Abstract
C1q/TNF-related protein 1 (CTRP1) is an endocrine factor with metabolic, cardiovascular, and renal functions. We previously showed that aged Ctrp1-knockout (KO) mice fed a control low-fat diet develop renal hypertrophy and dysfunction. Since aging and obesity adversely affect various organ systems, we hypothesized that aging, in combination with obesity induced by chronic high-fat feeding, would further exacerbate renal dysfunction in CTRP1-deficient animals. To test this, we fed wild-type and Ctrp1-KO mice a high-fat diet for 8 mo or longer. Contrary to our expectation, no differences were observed in blood pressure, heart function, or vascular stiffness between genotypes. Loss of CTRP1, however, resulted in an approximately twofold renal enlargement (relative to body weight), ∼60% increase in urinary total protein content, and elevated pH, and changes in renal gene expression affecting metabolism, signaling, transcription, cell adhesion, solute and metabolite transport, and inflammation. Assessment of glomerular integrity, the extent of podocyte foot process effacement, as well as renal response to water restriction and salt loading did not reveal significant differences between genotypes. Interestingly, blood platelet, white blood cell, neutrophil, lymphocyte, and eosinophil counts were significantly elevated, whereas mean corpuscular volume and hemoglobin were reduced in Ctrp1-KO mice. Cytokine profiling revealed increased circulating levels of CCL17 and TIMP-1 in KO mice. Compared with our previous study, current data suggest that chronic high-fat feeding affects renal phenotypes differently than similarly aged mice fed a control low-fat diet, highlighting a diet-dependent contribution of CTRP1 deficiency to age-related changes in renal structure and function.
Keywords: aging, heart, kidney, metabolism, obesity
INTRODUCTION
Aging and obesity adversely affect various organ systems through complex mechanisms that are not well understood (10, 35, 36, 45, 47, 66). Because of the integrative nature of physiology, deterioration of one organ compartment as a consequence of aging and/or obesity has cascading effects on the function of another. Altered secretion of endocrine factors is one possible factor that underlies obesity and aging-related decline (14, 25, 37, 52), leading to common cardiovascular, renal, and metabolic disorders.
C1q/TNF-related protein 1 (CTRP1), part of a novel family of endocrine factors with metabolic functions (41, 59, 60), plays an important role in tissue crosstalk. CTRP1 belongs to the CTRP subfamily (49, 60) and the larger C1q family (16) of secretory proteins, all of which are conserved from fish to human and share a signature globular C1q domain at the COOH-terminus of the protein (27). Adipose tissue and heart are among the major tissues that express CTRP1, with other tissues such as the kidney expressing CTRP1 at a lower level (26, 59, 60). Functional characterizations uncovered major roles of CTRP1 in regulating systemic glucose and lipid metabolism in vivo (41, 59) and were corroborated by subsequent studies (20, 43). Additional studies established an important role for CTRP in modulating cardiovascular (18, 24, 64) and renal functions (22, 44). Yet, the action of CTRP1 in the heart is context dependent. In a mouse model of ischemia/reperfusion injury, CTRP1 plays a protective role, and loss of the adipokine resulted in greater disease severity (64). In the context of atherosclerosis, however, CTRP1 plays an adverse role, and its deficiency reduces disease severity in apoE-knockout (KO) mice (30). In an LPS-induced sepsis model, overexpression of CTRP1 improved cardiovascular function and survival rate by dampening inflammation and oxidative damage in the heart; conversely, CTRP1 deficiency exacerbated cardiomyopathy and reduced survival in mice (23).
In our recent study, we observed renal hypertrophy and dysfunction in aged Ctrp1-knockout (KO) mice fed a control low-fat diet (LFD). Specifically, CTRP1 contributes to renal structural integrity and function in an age- and sex-dependent manner by reducing the expression of channels, transporters, and exchangers involved in sodium and potassium reabsorption, as well as reducing the expression of genes involved in inflammation, fibrosis, and oxidative stress (44). It is known that both aging and obesity are significant factors that contribute to functional deteriorations of various organ systems. Here, we test the hypothesis that aging, in combination with diet-induced obesity, would further exacerbate the detrimental effects of CTRP1 deficiency on renal function. Contrary to our expectation, although aged and Ctrp1-KO male mice had significant renal enlargement (relative to body weight), elevated urinary protein content, and pH, we did not observe an exacerbation of renal phenotypes in aged Ctrp1-KO mice that were chronically fed a high-fat diet (HFD). The different phenotypes we observed in similarly aged mice highlight the role of diet in influencing renal phenotypes in CTRP1-deficient mice.
MATERIALS AND METHODS
Mice.
Ctrp1/C1qtnf1 KO were as previously described (43). Both wild-type (WT) and KO mice were on a C57BL6/J genetic background. All Ctrp1-KO (−/−) and WT (+/+) littermate controls were generated by intercrossing Ctrp1 heterozygous (+/−) mice. Male and female Ctrp1-KO mice and WT littermate controls were housed in polycarbonate cages on a 12-h light/dark photocycle with ad libitum access to water and food, with no more than five adult mice per cage. Mice were fed a HFD (60% kcal derived from fat; D12492, Research Diets) beginning at 6 wk of age. For high-salt loading studies, WT and Ctrp1-KO male mice were transitioned to a high-salt diet (HSD; 4% NaCl; TD.92034, Envigo Teklad Diets) for 2 wk. At the termination of the study, mice were fasted for 2 h before euthanasia. All animal protocols were approved by the Institutional Animal Care and Use Committee of The Johns Hopkins University School of Medicine (Protocol No. MO16M431).
Serum and blood chemistry analysis.
Mouse blood chemistry was analyzed using an iSTAT Chem8+ cartridge with a handheld iSTAT system (Abbott Point of Care). For iSTAT analyses, blood samples were obtained from the superficial temporal vein using lithium-heparin-coated tubes. Mouse serum was harvested by retro-orbital bleeds at the time of euthanasia from 40-wk-old HFD-fed WT and Ctrp1-KO mice.
Tissue collection.
Whole kidneys were immediately harvested from euthanized mice, flash-frozen in liquid nitrogen, and kept at −80°C until analysis.
Quantitative real-time PCR analysis.
Total RNA was isolated from whole kidney tissue, reverse transcribed, and subjected to quantitative real-time PCR analyses using SYBR green as previously described (43). Data were normalized to 18S rRNA and expressed as relative mRNA levels using the ∆∆Ct method (48). Real-time PCR primers used in this study were previously published (40, 44).
RNA-sequencing and bioinformatics analysis.
RNA-sequencing (RNA-seq) and bioinformatics analyses of age-matched (40-wk-old) WT littermates (n = 4) and Ctrp1-KO (n = 3) male kidneys were performed by Novogen. In brief, data analysis was performed using a combination of programs, including STAR (12), HTseq (2), Cufflink (55), and Novogene wrapped scripts, wherein HTSeq v0.6.1 was used to generate fragments per kilobase of transcript per million mapped reads (FPKM) values. Transcript identifiers were then updated to current HGNC/National Center for Biotechnology Information nomenclature. The raw data files’ FPKM values of 0.0 were treated as nulls and the remaining values transformed into log2 notation. Quality control examination of the raw log2 signals in box plot, histogram, and principal component analysis showed technically consistent results across the samples, except for one outlier, which was excluded from further analysis. The remaining seven samples were quantile normalized to minimize experimental noise. The two biological classes underwent differential expression analysis with the two-tailed one-way t test ANOVA using the Partek GS 7.0 analytic platform (Partek, St. Louis, MO), yielding genes’ differential expression in fold change and log2-fold change and their statistical significance in uncorrected P values. A standard deviation analysis was performed for this class-class comparison using only those transcripts that had NCBI Entrez gene annotation and a mean FPKM log2 value >0.5 in the lower-expressed class to evaluate only genes of significantly high expression. On the order of 11.4k transcripts met these criteria. High-throughput sequencing data from this study were submitted to the NCBI Sequence Read Archive under accession number GSE151154.
Pathway analysis.
For pathway analysis, we focused on NCBI-annotated genes with >2 standard deviation (SD) difference between Ctrp1 KO and WT kidneys. These transcripts were considered to have significant differential expression. This SD corresponded to a linear fold-change of approximately ±1.16 for KO versus WT, which resolved to 727 upregulated and 496 downregulated transcripts between the two genotypes. These probeset transcripts were uploaded to the Ingenuity Pathway Analysis platform (www.ingenuity.com; Qiagen, Hilden, Germany) to evaluate their functional relevance in canonical pathways. Analysis was last performed on August 16, 2020. P values for pathway selection were calculated using Fisher’s exact test, and the top 20 pathways were analyzed for each two-way comparison. All listed pathways had P < 0.01 (right-tailed).
Histology.
Kidneys (40 wk old) and hearts (60 wk old) from HFD-fed WT and Ctrp1-KO mice were fixed immediately in 10% formalin at 4°C following dissection. Fixed tissues were embedded in paraffin, sectioned, and stained with hematoxylin and eosin at the Histology Reference Laboratory at The Johns Hopkins University School of Medicine.
Electron microscopy.
Small kidney pieces (1–2 mm3) were fixed by immersion with freshly prepared electron microscopy-grade 2% paraformaldehyde, 2% glutaraldehyde in 100 mM Sorenson’s phosphate buffer containing 3 mM MgCl2, pH 7.4, and 1,144 mOsmol overnight on a cold room slow rocker. Samples were rinsed in the same buffer containing 3% sucrose, 316 mOsmol, and osmicated in 2% osmium tetroxide reduced in 105% potassium ferrocyanide for 2 h at 4°C. Tissue was then rinsed in 100 mM maleate buffer containing 3% sucrose pH 6.2 and en bloc stained in 2% uranyl acetate in maleate buffer for 1 h at 4°C in the dark. Samples were dehydrated in a graded ethanol series, brought to room temperature in 70% ethanol, and completely dehydrated in 100% ethanol. Samples were resin-embedded (Epon 812, T. Pella) after a propylene oxide transition step and further infiltrated and cured the next day. Briefly, 80-nm-thin compression-free sections were obtained with a Diatome diamond knife (35 degree). Sections were picked up on 1 × 2-mm formvar-coated copper slot grids (Polysciences) and further stained with uranyl acetate followed by lead citrate. Grids were examined on a Hitachi H-7600 transmission electron microscope operating at 80 kV. Images were digitally captured with an AMT XR 50-5 megapixel CCD camera.
Quantification of glomeruli and podocyte parameters.
Glomeruli were quantified using HFD-fed 40-wk-old WT and Ctrp1-KO mice as previously described (44). Briefly, mice were subjected to 2-h food removal before kidneys were excised, fixed overnight in 10% formalin, and stored overnight at 4°C. Samples were paraffin-embedded and sectioned at the Histology Reference Laboratory at The Johns Hopkins University School of Medicine. A rabbit monoclonal anti-p57/KIP2 antibody (Abcam) was used to detect podocyte nuclei using tissue sections mounted on an automated stainer followed by periodic acid-Schiff (PAS) counterstaining. After whole slide images were generated, 50 randomly selected glomeruli in each cortical profile and p57-positive podocytes were counted. PAS counterstain was used as the delimiter of the glomerular area, glomerular area was measured, and the podocyte number was indexed to the glomerular area (µm2). Glomerular density was determined by calculating the number of glomeruli that were not globally sclerotic per total renal cortical area. Podocyte foot process effacement on the filtering surface of open capillary loops was evaluated using image processing software (ImageJ, Viewpoint Light). Length of the outer surface of the glomerular basement membrane (GBM) was measured. Overlying foot processes were manually counted. A ratio of foot process number to GBM length was computed for each image.
Urine collection and analysis.
For urine collection, one cohort of HFD-fed 50-wk-old WT and Ctrp1-KO were housed in diuresis cages with ad libitum access to water and food (HFD). A separate cohort of 52-wk-old HFD-fed WT and Ctrp1-KO mice were subjected to HSD for 2 wk before housing in diuresis cages and given ad libitum access to water and HSD during studies. Levels of urinary sodium, potassium, and chloride were measured using an EasyLyte electrolyte analyzer (Medica Corporation). Urine osmolality measurements were performed on an Advanced Instruments 3320 single-sample micro-osmometer (Advanced Instruments). Urinalysis of pH, creatinine, urea nitrogen, calcium, total protein, glucose, and albumin were measured at the Molecular and Comparative Pathobiology Core at The Johns Hopkins University School of Medicine.
