Abstract
Accumulation of unfolded proteins in the endoplasmic reticulum (ER) causes ER stress and activates a signaling network known as the unfolded protein response (UPR). Here we characterize how ER stress and the UPR inhibit insulin signaling. We find that ER stress inhibits insulin signaling by depleting the cell surface population of the insulin receptor. ER stress inhibits proteolytic maturation of insulin proreceptors by interfering with transport of newly synthesized insulin proreceptors from the ER to the plasma membrane. Activation of AKT, a major target of the insulin signaling pathway, by a cytosolic, membrane-bound chimera between the AP20187-inducible FV2E dimerization domain and the cytosolic protein tyrosine kinase domain of the insulin receptor was not affected by ER stress. Hence, signaling events in the UPR, such as activation of the JNK mitogen-activated protein (MAP) kinases or the pseudokinase TRB3 by the ER stress sensors IRE1α and PERK, do not contribute to inhibition of signal transduction in the insulin signaling pathway. Indeed, pharmacologic inhibition and genetic ablation of JNKs, as well as silencing of expression of TRB3, did not restore insulin sensitivity or rescue processing of newly synthesized insulin receptors in ER-stressed cells.
INTRODUCTION
In mammalian cells, most secreted proteins and proteins residing in the plasma membrane or the secretory pathway are transported into the endoplasmic reticulum (ER) while their polypeptide chains are being assembled by translating ribosomes (Walter and Lingappa, 1986). In the ER, newly synthesized proteins fold into their native three-dimensional structures and undergo multiple posttranslational modifications including asparagine (N)-linked glycosylation (Hubbard and Ivatt, 1981; Kornfeld and Kornfeld, 1985) and the formation of disulfide bonds (Fewell et al., 2001). Interaction of newly synthesized proteins with several chaperone systems facilitates their productive folding, but also serves as a quality control mechanism to retain newly synthesized proteins in the ER until they have completed their folding and maturation processes (Hebert and Molinari, 2007). Consequently, unfolded or only partially folded proteins are prevented from exiting the ER until they have completed their folding process or are targeted to degradation mechanisms if they fail to fold productively, such as in ER-associated degradation (ERAD) (Meusser et al., 2005) or ER-phagy (Bernales et al., 2006).
The accumulation of unfolded and partially folded proteins in the ER activates a signaling network termed the unfolded protein response (UPR) (Schröder and Kaufman, 2005; Walter and Ron, 2011). Three ER transmembrane proteins, the membrane-bound transcription factor ATF6 (Yoshida et al., 2000, 2001b), the protein kinase PERK (Shi et al., 1998, 1999; Harding et al., 1999), and the protein kinase-endoribonuclease (RNase) IRE1α (Tirasophon et al., 1998), initiate signaling in the UPR. After cleavage from the endomembrane system, ATF6 translocates to the nucleus and activates transcription of genes encoding ER-resident molecular chaperones and components of the ERAD machinery (Ye et al., 2000; Wu et al., 2007; Yamamoto et al., 2007). PERK transiently attenuates general translation in ER-stressed cells by phosphorylating the α subunit of eIF2 (Shi et al., 1998; Harding et al., 1999). Phosphorylation of eIF2α also promotes translation of mRNAs with several short upstream open reading frames leading to induction of the transcription factor CHOP (Harding et al., 2000) and the pseudokinase TRB3 (Ohoka et al., 2005).
IRE1α is a bifunctional protein kinase-RNase (Tirasophon et al., 1998, 2000). The IRE1α RNase activity initiates splicing of XBP1 mRNA, which encodes a basic leucine zipper (bZIP) transcription factor (Shen et al., 2001; Yoshida et al., 2001a; Calfon et al., 2002; Lee et al., 2002). Spliced XBP1 (XBP1s) is a more potent transcriptional activator than unspliced XBP1 (XBP1u) for genes encoding ER resident molecular chaperones, phospholipid biosynthetic enzymes, and proteins involved in ERAD (Shen et al., 2001; Yoshida et al., 2001a; Calfon et al., 2002; Lee et al., 2002). In addition, relaxed specificity of the RNase activity mediates decay of many mRNAs encoding proteins targeted to the secretory pathway (Hollien and Weissman, 2006; Hollien et al., 2009; Gaddam et al., 2013). Through association with the E3 ubiquitin ligase TRAF2, IRE1α activates the JNK family of mitogen-activated protein (MAP) kinases (Urano et al., 2000).
Insulin signaling is initiated by binding of insulin to the insulin receptor, activation of the protein tyrosine kinase domain and tyrosine autophosphorylation of the insulin receptor, and extensive tyrosine phosphorylation of insulin receptor substrate (IRS) proteins (reviewed in Saltiel and Kahn, 2001), including phosphorylation of Y612, Y632, Y896, Y941, Y1173, and Y1229 in human IRS1 (Shoelson et al., 1992; Sun et al., 1993; Rocchi et al., 1995; Xu et al., 1995; Esposito et al., 2001; Hers et al., 2002). Phosphorylated Y612, Y632, and Y941 are binding sites for the Src homology 2 (SH2) domain of the p85α subunit of phosphatidylinositol (PI) 3-kinase (PI3K) (Sun et al., 1993; Rocchi et al., 1995; Esposito et al., 2001). After formation of PI-3,4-bis- and PI-3,4,5-trisphosphate by PI3K, phosphoinositide-dependent kinases (PDKs) and isoforms of the protein serine/threonine kinase AKT are recruited to the plasma membrane. Colocalization of PDKs and AKT to the plasma membrane facilitates phosphorylation of AKT on T308 by PDK1 (Alessi et al., 1996) and on S473 by mTORC2 (Sarbassov et al., 2005; Guertin et al., 2006; Jacinto et al., 2006), PAK1 (Mao et al., 2008), and ILK (McDonald et al., 2008), leading to activation of AKT. Activated AKT facilitates glucose transport and protein and glycogen synthesis and inhibits gluconeogenesis. Recruitment of GRB2 to IRS1 phosphorylated at Y896 via its SH2 domain (Sun et al., 1993; Myers et al., 1994) activates MAP kinases, such as p42/p44, and contributes to generating a mitogenic signal in insulin-stimulated cells (Valverde et al., 2001).
Inhibition of signal transduction in the insulin signaling pathway is the cause for insulin resistance when cells fail to respond normally to insulin. Insulin resistance can be caused by a decrease in the number of insulin receptors or defects in signal transduction downstream of the insulin receptor. Activation of JNKs and TRB3 by the UPR has been implicated in the inhibition of insulin signaling downstream of the insulin receptor (Özcan et al., 2004; Koh et al., 2006, 2013). JNKs inhibit tyrosine phosphorylation of IRS1 by the insulin receptor by phosphorylating S307 in murine IRS1 and S312 in human IRS1 (Aguirre et al., 2000, 2002). Consequently, phosphorylation and activation of AKT by insulin is inhibited by JNKs (Lee et al., 2003; Nguyen et al., 2005; Emanuelli et al., 2008). TRB3 interacts with and inhibits phosphorylation of AKT (Du et al., 2003) and also interacts with IRS1 and inhibits its phosphorylation at Y612 by the insulin receptor (Koh et al., 2013).
JNKs become rapidly and transiently activated in ER-stressed cells to promote an adaptive response to ER stress (Brown et al., 2016). The motivation for this study was to characterize whether this rapid, initial JNK activation in the first 10–120 min of the ER stress response causes insulin resistance. However, we find no evidence for inhibition of insulin-stimulated AKT phosphorylation or IRS1 tyrosine phosphorylation in cells exposed to ER stress for up to ∼8–12 h despite activation of JNKs and induction of TRB3. Only ER stress lasting for more than ∼8–12 h inhibited insulin-stimulated AKT phosphorylation, but did so independent of JNKs and TRB3, and correlated with depletion of β chains of the mature insulin receptor, accumulation of unprocessed α-β precursors of the insulin receptor in the ER, and depletion of GFP-tagged insulin receptors from the plasma membrane. Moreover, phosphorylation of AKT at S473 in response to activation of a cytosolic, membrane-bound chimera between the AP20187-inducible FV2E dimerization domain (Clackson et al., 1998; Yang et al., 2000) and the protein tyrosine kinase domain of the insulin receptor is not affected by ER stress lasting for 24 h. We propose that inhibition of trafficking of newly synthesized insulin receptors to the plasma membrane suffices and is necessary to inhibit activation of AKT by insulin in ER-stressed cells by depleting the plasma membrane population of the insulin receptor.
RESULTS
ER stress for up to 8 h does not inhibit insulin-stimulated AKT activation
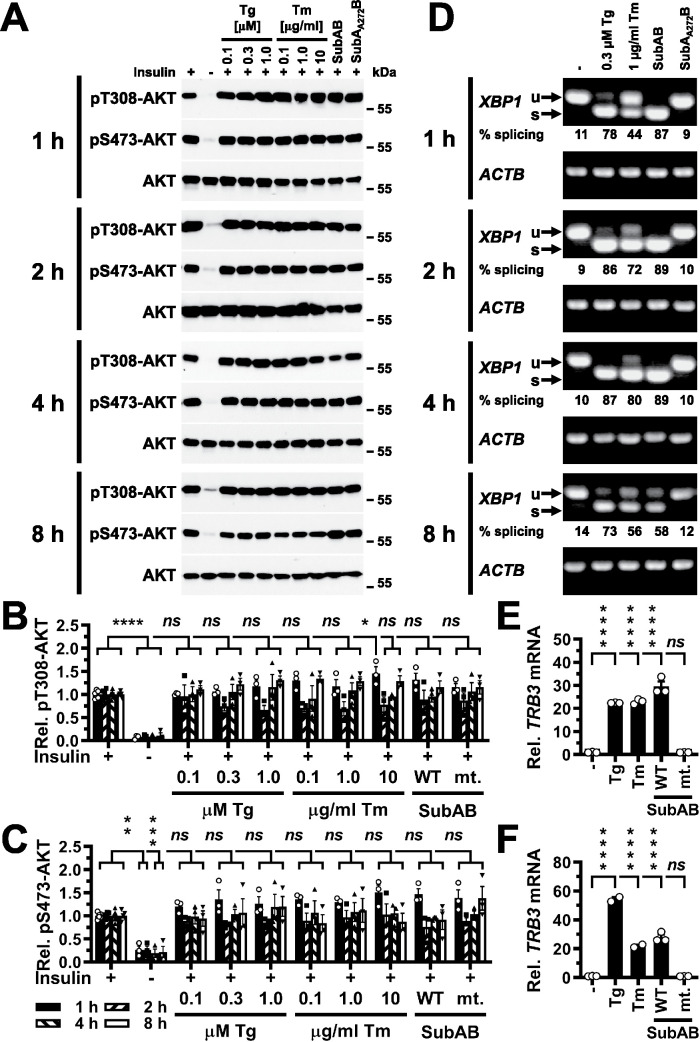
We used in vitro differentiated C2C12 myotubes, 3T3-F442A preadipocytes, and Hep G2 hepatoma cells to characterize the effects of ER stress on insulin signaling, because these cell types are cell culture models of the main tissues and organs contributing to glucose homeostasis, muscle, adipose tissue, and the liver (Saltiel and Kahn, 2001). We first characterized whether ER stress induced with different ER stressors for up to 8 h inhibits signal transduction downstream of the insulin receptor by monitoring insulin-stimulated phosphorylation of AKT at T308 and S473 in in vitro differentiated C2C12 myotubes. C2C12 myotubes were serum-starved for 18 h, treated with ER stressors for the last 1–8 h of serum starvation and then stimulated with 100 nM insulin for 15 min in the continued presence of ER stressors. Insulin (100 nM) was chosen because inhibition of insulin signaling downstream of the receptor manifests independent of insulin concentration (Olefsky and Kolterman, 1981). To exclude drug-specific effects on insulin signaling, we used three different ER stressors, the SERCA pump inhibitor thapsigargin (Thastrup et al., 1990), the N-glycosylation inhibitor tunicamycin (Kuo and Lampen, 1976; Lehle and Tanner, 1976), and the protease SubAB, which cleaves and inactivates the ER resident HSP70 class molecular chaperone BiP/GRP78 (Paton et al., 2006). We also titrated the concentrations of both thapsigargin and tunicamycin in the culture medium over a 10- or 100-fold concentration range, respectively. AKT phosphorylation was chosen as readout, because its dynamic range is larger than the dynamic ranges of many physiological responses to insulin such as translocation of GLUT4 to the plasma membrane (Hoehn et al., 2008), uptake of 2-deoxyglucose (Whitehead et al., 2001), or glucose oxidation (Kono and Barham, 1971). Induction of ER stress with 0.1–1.0 μM thapsigargin, 0.1–10 μg/ml tunicamycin, or SubAB for up to ∼8 h in C2C12 myotubes, however, did not decrease insulin-stimulated phosphorylation of AKT at T308 (Figure 1, A and B) or S473 (Figure 1, A and C).
FIGURE 1:
Acute ER stress does not inhibit phosphorylation of AKT on T308 or S473 in C2C12 myotubes stimulated with 100 nM insulin for 15 min. (A) C2C12 myotubes were serum-starved for 18 h and treated with the indicated concentrations of thapsigargin (Tg), tunicamycin (Tm), 1 μg/ml SubAB, or 1 μg/ml catalytically inactive SubAA272B during the last 1–8 h of serum starvation and then stimulated with 100 nM insulin for 15 min where indicated. Cell lysates were analyzed by Western blotting. Quantification of phosphorylation of AKT on (B) T308 and (C) S473. Bars represent SEs (n = 5 for S473 phosphorylation of AKT at 8 h in unstressed, insulin-stimulated cells, n = 6 for all other unstressed, insulin-stimulated samples, and n = 3 for all other treatments). p values for comparison of ER-stressed samples and samples not stimulated with 100 nM insulin to samples stimulated with 100 nM insulin were calculated by ordinary two-way ANOVA with Dunnett’s multiple comparisons test (Dunnett, 1955, 1964). (D) Detection of XBP1 splicing by reverse transcriptase PCR. PCR products derived from unspliced (u) and spliced (s) XBP1 mRNA are indicated by arrows. β-Actin (ACTB) was used as a loading control. (E, F) Induction of TRB3 in C2C12 cells by ER stress. C2C12 cells were treated with 300 nM thapsigargin, 1 μg/ml tunicamycin, 1 μg/ml SubAB (labeled “WT”), or 1 μg/ml SubAA272B (labeled “mt.”) for (E) 4 h and (F) 8 h. TRB3 mRNA levels were determined by reverse transcriptase-qPCR and standardized to the loading control ACTB. Bars represent SEs (n = 2 for the samples treated with thapsigargin or tunicamycin for 8 h, n = 3 for all other samples). p values for comparison of treated samples to the untreated sample (“–” ) were calculated by ordinary two-way ANOVA with Dunnett’s multiple comparisons test taking data shown in Figure 10F into account. Abbreviations in this and all other figures: ns, not significant, * or #, p < 0.05, ** or ##, p < 0.01, *** or ###, p < 0.001, and **** or ####, p < 0.0001.
To confirm that treatment of serum-starved C2C12 cells with ER stressors induces ER stress, we monitored XBP1 splicing using reverse transcriptase PCR. The IRE1α-initiated XBP1 splicing reaction removes a 26-nucleotide intron from XBP1 mRNA. Therefore, the appearance of a shorter reverse transcriptase PCR product on 2% (wt/vol) agarose gels indicates activation of the RNase activity of IRE1α. Upon exposure of serum-starved C2C12 cells to 0.3 μM thapsigargin, 1 μg/ml tunicamycin, or 1 μg/ml SubAB, a shorter reverse transcriptase PCR product appeared (Figure 1D), which represents spliced XBP1 mRNA. Strong induction of TRB3 mRNA after induction of ER stress for 4 or 8 h was also detected (Figure 1, E and F), which suggests that serum-starved C2C12 cells experience ER stress when challenged with thapsigargin, tunicamycin, or SubAB. Furthermore, serum starvation did not decrease XBP1 splicing in cells exposed to 1 μM thapsigargin for 1 h (Supplemental Figure S1A), which argues against the possibility that induction of ER stress is blunted by decreased protein synthesis rates in serum-starved cells. Thapsigargin-, tunicamycin-, or SubAB-induced ER stress for up to 12 h also did not inhibit insulin-stimulated AKT activation in 3T3-F442A adipocytes or Hep G2 hepatoma cells or over a period of 4 h in Fao rat hepatoma cells cultured in RPMI 1640 or Coon’s modification of Ham’s F12 medium (unpublished data).
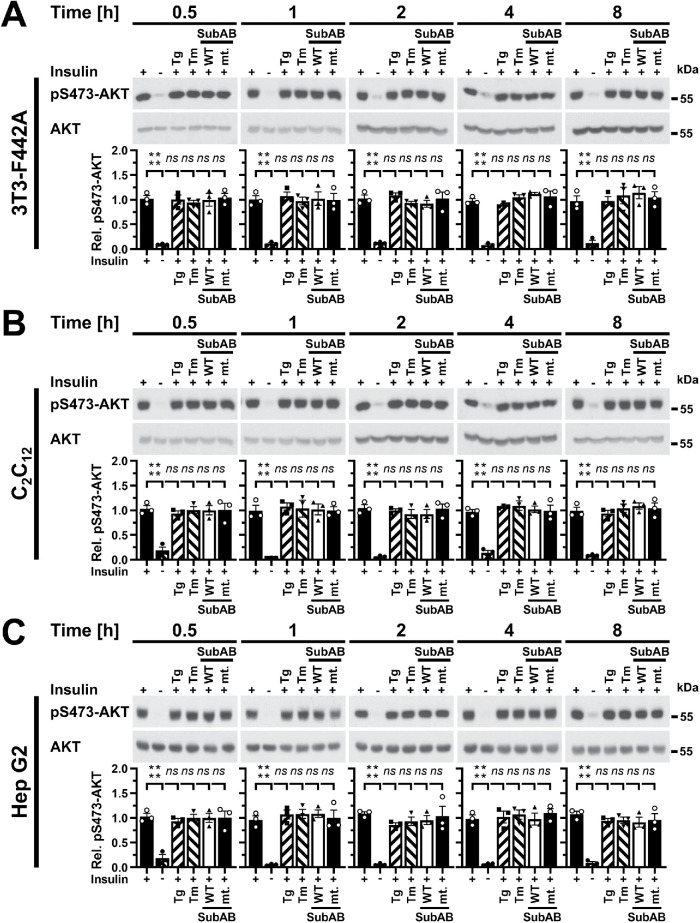
JNKs are activated as early as 10 min after induction of ER stress in C2C12 myotubes and 3T3-F442A adipocytes, and after 30 min in Hep G2 cells (Brown et al., 2016), which raises the possibility that ER stress may inhibit the insulin signaling pathway around these times in the ER stress response. However, 30 min of thapsigargin-induced ER stress did not decrease insulin-stimulated phosphorylation of AKT on S473 in 3T3-F442A adipocytes, C2C12 myotubes, or Hep G2 cells (Supplemental Figure S1, B and C). Induction of ER stress for 30 min with tunicamycin or SubAB also did not decrease insulin-induced phosphorylation of AKT (Supplemental Figure S1, B and C). These results suggest that activation of JNKs by ER stress does not inhibit signal transduction in the insulin signaling pathway. To characterise whether lower, more physiologic insulin concentrations (Cryer and Polonsky, 1998; Unger and Foster, 1998) unmask effects of ER stress on insulin-stimulated AKT phosphorylation, we stimulated cells with 10 nM insulin for 15 min. ER stress induced with 0.3 μM thapsigargin, 1 μg/ml tunicamycin or SubAB for 30 min up to 8 h had no effect on phosphorylation of AKT on S473 in 3T3-F442A, C2C12, or Hep G2 cells stimulated with 10 nM insulin for 15 min (Figure 2). In summary, these data establish that short periods of ER stress lasting for up to ∼8 h, in which JNKs are activated (Brown et al., 2016) and TRB3 is induced (Figure 1, E and F), do not inhibit insulin-stimulated AKT phosphorylation in 3T3-F442A, C2C12, Fao, and Hep G2 cells.
FIGURE 2:
Acute ER stress does not inhibit phosphorylation of AKT on S473 stimulated with 10 nM insulin for 15 min. (A) 3T3-F442A cells, (B) C2C12 myotubes, and (C) Hep G2 cells were serum-starved for 18 h and treated with 0.3 μM thapsigargin, 1 μg/ml tunicamycin, 1 μg/ml SubAB, or 1 μg/ml catalytically inactive SubAA272B during the last 30 min of serum starvation and then stimulated with 10 nM insulin for 15 min where indicated. Cell lysates were analyzed by Western blotting. Bars represent SEs (n = 3). p values for comparison of ER-stressed samples and samples not stimulated with 10 nM insulin to samples stimulated with 10 nM insulin were calculated by ordinary two-way ANOVA with Dunnett’s multiple comparisons test.
ER stress for up to 30 min does not inhibit IRS1 tyrosine phosphorylation
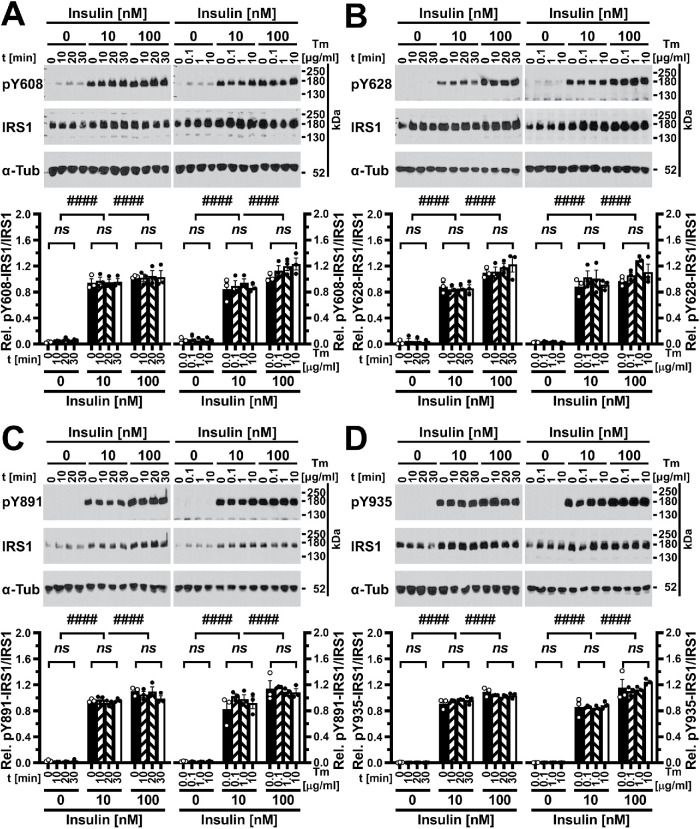
Phosphorylation of AKT is downstream of tyrosine phosphorylation of IRS1 by the activated insulin receptor in the insulin signaling pathway (Backer et al., 1992; Franke et al., 1995). The absence of effects of heterozygosity for IRS1 in lean mice on control of blood glucose levels (Shirakami et al., 2002) and the lack of effects of partial short hairpin RNA–mediated knockdown of IRS1 in skeletal muscle on local glucose clearance (Cleasby et al., 2007) suggest that IRS1 is available in excess over the amounts needed for full activation of downstream events in the insulin signaling pathway. To address the possibility that decreases in IRS1 tyrosine phosphorylation in ER-stressed cells are not reflected at the level of AKT phosphorylation, we directly examined the effects of ER stress on IRS1 tyrosine phosphorylation. First, we characterized whether within the initial 30 min time window after induction of ER stress, in which ER stress activates JNKs in 3T3-F442A, C2C12, and Hep G2 cells (Brown et al., 2016), a decrease in tyrosine phosphorylation of specific, well-characterized insulin-responsive tyrosine phosphorylation sites, such as Y608 (mouse)/Y612 (human; from here on abbreviated as Y608/612), Y628/632, Y891/896, and Y935/941 (Shoelson et al., 1992; Sun et al., 1993; Xu et al., 1995; Hers et al., 2002), could be observed. We could extract intact IRS1 from 3T3-F442A, C2C12, and Hep G2 cells only under strongly denaturing conditions such as 8 M urea, 2.5% (wt/vol) SDS, or 7 M urea, 2 M thiourea, 2.5% (wt/vol) SDS, or 8 M guanidinium hydrochloride, 1% (vol/vol) Triton X-100, or 4 M guanidinium thiocyanate, 1% (vol/vol) Triton X-100, or 10–20% (wt/vol) trichloroacetic acid (TCA; unpublished data). In addition, detection of full-length IRS1 by Western blotting required electrotransfer onto polyvinylidene difluoride (PVDF) membranes at pH ∼10 in the presence of SDS as described in Materials and Methods. Under these conditions, Western blots displaying one band at ∼180 kDa, the migration position of IRS1 in SDS–PAGE (Sun et al., 1991), were obtained with all four single tyrosine phosphorylation site antibodies and anti-IRS1 antibodies (Figure 3; Supplemental Figures S2 and S3). Antibodies against phosphorylated tyrosine phosphorylation sites gave much stronger signals on samples isolated from insulin-treated cells at ∼180 kDa (Figure 3; Supplemental Figures S2 and S3). Induction of ER stress with 1 μM thapsigargin for up to 30 min or with 0.1 μg/ml, 1 μg/ml, or 10 μg/ml tunicamycin for 30 min did not affect phosphorylation of Y608/612, Y628/632, Y891/896, or Y935/941 when cells were stimulated with 10 or 100 nM insulin for 5 min (Figure 3; Supplemental Figures S2 and S3). An approximately threefold increase in IRS1 levels upon insulin stimulation in 3T3-F442A cells (Supplemental Figure S2) and an approximately twofold increase in IRS1 levels in C2C12 myotubes and Hep G2 cells (Figure 3; Supplemental Figure S3) were also not affected by thapsigargin or tunicamycin.
