Abstract
Axonal dysfunction is a common phenotype in neurodegenerative disorders, including in amyotrophic lateral sclerosis (ALS), where the key pathological cell-type, the motor neuron (MN), has an axon extending up to a metre long. The maintenance of axonal function is a highly energy-demanding process, raising the question of whether MN cellular energetics is perturbed in ALS, and whether its recovery promotes axonal rescue. To address this, we undertook cellular and molecular interrogation of multiple patient-derived induced pluripotent stem cell lines and patient autopsy samples harbouring the most common ALS causing mutation, C9orf72. Using paired mutant and isogenic expansion-corrected controls, we show that C9orf72 MNs have shorter axons, impaired fast axonal transport of mitochondrial cargo, and altered mitochondrial bioenergetic function. RNAseq revealed reduced gene expression of mitochondrially encoded electron transport chain transcripts, with neuropathological analysis of C9orf72-ALS post-mortem tissue importantly confirming selective dysregulation of the mitochondrially encoded transcripts in ventral horn spinal MNs, but not in corresponding dorsal horn sensory neurons, with findings reflected at the protein level. Mitochondrial DNA copy number was unaltered, both in vitro and in human post-mortem tissue. Genetic manipulation of mitochondrial biogenesis in C9orf72 MNs corrected the bioenergetic deficit and also rescued the axonal length and transport phenotypes. Collectively, our data show that loss of mitochondrial function is a key mediator of axonal dysfunction in C9orf72-ALS, and that boosting MN bioenergetics is sufficient to restore axonal homeostasis, opening new potential therapeutic strategies for ALS that target mitochondrial function.
Supplementary Information
The online version of this article (10.1007/s00401-020-02252-5) contains supplementary material, which is available to authorized users.
Keywords: Amyotrophic lateral sclerosis, Axon, Energy metabolism, Frontotemporal dementia, Mitochondria, Motor neuron, Neurodegeneration
Introduction
Amyotrophic lateral sclerosis (ALS) is an incurable, rapidly progressive and fatal neurodegenerative disorder, characterised by loss of upper and lower motor neurons (MNs) [18]. Humans are the only species affected by sporadic ALS [8]. Approximately 10–20% of ALS cases are familial, of which the C9orf72 hexanucleotide repeat expansion mutation is the commonest cause [29, 81]. The finding that familial ALS is clinically and pathologically indistinguishable from sporadic ALS supports the study of monogenetic causes to better understand common pathogenic mechanisms [67]. Thus, human induced pluripotent stem cell (iPSC)-derived MN experimental platforms [84], combined with paired gene-edited isogenic control lines, provide a powerful approach, both to identify early-stage disease-driving mechanisms, and establish causality between a given mutation and phenotypes [31, 85, 91, 92]. Such in vitro disease modelling is complemented by in vivo approaches, particularly those that harness technologies interrogating molecular neuropathological signatures in human post-mortem tissue [40, 43, 92].
Although a clinical ALS phenotype that includes hypermetabolism and dyslipidaemia [32] is well recognised [103], there is limited understanding of the underlying mechanism and/or connection to MN metabolic state [13, 104, 109]. Neurons, in contrast to astrocytes (for example), are predominantly reliant upon mitochondrial oxidative phosphorylation [114]; this, coupled with the extraordinary length of MN axons—20,000 times longer than the diameter of their soma [47]—suggests a particular metabolic vulnerability of MNs to deficits in key processes involved in the maintenance of the form and function of the axon: dysfunctional axonal homeostasis. These processes include cytoskeletal dynamics and axonal transport. Both axonal growth, as well as transport of cargo driven by dynein and kinesin molecular motors, are energy demanding processes, with kinesin hydrolysing one ATP per 8 nm step [38]. Transport of mitochondrial cargo, in contrast to vesicular transport, depends on mitochondrial ATP, and not glycolysis [113]. Defective transport of cargo to the distal axon has been reported in several ALS causing mutations [1, 2, 9, 22, 28, 41, 48, 50, 64, 71, 78, 96], and not just in genes canonically involved in microtubule-based transport of cargo [70, 73, 98]. It is therefore perhaps surprising that, despite major advances in our understanding of the biology and consequence of the C9orf72 repeat expansion mutation on MNs [10], its specific impact on axonal homeostasis has been relatively understudied, particularly in human models [1, 64, 96]. It remains, for example, unknown whether C9orf72 MNs have a metabolic deficit, and whether this contributes to axonal dysfunction and selective MN vulnerability.
Against this background, we examined the cell-autonomous role of mitochondrial bioenergetics on C9orf72 MN axonal function using multiple patient-derived iPSCs combined with isogenic controls, transcriptomic analysis and human neuropathological study. We establish that C9orf72 MNs have dysfunctional axonal homeostasis, with aberrations in axonal morphology (reduced neurite length) and function (impaired fast axonal transport of mitochondrial cargo). We show that these axonal phenotypes are associated with concomitant metabolic dysfunction, owing to defective mitochondrial respiration. Unbiased RNA-sequencing revealed reduced expression of electron transport chain transcripts, encoded by mitochondrial DNA, the copy number of which was unaltered. Critically, we show, through neuropathological analysis of patient post-mortem tissue, that this transcriptomic dysregulation is selective to anterior horn spinal (motor) neurons, and is absent in dorsal horn spinal (sensory) neurons, with corresponding alterations reflected at the level of protein expression. Through manipulation of this molecular mitochondrial loss-of-function signature, leading to therapeutic rescue of the observed dysfunctional axonal homeostasis, we determine a novel causal relationship between axon dysfunction and contributory metabolic dysfunction in C9orf72-ALS.
Materials and methods
Cell culture and motor neuron generation from parental and gene-edited human iPSCs
Dermal fibroblasts from two unrelated healthy individuals (Con-1, Con-2) and three C9orf72 ALS/FTD patients (C9-1, C9-2, C9-3) harbouring the G4C2 repeat expansion in the C9orf72 gene were obtained and reprogrammed under full Ethical/Institutional Review Board approval at the University of Edinburgh, and isogenic controls were generated via CRISPR/Cas9 technology, as previously described by our group [92, 105] (Table 1). iPSCs were maintained in Matrigel® Growth Factor Reduced Basement Membrane Matrix (354230, Corning® Life Sciences)-coated plastic dishes in E8™ medium (A1517001, Gibco™ Thermo Fisher Scientific) at 37 °C and 5% CO2. Standard Giemsa banding chromosome analysis was periodically performed over the course of this study to exclude a karyotypic abnormality (The Doctors Laboratory Ltd, London), together with monthly testing of iPSC culture supernatants to exclude mycoplasma contamination using the Venor®GeM Classic detection kit (11-1050, Minerva Biolabs GmbH).
Table 1.
Details of human iPSC control and C9orf72 lines
| Line | Reprogramming method | Repeat Length | Sex | Ethnicity | Age of onset (years) | Age at skin biopsy (years) | Disease duration (months) | Diagnosis | Family history | Site of onset | Dementia |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Con-1 | Retroviral | N/A | Female | Unknown | N/A | 40 | N/A | N/A | N/A | No | |
| Con-2 | Episomal | N/A | Female | Caucasian | N/A | 56 | N/A | N/A | Psychiatric disorders | N/A | No |
| C9-1 | Sendai | c. 750 | Female | Dutch | 38 | 39 | 31 | ALS/FTD (pm confirmed) | ALS/FTD | Behavioural change, language loss, wasting of hand muscles | Yes |
| C9-2 | Retroviral | c. 638 | Male | Caucasian | 52 | 58 | 72 | ALS | Mother-dementia | Lower limb | Cognitive decline |
| C9-3 | Retroviral | c. 960 | Male | Dutch | 65 | 67 | 36 | ALS | ALS/FTD | Lower limb | No |
Spinal MN differentiation was performed using an established protocol [62] with minor modifications, yielding a highly enriched and electrophysiologically mature neuronal culture, devoid of glia, with circa 60% of cells being positive for islet-1 and islet-2 homeobox MN markers one week post platedown [92]. iPSCs were dissociated into single cells using 1X Accutase® (A6964, Sigma-Aldrich®), and neuralised as a suspension culture using dual-SMAD inhibition by SB-431542 (20 μM; 1614, Tocris® Bio-Techne), LDN-193189 (0.1 μM; S2618, Selleckchem) and potentiation with the Wnt-agonist, CHIR-99021 (3 μM; 4423, Tocris® Bio-Techne) in N2/B27 medium (0.5X Neurobasal™ [21103049, Gibco™ Thermo Fisher Scientific], 0.5X Advanced DMEM/F12 [12634028, Gibco™ Thermo Fisher Scientific], 1X Antibiotic–Antimycotic [15240062, Gibco™ Thermo Fisher Scientific], 1X GlutaMAX™ [35050061, Gibco™ Thermo Fisher Scientific], 100 μM beta-mercaptoethanol [31350010, Gibco™ Thermo Fisher Scientific], 1X B-27™ supplement [17504044, Gibco™ Thermo Fisher Scientific], 1X N-2 supplement [17502001, Gibco™ Thermo Fisher Scientific] and 10 μM l-ascorbic acid [A4403, Sigma-Aldrich®]). On day 2, neural spheres were simultaneously patterned to spinal cord identity by treating with retinoic acid (RA, 0.1 μM; R2625, Sigma-Aldrich®) and smoothened agonist (SAG, 0.5 μM; 566660, Sigma-Aldrich®), promoting caudalisation and ventralisation, respectively, along with SB-431542, LDN-193189 and CHIR-99021 in N2/B27 medium for an additional 5 days. On day 7, spheres were maintained in culture with RA and SAG, with the addition of recombinant human brain-derived neurotrophic factor (BDNF, 10 ng/ml; 248-BDB, R & D Systems® Bio-Techne) in N2/B27 medium to generate MN progenitors. From day 9, MN progenitors were cultured in day 7 medium with the addition of DAPT (10 μM; 2634, Tocris® Bio-Techne), an inhibitor of Notch signalling, for an additional 5–7 days. At day 14–16, MN spheres were dissociated using 0.05% Trypsin–EDTA (25300054, Gibco™ Thermo Fisher Scientific) and cells were plated as a monolayer onto laminin from Engelbreth-Holm-Swarm murine sarcoma basement membrane (5 μg/ml; L2020, Sigma-Aldrich®), fibronectin human plasma (10 μg/ml; F2006, Sigma-Aldrich®), and Matrigel® (1:20)-coated dishes that had been pre-treated with poly-l-ornithine bromide (100 µg/ml; P3655, Sigma-Aldrich®). Dissociated MNs were cultured in motor neuron neurotrophic factor (MN-NF) medium (1X Neurobasal, 1X Antibiotic–Antimycotic, 1X GlutaMAX™, 1X MEM Non-Essential Amino Acids solution [11140–035, Gibco™ Thermo Fisher Scientific], 100 μM beta-mercaptoethanol, 1X B-27™ supplement, 1X N-2 supplement, RA [1 μM], ascorbic acid [2.5 μM], BDNF [10 ng/ml], recombinant human glial-derived neurotrophic factor [GDNF, 10 ng/ml; 212-GD, R & D Systems® Bio-Techne], recombinant human ciliary neurotrophic factor [CNTF, 10 ng/ml; 257-NT, R & D Systems® Bio-Techne], and animal-free recombinant human insulin-like growth factor-1 [IGF-1, 10 ng/ml; AF-100–11, PeproTech®]). MN-NF medium was regularly changed every 2–3 days and supplemented with uridine/5-fluoro-2′-deoxyuridine (1 μM, U/FDU; U3003 and F0503, Sigma-Aldrich®) for at least 1 week post platedown to remove residual proliferating cells.
Overexpression of PGC1α
Briefly, FLAG-PGC1α-6xHIS (originated from Addgene [67637]) and P2A-eGFP (originated from pDONR-P2A-eGFP), were shuttled into a lentiviral backbone pLenti6-cppt-delta CMV-DEST-opre, to generate a FLAG-PGC1α-6xHIS-P2A-eGFP lentiviral construct. PGC1α overexpression lentivirus was generated as previously described [63, 76] and transduced as previously described by our group [56] in human iPSC-derived MNs at a multiplicity of infection (MOI) of 10. This achieved a mean transduction efficiency of circa 70% MNs. Vehicle control transduction was achieved by lentiviral transduction of emerald GFP in the equivalent plasmid backbone using a plasmid from Invitrogen™ (Thermo Fisher Scientific, V35520).
