Abstract
Long noncoding RNAs (lncRNAs) have emerged as diverse functional regulators involved in mammalian development; however, large-scale functional investigation of lncRNAs in mammalian spermatogenesis in vivo is lacking. Here, we delineated the global lncRNA expression landscape in mouse spermatogenesis and identified 968 germ cell signature lncRNAs. By combining bioinformatics and functional screening, we identified three functional lncRNAs (Gm4665, 1700027A15Rik, and 1700052I22Rik) that directly influence spermatogenesis in vivo. Knocking down Gm4665 hampered the development of round spermatids into elongating spermatids and disrupted key spermatogenic gene expression. Mechanistically, lncRNA Gm4665 localized in the nucleus of round spermatids and occupied the genomic regulatory region of important spermatogenic genes including Ip6k1 and Akap3. These findings provide a valuable resource and framework for future functional analysis of lncRNAs in spermatogenesis and their potential roles in other biological processes.
Spermatogenesis is a complex process involving a series of tightly regulated developmental programs, including mitosis, meiosis, and spermiogenesis (Griswold 2016). Each of these dynamic processes is driven by multiple gene products that undergo strict regulation in a testis- or germ cell–specific manner (Luk et al. 2014). It is well established that spermatogenesis follows well-defined steps during germ cell development at various time points, and as per this special pattern, distinct germ cell types can be isolated from the testis at different ages after birth (Gan et al. 2013). Many studies have been carried out based on the above theory and identified some key cellular and molecular regulators in spermatogenesis (da Cruz et al. 2016; Lesch et al. 2016; Naro et al. 2017). However, given the highly complex gene expression program underlying spermatogenesis, broadening the molecular basis and further functionally annotating them will facilitate a comprehensive understanding of the cellular and molecular regulation in spermatogenesis.
The testis has the highest transcriptional diversity among mammalian organs, which is particularly pronounced in the abundance of noncoding transcripts, especially long noncoding RNAs (lncRNAs) (Soumillon et al. 2013; Necsulea et al. 2014). LncRNAs are a class of RNA transcripts longer than 200 nucleotides in length with little protein-coding potential that have been revealed as a significant regulator of gene expression networks (Rinn et al. 2007; Quinn and Chang 2016). During cell differentiation and organ development, these molecules modulate nuclear architecture and transcription in the nucleus and cytoplasmic mRNA stability, translation, and posttranslational modifications (Rinn and Chang 2012; Batista and Chang 2013; Fatica and Bozzoni 2014; Kopp and Mendell 2018; Mattioli et al. 2019; Yin et al. 2020). Many lncRNAs have been identified in the mammalian testis, including in humans (Bao et al. 2013; Sun et al. 2013; Chalmel et al. 2014; Hammoud et al. 2014; Liang et al. 2014; Lin et al. 2016; Jan et al. 2017; Rolland et al. 2019; Gamble et al. 2020; Tan et al. 2020). Further in-depth genomic examination of spermatogenesis reveals individual features of lnRNAs, including germ cell specificity, longer exon length, or high representation of antisense (AS) lncRNAs, which have shown a potential relevance of lncRNAs for germ cell development (Chalmel et al. 2014; Rolland et al. 2019; Trovero et al. 2020). To date, several functionally characterized cases demonstrated that lncRNAs were essential for spermatogenesis in Caenorhabditis elegans (Wei et al. 2019), Drosophila (Wen et al. 2016), Danio rerio (Hosono et al. 2017), and Mus musculus (Wichman et al. 2017; Li et al. 2019). Despite these promising findings, the systematic functional screening of lncRNAs in spermatogenesis of mammalian models remains largely unexplored.
Here, we perform bulk RNA-seq to delineate the global lncRNA expression landscape in mouse spermatogenesis and then combine an in vivo functional screening approach with single-cell transcriptomics and chromatin genomics analysis to unveil the regulatory function of lncRNA during male germ cell development at the tissue and organismal levels.
Results
Global lncRNA expression dynamics during mouse spermatogenesis
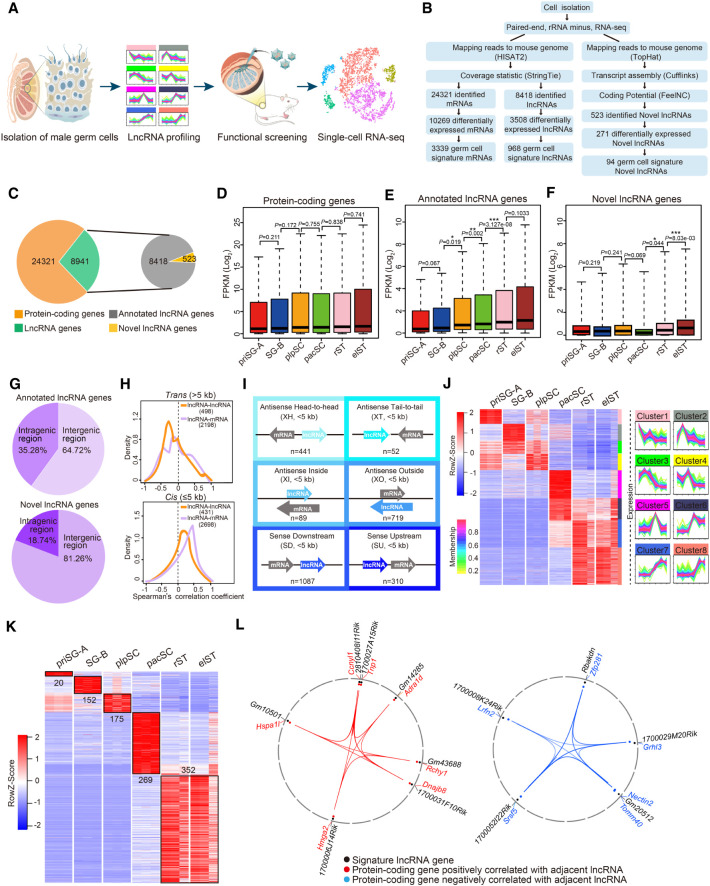
To define the lncRNA landscape during spermatogenesis, we performed bulk RNA-seq on six purified germ cell types that emerge successively during germ cell development: primitive type A spermatogonia (priSG-A), type B spermatogonia (SG-B), preleptotene spermatocytes (plpSC), pachytene spermatocytes (pacSC), round spermatids (rST), and elongating spermatids (elST) (Fig. 1A; Supplemental Fig. S1). By using unique whole-genome alignment (mouse genome [mm10]), a total of 24,321 protein-coding genes and 8941 lncRNA genes, including 8418 annotated and 523 novel lncRNAs, were detected as fragments per kilobase of exon per million reads mapped (FPKM) > 0.1 in at least three samples (Fig. 1B,C; Supplemental Table S2). According to bioinformatics analysis, both the annotated and novel lncRNAs largely had little protein-coding potential (Supplemental Fig. S2A). Additionally, both the transcript length and the number of exons per transcript of lncRNAs were less than that of protein-coding genes (Supplemental Fig. S2B,C). On the other hand, we found that annotated lncRNAs showed lower average expression levels than protein-coding genes and novel lncRNA genes (Supplemental Fig. S2D,E). Next, we observed that the average expression level of protein-coding genes (FPKM > 1) in each cell type along the spermatogenesis process was comparable between cell types, whereas the average expression levels of annotated and novel lncRNAs (FPKM > 1) underwent a gradual increase (Fig. 1D–F).
Figure 1.
Global lncRNA expression dynamics during mouse spermatogenesis. (A) Schematic workflow of dissecting lncRNA dynamics and identifying functional lncRNAs during mouse spermatogenesis. (B) Pipeline for transcriptome analysis of six distinct germ cell types. (C) The number of protein-coding and lncRNA genes expressed during spermatogenesis (left) and the proportion of lncRNA genes that were annotated versus novel (right). (D–F) Box plots showing differential expression characteristics of protein-coding (D), annotated (E), and novel (F) lncRNA genes in each germ cell type: priSG-A, SG-B, plpSC, pacSC, rST, and elST. P-values were calculated using Student's t-test. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. (G) Percentage of intergenic and intragenic lncRNA genes. (Top) Annotated lncRNA genes; (bottom) novel lncRNA genes. (H) Correlation of lncRNAs with neighboring genes in trans (top: pairwise with a distance >5 kb) or in cis (bottom: pairwise with a distance ≤5 kb). (I) Six locus biotypes of lncRNAs according to their genomic locations relative to the neighboring protein-coding genes: antisense lncRNA/mRNA gene pairs in the head-to-head position were designated divergent (XH); antisense lncRNA/mRNA gene pairs in the tail-to-tail position were designated convergent (XT); the gene body of an antisense lncRNA can be located within a protein-coding gene (XI) or can completely encompass a protein-coding gene (XO); the lncRNAs transcribed in the same direction can be located downstream (SD) or upstream (SU) of the neighboring coding genes. (J) Heat map showing dynamic expression pattern of lncRNAs using WGCNA (left) and STEM (right). (K) Heat map displaying relative expression level of germ cell developmental signature lncRNAs during spermatogenesis. (L) The positive correlation (left) and negative correlation (right) of germ cell signature lncRNAs with their neighboring protein-coding genes.