Water deprivation.
Water deprivation experiments were conducted on a separate cohort of HFD-fed 47-wk-old WT and Ctrp1-KO mice. On day 1, baseline bodyweights were measured before water was removed at 3 h before the start of the dark cycle (3:00 PM), when mice were active and feeding. The mice were then subjected to a 24-h water deprivation. On day 2, body weights were measured again at 3:00 PM, before mice were given access to water for the next 24 h. On day 3, body weights were measured again at 3:00 PM to assess the ability to recover from water depravation. Mice had ad libitum access to food during the experiment.
Blood pressure.
Tail-cuff blood pressure measurements were performed on HFD-fed 55-wk-old WT and Ctrp1-KO male mice using a BP-2000 blood pressure analysis system (Visitech Systems) as previously described (44). Briefly, mice underwent a 4-day training period before testing. During the testing week, mice had a 5-min acclimation period before blood pressure values were recorded 20 consecutive times. Only successful reads were accepted for analysis, and the results were averaged. Mice were tested daily between 3:00 and 5:00 PM for 4 days during the experiment.
Transthoracic echocardiography.
Transthoracic echocardiography was performed on 50-wk-old HFD-fed WT and Ctrp1-KO mice without sedation using a Vevo 2100 system (VisualSonics) equipped with a 40-MHz linear transducer as previously described (44).
Pulse-wave velocity.
Pulse-wave velocity was measured noninvasively on 48-wk-old HFD-fed Ctrp1-KO and WT mice with a high-frequency, high-resolution Doppler spectrum analyzer (DSPW, Indus Instruments) as previously described (7).
Multiplex quantification of serum cytokines.
Serum levels of 59 mouse cytokines were measured on 40-wk-old HFD-fed Ctrp1-KO and WT mice using a Luminex bead-based multiplex system according to the manufacturer’s protocol (MilliporeSigma). This method is sensitive and reproducible (13, 54) and was validated in multiple studies (33, 46). MCYTOMAG-70K, MECY2MAG-73K, and MSCRMAG-42K assay kits were used according to the manufacturer’s instructions (MilliporeSigma) and performed as previously described (29, 39). Standard curves were generated for each mouse cytokine, and all samples and standards were analyzed using a Luminex 200 instrument (Luminex) and XPonent 3.1 software (MilliporeSigma). Sample concentrations were determined for each of the 59 mouse cytokines relative to an appropriate six-point regression standard curve (pg/mL or ng/mL). All samples used for direct comparison or normalization were run on the same plate. For cytokines where not all samples were successfully detected in the multiplex analysis, data were analyzed when cytokine levels were detected in at least six of 10–12 samples. To enable unpaired t test analysis (where n ≥ 6), serum sample levels had to be detected in both WT and Ctrp1-KO mice. All data are reported as means ± standard error. Data were analyzed using GraphPad Prism v 7.00 for Windows (Graphpad Software).
Complete blood count analysis.
A complete blood count on blood samples from 51-wk-old HFD-fed Ctrp1-KO and WT mice was performed at the Pathology Phenotyping Core at the Johns Hopkins University School of Medicine. Blood samples were collected from the tail vein using EDTA-coated blood collection tubes (Sarstedt).
Statistical analysis.
Statistical analysis was performed with Prism 8 software (GraphPad Software, San Diego, CA). Comparisons between two groups of data were performed using two-tailed Student’s t tests with 95% confidence intervals, and ANOVA tests were used to make comparisons involving more than two groups. Values were considered statistically significant at P < 0.05 for all data. All data are presented as means ± SE.
RESULTS
No differences in cardiovascular function in aged and obese WT and Ctrp1-KO male mice.
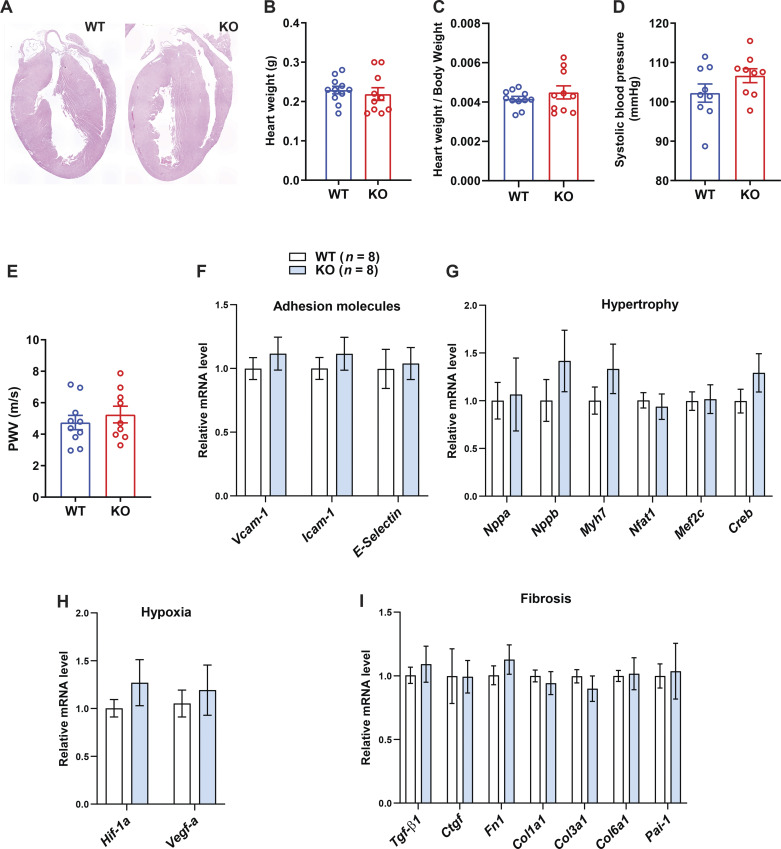
Neither heart histology, weight, relative heart weight (normalized to body weight), nor any of the functional parameters of the heart revealed by echocardiogram was significantly different between aged and obese WT and Ctrp1-KO mice (Fig. 1, A–C and Table 1). Furthermore, tail-cuff measurements of blood pressure did not reveal differences in systolic blood pressure between genotypes (Fig. 1D). Measurements of vascular stiffness by pulse wave velocity were not significantly different between WT and Ctrp1-KO male mice (Fig. 1E). None of the genes involved in cell adhesion, hypertrophy, hypoxia, and fibrosis in the heart showed significantly altered expression between genotypes (Fig. 1, F–I). To rule out potential compensation by other related CTRP family members, we measured the mRNA expression levels of CTRP2, -3, -5, -6, -7, -9, -12, and -15 in the heart and kidney and found no significant differences between genotypes (data not shown).
Figure 1.
Heart function, vascular stiffness, and blood pressure in aged and obese Ctrp1-knockout (KO) male mice. A: representative hematoxylin-eosin (H&E)-stained heart histology of aged and obese wild-type (WT) and Ctrp1-KO mice fed a high-fat diet. B: heart weight of WT (n = 11) and Ctrp1-KO (n = 10) male mice (10 mo old) fed a high-fat diet. C: relative heart weight (normalized to body weight) of WT and KO male mice. D: systolic blood pressure measured via tail-vein cuff. E: pulse wave velocity (PWV) measurements of vascular stiffness. F–I: expression of genes involved in cell adhesion (F), hypertrophy (G), hypoxia (H), and fibrosis (I) in the heart of WT (n = 8) and Ctrp1-KO (n = 8) male mice (10 mo of age) fed a high-fat diet. All data are expressed as means ± SE. Col1a1, collagen type I α 1 chain; Creb, cAMP-response element binding protein; Ctgf, connective tissue growth factor; Fn1, fibronectin 1; Hif-1a, hypoxia inducible factor 1α; Icam-1, intercellular adhesion molecule; Mef2c, myocyte enhancer factor 2C; Myh7, myosin heavy chain β; Nfat1, nuclear factor of activated T-cells 1; Nppa, natriuretic peptide A; Nppb, natriuretic peptide B; Pai-1, plasminogen activator inhibitor 1; Tgf-β1, transforming growth factor-β1; Vcam-1, vascular adhesion molecule; Vegf-a, vascular endothelial growth factor α.
Table 1.
Echocardiogram, tail-vein cuff blood pressure, and pulse-wave velocity analyses of aged and obese Ctrp1 WT and Ctrp1-KO male mice (12 mo old) fed a high-fat diet
| Variable | WT (n = 11) | KO (n = 12) | P |
|---|---|---|---|
| LVIDd, mm | 3.23 ± 0.083 | 3.295 ± 0.074 | 0.564 |
| LVIDs, mm | 1.639 ± 0.083 | 1.745 ± 0.073 | 0.35 |
| IVSd, mm | 1.234 ± 0.024 | 1.186 ± 0.016 | 0.119 |
| LVPWd, mm | 1.209 ± 0.019 | 1.259 ± 0.023 | 0.126 |
| HR, beats/min | 658.6 ± 14.96 | 640 ± 13.01 | 0.355 |
| FS, % | 49.54 ± 1.467 | 47.23 ± 1.284 | 0.248 |
| EF, % | 74.32 ± 1.483 | 71.97 ± 1.377 | 0.258 |
| LV mass | 157.3 ± 4.97 | 162.4 ± 5.683 | 0.515 |
Values are means ± SE; n, number of mice. EF, ejection fractionation; FS, fractional shortening; HR, heart rate; IVSD, inter-ventricular septal thickness at end diastole; LV mass, left ventricle mass; LVIDd, left ventricular internal dimension at end diastole; LVIDs, left ventricular internal dimension at end systole; LVPWd, left ventricular posterior wall thickness at end diastole.
Relative kidney enlargement in aged and obese Ctrp1-KO male, but not female mice.
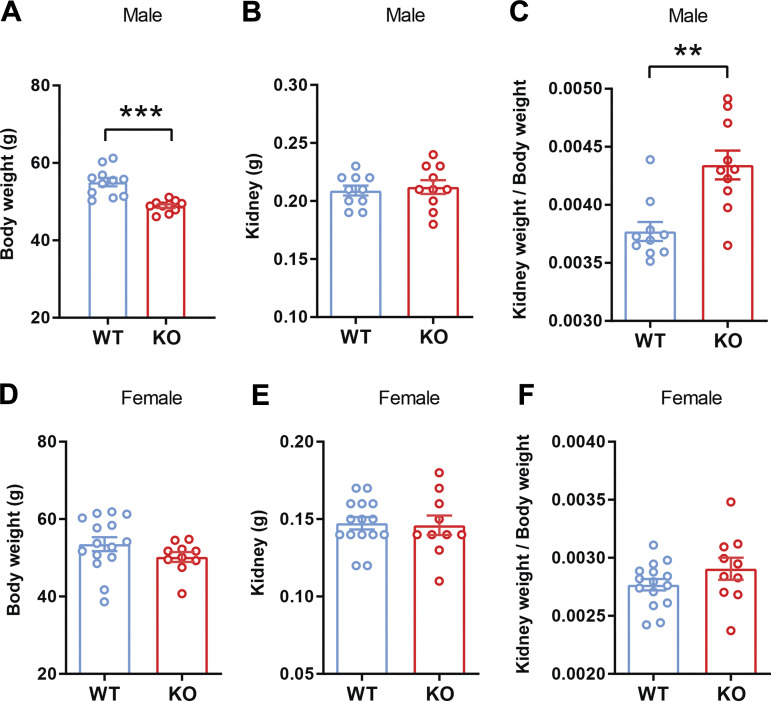
In a previous study (44), we reported that male Ctrp1-KO mice fed LFD develop renal hypertrophy due to an increase in kidney weight without a change in body weight. However, in this study, we found that the body weight of aged and obese Ctrp1-KO male mice fed HFD was lower compared with WT littermates (Fig. 2A). Although the absolute weight of the kidney was not different between genotypes (Fig. 2B), its relative weight (normalized to body weight) was significantly increased in aged and obese Ctrp1-KO male mice relative to WT controls (Fig. 2C). Relative kidney enlargement was consistently observed in multiple cohorts of aged and obese Ctrp1-KO male mice. In contrast to male mice, neither body weight, absolute kidney weight, nor relative kidney weight (normalized to body weight) was significantly different between aged and obese Ctrp1-KO female mice and WT littermates (Fig. 2, D–F).
