Abstract
Zika virus (ZIKV), a mosquito-transmitted Flavivirus, emerged in the last decade causing serious diseases and affecting human health globally. Currently, no licensed vaccines or antivirals are available to combat ZIKV, although several vaccine candidates are in the pipeline. In recent years, the presence of non-canonical G-quadruplex (GQ) secondary structures in viral genomes has ignited significant attention as potential targets for antiviral strategy. In this study, we identified several novel conserved potential GQ structures by analyzing published ZIKV genome sequences using an in-house algorithm. Biophysical and biochemical analysis of the RNA sequences containing these potential GQ sequences suggested the existence of such structures in the ZIKV genomes. Studies with known GQ structure-binding and -stabilizing ligands such as Braco-19 and TMPyP4 provided support for this contention. The presence of these ligands in cell culture media led to significant inhibition of infectious ZIKV yield, as well as reduced viral genome replication and viral protein production. Overall, our results, for the first time, show that ZIKV replication can be inhibited by GQ structure-binding and -stabilizing compounds and suggest a new strategy against ZIKV infection mitigation and control.
Keywords: Zika virus, G-quadruplex structure, Braco-19, TMPyP4, virus growth, RNA synthesis, E protein
Graphical Abstract
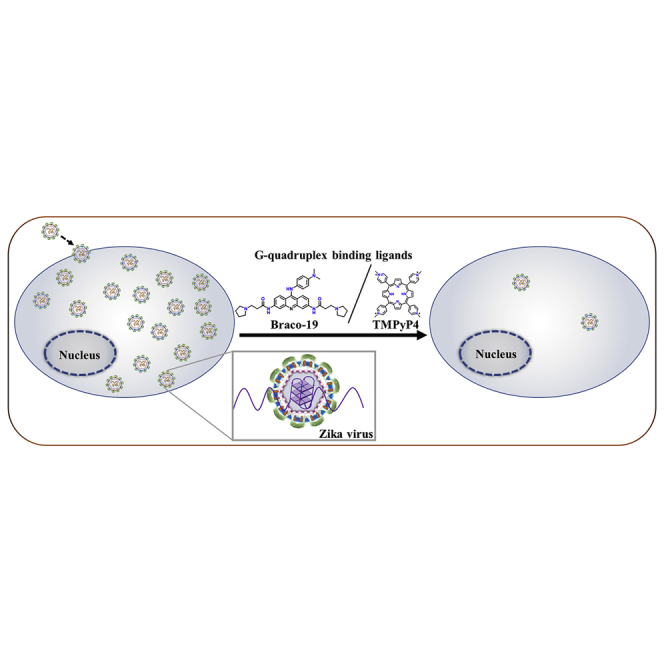
G-quadruplex structures are non-canonical secondary structures present in nucleic acid. In this manuscript, we targeted the Zika GQ structures with GQ-binding ligands like Braco-19 and TMPyP4 to inhibit ZIKV genome replication and transcription. Overall, 100 μM Braco-19 treatment produced >80-fold (96 hpi) reduction in viral titer production in cell culture, having a potential anti-viral effect.
Introduction
The emergence of the Zika virus (ZIKV) is of great concern worldwide in recent times. Though the virus was first isolated in 1947 in the Zika forest of Uganda,1 it came to prominence mainly after the 2007 outbreak for its association with serious congenital and neurological abnormalities.2, 3, 4, 5 The outbreaks, which occurred sporadically in parts of African and Asian countries, later manifested into major outbreaks in the Island of Yap, Federated States of Micronesia6 in 2007, French Polynesia7 in 2013, and the most explosive outbreak in Brazil8,9 in 2015. It is estimated that over 86 countries across the world have now witnessed ZIKV infections. The ZIKV infection may be asymptomatic or manifest as mild, self-limiting, flu-like febrile illnesses that are resolved within days. In certain cases, it can even give rise to congenital abnormalities and severe complications in children, including microcephaly, hydrocephaly, cerebral atrophy, ventriculomegaly, cortical malformations, intracranial calcifications, etc.2, 3, 4, 5,10 In adults, ZIKV infection may lead to fatal conditions involving neurological complications like acute auto-immune-mediated damage to peripheral nerves (Guillain-Barré syndrome), neuropathy, myelitis, meningoencephalitis, ischemic infarction, seizures, vasculopathy, etc.11,12
ZIKV, along with other mosquito-borne viruses like dengue virus (DENV), West-Nile virus (WNV), and Yellow fever virus (YFV), belongs to the Flaviviridae family. This enveloped virion consists of a positive-sense single-stranded RNA genome that codes for a single polyprotein that is further processed into three structural (capsid [C], precursor membrane [prM], and envelope [E] protein) and seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5).13 As there is no effective licensed vaccine or drug currently available, various strategies and approaches are being explored for the discovery of antiviral drugs against ZIKV. These include targeting viral proteins (structural and NS), repurposing the already approved drugs, etc.,14 and therefore further research on alternate strategies for the discovery of new and effective antiviral compounds is warranted.
The G-quadruplex (GQ)-forming sequences are being explored extensively as potential drug targets and have gained significant attention as potential antiviral strategies in recent years.15,16 The stretches of guanines in DNA or RNA sequences self-assemble to form a non-canonical secondary structure that can provide a specialized binding pocket for planar ligands.17 Targeting GQ structures for cancer treatment are being explored continuously for a very long time as they are reported to be extensively found in the telomeric region of the human chromosome.18 They are subsequently present in the promoter region of several proto-oncogenes like Bcl2 (B cell lymphoma 2)19 and c-Myc (cancer myelocytomatosis).20 Promisingly, researchers have designed ligands, which can specifically target these GQ sequences and inhibit the growth of cancer cells.21, 22, 23 These sequences also play significant role in neurodegenerative diseases like fragile X syndrome, Amyotrophic Lateral Sclerosis, and frontotemporal dementia.24,25 The search of biologically important GQ sequences has been ramified in other organisms as well, including yeast,26 bacteria,27, 28, 29 parasites,30 and viruses.15,16 Several viruses including ZIKV,31 human papillomaviruses (HPV),32 herpes simplex virus (HSV),33 and human immunodeficiency virus (HIV)34 are reported to bear GQ-forming sequences in their genome. Founded on this, the GQ-binding ligands such as Braco-19 and core-extended naphthalene diimides are reported to have a significant antiviral effect on HIV replication.35,36 Another potent GQ-binding ligand, TMPyP4, was found to stabilize the GQ structure in the Ebola virus polymerase (L) gene, thereby repressing its expression at the RNA transcription level.37 A similar effect of TMPyP4 was also observed in the case of the core gene of hepatitis C virus (HCV).38 These studies provide a framework for further research on the potential use of GQ-binding and -stabilizing ligands for antiviral strategies.15,16
In a recent report, Fleming et al.31 first reported the presence of putative GQ-forming sequences in the ZIKV genome that are largely conserved among the Flaviviridae family, although the functional consequences of the presence of such structures were not examined by the investigators. In this study, we have discovered a number of additional GQ-structure-forming sequences that are exclusively conserved in the ZIKV genome in addition to those reported by Fleming et al.31 Taking ZIKV genomic RNA sequences carrying several of these potential GQ structures, and by using various assays such as fluorescence titration assay, nuclear magnetic resonance (NMR) spectral analysis, isothermal titration calorimetric (ITC) assay, and thermal denaturation experiments, we show that (1) the GQ structures do in fact exist in these RNA sequences, (2) the GQ-binding ligands such as Braco-19 and TMPyP4 bind specifically to these structures and stabilize them, and (3) treatment of ZIKV-infected cells with these ligands results in significant reduction in infectious virus production, as well as inhibition of viral genome replication and viral protein production in a dose-dependent manner. Thus, GQ-ligand binding strategy displayed in this study stands as a proof of concept and appears promising toward the development of potential antiviral agents against ZIKV infection.