To classify the expressed lncRNA genes, we first determined their patterns of genomic distribution relative to protein-coding loci (Zhou et al. 2019). Among the 8418 annotated lncRNA genes, 64.72% were intergenic and 35.28% were intragenic, whereas for the 523 novel lncRNA genes, 81.26% were intergenic and 18.74% were intragenic (Fig. 1G). Pairwise Pearson's correlation analysis revealed that lncRNA showed a significantly higher expression correlation with the closest protein-coding gene compared to other distal genes (Supplemental Fig. S2F). Thus, to calculate pairwise expression correlations between lncRNA-mRNA gene pairs, we then focused on the coexpression patterns of lncRNA genes with their closest protein-coding genes. We defined two categories of lncRNA genes—those in trans correlations (the pairwise with a distance of >5 kb or located on different chromosomes) and those in cis correlations (the pairwise with a distance ≤5 kb)—and separately analyzed these categories of expression among lncRNA-lncRNA pairs and lncRNA-mRNA pairs. We found a higher proportion of positive correlations between cis pairs than trans pairs (Fig. 1H). A total of 2698 lncRNA/mRNA gene pairs involved in cis correlations were identified in at least one germ cell type during spermatogenesis and subsequently classified into six distinct biotypes; of these, “sense downstream” (SD, 40.3%) and “antisense outside” (XO, 26.6%) were the two largest (Fig. 1I; Supplemental Table S3). Also, we analyzed the evolutionary state of these lncRNAs using the reported lncRNA repertoires of 11 tetrapod species (Necsulea et al. 2014) and found that trans lncRNAs were expressed in more species than cis lncRNAs, suggesting trans lncRNAs were more conserved than cis lncRNAs (Supplemental Fig. S2G).
Functional prediction of lncRNAs by protein-coding genes via expression correlation analyses
We proceeded to identify lncRNAs of biological interest using weighted gene coexpression network analysis (WGCNA) (Casero et al. 2015). Of the 15 modules identified by WGCNA, we chose the top eight modules for further analysis (Supplemental Fig. S2H). For each module, we explored the dynamic expression pattern of lncRNA genes using Short Time-series Expression Miner (STEM) and annotated their functions through mRNAs in the same units by PANTHER. We identified 3508 differentially expressed lncRNAs and 271 differentially expressed novel lncRNAs whose expression was different between any two stages among six germ cell types (Fig. 1J; Supplemental Fig. S2I). Of note, we found that lncRNAs were preferentially expressed in clusters 5−6, with peak expression in pacSC, and in clusters 7−8, with peak expression in rST and elST (Fig. 1J).
Next, we identified 968 germ cell developmental signature lncRNAs that were expressed at FPKM > 10 in the corresponding germ cell types and 94 signature novel lncRNAs (Fig. 1K; Supplemental Fig. S2J). We further performed functional prediction of germ cell developmental signature lncRNAs based on their neighboring mRNAs. The priSG-A and SG-B signature lncRNAs were significantly enriched in the Gene Ontology (GO) terms of “stem cell proliferation” and “germ cell development,” whereas those in plpSC and pacSC were mainly enriched in “meiotic cell cycle” and “positive regulation of meiosis,” and those in rST and elST were significantly enriched in “acrosomal vesicle” and “sperm fibrous sheath” (Supplemental Fig. S2K). To further explain the association between germ cell signature lncRNAs and neighboring protein-coding genes, we built a noncoding to coding network using Circos (Krzywinski et al. 2009). We found that critical functional spermatogenic genes including Tnp1, Ccnyl1, Hmga2, and Hspa1l were located adjacent to the signature lncRNAs (1700027A15Rik, 2810408I11Rik, 1700006J14Rik, Gm10501), and their expression was positively correlated. In contrast, expression of other functional spermatogenic genes, such as Srsf5, Grhl3, and Nectin2, was negatively correlated with that of the signature lncRNAs (1700052I22Rik, 1700029M20Rik, Gm20512) (Fig. 1L). These results suggest that the germ cell developmental signature lncRNAs might play a regulatory role in spermatogenesis.
Characterization of lncRNAs with potential roles in spermatogenesis
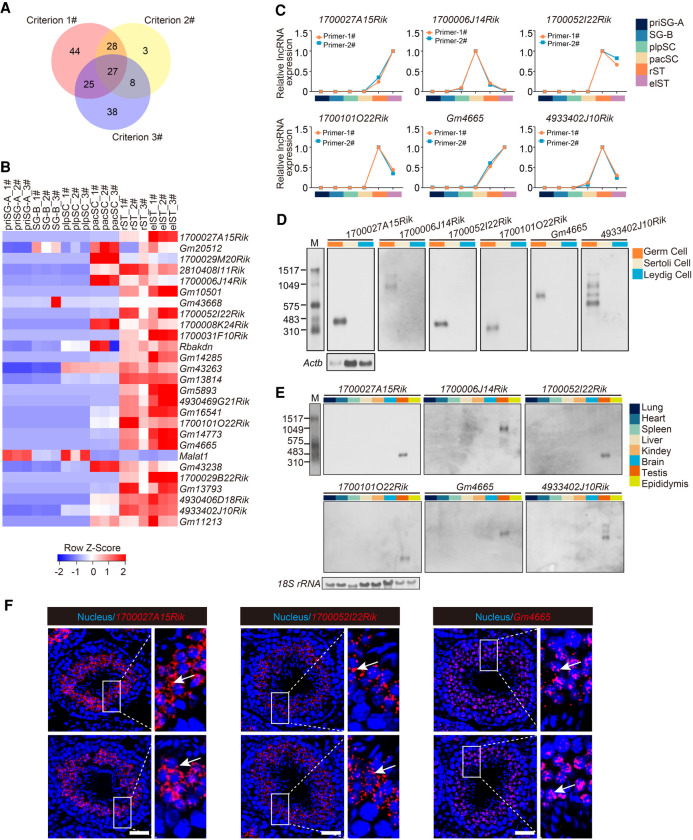
We next employed three criteria to systematically screen lncRNAs for potential functional roles in spermatogenesis: (1) their relation to spermatogenesis defined by WGCNA; (2) their differential expression between every two germ cell types (Max FPKM > 10 and fold change ≥5); and (3) having neighboring protein-coding genes involved in spermatogenesis. These screening criteria finally generated 27 candidate lncRNAs (Fig. 2A). Gene expression analysis showed that 22 of the 27 candidate lncRNAs were mainly detected in the late stage of spermatogenesis (Fig. 2B). To validate the observations from RNA-seq, we then measured the expression of candidate lncRNAs in each germ cell type by qRT-PCR. The results showed that the expression pattern of 20 of the 27 candidate lncRNAs (74%) was in accordance with the RNA-seq data set (Fig. 2C; Supplemental Fig. S3A). We next performed qRT-PCR with cDNA derived from germ cells, Sertoli cells, and Leydig cells to detect the expression pattern of the 20 candidate lncRNAs and observed that six candidate lncRNAs (1700027A15Rik, 1700006J14Rik, 1700052I22Rik, 1700101O22Rik, Gm4665, 4933402J10Rik) were germ cell–specific (Supplemental Fig. S3B,C). Among them, 1700027A15Rik and 1700101O22Rik have been previously identified in mouse testis but have yet to be functionally explored (Song et al. 2018; Trovero et al. 2020). We performed northern blots to validate the cell specificity of six candidate lncRNAs, and the results showed that they were specifically expressed in germ cells (Fig. 2D). We then used qRT-PCR and northern blots to detect the expression of the six lncRNAs across multiple tissues and found that all of them displayed a testis-specific pattern (Fig. 2E; Supplemental Fig. S3D). Additionally, we analyzed the transcriptome data generated by the Mouse ENCODE Project and found that all the six lncRNAs except 1700006J14Rik (not detected in ENCODE) were significantly testis-specific among the 30 mouse tissues (Supplemental Fig. S3E). The full-length transcript of the six lncRNAs was further identified by 5′ and 3′ rapid amplification of cDNA ends (RACE) (Supplemental Fig. S4A). Moreover, bioinformatics analysis indicated that the six lncRNAs had a very low coding potential (Supplemental Fig. S4B). We next performed an RNAscope assay to identify the subcellular location of three lncRNAs and found that 1700027A15Rik and 1700052I22Rik were mainly localized in the cytoplasm of spermatids at different stages of spermatogenesis, whereas Gm4665 was mainly localized in the nucleus of spermatids from stage I to stage VIII, and then localized in both nucleus and cytoplasm of spermatids from stage IX to stage XII. Further observation showed that these three lncRNAs underwent a significant increase of expression from stage VII to stage XII (step 7−12) (Fig. 2F; Supplemental Fig. S4C).
