Abstract
Adipogenesis is regulated by a cascade of signals that drive transcriptional reprogramming in adipocytes. Here, we report that nuclear actin regulates the chromatin states that establish tissue- specific expression during adipogenesis. To study the role of β-actin in adipocyte differentiation, we conducted RNA sequencing on wild-type and β-actin knockout mouse embryonic fibroblasts (MEFs) after reprograming to adipocytes. We found that β-actin depletion affects induction of several adipogenic genes during transcriptional reprograming. This impaired regulation of adipogenic genes is linked to reduced expression of the pioneer factor Cebpa and is rescued by reintroducing NLS-tagged β-actin. ATAC-Seq in knockout MEFs revealed that actin-dependent reduction of Cebpa expression correlates with decreased chromatin accessibility and loss of chromatin association of the ATPase Brg1. This, in turn, impairs CEBPB’s association with its Cebpa promoter-proximal binding site during adipogenesis. We propose a role for the nuclear β-actin pool in maintaining open chromatin for transcriptional reprogramming during adipogenic differentiation.
INTRODUCTION
In the nucleus, actin is required by all three eukaryotic RNA polymerases for both chromatin remodeling and transcription regulation (Visa and Percipalle, 2010; Percipalle, 2013; Virtanen and Vartiainen, 2017; Percipalle and Vartiainen, 2019). Actin-dependent epigenetic changes also significantly influence chromatin organization by facilitating heterochromatin segregation at the nuclear periphery and maintaining H3K9Me3-positive heterochromatin levels in the nuclear interior (Xie and Percipalle, 2018). We recently reported that these mechanisms have a significant impact on the expression of gene programs required during neurogenesis (Almuzzaini et al., 2016; Xie and Percipalle, 2018; Xie et al., 2018a). β-actin seems to perform these functions by regulating genomic deposition of Brg1 (Brahma-related gene 1), the ATPase subunit of the chromatin remodeling complex SWI/SNF or BAF (Brahma-associated factor). We found that β-actin-dependent loss of Brg1 binding at transcription start sites (TSS) of proneuronal and neuronal genes, along with promoter-specific H3K9me3 accumulation, accounts for loss of neuronal identity (Xie et al., 2018b). These findings suggest that β-actin may have a role in regulating activation of occluded genes during reprogramming to neurons. However, it remains unclear if β-actin-dependent Brg1 deposition is also required to maintain an open chromatin state during transcriptional reprogramming in other differentiation models such as adipogenesis.
During adipogenesis, preadipocytes differentiate into adipocytes, the main constituent of adipose tissue (Rosen et al., 2000; Rajala and Scherer, 2003; Kershaw and Flier, 2004; Rosen and Macdougald, 2006; Adamczak and Wiecek, 2013; McGown et al., 2014; Cinti, 2018). Adipocyte differentiation involves an interactive network of signaling cascades and transcription factors, including the peroxisome proliferator-activated receptor γ (PPARγ), members of the CCAAT/enhancer binding protein (CEBP) family (Spiegelman and Flier, 1996; Farmer, 2006; Lowe et al., 2011), and the BAF complex (Siersbaek et al., 2012; Shapira et al., 2017; Shapira and Seale, 2019). PPAR-γ is a master regulator of adipogenesis (Spiegelman, 1998), whereas members of CEBP family, specifically CEBPA (Linhart et al., 2001; Rosen et al., 2002) play a role in adipocyte subtype specificity, promoting white adipocyte over brown adipocyte differentiation (Linhart et al., 2001). During differentiation, adipocytes adopt a rounded morphology for maximal lipid storage (Smas and Sul, 1995; Rosen et al., 2002), a configuration modulated by the actin cytoskeleton (Jaffe and Hall, 2005; Sordella et al., 2003; Mcbeath et al., 2004; Noguchi et al., 2007; Tanegashima et al., 2008; Horii et al., 2009; Nobusue et al., 2014).
While investigating the potential role of actin in Brg1 recruitment to adipogenic gene loci, we discovered that loss of nuclear β-actin leads to changes in the expression of adipogenic gene programs, alterations in heterochromatin regulation, and down-regulation of the Cebpa gene. ATAC-Seq analysis of wild-type (WT) and knockout (KO) MEFs shows that this is due to loss of chromatin accessibility at the TSS and promoter-proximal region of Cebpa, consistent with increased H3K9Me3 accumulation and loss of Brg1 binding. Excessive chromatin compaction, in turn, impairs CEBPB binding to a specific regulatory motif upstream of the Cebpa TSS. Our results suggest that β-actin is required to maintain an open chromatin state during transcriptional reprogramming.
RESULTS AND DISCUSSION
Transcriptional reprograming to adipocytes is dysregulated in the absence of β-actin
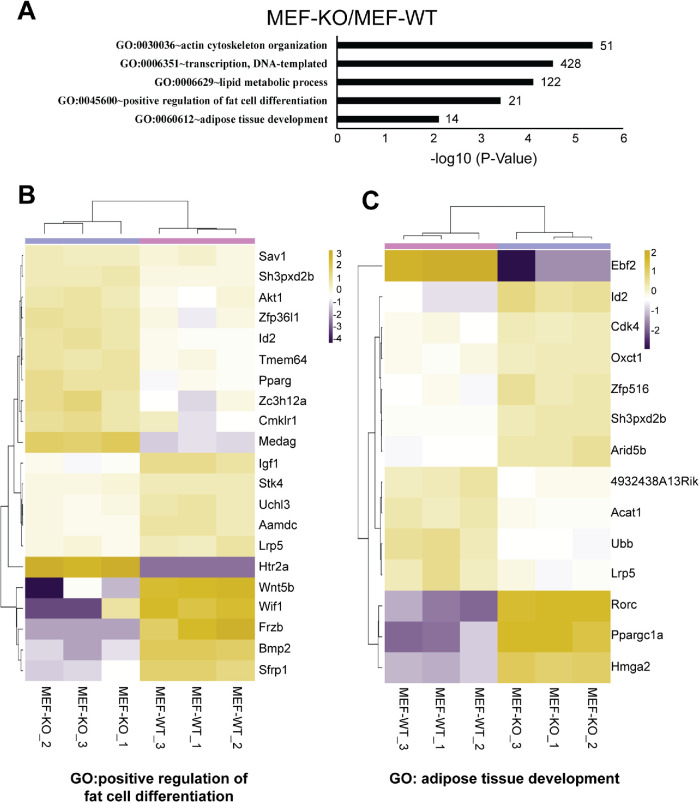
To find a potential role of β-actin in adipocyte differentiation, we performed gene ontology (GO) analysis on statistically significant genes (cutoff of p-value < 0.01) differentially expressed between WT and β-actin KO mouse embryonic fibroblasts (MEF-WT and MEF-KO; Xie et al., 2018a; Figure 1A). We identified GO terms for fat cell differentiation (GO:0045600) and adipose tissue development (GO:0060612; Figure 1A; Supplemental Table S1; Supplemental Table S2). Metric normalized (log count per million) heatmaps revealed down-regulation of Wif1, Frzb, Bmp2, Srfp1, and Wnt5b and up-regulation of Htr2a and Medag in the KO condition as compared with WT (Figure 1B). Among the genes involved in adipose tissue development, Ebf2 is heavily up-regulated, whereas genes such as Ppargc1a, Rorc, and Hmga2 are down-regulated in the KO condition (Figure 1C). These findings suggest that expression of genes related to adipocyte differentiation is dysregulated upon β-actin depletion.