Immunocytochemistry and neurite outgrowth quantification
MNs plated on 30-mm glass coverslips etched by the pre-treatment of nitric acid were washed once with phosphate-buffered saline (PBS; 10010, Gibco™ Thermo Fisher Scientific) and then incubated for 20 min at room temperature with 4% paraformaldehyde (AGR1026, Agar Scientific). Thereafter, cells were washed three times with PBS, followed by incubation for 10 min at room temperature with PBS containing 0.2% Triton X-100 (T8787, Sigma-Aldrich®) for permeabilisation, followed by a 1 h incubation at room temperature in 6% normal goat serum blocking buffer (S-1000, Vector Laboratories) in PBS. Primary antibodies diluted in blocking buffer were then added for overnight incubation at 4 °C. Cells were washed three times in phosphate-buffered saline plus 0.1% Tween® 20 (PBST; P9416, Sigma-Aldrich®) before the secondary antibodies coupled to fluorophores (Alexa Fluor® range, Thermo Fisher Scientific) were added diluted in PBST for 1 h at room temperature, with the samples protected from light. After washing with PBST, 4′,6-diamidino-2-phenylindole (DAPI, 1 µg/ml diluted in PBS; 10236276001, Roche) was applied for nuclear counterstaining for a further 15 min. Coverslips were then washed with PBST and distilled water, and then mounted onto glass slides (12362098, Thermo Fisher Scientific) with FluorSave™ Reagent (345789, Millipore) and imaged using an Axio Observer Z1 inverted motorised microscope (Carl Zeiss). Images were acquired from random positions of the coverslip. Primary antibodies used in this study are as follows: SMI-312 at 1:1000 (837904, BioLegend®), GFP at 1:1000 (ab13970, Abcam), and PGC1α at 1:300 (ab54481, Abcam). Neurite outgrowth was quantified 7 days post platedown (plating density of 5000 MNs per coverslip) blinded to sample genotype using the Simple Neurite Tracer plugin [58] within Fiji [89].
RNA extraction, RNA sequencing and transcriptomic analysis
Total RNA was extracted from MNs from two independent isogenic-corrected paired cell lines and two independent control cell lines (C9-1 and C9-2; Con-1 and Con-2) at day 21 post platedown using the RNeasy® Mini kit (74104, Qiagen®), according to the manufacturer’s instructions. RNA samples were assessed for concentration (NanoDrop ND-100 Spectrometer, NanoDrop Technologies) and quality (Agilent 2200 TapeStation, Agilent Technologies) before library preparation. Library preparation and sequencing were carried out by Edinburgh Genomics (Edinburgh, UK). For each sample, cDNA was converted to a sequencing library using the TruSeq stranded mRNA-seq library. Barcoded libraries were pooled and sequenced on an Illumina HiSeq 4000 using 75 base paired-end reads to generate at least 46 million raw reads per sample. The reads were mapped to the primary assembly of the human (hg38) reference genome contained in Ensembl release 90 [25]. Alignment was performed with STAR version 2.5.3a [30]. Tables of per-gene read counts were generated from the mapped reads with featureCounts version 1.5.2 [55]. Differential gene expression analysis, using DESeq2 version 1.16.1, specifically examined the intersection in commonly and concordantly differentially expressed genes between the two mutant-isogene pairs, using a false discovery rate of 20%, achieved by setting a Benjamini–Hochberg corrected p value threshold of 0.2 (genes with an average Fragments Per Kilobase of transcript per Million mapped reads [FPKM] < 1 were disregarded). Gene ontology (GO) [5, 100] analysis was performed on the differentially expressed genes to identify putatively altered pathways or processes using the GeneRanker module [11] within Genomatix proprietary software suite (Intrexon Bioinformatics Germany GmbH; threshold: p ≤ 0.01, Fisher’s exact test). Competitive gene set testing was performed using CAMERA [110]. Promoter analysis was performed using default parameters within Genomatix; first, the Gene2Promoter module was used to extract promoter sequences of all significantly dysregulated genes annotated in hg38. Overrepresented transcription factor binding sites (TFBS) were searched using MatBase within the input regions, and statistics on TFBSs were generated, together with overrepresentation values and Z-scores (the distance from the population mean in units of the population standard deviation) compared against genomic and promoter background. Promoter associations were defined as transcription factor families known to occur more than twice as often in promoters as in genomic sequence. A Z-score below -2 or above 2 can be considered statistically significant (it corresponds to a p value of ~ 0.05).
Quantitative reverse transcription PCR
Total RNA was extracted using the RNeasy® Mini kit (74104, Qiagen®), according to the manufacturer’s instructions. Genomic DNA was removed using the on-column DNase method (79254, Qiagen®). cDNA was synthesised using 250 ng of RNA using the DyNAmo™ cDNA Synthesis Kit (10748908, Thermo Fisher Scientific). Technical replicates, as well as no template and no RT negative controls, were included, and at least three biological replicates were studied in each case. Real-time reverse transcription quantitative PCR (RT-PCR) reactions were set up with DyNAmo ColorFlash SYBR Green qPCR kit (10442308, Thermo Fisher Scientific) and run on a CFX96 System (Bio-Rad). The data were visualised and exported using Maestro 1.1 software (Bio-Rad). Primers were synthesised by Sigma-Aldrich®. Primer sequences and annealing temperatures used in this study are provided in Supplementary Table 1, online resource.
Assessment of in vitro mitochondrial copy number
MNs, plated at a density of 2–4 million cells per 60 mm dish (430166, Corning® Life Sciences), were lysed at day 14 post platedown using 200 µl of Bradley Lysis buffer (10 mM Tris–HCl pH 7.5, 10 mM EDTA pH 8.0, 0.5% w/v SDS, 10 mM NaCl) containing 0.2 mg/ml of fresh Proteinase K (AM2546, Ambion™) and incubated in locking-lid microfuge tubes at 55 °C overnight on a ThermoMixer® comfort (Eppendorf AG) [80]. Subsequent steps are derived from classical techniques for genomic DNA extraction using phenol-choloroform [83]. An equal volume of phenol/choloroform/isoamyl alcohol solution (25:24:1; P2069, Sigma-Aldrich®) was added to each sample. After vortexing, organic and aqueous phases were separated by centrifugation at 16,000 rcf for 5 min. The upper aqueous phase was transferred to a new microfuge tube, and 200 µl of elution buffer (pH 8; 11–05-01–13, Integrated DNA Technologies, Inc) was added the original tube, which was vortexed and centrifuged as previously. The upper aqueous phase was transferred and combined with that obtained previously. An equal volume of chloroform/isoamyl alcohol solution (24:1; 25666, Sigma-Aldrich®) was added (choloroform back-extraction step). Following vortexing and centrifugation, the upper aqueous phase was transferred into a new microfuge tube, to which ammonium acetate (A2706, Sigma-Aldrich®) was added to a final concentration of 0.75 M, together with 20 µg of glycogen (9510, Ambion™). After mixing well, 2.5X volume of 100% ethanol was added. After a short 20 °C incubation on a ThermoMixer®, the samples were centrifuged at 4 °C at 16,000 rcf for 20 min. The supernatant was carefully decanted, and the pellet washed with 300 µl of cool 80% ethanol. After vortexing the samples, they were centrifuged at 16,000 rcf for 15 min at 4 °C. The supernatant was again carefully decanted and the pellet washed for a second time with 80% ethanol, followed by vortexing and centrifugation. The ethanolic solution was carefully aspirated and the pellet air dried at room temperature for 5 min followed by resuspension of the pellet in elution buffer, with gentle rocking overnight at 4 °C. Samples were assessed for concentration and quality using a NanoDrop ND-100 Spectrometer and stored at 4 °C for the duration of the study.
Mitochondrial DNA content was assessed by quantification of a unique (and rarely deleted) mitochondrial fragment in the minor arc of the mitochondrial genome (MT-ND1) relative to a single copy region of a nuclear gene (beta-2 microglobulin, B2M) using an established method [19, 39, 44, 61]. First, a customised double-stranded ND1–beta-2 microglobulin DNA fragment of 255 base pairs was synthesised as a gBlocks™ Gene Fragment (Integrated DNA Technologies, Inc.; sequence: 5′-TGC ATG ATC TAC GTG CGT CAC ATG CAG TAC CCA GCA GAG AAT GGA AAG TCA AAT TTC CTG AAT TGC TAT GTG TCT GGG TTT CAT CCA TCC GAC ATT GAA GTT GAC TTA CTG AAG AAT GGA GAG AGA CAC TAG CTC AGA TTC AGT AGA CCG CTG TTG CCC TAA AAC CCG CCA CAT CTA CCA TCA CCC TCT ACA TCA CCG CCC CGA CCT TAG CTC TCA CCA TCG CTC TAG TAA TGC AGA CAC TTG CGG TCC ATC TCG-3′). Serial dilutions were generated at 1:10 increments between 1 × 10–8 ng/µl and 10 ng/µl and stored at 4 °C for the duration of the study. Copy number was calculated as follows:
where C is the concentration of the gBlocks™ Gene Fragment in ng/µl, and M is the molecular weight (specified as 6.35 fmoles/ng).
A standard curve was generated. Real-time quantitative PCR (qPCR) reactions of the standards and the samples were set up in triplicate per experiment with DyNAmo ColorFlash SYBR Green qPCR kit (10442308, Thermo Fisher Scientific) and run on a CFX96 System (Bio-Rad). The data were visualised and exported using Maestro 1.1 software (Bio-Rad). Primers were synthesised by Sigma-Aldrich®. Primer sequences for ND1 and beta-2 microglobulin, and their annealing temperatures, are provided in Supplementary Table 1, online resource. To ensure the sample quantification cycles (Cq) for both reactions were within an optimal detection range of 17–33 [39], sample DNA concentrations differed by an order of magnitude between the mitochondrial (0.02X) and nuclear (1X) reactions. A standard curve of mean Cq value (ordinate) versus copy number (abscissa) was generated for both ND2 and beta-2 microglobulin, and copy numbers for each sample were interpolated using the semi-log line least squares fit function within Prism version 9.0.0 (GraphPad Software). The interpolated mitochondrial copy number was divided by the interpolated nuclear copy number to generate ratios of normalised values. Each experiment was performed at least three times (i.e., from at least three MN differentiations per cell line).
Western blot
MNs were cultured for 3 weeks. MN-NF medium was removed and cells were washed with ice-cold PBS. MNs were lysed with ice-cold RIPA buffer (150 mM NaCl, 50 mM Tris–HCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, pH 7.4; all constituents from Sigma-Aldrich®) supplemented with 1X cOmplete EDTA-free protease and phosphatase (PhosSTOP) inhibitor cocktail tablets (04693159001, 04906837001, Roche Diagnostics). MNs were gently scraped off the dish and samples were sonicated for 3 × 5 s at amplitude 60% using a Branson Digital Sonifier 450 (Branson Ultrasonics), keeping them on ice in between pulses. Samples were then centrifuged at 16,000 rcf at 4 °C for 10 min and the supernatant was retained for protein quantification using a bicinchoninic acid assay (23225, Pierce™ Thermo Fisher Scientific). 20 µg of protein was added to Laemmli buffer [51], whence samples were incubated at 37 °C for 30 min prior to being loaded onto a Novex™ Tris–glycine gel (XP00145BOX, Invitrogen™) and blotted to a polyvinylidene difluoride (PVDF) Immobilon®-FL membrane (05,317, Millipore) using 10 mM 3-(Cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer pH 11 (C2632, Sigma-Aldrich®) plus 10% methanol at 4 °C using a 150 mA transfer for 2 h. Total protein was quantified using Revert™ 700 Total Protein Stain (926–11,015, LI-COR Biosciences) as per manufacturer instructions, followed by blocking with Odyssey® blocking buffer in PBS (927–40,000, LI-COR Biosciences) for 1 h at room temperature. Thereafter, primary antibody incubation in blocking buffer plus 0.2% Tween® 20 (P9416, Sigma-Aldrich®) occurred at 4 °C overnight. The membrane was then washed with PBST three times and LI-COR IRDye® infrared dyes (1:15,000) added to blocking buffer plus 0.2% Tween® 20 and 0.01% SDS (L3771, Sigma-Aldrich®) for 1 h at room temperature. The membrane was then washed in PBST and imaged using an Odyssey CLx (LI-COR Biosciences). Blots were analysed using Empiria Studio 1.1 (LI-COR Biosciences). Primary antibodies used in this study are: Total OXPHOS Human WB Antibody Cocktail 1:1000 (ab110411, Abcam), VDAC1/Porin 1:2000 (ab15895, Abcam), and GAPDH 1:5000 (CB1001, Sigma-Aldrich®).