Figure 2.
Characterization of potential functional lncRNAs in mouse spermatogenesis. (A) Venn diagram showing the number of lncRNAs meeting the three screening criteria to determine potential functional lncRNAs. (B) Heat map showing relative expression level of the 27 candidate lncRNAs in each germ cell type. 1#, 2#, and 3# refer to three biological replicates per germ cell type. (C) Expression validation of six candidate lncRNAs in each germ cell type by qRT-PCR. Two independent pairs of primers were used for each candidate lncRNA (primer-1# and primer-2#); also see Supplemental Figure S3A. (D) Northern blotting analysis of six candidate lncRNAs in germ cells, Sertoli cells, and Leydig cells. Actb mRNA served as a loading control. (M) RNA molecular weight marker. (E) Northern blotting analysis of six candidate lncRNAs in multiple tissues. 18S rRNA served as a loading control. (M) RNA molecular weight marker. (F) Subcellular localization of candidate lncRNAs (red) from testis sections via RNAscope analysis. Nuclei were stained with DAPI (blue). Representative images with an enlargement of the white framed region are shown on the right. Scale bar, 50 μm.
Functional screening of candidate lncRNAs in spermatogenesis in vivo
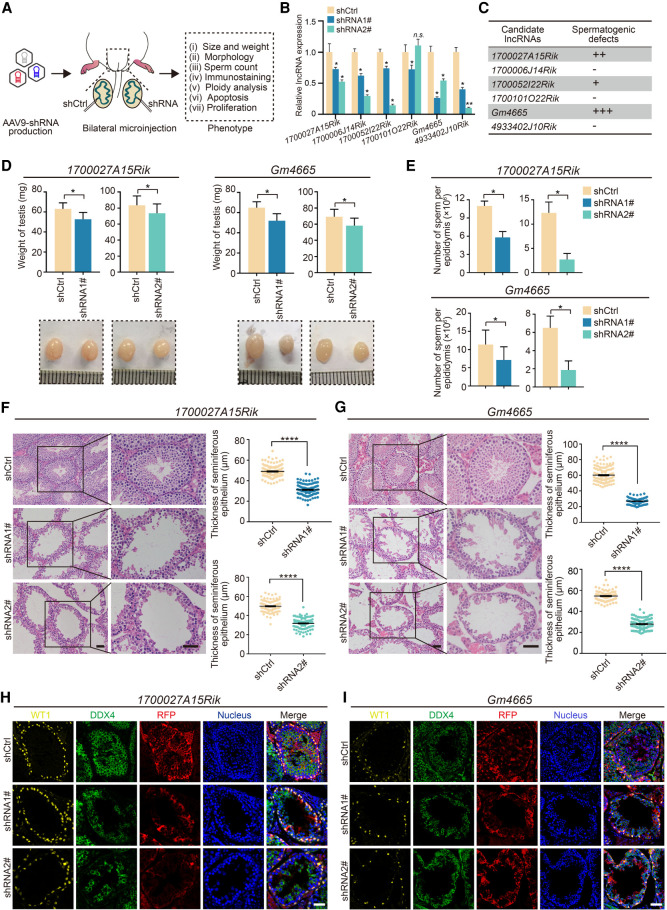
To determine the contribution of the six candidate lncRNAs to spermatogenesis in vivo, we performed shRNA-mediated knockdown of their expression in mouse testis by adeno-associated virus (AAV9). We selected two efficient shRNAs (shRNA1# and shRNA2#) for each lncRNA, with scrambled RNA as a negative control (shCtrl). To exclude differences between mice, we employed bilateral testicular microinjection, which meant that one testis was injected with AAV9-shCtrl-RFP and the contralateral testis was injected with AAV9-shRNA-RFP (Fig. 3A). As a positive control, we knocked down SYCP3 in mouse testis (Yuan et al. 2000), which led to severe developmental defects in spermatogenesis (Supplemental Fig. S5).
Figure 3.
Functional screening of candidate lncRNAs in mouse spermatogenesis in vivo. (A) Schematic overview of in vivo functional screening strategy. (B) qRT-PCR analysis of the efficacy of lncRNAs knockdown in vivo by AAV9-shRNA-RFP. Data represent the mean ± SEM for three biological replicates. (*) P < 0.05, (n.s.) P > 0.05 compared with shCtrl, Student's t-test. (C) Spermatogenesis phenotype profiles following lncRNAs knockdown. All results in this figure were collected 4 wk after microinjection with AAV9. (D) Testis morphology and weight from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice. (Top) The average weight of testes; (bottom) representative images of testes. Data represent mean ± SEM (n = 5). (*) P < 0.05 compared with shCtrl, Student's t-test. (E) Sperm counts in the cauda epididymis from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice. Data represent mean ± SEM (n = 5). (*) P < 0.05 compared with shCtrl, Student's t-test. (F,G) H&E staining of testis sections from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice. (Left) Representative staining images; (right) thickness of seminiferous epithelium. Data represent mean ± SEM of at least 100 seminiferous tubules from three mice. (****) P < 0.0001 compared with shCtrl, Student's t-test. Scale bar, 50 μm. (H,I) Immunostaining of testis sections from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice for WT1 (yellow), DDX4 (green), and RFP (red). Nuclei were stained with DAPI (blue). Scale bar, 50 μm.
Using this functional screening strategy, we observed successful reduction of the expression of the six lncRNAs by at least 40% in vivo relative to the shCtrl-injected testis (Fig. 3B). With repression of the candidate lncRNAs, we found that 1700027A15Rik- and Gm4665-depleted testes exhibited marked developmental defects in spermatogenesis as assessed by the loss of germ cells, whereas knockdown of 1700052I22Rik had only mild effects, and 1700006J14Rik, 1700101O22Rik, and 4933402J10Rik knockdown did not appear to affect spermatogenesis (Fig. 3C; Supplemental Fig. S6A,B). Therefore, we focused on 1700027A15Rik and Gm4665; the average weight of 1700027A15Rik- and Gm4665-depleted testes was ∼20% lower than that of shCtrl testes (Fig. 3D), and epididymal sperm number was reduced by 47.3%−78.1% in 1700027A15Rik-depleted testes and 50.5%–78.8% in Gm4665-depleted testes (Fig. 3E). We next examined tissue morphology via hematoxylin and eosin (H&E) staining and found that various stages of germ cells were presented in shCtrl testes, whereas both 1700027A15Rik- and Gm4665-depleted testes exhibited severe morphological defects. Compared to shCtrl testes, the seminiferous epithelium was up to ∼50% thinner, especially in Gm4665-depleted testes, and round spermatids and elongating spermatids were reduced significantly in the seminiferous tubules of both sets of testes (Fig. 3F,G). Immunolabeling for DDX4 also confirmed the shortage of germ cells in RFP-positive seminiferous tubules in 1700027A15Rik- and Gm4665-depleted testes (Fig. 3H,I).
