Significance
The ocean’s “biological pump” exports sinking particles containing carbon, nutrients, and energy to the deep sea, contributing centrally to the global carbon cycle. Here, we identify key organisms and biological processes associated with elevated carbon flux to the abyss. Our analyses reveal that, during summer export, specific populations of photosynthetic algae, heterotrophic protists, and bacteria reach the abyss on sinking particles. Deep-sea bacteria respond rapidly to this elevated nutrient delivery to the abyss in summer. During other seasons, different organisms and processes appear responsible for particle export to the deep sea. Our analyses reveal key biota and biological processes that interconnect surface productivity, particle export, and the deep-sea ecosystem, thereby influencing the function and efficiency of the ocean’s biological pump.
Keywords: carbon export, biological carbon pump, deep sea, marine microbes, open ocean
Abstract
In the open ocean, elevated carbon flux (ECF) events increase the delivery of particulate carbon from surface waters to the seafloor by severalfold compared to other times of year. Since microbes play central roles in primary production and sinking particle formation, they contribute greatly to carbon export to the deep sea. Few studies, however, have quantitatively linked ECF events with the specific microbial assemblages that drive them. Here, we identify key microbial taxa and functional traits on deep-sea sinking particles that correlate positively with ECF events. Microbes enriched on sinking particles in summer ECF events included symbiotic and free-living diazotrophic cyanobacteria, rhizosolenid diatoms, phototrophic and heterotrophic protists, and photoheterotrophic and copiotrophic bacteria. Particle-attached bacteria reaching the abyss during summer ECF events encoded metabolic pathways reflecting their surface water origins, including oxygenic and aerobic anoxygenic photosynthesis, nitrogen fixation, and proteorhodopsin-based photoheterotrophy. The abundances of some deep-sea bacteria also correlated positively with summer ECF events, suggesting rapid bathypelagic responses to elevated organic matter inputs. Biota enriched on sinking particles during a spring ECF event were distinct from those found in summer, and included rhizaria, copepods, fungi, and different bacterial taxa. At other times over our 3-y study, mid- and deep-water particle colonization, predation, degradation, and repackaging (by deep-sea bacteria, protists, and animals) appeared to shape the biotic composition of particles reaching the abyss. Our analyses reveal key microbial players and biological processes involved in particle formation, rapid export, and consumption, that may influence the ocean’s biological pump and help sustain deep-sea ecosystems.
As a central component of the ocean’s biological carbon pump (1–5), sinking particles mediate the export of photosynthetically derived organic matter, and the transport of carbon and energy to abyssal depths. At the open ocean time-series study site Station ALOHA (6, 7) in the North Pacific Subtropical Gyre (NPSG), prominent summertime elevated carbon flux (ECF) events called the “summer export pulse” (SEP) (8) typically occur from July through August. These summer ECF events, referred to henceforth as the SEP, are defined as those periods when summertime particulate carbon flux at 4,000 m exceeds the mean annual flux by 150% or more (8). Time-series measurements of particulate carbon, nitrogen, phosphorus, and silica flux have provided a long-term record of the amounts of carbon and nutrients reaching cold, well-oxygenated abyssal waters (8). These data also provide estimates of the amount of carbon sequestered in the deep ocean in the form of sinking particulate material that has escaped remineralization in the upper water column. Despite these critical datasets, the biological origins and composition of sinking particles, especially during periods of elevated flux, are less well known.
Historically, sinking particle-associated biota have been characterized and quantified in sediment trap collected samples via light microscopy (9, 10), or analyses of organic biomarkers like carotenoids or other photosynthetic pigments (11). Shell-bearing organisms like coccolithophorids, diatoms, tintinnids, foraminifera, radiolarians, or pelagic mollusk shells are the most readily identified by microscopic techniques (12). This approach cannot identify most microorganisms (including smaller protists, fungi, archaea, and bacteria), or soft-bodied pelagic metazoans like siphonophores or other hydrozoans. Additionally, the mineral-containing shells of some common pelagic organisms (like the aragonite shells of pteropods, or the strontium sulfate tests of Acantharea) dissolve at depths exceeding 1,000 m and so are difficult to quantify in deeper sediment traps (13, 14). New in situ optical techniques for identifying and quantifying sinking organisms have also been recently developed (15, 16), but these too suffer from the same difficulties encountered using traditional light microscopy. As a consequence, few comprehensive inventories of sinking particle-associated biota currently exist.
Recent studies are now beginning to apply nucleic acid-based taxonomic and genomic analyses to characterize particle-associated microorganisms. A few of these studies have used serial filtration of seawater to identify microbial taxa found in different particle size fractions (17–21). However, these approaches cannot identify or differentiate sinking from suspended particles (22). One approach to address this challenge employs large volume settling chambers, which can separate and differentiate suspended from sinking particles in seawater (22). Other oceanographic tools, like sediment traps that capture sinking particles in situ (23), are also being coupled with gene-based taxonomic analyses to determine the composition and variability of sinking particle-associated microbiota (22, 24–31). Different methods to preserve cells and nucleic acids in situ have been developed, including the use of non–cross-linking, precipitative fixatives (27, 29, 32) or poisons like mercuric chloride (31). These approaches have been leveraged to characterize microbial assemblages found on sinking particles and those responsible for particle degradation, using paired sediment traps deployed with or without preservatives (25, 27, 28, 33).
Despite great progress, few quantitative studies have linked elevated carbon flux events with their corresponding sinking particle-associated organisms reaching the deep ocean. In a recent sediment trap study off the California coast, Preston et al. (30) found that a single diatom species was associated with a bloom that led to elevated carbon flux to the abyss. A recent global ocean biogeographic survey of microbial metagenomes (from seawater sampled on filters) used optically estimated carbon flux at 150 m to infer associations of microorganisms with carbon flux (15). Another previous report, documenting the SEP events in the NPSG over several decades, postulated the involvement of diatom–diazotroph associations based on 1 y of biological data (8). Although a subsequent 1-y metagenomic study of sinking particle-associated microbes was conducted in 2014 in the NPSG (29), no elevated carbon flux events occurred that year, so no conclusions about SEP events could be inferred (29).
To better characterize the organisms and biological processes associated with ECF events in the NPSG, we conducted a 3-y metagenomic time-series study (21 time points per year) of particles sinking to abyssal depths (4,000 m) in the open ocean. We identified specific microbial populations whose abundances positively correlated with elevated carbon flux to the abyss. The temporal variability of carbon flux correlating microorganisms was analyzed, and distinguishing genomic features and inferred traits of sinking particle associated microbes were characterized. Our analyses provide perspective on microbial components and dynamics associated with the open ocean biological pump and inferred biological processes and variability associated with carbon transport to the deep ocean.
Results
Time-series analyses of sinking particle-associated microbial genes and genomes reaching 4,000-m depth were conducted from 2014 to 2016 at Station ALOHA in the NPSG (6–8). SEP events were observed in 2015 and 2016, each >150% of the 24-y mean annual particulate carbon flux (1992 to 2016; mean = 230.4 μmol⋅m−2, SE = 6.7, n = 394; Fig. 1, SI Appendix, Fig. S1, and Dataset S1). An additional ECF event was also observed in mid-May 2015. Particulate nitrogen flux patterns coincided with particulate carbon fluxes throughout the time series.