Figure 2.
Aged and obese Ctrp1-knockout (KO) male mice have increased kidney weight relative to body weight. A: body weight of wild-type (WT; n = 11) and Ctrp1-KO (n = 10) male mice (10-mo old). B: kidney weight of WT (n = 11) and Ctrp1-KO (n = 10) male mice (10-mo old) fed a high-fat diet (HFD). C: kidney weight-to-body weight ratio of HFD-fed aged male mice. D: body weight of WT (n = 15) and Ctrp1-KO (n = 10) female mice (10-mo old). E: kidney weight of WT (n = 15) and Ctrp1-KO (n = 10) female mice (10-mo old) fed an HFD. F: kidney weight-to-body weight ratio of HFD-fed aged female mice. All data are expressed as means ± SE. **P < 0.01; ***P < 0.001 (2-tailed Student’s t tests).
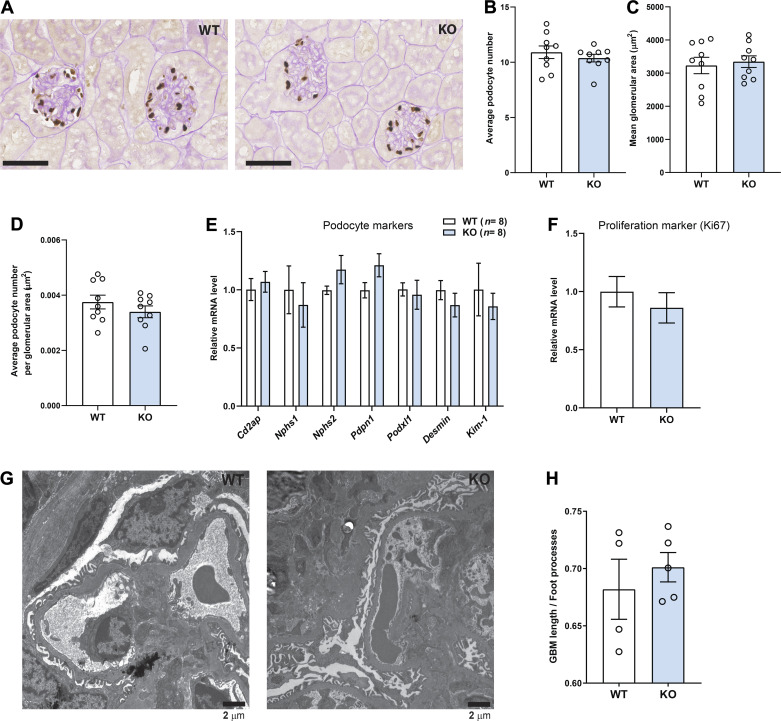
No differences in glomerular size, podocyte number, or podocyte foot process effacement in aged and obese WT and Ctrp1-KO male mice.
Histological quantification of renal sections revealed no difference in podocyte number, glomeruli size or area, or average number of podocytes per glomerular area between WT and Ctrp1-KO mice (Fig. 3, A–D). Expression of multiple podocyte marker genes did not differ between genotypes (Fig. 3E). Expression of Ki67 (a marker of cell proliferation) was also not different between genotypes (Fig. 3F). Podocyte foot process was indexed to the length of the GBM based on kidney electron micrographs of WT and Ctrp1-KO mice (Fig. 3G). A total of 49 measurements in WT (n = 4 mice) and 42 measurements in Ctrp1-KO (n = 5 mice) mice were obtained. Average length of GBM per podocyte foot process was not significantly different between genotypes (Fig. 3H).
Figure 3.
Kidney glomerular parameters in aged and obese Ctrp1-knockout (KO) male mice. A: representative kidney histology (×40 magnification) of Ctrp1-KO male mice and wild-type (WT) littermate (10-mo old) fed a high-fat diet. Tissue sections were stained with podocyte-specific marker p57/Kip2, followed by periodic acid-Schiff (PAS) counterstaining. B–D: quantification of kidney podocyte number (B), glomeruli area (C), and podocyte number per glomerular area (D) in WT and Ctrp1-KO male mice (based on 6 random fields per slide from each mouse; n = 9 mice per genotype). Measurements were made in a blinded fashion. E–F: quantitative real-time PCR analysis of podocyte marker genes (E) and Ki67 (cell proliferation marker) (F) in the kidney of WT (n = 8) and Ctrp1-KO (n = 8) male mice (∼10 mo of age) fed a high-fat diet. Cd2ap, CD2 associated protein; Kim-1, kidney injury molecule 1; Nphs1, Nephrin; Nphs2, podocin; Pdpn1, podoplanin; Podxl1, podocalyxin-like 1. G: representative electron micrograph of WT and Ctrp1-KO glomerulus showing the glomerulus basement membrane (GBM) and podocyte foot processes. H: indexed podocyte foot processes to length of GBM. Quantifications were based on a total of 49 measurements in WT (n = 4 mice) and 42 measurements in Ctrp1-KO (n = 5 mice). All data are expressed as means ± SE.
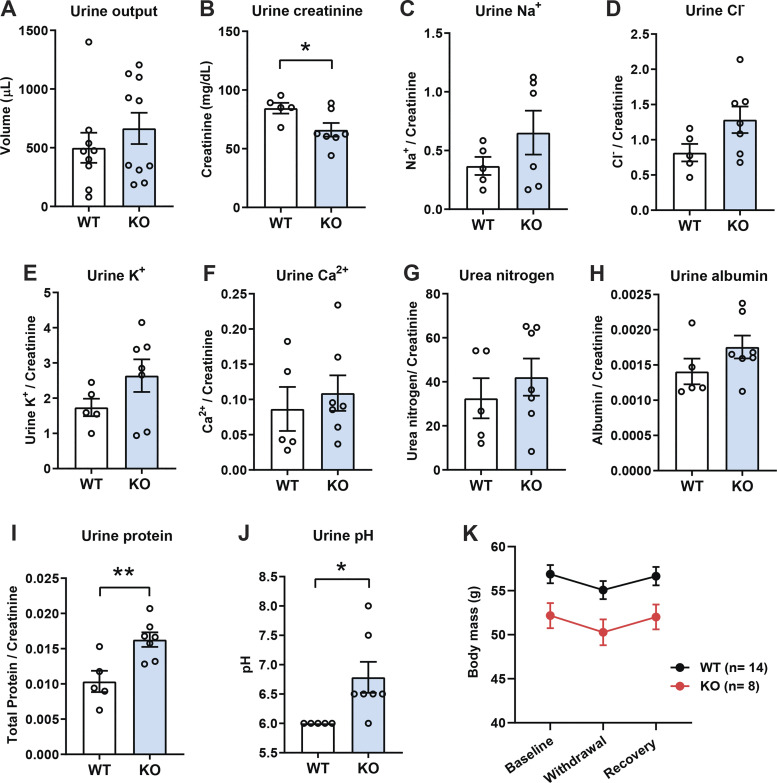
Elevated urinary total protein and pH in aged and obese Ctrp1-KO male mice.
Although urine output was not significantly different between genotypes (Fig. 4A), urine creatinine levels were reduced in aged and obese Ctrp1-KO mice relative to WT controls (Fig. 4B). Reduced urinary creatinine level likely reflects a lower body mass of the KO animals (Fig. 2A). When indexed against urine creatinine, urinary total protein content and pH were elevated in Ctrp1-KO mice whereas other analytes (Na+, Cl−, K+, Ca2+, urea nitrogen, and albumin) were not different between genotypes (Fig. 4, C–J). In contrast to urine, blood concentrations of glucose, carbon dioxide content (TCO2), Na+, K+, Cl−, ionized calcium (iCa2+), creatinine, hemoglobin, hematocrit, blood urea nitrogen, and anion gap were not different between WT and Ctrp1-KO mice (Table 2). When WT and KO mice were subjected to a 24-h water restriction followed by water replenishment, the magnitude of body mass reduction and rebound was not different between genotypes (Fig. 4K).
Figure 4.
Elevated urinary creatinine, total protein, and pH in aged and obese Ctrp1-knockout (KO) male mice. A–B: 24-h urine output (A) and urinary creatinine level (B) in wild-type (WT) and Ctrp1-KO male mice at ∼12 mo of age. C–J: 24-h urinary sodium (Na+) (C), chloride (Cl−) (D), potassium (K+) (E), calcium (Ca2+) (F), nitrogen/urea (G), albumin (H), total protein (I), and pH (J) in WT and Ctrp1-KO male mice at ∼12 mo of age. K: renal response to water deprivation. Body mass of aged and obese HFD-fed WT (n = 14) and Ctrp1-KO (n = 8) male mice before (baseline) and after a 24-h water deprivation (withdrawal), and 24-h after water was re-introduced (recovery). All data are expressed as means ± SE. WT (n = 5–8 mice) and Ctrp1-KO (n = 7–10 mice). *P < 0.05, **P < 0.01 (2-tailed Student’s t tests).
Table 2.
iSTAT blood analysis of aged and obese WT and Ctrp1-KO male mice (49 wk old) fed a high-fat diet
| Analyte | WT (n = 12) | KO (n = 14) | P |
|---|---|---|---|
| Na, mmol/L | 147.8 ± 0.505 | 147.6 ± 0.415 | 0.689 |
| K, mmol/L | 5.633 ± 0.060 | 5.757 ± 0.105 | 0.339 |
| Cl, mmol/L | 114.8 ± 0.561 | 115 ± 0.406 | 0.808 |
| iCa, mmol/L | 1.314 ± 0.009 | 1.287 ± 0.015 | 0.166 |
| TCO2, mmol/L | 24.17 ± 0.588 | 23.64 ± 0.452 | 0.48 |
| Glu, mg/dL | 207.8 ± 15.3 | 209.3 ± 9.207 | 0.933 |
| BUN, mg/dL | 23.5 ± 0.811 | 21.5 ± 0.761 | 0.085 |
| Creatinine, mg/dL | 0.2 | 0.2 | |
| Hct %PCV | 44.08 ± 0.856 | 44.71 ± 0.801 | 0.595 |
| Hb, g/dL | 14.99 ± 0.287 | 15.25 ± 0.271 | 0.514 |
| AnGap, mmol/L | 15.33 ± 0.541 | 15.62 ± 0.340 | 0.653 |
Values are means ± SE; n, number of mice. AnGAP, anion gap; BUN, blood urea nitrogen; Cl, chloride; Glu, glucose; Hct, hemotocrit; Hb, hemoglobin; iCa, ionized calcium; K, potassium; Na, sodium; TCO2, carbon dioxide content.