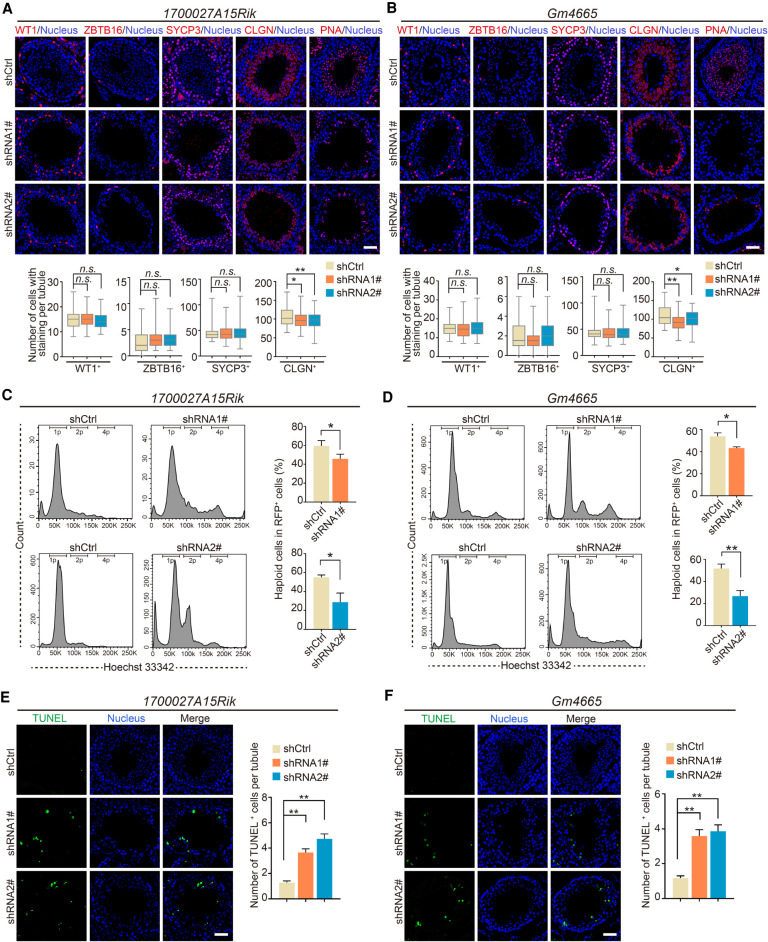
We further characterized these disrupted germ cell phenotypes via immunostaining for a set of germ cell–specific markers (ZBTB16, SYCP3, CLGN, and PNA). First, as Sertoli cells can be infected by AAV9-RFP, we conducted immunostaining for WT1, which showed that there were no significant differences in WT1+ cell number between shCtrl and 1700027A15Rik- or Gm4665-depleted testes; thus, AAV9-shRNA-mediated lncRNA knockdown had no effects on the maintenance of Sertoli cells (Fig. 4A,B). We also observed that ZBTB16+ and SYCP3+ germ cell numbers in 1700027A15Rik- and Gm4665-depleted testes were comparable to those of shCtrl testes, revealing that the mitotic and meiotic stages of spermatogenesis were not affected. In contrast, CLGN+ and PNA+ germ cell number was significantly reduced in the seminiferous tubules of 1700027A15Rik- and Gm4665-depleted testes, indicating that the late stage of spermatogenesis was damaged by the reduced levels of these specific lncRNAs (Fig. 4A,B). Accordingly, we went on to quantify the percentage of haploid cells in RFP+ germ cells using fluorescence-activated cell sorting (FACS). The proportion of haploid cells sorted from 1700027A15Rik- and Gm4665-depleted testes decreased by approximately 14%∼24% compared to those from shCtrl-testes (Fig. 4C,D).
Figure 4.
Knockdown of lncRNA candidates leading to defects in late spermatogenesis. (A,B) Immunostaining of testis sections from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice for marker proteins (red): WT1, ZBTB16, SYCP3, CLGN, PNA. Nuclei were stained with DAPI (blue). (Top) Representative images; (bottom) number of cells with staining per tubule. Data represent the mean ± SEM of at least 100 seminiferous tubules from three mice. (*) P < 0.05, (**) P < 0.01, (n.s.) P > 0.05 compared with shCtrl, Student's t-test. Scale bar, 50 μm. (C,D) Percentage of RFP+ haploid cells isolated from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) testes by FACS based on fluorescent labeling with Hoechst 33342. (Left) Representative flowcytometry histograms; (right) percentage of haploid cells. Data represent mean ± SEM (n = 4). (*) P < 0.05, (**) P < 0.01 compared with shCtrl, Student's t-test. (E,F) TUNEL assay of testis sections from the control (shCtrl), 1700027A15Rik and Gm4665 knockdown (shRNA1#, shRNA2#) mice. (Left) Representative images; (right) number of apoptotic cells per tubule. Data represent the mean ± SEM of at least 50 seminiferous tubules from three mice. (**) P < 0.01 compared with shCtrl, Student's t-test. Scale bar, 50 μm.
We then aimed to identify the main contributing factors explaining the decrease in the number of round spermatids and elongating spermatids by measuring levels of germ cell proliferation and apoptosis via immunolabeling for MKI67 and TUNEL assays, respectively. There were no significant differences in MKI67+ cell number between shCtrl and 1700027A15Rik- or Gm4665-depleted testes, indicating comparable levels of germ cell proliferation (Supplemental Fig. S6C,D). However, the TUNEL assay showed a significant increase in the number of apoptotic germ cells during late spermatogenesis in 1700027A15Rik- and Gm4665-depleted testes (Fig. 4E,F). Therefore, these results indicated that 1700027A15Rik and Gm4665 knockdown in part led to germ cell apoptosis in late spermatogenesis but had little effect on germ cell proliferation.
Single-cell RNA-seq analysis of regulatory role of Gm4665 in spermatid differentiation
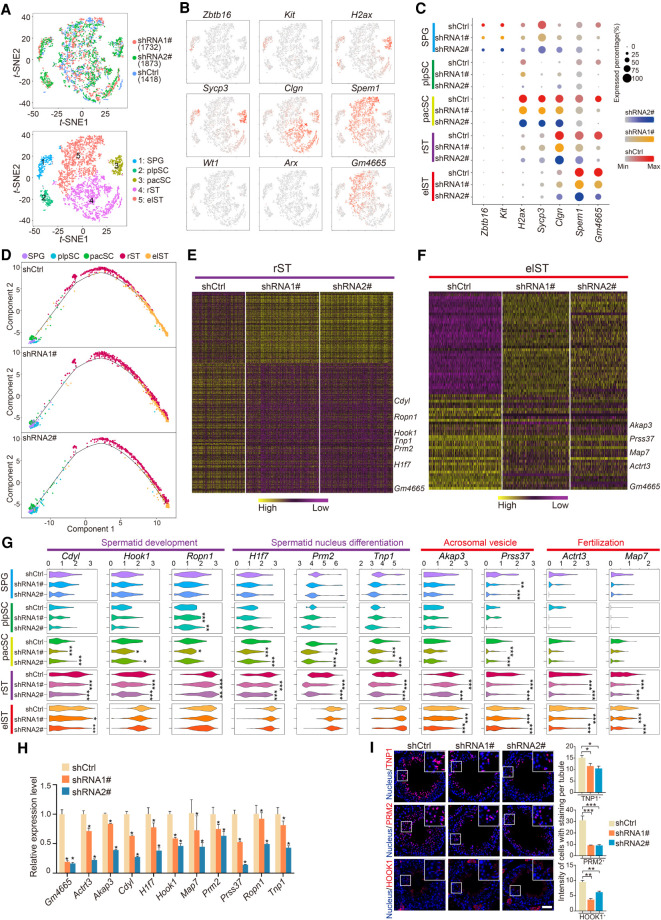
To understand the likely mechanisms behind the effects of candidate lncRNA knockdown, we applied single-cell RNA-seq (scRNA-seq) to decipher transcriptional alterations of affected germ cells induced by Gm4665 knockdown. We removed somatic cells and isolated RFP+ germ cells infected with AAV9-RFP from shCtrl or Gm4665-depleted testes by FACS, before performing scRNA-seq with the Chromium system (10x Genomics). Through stringent quality control filtering, 5023 single-cell (1418 shCtrl, 1732 Gm4665-shRNA1#, and 1873 Gm4665-shRNA2#) sequencing data sets with 3796 median genes and 21,092 mean confidently mapped reads per cell were retained for subsequent analysis (Supplemental Fig. S7A; Supplemental Table S1). We performed a t-distributed stochastic neighbor embedding (t-SNE) analysis to identify the cell types profiled: unsupervised clustering of the 5032 cells defined five distinct cell types (Fig. 5A), which expressed characteristic patterns of known marker genes, enabling us to identify them as spermatogonia (SPG), plpSC, pacSC, rST and elST (Fig. 5B). In addition, we calculated the cell numbers of each cluster and measured the expression of marker genes per cell of five distinct clusters (Supplemental Fig. S7B).
Figure 5.
High-resolution dissection of transcriptional alterations induced by Gm4665 knockdown in testis. (A) t-SNE plots of all 5023 RFP+ germ cells. (Top) Cells colored based on differential treatment with shCtrl, shRNA1#, or shRNA2#; (bottom) cells colored by the identified cell populations with the labeled numbers indicating the individual clusters. (B) Expression patterns of marker genes exhibited on t-SNE plots. Cluster 1 was SPG; cluster 2 was plpSC; cluster 3 was pacSC; cluster 4 was rST; and cluster 5 was elST; Wt1 and Arx, somatic cell marker genes, served as negative controls. Red indicates high expression and gray indicates low or no expression. (C) Expression of marker genes and Gm4665 in cell populations from differentially treated shCtrl, shRNA1#, and shRNA2#. (D) Pseudotime analysis of Gm4665 control and knockdown male germ cells during spermatogenesis. (E,F) Heat map showing differentially expressed genes in rST and elST populations upon Gm4665 knockdown according to defined clusters 4 and 5. The color key from yellow to purple indicates high to low gene expression levels. Differentially expressed genes were identified between two groups of cells using a Wilcoxon rank-sum test. (G) Violin plot displaying expression level of representative functional spermatogenic genes upon Gm4665 knockdown in indicated cell populations. P-values were calculated using a Wilcoxon rank-sum test. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 compared with shCtrl. (H) Expression validation of differentially expressed genes in rST and elST populations by qRT-PCR. Data represent the mean ± SEM for three biological replicates. (*) P < 0.05 compared with shCtrl, Student's t-test. (I) Immunostaining of testis sections from the shCtrl, shRNA1#, and shRNA2# mice for TNP1, PRM2, and HOOK1 (red). Nuclei were stained with DAPI (blue). (Left) Representative images; (right) intensity of cells with staining per tubule. Data represent the mean ± SEM of at least 50 seminiferous tubules from three mice. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 compared with shCtrl, Student's t-test. Scale bar, 50 μm.