Axonal transport studies
MNs, plated on laminin/Matrigel®/fibronectin-coated µ-slide 8-well ibidi dishes (80826, ibidi GmbH) at a density of 50,000–70,000 cells per well were sparsely transduced with lentivirus expressing mitoDsRed2 at platedown using an MOI of 0.5, optimised to visualise 1–2 labelled cells per field of view, as previously described by our group [37, 56]. Fluorescence live-cell imaging of mitochondrial axonal transport was performed at day 21 post platedown at 63X magnification (Plan-Apochrat 1.40 NA oil DIC M27 objective, Carl Zeiss) using an Axio Observer Z1 inverted motorised microscope (Carl Zeiss) equipped with a Cy3 FL filter-set (Carl Zeiss), Zen 2011 z-stack, time lapse and Definite Focus modules (Carl Zeiss), and an S1 Environmental System (Carl Zeiss) incubation chamber maintained at 37 °C and 5% CO2. Medium was changed 30 min prior to imaging to phenol red-free Neurobasal™ (12348017, Gibco™ Thermo Fisher Scientific) supplemented with 1X GlutaMAX™. Mitochondrial motility was recorded for 5 min using a 0.2 Hz capture of a ~ 100 µm stretch of axon and a small z-stack. ‘Proximal’ axon measurements were acquired from the axon immediately adjoining the cell body, and ‘distal’ were measured from the distal tip. At least four axons (n) were imaged per line per differentiation (N, where N = 3). MN genotype was counterbalanced and interleaved between experimental runs. Maximum intensity projections were computed in Fiji. Kymographs were generated and analysed using KymoToolBox [113] in Fiji [89] to determine the numbers of stationary (≤ 0.1 µm/s) versus bidirectionally motile mitochondria (either predominantly towards or away from the soma).
Metabolic profiling
MNs were plated on polyethyleinimine (2.2 mg/ml in 0.1 M borate buffer, pH 8.4; 408727 and B3545, Sigma-Aldrich®)-treated V28 Seahorse plates (100882-004, Agilent), that had been coated overnight with laminin/Matrigel®/fibronectin as described above, at a high density (200,000 cells per well) to ensure the presence of a confluent cellular monolayer. A modification of the standard Agilent Seahorse XF Cell Mito Stress Test protocol was performed at day 14 post platedown using a Seahorse XFe24 Analyzer (Agilent), providing a complete mitochondrial bioenergetic profile, revealing critical information not evident in basal metabolism measurements alone. Cells were transitioned from the MN-NF medium to low-buffering capacity Seahorse XF Base Medium (102,353, Agilent) supplemented with 1X Glutamax™, 10 mM glucose (Sigma-Aldrich®) and 2 mM pyruvate (Sigma-Aldrich®), pH 7.35 ± 0.5 at 37 °C and maintained in ambient CO2 for 30 min prior to the plate being inserted into the machine. Oxygen consumption rate (OCR), an indicator of oxidative phosphorylation and therefore mitochondrial respiration, and extracellular acidification rate (ECAR), an indicator of lactate export and anaerobic glycolysis, were measured during sequential injection of optimised concentrations of oligomycin (3 μM; Sigma-Aldrich®), carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP; Cayman Chemical, Cambridge Bioscience), antimycin/rotenone (3 μM; Sigma-Aldrich®), and 2-deoxy-d-glucose (1 M; D6134, Sigma-Aldrich®). FCCP dose was optimised for each cell line (low: 0.125 μM; medium: 0.25 μM; high: 0.5 μM) and the concentration adopted that maximised the OCR response. The protocol adopted a 3 min mixing, 2 min wait, 3 min measure cycle (i.e., cycle total duration of 8 min). Three measurements were taken basally (i.e., three cycles with a total duration of 24 min), and three measurements were taken after injection of each drug (in sequence: oligomycin for inhibiting ATP-linked respiration, FCCP for eliciting maximal uncoupled respiration, antimycin/rotenone for inhibiting the electron transport chain, and 2-deoxy-d-glucose for terminating glycolysis; see Supplementary Fig. 2a, 2b, online resource). The measured OCR was normalised to total protein [23] using the bicinchoninic acid assay (23225, Pierce™ Thermo Fisher Scientific). A minimum of four individual wells (n ≥ 4) per line were included per plate, with experiments repeated in three independent cultures from different differentiations (N = 3). Data were visualised in, and imported from, Seahorse Wave Desktop software (version 2.6.0.31, Agilent). Basal respiration was calculated by subtracting the minimum OCR value after antimycin/rotenone injection (measurement cycle 12) from the OCR value prior to injection of oligomycin (measurement cycle 3). FCCP-stimulated maximal uncoupled respiration was calculated by subtracting the minimum OCR after antimycin/rotenone injection (measurement cycle 12) from the maximum OCR after FCCP injection (measurement cycle 7). Basal glycolytic rate was estimated by subtracting the ECAR value immediately after injection of 2-deoxy-d-glucose (measurement cycle 13) from the ECAR value immediately prior to the addition of oligomycin (measurement cycle 3). Respiratory-inhibited maximal glycolysis was inferred by subtracting the ECAR value immediately after the injection of 2-deoxy-d-glucose (measurement cycle 13) from the ECAR value immediately prior to the injection of 2-deoxy-d-glucose (measurement cycle 12).
Post-mortem case identification
We identified a cohort of five C9orf72-ALS cases (from the Medical Research Council Edinburgh Brain Bank; Table 2) and five age- and sex-matched control cases (from the Sudden Death Brain Bank, with no neurological disorder during life and no significant neuropathology present at post-mortem; Supplementary Table 2, online resource) for neuropathological assessment of ventral and dorsal spinal cord, as adopted in our previously published work [40, 65, 92]. All clinical data were collected as part of Scottish Motor Neurone Disease Register (SMNDR) and Care Audit Research and Evaluation for Motor Neurone Disease (CARE-MND) platform (ethics approval from Scotland A Research Ethics Committee 10/MRE00/78 and 15/SS/0216). Additionally, all cases had corresponding whole-genome sequencing and diagnostic repeat prime PCR, demonstrating pathogenic repeat lengths in the C9orf72 locus [53]. The use of human tissue for post-mortem studies has been reviewed and approved by the Edinburgh Brain Bank ethics committee and the Academic and Clinical Central Office for Research and Development medical research ethics committee, in line with the Human Tissue (Scotland) Act 2006.
Table 2.
Clinical meta-data for C9orf72 cases used in post-mortem work
| Case ID (sex) | Age at death (years) | Disease duration (months) | Regions affected at death | Classification based on El Escorial criteria[60] | Past medical history | Smoker | Alcohol intake | Prescribed riluzole |
|---|---|---|---|---|---|---|---|---|
| Case 1 (F) | 63 | 25 | UL and LL | Amyotrophic lateral sclerosis (definite) | Migraine | Ex | Yes | No |
| Case 2 (M) | 50 | 29 | UL, LL and bulbar | Amyotrophic lateral sclerosis (probable) | Sarcoidosis | Ex | No | Yes |
| Case 3 (F) | 62 | 37 | UL, LL and bulbar | Amyotrophic lateral sclerosis (definite) | Lumbar discectomy | Yes | Yes | Yes |
| Case 4 (F) | 62 | 50 | UL, LL and bulbar | Amyotrophic lateral sclerosis (definite) | Nil known | No | Yes | No |
| Case 5 (M) | 58 | 87 | UL and LL | Amyotrophic lateral sclerosis (definite) | Lower back pain | Yes | Yes | Yes |
UL upper limb, LL lower limb, F female, M male
BaseScope™ RNA in situ hybridisation
Formalin-fixed paraffin embedded human post-mortem spinal cord tissue was sectioned at 4 µm thickness onto SuperFrost Plus™ slides (10149870, Fisher Scientific, Thermo Fisher Scientific) as previously reported [40], and BaseScope™ reagents (Advanced Cell Diagnostics™ Bio-Techne) were used as per manufacturer’s guidelines [106]. In brief, following deparaffinisation, tissue sections were incubated with hydrogen peroxide for 10 min at room temperature and target antigen retrieval was performed by submerging slides in BaseScope™ 1X target retrieval reagent at 99 °C in a Braun Multiquick FS 20 steamer for 15 min. The tissue was then permeabilised using BaseScope™ protease III at 40 °C for 30 min. Probe hybridisation was then performed by incubating the slides with four drops of custom-designed BaseScope™ probe: either BA-Hs-MT-ND2-3zz-st (853271, Advanced Cell Diagnostics™ Bio-Techne) or BA-Hs-MT-CO3-1zz-st (717961, Advanced Cell Diagnostics™ Bio-Techne), to recognise ND2 and CO3 mRNA transcripts, respectively, or negative control (l-2,3-dihydrodipicolinate reductase, DapB) or positive control (peptidyl-prolyl cis–trans isomerase, PPIB) probe for 2 hours at 40 °C. Following successive probe amplification steps, transcripts were detected using the BaseScope™ RED detection kit (322910, Advanced Cell Diagnostics™ Bio-Techne) and slides were counterstained using haematoxylin and lithium carbonate. The slides were subsequently cleared in xylene and mounted on a 24 × 50 mm coverslip using two drops of VectaMount® mounting medium. Sections were then imaged at 20 × magnification on a NanoZoomer slide scanner (Hamamatsu), and the relative neuronal abundance of transcripts (denoted by counting the number of spots per cell) was assessed by neuropathologists, blinded to the demographic and clinical metadata underlying each sample. Motor and sensory neurons were identified based on their anatomical location within the spinal cord, and according to established neuropathological criteria, including size and morphology.
BaseScope™ DNA in situ hybridisation
To examine for mitochondrial DNA copy number changes in post-mortem spinal cord neurons, we adapted a recently published, validated technique [21], permitting the application of BaseScope™ to achieve cell-type specific quantification. We optimised the BaseScope™ technique on control and diseased tissue to address probe specificity; thus, DNase treatment (optimised specifically to retain haematoxylin counterstaining, but eliminate probe binding, confirming that the probes were binding to DNA, and not RNA) and RNase treatment (optimised specifically to assess the extent of background probe binding to RNA, confirming that probes were binding to DNA, and not RNA) was carried out at 40 °C at a concentration of 800U/ml for DNase and RNase for 30 min immediately following the protease III tissue permeabilisation step, and prior to probe hybridisation. Probe hybridisation was then performed by incubating the slides with four drops of custom-designed BaseScope™ probe (BA-Hs-MT-CO1-3zz-st-sense; 891111, Advanced Cell Diagnostics™ Bio-Techne), targeting the antisense region of the MT-CO1 gene (i.e., the reverse-complement sequence of 273–402 of NC_012920.1:5904–7445) to bind to DNA, for 2 h at 40 °C. The method [21] elegantly profits on the fact that, although transcription of the circular mitochondrial DNA is bidirectional, meaning that theoretically probes could hybridise to either DNA or RNA (with resultant ambiguity in signal interpretation), most light strand transcripts are rapidly degraded, including those antisense to the MT-CO1 gene [12, 68]. The Fast RED chromogen incubation step was reduced from 10 to 8 min. Sections were imaged, anonymised, at 20 × magnification using a NanoZoomer slide scanner (Hamamatsu). Motor and sensory neurons were identified based on their anatomical location within the spinal cord and according to established neuropathological criteria, including size and morphology. The relative abundance of spots (denoting DNA) was quantified by neuropathologists, blinded to the demographic and clinical metadata underlying each sample, assessing the mean (± standard error) number of spots per cell across ten cells per condition. Notably, assuming that each motor neuron is circa 50 µm in maximum dimension, and that the photomicrograph captures only one 4 µm z-plane (range of spots per cell per 4 µm z-plane is between 1 and 8 dots), we anticipate that the average spots per cell equates to between 50 and 400 spots per cell.
Post-mortem tissue immunohistochemistry
Sections were dried overnight at 40 °C and immunostaining was performed, following epitope retrieval in citric acid buffer (pH 6) in a pressure cooker for 30 min, using the rabbit anti-MT-ND2 polyclonal antibody (PA5-103952, Invitrogen™ Thermo Fisher Scientific) at a 1:200 dilution and rabbit anti-MT-CO3 antibody (HPA042788, Sigma-Aldrich®) at a 1:50 dilution (both incubated for 30 min at room temperature). Immunohistochemical detection was performed using DAB chromogen counterstained with haematoxylin, according to standard operating procedures. The immunohistochemical staining pattern was noted to be dichotomous in nature (i.e., displaying either high intensity or low intensity); consequently, ND2 and CO3 mitochondrial staining intensity was graded semi-quantitatively, as low intensity (‘intensity 1’) or high intensity (‘intensity 2’), in line with other semi-quantitative staining patterns, such as the Allred oestrogen receptor (ER) histoscore used for prognostication in the breast cancer field [69, 101]. The percentage of neurons (n = 10 cells per case/control) falling into these staining intensity categories was recorded by neuropathologists, blinded to all demographic and clinical information. Statistical analysis was performed using a two-tailed chi-squared test with Yates’s correction (to reduce approximation error associated with small sample sizes) on integer means of immunoreactivity scores comparing ‘intensity 1′ with ‘intensity 2′ staining profiles across cases and controls (Supplementary Table 3, online resource).