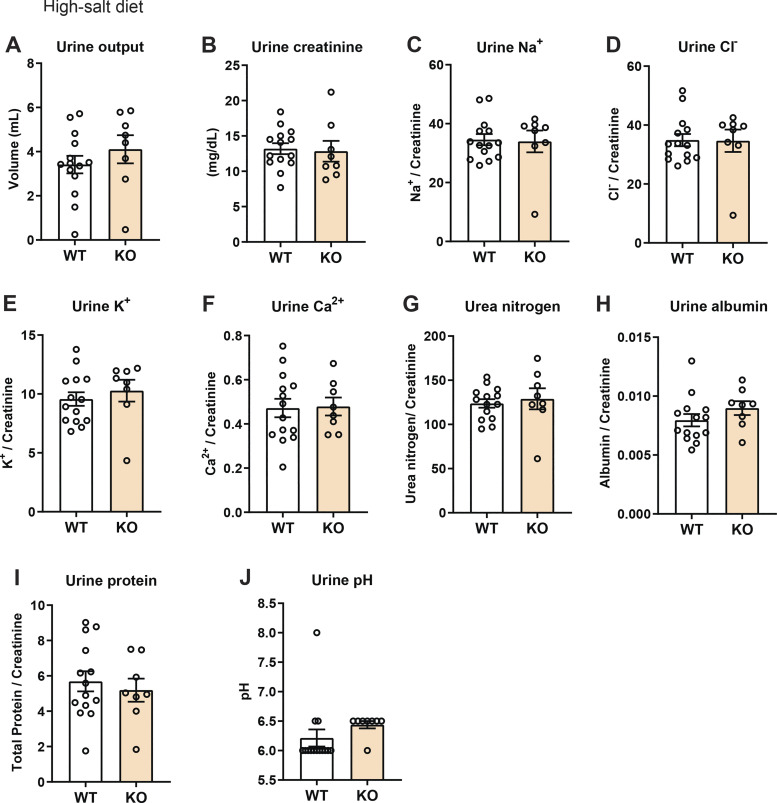
No difference in renal function after salt loading in aged and obese WT and Ctrp1-KO male mice.
Given the mild renal phenotype observed in aged and obese of WT and Ctrp1-KO male mice, we challenged the mice with HSD for 2 wk. As shown in Fig. 5, neither the urine output nor different urinary analyte levels (creatinine, Na+, Cl−, K+, Ca2+, urea nitrogen, albumin, total protein, and pH) were significantly different between WT and KO mice after HSD for 2 wk.
Figure 5.
Impact of salt loading (high-salt diet) on urine output and analytes in aged and obese Ctrp1-knockout (KO) male mice. A and B: 24-h urine output (A) and urinary creatinine level (B) in wild-type (WT) and Ctrp1-KO male mice (∼12 mo old) fed a high-salt diet for 2 wk. C–J: 24-h urinary sodium (Na+) (C), chloride (Cl−) (D), potassium (K+) (E), calcium (Ca2+) (F), nitrogen/urea (G), albumin (H), total protein (I), and pH (J) in WT and Ctrp1-KO male mice fed a high-salt diet for 2 wk. All data are expressed as means ± SE. WT (n = 14 mice) and Ctrp1-KO (n = 8 mice).
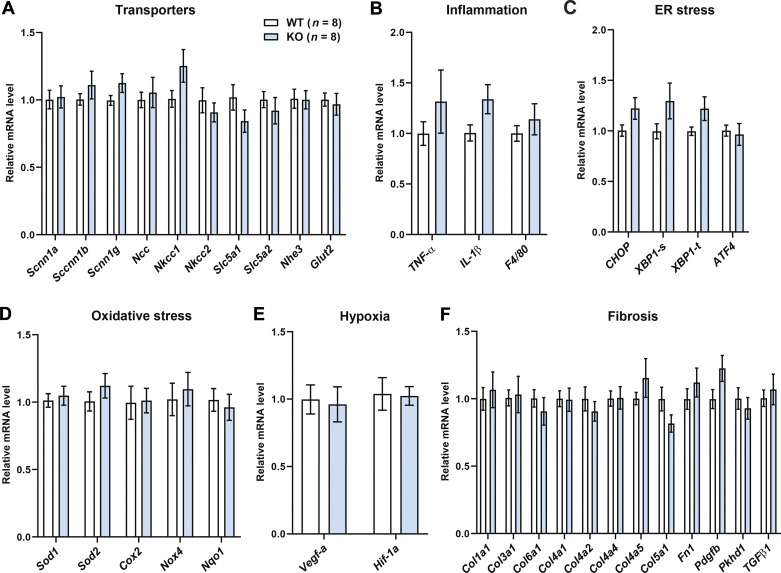
Aging and obesity affects renal gene expression in WT and Ctrp1-KO male mice.
None of the select genes involved in sodium and potassium transport, inflammation, endoplasmic reticulum and oxidative stress, hypoxia, and fibrosis in the kidney showed significantly altered expression between genotypes (Fig. 6). To further assess renal gene expression in a more global and unbiased manner, we performed RNA-seq analysis on whole kidneys isolated from aged and obese WT and Ctrp1-KO male mice. Principal component analysis revealed clear group differences between the three WT and four KO kidney samples. Of the 11,442 genes analyzed, 1,223 genes (∼10%) were differentially expressed between genotypes, with 727 genes (∼6%) significantly upregulated and 496 genes (∼4%) significantly downregulated in Ctrp1-KO kidney relative to WT controls. Tables 3 and 4 show some of the upregulated and downregulated genes involved in metabolism, signaling, transcription, solute and metabolite transport, and extracellular matrix remodeling. Pathway analysis further revealed that the top biological functions being altered based on differential gene expression between WT and KO kidney are related to metabolic disease, endocrine system, and inflammatory response, as well as to renal inflammation, proliferation, and glomerular injury (not shown).
Figure 6.
Renal gene expression in aged and obese Ctrp1-knockout (KO) male mice. Expression of genes involved in sodium and potassium reabsorption (A), inflammation (B), ER stress (C), oxidative stress (D), hypoxia (E), and fibrosis (F) in the kidney of wild-type (WT) (n = 8) and Ctrp1-KO (n = 8) male mice (∼10 mo of age) fed a high-fat diet. All data are expressed as means ± SE. Atf4, activating transcription factor 4; Chop, C/EBP homologous protein; Col, collagen; Cox2, cyclooxygenase 2; F4/80, also known as adhesion G protein-coupled receptor E1 (Adgre1); EGFn1, fibronectin 1; Glut2, glucose transporter 2 (Slc2a2); Hif-1α, hypoxia-inducible factor 1 alpha; Il-1β, interleukin 1 beta; Ncc, sodium chloride co-transporter (Slc12a3); Nhe3, Sodium hydrogen exchanger 3; Nkcc, Na-K-Cl cotransporter; Nox4, NADPH oxidase 4; Nqo1, NADPH-quinone oxidoreductase 1; Pdgfb, platelet-derived growth factor b; Scnn1, sodium channel epithelial 1; Slc5a1, solute carrier family 5 (sodium/glucose co-transporter) member 1; Sod, superoxide dismutase; Tgf-β, transforming growth factor beta; Tnf-α, tumor necrosis factor α; Vegf-a, vascular endothelial growth factor α; Xbp-1, X-box binding protein 1.
Table 3.
Genes significantly upregulated in Ctrp1 KO kidney relative to WT controls
| Gene Symbol | Protein Product | Function | Fold Change (KO/WT) | P Value |
|---|---|---|---|---|
| Pcdh10 | Protocadherin 10 | Cell adhesion | 5.846 | 0.0452 |
| Pcdhb18 | Protocadherin β-18 | Cell adhesion | 4.152 | 0.0444 |
| Ctnnd2 | Delta-catenin | Cell adhesion | 3.984 | 0.0116 |
| Megf11 | Multiple epidermal growth factor-like domains protein 11 | Cell adhesion | 3.637 | 0.0046 |
| Ceacam20 | CEA cell adhesion molecule 20 | Cell adhesion | 2.454 | 0.0383 |
| Kcna6 | Potassium voltage-gated channel subfamily A member 6 | Channel and transporter | 4.938 | 0.0383 |
| Slc39a4 | Solute carrier family 39 member 4 (Zinc transporter) | Channel and transporter | 4.311 | 0.0008 |
| Slc22a7 | Organic anion transporter 2 | Channel and transporter | 3.689 | 0.0239 |
| Slc6a12 | Sodium- and chloride-dependent betaine transporter | Channel and transporter | 2.594 | 0.0036 |
| Kcnj3 | Potassium inwardly rectifying channel subfamily J member 3 | Channel and transporter | 2.180 | 0.0442 |
| Kcnmb2 | Calcium-activated potassium channel subunit β-2 | Channel and transporter | 2.034 | 0.0375 |
| Slc6a14 | Sodium- and chloride-dependent neutral and basic amino acid transporter B(0+) | Channel and transporter | 1.997 | 0.0234 |
| Kcnk2 | Potassium two pore domain channel subfamily K member 2 | Channel and transporter | 1.910 | 0.0017 |
| Slc9a4 | Sodium/hydrogen exchanger 4 | Channel and transporter | 1.613 | 0.0283 |
| Slc14a1 | Urea transporter 1 | Channel and transporter | 1.522 | 0.0017 |
| Slc12a2 | Basolateral Na-K-Cl Symporter | Channel and transporter | 1.239 | 0.0394 |
| Alox15 | Arachidonate 15-lipoxygenase | Inflammation | 3.791 | 0.0165 |
| Mgst2 | Microsomal glutathione S-transferase 2 | Inflammation | 3.713 | 0.0175 |
| Cxcl11 | C-X-C motif chemokine 11 | Inflammation | 2.547 | 0.0457 |
| Csf3r | Granulocyte colony-stimulating factor receptor | Inflammation | 1.727 | 0.0246 |
| Il12b | Interleukin 12B | Inflammation | 1.585 | 0.0018 |
| Il17d | Interleukin 17D | Inflammation | 1.554 | 0.0037 |
| Ccr2 | C-C chemokine receptor type 2 | Inflammation | 1.472 | 0.0416 |
| Cxcr4 | C-X-C chemokine receptor type 4 | Inflammation | 1.337 | 0.0498 |
| Vcan | Versican | Matrix remodeling | 10.760 | 0.0115 |
| Lamb3 | Laminin subunit β-3 | Matrix remodeling | 3.774 | 0.0091 |
| Emilin2 | Elastin microfibril interfacer 2 | Matrix remodeling | 2.212 | 0.0200 |
| Col11a2 | Collagen type XI α 2 chain | Matrix remodeling | 1.707 | 0.0036 |
| Col6a6 | Collagen type VI α 6 chain | Matrix remodeling | 1.406 | 0.0431 |
| Col5a2 | Collagen type V α 2 chain | Matrix remodeling | 1.362 | 0.0401 |
| Aldh1a3 | Aldehyde dehydrogenase 1 family member A3 | Metabolism | 10.323 | 0.0248 |
| Hk2 | Hexokinase 2 | Metabolism | 1.992 | 0.0324 |
| Acsl6 | Acyl-CoA synthetase long-chain family member 6 | Metabolism | 1.751 | 0.0153 |
| Slc37a2 | Glucose-6-phosphate exchanger | Metabolism | 1.734 | 0.0033 |
| Fasn | Fatty acid synthase | Metabolism | 1.252 | 0.0209 |
| Adgrf1 | Adhesion G protein-coupled receptor F1 | Receptor | 4.421 | 0.0364 |
| Crhr2 | Corticotropin releasing hormone receptor 2 | Receptor | 4.265 | 0.0193 |
| Tspan11 | Tetraspanin 11 | Receptor | 3.962 | 0.0008 |
| Htr7 | 5-hydroxytryptamine receptor 7 | Receptor | 3.593 | 0.0184 |
| Tnfrsf19 | TNF receptor superfamily member 19 | Receptor | 3.218 | 0.0202 |
| Klrg2 | Killer cell lectin like receptor G2 | Receptor | 2.873 | 0.0099 |
| Gprc5a | G protein-coupled receptor class C group 5 member A | Receptor | 2.451 | 0.0244 |
| Olfr558 | Olfactory receptor 558 | Receptor | 2.010 | 0.0061 |
| Gpr183 | G protein coupled receptor 183 | Receptor | 1.899 | 0.0037 |
| Gpr27 | G protein-coupled receptor 27 | Receptor | 1.579 | 0.0220 |
| Tnfrsf14 | Tumor necrosis factor receptor superfamily member 14 | Receptor | 1.514 | 0.0438 |
| Tnfrsf23 | Tumor necrosis factor receptor superfamily member 23 | Receptor | 1.448 | 0.0351 |
| Tnfrsf25 | Tumor necrosis factor receptor superfamily member 25 | Receptor | 1.446 | 0.0279 |
| Hhip | Hedgehog interacting protein | Signaling | 10.145 | 0.0065 |
| Lrrn2 | Leucine-rich repeat neuronal protein 2 | Signaling | 9.445 | 0.0160 |
| Fzd9 | Frizzled-9 | Signaling | 6.384 | 0.0133 |
| Wnt4 | Wnt Family Member 4 | Signaling | 5.843 | 0.0077 |
| Crlf1 | Cytokine receptor-like factor 1 | Signaling | 5.264 | 0.0219 |
| Ahrr | Aryl-hydrocarbon receptor repressor | Signaling | 4.112 | 0.0063 |
| Rasal1 | RAS protein activator like 1 | Signaling | 3.958 | 0.0103 |
| Arhgap40 | Rho GTPase-activating protein 40 | Signaling | 3.523 | 0.0361 |
| Rgs1 | regulator of G protein signaling 1 | Signaling | 3.283 | 0.0138 |
| Wnt7b | Protein Wnt-7b | Signaling | 2.051 | 0.0394 |
| Lgr6 | Leucine rich repeat containing G protein-coupled receptor 6 | Signaling | 1.853 | 0.0460 |
| Styk1 | Serine/Threonine/tyrosine kinase 1 | Signaling | 1.837 | 0.0203 |
| Irs2 | Insulin receptor substrate 2 | Signaling | 1.802 | 0.0228 |
| Socs3 | Suppressor of cytokine signaling 3 | Signaling | 1.518 | 0.0212 |
| Grhl3 | Grainyhead-like transcription factor 3 | Transcription factor | 5.193 | 0.0010 |
| Foxa1 | Forkhead Box A1 | Transcription factor | 4.662 | 0.0160 |
| Npas4 | Neuronal PAS domain protein 4 | Transcription factor | 4.022 | 0.0092 |
| Zfp981 | Zinc finger protein 981 | Transcription factor | 3.279 | 0.0354 |
| Gli1 | Zinc finger protein | Transcription factor | 2.265 | 0.0253 |
| Tbx3 | T-Box transcription factor 3 | Transcription factor | 1.862 | 0.0221 |
| Cebpb | CCAAT enhancer binding protein β | Transcription factor | 1.584 | 0.0194 |
KO, knockout; WT, wild type.