FIGURE 3:
Acute ER stress does not inhibit insulin-stimulated phosphorylation of IRS1 at four specific tyrosine phosphorylation sites in C2C12 myotubes. Tyrosine phosphorylation at (A) Y608, (B) Y628, (C) Y891, and (D) Y935 was analyzed by Western blotting. C2C12 myotubes were serum-starved for 18 h before exposure to 1 μM thapsigargin for 10, 20, or 30 min (left side of figure panels) or to 0.1, 1.0, or 10 μg/ml tunicamycin for 30 min (right side of figure panels), followed by stimulation with the indicated concentrations of insulin for 5 min in the continued presence of thapsigargin or tunicamycin. Bars represent SEs (n = 3). p values for comparison of effects of thapsigargin or tunicamycin within one insulin concentration and for comparison of effects of different insulin concentrations were calculated by ordinary two-way ANOVA with Tukey’s multiple comparisons test (Tukey, 1949a). Abbreviation: α-Tub, α-tubulin.
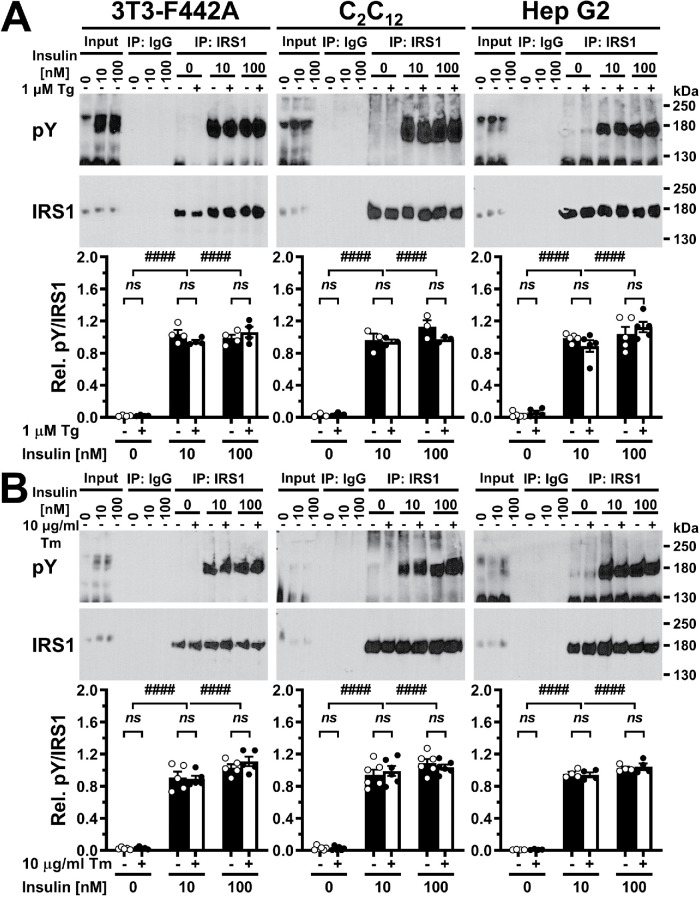
Human IRS1 has 32 tyrosyl residues, while murine IRS1 has 34. Phosphorylation of at least 19 tyrosines in human IRS1 and 13 tyrosines in murine IRS1 has been shown experimentally (Hornbeck et al., 2015). An additional eight tyrosines in human IRS1 and nine tyrosines in murine IRS1 feature at least one acidic amino acid in the six immediately upstream amino acids. Upstream acidic amino acids can be a feature of tyrosine phosphorylation sites (Neil et al., 1981; Smart et al., 1981; Baldwin et al., 1982, 1983; Hunter, 1982; Patschinsky et al., 1982; Pike et al., 1982) and are, for example, enriched in the experimentally confirmed tyrosine phosphorylation sites of human and murine IRS1 (human IRS1, upstream positions –1 to –3, χ2 p value < 0.05; murine IRS1, upstream positions –1 to –6, χ2 p value < 0.01). Given the large number of confirmed and putative tyrosine phosphorylation sites in IRS1, it is possible that individually surveying a subset of tyrosine phosphorylation sites may not uncover effects of ER stress on tyrosine phosphorylation of IRS1. To address this concern, we immunoprecipitated IRS1 from cell lysates prepared from unstressed cells or cells that were exposed to 1 μM thapsigargin or 10 μg/ml tunicamycin for 30 min, then stimulated with 10 or 100 nM insulin for 5 min in the continued presence of thapsigargin or tunicamycin, and Western blotted the immunoprecipitates with a pan-phosphotyrosine antibody (clone 4G10® Platinum) and an anti-IRS1 antibody (Figure 4). We observed a strong increase in tyrosine phosphorylation after stimulation with either 10 or 100 nM insulin for 5 min, but neither thapsigargin nor tunicamycin had any effect on the level of tyrosine phosphorylation of IRS1 (Figure 4). In summary, these data show that ER stress lasting for up to 30 min does not affect insulin-stimulated IRS1 tyrosine phosphorylation.
FIGURE 4:
Acute ER stress does not inhibit insulin-stimulated tyrosine phosphorylation of IRS1. Serum-starved cells were exposed to (A) 1 μM thapsigargin or (B) 10 μg/ml tunicamycin for 30 min, followed by stimulation with the indicated concentrations of insulin for 5 min in the continued presence of thapsigargin or tunicamycin. After immunoprecipitation of IRS1, Western blots were probed with an anti-phosphotyrosine (“pY”) and an anti-IRS1 antibody. Bars represent SEs (n = 3 for thapsigargin-treated C2C12 myotubes, n = 4 for thapsigargin-treated 3T3-F442A cells and tunicamycin-treated Hep G2 cells, n = 5 for tunicamycin-treated 3T3-F442A cells and thapsigargin-treated Hep G2 cells, and n = 6 for tunicamycin-treated C2C12 myotubes). p values for comparison of effects of thapsigargin or tunicamycin within one insulin concentration and for comparison of effects of different insulin concentrations were calculated by ordinary two-way ANOVA with Tukey’s multiple comparisons test. For 3T3-F442A cells and Hep G2 cells treated with tunicamycin, data were square root-transformed before statistical analyses.
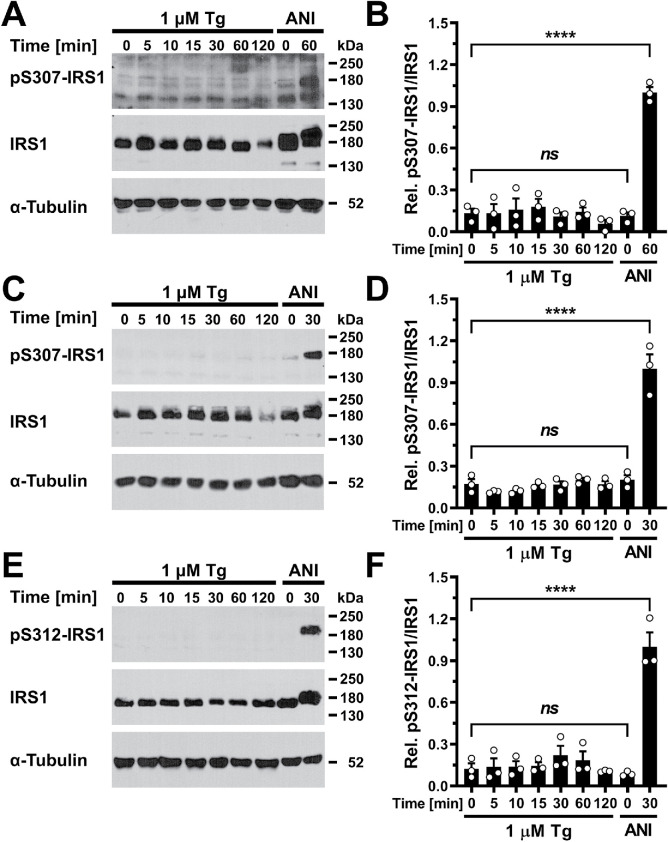
ER stress does not elicit serine 307/312 phosphorylation of IRS1
JNKs inhibit tyrosine phosphorylation of IRS1 by the activated insulin receptor by phosphorylating IRS1 at S307/312 (Aguirre et al., 2000, 2002). Unaltered tyrosine phosphorylation of IRS1 in ER-stressed cells (Figures 3 and 4; Supplemental Figures S2 and S3) suggested that JNKs, despite being activated by ER stress (Brown et al., 2016), do not phosphorylate IRS1 at S307/312 or that phosphorylation of IRS1 at S307/312 by JNKs does not inhibit tyrosine phosphorylation of IRS1 by the insulin receptor in ER-stressed cells. To distinguish between these possibilities, we measured IRS1 S307/312 phosphorylation in serum-starved cells and standardized the phospho-S307/S312 IRS1 signal to the signal for total IRS1 (Figure 5). We included treatment with 5 μg/ml anisomycin for 30–60 min, which has been reported to elicit phosphorylation of IRS1 at S307/312 (Aguirre et al., 2000, 2002; Werner et al., 2004), as a positive control, because we obtained only faint signals with the anti-pS307/S312 antibody with cell lysates prepared from cells exposed to 1 μM thapsigargin for up to 2 h (Figure 5). In 3T3-F442A and Hep G2 cells, anisomycin retarded migration of IRS1 in 7.5% SDS–PAGE gels (Figure 5, A and E). This suggests that IRS1 becomes phosphorylated at sites additional to those reported in the literature in response to anisomycin, S302/307 (Werner et al., 2004), S307/312 (Aguirre et al., 2000, 2002; Werner et al., 2004), and possibly S632/636 and S635/639 (Hiratani et al., 2005), to explain the shift in migration position. Retardation of IRS1 in SDS–PAGE was not seen in thapsigargin-treated cells (Figure 5). These data argue that ER stress induced with thapsigargin does not elicit S307/312 phosphorylation of IRS1 and that phosphorylation of IRS1 at other sites remains below the threshold necessary to affect retardation of IRS1 in SDS–PAGE.
FIGURE 5:
ER stress does not elicit IRS1 S307/S312 phosphorylation. (A, B) Serum-starved 3T3-F442A cells, (C, D) C2C12 myotubes, and (E, F) Hep G2 cells were treated with 1 μM thapsigargin or 5 μg/ml anisomycin (ANI) for the indicated times. (A, C, E) Cell lysates were analyzed by Western blotting. (B, D, F) Quantification of the Western blots shown in panels A, C, and E. Bars represent SEs (n = 3). p values for comparison of every sample to every other sample were calculated by ordinary one-way ANOVA with Tukey’s multiple comparisons test. Anisomycin-treated samples are positive controls in the phospho-S307/S312-IRS1 Western blots.
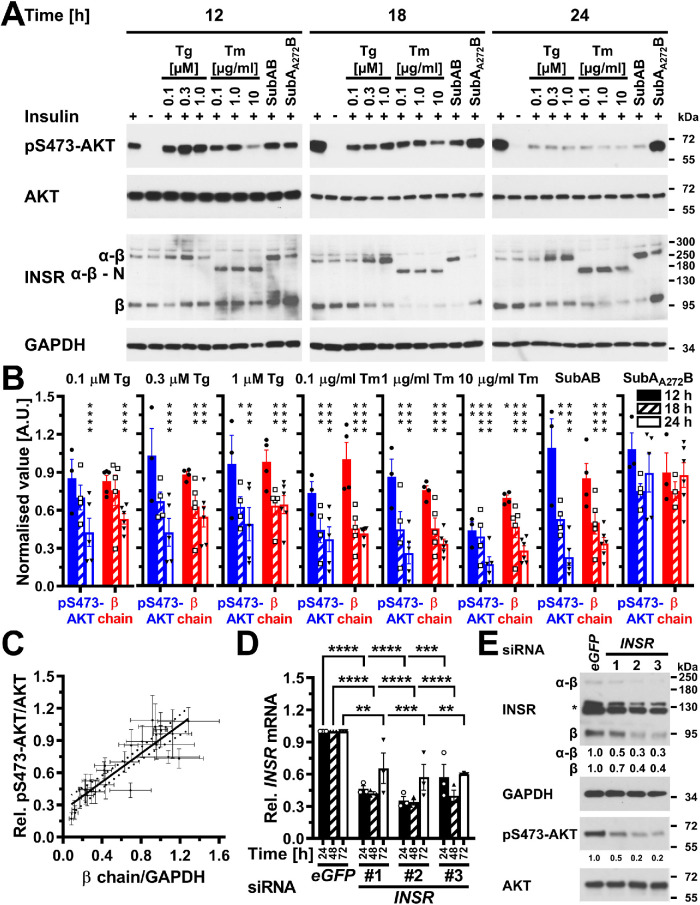
ER stress for ≥12 h inhibits insulin-stimulated AKT phosphorylation
Several studies have reported that ER stress lasting for 12 h or longer causes insulin resistance (Zhou et al., 2009; Avery et al., 2010; Xu et al., 2010; Tang et al., 2011; Hassan et al., 2012; Jung et al., 2013; Panzhinskiy et al., 2013). Such long periods of ER stress may cause insulin resistance by depleting the plasma membrane population of the insulin receptor, because the insulin receptor has a half-life of 7–13 h (Reed and Lane, 1980; Reed et al., 1981a, b; Kasuga et al., 1981; Capeau et al., 1985; Savoie et al., 1986; Grako et al., 1992). For this reason, we characterized whether ER stress for ≥12 h decreases insulin-stimulated phosphorylation of AKT. Twelve hours after induction of ER stress, insulin-stimulated S473 phosphorylation of AKT was decreased in C2C12 cells exposed to 10 μg/ml tunicamycin (Figure 6, A and B). Eighteen hours after induction of ER stress, several ER stressors and markedly lower tunicamycin concentrations decreased insulin-stimulated AKT phosphorylation at S473 (Figure 6, A and B). After 24 h of ER stress, all ER stress-inducing conditions decreased insulin-stimulated AKT phosphorylation at S473 in C2C12 cells. Twenty-four hours of ER stress induced with thapsigargin, tunicamycin, or SubAB decreased cell numbers (Supplemental Figure S4, A and B) but did not affect the activity of mitochondrial redox chains in the remaining cells (Supplemental Figure S4C), suggesting that the remaining cells were viable and that loss of viability cannot explain the decrease in S473 phosphorylation of AKT. We made similar observations in Hep G2 and 3T3-F442A cells. In Hep G2 cells, induction of ER stress for 18 h did not affect insulin-stimulated phosphorylation of AKT, except when cells were exposed to 10 μg/ml tunicamycin (Supplemental Figure S5, A and B). After 24 h of ER stress a 10-fold-lower tunicamycin concentration also reduced insulin-stimulated AKT phosphorylation, and after 36 h all ER stress-inducing conditions decreased insulin-stimulated AKT phosphorylation (Supplemental Figure S5, A and B). Thirty-six hours of ER stress decreased the number of Hep G2 cells remaining in culture dishes (Supplemental Figure S4, D and E), but did not affect the viability of the cells remaining in culture dishes (Supplemental Figure S4F). In 3T3-F442A cells, insulin-stimulated S473 phosphorylation of AKT started to decline after 12 h of ER stress and continued to decline over the next 12 h (unpublished data). These data confirm that periods of ER stress that exceed the half-life of the insulin receptor decrease insulin-stimulated AKT phosphorylation.
FIGURE 6:
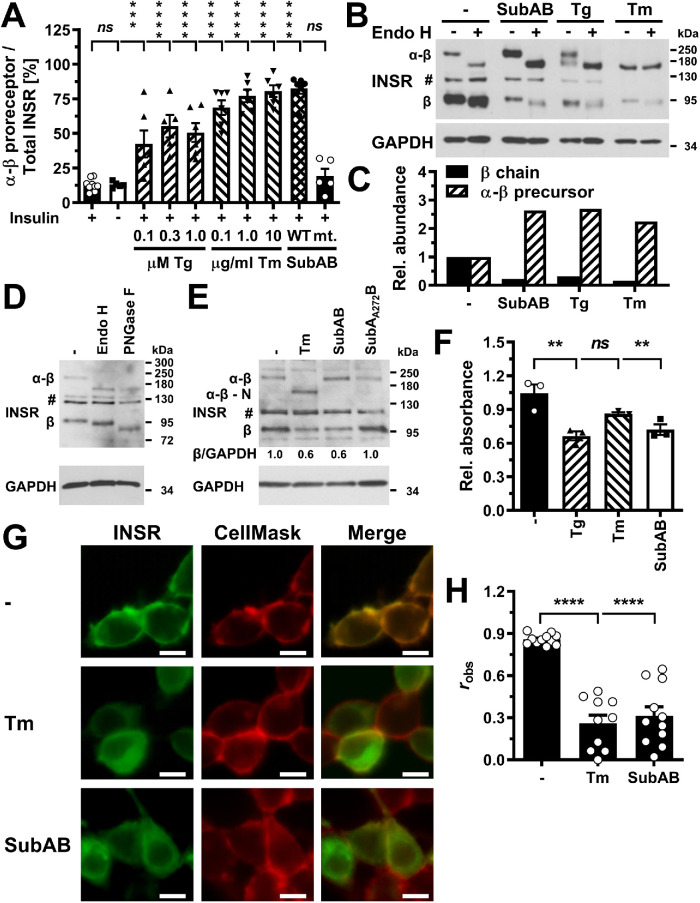
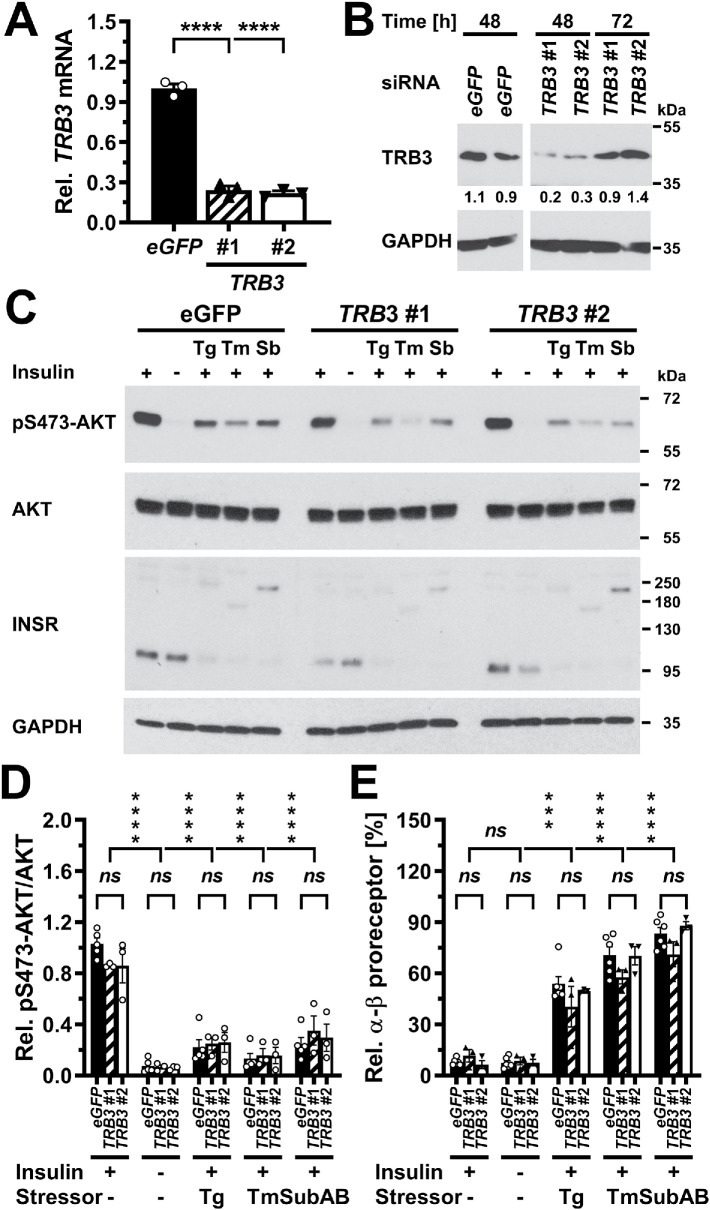
Depletion of insulin receptors coincides and correlates with decreased AKT S473 phosphorylation in ER-stressed C2C12 myoblasts and suffices to decrease AKT S473 phosphorylation. (A) Serum-starved C2C12 cells were treated with the indicated concentrations of thapsigargin, tunicamycin, 1 μg/ml SubAB, or 1 μg/ml SubAA272B for 12–24 h before stimulation with 100 nM insulin for 15 min. Cell lysates were analyzed by Western blotting for pS473-AKT, total AKT, the insulin receptor (INSR), and GAPDH. Bands representing the α-β proreceptor, the unglycosylated α-β proreceptor, and the β chain of the mature insulin receptor are labeled α-β, α-β - N, and β, respectively. (B) Quantification of the phosphorylation of AKT on S473 (“pS473-AKT”) and of the relative abundance of β chains of the insulin receptor (“β chain”). Bars represent SEs (AKT phosphorylation at S473: n = 6 for cells stimulated with 100 nM insulin at the 12 h time points, n = 7 and 9 for the 18 and 24 h time points; for all other samples n = 3 for the 12 h time point, n = 4 for the 18 h time point, and n = 5 for the 24 h time point; relative abundance of β chains: n = 9 for cells stimulated with 100 nM insulin at the 12 h time point, n = 10 for the 18 h time point, and n = 12 for the 24 h time point, 12 h: n = 5 for the unstimulated cells and the insulin-stimulated cells treated with 0.1 mM thapsigargin, n = 3 for the cells treated with SubAA272B, and n = 4 for all other samples, 18 h: n = 6 for the unstimulated cells, n = 3 for cells treated with SubAA272B, and n = 5 for all other samples, 24 h: n = 6 for all samples). Phosphorylation of AKT at S473 and the relative abundance of β chains are expressed relative to unstressed cells that were stimulated with 100 nM insulin for 15 min. p values for comparison of ER-stressed to unstressed samples were calculated using ordinary two-way ANOVA with Dunnett’s multiple comparisons test on the original data for AKT phosphorylation at S473 and square root–transformed data for the relative abundance of β chains. (C) Correlation of insulin-stimulated AKT phosphorylation with insulin receptor β chains (r2 = 0.80, two-tailed p < 0.0001 for a significantly nonzero slope, and p > 0.05 for deviation from linearity calculated by a runs test, n = 27). Dotted lines represent the 95% confidence interval of the linear regression line. The relative phosphorylation of AKT at S473 shown in panel B was plotted against the relative abundance of β chains shown in panel B. (D) Steady-state INSR mRNA levels in C2C12 cells transfected with 50 nM of the indicated siRNAs for 24, 48, or 72 h. Bars represent SEs (n = 3). p values for comparison of cells transfected with the three INSR siRNAs to the cells transfected with the eGFP siRNA were calculated with an ordinary two-way ANOVA with Tukey’s multiple comparisons test. Differences in INSR mRNA levels between different INSR siRNAs within individual time points or between different time points for individual siRNAs are not significant. (E) siRNA-mediated knockdown of expression of the insulin receptor inhibits insulin-stimulated phosphorylation of AKT. Serum-starved C2C12 cells were stimulated with insulin 48 h after transfection of 50 nM of the indicated siRNAs. Two unspecific bands are marked with an asterisk (*).
Decreased insulin-stimulated AKT phosphorylation correlates with depletion of the β chain of the mature insulin receptor in ER-stressed cells
The hypothesis, that ER stress for >12 h decreases insulin-stimulated S473 phosphorylation of AKT by depleting the insulin receptor at the plasma membrane, predicts a decrease in the number of mature β chains of the insulin receptor over the duration of ER stress. The insulin receptor is synthesized as a proreceptor of ∼190 or ∼210 kDa due to alternative glycosylation (Hwang and Frost, 1999). Cleavage of the proreceptor into mature α and β chains of ∼135 and ∼95 kDa in the trans-Golgi network by several proprotein convertases (Robertson et al., 1993; Bravo et al., 1994) yields the mature insulin receptor. Western blotting of cell lysates with an antibody against the β chain of the insulin receptor revealed bands representing the proreceptor at ∼190 and ∼210 kDa and the β chain at ∼95 kDa (Figure 6A; Supplemental Figure S5A). C2C12 cells stressed with 10 μg/ml tunicamycin displayed a decrease in the number of insulin receptor β chains 12 h after induction of ER stress, which is around the same time at which this condition decreases insulin-stimulated AKT S473 phosphorylation. After 18 and 24 h of ER stress, lower concentrations of ER stress and other ER stressors such as thapsigargin or SubAB also decreased the number of insulin receptor β chains (Figure 6, A and B). The decrease in the number of insulin receptor β chains correlated with the decrease in insulin-stimulated AKT phosphorylation (Figure 6C). Exposure of Hep G2 cells to 18 h of ER stress did not affect the abundance of β chains of the mature insulin receptor (Supplemental Figure S5A). By contrast, after 24 or 36 h of ER stress, a decline in β chain abundance coincided with decreased phosphorylation of AKT on S473 by insulin (Supplemental Figure S5, A and B). The decrease in S473 phosphorylation of AKT correlated with the decrease in β chain abundance in ER-stressed Hep G2 cells (Supplemental Figure S5C), and 36 h of ER stress significantly increased the number of unprocessed α-β proreceptors in Hep G2 cells (Supplemental Figure S5D). We also observed a correlation between a decrease in the number of insulin receptor β chains and decreased, insulin-stimulated AKT phosphorylation in 3T3-F442A cells (unpublished data). Insulin by itself, however, did not affect the abundance of β chains of the insulin receptor in any cell line (Figure 6, A and B; Supplemental Figure S5, A and B; unpublished data). In summary, these data establish that in ER-stressed cells a decrease in insulin-stimulated AKT phosphorylation correlates with a decrease in the number of mature insulin receptors.