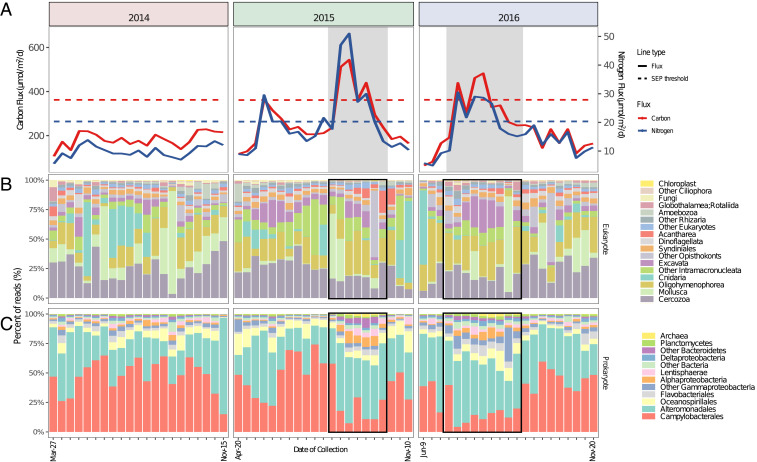
Fig. 1.
Particulate carbon and nitrogen flux and rRNA-based taxon abundances on sinking particles reaching 4,000 m from 2014 to 2016. (A) Particulate carbon (PC) and nitrogen (PN) throughout the time series. The dashed lines represent thresholds for the summer export pulse (SEP) for carbon and nitrogen flux (150% of the mean value). (B) Recovery of eukaryotic SSU rRNAs in the sediment trap time-series samples. (C) Recovery of bacterial and archaeal SSU rRNAs in the sediment trap time-series samples. Samples collected during the SEP ECF period are shaded in gray in A, and outlined in the black boxes in B and C.
To identify sinking particle-associated microbes reaching the abyss, total RNA and DNA were extracted from each sediment trap sample and the taxonomic representation of small subunit ribosomal RNA sequences (SSU rRNAs) was assessed (Fig. 1 and SI Appendix, Fig. S2). We focused most quantitative analyses on RNA-derived rRNAs (Fig. 1), rather than rRNA genes from metagenomic DNA (SI Appendix, Fig. S2 and Tables S1–S7), for several reasons: First, the total rRNA sequence reads per sample in the metatranscriptome dataset was ∼170-fold greater than the total rRNA sequence reads per sample in the metagenome dataset. As well, direct enumeration of rRNAs (as opposed to rRNA genes in DNA) is thought to better represent those microorganisms that are more metabolically active. The use of quantitative internal RNA standards for the metatranscriptome datasets also facilitated consistent count normalization across all RNA samples. On sinking particles, depending on the taxonomic group, rRNA abundances were either greater (Arcobacter, Colwellia, Intramacronucleata ciliates), approximately equal to (Moritella, Psychrobium, several Flavobacteriales genera), or less than (Ruegeria, Thalassobius, Tropicibacter, hydrozoans) corresponding rRNA gene abundances in DNA (SI Appendix, Tables S1–S7). Since deep-sea sinking particle-associated biota have diverse origins (e.g., attached microbes arriving from surface waters, colonizing and actively growing deep-sea microbes, or dead animals from various depths in the water column), these different abundance patterns reflect the different life histories of particle-associated groups.
Over the 3-y sampling period, particle-associated RNA-derived rRNAs (SI Appendix, Tables S1–S7) were dominated by bacteria affiliated with either the Epsilonproteobacteria (36.6% of all bacterial and archaeal SSU rRNAs were represented by Campylobacterales; Fig. 1) or Gammaproteobacteria (40.8% of all bacterial and archaeal SSU rRNAs were represented by Alteromonadales plus other Gammaproteobacteria; Fig. 1). The Epsilonproteobacteria order Campylobacterales was composed predominantly of Arcobacter spp. (Fig. 1 and SI Appendix, Table S1). Across the entire time series, the Alteromonadales clade was dominated by Colwellia, Moritella, and Shewanella (SI Appendix, Table S2), genera often associated with psychrophilic and piezophilic species (34, 35, 27–30). Alphaproteobacteria, Deltaproteobacteria, and Bacteroidetes each comprised a smaller percentage (1 to 3.6%) of the total rRNAs associated with sinking particles reaching abyssal depths across the time series (Fig. 1 and SI Appendix, Tables S3–S5). Archaeal rRNAs were found on sinking particles at much lower abundance, averaging 0.27% of the total rRNAs across all samples, and consisting primarily of Woesearchaeia, Nitrosopumilaceae, and Thermoplasmatota.
Sinking particle-associated eukaryotic rRNAs comprised 15.3% of the total rRNAs recovered across the 3-y time series. The three main supergroups represented in the eukaryotic SSU rRNA datasets included Rhizaria, Opisthokonta, and Alveolata. Among these, Cnidaria, Mollusca, and Cercozoa rRNAs were most abundant (accounting for 11.8%, 18.7%, and 19.7% of the eukaryotic SSU rRNAs, respectively). Among protists, ciliates, cercozoans, and acantharians were among the most well represented across the time series (Fig. 1 and SI Appendix, Table S6). Within the Cnidaria, siphonophores (Hydroidolina) were most abundant, while pteropods (Heterobranchia) were the most abundant molluscs (SI Appendix, Table S7).
To explore the genomic properties and functional traits of sinking particle-associated bacteria and archaea, a nonredundant dataset of 121 medium- to high-quality metagenome assembled genomes (MAGs) (≥70.7% complete and ≤8.9% contamination; Dataset S2) (36, 37) was prepared by analyzing all 63 time-series metagenomic assemblies. The taxonomic affiliation of these MAGs was assessed using GTDB-Tk (38), which classifies genomes based on their placement in a reference genome tree, relative evolutionary distance, and FastANI distance. The dereplicated MAGs encompassed 15 bacterial and 3 archaeal phyla (Dataset S2). Due partly to their larger genome sizes, greater complexity, and lower abundance in our samples, we did not recover any sinking particle-associated eukaryote MAGs.
To estimate strain diversity, read-based operational taxonomic unit (OTU) sequences of ribosomal proteins were identified in the metagenomes. Conservatively, we estimated that more than 6,908 strains were present in the sediment trap samples (Materials and Methods). To estimate the fraction of the bacterial and archaeal assemblage represented in the dereplicated MAG sets, comparison of the read-based OTU sequences to the ribosomal proteins in the dereplicated MAG set was conducted cumulatively for all MAGs, across all 63 samples. At the genus level, the dereplicated MAGs represented ∼40% of all bacterial and archaeal diversity present in sinking particle-associated metagenomes.
Mapping of metagenomic reads to dereplicated MAGs across the 3-y time series revealed significant shifts in the bacterial community during the 2015 and 2016 SEP events (Fig. 2, SI Appendix, Fig. S3, and Dataset S3). Individual MAGs were tested for differential abundance (via metagenomic read coverage profiles) during SEP versus non-SEP periods using the Mann–Whitney test. A subset of MAGs (40 of 121) was found to be significantly enriched (false-discovery rate-corrected value of P < 0.05) during the SEP compared to non-SEP periods. Some of the SEP-associated bacteria (14 of 40) belonged to Alteromonadaceae, Flavobacteriaceae, Rhodobacterales, Oceanospirillales, and Vibrio, presumptive copiotrophic particle-associated groups (Dataset S3 and 15, 39, 40). Nitrogen-fixing symbiotic or free-living cyanobacteria known to be prominent in surface waters of the NPSG (Richelia and Crocosphaera) (8, 9, 41, 42) were also well represented among the SEP-enriched bacterial genomes (Figs. 2A and 3 and SI Appendix, Fig. S3).
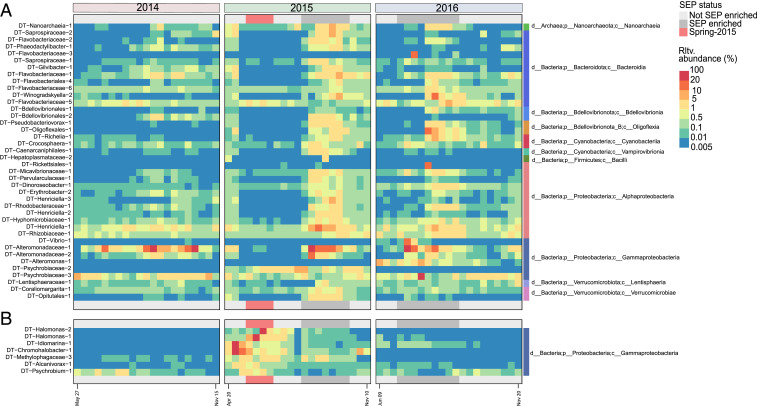
Fig. 2.
