Abstract
Classical osteogenesis imperfecta (OI) is an inherited rare brittle bone disease caused by dominant mutations in the COL1A1 or COL1A2 genes, encoding for the α chains of collagen type I. The definitive cure for the disease will require a gene therapy approach, aimed to correct or suppress the mutant allele. Interestingly, individuals lacking α2(I) chain and synthetizing collagen α1(I)3 homotrimers do not show bone phenotype, making appealing a bone specific COL1A2 silencing approach for OI therapy. To this aim, three different Col1a2-silencing RNAs (siRNAs), −3554, −3825 and −4125, selected at the 3′-end of the murine Col1a2 transcript were tested in vitro and in vivo. In murine embryonic fibroblasts Col1a2-siRNA-3554 was able to efficiently and specifically target the Col1a2 mRNA and to strongly reduce α2(I) chain expression. Its efficiency and specificity were also demonstrated in primary murine osteoblasts, whose mineralization was preserved. The efficiency of Col1a2-siRNA-3554 was proved also in vivo. Biphasic calcium phosphate implants loaded with murine mesenchymal stem cells were intramuscularly transplanted in nude mice and injected with Col1a2-siRNA-3554 three times a week for three weeks. Collagen α2 silencing was demonstrated both at mRNA and protein level and Masson's Trichrome staining confirmed the presence of newly formed collagen matrix. Our data pave the way for further investigation of Col1a2 silencing and siRNA delivery to the bone tissue as a possible strategy for OI therapy.
Abbreviations: BCP, biphasic calcium phosphate; D-MEM, Dulbecco-modified Eagle's medium; EDS, Ehlers Danlos syndrome; EGFP, enhanced green fluorescent protein; FBS, fetal bovine serum; MEF, murine embryonic fibroblast; MSC, mesenchymal stem cell; NMD, nonsense mediated RNA decay; OI, osteogenesis imperfecta; PBS, phosphate buffered saline; RNAi, RNA interference; SDS, sodium dodecyl sulphate; shRNA, short hairpin RNA; siRNA, small interfering RNA; TRAP, tartrate-resistant acid phosphatase
Keywords: Collagen, Osteogenesis imperfecta, siRNA, Silencing, Gene therapy
Highlights
-
•
Identification of a specific and efficient Col1a2 siRNA
-
•
Silencing of Col1a2 allows osteoblasts mineralization.
-
•
Col1a2 silencing is not impairing matrix deposition in vivo.
Introduction
Dominant mutations in the COL1A1 or COL1A2 genes, encoding for the two α chains of collagen type I, are responsible for the classical form of the rare bone disease osteogenesis imperfecta (OI). Individuals affected by OI are characterized by bone deformity, frequent fractures and growth delay [1,2]. No definitive treatment is so far available and the therapeutic options, based either on surgical interventions or drug administration, are for the most focused on ameliorating patient's quality of life [[3], [4], [5]]. Only a gene therapy approach, aimed to correct or suppress the expression of the mutant allele, could be effective to cure patients.
Interestingly, individuals heterozygous for null COL1A1 allele have a mild form of the disease [6] and null COL1A2 allele in heterozygosis where never described, suggesting an undetectable bone phenotype [1]. This observation points to allele specific silencing approach as an appealing strategy for the disease.
Indeed, for defects in COL1A1 or COL1A2, the specific silencing of the mutant allele could transform a severe phenotype into a mild osteoporotic form or even cure the disease. In classical dominant OI, over 85% of the mutations are single nucleotide substitutions changing one of the glycine, present every three residues in the helical domain and necessary for proper chains folding, with a bulkier amino acid [7]. RNA interference (RNAi) techniques based on small interfering RNA (siRNA) and short hairpin RNA (shRNA) were proved to be effective in allele specific down regulation of gene expression in several monogenic disorders [[8], [9], [10], [11], [12], [13], [14], [15]]. RNA therapeutics are easy to design, cost effective, generally stable and easy to combine with carriers [16]. The first therapy based on RNA interference gene silencing, termed Onpattro®, has been approved by the US Food and Drug Administration in 2018 [17]. This lipid complex suspension, for intravenous use, contains a transthyretin-directed siRNA and is indicated for the treatment of the polyneuropathy of hereditary transthyretin-mediated amyloidosis in adults [18].
The siRNAs are short chemical synthesized double strand RNAs, they can be delivered into the cells where they can be stabilized for weeks upon association to the RISC complex, although they are diluted during cell division. The shRNAs are short double strand RNA molecules with a hairpin structure transcribed upon their subcloning in a plasmid or viral vector used to transfect the host cells. shRNAs have the advantage to be continuously synthesized by the cell machinery upon integration in the host genome, but the templates are subjected to random DNA integration.
A single nucleotide difference in the highly repetitive and GC rich collagen sequence represents the first challenge in allele specific targeting of the collagen type I genes. Nevertheless, siRNA/shRNA were successfully used in OI. The transfection of allele specific siRNA specifically suppressing the mutant Col1a1 allele in primary fibroblasts from the OI murine model Brtl/+, carrying in heterozygosis the α1(I)-G349C substitution, led to about 60% down-regulation of gene expression, affecting only 20% the expression of the wild type allele. Mutant specific Col1a1-shRNA suppressed about 50% the gene expression, reducing to 60% the level of the mutated protein without compromising cells proliferation [19]. A limiting factor of these strategies is the requirement of specific silencing molecules for the hundreds of different mutations known to cause dominant OI (www.le.ac.uk/genetics/collagen/). To overcome this limitation, a mutation-independent approach in human bone derived cells using siRNA/shRNA targeting heterozygous single nucleotide polymorphisms in COL1A2 [20] or insertion/ deletion polymorphisms in both the COL1A1 and COL1A2 genes [21] were successfully attempted in vitro.
Interestingly, individuals carrying in homozygosis null COL1A2 allele are affected by a form of Ehlers Danlos syndrome (EDS) associated to vascular impairment, but they do not manifest bone phenotype [22]. Such observation suggests that a skeletal specific silencing of both mutant and normal COL1A2 allele could indeed ameliorate the OI skeletal outcome in these individuals.
In this report, we evaluate the effect of Col1a2 specific transcript silencing and α2 collagen protein suppression on osteoblast mineralization in vitro and on matrix formation in vivo in mice.
