Abstract
Dental caries, the most common chronic infectious disease worldwide, has a complex etiology involving the interplay of microbial and host factors that are not completely understood. In this study, the oral microbiome and 38 host cytokines and chemokines were analyzed across 23 children with caries and 24 children with healthy dentition. De novo assembly of metagenomic sequencing obtained 527 metagenome-assembled genomes (MAGs), representing 150 bacterial species. Forty-two of these species had no genomes in public repositories, thereby representing novel taxa. These new genomes greatly expanded the known pangenomes of many oral clades, including the enigmatic Saccharibacteria clades G3 and G6, which had distinct functional repertoires compared to other oral Saccharibacteria. Saccharibacteria are understood to be obligate epibionts, which are dependent on host bacteria. These data suggest that the various Saccharibacteria clades may rely on their hosts for highly distinct metabolic requirements, which would have significant evolutionary and ecological implications. Across the study group, Rothia, Neisseria, and Haemophilus spp. were associated with good dental health, whereas Prevotella spp., Streptococcus mutans, and Human herpesvirus 4 (Epstein-Barr virus [EBV]) were more prevalent in children with caries. Finally, 10 of the host immunological markers were significantly elevated in the caries group, and co-occurrence analysis provided an atlas of potential relationships between microbes and host immunological molecules. Overall, this study illustrated the oral microbiome at an unprecedented resolution and contributed several leads for further study that will increase the understanding of caries pathogenesis and guide therapeutic development.
Dental caries, the most prevalent chronic infectious disease globally, is caused by a dysbiotic oral microbiota that creates an acidic microenvironment adjacent to the tooth surface that demineralizes the enamel, which can lead to permanent damage to the tooth (Pitts et al. 2017). Although historically, Streptococcus mutans is the taxon that has received the major focus in regard to this disease, the true complexity of the caries-associated oral microbiota has only been realized following the relatively recent development of culture-independent detection methods (Banas and Drake 2018; Burne 2018). It is now understood that caries has a complex etiology and can occur in the absence of detectable levels of S. mutans, but a thorough understanding of what other taxa are involved has not been achieved. Furthermore, despite evidence that both the innate and adaptive arms of the immune system influence caries disease (Costalonga and Herzberg 2014; Meyle et al. 2017), the cross-talk between the oral microbiota and host immunological molecules during dental caries, compared to health, is not well characterized.
The majority of previous studies examining the caries-associated oral microbiome have used 16S rRNA gene amplicon sequencing (“16S sequencing”). However, 16S sequencing provides relatively low-resolution data that are biased, owing to PCR and amplicon choice, and overlooks crucial information regarding the true strain-level diversity and functional capabilities of the communities present (Hong et al. 2009; Pinto and Raskin 2012; Jovel et al. 2016; Hillmann et al. 2018). In contrast, metagenomic sequencing (i.e., shotgun, whole-genome sequencing) efforts are able to assemble large numbers of whole genomes, which can identify novel taxa and provide strain-level information for pangenomic analysis. Several recent landmark studies, which focused on the gut microbiome, have illustrated the power of this method (Almeida et al. 2019; Nayfach et al. 2019; Pasolli et al. 2019). Although sequencing is also used to quantify the abundances of resident taxa in microbial communities, sequencing provides only compositional data (i.e., relative abundances), which must be handled carefully to avoid generating spurious conclusions—a fact that is frequently neglected by microbiome studies (Gloor et al. 2017; Morton et al. 2017, 2019b; Knight et al. 2018). The major goals of this study were to (1) use metagenomics to identify novel taxa and strain-level differences that are likely to affect caries pathogenesis, (2) use recently developed compositional analysis tools to examine the oral microbiome during dental caries, and (3) survey host immunological markers and potential cross-talk between oral bacteria and these markers during caries. The data generated here illustrate the caries-associated oral microbiome at an unprecedented level of resolution and encourages several avenues of further study that will greatly increase the understanding of caries pathogenesis.
Results
Study design
In-depth details of the study design (Supplemental Fig. S1A), clinical sampling, and inclusion and exclusion criteria are provided in Methods and Supplemental Methods. A summary of the collected subject metadata is provided in Supplemental Table S1. Subjects were dichotomized into two groups: healthy or caries (two or more active caries lesions with penetration through the enamel into the underlying dentin; only lesions at least 2 mm in depth were considered). All subjects provided 2 mL of unstimulated saliva for analysis of host markers and 2 mL of stimulated saliva for microbiome analysis. The topic of whether saliva sampling is adequate to examine the caries-associated microbiota is one of debate. In this case, the choice to sample saliva instead of dental plaque from teeth was mainly because of the ease of collection (owing to noninvasiveness and patient compliance) and the ability to obtain sufficient sample volume for analysis (particularly for the case of the host markers). Although the various microenvironments of the oral cavity have distinct microbial residents, and an ideal sampling scenario would examine diversity at multiple sites, saliva bathes all oral tissues and is generally thought to represent the overall oral composition (Mira 2018). Furthermore, previous analysis showed that although incorporation of dental plaque data improved the ability of the oral microbiota to predict caries onset, saliva alone was generally sufficient to both distinguish and predict the onset of the disease (Teng et al. 2015). Several studies have also shown that caries impacts the microbiota of not just specific lesion sites but also that of other apparently healthy teeth, indicating that the oral microbiome as a whole may change significantly (Gross et al. 2010, 2012; Jiang et al. 2013, 2014). This is particularly true in the case of multiple, deep dentin lesions, such as those examined here, where the disease has progressed to a more systemic, rather than site-specific, state.
Assembly of metagenome-assembled genomes (MAGs) recovers 527 genomes, 42 representing novel taxa
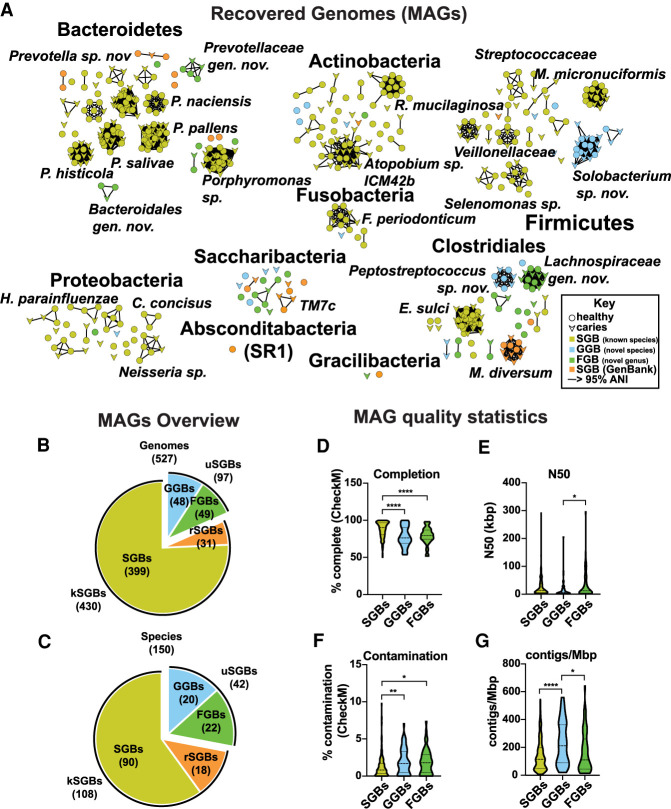
The metagenomics pipeline illustrated in Supplemental Figure S1B yielded 527 genomic bins that were of at least medium quality according to the guidelines set forth by the Genomic Standards Consortium (GSC) (>50% completeness, <10% contamination) (Supplemental Table S2; Bowers et al. 2017). Following dereplication of redundant species (≥95% ANI) across samples, there were 90 known species-level genome bins (kSGBs), representing 399 MAGs with ≥95% ANI to a RefSeq genome and 60 unknown species-level genome bins (uSGBs), representing 128 MAGs (Fig. 1A–G), with no genome in RefSeq with an ANI ≥95%. Individually comparing uSGBs against the wider GenBank database reassigned 18 uSGBs (rSGBs), representing 31 MAGs, to kSGBs, because they had ≥95% ANI match in GenBank (Supplemental Table S2). Twenty unknown species-level genome bins, representing 48 MAGs, that had 85%–95% ANI match to a GenBank genome were termed genus-level genome bins (GGBs), because the genus can be assigned with a fair amount of confidence, whereas the species appears to be not previously described. Twenty-two bins, representing 49 MAGs, had no match reference in GenBank with an ANI ≥85%. These were termed family-level genome bins (FGBs), because the family or higher-level taxa can be inferred, but the MAGs likely represent novel genera. These cutoffs for GGBs and FGBs were used and validated previously (Pasolli et al. 2019).