MAGs on sinking particles recovered at 4,000 m whose abundance positively correlates with either the SEP or Spring 2015 ECF periods. The heatmap displays the relative abundance of MAGs determined by read mapping from individual sediment trap sample time points (columns) during 2014 (orange), 2015 (green), and 2016 (blue). All MAGs displayed were significantly enriched (P < 0.05) during (A) SEP ECF periods (Top) or (B) the Spring 2015 ECF period (Bottom). SEP ECF periods are indicated by dark gray horizontal bars, and the Spring 2015 ECF period is indicated by the orange horizontal bar at the Top and Bottom of each panel.
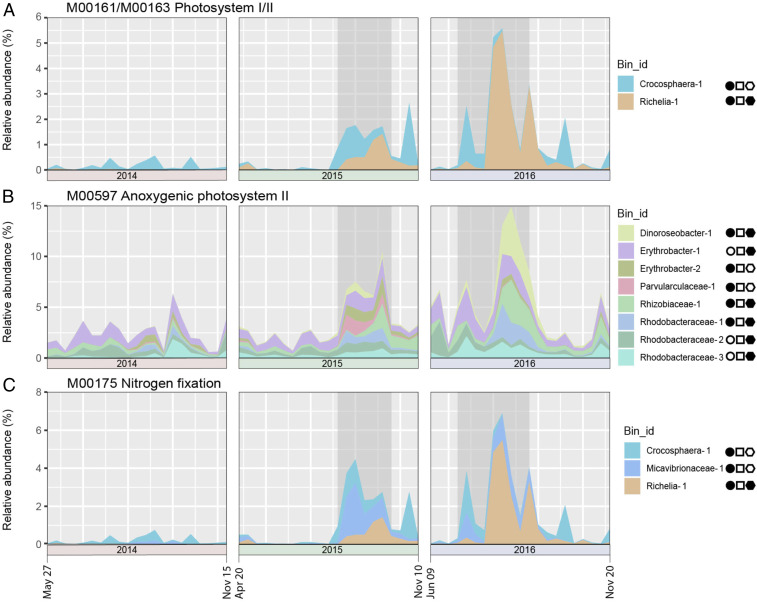
Fig. 3.
Changes in genome abundances and key metabolic pathways encoded by MAGs during the SEP. Key metabolic pathways found in different MAGs that encode oxygenic or anoxygenic photosynthesis, or nitrogen fixation. All MAGs found to encode these pathways (SEP, Spring 2015 ECF, or non-SEP) are shown, with respect to the total MAG relative abundance across the time series. (A) Oxygenic photosystem I/II MAGs; (B) anoxygenic photosystem II MAGs; (C) nitrogen fixation MAGs. The area chart displays the cumulative abundance of the specific MAGs that encoded each of the different metabolic pathways. The symbols in solid black to the Right of each panel indicate those MAGs that positively correlated with either the SEP time period (circles), the Spring 2015 ECF (squares), or carbon flux (polygon). Empty symbols indicate MAGs with no significant correlation in the test indicated. The 2015 and 2016 summer export pulse (SEP) periods are shaded in gray.
During the Spring 2015 ECF period, Mann–Whitney tests identified the enrichment of a different bacterial assemblage (7 of 121 MAGs) than was found in SEP events. This included representatives of Gammaproteobacteria belonging to the Halomonadaceae (Halomonas, Chromahalobacter), Idiomarinaceae (Idiomarina), Psychrobiaceae (Psychrobium), and Methylophagaceae (Fig. 2B and Dataset S3). Members of these genera are often found associated with surfaces, including abyssopelagic particles, or are known to inhabit abyssal and hadal plankton or sediments (17, 18, 43–45). Unlike SEP-enriched bacteria, none of the Spring 2015 ECF enriched bacteria encoded chlorophyll, bacteriochlorophyll, or proteorhodopsin-based photosystems. Different eukaryotic taxa were also enriched during the Spring 2015 ECF period compared to those found in SEP events (SI Appendix, Fig. S4 and Dataset S4). Polycystine radiolaria (Nassellaria, Polycystinea), fungi (Eurotiales), presumptive parasitic protists (Perkinsidae), and copepods (Misophrioida, Siphonostomatoida, Harpacticoida) in particular were enriched on deep-sea sinking particles during the Spring 2015 period (SI Appendix, Fig. S4 and Dataset S4). Consistent with the MAG results (Fig. 2 and Dataset S3), rRNA analyses associated Gammaproteobacteria belonging to the Halomonadaceae with the Spring 2015 ECF event as well (Dataset S4).
Both SSU rRNA and MAG analyses indicated a year-round prevalence of some groups Epsilonproteobacteria (Arcobacter spp.) and Gammaproteobacteria closely related to known deep-sea psychrophilic or piezophilic bacteria (e.g., Colwellia, Shewanella, Moritella spp.) (SI Appendix, Table S1, and Datasets S3 and S5) (29, 43). In contrast, other particle-associated bacteria found on deep-sea sinking particles were specifically associated with SEP events. The metabolic pathways of SEP-enriched MAGs recovered on deep-sea sinking particles, for example, revealed strong vertical connections between surface waters and the deep sea. In particular, SEP-enriched MAG-encoded metabolic pathways encompassed oxygenic and anoxygenic phototrophy, carbon fixation, and nitrogen fixation (Dataset S6), presumably as a consequence of the export of surface-dwelling microbes attached to fast-sinking particles that reach the deep sea.
As expected, the two SEP-enriched cyanobacterial MAGs (DT-Richelia-1 and DT-Crocosphaera-1) encoded the genes for oxygenic photosynthesis verifying their surface water origins. These cyanobacteria were significantly enriched during the SEP periods (value of P < 0.05; Fig. 3A). Additionally, eight Alphaproteobacteria MAGs in several different lineages encoded genes for bacterial anoxygenic photosynthesis (some with intact photosynthetic “superoperons”) (46) that were elevated in abundance during and after the SEP periods (Fig. 3B). Five (DT-Rhodobacteraceae-1,2,3, DT-Parvularculaceae-1, and DT-Rhizobiaceae-1) encoded genes for microaerobic cbb3-type cytochromes, and the other three were related to well-known aerobic anoxygenic phototrophs (DT-Dinoroseobacter-1, DT-Erythrobacter-1, and DT-Erythrobacter-2). During the SEP, these latter three aerobic anoxygenic phototrophs were the most abundant of all the anoxygenic phototrophs (Fig. 3B). A total of 70% of the SEP-associated MAGs on sinking particles reaching 4,000 m also encoded photolyases, supporting the surface water origins of many of these SEP-associated MAGs (Dataset S3; 47). Additionally, a total of 30% of all the SEP-associated MAGs encoded a proteorhodopsin gene (48), providing further evidence of the rapid export of bacterial photoheterotrophs to the deep sea during the SEP (Dataset S3).
A total of 10 MAGs encoded the Calvin–Benson–Bassham (CBB) cycle, which includes the key enzyme ribulose-1,5-bisphosphate carboxylase (RuBisCO). These included the cyanobacteria (DT-Richelia-1 and DT-Crocosphaera-1), which supports their contribution to SEP-associated export. Excluding the diazotrophic cyanobacteria, the genetic potential for the CBB cycle among sinking particle-associated MAGs was greatest between April and May of 2015 (SI Appendix, Fig. S5) and correlated with sinking particulate carbon fluxes above 150% of the average annual mean (Fig. 1 and SI Appendix, Fig. S1). Noncyanobacterial CBB encoding bacteria dominated by a Chromohalobacter species (DT-Chromohalobacter-1) were found to be enriched only during the Spring 2015 ECF event, suggesting functional differences between the Spring 2015 and summertime SEP events (Fig. 2 and SI Appendix, Fig. S5). An alternative carbon fixation pathway, the 3-hydroxyproprionate bicycle, was identified in four MAGs in the Rhodobacteraceae family (SI Appendix, Fig. S5), with two (DT-Rhodobacteraceae-2 and 3) encoding genes for anoxygenic photosynthesis. The 3-hydroxyproprionate bicycle encoding MAGs were enriched during and shortly following the SEP.