We next measured the expression level of germ cell markers and Gm4665 for each group (shCtrl, shRNA1#, and shRNA2#) and noted that Gm4665 was knocked down significantly in the pacSC, rST, and elST populations (Fig. 5C; Supplemental Fig. S7C). We then performed a pseudotime analysis to separately visualize the trajectory of the developmental germ cells for each group (shCtrl, shRNA1#, and shRNA2#) and found that the development of rST into elST was delayed, indicating Gm4665 knockdown hampered the process of spermiogenesis (Fig. 5D). We also calculated the proportion of each cell type in each group and found that of elST populations was reduced (Supplemental Fig. S7D). We next separately analyzed the differentially expressed genes (DEGs) induced by Gm4665 knockdown in rST and elST populations. This demonstrated that Gm4665 knockdown resulted in increased expression of 190 genes and decreased expression of 328 genes in rST (Fig. 5E; Supplemental Table S4) and increased expression of 135 genes and decreased expression of 35 genes in elST (Fig. 5F; Supplemental Table S4). Some representative functional spermatogenic genes involved in “spermatid development,” such as Hook1, Ropn1, and “spermatid nucleus differentiation,” such as Tnp1, Prm2, and H1f7, were significantly down-regulated following Gm4665 knockdown in rST and pacSC populations, whereas some representative functional spermatogenic genes involved in “acrosomal vesicle,” such as Akap3, Prss37, and “fertilization,” such as Map7 and Actrt3, were significantly down-regulated in elST and rST populations, indicating the critical role of Gm4665 in spermiogenesis (Fig. 5G; Supplemental Fig. S7E). Additionally, the pseudotime analysis of the functional spermatogenic genes suggested that Gm4665 knockdown delayed their expression, starting in the round spermatids (Supplemental Fig. S7F). We also validated the decreased expression of these functional spermatogenic genes by qRT-PCR from RFP+ germ cells and immunostaining for TNP1, PRM2, and HOOK1 (Fig. 5H,I; Supplemental Fig. S7G). Taken together, single-cell transcriptional analysis of male germ cells indicated the functional requirement of Gm4665 for multiple aspects of spermatid differentiation during spermiogenesis.
Gm4665 regulates important spermatogenic genes through binding to their genomic regulatory region
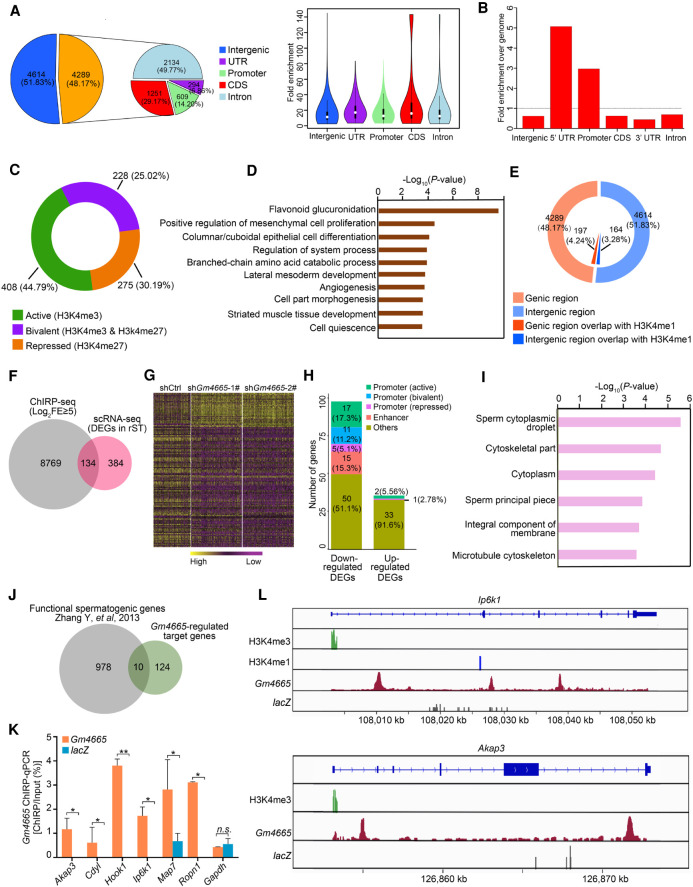
Because we observed Gm4665 nuclear localization in round spermatids, we hypothesized that this lncRNA might be regulating the expression of functional spermatogenic genes by binding to their genomic regulatory region and performed ChIRP-seq to test this. We first confirmed that the biotinylated probes targeting Gm4665 successfully enriched endogenous Gm4665, whereas the negative control probes targeting lacZ did not (Supplemental Fig. S8A). Analysis of the retrieved chromatin fragments by deep sequencing revealed Gm4665 binding sites distributed broadly across the genome, about half of which were intragenic. Among them, intronic binding sites were the most common (49.77%), but Gm4665 binding was also enriched significantly in the 5′ UTR and promoter region of genes relative to the whole genome, suggesting its potential role in regulating gene transcription (Fig. 6A,B; Supplemental Table S5). To further dissect the potential regulatory role of Gm4665 on these genes, we referenced the published epigenome data in spermatids and distinguished active promoters (marked with H3K4me3), repressed promoters (marked with H3K27me3), and bivalent promoters (coincidence of H3K4me3 with H3K27me3) (Hammoud et al. 2014). The results showed that Gm4665 was enriched in the active promoter region more frequently (44.79%) (Fig. 6C). These genes whose promoter was bound by Gm4665 participated in multiple developmental processes, including “flavonoid glucuronidation,” “epithelial cell differentiation.” “cell part morphogenesis,” and others (Fig. 6D). A small part of Gm4665 ChIRP peaks was also overlapped with a classic enhancer region (marked by H3K4me1), and these genes participated in multiple developmental processes, including “flavonoid glucuronidation,” “developmental growth,” and others (Fig. 6E; Supplemental Fig. S8B).
Figure 6.
ChIRP-seq analysis of chromatin interactions between Gm4665 and functional spermatogenic genes. (A) Pie chart showing the distribution of Gm4665-bound peaks in the genome. (B) Intragenic enrichment of ChIRP peaks over the genome. (C) The overlap of Gm4665 ChIRP-seq peaks within promoter regions with the known histone modification markers representative of promoter activities: H3K4me3, correlated with gene activation; H3K27me3, correlated with silencing; coincidence of H3K4me3 and H3K27me3, correlated with “bivalent.” (D) Enriched GO terms for the genes whose promoter was bound by Gm4665 and also marked with the above histone modification markers. (E) The overlap of Gm4665 ChIRP-seq peaks with the known histone modification marker representative of enhancer. (F) Venn diagram showing the number of target genes mediated by Gm4665. (FE) Fold enrichment. (G) Heat map showing 134 Gm4665-regulated target genes in rST populations induced by Gm4665 knockdown. The color key from yellow to purple indicates high to low gene expression levels. (H) The distribution of Gm4665 binding in the regulatory region of these down-regulated and up-regulated genes with Gm4665 knockdown. (I) Enriched GO terms for the 134 Gm4665-regulated target genes. (J) Ten functional spermatogenic genes identified by SpermatogenesisOnline 2.0 database. (K) ChIRP-qPCR validation of chromatin interaction of Gm4665 with six down-regulated functional spermatogenic gene loci. Gapdh gene locus was used as negative control. Data represent the mean ± SEM. (*) P < 0.05, (**) P < 0.01, (n.s.) P > 0.05, Student's t-test. (L) Graphical representation of Gm4665 ChIRP-seq peaks as well as the histone modification ChIP-seq peaks among Ip6k1 and Akap3 gene loci.
