Abstract
Conventional cancer cell lines and animal models have been mainstays of cancer research. More recently, human pluripotent stem cells (hPSCs) and hPSC-derived organoid technologies, together with genome engineering approaches, have provided a complementary platform to model cancer progression. Here, we review the application of these technologies in cancer modeling with respect to the cell-of-origin, cancer propagation, and metastasis. We further discuss the benefits and challenges accompanying the use of hPSC models for cancer research and discuss their broad applicability in drug discovery, biomarker identification, decoding molecular mechanisms, and the deconstruction of clonal and intra-tumoral heterogeneity. In summary, hPSC-derived organoids provide powerful models to recapitulate the pathogenic states in cancer and to perform drug discovery.
Keywords: Human pluripotent stem cells, Organoids, Cell-of-origin, Cancer propagation, Metastasis, Drug discovery
1. Introduction
Cancer is a highly complex disease and is the second leading cause of death worldwide. Over the past decades, the access to various human cancer cell lines and the use of animal models have greatly deepened our understanding of tumorigenesis and advanced the development of therapeutic targets. While human cancer cell lines are easy to handle and helpful for the evaluation of the efficacy of cytotoxic drugs, they tend to accumulate additional mutations over time, lack heterogeneity and poorly reflect early-stage cancer development (Mirabelli et al., 2019). Likewise, although animal models of cancer facilitate the study of basic cancer research, they cannot faithfully recapitulate the genetic alterations and heterogeneity in human cancers because of species-specific differences (Cheon and Orsulic, 2011). Patient-derived tumor xenografts (PDTXs) preserve cell–cell interactions and the tumor microenvironment, are better in representing tumor heterogeneity, and are more useful for preclinical drug screening applications. However, the generation of PDTX models is expensive and time-consuming, and engraftment efficiencies differ among distinct types of tumors (Murayama and Gotoh, 2019). Therefore, additional cancer modeling strategies need to be developed.
We have witnessed remarkable developments in pluripotent stem cell (PSC) technologies over the past few decades. The first human embryonic stem cell (hESC) lines were established in 1998 (Thomson et al., 1998), which marked a new era in the field of regenerative medicine. Several years later, the PSC field was revolutionized again by the generation of induced pluripotent stem cells (iPSCs) from mouse (Takahashi and Yamanaka, 2006) and human somatic cells (Takahashi et al., 2007). iPSCs have greatly advanced the establishment of new models of human disease, drug discovery, and regenerative medicine (Sharma et al., 2020). By introducing cocktails of small molecules and growth factors at defined concentrations and times, PSCs can be differentiated into various cell lineages that can recapitulate hallmarks of diseases. In recent years, PSC technologies, together with improved genome editing tools and differentiation protocols, including epigenetic programming, have opened up a new field of cancer modeling (Fig. 1).
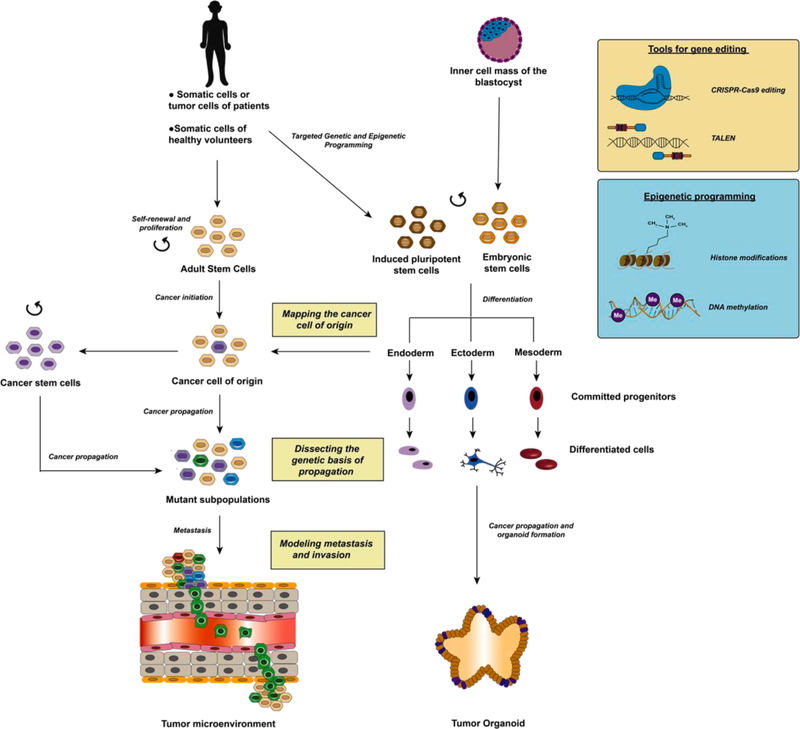
Fig. 1.
Cancer disease modeling using hPSC-derived organoids. Adult stem cells that undergo mutagenesis might give rise to the cancer cell-of-origin. Multistep accumulation of mutations then occurs to give rise to mutant subpopulations which escape cell senescence and acquire invasive properties to undergo metastasis. To model this progression, the somatic cells of cancer patients or healthy volunteers or the tumor cells of patients can be reprogrammed to form iPSCs. Additionally, ESCs can be obtained from the inner cell mass of the blastocyst. Some important tools for genetic engineering include CRISPR/Cas9 editing and TALEN. On the other hand, epigenetic programming can make use of methods that incorporate histone modifications or vary DNA methylation states including differential miRNA expression or expression of relevant transcription factors and genes. iPSCs and ESCs with self-renewal and proliferative capacity can be differentiated into three lineages to form committed progenitors, differentiated cells and subsequently organoids to mimic the process of cancer propagation.
There are three approaches to generate hPSC-based cancer models. Firstly, genetic alterations can be introduced to normal hPSCs by Transcription activator-like effector nucleases (TALENs) (Duan et al., 2015) or clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9) technologies (Terada et al., 2019). hPSC-derived cells, containing cancer-associated mutations, can be used to capture the earliest molecular events in cancer initiation, to trace cancer origins and to recapitulate cancer progression. Secondly, somatic cells can be reprogrammed from patients with a cancer predisposition syndrome. iPSC lines have been derived from patients with Li-Fraumeni syndrome (LFS) (TP53 mutation) (Lee et al., 2015), Fanconi anemia (FA) (FANCA and FANCC mutations) (Muller et al., 2012), Familial platelet disorder with associated myeloid malignancy (FPD/AML) (RUNX1 mutation) (Antony-Debre et al., 2015) and breast cancer predisposition (BRCA1 mutation) (Soyombo et al., 2013). Lastly, cancer cells can be directly reprogrammed into iPSCs. A variety of human cancer cell types have been successfully reprogrammed to iPSCs, including myeloid malignancies (Carette et al., 2010; Gandre-Babbe et al., 2013; Kotini et al., 2015), gastrointestinal cancers (Miyoshi et al., 2010), glioblastoma (Stricker et al., 2013), hepatocellular cancer (Kim et al., 2017), small cell lung cancers (SCLCs) (Chen et al., 2019) and pancreatic cancers (Kim et al., 2013; Khoshchehreh et al., 2019). A concerted approach including the expression or inhibition of relevant transcription factors and genes and/or epigenetic programming has been widely used in order to achieve this. For instance, the role of p53 as an inhibitor of reprogramming was previously studied in several liver cancer cell lines of varied genetic backgrounds and it was found that the p53-null Hep3B cell line was the most effective in attaining pluripotency upon the retroviral induction of pluripotency related genes (Kim et al., 2017). Moreover, several epigenetic reprogramming methodologies were adopted in a recent study to reprogram pancreatic ductal adenocarcinoma (PDAC) cell cultures including the induction of Yamanaka factors (Oct4, Sox2, Klf4 and c-Myc, termed OSKM), the expression of Oct4 and miR-302, and the use of episomal vectors (Khoshchehreh et al., 2019). After reprogramming, the established cancer iPSC lines can then be used to track different stages of cancer progression. Moreover, since patient-derived iPSCs and their differentiated tissues share patient-specific genetic traits, these tissues are able to phenocopy cancer progression after re-differentiation along the relevant lineage and can be potentially used for personalized therapy.
