The transfer of nutrients between cells, or cross-feeding, is a ubiquitous feature of microbial communities with emergent properties that influence our health and orchestrate global biogeochemical cycles. Cross-feeding inevitably involves the externalization of molecules.
KEYWORDS: biofilm, coculture, cross-feeding, exoenzymes, microbial ecology, mutualism, nanowires, quorum sensing, siderophores, synthetic ecology
SUMMARY
The transfer of nutrients between cells, or cross-feeding, is a ubiquitous feature of microbial communities with emergent properties that influence our health and orchestrate global biogeochemical cycles. Cross-feeding inevitably involves the externalization of molecules. Some of these molecules directly serve as cross-fed nutrients, while others can facilitate cross-feeding. Altogether, externalized molecules that promote cross-feeding are diverse in structure, ranging from small molecules to macromolecules. The functions of these molecules are equally diverse, encompassing waste products, enzymes, toxins, signaling molecules, biofilm components, and nutrients of high value to most microbes, including the producer cell. As diverse as the externalized and transferred molecules are the cross-feeding relationships that can be derived from them. Many cross-feeding relationships can be summarized as cooperative but are also subject to exploitation. Even those relationships that appear to be cooperative exhibit some level of competition between partners. In this review, we summarize the major types of actively secreted, passively excreted, and directly transferred molecules that either form the basis of cross-feeding relationships or facilitate them. Drawing on examples from both natural and synthetic communities, we explore how the interplay between microbial physiology, environmental parameters, and the diverse functional attributes of extracellular molecules can influence cross-feeding dynamics. Though microbial cross-feeding interactions represent a burgeoning field of interest, we may have only begun to scratch the surface.
INTRODUCTION
The sheer abundance of microbes on Earth is astounding. Globally, there are an estimated ∼1030 individual bacterial and archaeal cells (1, 2). Furthermore, most eukaryotes, including protists, microalgae, and some fungi, are unicellular and microscopic (3, 4). The abundance of microbes can be attributed, in part, to their unparalleled genomic and metabolic diversity, allowing them to inhabit nearly every imaginable niche (5–10). Within a given environment, microbes typically exist as multispecies communities. Some of the densest microbial communities are those found in the human gut and cow rumen, which are colonized by ∼1014 and ∼1015 microbes, respectively (10, 11). For the human colon, these numbers translate to about 1011 cells per ml (11). Other communities are comparatively dilute, such as those in seawater which range between 104 and 106 cells per ml (12, 13). In both dense and dilute communities, competition for scarce nutrients is the norm (14–21).
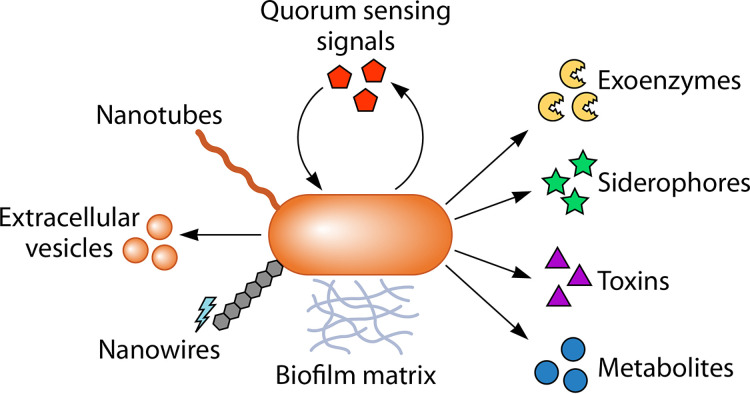
Competition for limiting resources has both physiological and ecological consequences. Most microbes spend at least part of their existence starving for nutrients and often enter a nongrowing state, called dormancy or growth arrest (22–24). In fact, dormant microbes represent a major fraction of the resident microbiota in many environments (22, 23). However, nongrowing microbes need not be metabolically inactive. Even dormant microbes typically exhibit a low metabolic rate to sustain critical activities, collectively termed maintenance metabolism, including DNA repair, protein and lipid turnover, osmoregulation, and nutrient transport (24–30). In addition to maintaining the cell, starved microbes combat nutrient limitation by engaging in behaviors that facilitate nutrient acquisition. These behaviors are commonly mediated by the production and release of molecules into the extracellular environment, such as metabolites, exoenzymes, siderophores, toxins, signals, and cell surface-associated factors (Fig. 1).
FIG 1.
Microbes release various molecules that promote cross-feeding. Microbially produced extracellular molecules such as quorum sensing signals, exoenzymes, siderophores, toxins, metabolites (e.g., sugars, organic acids, amino acids, etc.), biofilm matrix, nanowires, extracellular vesicles, and nanotubes can be consumed by neighboring microbes or can influence cross-feeding between cells.
Once outside the producing cell, these molecules can affect the survival and proliferation of the producer, neighboring microbes, and where applicable, host cells. Thus, ecological outcomes can be dictated by molecules that are actively secreted or passively excreted into the environment or, in some instances, directly transferred between cells. In many cases, externalized molecules instigate competition, but in other cases, externalized molecules facilitate synergistic cross-feeding (17, 31–41). If conditions prevail, cooperative cross-feeding relationships can be reinforced and ultimately influence community-wide processes of both global and societal importance, ranging from biogeochemical cycles that govern Earth’s climate to the progression of polymicrobial infections.
In this review, we highlight major classes of molecules produced and externalized by microbes that are either known to have important roles in cross-feeding or that have the potential to do so (Fig. 1). Our review thus adds a molecular perspective on cross-feeding interactions, supplementing other perspectives on cross-feeding available in other recent reviews (31–33, 36–44). While some externalized molecules promote a range of polymicrobial interactions, including exploitation and interbacterial warfare, we focus here on the establishment and maintenance of cross-feeding in both natural and synthetic communities. We also provide a nuanced perspective on cross-feeding as a dynamic phenomenon, highlighting conditions that affect the relative benefit of cross-feeding, including the unavoidable prospect of competition between otherwise cooperative partners. Given the current pace of discovery in microbial cross-feeding, we predict that the field has only begun to catalog the diverse range of cross-feeding interactions that exists in nature.
WHAT COUNTS AS CROSS-FEEDING?
This nonexhaustive review covers many examples of externalized molecules that can participate directly or indirectly in cross-feeding. Every microbe likely excretes a variety of molecules with the potential to impact the activity of a neighbor. How does one determine whether these molecules participate in cross-feeding?
We propose a broad definition of cross-feeding that should minimally be true to the literal notion of one producer cell or population feeding a recipient cell or population by meeting the following criteria.
(i) Material must be transferred from a producer to a recipient. Transferred material can include molecules but also subatomic particles like electrons and protons. For brevity we tend to refer to molecules in this review. We leave open the possibility for nonmaterial energy transfer. Arguably, the sharing of a proton motive force between cells is an example of energy cross-feeding since it is the shared electrochemical gradient that is important, rather than the protons themselves.
(ii) Transferred material must either be assimilated by the recipient or participate in energy transformation in the recipient and/or producer. This definition thus excludes some cases where a recipient benefits a producer through the detoxification of an inhibitory compound. While such detoxification can be critical to sustain a producer’s metabolism, the process would not count as cross-feeding unless the inhibitor was assimilated or used to derive energy. Otherwise, the metabolic activities would not meet the definition of “feeding.” Similarly, the degradation of intercellular signaling molecules, such as quorum quenching (45), would connect the metabolism of a producer and a recipient that degrades the molecule. However, unless the signal is assimilated or used for energy, this connection would not meet our definition of cross-feeding.
(iii) The fitness of the producer and/or the recipient must be altered as a result of assimilation or energy derived from the transferred material. Some assimilated or energy-yielding material will likely have a negligible impact on producer or recipient fitness due to either a low quantity of externalized material or an environmental context that diminishes the fitness effect of an externalized material. Assessing whether this criterion is met will not always be trivial. Measurements of growth rates and even competition assays are not always sensitive enough to detect small but important fitness effects that affect population frequencies over relatively long time scales.
(iv) Cross-feeding must involve different species or genotypically or phenotypically distinct populations. This criterion is a practical matter for studying the nature and consequences of cross-feeding. For example, a clonal population homogenously distributed in a test tube could involve nutrient transfer between cells. However, it would be a futile exercise to examine cross-feeding in this context because every cell would function equally as both a producer and a recipient. However, cross-feeding within a clonal population can be important in a mixed genotypic context because the amount of sharing between clones determines the extent to which externalized materials are privatized by a population versus available to other species or genotypes. Similarly, there are examples where a clonal population can differentiate into phenotypically distinct populations that engage in cross-feeding (46).
In this review, we cover a range of externalized molecules. Some of these molecules directly serve as cross-fed material that is assimilated or contributes to energy transformation. We also cover externalized molecules that can facilitate cross-feeding rather than serving as the transferred material. These facilitating molecules are thus not necessarily indicators of cross-feeding on their own since they can also have important physiological roles in the absence of cross-feeding. These facilitating molecules must act in concert with at least one directly cross-fed material or energy source.
There are a plethora of examples of nutrient transfer that meet the above criteria. Currently, the terminology used to describe different types of cross-feeding interactions lacks consensus, in part because of the expanding diversity of interactions and molecule functionalities being recognized. For instance, a catalog of 74 microbial interactions was recently created and applied to gain high-level comparative insights through hierarchical clustering (47). Rather than side with one proposed convention over another, we attempt here to use terminology that is sufficiently descriptive to be self-explanatory and inclusive of specific roles that externalized molecules can both directly and indirectly play in cross-feeding.
EXTRACELLULAR MOLECULES THAT PROMOTE CROSS-FEEDING
Metabolites
Microbes produce and release many small molecules, or metabolites. Here, we classify cross-fed metabolites as excreted molecules that primarily serve as biosynthetic intermediates and not as toxins or signals for cell-cell communication. However, this distinction is not always possible, and we point to cases throughout this review where metabolites might have inhibitory properties that allow them to act as toxins under certain situations. Common examples of cross-fed metabolites include sugars, organic acids, amino acids, vitamins, gasses, and reduced or oxidized inorganic elements and molecules.
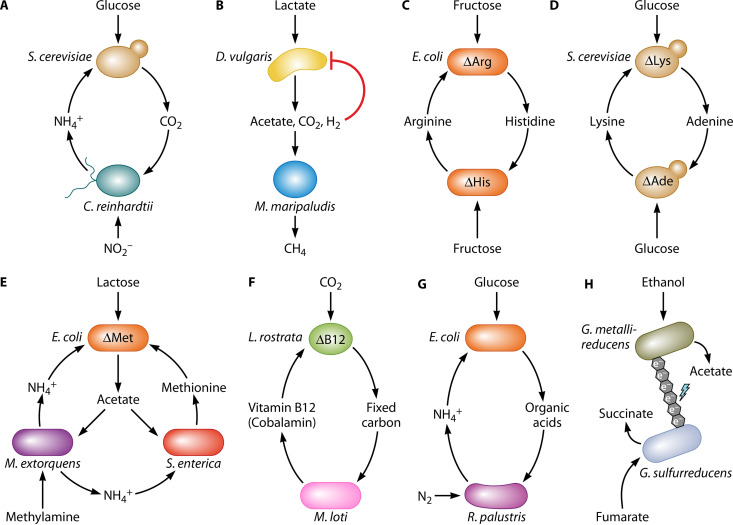
Metabolite cross-feeding is ubiquitous in microbial communities (31–41, 48–50). Furthermore, metabolite cross-feeding has been leveraged to establish a diverse array of synthetic communities, collectively spanning all three domains of life (Fig. 2). Such synthetic communities facilitate the study of microbial interactions by offering genetic and experimental tractability beyond what is possible with natural communities, while still retaining some of the complexity of natural communities (33, 42–44). In many cases, stable cross-feeding relationships can be established through genetic engineering and/or experimental evolution, while in other cases, careful design of environmental conditions is enough to enforce cross-feeding. The latter scenario was exemplified between NH4+-excreting Chlamydomonas reinhardtii and CO2-producing yeast, which established a cross-feeding relationship without any need for genetic engineering or evolution (Fig. 2A) (51).
FIG 2.
Synthetic microbial consortia featuring essential cross-feeding interactions. (A) Bidirectional cross-feeding of CO2 and NH4+ between Saccharomyces cerevisiae and Chlamydomonas reinhardtii (51). (B) Syntrophic cross-feeding of acetate, CO2, and H2 between Desulfovibrio vulgaris and Methanococcus maripaludis (61, 62). (C) Bidirectional cross-feeding of amino acids between Escherichia coli auxotrophs (64, 70). (D) Bidirectional cross-feeding of lysine and adenine between S. cerevisiae auxotrophs (75, 245). (E) Multidirectional cross-feeding of acetate, methionine, and NH4+ between auxotrophic E. coli, Salmonella enterica, and Methylobacterium extorquens (74, 246). (F) Bidirectional cross-feeding of fixed carbon and cobalamin between Lobomonas rostrata and Mesorhizobium loti (78). (G) Bidirectional cross-feeding of organic acids and NH4+ between E. coli and Rhodopseudomonas palustris (207). (H) Syntrophic cross-feeding of electrons between Geobacter metallireducens and Geobacter sulfurreducens (63). The “Δ” symbol indicates an auxotrophy for an incoming metabolite.
Metabolite cross-feeding can be unidirectional (with metabolites being transferred solely from one microbe to another), bidirectional, or even multidirectional, wherein metabolites are reciprocally exchanged between partners. Cross-fed metabolites can be either metabolic waste, which provide no further benefit to the producer after excretion, or communally valuable compounds, for which benefits can be reaped by whichever producer or recipient cell acquires the metabolite. Metabolite cross-feeding interactions within a microbial community can be numerous, creating a complex network involving either or both waste and communally valuable metabolites (Fig. 2).
Waste metabolites.
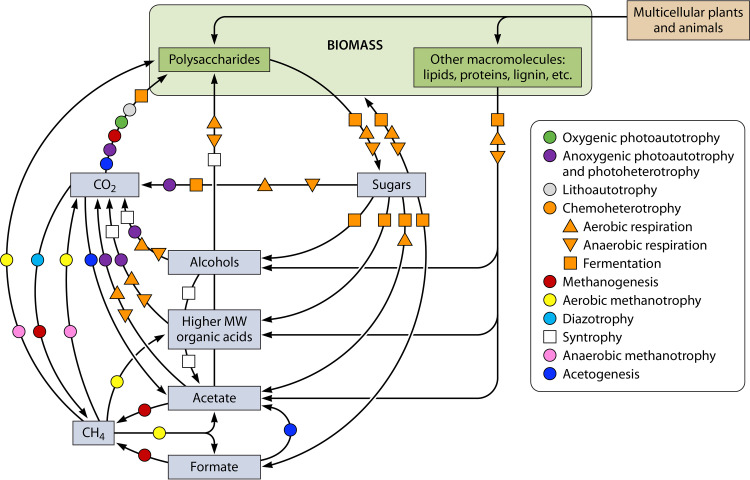
The vast metabolic diversity of microbes creates many scenarios for metabolite cross-feeding. For example, O2 waste from various photosynthetic cyanobacteria is collectively necessary for the aerobic respiration of diverse microbes. CO2 is a waste product of most heterotrophic lifestyles, including many fermentations and even photoheterotrophy (52), wherein organic substrates serve as an electron source for light-driven energy transformation reactions (Fig. 3). This CO2 in turn provides essential carbon for biosynthesis in diverse photo- and litho-autotrophs, and CO2 can even be used as a conditionally essential electron acceptor for photoheterotrophic lifestyles (53) (Fig. 3). CO2 also serves as an auxiliary carbon source for some chemoheterotrophic lifestyles, such as succinate-excreting fermentations which can exhibit net CO2 fixation (54). Fermentative microbes partition their carbon sources for biosynthesis and as disposable electron acceptors, resulting in the excretion of reduced compounds that serve as important carbon and/or electron sources for diverse lifestyles (Fig. 3). Anaerobically respiring microbes present even more variation in electron donor and acceptor couplings and thus establish cross-feeding of various inorganic metabolites. Inorganic reduced products of anaerobic respiration such as H2S, NH4+, and Fe2+ are utilized as electron sources in diverse lithoautotrophic and anoxygenic photoautotrophic lifestyles. The connections go on, eventually completing global biogeochemical cycles for carbon (Fig. 3) and other elements central to life such as N, S, Fe, and P. Beyond these classic examples, computational models involving relatively few species point to the possibility of even more cross-feeding interactions based on waste metabolites (55). Taken together, microbial cross-feeding of metabolic waste is a major variable that determines the course of life on Earth, as well as the Earth’s climate (56).