To further determine the target genes regulated by Gm4665, we compared the Gm4665 genomic occupancy data with our list of Gm4665-depletion-induced DEGs: this identified 134 candidate target genes that were differentially expressed with Gm4665 knockdown and also were bound by Gm4665 (Fig. 6F,G; Supplemental Table S6). Further analysis of the genomic distribution of Gm4665 and histone modifications on these 134 DEGs indicated that the percentage of Gm4665 binding with promoter (active or bivalent) or enhancer in the down-regulated genes was higher than that of up-regulated genes with Gm4665 knockdown, suggesting that Gm4665 mainly binds to the promoter or enhancer region to help to active these genes (Fig. 6H). A GO enrichment analysis showed that the candidate target genes were significantly associated with “sperm cytoplasmic droplet” and “sperm principal piece” pathways, which further supported the functional requirement of Gm4665 for expression of these important genes in spermiogenesis (Fig. 6I). In addition, we analyzed the overlap of Gm4665 genomic occupancy with genes preferentially expressed in early or late stages of spermatogenesis. The percentage of the overlapped genes in late stages including rST and elST (32.9%) was higher than that of early stages (25.9%), suggesting that Gm4665 might contribute to the preferentially expressed genes in rST and elST to a greater extent (Supplemental Fig. S8C).
To better understand the critical roles of the above Gm4665-regulated target genes in spermatogenesis, we subjected these candidate target genes to the SpermatogenesisOnline 2.0 database, which integrates the functional spermatogenic genes reported in the literature (Zhang et al. 2013). Of the 134 candidates, 10 corresponded to defined functional spermatogenic genes; of these, six were down-regulated (Ip6k1, Akap3, Hook1, Ropn1, Cdyl, Map7) and four were up-regulated (Rasl2-9, Acrbp, Ggnbp1, Dydc1) (Fig. 6J). We next performed ChIRP-qPCR to determine the chromatin interaction of Gm4665 with the six down-regulated genes and found that Gm4665 was highly enriched in genomic regions of the six genes (Fig. 6K). Further binding site analysis showed that the high-confidence Gm4665 ChIRP-seq peaks mapped to the active promoter region and enhancer region of Ip6k1 gene loci and the active promoter region of Akap3 gene loci and were overlapped with intronic regions or the “open state” of Hook1, Ropn1, Map7, and Cdyl gene loci. (Fig. 6L; Supplemental Fig. S8D). As previously reported, disruption of these genes, such as Ip6k1, Akap3, and Hook1, leads to development defects of spermatid (Mendoza-Lujambio et al. 2002; Bhandari et al. 2008; Xu et al. 2020). Our in vivo Gm4665 knockdown in mouse testes exhibited similar morphological disturbances in spermiogenesis as these gene-deficient mice. Taken together, these results suggested the potential role of Gm4665 in mediating chromatin state or transcription of target genes.
Discussion
Previous studies have identified numerous lncRNAs in testis, including human, suggesting their involvement in spermatogenesis (Sarropoulos et al. 2019). However, identifying functionally relevant lncRNAs is difficult due to their diversity and redundancy (Lee et al. 2019). Therefore applying strict screening criteria and a step-wise selection process (germ cell signature lncRNA, “guilt by association” with protein-coding genes, evolutionary analyses, etc.) gives the greatest probability of a genuine “hit” (Guttman and Rinn 2012; Sauvageau et al. 2013). Additionally, experimental scrutiny is further required to elucidate regulatory roles of these functionally relevant lncRNAs in spermatogenesis. In this study, we identified male germline-associated lncRNAs that are potentially important for spermatogenesis in vivo, based on several computational and experimental data sets. Meanwhile, our validated approach provides a useful framework for functional characterization of lncRNAs in spermatogenesis, as well as a potential template for their investigation in other tissues/processes.
Functional annotation of lncRNAs has been technically challenging in animal models in vivo. In the study of spermatogenesis, the majority of studies employ knockout animal models to investigate the molecular pathways involved, but this approach may not be suitable to target all lncRNAs, specifically regarding some special lncRNA loci (Goyal et al. 2017). For example, deletion of an entire lncRNA locus could have multiple off-target effects such as the loss of overlapping DNA regulatory elements or other functional gene bodies (Bassett et al. 2014; Ransohoff et al. 2018). Therefore, local lncRNA knockdown may be another complementary approach to study the in vivo functions of lncRNAs in animal models (Dai et al. 2017). In this study, we developed a relatively simple but efficient functional screening strategy to identify functional lncRNAs in spermatogenesis in vivo. Briefly, we introduced shRNA-mediated knockdown by AAV9 and then determined the contribution of lncRNAs to spermatogenesis based on metrics including weight of whole testes, number of sperm, tubule morphology, and apoptosis. Using this model, we successfully identified three functionally important lncRNAs (1700027A15Rik, 1700052I22Rik, and Gm4665) in mouse spermatogenesis. In particular, knockdown of 1700027A15Rik and Gm4665 resulted in a sharp decrease in the number of round spermatids and elongating spermatids in the abnormal seminiferous tubules. Taking into account their high abundance in late stages of spermatogenesis, we hypothesized that 1700027A15Rik and Gm4665 regulated spermiogenesis involving the development of round spermatids into elongating spermatids. Taken together, our functional screening strategy quickly and efficiently identified functional lncRNAs in mouse spermatogenesis in vivo, although it could be further optimized to, for example, increase germ cell infection efficiency.
Germ cell morphology is well understood from the histological perspective, but precisely deciphering the transcriptional alteration of different types of germ cell is difficult to achieve due to the technical limitations for germ cell culture in vitro, specifically late steps of meiosis and spermiogenesis (Witt et al. 2019). Additionally, regulatory roles of lncRNAs identified in vitro may not always translate to biological significance in vivo, as in the case of lncRNA Malat1 (Zhang et al. 2012; Wei et al. 2019). Therefore, here, we combined our in vivo lncRNA knockdown approach with scRNA-seq to decipher the transcriptional alteration of distinct germ cell types at single-cell resolution. We obtained highly purified RFP+ germ cells with somatic cell removal for single-cell sequencing and finally identified five distinct cell clusters: SPG, plpSC, pacSC, elST, and rST. This allowed us to conduct gene expression analysis of distinct germ cells, as well as dissecting the involvement of lncRNAs in cellular state transitions and, finally, to reconstitute the developmental trajectory during spermatogenesis following lncRNA knockdown (Guo et al. 2018, 2020; Wang et al. 2018, 2019). Our data showed that some critical regulatory genes were significantly down-regulated in rST and elST populations in which Gm4665 was knocked down. According to the developmental trajectory analysis, Gm4665 knockdown delayed expression of cell type–specific markers starting in rST, which coordinated with abnormal morphological transformation from the histological perspective. Taken together, the combination of our in vivo functional screening model and scRNA-seq will provide a potential strategy to investigate critical regulatory roles of lncRNAs or protein-coding genes during spermatogenesis.
It is generally acknowledged that the regulatory roles of lncRNAs are closely related to their subcellular localization (Chen 2016). Several well-characterized nuclear lncRNAs, such as Hotair and NRAD1, modulate gene transcription through chromatin interactions (Rinn et al. 2007; Tsai et al. 2010). Here, we performed ChIRP-seq to explore Gm4665-regulated target genes during germ cell development. Similar to previous reports on chromatin interaction of nuclear lncRNAs, the occupancy sites of Gm4665 were also genome-wide (Vidovic et al. 2020). Although not all binding events were functional, enrichment of Gm4665 in the 5′ UTR and promoter regions suggested its role in regulating gene transcription. The further comparative analysis with the histone modifications ChIP-seq data indicated Gm4665 was more frequently enriched in the promoters correlated with gene activation. Consistently, a high proportion of genes were down-regulated when Gm4665 was depleted. Both of these observations evidenced that Gm4665 was more inclined to active these genes. The colocalization of Gm4665 with the corresponding histone modifications indicated its greater regulatory possibilities on these genes, as the critical role of these regions in mediating gene transcription. The binding of Gm4665 in these regions may affect the relevant histone modifications or mediate the transcriptional factors’ binding or activity, even mediate the chromatin interaction or structures, as many lncRNAs do (Fatica and Bozzoni 2014; Yao et al. 2019). Additional experiments will be required to evaluate these hypotheses of gene regulation for Gm4665 in spermiogenesis. Overcoming existing technical limitations and generating an accurate cellular model for rST in vitro would greatly assist in functionally identifying Gm4665-specific interaction partners and thereby determine the regulatory mechanism of Gm4665 during spermiogenesis.
In summary, we have constructed a transcriptomic map of lncRNAs expressed during murine spermatogenesis using bulk RNA-seq and identified three functional lncRNAs using our in vivo functional screening strategy; finally, we provided mechanistic insights into the actions of one lncRNA Gm4665 through integrated analysis of scRNA-seq and ChIRP-seq data. Our findings offer genomic-level insights into the lncRNA dynamics involved spermatogenesis, and provide a promising strategy for identifying functional lncRNAs involved in the development of male germ cells at the tissue and organismal levels, which may also benefit lncRNA studies in other biological processes.