FIGURE 1:
Sets of adipocyte-related genes are differentially expressed between wild-type (MEF-WT) and β-knockout (MEF-KO) mouse embryonic fibroblasts. (A) Gene ontology on MEF-KO/MEF-WT pairwise comparison reveals enrichment of genes involved in adipose or fat development, actin cytoskeleton, and transcriptional mechanisms. GO enrichment terms are based on significance of p-value < 0.05 (normalized to -log10) cutoff. Numbers at the end of each bar reflect the numbers of genes identified in each GO-term. (B, C) Metric heat-map clustering of expression levels of genes associated with GO term: positive regulation of fat cell differentiation (GO:0045600) and adipose tissue development (GO:0060612). Genes are selected when they are differentially expressed by at least twofold in WT vs. KO comparisons. Clustering is based on the center values (CVs) of mean gene expression levels. Scale bar: log2 CPM. Data presented are based on three biological replicates (n = 3). See Materials and Methods for full description.
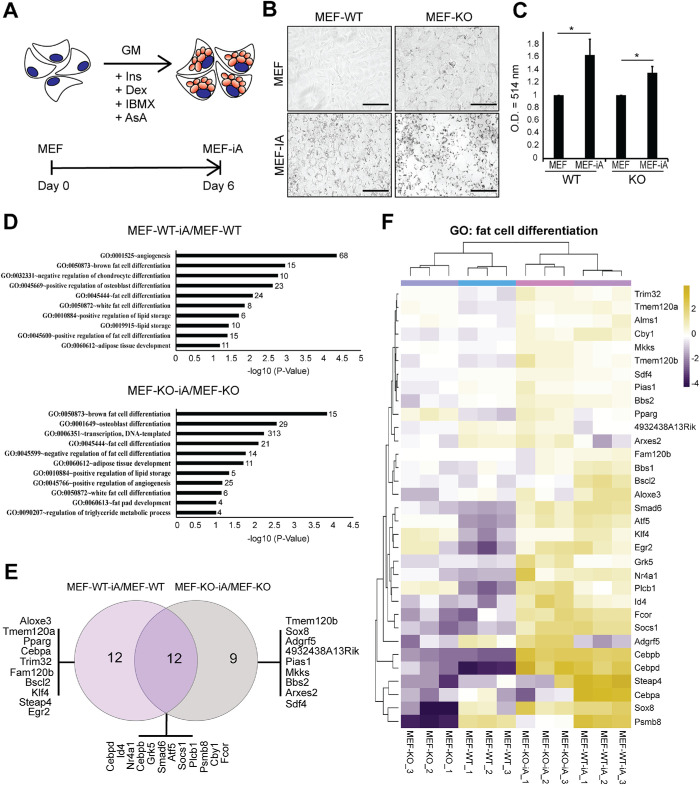
We next induced adipocytes from both WT and KO MEFs as recently described (Cuaranta-Monroy et al., 2014). Following a six-day induction (Figure 2A), the induced adipocytes (MEF-iA) were probed for the formation of lipid triglyceride droplets around cell nuclei (Ramirez-Zacarias et al., 1992). Using Oil Red O (ORO) staining, a triglyceride-specific stain (Ramirez-Zacarias et al., 1992), we found that both MEF-WT and MEF-KO cells were positive for lipid droplets (Figure 2, B and C) after induction with adipogenic differentiation medium. Interestingly, although the production of lipid droplets seems lower in KO cells with respect to WT condition, cells lacking β-actin showed the presence of lipid droplets even in the undifferentiated stage. This phenotype combined with differential expression of key genes involved in adipocyte differentiation between KO and WT suggests that loss of β-actin affects adipogenic gene programs during reprograming.
FIGURE 2:
Direct reprograming of MEFs into induced adipocytes. (A) Schematic illustration highlighting the differentiation protocol to induce MEFs (MEF-WT and MEF-KO) to adipocyte-like cells termed MEF-iA (MEF-WT-iA and MEF-KO-iA). GM = growth medium, Ins = insulin, DEX = dexamethasone, IBMX = 3-isobutyl-1-methylxanthine, AsA = ascorbic acid. (B) Lipid droplet detection in WT, KO MEFs and corresponding induced adipocytes by ORO staining at day 0 (basal condition) and day 6 (differentiation condition). Scale bar: 125 μm. (C) Quantification of oil red staining isolates via spectrophotometry comparing MEF and MEF-iA conditions for WT and KO with a wavelength optical density (O.D.) of 514 nm (n = 3; *p-value < 0.05) (D) Directional GO enrichment analysis on MEF-WT-iA/MEF-WT and MEF-KO-iA/MEF-KO pairs. Enrichment analysis of biological processes for up-regulated DEGs in MEF-WT-D/MEF-WT (Top) and MEF-KO-D/MEF-KO (Bottom) identifies GO terms associated with adipocyte-related biological processes. The p-value < 0.05 (normalized by -log10) was defined to be statistically significant and is plotted on the x-axis. Numbers at the ends of bars reflect the number of genes identified in each GO-term. (E) The Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) identifies common and unique DEGs under the GO term fat cell differentiation (GO:0045444) in the pairwise comparisons MEF-WT-D/MEF-WT and MEF-KO-D/MEF-KO. Common and unique genes are listed under each condition. (F) Heat-map plot illustration of DEG patterns under fat cell differentiation gene ontology. DESeq2 heatmap plot generated by TSAR with the expression patterns (scaled to a log2 count per million [log2cpm]) of DEGs (33 genes), displaying samples MEF-WT, MEF-KO, MEF-WT-iA, and MEF-KO-iA, along with their biological replicates, under the GO term fat cell differentiation (GO:0045444) identified in both MEF-WT-iA/MEF-WT and MEF-KO-iA/MEF-KO. Samples of each condition, along with three biological replicates, are displayed (n = 3). CVs for all three replicates are presented as mean values with error bars representing SD (SD). Significance t tests were performed based on a one-tailed hypothesis.
