Abstract
Ubiquitin-dependent proteolysis of cyclin B and securin initiates sister chromatid segregation and anaphase. The anaphase-promoting complex/cyclosome and its coactivator CDC20 (APC/CCDC20) form the main ubiquitin E3 ligase for these two proteins. APC/CCDC20 is regulated by CDK1-cyclin B and counteracting PP1 and PP2A family phosphatases through modulation of both activating and inhibitory phosphorylation. Here, we report that PP1 promotes cyclin B destruction at the onset of anaphase by removing specific inhibitory phosphorylation in the N-terminus of CDC20. Depletion or chemical inhibition of PP1 stabilizes cyclin B and results in a pronounced delay at the metaphase-to-anaphase transition after chromosome alignment. This requirement for PP1 is lost in cells expressing CDK1 phosphorylation–defective CDC206A mutants. These CDC206A cells show a normal spindle checkpoint response and rapidly destroy cyclin B once all chromosomes have aligned and enter into anaphase in the absence of PP1 activity. PP1 therefore facilitates the metaphase-to-anaphase transition by promoting APC/CCDC20-dependent destruction of cyclin B in human cells.
INTRODUCTION
Entry into and exit from mitosis is regulated by a conserved network of pathways controlling the activity and stability of the cyclin B–dependent protein kinase (CDK1-cyclin B). CDK1-cyclin B activation promotes entry into mitosis and the maintenance of the mitotic state until mitotic spindle formation and chromosome alignment have been completed (Nigg, 2001). Subsequent destruction of cyclin B once all chromosomes have aligned is the key event permitting exit from mitosis (Holder et al., 2019). Cyclin B stability is controlled by a specific ubiquitin E3 ligase known as the anaphase-promoting complex/cyclosome (APC/C) acting in concert with the ubiquitin–proteasome system (Sivakumar and Gorbsky, 2015; Alfieri et al., 2017; Watson et al., 2019). Anaphase onset is initiated by APC/C-dependent ubiquitylation of cyclin B and securin, marking them for destruction by the proteasome (Irniger et al., 1995; King et al., 1995; Murray, 1995). In addition to being a target of the APC/C, CDK1-cyclin B modulates APC/C activity toward different substrates through antagonistic phosphorylation of core APC/C subunits and the two coactivator subunits CDC20 and CDH1. CDK1-cyclin B phosphorylation of the core APC/C is necessary for CDC20-dependent ubiquitin ligase activity, whereas both CDC20 and CDH1 are inhibited by mitotic phosphorylation (Kramer et al., 2000; Yudkovsky et al., 2000; Kraft et al., 2003; Labit et al., 2012; Fujimitsu et al., 2016; Qiao et al., 2016; Zhang et al., 2016). Prior to mitosis when CDK1-cyclin B activity is low, the formation of active APC/CCDC20 complexes is unfavorable since the core APC/C subunits are not phosphorylated. Under these conditions an autoinhibitory segment within the APC1 subunit reduces binding of the CDC20 coactivator (Fujimitsu et al., 2016; Qiao et al., 2016; Zhang et al., 2016) and thus prevents ubiquitin–ligase activity toward cyclin B in late S-phase and G2. Phosphorylation of the APC1 and APC3 subunits by CDK1-cyclin B relieves this inhibition and is a prerequisite for CDC20 association with the core APC/C (Fujimitsu et al., 2016; Qiao et al., 2016; Zhang et al., 2016). Conversely, CDH1 binding to the APC/C does not require activating phosphorylation of these core subunits (Zhang et al., 2016). Activation of the APC/C by CDH1 is prevented until late during exit from mitosis by two means. CDH1 is sequestered by an inhibitory factor EMI1 throughout S-phase and G2 (Hsu et al., 2002; Miller et al., 2006). Additionally, CDK1-dependent phosphorylation of CDH1 prevents it from binding to the core APC/C during mitosis (Zachariae et al., 1998; Kramer et al., 2000). Thus, CDK1 promotes formation of active APC/CCDC20 while simultaneously inhibiting formation of the APC/CCDH1 complex. However, in the absence of further regulation, this arrangement would result in cyclin B destruction immediately upon entry into mitosis without a delay to allow for chromosome alignment and segregation, or cell division.
During mitosis, the core APC/C becomes phosphorylated and CDC20 would be expected to activate its ubiquitin ligase activity toward cyclin B immediately on mitotic entry. This is prevented by two further mechanisms. First, CDC20 is phosphorylated by CDK1 on entry into mitosis, reducing its affinity for the APC/C (Kramer et al., 2000; Yudkovsky et al., 2000). Second, CDC20 is sequestered into a diffusible inhibitor of the APC/C, the mitotic checkpoint complex (MCC), until the process of chromosome alignment has been completed (Musacchio, 2015; Hayward et al., 2019b). The MCC consists of four proteins, the checkpoint proteins MAD2, BUB3, and BUBR1 and CDC20 (Hardwick et al., 2000; Fraschini et al., 2001; Sudakin et al., 2001; Lara-Gonzalez et al., 2012; Musacchio, 2015). Formation of the MCC is triggered at unattached or incorrectly attached kinetochores through the action of the spindle checkpoint kinase MPS1 (Abrieu et al., 2001; Stucke et al., 2002; Musacchio, 2015). MPS1 promotes the accumulation of the MCC subunits at kinetochores through the phosphorylation of the outer kinetochore protein KNL1 (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012). MPS1-phosphorylated KNL1 acts as scaffold concentrating BUB1/BUB3 and BUBR1/BUB3 complexes at unattached kinetochores (Primorac et al., 2013; Overlack et al., 2015). BUB1 is further phosphorylated by MPS1, initiating the recruitment of MAD1 (Ji et al., 2017; Qian et al., 2017; Zhang et al., 2017). Crucially, MPS1 catalyzes the formation of the MCC through phosphorylation of the C-terminus of MAD1 (Faesen et al., 2017; Ji et al., 2017). MCC production is further strengthened by phosphorylation of CDC20 by CDK1-cyclin B, which biases CDC20 toward preferential incorporation into the MCC rather than association with the APC/C (Yudkovsky et al., 2000; D’Angiolella et al., 2003). The mechanism remains unclear, but the simplest explanation is that phosphorylation increases the free pool of CDC20 available for incorporation into the MCC. Completing this regulatory circuit, CDK1-cyclin B1 directly aids the production of MCC by promoting the activation and recruitment of the checkpoint kinase, MPS1, to kinetochores (Morin et al., 2012; Vazquez-Novelle et al., 2014; Alfonso-Perez et al., 2019; Hayward et al., 2019a, b). This CDK1 cyclin B–dependent cycle of MCC production and inhibition of APC/C thus prevents entry into anaphase until stable microtubule–kinetochore attachment has been achieved at all kinetochores.
CDK1-cyclin B therefore stabilizes the mitotic state in two ways and in doing so creates a crucial requirement for regulated phosphatase activity in mitotic exit. In mitosis, CDK1 inhibits APC/C activity through phosphorylation of CDC20 and CDH1 and by activating the spindle checkpoint pathway. Conversely, CDK1-cyclin B also promotes mitotic exit by phosphorylating and thereby activating APC/C (Kraft et al., 2003). Thus, CDC20 can only activate the APC/C in mitosis until the early stages of anaphase when APC/C phosphorylation is maintained. Maximal APC/CCDC20 activity in anaphase therefore requires differential dephosphorylation of the APC/C and CDC20. Simple logic dictates that CDC20 must be dephosphorylated before the core APC/C is dephosphorylated; otherwise active APC/CCDC20 complexes would not form. Later dephosphorylation of CDH1 would then explain the formation and activity of APC/CCDH1. The reported preference of the PP2A phosphatase for phosphorylated threonine compared with serine may explain differential dephosphorylation kinetics in mammalian cells (Cundell et al., 2016; Hein et al., 2017). For CDC20 a number of the important phosphorylated regulatory sites are on threonine residues, whereas those on the APC/C and CDH1 are serine (Hein et al., 2017; Fujimitsu and Yamano, 2020). Thus, CDC20 would become dephosphorylated before the core APC/C and CDH1, resulting in APC/CCDC20 activity. However, in other organisms there is good evidence that PP1 rather than PP2A dephosphorylates CDC20 to activate the APC/C at mitotic exit (Kim et al., 2017).
In general, the modulation of CDK1 cyclin B–mediated phosphorylation by counteracting phosphatases remains poorly understood, and this may reflect the involvement of multiple phosphatases acting on a diverse range of substrates. Both protein phosphatase 1 (PP1) and protein phosphatase 2A complexed to the B55 regulatory subunit (PP2A-B55) have been implicated as major CDK1-cyclin B opposing phosphatases (Wu et al., 2009; Schmitz et al., 2010; McCloy et al., 2015; Cundell et al., 2016; Godfrey et al., 2017). For PP2A-B55, unbiased proteomic screens have started identifying individual substrates as well as general motifs characterizing PP2A-B55 targets in anaphase cells (McCloy et al., 2015; Cundell et al., 2016). Indeed, the aforementioned phosphorylation of MPS1 by CDK1-cyclin B1 is removed by PP2A-B55 during anaphase (Hayward et al., 2019a). In mammalian cells, PP2A-B55 has also been suggested as the enzyme responsible for CDC20 dephosphorylation (Hein et al., 2017). However, depletion of PP2A-B55 does not affect progression through the metaphase-to-anaphase transition (Cundell et al., 2013; Hayward et al., 2019a), which relies on normal APC/CCDC20 activation, making it unlikely that PP2A-B55 is the only phosphatase carrying out this function. For PP1, which has three isoforms in human cells, α, β, and γ, substrates on chromatin and kinetochores have been identified (Francisco and Chan, 1994; Cohen, 2002; Wang et al., 2008; Yamashiro et al., 2008; Qian et al., 2011; London et al., 2012; Nijenhuis et al., 2014; Nilsson, 2019). In addition, the APC/C regulator and spindle checkpoint protein CDC20 has been reported to be a PP1 substrate in Caenorhabditis elegans (Kim et al., 2017); however whether this form of regulation is conserved in human cells has not been explored.
Here we investigate APC/C activation in mitotic exit and present evidence that PP1 is an important CDK1-counteracting CDC20 phosphatase promoting rapid cyclin B destruction in human cells. By controlling APC/CCDC20-dependent destruction of cyclin B downstream of the spindle checkpoint, PP1 accelerates the metaphase-to-anaphase transition and contributes to the timing of mitotic exit.
RESULTS
PP1 contributes to activation of APC/C during mitotic exit
Synchronous mitotic exit and entry into anaphase can be induced by chemical inhibition of either the spindle checkpoint kinase MPS1 or the mitotic master kinase CDK1 (Cundell et al., 2013). Despite the overall similar behavior, detailed biochemical analysis of mitotic exit reveals important differences between these two conditions (Supplemental Figure S1, A, CDK-i, and B, MPS1-i). Inhibition of CDK1 results in accelerated destruction of cyclin B1 when compared with inhibition of the checkpoint kinase MPS1 (Supplemental Figure S1C). This agrees with the notion that CDK1-cyclin B directly opposes APC/C activation independently of the MPS1-dependent spindle checkpoint pathway. In principle, this could be explained by CDK1-counteracting phosphatases of the PP1 and PP2A families. However, previous work showed that silencing of PP2A-B55 does not delay the metaphase-to-anaphase transition in the absence of chromosome segregation errors (Cundell et al., 2013; Hayward et al., 2019a). By contrast, PP2A-B56 is active throughout mitosis and is essential for chromosome alignment and checkpoint signaling, making it difficult to probe any downstream role at the metaphase-to-anaphase transition (Espert et al., 2014; Nilsson, 2019). We therefore focused on the role of PP1, in part because CDK1 inhibition results in accelerated dephosphorylation of the inhibitory T320 residue in PP1 with kinetics that parallel those for cyclin B destruction (Supplemental Figure S1, C and D). PP1 is phosphorylated and inhibited by CDK1 on a conserved C-terminal threonine, T320 in the PP1α isoform and the equivalent residues in the PP1β and PP1γ isoforms (Dohadwala et al., 1994; Goldberg et al., 1995; Kwon et al., 1997). The dephosphorylation of this residue is thought to be an autodephosphorylation event and can therefore be used as a surrogate measure of PP1 activity (Wu et al., 2009).
