelength-dependent DNA damage and human non-melanoma and melanoma skin cancer. Photochem Photobiol Sci Off J Eur Photochem Assoc Eur Soc Photobiol. 2012;11:90–97. doi: 10.1039/c1pp05144j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit NK, Robu M, Shah RG, Geacintov NE, Shah GM. Characterization of the interactions of PARP-1 with UV-damaged DNA in vivo and in vitro. Sci Rep. 2016;6:19020. doi: 10.1038/srep19020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puviani M, Barcella A, Milani M. Efficacy of a photolyase-based device in the treatment of cancerization field in patients with actinic keratosis and non-melanoma skin cancer Giornale italiano di dermatologia e venereologia : organo ufficiale. Societa italiana di dermatologia e sifilografia. 2013;148:693–698. [PubMed] [Google Scholar]
- Rastogi RP, Richa KA, Tyagi MB, Sinha RP. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucl Acids. 2010;1:592980. doi: 10.4061/2010/592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco V, Oppezzo O, Pizarro R, Sommaruga R, Ferraro M, Zagarese H. Ultraviolet damage and counteracting mechanisms in the freshwater copepod Boeckella poppei from the Antarctic Peninsula. Limnol Oceanogr. 2002;47:829–836. doi: 10.4319/lo.2002.47.3.0829. [DOI] [Google Scholar]
- Roldan-Arjona T, Ariza RR. Repair and tolerance of oxidative DNA damage in plants. Mutat Res. 2009;681:169–179. doi: 10.1016/j.mrrev.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Sancar A. Photolyase and cryptochrome blue-light photoreceptors. Adv Protein Chem. 2004;69:73–100. doi: 10.1016/s0065-3233(04)69003-6. [DOI] [PubMed] [Google Scholar]
- Sancar GB, Smith FW, Lorence MC, Rupert CS, Sancar A. Sequences of the Escherichia coli photolyase gene and protein. J Biol Chem. 1984;259:6033–6038. doi: 10.1016/S0021-9258(18)91118-X. [DOI] [PubMed] [Google Scholar]
- Sinha RP, Hader DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci Off J Eur Photochem Assoc Eur Soc Photobiol. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- Stege H, Roza L, Vink AA, Grewe M, Ruzicka T, Grether-Beck S, Krutmann J. Enzyme plus light therapy to repair DNA damage in ultraviolet-B-irradiated human skin. Proc Natl Acad Sci USA. 2000;97:1790–1795. doi: 10.1073/pnas.030528897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi L, Lee TH, Wahedi HM, Baek SH, Kim SY. Resveratrol-enriched rice attenuates UVB-ROS-induced skin aging via downregulation of inflammatory cascades. Oxid Med Cell Longev. 2017 doi: 10.1155/2017/8379539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagua VG, et al. Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc Natl Acad Sci USA. 2015;112:15130–15135. doi: 10.1073/pnas.1514637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Nakajima N, Saji H, Kondo N. Diurnal change of cucumber CPD photolyase gene (CsPHR) expression and its physiological role in growth under UV-B irradiation. Plant Cell Physiol. 2002;43:342–349. doi: 10.1093/pcp/pcf038. [DOI] [PubMed] [Google Scholar]
- Terashima M, Ohashi K, Takasuka TE, Kojima H, Fukui M. Antarctic heterotrophic bacterium Hymenobacter nivis P3(T) displays light-enhanced growth and expresses putative photoactive proteins. Environ Microbiol Rep. 2019;11:227–235. doi: 10.1111/1758-2229.12702. [DOI] [PubMed] [Google Scholar]
- Todo T, et al. Similarity among the Drosophila (6–4) photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–w303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang L, Zhong D. Photolyase: Dynamics and electron-transfer mechanisms of DNA repair. Arch Biochem Biophys. 2017;632:158–174. doi: 10.1016/j.abb.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Supplementary Fig. 1 Protein functional sites prediction of the amino acids sequence of Rhodococcus sp. NJ-530 PHR (TIF 73 KB)
Supplementary file2 Supplementary Fig. 2 Phylogenetic tree of CPF. PHR from Rhodococcus sp. NJ-530, shown in red box, belongs to CPD Class I PHRs (TIF 3408 KB)
Supplementary file3 Supplementary Fig. 3 Gene sequence of phr from Rhodococcus sp. NJ-530, before and after codon optimization for the next PHR expression (TIF 1723 KB)
Supplementary file4 Supplementary Fig. 4 Protein sequence of PHR from Rhodococcus sp. NJ-530. Peptides matched the results of mass spectrometry (MS) shown in bold red (TIF 356 KB)