Transcriptome analysis reveals differential gene expression in adipocytes lacking β-actin
We performed RNA-Seq analysis on induced KO and WT adipocytes (MEF-iA) and compared their transcriptomes with their MEF counterparts (Figure 2). Principal component analysis (PCA) of the RNA-Seq datasets showed 48% total variance in the first principal component (PC1) and clearly separated uninduced MEFs from induced adipocytes (Supplemental Figure S1A). Pairwise comparisons between MEFs and corresponding MEF-iA in KO and WT conditions based on significance (p-value < 0.01) and log 2-fold change of greater than 0 (FC > 0) among differentially expressed genes revealed major transcriptome differences. In the two pairwise comparisons, MEF-WT-iA/MEF-WT and MEF-KO-iA/MEF-KO, we identified 4098 and 3330 differentially expressed genes (DEGs), which were further analyzed for GO enrichment (biological processes) using DAVID (Figure 2D). We observed a strong enrichment of genes regulating fat cell differentiation and subtype specificity. The GO-term “fat cell differentiation” (GO:0045444) had 24 genes associated with it in the MEF-WT-iA/MEF-WT comparison and 21 genes in the MEF-KO-iA/MEF-KO comparison. Only 12 of these genes were common to both comparisons, with the remainder being exclusively enriched in one condition (Figure 2E). A heat map of the normalized read counts of these genes revealed that the majority of these genes were up-regulated upon adipogenesis induction in both WT and KO MEFs (Figure 2F).
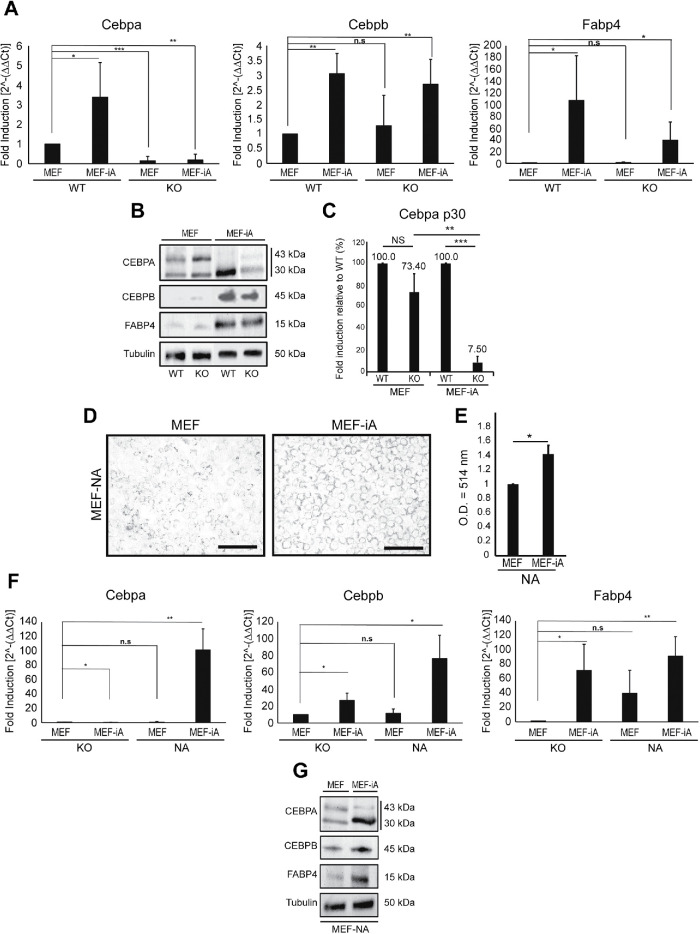
Members of the CCAAT/enhancer-binding protein beta (Cebpb) and delta (Cebpd) that function as transcription factors in cellular differentiation (Scott et al., 1992; Wu et al., 1996; Tanaka et al., 1997), metabolism (Cardinaux and Magistretti, 1996) and immune responses (Zannetti et al., 2010) were induced under both conditions (Figure 2F). Similarly, Socs1, Fcor, Sox8, Smad6, Atf5, Egr2, and Id4, generally involved at multiple stages of adipogenesis were also up-regulated under both conditions (Figure 2F). However, Cebpa, Steap4, and Psmb8 were not up-regulated in KO adipocytes, in contrast to WT cells (Figure 2F). Steap4 encodes the six-transmembrane epithelial antigen of the prostate-4, a plasma membrane protein that is associated with insulin sensitivity (Chen et al., 2010). Psmb8 encodes the proteasome subunit beta 8, an immunoproteasome that has been shown to be essential for adipocyte maturity during differentiation (Arimochi et al., 2016), whereas the Cebpa gene encodes the CCAAT/enhancer-binding protein alpha (CEBPA), a pioneer transcription factor known to induce adipogenesis through the PPARγ pathway (Rosen et al., 2002). qPCR analysis on total RNA from induced adipocytes confirmed a significant drop in Cebpa expression under the KO condition. In contrast, Cebpb and Fabp4, both of which are markers for mature adipocytes, showed significant up-regulation upon adipogenic induction (Moseti et al., 2016; Figure 3A). CEBPA is present in two alternatively spliced isoforms, CEBPA-p30 and CEBPA-p42 (Lane et al., 1996), and, in contrast to CEBPA-p42, during differentiation CEBPA-p30 is known to be up-regulated (Lin et al., 1993; Otto and Lane, 2005). While both WT and KO induced adipocytes showed CEBPA-p42 down-regulation, CEBPA-p30 expression was also significantly reduced in KO cells, consistent with the dysregulation of CEBPA expression in the absence of β-actin (Figure 3, B and C). In contrast, immunoblots of CEBPB and FABP4 did not reveal significant differences between WT and KO adipocytes.
FIGURE 3:
Expression of adipogenic genes requires nuclear β-actin. (A) Quantitative PCR analysis on total RNA isolated from WT and KO MEFs and the corresponding adipocytes using primers amplifying Cebpa (left chart), Cebpb (middle chart), and Fabp4 (right chart). The analysis was normalized against the housekeeping gene Nono. Conditions were normalized to MEF-WT to observe fold induction normalized to 1 (n = 3; *p-value < 0.05, **p-value < 0.01). (B) Immunoblots on lysates isolated from WT and KO MEFs and the corresponding induced adipocytes with antibodies against CEBPA (detecting p30 and p42 isoforms), CEBPB, FABP4, and α-tubulin as loading control. (C) Densitometric quantification of the immunoblot signals from three independent experiments (n = 3). Relative density values were generated via ImageJ software (n = 3; *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001). (D) Analysis of cell morphology and lipid droplet production in KO MEFs constitutively expressing β-actin in the cell nucleus (MEF-NA) and the corresponding induced adipocytes (MEF-NA-iA) by ORO staining at day 0 (basal condition) and day 6 (differentiation condition). Scale bar: 125 μm. (E) Quantification of oil red staining isolated via spectrophotometry comparing MEF-NA and MEF-NA-iA with wavelength optical density (O.D.) = 514 nm (n = 3; *p-value < 0.05) (F) Reintroduction of β-actin in the KO background rescues Cebpa expression during induction of adipocytes. Quantitative PCR analysis on total RNA isolated from MEF-KO and MEF-NA and the corresponding adipocytes using primers amplifying Cebpa (left chart), Cebpb (middle chart), and Fabp4 (right chart). The analysis was normalized against the housekeeping gene Nono. Conditions were normalized to MEF-KO to observe fold induction (n = 3; *p-value < 0.05, **p-value < 0.01). (G) Immunoblots on lysates isolated from KO MEFs and the corresponding induced adipocytes expressing an NLS-tagged β-actin construct with antibodies against CEBPA (detecting p30 and p42 isoforms), CEBPB, FABP4, and α-tubulin as loading control. All performed experiments consisted of three independent biological replicates (n = 3). CVs for all three replicates are presented as mean values with error bars representing SD. Significance t tests were performed based on a one-tailed hypothesis (p-values).