Results
ZIKV genome has potential GQ structures conserved across the strains
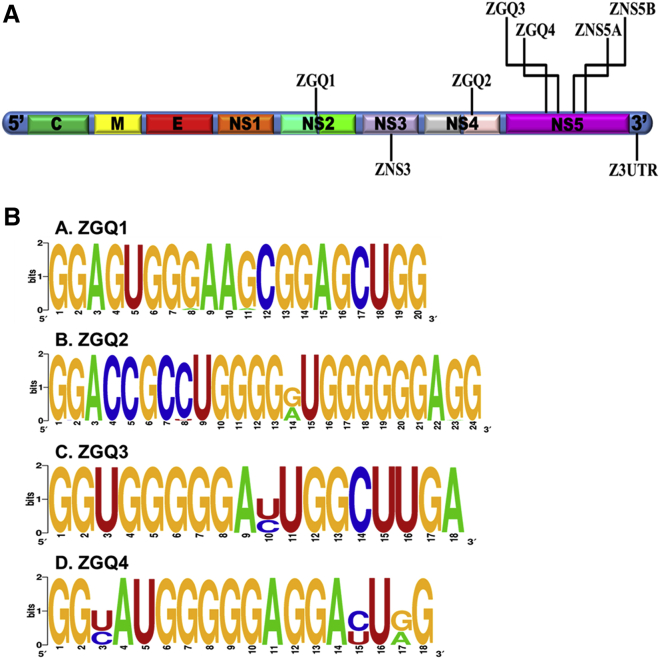
Earlier reports suggest that Flaviviruses including the ZIKV contain conserved GQ-forming sequences.31 By employing more stringent criteria where the number of guanine repeat was kept ≥2 and the loop length was restricted between 1 and 7, we identified four new ZIKV GQs (ZGQs) conserved in the NS genes such as NS2, NS4B, and NS5 genes of ZIKV genome in addition to the other ZGQs identified previously (Figure 1A).31 Importantly, the genome-wide mining of 247 ZIKV isolates revealed that these new ZGQs are conserved among >90% of the ZIKV strains (Figure 1B; Tables S1, S2, and S9). As Figure 1B displays, the WebLogo representation of the newly predicted four ZGQ sequences shows that the guanine residues involved in the GQ formation are highly conserved among the different evaluated strains and the residues present in the loop region are mostly divergent. Because ZIKV is an RNA virus, it is prone to a higher mutation rate,39 and thus, the conservation of these ZGQs across the isolates suggests functional importance associated with viral life cycle and pathogenesis. In addition, we have also identified highly conserved ZGQ sequence motifs in the viral anti-genomic (negative-sense) RNA (Figure S1; Table S3).
Figure 1.
G-quadruplex (GQ) structures are conserved in ZIKV genome
(A) Schematic diagram showing the location of the GQ sequences predicted in the ZIKV genome. The seven ZGQ sequences used in the study are distributed in the NS genes, namely, NS2, NS3, NS4B, and NS5, and one in the 3′ UTR region. (B) WebLogo representation of the newly predicted four ZGQ sequences showing conservation of the nucleotides among different strains. The height of the letter indicates the relative frequency of the particular nucleotides at that position, therefore reflecting the conservation of that particular nucleotide.
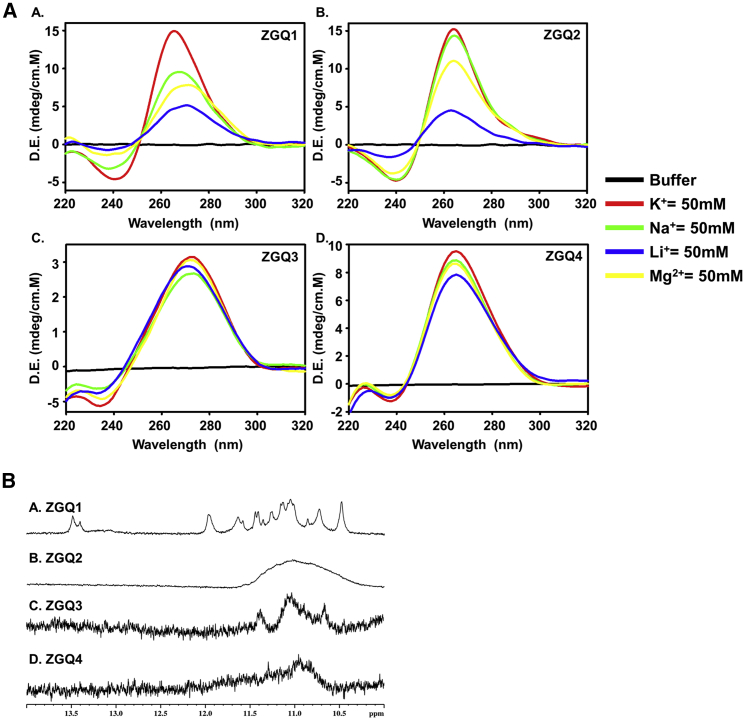
After identifying the ZGQ sequences in the positive-sense genomic RNA, we conducted circular dichroism (CD) spectroscopy and nuclear magnetic resonance (NMR) spectroscopy studies to determine whether they would form the GQ structures in vitro. It is well documented that these RNA GQ-forming sequences show less polymorphism in topology as compared to the DNA GQs and they are usually observed to form parallel topology, with one reported exception, i.e., Spinach, as it forms the anti-parallel topology.17 Consistent with this notion, we observed that the newly predicted ZGQs formed the signature pattern for parallel topology in the CD spectra, i.e., a negative peak at ∼240 nm and a positive peak at ∼265 nm in the presence of different cations (Figure 2A). Additionally, the 1H NMR spectra showed distinct chemical shift signals between 10.5 and 12 ppm, which depict the characteristic peaks of the imino region embracing the Hoogsteen base pairing in the GQ structures (Figure 2B). These results are consistent with the formation of ZGQ structures within the ZIKV RNA sequences examined here.
Figure 2.
Biophysical characterization of ZGQs
(A) CD spectra analysis of ZGQs showing parallel RNA topology in the presence of cations, namely, K+, Na+, Li+, and Mg2+. (B) 1H NMR spectra analysis of the ZGQs showing the signature imino proton peaks in the predicted four ZGQs, thus confirming the Hoogsteen bond formation in GQ structure.
