Abstract
The gut microbiota is a complex and plastic consortium of microorganisms that are intricately connected with human physiology. The liver is a central immunological organ that is particularly enriched in innate immune cells and constantly exposed to circulating nutrients and endotoxins derived from the gut microbiota. The delicate interaction between the gut and liver prevents accidental immune activation against otherwise harmless antigens. Work on the interplay between the gut microbiota and liver has assisted in understanding the pathophysiology of various liver diseases. Of immense importance is the step from high-throughput sequencing (correlation) to mechanistic studies (causality) and therapeutic intervention. Here, we review the gut microbiota, liver immunology, and the interaction between the gut and liver. In addition, the impairment in the gut–liver axis found in various liver diseases is reviewed here, with an emphasis on alcohol-associated liver disease (ALD), nonalcoholic fatty liver disease (NAFLD), and autoimmune liver disease (AILD). On the basis of growing evidence from these preclinical studies, we propose that the gut–liver axis paves the way for targeted therapeutic modalities for liver diseases.
Keywords: Gut–liver axis, Alcohol liver disease, Nonalcoholic fatty liver disease, Autoimmune liver disease, Microbiome
Subject terms: Adaptive immunity, Innate immunity, Autoimmunity
Introduction
Although the past decade has witnessed major efforts in fighting liver disease, it remains one of the top ten causes of death worldwide.1 The prevalence and incidence of nonalcoholic fatty liver disease (NAFLD) and alcohol-associated liver disease (ALD) continue to rise, which contribute to the burden of cirrhosis and liver cancer.2–4 Increasing epidemiological trends have also been observed in autoimmune liver disease (AILD), albeit less so in developing countries.5,6 Fine dissections of the pathophysiology of these liver diseases should be elucidated, which boosts the development of new therapeutic modalities and tackles the socioeconomic burden of these liver diseases.
The gut and liver communicate extensively through the biliary tract, portal vein and systemic circulation. This bidirectional crosstalk is called the gut–liver axis.7,8 The liver is a key and frontline immune organ. Positioned to receive various gut-derived signals (bacterial products, environmental toxins and food antigens), the liver’s default state is a balance between immunity and tolerance, which is essential to its function. Disruption of gut homeostasis leads to an altered immune state and various liver diseases—sterile liver inflammation caused by an excessive immune response even in the absence of pathogens; chronic infection and cancer caused by an insufficient immune response. Liver-derived factors, such as bile acids (BAs) and antibodies, also regulate the gut microbiota. The missing links in the gut–liver axis are being discovered piece by piece via well-designed experiments (detailed later). The identification of these altered elements in the gut–liver axis offers possibilities for intervention.9 Here, we first provide a basic overview of the gut microbiome (bacteria, fungi, viruses) and the microbiota-immune interaction. We then summarize the gut–liver axis with an emphasis on changes in liver immunology. Finally, we examined distinct microbial profiles and mechanistic studies in ALD, NAFLD, AILD, and other types of liver diseases.
The gut microbiome
The gut microbiota is a dense and diverse consortium of microorganisms composed of bacteria, fungi, viruses, archaea, and protozoa. The gut microbiome includes the assemblage of these microorganisms, their genome, and the environmental factors of a certain habitat. Following the first wave of intestinal microbiome studies mainly focusing on bacteria,10,11 knowledge of the gut microbiome has substantially increased. One could expect that ~1014 microbial cells reside in the human gut, comparable to human cells.12 Spatially, the majority (>99%) of the intestinal microbiome resides in the distal segments of the gastrointestinal tract (GIT), occupying different ecological niches. Temporally, an expansion in bacterial diversity is observed in infancy. It slows in early childhood and becomes stable in adults.13 Healthy adult intestinal bacteria are dominated by Bacteroidetes and Firmicutes, with smaller proportions of Proteobacteria, Actinobacteria, and Verrucomicrobia.14 To date, crosstalk between the gut and distal organs has been increasingly recognized. Physiologically, this diverse consortium has emerged as a potent modulator of the host’s immune system and metabolism. Pathologically, intestinal dysbiosis (i.e., qualitative and quantitative alterations of the gut microbiota) are associated with diseases including inflammatory bowel disease (IBD)15, type 1 diabetes (T1DM)16, obesity17, cardiovascular diseases18 and autism19. Methodologically, studies of the gut microbiota are divided into three stages: (1) descriptive/correlative studies to demonstrate the composition and diversity of the intestinal bacteria in the disease state, fueled by high-throughput sequencing methods, including 16S rRNA and metagenomics; (2) mechanistic/causal studies to elucidate the role of the intestinal bacteria in the pathogenesis of certain diseases, fueled by humanized animal models colonized by certain bacterial strains and/or bacterial communities isolated from patients or bacterial metabolites;20,21 and (3) microbe-based interventions to alleviate diseases, including fecal microbial transplantation (FMT), synthetic cocktails, and microbe-derived compounds.22
While intestinal bacteria have received considerable attention, there is a growing interest in gut-associated fungi and viruses. Fungi-relevant studies flourished with the advent of internal transcribed spacer (ITS) ribosomal sequencing. It is estimated that there are 105–106 fungal cells per gram of feces.12 The major phyla of fungi in the gut are Ascomycota and Basidiomycota. The “core mycobiota” within the human gut includes Candida, Saccharomyces, Penicillium, Aspergillus, and others.23 In the GIT, fungi interact with bacteria via mutualism, commensalism, or competition. Fungi elicit the host immune response by activating receptors, including Toll-like receptors (TLRs), C-type lectin receptors, NOD-like receptors (NLRs), and galectin 3, expressed on various immune cells, followed by the release of antimicrobial molecules.24 Recent studies have demonstrated the roles of the gut mycobiota in different diseases, including IBD25, colorectal cancer (CRC)26, irritable bowel syndrome27, ALD28, and AILD29. Bacteriophages (phages), which infect bacteria, constitute the majority of gut viruses (reviewed elsewhere30,31). The total counts of phages in human feces reach 109–1010 per gram of feces in the form of virus-like particles.32 Metagenomics has further revealed the complexity and richness of human intestinal phages.33 Morphologically, intestinal phages are mostly members of the Caudovirales order, comprising the Siphoviridae, Podoviridae, and Myoviridae families. Functionally, temperate phages, constituting at least 20–50% of free phages in the gut, play a major role in interacting with bacteria. CrAssphage is the most abundant phage in the human gut replicating on its host Bacteroides intestinalis.34 Ample evidence reveals the role of intestinal phages in diseases: (1) specific and durable changes in intestinal phages are observed in IBD35,36, CRC37, T1DM38, and rheumatic arthritis39; (2) selective engraftment of intestinal phages in FMT promotes therapeutic effects and safety;40 and (3) the delivery of phages targeting certain pathogens loaded with pharmaceutical nanoparticles could augment the effect of chemotherapy against cancer.41,42
The microbiota plays a significant role in the induction and education of the host immune system, shaping the functional diversity, and repertories of various immune cells.43 During hematopoiesis, intestinal bacterial metabolites (such as short-chain fatty acids (SCFAs)) can penetrate into the circulation and then tune immune cells (such as dendritic cell (DC) precursors) in the bone marrow.44 Commensals regulate both innate and adaptive immune systems to establish sustained tolerance to innocuous antigens. Innate lymphocytes are often located in peripheral tissues and are regulated by microbiota.45 Adaptive lymphocytes are also shaped by gut microbes, such as B cells generating IgA controlled by microbes,46 Th17 cells regulated by segmented filamentous bacteria,47 regulatory T (Treg) cells modulated by Clostridia,48 and T follicular helper cells influenced by Akkermansia muciniphila49. Translational investigations have been conducted, which permits the design of certain microbes that activate or suppress programs of the immune system to treat infections,50 autoimmunity,51 allergies52 and cancer53.
Gut microbiome and liver immunology
Immune cells in the liver communicating with the gut
The liver has been proposed as an innate immune organ54 because it is responsible for producing the majority of immune molecules in the circulation and contains a wealth of resident innate immune cells. Here, we highlight recent studies on those immune cells enriched in the liver that have a close relationship with gut microbiota.
Macrophages
Macrophages are key components of the innate immune system, and in the liver, they comprise subsets of different cell populations, including resident Kupffer cells (accounting for 80–90% of the total population of resident macrophages in the body) and recruited monocyte-derived macrophages.55 Located in the liver reticulo-endothelial system as the primary line of defense against invading microorganisms, macrophages regulate liver immune homeostasis via phagocytosis or serve as antigen-presenting cells.56 Traditionally, gut-derived endotoxins, such as LPS, sensed by TLRs expressed on liver macrophages, are one of the main factors contributing to macrophage activation. Physiologically, constitutive exposure to LPS educates liver macrophages, forming tolerance to LPS and downregulating TLRs.57 Pathologically, growing evidence suggests a key role for the excessive activation of liver macrophages induced by the LPS-TLR axis (in particular, TLR4) in the NAFLD process.58,59 Increased susceptibility of liver macrophages to LPS might be further mediated by lipids (called lipotoxicity) via enhanced release of proinflammatory cytokines60, recruitment of effective immune cells61, and ROS activation62. In addition, early colonization of the infant gut microbiota originating from obese mothers increases susceptibility to NAFLD via impaired phagocytosis of hepatic macrophages.63 By targeting the LPS-TLR axis, one could expect the potential to prevent inflammatory macrophage activation and alleviate liver diseases.64,65 In addition to LPS, some bacterial metabolites also regulate the immune state of hepatic macrophages. Tryptophan metabolites, including tryptamine and indole-3-acetate (I3A), reduce proinflammatory cytokines in macrophages by activating aryl hydrocarbon receptor66 or upregulating PFKFB3 (a key regulatory gene of glycolysis).67 Granisetron, a 5-hydroxytryptamine 3 (5-HT3) receptor antagonist derived from gut microbiota, suppresses the proinflammatory cytokine release by macrophages after LPS, alleviating sepsis-induced liver injury in mice.68 Recently, one study revealed that commensal-derived d-lactate could protect against pathogen dissemination by upregulating the phagocytic capability of Kupffer cells, thereby generating an intravascular immune firewall.69 BAs, including chenodeoxycholic acid and deoxycholic acid (DCA), could upregulate NLRP3 in hepatic macrophages, contributing to cholestatic liver diseases.70 In turn, hepatic macrophages could increase intestinal permeability and alter the microbiome composition by activating NLRP3, which aggravates the flux of endotoxin into the cholestatic liver.71
NKT cells
Natural killer T (NKT) cells are a heterogeneous group of innate-like T cells that recognize lipid antigens presented by a class I MHC-like molecule, CD1d (reviewed elsewhere72). NKT cells are predominantly found in the liver compared with other immune organs.73 On the basis of the diversity of the TCR repertoire, NKT cells are classified into two subtypes: type I or invariant NKT (iNKT) cells and type II NKT cells. Mice orally inoculated with Novosphingobium aromaticivorans, which expresses conserved PDC-E2 epitopes, could develop NKT cell-mediated destruction of small bile ducts resembling PBC, suggesting the role of NKT cells in the activation of organ-specific autoimmunity. One study also suggested that NKT cells facilitate ConA-induced hepatitis mediated by intestinal bacterial antigens presented by DCs.74 Another study demonstrated that ConA treatment failed to trigger the activation of hepatic NKT cells in GF mice, and liver injury could be restored by supplementation with killed intestinal bacteria, suggesting that gut-derived glycolipid antigens are necessary for NKT cell activation.75 Mice harboring the gut microbiota from a patient with severe ALD developed more severe liver inflammation with increased NKT cells, greater intestinal permeability and higher translocation of bacteria.76 In the scenario of liver cancer, one study suggested that the application of certain antibiotics targeting Gram-negative bacteria causes the primary/secondary BA ratio to reach a balance, which then increases the level of the chemokine CXCL16 of sinusoidal endothelial cells and thus promotes the accumulation of CXCR6-positive NKT cells in the liver, inhibiting tumor growth.77
γδ T cells
γδ T cells are enriched in the liver at a frequency of 3–5% among total hepatic lymphocytes.54 This group of cells is characterized by the expression of γβ TCRs that recognize lipid antigens resembling NKT cells and the rapid secretion of proinflammatory cytokines such as IL-17A upon stimulation.78 Growing evidence suggests that γδ T cells expand in response to invading bacterial pathogens and modulate tissue injuries.79,80 Recent studies further revealed γδ T cells at the nexus of the gut–liver interaction. Li et al. showed that hepatic γδ T cells are the major producers of IL-17A, which is quantitatively modulated by the commensal bacterial load.81 Tedesco et al. showed that in Mdr2−/− mice (an animal model resembling PSC), hepatic γδ T cells are activated by exposure to L. gasseri, leading to fibrosis and inflammation of the liver, suggesting that γδ T cells are a promising candidate for immunotherapies for liver diseases.82
MAIT cells
In a healthy adult, the frequency of mucosa-associated invariant T (MAIT) cells is 6–15% of the total T cells in the liver,83 serving as the most abundant innate-like T cells. Classically, MAITs can be activated by microbial riboflavin (vitamin B2) derivatives, which include 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), presented in the context of an MHC class I-related molecule (MR1).84 Two studies further suggested that the intrathymic development of MAITs in early life requires 5-OP-RU.85,86 MAITs can also be activated in an MR1-independent but cytokine-dependent manner, such as IL-12 and IL-18. Growing evidence suggests a wide range of functional options for MAITs in the context of different liver diseases (reviewed elsewhere83,87). In patients with acute hepatitis A, liver MAIT cells exert cytotoxicity and cause liver injury and are activated by IL-15 in the absence of TCR/MR1 engagement.88 In patients with decompensated liver cirrhosis, MAIT cells are enriched in ascites and display increased functional responses to stimulation with Escherichia coli and proinflammatory cytokines.89 MAITs could alleviate NAFLD by inducing anti-inflammatory macrophage polarization.90 In AILD, MAITs are regulated by cholic acid-induced IL-791 and promote HSC activation92, aggravating disease progression.