Recently developed techniques to construct three-dimensional (3D) spheroids and organoids have enabled the establishment of physiologically relevant cancer models and facilitated cancer drug discovery (Drost and Clevers, 2018; Zanoni et al., 2019). Tumor spheroids are self-assembled mixed cell aggregates cultured in an interactive 3D microenvironment. They have advantages over the conventional two-dimensional (2D) cell culture system in cancer modeling and drug screening. However, the widespread application of spheroids in high-throughput drug screening has been hampered by large variations in morphologies and sizes (Sant and Johnston, 2017). Organoids, derived from PSCs and adult stem cells (ASCs), contain multiple cell types and can self-organize to recapitulate the structures of in vivo organs (Clevers, 2016). Several hPSC-derived organoids have been successfully established, including brain (Lancaster et al., 2013), colon (Crespo et al., 2017; Munera et al., 2017), stomach (McCracken et al., 2014), kidney (Low et al., 2019), liver (Wu et al., 2019), lung (Miller et al., 2019; Chen et al., 2017), pancreas (Hohwieler et al., 2017) and small intestine (Holloway, et al., 2020) organoids. Tumor organoids can be generated directly from the tumor tissues of patients suffering from carcinomas (Fan et al., 2019). This allows for the recapitulation of the cancer genome changes of specific patients, paving the way for personalized cancer therapy. As tumor-derived organoids do not involve a complex differentiation scheme, it is easy to culture and maintain them in vitro. However, these tumor organoids do not accurately reflect the cancer microenvironment consisting of stromal cells and immune cells. Moreover, specific tumor organoids are only representative of certain cancer types, therefore making the establishment of large-scale biobanks of different organoids a necessity. Cancer-associated genetic alterations can be introduced into either ASC- or hPSC-derived organoids by CRISPR/Cas9-mediated gene editing. This method enables the generation of cancer-associated mutations under normal conditions, as well as in isogenic controls, facilitating the study of the dynamic events of cancer initiation and cancer evolution, but the application of stem cell-derived organoids requires widely accepted, standardized differentiation protocols for their generation. Thus far, hPSC-derived organoid systems have been used to model human brain cancer (Bian et al., 2018; Ballabio et al., 2020), colorectal cancer (CRC) (Crespo et al., 2017) and PDAC (Huang et al., 2015). The high culture efficiency and expandability of organoids reinforce the feasibility of the use of these systems to study tumor heterogeneity.
In this review, we will discuss recent studies that use hPSC-derived cells and organoids for the modeling of cancer progression. We will also highlight the applications of hPSC-based cancer modeling in the identification of biomarkers and therapeutic targets, the investigation of tumor heterogeneity and in drug discovery. Moreover, we will outline the potential challenges and opportunities of hPSC-based cancer modeling, which will further open up avenues for “cancer modeling in a dish”.
2. Tumor initiation in adult stem cells
ASCs, with their inherent self-renewal capacity, proliferative potential and multipotency, are deemed compelling candidates of oncogenesis as they continue to accumulate genetic mutations over their lifetime and respond to external stimuli such as inflammation. Acute myeloid leukemia (AML)-initiating cells that could reproduce the leukemia in non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice were first successfully isolated (Bonnet and Dick, 1997). Following the discovery of tumor-initiating cells for AML, tumor-initiating cells with stem cell properties have been isolated for a variety of cancers (Li et al., 2007; Eramo et al., 2008; Collins et al., 2005; Bapat et al., 2005; Singh et al., 2003; Yuan et al., 2004; Gibbs et al., 2005; Wu et al., 2007; Al-Hajj et al., 2003). In recent years, hPSCs have been extensively utilized to model a variety of ASC-related cancers including myeloid malignancies, brain tumors, and sarcomas, as well as ASC-related hereditary cancers including colon and breast cancer. However, the use of hPSC models can be extended to other cancers beyond those that are derived from ASCs, and hPSCs can serve as generally applicable models to investigate a wide spectrum of cancer types.
2.1. Brain cancer
Brain tumors remain among the most invasive and lethal forms of cancers. Neural stem cells (NSCs), characterized by their proliferative and multipotent potential, have been isolated from multiple germinal regions within the central nervous system and are posited to be the source of aggressive gliomas and medulloblastomas (Sanai et al., 2005; Fan and Eberhart, 2008). Deregulation of a number of growth-factor induced signaling cascades and developmental pathways are implicated in glioma and medulloblastoma formation, respectively. Modeling glioma has been made possible by the recapitulation of glioma tumor-initiating cells (GTICs) with iPSC-derived neural progenitor cells (NPCs) (Sancho-Martinez et al., 2016). iPSCs and iPSC-derived brain organoid models to study neurofibromatosis type 1 (NF1), a related cancer predisposition syndrome, have also been developed (Anastasaki et al., 2020). Bi-allelic inactivation of the NF1 gene gives rise to optic pathway gliomas (OPGs) in about 15–20% children with NF1 and additionally, other brainstem gliomas (Wegscheid et al., 2018). The prospects for the use of hPSCs in neuroblastoma modeling were expanded with the development of an iPSC model from Gorlin syndrome (GS) harboring heterozygous mutations in PTCH1 to investigate medulloblastoma occurrence in children (Tailor et al., 2018; Huang et al., 2019), as well as SMARCB1-deficient iPSC models for atypical teratoid/rhabdoid tumor (AT/RT) (Terada et al., 2019).
2.2. Myeloid malignancies
Myeloid malignancies are postulated to be the result of oncogenic transformation induced by mutations and epigenetic changes within hematopoietic stem cells (HSCs) or their committed progenitors. A broad range of myeloid cancers including myeloproliferative neoplasms (MPNs), myelodysplastic syndromes (MDS), and AML have been modeled using PSCs to dissect the mechanistic pathways underlying oncogenesis (Papapetrou, 2019). Many of these malignancies are characterized by chromosomal abnormalities. MDS-iPSCs (Kotini et al., 2017; Hsu et al., 2019), AML-iPSCs (Kotini et al., 2017; Chao et al., 2017), myelofibrosis-iPSCs (Hosoi et al., 2014), and 8p11 myeloproliferative syndrome (EMS)-iPSCs (Yamamoto et al., 2015), harboring chromosomal aberrations, have been generated. In addition, several studies have modeled chronic myeloid leukemia (CML) harboring the Philadelphia chromosome and the BCR/ABL oncogenic fusion gene (Miyauchi et al., 2018; Kumano et al., 2012; Suknuntha et al., 2015; Sloma et al., 2017; Amabile et al., 2015). Besides chromosomal aberrations, gene mutations in juvenile myelomonocytic leukemia (JMML) (Shigemura et al., 2019), secondary myelofibrosis (Hosoi et al., 2014), polycythemia vera (PV) (Stetka et al., 2017, 2019), CML (Sloma et al., 2017) and other myeloproliferative disorders (Ye et al., 2009) have also been studied using iPSC-derived models.
2.3. Sarcomas
Upon transformation, mesenchymal stem cells (MSCs) undergo sarcomagenesis to give rise to a variety of sarcomas, including bone sarcomas and soft tissue sarcomas. PSC models have been used in a variety of contexts to study the mechanisms associated with sarcomagenesis. For instance, the role of mutant TP53 in LFS, a cancer predisposition syndrome, has been explored using LFS-iPSCs (Lee et al., 2015; Lin et al., 2017; Zhou et al., 2017). Moreover, the contribution of the EWS-FLI1 oncogenic fusion transcript in inducing osteosarcoma has been explored using EWS-FLI1 expressing iPSCs (Komura et al., 2016; Moore et al., 2015).
2.4. Hereditary cancers
A number of hereditary cancers arise from ASCs, such as breast cancer and colon cancer. The existence of adult mammary epithelial stem cells within the normal breast has been previously corroborated with the discovery of rapidly cycling mammary stem cell populations within the basal mammary epithelium that give rise to progenitor populations, capable of forming adherent colonies (Stingl et al., 2006). Heterozygous mutations in BRCA1, a gene regulating DNA damage response, genetic stability and cell cycle progression, predisposes women – who typically have a familial history of cancer – to up to a 45% increase in lifetime risk of developing breast cancer (Griscelli et al., 2017). BRCA1-mutant iPSCs were derived and differentiated to recapitulate malignant breast cancer phenotypes (Soyombo et al., 2013; Griscelli et al., 2017). Although no differences were observed in the differentiation capacity of wild-type and BRCA1 mutants, an increase in protein kinase C-theta levels was observed in BRCA1-mutant iPSCs, as well as in cancer cell lines harboring BRCA1 mutations and hormone receptor negative breast cancers (Soyombo et al., 2013).
Analogous to mammary stem cells, steadily cycling intestinal stem cells, which give rise to transit-amplifying cells and other downstream differentiated cells, have been previously identified within intestinal crypts (Barker et al., 2007). Mutagenesis within crypt cells, subsequent cycles of niche succession, and clonal expansion promote colon cancer progression (Humphries and Wright, 2008). Traditionally, sequential progression of CRC occurs via a multi-step process involving heterozygous APC inactivation, KRAS oncogenic activation, loss of heterozygosity, and TP53 inactivation (Pino and Chung, 2010). iPSCs derived from patients diagnosed with hereditary familial adenomatous polyposis (FAP) have been differentiated toward colonic organoids to recapitulate the abnormal enhanced WNT activity caused by APC mutations (Crespo et al., 2017), and have revealed aberrations in cell identity, cell polarity, defective anaphase mechanisms and centrosome numbers as well (Sommer et al., 2018).
3. Mapping the cell-of-origin of cancer by directed differentiation of hPSCs
The cell-of-origin of cancer refers to normal cells that first acquire the mutations required for cancer initiation (Visvader, 2011). The cell-of-origin, in combination with its mutational profile, are key factors impacting tumor heterogeneity. A prerequisite to identify the cells of origin of cancers is to understand the normal lineage hierarchy during organ development, which originates from stem cells which divide to form committed progenitor cells and finally differentiated cells (the bulk of the organ). Certain cell types and differentiation states can be transformed by a given mutation, giving rise to specific disease states. For instance, a previous study of the cell-of-origin of CML indicated that the initiating oncogenic event, BCR/ABL translocation, occurred within HSCs (Tough et al., 1963). However, if this translocation occurred in committed progenitor cells, it gave rise to acute lymphoid leukemia (ALL) instead (Li et al., 1999). Likewise, different types of brain tumors are generated when mutations occur in NSCs, NPCs, oligodendrocyte progenitors (OPCs), or neuroepithelial stem cells (Huang et al., 2019; Alcantara Llaguno et al., 2009, 2015). Thus, mutations in either stem cells or progenitor cells give rise to distinct tumor types. Investigating the cell-of-origin of cancer will provide insights into cancer initiation and help to identify novel biomarkers for early cancer detection.