A diverse range of heterotrophic diazotrophs was recently reported on particles collected at relatively shallow depths (150 m) in the NPSG (49). This earlier study showed that a large proportion of PCR-amplified nifH gene sequences was derived from heterotrophic bacteria and diazotrophic cyanobacteria (Crocosphaera and Trichodesmium). In our study, in addition to SEP-associated diazotrophic cyanobacteria (DT-Richelia-1 and DT-Crocosphaera-1), one member of the alphaproteobacterial Micavibrionaceae family (DT-Micavibrionaceae-1) also encoded a nitrogen fixation gene cluster (nifHDK). Nitrogen fixation genes have not previously been reported in the Micavibrionaceae family, and this putative diazotroph was also selectively enriched during the SEP (Fig. 3C). To further explore the diversity of potential diazotrophs reaching seafloor depths, nifH gene sequences recovered from sediment trap metagenomic contigs were identified, revealing 15 additional nonredundant nifH genes (amino acid identity ≤95%) (Dataset S7 and SI Appendix, Fig. S6). Comparisons with the Genome Taxonomy Database (GTDB) (50) suggested that these nifH gene sequences were derived from Cyanobacteria (n = 10), including Trichodesmium, Gammaproteobacteria (n = 2), Alphaproteobacteria (n = 2), and Deferribacterota (n = 1). The nifH genes on deep-sea sinking particles were most abundant during the SEP events (SI Appendix, Fig. S6). A summer maximum of nifH genes was also observed in 2014, even though elevated carbon flux at 4,000 m was not detected in summer that year (SI Appendix, Fig. S6).
To test for potential relationships between SSU rRNA abundances and carbon flux measurements at 4,000 m, we performed weighted gene coexpression network analysis (WGCNA) (SI Appendix, Detailed Methods). WGCNA is a systems biology approach for identifying clusters (modules) of gene or taxon abundance profiles that are highly correlated with other external sample traits, in this case carbon flux values over time. Initial analyses identified specific, particle-associated rRNA-based taxa that clustered into one of four different modules (SI Appendix, Fig. S7 and Dataset S5). The rRNA module 1 correlated significantly with increased carbon flux (Fig. 4 A–C and Dataset S5). Similar analyses were also performed for bacterial and archaeal MAG taxa (Fig. 4D and Dataset S3).
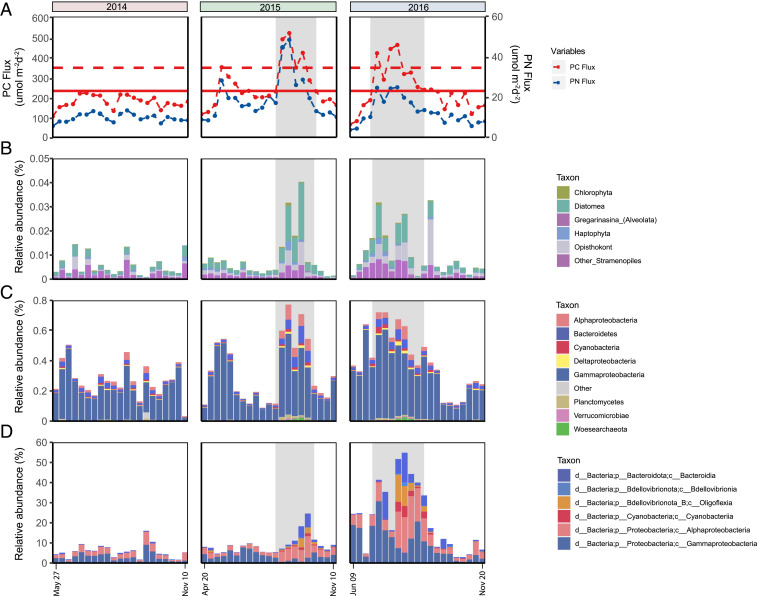
Fig. 4.
Carbon flux-correlating taxa found on sinking particles reaching 4,000 m. (A) Particulate carbon flux values across the time series are represented by the red data points connected by a dashed line. The average particulate carbon flux value is shown as a solid, horizontal red line. The SEP threshold (150% of the mean annual flux) is shown as a dashed horizontal red line. Dashed blue line plot: Particulate nitrogen flux at 4,000 m. Dashed red line plot: Particulate carbon flux at 4,000 m. (B) Abundance of eukaryote WGCNA module 1 SSU rRNAs relative to total SSU rRNAs. (C) Abundance of prokaryote WGCNA module 1 SSU rRNAs relative to total SSU rRNAs. (D) Abundance of carbon flux-associated bacterial and archaeal WGCNA module 1 MAGs relative to total MAG read counts across the time series. Specific taxa that positively correlate with elevated carbon flux are indicated in SI Appendix, Table S5 (rRNA abundances) and SI Appendix, Table S3 (MAG abundances). The 2015 and 2016 summer export pulse (SEP) periods are shaded in gray. WGCNA, weighted gene correlation network analysis. See SI Appendix, Detailed Methods for further detail.
The majority of taxa in SSU rRNA module 1 consisted of bacteria and archaea (85% of the total taxonomic diversity and >99% of all module 1 SSU rRNA reads). Only 2% of the bacterial taxonomic diversity across the time series was assigned to module 1, with a large percentage of SSU rRNAs falling within the Gammaproteobacteria order Alteromonadales (77.1% of all module 1 SSU rRNAs), including the family Colwelliaceae (Dataset S5). Other bacterial taxa in module 1 included Alphaproteobacteria (25% of bacterial taxonomic diversity assigned to module 1), Cyanobacteria (14% of module 1 bacterial taxonomic diversity, including a high representation of Nostocales, which contains nitrogen-fixing species like Richelia), and Bacteroidetes (13% of module 1 bacterial taxonomic diversity) (Fig. 4C and Dataset S5). A large proportion of eukaryote SSU rRNAs in module 1 were affiliated with Stramenopiles (68.4% of all module 1 eukaryote SSU rRNAs) and Diatomea (17.3% of the total module 1 eukaryotic SSU rRNAs; Fig. 4B and Dataset S5).
WGCNA analyses of MAG abundances similarly identified one module that correlated positively with elevated carbon flux (MAG module 1; Fig. 4D and Dataset S3). Carbon flux enriched bacterial genomes in MAG module 1 encompassed several members of Flavobacteriaceae (including Winogradskyella); the endosymbiotic diazotrophic cyanobacterium Richelia; Alphaproteobacteria associated with Rhizobiaceae and Rhodobacteraceae; members of Rickettsiales and Bdellovibrionales; and Gammaproteobacteria species affiliated with Halieaceae, Nitrincolaceae, Xanthomonadales, Pseudomonadales, Alteromonas, Pseudoalteromonas, Vibrio, and Colwellia (Dataset S3).
Most of the MAGs identified in ECF WGCNA analyses (Fig. 4D) were also positively correlated with summer SEP event time periods tested in Mann–Whitney tests (Fig. 2, SI Appendix, Fig. S3, and Dataset S3). These SEP-enriched MAGs included Nanoarchaeota; the diazotrophic cyanobacterium Crocosphaera; members of Chitinophagales (Saprospiraceae); Flavobacteriaceae (Gilvibacter spp.); Alphaproteobacteria affiliated with Hyphomonadaceae (Henricella spp.), Micavibrionaceae, Hyphomicrobiaceae, Parvularculaceae, and Sphingomonadaceae (Erythrobacter spp.); and members of Verrucomicrobiae (Datasets S2 and S3).