Figure 1.
Five hundred twenty-seven metagenome-assembled genomes (MAGs) were recovered. (A) Recovery of 151 species-level genome bins (SGBs), representing 527 MAGs and 42 novel taxa. Network representing an average nucleotide identity (ANI) distance matrix, generated by fastANI (Jain et al. 2018). Nodes represent MAGs, and edges represent an ANI > 95% (the cutoff chosen to designate species boundaries in this study). Circular nodes indicate MAGs recovered from healthy samples, and chevrons indicate MAGs recovered from caries samples. Nodes are colored based on bin designation: species-level (SGB: known species; yellow), genus-level (GGB: known genus, novel species; blue), family-level (FGB: novel genus and species; green), or reassigned to SGB (rSGB: orange). Subnetworks of interest are labeled with taxonomic names. (B,C) MAGs overview. Pie charts indicating the breakdown of bin types for genomes (B) and species (C). (D–G) Statistics indicating MAG quality. Violin charts illustrating the completion (D), N50 (E), contamination (F), and contigs/Mbp (G) of the SGBs, GGBs, and FGBs. Completion and contamination were determined by CheckM (Parks et al. 2015). Statistically significant differences between groups were determined using a Tukey's multiple comparisons posttest following a one-way ANOVA: (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001.
Twenty-five of the MAGs, including six genus-level genome bins and 11 family-level genome bins, represented Candidate Phyla Radiation (CPR) bacteria. This recently described supergroup is predicted to contain more than 35 phyla representing >15% of the diversity of all bacteria (Hug et al. 2016). CPR taxa have long been considered microbial “dark matter,” and only eight species have been cultivated thus far (He et al. 2015; Cross et al. 2019; Bor et al. 2020). CPR have reduced genomes and are thought to be obligate epibionts (He et al. 2015; Baker et al. 2017). In this data set, 22 CPR MAGs were Saccharibacteria (formerly, TM7), whereas two CPR MAGs were Gracilibacteria (formerly, GN02) and one was an Absconditabacteria (formerly, SR1). Because of their reduced genomes, CPR bacteria are missing many of the “essential” marker genes that are used when measuring genome completion, therefore completion percentages are highly underestimated for these genomes (Supplemental Table S2). For example, despite having a complete, closed, curated genome, TM7x has a completion of 65%, according to CheckM (McLean et al. 2020).
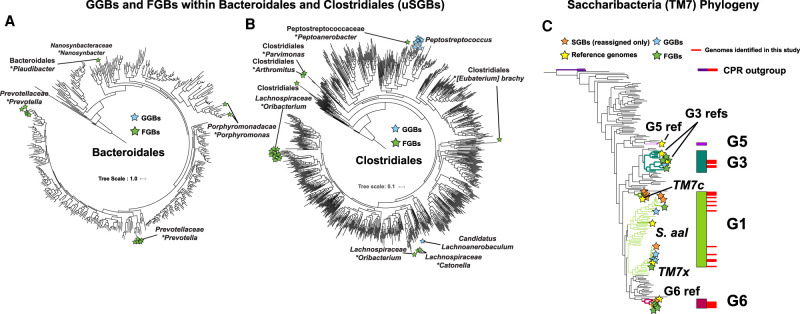
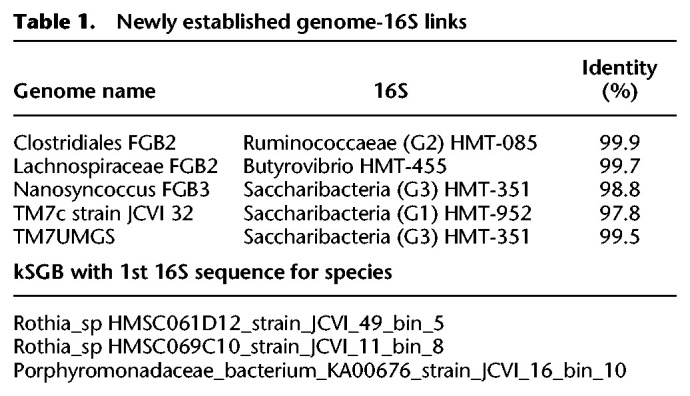
Because the majority of unknown genome bins were found within the clades Saccharibacteria, Bacteroidales, and Clostridiales, phylogenomics analysis was performed to place these genomes among available reference strains within these groups (Fig. 2A–C; Supplemental Fig. S2A–C). Many taxa have been identified exclusively by 16S sequencing and have no published genome. Conversely, there are taxa in which the only available genome sequences are not complete enough to include a 16S gene, which is notoriously difficult to obtain through de novo assembly because of the highly repetitive and conserved regions (Yuan et al. 2015). Thirty of the 527 genomes assembled in this study contained 16S sequences that were at least 80% complete. As a result, this study links five taxa that were previously known only by the16S sequence to their cognate genomes (four were unknown species-level genome bins) and three previously identified species with available genomes to a 16S sequence for the first time (Table 1).
Figure 2.
Phylogenetic trees of Bacteroidales (A), Clostridiales (B), and Saccharibacteria (C) reference genomes with placement of uSGBs. Reference genomes (C only), GGBs, FGBs, and rSGBs (C only) are denoted by stars of the indicated color. In A and B, the labels of GGBs indicate the genus of the uSGB, and the labels of FGBs indicate the family name and the name of the most closely related genus, denoted with the asterisk. In C, reference genomes and previously described oral Saccharibacteria clades G1, G3, G5, and G6 are labeled. The Bacteroidales and Clostridiales trees were constructed using PhyloPhlAn2, and the Saccharibacteria tree was constructed using Anvi'o. The trees with all leaves labeled are available in Supplemental Figure S2.
Table 1.
Newly established genome-16S links
Pangenomic analysis illustrates differences in functional potential between Saccharibacteria clades
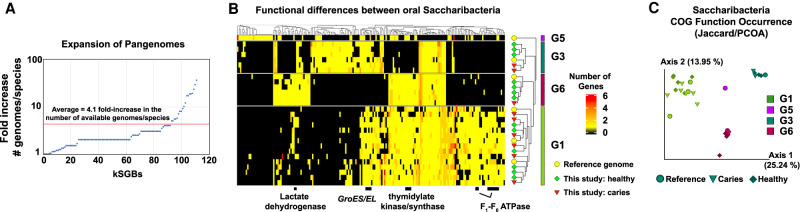
Another major advantage of the strain-level data provided by metagenomics is the ability to examine the pangenomes present in taxonomic clades. The large number of genomes assembled by this study significantly increased the available pangenomic information of many species (Fig. 3A). On average, the pangenome of each species with a MAG assembled in this study was increased by fourfold, and 60% of the taxa of known species-level genome bins had only one publicly available genome before this study. In a prime example of pangenome expansion, Saccharibacteria clade G6 (proposed family name of “Ca. Nanogingivalaceae”) had only one known representative genome (McLean et al. 2020). The phylogenomics performed here placed a second previously published genome into this clade (Espinoza et al. 2018; Shaiber and Eren 2019), added a second novel strain of that species, and four strains of a novel third species to clade G6. This study added a substantial amount of sequence data to Saccharibacteria clades G3 and G1 as well (Fig. 2C). The new information allowed for a detailed examination of the pangenome of the major human-associated Saccharibacteria clades, which revealed a significant variation in functional capability (Fig. 3B,C; Supplemental Fig. S3A,B). Many of the absent “essential” genes were unique to particular Saccharibacteria clades. For example, the F1-F0 ATPase appeared to be distinct to G1 Saccharibacteria, whereas most G1 and G3 genomes lacked lactate dehydrogenase. Because all Saccharibacteria cultured to-date are epibionts that depend on host bacteria, the diversity of critical functions present among the various Saccharibacteria clades suggests that different taxa may have different functional dependencies on their host species.