3.1. Brain cancer
It is controversial whether NPCs or pluripotent fetal cells are the cell-of-origin for AT/RT (Bouffard et al., 2004; Deisch et al., 2011). AT/RT is a highly aggressive central nervous system malignancy that mainly occurs in early childhood. The majority of AT/RTs harbor genome alterations in SMARCB1 (also known as INI1; SNF5, or BAF47) (Fruhwald et al., 2016). Mouse models of SMARCB1 ablation suggested that AT/RT may arise from NSCs or NPCs (Han et al., 2016). Recently, a group deconstructed the driving force of tumorigenesis by generating a human AT/RT model using SMARCB1- and TP53-deficient hiPSCs (hiPSCs SMARCB1−/−; TP53−/−) generated using the CRISPR/Cas9 system. hiPSCs SMARCB1−/−; TP53−/− were differentiated into neural progenitor-like cells (NPLCs) and transplanted into xenografts (Terada et al., 2019). The orthotopic transplantation of SMARCB1- and TP53-deficient NPLCs caused the development of aggressive tumors while control and TP53-deficient NPLCs gave rise to only microscopic tumors. Thus, SMARCB1 deficiency in NPLCs promotes tumor formation and the hPSC-based platform illustrates that NPLCs are the cell-of-origin for AT/RT. The use of hPSC-based models in tracing the cell-of-origin of cancer is also featured in another pediatric cancer, diffuse intrinsic pontine gliomas (DIPGs). DIPGs are highly aggressive pediatric brainstem tumors featuring the H3.3K27M mutation, which results in the global loss of H3K27me3 marks (Lewis et al., 2013; Bender et al., 2013). Funato et al. used a combination of lentiviruses, encoding H3.3 wt or H3.3K27M, PDGFRA, and sh-p53, to transform hESC-derived NPCs and found that the introduction of the H3.3K27M mutation in NPCs resulted in neoplastic transformation (Funato et al., 2014).
Interestingly, the suppression of a malignant phenotype was observed in glioblastoma iPSC (GiPSC) derived non-neural cell types, as compared to the parental glioblastoma-derived neural stem cells due to the reactivation of tumor suppressors (Stricker et al., 2013). However, GiPSC-derived neural progenitors remained malignant, although there was a widespread epigenetic resetting of common glioblastoma multiforme (GBM)-associated changes, including promoter regions of the TES, CDKN1C, and PRC2 target genes, which are normally hypermethylated (Stricker et al., 2013). Through these and other studies, a greater understanding of the putative cells of origin of various brain tumors and cancer-associated genetic lesions in cancer initiation have been obtained.
3.2. Small cell lung cancer (SCLC)
Pathways that increase the proportion of the cells of origin of SCLC via the conversion of hESCs into lung progenitor (LP) cells and eventually pulmonary neuroendocrine cells (PNECs) have been elucidated in a previous study (Chen et al., 2019). Specifically, inhibition of NOTCH signaling and the inhibition of the tumor suppressor genes, TP53 and RB, gave rise to an increase in the proportion of PNECs. Transcriptional profiles of the PNECs generated showed a remarkable similarity to those of early-stage human SCLC, with the upregulation of several genes associated with cell proliferation and inhibition of apoptosis. Moreover, tumors resembling early-stage SCLC developed upon the subcutaneous injection of PNECs with TP53 and RB inhibition in immunodeficient mice. In this context, hESCs serve as a useful model to decipher specific pathways that are key to cancer initiation via the development of cancer cells of origin.
3.3. Pancreatic cancer
Modeling changes in the epigenetic cancer landscape using hPSCs could provide further insights into the cancer cell-of-origin. This has been demonstrated in a study by Khoshchehreh et al. whereby multiple epigenetic reprogramming approaches were adopted in order to convert human PDAC cultures into a pluripotent state (Khoshchehreh et al., 2019). Three approaches, including the induction of the Yamanaka factors, the expression of Oct4 and miR-302, and the use of episomal vectors were explored. The pluripotent derivatives were found to be distinct from the parental counterparts, displaying reduced tumorigenicity in vivo and in vitro, especially upon the use of episomal vectors for reprogramming. Moreover, the reprogrammed cells readily differentiated in vivo. Such properties are indicative of a primitive state and as such, may enable the elucidation of epigenetic aberrations which play a role in these cancers.
4. Modeling cancer propagation and dissecting the genetic basis of cancer development
Point mutations, microsatellite instability (MIN), and chromosomal instability (CIN), initiate genomic instability, which leads to the development of cancer (Loeb et al., 2003). Cancer propagation is accompanied by a deregulation of key signaling pathways that offer a selective proliferative advantage and/or evasion from growth suppression. A selective proliferative advantage is acquired by cellular populations by one or all of the following mechanisms: receptor overexpression, constitutive receptor activation, production of oncogenic fusion proteins, or disruption of receptor recycling and degradation (Fouad and Aanei, 2017). Meanwhile, evasion from growth suppressors occurs via loss-of-function mutations in tumor suppressors, which results in the inhibition of critical cellular functions, including apoptosis, differentiation, maintenance of genomic integrity, DNA damage response, and intercellular interactions (Sun and Yang, 2010). Among the signaling pathways that are highly implicated in tumorigenesis, Ras and p53 signaling are the most predominant, with 30% and over 50% of cancers possessing the respective defective signaling networks (Fouad and Aanei, 2017; Zhou et al., 2019).
hPSC technologies have enabled investigations into proliferative pathways in cancer pathogenesis, especially in cancers with dysregulated RAS/MAPK and PI3K/AKT pathways. Among the various cancers, JMML has been extensively modeled using iPSCs. Gain of function mutations in PTPN11 modeled using iPSCs have been found to constitutively activate growth factor granulocyte macrophage colony-stimulating factor (GM-CSF) signaling and increase STAT5/ERK phosphorylation in JMML and Noonan syndrome (NS) (Gandre-Babbe et al., 2013; Shigemura et al., 2019; Tasian et al., 2019; Mulero-Navarro et al., 2015; Chan and Yoder, 2013). Recapitulation of aberrant myeloid cells in JMML using iPSC cells have provided insights on additional mechanisms underlying cancer progression, the study of which has been hampered by difficulties in obtaining specimens from very young children with the disease. iPSC-derived cancerous myeloid cells generated from patients harboring PTPN11 mutations have uncovered novel roles of miR-223 and miR-15a in activating the STAT5 pathway and inducing myeloid proliferation (Mulero-Navarro et al., 2015). In addition, specific mutations in PTPN11, including DG1H and G503R, were revealed to particularly promote hypersensitivity to GM-CSF. Graded differences in disease severity were found to be dependent on the extent of the destabilization of SHP-2, encoded by PTPN11 (Mulero-Navarro et al., 2015). A previous study modeling JMML using PTPN11 E76K or CBL Y371H mutant iPSCs brought the role of CBL, an E3 ubiquitin ligase, to light. iPSCs harboring PTPN11 mutations differentially activated RAS/MAPK signaling, while those that carried CBL mutations stimulated JAK/STAT signaling, indicating varied leukemogenic dependencies (Tasian et al., 2019). Sequencing studies on JMML iPSC clones have identified other single nucleotide variants including OSBP2 (c.389C > T), VPS13B (c.3379G > T), and SMC (c.2560A > G) and elucidated the co-occurrence of PTPN11 mutations with OSBP2 mutations (Shigemura et al., 2019). Although the role of the specific mutations was not extensively explored, some of the mutations identified may contribute to Ras-induced tumorigenesis.
Additionally, hPSCs have been used to deconstruct proliferative pathways in a number of other cancers. Recently, iPSCs were used to model a hereditary multiple endocrine neoplasia type 2A (MEN2A) cancer predisposing syndrome to evaluate the role of oncogenic RETC634Y and revealed the stimulation of the early growth response 1 (EGR1) transcriptional program through the RAS/MAPK signaling pathway (Hadoux et al., 2018). The constitutive activation of the HGF/c-MET axis and resultant altered gene expression has also been explored in the context of hereditary papillary renal cell carcinoma (PRCC) and glioblastoma using iPSCs (Hwang et al., 2019, 2020). Typically, stimulation of the HGF/c-MET axis leads to the activation of downstream signaling cascades, including PI3K/AKT, MAPK and STAT3 pathways, which enhance the survival and proliferation of tumorigenic cells (Zhang et al., 2018). Mutant c-MET iPSC-derived embryoid bodies, representative of PRCC, were generated and found to display a remarkably similar gene expression profile to primary human c-MET mutated PRCC, with overexpression of genes, such as KDM4C and BHLHE40 (Hwang et al., 2019). Analysis of the effect of c-MET mutants on glioblastoma formation were also evaluated using c-MET iPSC-derived organoids, which expressed elevated levels of glial fibrillary acidic protein (GFAP) and increased phospho-MET and phospho-STAT3 (Hwang et al., 2020). Activation of the OCT4/mTOR axis via MYCN overexpression was observed in another iPSC model of the Sonic Hedgehog (SHH)-subgroup medulloblastoma (Cancer et al., 2019). A number of genes, including KRAS, have also been activated to induce proliferation via Ras signaling during the generation of hPSCs and organoid models for the investigation of tumorigenesis (Kim et al., 2013; Huang et al., 2015). Moreover, iPSC-derived leukemic cell lines were applied to evaluate the response of varied KRAS mutants to Ras small molecule inhibitors (Kotini et al., 2017; Chao et al., 2017). The role of STAT signaling in oncogenesis has been explored in iPSC-derived models harboring JAK2 V617F mutations in a range of myeloid malignancies, including secondary myelofibrosis (Hosoi et al., 2014), PV (Stetka et al., 2017, 2019), CML (Sloma et al., 2017) and other myeloproliferative disorders (Ye et al., 2009).