Table 4.
Genes significantly downregulated in Ctrp1 KO kidney relative to WT controls
| Gene Symbol | Protein Product | Function | Fold Change (KO/WT) | P Value |
|---|---|---|---|---|
| Lilr4b | Leukocyte immunoglobulin-like receptor, subfamily B, member 4B | Immune response | −2.407 | 0.0037 |
| Ptx3 | Pentraxin 3 | Immune response | −2.387 | 0.0315 |
| Il23r | Interleukin 23 receptor | Immune response | −1.836 | 0.0142 |
| Cyp4a14 | Cytochrome P450 ω-hydroxylase 4A14 | Metabolism | −5.213 | 0.0003 |
| Hmgcs2 | 3-Hydroxy-3-methylglutaryl-CoA synthase 2 | Metabolism | −3.801 | 0.0009 |
| Slc2a7 | Glucose transporter 7 | Metabolism | −3.478 | 0.0071 |
| Ugt1a10 | UDP-glucuronosyltransferase 1–10 | Metabolism | −3.037 | 0.0209 |
| Kynu | Kynureninase | Metabolism | −1.848 | 0.0467 |
| Aldh1a1 | Aldehyde dehydrogenase 1 family, member A1 | Metabolism | −1.774 | 0.0073 |
| Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | Metabolism | −1.734 | 0.0009 |
| Acot3 | Acyl-CoA thioesterase 3 | Metabolism | −1.725 | 0.0032 |
| Cyp2j7 | Cytochrome P450, family 2, subfamily j, polypeptide 7 | Metabolism | −1.637 | 0.0079 |
| Cyp2c23 | Cytochrome P450, family 2, subfamily c, polypeptide 23 | Metabolism | −1.421 | 0.0166 |
| Aldh1a7 | aldehyde dehydrogenase family 1, subfamily A7 | Metabolism | −1.395 | 0.0170 |
| Acot1 | Acyl-CoA thioesterase 1 | Metabolism | −1.369 | 0.0149 |
| Cyp4a31 | Cytochrome P450, family 4, subfamily a, polypeptide 31 | Metabolism | −1.358 | 0.0007 |
| Lyplal1 | Lysophospholipase like 1 | Metabolism | −1.344 | 0.0167 |
| Pklr | Pyruvate kinase isozymes R/L | Metabolism | −1.331 | 0.0135 |
| Cox6b2 | Cytochrome c oxidase subunit 6B2 | Metabolism | −1.316 | 0.0211 |
| Slc25a20 | Mitochondrial carnitine/acylcarnitine carrier protein | Metabolism | −1.311 | 0.0086 |
| Fmo4 | Flavin containing dimethylaniline monoxygenase 4 | Metabolism | −1.300 | 0.0004 |
| Acsl1 | Acyl-CoA synthetase long chain family member 1 | Metabolism | −1.291 | 0.0294 |
| Pdk1 | Pyruvate dehydrogenase kinase 1 | Metabolism | −1.275 | 0.0123 |
| Acot2 | Acyl-CoA thioesterase 2 | Metabolism | −1.238 | 0.0414 |
| Plin5 | Perilipin 5 | Metabolism | −1.228 | 0.0095 |
| Acot6 | Acyl-CoA thioesterase 6 | Metabolism | −1.183 | 0.0227 |
| Acsl3 | Acyl-CoA synthetase long chain family member 3 | Metabolism | −1.160 | 0.0259 |
| Cd207 | Cluster of differentiation 207 (Langerin) | Receptor | −5.423 | 0.0029 |
| Cckbr | Cholecystokinin B receptor | Receptor | −4.259 | 0.0119 |
| Smim6 | Small integral membrane protein 6 | Receptor | −4.256 | 0.0001 |
| Olfr195 | Olfactory receptor 195 | Receptor | −2.942 | 0.0359 |
| Chrne | Acetylcholine receptor subunit ε | Receptor | −2.439 | 0.0332 |
| Gpr34 | G protein-coupled receptor 34 | Receptor | −2.016 | 0.0474 |
| Glp2r | Glucagon-like peptide 2 receptor | Receptor | −2.004 | 0.0191 |
| Cd200r4 | CD200 receptor 4 | Receptor | −1.965 | 0.0159 |
| Sirpb1c | Signal-regulatory protein β 1C | Signaling | −2.873 | 0.0441 |
| Crabp1 | Cellular retinoic acid-binding protein 1 | Signaling | −2.669 | 0.0330 |
| Ccn4 | Cellular communication network factor 4 | Signaling | −1.613 | 0.0050 |
| Pkmyt1 | Protein kinase, membrane associated tyrosine/threonine 1 | Signaling | −1.440 | 0.0466 |
| Nr1i3 | Nuclear receptor subfamily 1 group I member 3 | Transcription factor | −4.127 | 0.0184 |
| Hsf3 | Heat shock factor 3 | Transcription factor | −2.026 | 0.0183 |
| Utf1 | Undifferentiated embryonic cell transcription factor 1 | Transcription factor | −1.933 | 0.0180 |
| Pbx4 | Pre-B cell leukemia transcription factor 4 | Transcription factor | −1.918 | 0.0069 |
| Fosl1 | FOS-like 1, AP-1 transcription factor subunit | Transcription factor | −1.846 | 0.0390 |
| Hes2 | Hes family BHLH transcription factor 2 | Transcription factor | −1.724 | 0.0504 |
| Nr1i2 | Nuclear receptor subfamily 1 group I member 2 | Transcription factor | −1.480 | 0.0343 |
| Slc22a29 | Solute carrier family 22. member 29 | Transporter | −2.481 | 0.0206 |
| Slc25a18 | Mitochondrial glutamate transporter | Transporter | −1.568 | 0.0056 |
| Stra6 | Signaling receptor and transporter of retinol | Transporter | −1.557 | 0.0023 |
| Slc38a3 | Sodium-coupled neutral amino acid transporter 3 | Transporter | −1.449 | 0.0220 |
| Spns3 | Sphingolipid Transporter 3 | Transporter | −1.403 | 0.0275 |
| Slc23a3 | Solute carrier family 23 member 3 | Transporter | −1.309 | 0.0249 |
| Slc35f2 | Solute carrier family 35 member F2 | Transporter | −1.251 | 0.0249 |
| Slc7a9 | b(0, +)-type amino acid transporter 1 | Transporter | −1.180 | 0.0145 |
| Abcc2 | ATP binding cassette subfamily C member 2 | Transporter | −1.176 | 0.0324 |
| Abcc9 | ATP binding cassette subfamily C member 9 | Transporter | −1.172 | 0.0216 |
| Slc6a19 | Sodium-dependent neutral amino acid transporter B(0)AT1 | Transporter | −1.151 | 0.0345 |
| Slc25a13 | Solute carrier family 25 member 13 | Transporter | −1.146 | 0.0163 |
| Slc22a22 | Organic cation transporter | Transporter | −1.138 | 0.0092 |
KO, knockout; WT, wild type.
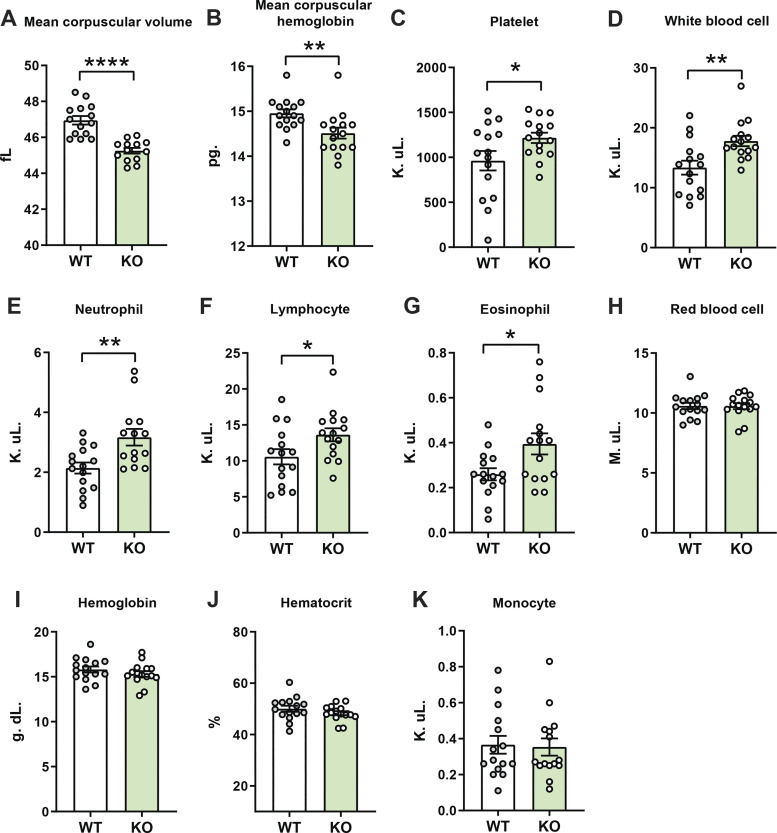
Elevated blood leukocytes in aged and obese Ctrp1-KO male mice.
Changes in blood cell count and composition are frequently observed in aging and obesity (11, 31, 56, 57, 62), and these systemic changes can contribute to altered local tissue homeostasis. For this reason, we also performed a complete blood count on aged and obese WT and Ctrp1-KO male mice. Our analyses revealed that mean corpuscular volume and hemoglobin of red blood cells were lower in Ctrp1-KO mice relative to WT controls (Fig. 7, A and B). In contrast, blood platelet, white blood cell, neutrophil, lymphocyte, and eosinophil count were significantly elevated in Ctrp1-KO mice relative to WT controls (Fig. 7, C–G). Red blood cell counts, monocytes, as well as hemoglobin and hematocrit, did not differ between genotypes (Fig. 7, H–K). Changes in circulating leukocyte levels prompted us to examine the circulating levels of cytokines, as these secretory proteins have diverse and pleiotropic effects on organ systems, including the kidney (1). We quantified the serum levels of 59 mouse cytokines, chemokines, and soluble cytokine receptors in aged (40-wk old) WT and Ctrp1-KO male mice (Table 5). Of these, only the circulating levels of chemokine CCL17 (TARC) and tissue inhibitor of metalloprotease (TIMP-1) were significantly increased in Ctrp1-KO male mice relative to WT controls (Table 5).
