Abstract
Mutations in ESR1 that confer constitutive estrogen receptor alpha (ER) activity in the absence of ligand are acquired by ≥40% of metastatic breast cancers (MBC) resistant to adjuvant aromatase inhibitor (AI) therapy. To identify targetable vulnerabilities in MBC, we examined steroid hormone receptors and tumor-infiltrating immune cells in metastatic lesions with or without ER mutations. ER and progesterone receptor (PR) were significantly lower in metastases with wild type (WT) ER compared to those with mutant ER, suggesting that metastases that evade AI therapy by mechanism(s) other than acquiring ER mutations lose dependency on ER and PR. Metastases with mutant ER had significantly higher T regulatory and T helper cells, total macrophages, and PD-L1 positive immune-suppressive macrophages than those with WT ER. BC cells with CRISPR Cas9-edited ER (D538G, Y537S, or WT) and patient derived xenografts (PDX) harboring mutant or WT ER revealed genes/proteins elevated in mutant ER cells, including androgen receptor (AR), Chitinase-3-Like Protein 1 (CHI3L1), and Interferon Stimulated Genes (ISG). Targeting these proteins blunted the selective advantage of ER mutant tumor cells to survive estrogen deprivation, anchorage independence, and invasion. Thus, patients with mutant ER MBC might respond to standard of care fulvestrant or other selective estrogen receptor degraders (SERD) when combined with AR or CHI3L1 inhibition, perhaps with the addition of immunotherapy.
Keywords: breast cancer, steroid hormone receptor, androgen receptor, estrogen receptor, ESR1 mutant, type-1 interferon, CHI3L1, IFITM3, IFITM1
Introduction
Two-thirds of all early (non-metastatic) primary breast cancers (BC) express estrogen receptor alpha (ER). First line adjuvant treatment for the majority of patients with ER+ disease is aromatase inhibitor (AI) therapy, which blocks conversion of androgens to estrogens by the enzyme aromatase and significantly extends time to recurrence (reviewed in (1)). However, all patients who recur will eventually develop AI resistant metastatic breast cancer (MBC) and acquired point mutations in the ESR1 gene often emerge under the selective pressure of this estrogen deprivation therapy. While other known mechanisms of endocrine therapy resistance exist, including amplification and fusions of ESR1 (2), point mutations in exon eight encoding the ER ligand binding domain (LBD) are the most prevalent. Indeed, ER mutations are detectable in circulating tumor cells or metastatic lesions in up to 40% of women with AI resistant MBC (reviewed in (3)). Specific mutations in this region result in amino acid alterations that confer constitutive receptor activity in the absence of estrogen (4-10) and consequent estrogen-independent gene regulation (11-16). In preclinical models, BC cells with mutant ER exhibit increased proliferation in vitro (13,14) and in vivo (11,12) compared to cells with wild type (WT) ER and have a greater propensity to metastasize with or without 17beta-estradiol (E2) (11,12).
Despite ER being constitutively active in the absence of E2, BC cells harboring mutant ER retain sensitivity to E2. Fulvestrant (Fulv), the selective ER degrader that serves as the current second line therapy for AI resistant disease, binds to mutant ER at lower affinity than to WT ER (reviewed in (17)). The poor bioavailability of Fulv necessitates intramuscular injections and higher doses are problematic. Hence, combination strategies to lower the effective dose of Fulv or alternative therapies are desirable. Cyclin dependent kinase-4/6 (CDK 4/6) inhibitors, such as Palbociclib (Palbo), improve survival for patients with ER+ MBC and Palbo+Fulv slightly improved survival in patients harboring mutant ER when compared to Fulv alone; however, CDK 4/6 inhibitors are not without side effects (18). Other therapies being tested in combination with Fulv include the CDK7 inhibitor THZ1 that showed enhanced efficacy in preclinical models (12) and second-generation selective ER degraders (SERDs), such as AZD9496 (19). However, there is clearly a need for novel therapeutic approaches for patients with AI refractory ER+ MBC.
Although both WT and mutant ER arise from ER+ BC that escapes adjuvant AI therapy, it is possible that due to differing biological properties, alternative therapeutic strategies may be required to treat these two forms of AI refractory MBC (11,20,21). In the companion manuscript, Arnesen et al molecularly characterize WT versus mutant ER BC cells, studying the most commonly occurring D538G and Y537S mutants associated with poor patient outcomes (4-7,22). Interestingly, concurrent analyses of new data from Arnesen et al and two other studies (12,13) indicated that many genes differentially expressed by mutant ER BC cells compared to WT are not E2 regulated. These results suggested that the selective advantages conferred by mutant ER are not solely attributable to constitutive ER activity. Since ER mutations represent an acquired adaptation that facilitates BC survival and recurrence as metastatic disease in women on AI therapy, we postulated that the selective advantages conferred by mutant ER might become more evident by modeling the clinical situation in which the mutations arise (E2-deprivation). Therefore, we examined specific proteins from mutant-enhanced pathways identified by genomics in Arnesen et al, under conditions that mimic the altered hormonal milieu that ensues upon AI therapy and anchorage independence, to simulate the selective pressure of treatment and early metastasis. This analysis led to identification of innate immune cell activating signatures that were confirmed by multiplex analysis of tumor infiltrating immune cells in MBC specimens. We also hypothesized that the long-term E2-deprivation induced by AI therapy blocking conversion of androgens to estrogens might cause tumor cells to shift to reliance on androgens/AR. The complement of steroid hormone receptor proteins including ER, progesterone receptor (PR), androgen receptor (AR), and glucocorticoid receptor (GR), had not previously been examined together in MBC with or without ESR1 mutations. Our approach combining data from preclinical models and biopsies of MBC, identified proteins that confer survival advantages relevant to AI refractory MBC that may serve as vulnerabilities and lead to new therapeutic approaches for this fatal form of the disease.
Materials and Methods
Metastatic Breast Cancer Patient Samples
Mutation analysis.
Core needle biopsies were acquired from patients, who gave their informed written consent, with ER+/Her2− measurable or evaluable metastatic breast cancer (MBC) without CNS disease enrolled in clinical trial NCT02953860 (COMIRB 16-1001). Median age of patients was 61 years (46-87); PS 1 (0-1); a median of 2 prior chemotherapy and 2 prior hormonal therapies for metastatic disease (including 7 with prior Fulvestrant), and 90% had visceral disease. An additional three biopsies were from NCT01597193 (COMIRB 12-0970). Formalin fixed paraffin embedded sections were analyzed for mutations in ESR1 exon 8 as well as 67 other gene hotspots frequently altered in cancer using a modified Archer VariantPlex Solid Tumor Assay through the CMOCO Laboratory (University of Colorado Department of Pathology). Mutation mutual exclusivity analyses were performed for commonly occurring mutations using cBioPortal (cBioPortal, RRID:SCR_014555) for our data set and data from the 2016 INSERM database and the 2020 Metastatic Breast Cancer Project database. The majority (67%) of biopsies were from the liver. The other sites included skin (3.7%) and the rest equally divided across lymph node, other soft tissue, breast and bone (at 7.4% each). Invasive ductal carcinomas represented 63% of the biopsies, invasive lobular carcinoma, 18.5%, and invasive mammary carcinoma (IMC) 14.8% and 3.7% unknown.
Immunohistochemistry (IHC).
IHC to detect ER (Agilent Cat# M7047, RRID:AB_2101946), PR (Agilent Cat# M3568, RRID:AB_2252608), AR (Agilent Cat# M3562, RRID:AB_2060174), GR (Cell Signaling Technology Cat# 3660, RRID:AB_11179215), Ki67 (Agilent Cat# M7240, RRID:AB_2142367), cleaved caspase 3 (Cell Signaling Technology Cat# 9661, RRID:AB_2341188), and IFITM3 (GeneTex Cat# GTX115407, RRID:AB_11172546) was performed on FFPE sections of the MBC biopsies described above. Antigen retrieval and detection were optimized for each antibody. IHC stained slides were scored for intensity and percent cells positive by breast pathologist Sharon B Sams, MD, who was blinded to ER mutation status. Pearson correlation values were calculated between the different receptors in MBC biopsies using percent positive cells.
Multiplex panel for tumor infiltrating immune cells.
FFPE sections were stained for immune markers using multiplex Opal™ TSA technology (Akoya Biosciences) along with the Vectra 3 Automated Quantitative Pathology Imaging System. TIL antibodies used were: CD4 (Agilent Cat# M7310, RRID:AB_2728838), Foxp3 (Abcam Cat# ab20034, RRID:AB_445284), CD8 (Agilent Cat# M7103, RRID:AB_2075537), CD20 (Abcam Cat# ab9475, RRID:AB_307267), and CD68 (Agilent Cat# GA60961-2, RRID:AB_2661840), and pan cytokeratin (Agilent Cat# M3515, RRID:AB_2132885) was used to identify tumor epithelium. PD-L1/CD68 Co-IF antibodies used were: PD-L1 (Abcam Cat# ab228462, RRID:AB_2827816) and CD68 (Agilent Cat# GA60961-2, RRID:AB_2661840). Dapi (Akoya Cat# FP1490) was used as a counterstain for each core needle biopsy, and positive cells in three to five 669um x 500um fields were scored using InForm software (Perkin Elmer) using either a pixel or cell-based algorithm including both tissue and cell segmentation.
Cell Culture
Cell culture conditions.
BC cell lines (MCF-7 and T-47D) were obtained from Dr. Kathryn Horwitz at the University of Colorado Anschutz Medical Campus. Cell lines were authenticated by Short Tandem Repeat DNA Profiling (Promega) and tested for mycoplasma in the University of Colorado Cancer Center (UCCC) Cell Technologies shared resource (September 2020). WT and mutant ER CRISPR cell lines were created by Dr. Jay Gertz at the Huntsman Cancer Institute using CETCH-seq to endogenously flag-tag WT or mutant (Y537S or D538G) ER using Cas9 mediated homologous recombination (see companion paper and (23)). MCF-7 cells were maintained in MEM with 5% FBS, 1% non-essential amino acids, 6.0 μg/mL insulin, and 1% penicillin/streptomycin. T-47D cells were cultured in RPMI-1640 media supplemented with 5% FBS and 1% penicillin/streptomycin. To mimic conditions resulting from AI therapy, where the ER mutations arise in patients, cells were also cultured in phenol red-free media (MEM for MCF-7 and RPMI-1640 for T-47D) supplemented with 5% dextran charcoal (DCC) stripped FBS for 6 months to generate long-term estrogen-deprived (LTED) cells. When appropriate, cell lines were cultured for 8 or fewer passages prior to experimentation.