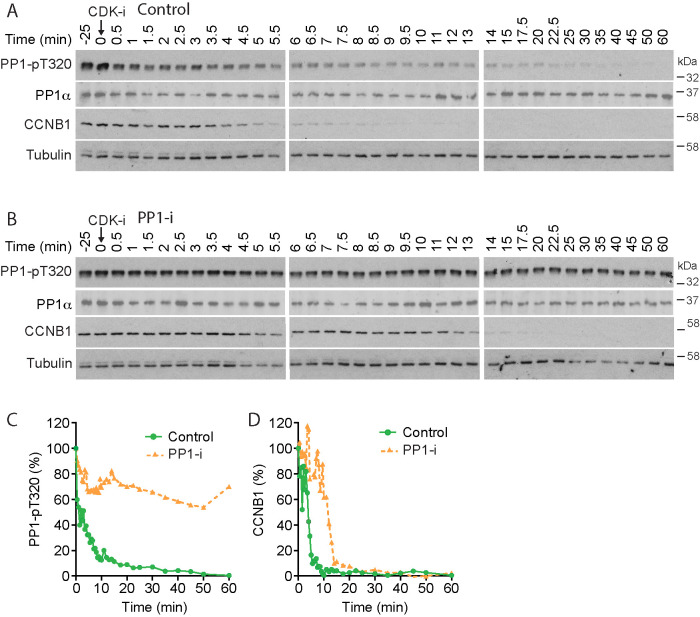
To test whether PP1 activity contributes to the accelerated destruction of cyclin B upon CDK1 inhibition, we performed a biochemical analysis of mitotic exit in the presence of the highly specific small molecule PP1 inhibitor (PP-i) tautomycetin, which inhibits all three PP1 isoforms but not PP2A (Choy et al., 2017; Hayward et al., 2019c). Tautomycetin prevented dephosphorylation of PP1-pT320 (Figure 1, A, control, and B, PP-i), and the half-life of pT320 increased from ∼2 min in the control to >60 min (Figure 1C). Cyclin B1 destruction was delayed in the presence of tautomycetin (Figure 1, A, control, and B, PP-i), and the half-life increased from 4 to 12 min (Figure 1D). This observation is consistent with the idea that PP1 plays a role in the regulation of cyclin B stability in the initial phase of mitotic exit.
FIGURE 1:
PP1 counteracts CDK1 regulation of APC/C during mitotic exit. (A, B) HeLa cells were arrested in mitosis and incubated for 25 min with either (A) DMSO (Control) or (B) 5 µM tautomycetin PP1 inhibitor (PP1-i). CDK1 was then inhibited (CDK-i) using 5 µM flavopiridol, and samples of the culture were taken at the times indicated and Western blotted for the indicated markers. (C) PP1α-pT320 and (D) CCNB1 levels were measured as a function of time and plotted in the line graphs for the representative experiments shown in A and B.
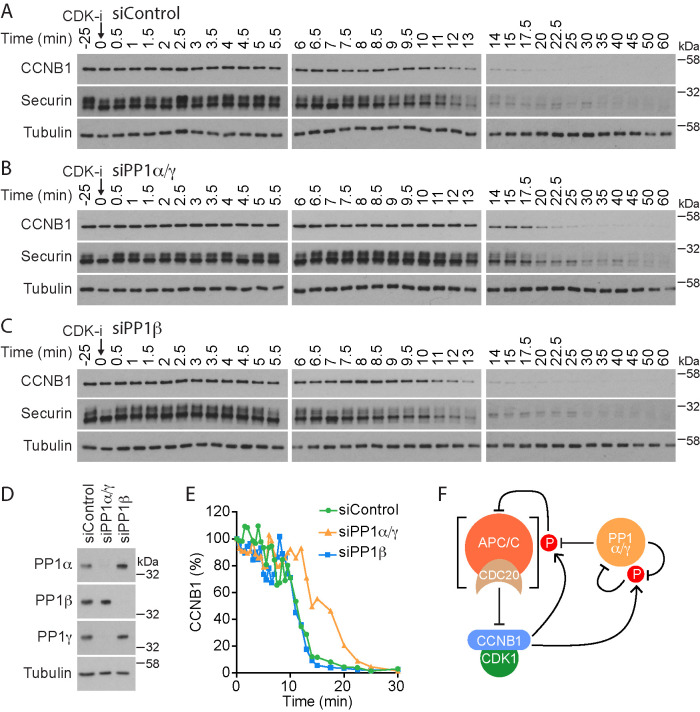
To identify which specific isoform of PP1 was responsible for this effect, HeLa cells were depleted of PP1 catalytic subunits, arrested in mitosis, and then treated with CDK1 inhibitors. Since PP1α and PP1γ are reported to have overlapping functions during mitosis (Trinkle-Mulcahy et al., 2006; Liu et al., 2010), these two catalytic subunits were depleted simultaneously (siPP1α/γ). PP1β, which has been shown to have a role distinct from PP1α and PP1γ (Yamashiro et al., 2008; Matsumura et al., 2011; Kiss et al., 2019), was knocked down separately. High-resolution mitotic exit time courses with cell samples taken every 30 s were then collected to monitor cyclin B destruction. In this assay, cyclin B1 and securin were rapidly degraded after CDK1 inhibition with a t1/2 of 11 min (Figure 2, A and E, control), whereas codepletion of PP1α and PP1γ slowed the rate of cyclin B1 destruction to a t1/2 of 17 min (Figure 2, B and E, siPP1α/γ). PP1β depletion did not alter the kinetics of cyclin B1 or securin destruction when compared with the control (Figure 2, C and E, siPP1β). Efficient PP1 depletion was confirmed for the different conditions by Western blotting (Figure 2D). We conclude that PP1 activity reverses CDK1-dependent phosphorylation of proteins normally limiting APC/CCDC20 activity toward cyclin B1 and securin at the metaphase-to-anaphase transition (Figure 2F).
FIGURE 2:
Identification of the major PP1 isoforms counteracting CDK1 regulation of APC/C. (A–C) HeLa cells were either (A) treated with control siRNA (siControl) or depleted for catalytic subunits of (B) PP1α/γ (PP1α/γ) or (C) PP1β (PP1β) and then arrested in mitosis. Mitotic exit was then triggered by CDK1 inhibition (CDK-i), and samples were taken for Western blot at the times indicated. (D) Western blot confirming efficient depletion of PP1 catalytic subunits. (E) CCNB1 levels were measured as a function of time in siControl, siPP1α/γ, and siPP1β for the representative experiments shown in A–C and plotted in the line graph. (F) Schematic depicting the potential role of PP1 in counteracting inhibitory CDK1-cyclin B phosphorylation of APC/CCDC20. CDK1-cyclin B phosphorylation also inhibits PP1 activity, and PP1 reactivates via an autocatalytic process. These are all freely reversible reactions. Activated APC/CCDC20 feeds back to irreversibly inhibit CDK1-cyclin B by promoting cyclin B destruction.
PP1 activity is required to trigger prompt cyclin B1 degradation
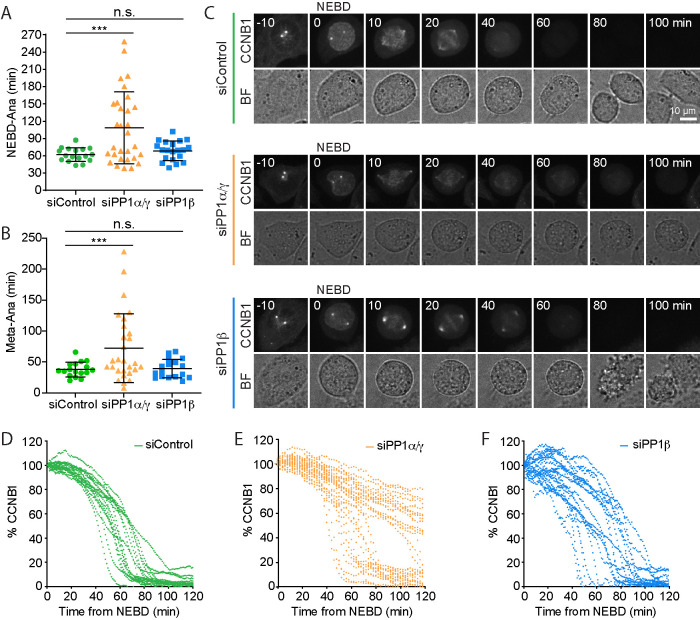
To investigate the role of PP1 at the metaphase-to-anaphase transition without the need to use CDK1 inhibition, we performed a single-cell analysis using HeLa cells expressing green fluorescent protein (GFP)-tagged cyclin B1 (CCNB1) from the endogenous promoter (Alfonso-Perez et al., 2019). This allowed us to follow cyclin B1 destruction in individual control cells passing through mitosis into anaphase and to compare control with PP1α/γ- or PP1β-depleted cells. In agreement with the biochemical data showing that PP1 is required for rapid cyclin B1 destruction, cells depleted for PP1α/γ showed delayed passage through mitosis. PP1α/γ-depleted cells took >100 min to reach anaphase from nuclear envelope breakdown (NEBD) compared with 60–70 min for control or PP1β-depleted cells (Figure 3A). This could be largely attributed to a delay in the time taken to proceed from the completion of the metaphase plate to anaphase (Figure 3B). In good agreement with previous observations (Clute and Pines, 1999), cyclin B1 was quickly degraded once chromosomes had aligned and established a metaphase plate in control cells, and this was followed by segregation of the chromosomes (Figure 3, C and D, siControl). However, codepletion of PP1α and PP1γ delayed cyclin B1 destruction (Figure 3, C and E, siPP1α/γ), resulting in a significantly increased overall length of mitosis (Figure 3A) and specifically an extended metaphase-to-anaphase transition (Figure 3B). A subpopulation of cells displayed wild-type kinetics for cyclin B1 destruction, possibly due to incomplete or variable levels of PP1α/γ depletion. Depletion of PP1β had no obvious effect on cyclin B1 destruction compared with the control condition (Figure 3, B and F, siPP1β).