Data analysis
Statistical analysis was performed using SPSS® Statistics for Windows version 25 (IBM® Corp) or Prism version 8.4.0 (GraphPad Software). Data are presented as mean ± standard error. Data were initially determined to be parametric or non-parametric before applying the appropriate statistical analysis, with false discovery rate (FDR) correction for multiple comparisons, as stated. *p < 0.05, **p < 0.01, ***p < 0.001, and ‘ns’ denotes a non-significant result (p ≥ 0.05).
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Results
C9orf72 MNs display dysfunctional axonal homeostasis associated with mitochondrial bioenergetic dysfunction
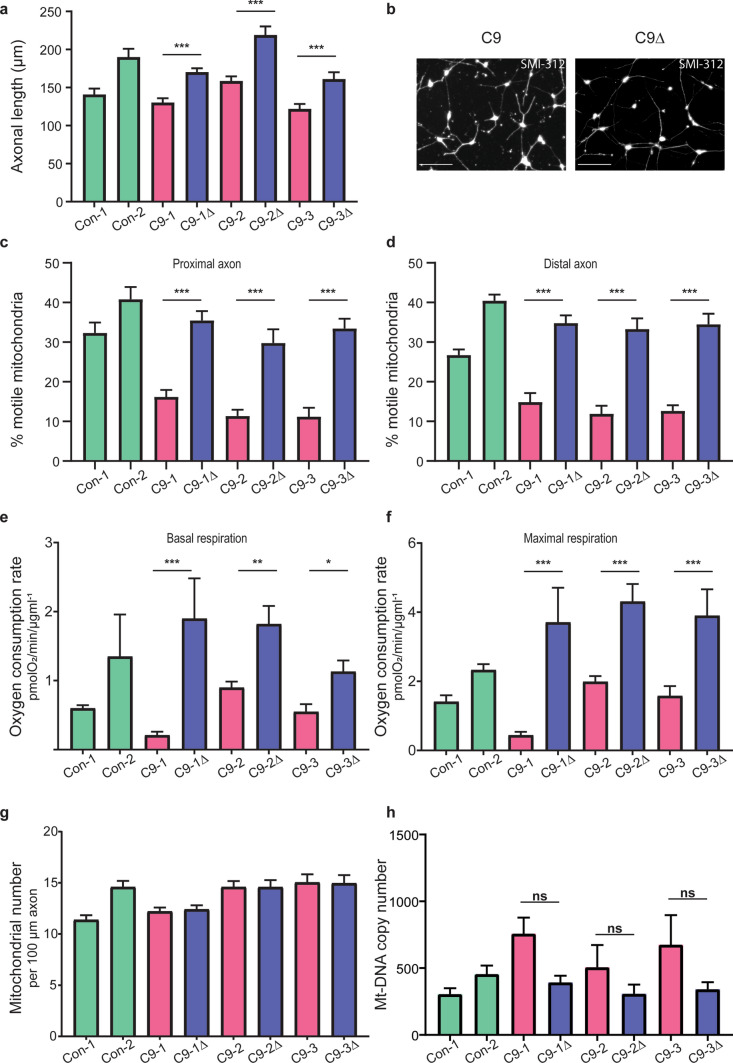
We generated highly enriched spinal cord MNs from three independent C9orf72 iPSC lines and their corresponding isogenic controls, as previously reported by our group [92]. To investigate the consequence of the C9orf72 repeat expansion mutation on MN axonal homeostasis, we first assessed axonal morphology and transport. We observed that C9orf72 MNs, when compared with their isogenic controls, have significantly shorter axons (Fig. 1a, 1b; Supplementary Fig. 1, online resource), and fewer motile mitochondria in both the proximal (Fig. 1c) and distal (Fig. 1d) axon, as assessed by live imaging of MNs transduced with mitoDsRed2 (Supplementary Video 1a, 1b, online resource). To investigate for the presence of associated deficits in MN cellular metabolism, we profiled C9orf72 MNs cellular energetics (Supplementary Fig. 2a, 2b, online resource). We found impaired basal (Fig. 1e) and maximal (Fig. 1f) mitochondrial respiration, despite normal axonal mitochondrial number (Fig. 1g) and mitochondrial copy number (Fig. 1h), implicating abnormalities in the electron transport chain machinery and/or mitochondrial substrate provision, necessary for maintenance of electron flow. Undifferentiated iPSCs did not demonstrate the mitochondrial deficit (Supplementary Fig. 2c, 2d, online resource), implying neuronal specificity. We also observed no change in the glycolytic function in the C9orf72 MNs, suggesting that glycolysis is neither involved in a negative nor compensatory fashion (Supplementary Fig. 2e, 2f, online resource).
Fig. 1.
C9orf72-dependent dysfunctional axonal homeostasis is associated with impaired mitochondrial bioenergetic function in human iPSC-derived MNs. a Quantification of SMI-312 antibody-labelled axonal length in MNs 7 days post platedown (n = 50 axons per independent MN differentiation, N = 3 independent differentiations; data are represented as mean ± SEM; genotypes used: two independent controls [Con-1, Con-2], three independent patient-derived C9orf72 lines [C9-1, C9-2, C9-3] with their corresponding isogenic controls [C9-1Δ, C9-2Δ, C9-3Δ]; statistical significance was evaluated using a mixed-effects negative binomial regression with fixed effects for mutant/isogene status and random effects for genotype, with paired mutant-isogene statistics performed via the Mann–Whitney test with FDR correction for multiple comparison testing). b Representative images depicting axonal staining via SMI-312 immunocytochemistry of day 7 post platedown C9orf72 (C9) and paired isogenic control (C9Δ) MNs (scale bars = 100 µm). Quantification of the percentage of motile mitochondria (labelled with mitoDsRed2) relative to the total number of mitochondria in a 100 µm stretch of proximal (c) and distal (d) axon in C9orf72-MNs and isogenic paired controls; two independent healthy controls are shown. Data are represented as mean ± SEM; n ≥ 4 axons per independent MN differentiation, N = 3 independent differentiations. Statistical significance was evaluated with the Kruskal–Wallis test with FDR correction. Quantification of the oxygen consumption rate (OCR) as measured by the Seahorse Analyzer, normalised to the amount of total protein, denoting basal (e) and maximal FCCP-uncoupled (f) mitochondrial respiration for C9orf72-MNs and isogenic paired controls; two independent healthy controls are also shown. Data are represented as mean ± SEM; n ≥ 4 wells per line per experiment, with experiments repeated in three independent cultures from different differentiations (N = 3). Statistical significance was evaluated with one-way ANOVA with post hoc mutant-isogene paired FDR-corrected t tests. g, Quantification of the axonal mitochondrial number in a 100 µm stretch of distal axon in C9orf72-MNs and isogenic paired controls; two independent healthy controls are also shown. Data are represented as mean ± SEM; n = 10 axons per line per experiment, with experiments repeated in three independent cultures from different differentiations (N = 3). Statistical significance was evaluated with one-way ANOVA. h Quantification of mitochondrial DNA copy number in C9orf72-MNs and isogenic paired controls; two independent healthy controls are also shown. Data are represented as mean ± SEM, with experiments repeated in at least three independent cultures from different differentiations (N ≥ 3). Statistical significance was evaluated with the Kruskal–Wallis test with FDR correction. *p < 0.05, **p < 0.01, ***p < 0.001, ‘ns’ denotes non-significant result (p ≥ 0.05)
Transcriptomic analysis of C9orf72 MNs reveals reduced expression of mitochondrially encoded transcripts of the mitochondrial respiratory chain
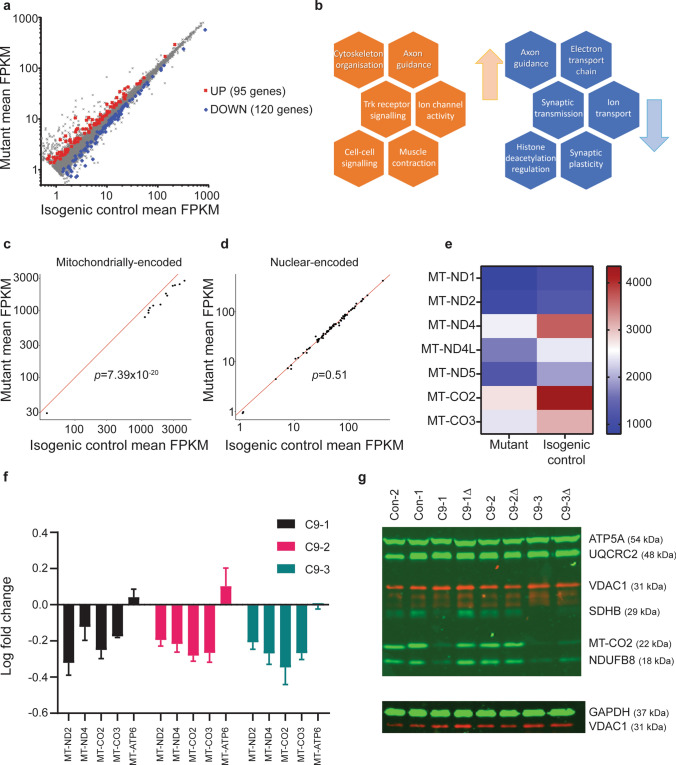
To identify putative dysregulated molecular pathways that may contribute to these axonal morphological and functional deficits, we next examined the C9orf72 MN transcriptome by performing RNA-seq on two pairs of C9orf72 and isogenic control MNs, and two independent controls. We observed a high degree of similarity between the transcriptome of mutant C9orf72 MNs and their corresponding isogenic control (Supplementary Fig. 3a, online resource). Differential gene expression analysis identified 215 dysregulated transcripts (95 upregulated and 120 downregulated) (Fig. 2a; Supplementary Data 1, online resource). Gene ontology analysis revealed, inter alia, dysregulation in pathways implicated in axonal homeostasis and oxidative phosphorylation (Fig. 2b; Supplementary Data 1, online resource).
Fig. 2.
Transcriptomic analysis revealed reduced gene expression of the mitochondrial electron transport chain in human iPSC-derived C9orf72-MNs. a Results of differential gene expression analysis, using DESeq2, examining the intersection in commonly and concordantly differentially expressed genes between the two mutant-isogene pairs, using an FDR of 20%, and discarding genes with an average FPKM < 1. The scatter plot shows the comparison of gene expression (as average FPKM). Red and blue data points denote the overlap of significantly up- and down-regulated genes in both mutant-correction pairs, respectively. b Summary of notable results from gene ontology analysis performed on the differentially expressed genes that identified putatively up (red) and down (blue) regulated altered pathways or processes. Average FPKM scatter plots comparing gene expression in C9orf72 mutant versus isogenic controls for two gene sets of interest (c, mitochondrially encoded mitochondrial transcripts and (d) nuclear-encoded mitochondrial transcripts). Competitive gene set testing using CAMERA, examining the differential gene expression for these two gene sets of interest compared with all other genes, showed that mitochondrially encoded transcripts were down-regulated (p = 7.39 × 10–20), whereas nuclear-encoded mitochondrial transcripts were not (p = 0.51). e Heatmap summarising the average gene expression (FPKM) of significantly dysregulated genes of complex I (MT-ND1, MT-ND2, MT-ND4, MT-ND4L, MT-ND5) and complex IV (MT-CO2, MT-CO3) subunits of the electron transport chain for C9orf72 versus isogenic control MNs. f Log10 mean fold change ± SEM of gene expression normalised to the respective isogenic control determined via RT-PCR for C9-1 pair (black bars), C9-2 pair (red bars), and C9-3 pair (green bars) for MT-ND2 and MT-ND4 subunits of complex I; MT-CO2 and MT-CO3 subunits of complex IV; and MT-ATP6 subunit of complex V. g Representative image of western blot of protein lysates from two independent controls [Con-1, Con-2], three independent patient-derived C9orf72 lines [C9-1, C9-2, C9-3] with corresponding isogenic controls [C9-1Δ, C9-2Δ, C9-3Δ] using a primary Total OXPHOS cocktail antibody against multiple mitochondrial electron transport chain subunits. Bands depict protein expression of complex I (NDUFB8; reduced), complex II (SDHB), complex III (UQCRC2), complex IV (MT-CO2; reduced) and complex V (ATP5A), and housekeeper proteins against GAPDH and mitochondrial outer membrane protein, VDAC-1
It is known that mitochondrial proteins are nuclear-encoded and mitochondrial DNA-encoded. Intriguingly, we found the mitochondrially encoded transcripts to be specifically downregulated in C9orf72 MNs using competitive gene set testing, where two gene sets of interest (for mitochondrial DNA-encoded mitochondrial proteins and nuclear-encoded mitochondrial proteins) were each compared with all other genes; mitochondrially encoded transcripts (p = 7.39 × 10–20) but not nuclear-encoded mitochondrial transcripts (p = 0.51) were significantly down-regulated (Fig. 2c, d). Furthermore, the nine mitochondrially encoded transcripts (contributing to complexes I and IV of the electron transport chain, and the mitochondrial small and large ribosomal subunits; Fig. 2e) whose expression was reduced are all present on the heavy strand of the mitochondrial DNA duplex, whereas the light strand transcripts, which notably encode for mitochondrial tRNAs, were unaltered (Supplementary Fig. 3b, online resource).