Interaction of potential ZGQs with GQ-binding ligand Braco-19 demonstrated by fluorescence-binding assay
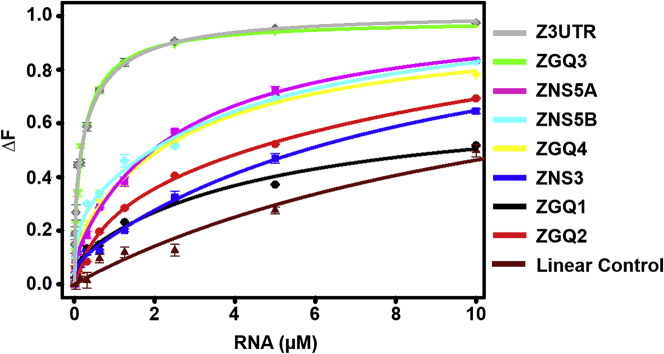
Along with the advent of predicting and analyzing GQ structures, the development of small-molecule ligands, which could specifically target these GQ structures, have also been intensified. GQ-specific molecules like Braco-19 and TMPyP4 are extensively used for targeting GQ structures in the telomere region,40, 41, 42 in viruses like in HIV35 and HSV.33 These particular molecules are immensely used to distinguish and characterize GQ forming sequences from the canonical DNA or RNA sequences. Thus, we evaluated the interaction ability of these GQ-specific ligands with the predicted ZGQs. We performed fluorescence-binding assays of Braco-19 with all the four newly predicted ZGQs along with the earlier reported GQs (ZNS3, ZNS5A, ZNS5B, and Z3UTR).31 This assay is a simple optical assay that makes use of the intrinsic fluorescence property of the ligand molecule. Fluorescence-binding assay helps to determine the binding affinity between two molecules (in our case, the GQ-specific ligand, Braco-19, and the RNA) and obtain a quantitative value in the form of dissociation constant (KD).43 The binding efficiency between the ligand and the biological molecules, i.e., RNA, is evaluated through the difference between the fluorescence intensity observed before the addition of the RNA and after its interaction with the ligand. The successful interaction results in either increase (enhancement) or decrease in fluorescence intensity (quenching). This change in fluorescence (ΔF) is plotted with respect to the concentration of RNA (GQs or canonical) using global curve fitting methods to obtain the KD values for each interaction. The respective KD value is used to analyze the binding affinity of the fluorescent molecule with each nucleic acid. In the present study, all the ZGQs showed higher binding affinity (>100-fold higher) for Braco-19 in comparison to the duplex control RNA sequence (Figure 3; Table S4). The variation in binding affinity of different ZGQs with Braco-19 is probably due to different polymorphic structures formed by the GQ-sequences and so the ligand molecules bind with the different RNA sequences with varying intensity. It should be noted that a higher affinity of Braco-19 toward the GQ sequences is attributed by the π-π interactions between the aromatic region of the ligand and the GQ-forming scaffolds of the ZGQs. Moreover, Braco-19 contains two pyrrolidine rings, which contribute to the cationic nature of the molecule and help to interact with the anionic core of the GQ structures. All these properties contribute to the mode of binding between the ligand and the biomolecule and thereby can exhibit different binding modes, for example, groove binding or end-stacking modes. In our study, all the sequences were best fitted in the two-mode binding model depicting more than one site being present in the RNA sequences for the ligand binding. The KD1 values were higher in each case, confirming preferential binding mode and better affinity of the ligand toward the ZGQ sequences.
Figure 3.
Fluorescence titration studies of Braco-19 with different ZGQ RNAs
The solid, colored lines represent the plot for fitting the data obtained due to the interactions of Braco-19 with the respective RNAs by two-mode binding. Table S4 lists the binding constant values (KD) obtained after fitting the plots.
The higher binding affinity of Braco-19 with ZGQs confirmed by ITC
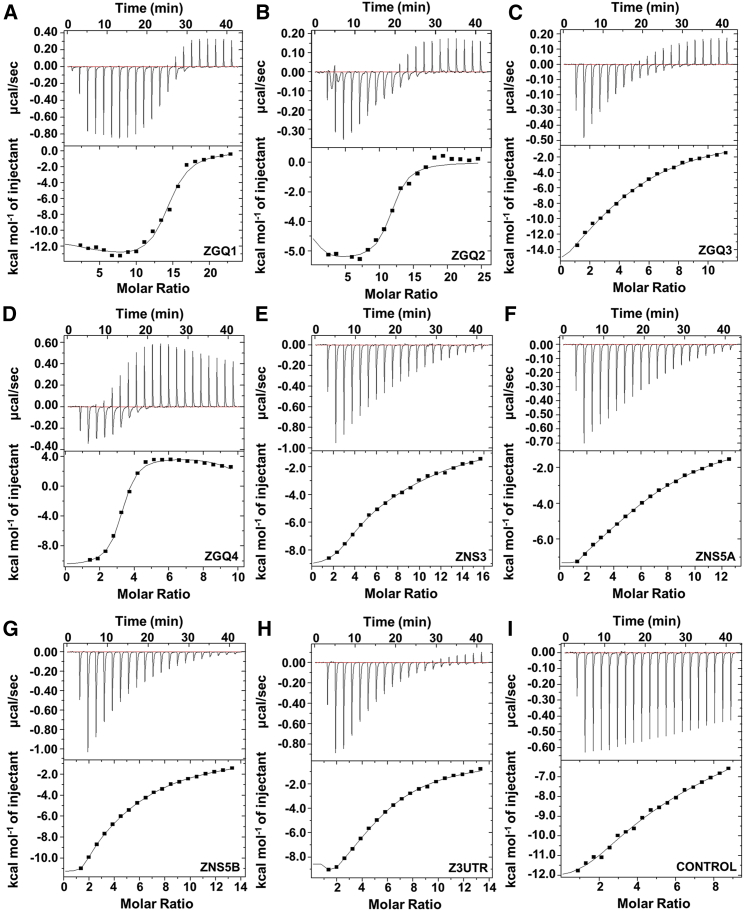
ITC is regarded as the gold standard technique for determining the thermodynamic properties of a bio-molecular interaction. Unlike the fluorescence-binding assay, it not only provides the binding constant values but also offers insight into the thermodynamic parameters including enthalpy changes, stoichiometry value, entropy changes, and Gibbs free energy. Thus, we performed ITC studies using the ZGQs and Braco-19 to have better insight regarding the interaction profile. Interestingly, the binding affinity values between Braco-19 and ZGQs were observed in μM scale, while with the control duplex RNA, the value ranged in mM scale confirming that the GQ specific ligand, Braco-19 has a higher selectivity for the ZGQ-forming structures over the non-GQ-forming sequences (Figure 4; Table S5). ITC was also performed for the ZGQs with TMPyP4 and in similar manner; TMPyP4 also showed a higher affinity for the GQ sequences (Figure S2; Table S7). In certain binding thermograms, for example in Figures 4A–4D, we find endothermic binding where the saturation level is low, while the reactions turn exothermic at higher levels of saturation. This implies that the stronger binding of the ligand to the RNA is entropically driven while those that show weaker binding are enthalpically directed.44
Figure 4.
Braco-19 has a higher binding affinity and thermal profile for ZGQs
Isothermal titration calorimetric experiment showing the binding affinity between the ZGQ RNAs and Braco-19 (A: ZGQ1, B: ZGQ2, C: ZGQ3, D: ZGQ4, E: ZNS3, F: ZNS5A, G: ZNS5B, H: Z3UTR and I: Duplex RNA-Control). The top panel represents the power in μcal unit versus time in s while the bottom panel depicts the thermogram of the integrated peak intensities plotted against the molar ratio. The binding constant values for all the ZGQ RNAs along with the control RNA are listed in Table S5.
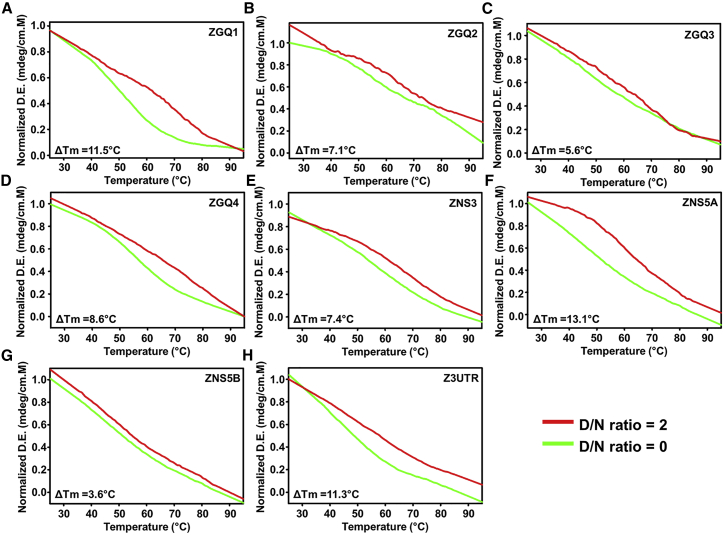
Thermal denaturation profile shows the stabilization of the ZGQs by Braco-19
The thermal denaturation experiments illustrate the stability of the GQ structures. The gradual increase in temperature leads to a change in conformation of the nucleic acid structure due to disruption of the bonds between the bases making it less stable.45 The higher melting temperature (Tm) signifies higher stability of the nucleic acid conformation. In our assays, when the ligand, Braco-19, was added in double the molar concentration to that of the ZGQ RNA solution, we observed a significant increase in Tm values. Tm differences of 3.6°C to 13.1°C were noted for different ZGQs on interaction with Braco-19 (Figure 5; Table S6). The ligand molecule interacts with the GQ structures, thereby stabilizing them by forming non-covalent bonds. On interaction with the ligand molecules, these GQ structures become relatively more stable at higher temperature, which in turn increases the melting temperature.