Methods
RNA extraction and qRT-PCR
Total RNA was extracted from cell or tissue using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. cDNAs were synthesized from equal amounts of total RNA using a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific). qRT-PCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems). The primer sequences are provided in Supplemental Table S7.
De novo assembly of novel lncRNAs
To identify novel lncRNAs, we referenced an algorithm that predicts multi-exonic transcribed loci based on transcribed island and splice junction coordinates, and we used StringTie to assemble transcripts from genomic read alignments (Zhou et al. 2019). LncRNAs were identified by an “ab initio” assembly pipeline. The final lncRNA annotation used throughout was composed of long (>200 base pairs) spliced transcripts with no sense exonic overlap with protein-coding genes. FlExible Extraction of LncRNAs (FEELnc) was used to evaluate the protein-coding potential of transcripts (Wucher et al. 2017).
LncRNA classification and neighboring gene correlation analysis
LncRNAs were classified into six locus biotypes based on their transcription orientation and the positions of their transcription start and end sites with respect to nearby protein-coding loci (5-kb distance cutoff). The observed fraction of protein-coding genes that are located in a defined genomic distance from a neighboring lncRNA or coding gene was compared with simulated distributions by random positioning.
Microinjection of AAV9
Virus particles were introduced into the seminiferous tubules via the rete testis in 3-wk-old ICR male mice. Briefly, mice were anesthetized with medical-grade oxygen and 3% isoflurane, then both testes were exposed under a dissecting microscope (Leica). Fifteen microliters of virus particles containing 0.04% Trypan blue solution were injected into the seminiferous tubules using a glass microcapillary pipette. One testis was injected with AAV9-shCtrl-RFP; the contralateral testis was injected with AAV9-shRNA-RFP. The testes were then returned to the abdominal cavity, and the abdominal wall and skin were closed with sutures. The testes were harvested 4 wk after microinjection. All animal experimentation protocols were performed with the approval of the Peking Union Medical College Animal Care and Use Committees. The sequences of siRNAs are provided in Supplemental Table S8.
Fluorescence-activated cell sorting and scRNA-seq
Germ cells were isolated from the AAV9-injected testes as previously described (Zhu et al. 2014). In brief, the testes were decapsulated and incubated with 0.5 mg/mL collagenase type IA (Sigma-Aldrich) at 37°C for 15 min. The suspensions, including interstitial cells, were then removed. The seminiferous tubules were resuspended in 0.5 mg/mL collagenase type IV for 5 min to remove the peritubular myoid cells. The tubules were cut into small pieces and then incubated with 1 mg/mL hyaluronidase with gentle pipetting to separate Sertoli cells and germ cells. The cell suspensions were cultured at 32°C for 2–6 h before the nonadherent germ cells were collected. Flow cytometry was used to sort these RFP+ germ cells. All FACS experiments were performed on an Aria III sorter (BD Immunocytometry Systems), and FACS data were analyzed using FlowJo software (TreeStar). The libraries were prepared using a Single Cell 3′ Library Gel Bead kit V2 (10x Genomics, 120237) following the manufacturer's instructions. For scRNA-seq, the gene cell barcode matrix was filtered based on the number of genes detected per cell (any cells with fewer than 500 or more than 4000 genes per cell were filtered out) and the percentage of mitochondrial UMI counts (any cells with more than 10% of mitochondrial UMI counts were filtered out). We used Cell Ranger version 1.3.1 to process raw sequencing data and Cell Ranger R kit version 2.0.0 and Seurat suite version 2.0.0 for downstream analysis (Butler et al. 2018). For clustering, principal component analysis was performed for dimension reduction. The top 10 principal components (PCs) were selected using a permutation-based test implemented in Seurat and passed to t-SNE for clustering visualization. Trajectory analysis was constructed by Monocle by pseudotemporal ordering of single cells (Trapnell et al. 2014).
ChIRP sample preparation, sequencing, and data analysis
ChIRP-seq was performed as previously described by Chu et al. (2011). Briefly, tiling antisense oligo probes spanning the Gm4665 sequence were generated using the Stellaris FISH Probe Designer (https://www.biosearchtech.com/support/tools/design-software/stellaris-probe-designer), and probes were synthesized and labeled with biotin at the 3′ end (Invitrogen). Germ cells isolated from the testis of wild-type mouse were cross-linked, lysed, and sonicated and then incubated with probesets. Following hybridization of biotinylated probesets to Gm4665 transcript, Gm4665-bound chromatin was retrieved. The purified DNA fragments were sequenced using high-throughput sequencing (Illumina HiSeq X Ten). Details about analysis of ChIRP-seq and the referenced published epigenome data are in Supplemental Methods.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE145189, GSE145130, and GSE161511. The processed sequencing data have been submitted to the ReproGenomics Viewer (RGV; https://rgv.genouest.org).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2018YFC1003500, 2019YFA0802600, 2019YFA0801800), the National Key Basic Research Program of China (2015CB943001), Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (2017-I2M-3-009, 2016-I2M-1-001, 2017-I2M-1-015), the State Key Laboratory Special fund from the Ministry of Science (2060204), the National Natural Science Foundation of China (31970794, 32000586, 81672472, 81530007, 31725013, 31900072), the CAMS (2016GH310001, 2017-I2M-B&R-04), and Medical Epigenetics Research Center, CAMS (2017PT31035). We thank Dr. Lucy Robinson of Insight Editing London for assistance in editing this manuscript.
Author contributions: W.S., K.L., J.Y., and Y.M. conceived and designed this study; R.H., Q.Z., and D.Z. isolated the germ cells; K.L. and S.Z. performed most of molecular detection experiments; Y.L. and K.L. performed in vivo functional screening experiments; J.X. performed computational analysis; X.M., M.L., Y.L., Y.S., P.L., C.J., and Z.W. provided some experimental support; S.M., L.W., B.W., and F.W. provided critical suggestions on manuscript preparation; W.S., K.L., Y.M., and J.X. prepared the manuscript with help from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.264333.120.
References
- Bao J, Wu J, Schuster AS, Hennig GW, Yan W. 2013. Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline. Biol Reprod 89: 107 10.1095/biolreprod.113.113308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al. 2014. Considerations when investigating lncRNA function in vivo. eLife 3: e03058 10.7554/eLife.03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. 2013. Long noncoding RNAs: cellular address codes in development and disease. Cell 152: 1298–1307. 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R, Juluri KR, Resnick AC, Snyder SH. 2008. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc Natl Acad Sci 105: 2349–2353. 10.1073/pnas.0712227105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero D, Sandoval S, Seet CS, Scholes J, Zhu YH, Ha VL, Luong A, Parekh C, Crooks GM. 2015. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat Immunol 16: 1282–1291. 10.1038/ni.3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F, Lardenois A, Evrard B, Rolland AD, Sallou O, Dumargne MC, Coiffec I, Collin O, Primig M, Jégou B. 2014. High-resolution profiling of novel transcribed regions during rat spermatogenesis. Biol Reprod 91: 5 10.1095/biolreprod.114.118166 [DOI] [PubMed] [Google Scholar]
- Chen LL. 2016. Linking long noncoding RNA localization and function. Trends Biochem Sci 41: 761–772. 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678. 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz I, Rodríguez-Casuriaga R, Santiñaque FF, Farías J, Curti G, Capoano CA, Folle GA, Benavente R, Sotelo-Silveira JR, Geisinger A. 2016. Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic- to postmeiotic-related processes at pachytene stage. BMC Genomics 17: 294 10.1186/s12864-016-2618-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Voloshin O, Potapova S, Camerini-Otero RD. 2017. Meiotic knockdown and complementation reveals essential role of RAD51 in mouse spermatogenesis. Cell Rep 18: 1383–1394. 10.1016/j.celrep.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15: 7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Gamble J, Chick J, Seltzer K, Graber JH, Gygi S, Braun RE, Snyder EM. 2020. An expanded mouse testis transcriptome and mass spectrometry defines novel proteins. Reproduction 159: 15–26. 10.1530/REP-19-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H, Wen L, Liao S, Lin X, Ma T, Liu J, Song CX, Wang M, He C, Han C, et al. 2013. Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat Commun 4: 1995 10.1038/ncomms2995 [DOI] [PubMed] [Google Scholar]