Quantitative RT-PCR of a representative panel of predicted dysregulated (MT-ND2 and MT-ND4 subunits of electron transport chain complex I, and MT-CO2 and MT-CO3 subunits of complex IV) and unaltered (MT-ATP6 subunit of complex V) mitochondrially encoded transcripts, and western blotting analysis of five OXPHOS complexes, were next undertaken from all three mutant-isogene C9orf72 MN pairs and two independent controls. This confirmed dysregulation of various subunits of the electron transport chain, most notably complex I and IV (Fig. 2f, g; Supplementary Fig. 3c, online resource).
We next performed promoter analysis on the dysregulated genes to identify putative common transcriptional factor family regulatory control (Supplementary Data 1, online resource). This showed an overrepresentation of potential transcription factor binding site motifs for the nuclear respiratory factor 1 (NRF1) transcription factor family, when compared with all other transcription factor families (Promoter Z score = 31.87).
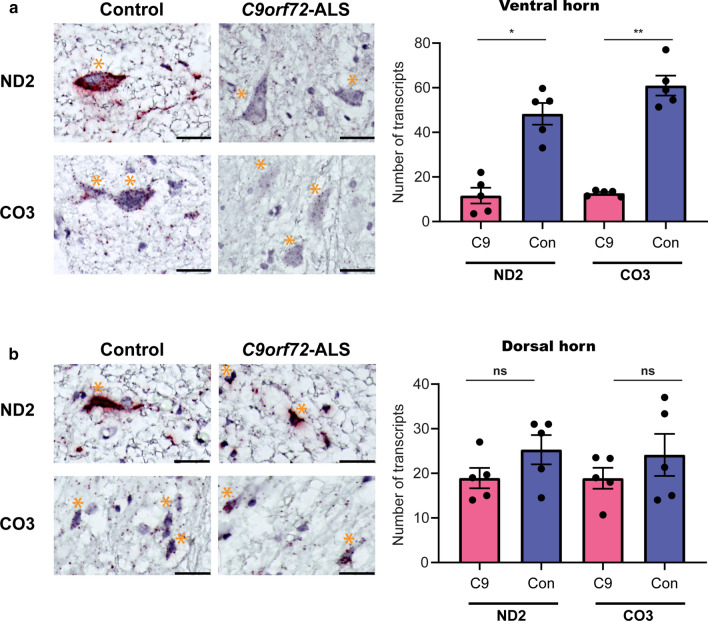
Examination of human C9orf72 post-mortem spinal cord tissue using BaseScope™ shows MN-selective reduced expression of complexes I and IV of the mitochondrial electron transport chain
To evaluate the in vivo relevance and selectivity of the observed in vitro transcriptional signature, we performed neuropathological analysis of human spinal cord motor neurons and sensory neurons. We used BaseScope™ to examine the expression of complex I (MT-ND2) and IV (MT-CO3) transcripts, in post-mortem samples from C9orf72-ALS cases (N = 5) and age- and sex-matched non-neurological controls (N = 5). We found significantly reduced expression of both MT-ND2 and MT-CO3 in ventral horn spinal motor neurons (Fig. 3a; Supplementary Fig. 4, online resource), but not in dorsal horn sensory neurons (Fig. 3b; Supplementary Fig. 4, online resource). Thus, these data confirm, in human autopsy cases, that the C9orf72 repeat expansion mutation results in down-regulation of genes associated with the mitochondrial electron transport chain, and that this dysregulation is selective to anterior horn (motor) neurons. Consistent with this transcriptional signature, we observed concomitant changes at the level of protein expression (Supplementary Fig. 5, online resource).
Fig. 3.
Examination of human C9orf72 post-mortem spinal cord tissue shows reduced expression of complexes I and IV of the mitochondrial electron transport chain in ventral horn motor, but not dorsal horn sensory, neurons. a Representative photomicrographs showing reduced expression of MT-ND2 (complex I) and MT-CO3 (complex IV) transcripts in ventral horn motor neurons of the post-mortem spinal cord from a C9orf72 hexanucleotide repeat expansion mutation case compared with its age- and sex-matched control. Red spots highlight individual mRNA molecules of MT-ND2 or MT-CO3. Tissue was counterstained with haematoxylin. Scale bars = 50 µm. Orange asterisk denotes an anterior horn motor neuron. Bar chart depicts the quantification of the number of MT-ND2 and MT-CO3 transcripts per ventral horn spinal motor neuron. Bars represent aggregate mean ± SEM, with each dot representing the mean count for between one and five cells for each of the five post-mortem specimens for C9orf72-ALS (red bars) and their age- and sex-matched controls (blue bars). Statistical significance was evaluated with the Kruskal–Wallis test with FDR correction. b Representative photomicrographs of the dorsal horn sensory neurons, examined using the same probes that recognise individual mRNA of MT-ND2 or MT-CO3, showing comparable expression between the same cases and controls. Scale bars = 50 µm. Orange asterisk denotes a dorsal horn sensory neuron. Bar chart depicts the quantification of the number of MT-ND2 and MT-CO3 transcripts per dorsal horn neuron. Bars represent aggregate mean ± SEM, with each dot representing the mean count for between one and five cells for each of the five post-mortem specimens for C9orf72-ALS (red bars) and their age- and sex-matched controls (blue bars). Statistical significance was evaluated with the Kruskal–Wallis test with FDR correction for multiple comparisons. *p < 0.05, **p < 0.01, ‘ns’ denotes non-significant result (p ≥ 0.05)
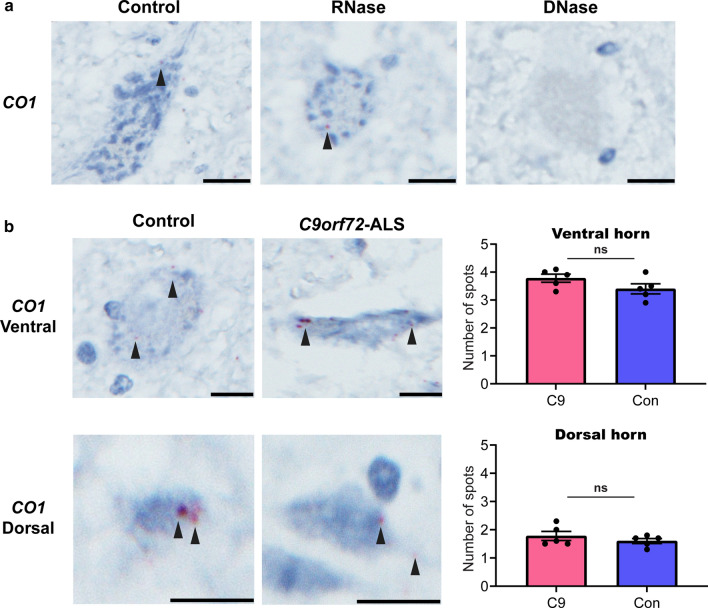
To exclude the possibility of reduced mitochondrial DNA driving the transcriptional signature in C9orf72-ALS MNs, we next quantified mitochondrial DNA copy number through a BaseScope™ DNA in situ hybridisation technique that uses a probe targeting the antisense strand of the MT-CO1 gene (see Materials and Methods). This showed unaltered mitochondrial copy number in neurons from cases versus controls, both in the ventral and dorsal horn spinal cord (Fig. 4).
Fig. 4.
Mitochondrial DNA copy number in human C9orf72 post-mortem spinal cord tissue is unaltered. a Representative photomicrographs showing DNA in situ hybridisation in control ventral horn spinal cord tissue. Red spots denote probe binding to MT-CO1—a surrogate of mitochondrial DNA copy number. RNase treatment resulted in no change in signal, while DNase treatment resulted in elimination of signal, confirming that the probe binds to DNA, and not RNA. Tissue was counterstained with haematoxylin and images were acquired at 40 × magnification. Scale bars = 20 µm. b Representative photomicrographs showing unaltered mitochondrial copy number in a C9orf72 hexanucleotide repeat expansion mutation case compared with its age- and sex-matched control, both in spinal cord ventral horn motor neurons (upper panel) and dorsal horn sensory neurons (lower panel). Red spots denote probe binding to MT-CO1. Tissue was counterstained with haematoxylin and images were acquired at 40 × magnification. Scale bars = 20 µm. Bar charts depict the quantification of the number of spots per neuron in either the ventral (upper panel) or dorsal horn (lower panel). Bars represent aggregate mean ± SEM, with each dot representing the mean count for ten cells for each of the five post-mortem specimens for C9orf72-ALS (red bars) and their age- and sex-matched controls (blue bars). Statistical significance was evaluated with the Mann–Whitney test; ‘ns’ denotes non-significant result (p ≥ 0.05)
C9orf72-dependent dysfunctional axonal homeostasis is caused by mitochondrial bioenergetic dysfunction
Peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1α) is a master regulator of mitochondrial energy metabolism and biogenesis [57], which are usually inextricably coupled [111]. PGC1α overexpression, through the modulation of the activity of multiple transcription factors (including NRF1), increases the expression of mitochondrial DNA-encoded and nuclear-encoded mitochondrial proteins [111], and it improves individual mitochondrial function [6], which, in turn, stimulates mitochondrial biogenesis [7, 88, 102]. We therefore hypothesised that overexpressing PGC1α, thereby increasing cellular bioenergetic function and mitochondrial biogenesis, may be sufficient to rescue the observed axonal phenotypes in C9orf72 MNs.
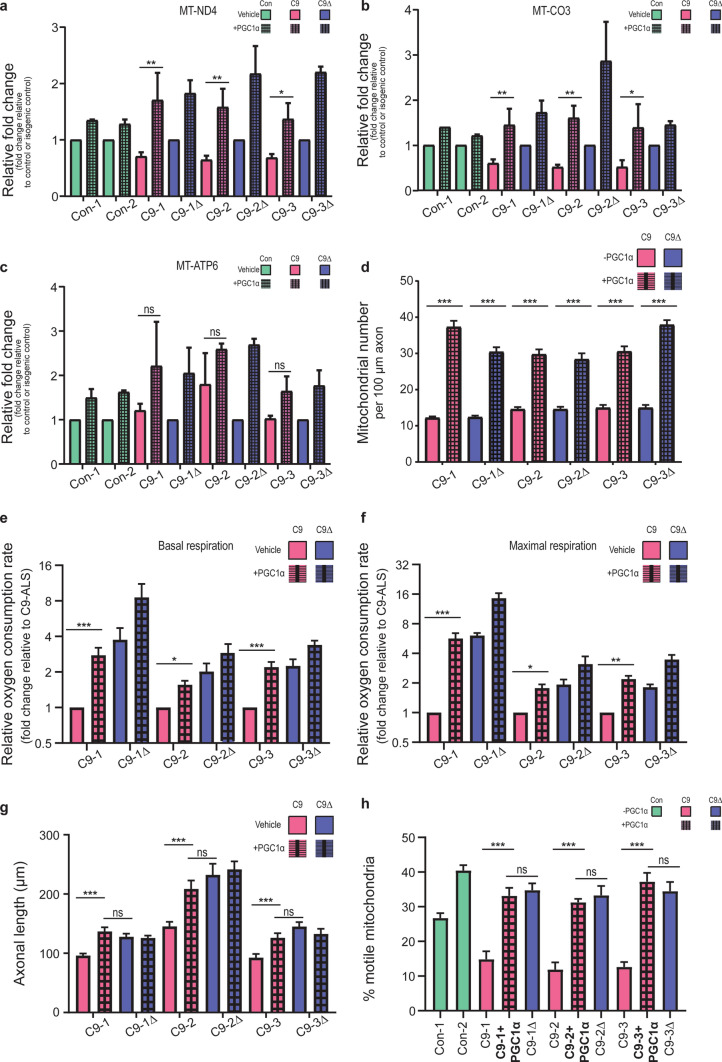
Accordingly, we performed lentiviral-mediated overexpression of PGC1α, the expression of which was itself unchanged basally between mutants and isogenic C9orf72 MNs (Supplementary Fig. 6, online resource). This resulted in the restoration of the expression of mitochondrial transcripts (complex I: MT-ND4 and complex IV: MT-CO3) of the electron transport chain (Fig. 5a–c), together with an increase in mitochondrial number in the distal axon (Fig. 5d). This in turn led to rescue of the C9orf72-dependent impairment in basal (Fig. 5e) and maximal (Fig. 5f) mitochondrial function. Critically, boosting mitochondrial bioenergetics was sufficient to rescue both the axonal length (Fig. 5g) and mitochondrial transport (Fig. 5h; Supplementary Video 2a, 2b, online resource) phenotypes observed in C9orf72 MNs. Taken together, these data establish a novel mechanistic link between mitochondrial bioenergetic dysfunction and axonal dysfunction in the pathophysiology of C9orf72-ALS.