Figure 3.
Expansion of pangenomes allows for discovery of large-scale functional differences between Saccharibacteria clades. (A) Expansion of pangenomes. Scatter plot illustrating the fold increase in the number of available genomes per species for each species in which a kSGB was recovered by this study. The red line denotes the mean of a 4.1-fold increase. (B) Differences in encoded COG functions pathways across human-associated Saccharibacteria. Heatmap illustrating the presence of various genes across four major clades of human-associated Saccharibacteria. Lactate dehydrogenase, GroES/EL, thymidylate kinase/synthase, and the F1-F0 ATPase are highlighted to illustrate differences. Rows and columns were clustered using the Jaccard distance and the “complete” clustering method. Only COG functions that were significantly different between clades are shown (adjusted Q-value < 0.05). The heatmap with all rows and columns labeled is available in Supplemental Figure S3. (C) Saccharibacteria COG function occurrence. PCoA plot illustrating a Jaccard distance matrix of COG functions across Saccharibacteria clades.
An increased ratio of Prevotella spp. to Rothia spp., Haemophilus spp., and Neisseria spp., as well as decreased functional diversity, were associated with caries
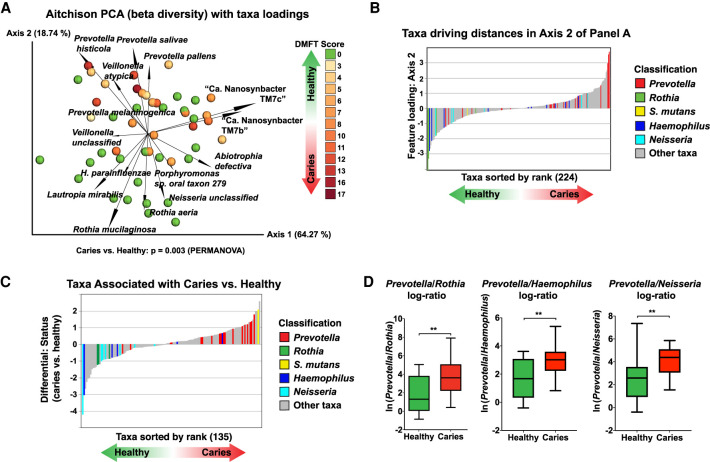
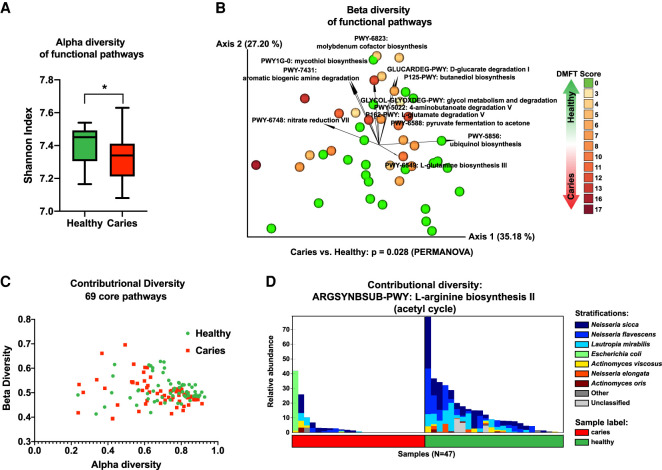
The taxa detected by MetaPhlAn2 abundance analysis in each sample are provided (Supplemental Figure S4A–E; Supplemental Table S3). Beta diversity, which illustrates differences in taxonomic diversity between samples/study groups, as well as correlation with caries, were examined using recently developed tools that are robust for investigating compositional data: DEICODE (Martino et al. 2019) and Songbird (Morton et al. 2019b). Both DEICODE and Songbird identified Prevotella as associated with caries and Rothia, Neisseria, and Haemophilus spp. as associated with health (Fig. 4A–D; Supplemental Table S4). According to Songbird, the taxa most correlated to disease was Human Herpesvirus 4 (Epstein-Barr virus [EBV]), which was detected in 10 subjects with caries and only one healthy subject (Supplemental Tables S3, S4). S. mutans, the classic caries pathogen, was the taxon with the second highest correlation to disease status (Fig. 4C), however S. mutans was only detected in seven subjects with caries and four healthy subjects. According to Songbird, both Saccharibacteria (formerly, TM7) species detected by MetaPhlAn2 were moderately associated with caries (Supplemental Table S4). Because of the compositional nature of sequencing data, log ratios are a preferable way to examine differences within these data sets (Morton et al. 2019b). Indeed, the log ratios of Prevotella to Rothia, Haemophilus, and Neisseria were significantly elevated in caries, indicating that the ratio of these taxa may have clinical significance and be a useful marker of disease (Fig. 4D). The functional pathways present in the oral microbiomes were examined in a similar manner (Fig. 5A–D; Supplemental Fig. S5A–D; Supplemental Tables S5, S6). Overall, there was a reduced diversity of functional pathways present in the caries-associated microbiomes, including the depletion of several pathways that were previously known to be health-promoting, such as biosynthesis of arginine (Nascimento et al. 2019), branched-chain amino acids (Santiago et al. 2012), and urea (Liu et al. 2012), and/or pathways dominated by taxa that were health-associated in this study (aerobic respiration by Neisseria) (Fig. 5A–D; Supplemental Fig. S5A–D). iRep (Brown et al. 2016) was used to calculate the replication rates of MAGs, but no difference in replication rates was detected between caries and health (Supplemental Fig. S6A,B). To examine the abundance of the specific genomes obtained by the assembly across the samples, sequencing reads were mapped back to the MAGs. This largely recapitulated the marker gene (MetaPhlAn2) taxonomic analysis, serving as a useful sanity check (Supplemental Fig. S6C).
Figure 4.
Significant taxonomic differences in the oral metagenome between healthy children and children with caries. (A) Beta diversity. Biplot generated using DEICODE (robust Aitchison PCA) (Martino et al. 2019). Data points represent individual subjects and are colored with a gradient to visualize DMFT score, indicating severity of dental caries. Feature loadings (i.e., taxa driving differences in ordination space) are illustrated by the vectors, which are labeled with the cognate species name. (B) Ranking of PCA Axis 2 taxonomic loadings. Qurro-produced bar chart illustrating the sorted ranks of the feature loadings of PCA Axis 2 from A, corresponding to the main PCA space separation between the healthy and caries groups. The indicated taxa are highlighted in the indicated color. (C) Differential rankings of taxa associated with disease status. Qurro-produced bar chart illustrating the sorted differential rankings of taxa associated with disease status determined by Songbird (Morton et al. 2019b). The indicated taxa are highlighted in the indicated color. (D) The log ratios of Prevotella spp. to Rothia, Haemophilus, and Neisseria spp. are significantly increased in caries. Bar chart illustrating the log2 ratios of Prevotella spp. to Rothia, Haemophilus, and Neisseria spp. across the healthy and caries sample groups. (**) Statistical significance based on a Welch's t-test (P = 0.001).
Figure 5.