Results
Col1a2-siRNA efficiently and specifically reduces gene expression in vitro in murine embryonic fibroblasts
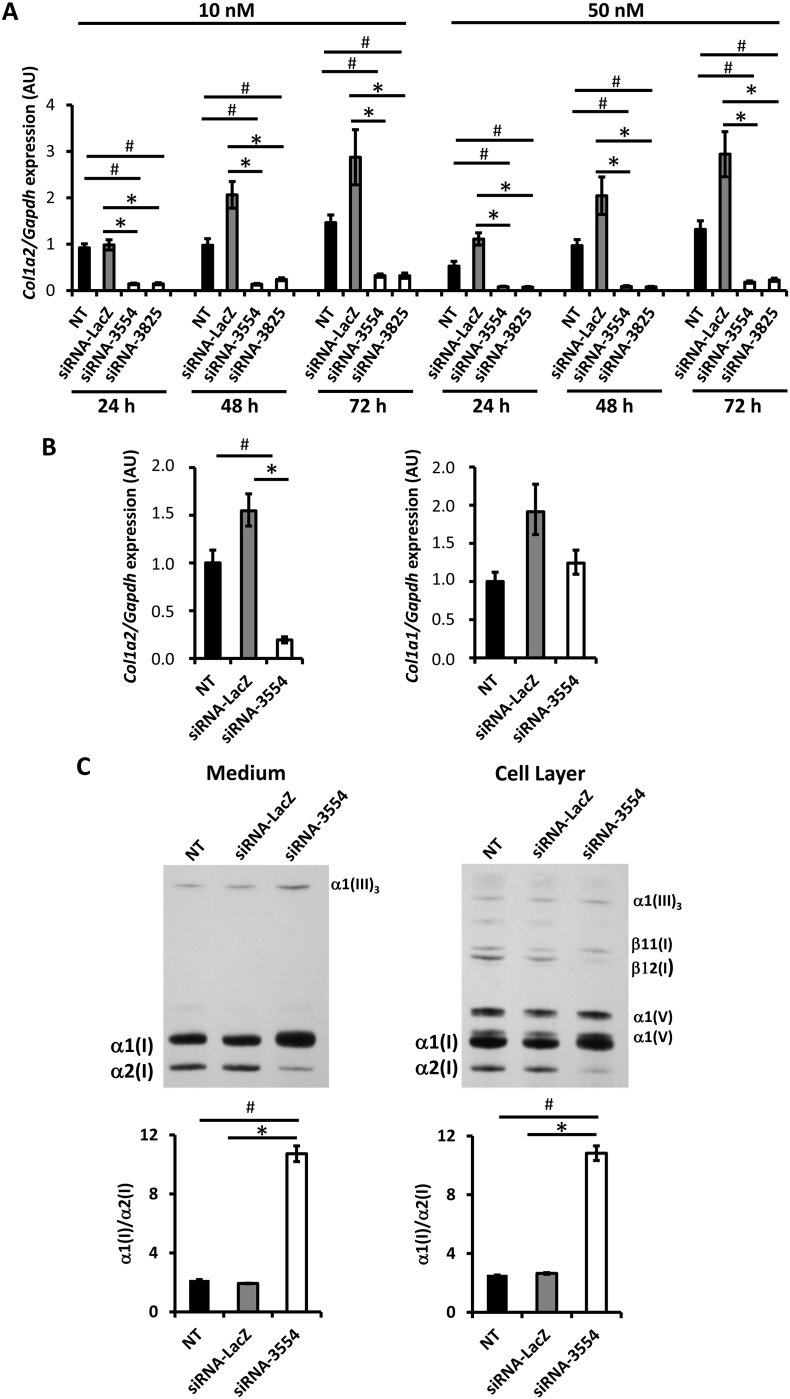
Three siRNAs targeting the murine Col1a2 gene were selected in silico at the 3′-end of the collagen I gene, encoding for the C-terminal propeptide domain and characterized by limited repetitive and GC rich sequence. The siRNAs target positions in the Col1a2 gene (NM_007743.3) were selected at the nucleotide 3554–3572 for siRNA-3554, 3825-3845 for siRNA-3825 and 4215-4233 for siRNA-4215. Their efficiency at two different concentrations, 10 nM and 50 nM, was first evaluated 48 h post transfection (hpt) in primary murine embryonic fibroblasts (MEFs). qPCR showed that siRNA-3554 silenced about 90% of the Col1a2 expression at both concentrations while siRNA-3825 63% and 81% at 10 and 50 nM, respectively (Supplementary Fig. 1A). On the contrary, siRNA-4215 did not significantly change Col1a2 expression, at least at the lower concentration, and its use was discontinued. The siRNA-LacZ, used as control, did not affect Col1a2 expression, as expected (Supplementary Fig. 1A).
To determine the temporal stability of the 2 most effective siRNAs, the Col1a2 gene expression was evaluated 24, 48 and 72 hpt. To this purpose two different cell densities, 1 × 104 and 2 × 104 cells/well, to mimic sub-confluent and confluent status, and two different siRNAs concentrations, 10 and 50 nM, were used. At the lowest cell density and at both siRNA concentrations, the Col1a2 expression was reduced about 85% at all time points (Fig. 1A).
Fig. 1.
Efficient and specific Col1a2-siRNAs suppress collagen α2 chain protein expression in primary murine embryonic fibroblasts.
(A) The stability of Col1a2 silencing of the 2 most effective siRNAs (siRNA-3554 and – 3825) at 10 or 50 nM was evaluated 24, 48 and 72 h post transfection in MEFs plated 1 × 104 cells/well. qPCR analyses showed Col1a2 relative expression. (B) The Col1a2 specificity was evaluated for the more promising molecule, siRNA-3554, analyzing the Col1a1 and Col1a2 expression by qPCR. (C) Representative images of SDS-Urea-PAGE analyses of 3H-proline labelled type I collagen extracted from medium and cell layer of transfected cells. The densitometric analyses of collagen type I α1 and α2 was performed and the α1/α2 ratio evaluated. In the collagen fraction extracted from cell layer a contribution of collagen type V in the α1(I) quantitation cannot be excluded. Not transfected cells (NT) and cells transfected with siRNA-LacZ were used as negative controls. A biological duplicate of the experiment was performed and each experiment was performed in triplicate. * p value <.05 vs siRNA-LacZ, # p value <.05 vs NT.
At the highest cell density only the 50 nM siRNAs concentration showed good efficiency for both siRNA molecules (Supplementary Fig. 1B).
Unfortunately, at the higher siRNA concentration the cell health was compromised and a large number of cells detached from the well, thus all the following experiments were performed using 10 nM and the 1 × 104 cell density was chosen. The siRNA-3554 was selected for its higher efficacy at the selected concentration.
Given the high similarity of the Col1a1 and Col1a2 genes, the siRNA-3554 specificity for Col1a2 was tested by qPCR. At 48 hpt siRNA-3554 suppressed Col1a2 without affecting the Col1a1 expression (Fig. 1B).
siRNA-3554 reduces α2(I) expression
To assess whether the silencing of the Col1a2 gene by siRNA-3554 leads to the α2 chain protein down-regulation, 48 hpt the 3H-proline labelled collagen was extracted from medium and cell layer at steady state. SDS-PAGE analysis demonstrated a strong reduction of the α2(I) chain in cells transfected with the siRNA-3554 compared to the cells transfected with siRNA-LacZ, used as control. The densitometric ratio of the α1 and α2 bands intensity was clearly higher in siRNA-3554 treated cells, with respect to the 2:1 expected value based on the collagen stoichiometry (2 α1 and 1 α2 chains). This result supported the successful α2(I) chain down regulation (Fig. 1C).
siRNA-3554 is efficient and specific in osteoblast cells
OI is mainly a bone disease and osteoblasts are the main affected cells, thus the efficiency and specificity of the siRNA-3554 were tested in primary murine osteoblasts. Cells were transfected in 24 well plates at cell density of 2 × 104 cells/well using 10 nM siRNA. The higher cell density was chosen based on the lower growth rate of osteoblasts compared to MEFs [23].