WGCNA analyses of rRNAs identified taxon correlations to carbon flux as a continuous variable. To further test for rRNA-based taxon association with SEP events only, rRNA temporal abundance profiles were classified as SEP-associated (or not) using a random forest (RF) classification and regression tree algorithm (SI Appendix, Detailed Methods) (51, 52). In the Eukaryote RF model, rhizosolenid diatoms were among the most important eukaryotic taxon variables, second only to the Filasterea family Capsasporidae (SI Appendix, Fig. S8 and Dataset S8). The Alphaproteobacteria family Sphingomonadaceae were the most important bacterial groups emerging from the Prokaryote RF models, along with cyanobacteria and other heterotrophic bacteria among the top 30 most highly ranked groups (SI Appendix, Fig. S8 and Dataset S9).
To further explore co-occurrence patterns among the sinking particle-associated microbes, a distribution-based clustering analyses on the rRNA data were also performed. Taxa that correlated with increased carbon flux in WGCNA analyses (Fig. 4B and Datasets S3 and S5) also emerged in the network analyses (SI Appendix, Fig. S9). Both phytoplankton and phytoplankton-associated bacteria were identified as central nodes in the network. In particular, eukaryote groups associated with Capsasporidae, Bicosoetida, and rhizosolenid diatoms appeared as central nodes connecting many bacterial nodes in the network. One archaeal Nanoarchaeota group, Woesearchaeia, was associated with rhizosolenid diatoms and Capsasporidae. The majority of network nodes were bacterial (89.5%), mostly composed of Alphaproteobacteria, Cyanobacteria, and Bacteroidetes, in concordance with WGCNA and RF results.
Discussion
To obtain a more thorough understanding of the ocean’s biological carbon pump, a deeper knowledge of the organisms that influence particle formation, sinking, and degradation is required. Here, time-series analyses of open-ocean particles and particle flux at 4,000 m allowed us to identify taxa associated with sinking particles over 3 y, in particular those that were differentially enriched during periods of elevated carbon flux. These observations provide a clearer picture of the specific organisms, and their variability, that are associated with open-ocean sinking particle flux (Fig. 5). The associated phylogenetic information and genome-enabled prokaryote trait predictions were also useful for inferring which organisms and metabolic processes might be important to the dynamic processes that promote particle formation, sinking, and degradation (Fig. 5).
Fig. 5.
Conceptual diagram of taxon temporal export dynamics, inferred traits, and predicted processes on particles collected at abyssal depths. Export dynamics line plots represent the temporal abundance profiles of representative taxonomic groups on sinking particles collected at 4,000 m. Groups shown correlated with elevated carbon flux and the summer export pulse (SEP) period (green oval), the Spring 2015 elevated carbon flux period (Spring 2015 ECF, blue oval), or neither period (Non-ECF, orange oval), in the 3-y time series. Inferred traits were derived from characteristics of known close relatives, or directly from genes and pathways present in the relevant particle-associated prokaryote MAGs. Predicted processes were derived from the export dynamics and inferred traits. Within each oval, the eukaryote Export dynamic temporal profiles are located in the topmost plot, with prokaryote temporal profiles below them. The gray area indicates the SEP time periods. The specific taxa associated with each Export dynamic plot are listed in Dataset S5 (eukaryotes) or Dataset S3 (prokaryotes). BChll, bacteriochlorophyll; Chll, chlorophyll.
Our analyses confirmed that symbiotic nitrogen-fixing cyanobacteria and their rhizosolenid diatom hosts are associated with SEP events, but also indicated that the free-living nitrogen-fixing cyanobacterium Crocosphaera (53–56) was also enriched on particles reaching the abyss during the SEP. In addition to diatom–diazotroph associations and free-living diazotrophic cyanobacterial groups, a wide variety of phototrophic and heterotrophic protists also appear to be key players in SEP events (Fig. 5). We did not find evidence of the abundant nitrogen-fixing cyanobacterial symbiont UCYN-A on sinking particles reaching the deep-sea during the SEP, or other time periods (53, 57).
In addition to rhizosolenid diatoms, a number of different photosynthetic protists correlated with elevated carbon flux during the SEP, which were also identified in RF classification analyses (Fig. 5 and SI Appendix, Tables S5 and S7). These carbon flux and SEP correlating photoautotrophic protists included other araphid and centric diatoms, green algae, coccolithophores, and other algal haptophyte and stramenopile lineages (Fig. 5 and SI Appendix, Table S5). Carbon flux correlating heterotrophic protists prevalent during the SEP included bicosoecids, filazoan species, MAST-9 and MAST-7 stramenopiles, parasitic protists, and thrausochytrids (Fig. 5 and SI Appendix, Table S5). The correlation of these groups with carbon flux suggests their importance in the grazing of particle-attached bacteria, and potential roles in particle aggregation and sinking, consistent with previous reports of protists occurring on sinking particles (15, 24, 25). The biomass of thrausochytrids (a heterotrophic fungus-like clade of Stramenopiles) was previously reported to rival that of bacteria on bathypelagic marine snow particles (58). Our results show that thrausochytrids also were correlated with elevated carbon flux, supporting important roles for them in the degradation of sinking particles.
The diversity and gene content of SEP-associated bacterial photoautotrophs and heterotrophs on particles reaching the abyss reflected rapid export from surface waters during SEP events. For example, the majority of the SEP-correlating, particle-associated MAGs reaching the abyss encoded photolyases (70% of all SEP-correlating MAGs), and many also encoded proteorhodopsins (30% of all SEP-correlating MAGs), indicative of their surface water origins (47, 48). In contrast, non-SEP MAGs encoded fewer photolyases (21% of the total non-SEP MAGs) and proteorhodopsins (11% of the total non-SEP MAGs), suggesting many of these represented deeper water colonizers with deep-sea origins.
A large cohort of bacterial photoheterotrophs on sinking particles reaching the abyss correlated significantly with the SEP. Among these, both bacteriochlorophyll-based anoxygenic phototrophs (46) and proteorhodopsin-based photoheterotophs (47) were well represented. SEP-associated heterotrophic bacteria were closely related to groups well known for their particle-associated lifestyles, physiologies, surface motility, predatory behaviors, or epiphytic associations (Fig. 5 and SI Appendix, Table S3) (19, 27, 59, 60). Heterotrophs included relatives of prosthecate bacteria (e.g., Phycisphaerae, Hyphomicrobiaceae, and Caulobacterales) known to attach to particles via stalks. The Flavobacteriaceae, well known for epiphytic associations, exopolymer degradation, and surface-associated gliding motility (59, 61), were also prevalent on sinking particles during the SEP. Other SEP-associated bacteria included Rhodobacteraceae, Alteromonadaceae, Vibrionaceae, and Saprospiraceae representatives, whose relatives are associated with biofilm formation, algal associations, or copiotrophic lifestyles. Bacteria with putative parasitic or predatory lifestyles were also associated with the SEP including Bdellovibrio and Micavibrio, the Caenarcaniphilales, and one archaeon within the class Nanoarchaeia.
In addition to surface-dwelling microbes exported to the abyss on rapidly sinking particles, SEP-correlating Gammaproteobacteria closely related to known autochthonous deep-sea adapted bacteria, were also prevalent (Figs. 2 and 5; SI Appendix, Table S3) (29, 30, 34, 35). Previous work has shown that long-term pressurization of deep-sea animal or sediment samples frequently recovers piezophilic bacterial species including Psychrobium, Colwellia, Moritella, Arcobacter, and Shewanella (34, 43). Notably, the positive correlation of some Colwellia spp. (a group not found in oligotrophic NPSG surface waters or shallow water sediment traps) (27, 28, 47) with the SEP, appears to reflect the rapid colonization and/or metabolic responses of deep-sea bacteria to elevated sinking particle inputs to the ocean’s interior.