Figure 5.
Thermal denaturation analysis showing stabilization ZGQs in the presence of Braco-19
The increase in Tm depicts the stable interaction of the ZGQs with Braco-19 (A: ZGQ1, B: ZGQ2, C: ZGQ3, D: ZGQ4, E: ZNS3, F: ZNS5A, G: ZNS5B and H: Z3UTR). The Tm difference (noted in the left bottom corner) is significant for all the ZGQ RNAs (also see Table S6).
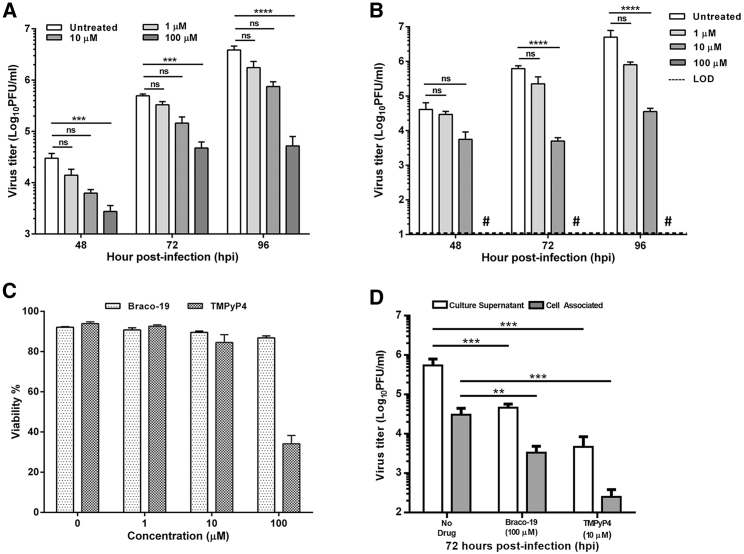
ZIKV growth is inhibited in vitro in the presence of GQ-binding compounds Braco-19 and TMPyP4
The studies described above suggest that the ZIKV genome contains GQ structures that can be stabilized through the binding of Braco-19, as well as TMPyP4. To determine whether the presence of such structures and stabilizing them through binding with Braco-19 could impact ZIKV growth in cultured cells, we performed experiments to assess ZIKV growth in Vero cells in the presence of varying concentrations of the drug. Vero cells infected with MR766 isolate of ZIKV were treated with Braco-19 and infectious virus yield in the supernatants at various time points was measured by plaque assay. In response to Braco-19 treatment, starting from 48 h post-infection, the total infectious virus yield showed a significant reduction in a dose-dependent manner, at each of the time points examined (Figure 6A). Strikingly, at 100 μM of Braco-19 (cell viability was greater than 85% (Figure 6C) at 100 μM concentration), inhibition of virus growth was most significant (>80-fold, p ≤ 0.0001) at 96 hpi compared to the untreated control (Figure 6A). We also investigated the effect of TMPyP4, another GQ-binding molecule on virus yield in a similar assay. Our results showed that TMPyP4 also inhibited virus growth significantly (Figure 6B). At a concentration of 10 μM, at which cell viability was over 80% (Figure 6C), the infectious virus yield in the supernatants at 72 hpi and 96 hpi was reduced nearly 120-fold and 170-fold, respectively, compared to the untreated control (Figure 6B). However, at 100 μM concentration, the infectious virus yield in the supernatant was below the limit of detection at any of the time points studied (Figure 6B), even though the cell viability was nearly 40% (Figure 6C). Overall, these results suggest that GQ-binding compounds Braco-19 and TMPyP4 inhibit ZIKV growth in vitro. To determine whether the lower levels of infectious virus in the culture supernatants in the presence of the Braco-19 or TMPyP4 were a result of increased accumulation of cell-associated infectious virus, we quantitated the amount of virus present in the cells. As can be seen in Figure 6D, on average, cell-associated virus represented about 17-fold less than the virus released in the supernatants of infected cells under no-drug treatment condition at 72 hpi. In the presence of 100 μM Braco-19 or 10 μM TMPyP4, cell-associated virus represented approximately 14- to 15-fold less than that in the corresponding supernatants. Importantly, the cell-associated virus in drug-treated cells were significantly lower than those in untreated cells (Figure 6D). These results show that both Braco-19 and TMPyP4 inhibit infectious ZIKV progeny production.
Figure 6.
Inhibition of ZIKV growth in cells treated with Braco-19 and TMPyP4
(A and B) Vero cells infected with ZIKV at an MOI of 1 were incubated in medium containing 0 (untreated), 1 μM, 10 μM, and 100 μM concentrations of Braco-19 (A) or TMPyP4 (B). Aliquots of culture supernatants were collected at various times (h post-infection, hpi) and infectious virus titers were determined by plaque assay. The graphs show mean values of virus titers, with error bars representing standard deviations from the results of four independent experiments. Two-way ANOVA was used to determine significant differences in virus titers between untreated and drug-treated samples. LOD, the limit of detection. # represent values below LOD. ∗∗∗∗p ≤ 0.0001; ∗∗∗p ≤ 0.001; ns, nonsignificant. (C) Cell viability in the presence of Braco-19 or TMPyP4. Vero cells were exposed to 0, 1, 10, and 100 μM Braco-19 or TMPyP4 for 4 days and percentage of viable of cells was measured. Mean values from three independent experiments with error bars representing standard deviations are shown. (D) Vero cells were infected with ZIKV at an MOI of 1 and treated with 100 μM Braco-19 or 10 μM TMPyP4 for 72 hpi. Infectious virus in the supernatants and cells were quantitated by plaque assay. The graphs show mean values of virus titers, with error bars representing standard deviations from the results of three independent experiments. Two-way ANOVA was used to determine significant differences in virus titers between untreated and drug-treated samples. ∗∗∗p ≤ 0.001; ∗∗p ≤ 0.01.
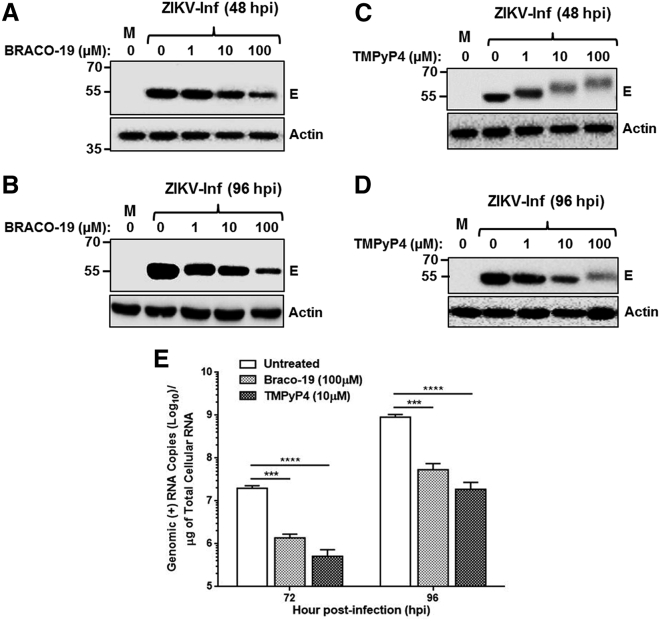
Inhibition of viral protein synthesis and genome replication in infected cells treated with Braco-19 and TMPyP4
Since Braco-19 and TMPyP4 treatment reduced infectious ZIKV yield, we next sought to find out whether this is due to lower viral protein expression in infected cells. We examined the E protein expression levels in virus-infected cells in the presence of Braco-19 or TMPyP4. Since the ZIKV genome is translated into a single large polyprotein, which is processed to yield 10 different proteins including the E protein, we picked the E protein as an indicator viral protein production. Vero cells, infected with MR766 ZIKV, were treated with varying concentrations of Braco-19 or TMPyP4 immediately after infection. Cell lysates were prepared from the infected cells at 48 or 96 hpi and analyzed for E protein levels by western blot, which showed a marked reduction in E protein expression with increasing concentrations of Braco-19 at both 48 hpi and 96 hpi (Figures 7A and 7B). The reduction of E protein levels was more pronounced at 100 μM Braco-19 and at 96 hpi. In response to TMPyP4 treatment, E protein expression showed observable reduction over time (Figure 7C). Surprisingly, the electrophoretic mobility of the E protein was demonstrably altered at 48 hpi time point (Figure 7C), with increasing dose of TMPyP4 resulting in increased molecular mass of the E protein. Interestingly at 96 hpi, the reduced electrophoretic mobility of E was only observed with 100 μM TMPyP4 (lane 5, Figure 7D) and was also less pronounced. The reason for the observed electrophoretic mobility shift in E protein in the presence of TMPyP4 is unclear at this time but it is possible that metabolic byproducts of TMPyP4, known to be generated in cells could bind to the E protein in a time- and concentration-dependent manner to reduce the mobility of the E protein (see Discussion for further details). Nevertheless, our results suggest that both Braco-19 and TMPyP4 inhibit viral protein expression in infected cells.