FIGURE 3:
PP1 is needed for rapid destruction of cyclin B at the metaphase–anaphase transition. (A, B) Comparison of the time spent (A) in mitosis or (B) to enter anaphase following completion of a metaphase plate in siControl, siPP1α/γ depletion, or siPP1β depletion for CRISPR-tagged CCNB1-GFP HeLa cells. Scatter plots of mean ± SD are shown for siControl (n = 17), siPP1α/γ (n = 30), and siPP1β (n = 20). ** denotes p < 0.001. (C) Live cell imaging of CCNB1-GFP with times shown in minutes. Percentage of CCNB1-GFP fluorescence following NEBD in (D) siControl, (E) siPP1α/γ, and (F) siPP1β cells. Brightfield images (BF) show the cell outline. CCNB1 images are maximum-intensity projections; quantification was carried out on sum intensity projections, and the single-cell traces were plotted as a function of time from NEBD in the graphs.
PP1 therefore promotes cyclin B destruction at the metaphase-to-anaphase transition in unperturbed mitosis, confirming the data obtained under CDK1-inhibited conditions.
PP1 activity promotes timely progression from metaphase to anaphase
The effects of PP1 on cyclin B destruction can be most simply explained by a direct effect on APC/C activity. Indeed, this is the case in C. elegans, where it has been found that PP1 promotes APC/C activation downstream of the spindle checkpoint (Kim et al., 2017). In human cells, PP1 together with PP2A-B56 has been reported to counteract the MPS1-dependent phosphorylation of MELT motifs in KNL1 (Espert et al., 2014; Nijenhuis et al., 2014). Removal of PP1 would therefore be expected to delay silencing of the spindle assembly checkpoint. This could also explain, either fully or in part, the delayed degradation of cyclin B1 upon PP1 inhibition and in PP1α/γ−depleted cells. To investigate both possibilities and define when PP1 is acting, we imaged HeLa cells stably coexpressing CCNB1-mCherry from its endogenous promoter and a GFP-tagged version of the spindle checkpoint protein MAD2. This approach enabled us to test whether the period from spindle checkpoint silencing to the onset of anaphase was increased.
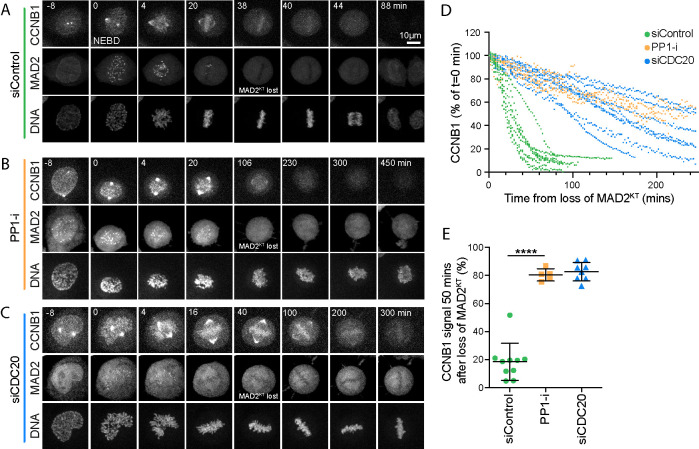
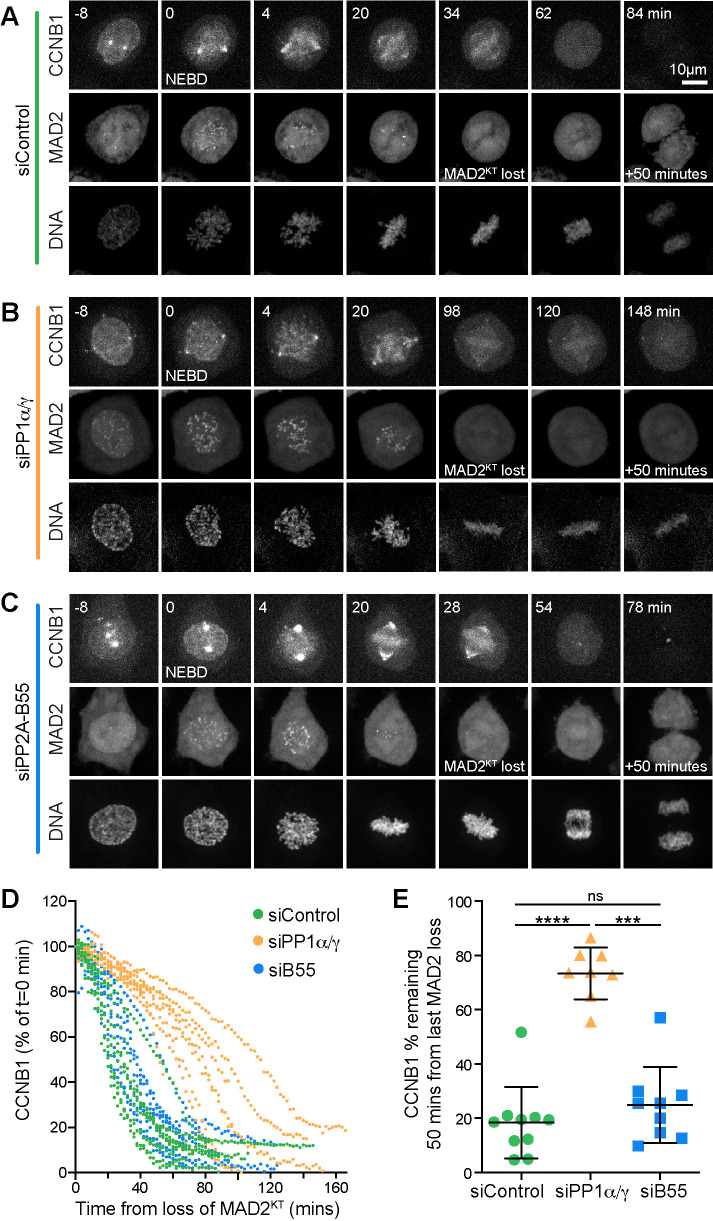
The defining hallmark of spindle checkpoint silencing is the loss of all MAD2-positive kinetochores. In control cells, complete chromosome alignment coincided with the disappearance of the last MAD2-positive kinetochore, rapidly followed by loss of the CCNB1-mCherry signal and chromosome segregation (Figure 4, A and D, siControl). Inhibition of PP1 with tautomycetin delayed cyclin B1 destruction following loss of the last MAD2-positive kinetochore (Figure 4, B and D, PP-i). The effect observed resembled a CDC20 depletion, in which spindle checkpoint silencing proceeded normally but cyclin B1 destruction was not initiated (Figure 4, C and D, and Supplemental Figure S4A, siCDC20). At 50 min after checkpoint silencing, monitored by the disappearance of GFP-MAD2 from kinetochores, PP1-inhibited cells retained the same level of cyclin B1 as CDC20-depleted cells, which are unable to target cyclin B for destruction (Figure 4E).
FIGURE 4:
PP1 inhibition prevents rapid cyclin B destruction downstream of the spindle checkpoint. (A–C) HeLa CCNB1-mCherry GFP-MAD2 cells treated with (A) control siRNA, (B) 5 µM PP1 inhibitor tautomycetin (PP1-i) for 30 min prior to the start of imaging, or (C) siCDC20 were imaged for 12 h at 2 min intervals. Chromosome congression was monitored using SiR-Hoechst, and checkpoint silencing was inferred from the loss of GFP-MAD2 from the kinetochore. Degradation of endogenously tagged CCNB1 indicated APC/C activation. Representative time course images are shown. (D) CCNB1 levels were measured as a function of time from the point at which the last MAD2-positive kinetochore (MAD2KT) was observed for siControl, PP1-i, and siCDC20 cells. CCNB1 levels were set to 100% at that point and plotted for single cells in the graph. (E) Scatter plots of the mean CCNB1 signal ± SD remaining at 50 min after loss of MAD2KT for siControl (n = 10), PP-i (n = 5), and siCDC20 (n = 8). *** denotes p < 0.001.
We then examined the relationship between checkpoint silencing and cyclin B destruction in PP1α/γ−depleted cells, focusing first on checkpoint silencing. The localization and subsequent efficient depletion of PP1 catalytic subunits was first confirmed by immunofluorescence analysis (Supplemental Figure S2, A and B) and Western blotting (Supplemental Figure S2C) of cells expressing GFP-tagged PP1 catalytic subunits. Next, the analysis of spindle checkpoint silencing revealed that MAD2-positive kinetochores were observed for a longer period in PP1-inhibited or PP1α/γ-depleted cells than in control cells (Supplemental Figure S3, A–C). Nevertheless, PP1α/γ-depleted or PP1-inhibited cells were able to silence the checkpoint and enter anaphase with a delay (Supplemental Figure S3, B, siPP1α/γ, and C, PP1-i). By contrast, PP2A-B56–depleted cells arrested with sustained MAD2-positive kinetochores and failed to enter anaphase (Supplemental Figure S3, D, siB56, and E). In agreement with the idea that checkpoint silencing and cyclin B destruction are independently delayed in the PP1-depleted cells, both the times taken for spindle checkpoint silencing to complete and from loss of the last MAD2 signal to the onset of anaphase were increased (Supplemental Figure S3F). These observations are consistent with a major role for PP2A-B56 in the spindle checkpoint and error correction pathways (Suijkerbuijk et al., 2012; Kruse et al., 2013; Xu et al., 2013; Espert et al., 2014), with an additional contribution by PP1 to the complete silencing of the spindle checkpoint (Nijenhuis et al., 2014).
We then measured the kinetics of cyclin B1 destruction in PP1-depleted cells to test whether this was delayed even after the spindle checkpoint had been silenced. In comparison to control cells, cyclin B1 destruction was delayed after the loss of MAD2-positive kinetochores in PP1α/γ−depleted cells (Figure 5, A, B, and D). At 50 min after the loss of the last MAD2-positive kinetochore, control cells had completed destruction of cyclin B1 (Figure 5, D and E, siControl). By contrast, PP1-depleted cells retained ∼80% of the maximal level of cyclin B1 measured at NEBD (Figure 5, D and E, siPP1α/γ). Importantly, PP2A-B55 depletion did not change the kinetics of the metaphase-to-anaphase transition or cyclin B1 destruction followed by live cell imaging (Figure 5, C–E, siB55) or cyclin B1 destruction and CDC20 dephosphorylation measured by biochemical analysis (Supplemental Figure S4, A–D).
FIGURE 5:
PP1α/γ are required for rapid cyclin B destruction downstream of the spindle checkpoint. (A–C) HeLa CCNB1-mCherry GFP-Mad2 cells treated with (A) control, (B) siPP1α/γ, or (C) siPP2A-B55 for 60 h were imaged for 12 h at 2 min intervals. DNA was visualized with SiR-DNA. (D) Quantitation of the CCNB1 levels of individual cells shown in A. (E) The CCNB1 signal remaining at 50 min in siControl, siPP1α/γ, or siPP2A-B55 was plotted as mean ± SD (siControl n = 10, siPP1α/γ n = 8, siPP2A-B55 n = 9). *** denotes p < 0.001; **** denotes p < 0.0001.