p53, regarded the guardian of the genome, has been implicated in a majority of human cancers. Several landmark studies have highlighted the critical role of the suppression of TP53 in bestowing malignant cell types with the ability to evade cell senescence and apoptosis, paving the way for the accumulation of genomic aberrations as the cells continue to replicate indefinitely (Greenblatt et al., 1994). In an ESC-based model of Ewing sarcoma (EWS), knockdown of p53, in addition to the doxycycline inducible expression of the oncogenic EWS-FLI1 transcript, was critical for the generation of embryoid bodies exhibiting a transformed phenotype and a strong EWS gene expression signature (Gordon et al., 2016). Likewise, aberrant p53 signaling and receptor tyrosine kinase signaling was found to be integral to the recapitulation of GTICs from NPCs (Sancho-Martinez et al., 2016). In addition, p53 suppression was also found to be key for the generation of several other hPSCs and organoid models recapitulating other cancer subtypes (Chen et al., 2019; Huang et al., 2015; Modrek et al., 2017). Typical loss of function mutations in other tumor suppressors and an enhancement of tumorigenic potential has been observed in multiple cancer types modeled using hPSCs. In one study, knockout of PTEN using TALEN in ESCs and subsequent differentiation to NSCs stimulated tumorigenesis and the formation of glioblastoma owing to an increase in PAX7 expression (Duan et al., 2015). Several other tumor suppressors such as Rb/P16(INK4A), BRCA1, CDKN1A, CDKN2A, and SMAD4 have also been implicated in various cancer states which have accordingly been modeled using hPSCs and organoids (Soyombo et al., 2013; Chen et al., 2019; Kim et al., 2013; Huang et al., 2015; Griscelli et al., 2017).
5. Modeling the tumor microenvironment using hPSC-derived organoids to evaluate invasiveness and metastatic capacity
Tumor invasion and metastasis, characterized by a multi-step cascade, is a bi-directional process governed by the interaction between tumor cells and the microenvironment (Chaffer and Weinberg, 2011). This process, supported by proliferative processes and angiogenesis, underlies the transformation of benign tumors to more aggressive forms. Transformation is characterized by invasion of tumor cells through the extracellular matrix (ECM) and stroma, followed by intravasation to enter the circulation. The circulating tumor cells (CTCs) which survive vasculature transport become sequestered at distant organ sites, extravasate and form micrometastases, and proliferate at secondary invasion sites. This process is accompanied by dynamic intercellular interactions and the development of a complex tumor microenvironment (TME), with stimuli acting on the tumor cells influenced by the neighboring environmental profiles of hypoxia, acidity, growth factors, and mechanical forces (Ivey et al., 2016).
Breakthroughs in the simulation of the tumor microenvironment occurred upon the successful generation of organoids from hPSCs. To date, several representative organoids have been generated from iPSCs which may facilitate the decoding of cancer etiology in specific contexts (Crespo et al., 2017; Ballabio et al., 2020; Linkous et al., 2019; Spence et al., 2011; Qu et al., 2017; Yucer et al., 2017). Some of the features associated with organoids include the constitution of multiple cell types typically present in the in vivo counterpart, the assembly of the tissue architecture in a manner that is archetypal of the primary tissue, and the presence of specific functions that are inherent to the organ (Xu et al., 2018). In particular, organoids are able to capture the intra- and inter-tumoral heterogeneity of the tumor microenvironment (Smith and Tabar, 2019). Moreover, they provide a greater understanding of not only intercellular interactions but also of cooperative interconnections between tumorigenic cells and the matrix and of the tumor niche. As such, microenvironment gradients and the molecular cues driving oncogenesis can be distinguished using organoid models. Furthermore, tumor-immune interactions can be dissected by introducing critical environmental factors to the organoid system and establishing co-cultures with stromal cells.
hPSC-derived organoids have provided insights into the acquisition of invasive and metastatic capacities by tumor cells, driving the progression of cancer to more malignant forms. The expression of metastasis promoting genes has been studied in iPSC-derived organoids. VIMENTIN, a marker of epithelial-mesenchymal transition (EMT), was found to be overexpressed in APC-mutant human intestinal organoids derived from iPSCs. VIMENTIN is upregulated in a variety of aggressive cancers with poor prognosis (Sommer et al., 2018). Another metastasis promoting gene, PTPRZ1, was identified upon the engraftment of primary GBMs into cortical organoids derived from iPSCs. PTPRZ1 has been proposed to reactivate mitotic somal translocation (MST) in outer radial glia-like cells derived from glioblastoma stem cells, thus promoting the invasive capacity of these cells (Bhaduri et al., 2020). Moreover, hPSC models have been used to model cerebral organoid gliomas (GLICOs), an aggressive tumor supported by an inter-connected network of tumor micro-tubes, facilitating infiltration into the normal host tissue (Linkous et al., 2019). The hPSC-derived GLICOs generated were found to be successful in fostering tumor growth and promoting the invasion of GLICOs.
Humanized animal models carrying xeno-transplanted organoids can further shed light on the metastatic potential of these organoids upon successful metastases formation in distinct sites in vivo (Bian et al., 2018; Ballabio et al., 2020). A neoplastic cerebral organoid generated from hESCs via CRISPR/Cas9- and transposon-mediated mutagenesis of candidate genes revealed that tumor cells within glioblastoma-like neoplastic cerebral organoids expressed invasiveness-relevant genes at higher levels compared to normal cells and breached the renal capsule in xenografts. An exciting prospect for PSC-derived organoids involves their use in constructing organ-on-a-chip technologies. Recently, iPSCs have been used to generate human intestinal organoids which have been integrated into a gut-on-a-chip technology (Richmond and Breault, 2018; Workman et al., 2018). The concerted use of micro-engineered chips and organoids enables the regulation of flow, mechanical stretch, and other culture conditions providing greater tunability than previous cancer models. Such technologies have further evolved to establish more representative models using a combination of iPSC-derived organoids within a closed perfusion system, which reproduce the integrated and dynamic physiological environment with greater accuracy (Skardal et al., 2017). The combination of organoids with 3D bioprinting may also result in better organized and functionally improved organoids (Ma et al., 2016; Reid et al., 2019). Hence, numerous techniques have been established which facilitate the modeling of the tumor microenvironment and tracking of cancer evolution.
6. Tracking malignancies along the spectrum of cancer progression
Cancer progression is marked by a continuum of disease progression with the accumulation of genetic mutations and acquisition of epigenetic changes within cellular populations. However, most tumor cell lines as well as xenografts of primary tumor cells recapitulate advanced stages of cancer, preventing the study of the early and discrete states of cancer evolution. hPSCs can address this limitation as the reprogramming of cancer cells to pluripotency reverts these cells to an earlier stage within the cancer spectrum and hence, hPSCs can be used to track the progression of cancer to more malignant states. Following the induction of pluripotency in human PDAC cells and subsequent differentiation into pancreatic tissue, pancreatic intraepithelial neoplasia (PanIN) precursors were generated that progressed to more aggressive states (Kim et al., 2013). The dynamic regulation of the HNF4α network during cancer progression, with HNF4α being expressed at intermediate stages, was also elucidated using PanIN precursors. In hematological cancers, iPSCs have allowed for the capture of a range of oncogenic states. For instance, normal cells, preleukemic cells, malignant clones and subclones in the spectrum of progression of MDS to AML were all recapitulated using iPSCs in a concerted approach combining cellular reprogramming and knowledge of cancer genetics (Kotini et al., 2017). The simulation of each stage confirmed the role of GATA2 mutations, additional mutational events and chromosomal aberrations in driving MDS to AML progression.
7. Applications of hPSC-based cancer modeling
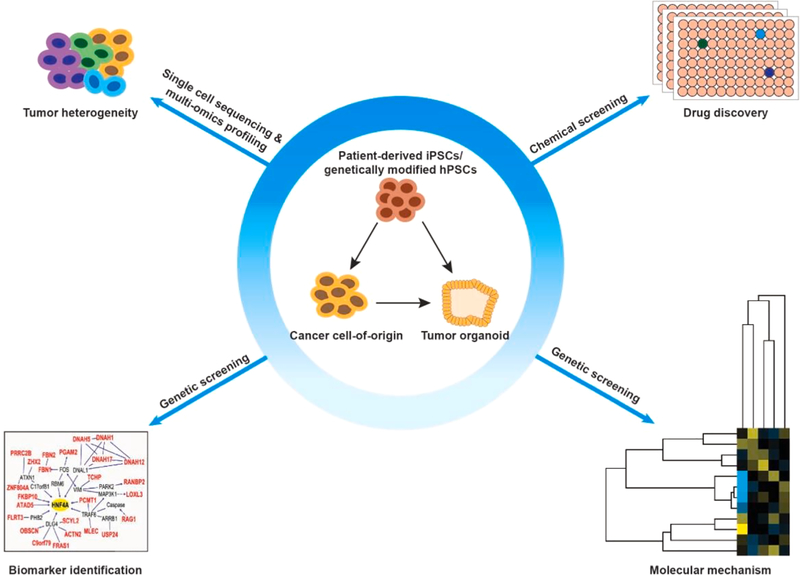
hPSC-based cancer modeling could benefit translational medicine as these models are powerful for drug discovery, the identification of biomarkers and molecular mechanisms, as well as tracking intra-tumoral heterogeneity (Fig. 2).