Profiling of functional pathways illustrates differences between health- and caries-associated oral microbiota. (A) A greater diversity of functional pathways is present in the healthy group. Bar chart illustrating the alpha diversity (Shannon Index) of the functional pathways present in the healthy and caries groups, as determined by HUMAnN2 (Franzosa et al. 2018) analysis. (*) Statistical significance based upon a Kruskal–Wallis test (P = 0.0136). (B) Beta diversity of functional pathways. Biplot generated using DEICODE (robust Aitchison PCA) (Martino et al. 2019). Data points represent individual subjects and are colored with a gradient to visualize DMFT score, indicating severity of dental caries. Feature loadings (i.e., functional pathways driving differences in ordination space) are illustrated by the vectors, which are labeled with the cognate pathway name. (C) Contributional diversity of 69 core pathways. Scatter plot indicating alpha and beta diversities of 69 functional pathways that were found across all samples. (D) Contributional diversity of arginine biosynthesis in caries versus health. Stacked bar chart illustrating the relative abundance and contributional diversity of arginine biosynthesis across the samples.
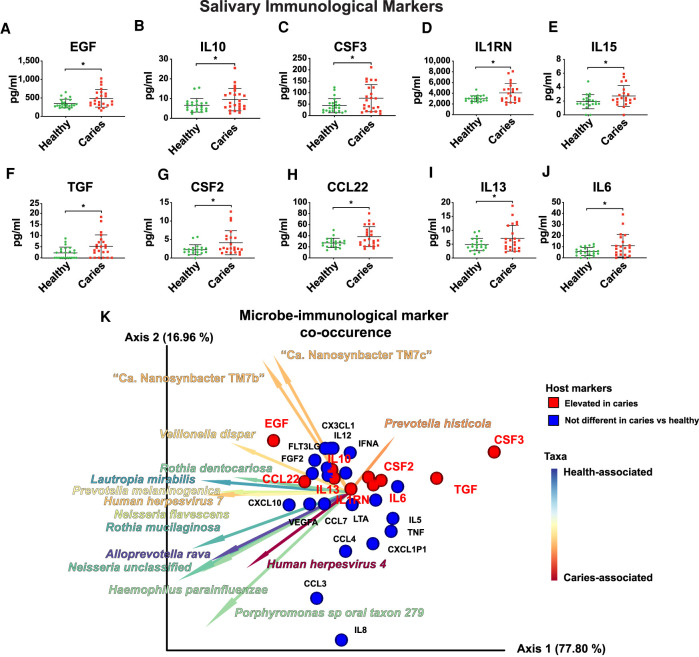
Ten host salivary immunological markers are more abundant in the saliva of children with caries than children with good dental health and co-occur with Prevotella histicola, Veillonella atypica, and TM7
Ten salivary immunological markers were found at significantly higher concentrations in the saliva of children with caries: epidermal growth factor (EGF), interleukin 10 (IL10), colony stimulating factor 3(CSF3), interleukin 1 receptor antagonist (IL1RN), colony stimulating factor 2 (CSF2), CCL22, interleukin 13 (IL13), interleukin 15 (IL15), and interleukin 6 (IL6) (Fig. 6A–J). MMvec (Morton et al. 2019a) is a recently developed tool that uses neural networks to address the statistical challenges confronting the inference of interactions across omics data sets. Here, MMvec was used to create microbe-metabolite vectors to examine co-occurrences between specific bacterial species and immunological markers (Supplemental Table S7). There was a noticeable similar trend in the directionality of many of the vectors representing taxa associated with caries (e.g., Prevotella histicola, Veillonella atypica, and the Saccharibacteria TM7b/TM7c) (Fig. 6K). These three vectors clearly indicated co-occurrence with several of the immunological markers that were elevated in caries, such as EGF and CSF2 (Fig. 6K). Additionally, IL13, CCL22, FGF2, IL7, CCL2, and FLT3LG formed a tight cluster in ordination space and had strong correlations with Rothia dentocariosa, Neisseria flavescens, Lautropia mirabilis, and Human herpesvirus 7 (Fig. 6K).
Figure 6.
Significant differences in the salivary immunological profile of healthy children and children with caries. (A–J) Scatter plots illustrating the 10 immunological markers: (A) EGF, (B) IL10, (C) CSF3, (D) IL1RN, (E) IL15, (F) TGF, (G) CSF2, (H) CCL22, (I) IL13, and (J) IL6, which were significantly different between healthy and caries subject groups. (*) P < 0.05, based on a Welch's t-test. (K) Microbe-immune marker co-occurrence. Biplot illustrating the co-occurrence of oral taxa with immune markers. The 31 detected immune markers are represented by spheres, and the bacterial taxa are represented by vectors. Red spheres indicate host markers that were elevated in caries, whereas blue spheres indicate host markers that were not significantly different between caries and health (based on the Welch's t-test described in A–J). Vectors are colored by Songbird ranks (Fig. 4C) indicating association with caries versus health.
Discussion
A major advantage of metagenomic sequencing is the ability to assemble MAGs, which allows the identification of novel taxa and analysis of pangenomes. Of the 527 MAGs reported in this study, 20% (98 MAGs) represented novel taxa that had no representative genome in public databases. Although many of these unknown taxa were likely observed previously by 16S sequencing, obtaining the cognate genomes is crucial to elucidate the ecology and possible pathogenesis of these species. The large number of genomes assembled by this study significantly increased the available pangenomic information for many oral species. These new genomes were particularly useful in the case of the Saccharibacteria clades G3 and G6, in which the number of available reference genomes was quite limited and estimates of genome completion are difficult owing to the absent “essential” genes in CPR bacteria (McLean et al. 2020). The large-scale differences observed in the metabolic pathways present among the Saccharibacteria clades suggests that these clades may have contrasting dependency requirements fulfilled by their host bacteria. For example, G3 and G6 Saccharibacteria may depend, in part, on a host bacterium for ATP production and/or pH homeostasis caused by a lack of an F1-F0 ATPase, whereas G1 Saccharibacteria may be independent in that regard because they still encode the enzyme complex. These major differences in functional capabilities support the hypothesis that loss of independence and acquisition of the various Saccharibacteria epibiont clades by the human host may have occurred in multiple, temporally separated events (McLean et al. 2020). Although this study and others (He et al. 2015) suggest that Saccharibacteria may be immunomodulatory, the role of this group in the ecology of dental caries is poorly understood, highlighting a significant need for more in-depth studies exploring the host-epibiont relationship of CPR at the metabolic level. In addition to the novel CPR taxa, novel taxa were also identified within more well-characterized clades including Prevotellaceae and Porphyromonadaceae. Several of these novel genomes, including unknown species-level genome bins of Peptostreptococcus, Solobacterium, and Lachnospiraceae were assembled and binned from a large number of subjects independently (8, 13, and 19 subjects, respectively), indicating that these unknown taxa may be widespread in the population, with roles in ecology, health, and disease waiting to be elucidated.
Examining the abundances of the taxa present in the caries- and health-associated microbiomes revealed that beta diversity of species-level taxonomy was significantly different between the caries and healthy groups. The importance of the canonical cariogenic species, S. mutans, in the development of dental caries has been the subject of recent debate (Banas and Drake 2018; Philip et al. 2018). Here, S. mutans, was associated with caries (second most correlated species-level taxa according to supervised methods), but was found in relatively low abundances and in only 11 of the 47 subjects. The fact that S. mutans is an exceptional biofilm former may contribute to this observation—it may be less likely to shed into the saliva, leading to its underrepresentation here, a hypothesis evidenced by several previous studies (Simón-Soro et al. 2013; Espinoza et al. 2018; Al-Hebshi et al. 2019). Alternatively, because this study examined deep dentin caries, where the tooth enamel has been dissolved and the underlying connective tissue is exposed, it is possible that new ecological niches were created, allowing for the growth of different species compared to earlier stages of the disease. On the other hand, the fact that S. mutans was strongly correlated with disease, despite having low abundance, supports the idea that, when present, S. mutans has a disproportionally large ability to influence disease and may be a keystone pathogen owing to its unique skill at generating insoluble glucans and resulting biofilms from sucrose (Bowen 2016; Banas and Drake 2018).