Next, we characterized whether the depletion of β chains in ER-stressed cells is sufficient to decrease insulin-stimulated AKT phosphorylation by silencing expression of the insulin receptor gene in C2C12 myoblasts by using three small interfering RNAs (siRNAs) and comparing insulin-stimulated AKT S473 phosphorylation to cells transfected with a siRNA against eGFP. All three siRNAs decreased insulin receptor mRNA steady-state levels by 50–70% (Figure 6D) and mature β chains to a similar extent (Figure 6E). Concomitant with the decrease in insulin receptor levels, insulin-stimulated AKT S473 phosphorylation was decreased by 50–80% (Figure 6E). These experiments suggest that an ∼50% decrease in insulin receptor levels suffices to decrease insulin-stimulated AKT S473 phosphorylation to a similar degree. In summary, these data suggest that depletion of β chains of the mature insulin receptor suffices to decrease insulin-stimulated AKT phosphorylation in ER-stressed cells.
Inhibition of protein synthesis and synthesis of α-β proreceptors cannot fully explain decreased insulin-stimulated S473 phosphorylation of AKT in ER stress lasting for 24 h
ER stress may decrease the number of mature insulin receptors by decreasing transcription of the insulin receptor gene (Örd and Örd, 2003; Jang et al., 2010), degrading the insulin receptor mRNA via the regulated IRE1α-dependent decay (RIDD) activity of IRE1α (Hollien and Weissman, 2006; Hollien et al., 2009), inhibiting translation of the insulin receptor mRNA, by interfering with folding and maturation of newly synthesized insulin receptors in the ER and transport of newly synthesized insulin receptors to the plasma membrane, or increasing the turnover of insulin receptors at the cell surface. Therefore, we decided to determine which of these processes contribute to lower levels of mature insulin receptors in ER-stressed cells.
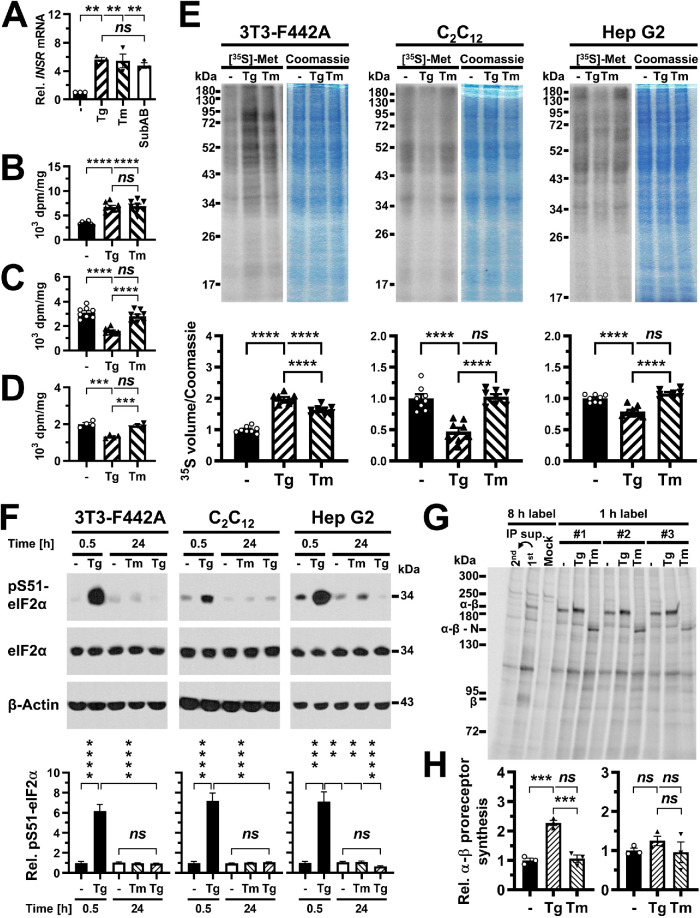
Reverse transcriptase-quantitative PCR (qPCR) showed that steady-state levels of the insulin receptor mRNA increase approximately sixfold in ER-stressed C2C12 cells (Figure 7A), thus making it unlikely that transcriptional effects or RIDD activity of IRE1α can explain the loss of insulin receptor β chains in ER-stressed cells.
FIGURE 7:
Inhibition of insulin receptor synthesis at the transcriptional or translational level cannot fully account for decreased insulin-stimulated AKT S473 phosphorylation. (A) INSR mRNA levels measured by reverse transcriptase-qPCR in C2C12 cells treated with 300 nM thapsigargin, 1 μg/ml tunicamycin, or 1 μg/ml SubAB for 24 h. Bars represent SEs (n = 3). p values for comparison of ER-stressed samples to unstressed samples were calculated using ordinary one-way ANOVA with Dunnett’s multiple comparisons test. (B–D) Protein synthesis rates in (B) 3T3-F442A (n = 8), (C) C2C12 (n = 8), and (D) Hep G2 (n = 4) cells treated with 0.1 μM thapsigargin or 0.1 μg/ml tunicamycin for 24 h measured by incorporation of [35S]-L-methionine into protein. Protein synthesis rates were determined as TCA-precipitable counts standardized to total protein. Bars represent SEs. p values were calculated by ordinary one-way ANOVA with Tukey’s multiple comparisons test. (E) Protein synthesis rates in cells treated for 24 h with 0.1 μM thapsigargin or 0.1 μg/ml tunicamycin measured by storage phosphor analysis of [35S]-L-methionine incorporation into protein. For each cell line, the storage phosphor image of the SDS–PAGE gel is shown to the left and an image of the Coomassie Brilliant Blue R-250–stained gel is shown to the right. Quantification of [35S]-L-methionine incorporation into protein, expressed as the volume of the 35S storage phosphor signal relative to the Coomassie Brilliant Blue R-250 staining intensity is shown in the bar graphs below the gel images. Bars represent SEs (n = 8). p values were calculated by ordinary one-way ANOVA with Tukey’s multiple comparisons test. (F) Phosphorylation of eIF2α at S51 in 3T3-F442A cells (n = 13 for 0.5 h and n = 12 for 24 h), C2C12 cells (n = 21), and Hep G2 cells (n = 13 for 0.5 h and n = 8 for 24 h) exposed for 0.5 or 24 h to 0.1 μg/ml tunicamycin or 0.1 μM thapsigargin. The treatment with 0.1 μM thapsigargin for 0.5 h is a positive control for the pS51-eIF2α Western blots. Bars represent SEs. For 3T3-F442A cells, p values were calculated by Welch’s ANOVA with Dunnett’s T3 multiple comparisons test. For C2C12 and Hep G2 cells, p values were calculated by a Kruskal–Wallis test with Dunn’s multiple comparisons test. (G) Immunoprecipitation of the insulin receptor after a 1 h label with [35S]-L-methionine in 3T3-F442A cells. Thapsigargin was used at 0.1 μM and tunicamycin at 0.1 μg/ml. Abbreviations: Mock, immunoprecipitation with nonimmune IgG; 1st, immunoprecipitation of the insulin receptor after an 8 h labeling period; 2nd, immunoprecipitation of insulin receptors remaining in the supernatant of the 1st immunoprecipitation. #1, #2, and #3 indicate three biological repeats. (H) Quantification of newly synthesized α-β proreceptors in 3T3-F442A cells (left) and Hep G2 cells (right). Bars represent SEs (n = 3). p values were calculated by ordinary one-way ANOVA with Tukey’s multiple comparisons test.
To explore whether a translational arrest can explain the loss of β chains, we labeled newly synthesized proteins for 30 min with a mix of [35S]-l-methionine and [35S]-l-cysteine and measured incorporation of [35S]-l-methionine/[35S]-l-cysteine into protein by scintillation counting of TCA precipitates (Figure 7, B–D) or after separating equal amounts of proteins on SDS–PAGE gels by storage phosphor imaging (Figure 7E). Storage phosphor signals were normalized to the intensity of Coomassie Brilliant Blue R-250 staining of the gels to account for small variations in loading of SDS–PAGE gels. Overall, scintillation counting of TCA precipitates and storage phosphor imaging of gels gave very similar results (Figure 7, B–E). In C2C12 and Hep G2 cells, 0.1 μM thapsigargin decreased protein synthesis rates measured by scintillation counting to 52 ± 4% and 66 ± 4% of untreated cells, respectively, or 55 ± 3% and 79 ± 2% when measured by storage phosphor imaging. In contrast, treatment with 0.1 μg/ml tunicamycin for 24 h did not affect protein synthesis rates (Figure 7, C–E). Surprisingly, both thapsigargin and tunicamycin increased protein synthesis rates in 3T3-F442A cells 1.97 ± 0.04- and 1.61 ± 0.03-fold when measured by storage phosphor imaging and 2.1 ± 0.1- and 2.1 ± 0.2-fold when measured by scintillation counting. These effects of both thapsigargin and tunicamycin on protein synthesis rates in 3T3-F442A cells were seen in four independent experiments. To investigate whether decreased protein synthesis in thapsigargin-treated C2C12 and Hep G2 cells may be caused by increased phosphorylation of eIF2α at S51, we examined phosphorylation of eIF2α at S51 by Western blotting. While treatment with 0.1 μM thapsigargin for 30 min led to a dramatic increase in phosphorylation of eIF2α in all three cell types, neither treatment with 0.1 μM thapsigargin or 0.1 μg/ml tunicamycin for 24 h affected phosphorylation of eIF2α at S51 (Figure 7F). Therefore, the inhibitory effect of 24 h of thapsigargin treatment of C2C12 and Hep G2 cells on protein synthesis rates is independent of phosphorylation of eIF2α at S51.
To directly establish whether ER stress affects synthesis of new insulin receptors, we measured synthesis rates of the α-β proreceptor by incorporation of [35S]-l-methionine/[35S]-l-cysteine into newly synthesized proteins and immunoprecipitation of the insulin receptor with an antibody against the β chain of the mature insulin receptor. Immunoprecipitates were resolved by SDS–PAGE after being boiled for 5 min in 10% (wt/vol) SDS and 2.5% (vol/vol) β-mercaptoethanol (see Materials and Methods for details). Initial experiments showed very faint bands for both the α-β proreceptor and β chain in cell lysates prepared from C2C12 cells labeled for 8 h with ∼70% [35S]-l-methionine/∼25% [35S]-l-cysteine (unpublished data). Much stronger signals obtained with lysates prepared from Hep G2 and especially 3T3-F442A cells labeled for 8 h allowed us to identify several bands that were not observed in a control immunoprecipitation with normal rabbit immunoglobulin G (IgG) (Figure 7G). The running positions of these bands in SDS–PAGE identified two of these bands as the α-β proreceptor and the β chain of the mature insulin receptor (Figure 7G). When cells were labeled for 1 h with [35S]-l-methionine/[35S]-l-cysteine, the band representing the β chain of the mature insulin receptor was no longer detected (Figure 7G). Quantification of storage phosphor signals revealed that in 3T3-F442A cells ≤2.6 ± 0.4% and in Hep G2 cells ≤9.0 ± 0.1% of α-β proreceptors were processed to mature insulin receptors in the 1 h label. These values likely represent an upper limit of α-β proreceptor processing in the 1 h label, because of the contribution of other comigrating, [35S]-l-methionine/[35S]-l-cysteine–labeled proteins to the storage phosphor signal at the migration position of the β chain. Hence, the number of α-β proreceptors synthesized in the 1 h labeling period is representative of the synthesis rate of the α-β proreceptor.
Consistent with total protein synthesis rates, 3T3-F442A cells treated for 24 h with 0.1 μM thapsigargin showed a 2.33 ± 0.04-fold increase in α-β proreceptor synthesis, while treatment of 3T3-F442A cells with 0.1 μg/ml tunicamycin or Hep G2 cells with either 0.1 μM thapsigargin or 0.1 μg/ml tunicamycin for 24 h did not affect α-β proreceptor synthesis (Figure 7, G and H). When cells were treated with tunicamycin, α-β proreceptors migrated faster in SDS–PAGE due to their decreased molecular weights caused by inhibition of N-glycosylation by tunicamycin (Figure 7G). In summary, these experiments reveal that conditions exist in which ER stress for >12 h decreases insulin-stimulated S473 phosphorylation of AKT without decreasing general protein synthesis or synthesis of the insulin proreceptor, for example exposure of C2C12 cells for 24 h to 0.1 μg/ml tunicamycin (Figures 6 and 7) and 24–36 h exposure of Hep G2 cells to low concentrations of thapsigargin or tunicamycin (Supplemental Figure S5; Figure 7).
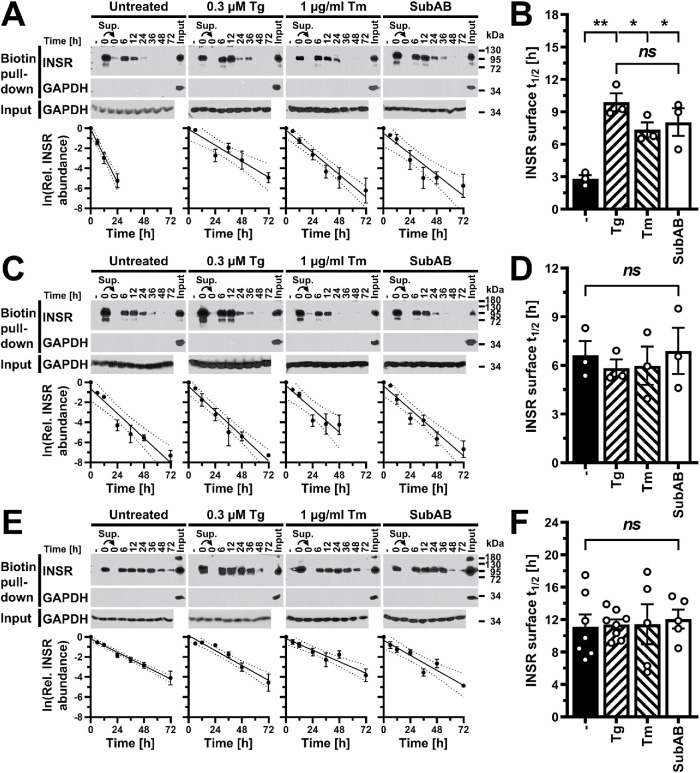
ER stress does not increase the rate of insulin receptor turnover at the cell surface
Another possibility for how ER stress may deplete β chains of the mature insulin receptor is that increased proteolytic activity associated with the secretory pathway, either lysosomal proteolytic activity (Chiang et al., 2012; Imanikia et al., 2019) or proteasomal activity associated with the ER (Casagrande et al., 2000; Friedlander et al., 2000; Termine et al., 2009; Ron et al., 2011; Chiang et al., 2012), results in increased turnover of mature insulin receptors in ER-stressed cells. To address this possibility, we determined the half-life of the insulin receptor at the cell surface in unstressed cells and in cells in which ER stress was induced with 0.3 μM thapsigargin, 1 μg/ml tunicamycin, or 1 μg/ml SubAB. We biotinylated surface exposed proteins with the membrane-impermeable biotinylation reagent sulfosuccinimidyl-6-(biotinamido)hexanoate and then continued to culture cells in the presence or absence of ER stressors for up to 72 h. At several times after biotinylation of cell surface proteins, we analyzed proteins isolated with streptavidin–agarose beads by SDS–PAGE and Western blotting. The stability of the streptavidin–biotin interaction in 6 M urea (Kurzban et al., 1991) allowed us to wash streptavidin–agarose beads twice with 6 M urea, 1% (vol/vol) Triton X-100 to remove nonbiotinylated proteins. Western blotting of proteins isolated on streptavidin–agarose beads for the abundant intracellular protein GAPDH revealed that GAPDH was not retained on streptavidin–agarose beads (Figure 8), showing that intracellular proteins were not biotinylated and that GAPDH was not retained on the beads via nonspecific interactions. Streptavidin–agarose beads also did not purify any insulin receptors from cells that were not exposed to the biotinylation reagent (Figure 8, A, C, and E, lanes labeled “–” ), showing that insulin receptors were retained on streptavidin–agarose beads only when they were biotinylated and, hence, surface exposed. When the supernatant of a pull-down reaction with streptavidin–agarose beads was used in a second pull-down reaction with a new aliquot of streptavidin–agarose beads (Figure 8, A, C, and E, lanes labeled “0” and marked with an arrowhead), no or only faint bands were obtained for the β chain of the insulin receptor, showing that the yield of the first pull-down reaction consistently was >96%. In all cell lines and under all conditions, the abundance of insulin receptors that bound to streptavidin–agarose beads decreased over time, indicating that initially surface exposed and biotinylated insulin receptor molecules were degraded. In all cases the decay of biotinylated insulin receptors followed a monoexponential relationship (Figure 8). This allowed us to calculate half-lives for the insulin receptor at the cell surface from the slopes of linearized relationships and to compare half-lives between unstressed cells and cells exposed to ER stressors (Figure 8). The calculated half-lives are similar to previously reported half-lives of total cellular insulin receptors (Reed and Lane, 1980; Reed et al., 1981a, b; Kasuga et al., 1981; Krupp and Lane, 1981; Grako et al., 1992) and insulin receptors at the cell surface (Rosen et al., 1979; Kasuga et al., 1981; Savoie et al., 1986). In C2C12 and Hep G2 cells, induction of ER stress with 0.3 μM thapsigargin, 1 μg/ml tunicamycin, or 1 μg/ml SubAB did not alter the half-life of the insulin receptor at the cell surface (Figure 8, D and F). By contrast, ER stress increased the half-life of insulin receptors at the surface of 3T3-F442A cells (Figure 8B). Therefore, increased turnover and degradation of cell surface–exposed insulin receptors cannot explain the loss of insulin receptors leading to a decrease in insulin-stimulated S473 phosphorylation of AKT in ER-stressed cells.
FIGURE 8:
ER stress does not increase turnover of insulin receptors at the cell surface. (A, C, E) Pull down of cell surface proteins with streptavidin–agarose after biotinylation with the cell-impermeable biotinylation reagent sulfosuccinimidyl-6-(biotinamido)hexanoate from (A) 3T3-F442A, (C) C2C12, and (E) Hep G2 cells. Cell extracts were prepared 0–72 h after labeling of cell surface proteins. Biotinylated proteins were isolated with streptavidin–agarose, separated by SDS–PAGE, and Western blotted for the insulin receptor and GAPDH. “–” refers to a pull-down reaction with unlabeled cell lysates. The arrows indicate that the supernatant of the pull-down reaction with the labeled 0 h sample was subjected to a second pull down with streptavidin–agarose. The lanes labeled “Input” serve as a positive control for the GAPDH Western blots on precipitates of the streptavidin–agarose pull-down reactions and themselves were not subjected to pull down with streptavidin–agarose. The rows labeled “Input” show Western blots for GAPDH on equal amounts of input protein for the streptavidin–agarose pull-down assays. The graphs show plots of the natural logarithm of the abundance of biotinylated insulin receptors over time, the line of linear regression (uninterrupted line), and the 95% confidence interval of the line of linear regression (dotted lines). (B, D, F) Comparison of the half-life, t1/2, of the insulin receptor at the cell surface of (B) 3T3-F442A (n = 3), (D) C2C12 (n = 3), and (F) Hep G2 (untreated n = 7, 0.3 μM thapsigargin n = 8, and 1 μg/ml tunicamycin and SubAB n = 5) cells. Half-lives were calculated from the slopes of linear regression lines obtained from plots of the natural logarithm of the abundance of biotinylated insulin receptors over time. Bars represent SEs. p values were calculated by ordinary one-way ANOVA with Tukey’s multiple comparisons test.
Unprocessed α-β proreceptors accumulate in the ER of ER-stressed cells
Since effects on transcription, translation, or degradation of insulin receptors cannot fully explain the loss of mature insulin receptors in ER-stressed cells, we characterized whether transport of newly synthesized insulin receptors to the plasma membrane is inhibited by ER stress. Consistent with this hypothesis is that while mature β chains decrease in ER-stressed cells, the levels of α-β proreceptors increase relative to the levels of the β chains (Figure 9A; Supplemental Figure S5D). This suggests that α-β proreceptors accumulate in an early compartment of the secretory pathway such as the ER or cis-Golgi, because cleavage of the proreceptor into α- and β chains occurs in the trans-Golgi network (Robertson et al., 1993; Bravo et al., 1994). To provide additional evidence that proreceptors accumulate in the ER or cis-Golgi, we digested protein extracts from un- and ER-stressed C2C12 cells with Endo H. Endo H releases high-mannose and some hybrid type N-linked oligosaccharides from glycoproteins by cleaving between the two N-acetylglucosamine units (Maley et al., 1989). High-mannose oligosaccharides are characteristic of proteins that have not been processed by enzymes in the Golgi complex. Endo H–digested α-β proreceptors migrated at the same position in SDS–PAGE as fully deglycosylated proreceptors synthesized in tunicamycin-treated cells (Figure 9, B and D) or obtained with PNGase F (Maley et al., 1989) (Figure 9, B–D), suggesting that none of the N-glycans on the majority of α-β proreceptors were exposed to processing enzymes of the Golgi complex. By contrast, mature α and β chains carry both Endo H–sensitive and Endo H–resistant N-linked oligosaccharides (Heidenreich and Brandenburg, 1986; Hwang and Frost, 1999, and Figure 9D). These data are consistent with the conclusion that α-β proreceptors accumulate in the ER or cis-Golgi of ER-stressed cells.
FIGURE 9:
Unprocessed α-β proreceptors accumulate in the ER of ER-stressed cells. (A) Quantification of the relative abundance of α-β precursors of the insulin receptor in C2C12 cells exposed to the indicated concentrations of thapsigargin, tunicamycin, 1 μg/ml SubAB, or 1 μg/ml SubAA272B for 24 h. Bars represent SEs (n = 12 for unstressed, insulin-stimulated cells, n = 5 for the samples treated with 0.3 μM thapsigargin and SubAA272B, and n = 6 for all other samples). p values for comparison of ER-stressed samples and samples not stimulated with 100 nM insulin to the sample stimulated with 100 nM insulin were calculated using ordinary two-way ANOVA with Dunnett’s multiple comparisons test including data for 12 h (n = 8 for unstressed, insulin-stimulated cells, n = 4 for all other samples) and 18 h (n = 10 for unstressed, insulin-stimulated cells, n = 5 for all other samples) time points. (B) Endo H digest of cell lysates prepared from C2C12 cells exposed to 1 μg/ml SubAB, 0.3 μM thapsigargin, or 1 μg/ml tunicamycin. (C) Quantification of the relative abundance of β chains and α-β proreceptors from panel B. (D) The mature β chain of the insulin receptor carries Endo H–sensitive N-linked oligosaccharides. Endo H and PNGase F digests of unstressed C2C12 cells were immunoblotted for the β chain of the insulin receptor. “#” indicates an unspecific band. (E) Steady-state insulin receptor levels in untreated HEK 293 cells or HEK 293 cells treated for 18 h with 0.1 μg/ml tunicamycin, 1 μg/ml SubAB or 1 μg/ml SubAA272B. (F) MTT activity of untreated HEK 293 cells and HEK 293 cells exposed for 18 h to 300 nM thapsigargin, 1 μg/ml tunicamycin, or 1 μg/ml SubAB. Bars represent SEs (n = 3). p values for comparison of treated samples to the untreated sample were calculated by ordinary one-way ANOVA with Dunnett’s multiple comparisons test. (G) Localization of GFP-tagged insulin receptors in transiently transfected HEK 293 cells after 18 h treatment with 1 μg/ml tunicamycin or 1 μg/ml SubAB. Scale bar = 10 μm. (H) Average Pearson’s correlation coefficient robs between the GFP-tagged insulin receptor and CellMask Deep Red fluorescence determined from 11 randomly chosen cells. Bars represent SEs (n = 10 for tunicamycin-treated samples and n = 11 for all other samples). p values for comparison of the treated to the untreated samples were calculated with an ordinary two-way ANOVA using Tukey’s multiple comparisons test on arcsine-transformed data. The Pearson correlation coefficients for the randomized images are –0.13 ± 0.08, –0.13 ± 0.07, and –0.33 ± 0.07 for the untreated, tunicamycin-treated, and SubAB-treated cells and are significantly different (p < 0.001, calculated with a two-way ANOVA using Šidák’s correction for multiple comparisons [ Šidák, 1967] on arcsine-transformed data) from the corresponding Pearson correlation coefficients for the nonrandomized images.