Fig. 2.
The applications of hPSC-based cancer modeling. iPSCs derived from patient cells or tumor samples, together with genetically engineered hPSCs with cancer-associated alterations undergo directed differentiation into cells of interest that represent the likely cancer cell-of-origin. These cells can be used to generate 3D tumor organoids under appropriate conditions. The hPSC-derived cancer models can be used in a variety of cancer studies including chemical screening for drug discovery, genetic screening to identify biomarkers and molecular mechanisms, as well as for the investigation of intra-tumoral heterogeneity by single cell sequencing and multi-omics profiling. The lower left image of biomarker identification was adapted from (Kim et al., 2013).
7.1. Drug discovery
hPSC modeling provides an opportunity for phenotype-based drug discovery (Fig. 2). The cells differentiated from hPSCs with genetic alterations could exhibit specific cellular and molecular phenotypes, which facilitates the identification of potential therapeutics by large-scale drug screens. FA is a rare inherited cancer predisposition syndrome, characterized by developmental abnormalities and bone marrow failure (Nalepa and Clapp, 2018). There is an urgent need for the discovery of novel drug candidates for the treatment of FA. Liu et al. generated FA-iPSCs/ESCs which can provide an unlimited source of differentiated, diverse hematopoietic cells. Subsequent screening of a panel of small molecules allowed for the discovery of several novel compounds that enhanced the hematopoietic differentiation of FA-iPSCs and FA patient bone marrow cells (Liu et al., 2014). Likewise, Chang et al. used MDS-iPSC-derived hematopoietic progenitor cells (iPSC-HPCs) to screen a library of 2000 compounds containing FDA-approved drugs, natural products, and other bioactive compounds, and identified Niflumic acid as a small molecule that selectively inhibited the proliferation of HPCs derived from del(7q) MDS iPSCs (Chang et al., 2018). The use of hPSC-based models as drug discovery platforms is not limited to hematopoietic disorders. By using transformed hiPSC/ESC-derived NPCs as well as NSCs to screen libraries of chemicals, several inhibitors of brain tumor growth have been identified, including MI-2, nelarabine, letrozole, capecitabine and MMC (Duan et al., 2015; Sancho-Martinez et al., 2016; Funato et al., 2014). Responses to various drugs depend on the specific stage of the disease as well. Chao et al. studied the impact of DOT1L inhibition on MLL-rearranged iPSCs and found that the DOT1L inhibitor, EPZ-5676, specifically inhibited the growth of hematopoietic cells differentiated from MLL-rearranged AML-iPSCs, while exhibiting a minimal growth inhibition of undifferentiated AML-iPSCs. Hence, cytotoxicity induced by EPZ-5676 was dependent on the specific differentiation state of AML cells (Chao et al., 2017).
The hPSC-derived organoid system also serves as an emerging platform for cancer drug discovery (Fig. 2). Initial drug screening studies used patient-derived organoids (PDOs) (Gao et al., 2014). PDO biobanks were later established which greatly expanded the types of patient samples. Organoid biobanks of colorectal cancer (van de Wetering et al., 2015), PDAC (Huang et al., 2015), bladder cancer (Lee et al., 2018), breast cancer (Sachs et al., 2018), gastric cancer (Yan et al., 2018), prostate cancer (Beshiri et al., 2018), ovarian cancer (Kopper et al., 2019), and glioblastomas (Jacob, 2020) have greatly facilitated high-throughput drug screening. Recently, colon organoids generated from FAP-iPSCs with mutations in APC were used as a platform for drug testing (Crespo et al., 2017). The authors found that a ribosome-binding antibiotic, geneticin, could rescue APC expression, reduce WNT overactivation, and block colonic epithelial cell hyperproliferation specifically in APC mutant FAP colon organoids. This highlights the powerful value of hPSC-derived organoids in drug development prior to clinical applications.
7.2. Identification of potential biomarkers and molecular mechanism
hPSC-based modeling represents an approach to identify disease biomarkers and therapeutic targets for clinical translation (Fig. 2). For example, Kim et al. used a PDAC-derived iPSC line and identified 107 proteins released or secreted from PanIN-like teratomas. Among these proteins, 43 fell into the category of TGFβ and integrin networks and 25 were components of the HNF4α network. The proteins discovered were found to be stable and may serve as potential biomarkers of early-stage PDAC (Kim et al., 2013). Further analysis of these 107 proteins revealed increased levels of THBS2 and CA19–9, which were considered potential markers of PDAC as well (Kim et al., 2017). In another study using stem cells derived from CML iPSCs, Miyauchi et al. identified ADAM8, a metalloproteinase, as an antigen of tyrosine kinase inhibitor (TKI)-resistant CML cells, and as a potential predictor of residual disease or relapse (Miyauchi et al., 2018).
hPSC systems also facilitate the identification of molecular mechanisms for various cancers. Terada et al. found that the activation of an ESC-like signature induced a rhabdoid phenotype in AT/RT recapitulated using SMARCB1-deficient NPLCs. Activation of an ESC-like gene expression signature is also a feature of other pediatric cancers, including neuroblastoma (NB), Wilms’ tumor, and hepatoblastoma (HB) (Terada et al., 2019). Previous studies using other models showed that poorly differentiated tumors exhibited an ESC-like gene expression signature (Ben-Porath et al., 2008; Wong et al., 2008). Moreover, the CRISPR/Cas9 system was used to screen for genes that are associated with ESC maintenance. Dozens of candidate genes can decrease cancer cell growth, including Enhancer of zeste homolog 2 (EZH2), which was previously reported as a target of AT/RT (Knutson et al., 2013). Similar genetic screens were performed for two neuroblastoma cell lines, SK-N-AS and SK-N-BE (2), and RAD21 knockout was found to inhibit hiPSC-derived cancer cell growth. Xenograft transplantation of hiPSC-derived cancer cell lines with EZH2 or RAD21 deletion significantly extended the overall survival of mice. Therefore, EZH2 and RAD21 are potential therapeutic targets for AT/RT (Terada et al., 2019). In addition, a phenotypic screen is another strategy to find therapeutic targets. Kotini et al. used Del(7q)-iPSCs to perform a phenotype rescue screen and identified candidate haplo-insufficient genes (HIPK2, ATP6V0E2, LUC7L2 and EZH2) underlying hematopoietic differentiation defects caused by del(7q) (Kotini et al., 2015).
7.3. Clonal evolution and intra-tumoral heterogeneity
Intra-tumoral heterogeneity is generated during the expansion of tumor cells and results in the formation of divergent clonal lineages. Intra-tumoral heterogeneity is a result of stochastic genetic variations (Heppner, 1984) and/or epigenetic alterations (Baylin and Jones, 2011), the presence of a minor subpopulation of cancer stem cells (CSCs) residing in tumors (Visvader, 2011), or the dynamic shift between CSCs and non-CSCs (Meacham and Morrison, 2013). Since iPSC clones are derived from single cells and can be clonally expanded, they can be used to trace tumor heterogeneity (Fig. 2). Chao et al. reported the use of AML-iPSCs to study clonal heterogeneity and cancer evolution. They identified two distinct AML-iPSC subclones (KRAS mutant or KRAS WT) from the same AML patient that shared all other leukemia-associated mutations, but exhibited differential cytokine (GM-CSF) dependence in vitro, differential leukemogenic properties in vivo, and differential sensitivity to MEK inhibitors. Interestingly, KRAS WT iPSC subclones showed greater chemotherapy resistance as compared to KRAS mutant subclones. Accordingly, clonal selection and evolution within the KRAS WT clone accounted for AML relapse in the patient. Moreover, the progression and relapse of myeloid neoplasms, including MDS, may be caused by the evolution of malignant cells harboring multiple genome lesions (Sperling et al., 2017). However, the sequence of accumulation of clonal mutations is in some cases unknown. In a recent study, Hsu and colleagues determined the discrete stages of preleukemic clonal evolution using MDS iPSCs reprogrammed by episomes. By using this platform, they identified t(4;12) to be an earlier event in a case of MDS, followed by mutations in SF3B1, EZH2, and del(5q) (Hsu et al., 2019). Thus, reprogramming provides a strategy to order genetic variants during clonal evolution.