Supervised methods also identified Human herpesvirus 4 (Epstein-Barr virus [EBV]) as the taxon that was most highly associated with disease. Although EBV has a known association with periodontitis (Imai and Ogata 2020), only one previous study has examined EBV during dental caries, which also reported elevated detection of EBV in caries subjects compared to healthy controls (Yildirim et al. 2010). It is possible that increased inflammation during severe dental caries leads to higher levels of viral shedding. In support of this hypothesis, there were strong co-occurrence relationships between EBV and several of the host immunological markers; however, EBV did not co-occur with the same cytokines/chemokines as other caries-associated taxa, such as Provotella histicola. Furthermore, EBV did not appear to be associated with changes in the oral microbiome independent of caries, as EBV positive subjects were broadly distributed through ordination space in the PCA analysis. The role of viruses and fungi in dental caries is likely to be of significance; however, the amount of research examining these relationships has been very limited compared to that of bacteria, mainly because they are not detected by 16S sequencing.
The significant elevation in the ratio of Prevotella to Haemophilus, Neisseria, and Rothia observed here may represent a useful novel biomarker for caries, but wider studies are needed because the present study group was rather homogenous in terms of host ethnicity and geography. Although Prevotella spp. were elevated in disease, they were highly abundant in all the samples, and this correlation was not as evident as that of Rothia, Neisseria, and Haemophilus with health, indicating that the positive effects of these three taxa may be more important than the negative effects of Prevotella (e.g., Prevotella may just have higher relative abundance because the health-associated taxa have lower relative abundance). Various Rothia and Prevotella species have been previously associated with either health (Agnello et al. 2017; Gomez et al. 2017) or caries (Nadkarni et al. 2004; Tanner et al. 2011; Teng et al. 2015; Jiang et al. 2016; Al-Hebshi et al. 2019; Hurley et al. 2019), although many prior studies did not use analysis methods that are robust for compositional data, sampled other oral sites, and/or did not examine advanced dentin caries, specifically, as was done here. The genera Rothia, Neisseria, and Haemophilus were recently documented to be important mediators of cell–cell interactions within the early biofilm derived from healthy individuals (Palmer et al. 2017), are among the first colonizers of the oral cavity after birth (Sulyanto et al. 2019), and may play a crucial, yet currently unrecognized, role in maintaining a health-associated oral microbiome. This hypothesis is supported by the fact that the diversity of metabolic pathways present in the microbiomes was reduced in the caries group. Many of the pathways depleted during caries were dominated by taxa that were also reduced, particularly Neisseria. This included several pathways with particular relevance to dental caries, including the BCAA (which produces ammonia) and arginine biosynthesis pathways, which serve to buffer the environment and prevent enamel demineralization via the production of alkaline molecules (Santiago et al. 2012; Nascimento et al. 2019).
A number of previous studies have illustrated that penetration of the dental plaque infection into the dentin is associated with elevation of a number of cytokines and host signaling molecules (Hahn et al. 2000; Sloan et al. 2000; Artese et al. 2002; McLachlan et al. 2004; Adachi et al. 2007; Kokkas et al. 2007; Horst et al. 2011). Several of these reports were supported by this study, in which 10 immunological factors were observed at significantly elevated concentrations in the caries group compared to the healthy group. These molecules have an array of functions and are likely to themselves influence the microbiota of the oral cavity (Chang et al. 2019). Similar to previous reports, we observed that higher IL10 and IL6 were associated with caries (McLachlan et al. 2004; Horst et al. 2011). However, unlike two previous studies examining pulpitis (McLachlan et al. 2004; Kokkas et al. 2007), TNF was not elevated in the caries group in this study. EGF was one of the markers most significantly elevated in the caries group and has been previously documented to be incorporated into dentin and released on orthodontic force (Derringer and Linden 2007). Microbe-host immunological marker co-occurrences have been characterized in periodontitis (Zhou et al. 2017; Arias-Bujanda et al. 2018; Lundmark et al. 2019), but this is the first study examining these co-occurrences in dental caries. Although the co-occurrences of various caries-associated microbes and host molecules presented an obvious chicken or egg dilemma (and it is likely that this cross-talk is bidirectional), it also provided an atlas of microbe-host metabolite interactions that are the most likely to be involved in caries pathogenesis and that deserve follow-up analysis.
Overall, this high-resolution survey of the oral microbiome and host immunological markers provides several important leads for future research. Because the role of CPR bacteria in dental caries is not well-understood, elucidating the basic lifestyle and metabolic requirements of the different Saccharibacteria clades will fill a significant knowledge gap in our understanding of oral microbial ecology and its relationship to pathogenesis. Confirmation that the ratio of Prevotella to Rothia, Neisseria, or Haemophilus represents a useful biomarker for caries-associated dysbiosis would afford a new diagnostic tool that is independent of S. mutans, which has historically been the target of such assays. Exploration into the mechanism behind the protective effect of the health-associated species and pathways will give further insights into how ecology affects caries pathogenesis and may lead to the development of novel therapeutics in the form of probiotics and/or targeted antimicrobials. Finally, investigation of the relationship between the caries-associated immunological molecules and specific taxa will improve the understanding of cross-talk between the oral microbiota and the host immune system and its function in disease.
Methods
Ethics statement
Child participants and parents understood the nature of the study, and parents/guardians provided informed consent before the commencement of the study. The Ethics Committees of the UCLA School of Dentistry, Los Angeles, California, and the J. Craig Venter Institute, La Jolla, California, approved the study design as well as the procedure for obtaining informed consent (IRB reference numbers: 13-001075 and 2016-226). All experiments were performed in accordance with the approved guidelines.
Study design
Detailed descriptions of the study design, study groups, oral examination, and saliva collection are provided in Supplemental Methods. Briefly, subjects were included in the study if the subject was 3 yr old or older, in good general health according to a medical history and clinical judgment of the clinical investigator, and had at least 12 teeth. Subjects were excluded from the study if they had generalized rampant dental caries, chronic systemic disease, or medical conditions that would influence the ability to participate in the proposed study. Health status was classified after a comprehensive oral examination, and subjects were dichotomized into two groups: caries free (dmft/DMFT = 0) and caries active (subjects with two or more active dentin lesions). Two milliliters of unstimulated and 2 mL of stimulated saliva were collected from subjects by drooling and/or spitting directly into a 50-mL conical tube. Then saliva samples were processed by centrifugation at 6000g for 5 min at 4°C, and the supernatants were transferred to cryotubes. The samples were immediately frozen in liquid nitrogen and stored at –80°C until analysis. Two milliliters of stimulated saliva was collected immediately following collection of unstimulated saliva.