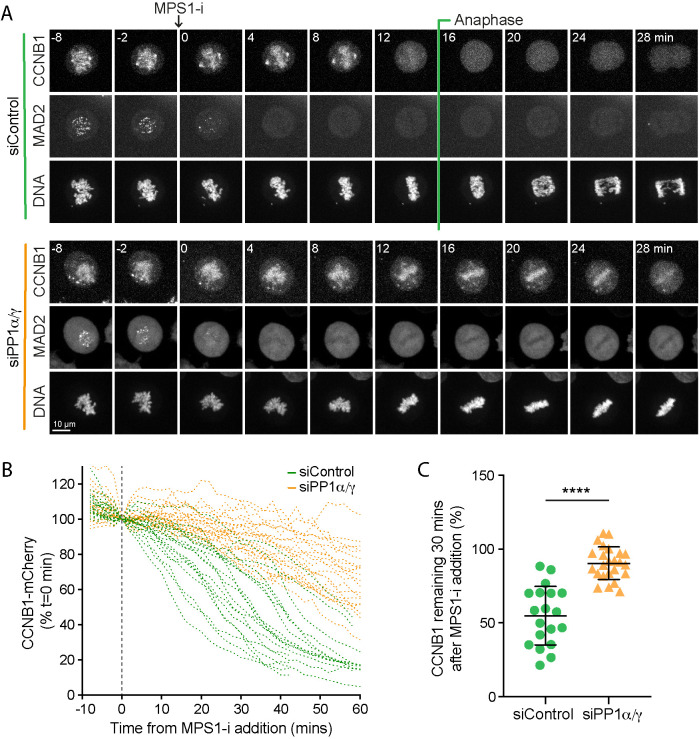
Because intermittent GFP-MAD2 signals at kinetochores are difficult to detect, it was possible that transient spindle checkpoint activation in PP1-depleted cells was the cause of delayed cyclin B destruction. For this reason, we sought further evidence that the delay in cyclin B destruction observed in cells depleted of PP1 was independent of ongoing spindle checkpoint signaling. To do this we used an MPS1 inhibitor to rapidly block spindle checkpoint signaling in live cells approaching metaphase and then measured the kinetics of cyclin B1 destruction in control and PP1-depleted cells. Addition of MPS1 inhibitor resulted in a rapid loss of kinetochore associated MAD2 within 2–4 min in all of the control and PP1-depleted cells examined (Figure 6A). Thus, in this assay MPS1 inhibition rapidly blocks checkpoint signaling in a PP1-independent manner (Espert et al., 2014; Hayward et al., 2019c). Under these conditions, cyclin B1 destruction occurred with a half-life of ∼35 min in the control cells (Figure 6, A and B, siControl) and >90 min in the PP1-depleted cells (Figure 6, A and B, siPP1α/γ). At 30 min after MPS1 inhibition, cyclin B1 levels were 54.8 ± 19.8% of the starting value in control cells, whereas in PP1-depleted cells they remained at 90.4 ± 11.0% (Figure 6C). Thus, the timing of cyclin B1 destruction but not silencing of MPS1-dependent checkpoint signaling is altered in PP1-depleted cells.
FIGURE 6:
In the absence of PP1α/γ, cyclin B destruction is delayed when the spindle checkpoint is inactivated by MPS1 inhibition. (A) HeLa CCNB1-mCherry GFP-MAD2 cells treated with control or siPP1α/γ were imaged progressing through mitosis and treated with MPS1 inhibitor AZ3146 (MPS1-i) when they approached metaphase. Loss of GFP-MAD2 and kinetics of CCNB1-mCherry degradation were monitored. (B) CCNB1-mCherry levels in single cells are plotted for siControl and siPP1α/γ cells undergoing mitotic exit. The time of MPS1-i addition (t = 0) is marked with a dashed line. (C) The percentage of cyclin B1-mCherry remaining at 30 min after MPS1-i addition in siControl (n = 19) and siPP1α/γ (n = 24) cells was plotted as mean ± SD in the line graph. A p value was calculated with an unpaired t test with Welch’s correction. **** denotes p < 0.0001.
Taken together, these results confirm that PP1 plays an important role in mediating the metaphase-to-anaphase transition, by contributing to spindle checkpoint silencing and through subsequent downstream regulation of cyclin B destruction.
PP1 dephosphorylates the N-terminus of CDC20
The results presented so far support the view that the activity of PP1 is required to trigger the rapid APC/C-dependent destruction of cyclin B downstream of the spindle checkpoint. The most parsimonious explanation for this observation is that one or more of the core components of the APC/C or an APC/C coactivator are dephosphorylated by PP1. Because CDC20 has been described as a PP1 target at the metaphase-to-anaphase transition in C. elegans (Kim et al., 2017), we decided to test the hypothesis that CDC20 may also be a critical PP1 target in human cells.
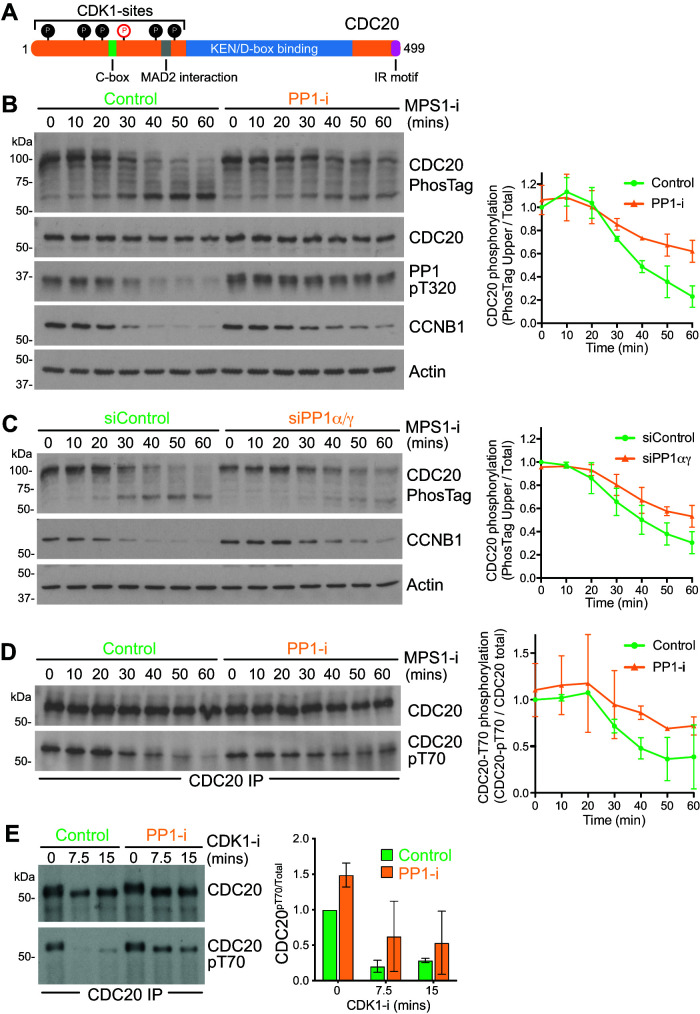
CDC20 is phosphorylated by CDK1 on six amino acids in the N-terminus of the protein (Figure 7A) (Yudkovsky et al., 2000; Labit et al., 2012). To investigate the kinetics of CDC20 dephosphorylation at the metaphase-to-anaphase transition in the presence and absence of PP1 activity, we first used PhosTag SDS–PAGE to visualize the phosphorylation status of CDC20. The PhosTag reagent binds to phosphate groups on proteins and thus results in reduced mobility and enhanced separation of the phosphorylated species away from nonphosphorylated forms on SDS–PAGE (Kinoshita et al., 2006). A pronounced downshift in CDC20 was seen as control cells progressed from mitosis into anaphase following MPS1 inhibition, consistent with the expected dephosphorylation of CDC20 at the metaphase-to-anaphase transition (Figure 7B, Control). Comparison with cells in which PP1 was inhibited revealed this downshift to be PP1-dependent (Figure 7B, PP-i). The PP1-dependent downshift was also seen when samples from control and PP1-depleted cell lysates were compared (Figure 7C, siControl and siPP1α/γ). Western blotting with a CDC20 antibody specific for phosphorylated T70 supported the view that the downshift in CDC20 was caused by PP1-dependent dephosphorylation during mitotic exit triggered by MPS1 inhibition (Figure 7D, Control and PP-i). To allow a direct comparison to the biochemical data shown in Figure 1, CDK1 inhibition was also used to trigger mitotic exit. Under these conditions, CDC20 T70 was dephosphorylated in control cells within 7.5 min (Figure 7E). As expected, pT70 dephosphorylation was more rapid than in the corresponding MPS1 inhibitor–treated cells (Figure 7D). In cells where PP1 was inhibited with tautomycetin, CDC20 pT70 was retained after 15 min (Figure 7E, Control and PP-i). Together, these results suggest that PP1 is an important CDC20 phosphatase at the metaphase-to-anaphase transition and identify T70 as a key diagnostic site for this regulation.
FIGURE 7:
PP1 dephosphorylates the N-terminal region of CDC20. (A) Schematic representation of CDC20 structure. Six N-terminal CDK1 sites are indicated as filled circles, and the pT70 site is marked in red in an open circle. (B) Synchronous progression of mitotic HeLa cells pretreated with DMSO or PP1 inhibitor into anaphase was triggered with MPS1 inhibitor and samples were collected every 10 min. Cell cycle progression was monitored by cyclin B1 Western blot, CDC20 phosphorylation state by PhosTag gel, and PP1 activation status by Western blot for PP1-pT320. Actin was used as a loading control. The mean level ± SD of phospho-CDC20 (top band in the PhosTag blot) relative to total CDC20 is plotted in the line graph as a function of time (n = 3). (C) Synchronous progression of control-depleted or PP1α/γ-depleted mitotic HeLa cells into anaphase was triggered with MPS1 inhibitor. The mean ± SD of phospho-CDC20 (top band in the PhosTag blot) relative to total CDC20 is plotted in the line graph (n = 3). (D) CDC20 was immunoprecipitated from synchronized cells treated with MPS1 inhibitor as in B, and samples were taken every 10 min. The immunoprecipitates were blotted for pT70-modified CDC20 and the total amount of CDC20. The mean level ± SD of pT70 relative to total CDC20 is plotted in the line graph (n = 2). (E) Synchronous progression of mitotic HeLa cells pretreated with DMSO or PP1 inhibitor into anaphase was triggered with CDK1 inhibitor, and samples were collected every 7.5 min. CDC20 was immunoprecipitated and the immunoprecipitates blotted for pT70. CDC20 pT70 levels relative to total CDC20 are plotted in the bar graphs as mean ± SD (n = 2).