FIG 3.
Cross-feeding of carbon waste metabolites creates a global carbon cycle. The indicated trophic categories only refer to possible effects on carbon transformation. For example, chemoheterotrophs are primarily responsible for the conversion of macromolecules from both microbes and multicellular organisms into diverse organic compounds that can serve as nutrients for other lifestyles but might also participate in the cycling of other elements. Acetogenesis arrows involve the excretion of acetate from the conversion of two CO2 to acetyl-CoA via energy-conserving Wood-Ljungdahl pathway activity, rather than referring to every lifestyle possible for a bacterium classified as an acetogen (247). Syntrophy arrows are for fermentative carbon transformations that require consumption by a partner to be thermodynamically feasible (31, 57, 58). The diazotrophy arrow represents one of the carbon transformations known to be carried out by nitrogenase in sufficient quantity to support the growth of a partner (236). In some cases, only certain organisms within a category might generate a given molecule. The figure does not necessarily capture every possible activity within a given lifestyle. The figure also does not include every carbon transformation known to be carried out by microbes.
At a physiological level, consumption of metabolic waste by a recipient often provides little benefit to the producer. In other cases, consumption of metabolic waste is essential for producer viability, such as for the lifestyle called syntrophy. Strictly and historical speaking, syntrophy is a form of cross-feeding wherein an energetically unfavorable metabolism of a producer is pulled forward by coupling to the metabolism of a recipient partner (31, 57, 58). The producer is typically a fermentative bacterium subsisting on electron-rich carbon sources, which quickly becomes thermodynamically unfeasible as products accumulate. However, these bacteria thrive in partnership with other microbes that remove fermentation products like H2, CO2, formate, and acetate (Fig. 3). In many cases, the recipient that consumes the fermentation products is an archaeal methanogen, though bacterial acetogens or sulfate or iron reducers can also fulfill this recipient role (31, 57, 58). In each case, removal of fermentation products by the recipient results in a thermodynamically favorable coupled metabolism (31, 57, 58). Syntrophies have been implicated as an important part of the carbon cycle and in the anaerobic degradation of electron-rich pollutants (58–60).
Syntrophic relationships have also been established and evolved in laboratories (61–63). A prominent example is that between the sulfate-reducing bacterium Desulfovibrio vulgaris, deprived of sulfate to force a fermentative lifestyle on lactate, and the methanogen Methanococcus maripaludis that consumes the fermentation products. Lactate fermentation by D. vulgaris on its own would be thermodynamically infeasible, but the coupled metabolism supports the growth of both species (Fig. 2B). Over time, evolved D. vulgaris populations frequently lost the ability to respire sulfate, suggesting that D. vulgaris was becoming specialized for obligate syntrophic growth with M. maripaludis (62). Such synthetic communities clearly illustrate how obligate cross-feeding, initially imposed by environmental conditions, can become fixed as genes for independent lifestyles decay.
Communally valuable metabolites.
Natural and synthetic cross-feeding of costly, communally valuable nutrients is similarly diverse. Communally valuable compounds include ammonia/ammonium (NH3/NH4+), amino acids, nucleobases, and vitamins. Many microbial taxa cannot produce certain essential metabolites, a trait referred to as auxotrophy. Auxotrophies for certain amino acids and vitamins are widespread in nature (32, 34, 36, 50, 64–67). The evolutionary origins of auxotrophies and cross-feeding are presumably also intertwined (65, 68). It seems plausible that the ubiquity of cross-feeding creates environments where costly metabolites are extracellularly available in enough abundance to select for the adaptive loss of biosynthetic genes, thus driving the emergence of auxotrophies (32, 35, 36, 65, 68, 69). Indeed, a genome-scale survey of 800 microbial communities noted a high potential for metabolic interdependencies between subcommunities (35). Amino acid auxotrophy is one of the most common strategies to engineer or evolve synthetic cross-feeding consortia (64, 70–75) (Fig. 2C to E). Even a single producer strain can potentially stabilize a community of otherwise competing auxotrophs (76).
There does not appear to be an energetic limitation to what valuable metabolites are cross-fed. Even vitamin B12, one of the most energetically costly small molecules to synthesize (77), is cross-fed between species (48, 49, 67, 78, 79) (Fig. 2F). Vitamin B12 cross-feeding likely has global importance as B12 is transferred from heterotrophic bacteria to major algal primary producers in the world’s oceans, such as diatoms (4, 48, 49, 67); marine diatoms are estimated to account for ∼40% of primary production in the oceans (56, 80) and up to 20% of primary production globally (81, 82). The exchange of B vitamins is also thought to be prevalent between gut microbiota members (32, 50, 66, 79, 83). Similar cross-feeding of metabolically expensive heme and quinones has been observed in emergent subpopulations of Staphylococcus aureus that relied on each other for the missing respiratory cofactor (84). The cross-feeding strains were more virulent together than the self-sufficient ancestor. What selective pressure enriched for this synergistic cross-feeding relationship and why this relationship resulted in a more severe infection remain open questions.
It is not obvious why microbes release costly compounds in sufficient quantities to support the survival or growth of other species. Part of the answer may be that excretion is accidental, occurring because cell membranes are leaky (68, 69, 85, 86). This is likely true at least for molecules wherein charge equilibrium is a function of the pH, as is the case with NH3/NH4+ (85, 86). Cell lysis is another possible mechanism of costly nutrient transfer. For example, nutrient release was associated with cell death in a synthetic mutualism between amino acid and nucleotide auxotrophs of yeast (Fig. 2D) (75). There can also be selection for excretion of valuable compounds if it stimulates enough reciprocation from a recipient to outweigh the cost of excretion, thus forming the basis of a mutualism (73, 87, 88). Reciprocal cross-feeding is thought to have been an important evolutionary driver of rapid metabolism and pyruvate and glycolate excretion by Prochlorococcus, one of the world’s most abundant marine cyanobacteria (89). Prochlorococcus metabolite excretion could have been reinforced in part by the coevolution of associated heterotrophs that utilize these compounds and reciprocate by excreting malate and citrate (89). The potential for positive selection for secretion raises the prospect for active processes of nutrient release, including transporters. Overall, the mechanisms by which costly metabolites are passively excreted or actively secreted are largely uncharacterized and could be as diverse as the molecules themselves.
For cooperative cross-feeding of communally valuable metabolites to evolve, it is critical that conditions allow for reciprocation to be confined to cooperative partners and avoid being outcompeted by exploitive community members (16, 90). The fact that the positive or negative effects of marine heterotrophs on Prochlorococcus growth is phylogenetically conserved suggests that some mechanism of directed reciprocation was involved in the evolution of cross-feeding in this case (91). One of the most effective mechanisms to achieve directed reciprocation is likely spatial orientation, an aspect that we raise at numerous points over this review.
The recipient itself can also drive cross-feeding simply through depleting a communally valuable cross-fed metabolite. Our group found that Escherichia coli mutants with a higher capacity for NH4+ acquisition greatly improved coculture growth with a poor NH4+-producing partner on which E. coli relied for nitrogen (92) (Fig. 2G). Though an example of waste metabolite cross-feeding, others have observed genetic and phenotypic differentiation of E. coli monocultures into subpopulations that established acetate cross-feeding, often cited as an example of division of labor. The acetate-consuming recipient subpopulation similarly evolved an increased affinity for acetate, whereas the acetate-excreting E. coli subpopulation showed higher growth rates on glucose (93, 94). The reciprocal benefit in this case could come from the removal of acetate as an inhibitory compound (95).
Exoenzymes
Extracellular enzymes, or exoenzymes, degrade large polymers into transportable monomers. Collectively, exoenzymes have diverse substrates. Prominent exoenzyme classes are glycosidases, proteases, nucleases, lipases, and lignin-cleaving laccases and peroxidases. Because exoenzymes release readily consumable nutrients into the extracellular milieu, their activities lend themselves to cross-feeding as the resulting monomers can be taken up by neighboring cells, be they clonal or unrelated. As a result, exoenzyme-secreting microbes are often viewed as keystone species and primary colonizers in establishing microbial communities on polymeric substrates (96–98). For example, host-associated cellulase-secreting microbes sustain communities of gut bacteria, as well as the cellulose-consuming animal hosts, such as ruminants (99) and termites (100), that otherwise have little to no native ability to access carbon in cellulose. At a broader scale, the breakdown of cellulose and lignin via exoenzymes is a key step in the global carbon cycle, driving the mineralization of plant polymers. The potential ease of cross-feeding from exoenzyme-secreting bacteria has also been embraced by industrial sectors. Lignocellulose-degrading enzymes are intensely researched with the goal of lowering the costs of liberating sugars for producing cellulosic ethanol and other biofuels (101). Cellulase-degrading microbes are also commonly cocultured with other microbes in proof-of-concept studies for biofuel production from cellulose based on cross-feeding relationships (102, 103).
In a given ecosystem, the potential for cross-feeding from exoenzymes is large. It was estimated that ∼99% of the glucose and fructose produced by digestion of sucrose via the exoenzyme invertase diffused away from the producer yeast cell before the monosaccharides could be imported under well-mixed conditions (104). Whereas this trait makes exoenzyme-producing microbes potentially cooperative, it also makes them vulnerable to exploitation, both from other species and by emergent “cheater” subpopulations that lose the ability to secrete exoenzymes but benefit from nearby exoenzyme-secreting cells without the cost of synthesis. In the case of invertase-secreting yeast, mutant cheaters that did not invest in invertase production could invade producer populations and coexist across a range of growth conditions (104). Likewise, Vibrio cholerae mutants that did not produce chitinase, which degrades insoluble chitin into N-acetylglucosamine oligomers and monomers, could acquire extracellular N-acetylglucosamine released by chitinase-producing strains and thus outcompete producers under well-mixed conditions (105).
How then do exoenzyme-secreting populations avoid being overrun by exploitive neighbors? Some polymer-degrading enzymes, such as bacterial cellulases, are often attached to the cell surface, requiring the exoenzyme-secreting cell be in close contact with the polymer substrate and likely increasing the chances of substrate acquisition after depolymerization (106, 107). In some cases, security comes from the use of exoenzymes that release soluble oligomers rather than monomers; oligomers are then imported into the periplasm before further depolymerization. While many microbes can access monomers, fewer have the necessary enzymes to degrade oligomers. This oligomer-cleaving and import strategy was shown to allow Bacteroides thetaiotaomicron from the human gut to utilize mannan without supporting the growth of a mannose monomer-utilizing cocultured strain (107). We suggest that the organization of chitin degradation pathways (108) hints at a similar strategy employed by chitin-degrading bacteria. While the use of oligomer-cleaving enzymes can be effective against unrelated competitors, the situation is less simple for combating related cheaters. Cheaters are more likely kept at bay, or policed, through group behaviors, including quorum sensing (QS) and clustering of producers in biofilms as detailed in later sections.
Siderophores
Siderophores are secreted small molecules that chelate iron, or in some cases other metals such as copper, manganese, and zinc (109, 110), and facilitate uptake through cell surface receptors. These trace metals are essential cofactors for numerous enzymes (110–112). Collectively, the activity of siderophores in the oceans makes them an important feature of global iron cycling, which in turn affects ocean productivity and nitrogen cycling (113). Iron is also frequently scarce in hosts, where there is competition for iron between host cells and their resident microbiota, as well as between microbes (111, 112, 114–117). Consequently, siderophore production and access can play crucial roles in determining the course of an infection (111, 112, 114–118). Some hosts actively try to block siderophore function as an innate immunity defense tactic, and pathogens have evolved “stealth siderophores” to evade detection by the immune system (112).
As secreted molecules, siderophores can participate in cross-feeding by enabling iron acquisition by not only producers but also by unrelated community members. Indeed, there are examples of siderophore-mediated iron cross-feeding between different bacterial taxa, and even between bacteria and eukaryotic microbes (18, 117, 119–121). Some bacteria might even specialize in scavenging siderophores produced by other organisms (122). However, the ability of a given microbe to use a foreign siderophore likely represents a snapshot of a molecular arms-race (112). Siderophore synthesis biochemistry permits a wide range of permutations on a given structure; nearly 60 pyoverdine siderophores alone have been described (123). Cell surface receptors that recognize siderophores can be species specific, restricting uptake by other community members (18, 116). As predicted by an eco-evolutionary model (124), siderophore specificity limits iron availability and decreases growth of other community members (18, 121).
Whereas siderophore specificity might protect against iron theft from unrelated organisms, siderophore-secreting bacteria are still prone to exploitation by cheaters. Siderophore cheating has been detected in soil, freshwater, and marine bacterial communities (18, 20). Nevertheless, siderophore cheating can be mitigated or policed. One example employs a secondary role of a pyoverdine siderophore in iron sequestration rather than acquisition. During oxidative stress, pyoverdine-producing Pseudomonas aeruginosa downregulates pyoverdine secretion, allowing pyoverdine to accumulate in the periplasm. There, pyoverdine sequesters iron and is thought to prevent Fe3+ from producing harmful OH radicals by the Fenton reaction (125). Cheaters that do not make pyoverdine would lose this protective role and be purged from the population during times of oxidative stress.
While there are many examples of siderophore exploitation (18, 20, 117, 120, 122, 124, 126), there are fewer known examples of siderophores forming the basis for mutualistic cross-feeding. In one known example, marine alga-associated bacteria produced a siderophore that enhanced algal iron acquisition (119). Both organisms appeared to benefit from the association, perhaps through the reciprocal release of consumable metabolites from the algae (119). Another intriguing exception is the finding that hosts might nutritionally benefit from siderophores secreted by their microbiota. Production of the siderophore enterobactin by E. coli was shown to promote iron uptake in mitochondria of Caenorhabditis elegans and mammalian cells, whereas siderophores, such as pyoverdine, from potential pathogens did not promote iron uptake (127).
Toxins
Broadly speaking, microbial toxins are secreted compounds that have likely been selected through evolution for their inhibitory, and sometimes lethal effects on other organisms, be it host cells or other microbes. Toxins come in a variety of chemical structures and sizes, from small molecules like antibiotics and cyanide to multisubunit proteins such as cholera toxin (14, 21, 118, 128, 129). Methods of toxin delivery also vary. Some are passively excreted or actively secreted into the extracellular milieu, whereas others are injected directly into host or microbial cells (14, 21).
Some small molecule examples of toxins can directly serve as cross-fed metabolites when toxins are at subinhibitory concentrations or when a recipient possesses sufficient tolerance to a toxin. For instance, certain bacteria can use cyanide as a nitrogen source for growth (130, 131). Other microbes might even consume antibiotics (132–135), although skepticism has been cast on such observations (136). However, we speculate that toxins can have a broader role by indirectly facilitating cross-feeding.
Toxins frequently facilitate nutrient acquisition by damaging or lysing cells and releasing the intracellular contents (118). It therefore seems intuitive that this toxin-mediated release of nutrients could also benefit neighboring microbes, thereby making toxins a facilitator of cross-feeding in a similar manner to that of exoenzymes. Synergistic interactions can occur between pathogens during polymicrobial infections and arguably these interactions are dependent on the hallmark toxins of an infection, though the role of toxin-mediated cross-feeding in these interactions remains obscure.