Fig. 5.
C9orf72-dependent dysfunctional axonal homeostasis is caused by mitochondrial bioenergetic dysfunction in human iPSC-derived MNs. a–c Mean fold change ± SEM of gene expression normalised to the respective isogenic control (or control) determined via RT-PCR; genotypes used: two independent controls [Con-1, Con-2], three independent patient-derived C9orf72 lines [C9-1, C9-2, C9-3; red bars] with their corresponding isogenic controls [C9-1Δ, C9-2Δ, C9-3Δ; blue bars], with (hatched bars) and without (unhatched bars) PGC1α overexpression, for transcripts: MT-ND4 (complex I; a), MT-CO3 (complex IV; b), and MT-ATP6 (complex V; c). d Quantification of the axonal mitochondrial number in a 100 µm stretch of distal axon in C9orf72-MNs and their respective isogenic controls, with (hatched bars) and without (unhatched bars) PGC1α overexpression. Data are represented as mean ± SEM; n ≥ 4 axons per line per experiment, with experiments repeated in three independent cultures from different differentiations (N = 3). Statistical significance was evaluated with one-way ANOVA with post hoc mutant-isogene paired FDR-corrected t tests. Quantification of the relative oxygen consumption rate (OCR) as measured by the Seahorse Analyzer, normalised to the amount of total protein, denoting basal (e) and maximal FCCP-uncoupled (f) mitochondrial respiration for C9orf72-MNs and isogenic paired controls, with (hatched bars) and without (unhatched bars) PGC1α overexpression. Data are represented as mean fold change (relative to C9orf72-ALS) ± SEM; n ≥ 4 wells per line per experiment, with experiments repeated in three independent cultures from different differentiations (N = 3). Statistical significance was evaluated with one-way ANOVA with post hoc mutant-isogene paired FDR-corrected t tests. g Quantification of the SMI-312 antibody-labelled axonal length in MNs 7 days post platedown, with (hatched bars) and without (unhatched bars) PGC1α overexpression (n > 15 axons per independent MN differentiation, N = 3 independent differentiations; data are represented as mean ± SEM; genotypes used: three independent patient-derived C9orf72 lines [C9-1, C9-2, C9-3] with their corresponding isogenic controls [C9-1Δ, C9-2Δ, C9-3Δ]; statistical significance was evaluated using the Kruskal–Wallis test with FDR correction). h Quantification of the percentage of motile mitochondria (labelled with mitoDsRed2) relative to the total number of mitochondria in a 100 µm stretch of axon in C9orf72-MNs, with (hatched bars) and without (unhatched bars) PGC1α overexpression, and isogenic paired controls; two independent healthy controls are also shown. Data are represented as mean ± S.E.M; n ≥ 4 axons per independent MN differentiation, N = 3 independent differentiations. Statistical significance was evaluated with the Kruskal–Wallis test with FDR correction. *p < 0.05, **p < 0.01, ***p < 0.001, ‘ns’ denotes non-significant result (p ≥ 0.05)
Discussion
Our human in vitro and neuropathological post-mortem findings establish, for the first time, the presence of metabolic deficits in C9orf72-ALS MNs, owing to loss of mitochondrial bioenergetic function. Through the use of isogenic controls, and pathway manipulation derived from unbiased transcriptomics, we further show a novel causal and mechanistic link between MN bioenergetic failure and axonal dysfunction. Given the particular vulnerability of MNs to metabolic stress [52], our findings suggest a cell-autonomous mechanism to explain the selective MN degeneration found in ALS [74], and further confirm that ameliorating mitochondrial function is an area of therapeutic promise [66].
It is well established from animal models of ALS that targeting the axon leads to delayed onset of disease and improved survival [90, 93, 95] and that preventing MN loss alone, but not axonal degeneration, is inadequate for promoting survival [82]. Given that importance of maintenance of axonal homeostasis, it is thus important to better understand the impact of the C9orf72 repeat expansion mutation on human MN axonal properties. In the present study, we found two axonal phenotypes underlying dysfunctional axonal homeostasis in C9orf72 MNs. First, we demonstrate a novel phenotype of impaired fast axonal transport of mitochondrial cargo in C9orf72 MNs. Second, we discovered a novel morphological deficit—of shorter MN axonal length—consistent with phenotypic observations across other, rarer, ALS mutations, such as SOD-1 [20, 48] and TDP-43 [17, 33, 34, 49], as well as sporadic ALS [36]. While to our knowledge this is the first report of these phenotypes in C9orf72 human iPSC-derived MNs, reduced mitochondrial trafficking has recently been shown in primary cortical excitatory neurons in an inducible mouse model of poly(GR) toxicity [22]. The precise underlying pathological mechanisms of C9orf72-ALS remain unclear, with non-mutually exclusive C9orf72 haploinsufficiency and toxic gain of function mechanisms implicated. Recent work from Sivadasan et al. showed modulation of C9orf72 protein levels in mouse primary MNs influences actin cytoskeletal dynamics and axon outgrowth [96]. However, in neurons, the primary mediator of long-range mitochondrial transport is via microtubule-based motor machinery. In addition, it has been recently showed that C9orf72 loss of function alone does not contribute to lysosomal fast axonal transport deficits in C9orf72 iPSC-derived MNs [1]. Previous C9orf72 human iPSC studies by ourselves [92, 115], and others [1], showing pathological dipeptide repeat proteins and RNA foci, but not reduction in expression of C9orf72 (both at transcriptomic and protein levels), suggest that the observed dysfunctional axonal homeostasis is due to toxic gain of function mechanisms [16].
A key component of either the dying backward [35, 71] or the dying forward model [15] of neurodegeneration is axonal transport [8]. Thus, perturbations in axonal transport are key mediators of neurodegeneration, warranting an improved understanding of upstream mechanisms. Mitochondria are transported via fast axonal transport to the distal neurites and provide the necessary energy for local axonal processes that shape neuronal wiring and axon maintenance. Microtubules are critical, as are two main motor proteins—kinesin (for anterograde transport) and dynein (for retrograde transport)—in conjunction with axonal actin filaments [45]. The processes involved are all highly energy consuming, raising the question of whether impairments in energy metabolism contribute to defective axonal transport. While impairments in microtubule machinery have been observed in a rare ALS-causing mutation, FUS, leading to axonal transport deficits [41], metabolic impairments were unexpectedly shown to be absent [104]. Indeed, until the present study, it has not been shown that ALS patient-derived MNs metabolically differ from healthy MNs. In contrast, we report a cell-autonomous bioenergetic functional deficit in ALS human iPSC-derived MNs. Mitochondrial metabolic function was impaired, with neither a concomitant difference in glycolysis, nor altered metabolic state in the parental iPSC lines. Unbiased bulk RNA-sequencing and transcriptomic analysis revealed a distinct mitochondrial signature, with reduction in expression of mitochondrial-DNA encoded transcripts relating to the electron transport chain. Importantly, this mitochondrial loss of function signature was recapitulated in patient spinal cord tissue in a selective manner; thus, ventral horn motor neurons—but not dorsal horn sensory neurons—displayed down-regulation of expression of transcripts relating to complexes I and IV of the electron transport chain. In addition to the observation of no difference in expression in the dorsal horn neurons (acting as an internal control), previous findings of an upregulation of the GRIA1 transcript in ventral spinal MNs from the same C9orf72-ALS cases [40, 92] suggest that the observed down-regulation of mitochondrial gene expression in ventral horn spinal motor neurons is not due to a non-specific post-mortem artifact. Furthermore, mitochondrial copy number is known to be unchanged in ALS cases in general compared to controls [108]. Indeed, our in vitro and post-mortem findings together also exclude the possibility of a reduction in mitochondrial copy number in C9orf72-ALS. Consistent with these findings, we did not observe significant changes in either axonal mitochondrial counts or transcript levels of mitochondrial tRNAs in C9orf72 MNs. The trend towards increased mitochondrial copy number observed in C9orf72-MNs may reflect a compensatory response to the mitochondrial transcriptional dysregulation. Taken together, we conclude that the observed reduced mitochondrial gene expression signature and concomitant metabolic deficit are not mechanistically driven by reduced mitochondrial DNA.
The molecular basis of how the C9orf72 repeat expansion mutation results in transcriptional dysregulation of mitochondrial electron transport chain genes is unclear. Future studies will also need to evaluate mitochondrial transcriptional status in non-C9orf72 cases, including sporadic ALS, as well as in other forms of motor neuron disease, such as spinal muscular atrophy [14], to begin to address whether our findings are directly related to C9orf72 pathogenic mechanisms, such as dipeptide repeat proteins and/or due to, for example, TDP-43 aggregation. Indeed, recent findings show that C9orf72 dipeptide repeat proteins can lead to cytoplasmic TDP-43 aggregation [24, 99]. Thus, does TDP-43 pathology, observed in the vast majority of ALS cases, including C9orf72 [4, 72], modulate mitochondrial homeostasis by regulating the processing of mitochondrial transcripts [46]? Indirect evidence for this idea is supported by recent findings of transcriptional dysregulation in pathways involved in mitochondrial function and energy production in TDP-43-depleted motor neurons [17]. Moreover, TDP-43 has been shown to exert toxicity by entering mitochondria and specifically impairing OXPHOS complex I via the preferential binding of mitochondrially transcribed ND3/6 mRNAs and by inhibiting their translation to cause mitochondrial dysfunction and neuronal loss [107], as well as triggering mitochondrial DNA release and neuroinflammatory cGAS/STING activation [112]. Finally, recent work by Onesto et al. showed that there is compensatory mitochondrial biogenesis, as evidenced by PGC1α upregulation, in dermal fibroblasts derived from patients with C9orf72-ALS [77]. This suggests that, despite carrying the pathological mutation, most cell types can compensate for the mitochondrial dysfunction by stimulating biogenesis, and selective vulnerability could arise partly from an inability of motor neurons to modulate this homeostatic mechanism.
Crucially, notwithstanding an unaltered NRF1-PGC1α axis basally in C9orf72 MNs, we were able to boost mitochondrial function (and biogenesis) through manipulation of its master regulator, PGC1α, leading to rescue of the axonal phenotypes, providing a novel mechanistic link between mitochondrial bioenergetics and axonal dysfunction. Fast axonal transport is a highly energy demanding process [97]. Chronic dysfunction of the mitochondrial electron transport chain could result in a reduction of global ATP levels in MNs. Consequently, motor proteins would arrest, given the cellular milieu being deprived of adequate ATP. The successful manipulation of a key pathway involved in mitochondrial energy metabolism adds to the emerging spinal cord injury literature that has shown that restoring cellular energetics and axonal transport promotes axonal outgrowth and regeneration [42]. Our data suggest that targeting this tractable pathway may thus also be highly relevant to neurodegeneration [56, 75], accepting the possibility of there being other competing mechanisms driving the dysfunctional axonal homeostasis observed in C9orf72 MNs.
Our findings add important new metabolic functional data to the existing ALS mitochondrial literature. Early, elegant post-mortem studies (well before the current rich genetic landscape of ALS had been determined) showed that patients with ALS have dense clusters of mitochondria in the anterior horn of the lumbar spinal cord [87] and presynaptic mitochondrial swelling in their motor neurons [94]. The cellular distribution of mitochondria was also shown to be affected, with the majority of mitochondria being located in the soma and proximal axon [86]. Moreover, the total amount of mitochondrial DNA, measured by Southern blot, was reduced in the spinal cord from sporadic ALS patients [109], associated with a decrease in the activity of the electron transport chain complexes in spinal cord mitochondria [109] and a reduction in the activity of key mitochondrial enzymes [13]. Our study provides a mechanistic link between the functional MN metabolic deficits to alterations in gene expression. More recently, astrocytic metabolic inflexibility [3] and mitochondrial aberrations in patient-derived fibroblasts [77] and iPSC models [27, 36, 59] have been demonstrated in C9orf72-ALS. C9orf72 patient-derived motor neurons have swollen mitochondria on electron microscopy [27], a reduced [27] or increased [59] mitochondrial membrane potential, associated with altered calcium buffering, contributing to glutamate excitotoxity [26], and elevated levels of cytochrome c [27]. They also show increased age-dependent oxidative stress associated with poly(GR) dipeptide repeat proteins binding to mitochondrial ribosomal proteins [59], and more recently in Drosophila muscle, poly(GR) has been shown to interact with components of the ‘mitochondrial contact site and cristae organising system’ impacting on mitochondrial ion homeostasis [54]. In our study, we show a distinct mitochondrial bioenergetic deficit in C9orf72 MNs. In addition to calcium buffering, mitochondrial bioenergetics could also contribute to neuronal vulnerability to excitotoxicity. PGC1α, which controls cellular energetics including mitochondrial biogenesis, has been shown to negatively regulate glutamate receptor mediated excitotoxicity in Huntington’s disease mouse models [79]. Indeed, we have previously shown that C9orf72 MNs display vulnerability to excitotoxicity, owing to dysregulation in glutamate receptors [92], and the present findings raise the possibility of there being positive feedback on the excitotoxic signalling pathways. Finally, a large screening study (> 1200 drugs) from 32 sporadic ALS human iPSC lines showed that ropinirole, a dopamine (D2) receptor agonist used to treat Parkinson’s disease motor symptoms, reverses their identified neurite length, cytotoxicity and abnormal protein aggregations phenotypes, by reducing the elevated reactive oxygen species, implicating ropinirole’s non-canonical effects on mitochondrial pathways [36].