Phosphorylation-defective CDC20 obviates the need for PP1
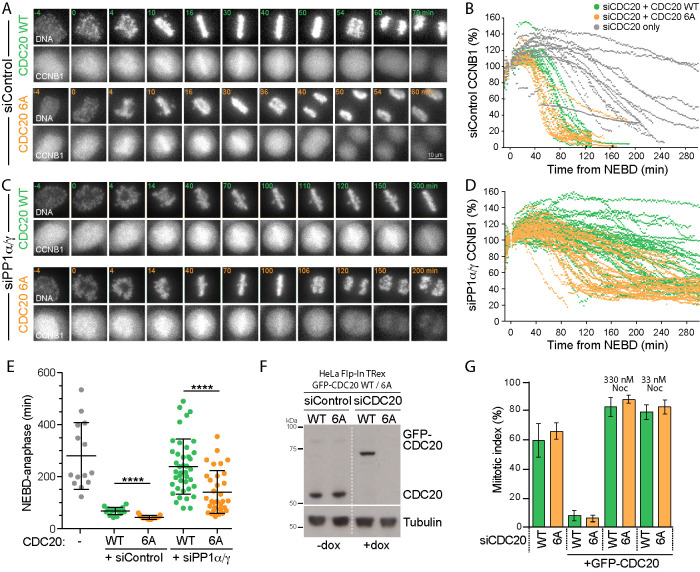
If CDC20 is a major PP1 target at the point of anaphase onset, then a CDK1 phosphorylation–resistant form of CDC20 should be able to rescue the cell cycle delay observed upon PP1 depletion or inhibition. To test this idea, endogenous CDC20 was depleted and replaced with either GFP-CDC20WT or GFP-CDC206A, lacking the N-terminal CDK1-phosphorylation sites. CDC206A localizes to kinetochores in checkpoint arrested cells, similar to the wild-type protein (Supplemental Figure S5A), but does not result in pT70 reactivity at kinetochores (Supplemental Figure S5B). In the presence of PP1, both GFP-tagged CDC20WT and CDC206A rescued the cell cycle arrest observed upon CDC20 depletion (Figure 8, A and B), indicating that APC/C activity toward cyclin B is restored. Cells expressing GFP-CDC206A passed through mitosis slightly faster than cells expressing CDC20WT (Figure 8E). In these cells, the half-life of cyclin B destruction was shortened from 80 to 60 min (Figure 8, B and D) and the time taken from NEBD to anaphase reduced from 67 to 42 min (Figure 8E). These effects were not due to differences in CDC20 levels, since these were comparable for all conditions (Figure 8F). The dominant effect on APC/C activity in the presence of endogenous wild-type CDC20 is consistent with the idea that inhibitory regulation is attenuated or lost for the GFP-CDC206A mutant, similar to results obtained with a phosphorylation-deficient CDC20 mutant in C. elegans (Kim et al., 2017). This may be due to the enhanced interaction with the APC/C detected using coimmune precipitation (Supplemental Figure S5C).
FIGURE 8:
CDK1 phosphorylation site mutant CDC206A bypasses the requirement for PP1 in rapid cyclin B destruction on mitotic exit. (A) HeLa Flp-In TRex cells expressing CCNB1-mCherry were depleted with siControl and siCDC20 and then induced to express either GFP-CDC20WT or GFP-CDC206A. Cell cycle progression and CCNB1-mCherry levels were followed by live cell imaging. DNA was visualized with SiR-DNA. (B) CCNB1 levels for individual cells are plotted in the line graph. Gray lines show CCNB1 levels in cells depleted with siCDC20 but without induction of the GFP-CDC20 transgene. (C) HeLa Flp-In TRex cells expressing CCNB1-mCherry were codepleted of siPP1α/γ and siCDC20 and then induced to express either GFP-CDC20WT or GFP-CDC206A. Cell cycle progression and CCNB1-mCherry levels were then followed by live cell imaging. DNA was visualized with SiR-DNA. (D) CCNB1 levels for individual cells are plotted in the line graph. (E) Scatter plots showing the mean time ± SD at which cells entered anaphase or the end of the movie was reached for siCDC20 uninduced (n = 12, gray), siControl with GFP-CDC20WT (n = 20, green) or GFP-CDC206A (n = 15, orange), and siPP1α/γ with GFP-CDC20WT (n = 41, green) or GFP-CDC206A (n = 33, orange). (F) Western blot of cells depleted of endogenous CDC20 and expressing GFP-CDC20 as in A, arrested with 330 nM nocodazole for 14 h. **** denotes p < 0.0001. (G) Cells depleted for endogenous CDC20 (siCDC20) and induced for GFP-CDC20WT (WT) or CDC206A (6A) were treated with nocodazole at the indicated concentrations for 14 h, and the mitotic index was plotted.
When PP1 was depleted, a cell cycle delay at the metaphase-to-anaphase transition was observed in cells rescued with CDC20WT (Figure 8, C and D). This delay was alleviated by replacement of the endogenous CDC20 with GFP-CDC206A (Figure 8, C and D). This supports the view that CDC20 is an important target for PP1 during mitotic exit. However, the half-life of mitosis was still slightly extended compared with cells expressing normal levels of PP1 and wild-type CDC20 (Figure 8E). This agrees with the notion that PP1 may have other targets in addition to CDC20 regulating the onset of anaphase.
Since CDC20 is a key part of the MCC, it was important to test whether CDC206A was defective for spindle checkpoint function. When GFP-CDC206A cells were treated with both high and low doses of nocodazole, the cells showed a normal cell cycle arrest (Figure 8G). Thus, the ability to trigger and sustain a spindle checkpoint response, that is, MCC formation and inhibition of APC/CCDC20, was not grossly compromised by expression of phosphorylation-resistant GFP-CDC206A. We therefore conclude that CDC20 is one of the key targets for dephosphorylation by PP1 at the metaphase-to-anaphase transition, independent of its role in the spindle checkpoint. The rescue of both rapid cyclin B destruction and cell cycle progression in PP1-depleted cells by CDC206A is thus a reflection of the need for PP1-mediated dephosphorylation of CDC20 to enable timely progression into anaphase.
DISCUSSION
CDC20 regulation by PP1
The metaphase-to-anaphase transition marks the point of no return for correction of errors in chromosome alignment and is a perilous stage during eukaryotic mitosis (Nasmyth, 2001). Mechanistically this is clearly understood since the release of APC/C inhibition at this point results in the activation of separase and consequently the loss of sister chromatid cohesion at all chromosomes simultaneously (Uhlmann, 2001). To ensure the synchronized separation and segregation of all the chromosomes, it is therefore crucial that the APC/C can be rapidly activated, yet is not triggered prematurely. Untimely APC/C activity at the metaphase–anaphase transition is prevented by both the spindle assembly checkpoint and phosphorylation of the APC/C coactivator CDC20 (Labit et al., 2012; Fujimitsu et al., 2016; Hein and Nilsson, 2016; Hein et al., 2017; Zhang et al., 2016). Phosphorylation of the N-terminal region of CDC20 has two notable consequences. First, to reduce the affinity of CDC20 for APC/C and second, to bias its incorporation into the MCC rather than APC/C (D’Angiolella et al., 2003; Hein and Nilsson, 2016). Thus, CDC20 has to be dephosphorylated in order to achieve full APC/C activity in exit from mitosis (Labit et al., 2012). On the basis of the data presented here, we conclude that in human cells PP1 is needed for timely CDC20 dephosphorylation at the metaphase-to-anaphase transition. Supporting the view that this is a conserved mechanism of APC/C regulation, PP1 has a similar role in C. elegans (Kim et al., 2017).
Our analysis thus sheds light on how PP1 regulates the metaphase-to-anaphase transition. However, some mechanistic details need to be explored further. There are two simple possibilities to explain how CDC20 dephosphorylation could promote cyclin B destruction. First, dephosphorylated CDC20 has already been shown to bind more readily to the APC/C (Kramer et al., 2000; Labit et al., 2012; Hein and Nilsson, 2016), as we confirm here (Supplemental Figure S5C). Second, the dephosphorylation of CDC20 at the metaphase-to-anaphase transition may also increase MCC turnover once the MPS1-dependent checkpoint signal stops and MCC production ceases. These possibilities are not mutually exclusive, and understanding the molecular consequences of CDC20 dephosphorylation is thus an important goal for future studies.
Local pools of PP1 in checkpoint signaling and APC/C regulation
A number of distinct pools of PP1 have been reported to contribute to the regulation of the metaphase-to-anaphase transition in mammalian cells. PP1 catalytic subunits bind to regulatory subunits via conserved short, linear motifs, the best characterized of which is the RVxF motif (Egloff et al., 1997; Terrak et al., 2004). Several RVxF-dependent PP1-interaction partners have been identified at attached kinetochores, including the outer kinetochore proteins KNL1 and Astrin and the mitotic motor proteins CENP-E and KIF18A (Kim et al., 2010; Liu et al., 2010; De Wever et al., 2014; Hafner et al., 2014; Conti et al., 2019). Additionally, the spindle and kinetochore–associated Ska complex also recruits PP1 in an RVxF-independent manner (Sivakumar et al., 2016). All of these PP1 complexes have been suggested to contribute to timely anaphase onset and would be good candidates to promote CDC20 dephosphorylation at attached kinetochores. Alternatively, it is conceivable that the bulk of CDC20 dephosphorylation takes place in the cytosol and involves a further, discrete, pool of PP1 or even the global pool of PP1. Consistent with this view, most of the checkpoint-dependent pool of CDC20 bound to BUB1 and BUBR1 will have already left the kinetochore prior to the metaphase-to-anaphase transition (Lischetti et al., 2014; Di Fiore et al., 2015; Vleugel et al., 2015). Identification of the PP1 pool relevant for CDC20 dephosphorylation is thus an important question to address in the future.
Timely roles for PP1 and PP2A in APC/C regulation and checkpoint signaling
PP2A-B55, PP2A-B56, and PP1 have all been reported to act on and regulate components of the APC/C and spindle checkpoint pathways (Hein et al., 2017; Kim et al., 2017; Fujimitsu and Yamano, 2020). Like PP1, PP2A-B55 is inhibited by CDK1-cyclin B and has also been suggested to be an important CDC20 phosphatase in mammalian cells (Mochida and Hunt, 2012; Hein et al., 2017). However, due to the temporal properties of its regulatory mechanism, PP2A-B55 becomes active in anaphase B only when cyclin B levels fall below a threshold level (Cundell et al., 2013, 2016). PP1 has been reported to be involved in the activation of PP2A-B55 through the dephosphorylation of the Gwl/MASTL kinase (Heim et al., 2015; Ma et al., 2016; Rogers et al., 2016; Ren et al., 2017) and could thus be considered an upstream regulator of PP2A-B55. PP2A-B55 action on CDC20 is therefore most likely restricted to a later point in anaphase, perhaps at the switch to APC/C regulation by CDH1 and destruction of late anaphase substrates. Consistent with this idea, and in agreement with previous work (Cundell et al., 2013; Hayward et al., 2019a), we did not see any impact on the kinetics of cyclin B destruction at the metaphase-to-anaphase transition when PP2A-B55 was depleted (Figure 5 and Supplemental Figure S4B).
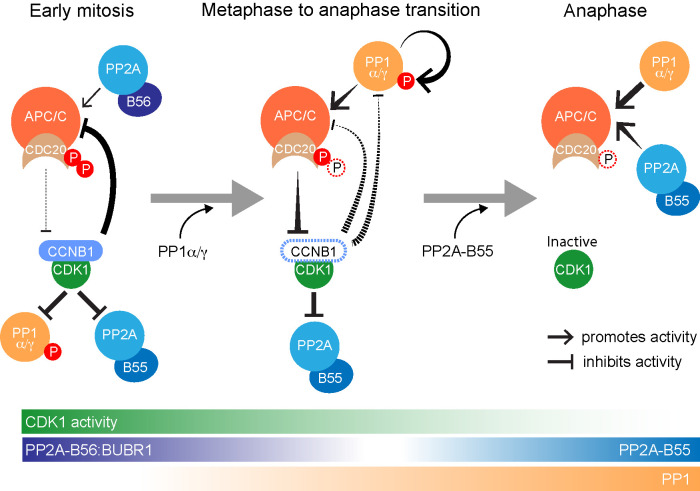
Unlike PP1 and PP2A-B55, PP2A-B56 is thought to retain full activity in mitosis, and CDK1-dependent phosphorylation of PP2A-B56 recognition sites may in fact increase its activity toward key substrates (Kruse et al., 2013; Smith et al., 2019). Interestingly, a specific pool of PP2A-B56, attached directly to the APC/C, has been implicated in vertebrate CDC20 dephosphorylation during mitosis (Lee et al., 2017; Fujimitsu and Yamano, 2020). Owing to the unique properties of PP2A-B56, we propose that this may promote the dynamic turnover of CDC20 phosphorylation during early mitosis, to create a limited level of APC/C activity prior to anaphase (Figure 9). One intriguing possibility is that PP2A-B56 is therefore important for the CDC20-dependent, spindle assembly checkpoint–independent destruction of cyclin A during the first few minutes of mitosis (Wolthuis et al., 2008; Di Fiore and Pines, 2010; van Zon and Wolthuis, 2010). By contrast, as the APC/C becomes active downstream of checkpoint silencing, PP1 promotes dephosphorylation of CDC20 and decisively tips the balance toward APC/CCDC20 activation, destabilization of the mitotic state, and consequently mitotic exit. The APC/CCDC20 active state is then consolidated by PP2A-B55, the activation of which is also initiated by PP1 (Heim et al., 2015; Ma et al., 2016; Rogers et al., 2016; Ren et al., 2017), ensuring that CDC20 is completely dephosphorylated in anaphase.