Figure 7.
Altered circulating levels of immune cells in aged and obese Ctrp1-knockout (KO) male mice. Complete blood count of wild-type (WT) (n = 15) and Ctrp1-KO (n = 15) male mice. At the time of sample collection, high-fat diet (HFD)-fed mice were ∼1-yr old. A: mean corpuscular volume. B: mean corpuscular hemoglobin. C: platelet. D: white blood cell. E: neutrophil. F: lymphocyte. G: eosinophil. H: red blood cell. I: hemoglobin. J: hematocrit. K: monocyte. All data are expressed as means ± SE. *P < 0.05, **P < 0.01, and ****P < 0.0001 (two-tailed Student’s t tests).
Table 5.
Summary of serum cytokine levels (pg/mL) in 40-wk-old male mice fed a high-fat diet
| Cytokine | WT (n = 11) | Ctrp1-KO (n = 9) | P |
|---|---|---|---|
| G-CSF | 555 ± 110 | 683 ± 240 | 0.613 |
| Eotaxin-1/CCL11 | 1,925 ± 228 | 2,038 ± 326 | 0.774 |
| GM-CSF | 77 ± 16 | 68 ± 15 | 0.727 |
| IFN-γ | 182 ± 16 | 174 ± 18 | 0.741 |
| IL-1α | 467 ± 89 | 408 ± 64 | 0.616 |
| IL-1β | 59 ± 14 | 75 ± 32 | 0.673 |
| IL-2 | 55 ± 22 | 46 ± 12 | 0.745 |
| IL-3 | 3.18 ± 0.85 | 2.08 ± 0.46 | 0.336 |
| IL-4 | ND | ND | ND |
| IL-5 | 57 ± 13 | 26 ± 5.4 | 0.059 |
| IL-6 | 14.7 ± 3.56 | 27 ± 15 | 0.41 |
| IL-7 | 294 ± 135 | 117 ± 46 | 0.272 |
| IL-9 | 684 ± 198 | 741 ± 298 | 0.89 |
| IL-10 | 85 ± 14 | 71 ± 8.3 | 0.446 |
| IL-11 | 109 ± 41 | 104 ± 50 | 0.953 |
| IL-12 (p40) | 63 ± 28 | 31 ± 12 | 0.394 |
| IL-12 (p70) | 59 ± 16 | 21 ± 3.17 | 0.051 |
| LIF | 4.88 ± 0.904 | 2.85 ± 0.186 | 0.25 |
| IL-13 | 132 ± 18 | 108 ± 14 | 0.342 |
| IL-15 | 199 ± 45 | 132 ± 15 | 0.248 |
| IL-16 | 1,080 ± 115 | 1,186 ± 171 | 0.602 |
| IL-17 | 9.39 ± 2.45 | 8.91 ± 1.85 | 0.9 |
| IL-17A/F | 1,208 ± 284 | 1,127 ± 370 | 0.862 |
| IL-20 | 468 ± 46 | 731 ± 129 | 0.053 |
| LIX/CXCL5 | 2,771 ± 715 | 3,494 ± 872 | 0.525 |
| IP-10/CXCL10 | 277 ± 25 | 366 ± 42 | 0.075 |
| KC/CXCL1 | 819 ± 159 | 644 ± 133 | 0.423 |
| MCP-1/CCL2 | 102 ± 14 | 129 ± 39 | 0.484 |
| MIP-1α/CCL3 | 98 ± 17 | 76 ± 4.42 | 0.353 |
| MIP-1β/CCL4 | 47 ± 6.63 | 40 ± 5.3 | 0.437 |
| M-CSF | 182 ± 56 | 138 ± 45 | 0.639 |
| MIP-2/CXCL2 | 268 ± 50 | 206 ± 37 | 0.367 |
| MIG/CXCL9 | 259 ± 19 | 383 ± 64 | 0.061 |
| RANTES/CCL5 | 30 ± 5.83 | 27 ± 5.13 | 0.731 |
| VEGF | 2.89 ± 0.36 | 2.98 ± 0.67 | 0.906 |
| TNF-α | 16.26 ± 2.51 | 37 ± 10 | 0.143 |
| EPO | 338 ± 36 | 304 ± 39 | 0.534 |
| Exodus-2/CCL21 | 35,266 ± 3,561 | 38,134 ± 2,678 | 0.543 |
| MCP-5/CCL12 | 308 ± 31 | 312 ± 31 | 0.921 |
| MIP-3β/CCL19 | 396 ± 33 | 371 ± 66 | 0.729 |
| MIP-3α/CCL20 | 128 ± 21 | 97 ± 11 | 0.25 |
| INF-b1 | 1,096 ± 321 | 1390 ± 895 | 0.792 |
| TARC/CCL17 | 48 ± 3.29 | 80 ± 16* | 0.048 |
| Fractalkine/CX3CL1 | 1,111 ± 280 | 999 ± 497 | 0.83 |
| MDC/CCL22 | 194 ± 19 | 240 ± 15 | 0.09 |
| TIMP-1 | 2,659 ± 204 | 3,846 ± 358* | 0.007 |
| sCD30 (sTNFRSF8) | 198 ± 23 | 155 ± 68 | 0.565 |
| sgp130 | 1,007 ± 103 | 1,088 ± 138 | 0.659 |
| sIL-1RI (sCD121a) | 510 ± 66 | 506 ± 79 | 0.97 |
| sIL-1RII (sCD121b) | 11,425 ± 251 | 11,321 ± 1321 | 0.932 |
| sIL-2Rα (sCD25) | 678 ± 35 | 639 ± 113 | 0.722 |
| sIL-4R (sCD124) | 2,852 ± 640 | 2,552 ± 355 | 0.704 |
| sIL-6R (sCD126) | 17,102 ± 675 | 17,601 ± 1,140 | 0.699 |
| sRAGE | 365 ± 90 | 329 ± 101 | 0.859 |
| sTNFRI (sTNFRSF1A) | 7,118 ± 247 | 7,273 ± 552 | 0.787 |
| sTNFRII (sTNFRSF1B) | 8,742 ± 612 | 7,831 ± 886 | 0.396 |
| sVEGFR1 (sFlt-1) | 2,024 ± 393 | 2,115 ± 609 | 0.904 |
| sVEGFR2 (sFlk-1) | 46,974 ± 3,913 | 46,296 ± 4,296 | 0.908 |
| sVEGFR3 (sFlt-4) | 77,965 ± 2,478 | 84,424 ± 6,867 | 0.351 |
Values are average means ± SE; n, number of mice. KO, knockout; ND, cytokines below the detection limit of the assay are not detected; WT, wild type.
Cytokines with statistically significant differences (P < 0.05) between WT and Ctrp1-KO mice.
DISCUSSION
Contrary to our hypothesis and expectation, the phenotypic outcomes of aged and obese Ctrp1-KO male mice fed an HFD were in fact different, and overall milder, than similarly aged mice fed a control LFD (44). Because aging and obesity adversely affect cardiovascular and renal functions (10, 35, 36, 45, 47, 66), these changes appear to partially mask the contributions of CTRP1 deficiency to age-related changes in renal structure and function.
In mouse models of ischemia reperfusion injury and atherosclerosis, loss of CTRP1 either protects or exacerbates cardiovascular disease outcomes in pathological contexts (30, 64). Circulating levels of CTRP1 are elevated in patients with metabolic (3, 6, 19, 38, 50, 61) and cardiovascular (18, 22, 30, 51, 53, 58, 63, 65) diseases. However, it is unclear whether CTRP1 plays a role in maintaining normal cardiovascular function over the life span in non-pathological contexts. In aged (∼1-yr-old) and obese male mice lacking CTRP1, we did not observe any significant differences in heart weight (either absolute or relative to body weight), heart function and contractility, or vascular stiffness compared with WT mice. Similarly aged Ctrp1-KO mice fed a control LFD also had normal cardiovascular function and vascular stiffness compared with WT mice (44). Although CTRP1 has important cardiovascular function in pathophysiological contexts, our current and previous studies suggest that CTRP1 deficiency likely has minimal impact on normal heart function in non-pathological settings. A limitation of our study is that we assessed mice between the ages of 8 and 13 mo old (considered midlife), and we do not know if older Ctrp1-KO mice closer to the end of the normal life span (∼2 yr old) would have altered heart function relative to WT mice. In contrast to our loss-of-function mouse model, Han et al. (18) reported structural (increased heart weight and left ventricular wall thickness) changes and reduced cardiac contractility and function in aged (∼1-yr-old) transgenic mice overexpressing CTRP1. A major caveat of this finding is that CTRP1 transgene expression is under the control of the ubiquitous chicken β-actin (CAG) promoter, and the circulating level of CTRP1 in the transgenic mouse is approximately eightfold above physiologic levels seen in WT mice. Nonetheless, these data collectively suggest that CTRP1 gain-of-function may be deleterious to the heart during aging, whereas its deficiency is dispensable for maintaining heart function in mice at midlife.
In LFD-fed mice, renal enlargement (both absolute and relative to body weight) and glomerular hypertrophy were observed in aged male, but not female or young, mice lacking CTRP1 (44). This age- and sex-dependent effect was also observed in the current study where mice were fed HFD. However, only relative kidney weight (normalized to body weight) was significantly increased in the HFD-fed aged and obese Ctrp1-KO male mice compared with WT littermates. Unlike LFD-fed male mice (44), we did not observe significant differences in the extent of glomerular hypertrophy or podocyte density between genotypes. Diet-induced obesity and aging frequently disrupt podocyte structural integrity as a consequence of podocyte foot process effacement (8, 34, 47). Ultrastructural analysis of podocyte foot process indexed to glomerular basement membrane (GBM) did not reveal any significant difference between WT and Ctrp1-KO mice. Thus, loss of CTRP1 in the context of aging and diet-induced obesity only affects renal structural integrity in a subtle manner (i.e., increase in relative kidney weight).
Aging, obesity, and diabetes each contribute to the deterioration of renal function (4, 10, 42). In the absence of CTRP1, urinary creatinine was reduced, and this likely reflects a lower body mass of the KO animals relative to WT control. Urinary total protein level (indexed against creatinine level) and pH were elevated in aged and obese Ctrp1-KO male mice relative to WT controls. Other aspects of renal function such as urine output, and the levels of sodium and potassium excretion, were not different between genotypes. These renal phenotypes are different compared with similarly aged Ctrp1-KO male mice fed a control LFD. In aged Ctrp1-KO male mice consuming an LFD, we observed elevated sodium and potassium excretion due to reduced expression of channels (Scnn1-α, Scnn1-β, Scnn1-γ), transporters (Ncc, Nkcc1, Nkcc2), and exchanger (Nhe3) genes involved in sodium and potassium reabsorption (44). In addition, we observed reduced expression of genes involved in inflammation, fibrosis, and oxidative stress in Ctrp1-KO mice fed an LFD (44). In contrast, none of the same renal transporters, channels, or exchanger gene expression were significantly altered in HFD-fed Ctrp1-KO male mice compared with WT controls. Nor did we see any altered expression of same select genes involved in inflammation, ER and oxidative stress, fibrosis, or hypoxia between genotypes.
A more global and unbiased profiling of gene expression using RNA-seq, however, revealed pathways altered in HFD-fed aged and obese Ctrp1-KO mice relative to WT controls. Among these are genes that encode proteins involved in metabolism, solute and metabolite transport, signaling, transcription, inflammation, and extracellular matrix remodeling. Pathway analysis also highlighted metabolic and endocrine disorders, and inflammatory response as being the top biological functions affected by the differential renal gene expression between WT and KO animals. One limitation of the observation, however, is that it is based on differential changes in mRNA levels; whether changes in transcript levels correspond to changes in protein levels remain to be determined. Although the specific sets of genes were different, RNA-seq revealed similar metabolism-related pathways altered in LFD-fed Ctrp1-KO male mice (44). Because CTRP1 is an endocrine hormone circulating in blood, it could act directly on different cell types within the kidney to affect renal gene expression or it could affect renal gene expression indirectly through either local or systemic effects on metabolism (20, 41, 43, 59). Although adipose and heart are the two major organs that express Ctrp1 transcript (59), podocytes within the kidney cortex also express CTRP1 transcript and protein (9, 44). Whether CTRP1 produced by podocytes acts locally in a paracrine manner within the kidney remains unclear and is a limitation of this study.