Notably, single-cell sequencing has impacted many areas of cancer research and provided a greater understanding of clonal evolution and intra-tumoral heterogeneity (Lim et al., 2020). Single-cell RNA sequencing (scRNA-seq) technology has been used to identify the subpopulations of a range of cancers, including hematopoietic (Jaitin et al., 2014), pancreatic (Peng et al., 2019), liver (Halpern et al., 2017), colorectal (Dai et al., 2019), and lung cancers (Kim et al., 2020). Moreover, single-cell DNA sequencing (scDNA-seq) is also useful for the discovery of copy-number (Gao et al., 2016; Laks et al., 2019) and branching evolution in cancers (Yu et al., 2014; Wang et al., 2017; Andor et al., 2020). Recently, single-cell-based transcriptomics or epigenomics have been applied to deconstruct heterogeneity present within hPSC-derived brain organoids (Tanaka et al., 2020) and kidney organoids (Wu and Humphreys, 2020; Subramanian et al., 2019). Since organoids contain a heterogeneous population of cells that exhibit different behaviors and responses, scRNA-seq can be used to comprehensively characterize distinct cell types and cell states and reconstruct lineage trajectories. Moreover, developments in computational analysis of tumor mutational profiles and large-scale biobanking of diverse patient-derived iPSCs have been achieved. The availability of such resources will further facilitate the capture of tumor heterogeneity and identification of predictive biomarkers using iPSC-derived differentiated cells or organoids.
8. Concluding remarks
Over the past decade, we have witnessed enormous leaps in PSC technologies which have facilitated the development of new models of human cancers. In this review, we have summarized the role of hPSC-based cancer modeling for the understanding of the initiation, propagation, and metastasis of cancer. Moreover, such models of cancer are also broadly applicable for the discovery of novel cancer drugs, biomarkers, and molecular mechanisms. One unique feature of this modeling approach is that it provides an opportunity to study early stages of cancer that are not well captured by cancer cell lines or xenografts. As early-stage cancers frequently evade detection, the understanding of early-stage cancer and the identification of potential biomarkers will ultimately result in a more timely and beneficial diagnosis and treatment of cancer.
When using this system, one should pay close attention to the line-to-line variations of iPSCs. Fortunately, such variations can be overcome by the generation of isogenic lines. With the help of the CRISPR/Cas9 technology, isogenic iPSC lines can be generated by the correction of mutations in patient-derived iPSCs. Another concern pertaining to the use of PSC systems is the accumulation of genetic aberrations during prolonged hPSC culture. This is an unavoidable phenomenon, but can be monitored by frequent karyotyping and routine checking of the mutational status of PSC lines (Assou et al., 2020). Maintaining early-passage cells, and avoiding unnecessary passaging and multiple freeze–thaw cycles are advisable. Moreover, the cellular immaturity of hPSC-derived cell types and the formation of fetal-like hPSC-derived organoids is another major challenge that has yet to be resolved. There is a need to optimize differentiation protocols for organoid generation. Vascularization or long-term culture may be additional options to improve organoid maturation (Lancaster and Huch, 2019). Additionally, recent developments in sophisticated 3D bioprinting and organs-on-a-chip systems may enable in vivo tumor pathology and interactions between the tumor and the microenvironment to be reflected with higher accuracy.
Combined with the development of genome editing, 3D organoids, single-cell sequencing, and multi-omics technologies, hPSC-based platforms for cancer modeling will become more powerful and tunable, especially with the manipulation of different stages of cancer development that cannot be achieved so far by other approaches. Large-scale biobanks of organoids or cells derived from hPSCs, together with large-scale small molecule library screens, will facilitate cancer drug development in the near future.
Acknowledgements
S.C is supported as Irma Hirschl Trust Research Award Scholars. L.A. L. is supported by an F32 post-doctoral fellowship from the National Institute of Health (1F32HD096810-01A1).
Abbreviations:
- ALL
acute lymphoid leukemia
- AML
acute myeloid leukemia
- ASCs
adult stem cells
- AT/RT
atypical teratoid/rhabdoid tumor
- CIN
chromosomal instability
- CML
chronic myeloid leukemia
- CRC
colorectal cancer
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeats/Cas9
- CSCs
cancer stem cells
- CTCs
circulating tumor cells
- DIPGs
diffuse intrinsic pontine gliomas
- ECM
extracellular matrix
- EGR1
early growth response 1
- EMS
8p11 myeloproliferative syndrome
- EMT
epithelial-mesenchymal transition
- EWS
ewing sarcoma
- EZH2
enhancer of zeste homolog 2
- FA
fanconi anemia
- FAP
familial adenomatous polyposis
- FPD/AML
familial platelet disorder with associated myeloid malignancy
- GBM
glioblastoma multiforme
- GFAP
glial fibrillary acidic protein
- GiPSC
glioblastoma induced pluripotent stem cells
- GLICOs
cerebral organoid gliomas
- GM-CSF
growth factor granulocyte macrophage colony-stimulating factor
- GS
gorlin syndrome
- GTICs
glioma tumor-initiating cells
- HB
hepatoblastoma
- hESCs
human embryonic stem cells
- HPCs
hematopoietic progenitor cells
- hPSCs
human pluripotent stem cells
- HSCs
hematopoietic stem cells
- iPSCs
induced pluripotent stem cells
- JMML
juvenile myelomonocytic leukemia
- LFS
Li-Fraumeni syndrome
- LP
lung progenitor cells
- MDS
myelodysplastic syndromes
- MEN2A
multiple endocrine neoplasia type 2A
- MIN
microsatellite instability
- MPNs
myeloproliferative neoplasms
- MSCs
mesenchymal stem cells
- MST
mitotic somal translocation
- NB
neuroblastoma
- NF1
neurofibromatosis type 1
- NOD/SCID
nonobese diabetic/severe combined immunodeficiency
- NPCs
neural progenitor cells
- NPLCs
neural progenitor-like cells
- NS
Noonan syndrome
- NSCs
neural stem cells
- OPCs
oligodendrocyte progenitors
- OPGs
optic pathway gliomas
- OSKM
OCT4, SOX2, KLF4 and c-MYC, also known as Yamanaka factors
- PanIN
pancreatic intraepithelial neoplasia
- PDAC
pancreatic ductal adenocarcinoma
- PDOs
patient-derived organoids
- PDTX
patient-derived tumor xenograft
- PNEC
pulmonary neuroendocrine cells
- PRCC
papillary renal cell carcinoma
- PSCs
pluripotent stem cells
- PV
polycythemia vera
- scDNA-seq
single-cell DNA sequencing
- scRNA-seq
single-cell RNA sequencing
- SHH
sonic hedgehog
- SCLC
small cell lung carcinoma
- TALENS
transcription activator-like effector nucleases
- TKI
tyrosine kinase inhibitor
- TME
tumor microenvironment
- 2D
two-dimensional
- 3D
three-dimensional
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Mirabelli P, Coppola L, Salvatore M, 2019. Cancer cell lines are useful model systems for medical research. Cancers (Basel) 11 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon DJ, Orsulic S, 2011. Mouse models of cancer. Annu. Rev. Pathol. 6, 95–119. [DOI] [PubMed] [Google Scholar]
- Murayama T, Gotoh N, 2019. Patient-derived xenograft models of breast cancer and their application. Cells 8 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, et al. , 1998. Embryonic stem cell lines derived from human blastocysts. Science 282 (5391), 1145–1147. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S, 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (4), 663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi K, et al. , 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5), 861–872. [DOI] [PubMed] [Google Scholar]
- Sharma A, et al. , 2020. Multi-lineage human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell 26 (3), 309–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, et al. , 2015. PTEN deficiency reprogrammes human neural stem cells towards a glioblastoma stem cell-like phenotype. Nat. Commun. 6, 10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y, et al. , 2019. Human pluripotent stem cell-derived tumor model uncovers the embryonic stem cell signature as a key driver in atypical teratoid/rhabdoid tumor. Cell Rep. 26 (10), 2608–2621 e6. [DOI] [PubMed] [Google Scholar]
- Lee DF, et al. , 2015. Modeling familial cancer with induced pluripotent stem cells. Cell 161 (2), 240–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller LU, et al. , 2012. Overcoming reprogramming resistance of Fanconi anemia cells. Blood 119 (23), 5449–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony-Debre I, et al. , 2015. Level of RUNX1 activity is critical for leukemic predisposition but not for thrombocytopenia. Blood 125 (6), 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyombo AA, et al. , 2013. Analysis of induced pluripotent stem cells from a BRCA1 mutant family. Stem Cell Rep. 1 (4), 336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, et al. , 2010. Generation of iPSCs from cultured human malignant cells. Blood 115 (20), 4039–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, et al. , 2013. Patient-derived induced pluripotent stem cells recapitulate hematopoietic abnormalities of juvenile myelomonocytic leukemia. Blood 121 (24), 4925–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini AG, et al. , 2015. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat. Biotechnol. 33 (6), 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N, et al. , 2010. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. U.S.A. 107 (1), 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker SH, et al. , 2013. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 27 (6), 654–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, et al. , 2017. Establishment of hepatocellular cancer induced pluripotent stem cells using a reprogramming technique. Gut Liver 11 (2), 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, et al. , 2019. Generation of pulmonary neuroendocrine cells and SCLC-like tumors from human embryonic stem cells. J. Exp. Med. 216 (3), 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. , 2013. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 3 (6), 2088–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshchehreh R, et al. , 2019. Epigenetic reprogramming of primary pancreatic cancer cells counteracts their in vivo tumourigenicity. Oncogene 38 (34), 6226–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, Clevers H, 2018. Organoids in cancer research. Nat. Rev. Cancer 18 (7), 407–418. [DOI] [PubMed] [Google Scholar]
- Zanoni M, et al. , 2019. Anticancer drug discovery using multicellular tumor spheroid models. Expert Opin. Drug Discov. 14 (3), 289–301. [DOI] [PubMed] [Google Scholar]