Figure 7.
Braco-19 and TMPyP4 inhibit viral protein synthesis and genome replication
(A–D) Vero cells were either mock-infected (M) or infected with ZIKV at an MOI of 1 and subsequently incubated in medium containing 0, 1 μM, 10 μM, and 100 μM concentrations of Braco-19 (A and B) or TMPyP4 (C and D). Cell extracts were prepared at 48 hpi (A and C) or 96 hpi (B and D) and 20 μg of proteins from each sample were analyzed by SDS-PAGE and western blotting with 4G2 antibody to detect viral E protein. Levels of actin detected in the blots using anti-actin antibody served as the loading control. Molecular mass markers (in kDa) are shown on the left. Representative data are shown in each panel. (E) Levels of viral genome in the presence of Braco-19 or TMPyP4. Vero cells infected as above were and incubated in medium without drug (untreated) or containing 100 μM Braco-19 or 10 μM TMPyP4. Viral genome copies in the cells at 72 and 96 hpi were quantitated and expressed as copies per μg of total cellular RNA from the cells. The graphs show mean values of genome copy numbers with error bars representing standard deviations from the results of three independent experiments. Two-way ANOVA was used to determine significant differences between untreated and drug-treated samples. ∗∗∗∗p ≤ 0.0001; ∗∗∗p ≤ 0.001.
We next sought to determine whether intracellular replication of the ZIKV genome is negatively affected by the GQ-binding ligands. Vero cells, infected with ZIKV MR766, were treated with Braco-19, TMPyP4, or left untreated and total RNA from the cells was extracted at 72 and 96 hpi. Total RNA was subjected to qRT-PCR with ZIKV-specific primers and probes to determine the copy numbers of the viral positive-sense genome. In these experiments, we used 100 μM Braco-19 because this concentration exhibited maximum inhibition without adversely affecting cell viability and 10 μM TMPyP4 because higher concentration of TMPyP4 was found to reduce cell viability substantially (Figure 6C). Results from the experiment showed that ZIKV genome copies were significantly lower in cells treated with Braco-19 or TMPyP4 at both 72 and 96 hpi (Figure 7E), suggesting that these GQ-binding molecules inhibit ZIKV genome replication in infected cells. Overall, from these studies we conclude that GQ-binding drugs inhibit both viral genome replication, as well as translation.
Discussion
Due to the unavailability of commercial vaccines or antiviral drugs, control and mitigation strategies for ZIKV infection remain a huge challenge to date. The unpredictable nature of ZIKV outbreaks and the global prevalence of its vector have further worsened the situation. Although attempts to repurpose the drugs like Sofosbuvir,46 Niclosamide,47 and Galidesivir48 have shown pronounced anti-Zika activity, these drugs also come with certain shortcomings such as poor bioavailability, poor liberation, and improper mode of administration. If infected during pregnancy, ZIKV can cause detrimental effects on the fetus and eventually on the newborn. Prognosis of newborn babies infected with ZIKV is unfavorable and infections could lead to permanent congenital defects. Thus, utmost precaution and precision should be taken while strategizing therapeutic countermeasures against ZIKV infection.
In recent years, GQ structures, which are non-canonical nucleic acid secondary structures, are being discovered in almost all organisms including many viral genomes and thus have been considered as potential targets for antiviral strategies.16 ZIKV and other Flaviviruses are earlier reported to possess conserved GQ structures.31 By expanding the analysis to all the available ZIKV genome sequences published in the NCBI Genome database and employing an in-house algorithm,49 we were able to predict four additional highly conserved GQ structures in ZIKV. Some GQ-forming sequences were also found to be conserved in the anti-genomic strand of the ZIKV (replication intermediate; Figure S1; Table S3). However, further studies will be required to validate whether they actually form GQs. The current study mainly focuses on the GQ structures formed in the genomic RNA (positive polarity) of the ZIKV. The observation that these GQ structures are highly conserved suggested to us that they may play a specific role in the biology and/or pathogenesis of the virus. Two of the GQs were found to be located in the coding region of NS5 gene, i.e., the RNA-dependent RNA polymerase coding gene, while the other two were present in the NS2 and NS4B gene. These GQs were characterized by CD and NMR spectra analysis and were found to form parallel GQ structures. Moreover, it has been previously shown that RNA GQ structures are more stable compared to their DNA counterparts due to the presence of the 2′ hydroxyl group in the ribose sugar. Following CD and NMR spectra studies, we evaluated whether these GQ structures would interact with the known GQ-binding ligands. GQ-binding ligands such as Braco-19, TMPyP4, etc. have been widely studied to measure the stability of the GQ structures and are presently being explored as an antiviral strategy. The ligands bear certain characteristics like basic aromatic rings, electron-deficient cationic core, and positively charged side chains, which facilitate the efficient interaction of these ligands with the GQ secondary structures.50 The mode of interaction of these ligands may be through end-stacking, groove binding, or interaction with the backbone or loop sequences. The molecules fit themselves within these GQ structures and bind through various non-covalent bonds thus stabilizing the structure. Further biophysical studies including the fluorescence titration assay, ITC, and thermal denaturation assay conclusively demonstrated that the Braco-19 and TMPyP4 bind selectively and preferentially to the GQ-forming sequences in comparison to the non-GQ linear RNA sequences. Our studies unequivocally demonstrated the existence and formation of GQ structures in the genome of ZIKV.
We reasoned that the binding of the Braco-19 or TMPyP4 could potentially impact the function(s) of the viral RNA containing the GQ structures. Our in vitro studies in cells infected with ZIKV and treated with Braco-19 or TMPyP4 suggested that both of these compounds inhibit the activities of the RNA template. Viral protein synthesis was substantially reduced in a dose-dependent manner as evidenced by the reduced levels of the E protein in the cells compared to control cells. Additionally, it is possible that binding of these compounds to the GQ structures inhibited the movement of the viral RNA polymerase, resulting in overall reduction of viral RNA copy numbers as we have observed in our experiments. Although our studies revealed the existence of multiple functional GQ structures in the viral genome, it would be interesting to examine whether a particular GQ structure functions more prominently than the other structures. In this regard, further studies are being planned to identify a specific contribution of these GQ structures in inhibiting viral genome replication and translation.