- Goyal A, Myacheva K, Groß M, Klingenberg M, Duran Arqué B, Diederichs S. 2017. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res 45: e12 10.1093/nar/gkw883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD. 2016. Spermatogenesis: the commitment to meiosis. Physiol Rev 96: 1–17. 10.1152/physrev.00013.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie X, Guo Y, Takei Y, Yun J, Cai L, et al. 2018. The adult human testis transcriptional cell atlas. Cell Res 28: 1141–1157. 10.1038/s41422-018-0099-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JT, Nie XC, Giebler M, Mlcochova H, Wang YQ, Grow EJ, Connect D, Kim R, Tharmalingam M, Matilionyte G, et al. 2020. The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell 26: 262–276.e4. 10.1016/j.stem.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346. 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Low DH, Yi C, Carrell DT, Guccione E, Cairns BR. 2014. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15: 239–253. 10.1016/j.stem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, Pitchiaya S, Tien J, Escara-Wilke J, Poliakov A, et al. 2017. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell 171: 1559–1572.e20. 10.1016/j.cell.2017.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan SZ, Vormer TL, Jongejan A, Röling MD, Silber SJ, de Rooij DG, Hamer G, Repping S, van Pelt AMM. 2017. Unraveling transcriptome dynamics in human spermatogenesis. Development 144: 3659–3673. 10.1242/dev.152413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, Mendell JT. 2018. Functional classification and experimental dissection of long noncoding RNAs. Cell 172: 393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Zhang ZL, Krause HM. 2019. Long noncoding RNAs and repetitive elements: junk or intimate evolutionary partners? Trends Genet 35: 892–902. 10.1016/j.tig.2019.09.006 [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Silber SJ, McCarrey JR, Page DC. 2016. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet 48: 888–894. 10.1038/ng.3591 [DOI] [PubMed] [Google Scholar]
- Li K, Zhong S, Luo Y, Zou D, Li M, Li Y, Lu Y, Miao S, Wang L, Song W. 2019. A long noncoding RNA binding to QKI-5 regulates germ cell apoptosis via p38 MAPK signaling pathway. Cell Death Dis 10: 699 10.1038/s41419-019-1941-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Li W, Tian H, Hu T, Wang L, Lin Y, Li Y, Huang H, Sun F. 2014. Sequential expression of long noncoding RNA as mRNA gene expression in specific stages of mouse spermatogenesis. Sci Rep 4: 5966 10.1038/srep05966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Han M, Cheng L, Chen J, Zhang Z, Shen T, Wang M, Wen B, Ni T, Han C. 2016. Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells. RNA Biol 13: 1011–1024. 10.1080/15476286.2016.1218588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk AC, Chan WY, Rennert OM, Lee TL. 2014. Long noncoding RNAs in spermatogenesis: insights from recent high-throughput transcriptome studies. Reproduction 147: R131–R141. 10.1530/REP-13-0594 [DOI] [PubMed] [Google Scholar]
- Mattioli K, Volders PJ, Gerhardinger C, Lee JC, Maass PG, Melé M, Rinn JL. 2019. High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Res 29: 344–355. 10.1101/gr.242222.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Lujambio I, Burfeind P, Dixkens C, Meinhardt A, Hoyer-Fender S, Engel W, Neesen J. 2002. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum Mol Genet 11: 1647–1658. 10.1093/hmg/11.14.1647 [DOI] [PubMed] [Google Scholar]
- Naro C, Jolly A, Di Persio S, Bielli P, Setterblad N, Alberdi AJ, Vicini E, Geremia R, De la Grange P, Sette C. 2017. An orchestrated intron retention program in meiosis controls timely usage of transcripts during germ cell differentiation. Dev Cell 41: 82–93.e4. 10.1016/j.devcel.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. 2014. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505: 635–640. 10.1038/nature12943 [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. 2016. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17: 47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, Khavari PA. 2018. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19: 143–157. 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland AD, Evrard B, Darde TA, Le Béguec C, Le Bras Y, Bensalah K, Lavoué S, Jost B, Primig M, Dejucq-Rainsford N, et al. 2019. RNA profiling of human testicular cells identifies syntenic lncRNAs associated with spermatogenesis. Hum Reprod 34: 1278–1290. 10.1093/humrep/dez063 [DOI] [PubMed] [Google Scholar]
- Sarropoulos I, Marin R, Cardoso-Moreira M, Kaessmann H. 2019. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571: 510–514. 10.1038/s41586-019-1341-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2: e01749 10.7554/eLife.01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Kyi-Tha-Thu C, Takizawa T, Naing BT, Takizawa T. 2018. 1700108J01Rik and 1700101O22Rik are mouse testis-specific long non-coding RNAs. Histochem Cell Biol 149: 517–527. 10.1007/s00418-018-1642-4 [DOI] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3: 2179–2190. 10.1016/j.celrep.2013.05.031 [DOI] [PubMed] [Google Scholar]
- Sun J, Lin Y, Wu J. 2013. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS One 8: e75750 10.1371/journal.pone.0075750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Song HW, Wilkinson MF. 2020. Single-cell RNAseq analysis of testicular germ and somatic cell development during the perinatal period. Development 147: dev183251 10.1242/dev.183251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovero MF, Rodríguez-Casuriaga R, Romeo C, Santiñaque FF, François M, Folle GA, Benavente R, Sotelo-Silveira JR, Geisinger A. 2020. Revealing stage-specific expression patterns of long noncoding RNAs along mouse spermatogenesis. RNA Biol 17: 350–365. 10.1080/15476286.2019.1700332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic D, Huynh TT, Konda P, Dean C, Cruickshank BM, Sultan M, Coyle KM, Gujar S, Marcato P. 2020. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ 27: 363–378. 10.1038/s41418-019-0362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu X, Chang G, Chen Y, An G, Yan L, Gao S, Xu Y, Cui Y, Dong J, et al. 2018. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell 23: 599–614.e4. 10.1016/j.stem.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Wang ZP, Xu XJ, Li JL, Palmer C, Maric D, Dean J. 2019. Sertoli cell-only phenotype and scRNA-seq define PRAMEF12 as a factor essential for spermatogenesis in mice. Nat Commun 10: 5196 10.1038/s41467-019-13193-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Chen H, Dzakah EE, Yu B, Wang X, Fu T, Li J, Liu L, Fang S, Liu W, et al. 2019. Systematic evaluation of C. elegans lincRNAs with CRISPR knockout mutants. Genome Biol 20: 7 10.1186/s13059-018-1619-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen K, Yang L, Xiong T, Di C, Ma D, Wu M, Xue Z, Zhang X, Long L, Zhang W, et al. 2016. Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res 26: 1233–1244. 10.1101/gr.199547.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman L, Somasundaram S, Breindel C, Valerio DM, McCarrey JR, Hodges CA, Khalil AM. 2017. Dynamic expression of long noncoding RNAs reveals their potential roles in spermatogenesis and fertility. Biol Reprod 97: 313–323. 10.1093/biolre/iox084 [DOI] [PubMed] [Google Scholar]
- Witt E, Benjamin S, Svetec N, Zhao L. 2019. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. eLife 8: e47138 10.7554/eLife.47138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucher V, Legeai F, Hédan B, Rizk G, Lagoutte L, Leeb T, Jagannathan V, Cadieu E, David A, Lohi H, et al. 2017. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res 45: e57 10.1093/nar/gkw1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Yang L, Zhang L, Qi H. 2020. Lack of AKAP3 disrupts integrity of the subcellular structure and proteome of mouse sperm and causes male sterility. Development 147: dev181057 10.1242/dev.181057 [DOI] [PubMed] [Google Scholar]
- Yao RW, Wang Y, Chen LL. 2019. Cellular functions of long noncoding RNAs. Nat Cell Biol 21: 542–551. 10.1038/s41556-019-0311-8 [DOI] [PubMed] [Google Scholar]
- Yin Y, Lu JY, Zhang X, Shao W, Xu Y, Li P, Hong Y, Cui L, Shan G, Tian B, et al. 2020. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 580: 147–150. 10.1038/s41586-020-2105-3 [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C. 2000. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 5: 73–83. 10.1016/S1097-2765(00)80404-9 [DOI] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung GN, Bhattacharjee G, Xiao XK, Booth CJ, Wu J, Zhang CL, et al. 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2: 111–123. 10.1016/j.celrep.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhong L, Xu B, Yang Y, Ban R, Zhu J, Cooke HJ, Hao Q, Shi Q. 2013. SpermatogenesisOnline 1.0: a resource for spermatogenesis based on manual literature curation and genome-wide data mining. Nucleic Acids Res 41: D1055–D1062. 10.1093/nar/gks1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xu J, Zhang L, Liu S, Ma Y, Wen X, Hao J, Li Z, Ni Y, Li X, et al. 2019. Combined single-cell profiling of lncRNAs and functional screening reveals that H19 is pivotal for embryonic hematopoietic stem cell development. Cell Stem Cell 24: 285–298.e5. 10.1016/j.stem.2018.11.023 [DOI] [PubMed] [Google Scholar]
- Zhu W, Liu P, Yu L, Chen Q, Liu Z, Yan K, Lee WM, Cheng CY, Han D. 2014. p204-initiated innate antiviral response in mouse Leydig cells. Biol Reprod 91: 8 10.1095/biolreprod.114.119396 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.