Contrary to the above expectations, there is some evidence to suggest that toxins lead to competition between populations. For example, several TnSeq experiments have shown that more genes are essential for a given pathogen during coinfection compared to monoinfection (137–139). These results might suggest that coinfection intensifies competition rather than improving conditions for the local community; intense competition could limit nutrient availability, causing more genes to be essential. However, the detection of competitive traits does not necessitate a net competitive relationship. As discussed below, competitive interactions are likely a common, and sometimes necessary, feature of cooperative relationships. While more genes were essential during coinfection, coinfection also alleviated the essentiality of some genes that were essential in monoculture (137), suggesting that some beneficial interactions occurred. As noted by Lewin et al., the increase in essential genes overall during coinfection could simply be indicative of increased environmental complexity due to partner interactions (138). Overall, the extent to which toxin-mediated cross-feeding occurs is unclear and merits further investigation.
Quorum-Sensing Signals
Quorum sensing (QS) is a microbial cell-cell communication system that coordinates gene expression based on population density through the production and subsequent recognition of diffusible signal molecules (140–144). Upon reaching a threshold population density, or quorum, bacterial populations synchronize expression of subsets of genes. Canonical QS systems consist of a signal synthase, which produces the signal, and a cytoplasmic receptor, which binds the signal and acts as transcriptional regulator to modulate gene expression, although two-component systems are also frequently involved in signal detection (141–145).
QS signals, also called autoinducers, can be used by some bacteria as a sole energy source in monoculture (146), thus raising the possibility of QS signals participating as cross-fed metabolites. However, catabolism of QS signals in communities might have greater physiological relevance in quorum quenching, alleviating the signaling effects of the molecules. To the best of our knowledge, the extent to which QS signals are cross-fed between organisms has not been tested. Better known is the role of QS signals in regulating the production of the aforementioned exoenzymes, siderophores, and toxins, in addition to other group behaviors (128, 129, 141–144, 147, 148). Thus, the role of QS in cross-feeding is typically auxiliary, functioning to control access to extracellular nutrients at the population level; a population is more likely to reap the benefits of an exoenzyme, siderophore, or toxin if those molecules are produced en masse.
QS often coordinates multiple and sometimes disparate activities. In this way, QS can also limit the loss of extracellular nutrients to cheaters. For example, expression of the casein-hydrolyzing exoenzyme elastase by the opportunistic human pathogen P. aeruginosa is regulated by the QS transcriptional regulator LasR (128, 129, 147, 149). lasR-null mutants that do not produce elastase rapidly emerge and cheat elastase-producing strains when casein is the sole carbon source (147, 149). However, the addition of adenosine to casein-containing medium decreased the relative fitness of lasR mutants in competition with the elastase-secreting strain because LasR is also required for activating adenosine consumption pathways (147). Thus, the coupled expression of exoenzymes with other physiological activities can help thwart cheaters by offsetting any benefit gained from the loss of exoenzyme secretion. Policing cheaters by LasR is even more multifaceted. LasR also regulates cyanide production and resistance via QS in P. aeruginosa such that any benefit gained from loss of elastase by a lasR mutant is negated by susceptibility to cyanide (128).
QS also controls access to nutrients through diversity in autoinducers. Autoinducers can be divided into broad categories; Gram-positive bacteria synthesize oligopeptide autoinducers, while Gram-negative bacteria generally produce acyl homoserine lactone autoinducers (141–145, 150). Within each of these categories there is further chemical and structural diversity. Although detection of a given autoinducer is usually confined to the producing species that encodes the cognate receptor, different QS receptors display various degrees of signal selectivity, with several receptors being able to recognize non-self-autoinducers, allowing for what is known as eavesdropping (143, 150). QS receptor promiscuity is widespread, suggesting that eavesdropping and interspecies cross talk between distinct QS systems could be common (143, 150). Along these lines, many Gram-positive and Gram-negative bacteria can produce an autoinducer distinct from those mentioned above, called autoinducer-2 (AI-2). AI-2 is a by-product of the activated methyl cycle in the methionine synthesis pathway (141, 151). As methionine synthesis is widespread, so is the capacity to produce AI-2. As a result, AI-2 has been proposed as an interspecies QS signal (141, 151). However, far fewer bacteria encode genuine AI-2 receptors, than are capable of generating AI-2 (151). Thus, while certain species might appraise the local polymicrobial density of their surroundings via detection of AI-2 and alter their behavior accordingly (152), this does not seem to be a widespread strategy.
The extent to which eavesdropping occurs and whether it coordinates the behaviors of multiple species in nature remains largely unknown. However, QS eavesdropping has been successfully exploited by synthetic biologists to coordinate behaviors between distinct bacterial populations, including between Gram-negative and Gram-positive bacteria (153, 154).
Extracellular Matrix Components of Biofilms
Microbes frequently attach to surfaces and each other to form multicellular biofilms. Biofilm formation is mediated by secretion of an extracellular matrix that can be composed of polysaccharides, nucleic acids, and/or protein (155). Matrix components can potentially contribute directly to cross-feeding in combination with exoenzymes that degrade matrix components into accessible monomers. For instance, isotopic tracer experiments have shown that some cyanobacteria store and later reacquire carbon from their extracellular matrix, and this material was also accessible to other bacteria within microbial mats (156). However, more is known about the role of biofilms in controlling access to externalized nutrients, a role similar to that of QS. In fact, QS and biofilm formation are often connected. QS commonly helps coordinate biofilm development by regulating the biosynthesis of biofilm matrix. Furthermore, biofilm-dwelling cells are densely packed, allowing smaller populations to a reach quorum compared to what would be necessary in dispersed, planktonic populations (16, 144).
The spatial structure provided by biofilms and other dense aggregations can promote cooperative cross-feeding of costly metabolites (16, 63, 71, 73, 87, 105, 157). Thus, clustering of partners is important to the evolutionary trajectory of certain cooperative cross-feeding relationships (71, 73, 87, 157–159). Spatially structured environments on agar plates were shown to select for costly methionine-excretion by a spontaneous Salmonella enterica mutant, enabling growth of nearby E. coli methionine auxotrophs (73). In return, E. coli evolved costly galactose secretion during lactose consumption when grown together with S. enterica as colonies on agar (87). Similarly, amino acid cross-feeding increased during long-term serial transfers in liquid cultures wherein cross-feeding partners were able to form clusters (157). These results indicate that certain spatially structured environments can select for of the evolution of mutualistic cross-feeding by favoring local retention of costly nutrients, thereby directing those nutrients to reciprocating neighbors and limiting diffusion to competing noncooperative populations. Beneficial cross-feeding interactions can also select for attachment between partners (31, 63, 160, 161). Attachment likely further enforces directed reciprocation between cooperative partners. The influence that spatial organization can have on cross-feeding communities has been utilized by synthetic ecologists. For example, cyanobacteria have been engineered to display surface epitopes that facilitate aggregations with yeast partners (162).
Clustering of cooperative cells is also thought to affect the ability of cheaters to exploit externalized metabolites, in some cases benefitting the cooperators but in other cases benefitting the cheaters. For example, in populations of V. cholerae, the production of a biofilm matrix concentrated chitinase-secreting cells on the chitin, limiting diffusion and restricting access of a cheater population to released monomers (105). As noted above, close spatial proximity between cross-feeding partners promotes efficient nutrient exchange and limits diffusion to nearby cheaters or competitors. Microfluidic and modeling experiments with closely packed amino acid cross-feeding auxotrophs showed that a single cell might only cross-feed to a relatively small neighborhood of one to four cell lengths (164). The distance over which cross-feeding can occur is likely further decreased in the presence of competitors. Recipients embedded in agarose beads with glucose-excreting producers, with an average distance of 15 µm between them, could not grow when competing cells were present in the surrounding medium (163). However, recipient growth was rescued in the presence of competitors when recipients and producers were allowed to form aggregates before being embedded in beads (163). There can also be circumstances where close proximity benefits cross-feeding partners but minimizing the distance between partners does not. In one tripartite synthetic community involving both cross-feeding and antibiotic detoxification, culturing all three members in a single well led to intense competition (165). However, when the three members were cultured in separate wells and allowed to exchange nutrients between wells, an optimal distance was identified that limited competition and led to synergistic effects on growth stemming from the complementary traits of the members (165).
Observations like those above have led to the general theory that close proximity of producer strains and/or cross-feeding partners is an important mechanism by which the impact of cheaters or nonproducers is minimized as they can only exploit producers at the edges of an aggregate or biofilm. However, the situation is likely more nuanced since there are also examples where high local concentrations of producers within multicellular aggregates can be susceptible to invasion by cheaters (20). Specifically, in marine Vibrio populations, nonproducing strains lacking siderophore biosynthesis genes were more frequently isolated from larger natural particles that were cocolonized by siderophore producers; it appears that nonproducer Vibrio populations cheat siderophore production by invading dense aggregations of producers (20). We propose that, in general, microbial cooperation seems to be more stable in biofilms that form through outward growth of clonemates or mutualistic partners, pushing nonproducers to the margins of a biofilm where some access to public goods is likely to be available (16). In contrast, biofilms that assemble through surface attachment of diverse species and strains from the surrounding environment are more likely to elicit competition and cheating (20). Thus, the exact form of spatial structure that maximizes cooperative nutrient acquisition and cross-feeding depends on many factors, including the biosynthetic cost and structure of the cross-fed compound(s), the physiochemical properties of the environment, the means by which spatial structuring is established, and the genetic composition of the local community.
Extracellular Vesicles
Like QS and biofilms, cross-feeding behavior can also be influenced through the release of extracellular vesicles. Extracellular vesicles have been reported for a variety of microscopic and multicellular eukaryotes, archaea, and both Gram-negative and Gram-positive bacteria (166–168). In Gram-negative bacteria, extracellular vesicle release is associated with a decrease in cross-linking between peptidoglycan and outer membrane proteins and might also be influenced by lipid and lipopolysaccharide composition (167). Mechanistic details for the release of vesicles from Gram-positive bacteria are currently obscure but could include being forced through weak points in the cell wall or facilitated via channels (168).
Some of the above details could suggest an accidental passive release of extracellular vesicles. However, a purposeful release of vesicles is suggested by the observed selectivity of vesicular cargo and the implication of vesicles in numerous microbial behaviors (166–168), some of which could directly participate in cross-feeding, or at least influence cross-feeding. For example, there are numerous examples of vesicle-mediated toxin delivery (166–168). In one case, the toxins were exoenzymes packaged in extracellular vesicles by Myxococcus xanthus to degrade macromolecules in E. coli prey cells (169). Extracellular vesicles have also been observed as a component of biofilm matrixes (168, 170). Extracellular vesicles can participate in QS, in one case by facilitating the diffusion of signals that would otherwise be too hydrophobic to be effective in an aqueous environment (171). In this case, the vesicle also imparted additional species specificity, thus limiting signaling to certain community members (171). Siderophores can also be secreted in association with extracellular vesicles. Vesicular association could help siderophores evade host immune responses that target bacterial iron acquisition (167, 172), though extracellular vesicles themselves can elicit immune responses in some cases (168). Perhaps extracellular vesicle-packaged siderophores also impart an additional layer of specificity for uptake of sequestered iron, similar to what was observed with QS signals (171).
Extracellular vesicles can also directly serve as a cross-fed nutrient. For example, vesicles released from Prochlorococcus cyanobacteria supported the growth of heterotrophic marine bacteria (173). Given that Prochlorococcus is one of the most abundant marine bacteria, extracellular vesicles from these cyanobacteria alone could account for the release of 104 to 105 metric tons of carbon into the oceans each day (173). While the vesicles and their cargo can likely serve as a cross-fed nutrient, vesicles also serve to protect their cargo (166). Thus, an important role of extracellular vesicles could instead be to combat unwanted cross-feeding by decreasing or delaying acquisition by competitors.
Contact-Dependent Cross-Feeding
QS, biofilms, and extracellular vesicles can all influence the fate and action of externalized molecules. However, there are also instances where little is left to chance through the use of contact-dependent mechanisms. Many contact-dependent interactions are antagonistic, directly delivering harmful toxins into neighboring cells (21, 174, 175), but beneficial contact-dependent cross-feeding also occurs.
In some cases, mechanisms of contact that facilitate cross-feeding are so intimate that one cell is housed within another (176–178). Less extreme, but still enclosed within a membrane, are examples of a shared periplasm, and thus likely a shared electrochemical gradient or proton motive force. For instance, multiple Pyrodictium archaea share a periplasm within tubular “cannulae” (179). This specific case of a shared periplasm between clones does not necessarily meet our definition of cross-feeding because there is no obvious phenotypic differentiation between cells. However, there are clear examples of cross-feeding between phenotypically distinct clones. Filaments of cable bacteria share a periplasm along with an electrically conductive intercellular appendage of unknown composition (180, 181). These filaments span redox gradients in sediments allowing separate cells to contribute oxidation and reduction reactions to a combined metabolism despite being centimeters apart (180, 181). Some filamentous cyanobacteria also share a periplasm and exhibit dramatic phenotypic differentiation into vegetative cells and nonvegetative heterocysts that are specialized for CO2 and N2 fixation (diazotrophy), respectively (182, 183). In this case, specialized connective structures participate in the transfer of nitrogen and carbon at the junction between vegetative cells and heterocysts (182, 183).
There are also examples of interspecific cross-feeding by direct contact. In some cases the mode of transfer is poorly defined, such as that behind observations of stable isotope transfer from cyanobacterial heterocysts to attached Rhizobia, both environmental isolates (184). In other cases, investigations into mechanistic features are revealing fascinating and unique physiological traits. The contact-dependent pairing of two archaea, Ignicoccus hospitalis and Nanoarchaeum equitans, likely involves transfer of phospholipids, amino acids, and perhaps even ATP from I. hospitalis to N. equitans (185). N. equitans does not appear to encode enough genes to make a complete ATP synthase. I. hospitalis, however, the only known organism to have an energized outer membrane, generates ATP in the periplasm via an outer membrane ATP synthase (186). The periplasmic generation of ATP has been speculated to be important for ATP cross-feeding to N. equitans (185).
Nanotubes.
Intimate contacts between cells also includes intercellular membranous connections called nanotubes (180). Nanotube connections have been reported as both intra- and interspecific (187), including between Gram-negative and Gram-positive bacteria (180), and potentially between bacteria and mammalian cells (188). Nanotubes were reported to enable the transfer of small metabolites, large proteins, and DNA molecules (187). How the inner membrane of a bacterium might be passed through the peptidoglycan layers of both donor and recipient cells is mechanistically obscure, though peptidoglycan hydrolases have been implicated, at least in part, in nanotube formation in B. subtilis (189, 190). Nanotube formation could also share mechanistic features with extracellular vesicle formation. Aside from the peptidoglycan hydrolases, a B. subtilis phosphodiesterase (191) and sigma factor (190) and pathogenic E. coli injectosome components (188) are the only other genetic factors that have been implicated in nanotube formation.
Nanotube function remains mysterious and at times controversial. The evidence for intercellular transfer of molecules via nanotubes is often correlative; nanotube structures are present, but molecule transfer via other mechanisms has not been ruled out (192). In some cases, intercellular transfer of fluorescent signatures was observed (190, 193–195). In these cases, frequencies of connections and transfer events within a population are rarely reported. In one relationship that depended on amino acid cross-feeding and in which nanotubes and fluorescent protein transfer were also observed, a subpopulation of the coculture was not fluorescent (194), suggesting that other mechanisms contributed to essential amino acid transfer, at least in part. There are also inconsistencies in the regulation of nanotubes. In one case, E. coli nanotube formation was stimulated by nutrient deprivation, supporting a role in nutrient acquisition from neighboring cells (194). However, nanotube formation by pathogenic E. coli (188) and B. subtilis (193) occurred in rich media.