To study if Cebpa expression is regulated by the nuclear β-actin pool during adipogenic induction, we differentiated MEF-KO cells constitutively expressing an NLS-tagged β-actin construct, referred to as MEF-NA (Almuzzaini et al., 2016; Xie et al., 2018a). We found that MEF-NA cells are also induced to adipocyte-like cells (MEF-NA-iA), as evidenced by the extensive formation of ORO-positive lipid droplets (Figure 3, D and E). We next performed qPCR analysis on total RNA from MEF-NA and MEF-KO cells before and after adipogenic induction and quantified the relative expression change of the Cebpa, Cebpb, and Fabp4 genes (Figure 3F). In both conditions, Cebpb and Fabp4 were differentially expressed upon induction to adipocytes. In contrast, the Cebpa gene, which does not exhibit differential expression between MEF-KO and the corresponding adipocyte condition, was significantly induced in MEF-NA after adipocyte induction (Figure 3F). In addition, while induced adipocytes from MEF-NA cells showed significantly higher levels of CEBPA-p30 expression compared with KO cells, (Figure 3G), immunoblots of CEBPB and FABP4 did not reveal significant differences between WT and KO adipocytes. These findings indicate that reintroduction of β-actin in the cell nucleus in MEF-KO cells rescues Cebpa expression.
β-actin regulates CEBPA expression by maintaining an open chromatin state
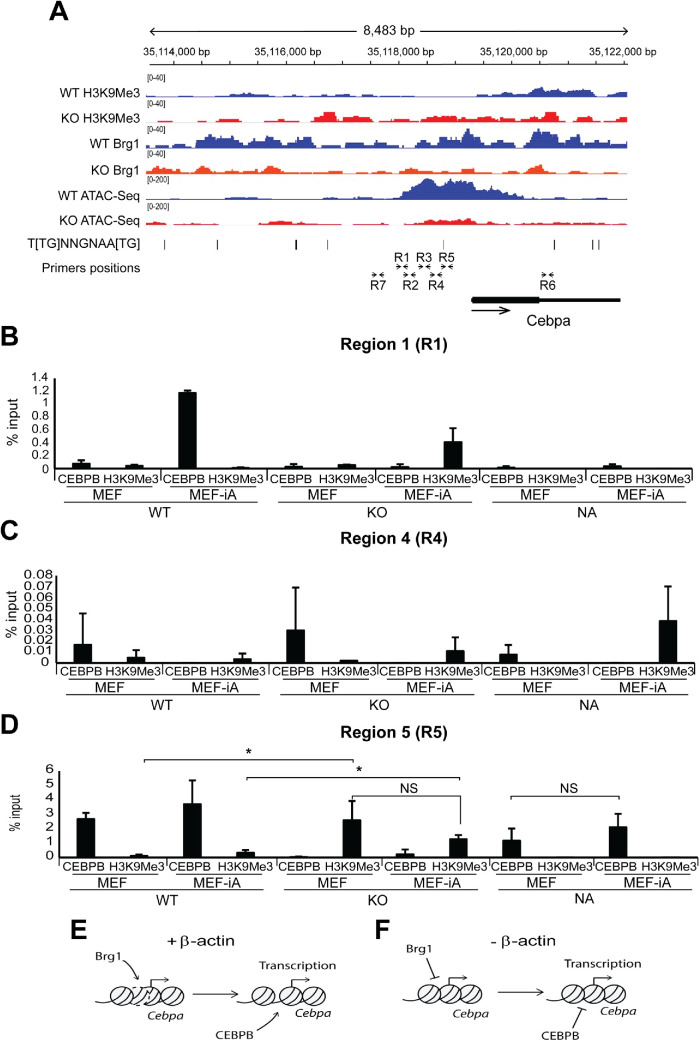
To investigate if β-actin-dependent chromatin remodeling influences adipogenic gene programs, we then used ATAC-Seq to analyze chromatin accessibility around the TSSs and promoter-proximal upstream regulatory regions of genes linked to adipocyte differentiation in WT and KO cells (Figure 2E). Average ATAC-Seq signal at TSS of adipogenic genes did not show a significant difference between WT and KO conditions (Supplemental Figure S2). Similarly, analysis of H3K9Me3, H3K27Me3, and Brg1 ChIP-Seq profiles, together with ATAC-Seq profiles of specific adipogenesis-related genes such as Cebpb, Fabp4, Psmb8, and Steap4, did not reveal a clear link between chromatin accessibility and differential expression (Supplemental Figure S2, A–D). However, we observed a significant reduction in chromatin accessibility at the TSS and promoter-proximal regulatory region of Cebpa (Figure 4; Supplemental Figure S2E). Furthermore, cells heterozygous for β-actin showed an accessibility profile identical to that of WT cells, suggesting that even reduced levels of β-actin can maintain Cebpa in an accessible state (Supplemental figure S2E). Remarkably, sequence analysis of the Cebpa promoter-proximal region also revealed that the loss of chromatin accessibility observed in KO cells correlated with a CEBP-binding motif present approximately 800 bp upstream of the Cebpa TSS (Figure 4A). To study if CEBPB binding to this region is compromised during adipogenic induction, we performed chromatin immunoprecipitation combined with RT-qPCR on chromatin isolated from WT and β-actin KO MEFs before and after induction to adipocytes (MEF-iA) with antibodies to CEBPB and H3K9Me3 (Figure 4, B–D). RT-qPCR analysis was performed with several primers targeting multiple regions (R1–R7) upstream of the Cebpa gene TSS, including a CEBPB binding motif, as well as a region (R6) located inside the Cebpa gene (Figure 4A). Our results show that under the WT condition, CEBPB is specifically enriched with primers amplifying R5, a unique region encompassing a putative CEBPB consensus binding site, but not in other regions (Figure 4, B–D; Supplemental Figure S2, F–I). Compatible with the fact that there is a putative CEBPB binding site (see Figure 4A), strong CEBPB association with R5 is maintained in differentiated adipocytes (MEF-iA) but is completely lost under the KO condition in both MEF and MEF-iA cells (Figure 4D). In addition, loss of CEBPB binding correlates with increased H3K9Me3 levels under both MEF and induced adipocyte conditions (Figure 4D), implying that an actin-dependent chromatin-based mechanism facilitates CEBPB access to a specific site to enhance Cebpa gene expression. This is consistent with the rescue of CEBPB association and loss of H3K9Me3 binding upon reintroduction of β-actin into nuclei of KO cells in both MEF and induced adipocytes (Figure 4D) and with enhanced CEBPA expression upon reintroduction of actin in the cell nucleus. Our results suggest that β-actin is required for the maintenance of chromatin accessibility in the promoter-proximal region of Cebpa during adipogenic reprogramming. This mode of regulation is compatible with the role of CEBPA as a pioneer transcription factor operating upstream of adipogenesis genes such as Psmb8 and Steap4.