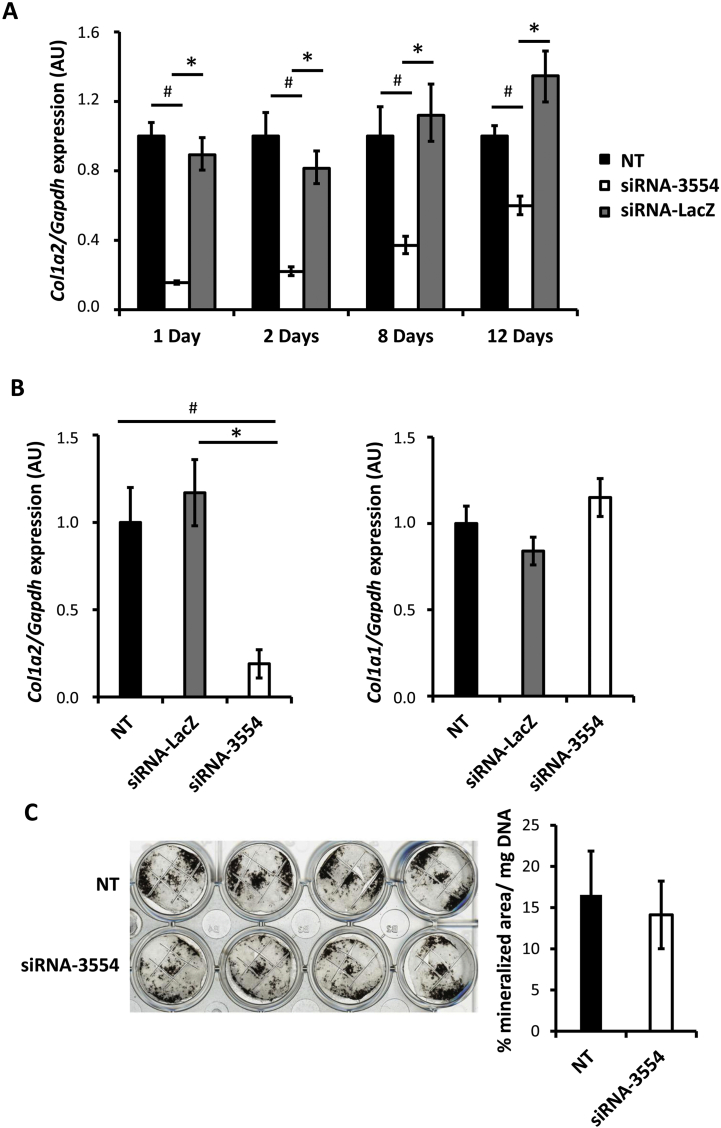
A silencing time course experiment with siRNA-3554 was performed. A down-regulation of Col1a2 expression of 82%, 73%, 67% and 56% with respect siRNA-LacZ transfection and 84%, 78%, 63% and 40% with respect siRNA-3554 transfection was obtained at 1, 2, 8, 12 days post transfection (dpt), respectively (Fig. 2A). At 2 dpt cells transfected with the siRNA-3554 showed a specific ~ 80% silencing of the Col1a2 expression, with no change in the Col1a1 expression (Fig. 2B). No effect on the expression of Col1a1 was detected at all time points in the time course experiment confirming the specificity of siRNA-3554 for Col1a2 also in primary osteoblasts (Supplementary Fig. 2A).
Fig. 2.
Efficient and specific Col1a2 silencing in primary murine osteoblasts does not impair mineralization.
(A) The stability of Col1a2 silencing was evaluated in murine osteoblasts untrasfected (NT) and upon 10 nM siRNA-3554 or siRNA-LacZ transfection at 1, 2, 8 and 12 days post transfection by qPCR quantitation of Col1a2 expression. (B) Similarly, the Col1a2 silencing specificity was evaluated by qPCR quantitation of Col1a2 and Col1a1 expression in murine osteoblasts NT and upon 10 nM siRNA-3554 or siRNA-LacZ transfection. (C) NT and transfected osteoblasts were stained by Von Kossa following 19 days growth in mineralization media. Mineral was quantified and normalized to DNA amount. A biological duplicate of the experiment was performed and each experiment was run at least in triplicate. * p value <.05 vs siRNA-LacZ, # p value <.05 vs NT.
Osteoblast mineralization is conserved following siRNA-3554 transfection
Mineralization is a fundamental step in bone formation [24], thus the mineralization of primary osteoblasts transfected with siRNA-3554 was evaluated. Osteoblasts mineralization was induced 24 hpt and after 19 days Von Kossa staining was performed to visualize the mineral nodules (Fig. 2C). Interestingly, transfected and control cells produced the same amount of mineralized area normalized to DNA content, indicating that the silencing of the Col1a2 is not affecting the process (Fig. 2C).
In vivo siRNA-3554 efficiency and specificity
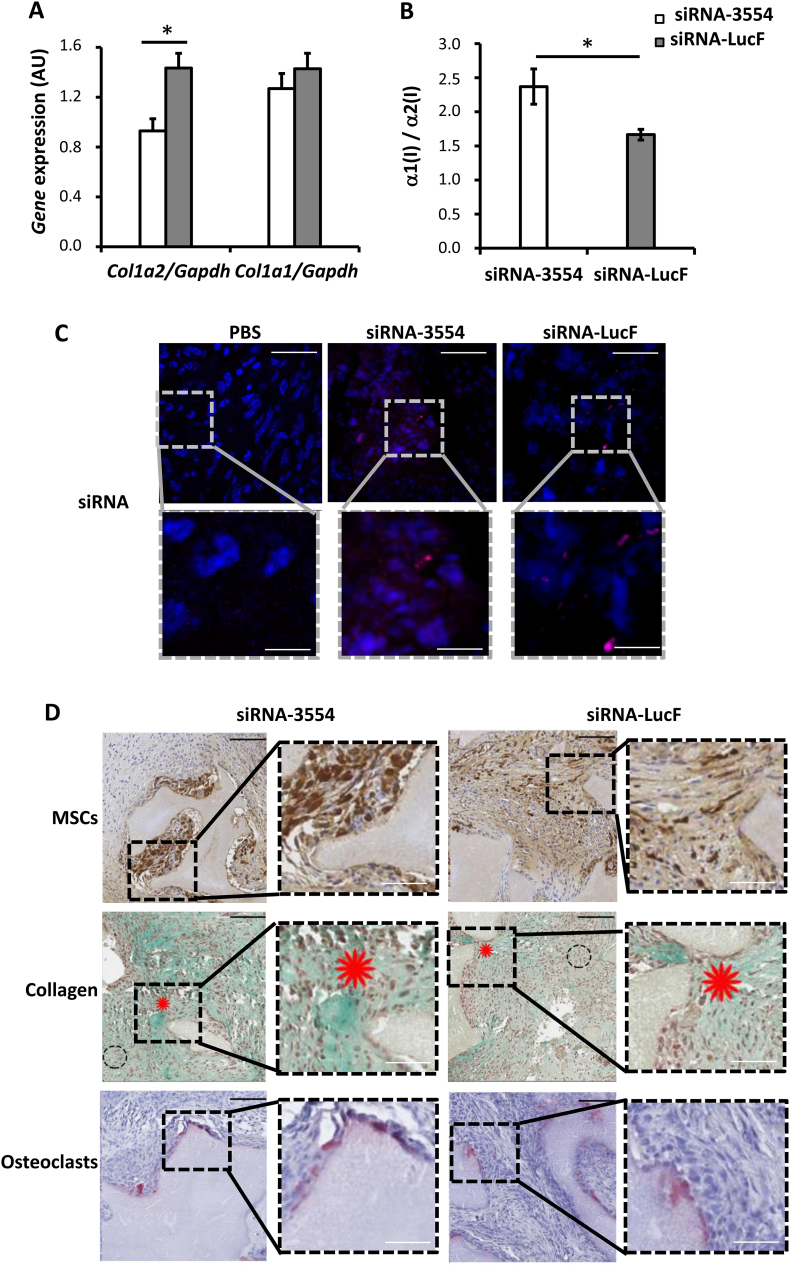
To have a proof-of-principle of the in vivo efficiency of siRNA-3554, biphasic calcium phosphate (BCP) implants loaded with mouse mesenchymal stem cells (MSCs) expressing the enhanced green fluorescent protein (EGFP) were prepared and transplanted intramuscularly in Rj:NMRI-nude mice. These implants were chosen based on their well known ability to favor bone formation [25,26]. Following implantation, siRNA-3554 and siRNA-LucF were injected at the implant site three times a week for three weeks, PBS injection was performed as control. The three last injections were performed with fluorescent siRNAs (ATTO 655-labelled siRNA). At the sacrifice the implants appeared as a compact mass of cells and particles (Supplementary Fig. 3A) and contained both EGFP positive cells and siRNAs in all siRNA injected samples (Fig. 3C,D).