Immunoblotting.
Cells were grown in full media under attached or forced-suspension conditions (on poly 2-hydroxyethyl methacrylate, poly-HEMA plates) for 24, 48, or 72 hours prior to harvest. Cells were then lysed in RIPA buffer (150 nM NaCl, 1% IPEGAL, 0.5% Na-Deoxycholate, 0.1% SDS, 50mM Tris, and 1.0mM EDTA) containing protease and phosphatase inhibitors. Whole cell protein lysates were separated by 10-12% SDS-PAGE gels, transferred to PVDF membranes, blocked in 3% BSA in Tris-buffered saline-Tween (TBST), and incubated with antibody overnight at 4°C. Primary antibodies used include: AR (Millipore Cat# 06-680, RRID:AB_310214), Flag (Sigma-Aldrich Cat# F1804, RRID:AB_262044), ER (Thermofisher Cat# RM9101-S), GR (Cell Signaling Technology Cat# 3660, RRID:AB_11179215), PR (Agilent Cat# M3568, RRID:AB_2252608), Tubulin (Sigma-Aldrich Cat# T5168, RRID:AB_477579), IFITM3 (GeneTex Cat# GTX115407, RRID:AB_11172546), and GAPDH (Sigma-Aldrich Cat# G8795, RRID:AB_1078991). Incubation in secondary antibody was followed by detection using an Odyssey CLx Imager (Odyssey CLx, RRID:SCR_014579). Densitometry was performed using the ImageJ software (ImageJ, RRID:SCR_003070) and reported as a ratio normalized to respective loading control (tubulin or GAPDH).
Immunohistochemistry on BC cell lines and patient-derived xenografts.
Cells were plated as described above and then were pelleted, fixed in 10% formalin, and paraffin-embedded by the University of Colorado Denver Tissue Biobanking and Processing Core. Slides were deparaffinized in xylene and ethanol and heat induced epitope retrieval was performed. Primary antibodies used were: AR (Abnova Cat# MAB10053, RRID:AB_10903299), ER (Thermofisher Cat# RM9101-S), PR (Agilent Cat# M3568, RRID:AB_2252608), IFITM3 (GeneTex Cat# GTX115407, RRID:AB_11172546), MUC1 (Cell Signaling Technology Cat# 4538, RRID:AB_2148549), and CHI3L1 (Abcam Cat# ab77528, RRID:AB_2040911). Representative images were taken at 200x, 400x, or 1000x magnification. Quantification of nuclear AR was completed using the Aperio microscope (Aperio ScanScope XT Leica, RRID:SCR_018457) and ImageScope (ImageScope, RRID:SCR_014311), to calculate percent nuclei positive and staining intensities of 0, 1+, 2+, or 3+. Data represents the nuclear intensity of all cells in each cell pellet (N>100 cells/pellet). For CHI3L1, IFITM3, and MUC1, ImageJ software (ImageJ, RRID:SCR_003070) was used to determine the average number of cells with 3+ staining in three representative images taken at 400x.
Soft Agar Colony Formation.
Soft agar colony formation assays were used to assay for anchorage-independent survival in WT and mutant ER MCF-7 and T-47D cells treated with vehicle (ethanol), 20μM Seviteronel (Sevi), or 20μM Enzalutamide (Enza). Experiments were performed in 6-well plates containing the following layers: 0.5% bottom agar, 0.3% top agar containing 20,000cells/well, and covered with the appropriate media. Cells were grown for approximately 21 days with bi-weekly media changes then fixed and stained with nitro blue tetrazolium at the time of harvest. Experiments were performed in triplicate (N=3) and the average colony number was quantified using ImageJ software (ImageJ, RRID:SCR_003070).
Invasion Assays.
Cells were plated at 50,000 cells/well in 1% FBS on the top of a Matrigel-coated invasion chamber with 8.0 micron holes (Corning) and normal growth media (with 5% FBS) was used as a chemoattractant in the bottom of each well. The following day 10μg/mL anti-CHI3L1 blocking antibody (Millipore Cat#MABC196) or IgG control (Thermo Fisher Scientific Cat# 31903, RRID:AB_10959891) was added to the top of each well. 72 hours post-treatment, cells were fixed in 10% formalin and stained with 1% crystal violet. Cell invasion was determined by analyzing the average area of crystal violet staining on the bottom of each insert using ImageJ software. Presented is the average area of two separate experiments (N=3/experiment).
Identification of mutant specific and AR regulated genes.
D538G and Y537S ER mutant-specific genes were identified by analyzing RNA-seq data from WT and mutant ER T-47D and MCF-7 cell lines generated by the Gertz lab (Arnesen et al, co-submitted), the Oesterreich lab [UPMC Hillman Cancer Center (13)], and the Brown lab [Dana Farber Cancer Institute groups (12)] using an adjusted p-value cutoff of <0.05. Pathway analysis of mutant-specific genes identified through the multivariate/multilab RNA-seq analysis (Arnesen et al) were analyzed using Illumina’s BaseSpace Correlation Engine to identify pathways and molecular and cellular processes that are significantly associated with mutant-specific gene expression changes. Gene sets were ranked by log2 fold change and p-value. Gene ontology enrichment p-values were calculated by Illumina’s BaseSpace Correlation Engine. A list of AR regulated genes was compiled from multiple sources including prostate and breast cancer, and cell lines derived from both types of cancer (24-26). This curated list of AR-regulated genes was overlapped with mutant-specific genes identified in our multi-lab RNA-seq analysis described above. Significant overlap between mutant specific genes and AR regulated genes was identified using a hypergeometric p-value cut off of <0.05.
Animal Studies
All animal experiments were performed in accordance with protocols approved by the University of Colorado Institutional Animal Care and Use Committee (IACUC) using humane procedures. 6 week old ovariectomized NOD scid gamma (NSG) mice (IMSR Cat# JAX:005557, RRID:IMSR_JAX:005557) were implanted subcutaneously at base of neck with pellets containing cellulose or pellets containing 2mg 17beta-estradiol (E2) in cellulose (N=10/group). Mice were injected via the tail vein with T-47DGFP/Luc cells containing either WT or D538G ER to generate 4 experimental groups of mice: cellulose pellets plus WT T47D cells, cellulose pellets plus D538G T-47D cells, E2 pellets plus WT T-47D cells, and E2 pellets plus D538G T-47D cells (N=5/group). Mice were imaged weekly by IVIS (Perkin Elmer IVIS Spectrum In-Vivo Imaging System, RRID:SCR_018621) beginning the day after tumor cell injection. Six weeks post cell injection, mice were imaged by IVIS, then were sacrificed, and excised lungs were imaged by IVIS ex vivo. Tumor burden at endpoint (6 weeks) is presented as total flux (photons/second) of luminescence from IVIS imaging. Patient derived xenografts (PDX) were kindly provided by Dr. Alana Welm, Huntsman Institute and Dr. Carol Sartorius, University of Colorado. PDX were passaged in cycling female NSG mice supplemented with E2 with the exception of HCI-013-EI tumors passaged in un-supplemented mice.
Statistical Analysis
Data are presented as the mean ± Standard Error of the Mean (SEM). When comparing two groups of data, an unpaired Student’s t-test was used. When heteroscedasticity was present or data were skewed, the non-parametric Mann-Whitney test was used. For statistical analysis between more than two groups a One way ANOVA or Two way ANOVA with a Tukey’s multiple comparison test was used. In the case of heteroscedasticity or skewed data, non-parametric Kruskal-Wallis or Friedman’s tests were used. When data were presented on a log-scale (Figure 4) data analysis using parametric tests was conducted on log transformed data to account for heteroscedasticity between groups. Statistical significance was defined as p<0.05 and all tests were two-sided. Analyses were done using GraphPad Prism (GraphPad Prism, RRID:SCR_002798, ver8.3). Power calculations for the animal study were conducted under the supervision of a collaborating biostatistician Dr. Kathleen Torkko. Animal numbers were calculated at 80% power to the expected difference at P<0.05 (two-tailed).
Results
Steroid hormone receptor status of metastases harboring ER mutations
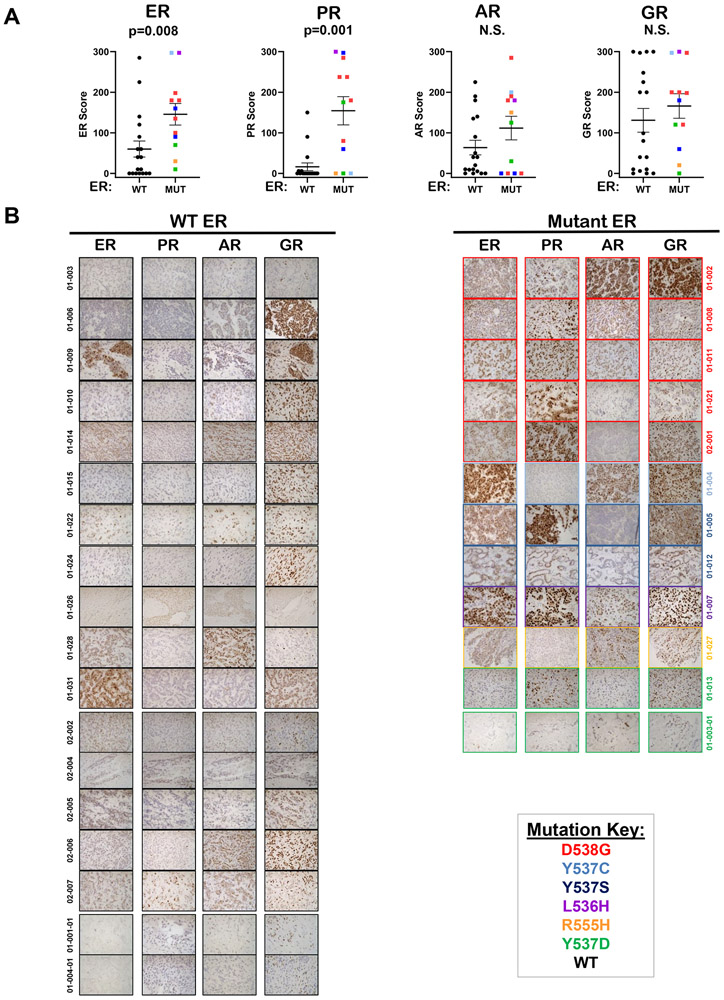
Core needle biopsies from accessible metastases were collected from consented patients primarily enrolled in NCT02953860 and 3 from NCT01597193. All patients had ER+HER2− BC at time of diagnosis. The steroid hormone receptor proteins, estrogen receptor alpha (ER), progesterone receptor (PR), androgen receptor (AR), and glucocorticoid receptor (GR), had not previously been examined together in the same cases of MBC harboring WT versus mutant ER. FFPE sections from core needle biopsies were stained for ER, PR, AR, and GR (Figure 1A-B), and Ki67 and cleaved caspase-3 (Sup Figure S1A-B). IHC was scored by breast pathologist (SBS) and graphed according to ER mutation status. ER and PR protein were significantly higher in biopsies harboring mutant ER as compared to metastases with no detectable mutations in exon 8 (designated as WT ER, Figure 1A). In contrast AR and GR were not significantly different in biopsies with WT ER compared to those with mutant ER (Figure 1A). In WT ER metastases, where ER and PR were often low, AR was frequently maintained (patients: 01-006, 01-010, 01-014, 01-022, 01-028, 02-006, 02-007: Figure 1B) and was therefore not significantly different between WT and mutant biopsies. Interestingly, some mutant ER cancers had a substantially higher AR than PR (patients: 01-002, 01-004 and 01-027: Figure 1B), and mutant ER metastases with high PR had low AR. Ki67 and cleaved caspase-3 were not significantly different between the two groups, although there was a trend towards higher Ki67 in metastases with mutant ER compared to WT ER (Sup Figure S1A-B). We also examined correlations between hormone receptors and as would be expected since ER regulates PR, these two receptors are positively correlated. ER is correlated with AR as well. It is evident that while many ER mutant containing biopsies (red) are high for ER and PR, the ER WT (black) have predominately lost PR, but often retain AR expression. ER does not correlate with GR and PR and AR do not correlate (Sup Figure S2A-D).
Figure 1. Steroid hormone receptor expression in biopsies of metastatic breast cancer with wild type or mutant ESR1.
A. Formalin fixed paraffin embedded (FFPE) sections of core needle biopsies (N=18 WT ER, N=12 mutant ER) were stained by IHC for ER, PR, AR, and GR. Depicted are the mean scores (intensity x percent cells staining) ± SEM. Mann-Whitney tests were performed for each receptor stained. B. Representative images for all WT ER metastases (left) and all mutant ER metastases (right) stained for ER, PR, and AR and GR are shown at 400X.
Mutant ER breast cancer cells upregulate AR expression and activity
To study mutant ER expressed from its native locus in MCF-7 and T-47D human BC cell lines, FLAG-tagged ER mutant models were engineered using CRISPR to introduce one allele of FLAG-tagged ESR1 containing the D538G or Y537S mutation or FLAG-tagged WT ESR1 (Arnesen et al, co-submitted). Immunoblot for FLAG confirmed CRISPR construct expression in two clones for each cell line (Sup Figure S3A-B). Further experiments were conducted using two clones pooled for each line.
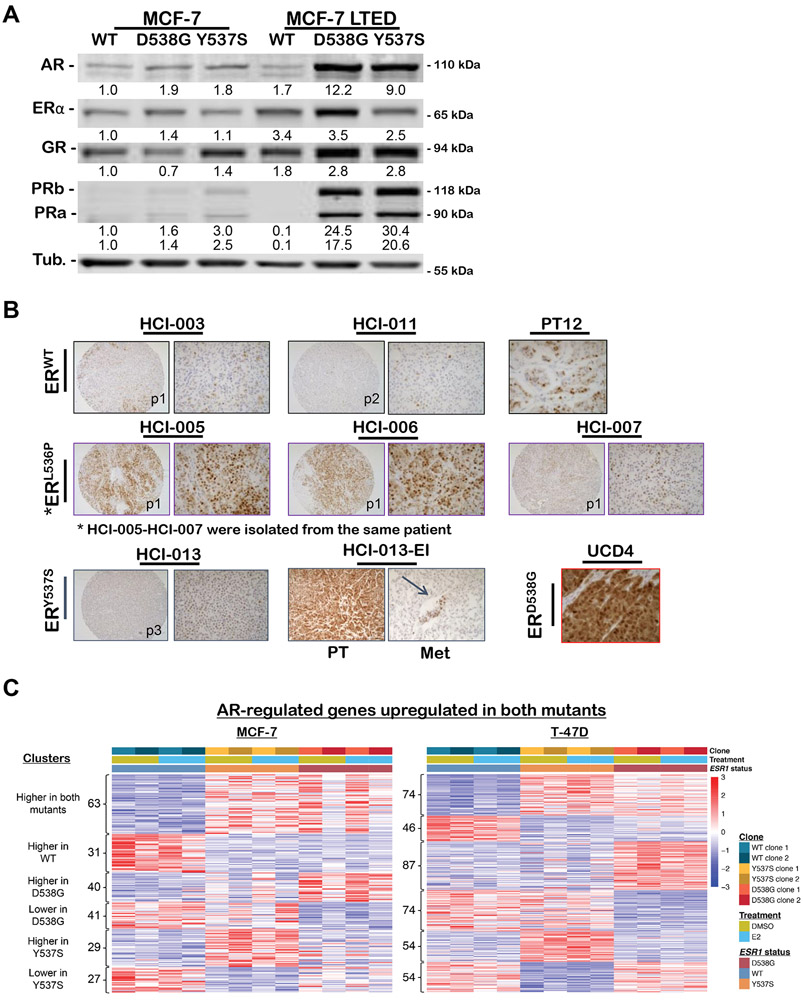
Steroid hormone receptor expression was assessed by immunoblot of WT, D538G, and Y537S MCF-7 and T-47D cells grown in culture media containing full serum (containing steroid hormones). However, to mimic conditions resulting from AI therapy, where the ER mutations arise in patients, the cells were also cultured in phenol red free media containing DCC stripped serum for 6 months to generate long-term estrogen-deprived (LTED) cells. While AR was slightly elevated in the MCF7 ER mutant compared to WT cells cultured in full serum, in the LTED conditions ER, PR, AR and GR were all increased. Interestingly AR and PR were dramatically increased in the LTED mutants compared to LTED WT. (Figure 2A). After LTED, mutant ER MCF-7 lines had dramatically increased ER, PR, AR and GR when compared to WT ER cells. In T-47D LTED resulted in increased ER, PR AR and GR compared to the cells cultured in full serum. AR and GR increased with LTED in both WT and Mutants, indicating that WT cells that survive LTED can also upregulate AR and GR. PR did not increase as a result of LTED in the WT (Sup Figure S4).
Figure 2. AR protein and gene regulation differs in WT versus mutant ER expressing MCF-7 breast cancer cells and patient derived xenografts (PDX), particularly after long-term estrogen deprivation (LTED).
A. Western blot of whole cell lysates from WT, D538G, and Y537S MCF7 cells grown in full serum media (left) or charcoal stripped serum containing media (LTED, right) were probed for AR, ER, GR, PR-A/B, and tubulin. B. ER+ patient-derived xenografts (PDX) tissue microarray from Dr. Alana Welm, Huntsman Cancer Institute (HCI) and ER+ PDX from University of Colorado (UCD) with ESR1 mutation status indicated, were stained for AR by IHC with representative images at 100X (left) and 400X (right). For the estrogen independent version of HCI-013, HCI-013-EI, an image representative of primary tumor (PT) and lung metastasis (Met) are shown. C. Heatmap of known AR regulated genes in MCF7 (left) and T47D (right) depicting relative expression of genes displayed as a Z-score across WT and mutant cells after 5 days of growth in hormone-depleted media followed by treatment with either 10nM E2 or vehicle (DMSO) for 8 hours. Significant overlap was determined using hypergeometric tests with a p-value cut off of <0.05: AR genes vs D538G up-regulated genes = 5.5x10−05; AR genes vs D538G down-regulated genes = 1.0x10−08; AR genes vs Y537S up-regulated genes = 3.2x10−08; AR genes vs Y537S down-regulated genes = 9.3x10−11.
Since AR was upregulated in ER mutant versus WT cells and by LTED, we evaluated AR by IHC in multiple ER+ patient-derived xenografts (PDX) (Figure 2B). Upon knowledge of the ER mutational status of the PDX, AR was found to be higher in PDX with ER mutations as compared to those with WT ER. Further, the Y537S mutant HCI-013-EI (estrogen independent) PDX, which was derived from the parental HCI-013 PDX passaged in mice without E2 supplementation (to mimic the E2 deprivation therapy in BC patients), exhibited elevated AR expression compared to parental HCI-013 tumors passaged in E2 supplemented mice. Human AR was also maintained in spontaneous metastases from the HCI-013-EI primary tumors to lung (Figure 2B).
Cross-referencing known AR target genes with genes differentially expressed in mutant ER BC cells revealed statistically significant enrichment (hypergeometric test p-values < 6x10-5) of AR target genes in mutant ER cells compared to WT (Figure 2C, genes identified in Sup Table S1). In this experiment, cells were cultured in phenol red free media containing DCC stripped serum for 5 days, then supplemented ± E2 for 8 hours. AR target genes were upregulated in mutant ER cell lines with or without E2, indicating that the mutant-specific alterations in known AR target genes occurred in an E2 independent manner.
AR inhibition abrogates the selective advantage of mutant ER BC cells for anchorage independent survival
We previously reported that AR mRNA, protein, and activity were increased in triple negative breast cancer (TNBC) cells that survive under anchorage independent conditions as compared to adherent cells, and AR protected against caspase-mediated cell death (27). To determine whether AR also increased in anchorage independent ER+ BC lines, parental MCF-7 cells were grown for 24-72 hours under adherent conditions (att) or forced suspension conditions on poly-HEMA coated plates (susp), which prevent cell adherence. AR was elevated at 24, 48, and 72 hours of anchorage independent growth as compared to adherent cells, while ER and GR were not (Sup Figure S5A). PR was slightly elevated at 24 hours of anchorage independent culture as compared to attached cells, but to a much lesser extent than AR. IHC on cell pellets harvested at 24, 48, and 72 hours demonstrated a striking increase in AR at each time point in the anchorage independent cells compared to attached cells, while ER and PR were not altered (Sup Figure S5B).
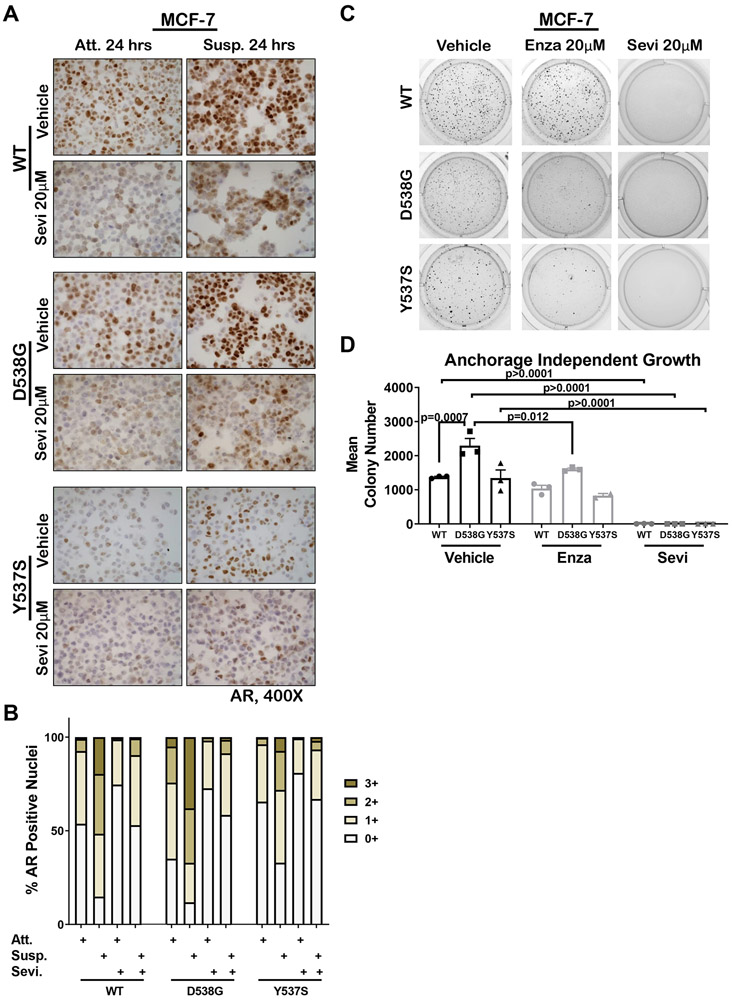
To determine if AR protein increased in mutant ER cells under anchorage independent conditions, AR was analyzed by IHC on FFPE sections of cell pellets generated from WT, D538G, and Y537S MCF-7 cells grown in attached or forced suspension conditions for 24 hours. AR protein increased in all three MCF-7 cell lines following suspension culture for 24 hours as compared to adherent cells and AR expression decreased with addition of the CYP17A1 lyase inhibitor Seviteronel (Sevi) that also acts as an AR antagonist (Figure 3A) (28). Quantification showed that strong (3+) nuclear AR staining in D538G cells was 5-fold higher at baseline than in WT MCF-7 cells (Figure 3B). The number of nuclei strongly positive for AR was increased by 19-fold for WT, 7.5-fold for D538G, and 21-fold for Y537S MCF-7 cells grown in suspension as compared to attached conditions. Sevi dramatically reduced the number of AR+ nuclei and the staining intensity (Figure 3A and B). Similar results were observed for WT, D538G, and Y537S T-47D cells (Sup Figure S6A-B). Furthermore, when grown on soft agar, D538G MCF-7 cells, which at baseline expressed the highest level of AR, formed significantly more colonies than WT counterpart (1.6-fold higher, Figure 3C-D). To determine if increased AR was indicative of a dependency on AR for survival in anchorage independent conditions, as in TNBC lines (27), MCF-7 cells were treated with Sevi, which completely abolished growth on soft agar of all MCF-7 lines. Since the AR inhibitor enzalutamide (Enza) showed clinical benefit in AR+ TNBC (as did bicalutamide) (29,30), we also tested Enza, which significantly inhibited D538G cell growth in soft agar. Importantly, Enza negated the survival advantage of D538G cells compared to WT MCF-7 in soft agar. Similar results were seen in D538G T-47D cells, where both Sevi and Enza inhibited growth on soft agar (Sup Figure S6C). WT and mutant ER MCF-7 and T-47D cells generated in the Oesterreich lab (13) were also studied on two-dimensional standard culture as compared to three-dimensional culture using ultra-low adhesion plates and three days of hormone deprivation (Sup Figure S7). AR was higher in both D538G and Y537S mutant ER MCF-7 lines compared to WT (in both 2D and 3D). While AR decreased in 3D compared to 2D in the WT cells, it remained elevated in 3D in the mutant ER lines, again demonstrating a potential role for AR in the ER mutants. Results in T47D mutants versus WT were similar.
Figure 3. AR increased in breast cancer cells grown in soft agar and AR inhibition abolished the selective advantage of mutant ER cells for anchorage-independent survival.
A. WT, D538G, and Y537S mutant MCF7 cells were grown in full serum media under attached (Att) or suspension conditions (on poly-HEMA plates, Susp) and treated ± 20μM Seviteronel (Sevi) for 24 hours. Cells were pelleted, FFPE, and AR IHC performed. Representative images are shown at 400x. B. Quantification of A with ImageScope software for percent positive nuclei for staining intensities of 0-3+ for all conditions attached (Att), suspended (Susp), and ± Sevi. C. WT, D538G, and Y537S cells plated in 0.3% agar and treated with 20μM Sevi, 20μM enzalutamide (Enza) or EtOH control and grown for three weeks with bi-weekly media and drug changes. Representative images of anchorage-independent growth are shown. D. Colony number quantified using ImageJ software for assay in C (N=3). Mean ± SEM, One way ANOVA with Tukey’s multiple comparison test is depicted. Two way ANOVA, interaction between cell line and treatment p = 0.003.
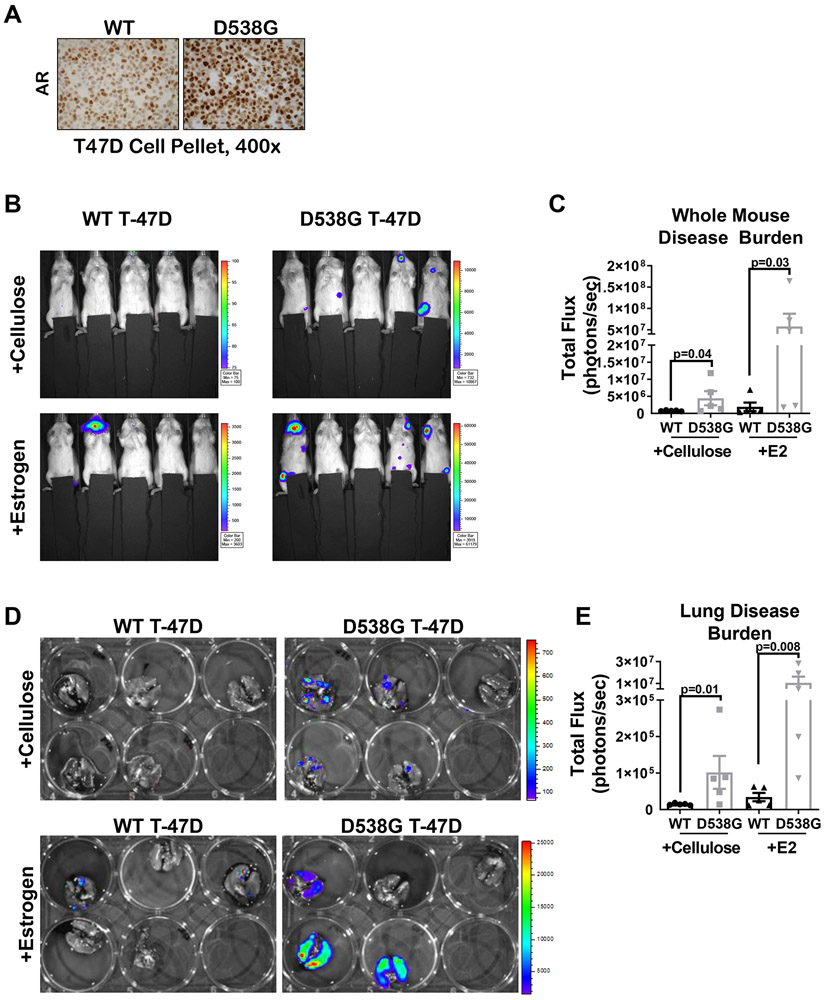
To determine whether mutant ER cells have a selective advantage for anchorage independent survival or metastatic outgrowth in vivo under conditions modeling the E2-depleted state, we introduced labeled WT or D538G T-47D cells, in which AR was elevated (Figure 4A), by tail vein injection into ovariectomized mice that were supplemented with either cellulose control or E2 pellets. Whole mouse IVIS imaging at the end of study showed significantly higher metastatic burden in mice injected with D538G T-47D cells compared to those with WT cells, both with and without E2 (Figure 4B-C). Ex vivo imaging of lungs completed immediately after necropsy also showed significantly higher metastatic burden in lungs harboring D538G cells as compared to those injected with WT T-47D with or without E2 (Figure 4D-E). In non-E2 supplemented mice, WT tumor cells were nearly undetectable when imaged in vivo or ex vivo.
Figure 4. Breast cancer cells harboring D538G ER mutation thrive at metastatic sites in an experimental metastasis model in estrogen-deprived mice, while WT ER cells do not.
A. WT and D538G ER T47DGFP/Luc cells were stained for AR and representative images shown at 400x. B. The same lines were delivered via tail vein injection into 6 week old ovariectomized NSG mice (N=5/group) with or without E2 or cellulose and six weeks post cell injection, mice were imaged by IVIS. C. Quantification of whole mouse luminescence signal (total flux), mean ± SEM. Raw data are presented with p-values from a Student’s unpaired two-tailed t test conducted on the log transformed data due to heteroscedasticity, one way ANOVA (p=0.006). D. At end of study mice were sacrificed, lungs excised and imaged ex vivo by IVIS. E. Quantification of luminescence in lungs ex vivo mean ± SEM. Raw data are presented with p-values from a Student’s unpaired two-tailed t test conducted on log transformed data due to heteroscedasticity, one way ANOVA (p=0.0003).
Mutant ER BC have elevated expression of pro-metastatic CHI3L1
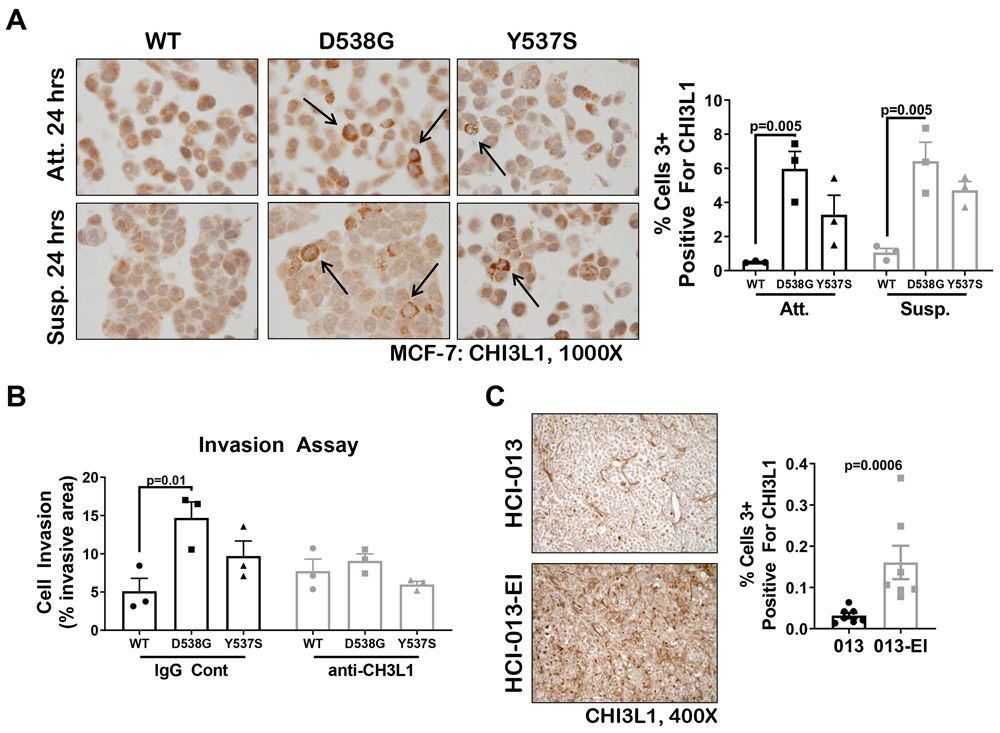
Analysis of RNA-seq data from three independent laboratories using mutant ER cell lines generated by different methods showed that CHI3L1 (also known as YKL-40) was elevated in Y537S MCF-7 and T-47D compared to WT (Sup Figure S8A-B and Arnesen et al, co-submitted). CHI3L1 is a well-established androgen/AR regulated gene in prostate cancer where it is detectable in tumor and patient serum and correlates with disease progression (31-33). Since we previously reported that CHI3L1 increased in TNBC lines under anchorage independent conditions (27), we evaluated CHI3L1 protein in WT, D538G, and Y537S MCF-7 cells in attached versus suspension culture for 24 hours (Figure 5A). CHI3L1 was significantly higher in both D538G (5-fold) and Y537S (3-fold) MCF-7 cells than WT; however, CHI3L1 was not further enhanced by 24 hours of anchorage independent culture. Since CHI3L1 promotes invasion in other cancer types, we compared the invasive capacity of D538G and Y537S MCF-7 to WT and found that D538G MCF-7, which had the highest expression of CHI3L1, were significantly more invasive than WT MCF-7 in Matrigel coated Boyden chamber assays (Figure 5B). Importantly, a CHI3L1 blocking antibody negated this significant invasive advantage of D538G MCF-7 cells compared to WT, suggesting that mutant ER BC cells can utilize CHI3L1 to promote invasion.
Figure 5. MCF-7 BC cells with mutant ER express increased CHI3L1 and blocking CHI3L1 decreased mutant ER BC cell invasion.
A. IHC staining for CHI3L1 protein in WT, D538G, and Y537S MCF-7 cells grown in full serum media under attached (Att) or suspension conditions (on poly-HEMA plates, Susp) for 24 hours. Representative images are shown at 1000x (left) and quantification of percent cells strongly positive (3+) for CHI3L1 is shown (right), mean ± SEM, N=3 photos/pellet, One way ANOVA with Tukey’s multiple comparison test is depicted. Two way ANOVA, interaction between cell line and treatment is N.S. B. Invasion through Matrigel was determined for WT, D538G, and Y537S MCF7 cells grown in full serum media. Cells were treated with 10μg/mL anti-CHI3L1 blocking antibody or IgG control 24 hours after plating and harvested 72 hours post-treatment. After crystal violet staining and imaging, the relative amount of invasion was determined using ImageJ software, mean ± SEM of two separate experiments, N=3 One way ANOVA with Tukey’s multiple comparison test is depicted. Two way ANOVA, interaction between cell line and treatment is N.S. C. CHI3L1 IHC was conducted on Y537S mutant HCI-013 grown in E2 supplemented mice and HCI-013-EI grown in ovariectomized mice. Representative images are shown at 400x (left) and quantification of strongly positive (3+) cells was conducted using the ImageScope Software (right), mean ± SEM, N=6-7, Mann-Whitney test.
We also examined CHI3L1 expression in HCI-013-EI (estrogen independent) PDX tumors, which harbor the Y537S mutant and were grown in the absence of E2, as compared to parental HCI-013 PDX tumors, which were grown in the presence of E2 (Figure 5C). HCI-013-EI tumors had an >3-fold increase in CHI3L1 expression compared to HCI-013 tumors, demonstrating that CHI3L1 is enhanced by propagating this PDX under conditions similar to the selective pressure of E2-depletion that selects for ER mutations in BC patients.
Mutant ER BC cells upregulated the immunomodulatory type-1 interferon pathway
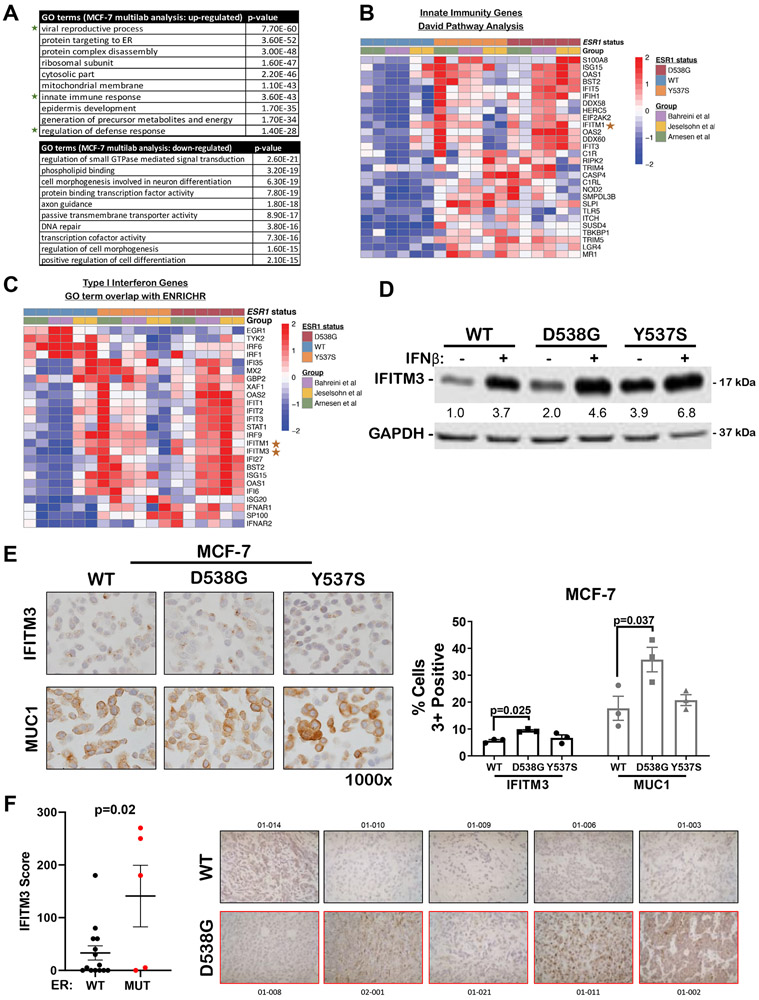
To identify additional mutant ER-specific vulnerabilities, we conducted pathway analysis of mutant-specific genes that revealed innate immune, viral recognition, and type-1 interferon among the top pathways preferentially upregulated in mutant versus WT ER MCF-7s (Figure 6A-C Arnesen et al, co-submitted). The interferon-induced transmembrane (IFITM) gene IFITM3 was consistently increased in D538G and Y537S MCF-7 cells in data from multiple laboratories (Sup Figure S8C). Both D538G and Y537S MCF-7 cells demonstrated enhanced levels of a number of interferon stimulated genes (ISGs) such as OAS1/2, IFIT1/2/3/5, IFITM1/3, and ISG15 compared to WT MCF-7 (Figure 6B-C and Sup Figure S8D-F), demonstrating robust activation of this pathway in mutant ER BC cells as compared to WT. Since IFITM proteins have been reported to play a role in AI-resistance (34,35), we explored the expression of IFITM3 in WT, D538G, and Y537S MCF-7 cells ± interferon-beta (IFNβ) stimulation, a key cytokine and activator of the type-1 interferon signaling pathway. Baseline IFITM3 levels were 2-4 fold higher in D538G and Y537S MCF-7 cells compared to WT and were further enhanced by IFNβ (Figure 6D). IHC quantification showed that IFITM3 was higher in D538G MCF-7 than WT (Figure 6E). Given that MUC1 drives IFITM1 overexpression in AI-resistant BC cells (36) and MUC1 was also on the list of genes significantly elevated in cells containing mutant versus WT ER, we also examined MUC1 protein by IHC and found it was elevated in D538G MCF-7 cells compared to WT (Figure 6E). Lastly, we performed IHC for IFITM3 in biopsies of five patients with MBC harboring the D538G ER mutation as compared to all of the WT biopsies presented in Figure 1 and found IFITM3 protein was significantly upregulated in the ER mutant versus WT biopsies (Figure 6F).
Figure 6. Mutant ER BC cells and specimens have increased type-1 interferon signaling pathway signature and increased IFITM3 protein compared to those with WT ER.
A. Illumina’s BaseSpace Correlation Engine was used to identify mutant-specific pathways from MCF7 mutant-specific genes identified through the multivariate/multilab RNA-seq analysis (Arnesen et al). B-C. Heatmap depicts relative expression, displayed as a Z-score, of genes associated with Innate Immunity (B) and Type I Interferon (C) identified by two different pathway analyses (David and ENRICHR) in ER WT and mutant MCF7 cells grown in hormone-depleted media without E2 for 3-5 days depending on the laboratory. D. WT, D538G, and Y537S MCF7 cells were treated with 1000 units of IFNβ for 24 hours, whole cell lysates were generated and analyzed by western blot for IFITM3. E. IHC for IFITM3 and MUC1 was performed on FFPE pellets generated from WT, D538G, and Y537S MCF-7 cells grown in media containing full serum, mean ± SEM, N=3 photos/pellet, One way ANOVA with Tukey’s multiple comparison test was conducted for each staining (right). Representative images are shown at 1000x (left). F. Formalin fixed paraffin embedded (FFPE) sections of core needle biopsies (N=14 WT ER, N=5 D538G mutant ER) were stained by IHC for IFITM3. Depicted are the mean scores (intensity x percent cells staining) ± SEM, Student’s unpaired two-tailed t test.
Metastases harboring mutant ER have significantly more CD4+ T cell and macrophage infiltrates
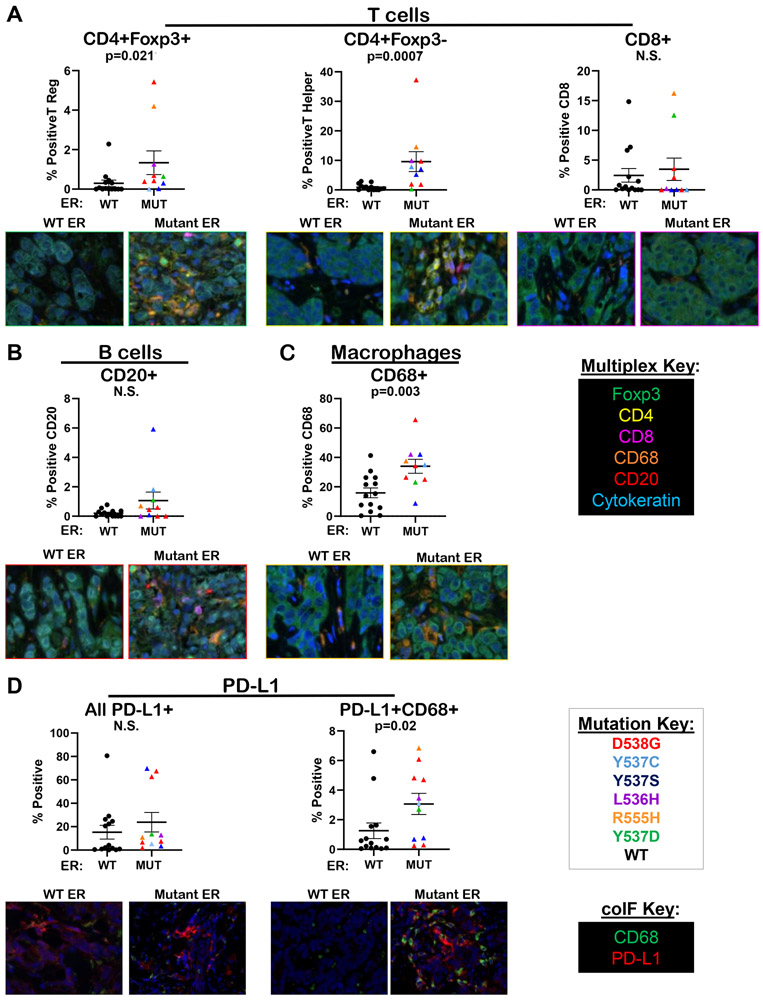
Given that mutant ER cell lines showed enhanced innate immune and interferon pathway signatures compared to WT ER, we tested biopsies of MBC with mutant versus WT ER for differences in tumor infiltrating immune cells. A multiplex panel consisting of antibodies that distinguish T cells (FoxP3+CD4+ T regulatory cells; FoxP3−CD4+ T helper cells; or CD8+ T effector cells), macrophages (CD68+), B cells (CD20+) and tumor cells (cytokeratin+) was utilized. InForm Software analysis demonstrated that the number of T regulatory (CD4+FoxP3+) and T helper cells (CD4+FoxP3−) were significantly increased in mutant ER BC metastases when compared to WT ER, although no difference in CD8+ T effector cells was observed (Figure 7A). Interestingly, in both our data (Sup Figure S9A) and data from two publicly available studies (Sup Figure S9B-C), ESR1 mutations were mutually exclusive with TP53 mutations. Further, patients with either TP53 or PIK3CA mutations contained significantly higher CD8+ cytotoxic T effector cells (Sup Figure S10A-B). There was no significant change in CD20+ B cells in mutant ER metastases compared to WT (Figure 7B). However, the number of CD68+ macrophages in mutant ER metastases was significantly increased when compared to biopsies with WT ER (Figure 7C). Finally, Programmed Death Ligand-1 (PD-L1) expression was analyzed in the same biopsies due to the recent testing of PD-L1 inhibitors in ER+ BC (Figure 7D) (37,38) and FDA approval in TNBC (39). Interestingly, while the overall level of PD-L1 did not differ between mutant and WT ER biopsies, PD-L1+ macrophages were significantly higher in mutant ER biopsies compared to WT, suggesting that macrophages infiltrating mutant ER metastases are more immunosuppressive. Interestingly, this finding was more significant when examining macrophages intimately associated with the tumor cells as compared to those found predominately in the intra-tumoral stroma (Sup Figure S11).
Figure 7. CD4+ T cells and PD-L1+ macrophages were significantly higher in biopsies of metastatic breast cancer harboring mutant ER as compared to WT ER.
Biopsies of the same metastases characterized in Figure 1 (N=14 with ER WT, N=10 with mutant ER) were stained for tumor infiltrating immune cells using Opal™ TSA technology (Akoya Biosciences) with the following colors indicating respective immune cell markers: CD4 (yellow), Foxp3 (green), CD8 (magenta), CD68 (orange), CD20 (red), and cytokeratin (cyan). Slides were scanned using Vectra 3 Automated Quantitative Pathology Imaging System (Perkin Elmer) technology and 3 to 5 representative 20x fields/tumor were analyzed using inform software (Perkin Elmer) and a pixel-based algorithm for percent positivity, mean percent positive cells ± SEM, Mann–Whitney test (top); representative images (20x, bottom) for: A. T regulatory cells (CD4+ FoxP3+), T helper cells (CD4+ FoxP3−), and cytotoxic T cells (CD8+), B. B cells (CD20+), and C. macrophages (CD68+). D. The same biopsies were stained for PD-L1 and CD68 using Opal™ TSA technology (Akoya Biosciences). Slides were scanned using a Vectra 3 Automated Quantitative Pathology Imaging System (Perkin Elmer), and up to 5 representative fields/tumor were analyzed for either total PD-L1 or dual expression of PD-L1 and CD68 using a cell phenotype-based algorithm for percent positive cells, mean positive cells ± SEM, Mann–Whitney test.
Discussion
Mutations in ESR1 that render ER constitutively active in the absence of ligand are a common mechanism of acquired resistance to AI therapy. While increased PR resulting from constitutively active mutant ER is well-documented, the four main steroid hormone receptor proteins (ER, PR, AR, and GR) had not been evaluated together in MBC biopsies harboring WT versus mutant ER. While E2 is extremely low in postmenopausal women with BC on AI, circulating and intra-tumoral androgens are increased (40). This, combined with the predominantly nuclear localization of AR in clinical samples (indicating ligand bound receptors), led us to postulate that long-term E2-deprivation via AI therapy, which blocks conversion of androgens to estrogens, might cause tumor cells to shift to reliance on androgens and AR. Indeed, in TNBC, where ER is absent, AR binds chromatin in a manner more similar to ER in ER+ BC cells than to AR in prostate cancer cells, suggesting that AR can replace ER in some TNBC (41) and confer the “luminal AR” designation (42). Our analysis of steroid hormone receptors suggests that ER+ MBC can become resistant to AI treatment by either 1) acquiring mutations that render ER constitutively active (retaining robust ER and PR positivity) or 2) by expressing little to no ER and PR in an apparent “subtype switch” to resemble TNBC, which often retain AR and/or GR.
Cross-referencing known AR regulated genes with genes differentially expressed by mutant ER versus WT ER BC cells revealed significant enrichment for AR regulated genes. Although AR protein had not specifically been examined in WT versus mutant ER MBC, it was elevated in both primary and MBC resistant to AI therapy when compared to responsive disease (43-48). AR was often maintained or elevated in MBC compared to patient-matched primary tumors, whereas ER was decreased (49). BC patient CTCs have been found to be AR+ in several studies and AR+ CTC associate with bone metastases (50) and AR mRNA was higher in CTC from BC PDX compared to primary tumors in mice (51). Furthermore, AR inhibition decreased metastatic burden of BC cells from an ER+/AR+ PDX (49) and conversely, ligand-activated AR potentiated metastasis in ER+/AR+ BC cell lines (52). These data, together with our analysis herein of hormone receptor status in clinical biopsies of AI resistant MBC and other data presented in the present study suggest that AR could be active in a subset of ER+ MBC, and might facilitate survival under conditions of E2 deprivation or anchorage independent survival. Indeed, MCF-7 and T-47D ER mutant BC lines, had higher ER, PR, and AR compared to WT, similar to results from the clinical biopsies of MBC. While all steroid receptors increased under long-term E2 deprivation (LTED) conditions, PR and AR were the most strikingly different in mutant compared to WT LTED cells. Historically, ER+ PDX were propagated in mice supplemented with E2 to increase take rate and growth rate for research studies regardless of ER mutational status. We now know that many of these ER+ PDX, derived from MBC in postmenopausal women on AI therapy (a long-term E2-deprived context) do harbor ER mutations. Placing these into mice supplemented with E2 could result in tumors that are “re-addicted” to E2 and therefore do not accurately model the selective pressure of long term E2-deprivation under which the mutations arise. We stained ER+ PDX for AR and found that PDX harboring ER mutations had higher AR than those with WT ER. Furthermore, in support of the theory that E2-deprivation enhances AR expression, the E2-independent HCI-013-EI PDX (with Y537S ER mutation) propagated in E2-deprived mice, expressed more AR protein than its parental counterpart propagated in mice supplemented with E2. Importantly, HCI-013-EI lung metastases originating from the orthotopic site still expressed AR. Thus, maintaining models of mutant ER disease under conditions similar to those in which they arise in the clinic (E2-deprived) may reveal important, clinically relevant molecular vulnerabilities.
In addition to E2-deprivation, we used anchorage independence as a model of early metastasis. Although AR significantly increased in both ER mutant and WT BC cells surviving anchorage independent conditions, anti-androgens abrogated the selective advantage of mutant ER cells for anchorage independent survival. This is consistent with previous studies in TNBC cells that demonstrated increased AR in anchorage independent cells, where it protected against apoptosis (27), rendering anchorage independent cells highly sensitive to AR inhibition (53). Therefore, AR targeting may limit AI-resistant MBC recurrence, perhaps for both patients with either WT or mutant ER, since some WT ER biopsies retained AR but had low ER and PR.
CHI3L1, encoding Chitinase 3-like 1, was preferentially expressed in BC cells harboring mutant ER. CHI3L1/YKL-40 is linked to chronic inflammation and cancer and is upregulated in the mammary gland during involution, where it contributes to a pro-tumorigenic wound-healing like environment via enhanced macrophage infiltration (54). In prostate cancer, CHI3L1 is upregulated by androgens/AR (33), and in breast cancer it is immune-suppressive (55), and correlates with M2 macrophage infiltration (56), enhanced metastasis (57,58), and poor prognosis (59). While CHI3L1 was noted previously as increased in mutant ER BC cells (13), it was not further explored. In the present study, CHI3L1 protein was higher in mutant ER MCF-7 compared to WT. Since CHI3L1 is associated with decreased E-cadherin and enhanced MMP-9 activity in the involuting mammary gland (54), we investigated its role in invasion and found that the selective advantage of D538G mutant ER MCF-7 cells for invasion was abolished by a CHI3L1 neutralizing antibody. While a gene signature suggestive of increased migration/invasive capacity was linked to mutant ER (12,13), to our knowledge this is the first demonstration that a specific protein can be targeted in mutant ER cells to decrease invasiveness.
Pathway analysis of genes significantly differentially expressed by mutant versus WT ER in data from three studies demonstrated that genes in the type-1 interferon (IFN) signaling and innate immune pathways were enhanced in mutant ER lines. In normal tissues, infectious pathogens are recognized by Pattern or Damage Recognition Receptors (PAMPs or DAMPs) that activate type-1 IFN production. IFNα/β signal through IFN receptors and STAT1 activate transcription of Interferon Stimulated Genes (ISGs) to facilitate an innate immune response against the pathogen. The cyclic GMP-AMP synthase (cGAS) is a known stimulator of ISGs via the STING signaling pathway that can be upregulated in cancer cells in response to cytoplasmic DNA or mitochondrial DNA (reviewed in (60)). We focused on the ISGs IFITM3 and M1 because they were elevated in mutant ER cells compared to WT in our mRNA profiling, correlated with AR positivity in TNBC (25), elevated in AI resistant BC cells (35), induced in therapy resistant prostate cancer models (61,62), and upregulated by MUC1 in BC cells (63). MUC1 also emerged as a mutant ER-specific gene increased in mutant ER cells at the protein level. We confirmed that both D538G and Y537S MCF-7 cells have elevated IFITM3 protein at baseline that was further elevated by IFNβ. Importantly, clinical biopsies harboring the D538G mutation had significantly more IFITM3 protein compared to metastatic lesions with WT ER.
Given the enhanced expression of the type-1 IFN pathway protein IFITM3 in mutant ER MBC specimens, we sought to determine if there was a difference in immune cell infiltrate in WT versus mutant ER biopsies. We found significantly elevated T helper and T regulatory cells, as well as enhanced macrophage infiltration in MBC with mutant compared WT ER. Androgens stimulated alternative activation (M2 pro-tumor polarization) of macrophages in an AR dependent manner in an allergic asthma model (64). Further, CHI3L1 stimulated by inflammation leads to M2 macrophage polarization during allergic reactions (65). Thus, enhanced macrophage infiltration in mutant ER MBC may be the result of elevated innate immune signaling pathways/proteins like type-1 IFN, active AR, and/or CHI3L1.
Although ER+ BC is more “immunologically cold” than TNBC, the recent efficacy of anti-PD-L1 antibody in high risk patients with ER+ disease (37), confirms that some ER+ BC may respond to immunotherapy. Indeed, we found that PD-L1 positive macrophages were significantly higher in mutant ER metastatic lesions than those with WT ER. Interestingly, the other two genes most commonly mutated in BC, TP53 and PIK3CA, did not correlate with altered macrophages, but did correlate with increased CD8+ cytotoxic T cells. These data suggest that enhanced immune suppression via PD-L1+ macrophages may be uniquely acquired by ER mutant MBC. Since expression of PD-L1 in the tumor microenvironment, primarily its expression on myeloid cells, predicts BC response to checkpoint inhibitors (39), future studies will explore checkpoint inhibition in WT versus mutant ER MBC.
Evidence presented here, and in Arnesen et al, suggest that mutant ER containing cells may be poised to survive the stress of E2-deprivation and anchorage independence by the constitutive activity of mutant ER and by changes in non-ER regulated genes. Our studies in BC cells, PDX, and patient biopsies revealed proteins/pathways that may serve as targetable vulnerabilities in ER mutant disease, including AR, CHI3L1, and ISGs. On the other hand, metastases without ESR1 exon 8 mutations expressed significantly less ER and PR than those with mutant ER, but clearly adapt to the same selective pressure of E2-deprivation (AI therapy) and the metastatic cascade. Adaptations may occur through shared targetable mechanisms like activated AR, or unique mechanisms that remain to be determined via preclinical modeling under clinically relevant conditions that mimic the selective pressures under which they evolve in patients. The differences in hormone receptor protein expression and tumor infiltrating immune cells in biopsies with ESR1 mutations versus ESR1 WT suggest that AI resistant MBC should be examined in light of the different pathways and adaptations that lead to resistance, which in turn result in new vulnerabilities to guide novel therapeutic strategies.
Supplementary Material
Significance statement:
Targetable alterations in MBC, including AR, CHI3L1, and ISG arise following estrogen-deprivation, and ER mutant metastases may respond to immunotherapies due to elevated PD-L1+ macrophages.
Acknowledgements
The authors acknowledge the use of the University of Colorado Cancer Center/NIH/NCI Cancer Core Support Grant P30 CA046934 for Tissue Biobanking and Histology, Molecular Pathology, Animal Imaging, Biostatistics and Bioinformatics and Cell Technologies shared resources. We thank Adrie Van Bokhoven, Kathryn Zolman, and Kurtis Davies in particular for their contributions. We would also like to thank the patients enrolled in NCT02953860 (COMIRB 16-1001) and patient advocates Dr. Karen Raines Hunt, Jane Perlmutter, and Vickie Tosher. This investigation was supported by DOD BCRP W81XWH-16-1-0422 BC151357 and BC151357P1 (JG and JKR); DOD BCRP BC120183 W81XWH-13-1-0090 (JKR and AE); NIH R01CA187733 (JKR), NIH R01CA221303 (SO), T32CA190216 and NIH NRSA F32 CA239436 (MMW) and DOD W81XWH-17-1-0615 BC161497 (RBR). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Disclosure of potential conflicts of interest: none
References
- 1.van Hellemond IEG, Geurts SME, Tjan-Heijnen VCG. Current Status of Extended Adjuvant Endocrine Therapy in Early Stage Breast Cancer. Curr Treat Options Oncol 2018;19:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartmaier RJ, Trabucco SE, Priedigkeit N, Chung JH, Parachoniak CA, Vanden Borre P, et al. Recurrent hyperactive ESR1 fusion proteins in endocrine therapy-resistant breast cancer. Ann Oncol 2018;29:872–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeselsohn R, De Angelis C, Brown M, Schiff R. The Evolving Role of the Estrogen Receptor Mutations in Endocrine Therapy-Resistant Breast Cancer. Curr Oncol Rep 2017;19:35. [DOI] [PubMed] [Google Scholar]
- 4.Karnik PS, Kulkarni S, Liu XP, Budd GT, Bukowski RM. Estrogen receptor mutations in tamoxifen-resistant breast cancer. Cancer Res 1994;54:349–53 [PubMed] [Google Scholar]
- 5.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep 2013;4:1116–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 2013;45:1439–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 2013;45:1446–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barone I, Iacopetta D, Covington KR, Cui Y, Tsimelzon A, Beyer A, et al. Phosphorylation of the mutant K303R estrogen receptor alpha at serine 305 affects aromatase inhibitor sensitivity. Oncogene 2010;29:2404–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herynk MH, Parra I, Cui Y, Beyer A, Wu MF, Hilsenbeck SG, et al. Association between the estrogen receptor alpha A908G mutation and outcomes in invasive breast cancer. Clin Cancer Res 2007;13:3235–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinert T, Saad ED, Barrios CH, Bines J. Clinical Implications of ESR1 Mutations in Hormone Receptor-Positive Advanced Breast Cancer. Front Oncol 2017;7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu L, Wang L, Mao C, Duraki D, Kim JE, Huang R, et al. Estrogen-independent Myc overexpression confers endocrine therapy resistance on breast cancer cells expressing ERalphaY537S and ERalphaD538G mutations. Cancer Lett 2019;442:373–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeselsohn R, Bergholz JS, Pun M, Cornwell M, Liu W, Nardone A, et al. Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer Cell 2018;33:173–86 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahreini A, Li Z, Wang P, Levine KM, Tasdemir N, Cao L, et al. Mutation site and context dependent effects of ESR1 mutation in genome-edited breast cancer cell models. Breast Cancer Res 2017;19:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin LA, Ribas R, Simigdala N, Schuster E, Pancholi S, Tenev T, et al. Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat Commun 2017;8:1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuqua SA, Fitzgerald SD, Chamness GC, Tandon AK, McDonnell DP, Nawaz Z, et al. Variant human breast tumor estrogen receptor with constitutive transcriptional activity. Cancer Res 1991;51:105–9 [PubMed] [Google Scholar]
- 16.McGuire WL, Chamness GC, Fuqua SA. Estrogen receptor variants in clinical breast cancer. Mol Endocrinol 1991;5:1571–7 [DOI] [PubMed] [Google Scholar]