In summary, our study establishes a novel mechanistic link between mitochondrial dysfunction and axonal dysfunction in ALS using human-based disease modelling. It is conceivable that boosting mitochondrial metabolism locally at the growth cone or synapses could improve the size of the motor unit by promoting terminal axonal sprouting and/or increasing stability of the neuromuscular junction. Accordingly, we believe that our findings highlight the importance of targeting mitochondrial dysfunction directly, to discover novel therapeutic agents for this devastating disease [66].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figures and Tables (PDF 2645 kb)
Acknowledgements
The authors thank James Cooper, Karen Gladstone, Judith O’Shaughnessy and Dr Ariane Willems, for technical assistance provided to Chandran Lab, Professor Michael Cousin and Dr James Longden for insightful discussions, and Dr Pamela Brown for preparing lentiviral vectors. The authors would also like to thank: (1) MRC Edinburgh Brain Bank for supplying all post-mortem brain material and the Scottish MND Register/CARE-MND Consortium for all clinical and demographic data; (2) Scottish MND clinical specialist team for discussing and obtaining consent from patients with ALS/MND for inclusion in these resources; and, (3) MND Scotland and the Sylvia Aitken Charitable Trust for funding C.S. to help establish the MND Tissue Bank.
Author contributions
ARM, GEH, SC and BTS conceived and designed the study. ARM, JMG and RNC performed data collection, analysis and interpretation. OD and ARM performed bioinformatics analysis. KB, JN and DS provided technical cell-culture and laboratory expertise. KM performed the post-mortem tissue work. CS, NMM, DJM, GEH, SC, and BTS contributed to interpretation of data. ARM drafted the manuscript, with critical input from DJM, GEH, SC, and BTS. All authors approved the final manuscript.
Funding
ARM is a Lady Edith Wolfson Clinical Fellow and is jointly funded by the Medical Research Council (MRC) and the Motor Neurone Disease Association (MR/R001162/1). He also acknowledges support from the Rowling Scholars scheme, administered by the Anne Rowling Regenerative Neurology Clinic (ARRNC), University of Edinburgh, and a seedcorn grant from The Chief Scientist Office and the RS Macdonald Charitable Trust via the Scottish Neurological Research Fund, administered by the University of St Andrews. JMG is funded by a starter grant for clinical lecturers from the Academy of Medical Sciences. CS is supported by a Medical Research Council grant (MR/L016400/1). NMM was funded by a Wellcome Trust New Investigator Award (100981/Z/13/Z). RNC and NMM are funded by a Diabetes UK grant (17/0005697). The Hardingham and Chandran laboratories are supported by the Euan MacDonald Centre for Motor Neurone Disease Research, and the UK Dementia Research Institute (DRI), which receives its funding from UK DRI Ltd, funded by the MRC, Alzheimer's Society and Alzheimer's Research UK. SC also acknowledges funding from the ARRNC, My Name’5 Doddie Foundation, and an MRC Dementias Platform UK Stem Cell Partnership grant (MR/N013255/1). BTS is a Rowling-DRI Fellow.
Compliance with ethical standards
Conflict of interest
The authors have no declarations to make.
Footnotes
Siddharthan Chandran and Bhuvaneish T. Selvaraj share co-senior authorship.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Siddharthan Chandran, Email: siddharthan.chandran@ed.ac.uk.
Bhuvaneish T. Selvaraj, Email: bthangar@ed.ac.uk
References
- 1.Abo-Rady M, Kalmbach N, Pal A, Schludi C, Janosch A, Richter T, et al. Knocking out C9ORF72 exacerbates axonal trafficking defects associated with hexanucleotide repeat expansion and reduces levels of heat shock proteins. Stem Cell Rep. 2020;14:390–405. doi: 10.1016/j.stemcr.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SSW, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen SP, Hall B, Woof R, Francis L, Gatto N, Shaw AC, et al. C9orf72 expansion within astrocytes reduces metabolic flexibility in amyotrophic lateral sclerosis. Brain. 2019;142:3771–3790. doi: 10.1093/brain/awz302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin S, Klimcakova E, St-Pierre J. Impact of PGC-1alpha on the topology and rate of superoxide production by the mitochondrial electron transport chain. Free Radic Biol Med. 2011;51:2243–2248. doi: 10.1016/j.freeradbiomed.2011.08.036. [DOI] [PubMed] [Google Scholar]
- 7.Austin S, St-Pierre J. PGC1 and mitochondrial metabolism—emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- 8.Baker MR. ALS—dying forward, backward or outward? Nat Rev Neurol. 2014;10:660. doi: 10.1038/nrneurol.2013.221-c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldwin KR, Godena VK, Hewitt VL, Whitworth AJ. Axonal transport defects are a common phenotype in Drosophila models of ALS. Hum Mol Genet. 2016;25:2378–2392. doi: 10.1093/hmg/ddw105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14:544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 12.Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013;41:1223–1240. doi: 10.1093/nar/gks1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borthwick GM, Johnson MA, Ince PG, Shaw PJ, Turnbull DM. Mitochondrial enzyme activity in amyotrophic lateral sclerosis: implications for the role of mitochondria in neuronal cell death. Ann Neurol. 1999;46:787–790. doi: 10.1002/1531-8249(199911)46:5<787::aid-ana17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Boyd PJ, Tu WY, Shorrock HK, Groen EJN, Carter RN, Powis RA, et al. Bioenergetic status modulates motor neuron vulnerability and pathogenesis in a zebrafish model of spinal muscular atrophy. PLoS Genet. 2017;13:e1006744. doi: 10.1371/journal.pgen.1006744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braak H, Brettschneider J, Ludolph AC, Lee VM, Trojanowski JQ, Del Tredici K. Amyotrophic lateral sclerosis—a model of corticofugal axonal spread. Nat Rev Neurol. 2013;9:708–714. doi: 10.1038/nrneurol.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braems E, Swinnen B, Van Den Bosch L. C9orf72 loss-of-function: a trivial, stand-alone or additive mechanism in C9 ALS/FTD? Acta Neuropathol. 2020;140:625–643. doi: 10.1007/s00401-020-02214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briese M, Saal-Bauernschubert L, Luningschror P, Moradi M, Dombert B, Surrey V. Loss of Tdp-43 disrupts the axonal transcriptome of motoneurons accompanied by impaired axonal translation and mitochondria function. Acta Neuropathol Commun. 2020;8:116. doi: 10.1186/s40478-020-00987-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377:162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 19.Campbell GR, Reeve A, Ziabreva I, Polvikoski TM, Taylor RW, Reynolds R, et al. Mitochondrial DNA deletions and depletion within paraspinal muscles. Neuropathol Appl Neurobiol. 2013;39:377–389. doi: 10.1111/j.1365-2990.2012.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Qian K, Du Z, Cao J, Petersen A, Liu H, et al. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell. 2014;14:796–809. doi: 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Zheng Q, Peiffer LB, Hicks JL, Haffner MC, Rosenberg AZ, et al. An in situ atlas of mitochondrial DNA in mammalian tissues reveals high content in stem and proliferative compartments. Am J Pathol. 2020;190:1565–1579. doi: 10.1016/j.ajpath.2020.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi SY, Lopez-Gonzalez R, Krishnan G, Phillips HL, Li AN, Seeley WW, et al. C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nat Neurosci. 2019;22:851–862. doi: 10.1038/s41593-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connolly NMC, Theurey P, Adam-Vizi V, Bazan NG, Bernardi P, Bolanos JP, et al. Guidelines on experimental methods to assess mitochondrial dysfunction in cellular models of neurodegenerative diseases. Cell Death Differ. 2018;25:542–572. doi: 10.1038/s41418-017-0020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cook CN, Wu Y, Odeh HM, Gendron TF, Jansen-West K, del Rosso G, Yue M, et al. C9orf72 poly(GR) aggregation induces TDP-43 proteinopathy. Sci Transl Med. 2020;12:eabb3774. doi: 10.1126/scitranslmed.abb3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dafinca R, Barbagallo P, Farrimond L, Candalija A, Scaber J, Ababneh NA, et al. Impairment of mitochondrial calcium buffering links mutations in C9ORF72 and TARDBP in iPS-derived motor neurons from patients with ALS/FTD. Stem Cell Rep. 2020;14:892–908. doi: 10.1016/j.stemcr.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dafinca R, Scaber J, Ababneh N, Lalic T, Weir G, Christian H, et al. C9orf72 hexanucleotide expansions are associated with altered endoplasmic reticulum calcium homeostasis and stress granule formation in induced pluripotent stem cell-derived neurons from patients with amyotrophic lateral sclerosis and frontotemporal dementia. Stem Cells. 2016;34:2063–2078. doi: 10.1002/stem.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Vos KJ, Hafezparast M. Neurobiology of axonal transport defects in motor neuron diseases: opportunities for translational research? Neurobiol Dis. 2017;105:283–299. doi: 10.1016/j.nbd.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolmetsch R, Geschwind DH. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupuis L, Pradat PF, Ludolph AC, Loeffler JP. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011;10:75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- 33.Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012;4:145–104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 34.Fiesel FC, Schurr C, Weber SS, Kahle PJ. TDP-43 knockdown impairs neurite outgrowth dependent on its target histone deacetylase 6. Mol Neurodegener. 2011;6:64. doi: 10.1186/1750-1326-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A, et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T, et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat Med. 2018;24:1579–1589. doi: 10.1038/s41591-018-0140-5. [DOI] [PubMed] [Google Scholar]
- 37.Fumagalli L, Young FL, Boeynaems S, Decker MD, Mehta AR, Swijsen A, et al. C9orf72-derived arginine-containing dipeptide repeats associate with axonal transport machinery and impede microtubule-based motility. bioRxiv. 2019 doi: 10.1101/835082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grady JP, Murphy JL, Blakely EL, Haller RG, Taylor RW, Turnbull DM, et al. Accurate measurement of mitochondrial DNA deletion level and copy number differences in human skeletal muscle. PLoS ONE. 2014;9:e114462. doi: 10.1371/journal.pone.0114462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory JM, Livesey MR, McDade K, Selvaraj BT, Barton SK, Chandran S, et al. Dysregulation of AMPA receptor subunit expression in sporadic ALS post-mortem brain. J Pathol. 2020;250:67–78. doi: 10.1002/path.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R, et al. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun. 2017;8:861. doi: 10.1038/s41467-017-00911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han Q, Xie Y, Ordaz JD, Huh AJ, Huang N, Wu W, et al. Restoring cellular energetics promotes axonal regeneration and functional recovery after spinal cord injury. Cell Metab. 2020;31(623–641):e628. doi: 10.1016/j.cmet.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawrot J, Imhof S, Wainger BJ. Modeling cell-autonomous motor neuron phenotypes in ALS using iPSCs. Neurobiol Dis. 2020;134:104680. doi: 10.1016/j.nbd.2019.104680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He L, Chinnery PF, Durham SE, Blakely EL, Wardell TM, Borthwick GM, et al. Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res. 2002;30:e68–e68. doi: 10.1093/nar/gnf067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–5419. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izumikawa K, Nobe Y, Yoshikawa H, Ishikawa H, Miura Y, Nakayama H, et al. TDP-43 stabilises the processing intermediates of mitochondrial transcripts. Sci Rep. 2017;7:7709. doi: 10.1038/s41598-017-06953-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jablonka S, Dombert B, Asan E, Sendtner M. Mechanisms for axon maintenance and plasticity in motoneurons: alterations in motoneuron disease. J Anat. 2014;224:3–14. doi: 10.1111/joa.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14:781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci. 2019;22:167–179. doi: 10.1038/s41593-018-0300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreiter N, Pal A, Lojewski X, Corcia P, Naujock M, Reinhardt P, et al. Age-dependent neurodegeneration and organelle transport deficiencies in mutant TDP43 patient-derived neurons are independent of TDP43 aggregation. Neurobiol Dis. 2018;115:167–181. doi: 10.1016/j.nbd.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 51.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Le Masson G, Przedborski S, Abbott LF. A computational model of motor neuron degeneration. Neuron. 2014;83:975–988. doi: 10.1016/j.neuron.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leighton D, Newton J, Colville S, Bethell A, Craig G, Cunningham L, et al. Clinical audit research and evaluation of motor neuron disease (CARE-MND): a national electronic platform for prospective, longitudinal monitoring of MND in Scotland. Amyotroph Lateral Scler Frontotemporal Degener. 2019;20:242–250. doi: 10.1080/21678421.2019.1582673. [DOI] [PubMed] [Google Scholar]