Different suites of organisms were enriched on abyssal sinking particles in the Spring 2015 ECF event than were found during SEP events (Fig. 5 and SI Appendix, Tables S3–S5). The Spring 2015 ECF period included different eukaryotic taxa (including radiolaria, copepods, and fungi) as well as different bacterial taxa (including members of the family Halomonadaceae) that were clearly distinct from those associated with the SEP (Fig. 5 and SI Appendix, Tables S3–S5). Unlike the SEP, fungi were specifically enriched during the Spring 2015 ECF event, a result consistent with a previous report of fungal prevalence on bathypelagic marine snow (58). These results suggest that distinct biological processes are at play at different times of year with respect to the formation, export, degradation, and recycling of sinking particles in the oligotrophic open ocean.
Some of the deep-sea sinking particle-associated taxa that we observed overlapped with those previously reported to correlate with optically inferred, near-surface (150-m) carbon export in a recent global ocean survey (15). These included thrausochytrids, and bacterial Arcobacter, Colwellia, Vibrio, Idiomarina, Halomonadaceae, Phycisphaeraceae, Piscirickettsiaceae, and Flavobacteriaceae species. In contrast, sinking particle-associated Alphaproteobacteria and Bdellovibrionales species we observed in SEP events, were not noted in this prior study (15). With respect to eukaryotes, we observed that siphonophores, copepods, foraminifera, radiolarians, and alveolate parasites were generally abundant on deep-sea sinking particles, a result consistent with this recent global survey (15). Notably, diatoms were not strongly correlated with carbon export in the global ocean survey reported by Guidi et al. (15), which the authors attributed to their sparse sampling of silicate replete regions. However, our study was conducted in a silicate-depleted region, and still revealed significant correlations of sinking diatoms, diatom-diazotrophic associations, and free-living diazotrophic cyanobacteria to directly measured carbon flux, and SEP events. These results support the utility of time-series analyses for documenting the intensity and variability of biological processes associated with deep-sea carbon export.
SEP-associated microbes on particles reaching the abyss are distinct from those found at other times of year, likely for several reasons. Flux coherence between sediment traps set at depths of 2,800 and 4,000 m indicate that particles sink rapidly, especially during the SEP (8). This period of rapid sinking appears to be the result of intense summer phytoplankton blooms and aggregation, which we show here to consist of nitrogen-fixing cyanobacteria, diatoms, haptophyte and stramenopile algae, protistan heterotrophs, and surface-attached Bacteria and Archaea. These faster sinking particles are predicted to reach the abyss less degraded during transit, better preserving the molecular signatures of their surface water origins (Fig. 5). The Spring 2015 ECF particle-associated assemblages, in contrast, were composed of different suites of algae, animals, heterotrophic protists, fungi, and bacteria, suggesting different modes of particle aggregation and processing in the ocean’s interior during these events (Fig. 5). In contrast to the SEP, at other times of year mid- and deep-water particle colonization, predation, degradation, and repackaging (by deep-sea bacteria, protists, and animals) appears to shape the biotic composition of particles reaching the abyss. Large sporadic pulses of dead meso- and bathypelagic animals (especially pteropods and siphonophores), and their associated microbiota, also appear to be important components of this open ocean, deep-sea carbon flux. SEP dynamics then reflect large, temporally constrained carbon flux events, during which SEP-associated microorganisms from surface waters augment other surface, mid-, and deep-water processes during their rapid export (Fig. 5).
Future studies focused on the nature of particle-forming microorganisms over different depth horizons, their diverse physiological activities, and their quantitative contributions to matter and energy flux will further clarify central processes of sinking particle formation, breakdown, and transformation. Controlled laboratory studies examining degradation of defined polymeric growth substrates using representative particle-associated bacterial cultivars also offer rich experimental testing grounds (62). Comparative genomic analyses of microbes on suspended versus sinking particles versus free-living bacterioplankton throughout the water column, can help to identify habitat-specific ecological and evolutionary trends (27, 28, 49). Along with oceanographic, biogeochemical, and process measurements and regionally oriented time-series efforts (8, 29, 30), the above approaches can provide a deeper understanding of biological mechanisms that help regulate the functioning and efficiency of the ocean’s biological pump.
Materials and Methods
Sample Collection.
Sinking particulate organic matter was collected and preserved in sequencing sediment traps deployed at 4,000 m over a 3-y time period as previously described (29). After sediment trap recovery and deployment, all samples were processed, and DNA and RNA extracted and purified as previously described (29). See SI Appendix, Detailed Methods for further information.
Sample Processing and Data Generation.
Metatranscriptomic and metagenomic sequencing libraries were prepared as previously described (29). Quality controlled sequences were compared to all rRNA models from RFAM release 12.1 (63) using cmsearch (parameters:–hmmonly) from infernal 1.1.2 (64). The taxonomic affiliations of the SSU rRNA sequences were assessed by homology to the SILVA SSU rRNA NR99 database release 132 (65) using BWA 0.7.15-r1140 (66) with matches limited to at least 97% identity over at least 70 bases. See SI Appendix, Detailed Methods for further information.
Recovery of the MAGs.
Mapping of quality reads was performed using CoverM v0.3.1 with default parameters (https://github.com/wwood/CoverM). For each assembled metagenome, MAGs were recovered using MetaBAT1 v0.32.5 (67) with all the sensitivity settings, MetaBAT2 v2.12.1 (68), and MaxBin2 v2.2.6 (69) using the 40 and 107 gene sets. The resulting MAGs from each assembly were assessed for completeness and dereplicated using DAStool (70). Completeness and contamination rates of the MAGs were assessed using CheckM v1.0.13 (71) with the “lineage wf” command. MAGs from all metagenomes were subsequently dereplicated using dRep v2.2.3 (72) using the dereplicate_wf at ≥97% average nucleotide identity over ≥70% alignment, and the best MAGs were chosen based on genome completeness. The dereplicated MAGs were further refined by reassembling the mapped quality trimmed reads with SPAdes (73) using the –careful and –trusted-contigs setting. Additional scaffolding and resolving ambiguous bases of the MAGs were performed using the “roundup” mode of FinishM v0.0.7 (https://github.com/wwood/finishm).
Taxonomic Inference of the MAGs.
Classification of the MAGs was determined using GTDB-Tk (74) v.0.3.3 implementing the classify_wf command (https://sciPy.com/Ecogenomics/GtdbTk). Briefly, marker genes were identified in each genome, aligned, concatenated, and classified with pplacer to identify the maximum-likelihood placement of each genome's concatenated protein alignment in the GTDB-Tk reference tree. GTDB-Tk classifies each genome based on its placement in the reference tree, its relative evolutionary distance, and FastANI distance. See SI Appendix, Detailed Methods for further details on MAG annotation and diversity analyses.
Calculation of MAG Relative Abundances.
To calculate the relative abundance, reads from each metagenomic dataset were mapped to the dereplicated MAGs using CoverM v0.3.1 with the “contig” command and a cutoff of 95% minimum identity and minimum aligned read length of 75% of each read. Coverage of each contig was calculated with the CoverM “trimmed_mean” option and the coverage for each MAG was calculated as the average of all contig coverages, weighted by their length. The relative abundance of MAGs in each metagenomic dataset was calculated as its coverage divided by the total coverage of all genomes in the dereplicated MAG dataset.
Genomes that Were Selectively Enriched during the SEP.
To determine which lineages were differentially abundant between the SEP and non-SEP samples, the relative abundance profiles from each group were compared. All statistical analysis was performed using sciPy (75) in the Python programming environment (76). The mean and statistical significance of the difference were analyzed using the Mann–Whitney U test. All P values that were affected by multiple testing were corrected for false discovery using the Benjamini–Hochberg procedure.