FIGURE 4:
β-actin KO cells show actin-dependent loss of chromatin accessibility around the TSS and upstream regulatory region of Cebpa, which impairs CEBPB binding for transcription activation. (A) ChIP-Seq profiles of H3K9Me3, H3K27Me3, Brg1, and ATAC-Seq profiles of WT and KO cells around the Cebpa gene. The y-axis data range represents RPKM (reads per kilobase of sequence range per million mapped reads) per bin. The y-axis of tracks in the same image were set to the same range. Gene body position (exon: box, intron: line), CEBPB consensus binding motif 5′-T[TG]NNGNAA[TG]-3′ (UniProtKB - P28033), and positions of the primers for qPCR analysis described in panel B are shown below the tracks. ChIP-Seq profile data for each assay are presented as individual points consisting of two biologically independent replicates (n = 2). (B–D) Results from ChIP experiments with antibodies against CEBPB and H3K9Me3 and qPCR analysis with primers across the Cebpa gene regulatory region (R1–R5) and control regions further upstream (R7) and inside the gene (R6) show that CEBPB binding is impaired at regions of decreased chromatin accessibility during adipogenesis. See A for the locations of the R1–R7 regions amplified by qPCR. The analysis was normalized against % input (n = 3; *p-value < 0.05, **p-value < 0.01). See Supplemental Figure S2 for ChIP qPCR analysis across regions R2, R3, R6, and R7. CV for all three replicates are presented as mean value with error bars representing SD. Significance t tests were performed based on a one-tailed hypothesis (p-values). (E, F) A speculative model suggesting that loss of nuclear β-actin impairs recruitment of Brg1, leading to compact chromatin with increased H3K9me3 and loss of chromatin accessibility that altogether impairs CEBPB binding required for Cebpa transcription activation.
An actin-based mechanism controls chromatin accessibility during adipogenesis
Establishment of an adipogenic phenotype requires a cohort of transcription factors including CEBPA, CEBPB, CEBPD, and PPARγ. Although they participate in a single pathway of fat cell development, PPARγ is the proximal effector, while CEBPA works as the pioneer factor needed for robust adipocyte-specific gene expression through binding at PPARγ-adjacent binding sites (Rosen et al., 2002; Lefterova et al., 2008). CEBPB and CEBPD are upstream of PPARγ and CEBPA. In the mouse 3T3-L1 preadipocyte cell line, PPARγ and CEBPA are regulated by CEBPB and CEBPD, both rapidly induced within 4 h postdifferentiation. PPARγ and CEBPA are thought to activate each other’s expression, inducing a number of genes that define terminally differentiated adipocytes (Rosen et al., 2002; Rosen and Macdougald, 2006). We found that loss of β-actin does not affect CEBPB, CEBPD, or PPARγ but leads to selective down-regulation of CEBPA at both mRNA and protein level. Reintroduction of β-actin in the nucleus of KO cells rescues CEBPA expression, suggesting that CEBPA is induced through a nuclear actin-based mechanism, fundamentally different from that activating PPARγ. Consistent with the selective impairment of CEBPA, GO terms on brown fat cell development in which CEBPA is involved (Kajimura et al., 2010; Saely et al., 2012; Zhang et al., 2018; Shapira and Seale, 2019) are enriched. CEBPA binds to DNA sites adjacent to PPARγ binding sites to regulate expression of genes that promote differentiation of preadipocytes to adipocytes (Lefterova et al., 2008; Madsen et al., 2014). Once bound, CEBPA functions as a pioneer factor, possibly by activating downstream effectors such as Psmb8 and Steap4 (Chen et al., 2010; Arimochi et al., 2016). Consistently, in β-actin KO cells, neither Psmb8 nor Steap4 is induced after adipogenic differentiation.
In preadipocytes, the CEBPA gene locus is maintained in a “poised” chromatin state ready for active transcription upon induction (Matsumura et al., 2015). Our ChIP-Seq and ChIP qPCR results suggest that this poised configuration at TSS and promoter-proximal region is achieved by maintaining low H3K9Me3 and high Brg1 levels in WT and is reversed in KO cells. Results from ATAC-Seq are compatible with these observations, showing loss of chromatin accessibility primarily at TSS and promoter-proximal region in the absence of β-actin. This indicates β-actin plays a key role in maintaining a chromatin state that ensures Cebpa gene expression during transcriptional reprograming of MEFs into adipocytes (see Figure 4, E and F, for a speculative model). Seeing that both ATPase activity and genomic deposition of Brg1 require β-actin, β-actin seems to contribute to chromatin accessibility by regulating the activity of the BAF complex at TSS and the promoter-proximal region of the Cebpa gene. We propose that maintenance of an open chromatin facilitates CEBPB binding to a specific motif located about 800 bp upstream from the TSS. This, in turn, allows the Cebpa gene to be in a poised configuration that is activated during transcriptional reprograming to adipocytes. The extent of chromatin accessibility correlates with transcriptional reprograming during oogenesis (Miyamoto et al., 2018). Impaired expression of Cebpa is rescued by the reintroduction of nuclear β-actin into KO cells. Although at this stage a contribution from the cytoplasmic actin cytoskeleton cannot be completely ruled out, we propose that maintenance of an open chromatin state at the Cebpa gene locus primarily requires the nuclear β-actin pool, highlighting an important role for nuclear β-actin in cell fate determination by contributing to the heterochromatin landscape.
MATERIALS AND METHODS
Antibodies
Mouse anti-CEBPA (catalogue number sc-365318) and rabbit anti-CEBPB (catalogue number sc-7962) antibodies were purchased from Santa Cruz Biotechnology and used at 1:200 and 1:400 dilution, respectively. The rabbit antibody to FABP4 (catalogue number ab66682) and the mouse antibody to α-tubulin (catalogue number ab6046) were purchased from AbCam and used at 1:500 dilutions. Horseradish peroxidase–conjugated goat anti-mouse and goat anti-rabbit secondary antibodies were obtained from JacksonImmunoResearch Laboratories and used at 1:2000 dilutions.
In vitro adipocyte differentiation
MEF-WT, MEF-KO, and MEF-NA were previously characterized (Almuzzaini et al., 2016; Xie et al., 2018a). They were maintained briefly in DMEM with high glucose supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (Pen/Strp growth media) in treated tissue culture T25 flasks (ThermoScientific NUNC) at 37°C with 5% CO2. An adipocyte differentiation protocol was adapted from a previous study (Cuaranta-Monroy et al., 2014). Cell lines were split with trypsin, counted, and plated at a density of 1 × 105 cells per well in six-well tissue culture plates (ThermoScientific NUNC). After 48 h, the medium was changed to an adipogenic differentiation cocktail, which included the growth medium supplemented with 10 μg/ml insulin (Ins), 2 μM dexamethasone (Dex), 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), and 25 μg/ml ascorbic acid (AsA; differentiation media; Cuaranta-Monroy et al., 2014). All supplements used were products of Sigma. Cells were allowed to differentiate for up to 6 d. The differentiation medium was changed every 2 d.