Recently, nanotubes were implicated as a by-product of cell death in E. coli, B. subtilis, and other Gram-positive bacteria (190). Although fluorescent proteins were observed in B. subtilis nanotubes, DNA transfer required well-described competence proteins, suggesting that nanotubes were not involved (190). In other bacteria that are not naturally competent, plasmid transfer via nanotubes is difficult to reconcile with common microbiology practices, even if occurring at a low frequency. For example, introduction of foreign DNA into noncompetent cells by nanotubes seems to trivialize long-used conjugation and other transformation techniques. Nanotube transfer of proteins and DNA between living cells could also call into question results from competition assays wherein genetically distinct populations are distinguished using drug resistance cassettes or fluorescent reporters. The cooccurrence of cell death with nanotube formation also invokes cell lysis, which could help explain correlative observations of cross-feeding (190). Overall, nanotubes are an intriguing mechanism for cross-feeding, but one for which critical mechanistic and functional details will likely remain a contentious topic of further investigation and debate.
Nanowires.
One of the most studied contact-dependent cross-feeding phenomena between species is that of direct electron transfer via structures often referred to as nanowires. Nanowires fulfill a role in anaerobic respiration, allowing the nanowire producer to deposit electrons on extracellular terminal electron acceptors that are often too large, insoluble, or toxic to reduce inside the cell (196). In this way, nanowires, as well as soluble extracellular electron shuttles, participate in cross-feeding via the generation of waste metabolites; the resulting reduced metals become electron sources for other lifestyles, like photo- or litho-autotrophs.
In other cases, nanowires enable the direct coupling of redox metabolisms between cells. Direct electron transfer via nanowires can sustain syntrophic partnerships by relieving thermodynamic limitations, much in the way that organic acids, H2, and other reduced compounds serve as intercellular electron shuttles in these partnerships (63). Nanowire-mediated electron transfer has been implicated in both natural and synthetic syntrophies (180). For example, a syntrophy was experimentally evolved between Geobacter metallireducens, forced to carry out an ethanol-fed fermentation, and fumarate-respiring Geobacter sulfurreducens (63) (Fig. 2H). Electrons were transferred from G. metallireducens to G. sulfurreducens, relieving the thermodynamic barrier on ethanol fermentation and providing electrons to drive fumarate respiration. The syntrophy was promoted through the formation of multicellular aggregates and was dependent on G. sulfurreducens nanowire components, namely, pili and an outer membrane cytochrome, but not on H2 oxidation by G. sulfurreducens (63). Syntrophic electron transfer has also been found to occur in natural aggregates formed between sulfate-reducing bacteria and archaeal anaerobic methanotrophs (197, 198) (Fig. 3; ANME), which are important players in controlling methane emissions from ocean sediments (199). Recent evidence also suggests that some H2 cross-feeding syntrophs can instead grow via direct electron transfer to a partner (200).
There is ongoing debate over the composition of nanowires. Some work suggests that electrons are transferred between aromatic amino acids in the pilus (201). More recently, a crystal structure of a pilus composed entirely of cytochrome subunits was solved (202), casting doubt on the role of pilin subunits in extracellular electron transfer. However, because G. sulfurreducens pilin was required to receive electrons from G. metallireducens in coculture (63), the pilus must still play a role in interspecies electron transfer even if it does not transfer electrons itself. It is also likely that nanowires are compositionally diverse. For example, Shewanella oneidensis nanowires were revealed to be extensions of the outer membrane containing electrically conductive cytochromes (203), a mechanism more akin to nanotubes or extracellular vesicles. Other mechanisms of direct electron transfer between species include cell surface cytochromes and transfer through abiotic surfaces, as well as the uncharacterized conductive structures that connect cells in filaments of cable bacteria (180, 204). Collectively, microbes that are capable of externalizing electron flow are also of interest for numerous applications ranging from, electricity generation, bioremediation, and electricity-driven production of reduced compounds (204, 205).
DYNAMIC ASPECTS OF MICROBIAL CROSS-FEEDING
The number of microbial relationships based on cross-feeding presents enough material for numerous reviews. Indeed, additional examples are reviewed elsewhere (31–33, 36–44). Here, we draw attention to some insights we have gained from our work on a synthetic cross-feeding community between E. coli and Rhodopseudomonas palustris (Fig. 2G) with supporting observations by others.
Cross-Feeding Rates Determine the Relative Benefit or Detriment of a Cross-Fed Metabolite
Many excreted molecules have toxic properties. Detoxification has emerged as an important aspect of cooperative microbial interactions, wherein one partner protects another by consuming a toxic metabolic product or by sharing the burden of processing a toxic intermediate (17, 95, 206). However, here we focus on a less obvious example wherein a cross-fed metabolite can shift from being a nutrient to a toxin.
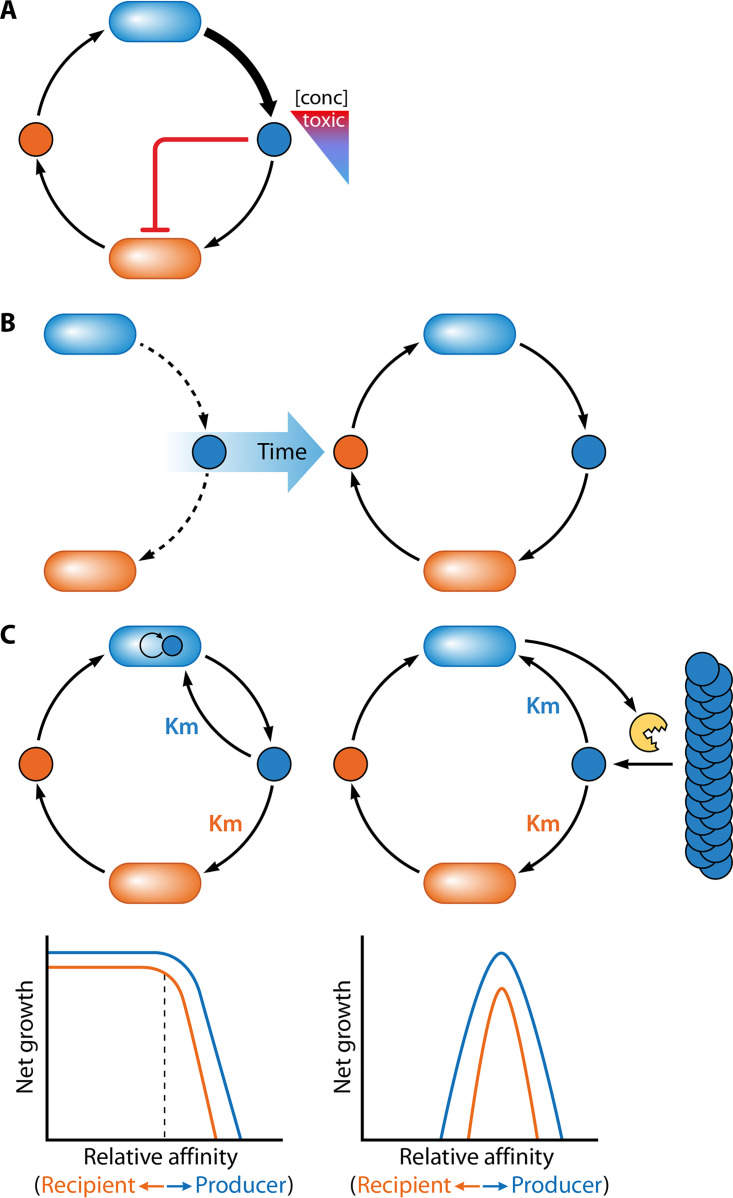
The scenario is similar to that noted above for cyanide and antibiotics, notoriously toxic compounds that can also serve as nutrients for some bacteria at certain concentrations. Similarly, fermentation products like alcohols and organic acids can be toxic but are commonly viewed as important shuttles of carbon and electrons in anaerobic food webs (31, 34, 58). Our group demonstrated that cross-feeding levels can determine the relative benefit of organic acids in a coculture featuring fermentative E. coli and a strain of N2-fixing R. palustris engineered to excrete NH4+ (207). In this coculture, R. palustris relies on E. coli for excreted organic acids as an essential source of carbon and E. coli relies on R. palustris for excreted NH4+ as an essential source of nitrogen (Fig. 2G). Under conditions with glucose and N2 as the sole carbon and nitrogen sources, one partner cannot grow without the other. When the NH4+ excretion level is low, the E. coli growth rate matches that of R. palustris, and R. palustris can consume organic acids as fast as E. coli excretes them. However, when R. palustris is engineered to excrete a higher level of NH4+, the growth rates become uncoupled; faster E. coli growth is stimulated, which is associated with a rate of organic acid excretion that exceeds the consumption rate by R. palustris. Organic acids accumulate and eventually acidify the medium, inhibiting R. palustris growth. Thus, although more cooperation by R. palustris led to more reciprocation by E. coli, the higher level of reciprocation was not beneficial to R. palustris since it shifted the role of organic acids from being nutrients to toxins (207) (Fig. 4A). We named this concept “dose-dependent toxicity.” At which concentration the accumulated compound becomes toxic is likely linked to local concentration and metabolite diffusion, both of which can vary with community spatial structure and environmental factors like flow rate. Unlike the dose-dependent toxicity of antibiotics and cyanide, for which a role as nutrients is likely limited to special cases, organic acids are a central feature of diverse anaerobic food webs. Thus, dose-dependent toxicity of organic acids has the potential to influence important ecosystems, including the mammalian gut and anaerobic sediments covering much of the Earth’s surface.
FIG 4.
Cross-feeding and its outcomes are dynamic. (A) A high level of cooperation by one partner can lead to excessive and harmful reciprocation by another. In this case, when production rate exceeds consumption rate (blue arrows), a metabolite (blue dot) can accumulate to toxic levels, for example by acidifying the environment (207). The “conc” triangle illustrates the effect of the metabolite on the recipient, ranging from beneficial (blue) to detrimental (red) as the concentration increases. (B) Growth-independent cross-feeding can rescue partners from starvation. Maintenance metabolism alone can lead to metabolite excretion under nongrowing conditions (left). Consumption of the metabolite by a recipient can stimulate recipient growth and reciprocation, creating a positive feedback loop and lifting both partners out of starvation (30). (C) The level of privatization influences the affinity that each partner must have for a communally valuable metabolite for cooperative coexistence to result (225). (Top) Cross-feeding of an intracellularly generated metabolite (left: high privatization) and an extracellularly generated metabolite liberated by an exoenzyme (right: low privatization). (Bottom) Simulated effect of the relative competition, in this case affinity (inverse of Km, which is the substrate concentration when the growth rate is at half the maximum), for the metabolite on the net growth of each partner under high and low privatization conditions. (Simulated trends are adapted from reference 225.)
Dose-dependent or conditional toxicity of transferred metabolites has been observed in other cross-feeding systems. For example, the toxicity of cross-fed nitrite (NO2–) varies based on pH such that NO2– is more inhibitory under more acidic conditions. Growth and colony expansion velocity of a denitrifying consortium of P. stutzeri strains decreased at lower pH when cross-fed NO2– was more toxic (206, 208). Dose-dependent toxicity of NO2– may similarly occur during the oxidation of NH4+ to nitrate (NO3–), known as nitrification, which can be carried out by two groups of bacteria: Nitrosomonus oxidizes NH4+ to NO2– and Nitrobacter oxidizes NO2– to NO3– (209). The NO2– consumer Nitrobacter is much more sensitive to NO2– than the producer Nitrosomonus (210), raising the possibility of Nitrosomonus harming Nitrobacter through high NO2– excretion rates.
Cross-Feeding Is Not Always Coupled to Growth
As noted in the introduction, most microbes likely exist in a state of nutrient deprivation (22–24). For microbes that are incapable of differentiating into specialized dormant structures like spores, a low level of metabolic activity is generally essential to produce maintenance energy for survival (24, 27, 28). While few studies have examined the role of cross-feeding on survival, one can envision that cross-feeding could be an important driver of maintenance metabolism in nutrient-limited environments. For instance, respiration is important for the stationary-phase survival of some lactic acid bacteria (211). Some of these lactic acid bacteria include group B Streptococcus (GBS) pathogens, for which respiration is important for infection, affecting both GBS growth and survival (212). GBS require exogenous sources of heme and naphthoquinones to carry out respiration. Because humans do not make napthoquinones, these respiratory cofactors must be acquired from other microbes. Indeed, several bacteria can support O2-dependent growth of GBS in coculture (213, 214). The reason why quinone cross-feeding favors stationary-phase GBS survival could simply be because aerobic respiration creates a more favorable pH by offsetting lactic acid accumulation. However, respiration could also help GBS maintain a proton motive force and thereby allow GBS to generate maintenance energy and persist under nongrowing conditions.
Metabolite cross-feeding is also likely important for the survival of the most abundant cyanobacteria on Earth, Synechococcus and Prochlorococcus, which together account for ∼25% of net primary productivity in the world’s oceans (215). Synechococcus survival through starvation was shown to be dependent on the supply of minerals such as NH4+ and phosphorous made available through the degradation of dissolved organic matter by heterotrophic Roseobacter partners (216). Degradation of dissolved organic matter by Roseobacter likely also played a detoxification role in this case (216). Similar prolonged survival during starvation was observed for Prochlorococcus when cocultured with heterotrophic Alteromonas (217). In this case, detoxification did not seem to play a major role since Prochlorococcus survival in monoculture was not improved when transferred to fresh media. In the absence of detoxification, the positive effect of Alteromonas on Prochlorococcus survival was likely through the mineralization of dissolved organic matter (217).
It is also possible that the low metabolic activity of nutrient-deprived cells can foster cross-feeding under certain conditions. Our lab demonstrated that nongrowing E. coli fermented glucose and excreted enough organic acids to support the phototrophic growth of R. palustris (30). In this case, the cross-feeding initially attributed to E. coli maintenance metabolism stimulated reciprocation from R. palustris and ultimately lifted both partners out of starvation (30) (Fig. 4B). Our lab has also shown that an obligate cross-feeding relationship can evolve naturally between E. coli and R. palustris (92). Initially, this relationship likely relied on growth-independent organic acid excretion by E. coli (92). Thus, maintenance metabolism could be important for both the initiation and maintenance of cross-feeding relationships.
Such growth-independent cross-feeding can also have medical relevance. For example, acetoin production by nongrowing S. aureus supported the growth and survival of P. aeruginosa isolated from the same cystic fibrosis patient samples (218). In turn, P. aeruginosa facilitated S. aureus survival by preventing acetoin accumulation to toxic levels (218). Thus, cross-feeding between nongrowing partners could support persistence during coinfection.
Cross-Feeding Can Be Facultative
While there is a tendency to think of cross-feeding as fulfilling an essential nutritional requirement of a recipient, cross-feeding can also act to beneficially augment a recipient’s metabolism. Cross-feeding of quinones to GBS is an example of facultative cross-feeding; GBS can grow by fermenting glucose when its electron transfer chain is incomplete, but the growth yield improves when aerobic respiration is enabled via quinone cross-feeding (212, 213).
With the exception of fermentations involved in syntrophic partnerships, most fermentations do not require removal of fermentation products by a partner to be thermodynamically feasible. Even so, product removal by a recipient can have thermodynamic benefits. We and others have observed that cross-feeding between fermentative and purple nonsulfur bacteria alters fermentation profiles; more formate typically accumulates in coculture than in monoculture (102, 207). Although the energetic benefits of cross-feeding have not been verified in this case, other cases are clear. For example, in monoculture, Ruminococcus albus ferments glucose to a mixture of acetate, ethanol, and H2. Acetate production is coupled to ATP generation, while ethanol and H2 production satisfy electron balance. However, when cocultured with fumarate-respiring Wolinella succinogenes, consumption of R. albus H2 waste by W. succinogenes overcomes a thermodynamic barrier, allowing electron balance in R. albus to be satisfied solely through H2 production (219). Without the need to produce ethanol, R. albus can divert more acetyl coenzyme A (acetyl-CoA) to acetate and maximize ATP production (219).