Read Mapping and Coverage Profile of the nifH Gene Sequences.
Coverage profile of the nifH gene sequences from the MAGs and metagenome generated using CoverM v0.3.1 with the “contig” command and a cutoff of 95% minimum identity and minimum aligned read length of 75% of each read. Coverage of each gene sequence was calculated with the CoverM “trimmed_mean” option.
WGCNA.
To identify assemblages of families highly correlating with carbon flux, WGCNA was done using the R package WGCNA (77). Transformations were performed using the R “DeSeq2” package variance stabilizing transformation function vst() (78, 79). Visualization of the resulting modules and their correlation with carbon flux was performed using Cytoscape (80). See SI Appendix, Detailed Methods for further information.
RF Models.
To further define rRNA-based taxon associations with SEP events, a RF classification and regression tree machine-learning model was built using the R package randomForest() (51). Each tree yielded a classification and the majority votes among all the trees were counted and a prediction was assigned. An out-of-bag-error-rate (OOB) was estimated to evaluate the accuracy of classifications. The final Prokaryote model was built with 10,001 trees and 32 variables were tried at each split. The OOB estimate was 4.76%, and our permutation test (n = 100) showed the model was significant (P < 0.05). The Eukaryote RF model was also built using 10,001 trees, with 29 variables tried at each split, and had an OOB estimate at 14.29%, and a cross-validation accuracy (85.7%), with Kappa values (15.8%); the permutation test (n = 100) yielded significant results (P < 0.05). See SI Appendix, Detailed Methods and Datasets S8 and S9, for further information.
Correlation Network Analyses of Sinking Particle-Associated Bacteria and Eukaryotes.
Correlation networks were inferred between the 2,259 families of bacteria, archaea, and eukaryotes using FastSpar(v0.0.10) (81), an efficient C++ implementation of the SparCC, which is based on the dbOTU3 algorithm (81–83). Correlations between bacteria, archaea, and eukaryotes were based on a distribution-based clustering method dbOTU3, 50 iterations, and a cutoff threshold of 0.40 (P < 0.05) (83). A permutation approach was used to assess correlation significance. From the original count data, 1,000 bootstrap counts were generated using the command fastspar_bootstrap. From the randomly permuted data, P values were calculated using fastspar_pvalues. Only interdomain correlation values calculated were retained. Using the same count table, a Kendall correlation test between taxa and carbon flux values was performed using the cor.test function in the R package v3.6.0 “Stats” (84). Only correlations with values of P < 0.05 were considered significant. The network was visualized using the Cytoscape (v3.7.2) software to identify the most significant positive correlations between groups and carbon flux (80).
Kendall Tau correlation tests were performed on SSU rRNA abundance data to identify taxa specifically enriched during the Spring 2015 ECF (SI Appendix, Fig. S4 and Dataset S4). Only correlation values found to be significant (P < 0.05) were retained. See SI Appendix, Detailed Methods and Datasets S8 and S9 for further information.
Data Visualization.
Figures were generated using pheatmap (85) and ggplot2 (86) packages in R (79) and further refined using Adobe Illustrator.
Supplementary Material
Acknowledgments
We thank Tara Clemente, Blake Watkins, and Eric Grabowski for their expert effort and leadership in sediment trap deployment, recovery, and biogeochemical analysis; the captain and crew of the RV Kilo Moana for able assistance at sea; Bethanie Edwards for assistance with nucleic acid extractions and purifications; and Paul Den Uyl and Anna Romano for assistance with metagenomic and metatranscriptomic library preparation and sequencing. This research was supported by grants from the Simons Foundation (329108 to E.F.D. and D.M.K., and 721223 to E.F.D.) and Gordon and Betty Moore Foundation Grants GBMF 3777 (to E.F.D.) and GBMF 3794 (to D.M.K.). This work is a contribution of the Simons Collaboration on Ocean Processes and Ecology and the Daniel K. Inouye Center for Microbial Oceanography: Research and Education.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018269118/-/DCSupplemental.
Data Availability.
Sequence read data are available from the National Center for Biotechnology Information (NCBI) short read archive (SRA) under Bioproject no. PRJNA482655. Individual MAGs are available under NCBI accession nos. SAMN14675689 to SAMN14675809. All other data products associated with this study are available from the corresponding author upon request.
References
- 1.Martin J. H., Knauer G. A., Karl D. M., Broenkow W. W., VERTEX: Carbon cycling in the northeast Pacific. Deep-Sea Res. 34, 267–285 (1987). [Google Scholar]
- 2.Karl D. M., Knauer G. A., Martin J. H., Downward flux of particulate organic matter in the ocean: A particle decomposition paradox. Nature 332, 438–441 (1988). [Google Scholar]
- 3.Grabowski E., Letelier R. M., Laws E. A., Karl D. M., Coupling carbon and energy fluxes in the North Pacific Subtropical Gyre. Nat. Commun. 10, 1895–1904 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruhl H. A., Ellena J. A., Smith K. L. Jr, Connections between climate, food limitation, and carbon cycling in abyssal sediment communities. Proc. Natl. Acad. Sci. U.S.A. 105, 17006–17011 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith K. L. Jr, Ruhl H. A., Huffard C. L., Messié M., Kahru M., Episodic organic carbon fluxes from surface ocean to abyssal depths during long-term monitoring in NE Pacific. Proc. Natl. Acad. Sci. U.S.A. 115, 12235–12240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karl D. M., Church M. J., Microbial oceanography and the Hawaii Ocean Time-series programme. Nat. Rev. Microbiol. 12, 699–713 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Karl D. M., Lukas R., The Hawaii Ocean Time-series (HOT) program: Background, rationale and field implementation. Deep Sea Res. Part II Top. Stud. Oceanogr. 43, 129–156 (1996). [Google Scholar]
- 8.Karl D. M., Church M. J., Dore J. E., Letelier R. M., Mahaffey C., Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc. Natl. Acad. Sci. U.S.A. 109, 1842–1849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venrick E. L., The distribution and significance of Richelia intracellularis Schmidt in the North Pacific Central Gyre. Limnol. Oceanogr. 19, 437–445 (1974). [Google Scholar]
- 10.Smetacek V., von Brockel K., Zeitzschel B., Zenk W., Sedimentation of particulate matter during a phytoplankton spring bloom in relation to the hydrographical regime. Mar. Biol. 47, 211–226 (1978). [Google Scholar]
- 11.Repeta D. J., Gagosian R. B., Carotenoid transformations in coastal marine waters. Nature 295, 51–54 (1982). [Google Scholar]
- 12.Bauerfeind E., et al. , Particle sedimentation patterns in the eastern Fram Strait during 2000–2005: Results from the Arctic long-term observatory HAUSGARTEN. Deep Sea Res. Part I Oceanogr. Res. Pap. 56, 1471–1487 (2009). [Google Scholar]
- 13.Bernstein R. E., et al. , Acantharian fluxes and strontium to chlorinity ratios in the North Pacific Ocean. Science 237, 1490–1494 (1987). [DOI] [PubMed] [Google Scholar]
- 14.Betzer P. R., et al. , The oceanic carbonate system: A reassessment of biogenic controls. Science 226, 1074–1077 (1984). [DOI] [PubMed] [Google Scholar]
- 15.Guidi L. et al.; Tara Oceans Coordinators , Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biard T., et al. , In situ imaging reveals the biomass of giant protists in the global ocean. Nature 532, 504–507 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Liu R., et al. , Depth-resolved distribution of particle-attached and free-living bacterial communities in the water column of the New Britain Trench. Front. Microbiol. 9, 625 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peoples L. M., et al. , Vertically distinct microbial communities in the Mariana and Kermadec trenches. PLoS One 13, e0195102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganesh S., Parris D. J., DeLong E. F., Stewart F. J., Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 8, 187–211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mestre M., et al. , Sinking particles promote vertical connectivity in the ocean microbiome. Proc. Natl. Acad. Sci. U.S.A. 115, E6799–E6807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz-González C., et al. , Major imprint of surface plankton on deep ocean prokaryotic structure and activity. Mol. Ecol. 29, 1820–1838 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Duret M. T., Lampitt R. S., Lam P., Prokaryotic niche partitioning between suspended and sinking marine particles. Environ. Microbiol. Rep. 11, 386–400 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Asper V. L., A review of sediment trap technique. Mar. Technol. Soc. J. 21, 18–25 (1987). [Google Scholar]
- 24.Amacher J., Neuer S., Anderson I., Massana R., Molecular approach to determine contributions of the protist community to particle flux. Deep Sea Res. Part I Oceanogr. Res. Pap. 56, 2206–2215 (2009). [Google Scholar]