Fig. 3.
Specific Col1a2 silencing in mouse mesenchymal stem cells injected in intramuscular biphasic calcium phosphate implants in nude mice allows collagen deposition.
(A)Col1a2 and Col1a1 qPCR analysis was performed on RNA obtained from biphasic calcium phosphate (BCP) implants extracted from nude mice 28 days after transplantation and following siRNA-3554 (n = 4) or siRNA-LucF (n = 4) injections for three weeks. (B) The α1/α2 collagen chains ratio was evaluated by quantification of collagen bands in SDS-Urea-PAGE of pepsin digested collagen from BCP implants extracted 28 days after transplantation following siRNA-3554 or siRNA-LucF injections. (C) siRNA presence was evaluated in intramuscular BCP implants injected with PBS, siRNA-3554 or siRNA-LucF extracted 28 days after transplantation. Fluorescent signal (pink) revealed siRNA-3554-ATTO 655 and siRNA-LucF-ATTO 655 that were used for the last three siRNA injections. (D) Histological analysis of intramuscular BCP implants injected with siRNA-3554 or siRNA-LucF extracted 28 days after transplantation. EGFP-MSCs are stained in brown following immunohistochemistry using anti-EGFP antibody. Masson's Trichrome staining revealed collagen in green, as indicated by red asterisks. Picnotic nuclei are indicated by dasher circles. TRAP staining revealed the presence of osteoclasts (pink) in the implants. * p value <.05. High magnification images are provided for all the panels in the inserts. Black scale bar = 100 μm, white scale bar = 30 μm.
Col1a2 expression was significantly silenced (48%) in the implants injected with the siRNA-3554, and the silencing was specific since no effect on Col1a1 was detectable (Fig. 3A).
The analyses of collagen type I extracted from the implants revealed an increased α1 / α2 chains ratio following injection with siRNA-3554, supporting α2 suppression (Fig. 3B, Supplementary Fig. 3B).
Bone formation upon siRNAs injection was evaluated by Masson's Trichrome histological staining. The presence of newly formed collagen matrix (red asterisks) was proved in all samples, demonstrating that the silencing of Col1a2 did not prevent collagen matrix formation in vivo (Fig. 3D). Of note, the area of the new collagen matrix was comparable in all samples (100 ± 17.14% in PBS-treated; 106.4 ± 24.5% in siRNA-3554; 117.5 ± 61% siRNA-LucF) (Fig. 3D, Supplementary Fig. 3C). Areas of picnotic nuclei were detectable (dashed circles) in siRNAs injected samples, suggesting a necrotic effect of lipoplex injections. Importantly, also osteoclasts were detected by TRAP staining, indicating that remodeling occurs upon siRNAs injection. Furthermore, the siRNAs did not affect the number of macrophages, markers of inflammation, as shown by immunohistochemistry using antibody against CD68 (Supplementary Fig. 4). Thus, data showed that the Col1a2 silencing was present in our in vivo model both at molecular and protein level.
Discussion
Type I collagen is a heterotrimer composed of two α1(I) and one α2(I) chains. While the complete deficiency of α1(I) chain appears to be incompatible with life [27], the synthesis of α1(I) homotrimers lacking α2(I) was described in several situations. α1(I)-trimers were isolated from normal human skin where they account for <5% of the total collagen [28], and their presence was also detected during embryogenesis [29,30], in tumors [31,32], in fibrotic tissues [33,34], in stressed mesangial cells [35], and in a variety of pathological conditions [[36], [37], [38]].
Homozygous null mutations in COL1A2 also lead to α1(I) homotrimers assembly and to a range of phenotypes with an effect on tissue properties still poorly understood. COL1A2 mutations impairing α2(I) chain assembly in the triple helix, without affecting gene transcription and translation, are associated to severe osteogenesis imperfecta phenotype both in humans and murine models [39,40]. On the contrary, COL1A2 defects affecting transcript stability and compromising α2(I) translation cause a mild EDS syndrome in patients, with hypermobility in childhood and cardiac valve disease in adulthood, but no bone phenotype [22]. To explain the very different effects of mutations in COL1A2, it is tempting to speculate that those mutations in the α2(I) C-propeptide associated to mutant α2(I) translation cause its intracellular accumulation inducing a cellular stress that we already described as a key player in OI skeletal outcome and a modulator of osteoblasts differentiation [41,42]. Of note, in the OI murine model oim/oim, in which mutant α2(I) translation is not impaired, but its assembly in collagen type I heterotrimer is compromised, the osteoblasts differentiation ability is affected [43].
The majority of the published silencing reports in OI comes from studies using in vitro approaches, only very limited data are available from in vivo studies in OI murine models [[44], [45], [46], [47]]. Indeed, the availability of both in vitro and in vivo models is particularly relevant in studies aimed to identify novel therapeutic approaches. The in vitro approach is an important tool that allows to dissect and analyze specific cellular pathways without the interference of the whole tissue/organ environment, on the other hand the use of in vivo models is a mandatory step before clinical trials.
Using Col1a2 silencing we demonstrated that in the absence of α2(I) the mineralization in vitro and collagen formation in vivo are not affected. Col1a2-siRNA transfection allowed osteoblasts mineralization, although we cannot exclude a rebound effect of Col1a2 gene expression level from day 12 to day 19 post transfection. Indeed, a local delivery of Col1a2-siRNA in vivo to BCP beads loaded with mesenchymal stem cells preserved collagen deposition [48].
The in vivo silencing of the Col1a2 expression, although specific, was less effective than in in vitro, and similarly, a lower reduction of the α2(I) chain was detected. A possible reason for this difference could be linked to some intrinsic limitations of the system, such as the difficulty to reach the cells inside the implant and the formation of a connective tissue capsule around the implant that could impair siRNAs entrance/uptake into the target cells. Nevertheless, collagen formation in the implant was not compromised by the Col1a2-siRNA injection. However, efficient in vivo model of new bone formation would need further optimization.
The ultimate cure for dominant OI would be the correction of the DNA mutation, but for the more severe forms the silencing of the mutant allele may represent an interesting and probably more feasible option. The highly repetitive primary sequence of type I collagen, along with over a thousand distinct single base substitutions identified in OI patients, presents a significant challenge to an allele-specific targeting approach to treat this disease [7]. In OI patients carrying over 600 point mutations in COL1A2, the bone phenotype could in principle be treated by a mutation independent silencing approach. Our data represent a proof of principle of the feasibility of the approach, but the issue of siRNA bone delivery remains to be addressed. Indeed, atelocollagen-mediated systemic administration of siRNAs successfully inhibited bone metastasis [49]. The complexation of siRNAs with polyethylenimine also was demonstrated to be effective following systemic administration to deliver the siRNAs into different organs, but no data on specific bone targeting are available yet [50,51].
Systemic delivery of siRNA is a quite promising therapeutic approach, but it should be taken into consideration that in clinical trial a high number of repeated siRNA injections could result in a decreased patient compliance and consequent impediment to patient treatment [52].
In conclusion, following the in silico design of three siRNAs molecules targeting the murine Col1a2, we successfully identified a siRNA that efficiently and specifically suppresses in vitro and in vivo the Col1a2 expression. The optimization of a bone specific delivery system will open a new era in the treatment of OI, at least for those forms with dominant mutations in the COL1A2 gene.
Materials and methods
Animals
C57Bl/6 control mice were purchased from Harlan Laboratories and maintained in the animal facility at the Department of Molecular Medicine of the University of Pavia according to the current laws on the animal care. All the experiments were approved by the Italian Ministry of Health (protocol n.1116/2015-PR, 21/10/2015), complied with the ARRIVE guidelines and carried out in accordance with the EU Directive 2010/63/EU for animal experiments. Rj:NMRI-nude mice were purchased from Elevages Janvier SAS and maintained in the animal facility at the faculty of Medicine of the University of Nantes. The local Ethic Committee (CEEA.2012.27) approved the animal experimentation protocol for the research.