Lipid droplet staining
Medium from each well plate was aspirated and washed with 1 ml of 1× phosphate-buffered saline (PBS). Paraformaldehyde (PFA) in PBS (4%; 2.4 ml) was added to each well for 5 min and then was changed to a fresh 2.4 ml 4%PFA for 1 h. Fixative solution was aspirated and cells were washed once with 2.4 ml of 60% isopropanol. After being washed, plates were allowed to dry completely before addition of 1 ml of diluted working ORO solution (six parts ORO stock solution [Sigma]: four parts H2O), after 10 min incubation at room temperature, staining solution was removed, and cells were washed four times with H2O to remove residual ORO. H2O was added (2 ml) to each well before the visualization under microscopy (EVOS FLOID Cell Imaging System Life Technologies). Visual assessment of ORO staining was conducted in four independent experiments. For quantification, ORO-stained particles were eluted with 100% isopropanol and analyzed using Thermo Scientific Varioskan Flash for spectrophotometry readings at 514 nm.
Protein isolation and Western blotting analysis
Cell lysates were prepared using in house–prepared radioimmune precipitation assay (1XRIPA) buffer, followed by incubation on ice for 30 min and vortexing at 5-min intervals. Lysates were then centrifuged at 15,000 rpm for 30 min and supernatant was collected. Protein concentrations were measured using a Pierce BCA protein assay kit (ThermoScientific). Samples were mixed with sodium dodecyl sulphate (SDS) loading buffer and boiled at 95°C for 5 min. From each sample, 20 µg was loaded and separated on 10% SDS–PAGE gels, followed by transferring proteins to polyvinylidene difluoride membranes at 90 V for 50 min. Membranes were then blocked for 1 h with 5% nonfat milk in PBS-T (0.05% Tween-20 in PBS). The membranes were incubated overnight at 4°C with primary antibodies in 1% nonfat milk in PBS. Membranes were washed with PBS and then incubated with secondary antibodies for 1 h at room temperature. Protein bands were developed using a Pierce ECL Western blot substrate kit (ThermoScientific) and were imaged using a Syngene GeneGnome system. Densitometry analysis was performed using ImageJ 1.x software (Schneider et al., 2012).
RNA isolation and purification
Total RNA was extracted using a combination of TriZol (Life Technologies) and an RNAeasy Mini Kit (Qiagen) with modifications from the manufacturer’s protocols. MEF cell lines in each well were washed once with 1 ml of 1XPBS before the addition of 1 ml of TriZol reagent. TriZol lysates were added to a fresh 1.5-ml tube. A volume of 0.2 ml of chloroform was added per ml of TriZol and was centrifuged at a speed of 12,000 × g at 4°C for 15 min. The upper aqueous phase (∼400 μl of RNA) was transferred and added to a fresh 1.5-ml tube. One volume of 70% ethanol was mixed with the RNA and the downstream purification was performed using an RNAeasy Mini spin column (Qiagen) according to the manufacturer’s protocol. RNA concentrations and integrity (RNA Integrity Number—RIN) were determined using NanoDrop (ThermoScientific) and an RNA Nanochip BioAnalyzer 2100 (Aligent), respectively.
cDNA synthesis and quantitative PCR
An amount of 0.1 μg of isolated RNA was used as a template for synthesizing complementary DNA (cDNA) using a 1st Strand Synthesis Kit (Invitrogen) according to the manufacturer’s protocol. To conduct quantitative PCR (qPCR), the cDNA product synthesized was diluted 1:50 before being used as a template for the SYBR-Green reaction (ThermoScientific). A total volume of 10 μl was prepared for the SYBR-Green qPCR reaction (2.5 μl diluted cDNA or nH2O, 2.5 μl 2 μM Forward+Reverse primers [Integrated DNA Technologies] mix, and 5 μl of SYBER-Green). The qPCR thermal cycle was run on an Applied Biosystems StepOnePlus real-time PCR System. The qPCR thermal cycle included a cycle of 50°C for 2 min and 95°C for 10 min followed by 44 cycles of 95°C for 15 s and 60°C for 1 min. Values of expression of gene targets were normalized to housekeeping gene Nono and determined via ΔΔCt. Primers for qPCR analysis were designed against the following genes: Cebpa (forward, AAACAACGCAACGTGGAGA; reverse, GCGGTCATTGTCACTGGTC), Cebpb (forward, ATCGACTTCAGCCCCTACCT; reverse, TAGTCGTCGGCGAAGAGG), Fabp4 (forward, GGATGGAAAGTCGACCACAA; reverse, TGGAAGTCACGCCTTTCATA), Nono (forward, GCCAGAATGAAGGCTTGACTAT; reverse, TATCAGGGGGAAGATTGCCCA).
RNA-sequencing library preparation and sequencing
Using a ThermoScientific NanoDrop 2000 and an Agilent BioAnalyzer 2100, the quantity and quality (RIN > 8) of total RNA were assessed before library preparation. RNA sequencing (RNA-Seq) libraries were prepared using an Illumina TruSeq Stranded mRNA prep kit. Using the manufacturer’s LS protocol, samples were bar coded, multiplexed, and sequenced (100 bp pair-end) in a single lane on the Illumina NextSeq 550 platform at the New York University Abu Dhabi (NYUAD) Genomic Core facility (Abu Dhabi, U.A.E).
Transcriptome data computational analysis
Transcriptional data from MEF-KO and MEF-WT cells were available from a previous study. The data sets are stored in the publicly available Gene Expression Omnibus (GEO) repository under GSE95830. The RNA-Seq data obtained on induced adipocytes were stored under GSE130765. For all the datasets, the DESeq2 computational pipeline was used to estimate the raw count reads aligned to the reference genome (Love et al., 2014). Mouse genome (GRCm38/mm10) from the University of California Santa Cruz Genome Browser (https://genome.ucsc.edu/) was utilized as a reference genome (Kent et al., 2002). Computing methods were run on a Linux-based command system on the NYUAD high-performance computing (HPC) server platform Dalma. Gene lists based on different comparative analyses (i.e., statistical significance [p-value < 0.01], fold inductions) and showing differential gene expression between two cell lines were identified using JMP genomics software (http://www.jmp.com/software/genomics/). Selected gene lists were subjected to Gene Ontology (GO) term enrichment analysis using the Database for Annotation, Visualization and Integrated Discover (DAVID) bioinformatics tool (https://david.ncifcrf.gov/home.jsp/). Heat maps and correlation analysis (i.e., principal component analysis—PCA, distance dendrogram) were generated by the RNA-Seq START (Shiny Transcriptome Analysis Resource Tool) application via the NYUAD Center of Genomic and Systems Biology (NYUAD-CGSB) Bioinformatics Online Analysis and Visualization Portal (http://tsar.abudhabi.nyu.edu/; Nelson et al., 2017).