A similar facultative cross-feeding interaction likely takes place in the human gut. Christensenella bacteria and the methanogen Methanobrevibacter smithii were found to co-occur in about 80% of >1,800 human gut samples (220). Characterization of metabolic traits in monoculture versus coculture revealed H2 cross-feeding from Christensenella species to M. smithii. The lower partial H2 pressure created by M. smithii allowed the Christensenella species to maintain electron balance by shifting electrons away from butyrate to H2 production, while still generating the same amount of ATP through acetate production (220). We speculate that this partial shift from butyrate to acetate and H2 benefits Christensenella by allowing it to achieve both electron balance and ATP production but with enzyme synthesis cost-savings by using shorter metabolic pathways. Human hosts might also benefit by receiving more acetate and less butyrate, which correlates with less obesity (220).
Facultative cross-feeding of vitamin B12 has also been noted between microbes that inhabit the human gut (83). In this case, Akkermansia muciniphila liberated sugars from mucus and produced propanediol as a fermentative end product, both of which could be used by Eubacterium hallii. E. hallii reciprocated with the release of vitamin B12, providing A. muciniphila with the necessary cofactor to shift its fermentation profile toward propionate instead of succinate excretion (83). Although the physiological benefit of propionate production on A. muciniphila was not examined, propionate can have potential benefits to the human host.
Mutualistic and Synergistic Relationships Contain Competitive Interactions
Cooperation between species can seem at odds with evolutionary theory; strong competitors are expected to dominate over species that invest in costly traits that benefit their neighbors. Indeed, competition is likely predominant in microbial interactions (15), including cooperative relationships. It has been proposed that truly selfless cooperative relationships are likely rare or nonexistent (221, 222). More likely, cooperative relationships arise as a necessity of environmental conditions. For example, cooperation could stem from a parasitic relationship if a host becomes reliant on the parasite for a function, such that the parasite can no longer be removed without detrimental consequences (221, 222).
There are several examples of competition between otherwise cooperative partners. Though not necessarily reflecting conditions in the ocean, laboratory cocultures suggested that the carbon requirement of the heterotrophic marine bacterium SAR11 could be largely met by a strain of Prochlorococcus that also proved to be a notable competitor for a key sulfur source (223). Competition was also noted in experimentally evolved syntrophic cocultures of D. vulgaris and M. maripaludis. Pairing different evolved partners resulted in growth rates and yields that were lower than what would be predicted from additive or multiplicative effects of evolved syntrophic mutations, suggesting that antagonistic interactions were at play even though conditions required syntrophic cross-feeding (61) to the extent that D. vulgaris lost traits required for an independent lifestyle (62). We thus suggest that it is useful to consider cooperation as the net sum of multiple interactions that microbial partners are likely to engage; inspection will invariably reveal competitive interactions between cooperative partners.
Cooperative cross-feeding can even occur while partners compete for a resource from which a cross-fed metabolite is derived. Long-term cultures of E. coli were noted to differentiate into distinct subpopulations, including a subpopulation that specialized in glucose uptake and excreted acetate, as well as another subpopulation that fed off that acetate (93). The relationship has been classified as cooperative because the acetate sustains the recipient, while the removal of acetate likely protects the producer from inhibition by acetate (95). However, the recipient can still consume glucose and actually exhibits a higher growth rate on glucose than the producer in comparative monocultures (93). Thus, the cross-feeding relationship likely hinges on the producer being more competitive for glucose than the acetate-consuming recipient. This scenario could also apply to other relationships based on cross-feeding of metabolic end products between species that might otherwise compete for a preferred carbon source. Cross-feeding of end products was identified as a generic stabilizing factor in communities derived from diverse environmental inocula, allowing potential competitors for a single carbon resource to coexist (224).
Perhaps counterintuitively, competition can even be at the heart of cooperative cross-feeding. Our group found that cross-feeding partners competed for the very nutrient that formed the basis of a cross-feeding relationship. As mentioned above, some cross-fed metabolites like NH4+, vitamins, nucleotides, and amino acids are communally valuable; any organism would take up these molecules, if available, rather than synthesize them. This logic extends to producers of communally valuable metabolites, who are likely to be among the contenders for valuable compounds they excrete. Through a combination of modeling and experimentation, we found that in cocultures wherein coexistence depended on cross-feeding of NH4+ from R. palustris to E. coli (Fig. 2G), the two partners competed for NH4+ (225). For the cooperative relationship to be maintained, E. coli had to be more competitive than R. palustris for excreted NH4+. Transcriptomic and proteomic analysis revealed that this competitive edge from E. coli relied on a nitrogen starvation response (226). If R. palustris were more competitive at reacquiring excreted NH4+, it would starve E. coli and in doing so cut itself off from reciprocal cross-feeding of essential carbon from E. coli (Fig. 4C). We later found that evolution of competitive NH4+ acquisition by E. coli was sufficient to establish cross-feeding with wild-type R. palustris, again involving mutations that likely further enhanced the nitrogen starvation response (92). Thus, not only can cross-feeding partners compete for a critical cross-fed nutrient, but a competitive trait can be required to establish and maintain the cooperative relationship. In other settings, enhanced NH4+ acquisition might have a net parasitic outcome, but in this case, cooperation resulted because the competitive trait was coupled to reciprocation. Importantly, we did not observe the spontaneous emergence of mutations that we knew through our previous engineering approaches would enhance NH4+ excretion by R. palustris (92). Such recipient-serving costly mutations are unlikely to arise naturally due to the likely negative impact on producer fitness. In support of this notion, the evolution of a seemingly partner-serving trait in a coculture of auxotrophic yeast (resembling Fig. 2D) was completely offset by a self-serving trait arising from the same mutation (227).
The example presented above required competitive bias toward the recipient. We anticipate that competitive bias toward the recipient versus the producer likely depends on the level of privatization on the communally valuable nutrient. When there is a high degree of privatization, such as for those metabolites that are generated intracellularly, competition must be biased in favor of the recipient; intracellular synthesis all but guarantees that the producer will have access to the majority of the metabolite. When there is a low degree of privatization, such as during metabolite release by exoenzymes or as is likely the case for some siderophores, the competitive bias required to maintain a cross-feeding relationship can be different. In this case, too much competitive bias for either the recipient or the producer can lead to a collapse of the relationship and even the extinction of both populations if obligate reciprocal cross-feeding is involved (Fig. 4C). A necessity of competitive bias for the producer has been observed in yeast populations that secrete the exoenzyme invertase to cleave sucrose (104, 228). As noted above, spatial organization in biofilms can be an important competitive factor to allow the producer to reap the benefits of exoenzymes (105), likely by increasing privatization through clustering of cooperative cells. Thus, the level of privatization dictates the level of competition required between the producer and the receiver to enable and maintain cross-feeding of communally valuable compounds.
THE ABUNDANCE OF CROSS-FEEDING RELATIONSHIPS IS LARGELY UNKNOWN
There is general recognition that cooperative cross-feeding is ubiquitous in microbial communities and thus is widespread in nature (37). This notion is potentially at odds with findings that pairings and mixtures of bacterial isolates often exhibited lower CO2 production rates than monocultures, suggesting a predominance for competitive interactions (15). However, as noted above, competition can be an integral part of cooperative cross-feeding and can subtract from its potential benefits (61, 225). Furthermore, cross-feeding can stabilize coexistence of competitive groups (224).
Computational models have also indicated a large solution space for cross-feeding, particularly when involving metabolites that incur little to no fitness cost on the producer (55). In many cases, cross-feeding was enough to offset competitive interactions, especially in anaerobic environments (55). Along these lines, we note that laboratory communities enriched from environmental samples and stabilized by cross-feeding were dominated by groups known to excrete waste metabolites (224). But metagenomic data have suggested that cross-feeding of not only costless metabolites, but costly metabolites that sustain auxotrophic partners is a widespread phenomenon by which microbial diversity is maintained (34, 35). The prevalence of these observations likely highlights the importance of physical clustering of cross-feeding partners to limit the benefit to parasitic groups.
Large-scale laboratory enrichments of microbial communities can overlook the molecular details of cross-feeding mechanisms. Similarly, model predictions on the abundance of cross-feeding in nature can only leverage existing biochemical knowledge. For example, in considering vitamin B12 alone, there are over a dozen analogs that could participate in cross-feeding, but this level of specificity cannot currently be gleaned from genomic data (79). Improved gene product annotations will undoubtedly lead to improved predictions on the abundance of cross-feeding. However, we also predict that the space for discovery in cross-feeding interactions extends well beyond more meaningful annotations of enzymes and transporters. We are in an era of renewed interest and discovery in microbial biochemistry. As classical definitions of the roles of molecules, enzyme activities, pathways, and lifestyles are blurred, new and unexpected possibilities for cross-feeding reactions are emerging. Thus, the abundance and nature of cross-feeding is potentially much greater than is currently appreciated.
A notable example is that of lactic acid bacteria, which have a long history in the production of fermented foods and are mostly regarded as being strictly fermentative. Yet, as noted above, some lactic acid bacteria such as GBS pathogens can respire when supplemented with heme and quinones, the latter of which are likely cross-fed from a bacterial partner (213, 214). We also note several Clostridia that, based on metabolomic analysis in monoculture, appear to operate central metabolism near the thermodynamic equilibrium (229, 230). Operating near equilibrium can provide a greater energetic efficiency by harnessing more of the energy released by central metabolism (229). However, operating central metabolism near equilibrium involves reversible reactions, translating to a slow metabolism and a low growth rate. These traits would leave a microbe susceptible to competition from microbes with less energetically efficient but faster metabolisms. Could a near-equilibrium metabolism instead hint at a cross-feeding lifestyle for these Clostridia in their natural environment? A fermentation product-consuming partner could shift thermodynamic landscapes similar to the facultative cross-feeding relationships noted above and allow these Clostridia to benefit from both a fast metabolism and efficient energy transformation mechanisms.
There are other examples of previously overlooked metabolic capabilities that could involve cross-feeding interactions. Aerobic methanotrophs, which normally couple aerobic respiration to CH4 oxidation, play an important role in influencing the amount of CH4 greenhouse gas that reaches the atmosphere (199, 231, 232). However, under low-O2 conditions, some methanotrophs can switch to a fermentative metabolism and convert methane into excreted organic acids (233, 234), setting the stage for cross-feeding (235) (Fig. 3). Similarly, it was recently discovered that Fe-nitrogenase generates CH4 from CO2 in vivo, alongside the enzyme’s better-known products, NH4+ and H2 (236) (Fig. 3). CH4 production via nitrogenase by R. palustris supported the growth of a methanotroph in a synthetic coculture (236). This alternative activity of nitrogenase implicates the involvement of N2-fixing diazotrophs in the carbon cycle with the potential for carbon cross-feeding (Fig. 3). What other undiscovered enzymatic activities could form the basis for microbial cross-feeding?
Even engineered cross-feeding relationships can involve unpredictable cross-feeding reactions. A multi-omics analysis of syntrophic cocultures between a sulfate reducer and a methanogen strongly suggested an unexpected transfer of alanine from the sulfate reducer to the methanogen (237). Our cocultures of E. coli and N2-fixing R. palustris were designed such that growth of both species is dependent on the reciprocal exchange of organic acids and NH4+ (207) (Fig. 2G). However, transposon sequencing analysis of an E. coli mutant library revealed that R. palustris unexpectedly also cross-feeds purines, since the growth of E. coli purine auxotrophs was fully supported in coculture with R. palustris but not in monoculture (238). While purine cross-feeding was not enough to satisfy the entire E. coli nitrogen requirement, as excreted NH4+ does, this finding nevertheless highlights the existence of covert cross-feeding, even in engineered consortia, and its potential to affect the ecology and evolution of microbial communities.
The potential for widespread cross-feeding has several societal implications, including new strategies for bioremediation (239), bioproduction (103), and combatting polymicrobial and chronic infections (84, 218, 240–244). For example, in a tripartite mutualistic consortium, the collective antibiotic tolerance of the cross-feeding community was dependent on the least tolerant member, or the weakest link, of the community (241). Thus, depending on how important cross-feeding is to the fitness of pathogens within a polymicrobial infection, it might be possible to identify and target the weakest link (242, 243). The viability and stability of a community can also hinge on a single producer strain, without which the rest of the community would fall into competition (76). The ability to identify such keystone species or strains could not only be key to disrupting harmful microbial community behavior but also advantageous for the design of synthetic communities that could benefit society.
As technologies advance, researchers will be able to better assess the functions of individual genes across conditions and better distinguish the activities of individual populations, and even individual cells, within communities. Such discoveries will invariably reveal a greater complexity of metabolic connections between microbes. This multivariable information will be challenging to interpret but will nevertheless be essential toward understanding how microbial cross-feeding relationships establish and evolve. This knowledge can in turn be leveraged to either harness or disrupt cross-feeding interactions in ways that will benefit our society.
ACKNOWLEDGMENTS
We were supported in part by U.S. Army Research Office grant W911NF-14-1-0411 and a National Science Foundation CAREER award, grant MCB-1749489. Financial support for A.L.M. also came from the Simons Foundation, Division of Life Sciences, Simons Postdoctoral Fellowships in Marine Microbial Ecology award 600755.
We are grateful to current and previous members of the McKinlay lab, to C. Fuqua and J. Lennon (Indiana University), and to G. M. Cook and K. Hards (University of Otago) for stimulating discussions on this topic.
We declare there are no conflicts of interest.
Biographies

Ryan K. Fritts obtained his Ph.D. in microbiology in 2019 from Indiana University. His doctoral research in the McKinlay lab focused on the evolution and molecular basis of metabolite cross-feeding interactions in a synthetic bacterial community. He is currently a postdoctoral researcher in Prof. Shelly Copley’s lab at the University of Colorado. His postdoctoral research examines the role of gene duplication during bacterial adaptation and enzyme evolution.

Alexandra L. McCully obtained her Ph.D. in microbiology in 2018 from Indiana University. During her doctoral work in the McKinlay lab she identified metabolic and physiological principles that drive mutualistic cross-feeding within a synthetic bacterial community. She is currently a postdoctoral researcher in Prof. Alfred Spormann’s lab at Stanford University. Her postdoctoral research focuses on the metabolism and physiology of sediment-associated marine microbes, including the capacity for direct electron uptake and competitive interspecies interactions. McCully’s research has been funded by a postdoctoral fellowship in Marine Microbial Ecology from the Simons Foundation and by the National Science Foundation’s Center for Dark Energy Biosphere Investigations.

James B. McKinlay obtained his Ph.D. in microbiology and molecular genetics from Michigan State University in 2006 for his characterization of a succinate-producing bacterium with mentors Prof. Claire Vieille and Prof. J. Gregory Zeikus. His postdoctoral research was conducted from 2007 to 2011 with Prof. Caroline Harwood at the University of Washington, where he studied the metabolism behind phototrophic H2 production. As an Associate Professor in the Department of Biology at Indiana University, McKinlay’s lab studies bacterial physiology and metabolism with an emphasis on cooperative cross-feeding between fermentative and phototrophic bacteria. McKinlay is also an artist and combines his illustrations with storytelling to help communicate detail-intensive subject matter in his teaching and outreach activities.