Bile acid-microbiota crosstalk and related signaling pathways
BA is a classic component in the gut–liver axis. The process of BA metabolism is thoroughly reviewed elsewhere.93 BA is involved in multiple processes connecting the gut–liver axis. First, the ratio of different BAs influences gut homeostasis and the immune system (reviewed in detail elsewhere94,95). Unconjugated BAs have stronger antibacterial activity than conjugated BAs.96 Gram-positive bacteria are more sensitive to BAs than Gram-negative bacteria, while some probiotics (such as Lactobacillus, Bifidobacterium and 7α-dehydroxy bacteria) show bile acid resistance related to glycolysis activation.96 Compared with patients with familial adenomatous polyposis, patients with ulcerative colitis (UC) have lower levels of lithocholic acid and DCA (the two most abundant secondary BAs in the intestinal tract), a reduction in the genes that convert primary BAs into secondary BAs, and a decrease in the abundance of the secondary BA-producing Ruminoccocus.97 Second, BAs have a bidirectional relationship with immune cells. BA can regulate regulatory T (Treg) cells in multiple ways, including agonizing VDR,98 antagonizing receptors expressed on DCs,99 and activating the reactive oxygen species (ROS) process.100 BA can also be regulated by immune cells. One study suggested that liver-infiltrating T cells could modulate the synthesis and metabolism of BAs in a TNF- and IFN-γ-dependent manner.101 Third, certain BA receptors have received attention. FXR (also known as NR1H4), which is located in intestinal epithelial cells, initiates the transcription process of fibroblast growth factor 19 (FGF19) after binding to BA. FGF19 enters the liver through the portal vein and downregulates the level of BA and maintains homeostasis by inhibiting the hepatocyte cholesterol 7α-monooxygenase (CYP7A1) enzyme, forming a feedback system for regulating BA production.102 FXR engagement mediates the restoration of homeostasis in the intestinal barrier, gut vascular barrier103 and energy metabolism104. Therapeutically, obeticholic acid, an FXR agonist, has proven useful in patients with PBC with poor response to UDCA.105 FGF19-52, an analog of FGF19, could also modulate the pool size and composition of BA and protect mice from intestinal inflammation.106 The probiotic Lactobacillus rhamnosus GG (LGG) could decrease hepatic BA by increasing intestinal FXR-FGF-15 signaling pathway-mediated suppression of BA synthesis and could prompt BA excretion, which alleviates excessive BA-induced liver injury and fibrosis in bile duct ligation and Mdr2−/− mice.107 Fourth, bile salt hydrolase (BSH) also has the potential to influence gut–liver crosstalk. The gut microbiota initiates BA metabolism via a critical first step catalyzed by BSH, that is, deconjugating glycine or taurine from primary BAs in preparation for various transformations into secondary BAs.108 Growing evidence shows the potency of BSH in modulating health and disease (beautifully reviewed elsewhere109), ranging from recurrent Clostridioides difficile infection (rCDI) to autism spectrum disorder. Therefore, BSH enzymes are promising for targeted modulation of the gut–liver axis.
The gut microbiome and ALD
Introduction of ALD
Chronic alcohol consumption leads in most cases to hepatic steatosis, which is reversible following a short period of abstinence. Approximately 10–20% of patients with alcohol-use disorder develop progressive liver disease, which is characterized by steatohepatitis, fibrosis and cirrhosis. Alcoholic hepatitis is an acute chronic form of ALD that develops mostly in patients with underlying cirrhosis and is associated with high mortality. Known factors that predispose to a more progressive type of ALD include the amount of consumed alcohol, female sex, the pattern of alcohol drinking (in particular, binge drinking and drinking outside of meals), cigarette smoking, obesity, concurrent other chronic liver diseases (CLDs) and genetic polymorphisms.110
The gut microbiome signature in ALD
Preclinical and clinical evidence supports an important role of the gut microbiota in ALD. Germ-free mice transplanted with stool from patients with severe alcoholic hepatitis develop more ethanol-induced liver inflammation (increased number of liver T lymphocyte subsets and NKT lymphocytes) and liver disease than germ-free mice colonized with fecal material from patients without alcoholic hepatitis.76 Steroid-ineligible patients with severe alcoholic hepatitis treated with daily fecal microbiota transplantation via a nasojejunal tube for 8 days had an increased survival rate compared with historical controls.111 Thus, the intestinal microbiota might be an additional factor that contributes to the progression of ALD, at least in a subset of patients.
Patients with alcohol-use disorder and liver disease show compositional changes in the bacterial fecal microbiota (Table 1). These changes are characterized by a decrease in bacterial diversity and lower proportions of several bacteria that are considered beneficial, including Lactobacillus spp., Bifidobacterium spp., Faecalibacterium prausnitzii112 and Akkermansia muciniphila113,114. In addition, patients with alcoholic cirrhosis have lower proportions of Bacteroidaceae and Prevotellaceae.115 Patients with alcoholic hepatitis show increased proportions of pathobionts, including Veilonella114 and Enterococcus faecalis.116 Quantitative changes are largely ignored when analyzing microbiota sequencing data, which focus on the relative proportions of bacteria. Patients with alcohol-use disorder show bacterial overgrowth in the luminal117 and mucosa-associated compartment of the small intestine118. Patients with alcohol-use disorder and ALD not only show bacterial dysbiosis but also changes in the gut mycobiome. They have a decrease in fungal diversity and a proportional and absolute increase in Candida spp. in the fecal mycobiota.28,119,120 We also recently described increased viral diversity in fecal samples from patients with ALD, with the most significant changes in samples from patients with alcoholic hepatitis. Escherichia-, Enterobacteria-, and Enterococcus phages were overrepresented in fecal samples from patients with alcoholic hepatitis, along with significant increases in mammalian viruses such as Parvoviridae and Herpesviridae.121 Taken together, changes in the fecal bacterial microbiota, mycobiota, and virome are observed in patients with alcohol-use disorder and ALD.
Table 1.
Studies of the gut microbiome in ALD patients
| Study | Country | Samples | Groups | Method | ALD-enriched Taxa | Controls-enriched Taxa |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| Bode et al.117 | Germany | Jejunal aspirate | Chronic alcoholics (27) vs. HC (13) | Culture | Coliform microorganisms, Gama-negative anaerobic bacteria, endospore-foming rod | / |
| Mutlu et al.115 | USA | Mucosa | ALD (19) vs. ALC (28) vs. HC (18) | 16S rRNA | Bacilli, Gammaproteobacteria | Clostridia, Bacteroidetes |
| Wang et al.118 | USA | Mucosa | Chronic alcoholics (8) vs. HC (5) | 16S rRNA | Mucosa-associated bacteria | / |
| Leclercq et al.112 | Belgium, Denmark and Sweden | Stool | AD high IP (26) vs. AD low IP (34) vs. HC (15) | 16S rRNA |
(At the family level) Lachnospiraceae, Incertae sedis XIV (At the genus level) Dorea, Blautia, Megasphaera |
(At the family level) Ruminococcaceae, Incertae sedis XIII (At the genus level) Faecalibacterium, Ruminococcus, Subdoligranulum, Oscillibacter, Anaerofilum, Clostridia, Bifidobacterium |
| Grander et al.113 | France | Stool | Severe ASH (15) vs. ASH (21) vs. HC (15) | 16S rRNA | / | Akkermansia muciniphila |
| Duan et al.116 | USA, Mexico, UK, France and Spain | Stool | Alcoholic hepatitis (75) vs. AUD (43) vs. HC (14) | 16S rRNA | Veillonella, Escherichia/Shigella, Megasphaera | Unclassified Ruminococcaceae, unclassified Lachnospiraceae, Clostridales, Ruminococcus |
| Lang et al.114 | Mutiple centers (USA, Mexico and Europe) | Stool | Alcoholic hepatitis in high MELD (54) vs. alcoholic hepatitis in low MELD (18) | 16S rRNA | (Comapred to alcoholic hepatitis in low MELD) Veillonella, Enterococcus | (Comapred to alcoholic hepatitis in high MELD) Akkermansia |
| Fungi | ||||||
| Yang et al.119 | Unclarified | Stool | AUD (10) vs. alcoholic hepatitis (6) vs. alcoholic cirrhosis (4) vs. HC (8) | ITS | Candida | Epicoccum, unclassified fungi, Galactomyces, Debaryomyces |
| Lang et al.28 | Multiple centers (USA, Mexico, Canada, UK, France and Spain) | Stool | AUD (15) vs. alcoholic hepatitis (59) vs. HC (11) | ITS | Candida | Penicillum |
| Chu et al.120 | Multiple centers (USA, Mexico, Canada, UK, France and Spain) | Stool | AUD (42) vs. alcoholic hepatitis (91) vs. HC (11) | Culture + qPCR | Candida | / |
| Viruses | ||||||
| Jiang et al.121 | Multiple centers (USA, Mexico, Canada, UK, France and Spain) | Stool | AUD (36) vs. alcoholic hepatitis (89) vs. HC (17) | Metagenomics | Escherichia phage, Enterobacteria phage, Enterococcus phage, Parvoviridae, Herpesviridae | / |
AD alcohol-dependent, IP intestinal permeability, ASH alcoholic steatohepatitis, MELD model for end-stage liver disease, ALC alcoholics without liver disease, AUD, alcoholic use disorder
The role of the gut microbiome in ALD
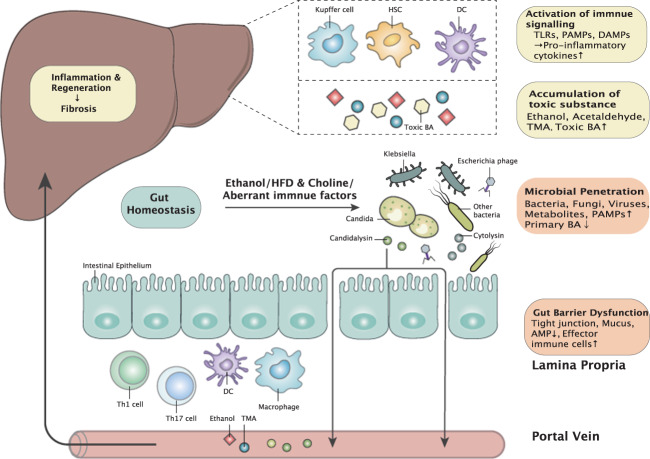
How does the gut microbiota increase susceptibility to ALD in patients? Dysbiosis-induced intestinal inflammation, acetaldehyde as a product of ethanol metabolism, changes in circadian rhythm, and alterations in intestinal bile acids and possibly other metabolites contribute to a disrupted gut barrier (Fig. 1).122–124 Since intestinal venous blood is drained into the portal vein, the first organ in our body that gut-derived microbial products and metabolites reach is the liver. Several pathogen-associated molecular pattern receptors, such as TLRs and NLRs, are expressed on parenchymal and nonparenchymal cells in the liver and recognize components of Gram-positive bacteria, the lipid A portion of LPS, flagellin and bacterial DNA. Once Kupffer cells and infiltrating macrophages are activated, they can produce a variety of inflammatory cytokines and chemokines, which contribute to disease progression.125 Chemokines such as CC-chemokine ligand 2 (CCL2) and IL8 recruit other immune cells, such as macrophages and neutrophils, to the liver. Another important cytokine is IL1β, which is induced by NF-kB following activation of TLR4 by LPS.126 Release and secretion of active IL1β require cleavage of pro–IL1β and production of mature IL1β by activating the canonical NOD-, LRR- and pyrin domain-containing 3 (NLRP3) and caspase-1 inflammasome pathways. In ALD, uric acid, and ATP levels are increased127,128, and fungal β-glucan activates the NLRP3 inflammasome in liver macrophages119. IL1β recruits other immune cells to the liver, induces lipid accumulation in hepatocytes and primes hepatocytes for cell death.3 Blocking IL1β reduces ethanol-induced liver disease in mice129 and is currently being evaluated in clinical trials for the treatment of alcoholic hepatitis (ClinicalTrials.gov Identifier: NCT04072822 and NCT03775109). Hepatic stellate cells undergo an activation process that can be triggered by microbial products and leads to liver fibrosis. Hepatocytes also recognize microbial components via TLRs, which could contribute to cell death.125 However, only ~50% of patients with alcohol-use disorder and mild liver disease show increased intestinal permeability.112 Future studies are required to determine whether the proportion of patients with increased intestinal permeability also shows progressive liver disease.
Fig. 1.
Interplay between the liver and gut microbiota in chronic liver diseases. Intestinal dysbiosis is the cornerstone of the impaired gut–liver axis, with different altered profiles in ALD, NAFLD, and AILD. Significant changes include (1) increased microbial invasion and impaired intestinal barrier and (2) activation of immune signaling and accumulation of toxins (especially in ALD and NAFLD) found in the liver. Changes in the gut and liver are connected by the portal vein, systemic circulation and bile acid metabolism. Finally, chronic inflammation and recurrent proliferation within the liver lead to cirrhosis. HFD high-fat diet, PAMP pathogen-associated molecular pattern, BA bile acid, AMP antimicrobial peptide, DC dendritic cell, TMA trimethylamine, TLR Toll-like receptor, DAMP damage-associated molecular pattern, HSC hepatic stellate cell
Several other mechanisms have been identified regarding how the gut microbiota contributes to ALD beyond increased intestinal permeability. We recently showed that patients with alcoholic hepatitis have elevated proportions of fecal E. faecalis compared with nonalcoholic subjects or patients with alcohol-use disorder.116 A higher severity of liver disease and mortality were seen in patients with alcoholic hepatitis who had cytolysin-positive (cytolytic) E. faecalis. Cytolysin is a two subunit toxin secreted by E. faecalis that can directly induce hepatocyte death presumably via pore formation.116 The importance of cytolytic E. faecalis for clinical outcome might be specific for alcoholic hepatitis, since the presence of cytolytic E. faecalis in patients with NAFLD was low and did not correlate with liver disease severity.130 In addition, we also discovered that the Candida albicans exotoxin candidalysin promotes ALD.120 Similar to cytolysin, candidalysin can damage primary hepatocytes and is associated with liver disease severity and mortality in patients with alcoholic hepatitis.120
In addition to toxins secreted by microbes, bacteria can metabolize primary bile acids into secondary bile acids in the gut. Alcohol-associated changes in the bacterial metagenome result in a dysregulation of bile acid metabolism, which is characterized by increased systemic bile acid levels and ethanol-induced liver disease in mice.131 The binding of bile acids to the nuclear receptor FXR impacts gut barrier function and host metabolic functions, including hepatic lipid metabolism. Modulation of intestinal bile acid signaling reduces ethanol-induced liver disease in mice.131 Total and conjugated bile acids are increased in patients with alcoholic hepatitis compared with controls.124
Bacteria can ferment nondigestible carbohydrates into SCFAs, with butyrate, propionate, and acetate being the most abundant in the intestine. Fecal levels of SCFAs are reduced in patients with chronic alcohol consumption.132 Similarly, the concentrations of SCFAs and numbers of SCFA-producing bacteria were lower in fecal samples from patients with alcoholic hepatitis than in samples from heavy drinkers.133 Supplementation with butyrate (tributyrin) protects mice from acute ethanol-induced gut injury.134
Taken together, changes in the gut microbiota contribute to ALD via different mechanisms, which include disruption of the intestinal barrier, secretion of toxins, and metabolism of microbial molecules. Alcohol-associated dysbiosis represents an attractive target to reduce ethanol-induced liver inflammation and hepatocyte damage.
The gut microbiome and NAFLD
The gut microbiome signature in NAFLD
Early studies have provided evidence that the gut microbiome might contribute to the development of fatty liver disease.135 In a landmark study from Anna Mae Diehl’s group, the authors observed that VSL#3, a multistrain probiotic, improved hepatic steatosis in ob/ob mice, suggesting that bacterial components contribute to liver steatosis. Whereas the exact pathomechanisms by which bacterial components might contribute to such a clinical phenotype remain unclear, many clinical studies from the last decade have convincingly demonstrated that patients with NAFLD exhibit a gut microbiome signature characterized mainly by reduced bacterial diversity.
The largest study on microbiome assessment in NAFLD was recently reported.136 Here, 472/1355 participants in the Rotterdam cohort exhibited evidence of hepatic steatosis, and steatosis was associated with lower microbial diversity and the presence of Coprococcus and Ruminococcus gnavus. Earlier smaller studies suggested that the gut microbiome in patients with NAFLD differed from that in healthy controls, with blooms of Proteobacteria, Enterobacteriaceae, and E. coli.137,138 Loomba et al. went a step further and demonstrated a gut microbiome signature in fibrotic NASH.139 In this study, patients with NASH exhibited a bacterial signature allowing us to differentiate between early and advanced liver fibrosis. Importantly, advanced liver fibrosis was paralleled by an increase in Proteobacteria and E. coli, whereas Firmicutes and F. prausnitzii were significantly decreased.139 Whereas several studies so far have assessed the gut microbiome, much less is known about the virome. A first report provided evidence from 73 patients with biopsy-proven NAFLD that more advanced disease shows a decrease in viral diversity and the respective proportion of bacteriophages.140 Overall, evidence is now substantial that the gut microbiome is affected even at the early stages of NAFLD and becomes more disturbed when this disease progresses towards a more advanced stage (Table 2).
Table 2.
Studies of the gut microbiome in NAFLD patients
| Study | Country | Samples | Groups | Method | NAFLD-enriched Taxa | Controls-enriched Taxa |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| Zhu et al.137 | USA | Stool | NASH (22) vs. obese (25) vs. HC (16) | 16S rRNA | Proteobacteria, Enterobacteriaceae, Escherichia | / |
| Mouzaki et al.138 | Canada | Stool | Simple steatosis (11) vs. NASH (22) vs. HC (17) | PCR | Clostridium coccoides | Bacteroidetes |
| Alferink et al.136 | Netherlands | Stool | No steatosis (883) vs. steatosis (472) | 16S rRNA | Ruminococcus gauvreauiigroup, Ruminococcus gnavusgroup | Coprococcus3 |
| Loomba et al.139 | USA | Stool | NAFLD (72) vs. advanced fibrosis (14) | Metagenomics | Proteobacteria, Escherichia coli | Fimicutes |
| Viruses | ||||||
| Lang et al.140 | Germany | Stool | NAFLD (73) vs. PBC (13) vs. HC (9) | 16S rRNA + Metagenomics | Escherichia phage, Enterobacteria phage, Lactobacillus phage | / |
NAS NAFLD activity score
What remains one of the most important questions is whether using certain probiotics could not only affect the gut microbiome but also influence hepatic steatosis. A large placebo-controlled study using a symbiotic (fructo-oligosaccharide plus Bifidobacterium animalis subspecies lactis BB-12) was recently reported.141 In this 10–14-month study, the symbiotic affected the fecal microbiome but failed to alter hepatic steatosis in subjects with NAFLD. A prebiotic-antibiotic strategy might also have the potential to affect NAFLD. When metronidazole was administered over 1 week followed by 11 weeks of inulin therapy, patients with NAFLD exhibited a reduction in ALT activity.142 Many studies have now been performed both preclinically and clinically studying the impact of various pre- and probiotics in NAFLD.143
Does an altered microbiota contribute to liver inflammation in NAFLD?
Whereas it seems well established that advanced liver disease with its complications, such as hepatic encephalopathy, is driven by microbial-derived products, it is still unclear whether dysbiosis observed at earlier stages of liver diseases might contribute to liver inflammation. Obviously, one of the most important and relevant parts protecting the host from bacterial invasion is an intact intestinal epithelial barrier (Fig. 1). It is now rather well established that the intestinal barrier is disrupted in many chronic inflammatory disorders, including NAFLD, and the gut microbiome might play a crucial role in the physiology of an intact epithelial barrier.8 Numerous factors might contribute to an impaired intestinal barrier,144 and various dietary factors, including a typical Western diet, might disturb this barrier. In mice with genetic impairment of the intestinal epithelial barrier, i.e., junctional adhesion molecule A-deleted mice, barrier disruption was accompanied by severe NASH.145 Targeting this defective barrier seems possible, and in addition to various beneficial diets, manipulation of the gut microbiome, e.g., by FMT, might reflect another possibility, as shown in a small clinical trial.146
Various bacteria-derived metabolites might play different roles at certain stages of liver disease.147 Several recent studies have identified gut bacteria-derived metabolites that might be involved in the development of hepatic steatosis. The serum levels of N,N,N-trimethyl-5-aminovaleric acid (TMAVA, a metabolite of gut bacteria) are increased in NAFLD, levels are reduced in mice treated with antibiotics or germ-free mice, and when administered to mice on a high-fat diet, this metabolite worsens hepatic steatosis.148 Gut microbes metabolize trimethyllysine to TMAVA, which inhibits butyrobetaine hydroxylase, thereby promoting steatosis. Hoyles et al. identified phenylacetate, a mainly bacteria-derived metabolite, as a molecule causing hepatic steatosis149, and fecal transfer from obese women into mice resulted in hepatic steatosis, as did feeding phenylacetate to mice.149 Goldstein et al. discovered another metabolite, imidazole propionate, a microbially produced histidine-derived metabolite,150 and serum and portal vein levels were increased in patients with type 2 diabetes. Furthermore, imidazole propionate regulated insulin signaling by activating p38 MAPK and phosphorylating p62, resulting in activation of mechanistic target of rapamycin.150
One important microbiota-derived driver of liver disease, especially in more advanced stages of disease, might be endotoxin. Endotoxin was considered a key pathogenetic factor in NAFLD more than two decades ago.151 Several bacteria, such as Enterobacter cloacae B29, E. coli PY102, and Klebsiella pneumonia A7, all endotoxin producers, induced NAFLD in germ-free mice on a high-fat diet.152 Patients with NASH show higher circulating endotoxin levels than patients with simple steatosis, and hepatocytes in NASH livers are positive for endotoxin accompanied by an increased number of TLR4 + hepatic macrophages.153 High-ethanol-producing K. pneumoniae strains isolated from patients with NAFLD may cause fatty liver disease in mice.154 All these studies are important, as they suggest that bacterial components such as endotoxin are of importance in various aspects of early and late liver disease, finally resulting in fibrosis and cirrhosis. As discussed in the chapter on ALD, similar mechanisms might take place in NAFLD, at least at a more advanced disease stage, when certain TLRs and NLRs expressed in the liver by various cell types are activated by gut-derived microbial components. Activation of TLRs and NLRs results in the production of numerous cytokines and chemokines driving liver inflammation. Microbially derived metabolites may especially affect patient outcomes in liver cirrhosis, as demonstrated by a large recent US study.155 Various metabolic pathways, such as bile acids, choline metabolism, aromatic amino acid metabolism, and xenobiotic pathways, correlated with the development of acute-on-chronic liver failure and death.155 Metabolomic approaches provide a large array of potentially involved molecules in the pathophysiology of liver diseases, many of which are released and regulated by gut microbes. Characterization of such key metabolites might have potential for future therapeutic targeting.
Extraintestinal microbiome signature in NAFLD: from adipose tissue to liver bacterial components
Whereas it had until recently been believed that bacterial signatures can only be observed in the intestine or other mucosal surfaces, there is increasing evidence that such signatures can be found in various tissues. The liver is continuously exposed to gut-derived bacteria and bacterial components, especially in advanced stages of disease. A circulating microbiome has been detected in central, hepatic, and portal venous blood and peripheral blood from patients with cirrhosis undergoing a transjugular portosystemic shunt.156 However, even in the early stages of liver disease, bacterial components might be present in the liver, as shown recently by Sookoian et al. when investigating liver tissue in NAFLD for the presence of bacterial DNA.157 In particular, liver Proteobacteria were increased in severe obesity, whereas in the case of moderate obesity, Gammaproteobacteria and Alphaproteobacteria as well as Deinococcus-Thermus dominated.157 This seems to be in accordance with other NAFLD studies assessing the gut microbiome where Proteobacteria also dominated.139 Another study investigated plasma and tissue (liver and 3 different adipose tissues) microbiomics in obese subjects + type 2 diabetes.158 In omental adipose tissue, the authors detected a high bacterial load with a decrease in certain Gram-positive bacteria, such as F. prausnitzii, and an increase in Enterobacteriaceae.158 Although all these “tissue microbiomic studies” are descriptive and probably detect only certain fragments of bacteria, such a phenomenon might contribute to local inflammatory processes in certain tissues, including the liver.
The gut microbiome and AILD
Introduction of AILD
AILD is a group of chronic and inflammatory liver diseases that is characterized by circulating autoantibodies and pathological inflammatory damage in the liver. On the basis of the involved cells (hepatocytes and/or biliary epithelial cells) in the liver, AILD can be categorized into four types: autoimmune hepatitis (AIH), primary biliary cholangitis (PBC), primary sclerosing cholangitis (PSC), and IgG4-related cholangitis (IgG4-SC). Variant forms of AILD (i.e., overlap syndromes) are not discussed here. Regarding pathogenesis, AILD is caused by environmental triggers in a genetically prone individual, involving the autoantigen, the major histocompatibility complex (MHC) and receptors expressed on various effective immune cells.5
The gut microbiome signature of AILD
Microbial composition and functional changes have been described in patients with AIH, PBC, and PSC (Table 3). For patients with AIH, our group revealed that Chinese patients with AIH are characterized by a decreased abundance of obligate anaerobes and increased potential pathobionts.159 Among the 11 changed bacterial genera, Veillonella dispar displays the strongest correlation with disease severity.159 Another study confirmed disease-specific fecal microbial alterations in patients with AIH in Germany.160 Moreover, a decreased abundance of Bifidobacterium is observed in this group of patients with AIH and is associated with increased disease activity. For patients with PBC, one cross-sectional study demonstrated correlations of the gut microbiome, metabolism, and immune changes in patients with PBC in China.161 Our group then revealed that the bacterial diversity decreased significantly in patients with PBC.162 The increased abundance of eight bacterial genera and the decreased abundance of four genera were also observed. This longitudinal study also revealed that gut microbial alterations in patients with PBC are associated with treatment with ursodeoxycholic acid (UDCA).162 Our group further analyzed serum and fecal BA profiles in patients with PBC and the association between BA and intestinal microbiota.163 This study demonstrated abnormal BA metabolism in patients with PBC, characterized by blockage of the conversion from conjugated to unconjugated and primary to secondary BAs. UDCA treatment leads to a decline in the level of certain conjugated BAs, therefore reversing the conjugated/unconjugated ratio in PBC. Moreover, the level of certain secondary BAs inversely correlates with upregulated bacteria in patients with PBC (e.g., Veillonella and Klebsiella) but positively correlates with upregulated bacteria in healthy controls (e.g., Faecalibacterium and Oscillospira).163 Interestingly, one study suggested that Veillonella might be a bile acid-sensitive bacterium and enable aldafermin (an FGF19 analog)-mediated suppression of bile acid synthesis, indicating its role as a novel marker for treatment response in patients with NASH.164 Recently, one study elucidated the relationships among clinical profiles, response to UDCA treatment, and gut microbiome composition in patients with PBC,165 indicating a decrease in Faecalibacterium as a novel prognostic factor in PBC. For patients with PSC, various studies show coherent changes in fecal and biliary bacteria in patients with PSC independent of IBD, characterized by low diversity and higher abundances of Enterococcus, Fusobacterium, and Lactobacillus.29,166–176 These studies suggested specific alterations in gut bacteria and bacterial metabolites in patients with AILD, suggesting the potential for using these gut microbiomes as novel biomarkers and targets in the diagnosis and treatment of AILD.
Table 3.
Studies of the gut microbiome in AILD patients
| Study | Country | Samples | Groups | Method | Certain AILD-enriched Taxa | Controls-enriched Taxa |
|---|---|---|---|---|---|---|
| AIH | ||||||
| Wei et al.159 | China | Stool | AIH (91) vs. HC (98) | 16S rRNA | Veillonella, Klebsiella, Streptococcus, Lactobacillus | Clostridiales, RF39, Ruminococcaceae, Rikenellaceae, Oscillospira, Parabacteroides, Coprococcus |
| Liwinski et al.160 | Germany | Stool | AIH (72) vs. HC (95) vs. PBC (99) vs. UC (81) | 16S rRNA | Veillonella, Klebsiella, Streptococcus | Faecalibacterium and Bifidobacterium |
| PBC | ||||||
| Lv et al.161 | China | Stool | PBC (42) vs. HC (30) | 16S rRNA | Proteobacteria, Enterobacteriaceae, Neisseriaceae, Spirochaetaceae, Veillonella, Streptococcus, Klebsiella, Actinobacillus, Anaeroglobus, Enterobacter, Haemophilus, Megasphaera, Paraprevotella | Acidobacteria, Lachnobacterium, Bacteroides and Ruminococcus |
| Tang et al.162 | China | Stool |
Treatment-naïve PBC(60) vs. HC (80) Treatment-naïve PBC vs. UDCA-treated PBC (37, longitudinal) |
16S rRNA | Haemophilus, Veillonella, Clostridium, Lactobacillus, Streptococcus, Pseudomonas, Klebsiella, Enterobacteriaceae | Oscillospira, Faecalibacterium, Sutterlla and Bacteroides |
| Furukawa et al.165 | Japan | Stool |
PBC (76) vs. HC (23) UDCA responder (43) vs. UDCA non-responder (30) |
16S rRNA | Lactobacillales | Clostridiales |
| PSC | ||||||
| Rossen et al.170 | Netherland | Mucosa | PSC-IBD (12) vs. UC (11) vs. HC (9) | 16S rRNA | / | Clostridiales II |
| Kevans et al.171 | Canada and Norway | Mucosa | PSC-UC (31) vs. UC (56) | 16S rRNA | / | / |
| Torres et al.172 | USA | Mucosa | PSC (20, 19 with IBD) vs. IBD (15, 13 with UC and 2 with CD) vs. HC (9) | 16S rRNA | Barnesiellaceae, Blautia, Ruminococcus | / |
| Quraishi et al.173 | UK | Mucosa | PSC-IBD (11) vs. IBD (10) vs. HC (9) | 16S rRNA | Lachnospiraceae, Escherichia, Megasphera | Prevotella, Roseburia, Bacteroides |
| Pereira et al.169 | Finland | Bile | PSC (80) vs. controls (46) | 16S rRNA | Streptrococcus | Bacteroides |
| Kummen et al.166 | Norway | Stool | PSC (85, 55 with IBD) vs. UC (36) vs. HC (263) | 16S rRNA | Viellonella | Coproccoccus, Phascolarctobacterium, Lachnospiraceae, Christensenellaceae |
| Sabino et al.168 | Belgium | Stool | PSC (52, 39 with IBD) vs. UC (13) vs. CD (30) vs. HC (52) | 16S rRNA | Veillonella, Streptococcus, Enterococcus, Lactobacillus, Fusobacterium | / |
| Iwasawa et al.174 | Japan | Stool | PSC (13, all pediatric-onset) vs. UC (15) vs. HC (23) | 16S rRNA | Veillonella, Streptococcus, Enterococcus | / |
| Bajer et al.175 | Czech | Stool | PSC (43, 32 with IBD) vs. IBD (32) vs. HC (31) | 16S rRNA | Veillonella, Rothia, Streptococcus, Enterococcus | Coprococcus |
| Torres et al.176 | USA and Portugal | Stool | PSC-IBD (15) vs. IBD (15) | 16S rRNA | Ruminococcus, Fusobacterium | Blautia, Roseburia, Veillonella, Doria |
| Rühlemann et al.167 | Germany and Norway | Stool | PSC (73, 38 with IBD) vs. UC (88) vs. HC (98) | 16S rRNA | Veillonella, Streptococcus, Enterococcus, Lactobacillus, Parabacterioides, Gammaproteobacteria | Coprococcus |
| Lemoinne et al.29 | France | Stool | PSC (49, 27 with IBD) vs. IBD (33) vs. HC (30) | 16S rRNA and ITS | Exophiala (fungal), Veillonella, Sphingomonadaceae, Alphaproteobacteria, Rhizobiales | S. Cereviseae (fungal), Ruminococcus, Ruminiclostridium, Faecalibacterium, Lachnoclostridium, Blautia |
The role of the gut microbiome in AILD
The mechanisms of how the gut microbiota contributes to the predisposition, initiation, and progression of certain autoimmune diseases include the following (summarized elsewhere177,178): (1) molecular mimicry; (2) translocation of gut commensals to distal organs; and (3) migration of gut immune cells to distal organs. A comprehensive understanding of the role of gut microbiota in AILD paves the way for therapeutic avenues to restore immune tolerance and homeostasis (Fig. 1).
Expression of autoantigen orthologs by the gut microbiome
Molecular mimicry could induce an autoimmune response.179 Multiple studies have suggested that autoantibodies isolated from patients with AILD can react with specific microbial proteins. The anti-mitochondrial antibody from patients with PBC can bind to certain E. coli proteins.180 The IgG3 autoantibody from patients with PBC can interact with Lactobacillus delbrueckii β-galactosidase, and its similarity with the PBC autoantigen PDC-E2 subunit is up to 67%.181 The peripheral antineutrophil cytoplasmic antibody (p-ANCA) derived from the PSC antibody can bind to the bacterial cell division protein FtsZ.182
Translocation of gut commensals to the liver
Physiologically, intestinal epithelial cells and their neighbors (mucus layer, antimicrobial peptides, immunoglobulins, symbiotic bacteria, etc.) jointly construct the gut barrier, while the mesenteric lymph nodes and the liver in turn act as immune firewalls to limit the translocation of gut microbiota to distal organs. Pathologically (named “gut barrier dysfunction”), bacteria and/or their metabolites can enter the liver through the portal vein, causing inflammation and damage in the liver. One study pointed out that Enterococcus gallinarum can pass through the intestinal epithelium and reach multiple distal organs, including the mesenteric lymph nodes and the liver, thereby causing autoimmune diseases (such as AIH and systemic lupus erythematosus).183 The application of antibiotics or specific vaccines in vivo can inhibit the growth of E. gallinarum and alleviate the disease. Another study showed that K. pneumonia in the intestines of patients with PSC can also destroy the intestinal epithelium, prompting other bacteria, such as E. gallinarum and Proteus mirabilis, to jointly cross the intestinal barrier, leading to Th17 cell-mediated liver inflammation.184
Migration of immune cells from the gut to the liver
Gut lymphocyte homing can cause inflammation and damage in the liver. One study revealed that in the state of inflammatory liver disease including PSC, hepatic sinusoidal endothelial cells overexpress MAdCAM-1, and CCL25, which recruit intestinal-derived lymphocytes into the liver and cause damage.185 Mechanistically, liver vascular adhesion protein (VAP)-1 is associated with the overexpression of MAdCAM-1 in the liver. Cysteamin, the substrate of VAP-1, originating from intestinal bacterial metabolism and the diet, acts on VAP-1 in the liver after entering the liver through the portal vein.186 Another study revealed that the TCRβ chain expressed on T cells in the intestine and liver is clonally related and recognizes the same antigen.187 B cells also participate in the gut–liver axis. One study showed that the subgroup of B cells that produce IgA in the liver is derived from intestinal lymphoid tissue and can recognize intestinal commensal bacterial antigens.188
The gut microbiome and other types of liver diseases
In the scenario of etiologically different CLDs, the vicious cycle of liver injury (inflammation and regeneration that spans decades) drives cirrhosis and liver cancer as the final stage of the disease process. Given the growing body of literature, it is becoming increasingly clear that intestinal dysbiosis plays a key role in this progression. Here, we will briefly review the role of the gut microbiome in viral hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). More detailed reviews have recently been published elsewhere for further exploration.189–191
Viral hepatitis
Hepatitis B virus (HBV) is one of the most common causes of chronic viral hepatitis, especially in developing countries. Intestinal dysbiosis has been found in patients with HBV. Opportunistic pathogenic bacteria (e.g., E. coli, Clostridium difficile, and Clostridium perfringens)192 and some fungi (e.g., Candida)193 are enriched in patients with HBV and HBV-related cirrhosis. Other bacteria, such as Bifidobacteria, display a shift from beneficial species to pathogenic species.194 Studies also demonstrate the interaction between gut microbiota and the elimination of HBV within the host. Using an HBV-infected mouse model, Chou et al. found that the establishment of intestinal bacteria via a TLR4-dependent pathway promotes liver immunity, resulting in rapid HBV clearance.195 Given the potential role of gut microbiota in the pathogenesis of HBV, randomized clinical trials of FMT for the treatment of HBV have been conducted (NCT02689245 and NCT03429439). Recently, one nonrandomized pilot trial of FMP for HBV treatment displayed safety and efficacy in viral suppression and HBeAg clearance.196
Other types of viral hepatitis also exhibit changes in the gut microbiota. Studies focusing on patients with hepatitis C virus demonstrated a reduction in microbial diversity and composition in the patient group by 16S rRNA197,198 and high levels of urease gene expression by metagenomics199. Hepatitis E virus infection, as one type of acute viral hepatitis, might lead to acute liver failure. One study illustrated a perturbation of intestinal microbiota effector T lymphocytes, the serum international normalized ratio and the severity of hepatic encephalopathy.200
Cirrhosis
Changes in the composition and function of the gut microbiota have been observed in cirrhosis. Regarding composition, the fecal microbiome in patients with cirrhosis is characterized by a decrease in diversity, an increase in immunostimulatory pathogens (e.g., Enterococcaceae (the major causative organism releasing endotoxin) and Staphylococcaceae) and a decline in potentially beneficial Firmicutes (e.g., Lachnospiraceae and Ruminococcaceae).201,202 Similar changes can be detected in the colon mucosa203, serum204 and saliva205 of patients with cirrhosis. Interestingly, the change in the gut microbiota composition is associated with cirrhosis patient outcomes (i.e., compensated vs uncompensated206, inpatient vs outpatient, and noninfected and infected patients202), suggesting its potential as a novel biomarker. Regarding function, leaky gut is a well-established feature of patients with cirrhosis and relevant animal models, rendering potentially pathogenic bacteria and their metabolites permeable to the circulation.207 In addition to the physical destruction of the gut barrier, intestinal infiltration with effective immune cells is also observed in cirrhosis, exemplified by the expansion of TNF-α- and IFN-γ-expressing lymphocytes and the depletion of Th17 cells.208 Moreover, reduced bile flow209 and impaired FXR signaling pathways210 are also detected in cirrhosis and parallel the disease severity.
A common gut microbiome signature in cirrhosis independent of the etiology of liver disease might exist. Oh et al. characterized a gut microbiome signature when combined with age that precisely detected NAFLD-related cirrhosis.211 Importantly, by using a machine-learning-based approach and when comparing their findings with liver cohorts from Italy and China comprising various etiologies of liver cirrhosis, a similar microbiome signature was detected independent of the etiology of cirrhosis. These findings suggest that at least in advanced stages of liver disease, the etiology of liver disease seems less relevant regarding the gut microbiome disturbances.212 The authors proposed that such an approach might allow the noninvasive diagnosis of liver cirrhosis in the near future.211 More studies are needed to clarify whether gut microbiome signatures differ between various etiologies at earlier stages of liver diseases and whether, in advanced stages, such differences might disappear.
HCC
Sustained liver injury and regeneration promote HCC, which is the third leading cause of cancer mortality worldwide. Recent studies have revealed changes in the gut microbiota in patients with HCC, suggesting a correlation of specific microbial profiles in patients with HCC with different etiologies, geographical distributions, and nutritional states.213,214 The contribution of gut microbiota to HCC is multifaceted: (1) A disrupted gut barrier brings a series of TLR ligands (LPS215 and bacterial DNA216) to the liver and triggers inflammation. The TLR signaling pathways mediate hepatocarcinogenesis via downregulation of hepatocyte apoptosis and upregulation of hepatic stellate cell proliferation64. (2) Impaired immunosurveillance is associated with abnormal gut microbiota in HCC. As mentioned before, a negative correlation between BA-metabolizing Clostridium cluster XIV and CXCR6-positive antitumor NKT cells is observed in mice with liver cancer.77 Moreover, the composition of the gut microbiota could potentially predict the response rates in patients with liver cancer treated with certain immune checkpoint inhibitors (such as PD-L1 inhibitors),217 suggesting the potential to harness the microbiome for HCC immunotherapy.
Concluding remarks and perspectives
An accumulating body of research suggests the close relationship between the gut and the liver, called the gut–liver axis. They communicate with each other anatomically and functionally via the portal vein, biliary tract, and systemic circulation. Extensive interplay between the gut and the liver is exemplified in specialized bacteria and their metabolites, immune cells in the liver, and BA signaling pathways. Pathologically, preliminary correlative studies have revealed different microbiota (bacteria, fungi, and viruses) phenotypes in different liver diseases, including ALD, NAFLD, AILD, and other types of liver diseases. More longitudinal studies are anticipated to provide persistent and accessory microbiomes within individuals. Mechanistic studies are the second step to explore potential microbiota-mediated causality by moving from human data and samples to animal models. Culturomics fuels functional studies of the key microbial member(s) identified in the disease state218. To gain a complete picture of the gut–liver axis, multiomics provides high-dimensional measurements to capture a comprehensive profile of both the microbiota and the immune system.219,220 There is also a trend to integrate environmental factors with genetic factors to better understand pathogenesis221,222.
Advances in knowledge of the gut–liver axis promote the development of novel diagnostic, prognostic and therapeutic modalities for liver diseases. FMT with precise modulation based on the prior selection of beneficial microbial members could bring benefits with reduced risks. Synthetic biology (i.e., engineered probiotics and human microbial members) also provide a sustainable therapeutic approach.223,224 Bacteriophages, which target specific bacterial hosts, also show efficacy as novel therapeutic modalities to treat liver disease.116 Thus, as the role of the gut microbiome in liver disease is increasingly recognized, more effort should be put into translating our current knowledge of the disease-modulating role of the gut–liver axis into well-designed trials in patients.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (grants #81830016, 81771732, and 81620108002 to X.M.; #81922010 and 81873561 to R.T.). This study was supported in part by services provided by the NIH centers P30 DK120515 and P50 AA011999. H.T. is supported by the excellence initiative VASCage (Centre for Promoting Vascular Health in the Ageing Community), an R&D K-Centre (COMET program—Competence Centers for Excellent Technologies) funded by the Austrian Ministry for Transport, Innovation and Technology, the Austrian Ministry for Digital and Economic Affairs and the federal states Tyrol, Salzburg and Vienna.
Author contributions
B.S. was responsible for writing the chapter “Contribution of the gut microbiota to alcohol-associated liver disease”. H.T. wrote the chapter “NAFLD and the Microbiome”. R.W., R.T. and X.M. wrote the chapters “The Gut Microbiome”, “The Gut Microbiome and Liver Immunology”, “The Gut Microbiome and AILD”, and “The Gut Microbiome and Other Types of Liver Diseases”. B.L. was responsible for draft calibration.
Competing interests
B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics, Mabwell Therapeutics and Patara Pharmaceuticals. B.S.’s institution (UC San Diego) has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company and Axial Biotherapeutics. H.T. and X.M. have no competing interests.
Footnotes
These authors contributed equally: Rui Wang, Ruqi Tang
These authors jointly supervised this work: Xiong Ma, Bernd Schnabl, Herbert Tilg
Contributor Information
Xiong Ma, Email: maxiongmd@hotmail.com.
Bernd Schnabl, Email: beschnabl@ucsd.edu.
Herbert Tilg, Email: herbert.tilg@i-med.ac.at.
References
- 1.World Health Organization. The Top 10 Causes of Death. WHO, https://www.whoint/en/news-room/fact-sheets/detail/the-top-10-causes-of-death (2018).
- 2.Younossi Z, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 3.Seitz HK, et al. Alcoholic liver disease. Nat. Rev. Dis. Prim. 2018;4:16. doi: 10.1038/s41572-018-0014-7. [DOI] [PubMed] [Google Scholar]
- 4.Wong MCS, et al. The changing epidemiology of liver diseases in the Asia-Pacific region. Nat. Rev. Gastroenterol. Hepatol. 2019;16:57–73. doi: 10.1038/s41575-018-0055-0. [DOI] [PubMed] [Google Scholar]
- 5.Mieli-Vergani, G. et al. Autoimmune hepatitis. Nat. Rev. Dis. Primers. 4, 18018 (2018). [DOI] [PubMed]
- 6.Lleo A, Colapietro F. Changes in the epidemiology of primary biliary cholangitis. Clin. Liver Dis. 2018;22:429–441. doi: 10.1016/j.cld.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Tripathi A, et al. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopyk, D. M. & Grakoui, A. Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology159, 849–863 (2020). [DOI] [PMC free article] [PubMed]
- 9.Albillos A, et al. The gut-liver axis in liver disease: pathophysiological basis for therapy. J. Hepatol. 2020;72:558–577. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Human Microbiome Project Consortium. et al. Structure, function and diversity of the healthy human microbiome. Nature. 486, 207–214 (2012). [DOI] [PMC free article] [PubMed]
- 12.Sender R, et al. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng J, et al. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 2016;10:1002–1014. doi: 10.1038/ismej.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariat D, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni J, et al. Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siljander H, et al. Microbiome and type 1 diabetes. EBioMedicine. 2019;46:512–521. doi: 10.1016/j.ebiom.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes AC, et al. The human gut microbiota: metabolism and perspective in obesity. Gut Microbes. 2018;9:308–325. doi: 10.1080/19490976.2018.1465157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang WHW, et al. Intestinal microbiota in cardiovascular health and disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2019;73:2089–2105. doi: 10.1016/j.jacc.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fattorusso, A. et al. Autism spectrum disorders and the gut microbiota. Nutrients. 11, 521 (2019). [DOI] [PMC free article] [PubMed]
- 20.Gilbert JA, Lynch SV. Community ecology as a framework for human microbiome research. Nat. Med. 2019;25:884–889. doi: 10.1038/s41591-019-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanoue T, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 22.D’Haens GR, Jobin C. Fecal microbial transplantation for diseases beyond recurrent clostridium difficile infection. Gastroenterology. 2019;157:624–636. doi: 10.1053/j.gastro.2019.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nash AK, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2019;16:331–345. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 25.Standaert-Vitse A, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology. 2006;130:1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Coker OO, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68:654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botschuijver S, et al. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology. 2017;153:1026–1039. doi: 10.1053/j.gastro.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Lang S, et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology. 2020;71:522–538. doi: 10.1002/hep.30832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemoinne S, et al. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut. 2020;69:92–102. doi: 10.1136/gutjnl-2018-317791. [DOI] [PubMed] [Google Scholar]
- 30.Shkoporov AN, Hill C. Bacteriophages of the human gut: The “Known Unknown” of the microbiome. Cell Host Microbe. 2019;25:195–209. doi: 10.1016/j.chom.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Sausset R, et al. New insights into intestinal phages. Mucosal Immunol. 2020;13:205–215. doi: 10.1038/s41385-019-0250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoyles L, et al. Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 2014;165:803–812. doi: 10.1016/j.resmic.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Breitbart M, et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shkoporov AN, et al. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018;9:4781. doi: 10.1038/s41467-018-07225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo T, et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68:1169–1179. doi: 10.1136/gutjnl-2018-318131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gogokhia L, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 2019;25:285–99.e8. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakatsu G, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155:529–41.e5. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 38.Kramná L, et al. Gut virome sequencing in children with early islet autoimmunity. Diabetes Care. 2015;38:930–933. doi: 10.2337/dc14-2490. [DOI] [PubMed] [Google Scholar]
- 39.Międzybrodzki R, et al. In vivo studies on the influence of bacteriophage preparations on the autoimmune inflammatory process. Biomed. Res. Int. 2017;2017:3612015. doi: 10.1155/2017/3612015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo T, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2018;67:634–643. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong X, et al. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci. Adv. 2020;6:eaba1590. doi: 10.1126/sciadv.aba1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng DW, et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 2019;3:717–728. doi: 10.1038/s41551-019-0423-2. [DOI] [PubMed] [Google Scholar]
- 43.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 44.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 45.Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bunker JJ, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ansaldo E, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019;364:1179–1184. doi: 10.1126/science.aaw7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ducarmon, Q. R. et al. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol Mol Biol Rev. 83 (2019). [DOI] [PMC free article] [PubMed]
- 51.Brown EM, et al. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev. Immunol. 2019;37:599–624. doi: 10.1146/annurev-immunol-042718-041841. [DOI] [PubMed] [Google Scholar]
- 52.Shu SA, et al. Microbiota and food allergy. Clin. Rev. Allergy Immunol. 2019;57:83–97. doi: 10.1007/s12016-018-8723-y. [DOI] [PubMed] [Google Scholar]
- 53.Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat. Rev. Cancer. 2017;17:271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- 54.Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43:S54–S62. doi: 10.1002/hep.21060. [DOI] [PubMed] [Google Scholar]
- 55.Kolios G, et al. Role of Kupffer cells in the pathogenesis of liver disease. World J. Gastroenterol. 2006;12:7413–7420. doi: 10.3748/wjg.v12.i46.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 2017;17:306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 57.Wu X, et al. Oral ampicillin inhibits liver regeneration by breaking hepatic innate immune tolerance normally maintained by gut commensal bacteria. Hepatology. 2015;62:253–264. doi: 10.1002/hep.27791. [DOI] [PubMed] [Google Scholar]
- 58.Rivera CA, et al. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J. Hepatol. 2007;47:571–579. doi: 10.1016/j.jhep.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vespasiani-Gentilucci U, et al. Hepatic toll-like receptor 4 expression is associated with portal inflammation and fibrosis in patients with NAFLD. Liver Int. 2015;35:569–581. doi: 10.1111/liv.12531. [DOI] [PubMed] [Google Scholar]
- 60.Kawaratani H, et al. Innate immune reactivity of the liver in rats fed a choline-deficient L-amino-acid-defined diet. World J. Gastroenterol. 2008;14:6655–6661. doi: 10.3748/wjg.14.6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leroux A, et al. Toxic lipids stored by Kupffer cells correlates with their pro-inflammatory phenotype at an early stage of steatohepatitis. J. Hepatol. 2012;57:141–149. doi: 10.1016/j.jhep.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 62.Kudo H, et al. Lipopolysaccharide triggered TNF-alpha-induced hepatocyte apoptosis in a murine non-alcoholic steatohepatitis model. J. Hepatol. 2009;51:168–175. doi: 10.1016/j.jhep.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 63.Soderborg TK, et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat. Commun. 2018;9:4462. doi: 10.1038/s41467-018-06929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dapito DH, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazagova M, et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. Faseb j. 2015;29:1043–1055. doi: 10.1096/fj.14-259515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krishnan S, et al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 2018;23:1099–1111. doi: 10.1016/j.celrep.2018.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma, L. et al. Indole alleviates diet-induced hepatic steatosis and inflammation in a manner involving myeloid cell 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3. Hepatology72, 1191–1203 (2020). [DOI] [PMC free article] [PubMed]
- 68.Gong S, et al. Intestinal microbiota mediates the susceptibility to polymicrobial sepsis-induced liver injury by granisetron generation in mice. Hepatology. 2019;69:1751–1767. doi: 10.1002/hep.30361. [DOI] [PubMed] [Google Scholar]
- 69.McDonald, B. et al. Programing of an intravascular immune firewall by the gut microbiota protects against pathogen dissemination during infection. Cell Host Microbe28, 660–668 (2020). [DOI] [PubMed]
- 70.Hao H, et al. Farnesoid X receptor regulation of the NLRP3 inflammasome underlies cholestasis-associated sepsis. Cell Metab. 2017;25:856–67.e5. doi: 10.1016/j.cmet.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Isaacs-Ten, A. et al. Intestinal microbiome-macrophage crosstalk contributes to cholestatic liver disease by promoting intestinal permeability. Hepatology (2020). [DOI] [PMC free article] [PubMed]
- 72.Marrero I, et al. Complex network of NKT cell subsets controls immune homeostasis in liver and gut. Front. Immunol. 2018;9:2082. doi: 10.3389/fimmu.2018.02082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bandyopadhyay K, et al. NKT cell subsets as key participants in liver physiology and pathology. Cell Mol. Immunol. 2016;13:337–346. doi: 10.1038/cmi.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen J, et al. Natural killer T cells play a necessary role in modulating of immune-mediated liver injury by gut microbiota. Sci. Rep. 2014;4:7259. doi: 10.1038/srep07259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei Y, et al. Enterogenous bacterial glycolipids are required for the generation of natural killer T cells mediated liver injury. Sci. Rep. 2016;6:36365. doi: 10.1038/srep36365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Llopis M, et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65:830–839. doi: 10.1136/gutjnl-2015-310585. [DOI] [PubMed] [Google Scholar]
- 77.Ma, C. et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 360, eaan5931 (2018). [DOI] [PMC free article] [PubMed]
- 78.Bonneville M, et al. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010;10:467–478. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- 79.Martin B, et al. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 80.Paget C, et al. CD3bright signals on γδ T cells identify IL-17A-producing Vγ6Vδ1+ T cells. Immunol. Cell Biol. 2015;93:198–212. doi: 10.1038/icb.2014.94. [DOI] [PubMed] [Google Scholar]
- 81.Li F, et al. The microbiota maintain homeostasis of liver-resident γδT-17 cells in a lipid antigen/CD1d-dependent manner. Nat. Commun. 2017;7:13839. doi: 10.1038/ncomms13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tedesco D, et al. Alterations in intestinal microbiota lead to production of interleukin 17 by intrahepatic γδ T-cell receptor-positive cells and pathogenesis of cholestatic liver disease. Gastroenterology. 2018;154:2178–2193. doi: 10.1053/j.gastro.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Provine NM, Klenerman P. MAIT cells in health and disease. Annu. Rev. Immunol. 2020;38:203–228. doi: 10.1146/annurev-immunol-080719-015428. [DOI] [PubMed] [Google Scholar]
- 84.Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 85.Legoux F, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 2019;366:494–499. doi: 10.1126/science.aaw2719. [DOI] [PubMed] [Google Scholar]
- 86.Constantinides, M. G. et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 366, eaax6624 (2019). [DOI] [PMC free article] [PubMed]
- 87.Lin, Q. et al. The dialogue between unconventional T cells and the microbiota. Mucosal Immunol. 13, 867–876 (2020). [DOI] [PubMed]
- 88.Rha, M. S. et al. Human liver CD8(+) MAIT cells exert TCR/MR1-independent innate-like cytotoxicity in response to IL-15. J. Hepatol. 73, 640–650 (2020). [DOI] [PubMed]
- 89.Niehaus, C. E. et al. MAIT cells are enriched and highly functional in ascites of patients with decompensated liver cirrhosis. Hepatology72, 1378–1393 (2020). [DOI] [PubMed]
- 90.Li Y, et al. Mucosal-associated invariant T cells improve nonalcoholic fatty liver disease through regulating macrophage polarization. Front Immunol. 2018;9:1994. doi: 10.3389/fimmu.2018.01994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang X, et al. The immunobiology of mucosal-associated invariant T cell (MAIT) function in primary biliary cholangitis: regulation by cholic acid-induced Interleukin-7. J. Autoimmun. 2018;90:64–75. doi: 10.1016/j.jaut.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 92.Böttcher K, et al. MAIT cells are chronically activated in patients with autoimmune liver disease and promote profibrogenic hepatic stellate cell activation. Hepatology. 2018;68:172–186. doi: 10.1002/hep.29782. [DOI] [PubMed] [Google Scholar]
- 93.Wahlström A, et al. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 94.Jia W, et al. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018;15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y, et al. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun. Rev. 2017;16:885–896. doi: 10.1016/j.autrev.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 96.Tian Y, et al. The microbiome modulating activity of bile acids. Gut Microbes. 2020;11:979–996. doi: 10.1080/19490976.2020.1732268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sinha SR, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27:659–70.e5. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song X, et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Campbell C, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581:475–479. doi: 10.1038/s41586-020-2193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hang S, et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. 2019;576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Glaser F, et al. Liver infiltrating T cells regulate bile acid metabolism in experimental cholangitis. J. Hepatol. 2019;71:783–792. doi: 10.1016/j.jhep.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 102.Zarrinpar A, Loomba R. Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and non-alcoholic fatty liver disease. Aliment Pharmacol. Ther. 2012;36:909–921. doi: 10.1111/apt.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mouries J, et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 2019;71:1216–1228. doi: 10.1016/j.jhep.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Copple BL, Li T. Pharmacology of bile acid receptors: evolution of bile acids from simple detergents to complex signaling molecules. Pharmacol. Res. 2016;104:9–21. doi: 10.1016/j.phrs.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nevens F, et al. A placebo-controlled trial of obeticholic acid in primary biliary cholangitis. N. Engl. J. Med. 2016;375:631–643. doi: 10.1056/NEJMoa1509840. [DOI] [PubMed] [Google Scholar]
- 106.Gadaleta RM, et al. Fibroblast Growth Factor 19 modulates intestinal microbiota and inflammation in presence of Farnesoid X Receptor. EBioMedicine. 2020;54:102719. doi: 10.1016/j.ebiom.2020.102719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Y, et al. Probiotic Lactobacillus rhamnosus GG prevents liver fibrosis through inhibiting hepatic bile acid synthesis and enhancing bile acid excretion in mice. Hepatology. 2020;71:2050–2066. doi: 10.1002/hep.30975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fiorucci S, Distrutti E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol. Med. 2015;21:702–714. doi: 10.1016/j.molmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 109.Foley MH, et al. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019;15:e1007581. doi: 10.1371/journal.ppat.1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lang S, Schnabl B. Microbiota and fatty liver disease-the known, the unknown, and the future. Cell Host Microbe. 2020;28:233–244. doi: 10.1016/j.chom.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Philips CA, et al. Healthy donor fecal microbiota transplantation in steroid-ineligible severe alcoholic hepatitis: a pilot study. Clin. Gastroenterol. Hepatol. 2017;15:600–602. doi: 10.1016/j.cgh.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 112.Leclercq S, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl Acad. Sci. USA. 2014;111:E4485–E4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grander C, et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 2018;67:891–901. doi: 10.1136/gutjnl-2016-313432. [DOI] [PubMed] [Google Scholar]
- 114.Lang S, et al. Changes in the fecal bacterial microbiota associated with disease severity in alcoholic hepatitis patients. Gut Microbes. 2020;12:1785251. doi: 10.1080/19490976.2020.1785251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mutlu EA, et al. Colonic microbiome is altered in alcoholism. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Duan Y, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575:505–511. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bode JC, et al. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology. 1984;31:30–34. [PubMed] [Google Scholar]
- 118.Wang L, et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19:227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang AM, et al. Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Investig. 2017;127:2829–2841. doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chu H, et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J. Hepatol. 2020;72:391–400. doi: 10.1016/j.jhep.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiang, L. et al. Intestinal virome in patients with alcoholic hepatitis. Hepatology 2020. 10.1002/hep.31459. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 122.Bluemel S, et al. Precision medicine in alcoholic and nonalcoholic fatty liver disease via modulating the gut microbiota. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;311:G1018–g36. doi: 10.1152/ajpgi.00245.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brenner DA, et al. Role of gut microbiota in liver disease. J. Clin. Gastroenterol. 2015;49(Suppl 1):S25–S27. doi: 10.1097/MCG.0000000000000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brandl K, et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J. Hepatol. 2018;69:396–405. doi: 10.1016/j.jhep.2018.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hendrikx T, Schnabl B. Antimicrobial proteins: intestinal guards to protect against liver disease. J. Gastroenterol. 2019;54:209–217. doi: 10.1007/s00535-018-1521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Inokuchi S, et al. Toll-like receptor 4 mediates alcohol-induced steatohepatitis through bone marrow-derived and endogenous liver cells in mice. Alcohol Clin. Exp. Res. 2011;35:1509–1518. doi: 10.1111/j.1530-0277.2011.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iracheta-Vellve A, et al. Inhibition of sterile danger signals, uric acid and ATP, prevents inflammasome activation and protects from alcoholic steatohepatitis in mice. J. Hepatol. 2015;63:1147–1155. doi: 10.1016/j.jhep.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Petrasek J, et al. Metabolic danger signals, uric acid and ATP, mediate inflammatory cross-talk between hepatocytes and immune cells in alcoholic liver disease. J. Leukoc. Biol. 2015;98:249–256. doi: 10.1189/jlb.3AB1214-590R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Petrasek J, et al. IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice. J. Clin. Investig. 2012;122:3476–3489. doi: 10.1172/JCI60777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lang S, et al. Cytolysin-positive Enterococcus faecalis is not increased in patients with non-alcoholic steatohepatitis. Liver Int. 2020;40:860–865. doi: 10.1111/liv.14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hartmann P, et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology. 2018;67:2150–2166. doi: 10.1002/hep.29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Couch RD, et al. Alcohol induced alterations to the human fecal VOC metabolome. PLoS ONE. 2015;10:e0119362. doi: 10.1371/journal.pone.0119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smirnova E, et al. Fecal microbiome distinguishes alcohol consumption from alcoholic hepatitis but does not discriminate disease severity. Hepatology. 2020;72:271–286. doi: 10.1002/hep.31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cresci GA, et al. Tributyrin supplementation protects mice from acute ethanol-induced gut injury. Alcohol Clin. Exp. Res. 2014;38:1489–1501. doi: 10.1111/acer.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Z, et al. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology. 2003;37:343–350. doi: 10.1053/jhep.2003.50048. [DOI] [PubMed] [Google Scholar]
- 136.Alferink, L. J. et al. Microbiomics, metabolomics, predicted metagenomics and hepatic steatosis in a population-based study of 1355 adults. Hepatology 2020. 10.1002/hep.31417. Online ahead of print. [DOI] [PubMed]
- 137.Zhu L, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 138.Mouzaki M, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- 139.Loomba R, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017;25:1054–62.e5. doi: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lang, S. et al. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology159, 1839–1852 (2020). [DOI] [PMC free article] [PubMed]
- 141.Scorletti E, et al. Synbiotics alter fecal microbiomes, but not liver fat or fibrosis, in a randomized trial of patients with nonalcoholic fatty liver disease. Gastroenterology. 2020;158:1597–610.e7. doi: 10.1053/j.gastro.2020.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chong, C. Y. L. et al. Randomised double-blind placebo-controlled trial of inulin with metronidazole in non-alcoholic fatty liver disease (NAFLD). Nutrients12, 937 (2020). [DOI] [PMC free article] [PubMed]
- 143.Aron-Wisnewsky J, et al. Nonalcoholic fatty liver disease: modulating gut microbiota to improve severity? Gastroenterology. 2020;158:1881–1898. doi: 10.1053/j.gastro.2020.01.049. [DOI] [PubMed] [Google Scholar]
- 144.Tilg H, et al. The intestinal microbiota fuelling metabolic inflammation. Nat. Rev. Immunol. 2020;20:40–54. doi: 10.1038/s41577-019-0198-4. [DOI] [PubMed] [Google Scholar]
- 145.Gupta B, et al. Western diet-induced increase in colonic bile acids compromises epithelial barrier in nonalcoholic steatohepatitis. FASEB J. 2020;34:7089–7102. doi: 10.1096/fj.201902687R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Craven, L. et al. Allogenic fecal microbiota transplantation in patients with nonalcoholic fatty liver disease improves abnormal small intestinal permeability: a randomized control trial. Am. J. Gastroenterol. 115, 1055–1065 (2020). [DOI] [PubMed]
- 147.Chu H, et al. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut. 2019;68:359–370. doi: 10.1136/gutjnl-2018-316307. [DOI] [PubMed] [Google Scholar]
- 148.Zhao M, et al. TMAVA, a metabolite of intestinal microbes, is increased in plasma from patients with liver steatosis, inhibits γ-butyrobetaine hydroxylase, and exacerbates fatty liver in mice. Gastroenterology. 2020;158:2266–81.e27. doi: 10.1053/j.gastro.2020.02.033. [DOI] [PubMed] [Google Scholar]
- 149.Hoyles L, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 2018;24:1070–1080. doi: 10.1038/s41591-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Goldstein JL. What makes a piece of art or science a masterpiece? Cell. 2018;175:1–5. doi: 10.1016/j.cell.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 151.Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 152.Fei, N. et al. Endotoxin producers overgrowing in human gut microbiota as the causative agents for nonalcoholic fatty liver disease. mBio. 11, e03263–19 (2020). [DOI] [PMC free article] [PubMed]
- 153.Carpino, G. et al. Increased liver localization of lipopolysaccharides in human and experimental NAFLD. Hepatology159, 1715–1730 (2019). [DOI] [PubMed]
- 154.Yuan J, et al. Fatty liver disease caused by high-alcohol-producing Klebsiella pneumoniae. Cell Metab. 2019;30:675–88.e7. doi: 10.1016/j.cmet.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 155.Bajaj, J. S. et al. Serum levels of metabolites produced by intestinal microbes and lipid moieties independently associated with acute on chronic liver failure and death in patients with cirrhosis. Gastroenterology (2020). [DOI] [PMC free article] [PubMed]
- 156.Schierwagen R, et al. Circulating microbiome in blood of different circulatory compartments. Gut. 2019;68:578–580. doi: 10.1136/gutjnl-2018-316227. [DOI] [PubMed] [Google Scholar]
- 157.Sookoian S, et al. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut. 2020;69:1483–1491. doi: 10.1136/gutjnl-2019-318811. [DOI] [PubMed] [Google Scholar]
- 158.Anhê FF, et al. Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat. Metab. 2020;2:233–242. doi: 10.1038/s42255-020-0178-9. [DOI] [PubMed] [Google Scholar]
- 159.Wei Y, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut. 2020;69:569–577. doi: 10.1136/gutjnl-2018-317836. [DOI] [PubMed] [Google Scholar]
- 160.Liwinski T, et al. A disease-specific decline of the relative abundance of Bifidobacterium in patients with autoimmune hepatitis. Aliment Pharmacol Ther. 2020;51:1417–1428. doi: 10.1111/apt.15754. [DOI] [PubMed] [Google Scholar]
- 161.Lv LX, et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ. Microbiol. 2016;18:2272–2286. doi: 10.1111/1462-2920.13401. [DOI] [PubMed] [Google Scholar]
- 162.Tang R, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67:534–541. doi: 10.1136/gutjnl-2016-313332. [DOI] [PubMed] [Google Scholar]
- 163.Chen W, et al. Comprehensive analysis of serum and fecal bile acid profiles and interaction with gut microbiota in primary biliary cholangitis. Clin. Rev. Allergy Immunol. 2020;58:25–38. doi: 10.1007/s12016-019-08731-2. [DOI] [PubMed] [Google Scholar]
- 164.Loomba, R. et al. The commensal microbe veillonella as a marker for response to an FGF19 analog in nonalcoholic steatohepatitis. Hepatology 2020. 10.1002/hep.31523. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 165.Furukawa M, et al. Gut dysbiosis associated with clinical prognosis of patients with primary biliary cholangitis. Hepatol. Res. 2020;50:840–852. doi: 10.1111/hepr.13509. [DOI] [PubMed] [Google Scholar]
- 166.Kummen M, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. 2017;66:611–619. doi: 10.1136/gutjnl-2015-310500. [DOI] [PubMed] [Google Scholar]
- 167.Rühlemann MC, et al. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut. 2017;66:753–754. doi: 10.1136/gutjnl-2016-312180. [DOI] [PubMed] [Google Scholar]
- 168.Sabino J, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut. 2016;65:1681–1689. doi: 10.1136/gutjnl-2015-311004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pereira P, et al. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS ONE. 2017;12:e0182924. doi: 10.1371/journal.pone.0182924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rossen NG, et al. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J. Crohns Colitis. 2015;9:342–348. doi: 10.1093/ecco-jcc/jju023. [DOI] [PubMed] [Google Scholar]
- 171.Kevans D, et al. Characterization of intestinal microbiota in ulcerative colitis patients with and without primary sclerosing cholangitis. J. Crohns Colitis. 2016;10:330–337. doi: 10.1093/ecco-jcc/jjv204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Torres J, et al. The features of mucosa-associated microbiota in primary sclerosing cholangitis. Aliment Pharmacol. Ther. 2016;43:790–801. doi: 10.1111/apt.13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Quraishi MN, et al. The gut-adherent microbiota of PSC-IBD is distinct to that of IBD. Gut. 2017;66:386–388. doi: 10.1136/gutjnl-2016-311915. [DOI] [PubMed] [Google Scholar]
- 174.Iwasawa K, et al. Characterisation of the faecal microbiota in Japanese patients with paediatric-onset primary sclerosing cholangitis. Gut. 2017;66:1344–1346. doi: 10.1136/gutjnl-2016-312533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bajer L, et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J. Gastroenterol. 2017;23:4548–4558. doi: 10.3748/wjg.v23.i25.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Torres J, et al. The gut microbiota, bile acids and their correlation in primary sclerosing cholangitis associated with inflammatory bowel disease. United European Gastroenterol. J. 2018;6:112–122. doi: 10.1177/2050640617708953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ruff WE, et al. Host-microbiota interactions in immune-mediated diseases. Nat. Rev. Microbiol. 2020;18:521–538. doi: 10.1038/s41579-020-0367-2. [DOI] [PubMed] [Google Scholar]
- 178.Meng X, et al. Microbe-metabolite-host axis, two-way action in the pathogenesis and treatment of human autoimmunity. Autoimmun. Rev. 2019;18:455–475. doi: 10.1016/j.autrev.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 179.Cai W, et al. Intestinal microbiome and permeability in patients with autoimmune hepatitis. Best. Pract. Res Clin. Gastroenterol. 2017;31:669–673. doi: 10.1016/j.bpg.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 180.Fussey SP, et al. Reactivity of primary biliary cirrhosis sera with Escherichia coli dihydrolipoamide acetyltransferase (E2p): characterization of the main immunogenic region. Proc. Natl Acad. Sci. USA. 1990;87:3987–3991. doi: 10.1073/pnas.87.10.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bogdanos DP, et al. Primary biliary cirrhosis is characterized by IgG3 antibodies cross-reactive with the major mitochondrial autoepitope and its Lactobacillus mimic. Hepatology. 2005;42:458–465. doi: 10.1002/hep.20788. [DOI] [PubMed] [Google Scholar]
- 182.Terjung B, et al. p-ANCAs in autoimmune liver disorders recognise human beta-tubulin isotype 5 and cross-react with microbial protein FtsZ. Gut. 2010;59:808–816. doi: 10.1136/gut.2008.157818. [DOI] [PubMed] [Google Scholar]
- 183.Manfredo Vieira S, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156–1161. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nakamoto N, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol. 2019;4:492–503. doi: 10.1038/s41564-018-0333-1. [DOI] [PubMed] [Google Scholar]
- 185.Trivedi PJ, et al. Intestinal CCL25 expression is increased in colitis and correlates with inflammatory activity. J. Autoimmun. 2016;68:98–104. doi: 10.1016/j.jaut.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Trivedi PJ, et al. Vascular adhesion protein-1 is elevated in primary sclerosing cholangitis, is predictive of clinical outcome and facilitates recruitment of gut-tropic lymphocytes to liver in a substrate-dependent manner. Gut. 2018;67:1135–1145. doi: 10.1136/gutjnl-2016-312354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Henriksen EK, et al. Gut and liver T-cells of common clonal origin in primary sclerosing cholangitis-inflammatory bowel disease. J. Hepatol. 2017;66:116–122. doi: 10.1016/j.jhep.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 188.Moro-Sibilot L, et al. Mouse and human liver contain immunoglobulin A-secreting cells originating from Peyer’s patches and directed against intestinal antigens. Gastroenterology. 2016;151:311–323. doi: 10.1053/j.gastro.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 189.Bajaj JS, Khoruts A. Microbiota changes and intestinal microbiota transplantation in liver diseases and cirrhosis. J. Hepatol. 2020;72:1003–1027. doi: 10.1016/j.jhep.2020.01.017. [DOI] [PubMed] [Google Scholar]
- 190.Schwabe RF, Greten TF. Gut microbiome in HCC—mechanisms, diagnosis and therapy. J. Hepatol. 2020;72:230–238. doi: 10.1016/j.jhep.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 191.Sehgal R, et al. Role of microbiota in pathogenesis and management of viral hepatitis. Front. Cell Infect. Microbiol. 2020;10:341. doi: 10.3389/fcimb.2020.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lu H, et al. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb Ecol. 2011;61:693–703. doi: 10.1007/s00248-010-9801-8. [DOI] [PubMed] [Google Scholar]
- 193.Cui L, et al. The human mycobiome in health and disease. Genome Med. 2013;5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Xu M, et al. Changes of fecal Bifidobacterium species in adult patients with hepatitis B virus-induced chronic liver disease. Microb. Ecol. 2012;63:304–313. doi: 10.1007/s00248-011-9925-5. [DOI] [PubMed] [Google Scholar]
- 195.Chou HH, et al. Age-related immune clearance of hepatitis B virus infection requires the establishment of gut microbiota. Proc. Natl Acad. Sci. USA. 2015;112:2175–2180. doi: 10.1073/pnas.1424775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chauhan, A. et al. Fecal microbiota transplantation in hepatitis B e antigen-positive chronic hepatitis B patients: a pilot study. Dig. Dis. Sci. 2020. 10.1007/s10620-020-06246-x. Online ahead of print. [DOI] [PubMed]
- 197.Aly AM, et al. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016;8:42. doi: 10.1186/s13099-016-0124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Heidrich B, et al. Intestinal microbiota in patients with chronic hepatitis C with and without cirrhosis compared with healthy controls. Liver Int. 2018;38:50–58. doi: 10.1111/liv.13485. [DOI] [PubMed] [Google Scholar]
- 199.Inoue T, et al. Gut dysbiosis associated with hepatitis C virus infection. Clin. Infect. Dis. 2018;67:869–877. doi: 10.1093/cid/ciy205. [DOI] [PubMed] [Google Scholar]
- 200.Wu J, et al. Altered faecal microbiota on the expression of Th cells responses in the exacerbation of patients with hepatitis E infection. J. Viral Hepat. 2020;27:1243–1252. doi: 10.1111/jvh.13344. [DOI] [PubMed] [Google Scholar]
- 201.Qin N, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 202.Bajaj JS, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 2014;60:940–947. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bajaj JS, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Santiago A, et al. Alteration of the serum microbiome composition in cirrhotic patients with ascites. Sci. Rep. 2016;6:25001. doi: 10.1038/srep25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bajaj JS, et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260–1271. doi: 10.1002/hep.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bajaj JS, et al. Association between intestinal microbiota collected at hospital admission and outcomes of patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2019;17:756–65.e3. doi: 10.1016/j.cgh.2018.07.022. [DOI] [PubMed] [Google Scholar]
- 207.Albillos A, et al. Cirrhosis-associated immune dysfunction: distinctive features and clinical relevance. J. Hepatol. 2014;61:1385–1396. doi: 10.1016/j.jhep.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 208.Muñoz L, et al. Intestinal immune dysregulation driven by dysbiosis promotes barrier disruption and bacterial translocation in rats with cirrhosis. Hepatology. 2019;70:925–938. doi: 10.1002/hep.30349. [DOI] [PubMed] [Google Scholar]
- 209.Lorenzo-Zúñiga V, et al. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 2003;37:551–557. doi: 10.1053/jhep.2003.50116. [DOI] [PubMed] [Google Scholar]
- 210.Sorribas M, et al. FXR modulates the gut-vascular barrier by regulating the entry sites for bacterial translocation in experimental cirrhosis. J. Hepatol. 2019;71:1126–1140. doi: 10.1016/j.jhep.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 211.Oh, T. G. et al. A universal gut-microbiome-derived signature predicts cirrhosis. Cell Metab. (2020). [DOI] [PMC free article] [PubMed]
- 212.Tilg H, et al. Gut microbiome and liver diseases. Gut. 2016;65:2035–2044. doi: 10.1136/gutjnl-2016-312729. [DOI] [PubMed] [Google Scholar]
- 213.Ponziani FR, et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology. 2019;69:107–120. doi: 10.1002/hep.30036. [DOI] [PubMed] [Google Scholar]
- 214.Ren Z, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014–1023. doi: 10.1136/gutjnl-2017-315084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lin RS, et al. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J. Hepatol. 1995;22:165–172. doi: 10.1016/0168-8278(95)80424-2. [DOI] [PubMed] [Google Scholar]
- 216.Bellot P, et al. Bacterial DNA translocation is associated with systemic circulatory abnormalities and intrahepatic endothelial dysfunction in patients with cirrhosis. Hepatology. 2010;52:2044–2052. doi: 10.1002/hep.23918. [DOI] [PubMed] [Google Scholar]
- 217.Routy B, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 218.Lagier JC, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 219.Metwaly A, Haller D. Multi-omics in IBD biomarker discovery: the missing links. Nat. Rev. Gastroenterol. Hepatol. 2019;16:587–588. doi: 10.1038/s41575-019-0188-9. [DOI] [PubMed] [Google Scholar]
- 220.Zhang Q, et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut. 2019;68:2019–2031. doi: 10.1136/gutjnl-2019-318912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Burberry A, et al. C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature. 2020;582:89–94. doi: 10.1038/s41586-020-2288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hu, S. et al. Whole exome sequencing analyses reveal gene-microbiota interactions in the context of IBD. Gut 2020;gutjnl-2019-319706. 10.1136/gutjnl-2019-319706. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 223.Bober JR, et al. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu. Rev. Biomed. Eng. 2018;20:277–300. doi: 10.1146/annurev-bioeng-062117-121019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hendrikx T, et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68:1504–1515. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]