- 54.Li S, Wu Z, Li Y, Tantray I, De Stefani D, Mattarei A, et al. Altered MICOS morphology and mitochondrial ion homeostasis contribute to poly(GR) toxicity associated with C9-ALS/FTD. Cell Rep. 2020;32:107989. doi: 10.1016/j.celrep.2020.107989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 56.Licht-Mayer S, Campbell GR, Canizares M, Mehta AR, Gane AB, McGill K, et al. Enhanced axonal response of mitochondria to demyelination offers neuroprotection: implications for multiple sclerosis. Acta Neuropathol. 2020;140:143–167. doi: 10.1007/s00401-020-02179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 58.Longair MH, Baker DA, Armstrong JD. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- 59.Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, et al. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92:383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ludolph A, Drory V, Hardiman O, Nakano I, Ravits J, Robberecht W, et al. A revision of the El Escorial criteria—2015. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16:291–292. doi: 10.3109/21678421.2015.1049183. [DOI] [PubMed] [Google Scholar]
- 61.Malik AN, Shahni R, Rodriguez-de-Ledesma A, Laftah A, Cunningham P. Mitochondrial DNA as a non-invasive biomarker: accurate quantification using real time quantitative PCR without co-amplification of pseudogenes and dilution bias. Biochem Biophys Res Commun. 2011;412:1–7. doi: 10.1016/j.bbrc.2011.06.067. [DOI] [PubMed] [Google Scholar]
- 62.Maury Y, Come J, Piskorowski RA, Salah-Mohellibi N, Chevaleyre V, Peschanski M, et al. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol. 2015;33:89–96. doi: 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- 63.McCloskey C, Rada C, Bailey E, McCavera S, Berg HA, Atia J, et al. The inwardly rectifying K+ channel KIR 7.1 controls uterine excitability throughout pregnancy. EMBO Mol Med. 2014;6:1161–1174. doi: 10.15252/emmm.201403944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehta A, Selvaraj B, Dando O, Burr K, Hardingham G, Chandran S. 229 Dysregulated axonal homeostasis in C9orf72 iPSC-derived motor neurones. J Neurol Neurosurg Psychiatry. 2019;90:e57–e57. doi: 10.1136/jnnp-2019-ABN-2.193. [DOI] [Google Scholar]
- 65.Mehta AR, Selvaraj BT, Barton SK, McDade K, Abrahams S, Chandran S, et al. Improved detection of RNA foci in C9orf72 amyotrophic lateral sclerosis post-mortem tissue using BaseScope shows a lack of association with cognitive dysfunction. Brain Commun. 2020;2:fcaa009. doi: 10.1093/braincomms/fcaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mehta AR, Walters R, Waldron FM, Pal S, Selvaraj BT, Macleod MR, et al. Targeting mitochondrial dysfunction in amyotrophic lateral sclerosis: a systematic review and meta-analysis. Brain Commun. 2019;1:fcz009. doi: 10.1093/braincomms/fcz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mehta PR, Jones AR, Opie-Martin S, Shatunov A, Iacoangeli A, Al Khleifat A, et al. Younger age of onset in familial amyotrophic lateral sclerosis is a result of pathogenic gene variants, rather than ascertainment bias. J Neurol Neurosurg Psychiatry. 2019;90:268–271. doi: 10.1136/jnnp-2018-319089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mohammed ZM, Edwards J, Orange C, Mallon E, Doughty JC, McMillan DC, et al. Breast cancer outcomes by steroid hormone receptor status assessed visually and by computer image analysis. Histopathology. 2012;61:283–292. doi: 10.1111/j.1365-2559.2012.04244.x. [DOI] [PubMed] [Google Scholar]
- 70.Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, et al. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- 71.Naumann M, Pal A, Goswami A, Lojewski X, Japtok J, Vehlow A, et al. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun. 2018;9:335. doi: 10.1038/s41467-017-02299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 73.Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97(1268–1283):e1266. doi: 10.1016/j.neuron.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nijssen J, Comley LH, Hedlund E. Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol. 2017;133:863–885. doi: 10.1007/s00401-017-1708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O'Donnell KC, Vargas ME, Sagasti A WldS and PGC-1alpha regulate mitochondrial transport and oxidation state after axonal injury. J Neurosci. 2013;33:14778–14790. doi: 10.1523/JNEUROSCI.1331-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohno N, Chiang H, Mahad DJ, Kidd GJ, Liu L, Ransohoff RM, et al. Mitochondrial immobilization mediated by syntaphilin facilitates survival of demyelinated axons. Proc Natl Acad Sci USA. 2014;111:9953–9958. doi: 10.1073/pnas.1401155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Onesto E, Colombrita C, Gumina V, Borghi MO, Dusi S, Doretti A, et al. Gene-specific mitochondria dysfunctions in human TARDBP and C9ORF72 fibroblasts. Acta Neuropathol Commun. 2016;4:47. doi: 10.1186/s40478-016-0316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pal A, Glass H, Naumann M, Kreiter N, Japtok J, Sczech R, et al. High content organelle trafficking enables disease state profiling as powerful tool for disease modelling. Sci Data. 2018;5:180241. doi: 10.1038/sdata.2018.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Puddifoot C, Martel MA, Soriano FX, Camacho A, Vidal-Puig A, Wyllie DJ, et al. PGC-1alpha negatively regulates extrasynaptic NMDAR activity and excitotoxicity. J Neurosci. 2012;32:6995–7000. doi: 10.1523/JNEUROSCI.6407-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramírez-Solis R, Rivera-Pérez J, Wallace JD, Wims M, Zheng H, Bradley A. Genomic DNA microextraction: a method to screen numerous samples. Anal Biochem. 1992;201:331–335. doi: 10.1016/0003-2697(92)90347-A. [DOI] [PubMed] [Google Scholar]
- 81.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sagot Y, Tan SA, Hammang JP, Aebischer P, Kato AC. GDNF slows loss of motoneurons but not axonal degeneration or premature death of pmn/pmn mice. J Neurosci. 1996;16:2335–2341. doi: 10.1523/jneurosci.16-07-02335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sambrook J. Molecular cloning : a laboratory manual. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 84.Sances S, Bruijn LI, Chandran S, Eggan K, Ho R, Klim JR, et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat Neurosci. 2016;19:542–553. doi: 10.1038/nn.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sandoe J, Eggan K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nat Neurosci. 2013;16:780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- 86.Sasaki S, Horie Y, Iwata M. Mitochondrial alterations in dorsal root ganglion cells in sporadic amyotrophic lateral sclerosis. Acta Neuropathol. 2007;114:633–639. doi: 10.1007/s00401-007-0299-1. [DOI] [PubMed] [Google Scholar]
- 87.Sasaki S, Iwata M. Ultrastructural study of the synapses of central chromatolytic anterior horn cells in motor neuron disease. J Neuropathol Exp Neurol. 1996;55:932–939. doi: 10.1097/00005072-199608000-00009. [DOI] [PubMed] [Google Scholar]
- 88.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Selvaraj BT, Frank N, Bender FL, Asan E, Sendtner M. Local axonal function of STAT3 rescues axon degeneration in the pmn model of motoneuron disease. J Cell Biol. 2012;199:437–451. doi: 10.1083/jcb.201203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Selvaraj BT, Livesey MR, Chandran S. Modeling the C9ORF72 repeat expansion mutation using human induced pluripotent stem cells. Brain Pathol. 2017;27:518–524. doi: 10.1111/bpa.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Selvaraj BT, Livesey MR, Zhao C, Gregory JM, James OT, Cleary EM, et al. C9ORF72 repeat expansion causes vulnerability of motor neurons to Ca(2+)-permeable AMPA receptor-mediated excitotoxicity. Nat Commun. 2018;9:347. doi: 10.1038/s41467-017-02729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sendtner M, Schmalbruch H, Stockli KA, Carroll P, Kreutzberg GW, Thoenen H. Ciliary neurotrophic factor prevents degeneration of motor neurons in mouse mutant progressive motor neuronopathy. Nature. 1992;358:502–504. doi: 10.1038/358502a0. [DOI] [PubMed] [Google Scholar]
- 94.Siklos L, Engelhardt J, Harati Y, Smith RG, Joo F, Appel SH. Ultrastructural evidence for altered calcium in motor nerve terminals in amyotropic lateral sclerosis. Ann Neurol. 1996;39:203–216. doi: 10.1002/ana.410390210. [DOI] [PubMed] [Google Scholar]
- 95.Simon CM, Jablonka S, Ruiz R, Tabares L, Sendtner M. Ciliary neurotrophic factor-induced sprouting preserves motor function in a mouse model of mild spinal muscular atrophy. Hum Mol Genet. 2010;19:973–986. doi: 10.1093/hmg/ddp562. [DOI] [PubMed] [Google Scholar]
- 96.Sivadasan R, Hornburg D, Drepper C, Frank N, Jablonka S, Hansel A, et al. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat Neurosci. 2016;19:1610–1618. doi: 10.1038/nn.4407. [DOI] [PubMed] [Google Scholar]
- 97.Sleigh JN, Rossor AM, Fellows AD, Tosolini AP, Schiavo G. Axonal transport and neurological disease. Nat Rev Neurol. 2019;15:691–703. doi: 10.1038/s41582-019-0257-2. [DOI] [PubMed] [Google Scholar]
- 98.Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp S, Kenna KP, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84:324–331. doi: 10.1016/j.neuron.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Solomon DA, Stepto A, Au WH, Adachi Y, Diaper DC, Hall R, et al. A feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-α mediates C9orf72-related neurodegeneration. Brain. 2018;141:2908–2924. doi: 10.1093/brain/awy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.The Gene Ontology C The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thomas J, Hanby A, Pinder S, Ellis I, Macartney J, Clements K, et al. Implications of inconsistent measurement of ER status in non-invasive breast cancer: a study of 1,684 cases from the Sloane Project. Breast J. 2008;14:33–38. doi: 10.1111/j.1524-4741.2007.00523.x. [DOI] [PubMed] [Google Scholar]
- 102.Uittenbogaard M, Chiaramello A. Mitochondrial biogenesis: a therapeutic target for neurodevelopmental disorders and neurodegenerative diseases. Curr Pharm Des. 2014;20:5574–5593. doi: 10.2174/1381612820666140305224906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vandoorne T, De Bock K, Van Den Bosch L. Energy metabolism in ALS: an underappreciated opportunity? Acta Neuropathol. 2018;135:489–509. doi: 10.1007/s00401-018-1835-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vandoorne T, Veys K, Guo W, Sicart A, Vints K, Swijsen A, et al. Differentiation but not ALS mutations in FUS rewires motor neuron metabolism. Nat Commun. 2019;10:4147. doi: 10.1038/s41467-019-12099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vasistha NA, Johnstone M, Barton SK, Mayerl SE, Thangaraj Selvaraj B, et al. Familial t(1;11) translocation is associated with disruption of white matter structural integrity and oligodendrocyte-myelin dysfunction. Mol Psychiatry. 2019;24:1641–1654. doi: 10.1038/s41380-019-0505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang W, Wang L, Lu J, Siedlak SL, Fujioka H, Liang J, et al. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat Med. 2016;22:869–878. doi: 10.1038/nm.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wei W, Keogh MJ, Wilson I, Coxhead J, Ryan S, Rollinson S, et al. Mitochondrial DNA point mutations and relative copy number in 1363 disease and control human brains. Acta Neuropathol Commun. 2017;5:13. doi: 10.1186/s40478-016-0404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wiedemann FR, Manfredi G, Mawrin C, Beal MF, Schon EA. Mitochondrial DNA and respiratory chain function in spinal cords of ALS patients. J Neurochem. 2002;80:616–625. doi: 10.1046/j.0022-3042.2001.00731.x. [DOI] [PubMed] [Google Scholar]
- 110.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/s0092-8674(00)80611-x. [DOI] [PubMed] [Google Scholar]
- 112.Yu C-H, Davidson S, Harapas CR, Hilton JB, Mlodzianoski MJ, Laohamonthonkul P, et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020 doi: 10.1016/j.cell.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, Cordelieres FP, et al. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell. 2013;152:479–491. doi: 10.1016/j.cell.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao C, Devlin AC, Chouhan AK, Selvaraj BT, Stavrou M, Burr K, et al. Mutant C9orf72 human iPSC-derived astrocytes cause non-cell autonomous motor neuron pathophysiology. Glia. 2020;68:1046–1064. doi: 10.1002/glia.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Tables (PDF 2645 kb)
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.