- Sant S, Johnston PA, 2017. The production of 3D tumor spheroids for cancer drug discovery. Drug Discov. Today Technol. 23, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, 2016. Modeling development and disease with organoids. Cell 165 (7), 1586–1597. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, et al. , 2013. Cerebral organoids model human brain development and microcephaly. Nature 501 (7467), 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo M, et al. , 2017. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 23 (7), 878–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munera JO, et al. , 2017. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell 21 (1), 51–64 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, et al. , 2014. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516 (7531), 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low JH, et al. , 2019. Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell 25 (3), 373–387 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, et al. , 2019. Generation of hepatobiliary organoids from human induced pluripotent stem cells. J. Hepatol. 70 (6), 1145–1158. [DOI] [PubMed] [Google Scholar]
- Miller AJ, et al. , 2019. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 14 (2), 518–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YW, et al. , 2017. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19 (5), 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohwieler M, et al. , 2017. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 66 (3), 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway EM, et al. , Differentiation of human intestinal organoids with endogenous vascular endothelial cells. bioRxiv, 2020: p. 2020.03.15.991950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Demirci U, Chen P, 2019. Emerging organoid models: leaping forward in cancer research. J. Hematol. Oncol. 12 (1), 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S, et al. , 2018. Genetically engineered cerebral organoids model brain tumor formation. Nat. Methods 15 (8), 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio C, et al. , 2020. Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat. Commun. 11 (1), 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, et al. , 2015. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21 (11), 1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE, 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3 (7), 730–737. [DOI] [PubMed] [Google Scholar]
- Li CW, et al. , 2007. Identification of pancreatic cancer stem cells. Cancer Res. 67 (3), 1030–1037. [DOI] [PubMed] [Google Scholar]
- Eramo A, et al. , 2008. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 15 (3), 504–514. [DOI] [PubMed] [Google Scholar]
- Collins AT, et al. , 2005. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 65 (23), 10946–10951. [DOI] [PubMed] [Google Scholar]
- Bapat SA, et al. , 2005. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 65 (8), 3025–3029. [DOI] [PubMed] [Google Scholar]
- Singh SK, et al. , 2003. Identification of a cancer stem cell in human brain tumors. Cancer Res. 63 (18), 5821–5828. [PubMed] [Google Scholar]
- Yuan X, et al. , 2004. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 23 (58), 9392–9400. [DOI] [PubMed] [Google Scholar]
- Gibbs CP, et al. , 2005. Stem-like cells in bone sarcomas: implications for tumorigenesis. Neoplasia 7 (11), 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CL, et al. , 2007. Side population cells isolated from mesenchymal neoplasms have tumor initiating potential. Cancer Res. 67 (17), 8216–8222. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, et al. , 2003. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100 (7), 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanai N, Alvarez-Buylla A, Berger MS, 2005. Neural stem cells and the origin of gliomas. N. Engl. J. Med. 353 (8), 811–822. [DOI] [PubMed] [Google Scholar]
- Fan X, Eberhart CG, 2008. Medulloblastoma stem cells. J. Clin. Oncol. 26 (17), 2821–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Martinez I, et al. , 2016. Establishment of human iPSC-based models for the study and targeting of glioma initiating cells. Nat. Commun. 7, 10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasaki C, et al. , 2020. Human iPSC-derived neurons and cerebral organoids establish differential effects of germline NF1 gene mutations. Stem Cell Rep. 14 (4), 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegscheid ML, Anastasaki C, Gutmann DH, 2018. Human stem cell modeling in neurofibromatosis type 1 (NF1). Exp. Neurol. 299 (Pt B), 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor J, et al. , 2018. A human Ips cell-based model of medulloblastoma demonstrates co-operativity between shh signalling and mutation in an epigenetic modifier. Neuro-Oncology 20, 2–3.29126327 [Google Scholar]
- Huang M, et al. , 2019. Engineering genetic predisposition in human neuroepithelial stem cells recapitulates medulloblastoma tumorigenesis. Cell Stem Cell 25 (3), 433–446 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou EP, 2019. Modeling myeloid malignancies with patient-derived iPSCs. Exp. Hematol. 71, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini AG, et al. , 2017. Stage-specific human induced pluripotent stem cells map the progression of myeloid transformation to transplantable leukemia. Cell Stem Cell 20 (3), 315–328 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J, et al. , 2019. Reprogramming identifies functionally distinct stages of clonal evolution in myelodysplastic syndromes. Blood 134 (2), 186–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, et al. , 2017. Human AML-iPSCs reacquire leukemic properties after differentiation and model clonal variation of disease. Cell Stem Cell 20 (3), 329–344 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi M, et al. , 2014. Generation of induced pluripotent stem cells derived from primary and secondary myelofibrosis patient samples. Exp. Hematol. 42 (9), 816–825. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, et al. , 2015. Screening of drugs to treat 8p11 myeloproliferative syndrome using patient-derived induced pluripotent stem cells with fusion gene CEP110-FGFR1. PLoS ONE 10 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi M, et al. , 2018. ADAM8 is an antigen of tyrosine kinase inhibitor-resistant chronic myeloid leukemia cells identified by patient-derived induced pluripotent stem cells. Stem Cell Rep. 10 (3), 1115–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K, et al. , 2012. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood 119 (26), 6234–6242. [DOI] [PubMed] [Google Scholar]
- Suknuntha K, et al. , 2015. Discovery of survival factor for primitive chronic myeloid leukemia cells using induced pluripotent stem cells. Stem Cell Res. 15 (3), 678–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloma I, et al. , 2017. Whole-genome analysis reveals unexpected dynamics of mutant subclone development in a patient with JAK2-V617F-positive chronic myeloid leukemia. Exp. Hematol. 53, 48–58. [DOI] [PubMed] [Google Scholar]
- Amabile G, et al. , 2015. Dissecting the role of aberrant DNA methylation in human leukaemia. Nat. Commun. 6, 7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemura T, et al. , 2019. Essential role of PTPN11 mutation in enhanced haematopoietic differentiation potential of induced pluripotent stem cells of juvenile myelomonocytic leukaemia. Br. J. Haematol. 187 (2), 163–173. [DOI] [PubMed] [Google Scholar]
- Stetka J, et al. , 2017. JAK2 V617F Progenitors Utilize Adaptations to Cell-Autonomous and Microenvironment-Dependent Inflammatory Stress in Polycythemia Vera, Likely Exhibiting Barrier Against Rapid Transformation to Myelofibrosis. Blood 130. [Google Scholar]
- Stetka J, et al. , 2019. Addiction to DUSP1 protects JAK2V617F-driven polycythemia vera progenitors against inflammatory stress and DNA damage, allowing chronic proliferation. Oncogene 38 (28), 5627–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, et al. , 2009. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood 114 (27), 5473–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, et al. , 2017. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol. Med. 23 (8), 737–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, et al. , 2017. Li-fraumeni syndrome disease model: a platform to develop precision cancer therapy targeting oncogenic p53. Trends Pharmacol. Sci. 38 (10), 908–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura S, et al. , 2016. An EWS-FLI1-Induced Osteosarcoma Model Unveiled a Crucial Role of Impaired Osteogenic Differentiation on Osteosarcoma Development. Stem Cell Rep. 6 (4), 592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.B.t., et al. , Epigenetic reprogramming and re-differentiation of a Ewing sarcoma cell line. Front Cell Dev Biol, 2015. 3: p. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, et al. , 2006. Purification and unique properties of mammary epithelial stem cells. Nature 439 (7079), 993–997. [DOI] [PubMed] [Google Scholar]
- Griscelli F, et al. , 2017. Generation of induced pluripotent stem cell (iPSC) line from a patient with triple negative breast cancer with hereditary exon 17 deletion of BRCA1 gene. Stem Cell Res. 24, 135–138. [DOI] [PubMed] [Google Scholar]
- Barker N, et al. , 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449 (7165), 1003–1007. [DOI] [PubMed] [Google Scholar]
- Humphries A, Wright NA, 2008. Colonic crypt organization and tumorigenesis. Nat. Rev. Cancer 8 (6), 415–424. [DOI] [PubMed] [Google Scholar]
- Pino MS, Chung DC, 2010. The chromosomal instability pathway in colon cancer. Gastroenterology 138 (6), 2059–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, et al. , 2018. Modeling APC mutagenesis and familial adenomatous polyposis using human iPS cells. PLoS ONE 13 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, 2011. Cells of origin in cancer. Nature 469 (7330), 314–322. [DOI] [PubMed] [Google Scholar]
- Tough IM, et al. , 1963. Cytogenetic studies on bone-marrow in chronic myeloid leukaemia. Lancet 1 (7286), 844–846. [DOI] [PubMed] [Google Scholar]
- Li S, et al. , 1999. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 189 (9), 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno S, et al. , 2009. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15 (1), 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno SR, et al. , 2015. Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell 28 (4), 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouffard JP, et al. , 2004. Double immunolabeling of central nervous system atypical teratoid/rhabdoid tumors. Mod. Pathol. 17 (6), 679–683. [DOI] [PubMed] [Google Scholar]
- Deisch J, Raisanen J, Rakheja D, 2011. Immunohistochemical expression of embryonic stem cell markers in malignant rhabdoid tumors. Pediatr. Dev. Pathol. 14 (5), 353–359. [DOI] [PubMed] [Google Scholar]
- Fruhwald MC, et al. , 2016. Atypical teratoid/rhabdoid tumors-current concepts, advances in biology, and potential future therapies. Neuro Oncol 18 (6), 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han ZY, et al. , 2016. The occurrence of intracranial rhabdoid tumours in mice depends on temporal control of Smarcb1 inactivation. Nat. Commun. 7, 10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, et al. , 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340 (6134), 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S, et al. , 2013. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24 (5), 660–672. [DOI] [PubMed] [Google Scholar]