FIGURE 9:
PP2A-B56, PP1, and PP2A-B55 dephosphorylate CDC20 at different points in the cell cycle. Schematic drawing illustrating the dephosphorylation of CDC20 by the three main mitotic phosphatases PP2A-B56, PP1α/γ, and PP2A-B55 in early mitosis, at the metaphase-to-anaphase transition and in anaphase, respectively. In this schematic, PP2A-B56 refers to the pool docked to the checkpoint protein BUBR1. Other pools of PP2A-B56 are active in anaphase.
In summary, we suggest that the involvement of three major mitotic phosphatases, PP1 and PP2A-B55 and -B56, in modulation of CDK1-dependent phosphorylation of CDC20 is crucial for restricting APC/CCDC20 activity to a narrow window at the metaphase-to-anaphase transition. Each of these phosphatases has unique spatial and temporal regulatory properties necessary for its role, depicted schematically in the model shown in Figure 9. PP2A-B56 is not inhibited by CDK1 and is thus well-placed to initiate spindle checkpoint silencing and mitotic exit. Owing to its regulatory properties, notably direct inhibition by CDK1-cyclin B (Dohadwala et al., 1994), PP1 may be particularly important downstream of PP2A-B56 to promote APC/C activity and hence mitotic exit. PP2A-B55 then acts slightly later to trigger the events of late anaphase (Cundell et al., 2013, 2016). This provides a compelling reason why PP1 is required in addition to PP2A to counteract CDK1 activity and thus promote mitotic exit and the transition into anaphase.
MATERIALS AND METHODS
Chemicals and antibodies
General laboratory chemicals and reagents were obtained from Sigma-Aldrich and Thermo-Fisher Scientific. Drugs were dissolved in dimethyl sulfoxide (DMSO) unless specifically indicated. Inhibitors were obtained from Sigma-Aldrich (CDK1 inhibitor flavopiridol, 5 mM stock), Tocris Bioscience (MPS1-inhibitor AZ3146, 20 mM stock; PP1 and PP2A–inhibitor calyculin A, 1 mM stock; PP1-inhibitor tautomycetin, 2.5 mM stock), Insight Bioscience (proteasome inhibitor MG132, 20 mM stock), and Merck (microtubule polymerization inhibitor nocodazole, 0.66 mM stock). Thymidine (Sigma-Aldrich; 100 mM stock) and doxycycline (Invivogen; 2 mM stock) were dissolved in water.
Commercially available polyclonal (pAb) or monoclonal (mAb) antibodies were used for β-actin (horseradish peroxidase [HRP]-conjugated; mouse mAb, Abcam, [AC-15] ab49900), tubulin (mouse mAb; Sigma, [DM1A] T6199), PP1α (rabbit pAb, Bethyl, A300-904A), PP1α-pT320 (rabbit pAb, Abcam, Ab62334), PP1β (rabbit pAb, Bethyl, A300-905A), PP1γ (goat pAb, Santa Cruz, sc6108), B55α/PPP2R2A (mouse mAb, Cell Signalling, 5689S), securin (rabbit pAb, Abcam, Ab79546), CDC27/APC3 (mouse mAb clone C-4, Santa Cruz, sc13154), CDC20 (mouse mAb clone E-7, Santa Cruz, sc-13162; mouse mAB, clone AR12, Millipore UK, MAB3775; mouse mAb, clone BA8, Bio-Techne (R&D Systems), NB100-2646—these three antibodies were mixed for immunoprecipitations. CDC20 (rabbit pAb, Santa Cruz, sc-8358) was used for Western blotting.
Antibodies against CDC20-pT70 were raised in rabbits using phospho-peptide CSKVQT(pT)PSKPG and affinity-purified using an immobilized form of the same peptide (Moravian). Secondary donkey antibodies against mouse, rabbit, guinea pig, or sheep and labeled with Alexa Fluor 488, Alexa Fluor 555, Alexa Fluor 647, Cy5, or HRP were purchased from Molecular Probes and Jackson ImmunoResearch Laboratories, respectively. Affinity-purified primary and HRP-coupled secondary antibodies were used at 1 µg/ml final concentration. For Western blotting, proteins were separated by SDS–PAGE and transferred to nitrocellulose using a Trans-blot Turbo system (Bio-Rad). Protein concentrations were measured by Bradford assay using Protein Assay Dye Reagent Concentrate (Bio-Rad). All Western blots were revealed using enhanced chemiluminescence reagent (GE Healthcare).
Molecular biology and siRNA reagents
Human CDC20, PP1α, and PP1γ were amplified from human testis cDNA (Marathon cDNA; Takara Bio) using Pfu polymerase (Agilent Technologies). CDC20 expression constructs were made using pcDNA5/FRT/TO vectors (Invitrogen) modified to encode the EGFP or FLAG reading frames; PP1 constructs were made using a modified pcDNA5/FRT/TO vector encoding a C-terminal GFPtag. Mutagenesis to introduce phospho-site mutations and resistance to CDC20 small interfering RNA (siRNA) oligo #14 was performed using the QuikChange method (Agilent Technologies). DNA primers were obtained from Invitrogen. For the knockdown of the catalytic subunits of PP1α and PP1γ, siRNA duplexes 5′-UGGAUUGAUUGUACAGAAAUU-3′ and 5′-GCGGUGAAGUUGAGGCUUAUU-3′ targeting the 3′-UTR of PPP1CA and PPP1CC, respectively, were used in Figures 2, 3, and 7 and Supplemental Figure S4. Duplexes targeting the ORFs 5′-CAUCUAUGGUUUCUACGAU-3′ and 5′-GAACGACCGUGGCGUCUCU-3′ for PPP1CA or 5′-GCGGAGAGUUUGACAAUGC-3′ and 5′-UAGAUAAACUCAACAUCGA-3′ for PPP1CC were used in Figures 5, 6, and 8 and Supplemental Figures S2 and S3. PP1β was depleted using a 3′-UTR duplex 5′-GGGAAGAGCUUUACAGACAUU-3′ targeting PPP1CB. CDC20 was depleted using siRNA duplex #14 5′-CGGAAGACCUGCCGUUACA-3′ (ThermoFisher). PP2A-B55 and PP2A-B56 were targeted with siRNA duplexes that have been described previously (Hayward et al., 2019a,c).
Cell culture and CRISPR procedures
HeLa cells were cultured in DMEM with 1% (vol/vol) GlutaMAX (Life Technologies) containing 10% (vol/vol) bovine calf serum at 37°C and 5% CO2. For plasmid transfection and siRNA transfection, Mirus LT1 (Mirus Bio LLC) and Oligofectamine (Invitrogen), respectively, were used. CCNB1-mCherry was integrated into the endogenous CCNB1 locus of the parental HeLa Flp-In TRex cell line (Invitrogen). Doxycycline-inducible, single copies of the GFP-CDC20 or GFP-CDC206A transgenes were then integrated into HeLa Flp-In TRex CCNB1-mCherry cells. CRISPR/Cas9-edited HeLa cells with an inserted GFP tag in the C-terminus of the CCNB1 gene product and HeLa cells stably expressing GFP-MAD2 have been described before (Alfonso-Perez et al., 2019; Hayward et al., 2019a). Doxycycline-inducible PP1α-GFP and PP1γ-GFP were integrated into HeLa Flp-IN TRex cells (Invitrogen) (Tighe et al., 2004).
High-resolution mitotic exit time courses for Western blotting
HeLa cells were seeded at 1,200,000 cells/dish onto 15 × 15 cm dishes per condition and grown for 72 h. Nocodazole, a microtubule depolymerization agent, was added to 100 ng/ml (330 nM) for 20 h to arrest the cells in mitosis. For experiments with siRNA knockdown, cells were seeded at 600,000 cells/dish and grown for 24 h before transfection with the appropriate siRNA duplexes for 72 h. Nocodazole was added to the dishes for the final 20 h of depletion. The mitotic cells were harvested by shake off and washed in 2 × 25 ml 1× phosphate-buffered saline (PBS) and 1 × 25 ml Opti-MEM, both of which had been preequilibrated to 37°C, 5% CO2. Wash centrifugations were carried out for 5 min at 200 × gav, 37C. The cells were then resuspended in preequilibrated Opti-MEM to give 15,000,000 cells/ml. The cells were incubated for 25 min (37°C, 5% CO2) to allow them to rebuild bipolar mitotic spindles, with gentle mixing every ∼5 min. While the cells were incubating, 500 μl of flavopiridol buffer (480 µl preequilibrated Opti-MEM + 20 µl, 5 mM flavopiridol) was made and prewarmed to 37°C. Where more than 500 µl of flavopiridol was required, this was scaled accordingly. Flavopiridol buffer was added to the cells at a 1:10 dilution, giving a final concentration of 20 µM. The cells were immediately mixed through inversion and pipetting before being split in half. Owing to the frequency of early timepoints, one 2 ml aliquot was kept incubating at 37°C, 5% CO2, and gently mixed every 5 min to ensure stable conditions. The other half was placed in a 37°C water bath and sampled accordingly. At each time point, 50 µl of cells was added to 25 µl of 3× sample buffer and boiled for 5 min. Samples were diluted 1:2 with 1× sample buffer prior to Western blotting. Typically, 6 µg was loaded on each Western blot but this was increased to 10 µg as required. Where necessary, drugs were added at the start of the 25 min incubation, to ensure maximal inhibition prior to the addition of flavopiridol. For MPS1 inhibition time courses (and for the comparative CDK1 inhibition time course in Supplemental Figure S1), drug treatments and washes for nocodazole release were performed in complete media (DMEM with 1% [vol/vol] GlutaMAX [Life Technologies] containing 10% [vol/vol] bovine calf serum) at 37°C and 5% CO2. MPS1 inhibitor (MPS1-i; AZ3146) was prediluted in complete media and prewarmed to 37°C before being added to cell suspensions (1.5 × 107 cells/ml) to a final concentration of 2 µM. For sample collection, cells were treated with 50 nM calyculin A to prevent further dephosphorylation events during centrifugation and 1× PBS wash before being resuspended in lysis buffer supplemented with phosphatase and protease inhibitors for snap-freezing.