Chromatin immunoprecipitation and qPCR analysis
Chromatin immunoprecipitation was previously described in Xie et al (2018a). Briefly, chromatin isolated from WT MEFs and β-actin KO MEFs was subjected to immunoprecipitations with an anti-CEBPB (catalogue number sc-7962) and anti-H3K9me3 (ab10812) antibodies. Analysis was performed by qPCR with primers amplifying overlapping regions covering approximately 2 kb upstream from the Cebpa gene TSS (regions R1–R5), a sequence within the gene coding region (R6), and a distant upstream sequence (R7). The following primers were used for qPCR analysis: R1 (forward, AAACTGTGTCTTCAGGCCCC; reverse, CTCGCGGAAAAGGACCCTAA), R2 (forward, AGCAATCCTATCGCTCTGGC; reverse, GCTCTTCAGAGTAGTAGGGCG), R3 (forward, CTCCCTAGTGTTGGCTGGAA; reverse, ACACGTGGTCCGTGGTTAG), R4 (forward, ACCGCCTTGGAAAGTCACAG; reverse, CGCGGGGACCGCTTTTATAG), R5 (forward, GGCCTGGCCATTCGC; reverse, ACTCCATGGGGGAGTTAGAGT), R6 (forward, AGAAAAGCCTTTCCCCACCC; reverse, CCATCAACCCAACCCTGTCT), R7 (forward, CGATCCTCTGCTCACACCAG; reverse TCGGTGCAGAGCGCATAAAA).
Chromatin immunoprecipitation sequencing
Data of chromatin immunoprecipitation sequencing (ChIP-Seq) for WT MEFs and β-actin KO MEFs using Brg1 and H3k9Me3 antibodies were from previous study in Xie et al. (2018a), which was deposited in the GEO repository with accession number GSE100096.
Statistical analysis
Statistical analysis was carried out using Student’s t tests analysis using Microsoft Excel. Results were represented as means of at least three independent experiments. A p-value < 0.05 was considered significant.
ATAC-Seq
50,000 cells per sample were used for the preparation of ATAC-Seq libraries. Cell samples in frozen medium (DMEM with 50% FBS and 10% DMSO) were shipped on dry ice to Novogene (Beijing, China). Subsequent processing, ATAC-seq library construction, and sequencing were performed by Novogene (Beijing, China). Briefly, cell nuclei were isolated from the cell samples and mixed with Tn5 Transposase with two adapters, and tagmentation was performed for 30 min at 37⁰C. The fragmented DNA was purified and amplified with a limited PCR cycle using index primers. Libraries were sequenced using Novaseq6000, paired-end 150 cycles. Raw reads were assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and quality trimmed using Trimmomatic (Bolger et al., 2014) to trim low-quality bases, systematic base calling errors, and sequencing adapter contamination. The specific parameters used were “trimmomatic_adapter.fa:2:30:10 TRAILING:3 LEADING:3 SLIDINGWINDOW:4:15 MINLEN:36.” Surviving paired reads were then aligned against the mouse reference genome (GRCm38) using a Burrows–Wheeler Aligner BWA-MEM 9 (Li and Durbin, 2010). The resulting BAM alignments were cleaned, sorted and deduplicated (PCR and Optical duplicates) with PICARD tools (http://broadinstitute.github.io/picard). Processed alignments were analyzed using the computeMatrix function of DeepTools2 (Ramirez et al., 2016) to plot the average signal around regions of interest. ChIP and ATAC-Seq profiles of genes of interest were visualized using IGV (Robinson et al., 2011). The ATAC-seq data are deposited in the GEO repository with accession number GSE133196 and are publicly available.
Supplementary Material
Acknowledgments
We thank the NYUAD Center for Genomics and Systems Biology, in particular Marc Arnoux and Mehar Sultana, for technical help, as well as Core Technology Platform Resources. We appreciate the computational platform provided by the NYUAD HPC team and are especially thankful to Yousif Ayman and Nizar Drou for technical help. This work is supported by individual grants provided by NYUAD to M.A.A. and P.P. as well as by grants from the Swedish Research Council (Vetenskapsrådet) and the Swedish Cancer Society (Cancerfonden) to P.P.
Abbreviations used:
- ATAC-Seq
assay for Transposase-Accessible Chromatin using sequencing
- BAF
Brahma-related gene (Brg)/Brahma-associated factor
- Brg1
Brahma-related gene 1
- ChIP
chromatin immunoprecipitation
- ChIP-Seq
ChIP and deep sequencing
- FBS
fetal bovine serum
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- H
histone
- HET
heterozygous
- HRP
horseradish peroxidase
- KO
knockout
- Me3
trimethylation
- MEF
mouse embryonic fibroblast
- MEF-iA
induced adipocytes from mouse embryonic fibroblast
- MEF-KO
knockout mouse embryonic fibroblast
- MEF-KO-iA
induced adipocytes from knockout mouse embryonic fibroblast
- MEF-NA
knockout mouse embryonic fibroblast expressing NLS-tagged actin
- MEF-NA-iA
induced adipocytes from knockout mouse embryonic fibroblast expressing NLS-tagged actin
- MEF-WT
wild-type mouse embryonic fibroblast
- NLS
nuclear localization factor
- Nono
non-POU domain-containing octamer-binding protein
- NYUAD
New York University Abu Dubai
- O.D.
optical density
- qPCR
quantitative PCR
- RNA-Seq
RNA deep sequencing
- RPKM
reads per kilobase per million mapped reads
- TSS
transcription start site
- WT
wild-type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-11-0628) on September 2, 2020.