In silico siRNA selection and synthesis
The Basic Local Alignment Search Tool (BLAST) was used to identify three siRNAs 19–21 nucleotides long, specifically targeting the 3′-end of the murine Col1a2 (NM_007743.3) gene encoding for the C-propeptide domain of the α2 chain of type I collagen: siRNA-3554 (5’-GGACUAUGAAGUUGAUGCA-3′) targeting exon 49 (NM_007743.3, 3633–3652), siRNA-3825 (5’-GCCAACAAGCATGTCTGGTTA-3′) targeting exon 50 (NM_007743.3, 3904–3925), and siRNA-4215 (5’-GAATTCCGTGTGGAGGTTG-3′) targeting exon 52 (NM_007743.3, 4303–4322). The three siRNAs targeting murine Col1a2 and two negative controls for in vitro and in vivo experiments: siRNA-LacZ (5’-GUGACCAGCGAAUACCUGU-3′) and siRNA-LucF (5’-CUUACGCUGAGUACUUCGA-3′) respectively, were purchased from Eurogentec (Seraing, Belgio) with the addition of a dTdT nucleotide at their 3′-end.
Isolation and culture of primary murine embryonic fibroblasts
Murine embryonic fibroblasts were obtained from E13.5–14.5 C57Bl/6 embryos. Briefly, upon sacrifice the body was transferred in a sterile tube, minced and sequentially digested with 1 g/l trypsin in 0.4 g/l EDTA, 1.7 g/l NaCl for 5 min at 37 °C. Cells obtained from four successive digestions were passed through a 40 μm polypropylene mesh (Millipore, Burlington, Massachusetts, USA) added with Dulbecco modified Eagle's medium (D-MEM, Lonza, Basilea, Switzerland) supplemented with 10% fetal bovine serum (FBS), 4 mM glutamine, 100 μg/ml penicillin and 0.1 mg/ml streptomycin and pooled. The cells obtained from each embryo were plated in 10 cm petri dish and incubated at 37 °C. Cells at passages P3-P6 were used for all the experiments.
Isolation and culture of primary murine calvarial osteoblasts
Murine calvarial osteoblasts were isolated from 1 to 2 days old mice. Pups were euthanized, the cranium was dissected, cleaned from the surrounding connective tissue, cut in half and collected in α-Minimal Essential Medium (α –MEM, Sigma Aldrich, St. Louis, Missouri, USA) supplemented with 10% FBS, 4 mM glutamine, 100 μg/ml penicillin and 0.1 mg/ml streptomycin and 25 μg/ml sodium ascorbate (Fisher Scientific, Hampton, New Hampshire, USA). Calvaria were then pooled and sequentially digested with 200 U/ml collagenase type II (Thermo Fisher Scientific, Waltham, Massachusetts, USA) at 37 °C in an oscillating water bath for 20 min. Cells obtained from the first two digestions were discharged, while cells from the following three digestions were passed through a 70 μm polypropylene mesh, added with α -MEM containing 10% FBS and pooled. Cells were plated in 10 cm petri dish at a density of 2 × 105 cells/dish. The medium was changed twice a week and cells were used at passage P1.
Culture of murine mesenchymal stem cells
Mouse mesenchymal stem cells (MSCs) were obtained from Invitrogen (GIBCO® Mouse C57BL/6 mesenchymal stem cells; Catalog no. S1502–100) and cultured as specified in the guidelines. They have been modified to express the enhanced green fluorescent protein (EGFP) using lentivirus as previously described [53].
Cell transfection
For MEFs transfection experiments cells were plated in 24 well plate at a density of 1 × 104 or 2 × 104 cells/well. After 24 h the cells were transfected using Interferin (PolyPlus transfection, Illkirch, France) following manufacturer's recommendation. For each siRNA two concentrations were tested, 10 and 50 mM respectively, and the RNA was collected 48 h after the transfection. For the two more efficient and specific siRNAs, siRNA-3554 and siRNA-3825 and the negative control siRNA-LacZ a time course was performed and the RNA was collected 24, 48 and 72 h after transfection with 10 nM siRNA.
For primary osteoblasts, cells were plated in 24 well plate at a density of 2 × 104 cells/well and transfected after 24 h. siRNA-3554 was used at a concentration of 10 nM and the RNA was collected 1, 2, 8, 12 days post transfection. Each experiment was performed in duplicate.
Biphasic calcium phosphate implants
The biphasic calcium phosphate (BCP) implant was composed by 40 mg of synthetic bone graft substitute bioactive calcium phosphate 0.5–1 mm granules (MBCP®, Biomatlante, Vigneux-de-Bretagne, France) incubated for 1 h at RT with 1.4 × 106 GIBCO® Mouse MSCs modified to express EGFP (EGFP-MSCs) in 60 μl of phosphate buffered saline (PBS). The lipoplex complex for the siRNA delivery was obtained as follow: a solution containing 0.67 μg/μl of siRNAs and 0.67 μg/μl of DNA plasmid pSL301 was mixed with 8 mM cationic liposome DMAPAP/DOPE (charge ratio 1:6) and incubated 20 min at room temperature to allow the siRNA encapsulation [54].
Rj:NMRI-nude female mice 5 week old were used. The mice were anesthetized by inhalation of 2% isoflurane and injected with 10 μg Buprécare intramuscularly. The leg was incised at the level of the tibia and a pocket in the muscle tissue was created. The implant was then inserted in both legs, and the cut was sutured with a non-resorbable suture thread. The week after, the siRNA lipoplex complex was injected on the implants three times a week for three weeks. Fluorescent labelled (ATTO655) siRNAs were injected the last week. The mice were injected with 10 μg of siRNA-3554 or siRNA-LucF or with PBS (n = 4/ group). The animals were then sacrificed 28 days after the implantation. The BCP implants were removed from the mice following euthanasia. The implants were used to evaluate collagen type I gene expression and protein levels and for histological analysis.
Gene expression analysis
The RNA from MEFs and osteoblasts was collected at the different time points detailed above using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer instruction.
The RNA from BCP implants was extracted, following removal of the surrounding connective tissue, using Tri-Reagent (Sigma Aldrich). Briefly, the implants were crushed with the use of a pestle in 1.5 ml tube (Argos EW-44468-19) in a final volume of 1 ml of Tri-Reagent and the extraction followed the manufacturer instruction. After DNase treatment, cDNA was synthesized from 100 ng of RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystem, Foster City, California, USA). qPCR analysis was performed using TaqMan probes, and Universal PCR Master Mix (Applied Biosystems) using the MX3000P qPCR (Stratagene, San Diego, California, USA). All samples were run in triplicate. Expression levels for Col1a1 (Mm00801666_g1) and Col1a2 (Mm00483888_m1) were evaluated. Gapdh (Mm99999915_g1) was used as housekeeping. Relative expression levels were calculated using the ΔΔCt method.