Examination of E protein in ZIKV-infected cells treated with TMPyP4 revealed slower migrating forms of the protein (Figures 7C and 7D). It appears to be specific for E protein since such mobility differences were not observed with another viral protein, the NS1 protein (data not shown). The molecular basis for the differences in mobility of E compared to that in TMPyP4-untreated cells is not clear at this time. However, it is known that TMPyP4 becomes metabolized51 in cells through oxidation by hemeoxygenase enzymes (HO-1 and HO-2) to produce 4-formyl-1-methylpyridinium (4F-MP) and 4-carboxyl-1-methylpyridinium (4C-MP). Since these metabolites can bind to certain cellular proteins such as acetylcholinesterase,51 it is possible that 4F-MP and/or 4C-MP bind specifically to the viral E protein in a time- and concentration-dependent manner to reduce its mobility. Although our results are consistent with such an interpretation, further research is needed to determine the molecular basis of the observed mobility shift in the E protein.
Taken together, our studies provide the first evidence that highly conserved GQ structures present in ZIKV genome and the ligands that bind and stabilize these GQ structures can potently inhibit ZIKV growth, genome replication, and viral protein expression in cultured cells. However, the two ligands evaluated here are the most common GQ-specific ligands used regularly to characterize these secondary structures and not highly specific to the particular ZIKV GQs. Moreover, the toxicity and bioavailability of these ligands pose a hindrance for their potential use as antiviral drugs. Though this piece of research stands as a proof of concept for using GQ-binding ligands as anti-ZIKV drugs, development of sequence-specific ligands needs further research.16 Moreover, the availability of ligands that can potentially target ZIKV GQ sequences and that have the ability to distinguish them from the host GQs can address the problem of off-target effects of the ligands. Thus, these results open the possibility for understanding the role of GQ structures in ZIKV life cycle and pathogenesis, as well as for use of these structures as targets for development of innovative and efficacious antiviral strategies.
Materials and methods
Oligonucleotides and reagents
Small-molecule ligands, namely Braco-19 and TMPyP4, were purchased from Sigma Aldrich Chemicals, dissolved in water or DMSO, respectively, for preparation of stock solutions, and stored at 4°C until further use. Other reagents and chemicals including KCl, KH2PO4, K2HPO4, DMSO, and D2O were procured from Sigma Aldrich Chemicals or HiMedia Laboratories. The DNA oligonucleotides and the RNA oligonucleotides used in the studies were obtained from Sigma Aldrich Chemicals and IDT Technologies, respectively (Table S8).
Prediction of putative GQ-forming sequences from the ZIKV genome
A total of 247 complete genome sequences of ZIKV were retrieved from the NCBI Genome database (https://www.ncbi.nlm.nih.gov/genomes). The FASTA sequences of the ZIKV genomes were then used for the prediction of the putative GQ-forming sequences with the help of our in-house GQ prediction tool.48 The search was based on the following algorithm:
where G corresponds to guanine and N corresponds to any base including guanine. Though the ZIKV is a positive-sense RNA virus, we also sought to find the probable GQ sequences in the anti-sense strand of the viral genome. Further, the consensus sequences for each of the ZGQs studied were generated using the WebLogo software.52
CD melting analysis
J-815 spectropolarimeter (JASCO) attached with a Peltier junction temperature controller was used to perform the CD experiment. With continuous supply of nitrogen gas, the required temperature was maintained to avoid condensation of water along the cuvette. 20 μM RNA sample dissolved in Tris buffer (10 mM, pH = 7.4, supplemented with the required cation) was used for the CD spectra analysis. For the CD spectra analysis, the wavelength range of 220–320 nm was scanned at the speed of 20 nm/min while the thermal denaturation experiments were carried out at ∼265 nm at a temperature range of 25°C to 98°C. The ligand was added gradually in increasing concentration to the ZGQ RNA solution. The experiments were done in triplicate and data were obtained and normalized by buffer subtraction. Following this, the result was plotted using SigmaPlot 12.0 software (Systat Software, Chicago, IL, USA).
NMR spectroscopy
The NMR experiment was carried on in the AVANCE 400 MHz (BioSpin International AG, Switzerland) spectrometer supported with the 5 mm broadband inverse (BBI) probe as reported earlier.28
Fluorescence-binding assay
The fluorescence titration experiment was performed using the Synergy H1 multi-mode plate reader. 2 nM of ligand was added to each of the 12 wells of a 96-well plate and gradually the target RNA was titrated in each of the wells (0–10 μM). Precisely, the highest concentration of RNA, i.e., 10 μM, was added to first well, followed by dilution in the subsequent well; for example, the second well then contained 5 μM, third well contained 2.5 μM, and so on until the eleventh well while no RNA was added in the last well (twelfth well) serving as blank. The enhancement in emission fluorescence was observed at specific excitation of the ligand molecule (λex = 360 nm and λem = 440 nm for Braco-19) on addition of RNA and the change in emission fluorescence (ΔF) for all datasets was normalized to the last well (containing no RNA). The two-site saturation using global curve fitting was used to extrapolate the curve between ΔF and concentration of RNA (μM) using SigmaPlot 12.5 software (Systat Software, Chicago, IL, USA) according to the following equation that accounts for two receptor binding sites with two different affinities, KD1 and KD2:
where Bmax is the maximum number of binding sites and KD is the dissociation equilibrium constant. Each binding study was performed in duplicate at room temperature (RT) and duplex RNA was used as the control.
ITC experiment
The ITC experiments were performed using the MicroCal iTC200 isothermal titration calorimeter (GE Healthcare) instrument. The RNA sample was prepared in potassium phosphate buffer and loaded in the cell while the ligand was loaded in the syringe. A total of 21 injections were performed where each injection delivered 1.78 μL of ligand and was of 3.56 s duration with a gap of 120 s between two. An initial injection of 0.4 μL having 60 s initial equilibrium delays was maintained during each experiment. Throughout the experimental period, 25°C temperature and 750 rpm speed were sustained. The data obtained were fitted to the two-site binding model and analyzed using the Origin scientific software version 7 (Microcal Software).
Cells and viruses
Vero (Cercopithecus aethiops, CCL-81) cells were obtained from American Type Culture Collection (ATCC). The cells were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS) and penicillin/streptomycin (PS) in humidified chamber with 5% CO2 at 37°C. The infectious clone-derived MR766 strain of ZIKV53 generated in our laboratory was used in this study. For determination of virus growth in the presence of Braco-19 or TMPyP4, Vero cells were infected with the virus at a multiplicity of infection (MOI) of 1 plaque-forming unit per cell (PFU/cell). Following adsorption at 37°C for 1 h, the inoculum was removed and replaced with virus growth medium as described before53,54 containing appropriate concentrations of Braco-19 or TMPyP4. Infected cells were incubated at 37°C and small aliquots of infected cell culture supernatants were collected at various hpi, clarified, and stored at −80°C for virus titration at a later time. To determine the amount of cell-associated infectious virus, after collecting culture supernatants, we subjected the cells to two cycles of freeze-thaw at −80°C in the presence of the same volume of growth medium followed by centrifugation at 17,000 × g for 10 min at 4°C. The clarified supernatants were then used to quantify the cell-associated virus. Virus quantitation was performed by plaque assay on Vero cells as described previously.53
Western blotting for E protein
ZIKV E protein was detected in infected cells treated without or with the GQ-binding ligands at appropriate concentrations. Cell extracts were prepared, and the viral E protein was detected by western blotting as described before.53 Anti-ZIKV E protein antibody was obtained from Genetex (Cat. # GTX1333325) and goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP)-conjugated (Cat. # A6154) was procured from Sigma Aldrich. Mock-infected or virus-infected cells were washed with cold PBS, trypsinized, and harvested at 500 × g for 5 min at 4°C. The cell pellet was resuspended in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) supplemented with the protease and phosphatase inhibitor cocktail and incubated on ice for 15 min with intermittent vortexing after which the lysates were clarified by centrifugation at 10,000 × g for 5 min, 4°C. Total protein quantification was done using the bicinchoninic acid (BCA) assay kit. 20 μg of total protein was separated by electrophoresis on a required percentage of polyacrylamide gel (PAGE) containing sodium dodecyl sulfate (SDS). The separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes using a Bio-Rad semi-dry transfer system. Non-specific protein binding was blocked with 5% non-fat milk or with bovine serum albumin (for phosphorylated proteins) in Tris-buffered saline containing 0.2% Tween 20 (TBS-T) and incubated with the appropriate primary antibodies (dilution: 1:5,000 for β-actin and 1:1,000 for all others) at 4°C overnight. Membranes were washed with TBS-T and incubated with appropriate HRP-conjugated secondary antibodies (dilution: 1:5,000) at RT for 2 h. Bands were detected using enhanced chemiluminescence (ECL) western blotting substrate using a BioRad imager.