Analysis of CDC20 phosphorylation
For analysis of CDC20 phosphorylation 10% (wt/vol) polyacrylamide separating gels were prepared with 100 µM MnCl2 and 25 µM PhosTag reagent (Wako Chemicals; AAL-107S1). Double concentrations of ammonium persulfate and N,N,N’,N’-tetramethylethane-1,2-diamine were used to aid polymerization. Typically, 10 µg of lysate was loaded for PhosTag gels. Prior to transfer, gels were equilibrated with 3 × 3 min washes in transfer buffer (20 mM Tris, 150 mM glycine, 0.1% [wt/vol] SDS, 20% [vol/vol] MeOH, 20 mM EDTA) and subsequently 3 × 3 min washes in transfer buffer without EDTA (20 mM Tris, 150 mM glycine, 0.1% [wt/vol] SDS, 20% [vol/vol] MeOH).
Analysis with anti–CDC20-pT70 antibodies was carried out on CDC20 immunoprecipitates. For CDC20 immunoprecipitations, 1.8 × 106 cells per sample were lysed for 15 min on ice, with vortexing briefly every 5 min, in lysis buffer supplemented with phosphatase and protease inhibitors (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% [vol/vol] IGEPAL® CA-630, 0.5 M β-glycerol phosphate, 10 mM NaF, 100 nM okadaic acid, 100 nM calyculin A, 1 mM phenymethylsulfonyl fluoride, phosphatase inhibitor cocktail 1:100 [Sigma-Aldrich] or protease inhibitor cocktail 1:250 [Sigma-Aldrich]). Volumes of lysis buffer were calculated during sample collection to give lysate concentrations of ∼1 mg/ml. Cells were then centrifuged at 14,000 × gav for 15 min at 4°C to produce a cleared lysate, which was analyzed by Bradford assay. For the experiment in Figure 6C, CDC20 was isolated from 0.35 mg of lysate by 90 min incubation at 4°C with 20 µl Protein-G Dynabeads (ThermoFisher; 10004D) and 2 µg of each of three anti-CDC20 mAbs: clone E-7, clone BA8, clone AR-12. For the experiment in Figure 7E, CDC20 was isolated from 1 mg of lysate by 1 h incubation at 4°C with 40 µl Protein-G Dynabeads and 3 µg mAb clone E-7. Dynabeads were washed 3× with lysis buffer and 3× with wash buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% [vol/vol] IGEPAL® CA-630) and resuspended in 100 µl 2.5× Laemmli sample buffer. For immunoprecipitation of GFP-CDC20, 20 µl Protein-A Dynabeads was used with 2.5 µg anti-GFP (rb pAb; Abcam ab290) or anti-mCh (rb pAb; Abcam ab167453) for control.
Functional analysis of CDC20 and CDC20 6A
For CDC20 siRNA rescue experiments, HeLa Flp-In TRex cells expressing CCNB1-mCherry from the endogenous promoter and GFP-CDC20WT or GFP-CDC206A from the Flp-In site were used. CDC20 siRNA rescue was performed by induction with 2 µM doxycycline of GFP-CDC20 transgenes (WT and 6A) resistant to siRNA oligo #14 for 6 h prior to 48 h siRNA depletion of endogenous CDC20 using oligo #14. A second induction was performed 18 h later. For live cell imaging and sample collection for immunoprecipitation, cells were treated for 18 h with 2 mM thymidine 18 h after transfection with the siRNA duplexes. The thymidine was removed by three washes with DMEM, with 2 µM doxycycline re-added in the final wash. For immunoprecipitation samples, 330 nM nocodazole was added 8 h after thymidine release for 4 h, and cells were collected by mitotic shake-off. For live cell imaging SiR-DNA (Spirochrome) was added to the final wash of the thymidine release at a concentration of 100 nM, and imaging commenced 9 h later.
Time lapse imaging of MAD2 and CCNB1
For the live cell imaging of spindle checkpoint silencing and cyclin B destruction, ∼50,000 GFP-MAD2 CCNB1-mCherry HeLa cells were seeded into 35-mm dishes with 1.5-thickness cover glass bottom (Fluorodishes; Applied Precision). After 24 h, cells were then transfected with siRNA duplexes. After 35 h of transfection, 2 mM thymidine was added to the media. After a further 18 h, the thymidine was removed by three washes with 2 ml of DMEM and imaging was started 9 h later. SiR-DNA (Spirochrome) was added to the final wash at a concentration of 100 nM. For experiments using a PP1 inhibitor, 5 µM tautomycetin was added to cells 30 min before imaging started.
Time lapse imaging was performed using a spinning disk confocal system (Ultraview Vox; PerkinElmer) mounted on an inverted microscope (IX81; Olympus) equipped with an EM charge-coupled device (CCD) camera (C9100-13; Hamamatsu Photonics) and controlled by Volocity software. CDC20 rescue assays were imaged on a Deltavision Elite system using an inverted microscope (IX81; Olympus) and equipped with a QuantEM EMCCD camera (Photometrics). Cell were placed in a 37°C and 5% CO2 environmental chamber (Tokai Hit) on the microscope stage with lens heating collar. Imaging was performed using a 60× NA 1.4 oil immersion objective lens.
To monitor chromosome congression, checkpoint silencing, and cyclin B1 destruction, HeLa CCNB1-mCherry GFP-MAD2 cells were treated with 100 nM SiR-DNA to allow live visualization of DNA. The cells were then imaged using 8% 561 nm laser power with 100 ms exposure for CCNB1-mCherry, 6% 488 nm laser power with 80 ms exposure for GFP-MAD2, and 2% 647 nm laser power with 20 ms exposure for SiR-DNA. Nineteen axial planes were captured 0.6 µm apart (for each wavelength) at an interval of 2 min for 10 h. These images were then used to determine the point of last chromosome congression from the SiR-DNA and the time at which the last GFP-MAD2 foci disappeared. CCNB1-mCherry fluorescence intensity was measured over time using ImageJ. Images were Z-projected to sum the intensity across the volume of the whole cell. A region of interest (ROI) was drawn around each cell, and the integrated density was then measured in these regions over time and normalized by dividing by the area. Background levels were measured by collecting the integrated density of a 4-µm-diameter circular ROI adjacent to the cell and again normalized by dividing by the area of the ROI; this value was then subtracted from the normalized signal.
For live cell imaging with the addition of the MPS1 inhibitor AZ3146, HeLa cells expressing GFP-MAD2 and CCNB1-mCherry were treated as above but seeded into imaging dishes covered with lids containing a preformed hole to facilitate drug addition. Cells were imaged at intervals of 2 min, and cells in mitosis were visually scanned to identify cells approaching metaphase. After four captured time points, MPS1-i diluted in 200 µl DMEM was added to the cells to a final concentration of 2 µM. After drug addition, imaging was continued for 1–2 h.
Statistical analysis
Statistical analysis of live cell imaging data and intensity measurements was carried out in Excel and GraphPad Prism. Statistical tests in Figure 3 were unpaired t tests with Welch’s correction for unequal SD and Mann–Whitney tests in Figures 4, 5, and 8. Unless stated otherwise, the measurements for graphs are derived from a compilation of three independent experiments. For Western blot analysis, representative examples of two to three independent repeats are shown.
Supplementary Material
Acknowledgments
J.B. and Z.G. were supported by a Medical Research Council Senior Non-Clinical Research fellowship awarded to U.G. (MR/K006703/1). T.A.-P. was supported by a Biotechnology and Biological Sciences Research Council Strategic LoLa grant (BB/M00354X/1). J.H. was supported by a Wellcome Trust PhD award. F.A.B. was funded by a Cancer Research UK program grant award (C20079/A15940). We acknowledge Catherine Wormald and Flavia Scialpi for initial observations, Fengying Liu for help with the generation of CDC20 Flp-In TRex cell lines, and Renaud Caous and Emile Roberts for help with live cell imaging analysis. We thank Elena Poser and Dan Hayward for critical reading of the manuscript.
Abbreviations used:
- APC/C
anaphase promoting complex/cyclosome core complex
- APC/C CDC20
APC/C with the CDC20 coactivator
- APC/C CDH1
APC/C with the CDH1 coactivator
- GFP
green fluorescent protein
- MCC
mitotic checkpoint complex
- NEBD
nuclear envelope breakdown
- PP1
protein phosphatase 1
- PP2A-B55
protein phosphatase 2A with the B55 regulatory subunit
- PP2A-B56
protein phosphatase 2A with the B56 regulatory subunit.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-04-0252) on August 5, 2020.