Type I collagen analysis
MEFs were plated in 6 well plate at a density of 1.2 × 105 cells/well. After 24 h the cells were transfected with 10 nM siRNA-3554 or siRNA-LacZ as described above. The day after transfection the cells were pre-labelled with D-MEM, 1% FBS containing 0.1 mg/ml ascorbic acid for 2 h to stimulate collagen production. The labelling was performed for 18 h in D-MEM pre-labelling medium using 20 μCi of 3H-Pro/well. Collagen was then extracted and analyzed as previously described [55]. For the analysis of collagen from biphasic calcium phosphate, dissected implants were washed in PBS and decalcified in 0.5 M EDTA pH 7.1. Collagen was extracted by pepsin digestion as described previously and lyophilized [55]. The pellet was resuspended in Laemmli buffer (62.5 mM Tris HCl, pH 6.8, 10% glycerol, 2% sodium dodecyl sulphate (SDS), 0.02% bromophenol blue), denatured at 80 °C for 5 min and separated on 6% SDS-PAGE gels in presence of 0.5 M urea. The gels were stained with Coomassie Picric Staining [56], and digitalized by Versadoc (Biorad, Hercules, California, USA). The bands intensity was measured using Quantity one software (Biorad).
Osteoblasts mineralization assay
24 h after transfection, the mineralization media α-MEM, 10% FBS, 100 μg/ml penicillin, 0.1 mg/ml streptomycin, 5 × 10−8 M dexamethasone (Sigma Aldrich), 0.2 mM ascorbic acid (Fisher Scientific) and 10 mM β-glycerophosphate (Sigma Aldrich) was added for 19 days, changing the media three times a week. The cells were then fixed in 10% formalin solution neutral buffer (Sigma Aldrich) for 30 min at RT and Von Kossa staining was performed. Images were acquired with a digital scanner at 2400 dpi resolution and analyzed with the Leica application suite V4.5 software. The percentage of mineralized area was calculated on the total well area and normalized on total DNA per well.
Histological and immunohistochemical analysis
The BCP implants were fixed for 24 h in 4% buffered paraformaldehyde (PFA), decalcified in 4% EDTA (Alfa Aesar, Haverhill, Massachusetts, USA) dehydrated and included in paraffin as previously described [57]. 3 μm sections were obtained using microtome RM2265 (Leica, Wetzlar, Germania, USA). Slides were stained with Masson's Trichrome combining hematoxylin for cell nuclei (blue/black), fuchsine for cytoplasm, muscle and erythrocytes (red) and light green solution for collagen (green), and with Tartrate-Resistant Acid Phosphatase (TRAP) commercial staining kit (Phosphatase Leukocyte Staining Kit, Sigma Aldrich) for osteoclasts.
Immunohistochemistry was performed to evaluate the presence of both EGFP expressing cells and macrophages. Briefly, after dewaxing and rehydration, heat-induced epitope retrieval was performed using 10 mM Tris HCl, 1 mM EDTA pH 9.0. Sections were treated with 3% H2O2 for 15 min at room temperature and the non-specific antibody binding was blocked using 2% normal donkey serum and 1% bovine serum albumin in 1× TBS Tween for 25 min at room temperature. Primary antibodies rat anti-mouse CD68 (1:100, FA-11, Biorad, France) and rabbit anti-EGFP (1:2000, ab290, Abcam, France) were incubated over night at 4 °C to detect macrophages and mouse EGFP-MSCs implanted with the BCP particles, respectively. Finally, a biotinylated goat anti-mouse IgG secondary antibody (Dako) and streptavidin/ horse radish peroxydase complexes (Dako) were used and revealed by a short incubation with 3,3′- diaminobenzidine (K3468, Agilent, Denmark). Nuclei were counterstained with a Gill-Hematoxylin solution. After dehydration and mounting with petex, slides were acquired using Nanozoomer 2.0 Hamamatsu slide scanner. Quantitative analysis of the collagen tissue stained by Masson's Trichrome was then performed. The collagen area and the total area of the implants were manually defined and quantified using Leica application suite V 4.5 (n = 4). The collagen amount was expressed as percentage of the total area.
Statistical analysis
All results were expressed as mean ± standard deviation. Statistical comparisons were based on Student's t-test. A p < .05 was considered significant.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank Dr. Marco Manca, Department of Molecular Medicine, Biochemistry Unit, University of Pavia, Italy, for initial experiments on murine embryonic fibroblasts and the OPBA of the University of Pavia for support in animal protocol drawing up.
Funding
The work was supported by AFM-Telethon, France (2012-1607) to AF and VT, Care4BrittleBone, The Netherlands (2015-0003) to AF and the European Community, FP7, ‘Sybil’ project, Europe (Grant No. 602300) and Italian Ministry of Education, University and Research (MIUR), Italy [Dipartimenti di Eccellenza (2018–2022)] to AR and AF.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2020.100028.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Forlino A., Marini J.C. Osteogenesis imperfecta. Lancet. 2016;387:1657–1671. doi: 10.1016/S0140-6736(15)00728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besio R., Chow C.W., Tonelli F., Marini J.C., Forlino A. Bone biology: insights from osteogenesis imperfecta and related rare fragility syndromes. The FEBS Journal. 2019;286:3033–3056. doi: 10.1111/febs.14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besio R., Forlino A. Treatment options for osteogenesis imperfecta. Expert Opinion on Orphan Drugs. 2015;3:165–181. [Google Scholar]
- 4.Rossi V., Lee B., Marom R. Osteogenesis imperfecta: advancements in genetics and treatment. Current Opinion in Pediatrics. 2019;31:708–715. doi: 10.1097/MOP.0000000000000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morello R. Osteogenesis imperfecta and therapeutics. Matrix Biology: Journal of the International Society for Matrix Biology. 2018;71–72:294–312. doi: 10.1016/j.matbio.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willing M.C., Deschenes S.P., Slayton R.L., Roberts E.J. Premature chain termination is a unifying mechanism for COL1A1 null alleles in osteogenesis imperfecta type I cell strains. American Journal of Human Genetics. 1996;59:799–809. [PMC free article] [PubMed] [Google Scholar]
- 7.Marini J.C., Forlino A., Cabral W.A., Barnes A.M., San Antonio J.D., Milgrom S., Hyland J.C., Korkko J., Prockop D.J., De Paepe A., Coucke P., Symoens S., Glorieux F.H., Roughley P.J., Lund A.M., Kuurila-Svahn K., Hartikka H., Cohn D.H., Krakow D., Mottes M., Schwarze U., Chen D., Yang K., Kuslich C., Troendle J., Dalgleish R., Byers P.H. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Human Mutation. 2007;28:209–221. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiroli D., Gomara M.J., Maurizi E., Atkinson S.D., Mairs L., Christie K.A., Cobice D.F., McCrudden C.M., Nesbit M.A., Haro I., Moore T. Effective in vivo topical delivery of siRNA and gene silencing in intact corneal epithelium using a modified cell-penetrating peptide. Molecular Therapy--Nucleic Acids. 2019;17:891–906. doi: 10.1016/j.omtn.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaleta-Rivera K., Dainis A., Ribeiro A.J.S., Cordero P., Rubio G., Shang C., Liu J., Finsterbach T., Parikh V.N., Sutton S., Seo K., Sinha N., Jain N., Huang Y., Hajjar R.J., Kay M.A., Szczesna-Cordary D., Pruitt B.L., Wheeler M.T., Ashley E.A. Allele-specific silencing ameliorates restrictive cardiomyopathy attributable to a human myosin regulatory light chain mutation. Circulation. 2019;140:765–778. doi: 10.1161/CIRCULATIONAHA.118.036965. [DOI] [PubMed] [Google Scholar]