Salivary immunological biomarker analysis
Frozen unstimulated saliva samples were thawed and processed through high-speed ultracentrifugation to precipitate cells and mucin for extraction of proteins. Host immunological marker profiles were determined by a Luminex Human Magnetic Assay using the Human Cytokine/Chemokine Panel (performed by Westcoast Biosciences). A total of 38 analytes were measured, the specific immune biomarkers that were studied in saliva samples included epidermal growth factor (EGF), fibroblast growth factor 2 (FGF2), C-C motif chemokine ligand 11 (CCL11, formerly, eotaxin), transforming growth factor alpha (TGFA), colony stimulating factor 3 (CSF3, formerly, granulocyte colony stimulating factor), colony stimulating factor 2 (CSF2, formerly, granulocyte-macrophage colony stimulating factor), fms related receptor tyrosine kinase 3 ligand (FLT3LG), vascular endothelial growth factor A (VEGFA), C-X3-C motif chemokine ligand 1 (CX3CL1, formerly, fractalkine), C-X-C motif chemokine ligand 1 pseudogene 1 (CXCL1P1, formerly, growth-regulated oncogene), C-C motif chemokine ligand 7 (CCL7, formerly, monocyte chemotactic protein 3), C-C motif chemokine ligand 22 (CCL22, formerly, macrophage derived chemokine), C-X-C motif chemokine ligand 8 (CXCL8, formerly, interleukin 8), C-X-C motif chemokine ligand 10 (CXCL10, formerly, IP-10), C-C motif chemokine ligand 2 (CCL2, formerly, monocyte chemoattractant protein-1), C-C motif chemokine ligand 3 (CCL3, formerly, macrophage inflammatory protein-1 alpha), C-C motif chemokine ligand 4 (CCL4, formerly, macrophage inflammatory protein-1 beta), interferon alpha 2 (IFNA2), interferon gamma (IFNG), interleukin 1 alpha (IL1A), interleukin 1 beta (IL1B), interleukin 1 receptor antagonist (IL1RN), interleukin 2 (IL2), interleukin 3 (IL3), interleukin 4 (IL4), interleukin 5 (IL5), interleukin 6 (IL6), interleukin 7 (IL7), interleukin 9 (IL9), interleukin 10 (IL10), interleukin 12B (IL12B), interleukin 12 (p70, IL12), interleukin 13 (IL13), interleukin 15 (IL15), interleukin 17 (IL17), CD40 ligand (CD40LG), tumor necrosis factor (TNF), and lymphotoxin alpha (LTA, formerly, tumor necrosis factor beta). Quantities of each host marker were compared between healthy and caries groups. In the cases of CCL11, CD40LG, IL17, IL9, IL2, IL3, and IL4, the majority of samples contained levels of the respective molecule below the limit of detection for the assay. Therefore, these salivary immunological markers were not analyzed subsequently. After removal of outliers using the ROUT method with a Q = 1%, a Welch's t-test was used to determine significantly differentially abundant immunological markers.
DNA extraction and sequencing
The DNA extraction and subsequent sequencing of these samples was reported in Aleti et al. (2019). Briefly, frozen stimulated saliva samples were thawed on ice. DNA was extracted and purified from the supernatant by using QIAmp Microbiome (Qiagen) and DNA Clean & Concentrator (Zymo Research) kit procedures in which host nucleic acid depletion step was skipped to maximize bacterial DNA recovery. Libraries were prepared using Illumina Nextera XT DNA library preparation kit (Illumina) (150-bp paired end reads) according to the manufacturer's instructions. Sequencing was carried out at the J. Craig Venter Institute (JCVI) Joint Technology Center (JTC) using an Illumina NextSeq 500 platform. DNA sample concentrations were normalized before sequencing. For 45 of the 47 samples, sequencing depth was 5–31 million reads per sample. Two samples, SC40 (caries) and SC33 (healthy) were sequenced ultradeep, to 366 and 390 million reads, respectively. The number of reads for each sample is listed in Supplemental Table S1. The raw sequencing data are available at the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA478018 with Sequence Read Archive (SRA) accession number SRP151559, as described in Aleti et al. (2019).
Sequencing analysis
A full description of the bioinformatics pipeline used in this study is provided in the Supplemental Methods and is also displayed in Supplemental Figure S1. Sequencing read quality control was performed by KneadData v0.5.4 (available at https://github.com/biobakery/kneaddata). De novo assembly of the metagenomes was performed using metaSPAdes (Nurk et al. 2017), and a separate assembly was performed for each sample. The resulting assemblies were binned using MetaWRAP (Uritskiy et al. 2018). fastANI (Jain et al. 2018) was used to dereplicate highly similar bins (≥95% ANI), likely representing the same species across different samples. To taxonomically identify bins, Mash v2.1 (Ondov et al. 2016) was used to compare each bin to the entire RefSeq database using a cutoff ≥95% ANI. This approach yielded 90 known species-level genome bins (kSGBs), representing 399 MAGs with ≥95% ANI to a RefSeq genome (based on Mash), and 60 unknown SGBs (uSGBs), representing 128 MAGs (Fig. 5), with no genome in RefSeq with an ANI ≥95% (Fig. 4). Here, a strategy for classifying uSGBs into genus-level genome bins (GGBs), which have an 85%–95% ANI to a GenBank genome, and family-level genome bins (FGBs), which have no match ≥85% ANI to a GenBank genome, was used, similar to the method described in Pasolli et al. (2019). The predicted family for each MAG was first inferred using the CheckM (Parks et al. 2015), Kraken (Wood and Salzberg 2014), and classify_bins tools from within the MetaWRAP pipeline. Next, because there are publicly available and, in many cases, described genomes in GenBank that do not appear in the RefSeq database used by Mash, each uSGB was compared against all GenBank genomes in its predicted family using fastANI. This process reassigned 18 uSGBs (rSGBs), representing 31 MAGs, to kSGBs, because they had ≥95% ANI match in GenBank (Supplemental Table S2). For the remaining “true” uSGBs, 20 uSGBs, representing 48 MAGs, that had 85%–95% ANI match to a GenBank genome were termed genus-level genome bins (GGBs), because the genus can be assigned with a fair amount of confidence although the species appears to be not previously described. The final 22 bins, representing 49 MAGs, had no matching reference in GenBank with an ANI ≥85%. These were termed family-level genome bins (FGBs), because the family or higher-level taxa can be inferred but the MAGs likely represent novel genera. When uSGBs contained multiple MAGs, the MAG with the best quality score according to the formula [completion – (2× contamination)] was used to find the best hit. Anvi'o (Eren et al. 2015) and PhyloPhlAn2 (Pasolli et al. 2019) were used to phylogenetically place uSGBs within predicted taxonomic groups. Individual assembled genomes were annotated with Anvi'o (COG Functions) and eggnog-mapper v2 (Huerta-Cepas et al. 2017, 2019). Anvi'o was used to perform pangenomics analysis (Delmont and Eren 2018). iRep was used (Brown et al. 2016) to estimate which taxa identified in the metagenomics analysis were alive and metabolically active and to compare these data between (Martino et al. 2019) health- and caries-associated microbiomes. Taxonomic abundance analysis based upon the assembled genomes was performed using BWA-MEM (Li et al. 2009; Li 2014), DEICODE (Martino et al. 2019), and QIIME2 (Bolyen et al. 2019). Read-based taxonomy was performed using MetaPhlAn2 v2.7.5 (Truong et al. 2015). QIIME2 (Bolyen et al. 2019) was used to calculate alpha diversity, whereas DEICODE was used to calculate beta diversity with feature loadings. The resulting PCA ordination was visualized with EMPeror (Vázquez-Baeza et al. 2013), and the feature loadings were visualized with Qurro (Fedarko et al. 2020). Songbird (Morton et al. 2019b) was used to rank species in regard to their association with disease, and the ranks were visualized using Qurro. HUMAnN2 (Franzosa et al. 2018) was used to provide abundance information regarding the functional pathways present in the metagenomes. Co-occurrences of species and immunological markers was determined using mmvec (Morton et al. 2019a) and visualized with EMPeror.
Data access
The genome sequences generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA624185.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health, National Institute of Dental and Craniofacial Research (NIH/NIDCR) grants F32-DE026947 (J.L.B.), K99-DE029228 (J.L.B.), and R00-DE024543 (A.E.). We thank Jeffrey S. McLean, R. Alexander Richter, Semar Petrus, Josh Espinoza, Drishti Kaul, Marcus Fedarko, Clarisse Marotz, and Cameron Martino for very helpful discussions.
Author contributions: Conception and design, J.L.B. and A.E.; sample collection and processing, M.D., R.A., N.C.T., and A.E.; public data curation, J.L.B.; software, J.L.B. and J.T.M.; data analysis, J.L.B., J.T.M., and M.D.; data interpretation, J.L.B., J.T.M., M.D., R.A., N.C.T., R.K., and A.E.; writing—original draft, J.L.B.; writing—review and editing, J.L.B., J.T.M., M.D., N.C.T., R.K., and A.E. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.265645.120.
References
- Adachi T, Nakanishi T, Yumoto H, Hirao K, Takahashi K, Mukai K, Nakae H, Matsuo T. 2007. Caries-related bacteria and cytokines induce CXCL10 in dental pulp. J Dent Res 86: 1217–1222. 10.1177/154405910708601215 [DOI] [PubMed] [Google Scholar]
- Agnello M, Marques J, Cen L, Mittermuller B, Huang A, Chaichanasakul Tran N, Shi W, He X, Schroth RJ. 2017. Microbiome associated with severe caries in Canadian First Nations children. J Dent Res 96: 1378–1385. 10.1177/0022034517718819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleti G, Baker JL, Tang X, Alvarez R, Dinis M, Tran NC, Melnik AV, Zhong C, Ernst M, Dorrestein PC, et al. 2019. Identification of the bacterial biosynthetic gene clusters of the oral microbiome illuminates the unexplored social language of bacteria during health and disease. mBio 10: e00321–19 10.1128/mBio.00321-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hebshi NN, Baraniya D, Chen T, Hill J, Puri S, Tellez M, Hasan NA, Colwell RR, Ismail A. 2019. Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children. J Oral Microbiol 11: 1557986 10.1080/20002297.2018.1557986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. 2019. A new genomic blueprint of the human gut microbiota. Nature 568: 499–504. 10.1038/s41586-019-0965-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Bujanda N, Regueira-Iglesias A, Alonso-Sampedro M, González-Peteiro MM, Mira A, Balsa-Castro C, Tomás I. 2018. Cytokine thresholds in gingival crevicular fluid with potential diagnosis of chronic periodontitis differentiating by smoking status. Sci Rep 8: 18003 10.1038/s41598-018-35920-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artese L, Rubini C, Ferrero G, Fioroni M, Santinelli A, Piattelli A. 2002. Vascular endothelial growth factor (VEGF) expression in healthy and inflamed human dental pulps. J Endod 28: 20–23. 10.1097/00004770-200201000-00005 [DOI] [PubMed] [Google Scholar]
- Baker JL, Bor B, Agnello M, Shi W, He X. 2017. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol 25: 362–374. 10.1016/j.tim.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA, Drake DR. 2018. Are the mutans streptococci still considered relevant to understanding the microbial etiology of dental caries? BMC Oral Health 18: 129 10.1186/s12903-018-0595-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B, Collins AJ, Murugkar PP, Balasubramanian S, To TT, Hendrickson EL, Bedree JK, Bidlack FB, Johnston CD, Shi W, et al. 2020. Insights obtained by culturing Saccharibacteria with their bacterial hosts. J Dent Res 99: 685–694. 10.1177/0022034520905792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH. 2016. Dental caries—not just holes in teeth! A perspective. Mol Oral Microbiol 31: 228–233. 10.1111/omi.12132 [DOI] [PubMed] [Google Scholar]
- Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35: 725–731. 10.1038/nbt.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Olm MR, Thomas BC, Banfield JF. 2016. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol 34: 1256–1263. 10.1038/nbt.3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA. 2018. Getting to know “the known unknowns”: heterogeneity in the oral microbiome. Adv Dent Res 29: 66–70. 10.1177/0022034517735293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AM, Liu Q, Hajjar AM, Greer A, McLean JS, Darveau RP. 2019. Toll-like receptor-2 and -4 responses regulate neutrophil infiltration into the junctional epithelium and significantly contribute to the composition of the oral microbiota. J Periodontol 90: 1202–1212. 10.1002/JPER.18-0719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costalonga M, Herzberg MC. 2014. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett 162: 22–38. 10.1016/j.imlet.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross KL, Campbell JH, Balachandran M, Campbell AG, Cooper SJ, Griffen A, Heaton M, Joshi S, Klingeman D, Leys E, et al. 2019. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat Biotechnol 37: 1314–1321. 10.1038/s41587-019-0260-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, Eren AM. 2018. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 6: e4320 10.7717/peerj.4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derringer K, Linden R. 2007. Epidermal growth factor released in human dental pulp following orthodontic force. Eur J Orthod 29: 67–71. 10.1093/ejo/cjl059 [DOI] [PubMed] [Google Scholar]
- Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi'o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3: e1319 10.7717/peerj.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza JL, Harkins DM, Torralba M, Gomez A, Highlander SK, Jones MB, Leong P, Saffery R, Bockmann M, Kuelbs C, et al. 2018. Supragingival plaque microbiome ecology and functional potential in the context of health and disease. mBio 9: e01631-18 10.1128/mBio.01631-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedarko MW, Martino C, Morton JT, González A, Rahman G, Marotz CA, Minich JJ, Allen EE, Knight R. 2020. Visualizing ‘omic feature rankings and log-ratios using Qurro. NAR Genom Bioinform 2: lqaa023 10.1093/nargab/lqaa023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, et al. 2018. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15: 962–968. 10.1038/s41592-018-0176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. 2017. Microbiome datasets are compositional: and this is not optional. Front Microbiol 8: 2224 10.3389/fmicb.2017.02224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Espinoza JL, Harkins DM, Leong P, Saffery R, Bockmann M, Torralba M, Kuelbs C, Kodukula R, Inman J, et al. 2017. Host genetic control of the oral microbiome in health and disease. Cell Host Microbe 22: 269–278.e3. 10.1016/j.chom.2017.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. 2010. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol 48: 4121–4128. 10.1128/JCM.01232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. 2012. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7: e47722 10.1371/journal.pone.0047722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CL, Best AM, Tew JG. 2000. Cytokine induction by Streptococcus mutans and pulpal pathogenesis. Infect Immun 68: 6785–6789. 10.1128/IAI.68.12.6785-6789.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, et al. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci 112: 244–249. 10.1073/pnas.1419038112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, Knight R, Knights D. 2018. Evaluating the information content of shallow shotgun metagenomics. mSystems 3: e00069-18 10.1128/mSystems.00069-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Bunge J, Leslin C, Jeon S, Epstein SS. 2009. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J 3: 1365–1373. 10.1038/ismej.2009.89 [DOI] [PubMed] [Google Scholar]
- Horst OV, Horst JA, Samudrala R, Dale BA. 2011. Caries induced cytokine network in the odontoblast layer of human teeth. BMC Immunol 12: 9 10.1186/1471-2172-12-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34: 2115–2122. 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47: D309–D314. 10.1093/nar/gky1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, et al. 2016. A new view of the tree of life. Nat Microbiol 1: 16048 10.1038/nmicrobiol.2016.48 [DOI] [PubMed] [Google Scholar]
- Hurley E, Barrett MPJ, Kinirons M, Whelton H, Ryan CA, Stanton C, Harris HMB, O'Toole PW. 2019. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health 19: 13 10.1186/s12903-018-0693-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Ogata Y. 2020. How does Epstein–Barr virus contribute to chronic periodontitis? Int J Mol Sci 21: 1940 10.3390/ijms21061940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9: 5114 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhang J, Chen H. 2013. Pyrosequencing analysis of oral microbiota in children with severe early childhood dental caries. Curr Microbiol 67: 537–542. 10.1007/s00284-013-0393-7 [DOI] [PubMed] [Google Scholar]
- Jiang W, Ling Z, Lin X, Chen Y, Zhang J, Yu J, Xiang C, Chen H. 2014. Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb Ecol 67: 962–969. 10.1007/s00248-014-0372-y [DOI] [PubMed] [Google Scholar]