REFERENCES
- Adamczak M, Wiecek A (2013). The adipose tissue as an endocrine organ. Semin Nephrol , 2–13. [DOI] [PubMed] [Google Scholar]
- Almuzzaini B, Sarshad AA, Rahmanto AS, Hansson ML, Von Euler A, Sangfelt O, Visa N, Farrants AK, Percipalle P (2016). In β-actin knockouts, epigenetic reprogramming and rDNA transcription inactivation lead to growth and proliferation defects. FASEB J , 2860–2873. [DOI] [PubMed] [Google Scholar]
- Arimochi H, Sasaki Y, Kitamura A, Yasutomo K (2016). Differentiation of preadipocytes and mature adipocytes requires PSMB8. Sci Rep , 26791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics , 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinaux JR, Magistretti PJ (1996). Vasoactive intestinal peptide, pituitary adenylate cyclase-activating peptide, and noradrenaline induce the transcription factors CCAAT/enhancer binding protein (C/EBP)-beta and C/EBP delta in mouse cortical astrocytes: involvement in cAMP-regulated glycogen metabolism. J Neurosci , 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhu C, Ji C, Zhao Y, Zhang C, Chen F, Gao C, Zhu J, Qian L, Guo X (2010). STEAP4, a gene associated with insulin sensitivity, is regulated by several adipokines in human adipocytes. Int J Mol Med , 361–367. [DOI] [PubMed] [Google Scholar]
- Cinti S (2018). Adipose organ development and remodeling. Comp Physiol , 1357–1431. [DOI] [PubMed] [Google Scholar]
- Cuaranta-Monroy I, Simandi Z, Kolostyak Z, Doan-Xuan QM, Poliska S, Horvath A, Nagy G, Bacso Z, Nagy L (2014). Highly efficient differentiation of embryonic stem cells into adipocytes by ascorbic acid. Stem Cell Res , 88–97. [DOI] [PubMed] [Google Scholar]
- Farmer SR (2006). Transcriptional control of adipocyte formation. Cell Metab , 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T, Morita S, Kimura M, Hatada I (2009). Epigenetic regulation of adipocyte differentiation by a Rho guanine nucleotide exchange factor, WGEF. PLoS One , e5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A (2005). Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol , 247–269. [DOI] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Spiegelman BM (2010). Transcriptional control of brown fat development. Cell Metab , 257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002). The human genome browser at UCSC. Genome Res , 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw EE, Flier JS (2004). Adipose tissue as an endocrine organ. J Clin Endocrinol Metab : 2548–2556. [DOI] [PubMed] [Google Scholar]
- Lane MD, Lin FT, Macdougald OA, Vasseur-Cognet M (1996). Control of adipocyte differentiation by CCAAT/enhancer binding protein alpha (C/EBP alpha). Int J Obes Relat Metab Disord , S91–96. [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ Jr, Liu XS, Lazar MA (2008). PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev , 2941–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2010). Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics , 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FT, Macdougald OA, Diehl AM, Lane MD (1993). A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci USA , 9606–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Ishimura-Oka K, Demayo F, Kibe T, Repka D, Poindexter B, Bick RJ, Darlington GJ (2001). C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc Natl Acad Sci USA , 12532–12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol , 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CE, O’rahilly S, Rochford JJ (2011). Adipogenesis at a glance. J Cell Sci , 2681–2686. [DOI] [PubMed] [Google Scholar]
- Madsen MS, Siersbaek R, Boergesen M, Nielsen R, Mandrup S (2014). Peroxisome proliferator-activated receptor gamma and C/EBPalpha synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol , 939–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Nakaki R, Inagaki T, Yoshida A, Kano Y, Kimura H, Tanaka T, Tsutsumi S, Nakao M, Doi T, et al. (2015). H3K4/H3K9me3 bivalent chromatin domains targeted by lineage-specific DNA methylation pauses adipocyte differentiation. Mol Cell , 584–596. [DOI] [PubMed] [Google Scholar]
- Mcbeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS (2004). Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell , 483–495. [DOI] [PubMed] [Google Scholar]
- McGown C, Birerdinc A, Younossi ZM (2014). Adipose tissue as an endocrine organ. Clin Liver Dis , 41–58. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Nguyen KT, Allen GE, Jullien J, Kumar D, Otani T, Bradshaw CR, Livesey FJ, Kellis M, Gurdon JB (2018). Chromatin accessibility impacts transcriptional reprogramming in oocytes. Cell Rep , 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseti D, Regassa A, Kim WK (2016). Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. Int J Mol Sci DOI: 10.3390/ijms17010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sklenar J, Barnes AP, Minnier J (2017). The START app: a Web-based RNAseq analysis and visualization resource. Bioinformatics , 447–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusue H, Onishi N, Shimizu T, Sugihara E, Oki Y, Sumikawa Y, Chiyoda T, Akashi K, Saya H, Kano K (2014). Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nat Commun , 3368. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Hosoda K, Fujikura J, Fujimoto M, Iwakura H, Tomita T, Ishii T, Arai N, Hirata M, Ebihara K, et al. (2007). Genetic and pharmacological inhibition of Rho-associated kinase II enhances adipogenesis. J Biol Chem , 29574–29583. [DOI] [PubMed] [Google Scholar]
- Otto TC, Lane MD (2005). Adipose development: from stem cell to adipocyte. Crit Rev Biochem Mol Biol , 229–242. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Vartiainen M (2019). Cytoskeletal proteins in the cell nucleus: a special nuclear actin perspective. Mol Biol Cell , 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P (2013). Co-transcriptional nuclear actin dynamics. Nucleus , 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala MW, Scherer PE (2003). Minireview: The adipocyte—at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology , 3765–3773. [DOI] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T (2016). deepTools2: a next generation Web server for deep-sequencing data analysis. Nucleic Acids Res , W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Zacarias JL, Castro-Munozledo F, Kuri-Harcuch W (1992). Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil Red O. Histochem , 493–497. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011). Integrative genomics viewer. Nat Biotechnol , 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM (2002). C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev , 22–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Macdougald OA (2006). Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol , 885–896. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM (2000). Transcriptional regulation of adipogenesis. Genes Dev , 1293–1307. [PubMed] [Google Scholar]
- Saely CH, Geiger K, Drexel H (2012). Brown versus white adipose tissue: a mini-review. Gerontology , 15–23. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LM, Civin CI, Rorth P, Friedman AD (1992). A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood , 1725–1735. [PubMed] [Google Scholar]
- Shapira SN, Lim HW, Rajakumari S, Sakers AP, Ishibashi J, Harms MJ, Won KJ, Seale P (2017). EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev , 660–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SN, Seale P (2019). Transcriptional control of brown and beige fat development and function. Obesity , 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, Mandrup S (2012). Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab , 56–64. [DOI] [PubMed] [Google Scholar]
- Smas CM, Sul HS (1995). Control of adipocyte differentiation. Biochem J , 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordella R, Jiang W, Chen GC, Curto M, Settleman J (2003). Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell , 147–158. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM (1998). PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes , 507–514. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Flier JS (1996). Adipogenesis and obesity: rounding out the big picture. Cell , 377–389. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yoshida N, Kishimoto T, Akira S (1997). Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J , 7432–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanegashima K, Zhao H, Dawid IB (2008). WGEF activates Rho in the Wnt-PCP pathway and controls convergent extension in Xenopus gastrulation. EMBO J , 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen JA, Vartiainen MK (2017). Diverse functions for different forms of nuclear actin. Curr Opin Cell Biol , 33–38. [DOI] [PubMed] [Google Scholar]
- Visa N, Percipalle P (2010). Nuclear functions of actin. Cold Spring Harb Perspect Biol , a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Bucher NL, Farmer SR (1996). Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol Cell Biol , 4128–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Almuzzaini B, Drou N, Kremb S, Yousif A, Farrants AO, Gunsalus K, Percipalle P (2018a). Beta-actin-dependent global chromatin organization and gene expression programs control cellular identity. FASEB J , 1296–1314. [DOI] [PubMed] [Google Scholar]
- Xie X, Jankauskas R, Mazari AMA, Drou N, Percipalle P (2018b). Beta-actin regulates a heterochromatin landscape essential for optimal induction of neuronal programs during direct reprograming. PLoS Genet , e1007846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Percipalle P (2018). An actin-based nucleoskeleton involved in gene regulation and genome organization. Biochem Biophys Res Commun , 378–386. [DOI] [PubMed] [Google Scholar]
- Zannetti C, Bonnay F, Takeshita F, Parroche P, Menetrier-Caux C, Tommasino M, Hasan UA (2010). C/EBP{delta} and STAT-1 are required for TLR8 transcriptional activity. J Biol Chem , 34773–34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu H, Ma S, Jing F, Yu C, Gao L, Zhao J (2018). Transcription regulators and hormones involved in the development of brown fat and white fat browning: transcriptional and hormonal control of brown/beige fat development. Physiol Res , 347–362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.