- 10.Giorgio E., Lorenzati M., Rivetti di Val Cervo P., Brussino A., Cernigoj M., Della Sala E., Bartoletti Stella A., Ferrero M., Caiazzo M., Capellari S., Cortelli P., Conti L., Cattaneo E., Buffo A., Brusco A. Allele-specific silencing as treatment for gene duplication disorders: proof-of-principle in autosomal dominant leukodystrophy. Brain. 2019;142:1905–1920. doi: 10.1093/brain/awz139. [DOI] [PubMed] [Google Scholar]
- 11.Keire P.A., Bressler S.L., Mulvihill E.R., Starcher B.C., Kang I., Wight T.N. Inhibition of versican expression by siRNA facilitates tropoelastin synthesis and elastic fiber formation by human SK-LMS-1 leiomyosarcoma smooth muscle cells in vitro and in vivo. Matrix Biology: Journal of the International Society for Matrix Biology. 2016;50:67–81. doi: 10.1016/j.matbio.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuppan D., Ashfaq-Khan M., Yang A.T., Kim Y.O. Liver fibrosis: direct antifibrotic agents and targeted therapies. Matrix Biology: Journal of the International Society for Matrix Biology. 2018;68–69:435–451. doi: 10.1016/j.matbio.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Molokanova O., Schonig K., Weng S.Y., Wang X., Bros M., Diken M., Ohngemach S., Karsdal M., Strand D., Nikolaev A., Eshkind L., Schuppan D. Inducible knockdown of procollagen I protects mice from liver fibrosis and leads to dysregulated matrix genes and attenuated inflammation. Matrix Biology: Journal of the International Society for Matrix Biology. 2018;66:34–49. doi: 10.1016/j.matbio.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z., Velazquez-Quesada I., Murdamoothoo D., Ahowesso C., Yilmaz A., Spenle C., Averous G., Erne W., Oberndorfer F., Oszwald A., Kain R., Bourdon C., Mangin P., Deligne C., Midwood K., Abou-Faycal C., Lefebvre O., Klein A., van der Heyden M., Chenard M.P., Christofori G., Mathelin C., Loustau T., Hussenet T., Orend G. Tenascin-C increases lung metastasis by impacting blood vessel invasions. Matrix Biology: Journal of the International Society for Matrix Biology. 2019;83:26–47. doi: 10.1016/j.matbio.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Melero-Fernandez de Mera R.M., Arasu U.T., Karna R., Oikari S., Rilla K., Vigetti D., Passi A., Heldin P., Tammi M.I., Deen A.J. Effects of mutations in the post-translational modification sites on the trafficking of hyaluronan synthase 2 (HAS2) Matrix Biology: Journal of the International Society for Matrix Biology. 2018;80:85–103. doi: 10.1016/j.matbio.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Coutinho M.F., Matos L., Santos J.I., Alves S. RNA therapeutics: how far have we gone? Adv. Exp. Med. Biol. 2019;1157:133–177. doi: 10.1007/978-3-030-19966-1_7. [DOI] [PubMed] [Google Scholar]
- 17.Weng Y., Xiao H., Zhang J., Liang X.J., Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnology Advances. 2019;37:801–825. doi: 10.1016/j.biotechadv.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., Lin K.P., Vita G., Attarian S., Plante-Bordeneuve V., Mezei M.M., Campistol J.M., Buades J., Brannagan T.H., 3rd, Kim B.J., Oh J., Parman Y., Sekijima Y., Hawkins P.N., Solomon S.D., Polydefkis M., Dyck P.J., Gandhi P.J., Goyal S., Chen J., Strahs A.L., Nochur S.V., Sweetser M.T., Garg P.P., Vaishnaw A.K., Gollob J.A., Suhr O.B. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. The New England Journal of Medicine. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 19.Rousseau J., Gioia R., Layrolle P., Lieubeau B., Heymann D., Rossi A., Marini J.C., Trichet V., Forlino A. Allele-specific Col1a1 silencing reduces mutant collagen in fibroblasts from Brtl mouse, a model for classical osteogenesis imperfecta. European Journal of Human Genetics. 2013;22:667–674. doi: 10.1038/ejhg.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindahl K., Rubin C.J., Kindmark A., Ljunggren O. Allele dependent silencing of COL1A2 using small interfering RNAs. International Journal of Medical Sciences. 2008;5:361–365. doi: 10.7150/ijms.5.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindahl K., Kindmark A., Laxman N., Astrom E., Rubin C.J., Ljunggren O. Allele dependent silencing of collagen type I using small interfering RNAs targeting 3′UTR Indels - a novel therapeutic approach in osteogenesis imperfecta. International Journal of Medical Sciences. 2013;10:1333–1343. doi: 10.7150/ijms.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarze U., Hata R., McKusick V.A., Shinkai H., Hoyme H.E., Pyeritz R.E., Byers P.H. Rare autosomal recessive cardiac valvular form of Ehlers-Danlos syndrome results from mutations in the COL1A2 gene that activate the nonsense-mediated RNA decay pathway. American Journal of Human Genetics. 2004;74:917–930. doi: 10.1086/420794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed H., Taipaleenmaki H., Aldahmash A.M., Abdallah B.M., Kassem M. Mouse embryonic fibroblasts (MEF) exhibit a similar but not identical phenotype to bone marrow stromal stem cells (BMSC) Stem Cell Reviews and Reports. 2011;8:318–328. doi: 10.1007/s12015-011-9315-x. [DOI] [PubMed] [Google Scholar]
- 24.Young M.F. Skeletal biology: where matrix meets mineral. Matrix Biology. 2016;52–54:1–6. doi: 10.1016/j.matbio.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim B.S., Choi M.K., Yoon J.H., Lee J. Evaluation of bone regeneration with biphasic calcium phosphate substitute implanted with bone morphogenetic protein 2 and mesenchymal stem cells in a rabbit calvarial defect model. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology. 2015;120:2–9. doi: 10.1016/j.oooo.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Gamblin A.L., Brennan M.A., Renaud A., Yagita H., Lezot F., Heymann D., Trichet V., Layrolle P. Bone tissue formation with human mesenchymal stem cells and biphasic calcium phosphate ceramics: the local implication of osteoclasts and macrophages. Biomaterials. 2014;35:9660–9667. doi: 10.1016/j.biomaterials.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 27.Schnieke A., Harbers K., Jaenisch R. Embryonic lethal mutation in mice induced by retrovirus insertion into the alpha 1(I) collagen gene. Nature. 1983;304:315–320. doi: 10.1038/304315a0. [DOI] [PubMed] [Google Scholar]
- 28.Uitto J. Collagen polymorphism: isolation and partial characterization of alpha 1(I)-trimer molecules in normal human skin. Archives of Biochemistry and Biophysics. 1979;192:371–379. doi: 10.1016/0003-9861(79)90105-x. [DOI] [PubMed] [Google Scholar]
- 29.Narayanan A.S., Meyers D.F., Page R.C., Welgus H.G. Action of mammalian collagenases on type I trimer collagen. Collagen and Related Research. 1984;4:289–296. doi: 10.1016/s0174-173x(84)80036-9. [DOI] [PubMed] [Google Scholar]
- 30.Jimenez S.A., Bashey R.I., Benditt M., Yankowski R. Identification of collagen alpha1(I) trimer in embryonic chick tendons and calvaria. Biochemical and Biophysical Research Communications. 1977;78:1354–1361. doi: 10.1016/0006-291x(77)91441-3. [DOI] [PubMed] [Google Scholar]
- 31.Rupard J.H., Dimari S.J., Damjanov I., Haralson M.A. Synthesis of type I homotrimer collagen molecules by cultured human lung adenocarcinoma cells. The American Journal of Pathology. 1988;133:316–326. [PMC free article] [PubMed] [Google Scholar]