- 17.Katzenellenbogen JA, Mayne CG, Katzenellenbogen BS, Greene GL, Chandarlapaty S. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer 2018;18:377–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner NC, Slamon DJ, Ro J, Bondarenko I, Im SA, Masuda N, et al. Overall Survival with Palbociclib and Fulvestrant in Advanced Breast Cancer. N Engl J Med 2018;379:1926–36 [DOI] [PubMed] [Google Scholar]
- 19.Hamilton EP, Patel MR, Armstrong AC, Baird RD, Jhaveri K, Hoch M, et al. A First-in-Human Study of the New Oral Selective Estrogen Receptor Degrader AZD9496 for ER(+)/HER2(−) Advanced Breast Cancer. Clin Cancer Res 2018;24:3510–8 [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Levine KM, Bahreini A, Wang P, Chu D, Park BH, et al. Upregulation of IRS1 Enhances IGF1 Response in Y537S and D538G ESR1 Mutant Breast Cancer Cells. Endocrinology 2018;159:285–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelsomino L, Gu G, Rechoum Y, Beyer AR, Pejerrey SM, Tsimelzon A, et al. Erratum to: ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res Treat 2017;163:639–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol 2016;2:1310–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savic D, Partridge EC, Newberry KM, Smith SB, Meadows SK, Roberts BS, et al. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res 2015;25:1581–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12:245–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, et al. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 2006;25:3994–4008 [DOI] [PubMed] [Google Scholar]
- 26.Jin HJ, Kim J, Yu J. Androgen receptor genomic regulation. Transl Androl Urol 2013;2:157–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barton VN, Christenson JL, Gordon MA, Greene LI, Rogers TJ, Butterfield K, et al. Androgen Receptor Supports an Anchorage-Independent, Cancer Stem Cell-like Population in Triple-Negative Breast Cancer. Cancer Res 2017;77:3455–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norris JD, Ellison SJ, Baker JG, Stagg DB, Wardell SE, Park S, et al. Androgen receptor antagonism drives cytochrome P450 17A1 inhibitor efficacy in prostate cancer. J Clin Invest 2017;127:2326–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, Carey LA, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res 2013;19:5505–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traina TA, Miller K, Yardley DA, Eakle J, Schwartzberg LS, O'Shaughnessy J, et al. Enzalutamide for the Treatment of Androgen Receptor-Expressing Triple-Negative Breast Cancer. J Clin Oncol 2018;36:884–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brasso K, Christensen IJ, Johansen JS, Teisner B, Garnero P, Price PA, et al. Prognostic value of PINP, bone alkaline phosphatase, CTX-I, and YKL-40 in patients with metastatic prostate carcinoma. Prostate 2006;66:503–13 [DOI] [PubMed] [Google Scholar]
- 32.Johansen JS, Brasso K, Iversen P, Teisner B, Garnero P, Price PA, et al. Changes of biochemical markers of bone turnover and YKL-40 following hormonal treatment for metastatic prostate cancer are related to survival. Clin Cancer Res 2007;13:3244–9 [DOI] [PubMed] [Google Scholar]
- 33.Jeet V, Tevz G, Lehman M, Hollier B, Nelson C. Elevated YKL40 is associated with advanced prostate cancer (PCa) and positively regulates invasion and migration of PCa cells. Endocr Relat Cancer 2014;21:723–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogony J, Choi HJ, Lui A, Cristofanilli M, Lewis-Wambi J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast Cancer Res 2016;18:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi HJ, Lui A, Ogony J, Jan R, Sims PJ, Lewis-Wambi J. Targeting interferon response genes sensitizes aromatase inhibitor resistant breast cancer cells to estrogen-induced cell death. Breast Cancer Res 2015;17:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Escher TE, Lui AJ, Geanes ES, Walter KR, Tawfik O, Hagan CR, et al. Interaction Between MUC1 and STAT1 Drives IFITM1 Overexpression in Aromatase Inhibitor-Resistant Breast Cancer Cells and Mediates Estrogen-Induced Apoptosis. Mol Cancer Res 2019;17:1180–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nanda R, Liu MC, Yau C, Asare S, Hylton N, Veer LVt, et al. Pembrolizumab plus standard neoadjuvant therapy for high-risk breast cancer (BC): Results from I-SPY 2. Journal of Clinical Oncology 2017;35:506-28029304 [Google Scholar]
- 38.Kyte JA, Andresen NK, Russnes HG, Fretland SO, Falk RS, Lingjaerde OC, et al. ICON: a randomized phase IIb study evaluating immunogenic chemotherapy combined with ipilimumab and nivolumab in patients with metastatic hormone receptor positive breast cancer. J Transl Med 2020;18:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 2018;379:2108–21 [DOI] [PubMed] [Google Scholar]
- 40.Gallicchio L, Macdonald R, Wood B, Rushovich E, Helzlsouer KJ. Androgens and musculoskeletal symptoms among breast cancer patients on aromatase inhibitor therapy. Breast Cancer Res Treat 2011;130:569–77 [DOI] [PubMed] [Google Scholar]
- 41.Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J 2011;30:3019–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dees EC, Carey LA. Improving Endocrine Therapy for Breast Cancer: It's Not That Simple. Journal of Clinical Oncology 2013;31:171–3 [DOI] [PubMed] [Google Scholar]
- 44.Giuliano M, Schiff R, Osborne CK, Trivedi MV. Biological mechanisms and clinical implications of endocrine resistance in breast cancer. The Breast 2011;20:S42–S9 [DOI] [PubMed] [Google Scholar]
- 45.Harvell DME, Spoelstra NS, Singh M, McManaman JL, Finlayson C, Phang T, et al. Molecular signatures of neoadjuvant endocrine therapy for breast cancer: characteristics of response or intrinsic resistance. Breast Cancer Research and Treatment 2008;112:475. [DOI] [PubMed] [Google Scholar]
- 46.Harvell DME, Richer JK, Singh M, Spoelstra N, Finlayson C, Borges VF, et al. Estrogen regulated gene expression in response to neoadjuvant endocrine therapy of breast cancers: tamoxifen agonist effects dominate in the presence of an aromatase inhibitor. Breast Cancer Research and Treatment 2008;112:489–501 [DOI] [PubMed] [Google Scholar]
- 47.De Amicis F, Thirugnansampanthan J, Cui Y, Selever J, Beyer A, Parra I, et al. Androgen Receptor Overexpression Induces Tamoxifen Resistance in Human Breast Cancer Cells. Breast cancer research and treatment 2010;121:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cochrane DR, Bernales S, Jacobsen BM, Cittelly DM, Howe EN, D’Amato NC, et al. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Research : BCR 2014;16:R7–R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Amato NC, Gordon MA, Babbs B, Spoelstra NS, Carson Butterfield KT, Torkko KC, et al. Cooperative Dynamics of AR and ER Activity in Breast Cancer. Mol Cancer Res 2016;14:1054–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aceto N, Bardia A, Wittner BS, Donaldson MC, O'Keefe R, Engstrom A, et al. AR Expression in Breast Cancer CTCs Associates with Bone Metastases. Mol Cancer Res 2018;16:720–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015;526:131–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng J, Li L, Zhang N, Liu J, Zhang L, Gao H, et al. Androgen and AR contribute to breast cancer development and metastasis: an insight of mechanisms. Oncogene 2017;36:2775–90 [DOI] [PubMed] [Google Scholar]
- 53.Barton VN, D'Amato NC, Gordon MA, Lind HT, Spoelstra NS, Babbs BL, et al. Multiple molecular subtypes of triple-negative breast cancer critically rely on androgen receptor and respond to enzalutamide in vivo. Mol Cancer Ther 2015;14:769–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes K, Wickenden JA, Allen JE, Watson CJ. Conditional deletion of Stat3 in mammary epithelium impairs the acute phase response and modulates immune cell numbers during post-lactational regression. J Pathol 2012;227:106–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen N, Shani O, Raz Y, Sharon Y, Hoffman D, Abramovitz L, et al. Fibroblasts drive an immunosuppressive and growth-promoting microenvironment in breast cancer via secretion of Chitinase 3-like 1. Oncogene 2017;36:4457–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steenbrugge J, Breyne K, Demeyere K, De Wever O, Sanders NN, Van Den Broeck W, et al. Anti-inflammatory signaling by mammary tumor cells mediates prometastatic macrophage polarization in an innovative intraductal mouse model for triple-negative breast cancer. J Exp Clin Cancer Res 2018;37:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Zhang S, Wang Q, Zhang X. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol 2017;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma B, Herzog EL, Lee CG, Peng X, Lee CM, Chen X, et al. Role of chitinase 3-like-1 and semaphorin 7a in pulmonary melanoma metastasis. Cancer Res 2015;75:487–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan G, Xiang L, Sun X, Wang X, Li H, Ge W, et al. Elevated YKL-40 expression is associated with a poor prognosis in breast cancer patients. Oncotarget 2017;8:5382–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loo TM, Miyata K, Tanaka Y, Takahashi A. Cellular senescence and senescence-associated secretory phenotype via the cGAS-STING signaling pathway in cancer. Cancer Sci 2020;111:304–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borghesan M, Fafian-Labora J, Eleftheriadou O, Carpintero-Fernandez P, Paez-Ribes M, Vizcay-Barrena G, et al. Small Extracellular Vesicles Are Key Regulators of Non-cell Autonomous Intercellular Communication in Senescence via the Interferon Protein IFITM3. Cell Rep 2019;27:3956–71 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Civenni G, Bosotti R, Timpanaro A, Vazquez R, Merulla J, Pandit S, et al. Epigenetic Control of Mitochondrial Fission Enables Self-Renewal of Stem-like Tumor Cells in Human Prostate Cancer. Cell Metab 2019;30:303–18 e6 [DOI] [PubMed] [Google Scholar]
- 63.Provance OK, Lewis-Wambi J. Deciphering the role of interferon alpha signaling and microenvironment crosstalk in inflammatory breast cancer. Breast Cancer Res 2019;21:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Becerra-Diaz M, Strickland AB, Keselman A, Heller NM. Androgen and Androgen Receptor as Enhancers of M2 Macrophage Polarization in Allergic Lung Inflammation. J Immunol 2018;201:2923–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwak EJ, Hong JY, Kim MN, Kim SY, Kim SH, Park CO, et al. Chitinase 3-like 1 drives allergic skin inflammation via Th2 immunity and M2 macrophage activation. Clin Exp Allergy 2019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.