- 25.Amacher J., Neuer S., Lomas M., DNA-based molecular fingerprinting of eukaryotic protists and cyanobacteria contributing to sinking particle flux at the Bermuda Atlantic time-series study. Deep Sea Res. Part II Top. Stud. Oceanogr. 93, 71–83 (2013). [Google Scholar]
- 26.LeCleir G. R., DeBruyn J. M., Maas E. W., Boyd P. W., Wilhelm S. W., Temporal changes in particle-associated microbial communities after interception by nonlethal sediment traps. FEMS Microbiol. Ecol. 87, 153–163 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Fontanez K. M., Eppley J. M., Samo T. J., Karl D. M., DeLong E. F., Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front. Microbiol. 6, 469–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelve E. A., Fontanez K. M., DeLong E. F., Bacterial succession on sinking particles in the ocean’s interior. Front. Microbiol. 8, 2269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeuf D., et al. , Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proc. Natl. Acad. Sci. U.S.A. 116, 11824–11832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preston C. M., Durkin C., Yamahara K., DNA metabarcoding reveals organisms contributing to particulate matter flux to abyssal depths in the North East Pacific Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 173, 104708 (2020). [Google Scholar]
- 31.Metfies K., et al. , Protist communities in moored long-term sediment traps (Fram Strait, Arctic)—preservation with mercury chloride allows for PCR-based molecular genetic analyses. Front. Mar. Sci. 4, 301 (2017). [Google Scholar]
- 32.Ottesen E. A., et al. , Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 5, 1881–1895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutierrez-Rodriguez A., et al. , High contribution of Rhizaria (Radiolaria) to vertical export in the California Current Ecosystem revealed by DNA metabarcoding. ISME J. 13, 964–976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLong E. F., Franks D. G., Yayanos A. A., Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl. Environ. Microbiol. 63, 2105–2108 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauro F. M., Chastain R. A., Blankenship L. E., Yayanos A. A., Bartlett D. H., The unique 16S rRNA genes of piezophiles reflect both phylogeny and adaptation. Appl. Environ. Microbiol. 73, 838–845 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albertsen M., et al. , Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Sharon I., et al. , Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 23, 111–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaumeil P.-A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database (Oxford University Press, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang H., Li T., Chen M., Huang G., Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchan A., LeCleir G. R., Gulvik C. A., González J. M., Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Foster R. A., et al. , Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J. 5, 1484–1493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villareal T. A., Evaluation of nitrogen fixation in the diatom genus Rhizosolenia Ehr. in the absence of its cyanobacterial symbiont Richelia intracellularis Schmidt. J. Plankton Res. 9, 965–971 (1987). [Google Scholar]
- 43.Peoples L. M., et al. , Microbial community diversity within sediments from two geographically separated hadal trenches. Front. Microbiol. 10, 347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunoura T., et al. , Hadal biosphere: Insight into the microbial ecosystem in the deepest ocean on Earth. Proc. Natl. Acad. Sci. U.S.A. 112, E1230–E1236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salazar G., et al. , Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 10, 596–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Béjà O., et al. , Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415, 630–633 (2002). [DOI] [PubMed] [Google Scholar]
- 47.DeLong E. F., et al. , Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311, 496–503 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Pinhassi J., DeLong E. F., Béjà O., González J. M., Pedrós-Alió C., Marine bacterial and archaeal ion-pumping rhodopsins: Genetic diversity, physiology, and ecology. Microbiol. Mol. Biol. Rev. 80, 929–954 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farnelid H., et al. , Diverse diazotrophs are present on sinking particles in the North Pacific Subtropical Gyre. ISME J. 13, 170–182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parks D. H., et al. , A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 52.Liaw A., Wiener M., Classification and regression by randomForest. R. News 2, 18–22 (2002). [Google Scholar]
- 53.Foster R. A., Zehr J. P., Diversity, genomics, and distribution of phytoplankton-cyanobacterium single-cell symbiotic associations. Annu. Rev. Microbiol. 73, 435–456 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Wilson S. T., et al. , Coordinated regulation of growth, activity and transcription in natural populations of the unicellular nitrogen-fixing cyanobacterium Crocosphaera. Nat. Microbiol. 2, 17118 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Dugenne M., Henderikx-Freitas F., Wilson S., Karl D. M., Life and death of Crocosphaera sp. in the Pacific Ocean: Fine scale predator-prey dynamics. Limnol. Oceanogr. 10.1002/lno.11473 (2020). [DOI] [Google Scholar]
- 56.Barone B., et al. , The ecological and biogeochemical state of the North Pacific Subtropical Gyre is linked to sea surface height. J. Mar. Res. 77, 215–245 (2019). [Google Scholar]
- 57.Thompson A. W., et al. , Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1550 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Bochdansky A. B., Clouse M. A., Herndl G. J., Eukaryotic microbes, principally fungi and labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME J. 11, 362–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeLong E. F., Franks D. G., Alldredge A. L., Phylogenetic diversity of aggregate-attached versus free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934 (1993). [Google Scholar]
- 60.Simon M., Grossart H. P., Schweitzer B., Ploug H., Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211 (2002). [Google Scholar]
- 61.McBride M. J., “The family Flavobacteriaceae” in The Prokaryotes, Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F., Eds. (Springer, Berlin, 2014), pp. 643–676. [Google Scholar]
- 62.Enke T. N., et al. , Modular assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. 29, 1528–1535.e1526 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Nawrocki E. P., et al. , Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 43, D130–D137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nawrocki E. P., Eddy S. R., Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quast C., et al. , The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN] (16 March 2013).
- 67.Kang D. D., Froula J., Egan R., Wang Z., MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang D. D., et al. , MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Y. W., Simmons B. A., Singer S. W., MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Sieber C. M. K., et al. , Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olm M. R., Brown C. T., Brooks B., Banfield J. F., dRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaumeil P.-A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics, btz848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Virtanen P. et al.; SciPy 1.0 Contributors , SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Rossum G., Drake F. L. Jr, Python Reference Manual (Centrum voor Wiskunde en Informatica, Amsterdam, 1995). [Google Scholar]
- 77.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2013).
- 80.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watts S. C., Ritchie S. C., Inouye M., Holt K. E., FastSpar: Rapid and scalable correlation estimation for compositional data. Bioinformatics 35, 1064–1066 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Friedman J., Alm E. J., Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8, e1002687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olesen S. W., Duvallet C., Alm E. J., dbOTU3: A new implementation of distribution-based OTU calling. PLoS One 12, e0176335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.R Core Team , R: a language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2016). [Google Scholar]
- 85.Kolde R. pheatmap v.1.0.12 (2015). https://cran.r-project.org/package=pheatmap. Accessed 8 March 2019.
- 86.Wickham H., ggplot2: Elegant Graphics for Data Analysis (Springer, 2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence read data are available from the National Center for Biotechnology Information (NCBI) short read archive (SRA) under Bioproject no. PRJNA482655. Individual MAGs are available under NCBI accession nos. SAMN14675689 to SAMN14675809. All other data products associated with this study are available from the corresponding author upon request.