To directly establish whether insulin receptors deplete at the plasma membrane and accumulate in intracellular compartments, we compared the localization of C-terminally GFP-tagged insulin receptors expressed in HEK 293 cells treated for 18 h with 0.1 μg/ml tunicamycin or 1 μg/ml SubAB to untreated HEK 293 cells. HEK 293 cells were chosen for these experiments because they can be easily transfected, in contrast to Hep G2 cells, do not grow in clumps, and adhered better to culture vessels when treated with ER stressors than C2C12 cells. ER stress lasting for 18 h depletes insulin receptor β chains in HEK 293 cells (Figure 9E) and slightly decreases cell numbers (Figure 9F). In unstressed cells, the GFP-tagged insulin receptor predominantly localized to the plasma membrane (Figure 9G), which is supported by the high Pearson’s correlation coefficient, robs, for the GFP fluorescence and the fluorescence of the CellMask Deep Red plasma membrane stain (Figure 9H). By contrast, ER-stressed HEK 293 cells displayed intracellular GFP fluorescence and decreased colocalization of GFP and CellMask Deep Red fluorescence (Figure 9, G and H). These observations are consistent with the conclusion that ER stress depletes the population of insulin receptors at the plasma membrane by interfering with trafficking of newly synthesized insulin receptors from the ER to the plasma membrane.
AKT activation by a cytosolic F V2E-insulin receptor chimera is not affected by ER stress
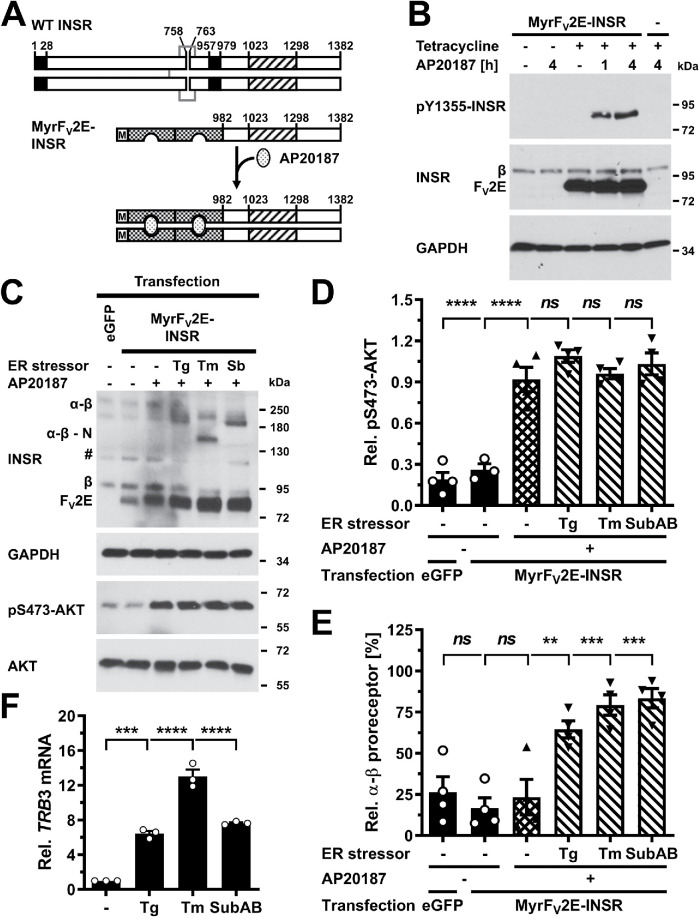
To establish whether inhibition of transport of insulin receptors in the secretory pathway is necessary for ER stress to cause insulin resistance, we bypassed the secretory pathway in synthesis of functional insulin receptor protein tyrosine kinase domains by creating a chimera in which the signal peptide, extracellular and transmembrane domains of the insulin receptor have been replaced by an N-terminal myristoylation signal and the FV2E domain (Figure 10A). The myristoylation signal mediates N-terminal myristoylation of the protein and its anchoring to intracellular membranes (Maurer-Stroh et al., 2002a, b). The FV2E domain contains two binding sites for the bivalent macrolide AP20187 and binds AP20187 with subnanomolar affinities (Clackson et al., 1998; Yang et al., 2000). Binding of AP20187 to the FV2E domain induces dimerization of the chimeric protein. Dimerization of the FV2E-insulin receptor chimera with AP20187 in stably transfected Flp-In T-Rex 293 cells increased phosphorylation of the chimera at Tyr-610, which is equivalent to Tyr-1355 in the long isoform of the human insulin receptor, showing that the chimera possesses tyrosine autophosphorylation activity (Figure 10B). Because AKT S473 phosphorylation was unresponsive to serum starvation in Flp-In T-Rex 293 cells (unpublished data), we transiently transfected the chimera into C2C12 myoblasts. In C2C12 cells, AP20187 stimulated AKT S473 phosphorylation ∼4.5 fold (Figure 10, C and D). Thus, activation of the FV2E-insulin receptor chimera recapitulates several events in insulin signaling. In transiently transfected C2C12 cells ER stress induced for 24 h with thapsigargin, tunicamycin, or SubAB reduced endogenous β chains by ∼40% (Figure 10C) but did not affect activation of AKT by the chimera (Figure 10, C and D). Furthermore, ER stress induced with thapsigargin, tunicamycin, or SubAB increased the abundance of unprocessed endogenous α-β proreceptors (Figure 10E) and induced TRB3 (Figure 10F). Hence, activation of AKT by the cytosolic, AP20187-stimulated insulin receptor chimera is not affected by ER stress despite induction of TRB3, depletion of β chains of the mature, endogenous insulin receptor, and accumulation of unprocessed endogenous α-β proreceptors.
FIGURE 10:
Bypass of the ER in insulin receptor synthesis abrogates ER stress–induced insulin resistance. (A) Schematic of the WT insulin receptor, the myristoylated FV2E-insulin receptor chimera, and activation of the chimera by AP20187. Black boxes represent the signal peptide sequence and transmembrane domain of the insulin preproreceptor, striped boxes the protein tyrosine kinase domain of the insulin receptor, and checkered boxes individual FV domains. Disulfide bonds that link α to β chains and two insulin receptor monomers are shown as gray lines. Abbreviations: M, Myr, myristoylation signal. (B) Autophosphorylation of the FV2E-insulin receptor chimera in stably transfected Flp-In T-Rex 293 cells. Expression of the chimera was induced for 27 h with 1 μg/ml tetracycline, followed by dimerization of the chimera with 100 nM AP20187 for 1 or 4 h. (C) C2C12 cells were transiently transfected with pmaxGFP or pcDNA5/FRT/TO-MyrFV2E-INSR. Twenty-four hours after transfection, ER stress was induced for 24 h with 0.1 μM thapsigargin, 0.1 μg/ml tunicamycin, or 1 μg/ml SubAB followed by dimerization of the receptor chimera with 100 nM AP20187 for 4 h. “#” indicates an unspecific band. Quantification of (D) S473 AKT phosphorylation and (E) the relative abundance of α-β proreceptors in panel C. Bars represent SEs (n = 3-4). p values for comparison of the treated to the untreated samples were calculated by ordinary one-way ANOVA using Dunnett’s multiple comparisons test. (F) Induction of TRB3 in C2C12 cells by ER stress. C2C12 cells were treated with 300 nM thapsigargin, 1 μg/ml tunicamycin, 1 µg/ml SubAB, or 1 µg/ml SubAA272B for 24 h. TRB3 mRNA levels were determined by reverse transcriptase-qPCR and standardized to the loading control ACTB. Bars represent SEs (n = 3). p values for comparison of treated samples to the untreated sample (“–”) were calculated by ordinary two-way ANOVA with Dunnett’s multiple comparisons test, taking data shown in Figure 1, E and F, into account.
Pharmacologic inhibition of JNKs does not rescue insulin-stimulated S473 phosphorylation of AKT in ER-stressed cells
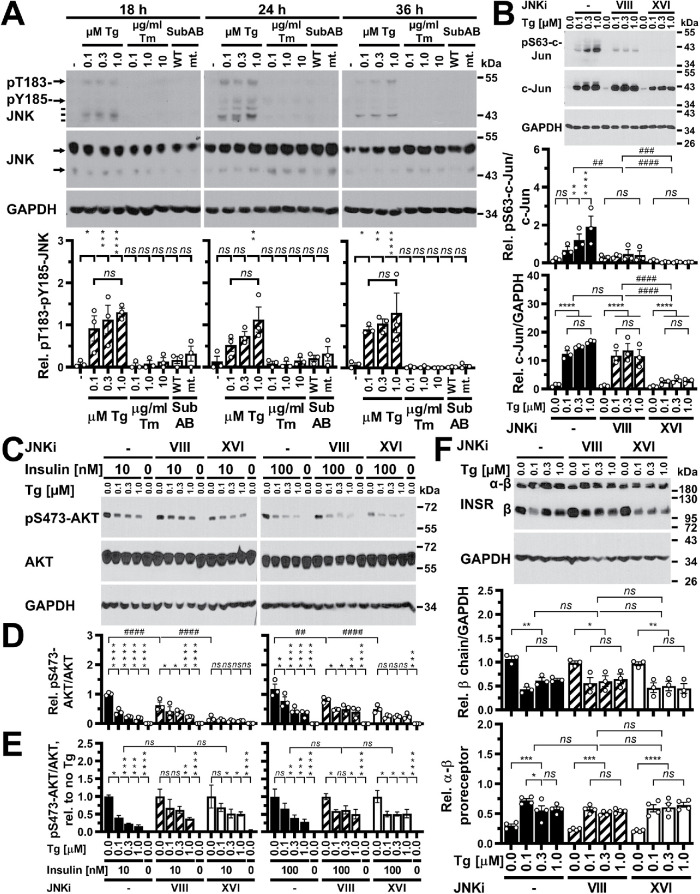
Previous work has suggested that activation of both the family of JNK MAP kinases and TRB3 by ER stress leads to a decrease in insulin-stimulated AKT phosphorylation in ER-stressed cells (Özcan et al., 2004; Koh et al., 2006, 2013). The lack of effects of 24 h of ER stress on the FV2E-insulin receptor chimera (Figure 10) prompted us to characterize whether the decrease in insulin-stimulated AKT phosphorylation in cells experiencing ER stress for >12 h manifests independent of activation of JNKs or TRB3. We first characterized JNK activation in C2C12 and Hep G2 cells exposed to ER stress for 12–36 h using phosphorylation of JNKs at T183 and Y185 in their T-loops as a marker for their activation (Lawler et al., 1998; Fleming et al., 2000). No signals for JNKs phosphorylated at T183 and Y185 were obtained for C2C12 cells exposed to 0.1–1.0 μM thapsigargin, 0.1–10 μg/ml tunicamycin, or 1 μg/ml SubAB for 12, 18, or 24 h or Hep G2 cells exposed to 0.1–10 μg/ml tunicamycin or 1 μg/ml SubAB for 18, 24, or 36 h in Western blots, despite detection of very strong signals in cells irradiated with UV light (400 J/m2, λ = 254 nm; Figure 11A and unpublished data). Thus, if JNKs are activated in these conditions, the levels of JNK species phosphorylated at T183 and Y185 are below levels that can be detected by Western blotting. By contrast, exposure of Hep G2 cells to 0.1–1.0 μM thapsigargin for 18–36 h revealed activation of JNKs (Figure 11A). Additional bands detected with the anti–phospho-T183-phospho-Y185-JNK antibody that migrate below the migration position of the 46 and 54 kDa isoforms of JNKs may represent phosphorylated species of other MAP kinases, such as p38, p42, and p44 (Figure 11A), suggesting that exposure to thapsigargin for ≥18 h may activate several MAP kinases in Hep G2 cells.
FIGURE 11:
JNKs are not required for insulin resistance in ER-stressed cells. (A) Activation of JNK in Hep G2 cells exposed to different concentrations of thapsigargin, but not tunicamycin or SubAB, for 18–36 h. The arrows indicate the p46 and p54 isoforms of JNKs; the three lines phosphorylated species of p38, p42, and p44 MAP kinases. Bars represent SEs (n = 3). p values for comparison of treated samples to the untreated sample (“–”) were calculated by ordinary two-way ANOVA with Tukey’s multiple comparisons test. (B) JNKi VIII and XVI inhibit phosphorylation of c-Jun at S63 in Hep G2 cells exposed to the indicated concentrations of thapsigargin for 36 h. Hep G2 cells were treated with 8 μM JNKi VIII or XVI for 0.5 h before exposure to thapsigargin in the presence of 8 μM JNKi VIII or XVI or no JNK inhibitor (“–”) for 36 h. The bar graphs show S63 phosphorylation of c-Jun standardized to c-Jun levels and c-Jun levels standardized to GAPDH levels. Bars represent SEs (n = 3). p values were calculated by ordinary two-way ANOVA with Tukey’s multiple comparisons test on square root or arctangent-transformed data, respectively. (C–E) JNKi VIII and XVI do not reverse inhibition of insulin-stimulated phosphorylation of AKT at S473 by thapsigargin in Hep G2 cells. Hep G2 cells were treated for 0.5 h with 8 μM JNKi VIII or XVI, followed by exposure to the indicated concentrations of thapsigargin in the presence of 8 μM JNKi VIII or XVI or no JNK inhibitor (“–” ) for 36 h. Cells were serum-starved in the last 18 h of thapsigargin treatment and then stimulated with 10 or 100 nM insulin for 15 min in the continued presence of thapsigargin and JNK inhibitors, where applicable. (C) Western blots for pS473-AKT, AKT, and GAPDH. (D) Quantification of the Western blots in panel C. Bars represent SEs (n = 3). p values were calculated by ordinary two-way ANOVA with Dunnett’s multiple comparisons test. (E) Reanalysis of the data in panel D after normalization of data for each condition of JNK inhibition to the insulin-stimulated sample not exposed to thapsigargin for the corresponding condition. p values were calculated by ordinary two-way ANOVA with Dunnett’s multiple comparisons test. (F) JNKi VIII and XVI do not restore levels of insulin receptor β chains or restore processing of α-β proreceptors to levels of untreated cells. Hep G2 cells were treated with 8 μM JNKi VIII or XVI for 0.5 h before exposure to thapsigargin in the presence of 8 μM JNKi VIII or XVI or no JNK inhibitor (“–”) for 36 h. Bars represent SEs (n = 3 for β chains and n = 4 for α-β proreceptors). p values were calculated by ordinary two-way ANOVA with Tukey’s multiple comparisons test.
To evaluate whether the JNK activation brought about by thapsigargin in Hep G2 cells contributes to the loss of insulin-stimulated S473 phosphorylation of AKT, we employed two selective JNK inhibitors, N-(4-amino-5-cyano-6-ethoxypyridin-2-yl)-2-(2,5-dimethoxyphenyl)acetamide (JNKi VIII) and (E)-3-(4-(dimethylamino)but-2-enamido)-N-(3-methyl-4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)benzamide (JNKi XVI). Both inhibitors inhibited phosphorylation of the JNK substrate c-Jun in Hep G2 cells stimulated with UV or 5 μg/ml anisomycin for 30 min with submicromolar EC50 values that are comparable to previously reported EC50 values for inhibition of c-Jun phosphorylation in tumor necrosis factor-α (TNF-α) treated Hep G2 or HeLa cells (Szczepankiewicz et al., 2006; Zhang et al., 2012) (Supplemental Figure S6). Both inhibitors also inhibited c-Jun phosphorylation at S63 in Hep G2 cells exposed to 0.1–1.0 μM thapsigargin for 36 h (Figure 11B). Thapsigargin increased steady-state levels of c-Jun (Figure 11B), which is consistent with c-Jun autoregulating its own expression (Angel et al., 1988). JNKi XVI at 8 µM decreased the increase in c-Jun levels, while the same concentration of JNKi VIII had no effect on the increase in c-Jun levels in thapsigargin-treated Hep G2 cells (Figure 11B), which correlates with the stronger inhibition of c-Jun S63 phosphorylation elicited by thapsigargin by JNKi XVI. Both JNK inhibitors, however, did not reverse inhibition of phosphorylation of S473 of AKT in Hep G2 cells that were exposed to 0.1–1.0 μM thapsigargin for 36 h and then stimulated with 10 or 100 nM insulin for 15 min in the continued presence of thapsigargin (Figure 11, C–E). Both JNK inhibitors decreased the insulin-stimulated phosphorylation of AKT at S473 (Figure 11, C and D). Normalization of S473 phosphorylation of AKT to the sample not exposed to thapsigargin within each group (no JNKi, treatment with JNKi VIII or XVI) reinforced the conclusion that both inhibitors do not reverse the inhibitory effect of thapsigargin on insulin-stimulated S473 phosphorylation of AKT (Figure 11E). Consistent with this observation and the proposed role for depletion of insulin receptors in ER-stressed cells as the cause for inhibition of insulin-stimulated S473 phosphorylation of AKT, both JNK inhibitors also did not rescue levels of β chains of the mature insulin receptor or α-β proreceptor processing in Hep G2 cells exposed to 0.1–1.0 μM thapsigargin for 36 h (Figure 11F).
Genetic ablation of JNK1 and JNK2 does not protect MEFs from inhibition of insulin-stimulated AKT phosphorylation by ER stress
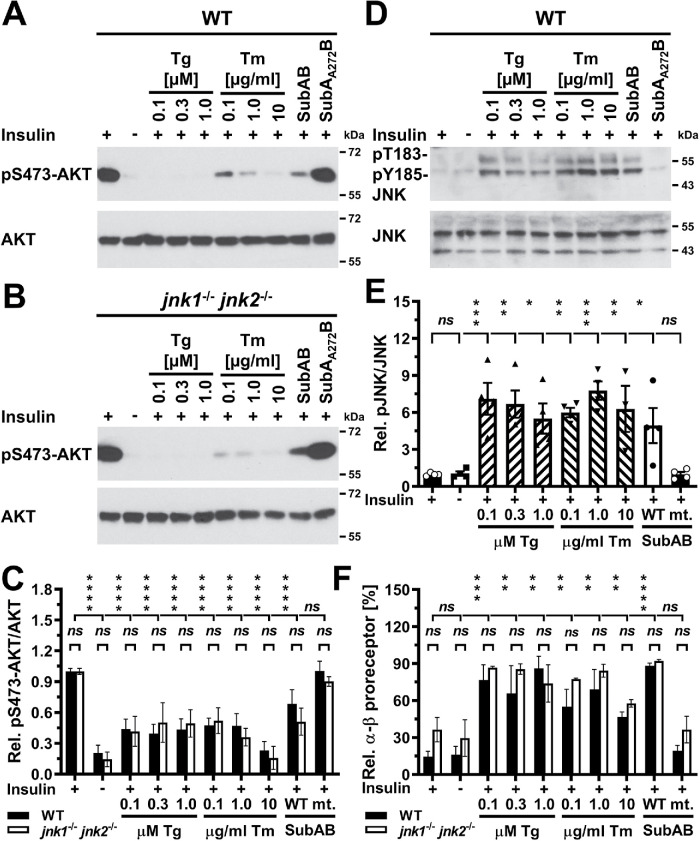
To confirm the results obtained with pharmacologic inhibitors of JNKs, we compared the effects of 24 h of ER stress on insulin-stimulated S473 phosphorylation of AKT in wild-type (WT) mouse embryonic fibroblasts (MEFs) and MEFs deficient in the JNK1 and JNK2 isoforms of the JNK kinases. These jnk1–/– jnk2–/– MEFs lack detectable JNK activity because these cells do not express the neuronal isoform of the JNKs, JNK3 (Tournier et al., 2000). Exposure of WT and jnk1–/– jnk2–/– MEFs to thapsigargin, tunicamycin, or SubAB for 24 h to elicit ER stress decreased insulin-stimulated AKT phosphorylation in both cell types to the same degree (Figure 12, A–C), despite a 6.1 ± 0.6 increase in phosphorylation of JNKs in ER-stressed WT MEFs (p < 0.0001 in an ordinary one-way analysis of variance [ANOVA]; Figure 12, D and E). ER stress also caused similar increases in the abundance of unprocessed α-β insulin proreceptors in WT and jnk1–/– jnk2–/– MEFs (Figure 12F), which suggests that activation of the JNK kinases in the ER stress response does not affect the protein folding capacity of the stressed ER.
FIGURE 12:
JNK1 and JNK2 are not required for development of insulin resistance in ER-stressed cells. (A) Serum-starved WT and (B) jnk1–/– jnk2–/– MEFs were treated for 24 h with the indicated concentrations of thapsigargin, tunicamycin, 1 μg/ml SubAB, or 1 μg/ml SubAA272B before stimulation with 100 nM insulin for 15 min. (C) Quantification of phosphorylation of AKT on S473 relative to total AKT levels in WT and jnk1–/– jnk2–/– MEFs exposed to thapsigargin, tunicamycin, and 1 μg/ml SubAB or 1 µg/ml SubAA272B. Bars represent SEs (n = 12 for unstressed, insulin-stimulated WT MEFs, n = 10 for unstressed, insulin-stimulated jnk1–/– jnk2–/– MEFs, n = 6 for all other samples for WT MEFs, and n = 5 for all other samples for jnk1–/– jnk2–/– MEFs). p values for comparison of the relative AKT S473 phosphorylation between WT and jnk1–/– jnk2–/– MEFs were calculated using ordinary two-way ANOVA with Šidák’s correction for multiple comparisons. p values for comparison of ER-stressed samples and samples not stimulated with 100 nM insulin to samples stimulated with 100 nM insulin were calculated using ordinary two-way ANOVA with Dunnett’s multiple comparisons test. (D) Activation of JNK in serum-starved WT MEFs exposed to the indicated concentrations of thapsigargin or tunicamycin, 1 μg/ml SubAB, or 1 µg/ml SubAA272B for 24 h before stimulation with 100 nM insulin for 15 min. (E) Quantification of the Western blots in panel D. Bars represent SEs (n = 8 for insulin-stimulated cells, n = 4 for all other samples). p values for comparison of ER-stressed samples and samples not stimulated with 100 nM insulin to samples stimulated with 100 nM insulin were calculated using ordinary one-way ANOVA with Dunnett’s multiple comparisons test. (F) JNK deficiency does not protect from the effects of ER stress on insulin receptor processing in the secretory pathway. The relative abundance of α-β proreceptors was determined by Western blotting of lysates of serum-starved WT and jnk1–/– jnk2–/– MEFs exposed for 24 h to the indicated concentrations of thapsigargin and tunicamycin and 1 μg/ml SubAB or 1 µg/ml SubAA272B followed by stimulation with 100 nM insulin for 15 min where indicated. Bars represent SEs (n = 4 for unstressed, insulin-stimulated cells, n = 2 for all other samples). p values for comparison of the relative abundance of α-β proreceptors between WT and jnk1–/– jnk2–/– MEFs were calculated by Welch’s test (Welch, 1947) followed by a Games–Howell post hoc test (Games and Howell, 1976). p values for comparisons of treatments were calculated by ordinary two-way ANOVA with Dunnett’s multiple comparisons test.
siRNA-mediated silencing of expression of TRB does not protect from inhibition of insulin-stimulated AKT phosphorylation by ER stress
To characterize whether TRB3 contributes to the decrease in insulin-stimulated AKT phosphorylation in cells that experience ER stress for 24 h, we used two siRNAs to knock down expression of TRB3. Forty-eight hours after transfection of C2C12 myoblasts, both siRNAs decreased TRB3 mRNA and protein levels to similar degrees (Figure 13, A and B). However, the knockdown of TRB3 mRNA and protein levels did not affect insulin-stimulated phosphorylation of AKT (Figure 13, C and D) or the accumulation of unprocessed α-β insulin proreceptors (Figure 13E) in cells exposed to thapsigargin, tunicamycin, or SubAB to elicit ER stress. These data argue that TRB3 does not contribute to decreased insulin-stimulated AKT phosphorylation or restoration of the protein folding capacity of cells that experience ER stress for 24 h. Taken together, the dispensability for JNKs and TRB3, as well as the absence of effects of ER stress on activation of AKT by the FV2E-insulin receptor chimera, argue that ER stress decreases insulin-stimulated AKT phosphorylation independent of signal transduction events.