REFERENCES
- 1.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burki F. 2014. The eukaryotic tree of Lile from a global phylogenomic perspective. Cold Spring Harb Perspect Biol 6:a016147. doi: 10.1101/cshperspect.a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazamia E, Helliwell KE, Purton S, Smith AG. 2016. How mutualisms arise in phytoplankton communities: building eco-evolutionary principles for aquatic microbes. Ecol Lett 19:810–822. doi: 10.1111/ele.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 6.Ettema TJG, Andersson SGE. 2009. The α-proteobacteria: the Darwin finches of the bacterial world. Biol Lett 5:429–432. doi: 10.1098/rsbl.2008.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 8.Cordero OX, Polz MF. 2014. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12:263–273. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- 9.Castelle CJ, Banfield JF. 2018. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 172:1181–1197. doi: 10.1016/j.cell.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Locey KJ, Lennon JT. 2016. Scaling laws predict global microbial diversity. Proc Natl Acad Sci U S A 113:5970–5975. doi: 10.1073/pnas.1521291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd KG, May MK, Kevorkian RT, Steen AD. 2013. Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl Environ Microbiol 79:7790–7799. doi: 10.1128/AEM.02090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arístegui J, Gasol JM, Duarte CM, Herndld GJ. 2009. Microbial oceanography of the dark ocean’s pelagic realm. Limnol Oceanogr 54:1501–1529. doi: 10.4319/lo.2009.54.5.1501. [DOI] [Google Scholar]
- 14.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster Kevin R, Bell T. 2012. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol 22:1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Nadell CD, Drescher K, Foster KR. 2016. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol 14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 17.Estrela S, Trisos CH, Brown SP. 2012. From metabolism to ecology: cross-feeding interactions shape the balance between polymicrobial conflict and mutualism. Am Nat 180:566–576. doi: 10.1086/667887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butaitė E, Baumgartner M, Wyder S, Kümmerli R. 2017. Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat Commun 8:414. doi: 10.1038/s41467-017-00509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright ES, Vetsigian KH. 2016. Inhibitory interactions promote frequent bistability among competing bacteria. Nat Commun 7:11274. doi: 10.1038/ncomms11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordero OX, Ventouras L-A, DeLong EF, Polz MF. 2012. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci U S A 109:20059–20064. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granato ET, Meiller-Legrand TA, Foster KR. 2019. The evolution and ecology of bacterial warfare. Curr Biol 29:R521–R537. doi: 10.1016/j.cub.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 22.Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9:119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 23.Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci U S A 107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergkessel M, Basta DW, Newman DK. 2016. The physiology of growth arrest: uniting molecular and environmental microbiology. Nat Rev Microbiol 14:549–562. doi: 10.1038/nrmicro.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirt SJ, Hinshelwood CN. 1965. The maintenance energy of bacteria in growing cultures. Proc Royal Soc B 163:224–231. [DOI] [PubMed] [Google Scholar]
- 26.Hoehler TM, Jørgensen BB. 2013. Microbial life under extreme energy limitation. Nat Rev Microbiol 11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 27.Basta DW, Bergkessel M, Newman DK. 2017. Identification of fitness determinants during energy-limited growth arrest in Pseudomonas aeruginosa. mBio 8:e01170-17. doi: 10.1128/mBio.01170-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pechter KB, Yin L, Oda Y, Gallagher L, Yang J, Manoil C, Harwood CS. 2017. Molecular basis of bacterial longevity. mBio 8:e01726-17. doi: 10.1128/mBio.01726-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinlay JB, Oda Y, Rühl M, Posto AL, Sauer U, Harwood CS. 2014. Non-growing Rhodopseudomonas palustris increases the hydrogen gas yield from acetate by shifting from the glyoxylate shunt to the tricarboxylic acid cycle. J Biol Chem 289:1960–1970. doi: 10.1074/jbc.M113.527515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCully AL, LaSarre B, McKinlay JB. 2017. Growth-independent cross-feeding modifies boundaries for coexistence in a bacterial mutualism. Environ Microbiol 19:3538–3550. doi: 10.1111/1462-2920.13847. [DOI] [PubMed] [Google Scholar]
- 31.Morris BEL, Henneberger R, Huber H, Moissl-Eichinger C. 2013. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 32.Seth EC, Taga ME. 2014. Nutrient cross-feeding in the microbial world. Front Microbiol 5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponomarova O, Patil KR. 2015. Metabolic interactions in microbial communities: untangling the Gordian knot. Curr Opin Microbiol 27:37–44. doi: 10.1016/j.mib.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Embree M, Liu JK, Al-Bassam MM, Zengler K. 2015. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc Natl Acad Sci U S A 112:15450–15455. doi: 10.1073/pnas.1506034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. 2015. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci U S A 112:6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zengler K, Zaramela LS. 2018. The social network of microorganisms: how auxotrophies shape complex communities. Nat Rev Microbiol 16:383–390. doi: 10.1038/s41579-018-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pande S, Kost C. 2017. Bacterial unculturability and the formation of intercellular metabolic networks. Trends Microbiol 25:349–361. doi: 10.1016/j.tim.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Hillesland KL. 2018. Evolution on the bright side of life: microorganisms and the evolution of mutualism. Ann N Y Acad Sci 1422:88–103. doi: 10.1111/nyas.13515. [DOI] [PubMed] [Google Scholar]
- 39.D’Souza G, Shitut S, Preussger D, Yousif G, Waschina S, Kost C. 2018. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep 35:455–488. doi: 10.1039/c8np00009c. [DOI] [PubMed] [Google Scholar]
- 40.Smith NW, Shorten PR, Altermann E, Roy NC, McNabb WC. 2019. The classification and evolution of bacterial cross-feeding. Front Ecol Evol 7:153. doi: 10.3389/fevo.2019.00153. [DOI] [Google Scholar]
- 41.Douglas AE. 2020. The microbial exometabolome: ecological resource and architect of microbial communities. Philos Trans R Soc Lond B Biol Sci 375:20190250. doi: 10.1098/rstb.2019.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Momeni B, Chen C-C, Hillesland KL, Waite A, Shou W. 2011. Using artificial systems to explore the ecology and evolution of symbioses. Cell Mol Life Sci 68:1353–1368. doi: 10.1007/s00018-011-0649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mee MT, Wang HH. 2012. Engineering ecosystems and synthetic ecologies. Mol Biosyst 8:2470–2483. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Widder S, Allen RJ, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, Cordero OX, Brown SP, Momeni B, Shou W, Kettle H, Flint HJ, Haas AF, Laroche B, Kreft J-U, Rainey PB, Freilich S, Schuster S, Milferstedt K, van der Meer JR, Groβkopf T, Huisman J, Free A, Picioreanu C, Quince C, Klapper I, Labarthe S, Smets BF, Wang H, Soyer OS, Isaac Newton Institute Foundation. 2016. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J 10:2557–2568. doi: 10.1038/ismej.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grandclement C, Tannieres M, Morera S, Dessaux Y, Faure D. 2016. Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev 40:86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 46.Ackermann M. 2015. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13:497–508. doi: 10.1038/nrmicro3491. [DOI] [PubMed] [Google Scholar]
- 47.Pacheco AR, Segrè D. 2019. A multidimensional perspective on microbial interactions. FEMS Microbiol Lett 366:fnz125. doi: 10.1093/femsle/fnz125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. 2005. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 49.Gómez-Consarnau L, Sachdeva R, Gifford SM, Cutter LS, Fuhrman JA, Sañudo-Wilhelmy SA, Moran MA. 2018. Mosaic patterns of B-vitamin synthesis and utilization in a natural marine microbial community. Environ Microbiol 20:2809–2823. doi: 10.1111/1462-2920.14133. [DOI] [PubMed] [Google Scholar]
- 50.Sharma V, Rodionov DA, Leyn SA, Tran D, Iablokov SN, Ding H, Peterson DA, Osterman AL, Peterson SN. 2019. B-vitamin sharing promotes stability of gut microbial communities. Front Microbiol 10:1485. doi: 10.3389/fmicb.2019.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hom EFY, Murray AW. 2014. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science 345:94–98. doi: 10.1126/science.1253320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKinlay JB, Harwood CS. 2011. Calvin cycle flux, pathway constraints, and substrate oxidation state together determine the H2 biofuel yield in photoheterotrophic bacteria. mBio 2:e00323-10. doi: 10.1128/mBio.00323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erb TJ. 2011. Carboxylases in natural and synthetic microbial pathways. Appl Environ Microbiol 77:8466–8477. doi: 10.1128/AEM.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKinlay JB, Vieille C, Zeikus JG. 2007. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol 76:727–740. doi: 10.1007/s00253-007-1057-y. [DOI] [PubMed] [Google Scholar]
- 55.Pacheco AR, Moel M, Segrè D. 2019. Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat Commun 10:103. doi: 10.1038/s41467-018-07946-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cavicchioli R, Ripple WJ, Timmis KN, Azam F, Bakken LR, Baylis M, Behrenfeld MJ, Boetius A, Boyd PW, Classen AT, Crowther TW, Danovaro R, Foreman CM, Huisman J, Hutchins DA, Jansson JK, Karl DM, Koskella B, Mark Welch DB, Martiny JBH, Moran MA, Orphan VJ, Reay DS, Remais JV, Rich VI, Singh BK, Stein LY, Stewart FJ, Sullivan MB, van Oppen MJH, Weaver SC, Webb EA, Webster NS. 2019. Scientists’ warning to humanity: microorganisms and climate change. Nat Rev Microbiol 17:569–586. doi: 10.1038/s41579-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 58.McInerney MJ, Sieber JR, Gunsalus RP. 2009. Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol 20:623–632. doi: 10.1016/j.copbio.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Struchtemeyer CG, Duncan KE, McInerney MJ. 2011. Evidence for syntrophic butyrate metabolism under sulfate-reducing conditions in a hydrocarbon-contaminated aquifer. FEMS Microbiol Ecol 76:289–300. doi: 10.1111/j.1574-6941.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 60.Sieber JR, Le HM, McInerney MJ. 2014. The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism. Environ Microbiol 16:177–188. doi: 10.1111/1462-2920.12269. [DOI] [PubMed] [Google Scholar]
- 61.Hillesland KL, Stahl DA. 2010. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci U S A 107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hillesland KL, Lim S, Flowers JJ, Turkarslan S, Pinel N, Zane GM, Elliott N, Qin Y, Wu L, Baliga NS, Zhou J, Wall JD, Stahl DA. 2014. Erosion of functional independence early in the evolution of a microbial mutualism. Proc Natl Acad Sci U S A 111:14822–14827. doi: 10.1073/pnas.1407986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS, Lovley DR. 2010. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330:1413–1415. doi: 10.1126/science.1196526. [DOI] [PubMed] [Google Scholar]
- 64.Mee MT, Collins JJ, Church GM, Wang HH. 2014. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci U S A 111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D’Souza G, Waschina S, Pande S, Bohl K, Kaleta C, Kost C. 2014. Less is more: selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68:2559–2570. doi: 10.1111/evo.12468. [DOI] [PubMed] [Google Scholar]
- 66.Degnan PH, Taga ME, Goodman AL. 2014. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab 20:769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cooper MB, Kazamia E, Helliwell KE, Kudahl UJ, Sayer A, Wheeler GL, Smith AG. 2019. Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J 13:334–345. doi: 10.1038/s41396-018-0274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Estrela S, Morris JJ, Kerr B. 2016. Private benefits and metabolic conflicts shape the emergence of microbial interdependencies. Environ Microbiol 18:1415–1427. doi: 10.1111/1462-2920.13028. [DOI] [PubMed] [Google Scholar]
- 70.Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, Kaleta C, Kost C. 2014. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J 8:953–962. doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pande S, Kaftan F, Lang S, Svatoš A, Germerodt S, Kost C. 2016. Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J 10:1413–1423. doi: 10.1038/ismej.2015.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziesack M, Gibson T, Oliver JKW, Shumaker AM, Hsu BB, Riglar DT, Giessen TW, DiBenedetto NV, Bry L, Way JC, Silver PA, Gerber GK. 2019. Engineered interspecies amino acid cross-feeding increases population evenness in a synthetic bacterial consortium. mSystems 4:e00352-19. doi: 10.1128/mSystems.00352-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harcombe W. 2010. Novel cooperation experimentally evolved between species. Evolution 64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 74.Harcombe WR, Betts A, Shapiro JW, Marx CJ. 2016. Adding biotic complexity alters the metabolic benefits of mutualism. Evolution 70:1871–1881. doi: 10.1111/evo.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shou W, Ram S, Vilar JMG. 2007. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci U S A 104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hammarlund SP, Gedeon T, Carlson RP, Harcombe W. 2020. Limitation by a shared mutualist promotes coexistence of multiple competing partners. bioRxiv doi: 10.1101/2020.04.22.055517. [DOI] [PMC free article] [PubMed]
- 77.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat Prod Rep 19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 78.Grant MAA, Kazamia E, Cicuta P, Smith AG. 2014. Direct exchange of vitamin B12 is demonstrated by modeling the growth dynamics of algal-bacterial cocultures. ISME J 8:1418–1427. doi: 10.1038/ismej.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sokolovskaya OM, Shelton AN, Taga ME. 2020. Sharing vitamins: cobamides unveil microbial interactions. Science 369:eaba0165. doi: 10.1126/science.aba0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nelson D, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B. 1995. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycles 9:359–372. doi: 10.1029/95GB01070. [DOI] [Google Scholar]
- 81.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 82.Tréguer P, Bowler C, Moriceau B, Dutkiewicz S, Gehlen M, Aumont O, Bittner L, Dugdale R, Finkel Z, Iudicone D, Jahn O, Guidi L, Lasbleiz M, Leblanc K, Levy M, Pondaven P. 2018. Influence of diatom diversity on the ocean biological carbon pump. Nat Geosci 11:27–37. doi: 10.1038/s41561-017-0028-x. [DOI] [Google Scholar]
- 83.Belzer C, Chia LW, Aalvink S, Chamlagain B, Piironen V, Knol J, de Vos WM. 2017. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. mBio 8:e00770-17. doi: 10.1128/mBio.00770-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hammer N, Cassat J, Noto MJ, Lojek L, Chadha A, Schmitz J, Creech C, Skaar E. 2014. Inter- and intraspecies metabolite exchange promotes virulence of antibiotic-resistant Staphylococcus aureus. Cell Host Microbe 16:531–537. doi: 10.1016/j.chom.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walter A, Gutknecht J. 1986. Permeability of small nonelectrolytes through lipid bilayer membranes. J Membr Biol 90:207–217. doi: 10.1007/BF01870127. [DOI] [PubMed] [Google Scholar]