Quantitative real-time PCR
Total RNA from infected cells at 72 or 96 hpi were isolated using TRIzol RNA isolation reagent (Thermo Fisher Scientific) and viral genomic copy numbers were determined using specific probes and primers as described in detail previously53 and expressed as log10 copies per μg of total cellular RNA. The forward primer (5′-GTCGTTGCCCAACACAAG-3′) and the reverse primer (5′-CCACTAATGTTCTTTTGCAGAC-3′) along with the fluorescent probe 5′-/56-FAM (5′-6-carboxyfluorescein)/AGCCTACCT/ZEN/TGACAAGCAATCAGACACTCAA/3IABkFQ (3′-Iowa black fluorescent quencher)/-3′ were used to determine the standard curve. The real-time PCR assays involved two steps. The first step included the cDNA synthesis from the viral RNA using the SuperScript II (Thermo Fisher Scientific, Waltham, MA, USA). The synthesized cDNAs were then used as templates in the qPCR. The qPCR cycling conditions used were as follows: initial denaturation at 95°C for 10 mins; 35 cycles of 95°C for 30 s, 55.3°C for 30 s, and 72°C for 30 s; and a melt curve from 65°C to 95°C with 0.5°C increments for 5 s. The reactions were run in CFX Connect instrument (Bio-Rad, Hercules, CA, USA). Viral-genome copy numbers were determined in parallel reactions using RNA samples in duplicate and expressed as copy numbers per μg of total cellular RNA.
Determination of cell viability by flow cytometry
Cell viability was determined by simultaneous determination of propidium iodide (PI, 1 μg/mL, Sigma Aldrich, P4170) uptake as a marker of plasma membrane integrity loss and changes in the forward scatter properties of the cell (cell size) using flow cytometry (fluorescence-activated cell sorting [FACS]). Flow cytometry and data analysis were performed as described previously.55
Statistical analysis
Data were analyzed using GraphPad Prism software version 6.0. Two-way ANOVA was used to determine significant differences between untreated and drug-treated samples for viral loads (RNA copies, infectious titer). Data were represented as either mean ± SEM or mean ± SD.
Acknowledgments
The research was supported in part by funds from the University of Nebraska-Lincoln, USA. P.M. and U.S. are thankful to the Ministry of Human Resource Development (MHRD), Government of India, New Delhi, India for their individual fellowship. We are thankful to Sophisticated Instrument Centre, IIT Indore for the CD and NMR facilities.
Author contributions
P.M. conducted biophysical and biochemical characterization and wrote the manuscript; A.P. and B.R.S. performed antiviral assays; U.S. performed NMR analysis; A.K., D.N., and A.K.P. conceptualized and executed the project and edited the manuscript; and A.K.P. acquired funding.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.12.030.
Contributor Information
Asit K. Pattnaik, Email: apattnaik2@unl.edu.
Amit Kumar, Email: amitk@iiti.ac.in.
Debasis Nayak, Email: nayakdn@iiti.ac.in.
Supplemental information
References
- 1.Dick G.W.A., Kitchen S.F., Haddow A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Chan J.F.W., Choi G.K.Y., Yip C.C.Y., Cheng V.C.C., Yuen K.-Y. Zika fever and congenital Zika syndrome: An unexpected emerging arboviral disease. J. Infect. 2016;72:507–524. doi: 10.1016/j.jinf.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musso D., Gubler D.J. Zika Virus. Clin. Microbiol. Rev. 2016;29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazear H.M., Diamond M.S. Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J. Virol. 2016;90:4864–4875. doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coyne C.B., Lazear H.M. Zika virus - reigniting the TORCH. Nat. Rev. Microbiol. 2016;14:707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- 6.Duffy M.R., Chen T.-H., Hancock W.T., Powers A.M., Kool J.L., Lanciotti R.S., Pretrick M., Marfel M., Holzbauer S., Dubray C. Zika virus outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 7.Musso D., Bossin H., Mallet H.P., Besnard M., Broult J., Baudouin L., Levi J.E., Sabino E.C., Ghawche F., Lanteri M.C., Baud D. Zika virus in French Polynesia 2013-14: anatomy of a completed outbreak. Lancet Infect. Dis. 2018;18:e172–e182. doi: 10.1016/S1473-3099(17)30446-2. [DOI] [PubMed] [Google Scholar]
- 8.Lowe R., Barcellos C., Brasil P., Cruz O.G., Honório N.A., Kuper H., Carvalho M.S. The Zika Virus Epidemic in Brazil: From Discovery to Future Implications. Int. J. Environ. Res. Public Health. 2018;15:96. doi: 10.3390/ijerph15010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro M.C., Han Q.C., Carvalho L.R., Victora C.G., França G.V.A. Implications of Zika virus and congenital Zika syndrome for the number of live births in Brazil. Proc. Natl. Acad. Sci. USA. 2018;115:6177–6182. doi: 10.1073/pnas.1718476115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrixson D.T., Newland J.G. Zika Virus Infection in Children. Infect. Dis. Clin. North Am. 2018;32:215–224. doi: 10.1016/j.idc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Carod-Artal F.J. Neurological complications of Zika virus infection. Expert Rev. Anti Infect. Ther. 2018;16:399–410. doi: 10.1080/14787210.2018.1466702. [DOI] [PubMed] [Google Scholar]
- 12.Petersen L.R., Jamieson D.J., Powers A.M., Honein M.A. Zika Virus. N. Engl. J. Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 13.Wang A., Thurmond S., Islas L., Hui K., Hai R. Zika virus genome biology and molecular pathogenesis. Emerg. Microbes Infect. 2017;6:e13. doi: 10.1038/emi.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou J., Shi P.-Y. Strategies for Zika drug discovery. Curr. Opin. Virol. 2019;35:19–26. doi: 10.1016/j.coviro.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Lavezzo E., Berselli M., Frasson I., Perrone R., Palù G., Brazzale A.R., Richter S.N., Toppo S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006675. e1006675–e1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruggiero E., Richter S.N. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018;46:3270–3283. doi: 10.1093/nar/gky187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fay M.M., Lyons S.M., Ivanov P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017;429:2127–2147. doi: 10.1016/j.jmb.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 19.Sun H., Xiang J., Shi Y., Yang Q., Guan A., Li Q., Yu L., Shang Q., Zhang H., Tang Y., Xu G. A newly identified G-quadruplex as a potential target regulating Bcl-2 expression. Biochim. Biophys. Acta. 2014;1840:3052–3057. doi: 10.1016/j.bbagen.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Stump S., Mou T.C., Sprang S.R., Natale N.R., Beall H.D. Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene. PLoS ONE. 2018;13:e0205584. doi: 10.1371/journal.pone.0205584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal P. Recent Advances in Anticancer Drugs Development: G-Quadruplex as New Drug Target. J. Pharmacovigil. 2015;3:e134. [Google Scholar]
- 22.Chen B.-J., Wu Y.-L., Tanaka Y., Zhang W. Small molecules targeting c-Myc oncogene: promising anti-cancer therapeutics. Int. J. Biol. Sci. 2014;10:1084–1096. doi: 10.7150/ijbs.10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang D., Okamoto K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simone R., Balendra R., Moens T.G., Preza E., Wilson K.M., Heslegrave A., Woodling N.S., Niccoli T., Gilbert-Jaramillo J., Abdelkarim S. G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 2018;10:22–31. doi: 10.15252/emmm.201707850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simone R., Fratta P., Neidle S., Parkinson G.N., Isaacs A.M. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015;589:1653–1668. doi: 10.1016/j.febslet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Hershman S.G., Chen Q., Lee J.Y., Kozak M.L., Yue P., Wang L.-S., Johnson F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., He X., Yuan G. Formation and recognition of G-quadruplex relevant for pilin antigenic variation in Neisseria gonorrhoeae. Can. J. Chem. 2011;90:34–38. [Google Scholar]