FIGURE 13:
TRB3 is not required for development of insulin resistance in ER-stressed C2C12 cells. (A) siRNA-mediated knockdown of TRB3 at the mRNA level 48 h after transfection of C2C12 cells with 50 nM of the indicated siRNAs. TRB3 mRNA was determined by reverse transcriptase-qPCR and normalized to ACTB. Bars represent the SE from three technical replicates. p values were calculated by ordinary one-way ANOVA with Dunnett’s multiple comparisons test. (B) siRNA-mediated knockdown of TRB3 at the protein level 48 and 72 h after transfection of C2C12 cells with 50 nM of the indicated siRNAs. (C) Twenty-four hours after transfection with the indicated siRNAs, C2C12 cells were exposed to 300 μM thapsigargin, 1 μg/ml tunicamycin, or 1 µg/ml SubAB for 24 h and serum-starved during the last 18 h of exposure to ER stressors before being stimulated with 100 nM insulin for 15 min. Phosphorylation of AKT at S473 and insulin receptors were analyzed by Western blotting. (D) Quantification of the relative phosphorylation of AKT at S473 in the Western blots of panel C. Bars represent SEs (n = 5 for cells transfected with siRNAs against eGFP, n = 3 for all other samples). p values were calculated by ordinary two-way ANOVA with Dunnett’s correction for multiple comparisons. (E) Quantification of the relative abundance of α-β proreceptors in the Western blots of panel C. Bars represent SEs (n = 6 for cells transfected with siRNAs against eGFP, n = 3 for all other samples). p values were calculated by ordinary two-way ANOVA with Dunnett’s multiple comparisons test to compare of samples to the insulin-stimulated, unstressed sample and Tukey’s multiple comparisons test to compare different siRNAs.
DISCUSSION
The data presented here establish that ER stress interferes with insulin-stimulated phosphorylation of AKT by inhibiting the processing of newly synthesized insulin receptors in the secretory pathway, which will deplete the cell surface population of the receptor over time (Figure 14A). Several lines of evidence support this conclusion. ER stress extending over several half-lives of the insulin receptor at the plasma membrane inhibits insulin-stimulated AKT phosphorylation (Figure 6; Supplemental Figure S5). Periods of ER stress shorter than the half-life of 6–12 h for the insulin receptor at the plasma membrane in ER-stressed cells (Figure 8) do not affect S473 phosphorylation of AKT by insulin (Figures 1 and 2; Supplemental Figure S1) or IRS1 tyrosine phosphorylation (Figures 3 and 4; Supplemental Figures S2 and S3). The inhibition of insulin-stimulated AKT phosphorylation in ER-stressed cells coincides with a decrease in the abundance of mature insulin receptors (Figure 6, A–C; Supplemental Figure S5, A–C). Colocalization experiments revealed that while GFP-tagged insulin receptors localize to the plasma membrane of unstressed cells, this localization of insulin receptors to the plasma membrane decreases in ER-stressed cells (Figure 9, G and H). At the same time, GFP-tagged insulin receptors accumulate within the cell (Figure 9G). Unprocessed α-β proreceptors, whose N-glycans remained sensitive to Endo H (Figure 9B), accumulate in ER-stressed cells (Figure 9A; Supplemental Figure S5D), suggesting that unprocessed α-β proreceptors do not reach the trans-Golgi where they are processed to mature receptors (Robertson et al., 1993; Bravo et al., 1994). siRNA-mediated knockdown of expression of the insulin receptor confirmed that an ∼50% decrease in insulin receptor levels suffices to cause a similar decrease in insulin-stimulated AKT phosphorylation (Figure 6E). Finally, the absence of effects of ER stress on phosphorylation by the activated chimera of the FV2E domain and protein tyrosine kinase domain of the insulin receptor (Figure 10) showed that processing of insulin receptors in the secretory pathway is necessary for ER stress to inhibit insulin signaling. These experiments also suggested that signaling events in the UPR, such as activation of JNKs or TRB3, do not affect insulin signaling downstream of the insulin receptor. Pharmacologic inhibition and genetic ablation of JNKs (Figures 11 and 12) and siRNA-mediated silencing of TRB3 (Figure 13) confirmed these conclusions.
FIGURE 14:
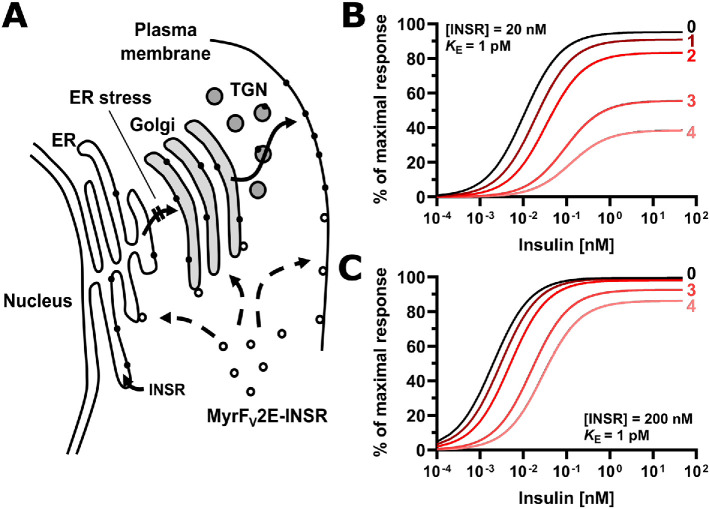
ER stress decreases insulin sensitivity by decreasing the plasma membrane population of the insulin receptor. (A) Inhibition of insulin signaling by ER stress requires transport of newly synthesized insulin receptors from the ER to the cell surface. The signal peptide sequence targets ribosomes translating the insulin receptor mRNA to the ER, where the newly synthesized polypeptide chain folds into molecules with insulin-binding activity. ER stress interferes with folding of newly synthesized insulin receptor molecules, preventing its transport to the Golgi complex. The Myr-FV2E-insulin receptor chimera is not affected by ER stress because it is translated by cytoplasmic ribosomes and folds in the cytosol into active molecules thus bypassing the ER. Abbreviation: TGN, trans-Golgi network. (B, C) Modeling of the response to insulin as a function of insulin and insulin receptor concentration. The response to insulin, R, is defined by the equation R = Rmax·[INS·INSR]/(KE + [INS·INSR]), with KE the concentration of insulin (INS)-insulin receptor (INSR) complexes at which R = 0.5·Rmax, and Rmax the maximal response to insulin. The concentration of insulin–insulin receptor complexes is calculated from the equilibrium INS + INSR ↔ INS·INSR, considering only high affinity binding of insulin to the insulin receptor with a dissociation constant of 200 pM (Bass et al., 1996). KE varies widely for different physiological responses to insulin (Gammeltoft and Gliemann, 1973; Hofmann et al., 1980; Crettaz and Kahn, 1984) and has been assumed to be equal to 1 pM for illustrative purposes only. Numbers represent the half-lives of the insulin receptor at the cell surface that have elapsed since ER stress was induced.
In previous research, tunicamycin was nearly exclusively used to inhibit trafficking of newly synthesized insulin receptors (Keefer and De Meyts, 1981; Reed et al., 1981b; Ronnett and Lane, 1981; Ronnett et al., 1984; Boyd and Raizada, 1983; Ercolani et al., 1984; Kadle et al., 1984; Capeau et al., 1985; Goldstein and Kahn, 1988), which led to the conclusion that the underlying cause for the trafficking defect is the lack of N-glycosylation of newly synthesized insulin receptors. We show that two other ER stressors, thapsigargin and SubAB, also inhibit processing of α-β proreceptors to mature receptors (Figure 9A; Supplemental Figure S5A), suggesting that newly synthesized α-β proreceptors do not reach the trans-Golgi to be cleaved into mature α- and β chains. This conclusion is supported by the observation that all N-glycans of α-β proreceptors in cells treated with thapsigargin or SubAB remain sensitive to Endo H (Figure 9B). The migration of α-β proreceptors synthesized in the presence of thapsigargin or SubAB in SDS–PAGE suggests that these proreceptors are N-glycosylated to the same extent as proreceptors synthesized by unstressed cells (Figures 6A, 7G, and 9B; Supplemental Figure S5A), suggesting that both thapsigargin and SubAB do not inhibit N-glycosylation of newly synthesized insulin receptors. Therefore, events other than lack of N-glycosylation can inhibit transport of newly synthesized insulin receptors from the ER to the trans-Golgi. Cleavage and inactivation of the ER luminal HSP70 chaperone BiP by SubAB (Paton et al., 2006) may interfere with correct folding of newly synthesized insulin receptors, leading to their retention in the ER by the quality control machinery of the ER (Bass et al., 1998) and, to some extent, targeting of unfolded proreceptors to ERAD machinery for degradation. The effects of thapsigargin on protein folding in the ER are less well understood, but depletion of the ER luminal Ca2+ store by thapsigargin (Thastrup et al., 1990) may inhibit several molecular chaperones of the ER, because many of them bind Ca2+ ions with high capacities (Macer and Koch, 1988; Fliegel et al., 1989; Wada et al., 1991). Indirect effects resulting from depletion of proteins functioning in vesicular trafficking and sorting may also account for some of the effects on insulin receptor trafficking and may, for example, explain the increased half-life of the insulin receptor at the plasma membrane in ER-stressed 3T3-F442A cells (Figure 8, A and B).
Insulin resistance can be a consequence of decreased insulin receptor levels, inhibition of transduction of the insulin signal downstream of the insulin receptor (sometimes referred to as “postreceptor events”), or a combination of the two (Olefsky and Kolterman, 1981). The experiments with the FV2E-insulin receptor chimera (Figure 10) show that ER stress does not affect insulin signaling downstream of the receptor. Our work also establishes that plasma membrane levels of the receptor decrease (Figure 9, G and H). Therefore, we propose that ER stress primarily causes insulin resistance by decreasing the levels of insulin receptors at the cell surface. Decreases in the number of insulin receptors affect insulin sensitivity of cells, in other words, shift the response curve to insulin toward higher insulin concentrations, but decrease the response to insulin only when receptors become severely depleted (see, for example, Figure 14, B and C, curves labeled “0” and “1” in B) (Olefsky and Kolterman, 1981). Elevated insulin concentrations can compensate for decreased insulin sensitivity, because many cell types possess “spare” receptors, which allows them to mount maximal responses to insulin even when only a small fraction of receptors have bound to insulin, possibly as low as a few percent (Kono and Barham, 1971; Olefsky, 1975; Le Marchand-Brustel et al., 1978; Hofmann et al., 1980; Frank et al., 1981). A decrease in the number of insulin receptors will diminish the response to insulin at one insulin concentration, but because insulin, the insulin receptor, and insulin–insulin receptor complexes are in a dynamic equilibrium, increases in the insulin concentration will compensate for the decrease in insulin receptor concentration and allow for formation of a sufficient number of insulin–insulin receptor complexes to elicit a maximal response to insulin. By contrast, a decrease in the response to insulin at all insulin concentrations is often indicative of the inhibition of signal transduction downstream of the insulin receptor (Olefsky and Kolterman, 1981). Hence, when high insulin concentrations are employed, as is often done in in vitro studies, this is expected to primarily reveal postreceptor events on insulin signaling because they affect insulin signaling independent of insulin concentration, and the high insulin concentration is thought to compensate for decreases in insulin sensitivity.
This, however, changes when insulin receptor levels decrease to such an extent that the concentration of insulin–insulin receptor complexes approaches the value of KE, the concentration of the hormone, or more precisely hormone-receptor complexes, at which the response to the hormone is half-maximal. At this point, the response to insulin will decrease over the whole insulin concentration range (Figure 14, B and C), and complete compensation by increasing insulin concentrations may, depending on the magnitude of the decrease in insulin receptor levels, no longer be possible. It then also becomes difficult, if not impossible, to distinguish between an effect on insulin sensitivity (due to decreased receptor levels) and postreceptor events, even if dose–response curves are recorded or high insulin concentrations are employed. Therefore, while previous studies have largely interpreted the effects of ER stress for >12 h as postreceptor events (Avery et al., 2010; Xu et al., 2010; Tang et al., 2011; Hassan et al., 2012; Jung et al., 2013; Panzhinskiy et al., 2013), it is also possible that the effects of long periods of ER stress are, at least in part, the consequence of profound decreases in the number of insulin receptors and subsequently insulin sensitivity. The FV2E-insulin receptor chimera described in this work may prove to be a valuable tool to distinguish more rigorously between receptor and postreceptor events in future work.
The interpretation of effects of ER stress on insulin signaling is further complicated by the gradual decrease in the number of insulin receptors at the plasma membrane over time until receptor levels fall below the critical level, at which cells no longer retain their complete responsiveness to insulin. The time it takes for insulin receptor levels to fall to this critical level depends on 1) the half-life of the receptor at the cell surface, which is affected by cell type and can be affected by ER stress (Figure 8), 2) the number of cell surface insulin receptors in unstressed cells, which can vary widely between cell types (Bar et al., 1976; Reed et al., 1977; Capeau et al., 1978), 3) the sizes of intracellular insulin receptor pools, including, for example, receptors that are transiting through the Golgi complex and receptors that are recycled at the plasma membrane at the time ER stress is induced, 4) the degree of completeness of inhibition of transport of newly synthesized receptors from the ER to the cell surface, and 5) the fraction of newly synthesized receptors that is targeted for degradation by ERAD in ER-stressed cells. For example, short periods of ER stress (<1–2 half-lives) will not deplete receptors to levels where the response to insulin decreases measurably, while long periods (>2 half-lives) will cause severe receptor depletion, severe loss of insulin sensitivity, and a loss in insulin responsiveness (Figure 14B). Likewise, an ∼10-fold-smaller plasma membrane insulin receptor population may result in measurable loss of insulin responsiveness after ER stress lasting for approximately three half-lives of the receptor at the plasma membrane, while in cells that possess 10 times more receptors, ER stress has to last for several additional half-lives before similar effects on insulin responsiveness manifest (Figure 14, B and C).
ER stress is present in muscle, liver, and adipose tissue of obese individuals (Özcan et al., 2004, 2006; Hosogai et al., 2007; Boden et al., 2008; Sharma et al., 2008; Sreejayan et al., 2008). Patients with impaired glucose tolerance or overt type II diabetes show an ∼50% decrease in the number of insulin receptors (Olefsky and Reaven, 1974, 1977; Olefsky, 1976; Goldstein et al., 1975; Pagano et al., 1977; Robinson et al., 1979; Helderman and Raskin, 1980; Kobayashi et al., 1980; Kolterman et al., 1980, 1981). This ∼50% decrease in the number of insulin receptors at the cell surface accounts for the decreased insulin sensitivity and abnormal glucose tolerance of patients with impaired glucose tolerance (Olefsky and Reaven, 1977; Pagano et al., 1977; Kolterman et al., 1980, 1981). ER stress may decrease the fraction of newly synthesized receptors that reach the plasma membrane or increase the transit time through the secretory pathway for all or some of the newly synthesized insulin receptors and through this may contribute to the decrease in the size of the steady-state cell surface population of receptors in obesity. Internalization of the insulin receptor is necessary for its degradation (Desbuquois et al., 1982; Knutson et al., 1983; Wang et al., 1983; Draznin et al., 1984). A decrease in the insulin receptor population at the cell surface is expected to decrease its internalization and degradation rates, and together with decreased synthesis rates may establish a new, smaller steady-state population of the receptor at the plasma membrane. Hyperinsulinemia in obesity may further aggravate decreases of the insulin receptor at the plasma membrane, because insulin stimulates internalization and degradation of its receptor (Kasuga et al., 1981; Knutson et al., 1982; Heidenreich et al., 1983; Ronnett et al., 1983; Freychet, 1984; Reed et al., 1984) and, in adipocytes, induces ER stress (Boden et al., 2014). Thus, the effects of ER stress on delivery of newly synthesized insulin receptors to the plasma membrane may contribute to decreased steady-state plasma membrane levels of the receptor in obesity.
In conclusion, we provide evidence that ER stress requires processing of insulin receptors in the secretory pathway to inhibit signal transduction in the insulin signaling pathway (Figure 14A). The effects of ER stress on trafficking of newly synthesized insulin receptors to the cell surface may account for the decrease in the number of insulin receptors in patients with impaired glucose tolerance and in patients with overt type II diabetes and underlie the decreased insulin sensitivity of patients with impaired glucose tolerance. Depending on the half-lives of individual plasma membrane receptors, analogous effects of ER stress on other plasma membrane receptors and transporters may exist.
MATERIALS AND METHODS
Antibodies and reagents
The mouse anti–β-actin antibody (clone 8F10-G10, cat. no. ab170325, batches GR184354-8 and GR327417-1) was purchased from Abcam (Cambridge, UK) and the rat anti–α-tubulin antibody (clone YOL1/34, cat. no. MCA78G, batch 1703) from Bio-Rad Laboratories (Hemel Hempstead, UK). Rabbit anti–phospho-T308-AKT (clone 244F9, cat. no. 4056S, batches 13 and 17), anti–phospho-S473-AKT (clone D9E, cat. no. 4060S, batches 16, 23, and 24), rabbit anti-AKT (clone C67E7, cat. no. 4691S, batches 11, 17, and 20), rabbit anti–phospho-S63-c-Jun (clone 54B3, cat. no. 2361S, batch 7), rabbit anti–c-Jun (clone 60A8, cat. no. 9165S, batch 11), rabbit anti–phospho-S51-eIF2α (cat. no. 9721S, batch 21), mouse anti-eIF2α (clone L57A5, cat. no. 2103S, batch 5), rabbit anti-insulin receptor β chain phosphorylated at Y1355 (clone 14A4, cat. no. 3026S, batch 1), rabbit anti-insulin receptor β chain (clone 4B8, cat. no. 3025S, batches 8 and 10), rabbit anti–phospho-Y891 (mouse)/Y896 (human)-IRS1 (cat. no. 3070S, batch 4), rabbit anti-IRS1 (clone D23G12, cat. no. 3407S, batch 6; clone D5T8J, cat no. 95816S, batch 1), rabbit anti–phospho-T183-phospho-Y185-JNK (clone 81E11, cat. no. 4668S, batches 9, 12 and 15), and rabbit anti-JNK (cat. no. 9252S, batches 15 and 17) antibody were purchased from Cell Signaling Technology (Danvers, MA). The mouse anti–phospho-S307 (mouse)/S312 (human)-IRS1 (clone 24.6.2, cat. no. 05-1087, batch 3063387), rabbit anti–phospho-Y608 (mouse)/Y612 (human)-IRS1 (cat. no. 09-432, batch 3018885), rabbit anti–phospho-Y628 (mouse)/Y632 (human)-IRS1 (cat. no. 09-433, batch 3023592), rabbit anti–phospho-Y935 (mouse)/941 (human)-IRS1 (cat. no. 07-848-I, batch Q2766987), mouse anti-phosphotyrosine (clone 4G10 Platinum, cat. no. 05-1050X, batches 2967319 and 3256630), and the rabbit anti-TRB3 (cat. no. 07-2160, batch 2716134) antibody were purchased from Merck Millipore (Watford, UK). Anti-insulin receptor β chain antibody (cat. no. sc-711, batch G0511) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and the mouse anti-GAPDH antibody (clone GAPDH-71.1, cat. no. G8795, batches 092M4820V and 067M4785V) from Sigma-Aldrich (Gillingham, UK).
Normal rabbit IgG was purchased from Fisher Scientific (Loughborough, UK; cat. no. 10312573, batch UA276761) and Santa Cruz Biotechnology (cat. no. sc-2027, batches C2712 and D1415). SureBeads protein A magnetic beads were purchased from Bio-Rad Laboratories (cat. no. 1614013) and streptavidin–agarose from Fisher Scientific (cat. no. 10302384, batch SJ523686). Goat anti-mouse IgG(H+L)-peroxidase (cat. no. 10330974, batch OE17149612) and goat anti-rat IgG(H+L)-peroxidase (cat. no. 11889140, batch PK209942) were purchased from Fisher Scientific, and goat anti-rabbit IgG(H+L)-peroxidase from Cell Signaling Technology (cat. no. 7074S, batches 26–28).
Sulfosuccinimidyl-6-(biotinamido)hexanoate (EZ-link-sulfo-NHS-biotin, cat. no. 11851185) was purchased from Fisher Scientific. JNK inhibitor VIII (N-(4-amino-5-cyano-6-ethoxypyridin-2-yl)-2-(2,5-dimethoxyphenyl)acetamide, cat. no. 420135-5MG), JNK inhibitor XVI ((E)-3-(4-(dimethylamino)but-2-enamido)-N-(3-methyl-4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)benzamide, cat. no. 420150-10MG), and tunicamycin (cat. no. 654380) were purchased from Merck Millipore. Endoglycosidase H (EndoH) and peptide-N-glycosidase (PNGase) F were obtained from New England Biolabs (Hitchin, UK). Bovine insulin (cat. no. I0516), bovine serum albumin (BSA; cat. no. A2153), dexamethasone, 3-isobutyl-1-methylxanthine, and thapsigargin (cat. no. T9033) were purchased from Sigma-Aldrich. Subtilase cytotoxin (SubAB) and catalytically inactive SubAA272B were purified from Escherichia coli as described before (Paton et al., 2004; Talbot et al., 2005). siRNAs against murine INSR mRNA and Aequora victoria enhanced green fluorescent protein (eGFP) were purchased from Sigma-Aldrich and siRNAs against murine TRB3 from Fisher Scientific. siRNA sequences are listed in Table 1.
TABLE 1:
siRNAs.
| Species | Gene | # | Sequence, sense strand | Sequence, antisense strand |
|---|---|---|---|---|
| Mus musculus | INSR | 1 | GAGAUCUCCUGGGAUUCAUdTdT | AUGAAUCCCAGGAGAUCUCdTdT |
| M. musculus | INSR | 2 | CCUUAUCAAGGCCUGUCUAdTdT | UAGACAGGCCUUGAUAAGGdTdT |
| M. musculus | INSR | 3 | GAAACUCUGCUUGUCUGAAdTdT | UUCAGACAAGCAGAGUUUCdTdT |
| M. musculus | TRB3 | 1 | GGCAGAAGCUGUGUGGAGdTdT | CUCCACACAGGCUUCUGCdTdT |
| M. musculus | TRB3 | 2 | GGACAAUCCCUUUCACAAAdTdT | UUUGUGAAAGGGAUUGUCCdTdT |
| Aequora victoria | eGFP | GCAAGCUGACCCUGAAGUUCAU | GAACUUCAGGGUCAGCUUGCCG |
Plasmids
Plasmids were maintained in E. coli XL10-Gold cells (Agilent Technologies, Stockport, UK; cat. no. 200314). Standard protocols for plasmid constructions were used (Ausubel et al., 2017). Plasmid pmaxGFP was obtained from Lonza Cologne AG (Cologne, Germany). Plasmid pEGFP-N2-hINSR encodes a fusion of the human insulin receptor to eGFP (Bass et al., 2000) and was obtained from Addgene (Cambridge, MA; Addgene ID 22286). Plasmid pcDNA5/FRT/TO-FV2E-INSRβ was generated by cloning the 1430-base-pair BsiWI-XmaI fragment of pCLFv2IRE (Cotugno et al., 2004) into BsiWI- and XmaI-digested pcDNA5/FRT/TO-FV2E-C’hIRE1α (D. Cox and M. Schröder, Durham University, Durham, UK, unpublished data). Plasmid pcDNA5/FRT/TO-MyrFV2E-INSRβ was generated by cloning the 501-base-pair EcoRI-XmaI fragment of pC4M-FV2E (Ariad Pharmaceuticals, Cambridge, MA) into HindIII- and XmaI-digested pcDNA5/FRT/TO-FV2E-INSRβ after blunting the EcoRI and HindIII sites with Klenow enzyme.
Cell culture
WT, ire1α–/– (Lee et al., 2002), jnk1–/– jnk2–/– (Tournier et al., 2000), and traf2–/– (Yeh et al., 1997) MEFs were obtained from Randal J. Kaufman (Sanford Burnham Medical Research Institute, La Jolla, CA), Roger Davis (University of Massachusetts, Worchester, MA), and Tak Mak (University of Toronto, Ontario Cancer Institute, Toronto, Ontario, Canada), respectively. 3T3-F442A preadipocytes (Green and Kehinde, 1976), C2C12 myoblasts (Blau et al., 1985), HEK 293 cells (Graham et al., 1977, 1978; Harrison et al., 1977), and Hep G2 cells (Knowles et al., 1980) were obtained from C. Hutchison (Durham University), R. Bashir (Durham University), M. Cann (Durham University), and A. Benham (Durham University), respectively. Fao rat hepatoma cells (Deschatrette and Weiss, 1974) were obtained from Public Health England (Salisbury, UK). The Flp-In T-Rex 293 cell line was obtained from Life Technologies (Paisley, UK). All cell lines were tested for mycoplasma contamination upon receipt in the laboratory with the EZ-PCR mycoplasma test kit from Geneflow (cat. no. K1-0210; Lichfield, UK). Mycoplasma testing was repeated approximately every 3 mo with all cells in culture at the time. Contaminated cultures were discarded.
All cell lines were grown in an atmosphere of 95% (vol/vol) air, 5% (vol/vol) CO2, and 95% humidity. Fao rat hepatoma cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Moore et al., 1967) or in Coon’s modification of Ham’s F12 medium (Coon and Weiss, 1969) containing 10% (vol/vol) fetal bovine serum (FBS) and 2 mM L-glutamine. Hep G2 cells were cultured in MEM (Eagle, 1959) supplemented with 10% (vol/vol) FBS and 2 mM L-glutamine. All other cell lines were cultured in DMEM containing 4.5 g/l D-glucose (Morton, 1970; Rutzky and Pumper, 1974), 10% (vol/vol) FBS, and 2 mM L-glutamine. The medium for ire1α–/– and corresponding WT MEFs was supplemented with 110 mg/l pyruvate (Lee et al., 2002). The medium for the Flp-In T-Rex 293 cells was supplemented with 100 μg/ml zeocin and 15 μg/ml blasticidin and the medium for Flp-In T-Rex 293 cells stably expressing the FV2E-insulin receptor chimera with 100 μg/ml hygromycin B and 15 μg/ml blasticidin. For immunoprecipitation of IRS1, [35S]-l-methionine pulse labeling experiments, and determination of the half-life of the insulin receptor at the cell surface, 3T3-F442A cells were cultured in gelatinized tissue culture dishes (Schröder and Friedl, 1997b).
siRNAs and plasmids were transfected with INTERFERin or jetPRIME transfection reagent (Polyplus Transfection, Illkirch, France) following the manufacturer’s instructions, respectively. The stably transfected Flp-In T-Rex 293 cell lines expressing a fusion of the FV2E drug-inducible dimerization domain (Clackson et al., 1998) to the β chain of the human insulin receptor with an N-terminal myristoylation signal were generated by transfection of the Flp-In T-Rex 293 cell line with pOG44 (Life Technologies) and pcDNA5/FRT/TO-MyrFV2E-INSRβ. Selection of stably transfected clones was initiated 24 h after transfection by using 50 μg/ml hygromycin B. After 2 d, the hygromycin B concentration was increased to 100 μg/ml.
C2C12 myoblasts were differentiated into myotubes and 3T3-F442A preadipocytes into adipocytes as described before (Mihai and Schröder, 2015; Brown et al., 2016). ER stress was induced with 0.1–1 μM thapsigargin, 0.1–10 μg/ml tunicamycin, or 1 μg/ml SubAB. To stimulate cells with insulin, cells were serum-starved for 18 h, followed by the addition of fresh serum-free culture medium containing 10 or 100 nM insulin for 5 or 15 min. Cells were serum-starved during the last 18 h of treatments with ER stressors lasting for more than 18 h. When cells were ER-stressed for shorter periods, the ER stressors were applied toward the end of the serum starvation, for example for the last 12 h of serum starvation in the case of treatment with ER stressors for 12 h. Expression of the FV2E-insulin receptor chimera was induced for 24 h with 1 μg/ml tetracycline in stably transfected Flp-In T-Rex 293 cells. The chimera was dimerized by treating cells with 100 nM AP20187 (B/B homodimeriser; TaKaRa Bio Europe, Saint-Germain-en-Laye, France; cat. no. 635058) for the times indicated in the text.
Crystal violet staining was used as a proxy to determine the number of cells remaining in culture dishes after exposure to ER stressors. Cultures were washed with phosphate-buffered saline (PBS; 4.3 mM Na2HPO4, 1.47 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4) at room temperature (RT) and stained for 10 min with 0.2% (wt/vol) crystal violet in 2% (vol/vol) ethanol at RT. After being washed with tap water to remove unbound crystal violet, the crystal violet was dissolved with 1% (wt/vol) SDS and the absorbance at 570 nm read in a Spectramax 190 microplate reader (Molecular Devices, San Jose, CA). The activity of mitochondrial redox chains was determined using thiazolyl blue tetrazolium bromide (MTT) as described previously (Mihai and Schröder, 2015), except that the absorbance at 690 nm was subtracted from the absorbance at 590 nm. Corrected MTT absorbances were normalized to the crystal violet absorbance of corresponding, identically treated cultures and expressed relative to the absorbance of cells exposed to 0.1% (vol/vol) dimethyl sulfoxide (DMSO), which was arbitrarily set to 1.0.
RNA extraction and reverse transcriptase PCRs
RNA was extracted and reverse transcribed as previously described (Cox et al., 2011). Protocols for quantification of splicing of XBP1 were described previously (Cox et al., 2011; Brown et al., 2016). qPCRs were run on a Rotorgene 3000 (Qiagen, Crawley, UK) using GoTaq G2 Flexi DNA polymerase (Promega, Southampton, UK; cat. no. M7801) and analyzed as described before (Brown et al., 2016). Primer sequences are listed in Table 2 or have been reported before (Brown et al., 2016). Amplification of a single PCR product was confirmed by recording the melting curves after each PCR. The amplification efficiencies for all qPCRs were ∼0.75 ± 0.05. Results represent the average and SE of three technical repeats. qPCR results were confirmed by at least one other biological replicate.
TABLE 2:
Oligodeoxynucleotides.
| Name | Purpose | Sequence |
|---|---|---|
| H8962 | TRB3 real time PCR, forward | TTTGGAACGAGAGCAAGGCA |
| H8963 | TRB3 real time PCR, reverse | CCACATGCTGGTGGGTAGG |
| H9044 | INSR real time PCR, forward | CTTCTCTTCCGTGTCTATGG |
| H0945 | INSR real time PCR, reverse | GACCATCTCGAAGATAACCA |
Cell lysis and Western blotting
Protein extracts for Western blotting except for extraction of IRS1 were produced as previously described (Brown et al., 2016). In brief, cells were washed three times with ice-cold PBS (pH 7.4) and lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, at 4°C, 150 mM NaCl, 0.5% [wt/vol] sodium deoxycholate, 0.1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS) containing Roche complete protease inhibitors (Roche Applied Science, Burgess Hill, UK; cat. no. 11836153001) or 10 mM EDTA, 2 mM PMSF, 6 mM 4-(2-aminoethyl)benzenesulfonyl flouride (AEBSF), 5 mM benzamidine, 10 μg/ml aprotinin, and each 1 μg/ml antipain dihydrochloride, chymostatin, leupeptin, and pepstatin A. To inhibit protein phosphatases, PhosSTOP phosphatase inhibitors (Sigma-Aldrich; cat. no. 04 906 837 001) were added when Roche complete protease inhibitors were used. When individual protease inhibitors were used, 1 mM sodium fluoride, 10 mM sodium β-glycerophosphate, 10 mM sodium pyrophosphate, and 200 nM okadaic acid were included to inhibit protein serine and threonine phosphatases. To preserve protein serine, threonine, and tyrosine phosphorylation when individual protease inhibitors were used, cells were lysed in 50 mM HEPES-NaOH (pH 8.0 at 4°C), 150 mM NaCl, 0.5% (wt/vol) sodium deoxycholate, 0.1% (vol/vol) Triton X-100, 0.1% (wt/vol) SDS, 6 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 1 mM sodium fluoride, 1 mM sodium vanadate, 10 mM sodium β-glycerophosphate, 10 mM sodium pyrophosphate, 2 mM PMSF, 6 mM AEBSF, 5 mM benzamidine, 10 μg/ml aprotinin, and each 1 μg/ml antipain dihydrochloride, chymostatin, leupeptin, and pepstatin A, 200 nM okadaic acid, 200 μM 2-bromo-4-methoxyacetophenone (Arabaci et al., 1999, 2002) and 20 μM ethyl 3,4-dephostatin (Watanabe et al., 2000; Suzuki et al., 2001). Protein concentrations were determined with the DC protein assay from Bio-Rad Laboratories (cat. no. 500-0116). Samples were denatured for 5 min at 100°C after addition of 1/6 volume of 6× SDS–PAGE sample loading buffer (350 mM Tris·HCl [pH 6.8 at RT], 30% [vol/vol] glycerol, 10% [wt/vol] SDS, 0.5 g/l bromophenol blue, 2% [vol/vol] β-mercaptoethanol). Total protein (10–100 μg) was separated by SDS–PAGE (Laemmli, 1970) and transferred onto PVDF membranes (Amersham HyBond-P, pore size 0.45 μm; GE Healthcare, Little Chalfont, UK; cat. no. RPN303F) by semidry blotting in 0.1 M Tris, 0.192 M glycine, 5% (vol/vol) methanol at 2 mA/cm2 for 75 min.
Protein extracts for Western blotting for IRS1 and phosphorylated species of IRS1 were prepared from cells serum-starved with media lacking phenol red. Media were aspirated and cells lysed in 8 M urea, 2.5% (wt/vol) SDS, 50 mM Tris·HCl (pH 7.5 at 4°C), 6 mM EDTA, 2 mM PMSF, 40 mM AEBSF, 5 mM benzamidine, and 10 μM E-64. Lysates were cleared by centrifugation at 627,000 × g for 2 h at 4°C. Protein concentrations were determined with bicinchoninic acid (Smith et al., 1985). Samples were denatured for 5 min at 70°C after addition of 1/6 volume of 6× SDS–PAGE sample loading buffer. Protein (50–200 μg) was separated on 7.5% SDS–PAGE gels and transferred to PVDF membranes by wet transfer in 10 mM NaHCO3, 3 mM NaCO3, 0.025% (wt/vol) SDS (Dunn, 1986) at 50 V for 18 h (for a 1-mm-thick gel) at 4°C after equilibration of gels for 3 × 20 min at 4°C in 10 mM NaHCO3, 3 mM NaCO3, 0.025% (wt/vol) SDS.
Membranes were blocked for 1 h at RT or overnight at 4°C in 5% (wt/vol) skimmed milk powder in TBST (20 mM Tris-HCl [pH 7.6 at RT], 137 mM NaCl, and 0.1% [vol/vol] Tween-20) or in TBST + 3% (wt/vol) skimmed milk powder when the anti-phosphotyrosine antibody was used. The anti–phospho-T308-AKT, anti–phospho-S473-AKT, anti-AKT, anti–phospho-S63-c-Jun, anti–c-Jun, anti–phospho-S51-eIF2α, anti-eIF2α, anti–phospho-Y1355-insulin receptor β chain, anti-insulin receptor β chain (clone 4B8), anti–phospho-Y891 (mouse)/Y896 (human)-IRS1, anti-IRS1 (clones D23G12 and D5T8J), anti–phospho-T183-phospho-Y185-JNK, and anti-JNK antibodies were incubated with membranes at a 1:1000 dilution in TBST + 5% (wt/vol) BSA overnight at 4°C with constant rotation. For the rabbit anti-IRS1 antibody, clone D23G12 was used to develop Western blots of cell lysates, while clone D5T8J was used to develop Western blots of immunoprecipitates of IRS1. The anti–phospho-Y608 (mouse)/Y612 (human)-IRS1 and anti–phospho-Y628 (mouse)/Y632 (human)-IRS1 antibodies were incubated with membranes at a 1:4000 dilution in TBST + 5% (wt/vol) skimmed milk power for 2 h at RT. The anti–phospho-Y935 (mouse)/941 (human)-IRS1 antibody was incubated with membranes at a 1:1000 dilution in TBST + 5% (wt/vol) skimmed milk powder for 2–4 h at RT or overnight at 4°C. The anti–phospho-S307 (mouse)/S312 (human)-IRS1 antibody was incubated with membranes at a 1:1000 dilution in TBST + 5% (wt/vol) skimmed milk powder overnight at 4°C and the anti-phosphotyrosine antibody at a 1:1000 dilution in TBST + 3% (wt/vol) skimmed milk powder for 4 h at RT. The polyclonal anti-INSR β chain antibody was used at a 1:200 dilution and the anti-TRB3 antibody at a concentration of 0.1 μg/ml in TBST + 5% (wt/vol) skimmed milk powder overnight at 4°C. The anti–β-actin antibody was used at a dilution of 1:10,000–1:20,000 in TBST + 5% (wt/vol) skimmed milk powder for 2 h at RT, the anti-GAPDH antibody at a dilution of 1:10,000–1:30,000 in TBST + 5% (wt/vol) skimmed milk powder for 2 h at RT or overnight at 4°C, and the anti–α-tubulin antibody at a dilution of 1:1000–1:2000 in TBST + 5% (wt/vol) skimmed milk powder for 2 h at RT. Membranes were washed four times with TBST for 5 min at RT and then probed with goat anti-mouse IgG(H+L)-peroxidase at a dilution of 1:5000 in TBST + 5% (wt/vol) skimmed milk power for 2 h at RT to detect the anti–phospho-S307 (mouse)/S312 (human)-IRS1 antibody and at a dilution of 1:10,000 in TBST + 5% (wt/vol) skimmed milk powder for 2 h at RT to detect all other murine antibodies. Goat anti-rabbit IgG(H+L)-peroxidase was used at a dilution of 1:2000 in TBST + 5% (wt/vol) skimmed milk powder for 2 h at RT and goat anti-rat IgG(H+L)-peroxidase at a dilution of 1:5000 in TBST + 5% (wt/vol) skimmed milk powder for 2 h at RT. Membranes were washed four times for 5 min at RT and then developed with Thermo Scientific Pierce ECL Western Blotting Substrate (Fisher Scientific; cat. no. 10455145), Thermo Scientific Pierce ECL 2 Western Blotting Substrate (Fisher Scientific; cat. no. 11517371), or enhanced chemiluminescence as described before (Schröder and Friedl, 1997a). Blots were exposed to Thermo Scientific CL-X Posure film (Fisher Scientific; cat. no. 10696384) or Thermo Scientific CL-X Posure film preflashed with a Sensitize preflash unit (GE Healthcare, Little Chalfont, UK; cat. no. RPN 2051) following the manufacturer’s instructions to capture weak signals. Exposure times were adjusted based on previous exposures to obtain exposures in the linear range of the film. Films were scanned on a CanoScan LiDE 600F scanner (Canon Europa, Amstelveen, The Netherlands) or a MFC-5320 DW all-in-one printer (Brother Industries, Manchester, UK) and saved as tif files. Bands were quantified with CLIQS 1.1 (Totallab, Newcastle upon Tyne, United Kingdom). Intensities for phosphorylated proteins were normalized to the intensity of the unphosphorylated species of the protein. Changes in protein abundance are expressed relative to the loading control, β-actin, GAPDH, or α-tubulin. Variation of normalizer samples was preserved for statistical analyses by normalizing all samples within one experiment to all samples with similar intensities or by normalizing all samples of one experimental repeat to the normalizer samples of all repeats for this type of experiment and using the average of these normalizations for graphing and statistical analyses.
To reprobe blots for detection of nonphosphorylated proteins and loading controls, membranes were stripped using Thermo Scientific Restore Western Blot Stripping Buffer (Fisher Scientific; cat. no. 10057103) or as described before (Armstrong et al., 2017), except that the pH of the100 mM glycine·HCl, 0.1% (vol/vol) Tween 20 solution was dropped to 1.5 and the incubation temperature with this buffer raised to 65°C. Membranes were then blocked with 5% (wt/vol) skimmed milk powder in TBST as described above.
Immunoprecipitation of IRS1
Protein extracts were prepared as described under Cell lysis and Western blotting for extraction of IRS1. Total protein (2 mg) was diluted with 50 mM Tris·HCl (pH 7.5 at 4°C), 1% (vol/vol) Triton X-100, 10 mM EDTA, 0.1% (wt/vol) BSA to a final SDS concentration of 0.1% (wt/vol). All immunoprecipitations in one experiment were adjusted to the same final volume and a SDS concentration of 0.1% (wt/vol). Lysates were precleared with 10 μl SureBeads protein A magnetic beads at 4°C with constant, slow rotation of the tubes. Samples were centrifuged at 500 × g and 4°C for 10 s, magnetic beads were collected in magnetic racks, and supernatants were centrifuged at 100,000 × g, 4°C, for 30 min and transferred to new tubes. After addition of 2 μl of anti-IRS1 antibody (clone D23G12), samples were incubated at 4°C overnight with constant, slow rotation. Samples were then centrifuged at 100,000 × g, 4°C, for 30 min, supernatants were transferred to new tubes, and immune complexes were collected by addition of 30 μl SureBeads protein A magnetic beads for 2 h at 4°C with constant, slow rotation. Samples were centrifuged at 500 × g and 4°C for 10 s and immunoprecipitates collected in magnetic racks. Immunoprecipitates were washed three times with 50 mM Tris·HCl (pH 7.5 at 4°C), 1% (vol/vol) Triton X-100, 10 mM EDTA, and 0.1% (wt/vol) BSA and once with 0.1% (wt/vol) BSA. The last wash solution was completely aspirated and the beads resuspended in 58.3 mM Tris·HCl (pH 6.8 at RT), 10% (wt/vol) SDS, 8 M urea, 2.5% (vol/vol) β-mercaptoethanol, 0.083 g/l bromophenol blue and denatured at 80°C for 5 min. Denatured samples were allowed to cool to RT for 5 min, centrifuged at 17,000 × g and RT for 5 min, and separated on 7.5% SDS–PAGE gels. Proteins were transferred onto PVDF membranes and membranes blotted with anti-phosphotyrosine and anti-IRS1 (clone D5T8J) antibodies as described under Cell lysis and Western blotting.
[ 35S]- l-methionine/[ 35S]- l-cysteine pulse labeling and immunoprecipitation of the insulin receptor
Confluent (70–80%) cultures were treated with 0.1 μM thapsigargin, 0.1 μg/ml tunicamycin, or 0.1% (vol/vol) DMSO for 22.5 h. Cultures were washed twice with methionine and cysteine-–free culture medium prewarmed to 37°C and incubated with methionine and cysteine-–free culture medium containing 2% (vol/vol) dialyzed FBS in the continued presence of 0.1% (vol/vol) DMSO, 0.1 μM thapsigargin, or 0.1 μg/ml tunicamycin for 20 min at 37°C. The medium was then replaced with fresh methionine and cysteine–free culture medium containing 2% (vol/vol) dialyzed FBS and DMSO, thapsigargin, or tunicamycin, and [35S]-l-methionine/[35S]-l-cysteine. 3T3-F442A cells were labeled with 250 μCi of 70% [35S]-l-methionine, 25% [35S]-l-cysteine (1000 Ci/mmol; Hartmann Analytic, Braunschweig, Germany; cat. no. SCIS-103) and Hep G2 cells with 125 μCi of 70% [35S]-l-methionine, 25% [35S]-l-cysteine (1000 Ci/mmol) per 10 cm dish for 1 or 8 h. Dishes were placed on ice, the medium aspirated, and cultures washed three times with ice-cold PBS (pH 7.4), and lysed in 125 μl of ice-cold 50 mM HEPES-NaOH (pH 7.5 at 4°C), 150 mM NaCl, 0.5% (wt/vol) sodium deoxycholate, 0.1% (vol/vol) Triton X-100, 0.1% (wt/vol) SDS, 10 mM EDTA, 2 mM PMSF, 6 mM AEBSF, 5 mM benzamidine, 10 μg/ml aprotinin, and each 1 μg/ml antipain dihydrochloride, chymostatin, leupeptin, and pepstatin A/10 cm dish. Lysates were cleared by centrifugation at 16,100 × g, 4°C, for 30 min and the protein concentration of lysates determined with the Bio-Rad Laboratories DC protein assay. For each immunoprecipitation 2.4 mg of total cell protein was diluted to a final concentration of 4.8 mg/ml with ice-cold 50 mM HEPES-NaOH (pH 7.5 at 4°C), 150 mM NaCl, 10 mM EDTA, 0.5% (wt/vol) sodium deoxycholate, 0.1% (vol/vol) Triton X-100, and 0.1% (wt/vol) SDS. 3T3-F442A cell lysates were precleared by incubation with 6.24 μl 100 ng/μl of normal rabbit IgG and 60 μl of SureBeads protein A magnetic beads for 2 h at 10°C with overhead rotation. For Hep G2 cell lysates, 3.12 μl 100 ng/μl of normal rabbit IgG and 30 μl of SureBeads protein A magnetic beads were used in the preclearing step. SureBeads protein A magnetic beads were collected in magnetic racks, supernatants centrifuged at 16,100 × g, 4°C, for 15 min, and supernatants transferred to new tubes. Anti-insulin receptor β chain antibody (clone 4B8) (6 μl of 52 ng/μl) or 3.12 μl 100 ng/μl normal rabbit IgG was added and samples incubated overnight at 10°C with overhead rotation. Samples were centrifuged at 16,100 × g, 4°C, for 15 min, supernatants transferred to new tubes, and 30 μl SureBeads protein A magnetic beads added to collect immune complexes while incubating samples for 2 h at 10°C with overhead rotation. Immune complexes were collected in magnetic racks. For immunoprecipitation of the insulin receptor from 3T3-F442A cell lysates, immunoprecipitates were washed three times with ice-cold 50 mM HEPES-NaOH (pH 7.5 at 4°C), 500 mM NaCl, 10 mM EDTA, 0.5% (wt/vol) sodium deoxycholate, 1% (vol/vol) Triton X-100, 0.1% (wt/vol) SDS, and 1 M urea and once with ice-cold water. For immunoprecipitation of the insulin receptor from Hep G2 cell lysates, immunoprecipitates were washed three times with ice-cold 50 mM HEPES-NaOH (pH 7.5 at 4°C), 2 M NaCl, 10 mM EDTA, 0.5% (wt/vol) sodium deoxycholate, 0.1% (vol/vol) Triton X-100, and 0.1% (wt/vol) SDS and once with ice-cold water. The water was completed aspirated and immunoprecipitates resuspended in 58.3 mM Tris·HCl (pH 6.8 at RT), 5% (vol/vol) glycerol, 10% (wt/vol) SDS, 2.5% (vol/vol) β-mercaptoethanol, 0.083 g/l bromophenol blue, and denatured at 100°C for 5 min. Denatured samples were allowed to cool to RT for 5 min, centrifuged at 17,000 × g, RT, for 15 min, and separated on 7.5% SDS–PAGE gels in which the SDS concentration had been raised to 1.25 g/l in both the stacking and separating gels. Gels were washed three times with water for 15 min at RT and dried for autoradiography with filter paper backing on a GelAir Drying System (Bio-Rad Laboratories; cat. no. 1651772) following the manufacturer’s instructions (Krishnan and Nguyen, 1990). Gels were exposed to Kodak storage phosphor SD230 screens for 12–16 d and scanned on a Typhoon 9400 system (GE Healthcare). The abundance of 35S-labeled α-β proreceptors was expressed relative to one randomly selected sample treated with 0.1% (vol/vol) DMSO.
Measurement of protein synthesis rates by [ 35S]- l-methionine/[ 35S]- l-cysteine pulse labeling
Cells were treated with 0.1% (vol/vol) DMSO, 0.1 μM thapsigargin, or 0.1 μg/ml tunicamycin and labeled with 70% [35S]-l-methionine, 25% [35S]-l-cysteine, and protein lysates were prepared as described in [35S]-l-methionine/[35S]-l-cysteine labeling and immunoprecipitation of the insulin receptor, except that cells were labeled in six-well plates with 0.35 μCi of 70% [35S]-l-methionine, 25% [35S]-l-cysteine (1000 Ci/mol) per cavity for 30 min and that lysates were prepared with 50 μl of lysis buffer per cavity. Total protein (50 μg) was denatured after the addition of 1/6 volume of 6× SDS–PAGE sample loading buffer and heating to 100°C for 5 min and resolved on 12% SDS–PAGE gels. Gels were stained with 0.1% (wt/vol) Coomassie Brilliant Blue R-250 in 50% (vol/vol) methanol, 10% (vol/vol) acetic acid for 30 min at RT, destained in 8% (vol/vol) acetic acid and 7% (vol/vol) methanol at RT, washed three times with water for 15 min at RT, and dried for autoradiography with filter paper backing as described above. Gels were exposed to Fujifilm BAS-IP multipurpose standard (MS) storage phosphor screens for 4 or 16 d and scanned on a Typhoon 9400 system (GE Healthcare) to record the 35S autoradiography signal and on a MFC-5320 DW all-in-one printer to record images of the Coomassie Brilliant Blue R-250–stained gels. Protein synthesis rates are expressed relative to the average of all cultures for one cell line treated with 0.1% (vol/vol) DMSO.
To measure [35S]-l-methionine/[35S]-l-cysteine incorporation rates by scintillation counting of TCA precipitates, 50 μg of protein lysate was added to 100 μl 1 mg/ml BSA on ice, followed by 1 ml of ice-cold 10% (wt/vol) TCA. Samples were incubated on ice for 30 min. TCA precipitates were collected on GF/C glass fiber filters (GE Healthcare; cat. no. 1822-025) prewetted with ice-cold 10% (wt/vol) TCA. The TCA precipitates were washed twice with ice-cold 10% (wt/vol) TCA and twice with ethanol and allowed to dry at RT. Precipitated radioactivity was measured by scintillation counting in a Tri-Carb 2200 CA Liquid Scintillation Analyzer (Canberra Packard, Pangbourne, UK) in 20 ml plastic scintillation vials with 10 ml Ecoscint A (National Diagnostics, Hessle, UK). Counts per minute were converted into disintegrations per minute by using a prerecorded 35S quench curve for the transformed spectral index of the external standard constructed by adding increasing amounts of chloroform to vials each containing 1·103 Bq of 70% [35S]-l-methionine, 25% [35S]-l-cysteine (1000 Ci/mol) in a total volume of 10 ml Ecoscint A/chloroform mixtures.
Measurement of the half-life of the insulin receptor at the cell surface
Confluent (70–80%) cultures were washed three times with ice-cold PBS (pH 8.0; 10 mM Na2HPO4, 2.0 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl), labeled for 5 min at 4°C in PBS (pH 8.0), 1 g/l D-glucose, 100 mg/l CaCl2, 100 mg/l MgCl2, 36 mg/l sodium pyruvate, and 2 mM sulfosuccinimidyl-6-(biotinamido)hexanoate, washed three times with PBS (pH 8.0), 100 mM glycine, at RT, three times with PBS (pH 7.4) at RT, replenished with culture medium containing 0.1% (vol/vol) DMSO, 0.3 μM thapsigargin, 1 μg/ml tunicamycin, or 1 μg/ml SubAB prewarmed to 37°C, and incubated at 37°C, 5% (vol/vol) CO2, and 95% relative humidity for up to 72 h. To prepare protein lysates, cultures were placed on ice, washed three times with ice-cold PBS (pH 7.4), and lysed in RIPA buffer containing 10 mM EDTA, 2 mM PMSF, 6 mM AEBSF, 5 mM benzamidine, 10 μg/ml aprotinin, and each 1 μg/ml antipain, chymostatin, leupeptin, and pepstatin A. Protein concentrations were determined with the Bio-Rad Laboratories DC protein assay. Biotinylated proteins were collected with 10 μl streptavidin–agarose beads from 100 μg total protein for 3T3-F442A cells, 200 μg total protein for C2C12 cells, and 50 μg total protein for Hep G2 cells for 1 h at 4°C. The streptavidin–agarose beads were washed once with ice-cold 10 mM HEPES (pH 7.5 at 4°C), 1% (vol/vol) Triton X-100, and 10 mM EDTA, twice with ice-cold 10 mM HEPES (pH 7.5 at 4°C), 1% (vol/vol) Triton X-100, 10 mM EDTA, and 6 M urea, and once with ice-cold water. The water was completed aspirated and beads resuspended in 15 μl of 6× SDS–PAGE sample loading buffer. After denaturation at 100°C for 5 min, purified proteins were resolved on 10% SDS–PAGE gels, transferred by semidry blotting onto PVDF membranes, developed with anti-insulin receptor β chain (clone 4B8) and anti-GAPDH antibodies, and chemiluminescence signals quantified as described under Cell lysis and Western blotting. To determine the half-life of the insulin receptor at the cell surface, the intensity of biotinylated insulin receptors purified on streptavidin–agarose beads immediately after labeling of cells with sulfosuccinimidyl-6-(biotinamido)hexanoate (“0 h time point”) was arbitrarily set to 1.0. Levels of biotinylated insulin receptors purified 6, 12, 24, 36, 48, and 72 h after labeling of cells with sulfosuccinimidyl-6-(biotinamido)hexanoate were expressed relative to the level of biotinylated insulin receptors purified at the 0 h time point. Half-lives were calculated from the slopes of linear decay curves of the natural logarithm of the relative abundance of biotinylated insulin receptors over time. For each condition, half-lives were determined in at least three independent experiments. Runs tests (Wald and Wolfowitz, 1940) did not reveal significant deviations from a linear relationship between the natural logarithm of the relative abundance of biotinylated insulin receptors and the time after the labeling reaction. Pilot experiments established that labeling of surface exposed insulin receptors is maximal and that biotinylation pull-down reactions are quantitative under the conditions described above (unpublished data).
Endo H and PNGase F digests
Protein (8 μg) was denatured in 0.5% (wt/vol) SDS, 40 mM 1,4-DL-dithiothreitol (DTT), at 100°C for 10 min. Samples were then incubated with 1000 U of Endo H in 50 mM sodium citrate, pH 5.5 (at 25°C), at 37°C for 2 h. For PNGase F digests, denatured samples were incubated with 1000 U of PNGase F in 50 mM sodium phosphate pH 7.5 (at 25°C), 1% (vol/vol) NP-40 at 37°C for 2 h.
Fluorescence microscopy
Images of GFP-tagged insulin receptors expressed in HEK 293 cells were taken on a Zeiss ApoTome microscope (Carl Zeiss, Cambridge, UK) equipped with a Zeiss 63x Plan-APOCHROMAT Oil PH3 objective with a numerical aperture of 1.4 18 h after induction of ER stress with 1 μg/ml tunicamycin or 1 μg/ml SubAB. The cell membrane was visualized by staining cells for 5 min at RT with 5 μg/ml CellMask Deep Red (Life Technologies). GFP fluorescence was observed using a band pass (BP) 450–490 filter (Carl Zeiss; FITC/GFP, filter set 9, cat. no. 488009-000) and a long pass (LP) 515 filter. CellMask Deep red fluorescence was observed using a BP546/12 filter (Carl Zeiss; rhodamine, filter set 15, cat. no. 488015-0000) and a LP 590 filter. To quantify colocalization of the GFP-tagged insulin receptors and CellMask Deep Red signals, individual cells were defined as regions of interest (ROIs) in ImageJ 1.47 (Schneider et al., 2012) and background-corrected for the intracellular fluorescence of CellMask Deep Red using the Background Subtraction from the ROI plug-in. The Pearson correlation coefficient between the INSR-GFP and CellMask Deep Red Fluorescence was determined in individual cells using the Colocalization Test plug-in and Costes’ image randomization (Costes et al., 2004) and a point spread function (PSF) width of 0.453 μm as a quantitative measure of colocalization of both fluorescence signals (Manders et al., 1992, 1993).
Statistical calculations
Experimental data are presented as means and SEs. For composite parameters, errors were propagated using the law of error propagation for random, independent errors (Ku, 1966). Data were analyzed for normality using the D’Agostino–Pearson omnibus normality test (D’Agostino and Pearson, 1973) or Shapiro–Wilk test (Shapiro and Wilk, 1965), equality of variances (homoscedasticity) using the Brown–Forsythe test (Brown and Forsythe, 1974), and, for additivity of means, treatment effects, and errors using Tukey’s test (Tukey, 1949b; Little and Hills, 1972) before ordinary one- or two-way ANOVA. Heteroscedastic data were transformed before ANOVA (Little and Hills, 1972) or examined by Welch’s test (Welch, 1947) and a Games–Howell post hoc test (Games and Howell, 1976) or Dunnett’s T3 multiple comparisons test (Dunnett, 1980). Kruskal–Wallis one-way ANOVA on ranks (Kruskal and Wallis, 1952) with Dunn’s post-hoc test (Dunn, 1964) was used to analyze data that are not normally distributed or heteroscedastic. Transformed data were reexamined for normality, equality of variances, and additivity. In all analyses, a familywise p value of <0.05 was considered statistically significant. Brown–Forsythe tests for equality of variances, Tukey’s test for additivity, and runs tests (Wald and Wolfowitz, 1940) were performed in Microsoft Excel 2010 or Microsoft 365 Excel (Microsoft Corporation, Redmond, WA) using the Real Statistics plug-in (www.real-statistics.com/). All other statistical calculations and linear and nonlinear regressions were performed in GraphPad Prism 6.0.7 or 8.4.3 (GraphPad Software, La Jolla, CA).
Supplementary Material
Acknowledgments
This work was supported by the European Community’s 7th Framework Programme (FP7/2007-2013) under grant agreement no. 201608, a PhD studentship from Diabetes UK (BDA 09/0003949), and a PhD studentship from Parkinson’s UK (H-1004). We thank R. Davis (University of Massachusetts), R. J. Kaufman (Sanford Burnham Medical Research Institute), T. Mak (University of Toronto), A. Benham (Durham University), R. Bashir (Durham University), M. Cann (Durham University), and C. Hutchison (Durham University) for providing cell lines. We thank A. Auricchio (Telethon Institute of Genetics and Medicine, Naples, Italy) for plasmid pCLFv2IRE and Arial Pharmaceuticals for providing plasmid pC 4M-F V2E.
Abbreviations used:
- ACTB
β actin
- AEBSF
4-(2-aminoethyl)benzenesulfonyl fluoride
- ANI
anisomycin
- ANOVA
analysis of variance
- AKT
AKR/J mice transforming retroviral oncogene
- AP20187
Arial Pharmaceuticals 20187
- ATF6
activating transcription factor 6
- ATP
adenosine triphosphate
- BAS-IP
BaFBr:Eu-sandwiched imaging plate
- BiP
heavy chain binding protein
- BP
band pass
- BSA
bovine serum albumin
- bZIP
basic leucine zipper
- CHOP
CCAAT/enhancer-binding protein homologous protein
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethyl sulfoxide
- DTT
1,4-DL-dithiothreitol
- E-64
trans-epoxysuccinyl- l-leucylamido(4-guanidino)butane
- ECL
enhanced chemiluminescence
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid
- Endo H
endoglycosidase H
- eGFP
enhanced green fluorescent protein
- eIF2
eukaryotic translation initiation factor 2
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- FK-506
Fujisawa Pharmaceutical Co. 506
- FKBP12
FK-506 binding protein of 12 kDa
- FV2E
P36V FKBP12 dimer-epitope
- GAPDH
D-glyceraldehyde 3-phosphate dehydrogenase
- GLUT4
glucose transporter type 4, insulin-responsive
- GRB2
growth factor receptor-bound protein 2
- GRP78
glucose-regulated protein of 78 kDa
- HEPES
4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
- IgG
immunoglobulin G
- ILK
integrin-linked kinase
- INSR
insulin receptor
- IP
immunoprecipitation
- IRE1α
inositol-requiring 1α
- IRS
insulin receptor substrate
- JNK
Jun N-terminal kinase
- JNKi
JNK inhibitor
- LP
long pass
- MAP
mitogen-activated protein
- MEF
mouse embryonic fibroblast
- MEM
minimum essential medium
- mRNA
messenger RNA
- mt.
mutant
- MS
multipurpose standard
- mTORC2
mammalian target of rapamycin complex 2
- PAGE
polyacrylamide gel electrophoresis
- PAK1
protein of 21 kDa (p21)-activated kinase 1
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PDK
phosphoinositide-dependent kinase
- PERK
PKR-like ER kinase
- PI
phosphatidylinositol
- PI3K
PI 3-kinase
- PKR
double-stranded RNA-activated protein kinase
- PMSF
phenylmethylsulfonyl fluoride
- PNGase F
peptide:N-glycosidase F
- PSF
point spread function
- PVDF
polyvinylidene difluoride
- pY
phosphotyrosine
- qPCRw
quantitative PCR
- quantitative PCR
regulated IRE1α-dependent decay
- RIPA
radioimmunoprecipitation assay
- RNA
ribonucleic acid
- RNase
ribonuclease
- ROI
region of interest
- RPMI
Roswell Park Memorial Institute
- RT
room temperature
- SDS
sodium dodecyl sulfate
- SERCA
Sarcoplasmic or endoplasmic reticulum Ca 2+-ATPase
- SH2
Src homology domain 2
- siRNA
small interfering RNA
- Src
Sarcoma formation
- SubAB
subtilase cytotoxin AB 5
- TBST
Tris-buffered saline, 0.1% (vol/vol) Tween-20
- TCA
trichloroacetic acid
- Tg
thapsigargin
- TGN
trans-Golgi network
- Tm
tunicamycin
- TNF
tumor necrosis factor
- TRAF2
TNF receptor-associated factor 2
- TRB3
Tribbles homolog 3
- Tris
Tris(hydroxymethyl)aminomethane
- α-Tub
α-tubulin
- UPR
unfolded protein response
- UV
ultraviolet
- WT
wild type
- XBP1
X-box-binding protein 1
- XBP1 s
spliced XBP1
- XBP1 u
unspliced XBP1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-01-0013) on September 2, 2020.
REFERENCES
- Aguirre V, Uchida T, Yenush L, Davis R, White MF (2000). The c-Jun NH2-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser307. J Biol Chem , 9047–9054. [DOI] [PubMed] [Google Scholar]
- Aguirre V, Werner ED, Giraud J, Lee YH, Shoelson SE, White MF (2002). Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J Biol Chem , 1531–1537. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA (1996). Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J , 6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Angel P, Hattori K, Smeal T, Karin M (1988). The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell , 875–885. [DOI] [PubMed] [Google Scholar]
- Arabaci G, Guo X-C, Beebe KD, Coggeshall KM, Pei D (1999). α-Haloacetophenone derivatives as photoreversible covalent inhibitors of protein tyrosine phosphatases. J Am Chem Soc , 5085–5086. [Google Scholar]
- Arabaci G, Yi T, Fu H, Porter ME, Beebe KD, Pei D (2002). α-Bromoacetophenone derivatives as neutral protein tyrosine phosphatase inhibitors: structure-activity relationship. Bioorg Med Chem Lett , 3047–3050. [DOI] [PubMed] [Google Scholar]
- Armstrong MC, Šestak S, Ali AA, Sagini HAM, Brown M, Baty K, Treumann A, Schröder M (2017). Bypass of activation loop phosphorylation by aspartate 836 in activation of the endoribonuclease activity of Ire1. Mol Cell Biol , e00655-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K (2017). Current Protocols in Molecular Biology, New York: John Wiley & Sons. [Google Scholar]
- Avery J, Etzion S, Debosch BJ, Jin X, Lupu TS, Beitinjaneh B, Grand J, Kovacs A, Sambandam N, Muslin AJ (2010). TRB3 function in cardiac endoplasmic reticulum stress. Circ Res , 1516–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM, Myers MG Jr, Shoelson SE, Chin DJ, Sun XJ, Miralpeix M, Hu P, Margolis B, Skolnik EY, Schlessinger J, White MF (1992). Phosphatidylinositol 3’-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J , 3469–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin GS, Burgess AW, Kemp BE (1982). Phosphorylation of a synthetic gastrin peptide by the tyrosine kinase of A431 cell membranes. Biochem Biophys Res Commun , 656–663. [DOI] [PubMed] [Google Scholar]
- Baldwin GS, Knesel J, Monckton JM (1983). Phosphorylation of gastrin-17 by epidermal growth factor-stimulated tyrosine kinase. Nature , 435–437. [DOI] [PubMed] [Google Scholar]
- Bar RS, Gorden P, Roth J, Kahn CR, De Meyts P (1976). Fluctuations in the affinity and concentration of insulin receptors on circulating monocytes of obese patients: effects of starvation, refeeding, and dieting. J Clin Invest , 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Chiu G, Argon Y, Steiner DF (1998). Folding of insulin receptor monomers is facilitated by the molecular chaperones calnexin and calreticulin and impaired by rapid dimerization. J Cell Biol , 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Kurose T, Pashmforoush M, Steiner DF (1996). Fusion of insulin receptor ectodomains to immunoglobulin constant domains reproduces high-affinity insulin binding in vitro . J Biol Chem , 19367–19375. [DOI] [PubMed] [Google Scholar]
- Bass J, Turck C, Rouard M, Steiner DF (2000). Furin-mediated processing in the early secretory pathway: sequential cleavage and degradation of misfolded insulin receptors. Proc Natl Acad Sci USA , 11905–11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P (2006). Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol , 2311–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC, Webster C (1985). Plasticity of the differentiated state. Science , 758–766. [DOI] [PubMed] [Google Scholar]
- Boden G, Cheung P, Salehi S, Homko C, Loveland-Jones C, Jayarajan S, Stein TP, Williams KJ, Liu ML, Barrero CA, Merali S (2014). Insulin regulates the unfolded protein response (UPR) in human adipose tissue. Diabetes , 912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden G, Duan X, Homko C, Molina EJ, Song W, Perez O, Cheung P, Merali S (2008). Increase in endoplasmic reticulum (ER) stress related proteins and genes in adipose tissue of obese, insulin resistant individuals. Diabetes , 2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd FT Jr, Raizada MK (1983). Effects of insulin and tunicamycin on neuronal insulin receptors in culture. Am J Physiol , C283–C287. [DOI] [PubMed] [Google Scholar]
- Bravo DA, Gleason JB, Sanchez RI, Roth RA, Fuller RS (1994). Accurate and efficient cleavage of the human insulin proreceptor by the human proprotein-processing protease furin. Characterization and kinetic parameters using the purified, secreted soluble protease expressed by a recombinant baculovirus. J Biol Chem , 25830–25837. [PubMed] [Google Scholar]
- Brown M, Strudwick N, Suwara M, Sutcliffe LK, Mihai AD, Ali AA, Watson JN, Schröder M (2016). An initial phase of JNK activation inhibits cell death early in the endoplasmic reticulum stress response. J Cell Sci , 2317–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MB, Forsythe AB (1974). Robust tests for the equality of variances. J Am Stat Assoc , 364–367. [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature , 92–96. [DOI] [PubMed] [Google Scholar]
- Capeau J, Lascols O, Flaig-Staedel C, Blivet MJ, Beck JP, Picard J (1985). Degradation of insulin receptors by hepatoma cells: insulin-induced down-regulation results from an increase in the rate of basal receptor degradation. Biochimie , 1133–1141. [DOI] [PubMed] [Google Scholar]
- Capeau J, Picard J, Caron M (1978). Insulin receptors in Zajdela rat ascites hepatoma cells and their sensitivity to certain enzymes and lectins. Cancer Res , 3930–3937. [PubMed] [Google Scholar]
- Casagrande R, Stern P, Diehn M, Shamu C, Osario M, Zúñiga M, Brown PO, Ploegh H (2000). Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell , 729–735. [DOI] [PubMed] [Google Scholar]
- Chiang W-C, Messah C, Lin JH (2012). IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol Biol Cell , 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, Stevenson LF, Magari SR, Wood SA, Courage NL, et al. (1998). Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci USA , 10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleasby ME, Reinten TA, Cooney GJ, James DE, Kraegen EW (2007). Functional studies of Akt isoform specificity in skeletal muscle in vivo; maintained insulin sensitivity despite reduced insulin receptor substrate-1 expression. Mol Endocrinol , 215–228. [DOI] [PubMed] [Google Scholar]
- Coon HG, Weiss MC (1969). A quantitative comparison of formation of spontaneous and virus-produced viable hybrids. Proc Natl Acad Sci USA , 852–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S (2004). Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J , 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotugno G, Pollock R, Formisano P, Linher K, Beguinot F, Auricchio A (2004). Pharmacological regulation of the insulin receptor signaling pathway mimics insulin action in cells transduced with viral vectors. Hum Gene Ther , 1101–1108. [DOI] [PubMed] [Google Scholar]
- Cox DJ, Strudwick N, Ali AA, Paton AW, Paton JC, Schröder M (2011). Measuring signaling by the unfolded protein response. Methods Enzymol , 261–292. [DOI] [PubMed] [Google Scholar]
- Crettaz M, Kahn CR (1984). Insulin receptor regulation and desensitization in rat hepatoma cells. Concomitant changes in receptor number and in binding affinity. Diabetes , 477–485. [DOI] [PubMed] [Google Scholar]
- Cryer PE, Polonsky KS (1998). Glucose homeostasis and hypoglycemia. In: Williams Textbook of Endocrinology, ed. Wilson JD, Foster DW, Kronenberg HM, Larsen PR, Philadelphia: W. B. Saunders Company, 939–972. [Google Scholar]
- D’Agostino R, Pearson ES (1973). Tests for departure from normality. Empirical results for the distributions of b2 and √b1. Biometrika , 613–622. [Google Scholar]
- Desbuquois B, Lopez S, Burlet H (1982). Ligand-induced translocation of insulin receptors in intact rat liver. J Biol Chem , 10852–10860. [PubMed] [Google Scholar]
- Deschatrette J, Weiss MC (1974). Characterization of differentiated and dedifferentiated clones from a rat hepatoma. Biochimie , 1603–1611. [DOI] [PubMed] [Google Scholar]
- Draznin B, Trowbridge M, Ferguson L (1984). Quantitative studies of the rate of insulin internalization in isolated rat hepatocytes. Biochem J , 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K, Herzig S, Kulkarni RN, Montminy M (2003). TRB3: A tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science , 1574–1577. [DOI] [PubMed] [Google Scholar]
- Dunn OJ (1964). Multiple comparisons using rank sums. Technometrics , 241–252. [Google Scholar]
- Dunn SD (1986). Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal Biochem , 144–153. [DOI] [PubMed] [Google Scholar]
- Dunnett CW (1955). A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc , 1096–1121. [Google Scholar]
- Dunnett CW (1964). New tables for multiple comparisons with control. Biometrics , 482–491. [Google Scholar]
- Dunnett CW (1980). Pairwise multiple comparisons in the unequal variance case. J Am Stat Assoc , 796–800. [Google Scholar]
- Eagle H (1959). Amino acid metabolism in mammalian cell cultures. Science , 432–437. [DOI] [PubMed] [Google Scholar]
- Emanuelli B, Eberle D, Suzuki R, Kahn CR (2008). Overexpression of the dual-specificity phosphatase MKP-4/DUSP-9 protects against stress-induced insulin resistance. Proc Natl Acad Sci USA , 3545–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercolani L, Brown TJ, Ginsberg BH (1984). Tunicamycin blocks the emergence and maintenance of insulin receptors on mitogen-activated human T lymphocytes. Metabolism , 309–316. [DOI] [PubMed] [Google Scholar]
- Esposito DL, Li Y, Cama A, Quon MJ (2001). Tyr612 and Tyr632 in human insulin receptor substrate-1 are important for full activation of insulin-stimulated phosphatidylinositol 3-kinase activity and translocation of GLUT4 in adipose cells. Endocrinology , 2833–2840. [DOI] [PubMed] [Google Scholar]
- Fewell SW, Travers KJ, Weissman JS, Brodsky JL (2001). The action of molecular chaperones in the early secretory pathway. Annu Rev Genet , 149–191. [DOI] [PubMed] [Google Scholar]
- Fleming Y, Armstrong CG, Morrice N, Paterson A, Goedert M, Cohen P (2000). Synergistic activation of stress-activated protein kinase 1/c-Jun N-terminal kinase (SAPK1/JNK) isoforms by mitogen-activated protein kinase kinase 4 (MKK4) and MKK7. Biochem J , 145–154. [PMC free article] [PubMed] [Google Scholar]
- Fliegel L, Burns K, MacLennan DH, Reithmeier RA, Michalak M (1989). Molecular cloning of the high affinity calcium-binding protein (calreticulin) of skeletal muscle sarcoplasmic reticulum. J Biol Chem , 21522–21528. [PubMed] [Google Scholar]
- Frank HJ, Davidson MB, Serbin PA (1981). Insulin binding and action in isolated rat hepatocytes: evidence for spare receptors. Metabolism , 1159–1164. [DOI] [PubMed] [Google Scholar]
- Franke TF, Yang SI, Chan TO, Datta K, Kazlauskas A, Morrison DK, Kaplan DR, Tsichlis PN (1995). The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell , 727–736. [DOI] [PubMed] [Google Scholar]
- Freychet P (1984). [Insulin resistance. Physiopathological and biochemical aspects]. Ann Endocrinol (Paris) , 107–114. [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T (2000). A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol , 379–384. [DOI] [PubMed] [Google Scholar]
- Gaddam D, Stevens N, Hollien J (2013). Comparison of mRNA localization and regulation during endoplasmic reticulum stress in Drosophila cells. Mol Biol Cell , 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Games PA, Howell JF (1976). Pairwise multiple comparison procedures with unequal N’s and/or variances: a Monte Carlo study. J Educ Stat , 113–125. [Google Scholar]
- Gammeltoft S, Gliemann J (1973). Binding and degradation of 125I-labelled insulin by isolated rat fat cells. Biochim Biophys Acta , 16–32. [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Kahn CR (1988). Initial processing of the insulin receptor precursor in vivo and in vitro . J Biol Chem , 12809–12812. [PubMed] [Google Scholar]
- Goldstein S, Blecher M, Binder R, Perrino PV, Recant L (1975). Hormone receptors, 5. Binding of glucagon and insulin to human circulating mononuclear cells in diabetes mellitus. Endocr Res Commun , 367–376. [DOI] [PubMed] [Google Scholar]
- Graham FL, Harrison T, Williams J (1978). Defective transforming capacity of adenovirus type 5 host-range mutants. Virology , 10–21. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R (1977). Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol , 59–74. [DOI] [PubMed] [Google Scholar]
- Grako KA, Olefsky JM, McClain DA (1992). Tyrosine kinase-defective insulin receptors undergo decreased endocytosis but do not affect internalization of normal endogenous insulin receptors. Endocrinology , 3441–3452. [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O (1976). Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell , 105–113. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM (2006). Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev Cell , 859–871. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell , 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature , 271–274. [DOI] [PubMed] [Google Scholar]
- Harrison T, Graham F, Williams J (1977). Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology , 319–329. [DOI] [PubMed] [Google Scholar]
- Hassan RH, Hainault I, Vilquin J-T, Samama C, Lasnier F, Ferré P, Foufelle F, Hajduch E (2012). Endoplasmic reticulum stress does not mediate palmitate-induced insulin resistance in mouse and human muscle cells. Diabetologia , 204–214. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Molinari M (2007). In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev , 1377–1408. [DOI] [PubMed] [Google Scholar]
- Heidenreich KA, Berhanu P, Brandenburg D, Olefsky JM (1983). Degradation of insulin receptors in rat adipocytes. Diabetes , 1001–1009. [DOI] [PubMed] [Google Scholar]
- Heidenreich KA, Brandenburg D (1986). Oligosaccharide heterogeneity of insulin receptors. Comparison of N-linked glycosylation of insulin receptors in adipocytes and brain. Endocrinology , 1835–1842. [DOI] [PubMed] [Google Scholar]
- Helderman JH, Raskin P (1980). The T lymphocyte insulin receptor in diabetes and obesity: an intrinsic binding defect. Diabetes , 551–557. [DOI] [PubMed] [Google Scholar]
- Hers I, Bell CJ, Poole AW, Jiang D, Denton RM, Schaefer E, Tavaré JM (2002). Reciprocal feedback regulation of insulin receptor and insulin receptor substrate tyrosine phosphorylation by phosphoinositide 3-kinase in primary adipocytes. Biochem J , 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani K, Haruta T, Tani A, Kawahara J, Usui I, Kobayashi M (2005). Roles of mTOR and JNK in serine phosphorylation, translocation, and degradation of IRS-1. Biochem Biophys Res Commun , 836–842. [DOI] [PubMed] [Google Scholar]
- Hoehn KL, Hohnen-Behrens C, Cederberg A, Wu LE, Turner N, Yuasa T, Ebina Y, James DE (2008). IRS1-independent defects define major nodes of insulin resistance. Cell Metab , 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Marsh JW, Miller B, Steiner DF (1980). Cultured hepatoma cells as a model system for studying insulin processing and biologic responsiveness. Diabetes , 865–874. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS (2009). Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol , 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science , 104–107. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res , D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosogai N, Fukuhara A, Oshima K, Miyata Y, Tanaka S, Segawa K, Furukawa S, Tochino Y, Komuro R, Matsuda M, Shimomura I (2007). Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes , 901–911. [DOI] [PubMed] [Google Scholar]
- Hubbard SC, Ivatt RJ (1981). Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem , 555–583. [DOI] [PubMed] [Google Scholar]
- Hunter T (1982). Synthetic peptide substrates for a tyrosine protein kinase. J Biol Chem , 4843–4848. [PubMed] [Google Scholar]
- Hwang JB, Frost SC (1999). Effect of alternative glycosylation on insulin receptor processing. J Biol Chem , 22813–22820. [DOI] [PubMed] [Google Scholar]
- Imanikia S, Özbey NP, Krueger C, Casanueva MO, Taylor RC (2019). Neuronal XBP-1 activates intestinal lysosomes to improve proteostasis in C. elegans . Curr Biol , 2322–2338.e2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B (2006). SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell , 125–137. [DOI] [PubMed] [Google Scholar]
- Jang YY, Kim NK, Kim MK, Lee HY, Kim SJ, Kim HS, Seo HY, Lee IK, Park KG (2010). The effect of tribbles-related protein 3 on ER stress-suppressed insulin gene expression in INS-1 cells. Korean Diabetes J , 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DY, Chalasani U, Pan N, Friedline RH, Prosdocimo DA, Nam M, Azuma Y, Maganti R, Yu K, Velagapudi A, et al. (2013). KLF15 is a molecular link between endoplasmic reticulum stress and insulin resistance. PLoS One , e77851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadle R, Fellows RE, Raizada MK (1984). The effects of insulin and tunicamycin on insulin receptors of cultured fibroblasts. Exp Cell Res , 533–541. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Kahn CR, Hedo JA, Van Obberghen E, Yamada KM (1981). Insulin-induced receptor loss in cultured human lymphocytes is due to accelerated receptor degradation. Proc Natl Acad Sci USA , 6917–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefer LM, De Meyts P (1981). Glycosylation of cell surface receptors: tunicamycin treatment decreases insulin and growth hormone binding to different levels in cultured lymphocytes. Biochem Biophys Res Commun , 22–29. [DOI] [PubMed] [Google Scholar]
- Knowles BB, Howe CC, Aden DP (1980). Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science , 497–499. [DOI] [PubMed] [Google Scholar]
- Knutson VP, Ronnett GV, Lane MD (1982). Control of insulin receptor level in 3T3 cells: effect of insulin-induced down-regulation and dexamethasone-induced up-regulation on rate of receptor inactivation. Proc Natl Acad Sci USA , 2822–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson VP, Ronnett GV, Lane MD (1983). Rapid, reversible internalization of cell surface insulin receptors. Correlation with insulin-induced down-regulation. J Biol Chem , 12139–12142. [PubMed] [Google Scholar]
- Kobayashi M, Ohgaku S, Iwasaki M, Harano Y, Maegawa H, Shigeta Y (1980). Evaluation of the method of insulin binding studies in human erythrocytes. Endocrinol Jpn , 337–342. [DOI] [PubMed] [Google Scholar]
- Koh HJ, Arnolds DE, Fujii N, Tran TT, Rogers MJ, Jessen N, Li Y, Liew CW, Ho RC, Hirshman MF, et al. (2006). Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol Cell Biol , 8217–8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh HJ, Toyoda T, Didesch MM, Lee MY, Sleeman MW, Kulkarni RN, Musi N, Hirshman MF, Goodyear LJ (2013). Tribbles 3 mediates endoplasmic reticulum stress-induced insulin resistance in skeletal muscle. Nat Commun , 1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolterman OG, Gray RS, Griffin J, Burstein P, Insel J, Scarlett JA, Olefsky JM (1981). Receptor and postreceptor defects contribute to the insulin resistance in noninsulin-dependent diabetes mellitus. J Clin Invest , 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolterman OG, Insel J, Saekow M, Olefsky JM (1980). Mechanisms of insulin resistance in human obesity: evidence for receptor and postreceptor defects. J Clin Invest , 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T, Barham FW (1971). The relationship between the insulin-binding capacity of fat cells and the cellular response to insulin. Studies with intact and trypsin-treated fat cells. J Biol Chem , 6210–6216. [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S (1985). Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem , 631–664. [DOI] [PubMed] [Google Scholar]
- Krishnan M, Nguyen HT (1990). Drying acrylamide slab gels for fluorography without using gel drier and vacuum pump. Anal Biochem , 51–53. [DOI] [PubMed] [Google Scholar]
- Krupp M, Lane MD (1981). On the mechanism of ligand-induced down-regulation of insulin receptor level in the liver cell. J Biol Chem , 1689–1694. [PubMed] [Google Scholar]
- Kruskal WH, Wallis WA (1952). Use of ranks in one-criterion variance analysis. J Am Stat Assoc , 583–621. [Google Scholar]
- Ku HH (1966). Notes on the use of propagation of error formulas. J Res Nat Bureau Standards Sect C–Eng Instrumentat , 263–273. [Google Scholar]
- Kuo SC, Lampen O (1976). Tunicamycin inhibition of [3H] glucosamine incorporation into yeast glycoproteins: binding of tunicamycin and interaction with phospholipids. Arch Biochem Biophys , 574–581. [DOI] [PubMed] [Google Scholar]
- Kurzban GP, Bayer EA, Wilchek M, Horowitz PM (1991). The quaternary structure of streptavidin in urea. J Biol Chem , 14470–14477. [PubMed] [Google Scholar]
- Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature , 680–685. [DOI] [PubMed] [Google Scholar]
- Lawler S, Fleming Y, Goedert M, Cohen P (1998). Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr Biol , 1387–1390. [DOI] [PubMed] [Google Scholar]
- Le Marchand-Brustel Y, Jeanrenaud B, Freychet P (1978). Insulin binding and effects in isolated soleus muscle of lean and obese mice. Am J Physiol , E348–E358. [DOI] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ (2002). IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev , 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Giraud J, Davis RJ, White MF (2003). c-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J Biol Chem , 2896–2902. [DOI] [PubMed] [Google Scholar]
- Lehle L, Tanner W (1976). The specific site of tunicamycin inhibition in the formation of dolichol-bound N-acetylglucosamine derivatives. FEBS Lett , 167–170. [DOI] [PubMed] [Google Scholar]
- Little TM, Hills FJ (1972). Statistical Methods in Agricultural Research, Davis: University of California Agricultural Extension. [Google Scholar]
- Macer DR, Koch GL (1988). Identification of a set of calcium-binding proteins in reticuloplasm, the luminal content of the endoplasmic reticulum. J Cell Sci , 61–70. [DOI] [PubMed] [Google Scholar]
- Maley F, Trimble RB, Tarentino AL, Plummer TH Jr (1989). Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem , 195–204. [DOI] [PubMed] [Google Scholar]
- Manders EMM, Stap J, Brakenhoff GJ, van Driel R, Aten JA (1992). Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci , 857–862. [DOI] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ, Aten JA (1993). Measurement of co-localization of objects in dual-colour confocal images. J Microsc , 375–382. [DOI] [PubMed] [Google Scholar]
- Mao K, Kobayashi S, Jaffer ZM, Huang Y, Volden P, Chernoff J, Liang Q (2008). Regulation of Akt/PKB activity by P21-activated kinase in cardiomyocytes. J Mol Cell Cardiol , 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F (2002a). N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J Mol Biol , 541–557. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F (2002b). N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J Mol Biol , 523–540. [DOI] [PubMed] [Google Scholar]
- McDonald PC, Oloumi A, Mills J, Dobreva I, Maidan M, Gray V, Wederell ED, Bally MB, Foster LJ, Dedhar S (2008). Rictor and integrin-linked kinase interact and regulate Akt phosphorylation and cancer cell survival. Cancer Res , 1618–1624. [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T (2005). ERAD: the long road to destruction. Nat Cell Biol , 766–772. [DOI] [PubMed] [Google Scholar]
- Mihai AD, Schröder M (2015). Glucose starvation and hypoxia, but not the saturated fatty acid palmitic acid or cholesterol, activate the unfolded protein response in 3T3-F442A and 3T3-L1 adipocytes. Adipocyte , 188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore GE, Gerner RE, Franklin HA (1967). Culture of normal human leukocytes. J Am Med Assoc , 519–524. [PubMed] [Google Scholar]
- Morton HJ (1970). A survey of commercially available tissue culture media. In Vitro , 89–108. [DOI] [PubMed] [Google Scholar]
- Myers MG Jr, Wang LM, Sun XJ, Zhang Y, Yenush L, Schlessinger J, Pierce JH, White MF (1994). Role of IRS-1-GRB-2 complexes in insulin signaling. Mol Cell Biol , 3577–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil JC, Ghysdael J, Vogt PK, Smart JE (1981). Homologous tyrosine phosphorylation sites in transformation-specific gene products of distinct avian sarcoma viruses. Nature , 675–677. [DOI] [PubMed] [Google Scholar]
- Nguyen MTA, Satoh H, Favelyukis S, Babendure JL, Imamura T, Sbodio JI, Zalevsky J, Dahiyat BI, Chi NW, Olefsky JM (2005). JNK and tumor necrosis factor-α mediate free fatty acid-induced insulin resistance in 3T3-L1 adipocytes. J Biol Chem , 35361–35371. [DOI] [PubMed] [Google Scholar]
- Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H (2005). TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J , 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM (1975). Effect of dexamethasone on insulin binding, glucose transport, and glucose oxidation of isolated rat adipocytes. J Clin Invest , 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM (1976). Decreased insulin binding to adipocytes and circulating monocytes from obese subjects. J Clin Invest , 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM, Kolterman OG (1981). Mechanisms of insulin resistance in obesity and noninsulin-dependent (type II) diabetes. Am J Med , 151–168. [DOI] [PubMed] [Google Scholar]
- Olefsky JM, Reaven GM (1974). Decreased insulin binding to lymphocytes from diabetic subjects. J Clin Invest , 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM, Reaven GM (1977). Insulin binding in diabetes. Relationships with plasma insulin levels and insulin sensitivity. Diabetes , 680–688. [DOI] [PubMed] [Google Scholar]
- Örd D, Örd T (2003). Mouse NIPK interacts with ATF4 and affects its transcriptional activity. Exp Cell Res , 308–320. [DOI] [PubMed] [Google Scholar]
- Özcan U, Cao Q, Yilmaz E, Lee A-H, Iwakoshi NN, Özdelen E, Tuncman G, Görgün C, Glimcher LH, Hotamisligil GS (2004). Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science , 457–461. [DOI] [PubMed] [Google Scholar]
- Özcan U, Yilmaz E, Özcan L, Furuhashi M, Vaillancourt E, Smith RO, Görgün CZ, Hotamisligil GS (2006). Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science , 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano G, Cassader M, Lenti G (1977). Insulin receptors in adipocytes of non-diabetic and diabetic subjects. Preliminary report. Acta Diabetol Lat , 164–169. [DOI] [PubMed] [Google Scholar]
- Panzhinskiy E, Hua Y, Culver B, Ren J, Nair S (2013). Endoplasmic reticulum stress upregulates protein tyrosine phosphatase 1B and impairs glucose uptake in cultured myotubes. Diabetologia , 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton AW, Beddoe T, Thorpe CM, Whisstock JC, Wilce MC, Rossjohn J, Talbot UM, Paton JC (2006). AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature , 548–552. [DOI] [PubMed] [Google Scholar]
- Paton AW, Srimanote P, Talbot UM, Wang H, Paton JC (2004). A new family of potent AB5 cytotoxins produced by Shiga toxigenic Escherichia coli . J Exp Med , 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patschinsky T, Hunter T, Esch FS, Cooper JA, Sefton BM (1982). Analysis of the sequence of amino acids surrounding sites of tyrosine phosphorylation. Proc Natl Acad Sci USA , 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LJ, Gallis B, Casnellie JE, Bornstein P, Krebs EG (1982). Epidermal growth factor stimulates the phosphorylation of synthetic tyrosine-containing peptides by A431 cell membranes. Proc Natl Acad Sci USA , 1443–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed BC, Glasted K, Miller B (1984). Direct comparison of the rates of internalization and degradation of covalent receptor-insulin complexes in 3T3-L1 adipocytes. Internalization of occupied receptors is not the rate-limiting step in receptor-hormone complex degradation. J Biol Chem , 8134–8143. [PubMed] [Google Scholar]
- Reed BC, Kaufmann SH, Mackall JC, Student AK, Lane MD (1977). Alterations in insulin binding accompanying differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA , 4876–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed BC, Lane MD (1980). Insulin receptor synthesis and turnover in differentiating 3T3-L1 preadipocytes. Proc Natl Acad Sci USA , 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed BC, Ronnett GV, Clements PR, Lane MD (1981a). Regulation of insulin receptor metabolism. Differentiation-induced alteration of receptor synthesis and degradation. J Biol Chem , 3917–3925. [PubMed] [Google Scholar]
- Reed BC, Ronnett GV, Lane MD (1981b). Role of glycosylation and protein synthesis in insulin receptor metabolism by 3T3-L1 mouse adipocytes. Proc Natl Acad Sci USA , 2908–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson BJ, Moehring JM, Moehring TJ (1993). Defective processing of the insulin receptor in an endoprotease-deficient Chinese hamster cell strain is corrected by expression of mouse furin. J Biol Chem , 24274–24277. [PubMed] [Google Scholar]
- Robinson TJ, Archer JA, Gambhir KK, Hollis VW Jr, Carter L, Bradley C (1979). Erythrocytes: a new cell type for the evaluation of insulin receptor defects in diabetic humans. Science , 200–202. [DOI] [PubMed] [Google Scholar]
- Rocchi S, Tartare-Deckert S, Mothe I, Van Obberghen E (1995). Identification by mutation of the tyrosine residues in the insulin receptor substrate-1 affecting association with the tyrosine phosphatase 2C and phosphatidylinositol 3-kinase. Endocrinology , 5291–5297. [DOI] [PubMed] [Google Scholar]
- Ron E, Shenkman M, Groisman B, Izenshtein Y, Leitman J, Lederkremer GZ (2011). Bypass of glycan-dependent glycoprotein delivery to ERAD by up-regulated EDEM1. Mol Biol Cell , 3945–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnett GV, Knutson VP, Kohanski RA, Simpson TL, Lane MD (1984). Role of glycosylation in the processing of newly translated insulin proreceptor in 3T3-L1 adipocytes. J Biol Chem , 4566–4575. [PubMed] [Google Scholar]
- Ronnett GV, Lane MD (1981). Post-translational glycosylation-induced activation of aglycoinsulin receptor accumulated during tunicamycin treatment. J Biol Chem , 4704–4707. [PubMed] [Google Scholar]
- Ronnett GV, Tennekoon G, Knutson VP, Lane MD (1983). Kinetics of insulin receptor transit to and removal from the plasma membrane. J Biol Chem , 283–290. [PubMed] [Google Scholar]
- Rosen OM, Chia GH, Fung C, Rubin CS (1979). Tunicamycin-mediated depletion of insulin receptors in 3T3-L1 adipocytes. J Cell Physiol , 37–42. [DOI] [PubMed] [Google Scholar]
- Rutzky LP, Pumper RW (1974). Supplement to a survey of commercially available tissue culture media (1970). In Vitro , 468–469. [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR (2001). Insulin signalling and the regulation of glucose and lipid metabolism. Nature , 799–806. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science , 1098–1101. [DOI] [PubMed] [Google Scholar]
- Savoie S, Rindress D, Posner BI, Bergeron JJ (1986). Tunicamycin sensitivity of prolactin, insulin and epidermal growth factor receptors in rat liver plasmalemma. Mol Cell Endocrinol , 241–246. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder M, Friedl P (1997a). Overexpression of recombinant human antithrombin III in Chinese hamster ovary cells results in malformation and decreased secretion of the recombinant protein. Biotechnol Bioeng , 547–559. [DOI] [PubMed] [Google Scholar]
- Schröder M, Friedl P (1997b). A protein-free solution as replacement for serum in trypsinization protocols for anchorage-dependent cells. Methods Cell Sci , 137–147. [Google Scholar]
- Schröder M, Kaufman RJ (2005). The mammalian unfolded protein response. Annu Rev Biochem , 739–789. [DOI] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB (1965). An analysis of variance test for normality (complete samples). Biometrika , 591–611. [Google Scholar]
- Sharma NK, Das SK, Mondal AK, Hackney OG, Chu WS, Kern PA, Rasouli N, Spencer HJ, Yao-Borengasser A, Elbein SC (2008). Endoplasmic reticulum stress markers are associated with obesity in non-diabetic subjects. J Clin Endocrinol Metab , 4532–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu C-Y, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell , 893–903. [DOI] [PubMed] [Google Scholar]
- Shi Y, An J, Liang J, Hayes SE, Sandusky GE, Stramm LE, Yang NN (1999). Characterization of a mutant pancreatic eIF-2α kinase, PEK, and co-localization with somatostatin in islet delta cells. J Biol Chem , 5723–5730. [DOI] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC (1998). Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol Cell Biol , 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakami A, Toyonaga T, Tsuruzoe K, Shirotani T, Matsumoto K, Yoshizato K, Kawashima J, Hirashima Y, Miyamura N, Kahn CR, Araki E (2002). Heterozygous knockout of the IRS-1 gene in mice enhances obesity-linked insulin resistance: a possible model for the development of type 2 diabetes. J Endocrinol , 309–319. [DOI] [PubMed] [Google Scholar]
- Shoelson SE, Chatterjee S, Chaudhuri M, White MF (1992). YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase. Proc Natl Acad Sci USA , 2027–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šidák Z (1967). Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc , 626–633. [Google Scholar]
- Smart JE, Oppermann H, Czernilofsky AP, Purchio AF, Erikson RL, Bishop JM (1981). Characterization of sites for tyrosine phosphorylation in the transforming protein of Rous sarcoma virus (pp60v-src) and its normal cellular homologue (pp60c-src). Proc Natl Acad Sci USA , 6013–6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985). Measurement of protein using bicinchoninic acid. Anal Biochem , 76–85. [DOI] [PubMed] [Google Scholar]
- Sreejayan N, Dong F, Kandadi MR, Yang X, Ren J (2008). Chromium alleviates glucose intolerance, insulin resistance, and hepatic ER stress in obese mice. Obesity , 1331–1337. [DOI] [PubMed] [Google Scholar]
- Sun XJ, Crimmins DL, Myers MG Jr, Miralpeix M, White MF (1993). Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol Cell Biol , 7418–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, Cahill DA, Goldstein BJ, White MF (1991). Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature , 73–77. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Hiroki A, Watanabe T, Yamashita T, Takei I, Umezawa K (2001). Potentiation of insulin-related signal transduction by a novel protein-tyrosine phosphatase inhibitor, Et-3,4-dephostatin, on cultured 3T3-L1 adipocytes. J Biol Chem , 27511–27518. [DOI] [PubMed] [Google Scholar]
- Szczepankiewicz BG, Kosogof C, Nelson LT, Liu G, Liu B, Zhao H, Serby MD, Xin Z, Liu M, Gum RJ, et al. (2006). Aminopyridine-based c-Jun N-terminal kinase inhibitors with cellular activity and minimal cross-kinase activity. J Med Chem , 3563–3580. [DOI] [PubMed] [Google Scholar]
- Talbot UM, Paton JC, Paton AW (2005). Protective immunization of mice with an active-site mutant of subtilase cytotoxin of Shiga toxin-producing Escherichia coli . Infect Immun , 4432–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Shen H, Chen J, Wang X, Zhang Y, Chen LL, Rukachaisirikul V, Jiang H-L, Shen X (2011). Activating transcription factor 6 protects insulin receptor from ER stress-stimulated desensitization via p42/44 ERK pathway. Acta Pharmacol Sin , 1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine DJ, Moremen KW, Sifers RN (2009). The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic downregulation of ER mannosidase I. J Cell Sci , 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, Dawson AP (1990). Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2+-ATPase. Proc Natl Acad Sci USA , 2466–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ (2000). The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev , 2725–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ (1998). A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev , 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ (2000). Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science , 870–874. [DOI] [PubMed] [Google Scholar]
- Tukey JW (1949a). Comparing individual means in the analysis of variance. Biometrics , 99–114. [PubMed] [Google Scholar]
- Tukey JW (1949b). One degree of freedom for non-additivity. Biometrics , 232–242. [Google Scholar]
- Unger RH, Foster DW (1998). Diabetes mellitus. In: Williams Textbook of Endocrinology, ed. Wilson J.D., Foster D.W., Kronenberg H.M., Larsen P.R., Philadelphia: W. B. Saunders Company, 973–1060. [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science , 664–666. [DOI] [PubMed] [Google Scholar]
- Valverde AM, Mur C, Pons S, Alvarez AM, White MF, Kahn CR, Benito M (2001). Association of insulin receptor substrate 1 (IRS-1) y895 with Grb-2 mediates the insulin signaling involved in IRS-1-deficient brown adipocyte mitogenesis. Mol Cell Biol , 2269–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I, Rindress D, Cameron PH, Ou WJ, Doherty JJ 2nd, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJ (1991). SSRα and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem , 19599–19610. [PubMed] [Google Scholar]
- Wald A, Wolfowitz J (1940). On a test whether two samples are from the same population. Ann Math Stat , 147–162. [Google Scholar]
- Walter P, Lingappa VR (1986). Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol , 499–516. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science , 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang CC, Sonne O, Hedo JA, Cushman SW, Simpson IA (1983). Insulin-induced internalization of the insulin receptor in the isolated rat adipose cell. Detection of the internalized 138-kilodalton receptor subunit using a photoaffinity 125I-insulin. J Biol Chem , 5129–5134. [PubMed] [Google Scholar]
- Watanabe T, Suzuki T, Umezawa Y, Takeushi T, Otsuka M, Umezawa K (2000). Structure-activity relationship and rational design of 3,4-dephostatin derivatives as protein tyrosine phosphatase inhibitors. Tetrahedron , 741–752. [Google Scholar]
- Welch BL (1947). The generalisation of Student’s problems when several different population variances are involved. Biometrika , 28–35. [DOI] [PubMed] [Google Scholar]
- Werner ED, Lee J, Hansen L, Yuan M, Shoelson SE (2004). Insulin resistance due to phosphorylation of insulin receptor substrate-1 at serine 302. J Biol Chem , 35298–35305. [DOI] [PubMed] [Google Scholar]
- Whitehead JP, Molero JC, Clark S, Martin S, Meneilly G, James DE (2001). The role of Ca2+ in insulin-stimulated glucose transport in 3T3-L1 cells. J Biol Chem , 27816–27824. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD-Y, Kaufman RJ (2007). ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell , 351–364. [DOI] [PubMed] [Google Scholar]
- Xu B, Bird VG, Miller WT (1995). Substrate specificities of the insulin and insulin-like growth factor 1 receptor tyrosine kinase catalytic domains. J Biol Chem , 29825–29830. [DOI] [PubMed] [Google Scholar]
- Xu L, Spinas GA, Niessen M (2010). ER stress in adipocytes inhibits insulin signaling, represses lipolysis, and alters the secretion of adipokines without inhibiting glucose transport. Horm Metab Res , 643–651. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K (2007). Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell , 365–376. [DOI] [PubMed] [Google Scholar]
- Yang W, Rozamus LW, Narula S, Rollins CT, Yuan R, Andrade LJ, Ram MK, Phillips TB, van Schravendijk MR, Dalgarno D, et al. (2000). Investigating protein-ligand interactions with a mutant FKBP possessing a designed specificity pocket. J Med Chem , 1135–1142. [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell , 1355–1364. [DOI] [PubMed] [Google Scholar]
- Yeh W-C, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa JL, Ferrick D, Hum B, Iscove N, et al. (1997). Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity , 715–725. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001a). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell , 881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K (2000). ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol , 6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K (2001b). Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol Cell Biol , 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Inesta-Vaquera F, Niepel M, Zhang J, Ficarro SB, Machleidt T, Xie T, Marto JA, Kim N, Sim T, et al. (2012). Discovery of potent and selective covalent inhibitors of JNK. Chem Biol , 140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang J, Fang Q, Liu M, Liu X, Jia W, Dong LQ, Liu F (2009). Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol Pharmacol , 596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.