- 86.Kim M, Zhang Z, Okano H, Yan D, Groisman A, Hwa T. 2012. Need-based activation of ammonium uptake in Escherichia coli. Mol Syst Biol 8:616. doi: 10.1038/msb.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harcombe WR, Chacón JM, Adamowicz EM, Chubiz LM, Marx CJ. 2018. Evolution of bidirectional costly mutualism from byproduct consumption. Proc Natl Acad Sci U S A 115:12000–12004. doi: 10.1073/pnas.1810949115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Germerodt S, Bohl K, Lück A, Pande S, Schröter A, Kaleta C, Schuster S, Kost C. 2016. Pervasive selection for cooperative cross-feeding in bacterial communities. PLoS Comput Biol 12:e1004986. doi: 10.1371/journal.pcbi.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Braakman R, Follows MJ, Chisholm SW. 2017. Metabolic evolution and the self-organization of ecosystems. Proc Natl Acad Sci U S A 114:E3091–E3100. doi: 10.1073/pnas.1619573114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Werner GD, Strassmann JE, Ivens AB, Engelmoer DJ, Verbruggen E, Queller DC, Noe R, Johnson NC, Hammerstein P, Kiers ET. 2014. Evolution of microbial markets. Proc Natl Acad Sci U S A 111:1237–1244. doi: 10.1073/pnas.1315980111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sher D, Thompson JW, Kashtan N, Croal L, Chisholm SW. 2011. Response of Prochlorococcus ecotypes to coculture with diverse marine bacteria. ISME J 5:1125–1132. doi: 10.1038/ismej.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fritts RK, Bird JT, Behringer MG, Lipzen A, Martin J, Lynch M, McKinlay JB. 2020. Enhanced nutrient uptake is sufficient to drive emergent cross-feeding between bacteria in a synthetic community. ISME J 14:2816–2828. doi: 10.1038/s41396-020-00737-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rosenzweig RF, Sharp RR, Treves DS, Adams J. 1994. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137:903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Treves DS, Manning S, Adams J. 1998. Repeated evolution of an acetate-cross-feeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol 15:789–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- 95.Gudelj I, Kinnersley M, Rashkov P, Schmidt K, Rosenzweig F. 2016. Stability of cross-feeding polymorphisms in microbial communities. PLoS Comput Biol 12:e1005269. doi: 10.1371/journal.pcbi.1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Datta MS, Sliwerska E, Gore J, Polz MF, Cordero OX. 2016. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat Commun 7:11965. doi: 10.1038/ncomms11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pelve EA, Fontanez KM, DeLong EF. 2017. Bacterial succession on sinking particles in the ocean’s interior. Front Microbiol 8:2269. doi: 10.3389/fmicb.2017.02269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Enke TN, Datta MS, Schwartzman J, Cermak N, Schmitz D, Barrere J, Pascual-García A, Cordero OX. 2019. Modular assembly of polysaccharide-degrading marine microbial communities. Curr Biol 29:1528–1535. doi: 10.1016/j.cub.2019.03.047. [DOI] [PubMed] [Google Scholar]
- 99.Solden LM, Naas AE, Roux S, Daly RA, Collins WB, Nicora CD, Purvine SO, Hoyt DW, Schückel J, Jørgensen B, Willats W, Spalinger DE, Firkins JL, Lipton MS, Sullivan MB, Pope PB, Wrighton KC. 2018. Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nat Microbiol 3:1274–1284. doi: 10.1038/s41564-018-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brune A. 2014. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 101.Chandel AK, Chandrasekhar G, Silva MB, Silvério da Silva S. 2012. The realm of cellulases in biorefinery development. Crit Rev Biotechnol 32:187–202. doi: 10.3109/07388551.2011.595385. [DOI] [PubMed] [Google Scholar]
- 102.Odom JM, Wall JD. 1983. Photoproduction of H2 from cellulose by an anaerobic bacterial coculture. Appl Environ Microbiol 45:1300–1305. doi: 10.1128/AEM.45.4.1300-1305.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roell GW, Zha J, Carr RR, Koffas MA, Fong SS, Tang YJ. 2019. Engineering microbial consortia by division of labor. Microb Cell Fact 18:35. doi: 10.1186/s12934-019-1083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gore J, Youk H, van Oudenaarden A. 2009. Snowdrift game dynamics and facultative cheating in yeast. Nature 459:253–256. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. 2014. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol 24:50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Artzi L, Bayer EA, Moraïs S. 2017. Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat Rev Microbiol 15:83–95. doi: 10.1038/nrmicro.2016.164. [DOI] [PubMed] [Google Scholar]
- 107.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munōz-Munōz JL, Day A, Peña MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. 2015. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol 74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Behnsen J, Raffatellu M. 2016. Siderophores: more than stealing iron. mBio 7:e01906-16. doi: 10.1128/mBio.01906-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnstone TC, Nolan EM. 2015. Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans 44:6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen–host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barber MF, Elde NC. 2015. Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genet 31:627–636. doi: 10.1016/j.tig.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boyd PW, Ellwood MJ. 2010. The biogeochemical cycle of iron in the ocean. Nature Geosci 3:675–682. doi: 10.1038/ngeo964. [DOI] [Google Scholar]
- 114.Wilson BR, Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y. 2016. Siderophores in iron metabolism: from mechanism to therapy potential. Trends Mol Med 22:1077–1090. doi: 10.1016/j.molmed.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kortman GAM, Raffatellu M, Swinkels DW, Tjalsma H. 2014. Nutritional iron turned inside out: intestinal stress from a gut microbial perspective. FEMS Microbiol Rev 38:1202–1234. doi: 10.1111/1574-6976.12086. [DOI] [PubMed] [Google Scholar]
- 116.Kramer J, Özkaya Ö, Kümmerli R. 2020. Bacterial siderophores in community and host interactions. Nat Rev Microbiol 18:152–163. doi: 10.1038/s41579-019-0284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ford SA, Kao D, Williams D, King KC. 2016. Microbe-mediated host defence drives the evolution of reduced pathogen virulence. Nat Commun 7:13430. doi: 10.1038/ncomms13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rivera-Chávez F, Mekalanos JJ. 2019. Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 572:244–248. doi: 10.1038/s41586-019-1453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Amin SA, Green DH, Hart MC, Küpper FC, Sunda WG, Carrano CJ. 2009. Photolysis of iron–siderophore chelates promotes bacterial–algal mutualism. Proc Natl Acad Sci U S A 106:17071–17076. doi: 10.1073/pnas.0905512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Loper JE, Henkels MD. 1999. Utilization of heterologous siderophores enhances levels of iron available to Pseudomonas putida in the rhizosphere. Appl Environ Microbiol 65:5357–5363. doi: 10.1128/AEM.65.12.5357-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Joshi F, Archana G, Desai A. 2006. Siderophore cross-utilization amongst Rhizospheric bacteria and the Role of their differential affinities for Fe3+ on growth stimulation under iron-limited conditions. Curr Microbiol 53:141–147. doi: 10.1007/s00284-005-0400-8. [DOI] [PubMed] [Google Scholar]
- 122.Champomier-Vergès M-C, Stintzi A, Meyer J-M. 1996. Acquisition of iron by the non-siderophore-producing Pseudomonas fragi. Microbiology 142:1191–1199. doi: 10.1099/13500872-142-5-1191. [DOI] [PubMed] [Google Scholar]
- 123.Cezard C, Farvacques N, Sonnet P. 2015. Chemistry and biology of pyoverdines, Pseudomonas primary siderophores. Curr Med Chem 22:165–186. doi: 10.2174/0929867321666141011194624. [DOI] [PubMed] [Google Scholar]
- 124.Niehus R, Picot A, Oliveira NM, Mitri S, Foster KR. 2017. The evolution of siderophore production as a competitive trait. Evolution 71:1443–1455. doi: 10.1111/evo.13230. [DOI] [PubMed] [Google Scholar]
- 125.Jin Z, Li J, Ni L, Zhang R, Xia A, Jin F. 2018. Conditional privatization of a public siderophore enables Pseudomonas aeruginosa to resist cheater invasion. Nat Commun 9:1383. doi: 10.1038/s41467-018-03791-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sexton DJ, Schuster M. 2017. Nutrient limitation determines the fitness of cheaters in bacterial siderophore cooperation. Nat Commun 8:230. doi: 10.1038/s41467-017-00222-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Qi B, Han M. 2018. Microbial siderophore enterobactin promotes mitochondrial iron uptake and development of the host via interaction with ATP synthase. Cell 175:571–582. doi: 10.1016/j.cell.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 128.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. 2015. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci U S A 112:2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smalley NE, An D, Parsek MR, Chandler JR, Dandekar AA. 2015. Quorum sensing protects Pseudomonas aeruginosa against cheating by other species in a laboratory coculture model. J Bacteriol 197:3154–3159. doi: 10.1128/JB.00482-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rollinson G, Jones R, Meadows MP, Harris RE, Knowles CJ. 1987. The growth of a cyanide-utilizing strain of Pseudomonas fluorescens in liquid culture on nickel cyanide as a source of nitrogen. FEMS Microbiol Lett 40:199–205. doi: 10.1111/j.1574-6968.1987.tb02025.x. [DOI] [Google Scholar]
- 131.Kunz DA, Nagappan O, Silva-Avalos J, Delong GT. 1992. Utilization of cyanide as nitrogenous substrate by Pseudomonas fluorescens NCIMB 11764: evidence for multiple pathways of metabolic conversion. Appl Environ Microbiol 58:2022–2029. doi: 10.1128/AEM.58.6.2022-2029.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pramer D, Starkey RL. 1951. Decomposition of streptomycin. Science 113:127–127. doi: 10.1126/science.113.2927.127. [DOI] [PubMed] [Google Scholar]
- 133.Johnsen J. 1977. Utilization of benzylpenicillin as carbon, nitrogen, and energy source by a Pseudomonas fluorescens strain. Arch Microbiol 115:271–275. doi: 10.1007/BF00446452. [DOI] [PubMed] [Google Scholar]
- 134.Dantas G, Sommer MOA, Oluwasegun RD, Church GM. 2008. Bacteria subsisting on antibiotics. Science 320:100–103. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 135.Barnhill AE, Weeks KE, Xiong N, Day TA, Carlson SA. 2010. Identification of multiresistant Salmonella isolates capable of subsisting on antibiotics. Appl Environ Microbiol 76:2678–2680. doi: 10.1128/AEM.02516-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Walsh F, Amyes SGB, Duffy B. 2013. Challenging the concept of bacteria subsisting on antibiotics. Int J Antimicrob Agents 41:558–563. doi: 10.1016/j.ijantimicag.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 137.Ibberson CB, Stacy A, Fleming D, Dees JL, Rumbaugh K, Gilmore MS, Whiteley M. 2017. Coinfecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat Microbiol 2:17079. doi: 10.1038/nmicrobiol.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lewin GR, Stacy A, Michie KL, Lamont RJ, Whiteley M. 2019. Large-scale identification of pathogen essential genes during coinfection with sympatric and allopatric microbes. Proc Natl Acad Sci U S A 116:19685–19694. doi: 10.1073/pnas.1907619116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Armbruster CE, Forsyth-DeOrnellas V, Johnson AO, Smith SN, Zhao L, Wu W, Mobley HLT. 2017. Genome-wide transposon mutagenesis of Proteus mirabilis: essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog 13:e1006434. doi: 10.1371/journal.ppat.1006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Whiteley M, Diggle SP, Greenberg EP. 2017. Progress in and promise of bacterial quorum sensing research. Nature 551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Abisado RG, Benomar S, Klaus JR, Dandekar AA, Chandler JR. 2018. Bacterial quorum sensing and microbial community interactions. mBio 9:e02331-17. doi: 10.1128/mBio.01749-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mukherjee S, Bassler BL. 2019. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol 17:371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.LaSarre B, Federle MJ. 2013. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev 77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang JJ, Han J-I, Zhang L-H, Leadbetter JR. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 69:5941–5949. doi: 10.1128/aem.69.10.5941-5949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dandekar AA, Chugani S, Greenberg EP. 2012. Bacterial quorum sensing and metabolic incentives to cooperate. Science 338:264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Whiteley M, Lee KM, Greenberg EP. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sandoz KM, Mitzimberg SM, Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A 104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wellington S, Greenberg EP. 2019. Quorum sensing signal selectivity and the potential for interspecies cross talk. mBio 10:e00146-19. doi: 10.1128/mBio.00146-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rezzonico F, Duffy B. 2008. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol 8:154. doi: 10.1186/1471-2180-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bridges AA, Bassler BL. 2019. The intragenus and interspecies quorum-sensing autoinducers exert distinct control over Vibrio cholerae biofilm formation and dispersal. PLoS Biol 17:e3000429. doi: 10.1371/journal.pbio.3000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pai A, Tanouchi Y, Collins CH, You L. 2009. Engineering multicellular systems by cell–cell communication. Curr Opin Biotechnol 20:461–470. doi: 10.1016/j.copbio.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marchand N, Collins CH. 2013. Peptide-based communication system enables Escherichia coli to Bacillus megaterium interspecies signaling. Biotechnol Bioeng 110:3003–3012. doi: 10.1002/bit.24975. [DOI] [PubMed] [Google Scholar]
- 155.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. 2015. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stuart RK, Mayali X, Lee JZ, Craig Everroad R, Hwang M, Bebout BM, Weber PK, Pett-Ridge J, Thelen MP. 2016. Cyanobacterial reuse of extracellular organic carbon in microbial mats. ISME J 10:1240–1251. doi: 10.1038/ismej.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Preussger D, Giri S, Muhsal LK, Ona L, Kost C. 2020. Reciprocal fitness feedbacks promote the evolution of mutualistic cooperation. Curr Biol 30:3580–3590. doi: 10.1016/j.cub.2020.06.100. [DOI] [PubMed] [Google Scholar]
- 158.Momeni B, Waite AJ, Shou W. 2013. Spatial self-organization favors heterotypic cooperation over cheating. Elife 2:e00960. doi: 10.7554/eLife.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Momeni B, Brileya KA, Fields MW, Shou W. 2013. Strong inter-population cooperation leads to partner intermixing in microbial communities. Elife 2:e00230. doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marx CJ. 2009. Getting in touch with your friends. Science 324:1150–1151. doi: 10.1126/science.1173088. [DOI] [PubMed] [Google Scholar]
- 161.Marchal M, Goldschmidt F, Derksen-Müller SN, Panke S, Ackermann M, Johnson DR. 2017. A passive mutualistic interaction promotes the evolution of spatial structure within microbial populations. BMC Evol Biol 17:106. doi: 10.1186/s12862-017-0950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fedeson DT, Ducat DC. 2017. Cyanobacterial surface display system mediates engineered interspecies and abiotic binding. ACS Synthetic Biol 6:367–374. doi: 10.1021/acssynbio.6b00254. [DOI] [PubMed] [Google Scholar]
- 163.van Tatenhove-Pel RJ, Rijavec T, Lapanje A, van Swam I, Zwering E, Hernandez-Valdes JA, Kuipers OP, Picioreanu C, Teusink B, Bachmann H. 2020. Microbial competition reduces metabolic interaction distances to the low μm range. ISME J doi: 10.1038/s41396-020-00806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dal Co A, van Vliet S, Kiviet DJ, Schlegel S, Ackermann M. 2020. Short-range interactions govern the dynamics and functions of microbial communities. Nat Ecol Evol 4:366–375. doi: 10.1038/s41559-019-1080-2. [DOI] [PubMed] [Google Scholar]
- 165.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. 2008. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A 105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Woith E, Fuhrmann G, Melzig MF. 2019. Extracellular vesicles-connecting kingdoms. Int J Mol Sci 20:5695. doi: 10.3390/ijms20225695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. 2015. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Evans AGL, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. 2012. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology (Reading) 158:2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 170.Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, Hanawa T, Kamiya S. 2009. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol 9:197. doi: 10.1186/1471-2180-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, Inaba H, Schmitt-Kopplin P, Eberl L, Nomura N. 2017. Membrane vesicle-mediated bacterial communication. ISME J 11:1504–1509. doi: 10.1038/ismej.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rodriguez GM, Prados-Rosales R. 2016. Functions and importance of mycobacterial extracellular vesicles. Appl Microbiol Biotechnol 100:3887–3892. doi: 10.1007/s00253-016-7484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. 2014. Bacterial vesicles in marine ecosystems. Science 343:183–186. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- 174.Danka ES, Garcia EC, Cotter PA. 2017. Are CDI systems multicolored, facultative, helping greenbeards? Trends Microbiol 25:391–401. doi: 10.1016/j.tim.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Coulthurst S. 2019. The Type VI secretion system: a versatile bacterial weapon. Microbiology 165:503–515. doi: 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 176.Yarlett N, Hackstein JHP. 2005. Hydrogenosomes: one organelle, multiple origins. Bioscience 55:657–668. doi: 10.1641/0006-3568(2005)055[0657:HOOMO]2.0.CO;2. [DOI] [Google Scholar]
- 177.McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol 21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M, Tanaka M, Satoh N, Bachtrog D, Wilson ACC, von Dohlen CD, Fukatsu T, McCutcheon JP. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 179.Nickell S, Hegerl R, Baumeister W, Rachel R. 2003. Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryo-electron tomography. J Struct Biol 141:34–42. doi: 10.1016/S1047-8477(02)00581-6. [DOI] [PubMed] [Google Scholar]
- 180.Lovley DR. 2017. Happy together: microbial communities that hook up to swap electrons. ISME J 11:327–336. doi: 10.1038/ismej.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Meysman FJR. 2018. Cable bacteria take a new breath using long-distance electricity. Trends Microbiol 26:411–422. doi: 10.1016/j.tim.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 182.Herrero A, Stavans J, Flores E. 2016. The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol Rev 40:831–854. doi: 10.1093/femsre/fuw029. [DOI] [PubMed] [Google Scholar]
- 183.Flores E, Nieves-Morión M, Mullineaux CW. 2018. Cyanobacterial septal junctions: properties and regulation. Life 9:1. doi: 10.3390/life9010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Behrens S, Lösekann T, Pett-Ridge J, Weber PK, Ng W-O, Stevenson BS, Hutcheon ID, Relman DA, Spormann AM. 2008. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol 74:3143–3150. doi: 10.1128/AEM.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huber H, Küper U, Daxer S, Rachel R. 2012. The unusual cell biology of the hyperthermophilic Crenarchaeon Ignicoccus hospitalis. Antonie Van Leeuwenhoek 102:203–219. doi: 10.1007/s10482-012-9748-5. [DOI] [PubMed] [Google Scholar]
- 186.Küper U, Meyer C, Müller V, Rachel R, Huber H. 2010. Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic Archaeon Ignicoccus hospitalis. Proc Natl Acad Sci U S A 107:3152–3156. doi: 10.1073/pnas.0911711107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.John M, Trzcinski AP, Zhou Y, Ng WJ. 2017. Microbial stress mediated intercellular nanotubes in an anaerobic microbial consortium digesting cellulose. Sci Rep 7:18006. doi: 10.1038/s41598-017-18198-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pal RR, Baidya AK, Mamou G, Bhattacharya S, Socol Y, Kobi S, Katsowich N, Ben-Yehuda S, Rosenshine I. 2019. Pathogenic Escherichia coli extracts nutrients from infected host cells utilizing injectisome components. Cell 177:683–696. doi: 10.1016/j.cell.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 189.Baidya AK, Rosenshine I, Ben-Yehuda S. 2020. Donor-delivered cell wall hydrolases facilitate nanotube penetration into recipient bacteria. Nat Commun 11:1938. doi: 10.1038/s41467-020-15605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pospisil J, Vitovska D, Kofronova O, Muchova K, Sanderova H, Hubalek M, Sikova M, Modrak M, Benada O, Barak I, Krasny L. 2020. Bacterial nanotubes as a manifestation of cell death. Nat Commun 11:4963. doi: 10.1038/s41467-020-18800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dubey GP, Malli Mohan GB, Dubrovsky A, Amen T, Tsipshtein S, Rouvinski A, Rosenberg A, Kaganovich D, Sherman E, Medalia O, Ben-Yehuda S. 2016. Architecture and characteristics of bacterial nanotubes. Dev Cell 36:453–461. doi: 10.1016/j.devcel.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 192.Galan JE. 2019. Reconfiguration of a bacterial transport system generates a reverse gear. Nature 569:44–45. doi: 10.1038/d41586-019-01320-5. [DOI] [PubMed] [Google Scholar]
- 193.Dubey GP, Ben-Yehuda S. 2011. Intercellular nanotubes mediate bacterial communication. Cell 144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 194.Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, Bischofs IB, Kost C. 2015. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun 6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 195.Benomar S, Ranava D, Cárdenas ML, Trably E, Rafrafi Y, Ducret A, Hamelin J, Lojou E, Steyer J-P, Giudici-Orticoni M-T. 2015. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nat Commun 6:6283. doi: 10.1038/ncomms7283. [DOI] [PubMed] [Google Scholar]
- 196.Gralnick JA, Newman DK. 2007. Extracellular respiration. Mol Microbiol 65:1–11. doi: 10.1111/j.1365-2958.2007.05778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. 2015. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526:531–535. doi: 10.1038/nature15512. [DOI] [PubMed] [Google Scholar]
- 198.Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. 2015. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526:587–590. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- 199.Conrad R. 2009. The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep 1:285–292. doi: 10.1111/j.1758-2229.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- 200.Walker DJF, Nevin KP, Holmes DE, Rotaru A-E, Ward JE, Woodard TL, Zhu J, Ueki T, Nonnenmann SS, McInerney MJ, Lovley DR. 2020. Syntrophus conductive pili demonstrate that common hydrogen-donating syntrophs can have a direct electron transfer option. ISME J 14:837–846. doi: 10.1038/s41396-019-0575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lovley DR, Malvankar NS. 2015. Seeing is believing: novel imaging techniques help clarify microbial nanowire structure and function. Environ Microbiol 17:2209–2215. doi: 10.1111/1462-2920.12708. [DOI] [PubMed] [Google Scholar]
- 202.Wang F, Gu Y, O’Brien JP, Yi SM, Yalcin SE, Srikanth V, Shen C, Vu D, Ing NL, Hochbaum AI, Egelman EH, Malvankar NS. 2019. Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers. Cell 177:361–369. doi: 10.1016/j.cell.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pirbadian S, Barchinger SE, Leung KM, Byun HS, Jangir Y, Bouhenni RA, Reed SB, Romine MF, Saffarini DA, Shi L, Gorby YA, Golbeck JH, El-Naggar MY. 2014. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc Natl Acad Sci U S A 111:12883–12888. doi: 10.1073/pnas.1410551111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Shi L, Dong H, Reguera G, Beyenal H, Lu A, Liu J, Yu HQ, Fredrickson JK. 2016. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol 14:651–662. doi: 10.1038/nrmicro.2016.93. [DOI] [PubMed] [Google Scholar]
- 205.Reguera G. 2018. Harnessing the power of microbial nanowires. Microb Biotechnol 11:979–994. doi: 10.1111/1751-7915.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Goldschmidt F, Regoes RR, Johnson DR. 2018. Metabolite toxicity slows local diversity loss during expansion of a microbial cross-feeding community. ISME J 12:136–144. doi: 10.1038/ismej.2017.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.LaSarre B, McCully AL, Lennon JT, McKinlay JB. 2017. Microbial mutualism dynamics governed by dose-dependent toxicity of cross-fed nutrients. ISME J 11:337–348. doi: 10.1038/ismej.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lilja EE, Johnson DR. 2016. Segregating metabolic processes into different microbial cells accelerates the consumption of inhibitory substrates. ISME J 10:1568–1578. doi: 10.1038/ismej.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Daims H, Lücker S, Wagner M. 2016. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol 24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Park S, Bae W. 2009. Modeling kinetics of ammonium oxidation and nitrite oxidation under simultaneous inhibition by free ammonia and free nitrous acid. Process Biochem 44:631–640. doi: 10.1016/j.procbio.2009.02.002. [DOI] [Google Scholar]
- 211.Duwat P, Sourice S, Cesselin B, Lamberet G, Vido K, Gaudu P, Le Loir Y, Violet F, Loubière P, Gruss A. 2001. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J Bacteriol 183:4509–4516. doi: 10.1128/JB.183.15.4509-4516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yamamoto Y, Poyart C, Trieu-Cuot P, Lamberet G, Gruss A, Gaudu P. 2005. Respiration metabolism of group B Streptococcus is activated by environmental haem and quinone and contributes to virulence. Mol Microbiol 56:525–534. doi: 10.1111/j.1365-2958.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- 213.Franza T, Delavenne E, Derré-Bobillot A, Juillard V, Boulay M, Demey E, Vinh J, Lamberet G, Gaudu P. 2016. A partial metabolic pathway enables group B Streptococcus to overcome quinone deficiency in a host bacterial community. Mol Microbiol 102:81–91. doi: 10.1111/mmi.13447. [DOI] [PubMed] [Google Scholar]
- 214.Rezaïki L, Lamberet G, Derré A, Gruss A, Gaudu P. 2008. Lactococcus lactis produces short-chain quinones that cross-feed group B Streptococcus to activate respiration growth. Mol Microbiol 67:947–957. doi: 10.1111/j.1365-2958.2007.06083.x. [DOI] [PubMed] [Google Scholar]
- 215.Flombaum P, Gallegos JL, Gordillo RA, Rincon J, Zabala LL, Jiao N, Karl DM, Li WK, Lomas MW, Veneziano D, Vera CS, Vrugt JA, Martiny AC. 2013. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci U S A 110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Christie-Oleza JA, Sousoni D, Lloyd M, Armengaud J, Scanlan DJ. 2017. Nutrient recycling facilitates long-term stability of marine microbial phototroph-heterotroph interactions. Nat Microbiol 2:17100. doi: 10.1038/nmicrobiol.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Roth-Rosenberg D, Aharonovich D, Luzzatto-Knaan T, Vogts A, Zoccarato L, Eigemann F, Nago N, Grossart H-P, Voss M, Sher D. 2020. Prochlorococcus cells rely on microbial interactions rather than on chlorotic resting stages to survive long-term nutrient starvation. mBio 11:e01846-20. doi: 10.1128/mBio.01846-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Camus L, Briaud P, Bastien S, Elsen S, Doléans-Jordheim A, Vandenesch F, Moreau K. 2020. Trophic cooperation promotes bacterial survival of Staphylococcus aureus and Pseudomonas aeruginosa. ISME J 14:3093–3105. doi: 10.1038/s41396-020-00741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Iannotti EL, Kafkewitz D, Wolin MJ, Bryant MP. 1973. Glucose fermentation products of Ruminococcus albus grown in continuous culture with Vibrio succinogenes: changes caused by interspecies transfer of H2. J Bacteriol 114:1231–1240. doi: 10.1128/JB.114.3.1231-1240.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ruaud A, Esquivel-Elizondo S, de la Cuesta-Zuluaga J, Waters JL, Angenent LT, Youngblut ND, Ley RE. 2020. Syntrophy via interspecies H2 transfer between Christensenella and Methanobrevibacter underlies their global cooccurrence in the human gut. mBio 11:e03235-19. doi: 10.1128/mBio.03235-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Oliveira NM, Niehus R, Foster KR. 2014. Evolutionary limits to cooperation in microbial communities. Proc Natl Acad Sci U S A 111:17941–17946. doi: 10.1073/pnas.1412673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.De Mazancourt C, Loreau M, Dieckmann ULF. 2005. Understanding mutualism when there is adaptation to the partner. J Ecol 93:305–314. doi: 10.1111/j.0022-0477.2004.00952.x. [DOI] [Google Scholar]
- 223.Becker JW, Hogle SL, Rosendo K, Chisholm SW. 2019. Coculture and biogeography of Prochlorococcus and SAR11. ISME J 13:1506–1519. doi: 10.1038/s41396-019-0365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, Segrè D, Mehta P, Sanchez A. 2018. Emergent simplicity in microbial community assembly. Science 361:469–474. doi: 10.1126/science.aat1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.McCully AL, LaSarre B, McKinlay JB. 2017. Recipient-biased competition for an intracellularly generated cross-fed nutrient is required for coexistence of microbial mutualists. mBio 8:e01620-17. doi: 10.1128/mBio.01620-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.McCully AL, Behringer MG, Gliessman JR, Pilipenko EV, Mazny JL, Lynch M, Drummond DA, McKinlay JB. 2018. An Escherichia coli nitrogen starvation response is important for mutualistic coexistence with Rhodopseudomonas palustris. Appl Environ Microbiol 84:e00404-18. doi: 10.1128/AEM.00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hart SFM, Pineda JMB, Chen C-C, Green R, Shou W. 2019. Disentangling strictly self-serving mutations from win-win mutations in a mutualistic microbial community. Elife 8:e44812. doi: 10.7554/eLife.44812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Celiker H, Gore J. 2012. Competition between species can stabilize public-goods cooperation within a species. Mol Syst Biol 8:621. doi: 10.1038/msb.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Park JO, Tanner LB, Wei MH, Khana DB, Jacobson TB, Zhang Z, Rubin SA, Li SH-J, Higgins MB, Stevenson DM, Amador-Noguez D, Rabinowitz JD. 2019. Near-equilibrium glycolysis supports metabolic homeostasis and energy yield. Nat Chem Biol 15:1001–1008. doi: 10.1038/s41589-019-0364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jacobson TB, Korosh TK, Stevenson DM, Foster C, Maranas C, Olson DG, Lynd LR, Amador-Noguez D. 2020. In vivo thermodynamic analysis of glycolysis in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum using 13C and 2H tracers. mSystems 5:e00736-19. doi: 10.1128/mSystems.00736-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Conrad R, Rothfuss F. 1991. Methane oxidation in the soil surface layer of a flooded rice field and the effect of ammonium. Biol Fert Soils 12:28–32. doi: 10.1007/BF00369384. [DOI] [Google Scholar]
- 232.Kuivila KM, Murray JW, Devol AH, Lidstrom ME, Reimers CE. 1988. Methane cycling in the sediments of Lake Washington. Limnol Oceanogr 33:571–581. doi: 10.4319/lo.1988.33.4.0571. [DOI] [Google Scholar]
- 233.Kalyuzhnaya MG, Yang S, Rozova ON, Smalley NE, Clubb J, Lamb A, Gowda GAN, Raftery D, Fu Y, Bringel F, Vuilleumier S, Beck DAC, Trotsenko YA, Khmelenina VN, Lidstrom ME. 2013. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun 4:2785. doi: 10.1038/ncomms3785. [DOI] [PubMed] [Google Scholar]
- 234.Gilman A, Fu Y, Hendershott M, Chu F, Puri AW, Smith AL, Pesesky M, Lieberman R, Beck DAC, Lidstrom ME. 2017. Oxygen-limited metabolism in the methanotroph Methylomicrobium buryatense 5GB1C. PeerJ 5:e3945. doi: 10.7717/peerj.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Krause SMB, Johnson T, Samadhi Karunaratne Y, Fu Y, Beck DAC, Chistoserdova L, Lidstrom ME. 2017. Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc Natl Acad Sci U S A 114:358–363. doi: 10.1073/pnas.1619871114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zheng Y, Harris DF, Yu Z, Fu Y, Poudel S, Ledbetter RN, Fixen KR, Yang Z-Y, Boyd ES, Lidstrom ME, Seefeldt LC, Harwood CS. 2018. A pathway for biological methane production using bacterial iron-only nitrogenase. Nat Microbiol 3:281–286. doi: 10.1038/s41564-017-0091-5. [DOI] [PubMed] [Google Scholar]
- 237.Walker CB, Redding-Johanson AM, Baidoo EE, Rajeev L, He Z, Hendrickson EL, Joachimiak MP, Stolyar S, Arkin AP, Leigh JA, Zhou J, Keasling JD, Mukhopadhyay A, Stahl DA. 2012. Functional responses of methanogenic archaea to syntrophic growth. ISME J 6:2045–2055. doi: 10.1038/ismej.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.LaSarre B, Deutschbauer AM, Love CE, McKinlay JB. 2020. Covert cross-feeding revealed by genome-wide analysis of fitness determinants in a synthetic bacterial mutualism. Appl Environ Microbiol 86:e00543-20. doi: 10.1128/AEM.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sousa DZ, Smidt H, Alves MM, Stams AJ. 2009. Ecophysiology of syntrophic communities that degrade saturated and unsaturated long-chain fatty acids. FEMS Microbiol Ecol 68:257–272. doi: 10.1111/j.1574-6941.2009.00680.x. [DOI] [PubMed] [Google Scholar]
- 240.Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, O’Toole GA. 2015. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol 197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Adamowicz EM, Flynn J, Hunter RC, Harcombe WR. 2018. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J 12:2723–2735. doi: 10.1038/s41396-018-0212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Adamowicz EM, Harcombe WR. 2020. Weakest link dynamics predict apparent antibiotic interactions in a model cross-feeding community. Antimicrob Agents Chemother 64:e00465-20. doi: 10.1128/AAC.00465-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Flynn JM, Cameron LC, Wiggen TD, Dunitz JM, Harcombe WR, Hunter RC. 2020. Disruption of cross-feeding inhibits pathogen growth in the sputa of patients with cystic fibrosis. mSphere 5:e00343-20. doi: 10.1128/mSphere.00343-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ramsey MM, Rumbaugh KP, Whiteley M. 2011. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog 7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Waite AJ, Shou W. 2012. Adaptation to a new environment allows cooperators to purge cheaters stochastically. Proc Natl Acad Sci U S A 109:19079–19086. doi: 10.1073/pnas.1210190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AH, Bonilla G, Kar A, Leiby N, Mehta P, Marx CJ, Segrè D. 2014. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep 7:1104–1115. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Schuchmann K, Müller V. 2016. Energetics and application of heterotrophy in acetogenic bacteria. Appl Environ Microbiol 82:4056–4069. doi: 10.1128/AEM.00882-16. [DOI] [PMC free article] [PubMed] [Google Scholar]