- 32.Makareeva E., Han S., Vera J.C., Sackett D.L., Holmbeck K., Phillips C.L., Visse R., Nagase H., Leikin S. Carcinomas contain a matrix metalloproteinase-resistant isoform of type I collagen exerting selective support to invasion. Cancer Research. 2010;70:4366–4374. doi: 10.1158/0008-5472.CAN-09-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojkind M., Giambrone M.A., Biempica L. Collagen types in normal and cirrhotic liver. Gastroenterology. 1979;76:710–719. [PubMed] [Google Scholar]
- 34.Ehrlich H.P., Brown H., White B.S. Evidence for type V and I trimer collagens in Dupuytren’s contracture palmar fascia. Biochemical Medicine. 1982;28:273–284. doi: 10.1016/0006-2944(82)90080-1. [DOI] [PubMed] [Google Scholar]
- 35.Haralson M.A., Jacobson H.R., Hoover R.L. Collagen polymorphism in cultured rat kidney mesangial cells. Laboratory Investigation. 1987;57:513–523. [PubMed] [Google Scholar]
- 36.Yamagata S., Yamagata T. FBJ virus-induced osteosarcoma contains type I, type I trimer, type III as well as type V collagens. Journal of Biochemistry. 1984;96:17–26. doi: 10.1093/oxfordjournals.jbchem.a134809. [DOI] [PubMed] [Google Scholar]
- 37.Moro L., Smith B.D. Identification of collagen alpha1(I) trimer and normal type I collagen in a polyoma virus-induced mouse tumor. Archives of Biochemistry and Biophysics. 1977;182:33–41. doi: 10.1016/0003-9861(77)90280-6. [DOI] [PubMed] [Google Scholar]
- 38.Narayanan A.S., Page R.C., Meyers D.F. Characterization of collagens of diseased human gingiva. Biochemistry. 1980;19:5037–5043. doi: 10.1021/bi00563a016. [DOI] [PubMed] [Google Scholar]
- 39.Chipman S.D., Sweet H.O., McBride D.J., Jr., Davisson M.T., Marks S.C., Jr., Shuldiner A.R., Wenstrup R.J., Rowe D.W., Shapiro J.R. Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: a model of human osteogenesis imperfecta. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:1701–1705. doi: 10.1073/pnas.90.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pace J.M., Wiese M., Drenguis A.S., Kuznetsova N., Leikin S., Schwarze U., Chen D., Mooney S.H., Unger S., Byers P.H. Defective C-propeptides of the proalpha2(I) chain of type I procollagen impede molecular assembly and result in osteogenesis imperfecta. The Journal of Biological Chemistry. 2008;283:16061–16067. doi: 10.1074/jbc.M801982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Besio R., Garibaldi N., Leoni L., Cipolla L., Sabbioneda S., Biggiogera M., Mottes M., Aglan M., Otaify G.A., Temtamy S.A., Rossi A & Forlino A Cellular stress due to impairment of collagen prolyl hydroxylation complex is rescued by the chaperone 4-phenylbutyrate. Disease Models & Mechanisms. 2019;12 doi: 10.1242/dmm.038521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Besio R., Iula G., Garibaldi N., Cipolla L., Sabbioneda S., Biggiogera M., Marini J.C., Rossi A., Forlino A. 4-PBA ameliorates cellular homeostasis in fibroblasts from osteogenesis imperfecta patients by enhancing autophagy and stimulating protein secretion. Biochimica et Biophysica Acta - Molecular Basis of Disease. 1864:1642–1652. doi: 10.1016/j.bbadis.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Jiang X., Delaney J., Franceschetti T., Bilic-Curcic I., Kalinovsky J., Lorenzo J.A., Grcevic D., Rowe D.W., Kalajzic I. Immature osteoblast lineage cells increase osteoclastogenesis in osteogenesis imperfecta murine. The American Journal of Pathology. 2010;176:2405–2413. doi: 10.2353/ajpath.2010.090704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khillan J.S., Li S.W., Prockop D.J. Partial rescue of a lethal phenotype of fragile bones in transgenic mice with a chimeric antisense gene directed against a mutated collagen gene. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:6298–6302. doi: 10.1073/pnas.91.14.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson P.A., Marini J.C. Hammerhead ribozymes selectively suppress mutant type I collagen mRNA in osteogenesis imperfecta fibroblasts. Nucleic Acids Research. 2000;28:4013–4020. doi: 10.1093/nar/28.20.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grassi G., Forlino A., Marini J.C. Cleavage of collagen RNA transcripts by hammerhead ribozymes in vitro is mutation-specific and shows competitive binding effects. Nucleic Acids Research. 1997;25:3451–3458. doi: 10.1093/nar/25.17.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toudjarska I., Kilpatrick M.W., Niu J., Wenstrup R.J., Tsipouras P. Delivery of a hammerhead ribozyme specifically downregulates mutant type I collagen mRNA in a murine model of osteogenesis imperfecta. Antisense & Nucleic Acid Drug Development. 2001;11:341–346. doi: 10.1089/108729001753231722. [DOI] [PubMed] [Google Scholar]
- 48.Brennan M.A., Renaud A., Amiaud J., Rojewski M.T., Schrezenmeier H., Heymann D., Trichet V., Layrolle P. Pre-clinical studies of bone regeneration with human bone marrow stromal cells and biphasic calcium phosphate. Stem Cell Research & Therapy. 2014;5:114. doi: 10.1186/scrt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeshita F., Minakuchi Y., Nagahara S., Honma K., Sasaki H., Hirai K., Teratani T., Namatame N., Yamamoto Y., Hanai K., Kato T., Sano A., Ochiya T. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urban-Klein B., Werth S., Abuharbeid S., Czubayko F., Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Therapy. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 51.Aigner A. Delivery systems for the direct application of siRNAs to induce RNA interference (RNAi) in vivo. Journal of Biomedicine & Biotechnology. 2006;2006:71659. doi: 10.1155/JBB/2006/71659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka T., Mangala L.S., Vivas-Mejia P.E., Nieves-Alicea R., Mann A.P., Mora E., Han H.D., Shahzad M.M., Liu X., Bhavane R., Gu J., Fakhoury J.R., Chiappini C., Lu C., Matsuo K., Godin B., Stone R.L., Nick A.M., Lopez-Berestein G., Sood A.K., Ferrari M. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Research. 2010;70:3687–3696. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rousseau J., Escriou V., Perrot P., Picarda G., Charrier C., Scherman D., Heymann D., Redini F., Trichet V. Advantages of bioluminescence imaging to follow siRNA or chemotherapeutic treatments in osteosarcoma preclinical models. Cancer Gene Therapy. 2010;17:387–397. doi: 10.1038/cgt.2009.89. [DOI] [PubMed] [Google Scholar]
- 54.Khoury M., Louis-Plence P., Escriou V., Noel D., Largeau C., Cantos C., Scherman D., Jorgensen C., Apparailly F. Efficient new cationic liposome formulation for systemic delivery of small interfering RNA silencing tumor necrosis factor alpha in experimental arthritis. Arthritis and Rheumatism. 2006;54:1867–1877. doi: 10.1002/art.21876. [DOI] [PubMed] [Google Scholar]
- 55.Forlino A., Tonelli F., Besio R. Steady-state and pulse-chase analyses of fibrillar collagen. Methods in Molecular Biology. 2019;1952:45–53. doi: 10.1007/978-1-4939-9133-4_4. [DOI] [PubMed] [Google Scholar]
- 56.Stephano J.L., Gould M., Rojas-Galicia L. Advantages of picrate fixation for staining polypeptides in polyacrylamide gels. Analytical Biochemistry. 1986;152:308–313. doi: 10.1016/0003-2697(86)90414-8. [DOI] [PubMed] [Google Scholar]
- 57.Paganini C., Monti L., Costantini R., Besio R., Lecci S., Biggiogera M., Tian K., Schwartz J.M., Huber C., Cormier-Daire V., Gibson B.G., Pirog K.A., Forlino A., Rossi A. Calcium activated nucleotidase 1 (CANT1) is critical for glycosaminoglycan biosynthesis in cartilage and endochondral ossification. Matrix Biology: Journal of the International Society for Matrix Biology. 2018;81:70–90. doi: 10.1016/j.matbio.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures