Bioengineered matrices from self-assembling peptides and extracellular matrix proteins can model the tumor microenvironment.
Abstract
Bioengineered three-dimensional (3D) matrices expand our experimental repertoire to study tumor growth and progression in a biologically relevant, yet controlled, manner. Here, we used peptide amphiphiles (PAs) to coassemble with and organize extracellular matrix (ECM) proteins producing tunable 3D models of the tumor microenvironment. The matrix was designed to mimic physical and biomolecular features of tumors present in patients. We included specific epitopes, PA nanofibers, and ECM macromolecules for the 3D culture of human ovarian cancer, endothelial, and mesenchymal stem cells. The multicellular constructs supported the formation of tumor spheroids with extensive F-actin networks surrounding the spheroids, enabling cell-cell communication, and comparative cell-matrix interactions and encapsulation response to those observed in Matrigel. We conducted a proof-of-concept study with clinically used chemotherapeutics to validate the functionality of the multicellular constructs. Our study demonstrates that peptide-protein coassembling matrices serve as a defined model of the multicellular tumor microenvironment of primary ovarian tumors.
INTRODUCTION
There is a need for improved three-dimensional (3D) cancer models to study tumor growth and progression as seen in patients and to test responses to new treatments. At present, 90% of successful cancer treatments tested preclinically fail in the early phases of clinical trials, and less than 5% of oncology drugs are successful in clinical trials (1). Preclinical tests rely on a combination of 2D in vitro cell cultures and in vivo models to predict responses to treatment. Conventional 2D cell cultures fail to recapitulate key features of tumor tissues, such as cellular heterogeneity, presence of spheroids, matrix stiffness, cell-matrix interactions, and cross-talk with other cell types (2). On the other hand, murine models allow for cell-matrix and cell-cell interactions, but interspecies differences result in many successful treatments in murine hosts being ineffective in humans. Thus, novel experimental 3D cancer models are needed to better recapitulate the human tumor microenvironment (TME) and incorporate patient-specific differences.
The TME is a rich mixture of malignant and nonmalignant cells, such as fibroblasts, endothelial cells, and immune cells, embedded in an extracellular matrix (ECM) (3). The TME affects tumor growth and undergoes changes in response to cancer progression, such as stiffening of the ECM, shifts in chemical signaling, and tumor angiogenesis (4). Tumor angiogenesis arises when the tumor core undergoes hypoxia and triggers the secretion of proangiogenic proteins and the migration of endothelial cells toward the tumor. The resulting vessels are leaky and disorganized, consequently influencing the components of the TME. Collectively, these changes in the local microenvironment affect both cancer progression and responses to treatments. Thus, modeling the TME is vital and must recapitulate cell-cell and cell-matrix interactions as well as key components of the ECM, such as proteins, growth factors (GFs), and chemical gradients. Furthermore, the model should integrate multiple cell types that enable the growth of tumor spheroids.
Currently, the gold standard for 3D cancer models is the commercially available Matrigel (5), a solubilized basement membrane extracted from mouse sarcoma that consists of an undefined mixture of ECM proteins and GFs. A major reason for Matrigel’s popularity is its capacity to enable cell-matrix interactions, which promote the growth of malignant and nonmalignant cells as well as spheroid formation (6). However, it lacks control in mimicking the TME because of its high batch variability, undefined composition, and murine origin. These features are important limitations to effectively screen and develop new treatments for cancer. To address these drawbacks, natural and synthetic materials are being explored as alternatives to generate 3D cancer models, such as hyaluronic acid hydrogels (7), chitosan/gelatin composites (8), and polyethylene glycol (PEG) (9). Natural materials recapitulate structural aspects of the ECM, but they exhibit limited control over their mechanical properties. While synthetic materials can overcome the limitations in mechanical control, the trade-off is bioactivity. Thus, hybrid systems, such as PEG modified with fibrinogen (10), heparin (11), isoleucine-lysine-valine-alanine-valine (IKVAV) or arginine-glycine-aspartate (RGD) peptides (12), are being studied. However, while these composites greatly improve cellular responses, they do not capture the fibrous ECM nanostructure and heterogeneous composition and tend to require the use of potentially cytotoxic cross-linking components.
Self-assembling peptide-based materials can be programmed to assemble into well-defined nanostructures, such as micelles, nanofibers, or ribbons, while generating porosity and displaying multiple bioactive epitopes (13). These materials have been optimized to enhance their capacity to recreate the ECM, for example, by enabling alignment (14), tunability of mechanical properties (15), and the incorporation of protein-mimetic sequences to promote angiogenesis (16). Peptide amphiphiles (PAs) are promising self-assembling materials, which are capable of assembling into well-defined nanofibers (17) with the capacity to display cell-instructive motifs that mimic ECM proteins or GFs, such as laminin, collagens, and fibronectin (FN) (18). This approach is increasingly being used to coassemble peptides with a variety of molecules, such as hyaluronic acid (19), heparin (20), and proteins (21, 22), to develop biologically relevant 3D cell cultures.
Here, we report the design and validation of a peptide-protein coassembling hydrogel-based multicellular 3D model for ovarian cancer. We demonstrate the capacity of the system to serve as a complex, yet controllable, alternative to Matrigel. The hydrogel supported the spheroid formation from single ovarian cancer cells reaching comparative sizes to those grown in Matrigel. When cocultured with human umbilical vein endothelial cells (HUVECs) and human mesenchymal stem cells (hMSCs), the hydrogel enabled cellular interactions between the different cell types. The model demonstrated an increase in spheroid size as a result of increased cell-cell interactions. We also tested the effect of different peptide sequences and proteins within the multicomponent hydrogel, indicating the tunability of the system. In a proof-of-concept study, we determined tumor cell responses to different chemotherapeutics.
RESULTS
Rationale of the design
The herein reported coassembling hydrogel is based on our recently developed PA/keratin (KN) bioink (22), which served as a supramolecular platform to incorporate and optimize structural, molecular, and cellular components of the TME of ovarian cancer (Fig. 1A). We synthesized three new PA sequences based on the previously established PA-H (C16VVVAAAH2K), which includes an histidine-histidine-lysine (H2K) motif to facilitate coassembly with ECM proteins (22): PA-VH (C16VVVAAAVPGIGH2K), which contains an additional elastin-mimetic segment that enhances recreation of the ECM and was found to improve hydrogel stability; PA-RGDS (C16VVVAAAH2KRGDS), which incorporates an additional arginine-glycine-aspartic acid-serine (RGDS) sequence to promote cell adhesion; and PA-GHK (C16VVVAAAH2KGHK), which includes an additional glycine-histidine-lysine (GHK) motif known to promote cell proliferation and to serve as an angiogenic factor (Figs. 1B and 2A and fig. S1) (23). The PAs were designed to gel by coassembling with proteins through charge screening using the base PA (PA-H) as a control. We included FN, which is up-regulated in the ECM of the ovarian TME (24), and hair KN, which supports cell proliferation and tissue regeneration (25). Specific KNs are involved in the regulation of inflammation and immunity in epithelial layers (26, 27). Furthermore, KN provides structural stability to tissues and is expressed in normal and cancerous ovaries (Fig. 2B) as well as in ovarian cancer cell monolayers (fig. S2A).
Fig. 1. Peptide/protein coassembling hydrogel for 3D cancer models.
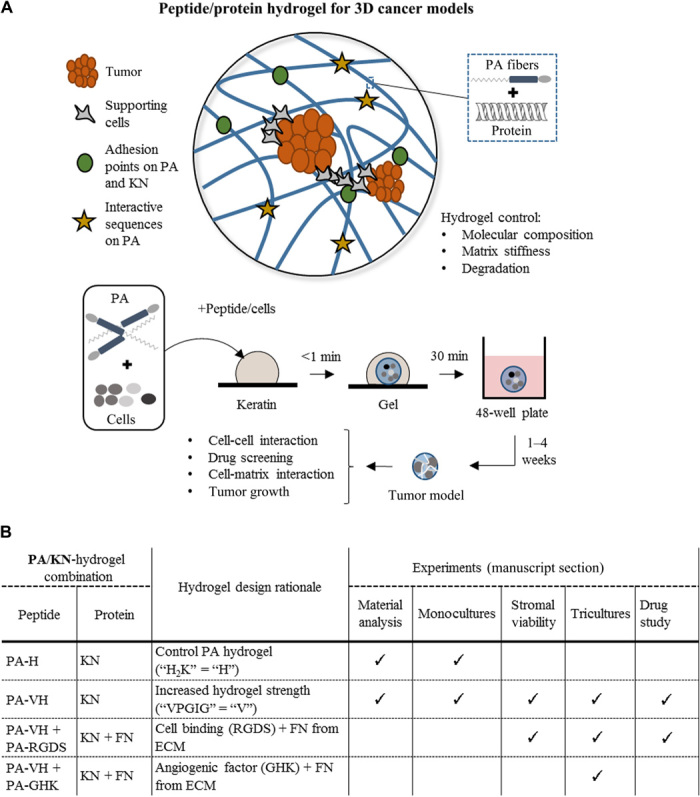
(A) Schematic of the peptide/protein coassembling hydrogel with key design features highlighted and an illustration of the process of cell encapsulation within the hydrogel. The hydrogel is made using solutions of PAs and a protein mixture [e.g., keratin (KN) and FN]. Cells are suspended in the PA solution leading to encapsulation. Cell-containing hydrogels are monitored over several weeks to observe cell-cell and cell-matrix patterns, cell growth, and drug response. (B) Table of the PA/KN hydrogel combinations used for the material analysis (see the “Hydrogel formation and characterization at the nano- and macroscale,” “Hydrogel stiffness characterization,” and “Hydrogel stability characterization” sections in Results), 3D monocultures (see the “Cell monocultures within PA/KN hydrogels generate tumor spheroids” section in Results), stromal-cell viability study (see the “PA/KN hydrogels support 3D monocultures of HUVECs and hMSCs” section in Results), 3D tricultures (see the “3D cocultures of cancer cells with stromal cells in PA/KN hydrogels promote intercellular network formation” section in Results), and treatment study (see the “Responses to treatment in spheroid-containing PA/KN hydrogels” section in Results).
Fig. 2. PA/KN hydrogel combinations and material analysis.
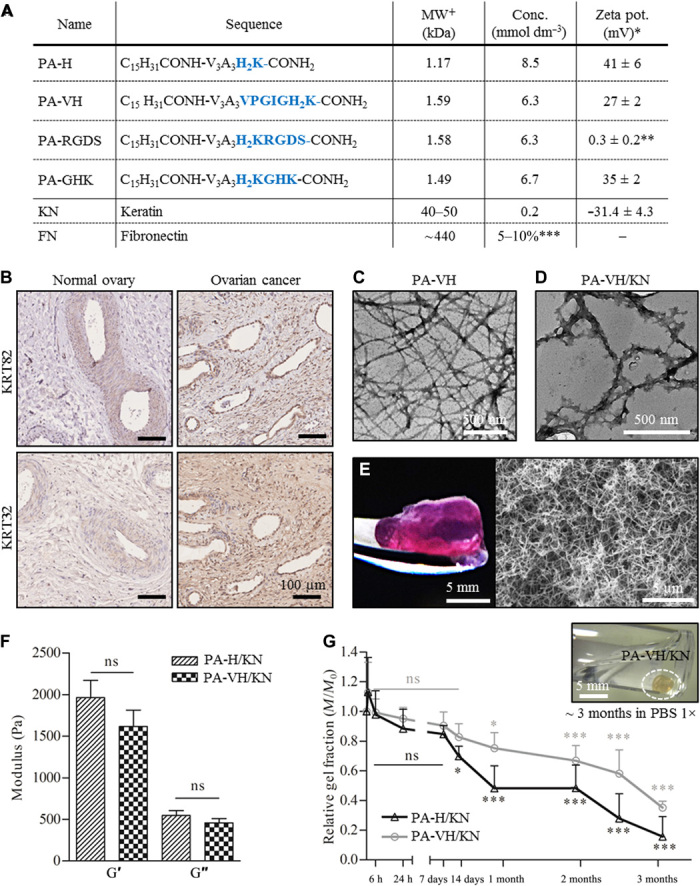
(A) Table of PA sequences and proteins used in this study including the chemical formula, molecular weight (MW), concentration used, and zeta potential at pH 7.5 (*in HEPES (pH 7.5), **in water, ***relative to keratin volume, +theoretical). (B) Immunohistochemical (IHC) staining for keratin type I (KRT32) and type II (KRT82) in normal (healthy) ovary and ovarian cancer tissue samples, with IHC controls in fig. S2. (C) TEM image of self-assembled PA fibers (PA-VH) in HEPES [0.1 mg/ml (pH 7.5)]. (D) TEM image of PA fibers interacting with KN (PA-VH/KN). Pure solutions of PA-VH and KN in HEPES [0.1 mg/ml (pH 7.5)] were mixed in ratio 1:1 before analysis. (E) Photograph of a PA-H/KN hydrogel after 14 days in culture medium at 37°C and a representative scanning electron microscopy image of the internal heterogeneous nanofibrous structure of PA/KN hydrogels. (F) Rheological characterization of the hydrogels, measuring the storage modulus (G′) and loss modulus (G″) of PA-H/KN and PA-VH/KN hydrogels (mean ± SEM). (G) Graphical plot of the hydrogel mass (Mt/M0) against time, indicating the slow degradation of PA-H/KN and PA-VH/KN hydrogels in PBS 1× at 37°C (mean ± SEM). Photo credit: Clara Hedegaard, Queen Mary University of London. ns, not significant.
A range of PA/KN hydrogel formulations were used throughout this work (Fig. 1B). In material characterization studies and 3D monocultures, PA-VH was tested using PA-H as a control. In a previous study, we used both PA-H and PA-VK sequences, showing favorable results for PA-VK in terms of hydrogel strength and PA-H for cell interaction. Thus, here, we wanted to build on our previous work by designing the sequence PA-VH and keeping PA-H as control for the influence of “V” (22). The hydrophobic section V (= VPGIG) was taken from elastin and has been used as part of biocompatible and biodegradable elastin-like polypeptides (28). We expected V to facilitate self-assembly into fibers and stronger interactions with KN. For 3D tricultures, PA-VH was mixed with PA-RGDS or PA-GHK at 10:1 (vol/vol) ratios before coassembly with the proteins. The rational here was to use the PA’s ability to coassemble between different PA molecules and present epitopes on the fiber surfaces. The PA ratio was selected to optimize the presentation and cellular recognition of the bioactive epitopes of PA-RGDS and PA-GHK. Previous studies have shown that epitope recognition can be maximized by spacing them within PA fibers (29). Last, in addition to the incorporation of multiple proteins and bioactive epitopes, the coassembling hydrogel was designed for encapsulation of three major cell types of the ovarian TME, including epithelial cancer cells, HUVECs, and hMSCs.
Hydrogel formation and characterization at the nano- and macroscale
Hydrogels were formed upon mixing PAs with the ECM proteins by injecting 15 μl of PA solution (10 mg/ml ± cells) into 100 μl of protein solution (10 mg/ml ± cells). PAs were synthesized as previously described (30), and their assembly assessed via circular dichroism (CD) and transmission electron microscopy (TEM). We found that all individual PAs and their mixtures exhibited the characteristic β sheet secondary structure (fig. S3A) and the classical nanofibrous architecture of self-assembled PAs (Fig. 2C and fig. S3C). The control sequence C16VVVAAAH2K of all tested PAs dictated that the inter-PA interaction mechanism and the different bioactive epitopes of the PAs did not interfere with the assembly process (fig. S3B). This result was further confirmed by TEM, which revealed that the PA-VH/KN coassembled nanofibers exhibited similar morphologies as those formed by the control PA-H/KN, as previously reported (Fig. 2D and fig. S3, C to E) (22). In addition, upon PA-protein coassembly, hydrogels formed immediately and exhibited a translucent appearance that did not appear to change 30 min after coassembly, at which point they were easily handled (Fig. 2E). However, PA-based hydrogels, like many self-assembling hydrogels, are fragile structures that can be difficult to manipulate without altering their macroscopic shape, despite retaining their inherent nanofibrous architecture (Fig. 2E). The moldability of this nanofibrous network (i.e., ability to expand, contract, and shift) is an important feature for cell encapsulation, creating a porous interactive structure. These characteristics facilitate cell migration, cell-cell communication, and spheroid growth, which are critical to recreate the TME. Nonetheless, the structural integrity of self-assembling hydrogels can be a limiting factor. To improve on this, our PA-protein coassembling strategy is expected to enhance stability over PA hydrogels and enable higher control over their stiffness (e.g., by varying PA sequences, PA-protein affinities, and PA and protein concentrations), as explored in the subsequent two sections.
Hydrogel stiffness characterization
Ovarian cancer spheroids are highly dependent on matrix stiffness (24). While soft hydrogels [storage modulus (G′) ~ 0.5 kPa] lead to loose cell aggregation, stiff hydrogels (G′ ≥ 7 kPa) inhibit cell proliferation, leading to smaller inhomogeneously shaped spheroids with decreased metabolic activity (31). A similar stiffness dependence is reported for prostate cancer spheroids (32). We tested the hydrogel stiffness by rheometry on acellular PA-VH/KN hydrogels (prepared as described in the “Hydrogel formation and characterization at the nano- and macroscale” section) in either cell culture medium or 10 mM HEPES after 24 hours in comparison to the control hydrogel PA-H/KN. Culture medium was used to assess the behavior of the hydrogels during cell cultures, while HEPES served as control.
In HEPES, no significant stiffness differences were observed between PA-VH/KN (G′ = 1616 ± 198 Pa) and the control PA-H/KN (G′ = 1965 ± 633 Pa) hydrogels (Fig. 2F). Comparing PA-VH/KN hydrogels in culture medium and in HEPES, a significant increase in stiffness was observed for hydrogels in medium (G′ = 2133 ± 111 Pa, P < 0.05; fig. S4A). When leaving the PA-VH/KN hydrogels for 15 days in culture medium, a further increase (G′ = 2633 ± 633 Pa) was observed compared to the 24-hour time point, although not statistically significant (fig. S4A). This kind of culture medium–induced increase in hydrogel stiffness has been reported (33). Overall, our findings show that the coassembling hydrogels are within the optimal stiffness range (G′ > 0.5 kPa and < 7 kPa) reported to support tumor spheroid formation from single-cell encapsulation (31).
Hydrogel stability characterization
Another critical parameter of the ECM is its capacity to degrade and to enable cell and spheroid growth (24). Peptide hydrogels are known to degrade enzymatically through peptide cleavage (34). Conversely, an ideal hydrogel needs to remain sufficiently stable for spheroid formation while enabling ECM secretion and remodeling (35, 36). We tested the hydrogel stability by incubating acellular PA-VH/KN hydrogels and control PA-H/KN hydrogels in cell diluents phosphate-buffered saline (PBS) and HEPES for 3 months at 37°C mimicking cell culture conditions. The hydrogel weight was monitored daily for the first week and subsequently weekly to assess degradation, with the data reported as the weight fraction (Mt) of the weight recorded at the start of the experiment (M0). Similar results were observed for PBS (Fig. 2G) and HEPES (fig. S4B). For PA-VH/KN hydrogels in PBS, the first significant difference in weight compared to the starting weight (M0) was observed after 27 days (M27 = 0.75 ± 0.1 fraction of M0; Fig. 2G), with a weight fraction below 40% remaining after 3 months (M92 = 0.35 ± 0.04 fraction of M0). Conversely, the control PA-H/KN hydrogels degraded comparatively faster. However, both hydrogels remained visibly present after 3 months despite the loss in mass (Fig. 2G). Previous studies have shown that spheroid formation from single cancer cells occurs within ~2 weeks (9, 12, 37), indicating that the PA-VH/KN hydrogels are sufficiently stable to support spheroid formation. The discrepancy between the quantified weight loss and the unchanged hydrogel structure may be attributed to swelling effects of the hydrogels, with >90% water content (22). Overall, PA-VH/KN hydrogels exhibit a suitable level of degradation to enable ECM generation.
Cell monocultures within PA/KN hydrogels generate tumor spheroids
Next, we sought to determine whether the encapsulation of ovarian cancer cells leads to tumor spheroid formation from a single-cell suspension within PA-VH/KN hydrogels compared to cells grown within Matrigel. Spheroid formation occurred over 21 days in PA-VH/KN hydrogels and Matrigel as detected by bright-field and fluorescence microscopy, with a higher spheroid density in Matrigel (Fig. 3, A and B, and fig. S5A) bright-field control of PA/KN. Assaying the metabolic activity as an indicator of cell proliferation, we found that PA-VH/KN hydrogels follow the same trend as Matrigel over 21 days, albeit with slightly lower cell numbers (Fig. 3C). The readout fluorescence count for Matrigel rose to 7600 ± 2037, whereas the PA/KN ended at 2961 ± 491 after 21 days (readout fluorescence values in fig. S5C). Other studies have shown that Matrigel artificially increases the growth rate of tumor cells (38). However, extensive cell detachment and hydrogel fragmentation were observed in Matrigel cultures on days 14 to 21 (fig. S5B), which was not the case for PA-VH/KN hydrogels. This enhanced stability is also evident, for example, in the metabolic activity studies where we retained all replicate PA/KN hydrogels for 21 days, compared to a 40% loss for Matrigel. These results are in alignment with previous studies that have also reported on the poor mechanical properties of Matrigel (38). We speculate that this characteristic may result from the limited mechanical stability of Matrigel, leading to uncontrolled cell migration and matrix digestion and subsequent collagen-induced shrinkage.
Fig. 3. Monoencapsulation of ovarian cancer cells in PA/KN hydrogels and Matrigel (500 cells/μl gel).
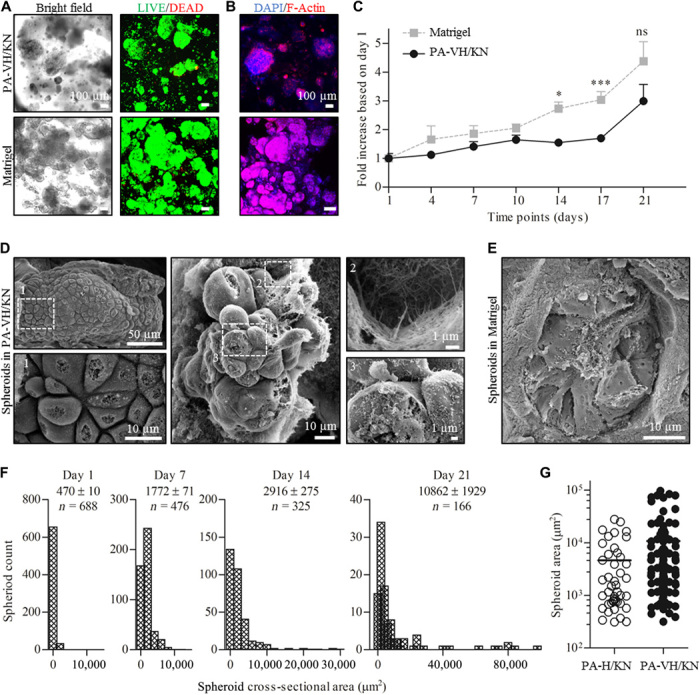
(A) Bright-field and Live (green)/Dead (red) images of ovarian cancer OVCAR-4 cells in PA-VH/KN hydrogels and Matrigel after 21 days of encapsulation. (B) Immunofluorescent (IF) images of F-actin network (phalloidin/red) and nuclei [4′,6-diamidino-2-phenylindole (DAPI)/blue] of ovarian cancer cells in PA-VH/KN hydrogels and Matrigel on day 21. (C) Comparison of the metabolic activity of ovarian cancer cells over time in PA-VH/KN hydrogels and Matrigel using an alamarBlue assay. The plot shows the values normalized to day 1 against time. (D) Scanning electron microscopy images of tumor spheroids grown within PA-VH/KN hydrogels on day 14. The images show the cobble structure of ovarian cancer and the anchoring points to the nanofibrous hydrogel matrix. (E) Scanning electron microscopy images of tumor spheroids grown in Matrigel on day 7, exhibiting comparative cobble structure. (F) IF images were used to measure the spheroid cross-sectional area (μm2), shown here as a Gaussian distribution on days 1, 7, 14, and 21. The graphical plots reveal the shift from single cells at ~500 μm2 to clusters up to 97,500 μm2. (G) PA-H/KN and PA-VH/KN hydrogels were compared by plotting the size distribution of tumor spheroids at day 21, demonstrating a wider distribution for PA-VH/KN hydrogels with the mean shifted above 104 μm2.
To investigate these differences, scanning electron microscopy confirmed the more compact spheroid formation in PA-VH/KN hydrogels compared to Matrigel (Fig. 3D). In PA-VH/KN hydrogels, the typical cobblestone morphology of epithelial cancer cells was observed [Fig. 3D; (1)], with varying degrees of spheroid development through continued proliferation and with defined cell-cell junctions. Cell-matrix interactions between cancer cells and the PA-VH/KN hydrogel were visible, exhibiting cell anchorage to the nanofibrous PA/KN network [Fig. 3D, (2)]. In addition, the spheroids in these hydrogels comprised surfaces covered in microvilli [Fig. 3D, (3)], which is characteristic of epithelial cancer cells and affects the sensitivity to cytotoxic compounds, such as chemotherapeutics (39). Comparatively, cells grown in Matrigel displayed both the cobblestone morphology and homogeneous spreading across cell aggregates (Fig. 3E). In the later stages of 3D cell culture (~day 21), individual spheroids were only distinguishable in PA-VH/KN hydrogels, suggesting less cell migration compared to Matrigel.
Further characterization was conducted by measuring the cross-sectional area of spheroids at several time points (days 1, 7, 14, and 21) using PA-H/KN hydrogels as control. The average spheroid size in PA-VH/KN hydrogels at day 21 (10,862 ± 1929 μm2; Fig. 3F) was comparable to those previously reported using prostate (11) and breast (12) cancer cells at day 14. Some spheroids grown within our PA-VH/KN hydrogels did reach similar sizes to those reported in the literature over 14 days in 3D cell culture. Spheroids formed from single cells and grew over time (Fig. 3F), with individual cancer cells giving way to larger spheroids leading to an overall reduced spheroid count at day 21 compared to day 1. Comparatively, PA-H/KN hydrogels led to smaller spheroids at day 21 (4623 ± 1002 μm2) with a significant decrease in spheroid count across all time points tested (fig. S5D). PA-VH/KN hydrogels exhibited higher propensity for spheroid formation compared to PA-H/KN (Fig. 3G), which was confirmed by microscopy of both cell-containing PA/KN hydrogels over 21 days, with PA-VH/KN hydrogels supporting comparable spheroid growth and size to Matrigel (Fig. 3 and fig. S5E). These findings demonstrate the feasibility of PA/KN hydrogels in supporting tumor spheroid formation from single cells, with PA-VH/KN being favorable over PA-H/KN. Stable spheroid formation occurred in PA-KN hydrogels over 21 days, representing the key advantage over Matrigel-based 3D cell cultures.
PA/KN hydrogels support 3D monocultures of HUVECs and hMSCs
The ovarian TME contains not only malignant cells but also nonmalignant cells that support tumor survival and growth through processes like angiogenesis. As tumors grow, their cores become hypoxic, which, in turn, leads to the expression of proangiogenic proteins and GFs, such as vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) (40). In previous 3D cocultures of HUVECs and cancer cells, HUVECs migrated toward the tumor spheroids (41). We sought to test the suitability of PA-VH/KN hydrogels to support the formation of capillary-like structures by encapsulating HUVECs and hMSCs. First, we explored the compatibility of PA/KN hydrogels with each cell type. Encapsulation of either HUVECs or hMSCs in PA-VH/KN hydrogels showed a high viability of 85 and 75%, respectively, over 7 days (fig. S6, A and B). Thus, PA/KN hydrogels are suitable for multicellular cultures of malignant with nonmalignant cells.
Next, the propensity of HUVECs to form endothelial networks was explored by comparing 3D monocultures of HUVECs in PA-VH/KN hydrogels to Matrigel. At day 2, limited cell spreading and networks were observed in cells grown within PA-VH/KN hydrogels. HUVECs in Matrigel seemed to form networks at the edges; however, these networks disappeared over time and were dissolved after day 5 (fig. S6C). Conversely, when HUVECs were instead seeded on top of PA-VH/PA-RGDS/KN hydrogels and Matrigel, they adhered (Fig. 4A) and interacted with both matrices (fig. S6D). It was not possible to process Matrigel samples for immunostaining because of the gels disintegrating when immersed in paraformaldehyde (PFA), a well-documented issue (Fig. 4B) (6). Using PA-VH/PA-RGDS/KN hydrogels, endothelial cell networks were formed 5 days after seeding (Fig. 4, A and B) and CD31 was detected on day 9 in proximity to tight junctions between the cells [Fig. 4, C (control) and D (sample)]. In comparison to the monolayer control, the CD31 staining of the hydrogels was relatively weak. Nonetheless, the network was confirmed by scanning electron microscopy, which depicted complete HUVEC coverage on top of the PA-VH/PA-RGDS/KN hydrogels (Fig. 4E). As with the encapsulation, networks did not remain intact and started to disappear with no CD31 staining evident after day 9. These results are in line with other studies reporting the formation of a HUVEC monolayer on collagen/chitosan hydrogels (42). Comparatively, cells grown on Matrigel aligned directionally to form tubular polygonal networks, which remained present for 8 days (Fig. 4A and fig. S6E). The network was not verified beyond day 8, as previous studies have shown this timeline to be sufficient (12). Because of the difficulties in fixing Matrigel with PFA (5), we were unable to use these samples for immunofluorescent (IF) analysis. Thus, while neither matrix supported cell spreading in 3D, both matrices promoted adhesion and proliferation when cells were seeded on top.
Fig. 4. HUVEC seeding on top of Matrigel and PA/KN hydrogels.
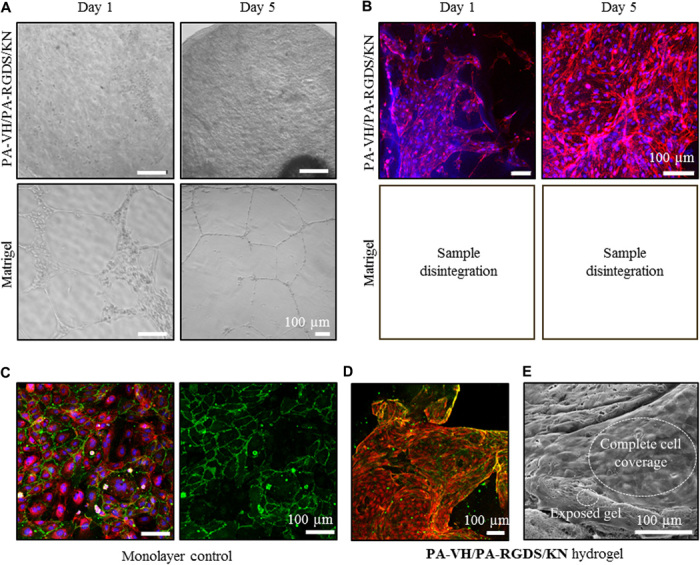
(A) Bright-field images of HUVECs seeded on top of Matrigel and PA-VH/PA-RGDS/KN hydrogels on days 1 and 5, with a seeding density of 1 × 105 cells per gel. (B) IF images of F-actin network (phalloidin/red) and nuclei (DAPI/blue) of HUVECs seeded on top of PA-VH/PA-RGDS/KN hydrogels. Samples from Matrigel disintegrated during sample preparation. (C) Monolayer of HUVECs stained for CD31 (green), nuclei (DAPI/blue), and F-actin network (phalloidin/red) as a positive staining control. In addition, the control was used as references for the signal strength when analyzing the CD31 stain in the PA/KN hydrogels. (D) IF images of CD31 (green) and F-actin network (phalloidin/red) of HUVECs grown on top PA-VH/PA-RGDS/KN hydrogels on day 9. (E) Scanning electron microscopy images of HUVECs covering the PA-VH/PA-RGDS/KN hydrogel surface on day 5.
Overall, our results demonstrate that, while PA/KN hydrogels support encapsulation of HUVECs, additional factors are needed to promote endothelial network structures within these hydrogels. Studies have indicated a role of hMSCs and ECM proteins, such as FN, in promoting capillary-like network formation (43). Thus, the inclusion of bioactive peptides (PA-RGDS or PA-GHK) and proteins in combination with 3D cocultures including hMSCs may further support HUVEC behavior.
3D cocultures of cancer cells with stromal cells in PA/KN hydrogels promote intercellular network formation
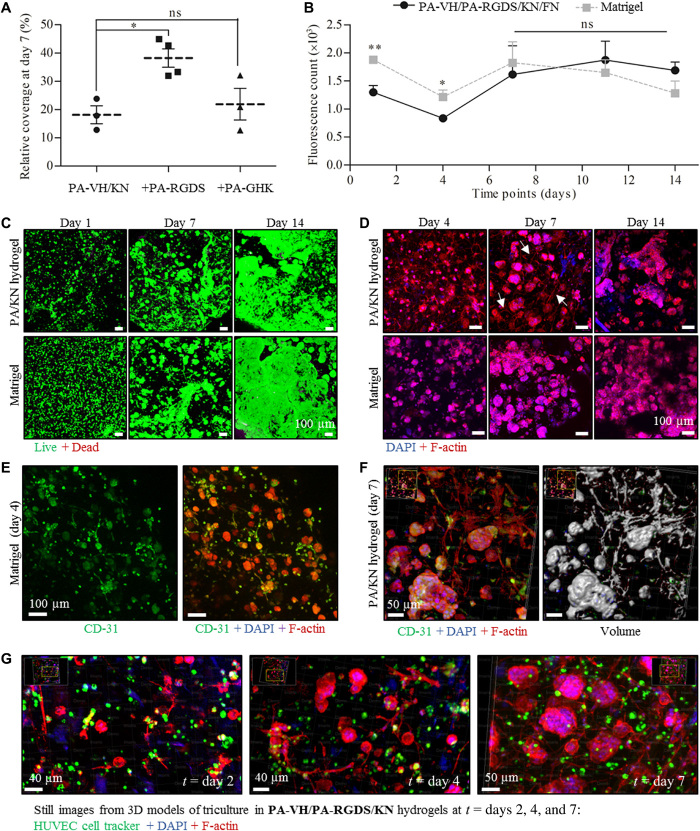
The TME mediates important interactions between tumor and endothelial cells (12). To investigate the potential of PA-VH/KN hydrogels to support cell-cell interactions within a controlled microenvironment, we conducted 3D tricultures of ovarian cancer cells together with HUVECs and hMSCs. Cell spreading was observed in PA-VH/KN hydrogels, which was not the case in 3D monocultures of HUVECs or hMSCs, suggesting a synergistic effect between the three cell types in 3D tricultures. Before in-depth analysis of the cell behavior in 3D tricultures, we first assessed the effect of the three different peptides PA-VH/PA-RGDS (10:1), PA-VH/PA-GHK (10:1), and PA-VH (1:0) and compared the extent of the F-actin networks. Therefore, all three cell types were encapsulated in PA-VH/KN, PA-VH/PA-RGDS/KN, and PA-VH/PA-GHK/KN hydrogels and F-actin stained using rhodamine-conjugated phalloidin (Figs. 1B and 5A). The immunofluorescent (IF) images were analyzed using AngioTool to calculate the area covered by the F-actin network (fig. S7A). A significant increase in F-actin network coverage was observed for PA-VH/PA-RGDS/KN (38 ± 7%) compared to PA-VH/KN (18 ± 6%) hydrogels (P < 0.05). On the other hand, PA-VH/PA-GHK/KN hydrogels did not yield a significant difference in F-actin network formation compared to PA-VH/KN or PA-VH/PA-RGDS/KN hydrogels due to high sample variability as a result of hydrogel instability and small sample range (n ≤ 3). Conversely, consistent results with smaller variability were obtained for PA-VH/PA-RGDS/KN hydrogels over a large sample range (n ≥ 6). When comparing the use of GHK to RGDS, cells grown within PA-VH/PA-GHK/KN hydrogels exhibited a similar level of spreading compared to cells grown within PA-VH/PA-RGDS/KN. This may be because of the angiogenic potential of GHK (44). Considering the variability in results obtained with PA-VH/PA-GHK, the peptide mixture PA-VH/PA-RGDS was found most favorable. Consequently, we used PA-VH/PA-RGDS/KN with and without FN to evaluate cell behaviors in comparison to Matrigel (Fig. 1B).
Fig. 5. Triencapsulation of ovarian cancer cells, HUVECs, and hMSCs in PA/KN hydrogels and Matrigel (ovarian cancer/HUVEC/hMSC ratio of 560:6000:600).
(A) Quantification of F-actin network coverage as percentage area coverage for different PA/KN hydrogel combinations. (B) Metabolic activity of tricultures over time in PA-VH/PA-RGDS/KN/FN hydrogels and Matrigel (values normalized to day 1). (C) Live (green)/Dead (red) images of tricultures in PA-VH/PA-RGDS/KN hydrogels and Matrigel on days 1, 7, and 14. (D) IF staining of F-actin network (phalloidin/red) and nuclei (DAPI/blue) of tricultures grown in PA-VH/PA-RGDS/KN/FN hydrogels and Matrigel on days 1, 7, and 14. (E) IF staining for CD31 (green), F-actin network (phalloidin/red), and nuclei (DAPI/blue) in Matrigel on day 4. (F) IF staining for CD31 (green), F-actin network (phalloidin/red), and nuclei (DAPI/blue) in PA-VH/PA-RGDS/KN hydrogel on day 7 with a 3D volume reconstruction (right). (G) Still video images of cell proliferation and spreading in PA-VH/PA-RGDS/KN hydrogels from days 2 to 7 (HUVECs labeled with a green cell tracker, F-actin network with red phalloidin, and nuclei with blue DAPI).
First, we determined the cell proliferation in 3D tricultures in PA-VH/PA-RGDS/KN(/FN) hydrogels compared to Matrigel. On the basis of the metabolic activity, a similar cell proliferation was observed in both matrices (Fig. 5B; readout fluorescence values in fig. S7B). There was an initial decrease in metabolic activity on day 4, most likely as a result of minor HUVEC and hMSC death, followed by a steady increase in metabolic activity. While cells in the PA-VH/PA-RGDS/KN(/FN) hydrogels showed an initial lower metabolic activity compared to Matrigel, no difference was observed from days 8 to 14. Whereas the cell monocultures in Matrigel were prone to gel fragmentation, the tricultures in Matrigel contracted in size over the 14 days. We speculate that this contractile behavior was due to the relatively high cell density in these cultures, increasing the number of cell-cell interactions.
Next, we compared spheroid formation in the 3D tricultures in PA-VH/PA-RGDS/KN(/FN) hydrogels and Matrigel to the spheroid data from the 3D monocultures (see the “Cell monocultures within PA/KN hydrogels generate tumor spheroids” section). In both matrices, we observed an immediate increase in spheroid formation and size compared to the monoculture counterparts, as detected by IF microscopy (Fig. 5, C and D; Live/Dead assay and bright-field images in fig. S7C). Comparing 3D mono- and tricultures within PA/KN hydrogels on day 14, we detected an increase in spheroid size in the tricultures (Fig. 5C versus Fig. 3B). Spheroid sizes observed in 3D monocultures in PA-VH/KN on day 21 (discussed in the “Hydrogel formation and characterization at the nano- and macroscale” section) were now observed by day 14 in the tricultures using PA-VH/PA-RGDS/KN(/FN) hydrogels. This accelerated growth may be the result of enhanced intercellular interactions between the different cell types, which have been associated with spheroid growth (45). No significant difference was detected between PA-VH/PA-RGDS/KN and PA-VH/PA-RGDS/KN/FN hydrogels on the basis of the fluorescent micrographs and proliferation data. We hypothesize that this may be a reflection of keratin-containing adhesion sequences that mask the effect of FN. However, to determine the influence of FN in our system, a more detailed analysis of the cell expression is required. Thus, on the basis of the comparative results without FN, our subsequent work focused on PA-VH/PA-RGDS/KN. Overall, we conclude that not only 3D tricultures in PA-VH/PA-RGDS/KN exhibit enhanced spheroid size and growth compared to 3D monocultures but also they are comparable to Matrigel.
Having evaluated the changes in spheroid formation between 3D tri- and monocultures, next we assessed the F-actin network formation in our PA/KN hydrogels. As discussed, 3D tricultures in PA-VH/PA-RGDS/KN led to an extensive F-actin network formation, which was absent in our stromal cell monocultures (see the “Cell monocultures within PA/KN hydrogels generate tumor spheroids” section). Using IF images, we performed a detailed image analysis on our 3D tricultures to determine the origin of the F-actin networks within PA-VH/PA-RGDS/KN hydrogels. The presence of hMSCs was confirmed by staining for 𝛼-smooth muscle actin (𝛼-SMA), which was aligned with the F-actin network (fig. S8A). We hypothesize that the presence of the F-actin network corresponds to the beginning of angiogenesis. To test this, we investigated the position of HUVECs within the hydrogels. Cells grown within PA-VH/PA-RGDS/KN hydrogels and Matrigel were stained with CD31, phalloidin/F-actin and 4′,6-diamidino-2-phenylindole (DAPI)/nuclei (Fig. 5E and fig. S8B). CD31 was stained positive in both matrices; however, unspecific staining was observed in the tumor spheroids. Thus, from the immunostaining alone, the F-actin network cannot be interpreted as the beginning of angiogenesis from HUVEC spreading. However, from the IF images, we created pseudo-volume filled images (Fig. 5F and fig. S8C). These images revealed that F-actin networks were integrated between adjacent spheroids, confirming that intracellular interactions between stromal and cancer cells play a role in the increased spheroid size observed in 3D tricultures compared to monocultures.
In a second attempt to elucidate the HUVEC behavior within the PA/KN hydrogel, we used a cell tracker dye to label HUVECs, allowing cell position to be tracked in relation to cell spreading over 14 days (Fig. 5G and fig. S8D). The image sequence confirmed that the F-actin network formation took place between days 2 and 7 (Fig. 5G), with a decreasing network by day 14, when instead large spheroids were visible (fig. S8E). Analyzing the cell tracker staining, some of the HUVECs coincide with the F-actin network. However, the majority was outside the F-actin network, suggesting that it did not originate from HUVECs. Thus, further optimization of the hydrogel formulation is required to promote tumor angiogenesis within PA/KN hydrogels. However, the images demonstrate the continuous cell spreading as evident by the buildup of F-actin networks. In summary, our 3D tricultures promote intercellular interactions in PA/KN hydrogels, while highlighting the positive impact of stromal cells on spheroid size and growth rate.
Responses to treatment in spheroid-containing PA/KN hydrogels
We have demonstrated the consistent tumor spheroid formation in our PA/KN hydrogels, as well as the benefits and added accuracy in modeling the TME by incorporating different cell populations. We have shown the versatility of the PA/KN hydrogels by changing PA sequences and protein composition to tailor the model’s applicability to the ovarian TME. To assess the functionality of our hydrogels, we tested the responses of tumor spheroids grown in PA/KN hydrogels to three different treatments: the matrix metalloproteinase inhibitor and antiangiogenic agent GM6001 and the first-line chemotherapeutics paclitaxel (TXL) and carboplatin (PLT) (Fig. 6A) (9, 31). The dosages used in this study for GM6001 (20 μM), TXL (100 μM), and PLT (100 μM) are commonly used for in vitro drug testing (6, 9, 31) and thus allow comparison with previous studies. For the treatment studies, the fold change in metabolic activity was normalized to day 1 to allow comparison between the different treatments. The readout values are in fig. S10.
Fig. 6. Treatment study with TXL, PLT, and GM6001 using cells grown in PA-VH/KN hydrogels and Matrigel.
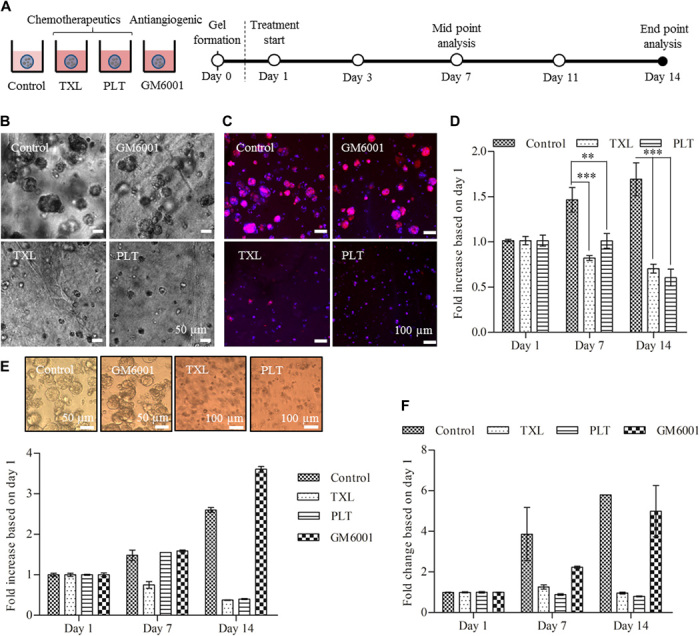
(A) Schematic showing the experimental setup for the treatment studies. Ovarian cancer cells (OVCAR-4) were encapsulated at a density of 500 cells/μl with treatment starting 1 day after encapsulation. (B) Bright-field images of the tumor spheroid sizes after 14 days for the control and three treatments. (C) IF images of the F-actin network (phalloidin/red) and nuclei (DAPI/blue) of tumor spheroids on day 14 for all treatment conditions. The IF images correspond to the bright-field images in (B). (D) Metabolic activity of cells grown in PA-VH/KN hydrogels for all treatments over 14 days (values normalized to day 1), measured using an almarBlue assay. (E) Metabolic activity of cells grown in Matrigel for all treatments over 14 days (values normalized to day 1), measured using an almarBlue assay. (F) Metabolic activity of tricultures (ovarian cancer cell/HUVEC/hMSC ratio of 560:6000:600) in PA/KN hydrogels for all treatments over 14 days (values normalized to day 1), measured using almarBlue assay.
Ovarian cancer cells were encapsulated in PA-VH/KN hydrogels and Matrigel and treated 1 day after encapsulation for 14 days, with treatment changes every 2 to 3 days (control hydrogels at day 1; fig. S9A). While GM6001 did not affect spheroid size and metabolic activity (fig. S9B), as reported previously (9), TXL and PLT prevented spheroid formation as detected by bright-field and fluorescence microscopy (Fig. 6, B and C). A small number of cancer cells were still present after 14 days of treatment, although with lower metabolic activity compared to controls on day 1 (TXL, 0.70 ± 0.05; PLT, 0.61 ± 0.09; control, 1.70 ± 0.18; Fig. 6D). There was no difference in the response to both chemotherapeutics. When comparing the response in spheroid-containing Matrigel (Fig. 6E), TXL and PLT treatment appeared more effective than in PA-VH/KN hydrogels, with a very low metabolic activity observed at day 14 compared to day 1 (TXL, 0.38 ± 0.02; PLT, 0.40 ± 0.02; control, 2.60 ± 0.05). However, previous studies have shown that drug uptake within Matrigel is unrealistically high, resulting in uptake values similar to those in 2D cell cultures (38). Thus, we conclude that cellular responses in PA/KN hydrogels are more physiological than Matrigel.
To further explore the effect of treatment, we conducted a brief study using PA/KN tricultures. Our preliminary results indicated a similar behavior to that of 3D monocultures, with reduced metabolic activity upon TXL and PLT treatment, however, with minor effects of GM6001 (Fig. 6E and fig. S9D). Interference from GM6001 may suggest inhibition of the network structures formed by HUVECs and hMSCs as previously reported (46). This aspect may be explored in future experiments. A follow-up study may also include the separation of multiple cell types after 3D culture for separate analysis to measure the effect of treatment. Within the scope of this study, the experiments with chemotherapeutics have shown the anticipated response in ovarian cancer cells.
DISCUSSION
There is a need to advance 3D cancer models to capture the key components of the TME and to screen improved or personalized therapeutics. Supramolecular hydrogels offer an opportunity to replicate the nanofibrous features of the extracellular microenvironment as well as include desired signaling epitopes. These self-assembling systems can also be engineered to interact with and incorporate key ECM components. Here, we demonstrate that PA/protein coassembling bioinks can model the TME of ovarian cancer to a level that is similar to, yet more controlled than, that provided by Matrigel.
Ovarian cancer spheroids were grown over 21 days and reached a similar cross-sectional size compared to previous studies (9). We did not observe a hypoxic core found in many spheroids or tumors. However, this phenomenon is only observed when the spheroids or tumor mass reaches a diameter of >200 μm, at which point diffusion is inhibited, eventually leading to a hypoxic core (47). We have shown the PA/KN hydrogel’s compatibility with multiple cell types and ability to selectively modify the peptide sequences to optimize cellular responses. In 3D tricultures, tumor spheroids interacted with HUVECs and hMSCs as well as the hydrogel matrix and their proliferation were enhanced. F-actin networks were formed throughout the hydrogel and intercalated between adjacent spheroids. The extent of network formation was enhanced by using the bioactive sequences RGDS and GHK. Many other sequences, for example, IKVAV and GFOGER (12), have been shown to specifically promote angiogenesis, which may be used instead of GHK. In addition, GFs, such as VEGF or FGF, may be entrapped within the fibrous network to allow slow release. We validated the PA/KN hydrogels using cytotoxic drugs, with cellular responses observed for all tested treatments.
In this study, we primarily assess the performance of the PA/KN hydrogel while comparing it with Matrigel, showing similar results in most of the performed experiments. In spheroid monocultures, Matrigel led to an increased proliferation compared to PA/KN hydrogels, which may result from Matrigel’s undefined composition of ECM proteins (primarily collagen, laminin, and proteoglycans) and GFs. However, the PA/KN hydrogels provide adaptability by permitting the inclusion of keratins that are found in ovarian cancer tissue (KRT32 and KRT82). In 3D tricultures, we did not detect a significant difference in proliferation between Matrigel and PA/KN hydrogels. Furthermore, the two materials exhibited comparative levels of intracellular networks. Major advantages of our PA/KN hydrogel over Matrigel is the possibility to improve reproducibility and minimize potential batch-to-batch variations, while providing an ECM-like nanofibrous network able to maintain comparative cell viability to the current gold standard, Matrigel. As a result, these hydrogels offer an exciting possibility for the design of 3D disease models.
Similarly, other studies assessing the advantages and disadvantages of using basement membrane–derived materials compared with synthetic materials have found that synthetic hydrogels offer more control, reproducibility, and versatility. For example, Bray et al. (11) reported comparative results between starPEG-heparin hydrogels and Matrigel, with the polymer hydrogels offering controlled mechanical properties, integrin-binding sequences, and quantity of GFs, which enabled testing of a range of therapeutics and inhibitors. Along with PEG-based hydrogels, gelatin methacryloyl (GelMA) has been used as a 3D ovarian cancer model. For example, Kaemmerer et al. (31) reported that GelMA supports tumor spheroid formation and sufficient porosity for treatment with anticancer drugs. These studies also highlight similar Matrigel limitations as we have reported here. Furthermore, by cross-comparing GelMA/PEG versus Matrigel studies to our PA/KN hydrogel versus Matrigel study, we see that the PA/KN hydrogels yield comparative performance. For example, the tumor sizes observed in our tricultures match those in studies using PEG (11) and GelMA (12) within a similar timeline (100 to 200 μm in diameter after 14 days). Taubenberger et al. reported the formation of endothelial networks in PEG-heparin gels sustainable over 10 days grown in cocultures of HUVECs and MSCs. Moreover, they observed that the inclusion of pregrown tumors enhanced angiogenesis. Similarly, we observed that PA/KN hydrogels support endothelial network formation in cocultures of MSCs and ovarian cancer cells, albeit not sustainable for 10 days. Last, the advantages that the porosity of GelMA offers for drug treatment (31) are also present in our PA/KN hydrogels as shown in Fig. 6. While PEG- and GelMA-based models are established hydrogels used normally in 3D cancer models, PA/KN hydrogels offer additional benefits. For example, PA/KN hydrogels exhibit both a nanofibrous network that resembles that of the native ECM, which neither PEG-based nor GelMA-based hydrogels recreate. Moreover, the peptide/protein coassembly strategy reported here permits incorporation of naturally occurring proteins in a manner that enables them to play both a structural and a signaling role. This emulates the way these macromolecules function in the native ECM. Overall, these hybrid hydrogels enable the design of 3D matrixes with controlled of molecular composition, nanofibrous architecture, and the capacity to present both specific bioactive epitopes and macromolecules.
Some of the limitations of PA-based hydrogels are cost of material and scalability. However, peptide-based self-assembling hydrogels offer attractive opportunities for in vitro applications, which is evidenced by a number of recent commercially available materials. Examples include Biogelx, which is currently being used in 3D breast cancer models for screening of new drugs; PeptiGels from Manchester Biogel, which have been used for postendoscopic treatment; and PuraStat from 3D Matrix, which has been developed for hemostasis. While these systems demonstrate that peptide-based self-assembling hydrogels are viable products for 3D culture applications, Matrigel remains the material of choice. Our study demonstrates that self-assembling materials can be rationally coassembled with ECM macromolecules to improve molecular and structural complexity while maintaining a high degree of control. Hence, it is possible to easily explore more complex hydrogels in a systematic manner, enabling the design of 3D microenvironments with high molecular diversity and reproducibility.
It is important to mention that the PA/KN coassembling system can be easily integrated with 3D bioprinting to provide macroscale precision and reproducibility (22). Bioprinting has already been explored to build 3D cancer models (48, 49). However, the nanofibrous network and the selective presentation of bioactive epitopes are unique properties to self-assembling materials and are known to directly affect cell behavior. While 3D bioprinting was not used in this study, the potential of the system to be used as a bioink offers additional opportunities for building more complex, yet controlled and reproducible, 3D cancer models (50). Nonetheless, in this study, we have demonstrated the possibility to use our PA/KN system to engineer an innovative 3D ovarian cancer model with a high degree of molecular versatility and tunability, enabling more effective testing of anticancer therapeutics.
METHODS
PAs (synthesis and purification)
PA-H (purity, 97.8%), PA-VH (purity, 98.9%), PA-GHK (purity, 99.5%), and PA-RGDS (purity, 99.2%) were synthesized in-house (fig. S1) as previously described (22). Additional PA-H (purity, 98.1%) was purchased from Biomatik (ON, Canada). In-house synthesis was carried out using an automatic microwave assisted solid-phase peptide synthesizer (SPPS) (CEM Liberty Blue) using 4-methylbenzhydrylamine Rink amide resin (0.52 mmol/g, Novabiochem Corporation, UK) with the tail coupled subsequently. Purification of all peptides was carried out through reverse-phase high-performance liquid chromatography (HPLC) using a 2545 Binary Gradient Preparative HPLC (Waters 2767, Waters, USA), a C18 column (Atlantis Prep OBD T3 Column, Waters, USA) and a gradient of 2 to 100% acetonitrile in water [0.1% (v/v) trifluoroacetic acid] over 40 min. Detection was carried out concomitantly with 2489 UV/Vis and an electrospray ionization mass spectrometer (ESI-MS) detector (Waters, USA). Peptide purity was determined by analytical HPLC as well as ESI-MS (Thermo LXQ, Thermo Fisher Scientific, USA). Purity ≥95% was accepted suitable for usage. Peptides were freeze-dried to yield lyophilized powder. Peptide solutions were prepared using 10 mM HEPES, (pH 7.5).
Matrigel
Matrigel (Corning Matrigel Growth Factor Reduced Basement Membrane Matrix, LDEV-Free) was purchased from Scientific Laboratory Supplies (catalog no. 356230). To minimize the potential influence of batch-to-batch variations of Matrigel, only one batch of Matrigel was used for this study (LOT 7202001).
Proteins
Keratin was extracted from human hair and freeze-dried using a modified protocol (51), which yields mainly in intact hair keratin. Bovine-derived FN was purchased from R&D Systems (1030-FN) provided as 1 mg/ml in a 400 mM urea, 500 mM NaCl, and 50 mM tris-HCl solution (pH 7.5). Peptide solutions were prepared using 10 mM HEPES, (pH 7.5).
Zeta potential
Samples (n = 3 per condition) were prepared at a concentration of 1 mg/ml in 10 mM HEPES and filtered before measuring on a Malvern Nano ZS series Zetasizer, with each measurement repeated three times.
Circular dichroism
The experiments were run at concentration 0.1 mg/ml in 10 mM HEPES buffer for both PA and keratin. Measurements were carried out at 25°C using a 0.1-cm–path length and 300-μl-volume cuvette (Chirascan, Applied Photophysics, UK). Three scans were run per sample and at least three independent repeats per sample condition (n ≥ 3). The data were normalized to the baseline (blank control; 10 mM HEPES buffer) and the averaged trace smoothed to reduce noise without causing spectrum distortion. The data manipulation was carried out using the Chircascan trace manipulation software.
Transmission electron microscopy
Samples were prepared at a concentration of 0.01 mg/ml in 10 mM HEPES. Carbon copper grids (Agar Scientific, Stansted, UK) were used to mount the samples. Excess was removed using filter paper before incubation with 2% filtered uranyl acetate solution for 30 s. Grids were then washed with ultrapure water for 30 s, air-dried for 24 hours at room temperature (RT) and imaged using a JEOL 1230 transmission electron microscope operated at an acceleration voltage of 80 kV. All the images were recorded by a Morada charge-coupled device camera (Image Systems). At least 10 images were taken in random locations per condition.
Degradation study
Hydrogels were formed using PA-VH and PA-H by injecting 30 μl of PA into 70 μl of KN, incubated over 4 hours before being transferred into vials filled with PBS or 10 mM HEPES. Hydrogels were kept in excess buffer (PBS or HEPES) to prevent false negatives as a result of dehydration. At predefined time points (daily in the first week, then biweekly; PBS, n = 6 per time point; 10 mM HEPES, n = 4 per time point), the solution was removed from the vial and the vial with hydrogel weighed. Liquid removal was removed with a pipette and a small cotton swab. After 3 months, the final vial with hydrogel weight was recorded, and the vial was cleaned and weighed. Mass loss was calculated as “M0 − MTime point/M0,” where M0 is the weight at day zero minus the weight of the vial.
Rheology
Hydrogels were prepared as 30 μl of PA in 70 μl of KN and formed for 4 hours at RT before being transferred to a 10 mM HEPES (n ≥ 6) solution or medium [Dulbecco’s Modified Eagle’s Medium (DMEM); n ≥ 3] and subsequently stored overnight before measurement. Samples were analyzed using a Discovery Hybrid Rheometer (DHR-3, TA Instruments, USA) equipped with an 8-mm-diameter parallel plate geometry. Rheological characteristics were monitored by amplitude sweep and frequency sweep. G′ (storage modulus) and G″ (loss modulus) were measured at 25°C and at a constant frequency of 1 Hz in the 0.01 to 10% strain during the amplitude sweep, while the oscillation frequency experiments were carried out at a 0.1% fixed strain along 0.1 to 100 Hz.
Scanning electron microscopy
Samples were fixed using 4% PFA solution for 2 hours at RT. Then, the samples were dehydrated from a 100% water solution to a 100% ethanol solution in serial steps and dried using a critical point drier, K850 (Quorum Technologies), exchanging the ethanol for carbon dioxide. Carbon black tape was used to adhere the samples to metal stubs. Before imaging, samples were cut open to reveal the inside of the hydrogel, gold coated for 30 to 45 s, and imaged on a FEI Inspect F.
Cell culture
Human epithelial ovarian cancer cells (NIH:OVCAR-4) and hMSCs (PromoCell C-12974 hMSC-BM-c) were maintained in DMEM (4.5 g/liter d-glucose) with GlutaMAX, supplemented with 1% penicillin-streptomycin (P/S) and 10% fetal bovine serum (FBS). HUVECs were purchased from PromoCell (pooled donors, C-12203) and grown in endothelial cell growth medium (C-22010; PromoCell). HUVECs and hMSCs were used at passage 2 (P2) to passage 7 (P7) and NIH:OvCar-4 at < P20. Monocultures were placed in DMEM supplemented with 1% P/S and 10% FBS. Tricultures were placed in endothelial cell growth medium, supplemented with VEGF (2 ng/ml). Treatments were carried out using DMEM without P/S and FBS.
2D cell cultures
For cell seeding experiments with HUVECs, a density 10,000 cells per gel was used. The cells were suspended in cell media and added to a well plate with preformed hydrogels (Matrigel or PA/KN hydrogels). In the case of PA/KN gels, HUVECs were suspended in a KN solution (10 mg/ml), and 70 μl of the KN/cell solution (~140 cells/μl) was added per well in a 96-well plate. Subsequently, 15 μl of PA was injected into the KN/cell solution and incubated for 30 min at 37°C on a shaker. One-day post-seeding hydrogels were transferred into a 48-well plate. In the case of Matrigel, the HUVECs were suspended in media and added to the preformed gels (10,000 cells per gel) and incubated for 30 min at 37°C on a shaker. One-day post-seeding hydrogels were transferred into a 48-well plate.
3D cell cultures
For cell encapsulation, lyophilized PA and proteins were treated with ultraviolet light for 30 min before dissolving in 10 mM HEPES at a concentration of 10 mg/ml. Cells were suspended in PA solution or Matrigel using the following cell densities:
• Stromal cell viability: 1000 HUVECs/μl and 600 hMSCs/μl, respectively.
• HUVEC encapsulation study: 1000 cells/μl.
• Monoculture: 500 OVCAR-4/μl.
• Triculture: 6000 HUVECs/μl, 600 hMSCs/μl, and 560 OVCAR-4/μl.
Polydimethylsiloxane (PDMS) squares were prepared in petri dishes (PDMS precut and autoclaved before use), and 45 μl of protein solution was pipetted onto each PDMS square. Subsequently, 15 μl of PA (with/without cells) was injected into the protein droplet. Hydrogels were formed for 30 min at RT prior and transferred into a 48-well plate with culture medium. Peptide sequences were used as PA-H or PA-VH (10 mg/ml) or in combination 10:1 with either PA-GHK or PA-RGDS (10 mg/ml), i.e., 10 μl of PA-RGDS or PA-GHK (10 mg/ml) per 100 μl of PA-VH (10 mg/ml). KN was used at a concentration of 10 mg/ml either alone or with addition of FN (1 mg/ml) at 100 μl FN per 900 μl of KN. As controls, undiluted Matrigel (GF reduced; Corning) was mixed with cells, and 15 μl was pipetted onto each PDMS square. Samples were incubated for 30 min at 37°C and transferred into a 48-well plate with culture medium. All 3D cell culture experiments were performed in biological replicates with at least triplicate samples.
alamarBlue and Live/Dead assays
alamarBlue reagent was used at a concentration of 700 μl in 10-ml medium (7%). Therefore, culture medium was removed, and 300 μl of alamarBlue solution was added and incubated for 6 hours including three control samples. Subsequently, fluorescent signals of 100-μl samples were measured in triplicate on a FLUOstar OPTIMA (BMG Laboratory) plate reader (excitation, 544 nm; emission, 590 nm) using a black 96-well plate. Media without cell-laden hydrogels with alamarBlue reagent served as negative control for background reading. The signal of pure media (no alamarBlue) was subtracted from all samples. Subsequently, the readout values were divided by the background reading (Media + alamarBlue + no cells), following the simplified calculation from Thermo Fisher Scientific. The corrected fluorescent signals were normalized to day 1. The uncorrected values can be found in the Supplementary Materials (figs. S5C, S7B, and S10). For Live/Dead staining, 2-μl ethidium homodimer-1 (2 mM) and 1-μl calcein AM (4 mM) were added to 1 ml of culture medium, and samples were imaged after 15 min using a confocal microscope. A monolayer was used as a control to check the Live/Dead staining.
Immunohistochemistry
Slides of normal and cancerous ovarian tissue were prepared as serial formalin-fixed paraffin-embedded tissue sections (5 μm). Sections were deparaffinized in xylene and rehydrated in dilutions of ethanol and water. Antigen retrieval was performed using 1× citrate-based solution (pH 6; H-3300; Vector Laboratories) at 95°C for 10 min. Then, sections were washed with 1× tris-buffered saline–Tween (TBS-T), blocked with 5% bovine serum albumin (BSA)/PBS for 1 hour. at RT and treated with 3% H2O2. Antibodies (KRT32 ABIN517410 and KRT82 ABIN651240; Antibodies-online) were diluted 1:50 in 5% BSA/PBS and incubated overnight at 4°C, followed by biotinylated goat anti-mouse immunoglobulin G (BA-9200; Vector Laboratories) at 1:200 in 5% BSA/PBS for 45 min. Samples were washed with 1× TBS-T and stained using an ABC HRP Elite kit (PK-6100-NB; Vectastain) and hematoxylin (GHS116; Sigma-Aldrich), mounted, and imaged using a NanoZoomer S210 (Hamamatsu). Staining controls without the primary antibody, as well as without primary and secondary, were included for all experiments. Examples of the controls for no primary are in fig. S2.
IF staining and confocal microscopy
DAPI (D1306) and rhodamine phalloidin (R415) were purchased from Sigma-Aldrich. Primary (CD31 ab32457, 𝛼-SMA ab7817, KRT32 ABIN517410, and KRT82 ABIN651240) and secondary (Alexa Fluor 488/555/568/594) antibodies were purchased from Abcam, Antibodies-online, and Life Technologies, respectively. Samples were fixed in 4% PFA/PBS for 2 hours, permeabilized in 0.2% Triton X-100/PBS for 2 hours and blocked in 2% BSA/PBS overnight at RT. Primary antibodies were diluted 1:100 in 2% BSA/PBS and incubated for 48 hours at 4°C. Secondary antibodies were diluted 1:1000 in 2% BSA/PBS and incubated overnight at 4°C. An Alexa Fluor 488–conjugated antibody (303110, BioLegend) was used at 1:100 in 2% BSA/PBS overnight at 4°C. DAPI and rhodamine phalloidin were diluted 1:500 and 1:250, respectively, in PBS and incubated for 2 hours at RT. Samples were washed with PBS between each step. A green cell tracker dye (CMFDA C2925; Life Technologies) was also used to label HUVECs by adding 4 mM to the cell suspension for 30 min before cell encapsulation into the hydrogels. Fluorescent images were acquired using Leica TCS SP2 and Zeiss LSM710 confocal microscopes. Samples were imaged acquiring z-stacks ranging from 5 to 20 μm depending on the sample size. The PA/KN hydrogels were checked for autofluorescence before staining. In addition, bright-field images were taken of PA hydrogels without cells after 21 days of incubation.
Image analysis
ImageJ (version l.52i) was used to compile z-stacks and adjust colors. AngioTool (version 0.5a) was used the measure the density of F-actin networks (fig. S7A). Imaris (version x64 9.2.1) was used to create 3D videos of the z-stacks and volume images. Spheroid cross-sectional area was calculated using ImageJ, with cutoff points for days 1 and 4 at 100 μm2 and for days 7 and 14 at 300 μm2.
Treatment with cytotoxic drugs and inhibitor
3D cell monocultures were treated on day 1 using GM6001 (20 μM), TXL (100 μM), and PLT (100 μM). Treatment solutions were prepared in DMEM without supplements and exchanged every 2 to 3 days. As controls, medium changes were performed without treatment.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0 (version 8.0.2). One-way analysis of variance for multiple comparisons was used. Nonparametric statistics were used when the samples did not present a normal distribution (Mann-Whitney test). Statistical significance was set to P < 0.05 (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Acknowledgments
Funding: We would like to acknowledge the funding from the ERC Starting Grant (STROFUNSCAFF), Medical Research Council (UK Regenerative Medicine Platform Acellular/Smart Materials-3D Architecture, MR/R015651/1), and the Program for Innovation and Human Capital from the Ministry of Science, Technology, and Telecommunications of the Government of Costa Rica (MICITT-PINN-PED-014-2015-2). We are grateful to Barts Gynae Tissue Bank for collecting the tissue specimen and S. Sharma for assistance with the immunohistochemistry. Author contributions: C.L.H., D.L., and A.M. conceived the study. C.L.H. performed the experiments. C.R.-G. purified the PAs and assisted with the TEM and rheology study. D.L. and C.L.H. designed the cell study. K.W.N. and B.Y.T. provided the keratin and insights to its interaction. D.L. and A.M. provided facilities for experiments. C.L.H., D.L., and A.M. interpreted the data and wrote the manuscript. All authors discussed the results and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials (figs. S1 to S10). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/40/eabb3298/DC1
REFERENCES AND NOTES
- 1.Wong C. H., Siah K. W., Lo A. W., Corrigendum: Estimation of clinical trial success rates and related parameters. Biostatistics 20, 366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrmann D., Conway J. R. W., Vennin C., Magenau A., Hughes W. E., Morton J. P., Timpson P., Three-dimensional cancer models mimic cell-matrix interactions in the tumour microenvironment. Carcinogenesis 35, 1671–1679 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Mitchell M. J., Jain R. K., Langer R., Engineering and physical sciences in oncology: Challenges and opportunities. Nat. Rev. Cancer 17, 659–675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holle A. W., Young J. L., Spatz J. P., In vitro cancer cell–ECM interactions inform in vivo cancer treatment. Adv. Drug Deliv. Rev. 97, 270–279 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Stock K., Estrada M. F., Vidic S., Gjerde K., Rudisch A., Santo V. E., Barbier M., Blom S., Arundkar S. C., Selvam I., Osswald A., Stein Y., Gruenewald S., Brito C., van Weerden W., Rotter V., Boghaert E., Oren M., Sommergruber W., Chong Y., de Hoogt R., Graeser R., Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery. Sci. Rep. 6, 28951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M. E., Ordóñez-Morán P., Clevers H., Lutolf M. P., Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Suo A., Xu W., Wang Y., Sun T., Ji L., Qian J., Dual-degradable and injectable hyaluronic acid hydrogel mimicking extracellular matrix for 3D culture of breast cancer MCF-7 cells. Carbohydr. Polym. 211, 336–348 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Li W., Hu X., Wang S., Xing Y., Wang H., Nie Y., Liu T., Song K., Multiple comparisons of three different sources of biomaterials in the application of tumor tissue engineering in vitro and in vivo. Int. J. Biol. Macromol. 130, 166–176 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Loessner D., Stok K. S., Lutolf M. P., Hutmacher D. W., Clements J. A., Rizzi S. C., Bioengineered 3D platform to explore cell–ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 31, 8494–8506 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Pradhan S., Clary J. M., Seliktar D., Lipke E. A., A three-dimensional spheroidal cancer model based on PEG-fibrinogen hydrogel microspheres. Biomaterials 115, 141–154 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Bray L. J., Binner M., Holzheu A., Friedrichs J., Freudenberg U., Hutmacher D. W., Werner C., Multi-parametric hydrogels support 3D in vitro bioengineered microenvironment models of tumour angiogenesis. Biomaterials 53, 609–620 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Taubenberger A. V., Bray L. J., Haller B., Shaposhnykov A., Binner M., Freudenberg U., Guck J., Werner C., 3D extracellular matrix interactions modulate tumour cell growth, invasion and angiogenesis in engineered tumour microenvironments. Acta Biomater. 36, 73–85 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Radvar E., Azevedo H. S., Supramolecular peptide/polymer hybrid hydrogels for biomedical applications. Macromol. Biosci. 19, 1800221 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Zhang S., Greenfield M. A., Mata A., Palmer L. C., Bitton R., Mantei J. R., Aparicio C., de la Cruz M. O., Stupp S. I., A self-assembly pathway to aligned monodomain gels. Nat. Mater. 9, 594–601 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou M., Smith A. M., Das A. K., Hodson N. W., Collins R. F., Ulijn R. V., Gough J. E., Self-assembled peptide-based hydrogels as scaffolds for anchorage-dependent cells. Biomaterials 30, 2523–2530 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Kumar V. A., Taylor N. L., Shi S., Wang B. K., Jalan A. A., Kang M. K., Wickremasinghe N. C., Hartgerink J. D., Highly angiogenic peptide nanofibers. ACS Nano 9, 860–868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartgerink J. D., Beniash E., Stupp S. I., Peptide-amphiphile nanofibers: A versatile scaffold for the preparation of self-assembling materials. Proc. Natl. Acad. Sci. U.S.A. 99, 5133–5138 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sieminski A. L., Was A. S., Kim G., Gong H., Kamm R. D., The stiffness of three-dimensional ionic self-assembling peptide gels affects the extent of capillary-like network formation. Cell Biochem. Biophys. 49, 73–83 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Capito R. M., Azevedo H. S., Velichko Y. S., Mata A., Stupp S. I., Self-assembly of large and small molecules into hierarchically ordered sacs and membranes. Science 319, 1812–1816 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Fernández-Muiños T., Recha-Sancho L., López-Chicón P., Castells-Sala C., Mata A., Semino C. E., Bimolecular based heparin and self-assembling hydrogel for tissue engineering applications. Acta Biomater. 16, 35–48 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Inostroza-Brito K. E., Collin E., Siton-Mendelson O., Smith K. H., Monge-Marcet A., Ferreira D. S., Rodríguez R. P., Alonso M., Rodríguez-Cabello J. C., Reis R. L., Sagués F., Botto L., Bitton R., Azevedo H. S., Mata A., Co-assembly, spatiotemporal control and morphogenesis of a hybrid protein–peptide system. Nat. Chem. 7, 897–904 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Hedegaard C. L., Collin E. C., Redondo-Gómez C., Nguyen L. T. H., Ng K. W., Castrejón-Pita A. A., Castrejón-Pita J. R., Mata A., Hydrodynamically guided hierarchical self-assembly of peptide-protein bioinks. Adv. Funct. Mater. 28, 1703716 (2018). [Google Scholar]
- 23.Pickart L., The human tri-peptide GHK and tissue remodeling. J. Biomater. Sci. Polym. Ed. 19, 969–988 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Pearce O. M. T., Delaine-Smith R. M., Maniati E., Nichols S., Wang J., Böhm S., Rajeeve V., Ullah D., Chakravarty P., Jones R. R., Montfort A., Dowe T., Gribben J., Jones J. L., Kocher H. M., Serody J. S., Vincent B. G., Connelly J., Brenton J. D., Chelala C., Cutillas P. R., Lockley M., Bessant C., Knight M. M., Balkwill F. R., Deconstruction of a metastatic tumor microenvironment reveals a common matrix response in human cancers. Cancer Discov. 8, 304–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan B. Y., Nguyen L. T. H., Kim H.-S., Kim J.-H., Ng K. W., Cultivation of human dermal fibroblasts and epidermal keratinocytes on keratin-coated silica bead substrates. J. Biomed. Mater. Res. Part A 105, 2789–2798 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Sequeira I., Neves J. F., Carrero D., Peng Q., Palasz N., Liakath-Ali K., Lord G. M., Morgan P. R., Lombardi G., Watt F. M., Immunomodulatory role of Keratin 76 in oral and gastric cancer. Nat. Commun. 9, 3437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilandzic M., Rainczuk A., Green E., Fairweather N., Jobling T. W., Plebanski M., Stephens A. N., Keratin-14 (KRT14) positive leader cells mediate mesothelial clearance and Invasion by Ovarian Cancer Cells. Cancers 11, 1228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elsharkawy S., Al-Jawad M., Pantano M. F., Tejeda-Montes E., Mehta K., Jamal H., Agarwal S., Shuturminska K., Rice A., Tarakina N. V., Wilson R. M., Bushby A. J., Alonso M., Rodriguez-Cabello J. C., Barbieri E., del Río Hernández A., Stevens M. M., Pugno N. M., Anderson P., Mata A., Protein disorder–order interplay to guide the growth of hierarchical mineralized structures. Nat. Commun. 9, 2145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storrie H., Guler M. O., Abu-Amara S. N., Volberg T., Rao M., Geiger B., Stupp S. I., Supramolecular crafting of cell adhesion. Biomaterials 28, 4608–4618 (2007). [DOI] [PubMed] [Google Scholar]
- 30.A. Mata, L. Palmer, E. Tejeda-Montes, S. I. Stupp, in Nanotechnology in Regenerative Medicine: Methods and Protocols, M. Navarro, J. A. Planell, Eds. (Humana Press, 2012), pp. 39–49. [Google Scholar]
- 31.Kaemmerer E., Melchels F. P. W., Holzapfel B. M., Meckel T., Hutmacher D. W., Loessner D., Gelatine methacrylamide-based hydrogels: An alternative three-dimensional cancer cell culture system. Acta Biomater. 10, 2551–2562 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Sieh S., Taubenberger A. V., Rizzi S. C., Sadowski M., Lehman M. L., Rockstroh A., An J., Clements J. A., Nelson C. C., Hutmacher D. W., Phenotypic characterization of prostate cancer LNCaP cells cultured within a bioengineered microenvironment. PLOS ONE 7, e40217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H., Ding Y., Sun X. S., Nguyen T. A., Peptide hydrogelation and cell encapsulation for 3D culture of MCF-7 breast cancer cells. PLOS ONE 8, e59482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zha R. H., Sur S., Stupp S. I., Self-assembly of cytotoxic peptide amphiphiles into supramolecular membranes for cancer therapy. Adv. Healthc. Mater. 2, 126–133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyga A., Cheema U., Loizidou M., 3D tumour models: Novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 5, 239–248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worthington P., Pochan D. J., Langhans S. A., Peptide hydrogels – Versatile matrices for 3D cell culture in cancer medicine. Front. Oncol. 5, 92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks E. A., Gencoglu M. F., Corbett D. C., Stevens K. R., Peyton S. R., An omentum-inspired 3D PEG hydrogel for identifying ECM-drivers of drug resistant ovarian cancer. bioRxiv, 560482 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischbach C., Chen R., Matsumoto T., Schmelzle T., Brugge J. S., Polverini P. J., Mooney D. J., Engineering tumors with 3D scaffolds. Nat. Methods 4, 855–860 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Lange K., Fundamental role of microvilli in the main functions of differentiated cells: Outline of an universal regulating and signaling system at the cell periphery. J. Cell. Physiol. 226, 896–927 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Loessner D., Holzapfel B. M., Clements J. A., Engineered microenvironments provide new insights into ovarian and prostate cancer progression and drug responses. Adv. Drug Deliv. Rev. 79–80, 193–213 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Chiew G. G. Y., Fu A., Perng Low K., Qian Luo K., Physical supports from liver cancer cells are essential for differentiation and remodeling of endothelial cells in a HepG2-HUVEC co-culture model. Sci. Rep. 5, 10801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miklas J. W., Dallabrida S. M., Reis L. A., Ismail N., Rupnick M., Radisic M., QHREDGS enhances tube formation, metabolism and survival of endothelial cells in collagen-chitosan hydrogels. PLOS ONE 8, e72956 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Böhrnsen F., Schliephake H., Supportive angiogenic and osteogenic differentiation of mesenchymal stromal cells and endothelial cells in monolayer and co-cultures. Int. J. Oral Sci. 8, 223–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lane T. F., Iruela-Arispe M. L., Johnson R. S., Sage E. H., SPARC is a source of copper-binding peptides that stimulate angiogenesis. J. Cell Biol. 125, 929–943 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horino K., Kindezelskii A. L., Elner V. M., Hughes B. A., Petty H. R., Tumor cell invasion of model 3-dimensional matrices: Demonstration of migratory pathways, collagen disruption, and intercellular cooperation. FASEB J. 15, 932–939 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Aplin A. C., Zhu W. H., Fogel E., Nicosia R. F., Vascular regression and survival are differentially regulated by MT1-MMP and TIMPs in the aortic ring model of angiogenesis. Am. J. Physiol. Cell Physiol. 297, C471–C480 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carmeliet P., Jain R. K., Angiogenesis in cancer and other diseases. Nature 407, 249–257 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Swaminathan S., Hamid Q., Sun W., Clyne A. M., Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication 11, 025003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duarte Campos D. F., Bonnin Marquez A., O’Seanain C., Fischer H., Blaeser A., Vogt M., Corallo D., Aveic S., Exploring cancer cell behavior in vitro in three-dimensional multicellular bioprintable collagen-based hydrogels. Cancers (Basel) 11, 180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hedegaard C. L., Mata A., Integrating self-assembly and biofabrication for the development of structures with enhanced complexity and hierarchical control. Biofabrication 12, 10.1088/1758-5090/ab84cb (2020). [DOI] [PubMed] [Google Scholar]
- 51.Wang S., Wang Z., Foo S. E. M., Tan N. S., Yuan Y., Lin W., Zhang Z., Ng K. W., Culturing fibroblasts in 3D human hair keratin hydrogels. ACS Appl. Mater. Interfaces 7, 5187–5198 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/40/eabb3298/DC1