- 28.Mishra S.K., Jain N., Shankar U., Tawani A., Sharma T.K., Kumar A. Characterization of highly conserved G-quadruplex motifs as potential drug targets in Streptococcus pneumoniae. Sci. Rep. 2019;9:1791. doi: 10.1038/s41598-018-38400-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra S.K., Shankar U., Jain N., Sikri K., Tyagi J.S., Sharma T.K., Mergny J.-L., Kumar A. Characterization of G-Quadruplex Motifs in espB, espK, and cyp51 Genes of Mycobacterium tuberculosis as Potential Drug Targets. Mol. Ther. Nucleic Acids. 2019;16:698–706. doi: 10.1016/j.omtn.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhartiya D., Chawla V., Ghosh S., Shankar R., Kumar N. Genome-wide regulatory dynamics of G-quadruplexes in human malaria parasite Plasmodium falciparum. Genomics. 2016;108:224–231. doi: 10.1016/j.ygeno.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Fleming A.M., Ding Y., Alenko A., Burrows C.J. Zika Virus Genomic RNA Possesses Conserved G-Quadruplexes Characteristic of the Flaviviridae Family. ACS Infect. Dis. 2016;2:674–681. doi: 10.1021/acsinfecdis.6b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tlučková K., Marušič M., Tóthová P., Bauer L., Šket P., Plavec J., Viglasky V. Human papillomavirus G-quadruplexes. Biochemistry. 2013;52:7207–7216. doi: 10.1021/bi400897g. [DOI] [PubMed] [Google Scholar]
- 33.Artusi S., Nadai M., Perrone R., Biasolo M.A., Palù G., Flamand L., Calistri A., Richter S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antiviral Res. 2015;118:123–131. doi: 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piekna-Przybylska D., Sullivan M.A., Sharma G., Bambara R.A. U3 region in the HIV-1 genome adopts a G-quadruplex structure in its RNA and DNA sequence. Biochemistry. 2014;53:2581–2593. doi: 10.1021/bi4016692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrone R., Butovskaya E., Daelemans D., Palù G., Pannecouque C., Richter S.N. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014;69:3248–3258. doi: 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- 36.Perrone R., Doria F., Butovskaya E., Frasson I., Botti S., Scalabrin M., Lago S., Grande V., Nadai M., Freccero M., Richter S.N. Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes. J. Med. Chem. 2015;58:9639–9652. doi: 10.1021/acs.jmedchem.5b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S.R., Zhang Q.Y., Wang J.Q., Ge X.Y., Song Y.Y., Wang Y.F., Li X.D., Fu B.S., Xu G.H., Shu B. Chemical Targeting of a G-Quadruplex RNA in the Ebola Virus L Gene. Cell Chem. Biol. 2016;23:1113–1122. doi: 10.1016/j.chembiol.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Wang S.-R., Min Y.-Q., Wang J.-Q., Liu C.-X., Fu B.-S., Wu F., Wu L.-Y., Qiao Z.-X., Song Y.-Y., Xu G.-H. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti-hepatitis C target. Sci. Adv. 2016;2:e1501535. doi: 10.1126/sciadv.1501535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shrinet J., Agrawal A., Bhatnagar R.K., Sujatha Sunil S. Analysis of the genetic divergence in Asian strains of ZIKA virus with reference to 2015–2016 outbreaks. Bull World Health Organ E-pub. 2016;22 [Google Scholar]
- 40.Taetz S., Baldes C., Mürdter T.E., Kleideiter E., Piotrowska K., Bock U., Haltner-Ukomadu E., Mueller J., Huwer H., Schaefer U.F. Biopharmaceutical characterization of the telomerase inhibitor BRACO19. Pharm. Res. 2006;23:1031–1037. doi: 10.1007/s11095-006-0026-y. [DOI] [PubMed] [Google Scholar]
- 41.Burger A.M., Dai F., Schultes C.M., Reszka A.P., Moore M.J., Double J.A., Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 42.Zhou G., Liu X., Li Y., Xu S., Ma C., Wu X., Cheng Y., Yu Z., Zhao G., Chen Y. Telomere targeting with a novel G-quadruplex-interactive ligand BRACO-19 induces T-loop disassembly and telomerase displacement in human glioblastoma cells. Oncotarget. 2016;7:14925–14939. doi: 10.18632/oncotarget.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard T.D. A guide to simple and informative binding assays. Mol. Biol. Cell. 2010;21:4061–4067. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alniss H., Zamiri B., Khalaj M., Pearson C.E., Macgregor R.B., Jr. Thermodynamic and spectroscopic investigations of TMPyP4 association with guanine- and cytosine-rich DNA and RNA repeats of C9orf72. Biochem. Biophys. Res. Commun. 2018;495:2410–2417. doi: 10.1016/j.bbrc.2017.12.108. [DOI] [PubMed] [Google Scholar]
- 45.Mergny J.-L., Li J., Lacroix L., Amrane S., Chaires J.B. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni134. e138–e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrows N.J., Campos R.K., Powell S.T., Prasanth K.R., Schott-Lerner G., Soto-Acosta R., Galarza-Muñoz G., McGrath E.L., Urrabaz-Garza R., Gao J. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu M., Lee E.M., Wen Z., Cheng Y., Huang W.-K., Qian X., Tcw J., Kouznetsova J., Ogden S.C., Hammack C. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim S.-Y., Osuna C., Lakritz J., Chen E., Yoon G., Taylor R., MacLennan S., Leonard M., Giuliano E., Mathis A. Galidesivir, a Direct-Acting Antiviral Drug, Abrogates Viremia in Rhesus Macaques Challenged with Zika Virus. Open Forum Infect. Dis. 2017;4 S55–S55. [Google Scholar]
- 49.Mishra S.K., Tawani A., Mishra A., Kumar A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep. 2016;6:38144. doi: 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vy Thi Le T., Han S., Chae J., Park H.J. G-quadruplex binding ligands: from naturally occurring to rationally designed molecules. Curr. Pharm. Des. 2012;18:1948–1972. doi: 10.2174/138161212799958431. [DOI] [PubMed] [Google Scholar]
- 51.Fujiwara N., Mazzola M., Cai E., Wang M., Cave J.W. TMPyP4, a Stabilizer of Nucleic Acid Secondary Structure, Is a Novel Acetylcholinesterase Inhibitor. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0139167. e0139167–e0139167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Annamalai A.S., Pattnaik A., Sahoo B.R., Muthukrishnan E., Natarajan S.K., Steffen D., Vu H.L.X., Delhon G., Osorio F.A., Petro T.M. Zika Virus Encoding Nonglycosylated Envelope Protein Is Attenuated and Defective in Neuroinvasion. J. Virol. 2017;91 doi: 10.1128/JVI.01348-17. e01348–e01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pattnaik A., Palermo N., Sahoo B.R., Yuan Z., Hu D., Annamalai A.S., Vu H.L.X., Correas I., Prathipati P.K., Destache C.J. Discovery of a non-nucleoside RNA polymerase inhibitor for blocking Zika virus replication through in silico screening. Antiviral Res. 2018;151:78–86. doi: 10.1016/j.antiviral.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Garcia A., Anandhan A., Burns M., Chen H., Zhou Y., Franco R. Impairment of Atg5-dependent autophagic flux promotes paraquat- and MPP+-induced apoptosis but not rotenone or 6-hydroxydopamine toxicity. Toxicol. Sci. 2013;136:166–182. doi: 10.1093/toxsci/kft188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.