- Jiang S, Gao X, Jin L, Lo EC. 2016. Salivary microbiome diversity in caries-free and caries-affected children. Int J Mol Sci 17: 1978 10.3390/ijms17121978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovel J, Patterson J, Wang W, Hotte N, O'Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, et al. 2016. Characterization of the Gut microbiome using 16S or shotgun metagenomics. Front Microbiol 7: 459 10.3389/fmicb.2016.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, et al. 2018. Best practices for analysing microbiomes. Nat Rev Microbiol 16: 410–422. 10.1038/s41579-018-0029-9 [DOI] [PubMed] [Google Scholar]
- Kokkas AB, Goulas A, Varsamidis K, Mirtsou V, Tziafas D. 2007. Irreversible but not reversible pulpitis is associated with up-regulation of tumour necrosis factor-α gene expression in human pulp. Int Endod J 40: 198–203. 10.1111/j.1365-2591.2007.01215.x [DOI] [PubMed] [Google Scholar]
- Li H. 2014. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30: 2843–2851. 10.1093/bioinformatics/btu356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Nascimento M, Burne RA. 2012. Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int J Oral Sci 4: 135–140. 10.1038/ijos.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark A, Hu YOO, Huss M, Johannsen G, Andersson AF, Yucel-Lindberg T. 2019. Identification of salivary microbiota and its association with host inflammatory mediators in periodontitis. Front Cell Infect Microbiol 9: 216 10.3389/fcimb.2019.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino C, Morton JT, Marotz CA, Thompson LR, Tripathi A, Knight R, Zengler K. 2019. A novel sparse compositional technique reveals microbial perturbations. mSystems 4: e00016-19 10.1128/mSystems.00016-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan JL, Sloan AJ, Smith AJ, Landini G, Cooper PR. 2004. S100 and cytokine expression in caries. Infect Immun 72: 4102–4108. 10.1128/IAI.72.7.4102-4108.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Bor B, Kerns KA, Liu Q, To TT, Solden L, Hendrickson EL, Wrighton K, Shi W, He X. 2020. Acquisition and adaptation of ultra-small parasitic reduced genome bacteria to mammalian hosts. Cell Rep 32: 107939 10.1016/j.celrep.2020.107939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyle J, Dommisch H, Groeger S, Giacaman RA, Costalonga M, Herzberg M. 2017. The innate host response in caries and periodontitis. J Clin Periodontol 44: 1215–1225. 10.1111/jcpe.12781 [DOI] [PubMed] [Google Scholar]
- Mira A. 2018. Oral microbiome studies: potential diagnostic and therapeutic implications. Adv Dent Res 29: 71–77. 10.1177/0022034517737024 [DOI] [PubMed] [Google Scholar]
- Morton JT, Sanders J, Quinn RA, McDonald D, Gonzalez A, Vazquez-Baeza Y, Navas-Molina JA, Song SJ, Metcalf JL, Hyde ER, et al. 2017. Balance trees reveal microbial niche differentiation. mSystems 2: e00162–16 10.1128/mSystems.00162-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JT, Aksenov AA, Nothias LF, Foulds JR, Quinn RA, Badri MH, Swenson TL, Van Goethem MW, Northen TR, Vazquez-Baeza Y, et al. 2019a. Learning representations of microbe-metabolite interactions. Nat Methods 16: 1306–1314. 10.1038/s41592-019-0616-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R. 2019b. Establishing microbial composition measurement standards with reference frames. Nat Commun 10: 2719 10.1038/s41467-019-10656-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni MA, Caldon CE, Chhour KL, Fisher IP, Martin FE, Jacques NA, Hunter N. 2004. Carious dentine provides a habitat for a complex array of novel Prevotella-like bacteria. J Clin Microbiol 42: 5238–5244. 10.1128/JCM.42.11.5238-5244.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento MM, Alvarez AJ, Huang X, Hanway S, Perry S, Luce A, Richards VP, Burne RA. 2019. Arginine metabolism in supragingival oral biofilms as a potential predictor of caries risk. JDR Clin Trans Res 4: 262–270. 10.1177/2380084419834234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. 2019. New insights from uncultivated genomes of the global human gut microbiome. Nature 568: 505–510. 10.1038/s41586-019-1058-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27: 824–834. 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using minHash. Genome Biol 17: 132 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RJ Jr, Shah N, Valm A, Paster B, Dewhirst F, Inui T, Cisar JO. 2017. Interbacterial adhesion networks within early oral biofilms of single human hosts. Appl Environ Microbiol 83: e00407-17 10.1128/AEM.00407-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. Checkm: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, Beghini F, Manghi P, Tett A, Ghensi P, et al. 2019. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176: 649–662.e20. 10.1016/j.cell.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip N, Suneja B, Walsh L. 2018. Beyond Streptococcus mutans: clinical implications of the evolving dental caries aetiological paradigms and its associated microbiome. Br Dent J 224: 219–225. 10.1038/sj.bdj.2018.81 [DOI] [PubMed] [Google Scholar]
- Pinto AJ, Raskin L. 2012. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7: e43093 10.1371/journal.pone.0043093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts NB, Zero DT, Marsh PD, Ekstrand K, Weintraub JA, Ramos-Gomez F, Tagami J, Twetman S, Tsakos G, Ismail A. 2017. Dental caries. Nat Rev Dis Primers 3: 17030 10.1038/nrdp.2017.30 [DOI] [PubMed] [Google Scholar]
- Santiago B, MacGilvray M, Faustoferri RC, Quivey RG Jr. 2012. The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J Bacteriol 194: 2010–2019. 10.1128/JB.06737-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaiber A, Eren AM. 2019. Composite metagenome-assembled genomes reduce the quality of public genome repositories. mBio 10: e00725-19 10.1128/mBio.00725-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Soro A, Belda-Ferre P, Cabrera-Rubio R, Alcaraz LD, Mira A. 2013. A tissue-dependent hypothesis of dental caries. Caries Res 47: 591–600. 10.1159/000351663 [DOI] [PubMed] [Google Scholar]
- Sloan AJ, Perry H, Matthews JB, Smith AJ. 2000. Transforming growth factor-β isoform expression in mature human healthy and carious molar teeth. Histochem J 32: 247–252. 10.1023/A:1004007202404 [DOI] [PubMed] [Google Scholar]
- Sulyanto RM, Thompson ZA, Beall CJ, Leys EJ, Griffen AL. 2019. The predominant oral microbiota is acquired early in an organized pattern. Sci Rep 9: 10550 10.1038/s41598-019-46923-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner AC, Kent RL Jr, Holgerson PL, Hughes CV, Loo CY, Kanasi E, Chalmers NI, Johansson I. 2011. Microbiota of severe early childhood caries before and after therapy. J Dent Res 90: 1298–1305. 10.1177/0022034511421201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Yang F, Huang S, Bo C, Xu ZZ, Amir A, Knight R, Ling J, Xu J. 2015. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe 18: 296–306. 10.1016/j.chom.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. Metaphlan2 for enhanced metagenomic taxonomic profiling. Nat Methods 12: 902–903. 10.1038/nmeth.3589 [DOI] [PubMed] [Google Scholar]
- Uritskiy GV, DiRuggiero J, Taylor J. 2018. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6: 158 10.1186/s40168-018-0541-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. 2013. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2: 16 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15: R46 10.1186/gb-2014-15-3-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim S, Yildiz E, Kubar A. 2010. TaqMan real-time quantification of Epstein-Barr virus in severe early childhood caries. Eur J Dent 4: 28–33. 10.1055/s-0039-1697805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Lei J, Cole J, Sun Y. 2015. Reconstructing 16S rRNA genes in metagenomic data. Bioinformatics 31: i35–i43. 10.1093/bioinformatics/btv231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yao Y, Jiao K, Zhang J, Zheng X, Wu F, Hu X, Li J, Yu Z, Zhang G, et al. 2017. Relationship between gingival crevicular fluid microbiota and cytokine profile in periodontal host homeostasis. Front Microbiol 8: 2144 10.3389/fmicb.2017.02144 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.