REFERENCES
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe JC (2001). Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell , 83–93. [DOI] [PubMed] [Google Scholar]
- Alfieri C, Zhang S, Barford D (2017). Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol , 170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso-Perez T, Hayward D, Holder J, Gruneberg U, Barr FA (2019). MAD1-dependent recruitment of CDK1-CCNB1 to kinetochores promotes spindle checkpoint signaling. J Cell Biol , 1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MS, Swingle M, D’Arcy B, Abney K, Rusin SF, Kettenbach AN, Page R, Honkanen RE, Peti W (2017). PP1:tautomycetin complex reveals a path toward the development of PP1-specific inhibitors. J Am Chem Soc , 17703–17706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P, Pines J (1999). Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol , 82–87. [DOI] [PubMed] [Google Scholar]
- Cohen PT (2002). Protein phosphatase 1 — targeted in many directions. J Cell Sci , 241–256. [DOI] [PubMed] [Google Scholar]
- Conti D, Gul P, Islam A, Martin-Duran JM, Pickersgill RW, Draviam VM (2019). Kinetochores attached to microtubule-ends are stabilised by Astrin bound PP1 to ensure proper chromosome segregation. eLife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundell MJ, Bastos RN, Zhang T, Holder J, Gruneberg U, Novak B, Barr FA (2013). The BEG (PP2A-B55/ENSA/Greatwall) pathway ensures cytokinesis follows chromosome separation. Mol Cell , 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundell MJ, Hutter LH, Nunes Bastos R, Poser E, Holder J, Mohammed S, Novak B, Barr FA (2016). A PP2A-B55 recognition signal controls substrate dephosphorylation kinetics during mitotic exit. J Cell Biol , 539–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V, Mari C, Nocera D, Rametti L, Grieco D (2003). The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev , 2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wever V, Nasa I, Chamousset D, Lloyd D, Nimick M, Xu H, Trinkle-Mulcahy L, Moorhead GB (2014). The human mitotic kinesin KIF18A binds protein phosphatase 1 (PP1) through a highly conserved docking motif. Biochem Biophys Res Commun , 432–437. [DOI] [PubMed] [Google Scholar]
- Di Fiore B, Davey NE, Hagting A, Izawa D, Mansfeld J, Gibson TJ, Pines J (2015). The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev Cell , 358–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Pines J (2010). How cyclin A destruction escapes the spindle assembly checkpoint. J Cell Biol , 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohadwala M, da Cruz e Silva EF, Hall FL, Williams RT, Carbonaro-Hall DA, Nairn AC, Greengard P, Berndt N (1994). Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc Natl Acad Sci USA , 6408–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D (1997). Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J , 1876–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert A, Uluocak P, Bastos RN, Mangat D, Graab P, Gruneberg U (2014). PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J Cell Biol , 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Thanasoula M, Maffini S, Breit C, Muller F, van Gerwen S, Bange T, Musacchio A (2017). Basis of catalytic assembly of the mitotic checkpoint complex. Nature , 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L, Chan CS (1994). Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell Mol Biol Res , 207–213. [PubMed] [Google Scholar]
- Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S (2001). Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J , 6648–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K, Grimaldi M, Yamano H (2016). Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science , 1121–1124. [DOI] [PubMed] [Google Scholar]
- Fujimitsu K, Yamano H (2020). PP2A-B56 binds to Apc1 and promotes Cdc20 association with the APC/C ubiquitin ligase in mitosis. EMBO Rep , e48503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey M, Touati SA, Kataria M, Jones A, Snijders AP, Uhlmann F (2017). PP2A(Cdc55) phosphatase imposes ordered cell-cycle phosphorylation by opposing threonine phosphorylation. Mol Cell , 393–402.e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J, Huang HB, Kwon YG, Greengard P, Nairn AC, Kuriyan J (1995). Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature , 745–753. [DOI] [PubMed] [Google Scholar]
- Hafner J, Mayr MI, Mockel MM, Mayer TU (2014). Pre-anaphase chromosome oscillations are regulated by the antagonistic activities of Cdk1 and PP1 on Kif18A. Nat Commun , 4397. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW (2000). MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol , 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D, Alfonso-Perez T, Cundell MJ, Hopkins M, Holder J, Bancroft J, Hutter LH, Novak B, Barr FA, Gruneberg U (2019a). CDK1-CCNB1 creates a spindle checkpoint-permissive state by enabling MPS1 kinetochore localization. J Cell Biol , 1182–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D, Alfonso-Perez T, Gruneberg U (2019b). Orchestration of the spindle assembly checkpoint by CDK1-cyclin B1. FEBS Lett 593, 2889–2907 [DOI] [PubMed] [Google Scholar]
- Hayward D, Bancroft J, Mangat D, Alfonso-Perez T, Dugdale S, McCarthy J, Barr FA, Gruneberg U (2019c). Checkpoint signaling and error correction require regulation of the MPS1 T-loop by PP2A-B56. J Cell Biol , 3188–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A, Konietzny A, Mayer TU (2015). Protein phosphatase 1 is essential for Greatwall inactivation at mitotic exit. EMBO Rep , 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein JB, Hertz EPT, Garvanska DH, Kruse T, Nilsson J (2017). Distinct kinetics of serine and threonine dephosphorylation are essential for mitosis. Nat Cell Biol , 1433–1440. [DOI] [PubMed] [Google Scholar]
- Hein JB, Nilsson J (2016). Interphase APC/C-Cdc20 inhibition by cyclin A2-Cdk2 ensures efficient mitotic entry. Nat Commun , 10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder J, Poser E, Barr FA (2019). Getting out of mitosis: spatial and temporal control of mitotic exit and cytokinesis by PP1 and PP2A. FEBS Lett , 2908–2924. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Reimann JD, Sorensen CS, Lukas J, Jackson PK (2002). E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nat Cell Biol , 358–366. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K (1995). Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell , 269–278. [DOI] [PubMed] [Google Scholar]
- Ji Z, Gao H, Jia L, Li B, Yu H (2017). A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. eLife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Lara-Gonzalez P, Prevo B, Meitinger F, Cheerambathur DK, Oegema K, Desai A (2017). Kinetochores accelerate or delay APC/C activation by directing Cdc20 to opposing fates. Genes Dev , 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW (2010). Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell , 444–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW (1995). A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell , 279–288. [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics , 749–757. [DOI] [PubMed] [Google Scholar]
- Kiss A, Erdodi F, Lontay B (2019). Myosin phosphatase: unexpected functions of a long-known enzyme. Biochim Biophys Acta Mol Cell Res , 2–15. [DOI] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM (2003). Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J , 6598–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM (2000). Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell , 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Zhang G, Larsen MS, Lischetti T, Streicher W, Kragh Nielsen T, Bjorn SP, Nilsson J (2013). Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J Cell Sci , 1086–1092. [DOI] [PubMed] [Google Scholar]
- Kwon YG, Lee SY, Choi Y, Greengard P, Nairn AC (1997). Cell cycle-dependent phosphorylation of mammalian protein phosphatase 1 by cdc2 kinase. Proc Natl Acad Sci USA , 2168–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H, Fujimitsu K, Bayin NS, Takaki T, Gannon J, Yamano H (2012). Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J , 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS (2012). The spindle assembly checkpoint. Curr Biol , R966–R980. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Rodriguez-Bravo V, Kim H, Datta S, Foley EA (2017). The PP2A(B56) phosphatase promotes the association of Cdc20 with APC/C in mitosis. J Cell Sci , 1760–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischetti T, Zhang G, Sedgwick GG, Bolanos-Garcia VM, Nilsson J (2014). The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nat Commun , 5563. [DOI] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA (2010). Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol , 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Ceto S, Ranish JA, Biggins S (2012). Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol , 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Vigneron S, Robert P, Strub JM, Cianferani S, Castro A, Lorca T (2016). Greatwall dephosphorylation and inactivation upon mitotic exit is triggered by PP1. J Cell Sci , 1329–1339. [DOI] [PubMed] [Google Scholar]
- Matsumura F, Yamakita Y, Yamashiro S (2011). Myosin light chain kinases and phosphatase in mitosis and cytokinesis. Arch Biochem Biophys , 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy RA, Parker BL, Rogers S, Chaudhuri R, Gayevskiy V, Hoffman NJ, Ali N, Watkins DN, Daly RJ, James DE, et al. (2015). Global phosphoproteomic mapping of early mitotic exit in human cells identifies novel substrate dephosphorylation motifs. Mol Cell Proteomics , 2194–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JJ, Summers MK, Hansen DV, Nachury MV, Lehman NL, Loktev A, Jackson PK (2006). Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev , 2410–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S, Hunt T (2012). Protein phosphatases and their regulation in the control of mitosis. EMBO Rep , 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin V, Prieto S, Melines S, Hem S, Rossignol M, Lorca T, Espeut J, Morin N, Abrieu A (2012). CDK-dependent potentiation of MPS1 kinase activity is essential to the mitotic checkpoint. Curr Biol , 289–295. [DOI] [PubMed] [Google Scholar]
- Murray A (1995). Cyclin ubiquitination: the destructive end of mitosis. Cell , 149–152. [DOI] [PubMed] [Google Scholar]
- Musacchio A (2015). The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol , R1002–R1018. [DOI] [PubMed] [Google Scholar]
- Nasmyth K (2001). Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet , 673–745. [DOI] [PubMed] [Google Scholar]
- Nigg EA (2001). Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol , 21–32. [DOI] [PubMed] [Google Scholar]
- Nijenhuis W, Vallardi G, Teixeira A, Kops GJ, Saurin AT (2014). Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat Cell Biol , 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J (2019). Protein phosphatases in the regulation of mitosis. J Cell Biol , 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overlack K, Primorac I, Vleugel M, Krenn V, Maffini S, Hoffmann I, Kops GJ, Musacchio A (2015). A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. eLife , e05269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, Ciliberto A, Musacchio A (2013). Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife , e01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Garcia-Gimeno MA, Beullens M, Manzione MG, Van der Hoeven G, Igual JC, Heredia M, Sanz P, Gelens L, Bollen M (2017). An attachment-independent biochemical timer of the spindle assembly checkpoint. Mol Cell , 715–730.e715. [DOI] [PubMed] [Google Scholar]
- Qian J, Lesage B, Beullens M, Van Eynde A, Bollen M (2011). PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr Biol , 766–773. [DOI] [PubMed] [Google Scholar]
- Qiao R, Weissmann F, Yamaguchi M, Brown NG, VanderLinden R, Imre R, Jarvis MA, Brunner MR, Davidson IF, Litos G, et al. (2016). Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc Natl Acad Sci USA , E2570–E2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Fisher LA, Zhao J, Wang L, Williams BC, Goldberg ML, Peng A (2017). Cell cycle-dependent regulation of Greatwall kinase by protein phosphatase 1 and regulatory subunit 3B. J Biol Chem , 10026–10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Fey D, McCloy RA, Parker BL, Mitchell NJ, Payne RJ, Daly RJ, James DE, Caldon CE, Watkins DN, et al. (2016). PP1 initiates the dephosphorylation of MASTL, triggering mitotic exit and bistability in human cells. J Cell Sci , 1340–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, et al. (2010). Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol , 886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JB (2012). Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol , 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Gorbsky GJ (2015). Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol , 82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Janczyk PL, Qu Q, Brautigam CA, Stukenberg PT, Yu H, Gorbsky GJ (2016). The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. eLife [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Cordeiro MH, Davey NE, Vallardi G, Ciliberto A, Gross F, Saurin AT (2019). PP1 and PP2A use opposite phospho-dependencies to control distinct processes at the kinetochore. Cell Rep , 2206–2219.e2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke VM, Sillje HH, Arnaud L, Nigg EA (2002). Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J , 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ (2001). Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol , 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ (2012). Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell , 745–755. [DOI] [PubMed] [Google Scholar]
- Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R (2004). Structural basis of protein phosphatase 1 regulation. Nature , 780–784. [DOI] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Taylor SS (2004). Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci , 6339–6353. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andersen J, Lam YW, Moorhead G, Mann M, Lamond AI (2006). Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J Cell Biol , 679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F (2001). Secured cutting: controlling separase at the metaphase to anaphase transition. EMBO Rep , 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zon W, Wolthuis RM (2010). Cyclin A and Nek2A: APC/C-Cdc20 substrates invisible to the mitotic spindle checkpoint. Biochem Soc Trans , 72–77. [DOI] [PubMed] [Google Scholar]
- Vazquez-Novelle MD, Sansregret L, Dick AE, Smith CA, McAinsh AD, Gerlich DW, Petronczki M (2014). Cdk1 inactivation terminates mitotic checkpoint surveillance and stabilizes kinetochore attachments in anaphase. Curr Biol , 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleugel M, Hoek TA, Tromer E, Sliedrecht T, Groenewold V, Omerzu M, Kops GJ (2015). Dissecting the roles of human BUB1 in the spindle assembly checkpoint. J Cell Sci , 2975–2982. [DOI] [PubMed] [Google Scholar]
- Wang W, Stukenberg PT, Brautigan DL (2008). Phosphatase inhibitor-2 balances protein phosphatase 1 and aurora B kinase for chromosome segregation and cytokinesis in human retinal epithelial cells. Mol Biol Cell , 4852–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ER, Brown NG, Peters JM, Stark H, Schulman BA (2019). Posing the APC/C E3 ubiquitin ligase to orchestrate cell division. Trends Cell Biol , 117–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J (2008). Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell , 290–302. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Guo JY, Tang W, Yang CS, Freel CD, Chen C, Nairn AC, Kornbluth S (2009). PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat Cell Biol , 644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Raetz EA, Kitagawa M, Virshup DM, Lee SH (2013). BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biol Open , 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Yang CH, Tanno Y, Watanabe Y (2012). MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol , 746–752. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Yamakita Y, Totsukawa G, Goto H, Kaibuchi K, Ito M, Hartshorne DJ, Matsumura F (2008). Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Dev Cell , 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky Y, Shteinberg M, Listovsky T, Brandeis M, Hershko A (2000). Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem Biophys Res Commun , 299–304. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W (1998). Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science , 1721–1724. [DOI] [PubMed] [Google Scholar]
- Zhang G, Kruse T, Lopez-Mendez B, Sylvestersen KB, Garvanska DH, Schopper S, Nielsen ML, Nilsson J (2017). Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling. Nat Commun , 15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chang L, Alfieri C, Zhang Z, Yang J, Maslen S, Skehel M, Barford D (2016). Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature , 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.