When stressed with a high-salt diet, the kidney upregulates its capacity to handle excess salt to maintain normal electrolyte levels and blood pressure (15). We performed salt loading to assess whether loss of CTRP1 in the context of aging and obesity affects the kidney’s ability to respond appropriately to a rise in dietary salt content. Two weeks of HSD did not significantly alter urine output, urinary creatinine, Na+, K+, Cl−, Ca2+, urea nitrogen, albumin, total protein, or pH. In similarly aged male mice fed an LFD, salt loading had no effect on renal function compared with WT littermates (44). Independent of age and diet, our data suggest that CTRP1 is dispensable for proper renal response to excess dietary salt intake, at least not in a chronic pathological setting.
When confronted with water loss or dehydration, the kidney responds by increasing water reabsorption and decreasing urine output (21). Interestingly, CTRP1 is upregulated in skeletal muscle in response to water restriction; the upregulated expression of CTRP1 transcript and protein is thought to be caused by a dehydration-induced rise in stress hormone (18). Elevated CTRP1 in plasma in turn promotes an angiotensin II receptor (AT1R)-dependent pathway in the vasculature to prevent a drop in blood pressure in response to dehydration. Consequently, mice lacking CTRP1 have impaired ability to maintain normal blood pressure when dehydrated (18). Considering the central role of the kidney in maintaining blood volume, we assessed whether loss of CTRP1 in the context of aging and obesity alters renal response to dehydration. Water-deprived mice consistently lose body mass (due to water loss) and quickly regain their weight when water is replenished. These stereotypical changes are robust and easy to measure. Our results suggest that CTRP1 is not required for renal response to dehydration in aged and obese mice. In similarly aged Ctrp1-KO male mice fed either a control LFD or an HSD, we also did not observe any differences in renal response to dehydration despite a significant elevation of plasma CTRP1 levels induced by water restriction (44). Independent of diet, our data do not support a role for CTRP1 in blood volume control.
One interesting and unexpected finding was that mean corpuscular volume and hemoglobin were reduced and leukocyte and platelet counts were elevated in Ctrp1-KO mice fed HFD. A reduction in mean corpuscular (red blood cell) volume and mean corpuscular hemoglobin is generally associated with iron deficiency (28, 32). Since blood hemoglobin and hematocrit levels were not different between WT and Ctrp1-KO mice, the moderate but significant decrease in mean corpuscular volume and hemoglobin may reflect mild anemia since the prevalence of anemia increases with age (17), as well as with obesity (5). Future studies may be warranted to determine if CTRP1 plays a role in regulating iron metabolism over the life span and whether different types of anemia are associated with altered circulating levels of CTRP1 in humans. The counts of several types of leukocytes (neutrophil, lymphocyte, and eosinophil) were moderately but significantly elevated in aged and obese Ctrp1-KO male mice relative to WT controls. These data point to a potential alteration in the immune status of the Ctrp1-KO mice. Since various causes elevate leukocyte counts, we do not know the mechanism underlying the increase in neutrophil, lymphocyte, and eosinophil count in Ctrp1-KO mice. Nonetheless, we examined possible changes in serum cytokine levels. Measurements of 59 cytokines, chemokines, and secreted cytokine receptors only uncovered two significant changes, with CCL17 and TIMP-1 elevated in aged and obese Ctrp1-KO male mice relative to WT controls. These changes are unlikely to contribute to the generally mild renal phenotypes observed in the Ctrp1-KO mice.
Perspectives and Significance
In summary, loss of CTRP1 did not exacerbate renal dysfunction in aged and obese mice. Instead, we observed a milder renal phenotype compared with similarly aged Ctrp1-KO mice fed a low-fat diet. Relative kidney enlargement (normalized to body weight) is associated with moderate but significant reduction in urinary creatinine and increased urinary total protein and pH. One consistent phenotype revealed by RNA-seq is that CTRP1 deficiency affects genes in metabolic, solute transport, signaling, and inflammatory pathways in the kidney. Thus, depending on the diet, the contribution of CTRP1 to aging-associated changes in renal structure and function may be more pronounced or nuanced. Future studies will seek to further elucidate the context- and tissue-dependent functions of CTRP1.
GRANTS
This work was supported by grants from the National Institutes of Health (NIH) DK084171 (to G.W. Wong), DK107726 and HL128512 (to J.L. Pluznick), a postdoctoral fellowship from the American Diabetes Association 1-18-PMF-022 (to S. Rodriguez), a predoctoral fellowship from NIH F31DK116537 (to H.C. Little), and a mini-grant from the National Kidney Foundation of Maryland NKFMD2017 (to S. Rodriguez).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.R. and G.W.W. conceived and designed research; S.R., H.C.L., S.Y.T., D.C.S., M.D., and S.J. performed experiments; S.R., P.D., P.F., and C.C.T. analyzed data; S.R., A.Z.R., and G.W.W. interpreted results of experiments; S.R. prepared figures; S.R. and G.W.W. drafted manuscript; C.C.T. edited and revised manuscript; H.C.L., P.D., P.F., S.Y.T., D.C.S., M.D., C.C.T., S.J., D.E.B., J.L.P., and A.Z.R. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Djahida Bedja for help with echocardiogram studies and Andrew Wolfe for help with multiplex cytokine profiling.
Present addresses: H. C. Little, Genentech, South San Francisco, California; S. Y. Tan, Pfizer, Cambridge, Massachusetts; D. E. Berkowitz, Dept. of Anesthesiology and Perioperative Medicine, University of Alabama at Birmingham School of Medicine, Birmingham, Alabama.
REFERENCES
- 1.Akchurin OM, Kaskel F. Update on inflammation in chronic kidney disease. Blood Purif 39: 84–92, 2015. doi: 10.1159/000368940. [DOI] [PubMed] [Google Scholar]
- 2.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169, 2015. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai B, Ban B, Liu Z, Zhang MM, Tan BK, Chen J. Circulating C1q complement/TNF-related protein (CTRP) 1, CTRP9, CTRP12 and CTRP13 concentrations in type 2 diabetes mellitus: in vivo regulation by glucose. PLoS One 12: e0172271, 2017. doi: 10.1371/journal.pone.0172271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camici M, Galetta F, Abraham N, Carpi A. Obesity-related glomerulopathy and podocyte injury: a mini review. Front Biosci (Elite Ed) 4: 1058–1070, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cepeda-Lopez AC, Aeberli I, Zimmermann MB. Does obesity increase risk for iron deficiency? A review of the literature and the potential mechanisms. Int J Vitam Nutr Res 80: 263–270, 2010. doi: 10.1024/0300-9831/a000033. [DOI] [PubMed] [Google Scholar]
- 6.Chalupova L, Zakovska A, Adamcova K. Development of a novel enzyme-linked immunosorbent assay (ELISA) for measurement of serum CTRP1: a pilot study: measurement of serum CTRP1 in healthy donors and patients with metabolic syndrome. Clin Biochem 46: 73–78, 2013. doi: 10.1016/j.clinbiochem.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee S, Bedja D, Mishra S, Amuzie C, Avolio A, Kass DA, Berkowitz D, Renehan M. Inhibition of glycosphingolipid synthesis ameliorates atherosclerosis and arterial stiffness in apolipoprotein E−/− mice and rabbits fed a high-fat and -cholesterol diet. Circulation 129: 2403–2413, 2014. doi: 10.1161/CIRCULATIONAHA.113.007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen HM, Liu ZH, Zeng CH, Li SJ, Wang QW, Li LS. Podocyte lesions in patients with obesity-related glomerulopathy. Am J Kidney Dis 48: 772–779, 2006. doi: 10.1053/j.ajkd.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 9.Cho KJ, Yang Y, Lee YH, Kim S. Podocyte-specific expression of C1q/TNF-related protein 1 in mice. Korean J Phys Anthropol 26: 147–153, 2013. doi: 10.11637/kjpa.2013.26.4.147. [DOI] [Google Scholar]
- 10.Denic A, Glassock RJ, Rule AD. Structural and functional changes with the aging kidney. Adv Chronic Kidney Dis 23: 19–28, 2016. doi: 10.1053/j.ackd.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon JB, O’Brien PE. Obesity and the white blood cell count: changes with sustained weight loss. Obes Surg 16: 251–257, 2006. doi: 10.1381/096089206776116453. [DOI] [PubMed] [Google Scholar]
- 12.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21, 2013. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.duPont NC, Wang K, Wadhwa PD, Culhane JF, Nelson EL. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol 66: 175–191, 2005. doi: 10.1016/j.jri.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckel J. Myokines in metabolic homeostasis and diabetes. Diabetologia 62: 1523–1528, 2019. doi: 10.1007/s00125-019-4927-9. [DOI] [PubMed] [Google Scholar]
- 15.Esteva-Font C, Ballarin J, Fernández-Llama P. Molecular biology of water and salt regulation in the kidney. Cell Mol Life Sci 69: 683–695, 2012. doi: 10.1007/s00018-011-0858-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghai R, Waters P, Roumenina LT, Gadjeva M, Kojouharova MS, Reid KB, Sim RB, Kishore U. C1q and its growing family. Immunobiology 212: 253–266, 2007. doi: 10.1016/j.imbio.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Goodnough LT, Schrier SL. Evaluation and management of anemia in the elderly. Am J Hematol 89: 88–96, 2014. doi: 10.1002/ajh.23598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han S, Jeong AL, Lee S, Park JS, Buyanravjikh S, Kang W, Choi S, Park C, Han J, Son WC, Yoo KH, Cheong JH, Oh GT, Lee WY, Kim J, Suh SH, Lee SH, Lim JS, Lee MS, Yang Y. C1q/TNF-α-related protein 1 (CTRP1) maintains blood pressure under dehydration conditions. Circ Res 123: e5–e19, 2018. doi: 10.1161/CIRCRESAHA.118.312871. [DOI] [PubMed] [Google Scholar]
- 19.Han S, Kim JD, Lee S, Jeong AL, Park JS, Yong HJ, Boldbaatar A, Ka HI, Rhee EJ, Lee WY, Yang Y. Circulating CTRP1 levels in type 2 diabetes and their association with FGF21. Int J Endocrinol 2016: 5479627, 2016. doi: 10.1155/2016/5479627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S, Park JS, Lee S, Jeong AL, Oh KS, Ka HI, Choi HJ, Son WC, Lee WY, Oh SJ, Lim JS, Lee MS, Yang Y. CTRP1 protects against diet-induced hyperglycemia by enhancing glycolysis and fatty acid oxidation. J Nutr Biochem 27: 43–52, 2016. doi: 10.1016/j.jnutbio.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Hoenig MP, Zeidel ML. Homeostasis, the milieu intérieur, and the wisdom of the nephron. Clin J Am Soc Nephrol 9: 1272–1281, 2014. doi: 10.2215/CJN.08860813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon JH, Kim KY, Kim JH, Baek A, Cho H, Lee YH, Kim JW, Kim D, Han SH, Lim JS, Kim KI, Yoon DY, Kim SH, Oh GT, Kim E, Yang Y. A novel adipokine CTRP1 stimulates aldosterone production. FASEB J 22: 1502–1511, 2008. doi: 10.1096/fj.07-9412com. [DOI] [PubMed] [Google Scholar]