- Funato K, et al. , 2014. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346 (6216), 1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Loeb KR, Anderson JP, 2003. Multiple mutations and cancer. Proc. Natl. Acad. Sci. U.S.A. 100 (3), 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouad YA, Aanei C, 2017. Revisiting the hallmarks of cancer. Am J Cancer Res 7 (5), 1016–1036. [PMC free article] [PubMed] [Google Scholar]
- Sun W, Yang J, 2010. Functional mechanisms for human tumor suppressors. J Cancer 1, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hao Q, Lu H, 2019. Mutant p53 in cancer therapy-the barrier or the path. J. Mol. Cell. Biol. 11 (4), 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasian SK, et al. , 2019. Mutation-specific signaling profiles and kinase inhibitor sensitivities of juvenile myelomonocytic leukemia revealed by induced pluripotent stem cells. Leukemia 33 (1), 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Navarro S, et al. , 2015. Myeloid Dysregulation in a Human Induced Pluripotent Stem Cell Model of PTPN11-Associated Juvenile Myelomonocytic Leukemia. Cell Rep 13 (3), 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RJ, Yoder MC, 2013. JMML patient-derived iPSCs induce new hypotheses. Blood 121 (24), 4815–4817. [DOI] [PubMed] [Google Scholar]
- Hadoux J, et al. , 2018. Transcriptional landscape of a RET(C634Y)-mutated iPSC and its CRISPR-corrected isogenic control reveals the putative role of EGR1 transcriptional program in the development of multiple endocrine neoplasia type 2A-associated cancers. Stem Cell Res. 26, 8–16. [DOI] [PubMed] [Google Scholar]
- Hwang JW, et al. , 2019. iPSC-Derived Embryoid Bodies as Models of c-Met-Mutated Hereditary Papillary Renal Cell Carcinoma. Int. J. Mol. Sci 20 (19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JW, et al. , 2020. A novel neuronal organoid model mimicking glioblastoma (GBM) features from induced pluripotent stem cells (iPSC). Biochim. Biophys. Acta, Gen. Subj. 1864 (4), 129540. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. , 2018. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer 17 (1), 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer M, et al. , 2019. Humanized stem cell models of pediatric medulloblastoma reveal an oct4/mtor axis that promotes malignancy. Cell Stem Cell 25 (6), p. 855–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt MS, et al. , 1994. Mutations in the P53 tumor-suppressor gene - clues to cancer etiology and molecular pathogenesis. Cancer Res. 54 (18), 4855–4878. [PubMed] [Google Scholar]
- Gordon DJ, Motwani M, Pellman D, 2016. Modeling the initiation of Ewing sarcoma tumorigenesis in differentiating human embryonic stem cells. Oncogene 35 (24), 3092–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek AS, et al. , 2017. Low-Grade Astrocytoma Mutations in IDH1, P53, and ATRX Cooperate to Block Differentiation of Human Neural Stem Cells via Repression of SOX2. Cell Rep 21 (5), 1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA, 2011. A perspective on cancer cell metastasis. Science 331 (6024), 1559–1564. [DOI] [PubMed] [Google Scholar]
- Ivey JW, et al. , 2016. Improving cancer therapies by targeting the physical and chemical hallmarks of the tumor microenvironment. Cancer Lett. 380 (1), 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkous A, et al. , 2019. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep 26 (12), 3203–3211 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, et al. , 2011. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470 (7332), 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, et al. , 2017. Differentiation of Human Induced Pluripotent Stem Cells to Mammary-like Organoids. Stem Cell Rep. 8 (2), 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucer N, et al. , 2017. Directed differentiation of human induced pluripotent stem cells into fallopian tube epithelium. Sci. Rep. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, et al. , 2018. Organoid technology and applications in cancer research. J. Hematol. Oncol. 11 (1), 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RC, Tabar V, 2019. Constructing and Deconstructing Cancers using Human Pluripotent Stem Cells and Organoids. Cell Stem Cell 24 (1), 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A, et al. , 2020. Outer Radial Glia-like Cancer Stem Cells Contribute to Heterogeneity of Glioblastoma. Cell Stem Cell 26 (1), 48–63 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond CA, Breault DT, 2018. Move over caco-2 cells: human-induced organoids meet gut-on-a-chip. Cell. Mol. Gastroenterol. Hepatol. 5 (4), 634–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman MJ, et al. , 2018. Enhanced utilization of induced pluripotent stem cell-derived human intestinal organoids using microengineered chips. Cell. Mol. Gastroenterol. Hepatol. 5 (4), 669–677 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skardal A, et al. , 2017. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 7 (1), 8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, et al. , 2016. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. U.S.A. 113 (8), 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JA, et al. , 2019. A 3D bioprinter platform for mechanistic analysis of tumoroids and chimeric mammary organoids. Sci. Rep. 9 (1), 7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalepa G, Clapp DW, 2018. Fanconi anaemia and cancer: an intricate relationship. Nat. Rev. Cancer 18 (3), 168–185. [DOI] [PubMed] [Google Scholar]
- Liu GH, et al. , 2014. Modelling fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat. Commun. 5, 4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, et al. , 2018. Dissecting the contributions of cooperating gene mutations to cancer phenotypes and drug responses with patient-derived iPSCs. Stem Cell Rep. 10 (5), 1610–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, et al. , 2014. Organoid cultures derived from patients with advanced prostate cancer. Cell 159 (1), 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, et al. , 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161 (4), 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, et al. , 2018. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173 (2), 515–528 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N, et al. , 2018. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172 (1–2), 373–386 e10. [DOI] [PubMed] [Google Scholar]
- Yan HHN, et al. , 2018. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell 23 (6), 882–897 e11. [DOI] [PubMed] [Google Scholar]
- Beshiri ML, et al. , 2018. A PDX/organoid biobank of advanced prostate cancers captures genomic and phenotypic heterogeneity for disease modeling and therapeutic screening. Clin. Cancer Res. 24 (17), 4332–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopper O, et al. , 2019. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 25 (5), 838–849. [DOI] [PubMed] [Google Scholar]
- Jacob F, et al. , 2020. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 180 (1), 188–204 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. , 2017. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19–9 blood markers. Sci. Transl. Med. 9 (398). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, et al. , 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40 (5), 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, et al. , 2008. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2 (4), 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, et al. , 2013. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl. Acad. Sci. U.S. A. 110 (19), 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner GH, 1984. Tumor heterogeneity. Cancer Res. 44 (6), 2259–2265. [PubMed] [Google Scholar]
- Baylin SB, Jones PA, 2011. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer 11 (10), 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ, 2013. Tumour heterogeneity and cancer cell plasticity. Nature 501 (7467), 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling AS, Gibson CJ, Ebert BL, 2017. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat. Rev. Cancer 17 (1), 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Lin Y, Navin N, 2020. Advancing cancer research and medicine with single-cell genomics. Cancer Cell 37 (4), 456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, et al. , 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343 (6172), 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, et al. , 2019. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 29 (9), 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern KB, et al. , 2017. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542 (7641), 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, et al. , 2019. Single-cell transcriptional profiling reveals the heterogenicity in colorectal cancer. Medicine (Baltimore) 98 (34), e16916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, et al. , 2020. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11 (1), 2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, et al. , 2016. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 48 (10), 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laks E, et al. , 2019. Clonal decomposition and DNA replication states defined by scaled single-cell genome sequencing. Cell 179 (5), 1207–1221 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, et al. , 2014. Discovery of biclonal origin and a novel oncogene SLC12A5 in colon cancer by single-cell sequencing. Cell Res. 24 (6), 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. , 2017. Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res. 27 (8), 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andor N, et al. , 2020. Joint single cell DNA-seq and RNA-seq of gastric cancer cell lines reveals rules of in vitro evolution. NAR Genom Bioinform. 2 (2), p. lqaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, et al. , 2020. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 30 (6), 1682–1689 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Humphreys BD, 2020. Single cell sequencing and kidney organoids generated from pluripotent stem cells. Clin. J. Am. Soc. Nephrol. 15 (4), 550–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, et al. , 2019. Single cell census of human kidney organoids shows reproducibility and diminished off-target cells after transplantation. Nat. Commun. 10 (1), 5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, et al. , 2020. Recurrent genetic abnormalities in human pluripotent stem cells: definition and routine detection in culture supernatant by targeted droplet digital PCR. Stem Cell Rep. 14 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Huch M, 2019. Disease modelling in human organoids. Dis. Model Mech 12 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]