- 23.Jiang W, Li W, Hu X, Hu R, Li B, Lan L. CTRP1 prevents sepsis-induced cardiomyopathy via Sirt1-dependent pathways. Free Radic Biol Med 152: 810–820, 2020. doi: 10.1016/j.freeradbiomed.2020.01.178. [DOI] [PubMed] [Google Scholar]
- 24.Kanemura N, Shibata R, Ohashi K, Ogawa H, Hiramatsu-Ito M, Enomoto T, Yuasa D, Ito M, Hayakawa S, Otaka N, Murohara T, Ouchi N. C1q/TNF-related protein 1 prevents neointimal formation after arterial injury. Atherosclerosis 257: 138–145, 2017. doi: 10.1016/j.atherosclerosis.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Karsenty G, Olson EN. Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell 164: 1248–1256, 2016. doi: 10.1016/j.cell.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KY, Kim HY, Kim JH, Lee CH, Kim DH, Lee YH, Han SH, Lim JS, Cho DH, Lee MS, Yoon S, Kim KI, Yoon DY, Yang Y. Tumor necrosis factor-alpha and interleukin-1beta increases CTRP1 expression in adipose tissue. FEBS Lett 580: 3953–3960, 2006. doi: 10.1016/j.febslet.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 27.Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Reid KB, Sim RB, Arlaud GJ. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol 25: 551–561, 2004. doi: 10.1016/j.it.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Lam AP, Gundabolu K, Sridharan A, Jain R, Msaouel P, Chrysofakis G, Yu Y, Friedman E, Price E, Schrier S, Verma AK. Multiplicative interaction between mean corpuscular volume and red cell distribution width in predicting mortality of elderly patients with and without anemia. Am J Hematol 88: E245–E249, 2013. doi: 10.1002/ajh.23529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Little HC, Tan SY, Cali FM, Rodriguez S, Lei X, Wolfe A, Hug C, Wong GW. Multiplex quantification identifies novel exercise-regulated myokines/cytokines in plasma and in glycolytic and oxidative skeletal muscle. Mol Cell Proteomics 17: 1546–1563, 2018. doi: 10.1074/mcp.RA118.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu L, Zhang RY, Wang XQ, Liu ZH, Shen Y, Ding FH, Meng H, Wang LJ, Yan XX, Yang K, Wang HB, Pu LJ, Zhang Q, Chen QJ, De Caterina R, Shen WF. C1q/TNF-related protein-1: an adipokine marking and promoting atherosclerosis. Eur Heart J 37: 1762–1771, 2016. doi: 10.1093/eurheartj/ehv649. [DOI] [PubMed] [Google Scholar]
- 31.Mahlknecht U, Kaiser S. Age-related changes in peripheral blood counts in humans. Exp Ther Med 1: 1019–1025, 2010. doi: 10.3892/etm.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maner BS, Moosavi L. Mean corpuscular volume In: StatPearls [Internet]. Treasure Island, FL: StatPearls Publishing, 2020. https://www.ncbi.nlm.nih.gov/books/NBK545275/. [PubMed] [Google Scholar]
- 33.Moncunill G, Aponte JJ, Nhabomba AJ, Dobaño C. Performance of multiplex commercial kits to quantify cytokine and chemokine responses in culture supernatants from Plasmodium falciparum stimulations. PLoS One 8: e52587, 2013. doi: 10.1371/journal.pone.0052587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neal CR. Podocytes ... what’s under yours? (podocytes and foot processes and how they change in nephropathy). Front Endocrinol (Lausanne) 6: 9, 2015. doi: 10.3389/fendo.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.North BJ, Sinclair DA. The intersection between aging and cardiovascular disease. Circ Res 110: 1097–1108, 2012. doi: 10.1161/CIRCRESAHA.111.246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Rourke MF, Safar ME, Dzau V. The cardiovascular continuum extended: aging effects on the aorta and microvasculature. Vasc Med 15: 461–468, 2010. doi: 10.1177/1358863X10382946. [DOI] [PubMed] [Google Scholar]
- 37.Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol 11: 85–97, 2011. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan X, Lu T, Wu F, Jin L, Zhang Y, Shi L, Li X, Lin Z. Circulating complement-C1q TNF-related protein 1 levels are increased in patients with type 2 diabetes and are associated with insulin sensitivity in Chinese subjects. PLoS One 9: e94478, 2014. doi: 10.1371/journal.pone.0094478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen PS, Lei X, Seldin MM, Rodriguez S, Byerly MS, Wolfe A, Whitlock S, Wong GW. Dynamic and extensive metabolic state-dependent regulation of cytokine expression and circulating levels. Am J Physiol Regul Integr Comp Physiol 307: R1458–R1470, 2014. doi: 10.1152/ajpregu.00335.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen PS, Lei X, Wolf RM, Rodriguez S, Tan SY, Little HC, Schweitzer MA, Magnuson TH, Steele KE, Wong GW. CTRP7 deletion attenuates obesity-linked glucose intolerance, adipose tissue inflammation, and hepatic stress. Am J Physiol Endocrinol Metab 312: E309–E325, 2017. doi: 10.1152/ajpendo.00344.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson JM, Aja S, Wei Z, Wong GW. C1q/TNF-related protein-1 (CTRP1) enhances fatty acid oxidation via AMPK activation and ACC inhibition. J Biol Chem 287: 1576–1587, 2012. doi: 10.1074/jbc.M111.278333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest 124: 2333–2340, 2014. doi: 10.1172/JCI72271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez S, Lei X, Petersen PS, Tan SY, Little HC, Wong GW. Loss of CTRP1 disrupts glucose and lipid homeostasis. Am J Physiol Endocrinol Metab 311: E678–E697, 2016. doi: 10.1152/ajpendo.00087.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez S, Little HC, Daneshpajouhnejad P, Shepard BD, Tan SY, Wolfe A, Cheema MU, Jandu S, Woodward OM, Talbot CC Jr, Berkowitz DE, Rosenberg AZ, Pluznick JL, Wong GW. Late-onset renal hypertrophy and dysfunction in mice lacking CTRP1. FASEB J 34: 2657–2676, 2020. doi: 10.1096/fj.201900558RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safar ME. Arterial aging—hemodynamic changes and therapeutic options. Nat Rev Cardiol 7: 442–449, 2010. doi: 10.1038/nrcardio.2010.96. [DOI] [PubMed] [Google Scholar]
- 46.Scheler M, Irmler M, Lehr S, Hartwig S, Staiger H, Al-Hasani H, Beckers J, de Angelis MH, Häring HU, Weigert C. Cytokine response of primary human myotubes in an in vitro exercise model. Am J Physiol Cell Physiol 305: C877–C886, 2013. doi: 10.1152/ajpcell.00043.2013. [DOI] [PubMed] [Google Scholar]
- 47.Schmitt R, Melk A. Molecular mechanisms of renal aging. Kidney Int 92: 569–579, 2017. doi: 10.1016/j.kint.2017.02.036. [DOI] [PubMed] [Google Scholar]
- 48.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108, 2008. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 49.Seldin MM, Tan SY, Wong GW. Metabolic function of the CTRP family of hormones. Rev Endocr Metab Disord 15: 111–123, 2014. doi: 10.1007/s11154-013-9255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shabani P, Naeimi Khaledi H, Beigy M, Emamgholipour S, Parvaz E, Poustchi H, Doosti M. Circulating level of CTRP1 in patients with nonalcoholic fatty liver disease (NAFLD): is it through insulin resistance? PLoS One 10: e0118650, 2015. doi: 10.1371/journal.pone.0118650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen Y, Lu L, Liu ZH, Wu F, Zhu JZ, Sun Z, Zhang RY, Zhang Q, Hu J, Chen QJ, Wu ZG, Shen WF. Increased serum level of CTRP1 is associated with low coronary collateralization in stable angina patients with chronic total occlusion. Int J Cardiol 174: 203–206, 2014. doi: 10.1016/j.ijcard.2014.03.205. [DOI] [PubMed] [Google Scholar]
- 52.Stefan N, Häring HU. The role of hepatokines in metabolism. Nat Rev Endocrinol 9: 144–152, 2013. doi: 10.1038/nrendo.2012.258. [DOI] [PubMed] [Google Scholar]
- 53.Tang JN, Shen DL, Liu CL, Wang XF, Zhang L, Xuan XX, Cui LL, Zhang JY. Plasma levels of C1q/TNF-related protein 1 and interleukin 6 in patients with acute coronary syndrome or stable angina pectoris. Am J Med Sci 349: 130–136, 2015. doi: 10.1097/MAJ.0000000000000378. [DOI] [PubMed] [Google Scholar]
- 54.Tighe P, Negm O, Todd I, Fairclough L. Utility, reliability and reproducibility of immunoassay multiplex kits. Methods 61: 23–29, 2013. doi: 10.1016/j.ymeth.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578, 2012. [Erratum in Nat Protoc 9: 2513, 2014]. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valiathan R, Ashman M, Asthana D. Effects of ageing on the immune system: infants to elderly. Scand J Immunol 83: 255–266, 2016. doi: 10.1111/sji.12413. [DOI] [PubMed] [Google Scholar]
- 57.Veronelli A, Laneri M, Ranieri R, Koprivec D, Vardaro D, Paganelli M, Folli F, Pontiroli AE. White blood cells in obesity and diabetes: effects of weight loss and normalization of glucose metabolism. Diabetes Care 27: 2501–2502, 2004. doi: 10.2337/diacare.27.10.2501. [DOI] [PubMed] [Google Scholar]
- 58.Wang H, Wang R, Du D, Li F, Li Y. Serum levels of C1q/TNF-related protein-1 (CTRP-1) are closely associated with coronary artery disease. BMC Cardiovasc Disord 16: 92, 2016. doi: 10.1186/s12872-016-0266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, Revett T, Gimeno R, Lodish HF. Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem J 416: 161–177, 2008. doi: 10.1042/BJ20081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong GW, Wang J, Hug C, Tsao TS, Lodish HF. A family of Acrp30/adiponectin structural and functional paralogs. Proc Natl Acad Sci USA 101: 10302–10307, 2004. doi: 10.1073/pnas.0403760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xin Y, Lyu X, Wang C, Fu Y, Zhang S, Tian C, Li Q, Zhang D. Elevated circulating levels of CTRP1, a novel adipokine, in diabetic patients. Endocr J 61: 841–847, 2014. doi: 10.1507/endocrj.EJ14-0016. [DOI] [PubMed] [Google Scholar]
- 62.Xu X, Su S, Wang X, Barnes V, De Miguel C, Ownby D, Pollock J, Snieder H, Chen W, Wang X. Obesity is associated with more activated neutrophils in African American male youth. Int J Obes 39: 26–32, 2015. doi: 10.1038/ijo.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Y, Liu S, Zhang RY, Luo H, Chen L, He WF, Lei R, Liu MR, Hu HX, Chen M. Association between C1q/TNF-related protein-1 levels in human plasma and epicardial adipose tissues and congestive heart failure. Cell Physiol Biochem 42: 2130–2143, 2017. doi: 10.1159/000479915. [DOI] [PubMed] [Google Scholar]
- 64.Yuasa D, Ohashi K, Shibata R, Mizutani N, Kataoka Y, Kambara T, Uemura Y, Matsuo K, Kanemura N, Hayakawa S, Hiramatsu-Ito M, Ito M, Ogawa H, Murate T, Murohara T, Ouchi N. C1q/TNF-related protein-1 functions to protect against acute ischemic injury in the heart. FASEB J 30: 1065–1075, 2016. doi: 10.1096/fj.15-279885. [DOI] [PubMed] [Google Scholar]
- 65.Yuasa D, Ohashi K, Shibata R, Takeshita K, Kikuchi R, Takahashi R, Kataoka Y, Miyabe M, Joki Y, Kambara T, Uemura Y, Matsuo K, Hayakawa S, Hiramatsu-Ito M, Ito M, Ikeda N, Murohara T, Ouchi N. Association of circulating C1q/TNF-related protein 1 levels with coronary artery disease in men. PLoS One 9: e99846, 2014. doi: 10.1371/journal.pone.0099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou XJ, Rakheja D, Yu X, Saxena R, Vaziri ND, Silva FG. The aging kidney. Kidney Int 74: 710–720, 2008. doi: 10.1038/ki.2008.319. [DOI] [PubMed] [Google Scholar]